- 1Department of Microbiology, Odisha University of Agriculture and Technology, Bhubaneswar, India
- 2Department of Biosciences and Bioengineering, Indian Institute of Technology Bombay, Mumbai, India
- 3National Centre for Cell Science (NCCS), Pune, India
The indiscriminate and intensive use of agrochemicals in developing nations to enhance crop productivity has posed an alarming threat to soil quality, fertility, biodiversity, food safety, agricultural sustainability, and groundwater quality, thus critically affecting planetary health and food productivity. Additionally, both abiotic and biotic stresses and developmental disorders, i.e., disease susceptibility, hormonal imbalance, and nutritional deficiency, are the major constraints on crop productivity. In this context, the use of soil–plant associated microbiomes “phytomicrobiome,” especially rhizospheric microbiota, in combination with agronomic practices (nutrient, water, and resource management, as integrated management options: INM/IPM/IWM) is the most promising alternative for managing soil health and crop productivity. The global recognition of plant/soil-associated microbiome has generated substantial investment of public and private bodies to grow microbe-based food products. However, understanding the molecular, genetic, physiological, and ecological aspects of phytomicrobiome toward sustainable agriculture would require broad attention along with associated environmental/physico-chemical control points. The underpinning mechanisms of plant–microbe interactions are of immense significance for strategizing host selection (single culture/consortia) and its field application. Taxa such as Rhizobium, Pseudomonas, Alcaligenes, Burkholderia, Sphingomonas, Stenotrophomonas, Arthrobacter, Bacillus, and Rhodococcus have emerged as promising plant growth-promoting (PGP) candidates with diverse beneficial traits, such as, producing phyto-hormones, volatile organics, antibiotics for disease suppression, N2-fixation, Fe uptake, and extracellular enzymes, but several physico-chemical constraints/extremities limit the field application (on-site) of such microbes. Hence, a detailed overview on genomic, physiological, metabolic, cellular, and ecological aspects is necessitated. Thorough insights into nutrient acquisition (especially limiting nutrients like Fe and P) during abiotic stress are still under-studied, so the use OMICS, robust bioinformatics pipeline/tools, might greatly revolutionize the field of PGP microbial ecology (complex plant–microbe interactions) for application in agricultural sustainability, nutritional security, and food safety. This review focusses on critical aspects of mechanisms of Fe and P transport-uptake (nutrient acquisition) by various PGP microbes, and their metabolism, genetics, and physiology relevant for managing stress and better crop production.
Introduction
Recent surge in food demand, unavoidable and excessive use of chemical fertilizers during agricultural practices, increased industrialization-led discharge of synthetic pollutants, altered land use pattern, and extreme climatic factors are seriously affecting soil health and crop productivity (Alori et al., 2020; Lehmann et al., 2020). The fertilizer-based mono-cropping model is posing an extra layer of harmful effects on human, soil health, and biota (Kumar et al., 2017; Pattnaik et al., 2019). Additionally, ecological stresses (abiotic: soil salinity, drought, pH of soil, environmental temperature, ozone, toxic metals, and low nutrient concentration; biotic: disease and pest, singly or in combination) have become primary hurdles of crop production in different agro-ecologies (Zhu, 2016; Ahmed et al., 2020; Janni et al., 2020). It has been estimated that around 70 % and 30 % of crop losses are due to abiotic (drought/flood) and biotic (bacteria, viruses, fungi, nematodes, and herbivores) factors, respectively (Enebe and Babalola, 2018; Majeed et al., 2018). On the other hand, soil–plant microbiome (microbial communities) is ascribed to be the key component of agroecosystem that performs multipartite functions, e.g., changing crop yield (quantity and quality), timing of key developmental stages of plants, tolerance to biotic and abiotic stresses, improving soil health, etc. (Lareen et al., 2016; Fitzpatrick et al., 2020). Since 1904, when Lorenz Hiltner coined the term “rhizosphere” stating the plethora of microorganisms (bacteria, fungi, archaea) around and inside plant roots in response to plant secretions/root exudates, many researchers have defined the multipartite interactions among soil biota and plant hosts (Berendsen et al., 2012; Carvalhais et al., 2013; Fitzpatrick et al., 2020). Through various cultivation-dependent and metagenomics studies, the importance of soil-plant microbiome interactions governing major nutrient/biogeochemical cycles and beneficial/pathogenic attributes of plant has been established. Based on microbial abundance, root-rhizospheric microbiome has also been considered as secondary genome, thus, providing microbe-derived compounds and traits to a plant (Berendsen et al., 2012; Rout and Southworth, 2013). Simultaneously, the host genotype influences the overall composition of plant-associated/root microbial communities.
Mostly, plant-derived metabolites (root exudates) act as signaling factors and nutrients for microbial communities in rhizospheric niches (Sasse et al., 2018; Mohapatra et al., 2020). Secretory metabolites of plant roots (exudates) excreted to the root vicinity because of the rupturing/mechanical damage of the root cortex cells along with microbial grazing, fungal infections/tissue invasion, or emergence of lateral roots, which break through the root epidermis. These compounds belong to a complex mixture of soluble low molecular weight organic materials belonging to classes of sugars, amino acids, organic acids, enzymes, nucleotides, co-factors, etc. (Table 1), usually present in larger quantity in the rhizosphere compared with bulk soil (Kumar et al., 2017; Verma et al., 2019a). In addition, some plants secret enzymes (extracellular, rhizodeposition) such as oxidase, hydrolase, phosphatase, nitroreductase, laccase, and peroxidase in response to nutrient deficiency and chemical compounds/pollutants in soil/root zone. It has been estimated that a typical rhizospheric region of Zea mays L. (Corn) harbors total organic acid and amino acids in the range of 10–20 mmol L−1 (accounting to 1 to 4 % of total dry weight), followed by sugars (90 mmol L−1) (Baetz and Martinoia, 2014). The major organic acids are detected to be malic, malonic, acetic, citric, fumaric, succinic, lactic, tartaric, and oxalic, which are solely responsible for solubilization of minerals/metal nutrients. Compared with organic acids, amino acids and nucleic acids appear to function as signals for microbial recognition. In addition, flavonoid compounds (mostly released by legumes) have been found to attract rhizobial members (Rhozobium, Agrobacterium), which suppress the activity of pathogenic fungi (Cooper, 2004). Root cap cells (border cell) and mucilage also appear to play a significant role in the establishment of root-microbial interactions (Hawes et al., 2002; Sasse et al., 2018). The components of root exudates or lysates of decaying root tissues have been found to act as precursors for phytohormone production with rhizosphere microorganisms, thus stimulating plant growth; whereas, amino acids such as tryptophan and L-methionine are required as precursor for indole acetic acid (IAA) and ethylene (C2H4) biosynthesis with rhizospheric microbiome (Hayat et al., 2010). In addition to this, soil edaphic factors (pH, temperature, redox equivalents, cation exchange capacity, salinity, moisture, O2 availability, organic carbon/matter, soil biochemical activity, etc.) or selection/genotypes of plants are ascribed to be the key determinants of root microbiome assembly (Pattnaik et al., 2019).
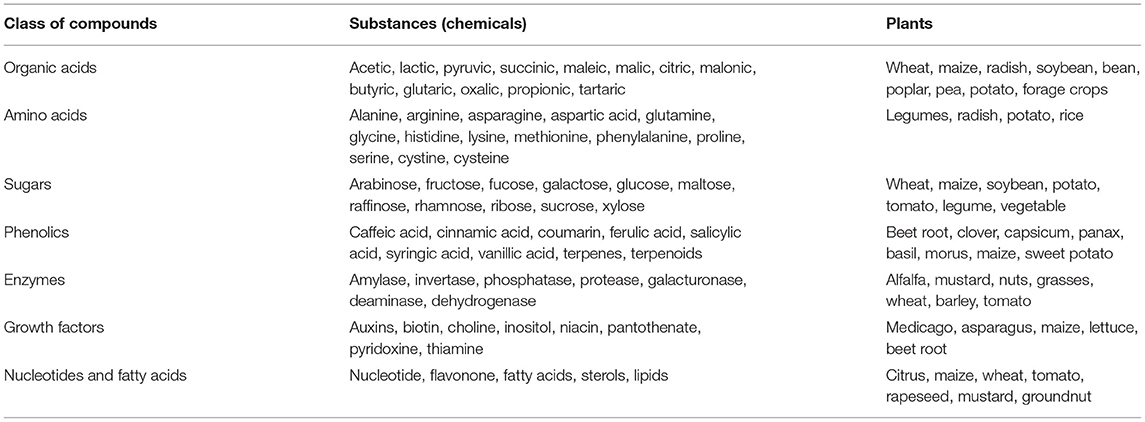
Table 1. Compounds secreted by root (as root exudates) by different plants into the rhizosphere-soil vicinity.
The complex interactions between plant host and microbiota, i.e., phytomicrobiome (the holobiont) are cautiously orchestrated with signaling crosstalk between microbe-microbe and microbe-host plant (Engelmoer et al., 2014). It has been established that microbe-associated molecular patterns (MAMPs) play a key role in elevating plant immune response (Smith et al., 2017), but various environmental stresses trigger metabolic, biochemical, and physiological alterations in plants that greatly affect the assembly and interaction of microbial communities with the host plant. Field crops face several biotic (living: viral, bacterial, and fungal, insects, nematodes, and parasitic weeds) and abiotic stresses (non-living/physico-chemical: excess water/flood, drought, heat, salinity, chilling, and oxidative damage), contributing to >50 % reduction in yield/productivity (Tardieu and Tuberosa, 2010; Augusto et al., 2017). During drought, decreased assimilation of photosynthetically active radiation (i.e., use of radiation capability) and diminished harvest index are the major factors of crop loss (Ashraf and Harris, 2013). Similarly, salinity affects more than 800 million hectares of cultivable land with suppression of photosynthesis, protein synthesis, and metabolism of lipids, thus affecting germination, phase transition, plant biomass, and yield (Ladeiro, 2012). Microbial members with plant growth-promoting (PGP) abilities are considered to be rescuers in such conditions to alleviate various stresses. Various beneficial traits such as phytohormone production, secretion of exopolysaccharides (EPS), volatile compounds, ACC deaminase, maintenance of osmolytes, antioxidants, regulation of stress-responsive genes, enhanced root and shoot growth, higher nutrient uptake, enhanced Na+/K+ ratio, production of enzymes and siderophore, chlorophyll content, and alteration of root morphology, help in mitigating stress in crops (Kumar and Verma, 2018; Jain et al., 2020). In addition, under severe stress, the rhizospheric microbiome can improve/modulate plant nutritional status by mineralizing bound or unavailable nutrients (limiting) at soil vicinity. Hence, in the context of fulfilling increased food demand within the framework of reduced cultivable land and climate change risks (stress), it is imperative to understand the basis of nutrient acquisition mechanisms of PGP microbiome for developing sustainable food production models and novel biotechnological solutions. This review comprehends the role of PGP microbes in nutrient acquisition, which (Fe, P, K, etc.) are limiting/redox active in soil system. The metabolic steps/pathways of siderophore-synthesis and -mediated Fe uptake in various bacteria, Fe oxidation-reduction processes, P (inorganic and organic) and K solubilization routes, associated genetic and physiological insights into these processes, are discussed, which are relevant for the management of stress in various crops.
PGP Microbes and Its Diversity
In rhizospheric niches, at any given time, wide microbial diversity is observed, i.e., 108−9 bacteria, 106−8 archaea, 105−6 fungi, 103−5 algae and protozoa, and 102 nematodes (Fierer, 2017; Dubey et al., 2019). Various hydrophobic and lipophilic root exudates or oxidative burst-mediated compounds exert varied cellular effects on rhizospheric microbiome (communities), e.g., act as carbon and nitrogen sources, uptake, signaling and quorum sensing, alter membrane fluidity (permeabilization), energy transduction (electron transport chain/proton motive force), changing of lipid composition, and activity of membrane-associated proteins (Qu et al., 2020; Zhou et al., 2020). The availability and abundance of such compounds in the root vicinity exert selection pressure on the microbial community to evolve as an efficient plant colonizer (Olanrewaju et al., 2019). In the context of plant-associated microbiome, in recent years, many rhizospheric colonizers have been found to display novel PGP activities and have emerged as a promising tool for agricultural application (Pattnaik et al., 2019). First, it is important to know plant-associated (ectosphere, endosphere, phyllosphere, and rhizosphere) microbial communities (who is/are there?) and their metabolic complexity and physiology (what are they doing?) (Mohapatra and Phale, 2021). The qualitative and quantitative aspects of growth-promoting activities largely depend on: (a) the nature of the plant-secreted compounds/substrates available; (b) the prevailing soil ecologies (nutrients and physico-chemical conditions); (c) microbial community composition; and (d) physiological activities of a community. Culture-based studies have characterized many plant-beneficial bacterial members from diverse habitat. Further non-cultivation-based studies (such as metagenomics) are providing new information on probable mutual beneficial interactions (such as pollutant degradation, bioremediation) within the population and with plant host(s) (Kanaly and Harayama, 2000; Olanrewaju et al., 2019). The PGP microbial communities belong to broad phylogenetic lineages, i.e., Bacteria, Archaea, Algae, and other Eukaryotes (filamentous fungi, yeast, protozoans), but bacterial members are significantly important (Figure 1A, Lugtenberg and Kamilova, 2009; Verma et al., 2019b; Khatoon et al., 2020). NCBI-Genebank and Ribosomal Database Project-II (RDP-II)-based data curation including nucleotides, rRNAs, and genomes (RefSeq), indicated that most PGP microbes belong to domain Bacteria (predominant) following Fungi and Archaea. Among bacterial domains (n = 7,594 reported taxa), Proteobacteria represented the highest [gamma- (2278), beta- (830), and alpha- (763) as abundant, up to 50%, n = 3,986)], followed by Firmicutes (n = 2,584), Actinobacteria (n = 844), Bacteroidetes (n = 191), Fungi (n = 165), Cyanobacteria, Verrucomicrobia, and Deinococcus-Thermus (Figure 1B). Depending on soil edaphic conditions, i.e., chemical composition of secretory pool, their concentration, temperature, pH, nutrients, O2, humidity, etc., the community composition changes at spatio-temporal scale. Amongst all, bacterial members such as Agrobacterium, Rhizobium, Azospirillum, Azotobacter, Burkholderia, Pseudomonas, Enterobacter, Erwinia, Flavobacterium, Microbacterium, Bacillus, Micrococcus, and Paenibacillus are recognized as promising PGP candidates (Table 2).
