
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 04 August 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00463
The growing consumption of snakes in China has led to a boom in edible snakes farming. Food producing reptiles, such as snakes can carry many pathogenic microbes and potentially infect humans. Here, we report the occurrence of multi drug resistant Salmonella enteritidis strains isolated from edible snakes in China. Our results showed that the isolated S. enteritidis was resistant to the majority of the tested drugs and sensitive to tetracycline and amikacin. Antimicrobial susceptibility test showed that the strains carried the blaTEM, qnrD, aadA1, catA1 o, sul I, and sul II genes. The pathogenicity testing of the S. enteritidis isolated strains showed that these strains were highly pathogenic (75% mortality, with LD50 at 107.7 CFU/mL). The chickens in the high-dose groups developed acute septicemia and died within 24 h. Results of the dissection showed extensive abdominal bleeding and swelling in the high dose groups, as well as hyperemia edema in the livers, lungs, kidneys, cecum, and bursa of the chickens, with spotty bleeding. In addition, rod-shaped bacterial aggregation was also seen in the visual field. A total of 23 virulence genes, mainly associated with pathogenicity island were tested, of which 8 genes including avrA, iacP, prgK, ssrA, siiD (spi4D), siiE, spi4H, and pipC were found positive. Altogether, our results provide useful information regarding edible snakes contaminated with S. enteritidis, which may have public health implications.
Salmonella is one of the most common Enterobacteriaceae. Since its discovery in 1885, more than 2,600 Salmonella serotypes have been reported (1), making it one of the most common zoonotic pathogens in the world. Salmonella is the leading cause of morbidity and mortality in children under 5 years of age in most developing countries (2–4). In China, 22.2% of food poisoning was caused by Salmonella, and the vast majority was caused by eating meat products (5). The strains that caused 99% human and animal infections belong to Salmonella enterica ssp. Enterica, of which S. typhimurium and S. enteritidis are the most common serotypes (6). And they are also important foodborne pathogens (7). One of the main characteristics of Salmonella enterica is that it can cause a variety of diseases of varying degrees in different hosts, such as human, swine, cows, birds and mice. In humans, S. enteritidis can cause a variety of foodborne diseases such as gastroenteritis and systemic or persistent diseases (8–10).
Poultry and poultry products are considered as the main reservoir of S. enteritidis (11), but it is worth noting that reptiles are also an important host of Salmonella. In the United States, about 6% of cases of human Salmonella infection were from reptiles. In southwest England, contacting with reptiles has been linked to 27.4% of Salmonella cases in children under 5 years of age (12). According to published reports, the majority of human cases of reptile-related salmonellosis come from non-toxic reptiles kept as pets (13). In China, snake meat has a huge number of consumers because of its high nutrition and good taste. Salmonella carried by snakes can be transmitted to humans through the food chain, causing harm to humans. Studies have found that vipers may be reservoirs of Salmonella serotypes associated with human salmonellosis (14). European Union has planned to establish food safety standards related to Salmonella contamination in reptile meat. The European Commission is proposing to limit Salmonella in reptile meat, which is another reminder that the presence of Salmonella in reptile meat could pose a potential threat to humans.
Salmonella enteritidis is causing serious complications in human since 1980. In 2011, the total number of cases reported to CED increased significantly and S. enteritidis became more common in South Africa (15, 16). Despite global efforts to constrain the spread of Salmonella, S. enteritidis infections continue to spread rapidly, which brings challenges to global health systems. In many developed countries, the development of resistance to commonly used antibiotics against zoonotic pathogens is almost the inevitable consequence of the abused use of antibiotics in food-producing animals. We have widely agreed that zoonotic strains acquired resistance from food animals first then infected human being through food chains. Salmonella from clinical practice and rapid spread of multi-drug resistance on a global scale have attracted the attention worldwide. Despite the development of national antimicrobial susceptibility testing programs, bacterial resistance remains a problem that needs to be addressed as soon as possible, given the severity of foodborne Salmonella resistance.
At present, there are only few reports about wild snakes or pet snakes carrying pathogenic bacteria. Limited reports about pathogens from food source snakes have been discussed. This study is the first report on S. enteritidis isolates from snake suffering from pneumonia. The aim of this study was to isolate and evaluate the pathogenicity of the S. enteritidis strains and determine the fundamental information for public health safety and further studies.
Under sterile conditions, the lungs of the snakes with pneumonia were collected. The internal structures of intestinal tissues were obtained with a sterile sterilizing ring, and the pathogens were separated by streaks on MAC agar (MacConkey). The culture medium was placed in a constant temperature incubator at 37°C for 18 h then the colony morphology was observed. Suspected bacterial colonies were selected by a sterile sterilizing ring and separated by streaking on a new MAC agar to be purified.
The pathogens were identified by Gram staining, biochemical examination, and PCR (Polymerase Chain Reaction) analysis. A pair of specific primers were designed for the specific gene sequence of invA, the specific gene of Salmonella, and the expected amplified target fragment size was 374 bp. The invA gene PCR primer sequence was: invA-F 5′-GCTCTTTCGTCTGGCATTA-3′. invA R-5′-CGGCATAGCGTCACCTT-3′. PCR reaction conditions were pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s. Annealed at 50°C and extended at 72°C for 1 min. There were 35 cycles of reaction and extended at 72°C for 10 min. The 7 μL PCR amplification product was subjected to 1% agarose gel electrophoresis, and the standard strain of S. enteritidis (NCTC13349) was used as the positive control. The strip was observed and preserved using a gel imaging system. Finally, PCR products were sent to Wuhan Jinkairui biological engineering co., LTD for sequencing.
The serotype of Salmonella was identified by slide agglutination tests. According to the Kauffmann-White scheme (17), Salmonella diagnostic serum kit (MicroFast; LR70602) was used for testing.
