
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Psychiatry , 17 July 2019
Sec. Child and Adolescent Psychiatry
Volume 10 - 2019 | https://doi.org/10.3389/fpsyt.2019.00473
This article is part of the Research Topic Advances in the Study of Pathogenic Mechanisms Responsible for Autism View all 14 articles
Autism spectrum disorder (ASD) is characterized by stereotyped behavior and deficits in communication and social interactions. Gastrointestinal (GI) dysfunction is an ASD-associated comorbidity, implying a potential role of the gut microbiota in ASD GI pathophysiology. Several recent studies found that autistic individuals harbor an altered bacterial gut microbiota. In some cases, remodeling the gut microbiota by antibiotic administration and microbiota transfer therapy reportedly alleviated the symptoms of ASD. However, there is little consensus on specific bacterial species that are similarly altered across individual studies. The aim of this study is to summarize previously published data and analyze the alteration of the relative abundance of bacterial genera in the gut microbiota in controls and individuals with ASD using meta-analysis. We analyzed nine studies, including 254 patients with ASD, and found that children with ASD had lower percentages of Akkermansia, Bacteroides, Bifidobacterium, and Parabacteroides and a higher percentage of Faecalibacterium in the total detected microflora compared to controls. In contrast, children with ASD had lower abundance of Enterococcus, Escherichia coli, Bacteroides, and Bifidobacterium and higher abundance of Lactobacillus. This meta-analysis suggests an association between ASD and alteration of microbiota composition and warrants additional prospective cohort studies to evaluate the association of bacterial changes with ASD symptoms, which would provide further evidence for the precise microbiological treatment of ASD.
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by stereotyped behavior and deficits in communication and social interactions. ASD is highly heterogeneous and its etiology is unclear. Previous studies have revealed several potential causes of this disease, such as genetic abnormalities, dysregulation of the immune system, inflammation, and environmental factors (1–5). Gastrointestinal (GI) problems, including constipation, abdominal pain, gaseousness, diarrhea, and flatulence, are common symptoms associated with ASD in a prevalence range from 23% to 70% (4, 6–8). Although there is no direct evidence that GI symptoms and ASD have a cause-effect relationship, studies have suggested that the gut has an important role in the etiology of ASD (9). Recently, interactions between the gut and the brain in ASD have received considerable attention (10–12). Over millennia, selected microbiota have become resident in the human GI tract, which is integrated with the immune system, metabolism, and nervous system (13, 14). These gut-adapted bacteria and their metabolites might have a critical role in the pathophysiology of ASD. Studies in rodents have indicated that the gut microbiota appears to influence the development of emotional behaviors and brain neurotransmitter systems, further suggesting the existence of a microbiota gut-brain axis (15–18). The gut microbiota has assumed its rightful position as a critical component of the brain-gut axis, highlighting its potential impact on behavior and mood at the level of the central nervous system (10). Furthermore, in some cases, remodeling the gut microbiota by antibiotic administration and microbiota transfer therapy reportedly alleviated the symptoms of ASD (19). The application of probiotics could influence microbiota composition and intestinal barrier function and alter mucosal immune responses (20). There are several possible microbial-related mechanisms implicated in ASD, such as dysbiosis-induced breakdown of gut integrity (21, 22), production of toxins (23), and immunological (24) and metabolic (23) abnormalities. A microbial shift within the gut of mice yields changes in serum metabolites and induces an autistic behavioral phenotype (25). Additionally, many studies have reported dysbiosis of the gut microbiota in individuals with ASD (26–28). However, different researchers reported various results. For example, Kang et al. reported a higher percentage of Bacteroides in the total detected microflora in children with ASD, whereas Strati et al. demonstrated a lower percentage of Bacteroides in children with ASD compared to controls (29, 30). Due to the currently available conflicting data, there is a need for a further investigation of the association between the gut microbiota and ASD. To better understand the effect of gut microbes on ASD, we carried out a meta-analysis to assess the differences in microbial populations between patients with ASD and age-matched controls. Such information is useful to design novel therapeutic strategies for modulating gut microbial populations in patients with ASD.
We performed a systematic literature search of PubMed, Web of Science, and Cochrane databases up to July 2017 using the following terms: “autism (autism spectrum disorder) and microbiota” or “microbiome” or “dysbiosis” according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (31). The abstracts identified in this search were screened to eliminate clearly irrelevant studies. The criteria for study inclusion were as follows: 1) observational prospective and retrospective studies, case–control studies, or cohort studies; 2) investigating gut bacteria in children diagnosed with autism or ASD; 3) including information about sample size and prevalence of the specific bacteria assessed; and 4) written in English. Studies about non-human subjects as well as reviews, case reports, and duplicate publications were excluded. All articles providing sufficient information about the relationship between the gut microbiota and ASD were included.
The outcome of interest was the association between ASD and the gut microbiota. The definition of ASD was based on a physician’s diagnosis according to the International Statistical Classification of Diseases and Related Health Problems, Tenth Revision or a history of ASD reported by the parents of the children. Assessments of the biodiversity and composition of microbiota were based on stool sample testing using culture-dependent methods (32, 33), real-time polymerase chain reaction (PCR) (34), fluorescence in situ hybridization (FISH) (35), and pyrosequencing for bacterial 16S ribosomal ribonucleic acid (rRNA) genes (23, 29, 30, 36, 37). To conduct the meta-analyses, at least three studies were used to assess the bacteria. To maintain consistency within the present meta-analysis, all bacterial information were reviewed and selected before the final analyses, including bacterial taxonomy, percentage, and relative abundance. In general, gut bacteria were classified at different taxonomic levels from phylum (high taxonomic level) to genus (low taxonomic level). For consistency, the included studies were analyzed at the genus level. We contacted the investigators of the eligible studies if we were unable to extract data on bacterial abundance from the published articles.
