
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron., 20 October 2021
Sec. Disease Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.676820
This article is part of the Research TopicNew Trends in Integrated Plant Disease ManagementView all 5 articles
 Ghulam Rasool1,2*†
Ghulam Rasool1,2*† Sumaira Yousaf1,3
Sumaira Yousaf1,3 Um e Ammara1,4
Um e Ammara1,4 Adnan Iqbal5
Adnan Iqbal5 Muhammad Saeed1,6
Muhammad Saeed1,6 Imran Amin1
Imran Amin1 Shahid Mansoor1
Shahid Mansoor1Cotton (Gossypium hirsutum L.) is the most essential fiber crop and is widely cultivated within the tropical regions of the world. It has a major impact on socio-economical get-together in Pakistan. Unfortunately, it has been tormented by cotton leaf curl disease (CLCuD) that is produced by distinct species of begomoviruses (Family; Geminiviridae) like Cotton leaf curl Kokhran virus-Burewala strain (CLCuKoV-Bu) associated with specific betasatellite—Cotton leaf curl Multan betasatellite (CLCuMuB). Host resistance against CLCuD has not been achieved yet. The present study employed the use of synthetic genes considering the phenomenon of pathogen derived resistance (PDR). The coat protein (CP) and replication associated protein (Rep) genes sequence from CLCuKoV-Bu were codon optimized, synthesized and used to control CLCuKoV-Bu and associated betasatellite related to CLCuD. The Nicotiana benthamiana plants were Agrobacterium mediated transformed with synthetic CP (CPsyn) and synthetic Rep (Repsyn) under the control of Cauliflower mosaic virus 35S promoter. The transgenic plants harboring CPsyn and Repsyn genes were infiltrated with CLCuKoV-Bu alone and CLCuKoV-Bu- CLCuMuB inoculums showed relatively milder symptoms as compared with wild sorts and low virus concentration as checked by southern hybridization. The results confirmed that CPsyn, Repsyn genes may be utilized for the resistance of CLCuKoV-Bu and associated betasatellites related to CLCuD and PDR mechanism may also be developed for the control of CLCuD.
The cotton leaf curl disease (CLCuD), a disease complex associated with diverse begomoviruses, is the most important limiting factor for the production of cotton across the sub-continent. Initially CLCuD was ignored as a major threat until it affected cotton production as during the first 3 years of the epidemics the disease incidence and severity were very high in the Punjab. About 97,580 ha were affected resulting in a loss of 543,294 bales of cotton during 1992–93 in Punjab (Amin et al., 2010). Since 2001 the resistance breaking “Burewala” strain of Cotton leaf curl Kokhran virus (CLCuKoV-Bu) has spread to all cotton producing areas of Pakistan (Mansoor et al., 2003a) which is still dominating.
During the past few decades the development of geminivirus resistant germplasm remains a challenge. The natural resistance of host plant is the most effective means of reducing the losses caused by these phytopathogens, however the lack of resistant germplasm has encouraged the interest of scientific community in the development of transgenic resistance. Pathogen derived resistance (PDR) is broadly divided into two classes depending upon prevailing molecular mechanisms protein mediated resistance (PMR) and nucleic acid-mediated resistance (RMR) (Smith et al., 2000; Niu et al., 2006). The PMR generally gives broad spectrum resistance whereas RMR offers highly specific resistance. The endeavors to control geminiviruses by PDR are not highly efficient as for RNA viruses suggestive of less susceptibility of these ssDNA viruses to the resistance mechanism (Lucioli et al., 2008). There is only one report of transgenic immunity against geminiviruses (Aragao and Faria, 2009) hence; exploring novel transgenic resistance approaches is highly desirable.
The effects of mutation in the coat protein (CP) gene was studied by Iqbal et al. (2012) by the introduction of premature stop codon which results the premature termination of translation of CP gene. The transient expression of CLCuKoV CP gene under the control of Cauliflower mosaic virus 35S promoter (35S-KoCP), in Nicotiana benthamiana plants was studied and in small number of plants viral DNA could be detected by PCR in tissues distal to the inoculation site.
The Cotton Leaf Curl Kokhran Virus-Dabawali recombinant coat protein (rCP) has conserved motif composed of C68, C72, H81, and H85 residues. The mutation in the H85A residue resulted in ~756-fold reduced the association rate and 3-fold decreased DNA binding affinity (Priyadarshini and Savithri, 2009). After the achievement of CP-mediated resistance (CPMR) it had greatly been developed for many other RNA viruses e.g., tobamoviruses (having CP closely related to TMV; Masuta et al., 1995; Bendahmane et al., 2007). It was also proposed that regulation of viral replication by CP is correlated to the relative strength of CPMR (Bendahmane et al., 2007). Different levels of protection by CPMR indicates the involvement of different mechanisms, broad-spectrum resistance/higher-level resistance was attributed to the co-existence PMR as well as RMR mediated interference (Prins et al., 2008).
First Rep protein-mediated resistance (RPMR) was achieved against TMV expressing TMV Rep gene sequence in transgenic tobacco (Golemboski et al., 1990). Mutations in conserved motifs of Rep acted as trans-dominant negative inhibitors of BGMV (Hanson and Maxwell, 1999) and similarly mutations in NTP-binding domain of Rep decreased ACMV but a high level of transgene expression was required to develop this resistance (Sangaré et al., 1999). The use of mutated and truncated Rep gene, resistance was successfully engineered to an African maize pathogen, Maize streak virus (MSV) in Digitaria sanguinalis (Shepherd et al., 2007a,b). The complete/partial sequence of Rep has been used by researchers to increase resistance in plants (Hanley-Bowdoin et al., 1990; Noris et al., 1996; Brunetti et al., 2001; Lucioli et al., 2003; Antignus et al., 2004). The 210amino acid (aa) from the N-terminal of Rep gene as (Rep210) and 130 aa as (Rep130) were used to develop Rep based resistance, however results exhibited that only 130 aa were sufficient to act as trans-dominant negative inhibitor (Lucioli et al., 2003).
