- Departamento de Biología Molecular de Plantas, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Cuernavaca, Mexico
Non-specific lipid transfer proteins (LTPs) constitute a large protein family in plants characterized by having a tunnel-like hydrophobic cavity, which allows them to transfer different lipid molecules. LTPs have been studied in various model plants including those of agronomic interest. Recent studies have demonstrated that LTPs play key functions in both biotic and abiotic stress. In plants-pathogen interaction, they act as either positive or negative regulators of defense responses. However, little is known about the roles of LTPs in symbiotic interactions, especially in root nodule symbiosis. Here, we performed a broad genome analysis of LTP family members in legumes and other important model plants, focusing on their possible roles in legume-rhizobium symbiosis. In silico analysis showed that legumes contain large LTP families, with at least 70 LTP members clustered into four clades. Although the structures of LTP genes and proteins are conserved among species, differences were observed between clades from different species. LTPs are widely expressed in different plant tissues. In general, genes of the LTP1 and LTP2 classes are highly expressed in shoot and reproductive tissues in all analyzed species. Furthermore, genes of the different classes are also expressed in roots inoculated with rhizobia and nodules of legumes. RT-qPCR expression profile analysis of seven PvLTP genes in common bean (Phaseolus vulgaris) revealed that these genes are differentially expressed during the early and late stages of nodulation and they are genetically regulated by PvRbohA. These findings provide insight into the putative roles of LTP family members in legume-rhizobium symbiosis and their possible interactions with RBOH-dependent ROS production.
Introduction
Lipid transfer proteins (LTPs) constitute a large family of plant proteins that bind to different lipids in a non-specific manner. As LTPs can transfer a wide range of lipid molecules, such as phospholipids, galactolipids, and free fatty acids, they are also known as non-specific LTPs (nsLTPs) (Wei and Zhong, 2014; Edqvist et al., 2018). LTPs have a low molecular weight and contain four or five α-helices connected by four disulfide bonds. These boundaries are formed by the conserved eight-cysteine motif (8CM) with the general structure C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C, where X represents any amino acid (Shin et al., 1995; Boutrot et al., 2008; Lascombe et al., 2008). LTPs are also characterized by the presence of a hydrophobic cavity that allows them to bind to lipid molecules (Lascombe et al., 2008).
LTP family members have been identified in various angiosperms, including rice (Oryza sativa L.), Arabidopsis thaliana (L.) Heynh, wheat (Triticum aestivum L.) (Boutrot et al., 2008), barley (Hordeum vulgare L.) (Zhang et al., 2019), maize (Zea mays L.) (Wei and Zhong, 2014), and potato (Solanum tuberosum L.) (Li et al., 2019). Also in non-vascular plants, such as Marchantia polymorpha L., Physcomitrium (Physcomitrella) patens (Hedw.) Bruch and Schimp., Selaginella moellendorffii Hieron, the fern Adiantum capillus-veneris L., and the conifer inus taeda L. (Edstam et al., 2011).
Plant LTPs are classified into 2 subgroups based on their structural organization: LTP1s, with a molecular weight of 9–10 kDa and ~90–95 amino acids; and LTP2s, with a molecular weight of 6–7 kDa and 65–70 amino acids (Finkina et al., 2016). LTPs have also been characterized based on the presence and positions of introns, the presence of a glycosylphosphatidylinositol (GPI) modification site, the Cys spacing in the 8CM, and the similarity of their amino acid sequences. Based on these features, LTPs are grouped into 10 subtypes: LTP1, LTP2, LTPc, LTPd, LTPe, LTPf, LTPg, LTPh, LTPj, and LTPk (Edstam et al., 2011).
LTPs participate in lipid barriers synthesis, facilitating the transportation of lipid polymers in the apoplastic space, as well as in the loosening of the cell wall (Edqvist et al., 2018). Under in vitro conditions, AsE246, an LTP from Chinese milk vetch (Astralagus sinicus L.), was shown to bind to different fatty acids, mainly those with chains 16–18 carbons in length. Under the same conditions, AsE246 also binds to phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), monogalactosyldiacylglycerol, (MGDG), and to digalactosyldiacylglycerol (DGDG) (Lei et al., 2014).
Genetic approaches have reveled LTPs functions in plant responses to pathogens. For instance, the Arabidopsis LTP3 gene transcript accumulated in response to the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 and its overexpression enhanced the susceptibility to this pathogen (Gao et al., 2016). Transcript accumulation of the LTP4 from Nicotiana tabacum L. (NtLTP4) also increased in response to wounding and infection with Ralstonia solanacearum (Xu et al., 2018). Moreover, salicylic acid (SA), a hormone that induces the plant systemic acquired resistance (SAR) of plants against biotrophic bacterial pathogens (Zhao, 2015), enhanced NtLTP4 transcription (Xu et al., 2018). Xu et al. (2018) also found that jasmonic acid (JA), a hormonal regulator of defense responses to wounding and pathogen attack (Halim et al., 2006), increased NtLTP4 transcription.
The Arabidopsis nsLPT-related gene DRN1 (disease related nonspecific lipid transfer protein) is suppressed in response to Pst DC3000 and Pseudomonas syringae pv. maculicola ES4326 (Psm ES4326). Interestingly, when plants were challenged with bacteria with a defective type three secretion system (TTSS), DRN1 was not suppressed. Additionally, drn1 mutants showed less resistance to both bacterial (Pst DC3000 and Psm ES4326) and fungal pathogens (Botrytis cinerea). Since DRN1 expression was also abrogated in SA, JA, and ethylene (ET)-mediated resistance, it is probably regulated by a pathway independent of these hormones. In this context, it was shown that DRN1 expression is apparently regulated by superoxide radicals (Dhar et al., 2020).
In root nodule symbiosis, LTP functions have also been reported. For instance, AsE246 was shown to bind DGDG from Chinese milk vetch nodule extract; interestingly, AsE246 knockdown reduced lipid abundance (PC, PE, PI, and DGDG) in the nodules. AsE246 knockdown also reduced number of nodules and infection threads (IT) and impaired the development of symbiosomes. Conversely, AsE246 overexpression increased the number of nodules and IT (Lei et al., 2014). Recently, AsE246 was observed to interact with the high temperature protein G (HtpG) of Mesorhizobium huakuii in infected nodule cells of A. sinicus and in symbiosome membranes (Zhou et al., 2019). Furthermore, downregulation of the LTP nodulin 5 from Medicago truncatula Gaertn. (MtN5) caused an increase in the curling of the root hair, but a reduction of invaded nodule primordia. On the contrary, MtN5 overexpression favored both root hair curling and invaded nodule primordia, also, the overexpression of MtN5 increased the content of lipids, MGDG, DGDG, PE and glucosyl ceramides, in infected roots (Santi et al., 2017).
Since LTPs bind to and transfer a variety of lipids, they are thought to play important roles in membrane remodeling. Both membrane remodeling and the formation of new membranes are key processes for the development of nodules during legume-rhizobium symbiosis. During this symbiotic interaction, rhizobium (soil bacteria capable of fixing atmospheric nitrogen) are internalized into nodule cells via a mechanism similar to endocytosis that encapsulates the bacteria within the plant membrane (Oldroyd and Downie, 2008; Roy et al., 2020). Inside the nodules, bacteria differentiate into bacteroids, which are surrounded by a membrane of plant origin. In these compartments, known as symbiosomes, bacteroids fix atmospheric nitrogen, and the plant-derived membrane acts as a symbiotic interface, allowing nutrient exchange (Oldroyd and Downie, 2008; Bapaume and Reinhardt, 2012).
Some legume species are widely exploited as crops, serving as a source of nutrients for human and animal consumption. The ability of legumes to develop symbiosis with nitrogen-fixing bacteria makes them suitable crops for the development of sustainable agriculture. However, little is known about the roles of LTPs in legume-rhizobium symbiosis. Therefore, in the current study, we examined the possible roles of these proteins in this process. We identified LTP family members in several model plant species, including seven species of legumes, using the criteria proposed by Edstam et al. (2011). To expand our knowledge of the evolutionary relationships of the LTPs, we constructed a phylogenetic tree based on the amino acid sequences of LTPs from legumes, cereals, and non-seed plants. Analysis of the gene structures of LTPs and their expression profiles in different organs of legumes and other species was performed based on several databases. Several LTPs are expressed in the roots and nodules of legumes. RT-qPCR assays showed that seven LTPs genes of common bean (Phaseolus vulgaris L.) were differential expressed in roots at different stages of nodule development and that some of these genes were regulated by downregulation of the NADPH oxidase PvRbohA. Therefore, we hypothesize that LTPs could regulate legume-rhizobium symbiosis under the regulation of ROS production, dependent on RBOHs.
Materials and Methods
Identification of LTP Family Members
To identify LTP genes in various plants (15 species, Supplementary Table 1), several databases were searched. LTPs from 12 species were identified in the Phytozome 12.1.6 database (https://phytozome.jgi.doe.gov) (Goodstein et al., 2012). LTPs from Lotus japonicus L. were identified by searching LotusBase (https://lotus.au.dk) (Mun et al., 2016). The LIS: Legume Information System database (https://legumeinfo.org) (Dash et al., 2016) was used to identify LTPs in the genomes of Pisum sativum L. and Lupinus albus. A. thaliana LTP amino acid sequences were used as queries in all BLASTP searches.
Gene/Protein Sequence Analysis, Chromosomal Localization, and Synteny Analysis of LTPs in Legume Species and A. thaliana
Exon-intron gene structure information was obtained from the corresponding databases (Supplementary Table 2). Gene structure mapping was carried out using Gene Structure Display Server 2.0 (http://gsds.gao-lab.org/Gsds_about.php). The conserved motifs and domains in the full-length amino acid sequences of all LTPs of legume species and A. thaliana were identified using the Multiple Expression motifs for Motif Elicitation version 5.2.0 tool (MEME) (Bailey et al., 2009). Chromosome localization was carried out with PhenoGram Plot (http://visualization.ritchielab.org/phenograms/plot) (Wolfe et al., 2013).
Synteny analysis was performed using the amino acid sequences of LTPs of L. japonicus, Glycine max L., P. vulgaris, M. truncatula, and A. thaliana. In all cases, a BLASTP file in m8 format and a simplified gff file were used as inputs for the MCScanx Collinearity Scanner Toolkit (http://chibba.pgml.uga.edu/mcscan2/) (Wang et al., 2012).
