- National Institute of Plant Genome Research, New Delhi, India
The sessile nature of plants has made them extremely sensitive and flexible toward the constant flux of the surrounding environment, particularly light and dark. The light is perceived as a signal by specific receptors which further transduce the information through the signaling intermediates and effector proteins to modulate gene expression. Signal transduction induces changes in hormone levels that alters developmental, physiological and morphological processes. Importance of light for plants growth is well recognized, but a holistic understanding of key molecular and physiological changes governing plants development under dark is awaited. Here, we describe how darkness acts as a signal causing alteration in hormone levels and subsequent modulation of the gene regulatory network throughout plant life. The emphasis of this review is on dark mediated changes in plant hormones, regulation of signaling complex COP/DET/FUS and the transcription factors PIFs which affects developmental events such as apical hook development, elongated hypocotyls, photoperiodic flowering, shortened roots, and plastid development. Furthermore, the role of darkness in shade avoidance and senescence is discussed.
Introduction
Light and dark, both act as environmental cues that regulate plant growth and development from seedling emergence till senescence. Plant development begins in the soil where darkness acts as a signal for etiolation which is characterized by elongated hypocotyl and shortened roots, apical hook of closed cotyledons covering shoot apical meristem and impaired chloroplast development (Arsovski et al., 2012; Mazzella et al., 2014; Gommers and Monte, 2018). This mode of plant growth is known as skotomorphogenesis. It is an evolutionary advanced program in angiosperms to safely get seedling through the soil to light (Seluzicki et al., 2017). Nonetheless, exposure of seeds to continuous light during early development causes seedling de-etiolation (photomorphogenesis) characterized by attenuated hypocotyl growth, root growth acceleration, apical hook straightening and chloroplast maturation.
Being photoautotrophs, plants have evolved with the diverse set of photoreceptors. Arabidopsis photoreceptors have been classified as phytochromes (phyA-E, red/far-red light), cryptochromes (cry1-2), phototropins (phot1-2) and zeitlupe family members (ZTL, blue/UV-A light and UVR8; UV-B light) (Galvao and Fankhauser, 2015; Mishra and Khurana, 2017; Podolec and Ulm, 2018). Light-mediated activation of different photoreceptors and subsequent release of the dark-mediated photomorphogenic repression leads to light-dependent plant development. Some of the major mechanisms that play important role in photomorphogenic development are phosphorylation of phytochrome interacting factors (PIFs), ubiquitin-mediated proteolysis (UMP), and modulation of CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) complex activity, organization and subcellular localization of positive regulators of light signaling like ELONGATED HYPOCTYL 5 (HY5), LONG AFTER FAR-RED LIGHT1 (LAF) and LONG HYPOCOTYL AFTER FAR-RED LIGHT1 (HFR1) (Huang et al., 2014; Pham et al., 2018). Dark germinated seedlings become sensitive to fluctuations of the day/night cycles when they are first exposed to light. Therefore, growth and development of seedling comes under the control of circadian clock components. Ample amount of information is available about how plants sense and respond to the complex light spectra, but the information is missing about their behavior under darkness, and light and dark signal interaction. Since, light simply reverses the dark-mediated development by activation of the photoreceptors, it has been speculated that the inactive photoreceptors might act as dark receptors and mediate the dark-triggered signal transduction (Seluzicki et al., 2017; Gommers and Monte, 2018; Armarego-marriott and Sandoval-ibañez, 2020). Nonetheless, exact mechanism of dark sensing and perception is still unknown, and the idea of darkness perception by the inactive light receptors is a matter of debate. Though, regulation of seedling establishment by the light and dark signaling in Arabidopsis was recently reviewed (Gommers and Monte, 2018), enough literature is not available for light and dark signal integration and consequent phenotypic alterations.
In this review, an update about how plants make sense of darkness and use it as a signal during different phases of skotomorphogenic development e.g., apical hook formation, hypocotyl elongation, shortened roots, photoperiodic flowering, and plastid development is provided. Moreover, how endogenous clock integrates plant growth and development with photoperiods, and the effect of darkness on the plant responses such as, shade avoidance and dark-mediated senescence is discussed.
Darkness as a Signal
As mentioned earlier, it is presumed that darkness might be perceived by the inactive light receptors that could activate the COP/DET/FUS complex proteins and the PIFs. The proteins of COP/DET/FUS complex are encoded by a group of pleiotropic genes. These proteins are assembled in three different functional complexes, i.e., SUPPRESSOR OF PHYA105 (COPI-SPA), COP10-DET1-DDB1 (CDD), and COP9 signalosome complex (CSN), all of them are connected by a scaffold protein CULLIN4 (CUL4) (Chen et al., 2010; Pokhilko et al., 2011; Dong et al., 2014; Huang et al., 2016). In darkness, COPI-SPA and CDD complex act together to degrade positive regulators of photomorphogenesis like HY5, LAF and HFR1 via UMP. However, these complexes also prevent degradation of PIFs (PIF3 and 4) by inhibiting a brassinosteroid (BR) signaling component BRASSINOSTEROID-INSENSITIVE 2 (BIN2) (Dong et al., 2014; Huang et al., 2016; Wang and Guo, 2018; Figure 1). The function of CSN complex is to derubylate and thus positively regulates the activity of CUL4 which is present on both COP1-SPA and CDD complexes (Chen et al., 2010; Dong et al., 2014). Additionally, DET1 interacts with PIFs under darkness to stabilize them (Dong et al., 2014; Pham et al., 2018). PIFs in turn activate a diverse set of hormone biosynthetic and signaling genes that promote etiolation and repression of photomorphogenic response (Leivar et al., 2008; Paik et al., 2017). In Arabidopsis, PIF1, 3, 4, and 5 are involved in skotomorphogenesis, as indicated by normal light-grown seedling like growth of quadruple mutant pif1pif3pif4pif5 (pifq) under complete darkness (Dong et al., 2014; Pfeiffer et al., 2014).
