- 1Plant Virology and Molecular Plant Pathology Laboratory, Division of Plant Pathology, SKUAST-Kashmir, Srinagar, India
- 2Directorate of Extension, SKUAST-Kashmir, Srinagar, India
- 3Division of Genetics and Plant Breeding, Faculty of Agriculture, SKUAST-Kashmir, Wadura, India
- 4Ambri Apple Research Centre, SKUAST-Kashmir, Srinagar, India
- 5Department of Plant Pathology, CSK HPKV, Palampur, India
Common bean (Phaseolus vulgaris L.) is an important legume crop of north-western (NW) Himalayan region and the major disease that causes catastrophic loss to the crop is anthracnose, which is caused by Colletotrichum lindemuthianum. The pathogen is highly diverse and most of the commercial cultivars are susceptible to different races prevalent in the region. The lack of information on the genomic regions associated with anthracnose resistance in NW Himalayan common bean population prompted us to dissect Quantitative Resistance Loci (QRLs) against major anthracnose races. In this study, 188 common bean landraces collected from NW region were screened against five important anthracnose races and 113 bean genotypes showed resistance to one or multiple races. Genotyping by sequencing (GBS) was performed on a panel of 192 bean lines (4 controls plus 188 Indian beans) and 22,589 SNPs were obtained that are evenly distributed. Population structure analysis of 192 bean genotypes categorized 188 Indian beans into two major clusters representing Andean and Mesoamerican gene pools with obvious admixtures. Many QRLs associated with anthracnose resistance to Indian C. lindemuthianum virulences (race 3, 87, and 503) are located at Pv04 within the gene models that encode typical resistance gene signatures. The QRLs associated with race 73 are located on Pv08 and overlaps with Co-4 anthracnose resistance gene. A SNP located at distal end of Pv11 in a gene model Phvul.011G202300 which encodes a LRR with a typical NB-ARC domain showed association with race 73 resistance. Common bean genomic regions located at Pv03, Pv09, and Pv11 showed association with resistance to anthracnose race 2047. The present study showed presence of many novel bean genomic regions associated with anthracnose resistance. The presence of Co-4 and Co-2 genes in our material is encouraging for breeding durable anthracnose resistant cultivars for the region.
Introduction
By 2050, the world population may increase by 2 billion and the important challenge before the humanity is to achieve food security and livelihood especially in developing countries. There is a need to increase the productivity of cheaper yet nutritionally rich crops. In this backdrop, Phaseolus vulgaris, a grain legume crop is a key commodity because of its rich nutrient profile (Broughton et al., 2003). Further, the nitrogen fixing capacity of bean crop enhances the soil fertility besides reduces environmental hazards of fertilizer application (Beaver et al., 2003; Rispail et al., 2010; Kamfwa et al., 2019). Phaseolus vulgaris is native to Andean and Mesoamerican regions where independent domestication resulted into two distinct major gene pools (Singh et al., 1991; Bitocchi et al., 2013; Bellucci et al., 2014; Pathania et al., 2014). Countries such as China, Africa, Europe, and Latin America are considered as secondary centers of beans (Duran et al., 2005; Asfaw et al., 2009; Pathania et al., 2014).
Common bean is an important legume crop of north-western (NW) Himalayan region, the largest producer of dry beans in India (Sofi et al., 2014; Rana et al., 2015). The Himalayan region is rich in common bean biodiversity and there is a scarcity of information about its dissemination in India (Pathania et al., 2014; Sofi et al., 2014). Europeans and Chinese who travelled to India for trading brought bright colored beans with them (Joshi and Thomas, 1987; Joshi and Mehra, 1993). Besides natural selection, farmers in the region selected bean genotypes according to their adaptability, taste and agronomic features that further enhanced bean diversity (Pathania et al., 2014). Common beans in India are grown over an area of 9.1 million ha with production of 3.63 million tons and yield of 399.0 kg/ha (Rana et al., 2015; Choudhary et al., 2018). Being nutritionally rich, it is socially an important crop in the NW states of India (Tiwari et al., 2005; Rana et al., 2012; Rana et al., 2015). The landraces particularly grown in the mountainous regions fetch higher market price (Rai et al., 2010; Rana et al., 2012). There are many famous bean landraces in the NW part of India such as “Chamba”, “Barot”, “Kinnauree”, “Auli”, “Munsiyari”, “Harshi”, “Bhaderwah”, and “Kashmiri” (Rana et al., 2015).
Many fungal, bacterial, and viral diseases infect common beans in northern India, but bean anthracnose caused by the hemibiotrophic fungus, Colletotrichum lindemuthianum, is a serious disease of P. vulgaris globally (Pastor-Corrales and Tu, 1989; Kumar et al., 1999; Pathania et al., 2006; Sharma et al., 2007; Sharma et al., 2008; Padder et al., 2010; Oblessuc et al., 2012). The pathogen is seed borne and planting of infected seed causes 80 to 100% yield loss (Sharma et al., 2008; Padder et al., 2017). Farmers in northern India plant their own saved seed that predisposes the crop to seed borne anthracnose infection. Further, the majority of the cultivated areas in the NW region of India experience frequent and high intensity rainfall during the cropping season, providing favorable conditions for the disease development; hence, anthracnose reoccurs regularly (Sharma et al., 1994; Sharma et al., 1999; Pathania et al., 2006; Sharma et al., 2007; Padder et al., 2009; Sharma et al., 2012). The pathogen is highly variable and 45 races are present in the NW region (Sharma et al., 1999; Pathania et al., 2006; Padder et al., 2007; Sharma et al., 2007; Padder et al., 2009; Padder et al., 2010; Sharma et al., 2019). Most of these races are different from those in the United States, Brazil, France, Canada, and many other countries. Being pathogenically variable, majority of commercially grown cultivars are susceptible to one or the other race(s) of the pathogen (Sharma et al., 2000; Pathania et al., 2006; Sharma et al., 2006; Sharma et al., 2012) which necessities the identification of durable anthracnose resistance cultivars.
Systematic studies on identification and mapping of anthracnose resistance genes in the United States, Canada, Brazil, and France resulted in the identification of around 25 anthracnose resistance genes. Most of the anthracnose resistance genes are dominant except the co-8 and are mapped to eight bean chromosomes (Burt et al., 2015; Meziadi et al., 2016; Zuiderveen et al., 2016). Seventeen of these genes are numbered from Co-1 to Co-17, however many anthracnose resistance genes have alphabets as suffix such as Co-x, Co-y, Co-w, Co-Pa, Co-z, Co-1HY, and are mapped to bean chromosomes where numbered genes are located (Zuiderveen et al., 2016). Co-1 and its alleles, Co-14, Co-x, Co-w, Co-1HY, Co-165-X, Co-173-X, and Co-Pa anthracnose resistance genes are mapped to the distal end of bean chromosome Pv01 (Melotto and Kelly, 2000; Geffroy et al., 2008; Gonçalves-Vidigal et al., 2011; Campa et al., 2014; Richard et al., 2014; Chen et al., 2017; de Lima Castro et al., 2017). Five anthracnose resistance genes (Co-u, CoPv02c3-X, CoPv02c7-X, CoPv02c19-X, and CoPv0c2449-X) are mapped to Pv02 (Kelly et al., 2003; Campa et al., 2014). Anthracnose resistance genes Co-13 and Co-17 are mapped to Pv03 (Goncalves-Vidigal et al., 2009; Lacanallo and Gonçalves-Vidigal, 2015; Trabanco et al., 2015). The anthracnose resistance gene mapped to Pv04 are Co-3 and its alleles, Co-10, Co-y, Co-z Co-15, Co-16, and Co-RVI (Geffroy et al., 2008; Coelho et al., 2013; Goncalves-Vidigal et al., 2013; Sousa et al., 2015; Coimbra-Gonçalves et al., 2016). Anthracnose resistance genes Co-5, Co-6, and Co-v map to Pv07 (Campa et al., 2009). Anthracnose resistance genes Co-4 and its alleles and Co-2 are located on Pv08 and Pv11, respectively (Young et al., 1998; Melotto and Kelly, 2001; Alzate-Marin et al., 2007; Rodriguez-Suarez et al., 2007). In addition to major resistance genes, studies showed that anthracnose resistance is quantitatively (QTLs) inherited like ANT02.1UC and ANT07.1UC (Oblessuc et al., 2014). Recently, a major QTL for anthracnose resistance against race 1545 was mapped to Pv05 with candidate gene Phvul.005G117900 that encodes a NBS-LRR protein (Gonzalez et al., 2015). Genome wide association studies (GWAS) showed association of a SNP and SSR marker located at Pv10 with anthracnose resistance (Zuiderveen et al., 2016; Wu et al., 2017). Access to P. vulgaris whole genome (Schmutz et al., 2014; Vlasova et al., 2016) and SNP Bean Chips (Hyten et al., 2010; Song et al., 2015) has resulted in the fine mapping of many economically important traits in beans including anthracnose resistance (Richard et al., 2014; Burt et al., 2015; Gonzalez et al., 2015; Oblessuc et al., 2015; Padder et al., 2016; Zuiderveen et al., 2016; Chen et al., 2017; de Lima Castro et al., 2017; Mungalu et al., 2020).
In India, a few studies identified anthracnose resistant sources to Indian races (Sharma et al., 1993; Kumar et al., 1997; Sharma et al., 1999; Pathania et al., 2006; Sharma et al., 2006; Sharma et al., 2012). Among the various sources of resistance, only a few genotypes were exploited to know the genetics of resistance (Sharma et al., 2000; Pathania et al., 2006; Katoch et al., 2019). For instance, inheritance pattern of broad spectrum anthracnose resistant cultivars such as KRC-5 and Baspa (KRC-8) showed dominant and recessive anthracnose resistance genes, respectively to a range of anthracnose races (Sharma et al., 2000; Pathania et al., 2006). The high diversity in common bean coupled with resistance to anthracnose races warrants identification of genomic regions associated with anthracnose. Mapping anthracnose resistance genes via bi-parental mapping populations is time consuming thus GWAS is the easiest approach to identify the genomic regions associated with anthracnose resistance genes in NW Himalayan common beans. Like the USA and other countries, India also need to exploit GWAS to its full potential to discern different agronomic and resistance traits in the diverse bean germplasm. Here we report a SNP based population structure of 188 common beans collected from NW Himalayan region. In the present study, genotyping by sequencing (GBS) approach was used with the aim to infer the population structure, to identify common beans with broad-spectrum resistance to anthracnose disease and to identify common bean genomic regions associated with anthracnose resistance to major anthracnose races present in the region.
Material and Methods
Common Bean Genotype Collection and Bean Anthracnose Race Material
Common beans cultivated in the NW Himalayan region are highly diverse (Rana et al., 2015) and we selected 188 common bean lines that were collected from SKUAST-K, Wadura, Himachal Pradesh Krishi Vishvavidyala (HPKV), Palampur, National Bureau of Plant Genetic Resources (NBPGR) located at Shimla and New Delhi. Four genotypes such as AND 277, Kaboon (Andean genotypes), TO, and Michelite (Mesoamerican genotypes) were used as checks for population structure study. Supplementary Table 1 contains the passport data of 192 lines that were used in the present study. All the bean genotypes were purified by a single seed decent method for three consecutive years.
We used five C. lindemuthianum races (3, 73, 87, 503, and 2047) to phenotype 188 bean genotype and choice of races was based on their virulence and geographic prevalence. Bean anthracnose race 87 is present in two states of NW India, whereas race 503 is a predominant across NW region (Sharma et al., 2019). Race 3 is present in major bean growing areas of Himachal Pradesh (Sharma et al., 2007). Anthracnose race 73 is an important race in the United States and Canada (Kelly et al., 1994; Rio et al., 2003a; Rio et al., 2003b; Conner et al., 2019). Race 2047 is highly virulent because it defeats most of the major anthracnose resistance genes (Supplementary Table 2) and only few genotypes show resistance against it (Zuiderveen et al., 2016). Professor James D. Kelly, Michigan State University, kindly provided race 73 and 2047. All C. lindemuthianum races are available in the Division of Plant Pathology, SKUAST-K, Shalimar and are maintained on sterilized Whatman filter paper discs and in glycerol stocks at -20°C for long time storage. To avoid virulence loss, races are inoculated to susceptible genotypes and pathogen is re-isolated from infected stem tissue.
Phenotype Screening
For screening bean genotypes to anthracnose races, six seeds of each line were planted in a tray containing coco-peat and vermiculite (2:1 ratio). The cotyledonary leaves were sprayed with a spore suspension that was prepared from seven days old culture by scraping the surface of culture with a spatula and few drops of 0.01% Tween 20 were added in sterile distilled water. Inoculum load was adjusted to 2 × 106 conidia/ml after counting the number of spores in an initial suspension with a haemocytometer. Inoculated plants were kept in plant growth chamber at 22°C with 90% humidity, 14:10-h day/night photoperiod, and light intensity of 120–150 µE/m2 (Mahiya-Farooq et al., 2019). The disease reaction was recorded with the 0–5 scale as described elsewhere (Drijfhout and Davis, 1989). Plants that were scored 0, 1, or 2 were categorized as resistant, whereas plants with scores of 3, 4, or 5 were graded as susceptible (Figure 1). The plant growth chamber was sterilized with 70% ethanol before performing screens of bean genotypes with other races. The phenotypic data was analyzed using R package and summary statistics such as density and frequency were calculated.

Figure 1 Screening of common bean population with anthracnose races (a, b). Plants showing disease score 5 were killed (c); whereas plants with 0 score showed no symptoms (d).
Genotyping by Sequencing
For GBS, the total genomic DNA from 192 bean genotypes was extracted from young leaf tissues using CTAB (Hexadecyltrimethyl ammonium bromide) protocol as described by Murray and Thompson (1980). DNA integrity and quality was checked by Agarose gel electrophoresis and quantification was performed on an Eppendroff nano-bio-spectrophotometer. The DNA samples were diluted to 25 ng/ml by adding nuclease free water. Approximately, 30 μl of each DNA sample from all the genotypes were shipped to Xcelris Labs Limited, Gujarat, India for GBS.
