Corrigendum: Heat Shock Protein HSP24 Is Involved in the BABA-Induced Resistance to Fungal Pathogen in Postharvest Grapes Underlying an NPR1-Dependent Manner
- 1College of Life and Food Engineering, Chongqing Three Gorges University, Chongqing, China
- 2College of Food Science and Technology, Nanjing Agricultural University, Nanjing, China
- 3College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo, China
- 4College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
Although heat shock proteins (HSPs), a family of ubiquitous molecular chaperones, are well characterized in heat stress-related responses, their function in plant defense remains largely unclear. Here, we report the role of VvHSP24, a class B HSP from Vitis vinifera, in β-aminobutyric acid (BABA)-induced priming defense against the necrotrophic fungus Botrytis cinerea in grapes. Grapes treated with 10 mmol L–1 BABA exhibited transiently increased transcript levels of VvNPR1 and several SA-inducible genes, including PR1, PR2, and PR5. Additionally, phytoalexins accumulated upon inoculation with the gray mold fungus B. cinerea, which coincided with the action of a priming mode implicated in pathogen-driven resistance. Intriguingly, electrophoretic mobility shift (EMSA), yeast two-hybrid (Y2H) and His pull-down assays demonstrated that the nuclear chaperone VvHSP24 cannot modulate the transcript of PR genes but does directly interact with VvNPR1 in vivo or in vitro. Furthermore, we found that VvHSP24 overexpression enhanced the transcript levels of NPR1 and SA-responsive genes (PR1, PR2, and PR5) and increased the resistance of transgenic Arabidopsis thaliana to B. cinerea compared with wildtype Col-0. An opposite trend between CRISPR mutants of AtHSFB1 (the orthologous gene of VvHSP24 in Arabidopsis) and wildtype plants was observed. Hence, our results suggest that VvHSP24 has a potential role in NPR1-dependent plant resistance to fungal pathogen. BABA-induced priming defense in grapes may require posttranslational modification of the chaperone VvHSP24 to activate VvNPR1 transcript, leading to PR gene expressions and resistance phenotypes.
Introduction
Grapes are cultivated worldwide with great functional importance in terms of their attractive flavor and constitution of natural phytonutrients (Dai et al., 2011). Nevertheless, the shelf life of postharvest grape berries is very limited, which is mainly ascribed to the invasion of bacterial and fungal pathogens. Indeed, gray mold disease by the fungal pathogen Botrytis cinerea has been recognized as one of the most severe epidemics threatening economic effectiveness in fruit crop farming (Feliziani et al., 2013). A variety of B. cinerea strains can secrete mycotoxins and extracellular cell wall-degrading enzymes to disintegrate and digest the host cell walls of vine crops (Gabler et al., 2003). Ongoing application of synthetic fungicides for controlling gray mold rot of grapevines is highly efficiently, but the occurrences of resistance as well as the negative impacts posed by the frequent use of fungicides have gradually threatened the environment. Hence, eco-compatible alternatives for harmful fungicides to suppress the postharvest spoilage of grapes have been urgently demanded (Mari et al., 2014).
Among a wide spectrum of emerging approaches, several phytohormones and a few organic elicitors have been described as high effectiveness against disease pressure in different kinds of agronomic fruits (Mengiste et al., 2009; Burketova et al., 2015; Romanazzi et al., 2016). To optimize defense against invading pathogens, plants gradually potentiate the systemic immunity driven by stress-induced signaling. This response has been defined as a “priming defense,” which suggests that systemic reactions against pathogen invasions can be triggered (van Hulten et al., 2006). β-aminobutyric acid (BABA), a non-proteinogenic amino acid, has been well documented as a predominant priming elicitor for effective resistance induction and is very desirable for disease management in agricultural fields (Cohen et al., 2016; Baccelli et al., 2017). The biobased resistance inducer of BABA employs its defense functions via abscisic acid (ABA)-responsive and/or phosphoinositide-responsive signaling transductions (Ton et al., 2005). A series of studies utilizing various Arabidopsis defense pathway mutants revealed that BABA not only primes the expressions of AtPR1, AtPR2, and AtPR5 through a salicylic acid (SA) signaling pathway but also intensifies the transcripts of a set of jasmonate acid (JA)- or ethylene (ET)-inducible genes, including AtPDFs, AtVSPs, and AtHELs (Zimmerli et al., 2001; Ton et al., 2002). Hence, BABA-induced resistance (BABA-IR) has been considered as an integrated disease alternative in plants and can be regulated by a signaling crosstalk involving SA, ABA, PI, JA, ET, and other hormone signaling pathways, which is dependent on the stress category (Cohen et al., 2016; AbuQamar et al., 2017; Sham et al., 2019). Similarly, the recent studies concluded that BABA-IR in peach, strawberry and grape occurs in a dose/concentration-dependent manner as a disease-dependent priming defense responding to BABA concentrations ranging from 10 to 50 mmol L–1, in contrast to a direct defense triggered by relatively high concentrations (≥100 mmol L–1 BABA) (Wang et al., 2016, 2018, 2019; Li et al., 2020b). Of note, there are metabolic costs involved in the activation of a certain defense, regardless of what type of BABA-IR is elicited (Perazzolli et al., 2011; Luna et al., 2014). Although the priming defense has some obvious ecological advantages by reducing the excessive cost of cellular resources unless plants suffer from severe hostile pressures (van Hulten et al., 2006), the critical node and its function underlying a priming defense have not been characterized in the specific plant-fungus interactions.
Biotic and abiotic stresses are environmental constraints that differ from natural or optimal conditions, impeding normal cellular functions and constituting a major hindrance to plant growth and development (Uyttewaal et al., 2010). Heat shock proteins (HSPs) are ubiquitous molecular chaperones that are recruited under diverse stressful conditions, including heat stress and other non-heat environmental stresses, such as osmotic, drought, salt, ultraviolet (UV) light, oxidative stresses, and pathogen infections (Swindell et al., 2007). HSPs/chaperones may serve as hubs in the proteostasis network by assisting in protein folding, protein assembly, compartment targeting, degradation, and signaling (Baniwal et al., 2004; Swindell et al., 2007). It has been evident that the expression of HSPs is constitutively regulated at the transcriptional level by heat shock transcription factors (HSFs), which are the terminal components of a signal transduction pathway modulating the transcript of genes responsive to distinct stimuli (von Koskull-Döring et al., 2007; Xin et al., 2010). Arabidopsis contains 21 HSF representatives that have been classified into three classes (A, B, and C) based on their characteristic cluster of DNA-binding domains (DBDs) and adjacent oligomerization domains (ODs, HR-A/B regions) (Nover et al., 2001; Scharf et al., 2012). Class B and Class C HSFs are characterized by the absence of the typical activator motif aromatic hydrophobic acidic (AHA), which is essential for the transcriptional activity of Class A HSFs (Guo et al., 2016). Along this line, HSF1a, HSF1b, HSF1d, and HSF1e are all responsible for basal thermotolerance, and the heat-acclimation phenotype fundamentally involves HSFA2, HSFA3, and HSFA7a (Bharti et al., 2000; Nishizawa et al., 2006; Larkindale and Vierling, 2008; Zheng et al., 2016). Expression of AtHSFA4a is transcriptionally elicited by numerous stresses, such as cold, drought and UV-B, and transgenic Arabidopsis overexpressing HSFA4a showed enhanced tolerance to those stresses (Pérez-Salamó et al., 2014). Enhancements of AtHSFA2 and AtHSFA7a transcripts have been monitored during virus infection, revealing the involvement of these two HSFs in the disease resistance of Arabidopsis (Sugio et al., 2009). Moreover, overexpression of HSP27 strongly augmented plant resistance to various apoptotic stimulis (Samali et al., 2001; Tanabe et al., 2010), and the multichaperone complex HSP70-HSP90 is implicated in resistance protein (R protein)-mediated immunity and is essential for proper defense signal transduction (Noël et al., 2007; Shirasu, 2009; Li et al., 2012).
For decades, HSFs/HSPs have been recognized as hallmarks of the environmental stress response, and research on most HSFs/HSPs has typically focused on their central roles in response to several abiotic stresses, in particular heat stress management, in model plants such as Arabidopsis thaliana, peruvian tomato (Solanum peruvianum), rice (Oryza sativa), and wheat (Triticum aestivum). Nonetheless, much less is known about the functional modulation of HSFs/HSPs against biotic stresses, especially against fungal pathogens, in horticultural crops. In this research, our focus extends the exploration of the chaperone function and potential regulation of HSFs/HSPs involved in the BABA-elicited priming response to fungal infection in grape fruit.
Materials and Methods
Plant and Fungi
Grapes (Vitis vinifera Cv ‘Kyoho’) from 6-year-old vines were purchased and hand picked from a local supplier with a standard orchard in Chuanshan District, Suining City, Sichuan Province, China. The grapes coincided with some of reliable quality parameters, including the berry diameter of 1.8–2.2 cm, total soluble solids (SCC) of 13.4–15.7%, titratable acidity (TA) of 0.64–0.72%, pH value of 3.14–3.27 and firmness of 4.7–6.3 N cm–2 were late-harvested at a commercial maturity stage that was identified according to the Balic et al. (2014) and Rosales et al. (2016). The harvested grapes were transported to our laboratory within 1 h by an air-condition vehicle (20°C).
Arabidopsis thaliana wildtype (WT) Columbia-0 (Col-0) plants were applied for genetic transformation, and WT and transgenic Arabidopsis lines were grown in a SPT-P500B illumination incubator (Darth carter Co., Hefei, China) equipped with an automatic stable controller for maintaining the R.H. at 60–70%, illumination at 2500–3000 lx and temperature at 22°C with a long-day photoperiod (light/dark cycle of 14 h/10 h) over 4 weeks.
Botrytis cinerea (Pers.: Fr.) B05.10 strain (phylogenetic analysis shown in Supplementary Figure 1) was isolated from decayed ‘Kyoho’ grape berries (Lovato et al., 2019) and cultured on PDA medium (Haibo Co., Qingdao, China) for 14 days at 26°C. The cultured medium was then rinsed with sterile water containing 0.5% (v/v) Tween-80 to produce B. cinerea suspensions and diluted to a final concentration of 1.0 × 105 spores mL–1, as counted with a Neubauer chamber.
Grape Treatments
Intact grape berries were picked and partitioned into four lots of 360 grapes each, which were superficially sterilized by 75% (v/v) alcohol and dried on sterile filter paper at 20°C for 2 h. Two symmetrical holes were then wounded (1.5-mm-deep × 1.5-mm-diameter) at the equator of each grape with a dissecting needle. BABA (purity ≥ 99%, Sigma Co., St. Louis, MO, United States), at 10 mmol L–1, was selected as the most effective concentration in basis of our current findings (Wang et al., 2019, 2020). Grapes in each lot were exposed to the four treatments following the description of Porat et al. (2003): (1) control, berries solely mock-inoculated with sterile water; (2) BABA, berries solely elicited with BABA solution; (3) inoculation, berries solely challenged with the B. cinerea suspensions; and (4) BABA + inoculation, berries elicited with BABA and subsequently challenged with B. cinerea suspensions. All treated grapes were then ventilated on stainless steel mesh for 6 h, transferred to PE boxes (60-μm-thick walls) and arranged in incubators for 5 days under 20 ± 1°C and 85 ± 5% R.H. A total of 30 g tissues from the healthy sarcocarp (3 mm distant from the diseased holes) of 60 grapes in each lot were sliced by a sterile scalpel before the incubation (0 h) and at 1-day intervals during the incubation. Tissues taken from each individual were rapidly frozen in liquid N2 and kept at –80°C until analysis. Each treatment was performed according to a completely randomized design and repeated three times, and the experiment was performed twice.
Assessment of Disease Development
If the width of the dot-inoculated area was beyond 1.5 mm, the grape could be defined to be diseased. Percentage of decaying grapes was calculated as disease incidence. The disease development of twenty berries from each triplicate was monitored continuously for 5 days at the intervals of 2 days.
Measurement of Phytoalexin Content
For phytoalexin extraction, approximately 5 g of frozen pulp was ground in liquid N2 and homogenized with 25 mL of 85% (v/v) methanol in ice bath. The homogenates were transferred into a dark incubator at 4°C overnight and centrifuged at 10,000 × g for 15 min (4°C). The supernatants were retained and evaporated under nitrogen, and the residue was further dissolved in absolute methanol before filtration. The individual phytoalexin content in the prepared extractions was measured by a HPLC analysis as described by Vitrac et al. (2005).
Classification and Sequence Analysis of HSFs and HSPs
Annotated complete lists of HSFs and HSPs in Vitis vinifera and Arabidopsis thaliana were obtained from the NCBI database. Furthermore, the phylogenetic relationship among the 37 identified Vitis vinifera HSFs/HSPs (VvHSFs/VvHSPs) and 20 Arabidopsis thaliana HSFs (AtHSFs) was conducted using MEGA 5.0 software with the neighbor-joining (NJ) method with 1000 bootstrap replicates and further visualized via the iTOL webtool. In addition, the online web resources SMART (Letunic and Bork, 2018) and TBtools (Chen et al., 2020) were used to visualize the conserved domains and sequence conservation, respectively.