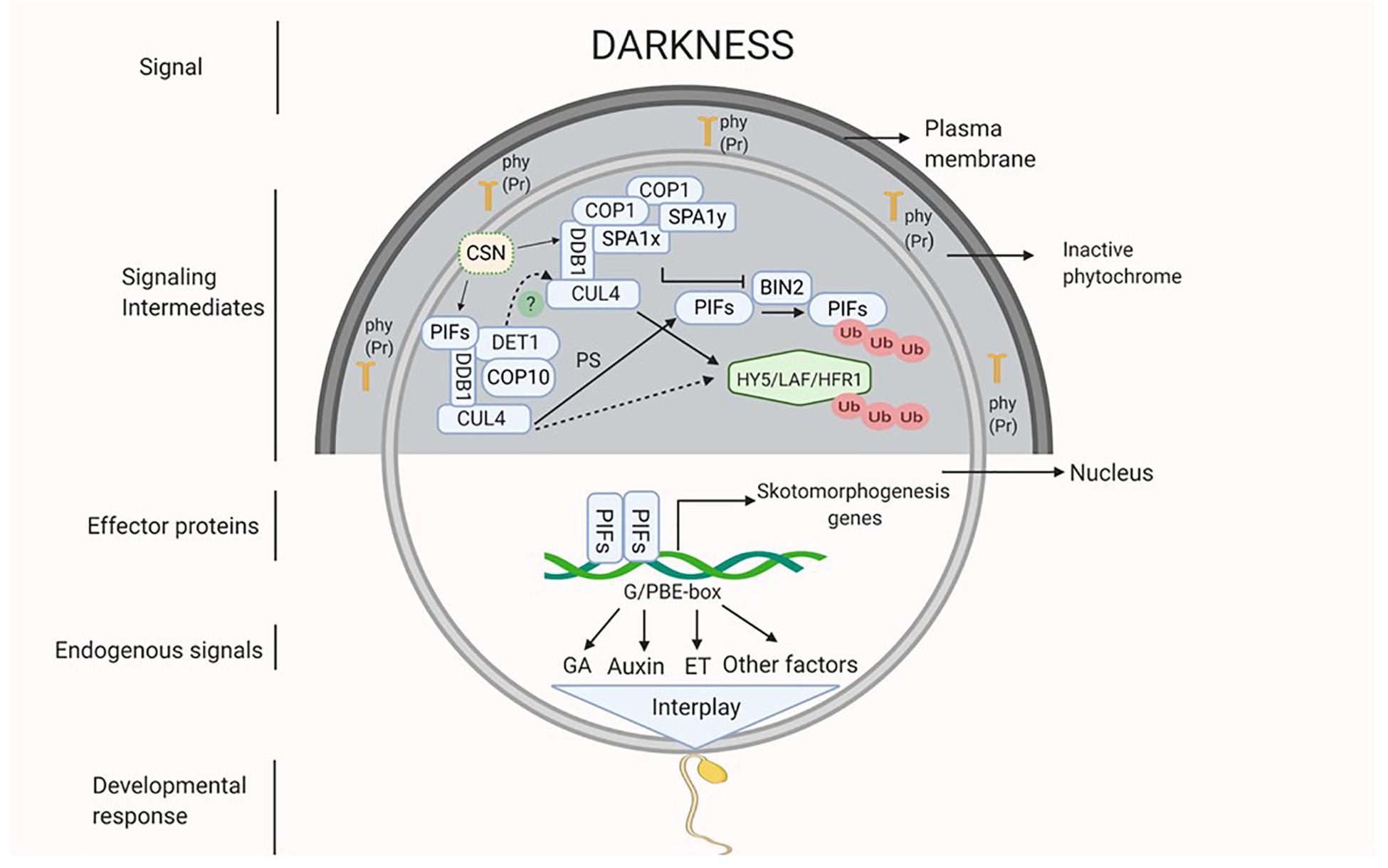
Figure 1. Darkness acts as a signal which might be perceived by inactive phytochromes (Pr form of phy) that activates the signaling intermediates COP1-SPA complex (CUL4-DDB1-COP1-SPA) and CDD complex (CUL4-DDB1-COP10-DET1) which interact with each other (dotted line). Active COP1-SPA complex lead to degradation of HY5 and inactivates BIN2 which causes degradation of PIFs by UMP and active CDD stabilizes PIFs. Active PIFs induces the expression of phytohormones (ET, GA and auxin) biosynthesis and signaling genes and of other skotomorphogenesis signals. Hormonal interplay along with many other factors integrate to generate skotomorphogenic developmental response.
In the forthcoming sections, how darkness regulates development of specific skotomorphogenic structures such as, apical hook, elongated hypocotyls, and shortened roots is discussed.
Hormonal Regulation of Plant Development in Dark
Apical Hook Development
Within 24 h of seed germination, after hypocotyl elongation, darkness induces the development of an apical hook (Mazzella et al., 2014) with curvature formation by modulating several hormonal pathways (de Wit et al., 2016). An asymmetrical distribution of auxin (auxin concentration increases at concave side and inhibit cell expansion) causes differential cell expansion and division at both the sides of the hypocotyl. This leads to faster growth at the outer side compared to inner side, which culminates in the hook formation (Béziat and Kleine-Vehn, 2018; Gommers and Monte, 2018; Wang and Guo, 2018). Asymmetric distribution of auxin is facilitated by the influx and efflux carriers on the cell membrane. In Arabidopsis, influx carriers AUXIN1 (AUX1) and LIKE AUXIN (LAX3) localize at the epidermal cells and vascular cylinder of the hook. These carriers facilitate the polar and basipetal flow of auxin from shoot apical meristem and cotyledons to hypocotyl. Efflux carrier PIN1 facilitates the outward flow of auxin through vascular cylinder and inner epidermis. PIN3 facilitates the auxin flow from vascular tissues to outer cortex and epidermis, whereas PIN4 and 7 support the auxin flow from vascular tissue to cortex and epidermis. In addition, ABCB transporter 1 and 19 which are localized at inner epidermal cells of the hook, steer the auxin transport to the convex side (Vandenbussche et al., 2010; Wu et al., 2010; Žádníkova et al., 2010, 2016; Farquharson, 2017; Wang and Guo, 2018). The maintenance of differential auxin gradient involves the co-ordination of auxin synthesis, transport and signaling. YUCs and TAR2 are the flavin monooxygenases and tryptophan aminotransferase related enzymes, which, respectively, catalyze two initial steps in the auxin biosynthesis pathway. These genes are differentially expressed in the apical hook region and their mutants (yuc1/2/4/6, tar2) display impaired hook phenotype due to developmental defects (Stepanova et al., 2008, 2011; Vandenbussche et al., 2010; Abbas et al., 2013; Cao et al., 2019). The auxin signaling mutants like iaa3, iaa12 and iaa13 lack apical hook, suggesting the involvement of auxin signaling in apical hook formation. Moreover, AUX/IAAs regulate auxin signaling effector proteins; auxin response factors (ARFs) (Žádníkova et al., 2010, 2016; Abbas et al., 2013).
The apical hook development is tightly regulated by signals from cell wall and root-hypocotyl interaction. The cell wall status affects apical hook bending by transcriptional regulation of PINs and AUX1 through ARF2 (Aryal et al., 2020; Sampathkumar and Eng, 2020). Recently, it was shown that auxin gradient formed at the root tip by PIN2 is required for root growth in response to gravity (Zhu and Gallem, 2019). The root auxin gradient gradually extends toward hypocotyl and might trigger the hypocotyl bending resulting in hook formation. A balanced concentration of hormones ABA and GA has been shown to be essential for a close interaction between root and hypocotyl (Baral et al., 2020).
PIFs have been involved in different aspects of gradient formation such as, auxin synthesis and polar auxin transport (PAT). In Arabidopsis, PIF4 and PIF5 induce the expression of YUCCA genes (Pfeiffer et al., 2014) and PIN localization regulatory kinase WAG2 (Willige et al., 2012; Mazzella et al., 2014). Darkness induced phytohormones such as, GA and ethylene (ET) affect PAT indirectly by modulating the expression of WAG2 (Willige et al., 2012) and ETHYLENE INSENSITIVE 3/EIN3-LIKE1 (EIN3/EIL1) (Wabnik et al., 2016; Wang and Guo, 2018). PIFs and EIN3/EIL1 co-regulate the curvature formation by directly binding to HOOKLESS 1 (HLS1) promoter. HLS1, which codes for a putative N-acetyltransferase and transcriptionally regulated by EIN3/EIL1 is a key regulator of apical hook development (Wang and Guo, 2018). Markedly, PIFs enhance the expression of HLS1 by binding to its promoter at a site different from that of EIN3/EIL1. In addition, PIFs work in tandem with EIN3/EIL1 to integrate hormonal signals such as, GA, JA, and physical factors including, light and mechanical pressure (Zhang et al., 2018). Besides acting along with PIFs, EIN3 also induces the expression PIF3 (Zhong et al., 2012). Also, divergent roles of ET in the light- and dark- mediated seedling growth are recently shown (Yu and Huang, 2017; Harkey et al., 2018, 2019; Gu et al., 2019).
In response to darkness, GA accumulates and binds to its receptor gibberellin insensitive dwarf 1 (GID1) and targets DELLAs (GAI, RGA), the negative regulators of GA signaling for UMP degradation (de Lucas et al., 2008). GA is essential for skotomorphogenesis as GA mutant ga1 seedlings and GA biosynthesis inhibitor Paclobutrazol (PAC) treated seedlings show light-grown phenotype and are unable to form hook even when grown in the complete darkness (Achard et al., 2007; Feng et al., 2008; Arana et al., 2011; Zhang et al., 2018). Antagonistic to auxin, GA promotes cell expansion and division on the convex side of hypocotyl (Alabadí et al., 2008; Arana et al., 2011).
