- 1Department of Preventive Veterinary Medicine, College of Veterinary Medicine, Shandong Agricultural University, Taian, China
- 2Shandong Provincial Key Laboratory of Animal Biotechnology and Disease Control and Prevention, Shandong Agricultural University, Taian, China
- 3China Animal Disease Control Center, Beijing, China
We investigated the prevalence of salmonellosis on 17 poultry breeding farms in nine Chinese provinces (Shandong, Jiangsu, Anhui, Zhejiang, Fujian, Guangdong, Yunnan, Sichuan, and Chongqing). Altogether, 3,508 samples from poultry breeding farms were collected in 2019, including 1,400 from cloaca swabs, 210 from feed, 1,688 from chicken embryos, and 210 from water. All the samples were subjected to bacterial isolation and culture, and bacterial species were identified by polymerase chain reaction. Serotyping, multilocus sequence typing (MLST), and drug-resistance phenotyping were performed on the isolates identified as Salmonella. Altogether, 126 Salmonella strains were detected in the 3,508 samples and the positivity rate for the samples was 3.59%. Among all the strains, 95 Salmonella isolates were selected for antimicrobial susceptibility test, resistance gene detection, serotyping, and genotyping. S. gallinarum-pullorum (57/95, 60.00%), S. enteritidis (22/95, 23.16%), and S. agona (16/95, 16.84%) serotypes were identified. The MLST classification showed that the 95 Salmonella strains fell into the following five sequence types (STs): ST92 (37/95, 38.95%), ST11 (22/95, 23.16%), ST2151 (19/95, 20.00%), ST13 (16/95, 16.84%), and ST470 (1/95, 1.05%). Apart from ST13, the other four STs shared close genetic relationships, and the genetic direction was ST11-ST470-ST92-ST2151. The resistance rates in the 95 isolates were 100% (95/95) for erythromycin, 68.42% (65/95) for tetracycline, and 53.68% (51/95) for streptomycin and ampicillin, respectively. The isolates were sensitive to polymyxin and sulfamethoxazole. Multi-drug resistance was seen in 70.53% (67/95) of the isolates. β-lactam-, aminoglycoside- and sulfonamide-encoding resistance genes were detected by PCR. The detection rate for blaTEM and sul3 was 100% (95/95), whereas sul2 and aaC4 had rates of 52.63 and 23.16%, respectively. These results indicate that some of the salmonellosis seen in Chinese breeding chicken farms may be caused by infection with S. gallinarum-pullorum, S. enteritidis, and S. agona. They also show that some Salmonella isolates have multi-drug resistance phenotypes and carry multi-drug resistance genes.
Introduction
Salmonella is a food-borne disease-causing zoonotic pathogen (1), and its occurrence has frequently been reported in recent years (2, 3). More than 2,600 Salmonella serotypes have been identified and recorded (4). According to the World Health Organization (WHO), with nearly one-tenth of the world's population becoming infected each year and around 33 million deaths (http://www.who.int/mediacentre/factsheets/fs139/en/). Most cases occur in older adults and in immunocompromised people (5, 6), and it is estimated that in 2017 Salmonella enterocolitis was responsible for 95.1 million cases and killed 50,771 people (7–9). In addition to diarrhea, 535,000 cases of non-typhoidal Salmonella invasive disease have occurred, and an estimated 77,500 people have died (10). At present, antibiotic therapy remains the main prevention and treatment method for salmonellosis. However, the long-term and unreasonable use of antibiotics has caused Salmonella to develop widespread and strong resistance to these agents (11). Drug resistant bacteria can also spread to humans, leading to public health problems (12).
Various molecular typing techniques have been widely used in the field of microbiology to track the origins of pathogenic bacteria (13), the most widely of which used are multilocus sequence typing (MLST) and pulsed field gel electrophoresis (PFGE). MLST is fast and convenient, producing reliable high resolution data, and easy real-time internet sharing (14). PFGE is recognized by laboratories around the world for its high resolution and repeatability. But PFGE does not have a strict and uniform international naming standard, making it difficult to achieve data sharing with it. In recent years, a large number of epidemiological surveys on Salmonella have emerged in China. However, researchers were more inclined to conduct their epidemiological Salmonella surveys on commercial broiler sales chains where sick birds appeared than on healthier chains (15–17). Therefore, data from epidemiological surveys on Salmonella in domestic poultry farms, especially chicken breeder flocks, are incomplete.
Here, we collected 3,508 samples from 17 poultry breeding farms in nine Chinese provinces in 2019. We investigated the drug-resistance genes, drug-resistance phenotypes, and genetic relationships among the Salmonella isolates we collected. We comprehensively and systematically studied the epidemiological characteristics of Salmonella, with the aim of improving the molecular typing network database in China and providing relevant data with which to support the prevention and control of Salmonella in China.
Materials and Methods
Sample Collection
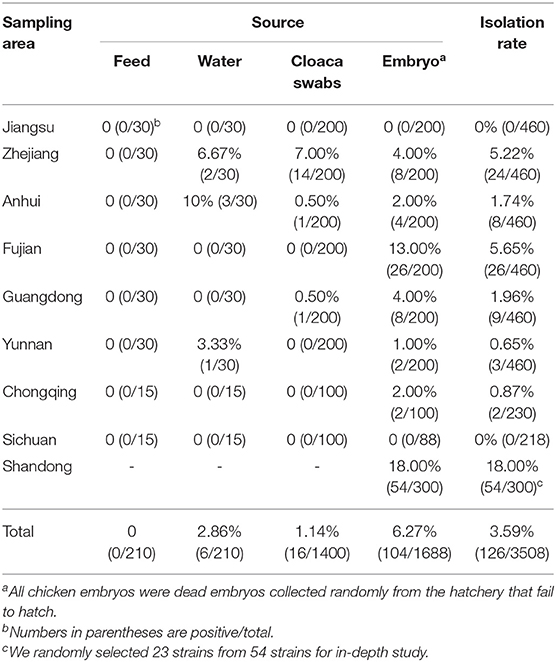
From May to July 2019, 3,208 samples were collected from 14 poultry breeding farms in Jiangsu, Anhui, Zhejiang, Fujian, Guangdong, Yunnan, Chongqing, and Sichuan Province, China. They included 1,400 cloacal swab samples, 210 animal feed samples, 1,388 embryo samples and 210 water samples. An additional 300 chicken embryo samples were obtained from three chicken breeding farms in Shandong Province, China (Figure 1A).
