- 1National Institute on Alcohol Abuse and Alcoholism, Bethesda, MD, United States
- 2National Institute on Drug Abuse, Bethesda, MD, United States
Opioid use in the United States has steadily risen since the 1990s, along with staggering increases in addiction and overdose fatalities. With this surge in prescription and illicit opioid abuse, it is paramount to understand the genetic risk factors and neuropsychological effects of opioid use disorder (OUD). Polymorphisms disrupting the opioid and dopamine systems have been associated with increased risk for developing substance use disorders. Molecular imaging studies have revealed how these polymorphisms impact the brain and contribute to cognitive and behavioral differences across individuals. Here, we review the current molecular imaging literature to assess how genetic variations in the opioid and dopamine systems affect function in the brain’s reward, cognition, and stress pathways, potentially resulting in vulnerabilities to OUD. Continued research of the functional consequences of genetic variants and corresponding alterations in neural mechanisms will inform prevention and treatment of OUD.
Introduction
Opioid use in the United States has steadily risen since the late 1990s, along with staggering increases in overdose fatalities (1). The use of illicit opioids such as heroin and fentanyl has increased dramatically, contributing to opioid-related morbidity and mortality (2). With approximately 115 Americans dying each day from an opioid overdose, this epidemic is now considered a public health emergency (3). The surge in prescription and illicit opioid abuse necessitates further investigation into the genetic risk factors and neuropsychological effects of opioid use disorder (OUD).
The roles of the opioid and dopamine (DA) systems in substance use disorders (SUDs) are well recognized (4). Drug reward and incentive salience develop during the acute effects of drug-taking and correspond to changes in opioid and DA signaling in the basal ganglia (5). Incentive salience is defined by the association of previously neutral stimuli with drug use, which promotes compulsive drug-seeking (4). Stress responses associated with withdrawal involve decreased DA signaling along reward pathways, increased dynorphin-mediated kappa opioid (KOP) receptor signaling, and increased corticotropin-releasing factor (CRF) signaling in the amygdala (4). These same principles apply to OUD. For example, Wang et al. (6) used positron-emission tomography (PET) imaging with [11C]raclopride to demonstrate lower dopamine receptor 2 (D2R) and 3 (D3R) availability in the striatum of opioid-dependent patients compared to controls. Another [11C]raclopride PET study found low striatal D2/3 receptor availability and low presynaptic DA in OUD patients compared to controls (7), which has also been found for other SUDs including cocaine, alcohol, methamphetamine, and cannabis [reviewed in Refs. (8, 9)]. Low D2R levels have also been associated with sleep deprivation (10–12) and lower socioeconomic status (13, 14). These factors may contribute to lower D2R availability found in SUDs, particularly since SUDs and sleep deprivation are highly comorbid (15). Other preclinical studies have found dynorphin-mediated KOP receptor signaling inhibits dopaminergic signaling and modulates aversive emotional states that maintain drug dependence (16–18). Based on these studies, both the opioid and DA signaling systems are implicated in OUD.
However, there are opposing views on these systems’ involvement in addiction. For example, there are studies that report no disruption of D2R in OUD, including no difference in baseline D2R availability in methadone-maintained OUD patients compared to controls (19). Moreover, PET studies of opioid-dependent patients on medications for OUD (MOUD) found no increase in striatal DA release in response to opioid administration (19, 20). Studies of other SUDs also present slight inconsistencies in their effects on the dopamine system. Imaging studies in individuals with alcohol use disorder (AUD) have reported marked reductions in dopamine release and in striatal D2R, and most preclinical studies have documented significant reductions in dopamine neuronal firing and tonic dopamine release (9, 21–27). However, studies in rodents have also reported dynamic changes in dopamine release with increases and decreases in accumbens at various days post alcohol withdrawal (28). The discrepancies in the preclinical studies are likely to reflect in part time at which the measurements were made (early versus late withdrawal) as well as the alcohol models used (active versus passive administration). Thus, further research is required to understand the complex relationship between opioid and DA systems in SUDs.
While it has long been postulated that genetics influence an individual’s susceptibility to addiction, there has been little success in pinpointing genes with well-defined, causal roles in SUDs (29). Nevertheless, OUD is highly heritable, with an estimated 50% genetic contribution (30–32). The use of candidate gene studies and genome-wide association studies has revealed several polymorphisms that reliably associate with SUDs; however, addiction is a polygenic disease with complex genetic interactions and therefore individual polymorphisms will likely only account for a fraction of the total genetic risk for OUD (33–35). Polymorphisms in the opioid signaling system have been associated with addiction, as well as addiction treatment response (29). For example, several studies have identified a single nucleotide polymorphism (SNP) in the OPRM1 gene that associates with improved response to naltrexone treatment in individuals with AUD (36–39). Other OPRM1 SNPs may also play a role in nicotine dependence and treatment response (40–42). Additionally, genetic variations in the DA system have been linked to various SUDs as DA modulates reward and aversion pathways central to addiction (29, 43). For example, polymorphisms in the genes coding for dopamine 1 receptor (D1R) and D2R are associated with OUD, cocaine use disorder (CUD), and AUD (6, 22, 44). In addition, polymorphisms in the gene DAT1, which codes for dopamine transporters (DAT), have been associated with CUD and AUD (45–47). In line with this, reduced striatal DAT availability has been associated with OUD (48–53) and DAT availability has been associated with various other SUDs (51, 54–62).
In this review, we compiled findings related to the genetics of the opioid and DA systems and corresponding changes in brain and behavior as evidenced by PET neuroimaging. Functional and structural magnetic resonance imaging (MRI) is another useful tool in examining altered neural circuits in individuals with SUDs, as well as in polymorphism carriers. However, we will limit the scope to molecular imaging as the literature on MRI in OUD was recently reviewed (63–66). Integrating genetics with regional changes in receptor binding may help uncover circuits relevant for the pathophysiology of OUD, and thereby inform precision-based prevention and treatment.
The Opioid Receptor System
OPRM1
OPRM1 Background
The OPRM1 gene codes for the MOP receptor, an inhibitory G-protein coupled receptor (GPCR) that binds endogenous opioid peptides such as β-endorphin and enkephalins as well as exogenous opioids such as morphine and heroin (67). MOP receptors are required to establish morphine place preference and physical dependence (68). MOP receptors are expressed throughout the brain’s reward pathways including the mesocorticolimbic network as illustrated in Figure 1; their proposed mechanism for positive reinforcement in OUD is through disinhibition of DA neurons that trigger drug reward upon DA release (69, 70). Originally it was thought that MOP receptor agonists hyperpolarize GABAergic interneurons of the ventral tegmental area (VTA), reducing GABA-mediated inhibitory input to DA neurons and thereby increasing DA signaling by disinhibition (69). However, most evidence now suggests that the rostromedial tegmental nucleus mediates opioid-induced disinhibition of DA neurons (71–73). There is preclinical evidence of DA-independent opioid-induced reward, but the mechanism is not well understood (74, 75).
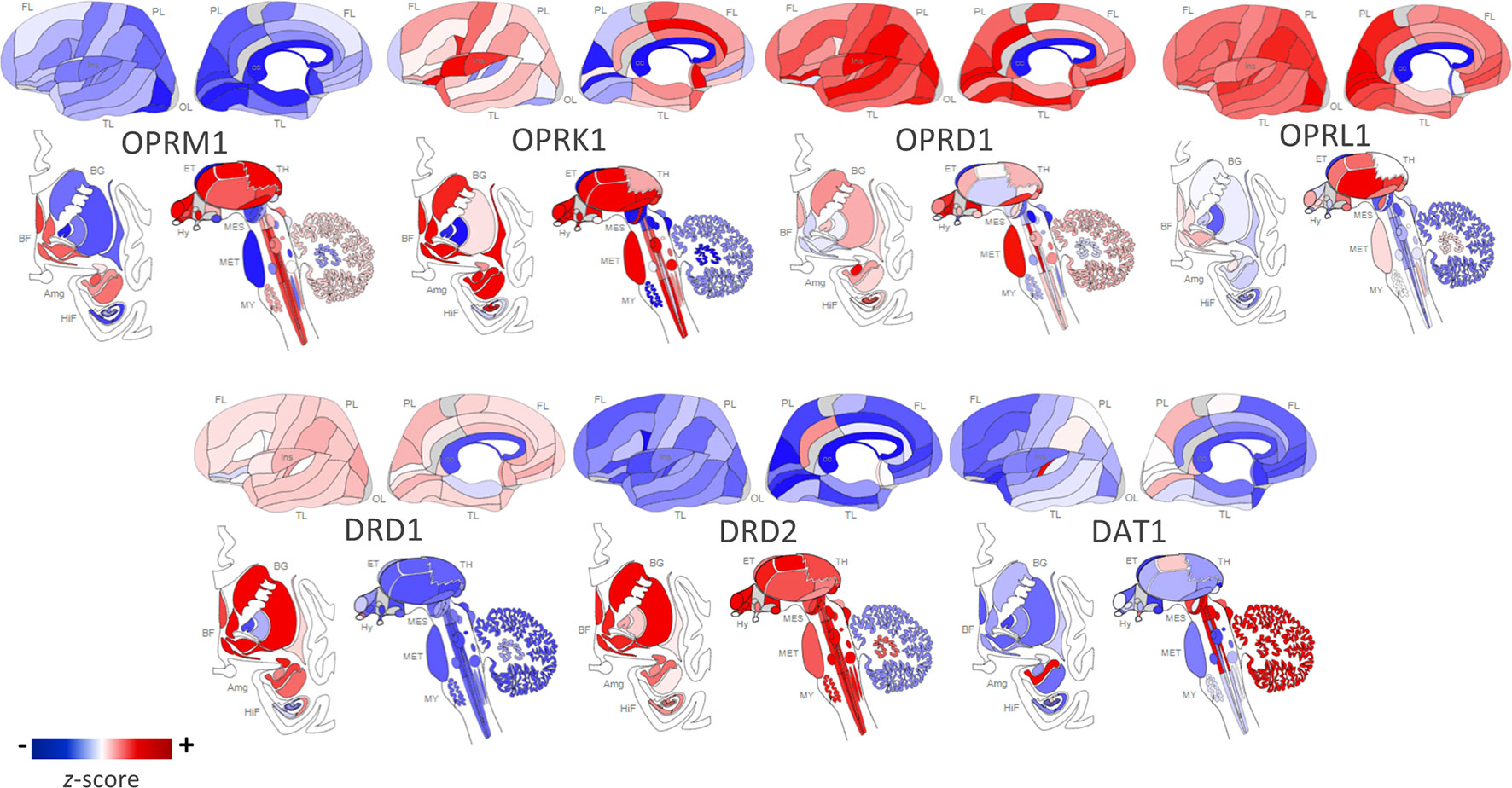
Figure 1 Regional distribution of receptor types in the human brain. Opioid and dopamine receptor gene expression in the human brain [Opioid Receptor Mu 1 (OPRM1), Opioid Receptor Kappa 1 (OPRK1), Opioid Receptor Delta 1 (OPRD1), Opioid Related Nociceptin Receptor 1 (OPRL1), Dopamine Receptor D1 (DRD1), Dopamine Receptor D2 (DRD2), Dopamine Active Transporter 1 (DAT1)]. Images constructed using Allen Human Brain Atlas. Data displayed are from one donor: H0351.2002, 39 years, M, Black or African American. The color bar displays expression values using z-score normalization. Color scale was altered to highlight regional differences in gene expression per receptor type; therefore, the absolute scale differs across each of the receptor subtypes. For quantitative results from all six postmortem donor brains, visit http://human.brain-map.org/static/brainexplorer.
The effects of prolonged opioid exposure on MOP receptors, whether in the context of chronic pain management or substance abuse, are not fully understood. Bolger et al. (76) demonstrated an upregulation in MOP receptor in rat brain after chronic heroin administration. However, several other studies have demonstrated that both morphine and buprenorphine administration downregulate MOP receptors in rat brain (76, 77) including striatum (78). Clinically, prolonged exposure to opioids results in tolerance and increased opioid dose requirements; several proposed mechanisms may explain this phenomenon, including phosphorylation and arrestin-driven uncoupling of the GPCR and receptor internalization and degradation (79–82). However, several studies cloned MOP receptors in human embryonic kidney cells and found that morphine does not promote MOP receptor endocytosis (80, 83–85), which results in protracted desensitization that could contribute to tolerance (86). Yet, several opioids including methadone, etorphine, and [D-Ala2, N-MePhe4, Gly-ol]-enkephalin (DAMGO) induced the expected receptor sequestration in cell line models (79, 80, 87, 88). A study in rats also showed MOP receptor internalization in the striatum and habenula after acute etorphine, but not morphine administration (80). These findings were replicated in the rat’s locus coeruleus where neurons showed MOP receptor internalization in response to DAMGO and methadone, but not morphine (89). Downregulation of MOP receptors is agent-specific as some opioids are more effective at activating the G-protein response than others (87). The concept of biased agonism explains differential activation patterns and intracellular signaling cascades based on ligand structure and GPCR conformations (90, 91). In the case of MOP receptors, ligands may preferentially activate G-protein coupling or β-arrestin recruitment (92). Schmid et al. (92) reported that fentanyl promotes bias toward β-arrestin recruitment, while morphine is relatively unbiased in mouse models and cell lines. Given that β-arrestin drives MOP receptor internalization and is associated with respiratory suppression and tolerance, these findings have clinical significance and may explain the differences in ligand-mediated MOP receptor internalization (92–95). Specifically, the increased lethality of fentanyl and structurally related synthetic opioids may not be due solely to greater potency, but also due to the preferential activation of an intracellular pathway that promotes respiratory depression (92, 96).
OPRM1 Polymorphisms
Genetic variations of OPRM1, the gene encoding for MOP, have been studied in the context of vulnerabilities to SUDs, treatment response, and relapse. Whole genome sequencing has identified 3,324 OPRM1 polymorphisms, the most commonly studied of which, rs1799971 (A118G), has a global minor allele frequency of 19% (97). Located on exon 1 of OPRM1, this SNP results in an asparagine replaced by an aspartate at position 40, which is in the amino-terminus of the receptor (98, 99). In preclinical studies, the G allele was associated with lower MOP receptor expression in transfected cell lines (100–103). In [11C]carfentanil PET scans, the G allele was also associated with lower global MOP receptor expression (104) and lower expression in anterior cingulate cortex (ACC), nucleus accumbens (NAc), and thalamus compared to the common genotype (105). One proposed mechanism suggests that the amino acid substitution removes an extracellular glycosylation site, potentially interfering with the protein’s folding or incorporation into the cell membrane (101). Other studies found that the G allele results in reduced levels of MOP receptor mRNA expression, although the underlying mechanism remains unknown (103). For example, a post-mortem study of heterozygotes for A118G found the wild-type A allele had twice the mRNA expression than the G variant in cortical and pons tissue samples (103). An in vitro study of G allele-transfected cells also showed reduced mRNA and lower receptor protein levels when compared to the wild-type allele (103). Oertel et al. (106) propose that rs1799971 creates a novel methylation site that suppresses transcription of OPRM1.
Interestingly, an initial in vitro study reported increased binding affinity of β-endorphin to the variant receptor (107); though subsequent in vitro studies were unable to replicate this finding (100, 108).
Genetic Association Studies: OPRM1 and OUD
Several studies have investigated the effects of genetic variations in OPRM1 on susceptibility to SUDs, including OUD. A systematic review and meta-analysis of 13 studies of the A118G polymorphism in OUD found significant associations of the G allele with CUD and OUD in Asian populations, but not in African American, Caucasian, or Hispanic populations (109). However, a behavioral study linked the G allele with increased addiction severity in Caucasian males with OUD (110). This could be attributable to the varying prevalence of the rs1799971 minor allele across ethnicities; for example, the G allele frequency is greater in Asian populations than in Caucasians (30–40% and 11–15%, respectively), and it is less than 5% in African American populations (107, 111, 112). Another study examined four low-frequency SNPs of OPRM1 in a cohort of European Americans and African Americans; only one polymorphism, rs62638690, was associated with both cocaine and heroin addiction in European Americans; however, it did not withstand correction for multiple testing (113). This may suggest that while OPRM1 polymorphisms alter vulnerability to OUD, the effects are race- and/or ethnicity-dependent. Finally, an intron 2 polymorphism, rs9479757, was not associated with OUD in a Chinese population, but OUD patients with the minor allele were found to consume higher levels of opioids (114). Further, Xu et al. (115) found the rs9479757 minor allele associated with addiction severity among Chinese OUD patients (115). These findings are outlined in Table 1.
Additionally, the A118G polymorphism may have relevance for OUD treatment. In a mouse model of A118G, the analgesic, anxiolytic, and hyperlocomotor effects of buprenorphine were attenuated in carriers of the minor G allele (162). In a study of opioid-dependent chronic pain patients, carriers of the minor G allele required higher morphine equivalent daily doses than AA homozygotes (163). This may be attributed to reduced MOP receptor functioning in carriers of the G allele that results in an increased opioid requirement for pain management (163, 164). However, a meta-analysis of the association between rs1799971 and methadone treatment response among OUD patients was inconclusive (165).
Several studies have examined associations between OPRM1 polymorphisms and stress response, as MOP receptors help regulate stress levels via tonic inhibition of the hypothalamic–pituitary–adrenal (HPA) axis (166). Naloxone is an opioid receptor antagonist with highest affinity for MOP receptors, thus eliciting an HPA axis stress response upon binding (167). Several studies demonstrate that healthy heterozygotes of A118G have increased stress response to naloxone compared to non-G allele carriers (168–170). Given the role of stress dysregulation in vulnerability to SUDs, this provides a potential mechanism for this SNP as a risk factor for OUD (167).
The A118G SNP has also been associated with personality traits relevant to SUDs (171). Several studies assessed participants with the five-factor NEO, a personality inventory that scores in domains of “Openness to Experience, Conscientiousness, Extraversion, Agreeableness, and Neuroticism” (172). High Neuroticism, low Conscientiousness, and low Agreeableness scores are associated with SUDs (173–176). Specifically, higher scores on Neuroticism and lower scores on Conscientiousness, Agreeableness, and Extraversion have been associated with OUD (177, 178). Compared to A118 homozygotes, carriers of the G allele scored lower on the Conscientiousness factor (170), which is associated with task organization and execution, and reflects control over impulsivity (179). Moreover, Pecina et al. (105) found that G carriers had higher Neuroticism scores than non-carriers, which negatively correlated with baseline MOP receptor availability in the anterior insula and subgenual ACC as assessed with [11C]carfentanil PET. However, Hernandez-Avila et al. (180) found no association between A118G and NEO personality dimensions in healthy and substance-dependent volunteers; thus, the role of this polymorphism in moderating personality is uncertain. Love et al. (181) used [11C]carfentanil PET in a study of healthy volunteers and assessed participants with the Revised NEO Personality Inventory, which includes domains “Impulsiveness” and “Deliberation,” that have been associated with negative risk-taking, including drug use (182, 183). Participants with high Impulsivity and low Deliberation scores showed higher baseline MOP receptor availability in several brain regions including the ACC and amygdala (181). Further, in response to a pain stress challenge, subjects with high Impulsivity/low Deliberation scores demonstrated a larger reduction in MOP receptor availability from baseline compared to low Impulsivity/high Deliberation scores in regions including the orbitofrontal cortex and amygdala (181). This suggests a possible mechanism for the role of personality traits in shaping vulnerabilities to SUDs.
Molecular Imaging: MOP Receptor and OUD
Several studies have used PET imaging to investigate MOP receptor availability in OUD patients receiving MOUD. The radioligand [11C]carfentanil is widely used in PET studies as it is a highly potent MOP-selective receptor agonist (184). [18F]cyclofoxy is less frequently used as it is both a MOP receptor and KOP receptor agonist, with some preliminary evidence of MOP receptor preference (185–188).
A number of studies have examined the effects of buprenorphine, a high-affinity MOP receptor partial agonist and KOP and delta opioid (DOP) receptor antagonist (189–191) in the treatment of OUD. Using [11C]carfentanil PET imaging, Greenwald et al. (192) investigated the duration of binding of buprenorphine at MOP receptor and the corresponding effects on withdrawal in 10 OUD patients. They found that 50–60% MOP receptor occupancy by buprenorphine was required for withdrawal suppression (192). At 28 h after buprenorphine, 46% of whole-brain MOP receptors were occupied, indicating inadequate withdrawal suppression (192). This may reflect the half-life of oral buprenorphine, which ranges from 28 to 37 h (193). Plasma concentrations of buprenorphine were time-dependent and correlated with levels of MOP receptor occupancy in brain (192, 194). Considering the minor allele of rs1799971 may lower MOP receptor expression, it stands to reason that this SNP may influence the dose of buprenorphine required to achieve adequate withdrawal suppression.
In two studies, heroin-dependent patients maintained on varying doses of buprenorphine underwent several [11C]carfentanil PET scans (194, 195). Buprenorphine was shown to reduce MOP receptor availability in a dose-dependent manner, and decreased MOP receptor availability correlated with decreased heroin craving and withdrawal symptoms (194, 195). After detoxification from buprenorphine, OUD participants demonstrated higher regional binding potential of MOP receptor particularly in the inferior frontal and anterior cingulate cortex compared to healthy controls (195). Yet, an animal study found buprenorphine maintenance down-regulates MOP receptor in rat brains (77). The higher MOP receptor binding potential among OUD participants found by Zubieta et al. (195) could reflect opioid or buprenorphine induced downregulation of enkephalins and β-endorphins in brain with a consequent reduced competition for [11C]carfentanil binding to MOP.
Another study used [18F]cyclofoxy PET scans in 14 methadone-maintained patients and 14 healthy controls (185). The methadone-maintained patients demonstrated 19–32% lower cyclofoxy binding than the controls in thalamus, caudate, anterior cingulate cortex, middle temporal cortex, and the middle frontal cortex (185). The lower [18F]cyclofoxy binding in the brain of OUD participants correlated with plasma methadone levels, likely reflecting the steady-state methadone occupancy of MOP receptors (185). These findings contrast with those obtained in OUD patients treated with buprenorphine who showed much greater levels of MOP occupancy consistent with the partial agonist effects of buprenorphine as compared to the full agonist effects of methadone (192). This discrepancy could also reflect less receptor internalization associated with a partial agonist and, therefore, greater levels of receptor occupancy by the radioligand.
PET studies have also investigated the effects of A118G on MOP receptor availability in individuals with SUDs. For example, the G allele has been associated with lower baseline MOP receptor binding potential in NAc and amygdala of smokers (146–148). Thus, A118G may shape predispositions to substance abuse by affecting MOP receptor availability, which could contribute to aberrant dopaminergic signaling. A [11C]raclopride PET study of tobacco smokers found that the G allele associated with greater DA release in the right caudate and ventral pallidum in response to smoking compared to the A allele (120). This is further evidence of the association between A118G and drug reward, which may increase vulnerability to SUDs (120). Longitudinal studies are needed to clarify the link between opioid receptor availability and SUDs.
OPRK1
OPRK1 Background
OPRK1 codes for the KOP receptor, an inhibitory GPCR that is implicated in the brain’s stress or anti-reward system (196). KOP receptors are the most abundant opioid receptors in the human brain and are highly expressed in key brain regions of the stress axis such as the prefrontal cortex and amygdala (197) as well as in reward-related regions including the VTA, NAc core, dorsal striatum, and substantia nigra as seen in Figure 1 (187, 198–201). KOP receptors are coupled with calcium channels and are localized in presynaptic terminals of dopaminergic cells; activation of KOP receptors inhibits adenylyl cyclase and calcium currents, thereby inhibiting DA release (199, 202–204). Prodynorphin (PDYN) codes for the precursor to the dynorphin peptide, which is the endogenous ligand to the KOP receptor. Using a phospho-selective antibody against KOP receptors, Land et al. (16) demonstrated that both stress paradigms and CRF injections elicit dynorphin-dependent KOP receptor activation in the basolateral amygdala, NAc, and hippocampus of mice. This indicates the key role KOP receptor signaling plays in stress and dysphoria. In general, KOP receptor agonists have anxiogenic properties in humans (205, 206) while KOR antagonists demonstrate anxiolytic properties in animal models (207, 208). However, there is evidence of dose-dependent effects; in a mouse study, KOP receptor agonist, U50,488H, was anxiolytic at high doses but anxiogenic in low doses (209). KOP receptor signaling may also influence stress responses associated with relapse; for example, heroin-dependent rats treated with KOP receptor antagonists show reduced anxiety- and stress-induced reinstatement of drug-seeking behavior (210, 211).
