
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Agron. , 15 December 2020
Sec. Pest Management
Volume 2 - 2020 | https://doi.org/10.3389/fagro.2020.608827
This article is part of the Research Topic New Applications of Insecticidal RNAi View all 9 articles
The sequence complementarity of the RNA interference (RNAi) pathway allows for targeted suppression of genes essential for insect survival, and enables development of pest management strategies specific to a given species while reducing the likelihood of adversely impacting non-target organisms (NTOs). The feasibility of manipulating the RNAi pathway to cause mortality in the highly invasive emerald ash borer (EAB) has been demonstrated. Here the spectrum of activity of three double stranded RNAs (dsRNAs) targeting the genes hsp, shi, and sn-rnp in EAB was evaluated in model insects representing five functional guilds including herbivore, predator, detritivore, pollinator, parasitoid; the last represented by the classical biological control agents currently deployed for EAB management in North America. All NTOs were exposed to EAB-specific dsRNAs in diet bioassays that measured potential lethal effects. Gene expression and in silico analysis were also assessed on NTOs for which gene sequences were publicly available. Bioassays demonstrated no lethal effects on our model insects, suggesting a narrow spectrum of activity for the three EAB-specific dsRNAs evaluated. The gene expression and in silico analyses suggest potential sublethal effects on our model pollinator; however we found no effects on insect survival. Overall, our results suggest no adverse effects of the RNAi strategy targeting EAB genes on the survival of the selected non-target organisms we evaluated. The results from this study provide guidance for future RNAi risk analyses that will allow this technology to move forward to a deployment stage.
RNA interference (RNAi) is a molecular mechanism triggered by the introduction of double-stranded RNA (dsRNA) designed to induce gene knock-down; it is a promising technology with powerful potential for insect pest control. The RNAi pathway disrupts target genes and can lead to insect mortality when essential genes are silenced and subsequent protein synthesis is interrupted (Huvenne and Smagghe, 2010; Zhang et al., 2017). The RNAi pathway involves the complementarity of ≥21 base pair sequences to the target genes in a given species or closely related species (Agrawal et al., 2003; Bachman et al., 2016). This specificity minimizes any detrimental effects of an RNAi strategy to non-target organisms (NTOs).
Implementation of crop protection strategies applying RNAi technology are under development (Poelchau et al., 2016; Vogel et al., 2019). Potential applications of environmental or dietary RNAi for pest control include applications of dsRNA in foliar sprays (San Miguel and Scott, 2016); systemic uptake through plant material (Hunter et al., 2012; Dalakouras et al., 2018; Pampolini et al., 2020), oral administration of dsRNA in baits (Zhou et al., 2008), embedded in nanoparticles (Zhang et al., 2010; Yan et al., 2020), expressed in genetically engineered microorganisms (Zhu et al., 2011; Joga et al., 2016), or expressed in transgenic plants (Baum et al., 2007; Mao et al., 2007; Ghag, 2017; Zhang et al., 2017).
In addition to its application for agricultural purposes, RNAi technology has been investigated in forest pests including the southern pine beetle, Dendroctonus frontalis (Coleoptera: Curculionidae) (Kyre et al., 2019), the congeneric mountain pine beetle, D. ponderosae (Kyre et al., 2020), the wood-boring Asian longhorned beetle, Anoplophora glabripennis (Coleoptera: Cerambycidae) (Rodrigues et al., 2017a), and the emerald ash borer (EAB), Agrilus planipennis Fairmaire (Coleoptera: Buprestidae) (Rodrigues et al., 2017b, 2018; Pampolini et al., 2020).
EAB is an exotic, highly invasive, phloem-feeding beetle native to Asia. Since its discovery in North America in 2002, EAB has killed millions of Fraxinus spp. throughout its invaded range (Cappaert et al., 2005; Poland and McCullough, 2006), devastating urban forests, posing a threat to forest ecosystems, reducing biodiversity, and causing unprecedented economic losses (Aukema et al., 2011). Fortunately, previous studies evaluating the susceptibility of EAB to RNAi (Zhao et al., 2015) and the effectiveness of this technology against this pest (Rodrigues et al., 2017b, 2018; Pampolini et al., 2020) shows promise, and suggests that RNAi could be a viable option when implemented as a component of an integrated management strategy for EAB.
To move a biopesticide using RNAi technology toward commercialization, its risk to the environment and possible adverse effects on non-target organisms must be evaluated (Vélez et al., 2016; Haller et al., 2019). Given the novel mode of action of dsRNA-based products, the regulatory requirements for their deployment have been discussed internationally (USEPA, 2014; Mendelsohn et al., 2020; Romeis and Widmer, 2020). The risk assessment framework currently in use for genetically engineered plants expressing insecticidal proteins, such as those expressing Cry proteins from Bacillus thuringiensis, has been suggested as a starting point to evaluate the potential hazards for RNAi products (Romeis et al., 2013; Romeis and Widmer, 2020). This framework tests the risk hypothesis that the stressor (dsRNA) does not adversely impact non-target arthropods in the field, and recommends investigating pest-specific dsRNA impacts in species that represent key functional groups of economic and ecological importance (Romeis et al., 2008; USEPA, 2013); this is the approach used here.
We evaluated model insects, including an herbivore, predator, detritivore, and pollinator, for any effects of the EAB-specific dsRNAs considered for future deployment. We then evaluated the effects of the same dsRNAs on classical biological control agents currently deployed for EAB management in North America. Our overall goal is to generate data that will allow the EAB-specific dsRNAs that silence genes and induce rapid mortality to move to the deployment stage.
Mortality of EAB neonate larvae and adults can be induced after oral ingestion of dsRNA targeting the genes heat shock 70-kDa protein (hsp), shibire (shi) (Rodrigues et al., 2018), and U1 small nuclear ribonucleoprotein (sn-rnp from hereafter referred as rnp) (Pampolini and Rieske, unpublished data), so these were selected to evaluate specific EAB-dsRNA effects in the non-target organisms (Supplementary Table 1). Green fluorescent protein (gfp) was used as the negative control, and a positive control included a dsRNA specific to the insect under evaluation (Supplementary Table 1) and/or potassium arsenate (KH2AsO4) (Sigma-Aldrich, St. Louis, MO USA), a stomach poison (Romeis et al., 2011) selected to confirm effectiveness of the feeding bioassay.
Total RNA is extracted using Trizole reagent (ThermoFisher, USA), and the quantity and quality of the RNA is checked by electrophoresis and spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). cDNA is synthesized from 3 μg of total RNA using a M-MLV reverse transcriptase kit (ThermoFisher, USA).
