- 1Laboratory of Food and Environmental Hygiene, Cooperative Department of Veterinary Medicine, Gifu University, Gifu, Japan
- 2Department of Food and Nutrition, Jumonji University, Saitama, Japan
- 3Education and Research Center for Food Animal Health, Gifu University (GeFAH), Gifu, Japan
- 4The United Graduate School of Veterinary Sciences, Gifu University, Gifu, Japan
- 5Joint Graduate School of Veterinary Sciences, Gifu University, Gifu, Japan
Orf virus is a prototype species of the genus Parapoxvirus, subfamily Chordopoxvirinae, family Poxviridae. Japanese orf viruses, infecting sheep and wild Japanese serows (Capricornis crispus), have been considered to be genetically closely related based on the sequence identities of the open reading frames (ORFs) 11, 20, and 132 in their genomes. However, since the genome size of orf viruses is about 140 kbp long, genetic variation among Japanese orf viruses remains unclear. In this study, we analyzed the sequences of ORFs 117, 119, 125, and 127 located in the 3′-proximal region of the viral genome using two strains from sheep and three strains from Japanese serows isolated from 1970 to 2007, and compared them with the corresponding sequences of reference orf viruses from other countries. Sequence analysis revealed that ORFs 125 and 127, which encode the inhibitor of apoptosis and viral interleukin (IL)-10, respectively, were highly conserved among the five Japanese orf viruses. However, high genetic variability with deletions or duplications was observed in ORFs 117 and 119, which encode granulocyte macrophage colony-stimulating factor and IL-2 inhibition factor (GIF), and inducer of cell apoptosis, respectively, in one strain from sheep and two strains from Japanese serows. Our results suggest that genetic variability exists in Japanese orf viruses even in the same host species. This is the first report of genetic variability of orf viruses in Japan.
Introduction
Orf virus is a prototype species of the genus Parapoxvirus, subfamily Chordopoxvirinae, family Poxviridae (1). Orf virus has a linear double-stranded DNA genome (134–139 kbp) with high GC content (~63–64%) and encodes 132 putative gene products (2). Orf virus is the causative agent of orf disease, also known as contagious pustular dermatitis, contagious ecthyma, or scabby mouth mainly in sheep and goats, and can be transmitted to humans (3). In Japan, the first reports of orf virus infections in sheep and wild Japanese serows (Capricornis crispus) were published in 1952 (4) and 1979 (5), respectively. Previously, we have reported nucleotide sequence homology in three open reading frames (ORFs) 11, 20, and 132 among 13 orf viruses isolated or polymerase chain reaction (PCR)-detected from sheep and wild Japanese serows (6). These ORFs encode viral envelope (7), virus interferon resistance (8), and viral vascular endothelial growth factor (VEGF) (9), respectively. The amino acid sequences derived from ORFs 11 and 20 were identical among the 13 orf viruses, and only one amino acid substitution was found in ORF 132 in an orf virus isolated from sheep (6). Therefore, the three viral genes of Japanese orf viruses are highly conserved. However, since only a part of the whole genome (~140 kbp) has been sequenced so far, the degree of genetic variation in other regions remains unclear.
To explore genetic differences between Japanese orf viruses, we conducted next-generation sequencing (NGS) of some strains of these orf viruses. However, whole genome sequences were not obtained, due to the large number of unmapped reads in the 3′-proximal region of viral genome (Figure 1). We hypothesized that the 3′-proximal region of a viral genome has genetic variation. Thus, in the present study, we characterized four ORFs 117, 119, 125, and 127, which are located in the 3′-proximal region of the viral genome. ORFs 117, 119, 125, and 127 encode granulocyte macrophage colony-stimulating factor and interleukin 2 (IL-2) inhibition factor (GIF) (11), inducer of cell apoptosis (12), inhibitor of apoptosis (13, 14), and viral IL-10 (15), respectively.
