- 1Department of Veterinary Medicine, School of Veterinary Medicine, College of Bioresources and Agriculture, National Taiwan University, Taipei, Taiwan
- 2Biology Division, Animal Health Research Institute, New Taipei City, Taiwan
- 3National Taiwan University Veterinary Hospital, Taipei, Taiwan
Extended-spectrum β-lactamases (ESBLs) are enzymes that mediate resistance to newer β-lactam antibiotics, including extended-spectrum cephalosporins and monobactams. The production of ESBL is primarily plasmid mediated, and such plasmids often comprise the genes that encode resistance to other classes of antimicrobials, such as aminoglycosides and fluoroquinolones. Therefore, ESBL-producing microorganisms leave clinicians with limited therapeutic options in both human and veterinary medicine. Compared with human medicine, information regarding ESBL-producing microorganisms is limited in veterinary medicine. We screened for ESBL-producing Escherichia coli in dogs and cats admitted to National Taiwan University Veterinary Hospital, Taipei, from 2014 to 2017 and further analyzed the genotypes and phylogenetic traits of these ESBL producers. Double disk tests specified by the Clinical and Laboratory Standards Institute were performed on 283 E. coli isolates and revealed a total of 65 E. coli (54 from dogs and 11 from cats) with the ESBL phenotype (22.8%). blaCTX−M−1 group and blaCTX−M−2group were the most commonly identified ESBL gene groups. blaCTX−M−55 was the main ESBL gene within the blaCTX−M−1group, whereas the blaCTX−M−2group contained only blaCTX−M−124. The ESBL-producing E. coli were all resistant to ampicillin. The resistance rate to ceftiofur, doxycycline, enrofloxacin, and ciprofloxacin was 93.8, 73.8, 80, and 78.5%, respectively. Of the antibiotics tested, greater sensitivity to imipenem and gentamicin was noted. Multilocus sequence typing indicated that ST457, ST131, and ST648 were the most common sequence types. Our study identified eight ST131/O25b isolates, which is a global zoonotic clone of public health concern. The major ESBL genes of these clones were blaCTX−M−174 and blaCTX−M−194. Because companion animals such as dogs and cats are in close contact with humans, the characterization of ESBL producers originating from them is crucial from the perspective of both public health and veterinary medicine.
Introduction
Escherichia coli, a type of Gram-negative bacteria is a ubiquitous inhabitant of the gastrointestinal tract of both humans and animals. This microorganism frequently causes urinary tract, skin, or soft tissue infections in cats and dogs (1). Commonly prescribed medications to treat E. coli infection in companion animals include ampicillin, amoxicillin-clavulanic acid, fluoroquinolones, or cephalosporins. However, the emergence of drug-resistant bacteria encountered in clinical practice decreases the therapeutic efficacy of these antimicrobial agents. One major mechanism of this drug resistance is the production of enzymes by microbes to inactivate antimicrobial agents. For example, β-lactam agents are widely used to treat bacterial infections in veterinary medicine, whereas extended-spectrum β-lactamases (ESBLs) are a group of enzymes that mediate resistance to most β-lactam antibiotics, including extended-spectrum cephalosporins and monobactams but excluding carbapenems and cephamycins (2). ESBLs are inhibited by clavulanic acid, sulbactam, and tazobactam; this fact is used as a criterion to classify β-lactamases and for ESBL diagnosis purposes (3). TEM, SHV, and CTX-M-group enzymes are examples of commonly encountered ESBLs (2). ESBL producers usually exhibit a multi-drug-resistant phenotype. In addition, the ESBL genes are mainly plasmid mediated, thus facilitating the transmission of drug-resistant genes to other bacteria. Such a situation poses a challenge for infection management in clinical practice. ESBLs have been previously documented primarily in human clinical cases (4). Because companion animals such as dogs and cats are in close contact with humans, they could contract ESBL-producing microorganisms from humans and then possibly transmit them back to humans, which represents a public health concern (5).
Information regarding the prevalence of ESBL producers or the genotypes of these clinical isolates from cats and dogs is limited in Taiwan. It is imperative to investigate related matters from both a veterinary medicine and public health perspective (6). The present study analyzed a collection of E. coli isolates obtained from National Taiwan University Veterinary Hospital (NTUVH), a university-based veterinary teaching hospital in Taipei, from 2014 to 2017 to determine the prevalence of ESBL-producing E. coli, assess their antimicrobial profile, and characterize the strains phylogenetically through multilocus sequence typing (MLST). The results obtained should provide insights into the role of ESBL-producing E. coli in companion animals. Some of the data herein have previously been reported at a conference (7).
Materials and Methods
Sample Collection
NTUVH is a teaching hospital affiliated with the College of Bioresources and Agriculture at National Taiwan University located in Taipei, Taiwan. Between 2014 and 2017, 283 E. coli isolates obtained from dogs (n = 224) and cats (n = 59) that were admitted to NTUVH were screened for ESBL producers. These E. coli isolates were cultured from different sources of the animals and identified using a Vitek 2 Compact (Biomérieux, Marcy-I'Etoile, France) to the species level and stored at −80°C. Urine and pus samples from the uterus or wounds comprised almost 70% (47 and 22%, respectively) of the E. coli sources. These samples were collected from the animals to facilitate diagnosis and treatment. An ethical review was not required for this study.