PCR templates for in vitro synthesis of dsRNA are generated using gene-specific primers containing T7 polymerase promoter sequence (TAATACGACTCACTATAGGG). PCR conditions are 4 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C, finishing with an extension step at 72°C for 10 min. The PCR template is purified using a PCR purification kit (Qiagen Inc., Valencia, CA, USA). After PCR purification, dsRNA synthesis is performed using the MEGAscript RNAi Kit (Ambion Inc., Foster City, CA USA) following manufacturer's instructions. The reaction is incubated for 14 h at 37 °C, followed by 15 min of DNase treatment. The dsRNA is precipitated by adding 0.1x volume of sodium acetate (3 M, pH 5.2) and 2.5x the volume of 100% ethanol, which is maintained at −20°C for at least 2 h followed by centrifugation at 4°C for 30 min. The dsRNA pellet is then rinsed with 750 μL of 75% ethanol and centrifuged again at 4°C for 15 min. The ethanol is removed and the dsRNA is diluted in ultrapure distilled water. The quality of the dsRNA is checked by electrophoresis and quantified using a spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). To attain the required concentration for each assay, dsRNA samples are vacuum concentrated using Concentrator plus (Eppendorf, Hauppauge, NY, USA).
Non-target organisms were chosen as models to represent multiple feeding guilds. The Colorado potato beetle (CPB), Leptinotarsa decemlineata (Coleoptera: Chrysomelidae), served as our model herbivore, and the spotted lady beetle, Coleomegilla maculata (Coleoptera: Coccinellidae), as our model predator. Our model detritivore was the eastern subterranean termite, Reticulitermes flavipes (Blattodea: Rhinotermitidae), and our model pollinator was the honeybee, Apis mellifera (Hymenoptera: Apidae). Finally, the classical biological control agents routinely deployed for EAB population management, adult Tetrastichus planipennisi (Hymenoptera: Eulophidae), and Spathius galinae (Hymenoptera: Braconidae), were evaluated.
Non-target effects were evaluated by oral delivery of EAB-specific dsRNAs incorporated into diets; insects were fed ad-libitum. Each NTO was exposed to three EAB-specific dsRNAs targeting the genes hsp, shi, and rnp, the negative control gfp, and a positive control appropriate for the assay (Supplementary Table 1). All NTOs were exposed to dsRNA treatments for 3 consecutive days, after which they were maintained on the appropriate species-specific diet until assays were terminated. On day 3, a subsample of dsRNA-treated NTOs were collected, placed individually into micro tubes and transferred immediately to −80°C for later analysis of gene expression using quantitative real time PCR analysis using the RT-qPCR primer sequences in Supplementary Table 2.
Following total RNA extraction, cDNA was synthesized using M-MLV Reverse Transcriptase (Life Technologies, Carlsbad, CA, USA), and was used as a template for gene expression studies. The expression analyses were conducted using SYBR Green PCR Master Mix containing 1 μl of cDNA, 0.2 μl of each primer (10 mM; Supplementary Table 2), 5 μl of the SYBR green PCR master mix and 3.6 μl of ddH2O, totaling 10 μl. The StepOnePlus Real-Time PCR system (Life Technologies, Carlsbad, CA, USA) was used to perform real-time quantitative PCR (RT-qPCR) under the following conditions: one cycle of 2 min at 50°C, one cycle of 2 min at 95°C, 40 cycles of denaturation of 1 s at 95°C, annealing and extension for 30 s at 60°C, ending with generation of a melting curve to confirm a single peak and rule out non-specific product and primer dimer formations. Reference genes (Supplementary Table 2) and the 2-ΔΔCt method was used to calculate the relative expression of the target gene compared to the control (Livak and Schmittgen, 2001). A two-tailed t-test was used to compare the means of a single variable.
Newly emerged second instar CPB were obtained from a laboratory reared colony in the UK Department of Entomology. Larvae were evaluated individually in 50 × 15 mm Petri dishes containing a 12 mm diameter potato leaf disc (Zhu et al., 2011) treated with 1 μL of 0.001% Triton x-100 (Sigma-Aldrich Co., St. Louis, MO USA) to increase permeability, followed by 2 μL of the EAB-specific dsRNA, dsGFP as the negative control, and dsRNA targeting the gene β-act for the positive control (Zhu et al., 2011). The dsRNA concentration was 5 μg/μL, resulting in a daily exposure of 10 μg of dsRNA per day for 3 days. The assay was maintained at room temperature (26°C +/−2°C; 10:14 L:D) for 12 days. On day 4 larvae were provided untreated leaf discs daily through day 12, after which the assay was terminated. Ten beetles per treatment were evaluated and the assay was repeated three times. Mortality was evaluated at 48 h intervals for the duration of the 12-days assay. A one-way analysis of variance (ANOVA) was used to evaluate differences in mortality among EAB-specific dsRNAs and the controls.
Following exposure to the EAB-specific dsRNAs, the gene expression analysis evaluated the relative mRNA levels of the target genes hsp, shi, and rnp, with gfp as the negative control and β-act as the positive control. The ribosomal protein genes rp18 and rp4 were selected as reference genes to normalize expression of the target genes (Zhu et al., 2011).
Eggs of C. maculata were obtained commercially (Insect Lore, Shafter, CA, USA) and neonate larvae were maintained on a lepidopteran egg diet (Ephestia kuehniella; Lepidoptera: Pyralidae) (Green Methods, Redding, CA, USA) until reaching the second instar, when they were used in bioassays. Newly emerged second instar larvae were starved for 24 h prior to being placed individually into wells of a 96 well, flat bottom plate (Haller et al., 2019). Each well-contained a small (~1.5 mm diameter) cotton ball soaked with 4 μL of a 15% sucrose solution + EAB-specific dsRNA at a concentration of 2.5 μg/μL, resulting in a daily exposure of 10 μg; dsGFP was our negative control and dsRNA targeting the gene vATPase was our positive control (Yang et al., 2015). dsRNA was replenished daily for 3 days. On day 4 the larvae were placed individually into 35 mm Petri dishes and maintained on a diet of E. kuehniella eggs until adult emergence (~20 days). A moistened cotton ball in each Petri dish provided humidity; the cotton balls and the food were replaced every other day. Mortality was evaluated at 48 h intervals for 20 days after ingestion of dsRNAs. The assay was maintained in a growth chamber at 25°C +/−2°C, 70 +/- 5% RH, and a 16:8 L:D. A total of 20 individuals were evaluated per treatment, and there were two biological replicates. A one-way ANOVA was used to evaluate differences in mortality among dsRNA treatments and controls.
The relative expression of the target gene hsp was compared to that of our gfp negative control and our vATPase positive control. The shi and rnp gene sequences were not publicly available at the time of our study, so they were not included in the gene expression analysis. The 18S ribosomal RNA (rp18S) and the 16S ribosomal RNA (rp16S) genes were selected as the reference genes (Yang et al., 2015).
Termites were obtained from laboratory reared colonies in the UK Department of Entomology; worker adults were selected for evaluation and were starved for 24 h prior to use in assays. Groups of 13–15 termites were placed into 35 mm culture Petri dishes (Fisher Scientific, Waltham, MA, USA) containing a 33 mm diameter filter paper treated with 20 μL of EAB-specific dsRNA solution at a concentration of 10 μg/cm2 (Zhou et al., 2008); dsGFP served as the negative control and the positive control consisted of potassium arsenate at 1%. The dsRNA and potassium arsenate solutions were applied to the filter paper and allowed to completely dry, after which the paper disks were moistened with an additional 50 μL of water and 1% blue food dye (Assorted Food Colors, Kroger Co., Cincinnati, OH, USA) to allow visual tracking of ingestion of the dsRNA in the termites' intestinal tract. Assays were maintained at 26°C +/- 2°C, and 0:24 L:D for 14 days, and the filter paper was moistened every other day with 50 μL of water and food dye. Three groups of 13–15 termites from three different colonies were evaluated per treatment, representing three biological replicates. A one-way ANOVA was used to evaluate differences in termite mortality among dsRNA treatments and controls.
