- 1Department of Agricultural and Food Science, University of Bologna, Bologna, Italy
- 2INRA, Laboratoire des Interactions Plantes-Microorganismes (LIPM), UMR441, Castanet-Tolosan, France
- 3CNRS, Laboratoire des Interactions Plantes-Microorganismes (LIPM), UMR2594, Castanet-Tolosan, France
From bacterial quorum sensing to the signals of bees, communication is the basis of biotic interactions. Frequently, more than two organisms can take part in the speeches, resulting in a complex network of cross-talks. Recent advances in plant-microbe interactions research have shown that communication, both inter-kingdom and intra-kingdom, is shaped by a broad spectrum of factors. In this context, the rhizosphere (i.e., the soil close to the root surface) provides a specific microhabitat where complex interactions occur. The complex environment that makes up the rhizosphere can select for certain microbial populations, which are adapted to this unique niche. Among them, rhizobia have emerged as an important component of the rhizospheric microbiome. The aim of this review is to explore the components of such a rhizospheric Talk Show in the frame of the rhizobium-legume interactions. This symbiosis is a complex process that involves several signals that can be shaped by plant rhizospheric exudates and microbiome composition. The relationship established by rhizobia with other rhizospheric organisms, together with the influence of the environmental factors, results in their beneficial role on host plant health. Here, we resume research accounting strategies, molecules, and organisms that influence the place of rhizobia in the rhizosphere. The focus is on the most recent approaches for the study and subsequent exploitation of the diversity of the organisms. Indeed, the study of plant-microbes communication and evolution is fundamental to develop highly efficient inoculants able to reduce the use of fertilizers in agriculture.
The Talk Show in the Rhizosphere
Prokaryotes and eukaryotes have interacted for millions of years, evolving and refining over time their communication systems. As proposed by Hauser in 1996 (Hauser, 1996), biological signals and the exchange of information are part of the definition of communication, while the signals themselves are considered as “every structure able to shape the behavior of the organisms” (Smith and Harper, 2003; Schott-Phillips, 2008). Consequently, the signals can evolve and persist thanks to the interaction between signals producers and receivers. Then, cooperation and fitness improvement are the basis of biological communication (Zahavi, 2008).
In a particular environment, individuals can communicate and interact with multiple partners, and the nature of interaction can determine variable costs and benefits to the partner, as a biological market (Werner et al., 2015). One of the most fascinating environments where complex biological interactions occur is the rhizosphere. Indeed, a large number of signals can be exchanged involving the plant itself, insects, fungi and microbes. This all take place in a high-density environmental niche. Usually, communication is the result of chemical responses of cells to signatory molecules coming from other cells. These signals affect both the metabolism and transcription of genes activating several regulatory mechanisms.
Frequently, in the rhizosphere more than two organisms (and species, across the tree of life) can take part in the communication, resulting in a complex network of interactions and cross-talks which can influence the fitness of all participating partners. Thus, this environment can be considered a hot spot for numerous inter-kingdom signals exchange, which involves plant-associated microbial communities (rhizobiome). The microbial community's composition is mainly shaped and recruited by hundreds of metabolites released in the soil by plant roots, which normally facilitate the interactions with the biotic and abiotic environment. Often the plant can modulate their diversity based on the benefits in terms of growth and health (Plant Growth Promoting, PGP) (Hartmann et al., 2009). Nevertheless, a large number of nutrients issued by the plant can be of interest for the pathogenic organisms, which can take advantage of plant products for their survival in the rhizosphere (Rasmann and Turlings, 2016).
It stands to reason that the plants play a fundamental role in the rhizosphere scene (Bending, 2017) (Figure 1). Indeed, because of the chemical signals conveyed by nutrient-rich exudates released by the plant roots, a large variety of microbes can first colonize the rhizosphere and then gradually penetrate the root and the overall plant tissue (endophytes) (Hardoim et al., 2008). Otherwise, they can colonize the host plant establishing a lasting and beneficial symbiotic relationship (Chi et al., 2005). To date, numerous investigations on root exudates composition have been performed (Hartmann et al., 2009; Bulgarelli et al., 2013; Venturi and Keel, 2016).
