- 1Student Research Committee, Afzalipour School of Medicine, Kerman University of Medical Sciences, Kerman, Iran
- 2Research Center for Hydatid Disease in Iran, Department of Parasitology, Kerman University of Medical Sciences, Kerman, Iran
- 3Dietary Supplements and Probiotic Research Center, Alborz University of Medical Sciences, Karaj, Iran
Cystic echinococcosis (CE) is one of the most widespread zoonotic diseases, with considerable public health and economic importance. Camels play a significant role in transmission cycle of Echinococcus granulosus especially, in the Middle East and North Africa (MENA). The present study aimed to identify the genetic variation and haplotype distribution of camel isolates of E. granulosus sensu lato using all existing E. granulosus mitochondrial DNA data from camels in different parts of the world. Sequence data from 1,144 camel isolates of E. granulosus s.l. available in the NCBI GenBank including 57 camel hydatid cysts collected in central Iran were used to analyze the nature of genetic variation within the camel isolates of E. granulosus s.l. in MENA region. Fifty-seven camel isolates were also PCR-sequenced on mitochondrial 12S rRNA gene. Haplotype network analysis revealed seven different haplotypes clustered into four major groups. E. intermedius G6 was identified as the most commonly represented genotype in camels followed by G1. Mitochondrial 12S rRNA gene sequence analysis on 57 camel isolates identified three different genotypes, including E. intermedius/G6 (35/57, 61.4%), E. granulosus sensu stricto/G1-G3 (21/57, 36.8%) as well as one isolate identified as E. ortleppi/G5 (1/57, 1.8%). The number of base substitutions per site over 420 positions of partial 12S rRNA gene sequences were shown as 0.000 and 0.004 for E. intermedius (G6) corresponding to the Middle East and sub-Saharan isolates, respectively. Camel isolates of E. granulosus in the MENA region present moderate genetic diversity (Hd = 0.5540–0.6050). The Middle East isolates demonstrated a more diverse population than the North/sub-Saharan isolates, where six out of seven 12S rRNA haplotypes were identified in the former region. E. intermedius (G6 genotype) was shown to be the most common species in the world camel population. In conclusion, camels showed to be an important intermediate host species in the MENA region with different patterns of genetic variation between the Middle East and Africa.
Introduction
Tapeworms of the genus Echinococcus, causing a spectrum of infections known as echinococcosis, are members of the family Taeniidae. Cystic echinococcosis (CE) caused by the larval stages of Echinococcus granulosus sensu lato represents serious zoonotic infections in human and animals with a cosmopolitan distribution (1, 2). The life cycle involves dogs and other canids as definitive and domestic and wild ungulates as the intermediate hosts (3).
Extensive intraspecific variations have been documented within this species with significant epidemiological and clinical implications. E. granulosus genotype variation, may affect parasite life cycle and transmission patterns, host range and pathogenicity to humans (4, 5). Based on the biological and molecular genetic analyses using nuclear and mitochondrial DNA sequences 10 distinct genotypes with different host preferences have been identified for E. granulosus sensu lato and new nomenclature has been adopted for several genotypes (6) i.e., E. granulosus sensu stricto (G1–G3) with a wide range of intermediate hosts particularly sheep, goat and buffaloes, E. equinus (G4) of horses, E. ortleppi (G5) of cattle, E. intermedius (G6–G7) of camels and pigs, E. canadensis (G8 and G10) of Fennoscandian and subarctic cervids and E. felidis of African wild felid population (2).
According to Alvarez Rojas et al. about 88 and 11% of human CE infections are due to E. granulosus sensu stricto and E. intermedius, respectively (7). Therefore, camels are one of the most important intermediate hosts of the parasite especially in endemic regions of the Middle East and North Africa (MENA) where they play an important role in the transmission of E. granulosus. CE prevalence in camels has been estimated as 8–36% in different endemic countries (2, 8). In camels the presence of three Echinococcus species have been documented in different regions of the world, i.e., E. granulosus sensu stricto, E. intermedius, and E. ortleppi (2, 9). Several studies identified 100% of the parasites isolated from camels as E. intermedius (G6 genotype), however E. granulosus sensu stricto (G1–G3) has been shown to perpetuate in the camel-dog cycle as well (1, 10). Different studies showed 17.0–88.4% of camels harbored E. granulosus G1 genotype (11, 12). The nature and significance of Echinococcus variation in camels is poorly understood (6). Genotype G6 is more widespread in camels as a common intermediate host in the Middle East, Asia, and Africa, while G7 distribution has been reported among pigs and wild boars in Europe, the highly endemic Mediterranean areas and Central America (6). Both genotypes have been found to co-exist in Turkey, Argentina and Peru.
Mitochondrial DNA has been widely used for molecular epidemiological studies on helminth parasites mainly because of its conserved structure, mode of inheritance, and relatively high evolutionary rate. Currently one of the accepted markers for investigation of genetics and characterization of helminth parasites is mitochondrial 12S ribosomal RNA (12S rRNA) gene (13). The present study was conducted to provide a global outlook on the nature of genetic variation and haplotype distribution of E. granulosus sensu lato infecting camels using mitochondrial 12S rRNA gene sequence analysis.
Materials and Methods
In this study all existing 12S rRNA gene sequences of camel isolates of E. granulosus sensu lato were retrieved form NCBI GenBank. To collect a dataset of 12S rDNA region, sequences data searched in and downloaded from NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) using keywords “Echinococcus,” “12S rRNA,” and “camel” with mitochondrial records filtration for sequences longer than 400 bp. The search strategy was performed as follows: ((((“Echinococcus”[Organism] OR Echinococcus[All Fields]) AND 12S[All Fields]) AND mitochondrion[filter]) AND (“Camelus dromedarius”[Organism] OR camel[All Fields])) AND (“400”[SLEN]: “14000”[SLEN]). All sequence features were manually checked to confirm the parasite host as camel.
In addition, liver and lung samples were collected from 57 camels slaughtered in the municipal abattoirs of Kerman, Qom, and Tehran provinces. The protoscoleces and/or germinal layer were aspirated from each cyst and were preserved in −20°C after three-time washing in sterile saline solution. Total genomic DNA (gDNA) was extracted from individual cyst samples using High Pure PCR template preparation kit (Roche Diagnostic, Germany) and was stored at −20°C for future molecular analysis.