Our gene expression analysis assessed relative expression of the target gene hsp compared to that of the gfp negative control; the genes NADH dehydrogenase (NADHdh) and β-actin (β-act) were selected as reference genes (Zhou et al., 2008). Sequences for shi and rnp were not publicly available when the study was conducted, so they were not included in the gene expression analysis.
Honeybees were obtained from a research apiary at the UK Department of Entomology. Hive frames were selected to ensure brood synchrony, and newly emerged adults were collected immediately from frames in early August for feeding bioassays (Tan et al., 2016; Vélez et al., 2016). Groups of 8–10 bees were placed directly into modified Petri dishes (150 × 20 mm) containing a microcentrifuge tube with access holes to a 50% sucrose solution (Figure 1), which contained dsRNA at a concentration of 10 μg per bee for dsHSP, dsSHI, dsRNP, and dsGFP. An additional treatment tested the combination of dsHSP+dsSHI at a concentration of 1 μg (500 ng of each) per bee. Potassium arsenate (0.05 μg/μL) was used as the positive control (Tan et al., 2016). Bees were offered 1 mL of 50% sucrose + dsRNA, and after consumption of the entire solution (~72 h), were maintained on a 50% sucrose lacking dsRNA. Assay dishes were maintained in a growth chamber at 34°C +/- 2°C, 75% relative humidity, and 0:24 L:D; honeybee mortality was evaluated for 30 days until all individuals were dead, and the mortality assessed on day 10 was compared among treatments. There were 8–9 dishes of 8–10 bees per dish evaluated for each treatment. Frames from two hives were selected, and bees from both frames were collected on 2 different days with a 14 days interval, representing four biological replicates. Honeybee mortality was evaluated using a one-way ANOVA to assess differences among dsRNA treatments and controls.
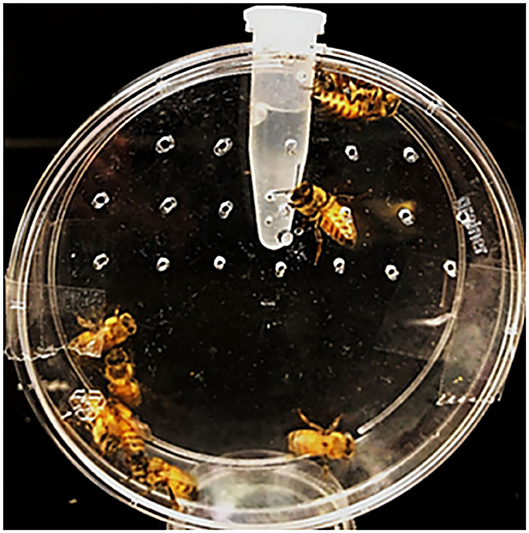
Figure 1. Honeybee bioassay chambers, in which 8–10 bees were placed in Petri dishes containing a microcentrifuge tube with holes allowing access to a 50% sucrose solution with the dsRNA treatments.
After 4 days of exposure to the EAB-specific dsRNAs, the relative mRNA levels of the target genes hsp, shi, and rnp were evaluated and compared to gfp, using act and ribosomal protein RP49 (rp49) as reference genes to normalize expression of the target genes (Lourenco et al., 2008).
Laboratory reared T. planipennisi and S. galinae adults were provided by the USDA APHIS PPQ Emerald Ash Borer Program laboratory (Brighton, MI). Immediately upon receipt, adult parasitoids were exposed to EAB specific dsRNAs using a droplet feeding bioassay (Rodrigues et al., 2017b). Droplets consisted of 2 μL of a 5% sucrose solution containing dsRNA at a concentration of 10 μg/μL, offered on parafilm in a 150 × 15 mm Petri dish containing a moistened filter paper and covered with an inverted transparent plastic cup (200 mL). There was one dsRNA-sucrose droplet for every two wasps in each cup. In groups of 8–10 individuals, adult wasps were exposed to the dsRNA treatments for 3 days; after day 4 they were maintained on a sucrose only solution, lacking dsRNA. Droplets were replenished every other day. Potassium arsenate at 0.05 μg/μL was the positive control and the assay was maintained at room temperature (26°C +/−2°C; 10:14 L:D) for 11 days. The experiment was repeated three times for S. galinae and twice for T. planipennisi. For T. planipennisi only, a second assay testing a dsRNA concentration of 1 μg/μL was also evaluated and repeated three times. A one-way analysis of variance (ANOVA) was used to evaluate differences in mortality among EAB-specific dsRNAs and the controls.
Because gene sequences for the classical biological control parasitoids T. planipennisi and S. galinae are not publicly available, we were not able to evaluate parasitoids for gene expression following EAB-specific dsRNA treatments.
Using publicly available sequences, an in silico analysis was performed to assess potential effects of the EAB-specific dsRNAs on our selected NTOs. BLAST searches were done using the NCBI nucleotide BLAST tool (BLASTn) to compare orthologous hsp, shi, and rnp sequences of EAB and each NTO. Alignments were analyzed to identify exact 21 or greater nucleotide (nt) matches. When coding sequences for a given species or a given gene were not available, they were replaced by predicted sequences derived from gene annotation of closely related species (Table 1).
Mortality of second instar larvae exposed to EAB-specific dsRNAs was <15% after 12 days, and did not differ among treatments and the negative control (p = 0.94), whereas mortality in the positive control (dsACT) reached 80% by day 6 and rose to 95% on day 8 (Figure 2). Our gene expression analysis demonstrated a significant reduction in gene expression for our positive control, dsACT, as expected, but there was no decrease in the relative expression of hsp, shi, and rnp (Figure 3). Further, our in silico analysis corroborates the findings of our bioassay and gene expression analysis; there were no 21 nucleotide alignments between the EAB sequences and the CPB coding sequences for the genes hsp, shi, and rnp (Table 1). Together, these results confirm that there are no adverse effects of EAB dsRNAs on survival of CPB, our model herbivore.
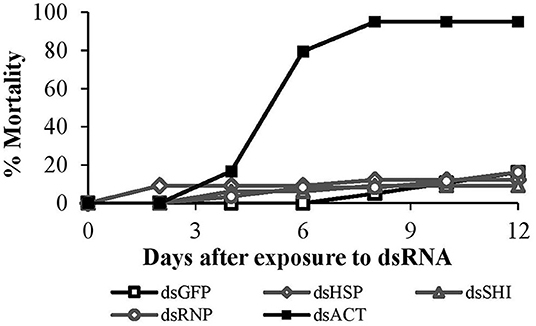
Figure 2. Second instar CPB mortality after exposure to 10 μg of EAB-specific dsRNAs targeting the genes hsp, shi, and rnp; 10 μg of dsGFP was used as negative control and 10 μg/μL of dsACT was used as positive control.