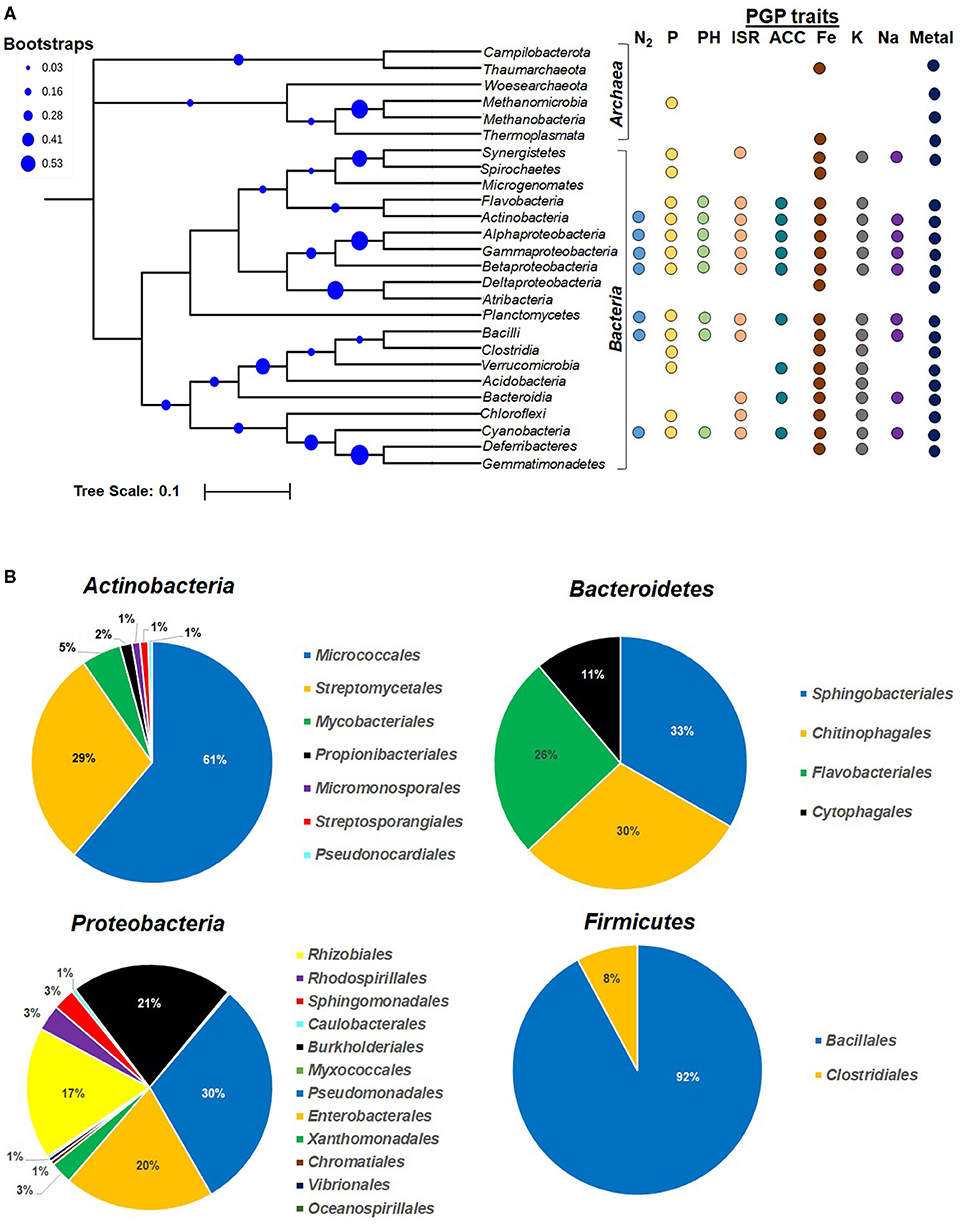
Figure 1. Phylogenetic diversity and percent abundance of prokaryotes involved in various plant growth promotion activities. (A) Phyla Proteobacteria, Firmicutes, and Euryarchaeota are further classified at class level. The root indicates the LUCA (last universal common ancestor). (B) Pie chart showing relative distribution (% abundance) of major PGP taxa reported in RDP II and NCBI taxonomy databases. The contributions of each PGP taxa in (A) are denoted with colored dots corresponding to each specific PGP traits. The scale bar 0.1 indicates substitution (10%) at nucleotide level of 16S rRNA gene, and blue circles indicate the bootstrap values (in %). N2, dinitrogen fixation; P, phosphate solubilization; PH, phyto-hormone production; ISR, induced systemic resistance; ACC, ACC deaminase; Fe, iron (siderophore) uptake; K, potasium uptake; Na, sodium uptake; Metal, heavy metal transformation and bioremediation.
Broad Mechanisms of Plant Growth-Promoting Attributes of PGP Taxa
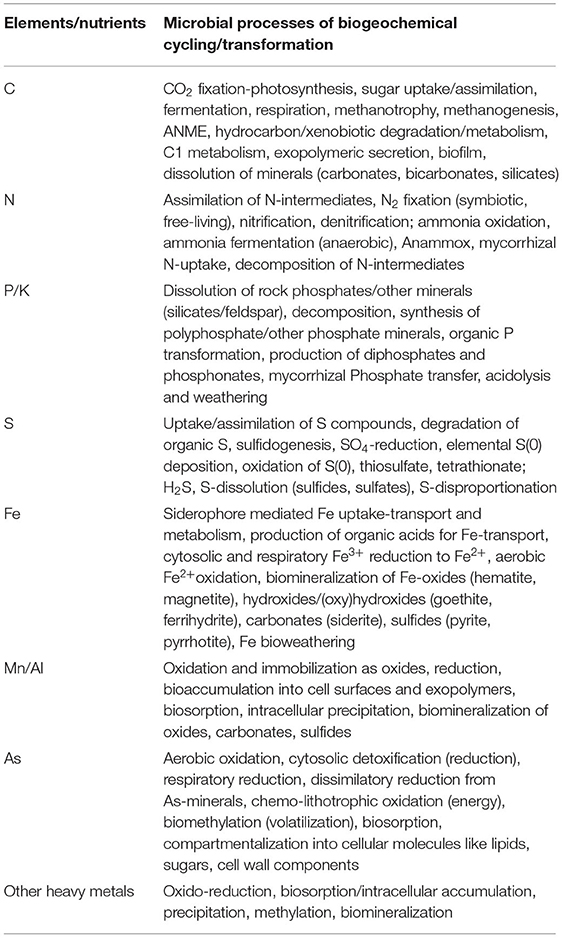
PGP microbes are ecologically and economically important and an expanded community with a significant role in enhancing the availability and uptake of nutrients through mobilization and fixation, or reducing the harmful effects of phyto-pathogens through direct and indirect modes of actions (Figure 2, Lugtenberg and Kamilova, 2009; Beneduzi et al., 2012; Olanrewaju et al., 2017). The direct mechanism comprises either facilitation of resource acquisition (N2 fixation, solubilization, or Fe-uptake through siderophore production/uptake) or modulation of plant hormone levels (production of IAA, cytokinin, ACC deaminase, or by decreasing the stress hormone). The indirect mechanisms include reduction of the inhibitory effects of various phytopathogens by niche exclusion, induced systemic resistance, and producing antibiotic or antiviral metabolites (Figure 2). In addition, these PGP microbes perform major biogeochemical functions, i.e., C, N, P, S, and Fe bio-geocycling (such as CO2 fixation, assimilation/fermentation, respiration, methano-trophy/genesis, ANME, hydrocarbon/xenobiotic degradation, nitrification, denitrification; anammox, rock phosphate solubilization, Fe-oxidation-respiration, dissolution, As detoxification, etc.) (Table 3) (Etesami and Adl, 2020; Basu et al., 2021a,b). Hence, specific group of PGP taxa are preferred corresponding to specific agricultural applications. Several Xanthomonads and Pseudomonads are reported for efficient nutrient solubilization (He et al., 2019; Basu et al., 2021a,b; Xiao et al., 2021). Inoculation of symbiotic N2-fixing Allorhizobium, Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium or associative fixers such as Azospirillum, Enterobacter, Klebsiella, Pseudomonas, and Xanthomonas resulted in improved growth, yield, and nutrient uptake in several crops by colonizing root surfaces (Glick, 1995). Achromobacter, Azospirillum, Bacillus, Enterobacter, Pseudomonas, and Rhizobium are recorded as potent genera of rhizobacteria with ACC deaminase activity to cut the level of ethylene in roots under severe stress (Glick et al., 2007). Azospirillum, Azotobacter, Bacillus, Enterobacter, Paenibacillus, Pseudomonas, Streptomyces, Glomus, and Gigaspora are documented as potential biocontrol microbes for exhibiting antagonistic activity against phytopathogens by producing antibiotics, β-1,3-glucanase, chitinases, cyanide, fluorescent pigment, and siderophores (Pathma et al., 2011; Bhattacharyya and Jha, 2012). Seed treatment with Agrobacterium, Alcaligenes, Bacillus, Comamonas, Paenibacillus, Pseudomonas, and Streptomyces has shown to induce the elongation rate of lateral roots. Strains of Bacillus, Pseudomonas, and Serratia have reported as agents of developing systemic resistance and promoting plant health. Plant hormones (IAA, gibberellins, cytokinins), extracellular substances produced by phyllospheric bacteria as well as rhizobacteria, are responsible for direct role in crop productivity (Glick et al., 2007). Abiotic stresses (such as extreme temperature, flooding, drought, salinity, and heavy metal contamination) are yield-limiting factors for plant growth and crop productivity, and also trigger a series of morpho-physiological, biochemical, and molecular defects in plants, thus microbial assemblages (Enebe and Babalola, 2018; Vaughan et al., 2018). The Application of several PGP microbial cultures or consortiums such as Serratia marcescens, Bacillus amyloliquefaciens, Bacillus subtilis, Brevibacillus brevis, Bacillus cereus, Pseudomonas putida, and Pseudomonas fluorescens has significantly improved the defense mechanism and modulated production of secondary metabolites. The induction of secondary metabolites by stress-tolerant PGP taxa is shown to improve conditions by producing antioxidants, osmolyte biosynthesis (soluble sugars, proline, glycine betaine, and amines), generating reactive oxygen species (ROS) scavengers, coenzymes, etc. (Sansinenea, 2019). Antibiotic compounds such as 2,4-diacetylphloroglucinol (DAPG), hydrogen cyanide, oomycin A, and phenazine secreted by some PGP bacteria suppress pathogens in soils, for example phenazines produced by Pseudomonas chlororaphis against Fusarium oxysporum (Raaijmakers and Mazzola, 2012). The presence of a wide array of antibiotic resistance genes (ARG) and their transmission night have important implications for agriculture in future.
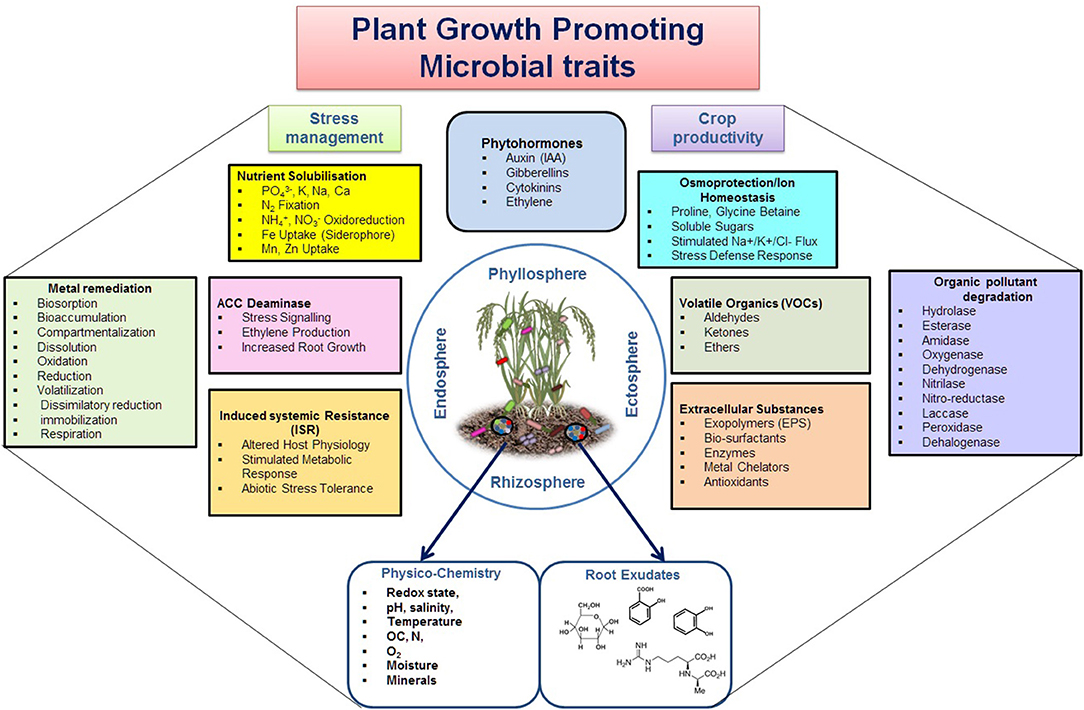
Figure 2. Multipartite growth promoting aspects of PGP microbes for enhancing crop yield and productivity under varied agro-ecologies. The rectangular boxes depict various aspects of plant growth activities, while circles indicate the soil physico-chemical and root exudate activities for influencing microbial colonization at root vicinity.
