- 1Plant Cell & Transformation Technologies, Research & Development, KWS SAAT SE & Co. KGaA, Einbeck, Germany
- 2OSU Plant Transformation Laboratory, College of Food, Agricultural and Environmental Sciences, Department of Horticulture and Crop Science, The Ohio State University, OH, United States
- 3BASF Corporation, Research Triangle Park, NC, United States
- 4Roche Tissue Diagnostics, Tucson, AZ, United States
- 5BASF Metabolome Solutions GmbH, Berlin, Germany
- 6BASF Belgium Coordination Center—Business Belux (Branch), Gent Zwijnaarde, Belgium
Successful regeneration of genetically modified plants from cell culture is highly dependent on the species, genotype, and tissue-type being targeted for transformation. Studies in some plant species have shown that when expression is altered, some genes regulating developmental processes are capable of triggering plant regeneration in a variety of plant cells and tissue-types previously identified as being recalcitrant to regeneration. In the present research, we report that developmental genes encoding GROWTH-REGULATING FACTORS positively enhance regeneration and transformation in both monocot and dicot species. In sugar beet (Beta vulgaris ssp. vulgaris), ectopic expression of Arabidopsis GRF5 (AtGRF5) in callus cells accelerates shoot formation and dramatically increases transformation efficiency. More importantly, overexpression of AtGRF5 enables the production of stable transformants in recalcitrant sugar beet varieties. The introduction of AtGRF5 and GRF5 orthologs into canola (Brassica napus L.), soybean (Glycine max L.), and sunflower (Helianthus annuus L.) results in significant increases in genetic transformation of the explant tissue. A positive effect on proliferation of transgenic callus cells in canola was observed upon overexpression of GRF5 genes and AtGRF6 and AtGRF9. In soybean and sunflower, the overexpression of GRF5 genes seems to increase the proliferation of transformed cells, promoting transgenic shoot formation. In addition, the transformation of two putative AtGRF5 orthologs in maize (Zea mays L.) significantly boosts transformation efficiency and resulted in fully fertile transgenic plants. Overall, the results suggest that overexpression of GRF genes render cells and tissues more competent to regeneration across a wide variety of crop species and regeneration processes. This sets GRFs apart from other developmental regulators and, therefore, they can potentially be applied to improve transformation of monocot and dicot plant species.
Introduction
Plants have an impressive capability to regenerate new tissues, organs, or even an entire plant. External signals can trigger differentiated somatic cells to reprogram their developmental fate to repair wounded organs and to regenerate new tissues or whole plants, allowing plants to cope with environmental threats (Ikeuchi et al., 2016). This natural competence for regeneration is fundamental in plant propagation and tissue culture techniques that regenerate plants through de novo organogenesis or somatic embryogenesis using exogenously applied plant hormones (Ikeuchi et al., 2016; Kareem et al., 2016; Méndez-Hernández et al., 2019). The study of these regeneration pathways and their developmental regulators has enabled the establishment of protocols for the in vitro production and propagation of many plant species under controlled conditions (George et al., 2008; Sugiyama, 2015).
Tissue culture-induced regeneration is also important for transformation protocols in crop species. For example, monocot transformation methods predominantly depend on plant regeneration through somatic embryogenesis, whereas the regeneration of transgenic dicot crops is often achieved through organogenesis (Kausch et al., 2019). Regeneration potential of plant cells and organs varies widely among plant species, which often restricts transformation methods to a limited set of plant varieties or genotypes as well as target explants for DNA delivery (Cheng et al., 2004). The phenomenon of limited regenerative capability in vitro, better known as recalcitrance, makes the recovery of transgenic lines difficult or impossible in many plant species. This hampers the advance of fundamental research as well as the application of technologies relying on regeneration, including plant transformation and genome editing (Altpeter et al., 2016).
Traditionally, tissue culture-based regeneration methods are based upon the application of plant hormone combinations (especially auxins and cytokinins) on explants that are amenable to regeneration (Sugiyama, 2015). The establishment of a successful regeneration or transformation protocol often requires customization of the plant hormone ratios along with other tissue culture factors for each genotype, which is laborious and very often fails. Recently, several lines of experimental evidence suggest that inherent developmental programs are activated in response to tissue culture, which can reprogram the cell fate or activate undifferentiated cells in cultured explants (Ikeuchi et al., 2019; Sugimoto et al., 2019). Observations gained from model organisms such as Arabidopsis have shown that specific transcription factors can integrate the signals leading to cell reprogramming and the reacquisition of an embryonic or a meristematic fate (Gordon et al., 2007; Duclercq et al., 2011; Ikeuchi et al., 2013; Gaillochet and Lohmann, 2015; Kareem et al., 2015; Perez-Garcia and Moreno-Risueno, 2018). Such transcription factors are referred to as developmental regulators since they coordinate the spatial distribution of cells in an organized way during the development of organs and embryos.
Several genes encoding developmental regulators have been described to improve the regeneration efficiency in various plant species (Heidmann et al., 2011; Horstman et al., 2017; Gordon-Kamm et al., 2019). For example, the constitutive expression of BABY BOOM (BBM), a transcription factor of the AP2/ERF family with diverse functions in plant development, can promote cell proliferation and ectopic embryo formation in cotyledons and leaves of Arabidopsis (Boutilier et al., 2002). In a study aimed at elucidating the molecular mechanism of somatic embryogenesis, gain-of-function mutations in the gene coding for the shoot apical meristem identity regulator WUSCHEL (WUS) were found to induce embryo formation from various vegetative tissues in Arabidopsis (Zuo et al., 2002; Somssich et al., 2016). These initial observations led researchers to develop an approach using ectopic co-expression of BBM and WUS to greatly improve in vitro transformation in a variety of monocots, including several recalcitrant maize inbred lines, rice and sorghum (Lowe et al., 2016; Mookkan et al., 2017; Lowe et al., 2018). The positive influence of these genes on the regeneration in tissue culture was also reported in studies conducted in several dicot plant species, including highly recalcitrant crop varieties (Srinivasan et al., 2007; Arroyo-Herrera et al., 2008; Deng et al., 2009; Solís-Ramos et al., 2009; Heidmann et al., 2011; Bouchabke-Coussa et al., 2013; Florez et al., 2015).
Additional developmental genes have been proposed as candidates to further improve regeneration and transformation technologies in monocots and dicots (Gordon-Kamm et al., 2019; Kausch et al., 2019). For example, overexpression of an AP2/ERF transcription factor, WOUND INDUCED DEDIFFERENTIATION1 (WIND1) increased callus-induction and shoot regeneration in rapeseed, tomato, and tobacco (Iwase et al., 2015). WIND1 directly binds the promoter of another member of the AP2/ERF transcription factor family, ENHANCER OF SHOOT REGENERATION1 (ESR1) gene, whose overexpression similarly increases callus formation and shoot regeneration in Arabidopsis (Banno et al., 2001; Iwase et al., 2007). However, in all these reports the continued overexpression of developmental genes caused severe growth defects during further plant development, such as aberrant development of vegetative and reproductive organs and infertility. Therefore, use of these developmental genes requires restricting or eliminating their expression in the plant by methods such as gene excision or the use of inducible or tissue-specific promoters (Gallois et al., 2002; Lowe et al., 2016).
Here, we report that ectopic expression of a developmental regulator, GROWTH-REGULATING FACTOR 5 (GRF5), has a positive effect in boosting regeneration and genetic transformation in various crop species using established organogenic or embryogenic regeneration systems. The GRF genes belong to a small plant-specific transcription factor family and they form a transcriptional complex with GRF-INTERACTING FACTOR (GIF) to regulate plant growth and development by providing cues to primordial cells of vegetative and reproductive organs (van der Knaap et al., 2000; Kim et al., 2003; Omidbakhshfard et al., 2015; Kim, 2019; Liebsch and Palatnik, 2020). Genetic analyses of plants with AtGRF5 overexpression and loss-of-function mutations in Arabidopsis revealed that this transcription factor is involved in the regulation of the cell division and expansion during leaf development (Horiguchi et al., 2005; Horiguchi et al., 2006; Debernardi et al., 2014). In addition, GRF5 overexpression in Arabidopsis increased chlorophyll content and chloroplast number per cell, and delayed leaf senescence (Vercruyssen et al., 2015). We observed that stable transformation of GRF5 genes in sugar beet accelerated shoot organogenesis and resulted in significant improvements in genetic transformation, including recalcitrant varieties. Ectopic expression of AtGRF5 and orthologs also resulted in significant increases in the formation and development of transgenic callus in canola and increases in the production of developing transgenic shoots in soybean and sunflower. In maize, transformation of AtGRF5 orthologs showed a higher rate of embryogenic callus growth leading to an increased recovery of transgenic plants. Unlike other reported developmental regulators, no detrimental pleiotropic effects were observed in transgenic events overexpressing GRF genes in the tested crops, other than possible reduction in de novo root initiation on soybean shoots. The role of GRF genes to improve transformation protocols based on either shoot organogenesis or somatic embryogenesis is discussed.
Material and Methods
KWS SAAT SE & Co. KGaA and BASF will provide the material described here to academic groups under a material transfer agreement for non-commercial use in research.
Agrobacterium Strains and Binary Plasmids
All bacterial strains and binary vectors used in this study were produced according to standard procedures (Russell and Sambrook, 2001). The Agrobacterium strains harboring the constructs used to transform sugar beet, canola, soybean, sunflower and maize, including the expression cassettes for the GRF5 genes or the fluorescent control reporters, are described in the Supplementary Table 1.
Sugar Beet Transformation
The multigerm inbred lines 9BS0448, 1RV6183, 7RV5706H and 8RV6921 were used in the sugar beet experiments. Agrobacterium-mediated transformation of sugar beet was performed by using the procedure and culture media as described elsewhere (Kishchenko et al., 2005) with minor modifications. The media composition is provided in the Supplementary Table 2. Briefly, to induce friable callus cultures, leaf explants from micropropagated shoots were incubated in callus induction medium at 28°C in the dark. The Agrobacterium culture was centrifuged and resuspended to an OD600 of 0.8 in liquid co-culture medium. Callus pieces were harvested and cultivated with Agrobacterium in co-culture medium solidified with 10 g/l agar for at least 2 days. Transformed calli were cultured in shoot regeneration medium for selection of transgenic shoots. Transgenic calli were transferred to fresh shoot regeneration medium every 3 weeks at least four times. Developing shoots were isolated and clonally propagated in shoot multiplication medium. Co-transformation experiments were performed by mixing Agrobacterium strains harboring the AtGRF5 construct and the tdTomato control construct in a 1:1 volume ratio. To regenerate untransformed plants, callus cultures were subjected to the same procedure by using medium without aminoglycoside antibiotic. Transformation efficiency was estimated as the frequency of transgenic plants normalized by the number of starting explants.
Canola Hypocotyl Transformation
Canola transformation was performed on the genotype BNS3 based on the hypocotyl transformation method (Radke et al., 1988) with modifications. Media components used are detailed in Supplementary Table 3. Seedlings were prepared by surface sterilizing seeds in 70% ethanol and sowing on germination medium in PlantCon™ boxes and growing for 4 days in the dark at 23°C. Agrobacterium rhizogenes strain SHA001 was used for delivery of the gene cassettes into hypocotyl segments of B. napus seedlings (Mankin et al., 2007). Two experiments were conducted. The first experiment had six replicates over time each with three operators testing AtGRF5, BnGRF5-LIKE, or DsRed control vectors, and the second had three replicates over time each performed by three operators testing AtGRF5, AtGRF6, AtGRF9, or DsRed control vectors. A. rhizogenes was grown to OD600 of 1.0 and subsequently diluted to 0.1 with liquid infection medium. Hypocotyl explants 7 to 10 mm in length were prepared from 4-day-old seedling. After cutting, the explant was dipped in the Agrobacterium suspension and placed on filter paper on solid co-culture medium in 15 mm x 100 mm plates. Plates containing ~50 explants were sealed with 3M Micropore™ tape and cultivated in 16 light/8 dark photoperiod at 23°C. After 3 days, all explants were transferred to recovery medium, sealed with tape, and cultivated for 7 days. Explants were then transferred to four rounds of selection, each cycle lasting 14 days. Shoots of at least 1 cm in size were removed to rooting medium. Explants were scored for callus growth and DsRed expression 38 days after transformation. Putative transformation efficiency was calculated as [(# events with either DsRed expression or rooting on selection/# explants transformed) *100].