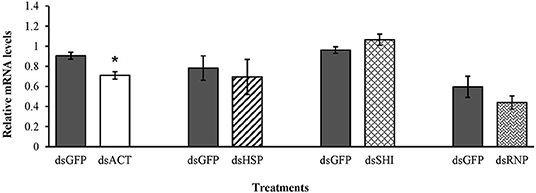
Figure 3. Transcript levels of the positive control act, hsp, shi, and rnp genes in second instar CPB after 3 days feeding on 10 μg of EAB-specific dsRNAs; 10 μg of dsGFP was used as control. Relative mRNA levels were normalized using rp4 and rp18 as reference genes. Means ± SE (N = 4–5) with asterisks indicate significant differences (t-test, one-tailed p < 0.05).
In our 20-days bioassay, mortality of larval C. maculata ranged from 2.5 to 7.5% for the EAB dsRNA treatments and the negative control, which did not differ (P = 0.37) (Figure 4A). On day 10, ~97% of the insects reached the pupal stage in all EAB dsRNAs treatments and the negative control, followed by adult emergence of 97.5% for dsHSP, 94.5% for dsSHI, and dsRNP, and 92.5% for dsGFP (Figure 4B). Larvae demonstrated a significant reduction in gene expression after exposure to the positive control, ds-vATPase in our gene expression analysis, but there was no decrease in relative expression of hsp in our model predator after exposure to EAB dsHSP (Figure 5). Additionally, the three EAB gene sequences were queried against spotted lady beetle and no similar sequences were found in the bioinformatics analysis (Table 1). Our results indicate that lady beetle larval survival, development, pupation, and adult emergence were unaffected by ingestion of EAB dsRNAs.
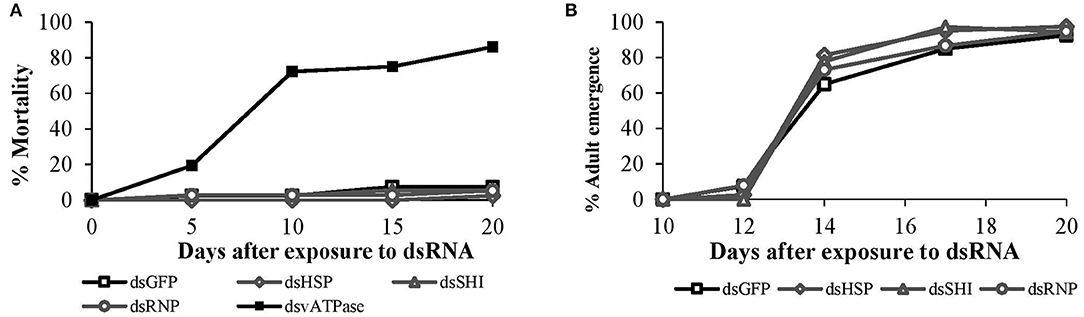
Figure 4. Spotted lady beetle (A) larval mortality after oral ingestion of EAB dsRNAs (10 μg) targeting the genes hsp, shi, and rnp; 10 μg of dsGFP was used as negative control and 10 μg/μL of dsvATPase was used as positive control, and (B) subsequent adult emergence from day 12 until day 20.
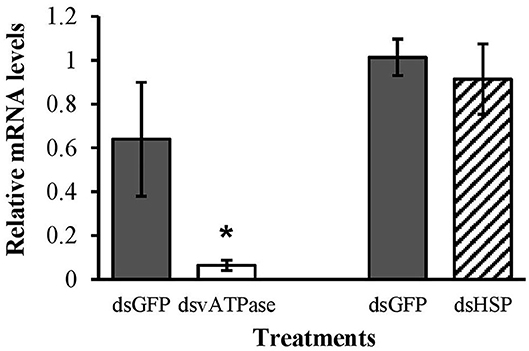
Figure 5. Transcript levels of the positive control vATPase, and hsp genes in second instar larvae of spotted lady beetle after 3 days feeding on 10 μg of EAB-specific dsRNAs; 10 μg of dsGFP was used as control. Relative mRNA levels were normalized using 16s and 18s as reference genes. Means ± SE (N = 4–5) with asterisks indicate significant differences (t-test, one-tailed p < 0.05).
Here we show that C. maculata second instar larvae are highly susceptible to orally delivered dsRNAs, whereas previous studies evaluating lady beetle response to dietary RNAi utilized neonates (Haller et al., 2019) or first instar larvae (Bachman et al., 2013). We found that ingestion of the positive control dsvATPase by second instars triggered gene silencing and induced significant mortality, but ingestion of the EAB-specific dsRNAs elicited no response. Collectively, our results confirm that there are no adverse effects of EAB dsRNAs on survival or gene expression in our model predator.
After 14 days, mortality of worker termites exposed to EAB specific dsRNA was 4.5–11%, and did not differ among treatments and the negative control (p = 0.89). After 4 days the positive control (potassium arsenate) exhibited 50% mortality; this increased to 86 and 98% on days 6 and 8, respectively (Figure 6).
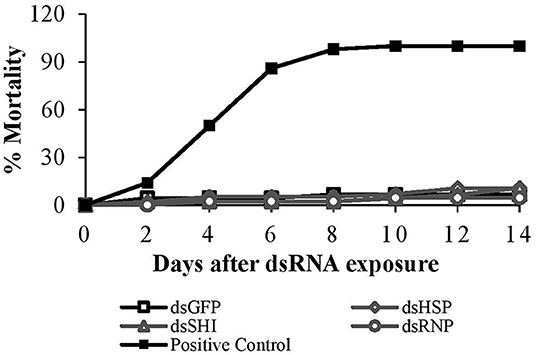
Figure 6. Worker termite mortality after exposure to EAB-specific dsRNAs (10 μg/cm2) targeting the genes hsp, shi, and rnp. 10 μg/cm2 of dsGFP was used as negative control and 1% potassium arsenate solution was used as positive control.
There was no significant decrease in mRNA levels of the hsp gene after exposure to the EAB dsRNA in our gene expression analysis (Figure 7); and the three EAB gene sequences were queried against Eastern subterranean termite and no similar sequences were found in the bioinformatics analysis (Table 1).
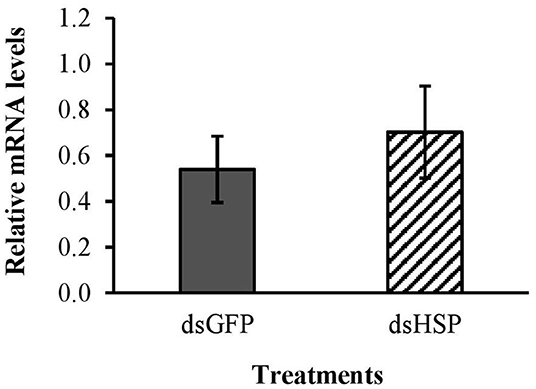
Figure 7. Relative expression of hsp gene in worker termites after 3 days feeding on 10 μg/cm2 dsHSP; 10 μg/cm2 of dsGFP was used as control. Relative mRNA levels were normalized using β-act and NADHdh as reference genes.
Termites are sensitive to ingested dsRNA and the RNAi response in termites has been characterized (Zhou et al., 2008). Although we used potassium arsenate as the positive control rather than a termite-specific dsRNA, our results demonstrate that the EAB dsRNAs had no impact on termite survival. This is supported by the in silico analysis, and by the gene expression study evaluating mRNA levels of the hsp gene.
