- 1Guangdong Provincial Key Laboratory of Gastroenterology, Department of Gastroenterology, Institute of Gastroenterology of Guangdong Province, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 2Department of General Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 3Department of Cell Biology, School of Basic Medicine, Southern Medical University, Guangzhou, China
- 4Guangzhou Weisengene Biological Technology Co., Ltd, Guangzhou, China
Intestinal probiotics are a primary focus area of current medical research. Probiotics such as bifidobacteria and lactobacilli can positively impact obesity and other metabolic diseases by directly or indirectly affecting lipid metabolism. However, the precise mechanisms of these effects remain unclear. In our previous work, the novel strain Lactobacillus reuteri HI120 was isolated and identified. HI120 expresses high levels of linoleic isomerase, resulting in the production of large amounts of conjugated linoleic acid (CLA) when mixed with linoleic acid (LA). As HI120 can efficiently transform LA into CLA, the effect of HI120 on the lipid metabolism in C57BL/6 obese mice was studied and the underlying molecular mechanism was explored in vitro. The results revealed no significant change in the diet, body weight, and serum triglyceride levels in mice. However, serum cholesterol levels were significantly decreased. The underlying mechanism may involve a CLA-mediated reduction in the gene expression levels of NPC1L1, SREBP-2, and HMG-CR, resulting in reduced cholesterol synthesis and absorption. Thus, HI120 can be developed as a potential probiotic formulation. After oral administration, LA from certain food sources can be converted into CLA in the human intestine to contribute to the prevention and treatment of obesity and hyperlipidemia.
Introduction
Obesity can cause a series of conditions such as hyperlipidemia, fatty liver, atherosclerosis, hypertension, type 2 diabetes, and even malignant tumors, all of which are harmful to human health (1–4).
Gut microbes can affect obesity, insulin resistance, diabetes, and metabolic syndrome, among other conditions (5). Gut microbes play important roles in metabolic diseases and can directly or indirectly affect lipid metabolism. However, the exact mechanism of these interactions remains unclear (6). As early as 1963, probiotics were found to have a cholesterol-lowering effect in humans (7, 8). The cholesterol level in the plasma of a high-fat-induced atherosclerosis animal model was decreased by administration of a lactobacillus-rich diet (9). Two strains of Lactobacillus plantarum, ATG-K2 and ATG-K6, can significantly reduce body weight, liver fat accumulation, and related gene expression in non-alcoholic fatty liver disease (NAFLD) rat models generated by a high-fat/high-fructose diet (10). Thus, certain probiotics may be useful as biological agents for preventing and controlling lipid metabolism-related diseases.
Certain lactobacillus strains, such as Lactobacillus reuteri, L. plantarum, and Lactobacillus acidophilus, contain linoleic acid isomerase (LAI), which can isomerize linoleic acid (LA) into two isomers of conjugated linoleic acid (CLA) containing conjugated double bonds (11). Lactobacillus reuteri has been reported to exist in the intestines of almost all vertebrates and mammals. Because of its beneficial effects, it is considered as a probiotic. In our previous work, we isolated a L. reuteri strain expressing high levels of LAI from the feces of healthy children and named this strain as HI120. Mixing L. reuteri HI120 with LA in vitro resulted in the generation of a large amount of CLA, as revealed by mass spectrometry. In this study, we performed sequencing and mass spectrometry analysis of HI120. In 2008, the FDA issued a food license for the use of CLA as a food additive (12). CLA shows effects against cancer, obesity, and atherosclerosis (13) as well as potent functions in weight and fat reduction. Sprague-Dawley rats fed CLA displayed lower body fat, weight, triglyceride, and cholesterol levels (14). The body weight of ob/ob mice was reduced after feeding of c9, t11-CLA (15). Furthermore, t10, c12-CLA reduced triglycerides and cholesterol levels in the liver of obese rats (16). Daily supplementation of 0.7–1.4 g CLA in patients with hyperlipidemia significantly reduced the concentrations of serum low-density lipoprotein-cholesterol, very low-density lipoprotein-cholesterol, and triglycerides (17). When 3.0% CLA was added to the diet of mice, the concentration of total cholesterol in the liver decreased by 41% (18). Based on these results, we fed C57BL/6 obese mice with HI120 and sunflower seed oil containing 60% LA to simulate the everyday human diet and observe whether any fat- and weight-reducing effects occurred. Lactobacillus reuteri DSM20016 is the standard strain of L. reuteri, which is often used as a control with other bacteria (19). In this study, L. reuteri DSM20016 was used as the control to compare the weight loss and lipid-lowering effects of HI120. Atorvastatin is a commonly used lipid-lowering drug in the clinic (20, 21). We also used atorvastatin in the positive control group to understand the lipid-lowering effect of HI120. NPC1L1, SREBP-2, HMG-CR, CYP7A1, FXR, and LXR (22–27) are important genes involved in cholesterol synthesis, absorption, and excretion. We examined these genes in vitro and in vivo to explore the molecular mechanism of HI120 in lowering cholesterol.
Materials and Methods
Ethics Statement
Animal experiments were performed in strict accordance with the National Institutes of Health guidelines and were approved by the Institutional Laboratory Animal Care and Use Committee of the South Medical University at Guangzhou (Approval No. L2017104, Guangzhou, Guangdong province, China).
Bacterial Strains, Animals, Cell Lines, and Culture Conditions
Lactobacillus reuteri HI120 is preserved at the culture preservation center of Guangdong Institute of Microbiology (DEPOSIT No.: GDWSD0600119, China). Lactobacillus reuteri DSM20016 (Deutsche Sammlung von Mikroorganismem und Zellkulturen'scollection, Braunschweig, Germany) was used in this study. Both species were verified by PCR amplification.
Male C57BL/6 mice (6–8-week-old) were obtained from the Southern Medical University animal facility (China) and reared under specific pathogen-free conditions. All animals were housed under controlled conditions (temperature 20 ± 2°C, humidity 55 ± 5%, 12/12 h light/dark cycle; 08:00–20:00 h) with free access to tap water and diet, and maintained in groups in cages with cottonwood sawdust pads. All mice were fed either a normal diet [32% (w/w) protein, 5% fat, 2% fiber, 1.8% calcium, 1.2% phosphorus, and 58% nitrogen free extract] or a high-fat diet [10% (w/w) lard, 1% cholesterol, 0.2% sodium cholate, and 88.8% normal diet].
The human colorectal adenocarcinoma cell line Caco-2 and hepatoma cell line HepG2 were provided by Southern Medical University (Guangzhou, China) and routinely cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) and DMEM high-glucose culture medium (Gibco), respectively, supplemented with 10% heat-inactivated fetal bovine serum (PAN, Aidenbach, Germany), penicillin (100 U/mL), and streptomycin (100 ng/mL). Cell cultures were maintained in an incubator at 37°C under 95% (v/v) humidified air and 5% (v/v) CO2.
Sequencing Identification of L. reuteri HI120 and Detection of Its LA–CLA Conversion Rate
HI120 was cultured and lysozyme was added to lyse the bacteria for genomic DNA extraction. 16S rRNA universal primers were used, and genomic DNA was used as template for PCR. Using the NCBI database, the 16S rRNA gene sequence of HI120 was compared with that of various L. reuteri strains using BLAST software. The LAI amino acid sequence of HI120 was also compared to that of other strains.
To detect the transformation rate of LA to CLA by HI120, several experimental groups were established. LA group: 1 mg/mL LA and 1 mg/mL Tween 80 MRS medium incubated at 37°C on a shaking table at 120 rpm for 48 h. DSM20016 + LA group: DSM20016 inoculated at a 1:100 ratio and cultured on a shaking table at 120 rpm and 37°C for 24 h. Subsequently, DSM20016 was inoculated at 5% v/v into MRS medium containing 1 mg/mL LA and 1 mg/mL Tween 80, followed by culture at 120 rpm and 37°C on a shaking table for 48 h. HI120 + LA group: HI120 inoculated at a 1:100 ratio, cultured on a shaking table at 120 rpm and 37°C for 24 h, and subsequently inoculated at 5% v/v into MRS medium containing 1 mg/mL LA and 1 mg/mL Tween 80, followed by culture on a shaking table at 120 rpm and 37°C for 48 h. The procedure for each group was carried out at least three times in parallel. The bacterial solution mixture was centrifuged at 5,000 rpm for 5 min, and the supernatant was collected. A volume of 1 mL culture medium + 2 mL isopropanol + 1.5 mL n-hexane was added for extraction for 3 min followed by centrifugation of the sample at 6,000 rpm for 5 min. The upper layer of n-hexane was absorbed, dried with anhydrous sodium sulfate, and placed for stratification. For gas chromatography-mass spectrometry (Agilent, Santa Clara, CA, USA) detection, the following parameters were used: sample inlet 280°C, constant temperature at 50°C for 2 min, and then the temperature was increased at 15°C/min up to 250°C. A constant temperature was maintained for 10 min with a sample volume of 0.2 μL. The external standard method was used for qualitative and quantitative detection, with c9, t11-CLA standard, t10, c12-CLA standard, and LA standard as standard samples and n-hexane as solvent.
Preparation of Bacterial Powder and Detection of Its Concentration
Powdered bacterial protective agent (100 mL deionized water with 4.5 g dissolved gelatin + 0.9 g sodium chloride + 3 g fructose + 3 g starch) was added to the high-concentration HI120/DSM20016 bacterial solution, and the mixture was suspended and precipitated again. This mixture was dried using a powder-spraying machine (drying conditions: rotation speed 15 rpm, drying temperature 115°C, fan frequency 50 Hz). The dry powder was collected, packed separately, marked, and dried at 4°C. Next, 100 mg of HI120 and DSM20016 bacterial powder was used to prepare 10 mL of solution. A volume of 100 μL of solution was collected and serially diluted by 10-fold at 107, 108, 109, and 1010. This diluted solution (100 μL) was applied to the solid medium. The mixtures were then labeled and cultured for 48 h at 37°C. According to the different dilution times, the above procedures were repeated until the plate counts reached 100–300 single colonies. The number of colonies was counted and multiplied by the dilution.