Four investigators independently carried out data extraction of the following items: author(s), publication year, study design, country, study population age, diagnosis of ASD, and effect size. Two reviewers completed the quality assessment independently. A set of structured criteria modified from previous studies was used to complete the quality assessment of publications. The total score ranged from 5 to 9 (with 9 as the highest), with a higher score indicating higher quality. In case of disagreement regarding the extracted data, discrepancies were resolved by consensus discussion.
A fixed-effects model and a random-effects model were used to report the most conservative result. Statistical heterogeneity was assessed using the I 2 value, which represents the percentage of total variation across different studies, owing to heterogeneity rather than chance. I 2 values of 25%, 50%, and 75% were related to low, moderate, and high heterogeneity (38). A random-effects model was applied when there was notable heterogeneity (I 2 index ≥ 50%); otherwise, a fixed-effects model was used. The standardized mean difference (SMD) measure of effect was used for the continuous variables (39). SMD > 0 indicates that participants with ASD have a higher bacterial abundance than controls, and SMD < 0 indicates participants with ASD have a lower bacterial abundance than controls. We also planned to analyze the influence of bias control in subgroup analyses as well as the evidence of publication bias and other small study effects using funnel plots and regression analyses. However, because of the limited number of studies, we only conducted subgroup analyses of studies that included participants with ASD or typically developing children. In our primary analysis, we included all published studies. The ratio of the bacterial percentage between children with ASD and controls was calculated to assess the relative abundance of bacteria in children with ASD compared to controls. All statistical analyses were carried out using Stata version 12.0 (Stata Corp, College Station, TX, USA).
Literature searches revealed 431 potentially eligible records (Figure 1). Three additional records were identified through a review of reference lists. After the review of the titles and abstracts and the removal of 246 duplicates, 112 publications were selected for a further review of the full texts. After the exclusion of records that were clearly irrelevant, involved nonhuman subjects, or have incomplete data, 30 full-text records were reviewed individually. Of these 30 articles, 9 studies were included in the present meta-analysis, as the remaining studies did not provide quantitative data about bacterial abundance or percentage in the report or after our request for essential details. In total, there were 254 participants with ASD and 167 age-matched typically developing controls with an age range from 6 to 11 years (Table 1). The diagnostic methods of ASD and comorbidity disease in the included studies are shown in Table 2. Gut microbiology was assessed using quantitative PCR (QPCR) or PCR, pyrosequencing, culture methods, or FISH (Table 3). Each study provided different types of bacteria for the meta-analysis (Table 3). The percentage and relative abundance of bacterial genera in the gut microbiota were used in the present analysis to avoid potential variation caused by different detection methods of the microbiota in the included papers. The absolute number of bacterial populations reported only in three studies was insufficient to perform a meta-analysis. The standard deviation of the mean was calculated for one study that only provided the mean and range (33) using a previously published method (40).
We analyzed the percentage of Akkermansia from five trials (Figure 2A). A fixed-effects meta-analysis showed that the percentage of Akkermansia in the total detected microflora was 0.1% in participants with ASD [95% confidence interval (CI): −0.005 to 0.007] compared to 0.2% in typically developing children (95% CI: −0.007 to 0.01). There was no evidence of between-study heterogeneity (I 2 = 0%; Figure 2A). However, its effect size (Z = 0.44, P = 0.658) was relatively small. The ratio of bacterial percentage between the ASD group (0.1%) and the control group (0.2%) was 0.5. The percentage of Akkermansia in patients with ASD was clearly lower compared to controls (Figure 5A).
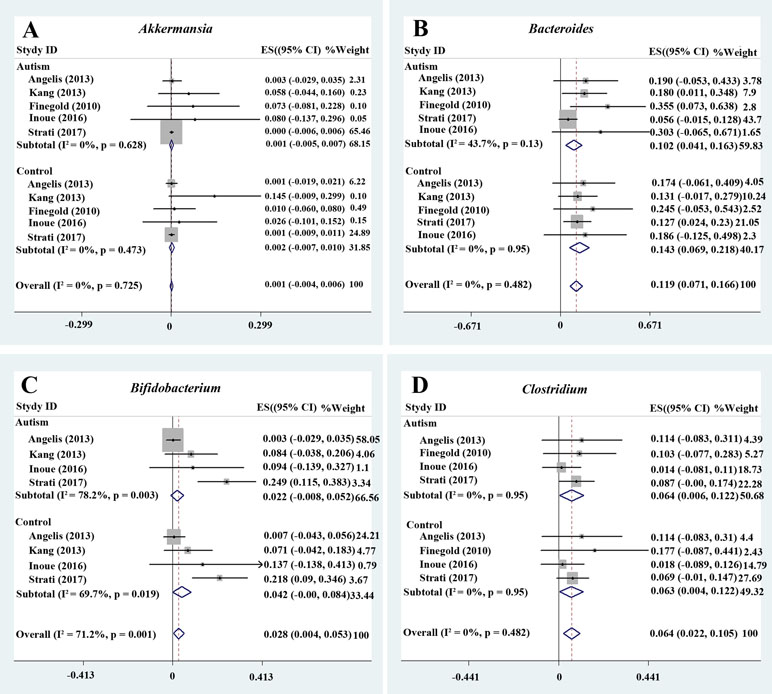
Figure 2 Forest plot of percentages of Akkermansia, Bacteroides, Bifidobacterium, and Clostridium in ASD. (A–D) Percentages of Akkermansia, Bacteroides, Bifidobacterium, and Clostridium in the total detected microflora, respectively. Fixed-effects models were used to assess Akkermansia, Bacteroides, and Clostridium. A random-effects model was used to analyze Bifidobacterium, contributing to higher between-study heterogeneity (I2 > 50%). The pooled percentages of Akkermansia, Bacteroides, Bifidobacterium, and Clostridium from the included studies were 0.1%, 11.9%, 2.8%, and 6.4%, respectively.
