- 1Department of Plant Pathology, University of Agriculture Faisalabad, Faisalabad, Pakistan
- 2Plant Protection Division, Nuclear Institute for Agriculture and Biology, Faisalabad, Pakistan
Brown leaf spot disease of rice is a dominant lethal disease, caused by the fungus Bipolaris oryzae. The pathogen is an obligate parasite and causes qualitative and quantitative damage to rice crop. The objective of the present study was to investigate what extent the defense related biochemical compounds reflect the distinct categories of resistance phenotypes in rice against brown spot disease. This was done by determining the concentration of Catalase (CAT), Phenylalanine ammonia-lyase (PAL), Polyphenol oxidase (PPO), Peroxidase (POD), and β-1,3-Glucanase enzymes in resistant, moderately resistant and susceptible rice genotypes. The disease resistant phenotypes in rice line (PARC-7) reflect the higher accumulation of CAT, PAL, PPO, POD, and β-1,3-Glucanase. The pattern of enzyme accumulation was similar in all resistant genotypes. The rice genotypes with moderately resistant phenotypes showed significant difference with respect to the concentration of biochemical defense-related compounds. The difference in accumulation of defense related enzymes reflected the level of disease severity (% leaf area covered) on resistant and moderately resistant genotypes. The susceptible rice genotypes showed the minimum concentration of these enzymes, with the lowest concentrations found in the rice variety Bas-2000 (80% Disease Index). The differential defense response in resistant and susceptible genotypes suggests that these enzymes can be used as biochemical markers for early detection of disease resistant genotypes. The study of enzyme accumulation at different time points and at different levels of disease severity helps to understand the resistance mechanisms against brown spot disease in rice.
Introduction
Rice (Oryza sativa) is a major food crop of Pakistan and staple food after wheat, and the third largest crop in terms of area sown (https://reliefweb.int/sites/reliefweb.int/files/resources/Pakistan_Food_Security_Bulletin_September_2016.pdf). Pakistani Basmati rice is famous and distinguished all around the world for its appearance and aroma, but unfortunately rice production is reduced because of the biotic and abiotic stresses. Among the biotic constraints, fungal and bacterial diseases play a vital role in damaging health of rice crops and lessening rice production. In Pakistan, reports reveal that there is a loss of $5 billion due to diseases of rice (Asghar et al., 2007). The three most important pathogens of rice are Pyricularia oryzae, Helminthosporium oryzae, and Xanthomonas oryzae, which causes blast, brown spot, and bacterial blight (Singh et al., 2013).
Brown leaf spot disease caused by Bipolaris oryzae is one of the distinctive and most damaging disease of rice in the world, because of the extensive distribution of numerous physiological races (Arshad et al., 2008). Brown spot disease is linked with many important epidemics in the world with the most destructive occurring in 1942 that resulted in the Great Bengal Famine. Reported yield losses vary broadly from 6 to 90% (Barnwal et al., 2013). The characteristic symptoms of brown spot on leaves and glumes include light reddish-brown lesions or lesions with a gray center surrounded by a dark to reddish-brown margin, and then by a bright yellow halo (Dallagnol et al., 2009). Spots have a distinctive shape and size on different parts of plant depending upon the environmental conditions (Don and Clayton, 2010). In adverse conditions, discoloration and poor grain filling occurs, resulting in a reduction of yield during the plant's reproductive stage (Huynh and Ashok, 2004). The symptoms are more apparent and severe when rice is sown in soil that is nutrient deficient (Nazari et al., 2015). The causal fungus Bipolaris oryzae remains in seeds for most of its lifecycle, but can also persist on infested rice stubble and straw. It spreads from plant to plant in the field by the airborne spores (Sato et al., 2008). Weeds that serve as alternative hosts and soils are important as sources of inoculum for this pathogen survival (Biswas et al., 2008).
Rice disease management strategies mainly relies on the deployment of host plant resistance and fungicides (Sarkar et al., 2014). Utilization of resistant varieties is the simple, harmless, effective, and the most economical approach for managing the brown spot disease. However, very few resistant cultivars are available for practical use (Biswas et al., 2011) and sometimes resistance is very unstable due to the presence of new and more virulent pathogen races (Katasntones et al., 2007). Therefore, it is necessary to develop and update resistant levels in each variety, every year and there is also a need to develop those strategies that will help in protecting the plant's resistance level over a longer period of time in a wide geographical area (Fengming and Robert, 2001). Host resistance is mediated by a complex network of molecular and biochemical events that determine a range between the susceptibility and resistance. Understanding of the host resistance mechanism and the biochemical interaction of host and pathogen plays a vital role in the identification of effective and durable resistance genotypes. Several defense-related enzymes are known to play a crucial role in the expression of rice disease resistance (Geetha et al., 2005). The most commonly studied defense compounds that act as first line of defense are mitogen activated protien kinases (MAPKs), peroxidase (POD), and phenylalanine ammonia lyase (PAL) (Eliahu et al., 2007). Together with the introduction and buildup of other pathogenesis-related (PR) proteins, like β-1,3-Glucanase (Kessmann et al., 1994) Catalase, Superoxide dismutases (Khan and Hemalatha, 2016), and PolyPhenol Oxidase (Canxing et al., 2013), these compounds contribute to systemic acquired resistance (SAR) by accumulating profusely at the site of infection. Most of these compounds accumulated in resistant rice genotypes under field conditions. However, the level and time of accumulation varied among different varieties. It is not well-known if the expression of specific defense compounds can act as biochemical marker for the detection of the resistance in diverse rice genotypes (Carella et al., 2019). Many previous studies investigated the role of defense related enzymes for understanding the resistance mechanisms (Sripriya et al., 2017; Minaeva et al., 2018; Aoki et al., 2020; Divya et al., 2020; Hoang Loc et al., 2020) but in Pakistan no such studies were carried out for the brown spot disease. The objective of the present study is to characterize the biochemical defense compounds with different levels of brown spot disease resistance in rice genotypes. The hypothesis investigated is to what extent the defense related compounds reflect specific phenotypes in rice against brown spot disease.
Materials and Methods
Isolation and Purification of Pathogen
The fungal pathogen of brown spot disease Bipolaris oryzae was isolated from the infected rice plant samples preserved in the rice pathology lab, Nuclear Institute for Agriculture and Biology (NIAB) Faisalabad. The infected samples were collected by the rice pathologist from different rice growing areas of Punjab during the previous surveys. For the isolation of pathogen from infected rice samples, Potato Dextrose Agar (potato starch 20 g, glucose 20 g, agar 20 g in a 1 L distilled water) media was prepared in 1 L flask. The solution was slowly shaken by hand and autoclaved at 121°C 15–20 psi for 15–20 min. The PDA solution was allowed to cool in a laminar flow cabinet for 24 h at room temperature and media was poured in it and kept for solidification. Small cuttings of infected leaf portions along with healthy rice leaves were made followed by the surface sterilization with 2% (v/v) solution of sodium hypochlorite (NaHCl) in three sterilized tubes. The leaves were soaked for 2 min in NaHCl in the first tube, followed by two washes in sterile distilled water in the second and third tubes. After drying on sterilized tissue paper, 3–4 leaf pieces were aseptically placed on PDA plates, and incubated at 30°C for 3–4 days. Patterns of fungal growth were observed on daily basis. For purification, sub-culturing was done. A minute portion of the greenish colonies was collected using a sterilized inoculating needle, placed on the center of the new solidified PDA plates and incubated for at 30°C. After 3–4 days, once the new colonies had appeared, slides were prepared from pure fungal colonies and mycelia and the shape of the spores were observed under microscope (Labomed, USA). The identifying features are the observation of gray to dark greenish gray mycelium showing fluffy and cottony growth, with septate gray-greenish hyphae. Conidia are brown, usually curved, widest at the middle and tapering to rounded ends.
