- 1Department of Crop, Soil, and Environmental Sciences, Auburn University, Auburn, AL, United States
- 2Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, United States
Whole genome duplication via polyploidization is a major driver of diversification within angiosperms and it appears to confer the most benefit during times of rapid environmental change. Polyploidization offers expanded access to novel phenotypes that facilitate invasion of new environments and increased resistance to stress. These new phenotypes can arise almost immediately through the novel interactions among or between transcription factors of the duplicated genomes leading to transgressive traits, and general heterosis, or they can occur more slowly through processes like neofunctionalization, and subfunctionalization. These processes are characterized by the changes within homologs of the duplicated genomes, homoeologs. It has been proposed that redundant homoeologs are released from selective constraints and serve as an additional source of adaptive genetic variation, particularly in neo and meso-polyploids. Current practices in weed management create rapid environmental change through the use of chemicals, practices that are meant to cause the extirpation of the designated weed, and represent a strong recurrent selective event—a scenario that should favor polyploidy species. Here we ask the question, “Do polyploids make better weeds?” It is our conclusion that such a question is impossible to answer at this time due to the lack of resources and understanding in weed genomics. The growing contingent of research in weed genomics, however, driven by herbicide resistance evolution is rapidly improving our understanding of weed molecular biology and will aid in improving understanding of the impacts of ploidy levels on weed evolution and adaptation in the future.
Introduction
All extant diploid angiosperms have been traced back to polyploid ancestors (Scarpino et al., 2014). Whole genome duplications (WGDs) are major drivers of adaptation and are responsible for the trajectory of flowering plant evolution. Phylogenetic analyses and molecular dating have traced an ancient genome-wide duplication event shared by all extant seed plants (Jiao et al., 2011). Ancient whole genome duplications (WGD), served as a major force in speciation and diversification in highly plastic angiosperm genomes. Compared to gymnosperms, angiosperms are more likely to endure the impact that polyploidy has on a genome, as <5% of gymnosperms are polyploid (Leitch and Leitch, 2008). While polyploidy is gaining traction as a viable and beneficial means of adaptation, polyploidization has previously been described and is still commonly referred to as an evolutionary “dead end,” as ancient WGD were seen scarce (Arrigo and Barker, 2012; Van De Peer et al., 2017). Polyploidy studies are continuing to rise in prevalence, and more cases of ancient and neopolyploid cases are being discovered and suggests that polyploidization via whole genome duplication is more common than previously thought (Hohmann et al., 2015; Barker et al., 2016; Yang et al., 2018). In rare instances polyploids could have had an evolutionary advantage on their non-polyploid competition, especially in times of stress or environmental upheaval, providing means to survive over their counterparts (Van De Peer et al., 2017). Recent studies provided evidence that there is an increased tolerance to genomic changes in polyploids relative to diploid progenitors, including how polyploid lineages were established and the rates at which this occurs, and the mechanisms they used to spread and maintain themselves (Schoenfelder and Fox, 2015; Shimizu-Inatsugi et al., 2017). Using a literature review and a survey of reported weeds and their ploidy level, we propose to show that polyploids make better weeds. The term weed is a generally vague description designated to virtually any plant deemed undesirable in the context of where it grows. This review focuses on the undesirable plants in an agricultural perspective that have adapted to the human condition (Harlan and de Wet, 1965). Polyploidization is especially important to understand within the study of weed science in the view of climate change and the ever-increasing size of highly managed tracts of land around the world, since both of these selective forces may favor polyploids. Here we ask if polyploidy confers an advantage in the evolution of glyphosate resistance in comparison to their diploid counterparts. Glyphosate resistance is an ideal trait to test since understanding its evolution has both practical and theoretical applications. As a herbicide it was highly effective and the evolution of target site and non-target site required decades (Pratley et al., 1999; Sammons and Gaines, 2014). Understanding if polyploidy confers an advantage in the case of glyphosate will elucidate how resistance to other herbicides will evolve and if special consideration needs to be given to polyploids in application of weed control.
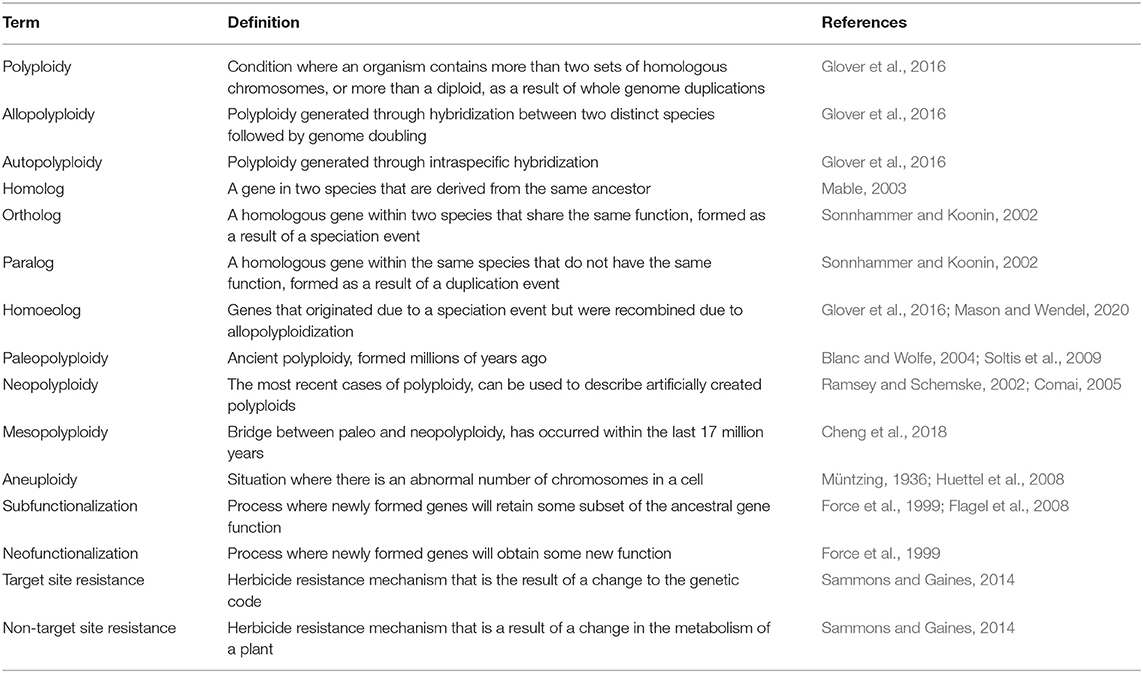
Definitions About Polyploidy
Polyploids are organisms that contain multiple copies of their chromosomes, or simply, a species that has more copies than diploids (Glover et al., 2016). Polyploidization itself is defined as whole genome duplication, where it has doubled in the form of either allopolyploidy or autopolyploidy, or as a combination of both forms (Table 1). Allopolyploids are generated through the hybridization of two or more different species each contributing unique subgenomes, while autopolyploids arise from the duplication of a single species' genome. On a gene level, the multiple copies of genes or chromosomes in allopolyploids are referred to as homoeologs. Not to be confused with homologs, homoeologs are related genes that lie in the different subgenomes of an allopolyploid (Mason and Wendel, 2020). Homologous genes share a common ancestor, while homoeologous genes have the same parental origin (Mable, 2003). Within homologous genes, there are orthologs and paralogs: orthologs are genes descended from a common ancestor in different species that share the same function or formed due to a speciation event. Paralogs are genes derived from a single gene as the result of a duplication event (Sonnhammer and Koonin, 2002). Homoeologs and orthologs can be construed as analogous, as homoeologs are orthologous genes within a polyploid species that occur on different subgenomes. Homoeologs originated through speciation and were recombined in the same genome through allopolyploidization (Glover et al., 2016). The correct usage of “homoeolog” has been debated and the sheer amount of different terms can lead to some confusion.
Paleopolyploidy is defined as polyploidy that occurred millions of years ago (Blanc and Wolfe, 2004; Soltis et al., 2009). Genes associated with paleopolyploidy can also be referred to as paleologs. Determining whether an organism is a paleopolyploid or used to be a difficult task because progenitor species could not be identified through cytological tools or DNA markers (Levy and Feldman, 2002). Advances in genomics has eased the process of identification with whole genome assemblies providing the necessary data for synteny plots, gene trees constructed from gene family analyses, and Ks plots from transcriptome assembles (Husemann and Stoye, 2009; Gao et al., 2018; Leebens-Mack et al., 2019). More recent polyploids have two different categories: mesopolyploid, if formed within the last 17 million years, or neopolyploids for the species that most recently experienced polyploidization (Ramsey and Schemske, 2002; Cheng et al., 2018). Neopolyploidy can also be described as a species that has experienced an artificially induced chromosome duplication (Comai, 2005). Aneuploidy is another term associated with polyploidy, as it signifies when there is an abnormal number of chromosomes compared to the wild type, which is commonly found in triploid (and sometimes pentaploid) populations (Müntzing, 1936; Huettel et al., 2008).