Disc diffusion test (K-B method) was used to conduct sensitivity tests for 19 antimicrobial drugs on isolated strains, and the whole process was developed, according to the standards of the American Association for Clinical and Laboratory Standards Institute (CLSI). Before antimicrobial resistant test, the purified strain was inoculated in TSB (tryptic soy broth) liquid culture for 18 h. One hundred micro-liters of fresh bacterial liquid were taken and placed on TSA (Tryptic Soy Agar) medium. The liquid was spread evenly with sterile cotton swabs. Each drug sensitive paper was clipped and attached to the medium with sterile tweezers by triplicate. All plates were placed upside down in an incubator at 37°C for 18–24 h. After the cultivation, the formation of bacteria-resistant zones was observed, the diameter of bacteria-resistant zone was measured, the average values were calculated, and judged as susceptible, intermediate or resistant (18, 19). The types and contents of selected antibacterial drugs are shown in Table 1.
Relevant gene sequences published on NCBI and literature were searched (20, 21), then primers were designed. They were synthesized by Wuhan Jinkairui biological engineering co. LTD. The categories, names, and primer sequences of resistant genes are shown in Table 2. The capacity of each PCR reaction system was 20 μL. It contains PCR Mix 10 μL. The upstream and downstream primers were 1 μL each. While DNA template and dd water was 2 and 6 μL, respectively. The reaction parameters of all genes amplified by PCR instruments were: pre-denaturation 95°C for 5 min, denaturation 95°C for 1 min, annealing at 54°C for 30 s, extension at 72°C for 1 min, a total of 35 cycles, extension at 72°C for 10 min. The amplified products were analyzed by 1% agarose gel electrophoresis and photographed by a gel imaging system. Samples with positive PCR results were selected for sequencing. The sequencing results were compared with the existing sequences in Blast software in GenBank.
The virulence gene sequence recorded on GenBank and related literature were searched (22–24), and 23 pairs of specific virulence gene primers were designed by Oligo6 software, as shown in Table 3. The primers were synthesized by Wuhan Jinkairui biological engineering co. LTD. Plasmid virulence genes: spvA and spvD. Virulence genes of SPI-1: invH, sipA, sopA, sopD, avrA, iacP, and prgK. Virulence genes of SPI-2: ssaB, ssaQ, sifA, sseL, ssrA, and ttrB. Virulence genes of SPI-3: misL, rmbA, rhuM, and sugR. Virulence genes of SPI-4: siiD (spi4D), siiE, and spi4H. Virulence genes of SPI-5: pipC. The volume of amplified virulence gene PCR system was 20 μL, PCR Mix 10 μL, upstream primer 1 μL, downstream primer 1 μL, DNA template 2 μL, and ddH2O 6 μL. PCR amplification procedures: pre-denaturation at 95°C for 5 min, denaturation at 94°C for 30–45 s, annealing at 52–58°C for 30 s, extension at 72°C for 1 min, reaction 35 cycles. Extend at 72°C for 10 min. The amplified products were analyzed by 1% agarose gel electrophoresis and photographed by a gel imaging system. Samples with positive PCR results were selected for sequencing. The sequencing results were compared with the existing sequences in Blast software in Genbank.
Fifteen three-day-old SPF (Specific pathogen Free) chickens were randomly divided into five groups evenly. Four experimental groups were set up, and 1 negative control group was set up. Each chicken in the test group was intraperitoneally injected with 0.2 mL of the bacterial solution at a concentration of 4 × 108 CFU/mL. The control group was intraperitoneally injected with the same amount of sterile PBS (phosphate buffer saline) solution. Death data were recorded after 7 consecutive days of observation.
Another 25 healthy 3-day-old SPF chicks were randomly divided into 5 groups of 5 in each group to avoid cross-infection. Four groups were taken as the experimental group. The concentration of bacterial suspensions was diluted into 4 × 109 CFU/mL, 4 × 108 CFU/mL, 4 × 107 CFU/mL and 4 × 106 CFU/mL, respectively, with sterile PBS.
The day of challenge was recorded as day 0. The onset time, symptoms and death time of the chickens were recorded twice a day. The dead chickens were dissected immediately, and the pathological changes were recorded. After 7 days of continuous observation, all chickens were euthanized. The bacteria were isolated and identified from dead chickens and the total number of deaths was recorded. The 50% lethal dose of bacteria (LD50) was calculated by the modified Kärber method.
“XK” means the maximum logarithmic dose, “d” means the difference of logarithmic dose between two adjacent groups, “Pi” means the mortality rate, and “i” means the group number.
The dead chickens were timely dissected. The pathological changes of tissues and organs were observed, and the hearts, lungs, livers, kidneys, spleens, bursa of fabricius and small intestine of the sick and dead chickens were collected, then fixed with 4% paraformaldehyde solution and stored at 4°C. Hematoxylin-eosin staining was performed on tissue samples. The histological changes were observed and analyzed under an optical microscope.
Intestines of sick snakes were aseptically sampled and streaked onto TSA solid medium for pathogen culture and isolation. The suspected colonies were streaked on MAC solid medium and SS (Salmonella Shigella agar) solid medium for identification. The isolated strains showed different morphology on different mediums as shown in Figure 1A. After Gram staining, the isolated bacteria could be seen under ordinary optical microscope (Figure 1B). The invA gene-specific to Salmonella was used to amplify the DNA of the isolated strain by PCR, and the product size of the target fragment was 374bp (Figure 1C). In summary, these characteristics mentioned above are consistent with Salmonella.
