- 1Istituto Zooprofilattico Sperimentale del Mezzogiorno, Portici, Italy
- 2Department of Veterinary Medicine and Animal Productions, Università degli Studi di Napoli “Federico II”, Naples, Italy
Babesiosis is an emerging tick-borne disease caused by apicomplexan parasites with widespread geographical distribution and various wildlife species as reservoir hosts. The aims of this study were to investigate the prevalence and assess the role of free-ranging canids and mustelids in the maintenance of Babesia spp. in southern Italy. PCR analysis of splenic samples targeting the 18S rRNA gene revealed the presence of Babesia spp. in 36 of 82 (43.9%) red foxes (Vulpes vulpes) including 29 (58%) from Campania region and seven (21.8%) from Calabria region, in seven of 13 (53.8%) Eurasian badgers (Meles meles), and in one of 13 (7.7%) gray wolves (Canis lupus). Samples from other host species including 9 Eurasian otters (Lutra lutra), 1 stone marten (Martes foina), 1 least weasel (Mustela nivalis), and 1 European polecat (Mustela putorius) tested Babesia spp. negative. Sequence analysis of the 18S rRNA gene demonstrated the presence of B. vulpes in the red fox and two sequence types of badger-associated Babesia spp. in the Eurasian badger. The Babesia sp. sequence detected in the gray wolf was identical to a badger-associated Babesia sp. This study shows that the number of Babesia spp. infecting free-ranging carnivores in Italy is higher than currently believed, and suggests that these hosts may play an important role in the maintenance of the sylvatic cycle of these parasites. It is the first report of badger-associated Babesia spp. in Italy and in a gray wolf.
Introduction
Babesiosis is a tick-borne disease caused by haemoprotozoan organisms of the genus Babesia (Apicomplexa: Piroplasma), and several wildlife species may represent important reservoir hosts. According to the most recent studies, Babesia spp. of carnivore mammals are divided into three main groups including the Babesia microti group infecting felids, canids, mustelids and procyonids; the prototheilerid group infecting felids, canids, herpestids and hyaenids; and Babesia sensu stricto group infecting canids, procyonids, and ursids (1–3).
In free-ranging carnivores in Europe, infection with at least three Babesia spp. has been documented including B. canis in the gray wolf (Canis lupus) (4), B. canis and B. vulpes (this latter reported in the literature as B. “Spanish dog isolate,” B. microti-like, B. annae, and Theileria annae) (5) in the red fox (Vulpes vulpes), and B. vogeli in a stone marten (Martes foina). Moreover, at least two genotypes named badger-associated Babesia spp. in the Eurasian badger (Meles meles) (1, 3, 6) and a genotype that clustered together to B. vulpes isolates [FJ225390.1 (7)] have been reported. An additional Babesia genotype in the European wildcat (Felis silvestris silvestris) from Bosnia and Herzegovina (8) has been also reported. B. vulpes and badger-associated Babesia spp. are included in the B. microti group; B. canis and B. vogeli are included in the Babesia sensu stricto group (1–3).
There are few studies focusing on the Babesia spp. in free-ranging carnivores other than red fox in Europe, and none in Italy (1, 3, 6, 8). Due to recent reports of DNA sequences than could represent new species and the overall lack of prevalence surveys in most carnivore species and geographic locations, it is likely that the number of Babesia spp. infecting free-ranging carnivores is higher that currently believed (1, 6–10).
Among Babesia species in free-ranging carnivores in Italy, only three recently studies from northern and central Italy reported the presence of B. vulpes in the red fox (11–13). All those studies suggested that regional factors as climatic and landscape variables linked to tick vector abundance and wild carnivore population densities may exist (11–13). Here we perform a molecular survey to study the occurrence of Babesia spp. in free-ranging canids and mustelids in the southern Italy where emerging tick-borne pathogens (i.e., B. canis, B. gibsoni, B. vogeli, Ehrlichia canis, Anaplasma spp., Borrelia burgdorferi spp., and Rickettia spp.) have been recently reported in domestic and free-ranging carnivores (14–18).