Preparation of GBS Library and Quantity/Quality Check
The 100-ng genomic DNA from 192 samples was digested using ApeK1 enzyme and the GBS library was prepared from the digested DNA fragments by ligating adaptors specific to the cut-site. The ligated products were pooled into 16 groups of 12 samples each. The pool was PCR amplified to generate the final library pool. The library pools were analyzed in Bioanalyzer 2100 (Agilent Technologies) using High Sensitivity (HS) DNA chip as per manufacturer’s instructions. Then next generation sequencing for all the samples was carried out independently on IlluminaHiSeq platform to generate up to 3.5–4.0 million reads.
Bioinformatics Analysis
The fastq files obtained from 192 samples in 16 different pools were first de-multiplexed and then we calculated overall amount of data and performed the QC. The high quality (HQ) data was extracted using Trimmomatic at Q20 quality values. The barcode tagged fastq files for both R1 and R2 (paired end) were condensed into single file and analyzed using TASSEL. The Tassel key file containing the information of each barcode corresponding to each sample was generated. Common bean reference genome (P. vulgaris v2.0) was downloaded from Phytozome (https://genome.jgi.doe.gov/portal/pages/dynamicOrganismDownload.jsf?organism=Pvulgaris). The genome has 11 chromosomes and 467 scaffolds with 537.22-Mb genome size. All reads from each sample were aligned to the common bean reference genome (v2.0).
We used TASSEL5 GBSv2 pipeline for identification of tags at cut sites and SNPs located across reference genome. The GBSv2 pipeline supports fastq files in the older format, which includes multiple taxa combined in a single file with barcodes attached to each read sequence. The ApekI tags were determined using GBSSeqToTagDBPlugin, which identifies tags from fastq input files then stores these tags and the taxa in which they appear into a local SQLite database. Next TagExportToFastqPlugin with default parameters was used to pull distinct tags from the database that were exported in the fastq format so that they can be aligned to the reference genome with various aligners. The fastq files were aligned against the common bean reference genome v.2.0 downloaded from phytozome using the Bowtie2 tool version 2.2.9 (Langmead and Salzberg, 2012). The sam file created from the Bowtie2 aligner program was used through SAMToGBSdbPlugin to store the position information for each aligned tag. We then used DiscoverySNPCallerPluginV2 to identify SNPs from the aligned tags. SNP positions for each allele and the tags associated with that allele were exported along with the number of times the tag appears in each taxa. SNP coverage, quality and genotypic statistics were calculated using SNPQualityProfilerPlugin. Finally, ProductionSNPCallerPluginV2 plugin was used for SNP calling. To acquire high quality SNPs with minimum missing data, filtration criteria of missing data was applied with following parameters; minor allele frequency (MAF >= 0.05, MAC >= 10) using VCF tools as per the procedure described by Mourad et al. (2018).
Population Structure, Kinship, and Linkage Decay (LD) Analysis
Structure threader software was used to assess the population genetic structure among 192 common bean (Pina-Martins et al., 2017). Population structure was estimated based on total SNP loci (22,589 SNPs) and K from 1 to 10 with 10 independent runs for each K. To determine the probable number of clusters based on genotypes we used 5,000 burnin and 50,000 MCMC (Markov Chain Monte Carlo) iteration. Structure output was then subjected to structure harvester for identification of effective number of clusters using the Evanno test implement in STRUCTURE HARVESTER (Earl, 2012). The principal component analysis was performed using TASSEL5.0 to determine the percentage of variation explained by top three principal components. For phylogenetic analysis, SNPhylo was run considering parameter of bootstrapping and multiple alignment by MUSCLE and the tree was plotted using figtree v1.4.1. To know the relatedness among 192 common bean genotypes based on shared alleles among individuals, we used GAPIT R package for generation of Kinship matrix. All the 22,589 filtered SNPs were used to perform the analysis because previous studies have shown that the complete set of markers control genome wide error rate better and is a good choice than pedigree based methods for kinship estimates. Linkage Disequilibrium (LD) was estimated using Genomic Association and Prediction Integrated Tool (GAPIT) and LD curve was fitted using a nonlinear model as described elsewhere (Marroni et al., 2011; Gao L. et al., 2016).
Genome Wide Association Analysis
After removing four controls (Kaboon, AND 277, Michelite, and TO) and eight genotypes that were phenotypically heterozygous, 180 genotypes were processed for GWAS analysis. GAPIT was used for GWAS analyses and SNP-trait association analysis was performed using Mixed Linear Model (MLM) (Zhang et al., 2010) implemented in GAPIT that include correction for both population structure and kinship. The qqman R package (Turner, 2014) was used to plot Manhattan and quantile-quantile (QQ) plots. A 5% significance level was used to identify SNPs significantly associated with anthracnose resistance. In the present study, we used linear scale for phenotypic measurements thus, smaller values represent resistance and larger values indicate susceptibility. Hence, SNPs with lower marker effect (Supplementary Tables 3-7) are linked to bean anthracnose resistance as per the procedure described by Mourad et al. (2018).
Candidate Gene Identification
Significant SNPs for anthracnose resistance obtained from MLM analysis were blasted against the P. vulgaris v 2.0 available at www.phytozome.jgi.doe.gov for candidate gene identification. Gene annotation and sequence flanking (200 bp) for SNPs was also obtained from the phytozome. The top 10 SNPs were also checked at phytozome for functional annotation.
Results
Identification of Resistant Sources to Five Anthracnose Races
Common bean genotypes (188) showed differential interaction when evaluated for anthracnose resistance with five anthracnose races. Genotypes that showed resistance against races were either immune or exhibited pinhead symptoms on leaves and stems after 3 days of inoculation (Figure 1D). Such cultivars showed hypersensitive response after 7 days of incubation. In contrary susceptible cultivars after 7 days showed severe infection on the cotyledons and the hypocotyls as small blackish sunken lesions that led to plant death (Figure 1C). Numerous anthracnose resistant lines were identified among the bean diversity panel. Out of 188 accessions, 113 were resistant to one or multiple races and 75 lines were susceptible to all races (Supplementary Table 8 and Figure 2). Common bean genotype WB-1634 and WB-967 collected from NBPGR, Shimla showed resistance to all the five races, whereas WB-716 (NBPGR, Shimla) was resistant to four races. WB-1637 was resistant to races 2047, 3, 87, and 503. Bean anthracnose race 2047 was highly virulent and only 12.5% of the cultivars exhibited resistance to it, followed by race 503 for which 14.36% of the genotypes showed resistance. Anthracnose races 73 and 87 were unable to infect 18.7% and 26.0% cultivars, respectively. In contrary to other races, anthracnose race 3 was less virulent, as 34.8% of the bean genotypes were resistant to it (Supplementary Table 8 and Figure 2). A number of anthracnose resistant bean genotypes were identified against the highly virulent anthracnose race 2047.
Population Structure, Kinship Matrix, and Linkage Disequilibrium (LD)
The lack of evidence about the common bean population structure and the gene pools to which the NW Himalayan beans belong to prompted us to undertake these studies. We used 192 bean genotypes (4 controls plus 188 Indian beans) for GBS and after SNP calling we obtained 250,526 SNPs (Supplementary Figure 1). The SNPs obtained varied from 18,198 to 28,005 on 11 bean pesudomolecules. In addition, 10,511 SNPs were present on various bean scaffolds (Supplementary Figure 1). After applying various quality-filtering parameters (MAF >= 0.05, MAC >= 10, Missing Data <= 50%) 22,589 SNPs were retained for downstream analysis and these SNPs were evenly distributed on 11 bean chromosomes (Figure 3). Linkage decay was observed after 1.0-Mb distance (Figure 4) that has a practical significance for identifying significant trait association even with a fewer number of markers. Population structure analysis of 192 bean genotypes (188 genotypes from India plus 4 controls) with STRUCTURE, GAPIT and SNPhylo categorized 188 Indian beans into two major clusters (Figures 5A–D) representing Andean and Mesoamerican gene pools. Among the 188 bean genotypes 114 (60.63%) belonged to Andean group and 60 (31.92%) belonged to Mesoamerican group. Both PCA and STRUCTURE analysis showed the presence of admixture within the 188 bean genotypes (Figures 5A, D). The two controls used in the present study helped us to categorize the NW beans into two distinct gene pools (5b). The first two principal components accounted for more than 40% of the variation. The kinship matrix also grouped NW Himalayan bean into two main groups belonging to Andean and Mesoamerican types (Figure 5C). However, subdivisions within the two main groups are evident and the genotypes showing admixture are located in the centre of the heat map (Figure 5C).
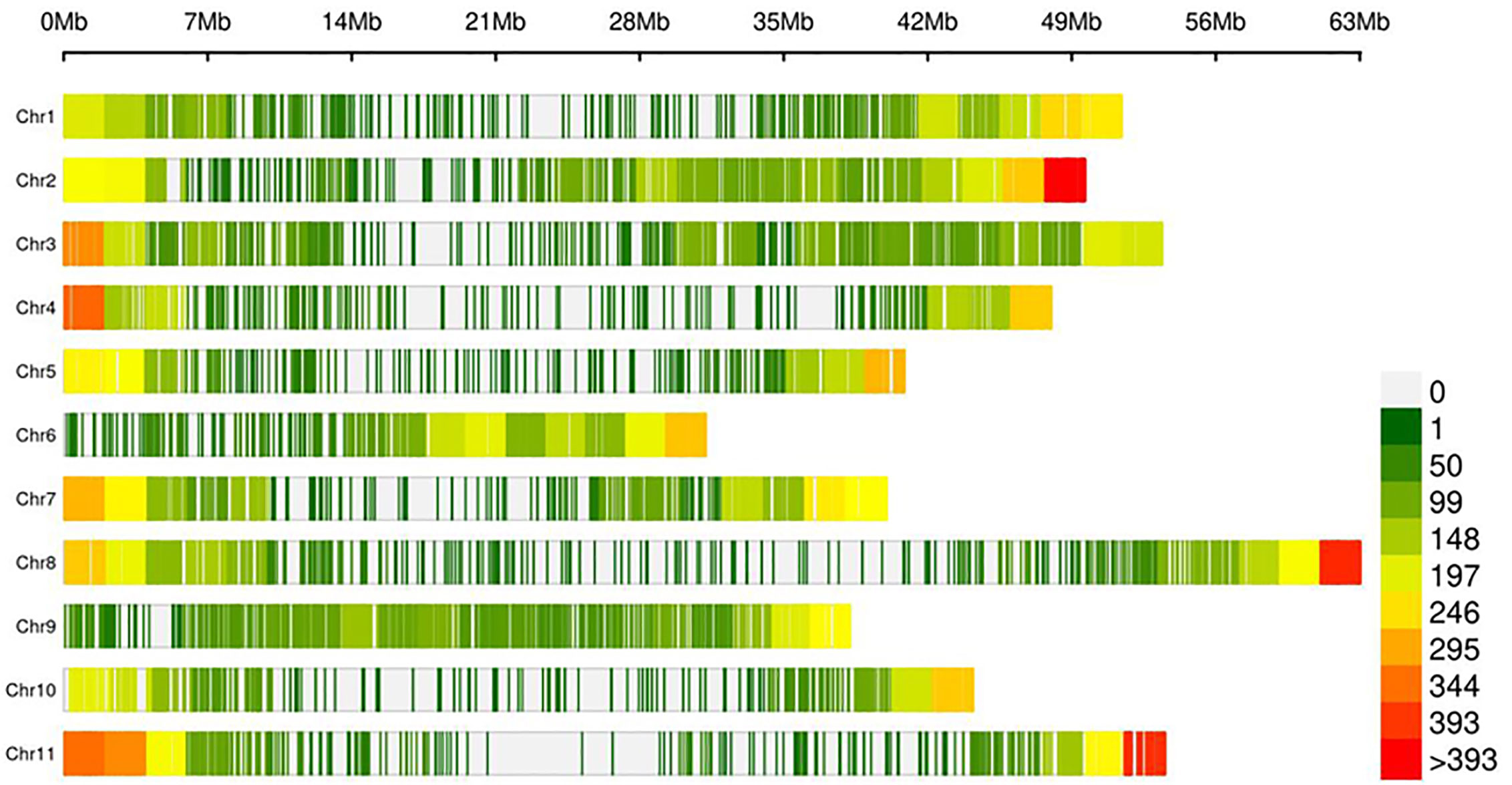
Figure 3 Density of 22,589 SNP within 1-Mb common bean genome window size. The different colors represent different density levels and “Chr” refers to common bean chromosomes.
