
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 16 October 2020
Sec. Animal Nutrition and Metabolism
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.552251
This article is part of the Research TopicNutrition and Management of Animals We Keep as CompanionsView all 17 articles
Canine atopic dermatitis (CAD) has a hereditary basis that is modified by interactions with the environment, including diet. Differentially expressed genes in non-lesional skin, determined by RNA sequencing before and after a dietary intervention, were compared between dogs with naturally occurring CAD (n = 4) and healthy dogs (n = 4). The dogs were fed either a common commercial heat-processed high carbohydrate food (kibble diet) (n = 4), or a non-processed high fat food (raw meat-based diet) (n = 4). At the end of the diet intervention, 149 differentially expressed transcripts were found between the atopic and healthy dogs. The main canonical pathways altered by the dysregulation of these genes were angiopoietin signaling, epidermal growth factor signaling, activation of angiogenesis, and alterations in keratinocyte proliferation and lipid metabolism. On the other hand, 33 differently expressed transcripts were found between the two diet groups, of which 8 encode genes that are annotated in the current version of the dog genome: immunoglobulin heavy constant mu (IGHM), immunoglobulin lambda-like polypeptide 5 (IGLL5), B-cell antigen receptor complex-associated protein beta chain (CD79B), polymeric immunoglobulin receptor (PIGR), cystathionine β-synthase (CBS), argininosuccinate synthase 1 (ASS1), secretory leukocyte peptidase inhibitor (SLPI), and mitochondrial ribosome recycling factor (MRRF). All genes were upregulated in the raw diet group. In conclusion the findings of this study suggest alterations in lipid and keratinocyte metabolism as well as angiogenesis in the skin of atopic dogs. Additionally, a possible enhancement of innate immunity and decrease in oxidative stress was seen in raw food fed dogs, which could have an important role in preventing hypersensitivities and disturbed immunity at young age.
Atopic dermatitis (AD) in humans is a complex hereditary disease characterized by its genetic pre- disposition, as well as how it causes immunologic hyperreactivity and a defective skin barrier. An impaired epidermal barrier is one of the typical features of AD (1–3). Also, environmental factors play a role in its pathogenesis (4). Canine atopic dermatitis (CAD) shares genetic, phenotypic, and histological similarities with human AD (5). CAD is estimated to affect 10–15% of the canine population and seems to be increasing over time (5, 6). It has been reported that pure bred dogs are more likely to develop atopy and allergic dermatitis than mixed breeds (7). Staffordshire bull terriers and similar breeds (bull terrier, American Staffordshire terrier, and pit bull terrier) have previously been shown to be particularly susceptible to CAD (8–13).
Diet has profound effects on the whole body, including its metabolism. People with skin conditions often modify their diet to influence their clinical symptoms (14–16), as do dog owners for their dogs (17). It has been previously suggested that clinical manifestations of CAD can be modulated by diet (18–21). It has been reported that certain diets can potentially affect skin symptoms positively both in humans (22–24) and in dogs (25–27). Research on how diets impact skin gene expression in dogs is still scarce. However, dietary changes are shown to change the gene expression in the skin of mice (28), rats (29), and humans (30, 31). The impact that the elimination diet has on the skin of dogs with cutaneous adverse food reactions has been studied using quantitative PCR (qPCR) (32). The diets influence on gene expression in the liver (33), skeletal muscle (34), adipose tissue (35), colonic mucosa (36), and brain tissue (37) of dogs has also been studied.
Previously published studies that have focused on altered gene expression in atopic canine skin have used mRNA microarrays (38–40) and (qPCR) (41–50). The objective of this study was to find differentially expressed genes between atopic and healthy dogs, as well as to compare the effects of two different diet types on the gene expression of the skin using RNA sequencing (RNA-Seq). As alterations in skin function of atopic patients are already well recognized, our hypothesis was that there would be differences in the gene expression between atopic and healthy dogs. In addition, we hypothesized that two different dietary choices would subsequently affect skin gene expression differently. Eight Staffordshire bull terriers were used, both atopic and healthy, equally distributed between diet groups. To our best knowledge, neither previous studies using RNA-Seq to compare atopic and healthy dogs, or studies regarding the effect of diet on canine skin gene expression have been conducted.
The study protocol was approved by the Animal Experiment Board in Finland (ELLA) (permit number: ESAVI/3244/04.10.07/2013). All owners filled in and signed a written consent form. Dogs were living in their home environment during the diet intervention trial. The eight study dogs used in the present study were part of a larger diet intervention trial studying CAD. The study was conducted at the department of equine and small animal medicine at the University of Helsinki.
At the baseline visit, dogs were evaluated by a veterinarian and blood samples were collected. Non-lesional skin biopsies were taken from the axillary area under anesthesia using an 8 mm biopsy punch, and samples were immediately stored at −80°C. The animals were sedated using dexmedetomidine (Dexdomitor, Orion Pharma) 5–10 μg/kg intramuscularly and intravenous propofol (PropoVet, Orion Pharma) as needed. Skin biopsies were taken only from dogs that had not taken oral glucocorticoids and cyclosporine for 4 weeks, or oral antihistamines, topical glucocorticoids, and medicated shampoos for 2 weeks prior to sample collection.
After the baseline visit, the dogs were divided into two diet groups, and fed either a commercial heat-processed, high carbohydrate (kibble) diet [Hill's Science PlanTM Canine Adult Sensitive Skin with Chicken (KD), detailed composition of food shown in Supplementary Table 1], or a commercial non-processed, high fat (raw meat-based) diet [MUSH BARF Vaisto® Pork-Chicken-Lamb and/or MUSH BARF Vaisto® Beef-Turkey-Salmon (RMBD), detailed compositions shown in Supplementary Table 2]. The diets differed by their fat and carbohydrate content, their ingredients (KD having chicken as a main animal protein source and RMBDs having three different animal protein sources each) and their manufacturing methods (the KD was heat-processed and the RMBDs were only ground and frozen). Owners were asked to feed their dogs at least 99.9% trial diet, giving portions as recommended by the manufacturer, adjusting if necessary, to maintain normal body weight. Water was allowed ad libitum. At the end visit, the same protocol was followed as the baseline visit, and non-lesional skin biopsies were again obtained from the same area and immediately stored at −80°C.
Skin biopsies were obtained from client-owned Staffordshire bull terriers (n = 8). They were stratified between diet cohorts by age, sex, and health status when possible, since the number of healthy dogs was very small. The dogs diagnosed with naturally occurring CAD (n = 4) were fed either the KD (n = 2) or RMBD (n = 2). The healthy control dogs were fed either the KD (n = 2) or the RMBD (n = 2). This allowed comparisons to be made between KD-fed (n = 4) and RMBD-fed (n = 4) dogs, as well as between atopic (n = 4) and healthy (n = 4) dogs. Information regarding the dogs chosen for RNA-Seq analyses (n = 8) are shown in Table 1.
Prior to RNA extraction, the skin biopsy samples were transferred to RNAlater®-ICE Frozen Tissue Transition Solution (Life Technologies, Carlsbad, CA, USA) and allowed to thaw overnight at −20°C. Subcutaneous fat was carefully trimmed from the skin biopsies, and the samples were homogenized using a tissue homogenizer (TissueRuptor, Qiagen, Hilden, Germany). Total RNA was extracted using Qiagen miRNAeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommended protocol. After RNA extraction, DNAse treatment using RNase-free DNase I (Thermo Fisher Scientific, Waltham, MA, USA) was performed. The total RNA concentration of samples was analyzed using a 260 nm ultraviolet spectrophotometer (NanoDrop ND-1000, Thermo Fisher Scientific, Wilmington, DE, USA). The integrity and quality of the RNA was analyzed using an Agilent 2100 Bioanalyzer (Agilent Biotechnologies Inc., Santa Clara, CA, USA) and only samples with RIN >7 and RNA amount higher than 1 μg were sent for analysis. Prior to being sent to the sequencing facility, the samples were stored at −80°C.
Next generation sequencing was performed on all of RNA skin samples of the dogs, taken at the baseline (n = 8) and end of the diet intervention (n = 8). One microgram (μg) of total RNA was ribodepleted and an RNA-Seq library was created using a ScriptSeq v2™ Complete kit for human/mouse/rat (Illumina, Inc., San Diego, CA, USA). Paired-end library creation and transcriptome sequencing were completed at the Institute for Molecular Medicine Finland (FIMM). Libraries were quality controlled by High Sensitivity chips by Agilent Bioanalyzer (Agilent) before being sequenced on an Illumina HiSeq platform (HiSeq 2000, Illumina, Inc., San Diego, CA, USA).
