- 1Department of Molecular Microbiology, John Innes Centre, Norwich, United Kingdom
- 2Institute of Plant Biology, Biological Research Center of the Eötvös Loránd Research Network, Szeged, Hungary
In nitrogen-fixing nodules of legumes such as pea (Pisum) and Medicago spp. the plant induces terminal differentiation in the rhizobial endosymbionts by targeting nodule-specific cysteine-rich defensin-like peptides into the bacteria. However, in nodules of other legumes such as soybean and Lotus spp. terminal bacterial differentiation does not occur; these legumes lack genes encoding equivalent peptides controlling rhizobial development. Here, we review the effects of some of these peptides on rhizobia and address the question as to how and why such peptides may have evolved to enslave rhizobia and become essential for nitrogen fixation in some clades of legumes but not in others.
Introduction
About 90% of 19,000 species of legumes have nitrogen-fixing nodules, which can have diverse structures in different legume genera (Sprent, 2001). There is a profound difference in the fate of nitrogen-fixing rhizobia in different types of legume nodules because in some, the rhizobia become terminally differentiated and cannot be cultured, whereas in others, the nitrogen-fixing rhizobia can return to the free-living state. These differences are caused by plant-encoded small peptides that can induce terminal bacterial differentiation.
Symbiotic nitrogen fixation has been most studied in legumes producing root nodules with either “determinate” or “indeterminate” meristems, that are usually infected by rhizobia entering roots via plant-made infection threads formed after rhizobial-legume signaling (Oldroyd et al., 2011). The infection threads are tunnel-like structures that allow the rhizobia to grow into the region of the root in which cell proliferation has initiated nodule organogenesis (Oldroyd et al., 2011).
“Indeterminate” nodules are cylindrical with a continuously active meristem, a bit like very stubby lateral roots. Such nodules, which e.g., are formed on Pisum, Medicago spp., have an age gradient along their length with a growing meristem at the tip, an infection zone, a nitrogen-fixation zone and a so-called interzone between the infection and nitrogen-fixation zones (Figure 1). Rapid bacterial differentiation occurs in the interzone and is initiated in the proximal infection zone after the bacteria are released from infection threads and are endocytosed into the cytoplasm surrounded by a plant-made membrane. Many of the legumes forming indeterminate nodules lack an ancient inverted repeat region indicating a deep phylogenetic split with other legumes (Wojciechowski et al., 2004); consequently, they are referred to as the “Inverted-Repeat-Lacking Clade” (IRLC) of legumes.
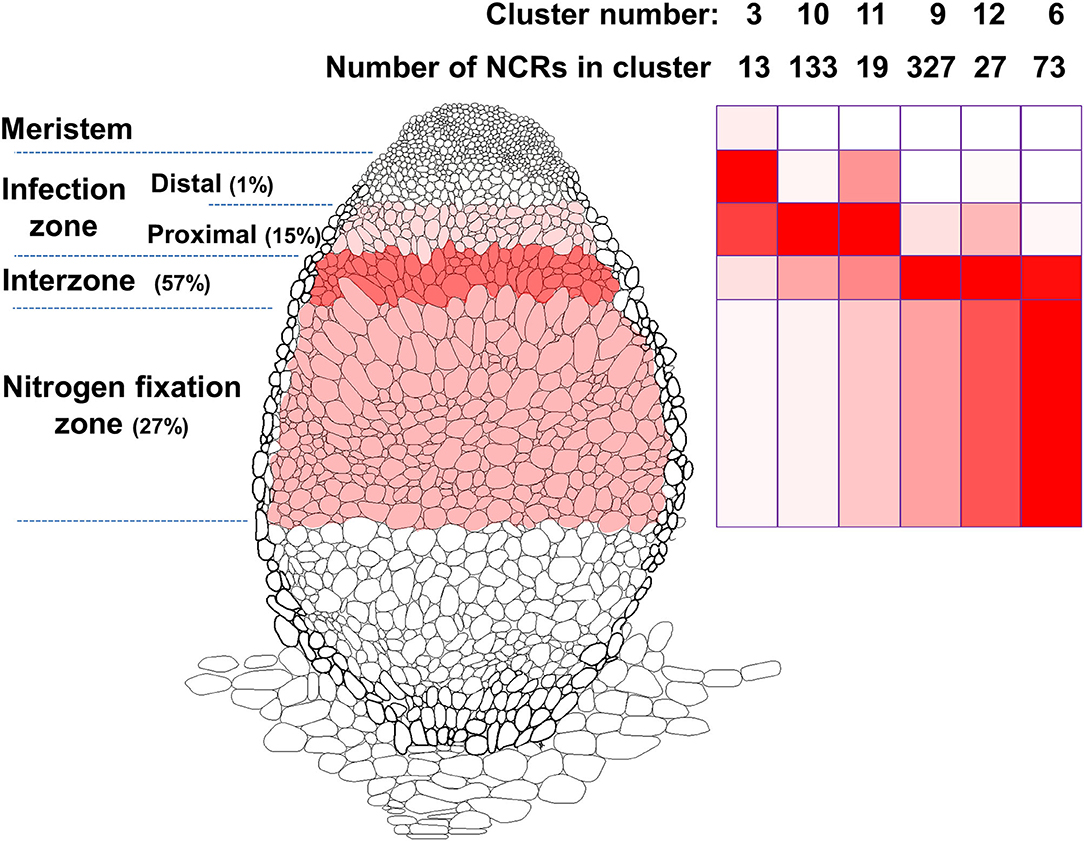
Figure 1. Model of a M. truncatula nodule illustrating the patterns of expression of NCR genes. The different stages of nodule development are named on the left. The intensity of color in the cells of these zones in the nodule reflects the relative expression of all the NCRs based on the data from Roux et al. (2014). NCRs were grouped into clusters by Roux et al. (2014) based on their expression patterns and the relative levels of expression of NCRs in the six clusters representing most of the NCRs are shown on the right panel. The color intensity illustrates their average patterns of expression in the different developmental stages with which they are aligned.