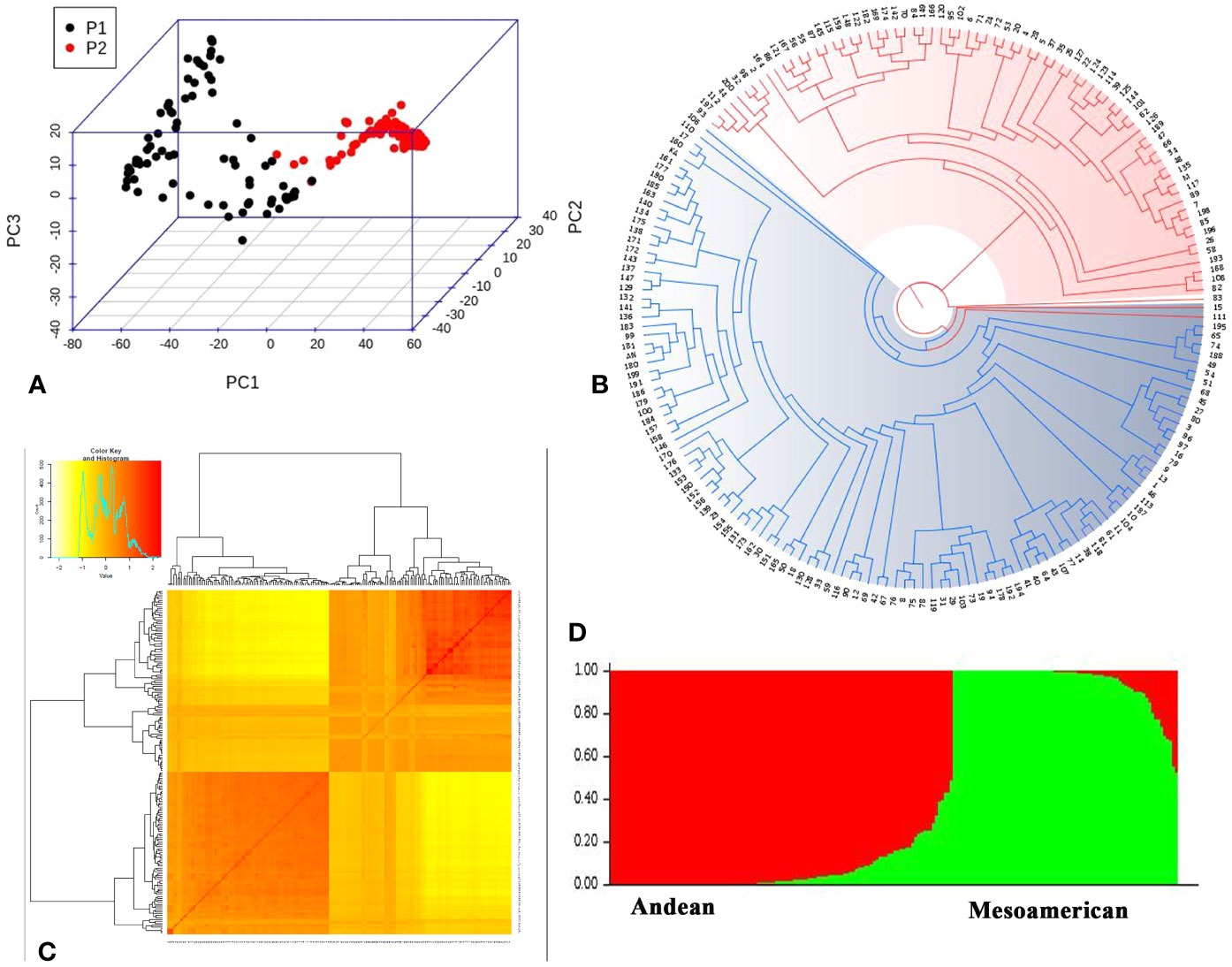
Figure 5 Population structure of 192 common beans inferred using different software’s. (A) Principal Component analysis performed using 22,598 evenly distributed SNPs along the bean genome. The two groups of accessions (P1 and P2) are depicted; one with black dots (Andean genotypes; P1) and dots with red color depict Mesoamerican genotypes (P2). (B) Phylogenetic tree generated using SNPhylo categorized 192 common bean genotypes into groups; Andean group (Blue) and Mesoamerican group (red). (C) Kinship matrix depicting pairwise similarities among 192 common bean genotypes; yellow and dark yellow color shows high similarities whereas red color depicts low similarity between the bean genotypes. (D) Genetic structure of 192 common bean genotypes based on model based approach implemented in STRUCTRE. Each genotypes is represented by a column divided into two colored segments (red and green) whose length indicates the proportions of the genome attributed to the two main groups (y axis); Andean (red) and Mesoamerican (green).
Genome Wide Association Study
After removing the controls and heterozygous lines, 180 common bean genotypes were retained for a GWAS analysis. The filtered SNPs were evenly distributed across all the 11 bean chromosomes and 22,589 SNPs were used to find the marker trait associations through GWAS. Our data set showed the presence of structure, so the MLM model implemented in GAPIT was used to find significant SNPs associated with five anthracnose races. The expected and observed –log(10)p values followed X = Y distribution along the diagonal line until the observed p values diverted from the expected p values, which further strengthens that MLM model fits the data set.
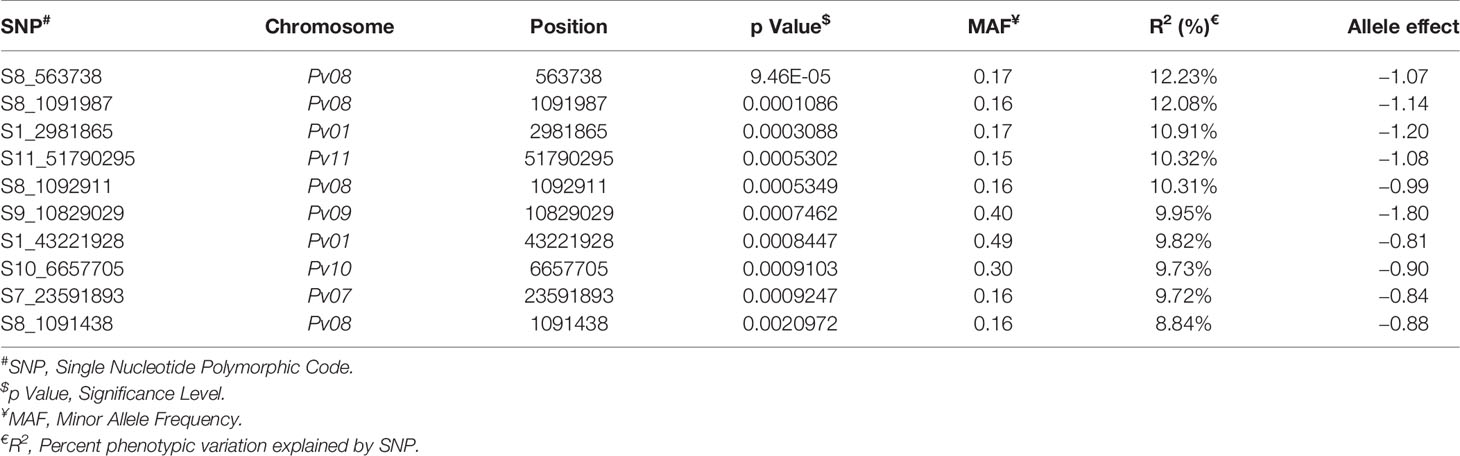
The significant SNPs associated with anthracnose race 3 are listed in supplementary Table 3 and the top 10 SNPs are listed in Table 1. There are many SNPs associated with anthracnose race 3 resistance at Pv04 (Figure 6) within the genomic region 1.31 to 2.03 Mb (Table 1). These SNPs contribute 16.71% to 19.57% of the phenotypic variation (Table 1). The other bean chromosome that contains a significant SNP associated with race 3 resistance is Pv03 and it contributes 16.62% of the phenotypic variation (Table 1 and Supplementary Table 3).
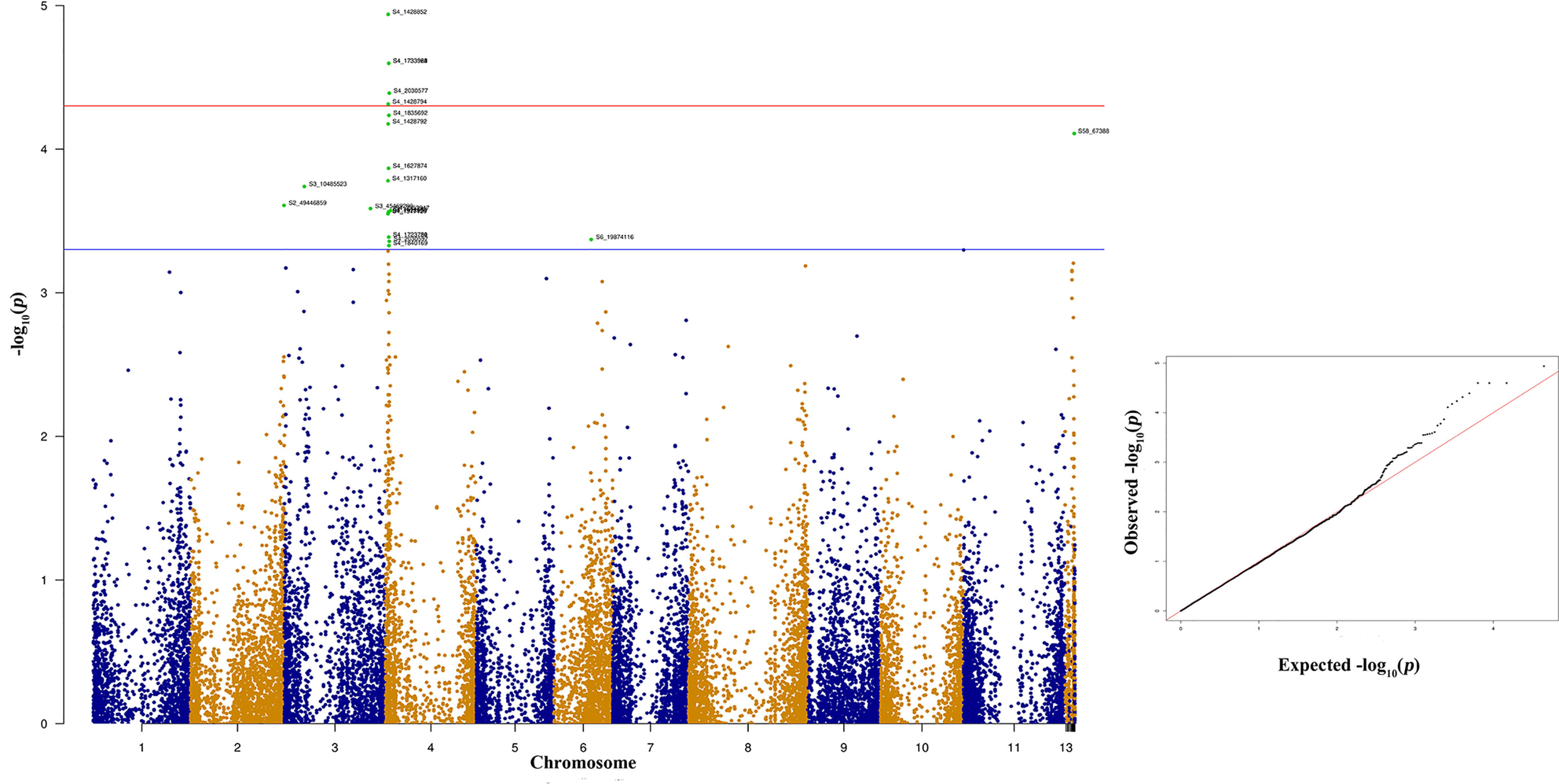
Figure 6 Manhattan and quantile-quantile (Q-Q) plots showing significant SNPs identified using MLM model that showed association with bean anthracnose race 3. Red line indicates Simple M Bonferroni corrected p-value threshold for significance and Green line indicates p-value of 1 × 103.
The top two significant SNPs that showed association with race 73 resistance are located between genomic region 0.5 to 1.09 Mb at Pv08 (Table 2). There is one more SNP (S8_1092911) on Pv08 at 1.09-Mb physical position and it contributes 10.31% of the phenotypic variation. These SNPs cumulatively contribute about 35% phenotypic variation. The significant SNPs such as S8_563938 and S8_1091987 (Figure 7 and Supplementary Table 4) indicate a major QTL associated with anthracnose race 73 resistance. The other significant SNPs among the top 10 SNPs associated with race 73 quantitative resistance are located at Pv01, Pv07, and Pv11 (Figure 7 and Table 2).
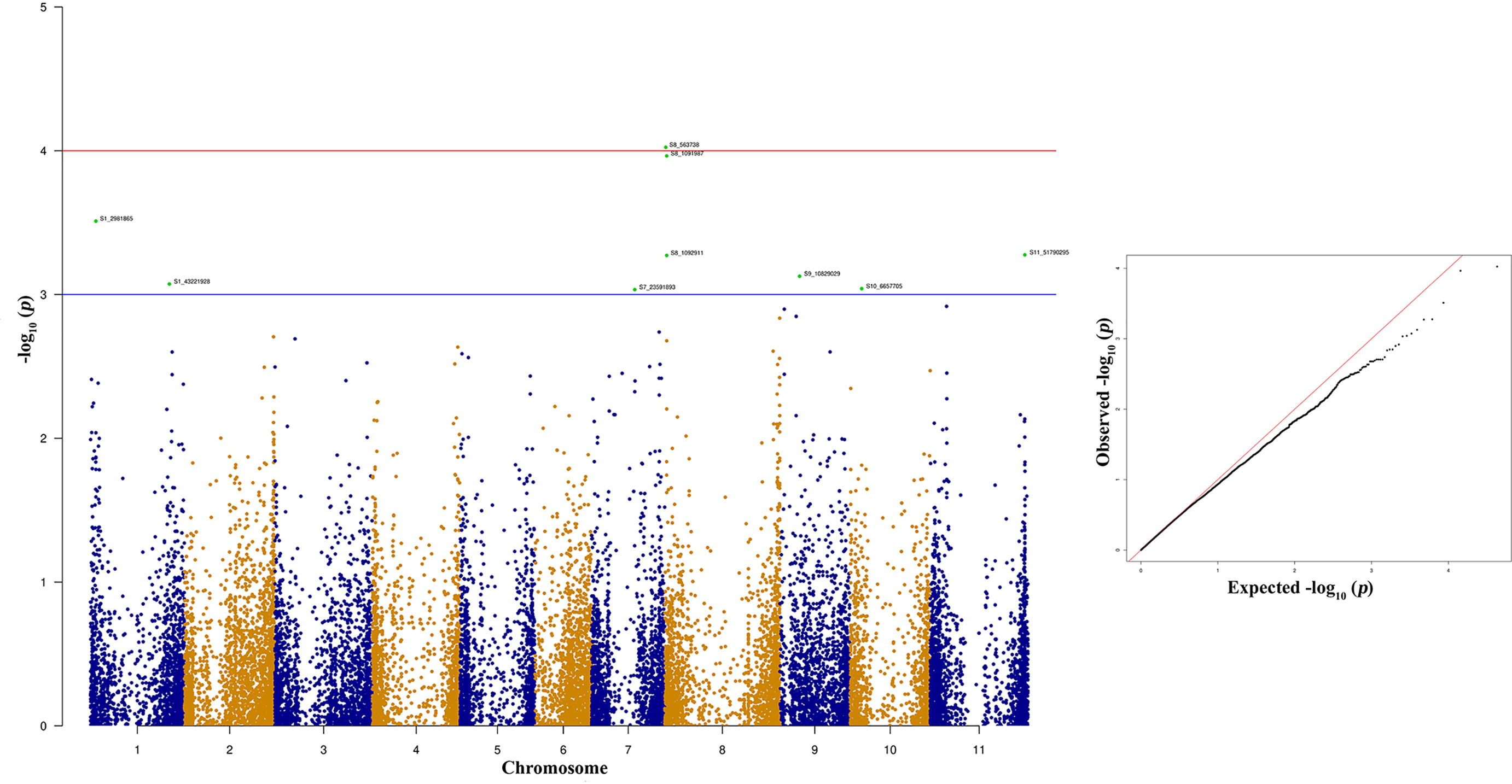
Figure 7 Manhattan and quantile-quantile (Q-Q) plots showing significant SNPs identified using MLM model that showed association with bean anthracnose race 73. Red line indicates Simple M Bonferroni corrected p-value threshold for significance and Green line indicates p-value of 1 × 103.