In silico Analysis of LTP Expression in P. vulgaris, G. max, M. truncatula, L. japonicus, and A. thaliana
The expression profiles of P. vulgaris LTPs were obtained from the Common Bean Gene Expression Atlas, PvGEA (https://plantgrn.noble.org/PvGEA/) (O'Rourke et al., 2014). The LTP expression profiles of G. max, A. thaliana, S. moellendorffii, Sorghum bicolor (L.) Moench, O. sativa, and Z. mays were retrieved from Bio-Analytic Resource for Plant Biology (BAR) (Schmid et al., 2005; Libault et al., 2010). The expression profiles of M. truncatula LTPs were downloaded from the M. truncatula Gene Expression Atlas (MtGEA) (Benedito et al., 2008; He et al., 2009). The expression profiles of L. japonicus LTPs were retrieved from LotusBASE (https://lotus.au.dk/) (Mun et al., 2016), and the expression profiles of P. patens LTPs were retrieved from PEATmoss (https://peatmoss.online.uni-marburg.de/) (Fernandez-Pozo et al., 2020). Heat maps of the transcript levels of the genes were constructed using the heatmap.2 function of the gplot package (Warnes et al., 2015) with R Project.
Phylogenetic Analysis of LTPs
All protein sequences were aligned with the MUSCLE algorithm and manually edited using MEGA version X (Kumar et al., 2018) to remove misaligned sequences. The phylogenetic tree was constructed with the IQ-TREE algorithm 1.6.12 (Nguyen et al., 2014) using the maximum-likelihood method based on a VT+R9 substitution model with 1,000 bootstraps.
Wild-Type Plant Growth and Generation of Composite Plants
Common been seeds (P. vulgaris cv. Negro Jamapa) were surface sterilized as described by Estrada-Navarrete et al. (2007) and incubated in a germination chamber for 2 days at 28°C in the dark. At 2 days postgermination (dpg), the seedlings were placed in pots with sterile vermiculite and inoculated with Rhizobium tropici (CIAT 899) at an OD600 of 0.05 or they were not inoculated (control plants). All plants were watered three times per week with Fahraeus medium; control plants were treated with KNO3 (10 mM) to avoid rhizobial contamination. At 7, 14, 21, and 30 days post-inoculation (dpi), the roots were harvested, frozen in liquid nitrogen, and stored at −75°C until RNA extraction.
To generate common bean composite plants (wild-type plants with transgenic hairy roots), 2 dpg seedlings were infected with Agrobacterium rhizogenes K599 strains carrying the PvRbohA-RNAi (Arthikala et al., 2017) and PvRbohB-RNAi (Montiel et al., 2012) constructs. As a control, composite plants carrying an inert RNAi vector were used (see details in Montiel et al., 2012); these plants are referred to as pTdT-SAC hereafter. All composite plants were generated as described by Estrada-Navarrete et al. (2007) with some modifications (Supplementary Material: protocol 1). Three to five days after removing the primary root, hairy roots of composite plants were observed under an epifluorescence microscope to confirm the presence of the reporter gene (RFP) and to remove non-transformed roots. Composite plants were potted in sterile vermiculite and inoculated with R. tropici (CIAT 899) as previously described. Hairy roots were harvested at 24 h after inoculation, frozen in liquid nitrogen, and stored at −75°C until RNA extraction.
RNA Extraction and qPCR Assays
Total RNA was extracted from frozen root tissue using TRIzol Reagent (Ambion®, Life Technologies™, USA) following the manufacturer's instructions. RNA integrity and concentration were verified by agarose gel electrophoresis (1%) and spectrometry (NanoDrop 2000/200c, Thermo Scientific, Waltham, MA, USA), respectively. DNA contamination was removed by incubation with RNase-free DNase (10 U/μl, Roche) at 37°C for 30 min. Complementary DNA (cDNA) was synthesized from 200 ng/μl of DNA-free RNA using RevertAid Reverse Transcriptase (200 U/μl, Thermo Scientific, Waltham, MA, USA), according to the manufacturer's instructions. Real-time quantitative PCR (RT-qPCR) assays were performed using Maxima SYBR Green/ROX qPCR Master Mix (2X) (Thermo Scientific, Waltham, MA, USA) on a RT-qPCR system (QuantStudio 5; Applied Biosystems, Waltham, MA, USA) as follows: 95°C for 10 min, 30 cycles of 95°C for 15 s, and 60°C for 60 s. The primer sequences used in the RT-qPCR assays are listed in Supplementary Table 1.
Relative transcript levels were calculated by the 2−ΔΔCT method using the transcript profile of the elongation factor 1α (EF1α) gene for normalization. The samples for the RT-qPCR assays were derived from three independent plants, with three biological replicates and three technical repeats performed per sample.
Statistical Analysis
The non-parametric Mann-Whitney test was performed using the wilcox.test function from the statistics package in R Project.
Results
Phylogenetic Analysis Indicates That LTPs Are an Ancient Protein Family
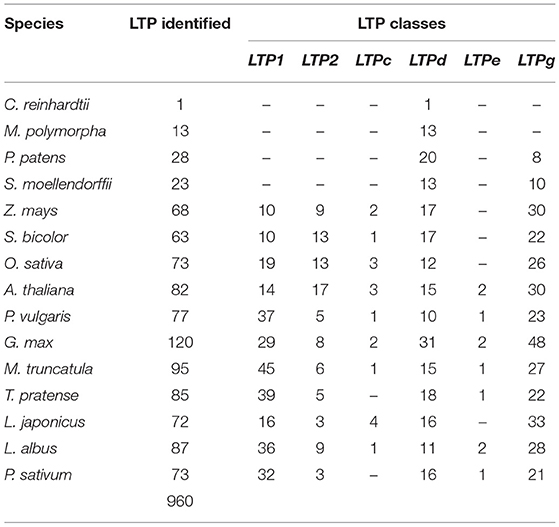
Previous studies aimed at identifying the members of the LTP family have focused on only a few model species, such as A. thaliana and a few cereal crops (Edstam et al., 2011; Salminen et al., 2016). To analyze the potential roles of LTP subfamily members in legume-rhizobium symbiosis, we identified these proteins in the most recent versions of the genomes of 14 plants and a green alga from various databases (Supplementary Table 1). We searched for LTP homologs in each species, followed by domain analysis of homologous proteins to verify the presence of the characteristic lipid transfer protein domain of the LTP family. We identified 960 LTPs in the 15 species analyzed (Table 1, Supplementary Table 3). Remarkably, the Chlamydomonas reinhardtii P. A. Dang. genome had one significant hit related to the LTP family, making it the first reported LTP in a chlorophyte species. In addition, we identified at least 13 LTPs encoded in each terrestrial plant genome, with fewer homologous LTPs present in the analyzed marchantiophyta, bryophyte, and lycophyte species (Supplementary Table 3).
A robust phylogenetic analysis of the amino acid sequences of these 960 LTPs divided them into 4 major clades (Figure 1, Supplementary Figure 1). Following the guidelines proposed by Edstam et al. (2011), the clades were named according to the content of the characteristic 8CM motif C-Xn-C-Xn-CC-Xn-CXC-Xn-C-Xn-C (Figure 1, Supplementary Table 3). One of these clades, consisting exclusively of the LTP1 class, was identified in all angiosperm species analyzed in this study (Figure 1, Supplementary Figure 1, light blue clade). Additionally, another clade consisting exclusively of the LTPg class was identified in all bryophyte and angiosperm species studied (Figure 1, Supplementary Figure 1, red clade). By contrast, two clades contained a mixture of classes, including one with classes LTP2, LTPc, and LTPd (Figure 1, Supplementary Figure 1, green clade) and the other with classes LTPd and LTPe (Figure 1, Supplementary Figure 1, lilac clade). At least one member of the LTPd class was present in all species analyzed. A few previously classified LTPs were reassigned based on a phylogenetic analysis and on the arrangement of 8CM motifs determined using the latest available versions of their genomes. These results suggest that LTPs are an ancient protein family that appeared before the first terrestrial plant; it was represented in the chlorophyte species by the LTPd class.
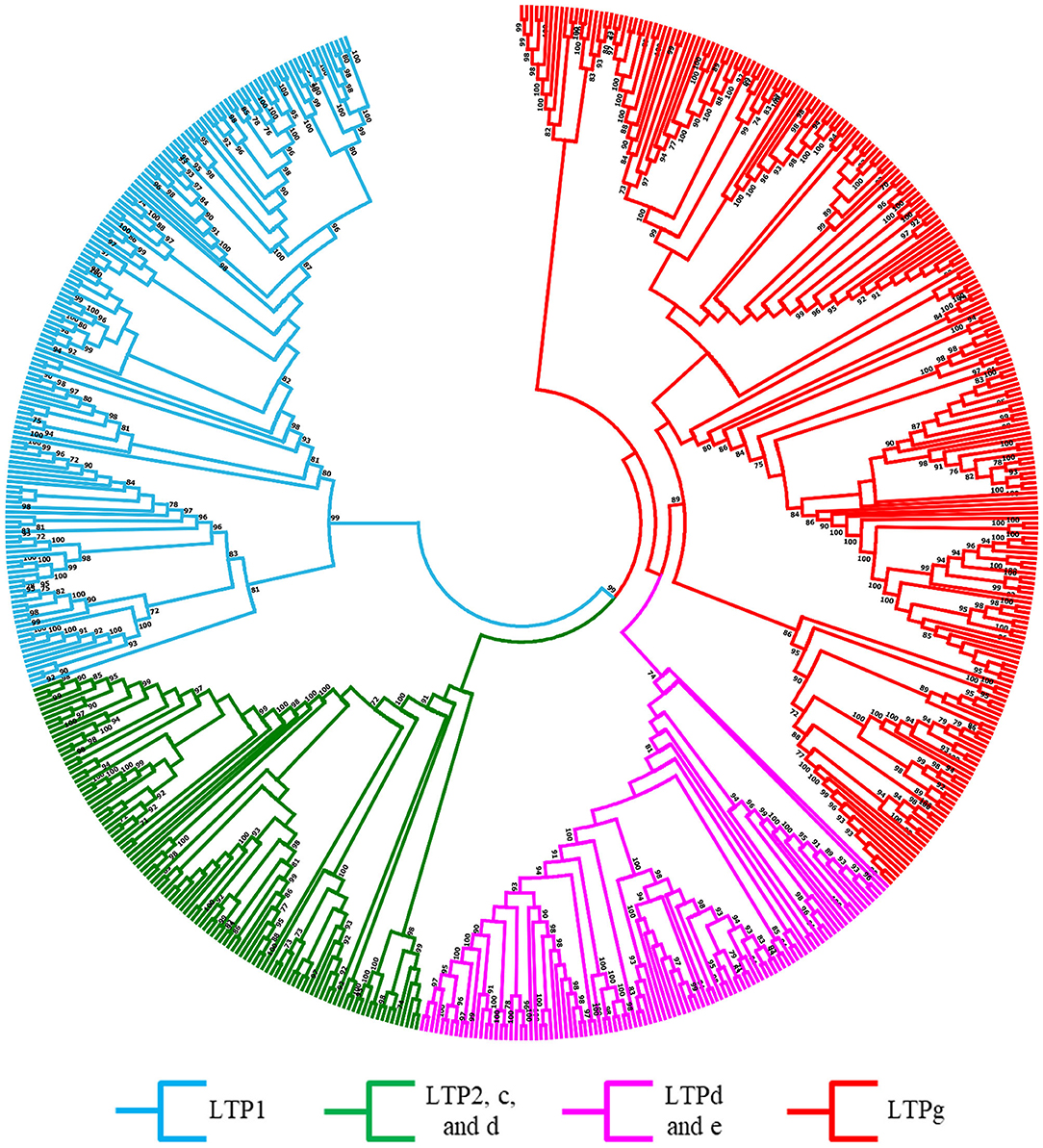
Figure 1. Evolutionary relationships of LTPs. Unrooted approximately maximum-likelihood phylogenetic tree inferred from 960 LTPs identified in 15 plant species: C. reinhardtii, P. patens, M. polymorpha, S. moellendorffii, O. sativa, S. bicolor, Z. mays, A. thaliana, G. max, P. vulgaris, L. japonicus, M. truncatula, L. albus, P. sativum, and T. pratense. The clades are shown in different colors according to the legend at the bottom. The phylogenetic tree was constructed using IQ-TREE software with the VT+R9 substitution model with 1,000 bootstrap iterations.
