- 1Graduate School of Bioresources, Mie University, Tsu, Japan
- 2Department of Plant and Soil Science, Texas Tech University, TX, United States
- 3Graduate School of Life Sciences, Tohoku University, Sendai, Japan
- 4Life Science Research Center, Mie University, Tsu, Japan
- 5School of Agriculture, Meiji University, Kawasaki, Japan
- 6Graduate School of Science, The University of Tokyo, Tokyo, Japan
- 7Department of Evolutionary Biology and Environmental Studies, University of Zurich, Zurich, Switzerland
- 8Kihara Institute for Biological Studies, Yokohama City University, Yokohama, Japan
- 9Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 10Division of Natural Science, Osaka Kyoiku University, Kashiwara, Japan
Self-compatibility in Arabidopsis thaliana represents the relatively recent disruption of ancestral obligate cross pollination, recognized as one of the prevalent evolutionary pathways in flowering plants, as noted by Darwin. Our previous study found that inversion of the male specificity gene (SP11/SCR) disrupted self-incompatibility, which was restored by overexpressing the SCR with the reversed inversion. However, SCR in A. thaliana has other mutations aside from the pivotal inversion, in both promoter and coding regions, with probable effects on transcriptional regulation. To examine the functional consequences of these mutations, we conducted reciprocal introductions of native promoters and downstream sequences from orthologous loci of self-compatible A. thaliana and self-incompatible A. halleri. Use of this inter-species pair enabled us to expand the scope of the analysis to transcriptional regulation and deletion in the intron, in addition to inversion in the native genomic background. Initial analysis revealed that A. thaliana has a significantly lower basal expression level of SCR transcripts in the critical reproductive stage compared to that of A. halleri, suggesting that the promoter was attenuated in inducing transcription in A. thaliana. However, in reciprocal transgenic experiments, this A. thaliana promoter was able to restore partial function if coupled with the functional A. halleri coding sequence, despite extensive alterations due to the self-compatible mode of reproduction in A. thaliana. This represents a synergistic effect of the promoter and the inversion resulting in fixation of self-compatibility, primarily enforced by disruption of SCR. Our findings elucidate the functional and evolutionary context of the historical transition in A. thaliana thus contributing to the understanding of the molecular events leading to development of self-compatibility.
Introduction
While a variety of mechanisms to promote outcrossing for diversification have evolved in angiosperms, selfing may nevertheless be favored in terms of reproductive assurance and inherent genetic transmission when these advantages outweigh a reduction in fitness (Darwin, 1876; Charlesworth and Vekemans, 2005; Franklin-Tong, 2008). In this evolutionary process, the switch from outcrossing to selfing mode is associated with the loss of self-incompatibility (SI) in many plant lineages (Stebbins, 1974; Goodwillie et al., 2005). SI is defined as the inability of a fertile hermaphrodite plant to produce zygotes after self-pollination, consequently promoting outcrossing, and diverse SI systems have evolved in angiosperms (de Nettancourt, 2001; Watanabe et al., 2012). The molecular basis of SI has been extensively studied in the family Brassicaceae, and it has been found to be controlled by a single highly polymorphic locus, the S-locus (Bateman, 1955; Watanabe et al., 2012), which comprises the male and female SI specificity genes SP11 (S-locus protein 11)/SCR (S-locus cysteine rich protein) and SRK (S-locus receptor kinase), respectively (Suzuki et al., 1999; Schopfer et al., 1999; Takayama et al., 2000; Takasaki et al., 2000; Shiba et al., 2001). SP11/SCR is a small cysteine-rich protein localized in the pollen coat that acts as a ligand, and SRK is a transmembrane serine/threonine receptor kinase on the stigma epidermis that functions as a receptor for SP11/SCR. Their physical interaction, in an allele-specific manner, initiates the subsequent incompatible reaction to reject self pollen (Takayama et al., 2001), through an as yet inadequately understood downstream system.
The model plant Arabidopsis thaliana is a self-compatible, predominantly selfing species of Brassicaceae, whereas its ancestral state is thought to have been an obligate outcrosser enforced by SI (Kusaba et al., 2001; Bechsgaard et al., 2006; Tang et al., 2007; Shimizu et al., 2008) through haplotype-specific interaction of SCR and SRK. Functional orthologues of SCR and SRK are found in A. lyrata and A. halleri, SI congeners, as shown by the successful recovery of SI in A. thaliana in heterologous experiments with introduction of the orthologues (Nasrallah et al., 2002; Nasrallah et al., 2004; Tsuchimatsu et al., 2010; Fujii et al., 2020). Although such experiments provided the insight that A. thaliana genomic and physiological background is compatible with the orthologues in recovering SI, heterologous experiments might not have captured the functional context of nucleotide variations in SCR and/or SRK. For example, A. lyrata SCR and SRK were of different haplotype origin (Nasrallah et al., 2002; Nasrallah et al., 2004) and the transgenic SCR construct was driven by the non-native anther-specific promoter (Tsuchimatsu et al., 2010), which might not align with the attempt to reverse the evolutionary course that occurred in this locus in A. thaliana. Indeed, even within A. thaliana the evolution of SI has been diverse with different paths leading to disruption of SCR and/or SRK (Kusaba et al., 2001; Sherman-Broyles et al., 2007; Shimizu et al., 2008; Tsuchimatsu et al., 2017; Fujii et al., 2020).
In this study our aim is to understand the functional and evolutionary contexts of the loss of SI in A. thaliana by segregating the primary and secondary mutations. For this we compared functional and disrupted SCRs between A. halleri and A. thaliana, including their transcriptional regulation, from the same haplotype origin and elucidated the historical transition in A. thaliana.
Materials and Methods
Plant Material
A. thaliana ecotype Oldenburg (Old-1) was obtained from the Arabidopsis Biological Resource Center (ABRC; http://abrc.osu.edu/). Old-1 plants were grown in a growth chamber (BIOTRON LH-240S, NK system, Osaka, Japan) at 22°C under 8 h light/16 h dark photoperiod for the vegetative stage and 16 h light/8 h dark photoperiod for the reproductive stage. A. halleri Tada-mine accession (W302) was originally collected in Japan and developed by self-fertilization for five generations. W302 plants were grown in a growth chamber at 22°C under 16 h light/8 h dark photoperiod.
Pollination Assay
Flower buds of Old-1 and W302 were emasculated at developmental stage 12 (Smyth et al., 1990) and incubated at 22°C on 1% agar medium until appropriate developmental stages. Each emasculated pistil at developmental stages 12, 13, 14, and 15 of W302 was self-pollinated. In Old-1, each emasculated pistil at developmental stages 12, 13, 14, and 15 was pollinated with pollen of W302 to test the female SI function of Old-1. After fixation and staining with aniline blue solution, pollen tubes were observed using fluorescence microscopy (Axio Imager A2, Carl Zeiss, Jena, Germany) according to Nou et al. (1991) and Watanabe et al. (1992).