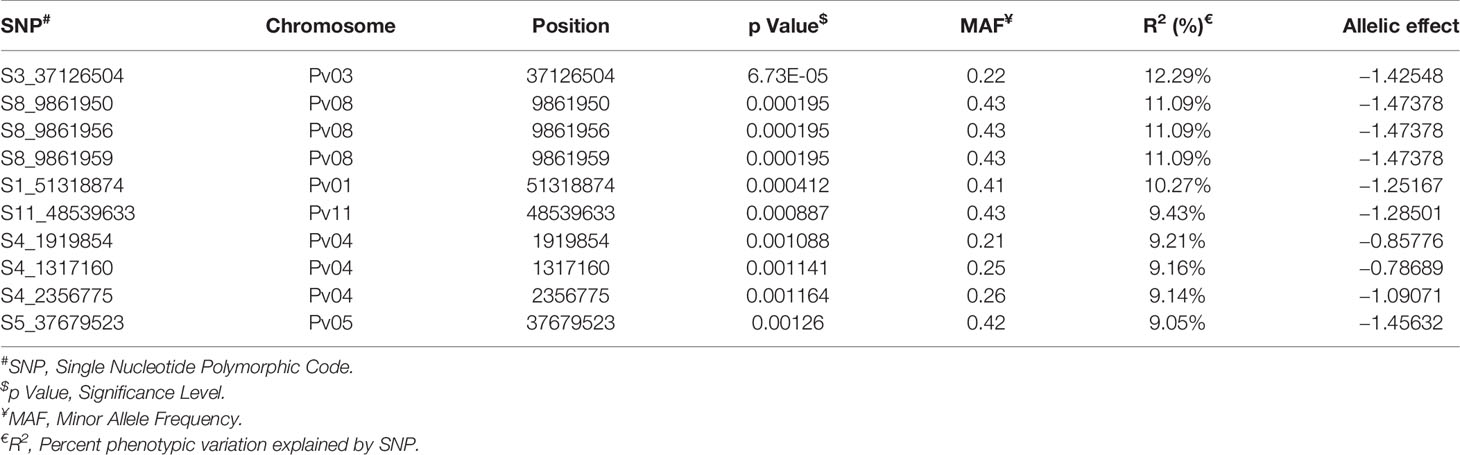
Among the top 10 SNPs associated with race 87 resistance, six SNPs are located at Pv08 and Pv04 (Figure 8 and Table 3). However, SNP, S3_37126504 located at Pv03, contributes 12.29% of the phenotypic variation. Three SNPs located at Pv08 explain 11.09% phenotypic variation. It was interesting to observe that the genomic region from 1.3 to 2.3 Mb at Pv04 is associated with race 87 and the region overlaps with genomic region associated with race 3. The distal end of Pv01 at 51.31 Mb contains another significant SNP that explains 10.27% phenotypic variation (Table 3). The distal end of Pv01 is known to contain many major anthracnose resistance genes and we hypnotize that this a most probable region for anthracnose resistance against race 87.
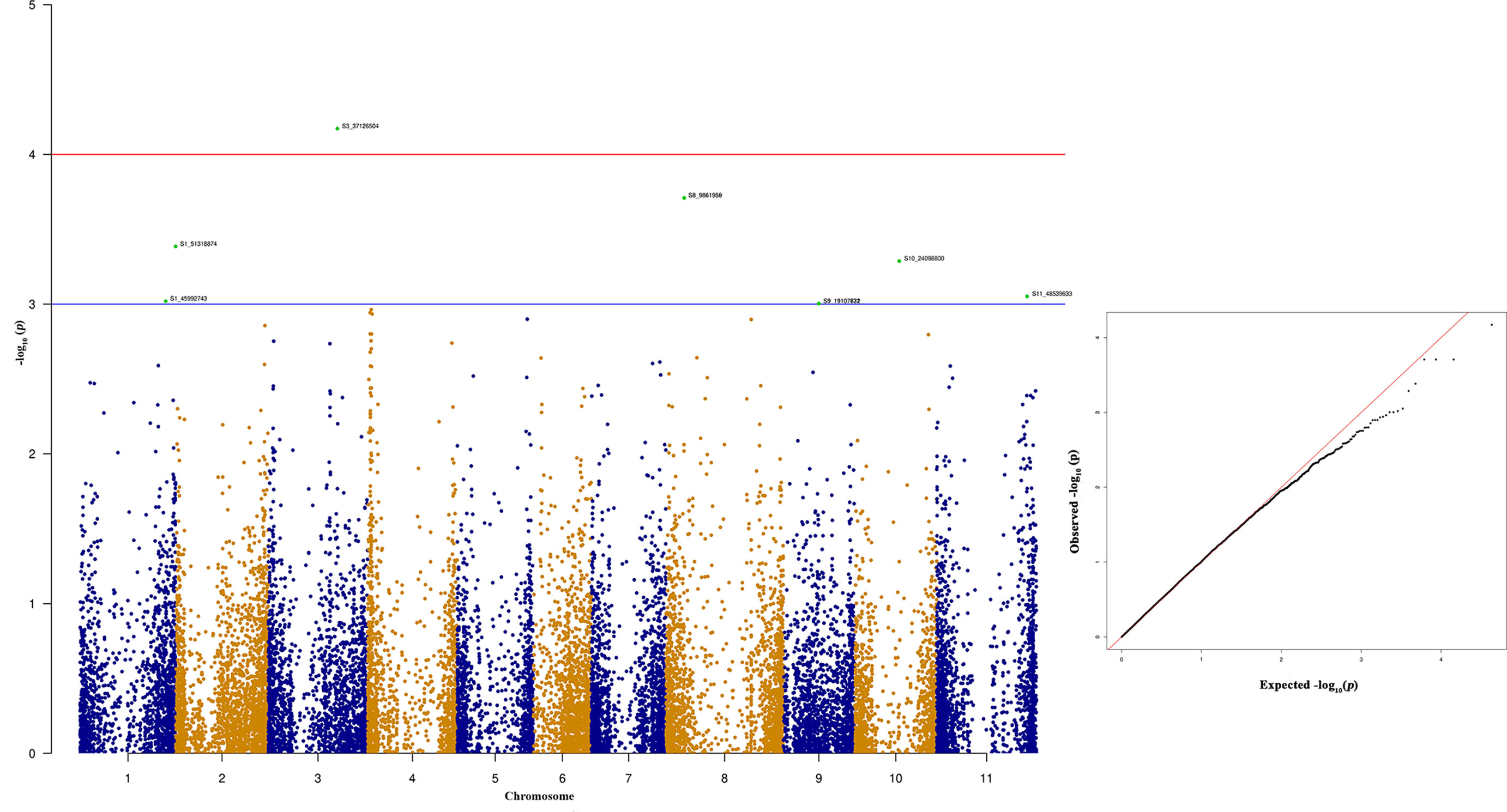
Figure 8 Manhattan and quantile-quantile (Q-Q) plots showing significant SNPs identified using MLM model that showed association with bean anthracnose race 87. Red line indicates Simple M Bonferroni corrected p-value threshold for significance and Green line indicates p-value of 1 × 103.
Seven out of ten of the most significant SNPs that showed strong association with quantitative anthracnose resistance against race 503 are located at Pv04 which explains above 12.30% of the phenotypic variation (Figure 9 and Table 4). Six of these SNPs are located between 1.35 to 2.72 Mb in the physical map of Pv04 and one is located at 47.62 Mb.
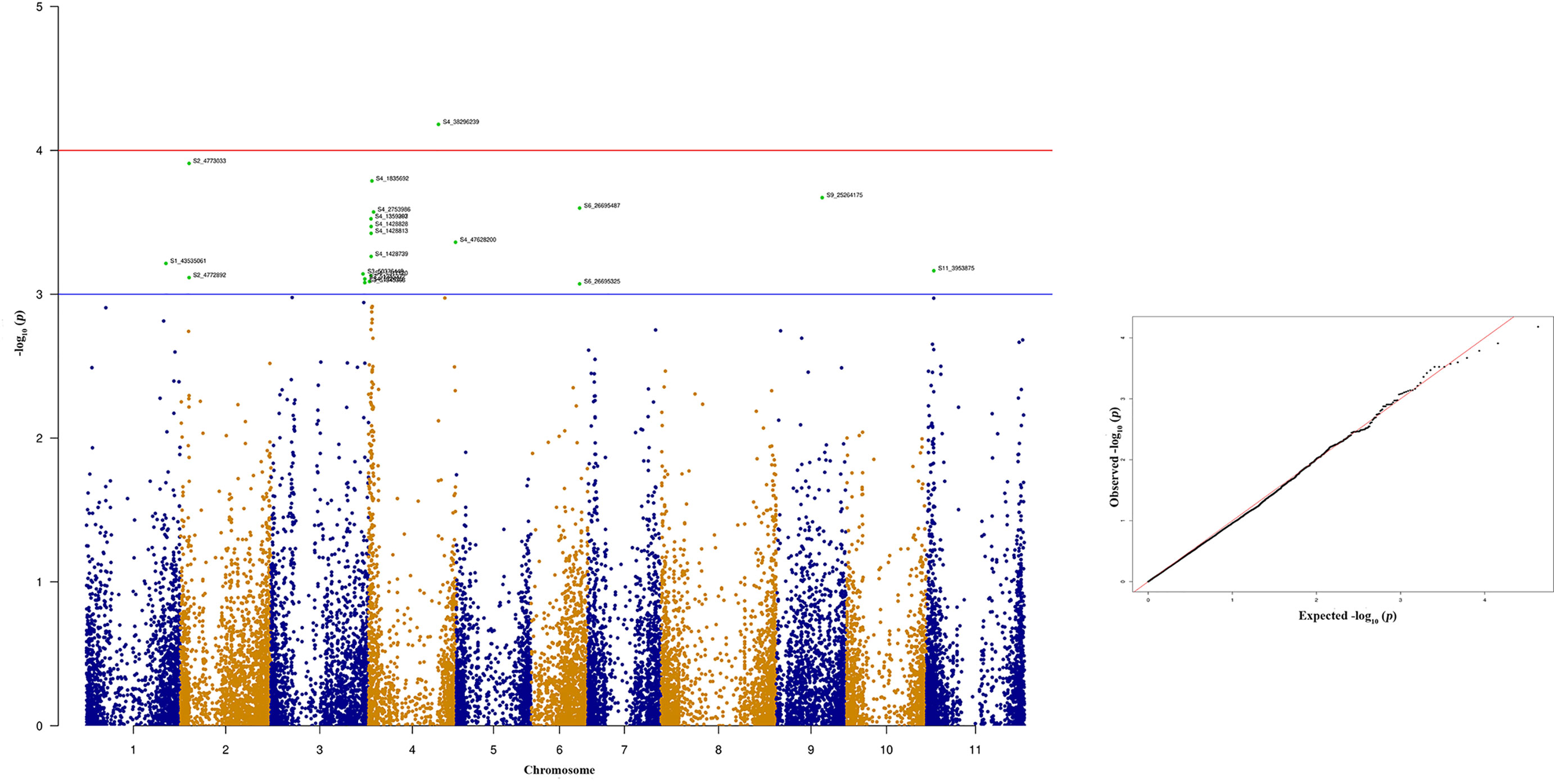
Figure 9 Manhattan and quantile-quantile (Q-Q) plots showing significant SNPs identified using MLM model that showed association with bean anthracnose race 503. Red line indicates Simple M Bonferroni corrected p-value threshold for significance and Green line indicates p-value of 1 × 103.
The top 10 significant SNPs associated with race 2047 resistance were distributed across the different bean chromosomes Pv03, Pv04, Pv06, and Pv09 (Figure 10 and Table 5). These top 10 significant SNPs explain phenotypic variation between 6.98% and 8.47% (Table 5). Bean genomic regions at Pv03 with physical positions of 28.62 and 27.317 Mb are associated with race 2047 resistance. The other two SNPs located at 6.18 Mb on Pv03 contribute 7.43% of the phenotypic variation (Table 5).
