
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 05 February 2019
Sec. Veterinary Infectious Diseases
Volume 6 - 2019 | https://doi.org/10.3389/fvets.2019.00017
Objectives: We investigated a collection of strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex obtained from a veterinary clinic with regard to their genetic relatedness, presence of antibiotic resistance genes and antimicrobial susceptibility profiles.
Methods: Fifty-eight ACB-complex strains from animals treated at a veterinary clinic between 2006 and 2017, and seven strains collected from the hospital environment during 2012 were analyzed. Assignment to sequence types (ST) and international complexes (IC) was done by multilocus sequence typing (MLST) according to the Pasteur scheme. Genes encoding carbapenemases, aminoglycoside-modifying enzymes, macrolide-, quinolone- and co-trimoxazole resistance genes, the ISAba1 element, virulence associated intI1 genes and plasmid associated toxin-antitoxin markers were identified by microarray. Genes encoding blaOXA−51-like carbapenemases were amplified by PCR and sequenced. Susceptibility profiles were determined by disc diffusion or by broth microdilution.
Results: Among 50 A. baumannii isolates from animals, two predominant clones were observed linked to CC1 (n = 27/54% of the isolates) and CC25 (n = 14/28%), respectively. Strains of IC I harbored blaOXA−69, aac(3′)-la, aadA1, sul1, intI1, and splA/T genes. Isolates belonging to CC25 possessed blaOXA−64. Six (12%) isolates belonging to CC2 and carrying blaOXA−66 were also noted. One isolate belonged to CC10 (blaOXA−68), one to CC149 (blaOXA−104), the remaining isolate was assigned to ST1220 and possessed blaOXA−116. Of six environmental A. baumannii, four (66.7%) belonged to CC25 (blaOXA−64), one (16.7%) to CC2 (blaOXA−66) and one to CC3 (blaOXA−71). Nine isolates (eight from animals and one environmental strain) were non-baumannii strains and did not harbor blaOXA−51-like genes. None of the isolates carried blaOXA−23, blaOXA−48, or blaOXA−58, and none were resistant to carbapenems.
Conclusions: Clonal lineages of the veterinary A. baumannii isolates in our collection are identical to those globally emerging in humans but do not harbor blaOXA−23. A. baumannii CC25 may be specific for this particular veterinary clinic environment.
The genus Acinetobacter is ubiquitous in diverse environments and as of today comprises 60 validly published species names (www.szu.cz/anemec/Classification.pdf). A list of Acinetobacter spp. also available at http://www.bacterio.net (1). In clinical settings, the species belonging to the Acinetobacter calcoaceticus- Acinetobacter baumannii (ACB) complex are of greatest importance (2). The ACB complex currently comprises Acinetobacter baumannii and its close relatives, A. calcoaceticus, A. dijkshoorniae (3), A. lactucae (4), A. nosocomialis, A. pittii, (5), and A. seifertii (6). Currently, A. dijkshoorniae and A. lactucae are considered conspecific (7). Hence, there exist to date six distinct ACB complex species with formal nomenclatural recognition.
A. baumannii is the most frequent Acinetobacter species isolated from patients in intensive care units (ICUs) and is the causative agent of ventilator associated pneumonia, catheter-related bloodstream infections, meningitis, and wound infection, often causing clonal outbreaks involving critically ill patients (8, 9). By constrast, A. calcoaceticus, although found sometimes in clinical specimens, seems to be more environmental, has unknown clinical significance and is usually well-susceptible to antibiotics (10).
The diversity of strains in epidemiology studies of A. baumannii is frequently investigated by multilocus sequence typing (MLST) using either the Oxford or the Pasteur scheme (11, 12). The majority of outbreak strains reported globally belong to IC I, IC II, and IC III, corresponding to clonal complexes (CC)1, CC2, and CC3 of the Pasteur scheme (9, 12–14).
Treatment of infections is frequently compromised due to the fact that ACB complex strains possess multiple intrinsic and acquired mechanisms that may result in antimicrobial resistance (10, 15). Overexpression of intrinsic ß-lactamases and of multidrug resistance efflux pumps, loss of outer membrane proteins and mutations in the quinolone resistance-determining regions (QRDRs) of the gyrA and parC are commonly detected in ACB isolates (10). Hence, following CLSI guidelines, ACB isolates are considered intrinsically resistant to antibiotics such as aminopenicillins, aztreonam, ertapenem, trimethoprim, chloramphenicol, macrolides, and fosfomycin (16).
Importantly, in addition to the presence of chromosomally located blaOXA−51-like genes encoding for naturally occurring carbapenemases, plasmid mediated carbapenem resistance genes including blaOXA−23, blaOXA−48, and blaOXA−58 have emerged globally in A. baumannii further restricting therapeutic options for treating infections in humans, with blaOXA−23 harboring A. baumannii representing one of the most problematic hospital-acquired human pathogens (17).
