
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 21 August 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00477
This article is part of the Research Topic FMD Research: Bridging the Gaps with Novel Tools View all 25 articles
Foot-and-mouth disease (FMD) is a devastating livestock disease caused by foot-and-mouth disease virus (FMDV). Outbreaks of this disease in a country always result in conspicuous economic losses to livestock industry and subsequently lead to serious socioeconomic damages due to the immediate imposition of trade embargo. Rapid and accurate diagnoses are imperative to control this infectious virus. In the current review, enzyme-linked immunosorbent assay (ELISA)-based methods used in FMD diagnosis are extensively reviewed, particularly the sandwich, liquid-phase blocking, and solid-phase competition ELISA. The differentiation of infected animals from vaccinated animals using ELISA-based methods is also highlighted, in which the role of 3ABC polyprotein as a marker is reviewed intensively. Recently, more studies are focusing on the molecular diagnostic methods, which detect the viral nucleic acids based on reverse transcription-polymerase chain reaction (RT-PCR) and RT-loop-mediated isothermal amplification (RT-LAMP). These methods are generally more sensitive because of their ability to amplify a minute amount of the viral nucleic acids. In this digital era, the RT-PCR and RT-LAMP are progressing toward the mobile versions, aiming for on-site FMDV diagnosis. Apart from RT-PCR and RT-LAMP, another diagnostic assay specifically designed for on-site diagnosis is the lateral flow immunochromatographic test strips. These test strips have some distinct advantages over other diagnostic methods, whereby the assay often does not require the aid of an external device, which greatly lowers the cost per test. In addition, the on-site diagnostic test can be easily performed by untrained personnel including farmers, and the results can be obtained in a few minutes. Lastly, the use of FMDV diagnostic assays for progressive control of the disease is also discussed critically.
Foot-and-mouth disease (FMD) is a contagious vesicular disease caused by foot-and-mouth disease virus (FMDV), a member of the Picornaviridae family. The virus infects a wide range of wild and domesticated cloven-footed mammals. An accidental introduction of FMDV in a susceptible population can result in an abrupt outbreak of the disease, leading to a massive economic loss.
Immediate actions are usually taken in response to an FMD outbreak to secure a differential and definitive diagnosis and to prevent further spread of the disease (Figure 1). To complement the vaccination and stamping out policies, early FMD detections in cloven-hoofed animals using current available diagnostic tools have been widely employed to counter this highly scrutinized agent.
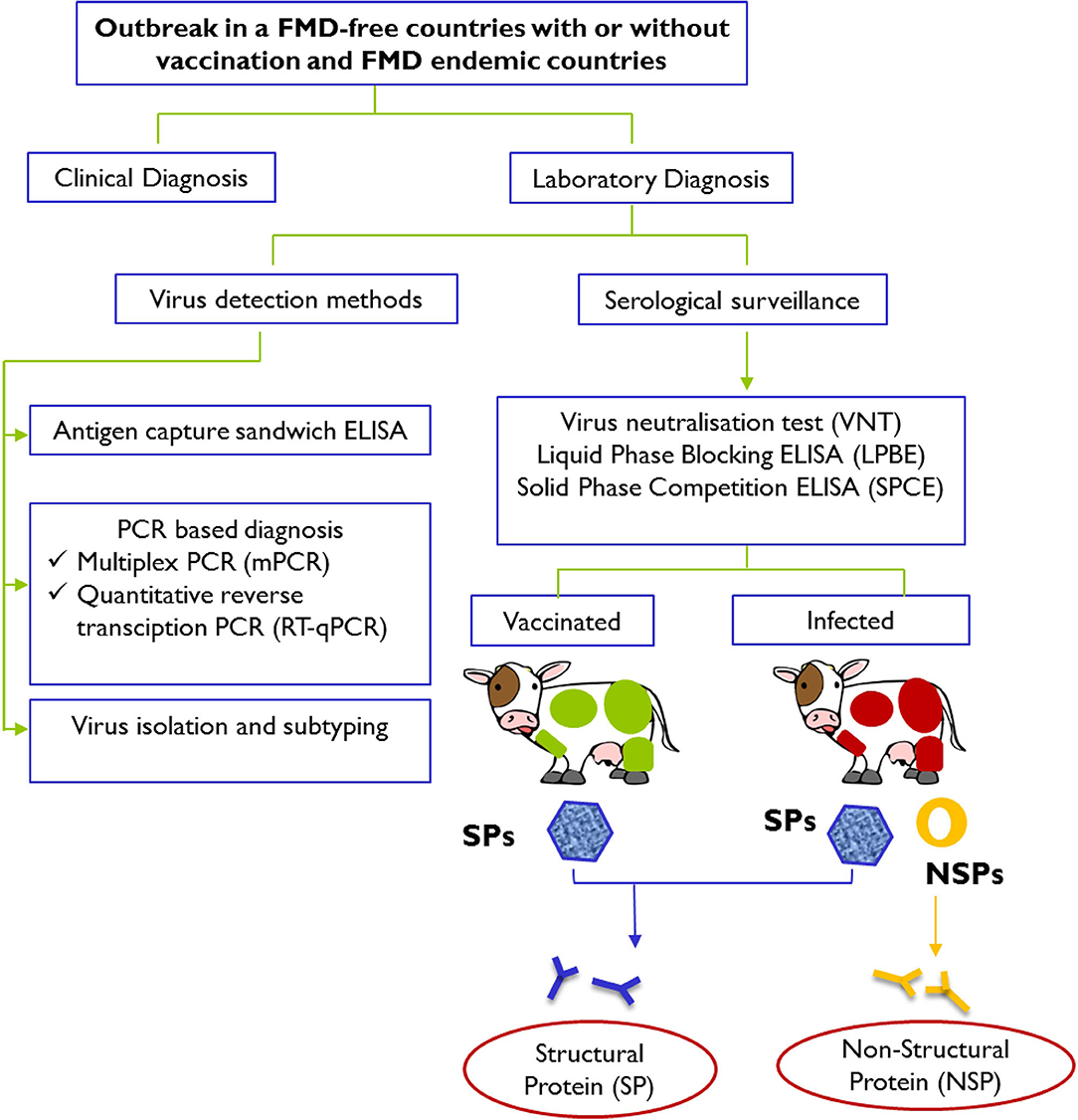
Figure 1. Schematic representation of laboratory tests for determining evidence of infection with FMDV after an outbreak in FMD-free countries with or without vaccination and FMD endemic countries. Laboratory confirmation of a presumptive diagnosis of FMD involves detection and identification of viral materials in animals' samples or presence of specific antibodies against structural proteins (SPs; presence in both vaccinated and infected animals) and specific antibodies against non-structural proteins (NSPs; presence in infected animals only) in serum samples. Diagnostic procedures for FMD can be found in the Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2019 (https://www.oie.int/standard-setting/terrestrial-manual/access-online/).
Generally, a suspected case of FMD can be identified based on observations of clinical signs. Severity of the symptoms in animals is affected by many factors, such as the species and age of the animals, virus strains, dosage of exposure to FMDV, and the host immunity. The symptoms are generally more severe in cattle and intensively reared swine (high-density indoor-rearing in straw-lined sheds or group-housing), as compared to goats and sheep (1, 2). Typically, FMDV-infected animals will develop lesions on the tongue, muzzle, oral cavity, coronary bands, and teats. Other symptoms frequently observed include fever, loss of appetite, weight loss, hypersalivation, depression, growth retardation, and severe decrease in milk production, which could persist after recovery (2). However, diagnoses based on clinical symptoms are highly unreliable, because several other diseases share similar symptoms as FMD, which include swine vesicular disease (SVD), vesicular stomatitis and vesicular exanthema. Swine are vulnerable to vesicular stomatitis, SVD, and FMD, whereas cattle are vulnerable to vesicular stomatitis and FMD, all of which could not be distinguished based on clinical symptoms (3–5). Hence, confirmatory laboratory diagnosis of any suspected FMD case is vital.
Conventional techniques such as complement fixation test (CFT), virus isolation test, virus neutralization test (VNT), and enzyme-linked immunosorbent assay (ELISA) are routinely used to detect FMDV in clinical samples. As virus isolation tests, CFT and VNT are well-established and often used as standards in development of new detection assays; thus, they will not be discussed in detail in this article. Advancement in molecular techniques accelerates rapid and accurate diagnoses of FMDV through detection of the viral RNA. In this article, the most recent advancements in reverse transcription-polymerase chain reaction (RT-PCR) and RT-loop-mediated isothermal amplification (RT-LAMP)-based methods are thoroughly reviewed. Lastly, the roles of lateral flow immunochromatographic (LFI) test strips in FMDV diagnosis are also discussed.
Nucleic acid detection methods are molecular-based techniques used to detect the presence of viral nucleic acids. As these methods involve amplifications of viral nucleic acids, they have higher sensitivity compared to serological methods. In addition to detection of FMDV, primers used for FMDV serotyping have also been developed (6). As FMDV is an RNA virus, RT is required before the targeted viral nucleic acid can be amplified. Two of the most common nucleic acid detection methods used to detect FMDV are RT-PCR and RT-LAMP. Although the detection and typing of FMDV using microarray was also reported (7–9), its usage is highly limited, possibly due to the high operating cost. Table 1 summarizes some recent studies on molecular diagnostic assays for the detection of FMDV.
Detection of FMDV using RT-PCR was first reported by Meyer et al. (32), in which a conserved region in the viral genome encoding the RNA polymerase was amplified and analyzed using agarose gel electrophoresis and further confirmed by restriction enzyme digestion or Southern blotting. Höfner et al. (33) also demonstrated the detection of FMDV in clinical samples using primers targeting the 1A and 2A/2B conserved regions, amplifying the whole viral capsid coding region. Nucleotide sequencing of the amplified region can directly aid in the study of viral epidemiology. In a separate study, Laor et al. (34) showed that the primers targeting the RNA polymerase coding region could detect FMDV of different isolates, whereas another primer set targeting the variable region of VP1 was capable of differentiation detection. Dill et al. (10) developed a universal RT-PCR, which is rapid and cost-effective in generating the genome sequences of all FMDV serotypes, allowing immediate virus genotyping, phylogenetic analysis, and epidemiological studies of FMDV. Most recently, a primer set, namely, FM8/9, which targets the conserved region of 3D domain has also been reported to be 100.6- to 103.8-fold more sensitive than the 1F/R primer set as suggested in the OIE manual (11).These agarose gel electrophoresis-based methods have since laid the basis for the modern RT-PCR and RT-LAMP detection methods.
To date, many improved versions of RT-PCR have been employed for the detection of FMDV. To simultaneously screen for the presence of multiple viruses, multiplex RT-PCR, which uses multiple primer sets in a single reaction, has been developed. Lung et al. (9) demonstrated the use of multiplex RT-PCR for simultaneous detection and differentiation of FMDV serotypes and other vesicular disease viruses, including vesicular stomatitis virus (VSV), swine vesicular disease virus (SVDV), and vesicular exanthema of swine virus (VESV). When the multiplex RT-PCR is used in conjunction with slide microarray, the sensitivity improved by at least one log unit. However, the sensitivity was reported to be comparable but no better than real-time RT-PCR (RT-qPCR). Similarly, Erickson et al. (22) also used multiplex RT-PCR coupled with a more advanced, automated electronic microarray assay for simultaneous detection and differentiation of several swine viruses, including FMDV and other viruses such as SVDV, African swine fever virus, porcine circovirus type 2, porcine respiratory and reproductive syndrome virus, VESV, and classical swine fever virus. Although it is convenient to detect multiple viruses of different serotypes in a single reaction, careful design, and testing of primers are needed to achieve desirable assay sensitivity and specificity, as multiplex RT-PCR assays have been reported to have better sensitivity upon removal of certain primers from the pool (22).
Real-time quantitative PCR (qPCR)-based analyses coupled with fluorescent-emitting compounds have been used to measure the number of amplicons during an amplification process in real time (35). Generally, there are two types of fluorescent-emitting compounds used in qPCR: (i) non-specific intercalating dye such as SYBR green and (ii) specific reporter probes with fluorochromes attached to specific oligonucleotide sequences, such as fluorescence resonance energy transfer (FRET) and Taqman probes. These compounds have been used in RT-qPCR for detection of FMDV [FRET- (36); TaqMan- (37, 38); SYBR green- RT-qPCR (17, 39)]. While SYBR green is more economic, TaqMan and FRET probes have the advantage as signals generated from unspecific PCR are negligible. The most common target sequences for the detection of FMDV with RT-qPCR include 3D and 5′ UTR sequences (37, 38), in which the addition of 5′-tails to the primers targeting 3D and 5′ UTR sequences was reported to enhance the detection of FMDV (16). As in RT-PCR, RT-qPCR has also been exploited for multiplex detection. Reid et al. (15) developed a multiplex RT-qPCR assay using primer/probe sets targeting the FMDV VP1 coding region for detection and differentiation of FMDV serotypes O, A, and Asia 1 circulating in the Middle East. Nonetheless, when compared to single RT-qPCR, multiplex RT-qPCR tends to produce false-negative results due to mismatch in the probe-binding regions, suggesting that a higher sequence identicality is required for application in multiplex system. This complexity has limited the usage of this technique.
Although highly reliable, RT-qPCR often requires laboratory setting with a qPCR thermocycler, a cost factor that limits its usage in the field. To overcome such limitation, on-site devices capable of performing FMDV diagnosis in the field have been developed, such as the fully automated cartridge-based RT-qPCR diagnostic system, Enigma MiniLab® (18). Another handheld RT-qPCR device, Biomeme two3™ (two3) has also been developed and evaluated as a field-deployable platform for FMDV diagnosis, in which the sensitivity was shown to be almost comparable to RT-qPCR using the ABI7500 platform (19). The RNA samples can be extracted with an on-site RNA extraction kit such as Biomeme M1 Sample Prep™ cartridge kit, a method that is dispensable of laboratory equipment and chemicals such as microcentrifuge, alcohol, phenol, and chloroform (19).
Another modified version of RT-PCR known as the RT-insulated isothermal PCR (RT-iiPCR) assay was also developed for qualitative detection of FMDV (12). Unlike traditional PCR that requires cycles of multiple temperatures, RT-iiPCR utilizes a temperature gradient generated from a thermal convection from a single heating source, which hastens the detection process (12). Additionally, RT-recombinase polymerase amplification (RT-RPA) has also been used for detection of FMDV (14, 40). The RT-RPA uses three specific proteins: recombinase allows primer annealing to double-stranded DNA; single-stranded DNA-binding protein stabilizes primer binding; and strand-displacing DNA polymerase. As for real-time detection, another method based on water-oil emulsion droplet technology for partition of nanoliter droplets containing viral cDNA, known as the RT-droplet digital PCR (RT-ddPCR) was established for the absolute quantitation of FMDV RNA in epithelial and esophageal-pharyngeal fluid samples from FMDV-infected cattle (13).