“Determinate” nodules such as those formed on soybean, Phaseolus bean and Lotus spp. are spherical and develop as a consequence of transient cell proliferation; so all infected cells in mature nodules are essentially at the same developmental stage.
NCR Peptides
Peptides Control Rhizobial Development in Some but Not Other Legumes
In the nodules of IRLC clade legumes such as pea and Medicago spp., nitrogen-fixing bacteria (bacteroids) become terminally differentiated and cannot be cultured (Mergaert et al., 2006), whereas with legumes like soybean and L. japonicus, nitrogen-fixing rhizobia do not terminally differentiate and can regrow (Gresshoff and Rolfe, 1978). Terminally differentiated nitrogen-fixing bacteroids show chromosomal endoreduplication, arrest of cell division, cell enlargement and changes in cell walls, membrane permeability, and patterns of gene expression (Batut et al., 2011; Kereszt et al., 2011; Haag et al., 2013; Kondorosi et al., 2013; Maroti and Kondorosi, 2014; Alunni and Gourion, 2016). Many of these changes are caused by plant-made small defensin-like peptides. This has been best characterized in M. truncatula in which there are two gene families, one (of about 650) encoding Nodule-specific Cysteine-Rich (NCR) peptides and one (of 24) encoding Glycine-Rich Peptides (GRPs). Similar genes have been identified in other members of the IRLC legumes (Kevei et al., 2002; Graham et al., 2004; Alunni et al., 2007; Montiel et al., 2017) but neither NCR nor GRP-encoding genes could be identified in the sequenced genomes of L. japonicus and soybean (Alunni et al., 2007). The NCRs are delivered through the plant-made membrane surrounding the bacteroids via a plant-determined secretion complex that recognizes the highly conserved N-terminal secretion signal on NCR peptides (Wang et al., 2010; Stonoha-Arther and Wang, 2018). The mature peptides (usually 35–55 residues) then enter the bacteroids, inducing the changes associated with terminal differentiation (Mergaert et al., 2006; Van de Velde et al., 2010; Tiricz et al., 2013; Farkas et al., 2014; Penterman et al., 2014). The effects of NCR peptides have been reviewed recently (Mergaert et al., 2020; Roy et al., 2020).
NCR Peptides: Activity and Rhizobial Protection Mechanisms
Most eukaryotes produce anti-microbial defensin peptides and these fall into different groups including cysteine-rich peptides that bind to microbial proteins. In such defensins, cysteine crosslinks hold together α-helical and β sheet regions such that highly variable “interactive” domains can bind efficiently to proteins (Shafee et al., 2017). NCRs are similar to, but different from most cysteine–rich defensins, usually having 4 or 6 cysteine residues rather than 8 or 10 seen in true defensins (Maroti et al., 2015). The sequences of NCR peptides are highly diverse and fall into cationic, anionic, and neutral groupings. The NMR-derived structure of NCR044 produced in Pichia pastoris revealed that it was mostly disordered, highly dynamic and internally cross-linked via two disulfide bonds that linked two antiparallel beta strands and linked one of these to a very short potential alpha helix. NCR044 entered cells of the fungal pathogen Botrytis cinerea via discrete membrane foci causing a loss of turgor and the production of reactive oxygen species (Velivelli et al., 2020). Another peptide (NCR247) can bind to multiple proteins in bacteroids (Farkas et al., 2014) and this is associated with inhibition of transcription, translation and cell division (Farkas et al., 2014; Penterman et al., 2014; Shabab et al., 2016). Cysteine crosslinking within NCR247 was essential for effects on transcription, was important but not essential for inhibition of translation and was not required for inhibition of cell division (Haag et al., 2011, 2012; Shabab et al., 2016). Rhizobia require some degree of protection against such potentially widespread disruptive effects. Some S. meliloti strains have a plasmid-encoded peptidase (HrrP), that suppresses nitrogen fixation on some Medicago species but not on others (Crook et al., 2012; Price et al., 2015). HrrP can degrade several NCR peptides resulting in premature nodule senescence and a defective symbiosis, similar to the phenotypes caused by mutations in NCR genes (see below). Another mechanism conferring protection against NCRs is mediated via rhizobial bacA (or bclA) genes that are essential for terminal bacteroid development in IRLC legumes but not in legumes in which nitrogen-fixing rhizobia can regrow (Glazebrook et al., 1993; Karunakaran et al., 2010; Maunoury et al., 2010; Guefrachi et al., 2015; Barriere et al., 2017). BacA may play a role in the transport of peptides, and the bacA mutation increased sensitivity to the stable folded form of NCR247 (Haag et al., 2011). BacA and BclA have been implied to act either by importing NCR peptides to remove them from the bacterial cell surface (a likely site of activity), or by exporting them to reduce their cytoplasmic toxicity (Haag et al., 2011, 2013; Barriere et al., 2017). Mutations affecting rhizobial cell envelope polysaccharides, inner and outer membrane proteins and transcriptional regulators affect the sensitivity to NCR247 (Arnold et al., 2017, 2018).