KOP receptor signaling is also involved in an array of physiological functions such as mood modulation, pain perception, learning and memory, and behavioral response to drugs of abuse (212, 213). Within the NAc, dynorphin signaling inhibits DA release, which leads to aversive effects on mood (214). In individuals with SUDs, KOP receptor-mediated dynorphin signaling drives negative affective states during drug withdrawal (215). One [11C]raclopride PET study showed blunted DA release with a methylphenidate challenge in recently detoxified OUD patients compared to healthy controls (7). This hypodopaminergic response may be explained by dynorphin-mediated withdrawal. This is consistent with a rodent study that found chronic exposure and subsequent withdrawal from morphine led to prolonged (15 day) decreases in spontaneous dopaminergic neuron activity (216). This hypodopaminergic state may underlie dysphoria that drives compulsive drug-seeking (216).
Interestingly, post-mortem brain samples of heroin abusers showed lower levels of PDYN mRNA expression in the amygdalar nucleus of the periamygdaloid cortex compared to controls (217). Further, a post-mortem study reported elevated dynorphin levels in heroin abusers with reduced striatal PDYN mRNA expression, suggesting upregulation of PDYN mRNA translation despite reduced PDYN mRNA levels (126). These results corroborate findings of reduced PDYN mRNA expression and elevated expression of the brain stress marker, CRF, in the periamygdaloid cortex of heroin-dependent rats that were euthanized following 24 h of abstinence (217). Increased CRF may reflect the dynorphin-mediated withdrawal response in the heroin-dependent rats despite seemingly reduced PDYN expression (217).
Preclinical studies have found that KOP receptor agonists, including salvinorin A, cause KOP receptor internalization in vitro (218, 219). A [11C]GR103545 PET study in rodents found that a dose of 0.60 mg/kg of salvinorin A resulted in a prolonged decrease in [11C]GR103545 binding that persisted even after salvinorin A had cleared from the brain, consistent with KOP receptor internalization (220). This study provides insight into the neurochemical adaptations to KOP receptor agonist exposure, which may contribute to opioid tolerance (18).
OPRK1 Polymorphisms
A few OPRK1 polymorphisms have been described in the context of SUDs, although the majority of them are silent and have no effect on gene expression (221). One example is rs1051660 (G36T), a synonymous SNP in exon 2 (153). These polymorphisms may affect KOP receptor signaling indirectly by altering mRNA stability or translation (222).
PDYN polymorphisms are associated with aberrant dynorphin expression and signaling (223) that may contribute to dysphoria and relapse during opioid withdrawal (155). Intronic variants may alter gene expression via splicing mechanisms or may be in linkage disequilibrium with neighboring variants that have more direct downstream effects (146, 224). Mutations within the 3’ tail of mRNA transcripts could alter important sequences like the polyadenylate tail and may disrupt transcription termination (225), translation, and stability of mRNA (226–228). For example, rs910080, a polymorphism in the 3’ untranslated region of PDYN, is in high linkage disequilibrium with two other 3’ untranslated region SNPs, rs910079 and rs2235749; in a post-mortem analysis, this haplotype block was associated with levels of PDYN expression in the striatum (229). Other polymorphisms may alter gene expression directly. The 68-base pair variable number tandem repeat (VNTR) polymorphism, rs35286281, ranges from two to five repeats in the promoter region of PDYN, with each repeat containing one binding site for a transcription factor (230, 231). Thus, high dynorphin expression alleles (H alleles) contain three or more repeats and are associated with higher PDYN transcription and translation compared to low dynorphin expression alleles (L alleles) with one or two repeats (230).
Genetic Association Studies: OPRK1, PDYN, and OUD
There has been little consensus regarding the role of OPRK1 polymorphisms in OUD. The minor alleles of two intronic polymorphisms, rs997917 and rs6985606, were reported as risk factors for OUD in an Iranian population (154) but were not associated with OUD in a European American population (136). These conflicting findings are likely explained by ethnicity-dependent effects. Interestingly, the rs6473797 minor allele was found to be protective against OUD in a Caucasian population (130), but not in an Iranian population (154). However, rs6473797 did associate with withdrawal severity among OUD patients who underwent naloxone-precipitated withdrawal in an American population of mixed ethnicities (147). Additionally, Wang et al. (232) found that two OPRK1 haplotype blocks associated with withdrawal symptoms such as joint aches, gooseflesh skin, and yawning in Taiwanese methadone-maintained OUD patients. Lastly, rs1051660 was initially linked to OUD (152), and this finding was replicated by Gerra et al. (153) in a Caucasian Italian population.
Given the critical role of dynorphin signaling in the negative emotion states of SUDs, several studies have examined PDYN polymorphisms in the context of OUD. One polymorphism, rs910080, has been associated with OUD across a wide range of ethnicities (139, 156–158). Additionally, there is evidence of sex effects on the association between another two PDYN polymorphisms and OUD. That is, both rs1997794 and rs1022563 were found to associate with OUD among European American females, but not males (158). In a prior study of Chinese females, Clarke et al. (159) found the rs1997794 minor allele associated with OUD. Further, these two polymorphisms were not associated with OUD in a study of Malaysian males (139). Together, these findings suggest sex- and ethnicity-specific effects of the PDYN genotype on susceptibility to OUD.
Two studies found that the H allele of the PDYN VNTR polymorphism was a risk factor for OUD in Chinese populations (155, 156). It was also associated with greater instances of withdrawal and subsequent relapse among heroin-dependent Chinese patients on methadone therapy (155). However, Hashemi et al. (157) did not find an association between the PDYN genotype and OUD in an Iranian population. While evidence exists that the H allele upregulates PDYN expression (230), further research is required to understand its functional consequences as it relates to OUD.
Despite preclinical and clinical evidence of KOP receptor signaling modulating anxiety and stress response (16, 205, 206, 210, 211, 233, 234), few studies have investigated the effects of OPRK1 polymorphisms on personality or behavior. One study using the five-factor NEO found the minor allele at rs963549, in exon 3 of OPRK1, was associated with higher Neuroticism scores among participants with SUDs but not among healthy controls (235). While this SNP was found to not be a risk factor for SUDs in an Indian population (116), its effects may be ethnicity-dependent or potentially mediated by opioid use. Future studies on the functional effects of OPRK1 polymorphisms and their associated changes in neurochemistry and behavior would clarify the link between KOP receptor signaling and OUD.
One study examining the effects of the PDYN VNTR polymorphism on behavior found that the L allele is associated with disinhibited behavior as assessed with the Zuckerman Sensation Seeking Scale (236). Given that higher scores on this scale correlate with a preference toward risky behavior, this finding suggests L allele carriers are at increased risk for SUDs, contradicting findings from genetic association studies described above (155, 156) but perhaps corroborating post-mortem findings of reduced PDYN expression in individuals with OUD (217).
Molecular Imaging: KOP Receptor and OUD
At this point, no studies have used PET to examine OPRK1 polymorphisms among patients with OUD. Only recently have radiotracers been developed to target KOP receptors, including the agonist tracers [11C]GR103545 and [11C]EKAP and the antagonist tracer [11C]LY2795050. These radiotracers have been evaluated in primates (237–240) and humans (241–244).
In a [11C]LY2795050 PET study, patients with AUD showed lower KOP receptor availability in the amygdala and pallidum compared to healthy controls (245). It is possible that the reduction in KOP receptor availability helps restore dopaminergic signaling and thus alleviates the aversive effects of drinking. However, reduced [11C]LY2795050 specific binding to KOP receptors in AUD could also reflect increased competition for radiotracer binding from upregulation of dynorphin. Another [11C]LY2795050 PET study found that healthy male subjects had greater KOP receptor availability in several brain regions including ACC, frontal cortex, insula, and ventral pallidum compared to females (246). According to the “simple occupation theory,” the robustness of a drug response is directly proportional to the number of receptors occupied by the drug (247). This is consistent with the finding by Vijay et al. (246) that greater KOP receptor availability may mediate stronger responses to KOP receptor antagonists such as naltrexone treatment. Among patients with co-occurring cocaine and alcohol dependence, one study showed that naltrexone treatment reduced cocaine and alcohol use in men, but increased substance use in women (248). While sex differences in KOP receptor availability were not examined by Pettinati et al. (248), the authors suggest that receptor bioavailability and naltrexone treatment response may be sex-dependent. A potential non-neurochemical basis for the poorer treatment response in women compared to men is that women report higher rates of naltrexone-induced nausea, which results in lower medication compliance (249). However, it is important to note that other clinical studies found no sex differences of naltrexone treatment response in AUD (250, 251). Overall, these findings suggest that KOP receptor availability is associated with alcohol use and could potentially mediate the efficacy of KOP-targeted pharmacotherapies for AUD (245). Given the high comorbidity between AUD and OUD (252–254), these findings might have implications for opioid-antagonist treatment response in OUD.
OPRD1
OPRD1 Background
OPRD1 codes for DOP receptors, which are also involved in the negative affect and withdrawal stage of addiction, albeit with inverse effects than KOP receptors. Specifically, greater DOP receptor signaling leads to improvements in negative emotional states (255). DOP receptor agonists have demonstrated antidepressant and anxiolytic effects in rodent models (256, 257). DOP receptors are highly expressed in cortical and limbic areas such as the hippocampus and amygdala, as well as basal ganglia and hypothalamus (258–260). DOP receptors are located on presynaptic terminals of GABAergic interneurons and have region-specific effects on cAMP production (261). While striatal DOP receptor activation is inhibitory and results in increased extracellular dopamine (262), DOP receptors located in the olfactory bulb, medial prefrontal cortex, and primary cultures of hippocampal neurons stimulate cAMP production thereby inhibiting dopamine release (263–265).
Studies suggest DOP receptors modulate the rewarding effects of drugs of abuse. Le Merrer et al. (197) report DOP receptor knockout has no effect on morphine self-administration but does impair place conditioning in mice. In another rodent study, DOP receptor knockout resulted in reduced morphine reward and tolerance (266). Further, DOP receptor antagonists block sensitization to conditioned rewarding effects of opioids (267), whereas agonists enhance conditioned place preference to morphine (268). In a mouse model of OUD, DOP receptor knockout was associated with increased anhedonia and dysphoria during heroin abstinence compared to the wild-type genotype (269). Thus, OPRD1 polymorphisms that alter DOP receptor signaling may influence opioid withdrawal-associated stress response and relapse.
OPRD1 Polymorphisms
Several polymorphisms of OPRD1 have been studied in the context of SUDs. One, rs1042114 (G80T), results in an amino acid substitution from cysteine to phenylalanine in the N-terminus of the DOP receptor, and is proposed to disrupt DOP receptor maturation, leading to increased internalization of the receptor compared to wild type (270). The coding-region variant rs2234918 (T921C) is a synonymous polymorphism, that is, it does not cause a change in the coding amino acid, and has conflicting evidence for a role in OUD. Finally, rs569356, located in the promoter region, has been implicated in altered OPRD1 expression; Zhang et al. (271) found the G allele increased OPRD1 transcription in transfected cell lines. Few other OPRD1 polymorphisms have been described in terms of their functional effects; however, several have been assessed in genetic association studies.
Genetic Association Studies: OPRD1 and OUD
Two polymorphisms in the coding region of OPRD1 have been associated with OUD. The rs1042114 polymorphism has been found to be a risk factor for OUD in Malaysian males (139) and in Caucasian populations (136, 140). However, Nelson et al. (137) did not replicate these findings in Australian OUD patients. Rs2234918, a synonymous OPRD1 polymorphism, has also been studied in OUD with conflicting findings. The minor C allele of this polymorphism was initially reported as a risk factor for OUD in a German (142) and Chinese population (272). However, several studies have failed to replicate this association (130, 136, 140, 143) including a study that examined a German population but used a family-based association approach to control for population stratification (144). Thus, it is uncertain what role, if any, these OPRD1 polymorphisms play in increasing vulnerability to OUD.
Several polymorphisms in intron 1 of OPRD1 have been studied in OUD, although their functional effects remain largely unknown. Two studies found an association between rs2236861 and OUD among Caucasian patients (137, 150). Levran et al. (130) also found that the rs2236861 minor allele increases the risk of heroin dependence; however, the association did not survive multiple testing, perhaps due to a small sample size. Another intron 1 polymorphism, rs2236857, was associated with OUD in Iranian- and European-descent populations (130, 137, 146). However, Zhang et al. (136) were unable to replicate this association in a study of European Americans. Interestingly, among Chinese OUD patients, carriers of the rs2236857 minor allele were found to have higher subjective stress responses than non-carriers as assessed with the Life Event Questionnaire (272). This suggests that OPRD1 polymorphisms may disrupt stress responses that increase addiction vulnerabilities. The minor allele of rs581111, located in intron 1, has also been reported as a risk factor for OUD among Australians (137) and African Americans but not European Americans (140). Additionally, the minor allele of rs581111 has been associated with poor response to buprenorphine treatment among Caucasian females, but not males, suggesting ethnicity- and sex-dependent influences on genetic associations (145). Lastly, the minor allele of an OPRD1 intron 1 polymorphism, rs3766951, was reported as a risk factor for OUD in Caucasian populations (130, 137).
In addition, several studies have investigated the effects of OPRD1 polymorphisms on treatment outcomes in OUD. For example, the major allele of rs678849 has been associated with higher relapse rates among African American OUD patients undergoing buprenorphine treatment, as indicated by positive opioid urine tests (148, 149). Interestingly, the major allele was initially associated with lower relapse rates among African American OUD patients on methadone treatment (148), but this association was not replicated (149). Jones et al. (147) reported an association between rs678849 and abstinence-induced opioid withdrawal severity; however, it did not withstand a multivariate analysis. While the mechanism of action is unknown, these findings suggest that rs678849 may affect OUD treatment outcomes by potentially mediating withdrawal symptoms.
Several other OPRD1 polymorphisms have been studied in association with OUD with conflicting results as seen in Table 1.
OPRD1 polymorphisms have also been associated with behaviors related to the negative affect and withdrawal stage of OUD. In one study of Pakistani OUD patients, the minor G allele of rs569356 was strongly associated with increased serum cortisol levels, a marker of stress response (273). Given the preclinical evidence that this minor allele may increase OPRD1 transcription (271), the minor G allele may affect DOP receptor expression and stress responses that could contribute to OUD. While Zhang et al. (136) found a nominally significant association between rs569356 and OUD in a European American population, no significant association was found in Australian and Pakistani populations (137, 273).
Molecular Imaging: DOP Receptor and OUD
No PET studies have examined neurochemical differences between carriers of OPRD1 polymorphisms in OUD. The only DOP-selective radiotracer that has been developed for PET imaging in humans is N1’-([11C]methyl)naltrindole ([11C]MeNTI) (274).
PET studies investigating DOP receptor availability in healthy controls and AUD patients may provide insight into the functional effects of OPRD1 polymorphisms in OUD. One [11C]MeNTl PET study found that patients with AUD had slightly greater DOP receptor availability compared to healthy controls in the cingulate, amygdala, insula, ventral striatum, putamen, caudate nucleus, globus pallidus, and thalamus; however, group differences did not reach statistical significance (275). Within the AUD group, DOP receptor availability in the caudate showed a positive association with recent alcohol drinking (275). However, Weerts et al. (275) did not report associations between DOP receptor availability and other behavioral measures of alcohol dependence or withdrawal. Another PET study in abstinent AUD patients demonstrated that while naltrexone completely blocked MOP receptor radioligand binding, it only partially blocked [11C]MeNTl binding and there was high interindividual variability in DOP receptor blockade (276). These findings could underlie interindividual differences in responses to naltrexone treatment in AUD that could translate to naltrexone treatment responses in OUD.
Additionally, one [11C]MeNTl PET study found a negative correlation between mesolimbic DOP receptor availability and total cortisol output over a 4-h period following naloxone in healthy controls, but not in recently abstinent AUD patients (277). Given that endogenous DOP receptor signaling improves negative emotional states (278), the dissociation of DOP receptor availability from naloxone-induced cortisol response in AUD may suggest that chronic alcohol abuse disrupts DOP-mediated stress signaling during alcohol withdrawal. Whether this is the case for OUD remains to be determined. Notably however, Lutz et al. (269) reported that DOP receptor signaling ameliorates opioid withdrawal in rodents, so together, these findings may suggest a shared mechanism for negative emotional states in opioid and alcohol withdrawal.
OPRL1
OPRL1 Background
The nociceptin opioid peptide (NOP) receptor is an inhibitory GPCR encoded by the Opioid Receptor-Like 1 gene (OPRL1) that has MOP, KOP, and DOP receptor structure homology and similar signaling cascades (279). However, the NOP receptor is pharmacologically distinct from classical opioid receptors. The NOP receptor is activated by nociceptin, and its effects are not blocked by the universal opioid antagonist naloxone (280, 281). NOP receptors are distributed throughout the amygdala, hippocampus, thalamus, and cortical processing areas (282) and have roles in both analgesia and hyperalgesia [reviewed in (283) and (284)]. NOP receptor signaling is also involved in processes including stress, anxiety, depression, cognition, and addiction (285–289).
Given the distribution of NOP receptors along the limbic region (290), it follows that NOP signaling is tied to stress signaling. For example, central injections of nociceptin in rats result in increased plasma stress hormone levels, reflecting activation of the HPA axis (291). However, there is also evidence that NOP receptors in extrahypothalamic brain regions exert anti-stress effects. For example, nociceptin injections in the central nucleus of the amygdala reduce anxiety behaviors in rodents exposed to restraint stress (292). Further, body restraint stress upregulates NOP receptor mRNA in the central nucleus of the amygdala and basolateral amygdala (292). In an electrophysiological study, nociceptin blocked CRF-induced GABAergic transmission in slices from the central nucleus of the amygdala; these effects were more pronounced in neurons from ethanol-dependent rodents (293). Additionally, nociceptin injections in the bed nucleus of the stria terminalis block CRF-induced anxiety behaviors in rodents (294, 295). Thus, the role of NOP receptors in stress is likely complex and may be relevant in OUD, particularly due to the high co-occurrence of anxiety and SUDs [reviewed in (296)].
NOP receptor signaling also seems to have an anti-reward effect. In microdialysis studies, nociceptin administration was found to decrease extracellular DA levels in the NAc of anesthetized mice (297) and to decrease morphine-induced DA release in the NAc of rats (298). Further, in several rodent studies, NOP receptor agonists reduced conditioned place preference to alcohol, amphetamines, cocaine, and morphine, suggesting NOP receptor signaling may reduce the rewarding effects of these substances (299–304). However, Walker et al. (305) found nociceptin administration failed to reduce heroin self-administration in rodents. There is also preliminary evidence that the NOP receptor antagonist, LY2940094, could be efficacious in treating AUD in rodents and humans, perhaps by blocking stress-induced relapse (306, 307). While an initial post-mortem analysis demonstrated individuals with AUD had lower OPRL1 expression in the central amygdala compared to controls (308), no difference in OPRL1 expression was detected in another post-mortem study in individuals with SUDs including AUD (309). Thus, the NOP receptor is likely implicated in substance abuse and poses a potential therapeutic target, but further research is required to clarify its roles in reward and stress-related behaviors.
OPRL1 Polymorphisms
The functional effects of several OPRL1 polymorphisms have been studied. For example, two adjacent SNPs in intron 1, rs6512305 and rs6090043, are in high linkage disequilibrium and there is evidence that variants in rs6090043 may alter transcription factor binding sites, which could affect OPRL1 gene expression (161). Further, the minor G allele at rs6090041, another intron 1 variant, and the minor C allele at rs6090043 provide additional transcription factor binding sites that could result in increased OPRL1 transcription and NOP receptor availability (161). Given that NOP receptor signaling has been implicated in decreasing drug reward, there may be a role of OPRL1 polymorphisms in susceptibility to SUDs.
Genetic Association Studies: OPRL1 and OUD
Xuei et al. (160) assessed correlations between SUDs and polymorphisms in OPRL1 as well as in the prepronociceptin gene (PNOC), which encodes the NOP receptor precursor, in a European American population; rs6512305 and rs6090043 were nominally associated with opioid dependence; however, no SNPs proved significant (160). Briant et al. (161) found that minor alleles at rs6090043 and rs6090041 were risk factors for OUD among Caucasians but not African Americans. One haplotype (AT) of these variants was found to be a risk factor in both Caucasians and African Americans, while another haplotype (GC) was a risk factor in Caucasians only (161). While there is preliminary evidence that OPRL1 may influence vulnerability to OUD, further analysis is required to determine the potential ethnicity-dependent effects.
Molecular Imaging: NOP Receptor and OUD
NOP receptor antagonist PET radioligands have been developed; [11C]NOP-1A has been tested in humans (290, 310, 311) and [18F]MK-0911 has been tested in rhesus monkeys (312). To date, no molecular imaging of NOP has been done in participants with OUD; however, studies of other SUDs may provide insight. Using [11C]NOP-1A, Narendran et al. (313) found no difference in NOP receptor availability between healthy controls and recently abstinent AUD subjects, nor did NOP receptor availability correlate with clinical measures of addiction severity. This conflicts with preclinical evidence that NOP receptor signaling is involved with AUD (289, 299, 300, 308). However, the subjects with AUD in this study were abstinent for 16 to 54 days before the PET scan, and there is preclinical evidence that prolonged abstinence may recover NOP receptor levels in rats (313, 314). In another PET study, recently abstinent CUD participants demonstrated a significant increase in [11C]NOP-1A distribution volume notably in the midbrain, ventral striatum, and cerebellum compared to healthy controls (315). This increased NOP receptor availability may reflect a compensatory response to increased CRF transmission or decreased endogenous nociceptin associated with CUD (315). Further studies are required to evaluate NOP in OUD, for while studies in CUD have shown upregulation in brain, studies in AUD showed no differences (313), which suggests that there might be differences between SUDs. Also, research is needed to clarify changes during the different stages of the addiction cycle and to assess if there is recovery of NOP receptor availability with treatment.
The Dopamine System
DRD2
DRD2 Background
The gene DRD2 codes for D2R, an inhibitory GPCR distributed throughout the brain. Expression of D2R is concentrated in the basal ganglia nuclei, including the caudate, putamen, NAc, substantia nigra, and VTA, as shown in Figure 1 (316). As such, D2R signaling plays an important role in cognition, reward, motivation, and drug addiction, including OUD (317, 318). MOP receptors are expressed on DA neurons in the reward pathway; thus, with opioid use, MOP receptor binding leads to a release of DA, which then binds striatal D2Rs, leading to a decrease in intracellular cAMP production (69, 319). This D2R signaling inhibits the indirect ventral striatal pathway, which is connected to punishment (320).
Ankyrin Repeat and Kinase Domain Containing 1 (ANKK1) is a gene directly downstream of DRD2 on chromosome 11 that expresses a serine/threonine kinase (321). The protein product of ANKK1 upregulates the expression of the transcription factor NF-κB (322). Increased NF-κB expression results in increased DRD2 transcription (323).
Several studies have shown that OUD is associated with a disruption of the mesolimbic dopaminergic pathway, which underlies the behavioral response to opioids (4). Koob and Volkow (4) suggest that D2Rs contribute to drug seeking behaviors, but not drug reward directly (324, 325). A conditioned place preference study of DRD2-null mice demonstrated that D2Rs are in part responsible for the reinforcing nature of morphine (326).
Lower D2R levels observed in SUDs may reflect a homeostatic downregulation of D2R after excessive drug use (29), and some evidence exists that D2R levels increase after pronounced abstinence (327). Alternatively, lower D2R availability may be an inherent risk factor for drug abuse, even before the initiation of drug taking (328, 329).
DRD2 Polymorphisms
A wide range of DRD2/ANKK1 polymorphisms have been studied in the context of SUDs. One of the most well studied of these SNPs is TaqIA, located on exon 8 of ANKK1, adjacent to DRD2 (321). Many studies have supported the role of TaqIA in addictive behaviors including various SUDs, obesity, and pathological gambling (330–333). Thus, the TaqIA1 variant, which alters ANKK1 substrate binding specificity, could lead to decreased D2R expression downstream (321). Indeed, [11C]raclopride and [11C]NMB PET studies have shown that minor alleles of ANKK1TaqIA and TaqIB, a linked DRD2 SNP, are associated with low D2R availability in healthy controls (334–336). However, TaqIA is in linkage disequilibrium with several functional DRD2 polymorphisms (337); thus, it is unclear if reduced D2R expression is associated with TaqIA directly.
