- 1Department of Animal Science, Division of Agriculture, University of Arkansas, Fayetteville, AR, United States
- 2United States Department of Agriculture, Agricultural Research Service, Booneville, AR, United States
- 3Department of Food Science and Center for Food Safety, University of Arkansas, Fayetteville, AR, United States
- 4Department of Food Science and Technology, Oregon State University, Corvallis, OR, United States
- 5Department of Animal Science, University of Tennessee, Knoxville, TN, United States
Tall fescue (Schedonorus arundinaceus) is a cool-season perennial grass that is widely used as a forage for many livestock species including sheep. An endophyte (Neotyphodium coenophialum) in tall fescue produces ergot alkaloids that enhance plant survival but produce toxicosis in animals. The objective of this study was to investigate the rumen microbiota from gestation and lactation in ewes grazing tall fescue pastures with high (HA) or moderate (MA) levels of endophyte infection, and their relationship with serum parameters. Data were collected at the beginning of the study (d1), the week before initiation of lambing (d51), and at the end of the trial (d115). The rumen microbiota was evaluated using 16S rRNA gene sequencing. Ewes grazing HA had greater serum non-esterified fatty acid (NEFA) (P = 0.024) levels compared with ewes in MA pasture at d115. Both the number of observed OTUs and Shannon diversity index tended (P = 0.08, P = 0.06) to be greater for HA than for MA on d115. At the genus level, Prevotella relative abundance increased with time in both MA and HA (on d1, d51, and d115: 15.17, 25.59, and 24.78% in MA; 14.17, 18.10, and 19.41% in HA). Taxa unclassified at the genus level including (unclassified) Lachnospiraceae, Coriobacteriaceae, and Veillonellaceae exhibited higher abundances in HA at d51 (3.72, 2.07, and 11.22%) compared with MA (2.06, 1.28, and 7.42%). The predictor microbiota for HA and MA were identified by a random forest classification model. The HA predictors included bacteria associated with unclassified Coriobacteriaceae and Ruminococcaceae. Other OTUs classified as Prevotella and Clostridiales could be microbial predictors for MA. The OTUs classified as Prevotella and Lachnospiraceae were negatively correlated with serum concentration of prolactin. Negative correlations with NEFA were observed in the microbiota such as species affiliated to unclassified Clostridiales and Prevotella. OTUs classified as Bacteroidetes and Coriobacteriaceae exhibited a positive correlation with NEFA. Our study confirmed that the rumen microbiota populations were affected by high levels of toxins in endophyte-infected tall fescue and were associated with host hormone and energy metabolism.
Introduction
Tall fescue [Schedonorus arundinaceus (Schreb.) Dumort, syn. Festuca arundinacea] is a cool-season perennial grass that has been widely used as a forage for livestock (1). A symbiotic association between tall fescue and the fungus Neotyphodium coenophialum has been detected in more than 90% of tall fescue pastures (2), which increases survival of tall fescue grass during extreme weather conditions (3, 4). However, the fungal endophyte produces a range of mycotoxins that can be detrimental to livestock performance (5, 6). One of the most common classes of mycotoxins associated with N. coenophialum include ergot alkaloids, in which toxicity is typically characterized within livestock by reduced weight gain and volatile fatty acid absorption, restricted blood flow, and reduced reproductive rates (7, 8). Ergovaline is the main ergopeptine alkaloid produced in the endophyte-infected tall fescue, and is considered the primary causative agent of fescue toxicosis in livestock (9, 10). It has been proposed that mechanistically, the structure of ergot alkaloids enables these compounds to act as sympathetic monoamine mimetics for host receptors (11). Ergot alkaloids contain a tetracyclic ergoline ring and are structurally similar to biogenic amines such as serotonin, dopamine, norepinephrine, and epinephrine (11). This structural confirmation enables the compound to ligate receptors for biogenic amines, which have been hypothesized to decrease serum prolactin (PRL) concentrations and induce vasoconstriction via downstream signaling events (12, 13). In addition, the EC50 dose of ergovaline is relatively low, and can induce a series of adverse reactions in animals, such as the reduction of feed efficiency and gastrointestinal passage rates (7).
Previous research has focused on elucidating the effects of endophyte-infected tall fescue on performance and other physiological parameters, but few studies have been conducted to evaluate its effects on the rumen microbiota. It is important to evaluate the effects of these bioactive alkaloids on the rumen and its microbial population, as the microbial population directly aids in providing nutrients to the ruminant host by forage fermentation (i.e., tall fescue) and also impacts lactation energy requirements (14, 15). Rumen microorganisms are able to degrade many of the ergot alkaloids consumed by the hosts; however, the identification of the organisms responsible for toxicity degradation is unknown (16). In an in vitro study conducted by Harlow et al. (17), it was determined that ruminal ammonia-hyperproducing bacteria Peptostreptococcus anaerobius and Clostridum species sporogenes, sticklandii, and aminophilum as well as Prevotella bryantii, albeit to a lesser extent, contribute to the breakdown and metabolism of ergovaline. However, there remains a substantial information gap between the in vitro studies done with the pure bacterial cultures mentioned above and how they may contribute to ergovaline detoxification in the animal. Moreover, little is known regarding potential associations between the in vivo rumen microbiota and the performance and well-being of pregnant ewes grazing tall fescue. Accordingly, the objectives of this study were to evaluate changes to the rumen microbiota from gestation to lactation in ewes grazing tall fescue with high or moderate levels of toxic endophyte infection, and its association with serum parameters.
Materials and Methods
This study was conducted from early February to late May 2016 at the USDA, ARS Dale Bumpers Small Farms Research Center (DBSFRC), Booneville, AR, USA. All experimental procedures and husbandry practices in this experiment were reviewed and approved by the University of Arkansas Institutional Animal Care and Use Committee (Protocol # 16046) and the USDA ARS Institutional Animal Care and Use Committee.
Animal Management and Samples Collection
Fifty Katahdin ewes (ages 2–9 years) confirmed as pregnant via ultrasound were selected from the DBSFRC flock, then stratified by body weight (BW) within age and the number of fetuses and were allocated randomly into three groups. Groups 1 and 3 contained 15 ewes each, with group 2 containing 20 ewes. Groups 1 and 3 were allocated to tall fescue pastures with a high level of endophyte infection (HA; 90% infection), and group 2 was allocated to a tall fescue pasture with moderate endophyte infection (MA; 58% infection). The numbers (58 and 90%) are derived from collecting individual tillers from each pasture (n = 50) and subjecting each individual tiller to the immunoblot test kit that is available commercially. This kit is a presence/absence test for endophyte infection and is widely accepted as being accurate in the determination of the presence of N. coenophialum. This infection was then verified by quantitative analysis of ergovaline and total ergot alkaloids to provide an indication of the level of toxicity of the different pastures. Within each of these larger groups, ewes were selected at random for rumen sampling. This resulted in five ewes selected from group 1 (HA) and three ewes from group 3 (HA) with an average initial BW of 51.6 ± 2.64 kg and an initial average body condition score (BCS; 1 = emaciated and 5 = obese) of 2.8 ± 0.18. Seven ewes were selected randomly from group 2 (MA) with an average BW of 40.6 ± 2.82 kg, and an initial BCS of 2.7 ± 0.19. All ewes had unrestricted access to water and free choice to trace mineralized salt (Supplementary Table 1).
Blood samples were taken from the jugular vein of each ewe directly prior to day 1 (d1), the week before initiation of lambing (d51), and at the end of the trial (d115), which corresponded to ~60 d post-parturition. Blood was collected into serum separator vacuum tubes (BD #367988 Vacutainer®, Becton Dickinson, Inc., Franklin Lakes, NJ, USA). Samples were transported on ice and stored in a standard refrigerator at 4°C overnight. The samples were centrifuged (3,000 × g for 20 min.), followed by transfer to plastic tubes, and stored frozen (−20 °C) for subsequent analyses.
Rumen fluid samples (~10 mL) were taken directly from the rumen from all ewes at d1, d51, and d115 by orogastric intubation using sterile 60 mL syringes and autoclaved tubes. Rumen samples from each ewe were kept separately in 50 mL centrifuge tubes and transported on dry ice to the University of Arkansas, and stored at −80°C until DNA extraction.