Features of LTPs Are Variable Among Plant Species
Our phylogenetic analysis showed that LTPs are a large family of conserved proteins in the phylum Plantae. However, some differences were detected in the physicochemical characteristics of LTPs in legumes vs. the other species. The angiosperm species contained more LTPs than the ancient lineages, suggesting that a major duplication event occurred during evolution. Although the total number of LTPs identified in legumes was similar to that in A. thaliana and cereals, the number of members in different classes varied. Our sequence analysis showed that LTPs in the legumes studied had a reduced number of members of the LTP2 class, with an average of 6 members, whereas A. thaliana and the cereal species had an average of 13 members (Table 1). By contrast, the LTP1 class was larger in legumes than in the other species, with an average of 33 members in the legume species studied and only an average of 13 members in the other angiosperm species (Table 1). These differential characteristics in the legumes could be related to their specific functions in this plant family, likely in root nodule symbiosis.
The 960 LTPs have variable amino acid length and molecular weight (MW), ranging from 55 to 793 amino acids and 8.8–20.8 kDa (Supplementary Table 4), respectively. In all species studied, the MW was closely related to the LTP class, with the highest MW in the LTPg class and the lowest in the LTPc class (Supplementary Figure 2A). Similarly, the isoelectric point (IP) showed some differences between LTP classes, ranging from 3.7 to 11.1 (Supplementary Table 4, Supplementary Figure 1B). The LTPc class showed more acidic IP values, while the remaining LTP classes tended to show basic IP values. As expected, the LTPd identified in the alga C. reinhardtii showed a slightly basic IP of 7.56, confirming this categorization of IP values by LTP class and the trend toward a basic IP for the LTPd class.
LTPs are characterized by the presence of a lipid transfer domain and an 8CM arrangement along the sequence (Figure 2B). To characterize these conserved motifs and examine their relationship to legume nodule symbiosis, we analyzed the LTP sequences of the legumes and A. thaliana using MEME software (Figure 2). In the 691 sequences examined, 15 different motifs with diverse cysteine arrangements were identified throughout the sequences (Figures 2A,C). The motifs were mostly class specific for all species analyzed, showing clear differential patterns between the LTP1 and LTP2 classes and the remaining classes. Classes LTP1 and LTPg showed five specific motifs, while classes LTPc and LTPd shared two or three motifs in some genes. The motif located in the LTP2 class (motif number two) was the longest and one of the most conserved motifs. Surprisingly, we identified a specific motif in the LTP1 class in legumes that form indeterminate nodules, suggesting it might play a specific role in this type of symbiotic nodule (Figures 2A,C). Our findings indicate that the physicochemical characteristics of LTPs in plants vary between classes, but also between species, suggesting specific functions for each species.
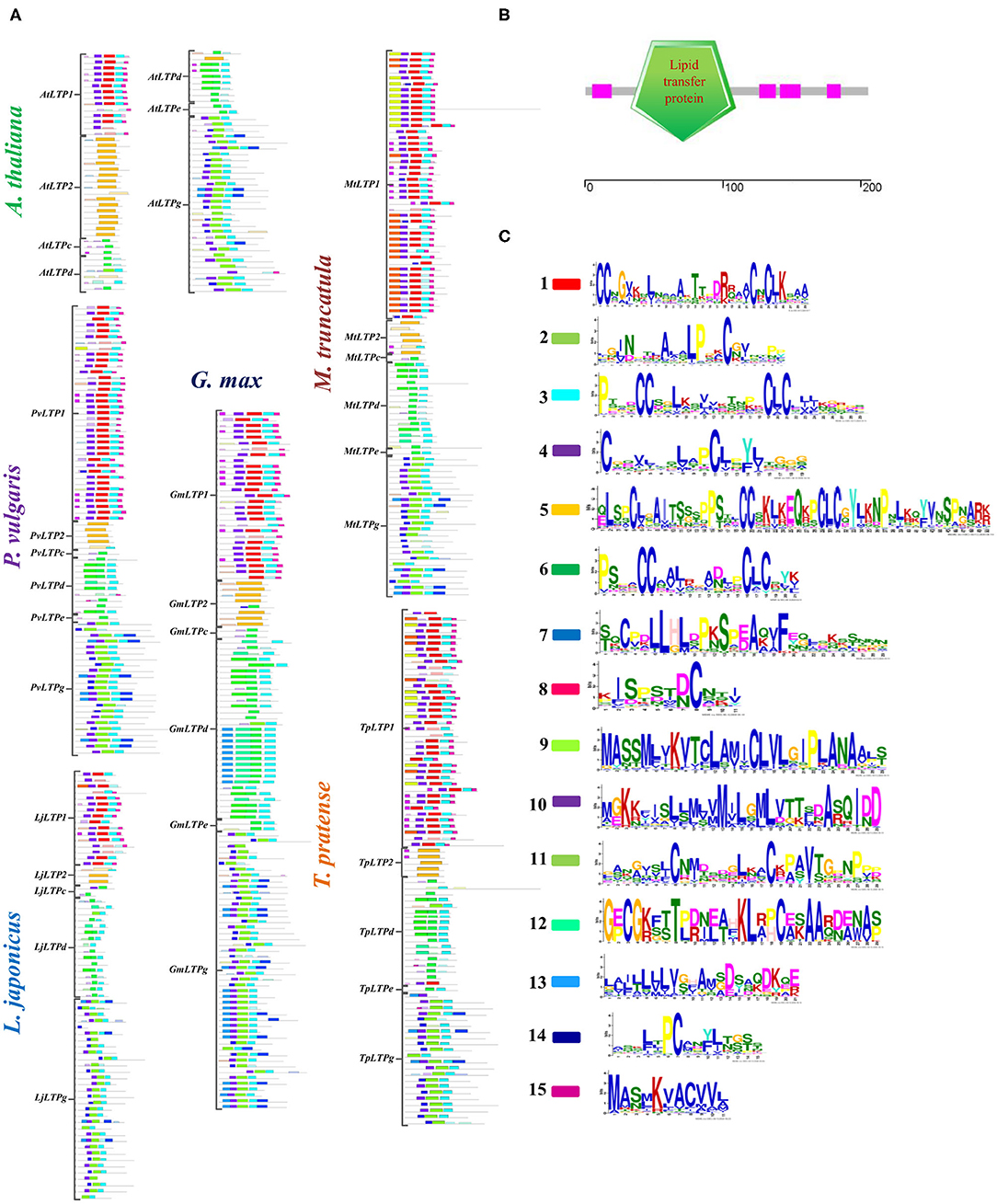
Figure 2. Conserved functional motifs in LTPs. (A) Schematic representation of the motifs identified in LTP amino acid sequences from P. vulgaris, G. max, L. japonicus, M. truncatula, T. pratense, and A. thaliana. Significantly overrepresented motifs are shown by bars in their predicted positions. (B) Characteristic lipid transfer domains in LTP sequences. (C) Logo of the identified overrepresented motifs; the color code is the same as in (A). The LTP classes on the left indicate the corresponding sequences for each class and species. MEME software was used to identify significantly overrepresented motifs.
Synteny of LTP Genes in Legumes and A. thaliana Showed a Duplicated Gene Family
To analyze the distribution of LTP genes in the genomes of the legumes and A. thaliana, we performed in silico mapping of the gene loci using PhenoGram Plot. The LTP gene family is distributed among all A. thaliana and legume chromosomes (Supplementary Figure 2). The LTP loci are clustered in a particular manner in some legume chromosomes by LTP class, which was not observed in A. thaliana. In P. vulgaris, 51% of the LTP1 genes are located on chromosome 9, while 40% of LTP1 genes are located on chromosome 7 in M. truncatula. Similarly, 50% of the LTPd genes are located on chromosome 18 in G. max, and 50% of LTPg genes are clustered on chromosomes 1 and 5 in L. japonicus.
We explored the evolutionary pathways of the LTP genes in the four legumes and A. thaliana individually and between species using MCScanx software. Individual syntenic analysis showed that A. thaliana, P. vulgaris, L. japonicus, and M. truncatula have a low percentage of LTP collinear genes in their genomes (10–17%), whereas G. max contains 52% of these genes (Supplementary Table 4, Supplementary Figure 3). These collinear genes represent less than half of the percentage of genetic synteny in the overall genomes of A. thaliana and P. vulgaris, which range from 27 to 29%. By contrast, in L. japonicus and M. truncatula, these collinear genes represent 1- or 2-fold that of their overall genomes, i.e., 5–13% (Supplementary Figure 4).
Syntenic analysis between species revealed 112 collinear LTP genes between P. vulgaris and G. max; 46 between P. vulgaris and L. japonicus; 54 between P. vulgaris and M. truncatula; 87 between L. japonicus and G. max; 93 between G. max and M. truncatula; and 49 between L. japonicus and M. truncatula (Figures 3A–C,E–G, Supplementary Table 4). Within these collinear genes, the LTPg class represents the highest percentage of syntenic genes (58%), followed by the LTP1 and LTPd classes (17 and 14%, respectively). Analysis of the collinearity between the LTPs of these four legumes against A. thaliana revealed 124 legume genes that are syntenic to those of the Brassicaceae. Of these 124 syntenic genes, 29 belong to P. vulgaris, 21 to L. japonicus, 52 to G. max, and 22 to M. truncatula (Figure 3D, Supplementary Table 5). G. max has the highest number of syntenic genes, and M. truncatula has the lowest number compared with A. thaliana, which are mainly represented by the LTPg class in all instances. These results indicate that LTP genes were subjected to various duplication events throughout the evolution of legumes.