As a consequence of viral attack, the pathogen derived transgenes which offer PMR may not work and counterbalanced in a short time due to post transcriptional gene silencing (PTGS)or virus induced gene silencing (VIGS) of the transgene (Lucioli et al., 2008). Furthermore, the modifications by reassortment and recombination of virus sequences (Padidam et al., 1999) put gene silencing (GS) and PDR together and therefore PDR which is a sequence specific mechanism may soon be compromised. To overcome the drawback of GS some other strategies like synthetic gene and non-pathogen derived resistance (NPDR) (Lucioli et al., 2008; Shepherd et al., 2009) are good options to evaluate against begomoviruses.
To overcome the phenomenon VIGS the silent point mutations were introduced in CP and truncated Rep130 gene sequence in a way that the protein sequence of CP and Rep130 was not disturbed. Considering these facts, the synthetic CP (CPsyn) and synthetic Rep130 (Repsyn130) were designed to check resistance against CLCuD by overcoming VIGS. The transient expression of CPsyn and Repsyn130 in Nicotiana benthamiana plants showed reduced symptoms and virus titer (Yousaf et al., 2013; Rasool et al., 2016b). The present study deals with the evaluation of transgenic resistance against CLCuKoV-Bu in N. benthamiana plants and assesses the resistance responses of CPsyn and Repsyn130 as well as resistance variations of independent transgenic lines.
The coat protein (CP) sequence of CLCuKoV-Bu was assembled and compared with the (accession number HF549183) CPs of other begomoviruses in the database, using BLAST (Mullineaux et al., 1988). The detailed mechanism of point mutations introduced in the CP sequence has been mentioned in Rasool et al. (2016a). The designed mutated CP construct was codon optimized for Gossypium hirsutum (http://www.kazusa.or.jp/codon) and synthesized commercially (GenScript, USA). The synthesized codon optimized CP (CPsyn) was cloned between SalI and EcoRI sites in pJIT60 vector under the control of cauliflower mosaic virus (CaMV) 35S promoter. The expression cassette of CPsyn (2.2 kb approx.) contained double CaMV 35S promoter and CaMV 35S terminator. The expression cassette along with promoter/terminator was digested and cloned in the binary vector pGreen0029 at KpnI and XhoI sites.
The 130 aa of Rep protein of (CLCuKoV-Bu: FR8197) was assembled and compared with the Rep of other begomoviruses in the database, using BLAST (Mullineaux et al., 1988). The detailed mechanism of point mutations introduced in the Rep sequence has been previously described. The designed Rep construct was codon optimized for Gossypium hirsutum (http://www.kazusa.or.jp/codon) and synthesized commercially (GenScript, USA) (Yousaf et al., 2013). The synthesized codon optimized Rep gene (Repsyn130) was cloned at HindIII and EcoRI restriction sites in plant expression vector pJIT60 under the control of the 35S Cauliflower mosaic virus (CaMV) promoter (Guerineau and Mullineaux, 1993). The Repsyn130 expression cassette was cloned in plant transformation vector pGreen0029 at KpnI and XhoI restriction sites (Hellens et al., 2000).
N. benthamiana was separately transformed with CPsyn and Repsyn130 expression cassette by the Agrobacterium tumefaciens-mediated leaf disc method (Horsch et al., 1985). All the primary transformants developed from tissue culture was confirmed by PCR using CaMV 35S promoter primers. These primary transformants were labeled as T0. All the T0 transgenic lines were phenotypically normal and kept in insect free glasshouse till flowering stage. The seeds were collected and germinated on MS medium supplemented with 500 mg/l kanamycin to get T1 generation.
Infectious clone CLCKoV-Bu/CLCuMuB (Mansoor et al., 2003b; Saeed et al., 2005) was cultured in LB medium along with appropriate antibiotics. The culture was centrifuged (4,000 rpm) for 15 min to harvest the cells, separately. The cells were resuspended in 10 mM MgCl2, 10 mM MES (pH 5.6) and 100 μM Acetosyringone was added in the culture after adjusting its OD600 to 0.6–1. The culture was left overnight or at least 2–3 h at room temperature before infiltration. Transgenic N. benthamiana plants at 5–6 leaves stage were chosen for agroinfiltration. The Agrobacterium suspension was infiltrated by 1 ml needleless syringe on the underside of 2–3 leaves. Plants were not watered 24 h before inoculation. After infiltration, plants were kept at 26–28°C with a 16 h photoperiod in an insect-free glasshouse and with 65% relative humidity.
Agro-inoculated transgenic plants were observed for percentage of symptoms severity, incidence of disease and systemic movement of CLCKoV-Bu/CLCuMuB to evaluate the resistance responses of two transgenic lines each of CPsyn and Repsyn130. The leaf samples were used to extract total genomic DNA by using the CTAB method (Doyle and Doyle, 1990) and quantified by Nanodrop spectrophotometer (NanoDrop 2000; Thermo Fisher Scientific, USA). The presence of inoculated CLCKoV-Bu was checked by PCR using primer set of CLCV1 and CLCV2 (Shahid et al., 2007) whilst CLCuMuB presence was confirmed by using primers β01 and β02 (Briddon et al., 2002; Saeed et al., 2005).
DNA samples were resolved in 1% agarose gels in 0.5X TAE buffer and blotted on nylon membranes (Roche GmbH, Germany). For the detection of virus replication and its systemic movement in the transgenic leaves were checked by Southern hybridization analysis using DIG labeled specific probes of CLCKoV-Bu DNA and CLCuKB DNA according to the manufacturer instructions (RocheGmbH, Germany).
In transgenic lines, for the detection of virus, 1,100-bp fragment of CP gene of CLCKoV-Bu was used. It was PCR amplified by usingCLCV1 and CLCV2 (Shahid et al., 2007) primers and labeled with digoxigenin using a PCR DIG Probe Synthesis Kit (Roche GmbH, Germany) as described by the manufacturer. For the detection of betasatellite, 380-bp fragment was amplified by using pair of β01 and β02 primers (Briddon et al., 2002; Saeed et al., 2005) and was labeled with digoxigenin using a DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche GmbH, Germany).
The PCR analysis of all putative (primary transformants) transgenic plants raised from tissue culture suggested 20 plants to be transformed with each CPsyn and Repsyn130 Constructs. However, the PCR analysis with CPsyn and Repsyn130 gene-specific primers showed only 13 and 11 plants to be transformed. All the primary transformants (T0) transgenic lines were phenotypically normal, allowed to grow till flowering stage and seeds were collected. The seeds (T1) of all the transgenic lines were germinated on MS growth medium supplemented with 500 mg/l kanamycin. On the selection medium, four lines of CPsyn (CP-2 and CP-4) and Repsyn130 (R1 and R2) constructs showed 3:1 of segregation pattern which is suggestive of a single insertion (data not shown). The seeds of all putative and singly inserted transformed CPsyn and Repsyn130 were collected and evaluated for resistance.
