
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 02 July 2021
Sec. Plant-Soil Interactions
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.666351
This article is part of the Research Topic Soil Fertility Management for Sustainable Food Production in Sub-Saharan Africa View all 11 articles
In Africa, intercropping and intra-hole cropping systems are common practices used by smallholder farmers to optimize land use and tap the benefits of plant-to-plant interactions. The aim of this study was to evaluate mineral nutrient concentrations, P-enzyme activity and changes in microbial communities in the rhizospheres of sole cropped and intra-hole planted cowpea (cvs. TVu 546 and PAN 311), maize (cv. ZM 521), and sorghum (cv. M48). Cowpea cv. TVu546 intra-hole planted with sorghum (i.e., TVu546+M48) produced the highest rhizosphere acid phosphatase (APase) activity (230.0 μg p-nitrophenol.g−1 soil.h−1). From 16S rRNA Miseq Illumina sequencing, the rhizosphere bacterial community structure was altered by intra-hole cropping, and was dominated by Actinobacteria, Acidobacteria, Bacteroidetes, Firmicutes, Planctomycetes, Proteobacteria, and Verrucomicrobia, which together accounted for about >95% of the total sequences. The Sphingobacteria phylum was the dominant microbial group in the rhizosphere soil of all the cropping systems. The Proteobacteria phylum was the second most abundant in this study, which included the beneficial bacteria in all the rhizosphere soils studied. In contrast, typical pathogens like Ralsotonia and Agrobacterium were completely absent, indicating that the intra-hole cropping system can provide protection against soil-borne diseases possibly through elimination by antibiotics and/or phytoalexins present in plant and microbial exudates in the rhizosphere. Mucilaginibacter and Flavobacterium were however selectively present with intra-hole cropping.
Intercropping, defined as the planting of two or more crops on the same piece of land within the same cropping season, is an ancient cultural practice common among smallholder farmers, especially in Africa. The intercropping system can vary from intra-row intercropping, inter-row intercropping to intra-hole cropping, which involves two or more different plant species. These intercropping systems are a common practice used by smallholder farmers in Africa to optimize land use as well as tap the benefits of plant-to-plant interaction. This practice is regarded as low-input and self-sustaining, in that, it enhances the amount of biomass and yield that is harvested per unit land area (Matusso and Mucheru-Muna, 2014). With this practice, yield reliability and insurance against crop failure are key, with intercrops tending to have higher yield stability than sole crops especially in areas prone to extreme weather conditions (Papastylianou, 2004; Wang et al., 2014). Where intercropped plants have different root architecture, soil fertility is improved as roots of such crops explore a larger soil volume through increased root length and density, as well as root distribution (Li et al., 2013). Where a legume is intercropped with non-legume, direct transfer of fixed-N from the legume to the non-legume can occur, leading to enhanced N nutrition of the companion crop (Xiao et al., 2004).
Intra-hole cropping is a system where two different crop species (e.g., legume and cereal) are planted in the same hole (Dakora et al., 2008). The intra-hole legume/cereal pairing is often practiced where soils are low in N. As a result, the legume's N-rich compounds excreted by root nodules, released through rhizodeposition, leached from leaves, as well as N transferred through mycorrhizal fungal mycelia become available to the associated cereal plant belowground, in addition to products of organic matter decomposition becoming more available to succeeding crops (Hamel and Smith, 1991; Fustec et al., 2010).
Enhanced P nutrition of cereals from intercropping with legumes has been attributed to P-solubilization in the rhizosphere by symbiotic rhizobia and the secretion of P compounds in root exudates (Dakora and Phillips, 2002; Maseko and Dakora, 2013). However, the root exudates also contain phosphatases that can decouple unavailable P from complexes (Gunes et al., 2007), organic acids that can lower soil pH and make P more available (Li et al., 2007), and protons released during nitrogen fixation that can readily acidify the rhizosphere for increased P availability (Shen et al., 2011). Where roots of the intercropped species are intermingled and/or in close proximity, the solubilized P can benefit the component non-legume plant. In fact, Makoi et al. (2010) have shown that a sorghum/cowpea intercrop increased APase and AlkPase activity, leading to enhanced P nutrition, greater plant growth, and higher grain yield. A soybean/sugarcane intercrop in the glasshouse also resulted in increased total soil microbe number when compared to the monocultures (Li et al., 2013).
Being indigenous to the African continent, both cowpea and sorghum are widely adapted to different soil ecologies, including tolerance to drought, and are used as food and cash crops by most smallholder and commercial farmers in Africa together with maize. During intra-hole cropping, where the legume and cereal are planted in the same hole, the roots of the two species can become interwoven, leading to direct nutrient acquisition and increased mobilization (Li et al., 2010). There are also reports that an increase in rhizosphere microbial biomass and P-enzymes can lead to improved nutrient supply to both partners compared to either sole crop (Inal et al., 2007; Makoi et al., 2010). However, soil and plant management practices can have much greater influence on soil microbial mass (Gupta and Germida, 1988; Dick et al., 1994; Alvey et al., 2003), with monocultures tending to show reduced microbial biomass and activity in comparison with mixed cultures (Moore et al., 2000). Furthermore, legumes generally accumulate greater microbial biomass in the soil than cereals (Walker et al., 2003).
Although there is considerable information on the relationship between soil management and soil microbial activity, little is known about it under mixed cultures commonly practiced by smallholder farmers in Africa (Dick et al., 1988; Deng and Tabatabai, 1996; Yang et al., 2015). Although rhizosphere microbial composition used to be studied by culturing the bacteria for single species interactions, now however, root-microbe interactions and the composition of root-associated microbial community have become easier to assess, with the development of next generation sequencing (Mendes et al., 2013; Hacquard et al., 2015). Furthermore, while several studies have been conducted on intercropping (Wahua, 1984; Li and Wu, 2018; Dang et al., 2020; Li et al., 2020), to our knowledge little is known about intra-hole cropping of legumes and cereals. We hypothesized that intra-hole cropping of cowpea-maize, and cowpea-sorghum would influence soil P-enzyme activity, mineral nutrient accumulation and microbial communities based on the partners used in the intra-hole cropping system. Therefore, the aim of this study was to evaluate P-enzyme activity, mineral nutrient accumulation and microbial community structure in the rhizosphere of sole and intra-hole cropped cowpea with cereals (namely, sorghum and maize).
The field experiment was carried out at the Marapyane Campus of the Lowveld College of Agriculture (24°57′53.777″S and 28°45′41.544″E), Mpumalanga Province of South Africa. The experimental field was grass fallow for over 30 years, but frequently grazed by livestock before the experiment was established in the 2013/2014 cropping season. The Mpumalanga Province is dominated by savanna vegetation and is semi-arid in climate with cold winter and warm summer seasons. The temperature and average rainfall can vary from 15.9 to 32.5°C and 0 to 34.4 mm, respectively, during planting summer season.
The experimental field layout consisted of plots measuring 4.2 m long and 2.4 m wide with an inter-row spacing of 80 cm, intra-row spacing of 30 cm and 90 cm between plots. The treatments included two cropping systems (monoculture and intra-hole cropping), two cowpea genotypes (PAN 311 and TVu 546), and two cereal species (maize cv. ZM521 and sorghum cv. M48). The experiment was laid out using a randomized complete block design with three replications. With sole cropping, two seeds were planted per hole for each crop species (cowpea, maize and sorghum), and later thinned to one plant per stand at 2 weeks after planting. With the intra-hole cropping system, four seeds (two of cereal and two of legume) were planted per hole, and later thinned to one plant of cowpea and one plant of cereal per stand at 2 weeks after sowing. The plants were naturally rain-fed without irrigation. Weeds were controlled manually, when necessary.
At 86 d after planting (DAP), samples of rhizosphere soil (defined here as soil adhering to the plant roots) were collected from each treatment, namely, sole cowpea, sole maize, sole sorghum, intra-hole cropped cowpea with maize and intra-hole cropped cowpea with sorghum. With the monocultures, soil adhering to the root system of each treatment was shaken off into a pre-labeled zipper plastic bag and sealed. With intra-hole planting, however, the soil around the intermingled roots of both cowpea and cereal was collected. The rhizosphere soil samples were immediately put into dry ice in a cooler box and transported to the laboratory where each sample was divided into three parts. One part was kept at −4°C for later use in APase and AlkPase assay, the second part stored at −80°C for later use in soil microbial DNA extraction, and the third part air-dried at room temperature, sieved (2 mm), and stored for mineral analysis.
To measure pH, 50 mL 0.01 M CaCl2 was added to 10 g of air-dried and sieved (2 mm) soil sample. The mixture was mixed thoroughly using an end-to-end shaker at 100 rpm for 3 h. After shaking, the suspension was allowed to settle for 30 min and the pH measured using a BT-675 pH benchtop meter [Boeckel + Co (GmbH + Co) KG, Hamburg, Germany].
APase and AlkPase activity in the rhizosphere soils was assayed as described by Tabatabai (1994). The solution was filtered through Whatman #2 filter paper and the absorbance of the supernatant was measured at 420 nm using a UV–VIS spectrophotometer (JENWAY 7300, Bibby Scientific Ltd, Stone, Staffs, UK). The absorbances of filtrates were compared with p-nitrophenol standards. For each assay, a control was included to account for non-enzymatic substrate hydrolysis, and the enzyme activity expressed as μg p-nitrophenol g−1 F wt soil h−1.
The rhizosphere soil samples that were processed and kept for mineral analysis were used to analyze for P, K, Ca, Mg, Cu, Zn, and Fe according to the citric acid method developed by Dyer (Dyer, 1894) and modified by the Division of Chemical Services (1956), and later by Du Plessis and Burger (1965). The acid digestion method was employed to determine soil concentrations of P, K, Ca, and Mg and the EDTA method for Cu, Zn, and Fe using plasma-mass spectrometer (ICP-MS) (IRIS/AP HR DUO Thermo Electron Corporation, Franklin, Massachusetts, USA).
The rhizosphere soils collected from each treatment plot were bulked together to obtain one composite sample. Genomic DNA was extracted from 0.5 g of the composite samples using PowerSoilTM DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA) according to the manufacturer's instructions. The DNA concentrations were measured using PicoGreen (Invitrogen, cat. # P7589) method and Victor 3 fluorometry. Fluorescence was measured for three cycles of 30 s at 25°C in 96 well plates, and a standard curve generated by means of the fluorescence results, was used to determine the DNA concentration and adjusted to a final level of 3.5 ng/μl. The DNA samples with known concentrations were sent to Macrogen (The Netherlands) for sequencing.
The sequencing library was prepared by random fragmentation of the DNA sample, followed by 5′ and 3′ adapter ligation. All samples of rhizosphere microbial DNA were subjected to PCR amplification in duplicates using primer pairs retaining the adapter-ligated fragments 5′TCGTCGGCAGCGT
CAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG3′ and 5′GTCTCGTGGGCT
CGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC3′ targeting variable regions V3-V4 of the 16S rDNA gene (Klindworth et al., 2013). The polymerase chain reaction (PCR) was carried out with about 10–15 ng DNA in 25 μl reaction volume containing 12.5 μl 2 × KAPA HiFi hotstart ready mix, and 5 μl of each primer (1 μM) with the following temperature profile: 95°C-30 s, 25 × (95°C-30 s, 55°C-30 s, 72°C-30 s), 72°C-5 min. To verify the size of PCR enriched fragments, we checked the template size distribution by running it on an Agilent Technologies 2100 Bioanalyzer using a DNA 1000 chip. Subsequently, PCR amplified products were cleaned using AMPure XP beads. The sample libraries were quantified using qPCR according to the Illumina qPCR Quantification Protocol Guide. The PCR amplified products were indexed by ligation of indexing adapters to the DNA fragments, and amplified by PCR. The amplified products were again purified using AMPure XP beads.