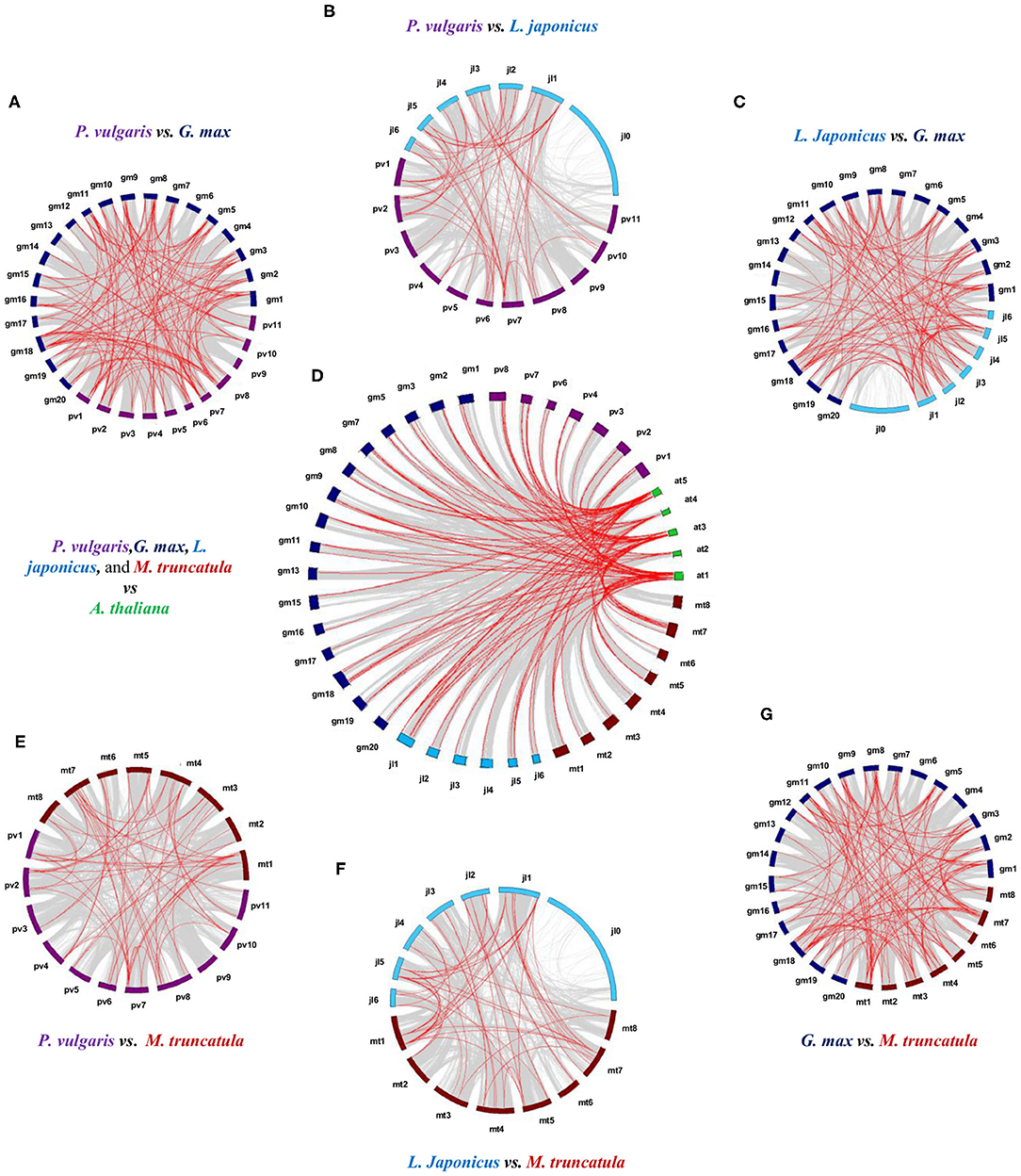
Figure 3. Synteny analysis of the LTP genes of P. vulgaris, L. japonicus, G. max, M. truncatula, and A. thaliana. Physical co-localizations of genetic loci in (A) P. vulgaris and G. max, (B) P. vulgaris and L. japonicus, (C) L. japonicus and G. max, (D) the four legumes against A. thaliana, (E) P. vulgaris and M. truncatula, (F) L. japonicus and M. truncatula, and (G) G. max and M. truncatula. Chromosomes are indicated by color code: purple, P. vulgaris; blue, L. japonicus; dark blue, G. max; burgundy, M. truncatula; and green, A. thaliana. The gray lines represent the remaining syntenic genes in the complete genome. MCScan software was used to analyze the syntenic relationship between species.
Conserved Exon-Intron Arrangement of LTP Genes in Legumes and A. thaliana
To analyze the functional roles of LTP genes, we assessed the exon-intron organization of the LTP genes in P. vulgaris, G. max, L japonicus, M. truncatula, T. pratense, and A. thaliana using Gene Structure Display Server 2.0 software. The positions of the introns are variable between all species analyzed and the LTP classes, but the number of introns showed little variation. In A. thaliana, four LTP genes lack introns (AtLTP2.8, AtLTP2.11, AtLTPe.2, and AtLTPg.18); the remaining genes of the LTP1, LTPc, and LTPd classes have one to three introns, with more genes containing three introns; genes of the LTP2 class have one or two introns; AtLTPe.1 has one intron, while LTPd genes have two to four introns, most with four introns (Figure 4A).
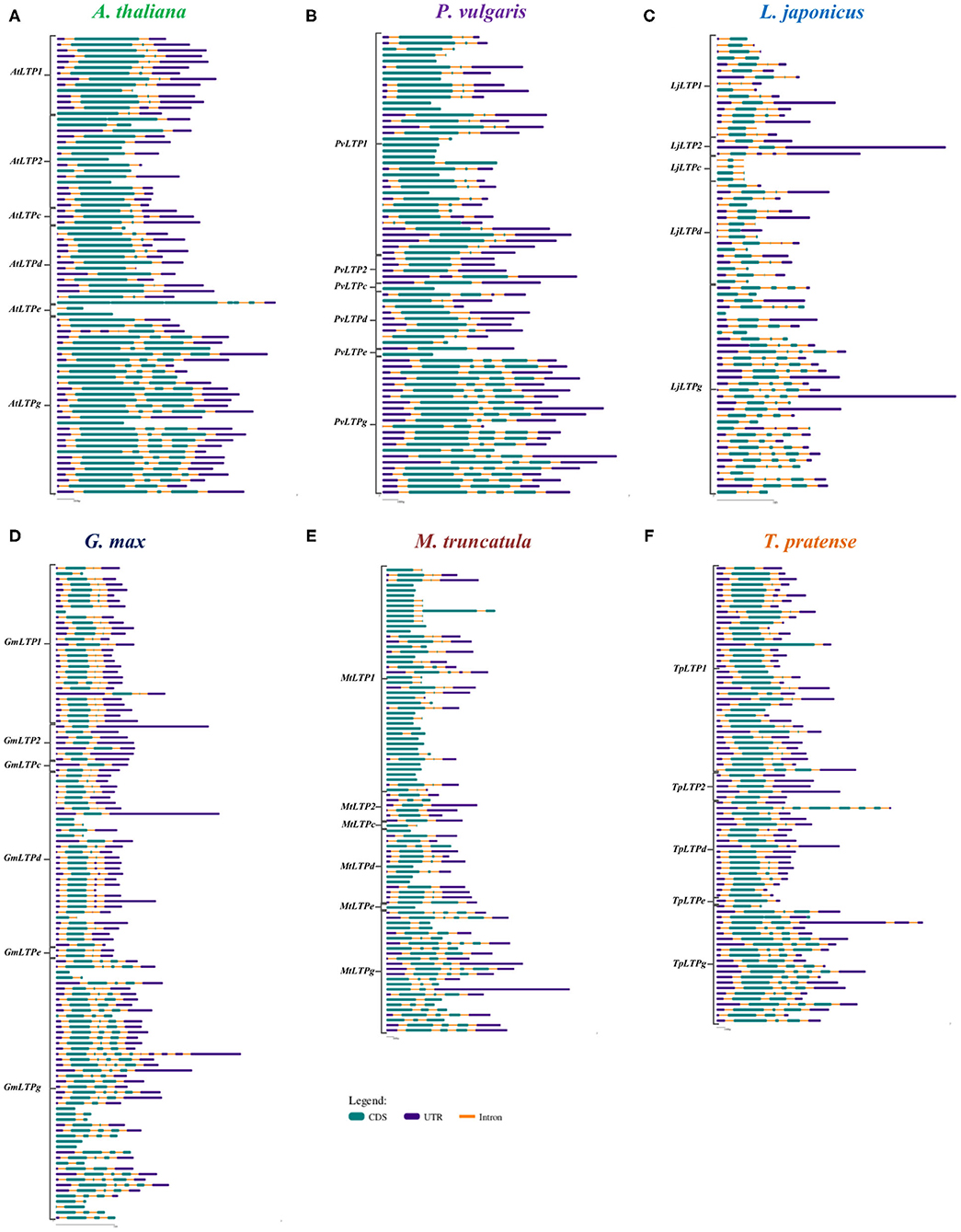
Figure 4. Structures of LTP genes. Exon-intron compositions of the LTP gene sequences of (A) A. thaliana, (B) P. vulgaris, (C) L. japonicus, (D) G. max, (E) M. truncatula, and (F) T. pratense. The LTP classes on the left indicate the corresponding sequences for each class and species. The exons (CDS), introns, and untranslated region (UTR) are represented according to the key. Gene Structure Display Server was used for analysis.
Among legumes, M. truncatula and P. vulgaris have the highest number of LTP genes without introns (20 and 11, respectively), followed by G. max and L. japonicus (6 and 1, respectively); all these genes are distributed among LTP1, LTPc, LTPd, LTPe, and LTPg (Figures 4B–F). Legumes that form determinate nodules have one to three introns in the LTP1 class (Figures 4B–D), while genes belonging to this class from legumes that form indeterminate nodules have up four introns (Figures 4E,F). Additionally, the classes LTP2, LTPd, and LTPg exhibit a homogeneous intron distribution, with two, one to three, and one to five introns, respectively, in almost all legumes. Finally, genes in the LTPc and LTPe classes contain one to three introns, depending on the legume species. Although some conservation of introns was observed among the six plants analyzed, more introns are present in LTP1 and LTPg genes in the legumes than in A. thaliana. However, more similarities between this non-legume dicot and legumes that form determinate vs. indeterminate nodules were observed.