History of Polyploid Evolution
The earliest concepts of polyploidy came about in the early 1900s. The independent rediscovery of Mendel's work by de Vries, Correns, and Tschermak was the beginning of a golden age of genetics (Corcos and Monaghan, 1990). Geneticists originally associated specific characteristics with morphological characteristics as opposed to genetic characteristics like karyotype (DeVries, 1915; Ramsey and Ramsey, 2014). Using morphological characteristics as a form identification was soon displaced by the acceptance of chromosomes as hereditary units (Roberts, 1929). While certain plants, like maize, had already been determined to be polyploid (Kuwada, 1911), the term polyploidy was coined by Winkler (1917), who created the first artificial polyploid. Winge (1917) had some of the most influential thoughts on the subject, proposing hybridization followed by the doubling of chromosomes (Harlan and De Wet, 1975; Soltis et al., 2014). Stebbins (1950) could be considered one of the most important thinkers on the importance of polyploidy, with fourteen chapters in his book Variation and Evolution in Plants dedicated to the subject. Scientists were tasked with the painstaking endeavor of manually counting chromosomes under a microscope using the squash method, until the genomics era eventually brought about flow cytometry, a more accurate way to measure cellular contents, including DNA and chromosomes (Kron et al., 2007; Windham et al., 2020).
Much of what is understood about the history of polyploidization has come from studying crops (Beasley, 1940; Mcfadden and Sears, 1946). Thus far, genomic studies on Triticum (wheat) and Gossypium (cotton) have contributed the most to the current knowledge (Flagel et al., 2008; Moshe Feldman and Levy, 2009). Cultivated wheat is a good example of how studying polyploidization can be useful. Cultivated wheat is classified in three different cytogenic categories: diploid, tetraploid, and hexaploid. While the wild type progenitors for the diploid and tetraploid varieties have been determined, studies have shown that the hexaploidy varieties, like bread wheat (T. aestivum) have formed as a byproduct of cultivated tetraploid and wild diploid progenitors as a result of polyploidization (Feldman, 2001; Feldman and Levy, 2005). In allohexaploid bread wheat, there are three identifiable subgenomes, A, B, and D, which is seen as an AABBDD genome. These subgenomes are known to have derived from diploid progenitors T. uratu (AA) and Aegilops tauschii (DD). The progenitor of the BB subgenome is extinct, but is likely derived from a diploid closely related to Aegilops speltoides (Dubcovsky and Dvorak, 2007; Gornicki et al., 2014). The ability to identify these subgenomes provides a history of polyploidization in wheat, visualizing its progenitors, its center of origin (likely in southwest Asia), and estimating when the polyploidization likely occurred (Vavilov and Love, 1992; Feldman, 2001).
Polyploidization is a seemingly irreversible process, but all polyploid plants eventually undergo the process of diploidization. The process of a polyploid becoming a diploid again is a result of genomic downsizing, where genomes have been significantly reduced as a result of loss of DNA fragments, segmental DNA loss, and gene silencing, mainly to stabilize the genome (Wendel and Adams, 2005; Bird et al., 2019). Genomic downsizing most likely occurs immediately following a chromosomal duplication event. Drastic alterations to the genome are referred to as genome shock; a plant might not be prepared for such intense changes to its genome and these stabilization events could possibly occur to counteract the shock (Mcclintock, 1983). There is a case to be made that there are no true extant diploids, and should be considered to be paleopolyploids (Levy and Feldman, 2002). Combined with the fact that all diploid angiosperms are descended from polyploid ancestors, genomic downsizing over the course of millions of years could contribute to this claim (Force et al., 1999; Feldman and Levy, 2005). An example of this is present in corn (Zea mays); it has paleopolyploid characteristics and has origins as a segmental allopolyploid, but its genome was so drastically altered and silenced that it is a cytogenic diploid (Gaut and Doebley, 1997; Soltis and Soltis, 1999).
Duplicate genes in polyploids have many different pathways they can take: they can develop a new function (neofunctionalization), retain the ancestral function (subfunctionalization), or accumulate deleterious mutations and decay (Force et al., 1999). In the process of trying to maintain its status as a diploid, some plants will undergo the process of instantaneous subfunctionalization, which occurs immediately following genomic merger in order to retain all duplicate genes (Flagel et al., 2008). Different loss-of-function mutations can develop in both copies, but both copies must be retained in order to keep its ancestral function (Cheng et al., 2018). Upland cotton (Gossypium hirsutum) demonstrates subfunctionalization in the reciprocal silencing of its adhA homoeolog; the homoeolog is silenced rather than deleted, retaining all copies present (Adams et al., 2003). Larger populations are more likely to experience neofunctionalization rather than subfunctionalization because the genetic drift in large populations is going to be so slow that parental alleles are likely going to be silenced by deleterious mutations before fixation can occur (Soltis et al., 2010).
Advantages of Polyploidy in Evolution
Polyploidization allows organisms to react and survive; by their very nature, polyploids have a much higher range of genetic diversity than diploids, which certain environmental factors, such as habitat disturbance, nutritional stress, physical stress, and climate changes, can trigger new phenotypes, like increased allelopathic effect (Hegarty and Hiscock, 2007; Ramsey, 2011; Te Beest et al., 2012; Omezzine et al., 2017). New phenotypes may arise through heterosis, gene redundancy, or the formation of transgressive traits (Comai, 2005; McCarthy et al., 2016; Wei et al., 2019). The effects of heterosis was first identified by Darwin, whose experimental crosses resulted in more vigorous hybrids, i.e., heterosis (Darwin, 1876). There are two main models involved in heterosis: the dominance model and overdominance model. The dominance model hypothesizes that the slightly deleterious recessive alleles are complemented by superior dominant ones in hybrids (Hochholdinger and Baldauf, 2018). The overdominance model is used to describe polyploidization, as the progressive heterosis associated with polyploids is more complex due to the increasing vigor with increasing number of genomes (Birchler et al., 2010). While heterosis generally results in polyploids with better phenotypic performance than its parent species, plants with transgressive traits display extreme phenotypes outside of the range of its progenitors (McCarthy et al., 2016). Heterosis and transgressive traits have been shown to be potential improvements for epigenetic mechanisms in allopolyploids, like histone modification or cytosine methylation (Renny-Byfield and Wendel, 2014). Gene redundancy acts as a protective feature, shielding polyploids from the effects of deleterious mutations with the numerous copies present (Wendel, 2000). Even allelopathy (the ability to suppress growth in another plant), which is present in both diploids and polyploids, has been shown to increase in polyploids compared to diploids (Colquhoun, 2006; Omezzine et al., 2017). Hexaploid barnyardgrass (Echinochloa crus-galli) shows considerable allelopathic tendencies and Omezzine et al. (2017) was able to show that allelopathy increased as ploidy increased in fenugreek (Trigonella foenum-graecum) (Omezzine et al., 2017).
Allopolyploids provide some evidence of increased fitness over their progenitors. When diploid parents are crossed, typically their offspring have an increase in performance; polyploids produced more viable seed in extreme heat and drought conditions and differences in stomatal pore sizes that improved drought survival over their diploids counterparts (Madlung, 2013; Godfree et al., 2017). For example, cultivated wheat (T. aestivum) is an allohexaploid that has managed to survive over its B genome donor (Feldman and Levy, 2009). Allopolyploids also have more potential for ecological adaptation over their diploid counterparts, as shown through diploid and allopolyploid species of Cardamine; while different diploid species had a tendency to prefer only one environment, the allopolyploid species was able to grow and survive in all the environments tested (Shimizu-Inatsugi et al., 2017). The ability to alter phenotypes, as in functional trait divergence or generalized trait plasticity is one of the leading hypotheses regarding overall increased fitness in polyploid species (Van De Peer et al., 2017; Wei et al., 2019). Polyploid crops have huge adaptation potential and further studies are necessary to show the role of genetic variation resulting from polyploidy in this potential (Ramsey and Ramsey, 2014; Schiessl et al., 2017).
The study of neopolyploids furnishes strong insights in the evolution of polyploid species. Spartina anglica (common cordgrass), is an invasive neoallopolyploid weed species that arose in the last 200 years (Baumel et al., 2002). The neo-dodecaploid weed arose at the end of the nineteenth century as a result of a genome duplication between the already hybrid species Spartina X townsendii, which is a cross between hexaploids Spartina alterniflora and Spartina maritima (Ainouche et al., 2004). The duplication of the two unique subgenomes in Spartina X townsendii cements S. anglica as an allopolyploid as opposed to an autopolyploid. Compared to its progenitors, S. anglica has been shown to have increased fitness with its prolific seed production, fertility, and extensive lateral clonal growth, which was not seen in its sterile progenitor Spartina X townsendii. Baumel et al. (2002) was able to demonstrate that rapid, non-Mendelian changes involving preferential sequence elimination or modification of methylation patterns may occur in the earliest stages of polyploid stabilization. Other neopolyploids, like Senecio and Tragopogon have also been established within the last 200 years (Abbott and Lowe, 2004; Soltis et al., 2004). The development new polyploids aids in understanding gene silencing, cytosine methylation, and parental “non-additivity” play an active role in polyploidization and improving overall understanding of the process (Adams and Wendel, 2005).
