- Department of Veterinary Clinical Sciences, College of Veterinary Medicine, China Agricultural University, Beijing, China
Diarrheagenic Escherichia coli is the causative agent of diarrhea in infants and animals worldwide. Many isolated strains recovered from pigs with postweaning diarrhea are multidrug resistance (MDR), and hybrids of E. coli are potentially more virulent, as enterotoxigenic E. coli (ETEC)/Shiga-toxigenic E. coli (STEC) hybrids. Here, we used whole-genome sequencing to analyze clinical isolates of the five colistin-resistant E. coli. The E. coli CAU15104, CAU15134, and CAU16060 belonged to ETEC/STEC hybrids, displaying the same serotype O3:H45 and sequence type ST4214. The E. coli CAU16175 and CAU16177 belonged to atypical enteropathogenic E. coli (aEPEC), display O4:H11 and O103:H2, ST29, and ST20, respectively. The E. coli CAU16175 carries six plasmids. An IncHI2-type plasmid, pCAU16175_1, harbors an IS26-enriched MDR region, which includes 16 antimicrobial-resistant genes. An IncFII-type plasmid, pCAU16175_3, harbors mcr-1.1, tet(M), and blaTEM−1B, whereas mcr-1.1 is located within a Tn2 derivative. Our findings indicate that the ETEC/STEC strains of the O3:H45 serotype as well as the aEPEC strains of the O4:H11 and O103:H2 serotypes are associated with postweaning diarrhea in swine and that some of diarrheagenic E. coli contains IS26-enriched MDR region and the mcr-1 gene located within a Tn2 derivative on IncFII plasmid.
Introduction
Diarrheagenic Escherichia coli (DEC) is a leading cause of infectious diarrhea in humans and animals around the world (1, 2). Diarrheagenic E. coli has six well-described pathotypes: enteropathogenic E. coli (EPEC), which is subdivided into typical EPEC (tEPEC) and atypical EPEC (aEPEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli, enteroaggregative E. coli, diffusely adherent E. coli, and enterohemorrhagic E. coli, which is subgroup of Shiga-toxigenic E. coli (STEC) (3, 4). All of these DECs possess diverse virulence factors, which are encoded by virulence genes and are responsible for their pathogenicity (5). Enterotoxigenic E. coli strains typically produce one or two toxins, heat-labile enterotoxin (LT) encoded by ltc, and heat-stable enterotoxin (ST) encoded by st (6). In pigs, STEC strains are characterized by producing the Shiga-like toxin variant Stx2e encoded by stx2e (7). Enteropathogenic E. coli strains are defined as forming the attaching and effacing (A/E) lesions mediated by genes located on the locus of enterocyte effacement (LEE) pathogenicity island (including eae) in the intestinal epithelium but not produce Shiga-like toxin (Stx) (8). Moreover, bfpA, which encodes the major subunit of bundle forming pili, is used to subdivide EPEC into tEPEC and aEPEC. Hence, aEPEC strains are defined by eae+, bfpA−, and stx− (9). Three pathotypes (ETEC, STEC, EPEC) are major DEC causing postweaning diarrhea (PWD) in pigs, and many hybrids of E. coli (ETEC/STEC, ETEC/STEC/EPEC) are present during PWD (6, 10). The recent data from The Global Enteric Multicenter Study showed that tEPEC and ETEC associated with a higher risk of fatal outcomes in children younger than 24 months with moderate to severe diarrhea (11). Atypical EPEC has outbreaks linked to diarrhea in children (12).
The occurrence of antimicrobial resistance and prevalence of multidrug resistance (MDR) Gram-negative Enterobacteriaceae has been increasing worldwide between humans and animals (13). In the cases of PWD, antimicrobial treatment has been widely used and caused severe drug resistance in DEC (1, 6, 14). In our previous study, among the 171 E. coli isolates, 94.15% of the strains were MDR, with antimicrobial resistance rates ranging from 2.34% for meropenem to 90.05% for nalidixic acid (6). Among the ETEC strains that cause PWD, resistance to apramycin, neomycin, trimethoprim sulfonamide, and colistin has been increasingly observed (1). Multidrug resistance strains may spread from animals to humans, causing antibiotics ineffective and increasing mortality and morbidity in developing countries (15).
Colistin as the last resort treatment against MDR bacterial infections may have been challenged by the mobile colistin-resistant gene (mcr-1), which has received widespread attention in different species of Enterobacteriaceae found in animals and humans around the world since it was first reported (16–19). So far, nine allelic variants of mcr-1 (mcr-2 to mcr-10) have been detected (20, 21). In addition, they have large number of variants, such as mcr-1 (mcr-1.1 to mcr-1.22), mcr-2 (mcr-2.1 to mcr-2.3), mcr-3 (mcr-3.1 to mcr-3.30), mcr-4 (mcr-4.1 to mcr-4.6), mcr-5 (mcr-5.1 to mcr-5.4), and mcr-8 (mcr-8.1 and mcr-8.2) (22). Our previous study found that the resistance rate to colistin was 20.47% in 171 E. coli isolates (6), and a recent article reported that direct sample testing rates of mcr-1 were higher than the rates of mcr-1–positive E. coli (64.6 vs. 49.2%) (23).
Insertion sequences (ISs) and transposons (Tns), arguably most numerous autonomous transposable elements, are crucial to shape their host genomes, particularly important in the bacterial antimicrobial resistance (24). IS26 and Tn2, which is a member of the IS6 family and Tn3 family, respectively, play a key role in the dissemination of antimicrobial-resistant genes in Gram-negative bacteria (25, 26). IS26 is often existed in MDR Gram-negative bacteria, which usually carry large regions containing several antimicrobial-resistant genes that are flanked by and interspersed with copies of IS26 (25). Tn2 is the most abundant in commensal E. coli. (27). ISSwi1-m2 in pNJST258C2 is a derivative of Tn2 that includes IS26 compared with Tn2 (28). ISApl1, which is an IS initially identified in Actinobacillus pleuropneumoniae, is a member of the IS30 family and is considered to be an essential element in the mobilization of mcr-1 (29, 30).
In the present study, we used whole-genome sequencing (WGS) to analyze virulence and resistant genes of the five clinical colistin-resistant E. coli isolates recovered from pigs with PWD and to characterize the complete sequence of a Tn2 derivative carrying mcr-1.1 on IncFII plasmid, pCAU16175_3, from a swine aEPEC isolate.
Materials and Methods
Bacterial Isolation and Identification
A total of 455 E. coli strains were obtained from feces samples or small intestinal content from pigs with diarrhea in China between 2014 and 2016 and identified with polymerase chain reaction (PCR) amplification of the uspA gene (6). Of these, 171 E. coli isolates were further screened by antimicrobial susceptibility testing, and the resistance rate of colistin was 20.47% (6). From these 35 colistin-resistant E. coli, we observed ST4214 was a major clone (6/35, 17.14%) based on the multilocus sequence typing and caused severe damage to IPEC-J2 cells (6). Then, ST4214 E. coli CAU15014, CAU15134, and CAU16060 were selected from different swine farms (one isolate per farm), and two MDR aEPEC isolates (E. coli CAU16175 and CAU16177) were randomly chosen for WGS.
