- School of Life Sciences, University of Essex, Colchester, United Kingdom
C4 photosynthesis is characterized by the compartmentalization of the processes of atmospheric uptake of CO2 and its conversion into carbohydrate between mesophyll and bundle-sheath cells. As a result, most of the enzymes participating in the Calvin-Benson-Bassham (CBB) cycle, including RubisCO, are highly expressed in bundle-sheath cells. There is evidence that changes in the regulatory sequences of RubisCO contribute to its bundle-sheath-specific expression, however, little is known about how the spatial-expression pattern of other CBB cycle enzymes is regulated. In this study, we use a computational approach to scan for transcription factor binding sites in the regulatory regions of the genes encoding CBB cycle enzymes, SBPase, FBPase, PRK, and GAPDH-B, of C3 and C4 grasses. We identified potential cis-regulatory elements present in each of the genes studied here, regardless of the photosynthetic path used by the plant. The trans-acting factors that bind these elements have been validated in A. thaliana and might regulate the expression of the genes encoding CBB cycle enzymes. In addition, we also found C4-specific transcription factor binding sites in the genes encoding CBB cycle enzymes that could potentially contribute to the pathway-specific regulation of gene expression. These results provide a foundation for the functional analysis of the differences in regulation of genes encoding CBB cycle enzymes between C3 and C4 grasses.
Introduction
C4 plants achieve higher photosynthetic efficiency by concentrating CO2 around RubisCO. In contrast with enzymes participating in C3 photosynthesis, C4-enzymes are compartmentalized to specific cell types, namely mesophyll (M) and bundle-sheath (BS) cells. Enzymes enriched in M cells include phosphoenolpyruvate carboxylase (PEPC) and pyruvate orthophosphate dikinase (Ppdk), whereas decarboxylating malic enzymes (NAD or NADP-Me) and RubisCO are enriched in the BS cells (Sheen and Bogorad, 1987; Hibberd and Covshoff, 2010; Berry et al., 2011).
During C4 evolution a change in localization of the enzymes involved in CO2 assimilation resulted in the compartmentalization of these reactions in either the M or BS cell types. A number of regulatory elements conferring a M or BS specific expression pattern have been identified in the regulatory sequences of the genes encoding PEPC, Ppdk; or NADP-ME, NAD-ME, and RubisCO (Nomura et al., 2000; Berry et al., 2011; Williams et al., 2016; Reyna-Llorens et al., 2018). To further interrogate those regulatory elements, a combination of comparative transcriptomics to identify differential expression of genes (Bräutigam et al., 2011; Aubry et al., 2014; Xu et al., 2016) and DNAse-seq to map differences in open chromatin regions between M and BS cells (Burgess et al., 2019) have been used. These studies have led to the identification of putative cis-regulatory elements and the trans-acting transcription factors binding to those elements, and have shown that the motifs conferring differences in expression in the C4 species have been recruited from pre-existing sequences in C3 species, rather than being generated de novo during the evolution of the C4 condition (Niklaus and Kelly, 2019).
Calvin Benson-Bassham (CBB) cycle enzymes, including RubisCO, are expressed in both C3 and C4 species. Similar to RubisCO, most of the CBB cycle enzymes are enriched in BS cells in C4 species (Sheen and Bogorad, 1987; John et al., 2014; Rao et al., 2016). Unlike RubisCO, little is known about the changes in the regulatory sequences of the other 10 genes encoding CCB cycle enzymes that enable such compartmentalization, limiting our ability to develop strategies to manipulate this pathway to improve photosynthetic efficiency. Here, we present a bioinformatics analysis of the regulatory sequences of genes encoding CBB cycle enzymes with the aim of identifying regulatory elements that are common to C3 and C4 species, or C4-specific regulatory elements that control photosynthesis and contribute to C4 compartmentalization. We selected four of the CBB cycle enzymes known to be redox-regulated by the ferredoxin/thioredoxin (Fd/TRX) system (Michelet et al., 2013) and that function exclusively in the CBB cycle: SBPase, FBPase (chloroplastic variant), PRK and GAPDH-B. Given the numerous independent origins of C4 photosynthesis that might have led to parallel evolution of cis-regulatory elements (Sage et al., 2012), in this paper we focus on a small subset of eight grasses from the Poaceae family whose genomes have been sequenced and annotated.
In this study we have identified putative regulatory elements that are common in both C3 and C4 species as well as C4-specific elements. We have also used existing data to explore the expression patterns of the trans-acting factors that have been shown or proposed to bind to these elements, suggesting a possible role in the compartmentalization of CBB cycle enzymes in C4 plants. The results presented here provide the basis for future functional studies.
Materials and Methods
DNA Sequences
Genomic sequences encoding CBB cycle enzymes of Oryza sativa (Ouyang et al., 2007), Hordeum vulgare (Beier et al., 2017; Mascher et al., 2017), Brachypodium distachyon (International Brachypodium Initiative, 2010), Zea mays (Schnable et al., 2009; Hirsch et al., 2016), Sorghum bicolor (McCormick et al., 2018), Setaria viridis (v2.1, DOE-JGI)1 and Panicum virgatum (v1.0, DOE-JGI, see footnote) were obtained from Phytozome12 (Goodstein et al., 2011). Arabidopsis thaliana genes (AT3G55800—SBPase, AT3G54050—chlFBPase, AT1G32060—PRK, and AT1G42970—GAPDHB) were used to identify orthologs in every species. For the genomic sequences encoding CBB cycle enzymes of Dichanthelium oligosanthes (Studer et al., 2016), the A. thaliana coding sequences were aligned against the D. oligosanthes genome using BLAST (Altschul et al., 1990) to find orthologous genes. Sequences used are included in Supplementary Material.
Motif Prediction in Conserved Non-coding Sequences (CNS)
Genomic sequences were aligned using mVISTA (Frazer et al., 2004) and aligned CNSs were used as input for motif prediction using MEME (v5.1.1; Bailey et al., 2009). Motif site distribution was set to zoops and maximum motif width to the size of the shorter CNS. Predicted motifs were used as input in FIMO (Grant et al., 2011) to scan the regulatory sequences of orthologous genes in other species.
Motif Scanning of Genomic Sequences
A collection of 529 plant transcription factor motifs validated in A. thaliana (O’Malley et al., 2016) were used to scan for motifs using FIMO (Grant et al., 2011) with default parameters.