A partial 450-bp fragment of mitochondrial 12S rRNA gene was amplified using specific primers as previously described by Rostami et al. 12SRF (5′-AGGGGATAGGACACAGTGCCAGC-3′) as the forward and 12SRR (5′-CGGTGTGTACATGAGCTAAAC-3′) as reverse primers (14). No template controls were included in each experiment. The PCR products were electrophoresed on 1% (w/v) agarose gel containing ethidium bromide. Moreover, a partial mitochondrial Cytochrome c oxidase subunit 1 (cox1) gene was amplified from individual isolates using the primer sets JB3/JB4.5 for an accurate distinction between genotypes within E. granulosus sensu stricto (15). All amplicons were sequenced by an ABI-3730XL capillary machine (Macrogen Inc., South Korea).
Reference sequences in this study were collected form reference nucleotide dataset (RefSeq) for representative Echinococcus species. Sequence data were trimmed, aligned, and edited manually in BioEdit software v.7.2 after equalizing each sample sequence length by the elimination of the PCR primers sequences and global sequence alignments using ClustalW algorithm (16). Phylogenetic analysis was performed with the best-fit model of nucleotide substitution on 12S rDNA sequences using the program Mega 7 software (17). Molecular phylogeny of E. granulosus isolates was determined by maximum likelihood method, and the corresponding phylogenetic tree was constructed. The heuristic tree search and tree topologies algorithm reliability test were estimated with bootstrap testing with 1,000 replicates.
Average evolutionary divergence of 12s rRNA sequences of all MENA camel E. granulosus sensu lato genotypes/haplotypes were also studied with MEGA7 software by using the Kimura 2-parameter model and the rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
Population genetics analysis of haplotypes was performed on a consensus string of 420 positions in the aligned sequences for nucleotide diversity (π) and haplotype diversity (Hd) using DnaSP software (18). Associations between each haplotype of Echinococcus species form camel and their corresponding genotypes were inferred by constructing the TCS network using PopART software (19). The individual nucleotide sequence data reported in this study were submitted to the GenBank database.
Results
Of 1,144 camel isolates of E. granulosus s.l. sequences retrieved from NCBI GenBank, 68 records for 12S rRNA gene were identified (Table 1). All 57 isolates were successfully PCR-sequenced on 12S rRNA gene and the sequences were submitted to the NCBI GenBank database under the accession numbers MH395757–MH395800 for 12sRNA and MH397251–MH397259 for cox1. Sequence analyses of 12S rRNA gene showed four genotypes including 35 G6 (61.4%), 15 G1 (26.3%), 6 G3 (10.5%), and one G5 (1.8%) representing three Echinococcus species, i.e., E. intermedius, E. granulosus sensu stricto, and E. ortleppi. Six camel isolates within G1-G3 complex were identified as G3 genotype using partial cox1 sequence analysis.
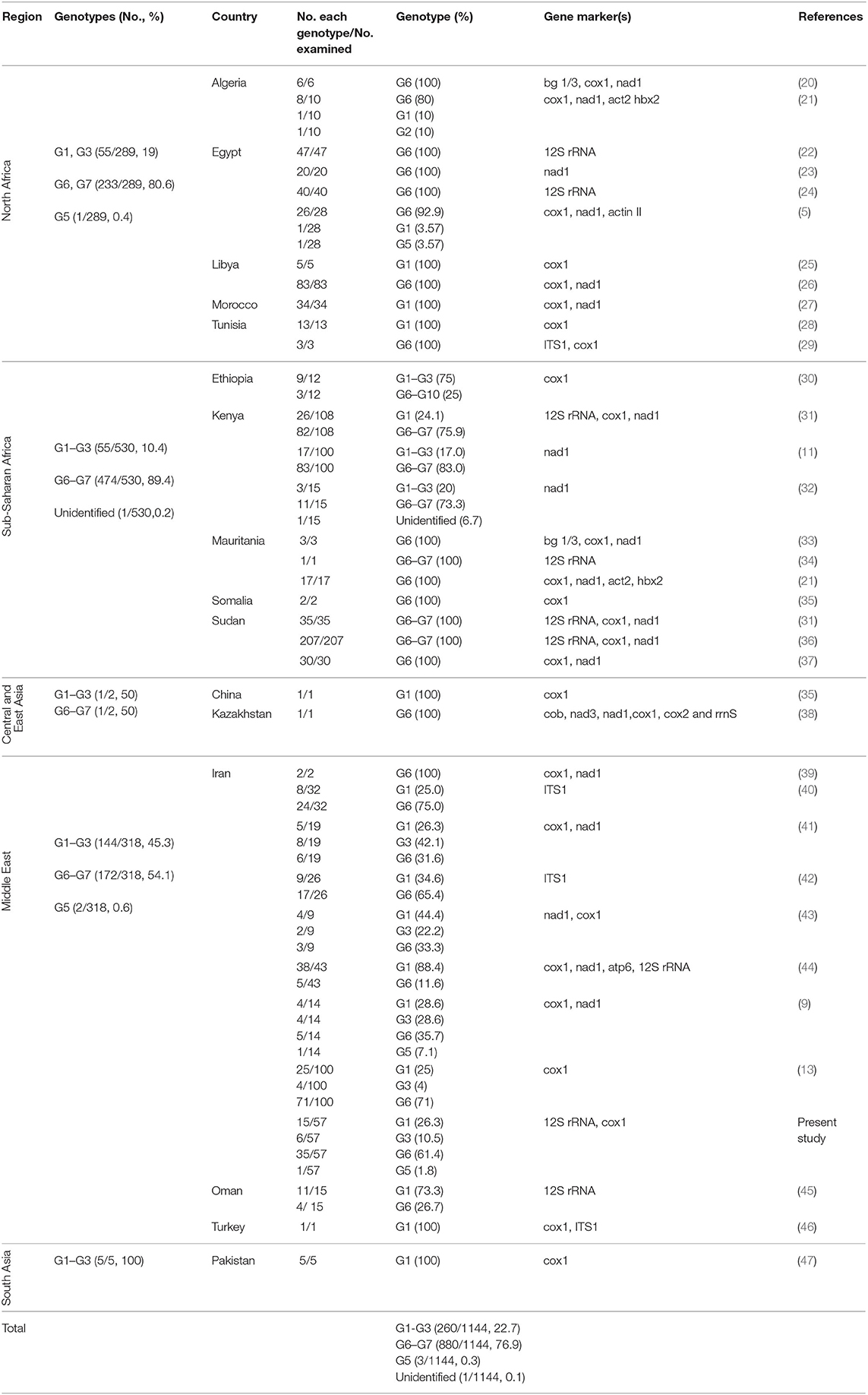
Table 1. The summary of global data on the frequency of E. granulosus sensu lato genotypes in camels (Camelus dromedarius) according to five major camel-rearing regions in the world.