Generally, the FMDV RNA for diagnostic purposes could be obtained from specimens, which include (i) swabs of oral, nasal, and lesion; (ii) epithelial tissue suspensions; and (iii) oral or vesicular fluid. In a recent study, Goller et al. (18) explored milk as a non-invasive sample for FMDV surveillance using RT-qPCR. They demonstrated that the RT-qPCR on milk sample was capable of detecting FMDV RNA 18 days after contact, which is later than the viral RNA detected in serum samples, suggesting milk as a feasible sample for FMD surveillance (18, 21). In addition, EDTA-treated blood samples have also been explored as a source of the viral RNA for the diagnosis of FMDV using RT-qPCR, owing to the samples' stability during transportation, as well as the ease of sample processing at the diagnostic laboratory (41). Fontél et al. (41) reported that the diagnostic assay using EDTA-treated blood samples was ~10 times less sensitive than that of serum samples. However, the study used double the volume of serum than that of EDTA-treated blood samples for RNA extraction, of which the serum contains almost double the virus concentration with the same sample volume as the red blood cells were spun off during centrifugation. This gave rise to an uneven amount of virus/volume in each sample type, with serum containing nearly four times the amount of virus, thereby resulting in a difference in cycle threshold (Ct) value of ~2, assuming that both types of samples were equally good for RT-qPCR detection. The results obtained by Fontél et al. (41) showed that the Ct value for serum was three to four times lower than that of EDTA-treated blood, in which the actual difference should be only 1- to 2-fold lower. Therefore, EDTA-treated blood samples, as initially proposed by Fontél et al. (41), may still remain a viable option for FMDV diagnosis.
Unlike PCR that requires cycles of different temperatures for amplification of DNA, LAMP is a method capable of amplifying DNA at a single temperature at around 60° to 65°C. It was first invented by Notomi et al. (42), and the method was demonstrated to be highly sensitive and specific. In general, the method involves the use of at least four primers and a DNA polymerase with high strand displacement activity. Two of the primers form loop structures at their respective 5′ ends, and the other two primers play the role of displacing the loop-forming strands from the template at the loop regions as they are being synthesized by DNA polymerase with high strand displacement activity. Once a double-stranded product with both ends capable of forming loop structures is synthesized, it then functions as a template for infinite amplification of the DNA, provided there are still loop-forming primers and dNTPs available. Loop-mediated isothermal amplification is capable of producing 109 copies of DNA in less than an hour (42), which probably exceeds the speed of PCR amplification, as no denaturation of double-stranded DNA is required. Another significant difference between LAMP and PCR is their amplification products. While PCR generates high copy numbers of identical products, LAMP generates a mixture of stem-loop DNAs, which can be observed through visual detection, based on the formation of insoluble magnesium pyrophosphate, which can be detected by simple turbidimeter or visual turbidity (43).
As FMDV is an RNA virus, RT-LAMP is needed for detecting the FMDV genomic sequence. Before the double-stranded DNA template with both loop-forming ends can be generated, a loop-forming primer targeting the positive-sense RNA template accompanied by reverse transcriptase is required to generate the RNA-DNA hybrid, which will then be displaced by another primer targeting the RNA template at the loop-forming region of the DNA strand through the displacement activity of the DNA polymerase. The antisense viral DNA will then function as a template for LAMP as described above.
RT-LAMP was first employed for FMDV diagnosis by Dukes et al. (44), targeting the FMDV 3D RNA polymerase gene, and the products of amplification can be visually inspected for turbidity, analyzed using agarose gel electrophoresis, or monitored in real time through addition of fluorescent dyes. Three years later, Li et al. (45) used RT-LAMP for detection of FMDV by targeting a conserved region within the FMDV polyprotein gene (3D), at positions 7,905–8,094 of FMDV O isolate o1bfs46 iso46 (GenBank accession no. AY593816). Thereafter, detections of FMDV serotypes C and Asia 1 using specific primers have also been reported (23, 24). Generally, for detection of FMDV regardless of serotypes, RT-LAMP requires a longer conserved region for primer design compared to RT-PCR and RT-qPCR, as RT-LAMP requires four to six primers to function. Although the target gene selected for detection is highly conserved, some degree of differences exists between different FMDV isolates (25), which will result in mismatch of nucleotides between primers and the target gene, thereby lowering the assay's sensitivity.
As FMDV genomic sequences vary widely between different serotypes, primers used for the detection of one particular serotype may not detect FMDV of another serotype. Therefore, Yamazaki et al. (25) developed a multiplex RT-LAMP, which contains multiple primer sets targeting the 3D conserved regions for the detection of FMDV regardless of serotypes, in which the assay's sensitivity and specificity were reported to be up to 98.0 and 98.1%, respectively. In each multiplex reaction tested, a combination of two primer sets was used. Unlike RT-PCR, RT-LAMP uses primer sets containing around six oligonucleotides with overlapping sequences. As multiplex RT-LAMP involves mixing of primer sets for simultaneous detection of different target genes, its application could be limited by the number of primer sets that can be used in a single reaction, as increase in varieties of primer sequences may increase the rate of unspecific binding, thereby affecting the assay's specificity. In addition, antigen capture RT-LAMP (Ag-RT/LAMP) assay has also been reported to be capable of detecting and serotyping FMDV. An anti-FMDV immunoglobulin G (IgG) that interacts with the VP1 epitope was immobilized on a tube to capture FMDV of various genotypes, and subsequently the viral 3D conserved region was amplified using RT-LAMP amplification (30). Although this method provides an alternative for the differential detection of FMDV using RT-LAMP, the method is heavily dependent on the efficiency of antibody used to capture the target virus. In addition, this method requires a longer time for completion compared to normal RT-LAMP, as extra incubation time is needed for capturing of target virus by antibody. In addition, the specificity of Ag-RT/LAMP has also been reported to be affected by high viral loads in samples of interest.
In a separate study, RT-LAMP was used together with a lateral flow device (LFD) for the detection of FMDV in minimally processed (without RNA extraction) samples (46). Although the coupling of LFD did not increase the assay's sensitivity, it allowed an easier interpretation of the test results compared to visual detection of turbidity from magnesium pyrophosphate, or color changes of double-stranded DNA-staining dyes. The combined use of RT-LAMP and LFD will be discussed in more detail under the subsection chromatographic strip test. For FMDV detection, RT-LAMP has been demonstrated to have an analytical sensitivity of 10-folds lower than that of the conventional RT-PCR, which is 10-folds lower than that of RT-qPCR method (26). To increase the sensitivity of RT-LAMP, a recent study that used swarm primers to improve the accessibility of DNA to standard LAMP primers was deployed for rapid and accurate diagnosis of FMDV from the pool-1 region (26). At a high concentration, swarm primers anneal to the double-stranded cDNA at higher rate, thereby exposing the inner primer annealing sites, facilitating the binding of the RT-LAMP primers, which results in a faster amplification rate, as well as more RT-LAMP products. Overall, the swarm primer-based RT-LAMP (sRT-LAMP) has been demonstrated to have analytical sensitivity of 10-folds higher than the conventional RT-PCR, which is comparable to RT-qPCR for FMDV detection.
To date, real-time RT-LAMP (qRT-LAMP) is available for the detection and differentiation of FMDV serotypes O, A, and Asia 1 (47). Unlike RT-qPCR, qRT-LAMP for FMDV detection could not produce accurate quantitative results, as measurements of magnesium pyrophosphate by turbidity or DNA by fluorogenic dye are directly proportional to the size of LAMP products. As LAMP products are of many different sizes, these methods could not truly reflect the number of replication cycle as in RT-qPCR. Recently, real-time detection and monitoring of LAMP using self-quenching and dequenching fluorogenic probes for direct quantification of LAMP products have been developed (48). However, this method has yet been applied for the detection of FMDV. RT-LAMP was used for detecting and serotyping FMDV in India and Pakistan, in which the results were reported to be comparable to RT-qPCR (27, 29, 31). Another study in Africa demonstrated that RT-LAMP outperformed RT-qPCR in the detection of FMDV (49), although further testing with a bigger sample size and a wide variety of serotypes is needed to support the finding. However, without the need of sophisticated devices as in RT-qPCR, RT-LAMP represents a potential method to be used for on-site diagnoses of FMDV. To further encourage the use of RT-LAMP in laboratory and field settings, Howson et al. (28) demonstrated the use of lyophilized reagents for RT-LAMP (Enigma Diagnostics Limited) and RT-qPCR (OptiGene Limited), in which lyophilization greatly improved the storage stability of test reagents without jeopardizing the assays' performance.
Serological methods detect the presence of viral antigens or antibodies in serum or other body fluid samples. Apart from detecting the viral genome 1 to 2 days postinfection in oral fluid and serum samples (50, 51), FMD diagnosis can also be confirmed through the detection of anti-FMDV antibodies in serum samples. As infection by FMDV will often result in the production of antibodies against the viral antigens (detectable ~4 days postinfection in cattle sera) (50), detection of these antibodies can therefore indicate the presence of current or past infection. Some of the serological detection methods include VNT, solid-phase competition (SPC) ELISA, and liquid-phase blocking (LPB) ELISA. When ELISA-based methods are used to detect antibodies against both the structural proteins (SPs) and non-structural proteins (NSPs), it is capable of differentiating the infected from vaccinated animals (DIVA), which will be discussed in detail in Enzyme-Linked Immunosorbent Assay. Table 2 summarizes recent studies on ELISA-based FMDV diagnostic assays.
Enzyme-linked immunosorbent assay, pioneered by Engvall and Perlmann (85), is an analytical method commonly used for qualitative and quantitative analyses. Current ELISA is a modified version of radioimmunoassay techniques, which was first described by Coons et al. (86), in which an antigen is immobilized on a solid phase either directly or indirectly to capture a targeted antibody, which is then reported through a secondary antibody conjugated to an enzyme, where signals will be generated in the presence of its corresponding substrate. In general, ELISA is categorized into direct, indirect, sandwich, and competitive ELISA (87). Currently, ELISA is one of the most common approaches in detection of FMDV in addition to the virus isolation, VNT, and PCR-based techniques (88). According to a report from the Regional Reference Laboratory for FMD in South East Asia, more than 13,000 ELISAs were performed compared to 304 VNTs and 790 PCR-based assays for diagnosis of FMD in 2017 (89).
Detection of FMDV specific antibodies in vaccinated bovine sera using an indirect ELISA was first reported by Abu Elzein and Crowther (90), in which the test sera from cattle reacted with the FMDV coated on the microtiter plate followed by detection with anti-bovine antiserum conjugated to an enzyme. Subsequently, the same research group demonstrated the capability of a sandwich ELISA in detecting and quantifying FMDV with 50 to 100 times higher sensitivity than CFT (91). A double-sandwich ELISA method developed by Roeder and Le Blanc Smith (92) further improved the sensitivity of FMDV detection with 125 times higher than that of CFT. Unlike CFT, specific detection of FMDV using the sandwich ELISA was reported to be unaffected by the presence of 12S antigen (93) and procomplementary or anticomplementary factors in the samples (88). Moreover, the sandwich ELISA allows direct assessment of samples without virus isolation, and it is generally more cost-effective than CFT because of the lower amount of sera required per test (88). In addition, ELISA is not affected by the variation in tissue culture susceptibility (88). Comparative studies via repeated testing of sera also indicated that ELISA was more reproducible than VNT, and their results could be generated within a day compared to VNT, which normally took more than 3 days (94).
Measurements of antibody titers using ELISA involve passive absorption of an antigen to a solid phase support, particularly the microtiter plate wells. Several studies have indicated that passive absorption of an antigen to a solid support either directly or indirectly via trapping antibodies may distort the conformation of the antigen. The conformation of FMDV antigen was previously reported to be altered with the exposure of internal viral proteins following a non-covalent binding to a PVC plate in an indirect ELISA (95–97). To resolve this problem, an LPB-ELISA was developed for the determination of antibody reactivity to the 146S antigen in its most native conformation (97). In this assay, the 146S antigen and test sera were mixed and incubated before being transferred to a plate precoated with serotype-specific anti-FMDV antisera (94). Attributed to the good reproducibility, faster results, and good correlation with VNT, LPB-ELISA quickly replaced VNT in FMD routine screening (98). Liquid-phase blocking ELISA was also reported to be the best strategy in differentiating the antigenic differences between FMDV strains (99). Nevertheless, the LPB-ELISA was shown to produce some degree of false-positive results and may require VNT for additional verification in some of the low-positive LPB-ELISA results (98). Conventionally, to measure the antibody titer by endpoint titration using LPB-ELISA requires the serum to be serially diluted, which is more laborious and more prone to error. To overcome this problem, a single-dilution LPB-ELISA was previously developed to measure the FMDV-specific antibody titer in serum. This method is based on a linear regression curve generated with reference standards to extrapolate the antibody titers of the sera tested (72, 73).
An SPC-ELISA was also developed for detecting FMDV (98). This method is based on the competition between the antibodies in the sera tested and the serotype-specific guinea pig anti-FMDV antibodies (98). Although both the LPB-ELISA and SPC-ELISA were shown to have similar sensitivity and limit of detection, the specificity of SPC-ELISA was reported to be higher, offering an improved FMDV-specific antibody detection method for mass screening (98). Paiba et al. (100) also demonstrated that SPC-ELISA was more sensitive than VNT for early serological detection of FMDV infection in cattle and sheep, although opposite findings were observed when tested in pigs. In addition, the sensitivity of SPC-ELISA is less affected by the strains of FMDV used in the assay, whereas VNT sensitivity could reduce significantly if heterologous virus is employed (100). Solid-phase competition ELISA was reported to be able to detect antibodies against six non-O serotypes of FMDV (A, C, SAT 1, SAT 2, SAT 3, and Asia 1) with specificity ranging from 99.4 to 99.9% and sensitivity comparable to LPB-ELISA and VNT (77). On the other hand, serotype specificity of the SPC-ELISA was evaluated against different reference sera representing six FMDV serotypes (O, A, Asia 1, SAT 1, SAT 2, and SAT 3). The SPC-ELISA detected all the reference sera correctly but not the FMDV serotype SAT 3-positive serum. Similarly, VNT also produced a borderline positive response on this sample, suggesting that the sample might be degraded. In addition, cross-reaction in SPC-ELISA between FMDV serotypes A and Asia 1-positive samples was observed (77). Solid-phase competition ELISA kits for detection of specific SPs of FMDV of different serotypes were commercially available. Nevertheless, one recent study reported that the sensitivity of the SPC-ELISA kit for specific detection of FMDV serotype O was lower, and it produced more false-positive results as compared to LPB-ELISA and VNT (78).