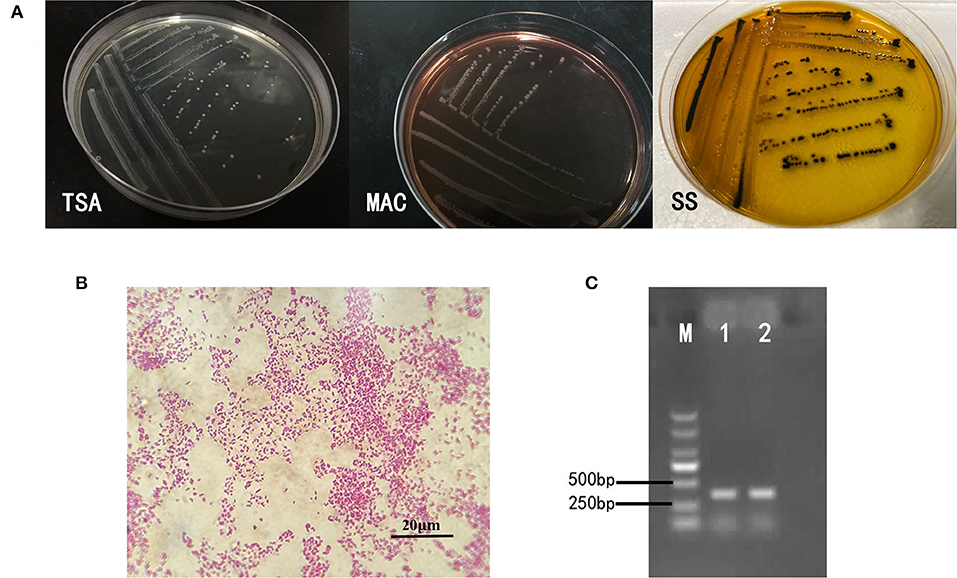
Figure 1. Isolation and identification of bacteria. (A) On the TSA medium, the bacteria presented a round, slightly raised, rounded and smooth surface, translucent colonies. Round translucent colonies with smooth edges appear on MAC identification medium. The strains on SS identification medium were round, slightly convex, with smooth and neat edges, and the center of the colony was black. (B) The bacteria were short shaped, without capsule and spores. Most of them are single. It was Gram-negative bacilli. (C) The invA gene-specific to Salmonella was used to amplify the DNA of the isolated strain by PCR, and the product size of the target fragment was 374 bp, which was the same as that of the reference strain of S. enteritidis.
The serotype of Salmonella identified by the method of glass slide agglutination test was S. enteritidis (-, 9, 12: g, m: 7).
The results of antimicrobial susceptibility tests are shown in Table 4. The results showed that the bacteria were resistant to most drugs, but sensitive to tetracycline and amikacin. The drug susceptibility results of ceftriaxone, kanamycin and neomycin were determined as intermediate.
DNA of the strains was extracted for PCR amplification of 17 drug-resistant genes. The amplification results of drug-resistant genes are shown in Figure 2A. Testing results show in Table 5. BlaTEM of β-lactams was found in the strains. The qnrD gene in 4-quinolones was found in the strains. The aadA1 gene in aminoglycosides was found in the strains. The catA1 in chloramphenicols was found in the strains. The genes sul I and sul II in sulfonamides were found in the strains. Sequencing results showed that the similarity between the six amplified drug-resistant gene sequences and the reference sequences in GenBank was more than 95% and they shared sequence homology.
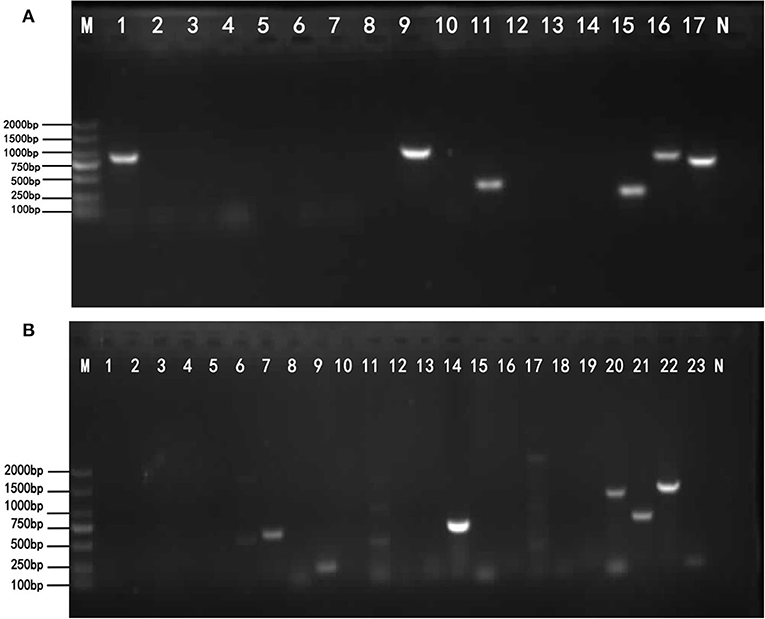
Figure 2. The results of drug-resistant genes and virulence genes. (A) The target resistant genes were amplified by PCR using the primers listed in Table 2. Lane M: DNA Marker DL2000 (from top to bottom is 2,000–1,500–1,000–750–500–250–100 bp). Lane 1–17: Resistance gene; Lane N: Negative control. The lane 1, lane 9, lane 11, lane 15, lane 16, and lane 17 show bright strips, which are, respectively, 643, 984, 343, 258, 822, and 722 bp. The results of the detected genes are shown in Table 5. (B) The target virulence genes were amplified by PCR using the primers listed in Table 3. Lane M: DNA Marker DL2000 (from top to bottom is 2,000–1,500–1,000–750–500–250–100 bp). Lane 1–23: virulence genes; Lane N: Negative control. The lane 7, 8, 9, 14, 20, 21, 22, and 23 show bright strips. The virulence genes corresponding to these positive bands are shown in Table 6.
The 23 pairs of virulence genes were amplified by PCR from isolated S. enteritidis. The amplification results of virulence genes are shown in Figure 2B. The Table 6 shows the virulence gene testing results. Virulence genes avrA, iacP, prgK, ssrA, siiD (spi4D), siiE, spi4H, and pipC were found in the strains. The sequences of these 8 virulence genes were highly homologous with the virulence genes logged on GenBank, and their similarity was more than 95.5%. Therefore, the PCR amplification products of the above 8 virulence genes belonged to the virulence genes of Salmonella.
