
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 04 September 2020
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.541968
In Arabidopsis shoot apical meristems (SAMs), a well-characterized regulatory loop between WUSCHEL (WUS) and CLAVATA3 (CLV3) maintains stem cell homeostasis by regulating the balance between cell proliferation and cell differentiation. WUS proteins, translated in deep cell layers, move into the overlaying stem cells to activate CLV3. The secreted peptide CLV3 then regulates WUS levels through a ligand-receptor mediated signaling cascade. CLV3 is specifically expressed in the stem cells and repressed in the deep cell layers despite presence of the WUS activator, forming an apical-basal polarity along the axis of the SAM. Previously, we proposed and validated a hypothesis that the HAIRY MERISTEM (HAM) family genes regulate this polarity, keeping the expression of CLV3 off in interior cells of the SAM. However, the specific role of each individual member of the HAM family in this process remains to be elucidated. Combining live imaging and molecular genetics, we have dissected the conserved and distinct functions of different HAM family members in control of CLV3 patterning in the SAMs and in the de novo shoot stem cell niches as well.
Pluripotent stem cells in plant shoot apical meristems (SAMs) can continuously divide and initiate new leaves and flowers. In the model plant Arabidopsis, the stem cells are located at the apical tip of the SAM, while cells that help to specify the stem cells are located more basally (Meyerowitz, 1997). Along the axis of the SAM, a regulatory feedback loop involving CLAVATA3 (CLV3) and WUSCHEL (WUS) controls stem cell homeostasis through cell-cell communication between these two cell types (Laux et al., 1996; Mayer et al., 1998; Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000). CLV3 mRNAs are specifically expressed in the stem cells at the central zone but not expressed in the rib meristem cells that are located beneath the stem cells (Fletcher et al., 1999; Brand et al., 2000). In contrast, WUS transcripts are confined to a small group of cells in the rib meristem that has been defined as the organizing center (OC) (Mayer et al., 1998; Schoof et al., 2000). Through plasmodesmata, WUS protein, a homeodomain transcription factor, can move from the cells at the OC into the stem cells in the central zone (Yadav et al., 2011; Daum et al., 2014), where it can directly activate CLV3 expression (Yadav et al., 2011). CLV3 encodes a secreted peptide that activates a ligand-receptor mediated signaling pathway (Clark et al., 1997; Fletcher et al., 1999; Kinoshita et al., 2010; Nimchuk et al., 2011; Nimchuk et al., 2015) to negatively regulate WUS levels and to limit stem cell proliferation. Thus, WUS and CLV3 form a negative feedback loop to maintain stem cell homeostasis (Schoof et al., 2000).
In previous work (Zhou et al., 2015; Zhou et al., 2018), we proposed that HAIRY MERISTEM (HAM, also known as LOST MERISTEMS – LOM, Schulze et al., 2010) family transcription factors regulate the CLV3-WUS feedback loop. HAM proteins together with WUS determine the apical-basal polarity of CLV3 expression in Arabidopsis SAMs and confine the CLV3 domain to the stem cells (Han et al., 2020a). Specifically, WUS protein moves upward and activates CLV3 in the central zone (Yadav et al., 2011; Daum et al., 2014), while HAM family members keep CLV3 off in the rib meristem by preventing WUS-dependent activation of CLV3 and/or repressing CLV3 transcription (Zhou et al., 2018). This hypothesis is supported by experimental results and shown plausible by a computational model (Zhou et al., 2018). The hypothesis also aligns with a number of results from earlier, independent studies (Brand et al., 2000; Schoof et al., 2000; Brand et al., 2002; Graf et al., 2010; Schulze et al., 2010; Biedermann and Laux, 2018; Zhou et al., 2018), including a computational model that features an essential role for HAM in control of CLV3 patterns (Gruel et al., 2018). Additionally, the concentration gradient of HAM has been shown to be essential in determining the CLV3 domain in both well-established SAMs and in initiating axillary stem cell niches (Biedermann and Laux, 2018; Zhou et al., 2018), suggesting the important roles of HAM family genes controlling both initiation and maintenance of patterns of gene expression in plant stem cell niches.
To date, the potentially overlapping and distinct roles of HAM family members in control of CLV3 patterning and meristem development remain unexplored. There are four HAM genes in Arabidopsis, which fall into two distinct subgroups—Type I and Type II (Engstrom et al., 2011). HAM1, HAM2, and HAM3 belong to the Type II clade, whereas HAM4 belongs to the Type I clade (Engstrom et al., 2011). Among them, HAM1 and HAM2 are the most closely related homologs (Schulze et al., 2010; Engstrom et al., 2011), and both HAM1 and HAM2 proteins physically interact with WUS as interacting cofactors (Zhou et al., 2015). Differently from HAM1-3 in the Type II clade that share similar N-terminal regions, the N terminus of HAM4 in the Type I clade is less similar to that in HAM1-3 (Engstrom et al., 2011). The transcripts of the Type II genes (HAM1-3) are targeted by the microRNA171, while the transcript of the Type I gene HAM4 lacks the microRNA171 target site (Engstrom et al., 2011). In addition, CLV3 is ectopically expressed in the rib meristem when all of the three Type II clade genes are nonfunctional in Arabidopsis, i.e. in the ham123 triple loss of function mutant (Schulze et al., 2010; Zhou et al., 2018). In contrast, HAM4 is specifically expressed in the provascular/vascular tissues (Zhou et al., 2015), and thus it is unlikely to be directly involved in CLV3 patterning at the center of the SAM. These findings suggest that HAM1, HAM2 and HAM3 in the Type II clade are potentially involved in CLV3 regulation. In this study, we aimed to define which member(s) of this clade is required and/or sufficient to control the CLV3 patterning and therefore meristem development. We performed confocal microscope live imaging and molecular genetic analyses. Our results demonstrate that HAM1 and HAM2, both expressed in the rib meristem, are necessary and sufficient to shape the CLV3-expression domain in established SAMs and in de novo-initiated axillary meristems. In contrast, HAM3 protein, naturally expressed only in the boundary between the meristem and primordia and a few cells of the peripheral zone of the SAM, does not contribute to CLV3 patterning. When the HAM3 protein is expressed in the HAM2 domain, it is able to shape the CLV3 expression pattern. These results uncover the different patterns and conserved functions of the Type II HAM proteins in Arabidopsis, and they suggest that HAM regulates CLV3 patterns cell-autonomously.
The ham123 triple mutant (Zhou et al., 2018) and ham12 double mutant (Engstrom et al., 2011) were previously described. The MIR171 overexpression lines were described previously (Zhou et al., 2018).