Rice Germplasm and Their Screening Against B. oryzae
Seventy-three (73) diverse genotypes of rice were screened against brown spot disease in this study. Among these 50 genotypes were obtained from International Rice Research Institute (IRRI), 16 types of rice germplasm from Pakistan Agriculture Research Council (PARC), 7 commercial varieties (Basmati-Super, Bas-370, Bas-385, Bas-198, Bas-2000, Bas-Pak, Bas-515, and Basmati C-622) were used. All genotypes were sown in the disease screening nursery at NIAB, according to the standard evaluation system of IRRI (2014). Seeds of each germplasm were sown on a raised bed of 30 cm in a single row and after two test rows, a highly susceptible variety (Basmati 2000) was sown in a single row to facilitate disease spread.
Each test genotype was sown in three replications with 10 cm distance between rows. The disease susceptible spreader variety was also sown in two parallel rows around the nursery. Seeds were covered by spreading thin layer of farm yard manure (FYM) and overhead watering took place three times per day. After 3 weeks, inoculum of the fungus B. oryzae was sprayed on the test plants. Inoculum was prepared by growing the fungus on rice seeds under laboratory conditions. For this, 1 kg of healthy rice seeds was placed in polythene bags, autoclaved and moisturized with distilled water. The top of the polythene bag was tightened by placing wooden disks, enclosed by cotton (Supplementary Figure 1) and incubated for 24 h at room temperature. The following day, fungal disks of 7-day old colonies were taken and 5 disks (5 mm) were added into seed bags and incubated for 7 days at room temperature. After this time, 250 ml distilled water was added into the seed bags and mixed well with hands covered with gloves, and then the resulting suspension was filtered through muslin cloth into a beaker. A slide was prepared from the spore suspension for microscopic observation and confirmed the presence of B. oryzae spores in suspension. The concentration of spores was adjusted to 1 × 106 using a haemocytometer. The spore suspension was added to a sterilized hand sprayer and sprayed on each rice plant. Disease data was recorded after 3 weeks of inoculation according to the standard disease rating scale of IRRI (2014) (Table 1). Ten plants of each genotype were observed for disease assessment and mean was used for statistical analysis. Similarly, disease data of 10 plants per genotype in each bed was recorded.
Biochemical Characterization
Leaf samples (20–25 leaves) of the 10 rice genotypes having resistant and moderately resistant phenotypes were collected from the diseased screening nursery of NIAB. In the same way, leaf samples of 10 susceptible genotypes were also collected for their biochemical profiling and stored at −1°C. The same number of leaves were collected from each replicate and 0.1 g leaf tissue per genotype was used for biochemical profiling.
Determination of the total soluble protein was done by following Bradford (1976) method. Suspension preparation was completed by weighing 0.1 g chilled leaves and cut into small pieces for grinding. These samples were ground in a mortar and pestle with 2 ml (0.1M) Sodium Phosphate Buffer, pH 6.0, and centrifuged at 13,000 rpm for 10 min. Supernatant was taken and stored at −18°C until use and residues were discarded. Extract (100 μl) of enzyme was taken in each test tube along with 5 ml of Coomassie blue agent (100 mg). In another test tube, a blank was prepared by adding 5 ml of Coomassie blue agent along with 100 μl Sodium Phosphate Buffer. Mixtures in the test tubes were vortexed for 5 min, then absorbance was measured by spectrophotometer at 595 nm (Bradford Protein assay, USA). For the preparation of protein standard, 1 mg/mL Bovine serum albumin was dissolved in the distilled water and utilized as the stock solution. The specific protein concentration was determined in the standard solution by noting its absorbance at 595 nm through spectrophotometer.
Catalase (CAT) activity was determined according to the method of Zhang et al. (2007). The consumption of H2O2. Leaf tissue (0.1 g) was homogenized with 2 ml (0.1M) Sodium Phosphate Buffer, pH 6.0. The homogenate was centrifuge at 13,000 rpm for 10 min. Supernatant was taken and stored at −18°C until use, and residues were discarded. Samples (100 μl) were treated with the reaction mixture containing 200 ml of H2O2 along with 30 ml Buffer (Sodium Phosphate) having pH 6.0 at room temperature (25°C). In a cuvette, 1.9 ml of distilled water was added, along with 1 ml substrate and 100 μl sample. The change in the absorbance was measured after 1 min at 240 nm. Utilization of H2O2 was quantified by its molar extinction coefficient (36 mol/1 cm). The activity of CAT enzyme was presented as CAT units min−1 mg−1 of protein.
For the estimation of peroxidase (POD), the sample suspension was prepared by weighing 0.1 g chilled leaves, grinding them in mortar and pestle with 2 ml (0.1M) Sodium Phosphate Buffer, pH 6.0, then centrifuging at 13,000 rpm for 10 min. Supernatant was retained for analysis, and stored at −18°C. The activity was estimated by a method described by Chance and Maehly (1955). The substrate buffer was prepared by adding 2.8 ml of (50 mM) Sodium Phosphate Buffer, 800 μl (40 mM) H2O2, 200 μl guaiacol and 60 ml of distilled water. Substrate (3 ml) and 100 ul sample were placed in cuvettes and absorbance at 470 nm was measured before and after 1 min. The enzyme activity was described by a variation of 0.01 unit min−1.
The enzyme phenylalanine ammonia-lyase (PAL) activity was estimated following the Zucker (1965) method. Leaf tissue (0.1 g) of its weight was homogenized with 1 ml (0.1M) buffer (Sodium borate) having pH 8.8. The homogenate was centrifuge at 13,000 rpm for 10 min. Supernatant was taken and stored at −18°C until use and residues were discarded. The 62.5 μl enzyme extract and 800 μl of sodium borate buffer were added in each test tube, along with 700 μl of (12 mM) phenylalanine, and tubes were incubated at 40°C in a water bath for 1 h. To stop the reaction, 200 μl of 5N HCL was added. Then, 0.5 ml of Trans-cinnamic acid (TCA) (1M) was added and estimation of the light absorbance was done at 290 nm.
For the estimation of polyphenol oxidase (PPO), 0.1 g chilled leaves were ground in mortar and pestle with 2 ml of 0.1M Sodium Phosphate Buffer, pH 6.0 and centrifuged at 13,000 rpm for 10 min. Supernatant was retained and stored at −18°C, and residues were discarded. The activity of PPO was estimated by following procedure described by Mayer et al. (1965). In the cuvette 1.0 ml of 0.1M sodium phosphate buffer, pH 6.5 and 1.0 ml of (0.01M) L-tyrosine in HCl was also added along with 0.9 mL of distilled water. Then 100 μl of sample was added and the absorbance was measured at 280 nm. The change in absorbance was determined at 30 s for 2 min and activity of PPO was expressed in min−1g−1 of fresh tissue.
The activity of β-1,3-Glucanase enzyme was estimated by following the method described by Liang et al. (1995). Chilled leaves (0.1 g) were ground in a taken in mortar and pestle with 1 ml of 0.05M Sodium Acetate Buffer, pH 5.0 and centrifuged at 13,000 rpm for 10 min. Supernatant was stored at −18°C and residues were discarded. In each test tube the reaction mixture comprised 0.1 mL of 50 mM Sodium Acetate Buffer, pH 5.0, 62.5 μl of 4% laminarin substrate and 62.2 μl enzyme extract followed by the incubation at 40°C for 10 min. The reaction was paused by introduction of 375 μl of dinitro salicylic acid and the mixture was boiled for 5 min in water bath and 4.5 ml of distilled water was added. Enzyme activity was then measured at 500 nm.
Statistical Analysis
The mean of disease and enzyme data was subjected to one-way ANOVA through software of SAS statistical (SAS institute, 1990). Rice genotypes and their enzymes activity means were compared using Least Significant Difference (LSD) at 5% level of significance.
