- 1Department of Sanità Animale, Istituto Zooprofilattico Sperimentale Della Sardegna, Sassari, Italy
- 2OIE Reference Laboratory for Bluetongue, Istituto Zooprofilattico Sperimentale Abruzzo e Molise, Teramo, Italy
Bluetongue (BT) is a major Office International des Epizooties (OIE)-listed disease of wild and domestic ruminants caused by several serotypes of Bluetongue virus (BTV), a virus with a segmented dsRNA genome belonging to the family Reoviridae, genus Orbivirus. BTV is transmitted through the bites of Culicoides midges. The aim of this study was to develop a new method for quantification of BTV Seg-10 by droplet digital RT-PCR (RTdd-PCR), using nucleic acids purified from complex matrices such as blood, tissues, and midges, that notoriously contain strong PCR inhibitors. First, RTdd-PCR was optimized by using RNAs purified from serially 10-fold dilutions of a BTV-1 isolate (105.43TCID50/ml up to 10−0.57 TCID50/ml) and from the same dilutions spiked into fresh ovine EDTA-blood and spleen homogenate. The method showed a good degree of linearity (R2 ≥ 0.995). The limit of detection (LoD) and the limit of quantification (LoQ) established were 10−0.67TCID50/ml (0.72 copies/μl) and 100.03TCID50/ml (3.05 copies/μl) of BTV-1, respectively. Second, the newly developed test was compared, using the same set of biological samples, to the quantitative RT-PCR (RT-qPCR) detecting Seg-10 assay widely used for the molecular diagnosis of BTV from field samples. Results showed a difference mean of 0.30 log between the two assays with these samples (p < 0.05). Anyway, the analysis of correlation demonstrated that both assays provided similar measurements with a very close agreement between the systems.
Introduction
Digital polymerase chain reaction (dPCR) is a recent technology enables an accurate absolute quantification of target nucleic acids. The principle of dPCR was first described in the 1990s (1, 2). The dPCR approach combines limiting dilutions, PCR, and Poisson distribution to quantitate the total number of amplifiable targets within a sample (1).
Digital technology is based on end-point PCR that provides the direct measure of nucleic acids without relying on a standard curve (3). In a dPCR assay, the sample is randomly partitioned into individual reactions, such that some contain no nucleic acid template and others contain one or more template copies (4). The very high number of partitions of the sample allows a significant precision on results (5).
After end-point PCR amplification, each partition is analyzed and distinct as positive (presence of PCR products) or negative (absence of PCR products). The fraction of amplification positive partitions is used to estimate the concentration of the initial target sequence using binomial Poisson statistics (4, 6). Nowadays, different dPCR commercial platforms are available as a useful tool for precise quantification of nucleic acids in a variety of basic research and clinical applications (4, 7–9).
Despite the fact that quantitative PCR or real-time PCR (qPCR) has been widely utilized to quantify nucleic acid in many areas of research and diagnostics tests, it has same disadvantages such as the necessity for a standard curve, the lack of universal standards of known quantity, and also the efficiency can be influenced by many factors including inhibitors. The qPCR is the gold standard for molecular quantitation in viral diagnostics; dPCR offers several potential advantages over qPCR (10). Digital PCR uses an amplification reaction system similar to a system of standard qPCR, but does not require the same level of calibration or controls as traditionally used in qPCR (5).
Digital PCR overcomes the need for a standard curve, and it is increasingly used for DNA/RNA viral quantification, in human and animal health (11–17). Bluetongue (BT) is an Office International des Epizooties (OIE)-listed infectious disease of domestic and wild ruminants (18), transmitted mainly through the bites of Culicoides midges. Bluetongue virus (BTV), which belongs to the genus Orbivirus of the family Reoviridae, causes significant economic losses due to mortality, decline in production, and restrictions on trade in animals from infected areas (19). BTV genome consists of 10 linear double strand segments (Seg-1 to Seg-10), encoding seven structural proteins (VP1-Vp7) and five non-structural proteins (NS1 to NS4 and S10-ORF2) involved in viral replication, morphogenesis, and assembly processes (20).
Currently, there are 24 classic serotypes of the virus, all capable of causing BT, plus a series of new serotypes, defined as atypical because infected animals are asymptomatic (21–27).
For molecular diagnostic laboratories, OIE recommends the use of a real-time RT-PCR (RT-PCRNS3) assay in order to confirm clinical cases, to establish uninfected animals before handling, to check the prevalence of infection, and for surveillance purposes.
The method real-time RT-PCRNS3 (21) allows to detect all circulating known BTV serotypes, by retro-transcription and amplification of a region of segment 10 of the viral RNA, coding for a non-structural NS3 protein, purified by blood-EDTA, biological liquid, and organ tissues taken from susceptible species and by hematophagous insects. RT-PCR targeting in Seg-2 coding for a least conserved virion outer capsid protein (VP2) identifies the specific BTV serotype (28, 29).