Lesser studied DRD2 variants may also contribute to OUD via a diminution of D2R expression (338). SNPs in the 5’ untranslated region of DRD2, including rs1799732, an insertion/deletion (Ins/Del) variant at position -141, have been shown to cause decreased promoter strength in an in vitro -141C Del luciferase construct (339). While one [11C]FLB-457 PET study found no association between rs1799732 and extrastriatal D2R in healthy volunteers (340), one [11C]raclopride PET study demonstrated higher striatal D2R availability in those with the combined minor variants of rs1799732, Ins/Del and Del/Del, compared to Ins/Ins (334). Until more studies are performed, the role of rs1799732 in D2R expression cannot be concluded.
Other DRD2 polymorphisms produce splicing errors of the DRD2 gene, resulting in altered D2R expression (341). For example, the minor allele of rs1076560, located in intron 6, is associated with a decreased ratio of short form D2 receptors (D2S) to long form receptors (D2L) (342). Preclinical studies have demonstrated that D2L knock-out mice have a loss of morphine preference in a conditioned place preference paradigm (343). Thus, this altered D2S/D2L ratio could help elucidate the mechanism of this SNP-OUD relationship. [123I]IBZM SPECT imaging revealed that in healthy volunteers, minor T allele carriers of this SNP showed lower levels of striatal D2R availability compared to G/G (344). However, another [123I]IBZM SPECT study in healthy volunteers did not replicate this finding (345). These findings may implicate DRD2/ANKK1 polymorphisms in the lower D2R levels observed in individuals with OUD (6).
Genetic Association Studies: DRD2 and OUD
Several polymorphisms in DRD2/ANKK1 have been suggested to predispose OUD, as outlined in Table 2. Indeed, a recent meta-analysis across 11 studies, with a total sample of 4,529 OUD patients and 4,168 healthy controls, found that the TaqIA1 allele is a risk factor for (OUD) (354). Further, several other minor alleles of TaqIA and TaqIB are more frequent among OUD patients compared to healthy controls (353, 355, 356, 360, 351).
There is less robust evidence for other DRD2 polymorphisms in OUD. For example, despite preclinical evidence that rs1076560 may alter D2R expression, genetic association studies between rs1076560 and OUD have been inconsistent (44, 341, 348, 352, 354). In contrast, while the role of rs1799732 on D2R expression is uncertain, subjects with the minor variant have shown to be at higher risk for OUD in the Jordanian Arabic population (352).
The extent to which DRD2 polymorphisms affect the response to MOUD in patients with OUD is inconsistent across studies. Lawford et al. (372) first reported that the TaqIA1 allele was associated with poorer treatment outcomes among Caucasian patients on methadone maintenance therapy. Since then, no group has replicated these findings in Caucasian populations (44, 134, 358, 361). Similarly, no association was found between TaqIB and methadone maintenance therapy response nor TaqIA and buprenorphine maintenance therapy response (44, 272). However, Crettol et al. (134) did report an association with rs6277 and patients’ response to methadone maintenance therapy; patients with the major CC genotype were more likely to abuse illicit opioids on methadone therapy than those with CT or TT genotype. Interestingly, in two [11C]raclopride PET studies, the major C allele of rs6277 was associated with lower striatal D2R availability in healthy volunteers (373, 374), while another [11C]FLB457 PET study found the C allele predicted high extrastriatal D2R availability across the cortex and hypothalamus (340). However, several studies found no association between rs6277 and OUD (44, 134). Further, Doehring et al. (44) found no relationship between rs6277 and methadone maintenance therapy response. Instead, this group found that minor allele carriers of a different polymorphism, rs6275, required greater methadone doses than non-carriers and took longer to reach their maximum methadone dose (44). Thus, genetic studies suggest a role of DRD2 polymorphisms in treatment response in OUD; however, they remain inconsistent and difficult to replicate.
Several studies have investigated the role of DRD2 variants on behaviors associated with OUD. The tridimensional personality questionnaire scores personality on harm avoidance, novelty seeking, and reward dependence (375). These scores are used to calculate a borderline index using the equation: borderline index = harm avoidance + novelty seeking − reward dependence (376). Borderline index reflects borderline personality trait, characterized by a fear of abandonment, self-injurious behaviors, and emotional dysregulation (376, 377) (DSM-5). A recent study found that OUD patients had higher harm avoidance and novelty seeking scores and lower reward dependence scores, and thus a higher borderline index, than healthy volunteers (356). Further, Huang et al. (272) found that borderline index scores are inversely correlated with methadone dose, indicating the relevance of borderline index score in OUD treatment. These personality scores have not shown associations with TaqIA or TaqIB polymorphisms (272, 356). However, the -141C Del polymorphism (rs1799732) is associated with higher harm avoidance scores among OUD patients (356). In contrast, Gerra et al. (377) found that OUD patients had lower harm avoidance scores compared to CUD patients and healthy volunteers. However, this study reported that both CUD and OUD patients had higher novelty seeking scores and lower reward dependence scores than healthy volunteers (378). Therefore, this difference in harm avoidance could be rooted in genetic differences between the groups, as -141C Del is associated with higher harm avoidance scores in OUD, though Gerra et al. (378) did not report the genetic composition of their cohort (356).
Molecular Imaging: D2R and OUD
[11C]raclopride and [123I]IBZM are widely used radiolabeled D2R antagonists differing via regioselectivity used to study D2R distribution, with additional affinity to D3Rs (D2-like inhibitory receptors) (379–381). [11C]NMB is another radiotracer used to study D2R availability with higher affinity for D2Rs over D3Rs than [11C]raclopride and [123I]IBZM (382, 383). Lastly, [11C]FLB-457 is a high-affinity radioligand that targets extrastriatal D2Rs and D3Rs (384).
In contrast to other SUDs, less is certain about D2R availability in OUD. In one [11C]raclopride PET study, OUD participants showed lower D2R availability compared to healthy controls (6). In this study, all OUD patients actively used heroin and most, but not all, were also on methadone therapy (6). In another [11C]raclopride PET study, recently detoxed OUD patients showed lower D2R availability than healthy controls (7). These patients also demonstrated lower levels of DA release in response to a methylphenidate challenge in comparison to healthy controls (7). In a [123I]IBZM SPECT study, the OUD patients were abstinent without maintenance therapy anywhere from 1 to 24 weeks (318). Zijlstra et al. (318) observed a negative correlation in length of opioid use history with striatal D2R availability. In contrast, two [11C]raclopride studies observed no differences in D2R availability between OUD patients receiving methadone therapy and healthy controls (19, 20). These findings suggest the potential therapeutic benefit of MOUD in restoring neurochemical imbalances resulting from substance abuse. These results demand further investigation into the relationship between OUD and D2R availability, particularly in the context of MOUD.
DRD1
DRD1 Background
The D1R is the most abundant DA receptor in the brain (380). Coded by DRD1, this excitatory GPCR is widespread, but most densely expressed in the dorsal striatum, hippocampus, amygdala, and neocortex, as illustrated in Figure 1 (385–388). D1Rs influence learning and memory via association with N-methyl-D-aspartate (NMDA)-mediated long-term potentiation as well as impact D2R-mediated events and regulate addiction-associated behaviors such as impulsivity (389–396). D1Rs are important mediators of several reward-related processes and there is evidence that D1Rs are required and sufficient for drug reward and conditioning (397, 398).
D1R function is relevant in OUD because DA release triggered by opioid-induced MOP receptor activation indirectly stimulates D1Rs and associated reward circuitry (69). While one post-mortem study showed lower D1R mRNA levels in the putamen and NAc shell in OUD subjects relative to controls (347), another postmortem analysis showed higher D1R mRNA and protein expression in VTA, NAc, and amygdala in the brains of opioid abusers compared to controls (399). This difference may be attributed to the difference in populations studied. Where Sadat-Shirazi et al. (399) studied patients who exclusively abused opioids, Jacobs et al. (347) included polysubstance users.
In addition, pharmacological manipulations of D1Rs in preclinical models of OUD demonstrate alterations in behaviors associated with dependence and withdrawal. For example, infusion of D1R agonist SKF 38393 into the NAc enhances, while antagonist SCH 23390 blunts, conditioned place preference in morphine-addicted rats (400). Additionally, infusions of SCH 23390 into the NAc core reduced cue-induced heroin-seeking in dependent rats (401). Furthermore, D1R agonist SKF 82958 relieved naloxone-precipitated withdrawal symptoms in morphine-dependent rats (402). These findings highlight the importance of D1Rs in OUD and correspond with other SUD models. For example, SCH 23390 infusion blocks reinstatement of cocaine-seeking in rats, while D1R agonist SKF 81297 reinstates cocaine-seeking (403, 404). In addition to pharmacological D1R blockade, D1R knock-out mice fail to self-administer cocaine (397). In models of AUD, NAc shell infusions of SCH 23390 blunt, while infusions of D1R agonist A-77636 enhance, ethanol self-administration in alcohol-preferring rats (405).
D1 and MOP receptors directly colocalize into hetero-oligomers in the rat cortex and striatum (including accumbens nucleus), regions of importance in reward and locomotor activity. Together, they promote locomotor sensitization in rats chronically treated with morphine, suggesting this association may be involved in the long-term neuronal changes associated with addiction (406, 407).
DRD1 Polymorphisms
While less attention has been given to variations in DRD1 than DRD2/ANKK1, there are several functional polymorphisms that have been studied in the context of SUDs. One study demonstrated that rs5326A, located in the 5’ untranslated region, correlated with decreased DRD1 promoter strength in an in vitro luciferase model (408). Other DRD1 polymorphisms may increase vulnerability to OUD by interacting with the glutamatergic system in the brain. Homer scaffold protein 1 (HOMER1) encodes HOMER1, a postsynaptic protein that facilitates glutamatergic transmission (409). Excitatory glutamatergic signaling has been shown to underlie the persistent compulsion to use drugs, suggesting SNPs disrupting this gene interaction may be relevant in OUD (410). In a post-mortem analysis of Caucasian samples, the DRD1 polymorphism rs265973 associated with HOMER1 expression in the striatum (347). Interestingly, the minor T allele associated with higher levels of striatal HOMER1 mRNA among the OUD cohort, but associated with lower levels of striatal HOMER1 mRNA in the control cohort (347). Thus, it is possible HOMER1-associated genetic variants disrupt glutamatergic and dopaminergic signaling and contribute to OUD.
Genetic Association Studies: DRD1 and OUD
Preliminary findings suggest a role of DRD1 SNPs in OUD, as outlined in Table 2. For example, Liu et al. (411) found that two SNPs located in the 5’ untranslated region of DRD1, major allele rs265981G and minor allele rs5326A, associated with OUD in a Han Chinese population. Levran et al. (348, 349) also found a trend toward an association between rs5326A and OUD in an African American sample. However, other groups were unable to replicate these findings (346, 350). Jacobs et al. (347) found a nominally significant association between DRD1 SNP rs265973 and OUD among Caucasians, but not African Americans. This provides further evidence of an association between HOMER1 and OUD, perhaps with ethnicity-dependent effects.
Several studies demonstrate that DRD1 variants associate with the duration of transition from the first use to dependence of opioids (346, 350). The duration of transition from the first use to dependence is of clinical significance; patients with a more rapid transition to dependence have poorer treatment outcomes and more severe SUDs (412, 413). Zhu et al. (346) found that the minor alleles of rs686 and rs4532 associated with a longer transition period. Peng et al. (350) were unable to replicate the rs4532 association, but found that homozygotes for the major alleles of rs5326 and rs6882300 had an accelerated transition to OUD. Interestingly, while these SNPS associated with the transition from first use to dependence, neither study found that they were associated with increased risk for OUD (346, 350).
DRD1 variants have also been implicated in subjective ratings of pleasure in response to opioids, both upon first use and after dependence (346). Typically, the pleasurable feeling associated with opioids increases with duration of use: most patients report a negative response upon their first use and a euphoric response after dependence (133, 346). This suggests that chronic opioid use induces changes to reward-related circuitry. One potential mechanism is through D1R-mediated phosphorylation of NMDA, contributing to long-term potentiation (414). DRD1 variants have been associated with this reward sensitization process in a Han Chinese population (346). This study revealed that DRD1 SNPs that modulate the subjective response to opioids upon first use are distinct from those that do so after dependence. Specifically, the minor alleles of rs5326, rs10063995, and rs10078866 are associated with a non-pleasurable first use of opioids, but are not associated with the subjective response after dependence. Conversely, the minor variants of rs686 and rs4532 are associated with less pleasurable responses to opioids after dependence, but are not associated with the initial response (346). Findings from a rat study indicate that there is a reward-switching mechanism in opioid response within the basolateral amygdala in which D1R signaling is associated with reward upon first use and D2R signaling with reward after dependence (415). Thus, it is possible that rs686 and rs4532 associate with less pleasurable opioid responses after dependence by modulating D2R activity.
Molecular Imaging: D1R and OUD
No molecular imaging studies have yet assessed D1R availability in OUD or in DRD1 polymorphism carriers. Few studies have examined the relationship between other SUDs and D1R levels. [11C]NNC 112 and [11C]SCH 23390 are radiolabeled D1R antagonists that differentially distribute throughout the brain; however, both display high affinity in the striatum and extrastriatal regions (416–418). In one [11C]NNC 112 study, D1R availability in CUD patients was not significantly different than in healthy controls (419). In contrast, studies utilizing [11C]SCH 23390 PET reveal individuals with tobacco use disorder have lower D1R availability than healthy controls (420, 421). These limited findings highlight the need for increased investigation into D1R availability in addiction.
DAT1
DAT1 Background
DAT are plasma membrane proteins essential for the clearance of DA from the synapse; they play a critical role in regulating DA neurotransmission, especially in the striatum (422–426). DAT harness the electrochemical gradient to transport two sodium ions with a DA molecule into the cell, thus regulating extracellular DA concentrations (423). DAT are coded by DAT1, a gene widely studied for its role in substance abuse (427).
DAT1 Polymorphisms
The most studied polymorphisms of DAT1 are VNTRs in the 3’ untranslated region, which may affect DAT expression (428–431). The most common variants are those with 9 or 10 repeats of the 40 base pair sequence (432) and multiple molecular imaging studies have investigated their functional effects. In several [123I]β-CIT SPECT studies, 9-repeat VTNR carriers demonstrated higher striatal DAT availability than the 10-repeat homozygotes (363–365). In contrast, two [123I]β-CIT SPECT studies found those homozygous for the 10-repeat allele had higher striatal DAT density compared to non-10-repeat carriers (370, 371). Finally, Martinez et al. (366) and Lynch et al. (367) found no effect of VNTR polymorphisms on striatal DAT expression in a [123I]β-CIT SPECT and [99mTc]TRODAT-1 study, respectively. Lastly, Guindalini et al. (433) found that the rare 6-repeat VNTR genotype reduced DAT1 expression in vitro, particularly when cocaine was added to the culture. However, the effects of the 6-repeat VNTR polymorphism on DAT1 availability has not been assessed in vivo with PET methodology. Thus, further research is required to determine these polymorphisms’ functional effects on DAT expression and availability.
Genetic Association Studies: DAT1 and OUD
Genetic association studies of DAT1 and OUD have yielded inconsistent results. While Galeeva et al. (362) found an association between 9-repeat VNTR allele and OUD in an ethnic Russian and Tartar male population, later studies in Han Chinese populations did not observe any association (360, 368). Ornoy et al. (369) examined the heritability of DAT1 ADHD risk alleles in Sephardic and Ashkenazi Jewish heroin-dependent individuals and their children. They found that mothers with OUD were more likely to be carriers of the DAT1 10-repeat allele than mothers without OUD. This association was not seen in fathers and was not explained by prevalence of ADHD among mothers with the polymorphism. Further, the children of heroin-dependent parents were more likely to inherit the 10-repeat allele than children of healthy volunteers (369). However, it is unclear how these VNTR polymorphisms impact DAT availability and thus vulnerability to OUD, as molecular imaging studies have conflicting results (363–367, 370, 371).
Polymorphisms in DAT1 have been associated with other SUDs, which may provide insight into their functional effects on DA signaling in addiction. DAT1 VNTR has been associated with OUD (362) as well as AUD in Western European and Japanese populations (47, 434). A meta-analysis also found that the 9-repeat VNTR was associated with increased withdrawal severity in AUD (435). The 6-repeat VNTR genotype was found to be a risk factor for CUD, but this variant has not yet been studied in OUD (45). Thus, it seems that DAT1 VNTR polymorphisms may affect DAT expression and contribute to SUDs.
Evidence suggests that the number of VNTR in patients with OUD influences their response to treatment. In each study, a “poor” treatment outcome indicates continued heroin use or treatment drop-out, whereas a “successful” outcome indicates cessation of illicit opioid use. In patients receiving buprenorphine therapy, carriers of the 10-repeat VNTR allele had poor outcomes more often than successful outcomes (436). Conversely, 6-, 7-, and 11-repeat VNTR allele carriers had successful outcomes in response to buprenorphine therapy more often than not (436). Gerra et al. (436) suggest that these variations in DAT1 may modulate buprenorphine-associated DA transmission and thus affect treatment success. In a study of both oral and implanted naltrexone therapy, Krupitsky et al. (437) found that OUD patients with the 9-repeat VNTR allele had poor outcomes more often than successful ones on both forms of naltrexone. Thus, genotyping DAT1 VNTR could be useful in OUD therapy selection.
While van Gestel et al. (438) reported an association between DAT1 VNTR polymorphisms and novelty seeking, a personality trait associated with SUDs (439), other studies have failed to replicate this finding (440, 441).
Molecular Imaging: DAT and OUD
Several molecular imaging studies have assessed DAT availability in SUDs utilizing DAT-sensitive tracers including [99mTc]TRODAT-1, [123I]β-CIT, [11C]WIN 35,428, [11C]cocaine, and [11C]CFT. There is evidence from molecular imaging studies that DAT availability is altered in SUDs. For example, CUD is associated with higher striatal DAT concentrations compared to healthy controls (54, 55), while methamphetamine-dependent individuals demonstrate lower striatal DAT availability compared to healthy controls (51, 57, 58). Alcohol and tobacco dependence have also been associated with lower striatal DAT levels (59, 60–62); however, other studies have observed no association between DAT levels and alcohol and tobacco dependency (22, 442). Although varied, these results overall suggest that DAT plays a role in SUDs.
PET and SPECT studies suggest that OUD is associated with decreased DAT availability. Chronic heroin users, detoxed abstainers, and methadone-maintained patients all present lower striatal DAT levels than healthy controls (48–53). A [99mTc]TRODAT-1 SPECT study comparing DAT concentrations between recently detoxed heroin-dependent patients and recently detoxed methamphetamine-dependent patients showed that both had lower striatal DAT availability than healthy controls and had no differences between them (51). In contrast, Cosgrove et al. (443) utilizing [123I]β-CIT SPECT imaging, reported no differences in striatal DAT levels between heroin users and healthy controls, though they acknowledged the limitations of their small sample sizes (443).
DAT availability may also vary based on the use of MOUD. For example, one [11C]CFT PET study reported methadone-maintained OUD patients showed lower DAT availability in the bilateral putamen than abstinent OUD patients, with both presenting lower striatal DAT availability compared to healthy controls (49). Further, while methadone-maintained patients showed lower DAT availability in caudate and putamen compared to controls, abstinent OUD patients showed lower DAT availability in the caudate only, suggesting that abstinence from opioids may partially recover DAT availability (49). However, a [99mTc]TRODAT-1 SPECT study found similar striatal DAT availability between methadone-maintained and abstinent OUD patients (50). This discrepancy may be due to methodological differences; in one study, patients were at least 6 months abstinent (49), while in the other, patients were abstinent for only 3 months or less (50). In a within-subjects [99mTc]TRODAT-1 SPECT study, Liu et al. (53) observed a 14–17% increase in DAT levels in the caudate and putamen of 64 heroin-dependent patients after 6 months of treatment with traditional Chinese Jitai tablets, an herbal remedy associated with withdrawal mitigation. No significant increase in DAT levels was observed in the placebo-treated group. However, even among the medication group, DAT availability was not restored to that of healthy control levels (53). Thus, further studies are required to determine the effects of MOUD compared to sustained abstinence on DAT availability.
Conclusion
Preclinical and clinical studies have demonstrated the importance of the opioid and DA systems in SUDs, including OUD. Polymorphisms within these systems have functional consequences that may influence a number of modalities in addiction, including vulnerabilities, addiction severity, treatment response, and relapse rates. PET and SPECT methodology allow for the study of these receptor systems in both healthy and substance-dependent populations and provide insight into the neurobiology of OUD.
Within the opioid system, the MOP receptor has been most closely studied in the context of OUD. The minor allele of the OPRM1 rs1799971 SNP has been widely linked to a reduction in MOP receptor availability (100–105). The implications of this in OUD, however, remain elusive; findings from genetic association studies are varied and seem largely ethnicity-dependent (109). The KOP and NOP receptors have also been studied in relation to OUD; both play important roles in the dysphoric effects of drug abuse seen during withdrawal, including modulating activation of the HPA axis (16, 291, 294, 295, 444). A number of polymorphisms in OPRK1 have been associated with OUD and opioid withdrawal severity (147, 155, 156, 232). Similarly, VNTR polymorphisms in PDYN have been correlated with opioid withdrawal, suggesting the importance of dynorphin-KOP receptor signaling system in the mediation of stress-induced withdrawal and compulsive drug-seeking (155). Lastly, genetic variants in both PDYN and OPRL1 have been associated with personality traits and behaviors associated with SUDs, another indication of their roles in OUD (235, 236, 445). The DOP receptor has an inverse function to the KOP receptor, in that DOP receptor activation improves negative emotional states (255). While several OPRD1 polymorphisms correlated with heroin dependence (130, 136, 138–141, 146, 150), it is likely that the effects are ethnicity-dependent, as several other studies found no significant associations between OPRD1 polymorphisms and OUD (137, 140, 144).
The DA system has several well-studied polymorphisms that have been linked with OUD and other SUDs. For example, polymorphisms in DRD2/ANKK1, in particular the TaqIA and TaqIB SNPs, may result in lower D2R availability (321, 334, 335) and have been associated with addictive behaviors including OUD (330, 332, 354, 446, 447). Less studied DRD2 polymorphisms may also affect D2R expression (341, 344) but results have been varied. Additionally, DRD2 polymorphisms may associate with response to medications for OUD; however, there are conflicting reports and further research is required (44, 134, 272, 335, 358, 361, 372). Fewer conclusions can be drawn about DRD1; for example, several DRD1 polymorphisms were initially associated with a rapid transition from first opioid use to opioid dependence, but the results could not be replicated (346, 350). Lastly, lower DAT availability has also been associated with OUD (48–53). Both the 9- and 10-repeat VNTR alleles have been associated with lower DAT availability (363–365, 370, 351); thus, more studies are required to pinpoint the effects of the different repeat VNTR polymorphisms in OUD.
While there is strong preliminary evidence of the role of genetic variants in the DA and opioid systems in OUD, more molecular imaging studies are required in individuals with OUD. In particular, studies utilizing PET tracers that target the less-studied opioid receptors, D1R, D3R, and DAT, would greatly contribute to our understanding of the complex interplay between these receptors in opioid addiction. For instance, as of yet, no imaging studies have examined DOP, KOP, NOP, D1, or D3 receptors in individuals with OUD. One of the most important molecular imaging research questions in OUD is how the different MOUD may alter the dopamine and opioid receptor systems and if these changes are associated with higher rates of successful abstinence. Current imaging studies largely group abstinent and medication-maintained OUD participants together and compare to healthy controls; however, analyses between OUD subgroups would shed light on any neurochemical benefits of MOUD. This would help inform treatment and ultimately improve outcomes for those suffering from OUD. Additionally, opioid receptor antagonist challenge studies would help assess the interaction between drugs like naloxone and semi-synthetic or synthetic opioids, improving safety and efficacy of overdose reversal and prevention. Finally, molecular imaging studies examining the effects of polymorphisms in the DA and opioid systems would help elucidate the genetic components of OUD. The literature relating to genetic association studies in OUD does suggest that certain polymorphisms are risk factors for OUD or may affect treatment outcomes. However, given that these associations are largely ethnicity-dependent, it is important to replicate these findings. Finally, there seem to be sex effects both on genetic association studies and PET/SPECT findings; therefore, future studies could investigate the sex differences in development and outcome of OUD. Further investigation into the underlying genetic factors of OUD and treatment response is critical to help curb the opioid crisis by means of addiction prevention, novel pharmacological targets, and precision treatment.
Author Contributions
PM, CW, NV, and G-JW contributed to the conception and design of the study. JB, DK, DF, and CK conducted a literature search and wrote the first draft. All authors contributed to manuscript revision, and read and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was accomplished with support from the National Institute on Alcohol Abuse and Alcoholism (Y1AA-3009). We also thank Dr. Marta Pecina for her counsel in constructing receptor distribution images using the Allen Brain Atlas.
