
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 25 March 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00135
 Ksenia Fursova1*
Ksenia Fursova1* Anatoly Sorokin2
Anatoly Sorokin2 Sergey Sokolov3
Sergey Sokolov3 Timur Dzhelyadin2
Timur Dzhelyadin2 Irina Shulcheva1
Irina Shulcheva1 Margarita Shchannikova1
Margarita Shchannikova1 Daria Nikanova4
Daria Nikanova4 Olga Artem'eva4
Olga Artem'eva4 Natalia Zinovieva4
Natalia Zinovieva4 Fedor Brovko1,4
Fedor Brovko1,4Staphylococcus aureus is a causative agent of different infectious processes, food poisoning, and autoimmune disorders. The horizontal transfer of pathogenic strains can occur from animal to human under both house and farm conditions, and the spread of strains with antibiotic resistance is an existing problem. In addition to the spread of antibiotic-resistant strains in clinics, this problem also exists in veterinary medicine. It is especially important to monitor antibiotic resistance on farms where antibiotics are the standard treatment of animals, which may trigger the spread of antibiotic-resistant strains among animals and to the human population, and these strains can also be distributed in milk products produced by these farms (milk, cheese, and butter). In this work, we investigated 21 S. aureus isolates using whole-genome sequence analysis and tried to establish a relationship between these isolates with the development of bovine mastitis in seven regions of Western Russia. An S. aureus virulence profile was identified. We identified two groups of S. aureus associated with subclinical mastitis, namely, the enterotoxin-positive and enterotoxin-negative groups. The most prevalent factor associated with bovine mastitis in Russia was cytotoxins, including hemolysins and leukocidins. Multidrug resistance strains were investigated, and antibiotic resistance genes were identified. We identified S. aureus ST 97 type as the most common type in the regions in Western Russia. To the best of our knowledge, this is the first in-depth study of a range S. aureus isolates originating from cattle infections in Russia.
Bovine mastitis is a widespread infectious disease that lacks a fully investigated etiology. It is considered that various factors, such as the environment, defects in milking techniques, lactation stage, or different infections, can trigger mastitis development (1). Infectious mastitis has a viral or bacterial origin. The most common bacterial agents are Staphylococcus, Streptococcus, coliforms, Micrococcus, and Bacillus species. These are opportunistic microorganisms that can switch to a pathogenic form and use various virulence factors to cause various pathologies in both animals and humans. Antibiotic therapy is actively carried out to prevent infection, which in turn triggers the problem of antibiotic resistance in microorganisms. At present, the methicillin-resistant S. aureus (MRSA) strains, including those of both animal and human origins, are prevalent (2). According to recent data, the frequency of MRSA cases has decreased, whereas the cases associated with methicillin-susceptible Staphylococcus aureus (MSSA) have been detected with increasing frequency (3).
In recent years, the frequency of the confirmed transition of antibiotic-resistant strains from animal to human populations is rapidly growing. Therefore, the study of the microflora of animals in close contact with humans is relevant (4–7).
This work aimed to investigate the virulence profile of S. aureus isolates obtained from cow's milk in Russia and to determine their potential involvement in the induction of mastitis in cows.
In this report, the whole-genome sequencing (WGS) characteristics of 21 S. aureus isolates from raw cow's milk samples from different regions of Russia are presented. Furthermore, we investigated the virulence factors and antibiotic resistance properties of the isolates. All the isolates were identified as methicillin-susceptible S. aureus, and multiple drug resistance was determined for several samples.
This study confirms the need for a systematic approach in modern studies of animal microflora because it can spread to the human population.
Raw milk samples were collected from cow farms from different regions of Russia in 2018. The milk samples were taken from each cow during aseptic milking and placed in sterile containers. The samples were transported to the laboratory at a temperature of +4°C within 2 h or frozen at −20°C. Salt meat broth (HiMedia Laboratories Pvt., Ltd, Mumbai, India) was inoculated with the milk samples at a ratio of 1:9 and stored at 37°C for 18–24 h. Internationally recognized traditional phenotypic methods, such as Gram-stained colony microscopy, growth in Baird–Parker agar (HiMedia Laboratories Pvt., Ltd.), hemolysis on azide blood agar Pronadisa (Condalab, Madrid, Spain), plasma coagulation, and biochemical identification, were applied to all the isolates. The strains were stored in trypticase soy broth (TSB) (Merck, Darmstadt, Germany) with 30% sterile glycerine at −18°C.
Staphylococcus aureus isolates were cultivated in TSB at 37°C on an orbital shaker for 14–16 h. The culture liquid was centrifuged at 4,000 g for 5 min at 4°C, and the cell pellet was used for DNA extraction.
Twenty-one isolates were selected for WGS. DNA samples were extracted using the Lysostaphin and GenElute Bacterial Genomic DNA kit (Sigma–Aldrich, Merck). DNA was analyzed by electrophoresis in a 1% agarose gel stained with ethidium bromide (5 μg/mL).
Sequencing was performed using an Illumina inc., San Diego, USA with MiSeq Reagent Kit v3 chemicals. The quality of the sequencing results was assessed by FastQC, and the sequences were trimmed with Bjorn Usadel Lab, Aachen, Germany to keep quality above 25.
The trimmed reads were uploaded to the PATRIC server (8), and the genomes were assembled and annotated with the PATRIC Comprehensive Genome Analysis pipeline (9). Only the genomes placed close to S. aureus by PATRIC Phylogenetic Analysis tree were retained for further analysis. All the genomes were deposited to GenBank.
The presence and allele of the antimicrobial resistance and virulence genes were evaluated by SRST2 (10). Two resources, namely, the ARG database (11) and ResFinder (12), were used for antimicrobial resistance, and VFDB (13) was used to identify virulence and pathogenicity gene annotation. The antimicrobial resistance database files were downloaded together with SRST2 software, and the S. aureus subset of VFDB was prepared as described in the SRST2 manual. The default parameter values were used for analysis in accordance with suggestions in the SRST2 manual.