Because of its accuracy and precision, real-time quantitative RT-qPCR is the method of choice when quantitative analysis is required. However, there are no available reference certificated material (standard) for the quantification of bluetongue virus. Toussaint et al. (30) used recombinant plasmid obtained by inserting BTV PCR product into PCRII-TOPO vector by TA-cloning and RNA synthesized in vitro with Riboprobe system T7 (Promega). (31) employed RiboMax Large scale RNA production System (Promega) for transcription of standard RNA by bluetongue recombinant plasmid PGEM –T Easy Vector RNA. Maan et al. (32) transcribed from recombinant pGEMT plasmid RNA BT using the mMessage mMachine T7 Ultra Kit (Life Technologies). Overall, the use of different calibration standards in different performing assays can lead to non-reproducible results between laboratories, even when testing the same material (12, 33).
To overcome these limitations, we aimed to develop a new method for quantification of BTV Seg-10 by droplet digital RT-PCR (RT-dd-PCR), using nucleic acids purified from complex matrices such as blood, tissues, and midges, that notoriously contain strong PCR inhibitors. The RT-PCRNS3 method recommended by OIE was transferred to the digital platform, optimized by using RNAs purified from serially 10-fold dilutions of a BTV-1 isolate, spiked into fresh ovine EDTA-blood and spleen homogenate. The limit of detection (LoD) and the limit of quantification (LoQ) were established by using serial dilutions of BTV-1 RNA purified. Using RNAs purified from field samples, the newly developed assay was compared, with the RT-qPCR NS3 detecting the same target Seg-10.
Materials and Methods
Virus Strain, Spiked-In Samples, and Field Samples Collection
BTV-1/2006 strain, isolated from the spleen of an infected sheep that succumbed during the BTV-1 outbreak in Sardinia (Italy) during 2006, was employed for the study. The BTV-1/2006 strain was titrated by end-point onto VERO cells by Sperman/Karber method (105.43/TCID50/ml). Four 10-fold serial dilutions of BTV-1 suspensions (from 102.43 to 10−0.57 TCID50/ml) spiked into ovine blood samples and in ovine spleen homogenates (10% w/v) were used to evaluate possible inhibition caused by matrices. The optimized droplet digital RT-PCR (RT-ddPCR) was finally evaluated on a total of 44 field samples tested positive for real-time RT-PCRNS3, including 16 of Culicoides imicola and 28 of ovine blood EDTA. Culicoides samples were collected in farms of southern Sardinia, during entomological surveillance, by the national surveillance plan, in the years 2017 and 2018. The blood samples were collected from farms located in the same part of the region. Four negative blood samples were used as negative controls. The blood samples were refrigerated at 5° ± 3°C. C. imicola samples were stored at −70 ± 10°C until the time of processing and analysis. The results were compared with those obtained by the RT-qPCRNS3.
Nucleic Acid Purification
RNA purification from viral suspensions, spiked-in samples, and field samples, mosquitos included, was performed by MagMax Core Nucleic Acid Purification Kit (Applied Biosystems-ThermoFisher Scientific-USA) in automated sample preparation workstation MagMAX Express 96 (Applied Biosystems) according to the manufacturer's instructions. Primers and probe were the same published by OIEb (34), which amplify a portion of Seg-10. This RT-PCRNS3 assay is widely used for molecular diagnosis of BTV. In order to optimize RT-ddPCR assay, the same set of primers and probe were used in all experiments.
Droplet Digital RT-PCR (RT-ddPCR) Optimization
Purified nucleic acids were quantified using QX200™ Droplet Digital PCR System (BioRad Laboratories, USA). The assay was performed in 20 μl using the One-step RT-ddPCR Advanced Kit for Probe (Bio-Rad) consisting of: 5 μl supermix 4 X, 2 μl of reverse transcriptase (20 U/μl), 1 μl of 300 mM DTT, 2 μl of RNA template, and the primers and probe at final concentrations of 0.9 and 0.25 μM, respectively, according to the manufacturer's instructions. In order to optimize RT-ddPCR assay, primers and probe were further tested at different concentrations within the range of 0.4–0.9 μM and 0.15–0.25 μM by using RNA purified from different BTV-1 suspensions (titer 102.43, 101.43, 100.43, 10−0.57 TCID50/ml). RNA was denatured with primers for 5 min at 95°C, stabilized for 3 min at 4°C, and then added to the RT-ddPCR mixture reaction as indicated above. No template controls (NTC) were used for monitoring primer-dimer formation and contaminations. Twenty microliters RT-ddPCR mixture/sample were placed in each well of droplet generator DG8 cartridge (BioRad Laboratories, USA) with 70 μl of droplet generator oil (BioRad Laboratories, USA) and emulsified in QX-200 Droplet Generator (BioRad Laboratories, USA) partitioning in 20,000 water in oil nanoliter-size droplet. Then, a volume of 40 μl of emulsion/sample was transferred to a 96-well reaction plate (Eppendorf, Hauppauge, NY), heat-sealed with pierceable foil sheets by the PX1™ PCR Plate Sealer (BioRad Laboratories, USA), and amplified in C1000 Touch™ Thermal Cycler (BioRad Laboratories, USA). So as to allow an optimal distinction between positive and negative droplets, PCR annealing temperature was optimized by thermal gradient from 55 to 65°C. Cycling conditions were the following: 48°C for 30 min, 95°C for 2 min, followed by 50 cycles of 95°C for 15 s, 55C-65°C for 30 s, and 72°C for 30 s. At the end of amplification, the PCR plates were read by the QuantaSoft Droplet Reader (BioRad Laboratories, USA) that measures the fluorescence intensity of each droplet and detects the size and shape as droplets pass detector. The absolute concentration of each sample was automatically reported as copy number of Seg-10 of BTV/μl by the ddPCR QuantaSoft Software V.1.7.4.0917 (BioRad Laboratories, USA) by calculating the ratio of the positive droplets over the total droplets combined with Poisson distribution with 95% confidence interval.
