
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Agron. , 18 March 2021
Sec. Pest Management
Volume 3 - 2021 | https://doi.org/10.3389/fagro.2021.652981
This article is part of the Research Topic New Applications of Insecticidal RNAi View all 9 articles
Since the discovery of RNA interference (RNAi) in the nematode worm Caenorhabditis elegans in 1998 by Fire and Mello et al., strides have been made in exploiting RNAi for therapeutic applications and more recently for highly selective insect pest control. Although triggering mRNA degradation in insects through RNAi offers significant opportunities in crop protection, the application of environmental naked dsRNA is often ineffective in eliciting a RNAi response that results in pest lethality. There are many possible reasons for the failed or weak induction of RNAi, with predominant causes being the degradation of dsRNA in the formulated pesticide, in the field or in the insect once ingested, poor cuticular and oral uptake of the nucleic acid and sometimes the lack of an innate strong systemic RNAi response. Therefore, in the last 10 years significant research effort has focused on developing methods for the protection and delivery of environmental dsRNA to enable RNAi-induced insect control. This review focuses on the design and synthesis of vectors (vehicles that are capable of carrying and protecting dsRNA) that successfully enhance mRNA degradation via the RNAi machinery. The majority of solutions exploit the ability of charged polymers, both synthetic and natural, to complex with dsRNA, but alternative nanocarriers such as clay nanosheets and liposomal vesicles have also been developed. The various challenges of dsRNA delivery and the obstacles in the development of well-designed nanoparticles that act to protect the nucleic acid are highlighted. In addition, future research directions for improving the efficacy of RNA-mediated crop protection are anticipated with inspiration taken from polymeric architectures constructed for RNA-based therapeutic applications.
The world population is increasing rapidly, and food supply must equally rise to meet demand. It has been estimated however that ~18% of total crop production is destroyed by insect pests, with wheat and cotton hit the hardest with losses of up to 50 and 80%, respectively. This induces a financial penalty with the global crop loss due to insect pests valued at an estimated US$470 billion (Oerke, 2006; Chakraborty and Newton, 2011; Borel, 2017; Sharma et al., 2017). In addition, the effectiveness of synthetic chemical pesticides introduced since the 1940s is continually being undermined by the appearance of resistant insect populations. Indeed, incidences of resistance to one or more synthetic pesticide has been reported for over 586 arthropod species, with the Arthropod Pesticide Resistance Database1 actively growing at an alarming rate (Tabashnik et al., 2014; Sparks and Nauen, 2015; Mota-Sanchez and Wise, 2017). Consequently, growers are increasingly relying on Integrated Pesticide Management programmes in order to prolong the effectiveness of their arsenal of chemical pesticides (Borel, 2017). Moreover, the lack of specificity of synthetic pesticides is a critical issue, with increasing reports of the detrimental impacts of these chemicals on beneficial pollinator species, predators, and other animals occurring through bioaccumulation or leaching into the environment (Windley et al., 2012; Nakasu et al., 2014; Frank and Tooker, 2020). Thus, a variety of biological-based insecticides, such as the bacterial toxins of Bacillus thuringiensis (Bt) and Saccharopolyspora spinosa, have been developed as alternatives to the synthetic chemical pesticides. These have modes of action quite distinct from the traditional synthetic chemicals, and whilst this minimises the risk of cross-resistance, field resistance has been widely reported for both biologicals (McGaughey, 1994; Scott, 2008; Bravo et al., 2011; Borel, 2017; Abbas, 2018; Santos and Pereira, 2020). The evolution of resistance of pest insect species to RNAi-based formulations also needs to be considered appropriately when developing delivery vectors. Introducing a large selection pressure could lead to mutations causing changes to core RNAi machinery (Scott et al., 2013; Vogel et al., 2019; Christiaens et al., 2020; Yan et al., 2020). For example, a population of western corn rootworm, Diabrotica virgifera virgifera, with resistance to DvSnf7 dsRNA showed cross-resistance to other dsRNAs (Khajuria et al., 2018). Future research must focus on specific management strategies to slow the development of this resistance, particularly in species that are already refractory to RNAi.
In this context, RNA interference (RNAi) offers a promising additional solution capable of generating biological insecticides with high selectivity toward target pest species. Triggering RNAi is a well-controlled approach that can target pest-specific transcripts and stop specific protein production with low risk of off-target toxicity (Price and Gatehouse, 2008; Huvenne and Smagghe, 2010).
RNAi was first discovered in animals by Fire et al. (1998) and has since been widely adopted in the post-genomic era for functional gene analysis. RNAi is elicited when exogenous double stranded-RNA (dsRNA) is internalised by cells and is cleaved by cytoplasmic ribonuclease III enzyme (Dicer-2) into 21–25 nucleotide-long duplexes called short-interfering-RNA (siRNA), which bind to the RNA-induced silencing complex (RISC). Proteins of the complex unwind the loaded duplex into its sense (passenger) and antisense (guide) strands. The passenger strand is degraded, whilst the guide is used to locate the complimentary messenger-RNA (mRNA) sequence. Upon recognition, the mRNA is cleaved by the RISC, leaving the fragments to be subsequently degraded by cytoplasmic nucleases, see Figure 1. The degradation of the mRNA and failure to translate to protein thus results in sequence-specific effects. Within the field of insect control, exogenously supplied dsRNA can result in insect mortality if the target protein has an essential function. The likelihood of mortality is significantly increased if the RNAi response is systemic via the transportation of the RNAi signal throughout the insect's body. However, not all insect species generate systemic RNAi responses to exogenously applied dsRNA. The reason for species differences in the spreading of RNAi between insect tissues is not fully understood (Fire et al., 1998; Huvenne and Smagghe, 2010; Smith et al., 2011; Katoch et al., 2013; Joga et al., 2016; Kunte et al., 2020; Liu et al., 2020).
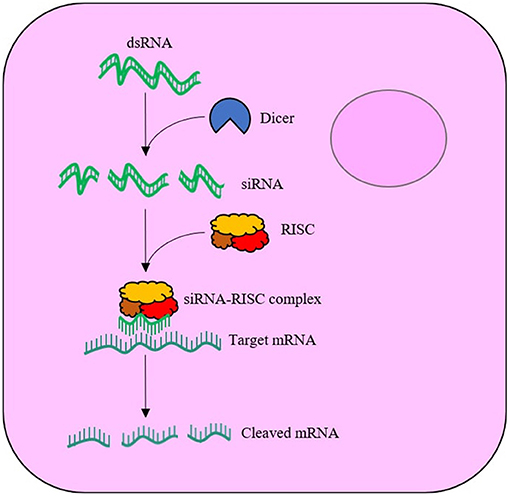
Figure 1. Schematic of the RNA interference (RNAi) mechanism within a cell. The exogenously supplied double stranded-RNA (dsRNA) enters the cell cytoplasm, where the enzyme, Dicer-2, cleaves the dsRNA into short interfering-RNA (siRNA) duplexes. The siRNA binds to the RNA-induced silencing complex (RISC), with the antisense (guide) strand maintained. The guide strand is used to locate complimentary messenger-RNA (mRNA), which is cleaved by RISC upon recognition.
The exploitation of RNAi for crop pest control requires efficient uptake of exogenously supplied dsRNA by the target insect. Various strategies have been employed in research environments to administer the nucleic acid to crop pest insects, ranging from direct injection to the in planta expression of dsRNA. The injection or soaking of the insect in a solution of dsRNA have often been shown to be more effective in comparison to topical application and oral administration (Whyard et al., 2009; Scott et al., 2013; Yu et al., 2013; Kang et al., 2018). However, there are some examples of the successful application of exogenous dsRNA by topical or oral administration in certain insect species (Baum et al., 2007; Pridgeon et al., 2008; Whyard et al., 2009, 2015; Killiny et al., 2014; Galdeano et al., 2017). Use of dsRNA as an insecticide for crop protection requires deposition by foliar or soil spraying followed by uptake either by contact with the insect's cuticle or by ingestion by the feeding insect (Webb and Green, 1945; Joga et al., 2016; Mamta and Rajam, 2017).
The failure to elicit a strong RNAi response by feeding or topical application of naked dsRNA in some insect species, despite success when administered by injection, highlights limitations in developing dsRNA as a general pest control solution (Whyard et al., 2009; Taning et al., 2016). The ineffective induction of RNAi by environmental naked dsRNA is often due to degradation of dsRNA (Christiaens et al., 2014; Dubelman et al., 2014; Shukla et al., 2016; Singh et al., 2017; Peng et al., 2018; Whitfield et al., 2018; Cooper et al., 2019; Zhu and Palli, 2020). There are several possible barriers to an effective insect response, as summarised in Figure 2. The most likely option for delivery of a formulated RNA-based insecticide is through sprayed deposition onto the crop followed by ingestion by the insect pest. Thus, it is paramount to take into account the requirements of the method of application in designing a delivery solution for the dsRNA, in particular the following considerations for successful pest control.
i. Prior to application, the insecticide must be designed as a dispersion with a homogenous distribution of dsRNA. Thus, dsRNA vectors (or carriers) must retain their colloidal stability throughout the various stages of the application, from the storage in the formulated pesticide, the dilution in water upon application, the spraying in the field, the deposition and drying on the crop and finally, during transport through the digestive tract of the insect and cellular uptake. In addition, the vectors should have low non-target cytotoxicity.
ii. Upon application of the formulation, the nucleic acid must survive deposition onto leaves or soil by withstanding harsh physical conditions and environmental biodegradation. Exogenously applied dsRNA can be fully degraded in living soil within 3 days (Dubelman et al., 2014; Whitfield et al., 2018).
iii. Following ingestion by the insect, dsRNA must survive attack by enzymatic nucleases and pH changes within the intestinal gut lumen before cellular internalisation by first traversing the inert peritrophic matrix that lines the gut and then the midgut epithelial cell membrane. The means of cellular uptake from the gut lumen appears to vary depending on species and might involve one or more mechanism, such as passive transport via a transmembrane protein, pinocytosis or receptor-mediated endocytosis (Cooper et al., 2019; Zhu and Palli, 2020).
iv. If internalised by endocytosis, the dsRNA must escape the endosomal compartment prior to lysosomal degradation (Yoon et al., 2017; Gurusamy et al., 2020b) before dsRNA may take part in the RNAi mechanism.
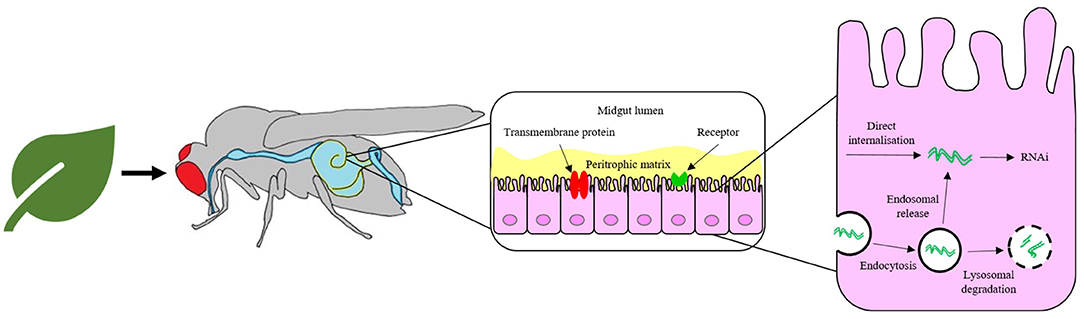
Figure 2. Pathway for the delivery of naked RNA to insect gut cells via foliar or soil application and oral feeding of the insect. The barriers to efficient delivery include metabolic instability in the gut lumen, inefficient uptake by gut epithelial cells and poor release of dsRNA from endosomes.
The species variability in the efficacy of exogenous dsRNA to elicit RNAi effects is well-documented (Terenius et al., 2011; Palli, 2014; Cooper et al., 2019; Zhu and Palli, 2020). Insects of the order Coleoptera, e.g., the red flour beetle Tribolium castaneum, respond efficiently to both injection and oral feeding of dsRNA. In contrast, many lepidopteran and dipteran species only respond to naked dsRNA through injection (Baum et al., 2007; Whyard et al., 2009; Terenius et al., 2011; Zhu et al., 2011). The life stage of the insect and the experimental protocol used have also been observed to impact on RNAi efficiency. Why there are these differences between species and at different stages of the life-cycle is not fully understood, but some contributing factors have been identified (Wynant et al., 2012, 2014; Christiaens and Smagghe, 2014; Christiaens et al., 2014, 2020; Joga et al., 2016; Mulot et al., 2016; Singh et al., 2017; Cooper et al., 2019; Vogel et al., 2019; Liu et al., 2020).
The degradation of dsRNA by RNases within the intestinal gut lumen, saliva or haemolymph of the insect species will reduce RNAi efficiency. Increased RNase activity at a particular life stage or differences in the chemical environment (e.g., gut pH) can explain some of the variability seen in the dsRNA responses of insects. Lepidopterans, in particular, can be refractory to RNAi via both injection and feeding with recent research by Singh et al. showing reduced processing of dsRNA to siRNA within this insect order (Shukla et al., 2016; Singh et al., 2017). The up56 gene, encoding for an additional RNAi efficiency-related nuclease (REase), could be responsible for the decrease in RNAi efficiency (Guan et al., 2018). In addition, ineffective endosomal escape within cells has also been suggested to contribute to lower RNAi efficiency in Lepidoptera (Shukla et al., 2016; Wang et al., 2016; Guan et al., 2018).
An in-depth discussion of the factors that contribute to varying RNAi efficiency in different insect species can be found in several recent review articles (Cooper et al., Scott et al., Terenius et al., Liu et al.) that also recommend strategies, such as the use of nanocarriers for the delivery and uptake of dsRNA, to improve the RNAi response for insect pest control (Zhou et al., 2008; Terenius et al., 2011; Scott et al., 2013; Mongelli and Saleh, 2016; Cooper et al., 2019; Liu et al., 2020).
The exciting potential of exogenous dsRNA to provide a new generation of highly selective insect control agents has focused attention on the development of delivery systems to counter the physical and metabolic instability of the nucleic acid in the field, which can result in the complete degradation of dsRNA within days (Dubelman et al., 2014; Whitfield et al., 2018). In the last decade, many novel nanoparticle delivery vehicles (also known as vectors) for dsRNA have been developed, including self-assembled block and branched copolymer nanoparticles, dendrimers, inorganic nanoparticles, and natural product-based nanoparticles, see Figure 3. It is worth noting at this stage that the majority of dsRNA delivery solutions utilise the electrostatic interaction between the anionic phosphate backbone of dsRNA and some cationic character expressed on the nanoparticle vector to assemble a system capable of protecting and transporting the dsRNA.