Antimicrobial Susceptibility Testing
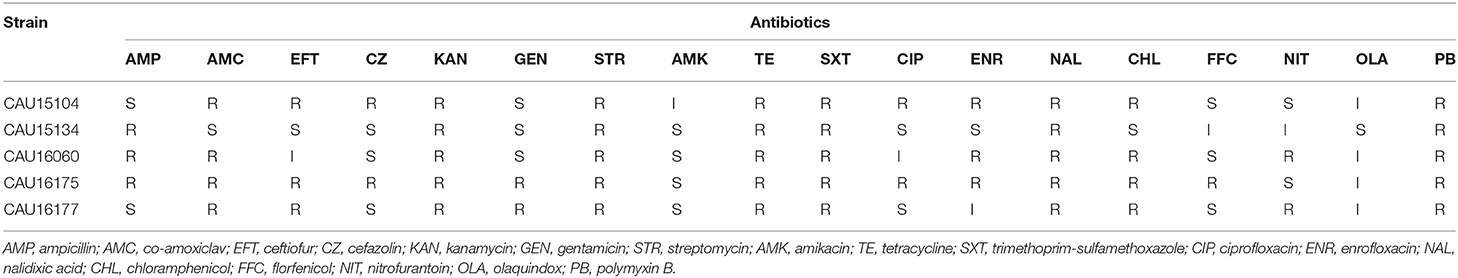
The susceptibility of the five isolates to 18 antimicrobials were tested by determining the minimum inhibitory concentration (MIC) using the US Clinical and Laboratory Standards Institute (CLSI) broth micro method (31). The results of MIC for ampicillin (AMP), co-amoxiclav (AMC), cefazolin (CZ), kanamycin (KAN), gentamicin (GEN), amikacin (AMK), tetracycline (TE), trimethoprim-sulfamethoxazole (SXT), ciprofloxacin (CIP), nalidixic acid (NAL), chloramphenicol (CHL), and nitrofurantoin (NIT) were interpreted according to guidelines of CLSI 2016 M100-S26 (31). The results of MIC for ceftiofur (EFT), enrofloxacin (ENR), and florfenicol (FFC) were interpreted according to CLSI VET01-A4 (32). In addition, the results with MIC values were defined resistant: streptomycin (STR) ≥64 μg/mL (33), olaquindox (OLA) ≥64 μg/mL (34), and polymyxin B (PB) >2 μg/mL (35). Escherichia coli ATCC 25922 was used as the quality control. According to the MIC determined for each antimicrobial, the isolates were defined as “susceptible,” “intermediate,” or “resistant.”
Whole-Genome Sequencing
Bacterial isolates were recultured from stock; DNA was extracted using a TIANamp Bacteria DNA kit (Tiangen Biotech Inc., Beijing, China). We used WGS by the Illumina Hiseq platform to get draft genome of the five E. coli isolates and further used WGS by the Oxford Nanopore Technologies MinION platform to get complete genome of the E. coli CAU16175.
The complete genome assembly was constructed from the two sequence data sets using Unicycler (Shanghai Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China). The serotype, multilocus sequence type, plasmid type, antimicrobial-resistant gene, and virulence gene detection were performed using the Center for Genomic Epidemiology server (https://cge.cbs.dtu.dk). Insertion sequence typing was carried out using the ISFinder database (https://www-is.biotoul.fr/). The complete genome sequences were initially annotated with Rapid Annotation using the Subsystem Technology server (http://rast.nmpdr.org) and curated manually using the BLAST server (https://blast.ncbi.nlm.nih.gov/Blast). The obtained plasmid sequences were aligned with homologous plasmid sequences from NCBI using the BRIG tool (36).
Conjugation Assay
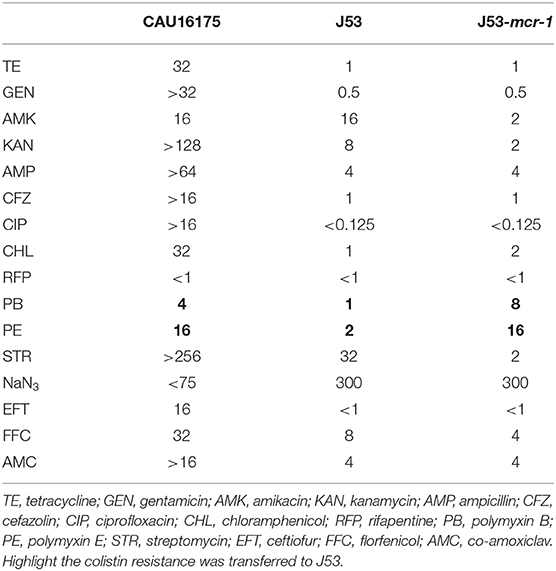
The conjugation experiment was carried out using the E. coli CAU16175 as the donor and the E. coli J53 (resistant to sodium azide) as the recipient. The transconjugant was screened on BHI agar plates containing sodium azide (150 mg/L) and colistin (4 mg/L). The presence of mcr-1 in the transconjugant was confirmed by PCR, and the MICs of antimicrobial agents for the transconjugant were determined using the agar dilution method. The conjugation experiment was repeated three times, and the conjugation frequencies were calculated as the number of transconjugants per recipient. The transconjugant strains were distinguished from donor occurring natural mutants of sodium azide–resistant E. coli by verifying eae, which is marker gene for E. coli CAU16175 donor.
GenBank Accession Number
The draft genome sequences of the E. coli CAU15104, CAU15134, CAU16060, and CAU16177 isolates have been deposited at GenBank under SRA accession no. SRR10828049, SRR10828048, SRR10828047, and SRR10828046, respectively. The complete genome sequences of the E. coli CAU16175 have been deposited at GenBank under SRA accession no. SRR10813965.
Results
Antimicrobial Susceptibility Testing of the Five E. coli Isolates
Antimicrobial susceptibility testing of the five isolates showed that they were MDR exhibiting resistant to at least three different classes of antimicrobials (Table 1). All the isolates showed MDR to KAN, STR, TE, SXT, NAL, and PB.
Characterization of the E. coli Isolates From the Draft Genome Sequences
The clinical isolates of the E. coli CAU15104, CAU15134, and CAU16060 belonged to ETEC/STEC (ltcA+, stb+, and stx+) hybrid strains, display the same serotype O3:H45 and sequence type ST4214, which are identical sequence type with the strains swine19 (LVMV00000000), swine22 (LVMY00000000), swine54 (LVMV00000000), and swine67 (LVOR00000000) from the NCBI database, all carrying fedA, fedF, iha, stb, ltcA, sepA, stx2A, and stx2B virulence genes and antimicrobial-resistant genes conferring resistant to KAN (aph(3′)-Ia), STR (aadA2, strA, and strB), TE [tet(A) and tet(M)], SXT (dfrA12, sul1, sul2, and sul3) and PB (mcr-1.1) (Tables 1, 2). Besides, the E. coli CAU15104, CAU15134, and CAU16060 carry cmlA1 and floR, which are associated to phenicol antibiotic (Table 2). The E. coli CAU15104 and CAU15134 carry qnrS2 (associated to quinolone antibiotic) and aac(6′)Ib-cr (associated to quinolone and aminoglycoside antibiotic). The E. coli CAU15134 and CAU16060 carry dfrA1 (associated to SXT), mcr-3.1 (associated to PB), oqxA, and oqxB, which are efflux pump conferring antibiotic resistance, and exhibit resistant to AMP associated to blaTEM−1B.