To improve the performance of ELISA in FMD diagnosis, many modifications have been made, primarily focusing on the development of new coating antigens and new monoclonal antibodies (mAbs) as trapping or detection antibodies. Majority of the ELISA-based assays involve inactivated FDMV antigens in the diagnostic process. However, the production of these inactivated antigens still requires handling of live virus in high-containment laboratories (101). Along with the advancement in recombinant DNA technology, coating antigens can be produced in a safer alternative. Recombinant SPs of FMDV serotypes O, Asia 1, and A were generated via the baculoviral expression system and used as diagnostic antigens in LPB-ELISA (61, 74). These recombinant LPB-ELISA assays exhibited specificity and sensitivity comparable to VNT (74). When this method was applied in the field during an FMDV serotype A outbreak in Korea in 2010, its specificity and sensitivity were reported to be 97 and 84%, respectively (75). The SPs VP1, VP2, VP3, and VP4 are the secondary cleavage products of a capsid precursor polyprotein (P1) of FMDV (102). Biswal et al. (79) produced a recombinant capsid polyprotein (rP1) and employed it as a diagnostic antigen in SPC-ELISA for detection of FMDV serotype O. Solid-phase competition ELISA based on rP1 demonstrated 100% specificity and 99% sensitivity. In addition, virus-like particles of FMDV serotype O (80) were also produced and used as a diagnostic antigen in SPC-ELISA. The specificity and sensitivity of this test were 100 and 96%, respectively (80). Interestingly, Wong et al. (52) genetically fused the capsid protein of T7 bacteriophage with the VP1 of FMDV and demonstrated that the recombinant protein, when served as the coating antigen in an indirect ELISA, could react with the vaccinated and positive infected bovine sera, suggesting its potential application in FMD diagnosis. Wong et al. (53) further delineated the VP1 sequence of FMDV to 12-amino-acid residues using amino acid sequence alignment, homology modeling, and phage display, in which the chimeric phage T7 displaying VP1159−170 epitope was demonstrated to have an improved sensitivity of 100% in a phage-based ELISA. Recently, a multiple-epitope protein (B4) comprising the G-H loops of VP1 from three topotypes of FMDV serotype O was developed as a potential vaccine candidate (54, 103). When the B4 was employed as a coating antigen in an indirect ELISA, it detected antibodies against FMDV serotype O in pigs with specificity and sensitivity up to 96.7 and 95.9%, respectively. These results were also reported to correlate well with the LPB-ELISA (55).
Serotyping and identification of FDMV based on sandwich ELISA normally use rabbit and guinea pig polyclonal antibodies as capture and detection antibodies, respectively. However, there are some disadvantages of using these polyclonal antibodies in ELISA, including batch-to-batch variation, inconsistent yield of antibodies, and limited serum samples collectable from individual animals (60). van Maanen (104) demonstrated the use of mAbs in ELISA for identification of three FMDV serotypes (A10, O1, C1). This mAb-based ELISA (mAb-ELISA) was shown to be sensitive, specific, and more reproducible than VNT. In the same year, Smitsaart et al. (105) developed a competition ELISA using an mAb that binds to the 12S protein subunit. This assay successfully detected six of the seven serotypes of FMDV with a sensitivity higher than that of CFT (105). More mAbs were later developed and utilized as trapping and/or detection antibodies in ELISA for FMDV detection (59, 62, 106–109). Veerasami et al. (60) also produced mAbs and chicken IgY specifically against the 146S antigen of three FMDV serotypes (O, Asia 1, and A) and used them in ELISA as capture and detection antibodies, respectively. There are several advantages in using chicken IgY in ELISA for FMD detection including minimal or no cross-reaction with mammalian IgG, complete absence of non-specific binding, and elimination of the need for cross-species immunoabsorptions due to the phylogenetic differences between birds and mammals (60). This method produced results comparable to the routine ELISA and RT-qPCR in FMDV serotyping (60). Another two mAbs that bind specifically to VP2 protein of FMDV serotype A were generated and employed as competing antibodies in SPC-ELISA. These mAbs interact with the VP2 protein, which is more conserved, thus offering a distinct advantage over another similar assay, which targets the more variable VP1 protein of FMDV serotype A (74, 81). This assay demonstrated specificity and sensitivity of 99.7 and 99.3%, respectively (81). In addition, two neutralizing mAbs, namely 72C1 and 65H6, which were raised against the FMDV O/JPN/2000 strain, were previously employed in LPB-ELISA as trapping and detection antibodies, respectively. This modified LPB-ELISA produced results that correlated well with VNT and demonstrated specificity of 100 and 99.7% in negative bovine and swine sera, respectively (71).
Apart from mAbs, recombinant antibody fragments such as the single-chain variable fragments (scFvs) were also used as detection antibodies in sandwich ELISA to detect FMDV-specific IgA in salivary samples from vaccinated and infected cattle (63). In addition, the variable heavy chain antibody fragments (VHHs) from camels have been explored for FMD diagnostic applications (76). The VHHs are composed of two heavy chains, but lack the light chains and CH1 domain present in conventional antibodies (110). Dash et al. (76) produced VHHs that bind specifically to 146S antigen of FMDV serotypes O, A, and Asia 1 and used them as trapping antibodies in LPB-ELISA. This modified LPB-ELISA yielded results that correlate well to routine LPB-ELISA, which uses coating antibodies from rabbits (76). The FMDV-specific VHHs could be produced with bacterial expression system, offering batch uniformity, and thus lower the production cost (111).
All field isolates of FMDV initiate infection using arginine-glycine-aspartic acid-binding integrins as the cell receptors (66). This knowledge was leveraged for the development of FMD diagnostic tools. A recombinant integrin αvβ6 was previously produced as an antigen-trapping reagent in a sandwich ELISA for FMDV diagnosis (64). When the serotype-specific polyclonal and mAbs were used as the detection antibody, the sensitivity of these methods was reported to be 98.1 and 97.9%, respectively. Nevertheless, the latter demonstrated superior serotypic specificity (96%) to that of the former (61.5%) (65). Later, Shimmon et al. (66) also generated a truncated bovine integrin αvβ6 as a universal trapping reagent in a sandwich ELISA for FMDV detection. Serotype specificity of sandwich ELISA assays based on the integrin αvβ6 (αvβ6-ELISA) was evaluated against FMDV-positive sera representing all seven serotypes. Depending on the serotype specificity of the mAb used for detection, little to no cross-reactivity was observed. Additionally, different sensitivities were observed when the αvβ6-ELISA was tested against different FMDV strains within the same serotypes (65, 66).
Exposure of animals to inactivated or live FMDV during vaccination or infection induces antibodies specific to the SPs. Therefore, a detection method targeting the SPs of FMDV alone cannot differentiate between the infected and vaccinated animals. Although the SPs and NSPs of FMDV are immunogenic, only the SPs serve as the main immunogen for the induction of protective responses (112, 113). Thus, the elimination of the NSPs from the inactivated FMDV vaccine could enable DIVA via differential detection of NSP-specific antibodies in animals infected with FMDV (114, 115). With some modifications, conventional ELISA methods have been adopted for the detection of NSPs of FMDV. Different NSPs of FMDV including 3ABC, 3AB, 3A, 3B, 3D, 2C, and 2B proteins have been employed in the establishment of NSP-based ELISAs.
Among the NSPs of FMDV, 3ABC polyprotein is reported to be the most antigenic and the most reliable marker for DIVA. Various formats of ELISA based on the 3ABC polyprotein were developed, including the LPB-ELISA, SPC-ELISA, and direct/indirect sandwich ELISA, all of which demonstrated good sensitivity, specificity, and capability for DIVA in various animals (56, 67, 68, 116–125). Enzyme-linked immunosorbent assays based on 3ABC have some added advantages over other NSPs including superior longevity of anti-3ABC antibody in infected animals compared to 2C, 3A, 3D, and Lb, and all infected cattle were shown to develop 3ABC-specific antibody at some points following the infection. Seroconversion to 3ABC in infected cattle was observed at 11 days postinfection, and the antibody remains detectable to the end of the experiments (301 days postinfection). Furthermore, repeated vaccination (fewer than five vaccinations) of the cattle with FMDV vaccine did not induce any antibody response against 3ABC polyprotein, in contrary to 3D protein (126). An agar gel immunodiffusion test was previously developed to detect 3D-specific antibodies in the sera of cattle, sheep, goats, and pigs for DIVA (127), but was found to have low sensitivity and specificity and was later replaced with LPB-ELISA (128). The conventional ELISA based on NSPs of FMDV uses partially purified antigens from infected cell cultures as diagnostic antigens, which require handling of live virus, posing risk of accidental virus escape from laboratories, and these partially purified antigens often lack batch-to-batch uniformity (129). Enzyme-linked immunosorbent assay based on the NSPs produced from either bacterial or baculoviral expression systems overcomes these concerns without compromising the sensitivity and specificity of the test (126, 130). Virus-like particles such as the tymovirus-like particles were also engineered to display 3B1, 3B2, 3AB, 3D, and 3ABD of FMDV and used as coating antigens in an indirect ELISA for DIVA (131). Variable heavy chain antibody fragments were also employed as competing antibodies for NSPs in SPC-ELISA and demonstrated high diagnostic specificity and sensitivity in detecting NSP-specific antibodies (82).
To further simplify and speed up the ELISA process for detecting FMDV-infected animals, the microchip-based ELISA was developed. This assay involved immobilization of the 6x-His tagged recombinant 3ABC polyprotein to microbeads with nickel (II) chelating chemistry, followed by immunoreaction with the 3ABC-specific antibodies in the test sera and detection with thermal lens microscopy based on the enzymatically colorimetric reaction between HRP-labeled antibody and the corresponding substrate. This method was demonstrated to be capable of detecting anti-3ABC antibodies in infected swine and cattle sera with good sensitivity and reproducibility. This assay is much faster (within 25 min) and requires lower serum volume (83, 84). Apart from the microchip-based ELISA, a chemiluminescence immunoassay (CLIA) was also developed for rapid identification of the anti-NSP antibodies. Chemiluminescence immunoassay was reported to simultaneously detect antibodies against 3ABC and 2C proteins of FMDV in experimentally infected pigs with sensitivity and specificity comparable to the commercial kits. This method produced results within 15 min, a remarkably short analysis time compared to other standard ELISA methods (132). Chemiluminescence immunoassay was later applied in the field for DIVA in bovines by simultaneously detecting 3A- and 3B-specific antibodies in the serum samples. In this field test, CLIA was reported to have concordance rate of 88.1% with the commercial PrioCHECK® FMDV NSP ELISA kit and produced no false-positive result in sera collected from bovine that had been vaccinated less than five times and low false-positive results in sera collected from bovine that had been vaccinated up to 10 (<2.2%) and 15 times (<6%) (133). Chemiluminescence immunoassay that enables simultaneous detection of two different antibodies against different NSPs of FMDV is advantageous over ELISA method, which detects only a single anti-NSP antibody. To ensure accurate diagnosis, retesting positive samples by detecting other antibodies against NSPs is a preferred measure (134).
Foot-and-mouth disease virus vaccines based on inactivated virus may contain a trace amount of FMDV NSPs, which could lead to the production of antibodies against the NSPs upon multiple vaccinations, which affect DIVA diagnosis (114, 130, 135). Negative marker vaccines that protect animals from FMDV infection while allowing DIVA were developed via removal of NSPs, which were used as markers for DIVA (57, 58, 136, 137). Alternatively, non-replicating FMDV virus-like particle was explored by Grubman (138) as a marker vaccine. While most negative marker vaccine developments involve deletion of NSPs, a few studies deleted part of the SPs, particularly the VP1 G-H loop, as the antibodies against G-H loop were demonstrated to be inefficient to provide a good protection (69, 70, 139, 140).
Fast detection and accurate identification of FMDV allow effective FMD surveillance and responses by imposing suitable controls and prevention strategies in case of an FMD outbreak. To date, typical assays for FMDV diagnosis such as virus isolation combined with antigen ELISA and RT-qPCR have been employed in FMDV reference laboratories (141). Despite the reliable and accurate diagnoses of FMDV, these diagnostic assays rely heavily on the availability of high-throughput equipment and highly trained personnel. Furthermore, the poor quality of the samples that resulted from the transport of materials from a field to a laboratory may obstruct or delay the early diagnosis of the disease. Thus, alternatives such as isothermal assays and dipsticks assays (also known as chromatographic strip tests) could serve as promising diagnostic methods in the field for a prompt FMD detection to allow timely implemented control measures. Reverse transcription-RPA (14, 40), RT-LAMP (23, 25, 29, 46, 47), and nucleic-acid sequence-based amplification (142) have been used to detect FMDV. A combination of dipsticks assays with RT-LAMP and RT-RPA has also been used for virus serotyping in field samples (14, 28, 46). Nevertheless, a drawback of the LAMP assay is that it involves the use of a few sets of intricate primers, while the RPA products require an electrophoresis setup and a fluorescent probe. Hence, a portable, rapid, and accurate detection method is still prominent for initial diagnosis of FMDV.
A chromatographic strip test such as LFI is a well-established fast paper-based analytical platform for detection and quantification of analytes. It is a simple and inexpensive point-of-care (POC) diagnosis without the need of elaborating sample preparations and sophisticated instruments (143). This has led to the increased applications of LFI assay in multiple field conditions where rapid screening is required. Table 3 summarizes LFI assays for FMDV diagnosis. A typical LFI strip normally consists of overlapping membranes that are mounted on a backing card. A liquid sample containing the analyte of interest moves through the cellulose membrane by a capillary force and is captured by the attached molecules that interact with the analyte along the membrane. In this context, a colored or fluorescent particle conjugated with an antibody that interacts specifically with the target analyte is used as the tracer for the development of signal (157). This LFI assay has been widely used for the diagnosis of infectious diseases (158–161) and detection of bioactive molecules (162, 163). Without the need of specific instruments, LFI strip test is a low-cost diagnostic method, which is easy to perform, giving straightforward results in a very short time. Lateral flow immunochromatographic strip tests have been used intensively for the detection of serotype-specific FMDV such as type-O (144, 149), -A (144, 145), -Asia 1 (144, 150, 164), and -SAT 2 (147). Likewise, LFI strips used for the detection of non-serotype-specific FMDV have also been reported (147, 151, 165, 166). However, one of the drawbacks for this non–serotype-specific LFI assay is the restricted usage of these strips in endemic countries, where rapid identification is essential for disease control (167, 168).
