- World Organisation for Animal Health (OIE), National Reference Laboratory for Salmonellosis, Istituto Zooprofilattico Sperimentale delle Venezie, viale dell'Università, Legnaro, Italy
Different Salmonella serovars generally display different antigenic formulae, but there are some exceptions. For instance, the same antigenic formula, 6,7:c:1,5, is shared by Salmonella enterica serovar, Paratyphi C, Typhisuis, and Choleraesuis. Moreover, three biotypes have been described within the S. Choleraesuis serovar. A distinction among such biotypes can only be based on biochemical behaviors (biotyping) posing serious concerns when rapid characterization is required. The study of an outbreak of severe epizootic salmonellosis in wild boars occurred in Italy between 2012 and 2014 and the typing of the isolates recovered from the outbreak were used to test different approaches for serovar identification. A number of 30 S. Choleraesuis var. Kunzendorf isolates from the outbreak were typed by means of four different methods to derive serovar and biotype: (i) slide agglutination method followed by biochemical tests, (ii) suspension array xMAP® Salmonella Serotyping Assay (SSA), (iii) whole genome sequencing (WGS) and data analysis using SeqSero tool, and (iv) WGS and data analysis using Salmonella TypeFinder tool. Slide agglutination, xMAP® SSA and WGS, followed by SeqSero analysis, are methods that infer the serovars according to the White-Kauffmann-Le Minor (WKL) scheme, based exclusively on antigens. Using these methods, isolates with incomplete antigenic formulae could be misleadingly excluded from an outbreak. On the contrary, WGS followed by Salmonella TypeFinder data analysis, which predicts the serotype on the basis of Multilocus sequence typing (MLST), might be able to cluster together isolates belonging to the same outbreak irrespective of the antigenic formula. Results suggest the benefit of routine use of a combination of in silico MLST and antigenic formula analysis to solve specific ambiguous case studies for outbreak investigation purposes.
Introduction
The White-Kauffmann-Le Minor (WKL) scheme summarizes antigenic formulae of all known Salmonella serovars, on the basis of antigenic variability in the outer membrane lipopolysaccharides (O antigen), flagellar proteins (H1 and H2 antigens) and capsular polysaccharide (Vi antigen) (1). The most recent edition of the WKL scheme has identified over 2,500 serovars belonging to the five subspecies of Salmonella enterica (1, 2).
Traditional serotyping of Salmonella based on slide agglutination has been used for decades worldwide (3), and it is still considered the gold standard method for Salmonella serotyping. According to this phenotype-based approach, the surface antigens are detected by agglutination of bacterial cells using specific Salmonella antisera (3). Traditional serotyping is labor intensive, and it requires trained technicians to provide valuable data (3). Another limitation of this method, which leads to inconclusive results, is a possible loss of expression of antigens required for definitive serovar identification (for example rough strains) (4). For all these reasons, molecular methods for Salmonella serotyping have been developed (3). An example of molecular alternative methods for Salmonella serotyping is a multiplex bead-based suspension array developed to detect the most common serovars using Luminex technology (5). Moreover, the technological advancements of Whole Genome Sequencing (WGS) and the improved bioinformatic analyses are revolutionizing surveillance programs and WGS data could also be used to derive information about Salmonella characteristics, such as serotype antimicrobial resistance determinants, virulence genetic factors, plasmid types, and in silico Multi Locus Sequence Type (MLST) (6).
Different Salmonella serovars generally display different antigenic formulae, but there are also some exceptions. Historically, different names have been assigned to serovars showing the same antigenic formula but differing either by biochemical characteristics, pathogenicity, or habitats (1).
The antigenic formula 6,7:c:1,5 is shared by different serovar: Paratyphi C, Typhisuis, Choleraesuis (1). Furthermore, three biotyping subdivisions on the basis of H2S production and the utilization of mucate and dulcitol have been described within Choleraesuis serovar: Choleraesuis sensu stricto, Choleraesuis var. Kunzendorf, and Choleraesuis var. Decatur (7).