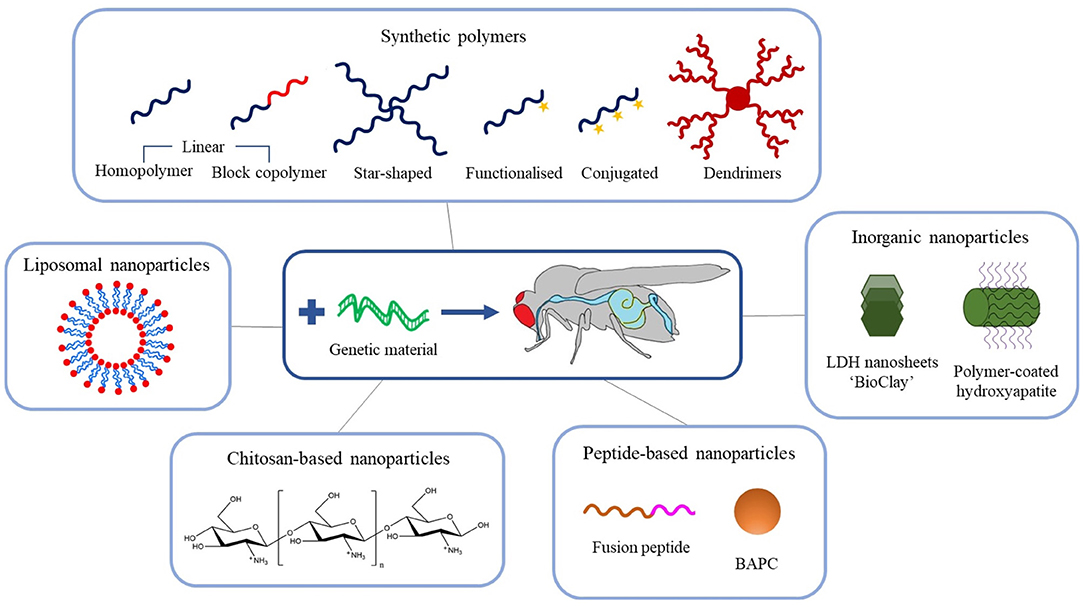
Figure 3. Schematic overview of the recent advances in engineered nanoparticles for RNA-mediated crop protection strategies.
The focus of this review is crop protection through the control of pest insect species. Whilst the majority of applications cited in this review target pest insect species specifically, some examples of dsRNA application to plant pathogens have been included to provide insight for potential future avenues for insecticidal applications. Further details of these systems are discussed in the following sections.
Synthetic polymeric delivery vectors often require well-defined polymer chains with precise molecular weight, narrow molar mass dispersity (Ð) and defined (co)polymer architecture (e.g., homo, block, star, or graft polymers). They are also tolerant to chemical functionalities, enabling the polymerisation of functional monomers without the need for protecting groups (Matyjaszewski, 2012). These highly desirable credentials have enabled not only the precise engineering of functional polymeric vectors; but also the ability to conduct highly systematic studies of the influence of the polymer construct on delivery efficiency.
The simplest polymeric architectures used for the delivery of dsRNA are linear homopolymers, such as poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA), poly(ethylene imine) (PEI), and poly(L-lysine) (PLL) (Wu and Wu, 1987; Boussif et al., 1995; Cherng et al., 1996; Klemm et al., 1998; Lungwitz et al., 2005; Agarwal et al., 2012). These polymers only comprise one monomer, which incorporates amine groups within the polymer chain backbone or on pendant groups which are positively charged below their pKa [e.g., 7.4–7.8 for PDMAEMA (van de Wetering et al., 1998), 8.2–9.5 for PEI (von Harpe et al., 2000; Ziebarth and Wang, 2010), and 9–11 for PLL (Choi et al., 2015)]. This cationic character facilitates complexation to the anionic dsRNA phosphate backbone through electrostatic attraction of the two macromolecules. This process is entropically favourable due to the release of counterions (Wu and Wu, 1987; Boussif et al., 1995; Cherng et al., 1996).
To date, linear homopolymers have not been efficient in their complexation and protection of dsRNA, with the additional issue of non-target cytotoxicity, which is unfortunate given their low cost. In particular, PEI and PDMAEMA homopolymer delivery vehicles are often subject to low cellular internalisation efficiency and can be highly toxic to mammalian cells (Godbey et al., 2001; Jones et al., 2004). This cytotoxicity might result from polymer interaction with cellular membranes and pore formation, leading to cell death (van de Wetering et al., 1997; Fischer et al., 2003; Prevette et al., 2010; Synatschke et al., 2011). Thus, to address these negative issues, many other, more complex, polymeric architectures, and functional groups have been designed to enhance RNA stability and efficiency of cellular uptake whilst reducing cytotoxicity. For example, the use of additional monomers to form diblock or triblock copolymers, or a change of architecture to form star-shaped polymers have been investigated as vehicles for effective delivery of dsRNA to a variety of insect species (Silva et al., 2010; Parsons et al., 2016, 2018; Christiaens et al., 2018; Cook et al., 2018; Whitfield et al., 2018). Variation in molecular weight, charge density, pKa, and ionic strength, that can be achieved by simply altering the chemistry of the polymeric vectors, have been considered in order to overcome the aforementioned delivery issues, with varying degrees of success (van de Wetering et al., 1997; Arigita et al., 1999; Cherng et al., 1999; Godbey et al., 1999; Kunath et al., 2003; Layman et al., 2009). For example, whilst higher molecular weight homopolymers complexed with dsRNA (or DNA) exhibit greater transfection efficiency, the cytotoxicity of the polyplexes increases with the increase in molecular weight (Layman et al., 2009). In the next sections, we describe recent advances in development and application of these various architectures as summarised in Table 1.
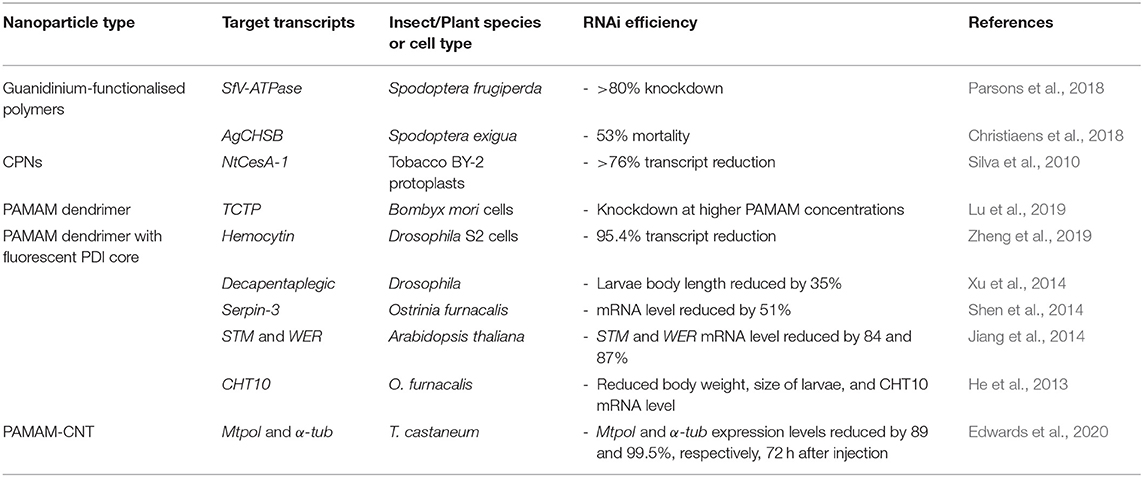
Table 1. Polymeric nanoparticles for RNA-mediated crop protection from the last 10 years that conducted in vitro or in vivo assay with the polyplexes.
More complex polymer architectures, such as branched polymers, have been investigated with the aim of improving transfection and RNAi efficiency, and reducing non-target cytotoxicity levels.
Synatschke et al. used ATRP to highlight the difference between linear PDMAEMA and 3 and 5-arm PDMAEMA star-shaped equivalents in terms of their non-target cytotoxicity and transfection efficiency in mammalian cells. Linear, 3- and 5-arm polymers were prepared with Ð ranging from 1.09 to 1.80. In this study, it was evident that as the complexity of branching increases (i.e. number of arms), cytotoxicity decreases as a result of the majority of the complexing nitrogen atoms being contained within the dense core of the star-shaped polymer. Consequently, interaction of the more branched polymers with membrane phospholipids and the resulting destabilisation of cell membranes is reduced. Transfection efficiency was comparable between the linear and star-shaped polymers, however a higher amount of polymer was required to fully stabilise the DNA in the case of the 3 and 5-arm polymers, as fewer nitrogen complexing moieties are available (Synatschke et al., 2011).
Whitfield et al. similarly compared the protection afforded to dsRNA by linear and star-shaped polymers, using poly(2-(dimethylamino)ethyl acrylate) (PDMAEA), instead. The aim of this work was to mitigate environmental biodegradation of dsRNA in soil (Dubelman et al., 2014). For this purpose, linear and 4-arm star-shaped PDMAEA were synthesised by ATRP with Ð-values of 1.18 and 1.14, respectively (Whitfield et al., 2018). The complexed dsRNA was more persistent in living soil, delaying metabolic degradation by an additional 7 and 14 days with linear and star-shaped PDMAEA, respectively. Molecular dynamics (MD) simulations indicated stronger binding of star-shaped PDMAEA with DNA, with more compact polyplexes formed from efficient bending around DNA. The reduced surface area of the more compact star-shaped PDMAEA polyplexes, limits contact of the nucleic acid with degrading nucleases, hence enhanced protection and slower release of dsRNA. In a different study, linear and 4-arm star-shaped PDMAEA have been synthesised by RAFT polymerisation, with Ð of 1.14–1.15 for the linear and 1.35–1.38 for the star-shaped polymers (Liao et al., 2017). These polymers were complexed with DNA and used to transfect mouse 3T3 fibroblast cells. The 4-arm star-shaped PDMAEA formed more compact DNA polyplexes, with less polymer required for equivalent stable binding in comparison to polyplexes made with the linear polymer (Liao et al., 2017; Whitfield et al., 2018).
It is important to note that PDMAEA, in contrast to PDMAEMA, is prone to self-catalysed hydrolysis, see Figures 4, 5 (van de Wetering et al., 1998; Cotanda et al., 2013; Schönemann et al., 2018), resulting from a phenomenon explained by Ros et al. (2020). This hydrolysis has been linked to more efficient release of complexed dsRNA for incorporation into the RNAi machinery and thus inducing the required effects more easily (Cook et al., 2018; Cooper et al., 2019). However, a potential drawback is that the complexes may be prone to premature release of dsRNA, prior to reaching the cells where RNAi would occur.
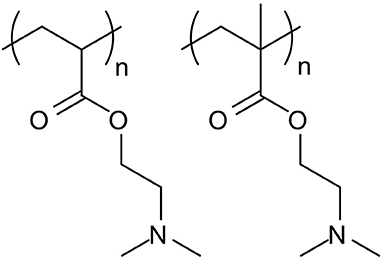
Figure 4. Structures of PDMAEA (left), used by Whitfield et al. (2018) in comparison to PDMAEMA (right), used by Synatschke et al. (2011). The additional methyl group on the PDMAEMA polymer backbone leads to a significant lowering of the polymer hydrolysis rate, which results in lower biodegradability but could potentially be used to tailor the release of the dsRNA at the point of action.
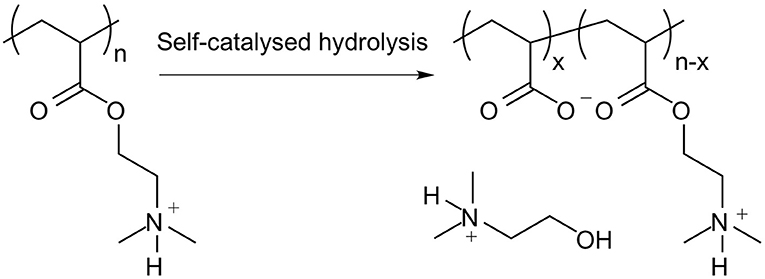
Figure 5. Outcome of the self-catalysed hydrolysis of PDMAEA to polyacrylic acid and 2-dimethylaminoethanol.
The recognised potential of RNAi in therapeutics has provided much of the impetus in the design and development of functionalised polymers as vectors of dsRNA. For example, recent developments in this field have included pH-responsive polymers that undergo a pH-dependent conformational change that releases the dsRNA from the complex in the acidic environment of the endosome compartment of the cell, over the physiological to endosomal pH transition (Auguste et al., 2008; Lin et al., 2008; Convertine et al., 2010; Manganiello et al., 2012; Cao et al., 2019; Liechty et al., 2019). However, the targeted pH transition range may not be applicable to insect control strategies. Lepidopteran pests, such as Spodoptera (armyworm) species, have an intestinal gut lumen pH of 10 to 11, which can increase dsRNA instability in the insect gut. The development of polymers designed to protect dsRNA in this strongly alkaline environment requires special consideration of the effect of pH on polymer complexation.
Guanidinium-based polymeric vectors have been designed for the protection of dsRNA over the alkaline pH range found in the gut of lepidopteran pests. These polymers bear similarity to arginine-rich cell-penetrating peptides (CPPs) that aid endocytic passage through cell membranes, as well as the escape of RNA from endosomes (Vivès et al., 2008; Cermenati et al., 2011; Chen et al., 2012; Gillet et al., 2017). The cationic homopolymer poly(N-(3-guanidinopropyl)methacrylamine) (PGPMA) made by RAFT polymerisation has a pKa of 12.5, ensuring protonation of the guanidinium functional groups even in the alkaline gut. Complexation of PGPMA with dsRNA was shown to occur at pH 10 with more compact polyplexes than at pH 7.4, see Figure 6. PGPMA/dsRNA complexes induced suppression of the CDC27 transcript in Sf9 cells after 48 h incubation, with ~90% reduction in CDC27 mRNA. Feeding second and third instar larvae of the fall armyworm, Spodoptera frugiperda, on PGMA/CDC27-dsRNA polyplex supplemented diet for 7 days, resulted in 80% transcript reduction, with ~30% larval/pupal mortality after 29 days (Parsons et al., 2018).