Bacteroides is a Gram-negative bacterium and is one of the earliest colonizing and most abundant constituents of the gut microbiota and may induce an anti-inflammatory milieu (41). A fixed-effects meta-analysis showed that the percentage of Bacteroides in the total detected microflora was 10.2% (95% CI: 0.041–0.163) in children with ASD but 14.3% in typically developing children (95% CI: 0.069–0.218). There was low between-study heterogeneity within the ASD group (I 2 = 43.7%; Figure 2B). The effect size (Z = 4.92, P = 0.000) was significant and large. The ratio of the bacterial percentage between the ASD group (10.2%) and the control group (14.3%) was 0.71 (Figure 5A). Furthermore, a random-effects model also showed a lower abundance of Bacteroides in participants with ASD compared to controls (SMD −0.35, 95% CI: −1.2 to 0.51; Figure 3D). However, its effect size (Z = 0.8, P = 0.427) was relatively small.
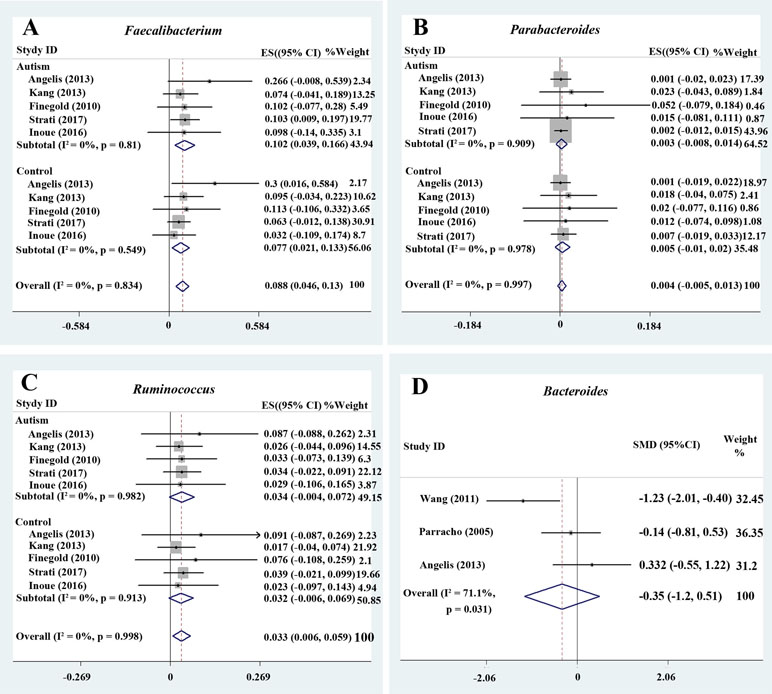
Figure 3 Forest plot of percentages of Faecalibacterium, Parabacteroides, Ruminococcus, and Bacteroides in autism spectrum disorder (ASD). (A–C) Percentages of Faecalibacterium, Parabacteroides, and Ruminococcus in the total detected microflora, respectively. Fixed-effects models were used to assess Faecalibacterium, Parabacteroides, and Ruminococcus. The pooled percentages of Faecalibacterium, Parabacteroides, and Ruminococcus from the included studies were 8.8%, 0.4%, and 3.3%, respectively. (D) Relative abundance of Bacteroides. A random-effects model was used to analyze Bacteroides, contributing to higher between-study heterogeneity (I2 > 50%).
Bifidobacterium has long been used as a probiotic to alleviate various diseases by changing the gut microbiota composition (34, 42). A random-effects meta-analysis showed 2.2% of Bifidobacterium in the total detected microflora of children with ASD (95% CI: −0.008 to 0.052), whereas the percentage in typically developing children was 4.2% (95% CI: −0.00 to 0.084) with moderate between-study heterogeneity (I 2 = 78.2% and 69.7%, respectively; Figure 2C). The effect size (Z = 2.27, P = 0.023) was significant and moderate. The ratio of the bacterial percentage between the ASD group (2.2%) and the control group (4.2%) was 0.52 (Figure 5A). The percentage of Bifidobacterium in patients with ASD was clearly lower compared to controls. Furthermore, a random-effects model showed a lower abundance of Bifidobacterium in children with ASD (SMD −1.05, 95% CI: −2.27 to 0.18; Figure 4A).
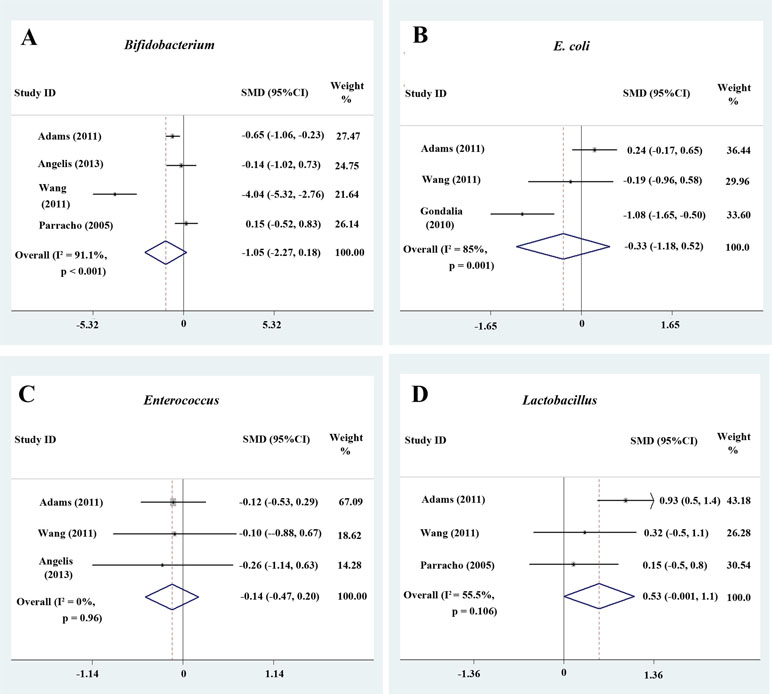
Figure 4 Forest plot of the relative abundance of Bifidobacterium, E. coli, Enterococcus, and Lactobacillus in ASD. (A–D) Relative abundance of Bifidobacterium, E. coli, Enterococcus, and Lactobacillus. Random-effects models were used to analyze Bifidobacterium, E. coli, and Lactobacillus, contributing to higher between-study heterogeneity (I2 > 50%), except Enterococcus.