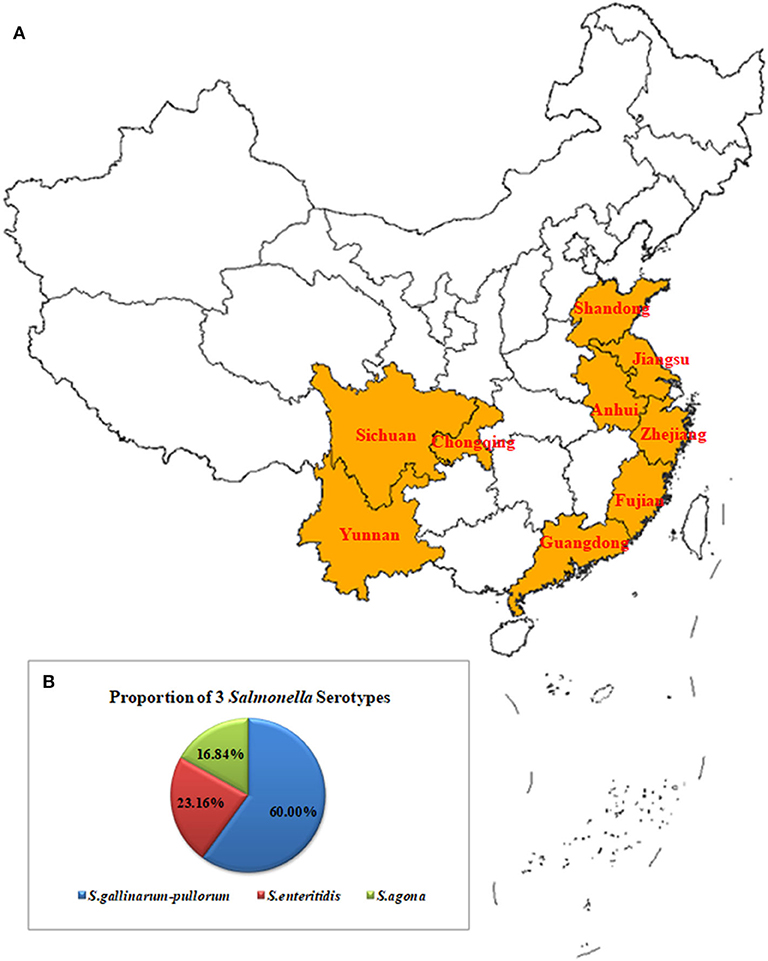
Figure 1. Geographical Map of the Chinese Sampling Sites and Proportions of the Serotypes for the Salmonella Isolates (A). A total of 3,208 samples were collected from 14 poultry breeding farms in Jiangsu, Zhejiang, Anhui, Fujian, Guangdong, Yunnan, Chongqing, and Sichuan Provinces, China. They included 1,400 cloacal swab samples, 210 feed samples, 1,388 embryo samples, and 210 water samples. An additional 300 chicken embryo samples were obtained from three chicken breeding farms in Shandong Province, China (B). The 95 isolates included 57 strains of S. gallinarum-pullorum (60.00%, 57/95), 22 strains of S. enteritidis (23.16%, 22/95), and 16 strains of S. agona (16.84 %, 16/95).
Isolating and Serotyping Salmonella
Buffered peptone water (4.5 mL) (BPW, Haibo Biotechnology, Qingdao, China) was added to each sample (0.5 g) for pre-enrichment, according to a previously described method (18). After incubation at 37°C for 8–12 h, 0.5 mL of each pre-enriched culture was incubated in 4.5 ml of Tetrathionate Broth Buffer (TTB, Haibo Biotechnology) at 37°C for 24 h. After selective enrichment, one loopful of each broth culture was streaked onto xylose lysine tergitol 4 (XLT4, Haibo Biotechnology) agar and the plates were incubated at 37°C for 24–36 h. The presumptive Salmonella colonies were identified by polymerase chain reaction (PCR) assays using invA primers (19). The invA gene has a conserved sequence in Salmonella species and can therefore be used to detect and validate Salmonella strains (20). Here, invA primers (19) (F: 5′-ACAGTGCTCGTTTACGACCTGAAT-3′, R: 5′-AGACGACTGGTACTGATCGATAAT-3′) were used for PCR. The PCR cycling conditions were as follows: 1 denaturation cycle at 95°C for 5 min, 30 cycles of denaturation at 95°C for 30 s, followed by annealing at 56°C for 30 s and elongation at 72°C for 1 min, and a final 10 min elongation cycle at 72°C. Positive colonies were subsequently confirmed using a microbial mass spectrometer (IVD MALDI Biotyper, Bruker Bremen, Germany).
Salmonella Serotyping
After being resuscitated on XLT4 medium, all the Salmonella isolates were serotyped according to the Kauffmann-White scheme by slide agglutination with O and H antigens (Tianrun Bio-Pharmaceutical, Ningbo, China). Normal saline was used as a negative control, and plate agglutination reactions were considered Salmonella positive. The Salmonella strains were PCR-identified using DNA as the template and Ipaj (F: 5′-TACCTGTCTGCTGCCGTGA-3′, R: 5′-ACCCTGCAAACCTGAAATC-3′) (21) as primers for detecting S. gallinarum-pullorum.
Antimicrobial Susceptibility Testing
The Kirby-Bauer disk diffusion method, as described by the Clinical and Laboratory Standards Institute (CLSI), was used to test the susceptibility of the Salmonella isolates to 14 commonly used antibiotics, including polymyxin B (PB, 300 IU), sulfamethoxazole (SXT, 25 μg), florfenicol (30 μg), ofloxacin (5 μg), cefoxitin (30 μg), doxycycline (30 μg), amoxicillin (20 μg), tetracycline (TE, 30 μg), streptomycin (STR, 10 μg), gentamycin (10 μg), erythromycin (EM, 15 μg), ceftazidime (CAZ, 30 μg), ampicillin (AM, 10 μg), and gatifloxacin (5 μg). The results were interpreted according to the standard guidelines from the CLSI (22). Salmonella isolates that were resistant to more than three antimicrobial classes were defined as multidrug-resistant (MDR) isolates.