Materials and Methods
Sample Collection
Carcasses of 120 free-ranging carnivores including members of two families (Canidae and Mustelidae) obtained from the southern Italy were used for this study (Table 1). Carcasses were all road-killed animals except for the red foxes obtained during the official hunting season from local hunters. Carcasses were all obtained between January 2017 and December 2018 except for six Eurasian otters (Lutra lutra) obtained between March 2004 and December 2016 [see also (16)].
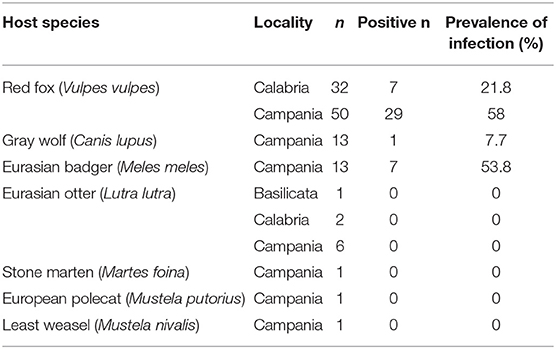
Table 1. Carnivore species, region of sampling and positive individuals for Babesia spp. in southern Italy.
Molecular Analyses
Spleen samples were collected at post-mortem examination and stored at −20°C for a week until DNA extraction. Genomic DNA was extracted from 25 mg of spleen with the QIAamp DNA mini KIT (QIAGEN) following manufacturer's instructions. All samples were screened for the presence of Babesia spp. by PCR targeting the 18S rRNA gene as described by Casati et al. (19). To assess the analytical sensitivity of the PCR assay, the DNA concentration was determined by biophotometry (BioPhotometer plus, Eppendorf, Italy). The extract was then serially diluted 10-fold in sterile water starting from 774 μg/ml, and each dilution was used as template for the PCR reaction. Briefly, reactions were performed in a total volume of 25 μL containing HotStar HiFidelity PCR Buffer 1X, 1 ul of HotStar HiFidelity DNA Polymerase, 0.6 μM of each primer, and 3.5 μL of DNA. The primers used were BJ1 (5′-GTC TTGTAATTGGAATGATGG-3′) and BN2 (5′-TAGTTTATGGTTAGGACTACG-3'). Amplification conditions included an initial denaturation/activation step at 95 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and extension at 72°C per 2 min, and a final extension step at 72°C for 5 min. Amplicons (486–520 bp) were resolved by automated capillary electrophoresis with the instrument QIAxcel (QIAGEN). Amplicons were subsequently purified and sequenced in both directions using the same primers as for PCR, employing the Big Dye Terminator Cycle Sequencing Kit v1.1 (Thermo Fischer Scientific, USA) in the automated sequencer 3500 Genetic Analyzer (ThermoFisher). Sequences were aligned using BIOEDIT software and compared with those available in GenBank (BLAST—http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic analyses were performed with the Neighbor-Joining method, using MEGA 6.0 (20, 21).
Results
Among DNA extracts, 36 of 82 (43.9%) samples of red fox, seven of 13 (53.8%) samples of Eurasian badger, and one of 13 (7.7%) samples of gray wolf were PCR-positive for Babesia spp. (Table 1). The amplicon detection limit for the PCR assay was 77.4 μg/ml corresponding to 1.9 μg/reaction.
Sequences from the red foxes (520 bp) were all identical and shared a similarity of 100% with B. vulpes 18S rRNA gene sequences found in GenBank (KY175167.1, KY175166.1). In the Eurasian badger and gray wolf we obtained six sequences belonging to two sequence types. The first type (515 bp) was found in four Eurasian badgers and a gray wolf (from MK742770 to MK742774) and showed a similarity of 100% with Babesia sp. badger type A 18S rRNA gene sequences found in China (MG799845.1, MH844530.1) and Spain (KT223484.1). The second one (486 bp) was found in a single Eurasian badger (MK742775), and showed a similarity of 100% with a Babesia sp. 18S rRNA gene sequence (JX679177.1) found in nymph individuals of the tick Ixodes canisuga collected from a red fox in Germany, and in an Eurasian badger in China (MG799847.1).