Antibiotic susceptibility testing was performed using the disc diffusion method for the following antibiotics (HiMedia Laboratories Pvt., Ltd.) in Mueller-Hinton Agar (HiMedia Laboratories Pvt., Ltd.): penicillin (10 IU), oxacillin (1 μg), gentamicin (10 μg), erythromycin (15 μg), and ciprofloxacin (5 μg). Methicillin-resistant S. aureus isolate screening was performed using oxacillin (1 μg/disc). The dishes with inoculated cultures were incubated under aerobic conditions at 37 ± 1°C for 18–24 h. The sensitivity was evaluated by the diameters of the zones of inhibition in accordance with the Performance Standards for Antimicrobial Disc Susceptibility tests (14); EUCAST, http://www.eucast.org.
The Single nucleotide polymorphism (SNP) tree was constructed using CSI Phylogeny 1.4 (Center for Genomic Epidemiology) using default settings and excluding heterozygous SNPs. To identify SNPs, all the input sequences were mapped to the S. aureus subspecies aureus LGA251 genome as the reference (NCBI accession no. FR821779.1) and screened for the relevant nucleotide variations as previously described (15). The following criteria for high-quality SNP calling and filtering were chosen: (i) a minimum depth of 10× at SNP positions; (ii) a minimum relative depth of 10% at SNP positions; (iii) a minimum distance of 10 bp between SNPs; (iv) a minimum SNP quality of 30; (v) a minimum read mapping quality of 25; and (vi) a minimum Z score of 1.96. Site validation for each SNP position was performed. SNPs that failed the necessary requirements were excluded in the final analysis. Based on the concatenated alignments of high-quality SNPs, maximum likelihood trees were created using MEGA7 (16).
SRST2 (10) software was used to map the trimmed reads to the multilocus sequence typing (MLST) database (17). Strain definition files for S. aureus were downloaded as described in the SRST2 manual, and the analysis was performed with SRST2 default parameter values.
The genomes of the S. aureus isolates, which had similar biochemical and genetic characteristics, were compared for a genome distance (18) using programs available online (http://ggdc.dsmz.de/). DNA-DNA hybridization values were measured based on formula 2.
All the animal manipulations were performed in accordance with the Instruction for Diagnostics, Therapy and Prevention of Mastitis (no. 13-5-2/1948; http://gov.cap.ru/home/65/aris/bd/vetzac/document/371.html) and approved by the Department of Veterinary, Ministry of Agriculture of the Russian Federation. In addition, all the procedures followed the Guidelines for the Microbiological Study of Milk and the Secretion of the Udder of Cows for the Diagnosis of Mastitis (https://standartgost.ru/g/pkey-14293740821), Ministry of Agriculture of the Russian Federation.
Raw milk samples were collected from farms in different regions of Russia (Figure 1). Pure S. aureus cultures were obtained from raw cow's milk. The purified DNA samples of 21 isolates (data not shown) were used for WGS.
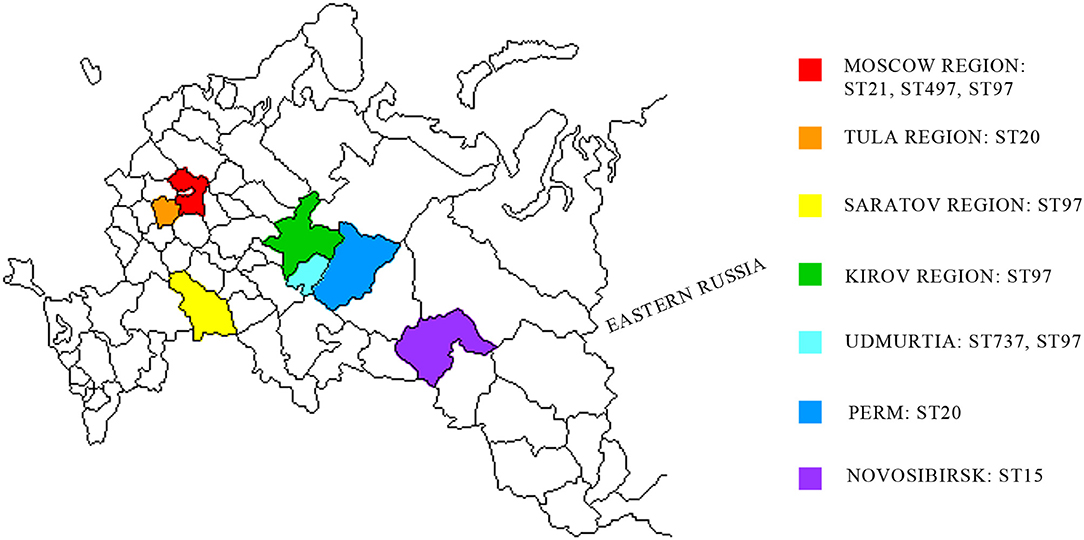
Figure 1. Geographic distribution of S. aureus strains associated with subclinical mastitis in the western region of Russia. Studied regions are shown in different colors. MLST distribution of S. aureus isolates are shown in studied regions.
The WGS data were analyzed by the following two separate pipelines: (i) SRST2 software was used to map the reads to special gene databases, such as MLST for strain annotation, ARG for antibiotic resistance genes, and VFDB for virulence factors; and (ii) the PATRIC Comprehensive Genome Analysis pipeline was used to assemble and annotate the genomes. The overall results of both approaches agreed with each other. For the analysis of virulence factors, however, we used SRST2 data, as these data provide depth and allele information.