For cluster generation, the library was loaded onto a flow cell where fragments were captured on a lawn of surface-bound oligos complementary to the library adapters. Each fragment was then amplified into distinct, clonal clusters through bridge amplification. After completion of clustering, the templates were sequenced.
Illumina SBS technology was used for paired-end-sequencing, a proprietary reversible terminator-based method that detects single bases as they get incorporated into DNA template strands. As all four reversible, terminator-bound dNTPs are present during each sequencing cycle, natural competition minimizes incorporation bias and greatly reduces raw error rates compared to other technologies. The results were a highly accurate base-by-base sequencing that virtually eliminated sequence-context-specific errors, even within repetitive sequence regions and homopolymers.
During data analysis and alignment, the newly identified sequence reads were aligned using FLASH to reference genome. The raw sequences were processed in CD-HIT-OTU for quality control (QC) assessment to remove noise data and to ultimately generate cluster files for each sample. All processed and QC passed cluster files were analyzed using Qunatitative Insights into Material Ecology (QIIME) pipeline (Caporaso et al., 2010). The representative reads from non-chimeric clusters were grouped using a greedy algorithm into OTUs at a user-specified OTU cutoff (e.g., 97% ID at species level). Genetic distance was calculated and sequences were clustered into operational taxonomic units (OTUs). The alpha diversity index (Chao1, Shanon, and Simpson) was calculated for each sample at both distances. The taxonomical abundance (%) of microbial communities of each rhizosphere soil sample was calculated using read files as queries against removed and de-replicated set of sequences from the small subunit (SSU) UCLUST (Edgar, 2010). The principal coordinate analysis (PCoA) was carried out using make_rd_plot.py of QIIME with output data of beta diversity (pairwise sample dissimilarity). Rarefaction analysis was performed at species level for each sample using alpha_rarefaction.py of QIIME. The data were submitted to NCBI Sequence Read Archive under the project Bioproject ID PRJNA397662. Soil APase/AklPase and nutrient data were statistically analyzed using STATISTICA software program version 10.1. The Duncan's multiple range test was used to separate means at p ≤ 0.05.
Rhizosphere soil acidity ranged from pH 6.05–7.07 for the test species. Sole planted maize (cv. ZM 521) had soil pH 7.0, and sorghum (cv. M48) pH 6.61. In contrast, the two cowpea genotypes showed much lower soil pH levels, with cv. TuV 546 recording pH 6.34, and cv PAN 311 pH 6.28. When planted with sorghum in a single hole, cowpea cv. PAN311 lowered soil pH to 6.05, in contrast to cv. TVu546, which increased rhizosphere soil pH to 7.07 when planted in the same hole with sorghum. Planting cowpea cv. TVu 546 with maize (cv. ZM 521) in the same hole produced a slightly acidic rhizosphere soil reaction (pH 6.43).
Available Fe in the soil ranged from 88.75 to 111.70 mg/kg for all treatments (Figure 1). Fe concentration in the rhizosphere of maize and sorghum monocultures (102.36 and 111.70 mg/kg, respectively) was much higher than that of the sole-grown cowpea cultivars. Cowpea cv. PAN 311 also showed greater Fe level in its rhizosphere (99.28 mg/kg) than cv. TVu 546 (88.75 mg/kg).
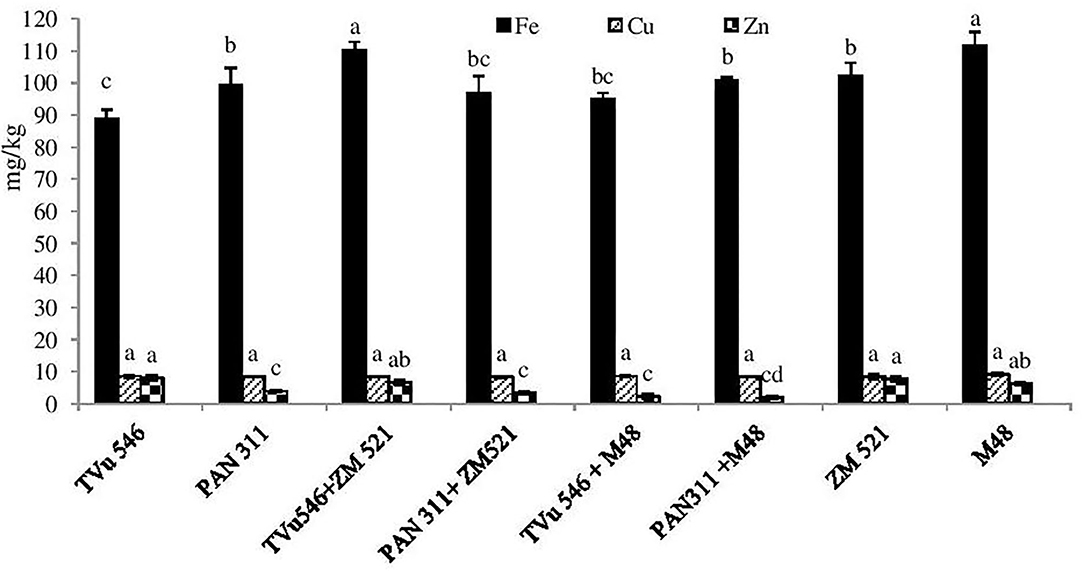
Figure 1. Available micro-element in rhizosphere of solo and intra-hole planted cowpea, sorghum, and maize. Bars with dissimilar letters are significantly different (P ≤ 0.05) for each element.
Intra-hole planting of sorghum with the two cowpea genotypes increased Fe levels in the rhizosphere soil relative to sole cowpea, but decreased it compared to sole sorghum (Figure 1). Planting maize with cowpea cv. TVu 546 also increased rhizosphere Fe relative sole cv. TVu 546.
Available Cu levels were generally similar in the rhizosphere soil of all the treatments, and ranged from 8.34 to 9.14 mg/kg. The concentration of Zn in the rhizosphere of all the treatments ranged from 1.96 to 8.06 mg/kg (Figure 1). The rhizosphere soil of cowpea cv. TVu 546 showed 2-fold higher Zn concentration (8.1 mg/kg) than cv. PAN 311 (3.9 mg/kg). However, the Zn level in the rhizosphere soil of the two cereals, sorghum (M 48) and maize (ZM 521), was much higher (6.30 and 7.74 mg/kg, respectively) relative to cowpea cv. PAN 311, but lower than cv. TVu 546. Intra-hole planting of sorghum or maize with the two cowpea cultivars decreased Zn levels relative to the monocultures of cowpea, sorghum, and maize (Figure 1).
The concentration of Ca in the rhizosphere soil of the test species ranged from 1012 to 11758 mg/kg (Figure 2), with the two cereals (maize and sorghum) registering the highest levels (11758 and 2118 mg/kg, respectively) relative to the legume. Cowpea cv. TVu 546 recorded 1287 mg/kg Ca and cv. PAN 311 1214 mg/kg. Intra-hole planting of maize with the two cowpea genotypes lowered Ca concentration in the rhizosphere relative to sole maize, but increased it when maize was planted in the same hole with cowpea cv. PAN 311 (Figure 2). Intra-hole planting of sorghum with each of the cowpea cultivars decreased rhizosphere Ca relative to monocultures of sorghum, cowpea cv. TVu 546 and cv. PAN 311.
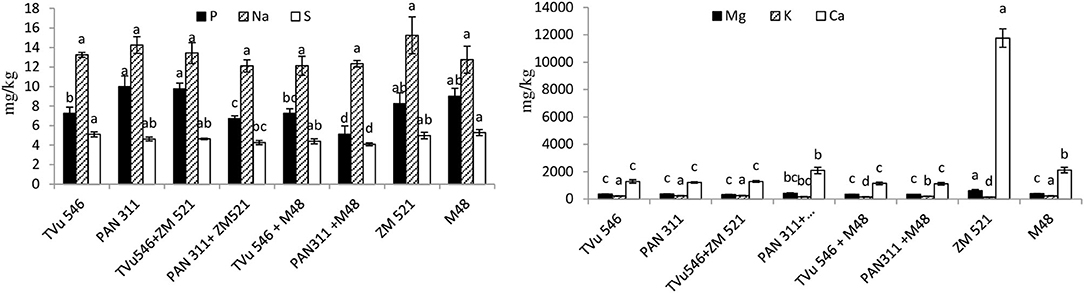
Figure 2. Presence of exchangeable macro-element in rhizosphere of solo and intra-hole planted cowpea, sorghum, and maize. Bars with dissimilar letters are significantly different (P ≤ 0.05) for each element.
The level of Mg in plant rhizospheres ranged from 324 to 605.10 mg/kg, with sole maize and sorghum exercising much greater concentration compared to sole cultures of the two cowpea genotypes (Figure 2). Planting maize with each of the two cowpea cultivars in same hole lowered Mg concentrations relative to monocultures of maize and cowpea cv. TVu 546, just as intra-hole planting of sorghum with the cowpea cultivars decreased the concentration of Mg in the rhizospheres of sorghum, cowpea cv. TVu 546 and cv. PAN 311 monocultures (Figure 2).
Potassium concentration ranged from 143.3 to 246.9 mg/kg in the rhizosphere soil of the test plants (Figure 2), with sole cowpea genotypes exhibiting greater K levels than the sole maize and sorghum. Intra-hole planting of maize with the cowpea genotypes increased rhizosphere K relative to sole-planted maize, as well as TVu 546 co-planted in one hole with maize. However, sowing sorghum seeds and cowpea seeds in one hole lowered rhizosphere K level when compared to sole-planted sorghum and cowpea (Figure 2).
Plant-available P in the rhizosphere soil of the test species ranged from 5.5 to 10.0 mg/kg, with cowpea cv. PAN 311 recording the highest P (10.00 mg/kg; Figure 2). Intra-hole planting of maize with cowpea increased the concentration of rhizosphere P relative to sole maize and cowpea cv. TVu 546, but decreased it when compared to sole maize and cowpea cv. PAN 311 (Figure 2). Similarly, planting sorghum and cowpea cv. PAN 311 in the same hole led to a decrease in P relative to monocultures of sorghum and cv. PAN 311, but had no effect with cv. TVu 546 (Figure 2).
Relative to APase, AlkPase activity was generally higher in the rhizosphere of the test species. Furthermore, the APase and AlkPase activity of sole-planted cowpea cultivars was much greater than that of their cereal counterparts (Figure 3). Intra-hole planting of maize with each of the two cowpea cultivars resulted in an increase in rhizosphere AlkPase activity relative to sole-planted maize, and a decrease relative to monocultured cowpea genotypes. However, planting maize in the same hole with cowpea cv. TVu 546 increased rhizosphere APase activity relative to sole-planted TVu 546, while with PAN 311, it decreased APase activity when compared with sole stands of maize and cv. PAN 311. Intra-hole planting of sorghum with cowpea cv. TVu 546 increased APase activity relative to sole-planted sorghum and cowpea cv. TVu 546, while with cv. PAN 311, it decreased it relative to sole stands of sorghum and cv. PAN 311. Planting sorghum in the same hole with cowpea cv. TVu 546 or PAN 311 decreased AlkPase activity relative to sole-planted cowpea cvs. TVu 546 and PAN 311, but increased AlkPase activity when compared with sole-planted sorghum (Figure 3).
