
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 02 April 2020
Sec. Comparative and Clinical Medicine
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00146
 Alba Gaspardo1
Alba Gaspardo1 Augusta Zannoni1,2
Augusta Zannoni1,2 Silvia Turroni3,4
Silvia Turroni3,4 Monica Barone3
Monica Barone3 Maria Chiara Sabetti1
Maria Chiara Sabetti1 Renato Giulio Zanoni1
Renato Giulio Zanoni1 Monica Forni1,2
Monica Forni1,2 Patrizia Brigidi3,4
Patrizia Brigidi3,4 Marco Pietra1*
Marco Pietra1*The increasing incidence of gastrointestinal tract pathologies in dogs and the worrisome topic of antibiotic resistance have raised the need to look for new therapeutic frontiers. Of these, the use of probiotics represents a potential therapeutic alternative. Lactobacillus kefiri (Lk) is a species of Lactobacillus isolated from kefir. Previous studies have demonstrated that its administration in mice downregulates the expression of proinflammatory mediators and increases anti-inflammatory molecules in the gut immune system. It also regulates intestinal homeostasis, incrementing immunoglobulin A (IgA) secretion. Since Lk has never been studied as a single probiotic in dogs, the aim of this study was to evaluate the safety of Lk in dogs, and its effect on IgA secretion and on intestinal microbiota composition. Ten healthy dogs without a history of gastrointestinal diseases were included. The dogs received Lk at a dose of 107 live microorganisms orally, once daily for 30 days. The fecal samples were tested before administration, in the middle, at the end, and 30 days after discontinuation. The IgA secretion concentration and the microbiota composition were evaluated on the fecal samples. The results in this study suggested that Lk did not influence the concentration of IgA, nor significant changes of the intestinal microbiota were observed during and after the treatment. Therefore, additional studies are needed to investigate if a higher daily dosage of Lk can influence the intestinal homeostasis of dogs.
In recent years, intestinal microbiota has become increasingly relevant for veterinary scientists and has been studied for its role on the welfare of the host (1). It is an ecosystem including mainly bacteria, but also archaea, fungi, protozoa, and viruses, which plays several roles in the host physiology by means of a range of metabolic and immunological interactions. In fact, this complex ecosystem helps in the digestion of food by assisting the absorption and metabolism of nutrients, and has trophic and protective functions (2). It defends the gastrointestinal tract (GIT) against pathogenic organisms, promotes mucus production and enterocyte turnover, and modulates host immune development and functionality (3). In particular, commensal bacteria provide intestinal immune protection by regulating, among other things, the secretion of IgA, the lack of which seems to be correlated with chronic enteropathies in dogs (4, 5).
Canine chronic enteropathies, categorized into four classes (food responsive; antibiotic responsive; immunosuppressant responsive; nonresponsive enteropathy) according to the response to treatment, are multifactorial diseases where host genetic factors, the immune system, and indigenous intestinal bacteria are supposed to be engaged in intricate interactions (6, 7).
Dogs affected by antibiotic responsive enteropathy (ARE) are generally young, predominantly belong to large breeds, and show remission of clinical signs following antimicrobial administration (metronidazole, tylosin, doxycycline, rifaximine) (8, 9). It is thought that antimicrobials are able to modify the intestinal microbial population by counteracting its imbalances (i.e., dysbiosis); however, although a short-term response to metronidazole and tylosin has been reported, very few papers have described the long-term control of ARE (9–11).
Moreover, there is now evidence that prolonged treatment with antibiotics, particularly with metronidazole, can lead to permanent unfavorable changes in the microbiota, promoting antimicrobial resistance, currently one of the most important threats to public health (12, 13).
Therefore, the increasing incidence of GIT pathologies and the worrisome topic of antibiotic resistance create the need for new therapeutic options (14), and toward replacing antibiotics with (a) probiotics (live microorganisms that confer a healthy benefit on the host); (b) prebiotics (a substrate selectively utilized by host microorganisms useful for reestablishing a eubiotic microbiota layout); (c) synbiotics (a mixture of probiotics and prebiotics having a synergistic action on host health); and (d) postbiotics (soluble factors or metabolic byproducts, secreted by live bacteria or released after bacterial lysis expressing biological activity in the host).
Lactic acid bacteria (LAB), including Lactococcus, Streptococcus, Enterococcus, Pediococcus, Leuconostoc, and Lactobacillus, are probiotic bacteria that are normally part of the intestinal microbiota of dogs and cats (2). Lactobacillus species are distributed throughout the canine intestinal tract in varying amounts (2). Several strains of Lactobacilli have specifically been studied for their ability to reduce the number of pathogenic bacteria in the canine intestine (15–20).
Among different naturally fermented foods and their potential probiotic properties, particular attention has recently been focused on kefir, a dairy product that could modulate canine intestinal microbiota if given regularly (21). Kefir has a complex composition of microbial organisms, which includes several species of LAB as well as acetic acid bacteria and yeast (22). Lactobacillus kefiri (Lk) is a Lactobacillus species that has been isolated from kefir (23). Previous studies have demonstrated that its administration in mice downregulates the expression of proinflammatory mediators and increases anti-inflammatory molecules in the gut immune system (24, 25). It also regulates intestinal homeostasis by increasing IgA secretion and mucin production (25). Probiotic properties of Lk have also recently been demonstrated in humans (26); however, to the best of the authors' knowledge, the use of Lk has never been evaluated in dogs.
The aim of this study was to evaluate the safety and ease of administration of Lk in healthy dogs and its ability to impact the intestinal microbiota composition and IgA secretion.
This study is based on the use of a commercial food supplement (Kefibios®), provided by the company Hulka S.r.L. (Rovigo, Italy), containing live lactic ferments of Lk (LKF01–DSM 32079), currently used as probiotics in human medicine. Copyright permission to publish the product name (Kefibios®) was given by the Chief Executive Officer of Hulka S.r.L. The product is marketed in capsules. Following the label of the product, five drops of the solution reconstituted with 6 ml of vegetable oil in prefilled bottles contain ≥109 active fluorescent units (AFUs) of live and viable Lk (ISO 19344:2015). The dose corresponds to the human one indicated by the company, regardless of age and body weight (BW). The study, involving 10 healthy privately owned dogs, was conducted at the Veterinary Teaching Hospital (VTH) of our department. The owners were carefully instructed as to how to use and mix the product, and then shake the bottle before each administration. The product was then stored at room temperature between 10°C and 25°C and away from direct light.