Tall fescue grass samples were collected monthly from each pasture throughout the study. Samples of tall fescue (n = 20/pasture) were harvested by walking in a zigzag pattern through each pasture and hand-clipping random samples to a 2.5 cm stubble height. The samples were placed in zip-lock plastic bags, stored on ice immediately, and then moved to a conventional freezer for at least 2 h. The grass samples were transported to the University of Arkansas Animal Science Department, and stored in an ultra-low freezer (−80°C), until they were lyophilized. Dried forage samples were ground to pass through 1-mm screen using a Thomas-Wiley laboratory mill model 4 (Arthur Thomas Co. Philadelphia, PA, USA) and stored in an ultra-low freezer except during analyses. Tillers for testing for the presence of endophyte fungus were collected on February 2nd by walking a zigzag pattern in the pastures, and collecting tall fescue tiller samples at random (50 per pasture). Stems were cut with a sharp knife to a height of 2.5 cm above of the soil surface. Tissue that contained the leaves was discarded; the tillers were rolled into moist paper towels to prevent drying, and then placed in plastic zip lock bags and stored in a refrigerator (1°C) pending endophyte analysis.
Measurements of Forage and Serum
Sequential analyses for forage neutral detergent fiber (NDF) and acid detergent fiber (ADF) were performed using the filter bag procedures described by Vogel et al. (18) using an Ankom 200 Fiber Analyzer (ANKOM Technology Corporation, Fairport, NY, USA). Nitrogen concentrations were determined via the Dumas total combustion method (Elmentar Americas, Mt. Laurel, NJ, USA; Method 990.03) (19). Crude protein (CP) was calculated by multiplying the total N concentration by 6.25. Total ergot alkaloids were determined for using Agrinostics Photo screen Ergot Alkaloids kit (Agrinostics Ltd. Co., Watkinsville, GA, USA) (20). Forage ergovaline concentrations were also determined using the HPLC procedure of (21). Pasture endophyte infection rates were analyzed using immunoblot test kits (Agrinostics Ltd. Co., Watkinsville, GA, USA).
Serum samples were analyzed for non-esterified fatty acid (NEFA) concentrations using an in vitro enzymatic colorimetric kit [NEFA-HR (2); Wako Chemicals, Inc., Richmond, VA, USA]. Serum prolactin (PRL) concentrations were determined at the University of Tennessee using their standard lab protocol following the procedures of Bernard et al. (22) with inter- and intra-assay coefficients of variability (CV) of 4.24 and 5.69%, respectively.
DNA Extraction and Next-Generation Sequencing
Total bacterial DNA was extracted using a PowerSoilTM DNA Isolation Kit (MO BIO Laboratories, Inc. Carlsbad, CA, USA) based on the manufacturer's instructions. Moreover, a bead beating step for rapid and thorough homogenization of bacterial cells was incorporated to yield high quantity and quality rumen microbial DNA. DNA concentrations were measured using a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). The V4 region of bacterial 16S rRNA gene was amplified for Illumina sequencing with the following primers 5′-GTGCCAGCMGCCGCGGTAA-3′ (forward) and 5′-GGACTACHVGGGTWTCTAAT-3′ (reverse) (23). PCR was performed using a T100 thermal cycler (Bio-Rad, Hercules, CA, USA) with the following conditions: 30 s initial denaturation at 95°C; 30 cycles at 95°C for 10 s annealing at 55°C for 30 s, and at 72°C for 1 min; at a 72°C final extension for 10 min. Two of microliters amplicons were examined on 1.0% agarose gel and remaining PCR products were subsequently normalized using a SequalPrepTM Normalization kit (Life Technology, Carlsbad, CA, USA) according to the manufacturer's recommendation. A 10 ng DNA aliquot was utilized to construct a sequencing library following a previous report (24). The concentration and quality of the library were measured using a KAPA Library Quantification Kit (Kapa Biosystem, Woburn, MA, USA) via a quantitative PCR (qPCR, Eppendorf, Westbury, NY, USA) assay and an Agilent 2100 Bioanalyser System (Agilent, Santa Clara, CA, USA), respectively. The library was mixed with PhiX control v3 (5%, v/v) (Illumina), loaded on a MiSeq v2 (2 × 250 bp, 500 cycle) reagent cartridge and then sequenced on an Illumina MiSeq sequencer (Illumina, San Diego, CA, USA) for next-generation sequencing.
Bioinformatics
The MOTHUR (v.1.39.1) software package was used to process raw sequences (25) following the standard procedures for MiSeq sequences (24). The forward and reverse sequences were first merged by using the make.contigs command in MOTHUR. Low-quality sequence reads were removed based on homopolymers longer than 8 bp, ambiguous bases, and raw reads of 500 bp or longer. The sequences were aligned with the SILVA database release 128 which contained the full-length 16S rRNA gene sequences (https://www.mothur.org/wiki/Silva_reference_files#Release_128). The chimeric sequences were removed using UCHIME (26). Also, singletons were removed to decrease the number of spurious sequences. High-quality sequences were clustered into operational taxonomic units (OTUs) having a similarity of 97% and classified using the RDP Classifier, with a naïve Bayesian algorithm (27). After normalization to the smallest number of reads (6946), alpha and beta diversity were estimated. Alpha diversity metrics consisted of the number of observed OTUs, and the Shannon diversity index. Bray-Curtis and Jaccard distance metrics were determined to compare the dissimilarities in community structure and membership, respectively. Principal Coordinate Analysis (PCoA) was utilized to visualize these distances. Analysis of similarity (ANOSIM) was also conducted in MOTHUR for testing the statistical significance of beta diversity distances. The raw sequences in the current study are available in the NCBI Sequence Read Archive repository (project number PRJNA577241).
Statistical Analyses
Based on a completely randomized design, BW, BCS, PRL, NEFA, and alpha diversities were tested using a linear mixed-effect model in R (v 3.6.0). Treatment and sampling time were included as fixed effects, while both individual dams, which served as experimental units, and the number of lambs born of each dam were included as nested random effects. The default unstructured covariance structure was used in linear mixed model when performing the “lmer” function (“lme4” package of R). P-values lower than 0.05 were considered as significant.
To analyze rumen microbiotas for differentiating high and moderate endophyte infection, a random forest model (area under the receiver operating characteristic curve (AUC) of the random forest, AUC-RF) with 10-fold cross-validation was performed (28). Thus, the model predicted the left-out subject and results were plotted as Receiver Operator Characteristic curves using the pROC package (v.1.13). The optimal predictors of AUCRF were listed based on their mean decrease accuracy (MDA). The boxplots portraying the relative abundance of optimal predictors was performed using the R “ggplot2” package (v.3.0) and labeled p-values of Wilcoxon test. To estimate the relationship between the microbiota and the animal phenotype (serum concentration of PRL and NEFA), a regression-based random forest model was performed using the package “randomForest” v.4.6.-7 in R with 10,000 trees. Top 50 predictors were selected and their relative abundances were then analyzed by using Pearson correlation with PRL and NEFA, respectively.
Results
Forage Characteristics and Ewes' Phenotype
The ergovaline concentration in both treatments ranged from 55 to 243 μg/kg during the experiment (February to May 2016, Booneville, AR, USA), which did not coincide with ambient temperatures and precipitation (Table 1 and Supplementary Table 2). Ergovaline concentrations were lowest in both treatments at the beginning of the study and increased gradually, resulting in the greatest concentrations at the end of the study in both HA and MA. The total ergot alkaloid concentrations also exhibited a similar pattern. While the concentration of ergovaline and total ergot alkaloids changed over time, they were greater in HA than MA throughout this study. Although ergovaline concentrations were low (<100 μg/kg) from both treatments on the first three sampling dates, the concentrations were 5.5, 1.5, 1.1, and 2.3 times greater from HA compared with MA on the February (2), March (22), April (12), and May (25) sampling dates, respectively. Total ergot alkaloids followed a somewhat similar pattern to ergovaline concentrations, excluding the fact that the lowest total ergot alkaloid concentrations occurred on the March (22) sampling date. Composition of other nutrients including crude protein and fiber was not appreciably different between the two treatments throughout this study.