Results
Symptomatology, Microscopic Identification of B. oryzae
Rice leaves infected by the brown spot disease exhibited lesions appeared on leaves that were originally dark brown in color ranging to purple-brown, with a light brown to grayish center and margins on the leaves were reddish-brown (Figure 1a, Supplementary Figure 2). Some leaves also showed spots with a typical yellow halo around the spot. Gray to dark greenish-gray, septate mycelia of brown spot pathogen were observed on PDA media (Figure 1b). The observed conidia were dark brown to olive-brown, curved or straight, fusiform with 6–16 transverse cell walls (Figure 1c, Supplementary Figure 3).
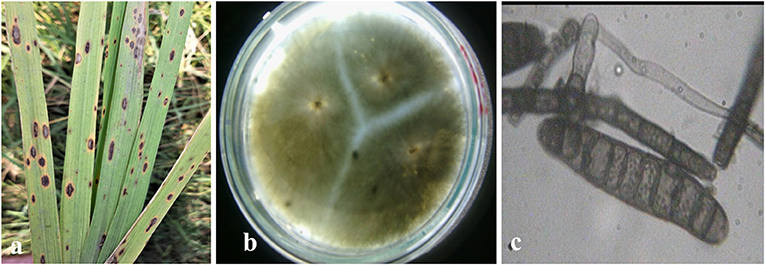
Figure 1. Symptomology and microscopic identification of Bipolaris oryzae the causal organism of brown spot disease in rice. Typical lesions appeared as dark brown in color with grayish center and margins and typical yellow halo around the spot on rice leaves (a). Isolation and purification of B. oryzae with gray to dark greenish septate mycelia observed on PDA media (b). Microscopic observation showed dark brown to olivaceous brown conidia, curved or straight, fusiform with 6–16 transverse cell walls (c).
Phenotypic Characterization of Rice Germplasm
The disease data revealed diverse phenotypic response of seventy-three (73) genotypes screened against the brown leaf spot disease (Table 2). None of the rice genotypes were found to be completely immune or highly resistant. Among the 50 genotypes developed by IRRI, only one resistant source (IRRI-43) was identified. However, 21 genotypes with moderately resistant phenotypes were found. The rest of the genotypes showed moderately susceptible to susceptible response (Table 2). Among the 16 genotypes from PARC, 4 genotypes were found to be resistant while five showed a moderately resistant response (Table 2). The screening of commercial varieties revealed the absence of resistance against brown spot disease. However, Basmati-Pak and Basmati-370 exhibited a moderately resistant phenotype. The spreader variety Basmati-2000 was highly susceptible with the disease index of 80.1%. Overall, the phenotypes of screened rice germplasm revealed the absence of resistance sources in exotic as well as local germplasm.
Biochemical Characterization of Disease Resistance
For biochemical characterization, five resistant genotypes, five moderately resistant (IRRI and PARC), and 10 susceptible genotypes were selected on the basis of their phenotypic response against the brown spot disease (Table 3). Defense related biochemical compounds were estimated and their expression was compared between resistant, moderately resistant and susceptible phenotypes (Table 3). The results showed that protein contents and defense related compounds were at maximum concentration in resistant genotypes, followed by the moderately resistant and susceptible rice lines. However, enzyme accumulation varied between the five resistant genotypes. The highest accumulation of total protein (2.45 mg/g.f.wt) and five defense related enzymes (CAT:1.72, POD:3.33, PAL:0.88, PPO: 13.30 and β-1,3-Glucanase: 3.70 U/mg protein) were recorded in resistant genotype PARC-7 followed by the PARC-13, PARC-8, PARC-1, and IRRI-43 (Table 3). All five genotypes showed a resistant phenotype, but had significantly different levels of the enzyme accumulation. The variation in the expression of defense enzymes suggest the induction of different biochemical pathways in each resistant genotype. Among the five moderately resistant genotypes, IRRI-11 showed highest enzyme accumulation (CAT:1.23, POD:1.70, PAL:0.78, PPO: 9.96, and β-1,3-Glucanase: 2.60 U/mg protein), followed by the PARC-14, IRRI-47, IRRI-36, and IRRI-27 (Table 3). The enzyme accumulation in all moderately resistant genotypes were significantly lower from the resistant lines. However, no significant difference in accumulation of CAT, POD, and β-1,3-Glucanase was observed between the IRRI-11, PARC-14, and IRRI-47 genotypes. The PAL and PPO accumulation were different in all five moderately resistant genotypes. Overall, the POD and PPO accumulation were ~2-fold lower in moderately resistant genotypes compared to the resistant lines (Table 3).
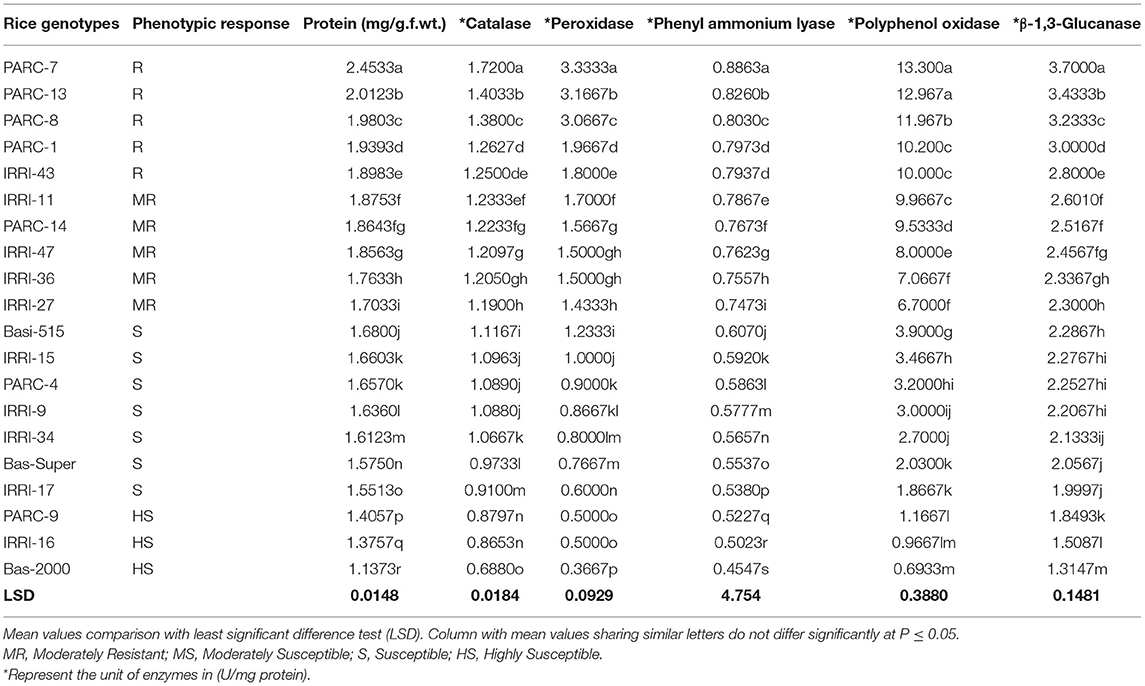
Table 3. Comparative analysis of total Protein and defense enzyme contents in disease resistant and susceptible rice germplasms.
The susceptible and highly susceptible genotypes of rice germplasm mostly showed minor difference in enzyme accumulation, with the maximum expression in Basmati-515 followed by IRRI-15, PARC-4, and IRRI-9. The lowest accumulation of defense related enzymes was observed in PARC-9, IRRI-16, and Basmati-2000, which expressed the highly susceptible phenotypes. The most significant difference between the susceptible and the resistant genotypes were reflected in the accumulation of POD and PPO enzymes. The POD accumulation in resistant genotype PARC-7 (3.33 U/mg protein) was 10-fold lower in the highly susceptible variety Basmati-2000 (0.366 U/mg protein) (Table 3). Likewise, the PPO expression was 20-fold lower in Basmati-2000 (0.69 U/mg protein) compared to the resistant genotype PARC-7 (13.30 U/mg protein). The accumulation of β-1,3-Glucanase was highest in the resistant genotype (2.8–3.7 U/mg protein) was reduced (2.3–2.6 U/mg protein) in moderately resistant genotypes and least expressed in highly susceptible rice variety Basmati-2000 (1.31 U/mg protein).