Determination of Full Structure of SCR-A
Genomic DNA and mRNA were extracted from Old-1 and W302 using Plant DNeasy and RNeasy kits (Qiagen, Hilden, Germany). First-strand cDNAs of anther and stigma were synthesized using a High Capacity RNA-to-cDNA Kit (Thermo Fisher Scientific, Waltham, MA, USA). The promoter and gene regions of SCR-A of Old-1 and W302 were amplified by genomic PCR (all primer information is summarized in Supplementary Table S1), subcloned into pCR2.1-TOPO vector (Thermo Fisher Scientific) and sequenced by an Applied Biosystems 3130 Genetic Analyzer (Thermo Fisher Scientific) according to Watanabe et al. (1994). The 5′- and 3′-UTRs of SCR-A were determined by First Choice RLM-RACE kit (Thermo Fisher Scientific), using anther cDNA of W302 based on Kakita et al. (2007).
Quantification of mRNA Expression of SCR-A and SRK-A
Expression levels of SCR-A and SRK-A in A. thaliana and A. halleri were quantified by qRT-PCR, using anther and stigma cDNAs of Old-1 and W302, respectively, by Step One Realtime PCR System (Thermo Fisher Scientific) according to Osaka et al. (2013). 18S rRNA was used as an internal control.
Transformation of A. thaliana
A 3,523-bp AhSCR-A fragment, consisting of 1,845-bp AhSCR promoter and 1,678-bp AhSCR gene, was amplified by genomic PCR and subcloned into pBI121 vector. Artificial chimeric SCR-A constructs, with one of the three major mutations of AtSCR, promoter fragments and former/latter parts of SCR-A coding region, were amplified from Old-1 and W302 by PCR with specific primers (Supplementary Table S1), respectively, connected and subcloned into pBI121 vector. Each vector was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Old-1 plants by the floral dip method according to Park et al. (2010).
Results and Discussion
Genetic and Functional Characteristics of SCR in A. thaliana and A. halleri
Thus far, the remnants of three haplotypes (haplogroups) at the S-locus have been identified in A. thaliana: haplogroup A is distributed predominantly in the northern hemisphere, haplogroup B is restricted to African islands, and haplogroup C is distributed mainly in Asia (Sherman-Broyles et al., 2007; Tang et al., 2007; Shimizu et al., 2008). In haplogroup A, disruption patterns in the S-locus are further classified into five variants, termed A-t1 to A-t5. While A-t2 and A-t3 groups have deleted the corresponding SCR genomic region, A-t1 and A-t5 still possess the SCR pseudogene (also known as ΨSCR1) together with the SRK pseudogene and A-t4 has the SCR pseudogene with a functional SRK, reflecting the complex evolutionary history of selfing in A. thaliana.
Previously we reported the pivotal role of the 213-bp inversion in the coding region of A. thaliana SCR, in which SI can be restored by reverting the inversion (Tsuchimatsu et al., 2010). In order to segregate the functional context of the numerous mutation that may be responsible for disruption (i.e. primary mutation) or not directly related (i.e. secondary mutation) in both coding and upstream regulatory regions, we used a comparative panel of self-compatible A. thaliana Oldenburg accession (Old-1) and outcrosser A. halleri Tada-mine accession (W302). This panel enables the comparative functional dissection of the S-locus in Old-1 and W302 because they have the identical S-haplotype, haplogroup A, trans-specifically, and Old-1 still retains the female SI function with an intact SRK-A coding region while the male SI function is disrupted (Tsuchimatsu et al., 2010). These two lines also have an identical flower developmental process and pollination behavior (Figure 1). The structure of SCR-A in W302 consists of two exons (67-bp exon 1 and 188-bp exon 2) separated by an intron of 1157 bp with 52-bp 5′ and 214-bp 3′ UTRs, and a total length of 1,678 bp (Figure 2A and Supplementary Figure S1). In comparison to the SCR-A of W302, as we reported previously (Tsuchimatsu et al., 2010), the nonfunctional form of SCR-A in Old-1 has variations including a 252-bp deletion in the intron, minor indels (≤36 bp) in the exons and a 213-bp inversion in exon 2 (Figure 2A and Supplementary Figure S1). Compared to the relatively well conserved SCR-A coding sequences, the upstream element has extensive alterations; there is no overall homology between upstream sequences of Old-1 and W302, except for regions -1 to -210 (Figure 2A and Supplementary Figure S1). Thus, to summarize, there are three major differences in SCR-A between Old-1 and W302: limited homology in the promoter, a 252-bp deletion in the intron and a 213-bp inversion in exon 2.