Soybean Primary-Node Transformation
Soybean transformation was performed using the cultivars, Jake and CD215, using the primary-node transformation method with modifications (Olhoft et al., 2007; Chang et al., 2012). Seedlings were grown for donor material by plating sterile soybean seeds on solid germination medium (see Supplementary Table 4) in PlantCons™ and placed under light (150 μm-2s-2) at 26°C for 7 to 8 days under 18 light/6 dark photoperiod. Agrobacterium rhizogenes strain SHA017 was grown and resuspended to OD600 1.5. Explants were prepared from seedlings by removing the roots, half of the hypocotyl, one cotyledon, and the epicotyl above the primary node. Approximately 50 explants were incubated in the Agrobacterium suspension for 30 min, then placed on co-cultivation medium and incubated at room temperature for 5 days. Explants were transferred to selection medium containing 3 μM imazapyr and cultivated at 26°C under light. After 3 weeks on selection, the explants were transferred to Oasis® growing media and subsequently scored for DsRed fluorescence on developing shoots at the primary node. Explants were regularly watered with 5.7 µM IAA for 3 weeks. Elongated shoots were removed, placed on Oasis® growing media, and watered with 5.7 µM IAA/2 µM imazapyr for 7 days. The number of rooted plants was recorded, and a subset was sent to the GH for molecular characterization of T0 and T1 materials.
Sunflower Shoot Induction and Agrobacterium-Mediated Transformation
Seeds of sunflower (cv. RHA280) were harvested from plants grown under greenhouse conditions and stored at 4°C in the dark for up to 1 year. Sunflower cotyledon explants, Agrobacterium inoculation and sunflower transformation were conducted using the low inoculum with long co-culture (LI/LC) method (Zhang and Finer, 2016) at inoculum density of 102 CFU/ml. After 2 weeks of co-culture on solid shoot induction medium (SIM), the explants were transferred onto solid shoot elongation medium (SEM). GFP expression in adventitious shoots was observed after 1 week on the SEM medium (3 weeks after inoculation). The GRF5s and GFP control vectors were transformed in triplicate (n ≥ 10).
Maize Transformation
Transgenic plants from the maize inbred line A188 were produced according to the method described by (Ishida et al., 2007). Transformation efficiency was calculated as the frequency of inoculated immature embryos that regenerated at least one transgenic plant.
Microscopy
Sugar beet and maize in vitro cultures were photographed using a SteREO Discovery.V12 stereomicroscope (Carl Zeiss Microscopy GmbH, Jena, Germany). To quantify the embryogenic callus area in maize, 36 randomly taken pictures of calli obtained from independent embryos for each construct were used. The callus area was quantified in the pictures by using the open source software Fiji (Schindelin et al., 2012). DsRed fluorescence signal in canola and soybean primary node transformation experiments was monitored by using a Zeiss SV11 stereomicroscope coupled with the mercury vapor short-arc illuminator HBO 100W (Carl Zeiss Vision International GmbH, Aalen, Germany). The rhodamine filter was used for DsRED2 (563 nm excitation max and 582 emission max) protein visualization. In sunflower transformation, tissue growth and GFP expression were monitored using a MZFLIII stereomicroscope (Leica, Heerbrugg, Switzerland) equipped with a “GFP-2” filter set (excitation 480 ± 40 nm; emission 510) and a pE-100 light-emitting diode (LED) lamp (Andover, Hampshire, UK) as an excitation light source.
Molecular Analysis
Sugar beet and maize: Genomic DNA was extracted by standard procedures from leaf tissue and used for Multiplex TaqMan assays to measure the copy number of integrated T-DNA in transgenic sugar beet and maize plants. In both cases detection of endogenous genes (BvEF1 of sugar beet and ZmEF1 of maize) were combined with the detection of either PAT or NPTII gene, respectively, in a duplex qPCR. Primers and probes were designed using the Primer Express 3.0 software for Real-Time PCR (Applied Biosystems Foster City, California, USA), selecting the option MGB TaqMan probes with amplicon length 60 to 80 bp, and using the default parameters of the software. The oligonucleotide sequences used in TaqMan assays are provided in the Supplementary Table 5. qPCRs were carried out on an Applied Biosystems QuantStudio 3 and 5 Real-Time PCR Systems (Applied Biosystems, Foster City, California, USA) using 80–150 ng of genomic DNA as template, Maxima Probe qPCR Master Mix (2X), with ROX (ThermoFisher Scientific, Waltham, Massachusetts, USA), 250 nM MGB probe each and 900 nM primer each in a total volume of 10 μl. Amplification and quantification were performed with the following cycling conditions: 2 min 50°C, 5 min at 95°C for initial DNA denaturation, followed by an amplification program of 40 cycles at 95°C for 15 s and 60°C for 30 s. The fluorescence threshold was manually set above the background level. Negative controls without DNA template and verified control samples with 1 copy and 2 copies of the T-DNA for either PAT or NPTII assay were run in parallel and used for absolute quantification and copy number determination.
Soybean: Genomic DNA was extracted from leaf tissue using a modified protocol for Wizard(R) Magnetic 96 DNA Plant System from Promega (Madison, Wisconsin, USA). The copy number of integrated T-DNA in transgenic soybean plants was measured in duplex TaqMan assays. Detection of endogenous one-copy genes (GmLectin) was combined with the detection of AHAS gene in a duplex qPCR. Primers and probes were designed using the Primer Express 3.0.1 software for Real-Time PCR (Applied Biosystems, Foster City, California, USA), selecting the option TaqMan® Quantification probes, and using the default parameters of the software. The oligonucleotide sequences used in TaqMan assays are provided in the Supplementary Table 5. qPCRs were carried out on an Applied Biosystems QuantStudio 7 Real-Time PCR Systems (Applied Biosystems, Foster City, California, USA) using isolated genomic DNA as template, Perfecta® qPCR ToughMix® Low ROX™ PCR master mix (QuantaBio, 2x), 200 nM Taqman probe and 900 nM forward and reverse primers in 5 μl. Amplification and quantification were performed with the following cycling conditions: 2 min 95°C for initial DNA denaturation, followed by an amplification program of 40 cycles at 95°C for 2 min and 60°C for 1 min. The fluorescence threshold was manually set above the background level and verified control samples with one copy of the T-DNA for AHAS assay were run in parallel and used for absolute quantification and copy number determination.
qRT-PCR Analysis
Total RNA from leaf samples was isolated with InviTrap® Spin Plant RNA Mini Kit (INVITEK Molecular, Berlin, Germany) and reverse transcribed using Invitrogen™ Oligo (dT) primers (Fisher Scientific GmbH, Schwerte, Germany) and Invitrogen™ SuperScript™ III Reverse Transcriptase (Fisher Scientific GmbH, Schwerte, Germany). 20 ng of reverse transcribed RNA was used as template in the real-time PCR using Applied Biosystems™ QuantStudio™ 6 Flex (Fisher Scientific GmbH, Schwerte, Germany). Specific primers binding the 3´-UTR of the NOS terminator were used for the detection of the transgene transcripts. The qPCR reactions for AtGRF5 and the endogenous reference (Beta vulgaris subsp. vulgaris CASEIN KINASE 1-LIKE PROTEIN 11) were performed in technical duplicates by using the following oligonucleotides: NOSt-F, 5´-AAC GTC ATG CAT TAC ATG TTA-3´; NOSt-R, 5´-CGG TCT TGC GAT GAT TAT CA-3´; S989-F, 5´-GAG GAA CTA GAC ATG GGG ATA CAT-3´; S990-R, 5´-GCG ATA CAA AGT AGA CAT TAG AAC TC-3´. ΔΔCt values were calculated and transcript levels given in log2 scale relative to the transgenic T0 event (L50) displaying the lowest AtGRF5 overexpression.
Data Analysis
In all experimental data sets, statistical significance of differences in average was determined with a Welch’s t-test.
Identification of GRF5 Homologs and Sequence Alignment
To identify the ortholog genes of AtGRF5 from sugar beet, canola, soybean, sunflower and maize, AtGRF5 amino acid sequence was used as the query to perform BLAST against respective plant-specific protein sequence databases (proteins from most actual annotated genome assemblies from NCBI and EnsemblPlants). Initial candidates were selected based on the BLAST results, and further analyzed by global, multiple alignments: true redundancy (= identical protein sequences) and outliers (= suspicious sequences, as revealed by manually checking using multiple alignments) were removed first. Then, the initial candidates were ranked based on true pairwise global identities by comparing with AtGRF5 (program NEEDLE from EMBOSS): ranking was based on global identity, thus also the less-conserved C-terminal region of the transcription factors is considered here. Based on this ranking, top candidates were selected, and confirmed to align best (global identity) with AtGRF5 when compared with all nine Arabidopsis GRF-family members in global alignments (back search). A summary of sequence identities found according to the blastp searches is shown in Supplementary Table 6.
GRF5 protein sequences were aligned using CLC DNA Workbench 8.0 (Qiagen) with the following settings: gap open cost, 10; gap extension cost, 1; end gap cost, as any other; alignment, very accurate. The conserved QLQ and WRC domains in the GRF crop orthologs were identified by using the Arabidopsis consensus amino acid sequences (Kim and Tsukaya, 2015) (Supplementary Figure 3).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers AY102638 (AtGRF5, At3g13960), XP_010666262 (BvGRF5-LIKE), XM_022719744.1 (BnGRF5-LIKE), XP_006589198.1 (GmGRF5-LIKE), XM_022128855.1 (HaGRF5-LIKE), GRMZM2G105335 (ZmGRF5-LIKE1), and GRMZM5G893117 (ZmGRF5-LIKE2).
Results
Overexpression of GRF5 Promotes Shoot Organogenesis and Increases Transformation Efficiency in Sugar Beet
During an Agrobacterium-mediated transformation experiment in sugar beet line 9BS0448, we noticed that a large number of shoots was unexpectedly regenerated from transgenic calli expressing the AtGRF5 gene under the CaMV 35S promoter (2x35S::AtGRF5). That observation prompted us to hypothesize that overexpression of AtGRF5 in callus cells positively influences shoot organogenesis. To test, we transformed leaf-derived callus from the same line with Agrobacterium containing 2x35S::AtGRF5 and compared to the control construct 2x35S::tdT, which expresses the tdTomato fluorescent protein. Compared to the control experiment, the calli transformed with the AtGRF5 transgene showed enhanced and rapid shoot formation in all experiments, confirming our initial observation (Figure 1A). The number of regenerated shoots was significantly greater from callus expressing AtGRF5 at the end of the second (S2), third (S3), and fourth (S4) round of selection compared to the control (Figure 1B). Interestingly, an average of 30 shoots was already formed from AtGRF5-transformed callus at the S2 round in each experiment, whereas in the control experiment, shoot formation was only visible at the selection round S3 (Figure 1B). Next, qPCR analyses confirmed that 97.6% of the regenerated shoots were transgenic in all experiments. Strikingly, callus transformed with 2x35S::AtGRF5 produced more transgenic events per experiment than the 2x35S::tdT control cultures, resulting in a significant 6-fold increase on transformation efficiency (Figure 1C). Similarly, transformation efficiency increased when sugar beet calli were inoculated with a mixture of Agrobacterium cultures containing either the 2x35S::AtGRF5 or the 2x35S::tdT construct. Transgenic plants containing both constructs were regenerated, resulting in an averaged co-transformation efficiency of 19.6% (Supplementary Figure 1). Compared to the experiments performed with the tdTomato construct alone, transgenic plants containing the 2x35S::tdT cassette were more efficiently produced in the co-transformation experiments, suggesting that AtGRF5 overexpression improves the recovery of transgenic events containing a construct of interest (Supplementary Figure 1). In addition, leaf explants from four out of five randomly selected T0 events produced callus cultures with enhanced shoot formation capacity compared to non-transgenic callus cultures (Figure 2). This correlated with the expression level of AtGRF5 of the lines (Figure 2). Sugar beet plants that were transformed with 2x35S::AtGRF5 grew well in vitro and showed no obvious differences in their phenotype compared with the transgenic control plants growing under the same conditions (Supplementary Figure 2). Collectively, our results indicate that the ectopic expression of AtGRF5 boosts the switch from callus phase to organogenesis and therefore, efficiently improves the transformation in sugar beet.