All the genomes were deposited to GenBank with the following GenBank accession numbers: WIPL00000000, WIPM00000000, WIPN00000000, WIPO00000000, WIPP00000000, WIPQ00000000, WIPR00000000, WIPS00000000, WIPT00000000, WIPU00000000, WIPV00000000, WIPW00000000, WIPX00000000, WIPY00000000, WIPZ00000000, WIQA00000000, WIQB00000000, WIQC00000000, WIQD00000000, WOUL00000000, and WNKR00000000.
Twenty-one S. aureus isolates were analyzed. We identified genes from the following seven virulence families: enterotoxins, enterotoxin-like proteins, superantigen-like proteins, cytotoxins (including hemolysins and leukocidins), adherence, immune evasion, and exfoliatins (Supplementary Table 1). The occurrence of virulence genes is presented in Figure 2.
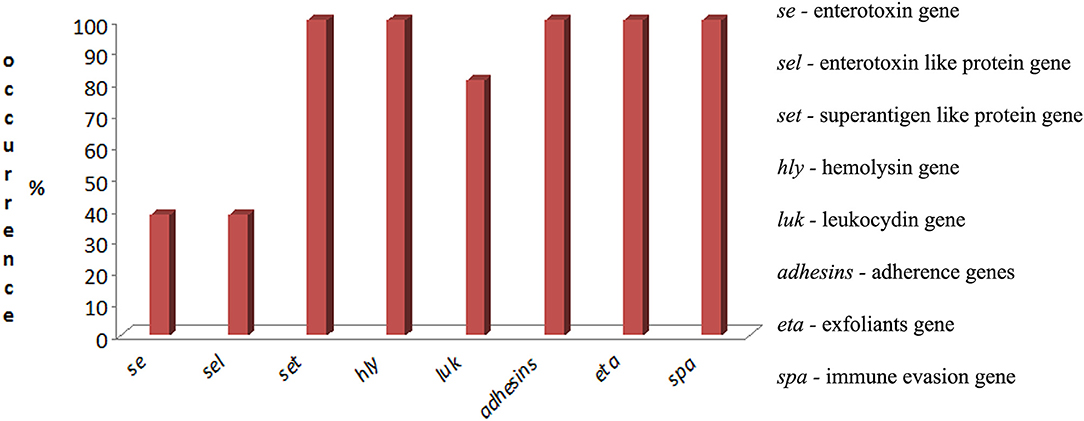
Figure 2. Occurrence of virulence genes of S. aureus isolated from raw cow's milk with subclinical mastitis in the western region of Russia.
None of the investigated strains contained genes encoding the classic enterotoxins from sea to see. Moreover, 38% of the isolates (n = 8) were classified as toxicogenic by enterotoxins seg and sei. Genes encoding the staphylococcal enterotoxin-like proteins (sel), namely, selm, seln, selo, and selv, were identified in this group, which consisted of 8 isolates (38%).
Genes encoding superantigen-like proteins (set) were detected in all the isolates (n = 21). All the isolates (n = 21) harbored genes encoding hemolysins (hla, hlb, hlg, and hld). Leukocidin genes, including lukE/D and lukM/F, were detected in 80.9% (n = 17) and −4.7% (n = 1) of isolates, respectively, but the luk S/F genes were not identified.
Genes encoding adhesins, namely, icaA, icaB, icaC, and icaD, were identified in 100% of the isolates (n = 21); and fnbA and fnbB were identified in 42.8% (n = 9) of the isolates. Spa genes were characteristic for all the S. aureus isolates (n = 21). Surprisingly, all the isolates (n = 21) had eta genes, but etb and etc were not identified.
Eight isolates that harbored staphylococcal enterotoxin genes (seg and sei) were selected for the study of antibiotic resistance. The antibiotic resistance results are shown in Table 1. The investigated isolates were resistant to erythromycin (100%), gentamicin (75%), penicillin (62.5%), and ciprofloxacin (25.5%). Among all the strains, the highest susceptibility was observed with respect to oxacillin (100%). Strains 615 and 1703 were resistant to gentamicin and erythromycin. Strains 88, 1838, and 1839 showed simultaneous penicillin, gentamicin, and erythromycin resistance. Strains 74 and 8656 had multiple resistance.
The following genes encoding antibiotic resistance proteins were identified: blaZ, ermC, aac (3), aph (2), and norA (Table 1).
SNP analysis (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) was conducted to study the differences in the genomes among the isolates and to determine their potential relationships (Figure 3). The studied isolates formed the following five clusters: (i) individual strain 8656; (ii) strains 1838 and 1839; (iii) strains 70, 74, 88, 615, and 1703; (iv) strains 812 and 724; and (v) strains 1817, 1816, 1813, 1829M, 1819M, 1835M, 18131, 187M, 1709, 23, and 8. Isolate 8656 was the most closely related to the reference genome LGA251 identified from bovine population (GenBank accession no. FR821779.1).