Different Legumes Have Widely Differing Numbers of NCR Peptides
M. truncatula has >650 NCR peptides predicted from genome sequencing (Young et al., 2011) of which about 600 were identified in RNA sequencing of nodule tissue (Roux et al., 2014). The expression of NCR peptides in nodules was reduced by added nitrate (Liese et al., 2017; Schulze et al., 2020) in parallel with nitrate-induced decreases in other nodule proteins. Mutations in specific NCR genes (Horvath et al., 2015; Kim et al., 2015), or affecting the secretion complex that delivers the peptides to bacteroids, abolish symbiotic nitrogen fixation (Van de Velde et al., 2010; Wang et al., 2010).
IRLC legumes express different numbers of NCRs; 7 were identified in Glycyrrhiza uralensis (Chinese licorice), 63 in Cicer arietinum (chickpea), 353 in Pisum sativum (pea), and 469 in M. sativa (alfalfa). The degree of bacteroid differentiation in the tested legumes correlated with the number and composition of NCR peptides (Montiel et al., 2016, 2017). The ability of the legumes to induce bacteroid swelling (an aspect of differentiation) was predicted to be acquired independently in at least five independent lineages and bacteroid morphotypes even within a single legume can vary (Oono et al., 2010). Such differentiation is not necessarily imposed on all rhizobia nodulating legumes in the IRLC clade. For example, in G. uralensis nodules Sinorhizobium fredii strain HH103 did not show the signs of bacteroid differentiation (Crespo-Rivas et al., 2016) that were seen with Mesorhizobium tianenshenense isolated from G. uralensis nodules (Montiel et al., 2016, 2017). This fits with the observation that S. fredii HH103 showed little in-vitro sensitivity to NCR peptides (Crespo-Rivas et al., 2016), that are toxic to other rhizobia (Tiricz et al., 2013). However, only cationic NCR peptides with a pI > 9.5 have so far been confirmed to have antimicrobial activity (Van de Velde et al., 2010; Ordogh et al., 2014) and G. uralensis lacks such NCRs.
Phylogenetic analyses indicated there were many legume-species-specific NCRs (Montiel et al., 2017). Such independent evolution of NCR peptides is consistent with the conclusion that there has been convergent evolution of endosymbiont differentiation driven by NCR-like peptides in the Dalbergoid clade of legumes that evolved separately from the IRLC clade (Czernic et al., 2015). However, the diversity in number and sequences of NCR peptides also point toward rapid evolution and diversification of this group of genes; the chromosomal clustering of groups of NCR genes and pseudogenes with related sequences in M. truncatula (Young et al., 2011) indicates that this diversification occurs via gene duplication. A comparison of 26 mature NCR sequences from different accessions of M. truncatula revealed a relatively high pattern of diversifying selection consistent with recent and rapid evolution leading to new functions of NCRs (Nallu et al., 2013). A comparison of NCR peptides in pea and lentil also indicated parallel evolution of NCR peptides and NCR genes arising from gene duplication (Duran et al., 2021). These observations imply that a single rhizobial genotype can encounter different NCR peptides in nodules within the same cross-inoculation group of legumes.
NCR Genes Regulate Bacteroid Development and Rhizobial Strain Discrimination
The rapid evolution and selection of NCRs implies that they confer a strong selective advantage. However, the absence of NCR peptides in several nitrogen-fixing legumes shows that NCR peptides are not required per se for symbiotic nitrogen fixation. Therefore, IRLC legumes probably use NCRs to manipulate rhizobia to optimize nitrogen fixation. The distribution of NCRs and the effects of mutations in individual NCR genes raise a few paradoxes. Firstly, NCRs have antimicrobial activity in vitro and yet the loss of individual NCR genes causes rapid senescence of bacteroids (Horvath et al., 2015; Kim et al., 2015). Secondly, although mutations in individual NCRs such as NCR169 and NCR211 cause rapid bacteroid senescence, these NCRs are not required for nitrogen fixation in those legumes lacking all NCRs. Thirdly, why should some legumes such as G. uralensis manage with few (Montiel et al., 2017) when M. truncatula has >600 nodule-expressed NCR genes?