Grouping and Treatment of Animals
After 3 days of adaptive feeding, the mice were randomly assigned to five groups (n = 6), including the blank, negative control, atorvastatin, DSM20016, and HI120 groups. In the blank group, mice were fed a normal diet for 12 weeks. negative-control group mice were fed a high-fat diet for 12 weeks. After 4 weeks, sunflower seed oil (containing 60% LA) (20 μL) was administered by gavage daily (every 2 days after 8 weeks). Mice in the atorvastatin group were fed a high-fat diet for 12 weeks; after 4 weeks, sunflower seed oil (20 μL) + atorvastatin (0.18 mg/kg) was administered by gavage daily (every 2 days after 8 weeks). In the DSM20016 group, the mice were fed a high-fat diet for 12 weeks; after 4 weeks, sunflower seed oil (20 μL) + DSM20016 bacterial powder (1,000 mg/kg) was administered by gavage daily (every 2 days after 8 weeks). HI120 group mice were fed a high-fat diet for 12 weeks. After 4 weeks, sunflower seed oil (20 μL) + HI120 bacterial powder (1,000 mg/kg) was administered by gavage daily (every 2 days after 8 weeks). Because atorvastatin is most effective when administered at night, the last four groups of mice were treated at 20:00 h every night.
Grouping and Treatment of Cells
Cholesterol Synthesis-, Absorption-, Excretion-, and Expression-Related Genes
Control group HepG2/Caco-2 cells were not treated. In the LA group, HepG2/Caco-2 cells were incubated with LA (0.7 μL/mL) for 48 h. In the DSM20016 group, LA (0.7 μL/mL) was incubated with DSM20016 solution (number of viable bacteria was approximately 1.0 × 107 cfu/mL) for 48 h. After centrifugation at 5,000 rpm, the supernatant was extracted (ensuring that the initial concentration of LA in the solution was 0.7 μL/mL) and incubated with HepG2/Caco-2 cells for 48 h. In the HI120 group, LA (0.7 μL/mL) was incubated with HI120 solution (number of viable bacteria was ~1.0 × 107 cfu/mL) for 48 h. After centrifugation at 5,000 rpm, the supernatant was extracted (initial concentration of LA in the solution is 0.7 μL/mL) and then incubated with HepG2/Caco-2 cells for 48 h.
NPC1L1/HMG-CR Expression Following siRNA Interference Targeting SREBP-2
In the NC group, HepG2/Caco-2 cells were transfected with negative-control siRNA. In siRNA group 1, HepG2/Caco-2 cells were transfected with the siRNA of SREBP-2 gene fragment 1. In siRNA group 2, HepG2/Caco-2 cells were transfected with the siRNA of SREBP-2 gene fragment 2. SREBP-2 siRNA primer sequence: 5′-AGCAGCAGCAGCAGCAATGG-3′ (forward), 5′-GCCGCCGAGGGAGAGAAGG-3′ (reverse).
Each transfected sample was prepared as follows: (1) siRNA dilution: 1.25 μL 20 μM siRNA stock solution was diluted with 30 μL Ribofect TM CP buffer (RiboBio Co., Ltd., Wuhan, China) and mixed gently. (2) Mixed solution: 3 μL Ribofect TM CP reagent (RiboBio Co., Ltd.) was added. The solution was gently blown and mixed, incubated at room temperature for 0–15 min, and used to prepare the transfection complex. (3). The Ribofect TM CP transfection complex was added to a suitable amount of complete culture medium without double antibody and mixed gently. Transfection step: cells were inoculated and spread onto a 6-well plate at a density of 5 × 105 cells per well and transfected at ~70% confluence. The final concentration of siRNA was 50 nM. Subsequently, 1,863 μL medium, 120 μL buffer, 5 μL siRNA, and 12 μL Regent were added to each pore and then mixed. The cells were cultured in an incubator for 48 h. The efficiency of siRNA silencing after transfection was assessed.
Immunofluorescence Analysis of Cholesterol Absorption and NPC1L1/SREBP-2 Expression
In the NC group, Caco-2 cells were incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) for 48 h. In the LA group, Caco-2 cells were incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) and LA (0.7 μL/mL) for 48 h. In the DSM20016 group, LA (0.7 μL/mL) was incubated with DSM20016 solution (number of viable bacteria was ~1.0 × 107 cfu/mL) for 48 h. After centrifugation at 5,000 rpm, the supernatant was extracted (ensuring that the initial concentration of LA in the solution was 0.7 μL/mL). Caco-2 cells were then incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) for 48 h. In the HI120 group, LA (0.7 μL/mL) was added to HI120 solution (number of viable bacteria was ~1.0 × 107 cfu/mL) and incubated for 48 h, After centrifugation at 5,000 rpm, the supernatant was extracted (ensuring that the initial concentration of LA in the solution was 0.7 μL/mL). Caco-2 cells were then incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) for 48 h.
Detection of Cholesterol Absorption by Immunofluorescence and NPC1L1 Expression After siRNA Silencing of SREBP-2
In the NC group, Caco-2 cells were incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) for 48 h. In the SREBP-2 RNAi 1 group, Caco-2 cells were transfected with the siRNA of SREBP-2 gene fragment 1 and incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) for 48 h. In the SREBP-2 RNAi 2 group, Caco-2 cells were transfected with the siRNA of SREBP-2 gene fragment 2 and incubated with Cholesteryl BodipyTM FL C12 (20 μM/L) for 48 h. The transfection procedure was performed as described above.
Detection Index of Animal- and Cell-Based Experiments
After 28 days, the body weight, food intake, and water intake of the mice were measured every 7 days. At the end of the experiment, the mice were anesthetized by intraperitoneal injection of sodium pentobarbital (1%, 40 mg/kg), blood was collected from the heart, and the mice were sacrificed. Tissue samples including small intestinal and liver tissues were collected from the same segment or lobe of each mouse and stored at −80°C. The blood was incubated at room temperature for 1 h and centrifuged at 3,000 rpm for 15 min, after which the serum was separated. The serum total cholesterol and triglyceride levels (HITACHI 7150 automatic biochemical analyzer, Tokyo, Japan) were then measured.
Total RNA was extracted from the liver/small-intestinal tissue of animals as well as from the cell lines. In both sample types, extraction was performed by the TRIzol method. The expression of NPC1L1, SREBP-2, HMG-CR, CYP7A1, LXR, and FXR was measured by RT-qPCR using the following primers: 51v 5′-TGACAAGTTCCAGGTTGCGT-3′ (forward), 5′-TAGCTGACGGCAAAGACAGG-3′ (reverse); 5′-ATGGACGACAGCGGCGAGC-3′ (forward), 5′-CAGAAGAATCCGTGAGCGGTCTAC-3′ (reverse); 5′-GATCCAGGAGCGAACCAAGAGAG-3′ (forward), 5′-GCTACAGAAGCCCCAAGCACAA-3′ (reverse); 5′-CAAGACCGCACATAAAGCC-3′ (forward), 5′-GATGCCCAGAGGATCACG-3′ (reverse); 5′-GATGCCCAGAGGATCACG-3′ (forward), 5′-GCTGACTCCAACCCTATCCC-3′ (reverse) and 5′-GTGAGGGCTGCAAAGGTTTC-3′ (forward), 5′-CAAACCTGTATACATACATTCAGCC-3′ (reverse). Their mRNA levels were presented as the ratio of the expression of each gene to GAPDH, in which its primers were 5′-GGACCTCATGGCCTACATGG-3′ (forward) and 5′-TAGGGCCTCTCTTGCTCAGT-3′ (reverse).
Total protein was extracted both from the liver/small intestine tissue of animals and from cell lines. The protein levels of NPC1L1, SREBP-2, HMG-CR, CYP7A1, LXR, and FXR were measured by western blotting using the following antibodies: anti-NPC1L1(Cat: PA1-16800, Thermo Fisher Scientific, Waltham, MA, USA), anti-SREBP-2 (Cat: ab28482, Abcam, Cambridge, UK), anti-HMG-CR (Cat: 3952-100, Bio Vision, San Francisco, CA, USA), anti-CYP7A1 (Cat: sc14423, Santa Cruz Biotechnology, Dallas TX, USA), LXR (Cat: GTX89656, GeneTex, Irvine, CA, USA), FXR (Cat: GTX113867, GeneTex, CA, USA), anti-GAPDH (Cat: AC027, ABclonal Technology, Wuhan, China), anti-rabbit secondary antibody (Cat: orb229657, Biorbyt, Cambridge, UK), anti-goat secondary antibody (Cat: 023002, Kangwei, Beijing, China).
Cells in the middle pores of the confocal dish were fixed, permeabilized, sealed, incubated with primary and secondary antibodies, stained with DAPI, and observed under a confocal laser scanning microscope (28).
Statistical Analysis
All cell experiments were performed three times independently and each experiment was performed in triplicate. GraphPad Prism 7 software (GraphPad, Inc., La Jolla, CA, USA) was used to prepare various statistical charts. The results obtained were expressed as the mean ± SD. Statistical analysis was performed by one-way analysis of variance followed by Tukey's multiple comparisons test for multiple comparison. P < 0.05 was considered as statistically significant.
Results
Morphological Characteristics of L. reuteri HI120
Analysis of the morphological characteristics of HI120 under an optical microscope (Figures 1A,B) revealed that HI120 is a slightly irregular type of campylobacter with a rounded end.
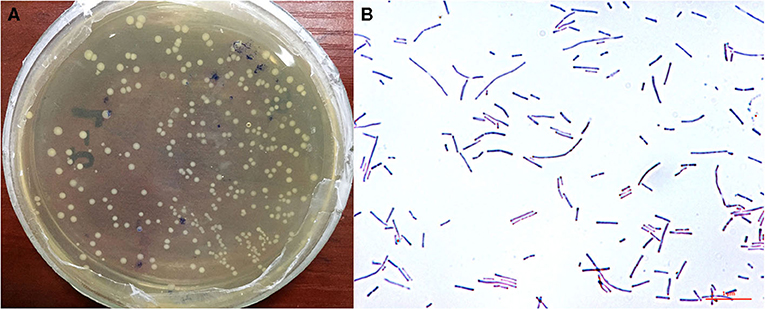
Figure 1. Morphological characteristics and observations of Lactobacillus reuteri HI120 under an optical microscope. (A) Morphological characteristics. (B) Observations after Gram staining (1,000× magnification).