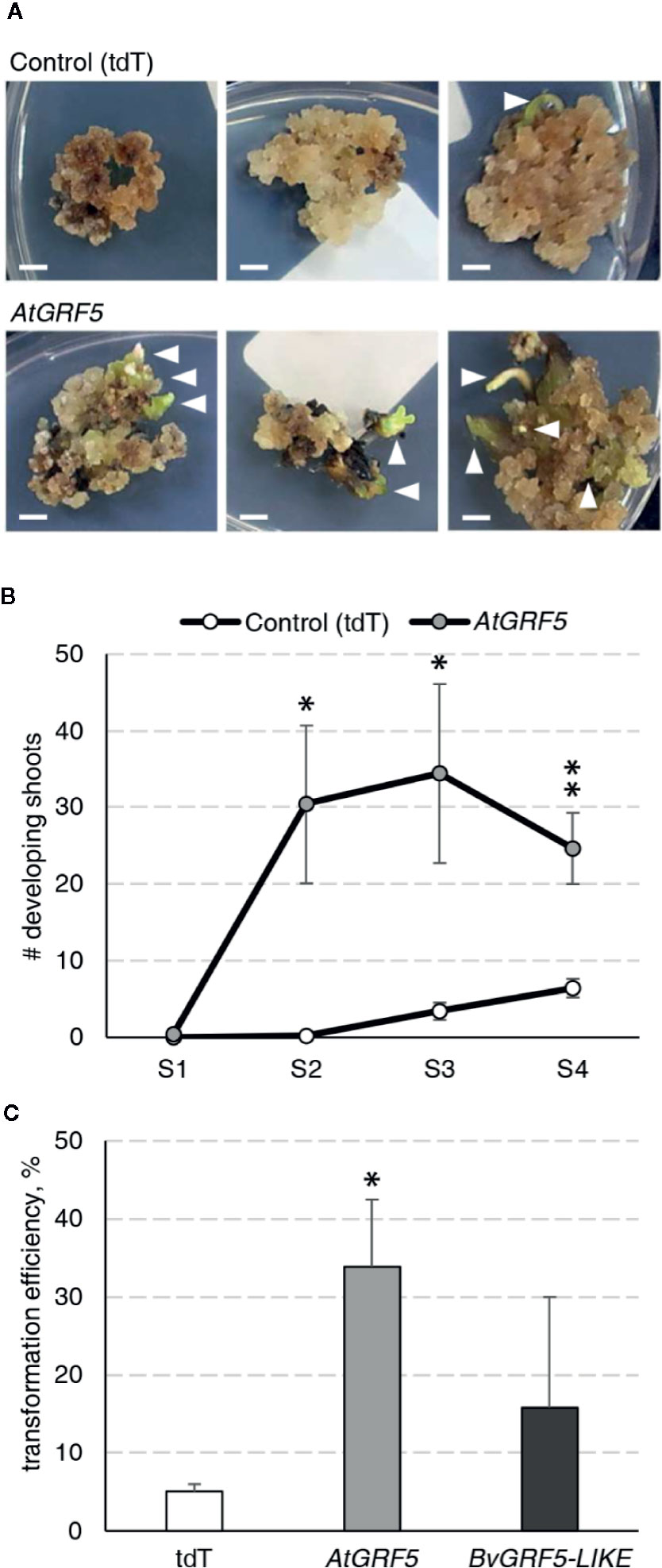
Figure 1 Detailed analysis of the AtGRF5 overexpression in sugar beet transformation experiments. Transformations with a construct expressing the tdTomato (tdT) were done in parallel as a control. (A) Representative pictures of transformed callus on selection medium. White arrow heads indicate developing shoots. Scale bar = 0.5 cm. (B) Number of shoots regenerated across four rounds of culture on selection medium (S1 to S4). The mean values ± SEM from five biological replicates for the tested constructs are represented. (C) Average transformation efficiency of the experiments performed with the construct to overexpress AtGRF5 or the putative sugar beet ortholog BvGRF5-LIKE construct and compared to the control construct. The transformation efficiency values are indicated as the mean ± SEM from at least three biological replicates per tested construct. In both graphs, differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; *p < 0.05; **p < 0.01.
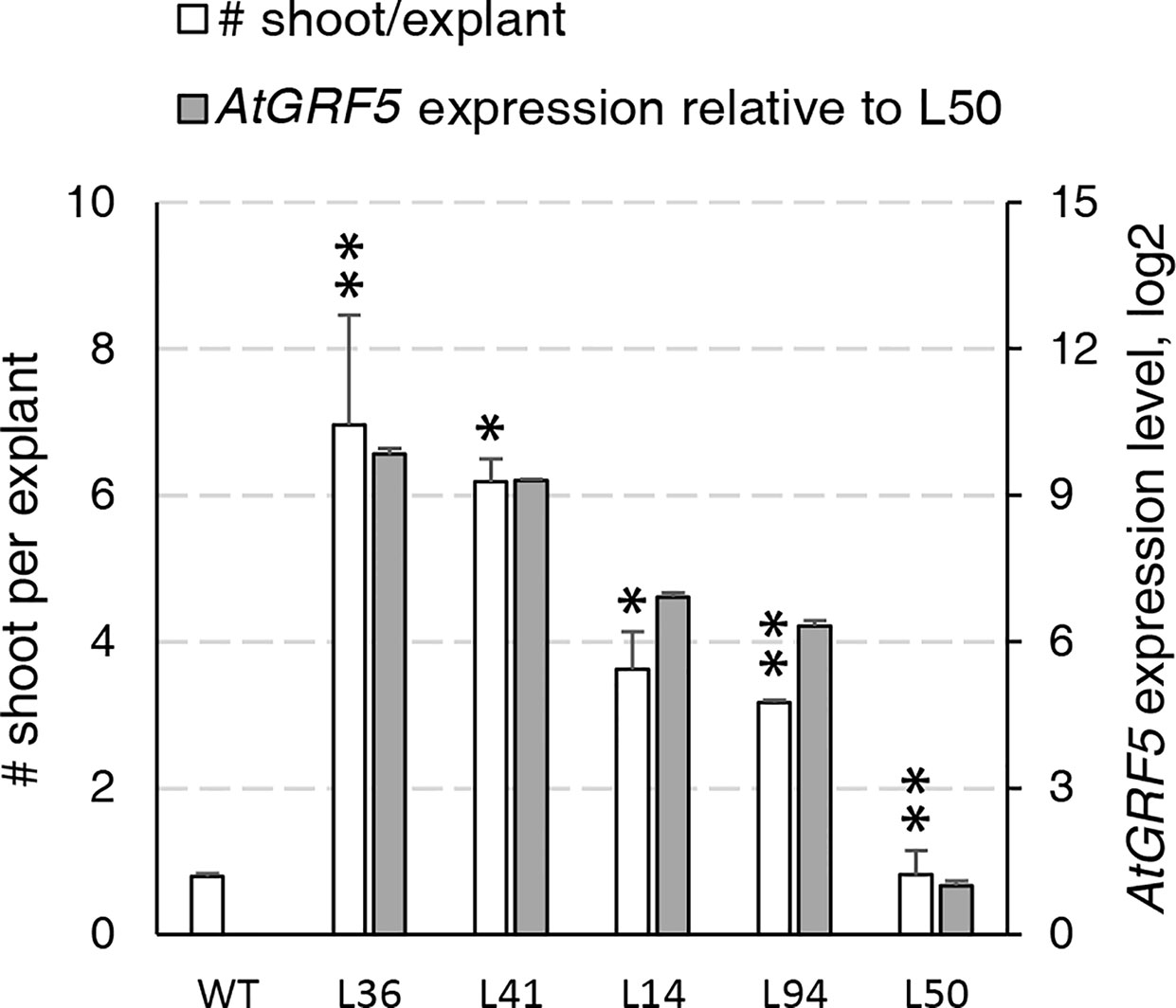
Figure 2 Characterization of the regeneration capacity in sugar beet T0 events expressing AtGRF5. Shoot regeneration capacity of friable callus cultures produced from five independent transgenic events containing the 2x35S::AtGRF5 construct (L36, L41, L14, L94, and L50) was compared with a non-transgenic plant regenerated from callus (WT) as a control (white bars). AtGRF5 expression level is represented with gray bars. The expression values were normalized to the low expressing event L50. The gene expression values are indicated as the mean ± SEM from two technical replicates, whereas the shoot/explant values are indicated as the mean ± SEM from at least 50 explants isolated from each transgenic event. Differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; *p < 0.05; **p < 0.01.
We suspected that the sugar beet GRF5 homolog might also influence shoot organogenesis. The putative ortholog in sugar beet was identified by blastp searches and named BvGRF5-LIKE (Supplementary Figure 3 and Supplementary Table 6). To test, we followed the same experimental design and transformed sugar beet callus of the line 9BS0448 with a construct to overexpress the BvGRF5-LIKE and compared with the control tdTomato. The transformation of BvGRF5-LIKE in sugar beet callus resulted in an increased transformation efficiency, although not statistically significant (Figure 1C).
GRF5 Promotes Shoot Organogenesis in Different Sugar Beet Varieties, Including Recalcitrant Ones
It is well known that sugar beet transformation is highly genotype dependent (Gurel et al., 2008). Consequently, we investigated the effect of AtGRF5 in the transformation efficiency of three other sugar beet varieties selected from the KWS germplasm. Leaf-derived callus obtained from the three inbred lines 1RV6183, 7RV5706H, and 8RV6921, with differing degrees of recalcitrance, was transformed with either the 2x35S::AtGRF5 construct or the 2x35S::tdT construct, respectively. The three lines showed enhanced transformability when the AtGRF5 construct was used (Figure 3A). The line 1RV6183 displayed transformation efficiencies with the control construct (6.2%) and the 2x35S::AtGRF5 construct (27.8%) that are comparable to the 9BS0448 variety (Figures 1C and 3A). The lines 7RV5706H and 8RV6921 showed an average transformation efficiency with the control construct of 1.8% and 0%, respectively, indicating a higher level of recalcitrance to transformation of these lines compared to 1RV6183 and 9BS0448 (Figures 1C and 3A). By contrast, the introduction of the 2x35S::AtGRF5 construct into the calli resulted in transformation efficiencies higher than the control experiment for the 7RV5706H (20.7%) and 8RV6921 (6.2%) sugar beet varieties (Figure 3A). Interestingly, all shoots that were regenerated from the recalcitrant 8RV6921 line contained the AtGRF5 gene, while the transformation of the 2x35S::tdT construct did not produce any shoot (Figures 3A, B). Overall, these results suggest that the overexpression of AtGRF5 can overcome recalcitrance to transformation in sugar beet.