The resistance evaluation of CPsyn transgenic N. benthamiana plants was observed in two lines (CP-2 and CP-4) at T2 generation. These lines were checked for resistance against the infectious clone of CLCKoV-Bu and CLCKoV-Bu/CLCuMuB. Twenty plants (confirmed by PCR for the presence of CPsyn) of each line and wild-type healthy N. benthamiana were agro-inoculated and kept under observation for 50 dpi. The periodic development of phenotypic symptoms was observed at three different intervals (7, 14, and 21 dpi). All the replicates of both CP-2 and CP-4 transgenic lines were remained asymptomatic at 7 dpi, however very mild symptoms (VMS) started to appear at 14 dpi (Table 1). The majority of replicates showed VMS at 21 dpi and few remained asymptomatic, the severe symptoms (SS) were observed in 4/20 in CP-2 replicates and 6/20 in CP-4 replicates (Table 1). The phenotypic data showed that the majority of CP-2 and CP-4 replicates were able to silence the CLCKoV-Bu along with the CLCuMuB. The data for evaluation of the resistance is presented in Table 2. Resistant phenotypes and symptom development as shown in Figure 1, panel were observed for all CPsyn transgenic replicates at the 21 dpi. The CP-2 and CP-4 transgenic lines replicates were inoculated with CLCuKoV-Bu alone showed mild to asymptomatic viral symptoms (12/20 and 10/20) whereas, the transgenic replicates (12/20 and 10/20) inoculated with CLCuKoV-Bu/CLCuMuB displayed mild to asymptomatic viral symptoms (Figure 1; Table 2). The PCR analysis showed the presence of CLCuKoV-Bu in all samples of CLCuKoV-Bu inoculated CP-2 and CP-4 transgenic replicates. Nevertheless, the transgenic replicates of both CP-2 and CP-4 transgenic lines showed the presence of both virus and betasatellite in all DNA samples when inoculated with of CLCuKoV-Bu/CLCuMuB (Table 2). The Southern blot analysis verified that when CP-2 and CP-4 transgenic replicates were inoculated with CLCuKoV-Bu showed moderate to lower level of CLCuKoV-Bu, respectively (Figure 2; Table 2). The same replicates when inoculated with CLCuKoV-Bu/CLCuMuB showed moderate to high level of CLCuKoV-Bu/CLCuMuB accumulation as compared to positive control plants by Southern analysis (Figure 2; Table 2).
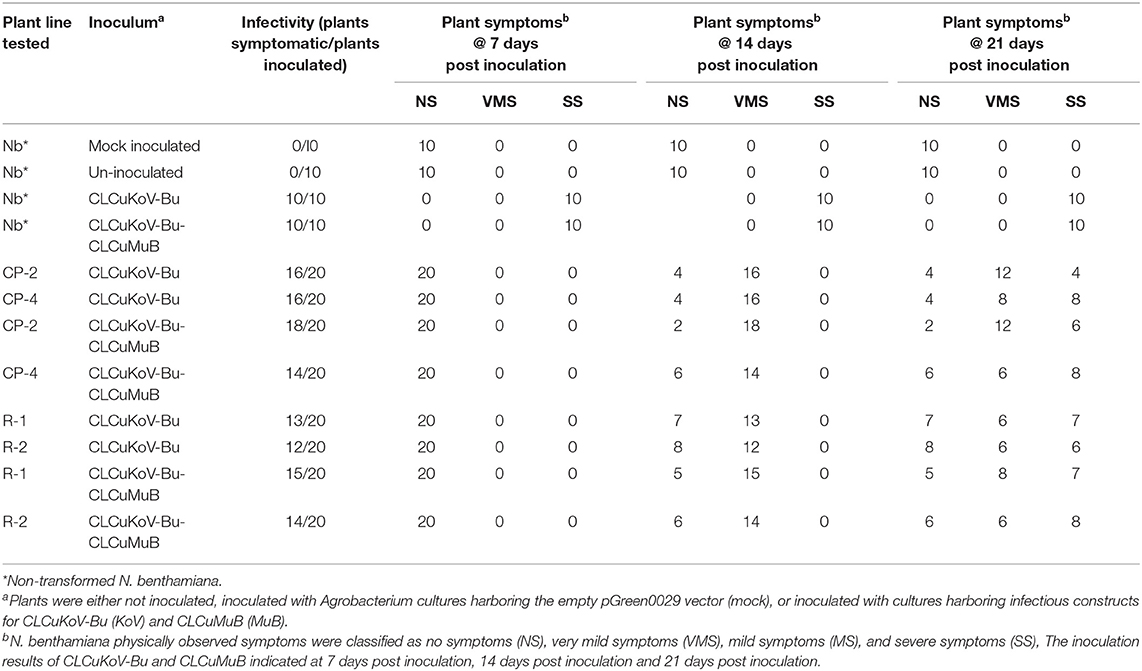
Table 1. Transgene response of transgenic plants expressing CPsyn and Repsyn130 at days post inoculations.