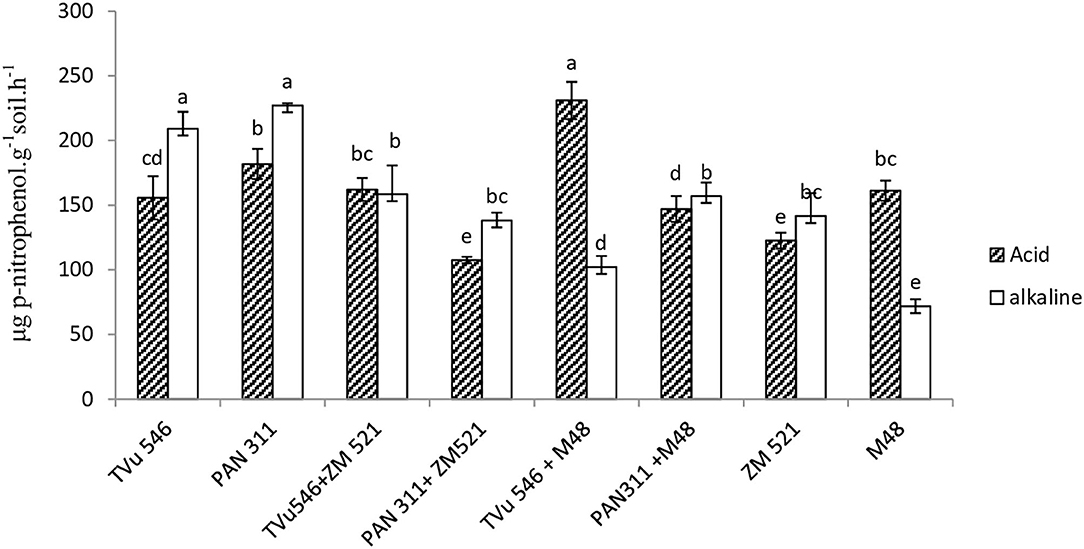
Figure 3. Phosphatase activity in the rhizosphere of solo and intra-hole planted cowpea, sorghum, and maize. Bars with dissimilar letters are significantly different (P ≤ 0.05) for each enzyme activity.
The results of sequencing microbial DNA extracted from rhizosphere soils showed that the intra-hole planting system exhibited higher nucleotide sequences when compared to sole planting. Intra-hole planting of cowpea cv. TVu 546 with sorghum (cv. M 48) revealed the highest sequence bases (>78 million), while sole-planted cowpea cv. PAN311 recorded the lowest (<54 million). The highest GC (55.6%) was found in the rhizosphere metagenome sequences of maize (ZM 521), but Q20 and Q30 were highest in the rhizosphere metagenome of intra-hole planted cowpea (TVu 546) with sorghum (M 48) (Table 1).
A total of 1,176,570 raw reads were generated from analysis of the eight rhizosphere soil samples collected from sole and intra-hole planted maize, sorghum and cowpea. There were variations in read output across the analyzed samples, with >100,000 reads obtained per sample (Table 1). With subsequent analysis, however, 107,171 quality sequences were added which resulted in 11,677 as a median per sample with a range of 9523–22907. The read length was 300 bp. All quality sequences were aligned at >97% sequence identity using QIIME to determine operational taxonomic units (OTUs). The reads were all successfully classified as belonging to the domain bacteria. The coverage was between 98 and 99% for each of the eight test samples. The highest OTU (780) was found in the rhizosphere metagenome of cowpea (PAN 311) co-planted in the same hole with maize (ZM 521), and the least (642) in sole-planted cowpea cv. PAN 311 (Table 1). Both the observed and estimated richness (Chao 1) were higher in the rhizosphere soil of cowpea (cv. PAN 311) planted in one hole with maize cv. ZM 521 (Table 1). The Shannon diversity index was 7.8 for rhizosphere soils from sole maize (cv. ZM 521) and intra-hole planted cowpea (cv. PAN 311) with maize, 7.7 for sole cowpea (cv. TVu 546) and cowpea (cv. PAN 311) co-planted with sorghum in one hole, 6.4 for rhizosphere soils of sole-planted cowpea (cv. PAN 311), 7.3 for sole sorghum, 5.9 for cowpea (cv. TVu 546) planted in the same whole with maize, and 6.8 for cowpea (cv. TVu 546) planted in one hole with sorghum (Figure 4).
The alpha rarefaction graph (Figure 5) for each soil sample did not reach the plateau phase, which indicates that more reads are required to capture all the diversity associated with the plateau part of the graph.
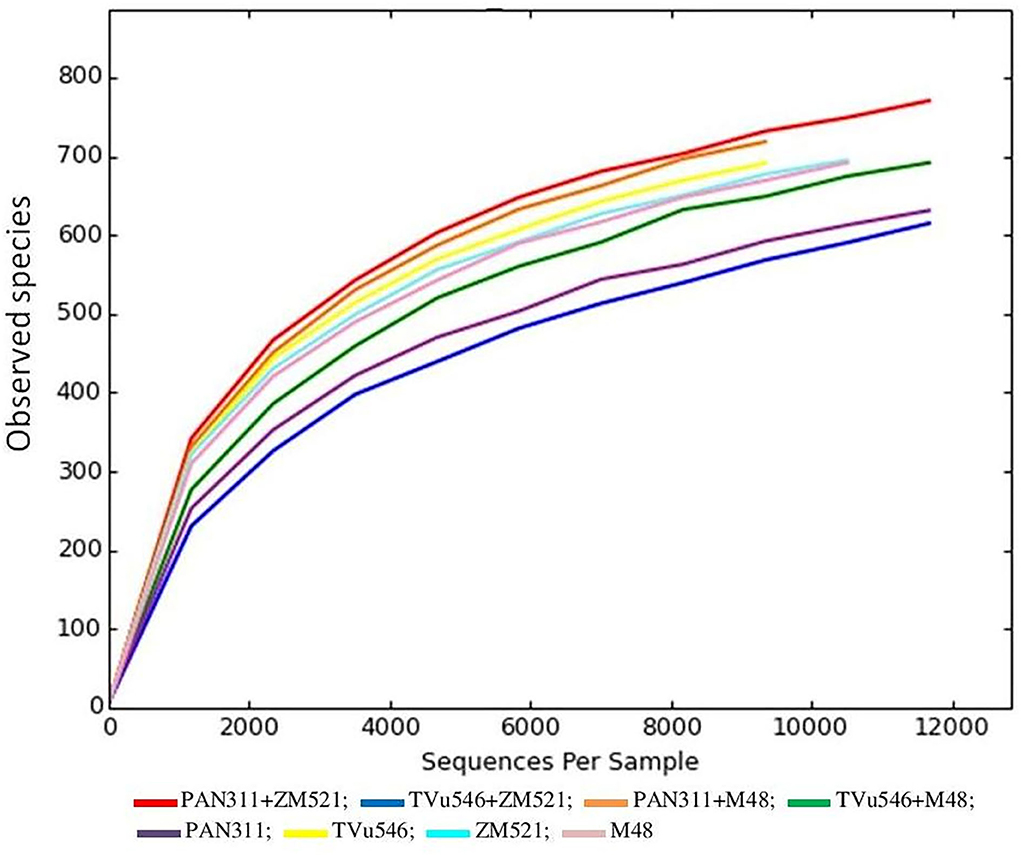
Figure 5. Rarefaction curves of all samples were generated for observed microbial species which contained unique sequences and were defined at 97% sequence similarities.
When the observed reads were assigned to high taxonomic ranks, the occurrence and relative abundance of different phyla suggested a strong dissimilarity among bacterial communities in all the rhizosphere soil samples tested. Except for cowpea (cv. PAN 311) co-planted with sorghum, the intra-hole planting system generally showed higher microbial phyla than sole cropping. Results showed that including undefined phylum the highest number of phyla (20) were found in the rhizosphere soil of intra-hole planted cowpea (cv. PAN 311) with maize, with the least number of phyla (16) detected in the rhizosphere soil of intra-hole planted cowpea (cv. PAN 311) with sorghum (Figure 6). The most dominant phyla in all the test samples were Actinobacteria, Acidobacteria, Bacteroidetes, Firmicutes, Planctomycetes, Proteobacteria, and Verrucomicrobia, as they accounted for >95% of the total bacteria observed. The microbial community profiles in the rhizosphere soils analyzed were not similar at the class, order and family levels due to little differences in the relative abundance of some bacteria. The rhizosphere soils of sole cowpea (cv. PAN 311), sole maize, and cowpea (cv. TVu 546) co-planted with maize showed 18 bacterial phyla, while sole cowpea (cv. TVu 546), sole sorghum and cowpea (cv. TuV546) intra-hole planted with sorghum revealed 17 bacterial phyla with some slight differences in relative presence and absence of certain bacterial phyla. For example, Thermomicrobia was absent in the rhizosphere soil of sole maize, as well as the two cowpea genotypes co-planted with sorghum (i.e., PAN311+M48 and TVu546+M48), but was present in the rhizosphere soils of sole-planted cowpea cultivars (i.e., cv. PAN 311 and TVu546), sole planted sorghum, and maize intra-hole planted with the two cowpea genotypes (i.e., PAN311+ZM521 and TuV546+ZM521). Similarly, Fibrobacteres was present in the rhizosphere soil of sole maize and maize intra-hole planted with the two cowpea cultivars (i.e., PAN311+ZM521, TVu546+M48), but absent in all other samples. Also, Parcubacteria was present in the rhizosphere soils of only sole sorghum and cowpea co-planted with maize. The phyla Aerophobetes and Tenericutes were present exclusively in the rhizosphere soils of intra-hole planted cowpea (cv. PAN 311) with maize, and sole-planted maize.