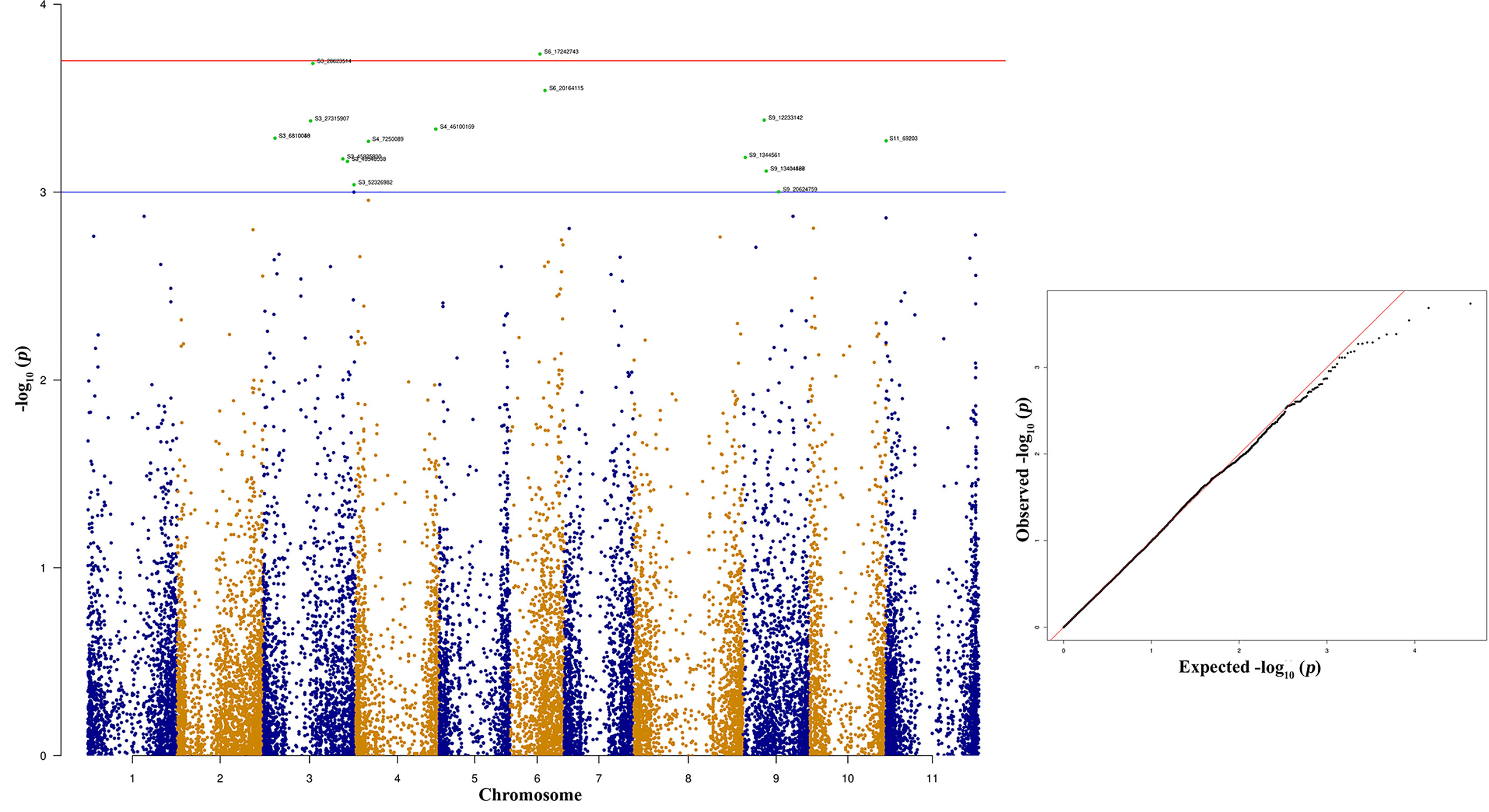
Figure 10 Manhattan and quantile-quantile (Q-Q) plots showing significant SNPs identified using MLM model that showed association with bean anthracnose race 2047. Red line indicates Simple M Bonferroni corrected p-value threshold for significance and Green line indicates p-value of 1 × 103.
Genomic Regions Associated With Anthracnose Resistance
It was interesting to observe that SNPs located at Pv04 are strongly associated with anthracnose resistance against races prevalent in NW Himalayan region. Many SNPs associated with race, 3, 87 and 503 are present up or downstream of genes having a strong role in disease resistance (Supplementary Table 9). For instance, common bean gene models Phvul.004G014800, Phvul.004G014801 and Phvul.004G015800 encode Leucine Rich Repeat (LRR) proteins. The common bean gene Phvul.004G015800 is having a typical NB-ARC domain (Table 6). A significant SNP (S9_13404457) located at Pv09 and associated with quantitative resistance against race 2047 encodes a serine threonine protein kinase (Table 6). In addition to the LRR region at Pv04 there is one more significant SNP (S9_25264175), which is located on Pv09 that encodes a zinc finger CCCH domain containing protein (Table 6). The other significant SNPs encode hypothetical proteins (Supplementary Table 9).
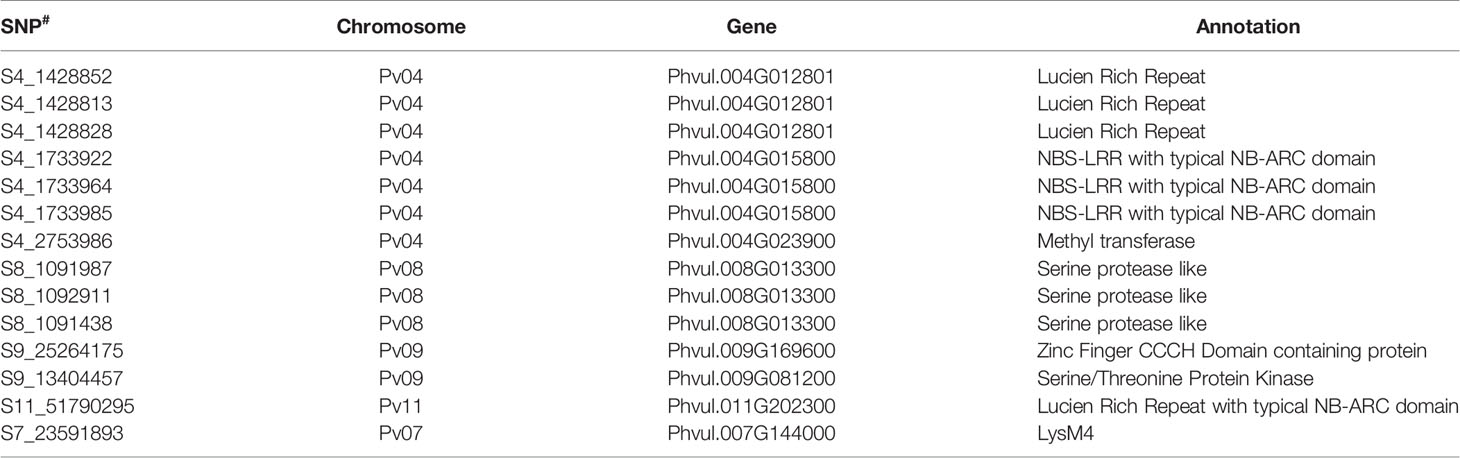
Table 6 Annotation of significant SNPs associated with different anthracnose resistance QTLs with typical role in disease resistance.
Discussion
Sources of Anthracnose Resistance
Phaseolus vulgaris – C. lindemuthianum pathosystem generally follows gene for gene interaction and most of the identified resistance genes in bean genotypes are dominant except the co-8 (Gonzalez et al., 2015). The pre-eminent approach to manage bean anthracnose is to identify broad spectrum resistant sources, exploit multitudinous genetic tools to unravel the underlying resistance mechanism in the resistant genotypes and then develop new resistant cultivars or deploy the identified cultivars spatially and temporally (Padder et al., 2010; Zuiderveen et al., 2016). In the present investigation, 188 bean genotypes from NW Himalayan region were screened against most prevalent five anthracnose races. Race 2047 is highly virulent (defeats all anthracnose resistance genes known in differentials except G2333) and only 4.4% genotypes from Andean diversity panel showed resistance to it (Zuiderveen et al., 2016). In NW region, 45 bean anthracnose races are reported and these are unable to infect G2333 and AB136 differential lines (Sharma et al., 2007; Sharma et al., 2019). Thus, it was anticipated that beans grown in northern India might show resistance against race 2047 because it is not reported in the bean growing areas but only 13.29% genotypes showed resistance, which is higher than the earlier reports (Zuiderveen et al., 2016). A good number of Andean beans such as WB967, WB-1634, WB-1637, and WB-716 showed resistance against multiple races and are good sources to start breeding programs for developing cultivars resistant to anthracnose race 2047. Similarly, we found a number of genotypes resistant to race 73, the most important race for the United States (Mahiya-Farooq et al., 2019). These genotypes can serve as a pool for breeding Andean and Mesoamerican genotypes. Anthracnose races 3, 87, and 503 are prevalent in the three northern states of India (Sharma et al., 2019) and the material resistant to these races can easily be exploited for the development of broad-spectrum anthracnose resistant cultivars for the region. The recent breakdown of the resistance gene present in AB136 by newly emerging races indicate coevolution of C. lindemuthianum in the region (Padder et al., 2009; Sharma et al., 2019) and warrants the effort to identify bean cultivars with resistance to multiple races. Although bean differential cultivars such as G2333, AB136 contain anthracnose resistance genes, their un-adaptability to NW Himalayan environment, hampers transfer of these genes to locally adapted cultivars. G2333 is photoperiod sensitive (Fernández et al., 2000; Ferreira et al., 2008; Goswami et al., 2011) similar to many anthracnose resistant genotypes identified by Fernández et al. (2000) and such cultivars are unsuitable for improvement of locally adapted beans. Alternatively, our material showed anthracnose resistance to race 2047 and resistance can easily be transferred to local commercially grown cultivars because these cultivars are adapted to the local environment and do not have limitations as those of G2333 and other differential cultivars. Over past few years, new anthracnose resistance genes were identified and mapped from diverse anthracnose resistant genotypes (Gonçalves-Vidigal et al., 2011; Richard et al., 2014; Lacanallo and Gonçalves-Vidigal, 2015; Trabanco et al., 2015; Chen et al., 2017; de Lima Castro et al., 2017). We expect our anthracnose resistant common bean material may contain diverse anthracnose resistance genes as the C. lindemuthianum virulence spectrum in NW region is different than the US, France and Canada (Padder et al., 2017).
Population Structure
Based on STRUCTURE, SNPhylo, and Kinship matrix, our common bean material (188 lines) along with the four control genotypes (each from Andean and Mesoamerican gene pools) helped us to categorize NW common bean populations into two distinct populations. This is the comprehensive study that shows common beans grown in India belong to two gene pools. Previously, Sharma et al. (2013) genotyped 67 common bean landraces of Himachal Pradesh with SSR markers and showed existence of two distinct gene pools in the state. It is well documented that common beans are native to Andean and Mesoamerican regions (Gepts and Debouck, 1991; McConnell et al., 2010; Bitocchi et al., 2012) and our study shows existence of two distinct gene pools in India that indicates probably multiple introductions of common bean occurred from their native places in to the region. We also observed admixture in our material similar to European common beans. Sharma et al. (2013) hypnotized that common beans grown in the NW region may have introduced either eastward or westward. This indicates that the common beans in India might be introduced from Europe (via Africa) or China and is strongly supported by earlier studies (Joshi and Thomas, 1987; Joshi and Mehra, 1993; Sharma et al., 2013). However, comprehensive studies similar to Bitocchi et al. (2013) are needed to answer single or multiple introductions into the region.
Association Mapping
A powerful tool currently available to scientific communities for QTL dissection is GWAS that provide higher resolution than QTL mapping (Muhammad et al., 2020). Hence, in recent years, use of GWAS for mapping economically important QTLs in agricultural crops has increased and P. vulgaris is not behind. Many QTLs having economic importance in common bean have been mapped using a GWAS approach (Moghaddam et al., 2016; Wu et al., 2017; Kelly and Bornowski, 2018; Kamfwa et al., 2019; Bisneta and Gonçalves-Vidigal, 2020; Bornowski et al., 2020; Gonçalves-Vidigal et al., 2020; Mungalu et al., 2020). In the present study, significant differences observed in NW Indian bean genotypes led to the identification of SNPs associated with the anthracnose resistance. Our findings resulted in the identification of new genomic regions associated with anthracnose resistance and to the best of our knowledge, it is the first study from India. Further, most of the GWAS studies explored bean chip to map economic traits in beans, however, present study explored GBS to find associations of SNPs with anthracnose resistance.
Many significant SNPs associated with anthracnose resistance to Indian C. lindemuthianum virulent races (3, 87, and 503) are located on Pv04. Most of these SNPs are either located up or down stream of gene models that encode LRR and have typical NB-ARC domains. Most of the plant disease resistance genes belong to the NBS-LRR class and play a significant roles in plant disease resistance (Jones and Dangl, 2006; Padder, 2014). The common bean genome also contains NBS-LRR regions that are distributed across all chromosomes (Ferrier-Cana et al., 2003; Richard et al., 2017; Wu et al., 2017) and a recent GWAS study showed associations between these NBS-LRR regions with anthracnose resistance (Wu et al., 2017). Anthracnose resistance genes Co-3 and its alleles, Co-16 are mapped to common bean Pv04 and this study also found association of SNPs on Pv04 with anthracnose resistance to races 3, 87, and 503 (Geffroy et al., 1999; David et al., 2008; Coimbra-Gonçalves et al., 2016; Murube et al., 2019). The Co-34 locus was previously mapped on the 3.36-Mb genomic position but it was recently mapped to the region between 0.49 to 0.58 Mb on Pv04 (Valentini et al., 2017) which indicates shift in the physical positions with better mapping tools (Kelly and Bornowski, 2018). A previous GWAS study with an Andean diversity panel by Zuiderveen et al. (2016) identified two SNPs located at 0.45 and 0.53 Mb on Pv04 against race 7 and 109, respectively. A recent study identified 21 kinases and 55 NBS-LRR encoding genes close to the major genes located on Pv04 (Bisneta and Gonçalves-Vidigal, 2020). In the current study, SNPs that showed strong association with race 3 and 503 were located between the physical positions of 1.3 to 2.75 Mb, which are close to the anthracnose resistance genes (Co-3 locus and Co-16) present on the proximal end of Pv04. We also found a gene model (Phvul.004G023900) that encodes a methyltransferase that impart quantitative resistance to race 503. The role of methyltransferases in plant disease resistance is well documented against an array of plant pathogens (Yang et al., 2017; Wang et al., 2018; Bulman et al., 2019). Besides Pv04, this study identified a SNP on Pv09 for quantitative resistance against race 503 in gene model Phvul.009G169600 that encodes a zinc finger protein. This gene model contains a CCCH domain and its involvement in plant disease resistance is well documented (Deng et al., 2012; Gao W. et al., 2016; Cui et al., 2018). In the common bean genome only two NBS-LRR genes are located at distal end of Pv09 (Richard et al., 2017) and identification of QTL against race 503 on Pv09 is interesting and warrants in silico identification of other plant disease resistance signatures such as zinc finger proteins from the common bean genomes.
The Co-1 locus is present on the distal end of Pv01 and contains five alleles that have been extensity used in breeding anthracnose resistant cultivars in the United States and Mexico (Melotto and Kelly, 2000; Goncalves-Vidigal and Kelly, 2006; Gonçalves-Vidigal et al., 2011). The Co-1 gene is present at a physical position of 50.30 Mb and we recently found high expression of gene model Phvul.001G243800 that encodes receptor like kinase following inoculation of a resistant NIL with race 73 (Zuiderveen et al., 2016; Mahiya-Farooq et al., 2019). Interestingly we were unable to find any significant SNPs among the list of top 10 SNPs associated with anthracnose resistance against race 73. However, this study identified a SNP associated with race 503 quantitative resistance. This SNP is located at 51.31 Mb and is close to the Co-1 locus and we hypothesize the involvement of Co-1 locus with race 503.