For Figures 4A–C, SAMs from Ler, ham123, and the pHAM1::YPET-HAM1 in ham123 transgenic line at 27 days after germination (DAG) were sampled and analyzed at the same time using an identical procedure. For Figures S1A, B Figure 4D, SAMs from Ler, ham123, and the pHAM2::YPET-HAM2 in ham123 transgenic line at 27 DAG were sampled and analyzed at the same time using an identical procedure. For Figures S2A, B and Figure 4E, SAMs from Ler, ham123, and the pHAM3::YPET-HAM3 in ham123 transgenic line at 27 DAG were sampled and analyzed at the same time using an identical procedure. For Figure 4F and Figures S3A–E, SAMs from ham123 and ham12 at 27 DAG were sampled and analyzed at the same time using an identical procedure. For Figures 5G–I, SAMs from Ler, ham123, and the pHAM2::YPET-HAM3 in ham123 transgenic line at 27 DAG were sampled and analyzed at the same time using an identical procedure. For Figure S4, the vegetative shoot apices including developing AMs from Ler and the MIR171 OE transgenic line at 27 DAG were sampled and analyzed at the same time using an identical procedure. For Figure 7, shoot apices including developing AMs from Ler, ham123, the pHAM1::YPET-HAM1 in ham123, the pHAM2::YPET-HAM2 in ham123, and the pHAM3::YPET-HAM3 in ham123 plants were grown in short days and analyzed using an identical procedure.
To generate the pHAM1::YPET-HAM1 in ham123, a YPET-HAM1 fusion was generated using overlapping PCR with the primers 5’- TACCGAGGGTATGAATGAATTGTACAAAAAATCTAGAATGCCCTTATCCTTTGAAAGGTTTCAAG - 3’, 5’- CTTGAAACCTTTCAAAGGATAAGGGCATTCTAGATTTTTTGTACAATTCATTCATACCCTCGGTA - 3’, 5’- CACCATGGCTGCAGCCAAGGGCGAGG - 3’, and 5’- CTAACATTTCCAAGCAGAGACAGTAACAAGT - 3’ following the published procedure (Heckman and Pease, 2007). A 3076 bp HAM1 promoter was PCR amplified using the primers 5’- ACAAgcggccgcGTTTTATATTTCAACTATCCCTAGATTTTAGC - 3’ and 5’- ACAAgcggccgcCGCCTCCTCAACAACACAGAGTAACTGTAAAAACA - 3’ (restriction enzyme sites are in lower case), and cloned to the 5’ of the YPET-HAM1 DNA fragment. A 1622 bp HAM1 3’ region was PCR amplified using the primers 5’ - ATAAggcgcgccACGAAGAAGAAACCACAAATCT - 3’ and 5’- ATAAggcgcgccAATCGGTGTATTCTTAATTAATGTCTAAAGTA - 3’ and cloned to the 3’ of the YPET-HAM1 DNA fragment. The whole fragment was then cloned into the pMOA34 binary vector (Barrell and Conner, 2006). pMOA series of binary vectors do not contain any 35S promoter elements and they have been used for generating the translational fusion fluorescence reporters (Nimchuk et al., 2011). The pMOA34 pHAM1::YPET-HAM1 plasmid was then introduced into ham123 triple homozygous plants through the floral dip method (Clough and Bent, 1998). To generate the pHAM2::YPET-HAM2 in ham123, the pMOA34 pHAM2::YPET-HAM2 plasmid that was described previously (Zhou et al., 2018) was introduced into ham123 triple homozygous plants through the Agrobacterium-mediated floral dip method (Clough and Bent, 1998).
To generate the pHAM3::YPET-HAM3 in ham123, a YPET-HAM3 fusion was generated using overlapping PCR with the primers 5’- TACCGAGGGTATGAATGAATTGTACAAAAAATCTAGAATGCCCTTACCCTTTGAAGAGTTTCAAGG- 3’, 5’- CCTTGAAACTCTTCAAAGGGTAAGGGCATTCTAGATTTTTTGTACAATTCATTCATACCCTCGGTA - 3’, 5’- CACCGTATTTTTACAACAATTACCAACAAC - 3’ and 5’- TCAGGAGGAGCGACATCTCCATGCT- 3’. A 3816 bp HAM3 promoter was PCR amplified using the primers 5’- ACAAgcggccgcTTTATAAGACTTGCTATGGTCGTGAG - 3’ and 5’- ACAAgcggccgcTGCAGACGATAAAAAATAGTGTATT - 3’ (restriction enzyme sites are in lower case), and cloned to the 5’ of the YPET-HAM3 DNA fragment. A 1755 bp HAM3 3’ region was PCR amplified using the primers 5’ - TACAggcgcgccTTTCCACCGGAGTTTCAATTATTAAA - 3’ and 5’- TACAggcgcgccTTAGTTGAAGGACAAATAACACCAAA - 3’ and cloned to the 3’ of the YPET-HAM3 DNA fragment. The whole fragment was then cloned into the pMOA34 binary vector (Barrell and Conner, 2006). The pMOA34 pHAM3::YPET-HAM3 plasmid was then introduced into ham123 triple homozygous plants through the floral dip method (Clough and Bent, 1998).
To generate the pHAM2::YPET-HAM3 in ham123, the HAM2 promoter was cloned to the 5’ of the YPET-HAM3 DNA fragment and the HAM2 3’ region was cloned to the 3’ of the YPET-HAM3 DNA fragment. The whole fragment was then cloned into the pMOA34 binary vector (Barrell and Conner, 2006). The pMOA34 pHAM2::YPET-HAM3 plasmid was introduced into ham123 triple homozygous plants through the floral dip method (Clough and Bent, 1998).
Independent transformants were first selected based on their hygromycin resistance. They were then imaged using the laser scanning confocal microscope to determine the expression of each HAM fusion protein in the SAMs. For each construct, multiple independent transgenic lines have been identified and used in the study. Specifically, four independent transgenic lines of the pHAM1::YPET-HAM1 in ham123 showed comparable expression patterns of YPET-HAM1 in the SAMs and displayed comparable growth phenotypes. Three independent transgenic lines of the pHAM2::YPET-HAM2 in ham123 showed comparable expression patterns of YPET-HAM2 in the SAMs and displayed comparable growth phenotypes. Four independent transgenic lines of the pHAM3::YPET-HAM3 in ham123 showed comparable expression patterns of YPET-HAM3 in the SAMs and displayed comparable growth phenotypes. Four independent transgenic lines of the pHAM2::YPET-HAM3 in ham123 showed comparable expression patterns of YPET-HAM3 in the SAMs and displayed comparable growth phenotypes. The results from one representative line for each construct were presented in the Figures.
All the plants for RNA in situ hybridization experiments were grown in short days at 22°C. Vegetative shoots were fixed with 4% paraformaldehyde overnight at 4°C. Tissues were embedded, sectioned, hybridized and washed as described previously (Krizek, 1999; Han et al., 2020b). The CLV3 probe was described previously (Zhou et al., 2018). At least three biological replicates were performed for each genotype and showed similar results.