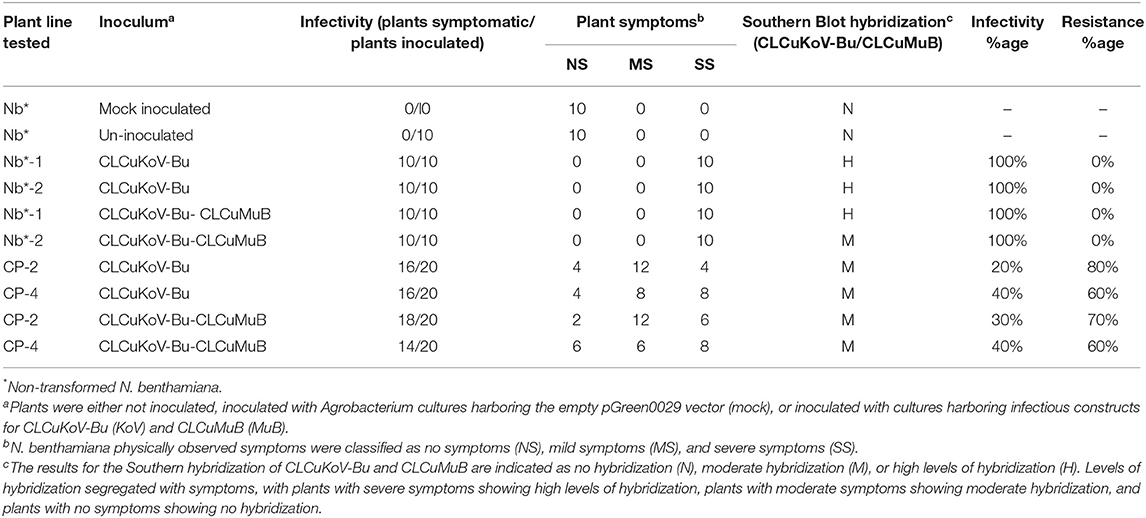
Table 2. Transgene response of transgenic plants expressing CPsyn to challenge with CLCuKoV-Bu/CLCuMuB.
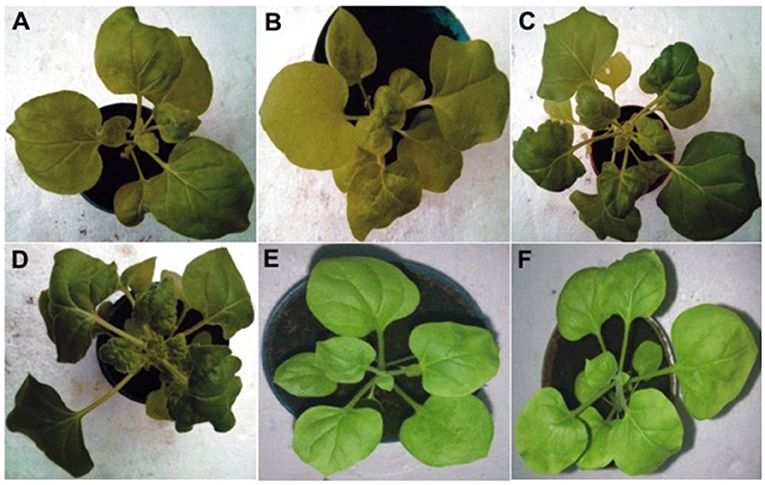
Figure 1. The photographs represents the phenotypic symptoms observed in the agro-inoculated two CPsyn transgenic N. benthamianalines compared with control plants. The CP-2 transgenic line was inoculated with CLCuKoV-Bu (A) and CLCuKoV-Bu/CLCuMuB showed mild leaf curl symptoms (B). The CP-4 line was inoculated with CLCuKoV-Bu (C) and inoculated with CLCuKoV-Bu/CLCuMuB showed severe symptoms (D). For mock inoculation the non-transgenic N. benthamiana plants were either inoculated with the empty pGreen0029 vector (E) or not inoculated (F). All the pictures were taken at 25 dpi.
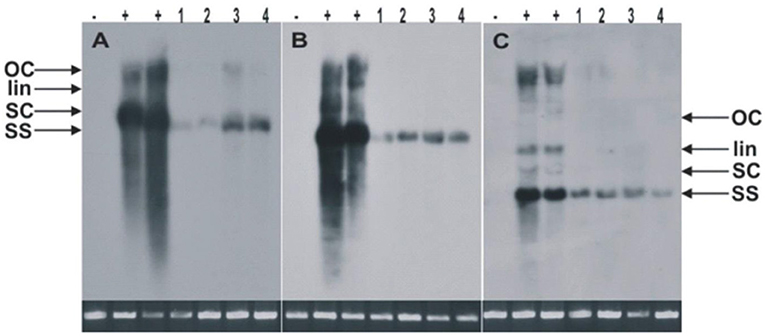
Figure 2. The Southern blots for the detection of CLCuKoV-Bu alone (A) or with the CLCuKo-Bu/CLCuMuB (B,C). The total genomic DNA samples were extracted from two transgenic replicates of CP-2 transgenic line (line 1 and 2 as indicated), and two replicates from CP-4 line as indicated in (line 3 and 4) for the expression of CPsyn. For all the blots the one negative uninoculated control plant in (- lanes), and two positive non-transgenic symptomatic control plants in (+ lanes). All the DNA samples were extracted from leaves at 21–25 dpi and ~10 μg of total genomic DNA was used in each case. The position of all replicative forms of DNA like single-stranded (ss), supercoiled (sc), linear (lin) and open circular (OC) have shown in all three blots. In each case, a photograph of the ethidium bromide stained genomic DNA bands on the agarose gel are shown below the blot to confirm.
The resistance evaluation of Repsyn130 transgenic plants was performed on two lines (R1 and R2) at T2 generation. Resistance against CLCKoV-Bu/CLCuMuB was checked on twenty plants (confirmed by PCR for the presence of Repsyn130) of each line and ten plants of wild-type healthy N. benthamiana. These plants were inoculated by CLCKoV-Bu and CLCKoV-Bu/CLCuMuB complex and kept under observation for 50 dpi. The data for evaluation of the resistance potential was recorded at three different intervals (7, 14, and 21 dpi) and presented in Table 1. All the transgenic replicates of lines R1 and R2 remained asymptomatic at 7 dpi, however the development of VMS was observed at 14 dpi. The severity in the symptom development was observed in 7/20 in R1 line and 6/20 in R2 line. The overall effect of CLCuMuB on the development of disease severity symptom was not very significant as 8/20 replicates developed SS whereas 6/20 exhibited SS without CLCuMuB. Resistant phenotypes as shown in Figures 3C,D were observed for Repsyn130 transgenic plants. The transgenic replicates 7/20 plants of R1 and 8/20 plants of R2, no symptoms were observed and these plants remained symptomless even at 50 dpi as well as till senescence (Table 3). The southern blot shown in Figure 4 is representing the titer of CLCuKoV-Bu in nine plants of R1 (lane 2–11) and 8 plants of R2 (lane 12–19). The lane 1 and 20 are representing the DNA from negative and positive control plants, respectively. The R1 and R2 transgenic replicates inoculated with CLCuKoV-Bu alone exhibited mild to asymptomatic viral symptoms (6/20 and 8/20) while plants (8/20 and 7/20) inoculated with CLCuKoV-Bu/CLCuMuB exhibited mild to asymptomatic viral symptoms (Figure 3; Table 3). A considerable decrease in the titer of CLCuKoV-Bu DNA was observed in both R1 and R2 lines compared to control (Figure 4 lane 20). All plants of R1 and four plants of line R2 also showed decrease in the level of CLCuKoV-Bu DNA.