Weeds and Unconscious Selection
Aside from polyploidy, one way weeds have been able to thrive in a world that strives to exterminate them is through unconscious selection, or actively breeding plants in an environment different from their wild habitat, usually human-made (Zohary, 2004). Crops once considered weeds have been domesticated through unconscious selection, as they could not be differentiated from the crop they were growing alongside. The phenomenon of weeds imitating desired crops was noticed and thoroughly discussed by Nikolai Vavilov, who covered the process in “Origin and Geography of Cultivated Plants” (Vavilov, 1922). Vavilov looked into the centers of origins of crops, determining that there is more diversity in a species in an area where a species originated (McElroy, 2014). The process of an undesirable species evolving to mimic desirable ones as a means of survival has become known as Vavilovian mimicry (Pasteur, 1982). This phenomenon was first noticed, and well-established, between rye (Secale cereale) and wheat (Triticum) (Vavilov, 1922).
Before the time of chemical management, the only way to eliminate weeds when harvesting was by hand or hand-held implements. Since rye is remarkably visually similar to wheat, farmers often could not differentiate the two. When harvesting, rye would be selected for alongside the wheat, and when the harvested seed was replanted, unwanted rye would grow as well. Eventually, rye evolving alongside a desirable crop made it desirable as well. Rye could also perform better in colder climates than weaker wheat species did, but its increased fitness could possibly be contributed to crossing between the weed species and the crop it grew alongside (Harlan, 1965). This same principle of mimicry can be applied to modern day crop and companion weeds with cultivated rice (Oryza) and barnyardgrass (Echinochloa crus-galli, specifically ssp. oryzicola). In the seedling stage, barnyardgrass growing in paddy fields are virtually indistinguishable from rice, which makes manual weeding extremely difficult (Barrett, 1983). Barnyardgrass has also demonstrated allelopathetic tendencies in rice paddies, which provides even more difficulty in its management (Khanh et al., 2007; Guo et al., 2017)?
Another subset of Vavilovian mimicry is seed mimicry, where weeds may or may not look morphologically similar to the crop they contaminate, but the seeds produced by both crop and weed are identical in size and shape. While this might not be a problem when weeding by hand, but once mechanical winnowing came into play, the weed seeds only need to trick the machine (Harlan, 1982). An example of this is evident between balloonvine (Cardiospermum halicacabum) and soybean (Glycine max), which are morphologically dissimilar, however their seeds are indistinguishable in size and shape by a machine, which aids in the weed survival (Johnston et al., 1979). One final way weeds are unconsciously selected is through genetic mimicry. As weeds continue to grow, it is likely that they will develop herbicide resistance as an adaptation to human management practices.
As chemical herbicides became the primary weed management practice, herbicide resistance developed as the weed's survival mechanism. The more herbicides are continuously sprayed on crops, the more likely resistance to such herbicides will result in those weeds (Powles and Yu, 2010). Gene flow works in both ways; crops and weeds don't grow independently, genetic material can be shared between them, which is what leads to hybridization between the two (Harlan and De Wet, 1975; Gould, 1991). While unconscious selection plays a major role in basically how all weeds are bred, the real question is are polyploids the better weed? Do more polyploids exhibit characteristics of Vavilovian mimicry within the genetic subset? Before we answer these questions, it is important to look further into modern day weed management practices, specifically regarding herbicide and the effect it has on crops and weeds in general.
Herbicide Resistance
In this modern era, herbicide resistance is the biggest problem currently faced with weeds. The two types of herbicide resistance typically dealt with are target site resistance (TSR) and non-target site resistance (NTSR). TSR develops directly against a mode of action, specifically as a mutation to the genetic code, as a single nucleotide polymorphism (SNP). NTSR relates to metabolism, as there are no direct changes to the genetic code (Sammons and Gaines, 2014). This can be seen as reduced absorption, translocation, or sequestration of the herbicide in the vacuole (Powles and Yu, 2010). To simplify our analysis of the effect of polyploidization on the formation of herbicide resistance traits, we have chosen to focus on well-described TSR mechanisms, specifically the glyphosate mechanism. Target site resistance provides a stronger case for increased weed survivability in polyploids, as multiple chromosome copies alone provide an environment for mutations to occur in at least one copy, and if that copy is expressed resistance should occur (Otto, 2007).
Polyploids develop glyphosate resistance at a faster rate than their diploid relatives. Glyphosate was first introduced by Monsanto in 1974 with the trade name “Roundup” and in the last decade has become the most used herbicide worldwide. It is a non-selective herbicide that targets the shikimic acid pathway and its ability produce folates and aromatic amino acids (Malik et al., 1989; Valavanidis, 2018). Target site resistance in glyphosate resistant plants results in mutations at the Pro106 location to either Thr, Leu, Ser, Ala, or a double substitution of Thr102 to Ile + Pro106 to Ser (TIPS) (Yu et al., 2015). With the introduction of Roundup Ready® (RR) crops as a staple in crop production, gene flow from RR crops has the potential to result in glyphosate resistant weeds, which presents a major problem for weed management (Mallory-Smith and Zapiola, 2008). Based on data from weedscience.org, weedy polyploid species like annual bluegrass (Poa annua), oat (Avena), junglerice (Echinochloa colona), and barnyardgrass (Echinochloa crus-galli) are just a few of the species reported resistant to glyphosate (Heap, 2019).
Should Polyploid Species Should Make Better Weeds and be More Common With Herbicide Resistance?
Theoretically, target site resistance should be more common in polyploid weeds, since theoretically polyploids have a more flexible expression profile that allows them to silence adaptive alleles or loci with fitness costs when the allele offers no adaptive advantage (Otto and Whitton, 2000; Otto, 2007). TSR fitness costs have been identified, but the level of costs varies among different plant species (Vila-Aiub et al., 2009) and modes of action. In general, fitness costs have been associated with ALS, ACCase, and photosystem II (PSII) herbicides, which is especially evident in PSII herbicides because of the reduced photosynthetic capacity (Jansen and Pfister, 1990). Fitness costs in ACCase inhibitors should have no association with polyploidy because ACCase inhibitors only affect the plastid isoform (Murphy and Tranel, 2019). Glyphosate's role as the most widely used herbicide provides the lens for the focus of herbicide resistance in polyploids compared to other herbicides due to the possibility of both target and non-target-site resistance. Studies have shown that herbicide resistance alleles do not universally endow some type of fitness cost, but there is more of a cost in diploid species over polyploid (Vila-Aiub et al., 2009; Yanniccari et al., 2016). A reduction in fitness has been identified in glyphosate resistant goosegrass (Eleusine indica), rigid ryegrass (Lolium rigidum), and perennial ryegrass (Lolium perenne) (Preston et al., 2009; Yanniccari et al., 2016; Han et al., 2017). However, fitness costs in glyphosate resistant biotypes seem to be present on a case-by-case basis. The TIPS double mutation in the E. indica population came at a very high resistance cost, resistant L. rigidum populations may or may not have a fitness penalty, depending on the resistance allele present, and the fitness cost in L. perenne is not associated with a target-sire mutation, but rather high EPSPS activity. The Pro-106-Ser mutation, the most common target-site in glyphosate resistant biotypes, endows a low-level glyphosate resistance and is seemingly negligible in fitness costs compared to mutations endowing high level resistance, like the TIPS mutation (Vila-Aiub et al., 2019). There has been no investigation comparing the fitness cost of herbicide resistant polyploid species to the cost seen in diploid species, or even delving into the costs of herbicide resistance in any polyploid species. While there have been reviews showing the fitness costs of different herbicides, all data and conclusions are drawn from diploid species (Vila-Aiub et al., 2009). More studies should be performed in order to ascertain whether polyploidy plays a role in reduced fitness in association with herbicide resistance.
In order to examine how polyploidy affects weed survival, a survey of weeds was done on those reported as glyphosate resistant (Heap, 2019). A list of weeds reported as glyphosate resistant was assembled and the polyploidy, chromosome counts, and C-values were compared in Table 2. Out of 48 species selected, 12 are known to be diploid, four tetraploid, two hexaploid, one octoploid, and the remaining 29 species have a ploidy level that has not been confirmed, based on data from the Kew C-value database and the Chromosome Counts Database (Rice et al., 2015; Pellicer and Leitch, 2020). A few assumptions can be drawn about the species with unconfirmed ploidy: they could be diploid, polyploid, or have mixed ploidy, but thus far no studies have been done to confirm their ploidy. Ploidy determination is just one part of a bigger issue; according to the genome database from NCBI, out of the 48 species surveyed, only nine have fully sequenced genomes and two have RefSeq genomes. So along with the fact that there is little to no data to draw conclusions from, the majority of studies reporting glyphosate resistant are lacking in research beyond spray trials. Target site resistance should be considered a major factor for resistance and studies that just ignore the benefits of sequencing data can be detrimental. When sequencing data is in play, one can more accurately confirm TSR or NTSR, rather than just claiming NTSR.