Data on molecular characteristics and antimicrobial resistance mechanisms of Acinetobacter of veterinary origin are still scarce compared to those of isolates from humans. However, it has been shown that A. baumannii isolated from animals may share clonal lineages and possess identical transmissible antibiotic resistance genes to those from humans, suggesting common pathways and/or sources of infection (15, 18). Furthermore, reports on the emergence of infections due to carbapenem resistant A. baumannii in hospitalized companion animals are of concern and emphasize the need for epidemiological studies and surveillance in order to maintain veterinary and public health (19–22).
The present study was designed to characterize clinical isolates belonging to the ACB complex originating from companion animals and horses hospitalized during 2006–2017 at a university veterinary clinic in Switzerland by (i) determining the genetic relatedness using multilocus sequence typing, (ii) performing genetic profiling using a microarray-based assay, and (iii) assessing their antimicrobial susceptibility profiles.
Between 2006 and 2017, a total of 93 non-duplicate Acinetobacter spp. isolated from hospitalized animals (one strain per animal) were obtained. Only isolates with clinical significance were collected. In addition, strains taken from the hospital environment during 2012 (n = 7) were included in the study. Strains were identified to the level of the genus Acinetobacter using the VITEK® 2 Compact system (Biomérieux, Nürtingen, Germany).
Species identification was performed by matrix-assisted laser desorption/ionization time-of- flight mass spectrometry (MALDI-TOF–MS, Bruker Daltronics, Bremen, Germany) and by amplification and sequencing of the 350 bp highly variable zone 1 of the rpoB gene (23, 24). Custom sequencing was done by Microsynth, Balgach, Switzerland.
In total, 65 ACB complex strains were identified, including 56 A. baumannii (50 isolates from animals, and six environmental strains), seven A. pittii (six animal, and one environmental isolate), and two animal A. calcoaceticus isolates.
The 58 animal strains originated from horses (n = 35), cats (n = 7), dogs (n = 6), chicken (n = 3), rabbits (n = 2), Andean bear (n = 1), cattle (n = 1), donkey (n = 1) reptile (n = 1), and rodent (n = 1) admitted to the veterinary clinic of the University of Zürich, Switzerland. The isolates were cultured from wounds (n = 20), abscesses (n = 19), urine (n = 4), synovial fluid aspirations (n = 2), tracheobronchial secretions (n = 2), abdominal aspiration (n = 1), alveolus (n = 1), bladder wall (n = 1), eye swab (n = 1), implant (n = 1), pus (n = 1), surgical sites (3), and other sites (n = 2).
In addition, strains collected from the hospital environment during 2012 (n = 7) were included in the study.
Non-ACB-complex strains (n = 35) comprising 21 A. lwoffii/‘A. pseudolwoffii’ (25), three A. guillouiae, three A. radioresistens, two A. beijerinckii, two A. towneri, one A. gandensis, one A. junii, one A. parvus, and one A. ursingii were not included in this study.
In accordance with local legislation, ethics approval was not required and no animal experiments were carried out for this study.
Multilocus sequence typing was performed according to the scheme developed by the Pasteur Institute (12). This scheme involves PCR amplification and sequencing of internal fragments of seven housekeeping genes (fusA, gltA, pyrG, recA, cpn60, rpoB, and rplB). Primers and PCR conditions are listed at the A. baumannii MLST database website http://pubmlst.org/abaumannii/. Sequencing of the amplification products was performed by Microsynth (Balgach, Switzerland). Sequences were uploaded to http://pubmlst.org/abaumannii/ to identify alleles and sequence types. The population structure of STs of the A. baumannii isolates was evaluated using the goeBURST software (http://www.phyloviz.net/goeburst/). CCs were defined as single-locus (SLVs) and double-locus variants (DLVs).
DNA was purified using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), according to manufacturer's protocol.
Isolates were genotyped using the oligonucleotide based microarray CarbDetect AS-2 Kit (Alere Technologies GmbH, Jena, Germany) to detect all currently known relevant carbapenemase genes, extended-spectrum ß-lactamase (ESBL) genes, aminoglycoside, macrolide, quinolone and co-trimoxazole resistance genes found in Enterobacteriaceae and Pseudomonadales (26). Additional markers included the ISAba1 element, integrase and transposase genes, and plasmid associated toxin-antitoxin (T/A) markers. An overview of the target genes and multiplex labeling, hybridization and data analysis has been described by Braun et al. (26). In brief, DNA was labeled internally with biotin-11-dUTP using a linear amplification protocol to generate single stranded (ss) DNA. Biotin labeled ssDNA was transferred and hybridized with DNA probes in oligonucleotide microarray strips. Hybridization was detected using streptavidin-horseradish peroxidase and a dye precipitation. The signals were detected using the platform ArrayMate Reader provided by Alere Technologies GmbH.
PCR and DNA sequencing analyses of blaOXA−51-like genes in A. baumannii isolates was carried out using custom synthesized primers (Microsynth, Balgach, Switzerland) and conditions published previously (27). Nucleotide sequences were analyzed with the CLC Main Workbench 8.0.1 and the BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov/blast/).
Screening for the plasmid-mediated colistin resistance genes mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 was performed by PCR using custom synthesized primers (Microsynth, Balgach, Switzerland) and conditions described previously (28).