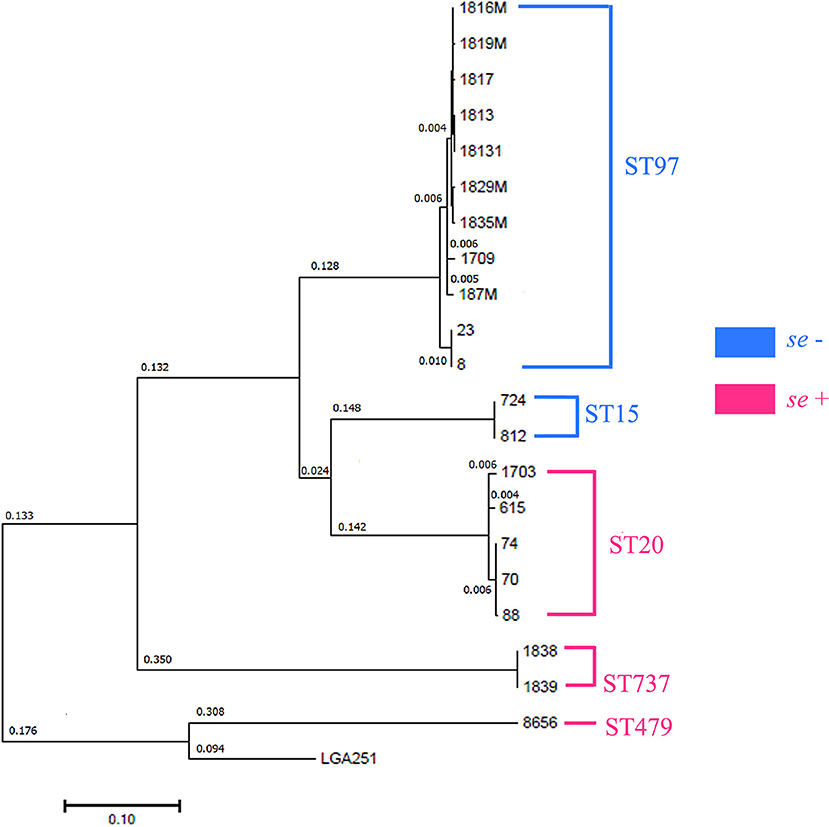
Figure 3. Phylogenetic analysis of S. aureus isolates by the maximum likelihood method. The log likelihood of the tree is (−233,811.0045). The tree is drawn to scale with branch lengths measured in the number of substitutions per site. All the positions containing gaps and missing data were eliminated. There were 39,835 positions in the final dataset. Evolutionary analyses were conducted in MEGA7. Distances >0.001 are shown. Sequence types (MLST) are shown in clusters.
Isolates 70, 74, and 88 differed with 0.01-nucleotide substitution per site, but they shared different phenotypes.
To determine the sequence type (ST) of the isolated strains, MLST analysis was performed in silico using the MLST database (https://cge.cbs.dtu.dk/services/MLST/).
Using MLST, we detected the following five STs: ST20, ST479, ST737, ST97, and ST15. ST97 was the most common type that was detected in the Moscow, Kirov, Udmurtia, and Saratov regions (Figure 1, Supplementary Table 1). The isolates were categorized in the following STs: isolates 70, 74, 88, 615, and 1703 belong to ST20; isolates 724 and 812 belong to ST15; isolates 1838 and 1839 belong to ST737; and isolate 8656 belongs to ST479.
Five SNP clusters were found to correlate both with genotypic (MLST) and with phenotypic (se) traits (Figure 3).
Genome-to-genome comparison was provided for the isolates with high identity according to their biochemical and genetic properties or pairs from one region.
The calculation of the intergenomic distance (Table 2) showed that genome pairs 1838–1839 (distance: 0.0001), 812–724 (distance: 0.0002), 70–88 (distance: 0.0000), and 23–8 (distance: 0.0000) belong to the same genomic subspecies. Pairs 70–74 (distance: 0.0014), 74–88 (distance: 0.0014), and 615–1703 (distance: 0.0008) were slightly spaced from each other.
Most often, bovine mastitis is associated with superantigens or enterotoxins produced by S. aureus (19–21) or coagulase-negative staphylococci (22). Staphylococcal enterotoxin C is the most common cause of bovine mastitis (23–25). According to other authors, one of the main enterotoxin genes of S. aureus isolated from cows with mastitis is sea. This gene occurs at rates ranging from 23.6 to 50% (21). Franck et al. (26) showed that mastitis in human females is also associated with the sec and sell genes, which are located in pathogenicity islands and are potentially transferred from cows to humans. For this reason, we conducted molecular characteristic analyses of the virulence potential of staphylococcal isolates from cow's milk to study mastitis in cows associated with S. aureus in Russia.
Twenty-one S. aureus samples were isolated from raw cow's milk with subclinical mastitis. By analyzing previous studies on S. aureus associated with intramammary infections in dairy cows of different geographical regions (4, 27–29) and using the virulence factor database (VFDB; http://www.mgc.ac.cn/cgi-bin/VFs/genus.cgi?Genus=Staphylococcus), we selected seven types of virulence genes because of their known pathogenic properties that may be associated with mastitis in Russia. Eight S. aureus isolates harbored the staphylococcal enterotoxin genes, seg and sei. Other genes encoding the classic se from sea to see were not identified. In previous studies, the occurrence of the seg and sei genes varied. Mello et al. (22) detected the seg gene only for 1% of S. aureus and 2% for central nervous system isolates, and sei was detected for 11 and 3% for S. aureus and coagulase-negative isolates, respectively. In contrast, Srinivasan et al. (30) detected seg and sei with higher frequencies (60.3% for sei and 24.4% for seg).
These seg/sei-positive isolates harbored genes encoding enterotoxin-like proteins, namely, selm, seln, selo, selu, and selv, which was not surprising because of their association with the enterotoxin gene cluster (egc) that is organized as an operon (31). The cluster consists of two enterotoxin genes (seg and sei), three enterotoxin-like genes with proven superantigenic activity but not emetic properties (selo, selm, and seln), and two pseudogenes (ϕent1 and 2). In turn, SEU is the fusion product of ϕent1 and ϕent2, which results from a 15-nucleotide deletion in ϕent1 (32).
The family of superantigen-like proteins (set) was identified as a common gene group in the analyzed isolates. These genes are located in the S. aureus genome by pathogenicity islands (33) and encode secreted virulence factors related to the host–pathogen interaction process (34).