In darkness, ET synthesis is enhanced by the transcriptional activation of ACS8 by PIF5 in GA dependent manner (Arana et al., 2011). In addition to darkness, physical factors such as, soil depth and compactness, mechanical pressure generated by hypocotyl against soil induce ET production (Zhong et al., 2014; de Wit et al., 2016). ET positively regulates skotomorphogenesis as dark-grown seedlings when treated with exogenous ET, produces exaggerated hooks (Mazzella et al., 2014). Similar to auxin and GA, ET also regulates cell division but at the apical basal parts of the hook. In addition, ET maintains auxin gradient by regulating its synthesis, transport and signaling (An et al., 2012; Chang et al., 2013; Wang et al., 2014; Wabnik et al., 2016; Zhang et al., 2018). JA and SA both are negative regulators of apical hook formation, and both of them act by disrupting ET signaling. JA activated MYC2 promotes the degradation of EIN3/EIL1. Also, MYC2 and NON-EXPRESSOR OF PR GENES1 (NPR1, SA signaling mediator) individually interact with EIN3 and inhibits its binding to the promoter of HLS1 (Zhang et al., 2018; Bertoni, 2020; Huang et al., 2020).
Hypocotyl Elongation
Another remarkable phenotype observed under darkness is an elongated hypocotyl. Darkness leads to this phenotype by modulating the levels of phytohormones such as, auxin, BR, ET, and GA (Reed et al., 2018). Irreversible increase in the plant organ size is primarily caused by cell expansion. Expansion of a cell is characterized by the vacuole enlargement and selective cell wall loosening, which releases the wall pressure and allows the water to flow inside (Cosgrove, 2016a,b). Consistently, the up-regulation of several auxin responsive cell wall loosening related genes such as, EXPANSINs (EXPA4,11), EXPANSIN- LIKE (EXLA3) and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE (XTH18,19) was found in dark-grown seedlings (Cosgrove, 2005; Orden et al., 2010; Majda, 2018). Auxin acidifies the wall matrix by stimulating the activity of epidermal cell H+-ATPases and K+ channels, thereby, generating the turgor pressure (Ivakov et al., 2017; Majda, 2018; Duman et al., 2020). Therefore, cell wall remodeling by auxin and cell wall generated signals promotes hypocotyl elongation. PIFs maintain auxin gradient and induce the expression of wall loosening enzymes (Rougemont et al., 2012; Leivar and Monte, 2014). PIF4 remains functional under darkness by binding directly to BR stabilized protein BZR1 (Wang et al., 2012). Active PIF4-BZR1 module positively regulates the synthesis of GA by targeting DELLAs. Thus, darkness acts through the functional PIF4-BZR1 by regulating the level of phytohormones. ABA has been found to be a negative regulator of hypocotyl elongation in the dark-grown seedlings. ABA induces the expression of DELLAs (GAI and RGA) and inhibits the expression of auxin biosynthetic genes and membrane H+-ATPases (Hayashi et al., 2014; Lorrai et al., 2018).
Shortened Roots
Like shoots, roots also sense light, consequently, root morphology is altered after light perception (Lee et al., 2017). Dark-germinated seedlings have short and thin primary roots with reduced lateral roots, whereas, the phenotype is reversed after light exposure (Dyachok et al., 2011). So, how the dark signaling represses root growth, and the light perceived at axial end of the plant alters root morphology is a curious question?
When light grown seedlings were decapitated or treated with PAT inhibitors, they showed inhibition of lateral root development (Salisbury et al., 2007), like the etiolated seedlings. This indicated that auxin synthesized in cotyledons in response to light acts as a positive regulator of lateral root emergence. Also, light facilitates auxin transport from apical part of shoot to root (Lee et al., 2016, 2017). In darkness, COP1 inhibits PIN1 gene expression in shoot. PIN1 is essential for shoot to root PAT therefore, repression of its expression leads to the root growth suppression. But, when shoot is exposed to light, COP1 moves out from the nucleus, relieving the suppression of PIN1 gene expression (Sassi et al., 2012; Gangappa and Botto, 2016). The HY5 deficient mutant also exhibits defects in lateral root elongation and growth, suggesting the involvement of HY5 in maintaining shoot to root continuum (Sibout et al., 2006). Light activates photoreceptors which interact with COP1, leading to its inactivation. Light-dependent inactivation of COP1 inhibits COP1 mediated degradation of HY5 thereby, promoting HY5 activity. Interestingly, light stabilized HY5 targets COP1 for degradation, thereby, shoot to root PAT is resumed that leads to altered root morphology (Mazzella et al., 2014). Moreover, HY5 translocation from shoot to root promotes root growth (Zhang et al., 2017). The dark-induced hormonal regulation of skotomorphogenic development is summarized in Figure 2.
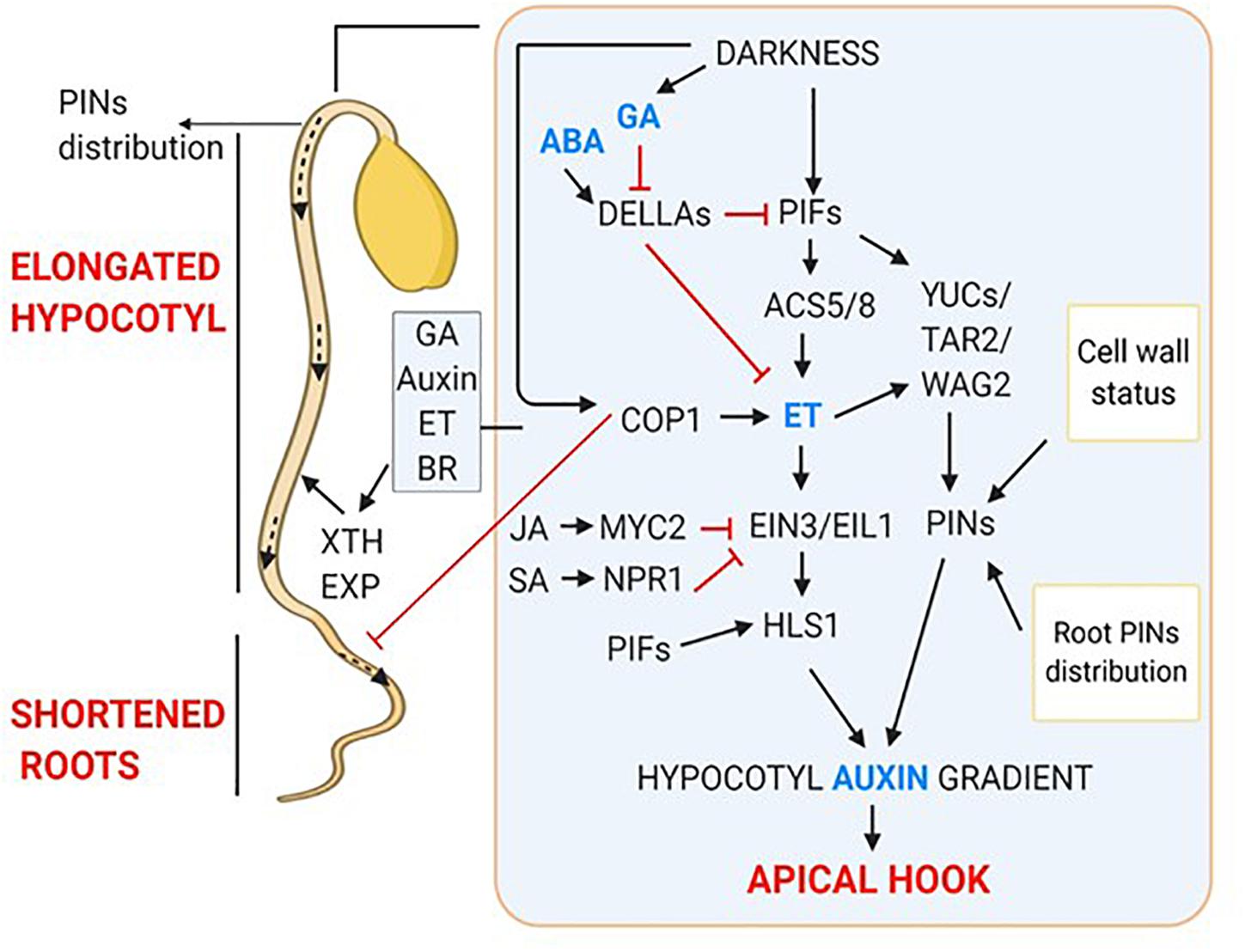
Figure 2. Hormonal regulation of skotomorphogenic organs; apical hook, elongated hypocotyls and shortened roots. Dark-induced signaling results in increased gradient of GA, ET and Auxin which together interplay to form apical hook. Outside inset showing hormonal regulation of hypocotyl elongation and short roots.