Most of the LFI strips detect FMDV SPs, but detection of specific antibodies against FMDV SPs (149) and NSPs (148, 152) has also been performed. Unlike strips that detect SPs, detections of antibodies against SPs are often performed to identify the vaccination status of animals, whereas detections of antibodies against NSPs are used to identify animals that have been infected by FMDV. Yang et al. (149) developed a lateral flow test strip using the recombinant VP1 protein for specific detection of antibodies against FMDV serotype O. Similar to ELISA, LFI test strips that are able to detect antibodies against SPs are unable to differentiate whether an animal that tested positive is vaccinated or infected. Therefore, test strips that detect antibodies against NSPs are required for the purpose of DIVA. Chen et al. (148) used recombinant 3ABC protein of FMDV serotype O for the detection of anti-NSPs antibodies in porcine. Although the NSPs of FMDV are highly conserved among all FMDV serotypes, only the samples of serotype O were tested. Later on, Wu et al. (152) developed an LFI test strip based on the recombinant 2C′3AB protein of FMDV serotype O, in which 3C was removed because of its low immunogenicity and replaced by part of 2C protein, which was fused to the N-terminus of 3AB. Despite the high sensitivity and specificity of the test, the serotypes of positive and vaccinated serum samples tested were not reported. The LFI strip technology has also been proposed for use in DIVA, but its practical usage in DIVA has yet been reported.
While most of the LFI strips utilize rabbit and guinea pig polyclonal sera, respectively, as the capture and detection antibodies, the usage of mAb as the capture and detection antibodies for FMDV detection in the LFI strip tests has also been developed to improve the efficiency of diagnosis (146, 165, 166, 169). For this purpose, strips are specifically designed for each antigen in order to increase the accuracy, sensitivity, and consistency of the assay. Reid et al. (165) reported that an equivalent sensitivity (100%) to the conventional antigen ELISA was observed in both the clinical samples from animals infected experimentally and in cell culture supernatant using the Clearview™ chromatographic strip test technology with mAb isotype IgG1, designated as Cla. The mAb Cla displayed high reactivity against FMDV serotypes O, A, C, and Asia 1 and no cross-reactivity with SVDV. Utilization of the mAb approach, in which specific mAbs were used as the capture antibody, and serotype-independent mAbs were employed as the detection antibody, produced a new generation of the generic Rapid Assay Device (gRAD) for the detection of FMDV serotypes O, A, and Asia 1 (153). The gRAD, which is currently commercially available, has been shown to achieve a sensitivity similar to that of the double antibody sandwich ELISA for viral antigen detection with a detection limit of 2.55 to 6.3 log10 TCID50/mL of 10% tissue suspension from epithelial lesions in a process that took only 10 min (153). Another commercially available LFI strip known as the Svanodip FMDV-Ag LFD by Boehringer Ingelheim Svanova (Sweden) can also be used to detect all the seven serotypes of FMDV antigens based on IF10 mAbs.
As serotype-specific LFI strips can only detect one FMDV serotype at a time (170), thus development of a multiplex platform for simultaneous detection of multiple FMDV serotypes will undoubtedly enhance the usage of the LFI strips in the field. A multiplex-LFI strip test for detecting Hantavirus in humans was developed by Amada et al. (171). The first study describing the development of a multiplex-LFI strip test for detecting FMDV serotypes O, A, and Asia 1 was reported by Yang et al. (151). Following this report, Morioka et al. (154) successfully developed another multiplex FMDV LFI strip based on mAbs that can detect all the seven serotypes and concurrently distinguish serotypes O, A, C, and Asia 1. The developed multiplex-LFI strip had a sensitivity ranging from 103 to 104 of a 50% tissue culture infectious dose (TCID50) of each FMDV strain, comparable to the commercial product, Svanodip FMDV-Ag LFD, which can detect all the seven serotypes of FMDV, but is not able to serotype them.
Recently, a combination of LFI assay and other technologies, such as PCR (172), RT-LAMP (173), RT-RPA (155, 174–176), and quantum dots (177), for the diagnosis of animal pathogens has also been explored. Therefore, the current approach in the development of a desirable FMD diagnostic test typically involves the incorporation of two assays such as RT-LAMP-LFD (46) and RT-RPA-LFD (155, 156). Waters et al. (46) modified an existing FMDV RT-LAMP assay to allow detection of LAMP products with LFD by labeling the FIP/BIP at the 5′ terminus with fluorescein (Flc) and biotin (Btn). This RT-LAMP-LFD assay produced concordant results as compared to those obtained using RT-qPCR with a positive detection of FMDV RNA when the FMDV spiked 10% epithelium suspensions diluted to a range of 10−5. The RT-LAMP-LFD assay also showed 104 times more sensitive in detecting FMDV than most of the FMD-specific antigen lateral flow devices. Hence, this assay not only resolved the problem of relatively low analytical sensitivity encountered by most LFD used in the field, but it also detected FMDV RNA in the raw epithelial suspension (in the absence of RNA extraction) by only diluting the samples with nuclease-free water and incubating the mixture using a water bath set at 60°C for RT-LAMP amplification. With its ideal characteristics, this LFD assay serves as a “proof of concept” for the future use of LAMP in the development of a pen-side assay for FMDV. However, the difficulty in designing the four to six primers needed in RT-LAMP, especially in a virus like FMDV that exhibits a high mutation rate during its replication, hinders the usage of RT-LAMP-LFD assay. In addition, the incubation for RT-LAMP for 45 to 60 min is disadvantageous compared to an RT-RPA approach with a run time of only 4 to 10 min (14). As described earlier, as an isothermal DNA amplification method, RPA has been widely used in the detection of different pathogens. Wang et al. (155) established a combination method of RT-RPA and lateral flow dipstick (RT-RPA-LFD) for detecting and serotyping of FMDV in the field. They constructed a recombinant vector, pcDNA3.1-2B, containing the 2B gene of FMDV, and amplified it with RT-RPA using specific primers and a probe within 20 min. The newly established FMDV RT-RPA-LFD assay has a higher sensitivity, up to 10 copies as compared with the previous FMDV RT-RPA assays (14, 40) with sensitivity limited to 100 RNA copies. Furthermore, this RT-RPA-LFD assay only requires a thermos metal bath at 38°C unlike other previous RT-RPA assays, which need a sophisticated instrumentation, and the RPA amplicons can be detected by LFD within 5 min. The RT-RPA-LFD is a promising POC diagnostic test for FMDV as it reacts with the FMDV reference strains, including serotypes O, A, and Asia 1, and with no cross-reactivity with other viral pathogens from cattle, which had similar vesicular lesions and clinical symptoms. At the same time, another FMDV RPA-LFD assay that targets the VP1 gene was also developed by the same research group (178). VP1 protein has been widely used to determine the genetic relationships between different strains of FMDV because of its high genetic heterogeneity (179). Therefore, primers and probes specific for serotypes O, A, and Asia 1 of FMDV were designed based on the alignment of the VP1 nucleotide sequences. The detection limits of these assays were three copies of plasmid DNA or 50 copies of viral RNA with 98.41% concordance between the RT-RPA-LFD and RT-qPCR assays. The development of this serotype-specific RT-RPA-LFD assay provides a rapid, sensitive, and specific method for differentiation of FMDV serotypes A, O, or Asia 1. On the other hand, an equipment-free FMDV RPA-LFD specifically designed for the 3D gene was also developed by Liu et al. (156). They performed the assay by incubating the reaction tubes in a closed fist using body heat for 15 min. The developed RPA-LFD was capable to detect FMDV serotypes O, A, and Asia 1 using 10 ng viral RNA and DNA as templates with no cross-detections observed. The analytical sensitivity was equivalent to RT-qPCR with 100 copies of in vitro–transcribed FMDV RNA per reaction. One of the benefits in their work is the instant utilization of FMDV RNA as the template in the RPA-LFD without the need to reverse-transcribe the viral RNA into cDNA as required in other RPA assays. This rapid, visible and equipment-free method makes FMDV RPA-LF assay ideal for reliable detection of FMDV in an underequipped laboratory and at point of need, especially in low-resource settings.
Over the past decades, livestock industry has developed remarkably, contributing 40% of the global value of agricultural output and, sustaining the food security of almost 1.3 billion people (180, 181). However, outbreaks of animal diseases remain a major concern that threatens the livestock industry. Foot-and-mouth disease, as one of the most significant animal diseases, poses a severe constraint on the reduction of poverty in countries where this disease is endemic and more prone to food insecurity. Contingency plans for an FMD emergency enable rapid detection of the virus before it progresses to an epidemic outbreak (182). Current laboratory approaches for FMD diagnosis are generally based on assays that exploit the clinical windows of infected animals. The diagnostic window is typically 2 to 14 days with an early observation of clinical signs from vesicular lesions. Rapid confirmation includes assays that aim to detect FMDV in vesicular epithelium and vesicular fluid from clinical lesions, as well as in the blood and mucosal swabs from the active surveillance of infected animals in preclinical cases. Furthermore, FMDV-specific antibody responses can also be detected by serological assays in animals exposed to and recovered from FMDV.
Foot-and-mouth disease diagnosis is performed at two levels: (i) in the field/local and (ii) in the central laboratory. If there is a suspected case of FMD in the field, a quick diagnosis is performed by the FMD diagnostic specialist in order to implement immediate control or biosecurity measures. Clinical examinations and collection of suspected animal's history are performed for epidemiological and disease prevalence investigations. In addition, a range of specimens that might be included in the differential diagnosis is collected and transported back to the regional or central laboratory for further examination. These specimens consist of (i) oral swabs from ruptured lesions; (ii) nasal swabs from lesion less than a week old, where vesicular material is not available; (iii) vesicular fluid from unruptured vesicles; (iv) epithelium from ruptured tissues, placed in a neutral buffer phosphate saline with 50% glycerol; and (v) blood specimens from suspected cases. Although proband samples are not recommended for the first-line diagnostic tests, the oropharyngeal fluid is collected if no fresh lesions are detected. All samples in the ideally leakproof transport containers are labeled and stored in an insulated cool box with a submission form with case history sealed in an external disinfectant see-through bag with photographs of infected animals. The assessment of the situation on the field, and steps taken to secure a confirmatory diagnosis must be immediately reported to the state or regional and central veterinary officers for further advices regarding the disease control strategies.
The subsequent diagnosis of FMD generally depends on the laboratory testing, which includes live virus isolation from tissue culture coupled with the identification of the viral antigen by ELISA or detection of the viral nucleic acid by RT-PCR. Detection of elevated FMD-specific antibodies by ELISA or VNT may also aid in indicating a recovery from the virus infection. These diagnostic assays were performed at a regional or central laboratory to prescribe appropriate control measures based on the confirmation of a definitive diagnosis (168). Hence, these tests should be highly sensitive and specific to provide a differential diagnosis. In the central laboratory, virus and its viral components can be detected with various diagnostic assays. These assays include VI, Ag-ELISA, multiplex RT-PCR, RT-qPCR, and nucleotide sequencing. In addition, antiviral antibodies against SPs can also be detected using VNT, LPB sandwich ELISA, and SPCE-ELISA, whereas antibodies against NSPs can be detected using 3ABC-ELISA. The detailed diagnoses performed at the central laboratory enable the confirmation of disease, serotyping of virus, molecular epidemiology, and phylogenetic analysis and lastly determined the most relevant vaccine matching strains to control an outbreak (183). The performance of all these assays varies in terms of sensitivity, specificity, and time required. The speed of a definitive diagnosis would vary depending on the distance of the samples being transported from the field to an appropriate laboratory. Thus, a network of international reference laboratories and collaborating centers is essential for handling of specimens in the event of a large outbreak, for the purpose of both surveillance and rapid diagnosis. Scalability and cost of each assay must also be taken into consideration especially in FMD endemic and underdeveloped countries. The establishment of centralized facilities for testing, together with the implementation of quality control systems, have improved significantly the assays for routine diagnostic purposes.
More recently, portable tests or POC diagnostics, such as LFD, mobile PCR, and isothermal assays, have been developed to increase the applicability of these assays in multiple field conditions where rapid screening is of paramount importance. Even though LFD can be operated by “non-specialist,” the usage of this portable test may be restricted by its low-throughput assay performance. A commercially available LFI strip known as the Svanodip FMDV-Ag LFD by Boehringer Ingelheim Svanova (Sweden) was reported to show similar assay performance to laboratory-based Ag-ELISA when it was applied on the field during the 2007 UK outbreaks (166). The deployment of LFD on field remains advantageous for FMD endemic countries as compared with portable RT-qPCR in terms of production cost (25, 184). Although these simple-to-use POC tests offer a rapid result that can support the local decisions, they are also limited by the cost-benefit analysis. In conclusion, the deployment of these portable tests on the field will be taken into consideration after their characteristics have been thoroughly evaluated in terms of test performance, speed, cost, simplicity, and robustness.
The control of FMD varies among countries, depending on the FMD status. As the FMD control in FMD-free countries emphasizes on reducing the risk and impact of the virus incursions from both neighboring and trade-partner countries, the control policies in FMD-free countries have been based on depopulation of infected and in-contact animals, together with restrictions on movement of animals and their products. Early detection followed by surveillance is crucial. In order to regain the international trading rights, FMD-free countries are required to identify the remaining sources of infection and to demonstrate that they are free of the disease. On the other hand, FMD control in endemic countries is implemented by diagnoses, surveillance, and regular mass vaccinations. Most importantly, there is a continuous need for an up-scaling of improved quality vaccines with longer-lasting protection at a lower cost (185). In this context, serological assays including ELISA for detection of antibodies against FMDV SPs and NSPs are used. The former is useful to measure the vaccine efficacy, and the latter is generally used to establish prevalence and to monitor virus circulation as it can detect the presence of the infection regardless of the vaccination status of the animals (186, 187). The SP tests are serotype specific. Therefore, virus and antigen closely related to the field strain are selected to be used in ELISA for optimal sensitivity. To date, the commercially available PrioCHECK® FMDV type-specific products by Prionics can only detect anti-SP antibodies of 3 FMDV serotypes: O, A, and Asia. Hence, determination of the serotype involved in field outbreaks is important for a proper control of the disease. On the other hand, the use of NSP tests in FMD endemic countries is complicated by the fact that the vaccinated animals may seroconvert after repeated vaccinations. Anti-NSP antibody responses may also be delayed in cases of subclinical or mild clinical infections following routine vaccinations. Moreover, anti-NSP antibodies can persist for a long period and may not indicate a recent FMDV infection (134, 188).