The bioinformatic analysis was performed at the Plataforma Andaluza de Bioinformática (University of Málaga, Spain). Quality control and initial pre-processing was performed using SeqTrimNext (v. 2.0.53), where low quality, ambiguous, low complexity stretches, adaptor, organelle DNA, polyA/polyT tails, and contaminating sequences were removed (51). Mapping was performed with restrictive conditions of bowtie2 (v.2.2.2), including parameters for rejecting discordant alignments and optimized for paired-end reads (52). Samtools (v.0.1.19) quantified known transcripts (count reads per transcripts) (53), and transcripts were annotated using Full-Lengther-Next (54). Statistical comparisons of transcript expression between diet groups and between atopic and healthy dogs were performed using DEgenes-Hunter (v.2.0.11) (55), a tool that imputes raw read counts generated by Bowtie2/Samtools into the EdgeR (56) and DESeq2 (57) algorithms. Fold change (FC) ≥2, and a false discovery rate (FDR) corrected p < 0.05 were set as thresholds. Differentially expressed transcripts common to both analyses (EdgeR and DESeq) were also identified. RNA-Seq can detect a higher percentage of differentially expressed genes compared to expression arrays, especially genes with low abundance. Typically, in many controlled experiments which utilize RNA-Seq, the number of biological samples is a limiting factor (57) which ultimately restricts the statistical inference to the largest gene expression differences between the groups. Specific algorithms have been developed to improve statistical inference in RNA-Seq datasets with small sample sizes (56–58). EdgeR and DESeq2 are shown to have the highest sensitivity to detect true differences between group means in read count datasets produced by RNA-Seq methods (57, 58). These tools utilize different approaches in calculation of dispersion estimates for read count datasets, and algorithms are differently affected by outlying observations (58).
The comparisons related to dogs' characteristics between the two diet groups were analyzed using the Mann-Whitney U test, Fisher's Exact test and the Wilcoxon signed ranks test. SPSS software (version 25, IBM SPSS Statistics. Chicago, Ill., USA) was used for the statistical analyses. The statistical significance threshold was set at P < 0.05.
There were no statistical differences between the comparison groups (n = 8) in any of the recorded descriptive data of the dogs (Tables 2, 3). The mean weight of the dogs in the atopic (n = 4) and healthy cohorts (n = 4) did not change significantly during the trial (p = 0.180 and 0.141, respectively; Table 2), or in the KD-fed and RMBD-fed cohorts (p = 0.285 and 0.102, respectively; Table 3). The diet intervention lasted 84–147 days (median 137 days).
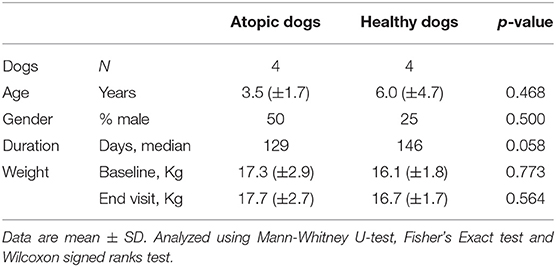
Table 2. Descriptive data and statistical analysis of the atopic and healthy dogs used in the study comparing the skin gene expression profiles (n = 8).
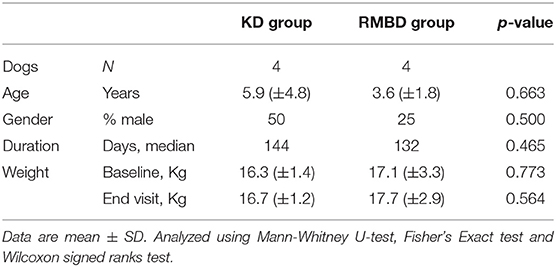
Table 3. Descriptive data and statistical analysis of the kibble diet (KD) and raw meat-based diet (RMBD) fed dogs used in the study comparing the skin gene expression profiles (n = 8).
An average of 52.2 million sequencing reads were obtained per sample, ranging from 40.7 to 69.4 million reads. Mean read length obtained from sequencing was 93.8 bp after pre-processing quality trimming. An average of 93.47% of the reads were mapped to the canine reference genome. The obtained read counts were then uploaded into the DEgenes-Hunter tool, where comparisons using two different software (DESeq2 and EdgeR) were performed.
At the baseline visit, three downregulated genes were found in atopic dogs compared to healthy dogs by EdgeR. Four downregulated genes and one upregulated gene were found by DESeq2 in atopic dogs compared to healthy dogs (Table 4).
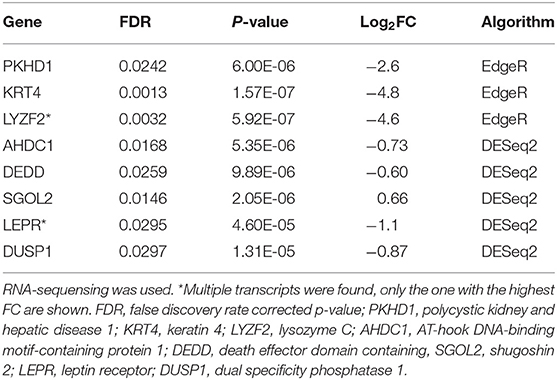
Table 4. Differentially expressed genes in the skin of atopic dogs (n = 4) compared to healthy dogs (n = 4) at the baseline visit.
After the diet intervention, EdgeR found 200 transcripts, and DeSeq found 451 transcripts that were differentially expressed between atopic and healthy groups after the diet intervention (S3), of which 149 differentially expressed transcripts between groups were found by both EdgeR and DESeq2 (Figure 1A; Supplementary Table 3). According to the analyses of their biological function, 69 of the differentially expressed transcripts were involved in dermatological and inflammatory conditions, of which 8 were associated with atopic dermatitis based on the analysis of biological functions (Figure 1B). Signaling pathways affected by gene dysregulation in the atopic group included angiopoietin signaling, epidermal growth factor signaling, AMP-activated protein kinase signaling, retinoid X receptor (RXR)/farnesoid X receptor (FXR) signaling, and leptin signaling in obesity. Also, upregulation of EGF, AKT3, KLB, ANGPT1, and TEK led to the predicted activation (z-score = 2.23) of the IL-8 inflammatory pathway (Figure 1C).
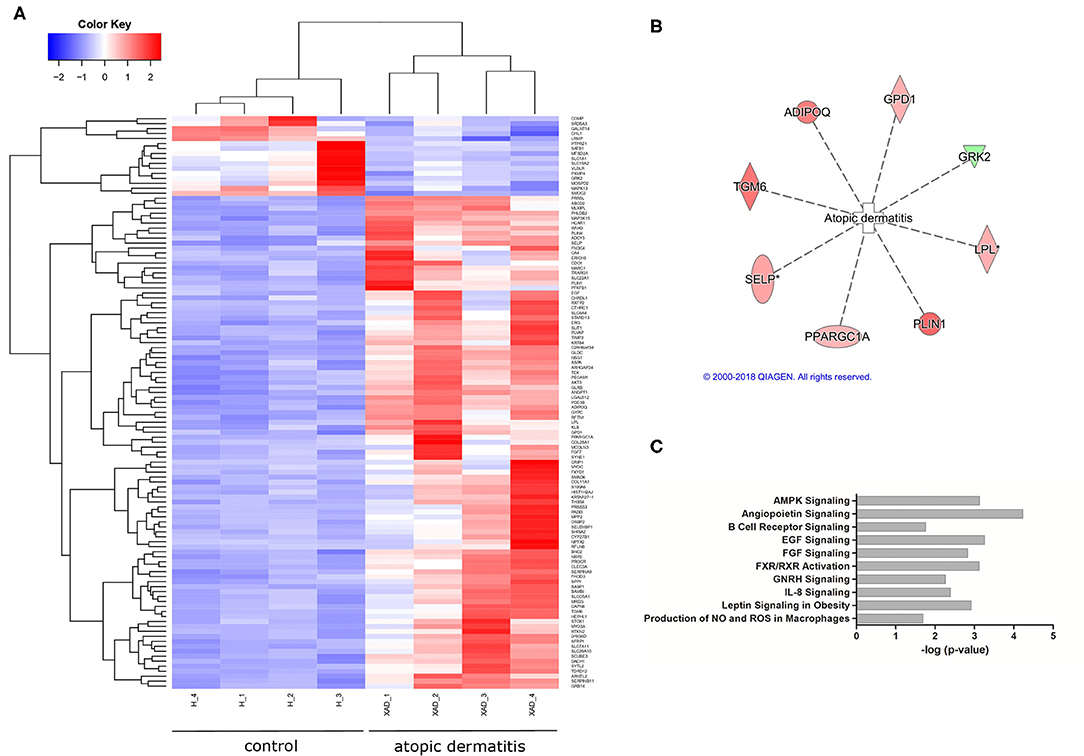
Figure 1. Differential gene expression in atopic vs. healthy dogs (n = 8). (A) A heatmap showing differentially expressed genes in atopic and healthy dogs (downregulated in blue, upregulated in red). (B) A total of eight genes associated with atopic dermatitis were found differentially expressed in atopic dogs after the diet trial (green indicates downregulation, red indicates increased expression). (C) Signaling pathways affected by gene dysregulation in dogs with atopic dermatitis compared to healthy dogs.