The biochemical profile of 20 rice genotypes clearly can be used to differentiate the resistant, moderately resistant, susceptible, and highly susceptible phenotypes. The pattern of total protein and five defense related enzymes showed a gradual decrease from resistant to highly susceptible phenotypes. However, individual enzyme accumulation varied with respect to the host genotype and its phenotypic response. The range of the enzyme accumulation representing resistant phenotypes was 1.25–1.72, 1.8–3.3, 0.79–0.80, 10.0–13.3, and 2.8–3.7 U/mg protein for catalase, Peroxidase, Phenyl ammonium lyase, Polyphenol oxidase, and β-1,3-Glucanase, respectively. The enzymes spectrum reflecting moderately resistant phenotypes was 1.23–1.19, 1.7–1.4, 0.78–0.74, 9.96–6.7, and 2.6–2.3 U/mg protein for catalase, Peroxidase, Phenyl ammonium lyase, Polyphenol oxidase, and β-1,3-Glucanase, respectively.
Discussion
The objective of the current study was the biochemical characterization of rice germplasm having resistant, moderately resistant and susceptible phenotypes against B. oryzae causing brown spot disease. It was achieved by determining the activity of five defense related enzymes at seedling stage after inoculation with prevailing population of B. oryzae. Increased activity of defense-related enzymes (CAT, PAL, PPO, POD, and beta-1,3-glucanase), correlated with higher levels of resistance to brown spot disease in rice cultivars, suggesting that these enzymes could be used as biochemical markers for early detection of disease-resistant genotypes. In addition, resistant and moderately resistant genotypes were also identified among the local and exotic rice germplasm against B.oryzae.
Brown spot (BS), caused by Bipolaris oryzae affects millions of acres of rice worldwide causing great yield losses yearly (~10% average) and reducing grain quality (Barnwal et al., 2013). The current study was carried out at rice seedlings on which small brownish lesions, typical of brown spot, were observed and leaf samples were collected for microscopic and biochemical characterization. The microscopic observation revealed the presence of fusiform and slightly curved bipolar conidia and these morphological observations are in line with the previous study (Wang et al., 2016). Bipolaris oryzae belongs to the ascomycetes family which is a very large group of fungi and Bipolaris has a broad host range. Although we isolated the fungus from rice and confirmed it through pathogenicity test and microscopy. The molecular confirmation of pathogen and genetic diversity in pathogen population through molecular markers is suggested in Pakistan as conducted earlier in Japan, Philippine and Iran (Ahmadpour et al., 2018). Such information will help to interpret the phenotypic diversity and variation in pathogen latent period and growth at multiple growth stages of rice.
Use of resistant varieties is an effective and durable way to protect crop from brown spot disease. Resistant varieties help to stabilize yield, but approaches for brown spot control through resistance are very scarce and not stable due to the presence of more severe races of pathogens (Katasntones et al., 2007). In the present study, 73 diverse and both local and exotic rice genotypes were evaluated against brown spot disease under inoculated field conditions. Five genotypes representing International Rice Research Institute (IRRI) and PARC showed resistant phenotype. The 50 genotypes provided by IRRI were high yielding yet were not tested at IRRI against brown spot disease. Out of 50, only 21 showed a moderately resistant phenotype. The result suggests the different genetic background of the germplasm. Another reason of contrasting phenotype could be the very different environmental conditions in Pakistan because the germplasm was not challenged against the prevailing B. oryzae pathogen population. The primary source of inoculum is likely through infected seeds and is perhaps a dominant process in many BS epidemics. There is the possibility that the rice seed provided by the IRRI is treated with fungicide which may inhibit seed infection. Four genotypes showed a resistant response and there were five moderately resistant phenotypes among the PARC material. Most of the genotypes were developed by crossing of the local varieties and exotic germplasm. However, the study for the confirmation of genes and/or QTLs in these genotypes for disease resistance has not be conducted yet. The results emphasize to adopt integrated approaches for the management of the brown spot disease.
Several previous studies identified resistant genotypes in rice collection under field conditions however very few genes or QTLs were identified against brown spot disease. Satija et al. (2005) identified 15 Oryza sativa entries out of 124 that were classified as resistant (<5% severity). Conversely, Hossain et al. (2004) identified one resistant variety out of 29 entries. Three quantitative trait loci (QTL) were detected in cultivar Tadukan (qBS2, qBS9, qBS11) on chromosomes 2, 9, and 11, respectively (Sato et al., 2008), and qBS11 is considered as having a major effect. Based on the phenotypic results, we can postulate that the resistant rice genotype may contain resistance genes, but validation through molecular markers or specific primers is required. The moderately resistant and moderately susceptible genotypes might carry minor QTLs but to support this hypothesis the germplasm needs to be evaluated at adult plant stages. It is generally accepted that the partial resistance is governed by the QTLs and is mostly expressed at adult plant stages. Our results also highlighted that none of the rice commercial variety showed a resistant reaction against brown spot disease. We are not certain whether the seed of the varieties used in this study was pathogen free. If seed of the commercial varieties carried Bipolaris oryzae spores, this could have increased the inoculum pressure resulting in the susceptible reaction of the rice varieties. Based on the results of our screening, the selected resistant germplasms can further be used in the breeding program as a source of resistance against brown leaf spot of rice. For future studies, it is suggested that the phenotyping of rice germplasm at multiple growth stages and against the diverse race of pathogen should be executed. It will not only improve our understanding of host-pathogen interaction but will also increase the resolution of rice resistance under field conditions.
Plants have evolved a sophisticated defense system which includes constitutive or induced, biochemical or structural mechanisms, to ward off pathogen attack. Among such mechanisms, some enzymes and compounds have been shown to be involved in rice defense against fungal infection. In the current study, accumulation of biochemical defense related enzymes was determined in resistant, moderately resistant and susceptible rice genotypes against brown spot disease. The results showed that the accumulation of total protein, catalase, peroxidase, phenyl ammonium lyase, polyphenol oxidase and β-1,3-Glucanase were higher in resistant genotypes followed by the moderately resistant rice lines. The results are in line with the previous studies in which the rice defense to B. oryzae was found to be related to higher activities of peroxidase (POD) (Dallagnol et al., 2011). In a similar study rice plants that displayed higher concentrations of POD, polyphenol oxidase (PPO), phenylalanine ammonia-lyase (PAL), and lipoxygenase (LOX) were more resistant to leaf scald (Tatagiba et al., 2014).
Involvement of protein components in plant disease resistance has been documented in many plants' pathogenic interactions (Tornero et al., 2002; Carvalho et al., 2006). In this study, we found that the higher amount of total protein in rice leaves correlated with lower disease incidence. However, the parameter of total protein cannot explain the total variation in phenotype because proteins are major constituents of the plant cell and are involved in many biochemical functions including host defense against diseases. Association of the total protein with plant defense against fungi and bacteria was earlier reported in wheat (Biswas et al., 2003), rice (Kumawat et al., 2008; Biswas et al., 2011), oat (Fink et al., 1988), maize (Nasser et al., 1990), and barley (Hoj et al., 1989).
We observed the reduction in the accumulation of all enzymes (CAT, POD, PAL, PPO, and β-1,3-Glucanase) as resistance level decreases. However, the reduction in POD and PPO in susceptible rice genotypes was very rapid and many-fold lower in comparison to the resistant ones. Supporting the data provided by Cruz et al. (2010), the present study brings novel evidence of efficient activation of defense enzymes in the leaves of resistant and the moderately resistance genotypes that removes the excess ROS, may be linked to the resistance. Among the enzymes involved in the removal of the excess ROS generated spontaneously, CAT also plays a key role (Mittler, 2002).