The mcr-1 harboring strain OW3E1 (29) and plasmid “Plasmid-MCR2-Positivkontrolle” (P. Keller, personal communication) were used as positive controls.
Antimicrobial susceptibility testing was carried out according to Clinical and Laboratory Standards Institute (CLSI) performance standards (16), using the disk-diffusion method and the antibiotics cefotaxime (CTX), cefepime (FEP), ciprofloxacin (CIP), sulfamethoxazole/trimethoprim (SXT), gentamicin (GM), and tetracycline (TE). Minimal inhibitory concentrations (MIC) of imipenem were determined using the E-Test® (bioMérieux, Marcy L'Etoile, France) according to manufacturer's protocol. Determination of the MIC of colistin was performed by broth microdilution according to the European Committee on Antimicrobial Susceptibility Testing EUCAST (eucast.org).
The degrees of antimicrobial resistance among the ACB complex isolates was defined in accordance to Falagas and Karageorgopoulos (30).
Multidrug resistance (MDR) was thereby defined as resistance to three or more classes of antimicrobial agents including β-lactams, fluoroquinolones, sulfonamides, aminoglycosides, tetracyclines, and polymyxins, and excluding those which cannot be regarded as potentially effective, i.e., to which ACB complex strains are intrinsically resistant: aminopenicillins, aztreonam, ertapenem, trimethoprim, chloramphenicol, macrolides and fosfomycin (16).
An overview of the isolates, their origins, molecular and phenotypic characteristics is shown in Figure 1.
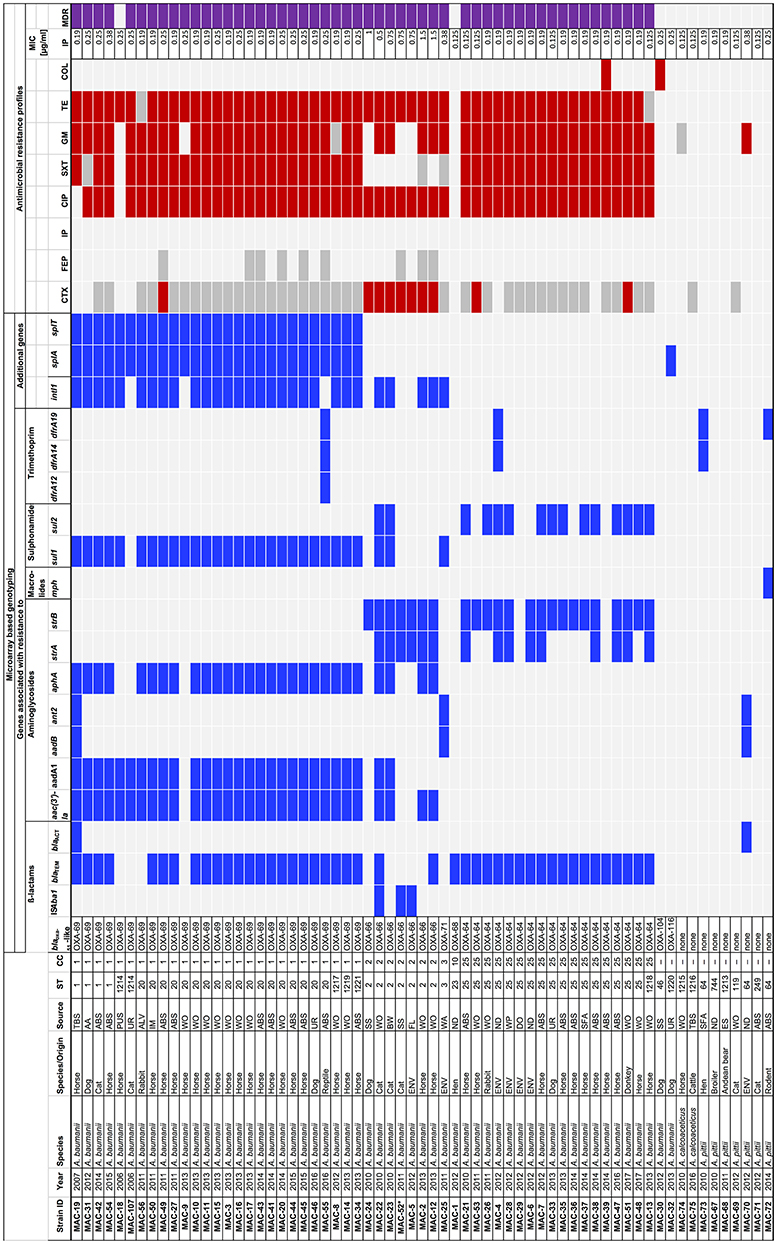
Figure 1. Molecular and phenotypic characterization of strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex isolated from animals and the environment at a veterinary hospital in Switzerland, 2006–2017. AA, abdominal aspiration; ABS, abscess; ALV, alveolus; AZM, azithromycin; BW, bladder wall; CC, clonal complex; CIP, ciprofloxacin; COL, colistin; CTX, cefotaxime; DO, door; ES, eye swab; ENV, environment; FEP, cefepime; FL, floor; GM, gentamicin; IM, implant; IP, imipenem; MIC, minimal inhibitory concentration; MDR, multidrug resistant; SS, surgical site; SFA, synovial fluid aspiration; ST, sequence type; SXT, sulfamethoxazole/trimethoprim; TBS, tracheobronchial secretion; TE, tetracycline; UR, urine; WA, water; WP, water pipe; WO, wound. * In isolate MAC-52, the ISAba1 element was not associated with blaOXA−66. Blue squares, positive result; red squares, resistant to a specific antimicrobial; gray squares, intermediately resistant to a specific antimicrobial; light gray squares, negative result or susceptible to a specific antimicrobial; purple squares, multidrug resistant.