The breakthrough of molecular diagnostics along with the development of pen-side devices has allowed the determination of the FMDV serotypes. For endemic countries, the use of LFD is more favorable. A routine screening with an LFD device has been viewed as a rapid and economical tool to determine incidences of the infection in countries where the emergence rate of FMDV is high, under limited-resources veterinary settings. Therefore, rapid action is needed to minimize the virus spread. As mentioned earlier, serotype-specific LFI test strips can be used for rapid detection and identification of various FMDV serotypes (144, 145, 147, 149, 150, 164). By identifying these FMDV serotypes, appropriate commercial FMD vaccine can be applied to the animal population to minimize the loss of productivity in majority of the smallholder and commercial farmer settings in endemic countries. As for FMD-free countries, confirmatory tests such as ELISA and RT-qPCR are more desirable. Due to the occurrence of FMDV is relatively much lower in these countries, confirmation of the disease is more important than rapid identification of the virus to avoid unnecessary culling of suspected animals. Setup of RT-qPCR in the regional laboratories in these more developed countries, which are typically FMD-free, can increase the diagnostic capacity and subsequently reduce the sample shipping times during a sudden outbreak. The FMD outbreak confirmation along with the virus typing and characterizations enables the study of the virus lineage and routes of transmission, which will provide substantial information for epidemiology study in the effort to control the spread of FMD.
The deployment of diagnostic tools to rapidly identify and confirm initial clinical symptoms of an infection is prerequisite in any epidemic disease control strategy, particularly when it comes to the prevalence of the FMDV in a livestock population. As the FMDV infection is clinically indistinguishable from infections resulting from other similar vesicular disease viruses, early diagnosis is critical for efficient disease control. Various diagnostic methods ranging from conventional such as virus isolation and competitive- and blocking-antigen ELISA to molecular-based methods such as RT-PCR and RT-LAMP have been developed over the years. Although ELISA-based methods have good diagnostic sensitivity and specificity, molecular detection methods have the advantage of higher analytical sensitivity for the detection of minimal viral RNA. Despite these accurate and reliable FMDV assays, researchers have been developing alternatives methods that allow for pen-side testing in an attempt to overcome some of the practical challenges such as tedious procedures and the availability of an equipped laboratory setting with trained field personnel. Development of lateral flow devices and integration of the portable RT-PCR, RT-LAMP, and RT-RPA with LF technologies have been made to increase the sensitivity of FMDV detection. Nevertheless, translations of these assays from laboratories to practical applications in the field remain limited, and various technical and cost issues need to be addressed to develop a more flexible and affordable diagnostic tools that can be widely used for FMDV detection.
CW, CY, and HO wrote the manuscript. CW, CY, HO, KH, and WT approved its final version. All authors contributed to the article and approved the submitted version.
This study was supported by the Ministry of Agriculture and Agro-based Industry, Malaysia (Grant no: 05-01-04-SF1149) and UPM Putra Grant (grant no: GP-IPS/2017/9539500) of Universiti Putra Malaysia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
CW was financially supported by MyPhD under the MyBrain 15 programme from the Ministry of Higher Education of Malaysia.
1. Kitching RP, Alexandersen S. Clinical variation in foot and mouth disease: pigs. Rev Sci Tech. (2003) 21:513–8. doi: 10.20506/rst.21.3.1367
2. OIE. Foot & Mouth Disease (FMD) [Online] (2018). Available online at: http://www.oie.int/en/animal-health-in-the-world/animal-diseases/Foot-and-mouth-disease/ (accessed June 13, 2020).
3. Mowat GN, Darbyshire JH, Huntley JF. Differentiation of a vesicular disease of pigs in Hong Kong from foot-and-mouth disease. Vet Rec. (1972) 90:618–21. doi: 10.1136/vr.90.22.618
4. Fernández J, Agüero M, Romero L, Sánchez C, Belák S, Arias M, et al. Rapid and differential diagnosis of foot-and-mouth disease, swine vesicular disease, and vesicular stomatitis by a new multiplex RT-PCR assay. J Virol Methods. (2008) 147:301–11. doi: 10.1016/j.jviromet.2007.09.010
5. Smith PF, Howerth EW, Carter D, Gray EW, Noblet R, Berghaus RD, et al. Host predilection and transmissibility of vesicular stomatitis New Jersey virus strains in domestic cattle (Bos taurus) and swine (Sus scrofa). BMC Vet Res. (2012) 8:183. doi: 10.1186/1746-6148-8-183
6. Vangrysperre W, De Clercq K. Rapid and sensitive polymerase chain reaction based detection and typing of foot-and-mouth disease virus in clinical samples and cell culture isolates, combined with a simultaneous differentiation with other genomically and/or symptomatically related viruses. Arch Virol. (1996)141:331–44.
7. Baxi MK, Baxi S, Clavijo A, Burton KM, Deregt D. Microarray-based detection and typing of foot-and-mouth disease virus. Vet J. (2006) 172:473–81. doi: 10.1016/j.tvjl.2005.07.007
8. Banér J, Gyarmati P, Yacoub A, Hakhverdyan M, Stenberg J, Ericsson O, et al. Microarray-based molecular detection of foot-and-mouth disease, vesicular stomatitis and swine vesicular disease viruses, using padlock probes. J Virol Methods. (2007) 143:200–6. doi: 10.1016/j.jviromet.2007.03.004
9. Lung O, Fisher M, Beeston A, Hughes KB, Clavijo A, Goolia M, et al. Multiplex RT-PCR detection and microarray typing of vesicular disease viruses. J Virol Methods. (2011) 175:236–45. doi: 10.1016/j.jviromet.2011.05.023
10. Dill V, Beer M, Hoffmann B. Simple, quick and cost-efficient: a universal RT-PCR and sequencing strategy for genomic characterisation of foot-and-mouth disease viruses. J Virol Methods. (2017) 246:58–64. doi: 10.1016/j.jviromet.2017.04.007
11. Nishi T, Kanno T, Shimada N, Morioka K, Yamakawa M, Fukai K. Reverse transcription-PCR using a primer set targeting the 3D region detects foot-and-mouth disease virus with high sensitivity. Transbound Emerg Dis. (2019) 66:1776–83. doi: 10.1111/tbed.13202
12. Ambagala A, Fisher M, Goolia M, Nfon C, Furukawa-Stoffer T, Ortega Polo R, et al. Field-deployable reverse transcription-insulated isothermal PCR (RT-iiPCR) assay for rapid and sensitive detection of foot-and-mouth disease virus. Transbound Emerg Dis. (2017) 64:1610–23. doi: 10.1111/tbed.12554
13. Pinheiro-de-Oliveira TF, Fonseca AA Jr, Camargos MF, Laguardia-Nascimento M, de Oliveira AM, Cottorello ACP, et al. Development of a droplet digital RT-PCR for the quantification of foot-and-mouth virus RNA. J Virol Methods. (2018) 259:129–34. doi: 10.1016/j.jviromet.2018.06.015
14. Abd El Wahed A, El-Deeb A, El-Tholoth M, Abd El Kader H, Ahmed A, Hassan S, et al. A portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PLoS ONE. (2013) 8:e71642. doi: 10.1371/journal.pone.0071642
15. Reid SM, Mioulet V, Knowles NJ, Shirazi N, Belsham GJ, King DP. Development of tailored real-time RT-PCR assays for the detection and differentiation of serotype O, A and Asia-1 foot-and-mouth disease virus lineages circulating in the Middle East. J Virol Methods. (2014) 207:146–53. doi: 10.1016/j.jviromet.2014.07.002
16. Vandenbussche F, Lefebvre DJ, De Leeuw I, Van Borm S, De Clercq K. Laboratory validation of two real-time RT-PCR methods with 5'-tailed primers for an enhanced detection of foot-and-mouth disease virus. J Virol Methods. (2017) 246:90–4. doi: 10.1016/j.jviromet.2017.04.014
17. Rios L, Perera CL, Coronado L, Relova D, Alvarez AM, Ganges L, et al. Multi-target strategy for pan/foot-and-mouth disease virus (FMDV) detection: a combination of sequences analysis, in silico predictions and laboratory diagnostic evaluation. Front Vet Sci. (2018) 5:160. doi: 10.3389/fvets.2018.00160
18. Goller KV, Dill V, Madi M, Martin P, Van der Stede Y, Vandenberge V, et al. Rapid and simple detection of foot-and-mouth disease virus: evaluation of a cartridge-based molecular detection system for use in basic laboratories. Transbound Emerg Dis. (2018) 65:578–84. doi: 10.1111/tbed.12744
19. Hole K, Nfon C. Foot-and-mouth disease virus detection on a handheld real-time polymerase chain reaction platform. Transbound Emerg Dis. (2019) 66:1789–95. doi: 10.1111/tbed.13227
20. Armson B, Mioulet V, Doel C, Madi M, Parida S, Lemire KA, et al. Detection of foot-and-mouth disease virus in milk samples by real-time reverse transcription polymerase chain reaction: optimisation and evaluation of a high-throughput screening method with potential for disease surveillance. Vet Microbiol. (2018) 223:189–94. doi: 10.1016/j.vetmic.2018.07.024
21. Armson B, Wadsworth J, Kibona T, Mshanga D, Fowler VL. Opportunities for enhanced surveillance of foot-and-mouth disease in endemic settings using milk samples. Transbound Emerg Dis. (2019) 66:1405–10. doi: 10.1111/tbed.13146
22. Erickson A, Fisher M, Furukawa-Stoffer T, Ambagala A, Hodko D, Pasick J, et al. A multiplex reverse transcription PCR and automated electronic microarray assay for detection and differentiation of seven viruses affecting swine. Transbound Emerg Dis. (2018) 65:e272–83. doi: 10.1111/tbed.12749
23. Chen HT, Zhang J, Liu YS, Liu XT. Rapid typing of foot-and-mouth disease serotype Asia 1 by reverse transcription loop-mediated isothermal amplification. Virol J. (2011) 8:489. doi: 10.1186/1743-422x-8-489
24. Ding Y-Z, Zhou J-H, Ma L-N, Qi Y-N, Wei G, Zhang J, et al. A reverse transcription loop-mediated isothermal amplification assay to rapidly diagnose foot-and-mouth disease virus C. J Vet Sci. (2014) 15:423–6. doi: 10.4142/jvs.2014.15.3.423
25. Yamazaki W, Mioulet V, Murray L, Madi M, Haga T, Misawa N, et al. Development and evaluation of multiplex RT-LAMP assays for rapid and sensitive detection of foot-and-mouth disease virus. J Virol Methods. (2013) 192:18–24. doi: 10.1016/j.jviromet.2013.03.018
26. Lim DR, Kim HR, Park MJ, Chae HG, Ku BK, Nah JJ, et al. An improved reverse transcription loop-mediated isothermal amplification assay for sensitive and specific detection of serotype O foot-and-mouth disease virus. J Virol Methods. (2018) 260:6–13. doi: 10.1016/j.jviromet.2018.06.017
27. Ranjan R, Kangayan M, Subramaniam S, Mohapatra JK, Biswal JK, Sharma GK, et al. Development and evaluation of a one step reverse transcription-loop mediated isothermal amplification assay (RT-LAMP) for rapid detection of foot and mouth disease virus in India. Virus Dis.(2014) 25:358–64. doi: 10.1007/s13337-014-0211-2
28. Howson ELA, Armson B, Madi M, Kasanga CJ, Kandusi S, Sallu R, et al. Evaluation of two lyophilized molecular assays to rapidly detect foot-and-mouth disease virus directly from clinical samples in field settings. Transbound Emerg Dis. (2017) 64:861–71. doi: 10.1111/tbed.12451
29. Farooq U, Latif A, Irshad H, Ullah A, Zahu AB, Naeem K, et al. Loop-mediated isothermal amplification (RT-LAMP): a new approach for the detection of foot-and-mouth disease virus and its sero-types in Pakistan. Iran J Vet Res. (2015) 16:331–4.
30. Guan H, Li Z, Yin X, Zhang Y, Gao P, Bai Y, et al. Rapid detection and differentiation of foot and mouth disease virus serotypes by antigen-capture reverse transcriptase loop-mediated isothermal amplification. Asian J Anim Vet Adv. (2013) 8:647–54. doi: 10.3923/ajava.2013.647.654
31. Madhanmohan M, Nagendrakumar SB, Manikumar K, Yuvaraj S, Parida S, Srinivasan VA. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid serotyping of foot-and-mouth disease virus. J Virol Methods. (2013) 187:195–202. doi: 10.1016/j.jviromet.2012.08.015
32. Meyer RF, Brown CC, House C, House JA, Molitor TW. Rapid and sensitive detection of foot-and-mouth disease virus in tissues by enzymatic RNA amplification of the polymerase gene. J Virol Methods. (1991) 34:161–72. doi: 10.1016/0166-0934(91)90096-I
33. Höfner MC, Carpenter WC, Donaldson AI. Detection of foot-and-mouth disease virus RNA in clinical samples and cell culture isolates by amplification of the capsid coding region. J Virol Methods. (1993) 42:53–61. doi: 10.1016/0166-0934(93)90176-R
34. Laor O, Torgersen H, Yadin H, Becker Y. Detection of FMDV RNA amplified by the polymerase chain reaction (PCR). J Virol Methods. (1992) 36:197–207. doi: 10.1016/0166-0934(92)90051-E
35. Smith CJ, Osborn AM. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol. (2009) 67:6–20. doi: 10.1111/j.1574-6941.2008.00629.x
36. Moonen P, Boonstra J, van der Honing RH, Leendertse CB, Jacobs L, Dekker A. Validation of a LightCycler-based reverse transcription polymerase chain reaction for the detection of foot-and-mouth disease virus. J Virol Methods. (2003) 113:35–41. doi: 10.1016/s0166-0934(03)00220-9
37. Callahan JD, Brown F, Osorio FA, Sur JH, Kramer E, Long GW, et al. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J Am Vet Med Assoc. (2002) 220:1636–42. doi: 10.2460/javma.2002.220.1636
38. Reid SM, Ferris NP, Hutchings GH, Zhang Z, Belsham GJ, Alexandersen S. Detection of all seven serotypes of foot-and-mouth disease virus by real-time, fluorogenic reverse transcription polymerase chain reaction assay. J Virol Methods. (2002) 105:67–80. doi: 10.1016/S0166-0934(02)00081-2
39. El-Shehawy L, Abu-Elnaga H, Abdel AM, Fawzy H, Al-Watany H, Azab A. Laboratory diagnosis of FMD using real-time RT-PCR in Egypt. Life Sci J. (2011) 8:384–7.