Detecting Drug-Resistance Genes
Bacterial DNA was extracted from the samples using a Bacterial Genome Kit (Bioteke, Beijing, China), according to the manufacturer's instructions. The β-lactam genes (blaTEM, blaSHV, blaPSE, blaCTX−M, and blaOXA) and other genes associated with resistance to aminoglycosides (aaC1, aaC3, and aaC4), quinolones (qnrB, qnrC, qnrD, qnrS, and aac(6′)-Ib-cr), tetracyclines (tetA, tetB), and sulfonamides (sul1, sul2, and sul3) were PCR-detected. The relevant primers are listed in Table 1.
Detecting Class I Integrons
Class I integron-targeting primers were designed based on previously described sequences (28). After DNA gel extraction kit (Vazyme Biotech, Nanjing, China) purification, the class I integron-positive DNA samples were sequenced (Sangon Biotech, Shanghai, China). The DNA sequences obtained were compared with those in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) using the Basic Local Alignment Search Tool to determine the gene cassettes within the variable region of the class I integrons.
MLST
Seven housekeeping genes (aroC, dnaN, hemD, hisD, purE, sucA, and thrA) were used to characterize the Salmonella isolates using MLST (http://mlst.warwick.ac.uk/mlst/). The protocols from the MLST homepage were used, including the PCR conditions and primer sequence information (http://mlst.warwick.ac.uk/mlst/dbs/Senterica). PCR products were purified and sequenced (Sangon Biotech). The nucleic acid sequences corresponding to the allele values of the seven pairs of Salmonella housekeeping genes were downloaded from Pubmlst (https://pubmlst.org/), and BioEdit v7.0.9 software was used to construct a local gene bank for them. Genes from the gene library were aligned to obtain the allele values of the isolated strains and their corresponding ST types. The sequence types were assigned according to the MLST online scheme (http://mlst.warwick.ac.uk/mlst/dbs/Senterica), and the results of the eBURST map were constructed by Phyloviz (https://online.phyloviz.net/index).
Results
Isolation and Salmonella Serotyping
As described in the methods section, samples of the diseased materials were subjected to two-step enrichment using BPW and TTB, and a single colony was obtained by a three-line method on XLT4 medium. Colorless, translucent small colonies were observed on the plates, and some had smooth black cores and very narrow transparent bands around them. DNA was extracted from the isolated bacterial colonies, and PCR was performed with invA primers for Salmonella detection. After the PCR detection and the microbiological mass spectrometer verification, 126 Salmonella strains were isolated, with the detection rate of 3.59% (126/3508; Table 2). The isolating rates among different sample source were shown as: 6.27% (104/1688) in chicken embryo samples, 2.86% (6/210) in water samples, 1.14% (16/1400) in cloaca swabs, 0% (0/210) in feed samples. Salmonella was not detected in samples from Jiangsu and Sichuan provience. Among all the isolates, 54 sratins were recovered from samples of Shandong province, with the highest isolation rate of 18% (54/300), while a total of 72 Salmonella isolates were isolated from the other 6 provinces. In this study, a total of 95 strains, consisted of 23 randomly selected Shandong isolates and the aforementioned 72 isolates, were collected for subsequent tests.
Salmonella Serotyping Results
The Ningbo Tianrun Serum Diagnostic Kit test results showed that the O antigens of the 95 Salmonella strains were O9, O12 (79/95) and O4, O12 (16/95). The results from the semi-solid puncture tests showed that 38/95 isolates had flagella and 57 had no flagella. The serum diagnostic kit results showed that the H antigens from the 38 isolates were Hg, m (22/38) and Hs (16/38). The kit's results when combined with the PCR results showed that the 95 isolates included 57 S. gallinarum-pullorum strains (60.00%, 57/95), 22 S. enteritidis strains (23.16%, 22/95), and 16 S. agona strains (16.84 %, 16/95) (Figure 1B).
Antibiotic Resistance and MDR Profiles
The KB method was used to count the diameters of the inhibition zones and to determine growth inhibition to 14 common antibacterial drugs in the 95 Salmonella strains according to the 2018 edition of the CLSI (22) standards. The resistance rates in the 95 isolates were 100% (95/95) for erythromycin, 68.42% (65/95) for tetracycline, and 53.68% (51/95) for STR and AM, respectively. The AM resistance rate for the 95 isolates was 53.68% (51/95), and they were all sensitive to PB and SXT (Figure 2A).