The next antigenic target of our analysis was a group of cytotoxins, including hemolysins and leukocidins. We observed high occurrence of these genes. Cytotoxin genes were identified at high levels. Namely, genes of hemolysins (hla, hlb, hld, hlg) were detected in all investigated regions of Russia. Genes of leukocydins lukE/D in 80.9% of isolates and lukM/F in only one isolate (4.7%), although lukM/F is associated with zoonotic infections, and the LukM/F produced during the course of infection and the LukM/F levels in milk are associated with the severity of mastitis (27). In our previous work, we detected the following cytotoxin genes with a high frequency: lucS (46.6%), hla (70%), and hld (78.3%) (35). The high level of hly detection (up to 93%) has been observed by others (29). These agents are known for their role in the development of mastitis. Leukocidins may cause the lysis of leucocytes in bovine mastitis (36) and provide a protective masking effect for S. aureus in the host (37). α-Hemolysin induces gangrenous mastitis, which damages smooth muscle, restricting blood supply to mammary tissue (38).
Interestingly, 100% of the isolates from different regions of Russia harbored the eta gene. However, this finding requires verification by further analysis. In recent work, Schmidt and coauthors did not identify eta, etb, or etd in bovine isolates, and they reported that only one human S. aureus isolate tested positive for eta (4). It is possible that our finding in the current study may be a result of the coincidence of mastitis with different dermatitis conditions, for example, hock lesions (39) or udder cleft dermatitis (40, 41). Udder cleft dermatitis is a pathology lacking a clear etiology, but one of the bacterial inducers can be S. aureus. Moreover, some cases of sheep mastitis have been associated with the exfoliative toxin D–like protein (42).
We identified genes of two protein families with high occurrence. All the isolates harbored genes encoding intercellular adhesins encoding intercellular adhesion operon (IcaADBC) (43), and 42.8% of isolates harbored genes encoding fibronectin-binding proteins (44, 45). Usually, these genes are associated with biofilm formation, which is associated with antibiotic resistance. In particular, fibronectin-binding proteins A and B mediate the adhesion of S. aureus to fibrinogen, elastin, and fibronectin, thus providing S. aureus the ability to invade endothelial cells without requiring additional factors, although slowly and inefficiently, through actin rearrangements in host cells (46) (https://www.uniprot.org/uniprot/P14738). Furthermore, S. aureus surface protein A may participate in biofilm formation (47).
Investigation of antibiotic resistance showed that eight of the studied isolates were resistant to different classes of antibiotics. Moreover, isolates 74, 88, 1838, 1839, and 8656 were identified as Multiple drug resistance (MDR), as they were resistant to three or more antimicrobial classes according to the guidelines recommended by the joint initiative of the European Center for Disease Prevention and Control and the Centers for Disease Control and Prevention (48). Whole-genome sequencing analysis confirmed the diversity of antibiotic resistance genes.
Antibiotic treatment of bacterial infections may cause the development of MDR. This problem has become widespread both in veterinary and human medicine. The drug resistance of S. aureus has multiple origins, such as the use of efflux pumps, increased expression of target proteins, or mutation of target proteins (49). In the present study, neither the mecA nor mecC genes were detected in any isolate, whereas all the isolates (excepted 8656) harbored the blaZ gene. Multiple and methicillin resistance can be associated with biofilm formation of S. aureus. Previous studies have reported that biofilm formation is a common characteristic of MRSA strains (50). Isolates 70, 74, and 88 also had the ermC gene, which is responsible for their resistance to erythromycin. We observed the presence of aac (3) and aph (2), which confirmed resistance to gentamicin (51). The norA gene was detected in the common group, and it may provide non-specific resistance to different antibiotics.
As described in many studies, the specific strains of S. aureus that are associated with mastitis vary across herds (28, 52, 53). Strains with identical genotypes may possess characteristics that provide some advantages for their survival in the environment and to colonize the udder (22). We performed phylogeny analysis and multilocus typing of isolates, and we detected five clusters. As shown at Figure 1, ST97 was identified to be geographically close to other regions, such as Kirov and Udmurtia, but not close to outlying regions, such as Moscow and Saratov. In our previous work, ST97 isolates were identified in farms of Central Russia (54). Based on the data presented on the MLST resource, ST97 was present in the remainder of the group. This type is quite common, as it is found among isolates of China, Spain, Japan, the Netherlands, Brazil, and other countries (https://pubmlst.org/bigsdb?db=pubmlst_saureus_isolates). In addition to being detected in the present study, ST20 is the most widely represented among the isolates of China and Japan followed by ST737 in Turkey and ST479 in the Netherlands and USA (https://pubmlst.org/bigsdb?db=pubmlst_saureus_isolates).
By summarizing these phylogeny results with virulence data, we identified the following two groups: se-negative (formed by ST15 and ST97) and se-positive (formed by ST20, ST737, and ST479). The se-negative group was composed of the samples isolated from regions located in different geographic and climatic regions (Kirov, Saratov, Novosibirsk, and partially Moscow). The se-positive group was isolated from regions that geographically pairwise border Udmurtia and Perm and Moscow and Tula and that belong to the same climatic zone. In addition, an animal transfer between farms of these neighboring regions is possible. These factors may account for the spreading of enterotoxigenic strains in these regions.
According to Grunert, infection in a herd can be linked to specific strain genotype and often results in one or a few dominant clones within a herd, indicating transmissibility between animals and the preference of particular pathotypic traits (55). We detected high genetic similarity within groups and the similarity of these groups to ST types, which allowed us to hypothesize that isolates of the phylogenetic group with one ST type may belong to the one genomic subspecies. In addition, such isolate pairs, namely, 1838–1839 (Udmurtia) and 615 (Perm)−1703 (Moscow), had similar antibiotic resistance in their pairs. A genome-to-genome comparison showed that 1838–1839 belonged to the same genomic subspecies but that 615–1703 was identified as different but closely related subspecies.