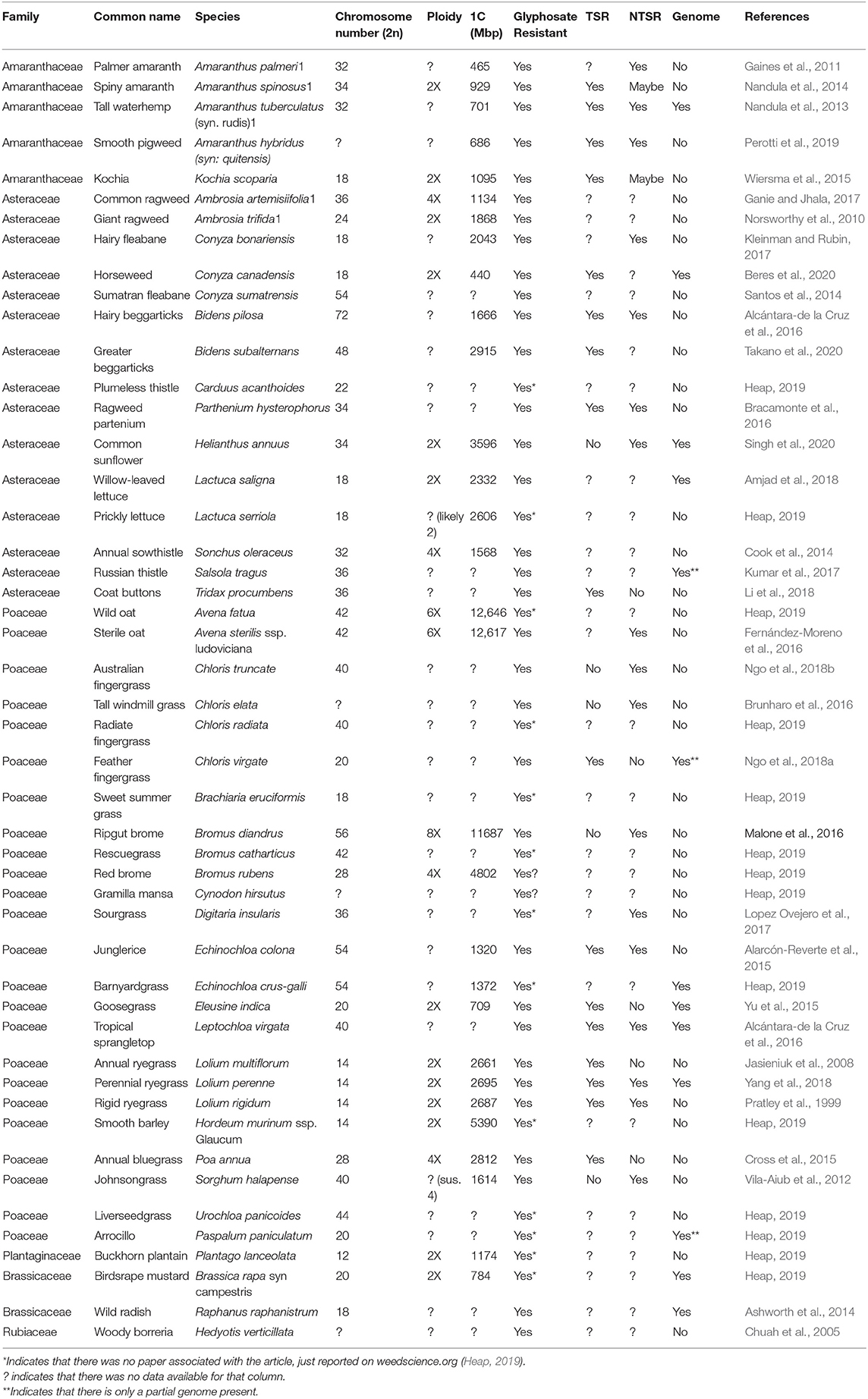
Table 2. Weed species currently (9 June 2020) reported as resistant to glyphosate according to The International Survey of Herbicide Resistant Weeds and corresponding reported genomic data.
But, do Polyploids Make Better Weeds?
Two questions need to be asked: “Are polyploids better weeds?” and “Are polyploids more likely to be resistant to herbicides?” The weeds historically referred to as the “world's worst weeds,” has had relatively no changes, or at least no reported changes, to this list since first reported in 1969, and last updated in 1977 (referenced in Table 3) (Holm and Herberger, 1969; Holm et al., 1977). Based on data from the Plant C-Value database and the International Herbicide-Resistant Weed database, little can be determined whether ploidy or glyphosate resistance has any determining factor on what makes a better weed (Heap, 2019; Pellicer and Leitch, 2020). Barnyardgrass (E. crus-galli), which is known to be glyphosate resistant, a possibly hexaploid weed, would support the case for polyploidy providing better weeds; barnyardgrass has easily adapted to human activity, along with other polyploids like rye (S. cereale) and annual bluegrass (Poa annua) (Ye et al., 2014; Cross et al., 2015). A case could also be supported with the glyphosate resistant diploid species goosegrass (Eleusine indica), as there is a fitness cost associated with the double EPSPS gene mutation TIPS endowing glyphosate resistance (Han et al., 2017). Polyploids have adapted and survived due to polygenic selection over millennia (Stebbins, 1950). Comparatively herbicide resistance, both TSR and NTSR, is more often than not monogenic: this can be seen as a divergent single nucleotide polymorphism (SNP), a single unregulated metabolic enzyme, or even as a widely duplicated gene (Délye, 2013; Jugulam and Shyam, 2019).
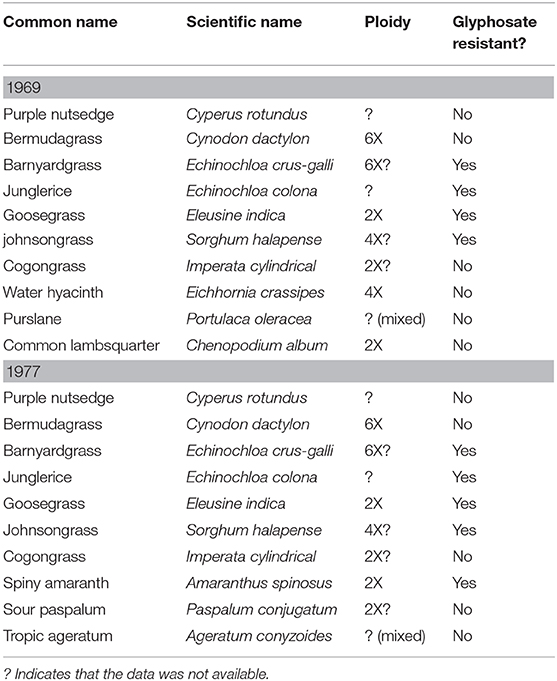
Table 3. Ploidy and glyphosate resistance status of the world's worst weeds as described by Holm and Herberger (1969), Holm et al. (1977).
Applying what is known about polyploidization in crops to weeds, one could try to assume an advantage for polyploid weeds. Factors that favor polyploid crop domestication should translate over to polyploid weed species. Heterosis, gene redundancy, and high genetic variability discussed above should act the same in weeds as they do in crops. Even in older, more established polyploid populations, you can see the benefit compared to diploids. Studies have shown that polyploids in general have the capacity to be more invasive over diploid species. Stevens et al. (2020) showed that tetraploid seeds tended to be larger than those of diploids, which contributed to tetraploid seeds being more dormant than the diploid seeds, less likely to germinate in stressful environments and therefore better adapted to said environments. Polyploid species have also been found to be more fecund and competitive than diploids, as seen in studies done with spotted knapweed (Centaurea stoebe) (Broz et al., 2009; Rosche et al., 2017). Studies on the three geo-cytotypes of C. stoebe, a native Eurasian diploid, a native Eurasian tetraploid, and an introduced North American tetraploid provided evidence that the polycarpic nature of the tetraploid biotypes allowed these biotypes to survive after the initial flowering and flower more than the monocarpic diploid biotype, and the introduced biotype even more than the native (Broz et al., 2009). Pandit et al. (2011) showed in an extensive study on rarity and invasiveness that diploid plants were more likely to be rare, while polyploids were more likely to be invasive. It has also been determined that polyploid species are less likely to experience inbreeding depression, due to the balancing effect of the presence of multiple gene copies (Rosche et al., 2017). The combination of higher seedling growth rates and diminished inbreeding depression creates an argument that polyploids are more invasive and therefore more competitive than diploids. Annual bluegrass (P. annua) has established itself on every continent and barnyardgrass continue to invade rice paddies, and when combined with the likelihood for developing herbicide resistance, namely glyphosate resistance, it creates some formidable weed species. And as mentioned before, there is even evidence that target site resistance in diploid plants significantly reduces fitness levels (Preston et al., 2009; Yanniccari et al., 2016; Han et al., 2017).
However, there are currently more reported diploid species resistant to glyphosate than there are polyploid. There are diploid species that are considered to be some of the most widespread and difficult weed species to manage, like smooth pigweed (Amaranthus hybridus) and horseweed (Conyza canadensis), that have high levels of glyphosate resistance with no fitness cost reported (Beres et al., 2018, 2020; Perotti et al., 2019). Purple nutsedge (Cyperus rotundus) and yellow nutsedge (Cyperus esculentus), whose ploidy levels (according to the Plant C-Value database) have not been reported, are considered some of the worst weeds on the planet because of control difficulty (Pereira et al., 1987; Arias et al., 2011; Pellicer and Leitch, 2020). And although its ploidy has, surprisingly, not yet been reported, Palmer amaranth (Amaranthus palmeri), is increasing one of the most difficult weeds to control (Rayburn et al., 2005; Gaines et al., 2011). Such fitness costs have not yet been reported in polyploid species and further investigation is required to determine if herbicide resistance has any negative effects in polyploids.