Altogether 33 differently expressed transcripts were found between the diet groups, of which 8 genes are annotated in the current version of the dog genome: immunoglobulin heavy constant mu (IGHM), immunoglobulin lambda-like polypeptide 5 (IGLL5), B-cell antigen receptor complex-associated protein beta chain (CD79B), polymeric immunoglobulin receptor (PIGR), cystathionine β-synthase (CBS), arginosuccinate synthase 1 (ASS1), secretory leukocyte peptidase inhibitor (SLPI), and mitochondrial ribosome recycling factor (MRRF). These genes (n = 8) were all upregulated in the RMBD group compared to the KD group (Table 5). They were then studied regarding their biological implication. Activation of ASS1 and CBS in the raw diet group indicated upregulation of cystein biosynthesis and methionine degradation, the citrulline-nitric oxide cycle, the urea cycle, and arginine biosynthesis (Figure 2). Biological functions activated by CBS included for example concentration of glutathione, conversion of homocysteine, and concentration of phospholipids, and inhibited by CBS included for example oxidative stress, hyperkeratosis, and accumulation of reactive oxygen species (Figure 3). In addition, the expression of IGHM, IGLL5, and CD79B activates the differentiation of B lymphocytes and the memory immune response, determines the quantity of different immunoglobulins, inhibits hypoplasia of the lymphoid organ, determines the quantity and secretion of autoantibody, inflammation of secretory structure, and lesioning of skin (Figure 4).
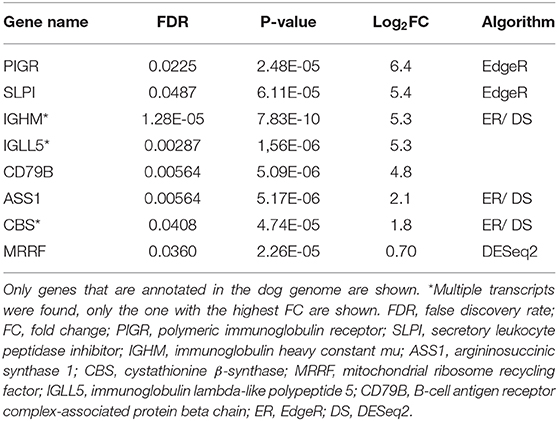
Table 5. Differentially expressed genes in raw meat-based diet fed dogs compared to kibble diet fed dogs after the diet intervention (n = 8).
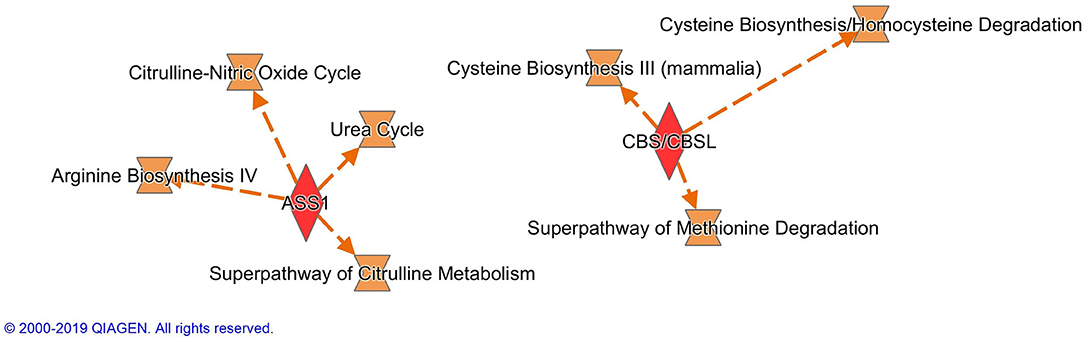
Figure 2. Canonical pathways activated by the expression of ASS1 and CBS in the raw meat-based diet fed dogs. ASS1, argininosuccinate synthase 1; CBS, cystathionine β-synthase.
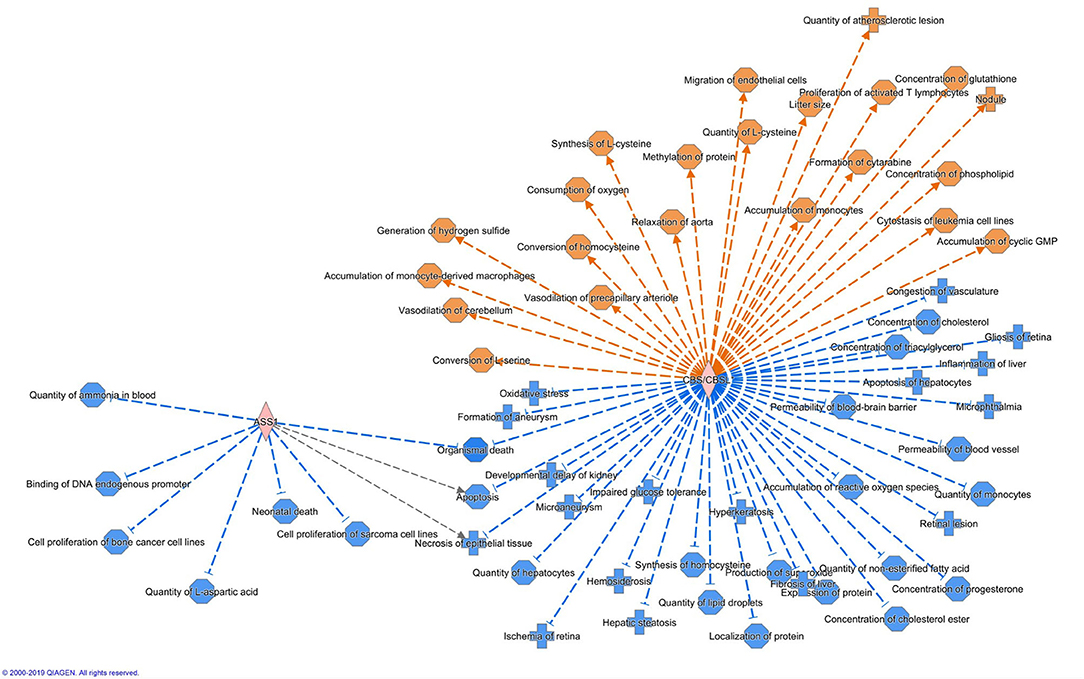
Figure 3. Biological functions activated (in orange) or inhibited (in blue) by the expression of ASS1 and CBS in the raw meat-based diet fed dogs. ASS1, argininosuccinate synthase 1; CBS, cystathionine β-synthase.