Although several workers have formulated and applied PGP microbial members on field (as biofertilizers), the effectiveness of such microbes are mainly focused on laboratory studies and yielded inefficient results at a field scale. Such failures mainly ascribed to: (a) lack of information on microbiota composition of specific plant genotypes; (b) assembled metabolic networks of microbes; (c) properties of their enzymes/activities; (d) crosstalk between host(s)-microbial metabolic routes; and (e) stress responses and cellular changes (Gopalakrishnan et al., 2015; Rilling et al., 2019). In this context, the advent of recent molecular approaches, i.e., “OMICS” (combination of genomic, proteomic, transcriptomic, and metabolomics) contributed markedly to avoid such hurdles and rationalize the application of PGP microbes for better crop growth. The use of high-throughput next-generation sequencing (for DNA/RNA), and its related algorithms/computational tools, and proteomic and metabolomic datasets allow us to understand dynamic microbial interactions and the physiological basis of plant colonization and would further enable to gain deeper insights into the system level understanding of such processes (Figure 3). As a consequence, it is important to analyze and understand the OMICS of PGP microbes to reduce the gap, controlling the timing, tissue-specificity, and expression level of genes for their optimal use in agriculture.
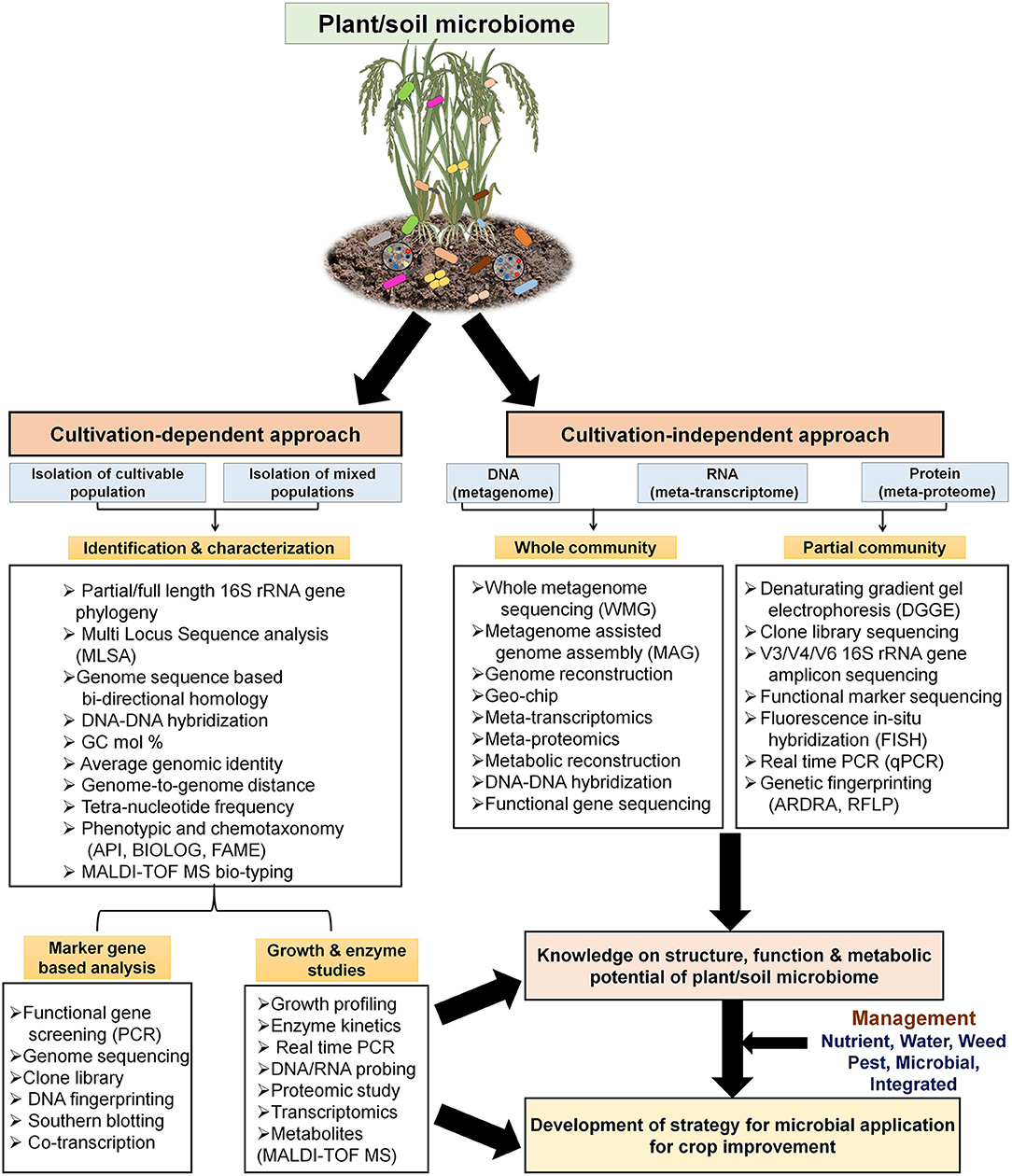
Figure 3. Use of cultivation-dependent and –independent (metagenomics, other OMICS) approaches for studying PGP microbial ecology, physiology, and biochemical functions at genetic and molecular levels. The steps and workflow are depicted for better understanding of structure-function of soil PGP microbiome together with application of various management strategies for sustainable crop production.
PGP Microbial Role in Acquisition of Limiting/Redox Active Nutrients
Out of 17 essential nutrients (some of them are limiting) [carbon (C), hydrogen (H), oxygen (O), nitrogen (N), phosphorus (P), potassium (K), sulfur (S), magnesium (Mg), calcium (Ca), iron (Fe), manganese (Mn), zinc (Zn), copper (Cu), molybdenum (Mo), nickel (Ni), chlorine (Cl), and boron (B)], paucity of any single nutrient leads to nutritional deficiency, thus resulting in low performance of crop, yield (crop loss), and quality of crop (Oldroyd and Leyser, 2020). Since many of the nutrients are found to be soil associated, i.e., bound to inorganic and organic soil constituents/insoluble precipitates, root absorption often becomes difficult. In such case, PGPR microbes play a pivotal role in releasing nutrients from soil minerals and organic complexes for own cellular metabolism, which aid in absorption by plant root (Uroz et al., 2009; Etesami and Adl, 2020). The application of such PGPR as bio-inoculants has become a swift alternative for increased availability of nutrients to various field crops, minimizing the use of chemical fertilizers, and bioremediation (metal remediation) purposes. Additionally, increased climatic variation and environmental pollution have led to incidence of various abiotic stresses on diverse crops like drought/extreme temperature, salinity, flooding (flash), UV irradiation, heavy metal(s) and synthetic organic pollution, which ultimately lead to nutritional imbalance (P, K, Fe, Mn, Ca, etc.) and serious reductions in growth and yield (Bukhari et al., 2019). Availability of nutrients and its acquisition by crops are affected by both internal or genetic factors (host genotype-phenotype) and external factors (soil physico-chemistry). It has been shown that, under slightly acidic conditions (pH = 6–6.5), most nutrients become available in soil solution. For example, at acidic pH, phosphate reacts with aluminum (Al) and Fe to form precipitates (sparingly soluble to insoluble), whereas at pH > 7.5, it reacts with Ca and Mg to form soluble complexes. In addition, the availability of Fe, Mn, Al, and S is also affected by redox conditions of the soil. Pertaining to this, the involvement of PGPR in mineral solubilization, nutrient acquisition, abiotic stress management, and enhancing defense responses has been effectively known (Majeed et al., 2018; Numan et al., 2018; Kumar et al., 2020). One of the key strategies of PGPR includes enabling higher contact/access of the root system to the nutrients in soil solution by increasing root growth (root interception) through different mechanisms. Root elongation by PGPR by production of IAA and ACC deaminase is the most important. It has been established that PGPR helps in reducing growth of primary root, increasing the number and length of lateral roots, and stimulating root hair elongation (Dobbelaere et al., 1999; Chamam et al., 2013; Santoyo et al., 2021). In literature, many researchers have systematically reviewed/highlighted various direct and indirect plant growth related activities of PGP microbes, but thorough insights into nutrient acquisition (especially limiting nutrients such as Fe and P) during abiotic stress are still under-studied and remained un-highlighted, thus necessitating a deeper study on metabolism, genetics, and eco-physiology of nutrient acquisition by PGP microbes.
PGP Microbes, Fe Acquisition, and Plant Growth
Supplementation of available forms of iron (Fe) (mostly through siderophore) to plants is one of the key bacterial functions under stress conditions (Kramer et al., 2020). Fe forms the integral component of cell metabolism including synthesis of chlorophyll, maintenance of chloroplast structure and function, DNA synthesis, respiration (electron transport chain proteins, ferredoxin), prosthetic group of many metallo-enzymes (oxido-reductases, cytochromes and non-heme oxidases) involved in oxido-reductive reactions (Rout and Sahoo, 2015). Despite Fe being the 4th most abundant (1 % by mass) on crust of the Earth (lithosphere), bio-availability is very limited because of its low solubility, mostly in the form of insoluble Fe (II/III)-oxides, -hydroxides, -oxy(hydroxides), and sulphidic complexes (hematite, magnetite, ferrihydrite, goethite, pyrite, pyrrhotite, etc.). This leads to very low (10−9 to 10−18 M) free Fe(II/III) concentrations in soil/rhizosphere (Hider and Kong, 2010; Zhang et al., 2019). In the soil and plant root vicinity, Fe availability and its release (weathering, dissolution, leaching, mineralization, etc.) from soil mineral sources are found to be a complex process, mostly regulated by various physico-chemical factors such as pH, O2 concentration, conductivity, redox, and dissolution-precipitation phenomena (Figure 4) (Colombo et al., 2014).
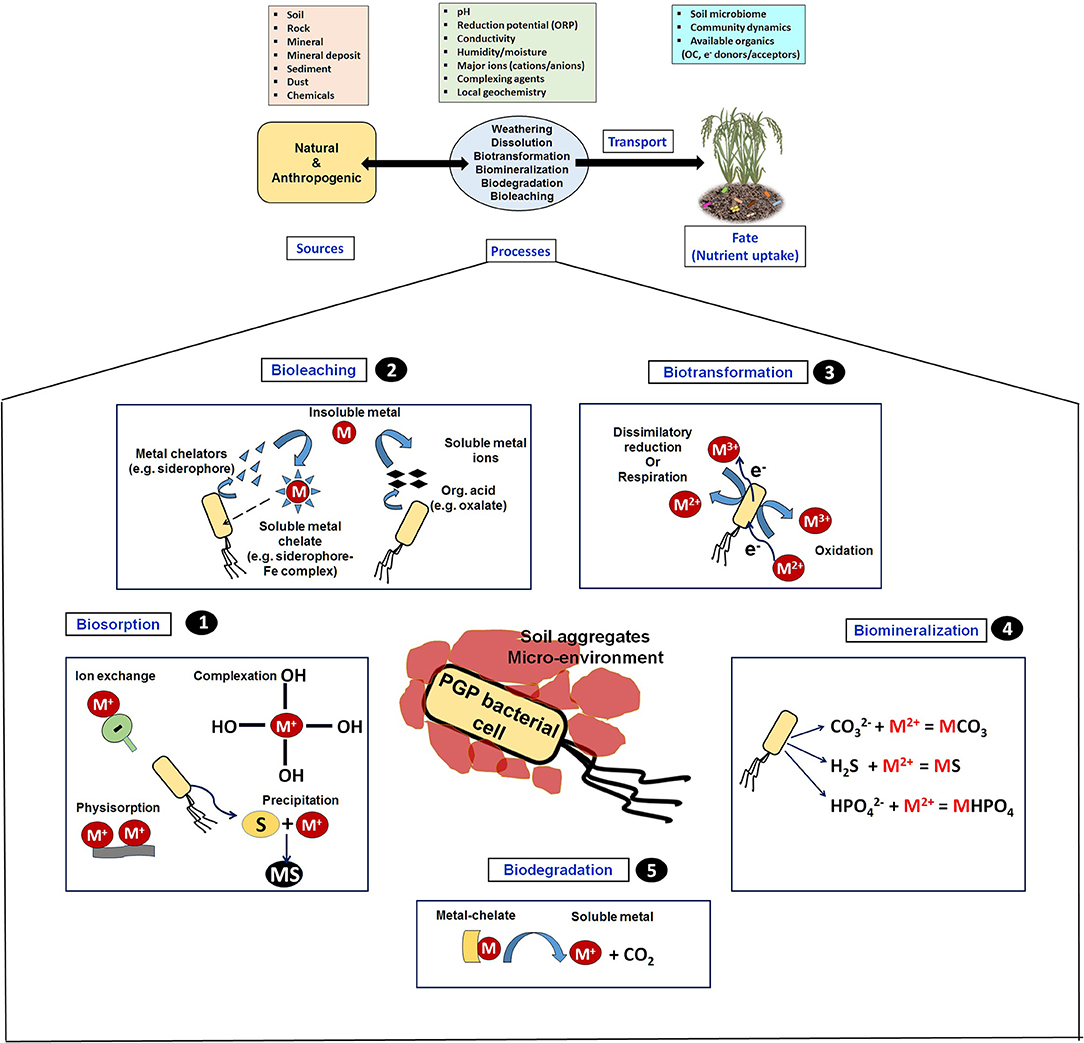
Figure 4. Geochemical pathways involving sources, processes, and transport factors influencing the fate of nutrient availability at soil solution and its uptake by plants (upper panel) and diverse types of geomicrobiological interactions that occur between microbe and solid mineral phase for metal (Fe, P, K) solubilization (lower panel in pentagon box).