Overall, 58 clinical isolates belonging to the ACB complex were collected from animals admitted to the veterinary hospital of Zürich, Switzerland between 2006 and 2017. The majority thereof (50/86.2%) were A. baumannii isolates collected from horses (n = 34), dogs (n = 6), cats (n = 5), rabbits (n = 2), and from one chicken, one donkey and one reptile, respectively. Six (10.3%) of the clinical isolates were A. pittii collected from cats (n = 2), chicken (n = 2), and from one Andean bear and one rodent, respectively. Two isolates (3.4%) were A. calcoaceticus from a cow and from a horse, respectively.
Environmental isolates collected from the premises of the veterinary hospital during 2012 included six A. baumannii and one A. pittii (Figure 1).
Multilocus sequence typing revealed that the majority (27/48.2%) of the 56 A. baumannii isolates belonged to ST20 and its SLVs ST1, ST1217, and ST1219, as well as its DLV ST1214. eBurst analysis assigned these sequence types to clonal complex CC1 (Figure 2). Fourteen (25%) of the A. baumannii strains belonged to ST25 and its SLV ST1218 and were assigned to CC25. Of the remaining isolates, six (10.7%) belonged to ST2 (CC2), and one (1.8%) to ST23 (CC10) and one to ST46 (CC149). One isolate typed and ST1220 and was not assigned to any CC (Figure 2).
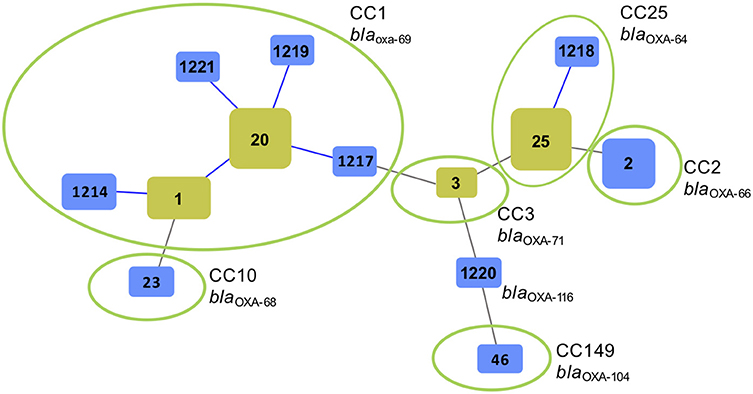
Figure 2. Genetic relatedness of Acinetobacter baumannii isolated from animals and the environment at a veterinary hospital in Switzerland during 2006–2017 using multilocus sequence typing (MLST) and goeBurst (Phylowiz). The sizes of the squares reflect the number of strains belonging to a particular sequence type (ST). Blue links show single locus variants (SLVs). Founder STs are highlighted in yellow. Green circles indicate isolates grouped into a clonal complex (CC) harboring a particular blaOXA−51-like allele.
Microarray based genotyping revealed the presence of blaOXA−51-like genes in all A. baumannii isolates. None of the A. pittii or A. calcoaceticus isolates tested positive for blaOXA−51-like genes (Figure 1).
Sequencing analysis of the blaOXA−51-like genes revealed the presence of blaOXA−69 in all 27 A. baumannii belonging to CC1 (Figures 1, 2). The blaOXA−64 gene was detected in the 18 A. baumannii isolates belonging to CC25, and blaOXA−66 was identified in the seven strains belonging to CC2. Other alleles included blaOXA−71, blaOXA−104, and blaOXA−116 (Figures 1, 2).
The ß-lactamase genes blaTEM and blaACT were found in 44 and 2 of the isolates, respectively (Figure 1).
Other bla genes encoding for carbapenemases (e.g., OXA-23, OXA-48, OXA-58, KPC, or NDM), or for acquired ESBLs (e.g., PER, VEB, or CTX-M types) were not detected.
The ISAba1 element which accounts for enhanced expression of blaOXA−51-like genes was detected adjacent to blaOXA−66 in two of the isolates (MAC5 and MAC22, Figure 1). In one isolate (MAC-52) the element was not associated with blaOXA−66 (Figure 1).
Genes associated with resistance to aminoglycosides, macrolides, sulphonamides and trimethoprim were detected as shown in Figure 1. The aac(3′)-Ia, aadA1, aphA, and sul1 genes occurred predominantly in association with the presence of the class 1 integrase gene intI1 in A. baumannii belonging to CC1 and CC2, whereas the majority of strains that lacked the intI1 gene harbored strA and/or strB and sul2 (Figure 1).