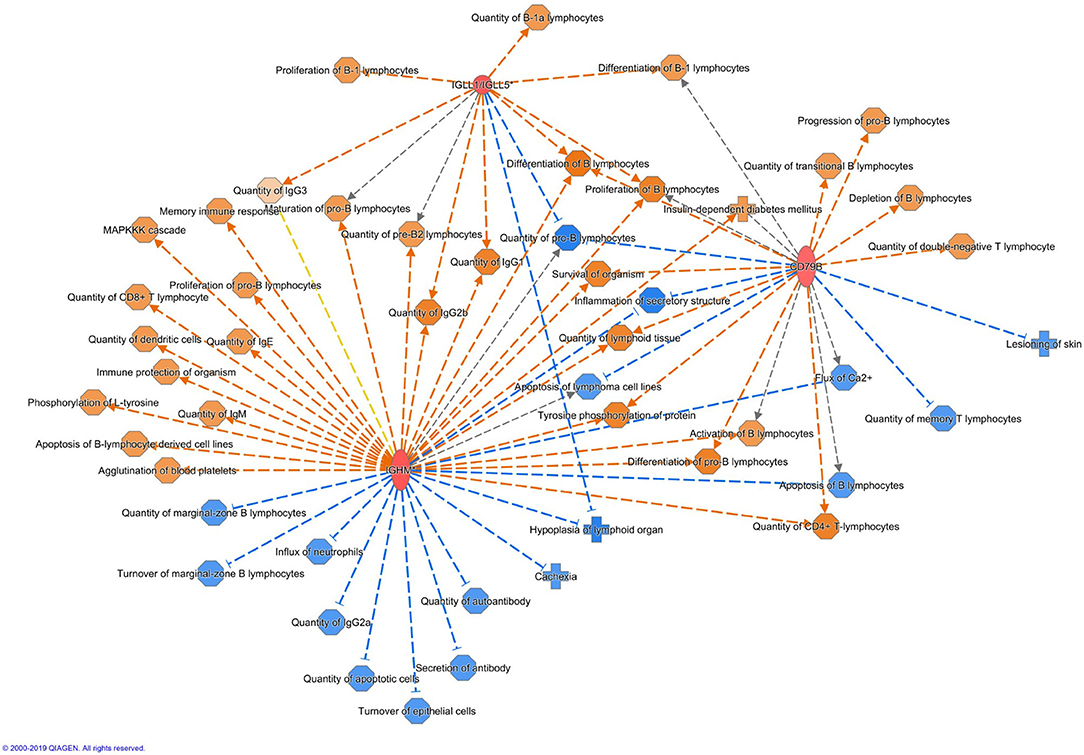
Figure 4. Biological functions activated (in orange) or inhibited (in blue) by the expression of IGHM, IGLL5, and CD79B in the raw meat-based diet fed dogs. IGHM, immunoglobulin heavy constant mu; IGLL5, immunoglobulin lambda-like polypeptide 5; CD79B, B-cell antigen receptor complex-associated protein beta chain.
The activation of immunological pathways such as IL-7 signaling, and B-cell related pathways were also found. Upregulation of (natural) IgM antibodies can result from enhanced IL-7 production by epithelial cells in the gastro-intestinal tract, where they are then shuttled across the epithelium into the lumen as a result of upregulated poly Ig receptor expression (59).
To our best knowledge, the present study is the first RNA-Seq study of altered skin gene expression between dogs fed two different diets. Although the sample sizes in this study was limited, differences were found both between the atopic and healthy groups and between the diet groups. There were many more differentially expressed genes found between the same atopic and healthy dogs at the end of the diet intervention than at baseline. This suggests that the diet is an important background factor which should be considered when studying the gene expression of animals with skin diseases.
In dogs suffering from CAD, the defective skin barrier is believed to facilitate the penetration of allergens into the skin. This then leads to sensitization against environmental allergens and subsequent cutaneous inflammation, which then further aggravates the impairment of the skin barrier (60). Lipids in the skin are important to its barrier function as they help prevent transepidermal water loss (TEWL) (61). Expression of galectin 12 (LGALS12), hydroxycarboxylic acid receptor 1 (HCAR1) and ATP Binding Cassette Subfamily D Member 2 (ABCD2) were upregulated in the atopic dogs in the present study. LGALS12 and HCAR1 suppress lipolysis (62, 63). As part of the barrier formation process, keratinocytes secrete lipids through lipolysis (64). Very-long chain fatty acids (VLCFA) act as a water barrier and are important in forming a cornified envelope, both of which help maintain the integrity and functionality of the skin (65, 66). ABCD2 is suggested to have a role in a transporting VLCFAs to peroxisomes for degradation by β-oxidation (67). These results suggest that the lipid metabolism in the skin of atopic dogs might have been impaired, detrimentally affecting skin barrier function.
The cornified envelope formation of the skin also requires the differentiation of keratinocytes (61, 68). This differentiation is regulated by the epidermal differentiation complex (EDC). Another gene cluster located in the EDC is the S100 gene family. A higher expression of S100 calcium binding protein A6 (S100A6) was found in the non-lesional skin of atopic dogs compared to healthy dogs. One member of the S100 gene family, S100A8, has been previously shown to be associated with lesional skin of atopic dogs (38–50). A recent in vitro study showed that the overexpression of S100A6 results in a less differentiated keratinocyte phenotype and thus disturbs the differentiation process (69). Additionally, one of the epigenetic regulators needed for the terminal differentiation of keratinocytes, special AT-rich sequence-binding protein 1 (SATB1) (70), was downregulated in the skin of atopic dogs in the present study. These results indicate that the differentiation of keratinocytes in the skin of atopic dogs might be dysfunctional. Altered gene expression of keratins (KRTs) are found in both the skin of atopic dogs (39, 40, 71) and human AD patients (39, 72). In the present study, a transcript of the KRT4 gene was highly downregulated, and the KRT84 gene was overexpressed in all atopic dogs compared to healthy dogs at the diet intervention baseline. An association of KRT84 with CAD has not been previously reported.
In the present study, angiopoietin signaling was the most upregulated canonical pathway found in the atopic dogs. Angiogenesis has been reported to play a role in atopic dermatitis (73–75). In the present study AKT serine/threonine kinase 3 (AKT3), angiopoietin 1 (ANGPT1), tyrosine receptor kinase (TEK), and secreted phosphoprotein 1 (SPP1) were upregulated in the skin of atopic dogs, all of which are known to regulate angiogenesis (76–78). ANGPT1 has been shown to be upregulated in the skin of the AD mouse model (75). Together with TEK, ANGPT1 has also been reported to be upregulated in psoriatic skin in humans (79). Elevated expression of SPP1 in the skin of psoriatic patients has been reported, but the expression of SPP1 in lesional skin of AD patients was not observed (80). Hence the upregulated angiogenesis found in the present study comports with previous research.
The upregulation of several immunity-related genes was found in the RMBD fed dogs at the end of the diet intervention. The IGHM gene encodes the C region of the mu heavy chain, which defines the IgM isotype. As immunoglobulin M's (IgM's) are the first antibodies to be produced in an ongoing immune response to infection or immunization (81), the upregulation of IGHM in the RMBD fed group may indicate activation of humoral immune mechanisms. IgM antibodies are generally polyspecific and have low binding affinities and reflect an increased innate immune defense. Together with IGLL5, IGHM regulates both the quantity of different immunoglobulins and inhibits the hypoplasia of lymphoid organs. Together with CD79B, IGHM increases lymphoid tissue quantity. CD79B inhibits biological functions such as skin lesioning and inflammation of secretory structures. Activation of these three genes also increases the proliferation of B lymphocytes. Playing an important role in the mucosal immune system, PIGR transports polymeric immunoglobulins to the apical surfaces of epithelia (82). While IgM and IgA antibodies only have a limited antigen specificity, they generally show a large bystander response. Thus, secretory IgMs and IgAs in the gastrointestinal tract are polyreactive against primarily commensal bacteria and most of these “natural” anti-commensal secretory Igs (sIgs) are made through T cell-independent B cell responses (83). Secretions collected at mucosal surfaces contain significant proportions of IgA due to passive transudation, reflecting the degree of mucosal inflammation. The sIgA is generated at the cleavage site of PIGR and acts as an inhibitory factor against bacteria on the skin surface. In human AD, abnormalities in sIgA have been reported (84). Previous studies have indicated that PIGR and its secretory component have an anti-inflammatory role in inflammatory skin diseases (85–87).
In the citrulline-nitric oxide (NO) cycle, found in many cells, ASS1 is a rate-limiting enzyme for nitric oxide synthesis (88). The expression of ASS1 in macrophages and neutrophils can be upregulated in response to bacterial lipopolysaccharides, and hence also contributes to the innate immune defense (89). Although bacteria present in the RMBD may have enhanced the expression of ASS1, its expression is also necessary for optimal control of persistent pathogens (90), which may be beneficial to dogs suffering from CAD. NO functions as an antimicrobial (91) and can initiate human keratinocyte differentiation (92), which is necessary for the proper development of a functional skin barrier (93). It has been suggested that high concentrations of NO are functionally important for the resolution of chronic inflammatory processes (42, 94).
SLPI, which is a serine protease inhibitor and an antimicrobial peptide, was upregulated in the RMBD group. Lancto et al. (42) found a lower expression of SLPI both in the lesional and non-lesional skin of atopic dogs compared to healthy dogs. Considering the roles of IGHM, PIGR, ASS1, and SLPI, the RMBD may have both enhanced the dogs' innate immunity and improved barrier function of the skin. Consumption of certain dietary constituents found in the RMBD, namely water-soluble vitamins and amino acids, have been previously shown to positively affect the skin barrier by decreasing TEWL in dogs (93). It remains unclear whether the effect seen in the present study was due to the quantity of certain nutrients, or due to the differences in fat or moisture content of the diets.
