- 1Department of Entomology, Plant Pathology and Nematology, University of Idaho, Moscow, ID, United States
- 2Department of Crop and Soil Sciences, Washington State University, Pullman, WA, United States
- 3Statistical Programs, College of Agricultural and Life Sciences, University of Idaho, Moscow, ID, United States
Quantification of variation for phenotypic traits within and among weed populations facilitate understanding of invasion mechanisms and management tactics. In the Pacific Northwest (PNW), USA, in response to climate change and to improve sustainability, producers are increasingly adopting broadleaf crops and cover crops, but Mayweed chamomile (Anthemis cotula L.) is a significant barrier to diversifying cropping systems because of its abundance and lack of herbicide options for its control. To quantify within-population phenotypic trait variation and heritability, plants (n = 300) from six half-sib families (i.e., seed source plants or mother plants) from each of 10 A. cotula populations (infested farms or sites) in the PNW were grown from seed through the flowering stage in the greenhouse common garden experiment. We measured percent seedling emergence, the initial date of flowering, flowering duration, plant biomass, number of flower heads, floral scent profiles, and other traits on individual plants. Trait variation was high among half-sib families within each population. For example, in two of the populations, percent seedling emergence within 30 days of planting ranged from 5 to 41% and 3 to 53%, respectively. As another example, initial date of flowering in two other populations ranged from 61 to 93 days and 58 to 92 days, respectively. Differences among half-sib families were greatest for flowering period, which differed by a month in most populations, and floral scent profiles. Heritability estimates were higher than 1.0 for most phenotypic traits, indicating that the study plants were more closely related than half-sibs (i.e., included full-sibs or products of selfing). These patterns of phenotypic trait variation are potentially caused by local edaphoclimatic factors and within-field farm management practices, suggesting that management of A. cotula might be challenging and differ within and across farms.
Introduction
Mayweed chamomile (Anthemis cotula L.; Asteraceae) is an annual, bushy, ill-scented, and globally invasive weed that originated in Eurasia to have a wide geographic range and long invasion history (Kay, 1971; Adhikari S. et al., 2020). It has been introduced worldwide, presumably as a contaminant of crop seed and other plant materials (CABI, 2018), and has become abundant especially in Mediterranean-like climates such as in the Pacific Northwest (PNW), USA. It can be aggressively weedy in croplands and pastures, significantly reducing crop yields and forage quality (Kay, 1971; Adhikari S. et al., 2020). Anthemis cotula has been present in the PNW for more than 143 years, but it has recently become much more problematic (Adhikari S. et al., 2020). Despite increases or stable crop production in recent decades, uncertainties caused by climate change and pest challenges have threatened low-input cereal-based cropping systems and, progressive farmers are responding by adopting climate-change-resilient practices including intercropping and cover crops, fall- and spring-seeded pulses and oilseed Brassica crops into cereal rotations (Eigenbrode et al., 2013; O'Leary et al., 2018). These new crops, however, are vulnerable to A. cotula because it can outcompete them and there is a lack of compatible herbicides for its management (Lyon et al., 2017). As a result, PNW growers indicate anecdotally that A. cotula is more severe in broadleaf cover crops than in cereal crops and presents a substantial barrier to diversifying their cropping systems. To help understand A. cotula and the management challenges it presents, there is a need to assess systematically the phenotypic traits that contribute to its invasiveness and weediness.
Invasive species like A. cotula have adapted and colonized habitats with diverse ecological conditions worldwide and are a threat to native biodiversity, a threat which is expected to be exacerbated by anthropogenic disturbance and global environmental change (Lee and Gelembiuk, 2008; Richardson and Pyšek, 2012; Downey and Richardson, 2016). Cropping systems are particularly vulnerable to plant invasion. Modern monoculture-based intensive farming practices fragment agricultural landscapes and disturb or displace native flora (Tilman et al., 2011; Chaudhary et al., 2016; Adhikari et al., 2019), enabling invasion by plant species that colonize and evolve rapidly (Williams et al., 2016). Under climate change, not only are the adverse effects of modern agriculture expected to intensify (Bradley et al., 2010; Giejsztowt et al., 2020) but widely distributed invasive weeds with long invasion histories are expanding their range and becoming more invasive (Adhikari A. et al., 2020; Sharma et al., 2020). These ecological challenges necessitate additional study to understand invasion drivers and avenues to improve weed management. Invasive species, usually with limited population sizes when initially colonizing novel environments, frequently adapt, accumulating phenotypic traits that facilitate their success in the new environment (Baker, 1974). For agricultural weeds, abiotic and biotic conditions created by edaphoclimatic variables and within-field management practices may exert localized selection pressure, causing adaptation to the site with implications for management practices (Gunton et al., 2011; Fried et al., 2012; Gaba et al., 2017; Yvoz et al., 2020). Traits such as seed germination, duration of flowering, and fecundity (i.e., abundant flowers or seeds) are known to contribute to invasiveness of plant populations (Pyšek and Richardson, 2008). Variation in traits such as seedling emergence rates within a weedy plant population could complicate management decisions (Sterling et al., 2004). Hence, identifying and quantifying variation in traits of adaptive significance could help improve management, reducing the potential long-term economic and environmental impacts of invasive weeds (Kueffer et al., 2013; Travlos, 2013).
Traits must be heritable for adaptation to occur, and heritability estimates help explain the phenotypic (i.e., underlying genetic) trait variation within a population. Heritability is a useful concept for evaluating and understanding invasive populations' evolutionary dynamics (Visscher et al., 2008; Zas and Sampedro, 2015). Heritability is the proportion of the total phenotypic variance due to genetic variance (Falconer and Mackay, 1989; Zhao et al., 2018) relative to the total phenotypic variance, so heritable traits are less controlled by the environment. Narrow-sense heritability is the proportion of phenotypic variance due to additive genetic variance only.
Anthemis cotula is an important, model, globally-invasive annual agricultural weed that has yet to be assessed for its phenotypic variation in any part of its invaded range. We delineated phenotypic trait variation among half-sib families (i.e., pooled seed from individual mother plants) collected from 10 A. cotula PNW populations using a common garden greenhouse experiment. We compared the traits seedling emergence, number of branches, initial day of flowering, number of flower heads, biomass production, plant height, flowering duration, and floral scent volatile organic compound (VOC) profiles (abundance, richness, diversity, evenness) among half-sib families from each of the 10 A. cotula populations. The study objectives were: (1) To assess the extent of phenotypic trait variation within A. cotula populations and determine if half-sib families explain more variation in traits than populations; and (2) Evaluate the extent of genetic variation (as estimated by heritability) of phenotypic traits within populations. We expected to find within-population (i.e., among half-sib families) variation in phenotypic traits in all of the populations.
Materials and Methods
Seed Collection and Greenhouse Common Garden Experiment
In the summer of 2018, seeds were harvested from six different individual plants from each of 10 A. cotula infested farms (names are based on nearest towns or farm name: Genesee, Kambitsch, Palouse, Parker, Potlatch, Spillman, St. John, Tensed, Thornton, and Troy) across a precipitation gradient (see Supplementary Table 1 for farm details) in the PNW. Seeds from each individual were kept separate, each constituting a half-sib family. Anthemis cotula is an obligate outcrossing species (Adhikari S. et al., 2020), and the seed from individual plants were assumed to be half-sibs for this analysis.