Phylogenetic analysis of 12S rRNA sequence data from 57 camel isolates compared with reference genotypes of E. granulosus s.l. is shown in Figure 1. Four genotypes, G1, G3, G6, and G5 exhibit distinct clusters along with corresponding reference sequences representing all of the sequences determined in the present study. The haplotype diversity for 57 camel isolates from Iran (categorized into six haplotypes) was calculated as Hd = 0.6050 (±0.063). However, the haplotype diversity for 68 camel records from MENA region (categorized into seven haplotypes) was estimated at Hd = 0.5540 (±0.064).
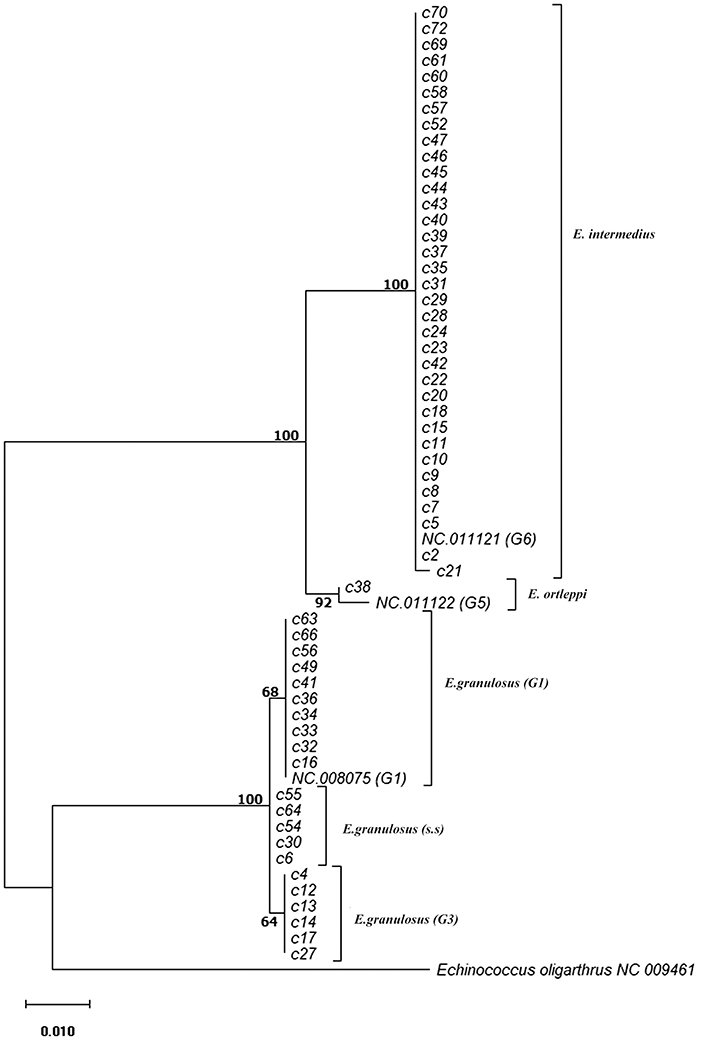
Figure 1. Phylogenetic relationships of camel isolates of Echinococcus granulosus sensu lato based on 12S mitochondrial rRNA sequences based on Hasegawa-Kishino-Yano model. The heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The percentage of trees in which the associated samples clustered together is shown next to the branches. The tree is drawn to scale with branch lengths based on the number of substitutions per site. The analysis involved 60 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 420 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.
Global data on the frequency of E. granulosus s.l. genotypes in camels according to five major camel-rearing regions in the world, i.e., the Middle East, North Africa, sub-Saharan Africa, East/Central Asia, and South Asia are summarized in Table 1. As it is shown, three different genotype patterns could be obtained in camels infected by E. granulosus sensu lato. E. intermedius (G6, camel strain) retained its dominance in camels from sub-Saharan Africa, so that about 90% of Echinococcus species isolated from camels were E. intermedius. In the Middle East, E. granulosus sensu stricto replaced E. intermedius, interestingly half of Echinococcus species isolated from camels in this region have been identified as E. granulosus sensu stricto. The situation in North Africa is something in between, ~80% of the parasites are G6 camel strain (Figure 2, Table 1).