Serovar Paratyphi C is associated with enteric fever in humans; serovar Typhisuis is associated with chronic paratyphoid/caseous lymphadenitis in swine (8) and serovar Choleraesuis may cause serious outbreaks of salmonellosis and paratyphoid in pigs (9), with clinical outcomes, such as enterocolitis and septicemia (10), often resulting in fatal systemic disease (11). This serovar is currently highly prevalent in North America and Asia, but it is rare in Australia and the European Union (EU) (12).
An unexpected and sudden outbreak of severe epizootic salmonellosis due to S. Choleraesuis occurred in wild boars in Italy between 2012 and 2014; recovered isolates were typed for outbreak investigation purposes (13). A total of 30 isolates belonging to the outbreak were serotyped using the slide agglutination method, and biochemical tests were performed to identify the biotype. Where serotyping didn't work, additional tests were used to determine the serotype.
Results suggest the benefit of a combination of in silico MLST and antigenic formula detection to deep insight into a specific case of uncertainty in Salmonella serovar attribution.
Materials and Methods
Bacterial Isolates
A panel of thirty Salmonella wild boar isolates was included in this study (Table 1). All the isolates were collected by the Istituto Zooprofilattico Sperimentale delle Venezie, Legnaro, Italy between 2012 and 2014. Isolation was performed according to ISO 6579:2002/Amd 1: 2007.
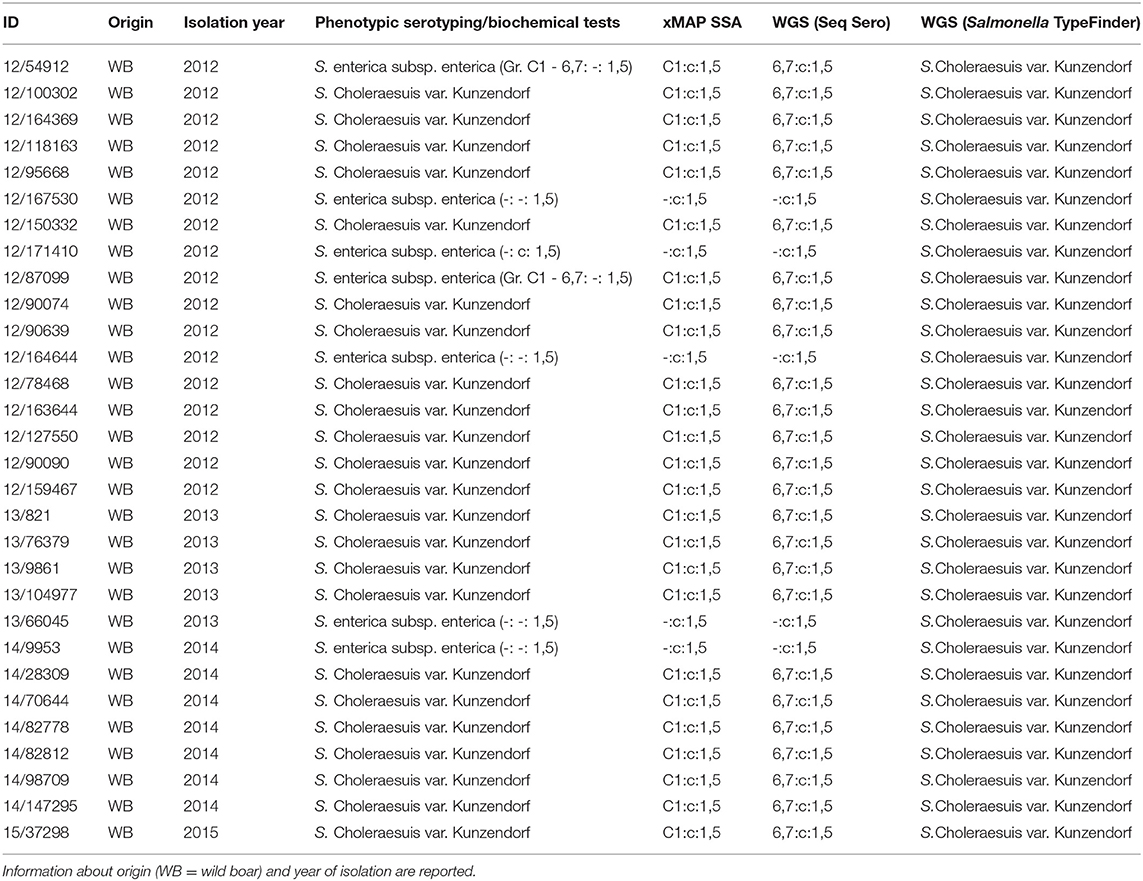
Table 1. Entire panel of analyzed isolates and serotyping results using different approaches: phenotypic serotyping and biochemical tests, xMAP® Salmonella Serotyping Assay (SSA), WGS (data analysis with SeqSero tool) and WGS (data analysis with Salmonella TypeFinder tool).