To measure phenotypic traits of individuals among half-sib families of 10 populations (i.e., sampling locations or farms), an experiment was conducted from February 27 to August 25, 2019. Anthemis cotula plants were grown in a common garden greenhouse under a 15-h photoperiod of sunlight and supplemental artificial light (high-pressure sodium lights of photosynthetic photon flux = 595 μmol m−2 s−1; model MVR1000/U, General Electric Company, Cleveland, OH) with an average temperature of 22.9 ± 0.26 (mean ± SE) °C and ambient humidity of 43.7 ± 9.5% (mean ± SE). Individual plants were grown in 2.3 L pots filled with a commercial greenhouse soil mix (PRO-MIX BX containing 75–80% Canadian sphagnum peat moss along with perlite and vermiculite; produced by Premier Tech Horticulture Ltd, Alberta, CA). From each of six families of the 10 populations (i.e., 60 seed source plants), 20 seeds were planted in each pot for a total of 300 pots, which were distributed on the greenhouse bench in a randomized complete block design with five replications. Pots were regularly watered as needed and not fertilized. Thirty days after seeding, seedlings were either thinned to a single plant per pot. If there was no emergence from any pot within a family, extra seedlings were transplanted into those pots to ensure within-family replication.
Phenotypic Traits
Data on 13 phenotypic traits were collected from plants in the study. The phenotypic traits assessed were percent seedling emergence in the first 30 days, days to first bud (time spent in vegetative growth), anthesis (days from the first bud to first opening or early flowering), the initial date of flowering (days to the first opening of bud), plant height (cm), number of branches, the final date of flowering (days to flower mature and wilting), flowering duration, the total number of flower heads, plant life (days from emergence to harvest), aboveground dry biomass (g) at harvest, and floral scent profiles. All traits were measured on every plant in the study except for floral scent characterization (see below).
Floral Scent Analysis
Four populations (Genesee, Kambitsch, Palouse Farm, and Thornton), that had at least three half-sib families each with three plants with five flowers blooming at the time of volatile collection, were selected for floral scent characterization among half-sibs of a subset of families. As an obligate out-crosser and a generalist species (Kay, 1971; Adhikari S. et al., 2020), A. cotula floral scents could be important to attract diverse pollinators and maximize seed production, contributing to invasiveness. When the plants were blooming abundantly, solid-phase microextraction [“SPME”: field sampler with 100-μm polydimethylsiloxane coated fiber; Supelco (Sigma-Aldrich) Bellefonte, PA, USA] was used to collect VOCs (volatile organic compounds) from eight or nine individual plants of each population. To collect VOCs, five live flowers, still attached to plants, were carefully inserted into a 90 mL (30 mm diameter) glass tube, and cotton was pushed in around the stems at the base of the tube to contain the airspace (Supplementary Figure 1). SPME fibers were then inserted through a 1 mm hole of a septum (PTFE silicon septum; 10 mm diameter) in the top of the tube and exposed for an hour. After exposure, the fibers were retracted and sampler was held on ice for 5 min while samples were transported to the laboratory for gas chromatography/mass spectrometry (GC/MS). An Agilent 7890A gas chromatograph/5973 Mass Selective Detector (Santa Clara, CA, USA) equipped with an EC-WAX polar column (30 m long, 0.25 mm × 0.25 μm film thickness: Grace, Deerfield, IL, USA) was used for analysis. Helium was used as the carrier gas at a constant flow rate of 1 mL min−1, and the injector temperature was held at 250°C. While analyzing samples, starting oven temperature was 60°C with a 3-min holding period, which later reached to a maximum temperature of 260°C after ramping 10°C per minute for 20 min. Compounds were provisionally identified based on match (≥80%) between their spectra and NIST-11 spectral library (National Institute of Standards and Technology, Gaithersburg, MD, USA) linked to Agilent Mass Hunter software. Compounds present in empty-tube controls were excluded as contaminants. Along with the abundance (total chromatogram m/z; mass-to-charge ratio) and richness (expressed as the number of compounds), Simpson's diversity (Simpson, 1949) and Pielou's evenness (Pielou, 1966) were calculated for VOCs.
Statistical Analysis
To assess phenotypic trait variation among half-sib families within each A. cotula population, linear mixed models were used for plant height and dry biomass:
Where yijk and yij are the response variables, μ is the mean, α(η)ik is the random effect for half-sib family within the population, ηk is the random effect for population, αi is a random effect due to half-sib family, βj is a random effect of the block, and ϵij is the error term. The R package “lme4” was used for analysis (Bates et al., 2015). Residuals were examined for homogeneity of variance, and Q-Q-plots were used to check for normality. For traits quantified as count variables, a generalized linear mixed model with a Poisson distribution and log link function was used, following the model framework described above. The count variables were days to seedling emergence, days to first budding, days to anthesis, days to first and last flowering, flowering duration (days), number of branches, number of flower heads, and plant life (days). For generalized linear models, the residual plots were also examined for homogeneity of variance. For percent emergence data, mixed-model beta regression with function “glmmTMB (generalized linear mixed model using template model builder)” was used (Brooks et al., 2017). In addition to the univariate analysis, using populations and half-sib families in the models, we also performed the variance partitioning to estimate the percentage of variation explained by half-sib families within populations for each trait, and the significance of variation was tested by using a likelihood ratio test. For floral VOC diversity and evenness, simple linear models were used and for VOC abundance and richness generalized linear models with a Poisson distribution and log link function were used. All data analyses were performed using R 3v.5.1 (R Development Core Team, 2018).
To assess if the overall composition of phenotypic traits were different among half-sib families in each population, we conducted a permutational multivariate analysis of variance (PERMANOVA) on a Bray-Curtis dissimilarity matrix (Bray and Curtis, 1957) of 10 phenotypic traits [plant height, the total number of branches, first budding date, first flowering date, anthesis, plant life, flowering period, last day of flowering, the total number of flower heads, and plant dry biomass (g)]. The results were visualized with Non-metric Multidimensional Scaling (NMDS) ordination. The package “vegan” was used for conducting PERMANOVA and NMDS (Oksanen, 2019).
Heritability
We calculated narrow-sense heritability (h2) on each phenotypic trait of 10 populations using mixed-effect models [Equation (2); see details in Statistical Analysis], with half-sib family and block as random effects.
Because half-sibs are expected to share ¼ of their genetic information by descent, four times the intraclass correlation of half-sib families can be used as an estimate of narrow-sense heritability (Equation 3) although maternal half-sib families may have an upward bias due to maternal environmental effects (Falconer and Mackay, 1989). In plants, maternal environmental effects are especially prevalent in seed traits such as seed weight or germination rate (Hendrix, 1984).
Results
Phenotypic Traits
The first day of budding and flowering periods varied among and within half-sib families within each of the 10 A. cotula populations or sampling locations (Table 1, Supplementary Table 2). Phenotypic variation among half-sib families was observed in nine populations for first flowering day and total flower heads, eight populations for seedling emergence within 30 days of planting and last day of flowering, six populations for plant life, four populations for plant height, two populations for total branches and plant dry biomass, and zero populations for anthesis (Table 1, Supplementary Table 2). For example, percent seedling emergence within 30 days of planting among half-sib families ranged from 2 to 35% in Parker Farm, 5 to 41% in Thornton, and 3 to 53% in Troy populations. Similarly, the initial date of flowering among half-sib families ranged from 61 to 93 days in Genesee and 58 to 92 days in Parker populations. Also, half-sibs of some individual families in Kambitsch, Palouse, St. John, and Thornton populations flowered for more than 1 month longer than the half-sibs of other families within these populations. Variance partitioning calculations indicated that half-sib families explained a higher proportion of variance than the population for all traits, except for anthesis and plant dry biomass (Table 2).