To summarize, WGS of 21 S. aureus isolates from different climatic regions in Russia showed the high virulence potential of S. aureus in all regions. We observed two groups of S. aureus isolates associated with subclinical mastitis, namely, the enterotoxin-positive and enterotoxin-negative groups. The most prevalent factor associated with bovine mastitis in Russia was cytotoxins, including hemolysins and leukocidins. Furthermore, we observed a potential connection of bovine mastitis in Russia with eta-positive S. aureus. We did not identify MRSA, but the investigated strains may be strongly associated with bovine mastitis due to their multiple combinations of virulence genes, including enterotoxin genes, cytotoxin genes, and bacterial adhesion mediator genes. The limited diversity of enterotoxin genes, which trigger inflammation in the host immune system, in combination with MDR may indicate a long-term chronic form of mastitis (56, 57). Previous antibiotic treatment may transform infection into a chronic subclinical form. MDR may be associated with biofilm formation, which in turn promotes bacterial cell survival during antibiotic treatment and host immune defense mechanisms (58, 59). In previous studies, authors have discussed the problem of long-term, subclinical, and persistent mastitis and its dependence on different virulence factors (60, 61). Grunert et al. (55) showed that high cellular invasiveness of S. aureus is associated with chronic infection. Furthermore, high biofilm formation and low level of cellular toxicity may be a supportive mechanism of S. aureus to successfully persist and spread in bovines (55).
Although the diversity of virulence factors was investigated in the present study, we did not show expression levels of these factors, which was a limitation of the present study. The expression levels may provide more information about mastitis in Russia. Therefore, our future work will investigate this next step. Our previous study showed that the expression level of some enterotoxins associated with subclinical mastitis in Russia was pure (35).
Studies using genome analysis have facilitated a comprehensive understanding of the epidemiology and genomic repertoire of S. aureus from the bovine population and associated this form of mastitis with S. aureus as one of the major mastitis pathogens in Russia.
The datasets generated for this study can be found in the GenBank: WIPL00000000, WIPM00000000, WIPN00000000, WIPO00000000, WIPP00000000, WIPQ00000000, WIPR00000000, WIPS00000000, WIPT00000000, WIPU00000000, WIPV00000000, WIPW00000000, WIPX00000000, WIPY00000000, WIPZ00000000, WIQA00000000, WIQB00000000, WIQC00000000, WIQD00000000, WOUL00000000, and WNKR00000000.
All animal manipulation were provided in accordance with the Instruction for Diagnostics, Therapy and Prevention of mastitis # 13-5-2/1948 (http://gov.cap.ru/home/65/aris/bd/vetzac/document/371.html) and approved by the Department of Veterinary, Ministry of Agriculture of the Russian Federation; and with the Guidelines for the Microbiological Study of Milk and the Secretion of the Udder of Cows for the Diagnosis of Mastitis (https://standartgost.ru/g/pkey-14293740821), Ministry of Agriculture of the Russian Federation.
KF and FB designed the study. AS and TD generated the genome assembly and annotation. SS analyzed the phylogeny of isolates. KF, OA, DN, IS, and MS performed the laboratory experiments. KF analyzed the genetic and bioinformatic materials and wrote the paper. All authors read and approved the final manuscript. FB and NZ are supervisors of the project.
This work was carried out under an agreement with the Russian Ministry of Science and Higher Education, Grant No. 0445-2019-0023 AAAA-A18-118021590136-7 and AAAA-A19-119050790041-1.
Frontiers Media Ltd. remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the GenoMed (Moscow, Russia) for sequencing, and we also thank Denis Pyankov (Head of Laboratory).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00135/full#supplementary-material
1. Zhang Z, Li XP, Yang F, Luo JY, Wang XR, Liu LH, et al. Influences of season, parity, lactation, udder area, milk yield, and clinical symptoms on intramammary infection in dairy cows. J Dairy Sci. (2016) 99:6484–93. doi: 10.3168/jds.2016-10932
2. Monaco M, Pimentel de Araujo F, Cruciani M, Coccia EM, Pantosti A. Worldwide epidemiology and antibiotic resistance of Staphylococcus aureus. Curr Top Microbiol Immunol. (2017) 409:21–56. doi: 10.1007/82_2016_3
3. Kourtis AP, Hatfield K, Baggs J, Mu Y, See I, Epson E, et al. Vital signs: epidemiology and recent trends in methicillin-resistant and in methicillin-susceptible Staphylococcus aureus bloodstream infections - United States. MMWR Morb Mortal Wkly Rep. (2019) 68:214–9. doi: 10.15585/mmwr.mm6809e1
4. Schmidt T, Kock MM, Ehlers MM. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African Dairy Herds: genetic diversity and inter-species host transmission. Front Microbiol. (2017) 8:511. doi: 10.3389/fmicb.2017.00511
5. Murray S, Pascoe B, Méric G, Mageiros L, Yahara K, Hitchings MD, et al. Recombination-mediated host adaptation by Avian Staphylococcus aureus. Genome Biol Evol. (2017) 9:830–42. doi: 10.1093/gbe/evx037
6. Islam MZ, Espinosa-Gongora C, Damborg P, Sieber RN, Munk R, Husted L, et al. Horses in Denmark are a reservoir of diverse clones of methicillin-resistant and–susceptible Staphylococcus aureus. Front Microbiol. (2017) 8:543. doi: 10.3389/fmicb.2017.00543
7. Miragaia M. Review factors contributing to the evolution of mecA-mediated β-lactam resistance in Staphylococci: update and new insights from whole genome sequencing (WGS). Front Microbiol. (2018) 9:2723. doi: 10.3389/fmicb.2018.02723
8. Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. (2017) 45:D535–42. doi: 10.1093/nar/gkw1017
9. Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. (2015) 5:8365. doi: 10.1038/srep08365
10. Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. (2014) 6:90–16. doi: 10.1186/s13073-014-0090-6
11. Liu B, Pop M. ARDB-antibiotic resistance genes database. Nucleic Acids Res. (2009) 37(Database issue):D443–7. doi: 10.1093/nar/gkn656
12. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. (2012) 67:2640–4. doi: 10.1093/jac/dks261
13. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. (2018) 47:D687–92. doi: 10.1093/nar/gky1080
14. CLSI. Performance Standards for Antimicrobial Susceptibility Tests; Approved Standard - Twelfth Edition. CLSI document M02-A12. Wayne, PA. Clinical and Laboratory Standards Institute, USA (2015).
15. Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. (2014) 9:e104984. doi: 10.1371/journal.pone.0104984
16. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
17. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. (2018) 3:124–0. doi: 10.12688/wellcomeopenres.14826.1
18. Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. (2013) 14:60. doi: 10.1186/1471-2105-14-60
19. Schabauer A, Pinior B, Gruber CM, Firth CL, Käsbohrer A, Wagner M, et al. The relationship between clinical signs and microbiological species, spa type, and antimicrobial resistance in bovine mastitis cases in Austria. Vet Microbiol. (2018) 227:52–60. doi: 10.1016/j.vetmic.2018.10.024
20. Artursson K, Söderlund R, Liu L, Monecke S, Schelin J. Genotyping of Staphylococcus aureus in bovine mastitis and correlation to phenotypic characteristics. Vet Microbiol. (2016) 25:156–61. doi: 10.1016/j.vetmic.2016.08.012
21. Rall VL, Miranda ES, Castilho IG, Camargo CH, Langoni H, Guimarães FF, et al. Diversity of Staphylococcus species and prevalence of enterotoxin genes isolated from milk of healthy cows and cows with subclinical mastitis. J Dairy Sci. (2014) 97:829–37. doi: 10.3168/jds.2013-7226
22. Mello PL, Moraes Riboli DF, Pinheiro L, de Almeida Martins L, Vasconcelos Paiva Brito MA, Ribeiro de Souza da Cunha Mde L. Detection of enterotoxigenic potential and determination of clonal profile in Staphylococcus aureus and coagulase-negative Staphylococci isolated from bovine subclinical mastitis in different Brazilian States. Toxins. (2016) 8:104. doi: 10.3390/toxins8040104
23. Fang R, Cui J, Cui T, Guo H, Ono HK, Park CH, et al. Staphylococcal enterotoxin C is an important virulence factor for mastitis. Toxins. (2019) 11:141. doi: 10.3390/toxins11030141
24. Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, Meaney WJ, et al. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J Bacteriol. (2001) 183:63–70. doi: 10.1128/JB.183.1.63-70.2001
25. Ebling TL, Fox LK, Bayles KW, Bohach GA, Byrne KM, Davis WC, et al. Bovine mammary immune response to an experimental intramammary infection with a Staphylococcus aureus strain containing a gene for staphylococcal enterotoxin C1. J Dairy Sci. (2001) 84:2044–50. doi: 10.3168/jds.S0022-0302(01)74648-6
26. Franck KT, Gumpert H, Olesen B, Larsen AR, Petersen A, Bangsborg J, et al. Staphylococcal aureus enterotoxin C and enterotoxin-like L associated with post-partum mastitis. Front Microbiol. (2017) 8:173. doi: 10.3389/fmicb.2017.00173
27. Vrieling M, Boerhout EM, van Wigcheren GF, Koymans KJ, Mols-Vorstermans TG, de Haas CJ, et al. LukMF' is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci Rep. (2016) 6:37759. doi: 10.1038/srep37759
28. Cremonesi P, Pozzi F, Raschetti M, Bignoli G, Capra E, Graber HU, et al. Genomic characteristics of Staphylococcus aureus strains associated with high within-herd prevalence of intramammary infections in dairy cows. J Dairy Sci. (2015) 98:6828–38. doi: 10.3168/jds.2014-9074
29. Wang D, Zhang L, Zhou X, He Y, Yong C, Shen M, et al. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J Dairy Sci. (2016) 99:9560–9. doi: 10.3168/jds.2016-11625
30. Srinivasan V, Sawant AA, Gillespie BE, Headrick SJ, Ceasaris L, Oliver SP. Prevalence of enterotoxin and toxic shock syndrome toxin genes in Staphylococcus aureus isolated from milk of cows with mastitis. Foodborne Pathog Dis. (2006) 3:274–83. doi: 10.1089/fpd.2006.3.274
31. Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, et al. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. (2001) 166:669–77. doi: 10.4049/jimmunol.166.1.669
32. Letertre C, Perelle S, Dilasser F, Fach P. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J Appl Microbiol. (2003) 95:38–43. doi: 10.1046/j.1365-2672.2003.01957.x
33. Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. (2001) 357:1225–40. doi: 10.1016/S0140-6736(00)04403-2
34. Arcus VL, Langley R, Proft T, Fraser JD, Baker E. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J Biol Chem. (2002) 277:32274–81. doi: 10.1074/jbc.M203914200
35. Fursova KK, Shchannikova MP, Loskutova IV, Shepelyakovskaya AO, Laman AG, Boutanaev AM, et al. Exotoxin diversity of Staphylococcus aureus isolated from milk of cows with subclinical mastitis in Central Russia. J Dairy Sci. (2018) 101:4325–31. doi: 10.3168/jds.2017-14074
36. Vrieling M, Koymans KJ, Heesterbeek DA, Aerts PC, Rutten VP, de Haas CJ, et al. Bovine Staphylococcus aureus secretes the leukocidin LukMF' to kill migrating neutrophils through CCR1. MBio. (2015) 6:e00335. doi: 10.1128/mBio.00335-15
37. Spaan AN, van Strijp JAG, Torres VJ. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol. (2017) 15:435–47. doi: 10.1038/nrmicro.2017.27
39. Capurro A, Aspán A, Ericsson Unnerstad H, Persson Waller K, Artursson K. Identification of potential sources of Staphylococcus aureus in herds with mastitis problems. J Dairy Sci. (2010) 93:1. doi: 10.3168/jds.2009-2471
40. Persson Waller K, Bengtsson M, Nyman AK. Prevalence and risk factors for udder cleft dermatitis in dairy cattle. J Dairy Sci. (2014) 97:1. doi: 10.3168/jds.2013-7186
41. Ekman L, Nyman AK, Landin H, Magnusson U, Waller KP. Mild and severe udder cleft dermatitis-prevalence and risk factors in Swedish dairy herds. J Dairy Sci. (2018) 101:1. doi: 10.3168/jds.2017-13133
42. Mariutti RB, Souza TA, Ullah A, Caruso IP, de Moraes FR, Zanphorlin LM, et al. Crystal structure of Staphylococcus aureus exfoliative toxin D-like protein: structural basis for the high specificity of exfoliative toxins. Biochem Biophys Res Commun. (2015) 467:171–7. doi: 10.1016/j.bbrc.2015.08.083
43. Avila-Novoa MG, Iñíguez-Moreno M, Solís-Velázquez OA, González-Gómez JP, Guerrero-Medina PJ, Gutiérrez-Lomelí M. Biofilm formation by Staphylococcus aureus isolated from food contact surfaces in the dairy industry of Jalisco, Mexico. J Food Qual. (2018). doi: 10.1155/2018/1746139. [Epub ahead of print].
44. Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. (1999) 67:2627–32. doi: 10.1128/IAI.67.5.2627-2632.1999
45. Vergara-Irigaray M, Valle J, Merino N, Latasa C, García B, Ruiz de Los Mozos I, et al. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. (2009) 77:3978–91. doi: 10.1128/IAI.00616-09
46. Murai M, Moriyama H, Hata E, Takeuchi F, Amemura-Maekawa J. Variation and association of fibronectin-binding protein genes fnbA and fnbB in Staphylococcus aureus Japanese isolates. Microbiol Immunol. (2016) 60:312–25. doi: 10.1111/1348-0421.12377
47. Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. (2009) 191:832–43. doi: 10.1128/JB.01222-08
48. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
49. Mayrink AL, Nedeljković M, Dessen A. New strategies for targeting and treatment of multi-drug resistant Staphylococcus aureus. Drug Resist Updat. (2017) 31:1–14. doi: 10.1016/j.drup.2017.03.001
50. Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int J Gen Med. (2018) 11:25–32. doi: 10.2147/IJGM.S153268
51. Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. (2010) 13:151–71. doi: 10.1016/j.drup.2010.08.003
52. van Leeuwen WB, Melles DC, Alaidan M, Al-Ahdal M, Boelens HAM, Snijders SV, et al. Host-and tissue-specific pathogenic traits of Staphylococcus aureus. J Bacteriol. (2005) 187:4584–91. doi: 10.1128/JB.187.13.4584-4591.2005
53. Zecconi A, Binda E, Borromeo V, Piccinini R. Relationship between some Staphylococcus aureus pathogenic factors and growth rates and somatic cell counts. J Dairy Res. (2005) 72:203–8. doi: 10.1017/S0022029905000841
54. Fursova KK, Artem'eva OA, Nikanova DA, Larin AK, Zinovieva NA, Brovko FA. Draft genome sequences of five Staphylococcus aureus strains Isolated from clinically healthy cows in the Russian Federation. Genome Announc. (2018) 6:16. doi: 10.1128/genomeA.00275-18
55. Grunert T, Stessl B, Wolf F, Sordelli DO, Buzzola FR, Ehling-Schulz M. Distinct phenotypic traits of Staphylococcus aureus are associated with persistent, contagious bovine intramammary infections. Sci Rep. (2018) 8:15968. doi: 10.1038/s41598-018-34371-1
56. Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, et al. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect Immun. (2007) 74:4724–34. doi: 10.1128/IAI.00132-06
57. de Freitas MFL, da Luz IS, da Silveira-Filho MV, Júnior JWP, Stamford TLM, Mota RA, et al. Staphylococcal toxin genes in strains isolated from cows with subclinical mastitis. Pesq Vet Bras. (2008) 28:617–21. doi: 10.1590/S0100-736X2008001200010
58. Scali F, Camussone C, Calvinho LF, Cipolla M, Zecconi A. Which are important targets in development of S. aureus mastitis vaccine? Res Vet Sci. (2015) 100:88–99. doi: 10.1016/j.rvsc.2015.03.019
59. Melchior MB, Vaarkamp H, Fink-Gremmels J. Biofilms: a role in recurrent mastitis infections? Vet J. (2006) 171:398–407. doi: 10.1016/j.tvjl.2005.01.006
60. Marbach H, Mayer K, Vogl C, Lee JYH, Monk IR, Sordelli DO, et al. Within-host evolution of bovine Staphylococcus aureus selects for a SigB-deficient pathotype characterized by reduced virulence but enhanced proteolytic activity and biofilm formation. Sci Rep. (2019) 9:13479. doi: 10.1038/s41598-019-49981-6
Keywords: S. aureus, virulence genes, enterotoxins, cytotoxins, multidrug resistance, biofilms, mastitis
Citation: Fursova K, Sorokin A, Sokolov S, Dzhelyadin T, Shulcheva I, Shchannikova M, Nikanova D, Artem'eva O, Zinovieva N and Brovko F (2020) Virulence Factors and Phylogeny of Staphylococcus aureus Associated With Bovine Mastitis in Russia Based on Genome Sequences. Front. Vet. Sci. 7:135. doi: 10.3389/fvets.2020.00135
Received: 09 December 2019; Accepted: 24 February 2020;
Published: 25 March 2020.
Edited by:
Michael Kogut, Agricultural Research Service, United StatesReviewed by:
Bo Han, China Agricultural University, ChinaCopyright © 2020 Fursova, Sorokin, Sokolov, Dzhelyadin, Shulcheva, Shchannikova, Nikanova, Artem'eva, Zinovieva and Brovko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ksenia Fursova, cGh1cnNvdmFfa0ByYW1ibGVyLnJ1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.