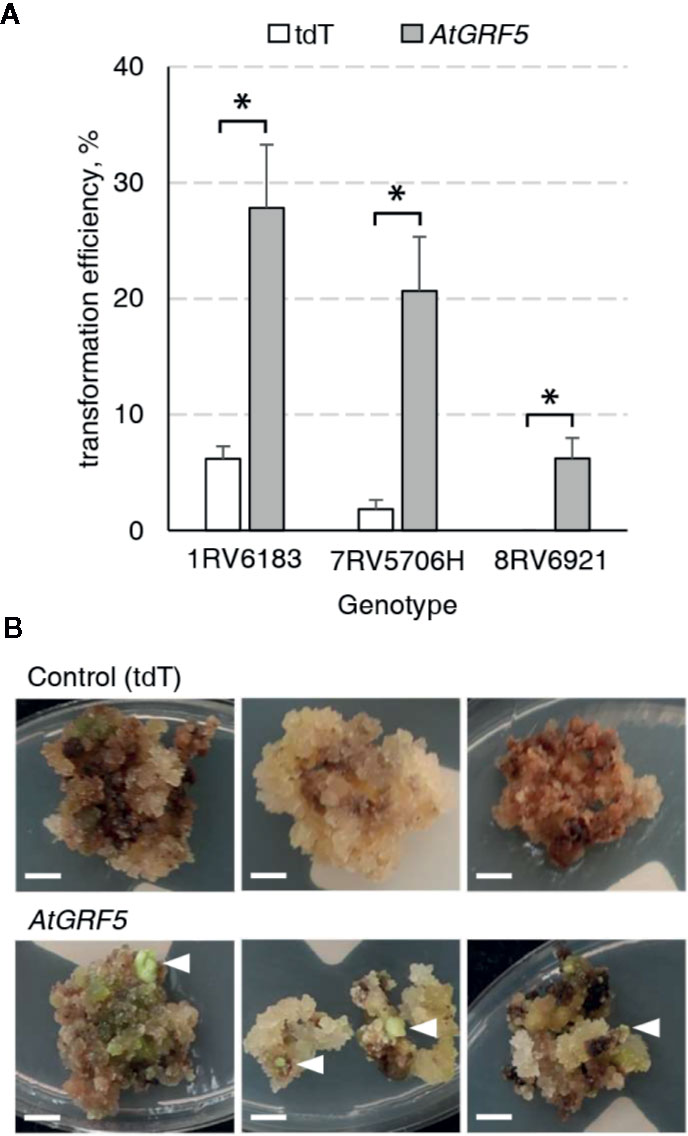
Figure 3 Overexpression of AtGRF5 enables transformation of recalcitrant sugar beet varieties. Transformations done with the tdTomato (tdT) and the AtGRF5 constructs were compared. (A) Average transformation efficiency of three sugar beet genotypes. Across the tested genotypes, the level of recalcitrance to shoot regeneration increases from left to right, the genotype 8RV6921 being the most recalcitrant. The transformation efficiency values are indicated as the mean ± SEM from at least three biological replicates per construct and genotype. Differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; *p < 0.05. (B) Representative pictures of transformed calluses of the recalcitrant 8RV6921 genotype in selection medium. White arrow heads indicate developing shoots. Scale bar = 0.5 cm.
Overexpression of GRF5, GRF6, and GRF9 Increases Transgenic Callus Production in Canola
Next, we explored whether ectopic expression of GRF orthologs and paralogs could result in increased transgenic callus and shoot formation in canola. In addition to AtGRF5 and its canola ortholog BnGRF5-LIKE, two further members of the AtGRF transcription factor family, i. e. AtGRF6 and AtGRF9, were also selected in order to test whether positive effects on transformation are specific to GRF5 or can be more generally attributed to the GRF gene family. The rationale to test the aforementioned two candidate genes was based on sequence homology with AtGRF6 being the most similar and AtGRF9 being the most distant GRF paralog compared to AtGRF5. In addition, AtGRF5 and AtGRF9 are both strongly expressed in the lower half of 6-day-old leaf primordia while AtGRF6 is expressed mainly near the midvein in Arabidopsis (Horiguchi et al., 2005).
The GRF ortholog and paralog expression cassettes were introduced into the canola genome using a hypocotyl transformation method (Radke et al., 1988). Although shoot production occurs through organogenesis in this method, the shoot primordia arise from callus tissue induced from hypocotyl segments that are targeted for T-DNA integration. To test whether overexpression of GRF genes could increase transgenic callus or shoot production, explants from cultivar BNS3 were transformed with AtGRF5, BnGRF5-LIKE, AtGRF6, AtGRF9, or DsRed control vectors in two experiments. Overexpression of all the GRF5 orthologs and paralogs resulted in significant increases in explants with DsRed fluorescing sectors than compared to the control vector (Figure 4A). At this step in the process, the hypocotyl has prominent callus formation, especially at the ends of the hypocotyls. To better understand if the DsRed fluorescence was localized to developing callus tissues, DsRed expression was further characterized in one experiment. The percent of explants with DsRed callus was found to be significantly greater on explants transformed with AtGRF5 (x = 53.3; SE = 3.4; p = 1.1 * 10-4) or BnGRF5-LIKE (x = 66.7; SE = 4.0; p = 2.7 * 10-6) compared to DsRed control (x = 28.7; SE = 3.5), indicating that the majority of DsRed sectors were in callus tissues. DsRed fluorescence was also notably stronger in those explants with callus overexpressing AtGRF5 or BnGRF5-LIKE (Figure 4B). The high DsRed expression suggests the callus is undergoing rapid cell proliferation compared to the control. Shoots that formed were transplanted onto rooting media and scored for rooting or DsRed fluorescence on the plantlet. Although there was a significant increase in DsRed expressing callus, no significant difference was found in the number of rooted and/or DsRed expressing shoots between GRF5 transformed events compared to DsRed control events. These results could be interpreted as either GRF5 overexpression in canola was unfavorable for shoot formation or the high DsRed expression is inhibiting further plant development.
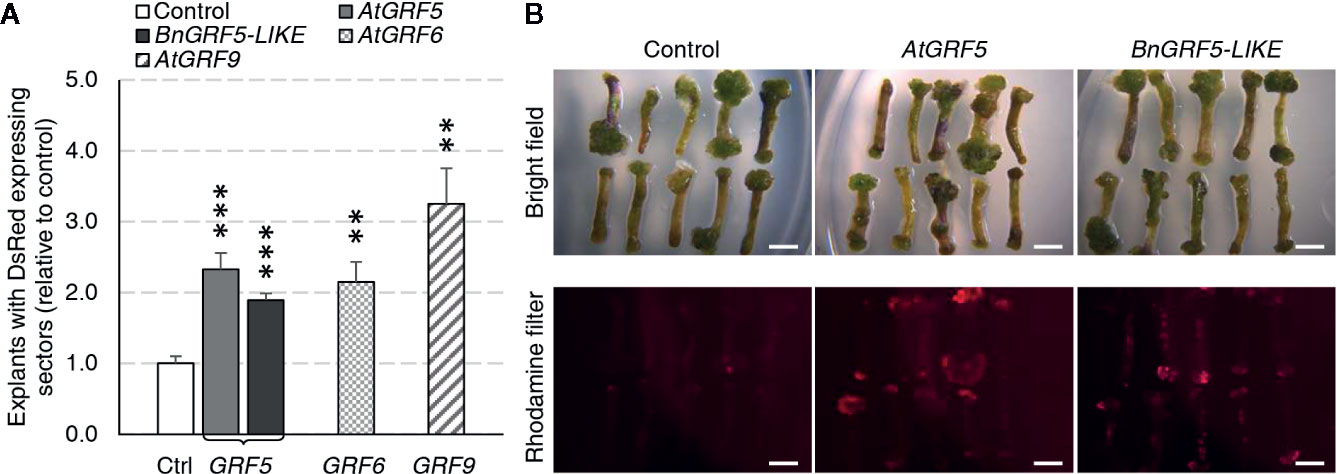
Figure 4 Canola hypocotyl sections producing callus increased in explants transformed with AtGRF5, BnGRF5-LIKE, AtGRF6, and AtGRF9 relative to DsRed control vectors. (A) Explants with DsRed expressing sectors (relative to control) were analyzed in two experiments where the first experiment had 6 biological replicates over time testing AtGRF5, BnGRF5-LIKE, or DsRed control vectors, and the second had three biological replicates over time testing AtGRF5, AtGRF6, AtGRF9 or DsRed control vectors. Explants overexpressing AtGRF5 or BnGRF5-LIKE resulted in a significant increase in DsRed sectors 38 days after transformation relative to DsRed control vector. In all graphs, differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; **p<0.01, ***p<0.001. (B) Representative pictures of explants 21 days after transformation transformed with vectors containing DsRed control, AtGRF5, and BnGRF5-LIKE show a relative increase in size and intensity of DsRed in overexpression cassettes of both GRF5 orthologs compared to DsRed control. Callus development on the hypocotyl segments under white light is shown on the top panel and DsRed fluorescence under a rhodamine filter on explants in the lower panel. Scale bar=5mm.
Overexpression of GRF5 Promotes Shoot Regeneration Directly From Axillary Meristems via Organogenesis in Soybean
We first wanted to address whether overexpression of GRF5 orthologs from Arabidopsis and soybean can increase transgenic shoot production in soybean directly from axillary meristematic cells. To answer this question, a series of experiments were initiated using two cultivars, Jake and CD215, and assayed for the presence of DsRed fluorescence in shoots developing at the seedling’s primary node 27 days after transformation. In the first experiment, prepared explants of the cultivar Jake were transformed with the DsRed control, AtGRF5, or GmGRF5-LIKE. In three experiments, explant material from the cultivar CD215 was transformed with GmGRF5-LIKE, AtGRF5, or DsRed control vectors. Significantly more explants developed shoots expressing DsRed when transformed with AtGRF5 or GmGRF5-LIKE compared to the DsRed control vector for both cultivars (Figure 5A). In addition, the explants transformed with GRF5 genes, especially GmGRF5-LIKE, frequently had more shoot development per explant giving it a swollen appearance at the primary node. Interestingly, DsRed fluorescence was also notably stronger in shoots developing from GRF5 transformed tissue compared to the DsRed control (Supplementary Figure 4A). The appearance of more meristem initials on the primary node suggests that overexpression of GRF5 is increasing meristem and shoot formation at the axillary nodes.
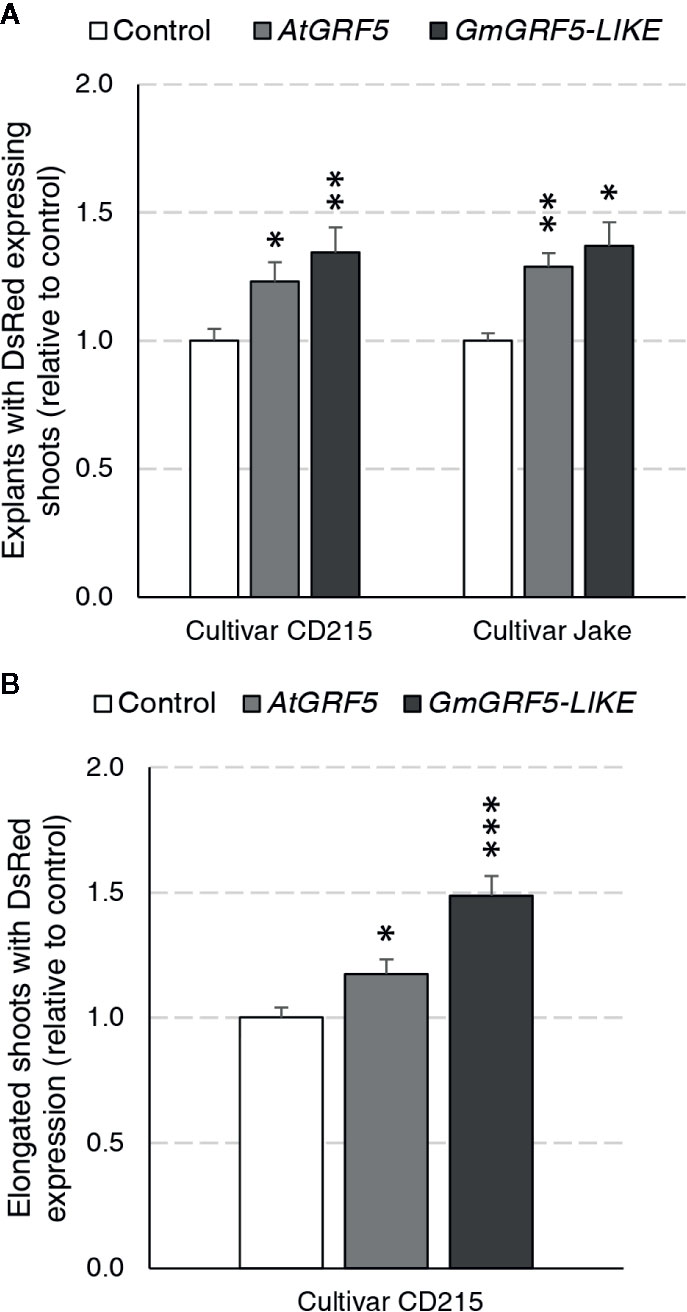
Figure 5 Soybean regeneration potential is increased in two cultivars expressing AtGRF5 or GmGRF5-LIKE transcription factors relative to control. Explants of cultivar CD215 were transformed across three experiments each with three biological replicates testing AtGRF5 or DsRed control vectors, GmGRF5-LIKE or DsRed control vectors, and GmGRF5-LIKE, AtGRF5, or DsRed control vectors. In a fourth experiment, prepared explants of the cultivar Jake were transformed with the DsRed control, AtGRF5, or GmGRF5-LIKE across four biological replicates. (A) Percent of explants with DsRed expressing shoots (normalized to control) significantly increased in explants transformed with either GRF5 ortholog in both cultivars tested 27 days after transformation. (B) For CD215 experiments, elongated shoots were removed, rooted on imazapyr selection, and scored for DsRed expression over a 5-week period. Overexpression of AtGRF5 and GmGRF5-LIKE resulted in the production of more DsRed expressing and/or rooted shoots. In both graphs, differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; *p < 0.05; *p < 0.01, ***p < 0.001.
