
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 08 July 2021
Sec. Pest Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.675247
This article is part of the Research Topic New Applications of Insecticidal RNAi View all 9 articles
 Wayne Brian Hunter1*†
Wayne Brian Hunter1*† William Rodney Cooper2*
William Rodney Cooper2* Andres F. Sandoval-Mojica3
Andres F. Sandoval-Mojica3 Greg McCollum1
Greg McCollum1 Veenu Aishwarya4
Veenu Aishwarya4 Kirsten S. Pelz-Stelinski3*†
Kirsten S. Pelz-Stelinski3*†We report on the development, evaluation, and efficient delivery of antisense oligonucleotide FANA (2′-deoxy-2′-fluoro-arabinonucleotide) RNA-targeting technology into citrus trees and potato plants for management of bacterial pathogens and arthropod pests. The FANA ASO technology is a single nucleotide strand of 20–24 nt in length that incorporates 2′F- chemically modifications of nucleotides, along with a phosphorothioate backbone and modified flanking nucleotides, in their structure called “gapmers,” produced by AUM LifeTech., Inc. These unique modified structures of FANA “triggers” enables gymnotic activity that self-delivers into cells, moving systemically in treated plants and insects, with significant suppression of their RNA targets. Reported is the FANA suppression of two plant-infecting bacterium Candidatus Liberibacter asiaticus, CLas (in citrus trees), and C. Liberibacter solanacearum, CLso (in potato and tomato). The CLas pathogen is associated with huanglongbing (a.k.a. Citrus Greening Disease), which causes severe loss of citrus trees, threatening global citrus production. The CLas bacterium is transmitted during feeding by the Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae). CLso causes Zebra-Chip disease in potato and is transmitted by the potato psyllid, Bactericera cockerelli (Hemiptera: Triozidae). Infected citrus trees or potato plants were treated with aqueous FANA solutions applied as a soil drench, root-infusion, topical spray, tree trunk injection or by absorption into cuttings, detached leaves, and leaf disks. Plants showed significant reduction of each pathogen or symptom development in response to FANA treatments. Similarly, ingestion of FANA solutions designed specifically to CLas by insects via artificial diets produced significant titer reductions in infected citrus psyllid adults that resulted in reduction of CLas transmission. The unique properties of FANA ASO solves many of the problems of stability, cell entry, and binding affinity that plagues exogenous RNAi strategies. Breakthroughs in production methods are reducing costs enabling these ASO to expand beyond medical applications into agricultural treatments. Thus, FANA ASO may provide viable treatments in the response to crop pandemics, like huanglongbing in citrus that threatens global food production.
Arthropod vectors of plant, animal, and human pathogens continue to threaten food security, and animal and human health on a global scale (Savary et al., 2019; He and Creasey Krainerm, 2020; Mesterházy et al., 2020). Many crop epidemics spread rapidly within an area in a country causing severe crop loss and economic hardship for local farmers. However, when these plant pathogens spread beyond international borders causing severe crop loss and starvation, they become pandemics (Savary et al., 2019; He and Creasey Krainerm, 2020). Crop pandemics, along with loss of food causes economic hardship with losses reported to be worth over $220 billion USD annually (Nicaise, 2014). Plant pathogens and pests are responsible for crop-yield losses up to 40% of maize, potato, rice, soybean, and wheat worldwide production, with another 30% loss in stored grains (Pimentel, 1991). Almost half of all plant diseases are caused by viruses, worth an annual global cost of $30 billion USD (Nicaise, 2014). Herbicides for weed control represented over half of all pesticides applied in the U.S. in 2012 and cost over $9 billion USD (Atwood and Paisley-Jones, 2017). The most severe crop pathosystems are from bacterial- and viral plant-pathogens vectored by plant-feeding hemipterans (psyllids, aphids, whitefly, leafhoppers, and planthoppers) (Nicaise, 2014; Sastry et al., 2019; Savary et al., 2019). These pathosystems, together with beetle pests and fungal pathogens, destroyed millions of tons of stored grains (Mesterházy et al., 2020), representing 30–40% loss of maize, potato, rice, soybean, and wheat crop-yield losses worldwide (Savary et al., 2019). Each year in response to these threats, ~500 million kilograms of pesticides are applied to plants in the United States. Studies show that only 0.1% of this reaches the desired target to eliminate pests (Pimentel and Levitan, 1986; Pimentel and Burgess, 2012), and the total pesticide expenditures in the US for agriculture showed a 5-fold increase (adjusted for inflation) between 1960 and 2008, at a cost of $12 billion USD (Fernandez Cornejo et al., 2014).
In the current global pandemic in citrus production, called Huanglongbing, HLB (a.k.a. “citrus greening disease” or “citrus yellows”), citrus trees infected with the bacterial pathogen (Candidatus Liberibacter asiaticus, CLas) produce small bitter fruits, eventually dropping all fruit, with infected trees ultimately dying (Da Graça and Korsten, 2004; Bové, 2006). Huanglongbing is the most serious threat to sustainable global citrus production (Halbert and Manjunath, 2004; Wang et al., 2017a,b). The economic losses in product and jobs in the Florida citrus industry alone is estimated to be over $4 billion USD (Hodges and Spreen, 2012; Stansly and Qureshi, 2020). Increased losses now threaten other U.S. citrus producing states (AZ, CA, TX) (Sétamou et al., 2016; Gottwald et al., 2018). Other countries that are citrus producers are also under threat from these psyllid-transmitted bacterial pathogens (Candidatus Liberibacter spp.) causing the global pandemic (Bové, 2006; Sétamou et al., 2016; Wang et al., 2017a,b; Kruse et al., 2019; Ammar et al., 2020a,b). The three known Liberibacter species associated with HLB are: Candidatus Liberibacter africanus, CLaf; Candidatus Liberibacter americanus, CLam; and Candidatus Liberibacter asiaticus, CLas (Bové, 2006; Haapalainen, 2014; Wang et al., 2017a,b; Coates et al., 2020). African countries are dominated by CLaf, which is transmitted by two psyllid species, the African citrus psyllid, Trioza erytreae (Hemiptera: Triozidae) and the Asian citrus psyllid (ACP) Diaphorina citri Kuwayama (Hemiptera: Liviidae), which can transmit both CLam and CLas bacteria (Lallemand et al., 1986). In Brazil, the CLam species is dominant, while CLas occurs globally in many citrus growing countries (Hall et al., 2013; Wang et al., 2017a,b; Ammar et al., 2020b). The pathogens replicate and circulate systemically in both the plant and the psyllid host tissues (Ammar et al., 2011, 2016, 2020a,b; Coy et al., 2014; Ramsey et al., 2015; Kruse et al., 2017, 2018), which facilitates their effective, rapid spread and persistence (Inoue et al., 2009; Ebert, 2019; Jiang et al., 2019; Huang et al., 2020).
A bacterium related to CLas, “Ca. L. solanacearum” (CLso) (syn. “Ca. L. psyllaurous”), causes disease symptoms in crops within the Solanaceae (Hansen et al., 2008; Liefting et al., 2009) in North America and New Zealand, and crops within the Apiaceae in Europe and North Africa (Munyaneza et al., 2010; Tahzima et al., 2014; Ahmed et al., 2015). CLso infection leads to rapid development of foliar symptoms in susceptible plants and is associated with the development of striped patterns in potato tubers known as zebra chip disease (Munyaneza, 2012, 2015). Multiple haplotypes of CLso have been identified (Nelson et al., 2011; Wen et al., 2013; Mauck et al., 2019; Swisher-Grimm and Garczynski, 2019). Haplotypes A and B appear to be most commonly associated Solanaceous crops in the United States (Dahan et al., 2017, 2019) with CLso haplotype B causing more severe symptoms than haplotype A (Mendoza-Herrera et al., 2018; Swisher-Grimm et al., 2018; Harrison et al., 2019; Tang et al., 2020). This pathogen infects multiple psyllid vectors within the Triozidae (Munyaneza et al., 2010; Tahzima et al., 2014; Antolinez et al., 2017; Borges et al., 2017), but is transmitted among Solanaceous plants by the potato psyllid (Bactericera cockerelli; Triozidae) in North America (Munyaneza et al., 2007; Hansen et al., 2008; Liefting et al., 2009). Like CLas, CLso is transmitted in a circulative and propagative manner, and colonization of the vector's salivary glands by CLso is required for transmission to new host plants (Cooper et al., 2014; Sengoda et al., 2014). CLso is transmitted by potato psyllids about 2–3 weeks after initial acquisition (Sengoda et al., 2014) and is known to affect psyllid fecundity (Nachappa et al., 2014; Ammar et al., 2020b).
Research efforts to develop effective management strategies for CLas and CLso have strongly focused on reducing the psyllid population, preventing acquisition and transmission of the bacterial pathogen by using several strategies including: RNAi-based suppression of the psyllid (Hajeri et al., 2014; Taning et al., 2016; Andrade and Hunter, 2017; Ghosh et al., 2017, 2018; NASEM, 2018; Dubrovina and Kiselev, 2019; Adeyinka et al., 2020; Dubrovina et al., 2020; Hunter et al., 2020; Yu and Killiny, 2020; Kiselev et al., 2021); antisense oligos, ASO (FANA and Morpholinos) targeting psyllids, their endosymbionts, and bacteria (Wesolowski et al., 2011, 2013; Pietri et al., 2014; Daly et al., 2017; Hunter et al., 2017a,b, 2018b, 2020; Metz et al., 2017a,b; Moulton and Moulton, 2017; Patel et al., 2017; Pelz-Stelinski et al., 2017; Hunter and Sinisterra-Hunter, 2018; Xue et al., 2018; Novopashina et al., 2019; Pelz-Stelinski, 2020; Sandoval-Mojica et al., 2020, 2021; Hunter and Pelz-Stelinski, 2021); gene–targeting and editing CRISPR/Cas9 systems (Chaverra-Rodriguez et al., 2018; Chaverra Rodriguez et al., 2020; Hunter et al., 2018a, 2019a; Hunter and Tomich, 2019); insecticides (Tiwari et al., 2011; Grafton-Cardwell et al., 2013; Qureshi et al., 2014; Chen and Stelinski, 2017; Chen et al., 2018; Della Vechia et al., 2019; Tian et al., 2019; Grafton-Cardwell, 2020; Monzo and Stansly, 2020) and biocontrol agents (Avery et al., 2009, 2011, 2013; Hoy et al., 2010; Chen and Stansly, 2014; Qureshi et al., 2014; Patt et al., 2015; Cicero et al., 2017; Pérez-Rodríguez et al., 2019). However, bacterial pathogens cannot be targeted directly using dsRNA strategies due to differences in cellular immune and replication processing pathways (Kruse et al., 2019; Blacutt et al., 2020). Bacteria also produce protective biofilms and have rapid replication that favors selection and development of resistance to chemical or antibiotic treatments, thus bacterial pathogens are very difficult to manage (Cansizoglu and Toprak, 2017; Jiang et al., 2019; Kumar et al., 2020; Naranjo et al., 2020; Puvača and de Llanos Frutos, 2021). The HLB pathosystem is the most difficult problem globally affecting the citrus industry (Bové, 2006; Wang and Trivedi, 2013; Wang et al., 2017a,b). A review by the national Academies of Sciences, of the past 10-year's research efforts, 2006–2018 summarized efforts, as “no significant solutions to stop HLB have yet been developed” (NASEM, 2018). Like many other plant pathogens that do not have effective treatments for infected hosts, the focus is on reduction of the insect vector population (Stansly and Qureshi, 2020), and/or targeting the pathogen using genetically modified plants (Puchta and Fauser, 2013; Saurabh et al., 2014; Younis et al., 2014; Aldemita et al., 2015; Huesing et al., 2016; Ricroch and Hénard-Damave, 2016; Ni et al., 2017; Sinisterra-Hunter and Hunter, 2018; ISAAA, 2019; Van Vu et al., 2019; Bramlett et al., 2020; Doyle et al., 2020; Kunte et al., 2020; Samada and Tambunan, 2020).
The purpose of this study was to assess FANA ASO as a novel strategy to manage Liberibacter species of bacterial pathogens in citrus, potato, tomato, and in the psyllid vector, and to suppress the psyllid vectors. We examined the potential of FANA ASO (2′-Deoxy-2′-Fluoro-β-D-Arabinonucleic Acid) antisense oligonucleotides to target bacterial pathogens inside of citrus trees and the psyllid vector. FANA are nucleic acids with a phosphorothioate backbone and modified flanking nucleotides, in which the 2′-OH group of the ribose sugar is substituted by a fluorine atom. The flank modifications increase the resistance of the ASO to degradation and enhances binding to targeted mRNA. The FANA/RNA duplex is recognized by ribonuclease H (RNase H), an enzyme that catalyzes the degradation of duplexed mRNA (Kalota et al., 2006; Teo et al., 2018; Gruenke et al., 2020; Ochoa and Milam, 2020). Therefore, antisense oligo modifications permit targeting of bacteria, like CLas, that cannot be suppressed with dsRNA strategies (Scott et al., 2013). The unique properties of the FANA designs make them more stable, enhance binding efficiency and increase specificity to the targeted organism (Gruenke et al., 2020; Ochoa and Milam, 2020), including the psyllid vectors (Hunter et al., 2017a,b; Pelz-Stelinski et al., 2017; Sandoval-Mojica et al., 2021). In the current study, FANA ASO were evaluated for plant delivery and systemic movement, insect delivery and activity to suppress psyllid and several bacteria: CLas, CLso, and psyllid endosymbionts. Confirmation analyses included (1) visualization using confocal microscopy of plant and insect tissues for their capacity to penetrate host cells, (2) silencing bacterial and insect essential genes and (3) evaluation on bacterial titer, and (4) insect mortality. The study demonstrated successful delivery of FANA with subsequent movement into target tissues in plants and psyllid, with reductions in CLas, and CLso titers within their perspective hosts (citrus, potato, tomato) and psyllid vector. Ultimately, the decreased bacteria titer, CLas in citrus and in the vector, ACP, translated into a biological significance of reduced CLas transmission by the vector, ACP1 (Pelz-Stelinski et al., 2017; Brown et al., 2020; Sandoval-Mojica et al., 2021).
To validate movement of the FANA into plant xylem and phloem, whole plants, cuttings, and detached leaves were treated with a fluorophore-tagged FANA and tissues prepared and examined with confocal microscopy. FANA bound to probe was observed in the xylem and phloem of citrus leaf midribs (Figure 1).
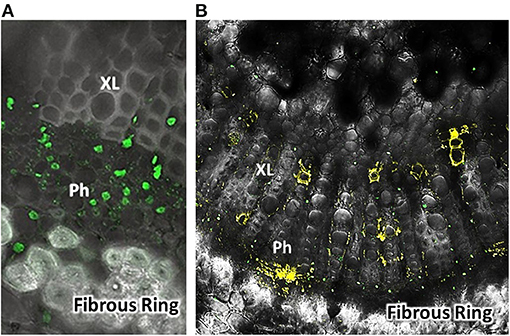
Figure 1. Fluorescent labeled FANA (yellow) after delivery into citrus leaves [Sweet orange group, Madam vinous, Citrus sinensis (L.) Osbeck] (B). Confocal microscope view as cross-section through leaf midrib near petiole. Control-Counter stain leaf cross-section. Green probe for nuclei used a tag fluorescence at 488 nm wavelength, was Nuclear-Green™ (A). Labels: Ph, Phloem; XL, Xylem; Fibrous Ring. Detection of FANA in Phloem and Xylem vessels. Sample image 7 days after treatment. FANA-probe treatment was a 6.93 nmol concentration in 500 μL water, absorbed into detached citrus leaves. Image view ~40X. Images were taken with a Zeiss LSM510 Confocal Microscope.
In lab and glasshouse trials, the FANA reduced CLas in infected citrus seedling trees, Madam vinous or Citrus sinensus. Methods as in Pelz-Stelinski et al. (2017), Andrade and Hunter (2016, 2017), Ghosh et al. (2018), and Sandoval-Mojica et al. (2020, 2021). Preliminary analyses at 3 weeks after FANA injections into infected citrus seedlings, showed a greater percentage of dead CLas bacteria (aver. 65%), over natural CLas mortality titers in control infected, untreated citrus trees (aver. 48.2%), method as in Cangelosi and Meschke (2014). However, at 4 months with no further treatments the CLas titers returned to pretreatment levels (data not shown). Thus, either more than a single-dose treatment would be required, or a higher concentrated dose would be needed to maintain suppression of CLas for longer than 3 months period.
Adult psyllids were fed on new citrus growth that absorbed solution of FANA labeled with a fluorescent probe psyllids and leaves were prepared and viewed with a confocal microscope (Figure 2). Feeding access periods were 48 h, 5 and 7 d (B). The tagged FANA were observed in a variety of tissues at 7 d after feeding, including the fat body, esophageal ganglia (brain), and vesicles in the psyllid body.