Data Processing and Visualization
Data processing and visualization were performed using R 3.6.0 (R Core Team, 2019). The dplyr package (Wickham et al., 2019) was used to filter the identified motifs by q < 0.05, genomic feature, and by species. The UpSetR package (Gehlenborg, 2019) was used to generate Figure 2A showing all possible interactions; and the ggplot2 (Wickham, 2016) and the gggenes packages (Wilkins, 2019) were used to generate Figure 2B.
Transcriptomics Analysis
Transcriptomic data from RNAseq experiments in which mesophyll and bundle sheath cells were separated in P. virgatum (Rao et al., 2016), S. viridis (John et al., 2014), Panicum hallii (Washburn et al., 2017), and Setaria italica (Washburn et al., 2017) were obtained from NCBI (BioProject accession numbers: PRJNA293441, PRJEB5074, PRJNA475365). A classification-based quantification was performed using kallisto (Bray et al., 2016) with the transcriptomes and genome annotation obtained from Phytozome 12 (Goodstein et al., 2011; Bennetzen et al., 2012). In short, a kallisto index was built with the reference transcriptome of each species, and kallisto quant was used to quantify abundance of pair-end reads with default parameters. Differential expression analysis was performed with R packages DESeq (Anders and Huber, 2010) using estimateSizeFactors, estimateispersions and nbinomTest functions; DESeq2 (Love et al., 2014) using DESeq function, and edgeR (Robinson et al., 2009; McCarthy et al., 2012) using estimateCommonDisp, estimateTagwiseDisp and exactTest functions. P-values were adjusted with the Hochberg method in the three analyses, and only genes with adjusted p < 0.05 in at least one of the analyses were included in Table 1. Parallel (Tange, 2018) was used at every step to run jobs in parallel.
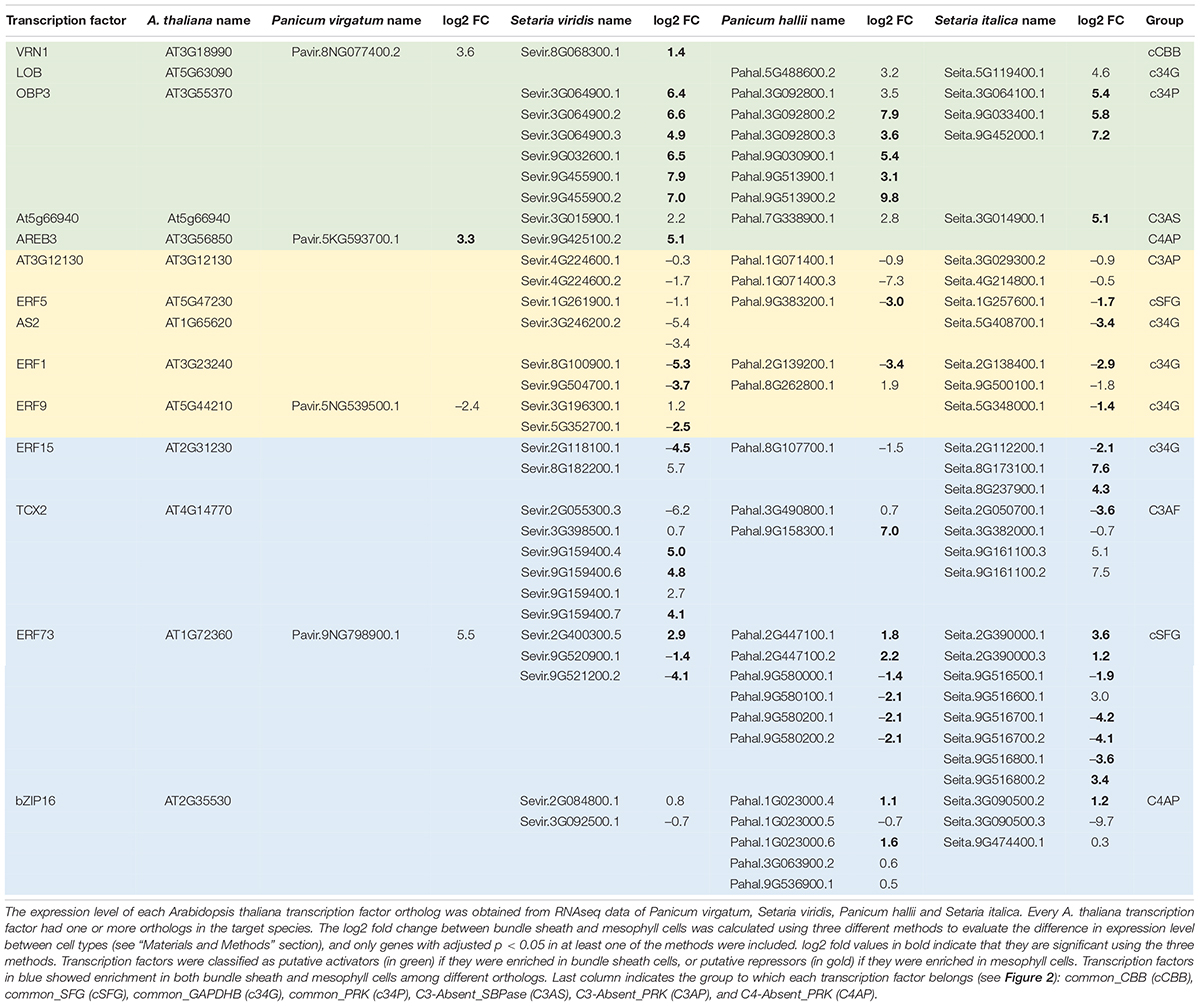
Table 1. Differential expression of trans-acting factors binding putative TFBS in bundle sheath and mesophyll cells of C4 species.
To construct Supplementary Table 1, we used the 57 A. thaliana transcription factors that have been shown to bind the identified transcription factor binding sites (TFBS, 50 shared by different orthologous genes, Figure 2; plus 7 absent from C3 or C4 species, Supplementary Figure S5). We identify the orthologous genes in grass species and evaluate their enrichment in M or BS cells using publicly available transcriptomic data for S. viridis (John et al., 2014), S. italica (Washburn et al., 2017), P. virgatum (Rao et al., 2016), P. halli (Washburn et al., 2017), Z. mays (Chang et al., 2012), and S. bicolor (Döring et al., 2016). All these databases separate M and BS cells from whole leaves. We identified 10 orthologous genes significantly enriched (adj. p < 0.05) in P. virgatum, which corresponded to 8 genes in A. thaliana. For P. halli we identified 21 orthologs corresponding to 11 A. thaliana genes. For S. viridis we identified 53 orthologs corresponding to 26 A. thaliana genes. For S. italica we identified 46 orthologs corresponding to 22 A. thaliana genes. For Z. mays we identified 10 orthologs corresponding to 4 A. thaliana genes. For S. bicolor we identified 2 orthologs corresponding to 2 A. thaliana genes. In Table 1, we only included the A. thaliana genes for which the log2 fold was at least 1, and with consistent data from at least two species. We also removed Z. mays and S. bicolor orthologous genes as their transcriptomic data did not add any information on the A. thaliana genes included on Table 1.