A Phylogenetic tree of the 18S rRNA gene of Babesia spp. of microti group using the Neighbor-Joining method, showed that B. vulpes sequences from this study clustered together with B. vulpes, B. sp. Spanish dog isolate, B. annae, T. annae, and B. microti-like piroplasm confirming the synonymy with those species. The first type sequence detected in the Eurasian badger and gray wolf clustered together with Babesia sp. badger type A and Babesia sp. Meles Hu1 found in European countries; the second type sequence detected in a Eurasian badger clustered together with a Babesia sp. found in Germany and China (Figure 1).

Figure 1. The evolutionary history of Babesia microti group DNA sequences was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 1,76412028 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown above the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances are in the units of the number of base substitutions per site. Sequences are presented by GenBank accession number, Babesia species, host species, and geographical location in brackets. Isolate from this study are in bold.
Discussion
In this study, the splenic samples of seven free-ranging carnivore species from the southern Italy were molecularly screened for the presence of Babesia spp. The DNA of three Babesia spp. all belonging to the B. microti group was detected.
Published data regarding Babesia spp. in free-ranging carnivores in Italy is limited to few studies that detected B. vulpes in the red fox only. In the red fox in Italy, B. vulpes prevalence was highly variable: 0.98% in north-western (11), 54% in north-eastern (13), and 22.88% in central Italy (12). PCR prevalence of B. vulpes in this survey differed depending on geographical locality. In Campania region, B. vulpes prevalence was almost three times higher than that reported in Calabria region (58 vs. 21.8%), but it was lower than prevalence found in Portugal [69.2% (22)] and Galicia, Spain [from 61 to 79.8% (23)].
Prevalence of badger-associate Babesia spp. in the Eurasian badger has been reported to range from 21.3 to 53.2% in Scotland (9), from 20 to 35.2% in Spain (6, 7), and it was 66.7% in Hungary (3). Prevalence of badger-associated Babesia spp. in the Eurasian badger in this study was among the higher prevalence values recorded to date.
Difference in Babesia spp. prevalence suggests regional differences in tick species diversity and abundance, and infection rate of the pathogen in ticks may influence the dynamic of Babesia spp. infection in mammalian hosts. In a recent study from Spain, Checa et al. (23) found significant differences in prevalence of B. vulpes infection among red fox populations of Galicia provinces suggesting that humid climates found in those provinces with highest prevalence rates are capable of sustaining higher burdens of ixodid ticks, which in turn may lead to the increased risk of tick-borne diseases. Differences in prevalence of B. vulpes between Campania and Calabria regions indicate a lower circulation of the parasite in Calabria region and could be related to the highest red fox density and tick vector abundance in those localities of the Campania region where the red foxes were obtained. A great abundance of potential hosts is an important factor for tick expansion and a potential for the spread of babesiosis (3, 4, 23). Southern Italy represents an endemic area for several tick-borne pathogens infecting domestic and free-ranging carnivores showing distribution corresponding to known distribution of its tick vectors (14–18). DNA of B. vulpes has been detected in many European countries in few tick species infecting the red fox including I. canisuga, I. hexagonus, I. ricinus, Rhipicephalus sanguineus s.l., and Dermacentor reticulatus; all these species are present in southern Italy except the last one. I. hexagonus is considered the most likely vector for B. vulpes (23, 24).
Despite the frequent contacts occurring between domestic dogs (as hunting dogs and sheepdogs) and red foxes, and the few ixodid tick vectors they share, infection of dogs and other free-ranging carnivores with B. vulpes occurred occasionally suggesting the red fox as its main natural host (1, 25). A recent survey of Babesia spp. in 1,311 hunting dogs in the Campania region of southern Italy revealed only the presence of B. canis, B. gibsoni, and B. vogeli (18). It suggests that at least in this region the red foxes and hunting dogs tend to acquire different Babesia spp.