Ten days after ingestion of dsRNAs, there were no differences in adult honeybee mortality among dsRNA treatments and the negative control; average mortality was 14% for dsHSP, 23% for dsSHI, and 15% for dsRNP and dsGFP (P = 0.83) (Figure 8A). Bees exposed to the potassium arsenate control experienced 100% mortality within 24 h, confirming the efficacy of oral delivery. Following evaluation of mortality on day 10, bees were kept for an additional 20 days when all treatments reached 100% mortality (P = 0.98) (Figure 8B). Our results indicate no adverse impact of EAB dsRNAs on honeybee survival and longevity; however, we did see substantial changes in gene expression.
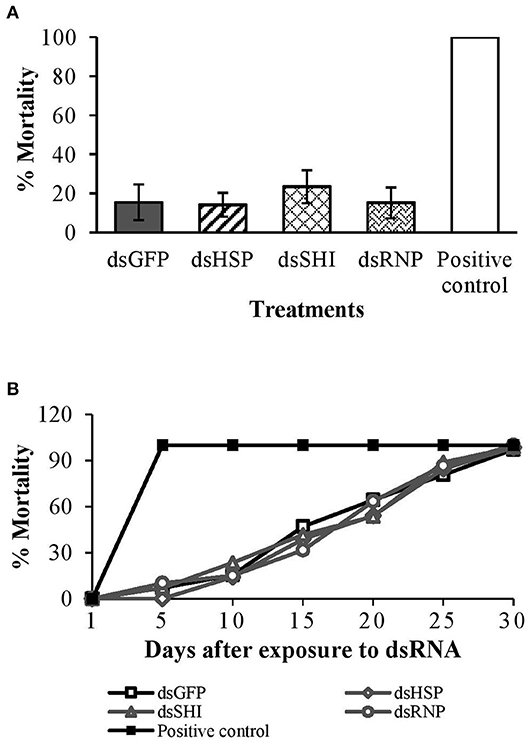
Figure 8. Worker honeybees <24 h after emergence were exposed to EAB-specific dsRNAs targeting the genes hsp, shi, and rnp at a concentration of 10 μg per individual; 10 μg per individual of dsGFP was used as negative control and 0.05 μg/μL of potassium arsenate was used as positive control. Adult mortality (A) after 10 days, and (B) over the 30-days assay.
Honeybees fed 10 μg per individual of dsHSP showed a decrease in the mRNA levels of hsp (47%), shi (70%), and rnp (75%) when compared to levels in honeybees fed dsGFP (Figure 9A). Honeybees fed 10 μg per individual of dsSHI also showed a decrease in the relative expression of hsp (38%), shi (75%), and rnp (80%) (Figure 9B). The treatment exposing bees to 10 μg per individual of dsRNP resulted in a decrease in relative expression of shi (70%), and rnp (80%) (Figure 9C), and bees exposed to 1 μg per individual of both dsHSP+dsSHI also showed a decrease in the relative expression of hsp (36%), shi (76%), and rnp (75%) (Figure 10).
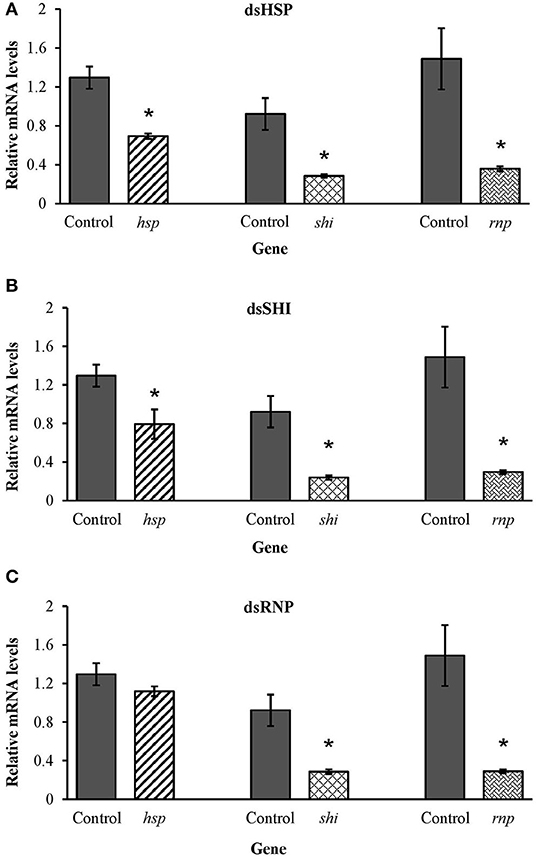
Figure 9. Relative expression of the genes hsp, shi, and rnp in worker honeybees after 3 days feeding on 10 μg per individual of (A) dsHSP, (B) dsSHI, and (C) dsRNP; 10 μg per individual of dsGFP was used as control. Relative mRNA levels were normalized using act and rp49 as reference genes. Means ± SE (N = 5) with asterisks indicate significant differences (t-test, one-tailed p < 0.05).
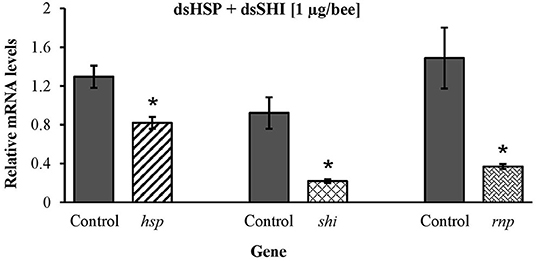
Figure 10. Transcript levels of hsp, shi, and rnp genes in worker bees after 3 days feeding on 1 μg per individual of combined dsRNAs (500 ng/individual each dsHSP and dsSHI) 10 μg per individual dsGFP was used as control. Relative mRNA levels were normalized using act and rp49 as reference genes. Means ± SE (N = 5) with asterisks indicate significant differences (t-test, one-tailed p < 0.05).
Our gene expression analysis demonstrated silencing of genes other than the targets, even at dsRNA concentrations an order of magnitude lower than previously used (1 μg per individual of dsHSP+dsSHI). However, these reductions in expression of hsp, shi, and rnp did not affect honeybee survival. One potential explanation for this is that, following exposure to a single high concentration of dsRNA, oversaturation of RNAi machinery may affect the insect immune response, but this oversaturation and the effects of silencing genes other than the target is not detectable in acute toxicity bioassays (Auer and Frederick, 2009; USEPA, 2013). Vélez et al. (2016), detected transient gene silencing in honeybees when comparing mRNA levels 48 and 96 h after dsRNA exposure. However, we evaluated gene expression only at a single time point (72 h after exposure), and so are unable to detect potential transient effects.