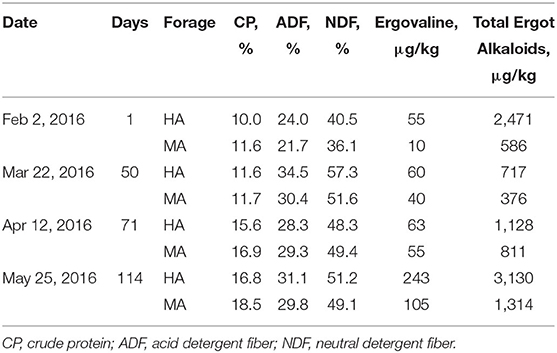
Table 1. Forage quality components and ergot alkaloid concentrations from tall fescue pastures that had moderate (MA) and high (HA) N. coenophialum infection levels and grazed by gestation and lactation ewes.
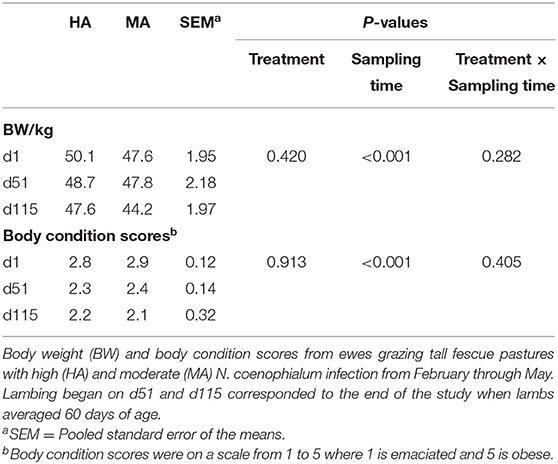
The BW and body condition scores of ewes during this study were not different (P > 0.10) between the HA and MA treatment groups, but these measurements decreased with sampling time (P < 0.05) (Table 2). Serum PRL and NEFA concentrations were not different (P > 0.10) between treatments throughout the study (Supplementary Table 3). However, they were affected by sampling time (P < 0.05). Moreover, serum NEFA displayed an interaction between treatment and sampling time (P < 0.05). NEFA concentration were higher on d51 than on d1 and d115 (P < 0.05). Serum NEFA was not different (P > 0.10) between treatments on d1 and d51, while ewes grazing HA had higher NEFA (P = 0.024) on d115 compared with ewes on MA pasture.
Alpha and Beta Diversities of the Rumen Microbiota
The microbial diversity in ruminal fluid between ewes grazing HA and MA endophyte tall fescue within different periods was initially investigated. Sampling time affected alpha diversity measures (P < 0.05) (Table 3), however, no treatment effects or interaction with sampling time was observed (P > 0.05). Overall, rumen microbial diversity decreased on lambing days (d51) and then reached the initial level on the lambs' weaning day (d115).
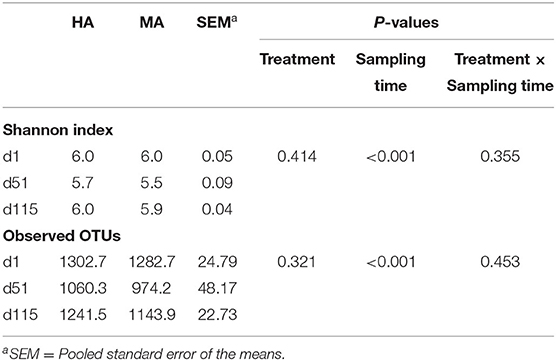
Table 3. Alpha diversities of rumen microbiota in ewes grazing tall fescue pastures with high (HA) and moderate (MA) N. coenophialum infection.
Bray-Curtis and Jaccard similarities were calculated for beta diversity (Figures 1A,B). No differences were observed on d1 based on Bray-Curtis and Jaccard similarities (ANOSIM: R = −0.03, 0.03; P = 0.58, 0.35). On d51, the rumen microbial community structure and membership tended to be distinct between HA and MA (ANOSIM: R = 0.30, 0.29; P = 0.06, 0.07). In the lactation period (d115), rumen microbiota were different between ewes grazing HA and MA tall fescue (ANOSIM: R = 0.28, 0.31; P = 0.02, 0.01). In addition, d1 samples were distinct from those of the other two time points (d51 and d115).
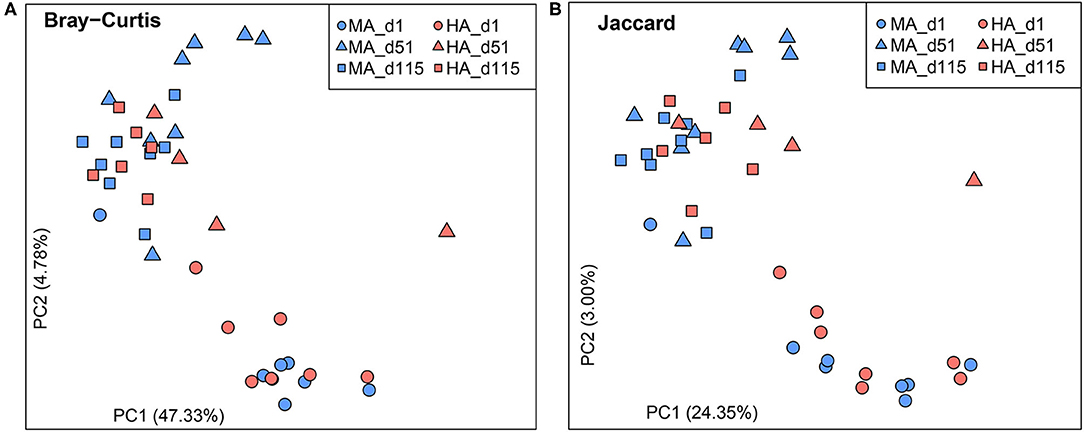
Figure 1. Beta diversity plots of rumen microbiota in ewes grazing tall fescue pastures with high (HA) and moderate (MA) N. coenophialum infection. (A,B) Principal coordinate analysis (PCoA) plots based on the Bray-Curtis and Jaccard similarities. The treatments of MA and HA are differentiated by colors (blue and red), and sampling days (d1, d51 and d115) are distinguished by shape (circle, triangle, and square, respectively).
Taxonomic Composition of the Rumen Microbiota
Bacteroidetes and Firmicutes (43.26 and 39.86% across all samples) were the dominant phyla in the rumen fluid (Figure 2). In this study, the relative abundances of the Bacteroidetes in MA were 42.51, 47.95, and 45.39% on d1, d51, and d115, while its relative abundances in HA were 44.68, 37.58, and 41.44% on d1, d51, and d115, respectively. On d51, the phylum Firmicutes was less relatively abundant in MA pasture (36.51%) compared with HA pastures (46.70%). The Actinobacteria phylum exhibited a similar pattern with Firmicutes, and its relative abundance on d51 were 3.94 and 6.16% in MA and HA, respectively.