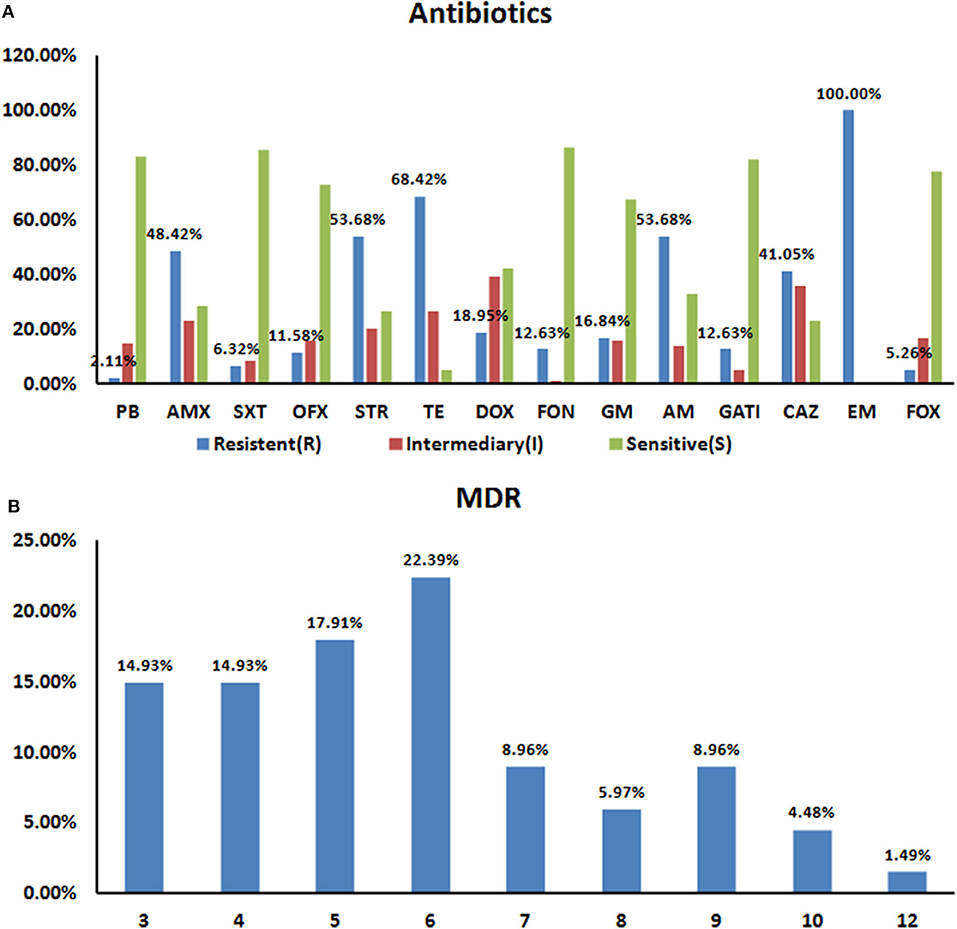
Figure 2. Resistance Characteristics of the 95 Salmonella Strains to 14 Antibiotics (A). Rates of antibiotic resistance amongst Salmonella isolates. The drug resistance rates for all 95 isolates were 100% for EM (95/95), 68.42% (65/95) for TE, and 53.68% (51/95) for STR and AM, respectively. All 95 strains showed polymyxin and sulfamethoxazole sensitivity (B). Prevalence of multidrug resistance amongst 95 Salmonella isolates. 70.53% (67/95) of the tested Salmonella isolates were resistant to at least three antibiotics. About 36.8% (35/95) strains resistant to 6–12 drugs.
We next statistically analyzed the drug resistance spectrum and multiple drug resistance in the isolated bacteria (Figure 2B). The MDR statistics showed that 70.53% (67/95) of the strains were resistant to three or more drug types, indicating that drug resistance in the isolated bacteria is prevalent with 38 different resistance patterns identified (Table 3). The drug-resistance spectrum was broad, and the most common pattern in seven of the isolates involved AMX-STR-TE-AM-CAZ-EM. The serotypes and resistance patterns were not significantly correlated. According to the resistance profile heat map (Figure 3), drug-resistant Salmonella isolated from the three chicken farms in Shandong Province had the highest drug-resistance rates.
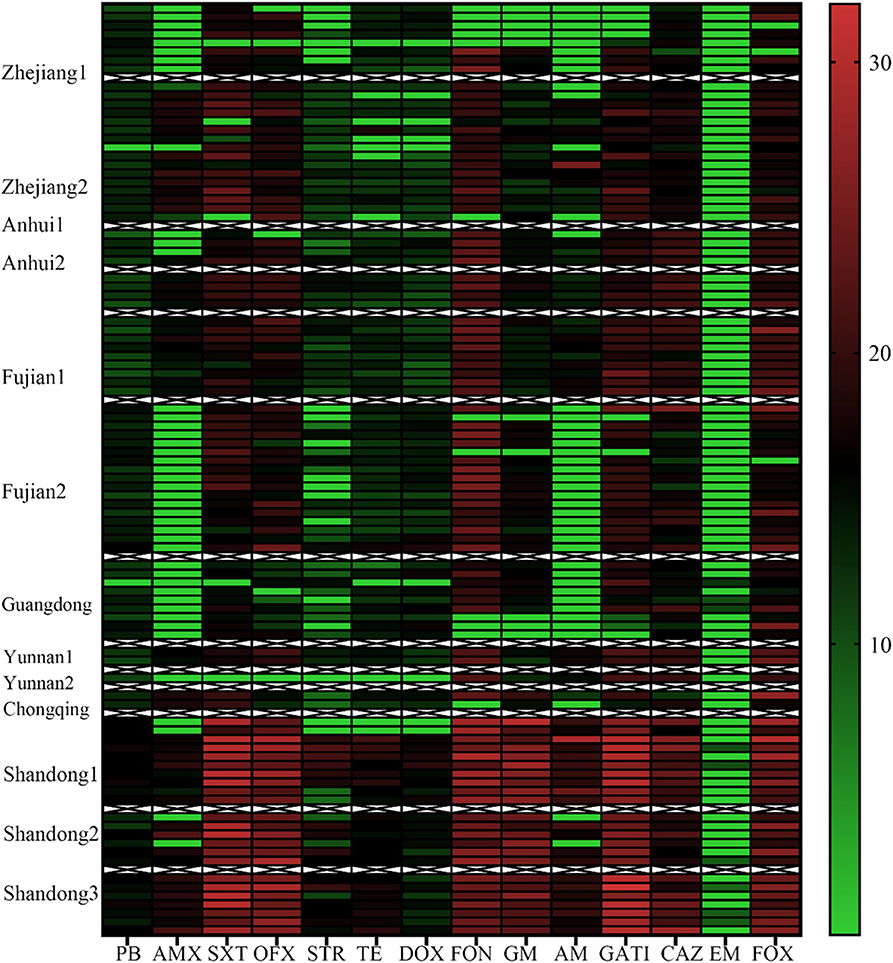
Figure 3. Antibiotic Resistance Heat Map for the 95 Salmonella Strains. Heat map showing the antibiotic-resistance distribution for the 95 Salmonella strains isolated from samples from broiler chicken farms in nine provinces to 14 antibiotic types. The color scale of the individual cells represents the resistance rate, as a percentage. PB, polymyxin B; AMX, amoxicillin; SXT, sulfamethoxazole; OFX, ofloxacin; STR, streptomycin; TE, Tetracycline; DOX, doxycycline; FON, florfenicol; GM, Gentamycin; AM, ampicillin; GATI, Gatifloxacin; CAZ, Ceftazidime; EM, erythromycin; and FOX, cefoxitin.