The recruitment of the dogs in the study was voluntary and at no cost to the owners. Written informed consent before enrollment in the study was obtained from the owners.
The trial was authorized by the Animal Welfare Committee of the University of Bologna (Protocol No. 3885 of 21/07/2017).
Privately owned dogs of various breeds, genders, and weight, over 1 year of age, were enrolled in the trial. Dogs with any disease in the previous 2 months before the start of the trial were excluded. The inclusion criterion was the absence of antimicrobial or immunosuppressive treatment up to 60 days prior to the start of and during the trial. For inclusion, each dog was evaluated with a clinical examination and a laboratory panel, which included a complete blood count, a serum biochemistry profile, and coprological examination for gastrointestinal parasites.
The body condition score (BCS) was calculated using the 1–9 score proposed by Royal Canine SAS, and the fecal score was evaluated according to the Fecal Score System (FSS 1-7) proposed by Nestlé-Purina Petcare. The BCS was determined as follows: 1–3 = too thin, 4–5 = ideal, and 6–9 = too heavy. The fecal samples were scored as follows: 1 = very hard and dry, leaves no residue on the ground when picked up; 2 = firm, but not hard, pliable, little or no residue on the ground when picked up; 3 = log-shaped, moist surface, leaves residue on the ground, but holds form when picked up; 4 = very moist and soggy, leaves residue on the ground and loses form when picked up; 5 = very moist but has a distinct shape, leaves residue on the ground and loses form when picked up; 6 = has texture, but no defined shape, leaves residue on the ground when picked up; and 7 = watery, no texture, present in flat patches.
Not being experimental animals but privately owned dogs, diet was not standardized during the trial but the previous diet (Table 1) was maintained, and water was supplied ad libitum.
During the entire experimental time, the dogs received five drops (≥109 AFUs) of Lk once daily for 30 days, administered directly in the mouth during the dinner meal.
Clinical examination, BCS, and FSS were performed at inclusion, and, during the trial, respectively, 15, 30, and 60 days after the start of Lk administration.
Samples of fresh feces were collected from each dog on three consecutive days: (a) at T0, before the start of Lk administration (days −3, −2, −1); (b) at T15, in the intermediate time of Lk administration (days 13, 14, 15); (c) at T30, at the end of Lk administration (days 28, 29, 30); (d) at T60, 1 month after the last Lk administration (days 58, 59, 60).
The samples were collected by the owner immediately after defecation, immediately stored at −20°C in the owner's household freezer until delivered frozen to the department where they were stored at −80°C until use.
All the samples were analyzed for IgA detection, while, for the gut microbiota, a pool of three samples from each dog at T0, T30, and T60 was prepared and analyzed.
Microbial DNA was extracted from about 250 mg of pooled fecal sample, derived from the experimental points (T0; T30; T60) for each dog, using the repeated bead-beating plus column method, as previously described (27). Briefly, samples were suspended in 1 ml of lysis buffer (500 mM NaCl, 50 mM Tris-HCl, pH 8, 50 mM EDTA, and 4% SDS) and bead-beaten three times in a FastPrep instrument (MP Biomedicals, Irvine, CA, USA) at 5.5 movements/s for 1 min, in the presence of four 3-mm glass beads and 0.5 g of 0.1-mm zirconia beads (BioSpec Products, Bartlesville, OK, USA). After incubation at 95°C for 15 min, samples were centrifuged at 13,000 rpm for 5 min. Two hundred and sixty microliters of 10 M ammonium acetate was added to the supernatant, followed by 5-min incubation on ice and 10-min centrifugation at 13,000 rpm. The supernatant was added with one volume of isopropanol, followed by incubation on ice for 30 min. Precipitated nucleic acids were washed with 70% ethanol, resuspended in 100 μl of TE buffer, and treated with 2 μl of 10 mg/ml DNase-free RNase at 37°C for 15 min. DNA was further purified using the QIAamp Mini Spin columns (QIAGEN, Hilden, Germany) following the manufacturer's instructions. DNA concentration and quality were evaluated using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
The V3–V4 hypervariable region of the 16S rRNA gene was amplified by using the 341F and 785R primers with Illumina adapter overhang sequences, as previously described (28). PCRs were performed in a final volume of 25 μl, containing 12.5 ng of genomic DNA, 200 nM of each primer, and 2X KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA, USA), in a Thermal Cycler T (Biometra GmbH, Göttingen, DE) with the following thermal cycle: initial denaturation at 95°C for 3 min, 25 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s, and a final extension step at 72°C for 5 min. Amplicons were purified using a magnetic bead-based system (Agencourt AMPure XP; Beckman Coulter, Brea, CA, USA). Indexed libraries were prepared by limited-cycle PCR using Nextera technology and further purified as described above. Final libraries were pooled at equimolar concentration, denatured with 0.2 N NaOH, and diluted to 6 pM before loading onto the MiSeq flow cell. Sequencing was performed on Illumina MiSeq platform with a 2 × 250 bp paired-end protocol, according to the manufacturer's instructions (Illumina, San Diego, CA, USA).
Sequencing Reads Were Deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA; BioProject ID PRJNA 592436).
Stool samples were thawed and subsequently freeze-dried (Modulyo EF4, 1044, Edwards, Apeldoorn, The Netherlands) for 16 h in order to eliminate the water contained and standardize the subsequent analysis, as reported by Grellet et al. (29). The lyophilized fecal samples were resuspended in 1X PBS (phosphate buffer saline) containing 0.5% Tween20 (Sigma-Aldrich, St. Louis, MO, USA), according to a weight/volume ratio of 100 mg/ml (1:10 dilution) by vortex (3') and then centrifuged at 1,500 × g for 10 min. After removing the supernatant, a further centrifugation was carried out at 10,000 × g for 20 min, then the aqueous phase was taken and frozen at −20°C until the processing moment. The determination of the IgA secretion amount was carried out by using a commercial kit (dog IgA ELISA Quantitation Set, Bethyl Laboratories Inc., TX, USA; Assay Range: 15.6–1,000 ng/ml), following the manufacturer's protocol.
In order to confirm the content of Lk declared by the manufacturer, we performed an independent assessment of Lk concentration in Kefibios® capsules (Bacteriological Laboratory, UNI EN ISO 9001:2015 registration number IT-15164).
Five different batches of the product, acquired in several drugstores, with expiration date from 1 to 2 years with respect to the time of the analysis, were analyzed, and enumeration of viable bacteria was conducted by the plate count method.