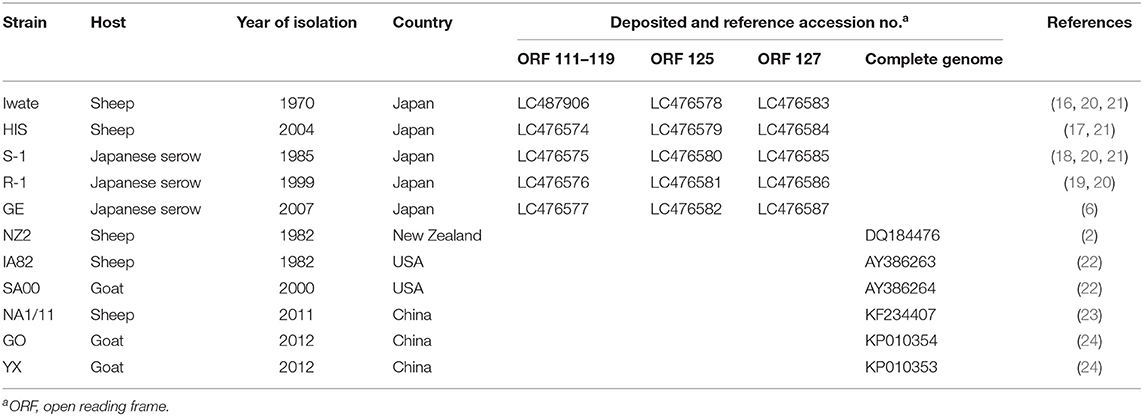
Figure 1. Visualization of next-generation sequencing (NGS) coverage of Japanese orf virus R-1 strain and the details of the open reading frame (ORF) analyzed in this study. NZ2 strain (GenBank accession no. DQ184476) was used as the reference strain. Binary version of the sequence alignment/map (bam) file was loaded onto the Integrated Genome Viewer (IGV) (10). The vertical axis shows the number of reads mapping to each location of the genome. Zoomed view of ORF 117 (A), ORF 119 (B), ORF 125 (C), and ORF 127 (D). Whole genome view (E).
Materials and Methods
Viruses
For epidemiologic and genetic characteristics of Japanese orf virus, five Japanese strains isolated from 1970 to 2007 were used. Two strains of Iwate (16) and HIS (17) were isolated from sheep and three strains of S-1 (18), R-1 (19), and GE (6) were isolated from wild Japanese serows (Table 1). Viruses were propagated in fetal lamb lung cells (kindly provided by Dr. H. Sentsui, Nihon University, Japan) at 37°C in Dulbecco's modified Eagle's medium (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (PAA Laboratories, Pasching, Austria).
Whole Genome Re-sequencing and Assembly
Total DNA extracted from virus-infected cells using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) were used for constructing the libraries using Nextera XT DNA sample Prep Kit (Illumina, San Diego, CA, USA), and sequenced using an Illumina MiSeq (Illumina). Obtained short read sequences collected in FASTQ files were aligned to the orf virus strain NZ2 (DQ184476) as the reference genome using Burrows–Wheeler transformation (BWA) ver 0.7.12-r103 software (25) and constructed binary version of the sequence alignment/map (bam) file using SAM tools ver. 0.1.19-96b5f2294a software (26).
Analysis of ORF
DNA was extracted from virus-infected cells using a DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer's instructions. Four different PCRs were carried out with GoTaq Hot Start Green Master Mix (Promega, Madison, WI, USA) using Veriti thermal cycler (Applied Biosystems, Foster City, CA, USA). The PCR conditions and analyzed ORFs are provided in Supplementary Table S1 and Figure 1, respectively. PCR products were purified using NucleoSpin Gel and PCR Clean-up (Macherey-Nagel, Duren, Germany), and the nucleotide sequences were determined by direct sequencing using a BigDye Terminator Cycle Sequencing Kit v3.1 (Applied Biosystems, Foster City, CA, USA). Sequence analysis was carried out using the software Genetyx-Win version 13 (Genetyx, Tokyo, Japan), and phylogenetic analysis was performed using the MEGA7 program (27). Phylogenetic trees were constructed using maximum likelihood methods, and the reliability of the branches was evaluated by 1,000 replicates. Sequence and phylogenetic analyses were compared with the reference orf viruses (Table 1).
Results
NGS was performed using total DNA extracted from the concentrated virus. However, whole genome sequences were not obtained, possibly due to the large number of deletion in the 3′-proximal region of viral genome (Figure 1).