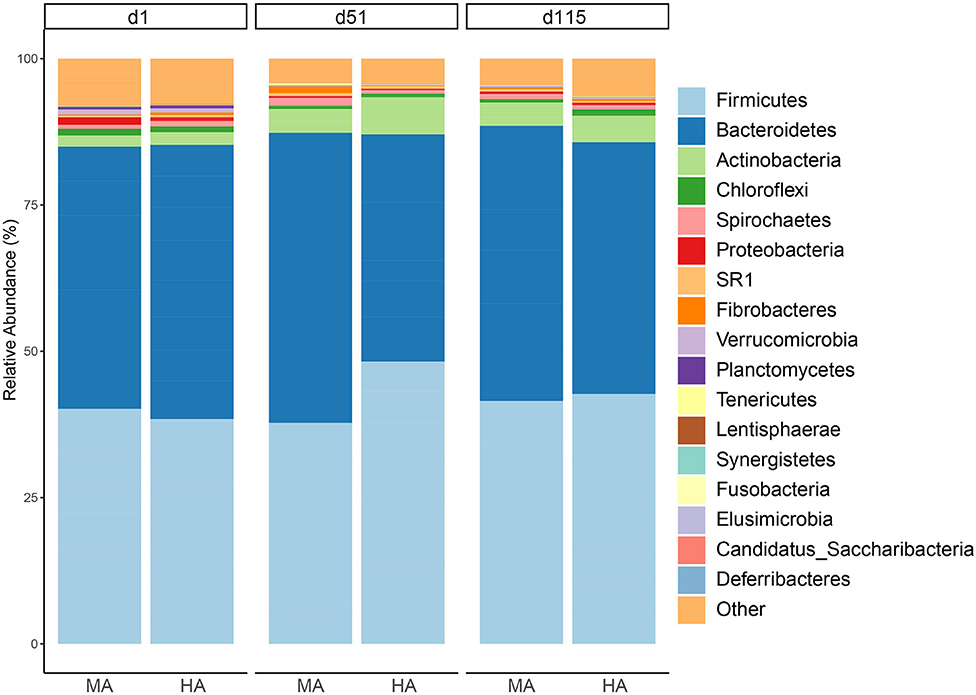
Figure 2. Relative abundance of rumen phyla from ewes consuming tall fescue infected with moderate (MA) or high (HA) levels of N. coenophialum on days 1 (d1), 51 (d51), and 115 (d115). Average relative abundance for each phylum by treatment and sampling time. Each color represents the relative abundance of a bacterial taxon on the stacked bar chart.
At the genus level (Figure 3), the dominant bacteria were Prevotella (19.54%) across all samples, followed by unclassified Lachnospiraceae (7.94%), unclassified Ruminococcaceae (7.62%), and unclassified Prevotellaceae (6.46%). The abundances of Prevotella increased with time in both MA and HA groups (on d1, d51 and d115: 15.17, 25.59, and 24.78% in MA; 14.17, 18.10, and 19.41% in HA). The unclassified Lachnospiraceae in HA were in higher abundance (11.22%) on d51 compared with MA (7.42%). Other minor genera such as unclassified Coriobacteriaceae and unclassified Veillonellaceae increased with time, and also had greater abundances in HA at d51 (3.72 and 2.07%) compared with MA (2.06 and 1.28%). Unclassified Ruminococcaceae on d115 were 9.78 and 8.13% in HA and MA groups, respectively.
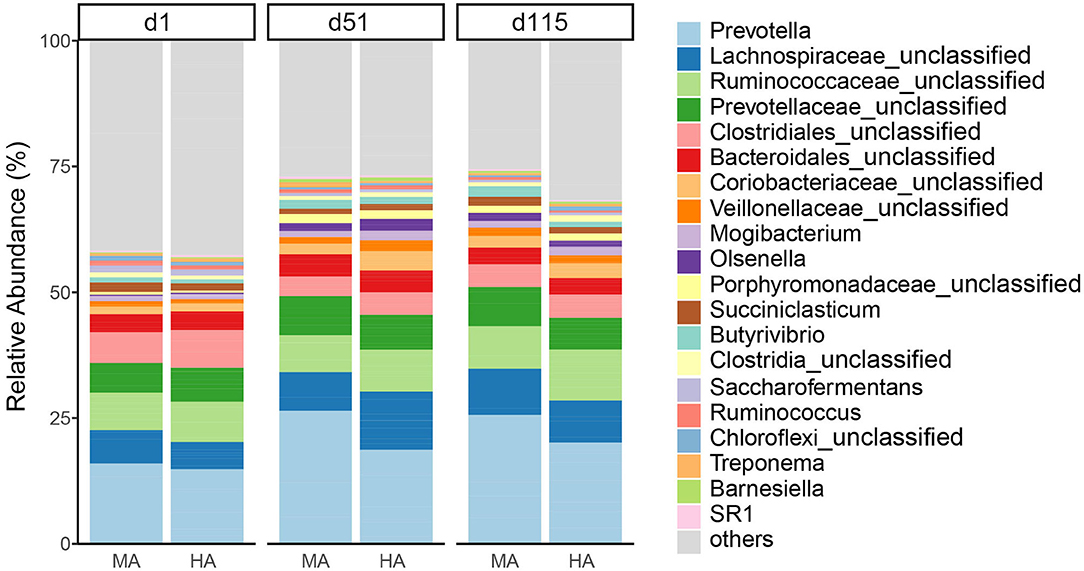
Figure 3. Relative abundance of rumen bacteria (genus) from ewes consuming tall fescue infected with moderate (MA) or high (HA) levels of N. coenophialum on days 1 (d1), 51 (d51), and 115 (d115). Each color represents the relative abundance of a bacterial taxon on the stacked bar chart.
Differential Rumen Microbiota Between Ewes Grazing HA and MA Tall Fescue Forage
Random Forest is able to analyze high dimensional microbiota data that usually has more variables (e.g., OTUs) than samples with complicated interactions between OTUs, and therefore, has been widely used to analyze human and animal microbiome data. Using random forest, a set of bacterial OTUs that differentiated HA and MA ewes were identified. The top 25 predictors are listed based on their ability to differentiate between ewes grazing HA and MA tall fescue (Supplementary Figures 2, 3). Six among the top 25 OTUs that differentiate HA and MA were listed as the most sensitive in distinguishing between the forages (Figures 4, 5).
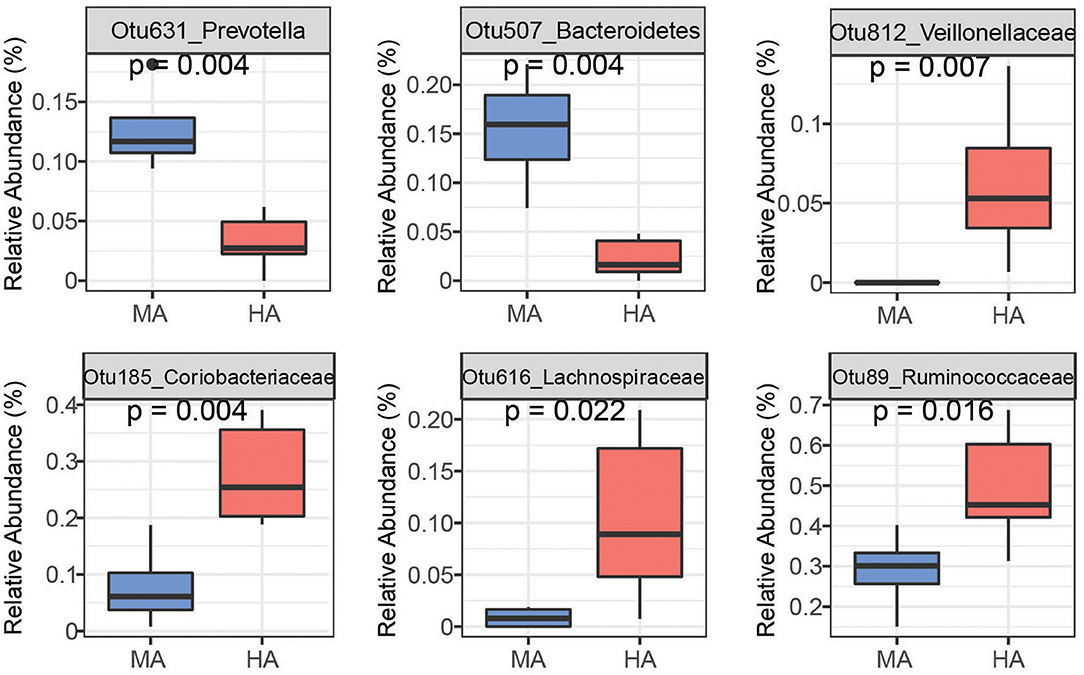
Figure 4. Bacteria OTUs determined using random forest differentiating MA and HA on d51. The rumen microbial data were tested using the Kruskal-Wallis test. MA and HA represent tall fescue with moderate or high levels of N. coenophialum, respectively.