Using the NCBI nucleotide BLAST tool (BLASTn), our in silico evaluation identified continuous matches of > 21 nucleotides for shi and rnp in honeybees when queried against the honeybee predicted sequences available (Table 1). However, the presence of sequence homologies between the dsRNA and the genome of an organism does not necessarily indicate sensitivity of that organism (Romeis and Widmer, 2020). Pan et al. (2016) identified six 21nt long matches between dsRNA targeting the vATPase A in D. virgifera and the springtail Sinella curviseta (Collembola: Entomobryidae), yet the springtail was not adversely affected in laboratory feeding assays. In our study, silencing genes other than the target indicates that the RNAi response was not a sequence-specific event, but a generalized immune response to dsRNA, leading Mendelsohn et al. (2020) to suggest that the use of bioinformatics may have limited value in assessing off-target effects due to interspecific variation in RNAi machinery and variability in environmental exposure across different organisms.
Two concentrations of EAB-specific dsRNAs, 1 μg/μL and 10 μg/μL, were evaluated for T. planipennisi, and there were no differences in parasite mortality between the dsRNA concentrations. Wasps exposed to dsRNA at the higher concentration (10 μg/μL) experienced 25% mortality for dsHSP, 5% for dsSHI, 9% for dsRNP, and 15% for dsGFP after 11 days, but there were no differences among dsRNA treatments (Figure 11A). Tetrastichus planipennisi exposed to the lower concentration (1 μg/μL) experienced 7% mortality for dsHSP, 12% for dsSHI, 10% for dsRNP, and 16% for dsGFP; again, there were no differences among treatments (Figure 11B). Mortality of S. galinae exposed to dsRNAs at a concentration of 10 μg/μL was 12% for dsHSP, 14% for dsSHI, 12% for dsRNP, and 9% for dsGFP (Figure 11C), again with no differences among treatments. Our results suggest that EAB dsRNAs have no impact on the survival of these classical biological control agents, but because of the lack of genetic sequences for the biological control species, a full evaluation is not possible.
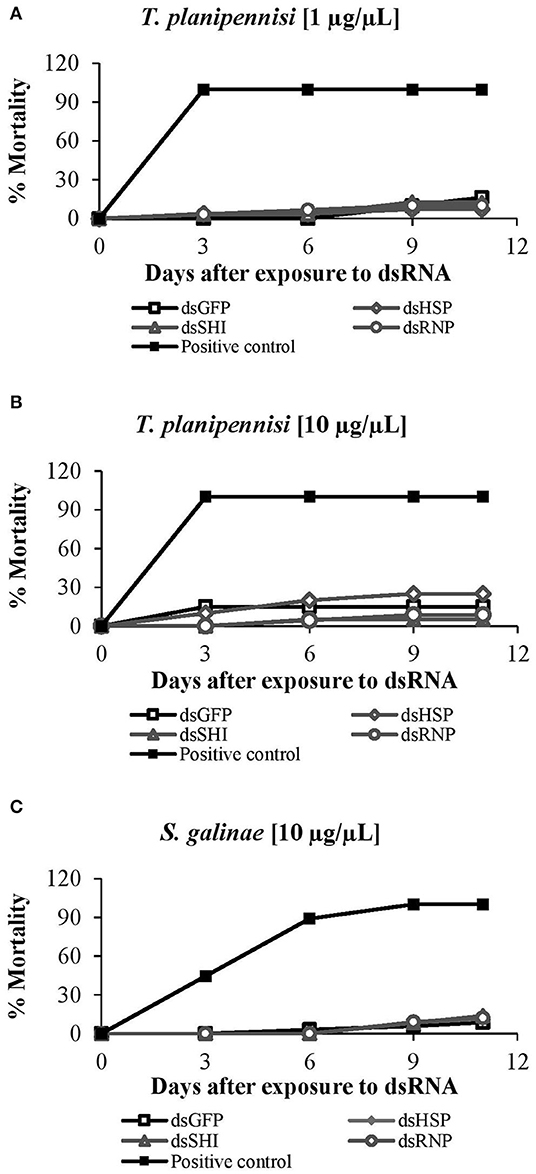
Figure 11. Mortality of classical biological control agents: T. planipennisi exposed to dsRNAs at (A) 1 μg/μL (P = 0.66) and (B) 10 μg/μL (P = 0.20); results did not differ between the two concentrations (two tailed t-test p > 0.05). Mortality of (C) S. galinae exposed to the EAB dsRNAs at 10 μg/μL (P = 0.91).
Sequence-specific gene silencing can suppress genes that are critical for insect survival or development, suggesting that dsRNAs can be used to develop pest control products that trigger the RNAi pathway, selectively targeting the pest species and reducing the likelihood of adversely affecting NTOs (Bachman et al., 2013). In addition to its efficacy, application of RNAi technology for insect control has the added advantage of extreme specificity to the target insect or to closely related species (Whyard et al., 2009). Characterization of the spectrum of activity for an insecticidal dsRNA should evaluate key species or guilds representing different ecological functions (Romeis et al., 2008). Our study evaluated the toxicity of dsRNAs targeting the genes hsp, shi, and rnp in the emerald ash borer, a highly destructive, invasive, tree killing pest, on model insects representing the herbivore, predator, detritivore, and pollinator guilds. In addition, we evaluated dsRNAs targeting the same genes for toxicity to the classical biological control agents currently deployed for EAB population management.
Safety protocols dictate that exposures used for toxicity studies with non-target arthropods must exceed the maximum amount of dsRNA expected to be available in the environment (EFSA European Food Safety Authority, 2014). Previous studies demonstrated that adult EAB experiences significant gene silencing and mortality when exposed to ash leaves treated with dsRNA solution at 70 ng/μl (Pampolini et al., 2020). In the present study, our model NTOs were exposed to EAB dsRNAs in diets at concentrations ~140 times higher than that inducing mortality in EAB delivered topically on plant foliage. The experimental diets in our study incorporated dsRNAs at 10 μg/μL (CPB, lady beetle, and classical biological control agents), 10 μg/cm2 (termites), and 10 ug per individual (honeybees) to evaluate non-target effects in representatives from six insect families (Chrysomelidae, Coccinellidae, Rhinotermitidae, Apidae, Eulophidae, Braconidae) in three orders (Coleoptera, Blattodea, and Hymenoptera), representing five functional guilds (herbivore, predator, detritivore, pollinator, parasitoid).
The efficiency of RNAi varies significantly among insect orders, but coleopterans are highly sensitive to ingested dsRNA (Joga et al., 2016), and our target species, emerald ash borer, is a coleopteran. Survival of the two additional coleopterans evaluated in this study were unaffected by ingestion of EAB-specific dsRNAs (Figures 2, 4A), and we found no differences in gene expression. Additionally, there were no sublethal effects on lady beetle larval development (Figure 4B).
Following ingestion of EAB-specific dsRNAs, our gene expression analysis demonstrated a reduction in expression of the honeybee genes hsp, shi, and rnp, but these did not affect adult survival. The responses we observed may be attributable to a generalized immune response to dsRNAs in honeybees. Previous studies evaluating off-target effects caused by dsRNA in honeybees (Jarosch and Moritz, 2012; Nunes et al., 2013) evaluated the expression of insect-specific genes after exposure to dsRNA targeting the gene gfp, which has no known honeybee homolog; they observed non-target effects characterized by the downregulation or upregulation of genes associated with a variety of biological processes. In general, dsRNAs can be recognized as a viral infection, culminating in the activation of immune genes, RNAi pathway, siRNA production and consequent off-target effects (Nunes et al., 2013). Vélez et al. (2016) conducted a toxicity assay to identify non-target effects of dsRNA targeting the Diabrotica virgifera virgifera gene vATPase in honeybees; our findings are similar to their results, with some effects on gene expression, but no effects on honeybee survival or longevity.