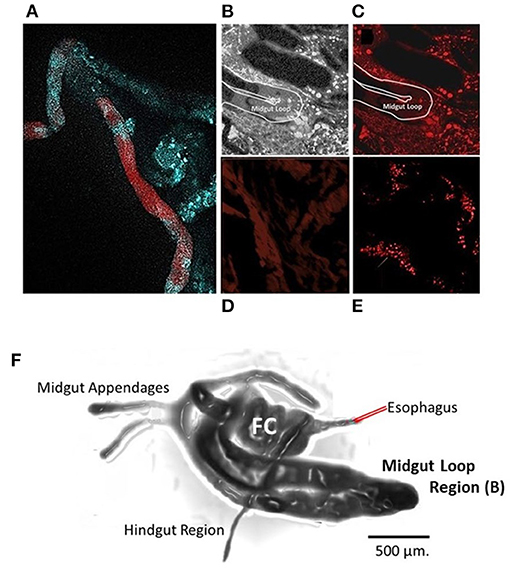
Figure 2. Citrus cutting feeding assay using FANA Fluorescence probe. Adult psyllid were fed on new citrus growth that absorbed solution of FANA with fluorescent probe or water. Feeding access periods were 48 h (A), 5 and 7 d (B,C). The midgut (A) of adult psyllid 48 h after feeding access on treated citrus leaf. The lumen of the midgut becomes void of fluorophores when examined after 7-day feeding access on untreated citrus leaf. (C,F) midgut (Loop). The tagged FANA were observed in a variety of tissues at 7 d after feeding, indicative of systemic movement into cells of tissues, and vesicles in the psyllid body (E) (Red). Control untreated psyllid sample shows level of background fluorescence (D). Drawing of dissected psyllid alimentary tract (F) provides perspective of location of the midgut loop shown in (C). Filter chamber (FC). Samples were viewed within 48 h after being mounted on glass slides, with a Zeiss LSM510 Confocal Microscope (Smithsonian Marine Station, Fort Pierce, FL). Image (A) separate sample preparation method as in Sandoval-Mojica et al. (2020).
CLas-infected adult psyllids fed on FANA DNA Ligase-A treated citrus leaves produced significant reduction of CLas titer. Four FANA_DNA Ligase-A (CLIBASIA-1,-2,-3,-4) designed to different regions were evaluated were evaluated in a citrus leaf feeding assay, as described in Sandoval-Mojica et al. (2021). Ingestion of FANA ASOs targeting the DNA Ligase-A, reduced CLas titers in infected-psyllid given a 6-day feeding access period on FANA treated citrus leaves (Figure 3). However, due to mRNA folding a FANA can be blocked from binding to the target region. Thus, not all selected target regions are equally accessible. Multiple FANA designs must be screened to find the best target regions to produce the greatest amount of suppression.
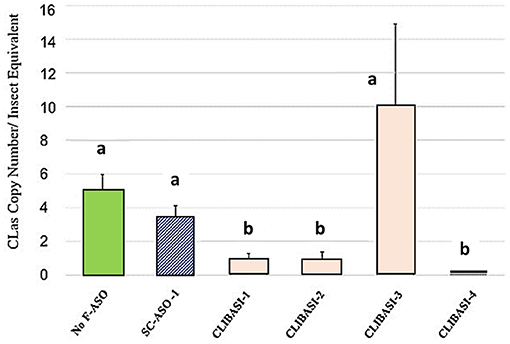
Figure 3. Psyllid ingestion of FANA ASO targeting DNA Ligase-A (CLIBASIA) reduced CLas titers in infected-psyllid given a 6-day feeding access period on FANA treated citrus leaves. Changes in CLas bacteria copy number in CLas-carrying psyllids after feeding on individual FANA leaf treatments shown. Error bars represent ±SE of the mean from six biological replicates, from three replicates. Bars with the same letter are not significantly different.
Psyllids that fed on FANA treated citrus seedlings had significantly reduced CLas titers. Two out of four FANA ASO designed to acetyl-CoA carboxylase biotin carboxylase subunit (CLas-BTIN), produced significant reductions of CLas titers in the CLas-infected adult psyllids given a 6-day feeding access period. LSD test post-hoc comparison of means (P ≤ 0.05) (Hayter, 1986). There was no significant difference observed for CLasBTIN-3 treatment with controls (Figure 4).
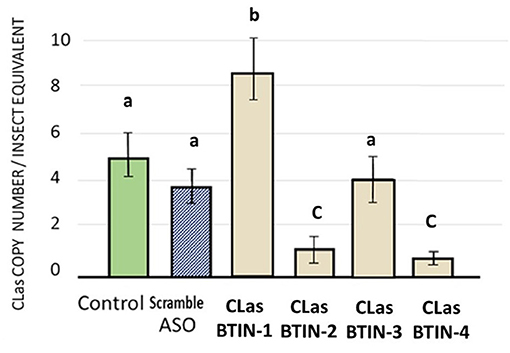
Figure 4. Effects of ingested FANA ASO by CLas-infected adult psyllid targeting CLas acetyl-CoA carboxylase biotin carboxylase subunit (CLas-BTIN). There was a statistically significant difference between group means as determined by one-way ANOVA [F(3,32) = 8.52, p = 0.0003], followed by post-hoc comparison of means used LSD test (P ≤ 0.05). Two out of four FANA ASO, designed to (CLas-BTIN) produced significant reduction of CLas in the CLas-infected adult psyllid given a 6-day feeding access period on citrus leaf disks. FANA CLasBTIN-2 and CLasBTIN-4 reduced bacteria concentrations by 3.5- to 3.7-fold less bacteria, respectively, compared to the untreated control. Error bars represent ±SE of the mean from six biological replicates, from three replicates. Bars with the same letter are not significantly different.
There was significant suppression of CLas titer in infected psyllid fed on the treatment with FANA_DNAB-1, out of the four FANA CLas_DNAB (replicative helicase DNA-B). The reduction of CLas represented a drop by 4.32 equivalents in CLas-infected psyllids fed on CLasDNAB-1 treated citrus leaves. There were no significant reductions in CLas in psyllids fed on FANA CLasDNAB-2 (0.48 CLas copy number), CLasDNAB-3 (2.37), or CLasDNAB-4 (3.7 log equivalents), as compared with the 4.8 CLas equivalents in the infected psyllids that fed on untreated control citrus leaves (Figure 5). The FANA_DNAB-1 was the most 5′ binding region within 107 nt from the “ATG” start codon (Supplementary Materials). Due to mRNA folding there can be regions that are blocked. Thus, not all FANA have equal activity. Individual FANA treatments may produce no effect, or no significant difference from the controls.
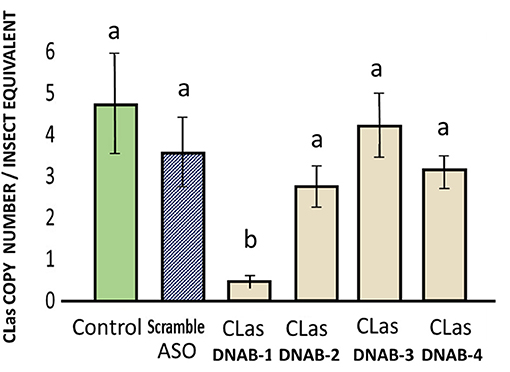
Figure 5. Suppression of CLas in infected adult psyllid given a 6-day feeding access period on FANA-CLasDNAB (replicative helicase DNA-B), treated citrus leaf tissue, which absorbed 300 μL of FANA at 1 μM concentration. Only the DNAB-1 treatment showed a significant decrease in CLas titer. One way ANOVA: F(5,48) = 5.77, P < 0.001, followed by post-hoc comparison of means using Tukey test (P ≤ 0.05). Error bars represent ±SE of the mean from six biological replicates, from three replicates. Bars with the same letter are not significantly different.
Adult psyllids were given a 12-day feeding access period on FANA-treated potted citrus seedlings to evaluate the effect of FANA targeting the Gyrase-A in the D. citri endosymbiont Profftella armatura. Reduction of the endosymbiont P. armatura significantly increased adult psyllid mortality by 76.3% (±SE 1.07) and 87.2% (±SE 0.89) after 8 and 12d of feeding on FANA-GyrA, respectively, compared to mortality in the control groups [15.7% (±SE 0.48) and 21.1% (±SE 0.38)]. The psyllid mortality between day 8 and 12 on FANA treated plants were not significantly different. There were no significance differences in mortality between psyllids on treated and control plants on day 2 or 5. Psyllid mortality reached 100% for six of 18 colonies on day 12 –feeding on FANA treated plants (33.33%), while 94% (17 of 18 colonies) had >60% mortality (Supplementary Figure 6B). The Eta Squared value (η2 = 0.92) supports a strong effect associated with the FANA treatment (Figure 6).
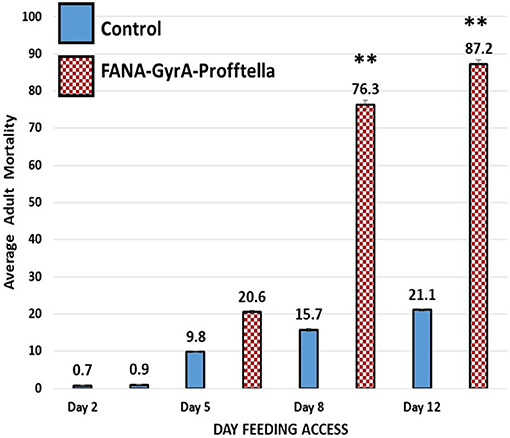
Figure 6. Average adult psyllid mortality on FANA treated citrus seedlings. Mortality of adult psyllids fed on citrus seedlings treated with FANA-GyrA to Profftella, was significantly greater than the control groups on days 8 and 12. Psyllid mortality associated with feeding on control and FANA-GyrA plants was not significantly different on days 2 or 5, reaching average of 0.8% (±SE 0.10) between treatments on day 2, and 9.81% (±SE 0.33) and 20.6% (±SE 0.24) in the control compared to the FANA-GyrA, respectively, on day 5. Single factor ANOVA, F(1,34) = 397.9, p < 0.001, η2 = 0.92, followed with Tukey post-hoc comparison of means (P ≤ 0.01). Error bars represent ±SE of the mean from 18 biological replicates, from two technical replicates. Symbol ** signifies significantly different from control.
Expression of Trehalase in psyllids given a 72 h-feeding access period on artificial diet containing FANA-TRS. Psyllids that ingested FANA DcTRS-2 showed a 0.73 log fold less of total Trehalase mRNA, while psyllids fed on DcTRS-3 had a 0.55 log fold less total Trehalase mRNA at 48 h of feeding. FANA DcTRS-1,−2, and−5, were not significantly different across all three sampled time periods (Figure 7).
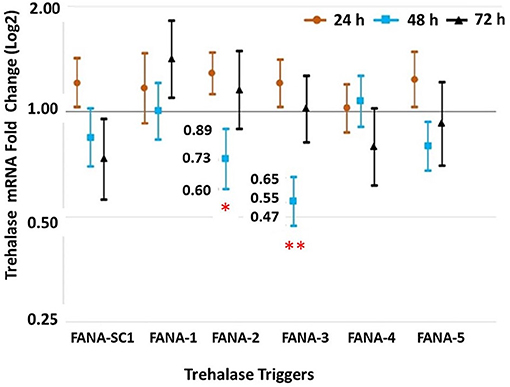
Figure 7. Feeding bioassay on diet targeting trehalase-of psyllid, D. citri. The qPCR analysis revealed that the treatments with FANA DcTRS-2 and DcTRS-3 significantly reduced mRNA Trehalase in the psyllids, D. citri at 48 h-post feeding (with reductions of 0.73 log fold and 0.55 log fold less total mRNA, respectively). Comparison of means post-hoc test using Tukey (P < 0.05). Error bars represent ±SE of the mean from six biological replicates, from three experimental replicates.
Foliar and tuber symptoms associated with CLso infection were not observed in potato plants that were not inoculated with CLso-infected potato psyllids (data not shown). Potato plants that were inoculated with CLso-infected potato psyllids did not exhibit foliar symptoms until about 5 weeks after inoculation, at which time the plants declined rapidly. Because foliar symptoms were not observed at the 2–4-wk observations, only data collected at the 6-wk observation was included in data analysis. Contingency table analyses indicated significant differences in Potato Foliar Symptom Scores of severity among treatments (row mean score = 10.5; d.f. = 3; P = 0.015). All untreated plants exhibited severe foliar symptoms associated with infection with CLso, but nearly all FANA treatments, except DNAB6, led to reduced or no foliar symptom development in at least some plant replicates (Figure 8A). There were also significant differences among treatments in Zebra Chip disease in tubers (row mean score = 5.9; d.f. = 1; P = 0.026). All tubers from untreated plants were symptomatic, while the proportion of symptomatic tubers was reduced by all FANA treatments (Figure 8B). GyraseA-4, DNAB2, and DNAB4 showed the largest reduction in tuber symptoms, and were significantly different from the untreated control (Figure 8B).
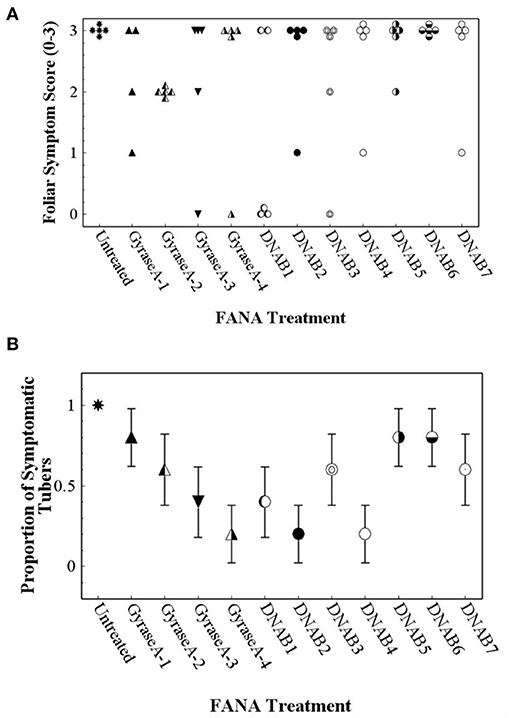
Figure 8. Foliar symptom score of “0” (no symptoms) to “3” (severe leaf yellowing) (A) and proportion of seed tubers exhibiting zebra chip disease (B) observed in potato plants 7 weeks after confining CLso-infected potato psyllids to plants, and 6 weeks after the initial treatment with water (untreated) or FANA treatments (A). Error bars represent ±SE; there is no error bar for the untreated control because all plants exhibited severe yellowing of leaves.
To confirm results of experiments conducted on potato, five treatments that showed the greatest reduction in foliar symptoms (GyraseA-2 and DNAB1) or tuber Zebra Chip disease (GyraseA-4, DNAB2, and DNAB4) were included in a second experiment using tomato plants. Foliar symptoms were first observed on untreated tomato plants just 2 weeks after removing infected psyllids, but were not observed in plants that were treated with FANA until at least the third weeks of the assay (Figure 9A). Contingency table analyses controlling for week indicated significant differences in symptom severity among treatments (row mean score = 19.4; d.f. = 5; P = 0.002). In general, plants that were treated with FANA had reduced or delayed foliar symptom development compared with the untreated plants (Figure 9A). CLso titers in leaves measured at the end of the experiment were not significantly different among treatments (F = 1.2; d.f. = 3, 42; P = 0.340), but were highly variable among plant replications ranging from nearly zero in four plants that were treated with FANA to over 100,000 copies in other plants (Figure 9B).
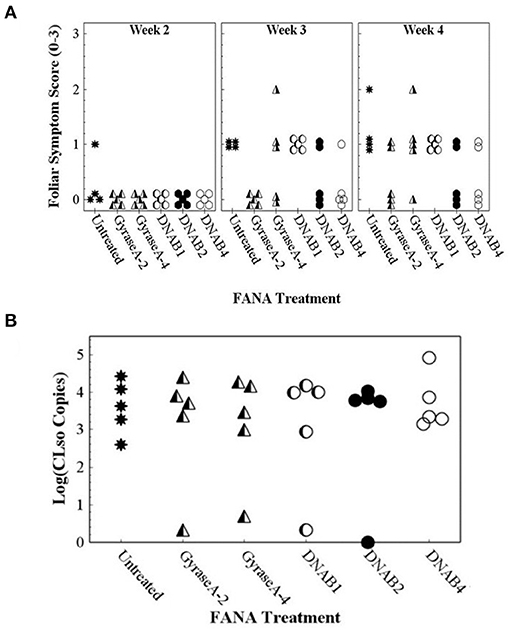
Figure 9. Foliar symptom score of “0” (no symptoms) to “3” (severe leaf yellowing) observed in tomato plants 2, 3, or 4 weeks after treating the plants with water (untreated control) or FANA products (A). Mean log number of copies of 16S gene of CLso 4 weeks after treating the tomato plants with water or FANA products (B). Error bars represent ±SE of the mean.
As crop pandemics continue to cause food shortages that threaten lives, there is a need to develop “stop-gap” emergency treatments like RNAi and FANA ASO to suppress invasive bacterial pathogens and insect vectors. FANA ASO easily enter cells without the need of additional carriers like peptides or nanoparticles. These additional traits combined with improved specificity to the RNA target, provide a therapeutic that could treat the most serious and difficult pathogens of fruit trees or crop plants, like bacteria (Kruse et al., 2019; Sastry et al., 2019; He and Creasey Krainerm, 2020). Bacteria pose some of the greatest problems due to rapid development of antibiotic resistance that limits traditional treatment effectiveness (Kumar et al., 2020; Puvača and de Llanos Frutos, 2021). Severe crop loss, stored grain loss, and starvation are associated with plant pathogens (Pimentel, 1991; He and Creasey Krainerm, 2020). Therefore, scientific research efforts are constantly working toward development of better, safer, more specific therapeutic treatments to manage pathogens and their insect vectors. Development of FANA ASO microbicides and biopesticides will provide improved targeting and binding affinity to increase pest specificity and management. These types of products could substantially reduce the amount of pesticides applied on crops and in urban settings each year. Estimates of pesticide costs for 2012, in the U.S.A. were over 380,000 tons used, at an annual cost of over 9 billion $USD (Fernandez Cornejo et al., 2014; Atwood and Paisley-Jones, 2017; Vega-Vásquez et al., 2020). Products with little to no negative impacts on biodiversity in ecosystems in and surrounding treated crops and homes would support greater numbers of beneficial organisms (Vogel et al., 2019; Lenard et al., 2020). These organisms in turn support plant growth and can increase plant yield (Mesterházy et al., 2020). Currently applied chemical pesticides and herbicides are more general in their toxicity, and broad in taxa that may be harmed (Sinisterra-Hunter and Hunter, 2018; ISAAA, 2019; Samada and Tambunan, 2020).