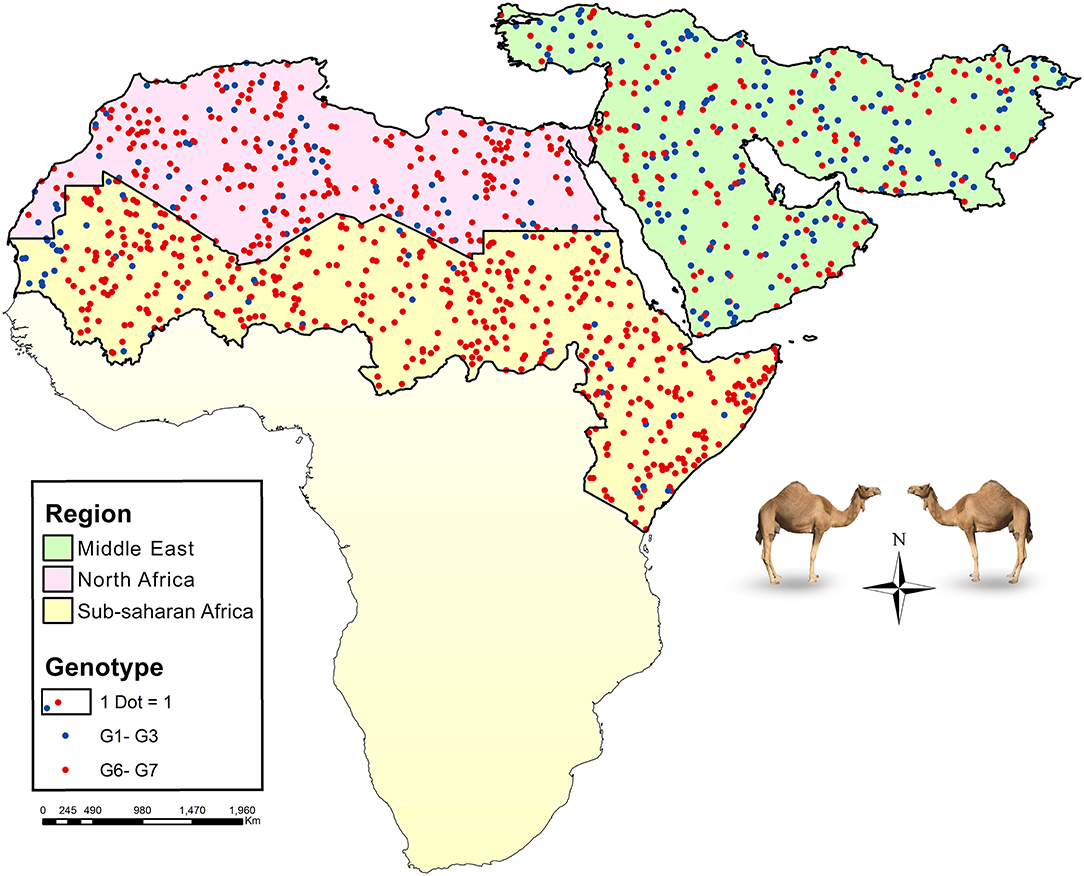
Figure 2. Map showing the relative frequency distribution of two major Echinococcus species infecting camels, E. granulosus sensu stricto (G1–G3) and E. intermedius (G6), in three main regions of camel CE transmission, the Middle East (green), North Africa (purple), and part of sub-Saharan Africa (yellow). Each dot represents a camel isolate.
The parsimony-based haplotype network analysis on all MENA records, indicated that E. granulosus sensu lato isolates were clustered into four major groups (Figure 3). The G6 genotype (NC011121) corresponds to the most commonly represented E. granulosus haplotype in camels originated from Central Asia, the Middle East and Africa, followed by G1 (NC008075) representing most of the isolates from Iran.
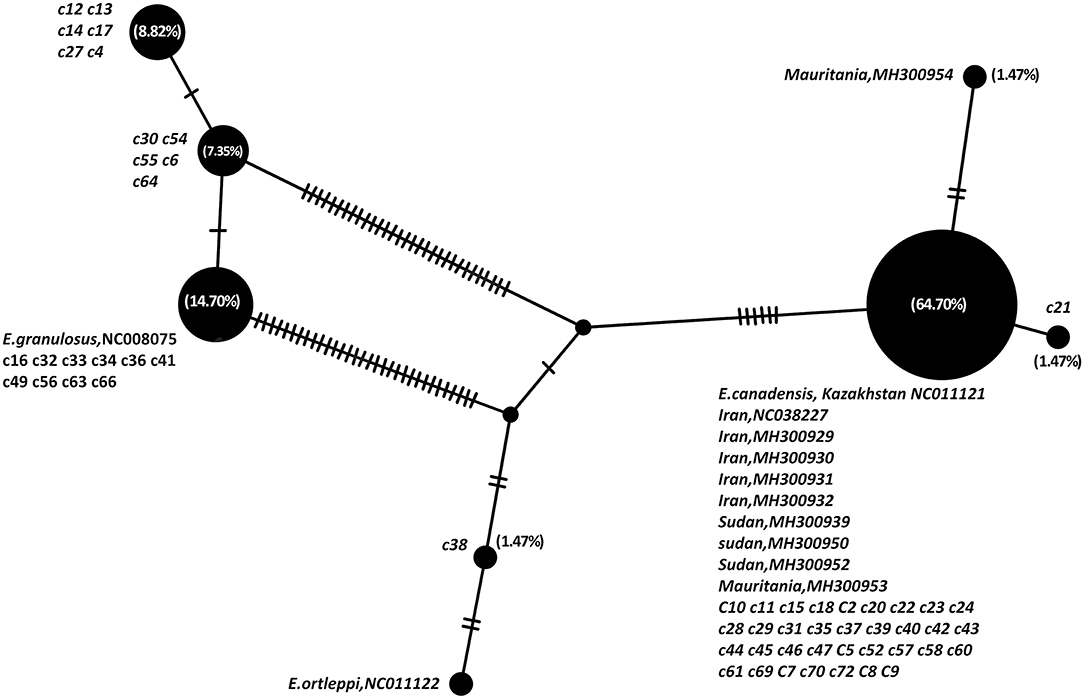
Figure 3. Haplotype network analysis of Echinococcus granulosus sensu lato of partial 420 bp of 12S rRNA gene on all available camel records from the Middle East and Africa, based on statistical parsimony. The size of the circles indicates the frequency of the haplotypes. The nucleotide accession numbers and each individual isolate code in the present study are indicated next to each circle. Numbers in parenthesis are the percent frequency of the isolates in each haplotype.
The number of base substitutions per site over all 420 positions of 12S rRNA partial sequence pairs were shown as 0.2–0.7% and 0.4% within the Middle Eastern and sub-Saharan E. intermedius (G6) isolates, respectively. The average overall distance of all MENA E. granulosus s.l. isolates was 0.6% (Figure 4).