Phenotipic Serotyping and Biochemical Tests
All Salmonella isolates were serotyped by slide agglutination with Salmonella antisera (Statens Serum Institut, Copenhagen, Denmark) and serovar names assigned according to the WKL; distinction between the biotypes of S. Choleraesuis was performed by biochemical tests (H2S production, mucate and dulcitol fermentation) (1).
xMAP® Salmonella Serotyping Assay
Salmonella isolates were serotyped by xMAP® Salmonella Serotyping Assay (SSA), Luminex Corp., Austin, TX, U.S. SSA is a molecular serotyping assay addressing a set of target genes involved in the expression of the most common Salmonella serotype-specific antigens (4, 5).
Whole Genome Sequencing and Data Analysis
Genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Valencia, CA) and quantified with a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA). Libraries for sequencing were prepared using Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA). High-throughput sequencing was performed on Illumina MiSeq with 2 × 250 paired-end reads. Raw sequence data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under accession number PRJEB27935.
Raw reads were assembled using SPAdes (version 3.9) (14), available online at the Center for Genomic Epidemiology (CGE) (www.genomicepidemiology.org). The serotyping was performed analyzing contigs with SeqSero (version 1.2) (15) and raw reads with Salmonella TypeFinder version 1.4 (https://cge.cbs.dtu.dk/services/SalmonellaTypeFinder/) (15–17).
Results
Based on phenotypic serotyping and biochemical tests, 23 out of 30 isolates were shown to be S. Choleraesuis var. Kunzendorf by phenotypic serotyping and biochemical tests. All these isolates were assigned to the antigenic formula 6,7: c: 1,5 and showed the following biochemical features: dulcitol (–), H2S (+) and mucate (–).
Seven additional isolates displayed incomplete antigenic formula, in particular, two isolates did not present the first flagellar antigen (c), one isolate did not display the somatic antigen (Serogroup C1–antigens 6,7) and four isolates presented neither somatic nor the first flagellar antigens. The second flagellar antigen (1,5) was always detected (Table 1). The isolates with incomplete antigenic formula couldn't be definitively typed as S. Choleraesuis according to the traditional serotyping and were thus classified as S. enterica subsp. enterica.
Salmonella isolates were serotyped by xMAP® SSA. Two out of the seven isolates harbored the entire panel of genes, which allowed to infer the complete antigenic formula (C1:c:1,5). Biochemical tests allowed the typing of these isolates as S. Choleraesuis var. Kunzendorf. The remaining five isolates did not display the genetic target of the relative somatic antigens.
On the basis of the WGS and data analysis with SeqSero tool, twenty-five isolates out of thirty presented a complete antigenic formula (6,7:c:1,5). The lack of somatic antigen sequence for the other five isolates was also confirmed by WGS analysis, leaving the typing incomplete.
Regarding the analysis with Salmonella TypeFinder tool, even though the antigenic formulae found with the preceding methods were confirmed for all tested isolates, it was possible to obtain only an indirect relationship for Sequence Type 145 with serovar Choleraesuis var. Kunzendorf.