Performance of RT-ddPCR Assay
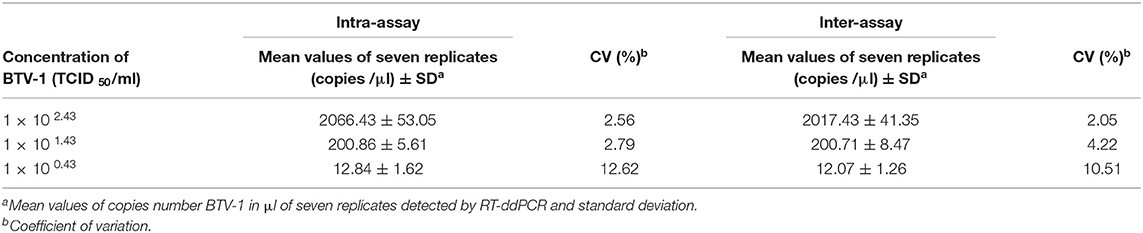
Linearity of the assay was defined by using 10-fold dilutions of BTV1-RNA purified (ranging from 102, 43 TCID50/ml up to 10−0, 57 TCID50/ml), analyzed in seven replicates, with optimized conditions of primers, probe, and amplification program. The range of linearity was defined by plotting the log value of titers (TCID50/ml) BTV-1 dilution against the absolute measured value (copies/μl). The LoD and LoQ were evaluated by using 20 replicates of 5-fold serial dilutions of BTV-1 RNA purified (ranging from 101.43 to 10−1.37 TCID50/ml). The LoD of RT-ddPCR was determined as the last serial dilution detected in 95% of replicates, whereas the LoQ was set at the lowest dilution showing a coefficient of variation percentage below the threshold (CV% = 25) for acceptance criteria of quantitative methods (35, 36). Furthermore, to evaluate the intra-assay and inter-assay repeatability, three different dilutions of BTV-1 (102.43, 101.43, 100.43 TCID50/ml) were tested in seven replicates in two different days; the CV% was then considered. pGEM T-easy vector (Promega, Milan-Italy) carrying Seg-10 of a BTV-1 strain in serial dilutions, from 2 × 104 to 2 × 101 copies, was also used to evaluate accuracy of the RT ddPCR for quantification purposes. Finally, matrix effect was evaluated comparing R2 values of RNA isolated from four BTV-1 dilutions (from 102.43 to 10−0.57 TCID50/ml), and RNA isolated from four blood and four spleen homogenates spiked (10% w/v) with the same BTV-1 TCID50/ml.
Quantitative Real Time Assay (RT-qPCRNS3)
RT-qPCRNS3 assay, as described above, was performed using the 7900HT Fast Real-Time PCR System (Applied Biosystems) and SuperScript™ III Platinum™ One-Step qRT-PCR Kit (ThermoFisher- Life Technologies). RNA was denatured with 0.4 μM primers for 5 min at 95°C, stabilized for 3 min at 4°C, and then added to the RT-qPCR mixture reaction as indicated above. The one step RT-qPCR mixture was prepared in 25 μl reaction volume, 12.5 μl 2X Reaction Mix, 0.5 μl of 50X ROX Reference Dye, 1 μl Mg2SO4 50 mM, 0.5 μl SSIII RT-Platinum™ Taq Mix, 0.2 μM probe, and 2 μl of RNA. Cycling conditions were the following: 48°C for 30 min, 95°C for 2 min, followed by 50 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s. In order to assess the standard curve the pGEM T-easy vector (Promega, Milan-Italy) carrying Seg-10 of a BTV-1 strain was employed, available at IZSAM (pGEM-BTV-NS3, 108 copies/μl). The standard curve was constructed placing Cq values of seven serial 10-fold dilutions of pGEM-BTV-NS3 (1 × 107 copies/μl up to 1 × 101 copies/μl), performed in triplicate wells, against the log value of the number of copies of BTV-1 Seg-10. BTV-1 Seg-10 copy number in each sample was determined by Cq value to the standard curve. Cq value was generated by 7900 Software SDS 2.4.1 (Applied Biosystems). Amplification Efficiency and R2 of the calibration curve were calculated.