40. Howson ELA, Kurosaki Y, Yasuda J, Takahashi M, Goto H, Gray AR, et al. Defining the relative performance of isothermal assays that can be used for rapid and sensitive detection of foot-and-mouth disease virus. J Virol Methods. (2017) 249:102–10. doi: 10.1016/j.jviromet.2017.08.013
41. Fontél KS, Botner A, Belsham GJ, Lohse L. Diagnostic comparison of serum and EDTA-stabilized blood samples for the detection of foot-and-mouth disease virus RNA by RT-qPCR. J Virol Methods. (2019) 270:120–5. doi: 10.1016/j.jviromet.2019.05.003
42. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. (2000) 28:E63. doi: 10.1093/nar/28.12.e63
43. Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. (2001) 289:150–4. doi: 10.1006/bbrc.2001.5921
44. Dukes JP, King DP, Alexandersen S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Arch Virol. (2006) 151:1093–106. doi: 10.1007/s00705-005-0708-5
45. Li J, Chen Q, Xiong W, Fang XE. [Establishment of RT- LAMP for rapid detection of foot-and-mouth disease virus]. Bing Du Xue Bao. (2009) 25:137–42.
46. Waters RA, Fowler VL, Armson B, Nelson N, Gloster J, Paton DJ, et al. Preliminary validation of direct detection of foot-and-mouth disease virus within clinical samples using reverse transcription loop-mediated isothermal amplification coupled with a simple lateral flow device for detection. PLoS ONE. (2014) 9:e105630. doi: 10.1371/journal.pone.0105630
47. Maryam S, Rashid T, Latif A, Zahra R, Bin Zahur A, Ahsan A, et al. One-step real-time loop-mediated isothermal amplification (RT-LAMP): evaluation and its application for the detection of foot-and-mouth-disease virus and its serotypes. Turk J Vet. Anim. Sci. (2017) 41:435–43. doi: 10.3906/vet-1611-10
48. Gadkar VJ, Goldfarb DM, Gantt S, Tilley PAG. Real-time detection and monitoring of loop mediated amplification (LAMP) reaction using self-quenching and de-quenching fluorogenic probes. Sci Rep. (2018) 8:5548. doi: 10.1038/s41598-018-23930-1
49. Kasanga CJ, Yamazaki W, Mioulet V, King DP, Mulumba M, Ranga E, et al. Rapid, sensitive and effective diagnostic tools for foot-and-mouth disease virus in Africa. Onderstepoort J Vet Res. (2014) 81:E1–5. doi: 10.4102/ojvr.v81i2.727
50. Pega J, Bucafusco D, Di Giacomo S, Schammas JM, Malacari D, Capozzo AV, et al. Early adaptive immune responses in the respiratory tract of foot-and-mouth disease virus-infected cattle. J Virol. (2013) 87:2489–95. doi: 10.1128/jvi.02879-12
51. Senthilkumaran C, Yang M, Bittner H, Ambagala A, Lung O, Zimmerman J, et al. Detection of genome, antigen, and antibodies in oral fluids from pigs infected with foot-and-mouth disease virus. Can J Vet Res. (2017) 81:82–90.
52. Wong CL, Sieo CC, Tan WS. Display of the VP1 epitope of foot-and-mouth disease virus on bacteriophage T7 and its application in diagnosis. J Virol Methods. (2013) 193:611–9. doi: 10.1016/j.jviromet.2013.07.053
53. Wong CL, Yong CY, Muhamad A, Syahir A, Omar AR, Sieo CC, et al. A 12-residue epitope displayed on phage T7 reacts strongly with antibodies against foot-and-mouth disease virus. Appl Microbiol Biotechnol. (2018) 102:4131–42. doi: 10.1007/s00253-018-8921-9
54. Cao Y, Lu Z, Li Y, Sun P, Li D, Li P, et al. Poly(I:C) combined with multi-epitope protein vaccine completely protects against virulent foot-and-mouth disease virus challenge in pigs. Antiviral Res. (2013) 97:145–53. doi: 10.1016/j.antiviral.2012.11.009
55. Cao Y, Zhou W, Xing X, Zhang J, Fu Y, Li K, et al. Indirect ELISA using a multi-epitope recombinant protein to detect antibodies against foot-and-mouth disease virus serotype O in pigs. J Virol Methods. (2018) 262:26–31. doi: 10.1016/j.jviromet.2018.09.008
56. Hosamani M, Basagoudanavar SH, Tamil Selvan RP, Das V, Ngangom P, Sreenivasa BP, et al. A multi-species indirect ELISA for detection of non-structural protein 3ABC specific antibodies to foot-and-mouth disease virus. Arch Virol. (2015) 160:937–44. doi: 10.1007/s00705-015-2339-9
57. Biswal JK, Subramaniam S, Ranjan R, Sharma GK, Misri J, Pattnaik B. Marker vaccine potential of foot-and-mouth disease virus with large deletion in the non-structural proteins 3A and 3B. Biologicals. (2015) 43:504–11. doi: 10.1016/j.biologicals.2015.07.004
58. Bhatt M, Mohapatra JK, Pandey LK, Mohanty NN, Das B, Prusty BR, et al. Mutational analysis of foot and mouth disease virus nonstructural polyprotein 3AB-coding region to design a negative marker virus. Virus Res. (2018) 243:36–43. doi: 10.1016/j.virusres.2017.10.010
59. Muller JD, McEachern JA, Bossart KN, Hansson E, Yu M, Clavijo A, et al. Serotype-independent detection of foot-and-mouth disease virus. J Virol Methods. (2008) 151:146–53. doi: 10.1016/j.jviromet.2008.03.011
60. Veerasami M, Singanallur NB, Thirumeni N, Rana SK, Shanmugham R, Ponsekaran S, et al. Serotyping of foot-and-mouth disease virus by antigen capture-ELISA using monoclonal antibodies and chicken IgY. New Microbiol. (2008) 31:549–54.
61. Ko YJ, Jeoung HY, Lee HS, Chang BS, Hong SM, Heo EJ, et al. A recombinant protein-based ELISA for detecting antibodies to foot-and-mouth disease virus serotype Asia 1. J Virol Methods. (2009) 159:112–8. doi: 10.1016/j.jviromet.2009.03.011
62. Chen H-T, Peng Y-H, Zhang Y-G, Liu X-T. Detection of foot-and-mouth disease serotype O by ELISA using a monoclonal antibody. Monoclon Antib Immunodiagn Immunother. (2013) 32:47–9. doi: 10.1089/mab.2012.0081
63. Sridevi NV, Shukra AM, Neelakantam B, Anilkumar J, Madhanmohan M, Rajan S, et al. Development of anti-bovine IgA single chain variable fragment and its application in diagnosis of foot-and-mouth disease. Eur J Microbiol Immunol. (2014) 4:34–44. doi: 10.1556/EuJMI.4.2014.1.3
64. Ferris NP, Abrescia NG, Stuart DI, Jackson T, Burman A, King DP, et al. Utility of recombinant integrin alpha v beta6 as a capture reagent in immunoassays for the diagnosis of foot-and-mouth disease. J Virol Methods. (2005) 127:69–79. doi: 10.1016/j.jviromet.2005.02.014
65. Ferris NP, Grazioli S, Hutchings GH, Brocchi E. Validation of a recombinant integrin alphavbeta6/monoclonal antibody based antigen ELISA for the diagnosis of foot-and-mouth disease. J Virol Methods. (2011) 175:253–60. doi: 10.1016/j.jviromet.2011.05.026
66. Shimmon G, Wood BA, Morris A, Mioulet V, Grazioli S, Brocchi E, et al. Truncated bovine integrin alpha-v/beta-6 as a universal capture ligand for FMD diagnosis. PLoS ONE. (2016) 11:e0160696. doi: 10.1371/journal.pone.0160696
67. Chen TH, Lee F, Lin YL, Dekker A, Chung WB, Pan CH, et al. Differentiation of foot-and-mouth disease-infected pigs from vaccinated pigs using antibody-detecting sandwich ELISA. J Vet Med Sci. (2011) 73:977–84. doi: 10.1292/jvms.10-0351
68. Chitray M, Grazioli S, Willems T, Tshabalala T, De Vleeschauwer A, Esterhuysen JJ, et al. Development and validation of a foot-and-mouth disease virus SAT serotype-specific 3ABC assay to differentiate infected from vaccinated animals. J Virol Methods. (2018) 255:44–51. doi: 10.1016/j.jviromet.2018.02.006
69. Fowler VL, Paton DJ, Rieder E, Barnett PV. Chimeric foot-and-mouth disease viruses: evaluation of their efficacy as potential marker vaccines in cattle. Vaccine. (2008) 26:1982–9. doi: 10.1016/j.vaccine.2008.02.012
70. Fowler VL, Knowles NJ, Paton DJ, Barnett PV. Marker vaccine potential of a foot-and-mouth disease virus with a partial VP1 G-H loop deletion. Vaccine. (2010) 28:3428–34. doi: 10.1016/j.vaccine.2010.02.074
71. Morioka K, Fukai K, Yoshida K, Yamazoe R, Onozato H, Ohashi S, et al. Neutralizing monoclonal antibody sandwich liquid-phase blocking enzyme-linked immunosorbent assay for detection of Foot-and-mouth disease virus type O antibodies. J Vet Diagn Invest. (2009) 21:499–503. doi: 10.1177/104063870902100411
72. Robiolo B, La Torre J, Duffy S, Leon E, Seki C, Torres A, et al. Quantitative single serum-dilution liquid phase competitive blocking ELISA for the assessment of herd immunity and expected protection against foot-and-mouth disease virus in vaccinated cattle. J Virol Methods. (2010) 166:21–7. doi: 10.1016/j.jviromet.2010.02.011
73. Sharma GK, Mahajan S, Matura R, Subramaniam S, Mohapatra JK, Pattnaik B. Quantitative single dilution liquid phase blocking ELISA for sero-monitoring of foot-and-mouth disease in India. Biologicals. (2015) 43:158–64. doi: 10.1016/j.biologicals.2015.03.003
74. Ko Y-J, Lee H-S, Jeoung H-Y, Heo E-J, Ko H-R, Chang B-S, et al. Use of a baculovirus-expressed structural protein for the detection of antibodies to foot-and-mouth disease virus type A by a blocking enzyme-linked immunosorbent assay. Clin Vaccine Immunol. (2010) 17:194–8. doi: 10.1128/CVI.00374-09
75. Ko YJ, Lee HS, Park JH, Lee KN, Kim SM, Cho IS. Field application of a recombinant protein-based ELISA during the 2010 outbreak of foot-and-mouth disease type A in South Korea. J Virol Methods. (2012) 179:265–8. doi: 10.1016/j.jviromet.2011.09.021
76. Dash L, Subramaniam S, Khulape SA, Prusty BR, Pargai K, Narnaware SD, et al. Development and utilization of VHH antibodies derived from camelus dromedarius against foot-and-mouth disease virus. Anim Biotechnol. (2019) 30:57–62. doi: 10.1080/10495398.2018.1433191
77. Li Y, Swabey KG, Gibson D, Keel PJ, Hamblin P, Wilsden G, et al. Evaluation of the solid phase competition ELISA for detecting antibodies against the six foot-and-mouth disease virus non-O serotypes. J Virol Methods. (2012) 183:125–31. doi: 10.1016/j.jviromet.2012.04.002
78. Kang YL, Jeong JY, Choi HY, Zhang Y, Liu Y, Lee HJ, et al. Evaluation and optimization of a conventional SPCE for FMD post-vaccination monitoring. BMC Vet Res. (2018) 14:371. doi: 10.1186/s12917-018-1686-7
79. Biswal JK, Bisht P, Mohapatra JK, Ranjan R, Sanya A, Pattnaik B. Application of a recombinant capsid polyprotein (P1) expressed in a prokaryotic system to detect antibodies against foot-and-mouth disease virus serotype O. J Virol Methods. (2015) 215–16, 45–51. doi: 10.1016/j.jviromet.2015.02.008
80. Ran X, Yang Z, Bai M, Zhang Y, Wen X, Guo H, et al. Development and validation of a competitive ELISA based on bacterium-original virus-like particles of serotype O foot-and-mouth disease virus for detecting serum antibodies. Appl Microbiol Biotechnol. (2019) 103:3015–24. doi: 10.1007/s00253-019-09680-8
81. Yang M, Xu W, Bittner H, Horsington J, Vosloo W, Goolia M, et al. Generation of mAbs to foot-and-mouth disease virus serotype A and application in a competitive ELISA for serodiagnosis. Virol J. (2016) 13:195. doi: 10.1186/s12985-016-0650-z
82. Gelkop S, Sobarzo A, Brangel P, Vincke C, Romao E, Fedida-Metula S, et al. The development and validation of a novel nanobody-based competitive ELISA for the detection of foot and mouth disease 3ABC antibodies in cattle. Front Vet Sci. (2018) 5:250. doi: 10.3389/fvets.2018.00250
83. Dong Y, Xu Y, Liu Z, Fu Y, Ohashi T, Tanaka Y, et al. Rapid screening swine foot-and-mouth disease virus using micro-ELISA system. Lab Chip. (2011) 11:2153–5. doi: 10.1039/c0lc00678e
84. Dong Y, Xu Y, Liu Z, Fu Y, Ohashi T, Mawatari K, et al. Determination of cattle foot-and-mouth disease virus by micro-ELISA method. Anal Sci. (2014) 30:359–63. doi: 10.2116/analsci.30.359
85. Engvall E, Perlmann P. Enzyme-linked immunosorbent assay, Elisa. 3. Quantitation of specific antibodies by enzyme-labeled anti-immunoglobulin in antigen-coated tubes. J Immunol. (1972) 109:129–35.
86. Coons AH, Creech HJ, Jones RN. Immunological properties of an antibody containing a fluorescent group. Proc Soc Exp Biol Med. (1941) 47:200–2. doi: 10.3181/00379727-47-13084P
87. Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides. (2015) 72:4–15. doi: 10.1016/j.peptides.2015.04.012
88. Ferris NP, Dawson M. Routine application of enzyme-linked immunosorbent assay in comparison with complement fixation for the diagnosis of foot-and-mouth and swine vesicular diseases. Vet Microbiol. (1988) 16:201–9.
89. Linchongsubongkoch W. OIE Reference Laboratory Reports Activities [Online] (2018). Available online at: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/reflabreports/2018/report_586_2018_Foot_and_mouth_disease_THAILAND.pdf?fbclid=IwAR2rn2ds53kkV0KCByhHuyDKVNGfUQcgOYz-sNRcevZqihoaj46N_FhLymA (accessed June 13, 2020).