CBS catalyzes a reaction in which serine and L-homocysteine (Hcy) are condensed to cystathionine and subsequently converted to cysteine, which is the limiting reagent in the production of glutathione, an important antioxidant (95–97). Inflammatory processes cause alterations to this pathway (97) and reduced glutathione production is associated with an increased vulnerability to oxidative stress (96). Elevated oxidative stress and immune dysfunction, which eventually leads to skin damage, appears to also play a role in the pathophysiology of atopic dermatitis. Clinical symptoms of atopic dermatitis can thus be mitigated by increasing antioxidant levels, as they help reduce oxidative damage. A study recently reported that cystathionine and cysteine inhibit the upregulation of proinflammatory mediators in human keratinocytes (98). Additionally, MRRF depletion results in elevated ROS production and cellular dysfunction (99). In our study, the expression of CBS and MRRF suggests that the RMBD may have inhibited ROS production in skin cells. Oxidative stress plays an important role in the pathogenesis of atopic dermatitis in mammals (100, 101), and the induction of oxidative stress is related to both excessive levels of ROS and to deficiencies within the antioxidant system (102). The possible effects of increased CBS and MRRF activity might offer therapeutic value to atopic dogs, since ROS production is increased in canine atopic dermatitis during the inflammatory process (103).
Anderson et al. (104) studied gene expression profiles of peripheral blood mononuclear cells from dogs fed either a kibble diet (n = 8) or a raw red meat diet (n = 7) using Agilent Canine 4 × 44 k microarrays. Their results indicated that a short-term (3 week) diet influenced gene expression at the system level, and that the kibble diet was proinflammatory and the raw red meat diet had anti-inflammatory effects. The comparison of the RMBD and KD in the present study showed that the differentially expressed genes mainly related to immune function, where CBS and PIGR also have anti-inflammatory effects when upregulated. Our findings support the results of Anderson et al. (104), demonstrating a similar effect over a longer diet intervention. However, more research is needed to verify this observation.
In the present study, genes upregulated in the skin of RMBD fed dogs were found to be related to innate immune function, inflammation and antioxidants, possibly indicating that their innate immunity was enhanced, and that there was less oxidative stress. Thus, RMBDs may have an important role in preventing hypersensitivities and disturbed immunity in puppyhood (105). Since the raw food was served raw, it may have enhanced the passive innate immunity more than the more sterile and processed KD. This has been suggested as a reason for the protective effect of raw meat-based diets in canine Toxocara canis infections (106). High-protein diets have been shown to be anti-inflammatory in the skin of mice and they might also prove to be beneficial for dogs suffering from allergic skin conditions (107). Secondary skin infections are common in atopic dogs, and antibiotics are often used when treating them. Antibiotic resistant bacteria are an increasing problem in veterinary medicine (108, 109). If the immunity of the skin could be enhanced through diet at a young age, it might decrease the frequency of antibiotic treatments, although further research is warranted.
Since this was a pilot study, the sample size was limited. To counteract this, two different algorithms were used to analyze the data. However, our results might still in part reflect the individual genetic differences between the dogs. Because these dogs were client-owned, it cannot be ruled out that different environmental factors may have affected the results. The KD and RMBDs had both very different macronutrient profiles and ingredients and their comparison was performed intentionally as a test between two common types of canine diet. Although this complicates the interpretation of the results it nevertheless shows the differential effect that diets had on gene expression. A larger sample size with more controlled diets should be used to validate the results of this study.
The present study showed that lipid metabolism and differentiation of keratinocytes were possibly altered in the skin of atopic dogs. Additionally, compared to the KD fed group, the gene transcription profile of dogs induced by the RMBD in this study is consistent with an enhancement of innate immunity and decreased oxidative stress and may have an important role in preventing hypersensitivities and a disturbed immunity. As there were two major factors differentiating the diets, processed vs. non-processed and high carbohydrate vs. high fat, further studies must be conducted to determine which, or to what extent these factors influenced the results seen in the present study.
The RNA-Seq data can be found in the SRA database under accession number SRP110851.
This animal study was reviewed and approved by Animal Experiment Board in Finland (ELLA) (permit number: ESAVI/3244/04.10.07/2013). Written informed consent was obtained from the owners for the participation of their animals in this study.
JA, AH-B, and KE contributed conception and design of the study. JA, SZ-L, and KE did laboratory work. JA, SZ-L, KE, AH-B, and HS analyzed and interpreted the data. JA wrote the first draft of the manuscript. SZ-L, KE, AH-B, and HS wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This study was funded by Svenska Kulturfonden (13/3307-1304). All study raw foods (MUSH Ltd., Pietarsaari, Finland) and part of the dry foods were given to the study for free (Hills via Berner Ltd., Helsinki, Finland). The companies had, however, no input on either planning the study, analyzing the results, or writing the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Institute for Molecular Medicine Finland FIMM Technology Centre and Biokeskus Suomi for the RNA sequencing. The authors thankfully acknowledge the PAB (Andalusian Bioinformatics Platform) center located at the University of Malaga (www.scbi.uma.es) for the bioinformatic analysis, computer resources, technical expertise and assistance provided. Robin Moore was thanked for the language revision.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.552251/full#supplementary-material
Supplementary Table 1. Composition and analytical constituent of the study kibble diet Hill's Science Plan Canine adult sensitive skin with chicken.
Supplementary Table 2. Composition and analytical constituent of the study raw meat-based food Mush Vaisto diets.
Supplementary Table 3. Differentially expressed transcripts, biological functions, and pathways after the diet intervention in atopic vs. healthy dogs.
1. Marsella R, Olivry T, Carlotti DN International Task Force on Canine Atopic Dermatitis. Current evidence of skin barrier dysfunction in human and canine atopic dermatitis. Vet Dermatol. (2011) 22:239–248. doi: 10.1111/j.1365-3164.2011.00967.x
2. Ramms L, Fabris G, Windoffer R, Schwarz N, Springer R, Zhou C, et al. Keratins as the main component for the mechanical integrity of keratinocytes. Proc Natl Acad Sci USA. (2013) 110:18513–8. doi: 10.1073/pnas.1313491110
3. Santoro D, Marsella R, Pucheu-Haston CM, Eisenschenk MN, Nuttall T, Bizikova P. Review: pathogenesis of canine atopic dermatitis: skin barrier and host-micro-organism interaction. Vet Dermatol. (2015) 26:84–e25. doi: 10.1111/vde.12197
4. Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. (2010) 125:16–29. doi: 10.1016/j.jaci.2009.11.008