ESBL Phenotype Testing
The ESBL producers of E. coli were tested using combination disk tests with cefotaxime and ceftazidime (30 μg), with and without clavulanic acid (10 μg), as specified by the Clinical and Laboratory Standards Institute (8). Briefly, the tested E. coli were plated on Muller–Hinton agar at a concentration of 0.5 McFarland standards and incubated at 35°C for 16–18 h. A difference of 5 mm or more in the inhibition zones for either cefotaxime or the ceftazidime–clavulanic acid combination vs. the corresponding cefotaxime or ceftazidime alone was defined as an ESBL-producing E. coli. Klebsiella pneumoniae ATCC 700603 and E. coli ATCC 25922 were used as the positive and negative controls, respectively.
Detection of bla Genes
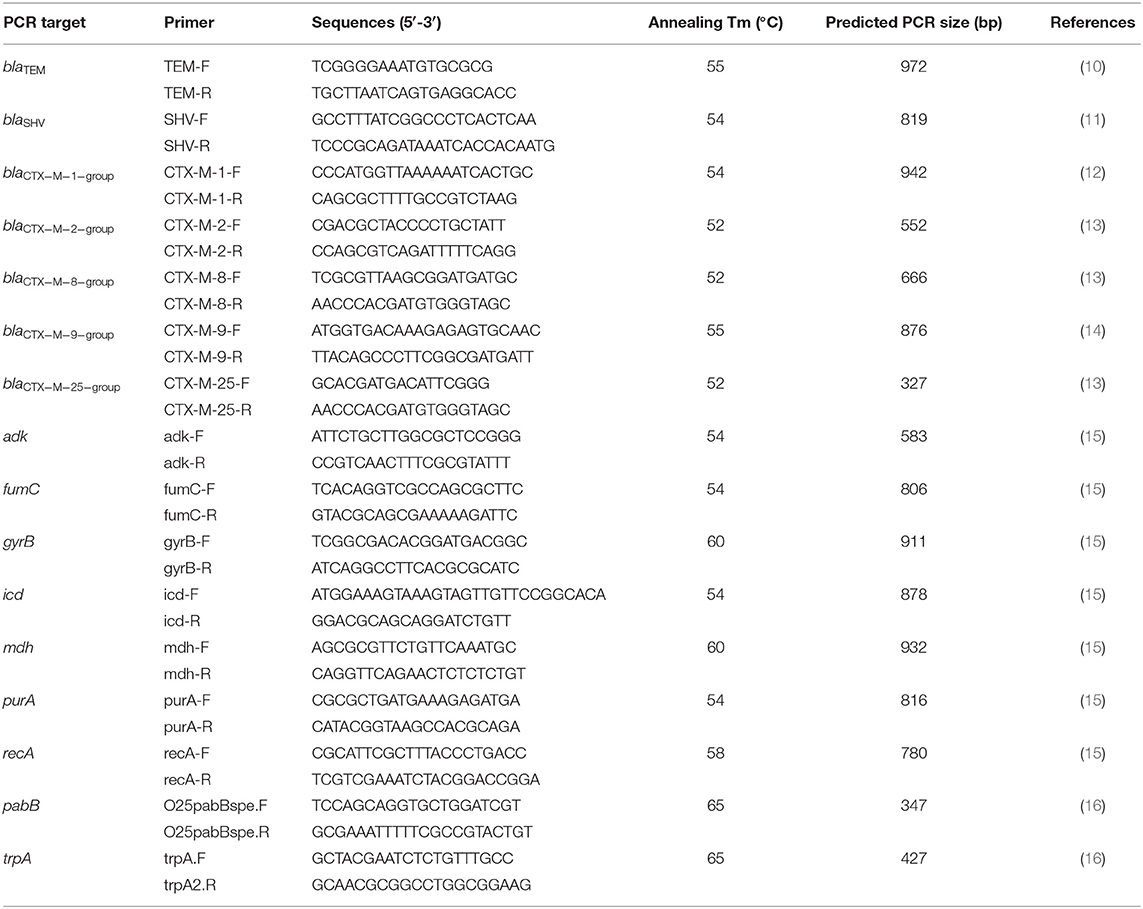
The E. coli isolates that were phenotypically ESBL producers were analyzed using polymerase chain reaction (PCR) to detect their bla genes. Bacterial DNA was extracted using the boiling method (9). Briefly, bacterial strains were cultured overnight at 37°C on tryptic soy agar plates (Difco/Becton Dickinson, Franklin Lakes, NJ), and a loopful of cells was boiled in 200 μL of ddH2O for 10 min. The supernatant was saved after centrifugation at 12,000 × g for 10 min and used as the source of template DNA for PCR. The primers used to amplify blaCTX−M−1−group, blaCTX−M−2−group, blaCTX−M−8−group, blaCTX−M−9−group, blaCTX−M−25−group, blaSHV, blaTEM, and the expected PCR product sizes are listed in Table 1. The PCR cycling conditions were as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, annealing at 52–55°C (as specified in Table 1) for 30 s, and a 72°C extension for 1 min. Ten microliters of each PCR sample were loaded onto a 1.5% agarose gel and electrophoresed at 100 V for 30 min. The gels were then stained with a fluorescent nucleic acid dye (Biotium, Hayward, CA) and examined under ultraviolet illumination. The PCR products were then purified using a GeneJet PCR purification kit (Thermo Fisher Scientific, Waltham, MA) according to the protocol provided by the manufacturer and subjected to sequencing (Mission Biotech, Taipei, Taiwan). The DNA sequences were examined using the Beta-Lactamase DataBase (www.bldb.eu) (17).