Analysis of the Expression Profiles of LTP Genes
To analyze the expression profiles of the LTP genes in various plant organs, we retrieved expression data for ancient species, cereals, legumes, and A. thaliana, available in different gene expression atlases. Comparison of expression data between plant species and between LTP classes was performed. The results are presented in the following sections.
Ancient Plants
Expression data for P. patens (a bryophyte) and S. moellendorffii (a lycophyte) were retrieved from the BAR for Plant Biology database. These species only possess the LTPd and LTPg classes, which exhibit differential expression patterns between species (Supplementary Figures 5A,B). In P. patens, the LTPd class (LTPd15, LTPd17, and LTPd.18) is highly expressed in whole gametophores, gametophores without rhizoids, and green sporophytes (Supplementary Figure 5A). Genes in the LTPg class are expressed 90% lower than LTPd genes, but are expressed in germinating spores and protonema. In S. moellendorffii, the LTPd class (LTPd.8 and LTPd.9) is mainly expressed in roots and rhizophores, while the LTPg class (LTPg.9) is 99% higher expressed in strobili than in other tissues (Supplementary Figure 5B). These data suggest that the LTPd class is more strongly associated with aboveground tissue in bryophytes and belowground tissue in lycophytes, but members of the LTPg class are more closely involved in reproductive organ activity in both species. However, additional comparative analyses with ancient species are needed to test this hypothesis.
Monocotyledons
To analyze the expression patterns of LTP family genes in monocot species, we also retrieved expression data for the cereal species, S. bicolor, O. sativa, and Z. mays from the BAR for Plant Biology database. The LTP genes are mainly expressed in the shoot systems (basically leaves, stems, flowers, and seeds) of the three monocot species, with LTP1 genes being the most highly represented class (Supplementary Figures 5C–E). In S. bicolor, the most highly expressed gene is SbLTP1.9 in the vegetative meristem, followed by SbLTP1.3 in shoots (Supplementary Figure 5C). In O. sativa, the most highly expressed gene is OsLTP1.4 in stems, followed by OsLTP2.4 in stems (Supplementary Figure 5D). The most highly expressed gene in Z. mays is ZmLTP1.8 in young seeds (Supplementary Figure 5E). In these cereal species, the LTP genes were expressed 70% lower in belowground vs. aboveground tissue, where genes in classes LTP1 and LTPd are mainly expressed (Supplementary Figures 5C–E). SbLTPd.10 and ZmLTPd.10 show the highest expression levels in S. bicolor and Z. mays, respectively, whereas 11 OsLTP1 genes and OsLTP2.3 show the highest expression levels in O. sativa. These data suggest that genes belonging to the LTP1 class could play various roles in shoot development and reproduction in cereals.
Non-legume Dicotyledons
LTP family proteins have been studied more extensively in A. thaliana than in any other species. In the present study, A. thaliana was included as a model non-legume dicotyledon to compare the expression profiles of LTPs in legumes against dicotyledons. We retrieved expression data for A. thaliana from the BAR for Plant Biology database. In this dicot species, AtLTP genes are expressed at their highest levels in siliques and flowers, followed by leaves and seeds (Figure 5A). The AtLTP1, AtLTP2, and AtLTPd classes are the most highly represented classes in these organs, whereas genes in the LTPg class are expressed 67% less. Despite the generally lower expression levels of AtLTP genes in roots compared to siliques and flowers, AtLTP2.5, AtLTP2.11, AtLTPd.4, and AtLTPg.19 are more highly expressed in roots than in these organs. These results suggest that members of the LTP family could play an important role in the reproductive process of this dicot species, as observed in the monocot species.
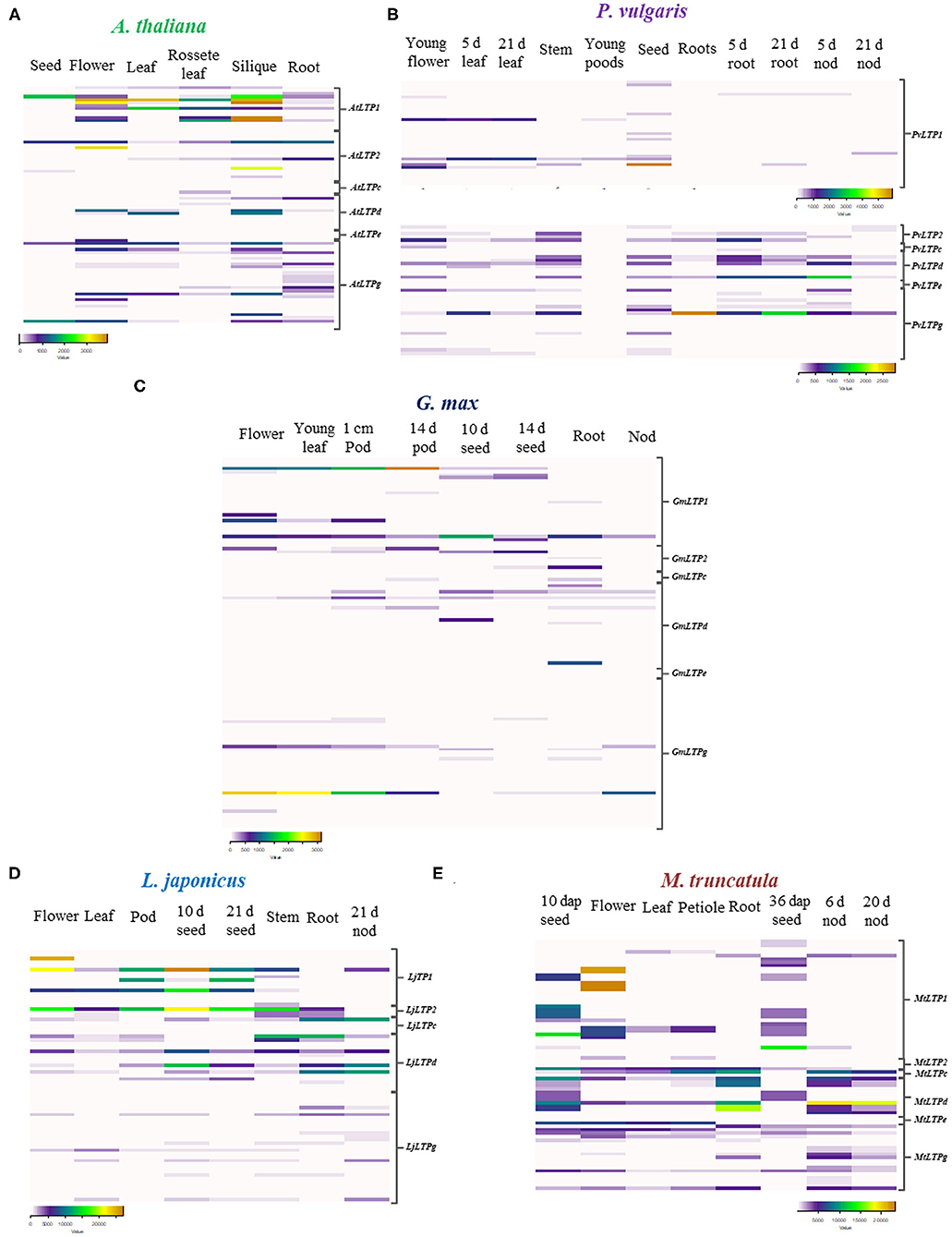
Figure 5. Expression patterns of LTP genes. Heat maps of LTP gene expression values in (A) A. thaliana, (B) P. vulgaris, (C) G. max, (D) L. japonicus, and (E) M. truncatula. Expression data were retrieved from the PvGEA, LotusBASE, MtGEA, and BAR databases. RPKM values are represented as color codes below each heat map. The LTP classes on the left indicate the corresponding sequences for each class and species. dap, days after pollination; d, days; nod, nodules.
Legumes
To analyze the possible roles of LTP family members in root nodule symbiosis, we evaluated the expression patterns of these genes in the following legumes: P. vulgaris, G. max, L. japonicus, and M. truncatula. We observed both similarities and differences in the expression patterns of LTP genes in these legumes compared to non-legumes species. Genes in classes LTP1 and LTP2 are highly expressed in leaves, flowers, and seeds at different stages of development (Figures 5B–E). Although some LTP genes are highly expressed in flowers and seeds in all four legumes, the different classes of LTPs are also highly expressed in roots and nodules of the legumes (Figures 5B–E).
In P. vulgaris, PvLTPg.11 is the most highly expressed gene in roots inoculated with rhizobia and in young nodules, followed by PvLTPd.4, PvLTPd.6, and PvLTPd.10 (Figure 5B), whereas in G. max, GmLTPd.1 and GmLTP1.1 are the most expressed genes in roots (Figure 5C). LjLTPd.1, LjLTPd.2, LjLTPd.3, and LjLTPc.1 are primarily expressed in roots and mature nodules of L. japonicus (Figure 5D). In M. truncatula, which forms indeterminate nodules, MtLTPd.1, MtLTPd.2, and MtLTP2.1 show high expression levels in young and mature nodules and roots (Figure 5E). Among legumes, the LTP1 and LTPd classes are the most commonly represented classes in roots and nodules, showing a differential expression pattern during early and late stages of nodulation.
These data suggest that some LTP classes are active in legume roots, roots inoculated with rhizobia, and nodules. To validate the expression patterns of LTP genes in the legumes analyzed in silico, we evaluated the expression profiles of seven P. vulgaris genes during the early and late stages of nodulation by RT-qPCR. We selected LTP genes that are potentially expressed under nodulation conditions in this legume that might also be expressed in the three other legumes examined. Also, we analyzed the expression profile of these genes in a home transcriptomic data of P. vulgaris roots inoculated and non-inoculated at 7 dpi with R. tropici (Fonseca-García et al., 2019), here they showed the highest fold changes with the highest read counts of the gene family in this legume (Supplementary Table 6). These seven genes (PvLTP1.6, PvLTP2.3, PvLTP2.5, PvLTPd.4, PvLTPd.6, PvLTPd.10, and PvLTPg20) presented two different expression profiles during nodulation in P. vulgaris (Figure 6).