The live imaging of pHAM1::YPET-HAM1, pHAM2::YPET-HAM2, pHAM3::YPET-HAM3 or pHAM2::YPET-HAM3 in the SAMs of ham123 mutants was performed using the ZEISS LSM880 confocal microscope. Plants were grown in short days for three weeks and then moved to continuous light to induce flowering. The inflorescence shoot apices were imaged when plants bolted at ~2 cm in height. At least four biological replicates were imaged for each pHAM::YPET-HAM reporter and showed similar results. The confocal imaging was performed with the similar methods previously described (Li et al., 2013; Zhou et al., 2015; Zhou et al., 2018; Geng and Zhou, 2019). Specifically, the shoot apices were stained with PI and imaged using the W plan-Apochromat 20x/1.0 water dipping lens (Zeiss). YPET and PI were excited by a 514 nm laser line and collected from the 520-560 nm and from the 600-675 nm, respectively. In general, the signal intensities of pHAM::YPET-HAM translational reporters are weaker than that of the previously reported HAM transcriptional reporters (Zhou et al., 2015; Zhou et al., 2018). In this study, all the confocal images were taken using the sum function as the averaging method in the Zeiss ZEN software. The confocal parameters of the laser power, gain and pinhole are different when imaging different pHAM::YPET-HAM translational reporters. The quantification for each confocal image was performed using the Fiji software with the Fire function as previously reported (Zhou et al., 2018).
All of the plants for the mPS-PI staining (Figure 6) were grown in short days for 28 days at 22°C. Old leaves were dissected out from vegetative shoots, and the vegetative shoot apices were fixed and stained following the procedure previously described (Truernit et al., 2008), except that Visikol (https://visikol.com/) instead of chloral hydrate was used. The confocal images were taken using a ZEISS LSM880, and the 3D projection view for each SAM was generated and analyzed using Image J.
As previously reported, a single mutation of HAM1, HAM2, or HAM3 does not result in any obvious defects (Engstrom et al., 2011) and the ham123 triple loss of function mutant leads to the ectopic activation of CLV3 in the rib meristem of SAMs (Schulze et al., 2010; Zhou et al., 2018), suggesting that HAM1-3 likely have shared functions. To precisely define the expression pattern of each HAM protein and to evaluate the role of each HAM in shaping the CLV3 domain, we generated HAM translational fluorescence reporters (pHAM::YPET-HAM) under the control of endogenous promoters and 3’ terminators, and introduced each reporter into ham123 mutants.
We first imaged a pHAM1::YPET-HAM1 translational reporter in the SAM from orthogonal and transverse optical section views, using confocal microscopy (Figures 1A–C). The signal intensity of the HAM1 translational reporter is weaker than the HAM1 transcriptional reporter we imaged previously (Zhou et al., 2015; Zhou et al., 2018), which we found is a common difference between translational and transcriptional reporters in live imaging. The expression of pHAM1::YPET-HAM1 shows a concentration gradient along the apical-basal axis of the SAM (Figures 1A–F), consistent with our previous observations on the pHAM1::2xYPET-N7mirS transcriptional reporter (Zhou et al., 2015; Zhou et al., 2018). Under the control of the endogenous HAM1 promoter and 3’ terminator, the YPET-HAM1 protein is not expressed in the epidermal L1 layer, barely expressed in the L2 layer, and highly expressed in the rib meristem and peripheral zone of the corpus (Figures 1A–F). In parallel, we imaged the pHAM2::YPET-HAM2 translational reporter (Zhou et al., 2015; Zhou et al., 2018; Han et al., 2020b) in the ham123 SAM (Figures 2A–C). pHAM2::YPET-HAM2 is highly expressed in the rib meristem and peripheral zone of the corpus, but it is repressed or completely off in the L1 and L2 layers (Figures 2A–F), a pattern comparable to that of pHAM1::YPET-HAM1 (Figures 1A–F). These results suggest that HAM1 and HAM2 proteins share similar expression patterns. We also generated a translational reporter for HAM3, and imaged the pHAM3::YPET-HAM3 in the ham123 background (Figures 3A–C). In contrast to pHAM1::YPET-HAM1 (Figures 1A–F) or HAM2::YPET-HAM2 (Figures 2A–F), we found that the pHAM3::YPET-HAM3 translational reporter is only expressed at the boundary between the meristem and primordia and in a few cells of the peripheral zone (PZ) of the SAM (Figures 3A–F). This pattern is not overlapping with the CLV3 or the WUS expression domain in either wild type or in the ham123 mutant (Schulze et al., 2010; Zhou et al., 2018). Hence, we hypothesized that in a wild type SAM, among three Type II HAM genes, HAM3 would be dispensable for the regulation of CLV3 expression due to its lack of expression in the WUS protein domain.
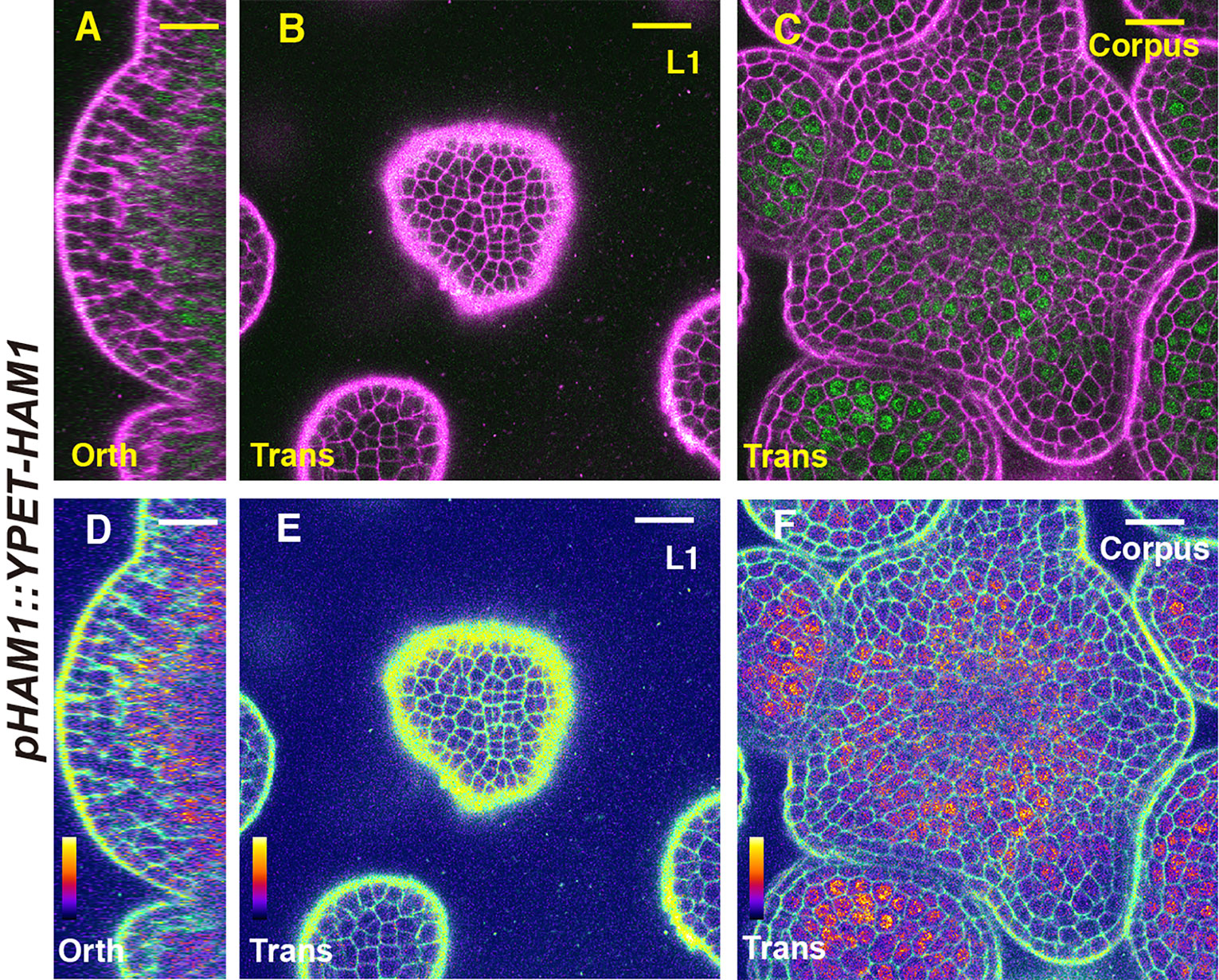
Figure 1 The expression of a HAM1 translational reporter in the SAM. (A–F) Confocal imaging of a pHAM1::YPET-HAM1 translational reporter in a SAM of a ham123 triple mutant, from the orthogonal view (A, D), transverse optical section view in L1 (B, E), and corpus (C, F). (A–C) merged channels from YFP (green) and PI (propidium iodide, purple). (D–F) merged channels from the quantified YFP (quantitatively indicated by color) and PI. Scale bar: 20 µm. Color bar: Fire quantification.