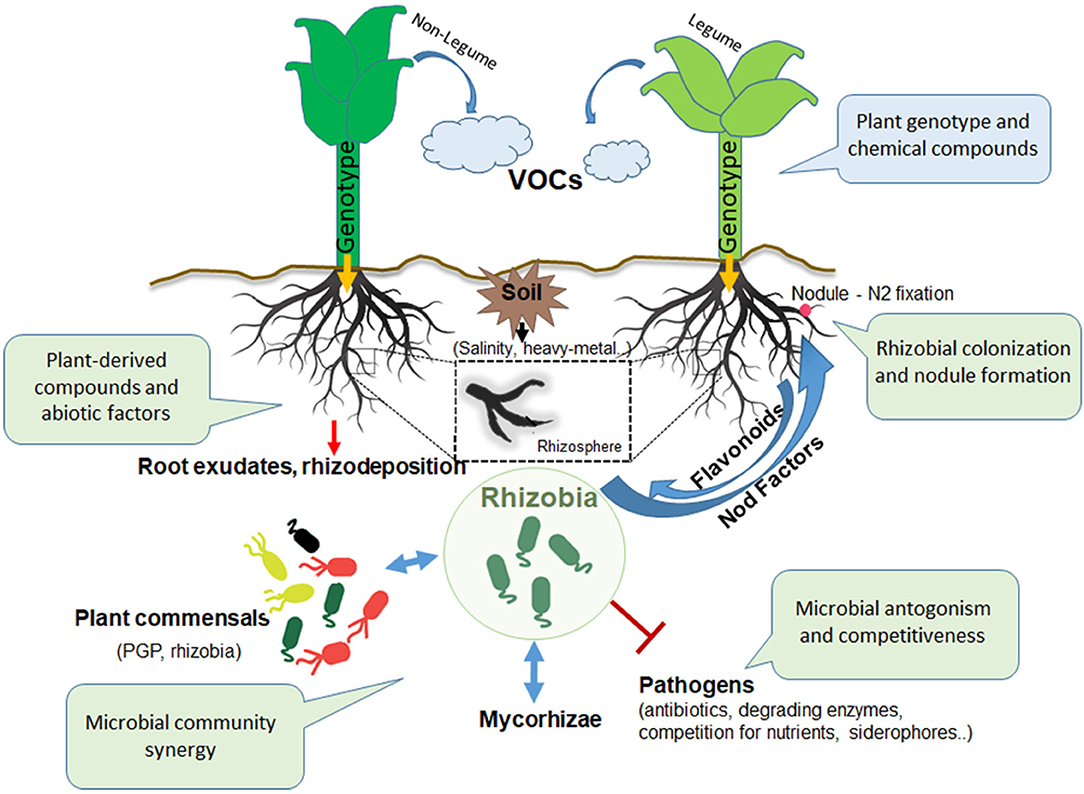
Figure 1. Actors and interactions in the rhizosphere. Inter-kingdom and intra-kingdom communication involving plants and microbes in the rhizosphere: the consistent role of rhizobia. VOCs, volatile organic compounds; PGP, plant growth promoting; AMF, arbuscular mycorrhizal fungi.
The most known plant-microbe dialogue on the rhizosphere scene, which determine direct and indirect advantages to the partners, was properly addressed as early as 1904 when Hiltner described the symbiotic interaction among legumes and rhizobia (Hartmann et al., 2008). This symbiosis is a highly specific process in which the genetic and chemical communication signals are strictly plant-bacterium specific. In this mutualistic interaction, rhizobia positively influence the host's growth thanks to the nitrogen fixation process and at the same time can benefit from the nutrients provided by the plant.
This symbiosis has been extensively studied over the last few decades, and many studies on the communication and the signaling between the two partners at different steps of the symbiosis (from root infection to nodules development) have been elucidated (Oldroyd et al., 2011; Oldroyd, 2013). However, the knowledge about the earlier steps of rhizosphere colonization, namely the opening line at the root surface, remains poorly characterized. Nonetheless, increasing data have shown the importance of intraspecies and multispecies communications among rhizospheric biotic components for the improvement of rhizobia–legumes interaction. In addition, it has been shown that rhizobia are part of the rhizosphere of a wide variety of non-legume plants where they can play a role as PGP components, recovering a central role in plant core microbiome (Yeoh et al., 2016).
This review provides an outline of the most studied intra- and inter-kingdom communication strategies in the rhizobium-legume cross talk, defining the most relevant mechanisms of rhizospheric communication. We conceived the exchange of signals between plants and microorganisms as an intricate Talk Show discussion, in which a large variety of dialogues can take place, regulated and coordinated by a defined line-up, but at the same time, free to influence each other.
The Place of Rhizobia: Setting Up the Scene
Rhizosphere complexity makes it difficult to identify and analyze the microbiome components and in particular, the laws governing the dynamics and the maintenance of plant associated microbial communities. Among factors able to influence those microbial communities, nutrients availability and soil composition have a strong influence (Naylor and Coleman-Derr, 2018). At this level, the root soil interface is rich in exudates that can differently alter gas and soil composition, thus selecting for certain microbial populations adapted to this unique niche. Root exudates contain a large variety of secondary metabolites, such as organic acids, amino acids and sugars, which are mainly used to attract microbes (PGP or pathogenic microorganisms) through chemotaxis process, and phenolic compounds such as flavonoids, fundamental for the signaling mechanism that allowst the symbiotic interaction with rhizobia and mycorrhizal fungi to start. Interestingly, several phytormones and antimicrobial compounds (phenolics and terpenoids) are secreted by roots and used by plant as defense weapons against soil-born pathogens, to select among beneficial microbes (Baetz and Martinoia, 2014). Additionally, roots are able to produce volatile organic compounds (VOCs), which fullfill the role of rhizosphere microbial growth regulator (as antimicrobial compounds) or as significant carbon source (Peñuelas et al., 2014). Therefore, the rhizodeposition makes microbial composition of the rhizophere significantly different from that found in bulk soil (de Oliveira et al., 2017).
The plant acts as a mediator in the rhizospheric stage, consistently contributing to the balance of soil microbiome composition (Hartmann et al., 2009). On the other hand, the soil microbial community actively participates in the composition of the rhizospheric environment, taking part in degradation and production of organic compounds, for its own benefit (Paterson et al., 2007).
Among bacteria living in the rhizosphere and in association with plants, rhizobia can impact on surrounding microbial population by affecting legumes rhizosphere's composition and structure of the soil microbiome (Nwoke et al., 2008; Fan et al., 2018). The role of rhizobia as the main actor is mediated by a plethora of chemical and molecular signals that regulate bacterial invasion and intracellular colonization, modulate host plant defense response and nutrient exchange (Gibson et al., 2008).