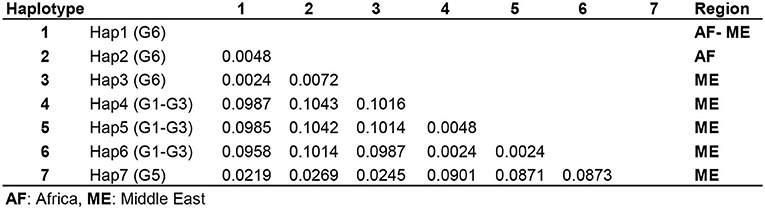
Figure 4. Pairwise comparison of nucleotide sequence differences (%) in mitochondrial 12S rRNA gene among seven available camel haplotypes within Echinococcus granulosus sensu lato.
Discussion
Control of cystic echinococcosis is complicated due to the fact that CE transmission occurs through different definitive/intermediate hosts systems (48). In this regard cystic echinococcosis of dromedaries has been received less attention among different livestock species. Camels play an essential role in the epidemiology and transmission of CE in the Middle East and North Africa. Little is known about the nature and significance of genetic variation of camel isolates of E. granulosus s.l. in the endemic areas. Most of the studies investigated a limited number of camel isolates which are not representatives of E. granulosus population in camels. In this study we present a global outlook of the significance of different species/genotypes of camel isolates of E. granulosus sensu lato in the molecular epidemiology of CE in the main camel breeding regions of the world using all available genetic markers including both nuclear and mitochondrial regions.
It has been already shown that 12S rRNA gene is a suitable marker for differentiating genotypes of E. granulosus, nonetheless few 12S rRNA nucleotide data are available in GenBank, particularly from camel CE. In the present study we investigated 12S rRNA gene diversity of 57 camel isolates of E. granulosus s.l. from Iran. The findings indicate that G6 genotype is the most common genotype (61.4%) in this Iranian dromedary population. The results also revealed the existence of three other genotypes including G1 (26.3%), G3 (10.5%), and G5 (1.8%).
The study showed that E. granulosus s.l. perpetuates in a camel-dog cycle mostly comprising of G6 genotype (E. intermedius) and several other less prevalent strains. Both species, E. granulosus s.s. and E. intermedius are believed to be overlapped in their cycle so that sheep and camels have contributed to the transmission of both species/genotypes. However, there are controversies on the contribution of the genotypes in each individual intermediate host species. Several studies showed the dominant species infecting camels is E. granulosus s.s. G1 genotype (10, 25, 28, 30, 43–45, 49), while several other studies reported higher frequency of E. intermedius in camels than E. granulosus s.s. (4, 9, 11, 20, 22, 25, 26, 29, 31, 33, 34, 36, 50, 51). In addition, sylvatic transmission of CE among camel populations should also be considered. Wild carnivores particularly wolves and golden jackals have long been considered as epidemiologically important definitive hosts in MENA region (52, 53). Regarding the nature of camel husbandry, the animals are usually roaming freely in vast geographical areas outside human dwellings during their long life span, therefore it is quite probable for camels to get involved in the sylvatic life cycle of E. granulosus sensu lato.
Assuming that G6 parasites had primarily perpetuated in wild camels across the old world, this is probably an indication of different forces of infection with G1 sheep strain in different camel-rearing areas from the Iranian plateau to the Maghreb and sub-Saharan Africa. With the Middle East has already been occupied by E. granulosus s.s. G1 genotype (sheep strain) as the dominant genotype in the region, exerting remarkable forces of infection on the camels regularly infected by G6 genotype.
The camel domestication took place some 8000 years later than that of sheep and goat (54).
It is believed that camels were first domesticated in the Fertile Crescent and Arabian Peninsula in 2000–1000 BCE. Increasing trade of basic commodities including incense, myrrh and frankincense mainly transported by the camels across the region, facilitated the dispersion of E. granulosus s.s. We could not find any significant different nucleotide diversity among isolates from Middle East and sub-Saharan Africa. This may be due the fact that the emergence of domestic camel-dog cycle of E. granulosus s.l. following camel domestication is a relatively recent event, therefore the development of significant haplotype diversity within the camel isolates is not expected in a short period of time. Similar molecular epidemiological picture has been demonstrated in sheep-dog cycle in which the universal sheep strain (G1 genotype) is the dominant variant, however other genotypes including G3 and G6 are perpetuating in the G1 endemic areas.
It should be noted that the sample size in several camel studies is small and the findings are not conclusive (Table 1). Mitochondrial gene sequences are highly informative markers for molecular taxonomy and phylogenetic investigation of helminth parasites. Cytochrome c oxidase subunit 1 and 12S ribosomal RNA genes have been extensively used for genetic classification of tapeworms. However, few studies on E. granulosus 12S rRNA sequences have been performed on African camel isolates and obviously more 12S rRNA data is required from this important endemic region. It has been shown that 12S rRNA gene presents higher sequence variability than cox1, therefore it is assumed that the camel isolates of E. granulosus s.l. in Africa present even more homogeneous population than those of the Middle East (13).
In addition, the majority of the studies have used small partial fragments of mitochondrial genes. Using longer sequences in phylogeographical studies provides more reliable information on the molecular epidemiology of CE in camels (6). This study presents the second evidence of G5 presence in the Middle East camels. This suggests that E. ortleppi, the common cattle strain, could also be transmissible through a camel-dog cycle in the region. Previous studies in MENA indicated that camels usually harbor highly fertile/highly viable hydatid cysts reaching >90% fertility and >80% viability (14, 28, 55, 56). However, interestingly in our study few protoscoleces were found in the camel hydatid cyst harboring G5 genotype (E. ortleppi) and DNA was extracted from the cyst germinal layer. This is an indication of low susceptibility of camels to E. ortleppi and can explain the low prevalence of G5 in camels, however, to obtain a more comprehensive picture of the global epidemiology of E. ortleppi, further studies are required on the molecular epidemiology of camel CE in the MENA region.
Conclusion
Global data on the frequency of E. granulosus s.l. genotypes in camel populations of major camel-rearing regions of the world revealed a changing pattern of genotype distribution between the Middle East and African isolates. E. intermedius (G6 genotype) was identified as the most common species in camels, however the contribution of this species in camel populations in various areas of MENA is significantly different across the region from the Iranian plateau to sub-Saharan Africa. Mitochondrial 12S rDNA study of camel isolates of E. granulosus s.l. revealed significant species/genotype diversity. Camel isolates of E. granulosus in the MENA region present moderate genetic diversity with the Middle East isolates demonstrating a more diverse population than the North/sub-Saharan isolates, where three species, four genotypes and six different 12S rRNA haplotypes were identified in the region.