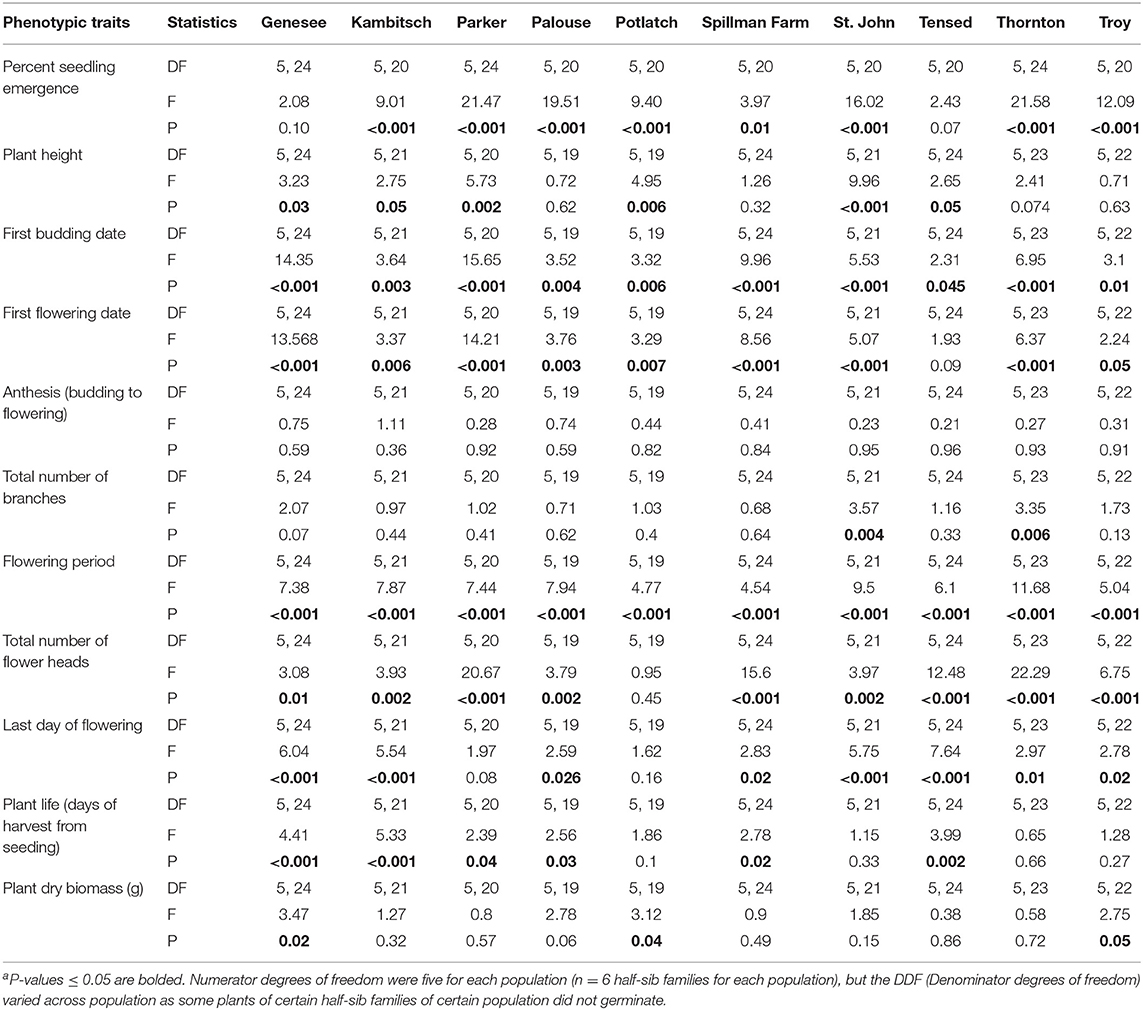
Table 1. Mixed effect model results for 11 phenotypic traits comparing among half-sib families across 10 Anthemis cotula populationsa.
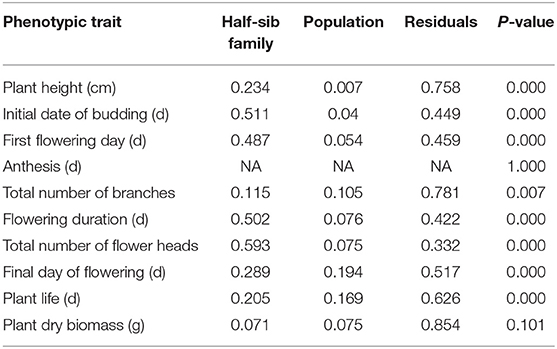
Table 2. Variation among half-sib families within population (i.e., variance partitioning) for each trait.
PERMANOVA on a Bray-Curtis dissimilarity matrix of 10 phenotypic traits [plant height, the total number of branches, first budding day, first flowering date, anthesis, plant life, flowering period, last day of flowering, the total number of flower heads, and plant dry biomass (g)] indicated that overall composition of traits in half-sibs among six families was dissimilar in five (Genesee, Parker Farm, Spillman, St. John, and Thornton), marginally dissimilar in one (Kambitsch), and similar in four (Palouse, Potlatch, Tensed, and Troy) populations (Figure 1, Table 3).
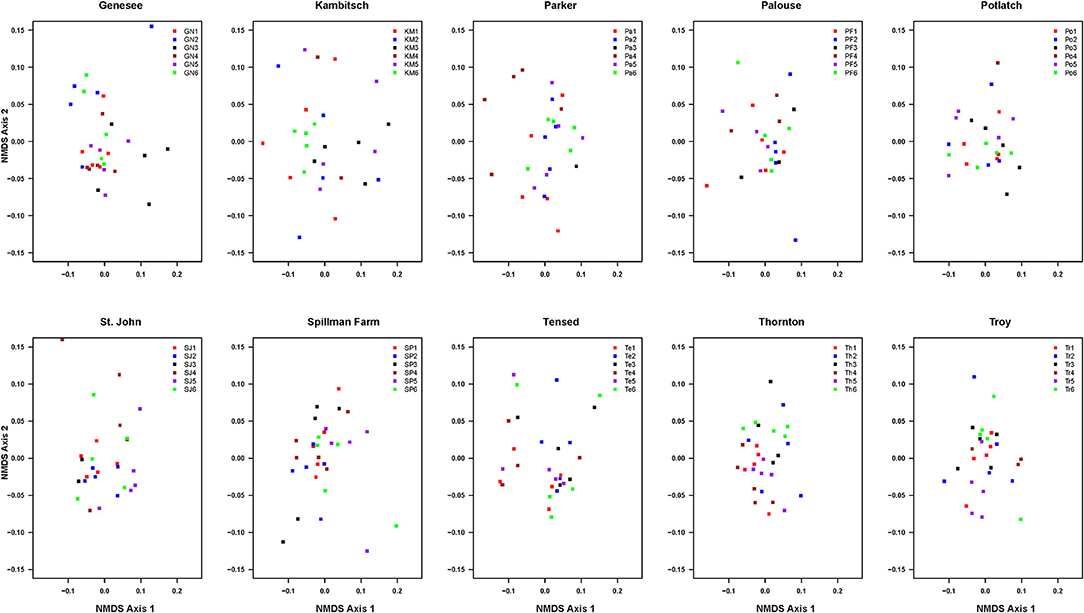
Figure 1. Non-metric multidimensional scaling (NMDS) ordination of selected phenotypic traits [plant height, the total number of branches, first budding date, first flowering date, anthesis, plant life, flowering period, last day of flowering, the total number of flower heads, and plant dry biomass (g)] of half-sibs among six families across 10 Anthemis cotula populations (See detail statistics for each population in Table 2).