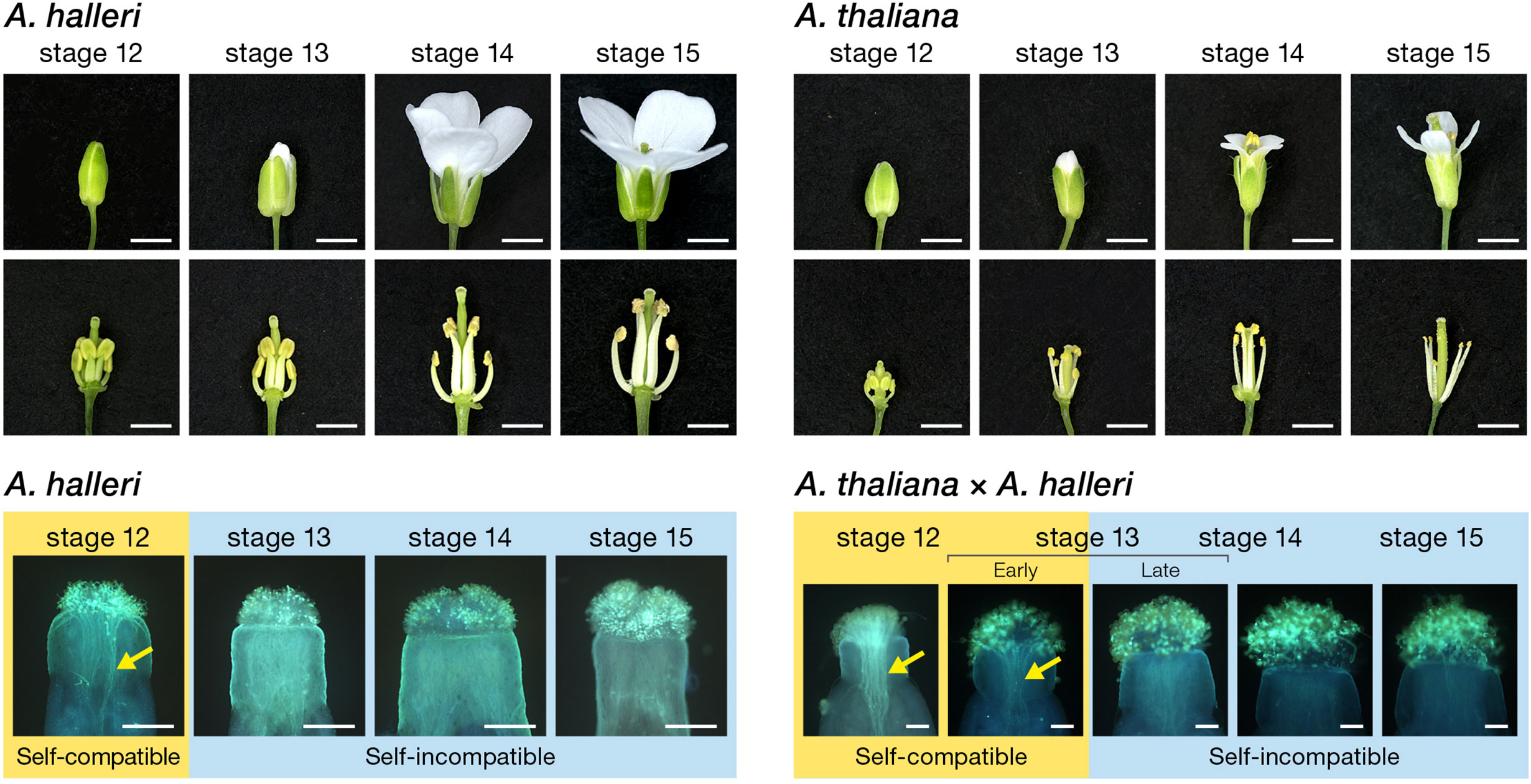
Figure 1 Floral morphology and pollination behaviour during flower development in A. thaliana and A. halleri. Appearance and inner morphological structure of flower buds, with floral developmental stages (Smyth et al., 1990). Flower buds of Old-1 and W302 were emasculated at developmental stage 12 and incubated at 22°C on 1% agar medium until appropriate developmental stages. Each emasculated pistil at developmental stages 12, 13, 14, and 15 was self-pollinated in W302 and cross-pollinated with pollen of W302 in Old-1 to test the female SI function of Old-1. Scale bar, 1 mm. Self-incompatible and self-compatible floral stages are highlighted with blue and yellow boarders, respectively, in self-pollinated pistils of A. halleri and A. thaliana pistils pollinated with A. halleri pollens. Arrows indicate growing pollen tubes. Scale bar, 0.1 mm.
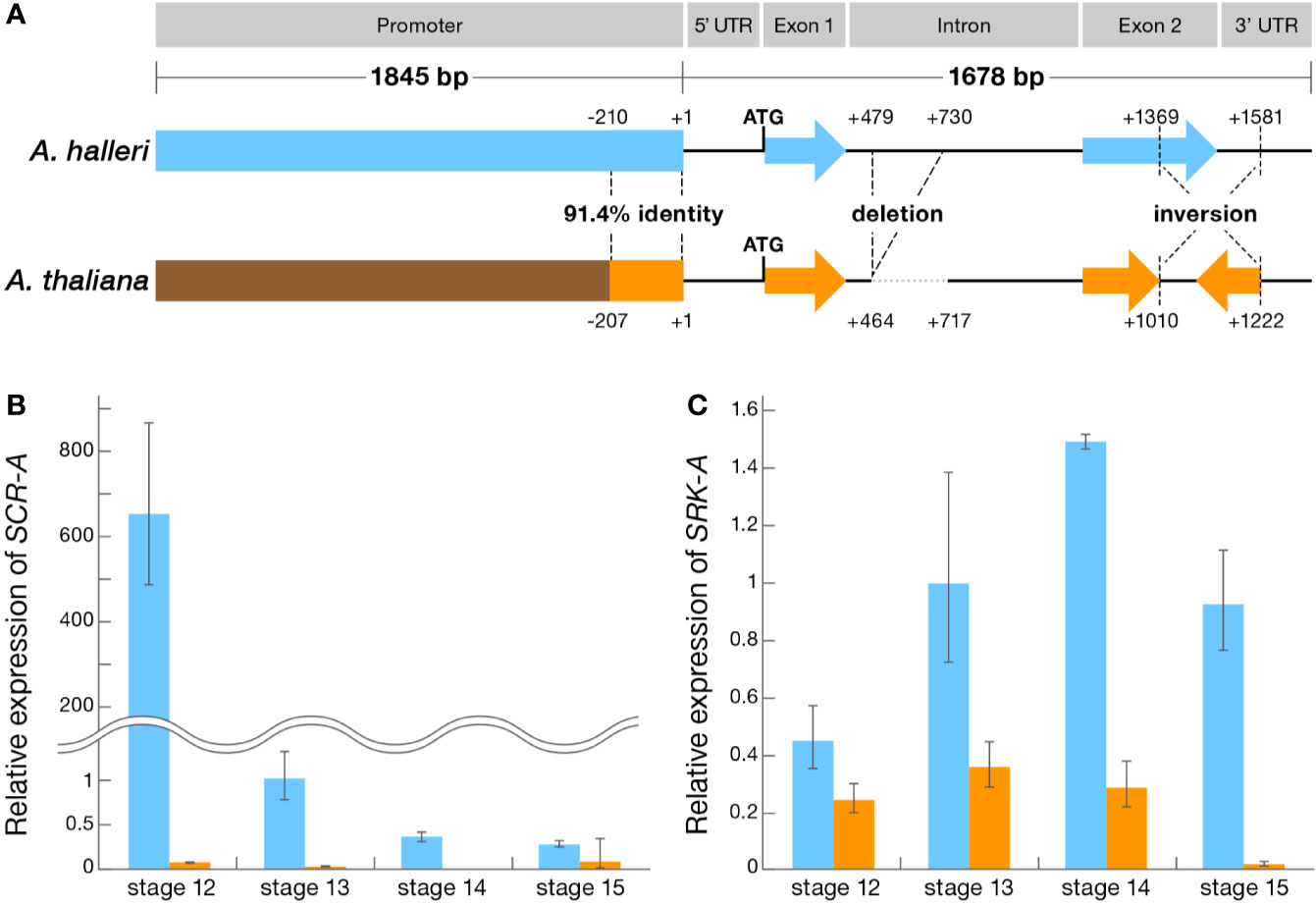
Figure 2 Genomic structure and expression profile of the SI genes in A. thaliana and A. halleri. (A) Schematic structure of SCR-A of A. thaliana and A. halleri. (B) Expression profile of SCR-A at flower developmental stages 12-15 in A. thaliana Old-1 (orange) and A. halleri W302 (blue). (C) Expression profile of SRK-A at flower developmental stages 12–15 in A. thaliana Old-1 and A. halleri W302. (B, C) Relative gene expression was determined by qRT-PCR and expression levels of transcripts at each stage are shown relative to A. halleri W302 at stage 13 (set to value of 1). Data from 4 biological replicates are shown. Error bars indicate ± SD.