Camels are an important intermediate host species in Iran, harboring different species and genotypes of E. granulosus s.l. throughout their long lifetime. More in-depth large-scale studies using multiple large fragments of mitochondrial/nuclear gene sequences are required to elucidate the significance and actual contribution of camels in CE epidemiology.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
This project was reviewed and approved by Research Review Committee of the National Institutes for Medical Research Development (NIMAD), No. 971237. The animals were slaughtered as part of the normal daily practice in the abattoirs.
Author Contributions
MF, MM, and SR: conceptualization and study design. MD, SH, SN, and SR: data curation. MD, MM, SN, and SR: data analysis and laboratory experiments. MF: funding acquisition. MM, SH, SR, and MF: data validation. MD, MM, SH, SN, SR, and MF: writing—original draft preparation. All authors contributed to the article and approved the submitted version.
Funding
Research reported in this publication was supported by Elite Researcher Grant Committee under award number 971237 from the National Institutes for Medical Research Development (NIMAD), Tehran, Iran.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AS declared a past co-authorship with one of the author MF to the handling editor.
Acknowledgments
This project was financially supported by the National Institutes for Medical Research Development (NIMAD), No. 971237. The authors wish to thank all the veterinary staff of different abattoirs in Kerman, Qom, and Tehran provinces that help collecting parasite specimens for this study.
References
1. Cardona GA, Carmena D. A review of the global prevalence, molecular epidemiology and economics of cystic echinococcosis in production animals. Vet Parasitol. (2013) 192:10–32. doi: 10.1016/j.vetpar.2012.09.027
2. Deplazes P, Rinaldi L, Alvarez Rojas CA, Torgerson PR, Harandi MF, Romig T, et al. Global distribution of alveolar and cystic echinococcosis. Adv Parasitol. (2017) 95:315–493. doi: 10.1016/bs.apar.2016.11.001
3. Harandi MF, Budke CM, Rostami S, Fasihi Harandi M, Budke CM, Rostami S, et al. The monetary burden of cystic echinococcosis in Iran. PLoS Negl Trop Dis. (2012) 6:e1915. doi: 10.1371/journal.pntd.0001915
4. Thompson RCA. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. (2008) 119:439–46. doi: 10.1016/j.exppara.2008.04.016
5. Amer S, Helal IB, Kamau E, Feng Y, Xiao L. Molecular characterization of Echinococcus granulosus sensu lato from farm animals in Egypt. PLoS ONE. (2015) 10:e0118509. doi: 10.1371/journal.pone.0118509
6. Laurimäe T, Kinkar L, Romig T, Omer RA, Casulli A, Umhang G, et al. The benefits of analysing complete mitochondrial genomes: deep insights into the phylogeny and population structure of Echinococcus granulosus sensu lato genotypes G6 and G7. Infect Genet Evol. (2018) 64:85–94. doi: 10.1016/j.meegid.2018.06.016
7. Alvarez Rojas CA, Romig T, Lightowlers MW. Echinococcus granulosus sensu lato genotypes infecting humans - review of current knowledge. Int J Parasitol. (2014) 44:9–18. doi: 10.1016/j.ijpara.2013.08.008
8. Ibrahim MM. Study of cystic echinococcosis in slaughtered animals in Al Baha region, Saudi Arabia: interaction between some biotic and abiotic factors. Acta Trop. (2010) 113:26–33. doi: 10.1016/j.actatropica.2009.08.029
9. Ebrahimipour M, Sadjjadi SM, Yousofi Darani H, Najjari M. Molecular studies on cystic echinococcosis of camel (Camelus dromedarius) and report of Echinococcus ortleppi in Iran. Iran J Parasitol. (2017) 12:323–31.
10. Sharbatkhori M, Mirhendi H, Harandi MF, Rezaeian M, Mohebali M, Eshraghian M, et al. Echinococcus granulosus genotypes in livestock of Iran indicating high frequency of G1 genotype in camels. Exp Parasitol. (2010) 124:373–9. doi: 10.1016/j.exppara.2009.11.020
11. Hüttner M, Siefert L, Mackenstedt U, Romig T. A survey of Echinococcus species in wild carnivores and livestock in East Africa. Int J Parasitol. (2009) 39:1269–76. doi: 10.1016/j.ijpara.2009.02.015
12. Rostami S, Shariat Torbaghan S, Dabiri S, Babaei Z, Ali Mohammadi M, Sharbatkhori M, et al. Genetic characterization of Echinococcus granulosus from a large number of formalin-fixed, paraffin-embedded tissue samples of human isolates in Iran. Am J Trop Med Hyg. (2015) 92:588–94. doi: 10.4269/ajtmh.14-0585
13. Rostami S, Salavati R, Beech RN, Sharbatkhori M, Babaei Z, Saedi S, et al. Cytochrome c oxidase subunit 1 and 12S ribosomal RNA characterization of Coenurus cerebralis from sheep in Iran. Vet Parasitol. (2013) 197:141–51. doi: 10.1016/j.vetpar.2013.07.001
14. Rostami S, Talebi S, Babaei Z, Sharbatkhori M, Ziaali N, Rostami H, et al. High resolution melting technique for molecular epidemiological studies of cystic echinococcosis: differentiating G1, G3, and G6 genotypes of Echinococcus granulosus sensu lato. Parasitol Res. (2013) 112:3441–7. doi: 10.1007/s00436-013-3523-z
15. Bowles J, Blair D, Mcmanus DPDPDPDPP, Bessho Y, Ohama T, Osawa S, et al. A molecular phylogeny of the genus Echinococcus. Parasitology. (1995) 110:317–28. doi: 10.1017/S0031182000080902