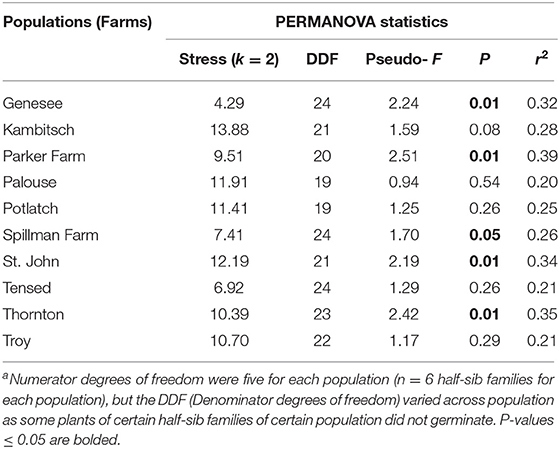
Table 3. Permutational Multivariate Analysis of Variance (PERMANOVA) statistics of half-sibs phenotypic traits among six mother plants across 10 Anthemis cotula populations (see Figure 1 for the ordination plots)a.
There was intrapopulation variation in floral scent VOCs within the four tested A. cotula populations. Differences among half-sib families occurred in VOC abundance (total chromatogram m/z) for all four populations (Figure 2, Table 4). VOC richness differed among half-sib families in the Kambitsch and Thornton populations, was marginally different in Genesee, and was not significant for the Palouse population (Figure 2, Table 4). Both Simpon's diversity and Pielou's evenness of floral VOCs differed among half-sib families within the Kambitsch population, but not within the Genesee, Palouse, or Thornton populations (Figure 2, Table 4).
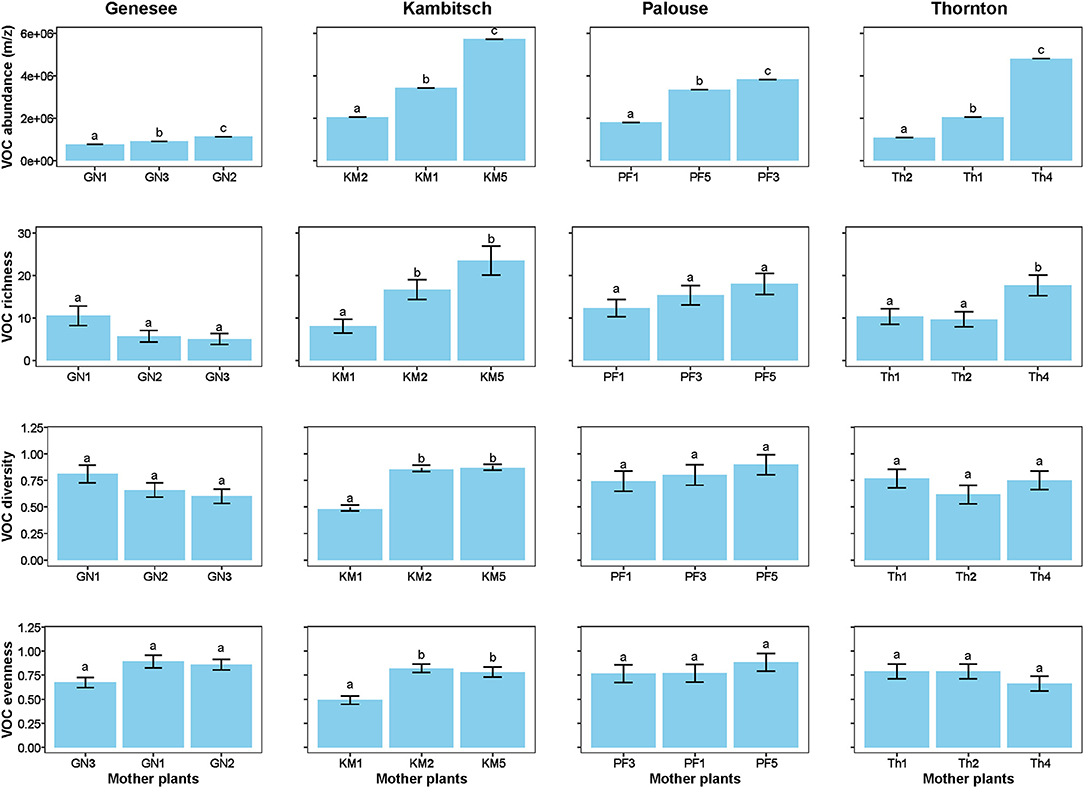
Figure 2. Genetic variation of half-sib families within each population (mean ± SE) floral VOCs abundance (m/z), richness, diversity, and evenness across four Anthemis cotula populations.
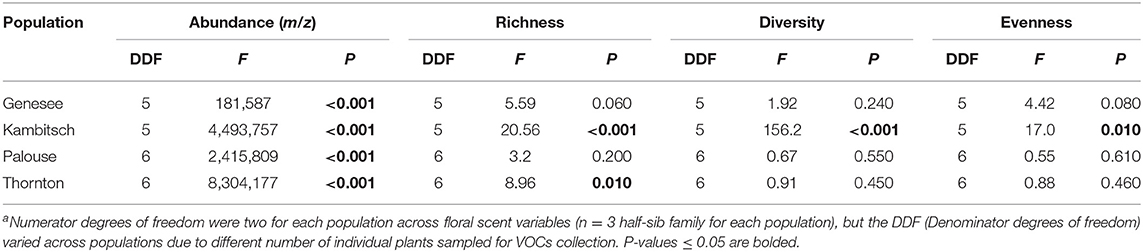
Table 4. Results from ANOVA for floral scent VOC abundance (m/z), richness, diversity, and evenness among half-sibs of three families across four Anthemis cotula populationsa.
Heritability
Considering the individual plants as half-sibs, heritability estimates for most phenotypic traits varied among populations and were high, occasionally reaching unusually high values greater than two (Table 5). Heritability estimates among tested populations ranged from 0.36 to 1.61 for seedling emergence, 0 to 2.44 for plant height, 0.52 to 2.56 for the initial date of budding, 0.39 to 2.53 for the initial date of flowering, 0 to 1.19 for total branches, 0 across all populations for anthesis, 0.67 to 2.37 for the flowering duration, 0.35 to 1.81 for the final date of flowering, 0 to 2.86 for total flower heads, 0 to 1.44 for plant life, and 0 to 1.23 for dry plant biomass (Table 5).
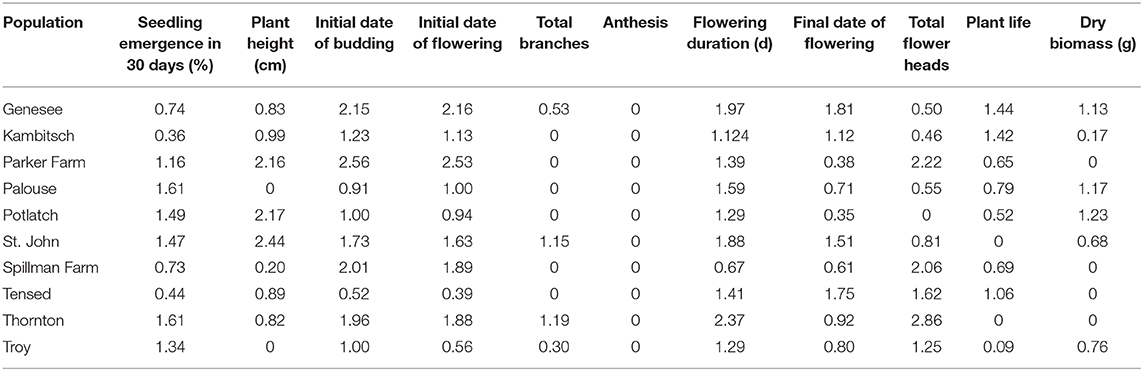
Table 5. Heritability (h2) estimates of 11 phenotypic traits for half-sib families from 10 Anthemis cotula populations.