Thus, there is a substantial need and willingness to try new therapeutic approaches for management of devastating plant pathogens, such as HLB and Zebra Chip disease. The proposed solutions using FANA oligonucleotide as therapeutics developed as antibacterial products targeting CLas and CLso pathogens and their respective insect vectors could prove to be an attractive strategy to maintain sustainable crop productivity. The rational design for specificity and relative speed of FANA screening and development provide a treatment option to fill the void, while development of other approaches, like plant breeding for tolerance or resistance to Liberibacter infection are completed (Stover et al., 2013; Saurabh et al., 2014; Younis et al., 2014).
The results from this study demonstrate a proof-of-concept for the activity and suitability of FANA ASO treatments as a novel strategy to reduce Liberibacter bacterial pathogens: HLB in citrus trees, Zebra-Chip pathogen in potato and tomato; and to reduce their psyllid vectors. Design of FANA to the same binding site in each bacterial specie, FANA DNAB-1 for CLas and CLso both produced significant reduction of bacteria, having a 2 SNP difference, while the FANA DNAB-3 targeting the same binding region in each bacterium had one SNP and also produce similar results in their perspective host plants (Supplementary Materials). Thus, these results provide evidence that when a FANA had good activity against the CLso in the Solanaceous plant, the corresponding FANA design would most likely have a similar activity against CLas in the citrus tree host. Therefore, Solanaceous plants may provide a rapid screening system, for FANA activity against plant pathogens that may have related pathogens in other crops (Liberibacter spp. in tomato comparatively to citrus trees). Solanaceous plants grow faster providing analyses of FANA activity in 5 weeks, while citrus trials take an average of 2–3 years. This study demonstrated that FANA ASO have the capacity for direct plant delivery, without any additional carrier or surfactants. FANA ASO applied in an aqueous solution (water), to potted potato, tomato plants and citrus seedlings resulting in systemic movement and availability to insect feeding from plant vascular tissues. Treatment to the root zones of citrus seedlings and trees in pots, from 20 cm to 2 m in height, and from 1 to 15 years of age in glasshouse conditions also successfully delivered the FANA ASO in a systemic manner (Hunter et al., 2017a,b, 2018b; Metz et al., 2017a,b). Effective systemic delivery of the FANA is due to their unique modifications (Scoles et al., 2019; Ochoa and Milam, 2020). The study of chemical modified nucleotides and structures have been reviewed elsewhere (Kurreck, 2003; Bennett and Swayze, 2010; Ludin et al., 2015; Crooke, 2017; Crooke et al., 2017, 2020; Hegarty and Stewart, 2018; Shen and Corey, 2018; Verma, 2018; Scoles et al., 2019; Shen et al., 2019; Gruenke et al., 2020; Ochoa and Milam, 2020). Incorporating the knowledge gained from studies over the past 30 years show chemical modification with 2′-Fluro- improves the stability and activity of “trigger” molecules, like ASO and dsRNA, supports their incorporation when possible (Malek-Adamian et al., 2019). Reports of improved RNAi activity by incorporating 2′Fluro-modified pyrimidines into dsRNA exogenous applied treatments show increased mortality of insects after feeding on treated plant tissues (Hunter and Sinisterra-Hunter, 2018; Hunter et al., 2020). Some of the credit in improved activity is the increased resistance to nuclease degradation, thus providing more “trigger” to the target within cells (Adeyinka et al., 2020; Crooke et al., 2020; Gruenke et al., 2020). One key trait is the gymnotic, “self-delivery” property of FANA oligos comes from the modifications of the phosphorothioate backbone, and modifications that produce a high affinity for proteins on the cell surface membrane promoting increase adsorptive endocytosis (Lebedeva et al., 2000; Souleimanian et al., 2012; Alves Ferreira-Bravo et al., 2015; Rose et al., 2019; Scoles et al., 2019; Ochoa and Milam, 2020). FANA are readily absorbed by the roots of citrus trees, potato, tomato, and okra, basil, and ornamental plants (sunflower, periwinkle, Orange jasmine, Murraya paniculata) and grapevines (see text footnote 1) (Hunter and Aishwarya, 2020; Pelz-Stelinski et al., 2020; Sandoval-Mojica et al., 2021). The movement of the FANA ASO through the plant xylem and phloem enables targeting and suppression of plant-infecting bacterial pathogens, like phloem-limited CLas in citrus trees; and CLso in potato and tomato plants (Hunter et al., 2017a,b, 2018b, 2019b, 2020; Pelz-Stelinski et al., 2017; Sandoval-Mojica et al., 2021). The reduction of the CLso bacterial titers with FANA soil applied treatments in infected potted potato and tomato plants decreased the foliar symptom severity and spud browning in potato, resulting in improved plant growth. Likewise, the reduction of CLas titers in infected citrus trees, and within the adult psyllid vector was reported to reduce CLas- transmission (Pelz-Stelinski et al., 2020; Sandoval-Mojica et al., 2021). Targeting one endosymbiont, Profftella armatura of the D. citri psyllid significantly increases psyllid mortality, providing another strategy for the control of plant-feeding hemipteran vectors (Ramsey et al., 2015).
The efficient absorption of a treatment is critical to both efficient activity on the target, and in reducing cost of treatments by reducing the amount of active ingredient required. Thus, the advantages of FANA ASO are the combined characteristics of increased stability, specificity, binding affinity, plus self-delivery (Scoles et al., 2019; Gruenke et al., 2020; Ochoa and Milam, 2020). Thus, FANA have the potential to change the face of agricultural production providing a greater variety of solutions across plant, animal or microbial agriculture. As society continues to look for safer biopesticides that are, more effective the development of FANA ASO looks bright. Emerging breakthroughs in production will further decrease the cost of FANA treatments (Jarvis et al., 2020). All these benefits make FANA ASO a leading transformative technology in therapeutics that also provide new alternative treatments for economically important arthropod pests and plant pathogens.
The results presented here support the potential of FANA ASO products to reduce plant bacterial pathogens and their insect vectors. However, preliminary analyses with single dose treatments only reduced bacteria in infected citrus seedlings for about 30–60 days, while providing significant improvement in potato and tomato for 55 days. Thus, further evaluation across a range of treatment doses and/or number of applications are still needed to determine how the effective dose needed to keep bacteria suppressed and maintain crop yields. The FANA solution was absorbed by the roots and transported systemically through the plants. Insects that fed on these treated plants also showed absorption into cells and tissues in a systemic manner, resulting in increased mortality. To extend the period of protection after as single FANA treatment, we conducted a pretrial using a clay absorbent product that successfully delivered a slow-release of FANA ASO to the plant root-zone. Our results show plant's are able to absorb FANA in water solution through the roots. Thus, when the FANA is absorbed into clay-Verge™ (OilDri, Inc.) and then added into the potting soil of plants as an amendment, the clay-Verge™ product2 acts as a “slow-release” system each time the soil is watered. With this method potted citrus seedlings provided delivery of FANA ASO for at least 90 days after treatment (Detected in plant material using BioTek H-1, Supplementary Materials). The slow-release clay system was previously reported by Hunter (2017) to extend delivery of dsRNA into plants with a single application mixed into the soil of potted citrus seedling trees providing RNAi activity for over 12 months (Ghosh et al., 2018; Hunter, 2019; Adeyinka et al., 2020; Hunter et al., 2020). This slow-release clay system was shown to deliver dsRNA into a variety of potted plants (citrus, grapevine, tomato, okra, and Murraya paniculata). Foliar applied clays, or penetrants were not evaluated in this study, however several studies report using topical sprays of clay with absorbed dsRNA that show extended delivery and period of activity for exogenous applied dsRNA for 20–30 days (San Miguel and Scott, 2016; Mitter et al., 2017). Preventing degradation of dsRNA from nucleases is not a problem with FANA ASO, but use of a method that provides a slow-sustained release would extend the period of plant protection and potentially reduce the cost of FANA treatments, making them more marketable.
Treatments with FANA reduced the bacterial pathogen CLas in citrus, and CLso in potato and tomato. Application of FANA ASOs in aqueous solutions applied to the soil of potted citrus, potato, and tomato plants was an effective method of delivery resulting in systemic movement throughout the plants. The bacterial pathogens where also reduced when targeted inside the psyllid vectors that fed on FANA-treated plants, or infected leaves. The endosymbionts of psyllids (Chu et al., 2016; Morrow et al., 2017; Hosseinzadeh et al., 2019) that are critical to psyllid survival were suppressed when insects fed on FANA-treated plants. The ability to target specific endosymbiotic microbes inside of insect vectors, like psyllids, makes FANA biopesticides an important control strategy that could be applied to many agricultural pathogens (Hunter et al., 2017a,b; Metz et al., 2017a,b; Pelz-Stelinski et al., 2017; Sandoval-Mojica et al., 2021).
Although FANA's are more stable, there needs to be more research on incorporating them within current agricultural practices such as with slow-release clays, chemical penetrants or surfactants similar to slow release fertilizers and pesticides (Ghosh et al., 2018; Hunter and Sinisterra-Hunter, 2018; Rudmin et al., 2020; Vega-Vásquez et al., 2020). Advances in medical applications includes nanoparticle-assisted delivery is rapidly developing a wide variety of molecules (Hammond et al., 2021; Mendonça et al., 2021). These novel materials may provide greater utility of biopesticide products in agriculture (Vega-Vásquez et al., 2020), aided by bioengineering and novel production methods to reduce costs or to add unique traits (Hochella et al., 2019), providing benefits across the environmental sciences (Lespes et al., 2020; Li et al., 2020; Slaveykova et al., 2020). Combined all these technological advances support the movement toward that day when FANA biopesticide are used to suppress invasive arthropod pests and insect vectors of pathogens to reduce threats to food security, or health of human and animal populations (Jiang et al., 2019; Adeyinka et al., 2020; Berber et al., 2020; Fletcher et al., 2020; He and Creasey Krainerm, 2020; Huang et al., 2020; Kleter, 2020; Mesterházy et al., 2020; Vega-Vásquez et al., 2020).
This study reports on FANA treatments that target and reduce CLas bacteria in infected citrus trees and the psyllid vector, while treatments also reduced symptoms in CLso infected potato and tomato. Repeat dose treatment trials were not conducted, therefore, additional field trials need to be completed to determine the effective dose, and number of applications needed to keep CLas titers below the economic threshold and improves plant health. With this information in hand, it will be determined if FANA ASO are an affordable and suitable treatment, to reduce bacterial pathogens of Huanglongbing, HLB, or Zebra Chip disease, ZC, in potato or other Solanaceous crop plants and their psyllid vectors.
FANA antisense oligonucleotides were synthesized by AUM Biotech (see text footnote 1) (Philadelphia, PA). Over 160 FANA ASO were produced for in vitro and in vivo experiments, the designed triggers averaged 22 nt long, single-stranded gapmers, with multiple binding regions in each RNA target. The DNA gyrase subunit A gene (GyrA) has been shown to be a good target in several bacteria (Wesolowski et al., 2011, 2013), and were used to identify GyrA in CLas, CLso, Wolbachia-Diaphorina (wDi) and Profftella armatura. On average, four individual FANA sequences were designed to different regions within each RNA target in bacteria and insects, with up to eight FANA designed for some RNA targets in insects. Multiple target regions and FANA are selected due to complex folding of RNA that can block or reduce the binding affinity and activity. Thus, as there is no guarantee that the FANA will be effective until they are screened in vivo. More information on design and methods of use are available at the AUM LifeTech website (AUM LifeTech, Inc., Philadelphia, PA) (see text footnote 1). For in vivo experiments targeting bacteria, the CLas NAD-dependent DNA Ligase A gene (LigA) [CLIBASIA], gyrase A [GyrA], and CLas acetyl-CoA carboxylase biotin carboxylase subunit [BTIN], were selected as targets. Previously, the psyllid endosymbiont, Wolbachia, was shown to be suppressed in in vitro cell culture experiments using FANA to demonstrate that bacteria like CLas could be suppressed (Sandoval-Mojica et al., 2021). Visualization of FANA in plants with fluorescent probe after absorption into leaves, stems, and roots used confocal microscopy. The plant tissue was then placed into a vial, 1.5 or 2.0 mL which held a range of treatment solutions from 10 to 50 nMoles of FANA in solution. The ASO solution was absorbed into the cuttings within 6–8 h, after which the vials were filled with Milli-Q filtered water and the top of the vials wrapped with parafilm loosely around the cut petiole stems to reduce water loss and prevent psyllids from entering.
The colonies of Asian citrus psyllid (ACP) were cultured at two different laboratory locations in FL [Univ. Florida, Citrus Research and Education Center (CREC), Lake Alfred, and USDA, ARS, Fort Pierce] about 180 km (112 miles) apart. B. cockerelli, cultures were reared on Russet Burbank cv. potato plants at the USDA-ARS laboratory, Wapato, WA, USA (Cooper et al., 2015). The USDA colony established in 2000 has been reared on Citrus macrophylla Wester since 2010 (Hall et al., 2015; Hall and Hentz, 2019) The colony was maintained using procedures similar to those described by Skelley and Hoy (2004), with no infusion of wild types. The insectary colony was subdivided among many cages (up to 78 at a time) containing psyllids at various different stages of growth and age. The University of Florida culture was established in 2005 from a field population collected in Polk Co., FL (28.0′ N, 81.9′ W), before the detection of HLB in the state. Psyllids not harboring CLas were maintained on uninfected Murraya koenigii (L.) Sprengel (Rutaceae) plants in a greenhouse not exposed to insecticides. Insects infected with CLas were collected from a subset of the uninfected D. citri culture reared on CLas-positive “Pineapple” sweet orange [Citrus sinensis (L.)] plants. Both psyllid colonies were maintained at 26 ± 1°C, 60–80% relative humidity and a photoperiod of 16:8 (L:D) h. To confirm the absence/presence of the bacterium in the colonies, random subsamples of both plants and insects were tested monthly using a quantitative real-time polymerase chain reaction procedure (Li et al., 2006; Chu et al., 2016, 2019). We produced four or more biological replicates for independent sample collection. Each treatment group was compared to the TE-Buffer control using students t-test (P ≤ 0.05).
CLso-infected and uninfected colonies of potato psyllid were maintained in separate environmentally controlled rooms at 25°C with a 16:8 (L:D) h photoperiod on potato Russet Burbank at the USDA laboratory in Wapato, WA. Both CLso haplotypes A and B were kept in colonies, but our experiments only used haplotype B. In addition, three biologically distinct haplotypes (western, central, and northwestern) of potato psyllid were kept in colonies. The colonies were regularly checked for the presence or absence of CLso using diagnostic PCR with primers OA2 and OI2c to amplify a ~1,200 bp region of 16S (Crosslin et al., 2011). Purity of CLso haplotypes was periodically assessed according to Wen et al. (2013), while purity of potato psyllid haplotypes was monitored based on variability in a region of the cytochrome oxidase 1 (CO1) gene (Swisher et al., 2012).
Total RNA extraction from untreated and treated samples psyllid (adults D. citri/B. cockerelli) and plants, were performed using Direct-Zol RNA MiniPrep (Zymo Research, Irvine, CA), following the manufacturer's instructions. The concentration and quality of RNA were measured by spectrophotometry (Nanodrop™ 2000; Thermo Scientific). cDNA was synthesized from total RNA (1 μg) using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific). Quantitative PCR assays were conducted using a QuantStudio™ 6 Flex Real-Time PCR Instrument (Thermo Fisher Scientific) and the Syber Green™ PCR Master Mix (Thermos Fisher Scientific). The data was analyzed using the comparative critical threshold (ΔΔCt) method as in Li et al. (2006) in which the expression level of the target mRNA in FANA-treated samples was compared to its expression in untreated samples. Genetic data available at the Citrusgreening working group (see text footnote 2) (Brown et al., 2020), International Psyllid genome consortium3 (Saha et al., 2012, 2017a,b, 2019; Reese et al., 2013; Hunter and Reese, 2014; Arp et al., 2016; Thomas et al., 2020); the AgData Commons (Leng et al., 2017; Saha et al., 2017c,d), and NCBI4 (www.ncbi.nlm.nih.gov). Primers for CLso extracted from plant material used the primers LsoF/HLB-r for qPCR (Levy et al., 2011). Primer pairs were designed using Primer3 (v. 0.4.0 software). PCR efficiencies of target and reference genes were confirmed to be within the range of 90–100% for all qPCR assays.