RNA Isolation and Real-Time qRT-PCR
Aliquots (5 g) of frozen tissues were ground in liquid N2, and total RNA was extracted using RNAprep Pure Kit for plants (Tiangen, China). Aliquots (1 μg) of RNA were prepared to synthesize first-strand cDNA using a PrimeScriptTM RT reagent kit (Takara, Japan). Quantitative real-time PCR (qRT-PCR) was conducted in triplicate using an ABI 7500 Fast Real-Time PCR system (Applied Biosystems). The 10-μL qPCR mixture contained 0.2 μL of each individual 10 μmol L–1 primer for VvHSP24, SA-dependent genes including VvNPR1, VvPR1, VvPR2, and VvPR5 as well as JA/ET-dependent genes including VvPDF1.2, VvVSP1, VvHEL, VvFAD3, VvTHI2, VvERS1, and VvERF1 (listed in Supplementary Table 1), 1 μL cDNA template, 5 μL 2 × ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) and 3.6 μL ddH2O. The thermal cycle for qRT-PCR was 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s with the ABI-specified hold and melt curve stages. Amplicon lengths were optimized to 90–150 bp to ensure optimal polymerization efficiency. qRT-PCR results were normalized by the cycle threshold value (CT) using the reference gene VvActin7 or AtActin2 according to the 2–Δ Δ CT method (Livak and Schmittgen, 2001). Relative expression levels of HSP/HSF and PR genes were calibrated, with the values for the control fruit/plant being set as 1.
Subcellular Localization
PCR-amplified VvHSP24 was cloned into the pEAQ-GFP vector to construct the pEAQ-VvHSP24-GFP fusion protein and the pEAQ-GFP empty vector was used as the positive control. Then, onion bulb epidermis cells were transformed by inoculation with A. tumefaciens strain EHA105 harboring pEAQ-GFP and the recombinant construction of pEAQ-VvHSP24-GFP via an Agrobacterium-mediated system, respectively. Briefly, the adaxial epidermises obtained from onion bulb were cultured on Murashige and Skoog (MS) medium for approximately 1 week and then immersed overnight in the suspension of EHA105 cells containing pEAQ-VvHSP24-GFP or pEAQ-GFP, respectively. Following 3 days of incubation on MS medium at 26°C in darkness, these epidermises were stained with the nuclear dye 4,6-diamidino-phenylindole (DAPI, 20 μg mL–1, Sigma, United States). Then, the fluorescence signal of the stained onion peels was captured by a Zeiss LSM 510 confocal laser scanning microscope with excitation wavelength set at 405 nm for DAPI fluorescence and 488 nm for GFP fluorescence and emission wavelength in the range of 450–550 nm at 3 h after DAPI staining.
Y2H Assay
Y2H assay was performed by employing the MatchmakerTM Gold Y2H System (Clontech). Full-length cDNA clones of VvHSP24 and VvNPR1 in BABA-treated, B. cinerea-infected berries were inserted into the pGBKT7 and pGADT7 plasmids, respectively. The recombinant plasmids, including BD-VvHSP24 and AD-VvNPR1, were cotransformed into yeast strain AH109 cells, which were further smeared on double dropout medium (DDO, SD/-Leu-Trp) and cultured for about 3 days at 29°C (Li et al., 2020b). Transformed colonies were cultured on triple dropout medium (TDO, SD/-Leu-Trp-His) and quadruple dropout medium (QDO, SD/-Leu-Trp-His-Ade) with or without 40 mg L–1 X-α-gal to determine possible interaction.
His Pull-Down Assay
The recombinant GST-VvNPR1 and His-VvHSP24 proteins were obtained through fusing with pGEX-4T-1 or pET-28a vector, respectively. The recombinant proteins were transformed into Escherichia coli BL21 (DE3) cells and expressed at 20°C for 3 h after the addition of 0.1 mmol L–1 IPTG (isopropyl β-D-galactopyranoside). Bacteria expressing the GST-VvNPR1 fusion protein were lysed by sonication and purified using glutathione agarose (Thermo Fisher Scientific, United States) following the instructions. E. coli harboring the His-VvHSP24 fusion protein were isolated and purified using a B-PER (R) 6 × His Spin Purification Kit (Pierce, Rockford, IL, United States). For His pull-down assay, the purified His-VvHSP24 fusion protein was immobilized with Ni sepharose 6 Fast Flow (GE Healthcare, code number: 17-5318-01) and mixed for 2 h at 4°C. Then, 5 g of GST-VvNPR1 fusion protein was incubated with 15 μL of prewashed resin overnight (4°C) before being washed three times with cold phosphate-buffered saline (PBS) based on standard protocols. Pulled down proteins (6 μL) were boiled in 2× Laemmli buffer and detected by Western blotting with an anti-GST antibody (TransGen Biotech Co., Ltd., Beijing, China, HT601); the His + GST-VvNPR1 fusion protein was used as a negative control.
EMSA Assay
The CDS fragment of VvHSP24 was synthesized and cloned into the pCzn1 vector to produce the His-VvHSP24 recombinant protein and introduced into ArcticExpress (DE3) chemically competent cells. Then, 0.5 mmol L–1 IPTG was applied to induce the expression of the target recombinant protein (His-VvHSP24) at 15°C for approximately 9 h and then purified using a Ni-NTA His-Bind resin column (Novagen, Gibbstown, NJ, United States). The heat stress elements (HSE: 5′-GAAnnTTC-3′), which could be recognized by HSFs/HSPs, were detected in the promoters of PR1, PR2 and PR5 (Supplementary Data 1), and their nucleotides adjacent to HSF motifs (approximately 50 bp) were adopted as probes, respectively. These probes were labeled at the 5′ end with biotin using Light Shift chemiluminescent EMSA kit (Pierce Biotechnology, Rockford, IL, United States) and incubated with the purified His-VvHSP24. The protein-DNA complexes formed during EMSA experiments were transferred electrophoretically to a nitrocellulose membrane after separated by SDS-PAGE. Then, the shift bands were visualized via a chemiluminescence reaction on X-ray films. The addition of 200-fold mutant probes with the mutant HSF motif served as the competitors for the 5′-labeled WT DNA probes to test for the specific of the binding assays; the biotin-EBNA control DNA + EBNA extract was used as a positive control.
Overexpression and Analysis of Vitis vinifera HSP24 in Transgenic Arabidopsis Plant
The coding region of VvHSP24 was amplified by PCR technology from a grape berry cDNA library with gene-specific primer listed in Supplementary Table 1. The construction of a plasmid for overexpression of VvHSP24 by introducing the VvHSP24 fragment into the binary vector pCambia 1305.1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Arabidopsis plant was transformed with A. tumefaciens GV3101 harboring the resulting 35S:VvHSP24 construct using the floral dip method (Clough and Bent, 1998). The harvested T1 seeds were surface sterilized in 2% (v/v) sodium hypochlorite for 10 min and stratification at 4°C for 3 days and then germinated on MS medium containing 50 mg L–1 kanamycin (kan) for more than 7 days. The kan-resistant seedlings were sown in pots containing a 1: 2 perlite-vermiculite mixture and cultured in illumination incubator with long-day photoperiod. Ultimately, homozygous plants of the T3 generation were used for subsequent experiments.
Generation of HSFB1 Mutants by CRISPR-Cas9
Mutations were introduced into Arabidopsis HSFB1 through the non-homologous end joining (NHEJ) method of RNA-guided endonuclease-mediated targeted mutagenesis with the clustered regularly interspersed short palindromic repeats (CRISPR)-Cas9 system (Lowder et al., 2015; Li et al., 2019). In brief, the gene-targeting binary vector M2CRISPR, with specific guide RNA (sgRNA) cassettes under the control of the U26:U29 promoter as well as an EC promoter driving Cas9 expression, was provided by Shanghai Weidi Biotechnology Co., Ltd. sgRNA cassettes were designed using sequences corresponding to the protospacer adjacent motif (PAM) sites (set as ‘NGG’) on the sense or antisense strand of the targeted AtHSFB1 gene, and the web server CRISPRdirect1 was used to check their off-target effects. Then, two sgRNA cassettes were annealed, and PCR products were further ligated into the BsaI-digested vector M2CRISPR to generate a U26 promoter-driven T1 sgRNA and U29 promoter-driven T2 sgRNA (Supplementary Figure 4). The primers used are provided in Supplementary Table 1. The resulting binary vector was used for A. tumefaciens GV3101-mediated genetic transformation of Arabidopsis. T1 generation transformants were selected on MS medium containing 50 mg L–1 hygromycin phosphotransferase (HYG), and the effectiveness of genome editing was evaluated using the HYG-resistant seedlings. Genomic DNA from true leaves was isolated, and the targeted genomic region was amplified by PCR using KOD FX (Toyobo Co. Ltd., Osaka, Japan) with the primers listed in Supplementary Table 1. The resulting PCR products were sequenced, and mutants were identified by comparison with WT sequences. INDELs (insertions and deletions) at the targeted sites were considered as mutations. The T1 mutant seedlings were transplanted into soil, and T2 generation seeds were collected. After the identification of homozygous mutants, the T2 seeds were used to produce T3 seedlings.
Inoculation of Arabidopsis Plant With Fungal Pathogen B. cinerea
Four- to five-week-old T3 transgenic and wildtype (Col-0) Arabidopsis were dot-inoculated with B. cinerea conidial suspensions (1.0 × 105 conidia mL–1) as previously described (Xiao and Chye, 2011). Detached leaf samples were used for morphological observations at 6 dpi and leaves picked at 0, 3, and 6 dpi were used to test for the transcript levels of disease resistance-related genes.
Trypan Blue Staining
Trypan blue staining was employed to assess the rate of cell death and fungal development of B. cinerea-infiltrated leaves from Arabidopsis WT (Col-0), VvHSP24-overexpressing plants and CRISPR mutants at 6 dpi, as described previously, with minor modifications (Cai et al., 2020). B. cinerea-infected Arabidopsis leaves were immersed and boiled in trypan blue staining solution [0.025% (m/v) trypan blue, 25% (m/v) phenol, 25% (v/v) lactic acid, and 25% (v/v) glycerol] for 5 min and then allowed to cool naturally and soak overnight. The stained leaves were immersed in 1.25 g mL–1 chloral hydrate solution for more than 24 h to eliminate chlorophyll; the samples were observed by binocular stereomicroscopy (Leica EZ4D) and photographed.
Electrolyte Leakage Measurement
Estimation of electrolyte leakage was carried out as previously described (Mackey et al., 2002). Leaf disks (8 mm diameter) of B. cinerea-infiltrated leaves from WT and VvHSP24-overexpressing Arabidopsis were washed in 50 mL ddH2O at least twice and then transferred to 10 mL ddH2O and floated for 8 h at 120 rpm and 25°C. Water conductivity was detected over time using a conductivity meter (MIK-TDS210-B).
Statistical Analysis
All data were subjected to t-test and one-way analysis of variance according to a Tukey’s test using SAS version 8.2 (SAS Inst., NC, United States); data were reported as the mean ± standard error of three replicates from one independent experiment. Different letters and asterisks indicated that the differences were statistically significant at the 0.05 or 0.01 level of the p-value.
Results
Inhibitory Effectiveness of BABA Treatment Against B. cinerea Infection in Grapes
As presented in Figure 1, grape berries are obviously susceptible to the necrotrophic pathogen B. cinerea and became deteriorated after infection at 20°C. 10 mmol L–1 BABA treatment caused a significant inhibition in disease development in postharvest grapes, and their lesion diameter and disease incidence were 32.16 and 32.32%, respectively, lower compared with the only B. cinerea-inoculated fruit at 5 dpi (Figure 1 and Supplementary Figure 2).
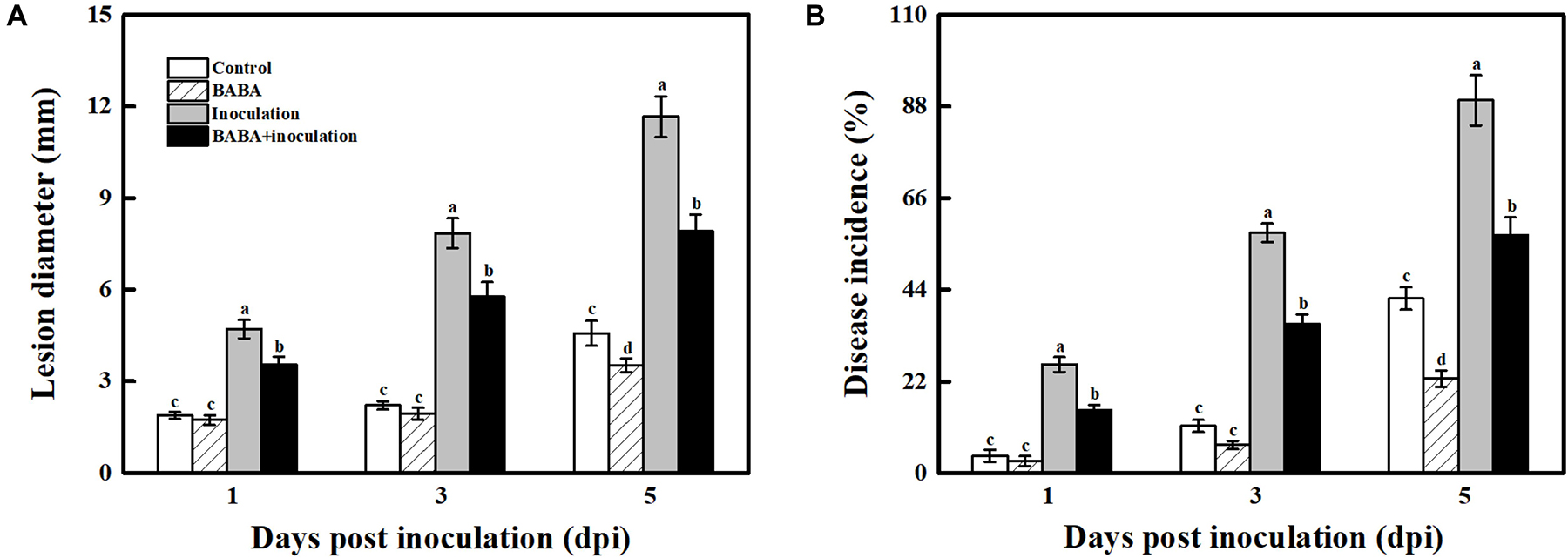
Figure 1. Impact of BABA on the disease development of grapes altered in their responses to the fungal pathogen B. cinerea. Lesion diameter (A) and disease incidence (B) measured at 1, 3, and 5 days post inoculation with B. cinerea. Grapes were pretreated with water (gray bars) or BABA (black bars) before inoculation. Disease development is expressed as the mean ± SE of nine assessments. Different letters above the bar indicate a significant difference (p < 0.05) between grapes with BABA-treated or B. cinerea-inoculated and untreated cases.