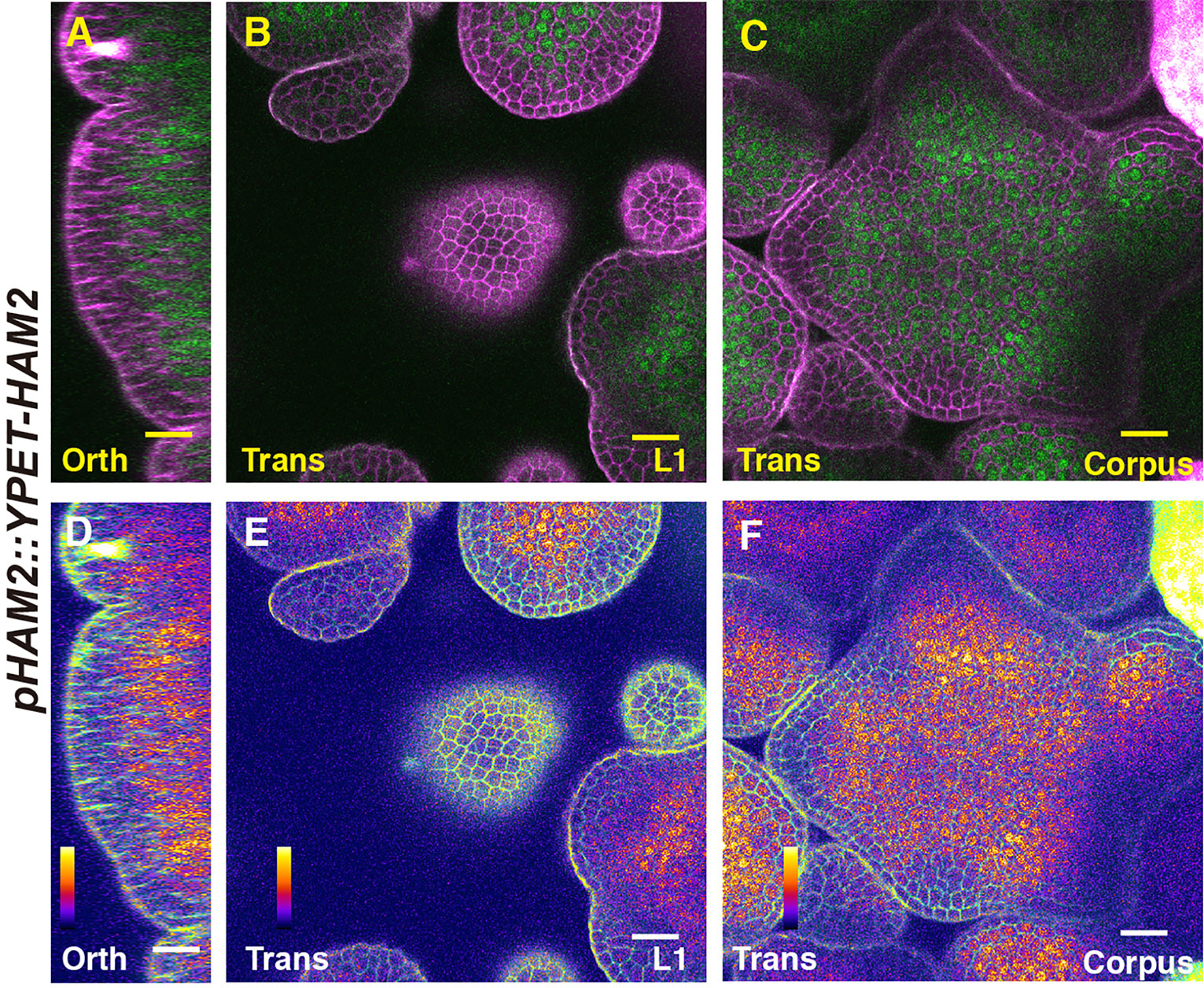
Figure 2 The expression of a HAM2 translational reporter in the SAM. (A–F) Confocal imaging of a pHAM2::YPET-HAM2 translational reporter in a SAM of a ham123 triple mutant, from the orthogonal view (A, D), transverse optical section view in L1 (B, E), and corpus (C, F). (A–C): merged channels from YFP (green) and PI (propidium iodide, purple). (D–F): merged channels from the quantified YFP (quantitatively indicated by color) and PI. Scale bar: 20 µm. Color bar: Fire quantification.