Briefly, each sample was firstly solubilized in MRS Broth (Oxoid CM359, Thermo Scientific, MA, USA) and gently shaken at room temperature for 15 min by using an orbital shaker, then serially diluted in the same broth and inoculated onto MRS Agar (Oxoid CM361, Thermo Scientific, MA, USA) plates. Plating was performed in triplicate.
Plates were incubated at 37°C for 72 h under anaerobic atmosphere, and the number of colony-forming units (CFUs) was determined.
The colonies obtained in the tests were identified by the API 50 CHL (BioMerieux, FI, Italy) test, and results are presented as the number of viable cells per capsule and per dose (five drops).
For the analysis of fecal microbiota composition and diversity, raw sequences were processed using a pipeline combining PANDAseq (30) and QIIME 1 (31). High-quality reads were binned into operational taxonomic units (OTUs) through an open-reference strategy at 0.97 similarity threshold by using UCLUST (32). Taxonomy was assigned using the RDP classifier and Greengenes as a reference database (release May 2013). All singleton OTUs and chimeras, identified by ChimeraSlayer (33), were discarded. Alpha rarefaction was performed using observed OTUs, Shannon, and the phylogenetic diversity (PD) whole tree metrics, while beta diversity was estimated by computing weighted and unweighted UniFrac distances. For the identification of Lk, OTUs assigned to the genus Lactobacillus were subjected to BLAST analysis (34).
All statistical analyses were performed in R (version 3.1.3) using the packages vegan, made4, and GraphPad Prism V.5.01 (GraphPad Software, La Jolla, CA, USA).
Assessment of data for normality was carried out by applying the D'Agostino and Pearson Omnibus normality test.
UniFrac distances were used for principal coordinates analysis (PCoA), and the significance of data separation was tested using a permutation test with pseudo-F ratios (function Adonis of vegan). Wilcoxon test for paired data was used to assess significant differences in alpha diversity and taxon relative abundance between groups.
A repeated-measures ANOVA (with Tukey post hoc test) was applied to evaluate the differences in BW and fecal IgA content between experimental points (T0, T15, T30, and T60). Friedman test (with Dunn's as post hoc test) was applied to evaluate the differences in BCS and FSS between experimental points (T0, T15, T30, and T60).
p < 0.05 was considered statistically significant.
Ten healthy adult dogs were included. Of those, one was male, three neutered males, two females, and four neutered females. Mean age was 4.9 ± 2.8 years (range 2–9). Breeds included were mixed breed dogs (n = 4), Border Collies (n = 3), Labrador Retriever (n = 1), Australian Shepherd (n = 1), and Dachshund (n = 1). Their BW ranged from 5 to 33 kg (18.3 ± 9.84 kg), while their average BCS was normal (range 4–6). Initial FSS was normal in all dogs (range 2–3; Table 1). All dogs had normal hematological and biochemical parameters, and the coprological examination was negative for parasites.
The liquid product containing Lk was spontaneously accepted by all the subjects. No clinical signs during the trial (with the exception of dog #6), and up to 1 month later, were reported by the owners.
There were no significant changes in BW and BCS during the study period, nor did the FSS of each animal changed.
Dog #6 developed a urinary tract infection at day 50 of the trial that required antibiotic treatment. This dog's last fecal sample was therefore excluded from the analysis.
A total of 1,536,903 high-quality reads (mean ± SD, 52,997 ± 16,302) were obtained and clustered into 2,716 OTUs at 97% similarity.
The PCoA of intersample diversity based on weighted and unweighted UniFrac distances showed no significant separation among the study groups (i.e., baseline, end of treatment, and follow-up; P = 0.7, permutation test with pseudo-F ratios; Figure 1A). Similarly, no significant differences were found in alpha diversity after Lk administration or in the follow-up compared to the baseline (P > 0.05, Wilcoxon test; Figure 1B).
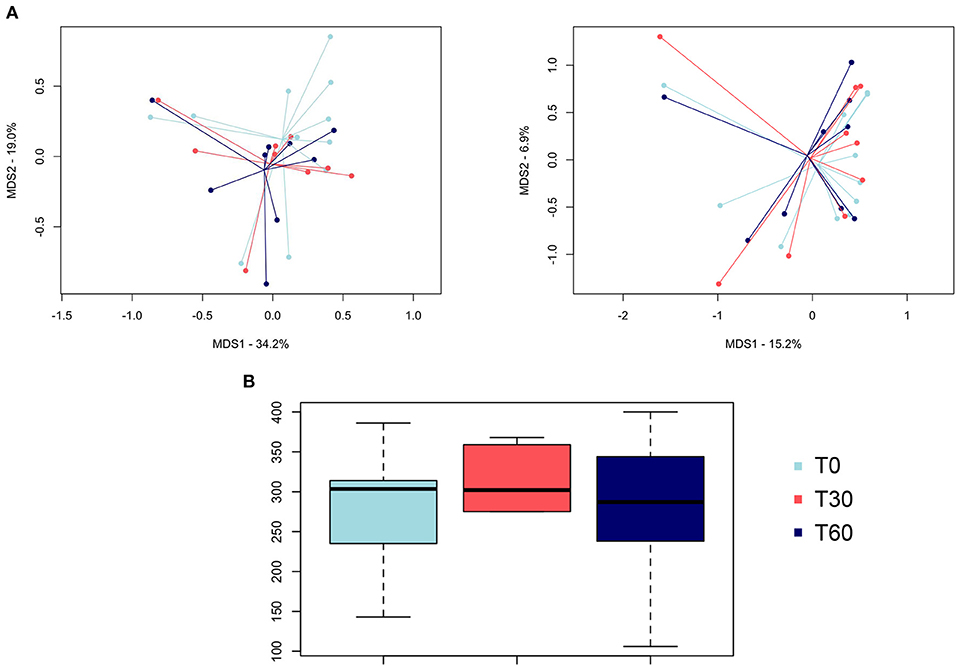
Figure 1. Gut microbiota diversity of healthy dogs following Lk administration. (A) Principal coordinates analysis of intersample diversity, based on weighted (left) and unweighted (right) UniFrac distances. (B) Alpha diversity computed with observed OTU metrics. T0, baseline; T30, after 30 days of Lk administration; T60, 1 month after the end of the treatment.