Five studies were used to evaluate the percentage of Faecalibacterium (Figure 3A). A fixed-effects meta-analysis showed that the percentage of Faecalibacterium in the total detected microflora of children with ASD was 10.2% (95% CI: 0.039–0.166), clearly higher than that of typically developing children (7.7%; 95% CI: 0.021–0.133). The effect size (Z = 4.12, P = 0.000) was significant and large. The ratio of the bacterial percentage between the ASD group (10.2%) and the control group (7.7%) was 1.32 (Figure 5A).
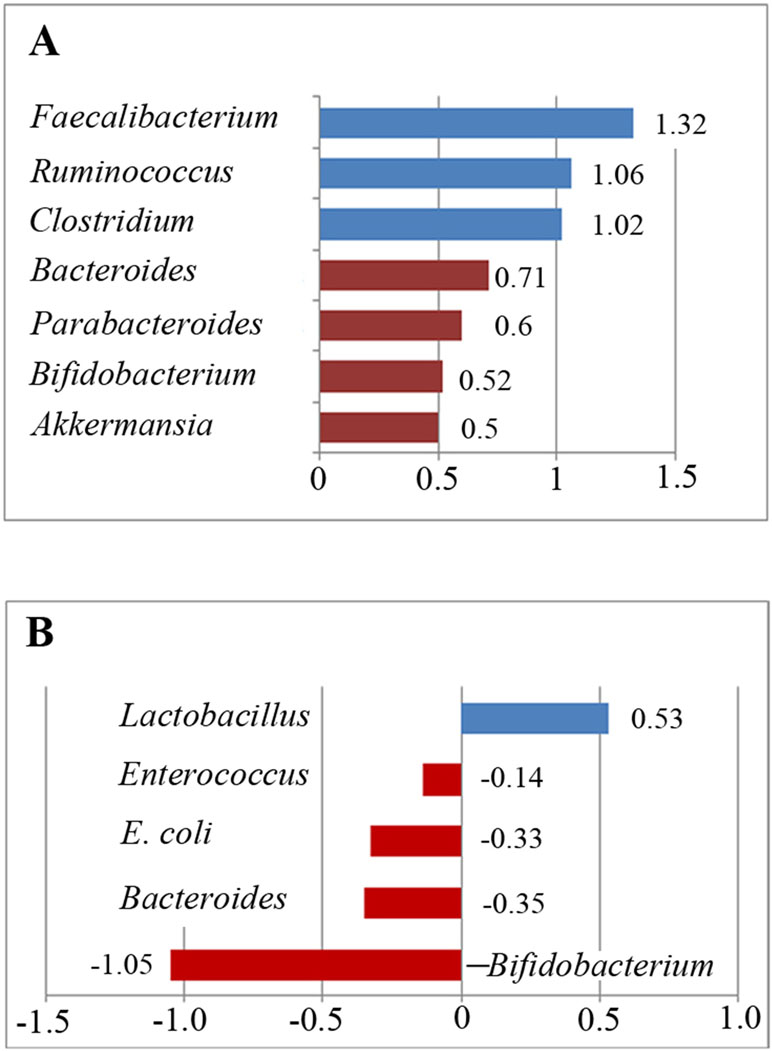
Figure 5 Relative abundance of the included bacteria in the meta-analysis. (A) Ratio of the bacterial percentages in children with ASD and typically developing children. A value greater than 1 indicates higher abundance in children with ASD (Faecalibacterium, Ruminococcus, and Clostridium), whereas a value less than 1 indicates lower abundance in children with ASD compared to controls. (B) Relative abundance of the gut microbiota in children with ASD. A positive value indicates higher abundance in children with ASD (Lactobacillus), whereas a negative value indicates lower abundance in children with ASD.
Ruminococcus is an anaerobic Gram-positive coccus that can be found in the GI tract (43, 44). The percentages of Ruminococcus in the total detected microflora were assessed. A fixed-effects meta-analysis showed 3.4% and 3.2% for children with ASD and typically developing controls, respectively (95% CI: −0.004 to 0.072 and −0.006 to 0.069, respectively). There was no evidence of between-study heterogeneity (I 2 = 0%; Figure 3C) with respect to Ruminococcus percentages. The effect size (Z = 2.42, P = 0.016) was significant and moderate. The ratio of the bacterial percentage between the ASD group (3.4%) and the control group (3.2%) was 1.06 (Figure 5A). The percentage of Ruminococcus in patients with ASD was slightly higher compared to controls.
A fixed-effects meta-analysis showed that the percentage of Clostridium in the total detected microflora of children with ASD was 6.4% (95% CI: 0.006–0.122), similar to that of typically developing children (6.3%; 95% CI: 0.004–0.122; Figure 2D). Additionally, the fixed-effects meta-analysis also showed that the percentage of Parabacteroides in the total detected microflora of children with ASD was 0.3% (95% CI: −0.008 to 0.014) compared to 0.5% in typically developing children (95% CI: −0.01 to 0.02; Figure 3B). The ratio of the bacterial percentage between the ASD group (0.3%) and the control group (0.5%) was 0.6, indicating a decreased percentage in children with ASD (Figure 5A).