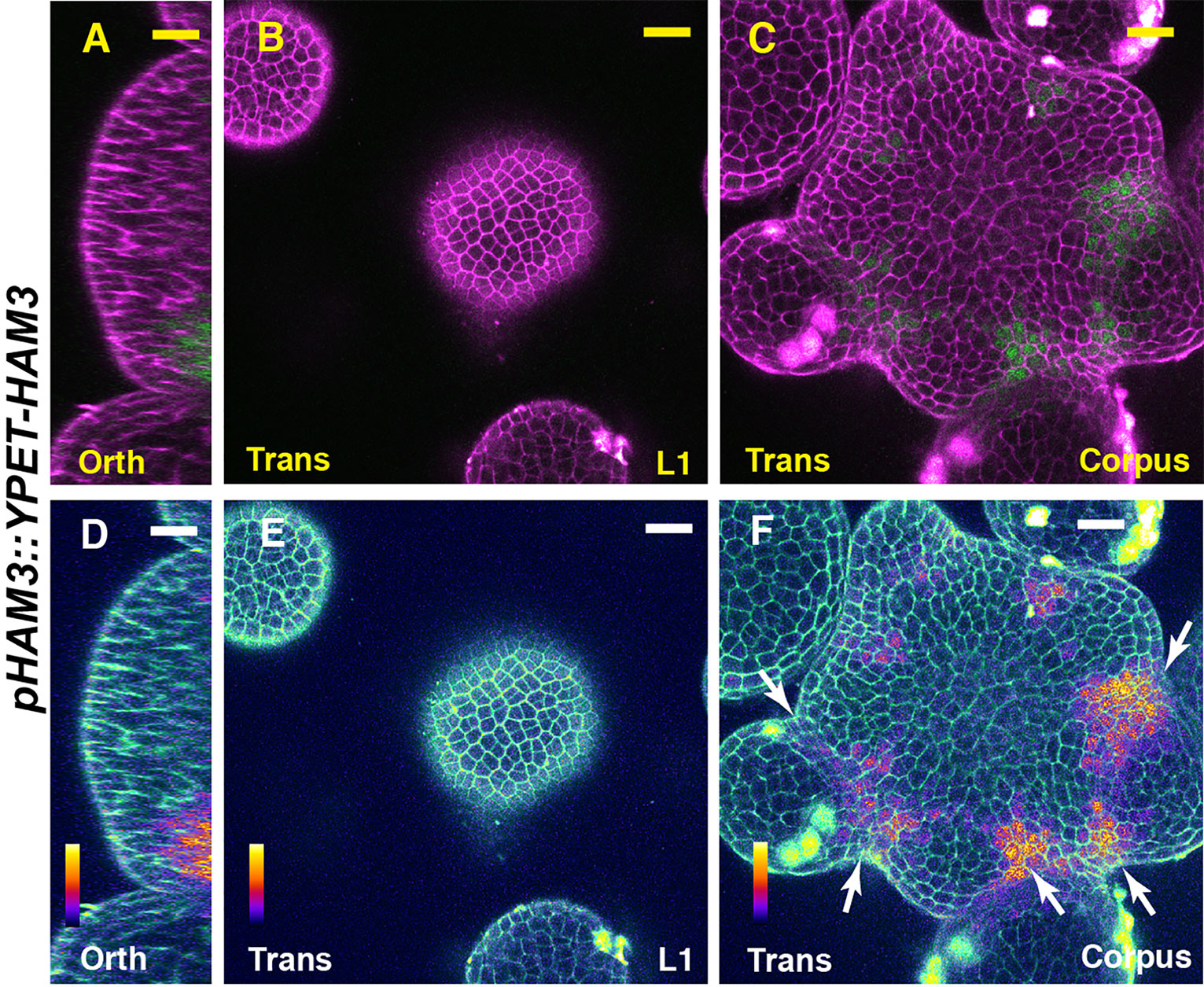
Figure 3 The expression of a HAM3 translational reporter in the SAM. (A–F) Confocal imaging of a pHAM3::YPET-HAM3 translational reporter in a SAM of ham123, from orthogonal view (A, D), transverse view in L1 (B, E), and corpus (C, F). (A–C): merged channels from YFP (green) and PI (purple). (D–F): merged channels from the quantified YFP (quantitatively indicated by color) and PI. Arrows indicate the boundary between shoot meristem and primordia. Scale bar: 20 µm. Color bar: Fire quantification.
We then examined the expression pattern of CLV3 using RNA in situ hybridization assays, in SAMs of the wild type, ham123, pHAM1::YPET-HAM1 ham123, pHAM2::YPET-HAM2 ham123, and pHAM3::YPET-HAM3 ham123 plants (Figures 4A–E). Differently from in the wild type control, CLV3 shows ectopic expression in the rib meristem of the ham123 SAM (Figures 4A, B) (Schulze et al., 2010; Zhou et al., 2018). We found that when the SAM only expresses HAM1, in a genotype with pHAM1::YPET-HAM1 in a ham123 background, the CLV3 expression domain is comparable to that in wild type (Figure 4A), showing a full complementation of the misregulated CLV3 expression domain of ham123 (Figure 4C). These results demonstrated that HAM1 is sufficient to keep CLV3 expression off in the interior layers of the SAM. In the SAM of the pHAM2::YPET-HAM2 ham123 line (Figure 4E), the CLV3 expression domain was specifically confined to the central zone, comparable to a wild type (Figure 4A, Figure S1) or a pHAM1::YPET-HAM1 ham123 plant (Figure 4C). The full complementation of the defective CLV3 patterning (Figures 4C, D) demonstrated that HAM2 and HAM1 share redundant function in their effect on CLV3 expression. The RNA in situ hybridization results also demonstrated that the CLV3 expression domain in pHAM3::YPET-HAM3 ham123 plants is distinct from that in wild type and comparable to that in ham123 (Figure 4E, Figure S2). Furthermore, we found that the expression pattern of CLV3 in the SAMs of the ham12 double mutants (Figure 4F) is largely comparable to that in the ham123 triple mutant (Figure 4B, Figures S3A–E).
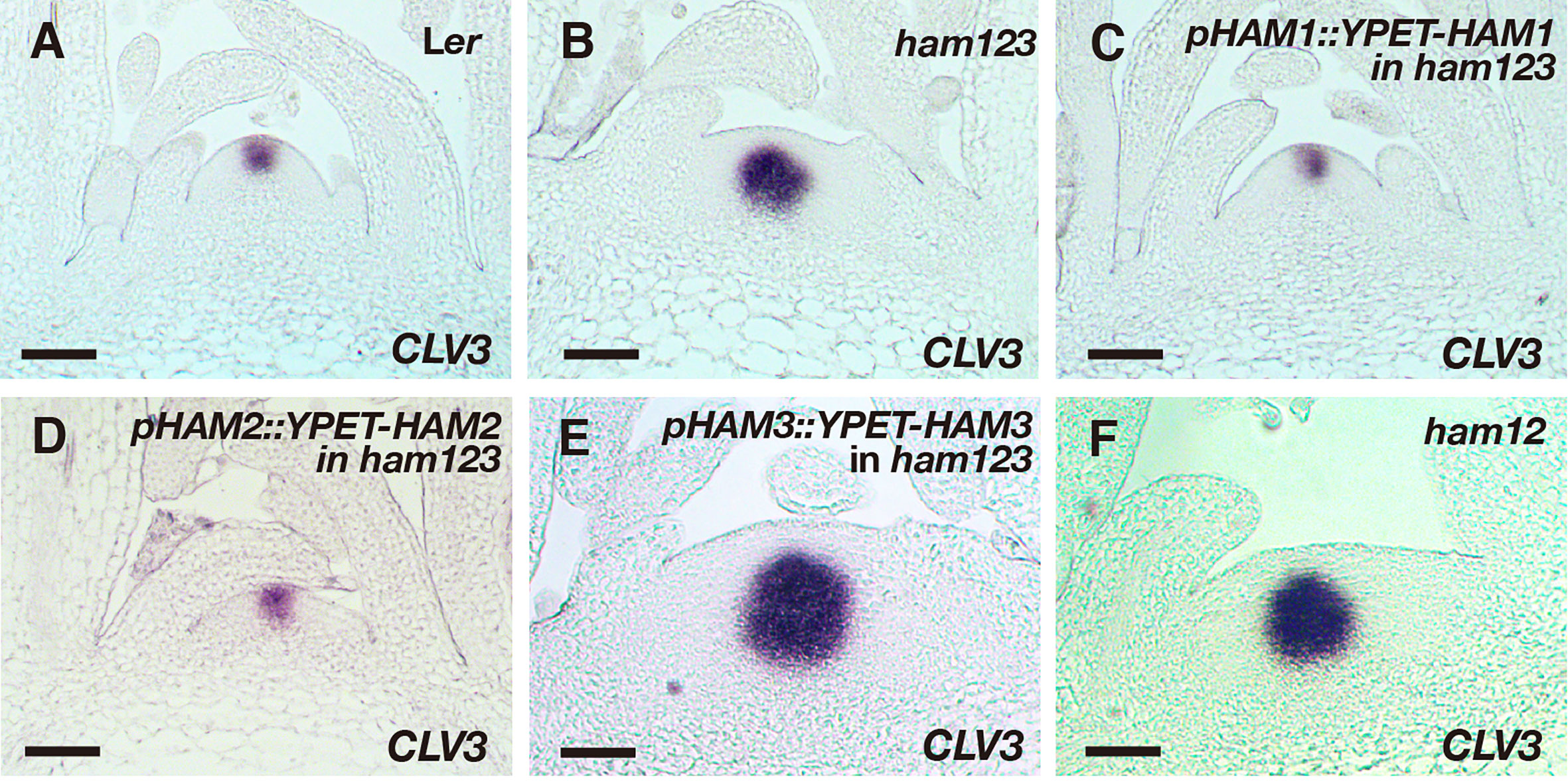