To address the consequences of extensive alteration in upstream sequences, we compared expression of SCR-A between Old-1 and W302 in anthers during flower development (Smyth et al., 1990). Real-time quantitative expression analysis revealed a significantly lower level of SCR-A expression in Old-1 compared to W302 (Figure 2B). The overall temporal patterns were similar in both Old-1 and W302, with peak expression observed prior to anthesis (i.e. stage 12) followed by a drastic reduction in later stages (i.e. stages 13–15). This indicates that the native SCR-A promoter in Old-1 seems to be attenuated in achieving the “optimal” expression level as seen in W302. From analysis of expression of the female-specificity gene SRK-A, again a lower expression in Old-1 stigmas was observed at stage 12 (Figure 2C). However, the degree of reduction was only about 10 times for SRK-A, unlike SCR-A which showed an ~6500 times reduction. Taken together, these results indicate that the S-locus transcriptional level in Old-1 was strongly silenced, especially in SCR-A and to a lesser extent in SRK-A, which may constitute a critical step in the loss of SI in A. thaliana.
Complete Experimental Reversal of the Historical Transition From Outcrossing to Selfing in A. thaliana
Given that our representative lines of A. halleri (functional SI) and A. thaliana (disrupted SI) have an identical S-haplotype with three major mutations in the SCR-A of A. thaliana Old-1 (AtSCR), namely a promoter with limited homology, a 252-bp deletion in the intron and a 213-bp inversion in exon 2, we regarded SCR-A of A. halleri W302 (AhSCR) as the functional, original form of AtSCR. The 3,523-bp AhSCR-A fragment (designated proAhSCR::AhSCR), consisting of a 1,845-bp AhSCR promoter and 1,678-bp AhSCR coding region, was introduced into Old-1. Expression of SCR-A in transgenic proAhSCR::AhSCR Old-1 plants was similar to that in W302, and transgenic Old-1 plants rejected self pollen on their stigmas, resulting in siliques with no or very few seeds (Figures 3A–F). In T2, T3, and T6 plants with proAhSCR::AhSCR, which were produced through bud self-pollination, developmentally stable SI was observed during autopollination at stages 12 to 15 (Figure 3G), while previously we observed that the SI reaction on the female side was attenuated at a relatively late stage (Tsuchimatsu et al., 2010). This indicates that the SI trait was restored in these transgenic Old-1 plants and this was heritable. Together with our previous results (Tsuchimatsu et al., 2010), this demonstrates a complete experimental reversal of the historical transition from outcrossing to selfing in A. thaliana, and is basically consistent with the heterologous SI in A. thaliana (Nasrallah et al., 2002; Nasrallah et al., 2004). This finding also indicates that inactivation of the S-locus was a primary step in the evolution of selfing in A. thaliana (Nasrallah et al., 2002; Sherman-Broyles et al., 2007; Boggs et al., 2009).
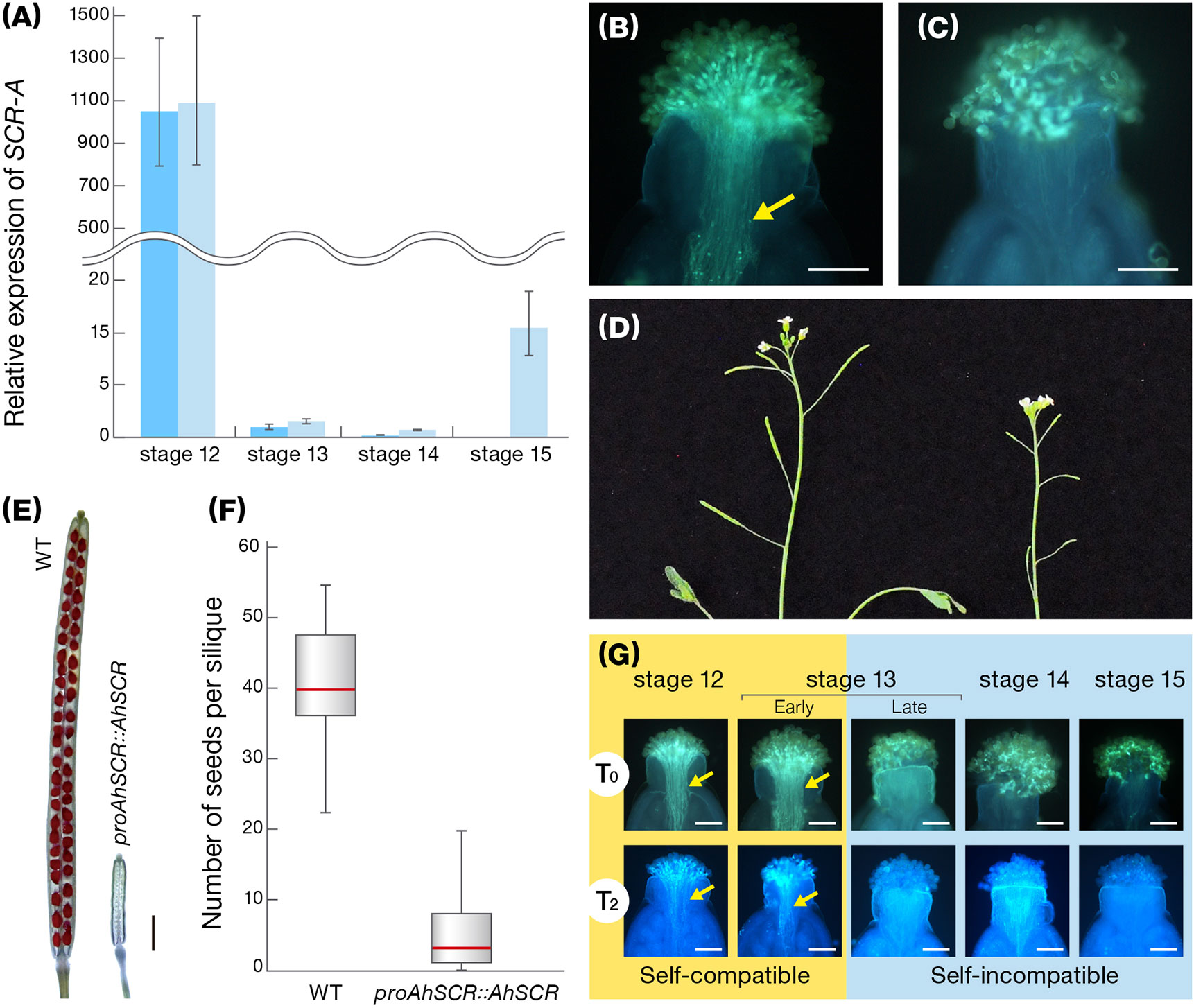