Specific PCR products of 799 bp for ORF 117 and 537–652 bp for ORF 119 were obtained from the four strains (Iwate, HIS, S-1, and GE) and from all five Japanese orf viruses, respectively. High genomic variability was seen in ORFs 117 and 119 in Japanese orf viruses. In ORF 117, 96.6–100% nucleotide identity was observed among four strains. Surprisingly, R-1 strain from Japanese serow completely lacked ORF 117 (Figure 2A). Partial deletion in ORF 117 was also observed in the amino acid sequences in reference orf virus strain NA1/11 isolated from sheep in China. In ORF 119, deletions were observed in the first half of the amino acid sequences in S-1 and R-1 strains as well as the reference Chinese NA1/11 strain (Figure 2B). Two and 12 amino acid deletion was observed in HIS and three reference strains from sheep and goat (NZ2, IA82, and YX), respectively. In the Iwate strain, 10 amino acids were found to be duplicated.
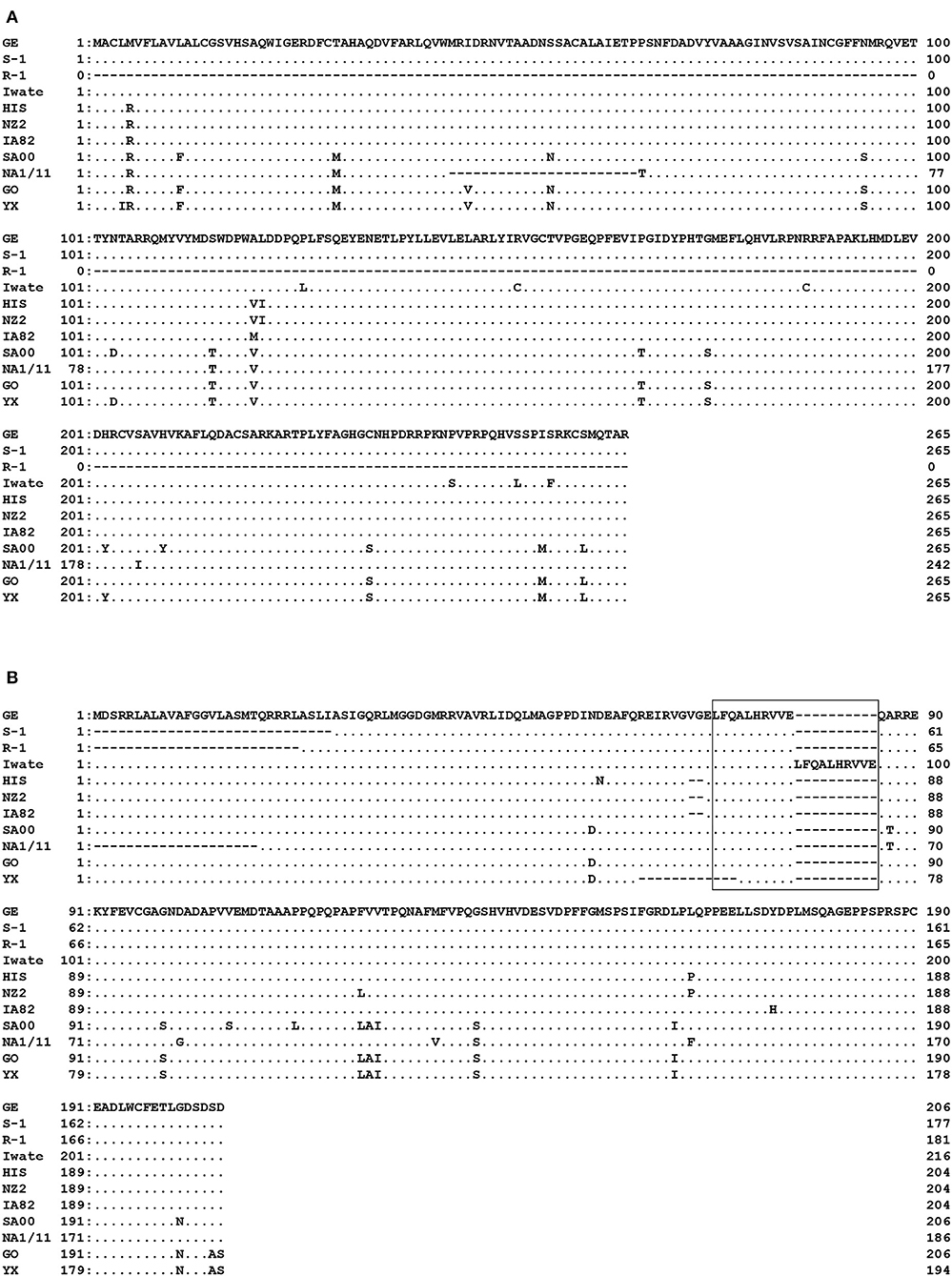
Figure 2. Alignment of the amino acid sequences derived from ORF 117 (A) and ORF 119 (B). Amino acids identical to GE strain at the given positions are represented by dots. R-1 strain completely lacked ORF 117. In ORF 119, deletions were observed in the first half of the nucleotide sequences in S-1 and R-1 strains. In the box, duplicate in Iwate strain is shown.