References
1. Kolodny A, Courtwright DT, Hwang CS, Kreiner P, Eadie JL, Clark TW, et al. The prescription opioid and heroin crisis: a public health approach to an epidemic of addiction. Annu Rev Public Health (2015) 36:559–74. doi: 10.1146/annurev-publhealth-031914-122957
2. Volkow ND, Jones EB, Einstein EB, Wargo EM. Prevention and treatment of opioid misuse and addiction: a review. JAMA Psychiatry (2019) 76(2):208–16. doi: 10.1001/jamapsychiatry.2018.3126
3. Centers for Disease Control and Prevention. Annual surveillance report of drug-related risks and outcomes—United States. surveillance special report 2. centers for disease control and prevention, U.S. Department of Health and Human Services. (2018).
4. Koob GF, Volkow ND. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry (2016) 3(8):760–73. doi: 10.1016/S2215-0366(16)00104-8
5. Volkow ND, Koob GF, McLellan AT. Neurobiologic advances from the brain disease model of addiction. N Engl J Med (2016) 374(4):363–71. doi: 10.1056/NEJMra1511480
6. Wang GJ, Volkow ND, Fowler JS, Logan J, Abumrad NN, Hitzemann RJ, et al. Dopamine D2 receptor availability in opiate-dependent subjects before and after naloxone-precipitated withdrawal. Neuropsychopharmacology (1997) 16(2):174–82. doi: 10.1016/S0893-133X(96)00184-4
7. Martinez D, Saccone PA, Liu F, Slifstein M, Orlowska D, Grassetti A, et al. Deficits in dopamine D(2) receptors and presynaptic dopamine in heroin dependence: commonalities and differences with other types of addiction. Biol Psychiatry (2012) 71(3):192–8. doi: 10.1016/j.biopsych.2011.08.024
8. Hou H, Wang C, Jia S, Hu S, Tian M. Brain dopaminergic system changes in drug addiction: a review of positron emission tomography findings. Neurosci Bull (2014) 30(5):765–76. doi: 10.1007/s12264-014-1469-5
9. Volkow ND, Wang GJ, Fowler JS, Tomasi D. Addiction circuitry in the human brain. Annu Rev Pharmacol Toxicol (2012b) 52:321–36. doi: 10.1146/annurev-pharmtox-010611-134625
10. Volkow ND, Wang GJ, Telang F, Fowler JS, Logan J, Wong C, et al. Sleep deprivation decreases binding of [11C]raclopride to dopamine D2/D3 receptors in the human brain. J Neurosci (2008) 28(34):8454–61. doi: 10.1523/JNEUROSCI.1443-08.2008
11. Volkow ND, Tomasi D, Wang GJ, Telang F, Fowler JS, Logan J, et al. Evidence that sleep deprivation downregulates dopamine D2R in ventral striatum in the human brain. J Neurosci (2012a) 32(19):6711–7. doi: 10.1523/JNEUROSCI.0045-12.2012
12. Wiers CE, Shumay E, Cabrera E, Shokri-Kojori E, Gladwin TE, Skarda E, et al. Reduced sleep duration mediates decreases in striatal D2/D3 receptor availability in cocaine abusers. Transl Psychiatry (2016) 6:e752. doi: 10.1038/tp.2016.14
13. Martinez D, Orlowska D, Narendran R, Slifstein M, Liu F, Kumar D, et al. Dopamine type 2/3 receptor availability in the striatum and social status in human volunteers. Biol Psychiatry (2010) 67(3):275–8. doi: 10.1016/j.biopsych.2009.07.037
14. Wiers CE, Shokri-Kojori E, Cabrera E, Cunningham S, Wong C, Tomasi D, et al. Socioeconomic status is associated with striatal dopamine D2/D3 receptors in healthy volunteers but not in cocaine abusers. Neurosci Lett (2016) 617:27–31. doi: 10.1016/j.neulet.2016.01.056
15. Berro LF, Frussa-Filho R, Tufik S, Andersen ML. Relationships between sleep and addiction: the role of drug-environment conditioning. Med Hypotheses (2014) 82(3):374–6. doi: 10.1016/j.mehy.2013.12.026
16. Land BB, Bruchas MR, Lemos JC, Xu M, Melief EJ, Chavkin C. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J Neurosci (2008) 28:407–14. doi: 10.1523/JNEUROSCI.4458-07.2008
17. Redila VA, Chavkin C. Stress-induced reinstatement of cocaine seeking is mediated by the kappa opioid system. Psychopharmacology (2008) 200:59–70. doi: 10.1007/s00213-008-1122-y
18. McLaughlin JP, Myers LC, Zarek PE, Caron MG, Lefkowitz RJ, Czyzyk TA, et al. Prolonged kappa opioid receptor phosphorylation mediated by G-protein receptor kinase underlies sustained analgesic tolerance. Biol Chem (2004) 279(3):1810–8. doi: 10.1074/jbc.M305796200
19. Daglish MRC, Williams TM, Wilson SJ, Taylor LG, Eap CB, Augsburger M, et al. Brain dopamine response in human opioid addiction. Br J Psychiatry (2008) 193(1):65–72. doi: 10.1192/bjp.bp.107.041228
20. Watson BJ, Taylor LG, Reid AG, Wilson SJ, Stokes PR, Brooks DJ, et al. Investigating expectation and reward in human opioid addiction with [11C]raclopride PET. Addict Biol (2014) 19:1032–40. doi: 10.1111/adb.12073
21. Rommelspacher H, Raeder C, Kaulen P, Bruning G. Adaptive changes of dopamine-D2 receptors in rat brain following ethanol withdrawal: a quantitative autoradiographic investigation. Alcohol (1992) 9:355–62. doi: 10.1016/0741-8329(92)90032-6
22. Volkow ND, Wang GJ, Fowler JS, Logan J, Hitzemann R, Ding YS, et al. Decreases in dopamine receptors but not in dopamine transporters in alcoholics. Alcohol Clin Exp Res (1996) 20(9):1594–8. doi: 10.1111/j.1530-0277.1996.tb05936.x
23. Syvälahti EK, Hietala J, Röyttä M, Grönroos J. Decrease in the number of rat brain dopamine and muscarinic receptors after chronic alcohol intake. Pharmacol Toxicol (1988) 62:4,210–2. doi: 10.1111/j.1600-0773.1988.tb01874.x
24. Martinez D, Gil R, Slifstein M, Hwang DR, Huang Y, Perez A, et al. Alcohol dependence is associated with blunted dopamine transmission in the ventral striatum. Biol Psychiatry (2005) 58(10):779–86. doi: 10.1016/j.biopsych.2005.04.044
25. Volkow ND, Wang GJ, Telang F, Fowler JS, Logan J, Jayne M, et al. Profound decreases in dopamine release in striatum in detoxified alcoholics: possible orbitofrontal involvement. J Neurosci (2007) 27:12700–6. doi: 10.1523/JNEUROSCI.3371-07.2007
26. Yoder KK, Albrecht DS, Dzemidzic M, Normandin MD, Federici LM, Graves T, et al. Differences in IV alcohol-induced dopamine release in the ventral striatum of social drinkers and nontreatment-seeking alcoholics. Drug Alcohol Depend (2016) 160:163–9. doi: 10.1016/j.drugalcdep.2016.01.001
27. Feltmann K, Borroto-Escuela DO, Rüegg J, Pinton L, de Oliveira Sergio T, Narváez M, et al. Effects of long-term alcohol drinking on the dopamine D2 receptor: gene expression and heteroreceptor complexes in the striatum in rats. Alcohol Clin Exp Res (2018) 42(2):338–51. doi: 10.1111/acer.13568
28. Hirth N, Meinhardt MW, Noori HR, Salgado H, Torres-Ramirez O, Uhrig S, et al. Convergent evidence from alcohol-dependent humans and rats for a hyperdopaminergic state in protracted abstinence. Proc Natl Acad Sci U S A (2016) 113(11):3024–9. doi: 10.1073/pnas.1506012113
29. Volkow ND, Wise RA, Baler R. The dopamine motive system: implications for drug and food addiction. Nat Rev Neurosci (2017) 18:12,741–752. doi: 10.1038/nrn.2017.130
30. Crist RC, Reiner BC, Berrettini WH. A review of opioid addiction genetics. Curr Opin Psychol (2019) 27:31–5. doi: 10.1016/j.copsyc.2018.07.014
31. Kreek MJ, Bart G, Lilly C, Laforge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev (2005) 57:1–26. doi: 10.1124/pr.57.1.1
32. Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, Fenton B, et al. Familial transmission of substance use disorders. Arch Gen Psychiatry (1998) 55:973–9. doi: 10.1001/archpsyc.55.11.973
33. Drgon T, Zhang PW, Johnson C, Walther D, Hess J, Nino M, et al. Genome wide association for addiction: replicated results and comparisons of two analytic approaches. PLoS One (2010) 5(1):e8832. doi: 10.1371/journal.pone.0008832
34. Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high density genome wide association study for nicotine dependence Hum. Mol Genet (2006) 16(1):24–35. doi: 10.1093/hmg/ddl441
35. Rutter JL. Symbiotic relationship of pharmacogenetics and drugs of abuse. Aaps J (2006) 8(1):E174–84. doi: 10.1208/aapsj080121
36. Chamorro AJ, Marcos M, Mirón-Canelo JA, Pastor I, González-Sarmiento R, Laso FJ. Association of µ-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systematic review and meta-analysis. Addict Biol (2012) 17(3):505–12. doi: 10.1111/j.1369-1600.2012.00442.x
37. Anton RF, Oroszi G, O’Malley S, Couper D, Swift R, Pettinati H, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry (2008) 65:135–44. doi: 10.1001/archpsyc.65.2.135
38. Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry (2007) 64(9):1069–77. doi: 10.1001/archpsyc.64.9.1069
39. Oslin DW, Leong SH, Lynch KG, Berrettini W, O’Brien CP, Gordon AJ, et al. Naltrexone vs placebo for the treatment of alcohol dependence: a randomized clinical trial. JAMA Psychiatry (2015) 72(5):430–7. doi: 10.1001/jamapsychiatry.2014.3053
40. Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang B-Z, Krupitsky E, et al. Association between two µ-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet (2006) 15(6):807–19. doi: 10.1093/hmg/ddl024
41. Lerman C, Berrettini W, Pinto A, Patterson F, Crystal-Mansour S, Wileyto EP, et al. Changes in food reward following smoking cessation: a pharmacogenetic investigation. Psychopharmacology (2004) 174:571–7. doi: 10.1007/s00213-004-1823-9
42. Ray R, Jepson C, Wileyto EP, Dahl JP, Patterson F, Rukstalis M, et al. Genetic variation in mu-opioid-receptor-interacting proteins and smoking cessation in a nicotine replacement therapy trial. Nicotine Tob Res (2007) 9:1237–41. doi: 10.1080/14622200701648367
43. Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: a comprehensive review. Behav Pharmacol (2009) 20(1):1–17. doi: 10.1097/FBP.0b013e3283242f05
44. Doehring A, Hentig NV, Graff J, Salamat S, Schmidt M, Geisslinger G, et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenet Genomics (2009) 19(6):407–14. doi: 10.1097/FPC.0b013e328320a3fd
45. Stolf AR, Müller D, Schuch JB, Akutagava-Martins GC, Guimaraes LSP, Szobot CM, et al. Association between the intron 8 VNTR polymorphism of the DAT1 gene and crack cocaine addiction. Neuropsychobiology (2017) 75(3):141–4. doi: 10.1159/000485128
46. Muramatsu T, Higuchi S. Dopamine transporter gene polymorphism and alcoholism. Biochem Biophys Res Commun (1995) 211:28–32. doi: 10.1006/bbrc.1995.1773
47. Sander T, Harms H, Podschus J, Finckh U, Nickel B, Rolfs A, et al. Allelic association of a dopamine transporter gene polymorphism in alcohol dependence with withdrawal seizures or delirium. Biol Psychiatry (1997) 41(3):299–304. doi: 10.1016/S0006-3223(96)00044-3
48. Jia SW, Wang W, Liu Y, Wu ZM. Neuroimaging studies of brain corpus striatum changes among heroin-dependent patients treated with herbal medicine, U’finerTM capsule. Addict Biol (2005) 10(3):293–7. doi: 10.1080/13556210500222456
49. Shi J, Zhao LY, Copersino ML, Fang YX, Chen Y, Tian J, et al. PET imaging of dopamine transporter and drug craving during methadone maintenance treatment and after prolonged abstinence in heroin users. Eur J Pharmacol (2008) 579(1–3):160–6. doi: 10.1016/j.ejphar.2007.09.042
50. Yeh TL, Chen KC, Lin SH, Lee IH, Chen PS, Yao WJ, et al. Availability of dopamine and serotonin transporters in opioid-dependent users-a two-isotope SPECT study. Psychopharmacology (2012) 220:1:55–64. doi: 10.1007/s00213-011-2454-6
51. Yuan J, Liu XD, Han M, Lv R, Bin, Wang YK, et al. Comparison of striatal dopamine transporter levels in chronic heroin-dependent and methamphetamine-dependent subjects. Addict Biol (2017) 22(1):229–34. doi: 10.1111/adb.12271
52. Zaaijer ER, Van Dijk L, De Bruin K, Goudriaan AE, Lammers LA, Koeter MWJ, et al. Effect of extended-release naltrexone on striatal dopamine transporter availability, depression and anhedonia in heroin-dependent patients. Psychopharmacology (2015) 232(14):2597–607. doi: 10.1007/s00213-015-3891-4
53. Liu Y, Han M, Liu X, Deng Y, Li Y, Yuan J, et al. Dopamine transporter availability in heroin-dependent subjects and controls: longitudinal changes during abstinence and the effects of Jitai tablets treatment. Psychopharmacology (2013) 230(2):235–44. doi: 10.1007/s00213-013-3148-z
54. Crits-Christoph P, Newberg A, Wintering N, Ploessl K, Gibbons MBC, Ring-Kurtz S, et al. Dopamine transporter levels in cocaine dependent subjects. Drug Alcohol Depend (2008) 98(1–2):70–6. doi: 10.1016/j.drugalcdep.2008.04.014
55. Malison RT, Best SE, Van Dyck CH, McCance EF, Wallace EA, Lamelle M, et al. Elevated striatal dopamine transporters during acute cocaine abstinence as measured by [123I]β-CIT SPECT. Am J Psychiatry (1998) 155(6):832–4. doi: 10.1176/ajp.155.6.832
56. Volkow ND, Chang L, Wang GJ, Fowler JS, Ding YS, Sedler M, et al. Low level of brain dopamine D2 receptors in methamphetamine abusers: association with metabolism in the orbitofrontal cortex. Am J Psychiatry (2001) 158(12):2015–21. doi: 10.1176/appi.ajp.158.12.2015
57. Volkow ND, Wang GJ, Smith L, Fowler JS, Telang F, Logan J, et al. Recovery of dopamine transporters with methamphetamine detoxification is not linked to changes in dopamine release. Neuroimage (2015) 121:20–8. doi: 10.1016/j.neuroimage.2015.07.035
58. McCann UD, Wong DF, Yokoi F, Villemagne V, Dannals RF, Ricaurte GA. Reduced striatal dopamine transporter density in abstinent methamphetamine and methcathinone users: evidence from positron emission tomography studies with [11 C]WIN-35,428. J Neurosci (1998) 18(20):8417–22. doi: 10.1523/JNEUROSCI.18-20-08417.1998
59. Laine TPJ, Ahonen A, Räsänen P, Tiihonen J. Dopamine transporter availability and depressive symptoms during alcohol withdrawal. Psychiatry Res Neuroimag (1999) 90(3):153–7. doi: 10.1016/S0925-4927(99)00019-0
60. Krause KH, Dresel SH, Krause J, Kung HF, Tatsch K, Ackenheil M. Stimulant-like action of nicotine on striatal dopamine transporter in the brain of adults with attention deficit hyperactivity disorder. Int J Neuropsychopharmacol (2002) 5(2):111–3. doi: 10.1017/S1461145702002821
61. Yang YK, Yao WJ, Yeh TL, Lee IH, Chen PS, Lu RB, et al. Decreased dopamine transporter availability in male smokers - A dual isotope SPECT study. Prog Neuropsychopharmacol Biol Psychiatry (2008) 32(1):274–9. doi: 10.1016/j.pnpbp.2007.08.018
62. Leroy C, Karila L, Martinot JL, Lukasiewicz M, Duchesnay E, Comtat C, et al. Striatal and extrastriatal dopamine transporter in cannabis and tobacco addiction: a high-resolution PET study. Addict Biol (2012) 17(6):981–90. doi: 10.1111/j.1369-1600.2011.00356.x
63. Moningka H, Lichenstein S, Worhunsky PD, DeVito EE, Scheinost D, Yip SW. Can neuroimaging help combat the opioid epidemic? a systematic review of clinical and pharmacological challenge fMRI studies with recommendations for future research. Neuropsychopharmacology (2019) 44(2):259–73. doi: 10.1038/s41386-018-0232-4
64. Fareed A, Kim J, Ketchen B, Kwak WJ, Wang D, Shongo-Hiango H, et al. Effect of heroin use on changes of brain functions as measured by functional magnetic resonance imaging, a systematic review. J Addict Dis (2017) 36(2):105–16. doi: 10.1080/10550887.2017.1280898
65. Ieong HF, Yuan Z. Resting-state neuroimaging and neuropsychological findings in opioid use disorder during abstinence: a review. Front Hum Neurosci (2017) 11:169. doi: 10.3389/fnhum.2017.00169
66. Wollman SC, Alhassoon OM, Hall MG, Stern MJ, Connors EJ, Kimmel CL, et al. Gray matter abnormalities in opioid-dependent patients: a neuroimaging meta-analysis. Am J Drug Alcohol Abuse (2017) 43:5,505–517. doi: 10.1080/00952990.2016.1245312
67. Kieffer BL, Gavériaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol (2002) 66(5):285–306. doi: 10.1016/S0301-0082(02)00008-4
68. Matthes HWD, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the µ-opioid-receptor gene. Nature (1996) 383:819–23. doi: 10.1038/383819a0
69. Johnson SW, North RA. Opioids excite dopamine neurons by hyperpolarization of local interneurons. J Neurosci (1992) 12(2):483–8. doi: 10.1523/JNEUROSCI.12-02-00483.1992
70. Gysling K, Wang RY. Morphine-induced activation of A10 dopamine neurons in the rat. Brain Res (1983) 277:119–27. doi: 10.1016/0006-8993(83)90913-7
71. Jalabert M, Bourdy R, Courtin J, Veinante P, Manzoni OJ, Barrot M, et al. Neuronal circuits underlying acute morphine action on dopamine neurons. Proc Natl Acad Sci U S A (2011) 108(39):16446–50. doi: 10.1073/pnas.1105418108
72. Jhou TC, Xu SP, Lee MR, Gallen CL, Ikemoto S. Mapping of reinforcing and analgesic effects of the mu opioid agonist endomorphin-1 in the ventral midbrain of the rat. Psychopharmacology (2012) 224(2):303–12. doi: 10.1007/s00213-012-2753-6
73. Matsui A, Jarvie BC, Robinson BG, Hentges ST, Williams JT. Separate GABA afferents to dopamine neurons mediate acute action of opioids, development of tolerance, and expression of withdrawal. Neuron (2014) 82(6):1346–56. doi: 10.1016/j.neuron.2014.04.030
74. Hnasko TS, Sotak BN, Palmiter RD. Morphine reward in dopamine-deficient mice. Nature (2005) 438(7069):854–7. doi: 10.1038/nature04172
75. Nader K, van der Kooy D. Deprivation state switches the neurobiological substrates mediating opiate reward in the ventral tegmental area. J Neurosci (1997) 17(1):383–90. doi: 10.1523/JNEUROSCI.17-01-00383.1997
76. Bolger GT, Skolnick P, Rice KC, Weissman BA. Differential regulation of mu-opiate receptors in heroin- and morphine-dependent rats. FEBS Lett (1988) 234(1):22–6. doi: 10.1016/0014-5793(88)81294-8
77. Belcheva MM, Barg J, McHale RJ, Dawn S, Ho MT, Ignatova E, et al. Differential down- and up-regulation of rat brain opioid receptor types and subtypes by buprenorphine. Mol Pharmacol (1993) 44:173–9.
78. Tempel A, Espinoza K. Morphine-induced downregulation of mu-opioid receptors in neonatal rat brain. Brain Res (1992) 469:129–33. doi: 10.1016/0165-3806(88)90176-9
79. Yu Y, Zhang L, Yin X, Sun H, Uhl GR, Wang JB. Mu opioid receptor phosphorylation, desensitization, and ligand efficacy. J Biol Chem (1997) 272:28869–74. doi: 10.1074/jbc.272.46.28869
80. Keith DE, Anton B, Murray SR, Zaki PA, Chu PC, Lissin DV, et al. mu-Opioid receptor internalization: opiate drugs have differential effects on a conserved endocytic mechanism in vitro and in the mammalian brain. Mol Pharmacol (1998) 53:377–84. doi: 10.1124/mol.53.3.377
81. Von Zastrow M, Svingos A, Haberstock-Debic H, Evans C. Regulated endocytosis of opioid receptors: cellular mechanisms and proposed roles in physiological adaptation to opiate drugs. Curr Opin Neurobiol (2003) 13:348–53. doi: 10.1016/S0959-4388(03)00069-2
82. Blanchet C, Sollini M, Luscher C. Two distinct forms of desensitization of G-protein coupled inwardly rectifying potassium currents evoked by alkaloid and peptide mu-opioid receptor agonists. Mol Cell Neurosci (2003) 24:517–23. doi: 10.1016/S1044-7431(03)00173-8
83. Arden JR, Segredo V, Wang Z, Lameh J, Sadée W. Phosphorylation and agonist-specific intracellular trafficking of an epitope-tagged mu-opioid receptor expressed in HEK 293 cells. J Neurochem (1995) 65:1636–45. doi: 10.1046/j.1471-4159.1995.65041636.x
84. Keith DE, Murray SR, Zaki PA, Chu PC, Lissin DV, Kang L, et al. Morphine activates opioid receptors without causing their rapid internalization. J Biol Chem (1996) 277:19021–4. doi: 10.1074/jbc.271.32.19021
85. Whistler JL, Chuang HH, Chu P, Jan LY, von Zastrow M. Functional dissociation of mu opioid receptor signaling and endocytosis: implications for the biology of opiate tolerance and addiction. Neuron (1999) 23:737–46. doi: 10.1016/S0896-6273(01)80032-5
86. Martini L, Whistler JL. The role of mu opioid receptor desensitization and endocytosis in morphine tolerance and dependence. Curr Opin Neurobiol (2007) 17(5):556–64. doi: 10.1016/j.conb.2007.10.004
87. Zaki PA, Keith DE Jr, Brine GA, Carroll FI, Evans CJ. The role of G protein activation in agonist-dependent mu-opioid receptor internalization. JPET (2000) 292(3):1127–34.