Prevalence of Antibiotic Resistance Genes
Using the DNA from each isolate as the template, PCR was used to detect the carriage status of the isolates to 18 drug resistance genes. Four different drug resistance genes were detected in DNA from the isolates, including those encoding β-lactams, aminoglycosides, and sulfonamides. The highest detection rates were seen for blaTEM and sul3 (100%, 95/95), followed by sul2 and aaC4 (52.63 and 23.16%, respectively) (Figure 4).
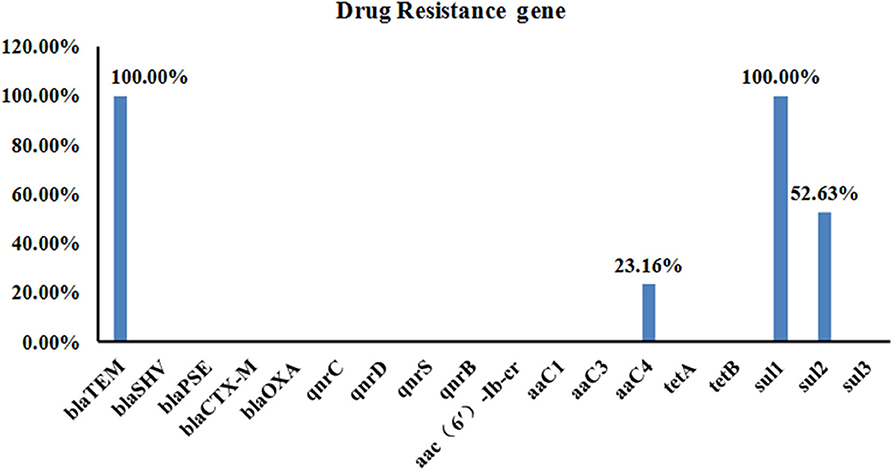
Figure 4. Carriage of 18 Drug Resistance Genes in the 95 Salmonella Strains. A total of four different drug resistance genes were detected in the DNA from the isolates, including those encoding β-lactams, aminoglycosides, and sulfonamides. The highest detection rate was for blaTEM and sul3 (both were 100%, 95/95), followed by sul2 and aaC4 with detection rates of 52.63 and 23.16%, respectively.
Detection of Class I Integrons
PCR testing detected no class I integrants in the DNA from the isolates; therefore none of the strains from this study carried class I integrants.
MLST
MLST classification showed that the 95 Salmonella strains fell into five ST types: ST92 (37/95, 38.95%), ST11 (22/95, 23.16%), ST2151 (19/95, 20.00%), ST13 (16/95, 16.84%), and ST470 (1/95, 1.05%) (Figure 5A). The results of the eBURST map showed that isolate ST11 was a cloned progenitor, isolates ST92 and ST2151 were subcloned groups from isolate ST11. Apart from ST13, the other four STs have close genetic relationships. The genetic direction was determined to be ST11-ST470-ST92-ST2151 (Figure 5B), with ST11 being the core sequence type.
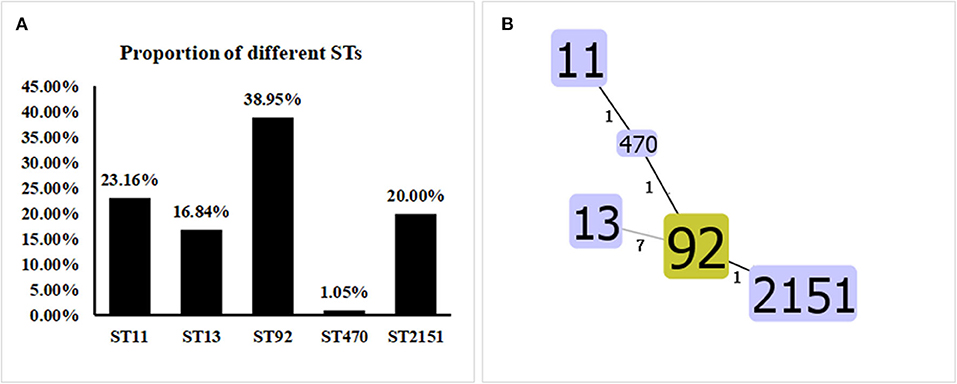
Figure 5. Proportions and Allelic Map of the Five STs in the 95 Salmonella Isolates. (A) MLST classification showed that the 95 Salmonella strains fell into the following five STs: ST92 (37/95, 38.95%), ST11 (22/95, 23.16%), ST2151 (19/95, 20.00%), ST13 (16/95, 16.84%), and ST470 (1/95, 1.05%). (B) Apart from ST13, the other four STs shared close genetic relationships. The genetic direction was found to be ST11-ST470-ST92-ST2151, and ST11 is the core sequence type.
Discussion
An important transmission route for Salmonella is vertical transmission. Therefore, chicken breeder farms are required to actively prevent, control, and work toward eliminating Salmonella infections on them. Once breeder chickens become infected with Salmonella, a “magnification effect” can occur as the generations expand. China has listed Salmonellosis as a priority disease control species in the National Medium and Long-Term Animal Disease Prevention and Control Plan (2012–2020) (http://www.gov.cn/zwgk/2012-05/25/content_2145581.htm), which requires all chicken farms across the country to meet the standards required to eliminate chicken salmonellosis by 2020. Hence, investigating salmonella epidemics on breeding farms is particularly important.
To investigate the prevalence and characteristics of Salmonella on Chinese poultry breeding farms requires information about drug-resistance genes, and the phenotypic characteristics of the isolates collected from them and their relationships. In this study, from May to July 2019, 3,508 samples from breeding farms were collected, including 1,400 cloaca swabs, 210 feed samples, 1,688 chicken embryo samples, and 210 water samples. The samples came from 14 breeder farms in eight provinces (Jiangsu, Anhui, Zhejiang, Fujian, Guangdong, Yunnan, Chongqing, and Sichuan) and three breeder flocks in Shandong Province. We detected 126 Salmonella strains in the 3,508 samples, a positivity rate of 3.59%. The positive isolation rate was very low, which may reflect the fact that the efforts made to prevent and control animal diseases in China are transitioning from effective control to gradual purification and elimination. In this investigation, especially in several breeder flocks in Jiangsu and Sichuan, Salmonella elimination has been done well. In contrast, the three local breed chicken breeder flocks in Shandong were more seriously contaminated with salmonella. It indicated that more attention should be paid on salmonella purification in local chicken breeds.