CAT is an oxygen-scavenging enzyme that protects cells from the toxic effects of substrates (H2O2) during development, which are otherwise lethal (Choodamani et al., 2009; Patel et al., 2011; Hameed and Iqbal, 2014). In the present study, the CAT activity only increased on leaves with resistant phenotype (<5% disease severity). Based on a previous study in tomato (Kuzniak and Sklodowska, 2005), CAT activities were increased during the initial stage of Botrytis cinerea infection and decreased after the necrotic lesion formation. The reduction in CAT activity in susceptible rice genotypes could be a consequence of enhanced proteolysis, which is induced during infection process (Palma et al., 2002), as is the case of Pyricularia orzyae infection. The investigation of CAT activity at multiple time points after inoculation is suggested for future studies for better understanding of the biochemical defense in rice against brown spot disease. The difference in the CAT activity between rice genotypes suggests that this enzyme plays a major role in the rice resistance to brown spot disease. This result is in agreement with Magbanua et al. (2007), who demonstrated that the CAT activity was higher in the leaves of maize lines resistant to Aspergillus flavus than in susceptible ones. The role of CAT in the plant–pathogen interaction seems to be more complex than for abiotic stress (Kuzniak and Sklodowska, 2005), which involves an association between CAT activity and plant tolerance (Miyagawa et al., 2000; Oksanen et al., 2003).
Peroxidase (PODs) is one of the first response enzymes which provides rapid defense against pathogen attack (Sulman et al., 2001). PODs are involved in the lignification, suberification, polymerization of hydroxy-proline-rich glycoproteins, and the resistance against pathogens in plants (Hammond-Kosack and Jones, 1996; Yoshida et al., 2003; Maksimov et al., 2014). The results of the present study showed that the peroxidase enzyme was accumulated in higher concentration in resistant and moderately resistant rice genotypes, with 10-fold less accumulation in highly susceptible rice variety Bas-2000. The results suggest that the POD played an important role in defense against brown spot disease. Infection with plant pathogens led to an induction in POD activity in plant tissues and a greater increase was recorded in resistant plants compared to the susceptible ones (Mydlarz and Harvell, 2006). A similar increase in POD activity was observed in tomato and bell pepper infected with tobacco mosaic virus and tomato mosaic tobamovirus (Madhusudhan et al., 2009) and tomato yellow leaf curl virus infected tomato plants (Dieng et al., 2011). In previous studies (Hong-xia et al., 2011) POD activity was found more important for resistant than for susceptible wheat cultivars when infected by Rhizoctonia cerealis. The higher POD activity in the leaves of wheat plants during the late stages of P.oryzae infection was detrimental to the resistance against blast (Xavier et al., 2011). These reports are in agreement with the data from the present study, which showed that the POD activity was maintained at a higher level in the leaves of resistant rice genotypes during seedling infection.
Phenyl ammonium lyase (PAL) is the primary enzyme in the phenylpropanoid metabolism and plays a significant role in the synthesis of several defense-related secondary compounds such as phenols and lignin (Hemm et al., 2004; Tahsili et al., 2014). In present study, PAL activity was higher in leaves of resistant genotypes and subsequently reduced as the resistance level decreased. PAL activity was found to change significantly in all four categories of host resistance. Similar results were reported by Ogawa et al. (2006) in tobacco leaves inoculated with tobacco mosaic virus. A previous study indicated that the presence of phenolic compounds in plants and their synthesis in response to infection is associated with disease resistance (Mandal et al., 2010). Another study highlighted that an increased accumulation of phenolics is due to increase in PAL activity that offered protection against diseases (Jayaraj et al., 2010).
Polyphenol oxidase (PPO) is important in the initial stage of plant defense where membrane damage causes release of phenols such as chlorogenic acid. PPO catalyzes the oxidation of phenolics to free radicals that can react with biological molecules, thus creating an unfavorable environment for pathogen development (Jockusch, 1966; Mohamed et al., 2012). PPO activity was significantly higher in resistant rice genotypes and rapidly decreased in susceptible genotype. Many studies have shown that PPO is induced in response to infection by different pathogens (Constabel et al., 2000; Stewart et al., 2001; Vanitha et al., 2009). Induction of PPO expression in resistant genotypes in the present study in response to Bipolaris oryzae infection might provide an additional line of defense to protect plants against further attack by pathogen and insects. There is a possibility that the resistant rice genotypes produce more secondary metabolites involved in plant defense mechanism than the susceptible genotypes. Similar findings were previously reported in other plant–pathogen interactions (Thipyapong et al., 1995; Stout et al., 1999).
β-1,3-Glucanase are the pathogenesis-related (PR) proteins associated with biotic stresses. They are coordinately induced, accumulate systemically and are linked to the development of systemic acquired response (SAR) (Ferreira et al., 2007). Plants produce enzymes such as β-1,3-glucanase and chitinase (Santos et al., 2004) that can break down the cell wall components of pathogens in a coordinated manner. These enzymes are important determinants of the resistance of plants to fungal diseases (Funnell et al., 2004). The results of this study indicated that the resistant genotypes displayed a significantly higher amount (3 times higher) of β-1,3-glucanase than the susceptible ones. The consistent and rapid increase in their activity in the resistant cultivar after pathogen infection suggests a possible role for β-1,3-glucanase in the defense mechanism of rice genotypes against B. oryzae. A direct role for β-1,3-glucanase in the defense of plants against pathogens has been proposed, because the substrate for the enzyme, β-1,3-glucan is a major component of the cell walls of many fungi (Wessels and Sietsma, 1981). However, β-1,3-glucanases can also play an indirect role in defense because they are known to release oligosaccharides from the walls of fungi, which in turn, act as signals in the elicitation of host defense responses (Poonam et al., 2013). In a previous study, enhanced activity of chitinase and β-1,3-glucanase was observed in rice plant infected with R. solani (Mondal et al., 2012). It has been reported earlier, that plants with elevated levels β-1,3-glucanase expression are more resistant to fungal pathogens (Datta et al., 2001). The results, therefore, suggest a possible role of β-1,3-glucanase in defense mechanism of Oryza sativa against the brown spot disease.
Conclusions
Overall, the results from the present study showed an elevated expression of defense enzyme activities in rice after the B. oryzae infection. Field based phenotyping identified the resistant and moderately resistant genotypes among the collection of 73 diverse rice germplasm. The biochemical profile of 20 diverse rice genotyped clearly reflect their phenotype. Our finding indicate that the elevated expression of various defense enzymes is an important feature of rice resistance to brown spot disease. We concluded that the total protein, CAT, POD, PAL, PPO, and β-1,3-Glucanase (PR proteins) play an active role in disease resistance against brown spot disease. However, the CAT, POD, and PPO were found to differ significantly in resistant and susceptible varieties after infection with B. oryzae. These enzymes can be used as biochemical markers for early detection of resistant during compatible and incompatible interactions. However, additional studies are needed to characterize these defense compounds at adult plant stages of the rice. On the basis of phenotypic and biochemical profiling of rice germplasm, the resistant and moderately resistance rice genotypes with elevated expression of defense enzymes can be used in rice breeding program for the development of disease resistant varieties.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
BA designed the study, carried out the experiment, and analyzed the data. HA contributed in execution of lab and field study and editing of manuscript. MA contributed to the statistical analysis. SY contributed in biochemical studies improved the text of manuscript. KS and AA wrote the manuscript. All authors have revised and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are highly grateful to Masood Babar and Umra Sadaf, NIAB Faisalabad for technical assistance during experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.675895/full#supplementary-material
References
Ahmadpour, A., Castell-Miller, C., Javan-Nikkhah, M., Naghavi, M. R., Dehkaei, F. P., Leng, Y., et al. (2018). Population structure, genetic diversity, and sexual state of the rice brown spot pathogen Bipolaris oryzae from three Asian countries. Plant Pathol. 67, 181–192. doi: 10.1111/ppa.12714
Aoki, S., Haga, S., and Suzuki, S. (2020). Direct antagonistic activity of chitinase produced by Trichoderma sp. SANA20 as biological control agent for grey mould caused by Botrytis cinerea. Cogent. Biol. 6:1747903. doi: 10.1080/23312025.2020.1747903
Arshad, H. M. I., Khan, J. A., and Jamil, F. F. (2008). Screening of rice germplasm against blast and brown spot diseases. Pak. J. Phytopathol. 20, 52–57.