Results
To account for the numerous independent origins of C4 photosynthesis, we focus on a small subset of eight grasses: Oryza sativa, Hordeum vulgare, Brachypodium distachyon, Dichanthelium oligosanthes, Zea mays, Sorghum bicolor, Panicum virgatum, and Setaria viridis. All of these plant species belong to the Poaceae family and shared a common ancestor around 50 million years ago. O. sativa, H. vulgare, B. distachyon, and D. oligosanthes perform C3 photosynthesis, whereas Z. mays, S. bicolor, P. virgatum, and S. viridis perform C4 photosynthesis. Notably, D. oligosanthes belongs to the PACMAD clade (Figure 1), to which all selected C4 species belong, and shares a common ancestor with them around 15 million years ago (Studer et al., 2016). To identify conserved regulatory regions in genes encoding CBB cycle enzymes of C3 and C4 grasses, we aligned each gene against its orthologous gene in a representative C3 species (B. distachyon; Figure 1A and Supplementary Figures S1A, S2A, S3A) and against its orthologous gene in a representative C4 species (S. bicolor; Figure 1C and Supplementary Figures S1C, S2C, S3B). The genomic sequence including potential promoters [2000 base pair (bp)] upstream from the annotated transcription start site (or start codon otherwise) and potential terminators (1,000 bp downstream from the end of 3′UTR or stop codon) was used to allow for the identification of putative regulatory regions outside coding sequences. Regions showing between 50 and 100% identity were plotted and conserved regions with over 70% identity were colored depending on the genomic feature (Figures 1A,C; coding sequences in purple, untranslated regions [UTRs] in cyan, and intergenic regions and introns in pink) As expected, most of the coding sequences were conserved among all orthologous genes, whereas only parts of the introns and intergenic sequences showed over 70% identity. We defined those regions as conserved non-coding sequences (CNS). For SBPase, we identified one CNS located at the last intron of most orthologs (Figures 1A,C), and two CNSs found only in SBPase orthologous genes from PACMAD species (C4 species + D. oligosanthes; Figure 1C). In addition, we found one CNS located at the 5′ intergenic region of all PRK genes (Supplementary Figure S1), and two CNSs located at the 5′ intergenic region of FBPase genes from PACMAD species (Supplementary Figure S2). To further characterize these CNSs, they were subjected to motif prediction using MEME (Bailey et al., 2009), which generated a position weight matrix for the predicted motifs (Figure 1B and Supplementary Figures S1B, S2B). We used these motifs to scan the orthologous genes of other species, and identified the PRK CNS in the intergenic regions of PRK orthologs in non-grasses species (Supplementary Dataset 1). These results indicate that there are conserved potential cis-regulatory sequences shared between C3 and C4 species. However, this alignment approach is based on sequence identity over at least 50 bp; so it was possible that smaller motifs, such as transcription factor binding sites (TFBS) could have been disregarded.
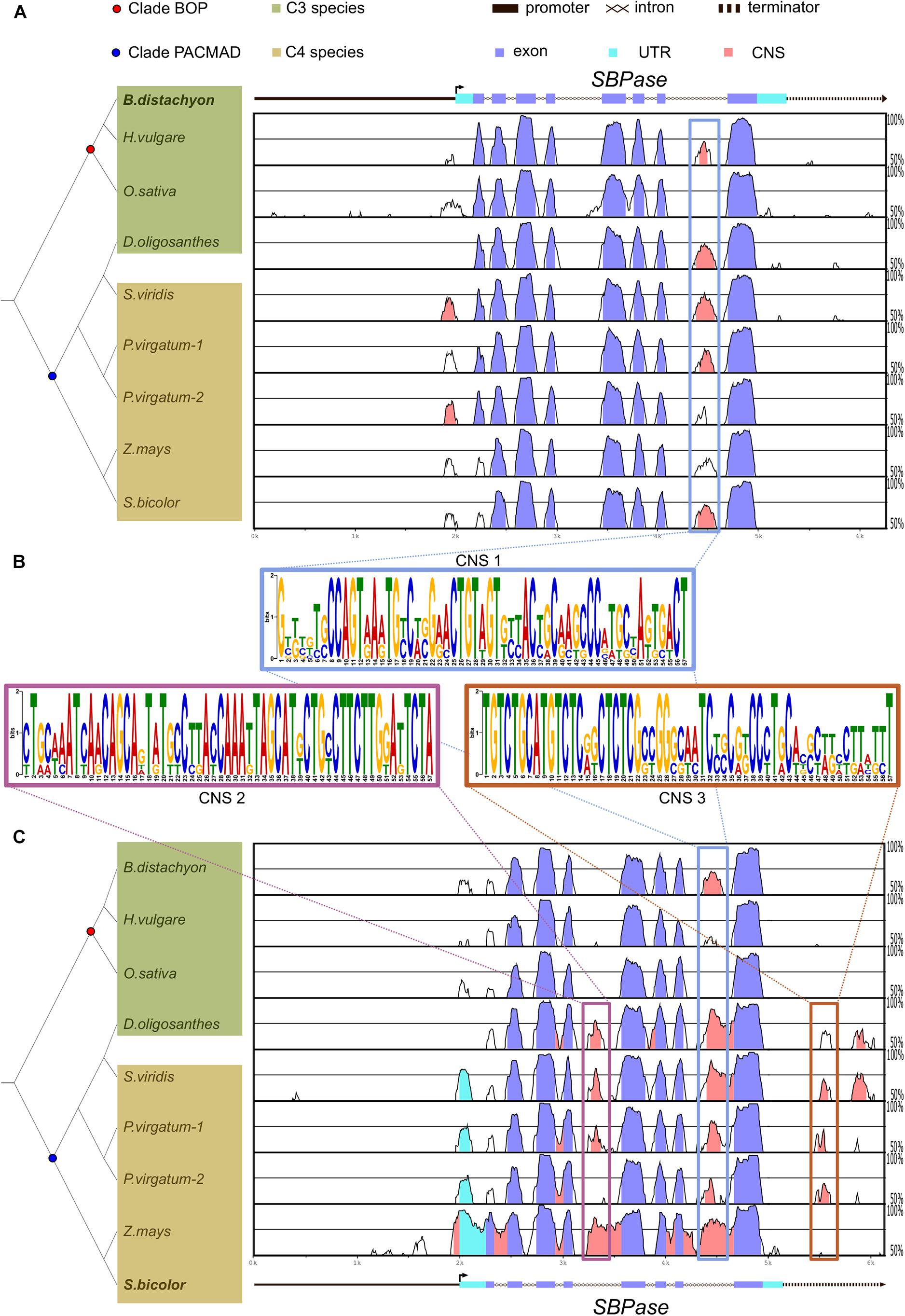
Figure 1. SBPase coding sequence is highly conserved among C3 and C4 grasses in comparison to putative regulatory regions. (A,C) mVISTA plots of Brachypodium distachyon (A) and Sorghum bicolor (C) SBPase aligned to SBPase orthologs in C3 and C4 grasses. Genomic region includes approximately 2 kb upstream from the transcription start site and 1 kb after the end of the 3′ untranslated region (UTR). UTRs, exons, and introns are annotated. The vertical line with a small perpendicular arrow indicates the transcription start site and the arrowhead the orientation of the gene. The graph shows sequences with 50–100% identity and regions with > 70% identity within 50 base pairs are highlighted in purple if they are located in exons, in cyan if they are located in UTRs, or in pink if they are located outside exons or UTRs. Boxes highlight conserved non-coding sequences (CNSs), and the predicted position weight matrix for each conserved sequence is included (B). On the left side, the phylogenetic relationship between C3 (in green) and C4 (in brown) grasses is shown. Common ancestor of BOP clade and PACMAD clade species are shown as a red and as a blue dot, respectively. Note that Dichanthelium oligosanthes is a C3 species within the PACMAD.