90. Abu Elzein EM, Crowther JR. Enzyme-labelled immunosorbent assay techniques in foot-and-mouth disease virus research. J Hyg. (1978) 80:391–9. doi: 10.1017/s0022172400024840
91. Crowther JR, Abu-el Zein EM. Detection and quantification of foot and mouth disease virus by enzyme labelled immunosorbent assay techniques. J Gen Virol. (1979) 42:597–602. doi: 10.1099/0022-1317-42-3-597
92. Roeder PL, Le Blanc Smith PM. Detection and typing of foot-and-mouth disease virus by enzyme-linked immunosorbent assay: a sensitive, rapid and reliable technique for primary diagnosis. Res Vet Sci. (1987) 43:225–32.
93. Elzein EM, Crowther JR. The specific detection of foot-and-mouth disease virus whole particle antigen (140S) by enzyme labelled immunosorbent assay. J Hyg. (1979) 83:127–34. doi: 10.1017/s0022172400025894
94. Hamblin C, Barnett IT, Hedger RS. A new enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies against foot-and-mouth disease virus. I. Development and method of ELISA. J Immunol Methods. (1986) 93:115–21.
95. Meloen RH, Rowlands DJ, Brown F. Comparison of the antibodies elicited by the individual structural polypeptides of foot-and mouth disease and polio viruses. J Gen Virol. (1979) 45:761–3. doi: 10.1099/0022-1317-45-3-761
96. Meloen RH, Briaire J. A study of the cross-reacting antigens on the intact foot-and-mouth disease virus and its 12S Subunits with antisera against the structural proteins. J Gen Virol. (1980) 51(Pt 1):107–16. doi: 10.1099/0022-1317-51-1-107
97. McCullough KC, Crowther JR, Butcher RN. Alteration in antibody reactivity with foot-and-mouth disease virus (FMDV) 146S antigen before and after binding to a solid phase or complexing with specific antibody. J Immunol Methods. (1985) 82:91–100.
98. Mackay DK, Bulut AN, Rendle T, Davidson F, Ferris NP. A solid-phase competition ELISA for measuring antibody to foot-and-mouth disease virus. J Virol Methods. (2001) 97:33–48. doi: 10.1016/s0166-0934(01)00333-0
99. Tekleghiorghis T, Weerdmeester K, van Hemert-Kluitenberg F, Moormann RJ, Dekker A. Comparison of test methodologies for foot-and-mouth disease virus serotype A vaccine matching. Clin Vaccine Immunol. (2014) 21:674–83. doi: 10.1128/cvi.00034-14
100. Paiba GA, Anderson J, Paton DJ, Soldan AW, Alexandersen S, Corteyn M, et al. Validation of a foot-and-mouth disease antibody screening solid-phase competition ELISA (SPCE). J Virol Methods. (2004) 115:145–58. doi: 10.1016/j.jviromet.2003.09.016
101. Valarcher JF, Leforban Y, Rweyemamu M, Roeder PL, Gerbier G, Mackay DK, et al. Incursions of foot-and-mouth disease virus into Europe between 1985 and 2006. Transbound Emerg Dis. (2008) 55:14–34. doi: 10.1111/j.1865-1682.2007.01010.x
102. Sangar DV, Black DN, Rowlands DJ, Brown F. Biochemical mapping of the foot-and-mouth disease virus genome. J Gen Virol. (1977) 35:281–97. doi: 10.1099/0022-1317-35-2-281
103. Cao Y, Lu Z, Li D, Fan P, Sun P, Bao H, et al. Evaluation of cross-protection against three topotypes of serotype O foot-and-mouth disease virus in pigs vaccinated with multi-epitope protein vaccine incorporated with poly(I:C). Vet Microbiol. (2014) 168:294–301. doi: 10.1016/j.vetmic.2013.11.023
104. van Maanen C. A complex-trapping-blocking (CTB) ELISA, using monoclonal antibodies and detecting specifically antibodies directed against foot-and-mouth disease types A, O and C. I. Method and characteristics. Vet Microbiol. (1990) 24:171–8.
105. Smitsaar EN, Saiz JC, Yedloutschnig RJ, Morgan DO. Detection of foot-and-mouth disease virus by competitive ELISA using a monoclonal antibody specific for the 12S protein subunit from six of the seven serotypes. Vet Immunol Immunopathol. (1990) 26:251–65.
106. Samuel AR, Knowles NJ, Samuel GD, Crowther JR. Evaluation of a trapping ELISA for the differentiation of foot-and-mouth disease virus strains using monoclonal antibodies. Biologicals. (1991) 19:299–310.
107. Smitsaart EN, Fernandez E, Maradei E, Morgan DO. Validation of an inhibition ELISA using a monoclonal antibody for foot-and-mouth disease (FMD) primary diagnosis. Zentralbl Veterinarmed B. (1994) 41:313–9.
108. Tosh C, Venkataramanan R, Pattnaik B, Hemadri D, Sanyal A. Monoclonal antibodies to an Indian strain of type A foot-and-mouth disease virus. Acta Virol. (1999) 43:219–25.
109. Morioka K, Fukai K, Yoshida K, Yamazoe R, Onozato H, Ohashi S, et al. Foot-and-mouth disease virus antigen detection enzyme-linked immunosorbent assay using multiserotype-reactive monoclonal antibodies. J Clin Microbiol. (2009) 47:3663–8. doi: 10.1128/jcm.00695-09
110. Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. (2005) 23:1126–36. doi: 10.1038/nbt1142
111. de Marco A. Biotechnological applications of recombinant single-domain antibody fragments. Microb Cell Fact. (2011) 10:44. doi: 10.1186/1475-2859-10-44
112. McCullough KC, De Simone F, Brocchi E, Capucci L, Crowther JR, Kihm U. Protective immune response against foot-and-mouth disease. J Virol. (1992) 66:1835–40.
113. Grubman MJ, Lewis SA, Morgan DO. Protection of swine against foot-and-mouth disease with viral capsid proteins expressed in heterologous systems. Vaccine. (1993) 11:825–9.
114. Mackay DK, Forsyth MA, Davies PR, Salt JS. Antibody to the nonstructural proteins of foot-and-mouth disease virus in vaccinated animals exposed to infection. Vet Q. (1998) 20(Suppl. 2):S9–11.
115. Paton DJ, de Clercq K, Greiner M, Dekker A, Brocchi E, Bergmann I, et al. Application of non-structural protein antibody tests in substantiating freedom from foot-and-mouth disease virus infection after emergency vaccination of cattle. Vaccine. (2006) 24:6503–12. doi: 10.1016/j.vaccine.2006.06.032
116. De Diego M, Brocchi E, Mackay D, De Simone F. The non-structural polyprotein 3ABC of foot-and-mouth disease virus as a diagnostic antigen in ELISA to differentiate infected from vaccinated cattle. Arch Virol. (1997) 142:2021–33.
117. Bronsvoort BM, Sorensen KJ, Anderson J, Corteyn A, Tanya VN, Kitching RP, et al. Comparison of two 3ABC enzyme-linked immunosorbent assays for diagnosis of multiple-serotype foot-and-mouth disease in a cattle population in an area of endemicity. J Clin Microbiol. (2004) 42:2108–14. doi: 10.1128/jcm.42.5.2108-2114.2004
118. Bruderer U, Swam H, Haas B, Visser N, Brocchi E, Grazioli S, et al. Differentiating infection from vaccination in foot-and-mouth-disease: evaluation of an ELISA based on recombinant 3ABC. Vet Microbiol. (2004) 101:187–97. doi: 10.1016/j.vetmic.2004.01.021
119. Clavijo A, Zhou EM, Hole K, Galic B, Kitching P. Development and use of a biotinylated 3ABC recombinant protein in a solid-phase competitive ELISA for the detection of antibodies against foot-and-mouth disease virus. J Virol Methods. (2004) 120:217–27. doi: 10.1016/j.jviromet.2004.05.007
120. Moonen P, van der Linde E, Chenard G, Dekker A. Comparable sensitivity and specificity in three commercially available ELISAs to differentiate between cattle infected with or vaccinated against foot-and-mouth disease virus. Vet Microbiol. (2004) 99:93–101. doi: 10.1016/j.vetmic.2003.12.003
121. Armstrong RM, Cox SJ, Aggarwal N, Mackay DJ, Davies PR, Hamblin PA, et al. Detection of antibody to the foot-and-mouth disease virus (FMDV) non-structural polyprotein 3ABC in sheep by ELISA. J Virol Methods. (2005) 125:153–63. doi: 10.1016/j.jviromet.2005.01.012
122. Foord AJ, Muller JD, Yu M, Wang LF, Heine HG. Production and application of recombinant antibodies to foot-and-mouth disease virus non-structural protein 3ABC. J Immunol Methods. (2007) 321:142–51. doi: 10.1016/j.jim.2007.01.014
123. Lu Z, Cao Y, Guo J, Qi S, Li D, Zhang Q, et al. Development and validation of a 3ABC indirect ELISA for differentiation of foot-and-mouth disease virus infected from vaccinated animals. Vet Microbiol. (2007) 125:157–69. doi: 10.1016/j.vetmic.2007.05.017
124. Bronsvoort BMDC, Parida S, Handel I, McFarland S, Fleming L, Hamblin P, et al. Serological survey for foot-and-mouth disease virus in wildlife in eastern Africa and estimation of test parameters of a nonstructural protein enzyme-linked immunosorbent assay for buffalo. Clin Vaccine Immunol. (2008) 15:1003–11. doi: 10.1128/CVI.00409-07
125. Ayebazibwe C, Mwiine FN, Balinda SN, Tjornehoj K, Alexandersen S. Application of the Ceditest(R) FMDV type O and FMDV-NS enzyme-linked immunosorbent assays for detection of antibodies against Foot-and-mouth disease virus in selected livestock and wildlife species in Uganda. J Vet Diagn Invest. (2012) 24:270–6. doi: 10.1177/1040638711435807
126. Mackay DKJ, Forsyth MA, Davies PR, Berlinzani A, Belsham GJ, Flint M, et al. Differentiating infection from vaccination in foot-and-mouth disease using a panel of recombinant, non-structural proteins in ELISA. Vaccine. (1998) 16:446–59. doi: 10.1016/S0264-410X(97)00227-2
127. McVicar JW, Sutmoller P. Foot-and-mouth disease: the agar gel diffusion precipitin test for antibody to virus-infection-associated (via) antigen as a tool for epizootiologic surveys1. Am J Epidemiol. (1970) 92:273–8. doi: 10.1093/oxfordjournals.aje.a121207
128. Alonso A, Gomes MPD, Martins MA, Sondahl MS. Detection of foot-and-mouth disease virus infection-associated antigen antibodies: comparison of the enzyme-linked immunosorbent assay and agar gel immunodiffusion tests. Prev Vet Med. (1990) 9:223–40. doi: 10.1016/0167-5877(90)90069-T
129. O'Donnell VK, Boyle DB, Sproat K, Fondevila NA, Forman A, Schudel AA, et al. Detection of antibodies against foot-and-mouth disease virus using a liquid-phase blocking sandwich ELISA (LPBE) with a bioengineered 3D protein. J Vet Diagn Invest. (1996) 8:143–50. doi: 10.1177/104063879600800201
130. Sørensen KJ, Madsen KG, Madsen ES, Salt JS, Nqindi J, Mackay DKJ. Differentiation of infection from vaccination in foot-and-mouth disease by the detection of antibodies to the non-structural proteins 3D, 3AB and 3ABC in ELISA using antigens expressed in baculovirus. Arch Virol. (1998) 143:1461–76. doi: 10.1007/s007050050390
131. Hema M, Nagendrakumar SB, Yamini R, Chandran D, Rajendra L, Thiagarajan D, et al. Chimeric tymovirus-like particles displaying foot-and-mouth disease virus non-structural protein epitopes and its use for detection of FMDV-NSP antibodies. Vaccine. (2007) 25:4784–94. doi: 10.1016/j.vaccine.2007.04.023
132. Liu Z, Shao J, Zhao F, Zhou G, Gao S, Liu W, et al. Chemiluminescence immunoassay for the detection of antibodies against the 2C and 3ABC nonstructural proteins induced by infecting pigs with foot-and-mouth disease virus. Clin. Vaccine Immunol. (2017) 24:e00153-17. doi: 10.1128/cvi.00153-17
133. Liu ZZ, Zhao FR. Development of a chemiluminescence immunoassay using recombinant non-structural epitope-based proteins to accurately differentiate foot-and-mouth disease virus-infected and vaccinated bovines. Transbound Emerg Dis. (2018) 65:338–44. doi: 10.1111/tbed.12811
134. Brocchi E, Bergmann IE, Dekker A, Paton DJ, Sammin DJ, Greiner M, et al. Comparative evaluation of six ELISAs for the detection of antibodies to the non-structural proteins of foot-and-mouth disease virus. Vaccine. (2006) 24:6966–79. doi: 10.1016/j.vaccine.2006.04.050
135. Lee F, Jong M-H, Yang D-W. Presence of antibodies to non-structural proteins of foot-and-mouth disease virus in repeatedly vaccinated cattle. Vet Microbiol. (2006) 115:14–20. doi: 10.1016/j.vetmic.2005.12.017
136. Henderson LM. Overview of marker vaccine and differential diagnostic test technology. Biologicals. (2005) 33:203–9. doi: 10.1016/j.biologicals.2005.08.006
137. Li P, Lu Z, Bai X, Li D, Sun P, Bao H, et al. Evaluation of a 3A-truncated foot-and-mouth disease virus in pigs for its potential as a marker vaccine. Vet Res. (2014) 45:51. doi: 10.1186/1297-9716-45-51
138. Grubman MJ. Development of novel strategies to control foot-and-mouth disease: marker vaccines and antivirals. Biologicals. (2005) 33:227–34. doi: 10.1016/j.biologicals.2005.08.009
139. Rieder E, Baxt B, Lubroth J, Mason PW. Vaccines prepared from chimeras of foot-and-mouth disease virus (FMDV) induce neutralizing antibodies and protective immunity to multiple serotypes of FMDV. J Virol. (1994) 68:7092–98.
140. Taboga O, Tami C, Carrillo E, Núñez JI, Rodríguez A, Saíz JC, et al. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: lack of solid protection in cattle and isolation of escape mutants. J Virol. (1997) 71:2606–14.