The sheer magnitude and complexity of polyploid genomes makes it difficult to perform large-scale genetics studies (Schiessl et al., 2017). While there have been polyploid genomes fully sequenced, the genomes sequenced have been relatively small, genome size wise, outside of the massive undertaking of sequencing the allohexaploid wheat genome (Zimin et al., 2017). Advances in genomics has made whole genome sequencing easier and cheaper as a whole, but it improving the possibility of sequencing polyploid genomes. Despite this, barnyardgrass remains the only polyploid weed genome sequenced (Kyriakidou et al., 2018). There is an obvious need for a well-established weed genomics database; while there are still challenges to this undertaking, it is a necessary step that needs to be taken in order to advance the understanding of polyploidy in weeds, and weed genomics in general (Patterson et al., 2019).
Conclusions
Based on the data that is available to us, no conclusions can be drawn that polyploids make better weeds than non-polyploids. At this point in time, there is no truly reliable database for genetic data on weed species, and even the available data on polyploid crops is lackluster. The Plant C-Value database is currently the most reliable, and while it offers data some data on polyploidy, including ploidy level and chromosome numbers, its purpose is to provide C-value data, not polyploid data. Even the list of the worst weeds in the world has not changed in the past 40 years, which should be highly unlikely, as the science is constantly changing. Research into weed genomics has room for improvement, and the development of weed genomics provides potential for greater understanding in how weed species evolve and the role polyploidy is playing and has played in weed evolution (Ravet et al., 2018; Patterson et al., 2019). The International Weed Genomics Consortium provides an outlet for collaborative research into weed genomics, with a growing genomics repository for weed species. The complexity of polyploids makes genomic work difficult; ploidy needs to be determined, chromosome copy number, and even then, certain genes might have more copies than are actually being tested. Next generation sequencing lends itself to providing more insight into polyploidy and its role in weed genomics. While there are more sources providing insight into weed genomics and a pathway for polyploid weeds, more extensive and in-depth research is required in order to fully comprehend the scale that polyploidy plays in understanding weeds.
Author Contributions
JM and NH conceived the idea, developed the outline for the paper, and edited the paper. CR wrote the paper. JM and CR developed the tables. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abbott, R. J., and Lowe, A. J. (2004). Origins, establishment and evolution of new polyploid species: senecio cambrensis and S. eboracensis in the British Isles. Biol. J. Linnean Soc. 82, 467–474. doi: 10.1111/j.1095-8312.2004.00333.x
Adams, K. L., Cronn, R., Percifield, R., and Wendel, J. F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. U.S.A. 100, 4649–4654. doi: 10.1073/pnas.0630618100
Adams, K. L., and Wendel, J. F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141. doi: 10.1016/j.pbi.2005.01.001
Ainouche, M. L., Baumel, A., and Salmon, A. (2004). Spartina anglica C. E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol. J. Linnean Soc. 82, 475–484. doi: 10.1111/j.1095-8312.2004.00334.x
Alarcón-Reverte, R., García, A., Watson, S. B., Abdallah, I., Sabaté, S., Hernández, M. J., et al. (2015). Concerted action of target-site mutations and high EPSPS activity in glyphosate-resistant junglerice (Echinochloa colona) from California. Pest Manag. Sci. 71, 996–1007. doi: 10.1002/ps.3878
Alcántara-de la Cruz, R., Fernández-Moreno, P. T., Ozuna, C. V., Rojano-Delgado, A. M., Cruz-Hipolito, H. E., Domínguez-Valenzuela, J. A., et al. (2016). Target and non-target site mechanisms developed by glyphosate-resistant hairy beggarticks (Bidens pilosa L.) populations from Mexico. Front. Plant Sci. 7:1492. doi: 10.3389/fpls.2016.01492
Amjad, M., Borger, C., Plan, E., Amjad, M., Borger, C., Development, R., et al. (2018). Willow-Leaved Lettuce (Lactuca saligna) Has Developed Resistance To Glyphosate In Western Australia. Grains and Research Development Corporation. Available online at: https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2018/02/willow-leaved-lettuce-resistance-to-glyphosate
Arias, R. S., Molin, W. T., Ray, J. D., Peel, M. D., and Scheffler, B. E. (2011). Isolation and characterisation of the first microsatellite markers for Cyperus rotundus. Weed Res. 51, 451–460. doi: 10.1111/j.1365-3180.2011.00861.x
Arrigo, N., and Barker, M. S. (2012). Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15, 140–146. doi: 10.1016/j.pbi.2012.03.010
Ashworth, M. B., Walsh, M. J., Flower, K. C., and Powles, S. B. (2014). Identification of the first glyphosate-resistant wild radish (Raphanus raphanistrum L.) populations. Pest Manag. Sci. 70, 1432–1436. doi: 10.1002/ps.3815
Barker, M. S., Arrigo, N., Baniaga, A. E., Li, Z., and Levin, D. A. (2016). On the relative abundance of autopolyploids and allopolyploids. N. Phytol. 210, 391–398. doi: 10.1111/nph.13698
Baumel, A., Ainouche, M., Kalendar, R., and Schulman, A. H. (2002). Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica C.E. Hubbard (Poaceae). Mol. Biol. Evol. 19, 1218–1227. doi: 10.1093/oxfordjournals.molbev.a004182
Beasley, J. O. (1940). The origin of American tetraploid gossypium species. Am. Nat. 74, 285–286. doi: 10.1086/280895
Beres, Z. T., Ernst, E. E., Ackley, B. A., Loux, M. M., Owen, M. D. K., and Snow, A. A. (2018). High levels of glyphosate resistance in conyza canadensis from agricultural and non-agricultural sites in Ohio and Iowa. Sci. Rep. 8, 1–8. doi: 10.1038/s41598-018-28163-w
Beres, Z. T., Giese, L. A., Mackey, D. M., Owen, M. D. K., Page, E. R., and Snow, A. A. (2020). Target-site EPSPS Pro-106-Ser mutation in Conyza canadensis biotypes with extreme resistance to glyphosate in Ohio and Iowa, USA. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-64458-7
Birchler, J. A., Yao, H., Chudalayandi, S., Vaiman, D., and Veitia, R. A. (2010). Heterosis. Plant Cell 22, 2105–2112. doi: 10.1105/tpc.110.076133
Bird, K., Niederhuth, C., Ou, S., Gehan, M., Pires, J. C., Xiong, Z., et al. (2019). Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. bioRxiv. doi: 10.1101/814491
Blanc, G., and Wolfe, K. (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678. doi: 10.1105/tpc.021345
Bracamonte, E., Fernández-Moreno, P. T., Barro, F., and de Prado, R. (2016). Glyphosate-resistant Parthenium hysterophorus in the Caribbean Islands: non target site resistance and target site resistance in relation to resistance levels. Front. Plant Sci. 7:1845. doi: 10.3389/fpls.2016.01845
Broz, A. K., Manter, D. K., Bowman, G., Müller-Schärer, H., and Vivanco, J. M. (2009). Plant origin and ploidy influence gene expression and life cycle characteristics in an invasive weed. BMC Plant Biol. 9, 1–13. doi: 10.1186/1471-2229-9-33
Brunharo, C. A., Patterson, E. L., Carrijo, D. R., de Melo, M. S., Nicolai, M., Gaines, T. A., et al. (2016). Confirmation and mechanism of glyphosate resistance in tall windmill grass (Chloris elata) from Brazil. Pest Manag. Sci. 72, 1758–1764. doi: 10.1002/ps.4205
Cheng, F., Wu, J., Cai, X., Liang, J., Freeling, M., and Wang, X. (2018). Gene retention, fractionation and subgenome differences in polyploid plants. Nature Plants 4, 258–268. doi: 10.1038/s41477-018-0136-7
Chuah, T. S., Noor-Zalila, M. R., Cha, T. S., and Ismail, B. S. (2005). Paraquat and glyphosate resistance in woody borreria growing in oil palm plantations in Terengganu, Malaysia. Malaysian Appl. Biol. 34, 43–49.
Colquhoun, J. B. (2006). “Allelopathy in weeds and crops: myths and facts,” in Wisconsin Fertilizer, Aglime and Pest Management Conference (Madison, WI). 45, 318–320.
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genetics 6, 836–846. doi: 10.1038/nrg1711
Cook, T., Davidson, B., and Miller, R. (2014). “A new glyphosate resistant weed species confirmed for northern New South Wales and the world: common sowthistle (Sonchus oleraceus),” in Nineteenth Australasian Weeds Conference, 206–209.