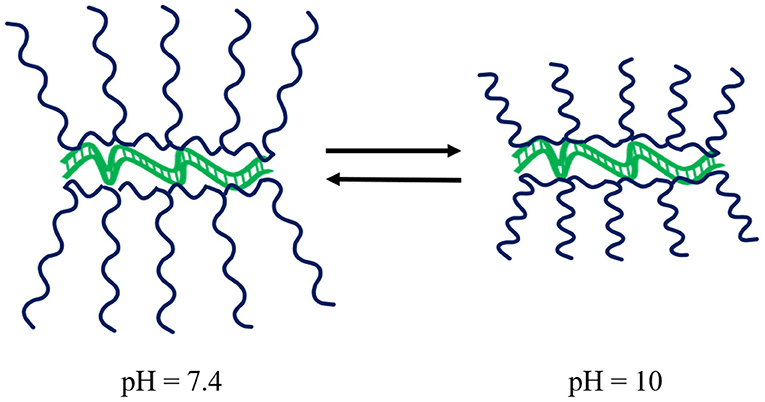
Figure 6. Schematic of the impact of pH on PGPMA polymers whilst complexed to dsRNA. Upon increasing pH, the polyplexes become more compact. Modified from work by Parsons et al. (2018).
Christiaens et al. synthesised a series of homopolymers and copolymers of poly(N-(2aminoethyl) methacrylate) (PAEMA) and PDMAEMA via free radical polymerisation, which were further functionalised by reacting part of the primary amine moieties of the PAEMA block with 1H-pyrazole-1-carboxamidine hydrochloride (HPC) to yield copolymers with guanidine (GUMA) (Ð of the copolymer ranged between 1.41 and 2.86) (Christiaens et al., 2018). The copolymer with the highest proportion of guanidine content [poly(AEMA-co-GUMA)] protected dsRNA to a greater extent than the other polyplexes when incubated with the midgut contents of Spodoptera exigua larvae, buffered at both pH 7.5 and 11. A low molecular weight poly(AEMA-co-GUMA) complexed with dsRNA of the vital chitin synthase B (ChSB) gene stabilised the dsRNA for at least 30 h in S. exigua gut juice at pH 11. Oral presentation to S. exigua larvae of 50 μg of poly(AEMA-co-GUMA)/ChSB dsRNA on cabbage leaves inhibited larval development and resulted in over a three-fold increase in mortality after 13 days when compared to feeding naked dsRNA. Confocal microscopy of cultured CF302 midgut cells incubated with Cy3-labelled dsRNA complexed with FITC-labelled poly(AEMA-co-GUMA) indicated greater cellular internalisation of polyplexes compared to naked non-complexed dsRNA (Christiaens et al., 2018).
These studies indicate that the incorporation of guanidine functional groups within the polymer structure provide enhanced protection for dsRNA at the alkaline pH conditions found in the gut of Spodoptera. They also show that the guanidine functionalised copolymers can greatly improve the in vivo efficacy of RNAi as an insect growth regulator and biocide.
Conjugated polymer nanoparticles (CPNs) are composed of polymers, the simplest example being polyacetylene, with a backbone of alternating single and double bonds that create a conjugated system of π-electrons. This induces a variety of useful properties such as conductivity and fluorescence (Abelha et al., 2020).
CPNs have been introduced for delivery of siRNA to HeLa cells (Moon et al., 2011). These nanoparticles comprised an amine-containing poly(phenylene ethynylene) (PPE) polymer, with a hydrophobic backbone to facilitate cellular internalisation and endosomal escape. These CPNs appear to form loosely aggregated particles, allowing effective complexation with siRNA due to their large, exposed surface area. A 94% knockdown of actin B expression was found when CPN/siRNA complexes were transfected into HeLa cells.
A similar PPE polymer was synthesised to form CPNs for use in plant protection against viral challenges and for the oral delivery of siRNA to insects. NtCes1-A siRNA, a cellulose synthase gene, complexed with the CPNs was incubated with protoplasts. A calcofluor white M2R staining assay revealed that following a 72 h incubation with the CPN-siRNA, protoplasts displayed 33–38% regeneration of their cell walls, in comparison to 51–54% in protoplasts that were either untreated or treated with naked siRNA or CPNs alone. Additionally, the transcription levels for NtCes1-A in CPN-siRNA treated protoplasts after a 48 h incubation were reduced by over 76%, compared to 17% NtCes1-A mRNA reduction with CPN treatment alone. Internalisation of nucleic acids by protoplasts has traditionally been achieved by using poly(ethylene glycol) (PEG) or by electroporation, which enhances the cell membrane permeability through short, high-intensity electrical pulses (Fisk and Dandekar, 2004; Davey et al., 2005). However, with use of both PEG and electroporation there was a detrimental impact on protoplast viability (Fromm et al., 1985; Fisk and Dandekar, 2004; Silva et al., 2010). In comparison, delivery of siRNA to plant protoplasts with CPNs significantly reduced cytotoxicity (Silva et al., 2010).
It is worth mentioning here a recent development of conjugated polymer/siRNA nanoparticle complexes for application in therapeutics, which may also hold promise for delivery of dsRNA for crop protection. Indeed, the recent design of conjugated polymer-dots that are formed by nanoprecipitation could be adapted to developing systems for delivery of dsRNA in other areas. These polymer-dots combine a core of conjugated polymer with a functional corona copolymer and a cationic lipid for complexation with siRNA. The resulting conjugated polymer-dot/siRNA complexes have shown comparable efficiency with lower cytotoxicity than Lipofectamine 2000® (Wang F. et al., 2019).
Dendrimers are highly branched polymer architectures defined by a central core, a branched interior structure of defined dimensions and a surface that contains functional groups. They can be used as delivery agents by retaining active species within the pores created by the branched structure and/or on their surface. So-called “Starburst” dendrimers were reported for gene transfection in 1993 (Haensler and Szoka, 1993) and such macromolecular species are now commonplace for therapeutic gene delivery within commercial DNA transfection kits such as SuperFect® (Yamano et al., 2010). However, the use of dendritic nanocarriers for delivery of dsRNA to insect species was not realised until 2013 by He et al., with the first report of the successful use of generation 5 (G5—referring to 5 cycles of repeated branching during the dendrimer synthesis) poly(amido amine) (PAMAM) dendrimers for dsRNA delivery to insect cells reported in 2019 (He et al., 2013; Kesharwani et al., 2015; Lu et al., 2019).
Dendrimers for the delivery of dsRNA are most commonly formed from PAMAM. They have well-defined nano-scale structures with a cationic surface charge that enables electrostatic capture of dsRNA. Lu et al. report the use of a G5 PAMAM dendrimer for the delivery of both dsRNA and plasmid DNA (pDNA) to two types of cultured cells from the lepidopteran insect, Bombyx mori (silkworm). G5-PAMAM/dsRNA dendriplexes were effective at interfering with expression of the translationally controlled tumour protein (TCTP) and the fluorescent red-ubiquitin (Ub) genes in BmE and BmN cell cultures (Lu et al., 2019).
Fluorescently-labelled PAMAM dendrimers have been synthesised with a perylenediimide (PDI) fluorophore core, see Figure 7, to aid visualisation of delivery via fluorescence microscopy. On topical application of the PDI-G2-PAMAM/dsRNA complexes, targeting the haemocytin transcript, to fourth instar Aphis glycines (soybean aphids), fluorescence from the PDI core was observed within 1 h in the circulating haemolymph and various internal tissues, demonstrating the systemic delivery of the dendriplexes. Haemocytin expression was reduced by 95.4% and a population density decrease of 80.5% was achieved by 5 days post-treatment (Zheng et al., 2019). A similar water-soluble PDI-G3-PAMAM nanocarrier, with extended dendritic arms (a higher generation dendrimer), was synthesised by Xu et al., and provided steric stability to the dendriplexes, preventing aggregation in aqueous solution due to the water insoluble PDI core. The nanocarrier was complexed with DNA and used to transfect Drosophila S2 cells. Fluorescence from the PDI core was seen in cultured cells 1 h post-incubation, with >90% cell viability. The same dendriplexes, targeting the developmental decapentaplegic transcript, fed to Drosophila melanogaster larvae, reduced body length of third instar larvae by 35%. This growth deficiency shows the potential of PDI-PAMAM as a dsRNA delivery vehicle. The high transfection capability of dendriplexes is in part due to the globular structure of the nanocarrier (Xu et al., 2014).
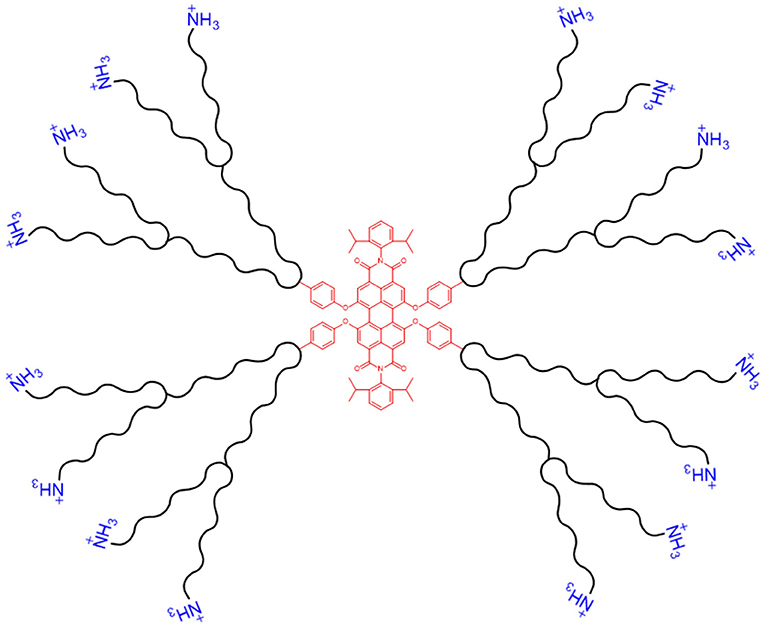
Figure 7. Schematic of a dendritic nanocarrier, with a PDI core (shown in red, centre) and NH3 groups for complexation with dsRNA (shown in blue, outer layer).
A dendritic nanocarrier with a PDI core has also been employed to deliver serpin-3 dsRNA by feeding to newly hatched larvae of the Asian corn borer, Ostrinia furnacalis. The cationic dendrimer was efficiently taken up by the midgut and fat body cells of the feeding larvae. When fed larvae were challenged with the bacterium Micrococcus luteus, the transcript of serpin-3, a critical immune-response gene, was reduced by 51% overall and more strikingly yet, serpin-3 protein was undetectable in the haemolymph. Similar fluorescent nanoparticle dendriplexes have been used for oral delivery of dsRNA targeting a vital chitinase-like transcript (CHT10) of O. furnacalis. Feeding first instar larvae with the nanoparticles stunted growth and after 5 days resulted in failure to moult and insect death (He et al., 2013; Shen et al., 2014).
The use of dendrimer-coated carbon nanotubes (PAMAN-CNTs) to deliver α-tubulin (α-tub) and mitochondrial RNA polymerase (mtpol) dsRNA to T. castaneum has recently been demonstrated. In both targets, fourth instar larvae injected with PAMAM-CNT-dsRNAs showed significantly greater reduction in gene expression after 72 h, in comparison to the naked dsRNA controls (Edwards et al., 2020).
Thus, several studies have shown that dendrimers based on PAMAM are effective polymeric vectors for dsRNA-mediated RNAi. These dendriplexes typically have high transfection efficiencies, low cytotoxicity and high water solubility, and show efficient RNAi induction in insect tissues through oral feeding. Further research on the effectiveness of these nanoparticle formulations to trigger RNAi in pest species from different orders is required to establish dendrimer/dsRNA formulations as a generic insect control solution, but the studies reviewed above have demonstrated the potential of such systems for specific insects.
A novel vector of nanometre dimensions that consist of layered double hydroxide clay nanosheets (LDH) for delivery of virus-specific dsRNA has also been developed to protect plants against virus infections (Mitter et al., 2017). The non-toxic LDH forms a positively charged layered lamellar structure with dsRNA (tested within the range of 300–1,800 bp) adsorbed to the surface, or sandwiched between multiple particles, forming what has been called “BioClay” formulations. The LDH material degrades in an atmosphere of CO2 and moisture due to the formation of carbonic acid with around 25% degradation after 7 days at 5% CO2 and 95% RH. Importantly, the BioClay formulation protects the dsRNA from leaf surface run-off and from metabolic breakdown for up to 30 days. The formulation provides sustained release of dsRNA on the leaf, which in turn results in long-term systemic protection against the targeted viral infection.
These properties are critical to overcoming the barriers of foliar sprayed application to crops, such as run-off and enzymatic degradation prior to ingestion by insect species, that must be reduced for efficient RNAi delivery and action. In particular, these recent studies demonstrated that Vigna unguiculata and Nicotiana tabacum plants were protected against viral challenges from Cucumber mosaic virus (CMV) and Pepper mild mottle virus (PMMoV), respectively, by the sprayed application of CMV2b and PMMoVIR54-specific BioClay formulations. A systemic RNAi response was observed in N. tabacum plants. Subsequently it was reported that bean common mosaic virus coat protein (BCMVCP) dsRNA incorporated within LDH BioClay nanosheets also protected Nicotiana benthamiana against virus transmission by the peach-potato aphid, Myzus persicae (Worrall et al., 2019). This recent work provides early evidence that BioClay has the potential to protect plants against viral challengers transmitted by pest insect species via a long-lasting integrated PTGS strategy (Mitter et al., 2017).
Here we specifically highlight studies where various inorganic nanoparticles are used as a template of known properties, particularly size and size distribution, to build hybrid polymer structures designed for efficient interaction with and retention of dsRNA. Indeed, the use of particles (or a well-known template particle) onto which appropriate chosen/designed polymers are coated allows for tight control of the carrier properties.
Biocompatible calcium phosphate (CaP) nanoparticles have been used in therapeutic applications for drug or gene delivery. CaP nanoparticles carrying dsRNA have been shown to enter cells via endocytosis and release their cargo within the endosome (Xu et al., 2014, 2016). In therapeutic applications the process of cellular internalisation and in vivo RNAi can be made more efficient for these systems by using polymeric coatings such as PEG and PEI to modify the nanoparticle surface (Roy et al., 2003; Xu et al., 2016).
In recent research by Elhaj Baddar et al., a hydroxyapatite (HA) inorganic nanocarrier with a polymer coating was used to induce RNAi in insect Sf9 cells. The HA nanoparticle template was coated in poly(acrylic acid) (PAA) through layer-by-layer electrostatic assembly. The addition of cationic amine groups is known to improve binding, therefore a layer of poly(arginine) (PLR10) was added to the nanoparticles to electrostatically interact with dsRNA (Das et al., 2015; Mostaghaci et al., 2015). When targeting the luciferase transcript in Sf9 cells, a 35% transcript reduction was achieved at a mass ratio of PLR10+PAA-coated HA nanoparticle to dsRNA of 5:1. In comparison, both naked dsRNA and the PAA-coated HA nanoparticle/dsRNA system did not lead to significant PTGS.