In this study four out of top 10 significant SNPs associated with race 73 quantitate resistance are located on Pv08. These SNPs are located between 0.56- and 1.09-Mb physical positions and encode hypothetical proteins. The role of hypothetical proteins cannot be completely ruled out. There is a single major anthracnose resistance gene (Co-4 and its alleles) present on Pv08 and a recent study fine mapped Co-4 between physical position of 0.49 to 2.84 Mb (Burt et al., 2015). Hence, it can be concluded that the SNPs overlap with the Co-4 anthracnose resistance gene. Identification of the Co-4 gene in the NW Himalayan bean lines is of paramount importance because this gene is highly effective against numerous anthracnose races (Melotto and Kelly, 2001; Oblessuc et al., 2015). Another source of Co-4, G2333 is photosensitive and does not flower under North Indian conditions, therefore hampering its usefulness as a source for introgression of Co-4 gene to Indian beans. The indication of presence of the Co-4 gene in our material is encouraging and breeders can easily introgress this gene into elite material for breeding anthracnose resistant cultivars. Additionally, we found a SNP (S11_51790295) located at distal end of Pv11 (51.79 Mb) in a gene model Phvul.011G202300 that encodes a LRR with typical NB-ARC domain. This region contains many NBS-LRR genes (Richard et al., 2017). The Co-2 gene was previously mapped at 39.73 Mb (Creusot et al., 1999; Zuiderveen et al., 2016), but a recent study fine mapped it to 51.52 Mb (Kelly and Bornowski, 2018; Miklas et al., 2020). There is a tight association between Co-2 and Ur-11, a bean rust resistance gene (Awale et al., 2008). The present study identified a SNP that overlaps with the Co-2 anthracnose resistance gene.
Different genomic regions are associated with quantitative resistance against race 2047. These regions belong to Pv03, Pv09, and Pv11 and the SNPs contribute less than 8.0% of the phenotypic variation. This may be because of the high virulence of race 2047 that defeats the entire major anthracnose resistance genes, except the three genes that are present in the differential cultivar, G2333. No significant SNP was found associated with anthracnose resistance against race 2047 in the Andean diversity panel (Zuiderveen et al., 2016) and the study also failed to identify any significant loci that was attributed to low level of resistance against it. The present study identified a significant SNP that is located on Pv11 and the gene model Phvul.009G081200 encodes a serine threonine protein kinase. Serine threonine protein kinases play significant role in plant disease resistance in many host pathogen interactions (Afzal et al., 2008; Juliana et al., 2018). There are a few effective anthracnose resistance genes against race 2047 such as Co-4, Co-12, Co-13, Co-14, and Co-15 (Goncalves-Vidigal et al., 2009; Goncalves-Vidigal et al., 2012; Sousa et al., 2015). Two of our significant SNPs are located on Pv03 at 27.31 and 28.62 Mb and both encode hypothetical proteins. The other genes present on Pv03 have not been fine mapped and we assume that the SNPs associated with quantitate resistance against race 2047 are different from Co-13 and Co-17 anthracnose resistance genes. The Co-17 gene was mapped to the extreme proximal end of Pv03 (Trabanco et al., 2015; Kelly and Bornowski, 2018; Bisneta and Gonçalves-Vidigal, 2020). Bisneta and Gonçalves-Vidigal (2020) found six genomic regions on Pv03 associated with bean anthracnose which are distant from the significant SNPs found associated with anthracnose resistance in the present study.
Conclusion
The present study revealed many sources of resistance to different anthracnose races prevalent in Northern India and to the two important anthracnose races 73 and 2047. A few genotypes are resistant to all the races among them, including a red colored Andean bean WB967, which will be a good source of resistance to develop durable anthracnose resistant cultivars. The present study showed that beans grown in the NW Himalayan region belong to Andean and Mesoamerican gene pools. Many novel QTLs particularly at Pv03, Pv08, Pv09, and Pv11 were identified in the gene models that encode typical plant disease resistance proteins. These novel genomic regions may contain major anthracnose resistance genes that needs to be elucidated using genetic studies. We found many SNPs that overlap with Co-4 anthracnose resistance gene and its presence in our material is encouraging for breeding durable anthracnose resistant cultivars for the region.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author Contributions
AB, AN, and RR multiplied common bean genotypes. M-F and A-N maintained the anthracnose race cultures. Phenotyping of bean genotypes was performed by AB, AN, RR, A-N, and MS. PAS and PNS provided the material 192 common bean genotypes. MA and HI helped in multiplication of genotypes at their respective stations. The whole work was supervised by BP. BP designed the study, analyzed SNP data set, and wrote/edited the MS. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JK declared a past co-authorship with one of the authors BP to the handling editor.
Acknowledgments
The corresponding author is highly thankful to Department of Biotechnology, Government of India for providing financial assistance (Grant No. BT/PR17344/AGII/106/1005/2016). The corresponding author is highly thankful to James D. Kelly, Michigan State University for providing anthracnose races 73 and 2047. The authors are also thankful to Xcelris Labs Limited, Ahmedabad-380015, India, for performing GBS of our material. The corresponding author is thankful to James D. Kelly and Halima E. Awale for editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.571618/full#supplementary-material
References
Afzal, A. J., Wood, A. J., Lightfoot, D. A. (2008). Plant Receptor-Like Serine Threonine Kinases: Roles in Signaling and Plant Defense. Mol. Plant-Microbe Interact. 21, 507–517. doi: 10.1094/MPMI-21-5-0507
Alzate-Marin, A. L., Souza, K. A., Silva, M. G. M., Oliveira, E. J., Moreira, M. A., Barros, E. G. (2007). Genetic Characterization of Anthracnose Resistance Genes Co-43 and Co-9 in Common Bean Cultivar Tlalnepantla 64 (PI207262). Euphytica 154, 1–8. doi: 10.1007/s10681-006-9253-x
Asfaw, A., Blair, M. W., Almekinders, C. (2009). Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces From the East African Highlands. Theor. Appl. Genet. 120, 1–12. doi: 10.1007/s00122-009-1154-7
Awale, H., Ismail, S. M., Vallejo, V. A., Kelly, J. D. (2008). SQ4 SCAR Marker Linked to the Co-2 Gene on B11 Appears to Be Linked to the Ur-11 Gene. Annu. Rep. Bean Improv. Coop. 51, 174–175.
Beaver, J. S., Rosas, J. C., Myers, J., Acosta, J., Kelly, J. D., Nchimbi-Msolla, S., et al. (2003). Contributions of the Bean/Cowpea CRSP to Cultivar and Germplasm Development in Common Bean. Field Crops Res. 82, 87–102. doi: 10.1016/S0378-4290(03)00032-7
Bellucci, E., Bitocchi, E., Rau, D., Rodriguez, M., Biagetti, E., Giardini, A., et al. (2014). “Genomics of Origin, Domestication and Evolution of Phaseolus vulgaris,” in Genomics of Plant Genetic Resources (Dordrecht: Springer), 483–507.
Bisneta, M. V., Gonçalves-Vidigal, M. C. (2020). Integration of Anthracnose Resistance Loci and RLK and NBS-LRR-Encoding Genes in the Phaseolus vulgaris L. Genome. Crop Sci. doi: 10.1002/Csc1002.20288
Bitocchi, E., Nanni, L., Bellucci, E., Rossi, M., Giardini, A., Zeuli, P. S., et al. (2012). Mesoamerican Origin of the Common Bean (Phaseolus vulgaris L.) Is Revealed by Sequence Data. Proc. Natl. Acad. Sci. 109, e788–e796. doi: 10.1073/pnas.1108973109
Bitocchi, E., Bellucci, E., Giardini, A., Rau, D., Rodriguez, M., Biagetti, E., et al. (2013). Molecular Analysis of the Parallel Domestication of the Common Bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 197, 300–313. doi: 10.1111/j.1469-8137.2012.04377.x
Bornowski, N., Song, Q., Kelly, J. D. (2020). QTL Mapping of Post-Processing Color Retention in Two Black Bean Populations. Theor. Appl. Genet. doi: 10.1007/S00122-00020-03656-00123
Broughton, W. J., Hernández, G., Blair, M., Beebe, S., Gepts, P., Vanderleyden, J. (2003). Beans (Phaseolus spp.)–Model Food Legumes. Plant Soil 252, 55–128. doi: 10.1023/a:1024146710611
Bulman, S., Richter, F., Marschollek, S., Benade, F., Jülke, S., Ludwig-Müller, J. (2019). Arabidopsis thaliana Expressing Pb BSMT, a Gene Encoding a SABATH-Type Methyltransferase From the Plant Pathogenic Protist Plasmodiophora brassicae, Show Leaf Chlorosis and Altered Host Susceptibility. Plant Biol. 21, 120–130. doi: 10.1111/plb.12728
Burt, A. J., William, H. M., Perry, G., Khanal, R., Pauls, K. P., Kelly, J. D., et al. (2015). Candidate Gene Identification With SNP Marker-Based Fine Mapping of Anthracnose Resistance Gene Co-4 in Common Bean. PLoS One 10, e0139450. doi: 10.1371/journal.pone.0139450
Campa, A., Giraldez, R., Ferreira, J. J. (2009). Genetic Dissection of the Resistance to Nine Anthracnose Races in the Common Bean Differential Cultivars MDRK and TU. Theor. Appl. Genet. 119, 1–11. doi: 10.1007/s00122-009-1011-8
Campa, A., Rodriguez-Suarez, C., Giraldez, R., Ferreira, J. J. (2014). Genetic Analysis of the Response to Eleven Colletotrichum lindemuthianum Races in a RIL Population of Common Bean (Phaseolus vulgaris L.). BMC Plant Biol. 14, 115. doi: 10.1186/1471-2229-14-115
Chen, M., Wu, J., Wang, L., Mantri, N., Zhang, X., Zhu, Z., et al. (2017). Mapping and Genetic Structure Analysis of the Anthracnose Resistance Locus Co-1HY in the Common Bean (Phaseolus vulgaris L.). PLoS One 12, e0169954. doi: 10.1371/journal.pone.0169954
Choudhary, N., Bawa, V., Paliwal, R., Singh, B., Bhat, M. A., Mir, J. I., et al. (2018). Gene/QTL Discovery for Anthracnose in Common Bean (Phaseolus vulgaris L.) From North-Western Himalayas. PLoS One 13, e0191700. doi: 10.1371/journal.pone.0191700
Coelho, R. T., Goncalves Vidigal, M. C., Vidigal Filho, P. S., Lacanallo, G. F., Darben, L. M., Silva, C. R., et al. (2013). Characterization of the Anthracnose Resistance Gene in the Mesoamerican Common Bean Cultivar Crioulo 159. Annu. Rep. Bean Improv. Coop. 56, 43–44.
Coimbra-Gonçalves, G. K., Gonçalves-Vidigal, M. C., Coelho, R. T., Valentini, G., Vidigal Filho, P. S., Lacanallo, G. F. (2016). Characterization and Mapping of Anthracnose Resistance Genes in Mesoamerican Common Bean Cultivar Crioulo 159. Crop Sci. 56, 2904–2915. doi: 10.2135/cropsci2015.10.0651
Conner, R. L., Boland, G. J., Gillard, C. L., Chen, Y., Shan, X., Mclaren, D. L., et al. (2019). Identification of Anthracnose Races in Manitoba and Ontario From 2005 to 2015 and Their Reactions on Ontario Dry Bean Cultivars. Can. J. Plant Sci. 100, 40–55. doi: 10.1139/cjps-2019-0003
Creusot, F., Macadré, C., Cana, E. F., Riou, C., Geffroy, V., Sévignac, M., et al. (1999). Cloning and Molecular Characterization of Three Members of the NBS-LRR Subfamily Located in the Vicinity of the Co-2 Locus for Anthracnose Resistance in Phaseolus vulgaris. Genome 42, 254–264. doi: 10.1139/g98-134
Cui, B., Pan, Q., Clarke, D., Villarreal, M. O., Umbreen, S., Yuan, B., et al. (2018). S-Nitrosylation of the Zinc Finger Protein SRG1 Regulates Plant Immunity. Nat. Commun. 9, 1–12. doi: 10.1038/s41467-018-06578-3
David, P., Sévignac, M., Thareau, V., Catillon, Y., Kami, J., Gepts, P., et al. (2008). BAC End Sequences Corresponding to the B4 Resistance Gene Cluster in Common Bean: a Resource for Markers and Synteny Analyses. Mol. Genet. Genomics 280, 521. doi: 10.1007/s00438-008-0384-8
de Lima Castro, S. A., Gonçalves-Vidigal, M. C., Gilio, T. A. S., Lacanallo, G. F., Valentini, G., Da Silva Ramos Martins, V., et al. (2017). Genetics and Mapping of a New Anthracnose Resistance Locus in Andean Common Bean Paloma. BMC Genomics 18, 306. doi: 10.1186/s12864-017-3685-7
Deng, H., Liu, H., Li, X., Xiao, J., Wang, S. (2012). a CCCH-Type Zinc Finger Nucleic Acid-Binding Protein Quantitatively Confers Resistance Against Rice Bacterial Blight Disease. Plant Physiol. 158, 876–889. doi: 10.1104/pp.111.191379
Drijfhout, E., Davis, J. (1989). Selection of a New Set of Homogeneously Reacting Bean (Phaseolus vulgaris) Differentials to Differentiate Races of Colletotrichum lindemuthianum. Plant Pathol. 38, 391–396. doi: 10.1111/j.1365-3059.1989.tb02158.x
Duran, L. A., Blair, M. W., Giraldo, M., Macchiavelli, R., Prophète, E., Nin, J. C., et al. (2005). Morphological and Molecular Characterization of Common Bean Landraces and Cultivars From the Caribbean. Crop Sci. 45, 1320–1328. doi: 10.2135/cropsci2004.0501
Earl, D. A. (2012). STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Fernández, M. T., Fernández, M., Casares, A., Rodriguez, R., Fueyo, M. (2000). Bean Germplasm Evaluation for Anthracnose Resistance and Characterization of Agronomic Traits: a New Physiological Strain of Colletotrichum lindemuthianum Infecting Phaseolus vulgaris L. in Spain. Euphytica 114, 143–149. doi: 10.1023/A:1003937812700
Ferreira, J. J., Campa, A., Perez-Vega, E., Giraldez, R. (2008). Reaction of a Bean Germplasm Collection Against Five Races of Colletotrichum lindemuthianum Identified in Northern Spain and Implications for Breeding. Plant Dis. 92, 705–708. doi: 10.1094/PDIS-92-5-0705
Ferrier-Cana, E., Geffroy, V., Macadre, C., Creusot, F., Imbert-Bollore, P., Sevignac, M., et al. (2003). Characterization of Expressed NBS-LRR Resistance Gene Candidates From Common Bean. Theor. Appl. Genet. 106, 251–261. doi: 10.1007/s00122-002-1032-z
Gao, L., Turner, M. K., Chao, S., Kolmer, J., Anderson, J. A. (2016). Genome Wide Association Study of Seedling and Adult Plant Leaf Rust Resistance in Elite Spring Wheat Breeding Lines. PLoS One 11, e0148671. doi: 10.1371/journal.pone.0148671
Gao, W., Long, L., Tian, X., Jin, J., Liu, H., Zhang, H., et al. (2016). Genome-Wide Identification and Expression Analysis of Stress-Associated Proteins (Saps) Containing A20/AN1 Zinc Finger in Cotton. Mol. Genet. Genomics 291, 2199–2213. doi: 10.1007/s00438-016-1252-6
Geffroy, V., Sicard, D., De Oliveira, J. C., Sevignac, M., Cohen, S., Gepts, P., et al. (1999). Identification of an Ancestral Resistance Gene Cluster Involved in the Coevolution Process Between Phaseolus vulgaris and Its Fungal Pathogen Colletotrichum lindemuthianum. Mol. Plant Microbe Interact. 12, 774–784. doi: 10.1094/MPMI.1999.12.9.774
Geffroy, V., Sévignac, M., Billant, P., Dron, M., Langin, T. (2008). Resistance to Colletotrichum lindemuthianum in Phaseolus vulgaris: a Case Study for Mapping Two Independent Genes. Theor. Appl. Genet. 116, 407–415. doi: 10.1007/s00122-007-0678-y
Gepts, P., Debouck, D. G. (1991). “Origin, Domestications, and Evolution of the Common Bean (Phaseolus vulgaris L.),” in Common Beans: Research for Crop Improvement. Eds. Schoonhoven, A., Voysest, O. (Cali: CIAT).