Photoperiodic Flowering
Plants sense diurnal variations which affect flowering and accordingly are classified as short- day, long-day and day neutral plants. Importance of darkness in short-day plants is evident from the fact that disruption of dark period with light significantly affects flowering (Andrés and Coupland, 2012; Johansson and Staiger, 2015; Cao et al., 2016). Under long-day condition, light inhibits the expression of flowering genes; HEADING DATE 3A (HD3A) and RICE FT-LIKE1 (RFT1) in rice by activating an inhibitor HEADING DATE 1 (HD1), whereas, under short-days, HD1 induces the expression of HD3A and RFT1 (Ishikawa et al., 2005). Interruption of long duration of darkness with short exposure of light induces PHOTOPERIOD1 (PPD1) mediated activation of FT1, which after moving from leaves into shoot apical meristem promotes accelerated flowering in wheat (Pearce et al., 2017). Moreover, a night break (NB) causes transcriptional up-regulation of PPD1 in wheat, levels of which increases with multiple NB and length of darkness. Thus, a period of darkness plays an important role in regulating photoperiodic flowering in plants.
Light- and Dark-Dependent Plastid Development
Development of plastids could be understood by the studies on the dark-grown seedlings. Plastids could be of various types like, proplastids, eoplasts, etioplasts, chloroplasts and chromoplasts based on their morphological characters, function and tissue location (Liebers et al., 2017). In the cotyledon of the dark-grown seedlings, eoplasts develop into etioplast and at this stage, the development of a prolamellar body (PLB) occurs (Bastien et al., 2016). A PLB is made up of regular arrangements of NADPH, the chlorophyll precursor protochlorophyllide (Pchlide), protochlorophyllide-oxido-reductase (POR), and the thylakoid membrane lipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG). In light, various nuclear genes which code for chloroplast biogenesis related proteins are expressed. This results in thylakoid membrane development and POR induced chlorophyll biosynthesis. The proplastids in the shoot apical meristem are directly converted to chloroplast during primary leaf development (Liebers et al., 2017).
Molecular Aspects of Plastid Development
TFs involved in early developmental process regulate the expression of plastid development related genes. PIF1 and PIF3 accumulate in dark-grown seedlings and repress the chlorophyll biosynthesis genes, whereas, in response to red light PIFs undergo active phy (Pfr) mediated degradation (Gommers and Monte, 2018; Hernández-Verdeja et al., 2020). Similarly, in light, EIN3 is directly targeted and degraded by crys and phys (Gommers and Monte, 2018).
In an early response to light the expression of HY5, which regulates the downstream components of chloroplast development, is induced. In dark COP1 mediates the degradation of HY5 and stabilization of PIF and EIN3. Under blue light, cryptochrome represses the expression of COP1 leading to enhanced HY5 activity, which is required for chloroplast development (Hernández-Verdeja et al., 2020). PIFs and HY5 act as negative and positive regulators, respectively, of chlorophyll and carotenoid synthesis. Therefore, PIFs and HY5 compete for the same binding site (G-box) on the promoter of common target genes, and eventually they regulate chloroplast development antagonistically (Toledo-Ortiz et al., 2014). In addition, HY5-PIF module controls the photosynthetic gene transcription by regulating PHYTOENE SYNTHASE (PSY) gene, which catalyzes a rate limiting step in carotenoid biosynthesis pathway (Toledo-Ortiz et al., 2014). Thus, the PIF-HY5 regulatory mechanism is crucial for the proplastid development. Golden2-Like (GLK) nuclear TFs which work independently of phytochromes and PIFs are also the key regulators of plastid biogenesis (Liu et al., 2020). GLKs strongly activate the chlorophyll biosynthesis enzymes and light-harvesting chlorophyll binding proteins including, GlutRNA reductase (HEMA1), magnesium chelatase (CHLH), pchlide oxidoreductase (POR-B) and chlidea oxygenase (CAO).
Two RNA polymerases; nuclear encoded RNA polymerase (NEP) and plastid encoded RNA polymerases (PEP) are involved in the proplastid biogenesis (Hernández-Verdeja et al., 2020). In dark, plastid gene transcription is driven by NEP whereas, upon light exposure, transition of NEP to PEP occurs leading to PEP driven transcription. In light, the main transcriptional activity is taken-up by PEP, thus, the transcription of PEP associated genes increases. PRIN2, a plastid localized redox regulated protein is required for PEP activity. In dark, PRIN2 forms a homo-dimer via di-sulfide bonds, however, upon exposure to light it get reduced to PRIN2 monomers, and contributes to PEP activity (Díaz et al., 2018). PEP mediated increase in transcription and light induced communication between developing chloroplast and nucleus through antero-retrograde signaling results in completion of chloroplast biogenesis (Pogson et al., 2015).
Hormonal Regulation of Plastid Development
Some phytohormones control the chlorophyll biosynthesis, thereby, regulate the chloroplast biogenesis. For example, in the dark-grown Arabidopsis seedlings cytokinin treatment induces the formation of prothylakoids whereas, these thylakoid membranes developed after about 6 h of light treatment in cytokinin untreated seedlings (Cortleven and Schmülling, 2015). The chlorophyll biosynthesis begins with the conversion of glutamate to 5-aminolevulinic acid (ALA), followed by the production of chlorophyll precursor, Pchlide. POR catalyzes the conversion of Pchlide into chlorophyllide which after esterification forms chlorophyll. Cytokinin promotes ALA synthesis and enhances POR activity to support the chloroplast biogenesis (Liu et al., 2017). GA regulated DELLA proteins are involved in the induction of PORA and PORB genes (Cheminant et al., 2011). Involvement of ET signaling has also been found in plastid development. In the double mutant of ET signaling; ein3eil1 expression of Pchlide/chlorophyll synthesis genes HEMA1, GUN4, and GUN5 was increased under darkness. PIF3 has been known to inhibit the expression of these genes (Stephenson et al., 2009; Shin et al., 2009), consistently, their expression was higher in pif3 mutant. Moreover, ein3eil1mutant but not pif3 had reduced expression of PORA and PORB genes. Thus, EIN3/EIL1 negatively regulate Pchlide/chlorophll biosynthesis through PIF3, but stimulate PORA and PORB independently of PIF3 (Zhong et al., 2014). These observations clearly suggest the crucial role of phytohormones in plastid development.
Shade Avoidance: An Intermediate Response Between Light and Dark
Due to their sessile nature, plants face shade arising from the neighboring vegetation canopies. In the shade, plants compete for their resources particularly red/far-red (R:FR) photon flux ratio. In response to shade, plants channelize their energy toward hypocotyl and stem elongation, enhanced apical dominance and early flowering, all of them collectively termed as shade avoidance syndrome (SAS) (de Wit et al., 2016). Along with photoreceptors, phytohormones like ET, GA, Auxin, and BR are implicated in shade-induced plant responses (Das et al., 2016; Yang and Li, 2017).
In the past decade, shade related research was carried out mainly in Arabidopsis, and indicated a crucial role of auxin in the shade avoidance response (Sessa et al., 2018). In the Arabidopsis seedlings, auxin accumulates in response to shade resulting in hypocotyl cells elongation (Ma and Li, 2019). A low R:FR ratio (as in case of shade) has been found to promote petiole elongation in Arabidopsis (Roig-Villanova and Martínez-García, 2016). Phytochrome B (phyB) is the key shade avoidance response regulating photoreceptor, whereas, phyD and phyE function redundantly in promoting shade-induced elongation (Franklin et al., 2003). In contrast, phyA represses the elongation response induced by low R/FR light (Martínez-García et al., 2014). phyB becomes active in the presence of light having high R:FR ratio. Upon activation, it translocates into the nucleus and interacts with PIFs. This interaction causes phosphorylation and subsequent inactivation of PIFs leading to their degradation via UMP (de Wit et al., 2016). Hence, PIF-dependent transcriptional activation of auxin homeostasis and cell wall remodeling related genes is inhibited (Courbier and Pierik, 2019). While, under light of low R:FR, phyB remains inactive, thus allowing PIFs to accumulate and induce the transcription of YUCCA genes (Müller-Moulé et al., 2016), resulting in enhanced auxin concentration in the cell. PIF4, 5, and 7 directly regulate several auxin biosynthesis and signaling genes in response to shade (Hornitschek et al., 2012; Li et al., 2012). A HomeoBox2 (ATHB2) TF acts as a positive regulator and Long Hypocotyl in Far-red 1/Slender In Canopy Shade 1 (HFR1/SICS1) (an atypical bHLH protein), acts as a negative regulator of PIFs controlled shade avoidance response (Sessa et al., 2018). Cryptochromes (cry1 and cry2) are involved in low blue light (LBL)-induced shade avoidance response. CRY1 and CRY2 physically interact with PIFs and regulates their activity for LBL induced hypocotyl growth (Ma et al., 2016; Sessa et al., 2018).