The type II T/A genes splA and splT were identified in all A. baumannii belonging to CC1.
In one strain (MAC-32), the toxin-antitoxin system was incomplete (Figure 1).
None of the isolates tested positive for mcr genes.
The antimicrobial susceptibility profiles of the isolates are summarized in Figure 1. The majority (52/92.9%) of the 56 A. baumannii strains was resistant to three or more classes of antimicrobials and were MDR according to Falagas and Karageorgopoulos (30). By contrast, among the A. calcoaceticus and A. pittii strains, one isolate (MAC-70) was resistant to gentamicin, the rest remained susceptible to all tested antimicrobials, and none were MDR.
Overall, 51 (91%) of the A. baumannii isolates were resistant to ciprofloxacin and to tetracycline, respectively, 47 (83.9%) were resistant to gentamicin, 43 (76.8%) were resistant to sulfamethoxazole-trimethoprim, and 10 (17.9%) were resistant to cefotaxime. Two (3.6%) isolates were resistant to colistin. All were susceptible to cefepime and imipenem, with MIC of imipenem ranging from 0.125 to 1.5 μg/ml (Figure 1).
There is growing concern that multidrug resistant, blaOXA−23 harboring A. baumannii in hospitalized companion animals and horses may be emerging as a threat to veterinary and public health (15). However, information on A. baumannii in veterinary medicine is still limited and there is a lack of comparable data to strains isolated from humans (15, 31). In this study, we provide a molecular and phenotypic analysis of strains belonging to the ACB complex isolated from diseased animals admitted to the veterinary hospital of the university of Zürich, Switzerland during 2006–2017. The main limitations of this study include its retrospective design and its restriction to a single center.
The two predominant lineages of A. baumannii comprised CC1, which is a globally distributed clade (9), and CC25, a lineage that has been responsible for epidemics in different European countries (32).
There are few reports on A. baumannii isolated from pets in Switzerland and overall, these isolates belonged primarily to CC1and CC2 (33, 34). Likewise, Ewers et al. (19) observed a prevalence of 26% of CC2 among A. baumannii recovered from animals hospitalized in various veterinary clinics in Germany, which is remarkably higher than the prevalence of 12% observed in this study. By contrast, A. baumannii ST25 (CC25) was not described in either of these studies. Its abundance in the collection of ACB isolates from the veterinary hospital of Zürich suggests that this clinical setting may be likely support the spread of this particular clonal lineage. Moreover, of the six environmental A. baumannii recovered during 2012, four (66.7%) belonged to ST25 (CC25), suggesting the existence of an environmental reservoir of this ST in or outside of the hospital setting. Its prevalence in the hospital environment may also be due to the elevated resistance to desiccation and high biofilm-forming capacity on abiotic surfaces, as demonstrated for this particular sequence type (35).
Notably, A. baumannii ST25 has recently been isolated from pets in France (20, 21). In both studies, the isolates possessed blaOXA−23 and were resistant to carbapenems, whereas the isolates analyzed in the current report possessed intrinsic blaOXA−51 –like carbapenemases only. Interestingly, the isolates from France were detected in companion animals in the community. Compared to clinical settings, little is known about Acinetobacter carriage in animals beyond these settings, but several studies during the last decade have detected A. baumannii in dogs in the community (21, 36), domestic birds (37), livestock (38) and other farmed animals such as mink (39). These reports indicate that community-acquired A. baumannii infections among animals may be increasing and that animals outside clinical settings may represent a reservoir for A. baumannii, including carbapenem resistant strains (15).
Overall, the blaOXA−51 alleles identified in the A. baumannii isolates correlated with their respective CCs, in accordance with previous observations for human isolates (27, 40).
Two isolates possessed the ISAba1 element upstream of blaOXA−66. As reported earlier, ISAba1 mediates overexpression of blaOXA−51-like enzymes, resulting in resistance to carbapenems (41, 42). However, there was no difference in the MICs of imipenem for these two isolates compared to those lacking the ISAba1 insertion, confirming recent observations that resistance to carbapenems is not guaranteed only by the presence of ISAba1, but depends on its orientation upstream of the bla gene (43, 44).
Aminoglycoside resistance genes were distributed unevenly among the A. baumannii isolates. The occurrence of aadA1 and aphA in association with intI1 among CC1, CC2, and CC3 is supportive of previous observations (45). By contrast, these genes were not prevalent among the CC25 isolates, among which strA and strB genes predominated. This may indicate that interclonal horizontal gene transfer plays a minor role in the dissemination of aminoglycoside resistance in the isolates analyzed in this study.
Of the two colistin resistant isolates, one belonged to CC25, which also occurs in humans. Colistin resistance in human Acinetobacter isolates is a source of great concern, although to date, there have been no reports of mcr positive Acinetobacter spp. (46).