Furthermore, water deficiency, salinity, heavy metals, acidity, as well as low nutrients levels are all abiotic factor that frequently can interfere with the role of rhizobia in the rhizosphere scene (Fagorzi et al., 2018; Bellabarba et al., 2019).
In addition, recent data have revealed that under identical soil conditions, the plant genotype, through its phenotypic features, can filter and modulate the microbial community structure and function, as well as the diversity of root associated bacteria (el Zahar Haichar et al., 2008; Berg and Smalla, 2009) carrying out a partner choice in rhizobia mutualism (Simms and Taylor, 2002).
Increasing field studies including large-scale rhizosphere samplings are now emerging with the aim to better understand the rhizospheric microbiome rules, focusing on the structure, the preservation and on the importance of communication occurring among the plants. In every case, the abiotic environmental factors (positive or negative), together with the communication among plants and rhizobial/non-rhizobial species are able to influence the rhizosphere Talk Show, giving order to the existing messy scenario.
Synergic Effects of Intra-Kingdom Bacterial Interaction
The communication of microorganisms in the rhizosphere is mediated by a large diversity of microbial metabolites and physical signals, as happens in a noisy Talk Show, in which every dialogue can influence the success of the performance. The soil bacteria attracted by root exudates (Figure 1), root cell lysates and mucilages, profusely colonize the rhizosphere, thus starting the rhizosphere dialogue. Quorum sensing (QS), the bacterial population-dependent regulatory mechanism, is involved in bacterial cell aggregation and in the first phases of rhizosphere colonization through biofilm formation (Ng and Bassler, 2009). Indeed, rhizobia typically produce and release into the surrounding environment diffusible N-acyl homoserine lactone (AHL) molecules, which are used as signals for the control of plant- microbe interaction. The recognition of AHL molecules by bacterial cells starts and controls several processes as the biofilm formation, Extracellular Polymeric Substances (EPS) production, bacterial motility and expression of bacterial genes relevant for symbiosis and nitrogen fixation (Loh et al., 2002; Sanchez-Contreras et al., 2007; Yang et al., 2009).
The rhizobial QS is based on the LuxR – LuxI type regulatory system. It is dependent on the threshold level of AHL which allows to induce expression of specific target genes (Veliz-Vallejos et al., 2020).
Actually, at least three LuxRI-type QS regulatory systems have been identified (Zheng et al., 2015b), and their role in interspecies communication also in co-inoculation experiments has been well demonstrated (Miao et al., 2018). Since several decades, extensive investigations of the role of the QS in rhizobial biofilms all along the root surface and inside the plants (i.e., endophytes) have been performed. Thus, strains with an effective roots colonizing ability through biofilm formation (Velmourougane et al., 2017) and flagellar-dependent chemotaxis ability (Zheng et al., 2015a) are always more advantaged than those strains without biofilm formation capabilities. Furthermore, a large variety of strategies are important for rhizobial roots colonization, such as the secretion of antibiotics and/or cell-wall degrading enzymes, which have been reported to be important for the protection against root phytopathogens (Siddiqui et al., 2000; Chandra et al., 2007) (Figure 1).
A wide spectrum of other physiological traits plays an important role in the rhizosphere by contributing to the complex phenomenon of nodulation competitiveness (Triplett and Sadowsky, 1992). These traits include swarming motility, type III secretion system, plasmid transfer, cell division, metabolism and transport (Calatrava-Morales et al., 2018). It is of note that their occurrence varies among species.
Microbial communities in the rhizosphere can compete for nutrients and for the plant roots colonization. Several rhizobia are able to interfere with the infecting capability of other stains through the production of bacteriocins, antimicrobial compounds that act on relative close phylogenetic species (Oresnik et al., 1999; Venter et al., 2001). Sinorhizobium meliloti, one of the most studied symbiotic species, is able to produce high-affinity siderophores which deprive antagonists of the available iron, thus limiting their plant growth (Arora et al., 2001).
Contrarily, in some cases, soil microbial communities can improve the infection capabilities of particular rhizobia and their communication with the host plant (Mehboob et al., 2013). The recent work by Miao et al. (2018) highlighted that strains of Rhizobium fabae are able to improve Rhizobium etli nodulation capabilities through a particular intra-species QS mechanism.
In this context, rhizosphere actors can participate in setting up the Talk Show by improving their performance. Synergic effects of specific co-inocula formulations should include a fair combination of PGP bacteria and rhizobia (Remans et al., 2008). Indeed, a range of PGP microbes can be used with rhizobia for the improvement of legumes growth and rhizobial mutalistic efficiency. The free - living diazotrophs Azospirillum, Azotobacter, Bacillus, Pseudomonas, Serratia, and Enterobacter are some of the genera that were successfully used with rhizobia as co-inoculants, potentiating growth and yield of several leguminous crops through different strategies, such as the production of phytohormones or biocontrol agents against rhizosphere phytopathogens (Tchebotar et al., 1998; Tilak et al., 2006; Remans et al., 2008). Presence of Azospirillum species can increase the size of the rhizobial infection site, providing space for infection and facilitating the nodulation ability of rhizobia (Tchebotar et al., 1998). Although there are many combinations of bacteria co-inoculation that have been explored for the improvement of rhizobia-legumes symbiosis, there is still the need for advanced comprehensive research in their communication systems. Indeed, a selection of the most appropriate and performing panels of PGP strains, might be used in the near future as inoculants replacing fertilizers by a more environmentally friendly agricultural practice.