Figure 3. The photographs represents the phenotypic symptoms in agro-inoculated Repsyn130transgenic N. benthamiana compared with control plants and were taken at 21 dpi. In (A) the transgenic N. benthamiana line R1 and in (B) R2 transgenic line inoculated with CLCKoV-Bu. The plant shown in (C,D) are R1 and R2 Repsyn130 transgenic plants, respectively, coinoculated with CLCKoV-Bu/CLCuMuB. (E,F) are the non-transgenic N. benthamiana plants were either not inoculated or with the empty pGreen0029 vector, respectively.
Table 3. Transgene response of transgenic plants expressing Repsyn130 to challenge with CLCuKoV-Bu/CLCuMuB.
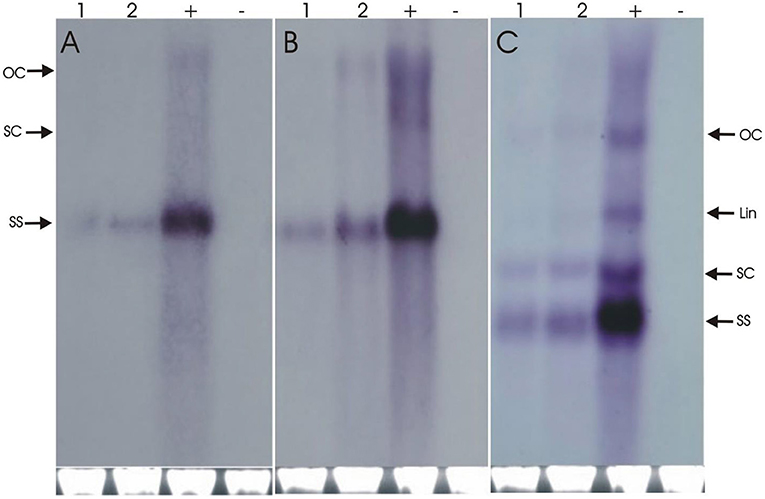
Figure 4. The Southern blot analysis for the detection of CLCuKoV-Bu alone (A) and CLCuKo-Bu/CLCuMuB (B,C). The total DNA was extracted from three transgenic replicates from R1 transgenic line in (lane R1-R3). For all three blots one negative uninoculated control plant in (H lanes), and one positive non-transgenic symptomatic control plants in (P lanes). The position of all replicative forms of DNA like single-stranded (ss), supercoiled (sc), linear (lin) and open circular (OC) have shown in all three blots. In each case, a photograph of the ethidium bromide stained genomic DNA bands on the agarose gel are shown below the blot to confirm equal loading of DNA samples.
The engineered resistance approaches are potentially offering an alternative and rapid method for the transfer of resistant genes to the traditional cultivars (Vanderschuren et al., 2007; Loriato et al., 2020). For engineered resistance the CP gene was selected in this study due to its function in assembly/dis-assembly of viral DNA during plant cell infection. The constitutive expression of CP elevated its levels that may protect the plant from secondary infection by the same virus (Powell et al., 1990). The similar mechanism of inhibition or interference may be involved in this study as the resistance response against CLCuKoV-Bu and CLCuMuB was observed. The resistance observed in transgenic plants supports the previous finding of Kunik et al. (1994) and Raj et al. (2005) who extended the use of CPMR from RNA to DNA viruses (geminiviruses). Similarly, codon optimized synthetic CP and RNAi construct has been used to develop resistance against Tomato yellow leaf curl virus Oman (TYLCV-OM) (Ammara et al., 2015, 2020). The CPsyn used in this study showed maximum aa sequence homology with different begomovirus strains associated with CLCuD complex. These results indicate that CPsynnt sequences have minimized the chances for VIGS (Rasool et al., 2016a,b). The resistance evaluation of transgenic N. benthamiana plants expressing CPsyn against CLCuKoV-Bu/CLCuMuB showed mild to delayed symptoms. The transgenic plants inoculated with CLCuKoV-Bu exhibited milder symptoms than the plants coinoculated with CLCuKoV-Bu and CLCuMuB. The southern hybridization results showed increased hybridization in plants coinoculated with CLCuKoV-Bu and CLCuMuB as compared to plants inoculated with CLCuKoV-Bu (Figures 1A–D). There has been many reports describing the role of betasatellite in relation to increased virus titer (Saunders et al., 2000). Some replicates when phenotypically observed displayed milder symptoms while some were asymptomatic (Table 1). These results have indicated that the presence of CPsyn have significantly reduced virus DNA accumulation however, the virus spread in the systemic leave was not reduced as shown by southern blot analysis (Figure 2).
The multifunctional property of Rep makes it well-studied for developing resistance against homologous as well as heterologous viruses and the mechanism of its action has also been described (Brunetti et al., 2001; Lucioli et al., 2003; Shivaprasad et al., 2006). However, it did not provide durable and enhanced resistance to DNA viruses as to RNA viruses (Ribeiro et al., 2007) because of GS. The Repsyn130 used in this study encompasses all the essential domains and its multifunctional property related with the oligomerization nature (Orozco et al., 2000).
The transgenic N. benthamiana plants constitutively expressing Repsyn130 controlled the accumulation of homologous CLCuKoV-Bu as well as its associated CLCuMuB by trans-dominant inhibition. Several reports of trans-dominant negative inhibition of both animal (Smith and Adeluca, 1992) and plant viruses particularly geminiviruses (Hou et al., 2000; Chatterji et al., 2001) have been described. The resistance response of transgenic N. benthamiana plants expressing Repsyn130 against CLCuKoV-Bu showed mild to delayed symptoms (Figure 3) as compared to plants coinoculated with CLCuKoV-Bu/CLCuMuB. However, the plants not showing the resistant phenotype may be due to some differences in the gene expression. The observed resistance is likely to be achieved as the expression of Repsyn130 might have acted as trans-dominant negative inhibitor and stopped the replication of attacking CLCuKoV-Bu as well as CLCuMuB. As the replication of CLCuMuB is dependent on helper begomovirus thus its titer was also reduced. Different studies had indicated that RPMR is positively correlated to the levels of transgene expression (Noris et al., 1996; Lucioli et al., 2003) whereas in some cases although a correlation between transcript level and resistance existed (Hong and Stanley, 1995).