Specific PCR products of 522 and 561 bp were obtained for ORFs 125 and 127 from all of five Japanese orf viruses. Amino acid sequences derived from these ORFs from four Japanese orf viruses (Iwate, S-1, R-1, and GE) were found to be 100% identical. The sequence from HIS strain revealed only two and seven amino acid substitutions in ORFs 125 and 127, respectively (Supplementary Tables S2, S3). The sequences of ORFs 125 and 127 were highly conserved among Japanese orf viruses. In the phylogenetic analysis, there were mainly two branches, and all Japanese orf viruses were classified into the same group (Figure 3). Our results indicate that Japanese orf viruses are closer to the IA82 and NZ2 strains isolated in the United States and New Zealand, respectively, than other reference strains. Sequences obtained in this study were submitted to DDBJ/EMBL/GenBank, and the accession numbers are given in Table 1.
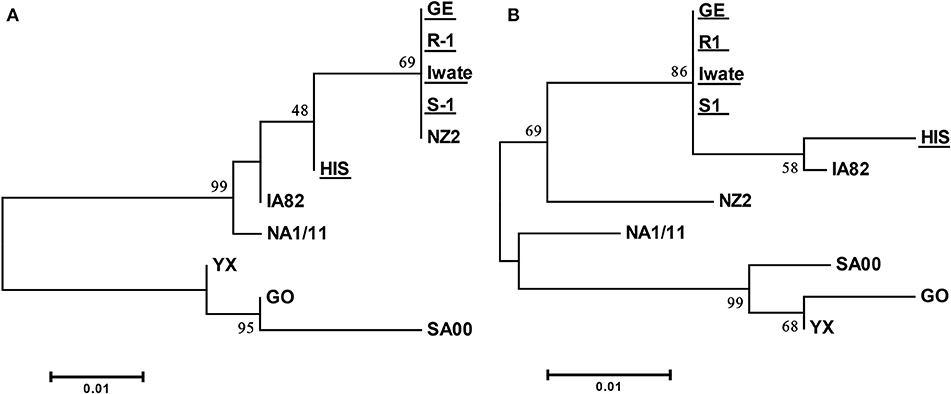
Figure 3. Phylogenetic tree based on deduced amino acid sequences derived from ORF 125 (A) and ORF 127 (B). Strains used in this study are underlined. The percentage bootstrap values calculated from 1,000 replicates are indicated above the internal nodes.
Discussion
In this study, we carried out ORF sequence analysis for five Japanese orf viruses, and our results revealed that the sequences of ORFs 125 and 127 were highly conserved. However, high genomic variability was seen in ORFs 117 and 119. Observed genetic variability was found to be the 3′-proximal region of Japanese orf viruses. To the best of our knowledge, this is the first report on the genetic variability of Japanese orf viruses.
In the phylogenetic analysis of ORF 125, Japanese orf viruses isolated from Japanese serows were classified into a group isolated from sheep. It has been reported that analyses of the phylogenetic tree of 47 ORFs including ORF 125 were found to assist in easily distinguishing between goat- and sheep-originated orf viruses (24). These results indicate a possibility that sheep orf virus may have infected Japanese serows. Furthermore, analyses of the phylogenetic tree of ORFs 125 and 127 clearly showed that the Japanese orf viruses were closer to IA82 and NZ2 strains than to other reference strains. In Japan, sheep are frequently imported from the United States and New Zealand for improved growth and to encourage breeding (28). Therefore, it is possible that these orf viruses came along with the imported animals and were introduced into breeding sheep and wild Japanese serows in Japan.