To cope up with very limiting Fe, plants and associated microbes employ complexation as key strategy, where soluble/secreted molecules (such as siderophore, organic acid) react directly with Fe mineral surfaces (silicates, oxy-hydroxides), thus weathering and mobilizing the micronutrient (Figure 4). In addition, these complexation agents are able to chelate Fe from organic complexes such as Fe-organic acids, phenols, humic substances, and proteins, via ligand exchange mechanism (Kostka et al., 1996; Cesco et al., 2000). With respect to complexation, plants have evolved dual Fe acquisition strategies (Zhang et al., 2019). In the reduction-based strategy (mostly for non-leguminous plants), protons and phenolic compounds are released by root cortical cells to the rhizospheric vicinity to lower the pH (acidify) and increase the Fe(III) solubility, thus uptaking it by apoplast Fe-regulated transporter (IRT1) and reducing it to more soluble Fe(II) mediated by ferric reduction oxidases (FRO) system (Olenska et al., 2020). Alternatively, leguminous plants employ chelation-based strategy. In this mechanism, phytosiderophores (PS) [mugineic acid (MA) family members: mugineic acid, 2′-deoxymugineic acid (DMA), 3-epihydroxymugineic acid (epi-HMA), and 3-epihydroxy 2′-deoxymugineic acid (epi-HDMA)] are produced having high affinity for binding to Fe(III). Three molecules of S-adenosyl-methionine serve to be the precursor for synthesis of such PS, and mostly they constitute of carboxyl, amine, and hydroxyl groups as ligand functional groups. The resulting binary complex [Fe(III)-PS] gets transported into the root epidermal cells through yellow stripe-like (YSL) transporters and metabolized (Ahmed and Holmstrom, 2014; Saha et al., 2016), but with various environmental stresses and physico-chemical conditions, the production of PS gets impaired, thus impacting Fe uptake by plants. In addition to PS, many organic molecules (oxalates, flavonoids) abundantly present in the rhizosphere are directly involved in efficient weathering/solubilization of Fe minerals (Colombo et al., 2014; Boiteau et al., 2018).
In contrast to plant PS, rhizospheric microbes (mainly bacteria and fungi) secret low molecular weight (400–1,500 D) siderophores (hydroxamate or phenolate groups) for scavenging Fe with higher efficiency (binding constant Kf > 1030) (Andrews et al., 2003; Ahmed and Holmstrom, 2014). Besides Fe, siderophores are found to form stable complexes with other trace metals such as Al, Cd, Cu, In, Pb, and Zn, thus helping in supplementing such elements for better growth (Yu et al., 2017). A total of 500 different siderophores are recognized today, out of which 270 are structurally characterized (Kramer et al., 2020). The up-to-date list of microbial (bacterial, archaeal, and fungal) siderophores identified is presented in Table 4. On the basis of their mode of biosynthesis, they are either non-ribosomal peptide synthetase (NRPS) multienzymes based or NRPS-independent mechanism, where larger groups are synthesized by non-ribosomal peptide synthetases (NRPSs) or polyketide synthase (PKS) (Carroll and Moore, 2018). These molecules form either octahedral (hexa-coordinate) complexes with Fe(III) where various functional groups act as bidentate ligands (hydroxamates, α-hydroxy carboxylates, and catecholates) or form polydentate phenolate/nitrogen heterocycle/carboxylate complexes (Hider and Kong, 2010).

Table 4. Microbial members responsible for producing/secreting several siderophores identified from various metabolite databases (SiderophoreBase, MetaCyc, BioCyc, and AntiSMASH).
Microbial (mainly bacteria and fungi) siderophores are broadly categorized into one; catecholate [C6H4(OH)2-1,2-dihydroxybenzene] or hydroxamate/phenolate [C6H5OH–hydroxybenzene] groups harboring backbone of polyamine, peptide, or macrocyclic lactone structural folds, where two [O] atoms chelate Fe(III), forming a hexadentate octahedral complex. Major members are enterobactin (Escherichia coli), pyoverdine (Pseudomonas aeruginosa), agrobactin (Agrobacterium tumefaciens), azotobactin (Azotobacter vinelandii), salmochelin (Salmonella enterica), malleobactin/ornibactin (Burkholderia cepacia, Burkholderia ambiferia, Burkholderia vietnamiensis), bacillibactin (Bacillus anthracis, B. subtilis), and parabactin (Paracoccus denitrificans) (Crosa and Walsh, 2002; Saha et al., 2016). Alternatively, hydroxamate siderophore contains C(=O)N(–OH), connected to the amino acid or its derivatives, where each group forms a bidentate ligand with Fe, thus forming a hexadentate octahedral symmetry. Major members of this group are ferribactin (Pseudomonas fluorescens), desferrioxamine (Streptomyces coelicolor). Members of Rhizobium, Staphylococcus, and some fungi (Mucorales) are found to produce (α-hydroxy)-carboxylates (complexones). More specifically, R. meliloti DM4 is reported to produce rhizobactin (amino poly-carboxylic acid) having ethylenediaminedicarboxyl and hydroxycarboxyl as Fe-chelating groups, whereas S. aureus synthesize staphylloferrin A (D-ornithine and two citric acid residues linked by two amide bonds) (Fukushima et al., 2014). Siderophore-mediated Fe uptake is different in both Gram-positive (Firmicutes, Actinobacteria) and Gram-negative (Proteobacteria) members. In Proteobacteria, Fe(III)-siderophore complex is recognized at receptors and targeted/bind to the outer membrane transporters (OMTs), followed by its entry through the periplasm into the cytoplasm using TonB-ExbBD complex, periplasmic siderophore-binding proteins (SBPs), and permease-ATPase system (Faraldo-Gómez and Sansom, 2003; Klebba, 2016). In Gram-positive taxa, lipoprotein SBPs anchored to the cell membrane bind the Fe(III)-siderophore complex and translocate into intracellular compartment through permease-ATPase system similar to the Gram-negative members (Fukushima et al., 2014).
Amongst all, catecholate are well-studied siderophores represented in broad phylogenetic lineages. The widely studied Enterobactin belongs to catecholate-type siderophore produced by E. coli, and related members are found to be composed of condensed three units of 2,3-Dihydroxybenzoylserine joined in a cyclic structure by lactone linkages. The biosynthetic precursor, i.e., 2,3-Dihydroxy-N-benzoylserine and its linear condensation products (dimer/trimer) are also found to transport Fe and are reported to serves as an exosiderophore for many other community members (siderophore deficient) such as P. aeruginosa, thus referred as strategy of siderophore piracy (Hantke, 1990; Barber and Elde, 2015). The biosynthesis starts with conversion of chorismate to 2,3-Dihydroxybenzoate, followed by condensation of three L-serine, three 2,3-Dihydroxybenzoate, and six ATP to form enterobactin catalyzed by different complex components of the non-ribosomal peptide synthase (NRPS), enterobactin synthase (Figure 5A). Similarly, pyoverdines (primary Fe uptake system) are produced by fluorescent Pseudomonads and with more than one type of siderophore molecules having a dihydroxyquinoline chromophore, a variable acyl side chain (dicarboxylic acid/amide) bound to the amino group of the chromophore, and a variable peptide chain linked by an amide group bound to the C1/C3 carboxyl group of the chromophore. The main chromophore is derived from L-tyrosine (Visca et al., 2007). Belonging to this class, glutamate-pyoverdine I (PVDI) produced by opportunistic Pseudomonas aeruginosa PAO1, has been studied well. The peptide chain consists of eight amino acids [D-serine- L-arginine- D-serine- N5-formyl-N5-hydroxy-L-ornithine-c(L-lysine- N5-formyl-N5-hydroxy-L-ornithine-L-threonine-L-threonine), and synthesis is carried out in the cytoplasm by genes pvdL, pvdI, pvdJ, and pvdD (non-ribosomal peptide synthase (NRPS) enzymes). Subsequent actions of L-Ornithine monooxygenase (PvdA), hydroxyl-L-ornithine formylase (PvdF), and ferribactin synthase (PvdDJIL) produce myristoylated ferribactin in the cytoplasm, which gets transported (PvdE) to periplasm. Further, the action of deacylase converts it to ferribactin, which, upon monooxygenation and dehydrogenation, yields glutamate-pyoverdine I (Figure 5B). The mature synthesized pyoverdines gets exported from the periplasm to outside by efflux systems, i.e., ABC-type exporter (PvdRT-OmpQ) and MdtABC-OmpB (Henríquez et al., 2019). After complexation with Fe(III), it is transported back into the cells via outer membrane FpvA-TonB ferric-pyoverdine transporter system (Schalk, 2008). In the periplasm, ferric-pyoverdine complex is scavenged by FpvC-FpvF complex, Fe(III) gets reduced by the FpvG ferric-pyoverdine reductase, and soluble Fe is released and transported into the cytoplasm by the ABC-type Fe2+ transporter (FpvDE), where sigma factor PvdS is essential for its maximal expression (Greenwald et al., 2009; Schalk et al., 2012). Similar biosynthetic pathway for bacillibactin, cyclic trimeric ester made of three units of 2,3-dihydroxybenzoate-glycine-threonine, joined in a cyclic structure by lactone linkages, is employed in Gram-positive Bacillus members.
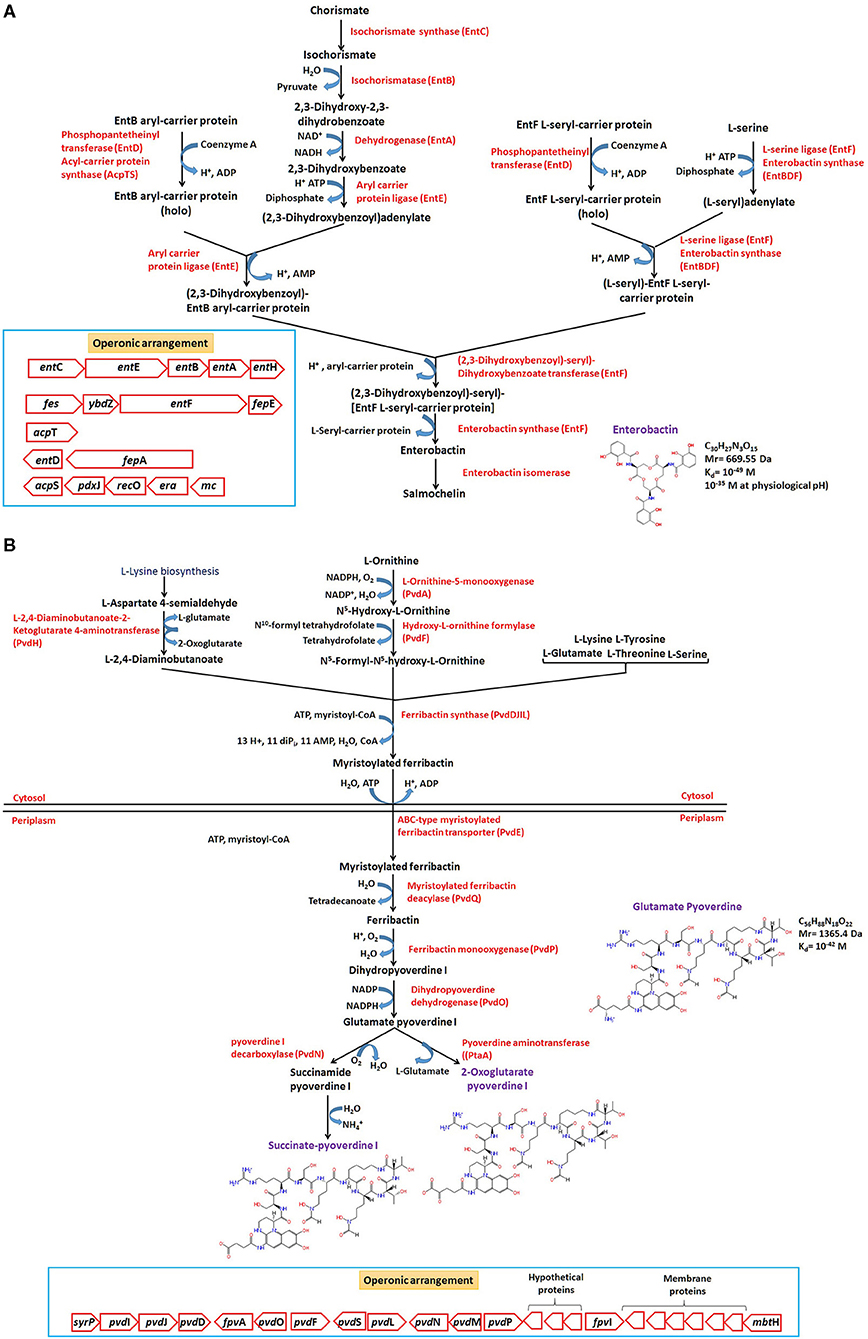
Figure 5. Biosynthetic pathways, reactions (KEGG, MetaCyc, Biocyc, and RAST-SEED based), and operonic (genetic) structure of predominant siderophores produced by E. coli (Enterobacteriaceae) and Pseudomonas members. The enzymes responsible for reactions (associated gene product name) are mentioned at each step (in red). The rectangular box in (A,B) denotes the operonic arrangement of the siderophore biosynthesis clusters.