The Most Acclaimed Rhizospheric Inter-Kingdom Communications
A large part of the rhizobia-host communication strategies in the rhizosphere influences the highly regulated and ongoing rhizobial interactions in the root tissue. In compatible legume cultivars, after the establishment in the rhizosphere, rhizobia detect plant-derived flavonoids inducing nod genes, which are involved in Nod factors (lipochitooligosaccharides) production (Poole et al., 2018).
Then, rhizobia gradually adhere to roots surfaces, penetrate the plant root tissues and address their lifestyle inside the newly formed root organ, the nodule. Here, rhizobia differentiate into bacteroids, the N2 fixing form of rhizobia, thus reducing atmospheric nitrogen to ammonia benefiting, in return, of protection and availability of nutrients from the plant (Kereszt et al., 2011). In this scenario, the accessibility to the symbiotic niche is naturally regulated and limited by the host plant to respond to nitrogen needs and by competition among rhizobial strains present in the neighboring soil (Triplett and Sadowsky, 1992).
The bi-directional inter-kingdom interaction between rhizobia and the compatible legume host involves a large panel of molecular signals as well as the exchange of metabolic resources, largely explored by scientist for decades (Oldroyd, 2013; Udvardi and Poole, 2013; Poole et al., 2018). The inter-kingdom molecular communication of these social interactions includes the volatile organic compounds (VOCs) and flavonoids issued by the plant and the bacterial Nod factors responsible for the nodulation signaling pathway. Bacteria cheaters among rhizospheric populations can profit of the plant permission state to escape plant sanctions and infect the plant tissue (Kiers et al., 2003; Sachs et al., 2010; Checcucci et al., 2016; diCenzo et al., 2018).
Representing more than 70% of all lands plants, legumes are able to interact with arbuscular mycorrhizal fungi (AMF) through the recognition of the mycorrhizal oligosaccharides' factors (Myc factors). Plant-fungi association allows the fungal elongation into the root cortex, where AMF can benefit the host mediating the nutrient uptake. It is largely demonstrated that in legumes, the Talk Show dialogue between fungi and rhizobia can take place at the root level (Barea and Pozo, 2005; Meghvansi et al., 2008; Kaschuk et al., 2010).
To our knowledge, Crush was the first researcher who observed that the presence of mycorrhizas stimulated nodulation and consequently plant growth (Crush, 1974). Then, many others reported additional effects produced by the tripartite interaction of Arbuscular Mycorrhizal Fungi – Rhizobia - Plant (ARP): it enhances symbiotic nitrogen fixation and effectiveness in drought soils (Hao et al., 2019; Laouane et al., 2019), increases number and dry weight of nodules (Antunes and Goss, 2005) and root soluble sugar contents (Hao et al., 2019; Tang et al., 2019). For more literature on the arguments, we refers to Anandakumar et al. (2019), Sakamoto et al. (2019), Sui et al. (2019) and Zhang et al. (2020).
Besides positive effects, fungi can act as antagonistic plant pathogens, which rhizobia can counteract as biocontrol agents for multiple plant species (Deshwal et al., 2013). Furthermore, it was demonstrated that the multiple inoculations of fungi, PGP bacteria can improve symbiotic behavior and plant nutrients availability, as was shown in semi-arid and alkaline soils (Requena et al., 1997; Abd-Alla et al., 2014).
It seems that communication which rhizobia can establish with the other components of the stage, can be a potential reserve for the improvement of rhizosphere interactions, and primarily those that contribute to host plant growth (Artursson et al., 2006). Recently, transcriptomic analysis in the presence of fungal exudates has shown their positive role in the transcription of rhizobial genes associated with the chemosensory (Zhang et al., 2020). In the future, further research should be addressed to the development of new technologies and applications to enable the optimization and the subsequent exploitation of ARP tripartite interactions. In particular, the monitoring of expression profiles of genes associated with communication mechanisms, the development of microcosm systems which simulate the natural condition, the analysis of the metabolic potential of microbial (and fungal) consortia in association with host plants, might be good starting points to better understand the multi-organism's rhizosphere communication. The most recent metagenomic approaches for the study of rhizobial microbial communities, as well as the determination of the role of each actor in the rhizosphere scene, will be essential for the understanding and the subsequent exploitation of organism diversity for sustainable agriculture (Faure et al., 2009; Arora et al., 2020).
Concluding Remarks
The current understanding of the rhizosphere is highlighting the complexity of the communication strategies taking place in this model environmental scenario. Increasing evidence is pointing out how in this scenario rhizobia occupy an important place that extends to non-legume plants. This niche is a consequence of the influence of several factors (biotic and abiotic), at different levels (intra- and inter-kingdom) and different degrees (competition and cooperation).
Every interaction and exchange of signals taking place in one of the most known complex environments cannot be conceived as uni or bidirectional reactions, but it can be imagined as a complex network of dialogues, a model of noisy discussion while remaining inexplicably tidy and regulated by the partners themselves. Therefore, the recent studies, focused on microbial community networks models (Melke et al., 2010; Barto et al., 2012; Succurro and Ebenhöh, 2018; Mai et al., 2019), have to be adapted to the complexity of the rhizosphere. The dissection of such rhizospheric communication is essential for the improvement of the benefic aspects of such communication (Mueller and Sachs, 2015; Checcucci et al., 2017; Mueller et al., 2019) with a view to agricultural applications.