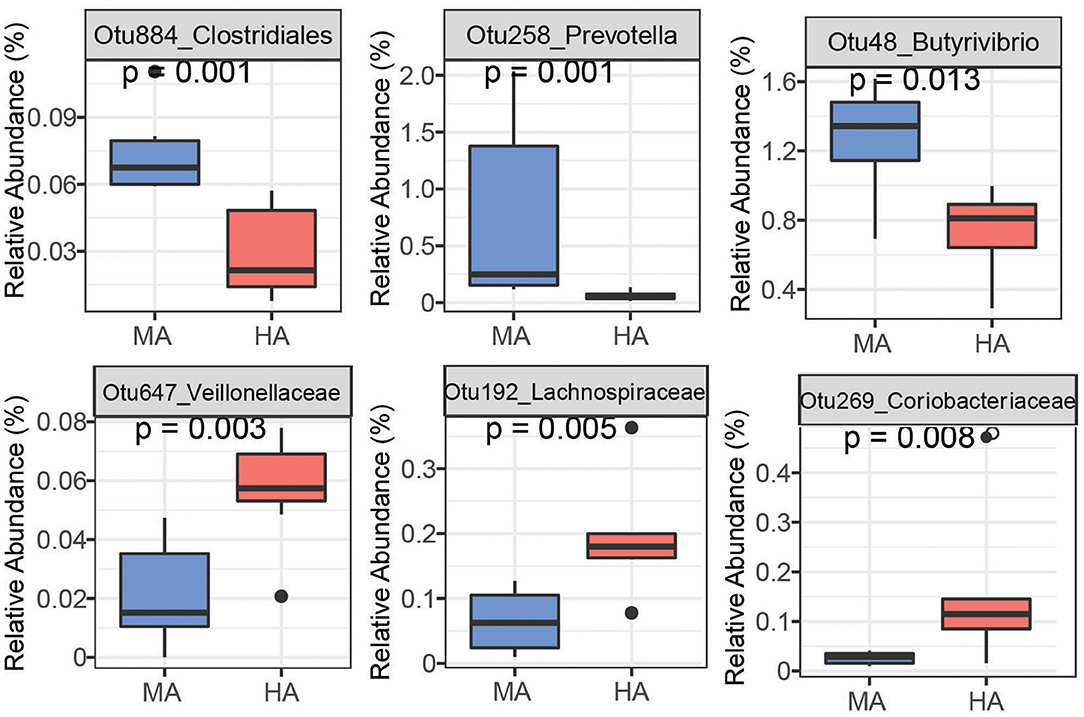
Figure 5. Bacteria OTUs determined using random forest differentiating MA and HA on d115. The rumen microbial data were tested using the Kruskal-Wallis test. MA and HA represent tall fescue with moderate or high levels of N. coenophialum, respectively.
On d51, the rumen species can differentiate MA and HA accurately by yielding AUC = 1 (sensitivity = 1, specificity = 1) (Supplementary Figure 1A). Several OTUs classified as Prevotella (Otu631, Otu134, OTU457, Otu266, and Otu11), Fibrobacter (Otu522), and unclassified Ruminococcaceae (Otu345) exhibited greater abundance in MA (Figure 4 and Supplementary Figure 2). Other OTUs including unclassified Veillonellaceae (Otu812), unclassified Coriobacteriaceae (Otu185, Otu269), unclassified Lachnospiraceae (Otu262, Otu616 and Otu23), and unclassified Ruminococcaceae (Otu89) were enriched in the HA treatment. Moreover, several OTUs classified as Prevotella occurred in higher abundance in HA, including Otu540, Otu717, and Otu272.
On d115, high accuracy in the random forest model was observed (Supplementary Figure 1B). Ewes grazing MA had high abundances of OTUs affiliated with unclassified Prevotella (Otu58, Otu171, Otu238), Clostridiales (Otu884), Bacteroidetes (Otu599 and Otu1102), Butyrivibrio (Otu48 and Otu215) and Succiniclasticum (Otu36) (Figure 5 and Supplementary Figure 3). Other OTUs enriched in HA were associated with unclassified Veillonellaceae (Otu647), unclassified Coriobacteriaceae (Otu269), unclassified Lachnospiraceae (Otu192), unclassified Ruminococcaceae (Otu49 and Otu345), and Mogibacterium (Otu10).
The Association Between the Ewe Rumen Microbiota and Serum PRL
To better understand the association between the rumen microbiota and serum PRL, a random forest regression model using PRL concentration as outcomes and the top 500 OTUs as independent variables was generated. Pearson correlations were subsequently estimated among the selected top 50 bacterial abundances and PRL. The bacteria with significant correlations are listed in Table 4. Some OTUs such as Lachnospiraceae (Otu896) exhibited a negative relation with PRL in both gestation and lactation periods (d51: r = −0.64, p = 0.03; d115: r = −0.52, p = 0.06).
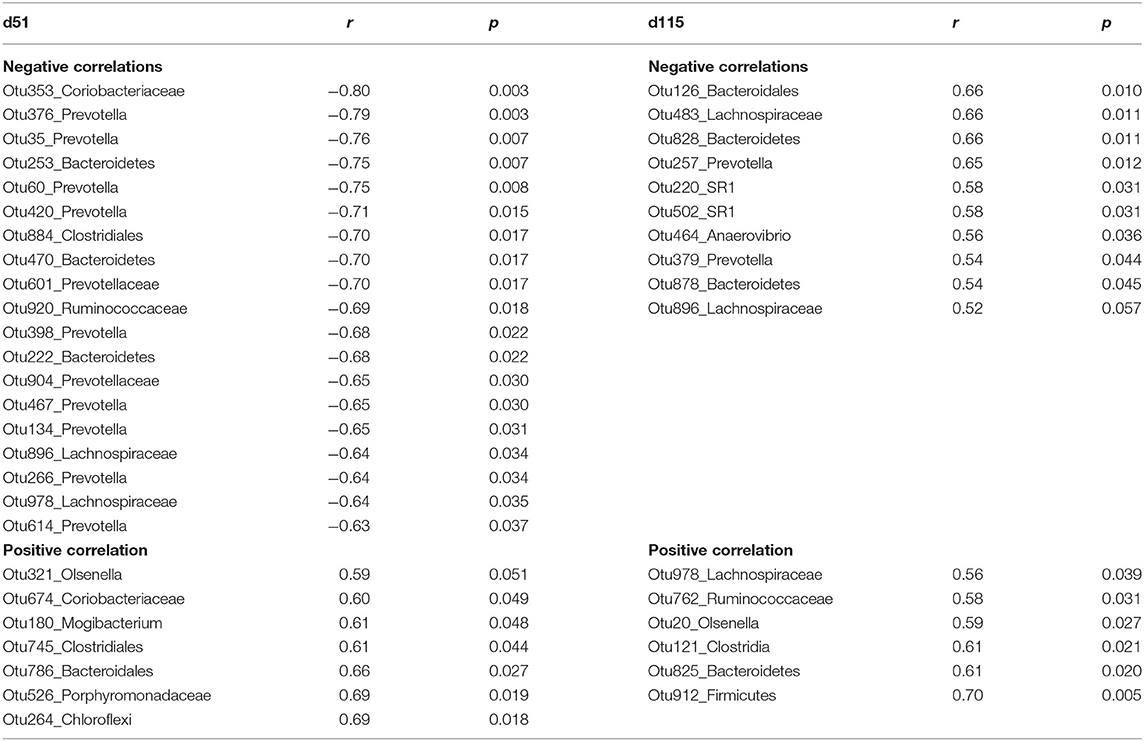
Table 4. The significant Pearson relationship between top bacteria OTUs identified by random forest regression model and serum prolactin (PRL) concentrations from ewes grazing N. coenophialum infected tall fescue pastures.
On d51, most OTUs classified as Prevotella were negatively correlated with PRL, e.g., Otu134 and Otu266 (r = −0.65, −0.64; p = 0.03, 0.03). Other OTUs belonging to unclassified Bacteroidetes (Otu253, Otu470, and Otu222), unclassified Clostridiales (Otu884), unclassified Ruminococcaceae (Otu920), and Coriobacteriaceae (Otu353) were also negatively correlated. In contrast, unclassified Coriobacteriaceae (Otu674), Mogibacterium (Otu180), Clostridiales (Out745), Bacteroidetes (Otu786), Porphyromonadaceae (Otu526), and unclassified Chloroflexi (Otu264) were positively correlated with PRL.