Sequencing Results of L. reuteri HI120
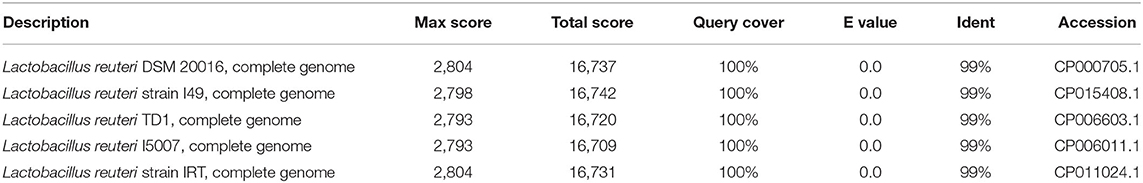
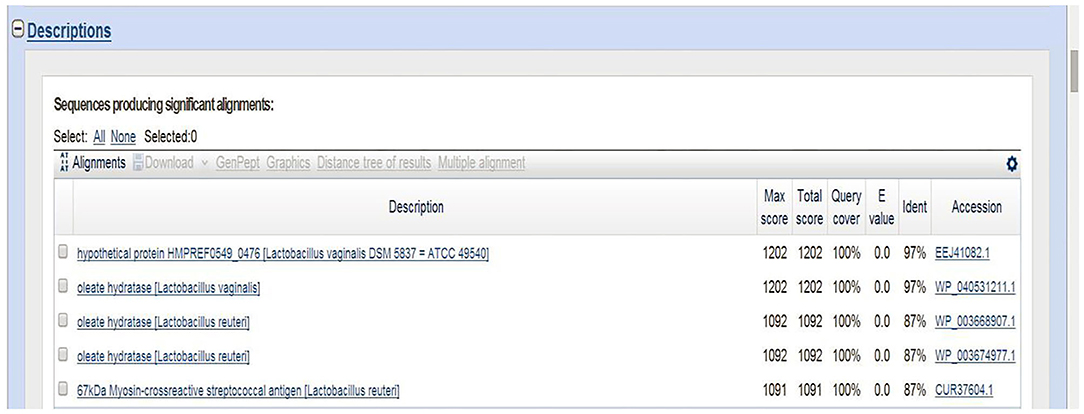
The 16S rRNA sequence of HI120 was 99% (Table 1) homologous with those of other L. reuteri strains. Thus, HI120 belongs to L. reuteri, which is supported by its biological characteristics. The LAI amino acid sequence of HI120 was 87% homologous with that of a standard strain of L. reuteri (DSM20016) and 97% homologous with that of Lactobacillus vaginalis (Figure 2). Therefore, HI120 may express high levels of LAI.
LA–CLA Conversion Rate of L. reuteri HI120
Gas chromatography (29) revealed that the main chemical structure of CLA generated by HI120 and DSM20016 was c9, t11-CLA (Figure 3). The conversion rate of LA to CLA was 27.63% for HI120 and 12.54% for DSM20016. The former rate was ~2.2-fold higher (Figure 4) than that of DSM20016.
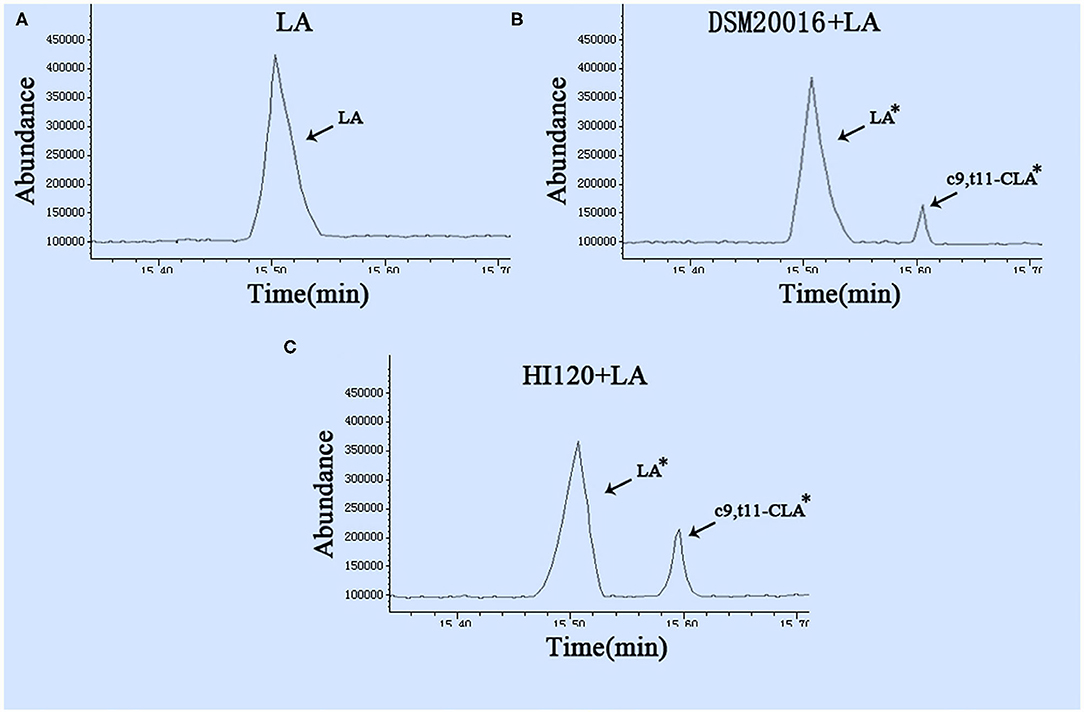
Figure 3. Gas chromatogram in all groups (A) LA group. (B) DSM20016 + LA group. (C) HI120 group + LA. *p < 0.05 compared to the LA group.
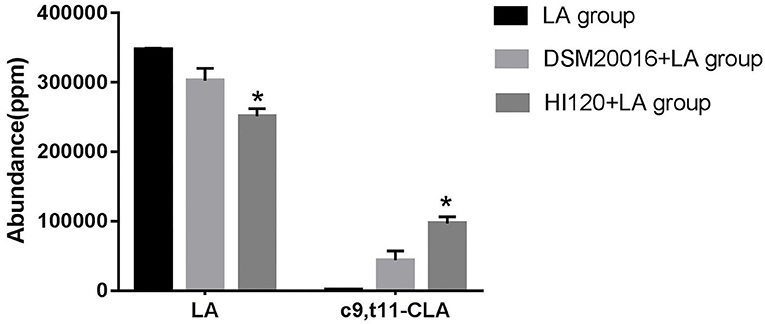
Figure 4. Abundance levels in the gas chromatogram of all groups. LA group. DSM20016 + LA group. HI120 + LA group. *p < 0.05 compared to the DSM20016+LA group.
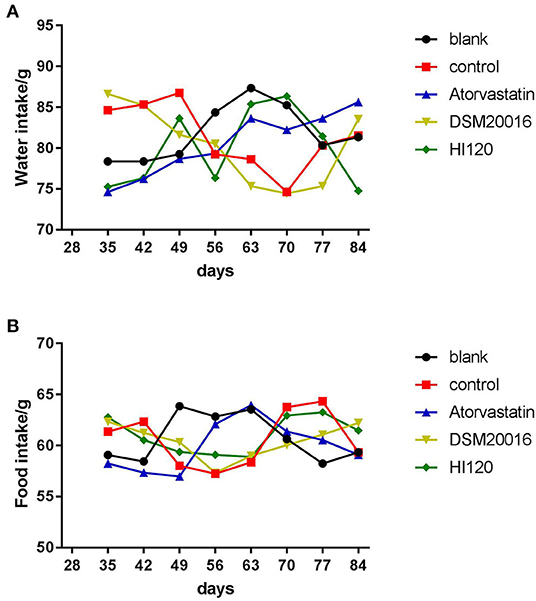
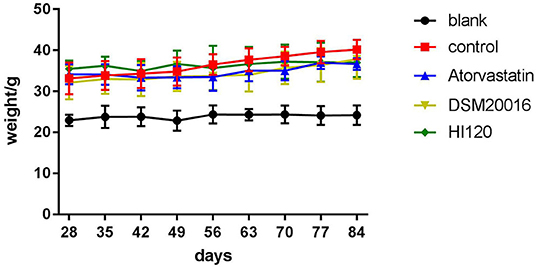
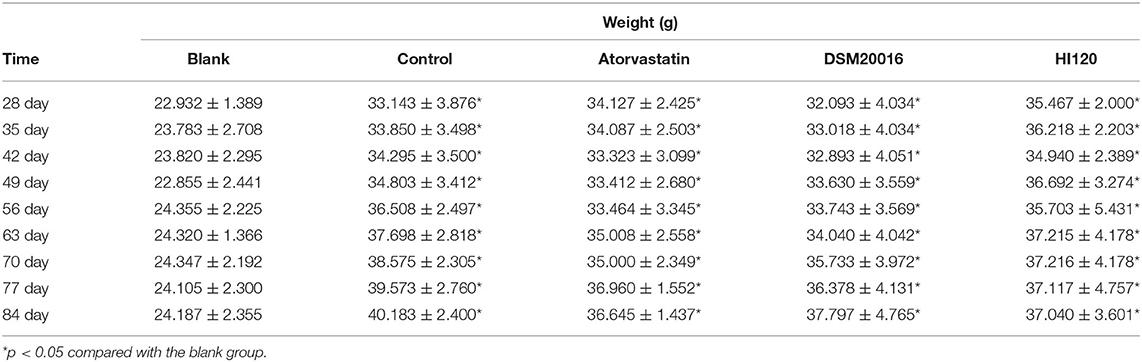
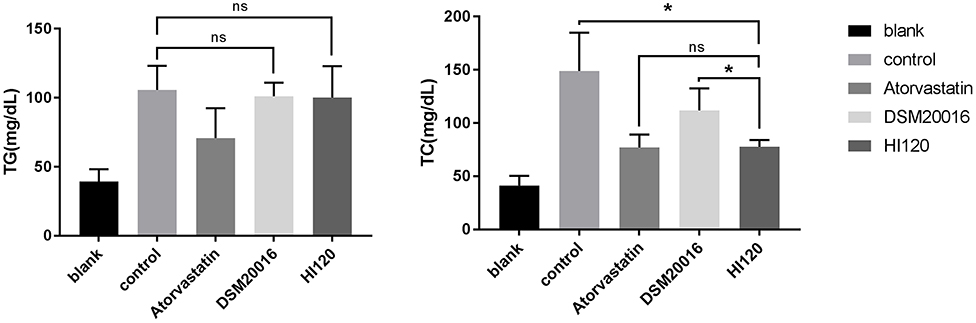
Food Intake, Water Intake, Body Weight, and Total Cholesterol and Triglyceride Serum Levels of Animals
There was no significant difference in the diet and water intake between experimental groups (Figures 5A,B). Additionally, no significant difference in body weight was observed between mice in the four experimental groups on a high-fat diet and body weight of mice on a regular diet (Figure 6, Table 2). The triglyceride levels of mice fed a high-fat diet were all increased; however, the levels in the mice fed with atorvastatin were decreased, whereas those fed HI120 and DSM20016 displayed no significant change. The cholesterol level of mice fed HI120 decreased as much as that of the level of the atorvastatin group mice and was significantly lower than that of DSM20016 group mice (Figure 7, Table 3). Mice fed HI120 did not lose weight or showed changes in triglycerides; however, cholesterol levels were decreased.