Comparison of RT-ddPCR and RT-qPCRNS3 Assays for Quantitation of BTV-1 Seg-10 in Field Samples
To evaluate the performance of RT-ddPCR against the RT-qPCRNS3, 44 field samples were tested in triplicate wells with both assays and the difference of log of quantification was evaluated to verify the agreement between two assays.
Statistical Analysis
Data were converted into a logarithmic format. Linear regression analyses were conducted using Microsoft Excel 2010. In order to compare quantification by RT-ddPCR and RT-qPCRNS3 statistical analysis was conducted by Statgraphics 18 Centurion Software (Version 18.1.06).
Results
Optimization of RT-ddPCR
The primers and probe concentrations were optimized by using RNA purified from dilution of BTV-1 from 102.43 to 10−0.57. As shown in Figure 1, the optimal primers and probe concentrations were 0.9 and 0.25 μM per reaction, respectively, i.e., the highest concentrations, among those tested, as recommended by the manufacturer (Biorad). PCR annealing temperature was optimized by thermal gradient from 55 to 65°C. The optimum annealing temperature was at 58.8°C, which resulted in the greatest difference of fluorescence amplitude between positive and negative droplets. The optimal cycling conditions were: 48°C for 30 min, 95°C for 2 min, followed by 50 cycles of 95°C for 15 s, 58.8°C for 30 s, and 72°C for 30 s, 1 cycle of 98°C for 10 min essential for droplet stabilization and infinite 12°C hold. It was used a 2.5°C/s ramp rate to ensure each droplet reached the correct temperature for each step during the cycling. The parameters above were used in following RT-ddPCR experiments of our study.
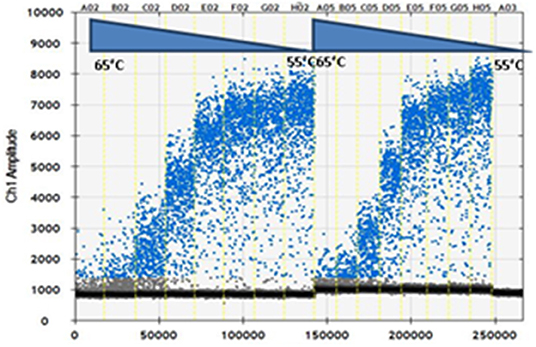
Figure 1. Optimization of droplet digital RT-PCR (RT-ddPCR). Fluorescence amplitude plotted against annealing temperature gradient for 0.7 μM primers (lanes A02-H02) and 0.9 μM primers (lanes A05-H05): 65, 64.3, 63, 61.3, 58.8, 56.9, 55.7, and 55°C; 0.25 μM probe. Lane A03 No Template Control (NTC).
Performance of RT-ddPCR Assay
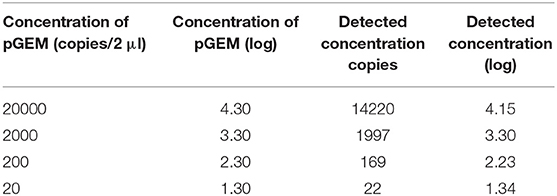
The trend line of detection BTV-1 concentration by RT-ddPCR exhibited a good degree of linearity (R2 ≥ 0.995) in the range from 102.43 TCID50/ml to 10−0.57 TCID50/ml (Figure 2). According to the definition, the LoD established was 10−0.67 TCID50/ml BTV1 corresponding to 0.72 copies/μl (Table 1). Conversely, the LoQ determined was 100.03 TCID50/ml of BTV-1 (3.05 copies/μl) with a CV% value 21 (Table 1). Results of the copy number obtained by RT-ddPCR relative to the four pGEM 10-fold dilutions give a good degree of linearity (R2 = 0.998), especially in the range 20–2000 copies as reported in Table 2 and Figure 3. The CV% values were also considered to assess repeatability. The analysis of the seven replicates for the three dilutions gave back CV% lower than the threshold (CV% = 25) in all cases for both intra- and inter-assay (Table 3).

Figure 2. Linear regression of RT-ddPCR assay using RNA extracted from 10-fold dilution of BTV-1/2006 from TCID50 102.43 to 10−0.57, analyzed in seven replicates, at the final optimized conditions.

Table 1. Limit of detection (LoD) and limit of quantification (LoQ) of droplet digital RT-PCR (RT-ddPCR).

Figure 3. Linear regression of RT-ddPCR assay using four 10-fold dilution of pGEM-BTV-NS3 from 2 × 104 to 2 × 101, analyzed in three replicates, at the final optimized conditions.