Phytohormones in Shade Avoidance Response
Upon low R: FR exposure, bioactive GA levels increases and the accumulated GA inhibits the DELLAs. The DELLA proteins directly interact with PIFs and this interaction prevents PIFs binding to DNA, thereby, negatively regulating the expression of cell elongation related genes (Li et al., 2016). Moreover, GA is implicated in shade-induced flowering, as silencing GA20ox2 expression results in delayed flowering in Arabidopsis under far-red light conditions (Yang and Li, 2017). In addition, ET is proposed to be a positive regulator of shade-induced petiole elongation, and is involved in organ specific shade avoidance response (Yang and Li, 2017). ABA biosynthesis mutants, nced3-2 and aba2-1 show increased branching under low R:FR suggesting that ABA suppresses branching under shade (Yang and Li, 2017). NPR1 also plays a crucial role in petiole elongation in shade (Nozue et al., 2018). Interestingly, elevated auxin and BR production in response to PIFs activation under low R:FR light costs both SA and JA based defenses (Martínez et al., 2018).
Day-Night Transitions
The rhythmic behavior of the biological processes is maintained by an endogenous oscillator/pacemaker called the circadian clock. This clock, under natural conditions maintains a period of 24 h, regulated by transitions from day to night (light/dark) and vice versa. As the clock is tightly coupled with diurnal cycles, it modulates many gene regulatory networks (GRNs) based on time of the day. When an organism is subjected to constant light or dark for a longer time, the rhythms dampen out and require transition from the existing environment. Since, plant life begins under complete darkness, the seedling growth during this period is clock independent, till it experiences light/dark (photoperiod) cycles (Salome et al., 2008). The fact that Arabidopsis seedling growth becomes photoperiod sensitive after de-etiolation was established by growing plants in continuous light and dark, separately. Until 12 h, similar hypocotyl length was observed in both conditions, and only prolonged exposure of darkness (> 12 h) resulted in elongated hypocotyls, suggesting the process to be short-day specific (Niwa et al., 2009; Seluzicki et al., 2017). Then, how clock is integrated with photoperiod after de-etiolation, was disclosed by the growth pattern of WT and clock mutants (CCA1-ox and elf3) seedlings in short-day conditions. WT seedlings elongated normally after prolonged darkness, but in both the clock mutants seedling elongation started at the beginning of the dark period and continued till it prolonged, indicating an inhibitory effect of clock genes during initial hours of darkness (Fankhauser et al., 2007). Interestingly, temporal transcriptional induction of PIF1, 3, 4, and 5 was also observed during late-night hours coinciding with the seedling etiolation (Leivar et al., 2012). PIFs are degraded during the day through PHY mediated photobodies, and are kept in check during early-night by clock evening complex (EC) genes (Huang et al., 2016). Another clock complex gene, TIMING OF CAB EXPRESSER 1 (TOC1) represses PIFs level during early- and mid-night (Soy et al., 2016; Paik et al., 2017). Therefore, clock exerts its effect by controlling the temporal expression of PIF genes under darkness.
Dark-Induced Senescence
Senescence is an age triggered developmental process characterized by an ordered and programmable degradation. The degradation involves the mobilization of building blocks at various levels of organization e.g., cell, tissue, organ and the organism, culminating in plant death (Lim et al., 2007). Its initiation, progression and completion are tightly linked to various external and internal cues (Kim et al., 2016; Law et al., 2018). Darkness is one of the external cues that positively regulate leaf senescence, and the process is called dark-induced senescence (DIS). DIS is physiologically quite different from an age-triggered leaf senescence (Kiddle et al., 2011). Interestingly, darkness only promotes senescence of individual plant organs, and inhibits senescence at the whole plant level (Keech et al., 2010; Sakuraba et al., 2014; Law et al., 2018), suggesting the involvement of other factors for death of the whole plant.
PIFs involvement in DIS became evident when PIF single and quadruple (pifq) mutants exhibited delayed leaf senescence, and their overexpressing plants showed the opposite phenotype (Song et al., 2014). Moreover, PIF3, 4 and 5 were significantly up-regulated in both age triggered senescence and DIS. Evidences indicate that PIFs mediate transcriptional activation of many SENESCENCE ASSOCIATED GENES (SAGs), i.e., STAY GREEN 1 (SGR1) and NON-YELLOW COLORING1 (NYC1) and other senescence promoting TFs such as WRKY22 and NAC (Zhang et al., 2015). Additionally, PIF4/5 have been shown to transcriptionally enhance the expression of ABSCISIC ACID 5 (ABI5) and ENHANCED EM LEVELS (EEL) (Qi et al., 2020). ABI5, EEL, and PIFs act together in a coherent feed forward loop to increase the expression of ORESARA1 (ORE1), a master regulator of senescence. Subsequently, PIFs, ORE1, ABI5, and EIN3 interplay to activate SAGs which finally lead to breakdown of chlorophyll, degradation of photosynthetic machinery culminating in leaf senescence (Sakuraba et al., 2014; Qiu et al., 2015; Liebsch and Keech, 2016).
Conclusion and Future Directions
Dark and light independently activate diverse signaling pathways which alter the levels of plant growth regulators consequently leading to a specific response. Efforts made in the past two decades in the area of skotobiology have advanced our understanding of how plant behaves and make sense of the dark period. The role of multifunctional dark signaling intermediates COP/DET/FUS and transcription factor PIF has been explored in diverse areas. We have provided the latest information about darkness acting as a signal during various plant growth processes such as, skotomorphogenesis, day-night transitions, shade avoidance, and senescence. Differential accumulation of several phytohormones, their regulatory effects on diverse molecular components and, in turn, the interplay of molecular players that determines the pattern of growth and development in dark has been elaborated. However, a complete understanding of the dark and light signaling integration needs exploration of the inter-organ communication mechanisms, necessary for establishment/transfer of hormonal gradients. Also, the hormonal interplay and regulatory mechanism underlying the integration of other subterranean environmental cues such as, soil compactness, temperature, biotic and abiotic factors with dark signaling, is still enigmatic and requires in-depth exploration in future.
Author Contributions
AS conceptualized and designed the study. Deepika, Ankit, and SS compiled the data. Deepika, Ankit, and AS wrote the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge DBT (Department of Biotechnology) – e-Library Consortium (DeLCON) for providing access to e-resources and financial support from National Institute of Plant Genome Research (NIPGR) core research grant. Deepika and SS are thankful to council of scientific and industrial research (CSIR), India for research fellowships.