Antibiotic Susceptibility Test
The ESBL-producing E. coli isolates were tested for susceptibility to antimicrobial agents used in clinical settings using the standard Kirby–Bauer disk diffusion method (8). The antimicrobial agents tested included β-lactams (amoxycillin/clavulanic acid, ampicillin, imipenem, and ceftiofur), tetracyclines (doxycycline), quinolones (enrofloxacin and ciprofloxacin), aminoglycosides (gentamicin), and sulfonamides (sulfamethoxazole/trimethoprim). The isolates were classified as susceptible, intermediate resistant, or resistant to the antimicrobial agents.
Genotyping and Phylogenetic Analysis
The ESBL-producing E. coli strains were genotyped using MLST (15). Internal fragments of adk, fumC, gyrB, icd, mdh, purA, and recA were amplified through a PCR by using the primers listed in Table 1 and sequenced. They were then uploaded to the EnteroBase MLST website (http://enterobase.warwick.ac.uk/) for comparison. Phylogenetic analysis of the strains was performed using BioNumerics version 7.0 (Applied Maths, Sint-Martens-Latem, Belgium).
E. coli ST131 O25b Detection
The PCR-based detection of E. coli ST131/O25b was based on the method described by Clermont et al. (16). The trpA and pabB primers and annealing temperature used are listed in Table 1. The PCR cycling conditions were as follows: initial denaturation at 94°C for 4 min, followed by 30 cycles at 94°C for 5 s, annealing at 65°C for 10 s, and 72°C extension for 5 min. Ten microliters of each PCR sample was loaded onto 2.0% agarose gel and electrophoresed at 100 V for 30 min. The gels were then stained with a fluorescent nucleic acid dye (Biotium) and examined under ultraviolet illumination.
Results
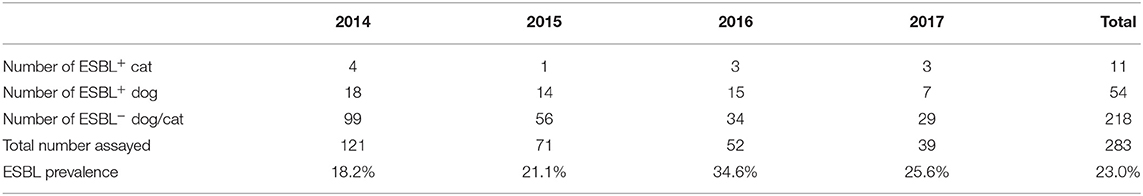
A total of 283 E. coli isolates (59 from cats and 224 from dogs) were obtained during our study period (2014–2017). Table 2 lists the prevalence of ESBL-producing E. coli from dogs and cats. In total, 65 ESBL-producing E. coli isolates, 54 from dogs and 11 from cats, were acquired from our assay. The prevalence of ESBL-producing E. coli isolates was 24.1% (54/224) in dogs and 18.6% (11/59) in cats, and the total prevalence for both animals was 23.0% (65/283).
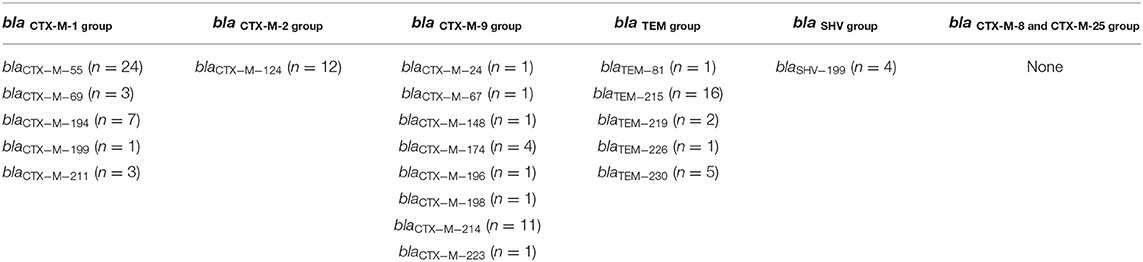
Table 3 lists the distribution of bla genes from the 65 ESBL-producing E. coli isolates. blaCTX−M−55 of the blaCTX−M−1group was the most prevalent bla gene encountered. The blaCTX−M−2group contained only blaCTX−M−124. The blaCTX−M−9group contained eight bla gene types, and blaCTX−M−214 was the most frequently observed. blaTEM−215 was the most common type encountered in the blaTEMgroup. We only detected blaSHV−199 in the blaSHV group. We did not detect blaCTX−M−8group or blaCTX−M−25group.