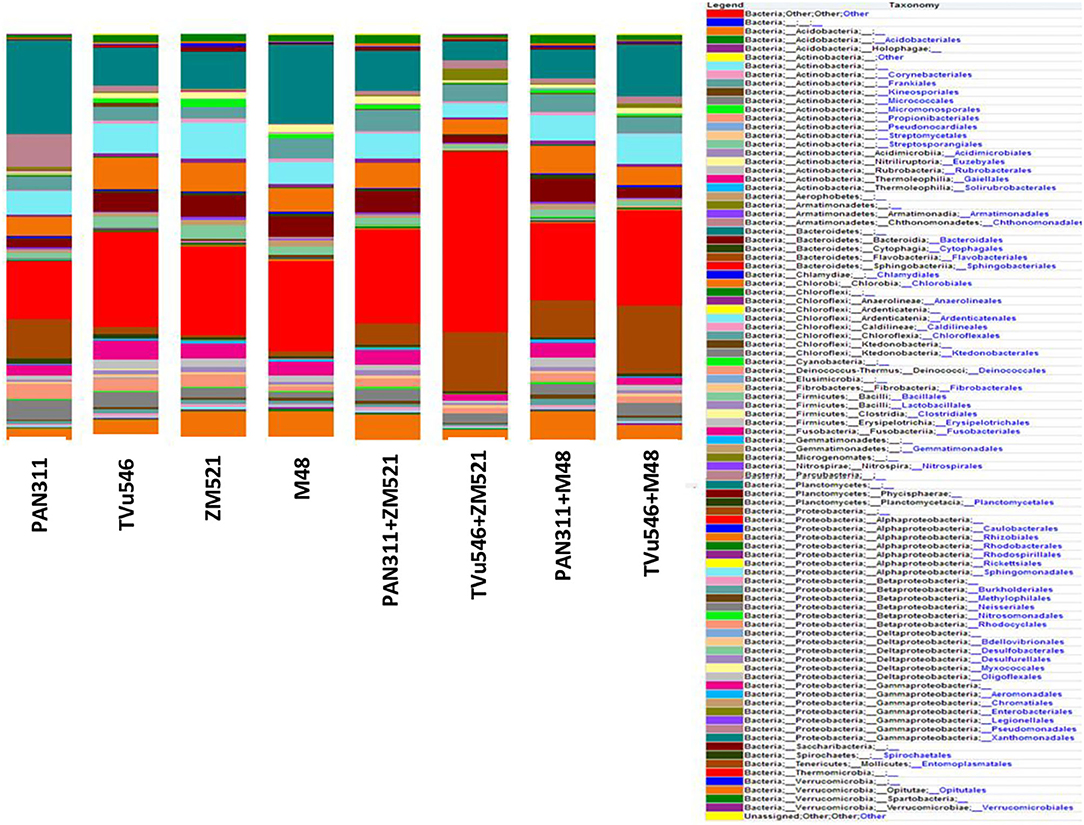
Figure 6. Average distribution of 16S rRNA sequences classified at order level in rhizosphere of solo and intra-hole planted cowpea, sorghum, and maize.
At the class and order level, the Sphingobacteria was the most dominant microbial community present in all rhizosphere soils (except for the rhizosphere of sole cowpea (cv. PAN 311) which was dominated by Gammaproteobacteria). Alphaproteobacteria was the second dominant microbial community present in the rhizosphere soils of sole cowpea (cv. TVu 546) (16.44%), sole maize (17.03%), intra-hole planted cowpea (cv PAN 311) with maize (13.7%) and intra-hole planted cowpea (cv. PAN 311 with sorghum (14.7%). By proportions, Sphingobacteria represented 14.2% of the microbial community in the rhizosphere of sole cowpea (cv. PAN 311), Gammaproteobacteria 19.9% in the rhizosphere of sole-planted sorghum and Flavobacteria 14.5% in the rhizosphere of intra-hole planted cowpea (cv. TVu546) with maize. The phylum Cyanobacteria was present in the rhizosphere soils of all treatments, except cowpea (cv. TVu 546) intra-hole planted with maize.
At the genus level, Lysobacter was dominantly present in the rhizospheres of monocultured cowpea (cv. PAN 311) (20.9%) and sorghum (15.5%), as well as cowpea (cv. PAN311) co-planted in one hole with maize (7.4%), while Sphingomonas was dominant (8.1%) in the rhizosphere soil of sole-planted maize. In contrast, Mucilaginibacter was the most abundant genus in the rhizosphere soils of intra-hole planted cowpea (cv. TVu 546) with maize (27.7%) and cowpea (cv. TVu 546) co-planted in one hole with maize (11.9%). The genus Flavobacterium was dominant in the rhizosphere soils of sole cowpea (cv. PAN311) (8.9%), sole sorghum (1.2%), intra-hole planted cowpea (cv. PAN 311) with maize (4.6%), cowpea (cv. TVu546) co-planted with maize (10.0%), and intra-hole planted cowpea genotypes TVu 546 and PAN 311 with sorghum (7.2 and 8.4%, respectively). An undefined genus of the Chitinophagaceae family was the most abundant in rhizosphere soil of sole-planted cowpea cv. TVu 546. Despite the presence of diverse bacterial species in the test rhizosphere soils, most of them were classified as unculturable.
To compare microbial community variation between samples, principal coordinate analysis (PCoA) was done using weighted UniFrac metric. The PCoA of the UniFrac metric illustrated clustering of soil microbial communities by number of sequences generated from each soil sample. Based on operational taxonomic units or OTUs (97% 16S rRNA identity), the results showed that the microbial communities were separated by rhizosphere soil of the test crop species and the cropping systems, indicating that there was a difference in the community composition (Figure 7). The two principal components (PC1 and PC2) accounted for 86.49% of the total microbial community variations in the rhizosphere soil samples tested. Despite this clear separation, the first axis in PC1 vs. PC2 explained 70.79% of the variance within the data, indicating that the relative abundance of the most OTUs was diverse between the rhizosphere soils of all the treatments.
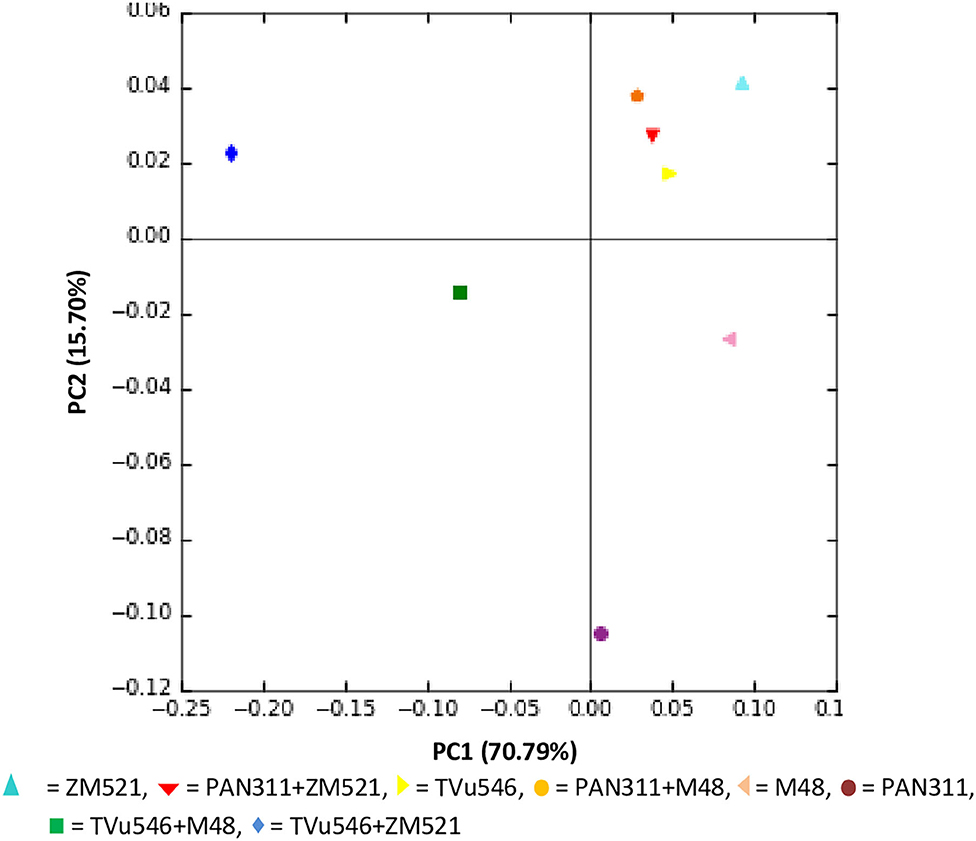
Figure 7. Principal Co-ordinate analysis (PCoA) of microbial communities based on OTUs for all samples from rhizosphere of solo and intra-hole planted cowpea, sorghum, and maize.
The number of sequences attributed to each taxon was compared between and among the rhizospheres of the test treatments based on distance matrix estimated with UPGMA algorithm represented in the phylogenetic tree (Figure 8). The results yielded one major cluster with two subclusters (Subcluster Ia and Ib). The microbial communities from the rhizosphere of sole-planted sorghum, sole cowpea (cv. PAN 311), and intra-hole planted cowpea (cv. TVu 546) with maize and sorghum stood separately, which clearly indicated the presence of diverse microbial community structures in the rhizosphere soils analyzed. The results further showed that the microbial communities in the rhizosphere soils of cowpea (cv. PAN 311) co-planted with sorghum or maize in one hole (Subcluster 1a), or sole-planted cowpea (cv. TVu546) and maize (Subcluster 1b) were very close when compared to the other cropping systems. The results of this study therefore suggest that the changes in microbial community structure in the rhizosphere soil samples were most distinct in intra-hole cropping systems relative to monocultures.
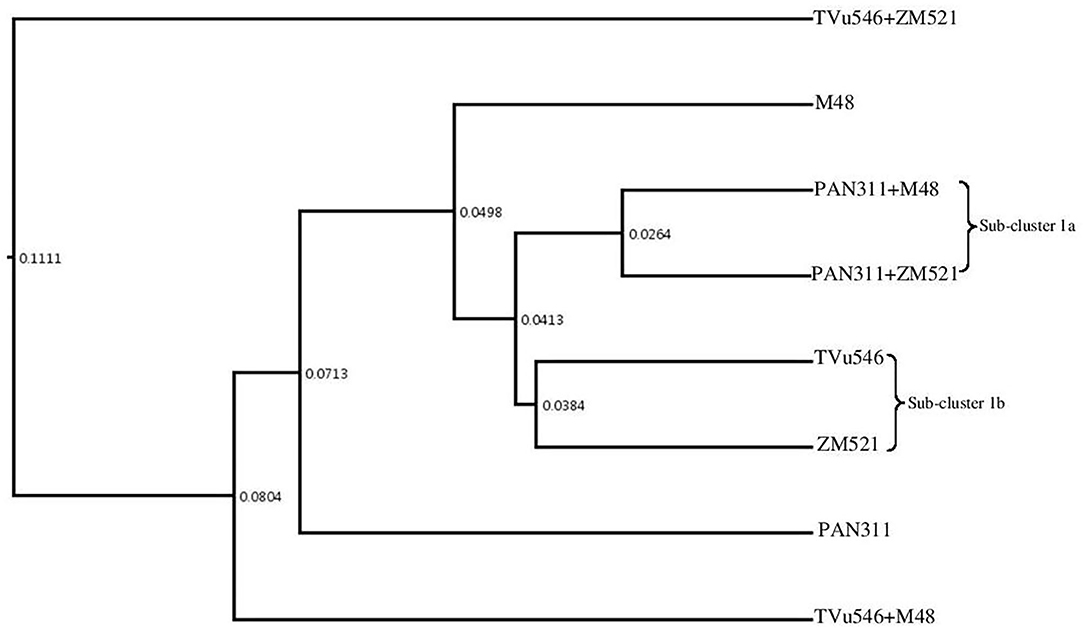
Figure 8. Hierarchical cluster analysis based on distance matrix with the UPGMA algorithm for eight rhizosphere metagenomes of solo and intra-hole planted cowpea, sorghum, and maize.
Rhizosphere functioning is plant species-dependent, but can be altered by many biological processes, including soil enzyme activity, microbial numbers, root border cells, mineral nutrient concentrations, and root exudates (Dakora and Phillips, 2002; Jaiswal et al., 2019). Soil nutrient profiles are often affected not only by plant species but also by soil enzymes and root border cells, and could be bio-indicators for soil fertility (Dick and Tabatabai, 1993; Dick et al., 2000). For example, soil enzymes such as APase and AlkPase, whether in plants or soils, are biomarkers for P nutrition, especially in nodulated legumes (Maseko and Dakora, 2013).