Given that more explants produced shoots expressing DsRed with GRF5 overexpression, we wanted to know if the increase in shoot production would also result in an increase of stable, transgenic plants. The experiments described above were continued until shoots elongated to 10–15 cm in length (Supplementary Figure 4B). Afterwards, one elongated shoot per explant was removed, placed on selection media for seven days and then scored for the presence or absence of roots. Despite the increase in shoot production on explants overexpressing GRF5 27 days after transformation, the observed differences in transgenic plant production between GRF5 and DsRed control experiments for both cultivars were not statistically significant due to high experimental variability. In CD215, an average of 51.3% and 51.2% of explants produced rooted shoots for AtGRF5 and GmGRF5-LIKE, respectively, compared to 44.7% of the control explants. Similarly, in Jake, an average of 50.2% of explants produced rooted shoots when transformed with AtGRF5, 47.2% with GmGRF5-LIKE, and 37.1% with DsRed control. To understand if the shoots expressing DsRed did not elongate or if the elongated shoots did not root, we scored all elongated shoots for DsRed expression after 1 week on rooting (Figure 5B). When compared to the control, explants transformed with GRF5 genes produced significantly more elongated shoots expressing DsRed in the CD215 cultivar (Figure 5B). These results suggest that GRF5 genes increased production of elongated, transgenic shoots, and reduced root formation negatively impacted the whole plant regeneration.
To confirm stable T-DNA integration in the T0 plant, copy number analysis of the AtAhasL gene in 126 independent events across all replicates was performed for CD215. The results confirmed that the transgene was indeed present in 99% of the rooted plants (Supplementary Table 7). Because overexpression of developmental genes has been associated with abnormal plant development and sterility, we continued to grow 42 events of CD215 until the R6 stage in the greenhouse. We observed no unusual morphological abnormalities in the developing plants and seeds of any of the events. Transgene inheritance into the T1 generation was then determined by a presence/absence assay for the AtAhasL gene in immature embryos using TaqMan (Supplementary Table 8). In all events, the AtAhasL gene was detected in the T1 generation in at least one progeny. A chi-square test of independence was performed on single copy T0 events to examine the relation between the genes, GmGRF5-LIKE, AtGRF5, and control cassette (DsRed) and the presence of the AtAhasL gene in the T1 generation. There was no significant relationship between the constructs and the ability to transmit the AtAhasL gene to the progeny, X2 (2, N = 405) = 5.2, p = 0.08, suggesting normal inheritance of the AtGRF5 and GmGRF5 genes. In several cases, the number of null segregants was greater than expected, which could be explained by small sample size and/or the occurrence of a chimeric T0 plant, which is common using organogenesis.
Overexpression of GRF5 Promotes Transgenic Shoot Production From Cotyledons via Organogenesis in Sunflower
Regeneration of transgenic sunflower plants remains problematic. Therefore, we wanted to test if overexpression of GRF5 could increase transgenic shoot production or regeneration of plants using a low inoculum/ long co-culture transformation method that targets sunflower cotyledons (Zhang and Finer, 2016). Although shoot induction directly from the cotyledons via organogenesis is high in sunflower, transformed cells are usually concentrated at the cut sides where shoot formation rarely occurs (Zhang and Finer, 2016). Mature cotyledons of the cultivar, RHA280, were prepared and transformed with overexpression cassettes of AtGRF5, a putative sunflower ortholog, HaGRF5-LIKE (Supplementary Figure 3), or a GFP control vector. Explants that were transformed with HaGRF5-LIKE produced significantly more (p=0.005) GFP expressing shoots per explant than the control vector, while the number of explants transformed with AtGRF5 was 2-fold greater, albeit not statistically significant (Figure 6A). In addition, GFP expressing sectors were more expansive, not only on the explants, but also on the shoots when compared to the control (Figure 6B).
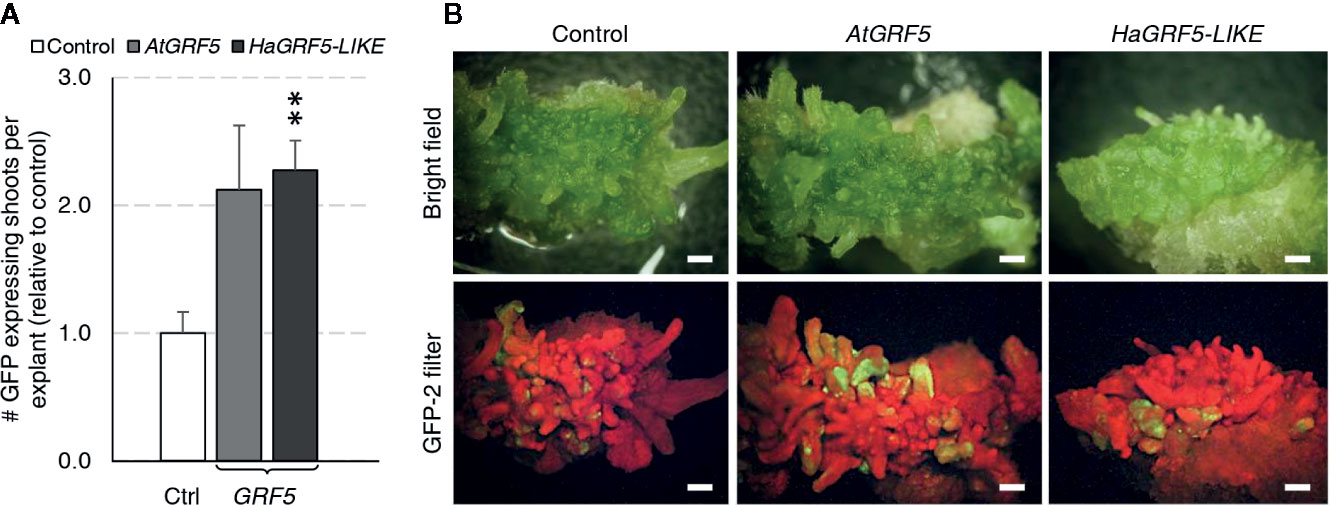
Figure 6 Overexpression of AtGRF5 and the ortholog HaGRF5-LIKE in sunflower (cv. RHA280) has a positive effect on the formation of GFP expressing shoots per explant. A Welch’s t-test was performed to determine significance of differences in averages from three experiments with seven biological replicates for GFP control, six biological replicates for AtGRF5, and 4 biological replicates for HaGRF5-LIKE vectors. (A) The number of GFP expressing shoots was significantly higher in HaGRF5-LIKE overexpressed explants relative to control, while a positive trend was observed on explants transformed with AtGRF5 (p = 0.079). Differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; **p < 0.01. (B) The size and frequency of GFP expressing shoots is more pronounced in explants transformed with GRF5. Representative pictures of explants with GFP expression under bright field (top) and GFP fluorescence (bottom) at 3 weeks post Agrobacterium inoculation with either GFP control, AtGRF5, HaGRF5-LIKE vectors are shown. Scale bar = 1 mm.
Overexpression of GRF5 Improves Somatic Embryogenesis-Based Transformation in Maize
To explore whether the GRF5 gene could boost the regeneration of transgenic plants through somatic embryogenesis, we employed a transformation protocol to introduce GRF5 constructs in the maize inbred line A188 (Ishida et al., 2007). To this end, the putative homologs ZmGRF5-LIKE1 and ZmGRF5-LIKE2 were identified as previously described for other crops (Supplementary Figure 3) and cloned under the BdEF1 promoter to ensure high expression level. Binary plasmids containing the expression cassette for either ZmGRF5-LIKE1, ZmGRF5-LIKE2, AtGRF5, and tdTomato were transformed into immature embryos. Over 50% of the embryos transformed with any of the two ZmGRF5-LIKE homologs developed type I embryogenic callus that were bigger than the control experiment at the end of the second selection round on callus induction medium (Supplementary Figure 5). Surprisingly, the size of the embryogenic calli in the transformation with the AtGRF5 construct remained comparable to the control at the same time point (Supplementary Figure 5). Quantitative analysis of callus area corroborated that the transformation of ZmGRF5-LIKE genes significantly increased the average embryogenic callus area compared to the control, whereas the transformation of the Arabidopsis counterpart had a very limited effect on the callus area (Figure 7A). These observations point out that, unlike AtGRF5, the maize GRF5 genes might intensify the proliferation of scutellum-derived embryogenic callus. In contrast to the control and the AtGRF5 transformations, a large amount of proliferating calli was obtained from the embryos transformed with ZmGRF5 at the end of the callus selection rounds (Figure 7B). Next, qPCR analysis confirmed the presence of the T-DNA in most regenerated maize plants. In general, the experiments done with the ZmGRF5-LIKE1 constructs showed a higher average transformation efficiency than the control (Figure 7C). Although a similar efficiency was observed when the ZmGRF5-LIKE2 was transformed, the effect of this ortholog was not statistically significant from controls due to a greater experimental variation. On the other hand, the transformation of the AtGRF5 gene in maize did not improve the efficiency of transgenic plant recovery (Figure 7C). The enhanced transformation efficiency is consistent with the increased embryogenic callus growth and revealed that the overexpression of ZmGRF5-LIKE genes improves transformation in maize.