Goncalves-Vidigal, M. C., Kelly, J. D. (2006). Inheritance of Anthracnose Resistance in the Common Bean Cultivar Widusa. Euphytica 151, 411–419. doi: 10.1007/s10681-006-9164-x
Goncalves-Vidigal, M. C., Vidigal Filho, P. S., Medeiros, A. F., Pastor-Corrales, M. A. (2009). Common Bean Landrace Jalo Listras Pretas Is the Source of a New Andean Anthracnose Resistance Gene. Crop Sci. 49, 133–138. doi: 10.2135/cropsci2008.01.0004
Gonçalves-Vidigal, M. C., Cruz, A. S., Garcia, A., Kami, J., Vidigal Filho, P. S., Sousa, L. L. (2011). Linkage Mapping of the Phg-1 and Co-14 Genes for Resistance to Angular Leaf Spot and Anthracnose in the Common Bean Cultivar AND 277. Theor. Appl. Genet. 122, 893–903. doi: 10.1007/s00122-010-1496-1
Goncalves-Vidigal, M. C., Meirelles, A. C., Poletine, J. P., Sousa, L. L. D., Cruz, A. S., Nunes, M. P., et al. (2012). Genetic Analysis of Anthracnose Resistance in ‘Pitanga’ Dry Bean Cultivar. Plant Breed. 131, 423–429. doi: 10.1111/j.1439-0523.2011.01939.x
Goncalves-Vidigal, M. C., Cruz, A. S., Lacanallo, G., Vidigal, P. S., Sousa, L. L., Pacheco, C. M. N. A., et al. (2013). Co-Segregation Analysis and Mapping of the Anthracnose Co-10 and Angular Leaf Spot Phg-ON Disease-Resistance Genes in the Common Bean Cultivar Ouro Negro. Theor. Appl. Genet. 126, 2245–2255. doi: 10.1007/s00122-013-2131-8
Gonçalves-Vidigal, M., Gilio, T., Valentini, G., Vaz-Bisneta, M., Vidigal Filho, P., Song, Q., et al. (2020). New Andean Source of Resistance to Anthracnose and Angular Leaf Spot: Fine-Mapping of Disease-Resistance Genes in California Dark Red Kidney Common Bean Cultivar. PLoS One 15, e0235215. doi: 10.1371/journal.pone.0235215
Gonzalez, A. M., Yuste-Lisbona, F. J., Rodino, A. P., De Ron, A. M., Capel, C., Garcia-Alcazar, M., et al. (2015). Uncovering the Genetic Architecture of Colletotrichum lindemuthianum Resistance Through QTL Mapping and Epistatic Interaction Analysis in Common Bean. Front. Plant Sci. 6, 141. doi: 10.3389/fpls.2015.00141
Goswami, R. S., Del Rio-Mendoza, L. E., Lamppa, R. S., Prischmann, J. (2011). Colletotrichum lindemuthianum Races Prevalent on Dry Beans in North Dakota and Potential Sources of Resistance. Plant Dis. 95, 408–412. doi: 10.1094/PDIS-06-10-0429
Hyten, D. L., Song, Q., Fickus, E. W., Quigley, C. V., Lim, J.-S., Choi, I.-Y., et al. (2010). High-Throughput SNP Discovery and Assay Development in Common Bean. BMC Genomics 11, 475. doi: 10.1186/1471-2164-11-475
Jones, J. D., Dangl, J. L. (2006). The Plant Immune System. Nature 444, 323–329. doi: 10.1038/nature05286
Joshi, B., Mehra, K. (1993). Adaptability in French Bean (Phaseolus vulgaris L.). Indian J. Plant Genet. Resour. 6, 73–77.
Joshi, B., Thomas, T. (1987). Genetic Resources in Temperate Grain Legumes. Plant Genet. Resources-Indian Perspect. NBPGR New Delhi, 255–260.
Juliana, P., Singh, R. P., Singh, P. K., Poland, J. A., Bergstrom, G. C., Huerta-Espino, J., et al. (2018). Genome-Wide Association Mapping for Resistance to Leaf Rust, Stripe Rust and Tan Spot in Wheat Reveals Potential Candidate Genes. Theor. Appl. Genet. 131, 1405–1422. doi: 10.1007/s00122-018-3086-6
Kamfwa, K., Cichy, K. A., Kelly, J. D. (2019). Identification of Quantitative Trait Loci for Symbiotic Nitrogen Fixation in Common Bean. Theor. Appl. Genet. 132, 1375–1387. doi: 10.1007/s00122-019-03284-6
Katoch, S., Katoch, A., Dhiman, S., Sharma, P., Rana, S., Sharma, P. (2019). Recitation of R Genes Identified in Common Bean Landrace KRC-5 and KRC-8 Native to Himachal Pradesh Against Colletotrichum lindemuthianum Virulences. Himachal J. Agric. Res. 45, 51–56.
Kelly, J. D., Bornowski, N. (2018). “Marker-Assisted Breeding for Economic Traits in Common Bean,” in Biotechnologies of Crop Improvement, Volume 3 (Cham: Springer), 211–238.
Kelly, J. D., Afanador, L., Cameron, L. S. (1994). New Races of Colletotrichum lindemuthianum in Michigan and Implications in Dry Bean Resistance Breeding. Plant Dis. 78, 892–894. doi: 10.1094/PD-78-0892
Kelly, J. D., Gepts, P., Miklas, P. N., Coyne, D. P. (2003). Tagging and Mapping of Genes and QTL and Molecular Marker-Assisted Selection for Traits of Economic Importance in Bean and Cowpea. Field Crops Res. 82, 135–154. doi: 10.1016/S0378-4290(03)00034-0
Kumar, A., Sharma, P. N., Sharma, O. P. (1997). Resistance to Colletotrichum lindemuthianum (Bean Anthracnose) in Kidney Bean Accessions of Diverse Origin in Himachal Pradesh. Indian Phytopathol. 50, 59–64.
Kumar, A., Sharma, P. N., Sharma, O. P., Tyagi, P. D. (1999). Epidemiology of Bean Anthracnose Colletotrichum lindemuthianum Under Sub-Humid Mid-Hills Zone of Himachal Pradesh. Indian Phytopathol. 52, 393–397.
Lacanallo, G. F., Gonçalves-Vidigal, M. C. (2015). Mapping of an Andean Gene for Anthracnose Resistance (Co-13) in Common Bean (Phaseolus vulgaris L.) Jalo Listras Pretas Landrace. Australas. J. Crop Sci. 9, 394–400.
Langmead, B., Salzberg, S. L. (2012). Fast Gapped-Read Alignment With Bowtie 2. Nat. Methods 4, 357. doi: 10.1038/nmeth.1923
Mahiya-Farooq, Padder, B. A., Bhat, N. N., Shah, M., Shikari, A. B., Awale, H. E., et al. (2019). Temporal Expression of Candidate Genes At the Co-1 Locus and Their Interaction With Other Defense Related Genes in Common Bean. Physiol. Mol. Plant Pathol. 108, 101424. doi: 10.1016/j.pmpp.2019.101424
Marroni, F., Pinosio, S., Zaina, G., Fogolari, F., Felice, N., Cattonaro, F., et al. (2011). Nucleotide Diversity and Linkage Disequilibrium in Populus nigra Cinnamyl Alcohol Dehydrogenase (CAD4) Gene. Tree Genet. Genomes 7, 1011–1023. doi: 10.1007/s11295-011-0391-5
McConnell, M., Mamidi, S., Lee, R., Chikara, S., Rossi, M., Papa, R., et al. (2010). Syntenic Relationships Among Legumes Revealed Using a Gene-Based Genetic Linkage Map of Common Bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 121, 1103–1116. doi: 10.1007/s00122-010-1375-9
Melotto, M., Kelly, J. D. (2000). An Allelic Series At the Co-1 Locus Conditioning Resistance to Anthracnose in Common Bean of Andean Origin. Euphytica 116, 143–149. doi: 10.1023/A:1004005001049
Melotto, M., Kelly, J. D. (2001). Fine Mapping of the Co-4 Locus of Common Bean Reveals a Resistance Gene Candidate, COK-4, That Encodes for a Protein Kinase. Theor. Appl. Genet. 103, 508–517. doi: 10.1007/s001220100609
Meziadi, C., Richard, M. M., Derquennes, A., Thareau, V., Blanchet, S., Gratias, A., et al. (2016). Development of Molecular Markers Linked to Disease Resistance Genes in Common Bean Based on Whole Genome Sequence. Plant Sci. 242, 351–357. doi: 10.1016/j.plantsci.2015.09.006
Miklas, P., Chilagane, L. A., Fourie, D., Nichimbi, S., Soler-Garzon, A., Hart, J., et al. (2020). QTL for Resistance to Angular Leaf Spot and Rust in Tanzania Vs South Africa for the Andean Panel & Rojo/Cal 143 RIL Population. Bean Improv. Coop. Annu. Rep. 63, 83–84.
Moghaddam, S. M., Mamidi, S., Osorno, J. M., Lee, R., Brick, M., Kelly, J., et al. (2016). Genome-Wide Association Study Identifies Candidate Loci Underlying Agronomic Traits in a Middle American Diversity Panel of Common Bean. Plant Genome 9, 1–21. doi: 10.3835/plantgenome2016.02.0012
Mourad, A. M., Sallam, A., Belamkar, V., Wegulo, S., Bowden, R., Jin, Y., et al. (2018). Genome-Wide Association Study for Identification and Validation of Novel SNP Markers for Sr6 Stem Rust Resistance Gene in Bread Wheat. Front. Plant Sci. 9, 380. doi: 10.3389/fpls.2018.00380
Muhammad, A., Hu, W., Li, Z., Li, J., Xie, G., Wang, J., et al. (2020). Appraising the Genetic Architecture of Kernel Traits in Hexaploid Wheat Using GWAS. Int. J. Mol. Sci. 21, 5649. doi: 10.3390/ijms21165649
Mungalu, H., Sansala, M., Hamabwe, S., Mukuma, C., Gepts, P., Kelly, J. D., et al. (2020). Identification of Race-Specific Quantitative Trait Loci for Resistance to Colletotrichum lindemuthianum in an Andean Population of Common Bean. Crop Sci. doi: 10.1002/Csc1002.20191
Murray, M., Thompson, W. F. (1980). Rapid Isolation of High Molecular Weight Plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Murube, E., Campa, A., Ferreira, J. J. (2019). Integrating Genetic and Physical Positions of the Anthracnose Resistance Genes Described in Bean Chromosomes Pv01 and Pv04. PloS One 14, e0212298. doi: 10.1371/journal.pone.0212298
Oblessuc, P. R., Borges, A., Chowdhury, B., Caldas, D. G., Tsai, S. M., Camargo, L. E., et al. (2012). Dissecting Phaseolus vulgaris Innate Immune System Against Colletotrichum lindemuthianum Infection. PLoS One 7, e43161. doi: 10.1371/journal.pone.0043161
Oblessuc, P. R., Baroni, R. M., Pereira, G. D., Chiorato, A. F., Carbonell, S. A. M., Brinez, B., et al. (2014). Quantitative Analysis of Race-Specific Resistance to Colletotrichum lindemuthianum in Common Bean. Mol. Breed. 34, 1313–1329. doi: 10.1007/s11032-014-0118-z
Oblessuc, P. R., Francisco, C., Melotto, M. (2015). The Co-4 Locus on Chromosome Pv08 Contains a Unique Cluster of 18 COK-4 Genes and Is Regulated by Immune Response in Common Bean. Theor. Appl. Genet. 128, 1193–1208. doi: 10.1007/s00122-015-2500-6
Padder, B. A., Sharma, P. N., Sharma, O. P., Kapoor, V. (2007). Genetic Diversity and Gene Flow Estimates Among Five Populations of Colletotrichum lindemuthianum Across Himachal Pradesh. Physiol. Mol. Plant Pathol. 70, 8–12. doi: 10.1016/j.pmpp.2007.05.003
Padder, B. A., Sharma, P. N., Sharma, O. P. (2009). Virulence and RAPD Data - A Tool to Study the Evolutionary Trends of Colletotrichum lindemuthianum Virulences in the North Western Himalayan Region of India. Arch. Phytopathol. Plant Prot. 42, 610–617. doi: 10.1080/03235400701287305
Padder, B. A., Sharma, P. N., Sharma, O. P. (2010). Distribution of Colletotrichum lindemuthianum Race Flora and Its Implication in Deployment of Resistant Sources Across Himachal Pradesh. Res. J. Agric. Sci. 1, 1–6.