Figure 3 Characteristics of transgenic A. thaliana with a functional SCR-A. (A) Expression of SCR-A at flower developmental stages 12–15 in proAhSCR::AhSCR transgenic A. thaliana plants (light blue) and A. halleri W302 plants (blue). Relative gene expression was determined by qRT-PCR and expression levels of transcripts at each stage are shown relative to A. halleri W302 at stage 13 (set to value of 1). Data from 4 biological replicates are shown. Error bars indicate ± SD. (B) Pollen tubes accepted in the stigma of a selfed pistil of wild-type A. thaliana Old-1. Arrow indicates growing pollen tubes. Scale bar, 0.1 mm. (C) Pollen tubes inhibited on the stigma of a selfed pistil of proAhSCR::AhSCR transgenic A. thaliana plant. Scale bar, 0.1 mm. (D) Inflorescences of proAhSCR::AhSCR transgenic A. thaliana plant (right) and wild-type A. thaliana Old-1 (left). (E) Silique resulting from selfing of proAhSCR::AhSCR transgenic A. thaliana (right) and wild-type A. thaliana Old-1 (left). Scale bar, 1 mm. (F) Comparison of number of seeds per silique resulting from selfing of the proAhSCR::AhSCR transgenic A. thaliana plant and wild-type A. thaliana Old-1. Thirty siliques were examined from each plant. Red bars, gray boxes, and black whiskers represent the median, the interquartile range, and 1.5 times extension of the interquartile range, respectively. (G) Pollination phenotype in T0 and progeny plants (T2 shown as a representative of T2, T3, and T6 plants) of proAhSCR::AhSCR transgenic A. thaliana, with flower development stage. Arrows indicate growing pollen tubes. Scale bar, 0.1 mm.
Discrimination of the Primary Inactivating Mutation and Decays for the Evolutionary Loss of SI in A. thaliana
To examine the functional consequences of the three SCR-A mutations in A. thaliana Old-1 (that is promoter alteration, deletion in the intron and inversion in exon 2), we conducted transgenic experiments in which chimeric constructs were introduced into Old-1 (Figure 4). As explained above, introduction of a W302-type SCR-A promoter and coding region (proAhSCR::AhSCR) achieved complete recovery of SI in all flowers as expected. However, to our surprise, the Old-1 promoter with W302 SCR-A coding region (designated proAtSCR::AhSCR) showed SI in 53 out of 76 flowers (Figures 4A, C), indicating the SCR-A promoter of Old-1 is partially functional in inducing transcription of SCR-A. The W302 promoter and coding sequences combined with the Old-1 deletion in the intron (designated proAhSCR::AhSCR Δintron) conferred SI in all flowers. The SI reaction was strongly prohibited by inversion of exon 2 regardless of the W302 promoter and exon 1 and intron (designated proAhSCR::AhSCR inv. exon2). This is consistent with the role of the inversion in exon 2 as the primary and strongest determining factor in the evolution of SI reaction, whereas the other two mutations of the deleted intron and attenuated expression are the result of decay. Hence, we conclude that the primary inactivating mutation for the evolutionary loss of SI in A. thaliana is the exon 2-disrupting inversion in SCR-A, which was a primary and definitive evolutionary event for the transition from outcrossing to selfing in A. thaliana.
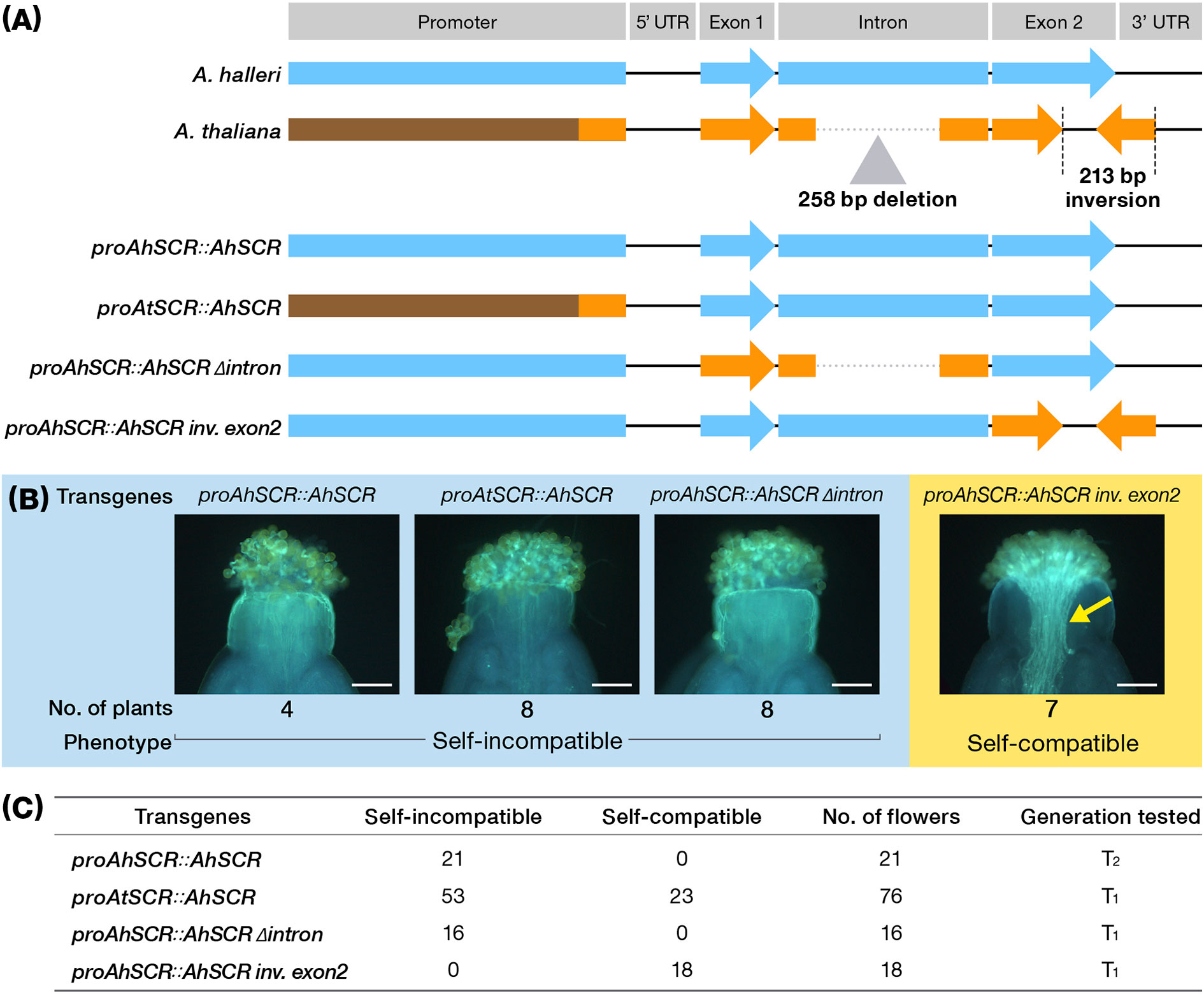
Figure 4 Schematic structure of artificial chimeric SCR-A and their effect on pollination in transgenic Arabidopsis plants. (A) Schematic structure of chimeric SCR-A. (B) Pollen tube behaviour on selfed pistils in each transgenic plant. Arrow indicates growing pollen tubes in the pistil. Scale bar, 0.1 mm. (C) Number of self-incompatible/self-compatible flowers in T1 or T2 generation of each transgenic plant.