The Shapiro–Wilk normality test and the Levene test of homogeneity of variances were employed to determine the type of distribution for the data obtained in each treatment. T-tests for independent samples or Mann–Whitney U-tests, depending on data distribution, were used to test for significant differences in expression levels (ΔCt values) of the target genes between the experimental and control. The differences in CLas copy number between the treatments were analyzed by Kruskal-Wallis H-test. One-way analysis of variance (ANOVA), with post-hoc analyses of means separated by Fisher's least significant difference (LSD) test (Hayter, 1986), considering P-values equal or less than 0.05 as statistically significant. The Software STATISTICA 13.3 (TIBCO Software Inc., Palo Alto, CA). Additionally, data were summarized as the mean ± SE (standard error). The data were analyzed using one-way analysis of variance (ANOVA) using Analyze-it® Statistical analysis add-in for Microsoft Excel, significance p ≤ 0.05. Post-hoc analyses of differences between means used Tukey (P ≤ 0.05) for differences to be considered statistically significantly different. Tukey Q Calculator (https://www.socscistatistics.com/pvalues/qcalculator.aspx). The severity of foliar symptoms and the proportion of symptomatic tubers were analyzed by contingency table analysis (PROC FREQ of SAS 9.4) using the Conchran-Mantel-Maenzel Row-Mean-Score statistic. CLso titers were analyzed using the GLIMMIX procedure of SAS 9.4 with the log number of copies as a dependent variable and treatment as the fixed effect.
DNA isolated from CLas bacterium from infected citrus trees was used to validate CLasP1 primers with positive control primers for RNA polymerase sigma 70 factor and 16S region. Each reaction consisted of 1 μL DNA, 0.5 μL of 10 μM of each primer, and 23 μL Platinum® PCR SuperMix (Invitrogen). PCR was performed in an MJ Research Peltier Thermal-cycler using the following parameters: 3 min at 92°C, followed by 45 cycles of 30 s at 92°C, 30 s at 60°, and 60 s at 72°C, and a single final cycle of 72°C for 10 min. All PCR reactions were performed in technical replicates. Ten microliter of each replicate was fractionated by electrophoresis for 45 min in a 2% agarose gel stained with ethidium bromide. Infected CLas-citrus seedlings were produced by psyllid transmission, at the USDA, ARS, U.S. Horticultural Research Lab, Fort Pierce, FL. Methods as in McCollum et al. (2018), McCollum et al. (2019), and McCollum (2021).
Reactions were performed to amplify the large region of the DNAB transcript following the previous method. A single edged blade was used to excise the amplified fragment from the agarose gel. A NucleoSpin® Gel and PCR Clean-up kit (Maherey-Nagel Ref. 740609.250) was used to purify the gel fragment according to manufacturer's protocol. The RNA was eluted using nuclease free water and quantified using a NanoDrop™ 8000 Spectrophotometer (Thermo Fisher Scientific).
Efficacy of target binding was analyzed by quantitative PCR. Samples were analyzed in three technical replicate reactions that consisted of: 2 μL sample, 0.5 μL of 10 μM of each CLasP1 primer, 1 μL ROX reference dye (diluted 1:10), 12.5 μL Platinum® SYBR® Green qPCR SuperMix-UDG, and 8.5 μL of nuclease free water. Quantitative PCR was performed in an Applied Biosystems 7500 Real-Time PCR System with the following parameters: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. Melting curve analysis was also obtained following completion of the final cycle. Analysis using qRT-PCR was performed using primers to CLas previously reported in the literature (Li et al., 2006), primers to the CLas Replicative DNAB helicase gene, and primers to the citrus dehydrin gene as the internal leaf control. Primers used for studying gene expression with reference to psyllids included the Elongation factor 1-alpha (EF1-a); wingless (Wg) and α-tubulin (Supplementary Materials). After an initial activation step at 95°C for 5 min, 40 cycles (95°C for 30 s, 57°C for 30 s, 72°C for 30 s) were performed. Cycle threshold (Ct) values were determined using the 7,500 Fast software supplied with the instrument. Levels of transcript expression were determined via the 2−ΔΔCt method (Livak and Schmittgen, 2001) by normalizing the amount of target transcript to the amount of the reference transcripts mentioned above.
Citrus cuttings were obtained and prepared using the method in Andrade and Hunter (2016, 2017). In brief, the new growth shoots from citrus trees (several varieties were tried: sweet orange, sour orange, grapefruit, and Carrizo rootstock variety), ornamentals, Murraya paniculata, periwinkle, were cut and washed in 0.2% hypochlorate solution, 10 min, then twice with deionized water, 10 min, each time. The cuttings had all but the most apical leaves removed (3 leaves, or leaflets), and the stem or petiole end was cut while held underwater at ~45 degrees with a new, clean razor. The cutting was then placed into a vial, 1.5 mL or 2.0 mL that held a range of treatment solutions from 10 to 50 nMoles, of FANA-ASO in solution. The ASO solution was absorbed into the cuttings within 6–8 h, after which the vials were filled with Milli-Q filtered water and the tops wrapped with parafilm to reduce water loss. For psyllid feeding assays the samples were maintained as needed from 2 to 40 days depending upon the bioassay by using a sterile needle and syringe to add filtered water, pH = 6.5–6.8 through the parafilm to fill each vial (~every 2–3 d).
The FANA-probe (either ATTO 530 or 633) was used at a concentration of 6.93 nmol in 500 μL water, absorbed into each detached citrus leaf. Image view ~40X, each leaf was cut into pieces and the tissue and whole insects were added to individual aliquots in four 1.5 mL vials of 4% paraformaldehyde for 24 h. Once fixed, the tissue was further dissected, midribs cross-section, and mounted onto glass slides. Control leaves with counter stain, Nuclear Green stain (NucGreen™ 488 nm, ThemoFisher Scientific) added prior to tissue before mounted in Fluoromount-G® (Fluoromount-G® Anti-Fade Cat. No. 0100-35 SouthernBiotech, Birmingham, AL, USA) and coverslip added. Images were taken with a Zeiss LSM510 Confocal Microscope (Figure 1). Methods as in Ammar et al. (2011).
Asian citrus psyllids were fed on cut citrus petioles with 3–4 young leaves placed into 200 μL volume of 6.93 nmol FANA in water. Cut petioles were from Citrus sinensis (L.) Osbeck (Sweet orange group: “Pineapple” cv. sweet orange, or Madam vinous sweet orange), Citrus macrophylla, or rootstock Carrizo citrange, or Citrus aurantifolia (Key Lime). Cut leaf pieces were prepared and observed on the same day by mounted in a drop of Fluoromount-G® (Fluoromount-G® Anti-Fade Cat. No. 0100-35 SouthernBiotech, Birmingham, AL, USA). Small pieces of filter paper (Whatman Grade #1) were placed around the edge of the coverslip to absorb excess mounting medium. The coverslip (Corning Optical Cover Glasses, 22 × 50 mm, 0.12–0.16 mm), edges were sealed with clear finger nail polish (Covergirl™). Sample were observed with a Zeiss LSM510 Confocal Microscope, Plant ASO delivery methods as in Hunter and Sinisterra-Hunter (2018) and Sandoval-Mojica et al. (2021).
The anatomy of the digestive system in Diaphorina citri and Bactericera cockerelli, along with methods of preparation for scanning and transmission electron microscopy (SEM and TEM, respectively) are reviewed in Cicero et al. (2009), Alba-Alejandre et al. (2018, 2020), and Alba-Tercedor et al. (2017, 2021). For fluorescent labeling adult psyllids fed on FANA treated citrus leaves for 3 d or more were dissected, placing dissected organs (i.e., alimentary tract) (Figure 2) into aliquots of fixative, 300 μL of 4% paraformaldehyde pH = 7 (6 h to overnight) (Electron Microscopy Sciences). Then samples were rinsed 3X in 1.0 M PBS buffer, pH = 8.5 (ThermoFisher Scientific). The counter-stains used were either one drop of Nuclear Green (NucGreen™ 488 nm, ThermoFisher Scientific), or DAPI; with some tissues submerged into a volume of the FANA solution (10 mM/1 mL) for a range of absorption times [1, 3, 8 h, overnight (~14 h)]. Tissue was rinsed twice, for 5 min each (1.0 M PBS buffer pH = 8.5), and twice in filtered water with the pH adjusted between 7.5 and 8.0. The excess liquid in the tissue was gently wicked away with a torn piece of Whatman® Grade #2 Filter Paper (Sigma-Aldrich), before placing on a glass slide. Mounted in a drop of Fluoromount-G® (Fluoromount-G® Anti-Fade Cat. No. 0100-35 SouthernBiotech, Birmingham, AL, USA). Small pieces of filter paper (Whatman Grade #1) were placed around the edge of the coverslip to absorb excess mounting medium. The coverslip (Corning Optical Cover Glasses, 22 × 50 mm, 0.12–0.16 mm), edges were sealed with clear finger nail polish (Covergirl™). Sample were observed with a Zeiss LSM510 Confocal Microscope, ~40X magnification. The control sample was observed first with settings place to just below background, then the treated sample observed at the same setting for above background signal (Microscope Lab at Smithsonian Oceanic Marine Station, Ft. Pierce, FL) (Figure 2).
Mfold web server was used to predict potential nucleic acid folding and hybridization to RNA targets by each FANA ASO. Fold prediction software, like MFOLD, can help show if a selected binding site is “open,” not blocked by complex folds and accessible for binding by the FANA ASO. In the example below the blue dots show the binding site prediction after folding of the FANA to DNAB-in CLas and CLso. Prediction shows region to be an open and suitable target for potential binding (http://unafold.rna.albany.edu/?q=mfold) (Zuker, 2003). Folding was performed with MFold program using standard conditions for RNA (Markham and Zuker, 2008) (Supplementary Figures 6.8A,B).
Five aliquots consisting of 5 ng purified DNAB were prepared in 0.2 mL microcentrifuge tubes. Each aliquot was treated with a single ASO and nuclease free water to dilute samples to final concentrations of: 0 μM (control), 0.34 μM FANA ASO, 0.67 μM FANA ASO, 1.01 μM FANA ASO, and 1.36 μM FANA ASO. Samples were vortexed gently and incubated at room temperature for 30 min. Relative binding of each single ASO to each transcript target was quantified using qPCR (Supplementary Materials).
Two to 3 years-old Citrus paradisi × Poncirus trifoliate, not treated with systemic insecticides, were used for plant assays. The trees were inoculated with CLas by exposing them to infected ACP for 1 month. After the inoculation access period, all developmental stages of ACP were eliminated from the trees by an insecticide treatment. The plants were maintained in a greenhouse for 4 months to allow for systemic infection of CLas.
The initial CLas titer (T0) of each tree was calculated by collecting three leaves per plant and extracting their genomic DNA, followed by quantitative real-time PCR. The Citrus trees were then treated with the LigA FANA ASO by root infusion (see text footnote 1) (Pelz-Stelinski et al., 2017, 2020). A single root from each tree was gently scraped under water with a razor blade and fitted into clear silicone tubing (1 m long, 6 mm diameter). The tubes were filled with 5 mL of either a 5 μM FANA ASO solution, a 5 mg/mL streptomycin sulfate solution or water (untreated control). Tubes were filled with water 24 h after treatment. From each tree, three leaves were removed from similar locations as the T0 samples at 2, 7, 14, and 30 days after treatment, to monitor the effect of the FANA ASO on the plants CLas titer. Five trees were used per treatment.
Leaf DNA was extracted as described in Pelz-Stelinski and Killiny (2016) and diluted to 15 ng μL-1. A multiplex TaqMan qPCR assay was performed as stated before, using probe and primers targeting CLas 16S rRNA gene and the citrus mitochondrial cytochrome oxidase gene (Cox) as internal control for DNA extractions. CLas copy number was quantified as previously reported (Coy et al., 2014). The standard curve obtained for CLas in the plant experiments was (y = −3.312x + 11.763; R2 = 0.99). The treatments effect was expressed as percent change in CLas titer [(mean titer after treatment - mean titer prior to treatment)/mean titer prior to treatment] × 100 (Cangelosi and Meschke, 2014). Initial and final CLas copy number in plants was compared by treatment using one-way analysis of variance (ANOVA). Means were separated by Fisher's least significant difference (LSD) test, considering P ≤ 0.05 as statistically significant.
Infected adult psyllids (≤3 days old) were placed in feeding arenas where they were exposed to a 5 μM FANA solution (LigA-FANA) or a 17% (w:v) sucrose solution, as described above. After seven days of continuous feeding, the psyllids were transferred to uninfected 9 months old Citrus macrophylla plants for a 15 d inoculation access period. Twenty adult psyllids (10 males and 10 females) were enclosed on individual plants using 1 L plastic deli containers equipped with mesh insect-proof panels for ventilation. Five plants were used per treatment. CLas-free psyllids were caged with healthy Citrus macrophylla seedlings as negative control. After the inoculation access period, every psyllid was collected and stored in 80% ethanol for subsequent CLas detection. The plants were maintained in an insect proof greenhouse and sampled after 6 months to quantify CLas infection. Three leaves were randomly sampled from each plant. DNA was extracted to determine the CLas copy number of each seedling as described before. Kruskal-Wallis H-test and Mann-Whitney U-test were used to compare CLas copy number between the treatments (P ≤ 0.05).
A 48-well leaf disc assay can be used to screen for efficacy of FANA ASO to reduce CLas in leaves of infected citrus trees {Valencia sweet orange [Citrus sinensis (L.) Osbeck]}. Valencia orange plants, previously validated to be CLas infected, were used as infection source material. Samples were assayed for CLas 16S rDNA target via qPCR. Results were considered CLas positive when mean Ct of three technical replicates of the same extract had a mean of ≤38 (CN = 1) (McCollum et al., 2018). Extracted nucleic acids were used for quantitative real-time PCR (qPCR) using primers and probe according to standard protocols. The method reduces variation and increases normalization of CLas copy number across citrus leaves from one tree. A set of 4–12 leaves per tree were collected and processed individually. The petiole or proximal end of the leaf attached to the tree was cut and collected (8.0–8.5 mm in length) and was used as a pretest to validate CLas positive leaves (positive if Ct value ≤ 36). Five to six disks were punched out along the midrib of CLas positive leaves starting from the proximal end (petiole). The leaf disks were floated on solutions containing individual FANA targeting CLas DNA Ligase (1–4), at ~50–100 nM concentration in basal salt mixture, M524 (PhytoTechnology Laboratories, Shawnee Missions, KS, 66282, USA). The individual leaf disks were collected and analyzed at 1 and 24 h after treatment. The extracted samples were run as technical replicate reactions in qPCR analyses. The Total CLas genome equivalents, gDNA, were compared to the CLas RNA Ct value to calculate CLas equivalents (Cangelosi and Meschke, 2014). A bacterial RNA/DNA ratio was calculated to get the indicator for live bacteria.
An in vivo assay was performed by collecting CLas infected leaves and validating infection (CLas copy number) by screening the petioles using real-time PCR, as previously described. Five leaf disks (10 mm diameter) were punched out with metal cork bore tool, down the midrib of each leaf and placed in a 48-well plate that contained 100 μL sterile water. The first two leaf disks closest the petiole were treated with individual FANA ASO targeting CLas-DNAB, replicative DNA B helicase (1–4). The leaf disc samples were collected at 1 and 24 h, then homogenized under liquid nitrogen, and stored in Trizol reagent. Total RNA isolation was performed using Zymo Research Direct-Zol™ RNA Microprep kit.
Infected adult psyllids (≤3 days old) were placed in feeding arenas containing an artificial diet solution consisting of 17% (w:v) sucrose solution plus (5 μM FANA ASO solution), or a 17% (w:v) sucrose solution. Sixteen adult psyllids (eight males and eight females) were placed in artificial feeding arenas for exposure to FANA ASO. The FANA ASO treatments targeting different C. Liberibacter asiaticus essential genes: CLas Gyrase A, CLas DNA Helicase, CLas biotin carboxylase subunit, or CLas Ligase. Two FANA ASO concentrations were used: 5 and 20 μM. RNA was extracted from each group of 16 psyllids, 6 days after treatment, followed by cDNA synthesis and quantitative reverse transcriptase PCR analysis. The copy number of the psyllid gene Wingless used as a reference gene.
The following experiments have shown successful suppression of psyllid mRNA transcripts using FANA ASO after ingestion. The enzyme trehalase controls the hydrolysis of trehalose, which has a role in the regulation of energy metabolism and glucose generation via trehalose catabolism (Shukla et al., 2015). The FANA ASO were designed to psyllid trehalase mRNA, DcTRS-1,−2,−3,−4,−5. A feeding bioassay was conducted and evaluated at 48 h after feeding. A 250 μL volume containing 1 μM of each FANA DcTRS molecule was added to artificial diet. This was repeated for the water control and Scramble SC-ASO control. Sixteen adult psyllids were introduced to each arena. The psyllid were given 24, 48, and 72 h of feeding access. Three biological replicates collected for each time period. Psyllid were sacrificed and total RNA was extracted from each sample. The RT-qPCR was performed in triplicates using primers for the target mRNA as well as a housekeeping gene (Elongation Factor, Accession No. XM_026827967.1 NCBI).
Insects were provided feeding access on citrus and other plants, which were treated with FANA ASO via soil treatments to potted plants, to investigate the effect of FANA ASOs targeting the psyllid endosymbiont C. Profftella armatura, and the effect on psyllid survival. Individual FANA were applied to potted plants either as 100 mL water solution (12.5 nmol FANA), or as a soil additive of montmorillonite clay pellets (see text footnote 2), mixed into the potting soil (35 g Clay-Verge™ in 25 mL water with 12.5 nmol FANA) (Ghosh et al., 2018; Hunter and Aishwarya, 2020). Both routes of soil treatment showed sufficient delivery for ingestion by insects with subsequent watering for root absorption and systemic movement of the FANA ASO throughout the plant tissues (Supplementary Materials).