Our results showed genetic variability in ORFs 117 and 119 in the Japanese orf viruses, suggesting that there is heterogeneity even in viruses infected with the same host species. In addition, deletions in ORF 119 were observed in Japanese (HIS, S-1, and R-1) and reference (NZ2, IA82, NA1/11, and YX) strains. Based on the previous comparative analysis, it is presumed that genes in the central region of the orf virus genome are more conserved, whereas those in the terminal region show remarkably high variability (29). Notably, this variability is accompanied by a high frequency of gene recombination and nucleotide deletions (23). The genetic analysis of ORFs 117 and 119 may help to characterize or differentiate strains that are otherwise shown to be identical by the envelope coding genes (30). A previous study demonstrated that viruses with high deletion in ORFs 114–120 showed low virulence in animal inoculation experiments and that genomic deletions attenuate virulence (24). At present, the relationship between the deletion of ORF 117 and virulence in the R-1 strain is unknown. Therefore, there is a need to analyze the correlation between genetic variability and virulence in more detail.
In this study, it was revealed that there were differences in conservation and variability among ORFs. Viral IL-10 encoded by ORF 127 shares remarkable similarity to mammalian IL-10. Mammalian IL-10 is highly conserved across all mammalian species (15). IL-10 is a multifunctional cytokine that has suppressive effects on inflammation, antiviral responses. Orf virus produces viral IL-10 by itself and avoids host's inflammatory and immune response by it (31). This suggests that viral IL-10 encoded by ORF 127 might require high conservation in orf virus. On the other hand, GIF encoded by ORF 117 does not resemble any known mammalian granulocyte-macrophage colony–stimulating factor (GM-CSF)- or IL-2-binding proteins, and indeed, there are no reports of any other protein capable of binding both GM-CSF and IL-2. In addition, human GM-CSF does not respond in sheep cells due to its inability to bind to ovine receptor (32). Therefore, GIF was thought to have evolved a unique binding specificity in sheep, the natural host of the orf virus (33). This suggests that GIF encoded by ORF 117 is gene whose necessity changes depending on the host species. It is thought that differences in necessity of gene may affect conservation and variability of the gene encoded by ORFs.
Japanese serows are wild animals and a natural monument that is endemic in Japan (34). Japanese serows are often witnessed in mountain villages and can come into contact with livestock sheep. There have been reports that a single strain of orf virus caused outbreak of proliferative dermatitis in various ruminant species at a zoo (35). Orf virus from Japanese serows can be spread to sheep or farmers, or orf virus from sheep can be spread to Japanese serows. It is important to know the characteristics of Japanese orf viruses in order to reduce the spread risk.
We tried NGS analysis, but it was unsuccessful. NGS results indicated the 3′-proximal region of the genome of Japanese orf viruses has genetic variation. Our results obtained by Sanger sequencing for variable region of Japanese orf viruses may be useful for understanding the characteristics of these viruses. However, we analyzed the limited region of the viral genomes, and sequencing other regions using improved methods for NGS might be required to better understand the characteristics of Japanese orf viruses.
Data Availability Statement
Sequence data obtained in this study is available in the DDBJ/EMBL/GenBank (accession nos. LC476574–LC476587 and LC487906).
Author Contributions
KS and YI analyzed all data and were major contributors in writing the manuscript. KS, AT, ST, and AO performed the nucleotide/amino acid sequencing and phylogenetic analysis. All authors read and approved the final manuscript.
Funding
This study was partly supported by the Grant for Joint Research Program of the Research Center for Zoonosis Control, Hokkaido University, from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This study was also supported, in part, by the Research Grant from The Okawa Foundation and by the Grant for Environmental Research Projects from The Sumitomo Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Dr. Hiroshi Sentsui (Nihon University, Japan) for providing fetal lamb lung cells.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00188/full#supplementary-material
Abbreviations
GIF, granulocyte-macrophage colony-stimulating factor and IL-2 inhibition factor; IL-2, interleukin 2; ORF, open reading frame; PCR, polymerase chain reaction; VEGF, vascular endothelial growth factor.
References
1. Skinner MA, Buller RM, Damon IK, Lefkowitz EJ, McFadden G, McInnes CJ, et al. Poxviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. New York, NY: Elsevier Academic Press (2012). p. 291–309. doi: 10.1016/B978-0-12-384684-6.00028-8
2. Mercer AA, Ueda N, Friederichs SM, Hofmann K, Fraser KM, Bateman T, et al. Comparative analysis of genome sequences of three isolates of Orf virus reveals unexpected sequence variation. Virus Res. (2006) 116:146–58. doi: 10.1016/j.virusres.2005.09.011
3. Knowles DP. Poxviridae. In: MacLachlan NJ, Dubovi EJ, editors. Fenner's Veterinary Virology. 4th ed. Amsterdam: Elsevier Academic Press. (2011) p. 151–65. doi: 10.1016/B978-0-12-375158-4.00007-9
4. Asakawa Y, Imaizum K, Tajima Y, Endo M. Studies on a contagious ecthyma-like disease observed among the sheep. Jpn J Med Sci Biol. (1952) 5:475–86. doi: 10.7883/yoken1952.5.475
5. Kumagai T, Shimizu M, Ito Y, Yamamoto S, Ito T, Ohyama S, et al. Contagious pustular dermatitis of Japanese serows in Akita Prefecture. In: Proceeding of the 87th Meeting of the Japanese Society of Veterinary Science. Kanagawa. (1979).