The rhizosphere scene can be considered as a highly suitable model for the application of a system biology study approach, including the large number of plants and microorganisms sequenced genomes, the studies on partners metabolic functionalities (Korenblum et al., 2020) and on the transcriptomic changes related to different partners interaction (Pathan et al., 2020). It is expected that future studies will continue to explore the selective forces that shape rhizosphere microbiome further elucidate the potential of the communication among the different rhizospheric partners.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This review was performed at the LIPM belonging to the Laboratoire d'Excellence (LABEX) entitled TULIP (ANR-10-LABX-41) and in the Laboratory of Microbiology, Department of Agricultural and Food Science, University of Bologna.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Prof. Lenie Dijkshoorn for the critical review and the English grammar editing of the manuscript.
References
Abd-Alla, M. H., El-Enany, A.-W. E., Nafady, N. A., Khalaf, D. M., and Morsy, F. M. (2014). Synergistic interaction of Rhizobium leguminosarum bv. viciae and arbuscular mycorrhizal fungi as a plant growth promoting biofertilizers for faba bean (Vicia faba L.) in alkaline soil. Microbiol. Res. 169, 49–58. doi: 10.1016/j.micres.2013.07.007
Anandakumar, S., Kalaiselvi, T., and Sivakumar, U. (2019). Arbuscular mycorrhizal fungi (Glomus intraradices) and diazotrophic bacterium (Rhizobium BMBS) primed defense in blackgram against herbivorous insect (Spodoptera litura) infestation. Microbiol. Res. 231:126355. doi: 10.1016/j.micres.2019.126355
Antunes, P. M., and Goss, M. J. (2005). Communication in the tripartite symbiosis formed by arbuscular mycorrhizal fungi, rhizobia, and legume plants: a review. Agronomy 48:199. doi: 10.2134/agronmonogr48.c11
Arora, N. K., Fatima, T., Mishra, I., and Verma, S. (2020). “Microbe-based inoculants: role in next green revolution,” in: Environmental Concerns and Sustainable Development, eds V. Shukla and N. Kumar (Singapore: Springer), 191–246.
Arora, N. K., Kang, S. C., and Maheshwari, D. K. (2001). Isolation of siderophore-producing strains of Rhizobium meliloti and their biocontrol potential against Macrophomina phaseolina that causes charcoal rot of groundnut. Curr. Sci. 81, 673–677.
Artursson, V., Finlay, R. D., and Jansson, J. K. (2006). Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 8, 1–10. doi: 10.1111/j.1462-2920.2005.00942.x
Baetz, U., and Martinoia, E. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 19, 90–98. doi: 10.1016/j.tplants.2013.11.006
Barea, J. M., and Pozo, M. J. (2005). Microbial co-operation in the rhizosphere. J. Exp. Bot. 56:e1778. doi: 10.1093/jxb/eri197
Barto, E. K., Weidenhamer, J. D., Cipollini, D., and Rillig, M. C. (2012). Fungal superhighways: do common mycorrhizal networks enhance below ground communication? Trends Plant Sci. 17, 633–637. doi: 10.1016/j.tplants.2012.06.007
Bellabarba, A., Fagorzi, C., diCenzo, G. C., Pini, F., Viti, C., and Checcucci, A. (2019). Deciphering the symbiotic plant microbiome: translating the most recent discoveries on rhizobia for the improvement of agricultural practices in metal-contaminated and high saline lands. Agronomy 9:529. doi: 10.3390/agronomy9090529
Bending, G. D. (2017). “The rhizopshere and its microorganisms,” in Encyclopedia of Applied Plant Sciences, 2nd edn, eds B. Thomas, B. G. Murray, and D. J. Murphy (Academic Press), 347–351. doi: 10.1016/B978-0-12-394807-6.00165-9
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bulgarelli, D., Schlaeppi, K., Spaepen, S., Van Themaat, E. V. L., and Schulze-Lefert, P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. doi: 10.1146/annurev-arplant-050312-120106
Calatrava-Morales, N., McIntosh, M., and Soto, M. (2018). Regulation mediated by N-Acyl homoserine lactone quorum sensing signals in the rhizobium-legume symbiosis. Genes 9:263. doi: 10.3390/genes9050263
Chandra, S., Choure, K., Dubey, R. C., and Maheshwari, D. K. (2007). Rhizosphere competent Mesorhizobiumloti MP6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of Indian mustard (Brassica campestris). Braz. J. Microbiol. 38, 124–130. doi: 10.1590/S1517-83822007000100026
Checcucci, A., Azzarello, E., Bazzicalupo, M., Galardini, M., Lagomarsino, A., Mancuso, S., et al. (2016). Mixed nodule infection in Sinorhizobium meliloti–Medicago sativa symbiosis suggest the presence of cheating behavior. Front. Plant Sci. 7:835. doi: 10.3389/fpls.2016.00835
Checcucci, A., DiCenzo, G. C., Bazzicalupo, M., and Mengoni, A. (2017). Trade, diplomacy, and warfare: the quest for elite rhizobia inoculant strains. Front. Microbiol. 8:2207. doi: 10.3389/fmicb.2017.02207
Chi, F., Shen, S.-H., Cheng, H.-P., Jing, Y.-X., Yanni, Y. G., and Dazzo, F. B. (2005). Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Env. Microbiol. 71, 7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005
Crush, J. R. (1974). Plant growth responses to vesicular-arbuscular mycorrhiza VII. Growth and modulation of some herbage legumes. New Phytol. 73, 743–749. doi: 10.1111/j.1469-8137.1974.tb01302.x
de Oliveira, M. C. F., Mafezoli, J., and Gunatilaka, A. A. L. (2017). Natural products of the rhizosphere and its microorganisms. Chem. Biol. Nat. Prod. 29, 285–331. doi: 10.1201/9781315117089-10
Deshwal, V. K., Singh, S. B., Kumar, P., and Chubey, A. (2013). Rhizobia unique plant growth promoting rhizobacteria: a review. Int. J. Life Sci. 2, 74–86.