In general, there was a good correlation between the presence of resistance genes detected by microarray and the phenotype of the isolates. There was however, a discrepancy between results of genotypic and phenotypic testing for three of isolates containing sulfonamide resistance genes and 4 isolates harboring aminoglycoside resistance genes, where the presence of resistance genes did not correspond with phenotypic resistance. Conversely, there was a lack of blaESBL genes such as blaGES, blaPER, or blaVEB that would explain the phenotypic resistance to cefotaxime observed in 10 isolates (10). On the other hand, ISAba1 and ISAba125 governed hyperexpression of the chromosomal blaADC cephalosporinase also leads to resistance to 3rd generation cephalosporins in A. baumannii (10, 47). Thus, further investigations targeting the genetic environment of blaADC in the cefotaxime resistant isolates are warranted to explain their phenotype.
In this study, we observed the presence of type II T/A genes splA and splT in A. baumannii CC1. Type II T/A systems are usually plasmid encoded and mediate plasmid maintenance through the post-segregational killing of plasmid-free daughter cells (48). The splA and splT genes are so far unique to A. baumannii and are encoded on small, ca. ~10 kb plasmids of the Rep-3 superfamily (49, 50). There is a lack of knowledge regarding these plasmids, however, some harbor blaOXA−24/blaOXA−40 or blaOXA−72 and are prevalent among carbapenem-resistant human clinical IC II isolates in Eastern Europe (49). The significance of the splA/T carrying plasmids identified among the CC1 isolates in this study remains to be investigated.
Finally, non-baumannii ACB complex species accounted for 13.8% of the animal, and 14.3% of the environmental isolates. These isolates were distinguished from A. baumannii strains by the lack of blaOXA−51-like genes, the low prevalence of acquired antibiotic resistance genes and high rate of susceptibility to antimicrobial agents, in agreement with observations from human isolates (8, 51).
In conclusion, this study provides a molecular and phenotypic analysis of ACB complex isolates obtained from animals admitted to a veterinary hospital in Switzerland during 2006–2017. Using established methods applied to isolates of human origin enabled the identification of clonal lineages and resistance determinants that occur globally among human isolates, including CC1 and CC25 A. baumannii. As opposed to A. baumannii CC1, CC25 isolates have infrequently been described in companion animals, but were prevalent among the isolates in this study. Contrasting to frequently occurring human clinical isolates worldwide, the veterinary ACB complex isolates in this study did not possess any known acquired carbapenemase genes. However, since A. baumannii, including CC25 isolates are emerging as blaOXA−23 carrying veterinary isolates in other countries, increased surveillance and targeted measures to prevent the dissemination of ACB complex strains are warranted.
RS designed the study. KZ, SP-S, and SS carried out the microbiological and molecular biological tests. SP-S, KZ, and MN-I analyzed and interpreted the data. MN-I drafted the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Marc Stevens for assistance with bioinformatics. This work was partly supported by the Swiss Federal Office of Public Health, Division Communicable Diseases.
1. Parte AC. LPSN-List of Prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol. (2018) 68:1825–29. doi: 10.1099/ijsem.0.002786
2. Towner KJ. Acinetobacter: an old friend, but a new enemy. J Hosp Infect. (2009) 73:355–63. doi: 10.1016/j.jhin.2009.03.032
3. Cosgaya C, Marí-Almirall M, Van Assche A, Fernández-Orth D, Mosqueda N, Telli M, et al. Acinetobacter dijkshoorniae sp. nov, a member of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int J Syst Evol Microbiol. (2016) 66:4105–11. doi: 10.1099/ijsem.0.001318
4. Rooney AP, Dunlap CA, Flor-Weiler LB. Acinetobacter lactucae sp. nov, isolated from iceberg lettuce (Asteraceae: Lactuca sativa). Int J Syst Evol Microbiol. (2016) 66:3566–72. doi: 10.1099/ijsem.0.001234
5. Nemec A, Krizova L, Maixnerova M, van der Reijden TJK, Deschaght P, Passet V, et al. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol. (2011) 162:393–404. doi: 10.1016/j.resmic.2011.02.006
6. Nemec A, Krizova L, Maixnerova M, Sedo O, Brisse S, Higgins PG. Acinetobacter seifertii sp. nov, a member of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex isolated from human clinical specimens. Int J Syst Evol Microbiol. (2015) 65:934–42. doi: 10.1099/ijs.0.000043
7. Dunlap CA, Rooney AP. Acinetobacter dijkshoorniae is a later heterotypic synonym of Acinetobacter lactucae. Int J Syst Evol Microbiol. (2017) 68:131–2. doi: 10.1099/ijsem.0.002470
8. Fitzpatrick MA, Ozer E, Bolon MK, Hauser AR. Influence of ACB complex genospecies on clinical outcomes in a U.S. hospital with high rates of multidrug resistance. J Infect. (2015) 70:144–52. doi: 10.1016/j.jinf.2014.09.004
9. Higgins PG, Dammhayn C, Hackel M, Seifert H. Global spread of carbapenem-resistant Acinetobacter baumannii. J Antimicrob Chemother. (2010) 65:233–8. doi: 10.1093/jac/dkp428
10. Doi Y, Murray GL, Peleg AY. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin Respir Crit Care Med. (2015) 36:85–98. doi: 10.1055/s-0034-1398388
11. Bartual SG, Seifert H, Hippler C, Luzon MAD, Wisplinghoff H, Rodríguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbio. (2005) 43:4382–90. doi: 10.1128/JCM.43.9.4382-4390.2005
12. Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE (2010) 5:e10034. doi: 10.1371/journal.pone.0010034
13. Tomaschek F, Higgins PG, Stefanik D, Wisplinghoff H, Seifert H. Head-to-head comparison of two multi-locus sequence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PLoS ONE (2016) 11:e0153014. doi: 10.1371/journal.pone.0153014
14. Zarrilli R, Pournaras S, Giannouli M, Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int J Antimicrob Agents. (2013) 41:11–9. doi: 10.1016/j.ijantimicag.2012.09.008
15. van der Kolk JH, Endimiani A, Graubner C, Gerber V, Perreten V. Acinetobacter in veterinary medicine with emphasis on A. baumannii J Glob Antimicrob Resist. (2018) 16:59–71. doi: 10.1016/j.jgar.2018.08.011
16. Clinical and Laboratory Standards Institute (2016). Performance Standards for Antimicrobial Susceptibility Testing, 26th Edn CLSI SupplementM100S.Wayne, PA: Clinical and Laboratory Standards Institute.
17. Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide Dissemination of the carbapenemase gene of blaOXA−23 carbapenemase gene of Acinetobacter baumannii. Emerg Infect Dis. (2009) 16:35–40. doi: 10.3201/eid1601.090852
18. Zordan S, Prenger-Berninghoff E, Weiss R, van der Reijden T, van den Broek P, Baljer G, et al. Multidrug-resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerg Infect Dis. (2011) 17:1751–4. doi: 10.3201/eid1709.101931
19. Ewers C, Klotz P, Leidner U, Stamm I, Prenger-Berninghoff E, Göttig S, et al. OXA-23 and ISAba1-OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int J Antimicrob Agents (2017) 49:37–44. doi: 10.1016/j.ijantimicag.2016.09.033
20. Lupo A, Châtre P, Ponsin C, Saras E, Boulouis H-J, Keck N, et al. Clonal spread of Acinetobacter baumannii sequence type 25 carrying blaOXA−23 in companion animals in France. Antimicrob Agents Chemother. (2017) 61:e01881–16. doi: 10.1128/AAC.01881-16
21. Hérivaux A, Pailhoriès H, Quinqueneau C, Lemarié C, Joly-Guillou M-L, Ruvoen N, et al. First report of carbapenemase-producing Acinetobacter baumannii carriage in pets from the community in France. Int J Antimicrob Agents (2016) 48:220–1. doi: 10.1016/j.ijantimicag.2016.03.012
22. Pomba C, Endimiani A, Rossano A, Saial D, Couto N, Perreten V. First report of OXA-23-mediated carbapenem resistance in sequence type 2 multidrug-resistant Acinetobacter baumannii associated with urinary tract infection in a cat. Antimicrob Agents Chemother. (2014) 58:1267–8. doi: 10.1128/AAC.02527-13
23. Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology (2009) 155(Pt 7):2333–41. doi: 10.1099/mic.0.026054-0
24. La Scola B, Gundi VAKB, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. (2006) 44:827–32. doi: 10.1128/JCM.44.3.827-832.2006
25. Nemec A, Radolfová-KríŽová L, Maixnerová M, Nemec M, Clermont D, Bzdil J, et al. Revising the taxonomy of the Acinetobacter lwoffii group: The description of Acinetobacter pseudolwoffii sp. nov. and emended description of Acinetobacter lwoffii. Syst Appl Microbiol. (2018) doi: 10.1016/j.syapm.2018.10.004. [Epub ahead of print].