In line with the available literature on the gut microbiota of healthy dogs (35, 36), the phylum-level microbial profiles at the baseline were dominated by Firmicutes (relative abundance, mean ± SEM, 73.1 ± 4.2%), with Actinobacteria (14.2 ± 3.1%), Bacteroidetes (7.8 ± 2.2%), and Fusobacteria (3.9 ± 1.2%) as minor components (Figure 2). Lachnospiraceae, Coriobacteriaceae, Clostridiaceae, and Erysipelotrichaceae were the major families of the baseline microbiota (relative abundance ≥ 10%; Figure 3A). Consistently, the most represented genera were Blautia, Clostridium, and Collinsella (Figure 3B). Following Lk administration, no significant differences in taxon relative abundance at these phylogenetic levels (i.e., phylum, family, and genus) were observed (Figures 2, 3). One month after the end of the treatment, these changes were no longer detectable.
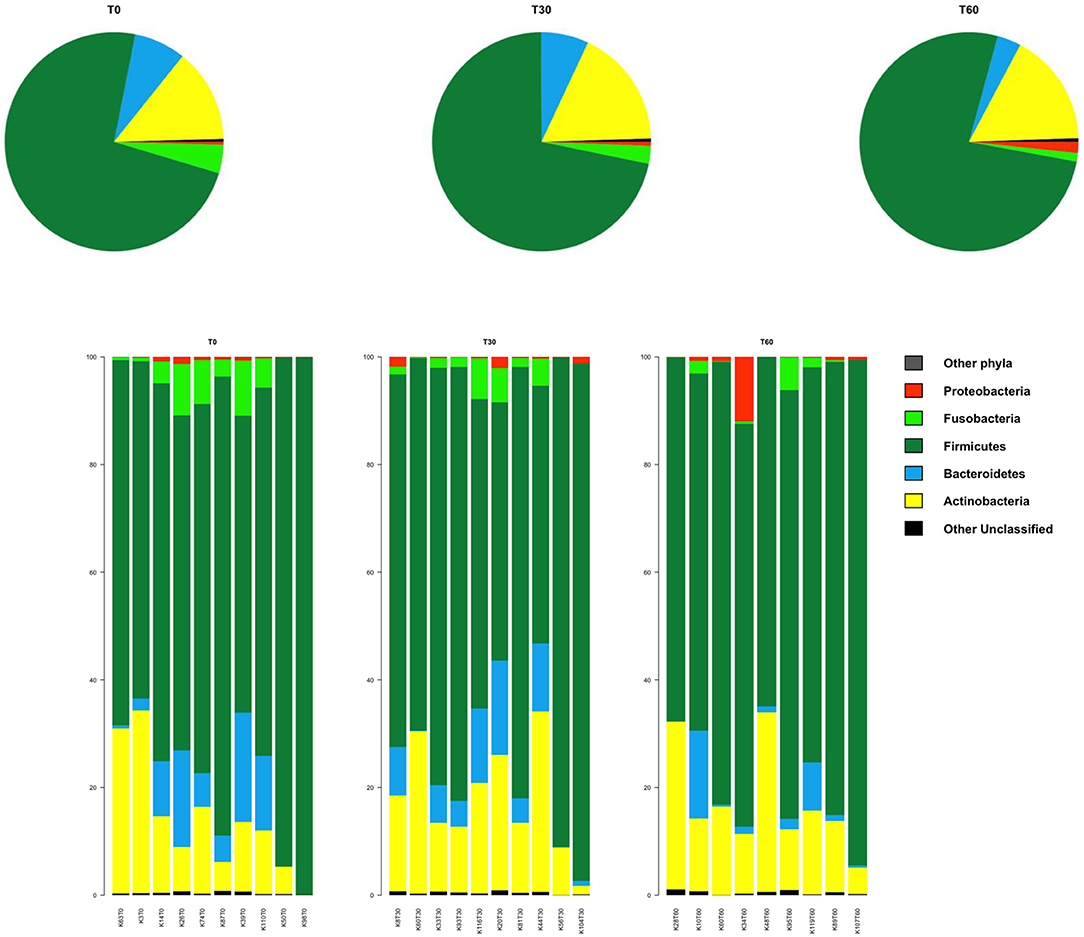
Figure 2. Impact of Lk administration on the phylum-level compositional structure of the gut microbiota of healthy dogs. Top: pie charts of mean values of relative abundance; Bottom: bar plots of individual profiles. T0, baseline; T30, after 30 days of Lk administration; T60, 1 month after the end of the treatment.
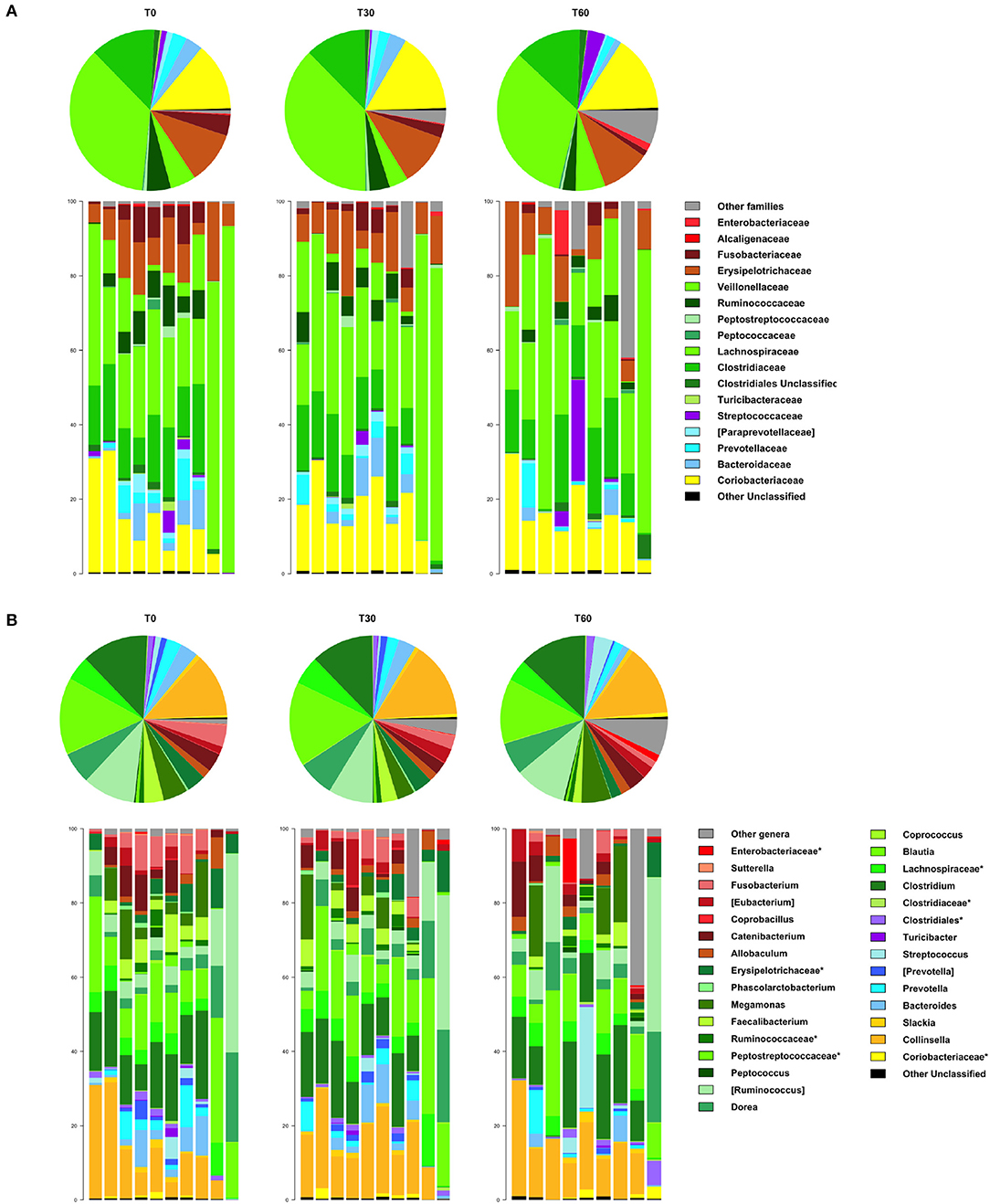
Figure 3. Impact of Lk administration on the family- and genus-level compositional structure of the gut microbiota of healthy dogs. Relative abundance profiles at family (A) and genus (B) level. Top: pie charts of mean values; bottom: bar plots of individual profiles. T0, baseline; T30, after 30 days of Lk administration; T60, 1 month after the end of the treatment.