From the 1990s onwards, pullorum disease has seriously threatened the poultry industry (29). S. pullorum infection is still threatening the poultry farming industry and related industries in developing countries including China (30), resulting in huge economic losses in these countries. Among the three serotypes identified in this study, S. gallinarum-pullorum (57/95, 60.00%) still dominates, with S. enteritidis (22/95, 23.16%) and S. agona (16/95, 16.84%) ranking second and third. In contrast, S. enteritidis, S. Indiana, and S. typhimurium are more common in the Chinese chicken retail industry.
After serotype identification, we used molecular typing to further analyze the genetic relationships among the isolates. Various molecular typing techniques are widely used in the microbiology field to track the origin of pathogenic bacteria (13), from which the most commonly used techniques are MLST and PFGE. The high resolution and repeatability of PFGE has been recognized by laboratories around the world, but PFGE lacks a strict and uniform international naming standard, so it is difficult to achieve data sharing with this technique. With its convenience, high resolution, reliable data, and easy real-time internet sharing, MLST is widely used (14). In this study, the MLST typing results showed that the 95 Salmonella strains we isolated fell into five ST types (ST92, ST11, ST2151, ST13, and ST47) (1/95, 1.05%). The following correlations between STs and Salmonella serovars were founded: ST13 with S. agona, ST11 with S. Enteritidis, and ST92, ST2151, and ST470 with S. gallinarum-pullorum. Among them, ST92 (37/95, 38.95%) was the most common, a finding consistent with the results of a study on S. enteritidis in China from 2011 to 2016 (30), thereby confirming that the current Salmonella type still dominates. In addition to ST13, the Salmonella isolates displayed four other close ST kinships, with the genetic direction being ST11-ST470-ST92-ST2151.
In this study, β-lactams, quinolones, aminoglycosides, tetracyclines, and sulfonamides were tested on the 95 isolates. Among them, β-lactam-, aminoglycoside-, and sulfonamide-resistance genes were detected by PCR. Four resistance genes were detected, among which the detection rate for the β-lactam blaTEMresistance gene and the sul3 sulfonamide resistance gene was 100% (95/95) for both, whereas those of sul2 and aaC4 were 52.63 and 23.16%, respectively. Quinolone resistance genes and tetracycline resistance genes were not detected. This result is consistent with the results of a study (31) on sulfonamide-resistance genes in Pasteurella multocida, where the detection rate for sul2 (and sul1) exceeded 97%. Furthermore, Zhu et al. (32) identified sulfonamide-resistance genes (sul1, sul2, and sul3) in 89 (97.8%) Salmonella isolates. These findings indicate that sulfonamide-resistance genes are relatively stable in the environment. Extended-spectrum β-lactamase (ESBL)-producing Salmonella is a significant clinical and food safety concern worldwide (33). Salmonella isolates that harbor ESBL-encoding genes are able to hydrolyze penicillin as well as most of the first, second, and third generation cephalosporins, and even carbapenems (34, 35). In the present study, all the Salmonella isolates carried blaTEM, a β-lactam resistance gene, highlighting their potential pathogenesis and the risk posed by this pathogen to Chinese poultry breeding farms.
The Chinese Veterinary Pharmacopeia report (30) pointed out that the drugs tested in the present study, including EM, TE, SXT, and AM, have been widely used in poultry production in China. The resistance rate of these 95 isolates to AM, STR, TE, and EM ranged from 53.68 to 100%. The Salmonella isolates from each province in this article were resistant to at least three common antibacterial drugs, and the proportion of MDR strains was 70.53% (67/95), which is consistent with the results from previous surveys (15, 36). This result is consistent with the findings of previous reports from Iran and the United States (37, 38). It is worth noting that should strains with the same MDR genotypes as those identified in the Salmonella isolates be transferred to humans via chickens and their derivatives and with different degrees of resistance to 14 antibacterial drugs as was shown for our isolates, this would constitute a major public health threat.
A recent study indicated that integrins and multiple drug resistance genes on mobile plasmids may be responsible for the stable spread of MDR genes in Salmonella (39). Previous studies on class I integrants in Salmonella in Hubei, Shandong, and Shanxi (18, 39, 40) provinces have found that class I integrants displaying the various antibiotic resistance gene cassettes that are commonly found in Salmonella isolates are associated with provinces, markets, and storage conditions differ. Multiple antibiotic resistance in isolates carrying class I integrons is significantly higher than that seen in isolates lacking these integrons, and they usually show a corresponding antibiotic resistance spectrum to the antibiotic resistance gene cassette contained in their class I integrants. In the present study, the detection rate for class I integrants was 0 (0/95), and the influence of gene mutations and other factors relating to drug resistance in Salmonella was not investigated. This may be related to the stricter control measures of breeding poultry farms compared to commercial chicken farms, bacterial infectious diseases are better controlled, and the application of antibiotics is greatly reduced. At the same time, it should be noted that poultry farming often uses antibiotics, which may explain the resistance patterns we observed with the tested antibiotics. Therefore, reducing antibiotic use is particularly important for limiting the emergence of super multidrug resistant organisms so that good health in humans and animals can be maintained.