On an overall basis, microbial siderophores influence host-plant Fe homeostasis under Fe limiting and stress conditions (such as salinity), metabolism, cell signaling, regulatory function, growth, and detoxification of toxic pollutants (Hesse et al., 2018). Studies have shown that the production of pyoverdine and its analog (apo-pyoverdine) by Pseudomonas fluorescens C7R12 modulates the expression of around 2,000 genes in Arabidopsis thaliana, related to development and Fe acquisition, as well as down-regulation of defense-related genes such as transcription factors (ERF, WRKY, MYB), salicylic acid (SA)-related genes (AT5G24210 lipase class 3 protein), and an abscisic acid (ABA)-related gene (lipid transfer protein LTP3) (Trapet et al., 2016). Apo-pyoverdine was impaired in Fe-regulated transporter 1 (IRT1) and ferric reduction oxidase 2 (FRO2) knockout mutants, an overexpression of the transcription factor HBI1, a key node for the crosstalk between growth and immunity, thus reflecting the increased susceptibility to pathogenic gray mold, Botrytis cinerea. P. fluorescens WCS417-colonized A. thaliana showed positive regulation of FIT, FRO2, IRT1, and MYB72 transcription factors involved in Fe mobilization and BGLU42, PDR9 for Fe metabolism (Verbon et al., 2017). Inoculation of three PGP microbes (Aneurinibacillus aneurinilyticus WBC1, Aeromonas sp. WBC4 and Pseudomonas sp. WBC10) having increased siderophore production showed higher growth promotion, biocontrol of Fusarium solani, and nutrient uptake by wheat growing in Punjab, India (Kumar et al., 2018). Increased Fe uptake and metabolism have been observed in A. thaliana in response to application of Paenibacillus polymyxa BFKC01. Increased Fe-biofortification has been observed in mungbean using siderophore producing plant growth promoting bacteria (Pantoea dispersa MPJ9 and Pseudomonas putida MPJ6) (Patel et al., 2018). Production of siderophore also showed decrease in toxic effects of several other metals (Cu, Cd) in plant root regions (Hesse et al., 2018). Inoculation of Micrococcus yunnanensis YIM 65004 and Stenotrophomonas chelatiphaga LPM-5 PGP organisms has shown significant increase in gain of weight and Fe content of roots and shoots of Canola (Ghavami et al., 2017). Similarly, Paenibacillus triticisoli BJ-18, a N2-fixing bacterium, has been shown to increase plant growth and antimicrobial properties (Zhang et al., 2020). Nevertheless, the molecular mechanisms that control synthesis of various yet to be uncharacterized siderophores such as tercoelichelin and fusachelin NRPSs have yet to be elucidated. The details of NRPS-independent siderophore biosynthetic pathways is still remains to be identified, and further research is needed to understand the substrate specificity, molecular mechanisms, and ecological considerations/implications of such molecules in crop productivity and development.
Alternatively, soil-/phyto-microbiome engage multiple redox (oxidation-reduction) mechanisms for mobilizing Fe in soil solution to the root vicinity (Figure 4). Particularly, under aerobic conditions and at pH 5-8, Fe (II) released from low-complex Fe minerals gets readily oxidized to Fe(III), while under anoxic conditions, Fe(III) gets reduced either by reducing equivalents (inorganic ions) or by Fe-reducing soil microbes (Li et al., 2019; Tang et al., 2019). Soil microbiota employ myriad of biogeochemical reactions to solubilize and uptake Fe (II/III), and they ultimately help in enhanced Fe acquisition by plants. Fe(III) gets reduced by direct or indirect mechanisms; where in direct, Fe(III) reduced to Fe(II) at the expense of energy (respiratory substrate). A variety of Fe(III) oxidic minerals was found to act as terminal electron acceptor (TEA) during microbial respiration, under anaerobic/anoxic conditions mediated by membrane bound respiratory reductase, termed as dissimilatory Fe reduction (DFeR) (Lovley et al., 2004). Both H2 and organic carbon are preferred as electron donor (oxidation) during the process by microbial taxa belonging to Geobacter, Shewanella, and other Bacillales members. Under anoxic paddy cultivation (waterlogging condition/wetlands: low redox potential Eh <0.4 V), this microbial reductive mechanism is attributed to derive high Fe(II) solubility (~3–5 orders of magnitude), thus helping in efficient Fe availability for plants (Lovley et al., 2004; Colombo et al., 2014). In addition, dissimilatory -reducing bacteria belonging to Desulfotomaculum and Desulfosporosinus further aid in reductive dissolution of poorly crystalline Fe (III)-hydroxides (ferrihydrite, goethite), thus releasing Fe(II) in soil solution. Alternatively, microbe-mediated change in extracellular pH and Eh promotes Fe solubilization at relatively lower pH, as in the case of acid mine drainage impacted soil/sediment (Gupta and Sar, 2020). On the other hand, under aerated/O2 saturated conditions (aerobic rice cultivation), soil microbes engage in oxidation of Fe(II) (Fe-oxidizing bacteria) in many minerals under both acidic and neutrophilic conditions. At lower pH, acidophilic taxa were found to couple Fe(II) oxidation (as electron donor and energy) to the reduction of various substrates, preferably nitrate or O2 (Acidothiobacillus ferrooxidans) (Baker and Banfield, 2003). At neutral pH, Fe(II) gets rapidly oxidized to Fe(III) spontaneously, which quickly hydrolyzes and precipitates as Fe(III)-hydroxides/oxyhydroxides, but recently many, neutrophilic taxa such as Pseudomonas, Gallionella, and Leptothrix spp. are ascribed for biotic Fe(II) oxidation under aerobic and anoxic soil conditions (Li et al., 2018; Liu et al., 2019). On an overall basis, the combined mechanisms of Fe solubilization by microbiome, i.e., proton/ligand-promoted dissolution of Fe-oxides/hydroxides, reductive dissolution of crystalline/amorphous Fe-complex minerals, redox reaction, weathering, sorption/desorption, and complexation processes (organic acids, siderophore, polysaccharides, etc.) occurring at the soil-rhizospheric (extracellular milieu) or cell surfaces mediate the acquisition under limiting Fe concentration.
PGP Microbes, P (Phosphate) Acquisition, and Plant Growth
Phosphorus (P) constitutes an important and is a key macroelement of cell having contribution in DNA, RNA, ATP, and lipids and critically influences various growth and developmental stages such as flowering, ripening of fruits/seeds, fertilization, embryo development, disease resistance, and shoot root system development (Razaq et al., 2017). But P availability in the soil solution is a limiting factor for uptake by plants. Earth's crust harbors total of average 1,200 mg/kg P (0.01–0.2% P2O5) (Olenska et al., 2020). Over 99 % of the naturally occurring phosphorus exists as inorganic P (Pi), mostly precipitated as insoluble sedimentary rocks phosphate (39 % P2O5), igneous (2 % P2O5), and metamorphic form (1.3 % P2O5), while the rest as organic phosphates. In soil, around 4 % of the total P remains as orthophosphate (35–70 % of total phosphorus) (Alori et al., 2017) in non-solubilization form. Variety of soil microbial taxa belonging to Aerobacter, Agrobacterium, Azotobacter, Burkholderia, Enterobacter, Achromobacter, Pseudomonas, Bradyrhizobium, Rhizobium, Erwinia, Flavobacterium, Bacillus, and Micrococcus dissolve dicalcium phosphate, tricalcium phosphate, or hydroxyl apatite to avail P, termed as phosphate solubilizing bacteria (PSB) (de Boer et al., 2019). This ability is of great interest in agro-ecologies due to their promising effect as bio-fertilizers on plant growth and maintaining soil fertility.
Bacterial members solubilize both inorganic and organic phosphates in several ways (Figure 6, Alori et al., 2017). In an acid-independent mechanism, bacteria were found to release H+ to the outer surface in exchange for cation uptake (Rodriguez and Fraga, 1999), but predominantly, the discharge/excretion of organic acid and soil acidification dissolves most of Pi. The carboxyl and hydroxyl residues of organic acid chelate cations bind to phosphate, resulting in a reduction of pH and release of phosphate anions. This process occurs in the periplasmic space mediated by direct oxidation (Lei and Zhang, 2015). Most secreted organic acids are gluconic, lactic, isovaleric, isobutyric, acetic, glycolic, oxalic, malonic, succinic, citric, and propionic, out of which many PSB (Burkholderia cepacia, Erwinia herbicola, Pseudomonas cepacia, Pseudomonas putida, Acinetobacter calcoaceticus, Rhizobium leguminosarum, Rhizobium meliloti, and Bacillus firmus) produce gluconic acid and 2-ketogluconic acid (Naraian and Kumari, 2018). The direct oxidation pathway of glucose (DOPG, non-phos-phorylating oxidation) leads to production of gluconic acid in the periplasmic space where glucose dehydrogenase (GCD/GDH, requiring pyrroloquinolinequinone (PQQ) cofactor) and gluconate dehydrogenase (GAD), oxidizes glucose to yield gluconate (Ge et al., 2015; Chen et al., 2016). PQQ cofactor is found to be encoded by pqqABCDEF in many PSB strains and any changes (loss)/mutation of any gene is shown to decrease/ablate phosphate release (Suleman et al., 2018). Besides Pi, 30–65 % of the total P of soil remains to be in organic phosphates (Po), which gets released from organophosphates by microbial mineralization reactions (Figure 6, Alori et al., 2017). Members of Gram-positive Arthrobacter, Bacillus, and Rhodococcus, and Gram-negative Citrobacter, Delftia, Phyllobacterium, Proteus, Pseudomonas, and Rhizobium hydrolyze Po to Pi by employing enzymes: a) non-specific acid phosphatases (NSAPs), e.g., acid and alkaline phosphomonoesterases (phosphatases) acting on phosphoester and phosphoanhydride; b) phytases cleaving phytate; and c) phosphonatases and C-P lyases, for cleaving organophosphonates (Sharma et al., 2013; Alori et al., 2017). Phytases, a class of myo-inositol phosphohydrolases, are an important class of enzymes for conversion of phytate (inositol hexakisphosphate) to Pi, which is subsequently taken up by plants (Azeem et al., 2015). Specific rhizospheric colonizers such as Citrobacter, Rhizobium, and Pseudomonas are the major phytase producers (Kumar et al., 2017). Production of phytase is pH and temperature-dependent, optimum being at pH 6–8 and 30–35°C (Farias et al., 2018; Valeeva et al., 2018).
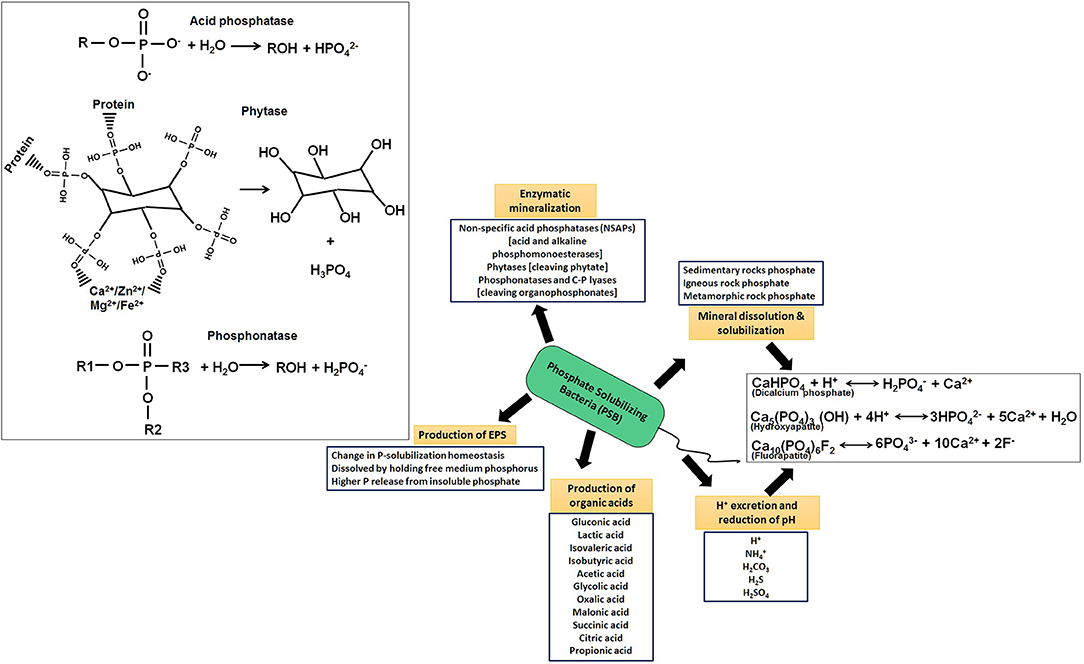
Figure 6. Various mechanistic aspects of phosphate solubilizing bacteria (PSB) showing important routes of solubilization of rock phosphate and organic phosphate for its metabolism as well as uptake by plant as plant growth beneficial traits.