Evaluation Matrix-Effect of RT-ddPCR
BTV-1 suspensions from 102.43 to 10−0.57 TCID50/ml spiked in blood and in spleen tissue showed good resilience to inhibitor blood and tissue factors compared to same viral suspensions without matrix, in linear range of the RT-ddPCR assay. As indicated in Figure 4 quantitative linearity analysis in matrices showed a good linearity with R2 close to 1.

Figure 4. RT-ddPCR Matrix effect. Linear regression analysis of 10-fold serial dilutions of BTV-1/2006, blood and spleen spiked with the same BTV-1/2006 TCID50/ml. Blue rhombus BTV-1/2006 viral suspension R2 = 0.9954; Green triangle BTV-1/2006 spiked-spleen R2 = 0.9964; Red square BTV-1/2006 spiked-blood R2 = 0.9996.
Comparison of RTdd-PCR and RT-qPCRNS3 Assays for Quantitation of Seg-10 of BTV in Field Samples
The two assays were compared calculating the difference between the logarithm of RT-qPCRNS3 quantification, i.e., copies of Seg-10 of BTV /μl of sample and the RT-ddPCR correspondent data. In field samples, the log difference average was 0.30 (p = 0.04968), in detail of 0.20 and 0.40, respectively, for blood EDTA samples and Culicoides midge as shown in Table 4 and Table S1. Figure 5 shows the correlations between the log copies of Seg-10 of BTV/μl of sample in RT-ddPCR with those in RT-qPCRNS3.

Table 4. Comparison of RT-qPCRNS3 and RT-ddPCR assays for quantitative detection of BTV in field samples.

Figure 5. Correlations between real-time RT-qPCRNS3 and ddPCR for field samples. Fit regression model (blue). 95% Confidence limits (gray). 95% Prediction limits (green).
Discussion
BTV is responsible for an important disease of ruminant that induces variable clinical signs, its pathogenicity depends on the host species. Seasonal incursions of the disease in parts of Europe (Mediterranean basin) during the summer cause economic losses due to direct impact on livestock and trade restriction. In the last few years, new serotypes have been identified, probably, originated under evolutionary dynamics and selection pressure (37); new potential vectors have been identified (38, 39); some field strains/serotypes proved to be able to transmit vertically or horizontally, to reassort their RNA, and to alter their pathogenicity, specificity, and spread capacity (37). Nowadays, the molecular diagnosis of BTV (real time RT-PCRNS3) by Hofmann allows to identify all circulating serotypes. These knowledges designate a complex contest in which it is necessary to have more sensitive and accurate methods, not only to assess the presence/absence of the virus but also to evaluate the RNA viral load in natural and/or experimental infected samples. In this study, we established a novel RT-ddPCR assay for the quantification of RNA BTV at low concentration of virus and in spiked and field samples including whole blood, tissues, and midges, showing the power and the potential of RT-ddPCR assay. This assay has been enabled to detect absolute target copy number in field samples without employing recombinant DNA or RNA plasmid to construct the standard curve. As a consequence, the amplification efficiency bias observed with quantitative RT-qPCR is minimized. A certified reference material (CRM) for BTV quantification is not available; therefore, further independent quantification methods are needed in order to quantify the BT virus as number of copies/μl sample, in the greatest way possible. NS3 gene primers and probe sequences used in RT-ddPCR assay were from the previously published BT reverse transcriptase real-time PCR (RT-PCRNS3) by Hofmann et al. (21) and OIE (34), mentioned above for diagnostic purposes. RT-qPCRNS3 is performed using degenerate primer and probe set that detect all serotypes of BTV; RT-ddPCRs were done with some specific modifications in order to optimize all parameters. RT-ddPCR assay exhibited a good degree of linearity (R2 ≥ 0.995) in the range from 102.43TCID50/ml to 10−0.57 TCID50/ml, overcoming the dependence on the availability of references or standards. BTV-1 102.43TCID50/ml was considered the first point of dynamic range. Evaluating the dynamic range of the digital assay, the 103.43 TCID50/ml viral dilution (corresponding to Cq 22.5 in RT-qPCRNS3) was excessively concentrated to be detectable because all of the droplets were positive (saturation of the reaction). In contrast, the last point of the range (BTV-1 10−1.57 TCID50/ml) was excessively diluted (data not shown); then the last point was 10−0.57 TCID50/ml viral dilution. As expected, the dynamic ranges of newly RT-ddPCR were lower (four orders of magnitude) than those obtained