Transgenic N. benthamiana harboring Repsyn130 showed that the gene has no toxic effect when constitutively expressed and do not cause any developmental abnormalities that are earlier described for complete Rep genes of several begomoviruses during plant transformation (Hong and Stanley, 1996). The Rep proteins are highly important and relatively conserved in begomoviruses, moreover due to codon optimization and introduction of point mutations Repsyn130 is viewed as a good candidate of trans-dominant negative inhibition of CLCuD. However, tomato plants expressing TYLCSV synthetic Rep have revealed some transcriptional changes similar to stress-related responses showing that this domain has interfered with viral infections (Lucioli et al., 2014). Resistance response of stably transformed Repsyn130 transgenic N. benthamiana plants was analogous to the transiently expressed Repsyn130 as previously described by the authors. Since synthetic Rep circumvented GS of the transgene on the attack of TYLCSV (Lucioli et al., 2008) in the same way this study confirmed the functioning of truncated Repsyn130 against CLCuKo-Bu. The resistance is correlated with the presence of transgene although in some cases the presence of the transgene was also associated with the infected plants expressing usual but mild symptoms e.g., some replicates of transgenic lines R1 and R2 showing tolerant behavior (Table 2) that might be due to differential expression of the transgene.
It may be predicted that CPsyn and Repsyn130 constructs can be used to achieve resistance against other viruses associated with CLCuD and its associated betasatellites. Moreover, further studies are required to check the combined resistance potential of both genes, CPsyn and Repsyn130, by gene staking or pyramiding that could offer broad spectrum resistance. Consequently, the synthetic gene approach alone, or in combination with other PDR or non-pathogen derived (NPDR) strategies, may provide effective control strategy for begomoviruses.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
This research article is a part of the Ph.D. thesis of GR and SY. SM and MS conceived the study and designed the experiments. SY and GR performed the experiments. SY, GR, UA, and AI analyzed the data. UA and AI wrote the first draft of the manuscript which was edited by SY and GR. All authors read and approved the final manuscript.
This work was supported by the Indigenous Ph.D. Fellowship Programme of the Higher Education Commission, Government of Pakistan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amin, I., Ilyas, M., Mansoor, S., Briddon, R. W., and Saeed, M. (2010). Emerging Geminiviral Diseases and their Management, eds P. Sharma, R. K. Gaur and M. Ikegami (Nova Science Publishers, Inc.), 1–36.
Ammara, U. E., Mansoor, S., Saeed, M., and Al-Sadi, A. M. (2020). Trans-dominant interference by synthetic coat protein of tomato yellow leaf curl virus expressed in transgenic tomato. Int. J. Aric. Bio. 6, 1513–1519. doi: 10.17957/IJAB/15.1589
Ammara, U. E., Mansoor, S., Saeed, M., Amin, I., Briddon, R. W., and Al-Sadi, A. M. (2015). RNA interference-based resistance in transgenic tomato plants against tomato yellow leaf curl virus-Oman (TYLCV-OM) and its associated betasatellite. Virol. J. 12, 38–49. doi: 10.1186/s12985-015-0263-y
Antignus, Y. R., Vunsh, R., Lachman, O., Pearlsman, M., Maslenin, L., et al. (2004). Truncated Rep gene originated from Tomato yellow leaf curl virus-Israel [Mild] confers strain-specific resistance in transgenic tomato. Ann. Appl. Biol. 144, 39–44. doi: 10.1111/j.1744-7348.2004.tb00314.x
Aragao, F. J. L., and Faria, J. C. (2009). First transgenic geminivirus-resistant plant in the field. Nat. Biotechnol. 27, 1086–1088. doi: 10.1038/nbt1209-1086
Bendahmane, M., Chen, I., Asurmendi, S., Bazzini, A. A., Szecsi, J., and Beachy, R. N. (2007). Coat protein-mediated resistance to TMV infection of Nicotiana tabacum involves multiple modes of interference by coat protein. Virol. J. 366, 107–116. doi: 10.1016/j.virol.2007.03.052
Briddon, R. W., Bull, S. E., Mansoor, S., Amin, I., and Markham, P. G. (2002). Universal primers for the PCR-mediated amplification of DNA β; a satellite associated with some monopartite begomoviruses. Mol. Biotechnol. 20, 315–318. doi: 10.1385/MB:20:3:315
Brunetti, A., Tavazza, R., Noris, E., Lucioli, A., Accotto, G. P., and Tavazza, N. (2001). Transgenically expressed T-Rep of tomato yellow leaf curl Sardinia virus acts as a trans-dominant-negative mutant, inhibiting viral transcription and replication. J. Virol. 75, 10573–10581. doi: 10.1128/JVI.75.22.10573-10581.2001
Chatterji, A., Beachy, R. N., and Fauquet, C. M. (2001). Expression of the oligomerization domain of the replication-associated protein (Rep) of Tomato leaf curl New Delhi virus interferes with DNA accumulation of heterologous geminiviruses. J. Biol. Chem. 276, 25631–25638. doi: 10.1074/jbc.M100030200
Golemboski, D. B., Lomonossoff, G. P., and Zaitlin, M. (1990). Plants transformed with a tobacco mosaic virus nonstructural gene sequence are resistant to the virus. Proc. Natl. Acad. Sci. U. S. A. 87, 6311–6315. doi: 10.1073/pnas.87.16.6311
Guerineau, F., and Mullineaux, P. (1993). “Plant transformation and expression vectors,” in Plant Molecular Biology Labfax, ed R. R. D. Croy (Oxford: Bios Scientific Publishers), 121–147.