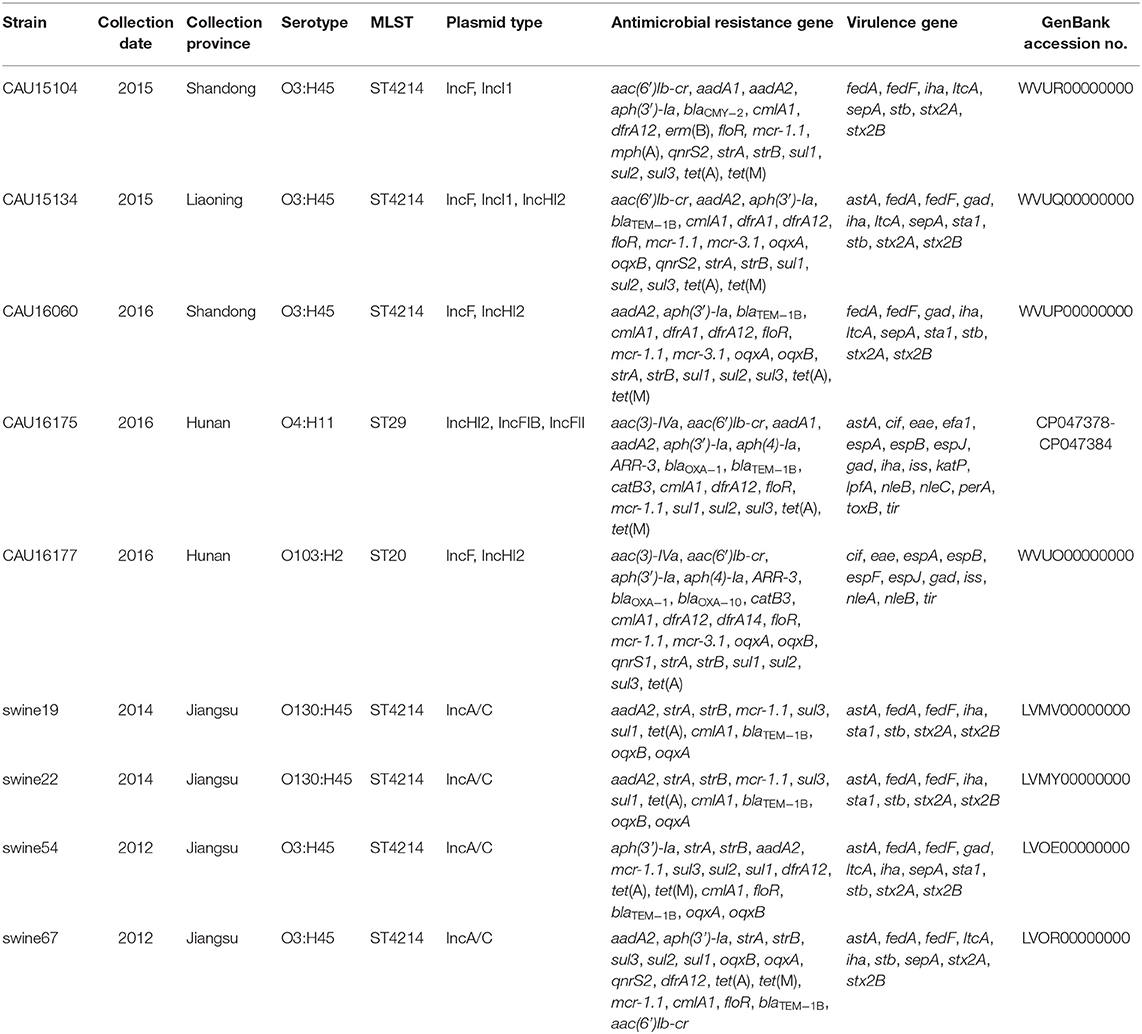
Table 2. Overview of the molecular typing, antimicrobial resistance genes and virulence genes of the five E. coli isolates from the whole-genome sequencing result by the Illumina Hiseq platform (CAU16175 Illumina/Nanopore) and four ST4214 E. coli strains from the NCBI database.
The clinical isolates of the E. coli CAU16175 and CAU16177, belonged to aEPEC (eae+, bfpA−, and stx−), display different serotype and sequence type, O4:H11 and O103:H2, ST29 and ST20, respectively. They both carry cif, eae, espA, espB, espJ, gad, iss, nleB, and tir virulence genes and antimicrobial-resistant genes conferring resistance to beta-lactam antibiotic (blaOXA−1), KAN (aph(3′)-Ia), GEN (aac(3)-IVa), STR (aadA1 or aadA2 or strA or strB), TE [tet(A) and/or tet(M)], SXT (dfrA12, sul1, sul2, sul3, and/or dfrA14), and PB (mcr-1.1 and/or mcr-3.1) (Tables 1, 2). In addition, the E. coli CAU16175 and CAU16177 both carry aac(6′)Ib-cr (associated to quinolone and aminoglycoside antibiotic), aph(4)-Ia (associated to aminoglycoside antibiotic), ARR-3 (associated to rifamycin antibiotic), catB3, cmlA1, and floR (associated to phenicol antibiotic) (Table 2). Interestingly, we observed that the mcr-1.1 gene and the blaTEM−1B gene existed in the same scaffold of the E. coli CAU16175 draft genome. Besides, partial sequences of this scaffold including mcr-1.1 and blaTEM−1B are highly homologous with Tn2 (KT002541).
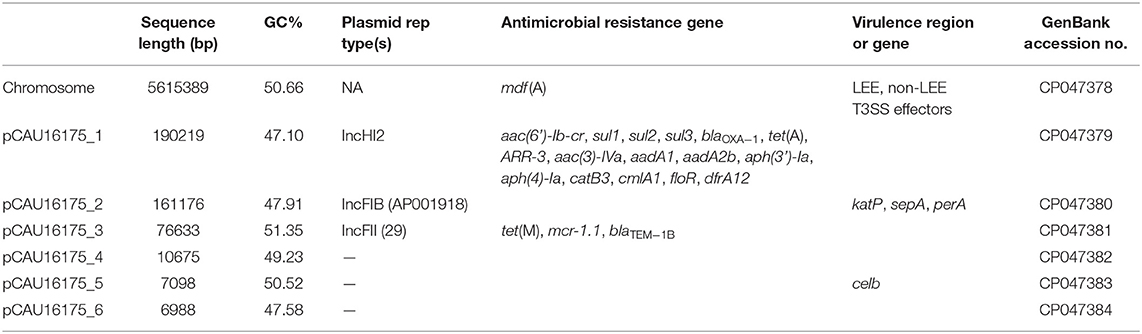
Complete Genome of the E. coli CAU16175
The 6.07-Mb complete genome of E. coli CAU16175 has a total GC content of 50.47% with a single chromosome and six plasmids. The 5.62 Mb chromosome has a GC content of 50.66%. The six plasmids sequences of E. coli CAU16175 range in length from 6.99 to 190.22 kb with a GC content from 47.10 to 51.35% (Table 3). The chromosome harbors LEE T3SS effectors, non-LEE T3SS effectors, and mdf (A) antimicrobial-resistant gene. The pCAU16175_1 (IncHI2 type) harbors an IS26-enriched MDR region, which includes blaOXA−1 and 15 additional antimicrobial-resistant genes. The pCAU16175_2 (IncFIB type) harbors katP, sepA, and perA virulence genes. The pCAU16175_3 (IncFII type) harbors mcr-1.1, tet(M), and blaTEM−1B antimicrobial-resistant genes. The pCAU16175_5 harbors celb virulence gene (Table 3).