Conclusions
The Salmonella detection rate for the 17 breeder farms in the Chinese provinces we surveyed was 3.59%. Five STs, ST92, ST11, ST2151, ST13, and ST470 were identified. Our results indicate that among the Salmonella prevalent in the poultry breeder farms in the various provinces, S. gallinarum, S. enteritidis, and S. agona still occupy the main positions. All of the Salmonella isolates carried blaTEM and sul3, and 64.21% were MDR strains, suggesting that antibiotic use in the poultry breeder farms still requires attention. The correlation between the serotypes and the molecular subtypes of the Salmonella isolates suggests that the results of a comprehensive analysis of different methods may provide more useful epidemiological information related to this infection. Our findings will update current knowledge on the prevalence of antibiotic resistance on poultry breeding poultry farms in China and provide information and technical support toward the long-term goal of eradicating salmonellosis.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SS, FW, and XG conceived and designed the experiments and analyzed the data. YanS, FZ, LZ, and FW performed the experiments and contributed the reagents. YL performed data validation. FW and YanyS wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the Fund of Shandong Agricultural Major Application Technology Innovation (SD2019XM009), the Key Research and Development Program of Shandong Province (Important Science and Technology Innovation Project, 2019JZZY010735), Collaborative Innovation Program of Shandong Higher Education Institutions (SDE [2017] 11), and the Funds of Shandong Double Tops Program (SYL2017YSTD11). These funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sandra Cheesman, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
1. Simpson KMJ, Hill-Cawthorne GA, Ward MP, Mor SM. Diversity of Salmonella serotypes from humans, food, domestic animals and wildlife in New South Wales, Australia. BMC Infect Dis. (2018) 18:623. doi: 10.1186/s12879-018-3563-1
2. Bae DH, Dessie HK, Baek HJ, Kim SG, Lee HS, Lee YJ. Prevalence and characteristics of Salmonella spp. Isolated from poultry slaughterhouses in Korea. J Vet Med Sci. (2013) 75:1193–200. doi: 10.1292/jvms.13-0093
3. Shao D, Shi Z, Wei J, Ma Z. A brief review of foodborne zoonoses in China. Epidemiol Infect. (2011) 139:1497–504. doi: 10.1017/S0950268811000872
4. Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PAD, et al. Supplement 2003–2007 (No. 47) to the white-kauffmann-le minor scheme. Res. Microbiol. (2010) 161:26–9. doi: 10.1016/j.resmic.2009.10.002
5. Lee LA, Puhr ND, Maloney EK, Bean NH, Tauxe RV. Increase in Antimicrobial-Resistant Salmonella Infections in the United States, 1989-1990. J Infect Dis. (1994) doi: 10.1093/infdis/170.1.128
6. Liang Z, Ke B, Deng X, Liang J, Ran L, Lu L, et al. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal Salmonella from human patients in Guangdong Province, China, 2009–2012. BMC Infect Dis. (2015) 15:53. doi: 10.1186/s12879-015-0784-4
7. Kyu HH, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2017. Lancet. (2018) 392:1859–922. doi: 10.1016/S0140-6736(18)32335-3
8. Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the global burden of disease study 2017. Lancet. (2018) 392:1736–88. doi: 10.1016/S0140-6736(18)32203-7
9. Spencer LJ, Degu A, Kalkidan HA, Solomon MA, Cristiana A, Nooshin A. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet. (2018) 392:1789–858. doi: 10.1016/S0140-6736(18)32279-7
10. Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, et al. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the global burden of disease study 2017. Lancet Infect Dis. (2019) 19:1312–24. doi: 10.1016/S1473-3099(19)30418-9
11. Kehrenberg C, Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother. (2006) 50:1156–63. doi: 10.1128/AAC.50.4.1156-1163.2006
12. Kehrenberg C, Schwarz S. Florfenicol-chloramphenicol exporter gene fexA is part of the novel transposon Tn558. Antimicrob Agents Chemother. (2005) 49:813–5. doi: 10.1128/AAC.49.2.813-815.2005
13. Achtman M, Wain J, Weill F, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLOS Pathog. (2012) 8:e1002776. doi: 10.1371/journal.ppat.1002776
14. Bilhere E, Lucas P, Claisse O, Lonvaudfunel A. Multilocus sequence typing of oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl Environ Microbiol. (2009) 75:1291–300. doi: 10.1128/AEM.02563-08
15. Yang X, Huang J, Zhang Y, Liu S, Chen L, Xiao C, et al. Prevalence, abundance, serovars and antimicrobial resistance of salmonella isolated from retail raw poultry meat in China. Sci Total Environ. (2020) 713:136385. doi: 10.1016/j.scitotenv.2019.136385
16. Xiong ZY, Wang SJ, Huang YM, Gao Y, Shen HY, Chen ZQ, et al. Ciprofloxacin-resistant Salmonella enterica Serovar Kentucky ST198 in broiler chicken supply chain and patients, China, 2010-2016. Microorganisms. (2020) 8:140. doi: 10.3390/microorganisms8010140
17. Zhu Z, Huang Q, Hong X, Chen X, Lu Y, Chen Z, et al. Isolation and characterization of salmonella in pork samples collected from retail and wholesale markets in each season from 2016 to 2018 in Wuhan, China. J Appl Microbiol. (2020) 128:875–83. doi: 10.1111/jam.14515
18. Yang J, Ju Z, Yang Y, Zhao X, Jiang Z, Sun S. Serotype, antimicrobial susceptibility and genotype profiles of salmonella isolated from duck farms and a slaughterhouse in Shandong province, China. BMC Microbiol. (2019) 19:202. doi: 10.1186/s12866-019-1570-z
19. Fardsanei F, Dallal MMS, Douraghi M, Salehi TZ, Mahmoodi M, Memariani H, et al. Genetic diversity and virulence genes of Salmonella enterica subspecies enterica serotype enteritidis isolated from meats and eggs. Microb Pathog. (2017) 107:451–6. doi: 10.1016/j.micpath.2017.04.026
20. Malorny B, Hoorfar J, Bunge C, Helmuth R. Multicenter validation of the analytical accuracy of salmonella PCR: towards an international standard. Appl Environ Microbiol. (2003) 69:290–6. doi: 10.1128/AEM.69.1.290-296.2003
21. Xu L, Liu Z, Li Y, Yin C, Hu Y, Xie X, et al. A rapid method to identify Salmonella enterica serovar Gallinarum biovar Pullorum using a specific target gene ipaJ. Avian Pathol. (2018) 47:238–44. doi: 10.1080/03079457.2017.1412084
22. CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth-third Informational Supplement M100-S23. Wayne: Clinical and Laboratory Standards Institute (2013).
23. Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, et al. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microb. (2007) 73:6686–90. doi: 10.1128/AEM.01054-07
24. Puah SM, Puthucheary SD, Liew F, Chua KH. Aeromonas aquariorum clinical isolates: antimicrobial profiles, plasmids and genetic determinants. Int J Antimicrob Agents. (2013) 41:281–4. doi: 10.1016/j.ijantimicag.2012.11.012
25. Navajasbenito EV, Alonso CA, Sanz S, Olarte C, Martinezolarte R, Hidalgosanz S, et al. Molecular characterization of antibiotic resistance in Escherichia coli strains from a dairy cattle farm and its surroundings. J Sci Food Agric. (2017) 97:362–5. doi: 10.1002/jsfa.7709
26. Aarestrup FM, Lertworapreecha M, Evans M, Bangtrakulnonth A, Chalermchaikit T, Hendriksen RS, et al. Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J Antimicrob Chemoth. (2003) 52:715–8. doi: 10.1093/jac/dkg426
27. Zhang A, Wang H, Tian G, Zhang Y, Yang X, Xia Q, et al. Phenotypic and genotypic characterisation of antimicrobial resistance in faecal bacteria from 30 Giant pandas. Int J Antimicrob Agents. (2009) 33:456–60. doi: 10.1016/j.ijantimicag.2008.10.030
28. Wang W, Wang L, Su J, Xu Z. Antibiotic susceptibility, biofilm-forming ability, and incidence of class 1 integron of salmonella spp., Escherichia coli, and Staphylococcus aureus isolated from various foods in a school canteen in China. Foodborne Pathog Dis. (2019) 17:269–75. doi: 10.1089/fpd.2019.2694
29. Zhang D, Zhuang L, Wang C, Zhang P, Zhang T, Shao H, et al. Virulence gene distribution of Salmonella pullorum isolates recovered from chickens in China (1953–2015). Avian Dis. (2018) 62:431–6. doi: 10.1637/11927-071318-ResNote.1
30. Guo X, Wang H, Cheng Y, Zhang W, Luo Q, Wen G, et al. Quinolone resistance phenotype and genetic characterization of Salmonella enterica serovar Pullorum isolates in China, during 2011 to 2016. BMC Microbiol. (2018) 18:225. doi: 10.1186/s12866-018-1368-4
31. Chen H, Cheng L, Deng HEA. Detection of sulfonamides sensitivity and analysis of resistant genes of Pasteurella multocida isolated from ducks. China J Anim Husb Vet Med. (2019) 46:223–30. doi: 10.16431/j.cnki.1671-7236.2019.01.026
32. Zhu Y, Lai H, Zou L, Yin S, Wang C, Han X, et al. Antimicrobial resistance and resistance genes in salmonella strains isolated from broiler chickens along the slaughtering process in China. Int J Food Microbiol. (2017) 259:43–51. doi: 10.1016/j.ijfoodmicro.2017.07.023
33. Seiffert SN, Hilty M, Perreten V, Endimiani A. Extended-spectrum cephalosporin-resistant gram-negative organisms in livestock: an emerging problem for human health? Drug Resist Update. (2013) 16:22–45. doi: 10.1016/j.drup.2012.12.001
34. Qiao J, Alali WQ, Liu J, Wang Y, Chen S, Cui S, et al. Prevalence of virulence genes in extended-spectrum β-lactamases (ESBLs)-producing salmonella in retail raw chicken in China. J Food Sci. (2018) 83:1048–52. doi: 10.1111/1750-3841.14111
35. Wang W, Liu F, Peng Z, Li F, Ma A. Complete genome sequence of Salmonella enterica subsp. enterica Serovar Indiana C629, a carbapenem-resistant bacterium isolated from chicken carcass in China. Genome Announc. (2016) 4:e00662–16. doi: 10.1128/genomeA.00662-16
36. Zhou X, Xu L, Xu X, Zhu Y, Suo Y, Shi C, et al. Antimicrobial resistance and molecular characterization of Salmonella enterica Serovar Enteritidis from retail chicken products in shanghai, China. Foodborne Pathog Dis. (2018) 15:346–52. doi: 10.1089/fpd.2017.2387
37. Fardsanei F, Dallal MMS, Douraghi M, Memariani H, Bakhshi B, Salehi TZ, et al. Antimicrobial resistance, virulence genes and genetic relatedness of Salmonella enterica serotype Enteritidis isolates recovered from human gastroenteritis in Tehran, Iran. J Glob Antimicrob Resist. (2017) 12:220–6. doi: 10.1016/j.jgar.2017.10.005
38. Velasquez C, Macklin KS, Kumar S, Bailey MA, Ebner PE, Oliver HF, et al. Prevalence and antimicrobial resistance patterns of salmonella isolated from poultry farms in southeastern United States. Poult Sci. (2018) 97:2144–52. doi: 10.3382/ps/pex449
39. Zhou M, Li X, Hou W, Wang H, Paoli GC, Shi X. Incidence and characterization of salmonella isolates from raw meat products sold at small markets in Hubei province, China. Front Microbiol. (2019) 10:2265. doi: 10.3389/fmicb.2019.02265
Keywords: Salmonella, multidrug resistance, MLST, phenotypes, chickens, breeder farms
Citation: Song Y, Wang F, Liu Y, Song Y, Zhang L, Zhang F, Gu X and Sun S (2020) Occurrence and Characterization of Salmonella Isolated From Chicken Breeder Flocks in Nine Chinese Provinces. Front. Vet. Sci. 7:479. doi: 10.3389/fvets.2020.00479
Received: 31 May 2020; Accepted: 29 June 2020;
Published: 05 August 2020.
Edited by:
Guillermo Tellez, University of Arkansas, United StatesReviewed by:
Roberto Senas Cuesta, University of Arkansas, United StatesXochitl Hernandez, National Autonomous University of Mexico, Mexico
Copyright © 2020 Song, Wang, Liu, Song, Zhang, Zhang, Gu and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuhong Sun, c3Vuc2h1aG9uZ0BzZGF1LmVkdS5jbg==; Xiaoxue Gu, Z3h4amFja2llQDE2My5jb20=
†These authors have contributed equally to this work
 Yan Song
Yan Song Fangkun Wang
Fangkun Wang Yang Liu3
Yang Liu3 Yanying Song
Yanying Song