Figure 4 Roles of different HAM genes in control of CLV3 patterning in SAMs. (A–F) RNA in situ hybridization of CLV3 in the SAMs of wild type (Ler) (A), ham123 (B), pHAM1::YPET-HAM1 in ham123 (C), pHAM2::YPET-HAM2 in ham123 (D), pHAM3::YPET-HAM3 in ham123 (E), and ham12 (F) grown in short days at the same developmental stage (27 DAG). Scale bar: 50 µm. At least three biological replicates were performed for each genotype with similar results.
The fact that pHAM3::YPET-HAM3 does not complement the defect of CLV3 patterning in ham123 led us to examine whether this is because expression of the HAM3 protein is not found in the center of the rib meristem. To test this possibility, we generated a pHAM2::YPET-HAM3 reporter in which YPET-HAM3 is under the control of the promoter and 3’ terminator of HAM2. We introduced this new reporter into a ham123 triple mutant and found that the expression pattern of pHAM2::YPET-HAM3 is largely comparable to that of pHAM2::YPET-HAM2, with broad expression in rib meristem and the peripheral zone in deep cell layers but reduced or absent expression in the central zone (Figures 5A–F). We then performed an RNA in situ hybridization experiment and we found that the CLV3 expression pattern in a pHAM2::YPET-HAM3 ham123 SAM became similar to that in the wild type plant (Figures 5G–I). This result suggests that the HAM3 protein maintains conserved function with HAM1/2 in regulating the CLV3 pattern, but the endogenous HAM3 gene alone cannot maintain correct CLV3 patterning due to its expression domain.
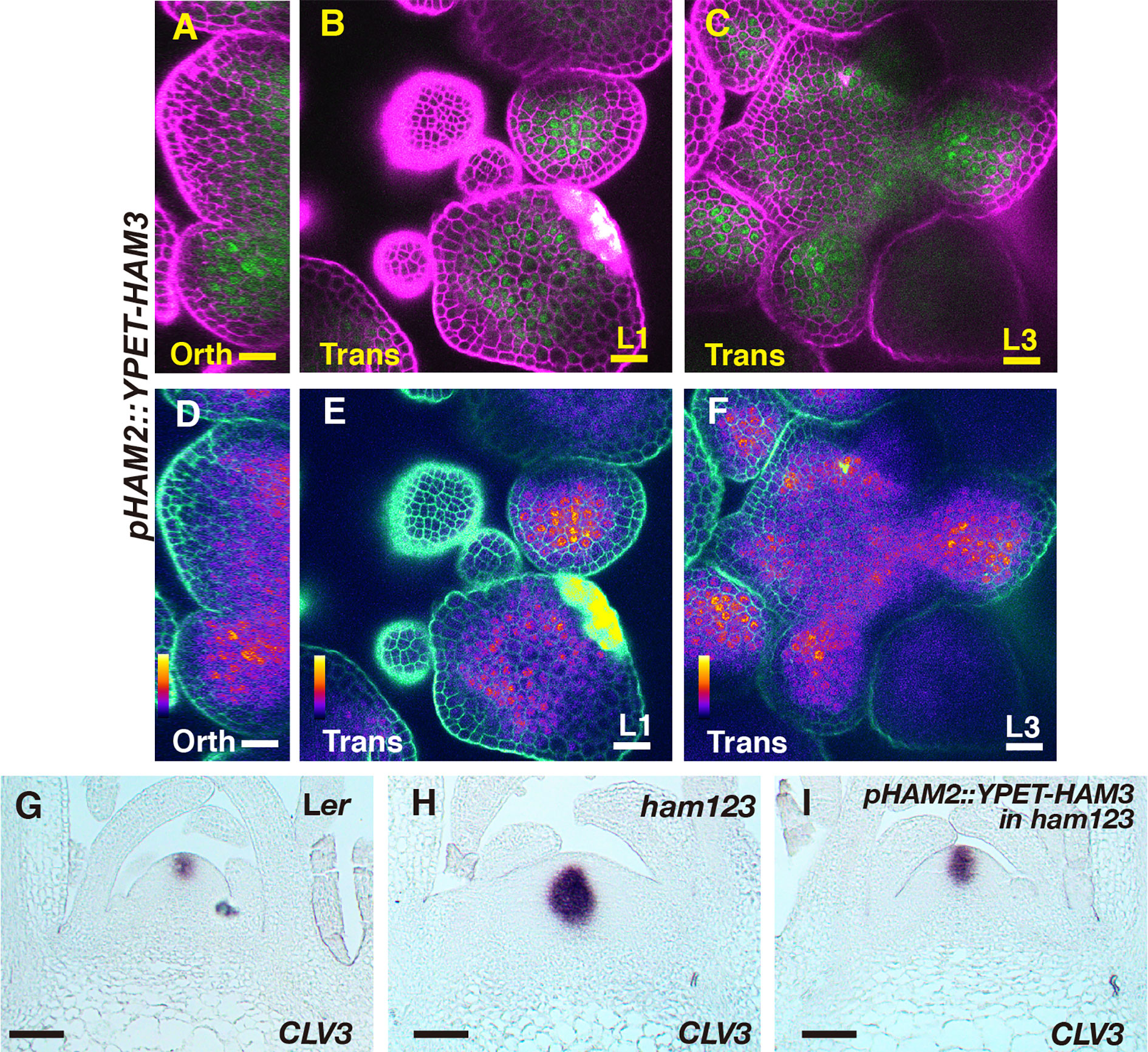
Figure 5 The expression and function of pHAM2::YPET-HAM3 in the SAM. (A–F) Confocal imaging of a pHAM2::YPET-HAM3 translational reporter in a SAM of ham123 from orthogonal view (A, D), transverse view in L1 (B, E), and corpus (C, F). (A–C): merged channels from YFP (green) and PI (purple). (D–F): merged channels from the quantified YFP (quantitatively indicated by color) and PI. Color bar: Fire quantification. Scale bar (A–F): 20 µm. (G–I) RNA in situ hybridization of CLV3 in the SAMs of wild type (Ler) (G), ham123 (H), and pHAM2::YPET-HAM3 in ham123 (I) at the same developmental stage (27 DAG). Scale bar (G–I): 50 µm. At least three biological replicates were performed for each genotype with similar results.