However, at T60, a decreasing trend in the relative abundances of Fusobacteriaceae and Ruminococcaceae was found (T0 vs. T60, P = 0.11 and 0.15, respectively; Figure 3A). With specific regard to Lk, OTUs assigned to this species were not present at T0 but accounted for 10.3% of Lactobacillus diversity (and 0.07% of the intestinal ecosystem) after 30 days of Lk administration, which then disappear again in the follow-up.
A total of 117 fecal samples were taken and analyzed. Three samples (T58, T59, and T60 from dog #6) were missing because dog #6 was excluded from the trial.
The water content of feces was very similar among all the samples with percentage values of 38.08 ± 4.97 (mean ± SD).
IgA was detected in all samples except for sample T15 from dog #3 (undetectable <15.6 ng/ml). All dogs have shown a huge intra-individual variability of fecal IgA content among the three following day samples at T0, T15, T30, and T60 (Figure 4A).
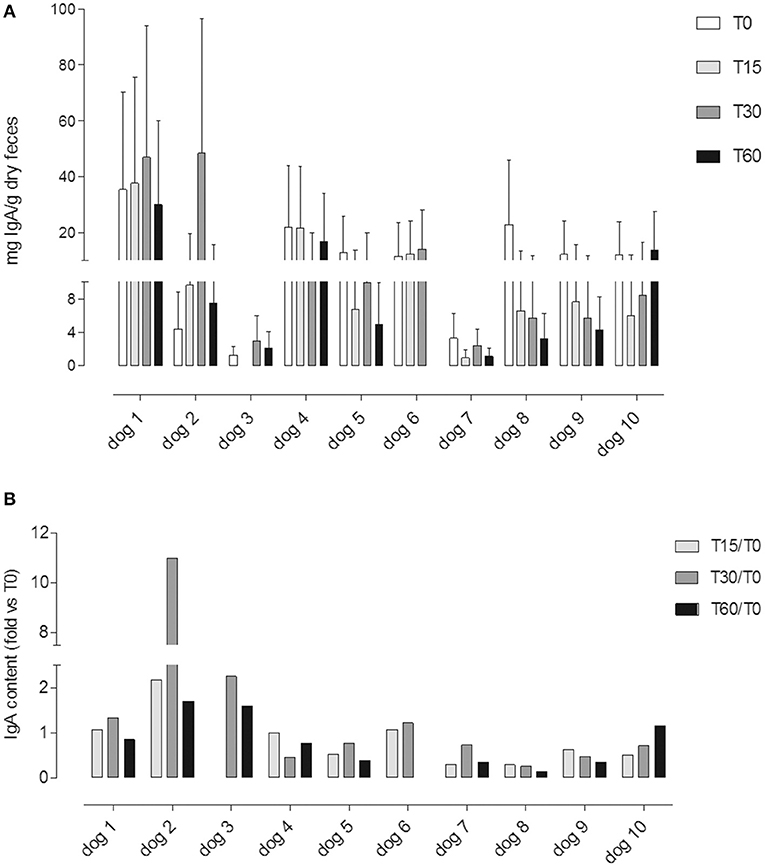
Figure 4. (A) Fecal IgA content at different time points (T0; T15; T30; T60); each value (mean ± SD) represents the average of the measurements of the three samples collected in three consecutive days for each experimental point: T0, before the administration, T15 and T30, after 15 and 30 days of Lk administration; and T60, 1 month after the last Lk administration. The T60 fecal samples of dog #6 were excluded from the analysis due to antibiotic therapy. (B) IgA fecal content evaluated by the ratio between the quantity at T15, T30, and T60 and the basal (T0) for each dog.
Only in four dogs it was possible to observe an increasing trend in the IgA fecal content (dogs 1, 2, 3, and 6; Figure 4A).
In Figure 4B, we showed a ratio between the quantity of IgA fecal content at T15, T30, and T60 and the basal (T0) for each dog.
Overall, the fecal IgA content between different time points did not show any significant differences (p = 0.1; Figure 5).
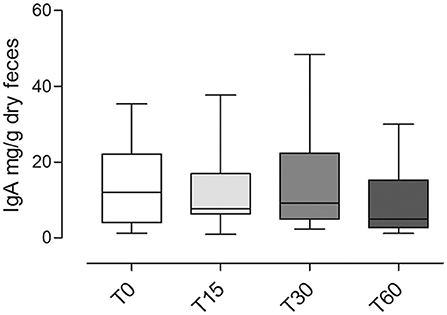
Figure 5. Fecal IgA content at different time points (T0; T15; T30; T60) in dogs included in the trial (mean ± SD). No significant statistical differences were observed (p = 0.1).