Discussion
Previous studies have reported phenotypic traits associated with the invasiveness of plant species (Bossdorf et al., 2005; Pyšek and Richardson, 2008; Murphy et al., 2016) but these traits have been measured at the species level, rarely among populations, and never among half-sib families within populations of invasive plants. In our common garden study, most of the individual traits and trait compositions of half-sibs differed among families across populations of A. cotula. For example, percent seedling emergence within 30 days of planting among half-sib families ranged from 2 to 35% in Parker Farm, 5 to 41% in Thornton, and 3 to 53% in Troy populations. Similarly, the initial date of flowering among half-sib families ranged from 61 to 93 days in Genesee and 58 to 92 days in Parker populations. Also, half-sibs of some individual families in Kambitsch, Palouse, St. John, and Thornton populations flowered for more than one month longer time than the half-sibs of other families within these populations.
The principal potential sources that contribute to intrapopulation trait variation in A. cotula populations are (1) varying local selection pressures across space and time, (2) original genetic variation of the introduced populations, (3) genetic drift of initial small populations, and (4) human-mediated inadvertent introductions and reintroductions of unrelated germplasm to existing populations. Regarding the first of these sources, spatial and temporal variation in abiotic and biotic factors associated with soil and climate, and farm management practices could select for trait variation (Menalled et al., 2016; Bagavathiannan and Davis, 2018). Factors such as soil properties (Metcalfe et al., 2019; Pätzold et al., 2020), tillage, crop rotation, the movement patterns of farm equipment, herbicide regimes (Gao et al., 2018), and crop stand (Mhlanga et al., 2016; Kaur et al., 2018) can all come into play, with their spatial patterns (Blank et al., 2019) driving within-site variation in phenotypic traits [Cardina et al., 1997; Baucom and Mauricio, 2008; Vigueira et al., 2013; Hovick et al., 2018, also see Miller et al. (1994)]. Different traits respond differently to one or more of these factors (Prunier et al., 2012). The variation we detected indicates more studies are required to assess this variation to describe spatiotemporal variation in selection pressure on specific traits.
The three remaining sources of intrapopulation variation also could account for the trait variation we detected. The original variation of the introduced population or populations, genetic drift, and the number and frequency of past introduction events are unknown. The agricultural practices or variable climate that favor different traits in different years could have maintained or amplified the genetic diversity associated with the initial introductions. Whatever the basis for the trait variation we have detected, its presence indicates a potential for continued adaptation in response to ongoing drivers (Menalled et al., 2016; Hovick et al., 2018) contributing to weed invasiveness (Pyšek and Richardson, 2008; Murphy et al., 2016).
Floral scent profiles also differed among the half-sib families across A. cotula populations. Our results are consistent with previous studies (Delle-Vedove et al., 2017; Campbell et al., 2019; Friberg et al., 2019), which indicated that VOCs could differ environmentally within and between populations based on diel period and other environmental drivers, or reflect responses to selection by pollinators and natural enemies, with a genetic basis. Our common garden data indicate that the floral scent traits of A. cotula are heritable with the potential to continue evolving to improve fitness of local populations. For pollination of an outcrossing invasive species such as A. cotula, floral scents could ensure immediate fitness of the plants, but also attract some floral visitors away from the native plants (Burkle and Runyon, 2017; Campbell et al., 2019). Variable timing and duration of flowering, flowering for extended periods, and producing floral scent diversity among individual plants within a population could contribute to greater visitation of local pollinators and contribute to success of invaders.
Heritability estimates differed for traits within populations with higher values for seedling emergence, plant height, first budding and flowering days, flowering duration, the final date of flowering, and total flower heads than those for other traits. Previous common garden and field studies have found that the heritability of traits in outcrossing species [reviewed in Geber and Griffen (2003)] and with larger range sizes (Hoffmann and Sgró, 2011; Zeng X. et al., 2017) tend to be higher than for species with some inbreeding or smaller range size. This seems consistent with our understanding of A. cotula ecology in the PNW. On the other hand, heritability estimates are generally higher or over-estimated in laboratory or greenhouse conditions (Conner et al., 2007), particularly for highly plastic traits such as seedling emergence, plant height, and phenological traits (e.g., first budding and flowering days; Dicenta et al., 1993; Geber and Griffen, 2003; Franks et al., 2007; Wright et al., 2020). Hence, high heritability in A. cotula could have occurred because the environmental variance component is reduced, and additive variance is increased in the controlled setting in the greenhouse common garden (Geber and Griffen, 2003; Conner et al., 2007).
High heritability estimates observed in many of the traits in our study indicate that the individuals were more related than expected for half-sibs, with perhaps some inbreeding (caused by selfing, producing full sibs) or maternal effects. Maternal effects are believed to influence phenotypes and the ability to adapt to the novel environmental conditions (Leiblein-Wild and Tackenberg, 2014; Albecker and McCoy, 2019). High narrow-sense heritability estimates in our study may also suggest that many traits have the potential for adaptive evolution (Zeng X. et al., 2017) in A. cotula, an understudied yet globally invasive species. While A. cotula occurring predominantly in agricultural fields might be under selection pressure of within-field farm management practices interacting with edaphoclimatic factors, heritable traits respond to selection pressure (Christoffers, 1999). We currently lack data to support these possible explanations for the observed heritability. Future work to assess these will require a larger sample size grown for multiple generations.
Results from our A. cotula study have several implications for management. As A. cotula in the PNW mainly occurs in agricultural fields where its range is apparently expanding, genetically based phenotypic variation in the species will allow it to respond to local differences in farm management practices such as types of crop planted (Ogg et al., 1994), continuous cropping (Murphy and Lemerle, 2006), fertilizer (Lemke et al., 2015; Zeng M. et al., 2017), tillage (Ghersa and Martínez-Ghersa, 2000), mowing and herbicides (Lyon et al., 2017; Hovick et al., 2018). Herbicides impose robust directional selection on herbicide resistance traits per se (Powles and Yu, 2010; Neve et al., 2018) with pleiotropic effects on traits such as seed dormancy and germination (Délye et al., 2013). Adaptation in response to herbicide regimes can introduce requirements for robust weed management tactics (Menalled et al., 2016; Bagavathiannan and Davis, 2018). All these selection pressures are likely to shift as producers adopt cropping systems and management regimes in response to climate change. A. cotula, given its genetically based phenotypic variation, is poised to evolve in response, creating continuing difficulties for its management (Murphy and Lemerle, 2006; Vigueira et al., 2013; Hovick et al., 2018). Given this potential, integrated weed management employing “many little hammers” (Liebman and Gallandt, 1997) could minimize strong directional selection pressure.