5. Marsella R, Girolomoni G. Canine models of atopic dermatitis: a useful tool with untapped potential. J Invest Dermatol. (2009) 129:2351–7. doi: 10.1038/jid.2009.98
6. Hillier A, Griffin CE. The ACVD task force on canine atopic dermatitis (I): incidence and prevalence. Vet Immunol Immunopathol. (2001) 81:147–51. doi: 10.1016/S0165-2427(01)00296-3
7. Bellumori TP, Famula TR, Bannasch DL, Belanger JM, Oberbauer AM. Prevalence of inherited disorders among mixed-breed and purebred dogs: 27,254 cases (1995-2010). J Am Vet Med Assoc. (2013) 242:1549–55. doi: 10.2460/javma.242.11.1549
8. Anturaniemi J, Uusitalo L, Hielm-Björkman A. Environmental and phenotype-related risk factors for owner-reported allergic/atopic skin symptoms and for canine atopic dermatitis verified by veterinarian in a Finnish dog population. PLoS ONE. (2017) 12:e0178771. doi: 10.1371/journal.pone.0178771
9. Mazrier H, Vogelnest LJ, Thomson PC, Taylor RM, Williamson P. Canine atopic dermatitis: breed risk in Australia and evidence for a susceptible clade. Vet Dermatol. (2016) 27:167–e42. doi: 10.1111/vde.12317
10. Nødtvedt A, Egenvall A, Bergvall K, Hedhammar A. Incidence of and risk factors for atopic dermatitis in a Swedish population of insured dogs. Vet Rec. (2006) 159:241–6. doi: 10.1136/vr.159.8.241
11. Oberbauer AM, Belanger JM, Bellumori T, Bannasch TL, Famula TR. Ten inherited disorders in purebred dogs by functional breed groupings. Canine Genet Epidemiol. (2015) 2:9. doi: 10.1186/s40575-015-0021-x
12. Picco F, Zini E, Nett C, Naegeli C, Bigler B, Rüfenacht S, et al. A prospective study on canine atopic dermatitis and food-induced allergic dermatitis in Switzerland. Vet Dermatol. (2008) 19:150–5. doi: 10.1111/j.1365-3164.2008.00669.x
13. Zur G, Ihrke PJ, White SD, Kass PH. Canine atopic dermatitis: a retrospective study of 266 cases examined at the University of California, Davis, 1992-1998. Part I Clinical features and allergy testing results. Vet Dermatol. (2002) 13:89–102. doi: 10.1046/j.1365-3164.2002.00285.x
14. Afifi L, Danesh MJ, Lee KM, Beroukhim K, Farahnik B, Ahn RS, et al. Dietary Behaviors in Psoriasis: Patient-Reported Outcomes from a U.S. National Survey. Dermatol Ther. (2017) 7:227–42. doi: 10.1007/s13555-017-0183-4
15. Johnston GA, Bilbao RM, Graham-Brown RA. The use of dietary manipulation by parents of children with atopic dermatitis. Br J Dermatol. (2004) 150:1186–9. doi: 10.1111/j.1365-2133.2004.05888.x
16. Nosrati A, Afifi L, Danesh MJ, Lee K, Yan D, Beroukhim K, et al. Dietary modifications in atopic dermatitis: patient-reported outcomes. J Dermatolog Treat. (2017) 28:523–38. doi: 10.1080/09546634.2016.1278071
17. Dell DL, Griffin CE, Thompson LA, Griffies JD. Owner assessment of therapeutic interventions for canine atopic dermatitis: a long-term retrospective analysis. Vet Dermatol. (2012) 23:228–e47. doi: 10.1111/j.1365-3164.2012.01054.x
18. Bizikova P, Pucheu-Haston CM, Eisenschenk MN, Marsella R, Nuttall T, Santoro D. Review: role of genetics and the environment in the pathogenesis of canine atopic dermatitis. Vet Dermatol. (2015) 26:95–e26. doi: 10.1111/vde.12198
19. Kawano K, Oumi K, Ashida Y, Horiuchi Y, Mizuno T. The prevalence of dogs with lymphocyte proliferative responses to food allergens in canine allergic dermatitis. Pol J Vet Sci. (2013) 16:735–9. doi: 10.2478/pjvs-2013-0104
20. Nødtvedt A, Bergvall K, Sallander M, Egenvall A, Emanuelson U, Hedhammar A. A case-control study of risk factors for canine atopic dermatitis among boxer, bullterrier and West Highland white terrier dogs in Sweden. Vet Dermatol. (2007) 18:309–15. doi: 10.1111/j.1365-3164.2007.00617.x
21. Popa I, Pin D, Remoué N, Osta B, Callejon S, Videmont E, et al. Analysis of epidermal lipids in normal and atopic dogs, before and after administration of an oral omega-6/omega-3 fatty acid feed supplement. A pilot study. Vet Res Commun. (2011) 35:501–9. doi: 10.1007/s11259-011-9493-7
22. Jaffary F, Faghihi G, Mokhtarian A, Hosseini SM. Effects of oral vitamin E on treatment of atopic dermatitis: a randomized controlled trial. J Res Med Sci. (2015) 20:1053–7. doi: 10.4103/1735-1995.172815
23. Kim G, Bae JH. Vitamin D and atopic dermatitis: a systematic review and meta-analysis. Nutrition. (2016) 32:913–20. doi: 10.1016/j.nut.2016.01.023
24. Mohajeri S, Newman SA. Review of evidence for dietary influences on atopic dermatitis. Skin Therapy Lett. (2014) 19:5–7.
25. Klinger CJ, Hobi S, Johansen C, Koch HJ, Weber K, Mueller RS. Vitamin D shows in vivo efficacy in a placebo-controlled, double-blinded, randomised clinical trial on canine atopic dermatitis. Vet Rec. (2018) 182:406. doi: 10.1136/vr.104492
26. Plevnik Kapun A, Salobir J, Levart A, Tavčar Kalcher G, Nemec Svete A, Kotnik T. Vitamin E supplementation in canine atopic dermatitis: improvement of clinical signs and effects on oxidative stress markers. Vet Rec. (2014) 175:560. doi: 10.1136/vr.102547
27. Saevik BK, Bergvall K, Holm BR, Saijonmaa-Koulumies LE, Hedhammar A, Larsen S, et al. A randomized, controlled study to evaluate the steroid sparing effect of essential fatty acid supplementation in the treatment of canine atopic dermatitis. Vet Dermatol. (2004) 15:137–45. doi: 10.1111/j.1365-3164.2004.00378.x
28. Kaikiri H, Miyamoto J, Kawakami T, Park SB, Kitamura N, Kishino S, et al. Supplemental feeding of a gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, alleviates spontaneous atopic dermatitis and modulates intestinal microbiota in NC/nga mice. Int J Food Sci Nutr. (2017) 68:941–51. doi: 10.1080/09637486.2017.1318116
29. Wang P, Sun M, Ren J, Aslam MN, Xu Y, Fisher GJ, et al. Dietary fish oil supplementation enhances expression of genes involved in cornified cell envelope formation in rat skin. J Invest Dermatol. (2018) 138:981–3. doi: 10.1016/j.jid.2017.11.019
30. Fabbrocini G, Bertona M, Picazo Ó, Pareja-Galeano H, Monfrecola G, Emanuele E. Supplementation with Lactobacillus rhamnosus SP1 normalises skin expression of genes implicated in insulin signalling and improves adult acne. Benef Microbes. (2016) 7:625–30. doi: 10.3920/BM2016.0089
31. Scott JF, Das LM, Ahsanuddin S, Qiu Y, Binko AM, Traylor ZP, et al. Oral vitamin D rapidly attenuates inflammation from sunburn: an interventional study. J Invest Dermatol. (2017) 137:2078–86. doi: 10.1016/j.jid.2017.04.040
32. Veenhof EZ, Knol EF, Schlotter YM, Vernooij JC, Rutten VP, Willemse T. Characterisation of T cell phenotypes, cytokines and transcription factors in the skin of dogs with cutaneous adverse food reactions. Vet J. (2011) 187:320–4. doi: 10.1016/j.tvjl.2010.02.005
33. Kil DY, Vester Boler BM, Apanavicius CJ, Schook LB, Swanson KS. Age and diet affect gene expression profiles in canine liver tissue. PLoS ONE. (2010) 5:e13319. doi: 10.1371/journal.pone.0013319
34. Middelbos IS, Vester BM, Karr-Lilienthal LK, Schook LB, Swanson KS. Age and diet affect gene expression profile in canine skeletal muscle. PLoS ONE. (2009) 4:e4481. doi: 10.1371/journal.pone.0004481
35. Swanson KS, Belsito KR, Vester BM, Schook LB. Adipose tissue gene expression profiles of healthy young adult and geriatric dogs. Arch Anim Nutr. (2009) 63:160–71. doi: 10.1080/17450390902733934
36. Kil DY, Vester Boler BM, Apanavicius CJ, Schook LB, Swanson KS. Gene expression profiles of colonic mucosa in healthy young adult and senior dogs. PLoS ONE. (2010) 5:e12882. doi: 10.1371/journal.pone.0012882
37. Swanson KS, Vester BM, Apanavicius CJ, Kirby NA, Schook LB. Implications of age and diet on canine cerebral cortex transcription. Neurobiol Aging. (2009) 30:1314–26. doi: 10.1016/j.neurobiolaging.2007.10.017
38. Merryman-Simpson AE, Wood SH, Fretwell N, Jones PG, McLaren WM, McEwan NA, et al. Gene (mRNA) expression in canine atopic dermatitis: microarray analysis. Vet Dermatol. (2008) 19:59–66. doi: 10.1111/j.1365-3164.2008.00653.x