Effects of various PSB bacterial members on plant growth and development have been observed. An increase in biomass production and P-uptake was reported in wheat (Triticum aestivum) inoculated with Pseudomonas spp., groundnut (Arachis hypogaea) inoculated with Pantoea spp., Castor (Ricinus communis), and sunflower (Helianthus annuus) inoculated with Psychrobacter (Ma et al., 2011). Transcriptomic study of PSB taxa Burkholderia multivorans WS-FJ9 has revealed differential expression of 446 genes involved in cell growth, P-solubilization; out of which 44 genes were up-regulated and 81 genes were down-regulated (Zeng et al., 2017). Inoculation of halo-tolerant PSB bacteria has shown to improve plant growth and suppress the effects of salts in salt-affected soil (Etesami and Beattie, 2018). Application of PSB microbes such as Arthrobacter, Bacillus, Azospirillum, and Oceanobacillus was shown to solubilize Ca3(PO4)2, AlPO4, and FePO4 in Avicennia marina, a halotolerant mangrove. Higher growth of Arabidopsis thaliana inoculated with Pseudomonas putida MTCC 5279 under salt stress and P-deficiency conditions has shown higher acidic and alkaline phosphatases activity, IAA and ABA levels, and up-regulation of several genes (At5g39610) encoding NAC-domain transcription factor and JAR1, At2g46370 for jasmonate, and AT3g32920 for DNA repair, leading to lowered senescence in leaves and stress adaptation (Srivastava and Srivastava, 2020). Addition of PSB members belonging to Klebsiella sp. RC3 and RCJ4, Stenotrophomonas sp. RC5, Serratia sp. RCJ6, and Enterobacter sp. RJAL6 exhibited high acid and alkaline phosphatase activity under P-starvation and increased Al toxicity (Barra et al., 2018). Inoculation of Arthrobacter nitroguajacolicus into Triticum aestivum seeds under salt stress gradient showed an increase in root-shoot length ration and overall biomass. The comparative transcriptome analysis showed involvement of 152 genes involved in biosynthesis of phenylpropanoid, metabolism of cysteine, methionine, flavonoids, and other secondary metabolites as well as induction of anti-oxidative enzymes cytochrome-P450, ascorbate peroxidase, nicotinamine, and ABC transporters (Safdarian et al., 2019). In addition, several researchers have attempted for phytoremediation of pollutants (metals) using PSB as bioinoculants in metalliferous soils. Various researchers have demonstrated the ameliorating effect of cheap organics, i.e., dry leaves, cellulose, manure, rice straw, distiller grain, saw dust etc. on precipitation of various metals and volatilization/methylation of As performed by microbes in acidic mine sites and impacted soil (Gupta and Sar, 2019, Mohapatra et al., 2020). Pseudomonas aeruginosa OSG41 has been used for Cr bioremediation under Chickpea cultivation (Oves et al., 2013). Other Pseudomonads have been used for Ni, Cu, Cd, and Zn bioremediation involving Brassicaceae family members, Black gram, and soybean (Rajkumar and Freitas, 2008; Ma et al., 2011). Other PSB members (Acinetobacter, Psychrobacter, and Bacillus spp.) have been used for multi-metal bioremediation involving various cereals and legumes (Pearl millet, Canola, Lycopersicon) (Ahemad, 2015). In severely As-polluted environment (contaminated groundwater irrigated for rice cultivation) of Bengal Basin, use of PGP bacteria for amelioration of As toxicity in soil and rice root/rhizosphere has become a swift alternative (Mohapatra et al., 2020). Although the solubilization of phosphates by indigenous PSB is very common under in-vitro conditions, the filed scale performance has been less satisfactory and, thus, greatly impacted the large-scale application of such microbes in sustainable agriculture. The molecular and physiological detailing of PSB microbes and real-time impact of phosphate solubilization using combined (OMICS) studies are still lacking, so further research in these aspects could largely benefit the farming community for its application along with suitable agronomic practices for better crop yield and maintenance of soil health.
In addition to Fe and P, potassium (K, as a part of NPK fertilizer dosing system) deficiency has become one of the major constraints for crop production because of introduction of high-yielding varieties, imbalanced K-fertilizer application, intensive cropping, run-off, leaching, and insoluble K minerals in soil (Sattar et al., 2019). Although, K is highly abundant, and it remains unavailable to plants because of its tightly bound nature in the constituent minerals (up to 90%) with non-exchangeable K, exchangeable K, mineral non-exchangeable K, and water-soluble K as major forms (Zorb et al., 2014). Hence, use of PGPR is gaining importance in enhancing K availability to plants under K deficiency. Many K-solubilizing bacterial (KSB) taxa, such as Pseudomonas, Burkholderia, Acidithiobacillus, Enterobacter, Paenibacillus, Arthrobacter, and Bacillus have shown to release K from insoluble K-bearing minerals (biotite, muscovite, feldspar, mica, vermiculite, orthoclase, illite, smectite) into soil solution, thus playing a key role in K biogeochemical cycling (Keshavarz Zarjani et al., 2013; Prajapati et al., 2013; Zhang and Kong, 2014; Sattar et al., 2019). Like P solubilization, KSB employs different mechanisms such as production of extracellular polysaccharides (chito-oligosachharides), ions/acids (acidolysis: citric, tartaric, 2-ketogluconic, oxalic, malic acid, propionic, fumaric, glycolic, and succinic acid), siderophores, organic ligands, and extracellular enzymes at soil-mineral interface to dissolute/release K to the root vicinity (Keshavarz Zarjani et al., 2013; Sattar et al., 2019). Amongst all, gluconic, oxalic, α-ketogluconic, and succinic acid are the most efficient organic acid for solubilization of K minerals by either a proton- or a ligand-mediated action or indirectly enhance dissolution by the forming complexes in solution with reaction products. Furthermore, bacterial IAA production also increases root growth and amount of root exudation, which ultimately enhances the surface area for reactivity (oxidize or complex) and K mobilization (Gahoonia et al., 1997). Microbial Fe(II) oxidation (biotite or silicates) has also been proposed as mechanism for microbial weathering of K minerals (Shelobolina et al., 2012). Overall, with K solubilization, KSB bacteria were reported to mediate exudation of soluble compounds, decomposition of soil organic matter, and mobilization of other nutrients (P, Fe, Mn), thus providing synergistic benefit to crop under field conditions (Zeng et al., 2012; Diep and Hieu, 2013; Zhang and Kong, 2014).
Conclusion and Future Perspective
Although PGP microbes are known for conferring multipartite plant-beneficial traits, their use/application under unfavorable environmental conditions (polluted sites, mine-impacted soil, saline/drought) is still limited. Therefore, a detailed study on plant–microbe–soil tripartite crosstalk must be investigate under both abiotic and biotic stress conditions. Use of real-time OMICS data with integrated informatics must be carried out to understand the eco-physiology of such microbes. Despite the marvelous progress in bioinformatics, transcriptome, proteome, and metabolome, adaptation system has been found to be ineffectual in the field of PGP microbial processes. Plant-beneficial microbes can change in response to several ecological factors and modify the cellular, biochemical, and molecular machineries in response to stress. Metagenomics/genomics and transcriptomics studies need to be performed for identifying the gene/transcript clusters, genomic plasticity, gene pool/flux, and pan-genomic detailing of PGP microbes. Alongside PGP traits, these microbes must be screened for novel insights into pesticide tolerance/degradation, anti-drought stress biomarkers, and development of biofungicides that will be surely helpful for their implication during harvest and storage of crops related to food quality and safety paradigm. In combination, system biology based metabolic engineering must be a priority to increase the metabolic diversity of PGP microbes (one microbe many function) for strain-specific and time-dependent metabolic fine-tuning.
Microbial approaches in agriculture have undergone a research transformation in recent years to straighten out the composition, diversity, and function for minimizing disease incidence and enhancing gross plant productivity. The scientific evidence supports the complexity of soil- microbial interactions, which frequently include microbial diversity and ecological covariates, and continues to challenge the discovery of PGP microbes. The general objective is to understand the use/selecetion of PGPB/R taxa, understanding their molecular mechanisms, a precise knowledge on key genes and use of molecular markers in including it in breeding programs. The emergent analytical toolsets of multi-omics integration will provide the prospect to resolve the PGPB/R functionally relating to specific bionetwork, which, in turn, would lead to another “Green Revolution” and help growing body of research to transform our ability in order to effectively target the right biomarkers for crop improvement. The future research must be prioritized in developing cost-effective and efficient PGP formulation with both public and farmers participation to propagate the use of these microbes in organic agriculture. Simultaneously, awareness on the use of PGP microbe might bring effective and sustainable crop framing in the near future.
Author Contributions
SP and BM conceptualized and organized the idea. SP, BM, and AG wrote, discussed, edited, and corrected the manuscript to its final form. All authors contributed equally to the article and approved the submitted version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
SP acknowledges OUAT-ICAR for providing research fellowship, and BM acknowledges IIT Bombay for providing the Institute Post-Doctoral fellowship. AG acknowledges NCCS, Pune for proving Research Associateship.
References
Ahemad, M. (2015). Phosphate-solubilizing bacteria-assisted phytoremediation of metalliferous soils: A review. 3 Biotech 5, 111–121. doi: 10.1007/s13205-014-0206-0
Ahmed, E., and Holmstrom, S. J. (2014). Siderophores in environmental research: roles and applications. Microb. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Ahmed, W., Xia, Y., Li, R., Bai, G., Siddique, K. H., and Guo, P. (2020). Non-coding RNAs: functional roles in the regulation of stress response in Brassica crops. Genomics 112, 1419–1424. doi: 10.1016/j.ygeno.2019.08.011
Alori, E. T., Adekiya, A. O., and Adegbite, K. A. (2020). Impact of agricultural practices on soil health. Soil Health 59, 89–98. doi: 10.1007/978-3-030-44364-1_5
Alori, E. T., Glick, B. R., and Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8, 971. doi: 10.3389/fmicb.2017.00971
Andrews, S. C., Robinson, A. K., and Rodríguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Ashraf, M. H. P. J. C., and Harris, P. J. (2013). Photosynthesis under stressful environments: an overview. Photosynthetica 51, 163–190. doi: 10.1007/s11099-013-0021-6
Augusto, L., Achat, D. L., Jonard, M., Vidal, D., and Ringeval, B. (2017). Soil parent material-a major driver of plant nutrient limitations in terrestrial ecosystems. Glob. Chang. Biol. 23, 3808–3824. doi: 10.1111/gcb.13691
Azeem, M., Riaz, A., Chaudhary, A. N., Hayat, R., Hussain, Q., Tahir, M. I., et al. (2015). Microbial phytase activity and their role in organic P mineralization. Arch. Agronomy Soil Sci. 61, 751–766. doi: 10.1080/03650340.2014.963796
Baetz, U., and Martinoia, E. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 19, 90–98. doi: 10.1016/j.tplants.2013.11.006
Baker, B. J., and Banfield, J. F. (2003). Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44, 139–152. doi: 10.1016/S0168-6496(03)00028-X
Barber, M. F., and Elde, N. C. (2015). Buried treasure: evolutionary perspectives on microbial iron piracy. Trends Genetics 31, 627–636. doi: 10.1016/j.tig.2015.09.001
Barra, P. J., Viscardi, S., Jorquera, M. A., Duran, P. A., Valentine, A. J., and de la Luz Mora, M. (2018). Understanding the strategies to overcome phosphorus-deficiency and aluminium-toxicity by ryegrass endophytic and rhizosphere phosphobacteria. Front. Microbiol. 9:1155. doi: 10.3389/fmicb.2018.01155