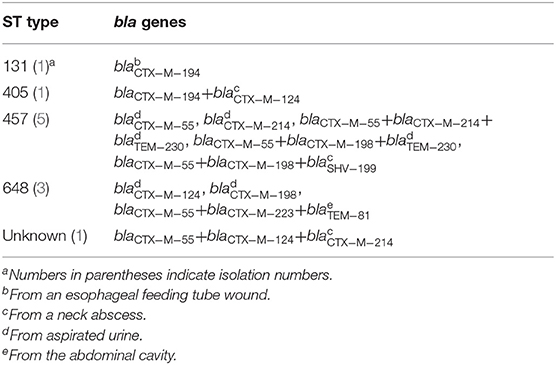
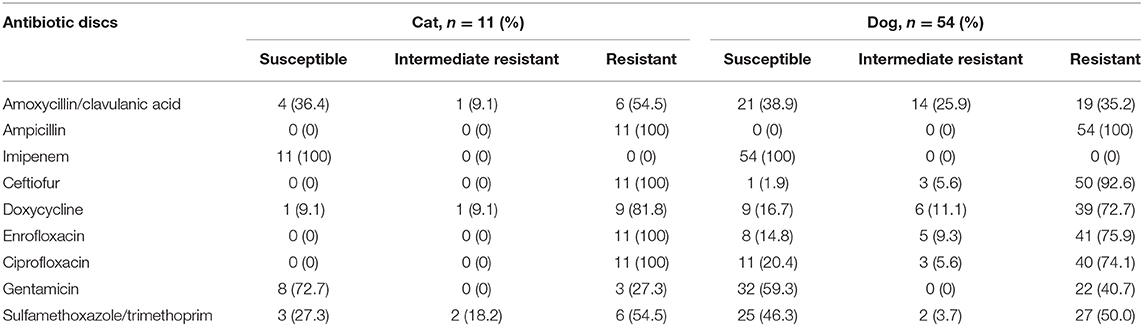
The sequence type (ST), bla genes, and the sampling sites of the ESBL-producing E. coli isolates from cats and dogs, respectively, are detailed in Tables 4, 5. MLST analysis identified 20 STs in our ESBL-producing E. coli isolates. In total, 16 E. coli isolates had STs that did not match any ST in the MLST databank. Combining the data of cats and dogs revealed that the commonest ST was ST457 (13/65, 20.0%), followed by ST131 (10/65, 15.4%), ST648 (6/65, 9.2%), ST38 (3/65, 4.6%), and ST405 (2/65, 3.1%); the other STs were encountered once. ESBL-producing E. coli were isolated from several sites but were principally observed in aspirated urine (44/65, 67.7%). Figure 1 reveals the minimal spanning tree of the 65 ESBL-producing E. coli STs according to the degree of allele sharing.
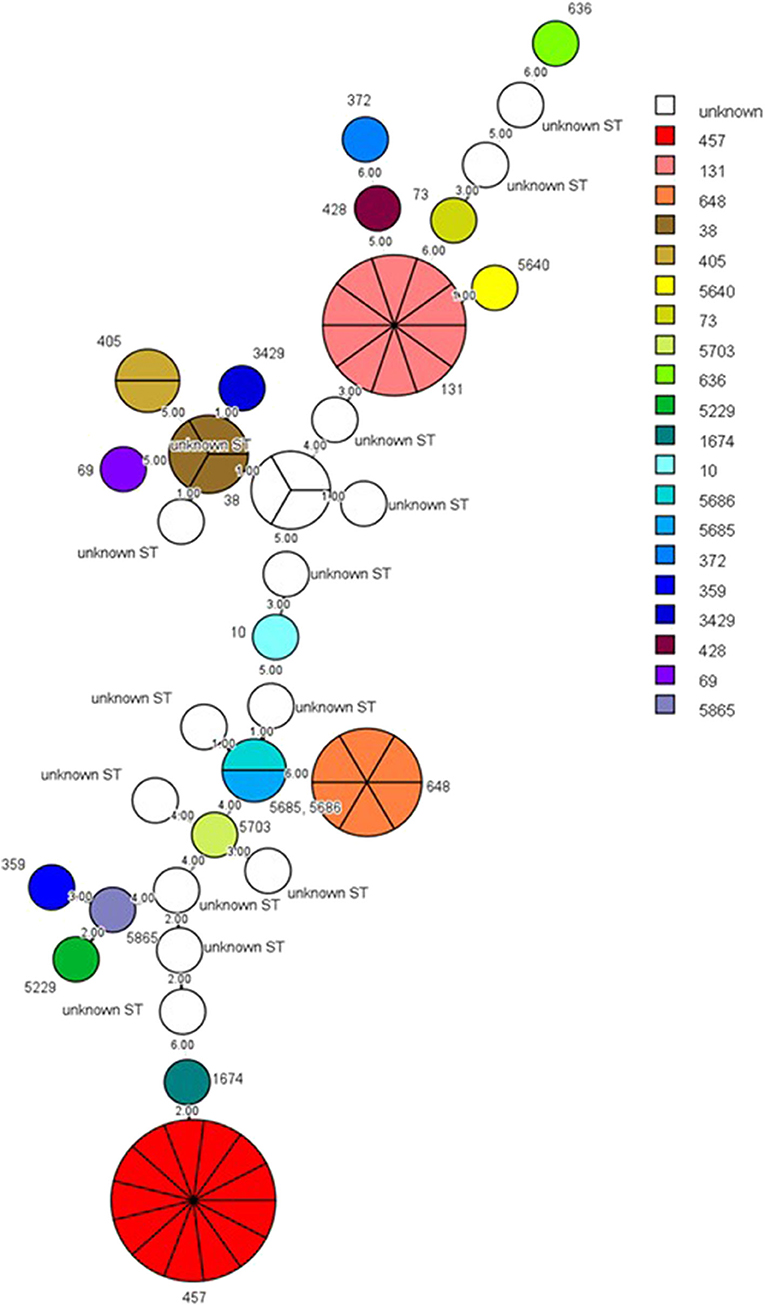
Figure 1. Minimal spanning tree of ESBL-producing E. coli. Each circle indicates one ST, subdivided into one sector for each isolate, and bordered by the ST number. White circles or sectors without an ST number denote a lack of comparison standard in the current databank. The numbers on the connecting line between STs within the MSTree indicate the number of different alleles. Solid lines represent an allele difference of three or fewer, whereas dotted lines and faint lines indicate an allele difference of four or more. ESBL, extended-spectrum β-lactamases; MSTree, minimal spanning tree; ST, sequence type.