Molecular Identification and Characterization of VvHSP24
Given that AtHSFB1—a class B-HSF from Arabidopsis, also referred to as AtHSF4 or AtTBF1—plays a positive role in the development of defense priming and systemic acquired resistance (SAR, Pajerowska-Mukhtar et al., 2012; Pick et al., 2012), we compared the amino acid sequences of HSFs/HSPs from Vitis vinifera specie and those of Arabidopsis thaliana HSFs by constructing a phylogenetic tree. According to the phylogenetic tree of Vitis vinifera HSFs/HSPs with AtHSFs, VvHSP24 and AtHSFB1 grouped into class B and a single conserved HSF_DNA-binding domain consisting of 94 amino acids is present at residue ranges of 11–104 and 6–99 in AtHSFB1 and VvHSP24, respectively (Supplementary Figure 3). Hence, the expression of VvHSP24, an AtHSFB1 orthologous gene in Vitis vinifera, might be related to grape defense. In addition, GFP-tagged VvHSP24 was exclusively detected in the nucleus of onion epidermal cells, contrarily the positive control, i.e., GFP alone, was found in both the cytoplasm and nucleus (Figure 2). As a result, VvHSP24 might be a nucleus-localized regulatory protein associated with the development of defense priming.
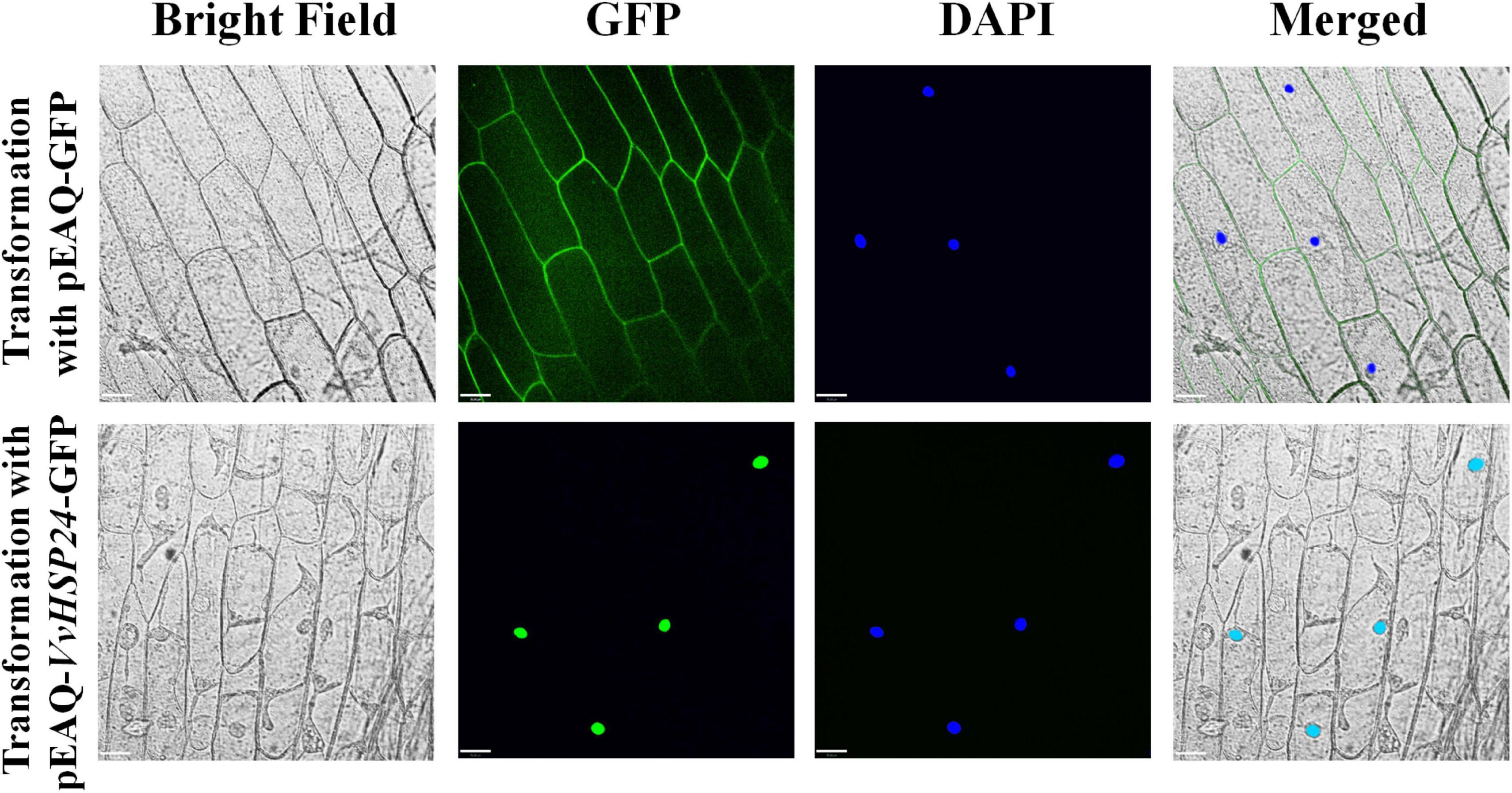
Figure 2. Nuclear localization of pEAQ-VvHSP24-GFP fusion protein. The fluorescence signals of pEAQ-GFP and pEAQ-VvHSP24-GFP fusion protein were observed with onion peels after transformation by Agrobacterium tumefaciens. Nuclei were visualized by staining onion peels with 4,6-diamino-2-phenylindole (DAPI), and GFP signals were captured using LSM (bars = 70 μm).
Effect of BABA Treatment on Transcript of VvHSP24 and SA-Dependent Genes in Grapes
For analyzing the intensity of BABA-triggered SAR defense, the qRT-PCR method was used to quantify the transcript levels of the VvHSP24 and SAR marker genes in grapes throughout 5 days of the inoculation. As shown in Figure 3A, treatment of grapes with 10 mmol L–1 BABA enhanced expression of the VvHSP24 gene within the incubation period of 5 days, similar to the trend in our previous RNA-seq database2 (Supplementary Figure 5). Thus, VvHSP24 may indeed participate in BABA-IR in grapes upon the perception of disease stress. In addition, although grapes reacted to BABA pretreatment with enhanced transcripts of VvNPR1 and SA-responsive genes such as VvPR1, VvPR2, and VvPR5 following mock inoculation with ddH2O, the reaction was much more pronounced in B. cinerea-inoculated grapes than in non-fungal-infected grapes. In BABA-treated and then B. cinerea-inoculated grapes, accumulation of VvNPR1 and SA-inducible genes transiently peaked at 1 day post inoculation (dpi) followed by a gradual decrease (Figures 3B–E). Thus, SA-dependent priming of pathogen responses was boosted in BABA-pretreated grapes because the transient accumulation of mRNA transcript levels detected in SA-dependent marker genes is a hallmark of the priming mode for BABA-IR (Ton et al., 2005).
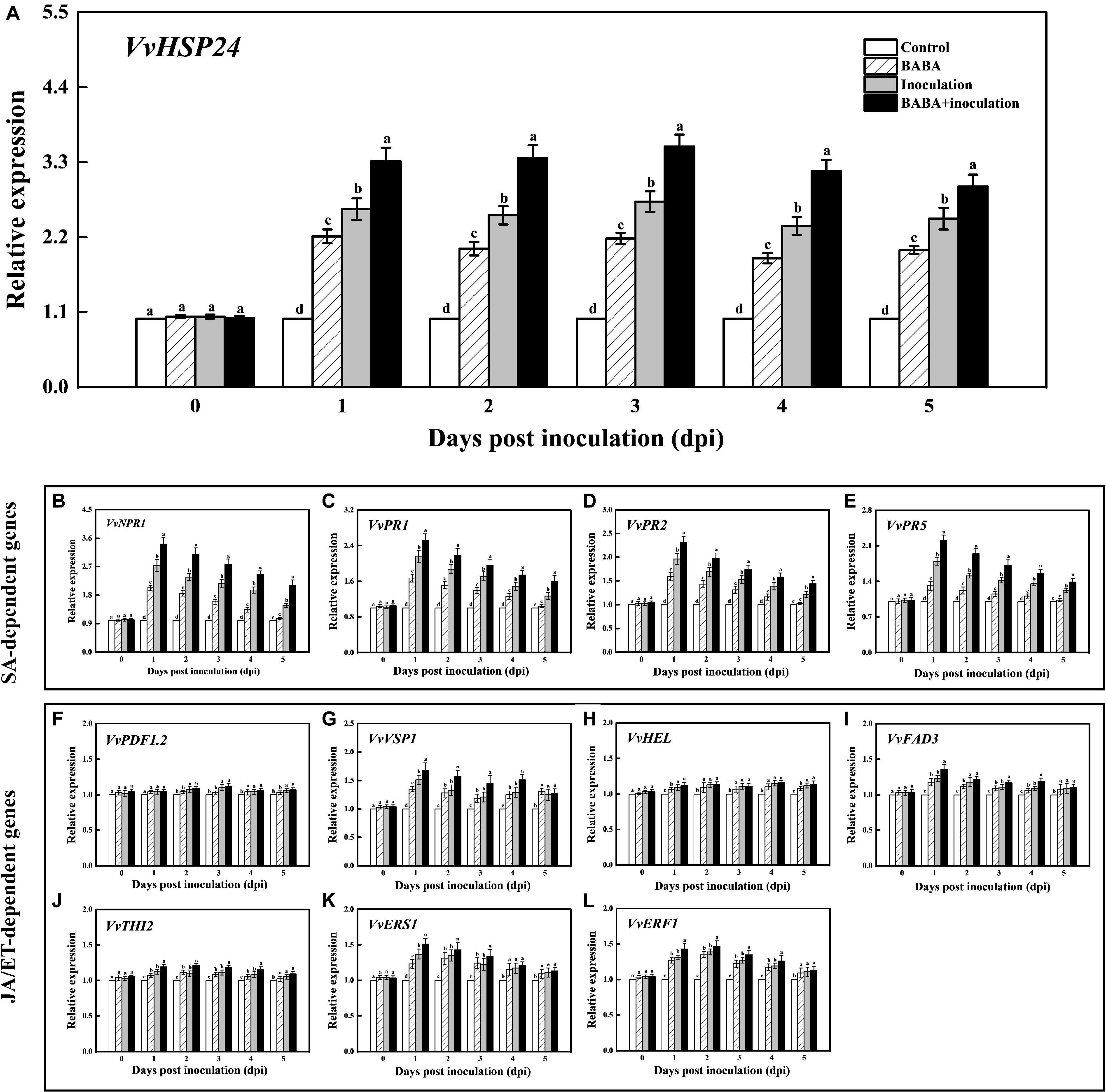
Figure 3. Transcript levels of VvHSP24 (A), SA-dependent VvNPR1 (B), VvPR1 (C), VvPR2 (D) and VvPR5 (E) genes and JA/ET-dependent VvPDF1.2 (F), VvVSP1 (G), VvHEL (H), VvFAD3 (I), VvTHI2 (J), VvERS1 (K), and VvERF1 (L) genes in grapes after inoculation with B. cinerea for 1-day intervals. Relative mRNA expression levels are expressed as the mean ± SE. Different letters above the bar represent a significant difference between treatments (p = 0.05).
Effect of BABA Treatment on Transcript of JA/ET-Dependent Genes in Grapes
It is well-documented that a subset of genes, including VvPDF1.2 (encoding a plant defensin), VvHEL (encoding a hevein-like protein), VvVSP1 (encoding an acid phosphatase), VvTHI2 (Encoding a antimicrobial thionin), VvFAD3 (encoding a fatty acid desaturase), VvERS1 (encoding an ethylene response sensor), and VvERF1 (encoding an ethylene response factor), are regulated by JA or ET signaling pathway and associated with a resistance against hostile threats (Zarate et al., 2007). Thus, the transcript levels of the above genes were assessed for determining the inductive effectiveness of BABA elicitation on JA/ET-dependent defense. Throughout the whole incubation period, the treatment of B. cinerea-inoculated grapes with 10 mmol L–1 BABA did not induce the significant change in the transcript levels of VvPDF1.2, VvHEL, and VvTHI2 compared with the only B. cinerea-inoculated samples (Figures 3F,H,J). On the contrary, an inductive effect of BABA on gene expressions of VvVSP1, VvFAD3, VvERS1, and VvERF1 could be observed in the BABA-stimulated and subsequent B. cinerea-challenged grapes (Figures 3G,I,K,L), indicating that the JA/ET-responsive genes were in part implicated in the BABA-induced priming defense.
Effect of BABA Treatment on Individual Phytoalexin Content in Grapes
To assess whether BABA-fueled defense was related to phytoalexin biosynthesis in grape berries, the contents of the dominant stilbenes in Vitis vinifera species, including trans-resveratrol and its oligomer ε-viniferin, were measured (Sarig et al., 1997). As depicted in Figure 4, the trans-resveratrol and ε-viniferin contents in the controls fluctuated within a very small range (from –23.72 to 17.06%) around their average values over 5 days at 20°C. BABA treatment alone was sufficient to promote stilbene synthesis in grapes, and subsequent B. cinerea inoculation resulted in an obvious enhancement of trans-resveratrol and ε-viniferin compared with the controls. It is worth noting that the amount of phytoalexin was higher in fruit treated with BABA elicitation in combination with B. cinerea suspension than that in non-B. cinerea-inoculated or non-BABA-treated grapes within 5 dpi, indicating the BABA elicitation exhibited a positive regulation on stilbene phytoalexin biosynthesis in grapes.