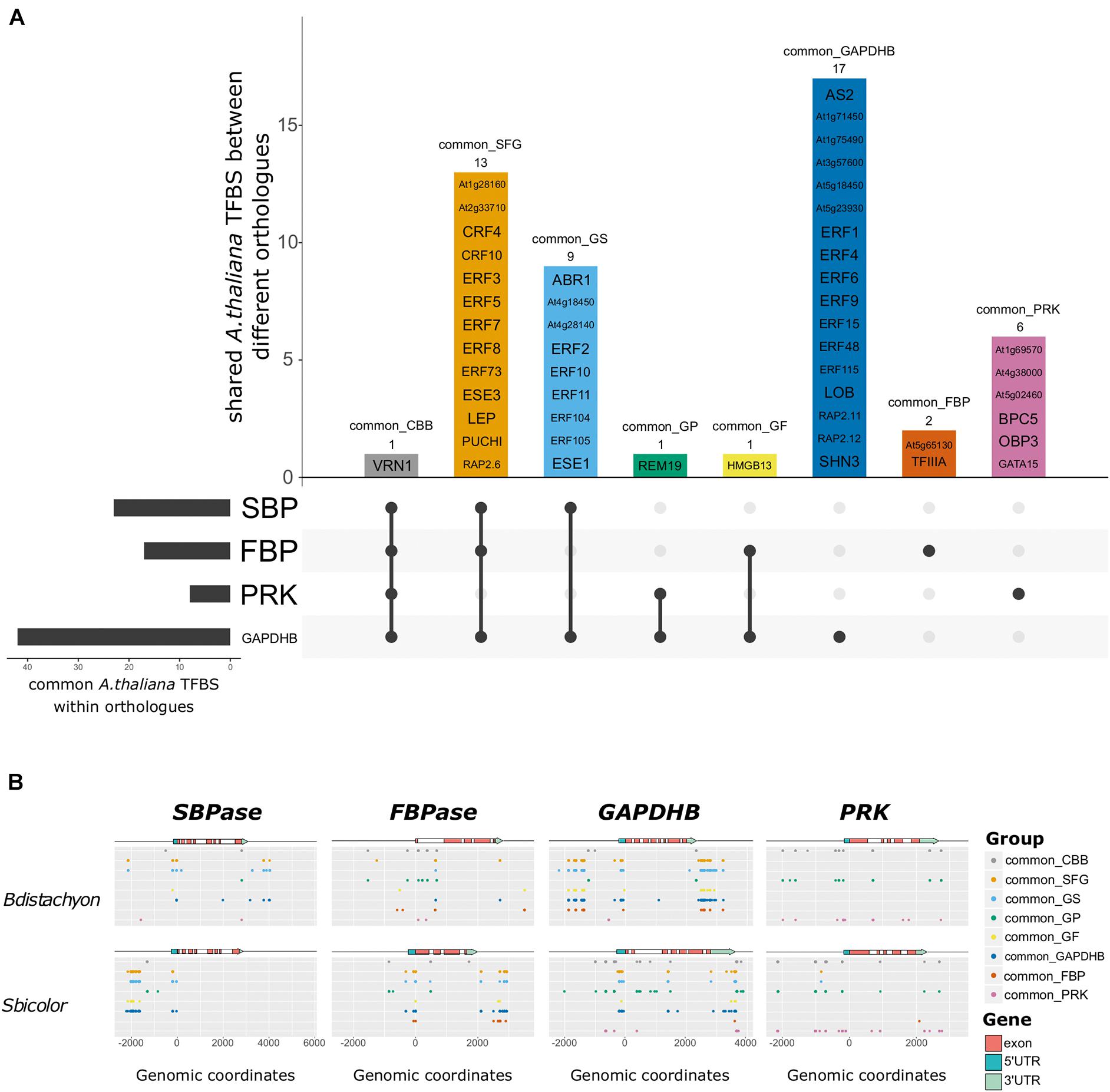
Figure 2. Arabidopsis thaliana transcription factor binding sites (TFBS) identified in the potential regulatory sequences of genes encoding C3 and C4 Calvin-Benson-Bassham (CBB) cycle enzymes. (A) Upset plot showing the identified A. thaliana TFBS and in which orthologous genes they are found. Horizontal bars represent the number of common motifs identified within orthologs, vertical bars represent the motifs shared between different orthologous genes, as indicated by the dots below. The name of the A. thaliana TFBS is included inside the vertical bars, and the number of motifs as well as the name of the gene group are indicated above. There is only one common motif shared across all orthologs (common_CBB: VRN1, gray bar), and most of the common motifs identified in GAPDHB are not common in the genes encoding other enzymes (common_GADPHB). In addition, many motifs are shared between GAPDHB, FBPase, and SBPase (common_SFG), whereas most PRK motifs are not shared with the genes encoding other enzymes (common_PRK). (B) Localization of each gene group in the genomic region around genes encoding CBB cycle enzymes in Brachypodium distachyon and Sorghum bicolor. The x-axis corresponds to the genomic coordinates with the start codon corresponding to the + 1 position. The colored arrow represents the gene structure with UTRs in blue, exons in red, and potential promoter and terminator as a black line. The dots represent the genomic coordinates of each of the motifs within each gene group. Different gene groups are separated along the y-axis. Despite being comprise by the same TFBS, the distribution of the dots changes between species. Notably, TFBS can be found at multiple coordinates in the same gene. Most of the trans-acting factors binding to the identified A. thaliana TFBS belong to the same family, and often bind to similar sequences. In fact, TFBS tend to cluster in discrete regions that might play a role in the regulation of the expression of the corresponding gene.