Conjugation Assay
The susceptibility testing of polymyxin B and E showed that the MIC values for the transconjugants (J53-mcr-1) increased to 8 and 16 mg/L, respectively (Table 4). The conjugation frequency was 2.1 × 10−8 transconjugants per recipient.
Genetic Characterization of the pCAU16175_1 Harboring the MDR Region
The 190.22-kb plasmid pCAU16175_1 was blasted against the GenBank nucleotide collection (nr/nt) database. An overall nucleotide sequence identity (99.66–99.86%) with query coverages of 90–99% to pSH16G4498 (MH522423), pSH16G2457 (MH522421), pHNYJC8 (KY019259), pHNLDF400 (KY019258), pHXY0908 (KM877269), and other 25 IncHI2-type plasmids were observed (Supplementary Table 1). We chose the five most similar plasmids for comparison analysis; these plasmids have MDR regions and many IS6 family insert sequences (IS26/IS15DI/IS15DIV/IS1006/ISEc59) (Figure 1). The pCAU16175_1 shows almost exactly the same sequence in MDR region with pSH16G4498 and pSH16G2457, which both existed in Salmonella typhimurium recovered from humans in China (Supplementary Table 1). In the pCAU16175_1, the MDR region locates at nucleotide location 121,521–178,916 (56,625 bp). The structure of the MDR region comprises twelve IS6 family insert sequence [including six copies IS26, three copies IS15DI (3 bp differ with IS26), one copy IS15DIV (1 bp differ with IS26), one copy IS1006, and one copy ISEc59] that flank containing different antimicrobial-resistant genes. The MDR region contains floR, sul2, aph(4)-Ia, aac(3)-IVa, aac(6')-Ib-cr, blaOXA−1, catB3, ARR-3, sul1 (two copies), dfrA12, aph(3')-Ia, sul3, aadA1, cmlA1, aadA2b, and tet(A). The MDR region is also interspersed with a number of different mobile elements including ΔTnAs3, ΔISVsa3, ISVsa3, ISAba1, ΔTn5393, ΔTn2, ΔIS15, and ΔTnAs1.
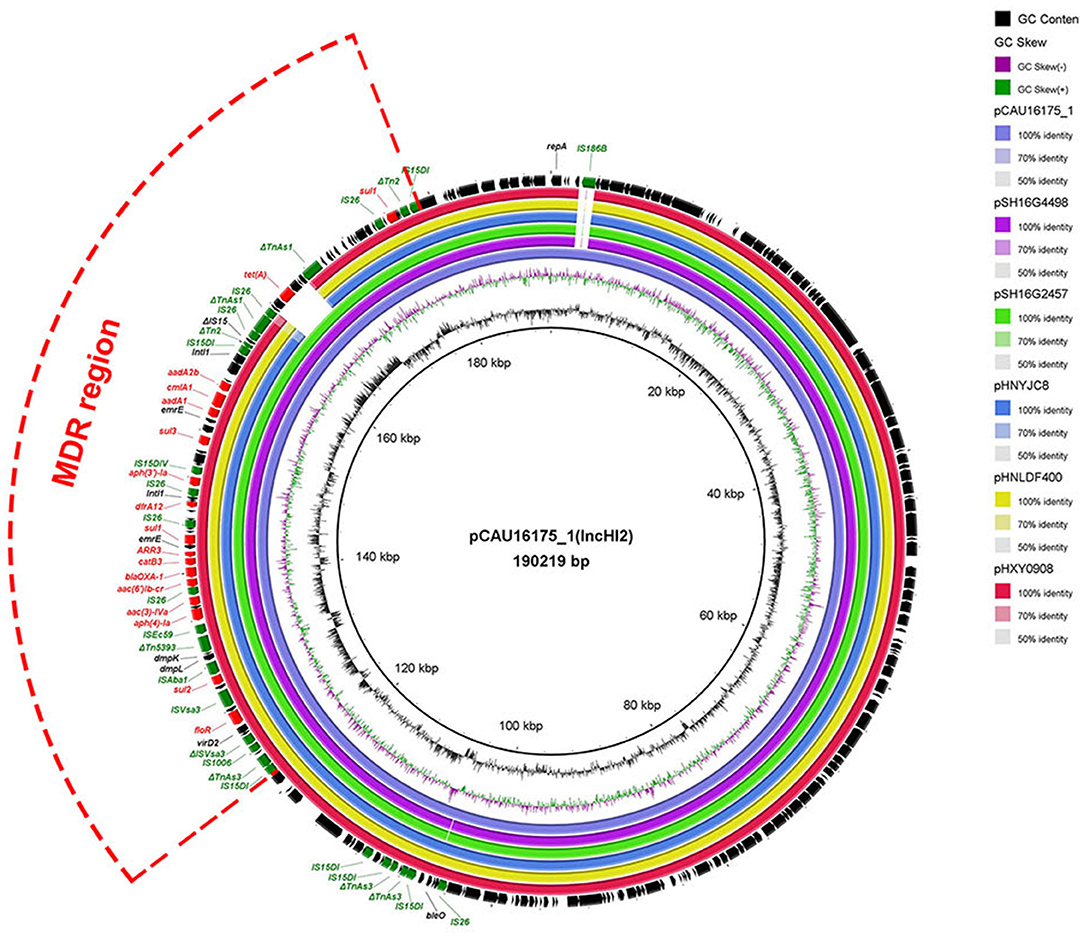
Figure 1. Comparative analysis of MDR-carrying plasmids pCAU16175_1, pSH16G4498, pSH16G2457, pHNYJC8, pHNLDF400, and pHXY0908 using the BLAST Ring Image Generator. The concentric rings show similarity compared the pCAU16175_1 as reference plasmid in inner ring with the other plasmids in outer rings. The varying color levels indicate a BLAST result with matched degree of shared regions, as shown to the right of the ring. The outer circle with red arrows and green arrows denotes antimicrobial resistance gene and transposable elements, respectively. Detailed information of the complete sequences of IncHI2 plasmids is described in Supplementary Table 1.
Genetic Characterization of the pCAU16175_3 Harboring the mcr-1.1 Gene
The 76.63-kb plasmid pCAU16175_3 was blasted against the GenBank Nucleotide collection (nr/nt) database. An overall nucleotide sequence identity (94.46–97.88%) with query coverages of 80–89% to pEC1515-3 (CP021847), pEC974-3 (CP021843), pH1038-142 (KJ484634), pFORC_081_1 (CP029058), plasmid R1 (KY749247), and other 27 IncFII-type plasmids were observed (Supplementary Table 2). We chose five most similar IncFII-type plasmids and reported IncFII-type plasmid-mcr-1 pKP81-BE (KU994859) for comparison analysis (Figure 2). Although the pCAU16175_3 demonstrates highly sequence homology with the other five IncFII-type plasmids, the mcr-1.1 gene only exists in pCAU16175_3 and pKP81-BE. Besides the mcr-1.1 gene, the pCAU16175_3 also carries the antimicrobial-resistant gene blaTEM−1B and tet(M), which are close to the mcr-1.1 gene.