Corcos, A. F., and Monaghan, F. V. (1990). Mendel's work and its rediscovery: a new perspective. CRC. Crit. Rev. Plant Sci. 9, 197–212. doi: 10.1080/07352689009382287
Cross, R. B., McCarty, L. B., Tharayil, N., McElroy, J. S., Chen, S., McCullough, P. E., et al. (2015). A pro 106 to ala substitution is associated with resistance to glyphosate in annual bluegrass (Poa annua). Weed Sci. 63, 613–622. doi: 10.1614/WS-D-15-00033.1
Darwin, C. (1876). The Effects of Cross and Self Fertilisation in the Vegetable kingdom. London: Murray. doi: 10.5962/bhl.title.110800
Délye, C. (2013). Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 69, 176–187. doi: 10.1002/ps.3318
DeVries, H. (1915). The coefficient of mutation in oenothera biennis L. Bot. Gazette. 59, 169–196. doi: 10.1086/331526
Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866. doi: 10.1126/science.1143986
Feldman, M. (2001). “Origin of cultivated wheat,” in The World Wheat Book: A History of Wheat Breading. Available online at: https://www.m-culture.go.th/mculture_th/download/king9/Glossary_about_HM_King_Bhumibol_Adulyadej's_Funeral.pdf
Feldman, M., and Levy, A. A. (2005). Allopolyploidy - a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109, 250–258. doi: 10.1159/000082407
Feldman, M., and Levy, A. A. (2009). Genome evolution in allopolyploid wheat-a revolutionary reprogramming followed by gradual changes. J. Genetics Genomics 36, 511–518. doi: 10.1016/S1673-8527(08)60142-3
Fernández-Moreno, P. T., Alcantara-De La Cruz, R., Cruz-Hipólito, H. E., Rojano-Delgado, A. M., Travlos, I., and De Prado, R. (2016). Non-target site tolerance mechanisms describe tolerance to glyphosate in Avena sterilis. Front. Plant Sci. 7:1220. doi: 10.3389/fpls.2016.01220
Flagel, L., Udall, J., Nettleton, D., and Wendel, J. (2008). Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6, 1–9. doi: 10.1186/1741-7007-6-16
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545.
Gaines, T. A., Shaner, D. L., Ward, S. M., Leach, J. E., Preston, C., and Westra, P. (2011). Mechanism of resistance of evolved glyphosate-resistant palmer amaranth (Amaranthus palmeri). J. Agric. Food Chem. 59, 5886–5889. doi: 10.1021/jf104719k
Ganie, Z. A., and Jhala, A. J. (2017). Glyphosate-resistant common ragweed (Ambrosia artemisiifolia) in Nebraska: Confirmation and response to postemergence corn and soybean herbicides. Weed Technol. 31, 225–237. doi: 10.1017/wet.2016.26
Gao, B., Chen, M., Li, X., Liang, Y., Zhu, F., Liu, T., et al. (2018). Evolution by duplication: paleopolyploidy events in plants reconstructed by deciphering the evolutionary history of VOZ transcription factors. BMC Plant Biol. 18, 1–19. doi: 10.1186/s12870-018-1437-8
Gaut, B. S., and Doebley, J. F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. U.S.A. 94, 6809–6814. doi: 10.1073/pnas.94.13.6809
Glover, N. M., Redestig, H., and Dessimoz, C. (2016). Homoeologs: what are they and how do we infer them? Trends Plant Sci. 21, 609–621. doi: 10.1016/j.tplants.2016.02.005
Godfree, R. C., Marshall, D. J., Young, A. G., Miller, C. H., and Mathews, S. (2017). Empirical evidence of fixed and homeostatic patterns of polyploid advantage in a keystone grass exposed to drought and heat stress. Royal Soc. Open Sci. 4:170934. doi: 10.1098/rsos.170934
Gornicki, P., Zhu, H., Wang, J., Challa, G. S., Zhang, Z., Gill, B. S., et al. (2014). The chloroplast view of the evolution of polyploid wheat. N. Phytol. 204, 704–714. doi: 10.1111/nph.12931
Guo, L., Qiu, J., Ye, C., Jin, G., Mao, L., Zhang, H., et al. (2017). Echinochloa crus-galli genome analysis provides insight into its adaptation and invasiveness as a weed. Nat. Commun. 8:1031. doi: 10.1038/s41467-017-01067-5
Han, H., Vila-Aiub, M. M., Jalaludin, A., Yu, Q., and Powles, S. B. (2017). A double EPSPS gene mutation endowing glyphosate resistance shows a remarkably high resistance cost. Plant Cell Environ. 40, 3031–3042. doi: 10.1111/pce.13067
Harlan, J. R. (1965). The possible role of weed races in the evolution of cultivated plants. Euphytica 14, 173–176. doi: 10.1007/BF00038984
Harlan, J. R. (1982). “Relationships between weeds and crops,” in Biology and Ecology of Weeds, (Dordrecht: Springer), 91–96. doi: 10.1007/978-94-017-0916-3_8
Harlan, J. R., and De Wet, J. M. (1975). On Ö. Winge and a prayer : the origins of polyploidy published by : springer on behalf of new york botanical garden press stable. Oecologia 41, 361–390. doi: 10.1007/BF02860830
Harlan, J. R., and de Wet, J. M. J. (1965). Some thoughts about weeds. Econ. Bot. 19, 16–24. doi: 10.1007/BF02971181
Heap, I. (2019). International Survey of Herbicide Resistant Weeds. International Survey of Herbicide Resistant Weeds. Available online at: http://www.weedscience.org/Home.aspx
Hegarty, M., and Hiscock, S. (2007). Polyploidy: doubling up for evolutionary success. Curr. Biol. 17, R927–R929. doi: 10.1016/j.cub.2007.08.060
Hochholdinger, F., and Baldauf, J. A. (2018). Heterosis in plants. Curr. Biol. 28, R1089–R1092. doi: 10.1016/j.cub.2018.06.041
Hohmann, N., Wolf, E. M., Lysak, M. A., and Koch, M. A. (2015). A time-calibrated road map of brassicaceae species radiation and evolutionary history. Plant Cell 27, 2770–2784. doi: 10.1105/tpc.15.00482
Holm, L. R. G., and Herberger, J. P. (1969). “The World's Worst Weeds,” 2nd Asian-Pacific Weed Science Society Conference (Laguna), 1–14.
Holm, L. R. G., Plucknett, D. L., Pancho, J. V., and Herberger, J. P. (1977). The World's Worst Weeds. Distribution and Biology. Honolulu, HI: University Press of Hawaii.
Huettel, B., Kreil, D. P., Matzke, M., and Matzke, A. J. M. (2008). Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet 4:10. doi: 10.1371/journal.pgen.1000226
Husemann, P., and Stoye, J. (2009). r2cat: Synteny plots and comparative assembly. Bioinformatics 26, 570–571. doi: 10.1093/bioinformatics/btp690
Jansen, M. A. k., and Pfister, K. (1990). Conserved kinetics at the reducing side of reaction-center ii in photosynthetic organisms; changed kinetics in triaiine-resistant weeds. Zeitschrift Fur Naturforschung 45, 441–445. doi: 10.1515/znc-1990-0522
Jasieniuk, M., Ahmad, R., Sherwood, A. M., Firestone, J. L., Perez-Jones, A., Lanini, W. T., et al. (2008). Glyphosate-resistant Italian ryegrass (Lolium multiflorum) in California: distribution, response to glyphosate, and molecular evidence for an altered target enzyme. Weed Sci. 56, 496–502. doi: 10.1614/ws-08-020.1
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Johnston, S. K., Murray, D. S., and Williams, J. C. (1979). Germination and Emergence of Balloonvine (Cardiospermum halicacabum). Cambridge: Cambridge University Press on Behalf of the Weed Science Society of America 27, 73–76. doi: 10.1017/S0043174500043526
Jugulam, M., and Shyam, C. (2019). Non-target-site resistance to herbicides. Plants 8:417. doi: 10.3390/plants8100417
Khanh, T. D., Xuan, T. D., and Chung, I. M. (2007). Rice allelopathy and the possibility for weed management. Annals Appl. Biol. 151, 325–339. doi: 10.1111/j.1744-7348.2007.00183.x
Kleinman, Z., and Rubin, B. (2017). Non-target-site glyphosate resistance in Conyza bonariensis is based on modified subcellular distribution of the herbicide. Pest Manag. Sci. 73, 246–253. doi: 10.1002/ps.4293
Kron, P., Suda, J., and Husband, B. C. (2007). Applications of flow cytometry to evolutionary and population biology. Annual Rev. Ecol. Evol. Systematics 38, 847–876. doi: 10.1146/annurev.ecolsys.38.091206.095504
Kumar, V., Spring, J. F., Jha, P., Lyon, D. J., and Burke, I. C. (2017). Glyphosate-resistant Russian-thistle (Salsola tragus) identified in Montana and Washington. Weed Technol. 31, 238–251. doi: 10.1017/wet.2016.32
Kuwada, Y. (1911). Maiosis in the pollen mother cells of zea mays L. Shokubutsugaku Zasshi 25, 405–415. doi: 10.15281/jplantres1887.25.298_405