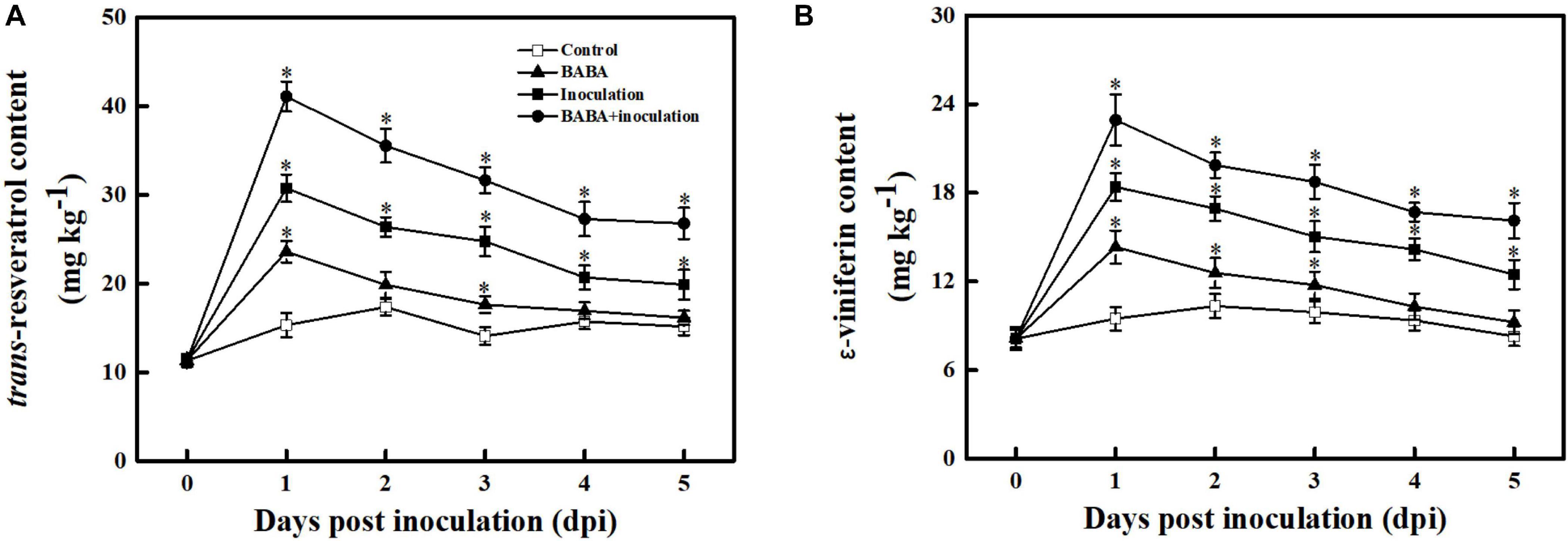
Figure 4. Changes in the amounts of trans-resveratrol (A) and ε-viniferin (B) in grapes after inoculation with B. cinerea at 1-day intervals. Phytoalexin contents are expressed as the mean ± SE. An asterisk represents a significant difference between pathogen- or BABA-treated and untreated grapes (p = 0.05).
Direct Interaction Between VvHSP24 and VvNPR1
The Y2H system was used to identify whether physical interaction occurs between VvHSP24 and VvNPR1 in vivo; meanwhile, the interaction between VvHSP24 and VvNPR1 in vitro was conducted by His pull-down. As shown in Figure 5A, colonies of competent yeast AH109 cotransfected with the BD-VvHSP24 and AD-VvNPR1 vectors grew within 4–6 days on synthetic dropout plates (SD/-Trp-Leu-His and SD/-Trp-Leu-His-Ade), and these cotransformants were blue on X-α-gal plates. Few or no colonies carrying BD-VvHSP24 plus the empty pGADT7 vector or AD-VvNPR1 plus the empty pGBKT7 vector were obtained. Therefore, the interaction between VvHSP24 and VvNPR1 was confirmed by the successful activated GAL4-responsive reporter genes.
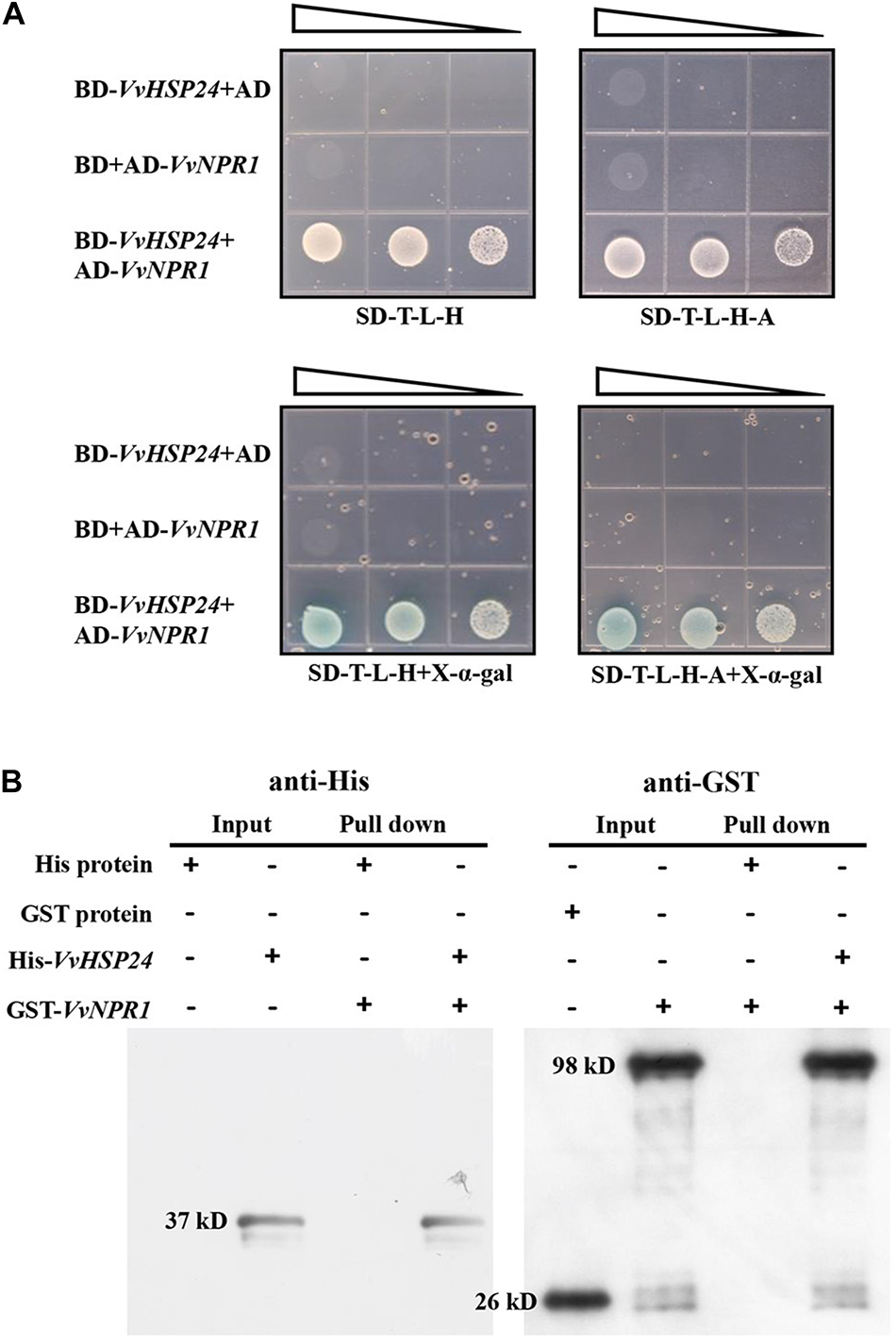
Figure 5. VvHSP24 interacts with VvNPR1 in vivo and in vitro. (A) Yeast two-hybrid analysis of the physical interaction between VvHSP24 and VvNPR1. SD-T-L-H, SD/-Trp-Leu-His agar medium; SD-T-L-H-A, SD/-Trp-Leu-His-Ade agar medium. The right-angled triangles on the top of the gridded Petri dishes represent the absorbance of yeasts at 600 nm in a 10-fold dilution series, from 1 to 10–2 abs. (B) The GST-fused VvNPR1 protein (1 mL) was incubated with 1 mL of preimmobilized His-VvHSP24 protein in a total volume of 25 mL at 4°C for more than 8 h. The pulled down proteins (6 μL) were analyzed by western blotting with anti-His or anti-GST antibodies.
Figure 5B illustrated that two intense bands observed at the position corresponding to the estimated sizes of His-VvHSP24 (37 kD) or GST-VvNPR1 (98 kD) when incubated with anti-His or anti-GST antibodies, respectively. For detection of a pulled down protein, a His-tagged VvHSP24 fragment protein immobilized on Ni sepharose 6 Fast Flow was used as bait against the GST-VvNPR1 fusion protein. As shown in lanes 7 and 8 of Figure 5B, GST-VvNPR1 was pulled down by the His-VvHSP24 fusion protein but not by the unfused His; hence, the VvHSP24 protein physically interacted with VvNPR1 in vitro.
VvHSP24 Cannot Directly Regulate the Transcript of PR Genes
qRT-PCR results showed that PR genes such as PR1, PR2, and PR5 display similar long-term potentiation (LTP) trends as VvHSP24 after BABA elicitation (Figure 3). In particular, previous studies have shown that HSFs/HSPs regulate the transcript of the target genes primarily by interacting with HSF sequence-binding elements (HSEs) (Nover et al., 2001; Guertin and Lis, 2010). HSEs (5′-GAAnnTTC-3′) were detected in the promoters of all PR genes (Supplementary Data 1). Hence, we tested the possibility whether VvHSP24 could directly control PR gene transcripts. Unexpectedly, the His-VvHSP24 fusion protein was unable to bind the biotin-labeled probes targeting the HSE motifs from the PR1, PR2, and PR5 promoters; indeed, mobility shifts were not observed, and the addition of mutated HSEs had no effect (Figure 6). However, a mobility shift was produced when biotin-EBNA control DNA was incubated with EBNA extract. Thus, the VvHSP24 could not directly regulate the transcript of PR genes.
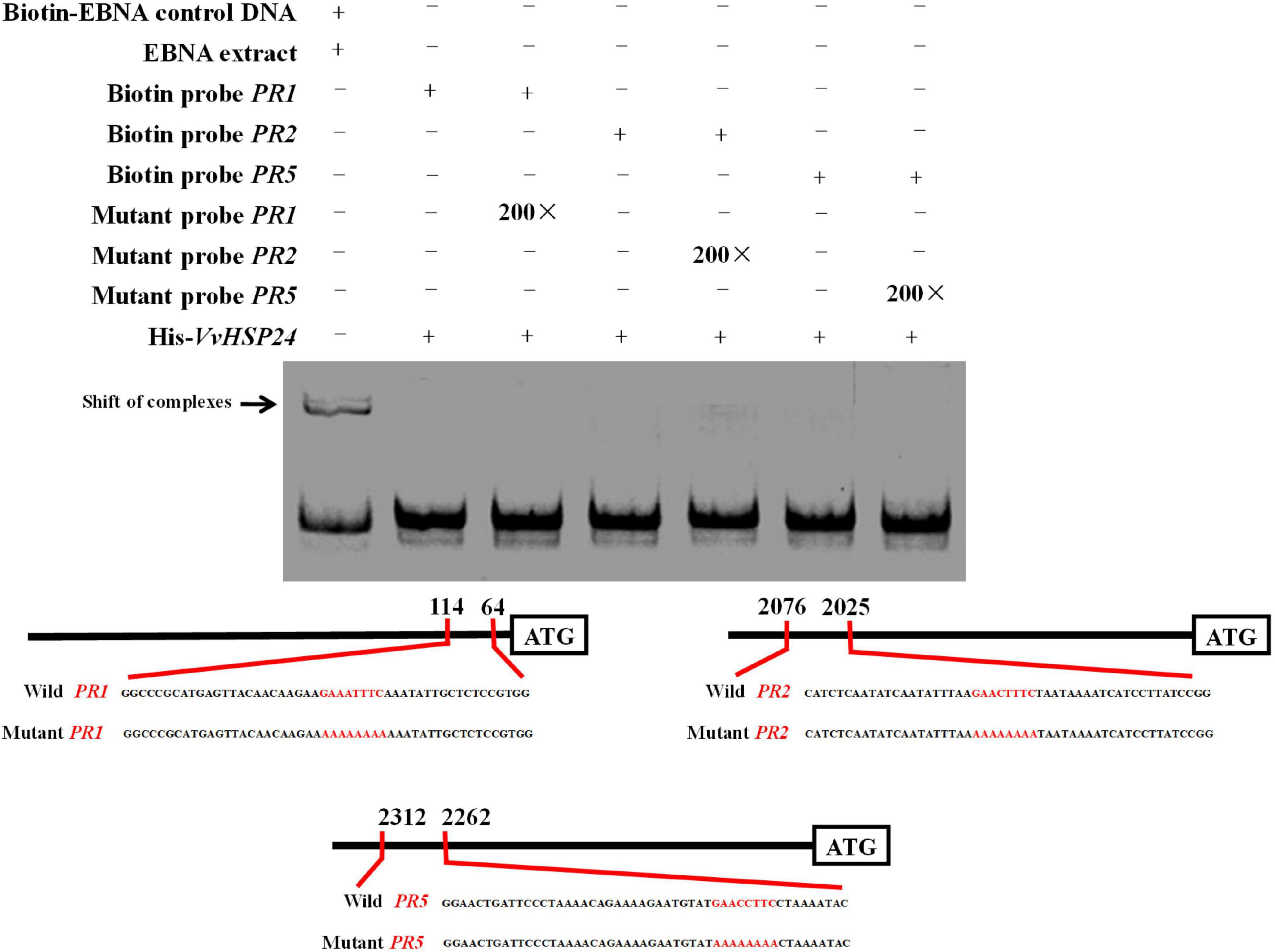
Figure 6. The binding patterns of VvHSP24 to the promoters of PR1-, PR2-, and PR5-containing HSE motifs were detected by EMSA. The HSE elements within the probe sequences are highlighted with red letters. The recombinant His-VvHSP24 protein was incubated with biotin-labeled probes and then the protein-DNA complexes were resolved on polyacrylamide gels and visualized by exposure of the dehydrated gels to X-ray films. The addition of 200-fold mutant probes was performed in the competitive binding assays to test for the specific of the binding assays between VvHSP24 and PR1, PR2, and PR5 promoters.