On d115, Otu257, and Otu379 classified as Prevotella were negatively correlated with PRL. Negative relationships were also observed in unclassified SR1 (Otu220 and Otu502), unclassified Bacteroidetes (Otu126, Otu828, Otu878), and Anaerovibrio (Otu464). Conversely, when serum PRL concentration increased, the abundances of unclassified Lachnospiraceae (Otu978), unclassified Ruminococcaceae (Otu762), Olsenella (Otu20), and unclassified Clostridia (Otu121) increased linearly.
The Correlation of Bacterial OTUs in the Ewe Rumen and Serum NEFA
To examine the relationship between serum NEFA and rumen microbiota we developed random forest regression models and performed Pearson correlations analysis, which shows the significant correlations between microbial abundances and serum NEFA in Table 5. Interestingly, the common OTU555 (unclassified Clostridiales) was negatively correlated with NEFA on d51 (r = −0.71, p = 0.01), while a positive correlation on d115 (r = 0.54, p = 0.05) was found.
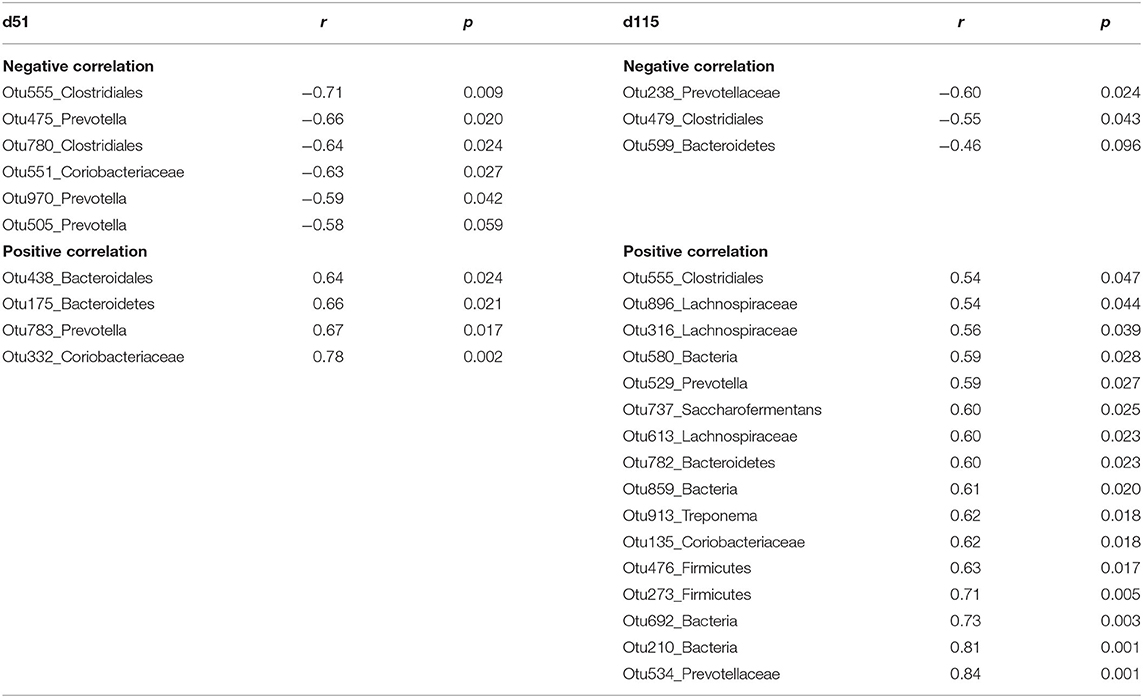
Table 5. The significant Pearson relationship between top bacteria OTUs identified by random forest regression model and serum non-esterified fatty acid (NEFA) from ewes grazing N. coenophialum infected tall fescue pastures.
On d51, negative correlations were observed in microbiota including unclassified Clostridiales (Otu555, Otu780), Prevotella (Otu475, Otu970, Otu505), and unclassified Coriobacteriaceae (Otu551). However, unclassified Coriobacteriaceae (Otu332), unclassified Bacteroidales (Otu438, Otu175), and Prevotella (Otu783) were positively correlated with NEFA.
On d115, the relative abundances of unclassified Clostridiales (Otu479), unclassified Prevotellaceae (Otu238), and unclassified Bacteroidetes (Otu599) decreased when serum NEFA increased. Rumen microbiota including unclassified Lachnospiraceae (Otu896, Otu316, Otu613), unclassified Coriobacteriaceae (Otu135), unclassified Prevotellaceae (Otu529, Otu534, Otu957), Clostridiales (Otu555), Saccharofermentans (Otu737), Bacteroidetes (Otu782), and Treponema (Otu913) linearly increased with increasing NEFA.
Discussion
This study was conducted to investigate changes within the rumen microbiota in gestating and lactating ewes that grazed tall fescue with different levels of endophyte infection. The toxicity (ergovaline) concentration in tall fescue was affected by seasonal effects, which is consistent with a previous study (6), suggesting that grazing at different growing seasons may have varying impacts on animals due to changing ergovaline intake. We observed that the fescue endophyte may negatively influence the health of lactating ewes more so than gestating ewes. Significant changes in serum NEFA and rumen microbial diversity were found at the end of lactation. A consortia of microbiota were linked to levels of endophyte-infection in gestation and lactation periods, and the microbiota responded to the step-wise increase in mycotoxins from the tall fescue. Moreover, the correlation between rumen bacteria and important reproductive parameters (serum PRL and NEFA) was measured that allowed for a better understanding of the host mechanism involving mycotoxin degradation.
Feed with mycotoxins or other potentially harmful substances have been shown to influence the rumen microbiota (16). In the current research, toxins in tall fescue affected alpha-diversity measures such as the number of observed OTUs and Shannon diversity index. Based on these results, there were no differences between d1 and d51, however, HA had greater alpha diversity than MA on d115. Similarly, beta diversity analysis also showed distinct clusters of microbial community structures between HA and MA groups on d115. We speculate that the lack of differences between d1 and51 could be explained by sheep consuming a forage that contained lower levels of toxins, allowing rumen bacteria to detoxify the mycotoxins. However, by d115, toxin concentrations had increased in both treatments resulting in significant impacts on alpha diversities. In a more recent study, Mote et al. (8) found that cattle grazing toxic tall fescue for 28 days (total ergot alkaloid levels 2357.7 ± 19.70 μg/kg) had a tendency to change the overall fecal microbiota community structure. Regarding the concentration of total ergot alkaloid in HA on May 25, 2016 (d114, 3,130 μg/kg), it was perhaps not surprising that significant results on d115 were observed compared with the results on d51.
The random forest algorithm was performed to classify the OTUs differentiating MA and HA treatments between d51 and d115, respectively. In gestation and lactation, only two shared OTUs differentiating high and moderate endophyte infection were observed. One of them, unclassified Coriobacteriaceae (Otu269), was enriched in the HA ewes at both time points. Importantly, it has been observed that members of the Coriobacteriaceae family can degrade mycotoxins to non-toxic metabolites (29). It was also consistently observed that OTUs associated with Coriobacteriaceae were more abundant in HA ewes and therefore could be viewed as an indicator of their contributions to ergovaline detoxification. Likewise, OTUs classified as Veillonellaceae and Lachnospiraceae were also observed with high abundances in HA ewes, which is in line with a recent study where high abundances of these bacteria were also found in cattle grazing toxic endophyte-infected tall fescue (8). Bacterial community structure has been shown to be affected by other mycotoxins (i.e., deoxynivalenol or fumonisin) in previous studies (30, 31). Therefore, these three rumen bacterial families may be associated with mycotoxin degradation. Lachnospiraceae can produce volatile fatty acids (VFA), whose metabolism may be influenced by changes in the bioavailability of the primary fermentation metabolites by mycotoxins (30, 32). Foote et al. (33) confirmed that ergovaline reduced rumen VFA concentrations, which provides further support that intake of highly endophyte-infected tall fescue impacts nutrient bioavailability, and potentially absorption, along with the resident microbiota (34). Moreover, members of the Prevotella genus and Clostridiales order relatively abundant in MA on d115 are reported as ergovaline degraders (17). Increased relative abundance of Prevotella were also observed when animals consumed another metabolic mycotoxin (deoxynivalenol) associated with a Fusarium fungi (35). Evidence from an ex vivo study demonstrated that Prevotellaceae and Clostridiaceae strains are not heavily involved in ergovaline degradation (17). We assumed that these two bacterial taxa associated with ergovaline degradation were affected by mycotoxin concentration. A hypothesis is that Prevotellaceae and Clostridiaceae can degrade ergovaline when its concentration was low, but these bacteria's activity decreased in environments containing high ergovaline concentration. Therefore, the concentration of ergovaline in tall fescue influences community structure of the rumen microbiota, with bacteria (e.g., Coriobacteriaceae, Veillonellaceae, and Lachnospiraceae) in high abundance in HA potentially playing roles in endophyte degradation.