In this study, APase and AlkPase activities in the rhizosphere of monocultures of the two cowpea genotypes were generally higher than those of the cereals, be it sorghum or maize (Figure 3). This is consistent with the findings by Li et al. (2004) which showed that chickpea secreted greater amount of APase than maize. Izaguirre-Mayoral et al. (2002) also found that phosphatase activity in plants can differ with crop species and varieties. The higher phosphatase activity in the rhizosphere of cowpea was not unexpected as nodulated legumes have a greater P requirement to support symbiotic functioning in root nodules when compared to non-legumes (Israel, 1987; Maseko and Dakora, 2013).
These enzyme activities were however altered by the cropping system. For example, co-planting of cowpea cv. TVu546 with sorghum in one hole significantly increased APase activity, whether compared with sole cowpea or sole sorghum, and is consistent with the results of other intercropping studies involving legumes and cereals (Li et al., 2004; Inal et al., 2007). It was further found that intra-hole cropping of cowpea cv. TVu546 with sorghum or maize showed greater enzyme activity relative to both sole legume and cereal. In contrast, co-planting cowpea cv. PAN 311 with any of the two cereals lowered APase activity relative to sole legume and cereal (Figure 3). Because APase activity of sole cowpea cv. TVU 546 was less than that of cv. PAN 311, the former would require higher APase activity to meet its P needs when compared to the latter. These results can therefore be interpreted to mean that intra-hole cropping of cowpea cv. TVU 546 with sorghum or maize optimizes P nutrition in both the cereal and cowpea cv. TVU 546 via plant-plant interaction in the rhizosphere. Other studies have also suggested that the legume facilitates P availability for uptake by the cereal partner in the cropping system (El Dessougi et al., 2003; Li et al., 2004).
Rhizosphere AlkPase activity was much higher than APase activity in the rhizospheres of monocultured cowpea cv. TVu 546 and cv. PAN 311 (Figure 3), implying that microbial biota which are responsible for secreting AlkPases in the rhizosphere contributed significantly to P nutrition of the cowpea plants. The AlkPase activity in the rhizosphere soil of intra-hole planted cowpea cv. TVu 546 with maize or sorghum was always lower than the sole legume, but higher than sole cereals, a finding consistent with a report by Makoi et al. (2010). As a result, co-planting of cowpea cv. PAN 311 in one hole with sorghum increased AlkPase activity relative to sole cereal, but decreased it when compared to sole cv. PAN 411.
In this study, there were strong species differences in mineral accumulation in the rhizosphere of cowpea, maize and sorghum. In general, the mineral concentrations (Na, Mg, Ca, S, Fe, and Cu) in the rhizosphere of sole cowpea were much lower than maize and sorghum. This is not unexpected as the symbiotic process in legumes imposes a greater demand on the host plant for enhanced mineral nutrition to support N2 fixation (O'Hara, 2001). It has also been shown that high N2 fixation in legumes generally results in greater mineral uptake from the soil compared to low-fixing symbiosis (Belane et al., 2014).
In this study, available P in the rhizosphere soil of the test species ranged from 5.5 to 10.0 mg/kg, with cowpea cv. PAN 311 recording the highest P (10.0 mg/kg; Figure 2). P supply in the rhizosphere is influenced by a number of factors, including the exudation of P-enzymes. As shown in Figure 1, cowpea cv. PAN 311 secreted more APases an AlkPases than all the test species, and this can explain the high P concentration in the rhizosphere of cv. PAN 311. However, the activity of P-solubilizing rhizobia has also been reported to increase P availability in rhizosphere soils of legumes (Rodríguez and Fraga, 1999). The fact that intra-hole planting of maize with cowpea increased P concentration in the rhizosphere relative to sole maize and sole cowpea cv. TVu 546, but decreased it when compared to sole maize and cowpea cv. PAN 311 (Figure 2) could suggest differences in microbial populations within the rhizospheres of the two systems. Any variation in the microbial community can affect P-solubilization and phosphatase secretion, and thus affect P availability.
The level of Ca and Mg were greater in the rhizospheres of sole maize and sorghum compared to monocultured cowpea cultivars TVu 546 and PAN 311. While this could be species-dependent, N2-fixing legumes tend to accumulate more mineral nutrients than cereals (Gardner and Boundy, 1983). Intra-hole planting of sorghum or maize with each of the cowpea cultivars decreased rhizosphere Ca relative to monocultures of sorghum, cowpea cv. TVu 546 and cv. PAN 311. This can be attributed to plant-plant competition by the species for nutrient uptake in the rhizosphere.
In this study, soil Cu levels were more than sufficient to support plant growth. Rhizosphere concentration of Fe and Zn varied among the treatments, but were generally higher in the rhizosphere of sole-planted maize and sorghum than the two cowpea genotypes, possibly due to greater uptake by the legume for symbiotic functioning. Intra-hole planting of sorghum or maize with cowpea cultivars TVu 546 and PAN 311 increased Fe levels in the rhizosphere relative to sole cowpea (Figure 1). While this could be attributed to increased secretion of phytosiderophores by the plant partners for Fe mobilization, the microsymbionts nodulating the legume probably also produced extra bacterial siderophores for increased Fe availability (Xue et al., 2016). Whatever the case, positive changes in mineral nutrient concentrations in the rhizosphere can be regarded as one of the benefits of legume/cereal intercropping systems.
Plants and microorganisms are intricately inter-linked for growth and survival. Plant interactions with rhizosphere microbial community play a crucial role in agricultural productivity, in addition to contributing to the resilience of the ecosystem to climatic stress. Understanding plant/microbe interactions could reveal novel ways of using these microbes to promote plant health and productivity, and thus change ecosystem functioning. In this study, we performed sequence analysis of the bacterial communities in the rhizospheres of sole and intra-hole planted cowpea and cereals using the MiSeq Illumina method. The results revealed the presence of many unculturable bacterial populations in the plant rhizospheres, as earlier reported by Jaiswal et al. (2019). There were distinct bacterial community members across the rhizosphere environments of both sole and intra-hole cropped plants, which probably denoted differences in bacterial community function.
Although the reads were applied to species level, the sequences were summarized and classified according to bacterial phylum to ascertain the overall composition of the microbial community at a high phylogenetic resolution. The results showed differences in microbial communities between rhizosphere soils of sole and intra-hole planted cowpea with maize and sorghum. The differences in OTU richness, both within and among phyla, increased non-linearly with test plant species which suggested that selection of rhizosphere microflora was highly sole and intra-hole specific (Micallef et al., 2009; Turner et al., 2013). Although intra-hole planted cowpea cv. PAN 311 with maize recorded the highest number of phyla (20), cv. PAN 311 with sorghum showed only 16 microbial phyla. This suggests that specific plant combinations define the bacterial community composition in the rhizosphere, and this is probably influenced by plant-related changes in the soil such as root exudation (Marschner et al., 2001; Micallef et al., 2009; Jaiswal et al., 2019). In fact, root exudates are a crucial determinant of rhizosphere microbial diversity (Lynch and Whipps, 1990; Barea et al., 2005; Badri and Vivanco, 2009). The results also suggest that microbial biomass is more likely to increase where intra-hole cropped plant partners share close root proximity and/or exhibit intermingling of their roots.
Though present in the rhizosphere soil of sole-planted cowpea cv. PAN 311 and cowpea cv. TVu 546 co-planted with maize in the same hole, the phylum Thermomicrobia was absent in the rhizosphere soil of sole maize. Similarly, Fibrobacteres was present in the rhizosphere of sole maize, but absent in the rhizosphere soil of monocultured cowpea cv. PAN 311 and cowpea cv. TVu 546 co-planted with maize. The phylum Parcubacteria was also present in the rhizosphere of sole sorghum, but absent in sole cowpea cv. TuV 546 and cv. TVu 546 co-planted with sorghum. However, the phyla Aerophobetes and Tenericutes were exclusively present only in the rhizosphere soils of intra-hole planted cowpea cv. PAN 311 with maize, and sole-planted maize. The presence or absence of microbial phyla in some plant rhizospheres, but not in others, can be interpreted in number of ways. Components of plant root exudates generally serve as a source of carbon for many bacteria, and as chemoattractants for mutualistic microbes (Aguilar et al., 1988; Compton et al., 2020), often leading to the establishment of symbiotic relationships. In contrast, some components of plant root exudates also serve as defense molecules for warding off soil-borne pathogens (Dakora et al., 1993a,b). Various studies (Dakora et al., 1993a,b) have shown that both pathogens and rhizobial symbionts can elicit phytoalexin exudation in the rhizosphere of N2-fixing legumes, and thus control populations of other microbes. Whether in this study any of these scenarios functioned to create the observed differences in rhizosphere microbial communities remains to be determined. Additionally, pioneer microbes in the rhizosphere can also produce antibiotics that reduce the populations of other microbial communities, but again, this was not assessed in this study. Similarly, the fact that the rhizospheres of the two test cowpea cultivars (cv. PAN 311 and cv. TVu 546) revealed different bacterial phyla can be attributed to differences in the profile of seed and root exudates produced (Tsamo et al., 2018, 2020). Root exudate composition can vary with plant taxa, and even within closely related plant species, as well as with genotypes and accessions of the same species (Czarnota et al., 2003; Warembourg et al., 2003; Micallef et al., 2009).
Cropping system per se has been reported to have an effect on rhizosphere microbial populations. For example, rhizosphere bacterial counts were found to increase with intercropping, and reportedly greater in intra-row than inter-row planting (Wahua, 1984). Because legumes and cereals release root exudates with different chemical profiles into their rhizospheres, intercropping and/or crop rotations are likely to significantly alter rhizosphere microbial communities (Alvey et al., 2003). Except for sole-planted cowpea cv. PAN 311, Sphingobacteria was the dominant microbial group in the rhizosphere soil of all the other cropping systems, followed by Alphaproteobacteria, which is home to the many rhizobial symbionts nodulating legumes. Similarly, all rhizosphere soils except that of cowpea cv. TVu 546 co-planted with maize showed high proportions of Cyanobacteria. These major groups were earlier identified in a PCR-based and metatranscriptomic study of both bulk and rhizosphere soils (Urich et al., 2008; Inceoglu et al., 2011; Lundberg et al., 2012; Turner et al., 2013). Despite the fact that soil remains the richest microbial ecosystems on earth, our study has revealed that only a few bacterial phyla (namely, Actinobacteria, Acidobacteria, Bacteriodetes, Firmicutes, Planctomycetes, Verrucomicrobia, and Proteobacteria) actually colonize the rhizosphere, a finding similar to the results of earlier studies (Peiffer et al., 2013; Figuerola et al., 2015; Shi et al., 2015; Zarraonaindia et al., 2015; Coleman-Derr et al., 2016; Jaiswal et al., 2019). In this study, the major contributors to microbial community diversity in the rhizosphere included the Flavobacteriales, Sphingobacteriales, Bacillales, Phycisphaerae, Rhizobiales, Sphingomonadales, Burkholderiales, and Xanthomonadales, which are well-known for their mutualistic interactions with plant roots in the rhizosphere (Lu et al., 2006). Recently, Yang et al. (2016) found that intercropping of groundnut with sorghum was beneficial to the composition of bacterial communities in the rhizosphere.