88. Kieffer BL, Evans CJ. Opioid tolerance—in search of a holy grail. Cell (2002) 108:5:587–90. doi: 10.1016/S0092-8674(02)00666-9
89. Bailey CP, Couch D, Johnson E, Griffiths K, Kelly E, Henderson G. μ-Opioid receptor desensitization in mature rat neurons: lack of interaction between DAMGO and morphine. J Neurosci (2003) 33:10515–20. doi: 10.1523/JNEUROSCI.23-33-10515.2003
90. Rankovic Z, Brust TF, Bohn LM. Biased agonism: an emerging paradigm in GPCR drug discovery. Bioorg Med Chem Lett (2016) 26:241–50. doi: 10.1016/j.bmcl.2015.12.024
91. Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, et al. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther (2007) 320:1–13. doi: 10.1124/jpet.106.104463
92. Schmid CL, Kennedy NM, Ross NC, Lovell KM, Yue Z, Morgenweck J, Bias factor and therapeutic window correlate to predict safer opioid analgesics. Cell (2017) 171(5):1165–1175.e13. doi: 10.1016/j.cell.2017.10.035
93. Raehal KM, Walker JK, Bohn LM. Morphine side effects in beta-arrestin 2 knockout mice. J Pharmacol Exp Ther (2005) 314(3):1195–201. doi: 10.1124/jpet.105.087254
94. Bohn LM, Lefkowitz RJ, Gainetdinov RR, Peppel K, Caron MG, Lin FT. Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science (1999) 286:2495–8. doi: 10.1126/science.286.5449.2495
95. Bu H, Liu X, Tian X, Yang H, Gao F. Enhancement of morphine analgesia and prevention of morphine tolerance by downregulation of β-arrestin 2 with antigene RNAs in mice. Int J Neurosci (2015) 125(1):56–65. doi: 10.3109/00207454.2014.896913
96. Beardsley PM, Zhang Y. Synthetic Opioids. Handb Exp Pharmacol (2018) 252:353–81. doi: 10.1007/164_2018_149
97. Crist RC, Berrettini WH. Pharmacogenetics of OPRM1. Pharmacol Biochem Behav (2014) 2014:25–33. doi: 10.1016/j.pbb.2013.10.018
98. Kasai S, Ikeda K. Pharmacogenomics of the human µ-opioid receptor. Pharmacogenomics (2011) 12:1305–20. doi: 10.2217/pgs.11.68
99. Yuferov V, Levran O, Proudnikov D, Nielsen DA, Kreek MJ. Search for genetic markers and functional variants involved in the development of opiate and cocaine addiction and treatment. Ann NY Acad Sci (2010) 1187:184–207. doi: 10.1111/j.1749-6632.2009.05275.x
100. Beyer A, Koch T, Schroder H, Schulz S, Höllt V. Effect of the A118G polymorphism on binding affinity, potency and agonist-mediated endocytosis, desensitization, and resensitization of the human mu-opioid receptor. J Neurochem (2004) 89:553–60. doi: 10.1111/j.1471-4159.2004.02340.x
101. Kroslak T, LaForge KS, Gianotti RJ, Ho A, Nielsen DA, Kreek MJ. The single nucleotide polymorphism A118G alters functional properties of the human mu opioid receptor. J Neurochem (2007) 103:1,77–87. doi: 10.1111/j.1471-4159.2007.04738.x
102. Robinson JE, Vardy E, DiBerto JF, Chefer VI, White KL, Fish EW, et al. Receptor reserve moderates mesolimbic responses to opioids in a humanized mouse model of the OPRM1 A118G polymorphism. Neuropsychopharmacology (2015) 40(11):2614–22. doi: 10.1038/npp.2015.109
103. Zhang Y, Wang D, Johnson AD, Papp AC, Sadée W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem (2005) 280(38):,32618–24. doi: 10.1074/jbc.M504942200
104. Weerts EM, McCaul ME, Kuwabara H, Yang X, Xu X, Dannals RF, et al. Influence of OPRM1 Asn40Asp variant (A118G) on [11C]carfentanil binding potential: preliminary findings in human subjects. Int J Neuropsychopharmacol (2013) 16(1):47–53. doi: 10.1017/S146114571200017X
105. Peciña M, Love T, Stohler CS, Goldman D, Zubieta JK. Effects of the Mu opioid receptor polymorphism (OPRM1 A118G) on pain regulation, placebo effects and associated personality trait measures. Neuropsychopharmacology (2015) 40(4):957–65. doi: 10.1038/npp.2014.272
106. Oertel BG, Doehring A, Roskam B, Kettner M, Hackmann N, Ferreiros N, et al. Genetic-epigenetic interaction modulates µ-opioid receptor regulation. Human Mol Genet (2012) 21(21):4751–60. doi: 10.1093/hmg/dds314
107. Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A (1998) 95:9608–13. doi: 10.1073/pnas.95.16.9608
108. Befort K, Filliol D, Decaillot FM, Gavériaux-Ruff C, Hoehe MR, Kieffer BL. A single nucleotide polymorphic mutation in the human mu-opioid receptor severely impairs receptor signaling. J Biol Chem (2001) 276:3130–7. doi: 10.1074/jbc.M006352200
109. Haerian BS, Haerian MS. OPRM1 rs1799971 polymorphism and opioid dependence: evidence from a meta-analysis. Pharmacogenomics (2013) 14(7):813–24. doi: 10.2217/pgs.13.57
110. Woodcock EA, Lundahl LH, Burmeister M, Greenwald MK. Functional mu opioid receptor polymorphism (OPRM1 A118G) associated with heroin use outcomes in Caucasian males: a pilot study. Am J Addict (2015) 24(4):329–35. doi: 10.1111/ajad.12187
111. Szeto CYK, Tang NLS, Lee DTS, Stadlin A. Association between µ opioid receptor gene polymorphisms and Chinese heroin addicts. Neuroreport (2001) 12:1103–6. doi: 10.1097/00001756-200105080-00011
112. Gelernter J, Kranzler H, Cubells J. Genetics of two µ opioid receptor gene (OPRM1) exon 1 polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjects. Mol Psychiatry (1999) 4:476–83. doi: 10.1038/sj.mp.4000556
113. Clarke TK, Crist RC, Kampman KM, Dackis CA, Pettinati HM, O’Brien CP, et al. Low frequency genetic variants in the μ-opioid receptor (OPRM1) affect risk for addiction to heroin and cocaine. Neurosci Lett (2013) 10(542):71–5. doi: 10.1016/j.neulet.2013.02.018
114. Shi J, Hui L, Xu Y, Wang F, Huang W, Hu G. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum Mutat (2002) 19:459–60. doi: 10.1002/humu.9026
115. Xu J, Lu Z, Xu M, Pan L, Deng Y, Xie X, et al. A heroin addiction severity-associated intronic single nucleotide polymorphism modulates alternative pre-mRNA splicing of the μ opioid receptor gene OPRM1 via hnRNPH interactions. J Neurosci (2014) 34(33):11048–66. doi: 10.1523/JNEUROSCI.3986-13.2014
116. Kumar D, Chakraborty J, Das S. Epistatic effects between variants of κ-opioid receptor gene and A118G of µ-opioid receptor gene increase susceptibility to addiction in Indian population. Prog Neuropsychopharmacol Biol Psychiatry (2012) 36:225–30. doi: 10.1016/j.pnpbp.2011.10.018
117. Nuechterlein EB, Ni L, Domino EF, Zubieta JK. Nicotine-specific and non-specific effects of cigarette smoking on endogenous opioid mechanisms. Biol Psychiatry (2016) 69:69–77. doi: 10.1016/j.pnpbp.2016.04.006
118. Domino EF, Hirasawa-Fujita M, Ni L, Guthrie SK, Zubieta JK. Regional brain [(11)C]carfentanil binding following tobacco smoking. Prog Neuropsychopharmacol Biol Psychiatry (2015) 59:100–4. doi: 10.1016/j.pnpbp.2015.01.007
119. Ray R, Ruparel K, Newberg A, Wileyto EP, Loughead JW, Divgi C, et al. Human mu opioid receptor (OPRM1 A118G) polymorphism is associated with brain mu-opioid receptor binding potential in smokers. Proc Natl Acad Sci U S A (2011) 108(22):9268–73. doi: 10.1073/pnas.1018699108
120. Domino EF, Evans CL, Ni L, Guthrie SK, Koeppe RA, Zubieta JK. Tobacco smoking produces greater striatal dopamine release in G-allele carriers with mu opioid receptor A118G polymorphism. Prog Neuropsychopharmacol Biol Psychiatry (2012) 38(2):236–40. doi: 10.1016/j.pnpbp.2012.04.003
121. Kapur S, Sharad S, Singh RA, Gupta AK. A118G polymorphism in µ opioid receptor gene (oprm1): association with opiate addiction in subjects of Indian origin. J Integr Neurosci (2007) 6(4):511–22. doi: 10.1142/S0219635207001635
122. Deb I, Chakraborty J, Gangopadhyay PK, Choudhury SR, Das S. Single-nucleotide polymorphism (A118G) in exon 1 of OPRM1 gene causes alteration in downstream signaling by µ-opioid receptor and may contribute to the genetic risk for addiction. J Neurochem (2010) 112:486–96. doi: 10.1111/j.1471-4159.2009.06472.x
123. Tan E-C, Tan C-H, Karupathivan U, Yap EPH. µ opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport (2003) 14:569–72. doi: 10.1097/00001756-200303240-00008
124. Nagaya D, Ramanathan S, Ravichandran M, Navaratnam V. A118G µ opioid receptor polymorphism among drug addicts in Malaysia. J Integr Neurosci (2012) 11:117–22. doi: 10.1142/S0219635212500082
125. Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, et al. Substantial attributable risk related to a functional µ-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry (2004) 9:547–9. doi: 10.1038/sj.mp.4001504
126. Drakenberg K, Nikoshkov A, Horváth MC, Fagergren P, Gharibyan A, Saarelainen K, et al. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc Natl Acad Sci U S A (2006) 103(20):7883–8. doi: 10.1073/pnas.0600871103
127. Luo X, Kranzlet HR, Zhao H, Gelernter J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am J Med Genet (2003) 120B:97–108. doi: 10.1002/ajmg.b.20034
128. Crowley JJ, Oslin DW, Patkar AA, Gottheil E, DeMaria PA Jr, O’Brien CP, et al. A genetic association study of the µ opioid receptor and severe opioid dependence. Psychiatr Genet (2003) 13:169–73. doi: 10.1097/00041444-200309000-00006
129. Franke P, Wang T, Nöthen MM, Knapp M, Neidt H, Albrecht S, et al. Nonreplication of association between µ-opioid-receptor gene (OPRM1) A118G polymorphism and substance dependence. Am J Med Genet (2001) 105(1):114–9. doi: 10.1002/1096-8628(20010108)105:1<114::AID-AJMG1074>3.3.CO;2-C
130. Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav (2008) 7:720–9. doi: 10.1111/j.1601-183X.2008.00410.x
131. Nikolov MA, Beltcheva O, Galabova A, Ljubenova A, Jankova E, Gergov G, et al. No evidence of association between 118A > G OPRM1 polymorphism and heroin dependence in a large Bulgarian case-control sample. Drug Alcohol Depend (2011) 117(1):62–5. doi: 10.1016/j.drugalcdep.2010.12.026
132. Li T, Liu X, Zhu ZH, Zhao J, Hu X, Sham PC, et al. Association analysis of polymorphisms in the µ opioid gene and heroin abuse in Chinese. Addict Biol (2000) 5:181–6. doi: 10.1080/13556210050003775
133. Zhang D, Shao C, Shao M, Yan P, Wang Y, Liu Y, et al. Effect of mu-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry (2007) 61(11):1244–51. doi: 10.1016/j.biopsych.2006.07.012
134. Crettol S, Besson J, Croquette-Krokar M, Hämmig R, Gothuey I, Monnat M, et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuropsychopharmacol Biol Psychiatry (2008) 32(7):1722–7. doi: 10.1016/j.pnpbp.2008.07.009
135. Levran O, Peles E, Randesi M, da Rosa JC, Adelson M, Kreek MJ. The μ-opioid receptor nonsynonymous variant 118A > G is associated with prolonged abstinence from heroin without agonist treatment. Pharmacogenomics (2017) 18(15):1387–91. doi: 10.2217/pgs-2017-0092
136. Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry (2008) 13:531–43. doi: 10.1038/sj.mp.4002035
137. Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol (2014) 19(1):111–21. doi: 10.1111/j.1369-1600.2012.00445.x
138. Gao X, Wang Y, Lang M, Yuan L, Reece AS, Wang W. Contribution of genetic polymorphisms and haplotypes in DRD2, BDNF, and opioid receptors to heroin dependence and endophenotypes among the Han Chinese. OMICS (2017) 21(7):404–12. doi: 10.1089/omi.2017.0057
139. Nagaya D, Zahari Z, Saleem M, Yahaya BH, Tan SC, Yusoff NM. An analysis of genetic association in opioid dependence susceptibility. J Clin Pharm Ther (2018) 43(1):80–6. doi: 10.1111/jcpt.12585
140. Crist RC, Ambrose-Lanci LM, Vaswani M, Clarke TK, Zeng A, Yuan C, et al. Case-control association analysis of polymorphisms in the delta-opioid receptor, OPRD1, with cocaine and opioid addicted populations. Drug Alcohol Depend (2013) 127:122–8. doi: 10.1016/j.drugalcdep.2012.06.023
141. Huang CC, Kuo SC, Yeh TC, Yeh YW, Chen CY, Liang CS, et al. OPRD1 gene affects disease vulnerability and environmental stress in patients with heroin dependence in Han Chinese. Prog Neuropsychopharmacol Biol Psychiatry (2018) 89:109–16. doi: 10.1016/j.pnpbp.2018.08.028
142. Mayer P, Rochlitz H, Rauch E, Rommelspacher H, Hasse HE, Schmidt S, et al. Association between a delta opioid receptor gene polymorphism and heroin dependence in man. Neuroreport (1997) 8:2547–50. doi: 10.1097/00001756-199707280-00025
143. Xu K, Liu XH, Nagarajan S, Gu XY, Goldman D. Relationship of the delta-opioid receptor gene to heroin abuse in a large Chinese case/control sample. Am J Med Genet (2002) 110:45–50. doi: 10.1002/ajmg.10374
144. Franke P, Nöthen MM, Wang T, Neidt H, Knapp M, Lichtermann D, et al. Human delta-opioid receptor gene and susceptibility to heroin and alcohol dependence. Am J Med Genet (1999) 88:462–4. doi: 10.1002/(SICI)1096-8628(19991015)88:5<462::AID-AJMG4>3.0.CO;2-S
145. Clarke TK, Crist RC, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ, et al. Genetic variation in OPRD1 and the response to treatment for opioid dependence with buprenorphine in European-American females. Pharmacogenomics J (2014a) 14(3):303–8. doi: 10.1038/tpj.2013.30
146. Sharafshah A, Fazel H, Albonaim A, Omarmeli V, Rezaei S, Mirzajani E, et al. Association of OPRD1 gene variants with opioid dependence in addicted male individuals undergoing methadone treatment in the north of Iran. J Psychoactive Drugs (2017) 49(3):242–51. doi: 10.1080/02791072.2017.1290303
147. Jones JD, Luba RR, Vogelman JL, Comer SD. Searching for evidence of genetic mediation of opioid withdrawal by opioid receptor gene polymorphisms. Am J Addict (2016) 25(1):41–8. doi: 10.1111/ajad.12316
148. Crist RC, Clarke TK, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ, et al. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology (2013) 38(10):2003–10. doi: 10.1038/npp.2013.99
149. Crist RC, Phillips KA, Furnari MA, Moran LM, Doyle GA, McNicholas LF, et al. Replication of the pharmacogenetic effect of rs678849 on buprenorphine efficacy in African-Americans with opioid use disorder. Pharmacogenomics J (2018) 19:260–68. doi: 10.1038/s41397-018-0065-x
150. Beer B, Erb R, Pavlic M, Ulmer H, Giacomuzzi S, Riemer Y, et al. Association of polymorphisms in pharmacogenetic candidate genes (OPRD1, GAL, ABCB1, OPRM1) with opioid dependence in European population: a case-control study. PLoS One (2013) 8(9):e75359. doi: 10.1371/journal.pone.0075359
151. Luo R, Li X, Qin S, Luo Z, Luo X, Hu P, et al. Impact of SNP–SNP interaction among ABCB1, ARRB2, DRD1 and OPRD1 on methadone dosage requirement in Han Chinese patients. Pharmacogenomics (2017) 18(18):1659–70. doi: 10.2217/pgs-2017-0072
152. Yuferov V, Fussella D, LaForgea KS, Nielsena DA, Gordonb D, Hoa A, et al. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenetics (2004) 14(12):793–804. doi: 10.1097/00008571-200412000-00002
153. Gerra G, Leonardi C, Cortese E, D’Amore A, Lucchini A, Strepparola G, et al. Human kappa opioid receptor gene (OPRK1) polymorphism is associated with opiate addiction. Am J Med Genet B Neuropsychiatr Genet (2007) 144B(6):771–5. doi: 10.1002/ajmg.b.30510
154. Albonaim A, Fazel H, Sharafshah A, Omarmeli V, Rezaei S, Ajamian F, et al. Association of OPRK1 gene polymorphisms with opioid dependence in addicted men undergoing methadone treatment in an Iranian population. J Addict Dis (2017) 36(4):227–35. doi: 10.1080/10550887.2017.1361724
155. Yuanyuan J, Rui S, Hua T, Jingjing C, Cuola D, Yuhui S, et al. Genetic association analyses and meta-analysis of Dynorphin-Kappa Opioid system potential functional variants with heroin dependence. Neurosci Lett (2018) 685:75–82. doi: 10.1016/j.neulet.2018.08.023
156. Wei SG, Zhu YS, Lai JH, Xue HX, Chai ZQ, Li SB. Association between heroin dependence and prodynorphin gene polymorphisms. Brain Res Bull (2011) 85:238–42. doi: 10.1016/j.brainresbull.2011.02.010
157. Hashemi M, Shakiba M, Sanaei S, Shahkar G, Rezaei M, Mojahed A, et al. Evaluation of prodynorphin gene polymorphisms and their association with heroin addiction in a sample of the southeast Iranian population. Mol Biol Res Commun (2018) 7(1):1–6. doi: 10.22099/mbrc.2017.27182.1294
158. Clarke T-K, Ambrose-Lanci L, Ferraro TN, Berrettini WH, Kampman KM, Dackis CA, et al. Genetic association analyses of PDYN polymorphisms with heroin and cocaine addiction. Genes Brain Behav (2012) 2012(11):415–23. doi: 10.1111/j.1601-183X.2012.00785.x
159. Clarke TK, Krause K, Li T, Schumann G. An association of prodynorphin polymorphisms and opioid dependence in females in a Chinese population. Addict Biol (2009) 14:366–70. doi: 10.1111/j.1369-1600.2009.00151.x
160. Xuei X, Flury-Wetherill L, Almasy L, Bierut L, Tischfield J, Schuckit M, et al. Association analysis of genes encoding the nociceptin receptor (OPRL1) and its endogenous ligand (PNOC) with alcohol or illicit drug dependence. Addict Biol (2008) 13(1):80–7. doi: 10.1111/j.1369-1600.2007.00082.x
161. Briant JA, Nielsen DA, Proudnikov D, Londono D, Ho A, Ott J, et al. Evidence for association of two variants of the nociceptin/orphanin FQ receptor gene OPRL1 with vulnerability to develop opiate addiction in Caucasians. Psychiatr Genet (2010) 20(2):65–72. doi: 10.1097/YPG.0b013e32833511f6
162. Browne CA, Erickson RL, Blendy JA, Lucki I. Genetic variation in the behavioral effects of buprenorphine in female mice derived from a murine model of the OPRM1 A118G polymorphism. Neuropharmacology (2017) 117:401–7. doi: 10.1016/j.neuropharm.2017.02.005
163. Muriel J, Margarit C, Planelles B, Serralta MJ, Puga C, Inda MD, et al. OPRM1 influence on and effectiveness of an individualized treatment plan for prescription opioid use disorder patients. Ann N Y Acad Sci (2018) 1425:82–94. doi: 10.1111/nyas.13735
164. Wu WD, Wang Y, Fang YM, Zhou HY. Polymorphism of the micro-opioid receptor gene (OPRM1 118A > G) affects fentanyl-induced analgesia during anesthesia and recovery. Mol Diagn Ther (2009) 13:331–7. doi: 10.1007/BF03256337
165. Oueslati B, Moula O, Ghachem R. The impact of OPRM1’s genetic polymorphisms on methadone maintenance treatment in opioid addicts: a systematic review. Pharmacogenomics (2018) 19(8):741–7. doi: 10.2217/pgs-2018-0017
166. Wand GS, Mangold D, El Deiry S, McCaul ME, Hoover D. Family history of alcoholism and hypothalamic opioidergic activity. Arch Gen Psychiatry (1998) 55:1114–9. doi: 10.1001/archpsyc.55.12.1114
167. Schluger JH, Ho A, Borg L, Porter M, Maniar S, Gunduz M, et al. Nalmefene causes greater hypothalamic–pituitary–adrenal axis activation than naloxone in normal volunteers: implications for the treatment of alcoholism. Alcohol Clin Exp Res (1998) 22:1430–6. doi: 10.1111/j.1530-0277.1998.tb03931.x
168. Chong RY, Oswald L, Yang X, Uhart M, Lin PI, Wand GS. The mu-opioid receptor polymorphism A118G predicts cortisol responses to naloxone and stress. Neuropsychopharmacology (2006) 31:204–11. doi: 10.1038/sj.npp.1300856
169. Hernandez-Avila CA, Wand G, Luo X, Gelernter J, Kranzler HR. Association between the cortisol response to opioid blockade and the Asn40Asp polymorphism at the mu-opioid receptor locus (OPRM1). Am J Med Genet (2003) 118:60–5. doi: 10.1002/ajmg.b.10054
170. Wand GS, McCaul M, Yang X, Reynolds J, Gotjen D, Lee S, et al. The mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacology (2002) 26:106–14. doi: 10.1016/S0893-133X(01)00294-9
171. Belcher AM, Volkow ND, Moeller GF, Ferre S. Personality traits and vulnerability or resilience to substance use disorders. Trends Cogn Sci (2014) 18(4):211–7. doi: 10.1016/j.tics.2014.01.010
172. McCrae RR, Costa PT Jr. Updating Norman’s “Adequate Taxonomy”: intelligence and personality dimensions in natural language and in questionnaires. J Pers Soc Psychol (1985) 49(3):710–21. doi: 10.1037//0022-3514.49.3.710
173. Martin ED, Sher KJ. Family history of alcoholism, alcohol-use disorders and the 5-factor model of personality. J Stud Alcohol (1994) 55:81–90. doi: 10.15288/jsa.1994.55.81
174. Ball SA, Tennen H, Poling JC, Kranzler HR, Rounsaville BJ. Personality, temperament, and character dimensions and the DSM-IV personality disorders in substance abusers. J Abnorm Psychol (1997) 106:545–53. doi: 10.1037/0021-843X.106.4.545
175. Terracciano A, Costa PT. Smoking and the five-factor model of personality. Addiction (2004) 99:472–81. doi: 10.1111/j.1360-0443.2004.00687.x
176. Raketic D, Barisic JV, Svetozarevic SM, Gazibara T, Tepavcevic DK, Milovanovic SD. Five-factor model personality profiles: the differences between alcohol and opiate addiction among females. Psychiatr Danub (2017) 29(1):74–80. doi: 10.24869/psyd.2017.74
177. Carter JA, Herbst JH, Stoller KB, King VL, Kidorf MS, Costa PT Jr, et al. Short-term stability of NEO-PI-R personality trait scores in opioid-dependent outpatients. Psychol Addict Behav (2001) 15(3):255–60. doi: 10.1037//0893-164X.15.3.255
178. Kornør H, Nordvik H. Five-factor model personality traits in opioid dependence. BMC Psychiatry (2007) 6(7):37. doi: 10.1186/1471-244X-7-37
179. McCrae RR, John OP. An introduction to the five-factor model and its applications. J Pers Disord (1992) 60(2):175–215. doi: 10.1111/j.1467-6494.1992.tb00970.x
180. Hernandez-Avila CA, Covault J, Gelernter J, Kranzler HR. Association study of personality factors and the Asn40Asp polymorphism at the mu-opioid receptor gene (OPRM1). Psychiatr Genet(2004) 14(2):89–92. doi: 10.1097/01.ypg.0000107931.32051.c7
181. Love TM, Stohler CS, Zubieta JK. Positron emission tomography measures of endogenous opioid neurotransmission and impulsiveness traits in humans. Arch Gen Psychiatry (2009) 66(10):1124–34. doi: 10.1001/archgenpsychiatry.2009.134
182. Smith GT, Fischer S, Cyders MA, Annus AM, Spillane NS, McCarthy DM. On the validity and utility of discriminating among impulsivity-like traits. Assessment (2007) 14(2):155–70. doi: 10.1177/1073191106295527
183. Fischer S, Anderson KG, Smith GT. Coping with distress by eating or drinking: role of trait urgency and expectancies. Psychol Addict Behav (2004) 18(3):269–74. doi: 10.1037/0893-164X.18.3.269
184. Frost JJ, Mayberg HS, Sadzot B, Dannals RF, Lever JR, Ravert HT, et al. Comparison of [11C]diprenorphine and [11C]carfentanil binding to opiate receptors in humans by positron emission tomography. J Cereb Blood Flow Metab (2001) 10(4):484–92. doi: 10.1038/jcbfm.1990.90
185. Kling MA, Carson RE, Borg L, Zametkin A, Matochik JA, Schluger J, et al. Opioid receptor imaging with positron emission tomography and [(18)F]cyclofoxy in long-term, methadone-treated former heroin addicts. J Pharmacol Exp Ther (2000) 295(3):1070–6.