Padder, B. A., Kamfwa, K., Awale, H. E., Kelly, J. D. (2016). Transcriptome Profiling of the Phaseolus vulgaris-Colletotrichum lindemuthianum Pathosystem. PLoS One 11, e0165823. doi: 10.1371/journal.pone.0165823
Padder, B. A., Sharma, P. N., Awale, H., Kelly, J. D. (2017). Colletotrichum lindemuthiam the Casual Agent of Bean Anthracnose. J. Plant Pathol. 99, 317–330. doi: 10.4454/jpp.v99i2.3867
Padder, B. (2014). “Plant Disease Resistance Genes: From Perception to Signal Transduction,” in Plant Signaling: Understanding the Molecular Crosstalk (New Delhi: Springer), 345–354.
Pastor-Corrales, M., Tu, J. C. (1989). “Anthracnose,” in Bean Production Problems in Tropics. Eds. Schwartz, H. F., Pastor-Corrales, M. A. (Cali, Colombia: Centro Internacional De Agricultura Tropical), 77–104.
Pathania, A., Sharma, P. N., Sharma, O. P., Chahota, R. K., Bilal, A., Sharma, P. (2006). Evaluation of Resistance Sources and Inheritance of Resistance in Kidney Bean to Indian Virulences of Colletotrichum lindemuthianum. Euphytica 149, 97–103. doi: 10.1007/s10681-005-9057-4
Pathania, A., Sharma, S. K., Sharma, P. N. (2014). “Common Bean,” in Broadening the Genetic Base of Grain Legumes (New Delhi: Springer), 11–50.
Pina-Martins, F., Silva, D. N., Fino, J., Paulo, O. S. (2017). Structure_Threader: an Improved Method for Automation and Parallelization of Programs Structure, Faststructure and Maverick on Multicore CPU Systems. Mol. Ecol. Resour. 16, 268–274. doi: 10.1111/1755-0998.12702
Rai, N., Singh, P., Verma, A., Yadav, P., Choubey, T. (2010). Hierarchical Analysis for Genetic Variability in Pole Type French Bean. Indian J. Hortic. 67, 150–153.
Rana, J., Dutta, M., Rathi, R. (2012). Plant Genetic Resources of the Indian Himalayan Region—An Overview. Indian J. Genet. Plant Breed. 72, 115.
Rana, J., Sharma, T., Tyagi, R., Chahota, R., Gautam, N., Singh, M., et al. (2015). Characterisation of 4274 Accessions of Common Bean (Phaseolus vulgaris L.) Germplasm Conserved in the Indian Gene Bank for Phenological, Morphological and Agricultural Traits. Euphytica 205, 441–457. doi: 10.1007/s10681-015-1406-3
Richard, M. M. S., Pflieger, S., Sevignac, M., Thareau, V., Blanchet, S., Li, Y. P., et al. (2014). Fine Mapping of Co-x, an Anthracnose Resistance Gene to a Highly Virulent Strain of Colletotrichum lindemuthianum in Common Bean. Theor. Appl. Genet. 127, 1653–1666. doi: 10.1007/s00122-014-2328-5
Richard, M. M. S., Gratias, A., Thareau, V., Kim, K. D., Balzergue, S., Joets, J., et al. (2017). Genomic and Epigenomic Immunity in Common Bean: the Unusual Features of NB-LRR Gene Family. DNA Res. 25, 161–172. doi: 10.1093/dnares/dsx046
Rio, L. E. D., Lamppa, R. S., Gross, P. L. (2003a). Characterization of the Reaction of North Dakota Dry Bean Cultivars to Three Races of Colletotrichum lindemuthianum. Plant Dis. 87, 263–265. doi: 10.1094/PDIS.2003.87.3.263
Rio, L. E. D., Lamppa, R. S., Gross, P. L., Brolley, B., Prischmann, J. (2003b). Identification of Colletotrichum lindemuthianum Race 73 in Manitoba, Canada. Can. J. Plant Pathol. 25, 104–107. doi: 10.1080/07060660309507055
Rispail, N., Kaló, P., Kiss, G. B., Ellis, T. H. N., Gallardo, K., Thompson, R. D., et al. (2010). Model Legumes Contribute to Faba Bean Breeding. Field Crops Res. 115, 253–269. doi: 10.1016/j.fcr.2009.03.014
Rodriguez-Suarez, C., Mendez-Vigo, B., Paneda, A., Ferreira, J. J., Giraldez, R. (2007). A Genetic Linkage Map of Phaseolus vulgaris L. and Localization of Genes for Specific Resistance to Six Races of Anthracnose (Colletotrichum lindemuthianum). Theor. Appl. Genet. 114, 713–722. doi: 10.1007/s00122-006-0471-3
Schmutz, J., Mcclean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A Reference Genome for Common Bean and Genome-Wide Analysis of Dual Domestications. Nat. Genet. 46, 707–713. doi: 10.1038/ng.3008
Sharma, P., Sugha, S., Panwar, K., Sagwal, J. (1993). Reaction of Landraces and Exotic Collections of Kidney Bean (Phaseolus vulgaris) to Anthracnose (Colletotrichum lindemuthianum). Indian J. Agric. Sci. 63, 456–457.
Sharma, P. N., Sharma, O. P., Tyagi, P. D. (1994). Status and Distribution of Bean Anthracnose in Himachal Pradesh. Himachal J. Agric. Res. 20, 91–96.
Sharma, P. N., Kumar, A., Sharma, O. P., Sud, D., Tyagi, P. D. (1999). Pathogenic Variability in Colletotrichum lindemuthianum and Evaluation of Resistance in Phaseolus vulgaris in the North-Western Himalayan Region of India. J. Phytopathol. 147, 41–45. doi: 10.1046/j.1439-0434.1999.147001041.x
Sharma, P. N., Kapila, R. K., Sharma, O. P., Deepika, S. (2000). Inheritance of Resistance in Two Indian Land Races of Phaseolus vulgaris to Colletotrichum lindemuthianum. Indian Phytopathol. 53, 83–86.
Sharma, T. R., Rana, J. C., Rajan, S., Rathour, R., Sharma, P. N. (2006). Genetic Diversity Analysis of Exotic and Indian Accessions of Common Bean (Phaseolus vulgaris L.) Using RAPD Markers. Indian J. Genet. Plant Breed. 66, 275–278.
Sharma, P. N., Padder, B. A., Sharma, O. P., Pathania, A., Sharma, P. (2007). Pathological and Molecular Diversity in Colletotrichum lindemuthianum (Bean Anthracnose) Across Himachal Pradesh, a North-Western Himalayan State of India. Australas. Plant Pathol. 36, 191–197. doi: 10.1071/AP07013
Sharma, P. N., Sharma, O. P., Padder, B. A., Kapil, R. (2008). Yield Loss Assessment in Common Bean Due to Anthracnose (Colletotrichum lindemuthianum) Under Sub-Temperate Conditions of North-Western Himalayas. Indian Phytopathol. 61, 323–330.
Sharma, P. N., Kajal, B., Rana, J. C., Ruby, N., Sharma, S. K., Anju, P. (2012). Screening of Common Bean Germplasm Against Colletotrichum lindemuthianum Causing Bean Anthracnose. Indian Phytopathol. 65, 99–101.
Sharma, P. N., Diaz, L. M., Blair, M. W. (2013). Genetic Diversity of Two Indian Common Bean Germplasm Collections Based on Morphological and Microsatellite Markers. Plant Genet. Resour. 11, 121–130. doi: 10.1017/S1479262112000469
Sharma, N., Kumari, N., Sharma, S. K., Padder, B. A., Sharma, P. N. (2019). Investigating the Virulence and Genetic Diversity of Colletotrichum lindemuthianum Populations Distributed in the North Western Himalayan Hill States. J. Plant Pathol. 101, 677–688. doi: 10.1007/s42161-019-00269-8
Singh, S. P., Gepts, P., Debouck, D. G. (1991). Races of Common Bean (Phaseolus vulgaris, Fabaceae). Econ. Bot. 45, 379–396. doi: 10.1007/BF02887079
Sofi, P., Rana, J., Bhat, N. (2014). Pattern of Variation in Common Bean (Phaseolus vulgaris L) Genetic Resources of Jammu and Kashmir. J. Food Legumes 27, 197–201.
Song, Q., Jia, G., Hyten, D. L., Jenkins, J., Hwang, E.-Y., Schroeder, S. (2015). SNP Assay Development for Linkage Map Construction, Anchoring Whole Genome Sequence and Other Genetic and Genomic Applications in Common Bean. G3: Genes Genomes Genet. 11, 2285–2290. doi: 10.1534/g3.115.020594
Sousa, L. L., Goncalves, A. O., Goncalves Vidigal, M. C., Lacanallo, G. F., Fernandez, A. C., Awale, H., et al. (2015). Genetic Characterization and Mapping of Anthracnose Resistance of Common Bean Landrace Cultivar Corinthiano. Crop Sci. 55, 1–11. doi: 10.2135/cropsci2014.09.0604
Tiwari, M., Singh, N., Rathore, M., Kumar, N. (2005). RAPD Markers in the Analysis of Genetic Diversity Among Common Bean Germplasm From Central Himalaya. Genet. Resour. Crop Evol. 52, 315–324. doi: 10.1007/s10722-005-5123-y
Trabanco, N., Campa, A., Ferreira, J. J. (2015). Identification of a New Chromosomal Region Involved in the Genetic Control of Resistance to Anthracnose in Common Bean. Plant Genome 8, 1–11. doi: 10.3835/plantgenome2014.10.0079
Turner, S. D. (2014). Qqman: An R Package for Visualizing GWAS Results Using QQ and Manhattan Plots. Biorxiv 005165. doi: 10.1101/005165
Valentini, G., Gonçalves-Vidigal, M. C., Hurtado-Gonzales, O. P., De Lima Castro, S. A., Cregan, P. B., Song, Q., et al. (2017). High-Resolution Mapping Reveals Linkage Between Genes in Common Bean Cultivar Ouro Negro Conferring Resistance to the Rust, Anthracnose, and Angular Leaf Spot Diseases. Theor. Appl. Genet. 130, 1705–1722. doi: 10.1007/s00122-017-2920-6
Vlasova, A., Capella-Gutiérrez, S., Rendón-Anaya, M., Hernández-Oñate, M., Minoche, A. E., Erb, I., et al. (2016). Genome and Transcriptome Analysis of the Mesoamerican Common Bean and the Role of Gene Duplications in Establishing Tissue and Temporal Specialization of Genes. Genome Biol. 17, 32. doi: 10.1186/s13059-016-0883-6
Wang, M., Zhu, X., Wang, K., Lu, C., Luo, M., Shan, T., et al. (2018). A Wheat Caffeic Acid 3-O-Methyltransferase Tacomt-3D Positively Contributes to Both Resistance to Sharp Eyespot Disease and Stem Mechanical Strength. Sci. Rep. 8, 1–14. doi: 10.1038/s41598-018-24884-0
Wu, J., Zhu, J., Wang, L., Wang, S. (2017). Genome-Wide Association Study Identifies NBS-LRR-Encoding Genes Related With Anthracnose and Common Bacterial Blight in the Common Bean. Front. Plant Sci. 8, 1398. doi: 10.3389/fpls.2017.01398
Yang, Q., He, Y., Kabahuma, M., Chaya, T., Kelly, A., Borrego, E., et al. (2017). A Gene Encoding Maize Caffeoyl-CoA O-Methyltransferase Confers Quantitative Resistance to Multiple Pathogens. Nat. Genet. 49, 1364. doi: 10.1038/ng.3919
Young, R., Melotto, M., Nodari, R., Kelly, J. (1998). Marker-Assisted Dissection of the Oligogenic Anthracnose Resistance in the Common Bean Cultivar,’G2333’. Theor. Appl. Genet. 96, 87–94. doi: 10.1007/s001220050713
Zhang, Z., Ersoz, E., Lai, C.-Q., Todhunter, R. J., Tiwari, H. K., Gore, M. A., et al. (2010). Mixed Linear Model Approach Adapted for Genome-Wide Association Studies. Nat. Genet. 42, 355. doi: 10.1038/ng.546
Keywords: Phaseolus vulgaris, SNPs, genome wide association study, anthracnose resistance, quantitative trait loci, northwestern Himalayan beans
Citation: Banoo A, Nabi A, Rasool RS, Mahiya-Farooq, Shah MD, Ahmad M, Sofi PA, Aasiya-Nabi, Itoo H, Sharma PN and Padder BA (2020) North-Western Himalayan Common Beans: Population Structure and Mapping of Quantitative Anthracnose Resistance Through Genome Wide Association Study. Front. Plant Sci. 11:571618. doi: 10.3389/fpls.2020.571618
Received: 08 July 2020; Accepted: 04 September 2020;
Published: 06 October 2020.
Edited by:
Roberto Papa, Marche Polytechnic University, ItalyReviewed by:
Karl Peter Pauls, University of Guelph, CanadaJames Kelly, Michigan State University, United States
Copyright © 2020 Banoo, Nabi, Rasool, Mahiya-Farooq, Shah, Ahmad, Sofi, Aasiya-Nabi, Itoo, Sharma and Padder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bilal A. Padder, YmFwYWRkZXJAcmVkaWZmbWFpbC5jb20=
 Aqleema Banoo1
Aqleema Banoo1 Mahiya-Farooq
Mahiya-Farooq Mehraj D. Shah
Mehraj D. Shah Parvaze A. Sofi
Parvaze A. Sofi Aasiya-Nabi
Aasiya-Nabi P. N. Sharma
P. N. Sharma Bilal A. Padder
Bilal A. Padder