The random-effects and fixed-effects models showed a lower relative abundance of E. coli and Enterococcus in children with ASD compared to controls (SMD −0.33, 95% CI: −1.18 to 0.52 and SMD −0.14, 95% CI: −0.47 to 0.20, respectively; Figure 4B and 4C). A random-effects meta-analysis showed a higher relative abundance of Lactobacillus in children with ASD (SMD 0.53, 95% CI: −0.001 to 1.1; Figure 4D). The pooled relative abundance of Lactobacillus, Enterococcus, E. coli, Bacteroides, and Bifidobacterium in children with ASD are shown in Figure 5B.
The gut microbiota plays a major role in human physiology and pathology (45–47). Both experimental and clinical cross-sectional studies showed that patients with ASD had alterations of the gut microbiota (48). These alterations were potentially relevant to behavioral and GI symptoms that are correlated with the severity of ASD (7, 43, 49–52), suggesting that the gut-brain axis participates in the pathogenesis of ASD (18, 53, 54).
Although several reviews suggested a microbiota alteration in patients with ASD (28, 30, 55–60), this is the first meta-analysis that systematically reviewed published data and examined microbiota alterations in patients with ASD. Standardized data collection, strict inclusion criteria, and multiple statistical tools were used to ensure the most accurate assessment. The present meta-analysis found that neither there were significant changes in the intestinal microbial diversity nor single microbial species may be perceived as “ASD-promoting microbes.”Our analyses showed that participants with ASD had a lower abundance of Akkermansia, Bacteroides, Bifidobacterium, E. coli, and Enterococcus, a higher abundance of Faecalibacterium and Lactobacillus, and a slightly increased abundance of Ruminococcus and Clostridium. It is possible that the reduced levels of beneficial bacteria combined with the increased levels of harmful bacteria contribute together to ASD symptoms. Our analysis is consistent with previous reviews (61), with one exception of Clostridium. Several studies showed there was a higher level of Clostridium in individuals with ASD compared to controls and hypothesized that Clostridium can produce neurotoxins and contribute to ASD (62, 63). The current analysis showed slightly increased levels of Clostridium and Ruminococcus, indicating that further studies should be performed to confirm these trends. In contrast, there is potentially a decrease in “beneficial” bacteria in patients with ASD (34, 64). This notion is further supported by a recent study showing that the supplementation of Bifidobacterium species-containing probiotics improves specific ASD symptoms (65).
The role of the gut microbiota in development and disease is not yet well understood. Potential mechanisms by which microbiota impacts the gut-brain axis and ASD progression involve inflammatory and metabolic pathways and alteration of epithelial barrier integrity. First, the abundance of Faecalibacterium may play a role in systemic immunity dysfunction. The abundance of Faecalibacterium was significantly higher in children with ASD compared to controls (30). The expression levels of interferon response factors 7 and 9 showed a strong correlation with the abundance of Faecalibacterium in fecal microbiota, which could produce substances that activate type I interferon signaling (66). In contrast, protective bacteria such as Bifidobacterium were decreased in abundance in individuals with ASD across the analyzed studies. Bifidobacteria are major producers of lactic acid, which suppress the growth of pathogens such as E. coli across the epithelium, reduce inflammation in the gut, and cooperate with the immune system (67, 68). In the present study, lower levels of Bifidobacterium and higher levels of Lactobacillus suggested an imbalance in beneficial bacteria. Decreased levels of Bifidobacterium and metabolites of free amino acids and short-chain fatty acids (SCFA) in the feces may also contribute to the development of ASD (23). A low level of SCFA was possibly related to probiotic usage, lower saccharolytic fermentation by beneficial bacteria, or increased gut permeability, subsequently exacerbating autistic symptoms (32). Akkermansia is a mucin-degrading bacterium present in the gut of typically developing adults. A lower abundance of Akkermansia in children with ASD could indicate a thinner GI mucus barrier in children with ASD compared to controls; the result might reflect an indirect evidence of impaired gut permeability in children with ASD (34). Second, animal studies have shown effects of the gut microbiota on neurodevelopment, suggesting that intestinally derived lipopolysaccharides can increase anxiety-like behavior in mice (69–71). Furthermore, gut microbial populations in ASD may produce toxic products, including neurotoxins that influence distal sites such as the brain, and exert systemic effects (35). Third, microbiota and their metabolites are essential in maintaining both white matter and epithelial barrier integrity, which is important for normal brain development and function (72). The development of the blood-brain barrier is now well established to be contingent upon the presence of commensal gut flora (10, 11). Additionally, diet-specific gut microbiota populations potentially influence white matter integrity in rats (59). These studies reveal a potential mechanism for the gut microbiota in influencing the brain-gut-enteric microbiota axis and contribute to the understanding of the role of the brain-gut axis in the pathogenesis of ASD.
Children with ASD also have a high rate of GI symptoms, which correlate with ASD severity (32, 73) and are associated with ASD-relevant emotional and behavioral problems (74, 75). More than 50% of GI symptoms may be due to dysbiotic gut microbiota, including increased Ruminococcaceae (76, 77). In our meta-analysis, only two studies enrolled children with ASD who had no GI symptoms (23, 37), and one study did not provide details about GI symptoms (33). Collectively, the studies included in the current analysis, however, indicate a high incidence of GI symptoms in children with ASD. The GI symptoms might be related to the ubiquity of food selectivity in this population, as the dietary patterns often associated with ASD involve a high intake of processed food and lack fiber-containing fruits and vegetables. Gastroesophageal reflux, gastroenteritis, food allergies, and inflammatory bowel disease are also more common in children with ASD, probably contributing to the development of GI symptoms (78).