Moreover, the same arginine-containing polymer (PLR10-PAA-HA) was complexed, in addition to the dsRNA, with a fluorescent probe, CypHer5E, that fluoresces specifically at the pH of late endosomes (pH 5) to test the endosomal escape of dsRNA following delivery to cells. It was found that, upon delivery of the complex, a significant reduction in the fluorescence within the acidic bodies of Sf9 cells occurred. In contrast, delivery of naked CypHer5E-dsRNA had a strong fluorescence in endosomal compartments. The enhanced RNAi response due to dsRNA complexation with PLR10-PAA-HA nanoparticles is thought to result from enhanced endosomal escape (Elhaj Baddar et al., 2020).
Gold (Au) is another inorganic nanoparticle that has been employed as a template for dsRNA delivery, due to its tuneable surface and low toxicity (Kozielski et al., 2013). In a recent report, a PLR10 polymer was covalently attached to the surface of Au nanoparticles (AuNPs) with hydroxysuccinimide (NHS) surface groups to which a bovine serum albumin (BSA) ligand was also attached to achieve a mixed surface layer of the polymer and BSA (Laisney et al., 2020). The BSA ligand, an amphiphilic peptide, is shown to improve endosomal escape in a similar mechanism to CPPs. The PLR10-Au-BSA/siRNA nanoparticles induced 31% reduction of the luciferase transcript in Sf9 cells, but in comparison, PLR10/siRNA nano-sized polyplexes elicited an efficiency of 58%, with the strong binding of PLR10 to the AuNPs hypothesised as the cause of the decreased transfection efficiency. However, the binding of PLR10 to citrate-stabilised gold nanoparticles through electrostatic layer-by-layer coating of PLR10, rather than covalent binding with NHS, led to less successful attachment of PLR10 and thus resulted in aggregation of the nanoparticles (Laisney et al., 2020).
These studies are important steps toward using polymer-coated inorganic nanoparticles for dsRNA delivery, as surface functional groups could improve cell-targeting and as a result enhance dsRNA uptake and endosomal escape (Laisney et al., 2020). Table 2 summarises the recent reports in this area.
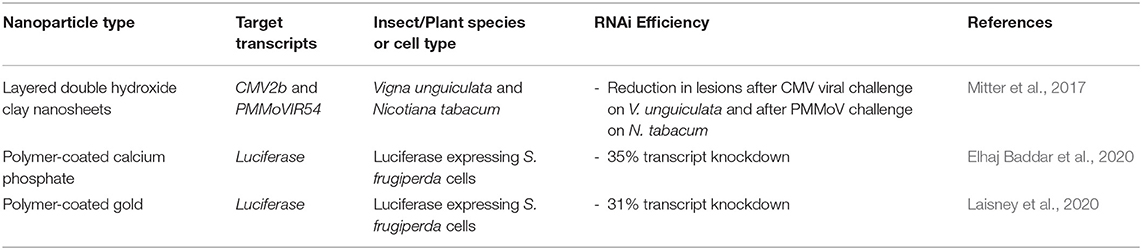
Table 2. Novel inorganic nanoparticles for RNA-mediated crop protection from the last 10 years that conducted in vitro or in vivo assay with the nanoparticle complexes.
There has been a drive to use natural product-based formulations in the design of nanoparticle vectors for dsRNA delivery, for example peptide-based systems, as they are generally non-toxic and biodegradable in comparison to synthetic polymeric materials, and therefore more eco-friendly (Wessel et al., 2019). Table 3 summarises the recent advances on this topic and the paragraphs below describe the main two systems developed.
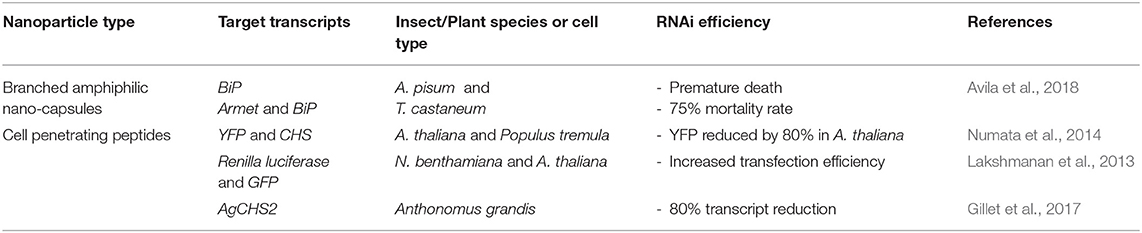
Table 3. Peptide-based nanoparticles for RNA-mediated crop protection from the last 10 years that conducted in vitro or in vivo assay with the peptiplexes.
In the past 10 years a new peptide-based nanoparticle (branched amphiphilic peptide capsules, BAPCs) has been developed for the nano-delivery of nucleic acids. These systems have a similar structure to liposomes, but have much greater stability, and self-assemble into bilayer delimited supramolecular nanovesicle structures. BAPCs have cationic lysine surface groups that form nanoparticle complexes with nucleic acids of ~80–200 nm in size (Barros et al., 2016, 2017, 2019; Tomich et al., 2019). BAPCs are biodegradable by a common soil fungus Aspergillus nidulans, thus lessening the potential impact of these nanoparticles on the environment (Wessel et al., 2019). Avila et al. reported the first use of BAPCs for the oral delivery of dsRNA to the pea aphid Acyrthosiphon pisum and the red flour beetle T. castaneum (Avila et al., 2018).
In this report, feeding BAPC/BiP-dsRNA complexes to adult A. pisum resulted in mortality 6–9 days earlier compared to aphids fed naked dsRNA alone. Two different T. castaneum transcripts, BiP, and Armet, were targeted by feeding dsRNA to early instar T. castaneum larvae, with mortality reaching 75% when BAPC/dsRNA complexes for both targets were fed simultaneously.
The potential for orally administered BAPC/dsRNA to generate RNAi effects in pest species other than aphids and beetles now needs to be demonstrated (Singh et al., 2017; Avila et al., 2018).
There are many naturally occurring proteins [pore/channel-forming proteins, fusion proteins, and cell-penetrating peptides (CPPs)] that facilitate the movement of molecules across cell membranes. These proteins might be exploited to enhance dsRNA uptake by cells and to assist endosomal escape within the cell (Yang and Hinner, 2015).
The discovery of the membrane traversal properties of the Tat protein (Trans-Activator of Transcription) from both the HIV-1 virus and the D. melanogaster Antennapedia homeodomain, led to the development of CPPs (Frankel and Pabo, 1988; Joliot et al., 1991; Vivès et al., 1997). The Tat peptide is an arginine-rich peptide, with a cationic guanidinium-based functionality that can interact with the anionic cell surface, provoking endocytic cellular uptake (Vivès et al., 2008; Cermenati et al., 2011; Yang and Hinner, 2015).
The combination of a CPP covalently bound to a polycationic peptide, forms a fusion peptide that can serve as a delivery vector for dsRNA or pDNA (Unnamalai et al., 2004; Lakshmanan et al., 2013; Numata et al., 2014). A copolymer of histidine and lysine, (KH)9, combined with the CPP Bp100, is an example of a fusion peptide that complexes with dsRNA (Numata et al., 2014). The (KH)9-Bp100/yellow fluorescent protein (YFP)-dsRNA complex, at a molar ratio of 2:1, targeting the transcript of YFP and the chalcone synthase genes in A. thaliana leaves, successfully affected both the exogenous and endogenous transcripts in <12 h post-application and the effect was sustained for at least a further 24 h. Similar fusion peptides: nona-arginine (R9)-Bp100, (KH)9-Bp100, and R9-Tat2 have been complexed with plasmid DNA. Rapid transfection was measured within the first 12 h of incubation of these fusion peptides complexed with Renilla luciferase pDNA with both N. benthamiana and A. thaliana leaves. Transfection efficiency then decreased for the remaining measurement time of 144 h (Lakshmanan et al., 2013; Numata et al., 2014).
A significant issue with the use of peptides for the delivery of dsRNA or pDNA, is the susceptibility of peptides to intracellular proteolysis which can attenuate the RNAi effect. The use of proteins/peptides (e.g., silk proteins) that are more resistant to metabolic degradation might provide a mechanism for the sustained release of dsRNA (Numata and Kaplan, 2010; Lakshmanan et al., 2013; Numata et al., 2014).
An example of a fusion peptide/dsRNA nanoparticle for an insect-specific application was recently reported (Gillet et al., 2017). In this work, a chimeric protein of a CPP fused to a DRBD (dsRNA binding domain) was synthesised, with the CPP including the fusogenic peptide haemagglutinin, that aids the endosomal escape of the complex following endocytosis into the cell by destabilising the endosomal membrane. The CPP-DRBD/dsRNA complexes, or ribonucleoprotein particles (RNPs), enhanced both the protection of dsRNA at pH 5.5 and the cellular uptake of Cy3-labelled dsRNA into Sf21 cells. Oral delivery of CPP-DRBD/chitin synthase II (Ag-ChSII) dsRNA complexes to cotton boll weevil (Anthonomus grandis), reduced Ag-ChSII transcript by 80%, in comparison to delivery of Ag-ChSII dsRNA alone that resulted in a reduction of only 30%. No significant mortality, however, was seen (Gillet et al., 2017).
Chitosan, the polysaccharide derived from crustacean shells, is a naturally-derived polymer that has been used for complexation with dsRNA to improve delivery for insect control. It is of particular interest to researchers due to its inherent biocompatibility and non-toxic nature (Serrano-Sevilla et al., 2019). Chitosan contains cationic groups along its polymer chain which electrostatically interact with dsRNA (Zhang et al., 2010). Table 4 summarises the recent advances in the use of chitosan/dsRNA nanoparticles for crop protection.
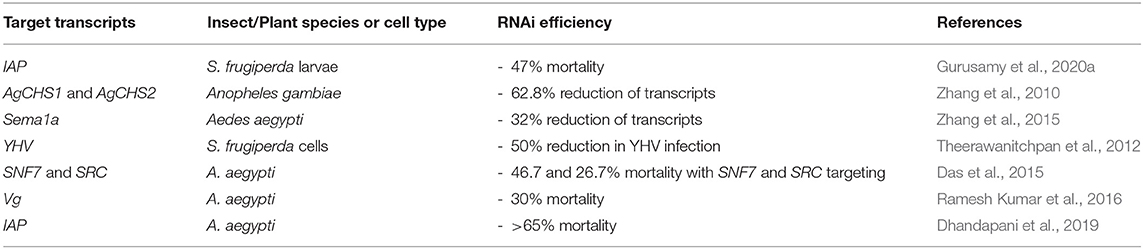
Table 4. Chitosan nanoparticles for RNA-mediated crop protection from the last 10 years that conducted in vitro or in vivo assay with the chitosan/dsRNA complexes.
Complexation of chitosan with dsRNA results in nanoparticles that can aid the endosomal escape of dsRNA and can consequently increase the RNAi efficiency. This has been illustrated in EGFP::Rab7 expressing Sf9 cells, with 60% reduction in the accumulation of CypHer-5E-labelled dsGFP in endosomal compartments when complexed with chitosan, in comparison to naked dsRNA. Upon oral feeding of the chitosan/dsIAP nanoparticles to third instar S. frugiperda larvae, a mortality rate of 47% was realised, in comparison to 25% mortality when larvae were fed naked dsIAP alone (Gurusamy et al., 2020a).
Chitosan/dsRNA complexes have been used to target the aquatic larval stages of mosquito pest species such as Anopheles gambiae and Aedes aegypti, with varying degrees of success. For example, chitosan/dsRNA nanoparticles targeting two different chitin synthase transcripts in A. gambiae reduced mRNA by 62.8% in comparison to the GFP equivalent control. However, the levels of chitin produced were only reduced by 33.8% and no insect mortality was observed (Zhang et al., 2010, 2015). The targeting of the A. aegypti vestigial (vg) transcript through delivery of chitosan/dsRNA complexes also did not significantly increase larval mortality (30% insect mortality compared to 20% mortality with the control group) (Ramesh Kumar et al., 2016). In a comparative study, chitosan/SNF7-dsRNA nanoparticles outperformed SNF7-dsRNA complexed with amine-functionalised silica nanoparticles (ASNP) in killing A. aegypti larvae and in knockdown of the A. aegypti SNF7 transcript. However, carbon quantum dot (CQD)/dsRNA complexes reduced mRNA levels for two different gene targets (SRC and SNF7), whereas chitosan/dsRNA complexes only gave significant RNAi response with SNF7-dsRNA (Das et al., 2015).
Chitosan-based dsRNA nanoparticles are insoluble at neutral and alkaline pH regimes, affecting their performance as effective delivery vectors of dsRNA (Das et al., 2015; Serrano-Sevilla et al., 2019). Consequently, recent developments have sought to improve chitosan-based nanoparticles by increasing the chitosan positive charge through chemical modification (quaternisation) (Theerawanitchpan et al., 2012). The greater charge density of the quaternised-chitosan (QCH) increases the polymer solubility and also improves stability of its complexes with dsRNA within cells. Typically, complexation of chitosan/dsRNA is achieved at a weight ratio of 1.5:1, with complexes 350–650 nm in size. In comparison, QCH/dsRNA achieves full complexation at 0.24:1 weight ratio, with lower sizes measured for the complexes between 150 and 350 nm (measurements made with a Zetasizer Nano ZS). The more compact complexes with QCH are due to a stronger electrostatic interaction with dsRNA, thus stronger binding and likely enhanced protection of the complex. The cytotoxicity of both complexes is lower than that of the commercial liposomal transfection agent, Cellfectin®, and both chitosan and QCH-based complexes with YHV-dsRNA were successful in reducing the viral infection of YHV (yellow head virus) in Sf9 cells by at least 50% 24 h post-viral challenge. Whilst QCH binds more strongly to dsRNA, due to the increased cationic charge, the difference in cell viability and RNAi efficiency was negligible (Theerawanitchpan et al., 2012).
The addition of sodium tripolyphosphate (TPP) to chitosan as an ionic cross-linking agent, prior to complexation with dsRNA (CS-TPP-dsRNA), has also been investigated to improve the stability of chitosan/dsRNA nanoparticles. The introduction of cross-links to the polyplexes caused an increase in the mortality rate in A. aegypti larvae of 65%, when targeting the IAP transcript, which was significantly more than the 35% mortality observed on feeding chitosan/dsIAP nanoparticles without TPP (Dhandapani et al., 2019).