The scramble control FANA-SC solution was prepared by mixing 200 nmoles (1.6 mg) of the oligonucleotide in 100 mL water for plants potted in pots or containers 2.6 L or larger in volume. The soil was not watered for 2 days prior, then the treatment was applied. Within 1 h of application to the soil, an additional volume of water alone, 100 mL, was applied to ensure the soil in the pot was adequately wetted. Plastic drip pans below the pots rarely showed excess runoff.
Clay pellet product, montmorillonite clay, used the same concentration 200 nmoles (1.6 mg) of FANA-SC-probe was mixed in 25 mL water. This solution was poured into an empty 50 mL centrifuge tube containing 35 g of granular dry clay particles (see text footnote 2). The cap was screwed onto the tube and shaken to mix the clay absorbent and FANA solution. The 35 g of wet clay was then mixed into half the volume of potting soil (damp potting mix, no water drip when squeezed in hand) that would fill the pot container. Each pot then had half the volume filled with regular potting soil, the citrus seedling was place into the pot and then the pot filled the rest of the way with the FANA+clay absorbent soil mixture. At 1 h after potting, 100 mL of water alone (no product) was then poured onto the soil to provide adequate water for root absorption. Plants were watered on a schedule of 100 mL water only, per potted citrus seedling, once or twice a week depending on evaporation for the duration of the experiment, method as in Ghosh et al. (2018). Analyses of the leaves and roots, plus insects that fed on the treated plants used confocal microscopy of plant and insect tissues revealed similar results to those previously shown for insects after fed on FANA+fluorophore absorbed into plant cuttings or topically applied foliar sprays (Hunter, 2019) (Supplementary Materials).
Citrus tissues were collected and processed for visualization using confocal microscopy at 5, 7, and 10 days after treatment. A second validation detection method used a BioTek™ Synergy H1, microplate fluorescence reader (Wavelength excitation scans were set from 300 to 700). Plants were sampled taking two leaves per plant, per treatment on days 2, 5, 10, and 30 after treatment. Tissue homogenized in water, centrifuged at 200 × g for 3 min, and the supernatant was added as a triplicated aliquot 300 μL per well of a 96 well plate specific for the spectrophotometric plate reader. Scan setting was set full to run 300–700 wv reads. Psyllid were collected live after feeding on plants or cuttings with FANA fluorophore-tagged (ATTO 633 or ATTO 530 wv), across a range of feeding access periods (3, 8, 24 h, and at 3, 7 d) to determine FANA absorption and systemic movement using both methods described above. The detection of the tagged-FANA in untreated leaves after soil applied treatments of potted citrus, tomato, and potato, along with detection in roots of potted citrus seedlings that had exogenously applied topical spray treatments to the leaves (Note: pots were wrapped in plastic to prevent contamination to soil and plant roots), both supports systemic movement of FANA ASO in plants (Supplementary Materials). Confocal microscopy showed tagged-FANA in xylem, and phloem when examined citrus leaf midribs (Figure 1). Furthermore, trials where the FANA where applied to the soil of potted citrus seedlings, potato, or tomato plants, provided for psyllid feeding-access resulted in RNA suppression in the psyllid, and of CLas or CLso in the infected plants.
Five potted potato plants (Russet Burbank cultivar) in 2.6 L (one gallon pots) (five per treatment) were inoculated by psyllid transmission with CLso haplotype “B” by confining three Liberibacter-infected psyllids to a single leaf for 1 week. Control-CLso infected plants were inoculated with psyllids, while Control-uninfected plants were challenged with uninfected potato psyllids (B. cockerelli). Following a 1-week inoculation access period, the psyllids were removed and confirmed to harbor CLso using PCR (Crosslin et al., 2011). Each plant was then treated with 100 mM FANA solution (one of 11 target sequences including 4 GyrA-FANA and 7 DNAB-FANA) or water (untreated controls) using a 100 ml solution as a soil applied drench. Two weeks (~14 days) following the initial treatment, the second dose of treatments were applied in the same manner as described. Development of Foliar Symptom Severity symptoms was monitored and photo-documented every 2 weeks for 6 weeks, and symptoms were ranked ordinals from “0” to “3,” where 0 = No symptoms, 1 and 2 = Leaf yellowing, and 3 = Dead. Seed tubers were harvested 6 weeks after the initial treatment and were sliced to observe the presence or absence of Zebra Chip symptoms. The severity of foliar symptoms and the proportion of symptomatic tubers was analyzed by contingency table analysis (PROC FREQ of SAS 9.4) using the Conchran-Mantel-Maenzel Row-Mean-Score statistic.
A second experiment was conducted on tomato “Moneymaker” cv. as described for potato, except that foliar symptoms were recorded every week, and the assay was terminated 4 weeks after removing infected psyllids. At the end of the experiment, three leaflets from separate leaves were collected from each plant to quantify pathogen titers (Levy et al., 2011). The severity of foliar symptoms was analyzed by contingency table analysis while controlling for week using the Conchran-Mantel-Maenzel Row-Mean-Score statistic. CLso titers were analyzed using the GLIMMIX procedure of SAS 9.4 with the log number of copies as a dependent variable and treatment as the fixed effect.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Materials.
WH, KP-S, AS-M, and WC: planned, designed, and conducted experiments. AS-M, WH, WC, GM, and VA: collected data. WH and KP-S: selected the target regions. WH and VA: FANA designs and production. WH, AS-M, and KP-S: processed and analyzed citrus data. WC and WH: experimental design, collected, and analyzed potato data. WH: wrote the first draft. WH, KP-S, and WC: wrote the final manuscript. All authors reviewed the manuscript.
This study was supported in-part by USDA, NIFA, National Institute of Food and Agriculture, Citrus Greening award #2015-70016-23028, Developing an Infrastructure and Product Test Pipeline to Deliver Novel Therapies for Citrus Greening Disease; USDA, NIFA-SCRI award #2015-10479, Targeting Microbes to Control Huanglongbing Disease of Citrus; and the USDA, NIFA, Western IPM Center award #1588 Novel control of the potato zebra chip pathogen and psyllid vector using FANA antisense oligonucleotides gene silencing. This research was supported in part by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE was managed by ORAU under DOE contract number DE-SC0014664.
Mention of trade names or commercial products herein is solely for the purpose of providing specific information and does not imply recommendation or endorsement, to the exclusion of other similar products or services by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer. All opinions expressed in this paper are the author's and do not necessarily reflect the policies and views of USDA, ARS, DOE, or ORAU/ORISE.
VA and AS-M are employees of AUM LifeTech, Inc., makers of the FANA products. No funding was received from AUM LifeTech, Inc., for these studies. Research was performed under cooperative research agreements (WH), USDA, ARS with (VA), AUM LifeTech, Inc. (KP-S), Univ. Florida from 2015 to 2019.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge and thank Maria T. Gonzalez, Salvador P. Lopez, Jennifer Wildonger Biological Technicians, USDA, ARS, Ft. Pierce, FL. Pauline Anderson, Jennifer Stout, Biological Technicians, USDA, ARS, Wapato, WA. Chris Holland, ORISE participant, BioMav, LLC, Ft. Pierce, FL. Jackie L. Metz, Univ. Florida/USDA, ARS; Dr. Michael Boyle, Smithsonian, Microscopy Facility, Oceanic Marine Station, Ft. Pierce, FL. Thanks to Tom Rutherford and Yasmith Bernal, for product VERGE™ Clay pellets (see text footnote 2).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.675247/full#supplementary-material
1. ^AUM LifeTech, Inc., More information on FANA products in this study at: www.aumlifetech.com.
2. ^Oil-Dri Corporation of America, Chicago, Illinois, USA. http://www.oildri.com.
3. ^Citrusgreening Working group/International Psyllid Genome annotation: www.citrusgreening.org.
4. ^National Center for biotechnology Information, NCBI, https://www.ncbi.nlm.nih.gov.
Adeyinka, O. S., Riaz, S., Tougiq, N., Yousaf, I., Bhatti, M. U., Batcho, A., et al. (2020). Advances in exogenous RNA delivery techniques for RNAi mediated pest control. Mol. Biol. Rep. 47, 6309–6319. doi: 10.1007/s11033-020-05666-2
Ahmed, M. Z., Li, S. J., Xue, X., Yin, J., Ren, S. X., Jiggins, F. M., et al. (2015). The intracellular bacterium Wolbachia uses parasitoid wasps as phoretic vectors for efficient horizontal transmission. PLoS Pathog. 11:e1004672. doi: 10.1371/journal.ppat.1004672
Alba-Alejandre, I., Alba-Tercedor, J., and Hunter, W. B. (2020). Anatomical study of the female reproductive system and bacteriome of Diaphorina citri Kuwayama, (Insecta: Hemiptera, Liviidae) using micro-computed tomography. Sci. Rep. 10:7161. doi: 10.1038/s41598-020-64132-y
Alba-Alejandre, I., Hunter, W. B., and Alba-Tercedor, J. (2018). Micro-CT study of male genitalia and reproductive system of the Asian citrus psyllid, Diaphorina citri Kuwayama, 1908 (Insecta: Hemiptera, Liviidae). PLoS ONE 13:e0202234. doi: 10.1371/journal.pone.0202234
Alba-Tercedor, J., Hunter, W. B., and Alba-Alejandre, I. (2021). Using micro-computed tomography to reveal the anatomy of adult Diaphorina citri Kuwayama (Insecta: Hemiptera, Liviidae) and how it pierces and feeds within a citrus leaf. Sci. Rep. 11:1358. doi: 10.1038/s41598-020-80404-z
Alba-Tercedor, J., Hunter, W. B., Cicero, J. M., Sáinz-Bariáin, M., and Brown, S. J. (2017). “Use of micro-CT to elucidate details of the anatomy and feeding of the Asian Citrus Psyllid Diaphorina citri Kuwayama, 1908 (Insecta: Hemiptera, Liviidae),” in Micro-CT User Meeting 2017 (Brussels: Royal Belgian Institute of Natural Science), 270–285. Available online at: https://www.citrusgreening.org/microtomography/index
Aldemita, R. R., Reaño, I. M., Solis, R. O., and Hautea, R. A. (2015). Trends in global approvals of biotech crops (1992-2014). GM Crops Food. 6, 150–166. doi: 10.1080/21645698.2015.1056972
Alves Ferreira-Bravo, I., Cozens, C., Holliger, P., and DeStefano, J. J. (2015). Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 43, 9587–9599. doi: 10.1093/nar/gkv1057
Ammar, E.-D., Ramos, J. E., Hall, D. G., Dawson, W. O., and Shatters, R. G. Jr. (2016). Acquisition, replication and inoculation of Candidatus Liberibacter asiaticus following various acquisition periods on huanglongbing-infected citrus by nymphs and adults of the Asian citrus psyllid. PLOS ONE 11:e0159594. doi: 10.1371/journal.pone.0159594
Ammar, E.-D., Shatters, R. G. Jr., and Hall, D. G. (2011). Localization of Candidatus Liberibacter asiaticus, associated with citrus huanglongbing disease, in its psyllid vector using fluorescence in situ hybridization. J. Phytopathol. 159, 726–734. doi: 10.1111/j.1439-0434.2011.01836.x
Ammar, E.-D., Shatters, R. G. Jr., and Heck, M. (2020b). “Huanglongbing pathogens: acquisition, transmission and vector interactions,” in Asian Citrus Psyllid, Chapter 8, Biology, Ecology and Management of the Huanglongbing Vector, eds P. Stansly and J. Qureshi [Wallingford: Commonwealth Agricultural Bureau International (CABI Press)], 113–139.
Ammar, E. D., George, J., Sturgeon, K., Stelinski, L. L., and Shatters, R. G. (2020a). Asian citrus psyllid adults inoculate huanglongbing bacterium more efficiently than nymphs when this bacterium is acquired by early instar nymphs. Sci. Rep. 10:18244. doi: 10.1038/s41598-020-75249-5
Andrade, E. A., and Hunter, W. B. (2017). RNAi feeding bioassay: development of a non-transgenic approach to control Asian citrus psyllid and other hemipterans. Entomol. Exp. Appl. 162, 389–396. doi: 10.1111/eea.12544
Andrade, E. C., and Hunter, W. B. (2016). “RNA interference – Natural gene-based technology for highly specific pest control (HiSPeC),” in RNA Interference, ed I. Y. Abdurakhmonov (Rijeka: InTech), 391–409.
Antolinez, C. A., Fereres, A., and Moreno, A. (2017). Risk assessment of 'Candidatus Liberibacter solanacearum' transmission by the psyllid Bactericera trigonica and B. tremblayi from Apiaceae crops to potato. Sci. Rep. 7:45534. doi: 10.1038/srep45534
Arp, A. P., Pelz-Stelinski, K., and Hunter, W. (2016). Annotation of the Asian citrus psyllid genome reveals a reduced innate immune system. Frontiers in cellular and infection microbiology. Front. Physiol. 7:570. doi: 10.3389/fphys.2016.00570
Atwood, D., and Paisley-Jones, C. (2017). Pesticides Industry Sales and Usage. United States Environmental Protection Agency, 24. Available online at: https://www.epa.gov/sites/production/files/2017-01/documents/pesticidesindustry-sales-usage-2016_0.pdf (accessed February 5, 2020).