The meta-analysis is inherently limited by the included studies. First, the study design, specificity, and sensitivity of the detection methods used in the included studies varied. The studies included in our analysis mainly used culture-based methods, PCR and pyrosequencing, to analyze the changes of particular bacterial groups, which might underestimate the complexity of the gut microbiota. Indeed, we found that suitable analytical and statistical methods are critical to detect the alterations of the abundance of some gut microbiota in patients with ASD. Second, many reports had relatively small sample sizes with only two of the nine studies recruiting more than 50 participants with ASD. Significant heterogeneity was found between studies when the data were pooled. Finally, our study only analyzed bacterial percentages and abundance at the genus level due to the insufficiency of data for various bacterial taxonomies. Further broad-based, longitudinal, unbiased studies of fecal microbial populations in patients with ASD and age-matched controls using next-generation sequencing will be more informative for clarifying ASD-associated dysbiosis.
Our review summarized the association between ASD and gut microbiota composition. Participants with ASD had a lower abundance of Akkermansia, Bacteroides, Bifidobacterium, E. coli, and Enterococcus, a higher abundance of Faecalibacterium and Lactobacillus, and a slightly increased abundance of Ruminococcus and Clostridium. There were important differences, such as the abundance of Akkermansia, Bifidobacterium, Bacteroides, E. coli, and Lactobacillus between the microbiota of children with ASD and typically developing children. Our analysis warrants additional prospective cohort studies to evaluate the influence of the microbiota in the pathogenesis of ASD and associated GI symptoms. A future impact of such studies could potentially guide the implementation of dietary/probiotic interventions impacting the gut microbiota in patients with ASD.
FL and JL designed and supervised the study. MX and XX performed the data analysis and interpretation and wrote the manuscript. All authors read and approved the final version to be published and agreed to be accountable for all aspects of the work.
This study was supported by grants from the National Natural Science Foundation of China (no. 81571031 and 8171101223), the Shanghai Committee of Science and Technology (no. 17XD1403200), the Shanghai Municipal Education Commission (Research Physician Project; no. 20152234), the Shanghai Municipal Commission of Health and Family Planning (no. GDEK201709, 2017ZZ02026, and 2017EKHWYX-02), the Shanghai Shenkang Hospital Development Center (no. 16CR2025B), and the Shanghai Municipal Health Commission (no. 2019SY068).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Fakhoury M. Autistic spectrum disorders: a review of clinical features, theories and diagnosis. Int J Dev Neurosci (2015) 43:70–7. doi: 10.1016/j.ijdevneu.2015.04.003
2. Jyonouchi H, Sun S, Itokazu N. Innate immunity associated with inflammatory responses and cytokine production against common dietary proteins in patients with autism spectrum disorder. Neuropsychobiology (2002) 46:76–84. doi: 10.1159/000065416
3. Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature (2012) 485:242–5. doi: 10.1038/nature11011
4. Newschaffer CJ, Croen LA, Daniels J, Giarelli E, Grether JK, Levy SE, et al. The epidemiology of autism spectrum disorders. Annu Rev Public Health (2007) 28:235–58. doi: 10.1146/annurev.publhealth.28.021406.144007
5. Rossignol DA, Frye RE. A review of research trends in physiological abnormalities in autism spectrum disorders: immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol Psychiatry (2012) 17:389–401. doi: 10.1038/mp.2011.165
6. Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord (2014) 44:1117–27. doi: 10.1007/s10803-013-1973-x
7. Holingue C, Newill C, Lee LC, Pasricha PJ, Daniele Fallin M. Gastrointestinal symptoms in autism spectrum disorder: a review of the literature on ascertainment and prevalence. Autism Res (2018) 11:24–36. doi: 10.1002/aur.1854
8. Myers SM, Johnson CP, American Academy of Pediatrics Council on Children With Disabilities. Management of children with autism spectrum disorders. Pediatrics (2007) 120:1162–82. doi: 10.1542/peds.2007-2362
9. Mayer EA, Padua D, Tillisch K. Altered brain-gut axis in autism: comorbidity or causative mechanisms? Bioessays (2014b) 36:933–9. doi: 10.1002/bies.201400075
10. Cussotto S, Sandhu KV, Dinan TG, Cryan JF. The neuroendocrinology of the microbiota-gut-brain axis: a behavioural perspective. Front Neuroendocrinol (2018) 51:80–101. doi: 10.1016/j.yfrne.2018.04.002
11. Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Front Physiol (2011) 2:94. doi: 10.3389/fphys.2011.00094
12. Stilling RM, Dinan TG, Cryan JF. Microbial genes, brain & behaviour — epigenetic regulation of the gut-brain axis. Genes Brain Behav (2014) 13:69–86. doi: 10.1111/gbb.12109
13. Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe (2015) 17:565–76. doi: 10.1016/j.chom.2015.04.011
14. Sommer F, Backhed F. The gut microbiota — masters of host development and physiology. Nat Rev Microbiol (2013) 11:227–38. doi: 10.1038/nrmicro2974
15. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A (2011) 108:16050–5. doi: 10.1073/pnas.1102999108
16. Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry (2014) 19:146–8. doi: 10.1038/mp.2013.65