The improved properties of chitosan-based dsRNA nanoparticles, by cross-linking or increased charge density, coupled with the low general toxicity adds to the attractiveness of this class of natural polymers for the delivery of dsRNA to insects. When such chitosan/siRNA nanoparticles have been used for therapeutic applications, the solubility and colloidal stability of complexes were improved by using PEG-modified chitosan or by combining chitosan with PEI and carboxymethyl dextran (Gutoaia et al., 2016; Rudzinski et al., 2016; Sun et al., 2016; Iranpur Mobarakeh et al., 2019). Enhanced cellular internalisation and endosomal escape were also achieved through grafting a CPP, e.g., Tat or nona-arginine peptides to chitosan (Malhotra et al., 2013; Park et al., 2013; Yang et al., 2013; Serrano-Sevilla et al., 2019). Such chitosan-based materials for dsRNA complexation and delivery may also of value for crop protection applications.
Liposomes are spherical vesicles formed from a phospholipid bilayer (resembling that of cell membranes) and are commonly used to encapsulate materials for drug delivery, with the first reported publication in literature in 1987 by Felgner et al. (1987) and Malone et al. (1989). Since then liposomes have been developed as non-viral vectors for dsRNA delivery, and mass-produced as commercial kits, such as Lipofectamine 2000® or Cellfectin® (Sparks et al., 2012; Zhang et al., 2012). Upon complexation of a cationic liposome with negatively charged dsRNA by electrostatic interaction, a lipoplex can be formed. It is thought that lipoplexes traverse the cell membrane via adsorptive endocytosis (Friedmann, 1992; Zuidam et al., 1999; Barenholz, 2001; Lungwitz et al., 2005; Joga et al., 2016), which exemplifies their appeal for delivery of dsRNA to cells (Xue et al., 2015).
Liposome encapsulated dsRNA has often been used to introduce nucleic acids to insect species that do not display systemic RNAi responses, such as D. melanogaster or Drosophila suzukii. For example, Taning et al. demonstrated that feeding rps13 and vha26 dsRNA, encapsulated in Lipofectamine 2000®, to D. suzukii larvae and adults, resulted in significant mortality after feeding, whereas naked dsRNA did not result in mortality (Taning et al., 2016). The lack of a SID-1-like transporter protein in Drosophila species means that the pathway for cellular uptake occurs via endocytosis (Saleh et al., 2006; Ulvila et al., 2006; Whyard et al., 2009), considered to be a slower uptake pathway in comparison to SID-1-like systems. The use of a liposome-based transfection agent improves cellular uptake and delivery of dsRNA in these recalcitrant species (Whyard et al., 2009; Joga et al., 2016).
Liposomes have also recently been shown to aid the release of dsRNA from endosomal compartments in S. frugiperda. Visualisation of CypHer-dsGFP dsRNA complexed to Cellfectin II® transfection reagent showed an 80% reduction in accumulation in the late endosomes, in comparison to the naked CypHer-dsGFP (Gurusamy et al., 2020b).
Liposomal encapsulation of dsRNA has been shown to be an effective method of delivery (as demonstrated below) in the tick species Rhipicephalus haemaphysaloides, the German cockroach Blattella germanica and the Neotropical stink bug Euschistus heros (Lin et al., 2017; Huang et al., 2018; Zhang et al., 2018; Castellanos et al., 2019).
In the case of R. haemaphysaloides, the Cy3-labelled dsRNA was encapsulated in three different commercial liposomal uptake facilitators (Lipofectamine 2000®, DMRIE-C®, and Cellfectin®) and the formulation was delivered via soaking of larvae, nymphs, and adults. All three lipoplexes induced more efficient RNAi than the dsRNA formulated in water alone (Zhang et al., 2018). The soaking of arthropods has been shown to be more effective than oral feeding, thus in vivo assay with artificial feeding would be beneficial, in particular as a SID-1 gene has not been found in ticks (Fuente et al., 2007; Whyard et al., 2009; Barry et al., 2013; Scott et al., 2013; Yu et al., 2013; Kang et al., 2018).
Blattella germanica are refractory to orally fed dsRNA due to enzymatic degradation by midgut nucleases, whereas micro-injection can induce an efficient RNAi response (Lin et al., 2017). The problem of metabolic instability in the cockroach midgut was overcome through encapsulation of the dsRNA in liposomes which resulted in 60% mortality over 40 days. Mortality rates from continuous oral feeding of lipoplexes increased from 8 days of feeding to 16 days of feeding, suggesting that modifications to liposomal nanoparticles could be introduced to shorten the time required for effective RNAi (Lin et al., 2017; Huang et al., 2018).
The efficacy of liposome encapsulated dsRNA to elicit RNAi has been improved by the addition of a chelating agent to the formulation and by modification of the lipid component. For example, in the stink bug E. heros, metal-dependent nuclease activity within the insect saliva is a limiting factor for the oral delivery of lipoplexes. A chelating-agent EDTA, which can isolate metal ions and prevent them from interacting with the liposomes, has been shown to improve mortality rate in these insects, when added in combination with lipoplexes (Castellanos et al., 2019).
Modifications of lipids for the encapsulation of dsRNA have been reported for therapeutic applications of RNAi. The 1,2-dioleoyl-3-dimethylammonium-propane (DAP) lipid was modified by anchoring PDMAEMA-b-PEG block copolymers to act as a pH-tuneable surface, see Figure 8. This polymer-coated liposome nanoparticle was shown to enhance protection of siRNA and to enhance endosomal escape. More efficient targeting of GFP in GFP-expressing HeLa cells was found with the polymer-coated lipoplexes than with the oligofectamine/siRNA control (Auguste et al., 2006, 2008). A similar modification has yet to be reported for RNAi triggering in either insects or plants. Table 5 summarises the recent reports of lipoplex nanoparticles for RNA-mediated crop protection.
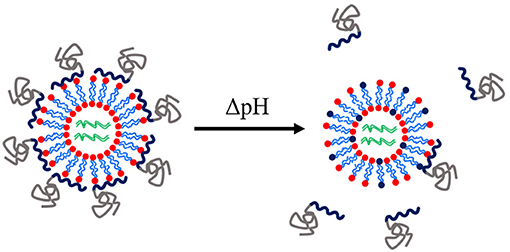
Figure 8. Schematic of a pH-tuneable liposome with adsorbed block copolymers that allow endosomal release upon decrease of pH. Modified from work by Auguste et al. (2008).
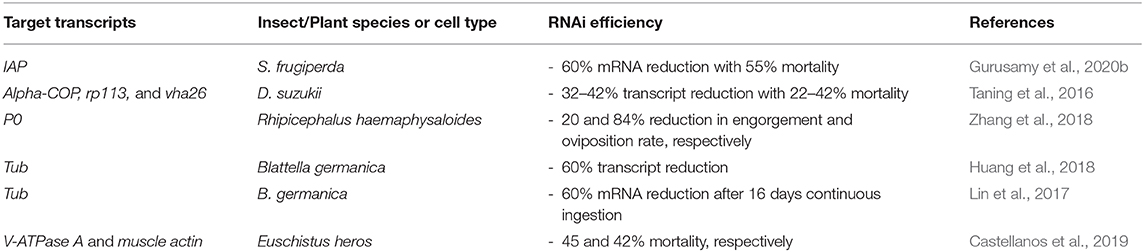
Table 5. Liposomal nanoparticles for RNA-mediated crop protection from the last 10 years that conducted in vitro or in vivo assay with the lipoplexes.
It is clear that there is a direct comparison to be drawn between engineered nanoparticles for therapeutic triggering of RNAi, and novel strategies for agricultural applications. Indeed, it has often been the case that strides in the field of therapeutics are swiftly followed by exploitation of these new routes in the area of crop protection. For example, Howard et al. reported the use of chitosan/siRNA nanoparticles in 2006, Eguchi et al. used CPP-DRBD/siRNA complexes in 2009 and Treat et al. synthesised guanidine-containing copolymers in 2012 (Howard et al., 2006; Eguchi et al., 2009; Treat et al., 2012), all for induction of RNAi in therapeutic applications, prior to reports of similar RNAi-based crop protection strategies. We anticipate that further insights and technological achievements in the clinical field will continue to be very valuable in developing improvements in the formulation of dsRNA for crop protection.
There has been significant progress in the design of responsive nanoparticle vectors for dsRNA delivery for application within therapeutics due to the in-depth knowledge of physiological temperature and pH. Whilst temperature-responsive nanoparticles may remain impractical for RNA-mediated insect control, pH-responsive nanoparticles have been the focus of recent developments. In reports by Parsons et al. and Christiaens et al., the insect midgut pH was characterised and exploited for a pH-tuneable delivery vehicle (Christiaens et al., 2018; Parsons et al., 2018). An example of this strategy within the field of lipid-based uptake facilitators has also been reported by Auguste et al. with a pH-tuneable liposome nanoparticle (Auguste et al., 2008).
Due to the revolutionary introduction of CRISPR/Cas9 as a gene editing biotechnology, there has been increasing recent interest in genetic material delivery. Thus, tailored polymer architectures have been synthesised to enable the improved efficacy of dsRNA/DNA cellular internalisation. Tan et al., for example, devised a triblock copolymer of PDMAEMA, hydrophobic PBMA, and PEO as a hydrophilic corona, capable of forming micelleplexes with sgRNA (single guide RNA) and Cas9 proteins for delivery to the cell nucleus (Tan et al., 2019). Recent advances in therapeutic applications of RNA delivery have also included reports of conjugated siRNA to nanoparticles, often with a brush-like polymer architecture. The use of cleavable disulfide linkages to the siRNA allows for a steady release mechanism within the cells (Bai et al., 2019; Wang D. et al., 2019). We expect that formulations with similar technology will soon be developed within the field of RNA-mediated crop protection.
The complexity of the pathway and barriers to induce RNAi, including enzymatic degradation, traversal of the cell membrane and subsequent endosomal escape, require tailored (often complex) nanoparticle designs to ensure that both the protection and delivery of dsRNA can be achieved. The recent developments reviewed in this article clearly demonstrate that more complex polymer architectures and nanoparticle modifications are possible and should provide future directions for enhancing efficacy of the use of dsRNA for crop protection. Additionally, it is evident that developing such polymer architectures will also need to take into account the more practical aspects of delivering the biopesticides to insects in a crop field. For example, a novel method of delivery through a non-invasive application has been developed to reduce the degradation of dsRNA. This high-throughput method integrates the dsRNA-nanoparticle complexes into aerosols, which could be a useful application tool for future pesticide formulation developments (Li-Byarlay et al., 2013; Thairu et al., 2017). Furthermore, any delivery system carried forward in the future will also need to show robustness when integrated within pesticide formulations (which vary depending on locations and types of crop for example) and when sprayed and dried on crop leaves. Sustainability issues will also have to be taken into account as the polymer particles should fully degrade in the environment to avoid long-term persistance. Additional concerns for the real-world application of dsRNA-based formulations include regulation of these products to ensure bio-safety, as well as the cost of large-scale production (Lundgren and Duan, 2013; Scott et al., 2013; Parker et al., 2019; Romeis and Widmer, 2020). These additional requirements will also bring about significant challenges to overcome.
CP led the writing of this review, with RI, NW, and OC responsible for editing. All authors conceptualised the format and topic of the article.
The authors would like to acknowledge the support of the EPSRC Centre for Doctoral Training in Soft Matter and Functional Interfaces (SOFI CDT - EP/L015536/1) and Syngenta for joint funding of Ph.D. studentship for CP. This work was supported by the University of Leeds Library UK Research and Innovation grant who provided funding for the open access publication fees.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge the support of Dr Rosa Domínguez-Espinosa and colleagues at Syngenta for reviewing the article and suggesting improvements. The authors also acknowledge help from Dr. Calum Ferguson in designing Figures 1, 3.
1. ^https://www.pesticideresistance.org/index.php (accessed May 7, 2020).