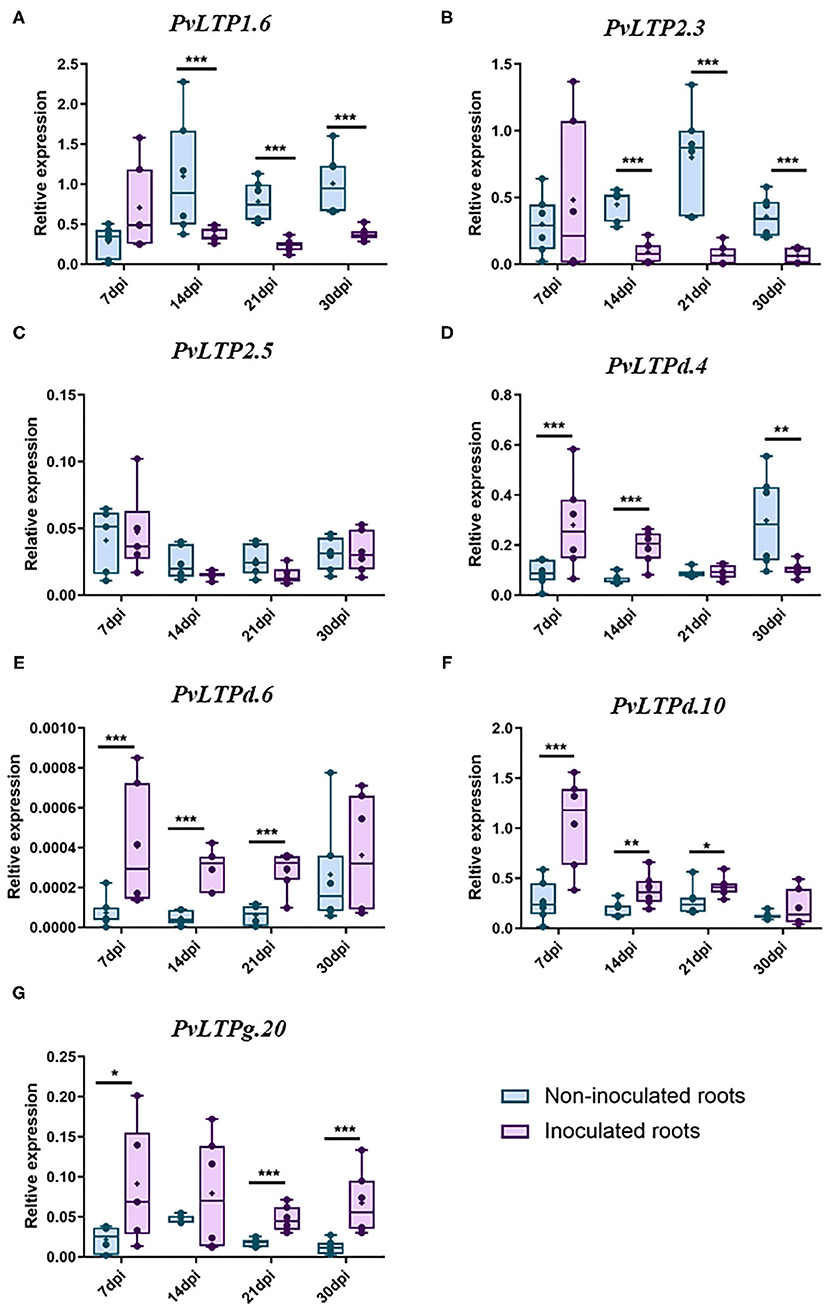
Figure 6. RT-qPCR analysis of selected P. vulgaris LTP genes. The relative expression profiles of seven LTP genes from wild-type P. vulgaris roots inoculated or non-inoculated with R. tropici: (A) PvLTP1.6, (B) PvLTP2.3, (C) PvLTP2.5, (D) PvLTPd.4, (E) PvLTPd.6, (F) PvLTPd.10, and (G) PvLTPg.20. The transcript abundance of the selected genes was evaluated by RT-qPCR and normalized according to the expression of the elongation factor 1α (EF1α) gene. The blue bars represent non-inoculated roots, and the purple bars represent roots inoculated with R. tropici. The borders of the boxes represent the first quartile to the third quartile, the horizontal line within the box represents the median, and the whiskers represent the smallest and the largest outlier point of the data set (n = 6). A non-parametric Mann-Whitney test was performed to evaluate significant differences, * p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
Two genes were downregulated during the late stages of nodulation: PvLTP1.6 was significantly suppressed at 14, 21, and 30 dpi, while PvLTP2.3 was clearly downregulated at 14 and 21 dpi (Figures 6A–C). By contrast, 4 genes were upregulated between the early and late stages of nodulation: PvLTPd.6, PvLTPd.10, and PvLTPg20 were upregulated at 7, 14, and 21 dpi, and PvLTPg20 was also upregulated at 30 dpi (Figures 6E–G). Similarly, PvLTPd.4 was upregulated at 7 and 14 dpi, but, intriguingly, it was downregulated at 30 dpi (Figure 6D). These results indicate that these seven LTP genes are differentially expressed during different stages of nodulation in common bean, suggesting that they are fine-tuning regulated in root nodule symbiosis.
Previously, our group demonstrated that ROS play an important role in common bean nodulation through RBOHs activity (Cárdenas et al., 2008; Montiel et al., 2012; Arthikala et al., 2017). To investigate the possible roles of these seven PvLTP genes in nodulation in more detail, we evaluated their transcript accumulation in transgenic roots expressing an RNA interference construct targeting PvRbohA following inoculation with R. tropici. PvRbohA encodes a NADPH oxidase required for the early stages of root nodule development in common bean (Arthikala et al., 2017). All three LTPd genes were downregulated with 23–54% less in the inoculated roots carrying the RbohA:RNAi construct at 1 dpi than in control roots, whereas LTPg.20 was upregulated with 38% more under the same conditions (Figure 7). Furthermore, the LTP2.5 gene presented a downregulation tendency with 44% less in the RbohA:RNAi roots than control, but LTP1.6 and LTP2.3 were expressed at similar levels in silenced and control roots. These results indicate that five of the seven genes analyzed could be regulated by ROS, produced by RbohA during the early stages of rhizobial symbiosis.
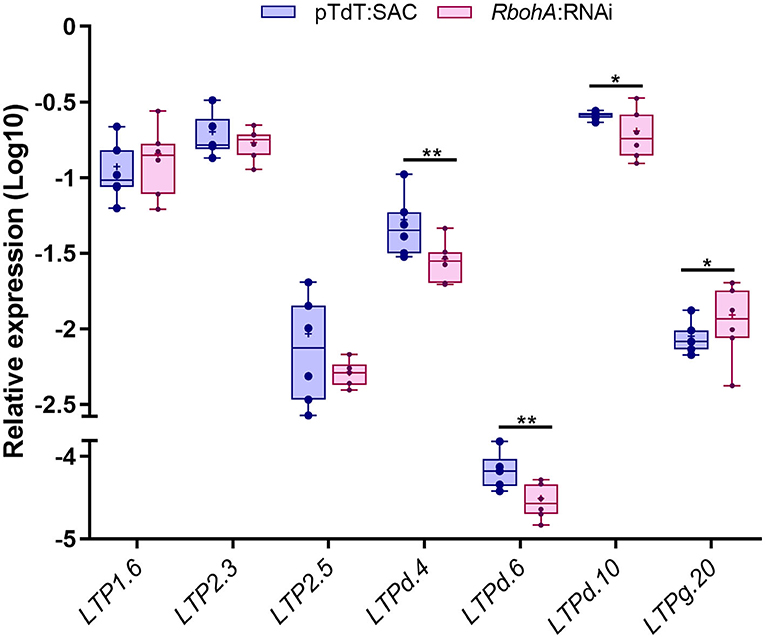
Figure 7. RT-qPCR analysis of selected P. vulgaris LTP genes in RbohA:RNAi transgenic roots. Relative expression profiles of the seven LTP genes in control (pTdT-SAC) and RbohA:RNAi transgenic roots of P. vulgaris inoculated with R. tropici. The transcript levels were normalized according to the expression of the elongation factor 1α (EF1α) gene. The blue bars represent control (pTdT-SAC) transgenic roots, and the purple bars represent RbohA:RNAi transgenic roots inoculated with R. tropici. The borders of the boxes represent the first quartile to the third quartile, the horizontal line within the box represents the median, and the whiskers represent the smallest and the largest outlier point of the data set (n = 6). A non-parametric Mann-Whitney test was performed to evaluate significant differences, *p ≤ 0.05, **p ≤ 0.01.
Discussion
Legumes are valuable crops that serve as sources of food and animal feed throughout the world. These plants interact with soil bacteria called rhizobia, which through a molecular dialog and a coordinated cellular reorganogenesis in the plant cortex, form a new organ, the nodule. In the nodules, rhizobia differentiate into bacteroids and are enclosed to form symbiosomes, in which atmospheric dinitrogen is transformed into ammonia, a source of nitrogen that is assimilable by the plant (Roy et al., 2020). In plants, LTP genes comprise an abundant family of proteins and perform various functions in their life cycle (Edqvist et al., 2018). This gene family has been identified in some important plant species such as, A. thaliana, rice, wheat (Boutrot et al., 2008), maize (Wei and Zhong, 2014), barley (Zhang et al., 2019) and potato (Li et al., 2019). However, little is known about LTPs in legumes and their role in the root-nodule symbiosis. In this study, we performed a genome identification of LPTs family in various legumes species and other important plant model, moreover, we analyzed the putative roles of LTPs in rhizobial symbiosis.
Through a robust phylogenetic analysis, we identified 960 LTP genes in 15 different plant species, which were grouped into 6 classes (LTP1, LTP2, LTPc, LTPd, LTPe, and LTPg), according to their phylogenetic features and the characteristic motif 8CM. We used the latest versions of the genomes of all species studied, allowing us to use the complete sequences of the putative LTP genes instead of the ESTs used in previous studies (Edstam et al., 2011); consequently, some previously reported LTP genes were reassigned in the current study. In addition, 30 genes showed incomplete 8CMs with at least 1 missing C; however, these sequences were considered to be LTPs because they all possess characteristic lipid transfer domains. This modification occurred in the 15 species studied and in the 6 classes of LTPs, suggesting that this feature was conserved during evolution. Notably, we identified the first reported LTP gene (LTPd) in an algae species, C. reinhardtii, indicating that LTPs are an ancient family of genes that has been present since the division of Chlorophyta, before the first land plant appeared.