The intrapopulation variation in traits and within a field could complicate management decisions (Sterling et al., 2004). For example, if some individuals in a field emerge and flower earlier a manager may choose to spray postemergence herbicide early. Later-emerging and flowering individuals from the same population will require additional treatment, or if not treated, lead to selection for later emergence increasing variability of that trait, exacerbating the problem. Understanding how phenotypic trait variation occurs within a field could allow targeted and more efficient herbicide treatments and reduce unnecessary applications (Cardina et al., 1997; Blank et al., 2019).
The degree of phenotypic trait variation in invading populations is thought to be determined by demographic history (Dlugosch and Parker, 2008; Hodgins and Rieseberg, 2011). The ongoing movement of A. cotula seeds among farms within or from outside in the PNW due to inadvertent yet common trade in crop seed, farming equipment and other inputs (Blanco-Moreno et al., 2004) should be a concern for farm managers, seed traders and others. These factors will contribute to maintaining genetic diversity of A. cotula, maintaining its potential to adapt to local environmental and management regimes. Hence, knowledge of anthropogenic A. cotula seed movement that could contribute to this diversity requires further study. Our study provides a baseline against which future assessments could be compared to assess ongoing changes in its phenotypic and genetic diversity.
Conclusions
Our study is the first of which we are aware that reports phenotypic trait variation among half-sib families in a globally invasive annual agricultural weed. In A. cotula, we found that half-sib families explained a higher proportion of variance than the population for most traits. Knowledge of plant functional traits among half-sibs across locally adapted plant populations can inform site-specific invasive species management and forecast their distributions in the context of global environmental changes. Farm management regime and several local abiotic (e.g., soil type, precipitation, and temperature) and biotic (e.g., natural enemies, flower visitors, symbiotic organisms, and co-occurring plant species) factors in agroecosystems could impose selection pressures affecting the response of A. cotula traits. As intrapopulation trait variation could favor different maternal lines under competitive local abiotic and biotic conditions, it could overall improve the invasion success of A. cotula. The current extensive (global) and intensive (local) distribution suggests that the climate niche of A. cotula is wide and its variable traits could help further spread during global environmental change, though further studies are required to confirm our hypothesis. Also, with the adoption of new crops, especially broadleaf crops and cover crops that are vulnerable to A. cotula because they are less competitive than traditional cereal crops or lack compatible herbicides for weed management, A. cotula could be more challenging to manage. To confirm whether the trait variation among half-sib families of A. cotula populations resulted from the adaptive responses to local selection pressure, how it is affected by maternal effects, selfing, or other factors, and how the trait variation could complicate within-farm weed management plans, requires additional study.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SA, ICB, and SDE designed the study. ICB and SDE acquired the funding and supervised the study. SA contributed to the seed collection, greenhouse experiment, data analysis, and manuscript preparation. SRR and JP contributed to data analysis. All authors contributed to the article, edited, and approved the submitted version.
Funding
This research is part of the LIT (Landscapes in Transition) project, supported by award #2017-68002-26819 and Hatch project 1017286 from the USDA National Institute of Food and Agriculture. Publication of this article was funded by the University of Idaho - Open Access Publishing Fund.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank PNW growers for allowing us to collect A. cotula seeds from their farms. We thank Philip Anderson for greenhouse space and support; Grace Overlie for seed counting; and Vincent Oliveras for data collection assistance. We are thankful to the Editor and Reviewers for their helpful comments on an earlier version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2021.640208/full#supplementary-material
References
Adhikari, A., Rew, L. J., Mainali, K. P., Adhikari, S., and Maxwell, B. D. (2020). Future distribution of invasive weed species across the major road network in the state of Montana, USA. Reg. Environ. Chang. 20, 1–14. doi: 10.1007/s10113-020-01671-0
Adhikari, S., Adhikari, A., Weaver, D. K., Bekkerman, A., and Menalled, F. D. (2019). Impacts of agricultural management systems on biodiversity and ecosystem services in highly simplified dryland landscapes. Sustainability 11:3223. doi: 10.3390/su11113223
Adhikari, S., Burke, I. C., and Eigenbrode, S. D. (2020). Mayweed chamomile (Anthemis cotula L.) biology and management—a review of an emerging global invader. Weed Res. 60, 313–322. doi: 10.1111/wre.12426
Albecker, M. A., and McCoy, M. W. (2019). Local adaptation for enhanced salt tolerance reduces non-adaptive plasticity caused by osmotic stress. Evolution 73, 1941–1957. doi: 10.1111/evo.13798
Bagavathiannan, M. V., and Davis, A. S. (2018). An ecological perspective on managing weeds during the great selection for herbicide resistance. Pest. Manag. Sci. 74, 2277–2286. doi: 10.1002/ps.4920
Baker, H. G. (1974). The evolution of weeds. Annu. Rev. Ecol. Syst. 5, 1–25. doi: 10.1146/annurev.es.05.110174.000245
Bates, D., Mächler, M., Bolker, B. M., and Walker, S. C. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67:i01. doi: 10.18637/jss.v067.i01
Baucom, R. S., and Mauricio, R. (2008). The evolution of novel herbicide tolerance in a noxious weed: the geographic mosaic of selection. Evol. Ecol. 22, 85–101. doi: 10.1007/s10682-007-9160-1
Blanco-Moreno, J. M., Chamorro, L., Masalles, R. M., Recasens, J., and Sans, F. X. (2004). Spatial distribution of Lolium rigidum seedlings following seed dispersal by combine harvesters. Weed Res. 44, 375–387. doi: 10.1111/j.1365-3180.2004.00412.x
Blank, L., Birger, N., and Eizenberg, H. (2019). Spatial and temporal distribution of Ecballium elaterium in almond orchards. Agronomy 9:751. doi: 10.3390/agronomy9110751
Bossdorf, O., Auge, H., Lafuma, L., Rogers, W. E., Siemann, E., and Prati, D. (2005). Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia 144, 1–11. doi: 10.1007/s00442-005-0070-z
Bradley, B. A., Blumenthal, D. M., Wilcove, D. S., and Ziska, L. H. (2010). Predicting plant invasions in an era of global change. Trends Ecol. Evol. 25, 310–318. doi: 10.1016/j.tree.2009.12.003
Bray, J. R., and Curtis, J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 325–349. doi: 10.2307/1942268
Brooks, M. E., Kristensen, K., Benthem, K. J., Magnusson, A., Berg, C. W., Nielsen, A., et al. (2017). glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R. J. 9, 378–400. doi: 10.32614/RJ-2017-066
Burkle, L. A., and Runyon, J. B. (2017). The smell of environmental change: using floral scent to explain shifts in pollinator attraction. Appl. Plant Sci. 5:1600123. doi: 10.3732/apps.1600123
CABI (2018). Anthemis cotula (dog fennel). Available online at: https://www.cabi.org/isc/datasheet/5672 (accessed January 5, 2020).