Abbas, M. S. T. (2018). Genetically engineered (modified) crops (Bacillus thuringiensis crops) and the world controversy on their safety. Egypt. J. Biol. Pest Control 28:52. doi: 10.1186/s41938-018-0051-2
Abelha, T. F., Dreiss, C. A., Green, M. A., and Dailey, L. A. (2020). Conjugated polymers as nanoparticle probes for fluorescence and photoacoustic imaging. J. Mater. Chem. B 8, 592–606. doi: 10.1039/C9TB02582K
Agarwal, S., Zhang, Y., Maji, S., and Greiner, A. (2012). PDMAEMA based gene delivery materials. Mater. Today 15, 388–393. doi: 10.1016/S1369-7021(12)70165-7
Arigita, C., Zuidam, N. J., Crommelin, D. J. A., and Hennink, W. E. (1999). Association and dissociation characteristics of polymer/DNA complexes used for gene delivery. Pharm. Res. 16, 1534–1541. doi: 10.1023/A:1015096302720
Auguste, D. T., Armes, S. P., Brzezinska, K. R., Deming, T. J., Kohn, J., and Prud'homme, R. K. (2006). pH triggered release of protective poly(ethylene glycol)-b-polycation copolymers from liposomes. Biomaterials 27, 2599–2608. doi: 10.1016/j.biomaterials.2005.08.036
Auguste, D. T., Furman, K., Wong, A., Fuller, J., Armes, S. P., Deming, T. J., et al. (2008). Triggered release of siRNA from poly(ethylene glycol)-protected, pH-dependent liposomes. J. Controll. Release 130, 266–274. doi: 10.1016/j.jconrel.2008.06.004
Avila, L. A., Chandrasekar, R., Wilkinson, K. E., Balthazor, J., Heerman, M., Bechard, J., et al. (2018). Delivery of lethal dsRNAs in insect diets by branched amphiphilic peptide capsules. J. Control. Release 273, 139–146. doi: 10.1016/j.jconrel.2018.01.010
Bai, Q., Liu, J., Tang, J., Li, Z., Zheng, X., and Chen, Q. (2019). Redox-responsive polymeric RNAi based on multivalent conjugation of siRNA for improved intracellular delivery. Bioconjugate Chem. 30, 2777–2781. doi: 10.1021/acs.bioconjchem.9b00680
Barenholz, Y. (2001). Liposome application: problems and prospects. Curr. Opin. Coll. Interface Sci. 6, 66–77. doi: 10.1016/S1359-0294(00)00090-X
Barros, S., de, M., Avila, L. A., Whitaker, S. K., Wilkinson, K. E., Sukthankar, P., et al. (2017). Branched amphipathic peptide capsules: different ratios of the two constituent peptides direct distinct bilayer structures, sizes, and DNA transfection efficiency. Langmuir 33, 7096–7104. doi: 10.1021/acs.langmuir.7b00912
Barros, S. M., Chandrasekar, R., Wilkinson-Nutsch, K. E., Yoonseong, P., Reeck, G., Tomich, J. M., et al. (2019). Delivery of lethal dsRNAs in insect diets by branched amphiphilic peptide capsules. FASEB J. 33:785.4 doi: 10.1096/fasebj.2019.33.1_supplement.785.4
Barros, S. M., Whitaker, S. K., Sukthankar, P., Avila, L. A., Gudlur, S., Warner, M., et al. (2016). A review of solute encapsulating nanoparticles used as delivery systems with emphasis on branched amphipathic peptide capsules. Arch. Biochem. Biophys. 596, 22–42. doi: 10.1016/j.abb.2016.02.027
Barry, G., Alberdi, P., Schnettler, E., Weisheit, S., Kohl, A., Fazakerley, J. K., et al. (2013). Gene silencing in tick cell lines using small interfering or long double-stranded RNA. Exp. Appl. Acarol. 59, 319–338. doi: 10.1007/s10493-012-9598-x
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Borel, B. (2017). CRISPR, microbes and more are joining the war against crop killers. Nature News 543:302. doi: 10.1038/543302a
Boussif, O., Lezoualc'h, F., Zanta, M. A., Mergny, M. D., Scherman, D., Demeneix, B., et al. (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301. doi: 10.1073/pnas.92.16.7297
Bravo, A., Likitvivatanavong, S., Gill, S. S., and Soberón, M. (2011). Bacillus thuringiensis: a story of a successful bioinsecticide. Insect. Biochem. Mol. Biol. 41, 423–431. doi: 10.1016/j.ibmb.2011.02.006
Cao, Z., Xiao, H., Li, L., Liu, M., Lin, G., Zhai, P., et al. (2019). The codelivery of siRNA and QDs by pH-responsive micelle for hepatoma cancer cells. Front. Pharmacol. 10:1194. doi: 10.3389/fphar.2019.01194
Castellanos, N. L., Smagghe, G., Sharma, R., Oliveira, E. E., and Christiaens, O. (2019). Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag. Sci. 75, 537–548. doi: 10.1002/ps.5167
Cermenati, G., Terracciano, I., Castelli, I., Giordana, B., Rao, R., Pennacchio, F., et al. (2011). The CPP Tat enhances eGFP cell internalization and transepithelial transport by the larval midgut of Bombyx mori (Lepidoptera, Bombycidae). J. Insect Physiol. 57, 1689–1697. doi: 10.1016/j.jinsphys.2011.09.004
Chakraborty, S., and Newton, A. C. (2011). Climate change, plant diseases and food security: an overview. Plant Pathol. 60, 2–14. doi: 10.1111/j.1365-3059.2010.02411.x
Chen, Y.-J., Liu, B. R., Dai, Y.-H., Lee, C.-Y., Chan, M.-H., Chen, H.-H., et al. (2012). A gene delivery system for insect cells mediated by arginine-rich cell-penetrating peptides. Gene 493, 201–210. doi: 10.1016/j.gene.2011.11.060
Cherng, J.-Y., van de Wetering, P., Talsma, H., Crommelin, D. J. A., and Hennink, W. E. (1996). Effect of size and serum proteins on transfection efficiency of poly ((2-dimethylamino)ethyl methacrylate)-plasmid nanoparticles. Pharm. Res. 13, 1038–1042. doi: 10.1023/A:1016054623543
Cherng, J. Y., Talsma, H., Verrijk, R., Crommelin, D. J., and Hennink, W. E. (1999). The effect of formulation parameters on the size of poly-((2-dimethylamino)ethyl methacrylate)-plasmid complexes. Eur. J. Pharm. Biopharm. 47, 215–224. doi: 10.1016/s0939-6411(98)00103-9
Choi, J.-H., Kim, S.-O., Linardy, E., Dreaden, E. C., Zhdanov, V. P., Hammond, P. T., et al. (2015). Influence of pH and surface chemistry on poly(l-lysine) adsorption onto solid supports investigated by quartz crystal microbalance with dissipation monitoring. J. Phys. Chem. B 119, 10554–10565. doi: 10.1021/acs.jpcb.5b01553
Christiaens, O., and Smagghe, G. (2014). The challenge of RNAi-mediated control of hemipterans. Curr. Opin. Insect Sci. 6, 15–21. doi: 10.1016/j.cois.2014.09.012
Christiaens, O., Swevers, L., and Smagghe, G. (2014). DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 53, 307–314. doi: 10.1016/j.peptides.2013.12.014
Christiaens, O., Tardajos, M. G., Martinez Reyna, Z. L., Dash, M., Dubruel, P., and Smagghe, G. (2018). Increased RNAi Efficacy in Spodoptera exigua via the formulation of dsRNA with guanylated polymers. Front. Physiol. 9:316. doi: 10.3389/fphys.2018.00316
Christiaens, O., Whyard, S., Vélez, A. M., and Smagghe, G. (2020). Double-stranded RNA technology to control insect pests: current status and challenges. Front. Plant Sci. 11:451. doi: 10.3389/fpls.2020.00451
Convertine, A. J., Diab, C., Prieve, M., Paschal, A., Hoffman, A. S., Johnson, P. H., et al. (2010). pH-responsive polymeric micelle carriers for siRNA drugs. Biomacromolecules 11, 2904–2911. doi: 10.1021/bm100652w
Cook, A. B., Peltier, R., Hartlieb, M., Whitfield, R., Moriceau, G., Burns, J. A., et al. (2018). Cationic and hydrolysable branched polymers by RAFT for complexation and controlled release of dsRNA. Polym. Chem. 9, 4025–4035. doi: 10.1039/C8PY00804C
Cooper, A. M., Silver, K., Zhang, J., Park, Y., and Zhu, K. Y. (2019). Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 75, 18–28. doi: 10.1002/ps.5126
Cotanda, P., Wright, D. B., Tyler, M., and O'Reilly, R. K. (2013). A comparative study of the stimuli-responsive properties of DMAEA and DMAEMA containing polymers. J. Polym. Sci. Part A Polym. Chem. 51, 3333–3338. doi: 10.1002/pola.26730
Das, S., Debnath, N., Cui, Y., Unrine, J., and Palli, S. R. (2015). Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Appl. Mater. Interfaces 7, 19530–19535. doi: 10.1021/acsami.5b05232
Davey, M. R., Anthony, P., Power, J. B., and Lowe, K. C. (2005). Plant protoplasts: status and biotechnological perspectives. Biotechnol. Adv. 23, 131–171. doi: 10.1016/j.biotechadv.2004.09.008
Dhandapani, R. K., Gurusamy, D., Howell, J. L., and Palli, S. R. (2019). Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti. Sci. Rep. 9:8775. doi: 10.1038/s41598-019-45019-z
Dubelman, S., Fischer, J., Zapata, F., Huizinga, K., Jiang, C., Uffman, J., et al. (2014). Environmental fate of double-stranded RNA in agricultural soils. PLoS ONE 9:e93155. doi: 10.1371/journal.pone.0093155
Edwards, C. H., Christie, C. R., Masotti, A., Celluzzi, A., Caporali, A., and Campbell, E. M. (2020). Dendrimer-coated carbon nanotubes deliver dsRNA and increase the efficacy of gene knockdown in the red flour beetle Tribolium castaneum. Sci. Rep. 10:12422. doi: 10.1038/s41598-020-69068-x
Eguchi, A., Meade, B. R., Chang, Y.-C., Fredrickson, C. T., Willert, K., Puri, N., et al. (2009). Efficient siRNA delivery into primary cells by a peptide transduction domain–dsRNA binding domain fusion protein. Nat. Biotechnol. 27, 567–571. doi: 10.1038/nbt.1541
Elhaj Baddar, Z., Gurusamy, D., Laisney, J., Tripathi, P., Palli, S. R., and Unrine, J. M. (2020). Polymer-coated hydroxyapatite nanocarrier for double-stranded RNA delivery. J. Agric. Food Chem. 68, 6811–6818. doi: 10.1021/acs.jafc.0c02182
Felgner, P. L., Gadek, T. R., Holm, M., Roman, R., Chan, H. W., Wenz, M., et al. (1987). Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U.S.A. 84, 7413–7417. doi: 10.1073/pnas.84.21.7413
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Fischer, D., Li, Y., Ahlemeyer, B., Krieglstein, J., and Kissel, T. (2003). In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials 24, 1121–1131. doi: 10.1016/S0142-9612(02)00445-3
Fisk, H. J., and Dandekar, A. M. (2004). “Electroporation: introduction and expression of transgenes in plant protoplasts,” in Transgenic Plants: Methods and Protocols Methods in Molecular BiologyTM, ed L. Peña (Totowa, NJ: Humana Press), 79–90. doi: 10.1385/1-59259-827-7:079
Frank, S. D., and Tooker, J. F. (2020). Opinion: neonicotinoids pose undocumented threats to food webs. Proc. Natl. Acad. Sci. U.S.A. 117, 22609–22613. doi: 10.1073/pnas.2017221117
Frankel, A. D., and Pabo, C. O. (1988). Cellular uptake of the tat protein from human immunodeficiency virus. Cell 55, 1189–1193. doi: 10.1016/0092-8674(88)90263-2
Fromm, M., Taylor, L. P., and Walbot, V. (1985). Expression of genes transferred into monocot and dicot plant cells by electroporation. Proc. Natl. Acad. Sci. U.S.A. 82, 5824–5828. doi: 10.1073/pnas.82.17.5824
Fuente, J., de la Kocan, K. M., Almazán, C., and Blouin, E. F. (2007). RNA interference for the study and genetic manipulation of ticks. Trends Parasitol. 23, 427–433. doi: 10.1016/j.pt.2007.07.002
Galdeano, D. M., Breton, M. C., Lopes, J. R. S., Falk, B. W., and Machado, M. A. (2017). Oral delivery of double-stranded RNAs induces mortality in nymphs and adults of the Asian citrus psyllid, Diaphorina citri. PLoS ONE 12:e0171847. doi: 10.1371/journal.pone.0171847
Gillet, F.-X., Garcia, R. A., Macedo, L. L. P., Albuquerque, E. V. S., Silva, M. C. M., and Grossi-de-Sa, M. F. (2017). Investigating engineered ribonucleoprotein particles to improve oral RNAi delivery in crop insect pests. Front. Physiol. 8:256. doi: 10.3389/fphys.2017.00256
Godbey, W. T., Wu, K. K., and Mikos, A. G. (1999). Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res 45, 268–275. doi: 10.1002/(SICI)1097-4636(19990605)45:3<268::AID-JBM15>3.0.CO;2-Q
Godbey, W. T., Wu, K. K., and Mikos, A. G. (2001). Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials 22, 471–480. doi: 10.1016/S0142-9612(00)00203-9
Guan, R.-B., Li, H.-C., Fan, Y.-J., Hu, S.-R., Christiaens, O., Smagghe, G., et al. (2018). A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 293, 6011–6021. doi: 10.1074/jbc.RA117.001553
Gurusamy, D., Mogilicherla, K., and Palli, S. R. (2020a). Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 104:e21677. doi: 10.1002/arch.21677
Gurusamy, D., Mogilicherla, K., Shukla, J. N., and Palli, S. R. (2020b). Lipids help double-stranded RNA in endosomal escape and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 104:e21678. doi: 10.1002/arch.21678
Gutoaia, A., Schuster, L., Margutti, S., Laufer, S., Schlosshauer, B., Krastev, R., et al. (2016). Fine-tuned PEGylation of chitosan to maintain optimal siRNA-nanoplex bioactivity. Carbohydr. Polym. 143, 25–34. doi: 10.1016/j.carbpol.2016.01.010
Haensler, J., and Szoka, F. C. (1993). Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjugate Chem. 4, 372–379. doi: 10.1021/bc00023a012
He, B., Chu, Y., Yin, M., Müllen, K., An, C., and Shen, J. (2013). Fluorescent nanoparticle delivered dsRNA toward genetic control of insect pests. Adv. Mater. 25, 4580–4584. doi: 10.1002/adma.201301201
Howard, K. A., Rahbek, U. L., Liu, X., Damgaard, C. K., Glud, S. Z., Andersen, M. Ø., et al. (2006). RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol. Ther. 14, 476–484. doi: 10.1016/j.ymthe.2006.04.010
Huang, J.-H., Liu, Y., Lin, Y.-H., Belles, X., and Lee, H.-J. (2018). Practical use of RNA interference: oral delivery of double-stranded RNA in liposome carriers for cockroaches. J. Vis. Exp. 135:57385. doi: 10.3791/57385
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