diCenzo, G. C., Debiec, K., Krzysztoforski, J., Uhrynowski, W., Mengoni, A., Fagorzi, C., et al. (2018). Genomic and biotechnological characterization of the heavy-metal resistant, arsenic-oxidizing bacterium Ensifer sp. M14. Genes 9:379. doi: 10.3390/genes9080379
el Zahar Haichar, F., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., Balesdent, J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2:1221. doi: 10.1038/ismej.2008.80
Fagorzi, C., Checcucci, A., diCenzo, G., Debiec-Andrzejewska, K., Dziewit, L., Pini, F., et al. (2018). Harnessing rhizobia to improve heavy-metal phytoremediation by legumes. Genes 9:542. doi: 10.3390/genes9110542
Fan, M., Xiao, X., Guo, Y., Zhang, J., Wang, E., Chen, W., et al. (2018). Enhanced phytoremdiation of Robinia pseudoacacia in heavy metal-contaminated soils with rhizobia and the associated bacterial community structure and function. Chemosphere 197, 729–740. doi: 10.1016/j.chemosphere.2018.01.102
Faure, D., Vereecke, D., and Leveau, J. H. J. (2009). Molecular communication in the rhizosphere. Plant Soil 321, 279–303. doi: 10.1007/s11104-008-9839-2
Gibson, K. E., Kobayashi, H., and Walker, G. C. (2008). Molecular determinants of a symbiotic chronic infection. Annu. Rev. Genet. 42, 413–441. doi: 10.1146/annurev.genet.42.110807.091427
Hao, Z., Xie, W., Jiang, X., Wu, Z., Zhang, X., and Chen, B. (2019). Arbuscular mycorrhizal fungus improves Rhizobium–Glycyrrhiza seedling symbiosis under drought stress. Agronomy 9:572. doi: 10.3390/agronomy9100572
Hardoim, P. R., van Overbeek, L. S., and van Elsas, J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–471. doi: 10.1016/j.tim.2008.07.008
Hartmann, A., Rothballer, M., and Schmid, M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hartmann, A., Schmid, M., van Tuinen, D., and Berg, G. (2009). Plant-driven selection of microbes. Plant Soil. 321, 235–257. doi: 10.1007/s11104-008-9814-y
Kaschuk, G., Leffelaar, P. A., Giller, K. E., Alberton, O., Hungria, M., and Kuyper, T. W. (2010). Responses of legumes to rhizobia and arbuscular mycorrhizal fungi: a meta-analysis of potential photosynthate limitation of symbioses. Soil Biol. Biochem. 42, 125–127. doi: 10.1016/j.soilbio.2009.10.017
Kereszt, A., Mergaert, P., and Kondorosi, E. (2011). Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims? Mol. Plant. Microbe Interact. 24, 1300–1309. doi: 10.1094/MPMI-06-11-0152
Kiers, E. T., Rousseau, R. A., West, S. A., and Denison, R. F. (2003). Host sanctions and the legume–rhizobium mutualism. Nature 425:78. doi: 10.1038/nature01931
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Laouane, R., Ben, M., eddich, A., Bechtaoui, N., Oufdou, K., and Wahbi, S. (2019). Effects of arbuscular mycorrhizal fungi and rhizobia symbiosis on the tolerance of Medicago sativa to salt stress. Gesunde Pflanz. 71, 135–146. doi: 10.1007/s10343-019-00461-x
Loh, J., Pierson, E. A., Pierson, I. I. I. L. S, Stacey, G., and Chatterjee, A. (2002). Quorum sensing in plant-associated bacteria. Curr. Opin. Plant Biol. 5, 285–290. doi: 10.1016/S1369-5266(02)00274-1
Mai, T. H., Schnepf, A., Vereecken, H., and Vanderborght, J. (2019). Continuum multiscale model of root water and nutrient uptake from soil with explicit consideration of the 3D root architecture and the rhizosphere gradients. Plant Soil 439, 273–292. doi: 10.1007/s11104-018-3890-4
Meghvansi, M. K., Prasad, K., Harwani, D., and Mahna, S. K. (2008). Response of soybean cultivars toward inoculation with three arbuscular mycorrhizal fungi and Bradyrhizobium japonicum in the alluvial soil. Eur. J. Soil Biol. 44, 316–323. doi: 10.1016/j.ejsobi.2008.03.003
Mehboob, I., Naveed, M., Zahir, Z. A., and Sessitsch, A. (2013). “Potential of rhizosphere bacteria for improving Rhizobium-legume symbiosis,” in: Plant Microbe Symbiosis: Fundamentals and Advances, ed N. K. Arora (New Delhi: Springer), 305–349.