Kyriakidou, M., Tai, H. H., Anglin, N. L., Ellis, D., and Strömvik, M. V. (2018). Current strategies of polyploid plant genome sequence assembly. Front. Plant Sci. 871, 1–15. doi: 10.3389/fpls.2018.01660
Leebens-Mack, J. H., Barker, M. S., Carpenter, E. J., Deyholos, M. K., Gitzendanner, M. A., Graham, S. W., et al. (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685. doi: 10.1038/s41586-019-1693-2
Leitch, A. R., and Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320, 481–483. doi: 10.1126/science.1153585
Levy, A. A., and Feldman, M. (2002). Levy2012 - ploidy grass evolution. Society 130, 1587–1593. doi: 10.1104/pp.015727
Li, J., Peng, Q., Han, H., Nyporko, A., Kulynych, T., Yu, Q., and Powles, S. (2018). Glyphosate resistance in Tridax procumbens via a novel EPSPS Thr-102-Ser substitution. J. Agr. Food Chem. 66, 7880–7888. doi: 10.1021/acs.jafc.8b01651
Lopez Ovejero, R. F., Takano, H. K., Nicolai, M., Ferreira, A., Melo, M. S. C., Cavenaghi, A. L., et al. (2017). Frequency and dispersal of glyphosate-resistant sourgrass (Digitaria insularis) populations across Brazilian agricultural production areas. Weed Sci. 65, 285–294. doi: 10.1017/wsc.2016.31
Mable, B. K. (2003). Breaking down taxonomic barriers in polyploidy research. Trends Plant Sci. 8, 582–590. doi: 10.1016/j.tplants.2003.10.006
Madlung, A. (2013). Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110, 99–104. doi: 10.1038/hdy.2012.79
Mallory-Smith, C., and Zapiola, M. (2008). Gene flow from glyphosate-resistant crops. Pest Manag. Sci. 64, 428–440. doi: 10.1002/ps.1517
Malone, J. M., Morran, S., Shirley, N., Boutsalis, P., and Preston, C. (2016). EPSPS gene amplification in glyphosate-resistant Bromus diandrus. Pest Manag. Sci. 72, 81–88. doi: 10.1002/ps.4019
Mason, A. S., and Wendel, J. F. (2020). Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 11:1014. doi: 10.3389/fgene.2020.01014
McCarthy, E. W., Chase, M. W., Knapp, S., Litt, A., Leitch, A. R., and Le Comber, S. C. (2016). Transgressive phenotypes and generalist pollination in the floral evolution of Nicotiana polyploids. Nat. Plants 2:16119. doi: 10.1038/nplants.2016.119
Mcclintock, B. (1983). The significance of responses of the genome to challenge. Science 226, 792–801. doi: 10.1126/science.15739260
McElroy, J. S. (2014). Vavilovian mimicry: nikolai vavilov and his little-known impact on weed science. Weed Sci. 62, 207–216. doi: 10.1614/WS-D-13-00122.1
Mcfadden, E. S., and Sears, E. R. (1946). The origin of triticum spelta and its free-threshing hexaploid relatives. J. Heredity. 37, 81–89. doi: 10.1093/oxfordjournals.jhered.a105590
Müntzing, A. (1936). The evolutionary significance of autopolyploidy. Hereditas 21, 363–378. doi: 10.1111/j.1601-5223.1936.tb03204.x
Murphy, B. P., and Tranel, P. J. (2019). Target-site mutations conferring herbicide resistance. Plants 8, 1–16. doi: 10.3390/plants8100382
Nandula, V. K., Ray, J. D., Ribeiro, D. N., Pan, Z., and Reddy, K. N. (2013). Glyphosate Resistance in Tall Waterhemp (Amaranthus tuberculatus) from Mississippi is due to both Altered Target-Site and Nontarget-Site Mechanisms. Weed Sci. 61, 374–383. doi: 10.1614/ws-d-12-00155.1
Nandula, V. K., Wright, A. A., Bond, J. A., Ray, J. D., Eubank, T. W., and Molin, W. T. (2014). EPSPS amplification in glyphosate-resistant spiny amaranth (Amaranthus spinosus): a case of gene transfer via interspecific hybridization from glyphosate-resistant Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 70, 1902–1909. doi: 10.1002/ps.3754
Ngo, T. D., Krishnan, M., Boutsalis, P., Gill, G., and Preston, C. (2018a). Target-site mutations conferring resistance to glyphosate in feathertop Rhodes grass (Chloris virgata) populations in Australia. Pest Manag. Sci. 74, 1094–1100. doi: 10.1002/ps.4512
Ngo, T. D., Malone, J. M., Boutsalis, P., Gill, G., and Preston, C. (2018b). EPSPS gene amplification conferring resistance to glyphosate in windmill grass (Chloris truncata) in Australia. Pest Manag. Sci. 74, 1101–1108. doi: 10.1002/ps.4573
Norsworthy, J. K., Jha, P., Steckel, L. E., and Scott, R. C. (2010). Confirmation and control of glyphosate-resistant giant ragweed (Ambrosia trifida) in Tennessee. Weed Technol. 24, 64–70. doi: 10.1614/wt-d-09-00019.1
Omezzine, F., De Biologie, D., and De Bizerte, S, and Haouala, R. (2017). Effect of Ploidy Level of Trigonella foenum-graecum on its Allelopathic Potential. Tunisian J. Plant Protect. 12, 11–18.
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. doi: 10.1016/j.cell.2007.10.022
Otto, S. P., and Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437. doi: 10.1146/annurev.genet.34.1.401
Pandit, M. K., Pocock, M. J. O., and Kunin, W. E. (2011). Ploidy influences rarity and invasiveness in plants. J. Ecol. 99, 1108–1115. doi: 10.1111/j.1365-2745.2011.01838.x
Pasteur, G. (1982). A classificatory review of mimicry systems. Annu. Rev. Ecol. Syst. 13, 169–199. doi: 10.1146/annurev.es.13.110182.001125
Patterson, E. L., Saski, C., Küpper, A., Beffa, R., and Gaines, T. A. (2019). Omics potential in herbicide-resistant weed management. Plants 1, 1–14. doi: 10.3390/plants8120607
Pellicer, J., and Leitch, I. J. (2020). The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. N. Phytol. 226, 301–305. doi: 10.1111/nph.16261
Pereira, W., Crabtree, G., and William, R. D. (1987). Herbicide action on purple and yellow nutsedge (Cyperus rotundus and C. esculentus). Weed Technol. 1, 92–98. doi: 10.1017/S0890037X00029201
Perotti, V. E., Larran, A. S., Palmieri, V. E., Martinatto, A. K., Alvarez, C. E., Tuesca, D., et al. (2019). A novel triple amino acid substitution in the EPSPS found in a high-level glyphosate-resistant Amaranthus hybridus population from Argentina. Pest Manag. Sci. 75, 1242–1251. doi: 10.1002/ps.5303
Powles, S. B., and Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annual Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Pratley, J., Urwin, N., Stanton, R., Baines, P., Broster, J., Cullis, K., et al. (1999). Resistance to glyphosate in Lolium rigidum. I. Bioevaluation. Weed Science, 47, 405–411. doi: 10.1017/S0043174500091992
Preston, C., Wakelin, A. M., Dolman, F. C., Bostamam, Y., and Boutsalis, P. (2009). A decade of glyphosate-resistant lolium around the world: mechanisms, genes, fitness, and agronomic management. Weed Sci. 57, 435–441. doi: 10.1614/WS-08-181.1
Ramsey, J. (2011). Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. U.S.A. 108, 7096–7101. doi: 10.1073/pnas.1016631108
Ramsey, J., and Ramsey, T. S. (2014). Ecological studies of polyploidy in the 100 years following its discovery. Philosophic. Transac. Royal Soc. 369, 15–19. doi: 10.1098/rstb.2013.0352
Ramsey, J., and Schemske, D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639. doi: 10.1146/annurev.ecolsys.33.010802.150437
Ravet, K., Patterson, E. L., Krähmer, H., Hamouzová, K., Fan, L., Jasieniuk, M., et al. (2018). The power and potential of genomics in weed biology and management. Pest Manag. Sci. 74, 2216–2225. doi: 10.1002/ps.5048
Rayburn, A. L., McCloskey, R., Tatum, T. C., Bollero, G. A., Jeschke, M. R., and Tranel, P. J. (2005). Genome size analysis of weedy Amaranthus species. Crop Sci. 45, 2557–2562. doi: 10.2135/cropsci2005.0163
Renny-Byfield, S., and Wendel, J. F. (2014). Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 101, 1711–1725. doi: 10.3732/ajb.1400119
Rice, A., Glick, L., Abadi, S., Einhorn, M., Kopelman, N. M., Salman-Minkov, A., et al. (2015). The Chromosome Counts Database (CCDB) - a community resource of plant chromosome numbers. N. Phytol. 206, 19–26. doi: 10.1111/nph.13191
Rosche, C., Hensen, I., Mráz, P., Durka, W., Hartmann, M., and Lachmuth, S. (2017). Invasion success in polyploids: the role of inbreeding in the contrasting colonization abilities of diploid versus tetraploid populations of Centaurea stoebe s.l. J. Ecol. 105, 425–435. doi: 10.1111/1365-2745.12670