6. Inoshima Y, Ito M, Ishiguro N. Spatial and temporal genetic homogeneity of orf viruses infecting Japanese serows (Capricornis crispus). J Vet Med Sci. (2010) 72:701–7. doi: 10.1292/jvms.09-0467
7. Inoshima Y, Morooka A, Sentsui H. Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J Virol Methods. (2000) 84:201–8. doi: 10.1016/S0166-0934(99)00144-5
8. McInnes CJ, Wood AR, Mercer AA. Orf virus encodes a homolog of the vaccinia virus interferon-resistance gene E3L. Virus Genes. (1998) 17:107–15.
9. Lyttle DJ, Fraser KM, Fleming SB, Mercer AA, Robinson AJ. Homologs of vascular endothelial growth factor are encoded by the poxvirus orf virus. J Virol. (1994) 68:84–92. doi: 10.1128/JVI.68.1.84-92.1994
10. The, Integrative Genomics Viewer. Available online at: http://software.broadinstitute.org/software/igv/ (accessed 7 January, 2020).
11. McInnes CJ, Deane D, Haig D, Percival A, Thomson J, Wood AR. Glycosylation, disulfide bond formation, and the presence of a WSXWS-like motif in the orf virus GIF protein are critical for maintaining the integrity of binding to ovine granulocyte–macrophage colony-stimulating factor and interleukin-2. J Virol. (2005) 79:11205–13. doi: 10.1128/JVI.79.17.11205-11213.2005
12. Li W, Chen H, Deng H, Kuang Z, Long M, Chen D, et al. Orf virus encoded protein ORFV119 induces cell apoptosis through the extrinsic and intrinsic pathways. Front Microbiol. (2018) 9:1056. doi: 10.3389/fmicb.2018.01056
13. Westphal D, Ledgerwood EC, Hibma MH, Fleming SB, Whelan EM, Mercer AA. A novel Bcl-2-like inhibitor of apoptosis is encoded by the parapoxvirus ORF virus. J Virol. (2007) 81:7178–88. doi: 10.1128/JVI.00404-07
14. Westphal D, Ledgerwood EC, Tyndall JD, Hibma MH, Ueda N, Fleming SB, et al. The orf virus inhibitor of apoptosis functions in a Bcl-2-like manner, binding and neutralizing a set of BH3-only proteins and active Bax. Apoptosis. (2009) 14:1317–30. doi: 10.1007/s10495-009-0403-1
15. Fleming SB, McCaughan CA, Andrews AE, Nash AD, Mercer AA. A homolog of interleukin-10 is encoded by the poxvirus orf virus. J Virol. (1997) 71:4857–61. doi: 10.1128/JVI.71.6.4857-4861.1997
16. Kumagai T, Shimizu M, Ito Y, Konno S, Nakagawa N, Sato K, et al. Contagious pustular dermatitis of sheep. In: Proceeding of the 71st Meeting of the Japanese Society of Veterinary Science. Tokyo. (1971).