17. Lim JS, Lim MY, Choi Y, Ko G. Modeling environmental risk factors of autism in mice induces IBD-related gut microbial dysbiosis and hyperserotonemia. Mol Brain (2017) 10:14. doi: 10.1186/s13041-017-0292-0
18. Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest (2015) 125:926–38. doi: 10.1172/JCI76304
19. Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome (2017) 5:10. doi: 10.1186/s40168-016-0225-7
20. Critchfield JW, van Hemert S, Ash M, Mulder L, Ashwood P. The potential role of probiotics in the management of childhood autism spectrum disorders. Gastroenterol Res Pract (2011) 2011:161358. doi: 10.1155/2011/161358
21. Navarro F, Pearson DA, Fatheree N, Mansour R, Hashmi SS, Rhoads JM. Are ‘leaky gut’ and behavior associated with gluten and dairy containing diet in children with autism spectrum disorders? Nutr Neurosci (2015) 18:177–85. doi: 10.1179/1476830514Y.0000000110
22. Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One (2011) 6:e24585. doi: 10.1371/journal.pone.0024585
23. De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One (2013) 8:e76993. doi: 10.1371/journal.pone.0076993
24. Onore C, Careaga M, Ashwood P. The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun (2012) 26:383–92. doi: 10.1016/j.bbi.2011.08.007
25. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell (2013) 155:1451–63. doi: 10.1016/j.cell.2013.11.024
26. Ding HT, Taur Y, Walkup JT. Gut microbiota and autism: key concepts and findings. J Autism Dev Disord (2017) 47:480–9. doi: 10.1007/s10803-016-2960-9
27. Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E, et al. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis (2002) 35:S6–S16. doi: 10.1086/341914
28. Rosenfeld CS. Microbiome disturbances and autism spectrum disorders. Drug Metab Dispos (2015) 43:1557–1. doi: 10.1124/dmd.115.063826
29. Kang DW, Park JG, Ilhan ZE, Wallstrom G, Labaer J, Adams JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One (2013) 8:e68322. doi: 10.1371/journal.pone.0068322
30. Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome (2017) 5:24. doi: 10.1186/s40168-017-0242-1
31. Shamseer L, Moher D, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ (2015) 350:g7647. doi: 10.1136/bmj.g7647
32. Adams JB, Johansen LJ, Powell LD, Quig D, Rubin RA. Gastrointestinal flora and gastrointestinal status in children with autism — comparisons to typical children and correlation with autism severity. BMC Gastroenterol (2011) 11:22. doi: 10.1186/1471-230X-11-22
33. Gondalia SV, Palombo EA, Knowles SR, Austin DW. Faecal microbiota of individuals with autism spectrum disorder. Electron J Appl Psychol (2010) 6:24–9. doi: 10.1002/aur.1253
34. Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol (2011) 77:6718–21. doi: 10.1128/AEM.05212-11
35. Parracho HM, Bingham MO, Gibson GR, McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol (2005) 54:987–91. doi: 10.1099/jmm.0.46101-0
36. Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe (2010) 16:444–53. doi: 10.1016/j.anaerobe.2010.06.008
37. Inoue R, Sakaue Y, Sawai C, Sawai T, Ozeki M, Romero-Perez GA, et al. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem (2016) 80:2450–8. doi: 10.1080/09168451.2016.1222267
38. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
39. Faraone SV. Interpreting estimates of treatment effects: implications for managed care. P T (2008) 33:700–11.
40. Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. (2005) 5:13. doi: 10.1186/1471-2288-5-13
41. Bolte ER. Autism and clostridium tetani. Med Hypotheses (1998) 51:133–44. doi: 10.1016/S0306-9877(98)90107-4
42. Mayer EA, Knight R, Mazmanian SK, Cryan JF, Tillisch K. Gut microbes and the brain: paradigm shift in neuroscience. J Neurosci (2014a) 34:15490–6. doi: 10.1523/JNEUROSCI.3299-14.2014
43. Prosperi M, Santocchi E, Balboni G, Narzisi A, Bozza M, Fulceri F, et al. Behavioral phenotype of ASD preschoolers with gastrointestinal symptoms or food selectivity. J Autism Dev Disord (2017) 47:3574–88. doi: 10.1007/s10803-017-3271-5
44. Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism (2013) 4:42. doi: 10.1186/2040-2392-4-42
45. Lei E, Vacy K, Boon WC. Fatty acids and their therapeutic potential in neurological disorders. Neurochem Int (2016) 95:75–84. doi: 10.1016/j.neuint.2016.02.014
46. Russo R, Cristiano C, Avagliano C, De Caro C, La Rana G, Raso GM, et al. Gut-brain axis: role of lipids in the regulation of inflammation, pain and CNS diseases. Curr Med Chem (2017) 25:3930–52. doi: 10.2174/0929867324666170216113756
47. Slattery J, MacFabe DF, Frye RE. The significance of the enteric microbiome on the development of childhood disease: a review of prebiotic and probiotic therapies in disorders of childhood. Clin Med Insights Pediatr (2016) 10:91–107. doi: 10.4137/CMPed.S38338
48. Ettinger G, MacDonald K, Reid G, Burton JP. The influence of the human microbiome and probiotics on cardiovascular health. Gut Microbes (2014) 5:719–28. doi: 10.4161/19490976.2014.983775
49. Bauerl C, Collado MC, Diaz Cuevas A, Vina J, Perez Martinez G. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett Appl Microbiol (2018) 66:464–71. doi: 10.1111/lam.12882
50. Karl JP, Margolis LM, Madslien EH, Murphy NE, Castellani JW, Gundersen Y, et al. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol Gastrointest Liver Physiol. (2017) 312:G559–G571. doi: 10.1152/ajpgi.00066.2017
51. Kong J, Fang J, Park J, Li S, Rong P. Treating depression with transcutaneous auricular vagus nerve stimulation: state of the art and future perspectives. Front Psychiatry (2018) 9:20. doi: 10.3389/fpsyt.2018.00020