The ESBL-producing E. coli isolates from cats were all resistant to ampicillin, ceftiofur, enrofloxacin, and ciprofloxacin, whereas those from dogs were all resistant to ampicillin. All the ESBL-producing E. coli were susceptible to imipenem, and more than 50% of the isolates were susceptible to gentamicin. Overall, most strains exhibited a multidrug resistant phenotype (Table 6).
PCR detection to target trpA and pabB was performed on 10 E. coli ST131 isolates, and 8 isolates were identified as E. coli ST131/O25b clones (Figure 2). The ESBL-producing E. coli possessed only the trpA specific DNA fragment, whereas the ESBL-producing E. coli ST131/O25b clones contained both the trpA and pabB DNA fragments. Among the 10 ESBL-producing E. coli, only one ST131/O25b clone was from a cat (E. coli 1942), whereas the others were from dogs. The two non-ST131/O25b clones were both from dogs.
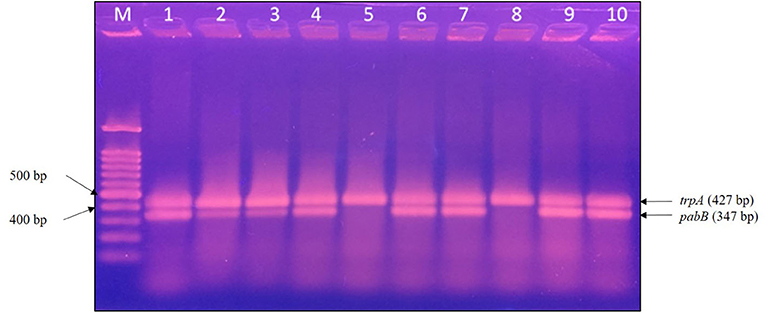
Figure 2. PCR detection of E. coli ST131/O25b clone. The trpA band corresponds to the positive control in all isolates, whereas the pabB band corresponds to the allele-specific amplification obtained only for the ST131/O25b clone. Eight isolates were confirmed to be ST131/O25b clones. M, molecular weight marker, 100 bp DNA ladder; lane 1, E. coli 1372; lane 2, E. coli 1933; lane 3, E. coli 1942; lane 4, E. coli 1972; lane 5, E. coli 2279; lane 6, E. coli 2289; lane 7, E. coli 2532; lane 8, E. coli 2588; lane 9, E. coli 2624; and lane 10, E. coli 2670. PCR, polymerase chain reaction.
Discussion
The overall prevalence of ESBL-producing E. coli in dogs and cats was 23.0% in our study. A comparable prevalence was also reported in Japan, China, and Switzerland (18–20). However, this prevalence is considerably higher than that reported in France (3.7%) and the Netherlands (2%) (21, 22). The medication strategy employed by first-line veterinarians from different countries or regions is a potential explanation for this difference. High prevalence of ESBL-producing E. coli threatens the efficacy of third-generation cephalosporins, such as cefovecin, approved for use in veterinary medicine (23).
The E. coli isolates were obtained from several sample types in cats and dogs. The most common source of ESBL-producing E. coli in cats and dogs was from aspirated urine samples, with prevalence's of 54.5% (6/11) and 68.5% (37/54), respectively. This is unsurprising because urinary tract infection (UTI) is a common diagnosis in companion animals (24). Moreover, UTIs in cats and dogs usually involve a single agent: E. coli (25).
The blaCTX−M−1 group was observed in 58.5% of the bla genes. This bla gene group is also commonly detected in Europe, the Middle East, and Asia (26). blaCTX−M−55 was the major bla gene in the blaCTX−M−1 group in our study. CTX-M-15 used to be common in human and animal isolates (27). CTX-M-55 was first identified in Thailand and is closely related to CTX-M-15 with only one amino acid substitution: Ala-77-Val (28). CTX-M-55 is a derivative of CTX-M-15. The presence of CTX-M-55 is widely reported in food and pets in China, and its geographic distribution is primarily in Asian countries (29–31). Notably, CTX-M-55 has rarely been encountered outside Asia. However, the recent emergence of CTX-M-55 in companion animals in Switzerland may indicate the spreading of this enzyme due to international food or animal trade, which warrants further attention (18). A study in the United Kingdom also revealed a decreased prevalence of CTX-M-15 producers over some years in favor of new variants, particularly CTX-M-55 (32). CTX-M-124 was another frequently observed β-lactamase in our study. CTX-M-124 was first detected in wild birds (33); the transmission of CTX-M-124 to other animals from the migratory behavior of wild birds may explain, in part, the presence of CTX-M-124 in ESBL-producing E. coli from pets (34).