Functional and Evolutionary Context of Transitional History in A. thaliana
The three major mutations in SCR-A are nearly fixed in a range of European A. thaliana accessions (Supplementary Figure S2), suggesting that these mutations in SCR-A had already occurred before differentiation of these accessions. Coupled with the prediction of a relatively recent origin of self-compatibility in A. thaliana (Nasrallah et al., 2002; Bechsgaard et al., 2006; Shimizu et al., 2008; Shimizu and Tsuchimatsu, 2015), it is conceivable that the ancestor of A. thaliana still retained SI after the split from the common ancestor of the sister species A. halleri and A. lyrata (Koch et al., 2000), in which all SI components were under selective constraint for the maintenance of SI. Interestingly, proAtSCR::AhSCR transgenic plants showed weakening of SI in some flowers, indicating the expression level of AhSCR driven by proAtSCR was on the threshold of the level required for an incompatibility reaction, resulting in partial SI (Shimizu and Tsuchimatsu, 2015) in proAtSCR::AhSCR transgenic plants. The partial recovery of SI could be attributed to two scenarios for the evolutionary loss of SI in A. thaliana (Figure 5): 1) a two-step scenario in which A. thaliana first evolved partial SI by depression of proAtSCR and then complete SC by inversion in SCR-A exon 2 (Castric et al., 2014; Shimizu and Tsuchimatsu, 2015), and 2) a direct scenario in which A. thaliana lost SI by inversion in SCR-A exon 2 together with a depression of proAtSCR as a secondary decay (Tsuchimatsu et al., 2010). Due to limitations in the methodology of calculating molecular clocks for large structural alterations compared to small mutations, it is not possible to reliably determine whether depression of proAtSCR proceeded the inversion or not. However, in either scenario, SCR-A provides the molecular basis of transition from outcrossing to selfing, leading to accumulation of mutations and decays following release from selective constraints.
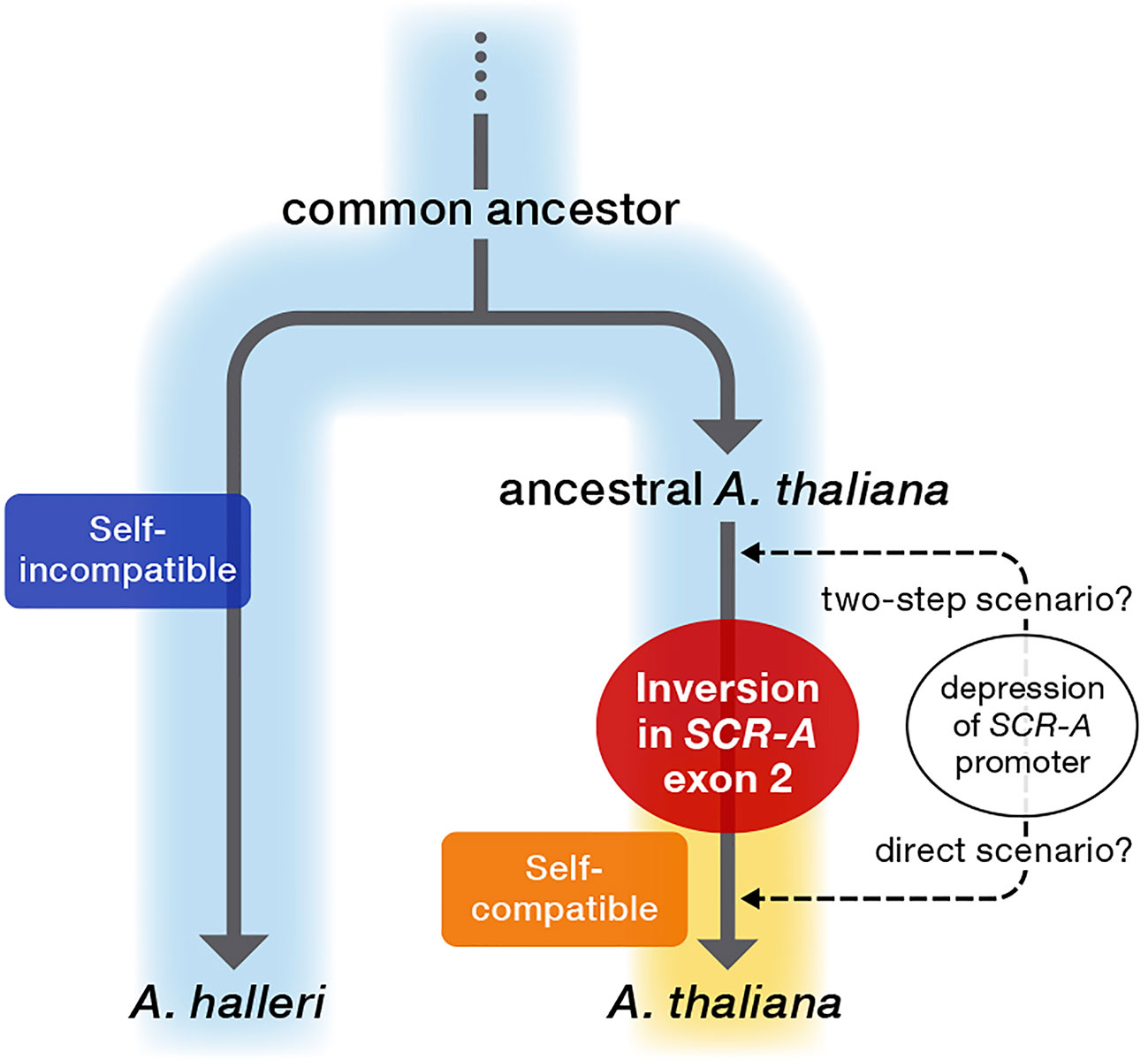
Figure 5 Proposed model of the evolutionary process of loss of self-incompatibility in genus Arabidopsis. After an evolutionary split from the common ancestor, ancestral A. thaliana still retained SI, as in the sister SI species A. halleri. The inactivating mutation resulting in the evolutionary loss of SI in A. thaliana occurred as an exon 2-disrupting inversion in SCR-A, which was a primary and definitive evolutionary event for the transition from outcrossing to selfing in A. thaliana. In addition, the SCR-A promoter was attenuated in this process, and thus two scenarios can be proposed for the evolutionary loss of SI in A. thaliana: 1) a two-step scenario in which A. thaliana first evolved partial SI by depression of proAtSCR and then complete SC by the inversion in SCR-A exon 2, and 2) a direct scenario in which A. thaliana lost SI by the inversion in SCR-A exon 2 together with a depression of proAtSCR as a secondary decay.