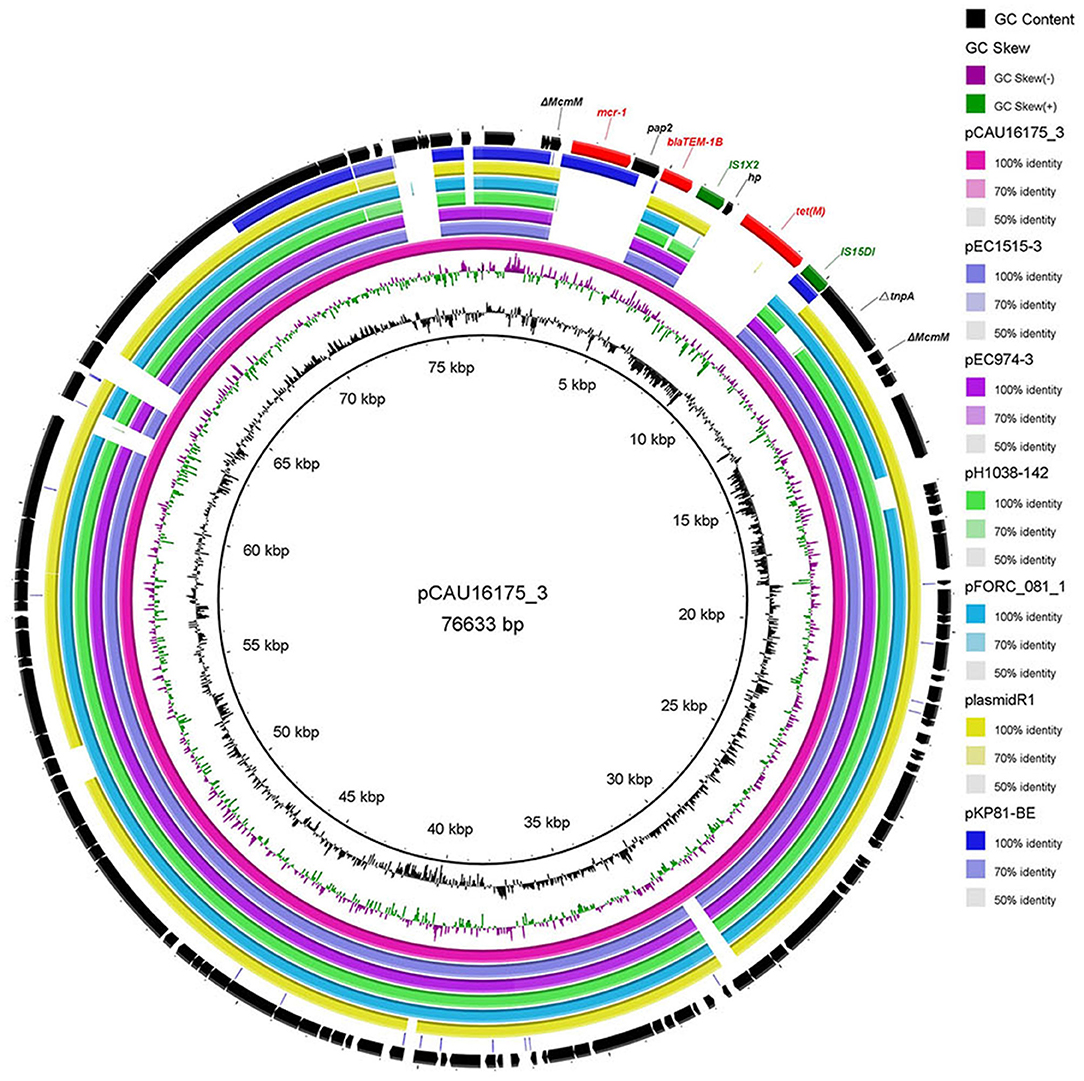
Figure 2. Comparative analysis of seven IncFII-type plasmids pCAU16175_3, pEC1515-3, pEC974-3, pH1038-142, pFORC_081_1, plasmid R1, and pKP81-BE (carrying mcr-1) using the BLAST Ring Image Generator. The concentric rings show similarity compared the pCAU16175_3 as reference plasmid in inner ring with the other plasmids in the outer rings. The varying color levels indicate a BLAST result with matched degree of shared regions, as shown to the right of the ring. The outer circle with red arrows and green arrows denotes antimicrobial resistance gene and transposable elements, respectively. Detailed information of the complete sequences of IncFII plasmids is described in Supplementary Table 2.
Noticeably, the nucleotide location of the pCAU16175_3 from 2,079 to 12,231 has high homology with the Tn2 derivative (Figure 3A). This structure is a unique identification compared with the NCBI database and was confirmed by conventional PCR (Supplementary Figure 1A). The genetic content of the structure analysis showed that the structure is located within mcmM (encoded microcin M). A five-nucleotide direct duplication (TATTT) was identified flanking the structure. Unlike the Tn2, this structure harbors mcr-1–pap2-ΔISApl1 and IS1X2-hp-tet(M)-IS15DI, deletes the resolvase (tnpR), and truncates the transposase (tnpA). Besides, the striking features of the insertion sites of ISApl1 were found, which includes high AT content and 2-bp direct repeats (DRs) (AA) (Figure 3B). From the result of conjugation experiment and PCR, the whole structure gene did not occur transposition (Table 4, Supplementary Figure 1B). The mcr-1 gene was transferred to J53 without the pap2 gene, whereas colistin resistance was transferred to J53. IS15DI was also transferred to J53.
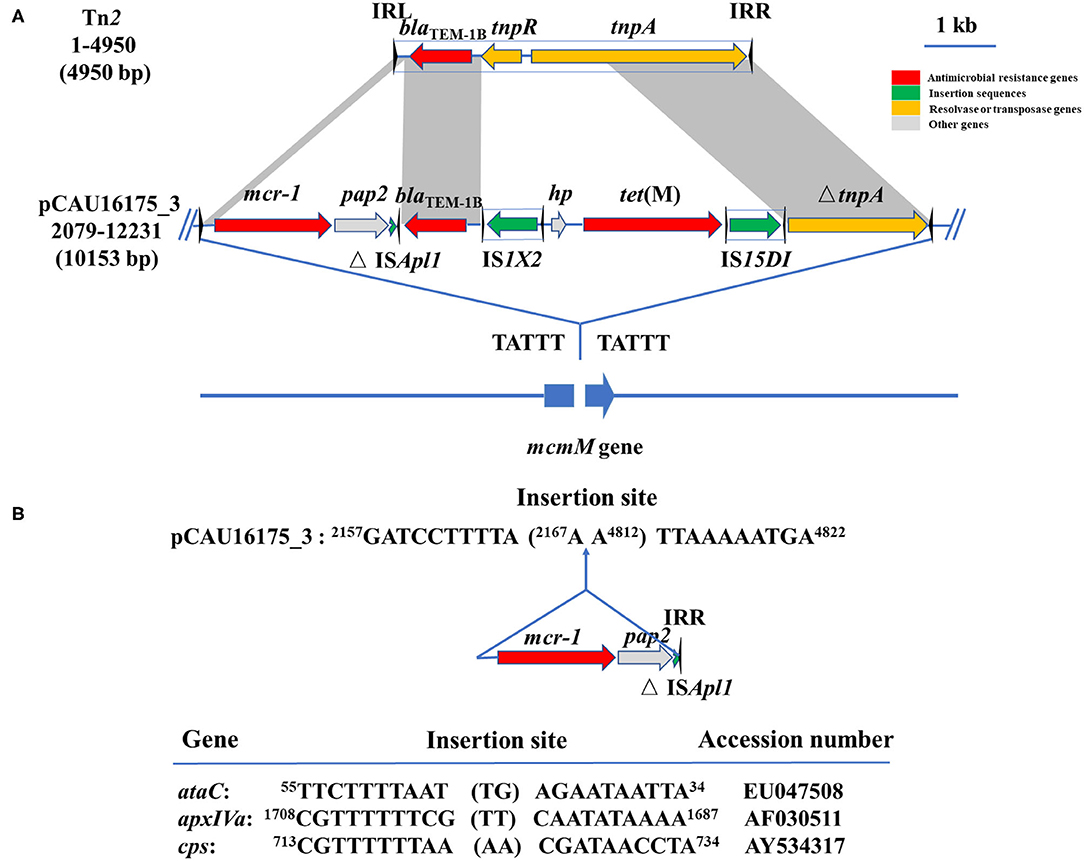
Figure 3. Genetic features of pCAU16175_3 (nucleotide location 2079-12231). (A) Linear sequence comparison of pCAU16175_3 (nucleotide location 2079-12231) with Tn2. Arrows indicate the positions and directions of the genes, Δ indicates a truncated gene. Regions with >99% homology are indicated by gray shading. Inverted repeat nucleotide sequences (IRL: left IR; IRR: right IR) of IS are marked by triangles. Antimicrobial resistance genes, insertion sequences, resolvase or transposase genes, and other genes are indicated by red, green, khaki, and gray, respectively. (B) DNA sequence of insertion sites of mcr-1-pap2-ΔISApl1 in pCAU16175_3 and ISApl1 in ataC, apxIVA, and cps gene (30). Shown sequences represent 10 base pairs upstream and downstream of insertion site, and 2-base-pair direct repeats in brackets. Numbers represent positions in the indicated sequences deposited in GenBank.