One reason for having different NCR genes could relate to their expression pattern. Figure 1 was made based on RNA sequencing and identification of gene expression clusters of NCR genes (Roux et al., 2014). It shows the pattern of expression of about 600 NCRs in different tissues of M. truncatula nodules; analysis of these data reveals that cationic NCR peptides with antimicrobial activity are restricted to the interzone and nitrogen fixing zone. Few (if any) NCR genes are expressed in the nodule meristem. Some NCRs are expressed in the distal part of the infection zone, in which many rhizobia remain in infection threads or some have recently been endocytosed into plant cells. Most rhizobia are released from infection threads in the proximal infection zone, where about 15% of the nodule-expressed NCR genes are induced. Most (56%) NCR genes are expressed in the interzone, which corresponds to the region of the nodule in which S. meliloti bacteroids are differentiating. Thus, over 70% of the NCR genes are induced before the onset of nitrogen fixation and many of these genes are switched off in the nitrogen fixation zone. The transient pattern of transcription of many NCR genes is correlated with the increasing ploidy levels of both the differentiating rhizobia (Mergaert et al., 2006) and the plant cells as they differentiate (Nagymihaly et al., 2017).
Different NCRs may enable legumes to discriminate against some rhizobia. Rhizobial-specific nitrogen fixation appears to be common among different accessions of M. truncatula (Liu et al., 2014) and discrimination between some rhizobial strains can be attributed directly to the presence of specific NCR genes (Wang et al., 2017, 2018; Yang et al., 2017). As observed in alfalfa and M. truncatula, different NCRs can be induced by different rhizobial strains (Burghardt et al., 2017; Kang et al., 2020). Mutation of some NCR genes conferred nitrogen fixation on a strain that was unable to fix nitrogen in plants carrying the NCR gene. One of these NCR genes was expressed in the proximal infection and transitional zones (Yang et al., 2017). This is an odd stage to apply sanctions to some rhizobia, because it follows the commitment to nodule development, but precedes an assessment of effectiveness of nitrogen fixation. Perhaps the effects of some NCRs could be an undesirable consequence of NCR function. A focus of future research will be to identify the modes of action of different NCR peptides.
Discussion
Why Are There So Many NCRs in Some Legumes?
To address this, we should consider the potential roles of NCR peptides. Some cationic peptides cause membrane depolarization and disintegration in vitro (Tiricz et al., 2013). This may be due to the relatively high concentrations used; at low concentrations labeled NCR247 could enter cells without causing membrane damage but at higher concentrations increased membrane permeability. It seems unlikely that M. truncatula would produce >600 proteins primarily causing membrane permeabilization and so these effects are probably not the primary role of most NCR peptides. Tagged NCR247 (Farkas et al., 2014) pulled down proteins associated with at least nine separate complexes (Ribosomal proteins, FtsZ, GroEL, Pyruvate dehydrogenase, transaldolase, RNA polymerase, Elongation factors, a Maf-like protein, and nitrogenase). Thus, some NCRs, like antimicrobial peptides, are probably “sticky” and can bind to many other proteins, possibly inhibiting or modulating their function. If even only 10% of the 600 or so NCRs expressed in M. truncatula nodules are “sticky,” then these NCRs have the potential to modulate activity of over a 100 rhizobial proteins. Why should some legumes evolve so many NCR peptides and to what end? Three reasons for having NCRs could be: (1) The NCRs may constrain rhizobial growth in nodules. (2) The NCRs may play a role in selection against cheaters (non-nitrogen fixing bacteria that get into nodules) or to select against rhizobia with poor nitrogen fixation. (3) Plants may use NCRs to take control of bacteroid development (and possibly aspects of metabolism) to optimize nitrogen fixation. Since each would induce positive selection, it is probable that some legumes combine aspects of each.
How might this have evolved? How can we explain why the loss of a single NCR can block nitrogen fixation, whereas other legumes fix nitrogen without that specific NCR? Let us assume that an early role of NCRs was to suppress growth in nodules of cheaters (or rhizobia that are poor at N-fixation). A nodule-expressed NCR may have evolved from a defensin to suppress growth of a cheater by targeting a protein that is absent from (or sufficiently different from that of) the rhizobial endosymbiont. This could confer symbiotic benefit and subsequent gene duplication and diversification could allow the acquisition of several related NCRs targeting different cheaters. However, the observation that a single NCR can bind to several targets implies that some side effects might negatively affect the symbiosis, for example by causing accumulation of some intermediate that limits symbiotic nitrogen fixation. If this did occur, the plant could resolve the problem by losing the NCR peptide. Alternatively, it could in theory, suppress the negative side effects by acquiring another NCR peptide that could down-regulate more of the pathway thereby decreasing accumulation of the problematic intermediate. It is possible that the acquisition of several NCRs could enable the plant to control different aspects of rhizobial development (and possibly metabolism) such that there would be selection against cheaters and optimization of nitrogen fixation. However, once such a control network had developed it could easily be associated with an interdependence of NCRs, such that e.g., if the first NCR was lost (e.g., by mutation) then an imbalance in the control network due to its loss, could result in a poor or ineffective symbiosis as seen with loss of NCR 169 or NCR211. Acquisition of NCRs that benefit some rhizobial strains could be detrimental to the symbiosis established with others. Therefore, there is the potential for both positive and negative selection, which could explain the observed diversifying selection (Nallu et al., 2013).