17. Kano Y, Inoshima Y, Shibahara T, Ishikawa Y, Kadota K, Ohashi S, et al. Isolation and characterization of a parapoxvirus from sheep with papular stomatitis. JARQ. (2005) 39:197–203. doi: 10.6090/jarq.39.197
18. Suzuki T, Minamoto N, Sugiyama M, Kinjo T, Suzuki Y, Sugimura M, et al. Isolation and antibody prevalence of a parapoxvirus in wild Japanese serows (Capricornis crispus). J Wildl Dis. (1993) 29:384–9. doi: 10.7589/0090-3558-29.3.384
19. Inoshima Y, Murakami K, Wu D, Sentsui H. Characterization of parapoxviruses circulating among wild Japanese serows (Capricornis crispus). Microbiol Immunol. (2002) 46:583–7. doi: 10.1111/j.1348-0421.2002.tb02738.x
20. Inoshima Y, Murakami K, Yokoyama T, Sentsui H. Genetic heterogeneity among parapoxviruses isolated from sheep, cattle and Japanese serows (Capricornis crispus). J Gen Virol. (2001) 82:1215–20. doi: 10.1099/0022-1317-82-5-1215
21. Inoshima Y, Ishiguro N. Molecular and biological characterization of vascular endothelial growth factor of parapoxviruses isolated from wild Japanese serows (Capricornis crispus). Vet Microbiol. (2010) 6:63–71. doi: 10.1016/j.vetmic.2009.07.024
22. Delhon G, Tulman ER, Afonso CL, Lu Z, de la Concha-Bermejillo A, Lehmkuhl HD, et al. Genomes of the parapoxviruses ORF virus and bovine papular stomatitis virus. J Virol. (2006) 78:168–77. doi: 10.1128/JVI.78.1.168-177.2004
23. Li W, Hao W, Peng Y, Duan C, Tong C, Song D, et al. Comparative genomic sequence analysis of Chinese orf virus strain NA1/11 with other parapoxviruses. Arch Virol. (2015) 160:253–66. doi: 10.1007/s00705-014-2274-1
24. Chi X, Zeng X, Li W, Hao W, Li M, Huang X, et al. Genome analysis of orf virus isolates from goats in the Fujian province of southern China. Front Microbiol. (2015) 6:1135. doi: 10.3389/fmicb.2015.01135
25. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. (2010) 26:589–95. doi: 10.1093/bioinformatics/btp698
26. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
27. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
28. Animal Quarantine Annual Report. Available online at: http://www.maff.go.jp/aqs/tokei/toukeinen.html. (accessed 7 January 2020).
29. Rziha HJ, Bauer B, Adam KH, Röttgen M, Cottone R, Henkel M, et al. Relatedness and heterogeneity at the near-terminal end of the genome of a parapoxvirus bovis 1 strain (B177) compared with parapoxvirus ovis (Orf virus). J Gen Virol. (2003) 84:1111–6. doi: 10.1099/vir.0.18850-0
30. Hosamani M, Bhanuprakash V, Scagliarini A, Singh RK. Comparative sequence analysis of major envelope protein gene (B2L) of Indian orf viruses isolated from sheep and goats. Vet Microbiol. (2006) 116:317–24. doi: 10.1016/j.vetmic.2006.04.028
31. Haig DM, McInnes CJ. Immunity and counter-immunity during infection with the parapoxvirus orf virus. Virus Res. (2002) 88:3–16. doi: 10.1016/S0168-1702(02)00117-X
32. McInnes CJ, Haig DM. Cloning and expression of a cDNA encoding ovine granulocyte-macrophage colony-stimulating factor. Gene. (1991) 105:275–9. doi: 10.1016/0378-1119(91)90163-6
33. Fleming SB, Wise LM, Mercer AA. Molecular genetic analysis of orf virus: a poxvirus that has adapted to skin. Viruses. (2015) 23:1505–39. doi: 10.3390/v7031505
34. Ohdachi SD, Ishibashi Y, Iwasa MA, Fukui D, Saitoh T. The Wild Mammals of Japan. 2th edn. Kyoto: Shoukadoh Book Sellers (2015) p. 314–7.
Keywords: genetic variability, Japanese serows, nucleotide sequence, orf virus, sheep
Citation: Shimizu K, Takaiwa A, Takeshima S-n, Okada A and Inoshima Y (2020) Genetic Variability of 3′-Proximal Region of Genomes of Orf Viruses Isolated From Sheep and Wild Japanese Serows (Capricornis crispus) in Japan. Front. Vet. Sci. 7:188. doi: 10.3389/fvets.2020.00188
Received: 20 January 2020; Accepted: 23 March 2020;
Published: 24 April 2020.
Edited by:
Satoshi Sekiguchi, University of Miyazaki, JapanReviewed by:
Eugene Ryabov, United States Department of Agriculture (USDA), United StatesShuhong Luo, Foshan University, China
Copyright © 2020 Shimizu, Takaiwa, Takeshima, Okada and Inoshima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuo Inoshima, aW5vc2hpbWFAZ2lmdS11LmFjLmpw
 Kaori Shimizu1
Kaori Shimizu1 Shin-nosuke Takeshima
Shin-nosuke Takeshima Ayaka Okada
Ayaka Okada Yasuo Inoshima
Yasuo Inoshima