Prolactin is known to play an important role for both lactation and mammogenesis (36). Prolactin homeostatic levels are sensitive to tall fescue toxins and are a measure of the degree of tall fescue mycotoxicosis (37). Since ergovaline has a ring structure similar to the major prolactin inhibitor (dopamine), this may significantly affect serum prolactin concentration (16). In the current study, serum prolactin levels were not statistically different between treatments; however, numerically lower values were observed in MA treatment on d115. Duckett et al. (38) found that serum PRL levels did not differ between ewes fed endophyte-infected tall fescue seed (0.8 μg of ergovaline/g diet DM) and free-infected seed on d 30 of gestation. Yet, significantly lower PRL levels occurred in ewes exposed to ergot alkaloids on d50 and d130 of gestation compared to an endophyte-free group (38). Additionally, another study did not find serum prolactin differences in goats grazing endophyte-infected tall fescue (87.5–95.8% infection) (39). It was speculated that the change of serum PRL was dependent on the concentrations of total ergot alkaloid in forage and the intake of tall fescue, as well as the ability of the rumen microbiota to degrade ergovaline. Consequently, we compared the correlation of PRL concentrations with the microbiota exposed under different levels of mycotoxins, which, in turn, could reflect the ability of rumen microbiota to degrade ergovaline. Lachnospiraceae exhibited a negative relation with PRL concentration in both gestation and lactation periods. Additionally, we observed that Clostridiales and Coriobacteriaceae were negatively correlated to serum prolactin concentration. Moreover, we found OTUs related to Prevotella that occurred in high abundance in MA were negatively correlated with PRL. Prevotella species were shown to induce a ruminal environment favorable to Prevotella and other hyper-ammonia-producing bacteria, potentially beneficial by reducing the prevalence of fescue toxicosis (16). Therefore, the intake of toxin could likely influence the rumen microbiota and ultimately impact serum prolactin.
There were several limitations of this study. First, while the baseline can be used as a control for endophyte infection, there was no control group that was exposed to non-infected tall fescue. However, endophyte-free tall fescue is not persistent in this area. Also, pastures seeded to newer non-toxic, novel-endophyte infected tall fescue are not located on the USDA station. Use of another forage as a control would confound the information by having different forages and different levels of toxicity. Second, this study did not measure the rumen alkaloid concentrations. Some measures of intake (based on weight gain), rumen volume and rate of passage would help estimate the extent of alkaloid degradation. Greater numbers of samples would be needed to measure the alkaloid concentration, but would have resulted in excessive stress on animals that are already stressed to a greater extent because of the toxins. Inserting a stomach tube causes considerable stress on the animal and the sampling days were hot, which would have caused excessive stress. Measuring rumen volume and rate of passage would require the use of pulse-dosed markers such as Yb then repeated and frequent sampling to develop the fecal excretion curve of the inserted marker. This again would have required a greater number of times the animals were handled and would not be allowed by the animal welfare group that oversees the animals at the USDA station.
To date, this is the first study to characterize the rumen microbiota and its relationship with serum prolactin and NEFA in gestating and lactating ewes grazing tall fescue with high or moderate levels of toxic endophyte infection. Since the concentration of total ergot alkaloids increased throughout the study, accurate levels of ergovaline in tall fescue that are shown to influence the rumen microbiota composition and ewes' phenotype cannot be confirmed. However, this study did identify rumen microbial changes affected by moderate and high levels of toxins in endophyte-infected tall fescue. Characteristics of the rumen microbiota associated with serum prolactin and NEFA were additionally identified. High endophyte infection exhibited increased abundances of Coriobacteriaceae, Lachnospiraceae, and Prevotellaceae families, which could be related with the endocrinology and metabolism of fatty acids. Future studies are needed to accurately identify the concentration of ergovaline significantly altering the rumen microbiota, and to further delineate the association between the rumen microbiota and host physiology.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by University of Arkansas Institutional Animal Care and Use Committee.
Author Contributions
KC, JB, and JZ conceived and designed the experiments. SA and JB performed the experiment and collected samples. JC, SP, and JZ performed next-generation sequencing. JC, SA, KC, JB, and JZ analyzed and interpreted the data. JC, SA, KC, JB, KF, SR, JE, and JZ drafted and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by grants from USDA National Institute for Food and Agriculture, Hatch Project (1005233).
Disclaimer
Mention of trade names or commercial products in this manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Robin Anderson (USDA-ARS, College Station, TX, USA) for his kind words and edits.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.544707/full#supplementary-material
References
1. Paterson J, Forcherio C, Larson B, Samford M, Kerley M. The Effects of fescue toxicosis on beef-cattle productivity. J Anim Sci. (1995) 73:889–98. doi: 10.2527/1995.733889x
2. Ball D, Schmidt S, Lacefield G, Hoveland C, Young W. Tall Fescue/Endophyte Concepts. Salem, MA: Oregon Tall Fescue Commission (2003).