To further define the role of each Type II HAM member in controlling the organization of SAMs, in addition to effect on CLV3 patterning, we imaged the SAMs of Landsberg erecta (Ler) wild type, ham123, pHAM1::YPET-HAM1 in a ham123 background, pHAM2::YPET-HAM2 in a ham123 background, pHAM3::YPET-HAM3 in a ham123 background, and ham12, all grown in short days (Figures 6A–F). Similarly to previous observations (Schulze et al., 2010; Zhou et al., 2018), the SAM of a ham123 triple mutant was flattened and broader than the dome-shaped wild type SAM (Figures 6A, B). Furthermore, consistent with the results from orthogonal sections in the RNA in situ hybridization experiments (Figures 4C–F), the 3D projection view of the confocal images showed that SAMs of the pHAM1::YPET-HAM1 ham123 strain and the pHAM2::YPET-HAM2 ham123 strain are comparable to wild type, whereas the SAMs of the pHAM3::YPET-HAM3 ham123 strain and of ham12 are very similar to that of ham123 (Figures 6C–F).
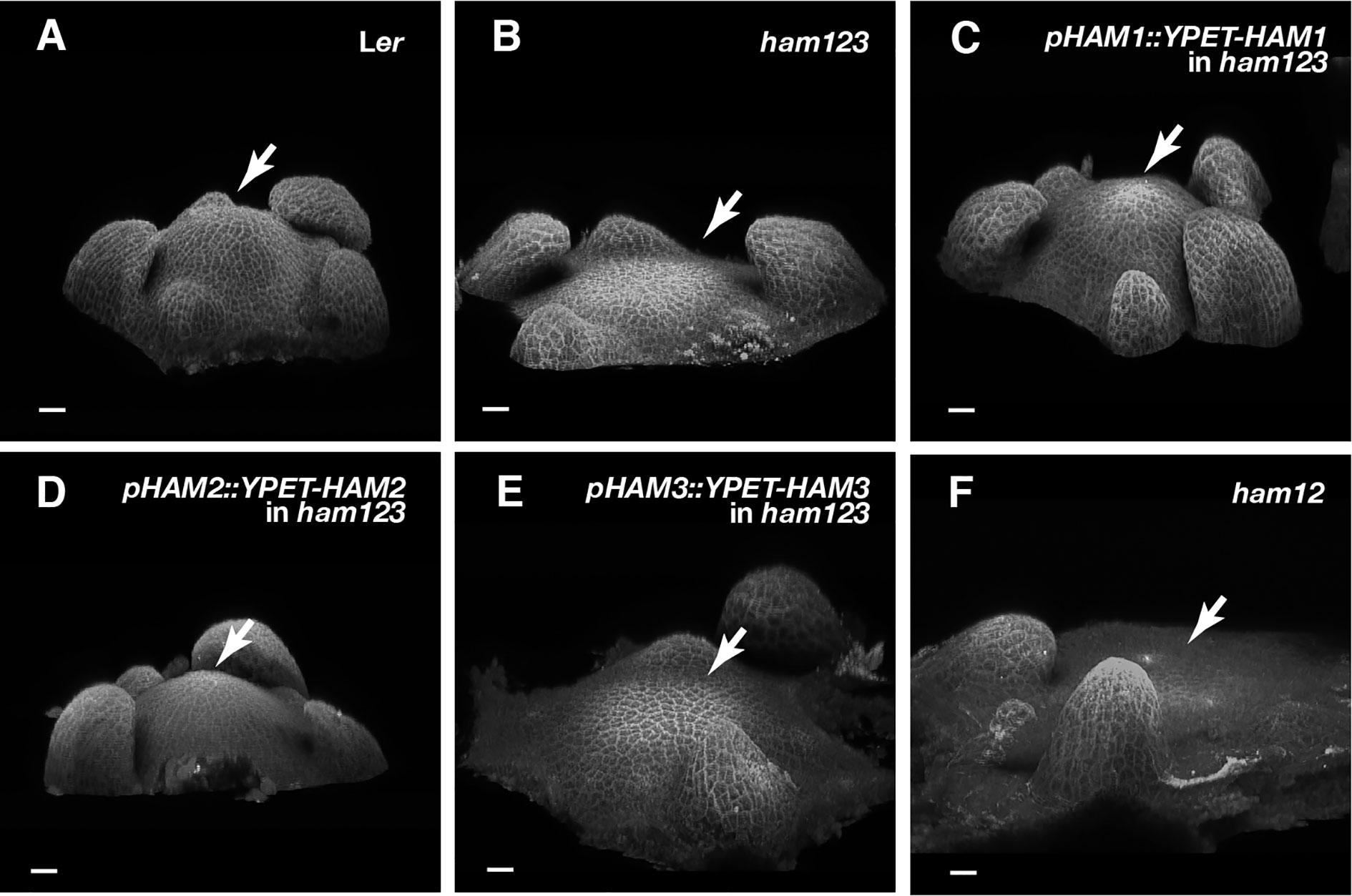
Figure 6 Roles of different HAM genes in control of vegetative SAM morphology. (A–E) 3D projection views of the vegetative SAMs of indicated genotypes are shown. Ler wild type (A), ham123 (B), pHAM1::YPET-HAM1 in ham123 (C), pHAM2::YPET-HAM2 in ham123 (D), pHAM3::YPET-HAM3 in ham123 (E), and ham12 (F) were grown in the short days and imaged at the same age (28 DAG). Four biological replicates were performed for each genotype with similar results. Arrows indicate center of the SAMs. Scale bar: 20 µm.