Our phylogenetic analysis clustered the 960 LTPs into 4 clades (Figure 1, Supplementary Figure 1). Ancient species here studied, P. patens (Rensing et al., 2020) and S. moellendorffii (Ferrari et al., 2020), only contained classes LTPd and LTPg, suggesting that these two classes are the oldest in this gene family. On the contrary, the LTPe class was present only in dicot species, indicating that the class is specific to these angiosperms and that these genes could be the youngest members of the LTP family (Table 1). Additionally, we detected notable differences in the number of LTP1 and LTP2 genes among monocotyledonous and dicotyledonous species. In particular, legumes had fewer LTP2 genes and more LTP1 genes than the other species (Table 1), which contained a specific motif in legumes that form indeterminate nodules (Figures 2A,C). These results suggest that these classes of genes exhibit specific gene duplication, as well as structural modifications in legumes that could have specific functions in these plants, which has also been suggested in other plant systems (Moore and Purugganan, 2005; Conant and Wolfe, 2008; Flagel and Wendel, 2009).
Genome duplication is a primary source of genetic novelty, providing increased functionality in angiosperms and has played a fundamental role in their evolution (Davies et al., 2004). Many reports have documented duplication events of LTP genes in various species, such as A. thaliana, rice, sorghum (Salminen et al., 2016), potato (Li et al., 2019), maize (Wei and Zhong, 2014), and wheat (Kouidri et al., 2018; Odintsova et al., 2019). Gene duplication and functional redundancy allow LTP gene sequences to gather mutations, that result in increased divergence and thus the expansion and evolution of the gene family (Conant and Wolfe, 2008). Our synteny analysis of LTP genes revealed a conserved homology between some genes in the four legumes and A. thaliana. We observed a low percentage of collinear LTP genes in the individual genomes of A. thaliana, P. vulgaris, L. japonicus, and M. truncatula, while more than half of the LTP genes in G. max showed collinearity (Supplementary Table 4, Supplementary Figure 3). This could be due to G. max is an allotetraploid species that originated from hybridization events involving two different diploid progenitors (Cannon and Shoemaker, 2012). Also, we observed a higher percentage of collinear genes between G. max and the other legumes than within A. thaliana, P. vulgaris, L. japonicus, and M. truncatula. The LTP class with the most syntenic genes was LTPg, which was generally observed in all species tested with an extended number of members., It could be because LTPg class regulate the export of cuticular wax and suberin (Kim et al., 2012; Lee and Suh, 2018), important processes for plant development.
Gene function has been associated with different structural characteristics such as exon-intron composition, which bear the marks of the evolution of a gene family (Moore and Purugganan, 2005; Flagel and Wendel, 2009). According to our data, the LTP family formed before the differentiation of terrestrial plants and generated new classes during their evolution. The dicotyledonous species analyzed have a variable arrangement of exon-introns among the LTP classes, and legume genes have more introns than A. thaliana genes (Figure 4). Also, legumes have more genes without introns in all LTP classes compared to A. thaliana. M. truncatula has the highest number of genes without introns (22, including 17 in the LTP1 class), followed by P. vulgaris (10 genes) and A. thaliana (only 5 genes). A compact genetic structure could be advantageous to plants responding to stimuli by allowing rapid gene expression (Jeffares et al., 2008). However, the expression patterns of intron less LTP genes in legumes were similar to those in A. thaliana (Figure 5).
Even though many potential LTP genes have been identified in some plant species using bioinformatics approaches, the functions of only a few LTPs have been revealed. However, some reports suggest that the a mechanism of action of LTPs is related to the transfer and deposition of the monomers required for the assembly of the waterproof lipid barriers (Charvolin et al., 1999; Han et al., 2001; Cheng et al., 2004). Root nodule symbiosis involve two complex processes that are simultaneously coordinated, rhizobial infection and nodule development, where the number of dividing cells increases dramatically, requiring membrane biosynthesis by the plant (Roy et al., 2020). Also, for the formation of symbiosomes the synthesis of the peribacteroidal membrane to enclose the bacteroids is required (Verma, 1992). The mentioned ability of LTPs to bind to and transfer a wide variety of lipids suggests that these proteins participate in membrane remodeling during root nodule symbiosis. In fact, the LTP AsE246 from Chinese milk vetch bids DGDG from nodule extract and its knockdown impairs symbiosomes development (Lei et al., 2014). In addition, in silico analysis of the expression profiles showed that many LTP genes, especially LTPd genes, are active in legume roots and nodules (Figure 5).
RT-qPCR assays confirmed that genes belonging to the PvLTPd and PvLTPg classes are highly expressed in P. vulgaris roots inoculated with rhizobia during the early and late stages of nodulation (Figures 6D–G). In addition, genes of the PvLTP1 and PvLTP2 classes are repressed during late stages of nodulation in this legume (Figures 6A–C). Indeed, a novel LTP gene in Chinese milk vetch might be involved in the establishment of rhizobial infection and the transport of plant-synthesized lipids to the symbiosome membrane (Lei et al., 2014). Another report suggests that an AtLTP1 gene is involved in regulating the ethylene response and signaling in A. thaliana (Wang et al., 2016), importantly, this phytohormone is a well-known negative regulator of nodulation. Our results suggest that genes of the PvLTPd and PvLTPg classes could mediate positively root nodule symbiosis, transporting plant-synthesized lipids. Whereas, members of the PvLTP1 and PvLTP2 classes might inhibit this process interacting with negatives regulators.
ROS function as early signals in root hairs during the establishment of root nodule symbiosis (Montiel et al., 2016). In root hair cells of P. vulgaris, a rapid, transient ROS burst was detected 15–30 s after treatment with Nod factors (Cárdenas et al., 2008). In plants, ROS are mainly produced by the activity of the NADPH oxidase enzymes RBOHs, which have been implicated in effective rhizobial infection and in promoting nodule organogenesis in P. vulgaris (Montiel et al., 2012; Arthikala et al., 2014) and M. truncatula (Marino et al., 2011). In the current study, we provided evidence that the expression of LTPd and LTPg class genes is regulated by the downregulation of a PvRBOH (PvRBOHA) in P. vulgaris roots during the early stages of nodulation. Our findings strongly suggest that LTP genes could play different roles during nodulation in P. vulgaris, and possibly interacting with RBOH-dependent ROS production since the early stages. ROS as early signals in this symbiotic process could activate the synthesis of lipids in the plasma membrane necessary for rhizobia infection and nodule organogenesis, however further functional analysis is needed to confirm this interaction.
Conclusions
In this study, we performed a comprehensive genome analysis of the LTP family in legumes and other important model plants. We identified 960 LTPs in 14 plant species and 1 green alga, which were grouped into 4 clades according to the LTP class. We report the first LTP gene identified in a chlorophyte species, C. reinhardtii, indicating that the LTP family appeared before the first terrestrial plant. Sequence analysis showed that the gene and protein structures of the LTP family members are highly conserved among species, but clear differences were observed between the LTP classes. Expression profiling analyses revealed that PvLTP genes are differentially expressed during the early and late stages of nodulation and that they might be regulated by RBOH-dependent ROS production. These findings shed light on the possible roles of LTP family members in the legume-rhizobium symbiosis, but functional analyses are necessary to increase our knowledge and, as a good start, PvLTP genes could be good candidates.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: Phytozome 12.1.6 database https://phytozome.jgi.doe.gov; LotusBase https://lotus.au.dk; LIS: Legume Information System database https://legumeinfo.org.
Author Contributions
CQ and CF-G conceived the study. CF-G and JS-M analyzed the data. RP and CF-G collected some of the plant tissues and performed RNA extraction and purification, cDNA synthesis, and RT-qPCR assays. CQ, CF-G, and RP drafted, edited, and revised the manuscript, which was approved by all authors. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Consejo Nacional de Ciencia y Tecnología in México (CONACyT) under Fronteras de la ciencia (FC) grants: FC-2016/1503 to CQ and CV200519 to Luis Cárdenas, with a postdoctoral fellowship to CF-G (268158) and doctoral fellowships to JS-M (463767) and RP (823333).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Noreide Nava for her technical support in the collection of plant tissues. In addition, we thank Dr. Paul Gaytan and MsC Jorge Arturo Yañez from Unidad de Síntesis y Secuenciación at Instituto de Biotecnología, UNAM for their technical contributions. We thank to Dr. Luis Cárdenas for providing the QuantStudio 5 equipment for the qPCR assays.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.660100/full#supplementary-material
References
Arthikala, M. K., Montiel, J., Sánchez-López, R., Nava, N., Cárdenas, L., and Quinto, C. (2017). Respiratory burst oxidase homolog gene a is crucial for rhizobium infection and nodule maturation and function in common bean. Front. Plant Sci. 8, 1–15. doi: 10.3389/fpls.2017.02003
Arthikala, M. K., Sánchez-López, R., Nava, N., Santana, O., Cárdenas, L., and Quinto, C. (2014). RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 202, 886–900. doi: 10.1111/nph.12714
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Bapaume, L., and Reinhardt, D. (2012). How membranes shape plant symbioses: signaling and transport in nodulation and arbuscular mycorrhiza. Front. Plant Sci. 3:223. doi: 10.3389/fpls.2012.00223
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Boutrot, F., Chantret, N., and Gautier, M. F. (2008). Genome-wide analysis of the rice and arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genom. 9, 1–19. doi: 10.1186/1471-2164-9-86
Cannon, S. B., and Shoemaker, R. C. (2012). Evolutionary and comparative analyses of the soybean genome. Breed. Sci. 61, 437–444. doi: 10.1270/jsbbs.61.437