Heterologous Expression of Vitis vinifera HSP24 Enhances the Resistance of Arabidopsis to B. cinerea and Mitigates Cell Death in Transgenic Leaves
As previously reported, AtHSFB1 is a critical player in the development of defense priming (Pajerowska-Mukhtar et al., 2012; Pick et al., 2012). To examine the effectiveness of VvHSP24 (an orthologous gene of AtHSFB1 from Vitis vinifera) in necrotrophic pathogen-induced defense responses, transcript levels of VvHSP24 and pathogenesis-related genes (NPR1, PR1, PR2, and PR5) in B. cinerea-inoculated WT plants and VvHSP24-overexpressing lines were monitored. qRT-PCR analysis showed that VvHSP24 gene expression was significantly enhanced in the VvHSP24-overexpressing lines, especially in OE-5, OE-7, and OE-10, in comparison with Col-0 WT (Figures 7A,B). As shown in Figure 7E, mRNA levels of AtNPR1, AtPR1, AtPR2, and AtPR5 were upregulated in the three VvHSP24-overexpressing lines (OE-5, OE-7, and OE-10) at 3 and 6 dpi compared with WT plants. To determine pathogen growth- and cell death-inducing activity, we further observed B. cinerea-inoculated leaves from WT and VvHSP24-transgenic plants by imaging after staining with lactophenol trypan blue. At 6 days after B. cinerea inoculation, almost all of the cells of WT leaves were basically dead and stained with trypan blue; in contrast, lesions and necrotic areas were visibly reduced in all transgenic lines with respect to dot-inoculated WT plants (Figures 7F,G). Coincidently, cell electrolyte leakage from transgenic leaves infiltrated with the B. cinerea conidial suspension was also significantly lower than that from WT plants (Figure 7H). Of note, heterologous expression of HSP24 from Vitis vinifera apparently retarded the growth of transgenic Arabidopsis plants, as displayed with dwarf leaves, smaller leaf area and less biomass in the transgenic lines than in the WT plants (Figures 7C,D). These data reflected that overexpression of VvHSP24 can obviously alleviate cell damage and disease progression.
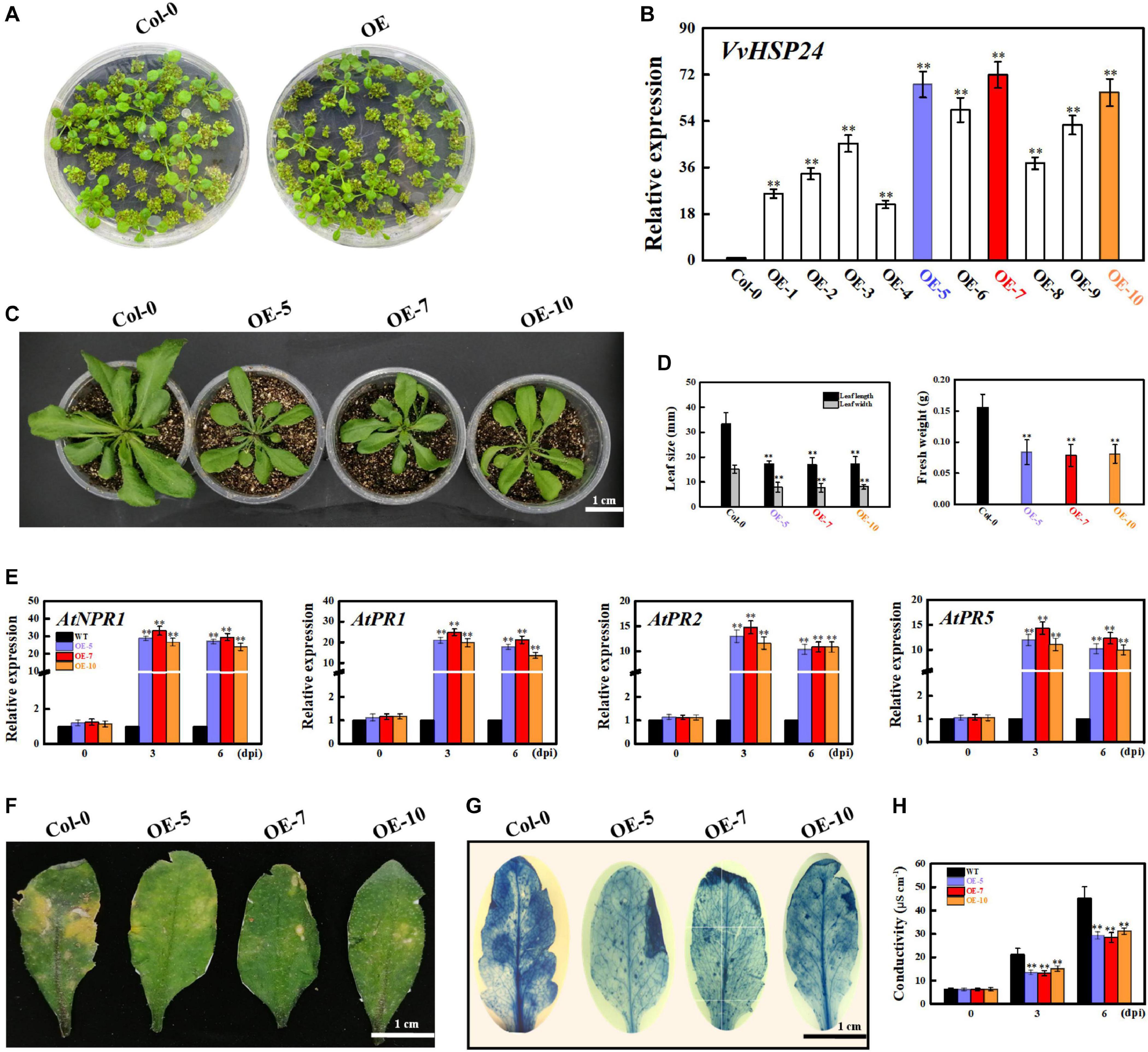
Figure 7. Overexpression of VvHSP24 activates PR gene expression in Arabidopsis leaves and increases the resistance of transgenic Arabidopsis to the fungal pathogen B. cinerea. (A) Phenotypes of Arabidopsis Col-0 wildtype and transgenic seedlings for 2 weeks. (B) Relative mRNA levels of VvHSP24 in Col-0 wildtype and transgenic Arabidopsis plants grown in soil for 4 weeks. Transcript abundances are expressed relative to the reference gene of AtActin2 and expressed as the mean ± SE. Two asterisks indicate significant differences between wildtype and transgenic plants (p < 0.01). (C,D) Overexpression of VvHSP24 in Arabidopsis inhibits growth, as exhibited by the shorter leaf length and lower biomass of the dominant VvHSP24-overexpressing Arabidopsis (OE5, OE7, and OE10) than of Col-0 wildtype. Bar = 1 cm. (E) qRT-PCR analysis of the transcript levels of defense-related genes (NPR1, PR1, PR2, and PR5) in Col-0 wildtype and VvHSP24-overexpressing Arabidopsis (OE5, OE7, and OE10) infected by B. cinerea at 0, 3, and 6 dpi. mRNA expression levels were quantified against the value of AtActin2; the data represent the mean ± SE of three separate replicates. Two asterisks indicate significant differences between wildtype and OE5, OE7, or OE10 (p < 0.01). (F) Disease symptoms of wildtype and VvHSP24-overexpressing (OE-5, OE-7, and OE-10) Arabidopsis leaves after B. cinerea infection at 6 dpi. Bar = 1 cm. (G) Necrotic areas in Col-0 wildtype and VvHSP24-overexpressing Arabidopsis leaves were determined by trypan blue staining after B. cinerea infection at 6 dpi, and images were captured with a digital camera. (H) Measurement of electrolyte leakage from wildtype and VvHSP24-overexpressing Arabidopsis leaves infected with B. cinerea at 6 dpi. Conductivities are expressed as the mean and SE of three independent biological replicates. Two asterisks indicate significant differences between wildtype and VvHSP24-overexpressing Arabidopsis at p = 0.01.
AtHSFB1 Mutants With Decreased Resistance in Response to Fungal Pathogen
Given that AtHSFB1 is a VvHSP24 orthologous gene in Arabidopsis, we harvested Arabidopsis AtHSFB1 mutant seedlings constructed via the CRISPR-Cas9 system to evaluate the disease resistance of HSP through mutant-based verification. As depicted in Figures 8A,B, AtHSFB1 transcript in all CRISPR mutants, especially in CRP-1, CRP-2, and CRP-6, were largely not expressed or drastically decreased compared with that in WT plants. Interestingly, the dominant CRISPR mutants CRP-1, CRP-2, and CRP-6 showed no apparent alteration in growth phenotypes compared with those in the WT (Figures 8C,D). In addition, CRISPR/Cas9-mediated AtHSFB1 knockout lines displayed similar kinetics and intensity of pathogenesis-related gene (NPR1, PR1, PR2, and PR5) transcript accumulations, and all were slightly or significantly decreased when compared with the WT at 3 and 6 dpi (Figure 8E). Consistent with this observation, the symptoms of fungal infection and staining level of the dot-inoculated mutants were stronger than those of WT leaves (Figures 8F,G). Thus, the AtHSFB1 mutation impaired the defense expression and led to a pathogenic sensitiveness.
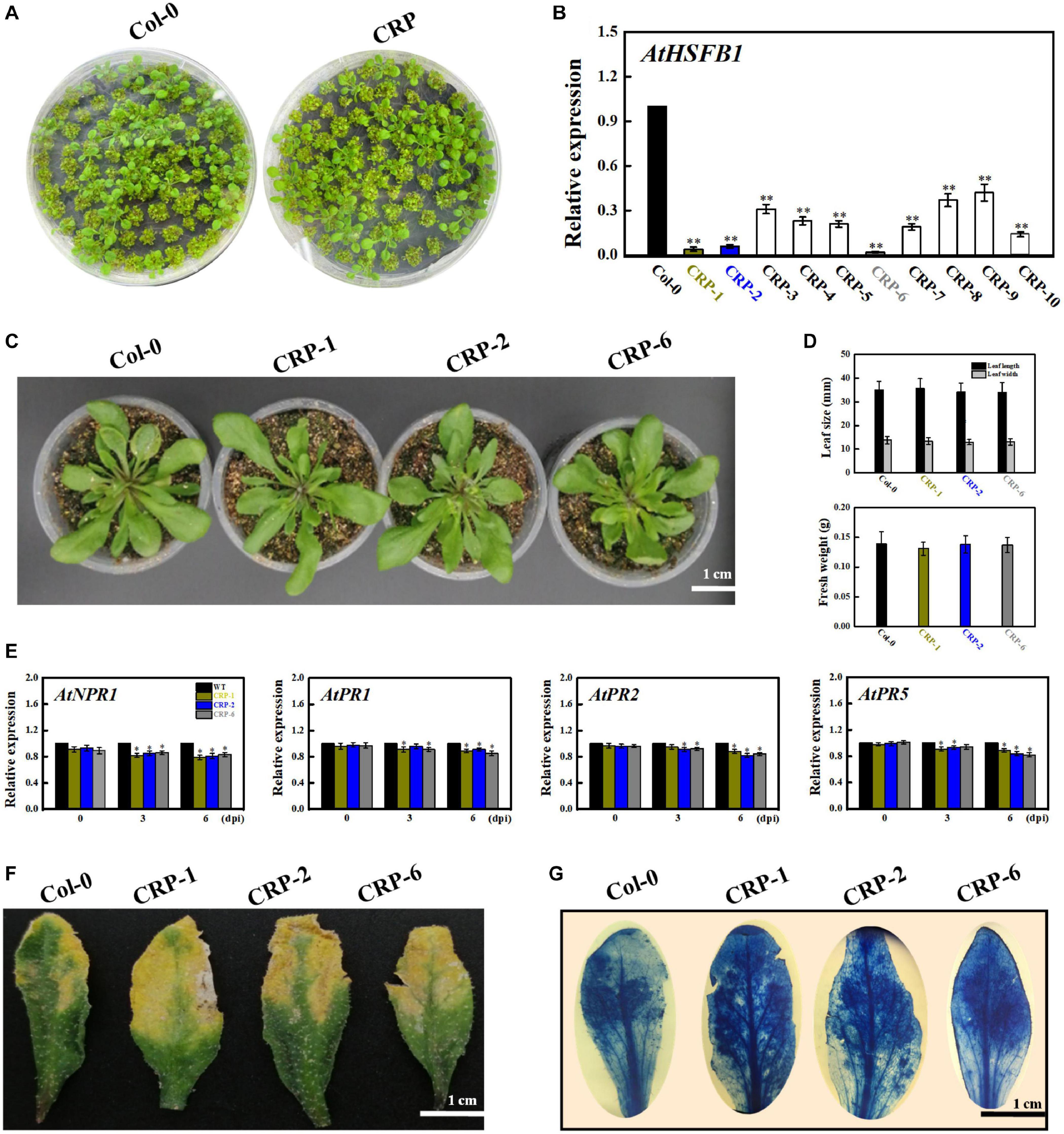
Figure 8. HSFB1 mutation mediated by the CRISPR/Cas9 system scarcely modifies the fungal resistance of Arabidopsis to B. cinerea. (A) Growth phenotypes of Arabidopsis Col-0 wildtype and mutant seedlings at 15 days after sowing on MS plants. (B) Transcript levels of HSFB1 from Col-0 wildtype and CRISPR mutants grown in soil for more than 4 weeks. Transcript amounts are normalized against the AtActin2 value, and the results are expressed as the mean ± SE. Asterisks indicate that the differences between wildtype and CRISPR mutants are statistically extremely significant at the p = 0.01 level. (C,D) CRISPR mutants (CRP1, CRP2, and CRP6) show unchanged levels of leaf length and biomass compared with Col-0 wildtype. Bar = 1 cm. (E) Relative mRNA levels of NPR1, PR1, PR2, and PR5 genes in Col-0 wildtype and CRISPR mutants (CRP1, CRP2, and CRP6) after inoculation with B. cinerea at 0, 3, and 6 days. Asterisks represent significant differences between wildtype and CRP1, CRP2, or CRP6 at p = 0.01. (F) Disease symptoms of Col-0 wildtype and CRISPR mutants (CRP1, CRP2, and CRP6) at 6 dpi. Bar = 1 cm. (G) Necrotic areas in wildtype and CRISPR mutants were stained with trypan blue and recorded by a camera mounted on a binocular stereomicroscope.