All the results are reported in Table 1.
Discussion
The wild boar epizootic mentioned in this work was caused by S. Choleraesuis var. Kunzendorf, which is considered the typical biotype of this serovar causing swine infections (9). The characterization of the Salmonella isolates responsible for wild boar's mortality provided us the opportunity to test different approaches to solve a specific ambiguous case study. The entire panel of isolates was serotyped with a phenotype-based approach at first, followed by biochemical tests. These analyses are labor intensive and quite long. Another limitation of traditional serotyping includes a possible loss of expression of one of the tested antigens (3). Seven isolates did not express one of the antigens required for serotyping, thus the result for the serovar assignment was incomplete (Salmonella enterica subsp. enterica). This would have misled the outbreak definition, as isolates from the same outbreak could have been assigned to different serovars.
Molecular serotyping methods offer a high-throughput alternative to traditional ones, which can strengthen the public health response capacity (18). In this study, the traditional serotyping was supported by three molecular approaches aiming at resolving the incomplete assignment of some isolates to a specific serovar. xMAP® SSA was a faster approach than the traditional serotyping, however, alone, it was not sufficient to discriminate S. Choleraesuis var. Kunzendorf for the entire panel of tested isolates. The two Salmonella isolates, showing absence of c flagellar antigen according to the phenotypic method, resulted to be S. Choleraesuis var. Kunzendorf by using xMAP® SSA. However, the lack of somatic antigen in the remaining five isolates was also confirmed by the xMAP® SSA, indicating the absence of the relative genetic target (rfb gene) (4).
The reads obtained from the entire panel of isolates by WGS were analyzed with two different tools. SeqSero tool, which assigns the serovar according to the antigenic formula, confirmed the results obtained by means of the molecular method.
Finally, Salmonella TypeFinder tool, which predicts the serovar using MLST typing (17), identified both serovar and biotype of the entire panel of the analyzed isolates as S. Choleraesuis var. Kunzendorf.
This study demonstrated that the antigenic formula detection might be not conclusive to cluster together isolates belonging to the same outbreak. The combined use of MLST and antigenic formula allowed, therefore, allocation of the investigated isolates to the same outbreak, irrespective of the antigenic formula. This suggests the perspective of integration of different data, both molecular and epidemiological, to provide deep insight into outbreak characterization in the presence of typing ambiguity.
Data Availability Statement
Raw sequence data were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under accession number PRJEB27935.
Author Contributions
AL, SP, AAL, LB, AR, CL, and VC contributed conception and design of the study. AL, SP, EM, and AT performed the analysis. AL and CL wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Grimont P, Weill FX. Antigenic Formulae of the Salmonella serovars. WHO Collab Cent Ref Res Salmonella. (2008). p. 1–166. Available online at: https://www.scacm.org/free/Antigenic%20Formulae%20of%20the%20Salmonella%20Serovars%202007%209th%20edition.pdf
2. Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, Nair S, et al. Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res Microbiol. (2014) 165:526–30. doi: 10.1016/j.resmic.2014.07.004
3. Shi C, Singh P, Ranieri ML, Wiedmann M, Moreno Switt AI. Molecular methods for serovar determination of Salmonella. Crit Rev Microbiol. (2015) 41:309–25. doi: 10.3109/1040841X.2013.837862
4. Fitzgerald C, Collins M, van Duyne S, Mikoleit M, Brown T, Fields P. Multiplex, bead-based suspension array for molecular determination of common Salmonella Serogroups. J Clin Microbiol. (2007) 45:3323–34. doi: 10.1128/JCM.00025-07
5. Dunbar SA, Ritchie VB, Hoffmeyer MR, Rana GS, Zhang H. Luminex® multiplex bead suspension arrays for the detection and serotyping of Salmonella spp. In: Schatten H, Eisenstark A, editors. Salmonella: Methods in Molecular Biology (Methods and Protocols), vol 1225. New York, NY: Humana Press.