In addition to identifying rhizobia and other beneficial bacterial taxa in this study, we also found the highest community of Lysobacter in the rhizosphere of sole-planted cowpea cv. PAN 311 (20.8%) and sorghum (15.5%), as well as in the rhizosphere of cowpea cv. TVu 546 co-planted with sorghum in one hole (10.8%) and cv. PAN 311 intra-hole planted with maize (7.4%). The genus Lysobacter is widely distributed in soil and has high P-solubilizing ability (Reichenbach, 2006). Its dominance in the rhizosphere of cv. PAN 311 probably explains the highest AlkPase activity observed in the rhizosphere soil of cv. PAN 311 (see Figure 3).
Furthermore, Lysobacter is reported to be an important source of new enzymes such as chitinases, glucanases, proteases, lipases, elastases, keratinases, phosphatases, endonucleases, endoamylases, and esterases (Zhang et al., 2001; Folman et al., 2003; Palumbo et al., 2005; Reichenbach, 2006; Stepnaya et al., 2008; Ko et al., 2009; Gökçen et al., 2014; Vasilyeva et al., 2014) that promote plant growth and enhance defense against pathogens (Gómez Expósito et al., 2015). Crop treatment with Lysobacter has been shown to enhance plant defense and decrease diseases in bean (Zhang et al., 2001), rice (Ji et al., 2008), pepper (Ko et al., 2009), cucumber (Folman et al., 2004; Postma et al., 2009) grapevine (Puopolo et al., 2014), and sugar beet, spinach (Islam et al., 2005).
The rhizosphere of sole-planted cowpea cv. PAN 311 was dominated by 8.9% Flavobacterium, and cv. PAN 311 co-planted with sorghum by 8.4%. The bacterium occurs in soil and other habitats (Bernardet and Bowman, 2006), and has extracellular macromolecular-degrading enzymes (amylase, cellulose, chitinase, peptidases and glycoside hydrolases) which enable it to digest degradable polymers like starch or chitin and therefore plays an important role in the degradation of bacteria, fungi, insects and nematodes (Peterson et al., 2006). Additionally, 70% of the rhizosphere carbon turnover is due to Flavobacterium (Johansen et al., 2009). As a result, Flavobacterium is a major member of the microbial community in the rhizosphere of pepper (Graber et al., 2010), lettuce (Cardinale et al., 2015), peanut (Haldar et al., 2011), and Arabidopsis thaliana (Lundberg et al., 2012; Bodenhausen et al., 2013). Recently, Qin et al. (2016) reported 0.9–8.5% Flavobacterium genera in wheat rhizosphere, and its presence correlated positively with plant biomass (Manter et al., 2010), plant disease resistance (Kolton et al., 2014), and bio-control against Phytophthora capsici in pepper (Sang et al., 2008). Flavobacterium also contains ACC deaminase activity for promoting plant growth (Maimaiti et al., 2007), in addition to the ability of some Flavobacterium isolates to elevate Rubisco gene expression and increase plant biomass in wheat and barley (Flynn et al., 2014).
The genus Mucilaginibacter, a member of the family Sphingobacteriaceae (Steyn et al., 1998; Pankratov et al., 2007) and phylum Bacteroidetes, was more abundant by 27.1 and 11.9%, respectively, in the rhizospheres of intra-hole planted cowpea cv. TVu546 with maize and sorghum. They are extracellular polysaccharide (EPS)-producing, plant-growth-promoting bacteria that were also isolated from cotton rhizosphere soil (Madhaiyan et al., 2010).
However, bacterial genera such as Pedobacter, Pseudomonas, Flavisolibacter, Bacillus, Rhizobium, Sphingomonas, and Streptomyces, which are beneficial microbes, were present in all the rhizosphere soils studied, thus indicating their general presence in the rhizosphere of crop species. A few studies have implicated Bacillus in the solubilization of Zn, Cu, and K in the rhizosphere (Pirhadi et al., 2016; Sunithakumari et al., 2016). Functionally, Sphingomonas is associated with IAA production in the rhizosphere of soybean for enhanced plant growth (Sugiyama et al., 2014), Pseudomonas, Bacillus, and Rhizobium with P solubilisation in the rhizosphere (Rodríguez and Fraga, 1999), and Streptomyces, Bacillus, and Pseudomonas with siderophore production for enhanced Fe mobilization (Matsuoka et al., 2013).
Although several studies have been undertaken on intercropping of legumes and cereals, few (if any) have addressed belowground processes involving critical analysis of the cowpea-sorghum intra-hole cropping system in terms of microbial community structure and mineral nutrition. This is possibly because data obtained from rhizosphere studies of mixed-cultured systems are often complex and difficult to interpret when compared to monocultures. The rhizosphere is home to millions of different microbial phyla, genera, families and species; and the complexity of this system increases with intercropping, especially with intra-hole planting where roots of legumes and cereals are closely interwoven and intermingled. In such an intimate system, there are plant-plant, plant-microbe and microbe-microbe interactions that are constantly occurring in the rhizosphere with different outcomes from mutualism and warfare. These interactions involve chemical products present in plant and microbial exudates. According to Dakora and Phillips (2002), plant root exudates consist of a complex mixture of enzymes, root border cells, phytosiderophores, phenolics, organic acids, gaseous molecules, organic and inorganic ions, as well as mineral nutrients, which are important for defense and promoting mineral uptake by roots. Soil microbes similarly secrete enzymes, organic acids and anions, siderophores and antibiotics for improving mineral nutrition and self-defense (Dakora and Phillips, 2002). For example, typical pathogens like Ralsotonia and Agrobacterium were completely absent in the rhizosphere of intra-hole cropping system possibly due to elimination by antibiotics and/or phytoalexins present in plant and microbial exudates (Dakora and Phillips, 1996). It is also therefore likely that the Pseudomonas and Xanthomonas species found in the Proteobacteria phylum in this study were the beneficial rather than pathogenic species, given the greater abundance of mutualistic microbes such as Flavobacteriales, Sphingobacteriales, Bascillales, Phycisphaerae, Rhizobiales, and Sphingomonadales found in this same study. Dang et al. (2020) found similar changes in some bacterial taxa, especially Proteobacteria, in a mung bean-millet intercropping system. Even with a tomato/potato-onion intercropping devoid of legumes, there was an alteration in soil microbial communities (Li et al., 2020), suggesting that changes in rhizosphere microflora is not unique to legume-cereal systems, but more likely an outcome of the composition of root exudates released by the intercropped partners.
In conclusion, intercropping of legumes with cereals is an old cultural practice perceived with the lens of “old science.” With new tools and techniques, however, more data can be generated on belowground processes that would advance our understanding of both plant-plant and plant-microbe interactions in the rhizosphere with potential for increasing crop yields. In this study, APase and AlkPase enzyme activity, mineral nutrient accumulation and microbial community structure in the rhizosphere were influenced by crop species (legume and cereals), as well as cropping system. The cropping system promoted an increase in the numbers and diversity of some functional and unculturable rhizosphere microbes. Intra-hole cropping significantly altered rhizosphere microbial community structure when compared with monoculture.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA397662.
SJ executed molecular work and drafted the manuscript. MM did soil analysis. FD conceived the idea, edited and approved the final version of the paper. All authors contributed to the article and approved the submitted version.
FD was grateful to the NRF, Tshwane University of Technology (TUT) and the South African Research Chair in Agrochemurgy and Plant Symbiosis for continued funding of his research. SJ and MM also respectively acknowledge the NRF and TUT for support of their postdoctoral and Master's fellowships through the Research Chair.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguilar, J. M. M., Ashby, A. M., Richards, A. J. M., Loake, G. J., Watson, M. D., and Shaw, C. H. (1988). Chemotaxis of Rhizobium leguminosarum biovar phaseoli towards Flavonoid Inducers of the Symbiotic Nodulation Genes. Microbiology 134, 2741–2746. doi: 10.1099/00221287-134-10-2741
Alvey, S., Yang, C.-H., Buerkert, A., and Crowley, D. E. (2003). Cereal/legume rotation effects on rhizosphere bacterial community structure in West African soils. Biol. Fertil. Soils 37, 73–82. doi: 10.1007/s00374-002-0573-2
Badri, D. V., and Vivanco, J. M. (2009). Regulation and function of root exudates. Plant. Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Barea, J. M., Werner, D., Azcón-Guilar, C., and Azcón, R. (2005). “Interactions of arbuscular mycorrhiza and nitrogen-fixing symbiosis in sustainable agriculture,” in Nitrogen Fixation in Agriculture, Forestry, Ecology, and the Environment (Berlin/Heidelberg: Springer-Verlag), 199–222.
Belane, A. K., Pule-Meulenberg, F., Makhubedu, T. I., and Dakora, F. D. (2014). Nitrogen fixation and symbiosis-induced accumulation of mineral nutrients by cowpea (Vigna unguiculata L. Walp.). Crop Pasture Sci. 65, 250–258. doi: 10.1071/CP13283
Bernardet, J.-F., and Bowman, J. P. (2006). “The genus flavobacterium,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 481–531.
Bodenhausen, N., Horton, M. W., Bergelson, J., Wolf, J., and van der, and Yanni, Y. (2013). Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 8:e56329. doi: 10.1371/journal.pone.0056329
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335. doi: 10.1038/nmeth.f.303
Cardinale, M., Grube, M., Erlacher, A., Quehenberger, J., and Berg, G. (2015). Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ. Microbiol. 17, 239–252. doi: 10.1111/1462-2920.12686
Coleman-Derr, D., Desgarennes, D., Fonseca-Garcia, C., Gross, S., Clingenpeel, S., Woyke, T., et al. (2016). Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 209, 798–811. doi: 10.1111/nph.13697
Compton, K. K., Hildreth, S. B., Helm, R. F., and Scharf, B. E. (2020). An updated perspective on Sinorhizobium meliloti chemotaxis to alfalfa flavonoids. Front. Microbiol. 11:581482. doi: 10.3389/fmicb.2020.581482
Czarnota, M. A., Rimando, A. M., and Weston, L. A. (2003). Evaluation of root exudates of seven sorghum accessions. J. Chem. Ecol. 29, 2073–2083. doi: 10.1023/A:1025634402071
Dakora, F. D., Chimphango, S. B. M., Valentine, A. J., Elmerich, C., and Newton, W. E. (eds.). (2008). Biological Nitrogen Fixation: Towards Poverty Alleviation Through Sustainable Agriculture. Dordrecht: Springer.