39. Plager DA, Torres SM, Koch SN, Kita H. Gene transcription abnormalities in canine atopic dermatitis and related human eosinophilic allergic diseases. Vet Immunol Immunopathol. (2012) 149:136–42. doi: 10.1016/j.vetimm.2012.06.003
40. Theerawatanasirikul S, Sailasuta A, Thanawongnuwech R, Suriyaphol G. Alterations of keratins, involucrin and filaggrin gene expression in canine atopic dermatitis. Res Vet Sci. (2012) 93:1287–92. doi: 10.1016/j.rvsc.2012.06.005
41. Klukowska-Rötzler J, Chervet L, Müller EJ, Roosje P, Marti E, Janda J. Expression of thymic stromal lymphopoietin in canine atopic dermatitis. Vet Dermatol. (2013) 24:54–9. doi: 10.1111/j.1365-3164.2012.01096.x
42. Lancto CA, Torres SM, Hendrickson JA, Martins KV, Rutherford MS. Altered expression of antimicrobial peptide genes in the skin of dogs with atopic dermatitis and other inflammatory skin conditions. Vet Dermatol. (2013) 24:414–21. doi: 10.1111/vde.12034
43. Mullin J, Carter S, Williams N, McEwan N, Nuttall T. Transcription of canine toll-like receptor 2, β-defensin 1 and β-defensin 103 in infected atopic skin, non-infected atopic skin, healthy skin and the CPEK cell line. Vet Microbiol. (2013) 162:700–6. doi: 10.1016/j.vetmic.2012.09.017
44. Nuttall TJ, Knight PA, McAleese SM, Lamb JR, Hill PB. Expression of Th1, Th2 and immunosuppressive cytokine gene transcripts in canine atopic dermatitis. Clin Exp Allergy. (2002) 32:789–95. doi: 10.1046/j.1365-2222.2002.01356.x
45. Roque JB, O'Leary CA, Kyaw-Tanner M, Duffy DL, Shipstone M. Real-time PCR quantification of the canine filaggrin orthologue in the skin of atopic and non-atopic dogs: a pilot study. BMC Res Notes. (2011) 4:554. doi: 10.1186/1756-0500-4-554
46. Santoro D, Marsella R, Ahrens K, Graves TK, Bunick D. Altered mRNA and protein expression of filaggrin in the skin of a canine animal model for atopic dermatitis. Vet Dermatol. (2013) 24:329–36. doi: 10.1111/vde.12031
47. Schamber P, Schwab-Richards R, Bauersachs S, Mueller RS. Gene expression in the skin of dogs sensitized to the house dust mite Dermatophagoides farinae. G3 (Bethesda). (2014) 4:1787–95. doi: 10.1534/g3.114.013003
48. Schlotter YM, Rutten VP, Riemers FM, Knol EF, Willemse T. Lesional skin in atopic dogs shows a mixed Type-1 and Type-2 immune responsiveness. Vet Immunol Immunopathol. (2011) 143:20–6. doi: 10.1016/j.vetimm.2011.05.025
49. van Damme CM, Willemse T, van Dijk A, Haagsman HP, Veldhuizen EJ. Altered cutaneous expression of beta-defensins in dogs with atopic dermatitis. Mol Immunol. (2009) 46:2449–55. doi: 10.1016/j.molimm.2009.05.028
50. Wood SH, Clements DN, Ollier WE, Nuttall T, McEwan NA, Carter SD. Gene expression in canine atopic dermatitis and correlation with clinical severity scores. J Dermatol Sci. (2009) 55:27–33. doi: 10.1016/j.jdermsci.2009.03.005
51. Falgueras J, Lara AJ, Fernández-Pozo N, Cantón FR, Pérez-Trabado G, Claros MG. SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinformatics. (2010) 11:38. doi: 10.1186/1471-2105-11-38
52. Hoeppner MP, Lundquist A, Pirun M, Meadows JR, Zamani N, Johnson J, et al. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS ONE. (2014) 9:e91172. doi: 10.1371/journal.pone.0091172
53. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
54. Supercomputing and Bioinnovation Center (SBC) (2018). Available online at: http://www.scbi.uma.es/site/scbi/downloads/313-full-lengthernext; (accessed February 15, 2018).
55. González Gayte I, Moreno RC, Zonjic RS, Claros MG. DEgenes Hunter - a flexible r pipeline for automated RNA-seq studies in organisms without reference genome. Genomics Comput Biol. 2365–7154. doi: 10.18547/gcb.2017.vol3.iss3.e31
56. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550doi: 10.1186/s13059-014-0550-8
57. Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res. (2014) 42:e91. doi: 10.1093/nar/gku310
58. Spencer J, Sollid LM. The human intestinal B-cell response. Mucosal Immunol. (2016) 9:1113. doi: 10.1038/mi.2016.59
59. Boguniewicz M, Leung DY. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev. (2011) 242:233–46. doi: 10.1111/j.1600-065X.2011.01027.x
60. Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. (2005) 6:328–40. doi: 10.1038/nrm1619
61. Cai TQ, Ren N, Jin L, Cheng K, Kash S, Chen R, et al. Role of GPR81 in lactate-mediated reduction of adipose lipolysis. Biochem Biophys Res Commun. (2008) 377:987–91. doi: 10.1016/j.bbrc.2008.10.088
62. Harrison WJ, Bull JJ, Seltmann H, Zouboulis CC, Philpott MP. Expression of lipogenic factors galectin-12, resistin, SREBP-1, and SCD in human sebaceous glands and cultured sebocytes. J Invest Dermatol. (2007) 127:1309–17. doi: 10.1038/sj.jid.5700743
63. Yang RY, Yu L, Graham JL, Hsu DK, Lloyd KC, Havel PJ, et al. Ablation of a galectin preferentially expressed in adipocytes increases lipolysis, reduces adiposity, and improves insulin sensitivity in mice. Proc Natl Acad Sci USA. (2011) 108:18696–701. doi: 10.1073/pnas.1109065108
64. Mizutani Y, Mitsutake S, Tsuji K, Kihara A, Igarashi Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie. (2009) 91:784–90. doi: 10.1016/j.biochi.2009.04.001
65. Fourcade S, Ruiz M, Camps C, Schlüter A, Houten SM, Mooyer PA, et al. A key role for the peroxisomal ABCD2 transporter in fatty acid homeostasis. Am J Physiol Endocrinol Metab. (2009) 296:E211–21. doi: 10.1152/ajpendo.90736.2008
66. Fürstenberger G, Epp N, Eckl KM, Hennies HC, Jørgensen C, Hallenborg P, et al. Role of epidermis-type lipoxygenases for skin barrier function and adipocyte differentiation. Prostaglandins Other Lipid Mediat. (2007) 82:128–34. doi: 10.1016/j.prostaglandins.2006.05.006
67. Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nature Rev Mol Cell Biol. (2003) 4:140–56. doi: 10.1038/nrm1014
68. Graczyk A, Leśniak W. S100A6 expression in keratinocytes and its impact on epidermal differentiation. Int J Biochem Cell Biol. (2014) 57:135–41. doi: 10.1016/j.biocel.2014.10.007
69. Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol. (2011) 194:825–39. doi: 10.1083/jcb.201101148
70. Fuchs E. Keratins and the skin. Annu Rev Cell Dev Biol. (1995) 11:123–53. doi: 10.1146/annurev.cb.11.110195.001011
71. Sugiura H, Ebise H, Tazawa T, Tanaka K, Sugiura Y, Uehara M, et al. Large-scale DNA microarray analysis of atopic skin lesions shows overexpression of an epidermal differentiation gene cluster in the alternative pathway and lack of protective gene expression in the cornified envelope. Br J Dermatol. (2005) 152:146–9. doi: 10.1111/j.1365-2133.2005.06352.x
72. Zhang Y, Matsuo H, Morita E. Increased production of vascular endothelial growth factor in the lesions of atopic dermatitis. Arch Dermatol Res. (2006) 297:425–9. doi: 10.1007/s00403-006-0641-9
73. Buroker NE, Ning XH, Zhou ZN, Li K, Cen WJ, Wu XF, et al. AKT3, ANGPTL4, eNOS3, and VEGFA associations with high altitude sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau. Int J Hematol. (2012) 96:200–13. doi: 10.1007/s12185-012-1117-7
74. Chen L, Marble DJ, Agha R, Peterson JD, Becker RP, Jin T, et al. The progression of inflammation parallels the dermal angiogenesis in a keratin 14 IL-4-transgenic model of atopic dermatitis. Microcirculation. (2008) 15:49–64. doi: 10.1080/10739680701418416
75. Varricchi G, Granata F, Loffredo S, Genovese A, Marone G. Angiogenesis and lymphangiogenesis in inflammatory skin disorders. J Am Acad Dermatol. (2015) 73:144–53. doi: 10.1016/j.jaad.2015.03.041
76. Kuroda K, Sapadin A, Shoji T, Fleischmajer R, Lebwohl M. Altered expression of angiopoietins and Tie2 endothelium receptor in psoriasis. J Invest Dermatol. (2001) 116:713–20. doi: 10.1046/j.1523-1747.2001.01316.x