Discussion
Postweaning diarrhea caused by DEC is an economically important disease for the swine industry around the world. In our previous study, we collected 455 E. coli isolates recovered from feces samples or small intestinal content from pigs with diarrhea in China between 2014 and 2016 to know the E. coli pathotype and the antimicrobial susceptibility of the isolates (6). Most of the isolates belonged to ETEC, followed by aEPEC, which is similar to the report in Spain; that is, most cases of PWD are significantly associated with ETEC (67%) and aEPEC (21.7%) (37). Our previous study showed that 95.91% of 171 E. coli isolates were MDR exhibiting resistance to at least three different classes of antimicrobials, and 20.47% E. coli isolates were resistant to colistin. It has been revealed that MDR isolates that existed in swine industry are associated with the widespread use of antibiotics (38, 39). Overuse of colistin is considered to contribute to the emergence and spread of mcr-1 (40). The 36 mcr-1–positive E. coli isolates recovered from pigs with PWD showed MDR (29). In this study, we used WGS to analyze the five MDR (including colistin resistance) of E. coli isolates from pigs with PWD and found that they demonstrate different serotype and sequence type and carry different antimicrobial-resistant genes and virulence genes.
The clinical isolates of the E. coli CAU15104, CAU15134, and CAU16060 belonged to ETEC/STEC, displaying the same serotype O3:H45 and sequence type ST4214. As far as we know, there are no reports about this clone strains, and only four have WGS of these clone strains. The E. coli swine19, swine22, swine54, and swine67 also belong to ETEC/STEC and display the same sequence type ST4214. The E. coli swine19 and swine22 display the same serotype O130:H45. The E. coli swine54 and swine67 display the same serotype O3:H45, similar to our collected the E. coli CAU15104, CAU15134, and CAU16060. Five ETEC/STEC isolates recovered from pigs with enteric colibacillosis in Spain display the serotype O141:H4 and sequence type ST10 (37). In the present study, all of the ST4214 ETEC/STEC we collected were MDR (at least three different classes of antimicrobials) and showed MDR to PB, TE, and NAL, carried 17–19 antimicrobial-resistant genes and 8–11 virulence genes. Besides, all the ST4214 ETEC/STEC harbored aminoglycoside-resistant genes (aadA2 and aph(3′)-Ia), phenicol-resistant genes (cmlA1 and floR), tetracycline-resistant genes [tet(A) and tet(M)], trimethoprim-resistant gene (dfrA12), sulfonamide-resistant genes (sul1, sul2, and sul3), streptomycin-resistant genes (strA and strB), and colistin-resistant genes (mcr-1.1). The ST4214 ETEC/STEC also harbored fimbrial adhesin genes (fedA and fedF), adherence gene (iha), heat-labile enterotoxin (LT) gene (ltcA), heat-stabile enterotoxin (ST) gene (stb), Shigella extracellular protein gene (sepA), and Shiga toxin 2 variant e genes (stx2A and stx2B). We observed the ST4214 strains were recovered from different provinces (Shandong, Jiangsu and Liaoning) and years (2012, 2014, 2015, and 2016) in China, which indicated this clone strains have spread in swine and should be brought to our attention.
In addition, the clinical isolates of the E. coli CAU16175 and CAU16177 belonged to aEPEC, displaying different serotype and sequence type, O4:H11 and O103:H2, ST29 and ST20, respectively. For aEPEC, serotypes O45 and O123 are frequently occurring in diarrheagenic pigs (41, 42). The E. coli CAU16175 and CAU16177 also showed MDR to AMC, KAN, PB, TE, NAL, and CHL and carried 19/23 antimicrobial-resistant genes and 17/11 virulence genes. They both harbored beta-lactamase-resistant genes (blaOXA−1), aminoglycoside-resistant genes (aac(3)-IVa, aac(6′)Ib-cr, aph(3′)-Ia and aph(4)-Ia), phenicol-resistant genes (cmlA1, catB3, and floR), tetracycline-resistant genes [tet(A)], trimethoprim-resistant gene (dfrA12), sulfonamide-resistant genes (sul1, sul2, and sul3), rifampicin-resistant gene (ARR-3), and colistin-resistant genes (mcr-1.1). Interestingly, E. coli CAU16177 also harbored mcr-3.1. Four mcr-1– and mcr-3-positive E. coli have been reported, and three E. coli isolates were recovered from pigs (23). The E. coli CAU16175 and CAU16177 both harbored type III secretion system-associated virulence genes (cif, espA, espB, and espJ), non–LEE-encoded effector gene (nleB), glutamate decarboxylase gene (gad), increased serum survival gene (iss), a marker gene for EPEC, intimin gene (eae), and translocated intimin receptor gene (tir).
The E. coli CAU16175 carries IS26-enriched MDR region in pCAU16175_1 (IncHI2 type). The IncHI2-type plasmids have been discovered as genetic elements mediating the transmission of MDR genes (16). IS26-flanked Tns play an increasingly critical role in the mobilization and development of antimicrobial-resistant genes (43–45). So far, there are 51 Tns related with IS26 from the Tn registry website. In an individual E. coli strain heterogeneous resistance-encoding plasmid, polymorphic MDR regions driven by IS26-flanked Tns have been detected (44). From the BLAST analysis, we found that there were abundant IS26-enriched MDR regions in Salmonella and E. coli, which were mostly isolated from humans and animals. The results showed that the plasmid carrying IS26-enriched MDR region has been widely distributed in humans and consumption animals (Supplementary Table 1). Combined with the antimicrobial susceptibility testing, we can deduce that the E. coli CAU16175 isolate has MDR and is difficult to control.