for the corresponding qPCRs for all samples. Several studies reported that digital PCR shows higher tolerance to inhibitors; as an end-point measurement, it can reduce the biases linked to matrix type often observed with qPCR (40), especially in clinical specimen (stool, sputum, tissue) (16, 17, 41). Pavšič et al. (42) suggested that it may be possible to perform ddPCR on samples without extraction of nucleic acid. Quantitative RT-ddPCR data obtained from blood and spleen showed a good level of tolerance to inhibitors when compared to the same TCID50/ml BTV-1 dilutions with a good linearity close to 1. Despite pGEM plasmid not being a properly standard reference, the use of serial dilutions of the plasmid allowed us to evaluate the precision of the RT-ddPCR for quantification purposes. These results suggest that RT-ddPCR assay provides an accurate quantification of BTV, unevenly distributed in different matrices, without precluding the quantitation efficiency due to impurities in the sample. The LoD and the LoQ were established at 10−0.67 TCID50/ml and 100.03 TCID50/ml of BTV-1 corresponding to 0.72 and 3.05 copies/μl, respectively. Data of intra- and inter-assay repeatability of the RT-ddPCR showed a good repeatability with a variability below the threshold (CV 25%) for acceptance criteria of quantitative methods (35, 36) and were assessed in two different days with <13% variability between the results. The good precision of RT-ddPCR is linked to the intrinsic characteristics of the method, enabling absolute quantification of the viral target at different work conditions. In the last part of this study, we compared the RT-ddPCR against RT-qPCRNS3 using 44 field samples that resulted positive to real-time RT-PCRNS3. Applicability of the technology has been tested on characteristic matrices of field and on the range of viral load generally distributed in routine samples. The collected whole blood samples, tested as positive with the official method, stored at 4°C, and selected for comparative study with both assays, showed the same Cq found in RT-qPCRNS3 during the diagnosis (data not shown). These results support a previous work in which the persistence of BTV in stored blood samples were observed (43). The main limitation of RT-ddPCR respect to RT-qPCRNS3 was that the higher concentration of template resulted in saturation of positive droplets, confirming the saturation at high concentration of RNA. Thus, the concentrated samples were diluted for viral load quantification in RT-ddPCR. The results showed a log difference average of 0.30 in field samples. A higher Seg-10 BTV-1 quantification by RT-qPCRNS3 could be addressed to the Cq values that are established on transcription and amplification efficiency. This could affect the RT-qPCR, but does not have any effects on digital PCR. Anyway, the analysis of correlation demonstrated that both assays provided similar measurements with a very close agreement between the systems. On the other hand, the main advantage of the RT-ddPCRNS3 is the ability to quantify any BTV serotype without the use of standard curves properly constructed for each specific group. Moreover, the implementation of the serotype-specific multiplexing system should be suitable to detect and quantify simultaneously different BTV serotypes in case of viral coinfection in areas that are circulating different BTV serotypes. Despite its general advantages, compared to qPCR, dPCR is more time consuming and labor intensive, but will certainly give further, in terms of applicability and throughput.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The study did not involve any animal experiment. Samples were collected from sheep with standard procedures by the Sardinian Veterinary Services A.T.S. and submitted to the Experimental Zooprophylactic Institute of Sardinia for BTV testing. Special authorization for sampling activities was not necessary; these actions are regulated by the Italian Ministry of Health and performed in case of BT outbreaks.
Author Contributions
MT, AR, and RB carried out experimental work. AR and MT wrote the first draft of the manuscript with support from GP and MD. All authors contributed to conception and design of the study, data analysis and interpretation, manuscript revision, and read and approved the submitted version.
Funding
This study was funded by the Italian Ministry of Health (grant # RC IZS SA 02/16), and by Experimental Zooprophylactic Institute of Sardinia—Sassari—Italy (grant #FNS 2016.11).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00170/full#supplementary-material