186. Rothman RB, McLean S. An examination of the opiate receptor subtypes labeled by [3H]cycloFOXY: an opiate antagonist suitable for positron emission tomography. Biol Psychiatry (1988) 23(5):435–58. doi: 10.1016/0006-3223(88)90016-9
187. Hiller JM, Fan LQ. Laminar distribution of the multiple opioid receptors in the human cerebral cortex. Neurochem Res (1996) 21(11):1333–45. doi: 10.1007/BF02532374
188. Cohen RM, Andreason PJ, Doudet DJ, Carson RE, Sunderland T. Opiate receptor avidity and cerebral blood flow in Alzheimer’s disease. J Neurol Sci (1997) 148(2):171–80. doi: 10.1016/S0022-510X(96)05315-4
189. Lewis JW, Husbands SM. The orvinols and related opioids—high affinity ligands with diverse efficacy profiles. Curr Pharm Des (2004) 10:717–32. doi: 10.2174/1381612043453027
190. Negus SS, Bidlack JM, Mello NK, Furness MS, Rice KC, Brandt MR. Delta opioid antagonist effects of buprenorphine in rhesus monkeys. Behav Pharmacol (2002) 13:557–70. doi: 10.1097/00008877-200211000-00005
191. Richards ML, Sadee W. In vivo opiate receptor binding of oripavines to mu, delta and kappa sites in rat brain as determined by an ex vivo labeling method. Eur J Pharmacol (1985) 114:343–53. doi: 10.1016/0014-2999(85)90379-6
192. Greenwald M, Johanson CE, Bueller J, Chang Y, Moody DE, Kilbourn M, et al. Buprenorphine duration of action: mu-opioid receptor availability and pharmacokinetic and behavioral indices. Biol Psychiatry (2007) 61(1):101–10. doi: 10.1016/j.biopsych.2006.04.043
193. Kuhlman JJ Jr, Lalani S, Magluilo J Jr, Levine B, Darwin WD. Human pharmacokinetics of intravenous, sublingual, and buccal buprenorphine. J Clin Pharmacol (1997) 37:31–7. doi: 10.1093/jat/20.6.369
194. Greenwald MK, Johanson CE, Moody DE, Woods JH, Kilbourn MR, Koeppe RA, et al. Effects of buprenorphine maintenance dose on mu-opioid receptor availability, plasma concentrations, and antagonist blockade in heroin-dependent volunteers. Neuropsychopharmacology (2003) 28(11):2000–9. doi: 10.1038/sj.npp.1300251
195. Zubieta J, Greenwald MK, Lombardi U, Woods JH, Kilbourn MR, Jewett DM, et al. Buprenorphine-induced changes in μ-opioid receptor availability in male heroin-dependent volunteers: a preliminary study. Neuropsychopharmacology (2000) 23(3):326–34. doi: 10.1016/S0893-133X(00)00110-X
196. Al-Hasani R, Bruchas MR. Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology (2011) 115(6):1363–81. doi: 10.1097/ALN.0b013e318238bba6
197. Le Merrer J, Becker JA, Befort K, Kieffer BL. Reward processing by the opioid system in the brain. Physiol Rev (2009) 89(4):1379–412. doi: 10.1152/physrev.00005.2009
198. Minami M, Satoh M. Molecular biology of the opioid receptors: structures, functions and distributions. Neurosci Res (1995) 23(2):121–45. doi: 10.1016/0168-0102(95)00933-K
199. Simonin F, Gavériaux-Ruff C, Befort K, Matthes H, Lannes B, Micheletti G, et al. kappa-Opioid receptor in humans: cDNA and genomic cloning, chromosomal assignment, functional expression, pharmacology, and expression pattern in the central nervous system. Proc Natl Acad Sci U S A (1995) 92(15):7006–10. doi: 10.1073/pnas.92.15.7006
200. Meng F, Xie GX, Thompson RC, Mansour A, Goldstein A, Watson SJ, et al. Cloning and pharmacological characterization of a rat kappa opioid receptor. Proc Natl Acad Sci U S A (1993) 90(21):9954–8. doi: 10.1073/pnas.90.21.9954
201. Mansour A, Fox CA, Akil H, Watson SJ. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci (1995) 18(1):22–9. doi: 10.1016/0166-2236(95)93946-U
202. Attali B, Saya D, Vogel Z. Kappa-opiate agonists inhibit adenylate cyclase and produce heterologous desensitization in rat spinal cord. J Neurochem (1989) 52(2):360–9. doi: 10.1111/j.1471-4159.1989.tb09130.x
203. Lawrence DM, Bidlack JM. The kappa opioid receptor expressed on the mouse R1.1 thymoma cell line is coupled to adenylyl cyclase through a pertussis toxin-sensitive guanine nucleotide-binding regulatory protein. J Pharmacol Exp Ther (1993) 266(3):1678–83.
204. Tallent M, Dichter MA, Bell GI, Reisine T. The cloned kappa opioid receptor couples to an N-type calcium current in undifferentiated PC-12 cells. Neuroscience (1994) 63(4):1033–40. doi: 10.1016/0306-4522(94)90570-3
205. Pfeiffer A, Brantl V, Herz A, Emrich HM. Psychotomimesis mediated by kappa opiate receptors. Science (1986) 233:774–6. doi: 10.1126/science.3016896
206. Gonzalez D, Riba J, Bouso JC, Gomez-Jarabo G, Barbanoj MJ. Pattern of use and subjective effects of Salvia divinorum among recreational users. Drug Alcohol Depend (2006) 85:157–62. doi: 10.1016/j.drugalcdep.2006.04.001
207. Knoll AT, Meloni EG, Thomas JB, Carroll FI, Carlezon WA Jr. Anxiolytic-like effects of kappa-opioid receptor antagonists in models of unlearned and learned fear in rats. J Pharmacol Exp Ther (2007) 323:838–45. doi: 10.1124/jpet.107.127415
208. Valdez GR, Harshberger E. κ opioid regulation of anxiety-like behavior during acute ethanol withdrawal. Pharmacol Biochem Behav (2012) 102:1,44–7. doi: 10.1016/j.pbb.2012.03.019
209. Wang YJ, Hang A, Lu YC, Long Y, Zan GY, Li XP, et al. κ Opioid receptor activation in different brain regions differentially modulates anxiety-related behaviors in mice. Neuropharmacology (2016) 110(Pt A):92–101. doi: 10.1016/j.neuropharm.2016.04.022
210. Schlosburg JE, Whitfield TW Jr, Park PE, Crawford EF, George O, Vendruscolo LF, et al. Long-term antagonism of κ opioid receptors prevents escalation of and increased motivation for heroin intake. J Neurosci (2013) 33(49):19384–92. doi: 10.1523/JNEUROSCI.1979-13.2013
211. Sedki F, Eigenmann K, Gelinas J, Schouela N, Courchesne S, Shalev U. A role for kappa-, but not mu-opioid, receptor activation in acute food deprivation-induced reinstatement of heroin seeking in rats. Addict Biol (2015) 20(3):423–32. doi: 10.1111/adb.12133
212. Shirayama Y, Ishida H, Iwata M, Hazama G, Kawahara R, Duman RS. Stress increases dynorphin immunoreactivity in limbic brain regions and dynorphin antagonism produces antidepressant-like effects. J Neurochem (2004) 90:1258–68. doi: 10.1111/j.1471-4159.2004.02589.x
213. Schwarzer C. 30 years of dynorphins—new insights on their functions in neuropsychiatric diseases. Pharmacol Therap (2009) 123:353–70. doi: 10.1016/j.pharmthera.2009.05.006
214. Massaly N, Morón JA, Al-Hasani R. A trigger for opioid misuse: chronic pain and stress dysregulate the mesolimbic pathway and kappa opioid system. Front Neurosci (2016) 10:480. doi: 10.3389/fnins.2016.00480
215. Koob GF. Addiction is a reward deficit and stress surfeit disorder. Front Psychiatry (2013) 4:72. doi: 10.3389/fpsyt.2013.00072
216. Diana M, Pistis M, Muntoni A, Gessa G. Profound decrease of mesolimbic dopaminergic neuronal activity in morphine withdrawn rats. J Pharmacol Exp Ther (1995) 272(2):781–5.
217. Anderson SA, Michaelides M, Zarnegar P, Ren Y, Fagergren P, Thanos PK, et al. Impaired periamygdaloid-cortex prodynorphin is characteristic of opiate addiction and depression. J Clin Invest (2013) 123(12):5334–41. doi: 10.1172/JCI70395
218. Liu-Chen LY. Agonist-induced regulation and trafficking of kappa opioid receptors. Life Sci (2004) 75(5):511–36. doi: 10.1016/j.lfs.2003.10.041
219. Wang Y, Tang K, Inan S, Siebert D, Holzgrabe U, Lee DY, et al. Comparison of pharmacological activities of three distinct kappa ligands (Salvinorin A, TRK-820 and 3FLB) on kappa opioid receptors in vitro and their antipruritic and antinociceptive activities in vivo. J Pharmacol Exp Ther (2005) 312(1):220–30. doi: 10.1124/jpet.104.073668
220. Placzek MS, de Bittner GC, Wey HY, Lukas SE, Hooker JM. Immediate and persistent effects of salvinorin A on the kappa opioid receptor in rodents, monitored in vivo with PET. Neuropsychopharmacology (2015) 40(13):2865–72. doi: 10.1038/npp.2015.159
221. Mayer P, Hollt V. Pharmacogenetics of opioid receptors and addiction. Pharmacogenet Genomics (2006) 16(1):1–7. doi: 10.1097/01.fpc.0000182781.87932.0d
222. Wei LN, Hu X, Bi J, Loh H. Post-transcriptional regulation of mouse kappa-opioid receptor expression. Mol Pharmacol (2000) 57(2):401–8.
223. Votinov M, Pripfl J, Windischberger C, Kalcher K, Zimprich A, Zimprich F, et al. A genetic polymorphism of the endogenous opioid dynorphin modulates monetary reward anticipation in the corticostriatal loop. PLoS One (2014) 9(2):e89954. doi: 10.1371/journal.pone.0089954
224. Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res (2018) 20(3):311–9. doi: 10.1101/gr.094151.109
225. Colgan D, Manley J. Mechanism and regulation of mRNA polyadenylation. Genes Dev (1997) 11:2755–66. doi: 10.1101/gad.11.21.2755
226. Ford LP, Bagga PS, Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol Cell Biol (1997) 17:398–406. doi: 10.1128/MCB.17.1.398
227. Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature (1998) 392:516–20. doi: 10.1038/33192
228. Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell (1997) 89:831–8. doi: 10.1016/S0092-8674(00)80268-8
229. Yuferov V, Ji F, Nielsen DA, Levran O, Ho A, Morgello S, et al. A functional haplotype implicated in vulnerability to develop cocaine dependence is associated with reduced PDYN expression in human brain. Neuropsychopharmacology (2008) 34:1185. doi: 10.1038/npp.2008.187
230. Zimprich A, Kraus J, Wöltje M, Mayer P, Rauch E, Höllt V. An allelic variation in the human prodynorphin gene promoter alters stimulus-induced expression. J Neurochem (2000) 74:472–7. doi: 10.1046/j.1471-4159.2000.740472.x
231. Saify K, Saadat I, Saadat M. Association between VNTR polymorphism in promoter region of prodynorphin (PDYN) gene and heroin dependence. Psychiatry Res (2014) 219(3):690–2. doi: 10.1016/j.psychres.2014.06.048
232. Wang SC, Tsou HH, Chung RH, Chang YS, Fang CP, Chen CH, et al. The association of genetic polymorphisms in the κ-opioid receptor 1 gene with body weight, alcohol use, and withdrawal symptoms in patients with methadone maintenance. J Clin Psychopharmacol (2014) 34:205–11. doi: 10.1097/JCP.0000000000000082
233. Bruchas MR, Land BB, Lemos JC, Chavkin C. CRF1-R activation of the dynorphin/kappa opioid system in the mouse basolateral amygdala mediates anxiety-like behavior. PLoS One (2009) 4:e8528. doi: 10.1371/journal.pone.0008528
234. Kastenberger I, Lutsch C, Herzog H, Schwarzer C. Influence of sex and genetic background on anxiety-related and stress-induced behaviour of prodynorphin-deficient mice. PLoS One (2012) 7:3,e34251. doi: 10.1371/journal.pone.0034251
235. Luo X, Zuo L, Kranzler H, Zhang H, Wang S, Gelernter J. Multiple OPR genes influence personality traits in substance dependent and healthy subjects in two American populations. Am J Med Genet B Neuropsychiatr Genet (2008) 147B(7):1028–39. doi: 10.1002/ajmg.b.30701
236. Flory JD, Pytte CL, Hurd Y, Ferrell RE, Manuck SB. Alcohol dependence, disinhibited behavior and variation in the prodynorphin gene. Biol Psychol (2011) 88(1):51–6. doi: 10.1016/j.biopsycho.2011.06.007
237. Talbot PS, Narendran R, Butelman ER, Huang Y, Ngo K, Slifstein M, et al. 11C-GR103545, a radiotracer for imaging kappa-opioid receptors in vivo with PET: synthesis and evaluation in baboons. J Nucl Med (2005) 46(3):484–94.
238. Tomasi G, Nabulsi N, Zheng MQ, Weinzimmer D, Ropchan J, Blumberg L, et al. Determination of in vivo Bmax and Kd for 11C-GR103545, an agonist PET tracer for κ-opioid receptors: a study in nonhuman primates. J Nucl Med (2013) 54(4):600–8. doi: 10.2967/jnumed.112.112672
239. Kim SJ, Zheng MQ, Nabulsi N, Labaree D, Ropchan J, Najafzadeh S, et al. Determination of the in vivo selectivity of a new κ-opioid receptor antagonist PET tracer 11C-LY2795050 in the rhesus monkey. J Nucl Med (2013) 54(9):1668–74. doi: 10.2967/jnumed.112.118877
240. Li S, Zheng MQ, Naganawa M, Kim S, Gao H, Kapinos M, et al. Development and in vivo evaluation of a novel kappa opioid receptor agonist as PET radiotracer with superior imaging characteristics. J Nucl Med (2019) 60(7):1023–30. doi: 10.2967/jnumed.118.220517
241. Naganawa M, Jacobsen LK, Zheng MQ, Lin SF, Banerjee A, Byon W, et al. Evaluation of the agonist PET radioligand [¹¹C]GR103545 to image kappa opioid receptor in humans: kinetic model selection, test-retest reproducibility and receptor occupancy by the antagonist PF-04455242. Neuroimage (2014a) 99:69–79. doi: 10.1016/j.neuroimage.2014.05.033
242. Naganawa M, Zheng MQ, Nabulsi N, Tomasi G, Henry S, Lin SF, et al. Kinetic modeling of (11)C-LY2795050, a novel antagonist radiotracer for PET imaging of the kappa opioid receptor in humans. J Cereb Blood Flow Metab (2014b) 34(11):1818–25. doi: 10.1038/jcbfm.2014.150
243. Naganawa M, Zheng MQ, Henry S, Nabulsi N, Lin SF, Ropchan J, et al. Test-retest reproducibility of binding parameters in humans with 11C-LY2795050, an antagonist PET radiotracer for the κ opioid receptor. J Nucl Med (2015) 56(2):243–8. doi: 10.2967/jnumed.114.147975
244. Matuskey D, Dias M, Naganawa M, Pittman B, Henry S, Li S, et al. Social status and demographic effects of the kappa opioid receptor: a PET imaging study with a novel agonist radiotracer in healthy volunteers. Neuropsychopharmacology (2019) 44:1714–19. doi: 10.1038/s41386-019-0379-7
245. Vijay A, Cavallo D, Goldberg A, de Laat B, Nabulsi N, Huang Y, et al. PET imaging reveals lower kappa opioid receptor availability in alcoholics but no effect of age. Neuropsychopharmacology (2018) 43(13):2539–47. doi: 10.1038/s41386-018-0199-1
246. Vijay A, Wang S, Worhunsky P, Zheng MQ, Nabulsi N, Ropchan J, et al. PET imaging reveals sex differences in kappa opioid receptor availability in humans, in vivo. Am J Nucl Med Mol Imaging (2016) 6:4,205–14.
248. Pettinati HM, Kampman KM, Lynch KG, Suh JJ, Dackis CA, Oslin DW, et al. Gender differences with high-dose naltrexone in patients with co-occurring cocaine and alcohol dependence. J Subst Abuse Treat (2007) 34(4):378–90. doi: 10.1016/j.jsat.2007.05.011
249. O’Malley SS, Krishnan-Sarin S, Farren C, O’Connor PG. Naltrexone-induced nausea in patients treated for alcohol dependence: clinical predictors and evidence for opioid-mediated effects. J Clin Psychopharmacol (2000) 20:69–76. doi: 10.1097/00004714-200002000-00012
250. Baros A, Latham P, Anton R. Naltrexone and cognitive behavioral therapy for the treatment of alcohol dependence: do sex differences exist? Alcohol Clin Exp Res (2008) 32:771–6. doi: 10.1111/j.1530-0277.2008.00633.x
251. Greenfield SF, Pettinati HM, O’Malley S, Randall PK, Randall CL. Gender differences in alcohol treatment: an analysis of outcome from the COMBINE study. Alcohol Clin Exp Res (2010) 34(10):1803–12. doi: 10.1111/j.1530-0277.2010.01267.x
252. Saxon AJ. The unmet challenges of co-occurring alcohol and opioid use. Alcohol Clin Exp Res (2018) 42:1406–7. doi: 10.1111/acer.13797
253. Hughes A, Williams MR, Lipari RN, Bose J, Copello EAP, Kroutil LA. Prescription drug use and misuse in the United States: Results from the 2015 National Survey on Drug Use and Health. NSDUH Data Rev (2016). Retrieved from http://www.samhsa.gov/data/.
254. Kerridge B. Nonmedical prescription opioid use and DSM-5 nonmedical prescription opioid use disorder in the United States. Drug Alcohol Depen(2016) 156:47–56. doi: 10.1016/j.drugalcdep.2015.08.026
255. Pradhan AA, Befort K, Nozaki C, Gavériaux-Ruff C, Kieffer BL. The delta opioid receptor: an evolving target for the treatment of brain disorders. Trends Pharmacol Sci (2011) 32(10):581–90. doi: 10.1016/j.tips.2011.06.008
256. Saitoh A, Kimura Y, Suzuki T, Kawai K, Nagase H, Kamei J. Potential anxiolytic and antidepressant-like activities of SNC80, a selective delta-opioid agonist, in behavioral models in rodents. J Pharmacol Sci (2004) 95:374–80. doi: 10.1254/jphs.FPJ04014X
257. Jutkiewicz EM, Rice KC, Traynor JR, Woods JH. Separation of the convulsions and antidepressant-like effects produced by the delta-opioid agonist SNC80 in rats. Psychopharmacology (Berl) (2005) 182:588–96. doi: 10.1007/s00213-005-0138-9
258. Simonin F, Befort K, Gavériaux-Ruff C, Matthes H, Nappey V, Lannes B, et al. The human delta-opioid receptor: genomic organization, cDNA cloning, functional expression, and distribution in human brain. Mol Pharmacol (1994) 46:6,1015–21.
259. Peckys D, Landwehrmeyer GB. Expression of mu, kappa, and delta opioid receptor messenger RNA in the human CNS: a 33P in situ hybridization study. Neuroscience (1999) 88(4):1093–135. doi: 10.1016/S0306-4522(98)00251-6
260. Peng J, Sarkar S, Chang SL. Opioid receptor expression in human brain and peripheral tissues using absolute quantitative real-time RT-PCR. Drug Alcohol Depend (2012) 124(3):223–8. doi: 10.1016/j.drugalcdep.2012.01.013
261. Rezaï X, Faget L, Bednarek E, Schwab Y, Kieffer BL, Massotte D. Mouse δ opioid receptors are located on presynaptic afferents to hippocampal pyramidal cells. Cell Mol Neurobiol (2012) 32:509–16. doi: 10.1007/s10571-011-9791-1
262. Hirose N, Murakawa K, Takada K, Oi Y, Suzuki T, Nagase H, et al. Interactions among mu- and delta-opioid receptors, especially putative delta1- and delta2-opioid receptors, promote dopamine release in the nucleus accumbens. Neuroscience (2005) 135(1):213–25. doi: 10.1016/j.neuroscience.2005.03.065
263. Onali P, Olianas MC. G protein activation and cyclic AMP modulation by naloxone benzoylhydrazone in distinct layers of rat olfactory bulb. Br J Pharmacol (2004) 143:638–48. doi: 10.1038/sj.bjp.0705951
264. Olianas MC, Dedoni S, Olianas A, Onali P. δ-Opioid receptors stimulate the metabolic sensor AMP-activated protein kinase through coincident signaling with G(q/11)-coupled receptors. Mol Pharmacol (2012) 81:154–65. doi: 10.1124/mol.111.075267
265. Yao L, Fan P, Jiang Z, Mailliard WS, Gordon AS, Diamond I. Addicting drugs utilize a synergistic molecular mechanism in common requiring adenosine and Gi-beta gamma dimers. Proc Natl Acad Sci U S A (2003) 100:14379–84. doi: 10.1073/pnas.2336093100
266. Chefer VI, Shippenberg TS. Augmentation of morphine-induced sensitization but reduction in morphine tolerance and reward in delta-opioid receptor knockout mice. Neuropsychopharmacology (2009) 34(4):887–98. doi: 10.1038/npp.2008.128
267. Shippenberg TS, Chefer VI, Thompson AC. Delta-opioid receptor antagonists prevent sensitization to the conditioned rewarding effects of morphine. Biol Psychiatry (2009) 65:2,169–74. doi: 10.1016/j.biopsych.2008.09.009
268. Suzuki T, Tsuji M, Mori T, Misawa M, Endoh T, Nagase H. Effect of the highly selective and nonpeptide delta opioid receptor agonist TAN-67 on the morphine-induced place preference in mice. J Pharmacol Exp Ther (1996) 279:1,177–85.