Avery, P. B., Hunter, W. B., Hall, D. G., Jackson, M. A., Powell, C. A., and Rogers, M. E. (2009). Diaphorina citri (Hemiptera: Psyllidae) infection and dissemination of the entomopathogenic fungus Isaria fumosorosea (Hypocreales: Cordycipitaceae) under laboratory conditions. Fla. Entomol. 92, 608–618. doi: 10.1653/024.092.0413
Avery, P. B., Pick, D. A., Aristizábal, L. F., Kerrigan, J., Powell, C. A., Rogers, M. E., et al. (2013). Compatibility of Isaria fumosorosea (Hypocreales: Cordycipitaceae) blastospores with agricultural chemicals used for management of the Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae). Insects 4, 694–711. doi: 10.3390/insects4040694
Avery, P. B., Wekesa, V. W., Hunter, W. B., Hall, D. G., McKenzie, C. L., Osborne, L. S., et al. (2011). Effects of the fungus Isaria fumosorosea (Hypocreales: Cordycipitaceae) on reduced feeding and mortality of the Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae). Biocontrol Sci. Technol. 21, 1065–1078. doi: 10.1080/09583157.2011.596927
Bennett, C. F., and Swayze, E. E. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293. doi: 10.1146/annurev.pharmtox.010909.105654
Berber, B., Aydin, C., Kocabas, F., Guney-Esken, G., Yilancioglu, K., Karadag-Alpaslan, M., et al. (2020). Gene editing and RNAi approaches for COVID-19 diagnostics and therapeutics. Gene Ther. 28, 290–305. doi: 10.1038/s41434-020-00209-7
Blacutt, A., Ginnan, N., Dang, T., Bodaghi, S., Vidalakis, G., Ruegger, P., et al. (2020). An in vitro pipeline for screening and selection of citrus-associated microbiota with potential anti-“Candidatus Liberibacter asiaticus” properties. Appl. Environ. Microbiol. 86, e02883–e02819. doi: 10.1128/AEM.02883-19
Borges, K. M., Cooper, W. R., Garczynski, S. F., Thinakaran, J., Jensen, A. S., Horton, D. R., et al. (2017). “Candidatus Liberibacter solanacearum” associated with the psyllid, Bactericera maculipennis (Hemiptera: Triozidae). Environ. Entomol. 46, 210–216. doi: 10.1093/ee/nvw174
Bové, J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37. doi: 10.4454/jpp.v88i1.828
Bramlett, M., Plaetinck, G., and Maienfisch, P. (2020). RNA-based biocontrols -a new paradigm in crop protection. Engineering 6, 522–527. doi: 10.1016/j.eng.2019.09.008
Brown, S. J., Shatters, R. S., Lupsky, S., Cilia, M., Saha, S., Mueller, L., et al. (2020). Developing an Infrastructure And Product Test Pipeline to Deliver Novel Therapies for Citrus Greening Disease (Final Report). USDA; National Institutes of Food and Agriculture, NIFA Award #2014-10154. Available online at: https://portal.nifa.usda.gov/web/crisprojectpages/1005600-developing-an-infrastructure-and-product-test-pipeline-to-deliver-novel-therapies-for-citrus-greening-disease.html
Cangelosi, G. A., and Meschke, J. S. (2014). Dead or alive: molecular assessment of microbial viability. Appl. Environ. Microbiol. 80, 5884–5891. doi: 10.1128/AEM.01763-14
Cansizoglu, M. F., and Toprak, E. (2017). Fighting against evolution of antibiotic resistance by utilizing evolvable antimicrobial drugs. Curr. Genet. 63, 973–976. doi: 10.1007/s00294-017-0703-x
Chaverra Rodriguez, D., Bui, M., Li, M., Raban, R., and Akbari, O. (2020). Developing genetic tools to control ACP. Citrus Research Board. Citrogr. Mag. 11, 64–68. Available online at: https://citrusresearch.org/citrograph/archive
Chaverra-Rodriguez, D., Macias, V. M., Hughes, G. L., Pujhari, S., Suzuki, Y., Peterson, D. R., et al. (2018). Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 9:3008. doi: 10.1038/s41467-018-05425-9
Chen, X., and Stansly, P. A. (2014). Biology of Tamarixia radiata (Hymenoptera: Eulophidae), parasitoid of the citrus greening disease vector Diaphorina citri (Hemiptera: Psylloidea): a mini review. Fla. Entomol. 97, 1404–1413. doi: 10.1653/024.097.0415
Chen, X. D., Gill, T. A., Ashfaq, M., Pelz-Stelinski, K. S., and Stelinski, L. L. (2018). Resistance to commonly used insecticides in Asian citrus psyllid: stability and relationship to gene expression. J. Appl. Entomol. 142, 967–977. doi: 10.1111/jen.12561
Chen, X. D., and Stelinski, L. L. (2017). Rapid detection of insecticide resistance in Diaphorina citri (Hemiptera: Liviidae) populations, using a bottle bioassay. Florida Entomol. 100, 124–133. doi: 10.1653/024.100.0118
Chu, C.-C., Gill, T. A., Hoffmann, M., and Pelz-Stelinski, K. S. (2016). Inter-population variability of endosymbiont densities in the Asian citrus psyllid (Diaphorina citri Kuwayama). Microb. Ecol. 71, 999–1007. doi: 10.1007/s00248-016-0733-9
Chu, C. C., Hoffmann, M., Braswell, W. E., and Pelz-Stelinski, K. S. (2019). Genetic variation and potential coinfection of Wolbachia among widespread Asian citrus psyllid (Diaphorina citri Kuwayama) populations. Insect Sci. 26, 671–682. doi: 10.1111/1744-7917.12566
Cicero, J. M., Adair, M. M., Adair, R. C., Hunter, W. B., Avery, P. B., and Mizell, R. F. (2017). Predatory behavior of long-legged flies (Diptera: Dolichopodidae) and their potential negative effects on the parasitoid biological control agent of the Asian citrus psyllid (Hemiptera: Liviidae). Fla. Entomol. 100, 485–487. doi: 10.1653/024.100.0243
Cicero, J. M., Brown, J. K., Roberts, P. D., and Stansly, P. A. (2009). The digestive system of Diaphorina citri and Bactericera cockerelli (Hemiptera: Psyllidae). Ann. Entomol. Soc. Am. 102, 650–665. doi: 10.1603/008.102.0410
Coates, L. C., Mahoney, J., Ramsey, J. S., Warwick, E., Johnson, R., MacCoss, M. J., et al. (2020). Development on Citrus medica infected with ‘Candidatus Liberibacter asiaticus' has sex-specific and -nonspecific impacts on adult Diaphorina citri and its endosymbionts. PLoS ONE 15:e0239771. doi: 10.1371/journal.pone.0239771
Cooper, W. R., Sengoda, V. G., and Munyaneza, J. E. (2014). Localization of 'Candidatus Liberibacter solanacearum' (Rhizobiales: Rhizobiaceae) in Bactericera cockerelli (Hemiptera: Triozidae). Ann. Entomol. Soc. Am. 107, 204–210. doi: 10.1603/AN13087
Cooper, W. R., Swisher, K. D., Garczynski, S. F., Mustafa, T., Munyaneza, J. E., and Horton, D. R. (2015). Wolbachia infection differs among divergent mitochondrial haplotypes of Bactericera cockerelli (Hemiptera: Triozidae). Ann. Entomol. Soc. Am. 108, 137–145. doi: 10.1093/aesa/sau048
Coy, M. R., Hoffmann, M., Gibbard, H. K., Kuhns, E. H., Pelz-Stelinski, K. S., and Stelinski, L. L. (2014). Nested-quantitative PCR approach with improved sensitivity for the detection of low titer levels of Candidatus Liberibacter asiaticus in the Asian citrus psyllid, Diaphorina citri Kuwayama. J. Microbiol. Methods 102, 15–22. doi: 10.1016/j.mimet.2014.04.007
Crooke, S., Wang, S., Vickers, T., Shen, W., and Liang, X.-h. (2017). Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 35, 230–237. doi: 10.1038/nbt.3779
Crooke, S. T. (2017). Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Therapeutics 27, 70–77. doi: 10.1089/nat.2016.0656
Crooke, S. T., Vickers, T. A., and Liang, X. H. (2020). Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Res. 48, 5235–5253. doi: 10.1093/nar/gkaa299
Crosslin, J. M., Lin, H., and Munyaneza, J. E. (2011). Detection of 'Candidatus Liberibacter solanacearum' in the potato psyllid, Bactericera cockerelli (Sulc), by conventional and real-time PCR. Southwestern Entomol. 36, 125–135. doi: 10.3958/059.036.0202
Da Graça, J. V., and Korsten, L. (2004). “Citrus huanglongbing: review, present status and future studies,” in Diseases of Fruits and Vegetables ed S. A. M. H. Naqvi (Dordrecht: Kluwer Academic Publishers), 1, 229–245. doi: 10.1007/1-4020-2606-4_4
Dahan, J., Wenninger, E. J., Thompson, B., Eid, S., Olsen, N., and Karasev, A. V. (2017). Relative abundance of potato psyllid haplotypes in southern Idaho potato fields during 2012 to 2015, and incidence of 'Candidatus Liberibacter solanacearum' causing zebra chip disease. Plant Dis. 101, 822–829. doi: 10.1094/PDIS-05-16-0668-RE
Dahan, J., Wenninger, E. J., Thompson, B. D., Eid, S., Olsen, N., and Karasev, A. V. (2019). Prevalence of ‘Candidatus Liberibacter solanacearum' haplotypes in potato tubers and psyllid vectors in Idaho from 2012 to 2018. Plant Dis. 103, 2587–2591. doi: 10.1094/PDIS-11-18-2113-RE
Daly, S. M., Sturge, C. R., and Greenberg, D. E. (2017). “Inhibition of bacterial growth by peptide-conjugated morpholino oligomers,” in Morpholino Oligomers, Vol. 1565, Methods in Molecular Biology, eds H. Moulton and J. Moulton (New York, NY: Humana Press), 115–122.
Della Vechia, J. F., Andrade, D. J., Azevedo, R. G., and Costa Ferreira, M. (2019). Effects of insecticide and acaricide mixtures on Diaphorina citri control. Rev. Bras. Frutic. Jaboticabal 41:e-076. doi: 10.1590/0100-29452019076
Doyle, C., Higginbottom, K., Swift, T. A., Winfield, M., Bellas, C., Benito-Alifonso, D., et al. (2020). A simple method for spray-on gene editing in planta. bioRxiv [preprint]. doi: 10.1101/805036
Dubrovina, A. S., Aleynova, O. A., Suprun, A. R., Ogneva, Z. V., and Kiselev, K. V. (2020). Transgene suppression in plants by foliar application of in vitro-synthesized small interfering RNAs. Appl. Microbiol. Biotechnol. 104, 2125–2135. doi: 10.1007/s00253-020-10355-y
Dubrovina, A. S., and Kiselev, K. V. (2019). Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 20:2282. doi: 10.3390/ijms20092282
Ebert, T. A. (2019). The probing behavior component of disease transmission in insect-transmitted bacterial plant pathogens. Insects 10:212. doi: 10.3390/insects10070212
Fernandez Cornejo, J., Wechsler, S., Livingston, M. J., and Mitchell, L. (2014). Genetically Engineered Crops in the United States (February 1). USDA-ERS Economic Research Report Number 162. doi: 10.2139/ssrn.2503388
Fletcher, S. J., Reeves, P. T., Hoang, B. T., and Mitter, N. (2020). A perspective on RNAi-based biopesticides. Front. Plant Sci. 11:51. doi: 10.3389/fpls.2020.00051
Ghosh, S. K., Gundersen-Rindal, D. E., Park, A. L., and Hunter, W. B. (2018). Double-stranded RNA oral delivery methods to induce RNA interference in phloem and plant-sap-feeding hemipteran insects. J. Vis. Exp. 135:57390. doi: 10.3791/57390
Ghosh, S. K. B., Hunter, W. B., Park, A. L., and Gundersen-Rindal, D. E. (2017). Double strand RNA delivery system for plant-sap-feeding insects. PLoS ONE 12:e0171861. doi: 10.1371/journal.pone.0171861
Gottwald, T., Luo, W., and McRoberts, N. (2018). Risk-based HLB surveys for California an analysis of the increasing risk of HLB over the last five years. CRB Research Project #5300-154. Citrograph 9, 38–43. Available online at: https://citrusresearch.org/citrograph
Grafton-Cardwell, E. E. (2020). “Management of Asian citrus psyllid in California,” in Asian Citrus Psyllid, Chapter 16, Biology, Ecology and Management of the Huanglongbing Vector, eds P. Stansly and J. Qureshi [Wallingford: Commonwealth Agricultural Bureau International (CABI Press)], 250–257.
Grafton-Cardwell, E. E., Stelinski, L. L., and Stansly, P. A. (2013). Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–432. doi: 10.1146/annurev-ento-120811-153542
Gruenke, P. R., Alam, K. K., Singh, K., and Burke, D. H. (2020). 2′-fluoro-modified pyrimidines enhance affinity of RNA oligonucleotides to HIV-1 reverse transcriptase. RNA 26, 1667–1679. doi: 10.1261/rna.077008.120
Haapalainen, M. (2014). Biology and epidemics of Candidatus Liberibacter species, psyllid-transmitted plant-pathogenic bacteria. Ann. Appl. Biol. 165, 172–198. doi: 10.1111/aab.12149
Hajeri, S., Killiny, N., El-Mohtar, C., Dawson, W. O., and Gowda, S. (2014). Citrus tristeza virus-based RNAi in citrus plants induces gene silencing in Diaphorina citri, a phloem-sap sucking insect vector of citrus greening disease (Huanglongbing). J. Biotechnol. 176, 42–49. doi: 10.1016/j.jbiotec.2014.02.010
Halbert, S. E., and Manjunath, K. L. (2004). Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: a literature review and assessment of risk in Florida. Fla. Entomol. 87, 330–353. doi: 10.1653/0015-4040(2004)087[0330:ACPSPA.0.CO;2]10.1653/0015-4040(2004)087[0330:ACPSPA]2.0.CO;2
Hall, D. G., and Hentz, M. G. (2019). Influence of light on reproductive rates of Asian citrus psyllid (Hemiptera: Liviidae). J. Insect Sci. 19:9. doi: 10.1093/jisesa/iey141
Hall, D. G., Hentz, M. G., and Patt, J. M. (2015). Behavioral assay on Asian citrus psyllid attraction to orange jasmine. J. Insect Behav. 28, 555–568. doi: 10.1007/s10905-015-9525-1
Hall, D. G., Richardson, M. L., Ammar, E. D., and Halbert, S. E. (2013). Asian citrus psyllid, Diaphorina citri, vector of citrus huanglongbing disease. Entomol. Exper. Appl. 146, 207–223. doi: 10.1111/eea.12025
Hammond, S. M., Aartsma-Rus, A., Alves, S., Borgos, S. E., Buijsen, R., Collin, R. W., et al. (2021). Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol. Med. 13:e13243. doi: 10.15252/emmm.202013243
Hansen, A. K., Trumble, J. T., Stouthamer, R., and Paine, T. D. (2008). A new huanglongbing species, “Candidatus Liberibacter psyllaurous,” found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Appl. Environ. Microbiol. 74, 5862–5865. doi: 10.1128/AEM.01268-08
Harrison, K., Tamborindeguy, C., Scheuring, D. C., Mendoza-Herrera, A., Silva, A., Badillo-Vargas, I. E., et al. (2019). Differences in zebra chip severity between ‘Candidatus Liberibacter solanacearum' haplotypes in Texas. Am. J. Potato Res. 96, 86–93. doi: 10.1007/s12230-018-9692-7
Hayter, A. J. (1986). The maximum familywise error rate of Fisher's least significant difference test. J. Amer. Stat. Assoc. 81, 1000–1004. doi: 10.1080/01621459.1986.10478364
He, S., and Creasey Krainerm, K. M. (2020). Pandemics of people & plants: which is the greater threat to food security? Mol. Plant. 13, 933–934. doi: 10.1016/j.molp.2020.06.007
Hegarty, J. P., and Stewart, E. B. (2018). Advances in therapeutic bacterial antisense biotechnology. Appl. Microbiol. Biotechnol. 102, 1055–1065. doi: 10.1007/s00253-017-8671-0
Hochella, M. F., Mogk, D. W., Ranville, J., Allen, I. C., Luther, G. W., Marr, L. C., et al. (2019). Natural, incidental, and engineered nanomaterials and their impacts on the Earth system. Science 363:eaau8299. doi: 10.1126/science.aau8299
Hodges, A., and Spreen, T. (2012). Economic Impacts of Citrus Greening (HLB) in Florida, 2006/07–2010/11, Electronic Data Information Source (EDIS) FE903. Gainesville, FL: University of Florida Gainesville, 32611.
Hosseinzadeh, S., Shams-Bakhsh, M., Mann, M., Fattah-Hosseini, S., Bagheri, A., Mehrabadi, M., et al. (2019). Distribution and variation of bacterial endosymbiont and “Candidatus Liberibacter asiaticus” titer in the huanglongbing insect vector, Diaphorina citri Kuwayama. Microb. Ecol. 78, 206–222. doi: 10.1007/s00248-018-1290-1
Hoy, M. A., Singh, R., and Rogers, M. E. (2010). Evaluations of a novel isolate of Isaria fumosorosea for control of the Asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae). Florida Entomol. 93, 24–32. doi: 10.1653/024.093.0103
Huang, W., Reyes-Caldas, P., Mann, M., Seifbarghi, S., Kahn, A., Almeida, R. P., et al. (2020). Bacterial vector-borne plant diseases: unanswered questions and future directions. Mol. Plant 13, 1379–1393. doi: 10.1016/j.molp.2020.08.010
Huesing, J. E., Andres, D., Braverman, M. P., Burns, A., Felsot, A. S., Harrigan, G. G., et al. (2016). Global adoption of genetically modified (GM) crops: challenges for the public sector. J. Agric. Food Chem. 64, 394–402. doi: 10.1021/acs.jafc.5b05116
Hunter, W. B. (2017). “Clay delivery of dsRNA into plants by root absorption,” in IPM Methods for Insect Pests of Orchard Crops. Subtropical Insects and Horticulture Research 2017 Annual Report. Available online at: https://www.ars.usda.gov/research/project/?accnNo=429787&fy=2017
Hunter, W. B. (2019). “Slow-release clay-delivery system, of dsRNA into plants beyond 12 months,” in IPM Methods for Insect Pests of Orchard Crops. Subtropical Insects and Horticulture Research 2019 Annual Report. Available online at: https://www.ars.usda.gov/research/project/?accnNo=429787&fy=2019
Hunter, W. B., and Aishwarya, V. (2020). Targeted Control of Pests and Pathogens by Plant Delivery of 2'FANA-Oligonucleotides. Patent Pending. AUM LifeTech, Inc. Available online at: www.aumlifetech.com
Hunter, W. B., Clarke, S. V., Sandoval Mojica, A. F., Paris, T., Metz, J. L., Holland, C. S., et al. (2020). “Advances in RNA suppression of the Asian Citrus Psyllid Vector of bacteria causing Huanglongbing (Hemipteran vector / Liberibacter pathogens),” in Asian Citrus Psyllid, Chapter 17, Biology, Ecology and Management of the Huanglongbing Vector, eds P. Stansly and J. Qureshi [Wallingford: Commonwealth Agricultural Bureau International (CABI Press)], 258–283.
Hunter, W. B., Gonzalez, M. T., and Tomich, J. (2018a). BAPC-assisted CRISPR/Cas9 system: Targeted delivery into adult ovaries for heritable germline gene editing (Arthropoda: Hemiptera). bioRxiv [preprint]. doi: 10.1101/478743
Hunter, W. B., Gonzalez, M. T., and Tomich, J. (2019a). BAPC-assisted-CRISPR-Cas9 delivery into nymphs and adults for heritable gene editing (Hemiptera). FASEB J. Biochem. Mol. Biol. 33:626.2. Available online at: https://www.fasebj.org/doi/abs/10.1096/fasebj.2019.33.1_supplement.626.2
Hunter, W. B., Metz, J. L., Lopez, S., Altman, S., Aishwarya, V., and Pelz-Stelinski, K. (2017b). “Method for detecting binding efficiencies of synthetic oligonucleotides: targeting bacteria and insects,” in Proceedings of 2017- 10th Arthropod Genomics Symposium, June 8–11 (Notre Dame, IN), 45. Available online at: http://globalhealth.nd.edu/assets/238736/ags_booklet.pdf
Hunter, W. B., Metz, J. L., Pelz-Stelinski, K., Sandoval Mojica, A. F., Altman, S., Boyle, M. J., et al. (2017a). “FANA and morpholino's, novel molecules for gene-targeting in plants and arthropods. W-56,” in Proceedings of XXV Int'l. Plant & Animal Conference, January 13-18 (San Diego, CA). Available online at: https://pag.confex.com/pag/xxv/webprogram/Paper25794.html
Hunter, W. B., Metz, J. L., Temeyer, K. B., Perez de Leon, B., Pelz-Stelinski, K., Sandoval-Mojica, A. F., et al. (2018b). “Antisense oligonucleotides, FANA, for treatment of microbes and arthropod pests of agricultural and medical importance. W069,” in Plant & Animal Genome XXVI Conference (San Diego, CA). Available online at: https://plan.core-apps.com/pag_2018/abstract/fff6e3855de940b33de03bc4efb11002
Hunter, W. B., Mojica, A. S., Miles, G., McCollum, G., Paris, T., Aishwarya, V., et al. (2019b). “Advances in treatments to manage huanglongbing: targeting Liberibacter bacteria and psyllid endosymbionts,” in Proceedings of Chemistry of Huanglongbing, AGFD, 179, 257th American Chemical Society, April 1-March 4, eds J. Manthey and R. Smitra (Orlando, FL). Available online at: https://plan.core-apps.com/acsorlando2019/abstracts
Hunter, W. B., and Pelz-Stelinski, K. S. (2021). Peptide Phosphorodiamidate Morpholino Oligomers Plant Delivery to Reduce Pathogens and Insect Pests. U.S. Patent No: US11,001,842_B2, Washington, DC, USA.