Importantly, we demonstrate no negative effects of the EAB-specific dsRNAs on survival of the classical biological control agents currently deployed for EAB management in the US. Our mortality assays with T. planipennisi and S. galinae showed no effects on parasitoid survival, regardless of the dsRNA concentration.
Overall, our results suggest no adverse effects of the RNAi strategy targeting EAB genes on the survival of the selected non-target organisms we evaluated. Additional research characterizing sub-lethal effects of dsRNA on the NTOs, and the persistence and fate of the molecule in the environment are needed; however, in our preliminary risk assessment study we identified high specificity for the target insect of the dsRNAs developed for EAB management. These results represent an important step to move this technology forward to a deployment stage.
Our study provides additional data demonstrating the feasibility of utilizing novel alternatives for EAB management that are ecofriendly and do not threaten the environment or beneficial and non-target organisms.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
FP conducted the experiments. FP and LR conceived the experiments, analyzed the results, and prepared the manuscript. All authors approve the manuscript.
This publication is from the Kentucky Agricultural Experiment Station and is published with the approval of the director. This work was supported by the University of Kentucky, the USDA APHIS Farm Bill, and the Kentucky Agricultural Experiment Station under McIntire-Stennis 2352657000.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Julian Dupuis (University of Kentucky) for bioinformatics support, Clare Rittschof, Xuguo Zhou, and Reddy Palli (University of Kentucky), and the USDA APHIS PPQ Biological Control Production Facility (Brighton, MI) for providing experimental insects. Hannah Hollowell, Holly Foster, Kylie Bickler, and Thèo Walden provided laboratory assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2020.608827/full#supplementary-material
ACT, actin; ANOVA, one-way analysis of variance; BLAST, Basic Local Alignment Search Tool; CPB, Colorado potato beetle; dsRNA, double-stranded RNA; EAB, emerald ash borer; GE plants, genetic engineered plants; GFP, green fluorescent protein; HSP, heat shock 70-kDa protein; NADHdh, NADH dehydrogenase; NCBI, National Center for Biotechnology Information; NTO, non-target organisms; RNAi, RNA interference; RNP, U1 small nuclear ribonucleoprotein; RP4, ribosomal protein4; RP16S, ribosomal RNA 16S; RP18, ribosomal protein 18; RP18S, ribosomal RNA 18S; RP49, ribosomal protein 49; SHI, shibire; vATPase, vacuolar ATPase.
Agrawal, N., Dasaradhi, P. V., Mohmmed, A., Malhotra, P., Bhatnagar, R. K., and Mukherjee, S. K. (2003). RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. doi: 10.1128/MMBR.67.4.657-685.2003
Auer, C., and Frederick, R. (2009). Crop improvement using small RNAs: applications and predictive ecological risk assessments. Trends Biotechnol. 27, 644–651. doi: 10.1016/j.tibtech.2009.08.005
Aukema, J. E., Leung, B., Kovacs, K., Chivers, C., Britton, K. O., Englin, J., et al. (2011). Economic impacts of non-native forest insects in the continental United States. PLoS ONE 6:e24587. doi: 10.1371/journal.pone.0024587
Bachman, P. M., Bolognesi, R., Moar, W. J., Mueller, G. M., Paradise, M. S., Ramaseshadri, P., et al. (2013). Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against western corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res. 22, 1207–1222. doi: 10.1007/s11248-013-9716-5
Bachman, P. M., Huizinga, K. M., Jensen, P. D., Mueller, G., Tan, J., Uffman, J. P., et al. (2016). Ecological risk assessment for DvSnf7 RNA: A plant-incorporated protectant with targeted activity against western corn rootworm. Regul. Toxicol. Pharmacol. 81, 77–88. doi: 10.1016/j.yrtph.2016.08.001
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Cappaert, D., McCullough, D. G., Poland, T. M., and Siegert, N. W. (2005). Emerald ash borer in North America: a research and regulatory challenge. Am. Entomol. 51, 152–163. doi: 10.1093/ae/51.3.152
Dalakouras, A., Jarausch, W., Buchholz, G., Bassler, A., Braun, M., Manthey, T., et al. (2018). Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petiole absorption. Front. Plant Sci. 9:1253. doi: 10.3389/fpls.2018.01253
EFSA European Food Safety Authority (2014). International Scientific Workshop “Risk Assessment Considerations for RNAi-based GM Plants” (4-5 June 2014, Brussels, Belgium).
Ghag, S. B. (2017). Host induced gene silencing, an emerging science to engineer crop resistance against harmful plant pathogens. Physiol. Mol. Plant Pathol. 100, 242–254. doi: 10.1016/j.pmpp.2017.10.003
Haller, S., Widmer, F., Siegfried, B. D., Zhuo, X., and Romeis, J. (2019). Responses of two ladybird beetle species (Coleoptera: Coccinellidae) to dietary RNAi. Pest Manag. Sci. 75, 2652–2662. doi: 10.1002/ps.5370
Hunter, W. B., Glick, E., Paldi, N., and Bextine, B. R. (2012). Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest suppression. Southwest Entomol. 37, 85–87. doi: 10.3958/059.037.0110
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
Jarosch, A., and Moritz, R. F. A. (2012). RNA interference in honeybees: off-target effects caused by dsRNA. Apidologie 43, 128–138. doi: 10.1007/s13592-011-0092-y
Joga, M. R., Zotti, M. J., Smagghe, G., and Christiaens, O. (2016). RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7:553. doi: 10.3389/fphys.2016.00553
Kyre, B. R., Bentz, B. J., and Rieske, L. K. (2020). Susceptibility of mountain pine beetle (Dendroctonus ponderosae Hopkins) to gene silencing through RNAi provides potential as a novel management tool. Forest Ecol. Manag. 473:118322. doi: 10.1016/j.foreco.2020.118322
Kyre, B. R., Rodrigues, T. B., and Rieske, L. K. (2019). RNA interference and validation of reference genes for gene expression analyses using qPCR in southern pine beetle, Dendroctonus frontalis. Sci. Rep. 9:5640. doi: 10.1038/s41598-019-42072-6
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lourenco, A. P., Mackert, A., Cristino, A. D. S., and Simoes, Z. L. P. (2008). Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39, 372–385. doi: 10.1051/apido:2008015
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Mendelsohn, M. L., Gathmann, A., Kardassi, D., Sachana, M., Hopwood, E. M., Dietz-Pfeilstetter, A., et al. (2020). Summary of discussions from the 2019 OECD conference on RNAi based pesticides. Front. Plant Sci. 11:740. doi: 10.3389/fpls.2020.00740
Nunes, F. M., Aleixo, A. C., Barchuk, A. R., Bomtorin, A. D., Grozinger, C. M., and Simões, Z. L. (2013). Non-target effects of green fluorescent protein (GFP)-derived double-stranded RNA (dsRNA-GFP) used in honey bee RNA interference (RNAi) assays. Insects 4, 90–103. doi: 10.3390/insects4010090
Pampolini, F., Rodrigues, T. B., Leelesh, R. S., Kawashima, T., and Rieske, L. K. (2020). Confocal microscopy provides visual evidence and confirms the feasibility of dsRNA delivery to emerald ash borer through plant tissues. J. Pest Sci. 93, 1143–1153. doi: 10.1007/s10340-020-01230-w
Pan, H., Xu, L., Noland, J. E., Li, H., Siegfried, B. D., and Zhou, X. (2016). Assessment of potential risks of dietary RNAi to a soil micro-arthropod, Sinella curviseta Brook (Collembola: Entomobryidae). Front. Plant Sci. 7:1028. doi: 10.3389/fpls.2016.01028
Poelchau, M. F., Coates, B. S., Childers, C. P., Peréz de León, A. A., Evans, J. D., Hackett, K., et al. (2016). Agricultural applications of insect ecological genomics. Curr. Opin. Insect Sci. 13, 61–69. doi: 10.1016/j.cois.2015.12.002
Poland, T. M., and McCullough, D. G. (2006). Emerald ash borer: invasion of the urban forest and the threat to north America's ash resource. J. Forest. 118–124.