Asghar, A., Rashid, H., Ashraf, M., Khan, M. H., and Chaudry, A. Z. (2007). Improvement of basmati rice, against fungal infection through gene transfer technology. Pak. J. Bot. 39, 1277–1283.
Barnwal, M. K., Kotasthane, A., Magculia, N., Mukherjee, P. K., Savary, S., Sharma, A. K., et al. (2013). A review on crop losses, epidemiology and disease management of rice brown spot to identify research priorities and knowledge gaps. Eur. J. Plant Pathol. 136, 443–457. doi: 10.1007/s10658-013-0195-6
Biswas, B., Scott, P. T., and Gresshoff, P. M. (2011). Tree legumes as feedstock for sustainable biofuel production: opportunities and challenges. J. Plant Physiol. 168, 1877–1884. doi: 10.1016/j.jplph.2011.05.015
Biswas, S. K., Ratan, V., Srivastava, S. S. L., and Singh, R. (2008). Influence of seed treatment with biocides and foliar spray with fungicides for management of brown leaf spot and sheath blight of paddy. Indian Phytopathol. 61, 55–59.
Biswas, S. K., Srivastava, K. D., Aggarwal, R., Praveen, S., and Singh, D. V. (2003). Biochemical changes in wheat induced by Chaetomium globosum against spot blotch pathogen. Indian Phytopathol. 56, 374–379.
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Canxing, D., Jiaojiao, Y., Jianyu, B., Zhendong, Z, and Xiaoming, W. (2013). Induced defense responses in rice plants against small brown planthopper infestation. Crop J. 2, 55–62. doi: 10.1016/j.cj.2013.12.001
Carella, P., Gogleva, A., Hoey, D. J., Bridgen, A. J., Stolze, S. C., Nakagami, H., et al. (2019). Conserved biochemical defenses underpin host responses to oomycete infection in an early-divergent land plant lineage. Curr. Biol. 29, 2282–2294. doi: 10.1016/j.cub.2019.05.078
Carvalho, D., Anastacio, Q., and Luciana, M. (2006). Proteins and isozymes electrophoresis in seeds of Desti (Leguminosae caesalpinioidea) artificially aged. Rev Arv. 30, 19–21. doi: 10.1590/S0100-67622006000100003
Chance, B., and Maehly, A. C. (1955). Assay of catalase and peroxidase. Meth. Enzymol. 2:7647775. doi: 10.1016/S0076-6879(55)02300-8
Choodamani, M. S., Hariprasad, P., Sateesh, M. K., and Umesha, S. (2009). Involvement of catalase in bacterial blight disease development of rice caused by Xanthomonas oryzae pv. oryza. Int. J. Pest Manage. 55, 121–127. doi: 10.1080/09670870802601076
Constabel, C. P., Yip, L., Patton, J. J., and Christopher, M. E. (2000). Polyphenol oxidase from hybrid poplar, cloning and expression in response to wounding and herbivory. Plant Physiol. 124, 285–295. doi: 10.1104/pp.124.1.285
Cruz, M. F. A., Prestes, A. M., Maciel, J. L. N., and Scheeren, P. L. (2010). Resistência parcial à brusone de genótipos de trigo comum e sintético nos estádios de planta jovem e de planta adulta. Trop. Plant Pathol. 35, 24–31. doi: 10.1590/S1982-56762010000100004
Dallagnol, L. J., Rodrigues, F. A., DaMatta, F. M., Mielli, M. V. B., and Pereira, S. C. (2011). Deficiency in silicon uptake affects cytological, physiological, and biochemical events in the rice–Bipolaris oryzae interaction. Phytopathology 101, 92–104. doi: 10.1094/PHYTO-04-10-0105
Dallagnol, L. J., Rodrigues, F. A., Mielli, M. V. B., Ma, J. F., and Datnoff, L. E. (2009). Defective active silicon uptake affects some components of rice resistance to brown spot. Phytopathology 99:1161121. doi: 10.1094/PHYTO-99-1-0116
Datta, K., Tu, J. M., Oliva, N., Ona, I., Velazhahan, R., Mew, T. W., et al. (2001). Enhanced resistance to sheath blight by constitutive expression of infection related rice chitinase in transgenic elite Indica rice cultivars. Plant Sci. 160, 405–414. doi: 10.1016/S0168-9452(00)00413-1
Dieng, H., Satho, T., Hassan, A. A., Aziz, A. T., Morales, R. E., Hamid, S. A., et al. (2011). Peroxidase activity after viral infection and whitefly infestation in juvenile and mature leaves of Solanum lycopersicum. J. Phytopathol. 159, 707–712. doi: 10.1111/j.1439-0434.2011.01830.x
Divya, K., Meenu, T., Smitha, V., Varghese, S., and Jisha, M. S. (2020). Induction of defence response in Oryza sativa L. against Rhizoctonia solani (Kuhn) by chitosan nanoparticles. Microb. Pathog. 149:104525. doi: 10.1016/j.micpath.2020.104525
Don, G., and Clayton, H. (2010). Brown Spot of Rice. Louisiana Plant pathology, Disease Identification and Management Series.
Eliahu, N., Igbaria, A., Rose, M. S., Horwitz, B. A., and Lev, S. (2007). Melanin biosynthesis in the maize pathogen Cochliobolus heterostrophus depends on two mitogen-activated protein kinases, Chk1 and Mps1, and the transcription factor Cmr1. Eukaryotic Cell 6, 421–429. doi: 10.1128/EC.00264-06
Fengming, S., and Robert, M. G. (2001). Molecular biology of disease resistance in rice. Physiol. Mol. Plant Pathol. 59:111. doi: 10.1006/pmpp.2001.0353
Ferreira, R. B., Monteiro, S., Freitas, R., Santos, C. N., Chen, Z., Batista, L. M., et al. (2007). The role of plant defense proteins in fungal pathogenesis. Mol. Plant Pathol. 8:677700. doi: 10.1111/j.1364-3703.2007.00419.x
Fink, W., Lifeland, M., and Mendgen, K. (1988). Chitinase and Beta 1,3 glucanase in the Apoplastic compartment of oat leaves (Avena sativa L.). Plant Physiol. 88, 270–275. doi: 10.1104/pp.88.2.270
Funnell, D. L., Lawrence, C. B., Pedersen, J. F., and Schardl, C. L. (2004). Expression of the tobacco b-1,3-glucanase gene, PR2d, following induction of SAR with Peronospora tabacina. Physiol. Mol. Plant Pathol. 65:285296. doi: 10.1016/j.pmpp.2005.02.010
Geetha, N. E., Amruthesh, K. N., Sharathchandra, R. G., and Shetty, H. S. (2005). Resistance to downy mildew in pearl millet is associated with increased phenylalanine ammonia-lyase activity. Funct. Plant Biol. 32, 267–275. doi: 10.1071/FP04068
Hameed, A., and Iqbal, N. (2014). Chemo-priming with mannose, mannitol and H2O2 mitigate drought stress in wheat. Cereal Res. Comm. 42, 450–462. doi: 10.1556/CRC.2013.0066
Hammond-Kosack, K. E., and Jones, J. D. G. (1996). Resistance gene dependent plant defense responses. Plant Cell 8, 1773–1791. doi: 10.1105/tpc.8.10.1773
Hemm, M. R., Rider, S. D., Ogas, J., Murry, D. J., and Chapple, C. (2004). Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38, 765–778. doi: 10.1111/j.1365-313X.2004.02089.x
Hoang Loc, N., Huy, N. D., Quang, H. T., Lan, T. T., and Thu Ha, T. T. (2020). Characterisation and antifungal activity of extracellular chitinase from a biocontrol fungus, Trichoderma asperellum PQ34. Mycology 11, 38–48. doi: 10.1080/21501203.2019.1703839
Hoj, P. B., Hartman, D., Morrice, N. A., Doan, D. N. P., and Finchar, G. B. (1989). Purification of β-1,3-glucan endohydrolase enzyme II from germinated barley and determination of the primary structure from a cDNA clone. Plant Biol. 13, 31–42. doi: 10.1007/BF00027333
Hong-xia, L., Zhi-yong, X., and Zeng-yan, Z. (2011). Changes in activities of antioxidant-related enzymes in leaves of resistant and susceptible wheat inoculated with Rhizoctonia cerealis. Agric. Sci. China 10, 526–533. doi: 10.1016/S1671-2927(11)60033-3
Hossain, M., Khalequzzaman, K. M., Mollah, M. R., Hussain, M. A., and Rahim, A. (2004). Reaction of breeding lines/ cultivars of rice against brown leaf spot of rice under field conditions. Asian J. Plant Sci. 3, 614–617. doi: 10.3923/ajps.2004.614.617
Huynh, V. N., and Ashok, G. (2004). Role of Bipolaris oryzae in producing abnormal seedlings of rice (Oryza sativa). Omonrice 12, 102-108. Identification of the causal agent fungi of rice brown spot disease in Guilan province. J. Agric. Sci. Nat. Resour. 12:136145.