To evaluate the presence of TFBS in the regulatory regions of genes encoding CBB cycle enzymes, a dataset containing validated TFBS in Arabidopsis thaliana (O’Malley et al., 2016) was used to scan the putative regulatory sequences (intergenic regions, untranslated regions, and introns) of orthologous genes, i.e., SBPase orthologs across the subset of eight grass species were scanned at the same time. We first determined the A. thaliana TFBS shared between orthologous genes, and used those to compare between the genes encoding the selected four CBB cycle enzymes (Figure 2A). This way, we identified one TFBS present in all of the potential regulatory sequences (common_CBB, in Figure 2A) that was bound by VRN1 in A. thaliana. This TFBS was also identified it in the putative regulatory regions of genes encoding photorespiratory (GDCH) and housekeeping proteins (CBP20) (Supplementary Dataset 2), suggesting that it might play a regulatory role not limited to photosynthetic genes. We also identified 13 putative TFBS shared between SBPase, FBPase, and GAPDHB orthologous genes (common_SFG), 9 TFBS shared between GAPDHB and SBPase orthologous genes (common_GS), one shared between GAPDHB and PRK orthologous genes (common_GP), and one TFBS shared between GAPDHB and FBPase orthologous genes (common_GF). In addition, 17, 2, and 6 putative TFBS were shared between GAPDHB orthologs (common_GAPDHB), FBPase orthologs (common_FBP), and PRK orthologs (common_PRK); but not between any other group of orthologous genes. Notably, these common sequences can be found in potential regulatory sequences of other orthologous genes from some but not in all of the species in the study. The fact that all the A. thaliana TFBS found in SBPase orthologs are shared with other genes (common_SFG, common_GS) whereas most of the TFBS found in PRK orthologs are not shared (common_PRK) suggests different mechanisms for the regulation of the expression of the genes encoding CBB cycle enzymes. Most of the trans-acting factors binding to the identified TFBS belong to the Apetala2/Ethylene-Response-Factor (AP2/ERF) family, and often recognize similar binding sites. A comparison between the location of the A. thaliana TFBS at the putative regulatory regions of the orthologs in the selected species (Figure 2B and Supplementary Figure S4) revealed that the TFBS tend to cluster together in discrete regions of the putative regulatory sequences, although the genomic coordinates of these clusters change between species.
We used a similar approach to identify C4-specific TFBS contributing to the difference in expression pattern between C3 and C4 species. After scanning together orthologous genes (i.e., all SBPase orthologous genes with the A. thaliana validated TFBS), we selected TFBS absent from the putative regulatory regions of genes encoding C3-enzymes (Supplementary Figure S5). Using this approach (choosing absent motifs from genes encoding C3 enzymes rather than present motifs in all genes encoding C4 enzymes), it was possible to account for the multiple independent origins of C4 photosynthesis. Three TFBS were found absent from SBPase C3-genes, bound by At5g66940, BZR1, and CEJ1 in A. thaliana; one absent from FBPase C3-genes (bound by TCX2) and one absent from PRK C3-genes (bound by At3g12130). In addition, using the same approach to identify TFBS absent from genes encoding C4 enzymes revealed two C3-specific motifs in the 5′ intergenic region of PRK (bound by AREB3 and bZIP16). These results suggest that there are C3- and C4-specific TFBS that might contribute to the compartmentalization of C4 CBB cycle enzymes.
To further understand how the identified TFBS might regulate the expression pattern of CBB cycle enzymes, we obtained the transcriptomic data from a collection of RNA-seq experiments on C4 species where samples were taken separately from mesophyll and bundle sheath cells (John et al., 2014; Rao et al., 2016; Washburn et al., 2017), and assessed the expression pattern of the trans-acting factors. Despite the complexities of using data from different experiments, and the limited validation of the interaction between trans-acting factors and the identified TFBS (i.e., only validated in the C3 species A. thaliana), we identified ten trans-acting factors differentially enriched in M and BS cell types (Table 1). These trans-acting factors were the orthologs of the validated trans-acting factors in A. thaliana, and could be classified into three categories in regard to BS-specific enrichment (Table 1): (1) putative activators, if all the orthologs were consistently enriched in BS over M cells, such as the orthologs of At5g66940, whose TFBS are only found in SBPase orthologs of C4 species; (2) putative repressors, if all the orthologs were consistently enriched in M over BS cells, for example the orthologs of At3g12130, whose TFBS are only found in PRK orthologs of C4 species; (3) broad regulators, if enrichment of orthologous genes was inconsistent within species (some were enriched in BS over M cells while others were enriched in M over BS cells), such as the orthologs of TCX2, which are found enriched in both M and BS cells, and whose TFBS are only found in FBPase orthologs of C4 species.
Discussion
In this study, we have used publicly available data to analyze putative regulatory regions of genes encoding a selected subset of CBB cycle enzymes (SBPase, FBPase, PRK, and GAPDHB) in C3 and C4 species. We used two different approaches to identify potential regulatory elements that might contribute to the compartmentalization of CBB enzymes in C4 species. The alignment of the genomic regions of the orthologs encoding the selected CBB cycle enzymes allowed us to identify conserved non-coding sequences (CNSs) shared by C3 and C4 orthologous genes, whereas the scanning of putative regulatory regions with TFBS validated in A. thaliana, allowed us to identify putative C4-specific regulatory elements. The results presented here provide new information on putative regulatory elements of the genes encoding SBPase, FBPase, PRK, and GAPDHB in both C3 and C4 species and although we do not provide experimental evidence in this paper the results form the basis for future functional studies.