Expression Analyses (Real-Time PCR and Western Blotting) in Animals
Compared with the control group parameters, the mRNA levels and protein expression of NPC1L1 and SREBP-2 were significantly decreased in the small intestine of mice in the HI120 and DSM20016 groups; however, these levels in the HI120 group were lower than those in the DSM20016 group (Figures 8, 9). Compared with control group parameters, the mRNA and protein expression levels of HMG-CR in the liver of mice in the HI120 and DSM20016 groups were significantly lower; however, HI120 group levels were lower than those in the DSM20016 group (Figure 10). Compared with control group parameters, the mRNA and protein expression levels of SREBP-2 in the liver of mice in the HI120 and DSM20016 groups were significantly lower, but there was no significant difference between the HI120 and DSM20016 groups (Figure 11). Compared with control group parameters, the mRNA and protein expression levels of CYP7A1, LXR, and FXR in the liver of mice in the HI120 and DSM20016 groups were not significantly different; there was also no significant difference observed between the HI120 and DSM20016 groups (Figures 12–14).
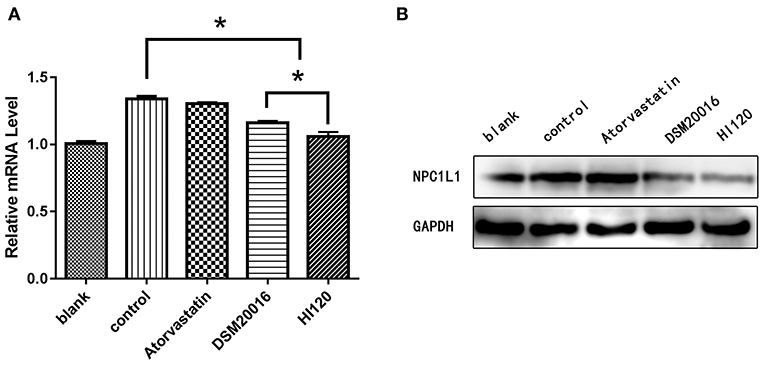
Figure 8. mRNA and protein expression levels of NPC1L1 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
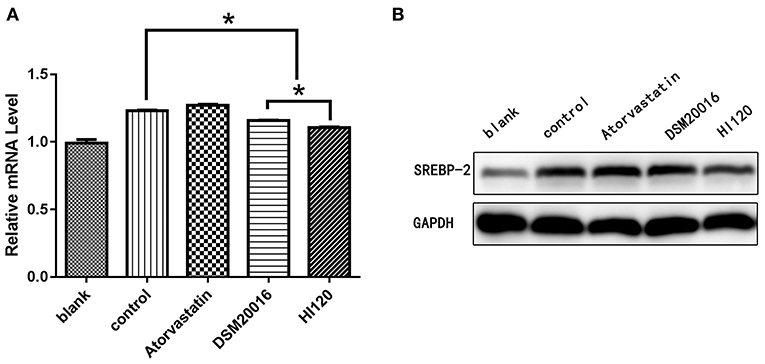
Figure 9. mRNA and protein expression levels of SREBP-2 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
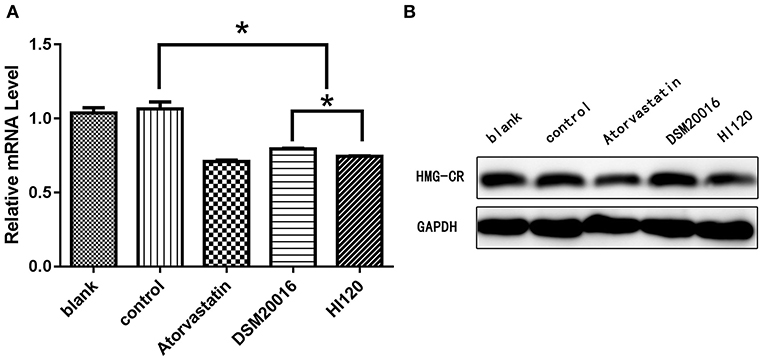
Figure 10. mRNA and protein expression levels of HMG-CR in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
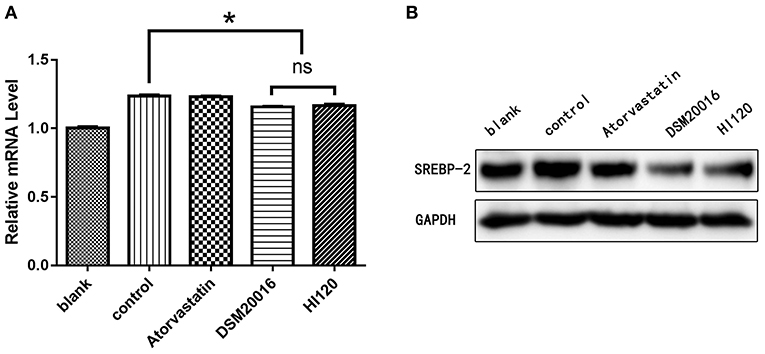
Figure 11. mRNA and protein expression levels of SREBP-2 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
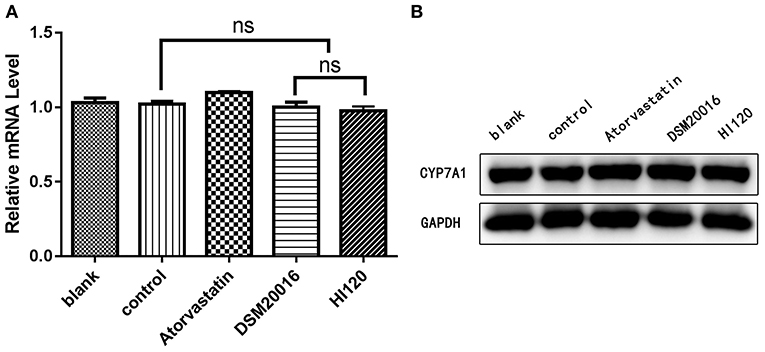
Figure 12. mRNA and protein expression levels of CYP7A1 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
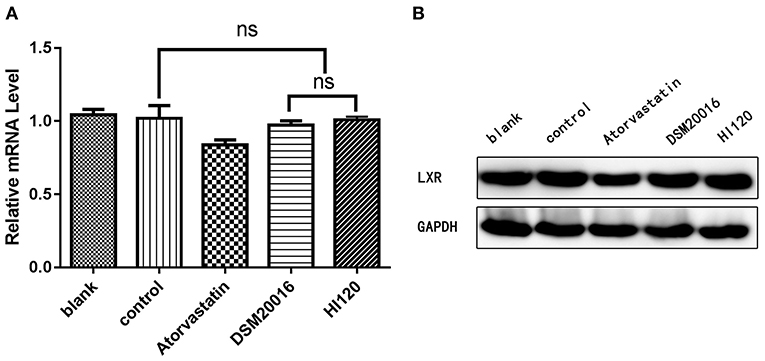
Figure 13. mRNA and protein expression levels of LXR in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
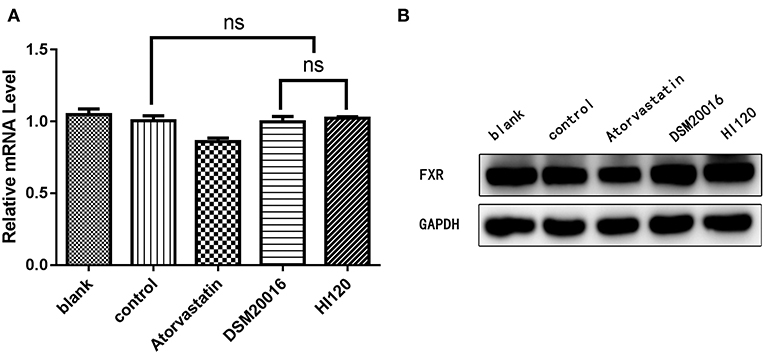
Figure 14. mRNA and protein expression levels of FXR in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
Expression Analyses (Real-Time PCR and Western Blotting) of Cells
Expression of Genes Related to Cholesterol Synthesis, Absorption, and Metabolism
Compared with the control group and LA group parameters, NPC1L1 and SREBP-2 mRNA levels and protein expression of Caco-2 cells from the HI120 group and DSM20016 group were lower; HI120 group levels were significantly lower than the corresponding DSM20016 levels (Figures 15, 16). Compared with the control group and LA group parameters, HMG-CR and SREBP-2 mRNA and protein expression levels in HepG2 cells from the HI120 group and DSM20016 group were lower; HI120 group levels were significantly lower than those in the DSM20016 group (Figures 17, 18). Compared with the control group and LA group parameters, CYP7A1, LXR, and FXR mRNA and protein expression levels of HepG2 cells in the HI120 group and DSM20016 group were not significantly different; there was also no significant difference between the HI120 and DSM20016 groups (Figures 19–21).