References
Abbas, M., Alabadí, D., and Blázquez, M. A. (2013). Differential growth at the apical hook: all roads lead to auxin. Front. Plant Sci. 4:441. doi: 10.3389/fpls.2013.00441
Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., et al. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172. doi: 10.1104/pp.106.092254
Alabadí, D., Gallego-Bartolomé, J., Orlando, L., García-Cárcel, L., Rubio, V., Martínez, C., et al. (2008). Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 53, 324–335. doi: 10.1111/j.1365-313X.2007.03346.x
An, F., Zhang, X., Zhu, Z., Ji, Y., He, W., Jiang, Z., et al. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22, 915–927. doi: 10.1038/cr.2012.29
Andrés, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonalcues. Nat. Publ. Gr. 13, 627–639. doi: 10.1038/nrg3291
Arana, V., Vandenbussche, F., Petra, Z., Gallego-bartolome, J., Guardiola, V., Van Der Straeten, D., et al. (2011). Hierarchy of hormone action controlling apical hook development in Arabidopsis. Plant J. 67, 622–634. doi: 10.1111/j.1365-313X.2011.04621.x
Armarego-marriott, T., and Sandoval-ibañez, O. (2020). Beyond the darkness: recent lessons from etiolation and de-etiolation studies. JXB 71, 1215–1225. doi: 10.1093/jxb/erz496
Arsovski, A. A., Galstyan, A., Guseman, J. M., and Nemhauser, J. L. (2012). Photomorphogenesis. Arabidopsis Book 10:e01475. doi: 10.1199/tab.0147
Aryal, B., Jonsson, K., Baral, A., Sancho-andres, G., Kierzkowska, A. R., Kierzkowski, D., et al. (2020). Interplay between cell Wall and Auxin mediates the control of differential cell elongation during apical hook development. Curr. Biol 30, 1733–1739.e3. doi: 10.1016/j.cub.2020.02.055
Baral, A., Morris, E., Aryal, B., Jonsson, K., Verger, S., Xu, T., et al. (2020). External mechanical cues reveal core molecular pathway behind tissue bending in plants. bioRxiv [Preprint]. doi: 10.1101/2020.03.05.978296
Bastien, O., Botella, C., Chevalier, F., Block, M. A., Jouhet, J., Breton, C., et al. (2016). New insights on thylakoid biogenesis in plant cells. Int. Rev. Cell Mol. Biol. 323, 1–30. doi: 10.1016/bs.ircmb.2015.12.001
Bertoni, G. (2020). Ethylene versus salicylic acid in apical hook formation. Plant Cell 32:531. doi: 10.1105/tpc.20.00031
Béziat, C., and Kleine-Vehn, J. (2018). The road to auxin-dependent growth repression and promotion in apical hooks. Curr. Biol. 28, R519–R525. doi: 10.1016/j.cub.2018.01.069
Cao, K., Cui, L., Ye, L., Zhou, X., Bao, E., Zhao, H., et al. (2016). Effects of red light and night break treatments on growth and flowering of tomato plants. Front. Plant Sci. 7:527. doi: 10.3389/fpls.2016.00527
Cao, M., Chen, R., Li, P., Yu, Y., Zheng, R., and Ge, D. (2019). Differential growth of the apical hook. Nature 568, 240–243. doi: 10.1038/s41586-019-1069-7
Chang, K. N., Zhong, S., Weirauch, M. T., Hon, G., Pelizzola, M., Li, H., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLIFE 2, e00675. doi: 10.7554/eLife.00675
Cheminant, S., Wild, M., Bouvier, F., Pelletier, S., Renou, J. P., Erhardt, M., et al. (2011). DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23, 1849–1860. doi: 10.1105/tpc.111.085233
Chen, H., Huang, X., Gusmaroli, G., Terzaghi, W., Lau, O. S., Yanagawa, Y., et al. (2010). Arabidopsis CULLIN4-Damaged DNA Binding Protein 1 Interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA Complexes to Regulate Photomorphogenesis and Flowering Time. Plant Cell 22, 108–123. doi: 10.1105/tpc.109.065490
Cortleven, A., and Schmülling, T. (2015). Regulation of chloroplast development and function by cytokinin. J. Exp. Bot. 66, 4999–5013. doi: 10.1093/jxb/erv132
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Cosgrove, D. J. (2016a). Catalysts of plant cell wall loosening. F1000Res. 5:119. doi: 10.12688/f1000research.7180.1
Cosgrove, D. J. (2016b). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall modifying enzymes. JXB 67, 463–476. doi: 10.1093/jxb/erv511
Courbier, S., and Pierik, R. (2019). Canopy light quality modulates stress responses in plants. iScience 22, 441–452. doi: 10.1016/j.isci.2019.11.035
Das, D., St. Onge, K. R., Voesenek, L. A. C. J., Pierik, R., and Sasidharan, R. (2016). Ethylene- and shade-induced hypocotyl elongation share transcriptome patterns and functional regulators. Plant Physiol. 172, 718–733. doi: 10.1104/pp.16.00725
de Lucas, M., Davière, J. M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
de Wit, M., Galvão, V. C., and Fankhauser, C. (2016). Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67, 513–537. doi: 10.1146/annurev-arplant-043015-112252
Díaz, M. G., Hernández-Verdeja, T., Kremnev, D., Crawford, T., Dubreuil, C., and Strand, Å. (2018). Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat. Commun. 9:50. doi: 10.1038/s41467-017-02468-2
Dong, J., Tang, D., Gao, Z., Yu, R., Li, K., He, H., et al. (2014). Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell 26, 3630–3645. doi: 10.1105/tpc.114.130666
Duman, Z., Eliyahu, A., Abu-abied, M., and Sadot, E. (2020). The contribution of cell wall remodeling and signaling to lateral organs formation. Isr. J. Plant Sci. 67, 110–127. doi: 10.1163/22238980-20191115
Dyachok, J., Zhu, L., Liao, F., He, J., Huq, E., and Blancaflor, E. B. (2011). SCAR mediates light-induced root elongation in arabidopsis through photoreceptors and proteasomes. Plant Cell 23, 3610–3626. doi: 10.1105/tpc.111.088823
Fankhauser, C., Nozue, K., Covington, M. F., Duek, P. D., Harmer, S. L., and Maloof, J. N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361. doi: 10.1038/nature05946
Farquharson, K. (2017). Division of labor during apical hook formation. Plant Cell 29, 917–918. doi: 10.1105/tpc.17.00357
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Franklin, K. A., Praekelt, U., Stoddart, W. M., Billingham, O. E., Halliday, K. J., and Whitelam, G. C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in arabidopsis. Plant Physiol. 131, 1340–1346. doi: 10.1104/pp.102.015487
Galvao, V. C., and Fankhauser, C. (2015). Sensing the light environment in plants: photoreceptors and early signaling steps. Curr. Opin. Neurobiol. 34, 46–53. doi: 10.1016/j.conb.2015.01.013
Gangappa, S. N., and Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gommers, C. M. M., and Monte, E. (2018). Seedling establishment: a dimmer switch-regulated process between dark and light signaling. Plant Physiol. 176, 1061–1074. doi: 10.1104/pp.17.01460
Gu, S. Y., Wang, L. C., Cheuh, C. M., and Lo, W. S. (2019). CHITINASE like1 regulates root development of dark-grown seedlings by modulating ethylene biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 10:600. doi: 10.3389/fpls.2019.00600
Harkey, A. F., Watkins, J. M., Olex, A. L., DiNapoli, K. T., Lewis, D. R., Fetrow, J. S., et al. (2018). Identification of transcriptional and receptor networks that control root responses to ethylene. Plant Physiol. 176, 2095–2118. doi: 10.1104/pp.17.00907
Harkey, A. F., Yoon, G. M., Seo, D. H., DeLong, A., and Muday, G. K. (2019). Light modulates ethylene synthesis, signaling, and downstream transcriptional networks to control plant development. Front. Plant Sci. 10:1094. doi: 10.3389/fpls.2019.01094
Hayashi, Y., Takahashi, K., Inoue, S. I., and Kinoshita, T. (2014). Abscisic acid suppresses hypocotyl elongation by dephosphorylating plasma membrane H+-ATPase in Arabidopsis thaliana. Plant Cell Physiol. 55, 845–853. doi: 10.1093/pcp/pcu028