Discussion
As sessile organisms, plants are frequently subjected to a fluctuating environment and exhibit a high degree of elasticity in survival under various stresses, suggesting broad crosstalk between plant defense responses and other biochemical and physiological processes (Khare et al., 2020). Of note, hyperactivation of resistance not only cannot terminate the wide variety of disease threats but also may entail an irreversible cost that might suppress the growth, fruit set, yield and quality of horticultural crops (Vos et al., 2013). Priming has been found to be an ecological approach of resistance owing to its capacity to respond promptly to a certain degree of biotic stress; thus, priming can balance the specific resistance and the resulting fitness costs or quality impairment in crops (Conrath et al., 2015; Ton et al., 2009). The induction of BABA on biotic resistance has been researching over half a century (Cohen et al., 2016). In this study, the resistance against B. cinerea fungal infection of postharvest grapes elicited by BABA at 10 mmol L–1 likely paralleled a priming mode. In fact, its definite role in the transient activation antimicrobial protein expression, particularly pathogenesis-related proteins (including VvNPR1, VvPR1, VvPR2, and VvPR5), and the synergistic stimulation of host-synthesized antifungal compounds such as trans-resveratrol and ε-viniferin occurred exclusively after combined therapy (BABA pretreatment with fungal infection; Figures 1, 3, 4; Thevenet et al., 2017). Although the infection process of the gray mold fungus B. cinerea could not be completely abolished by 10 mmol L–1 BABA, disease development was appreciably attenuated, leading to the smaller lesions and lower disease incidences during 5 days of the incubation at 20°C (Figure 1 and Supplementary Figure 1). Similarly, such priming resistance against pathogen stresses in Arabidopsis, peach and bayberry has been observed for other priming elicitors, such as methyl jasmonate (MeJA), 2,4-epibrassinolide (EBR), benzothiadiazole (BTH), and chitosan (Beckers et al., 2009; Wang et al., 2014; De Vega et al., 2020; Li et al., 2020a,b). In addition, Wang et al. (2016) reported an in vitro lethal activity of BABA on the cell membrane of B. cinerea. Thus, the combination of the enhanced host disease resistance elicited by BABA and its direct toxic effect on pathogens supports the inherent role of BABA as a defense inducer. Although the BABA conferred a positive resistance on disease development caused by B. cinerea, the combination strategy of BABA and other chemical or physical inducers should be tested for gaining a more satisfactory protection from fungal attack.
Specifically, primed grapes presented much stronger transcript levels of SA-responsive defense-related genes, such as VvPR1, VvPR2, and VvPR5, and stimulated disease resistance upon the artificial B. cinerea-inoculation over the observation period. We speculate that the mode of manner of BABA-elicited defense in grapes may be attributed to a SAR-like defense response. Although Nie et al. (2017) have recently pointed out that JA/ET-dependent signaling pathways have been shown to be involved in systemic resistance against B. cinerea in Arabidopsis, a set of JA/ET-dependent defensive genes (VvPDF1.2, VvHEL, and VvTHI2) were not promoted in B. cinerea-inoculated grapes after the BABA elicitation in our present study. In contrast, the transcriptional data exhibited a determinable induction of BABA on gene expressions of VvVSP1, VvFAD3, VvERS1, and VvERF1 in grapes. Noticeably, either of VvPDF1.2, VvTHI2, or VvHEL can directly participate in defense expression against biotic stress (Oh et al., 1999; Bertini et al., 2012), but the other JA/ET-dependent genes including VvVSP1, VvFAD3, VvERS1, and VvERF1 have been reported to be relevant to the plant development, ethylene perception, metabolic process or mechanical wounding healing, rather than the direct pathogenic response (Nath and Watson, 1980; Puttick et al., 2009; Liu and Wen, 2012). Hence, this BABA-IR in grapes is mainly associated with the expression of SA-dependent SAR. These results were in part consistent with our most current findings, showing that the BABA-activated resistance in grapes and peaches is dependent on SA signaling but not the JA/ET signaling pathways (Li et al., 2020c; Wang et al., 2021). Takahashi et al. (2004), Koornneef et al. (2008), and van der Does et al. (2013) also elucidated that the SA-dependent SAR reaction suppressed the transduction of JA signaling and the corresponding expression levels of JA-dependent defensive genes in model plant Arabidopsis thaliana. However, the obvious increases in the transcript levels of VvVSP1, VvFAD3, VvERS1, and VvERF1 in BABA-treated grapes provided a valuable clue that BABA may affect the fruit physiology and quality due to the genetic regulation of metabolic flow between the fruit defense and development, which will lead us to research the relationship between the induced resistance and its allocated metabolic influence. Coincidentally, the protection of Arabidopsis and tobacco plants by BABA against infection by the necrotrophic fungus B. cinerea, virulent pathogen tobacco mosaic virus (TMV) or avirulent pathogen Pseudomonas syringae pv. tomato (Pst) through the burst of the SA-dependent SAR pathway has been reported (Siegrist et al., 2000; Zimmerli et al., 2001; Slaughter et al., 2012). These results suggest that BABA-IR might be dependent on the SA-dependent defense pathway and several potential factors may propagate the SA signal.
Heat shock proteins are members of multigene families encoding molecular chaperones, and have long been recognized as heat stress-inducible proteins. However, a large number of evidence have pointed out that HSPs become activated when plants undergo other stresses, including drought, salt, cold, heavy metals, oxidative stresses and pathogen invasion, demonstrating extensive transcriptional overlap among heat and non-heat stress response pathways, and the essential role of HSPs/chaperones in the crosstalk of multistress resistance (Swindell et al., 2007; Jacob et al., 2017). A report by Pick et al. (2012) indicated that HSFB1, a class B heat shock factor, promotes primed expression of a subset of defense-related genes and is a potential candidate for SAR in Arabidopsis against Pseudomonas syringae. Windram et al. (2012) also implied a crosstalk mediating heat stress and B. cinerea infection. Similarly, a series of notable transcriptomic analysis by Sham et al. (2014, 2015, 2017) have emphasized that HSPs shared signaling pathways responding to biotic and abiotic stresses in Arabidopsis. In those cases, the specific HSP17.4 or HSP26.5-P was up- or down-regulated by the B. cinerea infection respectively, indicating that HSPs may be implicated in defense function. Hence, to explore whether a credible heat shock factor that also participates in the development of defense priming and SAR in grape berries might exist, we first comparatively analyzed the phylogenetic relationship between Vitis vinifera HSFs/HSPs and Arabidopsis thaliana HSFs (AtHSFs). A HSP (VvHSP24) with close sequence similarity to AtHSFB1 from Arabidopsis was identified and isolated (Supplementary Figure 3). Multiple alignment and conserved domain analysis of VvHSP24 and AtHSFB1 further revealed that both factors contain a highly conserved HSF_DNA-binding domain consisting of a total of 94 amino acids. Furthermore, VvHSP24 and AtHSFB1 group into class B, indicating that VvHSP24, as an ortholog of AtHSFB1, might exert a certain effect on SAR in grapes against the necrotrophic fungus B. cinerea (Supplementary Figure 3). Indeed, we found that treatment of grapes with a formulation of BABA increased the expression of VvHSP24 mRNA by several-fold, and the transcript level of VvHSP24 was significantly activated by B. cinerea fungal infection (Figure 3A). Together, these observations support our assumption that VvHSP24 is a candidate for BABA-induced systemic resistance in grape berries. Intriguingly, most npr1 (for non-expresser PR genes) alleles and NPR1 paralog null mutants exhibit enhanced disease susceptibility phenotype and are incapable of expressing PR genes (including PR1, PR2, and PR5) or developing SAR in their response to SA or its analog INA (2,6-dichloroisonicotinic acid) and benzothiadiazole (BTH); this reveals the positive function of NPR1 in SA-mediated activation of PR genes and SAR development (Canet et al., 2010). Because the dominant transgenic Arabidopsis plants overexpressing VvHSP24 exhibited potentiated resistance to necrotrophic fungal pathogens, including dozen-fold increased expression of VvNPR1 and the resulting constitutive expression of SA-responsive PR1, PR2, and PR5 genes and decreased levels in necrotic areas and cell death (Figures 7E–H), it can be deduced that VvHSP24 functions in the defense signaling pathway upstream of VvNPR1. This scenario would require the generation of the corresponding VvHSP24 mutation for confirmation. However, constitutive expression of NPR1 and SA-responsive genes in the CRISPR mutants of AtHSFB1 (an orthologous gene of VvHSP24 in Arabidopsis) was only slightly suppressed compared with that in the WT (Figure 8E). This finding elucidated that the NPR1-conferred resistance phenotype does not depend on HSP/chaperone; thus, HSP/chaperone is not a direct upstream regulator of NPR1.
If HSP/chaperone is not an upstream regulator of NPR1, where does it function in the defense signaling pathway? The presence of ankyrin repeats but the lack of a DNA-binding domain indicate that NPR1 may execute its regulatory role in SAR through interaction with other proteins (Dong et al., 2001). Therefore, it is possible that HSP24 physically interacts with NPR1 to stimulate the development of SAR against B. cinerea fungal infection in grape berries. As depicted in Figures 2, 5, VvHSP24 was identified as a nuclear chaperone that interacted directly with the VvNPR1 in both in vivo and in vitro systems, as expected. Additionally, the results of EMSA (Figure 6) show that VvHSP24 did not have the capacity of activating the transcript of the PR genes directly, implying that the protein binding activity of VvHSP24 to VvNPR1 is a prerequisite for PR expressions and consequent defense functions. In contrast to the resistance of transgenic Arabidopsis plants overexpressing VvHSP24 against fungal pathogens over 6 dpi, SA-responsive PR gene expressions and resistance to the necrotrophic fungus B. cinerea in the CRISPR mutants CRP1, CRP2 and CRP6 were not constitutively repressed. Moreover, an interesting observation of our study is the finding that unlike the limited influence of CRISPR mutation on transgenic plants, the high production of VvHSP24 in transgenic Arabidopsis overexpressing this gene can be beneficial for enhancing plant fungal resistance but can cause deleterious effects on plant growth (Figures 7C,D, 8C,D). Such negative influence of VvHSP24 overexpression on biomass and leaf length may occur because VvHSP24 initiates specific transcriptional reprogramming that prioritizes defense over growth-related costs. In fact, Pajerowska-Mukhtar et al. (2012) found that AtHSFB1, the VvHSP24 orthologous gene in Arabidopsis, plays a major role in the growth-toward-defense transition. Although overexpressing the stress protein HSP24 distinctly elevated the expression levels of NPR1 and SA-inducible genes, the CRISPR mutants only presented a slight decrease in NPR1, PR1, PR2, and PR5 transcript levels. Conversely, a more persuasive hypothesis is that HSPs/chaperones might not separately affect the defense signaling pathway downstream of VvNPR1, leading to PR gene expressions and resistance phenotypes. One probable explanation is that a second protein or factor, in addition to HSP/chaperone, is required for NPR1 activation in the defense signaling pathway. The CRISPR mutants might not totally bypass the positive function of this second protein or factor. In support of this possibility, Yu et al. (2001), Desveaux et al. (2002), Després et al. (2003), and Dong (2004) demonstrated that in addition to TGAs, the single-stranded DNA-binding protein Whirly1 and WRKY transcription factors are involved in the NPR1-mediated SAR network. Similarly, our previous studies showed that the involvement of TGA1 and WRKY transcription factors in the activation of fungal resistance progresses in peaches and grapes in an NPR1-dependent manner (Li et al., 2020c,d; Wang et al., 2020). Indeed, the NPR1 protein tends to form a large-scale oligomer under natural conditions, which seriously impedes its cellular functions (Tada et al., 2008), and the main chaperone function of HSPs is to modulate the depolymerization of bound substrates (Wang et al., 2004; Haslbeck et al., 2015). Thus, the stress protein HSP24 might control an important switch that, depending on fungal stress, may differentially disaggregate NPR1 into an active monomer to regulate NPR1-dependent SA signaling pathway. Similarly, Ramakrishna et al. (2003) found that the HSP viscosity 1 (visl) acts a positive function in pectin depolymerization. Moreover, BABA-IR, as exhibited in our and other studies, is a crosstalk mechanism through a convergence of several signaling pathways, such as the SA, ABA, PI, JA, and ET pathways, further suggesting the existence of shared components other than NPR1, TGAs, WRKYs and Whirly among these defense pathways. In terms of the SA signaling pathway, our current results suggest that BABA-IR in grapes can be fueled by the VvNPR1-VvHSP24 complex when VvNPR1 oligomers become disaggregated, resulting in producing active monomers that propagate the SA signal and induce PR gene expressions.