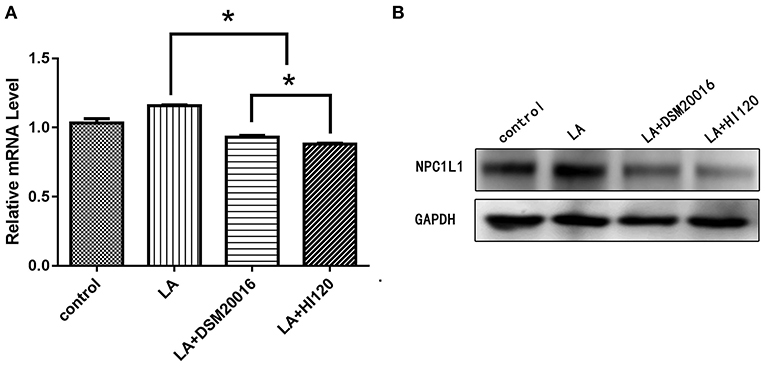
Figure 15. mRNA and protein expression levels of NPC1L1 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
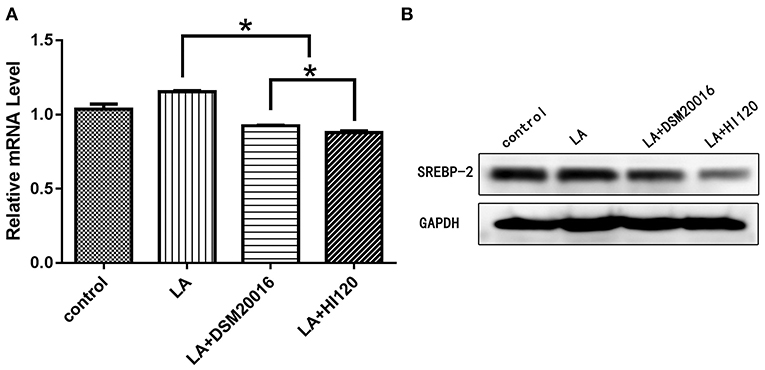
Figure 16. mRNA and protein expression levels of SREBP-2 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
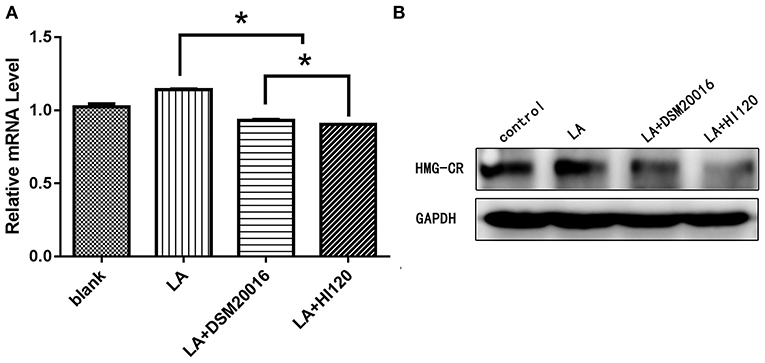
Figure 17. mRNA and protein expression levels of HMG-CR in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
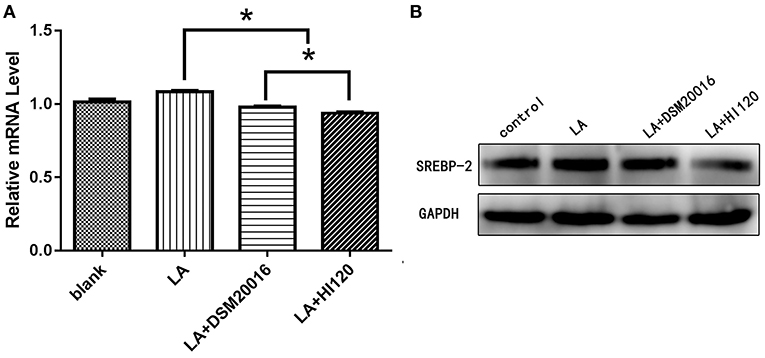
Figure 18. mRNA and protein expression levels of SREBP-2 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
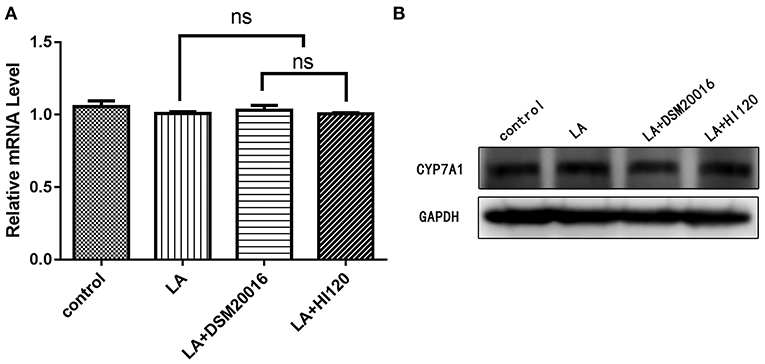
Figure 19. mRNA and protein expression levels of CYP7A1 in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
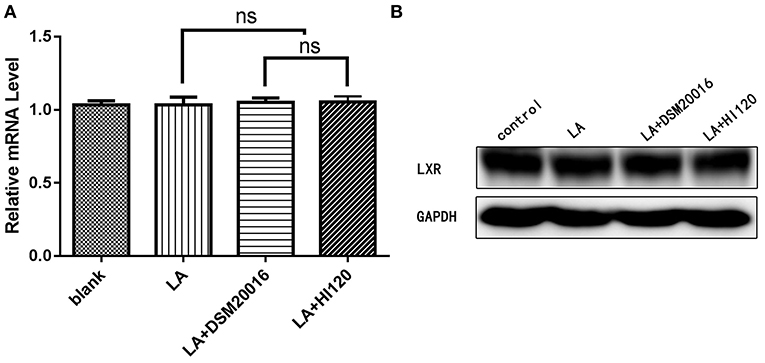
Figure 20. mRNA and protein expression levels of LXR in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
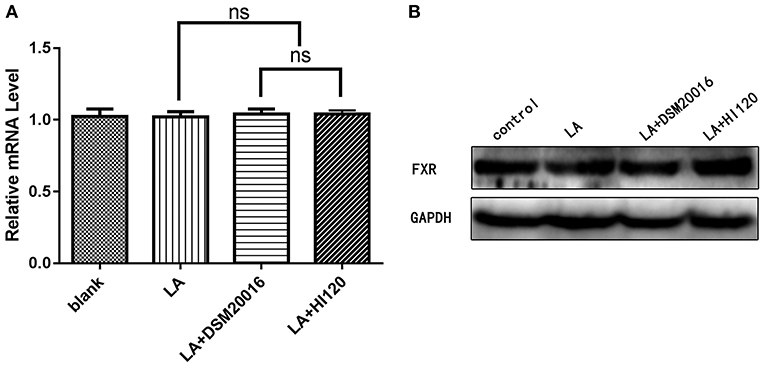
Figure 21. mRNA and protein expression levels of FXR in all groups. (A) qRT-PCR results. (B) Western blotting results. *p < 0.05.
NPC1L1/HMG-CR Expression After SREBP-2 siRNA Interference
After siRNA interference targeting SREBP-2, the mRNA and protein expression of NPC1L1 was significantly decreased in Caco-2 cells (Figure 22). HMG-CR mRNA and protein expression in HepG2 cells was decreased significantly (Figure 23).
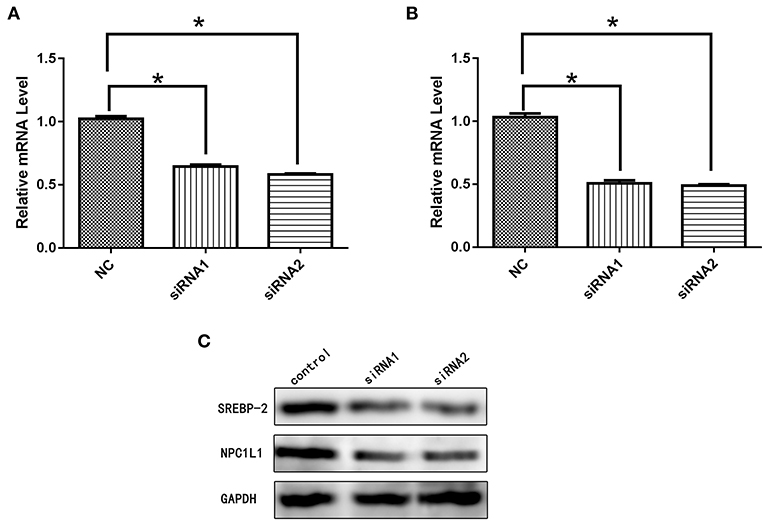
Figure 22. (A) mRNA expression level of SREBP-2 after interference. (B) mRNA expression level of NPC1L1 after SREBP-2 interference. (C) SREBP-2 and NPC1L1 protein expression levels after SREBP-2 interference. *p < 0.05.
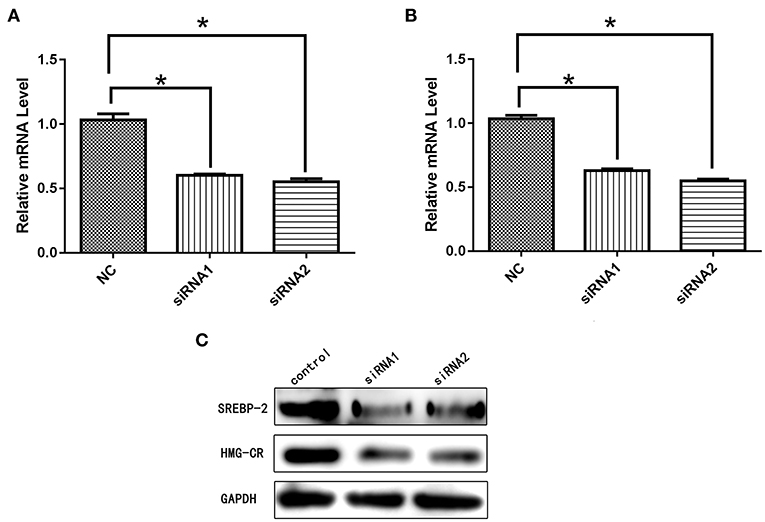
Figure 23. (A) mRNA expression level of SREBP-2 after interference. (B) mRNA expression level of HMG-CR after SREBP-2 interference. (C) SREBP-2 and HMG-CR protein expression levels after SREBP-2 interference. *p < 0.05.