Twelve chickens were intraperitoneally injected with 0.2mL of fresh bacterial solution at concentration of 4 × 108 CFU/mL. Within 4 h after challenge, some chickens died. Other chickens showed listlessness, loose fur, and body tremors after 48 h to challenge. The dead chickens had stiffened legs. Data were collected for 7 days of continuous observation. In the experimental group, 9 chickens died. While the control group had no obvious symptoms. So, the results showed that S. enteritidis had a 75% fatality rate in chickens.
The concentration of S. enteritidis administrated to chickens of each experimental group was 4 × 109 CFU/mL, 4 × 108 CFU/mL, 4 × 107 CFU/mL, 4 × 106 CFU/mL, respectively, corresponding to A1, A2, A3, and A4 groups. According to the data in Table 7, the modified Kärber method was used to calculate the 50% lethal dose of the strain.
The LD50 result was 107.7 CFU/mL. While, standard error SlogLD50 was 0.2449. Ninety-five percent and Confidence Interval 95% CI.
After the strain was cultured for 10 h, the bacterial quantity of the strain was examined, which was 4 × 109 CFU/mL by plate count method.
After intraperitoneal administration of S. enteritidis suspension with different concentrations into chickens, clinical signs showed as followed (Figure 3).
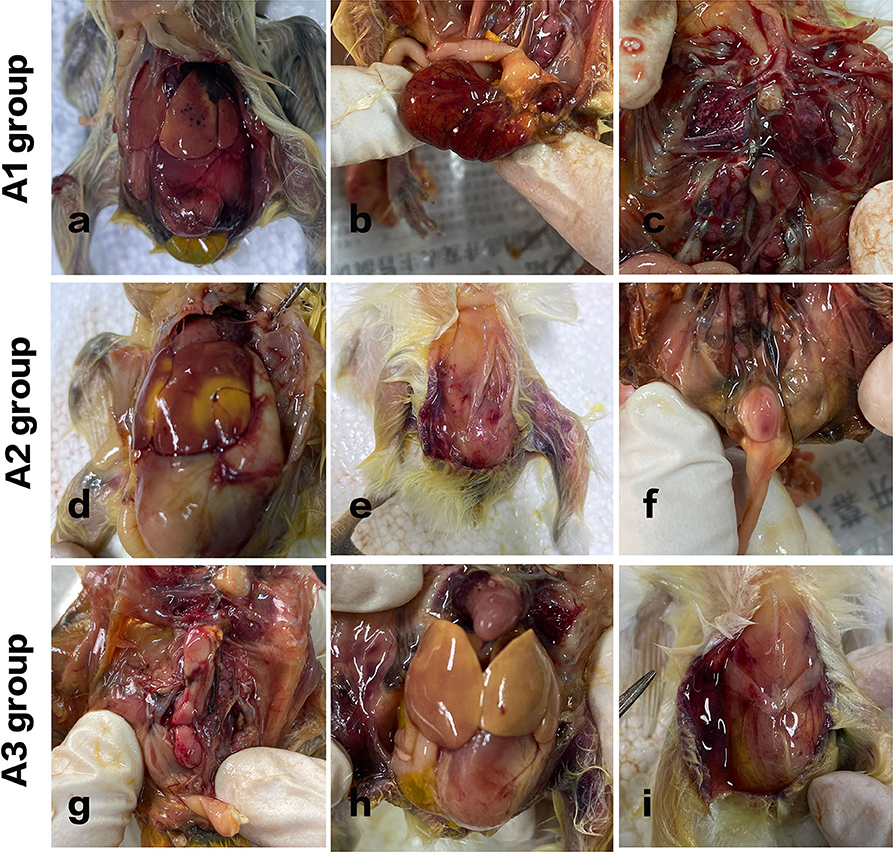
Figure 3. Pathological variation of chicks in different groups. A1 group: Acute septicemia, extensive abdominal bleeding, and redness. (a) Punctate bleeding points on the liver surface were seen. (b) The yolk sac was red and swollen. (c) Hyperemia in flamed lung tissues were seen. A2 group: (d) large yellow plaques and subcutaneous bleeding in the livers of the chickens. (e) Subcutaneous hemorrhage. (f) Enlarged bursa of Fabricius with hemorrhagic spots was observed. A3 group: (g) kidneys were swollen with spotty bleeding. (h) The liver turned yellow. (i) The peritoneum was in a viscous and gelatinous state.
In group A1, all the chickens died within 8 h after injection, and the course of the disease was urgent. Acute septicemia, extensive abdominal bleeding and redness, punctate bleeding points on the liver surface were seen. Kidneys were inflamed, and hyperemia in flamed lung tissues were seen. The control group of chickens showed no obvious redness and swelling in abdominal cavity.
In group A2, two chickens died 8 h after challenge, and the other three curled up in the corner, indolent, slow responsive, and anorexia. Another two died after 12 h administration. The rest of the chickens died within 24 h. And it turns out that large yellow plaques and subcutaneous bleeding in the livers of the chickens. Cecum was enlarged. Lungs were inflamed and congested. Enlarged bursa of Fabricius with hemorrhagic spots was also observed.
In group A3, one chicken died 2 days later after challenge, one chicken died on day 3. Figure 3 shows that the kidneys were swollen with spotty bleeding and the liver turned yellow. The intestinal tissue structure presented unclear boundaries and the peritoneum was in a viscous and gelatinous state.
In group A4, the chickens were bright, alert and responsive. They were euthanized on day 7. Compared with the A5 control group, the body weights were slightly lower.