The E. coli CAU16175 isolate also carries the mcr-1.1 gene in pCAU16175_3 (IncFII type). The emergence of the mcr-1 gene can be traced back to the E. coli isolated in the 1980s, and the outbreak of chicken-derived mcr-1–containing E. coli started in 2009 (46).The mcr-1 gene has been characterized in various genetic backgrounds and observed on a variety of plasmid type, including Incl2, IncX4, IncHI2, IncP, IncHI1, IncFII, IncFI, IncFIB, F18:A-:B+, IncY, IncK, and phage-like plasmid (29). Most of the recently reported the mcr-1 genes were primarily mobilized by an ISApl1 composite Tns Tn6330 and Tn6390 (47, 48). The ISApl1 would be lost over time, leading to the stability of the mcr-1 gene on plasmids (18, 49). Thus, only the truncated ISApl1 will lose the ability to transfer the mcr-1 gene.
Furthermore, the mcr-1.1 gene is located within the Tn2 derivative in pCAU16175-3. From the Tn registry website (https://transposon.lstmed.ac.uk/), we noticed the mcr-5 gene and the mcr-3.6 gene could be mobilized by Tn6452 and Tn6518 (belonged to Tn2 family). For the Tn6452, the mcr-5 gene was embedded within a Tn3-family Tn with 38-bp inverted repeats and flanked by 5-bp DRs, which were usually generated during the insertion (50, 51). Although DRs appear at the flanking site of the Tn2 derivative, the whole structure did not move by transposition because of truncated tnpA and deleted tnpR, which are essential to occur transposition for Tn2.
Conclusions
The current work shows the genetic characteristics of five DEC strains that exhibited MDR, including colistin resistance. Our data indicate that the ST4214 ETEC/STEC carried MDR and multivirulence genes and that the E. coli CAU16175 contains IS26-enriched MDR region and the mcr-1.1 gene, which is located within a Tn2 derivative. The coexistence of MDR and multivirulence in DEC may seriously compromise the effectiveness of clinical therapy, and heightened efforts are needed to control their dissemination.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR10813965; https://www.ncbi.nlm.nih.gov/, SRR10828049; https://www.ncbi.nlm.nih.gov/, SRR10828048; https://www.ncbi.nlm.nih.gov/, SRR10828047; and https://www.ncbi.nlm.nih.gov/, SRR10828046.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of the China Agricultural University under the protocol CAU20140616-1.
Author Contributions
LG and YZ conceived and designed the experiments. LG, JW, SW, JS, and XW performed the experiments. LG analyzed the sequencing data and wrote the manuscript. YZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (Project No. 2017YFD0502200) and the National Natural Science Foundation of China (Project Nos. 31873034 and 31672613).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Wang Yang (China Agricultural University) for providing the E. coli J53 strain.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00503/full#supplementary-material
Abbreviations
DEC, diarrheagenic Escherichia coli; WGS, whole-genome sequencing; MDR, multidrug resistance; ETEC, enterotoxigenic E. coli; STEC, Shiga-toxigenic E. coli; aEPEC, atypical enteropathogenic E. coli; tEPEC, typical enteropathogenic E. coli; PWD, postweaning diarrhea; IS, insertion sequence; Tn, transposon; PCR, polymerase chain reaction; DR, direct repeat.
References
1. Luppi A. Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porcine Health Manag. (2017) 3:16. doi: 10.1186/s40813-017-0063-4
2. Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. (1998) 11:142–201. doi: 10.1128/CMR.11.1.142
3. Bosak J, Hrala M, Pirkova V, Micenkova L, Cizek A, Smola J, et al. Porcine pathogenic Escherichia coli strains differ from human fecal strains in occurrence of bacteriocin types. Vet Microbiol. (2019) 232:121–7. doi: 10.1016/j.vetmic.2019.04.003
4. Hazen TH, Rasko DA. The complete genome of the atypical enteropathogenic Escherichia coli archetype isolate E110019 highlights a role for plasmids in dissemination of the type III secreted effector EspT. Infect Immun. (2019) 87:e00412–9. doi: 10.1128/IAI.00412-19
5. van Hattem JM, Cabal A, Arcilla MS, Alvarez J, de Jong MD, Melles DC, et al. Risk of acquisition of human diarrhoeagenic Escherichia coli virulence genes in intercontinental travellers: a prospective, multi-centre study. Travel Med Infect Dis. (2019) 31:101362. doi: 10.1016/j.tmaid.2018.12.005
6. Yang GY, Guo L, Su JH, Zhu YH, Jiao LG, Wang JF. Frequency of diarrheagenic virulence genes and characteristics in Escherichia coli isolates from pigs with diarrhea in China. Microorganisms. (2019) 7:308. doi: 10.3390/microorganisms7090308
7. Kusumoto M, Hikoda Y, Fujii Y, Murata M, Miyoshi H, Ogura Y, et al. Emergence of a multidrug-resistant Shiga toxin-producing enterotoxigenic Escherichia coli lineage in diseased swine in Japan. J Clin Microbiol. (2016) 54:1074–81. doi: 10.1128/JCM.03141-15
8. Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev. (2013) 26:822–80. doi: 10.1128/CMR.00022-13
9. Pearson JS, Giogha C, Wong Fok Lung T, Hartland EL. The genetics of enteropathogenic Escherichia coli virulence. Annu Rev Genet. (2016) 50:493–513. doi: 10.1146/annurev-genet-120215-035138
10. Shepard SM, Danzeisen JL, Isaacson RE, Seemann T, Achtman M, Johnson TJ. Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J Bacteriol. (2012) 194:395–405. doi: 10.1128/JB.06225-11
11. Levine MM, Nasrin D, Acacio S, Bassat Q, Powell H, Tennant SM, et al. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob Health. (2020) 8:e204–14. doi: 10.1016/S2214-109X(19)30541-8
12. Hu J, Torres AG. Enteropathogenic Escherichia coli: foe or innocent bystander? Clin Microbiol Infect. (2015) 21:729–34. doi: 10.1016/j.cmi.2015.01.015
13. Zajac M, Sztromwasser P, Bortolaia V, Leekitcharoenphon P, Cavaco LM, Zietek-Barszcz A, et al. Occurrence and characterization of mcr-1-positive Escherichia coli isolated from food-producing animals in Poland, 2011-2016. Front Microbiol. (2019) 10:1753. doi: 10.3389/fmicb.2019.01753
14. Gresse R, Chaucheyras-Durand F, Fleury MA, Van de Wiele T, Forano E, Blanquet-Diot S. Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. (2017) 25:851–73. doi: 10.1016/j.tim.2017.05.004
15. Iramiot JS, Kajumbula H, Bazira J, de Villiers EP, Asiimwe BB. Whole genome sequences of multi-drug resistant Escherichia coli isolated in a Pastoralist Community of Western Uganda: phylogenomic changes, virulence and resistant genes. PLoS ONE. (2020) 15:e0231852. doi: 10.1371/journal.pone.0231852
16. Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. (2017) 72:393–401. doi: 10.1093/jac/dkw411
17. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
18. Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. (2018) 9:1179. doi: 10.1038/s41467-018-03205-z
19. Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, et al. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio. (2016) 7:e00177. doi: 10.1128/mBio.00177-16
20. Anyanwu MU, Jaja IF, Nwobi OC. Occurrence and characteristics of mobile colistin resistance (mcr) gene-containing isolates from the environment: a review. Int J Environ Res Public Health. (2020) 17:1028. doi: 10.3390/ijerph17031028
21. Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerging Microbes Infect. (2020) 9:508–16. doi: 10.1080/22221751.2020.1732231
22. Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J. Antimicrob Chemother. (2020). doi: 10.1093/jac/dkaa205. [Epub ahead of print].