141. Knight-Jones TJ, Robinson L, Charleston B, Rodriguez LL, Gay CG, Sumption KJ, et al. Global foot-and-mouth disease research update and gap analysis: 4 - diagnostics. Transbound Emerg Dis. (2016) 63(Suppl. 1):42–8. doi: 10.1111/tbed.12523
142. Lau LT, Reid SM, King DP, Lau AM, Shaw AE, Ferris NP, et al. Detection of foot-and-mouth disease virus by nucleic acid sequence-based amplification (NASBA). Vet Microbiol. (2008) 126:101–10. doi: 10.1016/j.vetmic.2007.07.008
143. Alvarez I, Gutierrez G, Barrandeguy M, Trono K. Immunochromatographic lateral flow test for detection of antibodies to Equine infectious anemia virus. J Virol Methods. (2010) 167:152–7. doi: 10.1016/j.jviromet.2010.03.026
144. Jiang T, Liang Z, Ren W, Chen J, Zhi X, Qi G, et al. Development and validation of a lateral flow immunoassay using colloidal gold for the identification of serotype-specific foot-and-mouth disease virus O, A and Asia 1. J Virol Methods. (2011) 171:74–80. doi: 10.1016/j.jviromet.2010.10.002
145. Jiang T, Liang Z, Ren WW, Chen J, Zhi XY, Qi GY, et al. A simple and rapid colloidal gold-based immunochromatogarpic strip test for detection of FMDV serotype A. Virol Sin. (2011) 26:30–9. doi: 10.1007/s12250-011-3166-5
146. Oem JK, Ferris NP, Lee K-N, Joo Y-S, Hyun B-H, Park J-H. Simple and rapid lateral-flow assay for the detection of foot-and-mouth disease virus. Clin Vaccine Immunol. (2009) 16:1660. doi: 10.1128/CVI.00213-09
147. Ferris NP, Nordengrahn A, Hutchings GH, Paton DJ, Kristersson T, Brocchi E, et al. Development and laboratory validation of a lateral flow device for the detection of serotype SAT 2 foot-and-mouth disease viruses in clinical samples. J Virol Methods. (2010) 163:474–6. doi: 10.1016/j.jviromet.2009.09.022
148. Chen TH, Pan CH, Jong MH, Lin HM, Huang YL, Hsiung KP, et al. Development of a chromatographic strip assay for detection of porcine antibodies to 3ABC non-structural protein of foot-and-mouth disease virus serotype O. J Vet Med Sci. (2009) 71:703–8. doi: 10.1292/jvms.71.703
149. Yang S, Yang J, Zhang G, Wang X, Qiao S, Zhao D, et al. Development of an immunochromatographic strip for the detection of antibodies against foot-and-mouth disease virus serotype O. J Virol Methods. (2010) 165:139–44. doi: 10.1016/j.jviromet.2010.01.001
150. Lin T, Shao JJ, Du JZ, Cong GZ, Gao SD, Chang H. Development of a serotype colloidal gold strip using monoclonal antibody for rapid detection type Asia1 foot-and-mouth disease. Virol J. (2011) 8:418. doi: 10.1186/1743-422X-8-418
151. Yang M, Caterer NR, Xu W, Goolia M. Development of a multiplex lateral flow strip test for foot-and-mouth disease virus detection using monoclonal antibodies. J Virol Methods. (2015) 221:119–26. doi: 10.1016/j.jviromet.2015.05.001
152. Wu L, Jiang T, Lu Z-J, Yang Y-M, Sun P, Liang Z, et al. Development and validation of a prokaryotically expressed foot-and-mouth disease virus non-structural protein 2C'3AB-based immunochromatographic strip to differentiate between infected and vaccinated animals. Virol J. (2011) 8:186. doi: 10.1186/1743-422X-8-186
153. Yang M, Goolia M, Xu W, Bittner H, Clavijo A. Development of a quick and simple detection methodology for foot-and-mouth disease virus serotypes O, A and Asia 1 using a generic RapidAssay Device. Virol J. (2013) 10:125. doi: 10.1186/1743-422X-10-125
154. Morioka K, Fukai K, Yoshida K, Kitano R, Yamazoe R, Yamada M, et al. Development and evaluation of a rapid antigen detection and serotyping lateral flow antigen detection system for foot-and-mouth disease virus. PLoS ONE. (2015) 10:e0134931. doi: 10.1371/journal.pone.0134931
155. Wang H, Hou P, Zhao G, Yu L, Gao Y-W, He H. Development and evaluation of serotype-specific recombinase polymerase amplification combined with lateral flow dipstick assays for the diagnosis of foot-and-mouth disease virus serotype A, O and Asia1. BMC Vet Res. (2018) 14:359. doi: 10.1186/s12917-018-1644-4
156. Liu L, Wang J, Zhang R, Lin M, Shi R, Han Q, et al. Visual and equipment-free reverse transcription recombinase polymerase amplification method for rapid detection of foot-and-mouth disease virus. BMC Vet Res. (2018) 14:263. doi: 10.1186/s12917-018-1594-x
157. Koczula KM, Gallotta A. Lateral flow assays. Essays Biochem. (2016) 60:111–20. doi: 10.1042/EBC20150012
158. Sithigorngul P, Rukpratanporn S, Pecharaburanin N, Suksawat P, Longyant S, Chaivisuthangkura P, et al. A simple and rapid immunochromatographic test strip for detection of pathogenic isolates of Vibrio harveyi. J Microbiol Methods. (2007) 71:256–64. doi: 10.1016/j.mimet.2007.09.012
159. Cui S, Chen C, Tong G. A simple and rapid immunochromatographic strip test for monitoring antibodies to H5 subtype Avian Influenza Virus. J Virol Methods. (2008) 152:102–5. doi: 10.1016/j.jviromet.2008.06.011
160. Ferris NP, Clavijo A, Yang M, Velazquez-Salinas L, Nordengrahn A, Hutchings GH, et al. Development and laboratory evaluation of two lateral flow devices for the detection of vesicular stomatitis virus in clinical samples. J Virol Methods. (2012) 180:96–100. doi: 10.1016/j.jviromet.2011.12.010
161. Zhou SH, Cui SJ, Chen CM, Zhang FC, Li J, Zhou S, et al. Development and validation of an immunogold chromatographic test for on-farm detection of PRRSV. J Virol Methods. (2009) 160:178–84. doi: 10.1016/j.jviromet.2009.04.034
162. Zhang GP, Wang XN, Yang JF, Yang YY, Xing GX, Li QM, et al. Development of an immunochromatographic lateral flow test strip for detection of β-adrenergic agonist Clenbuterol residues. J Immunol Methods. (2006) 312:27–33. doi: 10.1016/j.jim.2006.02.017
163. Zhao Y, Zhang G, Liu Q, Teng M, Yang J, Wang J. Development of a lateral flow colloidal gold immunoassay strip for the rapid detection of enrofloxacin residues. J Agric Food Chem. (2008) 56:12138–42. doi: 10.1021/jf802648z
164. Jiang T, Liang Z, Chen J, He JJ, Lu L, Ma WM, et al. [Establishment of a colloid gold-immunochromatography assay for detection of type Asia I foot-and-mouth disease virus]. Chinese J Cell Mol Immunol. (2007) 23:1021–4.
165. Reid SM, Ferris NP, Bruning A, Hutchings GH, Kowalska Z, Akerblom L. Development of a rapid chromatographic strip test for the pen-side detection of foot-and-mouth disease virus antigen. J Virol Methods. (2001) 96:189–202. doi: 10.1016/s0166-0934(01)00334-2
166. Ferris NP, Nordengrahn A, Hutchings GH, Reid SM, King DP, Ebert K, et al. Development and laboratory validation of a lateral flow device for the detection of foot-and-mouth disease virus in clinical samples. J Virol Methods. (2009) 155:10–7. doi: 10.1016/j.jviromet.2008.09.009
167. Sammin D, Ryan E, Ferris NP, King DP, Zientara S, Haas B, et al. Options for decentralized testing of suspected secondary outbreaks of foot-and-mouth disease. Transbound Emerg Dis. (2010) 57:237–43. doi: 10.1111/j.1865-1682.2010.01141.x
168. Jamal SM, Belsham GJ. Foot-and-mouth disease: past, present and future. Vet Res. (2013) 44:116. doi: 10.1186/1297-9716-44-116
169. Yang M, Clavijo A, Suarez-Banmann R, Avalo R. Production and characterization of two serotype independent monoclonal antibodies against foot-and-mouth disease virus. Vet Immunol Immunopathol. (2007) 115:126–34. doi: 10.1016/j.vetimm.2006.10.002
170. Maradei E, Perez Beascoechea C, Malirat V, Salgado G, Seki C, Pedemonte A, et al. Characterization of foot-and-mouth disease virus from outbreaks in Ecuador during 2009-2010 and cross-protection studies with the vaccine strain in use in the region. Vaccine. (2011) 29:8230–40. doi: 10.1016/j.vaccine.2011.08.120
171. Amada T, Yoshimatsu K, Koma T, Shimizu K, Gamage CD, Shiokawa K, et al. Development of an immunochromatography strip test based on truncated nucleocapsid antigens of three representative hantaviruses. Virol J. (2014) 11:87. doi: 10.1186/1743-422x-11-87
172. Zhuang L, Ji Y, Tian P, Wang K, Kou C, Gu N, et al. Polymerase chain reaction combined with fluorescent lateral flow immunoassay based on magnetic purification for rapid detection of canine parvovirus 2. BMC Vet Res. (2019) 15:30. doi: 10.1186/s12917-019-1774-3
173. Yan G, Jun L, Kangchen Z, Yiyue G, Yang Y, Xiaoyu Z, et al. Rapid and visual detection of human enterovirus coxsackievirus A16 by reverse transcription loop-mediated isothermal amplification combined with lateral flow device. Lett Appl Microbiol. (2015) 61:531–7. doi: 10.1111/lam.12499
174. Yang M, Ke Y, Wang X, Ren H, Liu W, Lu H, et al. Development and evaluation of a rapid and sensitive EBOV-RPA test for rapid diagnosis of Ebola virus disease. Sci Rep. (2016) 6:26943. doi: 10.1038/srep26943
175. Yang Y, Qin X, Song Y, Zhang W, Hu G, Dou Y, et al. Development of real-time and lateral flow strip reverse transcription recombinase polymerase Amplification assays for rapid detection of peste des petits ruminants virus. Virol J. (2017) 14:24. doi: 10.1186/s12985-017-0688-6
176. Sun N, Wang W, Wang J, Yao X, Chen F, Li X, et al. Reverse transcription recombinase polymerase amplification with lateral flow dipsticks for detection of influenza A virus and subtyping of H1 and H3. Mol Cell Probes. (2018) 42:25–31. doi: 10.1016/j.mcp.2018.10.004
177. Cheng S, Sun J, Yang J, Lv J, Wu F, Lin Y, et al. A new immunoassay of serum antibodies against Peste des petits ruminants virus using quantum dots and a lateral-flow test strip. Anal Bioanal Chem. (2017) 409:133–41. doi: 10.1007/s00216-016-9972-3
178. Wang H-M, Zhao G-M, Hou P-L, Yu L, He C-Q, He H-B. Rapid detection of foot-and-mouth disease virus using reverse transcription recombinase polymerase amplification combined with a lateral flow dipstick. J. Virol. Methods. (2018) 261:46–50. doi: 10.1016/j.jviromet.2018.07.011
179. Samuel AR, Knowles NJ. Foot-and-mouth disease type O viruses exhibit genetically and geographically distinct evolutionary lineages (topotypes). J Gen Virol. (2001) 82(Pt 3):609–21. doi: 10.1099/0022-1317-82-3-609
180. Steinfeld H, Wassenaar T, Jutzi S. Livestock production systems in developing countries: status, drivers, trends. Rev Sci Tech. (2006) 25:505–16. doi: 10.20506/rst.25.2.1677
181. FAO. The State of Food Security and Nutrition in the World 2019 [Online]. (2019). Available online at: http://www.fao.org/state-of-food-security-nutrition/en/ (accessed June 14, 2020).
182. FAO. Preparation of Foot-and-Mouth Disease Contigency Plans: Early Warning Contingency Planning for FMD [Online] (2002). Available online at: http://www.fao.org/3/Y4382E/y4382e08.htm#bm08 (accessed June 14, 2020).
183. Paton DJ, Sumption KJ, Charleston B. Options for control of foot-and-mouth disease: knowledge, capability and policy. Philos Trans R Soc Lond Ser B Biol Sci. (2009) 364:2657–67. doi: 10.1098/rstb.2009.0100
184. Madi M, Hamilton A, Squirrell D, Mioulet V, Evans P, Lee M, et al. Rapid detection of foot-and-mouth disease virus using a field-portable nucleic acid extraction and real-time PCR amplification platform. Vet J. (2012) 193:67–72. doi: 10.1016/j.tvjl.2011.10.017
185. Knight-Jones TJ, Robinson L, Charleston B, Rodriguez LL, Gay CG, Sumption KJ, et al. Global foot-and-mouth disease research update and gap analysis: 1 - Overview of global status and research needs. Transbound Emerg Dis. (2016) 63(Suppl. 1):3–13. doi: 10.1111/tbed.12528
186. Gao M, Zhang R, Li M, Li S, Cao Y, Ma B, et al. An ELISA based on the repeated foot-and-mouth disease virus 3B epitope peptide can distinguish infected and vaccinated cattle. Appl Microbiol Biotechnol. (2012) 93:1271–9. doi: 10.1007/s00253-011-3815-0
187. Sharma GK, Mohapatra JK, Pandey LK, Mahajan S, Mathapati BS, Sanyal A, et al. Immunodiagnosis of foot-and-mouth disease using mutated recombinant 3ABC polyprotein in a competitive ELISA. J Virol Methods. (2012) 185:52–60. doi: 10.1016/j.jviromet.2012.05.029
Keywords: foot-and-mouth disease virus (FMDV) diagnosis, complement fixation test (CFT), virus neutralization test (VNT), enzyme-linked immunosorbent assay (ELISA), reverse transcription-polymerase chain reaction (RT-PCR), reverse transcription-loop-mediated isothermal amplification (RT-LAMP), reverse transcription-recombinase polymerase amplification (RT-RPA), lateral flow device (LFD)
Citation: Wong CL, Yong CY, Ong HK, Ho KL and Tan WS (2020) Advances in the Diagnosis of Foot-and-Mouth Disease. Front. Vet. Sci. 7:477. doi: 10.3389/fvets.2020.00477
Received: 24 March 2020; Accepted: 26 June 2020;
Published: 21 August 2020.
Edited by:
Mariano Pérez-Filgueira, National Agricultural Technology Institute, ArgentinaReviewed by:
Marie Rene Culhane, College of Veterinary Medicine, University of Minnesota, United StatesCopyright © 2020 Wong, Yong, Ong, Ho and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Siang Tan, d3N0YW5AdXBtLmVkdS5teQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.