Dakora, F. D., Joseph, C. M., and Phillips, D. A. (1993a). Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101, 819–824. doi: 10.1104/pp.101.3.819
Dakora, F. D., Joseph, C. M., and Phillips, D. A. (1993b). Common bean root exudates contain elevated levels of daidzein and coumestrol in response to Rhizobium inoculation. Mol. Plant Microbe Interact. 6, 665–668. doi: 10.1094/MPMI-6-665
Dakora, F. D., and Phillips, D. A. (1996). Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol. Mol. Plant Pathol. 49, 1–20. doi: 10.1006/pmpp.1996.0035
Dakora, F. D., and Phillips, D. A. (2002). Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245, 35–47. doi: 10.1023/A:1020809400075
Dang, K., Gong, X., Zhao, G., Wang, H., Ivanistau, A., and Feng, B. (2020). Intercropping alters the soil microbial diversity and community to facilitate nitrogen assimilation: a potential mechanism for increasing proso millet grain yield. Front. Microbiol. 11:2975. doi: 10.3389/fmicb.2020.601054
Deng, S. P., and Tabatabai, M. A. (1996). Effect of tillage and residue management on enzyme activities in soils. Biol. Fertil. Soils 22, 202–207. doi: 10.1007/BF00382513
Dick, R. P., Rasmussen, P. E., and Kerle, E. A. (1988). Influence of long-term residue management on soil enzyme activities in relation to soil chemical properties of a wheat-fallow system. Biol. Fertil. Soils 6, 159–164. doi: 10.1007/BF00257667
Dick, R. P., Sandor, J. A., and Eash, N. S. (1994). Soil enzyme activities after 1500 years of terrace agriculture in the Colca Valley, Peru. Agric. Ecosyst. Environ. 50, 123–131. doi: 10.1016/0167-8809(94)90131-7
Dick, W. A., Cheng, L., and Wang, P. (2000). Soil acid and alkaline phosphatase activity as pH adjustment indicators. Soil Biol. Biochem. 32, 1915–1919. doi: 10.1016/S0038-0717(00)00166-8
Dick, W. A., and Tabatabai, M. A. (1993). Significance and Potential Uses of Soil Enzymes:Soil Microbial Ecology: Application in Agricultural and Environmental Management. New York, NY: Marcel Dekker, 95–125.
Du Plessis, R. D. T., and Burger, R. F. (1965). A comparison of chemical extraction methods for the evaluation of phosphate availability of top soils. South African J. Agric. Sci. 8, 1113–1122.
Dyer, B. (1894). XV—On the analytical determination of probably available “mineral” plant food in soils. J. Chem. Soc. Trans. 65, 115–167. doi: 10.1039/CT8946500115
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
El Dessougi, H., zu Dreele, A., and Claassen, N. (2003). Growth and phosphorus uptake of maize cultivated alone, in mixed culture with other crops or after incorporation of their residues. J. Plant Nutr. Soil Sci. 166, 254–261. doi: 10.1002/jpln.200390037
Figuerola, E. L. M., Guerrero, L. D., Türkowsky, D., Wall, L. G., and Erijman, L. (2015). Crop monoculture rather than agriculture reduces the spatial turnover of soil bacterial communities at a regional scale. Environ. Microbiol. 17, 678–688. doi: 10.1111/1462-2920.12497
Flynn, B., Graham, A., Scott, N., Layzell, D. B., and Dong, Z. (2014). Nitrogen fixation, hydrogen production, and N2O emissions. Can. J. Plant Sci. 94, 1037–1041. doi: 10.4141/cjps2013-210
Folman, L., De Klein, M. J. E., Postma, J., and van Veen, J. (2004). Production of antifungal compounds by Lysobacter enzymogenes isolate 3.1T8 under different conditions in relation to its efficacy as a biocontrol agent of Pythium aphanidermatum in cucumber. Biol. Control 31, 145–154. doi: 10.1016/j.biocontrol.2004.03.008
Folman, L. B., Postma, J., and van Veen, J. A. (2003). Characterisation of Lysobacter enzymogenes (Christensen and Cook 1978) strain 3.1T8, a powerful antagonist of fungal diseases of cucumber. Microbiol. Res. 158, 107–115. doi: 10.1078/0944-5013-00185
Fustec, J., Lesuffleur, F., Mahieu, S., and Cliquet, J.-B. (2010). Nitrogen rhizodeposition of legumes. A review. Agron. Sustain. Dev. 30, 57–66. doi: 10.1051/agro/2009003
Gardner, W. K., and Boundy, K. A. (1983). The acquisition of phosphorus by Lupinus albus L. IV. The effect of interplanting wheat and white lupin on the growth and mineral composition of the two species. Plant Soil 70, 391–402. doi: 10.1007/BF02374894
Gökçen, A., Vilcinskas, A., and Wiesner, J. (2014). Biofilm-degrading enzymes from Lysobacter gummosus. Virulence 5, 378–387. doi: 10.4161/viru.27919
Gómez Expósito, R., Postma, J., Raaijmakers, J. M., and De Bruijn, I. (2015). Diversity and activity of lysobacter species from disease suppressive soils. Front. Microbiol. 6:1243. doi: 10.3389/fmicb.2015.01243
Graber, E. R., Meller Harel, Y., Kolton, M., Cytryn, E., Silber, A., Rav David, D., et al. (2010). Biochar impact on development and productivity of pepper and tomato grown in fertigated soilless media. Plant Soil 337, 481–496. doi: 10.1007/s11104-010-0544-6
Gunes, A., Bagci, E. G., and Inal, A. (2007). Interspecific facilitative root interactions and rhizosphere effects on phosphorus and iron nutrition between mixed grown chickpea and barley. J. Plant Nutr. 30, 1455–1469. doi: 10.1080/01904160701555648
Gupta, V. V. S. R., and Germida, J. J. (1988). Distribution of microbial biomass and its activity in different soil aggregate size classes as affected by cultivation. Soil Biol. Biochem. 20, 777–786. doi: 10.1016/0038-0717(88)90082-X
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Haldar, S., Roy Choudhury, S., and Sengupta, S. (2011). Genetic and functional diversities of bacterial communities in the rhizosphere of Arachis hypogaea. Antonie Van Leeuwenhoek 100, 161–170. doi: 10.1007/s10482-011-9570-5
Hamel, C., and Smith, D. L. (1991). Plant development in a mycorrhizal field-grown mixture. Soil Biol. Biochem. 23, 661–665. doi: 10.1016/0038-0717(91)90080-4
Inal, A., Gunes, A., Zhang, F., and Cakmak, I. (2007). Peanut/maize intercropping induced changes in rhizosphere and nutrient concentrations in shoots. Plant Physiol. Biochem. 45, 350–356. doi: 10.1016/j.plaphy.2007.03.016
Inceoglu, Ö., Al-Soud, W. A., Salles, J. F., Semenov, A. V., and van Elsas, J. D. (2011). Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE 6:e23321. doi: 10.1371/journal.pone.0023321
Islam, M. T., Hashidoko, Y., Deora, A., Ito, T., and Tahara, S. (2005). Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne Peronosporomycetes. Appl. Environ. Microbiol. 71, 3786–96. doi: 10.1128/AEM.71.7.3786-3796.2005
Israel, D. W. (1987). Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol. 84, 835–840. doi: 10.1104/pp.84.3.835
Izaguirre-Mayoral, M., Flores, S., and Carballo, O. (2002). Determination of acid phosphatase and dehydrogenase activities in the rhizosphere of nodulated legume species native to two contrasting savanna sites in Venezuela. Biol. Fertil. Soils 35, 470–472. doi: 10.1007/s00374-002-0477-1
Jaiswal, S. K., Mohammed, M., and Dakora, F. D. (2019). Microbial community structure in the rhizosphere of the orphan legume Kersting's groundnut [Macrotyloma geocarpum (Harms) Marechal andamp; Baudet]. Mol. Biol. Rep. 46, 4471–4481. doi: 10.1007/s11033-019-04902-8
Ji, G.-H., Wei, L.-F., He, Y.-Q., Wu, Y.-P., and Bai, X.-H. (2008). Biological control of rice bacterial blight by Lysobacter antibioticus strain 13-1. Biol. Control 45, 288–296. doi: 10.1016/j.biocontrol.2008.01.004
Johansen, J. E., Nielsen, P., and Binnerup, S. J. (2009). Identification and potential enzyme capacity of flavobacteria isolated from the rhizosphere of barley (Hordeum vulgare L.). Can. J. Microbiol. 55, 234–241. doi: 10.1139/W08-116
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks808
Ko, H.-S., Jin, R.-D., Krishnan, H. B., Lee, S.-B., and Kim, K.-Y. (2009). Biocontrol ability of lysobacter antibioticus HS124 against phytophthora blight is mediated by the production of 4-hydroxyphenylacetic acid and several lytic enzymes. Curr. Microbiol. 59, 608–615. doi: 10.1007/s00284-009-9481-0
Kolton, M., Frenkel, O., Elad, Y., and Cytryn, E. (2014). Potential role of flavobacterial gliding-motility and type IX secretion system complex in root colonization and plant defense. Mol. Plant Microbe Interact. 27, 1005–1013. doi: 10.1094/MPMI-03-14-0067-R
Li, H., Shen, J., Zhang, F., Marschner, P., Cawthray, G., and Rengel, Z. (2010). Phosphorus uptake and rhizosphere properties of intercropped and monocropped maize, faba bean, and white lupin in acidic soil. Biol. Fertil. Soils 46, 79–91. doi: 10.1007/s00374-009-0411-x
Li, L., Li, S.-M., Sun, J.-H., Zhou, L.-L., Bao, X.-G., Zhang, H.-G., et al. (2007). Diversity enhances agricultural productivity via rhizosphere phosphorus facilitation on phosphorus-deficient soils. Proc. Natl. Acad. Sci. U.S.A. 104, 11192–11196. doi: 10.1073/pnas.0704591104
Li, N., Gao, D., Zhou, X., Chen, S., Li, C., and Wu, F. (2020). Intercropping with potato-onion enhanced the soil microbial diversity of tomato. Microorganisms 8:834. doi: 10.3390/microorganisms8060834
Li, S., and Wu, F. (2018). Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front. Microbiol. 9:1521. doi: 10.3389/fmicb.2018.01521
Li, S. M., Li, L., Zhang, F. S., and Tang, C. (2004). Acid phosphatase role in chickpea/maize intercropping. Ann. Bot. 94, 297–303. doi: 10.1093/aob/mch140
Li, X., Mu, Y., Cheng, Y., Liu, X., and Nian, H. (2013). Effects of intercropping sugarcane and soybean on growth, rhizosphere soil microbes, nitrogen and phosphorus availability. Acta Physiol. Plant. 35, 1113–1119. doi: 10.1007/s11738-012-1148-y
Lu, Y., Rosencrantz, D., Liesack, W., and Conrad, R. (2006). Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ. Microbiol. 8, 1351–1360. doi: 10.1111/j.1462-2920.2006.01028.x
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Lynch, J. M., and Whipps, J. M. (1990). Substrate flow in the rhizosphere. Plant Soil 129, 1–10. doi: 10.1007/BF00011685
Madhaiyan, M., Poonguzhali, S., Lee, J.-S., Senthilkumar, M., Lee, K. C., and Sundaram, S. (2010). Mucilaginibacter gossypii sp. nov. and Mucilaginibacter gossypiicola sp. nov., plant-growth-promoting bacteria isolated from cotton rhizosphere soils. Int. J. Syst. Evol. Microbiol. 60, 2451–2457. doi: 10.1099/ijs.0.018713-0