269. Lutz PE, Ayranci G, Chu-Sin-Chung P, Matifas A, Koebel P, Filliol D, et al. Distinct mu, delta, and kappa opioid receptor mechanisms underlie low sociability and depressive-like behaviors during heroin abstinence. Neuropsychopharmacology (2014) 39(11):2694–705. doi: 10.1038/npp.2014.126
270. Leskela TT, Markkanen PM, Alahuhta IA, Tuusa JT, Petaja-Repo UE. Phe27Cys polymorphism alters the maturation and subcellular localization of the human delta opioid receptor. Traffic (2009) 10(1):116–29. doi: 10.1111/j.1600-0854.2008.00846.x
271. Zhang H, Gelernter J, Gruen JR, Kranzler HR, Herman AI, Simen AA. Functional impact of a single-nucleotide polymorphism in the OPRD1 promoter region. J Hum Genet (2010) 55:278. doi: 10.1038/jhg.2010.22\
272. Huang MW, Chiang TA, Lo PY, Huang CS. Relationship among methadone dose, polymorphisms of dopamine D2 receptor and tri-dimensional personality questionnaire in heroin-addicted patients. Behav Brain Funct (2016) 12(1):24. doi: 10.1186/s12993-016-0109-9
273. Rashid F, Zafar MM, Ahmed I, Jabeen S, Ahmad MS, Minhas NM, et al. Association of OPRD1 rs569356 SNP with stress response in opioid addicts. Presented at 20th European Congress of Endocrinology, Barcelona, Spain. Endocrine Abstracts (2018). 56 P669. doi: 10.1530/endoabs.56.P669
274. Madar I, Lever JR, Kinter CM, Scheffel U, Ravert HT, Musachio JL, et al. Imaging of δ opioid receptors in human brain by N1′- ([11C]methyl)naltrindole and PET. Synapse (1996) 24:19–28. doi: 10.1002/(SICI)1098-2396(199609)24:1<19::AID-SYN3>3.0.CO;2-J
275. Weerts EM, Wand GS, Kuwabara H, Munro CA, Dannals RF, Hilton J, et al. Positron emission tomography imaging of mu- and delta-opioid receptor binding in alcohol-dependent and healthy control subjects. Alcohol Clin Exp Res (2011) 35:2162–73. doi: 10.1111/j.1530-0277.2011.01565.x
276. Weerts EM, Kim YK, Wand GS, Dannals RF, Lee JS, Frost JJ, et al. Differences in δ- and μ-opioid receptor blockade measured by positron emission tomography in naltrexone-treated recently abstinent alcohol-dependent subjects. Neuropsychopharmacology (2007) 33:653. doi: 10.1038/sj.npp.1301440
277. Wand GS, Weerts EM, Kuwabara H, Wong DF, Xu X, McCaul ME. The relationship between naloxone-induced cortisol and delta opioid receptor availability in mesolimbic structures is disrupted in alcohol-dependent subjects. Addict Biol (2013) 18:181–92. doi: 10.1111/j.1369-1600.2011.00430.x
278. Nieto MM, Guen SLE, Kieffer BL, Roques BP, Noble F. Physiological control of emotion-related behaviors by endogenous enkephalins involves essentially the delta opioid receptors. Neuroscience (2005) 135:305–13. doi: 10.1016/j.neuroscience.2005.06.025
279. Mollereau C, Parmentier M, Mailleux P, Butour JL, Moisand C, Chalon P, et al. ORL1, a novel member of the opioid receptor family. FEBS Lett (1994) 341(1):33–8. doi: 10.1016/0014-5793(94)80235-1
280. Meunier JC, Mollereau C, Toll L, Suandeau C, Moisand C, Alvinerie P, et al. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature (1995) 377:532–5. doi: 10.1038/377532a0
281. Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, et al. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science (1995) 270(5237):792–4. doi: 10.1126/science.270.5237.792
282. Mollereau C, Mouledous L. Tissue distribution of the opioid receptor-like (ORL1) receptor. Peptides (2000) 21:907–17. doi: 10.1016/S0196-9781(00)00227-8
283. Mika J, Obara I, Przewlocka B. The role of nociceptin and dynorphin in chronic pain: implications of neuro-glial interaction. Neuropeptides (2011) 45(4):247–61. doi: 10.1016/j.npep.2011.03.002
284. Chiou LC, Liao YY, Fan PC, Kuo PH, Wang CH, Riemer C, et al. Nociceptin/orphanin FQ peptide receptors: pharmacology and clinical implications. Curr Drug Targets (2007) 8(1):117–35. doi: 10.2174/138945007779315605
285. Jenck F, Wichmann J, Dautzenberg FM, Moreau JL, Ouagazzal AM, Martin JR, et al. A synthetic agonist at the orphanin FQ/nociceptin receptor ORL1: anxiolytic profile in the rat. Proc Natl Acad Sci U S A (2000) 97(9):4938–43. doi: 10.1073/pnas.090514397
286. Gavioli EC, Vaughan CW, Marzola G, Guerrini R, Mitchell VA, Zucchini S, et al. Antidepressant-like effects of the nociceptin/orphanin FQ receptor antagonist UFP-101: new evidence from rats and mice. Arch Pharmacol (2004) 369:547–53. doi: 10.1007/s00210-004-0939-0
287. Martin-Fardon R, Zorrilla EP, Ciccocioppo R, Weiss F. Role of innate and drug-induced dysregulation of brain stress and arousal systems in addiction: focus on corticotropin-releasing factor, nociceptin/orphanin FQ, and orexin/hypocretin. Brain Res (2010) 1314:145–61. doi: 10.1016/j.brainres.2009.12.027
288. Nabeshima T, Noda Y, Mamiya T. The role of nociceptin in cognition. Brain Res (1999) 848(1–2):167–73. doi: 10.1016/S0006-8993(99)01906-X
289. Sakoori K, Murphy NP. Endogenous nociceptin (orphanin FQ) suppresses basal hedonic state and acute reward responses to methamphetamine and ethanol, but facilitates chronic responses. Neuropsychopharmacology (2008) 33:877–91. doi: 10.1038/sj.npp.1301459
290. Lohith TG, Zoghbi SS, Morse CL, Araneta MF, Barth VN, Goebl NA, et al. Brain and whole-body imaging of nociceptin/orphanin FQ peptide receptor in humans using the PET ligand 11C-NOP-1A. J Nucl Med (2012) 53(3):385–92. doi: 10.2967/jnumed.111.097162
291. Devine DP, Watson SJ, Akil H. Nociceptin/orphanin FQ regulates neuroendocrine function of the limbic–hypothalamic–pituitary–adrenal axis. Neuroscience (2001) 102(3):541–53. doi: 10.1016/S0306-4522(00)00517-0
292. Ciccocioppo R, de Guglielmo G, Hansson AC, Ubaldi M, Kallupi M, Cruz MT, et al. Restraint stress alters nociceptin/orphanin FQ and CRF systems in the rat central amygdala: significance for anxiety-like behaviors. J Neurosci (2014) 34(2):363–72. doi: 10.1523/JNEUROSCI.2400-13.2014
293. Cruz MT, Herman MA, Kallupi M, Roberto M. Nociceptin/orphanin FQ blockade of corticotropin-releasing factor-induced gamma-aminobutyric acid release in central amygdala is enhanced after chronic ethanol exposure. Biol Psychiatry (2012) 71(8):666–76. doi: 10.1016/j.biopsych.2011.10.032
294. Ciccocioppo R, Fedeli A, Economidou D, Policani F, Weiss F, Massi M. The bed nucleus is a neuroanatomical substrate for the anorectic effect of corticotropin-releasing factor and for its reversal by nociceptin/orphanin FQ. J Neurosci (2003) 23(28):9445–51. doi: 10.1523/JNEUROSCI.23-28-09445.2003
295. Rodi D, Zucchini S, Simonato M, Cifani C, Massi M, Polidori C. Functional antagonism between nociceptin/orphanin FQ (N/OFQ) and corticotropin-releasing factor (CRF) in the rat brain: evidence for involvement of the bed nucleus of the stria terminalis. Psychopharmacology (Berl) (2008) 196(4):523–31. doi: 10.1007/s00213-007-0985-7
296. Vorspan F, Mehtelli W, Dupuy G, Bloch V, Lépine JP. Anxiety and substance use disorders: co-occurrence and clinical issues. Curr Psychiatry Rep (2015) 17(2):4. doi: 10.1007/s11920-014-0544-y
297. Murphy NP, Ly HT, Maidment NT. Intracerebroventricular orphanin FQ/nociceptin suppresses dopamine release in the nucleus accumbens of anaesthetized rats. Neuroscience (1996) 75:1–4. doi: 10.1016/0306-4522(96)00322-3
298. Di Giannuario A, Pieretti S, Catalani A, Loizzo A. Orphanin FQ reduces morphine-induced dopamine release in the nucleus accumbens: a microdialysis study in rats. Neurosci Lett (1999) 272(3):183–6. doi: 10.1016/S0304-3940(99)00579-0
299. Ciccocioppo R, Panocka I, Polidori C, Regoli D, Massi M. Effect of nociceptin on alcohol intake in alcohol-preferring rats. Psychopharmacology (Berl) (1999) 141:220–4. doi: 10.1007/s002130050828
300. Kuzmin A, Sandin J, Terenius L, Ogren SO. Acquisition, expression, and reinstatement of ethanol-induced conditioned place preference in mice: effects of opioid receptor-like 1 receptor agonists and naloxone. J Pharmacol Exp Ther (2003) 304:310–8. doi: 10.1124/jpet.102.041350
301. Kotlinska J, Rafalski P, Biala G, Dylag T, Rolka K, Silberring J. Nociceptin inhibits acquisition of amphetamine-induced place preference and sensitization to stereotypy in rats. Eur J Pharmacol (2003) 474:233–9. doi: 10.1016/S0014-2999(03)02081-8
302. Kotlinska J, Wichmann J, Legowska A, Rolka K, Silberring J. Orphanin FQ/nociceptin but not Ro 65-6570 inhibits the expression of cocaine-induced conditioned place preference. Behav Pharmacol (2002) 13:229–35. doi: 10.1097/00008877-200205000-00006
303. Sakoori K, Murphy NP. Central administration of nociceptin/orphanin FQ blocks the acquisition of conditioned place preference to morphine and cocaine, but not conditioned place aversion to naloxone in mice. Psychopharmacology (Berl) (2004) 172:129–36. doi: 10.1007/s00213-003-1643-3
304. Ciccocioppo R, Angeletti S, Sanna PP, Weiss F, Massi M. Effect of nociceptin/orphanin FQ on the rewarding properties of morphine. Eur J Pharmacol (2000) 404:153–9. doi: 10.1016/S0014-2999(00)00590-2
305. Walker JR, Spina M, Terenius L, Koob GF. Nociceptin fails to affect heroin self-administration in the rat. Neuroreport (1998) 9:2243–7. doi: 10.1097/00001756-199807130-00017
306. Rorick-Kehn LM, Ciccocioppo R, Wong CJ, Witkin JM, Martinez-Grau MA, Stopponi S, et al. A novel, orally bioavailable nociceptin receptor antagonist, LY2940094, reduces ethanol self-administration and ethanol seeking in animal models. Alcohol Clin Exp Res (2016) 40(5):945–54. doi: 10.1111/acer.13052
307. Post A, Smart TS, Jackson K, Mann J, Mohs R, Rorick-Kehn L, et al. Proof-of-concept study to assess the nociceptin receptor antagonist LY2940094 as a new treatment for alcohol dependence. Alcohol Clin Exp Res (2016) 40(9):1935–44. doi: 10.1111/acer.13147
308. Kuzmin A, Bazov I, Sheedy D, Garrick T, Harper C, Bakalkin G. Expression of pronociceptin and its receptor is downregulated in the brain of human alcoholics. Brain Res (2009) 1305(Suppl):S80–5. doi: 10.1016/j.brainres.2009.05.067
309. Lutz PE, Zhou Y, Labbe A, Mechawar N, Turecki G. Decreased expression of nociceptin/orphanin FQ in the dorsal anterior cingulate cortex of suicides. Eur Neuropsychopharmacol (2015) 25:11,2008–14. doi: 10.1016/j.euroneuro.2015.08.015
310. Pike VW, Rash KS, Chen Z, Pedregal C, Statnick MA, Kimura Y, et al. Synthesis and evaluation of radioligands for imaging brain nociceptin/orphanin FQ peptide (NOP) receptors with positron emission tomography. J Med Chem (2011) 54(8):2687–700. doi: 10.1021/jm101487v
311. Lohith TG, Zoghbi SS, Morse CL, Araneta MD, Barth VN, Goebl NA, et al. Retest imaging of [11C]NOP-1A binding to nociceptin/orphanin FQ peptide (NOP) receptors in the brain of healthy humans. Neuroimage (2014) 87:89–95. doi: 10.1016/j.neuroimage.2013.10.068
312. Hostetler ED, Sanabria-Bohórquez S, Eng W, Joshi AD, Patel S, Gibson RE, et al. Evaluation of [¹⁸F]MK-0911, a positron emission tomography (PET) tracer for opioid receptor-like 1 (ORL1), in rhesus monkey and human. Neuroimage (2013) 68:1–10. doi: 10.1016/j.neuroimage.2012.11.053
313. Narendran R, Ciccocioppo R, Lopresti B, Paris J, Himes ML, Mason NS. Nociceptin receptors in alcohol use disorders: a positron emission tomography study using [11C]NOP-1A. Biol Psychiatry (2018) 84(10):708–14. doi: 10.1016/j.biopsych.2017.05.019
314. Lindholm S, Ploj K, Franck J, Nylander I. Nociceptin/orphanin FQ tissue concentration in the rat brain. Prog Neuropsychopharmacol Biol Psychiatry (2002) 26(2):303–6. doi: 10.1016/S0278-5846(01)00270-6
315. Narendran R, Tollefson S, Himes ML, Paris J, Lopresti B, Ciccocioppo R, et al. Nociceptin receptors upregulated in cocaine use disorder: a positron emission tomography imaging study using [11C]NOP-1A. Am J Psychiatry (2019) 176(6):468–76. doi: 10.1176/appi.ajp.2019.18081007
316. Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ Jr, et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science (1990) 250(4986):1429–32. doi: 10.1126/science.2147780
317. Trifilieff P, Feng B, Urizar E, Winiger V, Ward RD, Taylor KM, et al. Increasing dopamine D2 receptor expression in the adult nucleus accumbens enhances motivation. Mol Psychiatry (2013) 18(9):1025–33. doi: 10.1038/mp.2013.57
318. Zijlstra F, Booij J, van den Brink W, Franken IHA. Striatal dopamine D2 receptor binding and dopamine release during cue-elicited craving in recently abstinent opiate-dependent males. Eur Neuropsychopharmacol (2008) 18(4):262–70. doi: 10.1016/j.euroneuro.2007.11.002
319. Usiello A, Baik JH, Rougé-Pont F, Picetti R, Dierich A, LeMeur M, et al. Distinct functions of the two isoforms of dopamine D2 receptors. Nature (2000) 408:6809,199–203. doi: 10.1038/35041572
320. Volkow ND, Morales M. The brain on drugs: from reward to addiction. Cell(2015) 162(4):712–25. doi: 10.1016/j.cell.2015.07.046
321. Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANKK1: a novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat (2004) 23(6):540–5. doi: 10.1002/humu.20039
322. Meylan E, Tschopp J. The RIP kinases: crucial integrators of cellular stress. Trends Biochem Sci (2005) 30(3):151–9. doi: 10.1016/j.tibs.2005.01.003
323. Bontempi S, Fiorentini C, Busi C, Guerra N, Spano P, Missale C. Identification and characterization of two nuclear factor-kappaB sites in the regulatory region of the dopamine D2 receptor. Endocrinology (2007) 148(5):2563–70. doi: 10.1210/en.2006-1618
324. Norman AB, Tabet MR, Norman MK, Fey BK, Tsibulsky VL, Millard RW. The affinity of D2-like dopamine receptor antagonists determines the time to maximal effect on cocaine self-administration. J Pharmacol Exp Ther (2011) 338(2):724–8. doi: 10.1124/jpet.111.183244
325. Caine SB, Negus SS, Mello NK, Patel S, Bristow L, Kulagowski J, et al. Role of dopamine D2-like receptors in cocaine self-administration: studies with D2 receptor mutant mice and novel D2 receptor antagonists. J Neurosci (2002) 22(7):2977–88. doi: 10.1523/JNEUROSCI.22-07-02977.2002
326. Maldonado R, Saiardi A, Valverde O, Samad TA, Roques BP, Borrelli E. Absence of opiate rewarding effects in mice lacking dopamine D2 receptors. Nature (1997) 388:6642,586–9. doi: 10.1038/41567
327. Rominger A, Cumming P, Xiong G, Koller G, Boning G, Wulff M, et al. [18F]fallypride PET measurement of striatal and extrastriatal dopamine D 2/3 receptor availability in recently abstinent alcoholics. Addict Biol (2012) 17(2):490–503. doi: 10.1111/j.1369-1600.2011.00355.x
328. Volkow ND, Wang GJ, Fowler JS, Thanos PP, Logan J, Gatley SJ, et al. Brain DA D2 receptors predict reinforcing effects of stimulants in humans: replication study. Synapse (2002) 46:2,79–82. doi: 10.1002/syn.10137
329. Volkow ND, Wang GJ, Fowler JS, Logan J, Gatley SJ, Gifford A, et al. Prediction of reinforcing responses to psychostimulants in humans by brain dopamine D2 receptor levels. Am J Psychiatry (1999) 156(9):1440–3. doi: 10.1176/ajp.156.9.1440
330. Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, et al. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA (1990) 263(15):2055–60. doi: 10.1001/jama.263.15.2055
331. Noble EP, Blum K, Khalsa ME, Ritchie T, Montgomery A, Wood RC, et al. Allelic association of the D2 dopamine receptor gene with cocaine dependence. Drug Alcohol Depend (1993) 33(3):271–85. Erratum in: Drug Alcohol Depend. 34:1,83-4. doi: 10.1016/0376-8716(93)90113-5
332. Comings DE, Rosenthal RJ, Lesieur HR, Rugle LJ, Muhleman D, Chiu C, et al. A study of the dopamine D2 receptor gene in pathological gambling. Pharmacogenetics (1996) 6(3):223–34. doi: 10.1097/00008571-199606000-00004
333. Blum K, Braverman ER, Wood RC, Gill J, Li C, Chen TJ, et al. Increased prevalence of the Taq I A1 allele of the dopamine receptor gene (DRD2) in obesity with comorbid substance use disorder: a preliminary report. Pharmacogenetics (1996a) 6(4):297–305. doi: 10.1097/00008571-199608000-00003
334. Jönsson EG, Nöthen MM, Grünhage F, Farde L, Nakashima Y, Propping P, et al. Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol Psychiatry (1999) 4(3):290–6. doi: 10.1038/sj.mp.4000532
335. Eisenstein SA, Bogdan R, Love-Gregory L, Corral-Frías NS, Koller JM, Black KJ, et al. Prediction of striatal D2 receptor binding by DRD2/ANKK1 TaqIA allele status. Synapse (2016) 70(10):418–31. doi: 10.1002/syn.21916
336. Wiers CE, Towb PC, Hodgkinson CA, Shen PH, Freeman C, Miller G, et al. Association of genetic ancestry with striatal dopamine D2/D3 receptor availability. Mol Psychiatry (2018) 23:8,1711–1716. doi: 10.1038/mp.2017.208
337. Vereczkei A, Demetrovics Z, Szekely A, Sarkozy P, Antal P, Szilagyi A, et al. Multivariate analysis of dopaminergic gene variants as risk factors of heroin dependence. PLoS One (2013) 8(6):e66592. doi: 10.1371/journal.pone.0066592
338. Ritchie T, Noble EP. Association of seven polymorphisms of the D2 dopamine receptor gene with brain receptor-binding characteristics. Neurochem Res (2003) 28(1):73–82. doi: 10.1023/A:1021648128758
339. Arinami T, Gao M, Hamaguchi H, Toru M. A functional polymorphism in the promoter region of the dopamine D2 receptor gene is associated with schizophrenia. Human Mol Genet (1997) 6(4):577–82. doi: 10.1093/hmg/6.4.577
340. Hirvonen MM, Lumme V, Hirvonen J, Pesonen U, Någren K, Vahlberg T, et al. C957T polymorphism of the human dopamine D2 receptor gene predicts extrastriatal dopamine receptor availability in vivo. Prog Neuropsychopharmacol Biol Psychiatry (2009) 33(4):630–6. doi: 10.1016/j.pnpbp.2009.02.021
341. Clarke TK, Weiss AR, Ferarro TN, Kampman KM, Dackis CA, Pettinati HM, et al. The dopamine receptor D2 (DRD2) SNP rs1076560 is associated with opioid addiction. Ann Hum Genet (2014b) 78(1):33–9. doi: 10.1111/ahg.12046
342. Moyer RA, Wang D, Papp AC, Smith RM, Duque L, Mash DC, et al. Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology (2011) 36(4):753–62. doi: 10.1038/npp.2010.208
343. Smith JW, Fetsko LA, Xu R, Wang Y. Dopamine D2L receptor knockout mice display deficits in positive and negative reinforcing properties of morphine and in avoidance learning. Neuroscience (2002) 4:755–65:113. doi: 10.1016/S0306-4522(02)00257-9
344. Bertolino A, Taurisano P, Pisciotta NM, Blasi G, Fazio L, Romano R, et al. Genetically determined measures of striatal D2 signaling predict prefrontal activity during working memory performance. PLoS ONE (2010) 5:2. doi: 10.1371/journal.pone.0009348
345. Taurisano P, Romano R, Mancini M, Giorgio A, DiAntonucci LA, Fazio L, et al. Prefronto-striatal physiology is associated with schizotypy and is modulated by a functional variant of DRD2. Front Behav Neurosci (2014) 8:235. doi: 10.3389/fnbeh.2014.00235
346. Zhu F, Yan CX, Wen YC, Wang J, Bi J, Zhao YL, et al. Dopamine D1 receptor gene variation modulates opioid dependence risk by affecting transition to addiction. PLoS One (2013) 8(8):e70805. doi: 10.1371/journal.pone.0070805
347. Jacobs MM, Ökvist A, Horvath M, Keller E, Bannon MJ, Morgello S, et al. Dopamine receptor D1 and postsynaptic density gene variants associate with opiate abuse and striatal expression levels. Mol Psychiatry (2013) 18(11):1205–10. doi: 10.1038/mp.2012.140
348. Levran O, Randesi M, da Rosa JC, Ott J, Rotrosen J, Adelson M, et al. Overlapping dopaminergic pathway genetic susceptibility to heroin and cocaine addictions in African Americans. Ann Hum Genet (2015) 79(3):188–98. doi: 10.1111/ahg.12104
349. Levran O, Londono D, O’Hara K, Randesi M, Rotrosen J, Casadonte P, et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav (2009) 5:531–40. doi: 10.1111/j.1601-183X.2009.00501.x
350. Peng S, Du J, Jiang H, Fu Y, Chen H, Sun H, et al. The dopamine receptor D1 gene is associated with the length of interval between first heroin use and onset of dependence in Chinese Han heroin addicts. J Neural Transm (Vienna) (2013) 120(11):1591–8. doi: 10.1007/s00702-013-1029-6
351. Wang N, Zhang JB, Zhao J, Cai XT, Zhu YS, Li SB. Association between dopamine D2 receptor gene polymorphisms and the risk of heroin dependence. Genet Mol Res (2016) 15:4. doi: 10.4238/gmr15048772
352. Al-Eitan LN, Jaradat SA, Hulse GK, Tay GK. Custom genotyping for substance addiction susceptibility genes in Jordanians of Arab descent. BMC Res Notes (2012) 5:497. doi: 10.1186/1756-0500-5-497
353. Tsou CC, Chou HW, Ho PS, Kuo SC, Chen CY, Huang CC, et al. DRD2 and ANKK1 genes associate with late-onset heroin dependence in men. World J Biol Psychiatry (2017) 25:1–11. doi: 10.1080/15622975.2017.1372630
354. Zhang J, Yan P, Li Y, Cai X, Yang Z, Miao X, et al. A 35.8 kilobases haplotype spanning ANKK1 and DRD2 is associated with heroin dependence in Han Chinese males. Brain Res (2018) 1688:54–64. doi: 10.1016/j.brainres.2018.03.017
355. Xu K, Lichtermann D, Lipsky RH, Franke P, Liu X, Hu Y, et al. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry (2004) 61(6):597–606. doi: 10.1001/archpsyc.61.6.597
356. Teh LK, Izuddin AF MHFH, Zakaria ZA, Salleh MZ. Tridimensional personalities and polymorphism of dopamine D2 receptor among heroin addicts. Biol Res Nurs (2012) 14(2):188–96. doi: 10.1177/1099800411405030
357. Hung CC, Chiou MH, Huang BH, Hsieh YW, Hsieh TJ, Huang CL, et al. Impact of genetic polymorphisms in ABCB1, CYP2B6, OPRM1, ANKK1 and DRD2 genes on methadone therapy in Han Chinese patients. Pharmacogenomics (2011) 12(11):1525–33. doi: 10.2217/pgs.11.96
358. Bawor M, Dennis BB, Tan C, Pare G, Varenbut M, Daiter J, et al. Contribution of BDNF and DRD2 genetic polymorphisms to continued opioid use in patients receiving methadone treatment for opioid use disorder: an observational study. Addict Sci Clin Pract (2015) 10:19. doi: 10.1186/s13722-015-0040-7
359. Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. ANKK1, TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA Psychiatry (2013) 70(3):325–33. doi: 10.1001/jamapsychiatry.2013.282
360. Hou QF, Li SB. Potential association of DRD2 and DAT1 genetic variation with heroin dependence. Neurosci Lett (2009) 464(2):127–30. doi: 10.1016/j.neulet.2009.08.004
361. Barratt DT, Coller JK, Somogyi AA. Association between the DRD2 A1 allele and response to methadone and buprenorphine maintenance treatments. Am J Med Genet B Neuropsychiatr Genet (2006) 141B(4):323–31. doi: 10.1002/ajmg.b.30319
362. Galeeva AR, Gareeva AE, Iur’ev EB, Khusnutdinova EK. VNTR polymorphisms of the serotonin transporter and dopamine transporter genes in male opiate addicts. Mol Biol (Mosk) (2002) 36(4):593–8. doi: 10.1023/A:1019883806620
363. Jacobsen LK, Staley JK, Zoghbi SS, Seibyl JP, Kosten TR, Innis RB, et al. Prediction of dopamine transporter binding availability by genotype: a preliminary report. Am J Psychiatry (2000) 157(10):1700–3. doi: 10.1176/appi.ajp.157.10.1700
364. Van Dyck CH, Malison RT, Jacobsen LK, Seibyl JP, Staley JK, Laruelle M, et al. Increased dopamine transporter availability associated with the 9-repeat allele of the SLC6A3 gene. J Nucl Med (2005) 46:5,745–51.