Basu, A., Prasad, P., Das, S. N., Kalam, S., Sayyed, R. Z., Reddy, M. S., et al. (2021a). Plant Growth Promoting Rhizobacteria (PGPR) as green bioinoculants: recent developments, constraints, and prospects. Sustainability 13:1140. doi: 10.3390/su13031140
Basu, S., Kumar, G., Chhabra, S., and Prasad, R. (2021b). “Role of soil microbes in biogeochemical cycle for enhancing soil fertility,” in New and Future Developments in Microbial Biotechnology and Bioengineering (Elsevier). doi: 10.1016/B978-0-444-64325-4.00013-4
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bhattacharyya, P. N., and Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28, 1327–1350. doi: 10.1007/s11274-011-0979-9
Boiteau, R. M., Shaw, J. B., Pasa-Tolic, L., Koppenaal, D. W., and Jansson, J. K. (2018). Micronutrient metal speciation is controlled by competitive organic chelation in grassland soils. Soil Biol. Biochem. 120, 283–291. doi: 10.1016/j.soilbio.2018.02.018
Bukhari, S. A. H., Peerzada, A. M., Javed, M. H., Dawood, M., Hussain, N., and Ahmad, S. (2019). “Growth and development dynamics in agronomic crops under environmental stress,” in Agronomic Crops, eds M. Hasanuzzaman (Singapore: Springer). doi: 10.1007/978-981-32-9151-5_6
Carroll, C. S., and Moore, M. M. (2018). Ironing out siderophore biosynthesis: a review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit. Rev. Biochem. Mol. Biol. 53, 356–381. doi: 10.1080/10409238.2018.1476449
Carvalhais, L. C., Dennis, P. G., Fan, B., Fedoseyenko, D., Kierul, K., Becker, A., et al. (2013). Linking plant nutritional status to plant-microbe interactions. PLoS ONE 8:e68555. doi: 10.1371/journal.pone.0068555
Cesco, S., Römheld, V., Varanini, Z., and Pinton, R. (2000). Solubilization of iron by water-extractable humic substances. J. Plant Nutr. Soil Sci. 163, 285–290. doi: 10.1002/1522-2624(200006)163:3<285::AID-JPLN285>3.0.CO;2-Z
Chamam, A., Sanguin, H., Bellvert, F., Meiffren, G., Comte, G., Wisniewski-Dye, F., et al. (2013). Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry 87, 65–77. doi: 10.1016/j.phytochem.2012.11.009
Chen, W., Yang, F., Zhang, L., and Wang, J. (2016). Organic acid secretion and phosphate solubilizing efficiency of Pseudomonas sp. PSB12: effects of phosphorus forms and carbon sources. Geomicrobiol. J. 33, 870–877. doi: 10.1080/01490451.2015.1123329
Colombo, C., Palumbo, G., He, J. Z., Pinton, R., and Cesco, S. (2014). Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J. Soils Sediments 14, 538–548. doi: 10.1007/s11368-013-0814-z
Cooper, J. E. (2004). Multiple responses of rhizobia to flavonoids during legume root infection. Adv. Bot. Res. 41, 1–62. doi: 10.1016/S0065-2296(04)41001-5
Crosa, J. H., and Walsh, C. T. (2002). Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Re. 66, 223–249. doi: 10.1128/MMBR.66.2.223-249.2002
de Boer, M. A., Wolzak, L., and Slootweg, J. C. (2019). “Phosphorus: reserves, production, and applications,” in Phosphorus Recovery and Recycling, eds H. Ohtake, and S. Tsuneda (Singapore: Springer). doi: 10.1007/978-981-10-8031-9_5
Diep, C. N., and Hieu, T. N. (2013). Phosphate and potassium solubilizing bacteria from weathered materials of denatured rock mountain, Ha Tien, Kien Giang province Vietnam. Am. J. Life Sci. 1, 88–92. doi: 10.11648/j.ajls.20130103.12
Dobbelaere, S., Croonenborghs, A., Thys, A., Broek, A. V., and Vanderleyden, J. (1999). Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 212, 153–162. doi: 10.1023/A:1004658000815
Dubey, A., Malla, M. A., Khan, F., Chowdhary, K., Yadav, S., Kumar, A., et al. (2019). Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers. Conserv. 28, 2405–2429. doi: 10.1007/s10531-019-01760-5
Enebe, M. C., and Babalola, O. O. (2018). The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: a survival strategy. Appl. Microbiol. Biotechnol. 102, 7821–7835. doi: 10.1007/s00253-018-9214-z
Engelmoer, D. J. P., Behm, J. E., and Kiers, E. T. (2014). Intense competition between arbuscular mycorrhizal mutualists in an in-vitro root microbiome negatively affects total fungal abundance. Mol. Ecol. 23, 1584–1593. doi: 10.1111/mec.12451
Etesami, H., and Adl, S. M. (2020). Plant growth-promoting rhizobacteria (PGPR) and their action mechanisms in availability of nutrients to plants. Phyto-Microbiome in Stress Regulation 15, 147–203. doi: 10.1007/978-981-15-2576-6_9
Etesami, H., and Beattie, G. A. (2018). Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 9:148. doi: 10.3389/fmicb.2018.00148
Faraldo-Gómez, J. D., and Sansom, M. S. (2003). Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116. doi: 10.1038/nrm1015
Farias, N., Almeida, I., and Meneses, C. (2018). New bacterial phytase through metagenomic prospection. Molecules 23:448. doi: 10.3390/molecules23020448
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fitzpatrick, C. R., Salas-González, I., Conway, J. M., Finkel, O. M., Gilbert, S., Russ, D., et al. (2020). The plant microbiome: from ecology to reductionism and beyond. Annu. Rev. Microbiol. 74, 81–100. doi: 10.1146/annurev-micro-022620-014327
Fukushima, T., Allred, B. E., and Raymond, K. N. (2014). Direct evidence of iron uptake by the Gram-positive siderophore-shuttle mechanism without iron reduction. ACS Chem. Biol. 9, 2092–2100. doi: 10.1021/cb500319n
Gahoonia, T. S., Care, D., and Nielsen, N. E. (1997). Root hairs and phosphorus acquisition of wheat and barley cultivars. Plant Soil 191, 181–188. doi: 10.1023/A:1004270201418
Ge, X., Wang, W., Du, B., Wang, J., Xiong, X., and Zhang, W. (2015). Multiple pqqA genes respond differently to environment and one contributes dominantly to pyrroloquinoline quinone synthesis. J. Basic Microbiol. 55, 312–323. doi: 10.1002/jobm.201300037
Ghavami, N., Alikhani, H. A., Pourbabaei, A. A., and Besharati, H. (2017). Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J. Plant Nutr. 40, 736–746. doi: 10.1080/01904167.2016.1262409
Glick, B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Glick, B. R., Cheng, Z., Czarny, J., and Duan, J. (2007). Promotion of Plant Growth by ACC Deaminase-Producing Soil Bacteria. New perspectives and Approaches in Plant Growth-Promoting Rhizobacteria Research. Dordrecht: Springer. doi: 10.1007/978-1-4020-6776-1_8
Gopalakrishnan, S., Sathya, A., Vijayabharathi, R., Varshney, R. K., Gowda, C. L., and Krishnamurthy, L. (2015). Plant growth promoting rhizobia: challenges and opportunities. 3 Biotech 5, 355–377. doi: 10.1007/s13205-014-0241-x
Greenwald, J., Nader, M., Celia, H., Gruffaz, C., Geoffroy, V., Meyer, J. M., et al. (2009). FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter. Mol. Microbiol. 72, 1246–1259. doi: 10.1111/j.1365-2958.2009.06721.x
Gupta, A., and Sar, P. (2019). Role of cost-effective organic carbon substrates in bioremediation of acid mine drainage-impacted soil of Malanjkhand Copper Project, India: a biostimulant for autochthonous microbial populations. Environ. Sci. Pollut. Res. 27, 27407–27421. doi: 10.1007/s11356-019-06293-6
Gupta, A., and Sar, P. (2020). “Treatment options for acid mine drainage: remedial achievements through microbial-mediated processes,” in Combined Application of Physico-Chemical & Microbiological Processes for Industrial Effluent Treatment Plant, eds M. Shah, and A. Banerjee (Singapore: Springer). doi: 10.1007/978-981-15-0497-6_8
Hantke, K. (1990). Dihydroxybenzolyserine-a siderophore for E. coli. FEMS Microbiol. Lett. 67, 5–8. doi: 10.1111/j.1574-6968.1990.tb13826.x
Hawes, M. C., Bengough, G., Cassab, G., and Ponce, G. (2002). Root caps and rhizosphere. J. Plant Growth Regul. 21, 352–367. doi: 10.1007/s00344-002-0035-y
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
He, Y., Pantigoso, H. A., Wu, Z., and Vivanco, J. M. (2019). Co-inoculation of Bacillus sp. and Pseudomonas putida at different development stages acts as a biostimulant to promote growth, yield and nutrient uptake of tomato. J. Appl. Microbiol. 127, 196–207. doi: 10.1111/jam.14273
Henríquez, T., Stein, N. V., and Jung, H. (2019). PvdRT-OpmQ and MdtABC-OpmB efflux systems are involved in pyoverdine secretion in Pseudomonas putida KT2440. Environ. Microbiol. Rep. 11, 98–106. doi: 10.1111/1758-2229.12708
Hesse, E., O'Brien, S., Tromas, N., Bayer, F., Luján, A. M., van Veen, E. M., et al. (2018). Ecological selection of siderophore-producing microbial taxa in response to heavy metal contamination. Ecol. Lett. 21, 117–127. doi: 10.1111/ele.12878
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Jain, D., Phurailatpam, L., and Mishra, S. (2020). Microbes-mediated mitigation of drought stress in plants: recent trends and future challenges. Adv. Plant Microbio. Sustain. Agricult. 199–218. doi: 10.1007/978-981-15-3204-7_9
Janni, M., Gull,ì, M., Maestri, E., Marmiroli, M., Valliyodan, B., Nguyen, H. T., et al. (2020). Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J. Exp. Bot. 71, 3780–3802. doi: 10.1093/jxb/eraa034
Kanaly, R. A., and Harayama, S. (2000). Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182, 2059–2067. doi: 10.1128/JB.182.8.2059-2067.2000
Keshavarz Zarjani, J., Aliasgharzad, N., Oustan, S., Emadi, M., and Ahmadi, A. (2013). Isolation and characterization of potassium solubilizing bacteria in some Iranian soils. Arch. Agronomy Soil Sci. 59, 1713–1723. doi: 10.1080/03650340.2012.756977
Khatoon, Z., Huang, S., Rafique, M., Fakhar, A., Kamran, M. A., and Santoyo, G. (2020). Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manage. 273, 111–118. doi: 10.1016/j.jenvman.2020.111118
Klebba, P. E. (2016). ROSET model of TonB action in gram-negative bacterial iron acquisition. J. Bacteriol. 198, 1013–1021. doi: 10.1128/JB.00823-15
Kostka, J. E., Stucki, J. W., Nealson, K. H., and Wu, J. (1996). Reduction of structural Fe (III) in smectite by a pure culture of Shewanella putrefaciens strain MR-1. Clays Clay Miner. 44, 522–529.
Kramer, J., Ozkaya, O., and Kummerli, R. (2020). Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 18, 152–163. doi: 10.1038/s41579-019-0284-4
Kumar, A., Singh, S., Gaurav, A. K., Srivastava, S., and Verma, J. P. (2020). Plant growth-promoting bacteria: Biological tools for the mitigation of salinity stress in plants. Front. Microbiol. 11:1216. doi: 10.3389/fmicb.2020.01216
Kumar, A., and Verma, J. P. (2018). Does plant-microbe interaction confer stress tolerance in plants: a review? Microbiol. Res. 207, 41–52. doi: 10.1016/j.micres.2017.11.004
Kumar, P., Thakur, S., Dhingra, G. K., Singh, A., Pal, M. K., Harshvardhan, K., et al. (2018). Inoculation of siderophore producing rhizobacteria and their consortium for growth enhancement of wheat plant. Biocatal. Agric. Biotechnol. 15, 264–269. doi: 10.1016/j.bcab.2018.06.019
Kumar, U., Shahid, M., Tripathi, R., Mohanty, S., Kumar, A., Bhattacharyya, P., et al. (2017). Variation of functional diversity of soil microbial community in sub-humid tropical rice-rice cropping system under long-term organic and inorganic fertilization. Ecol. Indic. 73, 536–543. doi: 10.1016/j.ecolind.2016.10.014
Ladeiro, B. (2012). Saline agriculture in the 21st century: using salt contaminated resources to cope food requirements. J. Bot. 2012, 1–7. doi: 10.1155/2012/310705
Lareen, A., Burton, F., and Schafer, P. (2016). Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 90, 575–587. doi: 10.1007/s11103-015-0417-8
Lehmann, J., Bossio, D. A., Kögel-Knabner, I., and Rillig, M. C. (2020). The concept and future prospects of soil health. Nat. Rev. Earth Environ. 1, 544–553. doi: 10.1038/s43017-020-0080-8
Lei, Z. H. A. O., and Zhang, Y. Q. (2015). Effects of phosphate solubilization and phytohormone production of Trichoderma asperellum Q1 on promoting cucumber growth under salt stress. J. Integr. Agric. 14, 1588–1597. doi: 10.1016/S2095-3119(14)60966-7
Li, S., Li, X., and Li, F. (2018). Fe (II) oxidation and nitrate reduction by a denitrifying bacterium, Pseudomonas stutzeri LS-2, isolated from paddy soil. J. Soils Sediments 18, 1668–1678. doi: 10.1007/s11368-017-1883-1
Li, X., Mou, S., Chen, Y., Liu, T., Dong, J., and Li, F. (2019). Microaerobic Fe (II) oxidation coupled to carbon assimilation processes driven by microbes from paddy soil. Sci. China Earth Sci. 62, 1719–1729. doi: 10.1007/s11430-018-9329-3
Liu, T., Chen, D., Li, X., and Li, F. (2019). Microbially mediated coupling of nitrate reduction and Fe (II) oxidation under anoxic conditions. FEMS Microbiol. Ecol. 95:fiz030. doi: 10.1093/femsec/fiz030
Lovley, D. R., Holmes, D. E., and Nevin, K. P. (2004). Dissimilatory fe (iii) and mn (iv) reduction. Adv. Microb. Physiol. 49, 219–286. doi: 10.1016/S0065-2911(04)49005-5