Hanley-Bowdoin, L., Elmer, J. S., and Rogers, S. G. (1990). Expression of functional replication protein from tomato golden mosaic virus in transgenic tobacco plants. Proc. Natl. Acad. Sci. U. S. A. 87, 1446–1450. doi: 10.1073/pnas.87.4.1446
Hanson, S. F., and Maxwell, D. P. (1999). trans-Dominant inhibition of geminiviral DNA replication by bean golden mosaic geminivirus rep gene mutants. Phytopathology 89, 480–486. doi: 10.1094/PHYTO.1999.89.6.480
Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S., and Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/A:1006496308160
Hong, J., and Stanley, J. (1996). Virus resistance in Nicotiana benthamiana conferred by African cassava mosaic virus replication-associated protein (AC1) transgene. Mol. Plant Microbe Interact. 4, 219–225. doi: 10.1094/MPMI-9-0219
Hong, Y., and Stanley, J. (1995). Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J. Gen. Virol. 76, 2415–2422. doi: 10.1099/0022-1317-76-10-2415
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hou, Y. M., Sanders, R., Ursin, V. M., and Gilbertson, R. L. (2000). Transgenic plants expressing geminivirus movement proteins: abnormal phenotypes and delayed infection by Tomato mottle virus in transgenic tomatoes expressing the Bean dwarf mosaic virus BV1 or BC1 proteins. Mol. Plant Microbe Interact. 13, 297–308. doi: 10.1094/MPMI.2000.13.3.297
Iqbal, Z., Sattar, M. N., Kvarnheden, A., Mansoor, S., and Briddon, R. W. (2012). Effects of the mutation of selected genes of cotton leaf curl Kokhran virus on infectivity, symptoms and the maintenance of cotton leaf curl Multan betasatellite. Virus Res. 169, 107–116. doi: 10.1016/j.virusres.2012.07.016
Kunik, T., Salomon, R., Zamir, D., Navot, N., Zeidan, M., Michelson, I., et al. (1994). Transgenic tomato plants expressing the tomato yellow leaf curl virus capsid protein are resistant to the virus. Biotechnology 12, 500–504. doi: 10.1038/nbt0594-500
Loriato, V. A., Martins, L. G., Euclydes, N. C., Reis, P. A., Duarte, C. E., and Fontes, E. P. (2020). Engineering resistance against geminiviruses: a review of suppressed natural defenses and the use of RNAi and the CRISPR/Cas system. Plant Sci. 292:110410. doi: 10.1016/j.plantsci.2020.110410
Lucioli, A., Berardi, A., Gatti, F., Tavazza, R., Pizzichini, D., and Tavazza, M. (2014). Tomato yellow leaf curl Sardinia virus-resistant tomato plants expressing the multifunctional N-terminal domain of the replication-associated protein show transcriptional changes resembling stress-related responses. Mol. Plant Pathol. 15, 31–43. doi: 10.1111/mpp.12063
Lucioli, A., Noris, E., Brunetti, A., Tavazza, R., Ruzza, V., Castillo, A. G., et al. (2003). Tomato yellow leaf curl Sardinia virus Rep-derived resistance to homologous and heterologous geminiviruses occurs by different mechanisms and is overcome if virus-mediated transgene silencing is activated. J. Virol. 77, 6785–6798. doi: 10.1128/JVI.77.12.6785-6798.2003
Lucioli, A., Sallustio, D. E., Barboni, D., Beradni, A., Papacchioli, V., Tavazza, R., et al. (2008). A cautionary note on pathogen derived sequences. Nat. Biotechnol. 26, 617–619. doi: 10.1038/nbt0608-617
Mansoor, S., Amin, I., Iram, S., Hussain, M., Zafar, Y., Malik, K. A., et al. (2003a). The breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. New Dis. Rep. 52:784. doi: 10.1111/j.1365-3059.2003.00893.x
Mansoor, S., Briddon, R. W., Bull, S. E., Bedford, I. D., Bashir, A., Hussain, M., et al. (2003b). Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA b. Arch. Virol. 148, 1969–1986. doi: 10.1007/s00705-003-0149-y
Masuta, C., Tanaka, H., Uehara, K., Kuwata, S., Koiwai, A., and Noma, M. (1995). Broad resistance to plant viruses in transgenic plants conferred by antisense inhibition of a host gene essential in S-adenosylmethionine-dependent transmethylation reactions. Proc. Natl. Acad. Sci. U. S. A. 2:6117. doi: 10.1073/pnas.92.13.6117
Mullineaux, P. M., Boulton, M. I., Bowyer, P., van der Vlugt, R., Marks, M., Donson, J., et al. (1988). Detection of a non-structural protein of Mr 11000 encoded by the virion DNA of maize streak virus. Plant Mol. Biol. 11, 57–66. doi: 10.1007/BF00016014
Niu, Q. W., Lin, S. S., Reyes, J. L., Chen, K. C., Wu, H. W., Yeh, S. D., et al. (2006). Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24, 1420–1428. doi: 10.1038/nbt1255
Noris, E., Jupin, I., Accotto, G. P., and Gronenborn, B. (1996). DNA-binding activity of the C2 protein of tomato yellow leaf curl geminivirus. Virology 217, 607–612. doi: 10.1006/viro.1996.0157
Orozco, B. M., Kong, L. J., Batts, L. A., Elledge, S., and Hanley-Bowdoin, L. (2000). The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275, 6114–6122. doi: 10.1074/jbc.275.9.6114
Padidam, M., Sawyer, S., and Fauquet, C. M. (1999). Possible emergence of new geminiviruses by frequent recombination. Virology 265, 218–225. doi: 10.1006/viro.1999.0056
Powell, A. P., Sanders, P. R., Tumer, N., Fraley, R. T., and Beachy, R. N. (1990). Protection against tobacco mosaic virus infection in transgenic plants requires accumulation of coat protein rather than coat protein RNA sequences. Virol J. 175, 124–130. doi: 10.1016/0042-6822(90)90192-T
Prins, M., Laimer, M., Noris, E., Schubert, J., Wassenegger, M., and Tepfer, M. (2008). Strategies for antiviral resistance in transgenic plants. Mol. Plant Pathol. 9, 73–83. doi: 10.1111/j.1364-3703.2007.00447.x