Melke, P., Sahlin, P., Levchenko, A., and Jönsson, H. (2010). A cell-based model for quorum sensing in heterogeneous bacterial colonies. PLoS Comput. Biol. 6:e1000819. doi: 10.1371/journal.pcbi.1000819
Miao, J., Zhang, N., Liu, H., Wang, H., Zhong, Z., and Zhu, J. (2018). Soil commensal rhizobia promote Rhizobium etli nodulation efficiency through CinR-mediated quorum sensing. Arch. Microbiol. 200, 685–694. doi: 10.1007/s00203-018-1478-2
Mueller, C. W., Carminati, A., Kaiser, C., Subke, J.-A., and Gutjahr, C. (2019). Rhizosphere functioning and structural development as complex interplay between plants, microorganisms and soil minerals. Front. Environ. Sci: 7, 130. doi: 10.3389/fenvs.2019.00130
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Naylor, D., and Coleman-Derr, D. (2018). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8:2223. doi: 10.3389/fpls.2017.02223
Ng, W.-L., and Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Nwoke, O. C., Diels, J., Abaidoo, R., Nziguheba, G., and Merckx, R. (2008). Organic acids in the rhizosphere and root characteristics of soybean (Glycine max) and cowpea (Vigna unguiculata) in relation to phosphorus uptake in poor savanna soils. Afr. J. Biotechnol. 7, 3620–3627. doi: 10.5897/AJB08.438
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11:252. doi: 10.1038/nrmicro2990
Oldroyd, G. E. D., Murray, J. D., Poole, P. S., and Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Oresnik, I. J., Twelker, S., and Hynes, M. F. (1999). Cloning and characterization of a Rhizobium leguminosarum gene encoding a bacteriocin with similarities to RTX toxins. Appl. Environ. Microbiol. 65, 2833–2840. doi: 10.1128/AEM.65.7.2833-2840.1999
Paterson, E., Gebbing, T., Abel, C., Sim, A., and Telfer, G. (2007). Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610. doi: 10.1111/j.1469-8137.2006.01931.x
Pathan, S. I., Ceccherini, M. T., Sunseri, F., and Lupini, A. (2020). “Rhizosphere as hotspot for plant-soil-microbe interaction,” in: Carbon and Nitrogen Cycling in Soil, eds R. Datta, R. S. Meena, S. I. Pathan, and M. T. Ceccherini (Singapore: Springer), 17–43.
Peñuelas, J., Asensio, D., Tholl, D., Wenke, K., Rosenkranz, M., Piechulla, B., et al. (2014). Biogenic volatile emissions from the soil. Plant Cell Environ. 37, 1866–1891. doi: 10.1111/pce.12340
Poole, P., Ramachandran, V., and Terpolilli, J. (2018). Rhizobia: from saprophytes to endosymbionts. Nat. Rev. Microbiol. 16:291. doi: 10.1038/nrmicro.2017.171
Rasmann, S., and Turlings, T. C. J. (2016). Root signals that mediate mutualistic interactions in the rhizosphere. Curr. Opin. Plant Biol. 32, 62–68. doi: 10.1016/j.pbi.2016.06.017
Remans, R., Ramaekers, L., Schelkens, S., Hernandez, G., Garcia, A., Reyes, J. L., et al. (2008). Effect of Rhizobium–Azospirillum coinoculation on nitrogen fixation and yield of two contrasting Phaseolus vulgaris L. genotypes cultivated across different environments in Cuba. Plant Soil 312, 25–37. doi: 10.1007/s11104-008-9606-4
Requena, N., Jimenez, I., Toro, M., and Barea, J. M. (1997). Interactions between plant-growth-promoting rhizobacteria (PGPR), arbuscular mycorrhizal fungi and Rhizobium spp. in the rhizosphere of Anthyllis cytisoides, a model legume for revegetation in mediterranean semi-arid ecosystems. New Phytol. 136, 667–677. doi: 10.1046/j.1469-8137.1997.00786.x
Sachs, J. L., Ehinger, M. O., and Simms, E. L. (2010). Origins of cheating and loss of symbiosis in wild Bradyrhizobium. J. Evol. Biol. 23, 1075–1089. doi: 10.1111/j.1420-9101.2010.01980.x
Sakamoto, K., Ogiwara, N., Kaji, T., Sugimoto, Y., Ueno, M., Sonoda, M., et al. (2019). Transcriptome analysis of soybean (Glycine max) root genes differentially expressed in rhizobial, arbuscular mycorrhizal, and dual symbiosis. J. Plant Res. 132, 541–568. doi: 10.1007/s10265-019-01117-7
Sanchez-Contreras, M., Bauer, W. D., Gao, M., Robinson, J. B., and Allan Downie, J. (2007). Quorum-sensing regulation in rhizobia and its role in symbiotic interactions with legumes. Philos. Trans. R. Soc. B Biol. Sci. 362, 1149–1163.
Schott-Phillips, T. C. (2008). Defining biological communication. J. Evol. Biol. 21, 387–395. doi: 10.1111/j.1420-9101.2007.01497.x
Siddiqui, I. A., Ehteshamul-Haque, S., Zaki, M. J., and Abdul, G. (2000). Effect of urea on the efficacy of Bradyrhizobium Sp. and Trichoderma harzianum in the control of root infecting fungi in mungbean and sunflower. Sarhad J. Agric. 16, 403–406.