In Europe, at least two DNA sequences of badger-associated Babesia spp. that could be undescribed Babesia species (named Babesia sp. isolate badger type A or Babesia sp. isolate Meles Hu1, and Babesia sp. isolate badger type B) have been reported in the Eurasian badger in Spain (6), Scotland (9), and Hungary (3) (see phylogenetic tree in Figure 1). An additional sequence type available in GenBank corresponds to a Babesia sequence detected in a Eurasian badger in Xinjiang, China (Babesia sp. badger isolate Badger-1; MG799847.1), and in an I. canisuga tick collected in a red fox in Germany (Babesia sp. 4 NAN2012; JX679177.1). Four out of five badger-associated Babesia spp. sequences detected in the present study in the Eurasian badger were identical to type A sequences found in China and Spain; the other one (MK742775) was identical to those found in China (MG799847.1) and Germany (JX679177.1). The high homology among sequences here found and those deposited in GenBank suggests that the parasites may circulate among Eurasian badgers across Europe.
Contrary to that observed for B. vulpes, a recent study demonstrated that the infection of badger-associated Babesia spp. may occur in dogs showing clinical signs of babesiosis (3). The usage of dogs during badger hunting in Hungary was considered as a plausible explication for the dog infections (3), since few Babesia spp. within the microti group may be transmitted orally by direct contact with infected blood (26, 27). The absence of the badger-associated Babesia spp. infection in hunting dogs from southern Italy may be linked to two main facts: (a) in Italy the badger hunting is an unperformed practice; (b) the hunters treated regularly their dogs with ectoparasiticides, as a result of being informed of the risks of pathogen transmission by ticks and other vectors (17, 18).
To our knowledge there are only two previous reports of Babesia infection in the gray wolf (4, 28). Erdelyi et al. (28) described a fatal infection by B. canis in two male captive gray wolves in a private farm in Hungary. Beck et al. (4) reported the infection by B. canis in seven of 108 gray wolves (including one captive and six free-ranging) in Croatia. The Babesia positive gray wolf found in the present study was a road killed adult female collected in January 2017 in Montemarano (Avellino municipality). It represents the first molecular confirmed case of a badger associated Babesia infection in a free-ranging gray wolf. It is plausible to suppose that the gray wolf was infected via blood-sucking ticks or alternatively preying on an infected Eurasian badger similarly to that hypothesized by Hornok et al. (3) for hunting dogs in Hungary.
In conclusion, the red fox and the Eurasian badger in southern Italy exhibit a high prevalence of infection caused by B. vulpes and badger-associated Babesia spp., respectively. It suggests that these hosts may play an important role in the maintenance of the sylvatic cycle of these parasites. This represents the first detection of badger-associated Babesia spp. in Italy. The detection of a badger-associated Babesia sp. in a gray wolf deserves further studies to understand the gray wolf role in the maintenance of this Babesia strain in the wild.
Data Availability
Representative nucleotide sequences of Babesia spp. found in this study have been submitted to GenBank and are available with the following accession numbers: from MK742776 to MK742780 for B. vulpes; MK742775 for the Babesia sequence detected from a single Eurasian badger; from MK742770 to MK742774 for the Babesia sequences detected in four Eurasian badger and a gray wolf.
Ethics Statement
Ethical review and approval was not required for the animal study because The IZSM is accredited by the Italian Ministry of Health to perform systematic surveys on infectious diseases of domestic and free-ranging animals. Procedures for this study were performed in accordance with the guide for the care and use of animals by the Italian Ministry of Health.
Author Contributions
MS, GG, and GF: experimental conception and design. GS, ND, and VV: collection of samples. CA, ML, and GB: molecular analysis. MS: paper writing. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was partially funded by the grant from the Ministry of Health of the Italian Republic (IZS ME 05/16 RC).