Conclusion
We characterized a HSP, VvHSP24, as a potential candidate in NPR1-dependent plant resistance to the necrotrophic fungus B. cinerea. Furthermore, NPR1 is an essential regulator in the development of SAR and a key regulatory protein in activating transcripts of SA-inducible genes, but this capacity of VvNPR1 might rely on posttranslational modification of the VvHSP24 chaperone and their subsequent interaction after the perception of invading disease stress by plant cells. Our study reports the previously unclarified function of HSPs/chaperones and offers insight for uncovering the regulatory network of disease resistance in agronomic fruits. Given that the HSPs usually generate upon the plant suffers from various stresses, further researches should concentrate on the elaboration of the explicit molecular or metabolic modes of the cross-tolerance in postharvest fruit. Meanwhile, identification of stress-related HSPs and their functional co-activator under biotic stress may be definitely conducive to the studies on induced resistance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
KW, SC, YZ, and FX conceived the experiments. CLi, SC, KW, CLe, NJ, YJ, and LQ conducted the experiments. CLi and KW analyzed the data and wrote the manuscript. All the authors read, commented, revised, and approved the final revision manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31671913 and 31672209). The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Prof. Renhua Liu for his pivotal assistance for the transgenic trials. We are grateful of Ms. Yunxia Liao and Ms. Dongzhi Wu due to their help with sample preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.646147/full#supplementary-material
Supplementary Figure 1 | Tree analysis of the isolated Botrytis cinerea strain with the Botrytis cinerea family. Accession numbers of sequences: Botrytis cinerea B05.10, MH427722.1; Botryotinia fuckeliana T4, AJ422103.1; Botrytis cinerea Bo.CC166, MH427722.1; Botrytis cinerea strain B23, AJ422103.1; Botrytis cinerea BOT 40, KU145363.1; Botrytis cinerea BOT 5, KX772771.1; Botrytis cinerea BOT 16, KX781162.1; Botrytis cinerea BOT 61, KX781165.1; Botryotinia fuckeliana strain Minhang, AY694146.1; Botrytis cinerea BOT 74, KX781167.1; Botrytis cinerea strain SCB7-5, KR080285.1; Botrytis cinerea Bot 68, KU145391.1; Botrytis cinerea strain SCB2-2, KR080283.1; Botrytis cinerea strain SCB7-4, KP165498.1; Botrytis cinerea strain SCB5-2, KP165495.1; Botrytis cinerea strain BC-8, KP141792.1; Botryotinia fuckeliana species; AY674786.1; Botrytis cinerea strain KBC-13, KP141795.1; Botrytis cinerea strain SL, KU936083.1; Botrytis cinerea strain BC-10, KP141793.1; Botrytis cinerea strain KBC-14, KP141794.1; Botrytis cinerea strain BC-1, KP141790.1; Botrytis cinerea strain AC1, KU936079.1; Botrytis cinerea strain AR, KU936082.1; Botrytis cinerea strain SV, KU936085.1.
Supplementary Figure 2 | Grape fruit displays significant differences in visual appearances under four treatments (control, BABA, inoculation, and BABA + inoculation). BABA treatment and pathogen inoculation were conducted independently twice, with similar results. Representative grapes were photographed and compared after the 5 dpi at 20°C.
Supplementary Figure 3 | Phylogenetic and domain analyses of VvHSPs/AtHSFs. (A) Three classes of HSFs/HSPs are shown in the phylogenetic tree. VvHSP24 and AtHSFB1 are in bold in purple and group into class B. The sequences of 57 HSFs/HSPs from Vitis vinifera and Arabidopsis thaliana were aligned with Clustal W, and the phylogenetic analysis was made with MEGA (NJ method; bootstrap of 1000). (B) Identical amino acids between VvHSP24 and its closest related protein AtHSFB1 are shaded in black. In addition, a single conserved HSF_DNA-binding domain consisting of a total of 94 amino acids was visualized by TBtools software; it occurs at residue ranges of amino acids 6–99 in VvHSP24 and 11–104 in AtHSFB1.
Supplementary Figure 4 | Schematic diagram of the M2CRISPR/Cas9-AtHSFB1 vector.
Supplementary Figure 5 | Heat map of VvHSPs in pairwise comparisons of BABA-vs.-control, inoculation-vs.-control, BABA + inoculation-vs.-control, and BABA + inoculation-vs.-inoculation.
Supplementary Figure 6 | His-tagged protein was detected by a gel image analysis (un-cropped gels).
Supplementary Figure 7 | In the His pull-down assay, GST-VvNPR1 fusion protein was efficiently pull-downed by His-VvHSP24 fusion protein (un-cropped gels).
Supplementary Figure 8 | EMSA of VvHSP24 binding to the HSE elements of PR1, PR2, and PR5 (un-cropped blots).
Supplementary Table 1 | Primer sequence of the used genes.
Supplementary Data 1 | Promoter sequences of VvPR1, VvPR2, and VvPR5. HSEs (GAAnnTTC) and the translation start site (ATG) are marked in red and yellow boxed, respectively.
Footnotes
References
AbuQamar, S., Moustafa, K., and Tran, L. S. (2017). Mechanisms and strategies of plant defense against Botrytis cinerea. Crit. Rev. Biotechnol. 37, 262–274. doi: 10.1080/07388551.2016.1271767
Baccelli, I., Glauser, G., and Mauch-Mani, B. (2017). The accumulation of β-aminobutyric acid is controlled by the plant’s immune system. Planta 246, 791–796. doi: 10.1007/s00425-017-2751-3
Balic, I., Ejsmentewicz, T., Sanhueza, D., Silva, C., Peredo, T., Olmedo, P., et al. (2014). Biochemical and physiological study of the firmness of table grape berries. Postharvest Biol. Technol. 93, 15–23. doi: 10.1016/j.postharvbio.2014.02.001
Baniwal, S. K., Bharti, K., Chan, K. Y., Fauth, M., Ganguli, A., Kotak, S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. doi: 10.1007/BF02712120
Beckers, G. J., Jaskiewicz, M., Liu, Y., Underwood, W. R., He, S. Y., Zhang, S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21, 944–953. doi: 10.1105/tpc.108.062158
Bertini, L., Proietti, S., Aleandri, M. P., Mondello, F., Sandini, S., and Caporale, C. (2012). Modular structure of HEL protein from Arabidopsis reveals new potential functions for PR-4 proteins. Biol. Chem. 1, 1–14. doi: 10.1515/hsz-2012-0225
Bharti, K., Schmidt, E., Lyck, R., Heerklotz, D., Bublak, D., Scharf, K. D., et al. (2000). Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 22, 355–365. doi: 10.1046/j.1365-313x.2000.00746.x
Burketova, L., Trda, L., Ott, P. G., and Valentova, O. (2015). Bio-based resistance inducers for sustainable plant protection against pathogens. Biotechnol. Adv. 33, 994–1004. doi: 10.1016/j.biotechadv.2015.01.004
Cai, J. H., Chen, T., Wang, Y., Qin, G. Z., and Tian, S. P. (2020). SlREM1 triggers cell death by activating an oxidative burst and other regulators. Plant Physiol. 183, 717–732. doi: 10.1104/pp.20.00120
Canet, J. V., Dobón, A., Roig, A., and Tornero, P. (2010). Structure-function analysis of npr1 alleles in Arabidopsis reveals a role for its paralogs in the perception of salicylic acid. Plant Cell Environ. 33, 1911–1922. doi: 10.1111/j.1365-3040.2010.02194.x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cohen, Y., Vaknin, M., and Mauch-Mani, B. (2016). BABA-induced resistance: milestones along a 55-year journey. Phytoparasitica 44, 513–538. doi: 10.1007/s12600-016-0546-x
Conrath, U., Beckers, G. J. M., Langenbach, C. J. G., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Dai, Z. W., Ollat, N., Gomes, E., Decroocq, S., Tandonnet, J. P., Bordenave, L., et al. (2011). Ecophysiological, genetic, and molecular causes of variation in grape berry weight and composition: a review. Am. J. Enol. Vitic. 62, 413–425. doi: 10.5344/ajev.2011.10116
De Vega, D., Holden, N., Hedley, P. E., Morris, J., Luna, E., and Newton, A. (2020). Chitosan primes plant defence mechanisms against Botrytis cinerea, including expression of Avr9/Cf-9 rapidly elicited genes. Plant Cell Environ. 44, 290–303. doi: 10.1111/pce.13921
Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. doi: 10.1105/tpc.012849
Desveaux, D., Allard, J., Brisson, N., and Sygusch, J. (2002). A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat. Struct. Biol. 9, 512–517. doi: 10.1038/nsb814
Dong, X. N. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
Dong, X. N., Li, X., Zhang, Y. L., Fan, W. H., Kinkema, M., and Clarke, J. (2001). Regulation of systemic acquired resistance by NPR1 and its partners. Novartis. Found. Symp. 236, 165–173. doi: 10.1002/9780470515778.ch12
Feliziani, E., Smilanick, J. L., Margosan, D. A., Mansour, M. F., Romanazzi, G., Gu, S., et al. (2013). Preharvest fungicide potassium sorbate, or chitosan use on quality and storage decay of table grapes. Plant Dis. 97, 307–314. doi: 10.1094/PDIS-12-11-1043-RE
Gabler, F. M., Smilanick, J. L., Mansour, M., Ramming, D. W., and Mackey, B. E. (2003). Correlations of morphological, anatomical and chemical features of grape berries with resistance to Botrytis cinerea. Phytopathology 93, 1263–1273. doi: 10.1094/PHYTO.2003.93.10.1263
Guertin, M. J., and Lis, J. T. (2010). Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 6:e1001114. doi: 10.1371/journal.pgen.1001114
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., and Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7:114. doi: 10.3389/fpls.2016.00114
Haslbeck, M., Weinkauf, S., and Buchner, J. (2015). “Regulation of the chaperone function of small Hsps,” in The Big Book on Small Heat Shock Proteins. Heat Shock Proteins, Vol. 8, eds R. Tanguay and L. Hightower (Cham: Springer), doi: 10.1007/978-3-319-16077-1_6
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. doi: 10.1111/pbi.12659
Khare, S., Singh, N. B., Singh, A., Hussain, I., Niharika, K., Yadav, V., et al. (2020). Plant secondary metabolites synthesis and their regulations under biotic and abiotic constraints. J. Plant Biol. 11, 1–14. doi: 10.1007/s12374-020-09245-7
Koornneef, A., Leon-Reyes, A., Ritsema, T., Verhage, A., Den Otter, F. C., van Loon, L. C., et al. (2008). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147, 1358–1368. doi: 10.1104/pp.108.121392
Larkindale, J., and Vierling, E. (2008). Core genome responses involved in acclimation to high temperature. Plant Physiol. 146, 748–761. doi: 10.1104/pp.107.112060
Letunic, I., and Bork, P. (2018). 20 Years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, C. H., Du, M. Y., and Wang, K. T. (2020a). 2,4-Epibrassionolide activates priming resistance against Rhizopus stolonifer infection in peach fruit. Acta. Aliment. 49, 135–143. doi: 10.1556/066.2020.49.2.2
Li, C. H., Wang, J., Ji, N. N., Lei, C. Y., Zhou, D. X., Zheng, Y. H., et al. (2020b). PpHOS1, a RING E3 ubiquitin ligase, interacts with PpWRKY22 in the BABA-induced priming defense of peach fruit against Rhizopus stolonifer. Postharvest Biol. Technol. 159:111029. doi: 10.1016/j.postharvbio.2019.111029
Li, C. H., Wang, K. T., Lei, C. Y., and Zheng, Y. H. (2020c). Translocation of PpNPR1 is required for β-aminobutyric acid-triggered resistance against Rhizopus stolonifer in peach fruit. Sci. Hortic. 272:109556. doi: 10.1016/j.scienta.2020.109556
Li, C. H., Wang, K. T., and Zheng, Y. H. (2020d). Redox status regulates subcelluar localization of PpTGA1 associated with a BABA-induced priming defence against Rhizopus stolonifer. Mol. Biol. Rep. 47, 6657–6668. doi: 10.1007/s11033-020-05719-6
Li, J., Soroka, J., and Buchner, J. (2012). The Hsp90 chaperone machinery: conformational dynamics and regulation by cochaperones. Biochim. Biophys. Acta. 3, 624–635. doi: 10.1016/j.bbamcr.2011.09.003
Li, R., Liu, C. X., Zhao, R. R., Wang, L., Chen, L., Yu, W. Q., et al. (2019). CRISPR/Cas9-mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 19:38. doi: 10.1186/s12870-018-1627-4
Liu, Q., and Wen, C. K. (2012). Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes1[w]. Plant Physiol. 158, 1193–1207. doi: 10.1104/pp.111.187757
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lovato, A., Zenoni, S., Tornielli, G. B., Colombo, T., Vandelle, E., and Polverari, A. (2019). Specific molecular interactions between Vitis vinifera and Botrytis cinerea are required for noble rot development in grape berries. Postharvest Biol. Technol. 156, 110924–110938. doi: 10.1016/j.postharvbio.2019.05.025
Lowder, L. G., Zhang, D. W., Baltes, N. J., Paul, J. W. III, Tang, X., Zheng, X. L., et al. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. doi: 10.1104/pp.15.00636
Luna, E., López, A., Kooiman, J., and Ton, J. (2014). Role of NPR1 and KYP in long-lasting induced resistance by β-aminobutyric acid. Front. Plant Sci. 5:184. doi: 10.3389/fpls.2014.00184
Mackey, D., Holt, B. F. III, Wiig, A., and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
Mari, M., Di Francesco, A., and Bertolini, P. (2014). Control of fruit postharvest diseases: old issues and innovative approaches. Stewart Postharvest Rev. 10, 1–4. doi: 10.2212/spr.2014.1.1
Mengiste, T., Laluk, K., and AbuQamar, S. (2009). “Mechanisms of induced resistance against B. cinerea,” in Postharvest Pathology, eds D. Prusky and M. Gullino (Dordrecht: Springer), 13–30. doi: 10.1007/978-1-4020-8930-5_2
Nath, J., and Watson, C. V. (1980). Acid phosphatase changes associated with development of male sterile and fertile maize (Zea mays L.). Biochem. Genet. 18, 377–387. doi: 10.1007/BF00484250
Nie, P. P., Li, X., Wang, S. N., Guo, J. H., Zhao, H. W., and Niu, D. D. (2017). Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in Arabidopsis. Front. Plant Sci. 8:238. doi: 10.3389/fpls.2017.00238
Nishizawa, A., Yabuta, Y., Yoshida, E., Maruta, T., Yoshimura, K., and Shigeoka, S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48, 535–547. doi: 10.1111/j.1365-313X.2006.02889.x
Noël, L. D., Cagna, G., Stuttmann, J., Wirthmüller, L., Betsuyaku, S., Witte, C. P., et al. (2007). Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19, 4061–4076. doi: 10.1105/tpc.107.051896
Nover, L., Bharti, K., Doring, P., Mishra, S. K., Ganguli, A., and Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperon 6, 177–189.