Simms, E. L., and Taylor, D. L. (2002). Partner choice in nitrogen-fixation mutualisms of legumes and rhizobia. Integr. Comp. Biol. 42, 369–380. doi: 10.1093/icb/42.2.369
Succurro, A., and Ebenhöh, O. (2018). Review and perspective on mathematical modeling of microbial ecosystems. Biochem. Soc. Trans. 46, 403–412. doi: 10.1042/BST20170265
Sui, X., Zhang, T., Tian, Y., Xue, R., and Li, A. (2019). A neglected alliance in battles against parasitic plants: arbuscular mycorrhizal and rhizobial symbioses alleviate damage to a legume host by root hemiparasitic Pedicularis species. New Phytol. 221, 470–481. doi: 10.1111/nph.15379
Tang, T.-T., Xie, M.-M., Chen, S.-M., Zhang, S.-M., and Wu, Q.-S. (2019). Effects of arbuscular mycorrhizal fungi and rhizobia on physiological activities in white clover (Trifolium repens). Biotechnology 18, 49–54. doi: 10.3923/biotech.2019.49.54
Tchebotar, V. K., Kang, U. G., Asis Jr, C. A., and Akao, S. (1998). The use of GUS-reporter gene to study the effect of Azospirillum-Rhizobium coinoculation on nodulation of white clover. Biol. Fertil. Soils 27, 349–352. doi: 10.1007/s003740050442
Tilak, K., Ranganayaki, N., and Manoharachari, C. (2006). Synergistic effects of plant-growth promoting rhizobacteria and Rhizobium on nodulation and nitrogen fixation by pigeonpea (Cajanus cajan). Eur. J. Soil Sci. 57, 67–71. doi: 10.1111/j.1365-2389.2006.00771.x
Triplett, E. W., and Sadowsky, M. J. (1992). Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46, 399–422. doi: 10.1146/annurev.mi.46.100192.002151
Udvardi, M., and Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Veliz-Vallejos, D. F., Kawasaki, A., and Mathesius, U. (2020). The presence of plant-associated bacteria alters responses to N-acyl homoserine lactone quorum sensing signals that modulate nodulation in Medicago truncatula. Plants 9:777. doi: 10.3390/plants9060777
Velmourougane, K., Prasanna, R., and Saxena, A. K. (2017). Agriculturally important microbial biofilms: present status and future prospects. J. Basic Microbiol. 57, 548–573. doi: 10.1002/jobm.201700046
Venter, A. P., Twelker, S., Oresnik, I. J., and Hynes, M. F. (2001). Analysis of the genetic region encoding a novel rhizobiocin from Rhizobium leguminosarum bv. viciae strain 306. Can. J. Microbiol. 47, 495–502. doi: 10.1139/w01-043
Venturi, V., and Keel, C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Werner, G. D. A., Cornwell, W. K., Cornelissen, J. H. C., and Kiers, E. T. (2015). Evolutionary signals of symbiotic persistence in the legume–rhizobia mutualism. Proc. Natl. Acad. Sci. 112, 10262–10269. doi: 10.1073/pnas.1424030112
Yang, M., Sun, K., Zhou, L., Yang, R., Zhong, Z., and Zhu, J. (2009). Functional analysis of three AHL autoinducer synthase genes in Mesorhizobium loti reveals the important role of quorum sensing in symbiotic nodulation. Can. J. Microbiol. 55, 210–214. doi: 10.1139/W08-128
Yeoh, Y. K., Paungfoo-Lonhienne, C., Dennis, P. G., Robinson, N., Ragan, M. A., Schmidt, S., et al. (2016). The core root microbiome of sugarcanes cultivated under varying nitrogen fertilizer application. Environ. Microbiol. 18, 1338–1351. doi: 10.1111/1462-2920.12925
Zahavi, A. (2008). “The handicap principle and signalling in collaborative systems,” in: Sociobiology of Communication, eds P. d'Ettorre and D. P. Hughes (Oxford; New York, NY: Oxford University Press), 1–10.
Zhang, W., Li, X. G., Sun, K., Tang, M. J., Xu, F. J., Zhang, M., et al. (2020). Mycelial network-mediated rhizobial dispersal enhances legume nodulation. ISME J. 14, 1015–1029. doi: 10.1038/s41396-020-0587-5
Zheng, H., Mao, Y., Teng, J., Zhu, Q., Ling, J., and Zhong, Z. (2015a). Flagellar-dependent motility in Mesorhizobium tianshanense is involved in the early stage of plant host interaction: study of an flgE mutant. Curr. Microbiol. 70, 219–227. doi: 10.1007/s00284-014-0701-x
Keywords: rhizosphere, rhizobia, communication, microbial communities, plant-microorganisms interaction
Citation: Checcucci A and Marchetti M (2020) The Rhizosphere Talk Show: The Rhizobia on Stage. Front. Agron. 2:591494. doi: 10.3389/fagro.2020.591494
Received: 04 August 2020; Accepted: 16 November 2020;
Published: 02 December 2020.
Edited by:
Pilar Irisarri, Universidad de la República, UruguayReviewed by:
Ivan John Oresnik, University of Manitoba, CanadaLuciano Kayser Vargas, State Secretariat of Agriculture, Livestock and Irrigation, Brazil
Copyright © 2020 Checcucci and Marchetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alice Checcucci, YWxpY2UuY2hlY2N1Y2NpMkB1bmliby5pdA==
 Alice Checcucci
Alice Checcucci Marta Marchetti
Marta Marchetti