Priyadarshini, P. C., and Savithri, H. S. (2009). Kinetics of interaction of Cotton Leaf Curl Kokhran Virus-Dabawali (CLCuKV-Dab) coat protein and its mutants with ssDNA. Virology 386, 427–437. doi: 10.1016/j.virol.2009.01.016
Raj, S. K., Singh, R., Pandey, S. K., and Singh, B. P. (2005). Agrobacterium-mediated tomato transformation and regeneration of transgenic lines expressing tomato leaf curl virus coat protein gene for resistance against TLCV infection. Current Sci. 88, 1674–1679. Available online at: https://www.jstor.org/stable/24110495
Rasool, G., Yousaf, S., Akram, A., Mansoor, S., Briddon, R. W., and Saeed, M. (2016a). G5, a phage single-stranded DNA-binding protein, fused with a nuclear localization signal, attenuates symptoms and reduces begomovirus-betasatellite accumulation in transgenic plants. Mol. Biotechnol. 58, 595–602. doi: 10.1007/s12033-016-9959-5
Rasool, G., Yousaf, S., Mansoor, S., Briddon, R. W., and Saeed, M. (2016b). Transient expression of synthetic coat protein gene of Cotton leaf curl Burewala virus in tobacco (Nicotiana benthamiana). J. Agric. Res. 54, 21–34. Available online at: https://apply.jar.punjab.gov.pk/upload/1461838105_121_PAPER_NO._3.pdf
Ribeiro, S. G., Lohuis, H., Goldbach, R., and Prins, M. (2007). Tomato chlorotic mottle virus is a target of RNA silencing but the presence of specific short interfering RNAs does not guarantee resistance in transgenic plants. J. Virol. 81, 1563–1573. doi: 10.1128/JVI.01238-06
Saeed, M., Behjatania, S. A. A., Mansoor, S., Zafar, Y., Hasnain, S., and Rezaian, M. A. (2005). A Single complementrary-sense transcript 0f a geminiviral DNA β satellite is determinant of pathogenicity. Mol. Plant Microbe Interact. 18, 7–14. doi: 10.1094/MPMI-18-0007
Sangaré, A., Deng, D., Fauquet, C. M., and Beachy, R. N. (1999). Resistance to African cassava mosaic virus conferred by a mutant of the putative NTP-binding domain of the Rep Gene (AC1) in Nicotiana benthamiana. Mol. Plant Breed. 5, 95–102. doi: 10.1023/A:1009611403565
Saunders, K., Bedford, I. D., Briddon, R. W., Markham, P. G., Wong, S. M., and Stanley, J. (2000). A novel virus complex causes Ageratum yellow vein disease. Proc. Natl. Acad. Sci. U. S. A. 97, 6890–6895. doi: 10.1073/pnas.97.12.6890
Shahid, M. S., Mansoor, S., and Briddon, R. W. (2007). Complete nucleotide sequences of cotton leaf curl Rajasthan virus and its associated DNA beta molecule infecting tomato. Arch. Virol. 152, 2131–2134. doi: 10.1007/s00705-007-1043-9
Shepherd, D. N., Mangwende, T., Martin, D. P., Bezuidenhout, M., Kloppers, F. J., Carolissen, C. H., et al. (2007a). Maize streak virus-resistant transgenic maize: a first for Africa. Plant Biotechnol. J. 5, 759–767. doi: 10.1111/j.1467-7652.2007.00279.x
Shepherd, D. N., Mangwende, T., Martin, D. P., Bezuidenhout, M., Thomson, J. A., and Rybicki, E. P. (2007b). Inhibition of maize streak virus (MSV) replication by transient and transgenic expression of MSV replication-associated protein mutants. J. Gen. Virol. 88, 325–336. doi: 10.1099/vir.0.82338-0
Shepherd, D. N., Martin, D. P., and Thomson, J. A. (2009). Transgenic strategies for developing crops resistant to geminiviruses. Plant Sci. 176, 1–11. doi: 10.1016/j.plantsci.2008.08.011
Shivaprasad, P. V., Thillaichidambaram, P., Balaji, V., and Veluthambi, K. (2006). Expression of full-length and truncated Rep genes from Mungbean yellow mosaic virus-Vigna inhibits viral replication in transgenic tobacco. Virus Genes 33, 365–374. doi: 10.1007/s11262-006-0077-5
Smith, C. A., and Adeluca, N. (1992). Transdominant inhibition of herpes simplex virus growth in transgenic mice. Virol. J. 191, 581–588. doi: 10.1016/0042-6822(92)90233-F
Smith, N. A., P, S. S., Wang, M. B., Stoutjesdijk, P. A., Green, A. G., and Waterhouse, P. M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. doi: 10.1038/35030305
Vanderschuren, H., Stupak, M., Fütterer, J., Gruissem, W., and Zhang, P. (2007). Engineering resistance to geminiviruses – review and perspectives. Plant Biotechnol. J. 5, 207–220. doi: 10.1111/j.1467-7652.2006.00217.x
Yousaf, S., Rasool, G., Amin, I., Mansoor, S., and Saeed, M. (2013). Inference of a synthetic Rep protein to develop resistance against cotton leaf curl disease. Int. J. Agric. Biol. 15, 1140–1144. Available online at: http://www.fspublishers.org/published_papers/55250_.pdf
Keywords: cotton leaf curl disease, cotton leaf curl Kokhran virus-Burewala strain, cotton leaf curl Multan betasatellite, synthetic coat protein, synthetic replication associated protein, transgenic resistance
Citation: Rasool G, Yousaf S, Ammara Ue, Iqbal A, Saeed M, Amin I and Mansoor S (2021) Transgenic Expression of Synthetic Coat Protein and Synthetic Replication Associated Protein Produces Mild Symptoms and Reduce Begomovirus-Betasatellite Accumulation in Nicotiana benthamiana. Front. Agron. 3:676820. doi: 10.3389/fagro.2021.676820
Received: 06 March 2021; Accepted: 15 September 2021;
Published: 20 October 2021.
Edited by:
Tariq Mukhtar, Pir Mehr Ali Shah Arid Agriculture University, PakistanReviewed by:
Fasiha Qurashi, The Islamia University of Bahawalpu, PakistanCopyright © 2021 Rasool, Yousaf, Ammara, Iqbal, Saeed, Amin and Mansoor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ghulam Rasool, Z2h1bGFtcmFzb29sMzEyNkBnbWFpbC5jb21s
†Present address: Ghulam Rasool, Institute of Molecular Biology and Biotechnology (IMBB), Centre for Research in Molecular Medicine (CRIMM), University of Lahore, Lahore, Pakistan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.