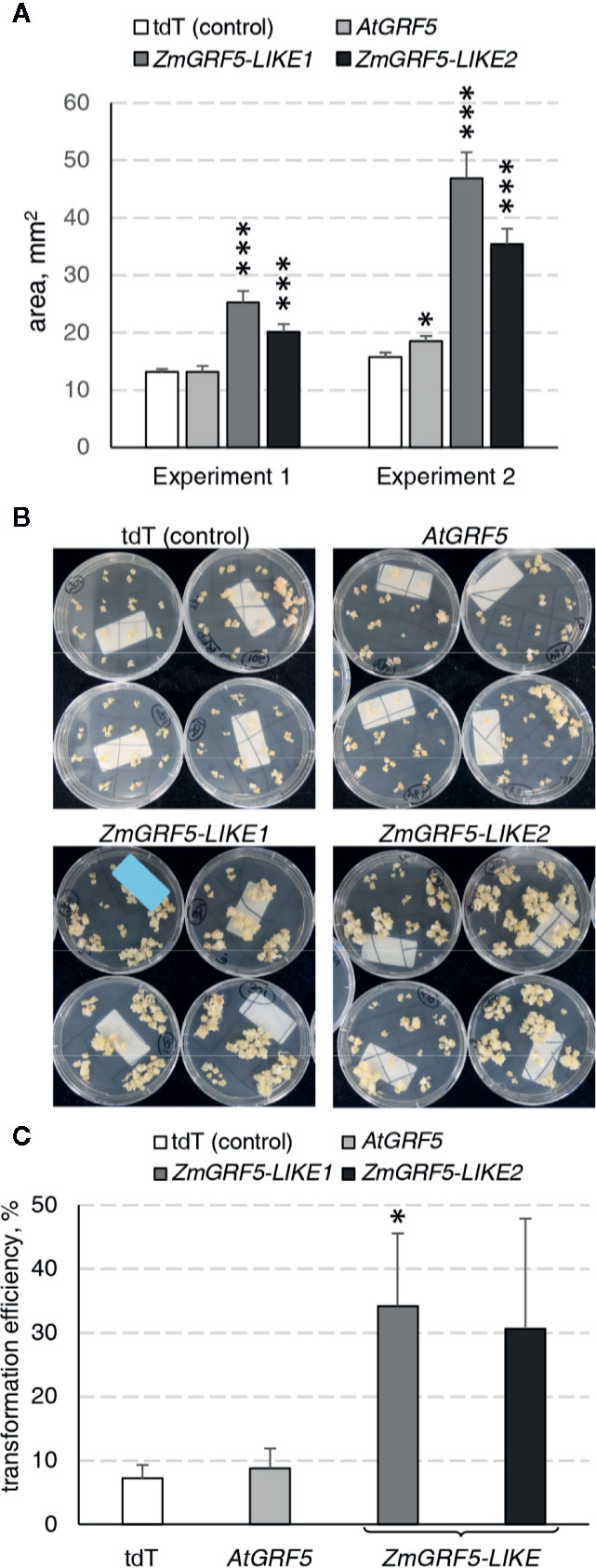
Figure 7 Analysis of the GRF5 genes in maize transformation experiments. In all experiments, constructs for either tdTomato (tdT), AtGRF5, ZmGRF5-LIKE1 or ZmGRF5-LIKE2 overexpression were used. (A) Quantification of the area of developing embryogenic calluses at 39 days after Agrobacterium inoculation. The area values are indicated as the mean ± SEM from 36 randomly selected calluses that were obtained from transformed immature embryos in each experiment. (B) Representative transgenic callus cultures at the end of the callus selection phase, before transferring to medium for regeneration of transformed plants. (C) Average transformation efficiency of the experiments performed in maize with the GRF5 and the tdT (control) constructs. The transformation efficiency values are indicated as the mean ± SEM from at least three biological replicates for tdTomato, AtGRF5 and ZmGRF5-LIKE1; and two biological replicates for ZmGRF5-LIKE2. In the graphs, differences as compared to the control are not significant unless indicated otherwise; error bars indicate standard error; *p < 0.05; ***p < 0.001.
Five randomly selected maize T0 plants for each ZmGRF5-LIKE construct were grown in the greenhouse until maturity and compared with plants transformed with the tdTomato control construct. All cultivated T0 maize plants showed normal organ morphology and produced a similar average number of seeds (Supplementary Figures 6 and 7). In addition, segregation analysis showed mendelian inheritance of the transgenes in all tested T1 progenies (Supplementary Table 9). In summary, these results demonstrate that the overexpression of ZmGRF5-LIKE genes does not impair the growth and fertility in maize and the transgene can be transmitted to the next generation.
Discussion
Despite decades of research on plant tissue culture and genetic engineering, the ability to efficiently regenerate fertile plants from transformed somatic tissues and cells is still limited. Even though there has been considerable progress enabling in vitro organogenesis and somatic embryogenesis in a variety of crops, transformation is often restricted to very few crop varieties, limiting the application of genome editing and transformation to crop improvement (Altpeter et al., 2016). Expression of developmental genes to boost regeneration has been reported as promising strategy to overcome this hurdle in transformation of plants of agricultural interest (Nagle et al., 2018; Gordon-Kamm et al., 2019). Here, we have discovered that the overexpression of the transcription factor GRF5 from Arabidopsis and/or its homologs increases transformation efficiency in sugar beet and maize. In addition, overexpression of GRF5 homologs improves transgenic shoot formation in soybean and sunflower, and transgenic callus cell proliferation in canola.
GRF5 Overexpression Boosts Transgenic Plant Production by Promoting Shoot Organogenesis in Sugar Beet
Creation of transgenic plants by shoot organogenesis from friable callus has been reported in sugar beet (Gurel et al., 2008). In these transformation protocols, transgenic callus cells were easily obtained, but the regeneration of transgenic plantlets was time consuming, difficult, and ultimately resulted in low transformation frequencies (Dhalluin et al., 1992; Kishchenko et al., 2005). In our study, we first discovered that the overexpression of AtGRF5 in sugar beet friable callus greatly promoted and accelerated shoot development, increasing the efficiency of transgenic plant production and, at the same time, decreasing its turnaround time (Figure 1). According to previous research, the recalcitrance in sugar beet is likely due to the low number of cells morphologically competent for regeneration, which are difficult to access for transformation as they are immersed within a large number of noncompetent cells within the callus (Gurel and Wren, 1995; Joersbo, 2007). The high transgenic shoot formation rates suggest that sugar beet callus cells overexpressing AtGRF5 might have acquired a regenerative advantage over the large population of noncompetent cells, which is in consonance with the function of GRF genes in regulating cell division and the pluripotent competence of cells and tissues in model plant species (Horiguchi et al., 2005; Lee et al., 2018). During de novo shoot organogenesis, cell division is required for both the generation of a new cell mass and the acquisition of shoot regeneration competency (Ikeuchi et al., 2019). In a genetic study, the grf1/2/3/4 quadruple Arabidopsis mutant was shown to produce a significant portion of seedlings lacking shoot apical meristem (Kim and Lee, 2006), indicating that GRF-mediated cell proliferation might regulate shoot meristem establishment. Thus, AtGRF5 overexpression might confer competence to the callus cells for de novo organogenesis facilitating the development of transgenic shoots. Further supporting that idea, friable callus obtained from stably AtGRF5-transformed leaf explants displayed enhanced de novo shoot formation compared to wild-type callus cultures in our experiments, which correlated with the AtGRF5 expression level (Figure 2).
More recently, a protocol for producing transgenic sugar beet plants from callus cells was published after 14 years of continuous optimization of tissue culture parameters whereby a 10% transformation efficiency was achieved (Kagami et al., 2015). By contrast, the overexpression of AtGRF5 led to transformation frequencies remarkably higher than 10%. Furthermore, the AtGRF5 overexpression also enabled efficient recovery of co-transformed events, being the first time that co-transformation is reported in this crop. Interestingly, the callus transformed with BvGRF5-LIKE showed more variable frequencies of transgenic plant production compared to the AtGRF5-expressing callus, as illustrated by the large standard error of the mean transformation efficiency. That could be attributed to a differential response of the transformed callus cells to the overexpression of AtGRF5 and BvGRF5-LIKE and their respective regulation or mode of action in sugar beet.
On the other hand, several reports have shown that sugar beet breeding lines and cultivars generally exhibit diverse in vitro responses to plant regeneration from friable callus, which illustrates strong genotype dependency of the regeneration in that crop (Dovzhenko and Koop, 2003; Ivic-Haymes and Smigocki, 2005; Tomita et al., 2013). Importantly, in our analysis AtGRF5 overexpression dramatically improved the recovery of transgenic events in several sugar beet inbred lines, including a genotype that was not amenable for transformation up to date (Figure 3). That demonstrates the potential of GRF genes in broadening the range of varieties that can be transformed in recalcitrant crops, such as sugar beet.
Overexpression of GRF5 Genes Promotes Transgenic Callus Proliferation in Canola
Next, we wondered whether GRF overexpression could improve indirect shoot organogenesis in other crops. Therefore, we decided to investigate the response of canola hypocotyl explants to the overexpression of AtGRF6 and AtGRF9, and compare with AtGRF5 and its putative Brassica napus ortholog BnGRF5-LIKE. In our analysis, the overexpression of AtGRF5 and BnGRF5-LIKE resulted in higher frequencies of callus with transgenic sectors. Likewise, AtGRF6 and AtGRF9 overexpression showed similar effects, indicating that GRF genes in general have a positive effect on promoting transgenic callus proliferation (Figure 4). Considering that, unlike several other members of the GRF family, AtGRF9 has been shown to have an opposite function in regulating cell proliferation and organ growth (Omidbakhshfard et al., 2018), it is very intriguing that the overexpression of this Arabidopsis homolog also promoted proliferation of transgenic callus cells in canola. Unlike sugar beet, there was no improvement in transgenic plant production in canola with any of the tested GRF genes. This could be attributed to differences in the transformation method where the hypocotyl explant is directly targeted for T-DNA delivery and callus is initiated from the explant. Shoot formation from the callus only occurs after several rounds of subcultures, which may explain some of the differences in plant production between canola and sugar beet.
GRF5 Overexpression Improves Transgenic Shoot Formation in Soybean and Sunflower
To further explore the effect of GRF overexpression in organogenesis, AtGRF5 and their respective soybean and sunflower orthologs were transformed using published methods, which are based on direct shoot organogenesis (Olhoft et al., 2007; Chang et al., 2012; Zhang and Finer, 2016). GRF5 overexpression in soybean resulted in visibly more meristematic initials at the axillary meristem, indicating increased cell division activity. Furthermore, transgenic shoot production at the axillary node was notably improved (Figure 5). Although most of the GRF5-expressing shoots developed well, a proportion of the elongating shoots that were expressing DsRed did not form roots, indicating that GRF5 or DsRed expression may negatively affect adventitious root initiation. By contrast, overexpression of GRF5 genes in sugar beet and canola plants did not impair root growth, suggesting that the effect of GRF5 on rooting might be crop specific, dependent on the promoter used, or due to the combination of GRF5 overexpression and the regeneration method. Similarly, the frequency of explants containing transgenic shoots significantly increased when GRF5 genes were transformed in sunflower, suggesting an effect on proliferation of transformed tissues (Figure 6). The results obtained in these two crops are again consistent with the notion that GRF genes positively regulate cell division in growing plant organs (Kim, 2019). Indeed, Arabidopsis plants overexpressing AtGRF5 develop enlarged leaves containing more cells, which was also accompanied with increased expression of cell cycle genes (Horiguchi et al., 2005; Gonzalez et al., 2010; Debernardi et al., 2014).
Overall, although GRF overexpression improved transgenic cell proliferation in canola, sunflower and soybean, transformation efficiency was not always increased. A possibility that cannot be ruled out is that differences in the transformation protocols, such as the target explant and phytohormones used for regeneration, might interact with the overexpression of GRFs. For instance, it was shown that the overexpression of AtGRF5 increases the sensitivity of Arabidopsis leaves to cytokinins and stimulated leaf growth when co-expressed with cytokinin catabolic enzymes (Vercruyssen et al., 2011; Vercruyssen et al., 2015). Several research groups also link the GRFs with other hormone pathways including brassinosteroids, gibberellins and auxin (Che et al., 2016; Gao et al., 2016; Lee et al., 2018; Zhang et al., 2018). It remains to be determined how GRF is linked to the hormone pathways related to plant regeneration.