Jayaraj, J., Bhuvaneswari, R., Rabindran, R., Muthukrishnan, S., and Velazhahan, R. (2010). Oxalic acid-induced resistance to Rhizoctonia solani in rice is associated with induction of phenolics, peroxidase and pathogenesis-related proteins. J. Plant Interact. 5, 147–157. doi: 10.1080/17429140903291012
Jockusch, H. (1966). The role of host genes, temperature and polyphenol oxidase in the necrotization of TMV infected tobacco tissue. Phytopathol Z. 55, 185–192. doi: 10.1111/j.1439-0434.1966.tb02222.x
Katasntones, G., Koutrovbas, S. D., Ntanos, D. A., and Lupoho, E. (2007). A comparison of three experimental designs for the field assessment of resistance to rice brown leaf spot of rice disease (Bipolaris oryzae). J. Phytopath. 155, 204–210. doi: 10.1111/j.1439-0434.2007.01218.x
Kessmann, H., Staub, T., Hofmann, C., Maetzke, T., Herzog, J., Ward, E., Uknes, S., and Ryals, J. (1994). Introduction of systemic acquired resistance in plants by chemicals. Annu. Rev. Phytopathol. 32, 439–459. doi: 10.1146/annurev.py.32.090194.002255
Khan, M. S., and Hemalatha, S. (2016). Biochemical and molecular changes induced by salinity stress in Oryae sativa L. Acta Physiol. Palnt 38:167. doi: 10.1007/s11738-016-2185-8
Kumawat, G. L., Biswas, S. K., and Srivastava, S. S. L. (2008). Biochemical evidence of defence response in plant induced by bio-agents against brown leaf spot pathogen. Indian Phytopath. 61, 197–203.
Kuzniak, E., and Sklodowska, M. (2005). Fungal pathogen-induced changes in the antioxidant systems of leaf peroxisomes from infected tomato plants. Planta 222, 192–200. doi: 10.1007/s00425-005-1514-8
Liang, Z. C., Hseu, R. S., and Wang, H. H. (1995). Partial purification and characterization of a 1,3-β-D-glucanase from Ganoderma tsugae. J. Ind. Microbiol. 14, 5–9. doi: 10.1007/BF01570058
Madhusudhan, K. N., Srikanta, B. M., Shylaja, M. D., Prakash, H. S., and Shetty, H. S. (2009). Changes in antioxidant enzymes, hydrogen peroxide, salicylic acid and oxidative stress in compatible and incompatible host tobamovirus interaction. J. Plant Interact. 4, 157–166. doi: 10.1080/17429140802419516
Magbanua, Z. V., De Moraes, C. M., Brooks, T. D., Williams, W. P., and Luthe, D. S. (2007). Is catalase activity one of the factors associated with maize resistance to Aspergillus flavus. Mol. Plant-Microbe Interact. 20, 697–706. doi: 10.1094/MPMI-20-6-0697
Maksimov, I., Troshina, N., Surina, O., and Cherepanova, E. (2014). Salicylic acid increases the defense reaction against bunt and smut pathogens in wheat calli. J. Plant Interact. 9, 306–314. doi: 10.1080/17429145.2013.832424
Mandal, S. M., Chakraborty, D., and Dey, S. (2010). Phenolic acids act as signaling molecules in plant-microbe. Plant Signal. Behav. 4, 359–368. doi: 10.4161/psb.5.4.10871
Mayer, A. M., Harel, E., and Shaul, R. B. (1965). Assay of catechol oxidase, a critical comparison of methods. Phytochemistry 5, 783–789. doi: 10.1016/S0031-9422(00)83660-2
Minaeva, O. M. E. E., Akimova, N. N., Tereshchenko, T. I., Zyubanova, M. V., and Apenysheva, A. V. K. (2018). Effect of Pseudomonas bacteria on peroxidase activity in wheat plants when infected with Bipolaris sorokiniana. Russ. J. Plant Physiol. 65, 717–725. doi: 10.1134/S1021443718040052
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Miyagawa, Y., Tamoi, M., and Shigeoka, S. (2000). Evaluation of defense system in chloroplasts to photooxidative stress caused by paraquat using transgenic tobacco plants expressing catalase from Escherichia coli. Plant Cell Physiol. 41, 311–320. doi: 10.1093/pcp/41.3.311
Mohamed, H., EL-Hady, A. A., Mansour, M., and El-Rheem ElSamawaty, A. (2012). Association of oxidative stress components with resistance to flax powdery mildew. Trop. Plant Pathol. 37, 386–392. doi: 10.1590/S1982-56762012000600002
Mondal, A., Dutta, S., Nandi, S., Das, S., and Chaudhuri, S. (2012). Changes in defence-related enzymes in rice responding to challenges by Rhizoctonia solani. Arch. Phytopathol. Plant Protect. 45, 1840–1851. doi: 10.1080/03235408.2012.712831
Mydlarz, L. D., and Harvell, C. D. (2006). Peroxidase activity and inducibility in the see fan coral exposed to a fungal pathogen. Comp. Biochem. Physiol. 10:1016. doi: 10.1016/j.cbpa.2006.09.005
Nasser, W., Tapia, M., and De Burkard, G. (1990). Maize pathogenesis related proteins: characterization and cellular distribution of beta 1,3 glucanase and chitinases indiced by brome mosaic virus infection. Physiol. Mol. Plant Pathol. 36, 1–14. doi: 10.1016/0885-5765(90)90087-E
Nazari, S., Javan Nikkhah, M., Khalil-Berdi, F., Khosravi, V., and Alizadeh, A. (2015). Bipolaris species associated with rice plant: pathogenicity and genetic diversity of Bipolaris oryzae using rep-PCR in Mazandaran province of Iran. J. Crop Prot. 4, 497–508.