The number of viable Lk per capsule from different product batches varied from 7.73 × 107 CFU to 152 × 107 CFU (81 ± 61 × 107 CFU, mean and standard deviation).
The number of viable Lk in the liquid formulation per dose of five drops varied from 0.39 × 107 CFU to 7.55 × 107 CFU (3.2 ± 2.4 × 107 CFU, mean and standard deviation) and was hence 2-log fold lower than the declared concentration (≥109 CFU) by the company.
In recent years, interest in characterizing the canine intestinal microbiota has soared, and therapeutical interventions that can positively influence the microbiota composition and function, specifically identification of novel probiotics, are sought (2, 3, 37).
Several potential probiotics have already been tested in dogs, including bacterial species belonging to the genera Enterococcus, Lactobacillus, Bifidobacterium, and Saccharomyces, demonstrating their role in the treatment of acute and chronic enteropathies (15–18, 38–40).
Among Lactobacillus strain, several species have been already studied (Lactobacillus acidophilus DSM13241; Lactobacillus fermentum CCM7421; Lactobacillus animalis LA4). However, the achieved results are difficult to compare as different dosages (107 CFU/daily dose; 109 CFU/daily dose; 3 to 3.6 × 109 CFU/daily dose) and different forms of application were used, and the duration of administration was also variable (15, 17, 18).
To date, no studies have evaluated the influence of Lk on the parameters of intestinal health in dogs.
Only one previous study has assessed the effect of kefir (a fermented dairy product containing Lk) on dogs (21). This study demonstrated a significant increase in the fecal LAB:Enterobacteriaceae ratio and a decrease in the fecal Firmicutes:Bacteroidetes ratio, which was interpreted as an improvement of the gut microbiota composition. However, this study did not use a single bacterial strain, but the mixture of more than 50 microorganisms contained in kefir, and only reported a dose for total LAB (9.32 ± 0.23 log10 CFU/ml) and yeast (7.12 ± 0.36 log10 CFU/ml) (21). It is therefore difficult to calculate and compare the precise concentration of Lk that was administered in this study, and to infer the changes observed to a single potentially probiotic strain, as it could be attributed to several microorganisms and their potential synergism.
In addition, the composition of microorganisms in kefir may vary depending on its origin, the substrate used in the fermentation process, and the culture maintenance methods (41).
The product used in our study is a commercial preparation registered as a probiotic for human medicine, containing Lk with 30-day stability of reconstituted product guaranteed by the producer. Currently, there are no recommendations for the dosage of Lk in dogs. Daily dosages used in mice and people were 108 and 1010 CFU, respectively (24, 26).
The dose administered to dogs in the present study was extrapolated from the dose for an adult person as recommended by the manufacturer. While the daily dose for people (five drops) should contain ≥109 AFUs of live and viable Lk, the results presented here indicate that the same dose, administered to the dogs of this study, was more equal to 3.2 ± 2.4 × 107 CFU.
The 30-day duration of the experiment was chosen based on available literature and a suspected washout time of 4 weeks after discontinuation of administration (42).
Fecal samples were chosen over other types of samples to evaluate the intestinal microbiota, as they can be collected in a noninvasive manner, raising no ethical concerns in comparison to, for example, intestinal mucosal biopsies. In addition, the ability of Lk to modulate microbiota composition has already been demonstrated using fecal samples in people and mice (25, 26). Similarly, concentrations of IgA from duodenal biopsies and fecal samples showed no difference in a previous study (4).
As for the gut microbiota, in line with what has already been reported for probiotic supplementations in healthy individuals (43), no changes were detected after Lk administration.
The IgA results showed a large interdog variability at T0 (from 1.33 to 35.35 mg/g of dehydrated feces), which, although all dogs appeared clinically healthy, could depend on the extreme variability in experimental dogs' signaling, age, life environment, and food taken.
Moreover, our IgA variability is in agreement with the data reported by other authors (29, 44).
Regardless of the basal value, we have noted the absence of a significant variation in the fecal IgA content comparing the experimental time points, i.e., pretreatment (T0), during (T15 and T30), and 30 days after Lk administration (T60).
Possible explanation of the results could be related to a poor immunomodulatory effect of Lk toward the canine GALT (gut associated lymphoid tissue) due to (a) poor LK viability in pharmaceuticals administered and consequent insufficient probiotic dose; (b) inadequate administration period; and (c) poor vitality in the gastrointestinal tract of the dog with lack of Lk probiotic activity in dogs.
Considering the first point, the differences found between the concentration declared by the company (Lk ≥ 25 × 109 AFU/capsule; ≥ 109 AFU/dose) and that found in our quality control analysis (81 ± 61 × 107 CFU/capsule; 3.2 ± 2.4 × 107 CFU/dose) must be emphasized.
It must be stressed that the gap between what was declared by the probiotic company and what was highlighted by an independent analysis is not a rare event. In fact, analyzing the literature, numerous papers, both in the field of human and veterinary medicine, evidence this gap, with several-folds reduction of live probiotics concentration with respect to which reported by the companies (45–47).
Though not much is known about the minimal dose and/or frequency of probiotics required for the probiotic effect, it seems to be dose-dependent (48). For this reason, it cannot be excluded that the absence of changes observed in fecal microbiota and IgA during the trial with Lk can be attributed to an insufficient dosage, corresponding to ~3% of the expected dose indicated for humans.
It is also true that an improvement in the enteric immune function in dogs was observed even after the administration of Lactobacillus fermentum at the dosage of 1 × 107 CFU/daily dose for 1 week, similar dose, and lower trial time, than the one actually used by us (18).
With respect to the treatment time, our study treatment is longer than employed in similar studies (15–17), with two intratreatment withdrawals in addition to pre- and post-treatment sampling, so this point can be excluded as a cause of poor response too.
Lastly, no previous studies have analyzed the Lk vitality in the canine gastrointestinal tract; therefore, it cannot be excluded that this may be the cause of the poor probiotic activity observed in our study.
Conversely, studies performed in human medicine have verified the gastrointestinal Lk vitality showing a high rate of adhesion of Lk to intestinal cells and strong resistance to gastric juice and intestinal bile salts (49).
Limitations of the study include the fact that dogs did not receive a uniform diet during the experiment, and the relatively low number of dogs included, further confounded by the exclusion of dog #6 from the analysis.
However, we believe that not artificially standardizing the dogs' diet might allow for results to be translated easier into real-life veterinary practice conditions and would showcase that Lk is able to impact the microbiota independent of the diet given. We also believe that our methods of analysis will, to a certain degree, be able to counteract any dietary effect, as the dogs could serve as their own controls.