References
1. Sykes PJ, Neoh SH, Brisco MJ, Hugues E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. (1992) 13:444–9.
2. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA. (1999) 16:9236–41. doi: 10.1073/pnas.96.16.9236
3. Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. (2015) 61:79–88. doi: 10.1373/clinchem.2014.221366
4. Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Analyt Chem. (2012) 84:1003–11. doi: 10.1021/ac202578x
5. Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. (2011) 83:8604–10. doi: 10.1021/ac202028g
6. Whale AS, Cowen S, Foy CA, Huggett JF. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS ONE. (2013) 8:e58177. doi: 10.1371/journal.pone.0058177
7. Quan PL, Sauzade M, Brouzes E. dPCR: a technology review. Sensors. (2018) 18:1271. doi: 10.3390/s18041271
8. Sedlak RH, Jerome KR. Viral diagnostics in the era of digital polymerase chain reaction. Diagn Microbiol Infect. Dis. (2013) 75:1–4. doi: 10.1016/j.diagmicrobio.2012.10.009
9. Doyle T, Smith C, Vitiello P, Cambiano V, Johnson M, Owen A, et al. Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy. Clin Infect Dis. (2012) 54:724–32. doi: 10.1093/cid/cir936
10. Sedlak RH, Jerome KR. The potential advantages of digital PCR for clinical virology diagnostics. Expert Rev Mol. Diagn. (2014) 14:501–7. doi: 10.1586/14737159.2014.910456
11. Varela MF, Monteiro S, Rivadulla E, Santos R, Romalde JL. Development of a novel digital RT-PCR method for detection of human sapovirus in different matrices. J Virol Methods. (2018) 254:21–4. doi: 10.1016/j.jviromet.2018.01.005
12. Jia P, Purcell MK, Pan G, Wang J, Kan S, Liu Y, et al. Analytical validation of a reverse transcriptase droplet digital PCR (RT-ddPCR) for quantitative detection of infectious hematopoietic necrosis virus. J Virol Methods. (2017) 245:73–80. doi: 10.1016/j.jviromet.2017.03.010
13. Abachin E, Convers S, Falque S, Esson R, Mallet L, Nougarede N. Comparison of reverse-transcriptase qPCR and droplet digital PCR for the quantification of dengue virus nucleic acid. Biologicals. (2018) 52:49–54. doi: 10.1016/j.biologicals.2018.01.001
14. Wu X, Lin H, Chen S, Xiao L, Yang M, An W, et al. Development of a reverse transcriptase droplet digital PCR (RT-ddPCR) for sensitive and rapid detection of Japanese encephalitis virus. J Virol Methods. (2017) 248:166–71. doi: 10.1016/j.jviromet.2017.06.015
15. Fraisse A, Coudray-Meunier C, Martin-Latin S, Hennechart-Collette C, Delannoy S, Fach P, et al. Digital RT-PCR method for hepatitis A virus and norovirus quantification in soft berries. Int J Food Microbiol. (2017) 243:36–45. doi: 10.1016/j.ijfoodmicro.2016.11.022
16. Sedlak RH, Nguyen T, Palileo I, Jerome R, Kuypers J. Superiority of digital Transcription-PCR(RT-PCR) over RealTime RT-PCR for quantitation of highly divergent human rhinoviruses. J Clin Microbiol. (2017) 55:442–9. doi: 10.1128/JCM.01970-16
17. Strain M, Lada S, Luong T, Rought SE, Gianella S, Terry VH, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE. (2013) 8:4:e55943. doi: 10.1371/journal.pone.0055943
18. OIEa. Chapter 8.3: Infection with bluetongue virus. In: OIE Terrestrial Animal Health Code. Paris: OIE (2019).
19. Rushton J, Lyons N. Economic impact of Bluetongue: a review of the effects on production. Vet Ital. (2015) 51, 401–6. doi: 10.12834/VetIt.646.3183.1
20. Patel A, Roy P. The molecular biology of Bluetongue virus replication. Virus Res. (2014) 182:5–20. doi: 10.1016/j.virusres.2013.12.017
21. Hofmann M, Griot C, Chaignat V, Perler L, Thür B. Bluetongue disease reaches Switzerland. Schweiz Arch. Tierheilk. (2008) 150:49–56. doi: 10.1024/0036-7281.150.2.49
22. Maan S, Maan NS, Nomikou K, Batten C, Antony F, Belaganahalli MN, et al. Novel bluetongue virus serotype from Kuwait. Emerg Infect Dis. (2011) 17:886–9. doi: 10.3201/eid1705.101742
23. Zientara S, Sailleau C, Viarouge C, Höper D, Beer M, Jenckel M, et al. Novel bluetongue virus in goats, Corsica, France, 2014. Emerg Infect Dis. (2014) 20:2123–5. doi: 10.3201/eid2012.140924
24. Schulz C, Breard E, Sailleau C, Jenckel M, Viarouge C, Vitour D, et al. Bluetongue virus serotype 27: detection and characterization of two novel variants in Corsica, France. J Gen Virol. (2016) 97:2073–83. doi: 10.1099/jgv.0.000557
25. Sun EC, Huang LP, Xu QY, Wang HX, Xue XM, Lu P, et al. Emergence of a novel bluetongue virus serotype, China 2014. Transbound Emerg Dis. (2016) 63:585–9. doi: 10.1111/tbed.12560
26. Savini G, Puggioni G, Meloni G, Marcaccia M, Di Domenico M, Rocchigiani AM, et al. Novel putative Bluetongue virus in healthy goats from Sardinia, Italy. Infect Genet Evol. (2017) 51:108–17. doi: 10.1016/j.meegid.2017.03.021
27. Marcacci M, Sant S, Mangone I, Goria M, Dondo A, Zoppi S, et al. One after the others: a novel Bluetongue virus strain related to Toggenburg virus detected in the Piemont region (North-western Italy), extends the panel of novel atypical BTV strains. Transbound Emerg Dis. (2018) 65:370–4. doi: 10.1111/tbed.12822
28. Huismans H, Erasmus BJ. Identification of the serotype-specific and groupspecific. Onderstepoort J Vet Res. (1981) 48:51–8.