77. Puxeddu I, Berkman N, Ribatti D, Bader R, Haitchi HM, Davies DE, et al. Osteopontin is expressed and functional in human eosinophils. Allergy. (2010) 65:168–74. doi: 10.1111/j.1398-9995.2009.02148.x
78. Thomas M, Augustin HG. The role of the angiopoietins in vascular morphogenesis. Angiogenesis. (2009) 12:125–37. doi: 10.1007/s10456-009-9147-3
79. Buommino E, Tufano MA, Balato N, Canozo N, Donnarumma M, Gallo L, et al. Osteopontin: a new emerging role in psoriasis. Arch Dermatol Res. (2009) 301:397–404. doi: 10.1007/s00403-009-0939-5
80. Petrušić V, Zivković I, Stojanović M, Stojićević I, Marinković E, Inić-Kanada A, et al. Antigenic specificity and expression of a natural idiotope on human pentameric and hexameric IgM polymers. Immunol Res. (2011) 51:97–107. doi: 10.1007/s12026-011-8236-8
81. Asano M, Komiyama K. Polymeric immunoglobulin receptor. J Oral Sci. (2011) 53:147–56. doi: 10.2334/josnusd.53.147
82. Turula H, Wobus CE. The role of the polymeric immunoglobulin receptor and secretory immunoglobulins during mucosal infection and immunity. Viruses. (2018) 10:237. doi: 10.3390/v10050237
83. Imayama S, Shimozono Y, Hoashi M, Yasumoto S, Ohta S, Yoneyama K, et al. Reduced secretion of IgA to skin surface of patients with atopic dermatitis. J Allergy Clin Immunol. (1994) 94:195–200. doi: 10.1053/ai.1994.v94.a55222
84. Nihei Y, Maruyama K, Zhang JZ, Kobayashi K, Kaneko F. Secretory component (polymeric immunoglobulin receptor) as an intrinsic inhibitor of biological functions of interferon gamma in keratinocytes. Arch Dermatol Res. (1995) 287:546–52. doi: 10.1007/BF00374074
85. Fernandez MI, Pedron T, Tournebize R, Olivo-Marin J-C, Sansonetti PJ, Phalipon A. Anti-inflammatory role for intracellular dimeric immunoglobulin A by neutralization of lipopolysaccharide in epithelial cells. Immunity. (2003) 18:739–49. doi: 10.1016/S1074-7613(03)00122-5
86. Husson A, Brasse-Lagnel C, Fairand A, Renouf S, Lavoinne A. Argininosuccinate synthetase from the urea cycle to the citrulline-NO cycle. Eur J Biochem. (2003) 270:1887–99. doi: 10.1046/j.1432-1033.2003.03559.x
87. Nihei Y, Maruyama K, Endo Y, Sato T, Kobayashi K, Kaneko F. Secretory component (polymeric immunoglobulin receptor) expression on human keratinocytes by stimulation with interferon-gamma and differences in response. J Dermatol Sci. (1996) 11:214–22. doi: 10.1016/0923-1811(95)00444-0
88. Hattori Y, Campbell EB, Gross SS. Argininosuccinate synthetase mRNA and activity are induced by immunostimulants in vascular smooth muscle. Role in the regeneration or arginine for nitric oxide synthesis. J Biol Chem. (1994) 269:9405–8.
89. Qualls JE, Subramanian C, Rafi W, Smith AM, Balouzian L, DeFreitas AA, et al. Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host Microbe. (2012) 12:313–23. doi: 10.1016/j.chom.2012.07.012
90. Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. (1992) 6:3051–64. doi: 10.1096/fasebj.6.12.1381691
91. Liebmann J, Born M, Kolb-Bachofen V. Blue-light irradiation regulates proliferation and differentiation in human skin cells. J Invest Dermatol. (2010) 130:259–69. doi: 10.1038/jid.2009.194
92. Watson AL, Fray TR, Bailey J, Baker CB, Beyer SA, Markwell PJ. Dietary constituents are able to play a beneficial role in canine epidermal barrier function. Exp Dermatol. (2006) 15:74–81. doi: 10.1111/j.0906-6705.2005.00385.x
93. Bruch-Gerharz D, Schnorr O, Suschek C, Beck KF, Pfeilschifter J, Ruzicka T, et al. Arginase 1 overexpression in psoriasis: limitation of inducible nitric oxide synthase activity as a molecular mechanism for keratinocyte hyperproliferation. Am J Pathol. (2003) 162:203–11. doi: 10.1016/S0002-9440(10)63811-4
94. Bogdan C. Nitric oxide and the immune response. Nat Immunol. (2001) 2:907–16. doi: 10.1038/ni1001-907
95. Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. (2004) 279:29871–4. doi: 10.1074/jbc.R400005200
96. Prudova A, Bauman Z, Braun A, Vitvitsky V, Lu SC, Banerjee R. S-adenosylmethionine stabilizes cystathionine beta-synthase and modulates redox capacity. Proc Natl Acad Sci USA. (2006) 103:6489–94. doi: 10.1073/pnas.0509531103
97. Rosado JO, Salvador M, Bonatto D. Importance of the trans-sulfuration pathway in cancer prevention and promotion. Mol Cell Biochem. (2007) 301:1–12. doi: 10.1007/s11010-006-9389-y
98. Lee E, Kim HJ, Lee M, Jin SH, Hong SH, Ahn S, et al. Cystathionine metabolic enzymes play a role in the inflammation resolution of human keratinocytes in response to sub-cytotoxic formaldehyde exposure. Toxicol Appl Pharmacol. (2016) 310:185–94. doi: 10.1016/j.taap.2016.09.017
99. Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. (2008) 36:5787–5579. doi: 10.1093/nar/gkn576
100. Niwa Y, Sumi H, Kawahira K, Terashima T, Nakamura T, Akamatsu H. Protein oxidative damage in the stratum corneum: evidence for a link between environmental oxidants and the changing prevalence and nature of atopic dermatitis in Japan. Br J Dermatol. (2003) 149:248–54. doi: 10.1046/j.1365-2133.2003.05417.x
101. Tsuboi H, Kouda K, Takeuchi H, Takigawa M, Masamoto Y, Takeuchi M, et al. 8-hydroxydeoxyguanosine in urine as an index of oxidative damage to DNA in the evaluation of atopic dermatitis. Br J Dermatol. (1998) 138:1033–5. doi: 10.1046/j.1365-2133.1998.02273.x
102. Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. (2007) 39:44–84. doi: 10.1016/j.biocel.2006.07.001
103. Kapun AP, Salobir J, Levart A, Kotnik T, Svete AN. Oxidative stress markers in canine atopic dermatitis. Res Vet Sci. (2012) 92:469–70. doi: 10.1016/j.rvsc.2011.04.014
104. Anderson RC, Armstrong KM, Young W, Maclean P, Thomas DG, Bermingham EN. Effect of kibble and raw meat diets on peripheral blood mononuclear cell gene expression profile in dogs. Vet J. (2018) 234:7–10. doi: 10.1016/j.tvjl.2018.01.005
105. Ji H, Li X-K. Oxidative stress in atopic dermatitis. Oxid Med Cell Longev. (2016) 2016:2721469. doi: 10.1155/2016/2721469
106. Nijsse R, Mughini-Gras L, Wagenaar JA, Ploeger HW. Recurrent patent infections with Toxocara canis in household dogs older than six months: a prospective study. Parasit Vectors. (2016) 9:531. doi: 10.1186/s13071-016-1816-7
107. Cui X, Kim E. Dual effects of high protein diet on mouse skin and colonic inflammation. Clin Nutr Res. (2018) 7:56–68. doi: 10.7762/cnr.2018.7.1.56
108. Hensel N, Zabel S, Hensel P. Prior antibacterial drug exposure in dogs with meticillin-resistant Staphylococcus pseudintermedius (MRSP) pyoderma. Vet Dermatol. (2016) 27:72–8e20. doi: 10.1111/vde.12292
109. Hillier A, Lloyd DH, Weese JS, Blondeau JM, Boothe D, Breitschwerdt E, et al. Guidelines for the diagnosis and antimicrobial therapy of canine superficial bacterial folliculitis (Antimicrobial Guidelines Working Group of the International Society for Companion Animal Infectious Diseases). Vet Dermatol. (2014) 25:163–75. doi: 10.1111/vde.12118
Keywords: atopic dermatitis, canine, diet, gene expression, RNAseq, skin, kibble diet, raw meat-based diet
Citation: Anturaniemi J, Zaldívar-López S, Savelkoul HFJ, Elo K and Hielm-Björkman A (2020) The Effect of Atopic Dermatitis and Diet on the Skin Transcriptome in Staffordshire Bull Terriers. Front. Vet. Sci. 7:552251. doi: 10.3389/fvets.2020.552251
Received: 15 April 2020; Accepted: 08 September 2020;
Published: 16 October 2020.
Edited by:
Emma Natasha Bermingham, AgResearch Ltd., New ZealandReviewed by:
Mohamed E. Abd El-Hack, Zagazig University, EgyptCopyright © 2020 Anturaniemi, Zaldívar-López, Savelkoul, Elo and Hielm-Björkman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johanna Anturaniemi, am9oYW5uYS5hbnR1cmFuaWVtaUBoZWxzaW5raS5maQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.