Hunter, W. B., and Reese, J. (2014). On behalf of ‘International Psyllid Genome Consortium'. The Asian citrus psyllid genome (Diaphorina citri, Hemiptera). 4.7P. 3rd International Research Conference on Huanglongbing - IRCHLB III. J. Citrus Pathol. Washington, DC, 1:143. doi: 10.5070/C411024833
Hunter, W. B., and Sinisterra-Hunter, X. (2018). Emerging RNA suppression technologies to protect citrus trees from citrus greening disease bacteria. Adv. Insect Phys. 55, 163–199. doi: 10.1016/bs.aiip.2018.08.001
Hunter, W. B., and Tomich, J. (2019). BAPC-Assisted-CRISPR-Cas9 Delivery Into Adult Psyllid Ovaries and Pupae for Heritable Gene Editing (Hemiptera). Citrus Greening Solutions. Boyce Thompson Institute. Res. Reports. Available online at: https://citrusgreening.org/disease/researchhighlights/index; https://www.researchgate.net/publication/336262798_BAPC-assisted-CRISPR-Cas9_Delivery_into_Adult_Psyllid_Ovaries_and_Pupae_for_Heritable_Gene_Editing_Hemiptera (accessed January 11, 2021).
Inoue, H., Ohnishi, J., Ito, T., Tomimura, K., Miyata, S., Iwanami, T., et al. (2009). Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann. Appl. Biol. 155, 29–36. doi: 10.1111/j.1744-7348.2009.00317.x
ISAAA (2019). International Service for the Acquisition of Agri-Biotech Applications, “Global Status of Commercialized Biotech/GM Crops in 2018: Biotech Crops Continue to Help Meet the Challenges of Increased Population and Climate Change.” ISAAA Brief No. 54. International Service for the Acquisition of Agri-Biotech Applications, Ithaca, NY.
Jarvis, B., Pavone, T., and Cedillo, I. (2020). Designing commercial-scale oligonucleotide synthesis. Pharm. Tech. 44, 30–34. Available online at: https://www.pharmtech.com/view/designing-commercial-scale-oligonucleotide-synthesis
Jiang, Y., Zhang, C. X., Chen, R., and He, S. Y. (2019). Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. U.S.A. 116, 23390–23397. doi: 10.1073/pnas.1915396116
Kalota, A., Karabon, L., Swider, C. R., Viazovkina, E., Elzagheid, M., Damha, M. J., et al. (2006). 2'-deoxy-2'-fluoro-beta-D-arabinonucleic acid (2'F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res. 34, 451–461. doi: 10.1093/nar/gkj455
Kiselev, K. V., Suprun, A. R., Aleynova, O. A., Ogneva, Z. V., and Dubrovina, A. S. (2021). Physiological conditions and dsRNA application approaches for exogenously induced RNA interference in Arabidopsis thaliana. Plants 10:264. doi: 10.3390/plants10020264
Kleter, G. A. (2020). Food safety assessment of crops engineered with RNA interference and other methods to modulate expression of endogenous and plant pest genes. Pest Manag. Sci. 76, 3333–3339. doi: 10.1002/ps.5957
Kruse, A., Fattah-Hosseini, S., Saha, S., Johnson, R., Warwick, E. R., Sturgeon, K., et al. (2017). Combining omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS ONE 12:e0179531. doi: 10.1371/journal.pone.0179531
Kruse, A., Fleites, L. A., and Heck, M. (2019). Lessons from one fastidious bacterium to another: what can we learn about Liberibacter species from Xylella fastidiosa. Insects 10:300. doi: 10.3390/insects10090300
Kruse, A., Ramsey, J. S., Johnson, R., Hall, D. G., MacCoss, M. J., and Heck, M. (2018). Candidatus Liberibacter asiaticus minimally alters expression of immunity and metabolism proteins in hemolymph of Diaphorina citri, the insect vector of huanglongbing. J. Proteome Res. 17, 2995–3011. doi: 10.1021/acs.jproteome.8b00183
Kumar, S. B., Arnipalli, S. R., and Ziouzenkova, O. (2020). Antibiotics in food chain: the consequences for antibiotic resistance. Antibiotics 9:688. doi: 10.3390/antibiotics9100688
Kunte, N., McGraw, E., Bell, S., Held, D., and Avila, L. A. (2020). Prospects, challenges and current status of RNAi through insect feeding. Pest Manag. Sci. 76, 26–41. doi: 10.1002/ps.5588
Kurreck, J. (2003). Antisense technologies: improvement through novel chemical modifications. Euro. J. Biochem. 270, 1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x
Lallemand, J., Fos, A., and Bové, J. M. (1986). Transmission de la bacteria associé à la forme africaine do la maladie du “greening” par le psylle asiatique Diaphorina citri Kuwayama. Fruits 41, 341–343.
Lebedeva, I., Benimetskaya, L., Stein, C. A., and Vilenchik, M. (2000). Cellular delivery of antisense oligonucleotides. Eur. J. Pharm. Biopharm. 50, 101–119. doi: 10.1016/S0939-6411(00)00088-6
Lenard, S. P., Powell, J. E., Perutka, J., Geng, P., Heckmann, L. C., Horak, R. D., et al. (2020). Engineered symbionts activate honey bee immunity and limit pathogens. Science 367, 573–576. doi: 10.1126/science.aax9039
Leng, N., English, A., Johnson, S., Richards, S., Hunter, W., and Saha, S. (2017). Diaphorina citri Genome Assembly DIACI 1.1. Washington, DC: Ag Data Commons. Available online at: https://data.nal.usda.gov/project/diaphorina-citri
Lespes, G., Faucher, S., and Slaveykova, V. I. (2020). Natural, nanoparticles, anthropogenic nanoparticles, where is the frontier? Front. Environ. Sci. 8:71. doi: 10.3389/fenvs.2020.00071
Levy, J. R. A., Gross, D., Tamborindeguy, T., and Pieson, E. (2011). Translocation of ‘Candidatus Liberibacter solanacearum', the zebra chip pathogen, in potato and tomato. Phytopathology 101, 1285–1291. doi: 10.1094/PHYTO-04-11-0121
Li, M., Liu, W., and Slaveykova, V. I. (2020). Effects of mixtures of engineered nanoparticles and metallic pollutants on aquatic organisms. Environments 7:27. doi: 10.3390/environments7040027
Li, W., Hartung, J. S., and Levy, L. (2006). Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods 66, 104–115. doi: 10.1016/j.mimet.2005.10.018
Liefting, L. W., Sutherland, P. W., Ward, L. I., Paice, K. L., Weir, B. S., and Clover, G. R. (2009). A new 'Candidatus Liberibacter' species associated with diseases of Solanaceous crops. Plant Dis. 93, 208–214. doi: 10.1094/PDIS-93-3-0208
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ludin, K. S., Gissberg, O., and Edvard Smith, C. I. (2015). Oligonucleotide therapies: the past and the present. Hum. Gene Ther. 26, 475–485. doi: 10.1089/hum.2015.070
Malek-Adamian, E., Fakhoury, J., Arnold, A. E., Martinez-Montero, S., Shoichet, M., and Damha, M. J. (2019). Effect of sugar 2′,4′-modifications on gene silencing activity of siRNA duplexes. Nucleic Acid Therapeutics 29, 187–194. doi: 10.1089/nat.2019.0792
Markham, N. R., and Zuker, M. (2008). “UNAFold,” in Bioinformatics, Vol. 453, Methods in Molecular Biology™, ed J. M. Keith (Humana Press), 453, 3–31. doi: 10.1007/978-1-60327-429-6_1
Mauck, K. E., Sun, P., Meduri, V., and Hansen, A. K. (2019). New Ca. Liberibacter psyllaurous haplotype resurrected from a 49-year-old specimen of Solanum umbelliferum: a native host of the psyllid vector. Sci. Rep. 9:9530. doi: 10.1038/s41598-019-45975-6
McCollum, G., Hall, D., and Luo, W. (2019). “CLas titer in ACP, not infected citrus, is the driving force for the spread of HLB,” in International Research Conference on HLB (Riverside, CA).
McCollum, G. (2021). To Find CLas in Citrus, Look for Where ACP Are of Have Been. CRB-funded Final Research Report. CRB Research Project #5300-176. Citrograph, Visalia, CA: Winter. 12, 46–51. Available online at: https://citrusresearch.org/citrograph
McCollum, G., Kunta, M., and Braswell, E. (2018). Improving early detection of CLas of HLB affected trees. Tracking citrus DNA modifications. Citrograph 9, 50–54. Visalia, CA. Available online at: https://citrusresearch.org/citrograph
Mendonça, M. C. P., Kont, A., Aburto, M. R., Cryan, J. F., and O'Driscoll, C. M. (2021). Advances in the design of (nano) formulations for delivery of antisense oligonucleotides and small interfering RNA: focus on the central nervous system. Mol. Pharm. 18, 1491–1506. doi: 10.1021/acs.molpharmaceut.0c01238
Mendoza-Herrera, A., Levy, J., Harrison, K., Yao, J., Ibanez, F., and Tamborindeguy, C. (2018). Infection by Candidatus Liberibacter solanacearum' haplotypes A and B in Solanum lycopersicum ‘Moneymaker'. Plant Dis. 102, 2009–2015. doi: 10.1094/PDIS-12-17-1982-RE
Mesterházy, A., Oláh, J., and Popp, J. (2020). Losses in the grain supply chain: causes and solutions. Sustainability 12:2342. doi: 10.3390/su12062342
Metz, J. L., Hunter, W. B., Boyle, M., and Aishwarya, V. (2017a). “Delivery of FANA ASOs by topical sprays to plants: a novel RNA silencing approach for arthropod pests control,” in Proceedings of XXV Int'l. Plant & Animal Conference, January 13-18 (San Diego, CA). Available online at: https://pag.confex.com/pag/xxv/webprogram/Paper25903.html
Metz, J. L., Hunter, W. B., Sandoval Mojica, A. F., Altman, S., Boyle, M. J., McCollum, G., et al. (2017b). “Delivery of gene biotechnologies to plants: pathogen and pest control,” in Proceedings of 2017- 10th Arthropod Genomics Symposium, June 8–11 (Notre Dame, IN), 44–45. Available online at: http://globalhealth.nd.edu/assets/238736/ags_booklet.pdf
Mitter, N., Worrall, E. A., Robinson, K. E., Li, P., Jain, R. G., Taochy, C., et al. (2017). Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 3, 1–10. doi: 10.1038/nplants.2016.207
Monzo, C., and Stansly, P. A. (2020). “Sampling and economic thresholds for Asian citrus psyllid,” in Asian Citrus Psyllid, Chapter 11, Biology, Ecology and Management of the Huanglongbing Vector, eds P. Stansly and J. Qureshi [Wallingford: Commonwealth Agricultural Bureau International (CABI Press)], 156–165.
Morrow, J. J., Hall, A. A., and Riegler, G. (2017). Symbionts in waiting: the dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 5:58. doi: 10.1186/s40168-017-0276-4
Moulton, H., and Moulton, J. (2017). “Morpholino oligomers,” in Methods in Molecular Biology, Vol. 1565, eds H. Moulton and J. Moulton (New York, NY: Humana Press), 277. doi: 10.1007/978-1-4939-6817-6_1
Munyaneza, J. E. (2012). Zebra chip disease of potato: biology, epidemiology, and management. Am. J. Potato Res. 89, 329–350. doi: 10.1007/s12230-012-9262-3
Munyaneza, J. E. (2015). Zebra Chip disease, Candidatus Liberibacter, and potato psyllid: a global threat to the potato industry. Am. J. Potato Res. 92, 230–235. doi: 10.1007/s12230-015-9448-6
Munyaneza, J. E., Crosslin, J. M., and Upton, J. E. (2007). Association of Bactericera cockerelli (Homoptera: Psyllidae) with “zebra chip,” a new potato disease in southwestern United States and Mexico. J. Econ. Entomol. 100, 656–663. doi: 10.1093/jee/100.3.656
Munyaneza, J. E., Fisher, T. W., Sengoda, V. G., Garczynski, S. F., Nissinen, A., and Lemmetty, A. (2010). First report of “Candidatus Liberibacter solanacearum” associated with psyllid-affected carrots in Europe. Plant Dis. 94, 639–639. doi: 10.1094/PDIS-94-5-0639A
Nachappa, P., Levy, J., Pierson, E., and Tamborindeguy, C. (2014). Correlation between “Candidatus Liberibacter solanacearum” infection levels and fecundity in its psyllid vector. J. Invertebr. Pathol. 115, 55–61. doi: 10.1016/j.jip.2013.10.008
Naranjo, E., Merfa, M. V., Santra, S., Ozcan, A., Johnson, E., Cobine, P. A., et al. (2020). Zinkicide is a ZnO-based nanoformulation with bactericidal activity against Liberibacter crescens in batch cultures and in microfluidic chambers simulating plant vascular systems. Appl. Environ. Microbiol. 86, e00788–e00720. doi: 10.1128/AEM.00788-20
NASEM (2018). “Committee on a Review of the Citrus Greening Research and Development Efforts Supported by the Citrus Research and Development Foundation: Fighting a Ravaging Disease,” Consensus Study Report. National Academies of Sciences, Engineering, and Medicine. National Academies Press, Washington, DC. Available online at: http://www.nap.edu; https://www.nap.edu/catalog/25026/a-review-of-the-citrus-greening-research-and-development-efforts-supported-by-the-citrus-research-and-development-foundation
Nelson, W. R., Fisher, T. W., and Munyaneza, J. E. (2011). Haplotypes of “Candidatus Liberibacter solanacearum” suggest long-standing separation. Euro. J. Plant Pathol. 130, 5–12. doi: 10.1007/s10658-010-9737-3
Ni, M., Ma, W., Wang, X., Gau, M., Dai, Y., Wei, X., et al. (2017). Next-generation transgenic cotton: pyramiding RNAi and Bt counters insect resistance. Plant Biotech. J. 15, 1204–1213. doi: 10.1111/pbi.12709
Nicaise, V. (2014). Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 5:660. doi: 10.3389/fpls.2014.00660
Novopashina, D., Vorobyeva, M., Nazarov, A., Davydova, A., Danilin, N., Koroleva, L., et al. (2019). Novel peptide conjugates of modified oligonucleotides for inhibition of bacterial RNase P. Front. Pharmacol. 10:813. doi: 10.3389/fphar.2019.00813
Ochoa, S., and Milam, V. T. (2020). Modified nucleic acids: expanding the capabilities of functional oligonucleotides. Molecules 25:4659. doi: 10.3390/molecules25204659
Patel, R. R., Sundin, G. W., Yang, C.-H., Wang, J., Huntley, R. B., Yuan, X., et al. (2017). Exploration of using antisense peptide nucleic acid (PNA) cell-penetrating peptide (CPP) as a novel bactericide against fire blight pathogen Erwinia amylovora. Front. Microbiol. 8:687. doi: 10.3389/fmicb.2017.00687
Patt, J. M., Chow, A., Meikle, W. G., Gracia, C., Jackson, M. A., Flores, D., et al. (2015). Efficacy of an autodisseminator of an entomopathogenic fungus, Isaria fumosorosea, to suppress Asian citrus psyllid, Diaphorina citri, under greenhouse conditions. Biol. Control 88, 37–45. doi: 10.1016/j.biocontrol.2015.04.014
Pelz-Stelinski, K. S. (2020). “Symbionts and pathogens of the Asian citrus psyllid,” in Asian Citrus Psyllid, Chapter 7, Biology, Ecology and Management of the Huanglongbing Vector, eds P. Stansly and J. Qureshi [Wallingford: Commonwealth Agricultural Bureau International (CABI Press)], 101–112.
Pelz-Stelinski, K. S., Hunter, W. B., and Aishwarya, V. (2020). Use of FANA Antisense Oligonucleotides for the Prevention and Management of Huanglongbing and Zebra-Chip Diseases. AUM LifeTech, Inc. Patent Pending [Footnote 5]. Available online at: www.aumlifetech.com
Pelz-Stelinski, K. S., Hunter, W. B., and Altman, S. (2017). SCRI Award #2015-10479 (Technical Report). USDA, NIFA. Available online at: https://portal.nifa.usda.gov/web/crisprojectpages/1008974-targeting-microbes-to-control-huanglongbing-disease-of-citrus.html
Pelz-Stelinski, K. S., and Killiny, N. (2016). Better together: association with ‘Candidatus Liberibacter asiaticus' increases the reproductive fitness of its insect vector, Diaphorina citri (Hemiptera: Liviidae). Ann. Entomol. Soc. Am. 109, 371–376. doi: 10.1093/aesa/saw007
Pérez-Rodríguez, J., Krüger, K., Pérez-Hedo, M., Ruiz-Rivero, O., Urbaneja, A., and Tena, A. (2019). Classical biological control of the African citrus psyllid Trioza erytreae, a major threat to the European citrus industry. Sci. Rep. 9:9440. doi: 10.1038/s41598-019-45294-w
Pietri, J. E., Cheung, K. W., and Luckhart, S. (2014). Knockdown of mitogen-activated protein kinase (MAPK) signalling in the midgut of Anopheles stephensi mosquitoes using antisense morpholinos. Insect Mol. Biol. 23, 558–565. doi: 10.1111/imb.12103
Pimentel, D. (1991). “World resources and food losses to pests,” in Ecology and Management of Food-Industry Pests, ed J. R. Gorham (Arlington, VA: Assoc. Analytical Chemists), 5–11.