6. Yachison CA, Yoshida C, Robertson J, Nash JHE, Kruczkiewicz P, Taboada EN, et al. The validation and implications of using whole genome sequencing as a replacement for traditional serotyping for a national Salmonella reference laboratory. Front Microbiol. (2017) 8:1044. doi: 10.3389/fmicb.2017.01044
7. Le Minor L, Beaud R, Laurent BMV. Salmonella possessing the 6,7:c:1,5 antigenic factors. Ann Inst Pasteur Microbiol. (1985)136B:225–34.
8. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. (2012) 8:e1002776. doi: 10.1371/journal.ppat.1002776
9. Pedersen K, Sørensen G, Löfström C, Leekitcharoenphon P, Nielsen B, Wingstrand A, et al. Reappearance of Salmonella serovar Choleraesuis var. Kunzendorf in danish pig herds. Vet Microbiol. (2015) 176:282–91. doi: 10.1016/j.vetmic.2015.01.004
10. Chiu CH, Su LH, Chu C. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin Microbiol Rev. (2004) 17:311–22. doi: 10.1128/cmr.17.2.311-322.2004
11. Boyen F, Haesebrouck F, Maes D, Van Immerseel F, Ducatelle R, Pasmans F. Non-typhoidal Salmonella infections in pigs: a closer look at epidemiology, pathogenesis and control. Vet Microbiol. (2008) 130:1–19. doi: 10.1016/j.vetmic.2007.12.017
12. Fedorka-Cray PJ, Gray JT, Wray C. Salmonella infections in pigs. In: Wray C, Wray A, editors. Salmonella in Domestic Animals. Wallingford: CAB International (2000). p. 191–207.
13. Longo A, Losasso C, Vitulano F, Mastrorilli E, Turchetto S, Petrin S, et al. Insight into an outbreak of Salmonella Choleraesuis var. Kunzendorf in wild boars. Vet Microbiol. (submitted).
14. Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, et al. Assembling genomes and mini-metagenomes from highly chimeric reads. J Comput Biol. (2013) 20:714–37. doi: 10.1089/cmb.2013.0084
15. Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, et al. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol. (2015) 53:1685–92. doi: 10.1128/JCM.00323-15
16. Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. (2014) 6:90. doi: 10.1186/s13073-014-0090-6
17. Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, et al. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ. (2016) 4:e1752. doi: 10.7717/peerj.1752
Keywords: Salmonella serotyping, antigenic formula, serovar Choleraesuis, biotype, xMAP® Salmonella serotyping assay, whole genome sequencing, MLST
Citation: Longo A, Petrin S, Mastrorilli E, Tiengo A, Lettini AA, Barco L, Ricci A, Losasso C and Cibin V (2019) Characterizing Salmonella enterica Serovar Choleraesuis, var. Kunzendorf: A Comparative Case Study. Front. Vet. Sci. 6:316. doi: 10.3389/fvets.2019.00316
Received: 28 June 2019; Accepted: 04 September 2019;
Published: 20 September 2019.
Edited by:
Bradley L. Bearson, United States Department of Agriculture (USDA), United StatesReviewed by:
Yves Millemann, INRA École Nationale Vétérinaire d'Alfort (ENVA), FranceNikki Shariat, University of Georgia, United States
Copyright © 2019 Longo, Petrin, Mastrorilli, Tiengo, Lettini, Barco, Ricci, Losasso and Cibin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmen Losasso, Y2xvc2Fzc29AaXpzdmVuZXppZS5pdA==
†These authors have contributed equally to this work
 Alessandra Longo
Alessandra Longo Sara Petrin
Sara Petrin Eleonora Mastrorilli
Eleonora Mastrorilli Alessia Tiengo
Alessia Tiengo Antonia Anna Lettini
Antonia Anna Lettini Lisa Barco
Lisa Barco Antonia Ricci
Antonia Ricci Carmen Losasso
Carmen Losasso Veronica Cibin
Veronica Cibin