Acute clinical symptoms developed in the chickens of group A1. Pathological slides of the livers showed that the hepatocytes were swollen with infiltrated inflammatory cells in the hepatic sinus. Fat droplets with varied sizes appeared in the cytoplasm of liver cells. The vacuoles in Figure 4a were small in size, indicating that steatosis of liver cells was less severe. There is also a field of view of vacuolar degeneration of hepatocytes showed in Figure 4b, in which the cellular matrix loosened and became an unstructured cystic space. The pathological changes of kidney in infected dead chicks showed numerous inflammatory cells oozing out in acute dead chickens' kidneys (Figure 4c), and the epithelial cells of the renal tubules were blurred, with bacterial accumulation in Figure 4d.
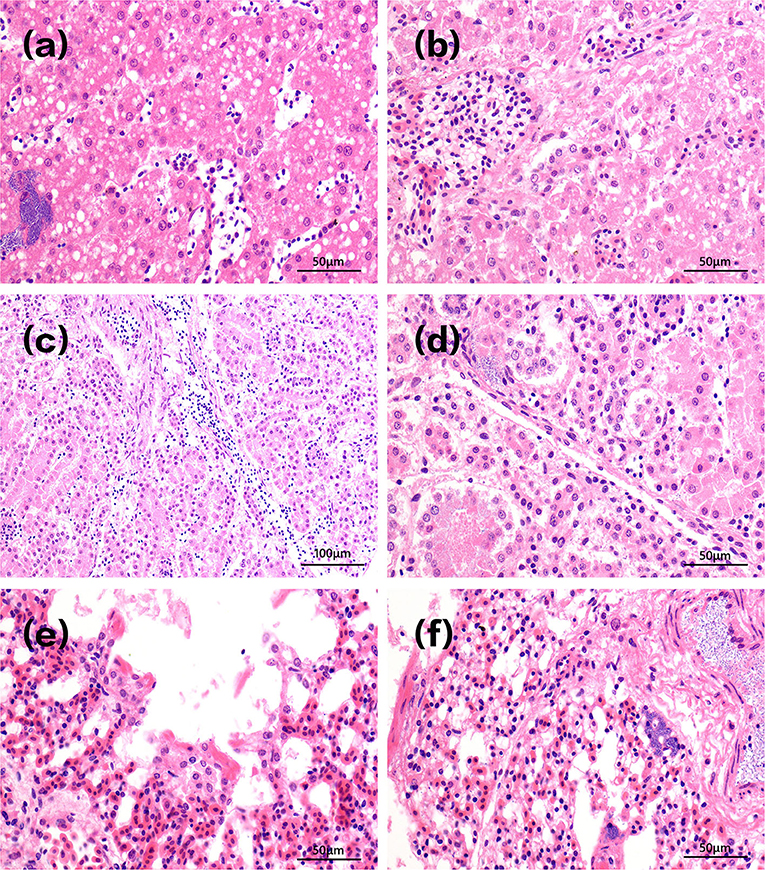
Figure 4. Pathological changes of tissues in infected dead chicks. Pathological changes of chick liver in acute A1 group are shown in (a,b). The vacuoles in (a) were small in size, indicating that steatosis of liver cells was less severe. There is also a field of view of vacuolar degeneration of hepatocytes showed in (b), in which the cellular matrix loosened and became an unstructured cystic space. Pathological changes of kidney in infected dead chicks were shown in (c,d). (c) Inflammatory cells infiltrate chicken kidneys. (d) The epithelial cells of renal tubules were swollen and their structures were blurred, bacterial accumulation. Pathological changes of the lung at different stages of infection were shown in (e,f). (e) Pulmonary capillary hyperemia, bacterial accumulation. (f) Alveolar serous fluid oozes out and alveolar epithelial cells proliferate and vacuoles appear in the cytoplasm.
Figure 4e shows infected lungs that congested pulmonary septum capillaries, a large number of neutrophils exudate, and rod-shaped bacteria in alveolar space can be seen in infected chickens that died at day 1. A large number of serous exudates from the alveolar cavity, mixed with more neutrophils, red blood cells, and exfoliated epithelial cells was seen 3 days after infection. After 10 days of injection, the alveolar structures were almost indistinguishable, alveolar epithelial cells proliferated, and alveolar cells showed vacuoles of different sizes in Figure 4f.
Salmonella enterica is considered an important zoonotic enterogenic pathogen. S. enterica can be divided into 6 subspecies and more than 2,600 serotypes, which can cause different degrees of infection (17). According to previous reports in other countries, frequent Salmonella infection in reptiles has been confirmed, suggesting that reptiles are also natural hosts for Salmonella (25–27). Reptiles are known to transmit Salmonella intermittently (28) and shed into the environment irregularly. In addition, bacteria-carrying fecal sprinkled on the surface of the earth will flow along with the rain and contaminate the soil, land crops, or residential water (29–31), which is difficult to eradicate through antibiotic treatment. Therefore, there may be risks for both carriers and people in close contact with these animals (32).
Contaminated water or food is thought to be the main source of Salmonella transmission. Currently, Salmonella prevention mainly focuses on water and food safety. Developed countries and areas such as Europe and the United States are able to implement procedures to prevent S. enteritidis transmission while developing countries and regions still need improvements. In addition to becoming Salmonella-contaminated food, edible snakes can also pollute water sources and transmit the bacteria to other animal farms. Infected chickens can also be the source for Salmonella infection in humans, leading to global public health concern (33).
Current published studies mostly focused on the increasing popularity of reptiles as pets and their possible role as hosts for pathogenic microorganisms. Frequent touch between reptiles and humans is considered a public health problem because these animals have been identified as carriers of zoonotic infectious diseases, especially Salmonella associated with human salmonellosis (32). In a Brazilian study, Salmonella isolated from reptiles was identified with a karyotype that has been reported to infect humans (34). Little is known about how reptiles carry pathogenic microorganisms (35). The information about major subspecies and serotypes of Salmonella isolated from reptiles is limited. Salmonella is isolated from snakes often more than other reptiles (36), possibly because of the snakes' dietary habits. Salmonella spreads fastest among carnivorous reptiles, such as snakes (37). This study investigated the incidence of pneumonia on a chicken-fed Chinese snake farm. It is likely that Salmonella from chicken meat accumulates in the snakes. Chicken was originally thought to be the main host of Salmonella (11). Chicken meats the potential source to infect this snake farm. Therefore, the pathogenicity of S. enteritidis isolated from snake pneumonia was studied in chickens.