With respect to the sample size, the current study is similar to other feeding trials performing similar analyses (15–18).
In conclusion, our study was unable to demonstrate a significant change in microbiota composition or function in healthy dogs administered with Lk at a dose of 3.2 ± 2.4 × 107 CFU/daily dose for 30 days. Further research will be necessary in order to assess the efficacy of a higher dose or of the combination of Lk with other potential probiotics.
The microbiota datasets generated for this study can be found in the Sequencing reads were deposited in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA; BioProject ID PRJNA 592436).
The trial was authorized by the Animal Welfare Committee of the University of Bologna (Protocol No. 3885 of 21/07/2017). Written informed consent was obtained from the owners for the participation of their animals in this study.
AG and MP were responsible for the conception of the study and data interpretation and wrote the manuscript. AZ and MS performed fecal IgA determination. MF reviewed the manuscript and provided critical suggestions and comments. ST, MB, and PB analyzed fecal microbiota. RZ performed quality control analysis. All authors discussed the results and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Lee WJ, Hase K. Gut microbiota–generated metabolites in animal health and disease. Nat Chem Biol. (2014) 10:416–24. doi: 10.1038/nchembio.1535
2. Grześkowiak Ł, Endo A, Beasley S, Salminen S. Microbiota and probiotics in canine and feline welfare. Anaerobe. (2015) 34:14–23. doi: 10.1016/j.anaerobe.2015.04.002
3. Redfern A, Suchodolski J, Jergens A. Role of the gastrointestinal microbiota in small animal health and disease. Vet Rec. (2017) 181:370. doi: 10.1136/vr.103826
4. Littler RM, Batt RM, Lloyd DH. Total and relative deficiency of gut mucosal IgA in German shepherd dogs demonstrated by faecal analysis. Vet Rec. (2006) 158:334–41. doi: 10.1136/vr.158.10.334
5. Fagarasan S. Evolution, development, mechanism and function of IgA in the gut. Curr Opin Immunol. (2008) 20:170–77. doi: 10.1016/j.coi.2008.04.002
6. Okanishi H, Sano T, Yamaya Y, Kagawa Y, Watari T. The characteristics of short- and long-term surviving Shiba dogs with chronic enteropathies and the risk factors for poor outcome. Acta Vet Scand. (2013) 55:32. doi: 10.1186/1751-0147-55-32
7. Dandrieux JRS. Inflammatory bowel disease versus chronic enteropathy in dogs: are they one and the same? J Small Anim Pract. (2016) 57:589–99. doi: 10.1111/jsap.12588
8. Menozzi A, Dall'Aglio M, Quintavalla F, Dallavalle L, Meucci V, Bertini S. Rifaximin is an effective alternative to metronidazole for the treatment of chronic enteropathy in dogs: a randomised trial. BMC Vet Res. (2016) 12:217. doi: 10.1186/s12917-016-0851-0
9. Makielski K, Cullen J, O'Connor A, Jergens AE. Narrative review of therapies for chronic enteropathies in dogs and cats. J Vet Intern Med. (2019) 33:11–22. doi: 10.1111/jvim.15345
10. Westermarck E, Skrzypczak T, Harmoinen J, Steiner JM, Ruaux CG, Williams DA, et al. Tylosin-responsive chronic diarrhea in dogs. J Vet Intern Med. (2005) 19:177–86. doi: 10.1892/0891-6640(2005)19<177:tcdid>2.0.co;2
11. Igarashi H, Maeda S, Ohno K, Horigome A, Odamaki T, Tsujimoto H. Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLoS ONE. (2014) 9:e107909. doi: 10.1371/journal.pone.0107909
12. Léger A, Stärk KDC, Rushton J, Nielsen LR. A one health evaluation of the University of Copenhagen Research Centre for control of antibiotic resistance. Front Vet Sci. (2018) 5:194. doi: 10.3389/fvets.2018.00194
13. Schmidt VM, Pinchbeck G, McIntyre KM, Nuttall T, McEwan N, Dawson S, et al. Routine antibiotic therapy in dogs increases the detection of antimicrobial-resistant faecal Escherichia coli. J Antimicrob Chemother. (2018) 73:3305–16. doi: 10.1093/jac/dky352
14. Watkins RR, Bonomo RA. Overview: global and local impact of antibiotic resistance. Infect Dis Clin North Am. (2016) 30:313–22. doi: 10.1016/j.idc.2016.02.001
15. Baillon ML, Marshall-Jones ZV, Butterwick RF. Effects of probiotic Lactobacillus acidophilus strain DSM13241 in healthy adult dogs. Am J Vet Res. (2004) 65:338–43. doi: 10.2460/ajvr.2004.65.338
16. Biagi G, Cipollini I, Pompei A, Zaghini G, Matteuzzi D. Effect of a Lactobacillus animalis strain on composition and metabolism of the intestinal microflora in adult dogs. Vet Microbiol. (2007) 124:160–65. doi: 10.1016/j.vetmic.2007.03.013
17. Pascher M, Hellweg P, Khol-Parisini A, Zentek J. Effects of a probiotic Lactobacillus acidophilus strain on feed tolerance in dogs with non-specific dietary sensitivity. Arch Anim Nutr. (2008) 62:107–16. doi: 10.1080/17450390801892583
18. Strompfová V, Lauková A, Gancarčíková S. Effectivity of freeze-dried form of Lactobacillus fermentum AD1-CCM7421 in dogs. Folia Microbiol. (2012) 57:347–50. doi: 10.1007/s12223-012-0139-0
19. Gómez-Gallego C, Junnila J, Männikkö S, Hämeenoja P, Valtonen E, Salminen S, et al. A canine-specific probiotic product in treating acute or intermittent diarrhea in dogs: A double-blind placebo-controlled efficacy study. Vet Microbiol. (2016) 197:122–28. doi: 10.1016/j.vetmic.2016.11.015
20. Suchodolski JS. Companion animals symposium: microbes and gastrointestinal health of dogs and cats. J Anim Sci. (2011) 89:1520–30. doi: 10.2527/jas.2010-3377
21. Kim DH, Jeong D, Kang IB, Lim HW, Cho Y, Seo KH. Modulation of the intestinal microbiota of dogs by kefir as a functional dairy product. J Dairy Sci. (2019) 102:3903–11. doi: 10.3168/jds.2018-15639