Iranpur Mobarakeh, V., Modarressi, M. H., Rahimi, P., Bolhassani, A., Arefian, E., Atyabi, F., et al. (2019). Optimization of chitosan nanoparticles as an anti-HIV siRNA delivery vehicle. Int. J. Biol. Macromol. 129, 305–315. doi: 10.1016/j.ijbiomac.2019.02.036
Jiang, L., Ding, L., He, B., Shen, J., Xu, Z., Yin, M., et al. (2014). Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 6, 9965–9969. doi: 10.1039/C4NR03481C
Joga, M. R., Zotti, M. J., Smagghe, G., and Christiaens, O. (2016). RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7:553. doi: 10.3389/fphys.2016.00553
Joliot, A., Pernelle, C., Deagostini-Bazin, H., and Prochiantz, A. (1991). Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 88, 1864–1868. doi: 10.1073/pnas.88.5.1864
Jones, R. A., Poniris, M. H., and Wilson, M. R. (2004). pDMAEMA is internalised by endocytosis but does not physically disrupt endosomes. J. Controll. Release 96, 379–391. doi: 10.1016/j.jconrel.2004.02.011
Kang, S., Shin, D., Noh, M. Y., Peters, J. S., Smartt, C. T., Han, Y. S., et al. (2018). Optimization of double-stranded RNAi intrathoracic injection method in Aedes aegypti. Entomol Res. 48, 269–278. doi: 10.1111/1748-5967.12300
Katoch, R., Sethi, A., Thakur, N., and Murdock, L. L. (2013). RNAi for insect control: current perspective and future challenges. Appl. Biochem. Biotechnol. 171, 847–873. doi: 10.1007/s12010-013-0399-4
Kesharwani, P., Banerjee, S., Gupta, U., Mohd Amin, M. C. I., Padhye, S., Sarkar, F. H., et al. (2015). PAMAM dendrimers as promising nanocarriers for RNAi therapeutics. Mater. Today 18, 565–572. doi: 10.1016/j.mattod.2015.06.003
Khajuria, C., Ivashuta, S., Wiggins, E., Flagel, L., Moar, W., Pleau, M., et al. (2018). Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS ONE 13:e0197059. doi: 10.1371/journal.pone.0197059
Killiny, N., Hajeri, S., Tiwari, S., Gowda, S., and Stelinski, L. L. (2014). Double-stranded RNA uptake through topical application, mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri. PLoS ONE 9:e110536. doi: 10.1371/journal.pone.0110536
Klemm, A. R., Young, D., and Lloyd, J. B. (1998). Effects of polyethyleneimine on endocytosis and lysosome stability. Biochem. Pharmacol. 56, 41–46. doi: 10.1016/S0006-2952(98)00098-7
Kozielski, K. L., Tzeng, S. Y., and Green, J. J. (2013). Bioengineered nanoparticles for siRNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 5, 449–468. doi: 10.1002/wnan.1233
Kunath, K., von Harpe, A., Fischer, D., Petersen, H., Bickel, U., Voigt, K., et al. (2003). Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Controll. Release 89, 113–125. doi: 10.1016/S0168-3659(03)00076-2
Kunte, N., McGraw, E., Bell, S., Held, D., and Avila, L.-A. (2020). Prospects, challenges and current status of RNAi through insect feeding. Pest Manag. Sci. 76, 26–41. doi: 10.1002/ps.5588
Laisney, J., Gurusamy, D., Baddar, Z. E., Palli, S. R., and Unrine, J. M. (2020). RNAi in Spodoptera frugiperda Sf9 cells via nanomaterial mediated delivery of dsRNA: a comparison of poly-l-arginine polyplexes and poly-l-arginine-functionalized Au nanoparticles. ACS Appl. Mater. Interfaces 12, 25645–25657. doi: 10.1021/acsami.0c06234
Lakshmanan, M., Kodama, Y., Yoshizumi, T., Sudesh, K., and Numata, K. (2013). Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules 14, 10–16. doi: 10.1021/bm301275g
Layman, J. M., Ramirez, S. M., Green, M. D., and Long, T. E. (2009). Influence of polycation molecular weight on poly(2-dimethylaminoethyl methacrylate)-mediated DNA delivery in vitro. Biomacromolecules 10, 1244–1252. doi: 10.1021/bm9000124
Liao, X., Walden, G., Falcon, N. D., Donell, S., Raxworthy, M. J., Wormstone, M., et al. (2017). A direct comparison of linear and star-shaped poly(dimethylaminoethyl acrylate) polymers for polyplexation with DNA and cytotoxicity in cultured cell lines. Eur. Polym. J. 87, 458–467. doi: 10.1016/j.eurpolymj.2016.08.021
Li-Byarlay, H., Li, Y., Stroud, H., Feng, S., Newman, T. C., Kaneda, M., et al. (2013). RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. U.S.A. 110, 12750–12755. doi: 10.1073/pnas.1310735110
Liechty, W. B., Scheuerle, R. L., Vela Ramirez, J. E., and Peppas, N. A. (2019). Cytoplasmic delivery of functional siRNA using pH-responsive nanoscale hydrogels. Int. J. Pharm. 562, 249–257. doi: 10.1016/j.ijpharm.2019.03.013
Lin, S., Du, F., Wang, Y., Ji, S., Liang, D., Yu, L., et al. (2008). An acid-labile block copolymer of PDMAEMA and PEG as potential carrier for intelligent gene delivery systems. Biomacromolecules 9, 109–115. doi: 10.1021/bm7008747
Lin, Y.-H., Huang, J.-H., Liu, Y., Belles, X., and Lee, H.-J. (2017). Oral delivery of dsRNA lipoplexes to German cockroach protects dsRNA from degradation and induces RNAi response. Pest Manag. Sci. 73, 960–966. doi: 10.1002/ps.4407
Liu, S., Jaouannet, M., Dempsey, D. A., Imani, J., Coustau, C., and Kogel, K.-H. (2020). RNA-based technologies for insect control in plant production. Biotechnol. Adv. 39:107463. doi: 10.1016/j.biotechadv.2019.107463
Lu, C., Li, Z., Chang, L., Dong, Z., Guo, P., Shen, G., et al. (2019). Efficient delivery of dsRNA and DNA in cultured silkworm cells for gene function analysis using PAMAM dendrimers system. Insects 11:12. doi: 10.3390/insects11010012
Lundgren, J. G., and Duan, J. J. (2013). RNAi-based insecticidal crops: potential effects on nontarget species. Bioscience 63, 657–665. doi: 10.1525/bio.2013.63.8.8
Lungwitz, U., Breunig, M., Blunk, T., and Göpferich, A. (2005). Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharmaceut. Biopharmac. 60, 247–266. doi: 10.1016/j.ejpb.2004.11.011
Malhotra, M., Tomaro-Duchesneau, C., and Prakash, S. (2013). Synthesis of TAT peptide-tagged PEGylated chitosan nanoparticles for siRNA delivery targeting neurodegenerative diseases. Biomaterials 34, 1270–1280. doi: 10.1016/j.biomaterials.2012.10.013
Malone, R. W., Felgner, P. L., and Verma, I. M. (1989). Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. U.S.A. 86, 6077–6081. doi: 10.1073/pnas.86.16.6077
Mamta, B., and Rajam, M. V. (2017). RNAi technology: a new platform for crop pest control. Physiol. Mol. Biol. Plants 23, 487–501. doi: 10.1007/s12298-017-0443-x
Manganiello, M. J., Cheng, C., Convertine, A. J., Bryers, J. D., and Stayton, P. S. (2012). Diblock copolymers with tunable pH transitions for gene delivery. Biomaterials 33, 2301–2309. doi: 10.1016/j.biomaterials.2011.11.019
Matyjaszewski, K. (2012). Atom transfer radical polymerization (ATRP): current status and future perspectives. Macromolecules 45, 4015–4039. doi: 10.1021/ma3001719
McGaughey, W. H. (1994). Problems of insect resistance to Bacillus thuringiensis. Agric. Ecosyst. Environ. 49, 95–102. doi: 10.1016/0167-8809(94)90027-2
Mitter, N., Worrall, E. A., Robinson, K. E., Li, P., Jain, R. G., Taochy, C., et al. (2017). Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nature Plants 3, 1–10. doi: 10.1038/nplants.2016.207
Mongelli, V., and Saleh, M.-C. (2016). Bugs are not to be silenced: small RNA pathways and antiviral responses in insects. Ann. Rev. Virol. 3, 573–589. doi: 10.1146/annurev-virology-110615-042447
Moon, J. H., Mendez, E., Kim, Y., and Kaur, A. (2011). Conjugated polymer nanoparticles for small interfering RNA delivery. Chem. Commun. 47, 8370–8372. doi: 10.1039/C1CC10991J
Mostaghaci, B., Susewind, J., Kickelbick, G., Lehr, C.-M., and Loretz, B. (2015). Transfection system of amino-functionalized calcium phosphate nanoparticles: in vitro efficacy, biodegradability, and immunogenicity study. ACS Appl. Mater. Interfaces 7, 5124–5133. doi: 10.1021/am507193a
Mota-Sanchez, D., and Wise, J. C. (2017). Field Evolved Resistance of Arthropods to Pesticides: An Analysis of Resistance Adaptation from 1914 to 2017. ESA. Available online at: https://esa.confex.com/esa/2017/meetingapp.cgi/Paper/126977 (accessed September 7, 2020).
Mulot, M., Boissinot, S., Monsion, B., Rastegar, M., Clavijo, G., Halter, D., et al. (2016). Comparative analysis of RNAi-based methods to down-regulate expression of two genes expressed at different levels in Myzus persicae. Viruses 8:316. doi: 10.3390/v8110316
Nakasu, E. Y. T., Williamson, S. M., Edwards, M. G., Fitches, E. C., Gatehouse, J. A., Wright, G. A., et al. (2014). Novel biopesticide based on a spider venom peptide shows no adverse effects on honeybees. Proc. R. Soc. B 281:20140619. doi: 10.1098/rspb.2014.0619
Numata, K., and Kaplan, D. L. (2010). Silk-based gene carriers with cell membrane-destabilizing peptides. Biomacromolecules 11, 3189–3195. doi: 10.1021/bm101055m
Numata, K., Ohtani, M., Yoshizumi, T., Demura, T., and Kodama, Y. (2014). Local gene silencing in plants via synthetic dsRNA and carrier peptide. Plant Biotechnol. J. 12, 1027–1034. doi: 10.1111/pbi.12208
Palli, S. R. (2014). RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci. 6, 1–8. doi: 10.1016/j.cois.2014.09.011
Park, S., Jeong, E. J., Lee, J., Rhim, T., Lee, S. K., and Lee, K. Y. (2013). Preparation and characterization of nonaarginine-modified chitosan nanoparticles for siRNA delivery. Carbohydr. Polym. 92, 57–62. doi: 10.1016/j.carbpol.2012.08.116
Parker, K. M., Barragán Borrero, V., van Leeuwen, D. M., Lever, M. A., Mateescu, B., and Sander, M. (2019). Environmental Fate of RNA interference pesticides: adsorption and degradation of double-stranded RNA molecules in agricultural soils. Environ. Sci. Technol. 53, 3027–3036. doi: 10.1021/acs.est.8b05576
Parsons, K. H., Holley, A. C., Munn, G. A., Flynt, A. S., and McCormick, C. L. (2016). Block ionomer complexes consisting of siRNA and aRAFT-synthesized hydrophilic-block-cationic copolymers II: the influence of cationic block charge density on gene suppression. Polym. Chem. 7, 6044–6054. doi: 10.1039/C6PY01048B
Parsons, K. H., Mondal, M. H., McCormick, C. L., and Flynt, A. S. (2018). Guanidinium-functionalized interpolyelectrolyte complexes enabling RNAi in resistant insect pests. Biomacromolecules 19, 1111–1117. doi: 10.1021/acs.biomac.7b01717
Peng, Y., Wang, K., Fu, W., Sheng, C., and Han, Z. (2018). Biochemical comparison of dsRNA degrading nucleases in four different insects. Front. Physiol. 9:624. doi: 10.3389/fphys.2018.00624
Prevette, L. E., Mullen, D. G., and Banaszak Holl, M. M. (2010). Polycation-induced cell membrane permeability does not enhance cellular uptake or expression efficiency of delivered DNA. Mol. Pharmaceutics 7, 870–883. doi: 10.1021/mp100027g
Price, D. R. G., and Gatehouse, J. A. (2008). RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400. doi: 10.1016/j.tibtech.2008.04.004
Pridgeon, J. W., Zhao, L., Becnel, J. J., Strickman, D. A., Clark, G. G., and Linthicum, K. J. (2008). Topically applied AaeIAP1 double-stranded RNA kills female adults of Aedes aegypti. J. Med. Entomol. 45, 414–420. doi: 10.1093/jmedent/45.3.414
Ramesh Kumar, D., Saravana Kumar, P., Gandhi, M. R., Al-Dhabi, N. A., Paulraj, M. G., and Ignacimuthu, S. (2016). Delivery of chitosan/dsRNA nanoparticles for silencing of wing development vestigial (vg) gene in Aedes aegypti mosquitoes. Int. J. Biol. Macromol. 86, 89–95. doi: 10.1016/j.ijbiomac.2016.01.030
Romeis, J., and Widmer, F. (2020). Assessing the risks of topically applied dsRNA-based products to non-target arthropods. Front. Plant Sci. 11:679. doi: 10.3389/fpls.2020.00679
Ros, S., Wang, J., Burke, N. A. D., and Stöver, H. D. H. (2020). A mechanistic study of the hydrolysis of poly[N,N-(dimethylamino)ethyl acrylates] as charge-shifting polycations. Macromolecules 53, 3514–3523. doi: 10.1021/acs.macromol.9b02272
Roy, I., Mitra, S., Maitra, A., and Mozumdar, S. (2003). Calcium phosphate nanoparticles as novel non-viral vectors for targeted gene delivery. Int. J. Pharm. 250, 25–33. doi: 10.1016/S0378-5173(02)00452-0
Rudzinski, W. E., Palacios, A., Ahmed, A., Lane, M. A., and Aminabhavi, T. M. (2016). Targeted delivery of small interfering RNA to colon cancer cells using chitosan and PEGylated chitosan nanoparticles. Carbohydr. Polym. 147, 323–332. doi: 10.1016/j.carbpol.2016.04.041
Saleh, M., Rij, R. P., van Hekele, A., Gillis, A., Foley, E., O'Farrell, P. H., et al. (2006). The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802. doi: 10.1038/ncb1439
Santos, V. S. V., and Pereira, B. B. (2020). Properties, toxicity and current applications of the biolarvicide spinosad. J. Toxicol. Environm. Health Part B 23, 13–26. doi: 10.1080/10937404.2019.1689878
Schönemann, E., Laschewsky, A., and Rosenhahn, A. (2018). Exploring the long-term hydrolytic behavior of zwitterionic polymethacrylates and polymethacrylamides. Polymers 10:639. doi: 10.3390/polym10060639
Scott, J. G. (2008). Unraveling the mystery of spinosad resistance in insects. J. Pestic. Sci. 33, 221–227. doi: 10.1584/jpestics.R08-04
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Serrano-Sevilla, I., Artiga, Á., Mitchell, S. G., De Matteis, L., and de la Fuente, J. M. (2019). Natural polysaccharides for siRNA delivery: nanocarriers based on chitosan, hyaluronic acid, and their derivatives. Molecules 24:2570. doi: 10.3390/molecules24142570