Ogawa, D., Nakajima, N., Seo, S., Mitsuhara, I., Kamada, H., and Oaci, Y. (2006). The phenylalanine pathway is the main route of salicylic acid biosynthesis in tobacco mosaic virus infected tobacco leaves. Plant Biotechnol. 23, 395–398. doi: 10.5511/plantbiotechnology.23.395
Oksanen, E., Häikkiö, E., Sober, J., and Karnosky, D. F. (2003). Ozone induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol. 161, 791–799. doi: 10.1111/j.1469-8137.2003.00981.x
Palma, J. M., Sandalio, L. M., Corpas, F. J., Romero-Puertas, M. C., McCarthy, I., and del Río, L. A. (2002). Plant proteases, protein degradation, and oxidative stress: role of peroxisomes. Plant Physiol. Biochem. 40, 521–530. doi: 10.1016/S0981-9428(02)01404-3
Patel, S. J., Subramanian, R. B., and Jha, Y. S. (2011). Biochemical and molecular studies of early blight disease in tomato. Phytoparasitica 39, 269–283. doi: 10.1007/s12600-011-0156-6
Poonam, G., Indu, R., and Vinay, S. (2013). Induction of β-1,3- glucanase and chitinase activity in the defense response of Eruca sativa plants against the fungal pathogen Alternaria brassicicola. J. Plant Interact. 8, 155–161. doi: 10.1080/17429145.2012.679705
Santos, I. S., Machado, O. L. T., Da Cunha, M., and Gomes, V. M. (2004). A chitinase from Adenanthera pavonina L. seeds: purification, characterization and immunolocalization. Plant Sci. 167:12031210. doi: 10.1016/j.plantsci.2004.04.021
Sarkar, D., Mandal, R., Roy, P., Taradar, J., and Dasgupta, B. (2014). Management of Brown Spot disease of rice by using safer fungicides and some bioagents. Bioscan 9, 437–441.
Satija, A., Chahal, S. S., and Pannu, P. P. S. (2005). Evaluation of rice genotypes against brown leaf spot disease. Plant Dis. Res. 20, 163–164.
Sato, H., Ando, I., Hirabayashi, H., Takeuchi, Y., Arase, S., and Kihara, J. (2008). QTL analysis of brown spot resistance in rice (Oryza sativa L.). Breed. Sci. 58, 93–96. doi: 10.1270/jsbbs.58.93
Singh, R. K., Singh, C. V., and Shukla, V. D. (2013). Phosphorus nutrition reduces brown spot incidence in rainfed upland rice. Int. Rice Res. Notes. 30, 31–32.
Sripriya, R., Parameswari, C., and Veluthambi, K. (2017). Enhancement of sheath blight tolerance in transgenic rice by combined expression of tobacco osmotin (ap24) and rice chitinase (chi11) genes. In Vitro Cell. Dev. Biol. Plant. 53, 12–21. doi: 10.1007/s11627-017-9807-8
Stewart, R. J., Sawyer, B. J. B., Bucheli, C. S., and Robinson, S. P. (2001). Polyphenol oxidase is induced by chilling and wounding in pineapple. Aust. J. Plant Physiol. 28, 181–191. doi: 10.1071/PP00094
Stout, M. J., Fidantsef, A. L., Duffey, S. S., and Bostock, R. M. (1999). Signal interactions in pathogen and insect attack: systemic plant mediated interactions between pathogens and herbivores of the tomato, Lycopersicon esculentum. Physiol. Mol. Plant Pathol. 54, 115–130. doi: 10.1006/pmpp.1998.0193
Sulman, M., Fox, G., Osman, A., Inkerman, A., Williams, P., and Michalowitz, M. (2001). “Relationship between total peroxidase activity and susceptibility to black point in mature grain of some barley cultivars,” in Proceeding of the 10th Australian Barley Technical Symposium (Canberra), 16–20.
Tahsili, J., Sharifi, M., Safaie, N., Esmaeilzadeh-Bahabadi, S., and Behmanesh, M. (2014). Induction of lignans and phenolic compounds in cell culture of Linum album by culture filtrate of Fusarium graminearum. J. Plant Interact. 9, 412–417. doi: 10.1080/17429145.2013.846419
Tatagiba, S. D., Rodrigues, F. A., Filippi, M. C., Silva, G. B., and Silva, L. C. (2014). Physiological responses of rice plants supplied with silicon to Monographella albescens infection. J. Phytopathol. 162, 596–606. doi: 10.1111/jph.12231
Thipyapong, P., Hunt, M. D., and Steffens, J. C. (1995). Systemic wound induction of potato (Solanum tuberosum) polyphenol oxidase. Phytochemistry 40, 673–676. doi: 10.1016/0031-9422(95)00359-F
Tornero, P., Chao, R., Luthin, W., Goff, S., and Dangl, J. (2002). Largescale structure, function, analysis, of Arabidopsis RPM1 disease resistance protein. Plant Cell 14, 435–450. doi: 10.1105/tpc.010393
Vanitha, S. C., Niranjana, S. R., and Umesha, S. (2009). Role of phenylalanine ammonia lyase and polyphenol oxidase in host. J. Phytopathol. 157, 552–557. doi: 10.1111/j.1439-0434.2008.01526.x
Wang, L., Al-Hatmi, A. M. S., Lai, X., Peng, L., Yang, C., Lai, H., et al. (2016). Bipolaris oryzae, a novel fungal opportunist causing keratitis. Diagn. Microbiol. Infect. Dis. 85, 61–65. doi: 10.1016/j.diagmicrobio.2015.11.020
Wessels, J. G. H., and Sietsma, J. H. (1981). “Fungal cell walls: a survey,” in Encyclopedia of Plant Physiology, eds W. Tanner and F. A. Louewus (New York, NY: Springer-Verlag), 352394. doi: 10.1007/978-3-642-68234-6_15
Xavier, F. M. S., Rodrigues, F. A., Domiciano, G. P., Oliveira, H. V., Silveira, P. R., and Moreira, W. R. (2011). Wheat resistance to leaf blast mediated by silicon. Aust. Plant Pathol. 40, 28–38. doi: 10.1007/s13313-010-0010-1
Yoshida, K., Kaothien, P., Matsui, T., Kawaoka, A., and Shinmyo, A. (2003). Molecular biology and application of plant peroxidase genes. Appl. Microbiol. Biotechnol. 60, 665–670. doi: 10.1007/s00253-002-1157-7
Zhang, F. Q., Wang, Y. S., Lou, Z. P., and Dong, J. D. (2007). Effect of heavy metal stress on antioxidative enzymes and lipid peroxidation in leaves and roots of two mangroves plant seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere 67, 44–50. doi: 10.1016/j.chemosphere.2006.10.007
Keywords: bipolaris, phenotype, host resistance, biochemical, susceptible
Citation: Ashfaq B, Arshad HMI, Atiq M, Yousaf S, Saleem K and Arshad A (2021) Biochemical Profiling of Resistant Phenotypes Against Bipolaris oryzae Causing Brown Spot Disease in Rice. Front. Agron. 3:675895. doi: 10.3389/fagro.2021.675895
Received: 04 March 2021; Accepted: 28 July 2021;
Published: 24 August 2021.
Edited by:
Tariq Mukhtar, Pir Mehr Ali Shah Arid Agriculture University, PakistanReviewed by:
Kirstin Verity Wurms, The New Zealand Institute for Plant and Food Research Ltd, New ZealandMuhammad Umar Shahbaz, Ayub Agriculture Research Institute, Pakistan
Copyright © 2021 Ashfaq, Arshad, Atiq, Yousaf, Saleem and Arshad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amna Arshad, YWFnaWxsXzUyQHlhaG9vLmNvbQ==
 Benish Ashfaq1
Benish Ashfaq1 Hafiz M. Imran Arshad
Hafiz M. Imran Arshad Sumaira Yousaf
Sumaira Yousaf Kamran Saleem
Kamran Saleem Amna Arshad
Amna Arshad