Conclusion
Our work has established the functional and evolutionary contexts of the major mutations in promoters, exons and introns of SCR and has suggested a transcriptional contribution to achieve the complete switch from outcrossing to selfing in A. thaliana. It is notable that the functionality of the pseudogene promoter has been retained. The findings from analysis of the fixed mutations in SCR-A further support the hypotheses of a recent origin and selection of selfing in A. thaliana (Bechsgaard et al., 2006; Shimizu et al., 2008) and the advantage of mutation in the male specificity gene (Tsuchimatsu et al., 2010), rather than the female specificity gene, for the evolution of self-compatibility (Uyenoyama et al., 2001; Busch and Schoen, 2008; Tsuchimatsu and Shimizu, 2013).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
KS, TT, KKS, ST, GS, and MW planned and designed the study, and wrote the paper. KN, EAW, CH, TO, MT, HM-S, and YK performed sequencing, expression profiling, crossing experiment and transgenic analysis. AK conducted nucleotide sequence analysis, and wrote the paper. KY conducted nucleotide sequence analysis. KS, KN, and EAW contributed equally to the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by MEXT KAKENHI (Grant Numbers 16H06467 to ST; 16H06469 to KKS; 17H05833, 19H04851 to TT; 19H04870 to KY; 16H06470, 16H06464, and 16K21727 to MW), JSPS KAKENHI (Grant Numbers 16H06380 to ST; 20K05982 to GS; 20H02956 to KS; 17H00821, 18KT0048, 19K22342 to MW) and JSPS Bilateral Programs (Grant Number 18032211-000481 to MW), by the Swiss National Science Foundation (31003A_159767 to KKS) and by the Research Funding for Computational Software Supporting Program from Meiji University to KY.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Kana Ito (Tohoku University) for technical assistances.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.576140/full#supplementary-material
Supplementary Figure 1 | Nucleotide sequence comparison of SCR-A between A. thaliana and A. halleri. Promoter and coding region of SCR-A aligned between A. thaliana and A. halleri. Identical nucleotide sequences between A. thaliana and A. halleri are highlighted with black background. In promoter region, open box outlined by red lines indicates a homologous region between A. thaliana and A. halleri. In coding region, two exons, the deletion in intron and the inversion in exon 2 are highlighted with red, yellow and blue backgrounds, respectively. Black arrows indicate the primer site for qRT-PCR.
Supplementary Figure 2 | Alignment of nucleotide sequences of SCR-A among A. thaliana accessions. Promoter and coding region of SCR-A aligned among 11 A. thaliana accessions in haplogroup A. Identical conserved sequences are highlighted with black background.
Supplementary Table 1 | List of Primers
References
Bateman, A. J. (1955). Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 9, 53–68. doi: 10.1038/hdy.1955.2
Bechsgaard, J. S., Castric, V., Charlesworth, D., Vekemans, X., Schierup, M. H. (2006). The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Ecol. 23, 1741–1750. doi: 10.1093/molbev/msl042
Boggs, N. A., Nasrallah, J. B., Nasrallah, M. E. (2009). Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PloS Genet. 5, e1000426. doi: 10.1371/journal.pgen.1000426
Busch, J. W., Schoen, D. J. (2008). The evolution of self-incompatibility when mates are limiting. Trends Plant Sci. 13, 128–136. doi: 10.1016/j.tplants.2008.01.002
Castric, V., Billiard, S., Vekemans, X. (2014). Trait transitions in explicit ecological and genomic contexts: plant mating systems as case studies. Adv. Exp. Med. Biol. 781, 7–36. doi: 10.1007/978-94-007-7347-9_2
Charlesworth, D., Vekemans, X. (2005). How and when did Arabidopsis thaliana become highly self-fertilising. Bioessays 27, 472–476. doi: 10.1002/bies.20231
Darwin, C. (1876). The Effect of Cross and Self Fertilisation in the Vegetable Kingdom (London: John Murray).
de Nettancourt, D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. 2nd edn (Berlin: Springer).
Franklin-Tong, V. E. (2008). Self-Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms (Berlin: Springer).