Immunofluorescence Detection of Cholesterol Absorption and NPC1L1/SREBP-2 Expression
Compared with the NC and LA groups, the HI120 and DSM20016 groups exhibited a weak green fluorescent signal (Cholesteryl BodipyTM FL C12) and red fluorescent signal (NPC1L1 and SREBP-2). The HI120 signal was weaker than that in the DSM20016 group (Figures 24, 25).
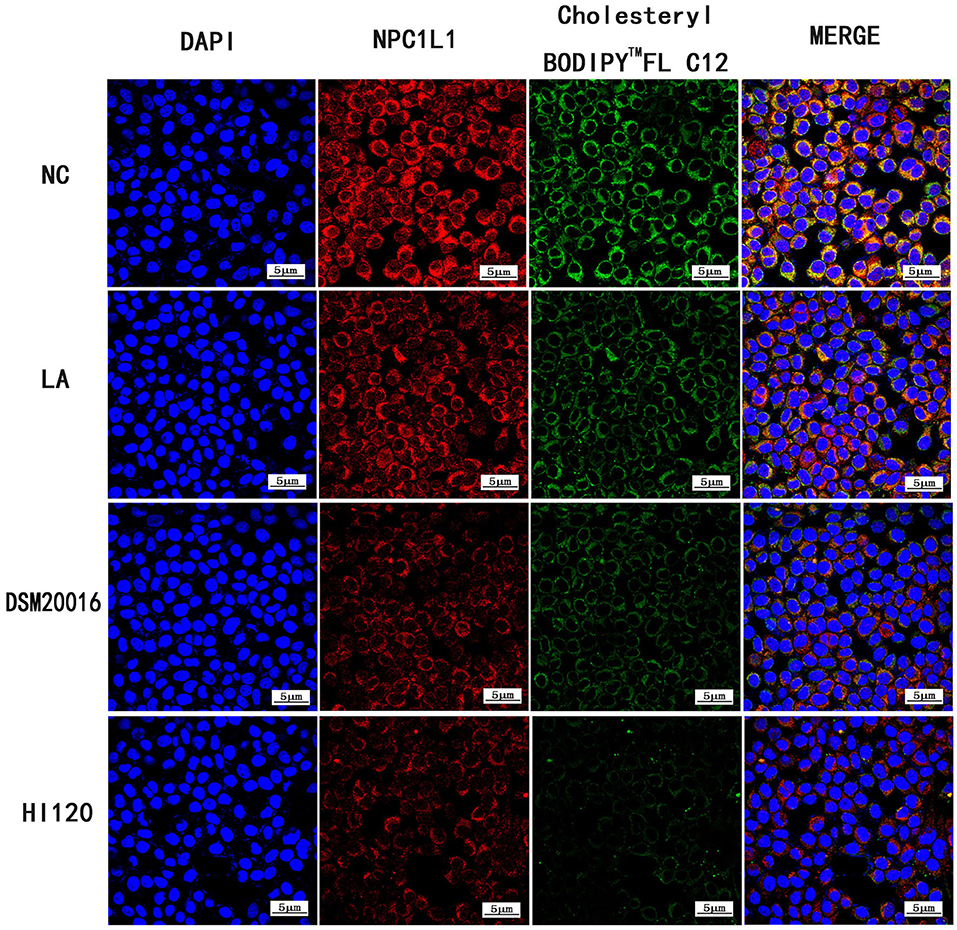
Figure 24. Expression of Cholesteryl BodipyTM FL C12 and NPC1L1 observed under confocal laser microscope (600× magnification).
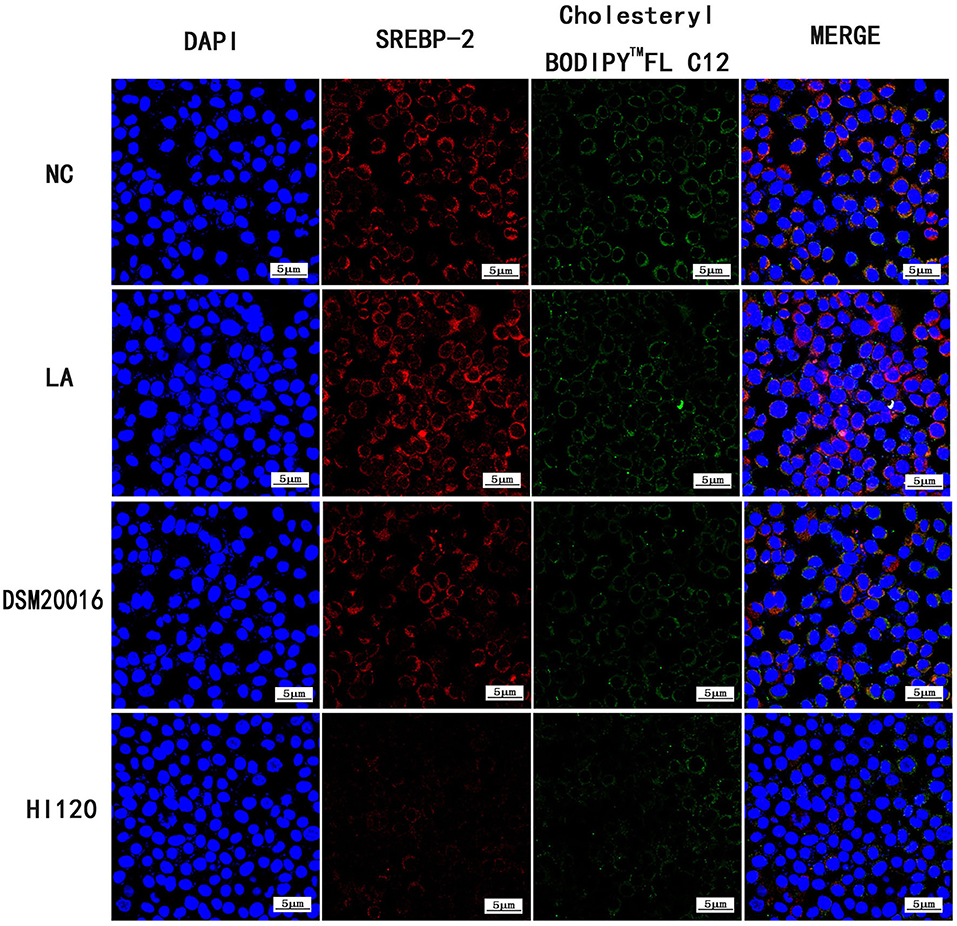
Figure 25. Expression of Cholesteryl BodipyTM FL C12 and SREBP-2 observed under confocal laser microscope (600× magnification).
Fluorescence Detection of Cholesterol Absorption and NPC1L1 Expression After SREBP-2 siRNA Interference
Following SREBP-2 siRNA interference, cholesterol absorption and NPC1L1 expression were decreased (Figure 26).
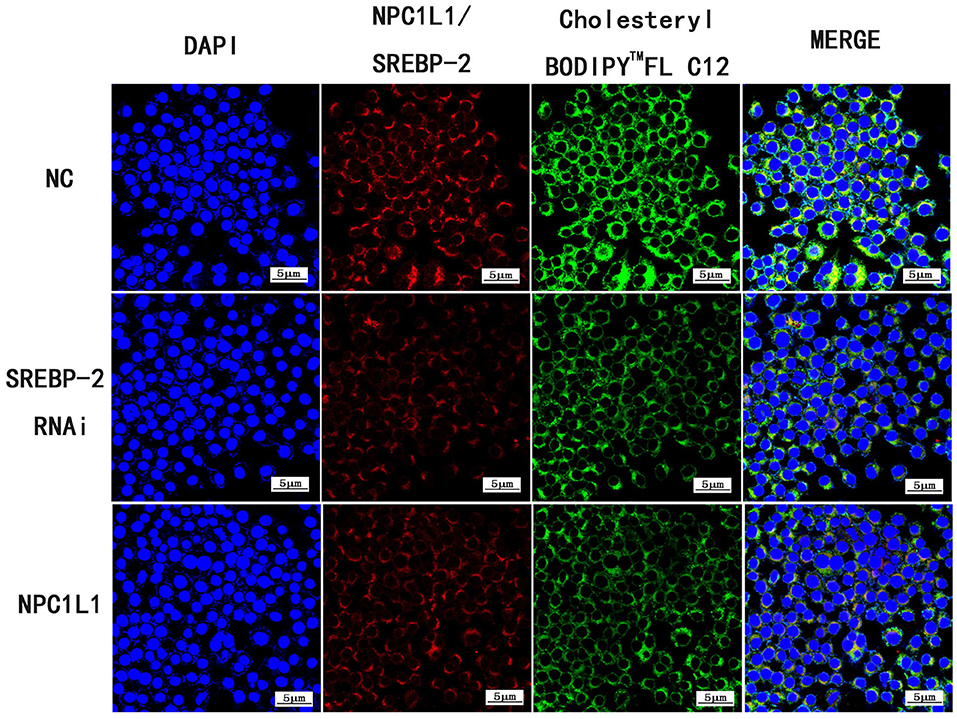
Figure 26. Expression of Cholesteryl BodipyTM FL C12, SREBP-2 RNAi and NPC1L1 under laser confocal microscope (600× magnification).
Discussion
Obesity is an increasingly widespread and detrimental health condition. Among cardiovascular-related deaths, 77% are attributed to elevated cholesterol (30). For every 10% reduction in serum cholesterol, the incidence of coronary heart disease is reduced by 15% (31). The prevention and treatment of lipid metabolism abnormalities, regulation of fatty acid transport, and reduction in lipid accumulation are key for alleviating obesity, diabetes, fatty liver, and other diseases. Therefore, suitable foods and drug must be identified to reduce obesity and improve lipid metabolism.
Obesity is closely related to gut microbes (32). Some bacterial strains can reduce macrophage lipid deposition and may improve atherosclerosis (33). Lactobacillus casei YRL577 have the potential effect of alleviating NAFLD, and significantly reduced liver weight and liver index and could regulate the levels of lipid metabolism (34). In addition, oral administration of a strain of human FGF21 expressing Lactobacillus significantly reduced the weight of mice and enhanced the activity of brown adipose tissue (35).