Overexpression of ZmGRF5 Genes Enhances Transformation Based on Somatic Embryogenesis in Maize
Besides the positive effect on organogenesis, further analyses uncovered that overexpression of GRF5 genes promotes regeneration in somatic embryogenesis-based transformation protocols in maize. Qualitative and quantitative analysis revealed that the overexpression of ZmGRF5-LIKE genes enhanced the growth of embryogenic callus, suggesting that ZmGRF5-LIKE homologs might have a positive influence on cell proliferation, thereby boosting the formation of somatic embryos and enhancing the transformation efficiency (Figure 7). According to the nomenclature used by RefSeq and adopted by Debernardi et al. (2020) the ZmGRF5 genes tested in our study are orthologous to the wheat GRF1 gene, which has also showed a positive effect on wheat transformation in a concurrent research (Debernardi et al., 2020). Like in dicot plants, GRF-promoting cell division and organ growth has also been described in monocots (Nelissen et al., 2015), which is again in accordance with the enhanced growth of transgenic embryogenic callus reported here. Interestingly, unlike the ZmGRF5-LIKE genes, overexpression of AtGRF5 in maize only slightly promoted proliferation of maize embryogenic callus and did not enhanced transformation efficiency, suggesting functional diversification of the monocot and dicot GRF genes.
Although little is known about the role of GRF genes in somatic embryogenesis, a recent publication demonstrated that the overexpression of AtGRF1 resulted in increased sensitivity of Arabidopsis explants to 2,4-D treatment and improved induction of somatic embryogenesis (Szczygiel-Sommer and Gaj, 2019). In our maize transformation protocol, the induction of somatic embryogenesis is achieved on 2,4-D-containing medium. Therefore, one can hypothesize that the overexpression of ZmGRF5-LIKE1 or ZmGRF5-LIKE2 might increase the sensitivity of the maize explants to 2,4-D, consequently improving their embryogenic response.
Finally, a technological advance employing Agrobacterium-mediated introduction of the BABYBOOM (BBM) and WUSCHEL (WUS) transcription factors greatly improved transformation efficiency in monocot crops, including maize recalcitrant varieties (Lowe et al., 2016; Lowe et al., 2018). However, constitutive expression of these transcription factors led to impaired plant development and sterility, which obliged researchers to develop strategies to limit expression to the first steps of the transformation process (Lowe et al., 2016). By contrast, the transgenic plants overexpressing either AtGRF5 or GRF5 orthologs in this study showed normal growth until maturity and produced viable seeds.
Concluding Remarks and Future Prospects
In summary, we have demonstrated that members of the GRF transcription factor family can be used to improve transformation protocols based on either organogenesis or embryogenesis. In light of our results, it appears that the GRF-mediated regeneration improves genetic transformation in various plant species, including dicotyledonous and monocotyledonous crops. Interestingly, high transformation efficiencies were achieved in recalcitrant sugar beet genotypes, revealing, that GRF genes can be applied to decrease genotype-dependency of transformation protocols.
This work presents the first evidence that members of the GRF gene family can trigger cell reprogramming to accomplish more efficient in vitro regeneration and transformation. We hypothesize that the effect of AtGRF5 and its counterparts on transformation in the crops analyzed in this study, can be mostly attributed to the well-known role of GRF transcription factors in regulating crucial developmental processes, such as cell proliferation. Accumulating evidence determined that the growth of numerous plant organs is controlled by a conserved gene network consisting of GRFs, GRF-INTERACTING FACTOR transcriptional co-regulators (GIF) and miR396 (Kim and Kende, 2004; Liebsch and Palatnik, 2020). Among other GRFs, AtGRF5 was also shown to interact with AtGIF1 (Horiguchi et al., 2005; Vercruyssen et al., 2014). Preliminary data suggest that co-expression of AtGIF1 and AtGRF5 induces a more pronounced effect on regeneration (data not shown). In line with this finding, researchers at UC Davis concurrently observed that overexpression of a chimeric protein containing GRF and its transcriptional co-activator GIF greatly increased the transformation efficiency in wheat, rice and citrus (Debernardi et al., 2020). In addition to the interaction with GRFs, GIF1 was shown to physically interact with SWI/SNF chromatin remodeling members to regulate transcription of various genes (Debernardi et al., 2014; Nelissen et al., 2015). The SWI/SNF complex is known to be antagonistic to the polycomb PRC2 complex (Wilson et al., 2011). Interestingly, mutations in the PRC2 complex subunits led to upregulation of downstream targets WIND3 and LEC2, which promote regeneration (Ikeuchi et al., 2015). One can hypothesize that GRF5 might interact with the GIF1-SWI/SNF complex in order to inhibit the polycomb PRC2 complex, thereby releasing its repression on other developmental regulators, consequently triggering cell reprogramming (Ikeuchi et al., 2015). Continuous efforts to unravel the function of GRF genes in the context of transcriptional modulation of key meristem or embryonic regulators would contribute to our fundamental understanding of the molecular pathways determining cell reprogramming during plant in vitro regeneration. In addition, further optimization of protocols would help to find specific conditions in which GRF-mediated regeneration boosting leads to higher transformation efficiencies in recalcitrant crops.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
Conceived and designed the experiments: JK, SM-O, DP-V, PO, JF, OS, and MS. Performed the experiments: JK, SM-O, NO, AG, JF, LB, DT, BR, and PO. Analyzed the data: OS and DP-V. Wrote the paper: JK, SM-O, MS, OS, PO, and DP-V.
Conflict of Interest
The authors declare that the reported results were in part filed in the following patent application: WO 2019/134884 A1. Authors JK, SM-O, and DP-V were employed by the company KWS SAAT SE & Co. KGaA. Authors LB, BR, OS, MS, and PO were employed by BASF. DT conducted research for this manuscript being employed by BASF and his current address is at Roche.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Linda Schwerdtfeger, Franziska Mense, Heike Lehmann, and Robin Pfeil for excellent technical assistance on transformation experiments at KWS; Benjamin Gruber for critical reading of the manuscript; Florian Schauwecker for conducting sequence analyses to derive GRF5 orthologs; James Silva Garcia for statistical analyses; Allan Wenck for critical review of manuscript; Fernando Ferreira for vector design and construction; Zhong Qi Wang, Shyam Barampuram, Harvinder Kaur, Minesh Patel, Yan Liu, Brandi Chappell and Mao Wang for executing and delivering on transformation experiments at BASF.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.572319/full#supplementary-material
References
Altpeter, F., Springer, N. M., Bartley, L. E., Blechl, A. E., Brutnell, T. P., Citovsky, V., et al. (2016). Advancing crop transformation in the era of genome editing. Plant Cell 28 (7), 1510–1520. doi: 10.1105/tpc.16.00196
Arroyo-Herrera, A., Gonzalez, A. K., Moo, R. C., Quiroz-Figueroa, F. R., Loyola-Vargas, V., Rodriguez-Zapata, L., et al. (2008). Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ Culture 94 (2), 171–180. doi: 10.1007/s11240-008-9401-1
Banno, H., Ikeda, Y., Niu, Q. W., Chua, N. H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13 (12), 2609–2618. doi: 10.1105/tpc.13.12.2609
Bouchabke-Coussa, O., Obellianne, M., Linderme, D., Montes, E., Maia-Grondard, A., Vilaine, F., et al. (2013). Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 32 (5), 675–686. doi: 10.1007/s00299-013-1402-9
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 (8), 1737–1749. doi: 10.1105/tpc.001941
Chang, Y.-F. V. A. S., Grist, L., Tuttle, H., Hong, H.-P., Olhoft, P. (2012). Plant transformation Method, US 2014237688 [A1] / WO 2013014585 [A1].
Che, R. H., Tong, H. N., Shi, B. H., Liu, Y. Q., Fang, S. R., Liu, D. P., et al. (2016). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2 (1), 1–7. doi: 10.1038/nplants.2015.195
Cheng, M., Lowe, B. A., Spencer, T. M., Ye, X., Armstrong, C. L. (2004). Factors influencing Agrobacterium-mediated transformation of monocotyledonous species. Vitro Cell. Dev. Biology-Plant 40 (1), 31–45. doi: 10.1079/IVP2003501
Debernardi, J. M., Mecchia, M. A., Vercruyssen, L., Smaczniak, C., Kaufmann, K., Inze, D., et al. (2014). Post-transcriptional control of GRF transcription factors by micro RNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79 (3), 413–426. doi: 10.1111/tpj.12567
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. in press.
Deng, W., Luo, K., Li, Z., Yang, Y. (2009). A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci. 177 (1), 43–48. doi: 10.1016/j.plantsci.2009.03.009
Dhalluin, K., Bossut, M., Bonne, E., Mazur, B., Leemans, J., Botterman, J. (1992). Transformation of Sugar-Beet (Beta-Vulgaris L) and Evaluation of Herbicide Resistance in Transgenic Plants. Bio-Technology 10 (3), 309–314. doi: 10.1038/nbt0392-309
Dovzhenko, A., Koop, H. U. (2003). Sugarbeet (Beta vulgaris L.): shoot regeneration from callus and callus protoplasts. Planta 217 (3), 374–381. doi: 10.1007/s00425-003-1006-7
Duclercq, J., Sangwan-Norreel, B., Catterou, M., Sangwan, R. S. (2011). De novo shoot organogenesis: from art to science. Trends Plant Sci. 16 (11), 597–606. doi: 10.1016/j.tplants.2011.08.004
Florez, S. L., Erwin, R. L., Maximova, S. N., Guiltinan, M. J., Curtis, W. R. (2015). Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol. 15 (1), 121. doi: 10.1186/s12870-015-0479-4
Gaillochet, C., Lohmann, J. U. (2015). The never-ending story: from pluripotency to plant developmental plasticity. Development 142 (13), 2237–2249. doi: 10.1242/dev.117614
Gallois, J.-L., Woodward, C., Reddy, G. V., Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129 (13), 3207–3217.
Gao, F., Wang, K., Liu, Y., Chen, Y., Chen, P., Shi, Z., et al. (2016). Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2 (1), 1–9. doi: 10.1038/nplants.2015.196
George, E. F., Hall, M. A., De Klerk, G.-J. (2008). Plant propagation by tissue culture 3rd Edition. Netherland Back Ground Springer 1, 1–502. doi: 10.1007/978-1-4020-5005-3
Gonzalez, N., De Bodt, S., Sulpice, R., Jikumaru, Y., Chae, E., Dhondt, S., et al. (2010). Increased leaf size: different means to an end. Plant Physiol. 153 (3), 1261–1279. doi: 10.1104/pp.110.156018
Gordon, S. P., Heisler, M. G., Reddy, G. V., Ohno, C., Das, P., Meyerowitz, E. M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134 (19), 3539–3548. doi: 10.1242/dev.010298
Gordon-Kamm, B., Sardesai, N., Arling, M., Lowe, K., Hoerster, G., Betts, S. (2019). Using Morphogenic Genes to Improve Recovery and Regeneration of Transgenic Plants. Plants 8 (2), 38. doi: 10.3390/plants8020038
Gurel, E., Wren, M. J. (1995). In-Vitro Development From Leaf Explants Of Sugar-Beet (Beta-Vulgaris L) - Rhizogenesis And The Effect Of Sequential Exposure To Auxin And Cytokinin. Ann. Bot. 75 (1), 31–38. doi: 10.1016/s0305-7364(05)80006-x
Gurel, E., Gurel, S., Lemaux, P. G. (2008). Biotechnology applications for sugar beet. Crit. Rev. Plant Sci. 27 (2), 108–140. doi: 10.1080/07352680802202000
Heidmann, I., De Lange, B., Lambalk, J., Angenent, G. C., Boutilier, K. (2011). Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 30 (6), 1107–1115. doi: 10.1007/s00299-011-1018-x
Horiguchi, G., Kim, G. T., Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43 (1), 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Horiguchi, G., Ferjani, A., Fujikura, U., Tsukaya, H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119 (1), 37–42. doi: 10.1007/s10265-005-0232-4
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017). The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175 (2), 848–857. doi: 10.1104/pp.17.00232
Ikeuchi, M., Sugimoto, K., Iwase, A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25 (9), 3159–3173. doi: 10.1105/tpc.113.116053
Ikeuchi, M., Iwase, A., Rymen, B., Harashima, H., Shibata, M., Ohnuma, M., et al. (2015). PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1 (7), 1–7. doi: 10.1038/nplants.2015.89
Ikeuchi, M., Ogawa, Y., Iwase, A., Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143 (9), 1442–1451. doi: 10.1242/dev.134668
Ikeuchi, M., Favero, D. S., Sakamoto, Y., Iwase, A., Coleman, D., Rymen, B., et al. (2019). Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 70, 377–406. doi: 10.1146/annurev-arplant-050718-100434
Ishida, Y., Hiei, Y., Komari, T. (2007). Agrobacterium-mediated transformation of maize. Nat. Protoc. 2 (7), 1614. doi: 10.1038/nprot.2007.241
Ivic-Haymes, S. D., Smigocki, A. C. (2005). Identification of highly regenerative plants within sugar beet (Beta vulgaris L.) breeding lines for molecular breeding. Vitro Cell. Dev. Biology-Plant 41 (4), 483–488. doi: 10.1079/ivp2005666
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Arai, T., Inoue, Y., et al. (2007). Functional analyses of an Arabidopsis transcription factor that is involved in callus formation. Plant Cell Physiol. 48, S120–S120. doi: 10.14841/jspp.2007.0.414.0
Iwase, A., Mita, K., Nonaka, S., Ikeuchi, M., Koizuka, C., Ohnuma, M., et al. (2015). WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J. Plant Res. 128 (3), 389–397. doi: 10.1007/s10265-015-0714-y
Joersbo, M. (2007). “Sugar Beet,” in Transgenic Crops IV. Eds. Pua, E.-C., Davey, M. R. (Berlin, Heidelberg: Springer Berlin Heidelberg), 355–379.