26. Braun SD, Monecke S, Thürmer A, Ruppelt A, Makarewicz O, Pletz M, et al. Rapid Identification of carbapenemase genes in Gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS ONE (2014) 9:e102232. doi: 10.1371/journal.pone.0102232
27. Pournaras S, Gogou V, Giannouli M, Dimitroulia E, Dafopoulou K, Tsakris A, et al. Single-locus-sequence-based typing of blaOXA−51-like genes for rapid assignment of Acinetobacter baumannii clinical isolates to international clonal lineages. J Clin Microbiol. (2014) 52:1653–7. doi: 10.1128/JCM.03565-13
28. Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveill (2018) 23:17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
29. Zurfluh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother. (2016) 60:2594–5. doi: 10.1128/AAC.00066-16
30. Falagas ME, Karageorgopoulos DE. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: need for international harmonization in terminology. Clin Infect Dis. (2008) 46:1121–2. doi: 10.1086/528867
31. Müller S, Janssen T, Wieler LH. Multidrug resistant Acinetobacter baumannii in veterinary medicine–emergence of an underestimated pathogen. Berl Munch Tierarztl Wochenschr. (2014) 127:435–46. doi: 10.2376/0005-9366-127-435
32. Sahl JW, Del Franco M, Pournaras S, Colman RE, Karah N, Dijkshoorn L, et al. Phylogenetic and genomic diversity in isolates from the globally distributed Acinetobacter baumannii ST25 lineage. Sci Rep. (2015) 5:15188. doi: 10.1038/srep15188
33. Endimiani A, Hujer KM, Hujer AM, Bertschy I, Rossano A, Koch C, et al. Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data. J Antimicrob Chemother. (2011) 66:2248–54. doi: 10.1093/jac/dkr289
34. Boerlin P, Eugster S, Gaschen F, Straub R, Schawalder P. Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet Microbiol. (2001) 82:347–59. doi: 10.1016/S0378-1135(01)00396-0
35. Giannouli M, Antunes LCS, Marchetti V, Triassi M, Visca P, Zarrilli R. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis. (2013) 13:282. doi: 10.1186/1471-2334-13-282
36. Pailhoriès H, Belmonte O, Kempf M, Lemarié C, Cuziat J, Quinqueneau C, et al. Diversity of Acinetobacter baumannii strains isolated in humans, companion animals, and the environment in Reunion Island: an exploratory study. Int J Infect Dis. (2015) 37:64–9. doi: 10.1016/j.ijid.2015.05.012
37. Klotz P, Jacobmeyer L, Stamm I, Leidner U, Pfeifer Y, Semmler T, et al. Carbapenem-resistant Acinetobacter baumannii ST294 harbouring the OXA-72 carbapenemase from a captive grey parrot. J Antimicrob Chemother. (2018) 73:1098–100. doi: 10.1093/jac/dkx490
38. Poirel L, Berçot B, Millemann Y, Bonnin RA, Pannaux G, Nordmann P. Carbapenemase-producing Acinetobacter spp. in cattle, France. Emerg Infect Dis. (2012) 18:523–5. doi: 10.3201/eid1803.111330
39. Molenaar RJ, van Engelen E. Pneumonia associated with Acinetobacter baumannii in a group of minks (Neovison vison). Vet Q. (2015) 35:174–6. doi: 10.1080/01652176.2015.1030714
40. Evans BA, Hamouda A, Towner KJ, Amyes SG. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin Microbiol Infect. (2008) 14:268–75. doi: 10.1111/j.1469-0691.2007.01919.x
41. Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural blaOXA−66 oxacillinase gene. Antimicrob Agents Chemother. (2009) 53:2657–9. doi: 10.1128/AAC.01663-08
42. Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. (2006) 258:72–7. doi: 10.1111/j.1574-6968.2006.00195.x
43. Nigro SJ, Hall RM. Does the intrinsic oxaAb (blaOXA−51-like) gene of Acinetobacter baumannii confer resistance to carbapenems when activated by ISAba1? J Antimicrob Chemother. (2018) 73:3518–20. doi: 10.1093/jac/dky334
44. Rodrigues-Costa F, Cayô R, Matos AP, Girardello R, Martins WMBS, Carrara-Marroni FE, et al. Temporal evolution of Acinetobacter baumannii ST107 clone: conversion of blaOXA−143 into blaOXA−231 coupled with mobilization of ISAba1 upstream occAB1. Res Microbiol (2018). doi: 10.1016/j.resmic.2018.07.001. [Epub ahead of print].
45. Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. (2004) 53:1233–40. doi: 10.1099/jmm.0.45716-0
46. Rahman A, Iza N, Ismail S, Alattraqchi AG, Cleary DW, Clarke SC, et al. Acinetobacter spp. infections in Malaysia: a review of antimicrobial resistance trends, mechanisms and epidemiology. Front Microbiol. (2017) 8:2479. doi: 10.3389/fmicb.2017.02479
47. Lopes BS, Amyes SG. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J Med Microbiol. (2012) 61(Pt 8):1103–8. doi: 10.1099/jmm.0.044156-0
48. Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science (2003) 301:1496–9. doi: 10.1126/science.1088157
49. Lean SS, Yeo CC. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: good, bad, who knows. Front Microbiol. (2017) 8:1547. doi: 10.3389/fmicb.2017.01547
50. Jurenaite M, Markuckas A, Suziedeliene E. Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J Bacteriol. (2013) 195:3165–72. doi: 10.1128/JB.00237-13
Keywords: Acinetobacter, blaOXA−51-like, genotypes, antimicrobial resistance, animals
Citation: Püntener-Simmen S, Zurfluh K, Schmitt S, Stephan R and Nüesch-Inderbinen M (2019) Phenotypic and Genotypic Characterization of Clinical Isolates Belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) Complex Isolated From Animals Treated at a Veterinary Hospital in Switzerland. Front. Vet. Sci. 6:17. doi: 10.3389/fvets.2019.00017
Received: 14 October 2018; Accepted: 17 January 2019;
Published: 05 February 2019.
Edited by:
Armanda Bastos, University of Pretoria, South AfricaReviewed by:
Lenie Dijkshoorn, Leiden University Medical Center, NetherlandsCopyright © 2019 Püntener-Simmen, Zurfluh, Schmitt, Stephan and Nüesch-Inderbinen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Magdalena Nüesch-Inderbinen, bWFnZGFsZW5hLm51ZWVzY2gtaW5kZXJiaW5lbkB1emguY2g=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.