In this study, a new L. reuteri HI120 was evaluated in detail, which revealed that the 16S rRNA sequence of HI120 was 99% homologous with other L. reuteri 16S rRNA genes, indicating that HI120 belongs to L. reuteri. Additionally, the LAI amino acid sequence of HI120 was 87% homologous with a L. reuteri standard strain. Therefore, HI120 may express high levels of LAI. After mixing L. reuteri HI120 with LA in vitro, a large amount of CLA was generated, as revealed by mass spectrometry. The produced CLA was mainly composed of C9, t11-CLA isomers.
However, unexpectedly, our in vivo experiments revealed no decrease in the body weight of mice fed HI120 and DSM20016. The triglyceride level of mice fed HI120 and DSM20016 also did not decrease significantly. However, the cholesterol level was decreased. This may be because the mice were given a high-fat diet for only 8 weeks, which may not have been long enough to cause large changes. However, the cholesterol level in mice fed HI120 was significantly lower than that of mice fed DSM20016, and the observed reduction was comparable to the reductions observed in the atorvastatin group. Currently, atorvastatin is the first-line treatment for reducing blood lipids, particularly cholesterol levels. Although the effect of HI120 on cholesterol reduction was as good as that of atorvastatin, this was likely related to the atorvastatin dosage. Based on these results, HI120 may be useful for preventing and treating obesity and hyperlipidemia.
To explore the causes of cholesterol reduction in vivo, we evaluated cholesterol absorption, synthesis, and excretion. In 2004, Altmann et al. identified the transmembrane protein NPC1L1, which is responsible for the efficient and specific transport of cholesterol into cells (22). NPC1L1 was highly expressed in the small intestine, with highest expression in the jejunum and ileum (36). NPC1L1 knockout resulted in a 70% reduction in cholesterol absorption efficiency, whereas the absorption of triglycerides and other lipids was not affected, indicating that NPC1L1 specifically participated in the absorption process of cholesterol in the small intestine (22). NPC1L1 is targeted by ezetimibe, the only cholesterol absorption inhibitor in use in the clinic (37). The cholesterol content of cells can regulate the transcription of NPC1L1 via the SREBP-2 pathway (23, 38, 39). Kumar et al. found that curcumin indirectly regulates the absorption and transport of cholesterol by NPC1L1 by inhibiting SREBP-2 expression (40, 41). Bisphenol A can increase cholesterol absorption of Caco-2 cells by enhancing NPC1L1 expression, an effect that may also be regulated via SREBP-2 (42). When Caco-2 cells were incubated with ω-3 polyunsaturated fatty acid, the expression of NPC1L1 mRNA decreased by 35–58% (43). LA, palmitic acid, and oleic acid exerted no such effect. ω-3 polyunsaturated fatty acid regulates the absorption of cholesterol by inhibiting NPC1L1 expression, potentially through the LXR or SREBP-2 pathway. HMG-CR is an important rate-limiting enzyme in cholesterol synthesis (24). SREBP-2 is mainly responsible for the transcriptional regulation of cholesterol synthesis genes and can maintain the relative stability of cholesterol in cells by regulating the expression of HMG-CR (44, 45). In mammalian cells, the HMG-CR mRNA level is regulated by SREBP-2. When the sterol level in cells increases, the HMG-CR mRNA level decreases significantly. When the sterol level is low, SREBP-2 expression is induced, resulting in transcription of HMG-CR (24). CYP7A1 is the key and rate-limiting enzyme of bile acid production (cholesterol excretion). By interfering with CYP7A1 expression and reducing the synthesis of bile acids in mice, the absorption efficiency of cholesterol can be significantly reduced (25). The positive regulatory factor of CYP7A1 is LXR, whereas FXR is the negative regulatory factor. LXR and FXR cooperate to control the level of bile acids (46). Bile acid feedback regulation is mediated via the transcription regulation of CYP7A1 by FXR protein (26). LXR can promote in vivo reverse cholesterol transport and upregulate CYP7A1 (27, 47, 48).
The current study examined the expression of genes related to cholesterol synthesis, absorption, and metabolism. In vivo and in vitro experiments revealed that the expression of NPC1L1, SREBP-2, and HMG-CR was decreased, whereas that of CYP7A1, FXR, and LXR was not changed significantly by probiotic treatment. CLA, generated by two L. reuteri strains, reduced the gene expression of SREBP-2 and NPC1L1, contributing to reduced cholesterol absorption. Furthermore, SREBP-2 and HMG-CR expression was reduced, potentially leading to lower cholesterol synthesis. HI120 exhibited a more prominent effect than DSM20016. However, whether cholesterol levels are affected via corresponding metabolic pathway remains unclear. We used BODIPY fluorescence (49) to label cholesterol to assess the molecular mechanism of absorption. The results revealed that CLA regulates NPC1L1 and cholesterol absorption through SREBP-2. By siRNA-mediated interference of SREBP-2, SREBP-2 was confirmed to regulate NPC1L1 and HMG-CR and thus affect the absorption and synthesis of cholesterol.
CLA of high purity is costly, and industrially produced CLA cannot be used in the daily diet, as food safety cannot be guaranteed. The edible oil in people's daily diet contains a considerable amount of LA. Long-term use of L. reuteri, a safe and reliable edible strain, may contribute to cholesterol reduction, supporting the prevention and treatment of high blood lipid–related disease. It is necessary to consider that the dosage used in the present study and duration of the experiments may need to be adjusted. Further investigation of whether HI120 can directly reduce cholesterol and the underlying mechanism is needed.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by Institutional Laboratory Animal Care and Use Committee of the South Medical University at Guangzhou.
Author Contributions
YS performed the experiments with bacteria, mice, cells, analyzed the data, and wrote the manuscript. XH and YT performed the experiments with mice. HW and LH performed the experiments with bacteria. JW provided the bacteria. HN and WZ designed the experiments, analyzed the data, and contributed to revising the manuscript. YB provided overall direction and contributed to revising the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81373315), National Science and Technology Major Special Fund (No. 2020zx09201017), Basic and Applied Basic Research Fund Project of Guangdong Province (No. 2019A1515012115), Science and Technology Planning Project of Guangdong Province (Nos. 201704020064, 2017b0209003, 2017b030314037, and 2015A020211013), and Science and Technology Planning Project of Guangzhou City (No. 2014J4100154).
Conflict of Interest
JW was employed by the Guangzhou Weisengene Biological Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professors Fachao Zhi, Side Liu, and Xiaochun Bai.
References
1. Reardon CA, Lingaraju A, Schoenfelt KQ, Zhou G, Cui C, Jacobs-El H, et al. Obesity and insulin resistance promote atherosclerosis through an IFNγ-regulated macrophage protein network. Cell Rep. (2018) 23:3021–30. doi: 10.1016/j.celrep.2018.05.010
2. Smith AG, Singleton JR. Obesity and hyperlipidemia are risk factors for early diabetic neuropathy. J Diabetes Comp. (2013) 27:436–42. doi: 10.1016/j.jdiacomp.2013.04.003
3. Aindelis G, Chlichlia K. Modulation of anti-tumour immune responses by probiotic bacteria. Vaccines. (2020) 8:329. doi: 10.3390/vaccines8020329
4. Stec DE, Gordon DM, Hipp JA, Hong S, Mitchell ZL, Franco NR, et al. Loss of hepatic PPARα promotes inflammation and serum hyperlipidemia in diet-induced obesity. Am J Physiol Regul Integr Comp Physiol. (2019) 317:R733–45. doi: 10.1152/ajpregu.00153.2019
5. Halmos T, Suba I. Alzheimer-kór és diabétesz mellitusz - a közös patomechanizmus [Alzheimer's disease and diabetes - the common pathogenesis]. Neuropsychopharmacol Hung. (2016) 18:5–19.