52. Wasilewska J, Klukowski M. Gastrointestinal symptoms and autism spectrum disorder: links and risks — a possible new overlap syndrome. Pediatric Health Med Ther (2015) 6:153–66. doi: 10.2147/PHMT.S85717
53. Beaudet AL. Brain carnitine deficiency causes nonsyndromic autism with an extreme male bias: a hypothesis. Bioessays (2017) 39:1700012(1–11). doi: 10.1002/bies.201700012
54. Evrensel A, Ceylan ME. Fecal microbiota transplantation and its usage in neuropsychiatric disorders. Clin Psychopharmacol Neurosci (2016) 14:231–7. doi: 10.9758/cpn.2016.14.3.231
55. Buie T. Potential etiologic factors of microbiome disruption in autism. Clin Ther (2015) 37:976–83. doi: 10.1016/j.clinthera.2015.04.001
56. Felice VD, O’Mahony SM. The microbiome and disorders of the central nervous system. Pharmacol Biochem Behav (2017) 160:1–13. doi: 10.1016/j.pbb.2017.06.016
57. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med (2018) 24:392–400. doi: 10.1038/nm.4517
58. Mulle JG, Sharp WG, Cubells JF. The gut microbiome: a new frontier in autism research. Curr Psychiatry Rep (2013) 15:337. doi: 10.1007/s11920-012-0337-0
59. Ong IM, Gonzalez JG, McIlwain SJ, Sawin EA, Schoen AJ, Adluru N, et al. Gut microbiome populations are associated with structure-specific changes in white matter architecture. Transl Psychiatry (2018) 8:6. doi: 10.1038/s41398-017-0022-5
60. Vuong HE, Hsiao EY. Emerging roles for the gut microbiome in autism spectrum disorder. Biol Psychiatry (2017) 81:411–23. doi: 10.1016/j.biopsych.2016.08.024
61. Li Q, Han Y, Dy ABC, Hagerman RJ. The gut microbiota and autism spectrum disorders. Front Cell Neurosci (2017) 11:120. doi: 10.3389/fncel.2017.00120
62. Argou-Cardozo I, Zeidan-Chulia F. Clostridium bacteria and autism spectrum conditions: a systematic review and hypothetical contribution of environmental glyphosate levels. Med Sci (Basel) (2018) 6:E29. doi: 10.3390/medsci6020029
63. Finegold SM, Summanen PH, Downes J, Corbett K, Komoriya T. Detection of Clostridium perfringens toxin genes in the gut microbiota of autistic children. Anaerobe (2017) 45:133–7. doi: 10.1016/j.anaerobe.2017.02.008
64. Tomova A, Husarova V, Lakatosova S, Bakos J, Vlkova B, Babinska K, et al. Gastrointestinal microbiota in children with autism in Slovakia. Physiol Behav (2015) 138:179–87. doi: 10.1016/j.physbeh.2014.10.033
65. Yang Y, Tian J, Yang B. Targeting gut microbiome: a novel and potential therapy for autism. Life Sci (2018) 194:111–9. doi: 10.1016/j.lfs.2017.12.027
66. Finsen B, Owens T. Innate immune responses in central nervous system inflammation. FEBS Lett (2011) 585:3806–12. doi: 10.1016/j.febslet.2011.05.030
67. Hashemi Z, Fouhse J, Im HS, Chan CB, Willing BP. Dietary pea fiber supplementation improves glycemia and induces changes in the composition of gut microbiota, serum short chain fatty acid profile and expression of mucins in glucose intolerant rats. Nutrients (2017) 9:E1236. doi: 10.3390/nu9111236
68. Mudd AT, Berding K, Wang M, Donovan SM, Dilger RN. Serum cortisol mediates the relationship between fecal Ruminococcus and brain N-acetylaspartate in the young pig. Gut Microbes (2017) 8:589–600. doi: 10.1080/19490976.2017.1353849
69. de Theije CG, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, et al. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun (2014) 37:197–206. doi: 10.1016/j.bbi.2013.12.005
70. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell (2016) 167:1469–80 e1412. doi: 10.1016/j.cell.2016.11.018
71. Tognini P. Gut microbiota: a potential regulator of neurodevelopment. Front Cell Neurosci (2017) 11:25. doi: 10.3389/fncel.2017.00025
72. Golubeva AV, Joyce SA, Moloney G, Burokas A, Sherwin E, Arboleya S, et al. Microbiota-related changes in bile acid & tryptophan metabolism are associated with gastrointestinal dysfunction in a mouse model of autism. EBioMedicine (2017) 24:166–78. doi: 10.1016/j.ebiom.2017.09.020
73. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature (2009) 461:1282–6. doi: 10.1038/nature08530
74. Fulceri F, Morelli M, Santocchi E, Cena H, Del Bianco T, Narzisi A, et al. Gastrointestinal symptoms and behavioral problems in preschoolers with autism spectrum disorder. Dig Liver Dis (2016) 48:248–54. doi: 10.1016/j.dld.2015.11.026
75. Vinolo MA, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients (2011) 3:858–76. doi: 10.3390/nu3100858
76. Mazefsky CA, Schreiber DR, Olino TM, Minshew NJ. The association between emotional and behavioral problems and gastrointestinal symptoms among children with high-functioning autism. Autism (2014) 18:493–501. doi: 10.1177/1362361313485164
77. Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, et al. Gastrointestional microbome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology (2011) 141:1782–91. doi: 10.1053/j.gastro.2011.06.072
Keywords: autism spectrum disorder, children, GI problems, gut microbiota, microflora, meta-analysis
Citation: Xu M, Xu X, Li J and Li F (2019) Association Between Gut Microbiota and Autism Spectrum Disorder: A Systematic Review and Meta-Analysis. Front. Psychiatry 10:473. doi: 10.3389/fpsyt.2019.00473
Received: 16 June 2018; Accepted: 13 June 2019;
Published: 17 July 2019.
Edited by:
David Cohen, Université Pierre et Marie Curie, FranceReviewed by:
Magdalena Romanowicz, Mayo Clinic, United StatesCopyright © 2019 Xu, Xu, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Li, ZmVpbGlAc2hzbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.