The alignment of the genomic regions of orthologous CBB genes revealed a number of CNSs shared between C3 and C4 species. We identified a highly conserved sequence in the 5′ intergenic region of every PRK gene (Supplementary Figure S1). This CNS stands out because of its length (113 bp) and the level of conservation, as it can be found in C3 species even outside of Poaceae (Supplementary Dataset S1). These attributes suggest that this region could have contributed to the regulation of PRK expression throughout evolutionary history. In contrast, the CNS identified in the last intron of SBPase orthologous genes (Figure 1) was only found in species belonging to Poaceae, suggesting that the possible contribution to the regulation of SBPase genes is limited to Poaceae species (Supplementary Dataset S1). Nevertheless, the location of this conserved region highlights the relevance of searching for regulatory elements outside of the up- and down-stream non-coding sequences of genes (Rose, 2019). Additionally, we also identified CNSs conserved only within the more closely related species of the PACMAD clade (D. oligosanthes, Z. mays, S. bicolor, P. virgatum, and S. viridis) but not within the more distant related species of the BOP clade (O. sativa, H. vulgare, and B. distachyon; Figure 1 and Supplementary Figure S2). The fact that these CNSs are only shared between the species of the PACMAD clade, including D. oligosanthes which performs C3 photosynthesis, suggests that these CNSs do not play a role in C4 compartmentalization and instead they are a result of shared evolutionary history. However, the significance of the contribution of these conserved regions to the levels or patterns of expression of these genes remains to be elucidated experimentally.
Using a different approach based on validated TFBS and their trans-acting factors in A. thaliana, we identified putative (i.e., non-validated in grasses) TFBS shared by the genes encoding CBB cycle enzymes in both C3 and C4 species (Figure 2A), as well as C4-specific (C3-absent) putative TFBS (Supplementary Figure S5). We found three putative TFBS absent from SBPase C3-genes, one absent from FBPase C3-genes, and one absent from PRK C3-genes. The identification of A. thaliana TFBS in genes encoding C4 CBB cycle enzymes supports the hypothesis that C4 genes co-opted regulatory elements of C3 genes to establish their restricted expression pattern (Brown et al., 2011; Xu et al., 2016; Borba et al., 2018; Reyna-Llorens et al., 2018). Notably, we did not identify any C3- or C4-specific putative TFBS in the regulatory regions of GAPDHB orthologs (Supplementary Figure S5) but found more shared TFBS between C3 and C4 GAPDHB orthologs (Figure 2A). Despite being expressed in BS cells, which should allow for CBB cycle function in those cells, GAPDHB is enriched in M cells (Majeran et al., 2005; Rao et al., 2016). The lack of C4-specific putative TFBS in GAPDHB regulatory regions suggests that its expression might be regulated similarly in both C3 and C4 plants. Most of the identified TFBS are recognized by members of the AP2/ERF family in A. thaliana, which supports the results of a recent study in which this family of TFBS was enriched in the regulatory regions of C4 photosynthetic genes (Burgess et al., 2019). We realized that these putative TFBS were often quite similar and cluster together at specific locations in the genome and this warrants further investigation to explore the functional significance. Despite the similarities, we only identified one TFBS, bound by VRN1 in A. thaliana, in the putative regulatory regions of every gene selected for this study, but its presence in other non-photosynthetic genes indicates that VRN1 is unlikely to be exclusive to the regulation of the expression of genes encoding CBB cycle enzymes. Furthermore, the variety in the putative TFBS identified in different sets of orthologous genes indicates differences in the regulatory networks controlling their expression. These results suggest that there is no “master” transcriptional regulator coordinating the expression of the genes encoding CBB cycle enzymes, in contrast to what has been reported in other metabolic pathways (Okada et al., 2009; Nützmann et al., 2018). In addition, the lack of a unique, “master” regulator would emphasize the importance of the simultaneous manipulation of multiple targets to increase CBB cycle efficiency (Simkin et al., 2015, 2017; López-Calcagno et al., 2020).
Based on data validated in the model plant A. thaliana, we used a computational approach to identify cis-regulatory elements whose putative trans-acting factors might play a role in C4 compartmentalization. These data have been used to investigate the putative role of orthologous genes in other crops (Capote et al., 2018; Moon et al., 2018; Burgess et al., 2019; Zeng et al., 2019; DeMers et al., 2020; Elzanati et al., 2020; Gray et al., 2020; Zhou et al., 2020), and allow us to generate a compelling hypothesis, as it is expected that similar DNA-binding domains of trans-acting factors would have similar DNA sequence preferences (Lambert et al., 2019). However, several complementary experimental approaches will be needed to provide evidence of functional significance in C4 plants. To confirm the TFBS in different species, transcription factor binding assays such as DAP-seq (O’Malley et al., 2016) could be developed in some of the grass species examined in this study. To assess the chromatin accessibility of potential regulatory regions, experiments such as DNAse-seq (Zhang and Jiang, 2015) or ATAC-seq (Buenrostro et al., 2015; Bajic et al., 2018; Maher et al., 2018), could be implemented. To enhance our ability to detect regulatory elements within coding sequences (Reyna-Llorens et al., 2018), functional assays that discriminate between conserved sites with a regulatory role and conserved sites with a coding sequence role could be developed. Finally, the generation of transcriptomic data from different species using a comparable sampling process, should allow us to unveil consistent pattern of expression among different species.
Taking all of our results together, we propose that the compartmentalization of the CBB cycle enzymes investigated in this study has occurred through the recruitment of TFBS whose trans-acting factors are enriched in either one of the C4 cell types. The expression pattern of any gene is determined by a combination of the TFBS present and the corresponding trans-acting factors binding to these regulatory regions at any given time. It then follows that the expression pattern of any gene can be changed either by recruiting new TFBS or by altering the expression pattern of the trans-acting factors. Thus, to enrich the expression of C4 enzymes in BS cells, new TFBS could be recruited into gene regulatory regions of C4 species to confer BS-specific expression. Alternatively, trans-acting factors could become enriched in BS cells to promote the expression of C4 enzymes in BS cells (as the predicted putative activators), or these factors could become enriched in M cells to repress the expression of C4 enzymes in M cells (predicted putative repressors). This transcriptional regulation would likely be complemented by regulation at post-transcriptional and/or post-translational level to achieve a precise regulation of the expression pattern of CBB cycle enzymes.