Rodrigues, T. B., Dhandapani, R. K., Duan, J. J., and Palli, S. R. (2017a). RNA interference in the Asian longhorned beetle: identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 7:8913. doi: 10.1038/s41598-017-08813-1
Rodrigues, T. B., Duan, J. J., Palli, S. R., and Rieske, L. K. (2018). Identification of highly effective target genes for RNAi-mediated control of emerald ash borer, Agrilus planipennis. Sci. Rep. 8:5020. doi: 10.1038/s41598-018-23216-6
Rodrigues, T. B., Rieske, L. K., Duan, J. J., Mogilicherla, K., and Palli, S. R. (2017b). Development of RNAi method for screening candidate genes to control emerald ash borer, Agrilus planipennis. Sci. Rep. 7:7379. doi: 10.1038/s41598-017-07605-x
Romeis, J., Bartsch, D., Bigler, F., Candolfi, M. P., Gielkens, M. M., Hartley, S. E., et al. (2008). Assessment of risk of insect-resistant transgenic crops to nontarget arthropods. Nat. Biotechnol. 26, 203–208. doi: 10.1038/nbt1381
Romeis, J., Hellmich, R. L., Candolfi, M. P., Carstens, K., Schrijver, A., Gatehouse, A. M. R., et al. (2011). Recommendations for the design of laboratory studies on non-target arthropods for risk assessment of genetically engineered plants. Transgenic Res. 20, 1–22. doi: 10.1007/s11248-010-9446-x
Romeis, J., Raybould, A., Bigler, F., Candolfi, M. P., Hellmich, R. L., Huesing, J. E., et al. (2013). Deriving criteria to select arthropod species for laboratory tests to assess the ecological risks from cultivating arthropod-resistant genetically engineered crops. Chemosphere 90, 901–909. doi: 10.1016/j.chemosphere.2012.09.035
Romeis, J., and Widmer, F. (2020). Assessing the risks of topically applied dsRNA-based products to non-target arthropods. Front. Plant Sci. 11:679. doi: 10.3389/fpls.2020.00679
San Miguel, K., and Scott, J. G. (2016). The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manage. Sci. 72, 801–809. doi: 10.1002/ps.4056
Tan, J., Levine, S. I., Bachman, P. M., Jensen, P. D., Mueller, G. M., Uffman, J. P., et al. (2016). No impact of DvSnf7 RNA on honey bee (Apis mellifera L.) adults and larvae in dietary feeding tests. Environ. Toxicol. Chem. 35, 287–294. doi: 10.1002/etc.3075
USEPA (2013). White Paper on RNAi Technology as a Pesticide: Problem Formulation for Human Health and Ecological Risk Assessment. (Washington, DC: FIFRA Science Advisory Panel). Available online at: https://www.thecre.com/premium/wp-content/uploads/2012/04/RNAi-White-Paper.pdf (accessed September, 2020).
USEPA (2014). Transmittal of the Meeting Minutes of the FIFRA SAP Meeting Held January 28, 2014 on the Scientific Issues Associated with the Use of ‘RNAi Technology as a Pesticide: Problem Formulation for Human Health and Ecological Risk Assessment’ Scientific Advisory Panel Minutes Number 2014–02. Washington, DC: USEPA. Available online at: http://www.epa.gov/scipoly/sap/meetings/2014/january/012814minutes.pdf
Vélez, A. M., Jurzenski, J., Matz, N., Zhou, X., Wang, H., Ellis, M., et al. (2016). Developing an in vivo toxicity assay for RNAi risk assessment in honey bees, Apis mellifera L. Chemosphere 144, 1083–1090. doi: 10.1016/j.chemosphere.2015.09.068
Vogel, E., Santos, D., Mingels, L., Verdonckt, T.-W., and Broeck, J. V. (2019). RNA Interference in insects: protecting beneficials and controlling pests. Front. Physiol. 9:1912. doi: 10.3389/fphys.2018.01912
Whyard, S., Singh, A. D., and Wong, S. (2009). Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 39, 824–832. doi: 10.1016/j.ibmb.2009.09.007
Yan, S., Ren, B., and Shen, J. (2020). Nanoparticle-mediated double-stranded RNA delivery system: a promising approach for sustainable pest management. Insect Sci. doi: 10.1111/1744-7917.12822. [Epub ahead of print].
Yang, C., Pan, H., Noland, J. E., Zhang, D., Zhang, Z., Liu, Y., et al. (2015). Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 5:18201. doi: 10.1038/srep18201
Zhang, J., Khan, S. A., Heckel, D. G., and Bock, R. (2017). Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 35, 871–882. doi: 10.1016/j.tibtech.2017.04.009
Zhang, X., Zhang, J., and Zhu, K. Y. (2010). Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 19, 683–693. doi: 10.1111/j.1365-2583.2010.01029.x
Zhao, C., Gonzales, M. A. A., Poland, T. M., and Mittapalli, O. (2015). Core RNAi machinery and gene knockdown in the emerald ash borer (Agrilus planipennis). J. Insect Physiol. 72, 70–78. doi: 10.1016/j.jinsphys.2014.12.002
Zhou, X., Wheeler, M. M., Oi, F. M., and Scharf, M. E. (2008). RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem. Mol. Biol. 38, 805–815. doi: 10.1016/j.ibmb.2008.05.005
Keywords: RNAi, specificity, Agrilus planipennis, dsRNA, environmental risk assessment
Citation: Pampolini F and Rieske LK (2020) Emerald Ash Borer Specific Gene Silencing Has No Effect on Non-target Organisms. Front. Agron. 2:608827. doi: 10.3389/fagro.2020.608827
Received: 01 October 2020; Accepted: 23 November 2020;
Published: 15 December 2020.
Edited by:
Jeremy Dean Allison, Canadian Forest Service, CanadaReviewed by:
Christian Stauffer, University of Natural Resources and Life Sciences Vienna, AustriaCopyright © 2020 Pampolini and Rieske. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynne K. Rieske, THJpZXNrZUB1a3kuZWR1; orcid.org/0000-0002-3049-4465
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.