References
1. Alvarado-Rybak M, Solano-Gallego L, Millán J. A review of piroplasmid infections in wild carnivores worldwide: importance for domestic animal health and wildlife conservation. Parasit Vectors. (2016) 9:538. doi: 10.1186/s13071-016-1808-7
2. Schreeg ME, Marr HS, Tarigo JL, Cohn LA, Bird DM, Scholl EH, et al. Mitochondrial genome sequences and structures aid in the resolution of Piroplasmida phylogeny. PLoS ONE. (2016) 11:e0165702. doi: 10.1371/journal.pone.0165702
3. Hornok S, Horváth G, Takács N, Kontschán J, Szőke K, Farkas R. Molecular identification of badger-associated Babesia sp. DNA in dogs: updated phylogeny of piroplasms infecting Caniformia. Parasit Vectors. (2018) 11:235. doi: 10.1186/s13071-018-2794-8
4. Beck A, Huber D, Polkinghorne A, Kurilj AG, Benko V, Mrljak V, et al. The prevalence and impact of Babesia canis and Theileria sp. in free-ranging grey wolf (Canis lupus) populations in Croatia. Parasit Vectors. (2017) 10:168. doi: 10.1186/s13071-017-2106-8
5. Baneth G, Florin-Christensen M, Cardoso L, Schnittger L. Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasit Vectors. (2015) 8:207. doi: 10.1186/s13071-015-0830-5
6. Barandika JF, Espí A, Oporto B, Del Cerro A, Barral M, Povedano I, et al. Occurrence and genetic diversity of piroplasms and other apicomplexa in wild carnivores. Parasitol Open. (2016) 2:1–7. doi: 10.1017/pao.2016.4
7. Gimenez C, Casado N, Criado-Fornelio A, de Miguel FA, Dominguez-Peñafiel G. A molecular survey of Piroplasmida and Hepatozoon isolated from domestic and wild animals in Burgos (northern Spain). Vet Parasitol. (2009) 162:147–50. doi: 10.1016/j.vetpar.2009.02.021
8. HodŽić A, Alić A, Duscher GG. High diversity of blood-associated parasites and bacteria in European wild cats in Bosnia and Herzegovina: a molecular study. Ticks Tick Borne Dis. (2018) 9:589–93. doi: 10.1016/j.ttbdis.2018.01.017
9. Bartley PM, Wilson C, Innes EA, Katzer F. Detection of Babesia DNA in blood and spleen samples from Eurasian badgers (Meles meles) in Scotland. Parasitology. (2017) 144:1203–10. doi: 10.1017/S0031182017000476
10. Liu X, Yang M, Liu G, Zhao S, Yuan W, Xiao R, et al. Molecular evidence of Rickettsia raoultii, “Candidatus Rickettsia barbariae” and a novel Babesia genotype in marbled polecats (Vormela peregusna) at the China-Kazakhstan border. Parasit Vectors. (2018) 11:450. doi: 10.1186/s13071-018-3033-z
11. Zanet S, Trisciuoglio A, Bottero E, de Mera IG, Gortazar C, Carpignano MG, et al. Piroplasmosis in wildlife: Babesia and Theileria affecting free-ranging ungulates and carnivores in the Italian Alps. Parasit Vectors. (2014) 7:70. doi: 10.1186/1756-3305-7-70
12. Ebani VV, Rocchigiani G, Nardoni S, Bertelloni F, Vasta V, Papini RA, et al. Molecular detection of tick-borne pathogens in wild red foxes (Vulpes vulpes) from Central Italy. Acta Trop. (2017) 172:197–200. doi: 10.1016/j.actatropica.2017.05.014
13. Da Rold G, Ravagnan S, Soppelsa F, Porcellato E, Soppelsa M, Obber F, et al. Ticks are more suitable than red foxes for monitoring zoonotic tick-borne pathogens in northeastern Italy. Parasit Vectors. (2018) 11:137. doi: 10.1186/s13071-018-2726-7
14. René-Martellet M, Lebert I, Chêne J, Massot R, Leon M, Leal A, et al. Diagnosis and incidence risk of clinical canine monocytic ehrlichiosis under field conditions in Southern Europe. Parasit Vectors. (2015) 8:3. doi: 10.1186/s13071-014-0613-4
15. Santoro M, Veneziano V, D'Alessio N, Di Prisco F, Lucibelli MG, Borriello G, et al. Molecular survey of Ehrlichia canis and Coxiella burnetii infections in wild mammals of southern Italy. Parasitol Res. (2016) 115:4427–31. doi: 10.1007/s00436-016-5213-0
16. Santoro M, D'Alessio N, Cerrone A, Lucibelli MG, Borriello G, Aloise G, et al. The Eurasian otter (Lutra lutra) as a potential host for rickettsial pathogens in southern Italy. PLoS ONE. (2017) 12:e0173556. doi: 10.1371/journal.pone.0173556
17. Piantedosi D, Neola B, D'Alessio N, Di Prisco F, Santoro M, Pacifico L, et al. Seroprevalence and risk factors associated with Ehrlichia canis, Anaplasma spp., Borrelia burgdorferi sensu lato, and D. immitis in hunting dogs from southern Italy. Parasitol Res. (2017) 116:2651–60. doi: 10.1007/s00436-017-5574-z
18. Veneziano V, Piantedosi D, Ferrari N, Neola B, Santoro M, Pacifico L, et al. Distribution and risk factors associated with Babesia spp. infection in hunting dogs from Southern Italy. Ticks Tick Borne Dis. (2018) 9:1459–63. doi: 10.1016/j.ttbdis.2018.07.005
19. Casati S, Sager H, Gern L, Piffaretti JC. Presence of potentially pathogenic Babesia spp. for human in Ixodes ricinus in Switzerland. Ann Agric Environ Med. (2006) 13:65–70.
20. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. (1987) 4:406–25.
21. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. (2013) 30:2725–9. doi: 10.1093/molbev/mst197
22. Cardoso L, Cortes HC, Reis A, Rodrigues P, Simões M, Lopes AP, et al. Prevalence of Babesia microti-like infection in red foxes (Vulpes vulpes) from Portugal. Vet Parasitol. (2013) 196:90–5. doi: 10.1016/j.vetpar.2012.12.060
23. Checa R, López-Beceiro AM, Montoya A, Barrera JP, Ortega N, Gálvez R, et al. Babesia microti-like piroplasm (syn. Babesia vulpes) infection in red foxes (Vulpes vulpes) in NW Spain (Galicia) and its relationship with Ixodes hexagonus. Vet Parasitol. (2018) 252:22–8. doi: 10.1016/j.vetpar.2018.01.011
24. Camacho AT, Pallas E, Gestal JJ, Guitián FJ, Olmeda AS, Telford SR, et al. Ixodes hexagonus is the main candidate as vector of Theileria annae in northwest Spain. Vet Parasitol. (2003) 112:157–63. doi: 10.1016/S0304-4017(02)00417-X
25. Nayyar Ghauri H, Ijaz M, Farooqi SH, Ali A, Ghaffar A, Saleem S, et al. A comprehensive review on past, present and future aspects of canine theileriosis. Microb Pathog. (2018) 126:116–22. doi: 10.1016/j.micpath.2018.10.033
26. Malagon F, Tapia JL. Experimental transmission of Babesia microti infection by the oral route. Parasitol Res. (1994) 80:645–8. doi: 10.1007/BF00932947
27. Jefferies R, Ryan UM, Jardine J, Broughton DK, Robertson ID, Irwin PJ. Blood, bull terriers and babesiosis: further evidence for direct transmission of Babesia gibsoni in dogs. Aust Vet J. (2007) 85:459–63. doi: 10.1111/j.1751-0813.2007.00220.x
Keywords: Vulpes vulpes, Meles meles, Canis lupus, Babesia vulpes, badger-associated Babesia spp., babesiosis, tick-borne disease
Citation: Santoro M, Auriemma C, Lucibelli MG, Borriello G, D'Alessio N, Sgroi G, Veneziano V, Galiero G and Fusco G (2019) Molecular Detection of Babesia spp. (Apicomplexa: Piroplasma) in Free-Ranging Canids and Mustelids From Southern Italy. Front. Vet. Sci. 6:269. doi: 10.3389/fvets.2019.00269
Received: 19 June 2019; Accepted: 05 August 2019;
Published: 23 August 2019.
Edited by:
Robert James Ossiboff, University of Florida College of Veterinary Medicine, United StatesReviewed by:
Steven Victor Kubiski, University of California, Davis, United StatesNikolaus Huber, University of Veterinary Medicine, Austria
Copyright © 2019 Santoro, Auriemma, Lucibelli, Borriello, D'Alessio, Sgroi, Veneziano, Galiero and Fusco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario Santoro, bWFyaW8uc2FudG9yb0BpenNtcG9ydGljaS5pdA==
 Mario Santoro
Mario Santoro Clementina Auriemma1
Clementina Auriemma1 Giovanni Sgroi
Giovanni Sgroi Vincenzo Veneziano
Vincenzo Veneziano