ST457, ST131, and ST648 are the three major STs of ESBL-producing E. coli detected in our study, with ST457 being the most prevalent. This ST has been associated with diseases in companion animals in other studies (21, 35). E. coli ST131 and ST648 with CTX-M have been reported worldwide in both human and animal samples. These two clones combine multidrug resistance and virulence; ST131, in particular, is a globally distributed uropathogenic E. coli lineage (36). E. coli ST131 O25b carrying CTX-M-15 is a globally spreading clone with a high virulence potential, making it a public health concern (37), whereas ST131 O25b with CTX-M-14 has predominated in Japan (38). By contrast, CTX-M-174 and CTX-M-194 were the two main β-lactamases in our E. coli ST131 O25b clones. An E. coli ST131 carrying CTX-M-174 was identified in humans in Korea (39). CTX-M-174 is a variant of CTX-M-14 with two amino acid substitutions (Glu-7-Leu and Asp-242-Gly). Regardless of the type of CTX-M present in our ST131 isolates, the presence of these clones in cats and dogs raises concerns about potential zoonotic risks. This finding also justifies the continued investigation of ESBL-producing E. coli to evaluate the persistence of these fast-spreading clones in companion animals in Taiwan. A study in Europe indicated that 1.6% of the diseased dogs and cats carried ESBL-producing Enterobacteriaceae but only 2 E. coli ST131 isolates were identified; therefore, companion animals may be a source of bla genes but may not be the major source of epidemic clones (40).
Previously, LeCuyer et al. (41) revealed a thought-provoking finding regarding uropathogenic E. coli in canines. They found that ST372 was the predominant ST in dogs, whereas ST372 was an infrequent human pathogen. The prevalence of ST372 observed in dogs was similar to that of ST131 in human uropathogenic E. coli and ST73 in feline E. coli that caused urinary tract infections. They therefore concluded that each host species may have a particular ST that comprises most of the E. coli uropathogens. A French study also reached a similar conclusion, identifying ST372 as the major pathogenic E. coli ST in dogs (42). Similar findings in two distinct geographic areas may indicate a dog-specific distribution of pathogenic E. coli clones instead of the effect of regional factors (42). In contrast to LeCuyer's and Valat's reports, ST372 was observed only once in our study. Different criteria for the screening of E. coli in the study design may have contributed to this discrepancy.
Some STs such as ST3429, ST5229, ST5640, ST5685, ST5686, ST5703, and ST5865, to the best of our knowledge, have not been reported before; therefore, the pathogenic potentials of these strains were unknown.
Imipenem reportedly remains relatively active against ESBL-producing bacteria (43), which is consistent with our results (Table 6). Nonetheless, the use of carbapenems in companion animals should be avoided, since the emergence of carbapenem resistance in companion animals has been reported (44).
The current study had some limitations. AmpC-β-lactamases, which also hydrolyze the third generation of cephalosporins, were not assayed for the E. coli isolates. In addition, resistant plasmids were not characterized using PCR-based replicon typing. Although the results obtained in this study originate from only one veterinary hospital, this university-based teaching hospital is the major referral hospital for local veterinary clinics in Taipei. We believe that the information regarding ESBL in cats and dogs reported herein could be helpful for infection management and prevention.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The purpose of collecting these samples from animals was for diagnosis and treatment. An ethical review process was not required for this study according to national/local guidelines.
Author Contributions
Y-HH conducted the characterization of the phenotype and genotype of the ESBL-producing E. coli and drafted the manuscript. N-LK analyzed the ESBL-producing E. coli through MLST. K-SY conceived and coordinated this research plan. All authors have read and approved the final manuscript.
Funding
This work was supported by National Taiwan University grant G049919. Some of the results has been reported in the Chinese Society of Veterinary Science Academic Conference in 2018.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. L. J. Teng from the Department of Clinical Laboratory Sciences and Medical Biotechnology, National Taiwan University, for providing Klebsiella pneumoniae ATCC 700603.
References
1. Koenig A. Gram-negative bacterial infection. In: Greene CE, editor. Infectious Diseases of the Dog and Cat. St. Louis, MO: Elsevier Saunders (2012). p. 349–59.
2. Bush K. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother. (2018) 62:e01076-18. doi: 10.1128/AAC.01076-18
3. Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. (2010) 54:969–76. doi: 10.1128/AAC.01009-09
4. Bradford PA. Extended-spectrum beta-lactamases in the 21 st centry: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. (2001) 14:933–51. doi: 10.1128/CMR.14.4.933-951.2001
5. Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L, et al. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev. (2010) 34:295–316. doi: 10.1111/j.1574-6976.2009.00198.x
6. Kuan NL, Chang CW, Lee CA, Yeh KS. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the urine of dogs and cats suspected of urinary tract infection in a veterinary teaching hospital. Taiwan Vet J. (2016) 42:143–8. doi: 10.1142/S1682648515500274
7. Huang YH, Yeh KS. Characteristics of the extended-spectrum β-lactamases-producing Escherichia coli isolated from the dogs and cats in National Taiwan University Veterinary Hospital from 2014 to 2017. In: Chinese Society of Veterinary Science Academic Conference. Taichung (2018).