29. Shaw AE, Ratinier M, Nunes SF, Nomikou K, Caporale M, Golder M, et al. Reassortment between two serologically unrelated bluetongue virus strains is flexible and can involve any genome segment. J Virol. (2013) 87:543–57. doi: 10.1128/JVI.02266-12
30. Toussaint JF, Sailleau C, Breard E, Zientara S, De Clercq K. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J Virol Methods. (2007) 140:115–23. doi: 10.1016/j.jviromet.2006.11.007
31. Yin HQ, Zhang H, Shi LJ, Yang S, Zhang GP, Wang SQ, et al. Detection and quantitation of bluetongue virus serotypes by a TaqMan probe-based real-time RT-PCR and differentiation from epizootic hemorrhagic disease virus. J Virol Methods. (2010) 168:237–41. doi: 10.1016/j.jviromet.2010.04.018
32. Maan S, Maan NS, Belaganahalli MN, Rao PP, Singh KP, Hemadri D, et al. Full-genome sequencing as a basis for molecular epidemiology studies of blue-tongue virus in India. PLoS ONE. (2015) 10:e0131257. doi: 10.1371/journal.pone.0131257
33. Pavšič J, Devonshire A, Blejec A, Foy CA, Van Heuverswyn F, Jones GM, et al. Inter-laboratory assessment of different digital PCR platforms for quantification of human cytomegalovirus DNA. Analyt Bioanalyt Chem. (2017) 409:2601–14. doi: 10.1007/s00216-017-0206-0
34. OIEb. Chapter 3.1.3: Bluetongue. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (2019). Available online at: www.oie.int
35. George D, Czech J, John B, Yu M, Jennings LJ. Detection and quantification of chimerism by droplet digital PCR. Chimerism. (2013) 4:102–8. doi: 10.4161/chim.25400
36. EURL-GMFF (European Union Reference Laboratory for GM Food and Feed). Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. (2015). Available online at: https://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm (accessed October 21, 2015).
37. Savini G. Bluetongue: a disease that does not speak ‘one tongue,' only. Vet Ital. (2015) 51:247–8.
38. Foxi C, Meloni G, Puggioni G, Manunta D, Rocchigiani A, Vento L, et al. Bluetongue virus detection in new Culicoides species in Sardinia, Italy. Vet Rec. (2019) 184:621. doi: 10.1136/vr.105118
39. Paslaru AI, Mathis A, Torgerson P, Veronesi E. Vector competence of pre-alpine Culicoides(Diptera:Ceratopogonidae) for bluetongue virus serotypes 1, 4 and 8. Parasites Vectors. (2018) 11:466. doi: 10.1186/s13071-018-3050-y
40. Burns MJ, Burrell AM, Foy C. The applicability of digital PCR for the assessment of detection limits in GMO analysis. Eur Food Res Technol. (2010) 231:353–62. doi: 10.1007/s00217-010-1277-8
41. Vynck M, Trypsteen W, Thas O, Vandekerckhove L, De Spiegelaere W. The future of the polymerase chain reaction in virology. Mol Diagn Ther. (2016) 20:437–47. doi: 10.1007/s40291-016-0224-1
42. Pavšič J, Zel J, Milavec M. Digital PCR for direct quantification of viruses without DNA extraction. Anal Bioanal Chem. (2016) 408:67–75. doi: 10.1007/s00216-015-9109-0
Keywords: Bluetongue, Reoviridae, RNA quantification, droplet digital RT-PCR, Quantitative Real Time -RT- PCR
Citation: Rocchigiani AM, Tilocca MG, Portanti O, Vodret B, Bechere R, Di Domenico M, Savini G, Lorusso A and Puggioni G (2020) Development of a Digital RT-PCR Method for Absolute Quantification of Bluetongue Virus in Field Samples. Front. Vet. Sci. 7:170. doi: 10.3389/fvets.2020.00170
Received: 29 November 2019; Accepted: 11 March 2020;
Published: 21 April 2020.
Edited by:
Jesus Hernandez, Centro de Investigación en Alimentación y Desarrollo (CIAD), MexicoReviewed by:
Faten Abdelaal Okda, St. Jude Children's Research Hospital, United StatesSemmannan Kalaiyarasu, ICAR-National Institute of High Security Animal Diseases (ICAR-NIHSAD), India
Copyright © 2020 Rocchigiani, Tilocca, Portanti, Vodret, Bechere, Di Domenico, Savini, Lorusso and Puggioni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria G. Tilocca, bWd0aWxvY2NhQGdtYWlsLmNvbQ==
 Angela M. Rocchigiani
Angela M. Rocchigiani Maria G. Tilocca
Maria G. Tilocca Ottavio Portanti
Ottavio Portanti Bruna Vodret
Bruna Vodret Roberto Bechere
Roberto Bechere Marco Di Domenico
Marco Di Domenico Giovanni Savini
Giovanni Savini Alessio Lorusso
Alessio Lorusso Giantonella Puggioni
Giantonella Puggioni