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Ma, Y., Prasad, M. N. V., Rajkumar, M., and Freitas, H. (2011). Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 29, 248–258. doi: 10.1016/j.biotechadv.2010.12.001
Majeed, A., Muhammad, Z., and Ahmad, H. (2018). Plant growth promoting bacteria: role in soil improvement, abiotic and biotic stress management of crops. Plant Cell Rep. 37, 1599–1609. doi: 10.1007/s00299-018-2341-2
Mohapatra, B., Bose, H., Saha, A., and Sar, P. (2020). Arsenic toxicity amelioration in rice soils by plant beneficial microbes. ORYZA-An Int. J. Rice 57, 70–78. doi: 10.35709/ory.2020.57.1.9
Mohapatra, B., and Phale, P. S. (2021). Microbial degradation of naphthalene and substituted naphthalenes: metabolic diversity and genomic insight for bioremediation. Front. Bioeng. Biotechnol. 9:144. doi: 10.3389/fbioe.2021.602445
Naraian, R., and Kumari, S. (2018). “Microbial production of organic acids,” in: Microbial Functional Foods and Nutraceuticals, eds V. K. Gupta, H. Treichel, V. Shapaval, L. A. de Oliveira, and M. G. Tuohy (New York, NY: John Wiley & Sons Ltd.) doi: 10.1002/9781119048961.ch5
Numan, M., Bashir, S., Khan, Y., Mumtaz, R., Shinwari, Z. K., Khan, A. L., et al. (2018). Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol. Res. 209, 21–32. doi: 10.1016/j.micres.2018.02.003
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi: 10.1007/s00253-018-9556-6
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33, 1–16. doi: 10.1007/s11274-017-2364-9
Oldroyd, G. E., and Leyser, O. (2020). A plant's diet, surviving in a variable nutrient environment. Science 368:6486. doi: 10.1126/science.aba0196
Olenska, E., Małek, W., Wójcik, M., Swiecicka, I., Thijs, S., and Vangronsveld, J. (2020). Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 743:140682. doi: 10.1016/j.scitotenv.2020.140682
Oves, M., Khan, M. S., and Zaidi, A. (2013). Chromium reducing and plant growth promoting novel strain Pseudomonas aeruginosa OSG41 enhance chickpea growth in chromium amended soils. Eur. J. Soil Biol. 56, 72–83. doi: 10.1016/j.ejsobi.2013.02.002
Patel, P., Trivedi, G., and Saraf, M. (2018). Iron biofortification in mungbean using siderophore producing plant growth promoting bacteria. Environ. Sustainabil. 1, 357–365. doi: 10.1007/s42398-018-00031-3
Pathma, J., Kennedy, R. K., and Sakthivel, N. (2011). “Mechanisms of fluorescent pseudomonads that mediate biological control of phytopathogens and plant growth promotion of crop plants,” in: Bacteria in Agrobiology: Plant Growth Responses, ed D. K. Maheshwari (Berlin: Springer). doi: 10.1007/978-3-642-20332-9_4
Pattnaik, S., Mohapatra, B., Kumar, U., Pattnaik, M., and Samantaray, D. (2019). “Microbe-mediated plant growth promotion: a mechanistic overview on cultivable plant growth-promoting members,” In: Biofertilizers for Sustainable Agriculture and Environment, eds B. Giri, R. Prasad, Q.-S. Wu, and A. Varma (Cham: Springer). doi: 10.1007/978-3-030-18933-4_20
Prajapati, K., Sharma, M. C., and Modi, H. A. (2013). Growth promoting effect of potassium-solubilizing microorganisms on okra (Abelmoscus Esculantus). Int. J. Agricult. Sci. Res. 1, 181–188. Available online at: http://s-o-i.org/1.15/ijarbs-2016-3-5-24
Qu, Q., Zhang, Z., Peijnenburg, W. J. G. M., Liu, W., Lu, T., Hu, B., et al. (2020). Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 68, 5024–5038. doi: 10.1021/acs.jafc.0c00073
Raaijmakers, J. M., and Mazzola, M. (2012). Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 50, 403–424. doi: 10.1146/annurev-phyto-081211-172908
Rajkumar, M., and Freitas, H. (2008). Influence of metal resistant-plant growth-promoting bacteria on the growth of Ricinus communis in soil contaminated with heavy metals. Chemosphere 71, 834–842. doi: 10.1016/j.chemosphere.2007.11.038
Razaq, M., Zhang, P., and Shen, H. L. (2017). Influence of nitrogen and phosphorous on the growth and root morphology of Acer mono. PLoS ONE 12:e0171321. doi: 10.1371/journal.pone.0171321
Rilling, J. I., Acuna, J. J., Nannipieri, P., Cassan, F., Maruyama, F., and Jorquera, M. A. (2019). Current opinion and perspectives on the methods for tracking and monitoring plant growth-promoting bacteria. Soil Biol. Biochem. 130, 205–219. doi: 10.1016/j.soilbio.2018.12.012
Rodriguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rout, G. R., and Sahoo, S. (2015). Role of iron in plant growth and metabolism. Rev. Agricult. Sci. 3, 1–24. doi: 10.7831/ras.3.1
Rout, M. E., and Southworth, D. (2013). The root microbiome influences scales from molecules to ecosystems: the unseen majority1. Am. J. Bot. 100, 1689–1691. doi: 10.3732/ajb.1300291
Safdarian, M., Askari, H., Shariati, V., and Nematzadeh, G. (2019). Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-018-38398-2
Saha, M., Sarkar, S., Sarkar, B., Sharma, B. K., Bhattacharjee, S., and Tribedi, P. (2016). Microbial siderophores and their potential applications: a review. Environ. Sci. Pollut. Res. 23, 3984–3999. doi: 10.1007/s11356-015-4294-0
Sansinenea, E. (2019). “Bacillus spp.: As plant growth-promoting bacteria,” in: Secondary Metabolites of Plant Growth Promoting Rhizo-Microorganisms, eds H. B. Singh, C. Keswani, M. S. Reddy, E. S. Royano, and C. García-Estrada (Singapore: Springer). doi: 10.1007/978-981-13-5862-3_11
Santoyo, G., Urtis-Flores, C. A., Loeza-Lara, P. D., Orozco-Mosqueda, M., and Glick, B. R. (2021). Rhizosphere colonization determinants by Plant Growth-Promoting Rhizobacteria (PGPR). Biology 10:475. doi: 10.3390/biology10060475
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Sattar, A., Naveed, M., Ali, M., Zahir, Z. A., Nadeem, S. M., Yaseen, M., et al. (2019). Perspectives of potassium solubilizing microbes in sustainable food production system: A review. Appl. Soil Ecol. 133, 146–159. doi: 10.1016/j.apsoil.2018.09.012
Schalk, I. J. (2008). Metal trafficking via siderophores in Gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. J. Inorg. Biochem. 102, 1159–1169. doi: 10.1016/j.jinorgbio.2007.11.017
Schalk, I. J., Mislin, G. L., and Brillet, K. (2012). Structure, function and binding selectivity and stereoselectivity of siderophore-iron outer membrane transporters. Curr. Top. Membr. 69, 37–66. doi: 10.1016/B978-0-12-394390-3.00002-1
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2, 1–14. doi: 10.1186/2193-1801-2-587
Shelobolina, E., Xu, H., Konishi, H., Kukkadapu, R., Wu, T., Blothe, M., et al. (2012). Microbial lithotrophic oxidation of structural Fe(II) in biotite. Appl. Environ. Microbiol. 78, 5746–5752. doi: 10.1128/AEM.01034-12
Smith, D. L., Gravel, V., and Yergeau, E. (2017). Signalling in the Phytomicrobiome. Front. Plant Sci. 8:611. doi: 10.3389/fpls.2017.00611
Srivastava, S., and Srivastava, S. (2020). Prescience of endogenous regulation in Arabidopsis thaliana by Pseudomonas putida MTCC 5279 under phosphate starved salinity stress condition. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-62725-1
Suleman, M., Yasmin, S., Rasul, M., Yahya, M., Atta, B. M., and Mirza, M. S. (2018). Phosphate solubilizing bacteria with glucose dehydrogenase gene for phosphorus uptake and beneficial effects on wheat. PLoS ONE 13:e0204408. doi: 10.1371/journal.pone.0204408
Tang, H. Y., Holmes, D. E., Ueki, T., Palacios, P. A., and Lovley, D. R. (2019). Iron corrosion via direct metal-microbe electron transfer. MBio 10, e00303–e00319. doi: 10.1128/mBio.00303-19
Tardieu, F., and Tuberosa, R. (2010). Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 13, 206–212. doi: 10.1016/j.pbi.2009.12.012
Trapet, P., Avoscan, L., Klinguer, A., Pateyron, S., Citerne, S., Chervin, C., et al. (2016). The Pseudomonas fluorescens siderophore pyoverdine weakens Arabidopsis thaliana defense in favor of growth in iron-deficient conditions. Plant Physiol. 171, 675–693. doi: 10.1104/pp.15.01537
Uroz, S., Calvaruso, C., Turpault, M.-P., and Frey-Klett, P. (2009). Mineral weathering by bacteria: ecology, actors and mechanisms. Trends Microbiol. 17, 378–387. doi: 10.1016/j.tim.2009.05.004
Valeeva, L. R., Nyamsuren, C., Sharipova, M. R., and Shakirov, E. V. (2018). Heterologous expression of secreted bacterial BPP and HAP phytases in plants stimulates Arabidopsis thaliana growth on phytate. Front. Plant Sci. 9:186. doi: 10.3389/fpls.2018.00186
Vaughan, M. M., Block, A., Christensen, S. A., Allen, L. H., and Schmelz, E. A. (2018). The effects of climate change associated abiotic stresses on maize phytochemical defenses. Phytochem. Rev. 17, 37–49. doi: 10.1007/s11101-017-9508-2
Verbon, E. H., Trapet, P. L., Stringlis, I. A., Kruijs, S., Bakker, P. A., and Pieterse, C. M. (2017). Iron and immunity. Annu. Rev. Phytopathol. 55, 355–375. doi: 10.1146/annurev-phyto-080516-035537
Verma, D. K., Pandey, A. K., Mohapatra, B., Srivastava, S., Kumar, V., Talukdar, D., et al. (2019a). “Plant growth-promoting rhizobacteria: An eco-friendly approach for sustainable agriculture and improved crop production,” in Microbiology for Sustainable Agriculture, Soil Health, and Environmental Protection, ed D. K. Verma (Apple Academic Press). doi: 10.1201/9781351247061-1
Verma, M., Mishra, J., and Arora, N. K. (2019b). “Plant growth-promoting rhizobacteria: diversity and applications,” in Environmental Biotechnology: For Sustainable Future, eds R. C. Sobti, N. K. Arora, and R. Kothari (Singapore: Springer). doi: 10.1007/978-981-10-7284-0_6
Visca, P., Imperi, F., and Lamont, I. L. (2007). Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15, 22–30. doi: 10.1016/j.tim.2006.11.004
Xiao, C., Guo, S., Wang, Q., and Chi, R. (2021). Enhanced reduction of lead bioavailability in phosphate mining wasteland soil by a phosphate-solubilizing strain of Pseudomonas sp., LA, coupled with ryegrass (Lolium perenne L.) and sonchus (Sonchus oleraceus L.). Environ. Pollut. 274:116572. doi: 10.1016/j.envpol.2021.116572
Yu, S., Teng, C., Bai, X., Liang, J., Song, T., Dong, L., et al. (2017). Optimization of siderophore production by Bacillus sp. PZ-1 and its potential enhancement of phytoextration of Pb from soil. J. Microbiol. Biotechnol. 27, 1500–1512. doi: 10.4014/jmb.1705.05021
Zeng, Q., Wu, X., Wang, J., and Ding, X. (2017). Phosphate solubilization and gene expression of phosphate-solubilizing bacterium Burkholderia multivorans WS-FJ9 under different levels of soluble phosphate. J. Microbiol. Biotechnol. 27, 844–855. doi: 10.4014/jmb.1611.11057
Zeng, X., Liu, X., Tang, J., Hu, S., Jiang, P., Li, W., et al. (2012). Characterization and Potassium-solubilizing ability of Bacillus circulans Z1-3. Adv. Sci. Lett. 10, 173–176. doi: 10.1166/asl.2012.3726
Zhang, C., and Kong, F. (2014). Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Agric. Ecosyst. Environ,. Appl. Soil Ecol. 82, 18–25. doi: 10.1016/j.apsoil.2014.05.002
Zhang, X., Zhang, D., Sun, W., and Wang, T. (2019). The adaptive mechanism of plants to iron deficiency via iron uptake, transport, and homeostasis. Int. J. Mol. Sci. 20:2424. doi: 10.3390/ijms20102424
Zhang, Y., Ren, J., Wang, W., Chen, B., Li, E., and Chen, S. (2020). Siderophore and indolic acid production by Paenibacillus triticisoli BJ-18 and their plant growth-promoting and antimicrobe abilities. PeerJ 8:e9403. doi: 10.7717/peerj.9403
Zhou, Y., Coventry, D. R., Gupta, V. V., Fuentes, D., Merchant, A., Kaiser, B. N., et al. (2020). The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Biol. 21, 1–19. doi: 10.1186/s13059-020-01999-0
Zhu, J. K. (2016). Abiotic stress signalling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: PGP microbes, soil-plant-microbe interaction, nutrient acquisition, siderophere-producing rhizobacteria, phosphate solubilisation, abiotic stress, crop productivity
Citation: Pattnaik S, Mohapatra B and Gupta A (2021) Plant Growth-Promoting Microbe Mediated Uptake of Essential Nutrients (Fe, P, K) for Crop Stress Management: Microbe–Soil–Plant Continuum. Front. Agron. 3:689972. doi: 10.3389/fagro.2021.689972
Received: 01 April 2021; Accepted: 30 June 2021;
Published: 09 August 2021.
Edited by:
Muhammad Arslan Ashraf, Government College University, Faisalabad, PakistanReviewed by:
Mehar Fatma, Aligarh Muslim University, IndiaBharati Kollah, Indian Institute of Soil Science (ICAR), India
Copyright © 2021 Pattnaik, Mohapatra and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Swati Pattnaik, cGF0dG5haWtzd2F0aTRAZ21haWwuY29t; Balaram Mohapatra, YmFsYXJhbW1vaGFwYXRyYTA5QGdtYWlsLmNvbQ==
 Swati Pattnaik
Swati Pattnaik Balaram Mohapatra
Balaram Mohapatra Abhishek Gupta
Abhishek Gupta