8. CLSI. M-100, Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Wayne, PA: CLSI (2020).
9. Shaheen BW, Oyarzabal OA, Boothe DM. The role of class 1 and 2 integrons in mediating antimicrobial resistance among canine and feline clinical E. coli isolates from the US. Vet Microbiol. (2010) 144, 363–70. doi: 10.1016/j.vetmic.2010.01.018
10. Sutcliffe JG. Nucleotide-sequence of ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. (1978) 75:3737–41. doi: 10.1073/pnas.75.8.3737
11. Chia JH, Chu C, Su LH, Chiu CH, Kuo AJ, Sun CF, et al. Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M beta-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J Clin Microbiol. (2005) 43:4486–91. doi: 10.1128/JCM.43.9.4486-4491.2005
12. Yu Y, Ji S, Chen Y, Zhou W, Wei Z, Li L, et al. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect. (2007) 54:53–7. doi: 10.1016/j.jinf.2006.01.014
13. Woodford N, Fagan EJ, Ellington MJ. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J Antimicrob Chemother. (2006) 57:154–5. doi: 10.1093/jac/dki412
14. Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis. (2014) 14:659–69. doi: 10.1186/s12879-014-0659-0
15. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. (2006) 60:1136–51. doi: 10.1111/j.1365-2958.2006.05172.x
16. Clermont O, Dhanji H, Upton M, Gibreel T, Fox A, Boyd D, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother. (2009) 64:274–7. doi: 10.1093/jac/dkp194
17. Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, et al. Beta-lactamase database (BLDB) - structure and function. J Enzyme Inhib Med Chem. (2017) 32:917–9. doi: 10.1080/14756366.2017.1344235
18. Zogg AL, Simmen S, Zurfluh K, Stephan R, Schmitt SN, Nuesch-Inderbinen M, et al. High prevalence of extended-spectrum beta-lactamase producing Enterobacteriaceae among clinical isolates from cats and dogs admitted to a veterinary hospital in Switzerland. Front Vet Sci. (2018) 5:62. doi: 10.3389/fvets.2018.00062
19. Sun Y, Zeng Z, Chen S, Ma J, He L, Liu Y, et al. High prevalence of bla(CTX-M) extended-spectrum beta-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin Microbiol Infect. (2010) 16:1475–81. doi: 10.1111/j.1469-0691.2010.03127.x
20. Tsuyuki Y, Kurita G, Murata Y, Takahashi T, Veterinary Infection Control Association Sepsis Working G. Bacteria isolated from companion animals in Japan (2014-2016) by blood culture. J Infect Chemother. (2018) 24:583–7. doi: 10.1016/j.jiac.2018.01.014
21. Dierikx CM, van Duijkeren E, Schoormans AHW, van Essen-Zandbergen A, Veldman K, Kant A, et al. Occurrence and characteristics of extended-spectrum-β-lactamase and AmpC-producing clinical isolates derived from companion animals and horses. J Antimicrob Chemother. (2012) 67:1368–74. doi: 10.1093/jac/dks049
22. Dahmen S, Haenni M, Chatre P, Madec JY. Characterization of blaCTX−M IncFII plasmids and clones of Escherichia coli from pets in France. J Antimicrob Chemother. (2013) 68:2797–801. doi: 10.1093/jac/dkt291
23. Stegemann MR, Passmore CA, Sherington J, Lindeman CJ, Papp G, Weigel DJ, et al. Antimicrobial activity and spectrum of cefovecin, a new extended- spectrum cephalosporin, against pathogens collected from dogs and cats in Europe and North America. Antimicrob Agents Chemother. (2006) 50:2286–92. doi: 10.1128/AAC.00077-06
24. Ling GV. Therapeutic strategies involving antimicrobial treatment of the canine urinary tract. J Am Vet Med Assoc. (1984) 185:1162–4.