Why Are NCR Genes Absent From Many Legumes?
If the acquisition of NCRs is of some benefit to legumes in the IRLC species, why are they not present in legumes such as soybean and Lotus spp? One possibility is that there may be a disadvantage for legumes to impose terminal differentiation on bacteroids in legumes like soybean, that lack persistent infection threads or other means of protecting some of the rhizobia from NCRs. Indeterminate nodules such as those in Medicago spp. have many persistent infection threads from which rhizobia are released, and these released rhizobia then enlarge mostly without undergoing cell division. Rhizobia within infection threads in M. truncatula do not differentiate, implying that these rhizobia are not exposed to NCRs (Mergaert et al., 2006). Therefore, the bacteria cultured from such nodules are probably those from within the infection threads, which are seen in sections of almost all infected cells.
In contrast, rhizobia released into cells of determinate nodules as in soybean do divide (Goodchild and Bergersen, 1966) and so infection threads can be much reduced and shorter compared with the complex architecture of infection threads seen e.g., in M. trunctula (Gage, 2004; Monahan-Giovanelli et al., 2006). Therefore, if a legume with determinate nodules acquired NCRs that induce terminal bacteroid differentiation, relatively few bacteria would be protected within infection threads. A successful symbiosis requires mutual benefit to both partners; if the legume were to impose constraints on bacteroid survival, such that there were insufficient rhizobia that could escape from senescing nodules, then a key aspect of the symbiosis, namely rhizobial benefit, would be lost. Thus, any advantage of controlling bacterial growth by inducing terminal differentiation could be outweighed by the disadvantage of having two few rhizobia surviving the symbiosis. It remains to be established whether, in diverse legumes, there is a correlation between protection of rhizobia within infection threads and acquisition of terminal differentiation of bacteroids induced by NCR peptides.
Future Perspectives
Key questions remain with regard to understanding the functions of NCR peptides acting alone and/or in combination. One problem is that they can bind many proteins (including other NCRs), but several of the observed interactions may be spurious and have no effect in nodules. Another issue is that the targets of NCRs may be expressed only in nitrogen-fixing bacteroids, making it difficult to use rhizobial genetics to identify their targets. So how can the analysis of the physiological functions of NCRs be addressed? One approach could be to analyze NCR function in IRLC legumes that express few NCRs, but getting the molecular genetics systems established for such legumes could be difficult. Another approach as suggested (Van de Velde et al., 2010) could be to introduce NCRs from IRLC legumes into a transformable non-IRLC legume such as L. japonicus or a promiscuous legume such as Phaseolus vulgaris. Although such a transgenic legume may be compromised for rhizobial survival in nodules, this would not be a problem with laboratory-maintained plants. Such an approach could be feasible using NCR genes from an IRLC legume that has relatively few NCRs that can induce terminal bacteroid differentiation. The technology is now in place to allow legume transformation using single constructs carrying multiple genes, so it should be possible to introduce multiple NCR genes into a transformable IRLC legume such as L. japonicus and determine effects on bacteroid differentiation and symbiotic nitrogen fixation.
Author Contributions
JAD and EK discussed the ideas and edited the manuscript prior to submission. JAD drafted the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
JAD was a recipient of a grant from the Hungarian Academy of Sciences to facilitate a working visit to the laboratory of EK. EK is a recipient of the NKFIH Frontline Research project KKP129924 and the Balzan research grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jeremy Murray for help in preparation of the figure and the colleagues of EK for helpful discussions. This article is dedicated to the memory of our friend and colleague Tomás Ruiz Argüeso.