16. Hall TAT. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series, London (1999). p. 95–98.
17. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
18. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. (2009) 25:1451–2. doi: 10.1093/bioinformatics/btp187
19. Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. (2015) 6:1110–6. doi: 10.1111/2041-210X.12410
20. Bardonnet K, Benchikh-Elfegoun MC, Bart JM, Harraga S, Hannache N, Haddad S, et al. Cystic echinococcosis in Algeria: Cattle act as reservoirs of a sheep strain and may contribute to human contamination. Vet Parasitol. (2003) 116:35–44. doi: 10.1016/S0304-4017(03)00255-3
21. Maillard S, Benchikh-Elfegoun MC, Knapp J, Bart JM, Koskei P, Gottstein B, et al. Taxonomic position and geographical distribution of the common sheep G1 and camel G6 strains of Echinococcus granulosus in three African countries. Parasitol Res. (2007) 100:495–503. doi: 10.1007/s00436-006-0286-9
22. Aaty HEA, Abdel-Hameed DM, Alam-Eldin YH, El-Shennawy SF, Aminou HA, Makled SS, et al. Molecular genotyping of Echinococcus granulosus in animal and human isolates from Egypt. Acta Trop. (2012) 121:125–8. doi: 10.1016/j.actatropica.2011.10.014
23. Khalifa N, F, Khater H, A, Fahmy H, E.I. Radwan M, S.A. Afify J. Genotyping and phylogenetic analysis of cystic Echinococcosis isolated from camels and humans in Egypt. Am J Epidemiol Infect Dis. (2014) 2:74–82. doi: 10.12691/ajeid-2-3-2
24. Alam-Eldin YH, Abdel Aaty HE, Ahmed MA. Molecular characterization of cystic echinococcosis: first record of G7 in Egypt and G1 in Yemen. Acta Parasitol. (2015) 60:662–5. doi: 10.1515/ap-2015-0094
25. Tashani OA, Zhang LH, Boufana B, Jegi A, McManus DP. Epidemiology and strain characteristics of Echinococcus granulosus in the Benghazi area of eastern Libya. Ann Trop Med Parasitol. (2002) 96:369–81. doi: 10.1179/000349802125000952
26. Abushhewa MH, Abushhiwa MHS, Nolan MJ, Jex AR, Campbell BE, Jabbar A, et al. Genetic classification of Echinococcus granulosus cysts from humans, cattle and camels in Libya using mutation scanning-based analysis of mitochondrial loci. Mol Cell Probes. (2010) 24:346–51. doi: 10.1016/j.mcp.2010.07.005
27. Azlaf R, Dakkak A. Epidemiological study of the cystic echinococcosis in Morocco. Vet Parasitol. (2006) 137:83–93. doi: 10.1016/j.vetpar.2006.01.003
28. Lahmar S, Debbek H, Zhang LH, McManus DP, Souissi A, Chelly S, et al. Transmission dynamics of the Echinococcus granulosus sheep-dog strain (G1 genotype) in camels in Tunisia. Vet Parasitol. (2004) 121:151–6. doi: 10.1016/j.vetpar.2004.02.016
29. M'rad S, Filisetti D, Oudni M, Mekki M, Belguith M, Nouri A, Sayadi T, et al. Molecular evidence of ovine (G1) and camel (G6) strains of Echinococcus granulosus in Tunisia and putative role of cattle in human contamination. Vet Parasitol. (2005) 129:267–72. doi: 10.1016/j.vetpar.2005.02.006
30. Hailemariam Z, Nakao M, Menkir S, Lavikainen A, Yanagida T, Okamoto M, et al. Molecular identification of unilocular hydatid cysts from domestic ungulates in Ethiopia: Implications for human infections. Parasitol Int. (2012) 61:375–7. doi: 10.1016/j.parint.2012.01.007
31. Dinkel A, Njoroge EM, Zimmermann A, Wälz M, Zeyhle E, Elmahdi IE, et al. A PCR system for detection of species and genotypes of the Echinococcus granulosus-complex, with reference to the epidemiological situation in eastern Africa. Int J Parasitol. (2004) 34:645–53. doi: 10.1016/j.ijpara.2003.12.013
32. Mbaya H, Magambo J, Njenga S, Zeyhle E, Mbae C, Mulinge E, et al. Echinococcus spp. in central Kenya: a different story. Parasitol Res. (2014) 113:3789–94. doi: 10.1007/s00436-014-4045-z
33. Bardonnet K, Piarroux R, Dia L, Schneegans F, Beurdeley A, Godot V, et al. Combined eco-epidemiological and molecular biology approaches to assess Echinococcus granulosus transmission to humans in Mauritania: occurrence of the ‘camel’ strain and human cystic echinococcosis. Trans R Soc Trop Med Hyg. (2002) 96:383–6. doi: 10.1016/S0035-9203(02)90369-X
34. Farjallah S, Busi M, Mahjoub MO, Slimane B Ben, Said K, D'Amelio S. Molecular characterization of Echinococcus granulosus in Tunisia and Mauritania by mitochondrial rrnS gene sequencing. Parassitologia. (2007) 49:239–46.
35. Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. (1992) 54:165–73. doi: 10.1016/0166-6851(92)90109-W
36. Omer RA, Dinkel A, Romig T, Mackenstedt U, Elnahas AA, Aradaib IE, et al. A molecular survey of cystic echinococcosis in Sudan. Vet Parasitol. (2010) 169:340–6. doi: 10.1016/j.vetpar.2010.01.004
37. Ibrahim K, Thomas R, Peter K, Omer RA. A molecular survey on cystic echinococcosis in Sinnar area, blue Nile state (Sudan). Chin Med J. (2011) 124:2829–33. doi: 10.3760/cma.j.issn.0366-6999.2011.18.006
38. Nakao M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology. (2007) 134:713–22. doi: 10.1017/S0031182006001934
39. Zhang L, Eslami A, Hosseini SH, McManus DP. Indication of the presence of two distinct strains of Echinococcus granulosus in Iran by mitochondrial DNA markers. Am J Trop Med Hyg. (1998) 59:171–4. doi: 10.4269/ajtmh.1998.59.171
40. Harandi MF, Hobbs RP, Adams PJ, Mobedi I, Morgan-Ryan UM, Thompson RCA. Molecular and morphological characterization of Echinococcus granulosus of human and animal origin in Iran. Parasitology. (2002) 125:367–73. doi: 10.1017/S0031182002002172
41. Sharbatkhori M, Fasihi Harandi M, Mirhendi H, Hajialilo E, Kia EB. Sequence analysis of cox1 and nad1 genes in Echinococcus granulosus G3 genotype in camels (Camelus dromedarius) from central Iran. Parasitol Res. (2011) 108:521–7. doi: 10.1007/s00436-010-2092-7
42. Shahnazi M, Hejazi H, Salehi M, Andalib AR. Molecular characterization of human and animal Echinococcus granulosus isolates in Isfahan, Iran. Acta Trop. (2011) 117:47–50. doi: 10.1016/j.actatropica.2010.09.002
43. Hajialilo E, Harandi MF, Sharbatkhori M, Mirhendi H, Rostami S. Genetic characterization of Echinococcus granulosus in camels, cattle and sheep from the south-east of Iran indicates the presence of the G3 genotype. J Helminthol. (2012) 86:263–70. doi: 10.1017/S0022149X11000320
44. Rostami Nejad M, Taghipour N, Nochi Z, Nazemalhosseini Mojarad E, Mohebbi SR, Fasihi Harandi M, et al. Molecular identification of animal isolates of Echinococcus granulosus from Iran using four mitochondrial genes. J Helminthol. (2012) 86:485–92. doi: 10.1017/S0022149X1100071X
45. Al Kitani FA, Al Riyami S, Al Yahyai S, Al Awahi AH, Al Aawali M, Hussain MH. Abattoir based surveillance of cystic echinococcosis (CE) in the Sultanate of Oman during 2010-2013. Vet Parasitol. (2015) 211:208–15. doi: 10.1016/j.vetpar.2015.06.011
46. Utuk AE, Simsek S, Koroglu E, McManus DP. Molecular genetic characterization of different isolates of Echinococcus granulosus in east and southeast regions of Turkey. Acta Trop. (2008) 107:192–4. doi: 10.1016/j.actatropica.2008.05.026
47. Latif AA, Tanveer A, Maqbool A, Siddiqi N, Kyaw-Tanner M, Traub RJ. Morphological and molecular characterisation of Echinococcus granulosus in livestock and humans in Punjab, Pakistan. Vet Parasitol. (2010) 170:44–9. doi: 10.1016/j.vetpar.2010.02.003
48. Tamarozzi F, Deplazes P, Casulli A. Reinventing the wheel of Echinococcus granulosus sensu lato transmission to humans. Trends Parasitol. (2020)36:427–34. doi: 10.1016/j.pt.2020.02.004
49. Sharbatkhori M, Mirhendi H, Jex AR, Pangasa A, Campbell BE, Kia EB, et al. Genetic categorization of Echinococcus granulosus from humans and herbivorous hosts in Iran using an integrated mutation scanning-phylogenetic approach. Electrophoresis. (2009) 30:2648–55. doi: 10.1002/elps.200900145
50. Schwenkenbecher JM, Wirth T, Schnur LF, Jaffe CL, Schallig H, Al-Jawabreh A, et al. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int J Parasitol. (2006) 36:237–46. doi: 10.1016/j.ijpara.2005.09.010
51. Knapp J, Bart JM, Glowatzki M-L, Ito A, Gerard S, Maillard S, et al. Assessment of use of microsatellite polymorphism analysis for improving spatial distribution tracking of Echinococcus multilocularis. J Clin Microbiol. (2007) 45:2943–50. doi: 10.1128/JCM.02107-06
52. Dalimi A, Motamedi G, Hosseini M, Mohammadian B, Malaki H, Ghamari Z, et al. Echinococcosis/hydatidosis in western Iran. Vet Parasitol. (2002) 105:161–71. doi: 10.1016/s0304-4017(02)00005-5
53. Gholami S, Jahandar H, Abastabar M, Pagheh A, Mobedi I, Sharbatkhori M. Echinococcus granulosus sensu stricto in Dogs and Jackals from Caspian Sea region, northern Iran. Iran J Parasitol. (2016) 11:186–94.
54. Orlando L. Back to the roots and routes of dromedary domestication. Proc Natl Acad Sci USA. (2016) 113:6588–90. doi: 10.1073/pnas.1606340113
55. Casulli A, Zeyhle E, Brunetti E, Pozio E, Meroni V, Genco F, et al. Molecular evidence of the camel strain (G6 genotype) of Echinococcus granulosus in humans from Turkana, Kenya. Trans R Soc Trop Med Hyg. (2010) 104:29–32. doi: 10.1016/j.trstmh.2009.08.001
Keywords: hydatid disease, Echinococcus intermedius, Echinococcus canadensis, 12S rDNA, genetic variation, Camelus dromedarius, strain, MENA
Citation: Dehghani M, Mohammadi MA, Hemmati S, Nasibi S, Rostami S and Fasihi Harandi M (2020) Cystic Echinococcosis of Camels: 12S rRNA Gene Variation Revealed Changing Pattern of Genetic Diversity Within Echinococcus granulosus sensu lato in the Middle East and North/Sub-Saharan Africa. Front. Vet. Sci. 7:618. doi: 10.3389/fvets.2020.00618
Received: 11 May 2020; Accepted: 29 July 2020;
Published: 17 September 2020.
Edited by:
Viliam Šnábel, Institute of Parasitology (SAS), SlovakiaReviewed by:
Adel Spotin, Tabriz University of Medical Sciences, IranAntonio Varcasia, University of Sassari, Italy
Copyright © 2020 Dehghani, Mohammadi, Hemmati, Nasibi, Rostami and Fasihi Harandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Ali Mohammadi, bWEubW9oYW1tYWRpQGttdS5hYy5pcg==; Majid Fasihi Harandi, ZmFzaWhpQGttdS5hYy5pcg==
 Mansoureh Dehghani
Mansoureh Dehghani Mohammad Ali Mohammadi
Mohammad Ali Mohammadi Sahel Hemmati2
Sahel Hemmati2 Majid Fasihi Harandi
Majid Fasihi Harandi