To our knowledge, and excluding the extensive work on RubisCO (discussed in Hibberd and Covshoff, 2010; Berry et al., 2011; Schlüter and Weber, 2020), this is the first study to focus specifically on the differences in the regulatory sequences of CBB cycle genes between C3 and C4 species. These results provide a hypothetical foundation for future functional analysis. Future experiments should include the in vivo validation of the trans-acting factors binding to cis-regulatory elements, and the resultant regulation of CBB cycle genes; the transfer of C4-specific transcription factors into C3 species to establish a C4-like expression pattern; or the precise genome editing of the cis-elements to evaluate their contribution to compartmentalization in C4 plants.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
CAR conceived the study and supervised the research with input from CA. CA performed the analysis and wrote the manuscript with input from CAR. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Realising Improved Photosynthetic Efficiency (RIPE) initiative awarded to CAR by the University of Illinois, United States. RIPE was possible through support from the Bill & Melinda Gates Foundation, FCDO and FFAR, grant no. OPP1172157.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.570436/full#supplementary-material
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106.
Aubry, S., Kelly, S., Kümpers, B. M. C., Smith-Unna, R. D., and Hibberd, J. M. (2014). Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10:e1004365. doi: 10.1371/journal.pgen.1004365
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208.
Bajic, M., Maher, K. A., and Deal, R. B. (2018). Identification of open chromatin regions in plant genomes using ATAC-Seq. Methods Mol. Biol. 1675, 183–201. doi: 10.1007/978-1-4939-7318-7_12
Beier, S., Himmelbach, A., Colmsee, C., Zhang, X. Q., Barrero, R. A., Zhang, Q., et al. (2017). Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Sci. Data 4:170044.
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 30, 555–561.
Berry, J. O., Patel, M., and Zielinski, A. (2011). “Chapter 12 C4 gene expression in mesophyll and bundle sheath cells,” in C4 Photosynthesis and Related CO2 Concentrating Mechanisms, eds A. S. Raghavendra and R. F. Sage (Dordrecht: Springer), 221–256. doi: 10.1007/978-90-481-9407-0_12
Borba, A. R., Serra, T. S., Górska, A., Gouveia, P., Cordeiro, A. M., Reyna-Llorens, I., et al. (2018). Synergistic binding of bHLH transcription factors to the promoter of the maize NADP-ME gene used in C4 photosynthesis is based on an ancient code found in the ancestral C3 state. Mol. Biol. Evol. 35, 1690–1705. doi: 10.1093/molbev/msy060
Bräutigam, A., Kajala, K., Wullenweber, J., Sommer, M., Gagneul, D., Weber, K. L., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155:142. doi: 10.1104/pp.110.159442
Bray, N. L., Pimentel, H., Melsted, P., and Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. doi: 10.1038/nbt.3519
Brown, N. J., Newell, C. A., Stanley, S., Chen, J. E., Perrin, A. J., Kajala, K., et al. (2011). Independent and parallel recruitment of preexisting mechanisms underlying C(4) photosynthesis. Science 331, 1436–1439. doi: 10.1126/science.1201248
Buenrostro, J. D., Wu, B., Chang, H. Y., and Greenleaf, W. J. (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.21–21.29.29.
Burgess, S. J., Reyna-Llorens, I., Stevenson, S. R., Singh, P., Jaeger, K., and Hibberd, J. M. (2019). Genome-wide transcription factor binding in leaves from C3 and C4 grasses. Plant Cell 31, 2297–2314. doi: 10.1105/tpc.19.00078
Capote, T., Barbosa, P., Usie, A., Ramos, A. M., Inacio, V., Ordas, R., et al. (2018). ChIP-Seq reveals that QsMYB1 directly targets genes involved in lignin and suberin biosynthesis pathways in cork oak (Quercus suber). BMC Plant Biol. 18:198. doi: 10.1186/s12870-018-1403-5
Chang, Y.-M., Liu, W.-Y., Shih, A. C.-C., Shen, M.-N., Lu, C.-H., Lu, M.-Y. J., et al. (2012). Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 160:165. doi: 10.1104/pp.112.203810
DeMers, L. C., Redekar, N. R., Kachroo, A., Tolin, S. A., Li, S., and Saghai Maroof, M. A. (2020). A transcriptional regulatory network of Rsv3-mediated extreme resistance against Soybean mosaic virus. PLoS One 15:e0231658. doi: 10.1371/journal.pgen.0231658
Döring, F., Streubel, M., Bräutigam, A., and Gowik, U. (2016). Most photorespiratory genes are preferentially expressed in the bundle sheath cells of the C4 grass Sorghum bicolor. J. Exper. Bot. 67, 3053–3064. doi: 10.1093/jxb/erw041
Elzanati, O., Mouzeyar, S., and Roche, J. (2020). Dynamics of the transcriptome response to heat in the moss, Physcomitrella patens. Int. J. Mol. Sci. 21:1512. doi: 10.3390/ijms21041512
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279.
Gehlenborg, N. (2019). UpSetR: a More Scalable Alternative to Venn and Euler Diagrams for Visualizing Intersecting Sets. Available online at: https://rdrr.io/cran/UpSetR/ (accessed May 21, 2020).
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186.
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Gray, S. B., Rodriguez-Medina, J., Rusoff, S., Toal, T. W., Kajala, K., Runcie, D. E., et al. (2020). Translational regulation contributes to the elevated CO2 response in two Solanum species. Plant J. 102, 383–397. doi: 10.1111/tpj.14632
Hibberd, J. M., and Covshoff, S. (2010). The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 61, 181–207.
Hirsch, C. N., Hirsch, C. D., Brohammer, A. B., Bowman, M. J., Soifer, I., Barad, O., et al. (2016). Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28, 2700–2714. doi: 10.1105/tpc.16.00353
International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
John, C. R., Smith-Unna, R. D., Woodfield, H., Covshoff, S., and Hibberd, J. M. (2014). Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 165, 62–75. doi: 10.1104/pp.114.238667
Lambert, S. A., Yang, A. W. H., Sasse, A., Cowley, G., Albu, M., Caddick, M. X., et al. (2019). Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 51, 981–989. doi: 10.1038/s41588-019-0411-1
López-Calcagno, P. E., Brown, K. L., Simkin, A. J., Fisk, S. J., Vialet-Chabrand, S., Lawson, T., et al. (2020). Stimulating photosynthetic processes increases productivity and water-use efficiency in the field. Nat. Plants 6, 1054–1063. doi: 10.1038/s41477-020-0740-1
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550.
Maher, K. A., Bajic, M., Kajala, K., Reynoso, M., Pauluzzi, G., West, D. A., et al. (2018). Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell 30, 15–36. doi: 10.1105/tpc.17.00581
Majeran, W., Cai, Y., Sun, Q., and Van Wijk, K. J. (2005). Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17:3111. doi: 10.1105/tpc.105.035519
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433.