Maimaiti, J., Zhang, Y., Yang, J., Cen, Y.-P., Layzell, D. B., Peoples, M., et al. (2007). Isolation and characterization of hydrogen-oxidizing bacteria induced following exposure of soil to hydrogen gas and their impact on plant growth. Environ. Microbiol. 9, 435–444. doi: 10.1111/j.1462-2920.2006.01155.x
Makoi, J. H. J. R., Chimphango, S. B. M., and Dakora, F. D. (2010). Elevated levels of acid and alkaline phosphatase activity in roots and rhizosphere of cowpea (Vigna unguiculata L. Walp.) genotypes grown in mixed culture and at different densities with sorghum (Sorghum bicolor L.). Crop Pasture Sci. 61:279. doi: 10.1071/CP09212
Manter, D. K., Delgado, J. A., Holm, D. G., and Stong, R. A. (2010). Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb. Ecol. 60, 157–166. doi: 10.1007/s00248-010-9658-x
Marschner, P., Yang, C.-H., Lieberei, R., and Crowley, D. (2001). Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 33, 1437–1445. doi: 10.1016/S0038-0717(01)00052-9
Maseko, S. T., and Dakora, F. D. (2013). Rhizosphere acid and alkaline phosphatase activity as a marker of P nutrition in nodulated Cyclopia and Aspalathus species in the Cape fynbos of South Africa. South African J. Bot. 89, 289–295. doi: 10.1016/j.sajb.2013.06.023
Matsuoka, H., Akiyama, M., Kobayashi, K., and Yamaji, K. (2013). Fe and P solubilization under limiting conditions by bacteria isolated from carex kobomugi roots at the hasaki coast. Curr. Microbiol. 66, 314–321. doi: 10.1007/s00284-012-0276-3
Matusso, J. M. M., and Mucheru-Muna. (2014). Potential role of cereal-legume intercropping systems in integrated soil fertility management in smallholder farming systems of Sub-Saharan Africa. Res. J. Agric. Environ. Manag. 3, 162–174. Available online at: https://www.scopus.com/record/display.uri?eid=2-s2.0-85013826983&origin=inward&txGid=a78dd0a5a92a9a0972582cfe425f111d
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Micallef, S. A., Shiaris, M. P., and Colón-Carmona, A. (2009). Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 60, 1729–1742. doi: 10.1093/jxb/erp053
Moore, J. M., Klose, S., and Tabatabai, M. A. (2000). Soil microbial biomass carbon and nitrogen as affected by cropping systems. Biol. Fertil. Soils 31, 200–210. doi: 10.1007/s003740050646
O'Hara, G. W. (2001). Nutritional constraints on root nodule bacteria affecting symbiotic nitrogen fixation: a review. Aust. J. Exp. Agric. 41, 417–433. doi: 10.1071/EA00087
Palumbo, J. D., Yuen, G. Y., Jochum, C. C., Tatum, K., and Kobayashi, D. Y. (2005). Mutagenesis of β-1,3-Glucanase genes in Lysobacter enzymogenes Strain C3 results in reduced biological control activity toward bipolaris leaf spot of tall fescue and pythium damping-off of sugar beet. Phytopathology 95, 701–707. doi: 10.1094/PHYTO-95-0701
Pankratov, T. A., Tindall, B. J., Liesack, W., and Dedysh, S. N. (2007). Mucilaginibacter paludis gen. nov., sp. nov. and Mucilaginibacter gracilis sp. nov., pectin-, xylan- and laminarin-degrading members of the family Sphingobacteriaceae from acidic Sphagnum peat bog. Int. J. Syst. Evol. Microbiol. 57, 2349–2354. doi: 10.1099/ijs.0.65100-0
Papastylianou, I. (2004). Effect of rotation system and N fertilizer on barley and vetch grown in various crop combinations and cycle lengths. J. Agric. Sci. 142, 41–48. doi: 10.1017/S0021859604004009
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Peterson, S. B., Dunn, A. K., Klimowicz, A. K., and Handelsman, J. (2006). Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl. Environ. Microbiol. 72, 5421–5427. doi: 10.1128/AEM.02928-05
Pirhadi, M., Enayatizamir, N., Motamedi, H., and Sorkheh, K. (2016). Screening of salt tolerant sugarcane endophytic bacteria with potassium and zinc for their solubilizing and antifungal activity. Agric. Commun. Biosci. Biotech. Res. Comm. 9, 530–538. doi: 10.21786/bbrc/9.3/28
Postma, J., Stevens, L. H., Wiegers, G. L., Davelaar, E., and Nijhuis, E. H. (2009). Biological control of Pythium aphanidermatum in cucumber with a combined application of Lysobacter enzymogenes strain 3.1T8 and chitosan. Biol. Control 48, 301–309. doi: 10.1016/j.biocontrol.2008.11.006
Puopolo, G., Giovannini, O., and Pertot, I. (2014). Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiol. Res. 169, 633–642. doi: 10.1016/j.micres.2013.09.013
Qin, Y., Fu, Y., Dong, C., Jia, N., and Liu, H. (2016). Shifts of microbial communities of wheat (Triticum aestivum L.) cultivation in a closed artificial ecosystem. Appl. Microbiol. Biotechnol. 100, 4085–4095. doi: 10.1007/s00253-016-7317-y
Reichenbach, H. (2006). “The genus lysobacter,” in The Prokaryotes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 939–957.
Rodríguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Sang, M. K., Chun, S.-C., and Kim, K. D. (2008). Biological control of Phytophthora blight of pepper by antagonistic rhizobacteria selected from a sequential screening procedure. Biol. Control 46, 424–433. doi: 10.1016/j.biocontrol.2008.03.017
Shen, J., Yuan, L., Zhang, J., Li, H., Bai, Z., Chen, X., et al. (2011). Phosphorus dynamics: from soil to plant. Plant Physiol. 156, 997–1005. doi: 10.1104/pp.111.175232
Shi, S., Nuccio, E., Herman, D. J., Rijkers, R., Estera, K., Li, J., et al. (2015). Successional trajectories of rhizosphere bacterial communities over consecutive seasons. MBIO 6:e00746. doi: 10.1128/mBio.00746-15
Stepnaya, O. A., Tsfasman, I. M., Chaika, I. A., Muranova, T. A., and Kulaev, I. S. (2008). Extracellular yeast-lytic enzyme of the bacterium Lysobacter sp. XL 1. Biochem 73, 310–314. doi: 10.1134/S0006297908030115
Steyn, P. L., Segers, P., Vancanneyt, M., Sandra, P., Kersters, K., and Joubert, J. J. (1998). Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobac. Int. J. Syst. Evol. Microbiol. 48, 165–177. doi: 10.1099/00207713-48-1-165
Sugiyama, A., Ueda, Y., Zushi, T., Takase, H., and Yazaki, K. (2014). Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE 9:e100709. doi: 10.1371/journal.pone.0100709
Sunithakumari, K., Devi, S. N. P., and Vasandha, S. (2016). Zinc solubilizing bacterial isolates from the agricultural fields of Coimbatore, Tamil Nadu, India. Curr. Sci. 110, 196–205. doi: 10.18520/cs/v110/i2/196-205
Tabatabai, M. A. (1994). “Soil enzymes,” in Methods of Soil Analysis, Part 2: Microbiological and Biochemical Properties, 775–833.
Tsamo, A. T., Mohammed, H., Mohammed, M., Papoh Ndibewu, P., and Dapare Dakora, F. (2020). Seed coat metabolite profiling of cowpea (Vigna unguiculata L. Walp.) accessions from Ghana using UPLC-PDA-QTOF-MS and chemometrics. Nat. Prod. Res. 34, 1158–1162. doi: 10.1080/14786419.2018.1548463
Tsamo, A. T., Ndibewu, P. P., and Dakora, F. D. (2018). Phytochemical profile of seeds from 21 Bambara groundnut landraces via UPLC-qTOF-MS. Food Res. Int. 112, 160–168. doi: 10.1016/j.foodres.2018.06.028
Turner, T. R., Ramakrishnan, K., Walshaw, J., Heavens, D., Alston, M., Swarbreck, D., et al. (2013). Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7:2248. doi: 10.1038/ismej.2013.119
Urich, T., Lanzén, A., Qi, J., Huson, D. H., Schleper, C., and Schuster, S. C. (2008). Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE 3:e2527. doi: 10.1371/journal.pone.0002527
Vasilyeva, N. V., Shishkova, N. A., Marinin, L. I., Ledova, L. A., Tsfasman, I. M., Muranova, T. A., et al. (2014). Lytic peptidase L5 of Lysobacter sp. XL1 with broad antimicrobial spectrum. J. Mol. Microbiol. Biotechnol. 24, 59–66. doi: 10.1159/000356838
Wahua, T. A. T. (1984). Rhizosphere bacterial counts for intercropped maize (Zea mays L.), cowpea (Vigna unguiculata L.) and ‘egusi’ melon (Colosynthis vulgaris L.). Field Crop. Res. 8, 371–379. doi: 10.1016/0378-4290(84)90084-4
Walker, T. S., Bais, H. P., Grotewold, E., and Vivanco, J. M. (2003). Root exudation and rhizosphere biology. Plant Physiol. 132, 44–51. doi: 10.1104/pp.102.019661
Wang, Z.-G., Jin, X., Bao, X.-G., Li, X.-F., Zhao, J.-H., Sun, J.-H., et al. (2014). Intercropping enhances productivity and maintains the most soil fertility properties relative to sole cropping. PLoS ONE 9:e113984. doi: 10.1371/journal.pone.0113984
Warembourg, F. R., Roumet, C., and Lafont, F. (2003). Differences in rhizosphere carbon-partitioning among plant species of different families. Plant Soil 256, 347–357. doi: 10.1023/A:1026147622800
Xiao, Y., Li, L., and Zhang, F. (2004). Effect of root contact on interspecific competition and N transfer between wheat and fababean using direct and indirect 15N techniques. Plant Soil 262, 45–54. doi: 10.1023/B:PLSO.0000037019.34719.0d
Xue, Y., Xia, H., Christie, P., Zhang, Z., Li, L., and Tang, C. (2016). Crop acquisition of phosphorus, iron, and zinc from soil in cereal/legume intercropping systems: a critical review. Ann. Bot. 117, 363–377. doi: 10.1093/aob/mcv182
Yang, A., Liu, N., Tian, Q., Bai, W., Williams, M., Wang, Q., et al. (2015). Rhizosphere bacterial communities of dominant steppe plants shift in response to a gradient of simulated nitrogen deposition. Front. Microbiol. 6:789. doi: 10.3389/fmicb.2015.00789
Yang, Z., Yang, W., Li, S., Hao, J., Su, Z., and Sun, M. (2016). Variation of bacterial community diversity in rhizosphere soil of sole-cropped versus intercropped wheat field after harvest. PLoS ONE 11:e0150618. doi: 10.1371/journal.pone.0150618
Zarraonaindia, I., Owens, S. M., Weisenhorn, P., West, K., Hampton-Marcell, J., Lax, S., et al. (2015). The soil microbiome influences grapevine-associated microbiota. MBio 6, e02527–e02514. doi: 10.1128/mBio.02527-14
Keywords: intra-hole and sole cropping, acid phosphatase activity, microbial community, 16S rRNA, Miseq Illumina sequencing
Citation: Jaiswal SK, Maredi MP and Dakora FD (2021) Rhizosphere P-Enzyme Activity, Mineral Nutrient Concentrations, and Microbial Community Structure Are Altered by Intra-Hole Cropping of Cowpea With Cereals. Front. Agron. 3:666351. doi: 10.3389/fagro.2021.666351
Received: 10 February 2021; Accepted: 04 June 2021;
Published: 02 July 2021.
Edited by:
Samuel Adjei-Nsiah, University of Ghana, GhanaReviewed by:
Bharati Kollah, Indian Institute of Soil Science (ICAR), IndiaCopyright © 2021 Jaiswal, Maredi and Dakora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Felix D. Dakora, ZGFrb3JhZmRAdHV0LmFjLnph
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.