Pimentel, D., and Burgess, M. (2012). Small amounts of pesticides reaching target insects. Environ. Dev. Sustain. 14, 1–2. doi: 10.1007/s10668-011-9325-5
Pimentel, D., and Levitan, L. (1986). Pesticides: amounts applied and amounts reaching pests. Bioscience 36, 86–91. doi: 10.2307/1310108
Puchta, H., and Fauser, F. (2013). Gene targeting in plants: 25 years later. Int. J. Dev. Biol. 57, 629–637. doi: 10.1387/ijdb.130194hp
Puvača, N., and de Llanos Frutos, R. (2021). Antimicrobial resistance in Escherichia coli strains isolated from humans and pet animals. Antibiotics 10:69. doi: 10.3390/antibiotics10010069
Qureshi, J. A., Kostyk, B. C., and Stansly, P. A. (2014). Insecticidal suppression of Asian citrus psyllid Diaphorina citri (Hemiptera: Liviidae) vector of Huanglongbing pathogens. PLoS ONE 9:e112331. doi: 10.1371/journal.pone.0112331
Ramsey, J. S., Johnson, R. S., Hoki, J. S., Kruse, A., Mahoney, J., Hilf, M. E., et al. (2015). Metabolic interplay between the Asian citrus psyllid and its Profftella symbiont: an Achilles' heel of the citrus greening insect vector. PLoS ONE 10:e0140826. doi: 10.1371/journal.pone.0140826
Reese, J., Christenson, M. K., Leng, N., Saha, S., Cantarel, B., Lindeberg, M., et al. (2013). The Asian citrus psyllid transcriptome. J. Genomics 2, 54–58. doi: 10.7150/jgen.7692
Ricroch, A. E., and Hénard-Damave, M. C. (2016). Next biotech plants: new traits, crops, developers and technologies for addressing global challenges. Crit. Rev. Biotechnol. 36, 675–690. doi: 10.3109/07388551.2015.1004521
Rose, K. M., Alves Ferreira-Bravo, I., Li, M., Craigie, R., Ditzler, M. A., Holliger, P., et al. (2019). Selection of 2′-deoxy-2′-fluoroarabino nucleic acid (FANA) aptamers that bind HIV-1 integrase with picomolar affinity. ACS Chem. Biol. 14, 2166–2175. doi: 10.1021/acschembio.9b00237
Rudmin, M., Banerjee, S., Yakich, T., Tabakaev, R., Ibraeva, K., Buyakov, A., et al. (2020). Formulation of a slow-release fertilizer by mechanical activation of smectite/glauconite and urea mixtures. Appl. Clay Sci. 196:105775. doi: 10.1016/j.clay.2020.105775
Saha, S., Hosmani, P. S., Flores-Gonzalez, M., Hunter, W., and D'Elia, T. (2017a). “Biocuration and improvement of the Diaphorina citri draft genome assembly with long reads, optical maps and long-range scaffolding,” in International Psyllid Annotation Consortium, Proceedings of 10th Arthropod Genomics Symposium, June 8–11, eds S. Brown and L. A. Mueller (Notre Dame, IN), 27–28. Available online at: http://globalhealth.nd.edu/assets/238736/ags_booklet.pdf
Saha, S., Hosmani, P. S., Villalobos-Ayala, K., Miller, S., Shippy, T., Brown, S. J., et al. (2019). Improved annotation of the insect vector of citrus greening disease: biocuration by a diverse genomics community. J. Biol. Database Curation. 2019:baz035. doi: 10.1093/database/baz035
Saha, S., Hosmani, PS., Villalobos-Ayala, K., Miller, S., Shippy, T., Flores, M., et al. (2017b). Improved annotation of the insect vector of citrus greening disease: biocuration by a diverse genomics community. Database 2017:bax032. doi: 10.1093/database/bax032
Saha, S., Hosmani, P. S., Villalobos-Ayala, K., Miller, S., Shippy, T., and Flores, M. (2017d). Diaphorina citri Official Gene Set v1.0. Washington, DC: Ag Data Commons. Available online at: https://data.nal.usda.gov/project/diaphorina-citri
Saha, S., Hunter, W., Mueller, L., and Brown, S. (2017c). Diaphorina citri Genome Assembly DIACI 1.9. Washington, DC: Ag Data Commons. Available online at: https://data.nal.usda.gov/project/diaphorina-citri
Saha, S., Hunter, W. B., Reese, J., Morgan, J. K., Marutani-Hert, M., Huang, H., and Lindeberg, M. (2012). Survey of endosymbionts in the Diaphorina citri metagenome and assembly of a Wolbachia wDi draft genome. PLoS ONE 7:e50067. doi: 10.1371/journal.pone.0050067
Samada, L. H., and Tambunan, U. S. F. (2020). Biopesticides as promising alternatives to chemical pesticides: a review of their current and future status. Online J. Biol. Sci. 20, 66–76. doi: 10.3844/ojbsci.2020.66.76
San Miguel, K., and Scott, J. G. (2016). The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manage. Sci. 72, 801–809. doi: 10.1002/ps.4056
Sandoval-Mojica, A. F., Altman, S., Hunter, W. B., and Pelz-Stelinski, K. S. (2020). Peptide conjugated morpholinos for management of the huanglongbing pathosystem. Pest Manag. Sci. 76, 3217–3224. doi: 10.1002/ps.5877
Sandoval-Mojica, A. F., Hunter, W. B., Aishwarya, V., Bonilla, S., and Pelz-Stelinski, K. S. (2021). Antibacterial FANA oligonucleotides as a novel approach for managing the huanglongbing pathosystem. Sci. Rep. 11:2760. doi: 10.1038/s41598-021-82425-8
Sastry, K. S., Mandal, B., Hammond, J., Scott, S. W., and Briddon, R. W. (2019). Encyclopedia of Plant Viruses and Viroids. Springer Nature.
Saurabh, S., Vidyarthi, A., and Prasad, D. (2014). RNA interference: concept to reality in crop improvement. Planta 239, 543–564. doi: 10.1007/s00425-013-2019-5
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Scoles, D. R., Minikel, E. V., and Pulst, S. M. (2019). Antisense oligonucleotides. Neurol. Genet. 5:e323. doi: 10.1212/NXG.0000000000000323
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Sengoda, V. G., Cooper, W. R., Swisher, K. D., Henne, D. C., and Munyaneza, J. E. (2014). Latent period and transmission of “Candidatus Liberibacter solanacearum” by the potato psyllid Bactericera cockerelli (Hemiptera: Triozidae). PLoS ONE 9:e93475. doi: 10.1371/journal.pone.0093475
Sétamou, M., Alabi, O. J., Kunta, M., Jifon, J. L., and da Graça, J. V. (2016). Enhanced acquisition rates of ‘Candidatus Liberibacter asiaticus' by the Asian citrus psyllid (Hemiptera: Liviidae) in the presence of vegetative flush shoot growth in citrus. J. Econ. Entomol. 109, 1973–1978. doi: 10.1093/jee/tow171
Shen, W., De Hoyos, C. L., Migawa, M. T., Vickers, T. A., Sun, H., Low, A., et al. (2019). Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 37, 640–650. doi: 10.1038/s41587-019-0106-2
Shen, X., and Corey, D. R. (2018). Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46, 1584–1600. doi: 10.1093/nar/gkx1239
Shukla, E., Thorat, L. J., Nath, B. B., and Gaikwad, S. M. (2015). Insect trehalase: physiological significance and potential applications. Glycobiology 25, 357–367. doi: 10.1093/glycob/cwu125
Sinisterra-Hunter, X., and Hunter, W. B. (2018). “Towards a holistic integrated pest management: lessons learned from plant-insect mechanisms in the field,” in Chapter 10, The Biology of Plant-Insect Interactions: A Compendium for the Plant Biotechnologist, ed C. Emani (Boca Raton, FL: CRC Press), 204–226.
Skelley, L. H., and Hoy, M. A. (2004). A synchronous rearing method for the Asian citrus psyllid and its parasitoids in quarantine. Biol. Control 29, 14–23. doi: 10.1016/S1049-9644(03)00129-4
Slaveykova, V. I., Li, M., Worms, I. A., and Liu, W. (2020). When environmental chemistry meets ecotoxicology: bioavailability of inorganic nanoparticles to phytoplankton. CHIMIA 74, 115–121. doi: 10.2533/chimia.2020.115
Souleimanian, N., Deleavey, G. F., Soifer, H., Wang, S., Tiemann, K., Damha, M. J., et al. (2012). Antisense 2′-deoxy, 2′-fluoroarabino nucleic acid (2′F-ANA) oligonucleotides: In vitro gymnotic silencers of gene expression whose potency is enhanced by fatty acids. 1:e43. doi: 10.1038/mtna.2012.35
Stansly, P. A., and Qureshi, J. A. (2020). “Management objectives and integration of strategies for the Asian citrus psyllid,” in Asian Citrus Psyllid, Chapter 11, Biology, Ecology and Management of the Huanglongbing Vector, eds P. Stansly and J. Qureshi [Wallingford: Commonwealth Agricultural Bureau International (CABI Press)], 166–179.
Stover, E., Stange, R. R., McCollum, T. G., Jaynes, J., Irey, M., and Mirkov, E. (2013). Screening antimicrobial peptides in vitro for use in developing transgenic citrus resistant to huanglongbing and citrus canker. J. Am. Soc. Hortic. Sci. 138, 142–148. doi: 10.21273/JASHS.138.2.142
Swisher, K. D., Munyaneza, J. E., and Crosslin, J. M. (2012). High resolution melting analysis of the cytochrome oxidase I gene identifies three haplotypes of the potato psyllid in the United States. Environ. Entomol. 41, 1019–1028. doi: 10.1603/EN12066
Swisher-Grimm, K. D., and Garczynski, S. F. (2019). Identification of a new haplotype of ‘Candidatus Liberibacter solanacearum' in Solanum tuberosum. Plant Dis. 103, 468–474. doi: 10.1094/PDIS-06-18-0937-RE
Swisher-Grimm, K. D., Mustafa, T., Cooper, W. R., and Munyaneza, J. E. (2018). Role of 'Candidatus Liberibacter solanacearum' and Bactericera cockerelli haplotypes in zebra chip incidence and symptom severity. Am. J. Potato Res. 95, 709–719. doi: 10.1007/s12230-018-9678-5
Tahzima, R., Maes, M., Achbani, E. H., Swisher, K. D., Munyaneza, J. E., and De Jonghe, K. (2014). First report of 'Candidatus Liberibacter solanacearum' on carrot in Africa. Plant Dis. 98, 1426–1426. doi: 10.1094/PDIS-05-14-0509-PDN
Tang, X. T., Longnecker, M., and Tamborindeguy, C. (2020). Acquisition and transmission of two ‘Candidatus Liberibacter solanacearum' haplotypes by the tomato psyllid Bactericera cockerelli. Sci. Rep. 10:14000. doi: 10.1038/s41598-020-70795-4
Taning, C. N. T., Andrade, E. C., Hunter, W. B., Christiaens, O., and Smagghe, G. (2016). Asian citrus psyllid RNAi pathway – RNAi evidence. Sci. Rep. Nat. 6:38082. doi: 10.1038/srep38082
Teo, R. D., Terai, K., Migliore, A., and Beratan, D. N. (2018). Electron transfer characteristics of 2'-deoxy-2'-fluoro-arabinonucleic acid, a nucleic acid with enhanced chemical stability. Phys. Chem. Chem. Phys. 20, 26063–26067. doi: 10.1039/C8CP04816A
Thomas, G. C., Dohmen, E., Hughes, D. T., Murali, S. C., Poelchau, M. F., Glastad, K., et al. (2020). Gene content evolution in the arthropods. Genome Biol. 21:15. doi: 10.1186/s13059-019-1925-7
Tian, F., Wang, Z., Li, C., Liu, J., and Zeng, X. (2019). UDP-Glycosyltransferases are involved in imidacloprid resistance in the Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae). Pesticide Biochem. Physiol. 154:23–31. doi: 10.1016/j.pestbp.2018.12.010
Tiwari, S., Mann, R. S., Rogers, M. E., and Stelinski, L. L. (2011). Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 67, 1258–1268. doi: 10.1002/ps.2181
Van Vu, T., Sung, Y. W., Kim, J., Hai Doan, D. T., Tran, M. T., and Kim, J.-Y. (2019). Challenges and perspectives in homology-directed gene targeting in monocot plants. Rice 12:95. doi: 10.1186/s12284-019-0355-1
Vega-Vásquez, P., Mosier, N. S., and Irudayaraj, J. (2020). Nanoscale drug delivery systems: from medicine to agriculture. Front. Bioeng. Biotechnol. 8:79. doi: 10.3389/fbioe.2020.00079
Verma, A. (2018). Recent advances in antisense oligonucleotide therapy in genetic neuromuscular diseases. Ann. Indian Acad. Neurol. 21, 3–8. doi: 10.4103/astrocyte.astrocyte_55_18
Vogel, E., Santos, D., Mingels, L., Verdonckt, T.-W., and Broeck, J. V. (2019). RNA Interference in insects: protecting beneficials and controlling pests. Front. Physiol. 9:1912. doi: 10.3389/fphys.2018.01912
Wang, N., Pierson, E. A., Setubal, J. C., Xu, J., Levy, J. G., Zhang, Y., et al. (2017a). The Candidatus Liberibacter-host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 55, 451–482. doi: 10.1146/annurev-phyto-080516-035513
Wang, N., Stelinski, L. L., Pelz-Stelinski, K. S., Graham, J. H., and Zhang, Y. (2017b). Tale of the huanglongbing disease pyramid in the context of the citrus microbiome. Phytopathology 107, 380–387. doi: 10.1094/PHYTO-12-16-0426-RVW
Wang, N., and Trivedi, P. (2013). Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103, 652–665. doi: 10.1094/PHYTO-12-12-0331-RVW
Wen, A. M., Johnson, C., and Gudmestad, N. C. (2013). Development of a PCR assay for the rapid detection and differentiation of 'Candidatus Liberibacter solanacearum' haplotypes and their spatiotemporal distribution in the United States. Am. J. Potato Res. 90, 229–236. doi: 10.1007/s12230-012-9293-9
Wesolowski, D., Alonso, D., and Altman, S. (2013). Combined effect of a peptide-morpholino oligonucleotide conjugate and a cell-penetrating peptide as an antibiotic. PNAS. 110, 8686–8689. doi: 10.1073/pnas.1306911110
Wesolowski, D., Tae, H. S., Gandotra, N., Llopis, P., Shen, N., and Altman, S. (2011). Basic peptide morpholino oligomer conjugate that is very effective in killing bacteria by gene-specific and nonspecific modes. PNAS 108, 16582–16587. doi: 10.1073/pnas.1112561108
Xue, X.-Y., Mao, X.-G., Zhou, Y., Chen, Z., Hu, Y., Hou, Z., et al. (2018). Advances in the delivery of antisense oligonucleotides for combating bacterial infectious diseases. Nanomed. NBM 14, 745–758. doi: 10.1016/j.nano.2017.12.026
Younis, A., Siddique, M. I., Kim, C. K., and Lim, K. B. (2014). RNA Interference (RNAi) induced gene silencing: a promising approach of hi-tech plant breeding. Int. J. Biol. Sci. 10, 1150–1158. doi: 10.7150/ijbs.10452
Yu, X., and Killiny, N. (2020). RNA interference-mediated control of Asian citrus psyllid, the vector of the huanglongbing bacterial pathogen. Trop. Plant Pathol. 45, 298–305. doi: 10.1007/s40858-020-00356-7
Keywords: antisense oligonucleotide, citrus huanglongbing, psyllid vectors, Liberibacter disease, FANA, biopesticide, microbicide, Solanaceae
Citation: Hunter WB, Cooper WR, Sandoval-Mojica AF, McCollum G, Aishwarya V and Pelz-Stelinski KS (2021) Improving Suppression of Hemipteran Vectors and Bacterial Pathogens of Citrus and Solanaceous Plants: Advances in Antisense Oligonucleotides (FANA). Front. Agron. 3:675247. doi: 10.3389/fagro.2021.675247
Received: 02 March 2021; Accepted: 04 June 2021;
Published: 08 July 2021.
Edited by:
Thais B. Rodrigues, GreenLight Biosciences, United StatesReviewed by:
Steve Whyard, University of Manitoba, CanadaCopyright © 2021 Hunter, Cooper, Sandoval-Mojica, McCollum, Aishwarya and Pelz-Stelinski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wayne Brian Hunter, d2F5bmUuaHVudGVyQHVzZGEuZ292; William Rodney Cooper, cm9kbmV5LmNvb3BlckB1c2RzYS5nb3Y=; Kirsten S. Pelz-Stelinski, cGVsenN0ZWxpbnNraUB1ZmwuZWR1
†Lead authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.