Hernández-Verdeja, T., Vuorijoki, L., and Strand, Å. (2020). Emerging from the darkness: interplay between light and plastid signaling during chloroplast biogenesis. Physiol. Plant. 169, 397–406. doi: 10.1111/ppl.13100
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., López-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Huang, H., Alvarez, S., Bindbeutel, R., Shen, Z., Naldrett, M. J., Evans, B. S., et al. (2016). Identification of evening complex associated proteins in arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15, 201–217. doi: 10.6019/PXD002606
Huang, P., Dong, Z., Guo, P., Zhang, X., Qiu, Y., Li, B., et al. (2020). Salicylic acid suppresses apical hook formation via NPR1-mediated repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 32, 612–629. doi: 10.1105/tpc.19.00658
Huang, X., Ouyang, X., and Deng, X. W. (2014). Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Biol. 21, 96–103. doi: 10.1016/j.pbi.2014.07.003
Ishikawa, R., Tamaki, S., Yokoi, S., Inagaki, N., Shinomura, T., Takano, M., et al. (2005). Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17, 3326–3336. doi: 10.1105/tpc.105.037028.1
Ivakov, A., Flis, A., Apelt, F., Scherer, U., Stitt, M., Kragler, F., et al. (2017). Cellulose synthesis and cell expansion are regulated by different mechanisms in growing Arabidopsis hypocotyls. Plant Cell 29, 1305–1315. doi: 10.1105/tpc.16.00782
Johansson, M., and Staiger, D. (2015). Time to flower: interplay between photoperiod and the circadian clock. JXB 66, 719–730. doi: 10.1093/jxb/eru441
Keech, O., Pesquet, E., Gutierrez, L., Ahad, A., Bellini, C., Smith, S. M., et al. (2010). Leaf senescence is accompanied by an early disruption of the microtubule network in arabidopsis. Plant Physiol. 154, 1710–1720. doi: 10.1104/pp.110.163402
Kiddle, S., Kim, Y., Penfold, C. A., Jenkins, D., Zhang, C., Morris, K., et al. (2011). High-resolution temporal profiling of transcripts during arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Physiol. 23, 873–894. doi: 10.1105/tpc.111.083345
Kim, J., Woo, H. R., and Nam, H. G. (2016). Toward systems understanding of leaf senescence: an integrated multi-omics perspective on leaf senescence research. Mol. Plant 9, 813–825. doi: 10.1016/j.molp.2016.04.017
Law, S. R., Chrobok, D., Juvany, M., Delhomme, N., Lindén, P., Brouwer, B., et al. (2018). Darkened leaves use different metabolic strategies for senescence and survival. Plant Physiol. 177, 132–150. doi: 10.1104/pp.18.00062
Lee, H., Park, Y., Ha, J., Baldwin, I. T., and Park, C. (2017). Multiple Routes of Light Signaling during Root Photomorphogenesis. Trends Plant Sci. 22, 803–812. doi: 10.1016/j.tplants.2017.06.009
Lee, H.-J., Ha, J.-H., Kim, S.-G., Choi, H.-K., Kim, Z. H., Han, Y.-J., et al. (2016). Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci. Signal. 9:ra106. doi: 10.1126/scisignal.aaf6530
Leivar, P., and Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cell 26, 56–78. doi: 10.1105/tpc.113.120857
Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., et al. (2008). Article multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18, 1815–1823. doi: 10.1016/j.cub.2008.10.058
Leivar, P., National, S., and Sentandreu, M. (2012). Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 71, 390–401. doi: 10.1111/j.1365-313X.2012.04992.x
Li, K., Yu, R., Fan, L. M., Wei, N., Chen, H., and Deng, X. W. (2016). DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7:11868. doi: 10.1038/ncomms11868
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Liebers, M., Grübler, B., Chevalier, F., Lerbs-Mache, S., Merendino, L., Blanvillain, R., et al. (2017). Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front. Plant Sci. 8:23. doi: 10.3389/fpls.2017.00023
Liebsch, D., and Keech, O. (2016). Dark-induced leaf senescence: new insights into a complex light-dependent regulatory pathway. New Phytol. 212, 563–570. doi: 10.1111/nph.14217
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf Senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Liu, L., Lin, N., Liu, X., Yang, S., Wang, W., and Wan, X. (2020). From chloroplast biogenesis to chlorophyll accumulation: the interplay of light and hormones on gene expression in Camellia sinensis cv. Shuchazao Leaves. Front. Plant Sci. 11, 256. doi: 10.3389/fpls.2020.00256
Liu, X., Li, Y., and Zhong, S. (2017). Interplay between light and plant hormones in the control of Arabidopsis seedling chlorophyll biosynthesis. Front. Plant Sci. 8:1433. doi: 10.3389/fpls.2017.01433
Lorrai, R., Boccaccini, A., Ruta, V., Possenti, M., Costantino, P., and Vittorioso, P. (2018). Abscisic acid inhibits hypocotyl elongation acting on gibberellins. DELLA proteins and auxin. AoB Plants 10:ply061. doi: 10.1093/aobpla/ply061
Ma, D., Li, X., Guo, Y., Chu, J., Fang, S., Yan, C., et al. (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. U.S.A. 113, 224–229. doi: 10.1073/pnas.1511437113
Ma, L., and Li, G. (2019). Auxin-dependent cell elongation during the shade avoidance response. Front. Plant Sci. 10:914. doi: 10.3389/fpls.2019.00914
Majda, M. (2018). The role of auxin in cell wall expansion. Int. J. Mol. Sci. 19:951. doi: 10.3390/ijms19040951
Martínez, C., Espinosa-Ruíz, A., Lucas, M., Bernardo-García, S., Franco-Zorrilla, J. M., and Prat, S. (2018). PIF 4-induced BR synthesis is critical to diurnal and thermomorphogenic growth. EMBO J. 37:e99552. doi: 10.15252/embj.201899552
Martínez-García, J. F., Gallemí, M., Molina-Contreras, M. J., Llorente, B., Bevilaqua, M. R. R., and Quail, P. H. (2014). The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLoS One 9:e109275. doi: 10.1371/journal.pone.0109275
Mazzella, M. A., Casal, J. J., Jorge, P., and Fox, A. R. (2014). Hormonal networks involved in apical hook development in darkness and their response to light. Front. Plant Sci. 5:52. doi: 10.3389/fpls.2014.00052
Mishra, S., and Khurana, J. P. (2017). Emerging roles and new paradigms in signaling mechanisms of plant cryptochromes. CRC Crit. Rev. Plant Sci. 36, 89–115. doi: 10.1080/07352689.2017.1348725
Müller-Moulé, P., Nozue, K., Pytlak, M. L., Palmer, C. M., Covington, M. F., Wallace, A. D., et al. (2016). YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 2016:e2574. doi: 10.7717/peerj.2574
Niwa, Y., Yamashino, T., and Mizuno, T. (2009). The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 1, 838–854. doi: 10.1093/pcp/pcp028
Nozue, K., Devisetty, U. K., Lekkala, S., Mueller-Moulé, P., Bak, A., Casteel, C. L., et al. (2018). Network analysis reveals a role for salicylic acid pathway components in shade avoidance. Plant Physiol. 178, 1720–1732. doi: 10.1104/pp.18.00920
Orden, V., Wolf, S., Vissenberg, K., Delacourt, J., Assoumou, Y., and Pelletier, S. (2010). A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark- grown Arabidopsis hypocotyls. New Phytol. 188, 726–739. doi: 10.1111/j.1469-8137.2010.03409.x
Paik, I., Kathare, P. K., Kim, J., and Huq, E. (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035–1046. doi: 10.1016/j.molp.2017.07.002
Pearce, S., Shaw, L. M., Lin, H., Cotter, J. D., Li, C., and Dubcovsky, J. (2017). Night-break experiments shed light on the photoperiod1-mediated flowering. Plant Physiol. 174, 1139–1150. doi: 10.1104/pp.17.00361
Pfeiffer, A., Shi, H., Tepperman, J. M., Zhang, Y., and Quail, P. H. (2014). Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis. Mol. Plant 7, 1598–1618. doi: 10.1093/mp/ssu087
Pham, V. N., Kathare, P. K., and Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038. doi: 10.1104/pp.17.01384