6. Singhvi N, Gupta V, Gaur M, Sharma V, Puri A, Singh Y, et al. Interplay of human gut microbiome in health and wellness. Ind J Microbiol. (2020) 60:26–36. doi: 10.1007/s12088-019-00825-x
7. Shaper AG, Jones KW, Jones M, Kyobe J. Serum lipids in three nomadic tribes of Northern Kenya. Am J Clin Nutr. (1963) 13:135–46. doi: 10.1093/ajcn/13.3.135
8. Mann GV. Studies of a surfactant and cholesteremia in the Maasai. Am J Clin Nutr. (1974) 27:464–9. doi: 10.1093/ajcn/27.5.464
9. Hennessy AA, Ross RP, Fitzgerald GF, Caplice N, Stanton C. Role of the gut in modulating lipoprotein metabolism. Curr Cardiol Rep. (2014) 16:515. doi: 10.1007/s11886-014-0515-2
10. Park EJ, Lee YS, Kim SM, Park GS, Lee YH, Jeong DY, et al. Beneficial effects of Lactobacillus plantarum strains on non-alcoholic fatty liver disease in high fat/high fructose diet-fed rats. Nutrients. (2020) 12:542. doi: 10.3390/nu12020542
11. Rosberg-Cody E, Johnson MC, Fitzgerald GF, Ross PR, Stanton C. Heterologous expression of linoleic acid isomerase from Propionibacterium acnes and anti-proliferative activity of recombinant trans-10, cis-12 conjugated linoleic acid. Microbiology. (2007) 153(Pt 8):2483–90. doi: 10.1099/mic.0.2006/001966-0
12. Diniz YS, Santos PP, Assalin HB, Souza GA, Rocha KK, Ebaid GM, et al. Conjugated linoleic acid and cardiac health: oxidative stress and energetic metabolism in standard and sucrose-rich diets. Eur J Pharmacol. (2008) 579:318–25. doi: 10.1016/j.ejphar.2007.11.008
13. den Hartigh LJ. Conjugated linoleic acid effects on cancer, obesity, and atherosclerosis: a review of pre-clinical and human trials with current perspectives. Nutrients. (2019) 11:370. doi: 10.3390/nu11020370
14. Stangl GI. Conjugated linoleic acids exhibit a strong fat-to-lean partitioning effect, reduce serum VLDL lipids and redistribute tissue lipids in food-restricted rats. J Nutr. (2000) 130:1140–6. doi: 10.1093/jn/130.5.1140
15. Rungapamestry V, McMonagle J, Reynolds C, Rucklidge G, Reid M, Duncan G, et al. Inter-organ proteomic analysis reveals insights into the molecular mechanisms underlying the anti-diabetic effects of cis-9, trans-11-conjugated linoleic acid in ob/ob mice. Proteomics. (2012) 12:461–76. doi: 10.1002/pmic.201100312
16. Wang YM, Nagao K, Inoue N, Ujino Y, Shimada Y, Nagao T, et al. Isomer-specific anti-obese and hypolipidemic properties of conjugated linoleic acid in obese OLETF rats. Biosci Biotechnol Biochem. (2006) 70:355–62. doi: 10.1271/bbb.70.355
17. Noone EJ, Roche HM, Nugent AP, Gibney MJ. The effect of dietary supplementation using isomeric blends of conjugated linoleic acid on lipid metabolism in healthy human subjects. Br J Nutr. (2002) 88:243–51. doi: 10.1079/BJN2002615
18. Stangl GI, Müller H, Kirchgessner M. Conjugated linoleic acid effects on circulating hormones, metabolites and lipoproteins, and its proportion in fasting serum and erythrocyte membranes of swine. Eur J Nutr. (1999) 38:271–7. doi: 10.1007/s003940050077
19. Bhushan B, Tomar SK, Chauhan A. Techno-functional differentiation of two vitamin B12 producing Lactobacillus plantarum strains: an elucidation for diverse future use. Appl Microbiol Biotechnol. (2017) 101:697–709. doi: 10.1007/s00253-016-7903-z
20. Schonewille M, de Boer JF, Mele L, Wolters H, Bloks VW, Wolters JC, et al. Statins increase hepatic cholesterol synthesis and stimulate fecal cholesterol elimination in mice. J Lipid Res. (2016) 57:1455–64. doi: 10.1194/jlr.M067488
21. Jiang R, Zhao S, Wang R, Feng H, Zhang J, Li X, et al. Safety and efficacy of atorvastatin for chronic subdural hematoma in chinese patients: a randomized clinical trial. JAMA Neurol. (2018) 75:1338–46. doi: 10.1001/jamaneurol.2018.2030
22. Altmann SW, Davis HR Jr, Zhu LJ, Yao X, Hoos LM, Tetzloff G, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. (2004) 303:1201–4. doi: 10.1126/science.1093131
23. Pramfalk C, Jiang ZY, Cai Q, Hu H, Zhang SD, Han TQ, et al. HNF1alpha and SREBP2 are important regulators of NPC1L1 in human liver. J Lipid Res. (2010) 51:1354–62. doi: 10.1194/jlr.M900274-JLR200
24. Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. (1990) 343:425–30. doi: 10.1038/343425a0
25. Schwarz M, Russell DW, Dietschy JM, Turley SD. Alternate pathways of bile acid synthesis in the cholesterol 7alpha-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding. J Lipid Res. (2001) 42:1594–603.
26. Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. (2000) 102:731–44. doi: 10.1016/S0092-8674(00)00062-3
27. Yasuda T, Grillot D, Billheimer JT, Briand F, Delerive P, Huet S, et al. Tissue-specific liver X receptor activation promotes macrophage reverse cholesterol transport in vivo. Arterioscler Thromb Vasc Biol. (2010) 30:781–6. doi: 10.1161/ATVBAHA.109.195693
28. Deng H, Yang S, Zhang Y, Qian K, Zhang Z, Liu Y, et al. Bacteroides fragilis prevents Clostridium difficile infection in a mouse model by restoring gut barrier and microbiome regulation. Front Microbiol. (2018) 9:2976. doi: 10.3389/fmicb.2018.02976
29. Ju H, Kim B, Kim J, Baek SY. Development of candidate reference method for accurate determination of four polycyclic aromatic hydrocarbons in olive oil via gas chromatography/high-resolution mass spectrometry using 13C-labeled internal standards. Food Chem. (2020) 309:125639. doi: 10.1016/j.foodchem.2019.125639
30. Critchley J, Liu J, Zhao D, Wei W, Capewell S. Explaining the increase in coronary heart disease mortality in Beijing between 1984 and 1999. Circulation. (2004) 110:1236–44. doi: 10.1161/01.CIR.0000140668.91896.AE
31. Pandian A, Arora A, Sperling LS, Khan BV. Targeting multiple dyslipidemias with fixed combinations–focus on extended release niacin and simvastatin. Vasc Health Risk Manag. (2008) 4:1001–9. doi: 10.2147/VHRM.S3460
32. Marzullo P, Di Renzo L, Pugliese G, De Siena M, Barrea L, Muscogiuri G, et al. From obesity through gut microbiota to cardiovascular diseases: a dangerous journey. Int J Obesity Suppl. (2020) 10:35–49. doi: 10.1038/s41367-020-0017-1
33. Liavonchanka A, Hornung E, Feussner I, Rudolph MG. Structure and mechanism of the Propionibacterium acnes polyunsaturated fatty acid isomerase. Proc Natl Acad Sci USA. (2006) 103:2576–81. doi: 10.1073/pnas.0510144103
34. Zhang Z, Zhou H, Zhou X, Sun J, Liang X, Lv Y, et al. Lactobacillus casei YRL577 ameliorates markers of non-alcoholic fatty liver and alters expression of genes within the intestinal bile acid pathway. Br J Nutr. (2020). doi: 10.1017/S0007114520003001. [Epub ahead of print].
35. Cao WY, Dong M, Hu ZY, Wu J, Li YC, Xu HD. Recombinant Lactococcus lactis NZ3900 expressing bioactive human FGF21 reduced body weight of Db/Db mice through the activity of brown adipose tissue. Benef Microbes. (2020) 11:67–78. doi: 10.3920/BM2019.0093
36. Xie C, Zhou ZS, Li N, Bian Y, Wang YJ, Wang LJ, et al. Ezetimibe blocks the internalization of NPC1L1 and cholesterol in mouse small intestine. J Lipid Res. (2012) 53:2092–101. doi: 10.1194/jlr.M027359
37. Rosenblum SB, Huynh T, Afonso A, Davis HRJr, Yumibe N, Clader JW, et al. Discovery of 1-(4-fluorophenyl)-(3R)-[3-(4-fluorophenyl)-(3S)-hydroxypropyl]-(4S)-(4 -hydroxyphenyl)-2-azetidinone (SCH 58235): a designed, potent, orally active inhibitor of cholesterol absorption. J Med Chem. (1998) 41:973–80. doi: 10.1021/jm970701f
38. Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. (1997) 89:331–40. doi: 10.1016/S0092-8674(00)80213-5
39. Alrefai WA, Annaba F, Sarwar Z, Dwivedi A, Saksena S, Singla A, et al. Modulation of human Niemann-Pick C1-like 1 gene expression by sterol: role of sterol regulatory element binding protein 2. Am J Physiol Gastrointest Liver Physiol. (2007) 292:G369–76. doi: 10.1152/ajpgi.00306.2006
40. Kumar P, Malhotra P, Ma K, Singla A, Hedroug O, Saksena S, et al. SREBP2 mediates the modulation of intestinal NPC1L1 expression by curcumin. Am J Physiol Gastrointest Liver Physiol. (2011) 301:G148–55. doi: 10.1152/ajpgi.00119.2011
41. Li Y, Li M, Wu S, Tian Y. Combination of curcumin and piperine prevents formation of gallstones in C57BL6 mice fed on lithogenic diet: whether NPC1L1/SREBP2 participates in this process? Lipids Health Dis. (2015) 14:100. doi: 10.1186/s12944-015-0106-2
42. Feng D, Zou J, Zhang S, Li X, Li P, Lu M. Bisphenol A promotes cholesterol absorption in Caco-2 cells by up-regulation of NPC1L1 expression. Lipids Health Dis. (2017) 16:2. doi: 10.1186/s12944-016-0395-0
43. Alvaro A, Rosales R, Masana L, Vallvé JC. Polyunsaturated fatty acids down-regulate in vitro expression of the key intestinal cholesterol absorption protein NPC1L1: no effect of monounsaturated nor saturated fatty acids. J Nutr Biochem. (2010) 21:518–25. doi: 10.1016/j.jnutbio.2009.02.010
44. Ford AC, Quigley EM, Lacy BE, Lembo AJ, Saito YA, Schiller LR, et al. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am J Gastroenterol. (2014) 109:1547–62. doi: 10.1038/ajg.2014.202
45. De Greef E, Vandenplas Y, Hauser B, Devreker T, Veereman G. The use of probiotics in IBD and IBS. Miner Pediatr. (2014) 66:491–500.
46. Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. (2000) 6:507–15. doi: 10.1016/S1097-2765(00)00050-2
47. Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. (1998) 93:693–704. doi: 10.1016/S0092-8674(00)81432-4
48. Lo Sasso G, Murzilli S, Salvatore L, D'Errico I, Petruzzelli M, Conca P, et al. Intestinal specific LXR activation stimulates reverse cholesterol transport and protects from atherosclerosis. Cell Metab. (2010) 12:187–93. doi: 10.1016/j.cmet.2010.07.002
Keywords: probiotics, obesity, cholesterol, lipid metabolism, conjugated linoleic acid, Lactobacillus reuteri
Citation: Sun Y, Tang Y, Hou X, Wang H, Huang L, Wen J, Niu H, Zeng W and Bai Y (2020) Novel Lactobacillus reuteri HI120 Affects Lipid Metabolism in C57BL/6 Obese Mice. Front. Vet. Sci. 7:560241. doi: 10.3389/fvets.2020.560241
Received: 20 May 2020; Accepted: 02 September 2020;
Published: 14 October 2020.
Edited by:
Stephen Brent Smith, Texas A&M University, United StatesReviewed by:
Elsa Lamy, University of Évora, PortugalF. Capela e Silva, Universidade de Évora, Portugal
Copyright © 2020 Sun, Tang, Hou, Wang, Huang, Wen, Niu, Zeng and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Bai, YmFpeWFuZzEwMzAmI3gwMDA0MDtob3RtYWlsLmNvbQ==; Weisen Zeng, emVuZ3dzJiN4MDAwNDA7c211LmVkdS5jbg==; Hongxin Niu, bmhvbmd4aW4mI3gwMDA0MDsxMjYuY29t
 Ye Sun
Ye Sun Yanqing Tang3
Yanqing Tang3