Sammons, R. D., and Gaines, T. A. (2014). Glyphosate resistance: state of knowledge. Pest Manag. Sci. 70, 1367–1377. doi: 10.1002/ps.3743
Santos, G., Oliveira, R. S., Constantin, J., Constantin Francischini, A., Machado, M. F. P. S., Mangolin, C. A., et al. (2014). Conyza sumatrensis: a new weed species resistant to glyphosate in the Americas. Weed Biol. Manag. 14, 106–114. doi: 10.1111/wbm.12037
Scarpino, S. V., Levin, D. A., and Meyers, L. A. (2014). Polyploid formation shapes flowering plant diversity. Am. Nat. 184, 456–465. doi: 10.1086/677752
Schiessl, S., Huettel, B., Kuehn, D., Reinhardt, R., and Snowdon, R. (2017). Post-polyploidisation morphotype diversification associates with gene copy number variation. Sci. Rep. 7, 1–18. doi: 10.1038/srep41845
Schoenfelder, K. P., and Fox, D. T. (2015). The expanding implications of polyploidy. J. Cell Biol. 209, 485–491. doi: 10.1083/jcb.201502016
Shimizu-Inatsugi, R., Terada, A., Hirose, K., Kudoh, H., Sese, J., and Shimizu, K. K. (2017). Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Mol. Ecol. 26, 193–207. doi: 10.1111/mec.13738
Singh, V., Etheredge, L., McGinty, J., Morgan, G., and Bagavathiannan, M. (2020). First case of glyphosate resistance in weedy sunflower (Helianthus annuus). Pest Manag. Sci. 76, 3685–3692. doi: 10.1002/ps.5917
Soltis, D. E., Albert, V. A., Leebens-Mack, J., Bell, C. D., Paterson, A. H., Zheng, C., et al. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348. doi: 10.3732/ajb.0800079
Soltis, D. E., Buggs, R. J. A., Doyle, J. J., and Soltis, P. S. (2010). What we still don't know about polyploidy. Taxon 59, 1387–1403. doi: 10.1002/tax.595006
Soltis, D. E., and Soltis, P. S. (1999). Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14, 348–352. doi: 10.1016/S0169-5347(99)01638-9
Soltis, D. E., Soltis, P. S., Pires, J. C., Kovarik, A., Tate, J. A., and Mavrodiev, E. (2004). Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linnean Soc. 82, 485–501. doi: 10.1111/j.1095-8312.2004.00335.x
Soltis, D. E., Visger, C. J., and Soltis, P. S. (2014). The polyploidy revolution then...and now: stebbins revisited. Am. J. Bot. 101, 1057–1078. doi: 10.3732/ajb.1400178
Sonnhammer, E. L. L., and Koonin, E. V. (2002). Orthology, paralogy and proposed classification for paralog subtypes. Trends Genetics. 18, 619–620. doi: 10.1016/S0168-9525(02)02793-2
Stebbins, G. L. (1950). Variation and Evolution in Plants. New York, NY: Columbia University Press. doi: 10.7312/steb94536
Stevens, A. V., Nicotra, A. B., Godfree, R. C., and Guja, L. K. (2020). Polyploidy affects the seed, dormancy and seedling characteristics of a perennial grass, conferring an advantage in stressful climates. Plant Biol. 22, 500–513. doi: 10.1111/plb.13094
Takano, H. K., Fernandes, V. N. A., Adegas, F. S., Oliveira, R. S., Westra, P., Gaines, T. A., et al. (2020). A novel TIPT double mutation in EPSPS conferring glyphosate resistance in tetraploid Bidens subalternans. Pest Manag. Sci. 76, 95–102. doi: 10.1002/ps.5535
Te Beest, M., Le Roux, J. J., Richardson, D. M., Brysting, A. K., Suda, J., Kubešová, M., et al. (2012). The more the better? The role of polyploidy in facilitating plant invasions. Annals Botany 109, 19–45. doi: 10.1093/aob/mcr277
Valavanidis, A. (2018). Glyphosate, the World's Most Widely Used Herbicide. Health and safety issues. Why scientists differ in their evaluation of its adverse health effects. Sci. Rev.
Van De Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genetics 18, 411–424. doi: 10.1038/nrg.2017.26
Vavilov, N. I. (1922). The law of homologous series in variation. J. Genetics Genomics 12, 47–89. doi: 10.1007/BF02983073
Vavilov, N. I., and Love, D. (1992). Origin and Geography of Cultivated Plants. Cambridge: Syndicate of the University of Cambridge. doi: 10.2307/215338
Vila-Aiub, M. M., Balbi, M. C., Distéfano, A. J., Fernández, L., Hopp, E., Yu, Q., et al. (2012). Glyphosate resistance in perennial Sorghum halepense (Johnsongrass), endowed by reduced glyphosate translocation and leaf uptake. Pest Manag. Sci. 68, 430–436. doi: 10.1002/ps.2286
Vila-Aiub, M. M., Neve, P., and Powles, S. B. (2009). Fitness costs associated with evolved herbicide resistance alleles in plants. N. Phyto. 184, 751–767. doi: 10.1111/j.1469-8137.2009.03055.x
Vila-Aiub, M. M., Yu, Q., and Powles, S. B. (2019). Do plants pay a fitness cost to be resistant to glyphosate? N. Phytol. 223, 532–547. doi: 10.1111/nph.15733
Wei, N., Cronn, R., Liston, A., and Ashman, T. L. (2019). Functional trait divergence and trait plasticity confer polyploid advantage in heterogeneous environments. N. Phytol. 221, 2286–2297. doi: 10.1111/nph.15508
Wendel, J. F. (2000). Genome evolution in polyploids. Plant Mol. Evol. 42, 225–249. doi: 10.1007/978-94-011-4221-2_12
Wendel, J. F., and Adams, K. L. (2005). Novel patterns of gene expression in polyploid plants. Trends Genetics 21, 539–543. doi: 10.1016/j.tig.2005.07.009
Wiersma, A. T., Gaines, T. A., Preston, C., Hamilton, J. P., Giacomini, D., Robin Buell, C., et al. (2015). Gene amplification of 5-enol-pyruvylshikimate-3-phosphate synthase in glyphosate-resistant Kochia scoparia. Planta, 241, 463–74. doi: 10.1007/s00425-014-2197-9
Windham, M. D., Pryer, K. M., Poindexter, D. B., Li, F. W., Rothfels, C. J., and Beck, J. B. (2020). A step-by-step protocol for meiotic chromosome counts in flowering plants: a powerful and economical technique revisited. Appl. Plant Sci. 8, 1–8. doi: 10.1002/aps3.11342
Winge, O. (1917). “The chromosomes: their numbers and general importance,” in Comptes Rendus des Travaux du Laboratoire Carlesberg, (Vol. 13), (Copenhagen: Carlesberg laboratoriet).
Winkler, H. (1917). Über die experimentelle Erzeugung yon Pflanzen mit abweichenden Chromosomenzahlen. Zeitschrift Für Induktive Abstammungs- Und Vererbungslehre. 17, 270–272. doi: 10.1007/BF01740617
Yang, Y., Moore, M. J., Brockington, S. F., Mikenas, J., Olivieri, J., Walker, J. F., et al. (2018). Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. N. Phytol. 217, 855–870. doi: 10.1111/nph.14812
Yanniccari, M., Vila-Aiub, M., Istilart, C., Acciaresi, H., and Castro, A. M. (2016). Glyphosate resistance in perennial ryegrass (Lolium perenne L.) is associated with a fitness penalty. Weed Sci. 64, 71–79. doi: 10.1614/WS-D-15-00065.1
Ye, C. Y., Lin, Z., Li, G., Wang, Y. Y., Qiu, J., Fu, F., et al. (2014). Echinochloa chloroplast genomes: insights into the evolution and taxonomic identification of two weedy species. PLoS ONE 9:e113657. doi: 10.1371/journal.pone.0113657
Yu, Q., Jalaludin, A., Han, H., Chen, M., Douglas Sammons, R., and Powles, S. B. (2015). Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 167, 1440–1447. doi: 10.1104/pp.15.00146
Zimin, A. V., Puiu, D., Hall, R., Kingan, S., Clavijo, B. J., and Salzberg, S. L. (2017). The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 6, 1–7. doi: 10.1093/gigascience/gix097
Keywords: polyploidy, herbicide resistance, evolution, genomics, glyphosate, weeds
Citation: Rutland CA, Hall ND and McElroy JS (2021) The Impact of Polyploidization on the Evolution of Weed Species: Historical Understanding and Current Limitations. Front. Agron. 3:626454. doi: 10.3389/fagro.2021.626454
Received: 05 November 2020; Accepted: 29 January 2021;
Published: 01 March 2021.
Edited by:
Mithila Jugulam, Kansas State University, United StatesReviewed by:
Ilias Travlos, Agricultural University of Athens, GreeceJamal Ragheb Qasem, The University of Jordan, Jordan
Copyright © 2021 Rutland, Hall and McElroy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph Scott McElroy, anNtMDAxMEBhdWJ1cm4uZWR1
 Claudia Ann Rutland
Claudia Ann Rutland Nathan D. Hall2
Nathan D. Hall2