365. Van de Giessen EM, de Win MML, Tanck MWT, van den Brink W, Baas F, Booij J. Striatal dopamine transporter availability associated with polymorphisms in the dopamine transporter gene SLC6A3. J Nucl Med (2008) 50(1):45–52. doi: 10.2967/jnumed.108.053652
366. Martinez D, Gelernter J, Abi-Dargham A, van Dyck CH, Kegeles L, Innis RB, et al. The variable number of tandem repeats polymorphism of the dopamine transporter gene is not associated with significant change in dopamine transporter phenotype in humans. Neuropsychopharmacology (2001) 24(5):553–60. doi: 10.1016/S0893-133X(00)00216-5
367. Lynch DR, Mozley PD, Sokol S, Maas NM, Balcer LJ, Siderowf AD. Lack of effect of polymorphisms in dopamine metabolism related genes on imaging of TRODAT-1 in striatum of asymptomatic volunteers and patients with Parkinson’s disease. Mov Disord (2003) 18(7):804–12. doi: 10.1002/mds.10430
368. Yeh YW, Lu RB, Tao PL, Shih MC, Lin WW, Huang SY. Neither single-marker nor haplotype analyses support an association between the dopamine transporter gene and heroin dependence in Han Chinese. Genes Brain Behav (2010) 9:6,638–47. doi: 10.1111/j.1601-183X.2010.00597.x
369. Ornoy A, Finkel-Pekarsky V, Peles E, Adelson M, Schreiber S, Ebstein PR. ADHD risk alleles associated with opiate addiction: study of addicted parents and their children. Pediatr Res (2016) 80(2):228–36. doi: 10.1038/pr.2016.78
370. Heinz A, Goldman D, Jones DW, Palmour R, Hommer D, Gorey JG, et al. Genotype influences in vivo dopamine transporter availability in human striatum. Neuropsychopharmacology (2000) 22:133–9. doi: 10.1016/S0893-133X(99)00099-8
371. Cheon KA, Ryu YH, Kim JW, Cho DY. The homozygosity for 10-repeat allele at dopamine transporter gene and dopamine transporter density in Korean children with attention deficit hyperactivity disorder: relating to treatment response to methylphenidate. Eur Neuropsychopharmacol (2005) 15:95–101. doi: 10.1016/j.euroneuro.2004.06.004
372. Lawford BR, Young RM, Noble EP, Sargent J, Rowell J, Shadforth S, et al. The D(2) dopamine receptor A(1) allele and opioid dependence: association with heroin use and response to methadone treatment. Am J Med Genet (2000) 96(5):592–8. doi: 10.1002/1096-8628(20001009)96:5<592::AID-AJMG3>3.0.CO;2-Y
373. Hirvonen M, Laakso A, Någren K, Rinne JO, Pohjalainen T, Hietala J. C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry (2004) 9(12):1060–1. doi: 10.1038/sj.mp.4001561
374. Smith CT, Dang LC, Buckholtz JW, Tetreault AM, Cowan RL, Kessler RM, et al. The impact of common dopamine D2 receptor gene polymorphisms on D2/3 receptor availability: C957T as a key determinant in putamen and ventral striatum. Transl Psychiatry (2017) 7:e1091. doi: 10.1038/tp.2017.45
375. Cloninger CR. A systematic method for clinical description and classification of personality variants. Arch Gen Psychiatry (1987) 44(6):573–88. doi: 10.1001/archpsyc.1987.01800180093014
376. Huang WL, Lin YH, Kuo TB, Chang LR, Chen YZ, Yang CC. Methadone-mediated autonomic functioning of male patients with heroin dependence: the influence of borderline personality pattern. PLoS One (2012) 7(5):e37464. doi: 10.1371/journal.pone.0037464
377. American Psychiatric Association. Substance use disorders. In: Diagnostic and statistical manual of mental disorders., 5th ed. American Psychiatric Publishing (2013). doi: 10.1176/appi.books.9780890425596
378. Gerra G, Bertacca S, Zaimovic A, Pirani M, Branchi B, Ferri M. Relationship of personality traits and drug of choice by cocaine addicts and heroin addicts. Subst Use Misuse (2008) 43(3–4):317–30. doi: 10.1080/10826080701202726
379. Hall H, Köhler C, Gawell L, Farde L, Sedvall G. Raclopride, a new selective ligand for the dopamine-D2receptors. Prog Neuropsychopharmacol Biol Psychiatry (1988) 12(5):559–68. doi: 10.1016/0278-5846(88)90001-2
380. Kung HF, Alavi A, Chang W, Kung MP, Keyes JW Jr, Velchik MG, et al.In vivo SPECT imaging of CNS D-2 dopamine receptors: initial studies with iodine-123-IBZM in humans. J Nucl Med (1990) 31(5):573–9.
381. Videbk C, Toska K, Scheideler MA, Paulson OB, Knudsen GM. SPECT tracer [123I]IBZM has similar affinity to dopamine D2 and D3 receptors. Synapse (2000) 38(3):338–42. doi: 10.1002/1098-2396(20001201)38:3<338::AID-SYN13>3.0.CO;2-N
382. Suehiro M, Dannals RF, Scheffel U, Stathis M, Wilson AA, Ravert HT, et al. In vivo labeling of the dopamine D2 receptor with N-11C-methyl-benperidol. J Nucl Med (1990) 31:12,2015–21.
383. Moerlein SM, Perlmutter JS, Markham J, Welch MJ. In vivo kinetics of [18F](N-methyl)benperidol: a novel PET tracer for assessment of dopaminergic D2-like receptor binding. J Cereb Blood Flow Metab (1997) 17(8):833–45. doi: 10.1097/00004647-199708000-00002
384. Halldin C, Farde L, Högberg T, Mohell N, Hall H, Suhara T, et al. Carbon-11-FLB 457: a radioligand for extrastriatal D2 dopamine receptors. J Nucl Med (1995) 36(7):1275–81.
385. Hall H, Sedvall G, Magnusson O, Kopp J, Halldin C, Farde L. Distribution of D1- and D2-dopamine receptors, and dopamine and its metabolites in the human brain. Neuropsychopharmacology (1994) 11(4):245–56. doi: 10.1038/sj.npp.1380111
386. Kebabian JW, Calne DB. Multiple receptors for dopamine. Nature (1979) 277:5692,93–6. doi: 10.1038/277093a0
387. Stoof JC, Kebabian JW. Opposing roles for D-1 and D-2 dopamine receptors in efflux of cyclic AMP from rat neostriatum. Nature (1981) 294(5839):366–8. doi: 10.1038/294366a0
388. Palacios JM, Camps M, Cortés R, Probst A. Mapping dopamine receptors in the human brain. J Neural Transm Suppl (1988) 27:227–35. doi: 10.1007/978-3-7091-8954-2_20
389. Beninger RJ, Miller R. Dopamine D1-like receptors and reward-related incentive learning. Neurosci Biobehav Rev (1998) 22(2):335–45. doi: 10.1016/S0149-7634(97)00019-5
390. Andrzejewski ME, Spencer RC, Kelley AE. Instrumental learning, but not performance, requires dopamine D1-receptor activation in the amygdala. Neuroscience (2005) 135(2):335–45. doi: 10.1016/j.neuroscience.2005.06.038
391. Tran AH, Tamura R, Uwano T, Kobayashi T, Katsuki M, Ono T. Dopamine D1 receptors involved in locomotor activity and accumbens neural responses to prediction of reward associated with place. Proc Natl Acad Sci(2005) 102(6):2117–22. doi: 10.1073/pnas.0409726102
392. van Gaalen MM, van Koten R, Schoffelmeer AN, Vanderschuren LJ. Critical involvement of dopaminergic neurotransmission in impulsive decision making. Biol Psychiatry (2006) 60(1):66–73. doi: 10.1016/j.biopsych.2005.06.005
393. Gurden H, Takita M, Jay TM. Essential role of D1 but not D2 receptors in the NMDA receptor-dependent long-term potentiation at hippocampal-prefrontal cortex synapses in vivo. J Neurosci (2000) 20(22):106RC–. doi: 10.1523/JNEUROSCI.20-22-j0003.2000
394. Jung E-Y, Shim I. Differential DAergic control of D1 and D2 receptor agonist over locomotor activity and GABA level in the striatum. Exp Neurobiol (2011) 20(3):153. doi: 10.5607/en.2011.20.3.153
395. Molloy AG, Waddington JL. Behavioural responses to the selective D1-dopamine receptor agonist R-SK&F 38393 and the selective D2-agonist RU 24213 in young compared with aged rats. Br J Pharmacol (1988) 95(2):335–42. doi: 10.1111/j.1476-5381.1988.tb11651.x
396. Eagle DM, Wong JCK, Allan ME, Mar AC, Theobald DE, Robbins TW. Contrasting roles for dopamine D1 and D2 receptor subtypes in the dorsomedial striatum but not the nucleus accumbens core during behavioral inhibition in the stop-signal task in rats. J Neurosci (2011) 31(20):7349–56. doi: 10.1523/JNEUROSCI.6182-10.2011
397. Caine SB, Thomsen M, Gabriel KI, Berkowitz JS, Gold LH, Koob GF, et al. Lack of self-administration of cocaine in dopamine D1 receptor knock-out mice. J Neurosci (2007) 27:13140–50. doi: 10.1523/JNEUROSCI.2284-07.2007
398. Zweifel LS, Parker JG, Lobb CJ, Rainwater A, Wall VZ, Fadok JP, et al. Disruption of NMDAR-dependent burst firing by dopamine neurons provides selective assessment of phasic dopamine-dependent behavior. Proc Natl Acad Sci U S A (2009) 106:7281–8. doi: 10.1073/pnas.0813415106
399. Sadat-Shirazi MS, Zarrindast MR, Daneshparvar H, Ziaie A, Fekri M, Abbasnezhad E, et al. Alteration of dopamine receptors subtypes in the brain of opioid abusers: a postmortem study in Iran. Neurosci Lett (2018) 687:169–76. doi: 10.1016/j.neulet.2018.09.043
400. Zarrindast MR, Azami BN, Rostami P, Rezayof A. Repeated administration of dopaminergic agents in the nucleus accumbens and morphine-induced place preference. Behav Brain Res (2006) 169(2):248–55. doi: 10.1016/j.bbr.2006.01.011
401. Bossert JM, Poles GC, Wihbey KA, Koya E, Shaham Y. Differential effects of blockade of dopamine D1-family receptors in nucleus accumbens core or shell on reinstatement of heroin seeking induced by contextual and discrete cues. J Neurosci (2007) 27(46):12655–63. doi: 10.1523/JNEUROSCI.3926-07.2007
402. Chartoff EH, Mague SD, Barhight MF, Smith AM, Carlezon WA. Behavioral and molecular effects of dopamine D1 receptor stimulation during naloxone-precipitated morphine withdrawal. J Neurosci (2006) 26(24):6450–7. doi: 10.1523/JNEUROSCI.0491-06.2006
403. Anderson SM, Bari AA, Pierce RC. Administration of the D1-like dopamine receptor antagonist SCH-23390 into the medial nucleus accumbens shell attenuates cocaine priming-induced reinstatement of drug-seeking behavior in rats. Psychopharmacology (Berl) (2003) 168(1-2):132–8. doi: 10.1007/s00213-002-1298-5
404. Bachtell RK, Whisler K, Karanian D, Self DW. Effects of intra-nucleus accumbens shell administration of dopamine agonists and antagonists on cocaine-taking and cocaine-seeking behaviors in the rat. Psychopharmacology (Berl) (2005) 183(1):41–53. doi: 10.1007/s00213-005-0133-1
405. Hauser SR, Deehan GA Jr, Dhaher R, Knight CP, Wilden JA, McBride WJ, et al. D1 receptors in the nucleus accumbens-shell, but not the core, are involved in mediating ethanol-seeking behavior of alcohol-preferring (P) rats. Neuroscience (2015) 295:243–51. doi: 10.1016/j.neuroscience.2015.03.030
406. Tao YM, Yu C, Wang WS, Hou YY, Xu XJ, Chi ZQ, et al. Heteromers of μ opioid and dopamine D1 receptors modulate opioid-induced locomotor sensitization in a dopamine-independent manner. Br J Pharmacol (2017) 174(17):2842–61. doi: 10.1111/bph.13908
407. Juhasz JR, Hasbi A, Rashid AJ, So CH, George SR, O’Dowd BF. Mu-opioid receptor heterooligomer formation with the dopamine D1 receptor as directly visualized in living cells. Eur J Pharmacol (2008) 581(3):235–43. doi: 10.1016/j.ejphar.2007.11.060
408. Tsang J, Fullard JF, Giakoumaki SG, Katsel P, Katsel P, Karagiorga VE, et al. The relationship between dopamine receptor D1 and cognitive performance. NPJ Schizophr (2015) 1:14002. doi: 10.1038/npjschz.2014.2
409. Thomas U. Modulation of synaptic signalling complexes by Homer proteins. J Neurochem (2002) 81(3):407–13. doi: 10.1046/j.1471-4159.2002.00869.x
410. Kalivas PW, Volkow ND. The neural basis of addiction: a pathology of motivation and choice. Am J Psychiatry (2005) 162(8):1403–13. doi: 10.1176/appi.ajp.162.8.1403
411. Liu JH, Zhong HJ, Dang J, Peng L, Zhu YS. Single-nucleotide polymorphisms in dopamine receptor D1 are associated with heroin dependence but not impulsive behavior. Genet Mol Res (2015) 14:2,4041–50. doi: 10.4238/2015.April.27.19
412. Behrendt S, Wittchen HU, Höfler M, Lieb R, Beesdo K. Transitions from first substance use to substance use disorders in adolescence: is early onset associated with a rapid escalation? Drug Alcohol Depend (2009) 99(1-3):68–78. doi: 10.1016/j.drugalcdep.2008.06.014
413. Sartor CE, Lynskey MT, Bucholz KK, Madden PA, Martin NG, Heath AC. Timing of first alcohol use and alcohol dependence: evidence of common genetic influences. Addiction (2009) 104(9s):1512–8. doi: 10.1111/j.1360-0443.2009.02648.x
414. Dudman JT, Eaton ME, Rajadhyaksha A, Macías W, Taher M, Barczak A, et al. Dopamine D1 receptors mediate CREB phosphorylation via phosphorylation of the NMDA receptor at Ser897-NR1. J Neurochem (2003) 87(4):922–34. doi: 10.1046/j.1471-4159.2003.02067.x
415. Lintas A, Chi N, Lauzon NM, Bishop SF, Gholizadeh S, Sun N, et al. Identification of a dopamine receptor-mediated opiate reward memory switch in the basolateral amygdala-nucleus accumbens circuit. J Neurosci (2011) 31(31):11172–83. doi: 10.1523/JNEUROSCI.1781-11.2011
416. Andersen PH, Grønvald FC, Hohlweg R, Hansen LB, Guddal E, Braestrup C, et al. NNC-112, NNC-687 and NNC-756, new selective and highly potent dopamine D1 receptor antagonists. Eur J Pharmacol (1992) 219(1):45–52. doi: 10.1016/0014-2999(92)90578-R
417. Abi-Dargham A, Martinez D, Mawlawi O, Simpson N, Hwang DR, Slifstein M, et al. Measurement of striatal and extrastriatal dopamine D1 receptor binding potential with [11C]NNC 112 in humans: validation and reproducibility. J Cereb Blood Flow Metab (2000) 20(2):225–43. doi: 10.1097/00004647-200002000-00003
418. Sedvall G, Farde L, Barnett A, Hall H, Halldin C. 11C-SCH 39166, a selective ligand for visualization of dopamine-D1 receptor binding in the monkey brain using PET. Psychopharmacology (Berl) (1991) 103(2):150–3. doi: 10.1007/BF02244195
419. Martinez D, Slifstein M, Narendran R, Foltin RW, Broft A, Hwang DR, et al. Dopamine D1 receptors in cocaine dependence measured with PET and the choice to self-administer cocaine. Neuropsychopharmacology (2009) 34(7):1774–82. doi: 10.1038/npp.2008.235
420. Yasuno F, Ota M, Ando K, Ando T, Maeda J, Ichimiya T, et al. Role of ventral striatal dopamine D1 receptor in cigarette craving. Biol Psychiatry (2007) 61(11):1252–9. doi: 10.1016/j.biopsych.2006.06.028
421. Dagher A, Bleicher C, Aston JA, Gunn RN, Clarke PB, Cumming P. Reduced dopamine D1 receptor binding in the ventral striatum of cigarette smokers. Synapse (2001) 42(1):48–53. doi: 10.1002/syn.1098
422. Dubol M, Trichard C, Leroy C, Sandu AL, Rahim M, Granger B, et al. Dopamine transporter and reward anticipation in a dimensional perspective: a multimodal brain imaging study. Neuropsychopharmacology (2018) 43(4):820–7. doi: 10.1038/npp.2017.183
423. Mulvihill KG. Presynaptic regulation of dopamine release: role of the DAT and VMAT2 transporters. Neurochem Int (2019) 122:94–105. doi: 10.1016/j.neuint.2018.11.004
424. Sotnikova TD, Beaulieu JM, Gainetdinov RR, Caron MG. Molecular biology, pharmacology and functional role of the plasma membrane dopamine transporter. CNS Neurol Disord Drug Targets (2006) 5(1):45–56. doi: 10.2174/187152706784111579
425. Schmitz Y, Benoit-Marand M, Gonon F, Sulzer D. Presynaptic regulation of dopaminergic neurotransmission. J Neurochem (2003) 87(2):273–89. doi: 10.1046/j.1471-4159.2003.02050.x
426. Sesack SR, Hawrylak VA, Guido MA, Levey AI. Cellular and subcellular localization of the dopamine transporter in rat cortex. Adv Pharmacol (1998) 42:171–4. doi: 10.1016/S1054-3589(08)60720-6
427. Sweitzer MM, Donny EC, Hariri AR. Imaging genetics and the neurobiological basis of individual differences in vulnerability to addiction. Drug Alcohol Depend (2012) 123(1):S59–71. doi: 10.1016/j.drugalcdep.2012.01.017
428. VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats element in DAT1 regulates in vitro dopamine transporter density. Genetics (2005) 6:1–11. doi: 10.1186/1471-2156-6-55
429. Fuke S, Suo S, Takahashi N, Koike H, Sasagawa N, Ishiura S. The VNTR polymorphism of the human dopamine transporter (DAT1) gene affects gene expression. Pharmacogenomics J (2001) 1(2):152–6. doi: 10.1038/sj.tpj.6500026
430. Bannon MJ, Michelhaugh SK, Wang J, Sacchetti P. The human dopamine transporter gene: gene organization, transcriptional regulation, and potential involvement in neuropsychiatric disorders. Neuropsychopharmacol (2001) 11:449–55. doi: 10.1016/S0924-977X(01)00122-5
431. Nakamura Y, Koyama K, Matsushima M. VNTR (variable number of tandem repeat) sequences as transcriptional, translational, or functional regulators. J Hum Genet (1998) 43:149–52. doi: 10.1007/s100380050059
432. Mitchell RJ, Howlett S, Earl L, White NG, McComb J, Schanfield MS, Distribution of the 3′ VNTR polymorphism in the human dopamine transporter gene in world populations. Hum Biol (2000) 72:295–304.
433. Guindalini C, Howard M, Haddley K, Laranjeira R, Collier D, Ammar N, et al. A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proc Natl Acad Sci U S A (2006) 103(12):4552–7. doi: 10.1073/pnas.0504789103
434. Ueno S, Nakamura M, Mikami M, Kondoh K, Ishiguro H, Arinami T, et al. Identification of a novel polymorphism of the human dopamine transporter (DAT1) gene and the significant association with alcoholism. Mol Psychiatry (1999) 4(6):552–7. doi: 10.1038/sj.mp.4000562
435. Du Y, Nie Y, Li Y, Wan YJ. The association between the SLC6A3 VNTR 9-repeat allele and alcoholism-a meta-analysis. Alcohol Clin Exp Res (2011) 35(9):1625–34. doi: 10.1111/j.1530-0277.2011.01509.x
436. Gerra G, Somaini L, Leonardi C, Cortese E, Maremmani I, Manfredini M, et al. Association between gene variants and response to buprenorphine maintenance treatment. Psychiatry Res (2014) 215(1):202–7. doi: 10.1016/j.psychres.2013.11.001
437. Krupitsky ЕМ, Kibitov АО, Blokhina ЕА, Verbitskaya ЕV, Brodyansky VМ, Alekseeva NP, et al. Stabilization of remission in patients with opioid dependence with naltrexone implant: a pharmacogenetic approach. Zh Nevrol Psikhiatr Im S S Korsakova (2015) 115(4 Pt 2):14–23. doi: 10.17116/jnevro20151154214-23
438. van Gestel S, Forsegren T, Claes S, Del-Favero J, van Duijn GM, Sluijs S, et al. Epistatic effect of genes from the dopamine and serotonin systems on the temperament traits of novelty seeking and harm avoidance. Mol Psychiatry (2002) 7:448–50. doi: 10.1038/sj.mp.4001005
439. Bardo MT, Donohew R, Harrington NG. Psychobiology of novelty seeking and drug seeking behavior. Behav Brain Res (1996) 77:23–43. doi: 10.1016/0166-4328(95)00203-0
440. Chmielowiec J, Chmielowiec K, Suchanecka A, Trybek G, Mroczek B, Małecka I, et al. Associations between the dopamine D4 receptor and DAT1 dopamine transporter genes polymorphisms and personality traits in addicted patients. Int J Environ Res Public Health (2018) 15(10):2076. doi: 10.3390/ijerph15102076
441. Kim SJ, Kim YS, Kim CH, Lee HS. Lack of association between polymorphism of the dopamine receptor D4 and dopamine transporter genes and personality in a Korean population. Yonsei Med J (2006) 47:787–92. doi: 10.3349/ymj.2006.47.6.787
442. Staley JK, Krishnan-Sarin S, Zoghbi S, Tamagnan G, Fujita M, Seibyl JP, et al. Sex differences in [123I]β-CIT SPECT measures of dopamine and serotonin transporter availability in healthy smokers and nonsmokers. Synapse (2001) 41(4):275–84. doi: 10.1002/syn.1084
443. Cosgrove KP, Tellez-Jacques K, Pittman B, Petrakis I, Baldwin RM, Tamagnan G, et al. Dopamine and serotonin transporter availability in chronic heroin users: A [123I]β-CIT SPECT imaging study. Psychiatry Res Neuroimag (2010) 184(3):192–5. doi: 10.1016/j.pscychresns.2010.08.001
444. Knoll AT, Carlezon WA Jr. Dynorphin, stress, and depression. Brain Res (2010) 1314:56–73. doi: 10.1016/j.brainres.2009.09.074
445. Andero R, Brothers SP, Jovanovic T, Chen YT, Salah-Uddin H, Cameron M, et al. Amygdala-dependent fear is regulated by Oprl1 in mice and humans with PTSD. Sci Transl Med (2013) 5(188):188ra73. doi: 10.1126/scitranslmed.3005656
446. Uhl G, Blum K, Noble E, Smith S. Substance abuse vulnerability and D2 receptor genes. Trends Neurosci (1993) 16(3):83–8. doi: 10.1016/0166-2236(93)90128-9
Keywords: opioid use disorder, neuroimaging, genetics, positron emission tomography, PET, polymorphism, opioid receptors, dopamine receptors
Citation: Burns JA, Kroll DS, Feldman DE, Kure Liu C, Manza P, Wiers CE, Volkow ND and Wang G-J (2019) Molecular Imaging of Opioid and Dopamine Systems: Insights Into the Pharmacogenetics of Opioid Use Disorders. Front. Psychiatry 10:626. doi: 10.3389/fpsyt.2019.00626
Received: 15 January 2019; Accepted: 05 August 2019;
Published: 18 September 2019.
Edited by:
Alexander Mario Baldacchino, University of St Andrews, United KingdomReviewed by:
Olivier George, The Scripps Research Institute, United StatesRoberto Ciccocioppo, University of Camerino, Italy
Annabelle Belcher, University of Maryland, United States
Diana Martinez, Columbia University, United States
Copyright © 2019 Burns, Kroll, Feldman, Kure Liu, Manza, Wiers, Volkow and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gene-Jack Wang, Z2VuZS1qYWNrLndhbmdAbmloLmdvdg==
 Jamie A. Burns
Jamie A. Burns Danielle S. Kroll
Danielle S. Kroll Dana E. Feldman
Dana E. Feldman Christopher Kure Liu
Christopher Kure Liu Peter Manza1
Peter Manza1 Corinde E. Wiers
Corinde E. Wiers Gene-Jack Wang
Gene-Jack Wang