Cárdenas, L., Martínez, A., Sánchez, F., and Quinto, C. (2008). Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). Plant J. 56, 802–813. doi: 10.1111/j.1365-313X.2008.03644.x
Charvolin, D., Douliez, J. P., Marion, D., Cohen-Addad, C., and Pebay-Peyroula, E. (1999). The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 A resolution. Eur. J. Biochem. 264, 562–568. doi: 10.1046/j.1432-1327.1999.00667.x
Cheng, H.-C., Cheng, P.-T., Peng, P., Lyu, P.-C., and Sun, Y.-J. (2004). Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa. Protein Sci. 13, 2304–2315. doi: 10.1110/ps.04799704
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Dash, S., Campbell, J. D., Cannon, E. K. S., Cleary, A. M., Huang, W., Kalberer, S. R., et al. (2016). Legume information system (LegumeInfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Res. 44, D1181–D1188. doi: 10.1093/nar/gkv1159
Davies, T. J., Barraclough, T. G., Chase, M. W., Soltis, P. S., Soltis, D. E., and Savolainen, V. (2004). Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc. Natl. Acad. Sci. U. S. A. 101, 1904–1909. doi: 10.1073/pnas.0308127100
Dhar, N., Caruana, J., Erdem, I., and Raina, R. (2020). An arabidopsis DISEASE RELATED NONSPECIFIC LIPID TRANSFER PROTEIN 1 is required for resistance against various phytopathogens and tolerance to salt stress. Gene 753, 1–13. doi: 10.1016/j.gene.2020.144802
Edqvist, J., Blomqvist, K., Nieuwland, J., and Salminen, T. A. (2018). Plant lipid transfer proteins: are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 59, 1374–1382. doi: 10.1194/jlr.R083139
Edstam, M. M., Viitanen, L., Salminen, T. A., and Edqvist, J. (2011). Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 4, 947–964. doi: 10.1093/mp/ssr019
Estrada-Navarrete, G., Alvarado-Affantranger, X., Olivares, J. E., Guillén, G., Díaz-Camino, C., Campos, F., et al. (2007). Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat. Protoc. 2, 1819–1824. doi: 10.1038/nprot.2007.259
Fernandez-Pozo, N., Haas, F. B., Meyberg, R., Ullrich, K. K., Hiss, M., Perroud, P. F., et al. (2020). PEATmoss (Physcomitrella Expression Atlas Tool): a unified gene expression atlas for the model plant Physcomitrella patens. Plant J. 102, 165–177. doi: 10.1111/tpj.14607
Ferrari, C., Shivhare, D., Hansen, B. O., Pasha, A., Esteban, E., Provart, N. J., et al. (2020). Expression atlas of Selaginella moellendorffii provides insights into the evolution of vasculature, secondary metabolism, and roots. Plant Cell 32, 853–870. doi: 10.1105/tpc.19.00780
Finkina, E. I., Melnikova, D. N., Bogdanov, I. V., and Ovchinnikova, T. V. (2016). Lipid transfer proteins as components of the plant innate immune system: structure, functions, and applications. Acta Nat. 8, 47–61. doi: 10.32607/20758251-2016-8-2-47-61
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Fonseca-García, C., Zayas, A. E., Montiel, J., Nava, N., Sánchez, F., and Quinto, C. (2019). Transcriptome analysis of the differential effect of the NADPH oxidase gene RbohB in Phaseolus vulgaris roots following Rhizobium tropici and Rhizophagus irregularis inoculation. BMC Genom. 20:800. doi: 10.1186/s12864-019-6162-7
Gao, S., Guo, W., Feng, W., Liu, L., Song, X., Chen, J., et al. (2016). LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 17, 412–426. doi: 10.1111/mpp.12290
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Halim, V. A., Vess, A., Scheel, D., and Rosahl, S. (2006). The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol. 8, 307–313. doi: 10.1055/s-2006-924025
Han, G. W., Lee, J. Y., Song, H. K., Chang, C., Min, K., Moon, J., et al. (2001). Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 308, 263–278. doi: 10.1006/jmbi.2001.4559
He, J., Benedito, V. A., Wang, M., Murray, J. D., Zhao, P. X., Tang, Y., et al. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinform. 10:a441. doi: 10.1186/1471-2105-10-441
Jeffares, D. C., Penkett, C. J., and Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Kim, H., Lee, S. B., Kim, H. J., Min, M. K., Hwang, I., and Suh, M. C. (2012). Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53, 1391–1403. doi: 10.1093/pcp/pcs083
Kouidri, A., Whitford, R., Suchecki, R., Kalashyan, E., and Baumann, U. (2018). Genome-wide identification and analysis of non-specific lipid transfer proteins in hexaploid wheat. Sci. Rep. 8:17087. doi: 10.1038/s41598-018-35375-7
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lascombe, M.-B., Bakan, B., Buhot, N., Marion, D., Blein, J.-P., Larue, V., et al. (2008). The structure of “defective in induced resistance” protein of Arabidopsis thaliana, DIR1, reveals a new type of lipid transfer protein. Protein Sci. 17, 1522–1530. doi: 10.1110/ps.035972.108
Lee, S. B., and Suh, M. C. (2018). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein 15 affects seed coat permeability in Arabidopsis. Plant J. 96, 1206–1217. doi: 10.1111/tpj.14101
Lei, L., Chen, L., Shi, X., Li, Y., Wang, J., Chen, D., et al. (2014). A nodule-specific lipid transfer protein AsE246 participates in transport of plant-synthesized lipids to symbiosome membrane and is essential for nodule organogenesis in Chinese milk vetch. Plant Physiol. 164, 1045–1058. doi: 10.1104/pp.113.232637
Li, G., Hou, M., Liu, Y., Pei, Y., Ye, M., Zhou, Y., et al. (2019). Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genom. 20:375. doi: 10.1186/s12864-019-5698-x
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D., et al. (2010). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. doi: 10.1111/j.1365-313X.2010.04222.x
Marino, D., Andrio, E., Danchin, E. G. J., Oger, E., Gucciardo, S., Lambert, A., et al. (2011). A Medicago truncatula NADPH oxidase is involved in symbiotic nodule functioning. New Phytol. 189, 580–592. doi: 10.1111/j.1469-8137.2010.03509.x
Montiel, J., Arthikala, M. K., Cárdenas, L., and Quinto, C. (2016). Legume NADPH oxidases have crucial roles at different stages of nodulation. Int. J. Mol. Sci. 17:680. doi: 10.3390/ijms17050680
Montiel, J., Nava, N., Cárdenas, L., Sánchez-López, R., Arthikala, M. K., Santana, O., et al. (2012). A Phaseolus vulgaris NADPH oxidase gene is required for root infection by Rhizobia. Plant Cell Physiol. 53, 1751–1767. doi: 10.1093/pcp/pcs120
Moore, R. C., and Purugganan, M. D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8, 122–128. doi: 10.1016/j.pbi.2004.12.001
Mun, T., Bachmann, A., Gupta, V., Stougaard, J., and Andersen, S. U. (2016). Lotus base: an integrated information portal for the model legume Lotus japonicus. Sci. Rep. 6, 1–18. doi: 10.1038/srep39447
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Odintsova, T. I., Slezina, M. P., Istomina, E. A., Korostyleva, T. V., Kovtun, A. S., Kasianov, A. S., et al. (2019). Non-specific lipid transfer proteins in Triticum kiharae dorof. Et migush.: Identification, characterization and expression profiling in response to pathogens and resistance inducers. Pathogens 8:221. doi: 10.3390/pathogens8040221
Oldroyd, G. E. D., and Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Ann. Rev. Plant Biol. 59, 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
O'Rourke, J. A., Iniguez, L. P., Fu, F., Bucciarelli, B., Miller, S. S., Jackson, S. A., et al. (2014). An RNA-Seq based gene expression atlas of the common bean. BMC Genom. 15, 1–16. doi: 10.1186/1471-2164-15-866
Rensing, S. A., Goffinet, B., Meyberg, R., Wu, S.-Z., and Bezanilla, M. (2020). The Moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell 32, 1361–1376. doi: 10.1105/tpc.19.00828
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Salminen, T. A., Blomqvist, K., and Edqvist, J. (2016). Lipid transfer proteins: classification, nomenclature, structure, and function. Planta 244, 971–997. doi: 10.1007/s00425-016-2585-4
Santi, C., Molesini, B., Guzzo, F., Pii, Y., Vitulo, N., and Pandolfini, T. (2017). Genome-wide transcriptional changes and lipid profile modifications induced by medicago truncatula N5 overexpression at an early stage of the symbiotic interaction with sinorhizobium meliloti. Genes 8:396. doi: 10.3390/genes8120396
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
Shin, D. H., Lee, J. Y., Hwang, K. Y., Kyu Kim, K., and Suh, S. W. (1995). High-resolution crystal structure of the non-specific lipid-transfer protein from maize seedlings. Structure 3, 189–199. doi: 10.1016/S0969-2126(01)00149-6
Verma, D. P. S. (1992). Signals in root nodule organogenesis and endocytosis of Rhizobium. Plant Cell 4, 373–382. doi: 10.2307/3869439
Wang, H., Sun, Y., Chang, J., Zheng, F., Pei, H., Yi, Y., et al. (2016). Regulatory function of Arabidopsis lipid transfer protein 1 (LTP1) in ethylene response and signaling. Plant Mol. Biol. 91, 471–484. doi: 10.1007/s11103-016-0482-7
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, 1–14. doi: 10.1093/nar/gkr1293
Warnes, G. R., Bolker, B., Huber, W., Lumley, T., Maechler, M., Magnusson, A., et al. (2015). gplots: Various R Programming Tools for Plotting Data, p. 70. Available online at: https://cran.r-project.org/web/packages/gplots/index.html
Wei, K., and Zhong, X. (2014). Non-specific lipid transfer proteins in maize. BMC Plant Biol. 14, 1–18. doi: 10.1186/s12870-014-0281-8
Wolfe, D., Dudek, S., Ritchie, M. D., and Pendergrass, S. A. (2013). Visualizing genomic information across chromosomes with PhenoGram. BioData Mining 6, 1–12. doi: 10.1186/1756-0381-6-18
Xu, Y., Zheng, X., Song, Y., Zhu, L., Yu, Z., Gan, L., et al. (2018). NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 8, 1–14. doi: 10.1038/s41598-018-27274-8
Zhang, M., Kim, Y., Zong, J., Lin, H., Dievart, A., Li, H., et al. (2019). Genome-wide analysis of the barley non-specific lipid transfer protein gene family. Crop J. 7, 65–76. doi: 10.1016/j.cj.2018.07.009
Zhao, J. (2015). Phospholipase D and phosphatidic acid in plant defence response: from protein-protein and lipid-protein interactions to hormone signalling. J. Exp. Bot. 66, 1721–1736. doi: 10.1093/jxb/eru540
Keywords: expression profile, legumes, lipid transfer proteins, nodule symbiosis, phylogenetic analysis
Citation: Fonseca-García C, Solis-Miranda J, Pacheco R and Quinto C (2021) Non-specific Lipid Transfer Proteins in Legumes and Their Participation During Root-Nodule Symbiosis. Front. Agron. 3:660100. doi: 10.3389/fagro.2021.660100
Received: 28 January 2021; Accepted: 22 March 2021;
Published: 22 April 2021.
Edited by:
Jose Palacios, Polytechnic University of Madrid, SpainReviewed by:
Estibaliz Larrainzar, Public University of Navarre, SpainKanika Khanna, Guru Nanak Dev University, India
Copyright © 2021 Fonseca-García, Solis-Miranda, Pacheco and Quinto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmen Quinto, Y2FybWVuLnF1aW50b0BpYnQudW5hbS5teA==
In Memoriam: This paper is dedicated to the memory of Dr. Tomás Ruiz-Argüeso.
 Citlali Fonseca-García
Citlali Fonseca-García Jorge Solis-Miranda
Jorge Solis-Miranda Carmen Quinto
Carmen Quinto