Sharma, S., Kooner, R., and Arora, R. (2017). “Insect pests and crop losses,” in Breeding Insect Resistant Crops for Sustainable Agriculture, eds R. Arora and S. Sandhu (Singapore: Springer), 45–66. doi: 10.1007/978-981-10-6056-4_2
Shen, D., Zhou, F., Xu, Z., He, B., Li, M., Shen, J., et al. (2014). Systemically interfering with immune response by a fluorescent cationic dendrimer delivered gene suppression. J. Mater. Chem. B 2, 4653–4659. doi: 10.1039/C4TB00411F
Shukla, J. N., Kalsi, M., Sethi, A., Narva, K. E., Fishilevich, E., Singh, S., et al. (2016). Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 13, 656–669. doi: 10.1080/15476286.2016.1191728
Silva, A. T., Nguyen, A., Ye, C., Verchot, J., and Moon, J. H. (2010). Conjugated polymer nanoparticles for effective siRNA delivery to tobacco BY-2 protoplasts. BMC Plant Biol. 10:291. doi: 10.1186/1471-2229-10-291
Singh, I. K., Singh, S., Mogilicherla, K., Shukla, J. N., and Palli, S. R. (2017). Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 7:17059. doi: 10.1038/s41598-017-17134-2
Smith, D., Holley, A. C., and McCormick, C. L. (2011). RAFT-synthesized copolymers and conjugates designed for therapeutic delivery of siRNA. Polym. Chem. 2, 1428–1441. doi: 10.1039/C1PY00038A
Sparks, J., Slobodkin, G., Matar, M., Congo, R., Ulkoski, D., Rea-Ramsey, A., et al. (2012). Versatile cationic lipids for siRNA delivery. J. Control. Release 158, 269–276. doi: 10.1016/j.jconrel.2011.11.006
Sparks, T. C., and Nauen, R. (2015). IRAC: mode of action classification and insecticide resistance management. Pestic. Biochem. Physiol. 121, 122–128. doi: 10.1016/j.pestbp.2014.11.014
Sun, P., Huang, W., Jin, M., Wang, Q., Fan, B., Kang, L., et al. (2016). Chitosan-based nanoparticles for survivin targeted siRNA delivery in breast tumor therapy and preventing its metastasis. Int. J. Nanomedicine 11, 4931–4945. doi: 10.2147/IJN.S105427
Synatschke, C. V., Schallon, A., Jérôme, V., Freitag, R., and Müller, A. H. E. (2011). Influence of polymer architecture and molecular weight of poly(2-(dimethylamino)ethyl methacrylate) polycations on transfection efficiency and cell viability in gene delivery. Biomacromolecules 12, 4247–4255. doi: 10.1021/bm201111d
Tabashnik, B. E., Mota-Sanchez, D., Whalon, M. E., Hollingworth, R. M., and Carrière, Y. (2014). Defining terms for proactive management of resistance to Bt crops and pesticides. J. Econ. Entomol. 107, 496–507. doi: 10.1603/EC13458
Tan, Z., Jiang, Y., Ganewatta, M. S., Kumar, R., Keith, A., Twaroski, K., et al. (2019). Block Polymer micelles enable CRISPR/Cas9 ribonucleoprotein delivery: physicochemical properties affect packaging mechanisms and gene editing efficiency. Macromolecules 52, 8197–8206. doi: 10.1021/acs.macromol.9b01645
Taning, C. N. T., Christiaens, O., Berkvens, N., Casteels, H., Maes, M., and Smagghe, G. (2016). Oral RNAi to control Drosophila suzukii: laboratory testing against larval and adult stages. J. Pest. Sci. 89, 803–814. doi: 10.1007/s10340-016-0736-9
Terenius, O., Papanicolaou, A., Garbutt, J. S., Eleftherianos, I., Huvenne, H., Kanginakudru, S., et al. (2011). RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 57, 231–245. doi: 10.1016/j.jinsphys.2010.11.006
Thairu, M. W., Skidmore, I. H., Bansal, R., Nováková, E., Hansen, T. E., Li-Byarlay, H., et al. (2017). Efficacy of RNA interference knockdown using aerosolized short interfering RNAs bound to nanoparticles in three diverse aphid species. Insect Mol. Biol. 26, 356–368. doi: 10.1111/imb.12301
Theerawanitchpan, G., Saengkrit, N., Sajomsang, W., Gonil, P., Ruktanonchai, U., Saesoo, S., et al. (2012). Chitosan and its quaternized derivative as effective long dsRNA carriers targeting shrimp virus in Spodoptera frugiperda 9 cells. J. Biotechnol. 160, 97–104. doi: 10.1016/j.jbiotec.2012.04.011
Tomich, J. M., Wessel, E., Choi, J., and Avila, L. A. (2019). “Nonviral gene therapy: peptiplexes,” in Nucleic Acid Nanotheranostics eds Filice, M. and Ruiz-Cabello, J. (Amsterdam: Elsevier), 247–276. doi: 10.1016/B978-0-12-814470-1.00008-3
Treat, N. J., Smith, D., Teng, C., Flores, J. D., Abel, B. A., York, A. W., et al. (2012). Guanidine-containing methacrylamide (Co)polymers via aRAFT: toward a cell-penetrating peptide mimic. ACS Macro Lett. 1, 100–104. doi: 10.1021/mz200012p
Ulvila, J., Parikka, M., Kleino, A., Sormunen, R., Ezekowitz, R. A., Kocks, C., et al. (2006). Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in drosophila S2 cells. J. Biol. Chem. 281, 14370–14375. doi: 10.1074/jbc.M513868200
Unnamalai, N., Kang, B. G., and Lee, W. S. (2004). Cationic oligopeptide-mediated delivery of dsRNA for post-transcriptional gene silencing in plant cells. FEBS Lett. 566, 307–310. doi: 10.1016/j.febslet.2004.04.018
van de Wetering, P., Cherng, J.-Y., Talsma, H., and Hennink, W. E. (1997). Relation between transfection efficiency and cytotoxicity of poly(2-(dimethylamino)ethyl methacrylate)/plasmid complexes. J. Controll. Release 49, 59–69. doi: 10.1016/S0168-3659(97)00059-X
van de Wetering, P., Zuidam, N. J., van Steenbergen, M. J., van der Houwen, O. A. G. J., Underberg, W. J. M., and Hennink, W. E. (1998). A mechanistic study of the hydrolytic stability of poly(2-(dimethylamino)ethyl methacrylate). Macromolecules 31, 8063–8068. doi: 10.1021/ma980689g
Vivès, E., Brodin, P., and Lebleu, B. (1997). A truncated HIV-1 tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 272, 16010–16017. doi: 10.1074/jbc.272.25.16010
Vivès, E., Schmidt, J., and Pèlegrin, A. (2008). Cell-penetrating and cell-targeting peptides in drug delivery. Biochim. Biophys. Acta 1786, 126–138. doi: 10.1016/j.bbcan.2008.03.001
Vogel, E., Santos, D., Mingels, L., Verdonckt, T.-W., and Broeck, J. V. (2019). RNA Interference in insects: protecting beneficials and controlling pests. Front. Physiol. 9:1912. doi: 10.3389/fphys.2018.01912
von Harpe, A., Petersen, H., Li, Y., and Kissel, T. (2000). Characterization of commercially available and synthesized polyethylenimines for gene delivery. J. Controll. Release 69, 309–322. doi: 10.1016/S0168-3659(00)00317-5
Wang, D., Lin, J., Jia, F., Tan, X., Wang, Y., Sun, X., et al. (2019). Bottlebrush-architectured poly(ethylene glycol) as an efficient vector for RNA interference in vivo. Sci. Adv. 5:eaav9322. doi: 10.1126/sciadv.aav9322
Wang, F., Chen, H., Liu, Z., Mi, F., Fang, X., Liu, J., et al. (2019). Conjugated polymer dots for biocompatible siRNA delivery. New J. Chem. 43, 14443–14449. doi: 10.1039/C9NJ03277K
Wang, K., Peng, Y., Pu, J., Fu, W., Wang, J., and Han, Z. (2016). Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 77, 1–9. doi: 10.1016/j.ibmb.2016.07.007
Webb, J. E., and Green, R. A. (1945). On the penetration of insecticides through the insect cuticle. J. Exp. Biol. 22, 8–20.
Wessel, E. M., Tomich, J. M., and Todd, R. B. (2019). Biodegradable drug-delivery peptide nanocapsules. ACS Omega 4, 20059–20063. doi: 10.1021/acsomega.9b03245
Whitfield, R., Anastasaki, A., Truong, N. P., Cook, A. B., Omedes-Pujol, M., Loczenski Rose, V., et al. (2018). Efficient binding, protection, and self-release of dsRNA in soil by linear and star cationic polymers. ACS Macro Lett. 7, 909–915. doi: 10.1021/acsmacrolett.8b00420
Whyard, S., Erdelyan, C. N., Partridge, A. L., Singh, A. D., Beebe, N. W., and Capina, R. (2015). Silencing the buzz: a new approach to population suppression of mosquitoes by feeding larvae double-stranded RNAs. Parasit. Vect. 8:96. doi: 10.1186/s13071-015-0716-6
Whyard, S., Singh, A. D., and Wong, S. (2009). Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 39, 824–832. doi: 10.1016/j.ibmb.2009.09.007
Windley, M. J., Herzig, V., Dziemborowicz, S. A., Hardy, M. C., King, G. F., and Nicholson, G. M. (2012). Spider-venom peptides as bioinsecticides. Toxins 4, 191–227. doi: 10.3390/toxins4030191
Worrall, E. A., Bravo-Cazar, A., Nilon, A. T., Fletcher, S. J., Robinson, K. E., Carr, J. P., et al. (2019). Exogenous application of RNAi-inducing double-stranded rna inhibits aphid-mediated transmission of a plant virus. Front. Plant Sci. 10:265. doi: 10.3389/fpls.2019.00265
Wu, G. Y., and Wu, C. H. (1987). Receptor-mediated in vitro gene transformation by a soluble DNA carrier system. J. Biol. Chem. 262, 4429–4432.
Wynant, N., Santos, D., and Vanden Broeck, J. (2014). Biological mechanisms determining the success of RNA interference in insects. Int. Rev. Cell Mol. Biol. 312, 139–167. doi: 10.1016/B978-0-12-800178-3.00005-1
Wynant, N., Verlinden, H., Breugelmans, B., Simonet, G., and Vanden Broeck, J. (2012). Tissue-dependence and sensitivity of the systemic RNA interference response in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 42, 911–917. doi: 10.1016/j.ibmb.2012.09.004
Xu, X., Li, Z., Zhao, X., Keen, L., and Kong, X. (2016). Calcium phosphate nanoparticles-based systems for siRNA delivery. Regen Biomater. 3, 187–195. doi: 10.1093/rb/rbw010
Xu, Z., He, B., Wei, W., Liu, K., Yin, M., Yang, W., et al. (2014). Highly water-soluble perylenediimide-cored poly(amido amine) vector for efficient gene transfection. J. Mater. Chem. B 2, 3079–3086. doi: 10.1039/C4TB00195H
Xue, H. Y., Guo, P., Wen, W.-C., and Wong, H. L. (2015). Lipid-based nanocarriers for RNA delivery. Curr. Pharm. Des. 21, 3140–3147. doi: 10.2174/1381612821666150531164540
Yamano, S., Dai, J., and Moursi, A. M. (2010). Comparison of transfection efficiency of nonviral gene transfer reagents. Mol. Biotechnol. 46, 287–300. doi: 10.1007/s12033-010-9302-5
Yan, S., Ren, B., Zeng, B., and Shen, J. (2020). Improving RNAi efficiency for pest control in crop species. BioTechniq. 68, 283–290. doi: 10.2144/btn-2019-0171
Yang, F., Huang, W., Li, Y., Liu, S., Jin, M., Wang, Y., et al. (2013). Anti-tumor effects in mice induced by survivin-targeted siRNA delivered through polysaccharide nanoparticles. Biomaterials 34, 5689–5699. doi: 10.1016/j.biomaterials.2013.03.047
Yang, N. J., and Hinner, M. J. (2015). Getting across the cell membrane: an overview for small molecules, peptides, and proteins. Methods Mol. Biol. 1266, 29–53. doi: 10.1007/978-1-4939-2272-7_3
Yoon, J.-S., Gurusamy, D., and Palli, S. R. (2017). Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem. Mol. Biol. 90, 53–60. doi: 10.1016/j.ibmb.2017.09.011
Yu, N., Christiaens, O., Liu, J., Niu, J., Cappelle, K., Caccia, S., et al. (2013). Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci. 20, 4–14. doi: 10.1111/j.1744-7917.2012.01534.x
Zhang, S., Zhi, D., and Huang, L. (2012). Lipid-based vectors for siRNA delivery. J. Drug Target. 20, 724–735. doi: 10.3109/1061186X.2012.719232
Zhang, X., Mysore, K., Flannery, E., Michel, K., Severson, D. W., Zhu, K. Y., et al. (2015). Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. J. Vis. Exp. 97:e52523. doi: 10.3791/52523
Zhang, X., Zhang, J., and Zhu, K. Y. (2010). Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 19, 683–693. doi: 10.1111/j.1365-2583.2010.01029.x
Zhang, Y., Cui, J., Zhou, Y., Cao, J., Gong, H., Zhang, H., et al. (2018). Liposome mediated double-stranded RNA delivery to silence ribosomal protein P0 in the tick Rhipicephalus haemaphysaloides. Ticks Tick Borne Dis. 9, 638–644. doi: 10.1016/j.ttbdis.2018.01.015
Zheng, Y., Hu, Y., Yan, S., Zhou, H., Song, D., Yin, M., et al. (2019). A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag. Sci. 75, 1993–1999. doi: 10.1002/ps.5313
Zhou, R., Hotta, I., Denli, A. M., Hong, P., Perrimon, N., and Hannon, G. J. (2008). Comparative analysis of argonaute-dependent small RNA pathways in drosophila. Mol. Cell 32, 592–599. doi: 10.1016/j.molcel.2008.10.018
Zhu, F., Xu, J., Palli, R., Ferguson, J., and Palli, S. R. (2011). Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 67, 175–182. doi: 10.1002/ps.2048
Zhu, K. Y., and Palli, S. R. (2020). Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 65, 293–311. doi: 10.1146/annurev-ento-011019-025224
Ziebarth, J. D., and Wang, Y. (2010). Understanding the protonation behavior of linear polyethylenimine in solutions through Monte Carlo simulations. Biomacromolecules 11:29. doi: 10.1021/bm900842d
Keywords: polymer, RNA interference, crop protection, insect control, nanoparticles
Citation: Pugsley CE, Isaac RE, Warren NJ and Cayre OJ (2021) Recent Advances in Engineered Nanoparticles for RNAi-Mediated Crop Protection Against Insect Pests. Front. Agron. 3:652981. doi: 10.3389/fagro.2021.652981
Received: 13 January 2021; Accepted: 24 February 2021;
Published: 18 March 2021.
Edited by:
Thais B. Rodrigues, GreenLight Biosciences, United StatesReviewed by:
Olivier Christiaens, Ghent University, BelgiumCopyright © 2021 Pugsley, Isaac, Warren and Cayre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charlotte E. Pugsley, ZnNjcEBsZWVkcy5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.