3. Hoveland CS, Schmidt SP, King CC, Odom JW, Clark EM, Mcguire JA, et al. Steer performance and association of Acremonium-coenophialum fungal endophyte on tall fescue pasture. Agron J. (1983) 75:821–4. doi: 10.2134/agronj1983.00021962007500050021x
4. Bouton JH, Gates RN, Belesky DP, Owsley M. Yield and persistence of tall fescue in the southeastern coastal-plain after removal of its endophyte. Agron J. (1993) 85:52–5. doi: 10.2134/agronj1993.00021962008500010011x
5. Hill NS, Thompson FN, Stuedemann JA, Rottinghaus GW, Ju HJ, Dawe DL, et al. Ergot alkaloid transport across ruminant gastric tissues. J Anim Sci. (2001) 79:542–9. doi: 10.2527/2001.792542x
6. Rogers WM, Roberts CA, Andrae JG, Davis DK, Rottinghaus GE, Hill NS, et al. Seasonal fluctuation of ergovaline and total ergot alkaloid concentrations in tall fescue regrowth. Crop Sci. (2011) 51:1291–6. doi: 10.2135/cropsci2010.07.0402
7. Klotz JL. Activities and effects of ergot alkaloids on livestock physiology and production. Toxins. (2015) 7:2801–21. doi: 10.3390/toxins7082801
8. Mote RS, Hill NS, Skarlupka JH, Turner ZB, Sanders ZP, Jones DP, et al. Response of beef cattle fecal microbiota to grazing on toxic tall fescue. Appl Environ Microbiol. (2019) 85:e00032-19. doi: 10.1128/AEM.00032-19
9. Klotz JL, Bush LP, Smith DL, Shafer WD, Smith LL, Arrington BC, et al. Ergovaline-induced vasoconstriction in an isolated bovine lateral saphenous vein bioassay. J Anim Sci. (2007) 85:2330–6. doi: 10.2527/jas.2006-803
10. Lea K, Smith L, Gaskill C, Coleman R, Smith SR. Ergovaline stability in tall fescue based on sample handling and storage methods. Front Chem. (2014) 2:76. doi: 10.3389/fchem.2014.00076
11. Strickland JR, Looper ML, Matthews JC, Rosenkrans CF Jr, Flythe MD, Brown KR. Board-invited review: St. Anthony's Fire in livestock: causes, mechanisms, and potential solutions. J Anim Sci. (2011) 89:1603–26. doi: 10.2527/jas.2010-3478
12. Aiken GE, Strickland JR, Looper ML, Bush LP, Schrick FN. Hemodynamics are altered in the caudal artery of beef heifers fed different ergot alkaloid concentrations. J Anim Sci. (2009) 87:2142–50. doi: 10.2527/jas.2008-1562
13. Klotz JL, Brown KR, Xue Y, Matthews JC, Boling JA, Burris WR, et al. Alterations in serotonin receptor-induced contractility of bovine lateral saphenous vein in cattle grazing endophyte-infected tall fescue. J Anim Sci. (2012) 90:682–93. doi: 10.2527/jas.2011-4323
14. Lima FS, Oikonomou G, Lima SF, Bicalho ML, Ganda EK, Filho JC, et al. Prepartum and postpartum rumen fluid microbiomes: characterization and correlation with production traits in dairy cows. Appl Environ Microbiol. (2015) 81:1327–37. doi: 10.1128/AEM.03138-14
15. Tong J, Zhang H, Yang D, Zhang Y, Xiong B, Jiang L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS ONE. (2018) 13:e0198225. doi: 10.1371/journal.pone.0198225
16. Melchior EA, Myer PR. Fescue toxicosis and its influence on the rumen microbiome: mitigation of production losses through clover isoflavones. J Appl Anim. (2018) 46:1280–8. doi: 10.1080/09712119.2018.1496920
17. Harlow BE, Goodman JP, Lynn BC, Flythe MD, Ji H, Aiken GE. Ruminal tryptophan-utilizing bacteria degrade ergovaline from tall fescue seed extract. J Anim Sci. (2017) 95:980–8. doi: 10.2527/jas.2016.1128
18. Vogel KP, Pedersen JF, Masterson SD, Toy JJ. Evaluation of a filter bag system for NDF, ADF, and IVDMD forage analysis. Crop Sci. (1999) 39:276–9. doi: 10.2135/cropsci1999.0011183X003900010042x
19. AOAC. Official Methods of Analysis. 16th ed. Gaithersburg, MD: Association of Official Analytical Chemists (1988).
20. Adcock R, Hill N, Bouton J, Boerma H, Ware G. Symbiont regulation and reducing ergot alkaloid concentration by breeding endophyte-infected tall fescue. J Chem Ecol. (1997) 23:691–704. doi: 10.1023/B:JOEC.0000006404.33191.60
21. Rottinghaus GE, Garner GB, Cornell CN, Ellis JL. HPLC method for quantitating ergovaline in endophyte-infested tall fescue: seasonal variation of ergovaline levels in stems with leaf sheaths, leaf blades, and seed heads. J Agr Food Chem. (1991) 39:112–5. doi: 10.1021/jf00001a022
22. Bernard J, Chestnut A, Erickson B, Kelly F. Effects of prepartum consumption of endophyte-infested tall fescue on serum prolactin and subsequent milk production of holstein cows. J Dairy Sci. (1993) 76:1928–33. doi: 10.3168/jds.S0022-0302(93)77526-8
23. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. (2011) 108:4516–22. doi: 10.1073/pnas.1000080107
24. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. (2013) 79:5112–20. doi: 10.1128/AEM.01043-13
25. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
26. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
27. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
28. Calle ML, Urrea V, Boulesteix AL, Malats N. AUC-RF: a new strategy for genomic profiling with random forest. Hum Hered. (2011) 72:121–32. doi: 10.1159/000330778
29. Sayyari A, Faeste CK, Hansen U, Uhlig S, Framstad T, Schatzmayr D, et al. Effects and biotransformation of the mycotoxin deoxynivalenol in growing pigs fed with naturally contaminated pelleted grains with and without the addition of Coriobacteriaceum DSM 11798. Food Addit Contam A Chem Anal Control Expo Risk Assess. (2018) 35:1394–409. doi: 10.1080/19440049.2018.1461254
30. Lucke A, Bohm J, Zebeli Q, Metzler-Zebeli BU. Dietary deoxynivalenol contamination and oral lipopolysaccharide challenge alters the cecal microbiota of broiler chickens. Front Microbiol. (2018) 9:804. doi: 10.3389/fmicb.2018.00804
31. Mateos I, Combes S, Pascal G, Cauquil L, Barilly C, Cossalter AM, et al. Fumonisin-exposure impairs age-related ecological succession of bacterial species in weaned pig gut microbiota. Toxins. (2018) 10:230. doi: 10.3390/toxins10060230
32. Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. (2015) 74:13–22. doi: 10.1017/S0029665114001463
33. Foote AP, Kristensen NB, Klotz JL, Kim DH, Koontz AF, Mcleod KR, et al. Ergot alkaloids from endophyte-infected tall fescue decrease reticuloruminal epithelial blood flow and volatile fatty acid absorption from the washed reticulorumen. J Anim Sci. (2013) 91:5366–78. doi: 10.2527/jas.2013-6517
34. Melchior EA, Smith JK, Schneider LG, Mulliniks JT, Bates GE, Mcfarlane ZD, et al. Effects of red clover isoflavones on tall fescue seed fermentation and microbial populations in vitro. PLoS ONE. (2018) 13:e0201866. doi: 10.1371/journal.pone.0201866
35. Saint-Cyr MJ, Perrin-Guyomard A, Houee P, Rolland JG, Laurentie M. Evaluation of an oral subchronic exposure of deoxynivalenol on the composition of human gut microbiota in a model of human microbiota-associated rats. PLoS ONE. (2013) 8:e80578. doi: 10.1371/journal.pone.0080578
36. Houdebine LM, Djiane J, Dusanterfourt I, Martel P, Kelly PA, Devinoy E, et al. Hormonal action controlling mammary activity. J Dairy Sci. (1985) 68:489–500. doi: 10.3168/jds.S0022-0302(85)80848-1
37. Hill NS, Agee CS. Detection of ergoline alkaloids in endophyte-infected tall fescue by immunoassay. Crop Sci. (1994) 34:530–4. doi: 10.2135/cropsci1994.0011183X003400020041x
38. Duckett SK, Andrae JG, Pratt SL. Exposure to ergot alkaloids during gestation reduces fetal growth in sheep. Front Chem. (2014) 2:68. doi: 10.3389/fchem.2014.00068
39. Junell BL, Harp DA, Bouton JH, Crenshaw DB. Goat performance and prolactin levels as affected by tall fescue toxicosis. Tex J Agric Nat Res. (2016) 24:88–96. Available online at: https://txjanr.agintexas.org/index.php/txjanr/article/view/55
Keywords: tall fescue, toxins, microbiota, rumen microbiota, sheep, ergot alkaloids, endophyte
Citation: Chai J, Alrashedi S, Coffey K, Burke JM, Feye K, Ricke SC, Park SH, Edwards JL and Zhao J (2020) Endophyte-Infected Tall Fescue Affects Rumen Microbiota in Grazing Ewes at Gestation and Lactation. Front. Vet. Sci. 7:544707. doi: 10.3389/fvets.2020.544707
Received: 26 March 2020; Accepted: 09 September 2020;
Published: 14 October 2020.
Edited by:
Alexandre Vieira Chaves, The University of Sydney, AustraliaReviewed by:
Devin Holman, Agriculture and Agri-Food Canada, CanadaPaul James Weimer, University of Wisconsin-Madison, United States
Copyright © 2020 Chai, Alrashedi, Coffey, Burke, Feye, Ricke, Park, Edwards and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiangchao Zhao, anpoYW83N0B1YXJrLmVkdQ==
†These authors have contributed equally to this work
 Jianmin Chai
Jianmin Chai Saleh Alrashedi1†
Saleh Alrashedi1† Ken Coffey
Ken Coffey Joan M. Burke
Joan M. Burke Kristina Feye
Kristina Feye Steven C. Ricke
Steven C. Ricke Si Hong Park
Si Hong Park Jiangchao Zhao
Jiangchao Zhao