Campbell, D. R., Sosenski, P., and Raguso, R. A. (2019). Phenotypic plasticity of floral volatiles in response to increasing drought stress. Ann. Bot. 123, 601–610. doi: 10.1093/aob/mcy193
Cardina, J., Johnson, G. A., and Sparrow, D. H. (1997). The nature and consequence of weed spatial distribution. Weed Sci. 45, 364–373. doi: 10.1017/S0043174500092997
Chaudhary, A., Pfister, S., and Hellweg, S. (2016). Spatially explicit analysis of biodiversity loss due to global agriculture, pasture and forest land use from a producer and consumer perspective. Environ. Sci. Technol. 50, 3928–3936. doi: 10.1021/acs.est.5b06153
Christoffers, M. J. (1999). Genetic aspects of herbicide-resistant weed management. Weed Technol. 13, 647–652. doi: 10.1017/S0890037X00046340
Conner, J. K., Franks, R., and Stewart, C. (2007). Expression of additive genetic variances and covariances for wild radish floral traits: comparison between field and greenhouse environment. Evolution 57, 487–495. doi: 10.1111/j.0014-3820.2003.tb01540.x
Delle-Vedove, R., Schatz, B., and Dufay, M. (2017). Understanding intraspecific variation of floral scent in light of evolutionary ecology. Ann. Bot. 120, 1–20. doi: 10.1093/aob/mcx055
Délye, C., Menchari, Y., Michel, S., Cadet, É., and Le Corre, V. (2013). A new insight into arable weed adaptive evolution: mutations endowing herbicide resistance also affect germination dynamics and seedling emergence. Ann. Bot. 111, 681–691. doi: 10.1093/aob/mct018
Dicenta, F., Garcia, J. E., and Carbonell, E. A. (1993). Heritability of flowering, productivity and maturity in almond. J. Hortic. Sci. 68, 113–120. doi: 10.1080/00221589.1993.11516334
Dlugosch, K. M., and Parker, I. M. (2008). Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449. doi: 10.1111/j.1365-294X.2007.03538.x
Downey, P. O., and Richardson, D. M. (2016). Alien plant invasions and native plant extinctions: a six-threshold framework. AoB Plants 8:plw047. doi: 10.1093/aobpla/plw047
Eigenbrode, S. D., Capalbo, S. M., Houston, L. L., Johnson-Maynard, J., Kruger, C., and Olen, B. (2013). “Agriculture: Impacts, adaptation, and mitigation,” in Dalton, M. M., Mote, P. W., Snover, A. K., eds Climate Change in the Northwest: Implications for Our Landscapes, Waters, and Communities (Island Press-Center for Resource Economics), 149–180. doi: 10.5822/978-1-61091-512-0_6
Falconer, D. S., and Mackay, T. F. C. (1989). Introduction to Quantitative Genetics. 4th ed. (Essex: Longman Scientific & Technical), 448.
Franks, S. J., Sim, S., and Weis, A. E. (2007). Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. U. S. A. 104, 1278–1282. doi: 10.1073/pnas.0608379104
Friberg, M., Schwind, C., Guimaraes, P. R., Raguso, R. A., and Thompson, J. N. (2019). Extreme diversification of floral volatiles within and among species of Lithophragma (Saxifragaceae). Proc. Natl. Acad. Sci. U. S. A. 116, 4406–4415. doi: 10.1073/pnas.1809007116
Fried, G., Kazakou, E., and Gaba, S. (2012). Trajectories of weed communities explained by traits associated with species' response to management practices. Agric. Ecosyst. Environ. 158, 147–155. doi: 10.1016/j.agee.2012.06.005
Gaba, S., Perronne, R., Fried, G., Gardarin, A., Bretagnolle, F., Biju-Duval, L., et al. (2017). Response and effect traits of arable weeds in agro-ecosystems: a review of current knowledge. Weed Res. 57, 123–147. doi: 10.1111/wre.12245
Gao, P., Zhang, Z., Sun, G., Yu, H., and Qiang, S. (2018). The within-field and between-field dispersal of weedy rice by combine harvesters. Agron. Sustain. Dev. 38, 1–10. doi: 10.1007/s13593-018-0518-2
Geber, M. A., and Griffen, L. R. (2003). Inheritance and natural selection on functional traits. Int. J. Plant. Sci. 164, 21–42. doi: 10.1086/368233
Ghersa, C. M., and Martínez-Ghersa, M. A. (2000). Ecological correlates of weed seed size and persistence in the soil under different tilling systems: implications for weed management. F. Crop Res. 67, 141–148. doi: 10.1016/S0378-4290(00)00089-7
Giejsztowt, J., Classen, A. T., and Deslippe, J. R. (2020). Climate change and invasion may synergistically affect native plant reproduction. Ecology 101:e02913. doi: 10.1002/ecy.2913
Gunton, R. M., Petit, S., and Gaba, S. (2011). Functional traits relating arable weed communities to crop characteristics. J. Veg. Sci. 22, 541–550. doi: 10.1111/j.1654-1103.2011.01273.x
Hendrix, S. D. (1984). Variation in seed weight and its effects on germination in Pastinaca sativa L. (Umbelliferae). Am. J. Bot. 71:795. doi: 10.1002/j.1537-2197.1984.tb14144.x
Hodgins, K. A., and Rieseberg, L. (2011). Genetic differentiation in life-history traits of introduced and native common ragweed (Ambrosia artemisiifolia) populations. J. Evol. Biol. 24, 2731–2749. doi: 10.1111/j.1420-9101.2011.02404.x
Hoffmann, A. A., and Sgró, C. M. (2011). Climate change and evolutionary adaptation. Nature 470, 479–485. doi: 10.1038/nature09670
Hovick, S. M., McArdle, A., Harrison, S. K., and Regnier, E. E. (2018). A mosaic of phenotypic variation in giant ragweed (Ambrosia trifida): local- and continental-scale patterns in a range-expanding agricultural weed. Evol. Appl. 11, 995–1009. doi: 10.1111/eva.12614
Kaur, S., Kaur, R., and Chauhan, B. S. (2018). Understanding crop-weed-fertilizer-water interactions and their implications for weed management in agricultural systems. Crop prot. 103, 65–72. doi: 10.1016/j.cropro.2017.09.011
Kueffer, C., Pyšek, P., and Richardson, D. M. (2013). Integrative invasion science: model systems, multi-site studies, focused meta-analysis and invasion syndromes. New Phytol. 200, 15–633. doi: 10.1111/nph.12415
Lee, C. E., and Gelembiuk, G. W. (2008). Evolutionary origins of invasive populations. Evol. Appl. 1, 427–448. doi: 10.1111/j.1752-4571.2008.00039.x
Leiblein-Wild, M. C., and Tackenberg, O. (2014). Phenotypic variation of 38 European Ambrosia artemisiifolia populations measured in a common garden experiment. Biol. Invasions 16, 2003–2015. doi: 10.1007/s10530-014-0644-y
Lemke, I. H., Kolb, A., Graae, B. J., De Frenne, P., Acharya, K. P., Blandino, C., et al. (2015). Patterns of phenotypic trait variation in two temperate forest herbs along a broad climatic gradient. Plant Ecol. 216, 1523–1536. doi: 10.1007/s11258-015-0534-0
Liebman, M., and Gallandt, E. (1997). “Many little hammers: ecological management of crop-weed interactions,” in LE Jackson, ed Ecology in Agriculture (San Diego, CA: Academic Press), 291–343. doi: 10.1016/B978-012378260-1/50010-5
Lyon, D., Burke, I. C., Hulting, A. G., and Campbell, J. M. (2017). Integrated Management of Mayweed Chamomile in Wheat and Pulse Crop Production Systems. Pullman, WA: Washington State University Extension Publications PNW695:1–6.