Oh, B. J., Ko, M. K., Kostenyuk, I., Shin, B., and Kim, K. S. (1999). Coexpression of a defensin gene and a thionin-like via different signal transduction pathways in pepper and Colletotrichum gloeosporioides interactions. Plant Mol. Biol. 41, 313–319. doi: 10.1023/a:1006336203621
Pajerowska-Mukhtar, K. M., Wang, W., Tada, Y., Oka, N., Tucker, C. L., Fonseca, J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112. doi: 10.1016/j.cub.2011.12.015
Perazzolli, M., Roatti, B., Bozza, E., and Pertot, I. (2011). Trichoderma harzianum T39 induces resistance against downy mildew by priming for defense without costs for grapevine. Biol. Control. 58, 74–82. doi: 10.1016/j.biocontrol.2011.04.006
Pérez-Salamó, I., Papdi, C., Rigó, G., Zsigmond, L., Vilela, B., Lumbreras, V., et al. (2014). The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol. 165, 319–334. doi: 10.1104/pp.114.237891
Pick, T., Jaskiewicz, M., Peterhänsel, C., and Conrath, U. (2012). Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 159, 52–55. doi: 10.1021/bi981774j
Porat, R., Vinokur, V., Weiss, B., Cohen, L., Daus, A., Goldschmidt, E. E., et al. (2003). Induction of resistance to Penicillium digitatum in grapefruit by β-aminobutyric acid. Eur. J. Plant. Pathol. 109, 901–907. doi: 10.1023/B:EJPP.0000003624.28975.45
Puttick, D., Dauk, M., Lozinsky, S., and Smith, M. A. (2009). Overexpression of a FAD3 desaturase increases synthesis of a polymethylene-interrupted dienoic fatty acid in seeds of Arabidopsis thaliana L. Lipids 44, 753–757. doi: 10.1007/s11745-009-3315-5
Ramakrishna, W., Deng, Z. P., Ding, C. K., Handa, A. K., and Ozminkowski, R. H. (2003). A novel small heat shock protein gene, visl, contributes to pectin depolymerization and juice viscosity in tomato fruit. Plant Physiol. 131, 725–735. doi: 10.1104/pp.012401
Romanazzi, G., Sanzani, S. M., Bi, Y., Tian, S. P., Martínez, P. G., and Alkan, N. (2016). Induced resistance to control postharvest decay of fruit and vegetables. Postharvest Biol. Technol. 122, 82–94. doi: 10.1016/j.postharvbio.2016.08.003
Rosales, R., Romero, I., Fernandez-Caballero, C., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2016). Low temperature and short-term high-CO2 treatment in postharvest storage of table grapes at two maturity stages: effects on transcriptome profiling. Front. Plant Sci. 7:1020. doi: 10.3389/fpls.2016.01020
Samali, A., Robertson, J. D., Peterson, E., Manero, F., van Zeijl, L., Paul, C., et al. (2001). Hsp27 protects mitochondria of thermotolerant cells against apoptotic stimuli. Cell Stress Chaperones 6, 49–58.
Sarig, P., Zutkhi, Y., Monjauze, A., Lisker, N., and Ben-Arie, R. (1997). Phytoalexin elicitation in grape berries and their susceptibility to Rhizopus stolonifer. Physiol. Mol. Plant Pathol. 50, 337–347. doi: 10.1006/pmpp.1997.0089
Scharf, K. D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta. 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Sham, A., Al-Ashram, H., Whitley, K., Iratni, R., El-Tarabily, K. A., and AbuQamar, S. F. (2019). Metatranscriptomic analysis of multiple environmental stresses identifies RAP2.4 gene associated with Arabidopsis immunity to Botrytis cinerea. Sci. Rep. 9:17010. doi: 10.1038/s41598-019-53694-1
Sham, A., Al-Azzawi, A., Al-Ameri, S., Al-Mahmoud, B., Awwad, F., Al-Rawashdeh, A., et al. (2014). Transcriptome analysis reveals genes commonly induced by Botrytis cinerea infection, cold, drought and oxidative stresses in Arabidopsis. PLoS One 9:e113718. doi: 10.1371/journal.pone.0113718
Sham, A., Moustafa, K., Al-Ameri, S., Al-Azzawi, A., Iratni, R., and AbuQamar, S. (2015). Identification of Arabidopsis candidate genes in response to biotic and abiotic stresses using comparative microarrays. PLoS One 10:e0125666. doi: 10.1371/journal.pone.0125666
Sham, A., Moustafa, K., Al-Shamisi, S., Alyan, S., Iratni, R., and AbuQamar, S. (2017). Microarray analysis of Arabidopsis WRKY33 mutants in response to the necrotrophic fungus Botrytis cinerea. PLoS One 12:e0172343. doi: 10.1371/journal.pone.0172343
Shirasu, K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Boil. 60, 139–164. doi: 10.1146/annurev.arplant.59.032607.092906
Siegrist, J., Orober, M., and Buchenauer, H. (2000). β-Aminobutyric acid-mediated enhancement of resistance in tobacco to tobacco mosaic virus depends on the accumulation of salicylic acid. Physiol. Mol. Plant Pathol. 56, 95–106. doi: 10.1006/pmpp.1999.0255
Slaughter, A., Daniel, X., Flors, V., Luna, E., Hohn, B., and Mauch-Mani, B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158, 835–843. doi: 10.1104/pp.111.191593
Sugio, A., Dreos, R., Aparicio, F., and Maule, A. J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21, 642–654. doi: 10.1105/tpc.108.062596
Swindell, W. R., Huebner, M., and Weber, A. P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. doi: 10.1186/1471-2164-8-125
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Takahashi, H., Kanayama, Y., Zheng, M. S., Kusano, T., Hase, S., Ikegami, M., et al. (2004). Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. doi: 10.1093/pcp/pch085
Tanabe, K., Matsushima-Nishiwaki, R., Dohi, S., and Kozawa, O. (2010). Phosphorylation status of heat shock protein 27 regulates the interleukin-1β-induced interleukin-6 synthesis in C6 glioma cells. Neuroscience 170, 1028–1034. doi: 10.1016/j.neuroscience.2010.08.014
Thevenet, D., Pastor, V., Baccelli, I., Balmer, A., Vallat, A., Neier, R., et al. (2017). The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. New Phytol. 213, 552–559. doi: 10.1111/nph.14298
Ton, J., Jakab, G., Toquin, V., Flors, V., Iavicoli, A., Maeder, M. N., et al. (2005). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17, 987–999. doi: 10.1105/tpc.104.029728
Ton, J., van der Ent, S., van Hulten, M., Pozo, M., van Oosten, V., van Loon, L. C., et al. (2009). Priming as a mechanism behind induced resistance against pathogens, insects and abiotic stress. IOBC/WPRS Bull. 44, 3–13. doi: 10.2307/3308631
Ton, J., van Pelt, J. A., van Loon, L. C., and Pieterse, C. M. J. (2002). Differential effectiveness of salicylate-dependent and jasmonate/ethylene-dependent induced resistance in Arabidopsis. Mol. Plant Microbe. Interact. 15, 27–34. doi: 10.1094/MPMI.2002.15.1.27
Uyttewaal, M., Traas, J., and Hamant, O. (2010). Integrating physical stress, growth, and development. Curr. Opin. Plant Biol. 13, 46–52. doi: 10.1016/j.pbi.2009.10.004
van der Does, D., Leon-Reyes, A., Koornneef, A., van Verk, M. C., Rodenburg, N., Pauwels, L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25, 744–761. doi: 10.1105/tpc.112.108548
van Hulten, M., Pelser, M., Loon, L. C. V., Pieterse, C. M. J., and Ton, J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 5602–5607. doi: 10.1073/pnas.0510213103
Vitrac, X., Bornet, A., Vanderlinde, R., Valls, J., Richard, T., Delaunay, J. C., et al. (2005). Determination of stilbenes (δ-viniferin, trans-astringin, trans-piceid, cis- and trans-resveratrol, ε-viniferin) in Brazilian wines. J. Agric. Food Chem. 53, 5664–5669. doi: 10.1021/jf050122g
von Koskull-Döring, P., Scharf, K. D., and Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant. Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Vos, I. A., Pieterse, C. M. J., and van Wees, S. C. M. (2013). Costs and benefits of hormone-regulated plant defences. Plant Pathol. 62, 43–55. doi: 10.1111/ppa.12105
Wang, J., Cao, S. F., Wang, L., Wang, X. L., Jin, P., and Zheng, Y. H. (2018). Effect of β-Aminobutyric acid on disease resistance against Rhizopus rot in harvested peaches. Front. Microbiol. 9:1505. doi: 10.3389/fmicb.2018.01505
Wang, K. T., Jin, P., Han, L., Shang, H. T., Tang, S. S., Rui, H. J., et al. (2014). Methyl jasmonate induces resistance against Penicillium citrinum in Chinese bayberry by priming of defense responses. Postharvest Biol. Technol. 98, 90–97. doi: 10.1016/j.postharvbio.2014.07.009
Wang, K. T., Li, C. H., Lei, C. Y., Jiang, Y. B., Qiu, L. L., Zou, X. Y., et al. (2020). β-aminobutyric acid induces priming defence against Botrytis cinerea in grapefruit by reducing intercellular redox status that modifies posttranslation of VvNPR1 and its interaction with VvTGA1. Plant Physiol. Biochem. 156, 552–565. doi: 10.1016/j.plaphy.2020.09.026
Wang, K. T., Li, C. H., Lei, C. Y., Zou, Y. Y., Li, Y. J., and Zheng, Y. H. (2021). Dual function of VvWRKY18 transcription factor in the β-aminobutyric acid-activated priming defense in grapes. Physiol. Plantarum doi: 10.1111/ppl.13341 [Epub ahead of print].
Wang, K. T., Liao, Y. X., Xiong, Q., Kan, J. Q., Cao, S. F., and Zheng, Y. H. (2016). Induction of direct or priming resistance against Botrytis cinerea in strawberries by β-Aminobutyric acid and their effects on sucrose metabolism. J. Agr. Food Chem. 64, 5855–5865. doi: 10.1021/acs.jafc.6b00947
Wang, K. T., Wu, D. Z., Bo, Z. Y., Chen, S., Wang, Z. R., Zheng, Y. H., et al. (2019). Regulation of redox status contributes to priming defense against Botrytis cinerea in grape berries treated with β-aminobutyric acid. Sci. Hortic. 244, 352–364. doi: 10.1016/j.scienta.2018.09.074
Wang, W. X., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Windram, O., Madhou, P., McHattie, S., Hill, C., Hickman, R., Cooke, E., et al. (2012). Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24, 3530–3557. doi: 10.1105/tpc.112.102046
Xiao, S., and Chye, M. L. (2011). Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv tomato DC3000. Plant Physiol. 156, 2069–2081. doi: 10.1104/pp.111.176933
Xin, H. B., Zhang, H., Chen, L., Li, X. X., Lian, Q. L., Yuan, X., et al. (2010). Cloning and characterization of HsfA2 from Lily (Lilium longiflorum). Plant Cell Rep. 29, 875–885. doi: 10.1007/s00299-010-0873-1
Yu, D. Q., Chen, C. H., and Chen, Z. X. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540. doi: 10.1105/TPC.010115
Zarate, S. I., Kempema, L. A., and Walling, L. L. (2007). Silver leaf white fly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 143, 866–875. doi: 10.1104/pp.106.090035
Zheng, X., Krakowiak, J., Patel, N., Beyzavi, A., Ezike, J., Khalil, A. S., et al. (2016). Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife 5:e18638. doi: 10.7554/eLife.18638
Keywords: β-aminobutyric acid, heat shock protein, priming resistance, NPR1, Botrytis cinerea, grape berries
Citation: Li C, Cao S, Wang K, Lei C, Ji N, Xu F, Jiang Y, Qiu L and Zheng Y (2021) Heat Shock Protein HSP24 Is Involved in the BABA-Induced Resistance to Fungal Pathogen in Postharvest Grapes Underlying an NPR1-Dependent Manner. Front. Plant Sci. 12:646147. doi: 10.3389/fpls.2021.646147
Received: 25 December 2020; Accepted: 08 February 2021;
Published: 08 March 2021.
Edited by:
Essaid Ait Barka, Université de Reims Champagne-Ardenne, FranceReviewed by:
Synan F. AbuQamar, United Arab Emirates University, United Arab EmiratesBrigitte Mauch-Mani, Université de Neuchâtel, Switzerland
Copyright © 2021 Li, Cao, Wang, Lei, Ji, Xu, Jiang, Qiu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaituo Wang, wangkaituo83@gmail.com
 Chunhong Li1,2
Chunhong Li1,2 Shifeng Cao
Shifeng Cao Kaituo Wang
Kaituo Wang Yonghua Zheng
Yonghua Zheng