In order to explore the pathogenicity of S. enteritidis isolated from snake pneumonia, the strain was administrated intraperitoneally in 3-day-old chicks, and half of the lethal dose of the strain was examined, as well as the pathological changes of each organ of the chicks. In the bacterial strain infection model, acute sepsis was observed in the highest dose group A1, which was consistent with previous research results (38–41). The natural infection of Salmonella enterica leads to systemic and persistent infection, which potentially causes bacteria to spread to other hosts, including in the poultry industry (42–44). Obvious redness and congestion were also seen in the lungs. Due to the rapid onset and short course of the disease, no respiratory symptoms were observed in the chickens. Bacteria solution was administrated intraperitoneally, and half of the lethal dose was 107.7 CFU/mL. For the high-dose A1 and A2 groups, the mortality rate was 100%, with all deaths occurring within 24 h. In addition, the lower dose groups A3 and A4 showed decreased weight gain after Salmonella infection, and the body weight began to decrease 5 days after infection. Chickens challenged with Salmonella showed symptoms of anorexia, diarrhea with high mortality. Chickens that had made through the infection also suffered stunted growth. Results of the pathogenicity test showed that this strain is highly pathogenic in warm-blooded animals with a low median lethal dose, and the potential human transmission route needs further study. Existing studies have shown that Salmonella from reptile sources is more likely to transmit to human beings than Salmonella from non-reptile sources. Therefore, reptile-related Salmonella should be considered to be a global threat to public health (12) and worthy of vigilance.
PCR detection of Salmonella virulence genes has been widely used as a predictive measurement of Salmonella virulence. It is known that there are significant genetic variations in virulence gene sequences between Salmonella serotypes. This variability in the genome sequence may translate into changes in function, leading to differences in pathogenicity. However, it must be noted that having one or more virulence genes does not confer pathogenicity on a strain unless the strain acquires the appropriate virulence gene combination to cause disease in a particular host species (45). The Salmonella type III secretory protein system, composed of proteins encoded by the virulence island gene, also plays a key role in the pathogenicity of Salmonella (46). The low dose of LD50 and the presence of multiple virulence genes indicate that the number of virulence genes in the strain is related to the pathogenicity.
Antimicrobial susceptibility test showed that the isolated Salmonella enteritidis was resistant to the vast majority of the 19 selected antimicrobials, and some do not appear bacteriostatic ring at all. It is only sensitive to tetracycline, amikacin, kanamycin, neomycin, and ceftriaxone. Currently, 4-quinolonesand β-lactams are mainly used to treat Salmonella infection. Notably, cephalosporin is an antimicrobial agent that can be used to treat human urinary tract infections (47, 48), and ciprofloxacin is a quinolone that is of great importance in human medicine and is mainly used to treat severe infections (49). However, in this study, isolated Salmonella were resistant to the selected third generation quinolones. The isolated strains also showed low sensitivity to β-lactams antibiotics. Potential drug resistance could occur in human beings with salmonellosis if such strains transmitted into people (50).
The antimicrobial resistance of bacteria has a certain relationship with the resistance genes carried by bacteria. The test results showed that the resistance phenotype of the strains was basically in consistent with the detection rate of antimicrobial resistance genes. For each selected drug-resistant gene category, the corresponding drug-resistant genes were detected in β-lactams, 4-quinolones, Aminoglycosides, Tetracyclines, Chloramphenicols, and Sulfonamides. As time goes on, bacterial resistance rates will also continue to rise, and antimicrobial resistance profiles will become more complex, perhaps leading to more intense foodborne salmonellosis. The chromosomal resistance can be transmitted to the next generation and also at the level of different genera, which greatly expands the number and range of drug-resistant bacteria, leading to the increasingly serious phenomenon of Salmonella resistance. At the same time, the resistance of pathogenic Salmonella strains of the same animal source isolated from different countries or regions in the same time period or in the same country or region in different time periods also varies, that is, the expression has certain locality and timeliness (20, 51). Pathogenic strains from different animal sources vary even more. Therefore, it is necessary to strengthen the study of antimicrobial resistance phenotype of Salmonella. The recombination of virulence genes and antimicrobial resistance gives Enterobacteriaceae a survival advantage when exposed to a drug environment that is unfavorable to growth, posing a serious threat to public health. This raises the potential risk to human health from these bacteria. In summary, this is the first report on Salmonella enteritis strains isolated from edible snakes with pneumonia in China. The isolated strains were extensively antimicrobial resistant, and potentially pathogenic. Thereby, rising public health concerns. Further studies should focus the underlying mechanisms involved in the possible spread of these strains to humans.
All datasets generated for this study are included in the article/supplementary material.
This animal study was reviewed and approved by Institution's Animal Care and Health and Supervisory Committee of Huazhong Agriculture University, Wuhan, China.
YS and YX: conceived and designed the experiments, analyzed the data, and wrote the paper. YS: provided the material and revised the article. YX and HL: performed the experiments. All authors contributed to the article and approved the submitted version.
This study was supported by the Fundamental Research Funds for the Central Universities, China (2662017QD015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
CFU, Colony-Forming Units; CLSI, Clinical and Laboratory Standards Institute; K-B, Kirby-Bauer; LD50, Median Lethal Dose; PBS, Phosphate Buffered Saline; S. enterica, Salmonella enterica ssp. Enterica; S. enteritidis, Salmonella enterica serovar Enteritidis; SPF, Specific Pathogen Free; SS, Salmonella Shigella Agar; S. typhimurium, Salmonella Typhimurium; TSA, Tryptose Soya Agar; TSB, Tryptic Soy Broth.
1. Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, Nair S, et al. Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res Microbiol. (2014) 165:526–30. doi: 10.1016/j.resmic.2014.07.004
2. Bern C, Martines J, de Zoysa I, Glass RI. The magnitude of the global problem of diarrhoeal disease: a ten-year update. Bull World Health Organ. (1992) 70:705–14.
3. Graham SM. Salmonellosis in children in developing and developed countries and populations. Curr Opin Infect Dis. (2002) 15:507–12. doi: 10.1097/00001432-200210000-00009
4. Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. (2003) 81:197–204. doi: 10.1590/S0042-96862003000300010
5. Wang S, Duan H, Zhang W, Li J-W. Analysis of bacterial foodborne disease outbreaks in China between 1994 and 2005. FEMS Immunol Med Microbiol. (2007) 51:8–13. doi: 10.1111/j.1574-695X.2007.00305.x
6. Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschäpe H, Adams LG, et al. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun. (2002) 70:2249–55. doi: 10.1128/IAI.70.5.2249-2255.2002
7. Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. (2004) 82:346–53. doi: 10.1590/S0042-96862004000500008
8. Wilks C, Parkinson G. International Review of Salmonella enteritidis Epidemiology and Control Policies. A Report for the Rural Industries Research and Development Corporation RIRDC Project No. DAV-146A (2000).
9. Sanchez-Vargas FM, Abu-El-Haija MA, Gomez-Duarte OG. Salmonella infections: an update on epidemiology, management, and prevention. Travel Med Infect Dis. (2011) 9:263–77. doi: 10.1016/j.tmaid.2011.11.001
10. Eguale T, Gebreyes WA, Asrat D, Alemayehu H, Gunn JS, Engidawork E. Non-typhoidal Salmonella serotypes, antimicrobial resistance and co-infection with parasites among patients with diarrhea and other gastrointestinal complaints in Addis Ababa, Ethiopia. BMC Infect Dis. (2015) 15:497. doi: 10.1186/s12879-015-1235-y
11. Chousalkar K, Gole VC. Salmonellosis acquired from poultry. Curr Opin Infect Dis. (2016) 29:514–19. doi: 10.1097/QCO.0000000000000296
12. Murphy D, Oshin F. Reptile-associated salmonellosis in children aged under 5 years in South West England. Arch Dis Child. (2015) 100:364–65. doi: 10.1136/archdischild-2014-306134
13. Whiley H, Gardner MG, Ross K. A review of Salmonella and squamates (lizards, snakes and amphibians): implications for public health. Pathogens. (2017) 6:38. doi: 10.3390/pathogens6030038
14. Pulford CV, Wenner N, Redway ML, Rodwell EV, Webster HJ, Escudero R, et al. The diversity, evolution and ecology of Salmonella in venomous snakes. PLoS Negl Trop Dis. (2019) 13:e0007169. doi: 10.1371/journal.pntd.0007169
15. Rodrigue DC, Tauxe RV, Rowe B. International increase in Salmonella enteritidis: a new pandemic? Epidemiol Infect. (1990) 105:21–7. doi: 10.1017/S0950268800047609
16. Von Mollendorf C, Cohen C, Tempia S, Meiring S, de Gouveia L, Quan V, et al. Epidemiology of serotype 1 invasive pneumococcal disease, South Africa, 2003-2013. Emerg Infect Dis. (2016) 22:261–70. doi: 10.3201/eid2202.150967
17. Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemühl J, Grimont PA, et al. Supplement 2003-2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. (2010)161:26–9. doi: 10.1016/j.resmic.2009.10.002
18. Clinical and Laboratory Standards Institute (CLSI). M31-A3. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Testing for Bacteria Isolated From Animal. Approved Standard-Third Edition (2008).
19. Clinical and Laboratory Standards Institute (CLSI). M100-S22. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Second Information Supplement (2012).
20. Lu Y, Wu C-M, Wu G-J, Zhao H-Y, He T, Cao X-Y, et al. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog Dis. (2011) 8:45–53. doi: 10.1089/fpd.2010.0605
21. Nde CW, Logue CM. Characterization of antimicrobial susceptibility and virulence genes of Salmonella serovars collected at a commercial turkey processing plant. J Appl Microbiol. (2008) 104:215–23. doi: 10.1111/j.1365-2672.2007.03535.x
22. Berndt A, Müller J, Borsi L, Kosmehl H, Methner U, Berndt A. Reorganisation of the caecal extracellular matrix upon Salmonella infection–relation between bacterial invasiveness and expression of virulence genes. Vet Microbiol. (2009) 133:123–37. doi: 10.1016/j.vetmic.2008.06.025
23. Foley SL, Johnson TJ, Ricke SC, Nayak R, Danzeisen J. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol Mol Biol Rev. (2013) 77:582–607. doi: 10.1128/MMBR.00015-13
24. Osman KM, Marouf SH, Erfan AM, Alatfeehy N. Salmonella enterica in imported and domestic day-old turkey poults in Egypt: repertoire of virulence genes and their antimicrobial resistance profiles. Rev Sci Tech. (2014) 33:1017–26. doi: 10.20506/rst.33.3.2338
25. de Sa IVA, Solari CA. Salmonella in Brazilian and imported pet reptiles. Braz J Microbiol. (2001) 32:293–7. doi: 10.1590/S1517-83822001000400007
26. Chen C-Y, Chen W-C, Chin S-C, Tung K-C, Chiou C-S, Hsu Y-M, et al. Prevalence and antimicrobial susceptibility of salmonellae isolates from reptiles in Taiwan. J Vet Diagn Invest. (2010) 22:44–50. doi: 10.1177/104063871002200107
27. Gay N, Le Hello S, Weill F-X, de Thoisy B, Berger F. Salmonella serotypes in reptiles and humans, French Guiana. Vet Microbiol. (2014) 170:167–71. doi: 10.1016/j.vetmic.2014.01.024
28. Burnham BR, Atchley DH, Defusco RP, Ferris KE, Zicarelli JC, Lee H, et al. Prevalence of fecal shedding of Salmonella organisms among captive green iguanas and potential public health implications. J Am Vet Med Assoc. (1998) 213:48–50.