Kagami, H., Kurata, M., Matsuhira, H., Taguchi, K., Mikami, T., Tamagake, H., et al. (2015). “Sugar beet (Beta vulgaris L.),” in Agrobacterium Protocols (New York: Springer) 335–347.
Kareem, A., Durgaprasad, K., Sugimoto, K., Du, Y., Pulianmackal, A. J., Trivedi, Z. B., et al. (2015). PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25 (8), 1017–1030. doi: 10.1016/j.cub.2015.02.022
Kareem, A., Radhakrishnan, D., Sondhi, Y., Aiyaz, M., Roy, M. V., Sugimoto, K., et al. (2016). De novo assembly of plant body plan: a step ahead of Deadpool. Regeneration 3 (4), 182–197. doi: 10.1002/reg2.68
Kausch, A. P., Nelson-Vasilchik, K., Hague, J., Mookkan, M., Quemada, H., Dellaporta, S., et al (2019). Edit at will: Genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci. 281, 186–205. doi: 10.1016/j.plantsci.2019.01.006
Kim, J. H., Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. 101 (36), 13374–13379. doi: 10.1073/pnas.0405450101
Kim, J. H., Lee, B. H. (2006). GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49 (6), 463–468. doi: 10.1007/Bf03031127
Kim, J. H., Tsukaya, H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66 (20), 6093–6107. doi: 10.1093/jxb/erv349
Kim, J. H., Choi, D. S., Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36 (1), 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, J. H. (2019). Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 52 (4), 227. doi: 10.5483/BMBRep.2019.52.4.051
Kishchenko, E. M., Komarnitskii, I. K., Kuchuk, N. V. (2005). Production of transgenetic sugarbeet (Beta vulgaris L.) plants resistant to phosphinothricin. Cell Biol. Int. 29 (1), 15–19. doi: 10.1016/j.cellbi.2004.11.003
Lee, S.-J., Lee, B. H., Jung, J.-H., Park, S. K., Song, J. T., Kim, J. H. (2018). GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 176 (1), 717–729. doi: 10.1104/pp.17.00960
Liebsch, D., Palatnik, J. F. (2020). MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 53, 31–42. doi: 10.1016/j.pbi.2019.09.008
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M.-J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28 (9), 1998–2015. doi: 10.1105/tpc.16.00124
Lowe, K., La Rota, M., Hoerster, G., Hastings, C., Wang, N., Chamberlin, M., et al. (2018). Rapid genotype “independent” Zea mays L.(maize) transformation via direct somatic embryogenesis. Vitro Cell. Dev. Biology-Plant 54 (3), 240–252. doi: 10.1007/s11627-018-9905-2
Mankin, S. L., Hill, D. S., Olhoft, P. M., Toren, E., Wenck, A. R., Nea, L., et al. (2007). Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. Vitro Cell. Dev. Biol. Plant 43 (6), 521–535. doi: 10.1007/s11627-007-9071-4
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., Avilez-Montalvo, J., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10, 77. doi: 10.3389/fpls.2019.00077
Mookkan, M., Nelson-Vasilchik, K., Hague, J., Zhang, Z. J., Kausch, A. P. (2017). Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 36 (9), 1477–1491. doi: 10.1007/s00299-017-2169-1
Nagle, M., Dejardin, A., Pilate, G., Strauss, S. H. (2018). Opportunities for Innovation in Genetic Transformation of Forest Trees. Front. Plant Sci. 9, 1443. doi: 10.3389/fpls.2018.01443
Nelissen, H., Eeckhout, D., Demuynck, K., Persiau, G., Walton, A., Van Bel, M., et al. (2015). Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27 (6), 1605–1619. doi: 10.1105/tpc.15.00269
Olhoft, P. M., Bernal, L. M., Grist, L. B., Hill, D. S., Mankin, S. L., Shen, Y., et al. (2007). A novel Agrobacterium rhizogenes-mediated transformation method of soybean [Glycine max (L.) Merrill] using primary-node explants from seedlings. Vitro Cell. Dev. Biol. Plant 43 (6), 536–549. doi: 10.1007/s11627-007-9050-9
Omidbakhshfard, M. A., Proost, S., Fujikura, U., Mueller-Roeber, B. (2015). Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8 (7), 998–1010. doi: 10.1016/j.molp.2015.01.013
Omidbakhshfard, M. A., Fujikura, U., Olas, J. J., Xue, G.-P., Balazadeh, S., Mueller-Roeber, B. (2018). GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PloS Genet. 14 (7), 1–31. doi: 10.1371/journal.pgen.1007484
Perez-Garcia, P., Moreno-Risueno, M. A. (2018). Stem cells and plant regeneration. Dev. Biol. 442 (1), 3–12. doi: 10.1016/j.ydbio.2018.06.021
Radke, S. E., Andrews, B. M., Moloney, M. M., Crouch, M. L., Kridl, J. C., Knauf, V. C. (1988). Transformation of brassica-napus l using agrobacterium-tumefaciens - developmentally regulated expression of a reintroduced napin gene. Theor. Appl. Genet. 75 (5), 685–694. doi: 10.1007/bf00265588
Russell, D. W., Sambrook, J. (2001). Molecular cloning: a laboratory manual (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory).
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9 (7), 676–682. doi: 10.1038/nmeth.2019
Solís-Ramos, L., González-Estrada, T., Nahuath-Dzib, S., Zapata-Rodriguez, L., Castaño, E. (2009). Overexpression of WUSCHEL in C. chinense causes ectopic morphogenesis. Plant Cell Tissue Organ Culture (PCTOC) 96 (3), 279–287. doi: 10.1007/s11240-008-9485-7
Somssich, M., Je, B. I., Simon, R., Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143 (18), 3238–3248. doi: 10.1242/dev.133645
Srinivasan, C., Liu, Z., Heidmann, I., Supena, E. D. J., Fukuoka, H., Joosen, R., et al. (2007). Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta 225 (2), 341. doi: 10.1007/s00425-006-0358-1
Sugimoto, K., Temman, H., Kadokura, S., Matsunaga, S. (2019). To regenerate or not to regenerate: factors that drive plant regeneration. Curr. Opin. Plant Biol. 47, 138–150. doi: 10.1016/j.pbi.2018.12.002
Sugiyama, M. (2015). Historical review of research on plant cell dedifferentiation. J. Plant Res. 128 (3), 349–359. doi: 10.1007/s10265-015-0706-y
Szczygiel-Sommer, A., Gaj, M. D. (2019). The miR396-GRF Regulatory Module Controls the Embryogenic Response in Arabidopsis via an Auxin-Related Pathway. Int. J. Mol. Sci. 20 (20), 18. doi: 10.3390/ijms20205221
Tomita, K.-i., Hiura, S., Tamagake, H. (2013). Evaluation of the potential for somatic embryogenesis in sugar beet (Beta vulgaris L.) breeding lines and improvement of regeneration efficiency. Plant Biotechnol. 30 (5), 479–487. doi: 10.5511/plantbiotechnology.13.0816a
van der Knaap, E., Kim, J. H., Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122 (3), 695–704. doi: 10.1104/pp.122.3.695
Vercruyssen, L., Gonzalez, N., Werner, T., Schmülling, T., Inzé, D. (2011). Combining enhanced root and shoot growth reveals cross talk between pathways that control plant organ size in Arabidopsis. Plant Physiol. 155 (3), 1339–1352. doi: 10.1104/pp.110.167049
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S.-K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26 (1), 210–229. doi: 10.1105/tpc.113.115907
Vercruyssen, L., Tognetti, V. B., Gonzalez, N., Van Dingenen, J., De Milde, L., Bielach, A., et al. (2015). GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 167 (3), 817–832. doi: 10.1104/pp.114.256180
Wilson, B. G., Wang, X., Shen, X., McKenna, E. S., Lemieux, M. E., Cho, Y.-J., et al. (2011). Epigenetic Antagonism between Polycomb and SWI/SNF Complexes during Oncogenic Transformation (vol 182010). Cancer Cell 19 (1), 153–153, pg 316. doi: 10.1016/j.ccr.2011.01.016
Zhang, Z., Finer, J. J. (2016). Low Agrobacterium tumefaciens inoculum levels and a long co-culture period lead to reduced plant defense responses and increase transgenic shoot production of sunflower (Helianthus annuus L.). Vitro Cell. Dev. Biol. Plant 52 (4), 354–366. doi: 10.1007/s11627-016-9774-5
Zhang, D., Sun, W., Singh, R., Zheng, Y., Cao, Z., Li, M., et al. (2018). GRF-interacting factor1 Regulates Shoot Architecture and Meristem Determinacy in Maize. Plant Cell 30 (2), 360–374. doi: 10.1105/tpc.17.00791
Keywords: transformation, GROWTH-REGULATING FACTOR, regeneration, organogenesis, embryogenesis, monocot, dicot, crop
Citation: Kong J, Martin-Ortigosa S, Finer J, Orchard N, Gunadi A, Batts LA, Thakare D, Rush B, Schmitz O, Stuiver M, Olhoft P and Pacheco-Villalobos D (2020) Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species. Front. Plant Sci. 11:572319. doi: 10.3389/fpls.2020.572319
Received: 13 June 2020; Accepted: 21 August 2020;
Published: 12 October 2020.
Edited by:
Ryozo Imai, National Agriculture and Food Research Organization (NARO), JapanReviewed by:
Lin Xu, Shanghai Institutes for Biological Sciences (CAS), ChinaSerena Varotto, University of Padua, Italy
Copyright © 2020 Kong, Martin-Ortigosa, Finer, Orchard, Gunadi, Batts, Thakare, Rush, Schmitz, Stuiver, Olhoft and Pacheco-Villalobos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paula Olhoft, cGF1bGEub2xob2Z0QGJhc2YuY29t; David Pacheco-Villalobos, ZGF2aWQucGFjaGVjb3ZpbGxhbG9ib3NAa3dzLmNvbQ==
 Jixiang Kong
Jixiang Kong Susana Martin-Ortigosa1
Susana Martin-Ortigosa1 Nuananong Orchard
Nuananong Orchard Andika Gunadi
Andika Gunadi Lou Ann Batts
Lou Ann Batts Dhiraj Thakare
Dhiraj Thakare Oliver Schmitz
Oliver Schmitz Maarten Stuiver
Maarten Stuiver David Pacheco-Villalobos
David Pacheco-Villalobos