Podolec, R., and Ulm, R. (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45, 18–25. doi: 10.1016/j.pbi.2018.04.018
Pogson, B. J., Ganguly, D., and Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta Bioenerg. 1847, 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Pokhilko, A., Ramos, J. A., Holtan, H., Maszle, D. R., Khanna, R., and Millar, A. J. (2011). Ubiquitin ligase switch in plant photomorphogenesis: a hypothesis. J. Theor. Biol. 270, 31–41. doi: 10.1016/j.jtbi.2010.11.021
Qi, L., Liu, S., Li, C., Fu, J., Jing, Y., Cheng, J., et al. (2020). PHYTOCHROME-INTERACTING FACTORS Interact with the ABA Receptors PYL8 and PYL9 to Orchestrate ABA Signaling in Darkness. Mol. Plant 13, 414–430. doi: 10.1016/j.molp.2020.02.001
Qiu, K., Li, Z., Yang, Z., Chen, J., Wu, S., Zhu, X., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 11:e1005399. doi: 10.1371/journal.pgen.1005399
Reed, J. W., Wu, M., Reeves, P. H., Hodgens, C., Yadav, V., Hayes, S., et al. (2018). Three auxin response factors promote hypocotyl elongation. Plant Physiol. 178, 864–875. doi: 10.1104/pp.18.00718
Roig-Villanova, I., and Martínez-García, J. F. (2016). Plant responses to vegetation proximity: a whole life avoiding shade. Front. Plant Sci. 7:236. doi: 10.3389/fpls.2016.00236
Rougemont, J., Ljung, K., Lo, I., Solano, R., Trevisan, M., Pradervand, S., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 1, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Sakuraba, Y., Jeong, J., Kang, M., Kim, J., Paek, N., and Choi, G. (2014). PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5:4636. doi: 10.1038/ncomms5636
Salisbury, F. J., Hall, A., Grierson, C. S., Halliday, K. J., Buildings, K., Road, M., et al. (2007). Phytochrome coordinates Arabidopsis shoot and root development. Plant J. 50, 429–438. doi: 10.1111/j.1365-313X.2007.03059.x
Salome, P. A., Xie, Q., and McClung, C. R. (2008). Circadian timekeeping during early Arabidopsis development. Plant Physiol. 147, 1110–1125. doi: 10.1104/pp.108.117622
Sampathkumar, A., and Eng, R. C. (2020). Plant biology: bending of plant organs. Curr. Biol. 30, R402–R405. doi: 10.1016/j.cub.2020.03.010
Sassi, M., Lu, Y., Zhang, Y., Wang, J., Dhonukshe, P., Blilou, I., et al. (2012). COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 3412, 3402–3412. doi: 10.1242/dev.078212
Seluzicki, A., Burko, Y., and Chory, J. (2017). Dancing in the dark: darkness as a signal in plants. Plant Cell Environ. 40, 2487–2501. doi: 10.1111/pce.12900
Sessa, G., Carabelli, M., Possenti, M., Morelli, G., and Ruberti, I. (2018). Multiple pathways in the control of the shade avoidance response. Plants 7:102. doi: 10.3390/plants7040102
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Sibout, R., Sukumar, P., Hettiarachchi, C., Holm, M., Muday, G. K., and Hardtke, C. S. (2006). Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2:e202. doi: 10.1371/journal.pgen.0020202
Song, Y., Yang, C., Gao, S., Zhang, W., Li, L., and Kuai, B. (2014). Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 7, 1776–1787. doi: 10.1093/mp/ssu109
Soy, J., Leivar, P., González-schain, N., Martín, G., Diaz, C., and Sentandreu, M. (2016). Molecular convergence of clock and photosensory pathways through PIF3 – TOC1 interaction and co-occupancy of target promoters. Proc. Natl. Acad. Sci. U.S.A. 113, 4870–4875. doi: 10.1073/pnas.1603745113
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D. Y., Doležal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Stepanova, A. N., Yun, J., Robles, L. M., Novak, O., He, W., Guo, H., et al. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973. doi: 10.1105/tpc.111.088047
Stephenson, P. G., Fankhauser, C., and Terry, M. J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 106, 7654–7659. doi: 10.1073/pnas.0811684106
Toledo-Ortiz, G., Johansson, H., Lee, K. P., Bou-Torrent, J., Stewart, K., Steel, G., et al. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10:e1004416. doi: 10.1371/journal.pgen.1004416
Vandenbussche, F., Petrášek, J., Žádníková, P., Hoyerová, K., Pešek, B., Raz, V., et al. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137, 597–606. doi: 10.1242/dev.040790
Wabnik, K., Abuzeineh, A., Gallemi, M., Van Der Straeten, D., Zádníková, P., Smith, R. S., et al. (2016). A model of differential growth-guided apical hook formation in plants. Plant Cell 28, 2464–2477. doi: 10.1105/tpc.15.00569
Wang, S., Zhang, S., Sun, C., Xu, Y., Chen, Y., Yu, C., et al. (2014). Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 201, 91–103. doi: 10.1111/nph.12499
Wang, Y., and Guo, H. (2018). Tansley insight on hormonal regulation of the dynamic apical hook development. New Phytol. 222, 1230–1234. doi: 10.1111/nph.15626
Wang, Z., Bai, M., and Oh, E. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Willige, B. C., Ogiso-tanaka, E., Zourelidou, M., and Schwechheimer, C. (2012). WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 139, 4020–4028. doi: 10.1242/dev.081240
Wu, G., Cameron, J. N., Ljung, K., and Spalding, E. P. (2010). A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 62, 179–191. doi: 10.1111/j.1365-313X.2010.04137.x
Yang, C., and Li, L. (2017). Hormonal regulation in shade avoidance. Front. Plant Sci. 8:1527. doi: 10.3389/fpls.2017.01527
Yu, Y., and Huang, R. (2017). Integration of ethylene and light signaling affects hypocotyl growth in Arabidopsis. Front. Plant Sci. 8:57. doi: 10.3389/fpls.2017.00057
Žádníkova, P., Petrášek, J., Marhavý, P., Raz, V., Vandenbussche, F., Ding, Z., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137, 607–617. doi: 10.1242/dev.041277
Žádníkova, P., Wabnik, K., Abuzeineh, A., Gallemi, M., Van Der Straeten, D., Smith, R. S., et al. (2016). A model of differential growth-guided apical hook formation in plants. Plant Cell 28, 2464–2477. doi: 10.1105/tpc.15.00569
Zhang, X., Ji, Y., Xue, C., Ma, H., Xi, Y., Huang, P., et al. (2018). Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 30, 1971–1988. doi: 10.1105/tpc.18.00018
Zhang, Y., Li, C., Zhang, J., Wang, J., Yang, J., Lv, Y., et al. (2017). Dissection of HY5/HYH expression in Arabidopsis reveals a root-autonomous HY5-mediated photomorphogenic pathway. PLoS One 12:e0180449. doi: 10.1371/journal.pone.0180449
Zhang, Y., Liu, Z., Chen, Y., He, J., and Bi, Y. (2015). Plant Science PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 237, 57–68. doi: 10.1016/j.plantsci.2015.05.010
Zhong, S., Shi, H., Xue, C., Wang, L., Xi, Y., Li, J., et al. (2012). A molecular framework of light-controlled phytohormone action in arabidopsis. Curr. Biol. 22, 1530–1535. doi: 10.1016/j.cub.2012.06.039
Zhong, S., Shi, H., Xue, C., Wei, N., Guo, H., and Deng, X. W. (2014). Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. U.S.A. 111, 3913–3920. doi: 10.1073/pnas.1402491111
Keywords: dark, signaling, hormone, growth, development
Citation: Deepika, Ankit, Sagar S and Singh A (2020) Dark-Induced Hormonal Regulation of Plant Growth and Development. Front. Plant Sci. 11:581666. doi: 10.3389/fpls.2020.581666
Received: 09 July 2020; Accepted: 16 September 2020;
Published: 07 October 2020.
Edited by:
M. Iqbal R. Khan, Jamia Hamdard University, IndiaReviewed by:
Sushma Mishra, Dayalbagh Educational Institute, IndiaJigang Li, China Agricultural University, China
Copyright © 2020 Deepika, Ankit, Sagar and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amarjeet Singh, YW1hcmplZXQuc2luZ2hAbmlwZ3IuYWMuaW4=
 Deepika
Deepika Ankit
Ankit Amarjeet Singh
Amarjeet Singh