Menalled, F. D., Peterson, R. K. D., Smith, R. G., Curran, W. S., Páez, D. J., and Maxwell, B. D. (2016). The eco-evolutionary imperative: revisiting weed management in the midst of an herbicide resistance crisis. Sustainability 8:1297. doi: 10.3390/su8121297
Metcalfe, H., Milne, A. E., Coleman, K., Murdoch, A. J., and Storkey, J. (2019). Modelling the effect of spatially variable soil properties on the distribution of weeds. Ecol. Modell. 396, 1–11. doi: 10.1016/j.ecolmodel.2018.11.002
Mhlanga, B., Chauhan, B. S., and Thierfelder, C. (2016). Weed management in maize using crop competition: a review. Crop Prot. 88, 28–36. doi: 10.1016/j.cropro.2016.05.008
Miller, T. E., Winn, A. A., and Schemske, D. W. (1994). The effects of density and spatial distribution on selection for emergence time in Prunella vulgaris (Lamiaceae). Am. J. Bot. 81, 1–6. doi: 10.1002/j.1537-2197.1994.tb15401.x
Murphy, C. E., and Lemerle, D. (2006). Continuous cropping systems and weed selection. Euphytica 148, 61–73. doi: 10.1007/s10681-006-5941-9
Murphy, J. E., Burns, J. H., Fougère-Danezan, M., and Drenovsky, R. E. (2016). Functional trait values, not trait plasticity, drive the invasiveness of Rosa sp. in response to light availability. Am. J. Bot. 103, 2058–2069. doi: 10.3732/ajb.1600235
Neve, P., Barney, J. N., Buckley, Y., Cousens, R. D., Graham, S., Jordan, N. R., et al. (2018). Reviewing research priorities in weed ecology, evolution and management: a horizon scan. Weed Res. 58, 250–258. doi: 10.1111/wre.12304
Ogg, A. G., Stephens, R. H., and Gealy, D. R. (1994). Interference between Mayweed chamomile (Anthemis cotula) and Pea (Pisum sativum) is affected by form of interference and soil water regime. Weed Sci. 42, 579–585. doi: 10.1017/S0043174500076980
Oksanen, J. (2019). CRAN - Package vegan. Available online at: https://cran.r-project.org/web/packages/vegan/index.html (accessed January 10, 2020).
O'Leary, G., Aggarwal, P., Calderini, D., Connor, D., Craufurd, P., Eigenbrode, S. D., et al. (2018). Challenges and responses to ongoing and projected climate change for dryland cereal production systems throughout the world. Agronomy 8:34. doi: 10.3390/agronomy8040034
Pätzold, S., Hbirkou, C., Dicke, D., Gerhards, R., and Welp, G. (2020). Linking weed patterns with soil properties: a long-term case study. Precis. Agric. 21, 569–588. doi: 10.1007/s11119-019-09682-6
Pielou, E. C. (1966). Species-diversity and pattern-diversity in the study of ecological succession. J. Theor. Biol. 10, 370–383. doi: 10.1016/0022-5193(66)90133-0
Powles, S. B., and Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Prunier, R., Holsinger, K. E., and Carlson, J. E. (2012). The effect of historical legacy on adaptation: do closely related species respond to the environment in the same way? J. Evol. Biol. 25, 1636–1649. doi: 10.1111/j.1420-9101.2012.02548.x
Pyšek, P., and Richardson, D. M. (2008). “Traits associated with invasiveness in alien plants: where do we stand?” in Nentwig, W., ed Biological Invasions (Berlin, Heidelberg: Springer, Berlin, Heidelberg), 97–125. doi: 10.1007/978-3-540-36920-2_7
R Development Core Team (2018). R Development Core Team. Available online at: https://cran.r-project.org/manuals.html (accessed May 20, 2020).
Richardson, D. M., and Pyšek, P. (2012). Naturalization of introduced plants: ecological drivers of biogeographical patterns. New Phytol. 196, 383–396. doi: 10.1111/j.1469-8137.2012.04292.x
Sharma, S., Walia, S., Rathore, S., Kumar, P., and Kumar, R. (2020). Combined effect of elevated CO2 and temperature on growth, biomass and secondary metabolite of Hypericum perforatum L. in a western Himalayan region. J. Appl. Res. Med. Aromat. Plants 16:100239. doi: 10.1016/j.jarmap.2019.100239
Sterling, T. M., Thompson, D. C., and Abbott, L. B. (2004). Implications of invasive plant variation for weed management. Weed Technol. 18, 1319–1324. doi: 10.1614/0890-037X(2004)018[1319:IOIPVF]2.0.CO;2
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U. S. A. 108:20260–20264. doi: 10.1073/pnas.1116437108
Travlos, I. S. (2013). Responses of invasive silverleaf nightshade (Solanum elaeagnifolium) populations to varying soil water availability. Phytoparasitica 41, 41–48. doi: 10.1007/s12600-012-0262-0
Vigueira, C. C., Olsen, K. M., and Caicedo, A. L. (2013). The red queen in the corn: agricultural weeds as models of rapid adaptive evolution. Heredity 110, 303–311. doi: 10.1038/hdy.2012.104
Visscher, P. M., Hill, W. G., and Wray, N. R. (2008). Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet. 9, 255–266. doi: 10.1038/nrg2322
Williams, J. L., Kendall, B. E., and Levine, J. M. (2016). Rapid evolution accelerates plant population spread in fragmented experimental landscapes. Science 353, 482–485. doi: 10.1126/science.aaf6268
Wright, T. I. C., Burnett, A. C., Griffiths, H., Kadner, M., Powell, J. S., Oliveira, H. R., et al. (2020). Identification of quantitative trait loci relating to flowering time, flag leaf and awn characteristics in a novel Triticum dicoccum mapping population. Plants 9:829. doi: 10.3390/plants9070829
Yvoz, S., Cordeau, S., Zuccolo, C., and Petit, S. (2020). Crop type and within-field location as sources of intraspecific variations in the phenology and the production of floral and fruit resources by weeds. Agric. Ecosyst. Environ. 302:107082. doi: 10.1016/j.agee.2020.107082
Zas, R., and Sampedro, L. (2015). Heritability of seed weight in Maritime pine, a relevant trait in the transmission of environmental maternal effects. Heredity 114, 116–124. doi: 10.1038/hdy.2014.76
Zeng, M., de Vries, W., Bonten, L. T. C., Zhu, Q., Hao, T., Liu, X., et al. (2017). Model-based analysis of the long-term effects of fertilization management on cropland soil acidification. Environ Sci. Technol. 51, 3843–3851. doi: 10.1021/acs.est.6b05491
Zeng, X., Durka, W., Welk, E., and Fischer, M. (2017). Heritability of early growth traits and their plasticity in 14 woody species of Chinese subtropical forest. J. Plant Ecol. 10, 222–231. doi: 10.1093/jpe/rtw086
Keywords: cropping system, floral scent volatile, half-sib family, heritability, phenotypic trait, selection pressure, weed management
Citation: Adhikari S, Burke IC, Revolinski SR, Piaskowski J and Eigenbrode SD (2021) Within-Population Trait Variation in a Globally Invasive Plant Species Mayweed Chamomile (Anthemis cotula): Implications for Future Invasion and Management. Front. Agron. 3:640208. doi: 10.3389/fagro.2021.640208
Received: 10 December 2020; Accepted: 20 January 2021;
Published: 10 February 2021.
Edited by:
Ali Bajwa, New South Wales Department of Primary Industries, AustraliaReviewed by:
Shane Douglas Campbell, The University of Queensland, AustraliaSajid Latif, Charles Sturt University, Australia
Simerjeet Kaur, Punjab Agricultural University, India
Copyright © 2021 Adhikari, Burke, Revolinski, Piaskowski and Eigenbrode. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subodh Adhikari, c3Vib2RoYUB1aWRhaG8uZWR1
 Subodh Adhikari
Subodh Adhikari Ian C. Burke2
Ian C. Burke2 Samuel R. Revolinski
Samuel R. Revolinski Sanford D. Eigenbrode
Sanford D. Eigenbrode