29. Harrison RA, Hargreaves A, Wagstaff SC, Faragher B, Lalloo DG. Snake envenoming: a disease of poverty. PLoS Negl Trop Dis. (2009) 3:e569. doi: 10.1371/journal.pntd.0000569
30. Warrell DA, Greenwood BM, Davidson NM, Ormerod LD, Prentice CR. Necrosis, haemorrhage and complement depletion following bites by the spitting cobra (Naja nigricollis). Q J Med. (1976) 45:1–22.
31. Ogunfowokan O, Jacob DA, Livinus OL. Relationship between bite-to-hospital time and morbidity in victims of carpet viper bite in North-Central Nigeria. West Afr J Med. (2011) 30:348–53.
32. Centers for Disease Control and Prevention (CDC). Reptile-associated salmonellosis–selected states, 1998-2002. MMWR Morb Mortal Wkly Rep. (2003) 52:1206–9.
33. Antunes P, Mourao J, Campos J, Peixe L. Salmonellosis: the role of poultry meat. Clin Microbiol Infect. (2016) 22:110–21. doi: 10.1016/j.cmi.2015.12.004
34. Ramos CP, Santana JA, Morcatti Coura F, Xavier RGC, Leal CAG, Oliveira C, et al. Identification and characterization of Escherichia coli, Salmonella spp., Clostridium perfringens, and C. difficile isolates from reptiles in Brazil. Biomed Res Int. (2019) 2019:9530732. doi: 10.1155/2019/9530732
35. Mermin J, Hutwagner L, Vugia D, Shallow S, Daily P, Bender J, et al. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin Infect Dis. (2004) 3:S253–61. doi: 10.1086/381594
36. Wheeler E, Hong P-Y, Bedon LC, Mackie RI. Carriage of antibiotic-resistant enteric bacteria varies among sites in Galapagos reptiles. J Wildl Dis. (2012) 48:56–67. doi: 10.7589/0090-3558-48.1.56
37. Chlebicz A, Slizewska K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as zoonotic foodborne diseases: a review. Int J Environ Res Public Health. (2018) 15:863. doi: 10.3390/ijerph15050863
38. Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. The Salmonella enterica pan-genome. Microbial Ecol. (2011) 62:487–504. doi: 10.1007/s00248-011-9880-1
39. Swart AL, Hensel M. Interactions of Salmonella enterica with dendritic cells. Virulence. (2012) 3:660–7. doi: 10.4161/viru.22761
40. Monack DM. Salmonella persistence and transmission strategies. Curr Opin Microbiol. (2012) 15:100–7. doi: 10.1016/j.mib.2011.10.013
41. Ilyas B, Tsai CN, Coombes BK. Evolution of -host cell interactions through a dynamic bacterial genome. Front Cell Infect Microbiol. (2017) 7:428. doi: 10.3389/fcimb.2017.00428
42. Guard-Petter J, Henzler DJ, Rahman MM, Carlson RW. On-farm monitoring of mouse-invasive Salmonella enterica serovar enteritidis and a model for its association with the production of contaminated eggs. Appl Environ Microbiol. (1997) 63:1588–93. doi: 10.1128/AEM.63.4.1588-1593.1997
43. Marin C, Martelli F, Rabie A, Davies R. Commercial Frozen mice used by owners to feed reptiles are highly externally contaminated with Salmonella Enteritidis PT8. Vector Borne Zoonotic Dis. (2018) 18:453–57. doi: 10.1089/vbz.2018.2295
44. Guard J, Cao G, Luo Y, Baugher JD, Davison S, Yao K, et al. Genome sequence analysis of 91 Salmonella enteritidis isolates from mice caught on poultry farms in the mid (1990s). Genomics. (2019) 112:528–544. doi: 10.1101/301432
45. Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, et al. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS ONE. (2013) 8:e58449. doi: 10.1371/journal.pone.0058449
46. Amavisit P, Lightfoot D, Browning GF, Markham PF. Variation between pathogenic serovars within Salmonella pathogenicity islands. J Bacteriol. (2003) 185:3624–35. doi: 10.1128/JB.185.12.3624-3635.2003
47. Gunduz S, Uludag Altun H. Antibiotic resistance patterns of urinary tract pathogens in Turkish children. Glob Health Res Policy. (2018) 3:10. doi: 10.1186/s41256-018-0063-1
48. Beetz R, Westenfelder M. Antimicrobial therapy of urinary tract infections in children. Int J Antimicrob Agents. (2011) 38(Suppl.):42–50. doi: 10.1016/j.ijantimicag.2011.09.006
49. World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine. 5th ed. Geneva: World Health Organization (2017).
50. Clinical and Laboratory Standards Institute [CLSI]. M100-S25. Performance Standards for Antimicrobial Susceptibility Testing, Twenty-Fi h Informational Supplement. Vol. 35. Wayne, PA: Clinical and Laboratory Standards Institute (2015).
Keywords: Salmonella enteritidis, antimicrobial resistance, edible snakes, foodborne pathogen, pathogenicity
Citation: Xia Y, Li H and Shen Y (2020) Antimicrobial Drug Resistance in Salmonella enteritidis Isolated From Edible Snakes With Pneumonia and Its Pathogenicity in Chickens. Front. Vet. Sci. 7:463. doi: 10.3389/fvets.2020.00463
Received: 03 April 2020; Accepted: 23 June 2020;
Published: 04 August 2020.
Edited by:
Guillermo Tellez, University of Arkansas, United StatesReviewed by:
Daniel Hernandez-Patlan, Universidad Nacional Autonóma de México, MexicoCopyright © 2020 Xia, Li and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoqin Shen, eXNoZW5AbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.