Fujii, S., Shimosato-Asano, H., Kakita, M., Kitanishi, T., Iwano, M., Takayama, S. (2020). Parallel evolution of dominant pistil-side self-incompatibility suppressors in Arabidopsis. Nat. Commun. 11, 1404. doi: 10.1038/s41467-020-15212-0
Goodwillie, C., Kalisz, S., Eckert, C. G. (2005). The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu. Rev. Ecol. Evol. Syst. 36, 47–79. doi: 10.1146/annurev.ecolsys.36.091704.175539
Kakita, M., Murase, K., Iwano, M., Matsumoto, T., Watanabe, M., Shiba, H., et al. (2007). Two distinct forms of M-locus protein kinase localize to the plasmamembrane and interact directly with S-locus receptor kinase to transduce self-incompatibility signaling in Brassica rapa. Plant Cell 19, 3961–3973. doi: 10.1105/tpc.106.049999
Koch, M. A., Haubold, B., Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248
Kusaba, M., Dwyer, K., Hendershot, J., Vrebalov, J., Nasrallah, J. B., Nasrallah, M. E. (2001). Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A.lyrata and its autogamous relative A. thaliana. Plant Cell 13, 627–643. doi: 10.1105/tpc.13.3.627
Nasrallah, M. E., Liu, P., Nasrallah, J. B. (2002). Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A.lyrata. Science 297, 247–249. doi: 10.1126/science.1072205
Nasrallah, M. E., Liu, P., Sherman-Broyles, S., Boggs, N. A., Nasrallah, J. B. (2004). Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. U.S.A. 101, 16070–16074. doi: 10.1073/pnas.0406970101
Nou, I. S., Watanabe, M., Isogai, A., Shiozawa, H., Suzuki, A., Hinata, K. (1991). Variation of S-alleles and S-glycoproteins in a naturalized population of self-incompatible Brassica campestris L. Jpn. J. Genet. 66, 227–239. doi: 10.1266/jjg.66.227
Osaka, M., Matsuda, T., Sakazono, S., Masuko-Suzuki, H., Maeda, S., Sewaki, M., et al. (2013). Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol. 54, 1894–1904. doi: 10.1093/pcp/pct133
Park, J.-I., Ishimizu, T., Suwabe, K., Sudo, K., Masuko, H., Hakozaki, H., et al. (2010). UDP-glucose pyrophosphoryalse is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51, 981–996. doi: 10.1093/pcp/pcq057
Schopfer, C. R., Nasrallah, M. E., Nasrallah, J. B. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. doi: 10.1126/science.286.5445.1697
Sherman-Broyles, S., Boggs, N., Farkas, A., Liu, P., Vrebalov, J., Nasrallah, M. E., et al. (2007). S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell 19, 94–106. doi: 10.1105/tpc.106.048199
Shiba, H., Takayama, S., Iwano, M., Shimosato, H., Funato, M., Nakagawa, T., et al. (2001). A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica. Plant Physiol. 125, 2095–2103. doi: 10.1104/pp.125.4.2095
Shimizu, K. K., Tsuchimatsu, T. (2015). Evolution of selfing: Recurrent patterns in molecular adaptation. Annu. Rev. Ecol. Evol. Syst. 46, 593–622. doi: 10.1146/annurev-ecolsys-112414-054249
Shimizu, K. K., Shimizu-Inatsugi, R., Tsuchimatsu, T., Purugganan, M. D. (2008). Independent origins of self-compatibility in Arabidopsis thaliana. Mol. Ecol. 17, 704–714. doi: 10.1111/j.1365-294X.2007.03605.x
Smyth, D. R., Bowman, J. L., Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. doi: 10.1105/tpc.2.8.755
Stebbins, G. L. (1974). Flowering Plants: Evolution Above the Species Level (Cambridge, Massachusetts: Harvard University Press).
Suzuki, G., Kai, N., Hirose, T., Fukui, K., Nishio, T., Takayama, S., et al. (1999). Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153, 391–400.
Takasaki, T., Hatakeyama, K., Suzuki, G., Watanabe, M., Isogai, A., Hinata, K. (2000). The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403, 913–916. doi: 10.1038/35002628
Takayama, S., Shiba, H., Iwano, M., Shimosato, H., Che, F.-S., Kai, N., et al. (2000). The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. U.S.A. 97, 1920–1925. doi: 10.1073/pnas.040556397
Takayama, S., Shimosato, H., Shiba, H., Funato, M., Che, F.-S., Watanabe, M., et al. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413, 534–538. doi: 10.1038/35097104
Tang, C., Toomajian, C., Sherman-Broyles, S., Plagnol, V., Guo, Y.-L., Hu, T. T., et al. (2007). The evolution of selfing in Arabidopsis thaliana. Science 317, 1070–1072. doi: 10.1126/science.1143153
Tsuchimatsu, T., Shimizu, K. K. (2013). Effects of pollen availability and the mutation bias on the fixation of mutations disabling the male specificity of self-incompatibility. J. Evol. Biol. 26, 2221–2232. doi: 10.1111/jeb.12219
Tsuchimatsu, T., Suwabe, K., Shimizu-Inatsugi, R., Isokawa, S., Pavlidis, P., Städler, T., et al. (2010). Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464, 1342–1346. doi: 10.1038/nature08927
Tsuchimatsu, T., Goubet, P. M., Gallina, S., Holl, A.-C., Fobis-Loisy, I., Bergès, H., et al. (2017). Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes. Mol. Biol. Evol. 34, 1878–1889. doi: 10.1093/molbev/msx122
Uyenoyama, M. K., Zhang, Y., Newbigin, E. (2001). On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics 157, 1805–1817. https://www.genetics.org/content/157/4/1805
Watanabe, M., Nou, I. S., Takayama, S., Yamakawa, S., Isogai, A., Suzuki, A., et al. (1992). Variations in and inheritance of NS-glycoprotein in self-incompatible Brassica campestris L. Plant Cell Physiol. 33, 343–351. doi: 10.1093/oxfordjournals.pcp.a078261
Watanabe, M., Takasaki, T., Toriyama, K., Yamakawa, S., Isogai, A., Suzuki, A., et al. (1994). A high degree of homology exists between the protein encoded by SLG and the S receptor domain encoded by SRK in self-incompatible Brassica campestris L. Plant Cell Physiol. 35, 1221–1229. doi: 10.1093/oxfordjournals.pcp.a078716
Keywords: Arabidopsis thaliana, artificial chimeric gene, evolutionary process, flower development, promoter activity, self-compatibility, S-locus protein 11/S-locus cysteine rich protein gene
Citation: Suwabe K, Nagasaka K, Windari EA, Hoshiai C, Ota T, Takada M, Kitazumi A, Masuko-Suzuki H, Kagaya Y, Yano K, Tsuchimatsu T, Shimizu KK, Takayama S, Suzuki G and Watanabe M (2020) Double-Locking Mechanism of Self-Compatibility in Arabidopsis thaliana: The Synergistic Effect of Transcriptional Depression and Disruption of Coding Region in the Male Specificity Gene. Front. Plant Sci. 11:576140. doi: 10.3389/fpls.2020.576140
Received: 25 June 2020; Accepted: 31 August 2020;
Published: 11 September 2020.
Edited by:
Tetsu Kinoshita, Yokohama City University, JapanReviewed by:
Tomohiko Kubo, Hokkaido University, JapanJong-In Park, Sunchon National University, South Korea
Copyright © 2020 Suwabe, Nagasaka, Windari, Hoshiai, Ota, Takada, Kitazumi, Masuko-Suzuki, Kagaya, Yano, Tsuchimatsu, Shimizu, Takayama, Suzuki and Watanabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keita Suwabe, c3V3YWJlQGJpby5taWUtdS5hYy5qcA==; Masao Watanabe, bmFiZUBpZ2UudG9ob2t1LmFjLmpw
†These authors have contributed equally to this work
 Keita Suwabe
Keita Suwabe Kaori Nagasaka1†
Kaori Nagasaka1† Ai Kitazumi
Ai Kitazumi Takashi Tsuchimatsu
Takashi Tsuchimatsu Kentaro K. Shimizu
Kentaro K. Shimizu Go Suzuki
Go Suzuki Masao Watanabe
Masao Watanabe