25. Byron JK. Urinary tract infection. Vet Clin North Am Small Anim Pract. (2019) 49:211–21. doi: 10.1016/j.cvsm.2018.11.005
26. Zeynudin A, Pritsch M, Schubert S, Messerer M, Liegl G, Hoelscher M, et al. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum beta-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect Dis. (2018) 18:524. doi: 10.1186/s12879-018-3436-7
27. Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. (2012) 18:646–55. doi: 10.1111/j.1469-0691.2012.03850.x
28. Kiratisin P, Apisarnthanarak A, Saifon P, Laesripa C, Kitphati R, Mundy LM, et al. The emergence of a novel ceftazidime-resistant CTX-M extended-spectrum beta-lactamase, CTX-M-55, in both community-onset and hospital-acquired infections in Thailand. Diagn Microbiol Infect Dis. (2007) 58:349–55. doi: 10.1016/j.diagmicrobio.2007.02.005
29. Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis. (2014) 14:659. doi: 10.1186/s12879-014-0659-0
30. Kawamura K, Sugawara T, Matsuo N, Hayashi K, Norizuki C, Tamai K, et al. Spread of CTX-type extended-spectrum beta-lactamase-producing Escherichia coli isolates of epidemic clone B2-O25-ST131 among dogs and cats in Japan. Microb Drug Resist. (2017) 23:1059–66. doi: 10.1089/mdr.2016.0246
31. Norizuki C, Kawamura K, Wachino JI, Suzuki M, Nagano N, Kondo T, et al. Detection of Escherichia coli producing CTX-M-1-group extended-spectrum beta-lactamases from pigs in Aichi Prefecture, Japan, between 2015 and 2016. Jpn J Infect Dis. (2018) 71:33–8. doi: 10.7883/yoken.JJID.2017.206
32. Bortolami A, Zendri F, Maciuca EI, Wattret A, Ellis C, Schmidt V, et al. Diversity, virulence, and clinical significance of extended-spectrum beta-lactamase- and pAmpC-producing Escherichia coli from companion animals. Front Microbiol. (2019) 10:1260. doi: 10.3389/fmicb.2019.01260
33. Poirel L, Potron A, De La Cuesta C, Cleary T, Nordmann P, Munoz-Price LS, et al. Wild coastline birds as reservoirs of broad-spectrum-beta-lactamase-producing Enterobacteriaceae in Miami Beach, Florida. Antimicrob Agents Chemother. (2012) 56:2756–8. doi: 10.1128/AAC.05982-11
34. Wang J, Ma ZB, Zeng ZL, Yang XW, Huang Y, Liu JH, et al. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool Res. (2017) 38:55–80. doi: 10.24272/j.issn.2095-8137.2017.024
35. Zogg AL, Zurfluh K, Schmitt S, Nuesch-Inderbinen M, Stephan R. Antimicrobial resistance, multilocus sequence types and virulence profiles of ESBL producing and non-ESBL producing uropathogenic Escherichia coli isolated from cats and dogs in Switzerland. Vet Microbiol. (2018) 216:79–84. doi: 10.1016/j.vetmic.2018.02.011
36. Shaik S, Ranjan A, Tiwari SK, Hussain A, Nandanwar N, Kumar N, et al. Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. mBio. (2017) 8:>e01596-17. doi: 10.1128/mBio.01596-17
37. Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. (2015) 28:565–91. doi: 10.1128/CMR.00116-14
38. Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, et al. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother. (2015) 70:1639–49. doi: 10.1093/jac/dkv017
39. Joo EJ, Kim SJ, Baek M, Choi Y, Seo J, Yeom JS, et al. Fecal carriage of antimicrobial-resistant Enterobacteriaceae in healthy Korean adults. J Microbiol Biotechnol. (2018) 28:1178–84. doi: 10.4014/jmb.1801.12060
40. Bogaerts P, Huang TD, Bouchahrouf W, Bauraing C, Berhin C, El Garch F, et al. Characterization of ESBL- and AmpC-producing Enterobacteriaceae from diseased companion animals in Europe. Microb Drug Resist. (2015) 21:643–50. doi: 10.1089/mdr.2014.0284
41. LeCuyer TE, Byrne BA, Daniels JB, Diaz-Campos DV, Hammac GK, Miller CB, et al. Population structure and antimicrobial resistance of canine uropathogenic Escherichia coli. J Clin Microbiol. (2018) 56:e00788-18. doi: 10.1128/JCM.00788-18
42. Valat C, Drapeau A, Beurlet S, Bachy V, Boulouis HJ, Pin R, et al. Pathogenic Escherichia coli in dogs reveals the predominance of ST372 and the human-associated ST73 extra-intestinal lineages. Front Microbiol. (2020) 11:580. doi: 10.3389/fmicb.2020.00580
43. Hsueh PR, Hoban DJ, Carmeli Y, Chen SY, Desikan S, Alejandria M, et al. Consensus review of the epidemiology and appropriate antimicrobial therapy of complicated urinary tract infections in Asia-Pacific region. J Infect. (2011) 63:114–23. doi: 10.1016/j.jinf.2011.05.015
Keywords: extended-spectrum-β-lactamases, Escherichia coli, CTX-M, multilocus sequence typing, multidrug resistance
Citation: Huang Y-H, Kuan N-L and Yeh K-S (2020) Characteristics of Extended-Spectrum β-Lactamase–Producing Escherichia coli From Dogs and Cats Admitted to a Veterinary Teaching Hospital in Taipei, Taiwan From 2014 to 2017. Front. Vet. Sci. 7:395. doi: 10.3389/fvets.2020.00395
Received: 02 March 2020; Accepted: 02 June 2020;
Published: 16 July 2020.
Edited by:
Magdalena Rzewuska, Warsaw University of Life Sciences, PolandReviewed by:
Jean-Yves Madec, Agence Nationale de Sécurité Sanitaire de l'Alimentation, de l'Environnement et du Travail (ANSES), FranceYves Millemann, INRA École Nationale Vétérinaire d'Alfort (ENVA), France
Copyright © 2020 Huang, Kuan and Yeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuang-Sheng Yeh, a3N5ZWgmI3gwMDA0MDtudHUuZWR1LnR3
 Yi-Hsuan Huang1
Yi-Hsuan Huang1 Nan-Ling Kuan
Nan-Ling Kuan Kuang-Sheng Yeh
Kuang-Sheng Yeh