References
Alunni, B., and Gourion, B. (2016). Terminal bacteroid differentiation in the legume-rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond. N. Phytolol. 211, 411–417. doi: 10.1111/nph.14025
Alunni, B., Kevei, Z., Redondo-Nieto, M., Kondorosi, A., Mergaert, P., and Kondorosi, E. (2007). Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol. Plant Microbe Interact. 20, 1138–1148. doi: 10.1094/MPMI-20-9-1138
Arnold, M. F. F., Penterman, J., Shabab, M., Chen, E. J., and Walker, G. C. (2018). Important late-stage symbiotic role of the Sinorhizobium meliloti exopolysaccharide succinoglycan. J. Bacteriol. 200, e00665–e00617. doi: 10.1128/JB.00665-17
Arnold, M. F. F., Shabab, M., Penterman, J., Boehme, K. L., Griffitts, J. S., and Walker, G. C. (2017). Genome-wide sensitivity analysis of the microsymbiont Sinorhizobium meliloti to symbiotically important, defensin-like host peptides. mBio 8, e01060–17. doi: 10.1128/mBio.01060-17
Barriere, Q., Guefrachi, I., Gully, D., Lamouche, F., Pierre, O., Fardoux, J., et al. (2017). Integrated roles of BclA and DD-carboxypeptidase 1 in Bradyrhizobium differentiation within NCR-producing and NCR-lacking root nodules. Sci. Rep. 7:9063. doi: 10.1038/s41598-017-08830-0
Batut, J., Mergaert, P., and Masson-Boivin, C. (2011). Peptide signalling in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 14, 181–187. doi: 10.1016/j.mib.2010.12.010
Burghardt, L. T., Guhlin, J., Chun, C. L., Liu, J. Q., Sadowsky, M. J., Stupar, R. M., et al. (2017). Transcriptomic basis of genome by genome variation in a legume-rhizobia mutualism. Mol. Ecol. 26, 6122–6135. doi: 10.1111/mec.14285
Crespo-Rivas, J. C., Guefrachi, I., Mok, K. C., Villaecija-Aguilar, J. A., Acosta-Jurado, S., Pierre, O., et al. (2016). Sinorhizobium fredii HH103 bacteroids are not terminally differentiated and show altered O-antigen in nodules of the Inverted Repeat-Lacking Clade legume Glycyrrhiza uralensis. Environ. Microbiol. 18, 2392–2404. doi: 10.1111/1462-2920.13101
Crook, M. B., Lindsay, D. P., Biggs, M. B., Bentley, J. S., Price, J. C., Clement, S. C., et al. (2012). Rhizobial plasmids that cause impaired symbiotic nitrogen fixation and enhanced host invasion. Mol. Plant Microbe Interact. 25, 1026–1033. doi: 10.1094/MPMI-02-12-0052-R
Czernic, P., Gully, D., Cartieaux, F., Moulin, L., Guefrachi, I., Patrel, D., et al. (2015). Convergent evolution of endosymbiont differentiation in Dalbergioid and Inverted Repeat-Lacking Clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 169, 1254–1265. doi: 10.1104/pp.15.00584
Duran, D., Albareda, M., Garcia, C., Marina, A.-I., Ruiz-Argueso, T., and Palacios, J.-M. (2021). Proteome analysis reveals a significant host-specific response in Rhizobium leguminosarum bv. viciae endosymbiotic cells. Mol. Cell Proteomics 20:100009. doi: 10.1074/mcp.RA120.002276
Farkas, A., Maroti, G., Durgo, H., Gyorgypal, Z., Lima, R. M., Medzihradszky, K. F., et al. (2014). Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc. Natl. Acad. Sci. U.S.A. 111, 5183–5188. doi: 10.1073/pnas.1404169111
Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. doi: 10.1128/MMBR.68.2.280-300.2004
Glazebrook, J., Ichige, A., and Walker, G. C. (1993). A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7, 1485–1497. doi: 10.1101/gad.7.8.1485
Goodchild, D. J., and Bergersen, F. J. (1966). Electron microscopy of the infection and subsequent development of soybean nodule cells. J. Bacteriol. 92, 204–213. doi: 10.1128/JB.92.1.204-213.1966
Graham, M. A., Silverstein, K. A., Cannon, S. B., and VandenBosch, K. A. (2004). Computational identification and characterization of novel genes from legumes. Plant Physiol. 135, 1179–1197. doi: 10.1104/pp.104.037531
Gresshoff, P. M., and Rolfe, B. G. (1978). Viability of Rhizobium bacteroids isolated from soybean nodule protoplasts. Planta 142, 329–333. doi: 10.1007/BF00385085
Guefrachi, I., Pierre, O., Timchenko, T., Alunni, B., Barriere, Q., Czernic, P., et al. (2015). Bradyrhizobium BclA is a peptide transporter required for bacterial differentiation in symbiosis with Aeschynomene legumes. Mol. Plant Microbe Interact. 28, 1155–1166. doi: 10.1094/MPMI-04-15-0094-R
Haag, A. F., Arnold, M. F., Myka, K. K., Kerscher, B., Dall'Angelo, S., Zanda, M., et al. (2013). Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol. Rev. 37, 364–383. doi: 10.1111/1574-6976.12003
Haag, A. F., Baloban, M., Sani, M., Kerscher, B., Pierre, O., Farkas, A., et al. (2011). Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 9:e1001169. doi: 10.1371/journal.pbio.1001169