22. Korsak N, Taminiau B, Leclercq M, Nezer C, Crevecoeur S, Ferauche C, et al. Evaluation of the microbiota of kefir samples using metagenetic analysis targeting the 16S and 26S ribosomal DNA fragments. J Dairy Sci. (2015) 98:3684–89. doi: 10.3168/jds.2014-9065
23. Pogačić T, Mancini A, Santarelli M, Bottari B, Lazzi C, Neviani E, et al. Diversity and dynamic of lactic acid bacteria strains during aging of a long ripened hard cheese produced from raw milk and undefined natural starter. Food Microbiol. (2013) 36:207–15. doi: 10.1016/j.fm.2013.05.009
24. Carasi P, Díaz M, Racedo SM, De Antoni G, Urdaci MC, Serradell Mde L. Safety characterization and antimicrobial properties of kefir-isolated Lactobacillus kefiri. Biomed Res Int. (2014) 2014:208974. doi: 10.1155/2014/208974
25. Carasi P, Racedo SM, Jacquot C, Romanin DE, Serradell MA, Urdaci MC. Impact of kefir derived Lactobacillus kefiri on the mucosal immune response and gut microbiota. J Immunol Res. (2015) 2015:361604. doi: 10.1155/2015/361604
26. Toscano M, De Grandi R, Miniello VL, Mattina R, Drago L. Ability of Lactobacillus kefiri LKF01 (DSM32079) to colonize the intestinal environment and modify the gut microbiota composition of healthy individuals. Dig Liver Dis. (2017) 49:261–67. doi: 10.1016/j.dld.2016.11.011
27. Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. (2004) 36:808–12. doi: 10.2144/04365ST04
28. Barone M, Turroni S, Rampelli S, Soverini M, D'Amico F, Biagi E, et al. Gut microbiome response to a modern Paleolithic diet in a Western lifestyle context. PLoS ONE. (2019) 14:e0220619. doi: 10.1371/journal.pone.0220619
29. Grellet A, Heilmann RM, Polack B, Feugier A, Boucraut-Baralon C, Grandjean D, et al. Influence of breed size, age, fecal quality, and enteropathogen shedding on fecal calprotectin and immunoglobulin A concentrations in puppies during the weaning period. J Vet Intern Med. (2016) 30:1056–64. doi: 10.1111/jvim.14255
30. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. (2012) 13:31. doi: 10.1186/1471-2105-13-31
31. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–36. doi: 10.1038/nmeth.f.303
32. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–61. doi: 10.1093/bioinformatics/btq461
33. Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. (2011) 21:494–04. doi: 10.1101/gr.112730.110
34. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
35. Sandri M, Dal Monego S, Conte G, Sgorlon S, Stefanon B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet Res. (2017) 13:65. doi: 10.1186/s12917-017-0981-z
36. Omatsu T, Omura M, Katayama Y, Kimura T, Okumura M, Okumura A, et al. Molecular diversity of the faecal microbiota of Toy Poodles in Japan. J Vet Med Sci. (2018) 80:749–54. doi: 10.1292/jvms.17-0582
37. Tizard IR, Jones SW. The microbiota regulates immunity and immunologic diseases in dogs and cats. Vet Clin North Am Small Anim Pract. (2018) 48:307–22. doi: 10.1016/j.cvsm.2017.10.008
38. Fenimore A, Martin L, Lappin MR. Evaluation of metronidazole with and without Enterococcus faecium SF68 in shelter dogs with diarrhea. Top Companion Anim Med. (2017) 32:100–03. doi: 10.1053/j.tcam.2017.11.001
39. Strompfová V, Kubašová I, Lauková A. Health benefits observed after probiotic Lactobacillus fermentum CCM 7421 application in dogs. Appl Microbiol Biotechnol. (2017) 101:6309–19. doi: 10.1007/s00253-017-8425-z
40. D'Angelo S, Fracassi F, Bresciani F, Galuppi R, Diana A, Linta N, et al. Effect of Saccharomyces boulardii in dogs with chronic enteropathies: double-blinded, placebo-controlled study. Vet Rec. (2018) 182:258. doi: 10.1136/vr.104241
41. Prado MR, Blandón LM, Vandenberghe LPS, Rodrigues C, Castro GR, Thomaz-Soccol V, et al. Milk kefir: composition, microbial cultures, biological activities, and related products. Front Microbiol. (2015) 30; 6:1177. doi: 10.3389/fmicb.2015.01177
42. Sanders ME. Impact of probiotics on colonizing microbiota of the gut. J Clin Gastroenterol. (2011) 45:S115–S119. doi: 10.1097/MCG.0b013e318227414a
43. Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr. (2019) 73:24–37. doi: 10.1038/s41430-018-0135-9
44. Delucchi L, Fraga M, Perelmuter K, Cella CD, Zunino P. Effect of native Lactobacillus murinus LbP2 administration on total fecal IgA in healthy dogs. Can J Vet Res. (2014) 78:153–55.
45. Hamilton-Miller JMT, Shah S. Deficiencies in microbiological quality and labelling of probiotic supplements. Int J Food Microbiol. (2002) 72:175–76. doi: 10.1016/s0168-1605(01)00703-6
46. Drago L, De Vecchi E, Nicola L, Colombo A, Gismondo MR. Microbiological evaluation of commercial probiotic products available in Italy. J Chemother. (2004) 16:463–67. doi: 10.1179/joc.2004.16.5.436
47. Weese JS, Martin H. Assessment of commercial probiotic bacterial contents and label accuracy. Can Vet J. (2011) 52:43–6.
48. Bertazzoni Minelli E, Benini A. Relationship between number of bacteria and their probiotic effects. Microb Ecol Health Dis. (2008) 20:180–83. doi: 10.1179/1973947813Y.0000000078
Keywords: dog, stool, gut microbiota, probiotic, IgA
Citation: Gaspardo A, Zannoni A, Turroni S, Barone M, Sabetti MC, Zanoni RG, Forni M, Brigidi P and Pietra M (2020) Influence of Lactobacillus kefiri on Intestinal Microbiota and Fecal IgA Content of Healthy Dogs. Front. Vet. Sci. 7:146. doi: 10.3389/fvets.2020.00146
Received: 28 November 2019; Accepted: 27 February 2020;
Published: 02 April 2020.
Edited by:
Thomas Schermerhorn, Kansas State University, United StatesReviewed by:
Jan S. Suchodolski, Texas A&M University, United StatesCopyright © 2020 Gaspardo, Zannoni, Turroni, Barone, Sabetti, Zanoni, Forni, Brigidi and Pietra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Pietra, bWFyY28ucGlldHJhQHVuaWJvLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.