23. Wang Z, Fu Y, Schwarz S, Yin W, Walsh TR, Zhou Y, et al. Genetic environment of colistin resistance genes mcr-1 and mcr-3 in Escherichia coli from one pig farm in China. Vet Microbiol. (2019) 230:56–61. doi: 10.1016/j.vetmic.2019.01.011
24. Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. (2014) 38:865–91. doi: 10.1111/1574-6976.12067
25. Harmer CJ, Moran RA, Hall RM. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio. (2014) 5:e01801–14. doi: 10.1128/mBio.01801-14
26. Nicolas E, Lambin M, Dandoy D, Galloy C, Nguyen N, Oger CA, et al. The Tn3-family of replicative transposons. Microbiol Spectrum. (2015) 3:693–726. doi: 10.1128/9781555819217.ch32
27. Bailey JK, Pinyon JL, Anantham S, Hall RM. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother. (2011) 66:745–51. doi: 10.1093/jac/dkq529
28. Partridge SR. What's in a name? ISSwi1 corresponds to transposons related to Tn2 and Tn3. mBio. (2015) 6:e01344–15. doi: 10.1128/mBio.01344-15
29. Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. (2018) 26:794–808. doi: 10.1016/j.tim.2018.02.006
30. Tegetmeyer HE, Jones SC, Langford PR, Baltes N. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet Microbiol. (2008) 128:342–53. doi: 10.1016/j.vetmic.2007.10.025
31. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-sixth Informational Supplement. CLSI document, Approved Standard M100-S26. Wayne, PA: Clinical and Laboratory Standards Institute (2016).
32. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 4th ed. Wayne, PA: Clinical and laboratory standards institute (2013).
33. Chen S, Zhao S, White DG, Schroeder CM, Lu R, Yang H, et al. Characterization of multiple-antimicrobial-resistant salmonella serovars isolated from retail meats. Appl Environ Microbiol. (2004) 70:1–7. doi: 10.1128/AEM.70.1.1-7.2004
34. Sorensen AH, Hansen LH, Johannesen E, Sorensen SJ. Conjugative plasmid conferring resistance to olaquindox. Antimicrob Agents Chemother. (2003) 47:798–9. doi: 10.1128/AAC.47.2.798-799.2003
35. EUCAST. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of Mics and Zone Diameters version 7.1. EU(European Union). (2017).
36. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. (2011) 12:402. doi: 10.1186/1471-2164-12-402
37. Garcia-Menino I, Garcia V, Mora A, Diaz-Jimenez D, Flament-Simon SC, Alonso MP, et al. Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front Microbiol. (2018) 9:2659. doi: 10.3389/fmicb.2018.02659
38. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
39. Rhouma M, Beaudry F, Theriault W, Letellier A. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and one health perspectives. Front Microbiol. (2016) 7:1789. doi: 10.3389/fmicb.2016.01789
40. Curcio L, Luppi A, Bonilauri P, Gherpelli Y, Pezzotti G, Pesciaroli M, et al. Detection of the colistin resistance gene mcr-1 in pathogenic Escherichia coli from pigs affected by post-weaning diarrhoea in Italy. J Glob Antimicrob Resist. (2017) 10:80–3. doi: 10.1016/j.jgar.2017.03.014
41. Malik A, Nagy B, Kugler R, Szmolka A. Pathogenic potential and virulence genotypes of intestinal and faecal isolates of porcine post-weaning enteropathogenic Escherichia coli. Res Vet Sci. (2017) 115:102–8. doi: 10.1016/j.rvsc.2017.02.002
42. Zhu C, Harel J, Jacques M, Desautels C, Donnenberg MS, Beaudry M, et al. Virulence properties and attaching-effacing activity of Escherichia coli O45 from swine postweaning diarrhea. Infect Immun. (1994) 62:4153–9. doi: 10.1128/IAI.62.10.4153-4159.1994
43. He D, Liu L, Guo B, Wu S, Chen X, Wang J, et al. Chromosomal location of the fosA3 and blaCTX−M genes in Proteus mirabilis and clonal spread of Escherichia coli ST117 carrying fosA3-positive IncHI2/ST3 or F2:A-:B- plasmids in a chicken farm. Int J Antimicrob Agents. (2017) 49:443–8. doi: 10.1016/j.ijantimicag.2016.12.009
44. He DD, Zhao SY, Wu H, Hu GZ, Zhao JF, Zong ZY, et al. Antimicrobial resistance-encoding plasmid clusters with heterogeneous MDR regions driven by IS26 in a single Escherichia coli isolate. J Antimicrob Chemother. (2019) 74:1511–6. doi: 10.1093/jac/dkz044
45. Roy Chowdhury P, McKinnon J, Liu M, Djordjevic SP. Multidrug resistant uropathogenic Escherichia coli ST405 with a novel, composite IS26 transposon in a unique chromosomal location. Front Microbiol. (2018) 9:3212. doi: 10.3389/fmicb.2018.03212
46. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. (2016) 16:293. doi: 10.1016/S1473-3099(16)00061-X
47. Liang B, Roberts AP, Xu X, Yang C, Yang X, Wang J, et al. Transferable plasmid-borne mcr-1 in a colistin-resistant Shigella flexneri isolate. Appl Environ Microbiol. (2018) 84:e02655–17. doi: 10.1128/AEM.02655-17
48. Snesrud E, McGann P, Chandler M. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio. (2018) 9:e02381–17. doi: 10.1128/mBio.02381-17
49. Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. (2016) 60:6973–6. doi: 10.1128/AAC.01457-16
50. Borowiak M, Hammerl JA, Deneke C, Fischer J, Szabo I, Malorny B. Characterization of mcr-5-harboring Salmonella enterica subsp. enterica serovar Typhimurium isolates from animal and food origin in Germany. Antimicrob Agents Chemother. (2019) 63:e00063–19. doi: 10.1128/AAC.00063-19
Keywords: Escherichia coli, whole-genome sequencing, multidrug resistance, mcr-1, Tn2, swine
Citation: Guo L, Wang J, Wang S, Su J, Wang X and Zhu Y (2020) Genome Characterization of mcr-1–Positive Escherichia coli Isolated From Pigs With Postweaning Diarrhea in China. Front. Vet. Sci. 7:503. doi: 10.3389/fvets.2020.00503
Received: 01 April 2020; Accepted: 02 July 2020;
Published: 25 August 2020.
Edited by:
Rafael Vignoli, University of the Republic, UruguayReviewed by:
José Alejandro Di Conza, University of Buenos Aires, ArgentinaNilton Lincopan, University of São Paulo, Brazil
Copyright © 2020 Guo, Wang, Wang, Su, Wang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaohong Zhu, emh1X3lhb2hvbmdAaG90bWFpbC5jb20=
 Liang Guo
Liang Guo Jing Wang
Jing Wang Yaohong Zhu
Yaohong Zhu