Haag, A. F., Kerscher, B., Dall'Angelo, S., Sani, M., Longhi, R., Baloban, M., et al. (2012). Role of cysteine residues and disulfide bonds in the activity of a legume root nodule-specific, cysteine-rich peptide. J. Biol. Chem. 287, 10791–10798. doi: 10.1074/jbc.M111.311316
Horvath, B., Domonkos, A., Kereszt, A., Szucs, A., Abraham, E., Ayaydin, F., et al. (2015). Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. U.S.A. 112, 15232–15237. doi: 10.1073/pnas.1500777112
Kang, W. J., Jiang, Z. H., Chen, Y. G., Wu, F., Liu, C., Wang, H. F., et al. (2020). Plant transcriptome analysis reveals specific molecular interactions between alfalfa and its rhizobial symbionts below the species level. BMC Plant Biol. 20:293. doi: 10.1186/s12870-020-02503-3
Karunakaran, R., Haag, A. F., East, A. K., Ramachandran, V. K., Prell, J., James, E. K., et al. (2010). BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 192, 2920–2928. doi: 10.1128/JB.00020-10
Kereszt, A., Mergaert, P., and Kondorosi, E. (2011). Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims? Mol. Plant Microbe Interact. 24, 1300–1309. doi: 10.1094/MPMI-06-11-0152
Kevei, Z., Vinardell, J. M., Kiss, G. B., Kondorosi, A., and Kondorosi, E. (2002). Glycine-rich proteins encoded by a nodule-specific gene family are implicated in different stages of symbiotic nodule development in Medicago spp. Mol. Plant Microbe Interact. 15, 922–931. doi: 10.1094/MPMI.2002.15.9.922
Kim, M., Chen, Y. H., Xi, J. J., Waters, C., Chen, R. J., and Wang, D. (2015). An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 15238–15243. doi: 10.1073/pnas.1500123112
Kondorosi, E., Mergaert, P., and Kereszt, A. (2013). A paradigm for endosymbiotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 67, 611–628. doi: 10.1146/annurev-micro-092412-155630
Liese, R., Schulze, J., and Cabeza, R. A. (2017). Nitrate application or P deficiency induce a decline in Medicago truncatula N2-fixation by similar changes in the nodule transcriptome. Sci. Rep. 7:46264. doi: 10.1038/srep46264
Liu, J. G., Yang, S. M., Zheng, Q. L., and Zhu, H. Y. (2014). Identification of a dominant gene in Medicago truncatula that restricts nodulation by Sinorhizobium memiloti strain Rm41. BMC Plant Biol. 14:167. doi: 10.1186/1471-2229-14-167
Maroti, G., Downie, J. A., and Kondorosi, E. (2015). Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Curr. Opin. Plant Biol. 26, 57–63. doi: 10.1016/j.pbi.2015.05.031
Maroti, G., and Kondorosi, E. (2014). Nitrogen-fixing Rhizobium-legume symbiosis: are polyploidy and host peptide-governed symbiont differentiation general principles of endosymbiosis? Front. Microbiol. 5:326. doi: 10.3389/fmicb.2014.00326
Maunoury, N., Redondo-Nieto, M., Bourcy, M., Van de Velde, W., Alunni, B., Laporte, P., et al. (2010). Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS ONE 5:e9519. doi: 10.1371/journal.pone.0009519
Mergaert, P., Kereszt, A., and Kondorosi, E. (2020). Gene expression in nitrogen-fixing symbiotic nodule cells in Medicago truncatula and other nodulating plants. Plant Cell 32, 42–68. doi: 10.1105/tpc.19.00494
Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235. doi: 10.1073/pnas.0600912103
Monahan-Giovanelli, H., Pinedo, C. A., and Gage, D. J. (2006). Architecture of infection thread networks in developing root nodules induced by the symbiotic bacterium Sinorhizobium meliloti on Medicago truncatula. Plant Physiol. 140, 661–670. doi: 10.1104/pp.105.072876
Montiel, J., Downie, J. A., Farkas, A., Bihari, P., Herczeg, R., Balint, B., et al. (2017). Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. U.S.A. 114, 5041–5046. doi: 10.1073/pnas.1704217114
Montiel, J., Szucs, A., Boboescu, I. Z., Gherman, V. D., Kondorosi, E., and Kereszt, A. (2016). Terminal bacteroid differentiation is associated with variable morphological changes in legume species belonging to the inverted repeat-lacking clade. Mol. Plant Microbe Interact. 29, 210–219. doi: 10.1094/MPMI-09-15-0213-R
Nagymihaly, M., Veluchamy, A., Gyorgypal, Z., Ariel, F., Jegu, T., Benhamed, M., et al. (2017). Ploidy-dependent changes in the epigenome of symbiotic cells correlate with specific patterns of gene expression. Proc. Natl. Acad. Sci. U.S.A. 114, 4543–4548. doi: 10.1073/pnas.1704211114
Nallu, S., Silverstein, K. A. T., Samac, D. A., Bucciarelli, B., Vance, C. P., and VandenBosch, K. A. (2013). Regulatory patterns of a large family of defensin-like genes expressed in nodules of Medicago truncatula. PLoS ONE 8:e60355. doi: 10.1371/journal.pone.0060355
Oldroyd, G. E. D., Murray, J. D., Poole, P. S., and Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Oono, R., Schmitt, I., Sprent, J. I., and Denison, R. F. (2010). Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. N. Phytol. 187, 508–520. doi: 10.1111/j.1469-8137.2010.03261.x