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. doi: 10.1111/tpj.13781
Michelet, L., Zaffagnini, M., Morisse, S., Sparla, F., Perez-Perez, M. E., Francia, F., et al. (2013). Redox regulation of the Calvin-benson cycle: something old, something new. Front. Plant Sci. 4:470. doi: 10.3389/fpls.2013.00470
Moon, S., Chandran, A. K. N., An, G., Lee, C., and Jung, K. H. (2018). Genome-wide analysis of root hair-preferential genes in rice. Rice 11:48.
Niklaus, M., and Kelly, S. (2019). The molecular evolution of C4 photosynthesis: opportunities for understanding and improving the world’s most productive plants. J. Exp. Bot. 70, 795–804. doi: 10.1093/jxb/ery416
Nomura, M., Katayama, K., Nishimura, A., Ishida, Y., Ohta, S., Komari, T., et al. (2000). The promoter of rbcS in a C3 plant (rice) directs organ-specific, light-dependent expression in a C4 plant (maize), but does not confer bundle sheath cell-specific expression. Plant Mol. Biol. 44, 99–106.
Nützmann, H.-W., Scazzocchio, C., and Osbourn, A. (2018). Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52, 159–183. doi: 10.1146/annurev-genet-120417-031237
Okada, A., Okada, K., Miyamoto, K., Koga, J., Shibuya, N., Nojiri, H., et al. (2009). OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of Diterpenoid Phytoalexins in rice. J. Biol. Chem. 284, 26510–26518. doi: 10.1074/jbc.m109.036871
O’Malley, R. C., Huang, S.-C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and Epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Ouyang, S., Zhu, W., Hamilton, J., Lin, H., Campbell, M., Childs, K., et al. (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887.
Rao, X., Lu, N., Li, G., Nakashima, J., Tang, Y., and Dixon, R. A. (2016). Comparative cell-specific transcriptomics reveals differentiation of C4 photosynthesis pathways in switchgrass and other C4 lineages. J. Exper. Bot. 67, 1649–1662. doi: 10.1093/jxb/erv553
Reyna-Llorens, I., Burgess, S. J., Reeves, G., Singh, P., Stevenson, S. R., Williams, B. P., et al. (2018). Ancient duons may underpin spatial patterning of gene expression in C4 leaves. Proc. Natl. Acad. Sci. U.S.A. 115:1931. doi: 10.1073/pnas.1720576115
Robinson, M. D., Mccarthy, D. J., and Smyth, G. K. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rose, A. B. (2019). Introns as gene regulators: a brick on the accelerator. Front. Genet. 9:672. doi: 10.3389/fgene.2018.00672
Sage, R. F., Sage, T. L., and Kocacinar, F. (2012). Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 63, 19–47.
Schlüter, U., and Weber, A. P. M. (2020). Regulation and evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 71, 183–215.
Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F., Pasternak, S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115.
Sheen, J. Y., and Bogorad, L. (1987). Regulation of levels of nuclear transcripts for C4 photosynthesis in bundle sheath and mesophyll cells of maize leaves. Plant Mol. Biol. 8, 227–238. doi: 10.1007/bf00015031
Simkin, A. J., Lopez-Calcagno, P. E., Davey, P. A., Headland, L. R., Lawson, T., Timm, S., et al. (2017). Simultaneous stimulation of sedoheptulose 1,7-bisphosphatase, fructose 1,6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnol. J. 15, 805–816. doi: 10.1111/pbi.12676
Simkin, A. J., Mcausland, L., Headland, L. R., Lawson, T., and Raines, C. A. (2015). Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. J. Exp. Bot. 66, 4075–4090. doi: 10.1093/jxb/erv204
Studer, A. J., Schnable, J. C., Weissmann, S., Kolbe, A. R., Mckain, M. R., Shao, Y., et al. (2016). The draft genome of the C3 panicoid grass species Dichanthelium oligosanthes. Genome Biol. 17:223.
Washburn, J. D., Kothapalli, S. S., Brose, J. M., Covshoff, S., Hibberd, J. M., Conant, G. C., et al. (2017). Ancestral reconstruction and C3 bundle sheath transcript abundance in the paniceae grasses indicate the foundations for all three biochemical c4 sub-types were likely present in the most recent ancestor. bioRxiv [Preprint]. doi: 10.1101/162644
Wickham, H., François, R., Henry, L., and Müller, K. (2019). dplyr: a Grammar of Data Manipulation. Available online at: https://CRAN.R-project.org/package=dplyr (accessed May 21, 2020).
Wilkins, D. (2019). gggenes: Draw Gene Arrow Maps in ‘ggplot2’. Available online at: https://rdrr.io/cran/gggenes/ (accessed May 17, 2020).
Williams, B. P., Burgess, S. J., Reyna-Llorens, I., Knerova, J., Aubry, S., Stanley, S., et al. (2016). An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra Mesophyll cells. Plant Cell 28:454. doi: 10.1105/tpc.15.00570
Xu, J., Bräutigam, A., Weber, A. P. M., and Zhu, X.-G. (2016). Systems analysis of cis-regulatory motifs in C4 photosynthesis genes using maize and rice leaf transcriptomic data during a process of de-etiolation. J. Exper. Bot. 67, 5105–5117. doi: 10.1093/jxb/erw275
Zeng, Z., Zhang, W., Marand, A. P., Zhu, B., Buell, C. R., and Jiang, J. (2019). Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 20:123.
Zhang, W., and Jiang, J. (2015). Genome-wide mapping of DNase I hypersensitive sites in plants. Methods Mol. Biol. 1284, 71–89. doi: 10.1007/978-1-4939-2444-8_4
Keywords: C4 photosynthesis, gene expression regulation, cis-regulatory elements, transcription factor binding sites, Calvin-Benson-Bassham cycle
Citation: Afamefule C and Raines CA (2020) Insights Into the Regulation of the Expression Pattern of Calvin-Benson-Bassham Cycle Enzymes in C3 and C4 Grasses. Front. Plant Sci. 11:570436. doi: 10.3389/fpls.2020.570436
Received: 07 June 2020; Accepted: 23 September 2020;
Published: 16 October 2020.
Edited by:
Martha Ludwig, University of Western Australia, AustraliaReviewed by:
Nelson J. M. Saibo, New University of Lisbon, PortugalThomas D. Sharkey, Michigan State University, United States
Copyright © 2020 Afamefule and Raines. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chidi Afamefule, Y2hpZGkuYWZhbWVmdWxlQGVzc2V4LmFjLnVr; Christine A. Raines, cmFpbmNAZXNzZXguYWMudWs=
 Chidi Afamefule
Chidi Afamefule Christine A. Raines
Christine A. Raines