Ordogh, L., Voros, A., Nagy, I., Kondorosi, E., and Kereszt, A. (2014). Symbiotic plant peptides eliminate Candida albicans both in vitro and in an epithelial infection model and inhibit the proliferation of immortalized human cells. Biomed. Res. Int. 2014, 320796. doi: 10.1155/2014/320796
Penterman, J., Abo, R. P., De Nisco, N. J., Arnold, M. F. F., Longhi, R., Zanda, M., et al. (2014). Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc. Natl. Acad. Sci. U.S.A. 111, 3561–3566. doi: 10.1073/pnas.1400450111
Price, P. A., Tanner, H. R., Dillon, B. A., Shabab, M., Walker, G. C., and Griffitts, J. S. (2015). Rhizobial peptidase HrrP cleaves host-encoded signaling peptides and mediates symbiotic compatibility. Proc. Natl. Acad. Sci. U.S.A. 112, 15244–15249. doi: 10.1073/pnas.1417797112
Roux, B., Rodde, N., Jardinaud, M. F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. doi: 10.1111/tpj.12442
Roy, P., Achom, M., Wilkinson, H., Lagunas, B., and Gifford, M. L. (2020). Symbiotic outcome modified by the diversification from 7 to over 700 nodule-specific cysteine-rich peptides. Genes 11:348. doi: 10.3390/genes11040348
Schulze, J., Liese, R., Ballesteros, G., Casieri, L., Salinas, G., and Cabeza, R. A. (2020). Ammonium acts systemically while nitrate exerts an additional local effect on Medicago truncatula nodules. Plant Sci. 292:110383. doi: 10.1016/j.plantsci.2019.110383
Shabab, M., Arnold, M. F., Penterman, J., Wommack, A. J., Bocker, H. T., Price, P. A., et al. (2016). Disulfide cross-linking influences symbiotic activities of nodule peptide NCR247. Proc. Natl. Acad. Sci. U.S.A. 113, 10157–10162. doi: 10.1073/pnas.1610724113
Shafee, T. M. A., Lay, F. T., Phan, T. K., Anderson, M. A., and Hulett, M. D. (2017). Convergent evolution of defensin sequence, structure and function. Cell. Mol. Life Sci. 74, 663–682. doi: 10.1007/s00018-016-2344-5
Stonoha-Arther, C., and Wang, D. (2018). Tough love: accommodating intracellular bacteria through directed secretion of antimicrobial peptides during the nitrogen-fixing symbiosis. Curr. Opin. Plant Biol. 44, 155–163. doi: 10.1016/j.pbi.2018.04.017
Tiricz, H., Szucs, A., Farkas, A., Pap, B., Lima, R. M., Maroti, G., et al. (2013). Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl. Env. Microbiol. 79, 6737–6746. doi: 10.1128/AEM.01791-13
Van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057
Velivelli, S. L. S., Czymmek, K. J., Li, H., Shaw, J. B., Buchko, G. W., and Shah, D. M. (2020). Antifungal symbiotic peptide NCR044 exhibits unique structure and multifaceted mechanisms of action that confer plant protection. Proc. Natl. Acad. Sci. U.S.A. 117, 16043–16054. doi: 10.1073/pnas.2003526117
Wang, D., Griffitts, J., Starker, C., Fedorova, E., Limpens, E., Ivanov, S., et al. (2010). A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327, 1126–1129. doi: 10.1126/science.1184096
Wang, Q., Liu, J. G., Li, H., Yang, S. M., Kormoczi, P., Kereszt, A., et al. (2018). Nodule-specific cysteine-rich peptides negatively regulate nitrogen-fixing symbiosis in a strain-specific manner in Medicago truncatula. Mol. Plant Microbe Interact. 31, 240–248. doi: 10.1094/MPMI-08-17-0207-R
Wang, Q., Yang, S. M., Liu, J. G., Terecskei, K., Abraham, E., Gombar, A., et al. (2017). Host-secreted antimicrobial peptide enforces symbiotic selectivity in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114, 6854–6859. doi: 10.1073/pnas.1700715114
Wojciechowski, M. F., Lavin, M., and Sanderson, M. J. (2004). A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91, 1846–1862. doi: 10.3732/ajb.91.11.1846
Yang, S. M., Wang, Q., Fedorova, E., Liu, J. G., Qin, Q. L., Zheng, Q. L., et al. (2017). Microsymbiont discrimination mediated by a host-secreted peptide in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 114, 6848–6853. doi: 10.1073/pnas.1700460114
Keywords: cysteine-rich peptides, nitrogen fixation, NCR, Rhizobium, Sinorhizobium, Medicago, Lotus
Citation: Downie JA and Kondorosi E (2021) Why Should Nodule Cysteine-Rich (NCR) Peptides Be Absent From Nodules of Some Groups of Legumes but Essential for Symbiotic N-Fixation in Others? Front. Agron. 3:654576. doi: 10.3389/fagro.2021.654576
Received: 16 January 2021; Accepted: 25 May 2021;
Published: 22 June 2021.
Edited by:
Juan Imperial, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Zhenhua Zhang, Hunan Agricultural University, ChinaEsther Menendez, University of Evora, Portugal
Copyright © 2021 Downie and Kondorosi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Allan Downie, YWxsYW4uZG93bmllJiN4MDAwNDA7amljLmFjLnVr
 J. Allan Downie
J. Allan Downie Eva Kondorosi
Eva Kondorosi