In addition to established SAMs, we also examined the roles of different HAM genes in controlling the initiating stem cell niches in leaf axils. We previously found that CLV3 expression is restricted to the basal part of initiating meristems in the ham123 complete loss of function mutant (Zhou et al., 2018). Our new results show that the partial loss of function of the Type II HAM genes in a MIR171 overexpression transgenic line is sufficient to disturb axillary meristem (AM) formation and de novo patterning of the CLV3 domain (Figures S4A–D), and the expression of CLV3 is also restricted to deep cell layers of the developing stem cell niches in the MIR171 OE line (Figure S4D). To further dissect the role of each HAM gene in controlling the initiation of stem cell niches, we examined CLV3 expression in the initiating axillary meristems from different genotypes (Figures 7A–E). We found that when only HAM1 (a pHAM1::YPET-HAM1 in ham123 background) or HAM2 (a pHAM2::YPET-HAM2 in ham123 background) is present, the CLV3 gene is expressed at the apical part of the initiating meristem (Figures 7C, D). In contrast, and similar to the phenotype of the ham123 triple mutant (Figure 7B), the pHAM3::YPET-HAM3 ham123 plant (Figure 6E) has a CLV3 expression domain confined to deeper layers. Furthermore, the projection of the new axillary meristem from the leaf does not occur—that is, the formation of the meristem is not completed. In addition to the axillary meristems, we characterized branches initiated from the axils of cauline leaves in each genotype (Figures 7F–J). We found that branches can normally initiate from the cauline leaves of the wild type, the pHAM1::YPET-HAM1 ham123 plant and the pHAM2::YPET-HAM2 ham123 plant (Figures 7F, H, I). However, branches were missing from the axils of the cauline leaves in ham123 or the pHAM3::YPET-HAM3 ham123 plant (Figures 7G, J). These results in de novo formed meristems (Figure 7) together with the characterization of the primary SAM (Figure 4) demonstrate that HAM1 and HAM2, a pair of closely related proteins (Engstrom et al., 2011), play major roles in de novo patterning of CLV3 expression in developing axillary meristems and in maintaining the CLV3 expression domain in established SAMs.
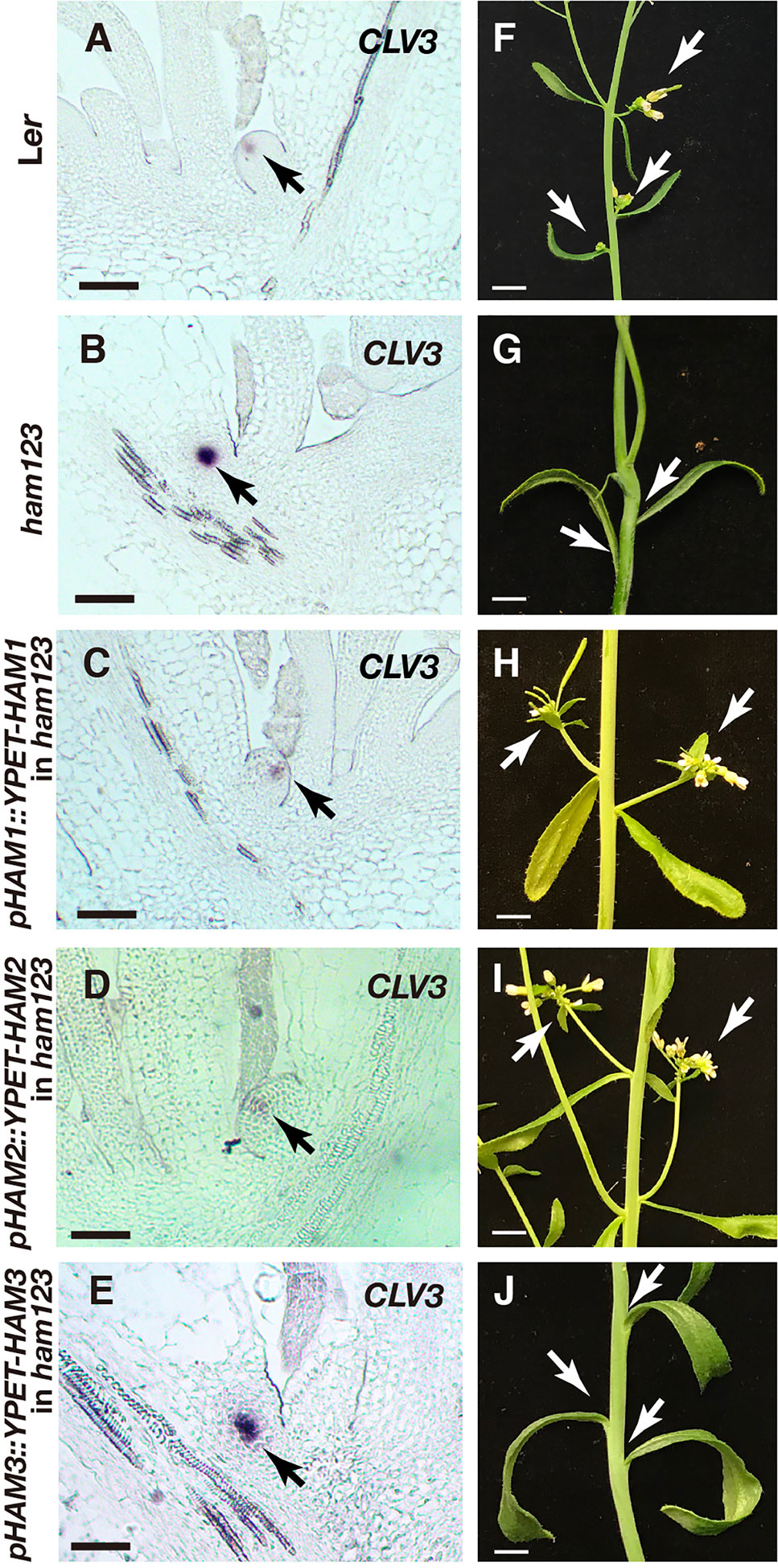
Figure 7 Roles of different HAM genes in control of CLV3 patterning in de novo stem cell niches and in branch development. (A–E) RNA in situ hybridization of CLV3 in the initiating stem cell niches of Ler wild type (A), ham123 (B), pHAM1::YPET-HAM1 in ham123 (C), pHAM2::YPET-HAM2 in ham123 (D), and pHAM3::YPET-HAM3 in ham123 (E). Scale bar: 50 µm. Arrows indicate the CLV3 expressing cells. (F–J) Images of branches initiated from base of the cauline leaves in different genotypes, which are grown in the same condition at the same age. Arrows indicate the branches initiated normally from the base of the cauline leaves of Ler wild type (F), pHAM1::YPET-HAM1 in ham123 (H) and pHAM2::YPET-HAM2 in ham123 (I), and they indicate the absence of branches initiated from the base of the cauline leaves in ham123 (G) and pHAM3::YPET-HAM3 in ham123 (J). Scale bar: 0.5cm. At least three biological replicates were performed for each genotype with similar results.
Although the defects of meristem initiation and organization of the CLV3 patterning in the ham123 mutant cannot be rescued by the pHAM3::YPET-HAM3 translational reporter at all (Figure 4E, Figure 6E, Figures 7E, J), the defects of retarded leaf growth in ham123 can be largely complemented by pHAM3::YPET-HAM3 (Figures S5A–E). These results suggest that HAM1/2 are necessary and sufficient for determining the CLV3 patterns in the meristems, whereas HAM3 appears to share redundant function with HAM1/2 in control of aspects of leaf development.
In the previous study, we proposed and provided evidence for a model that the apical-basal extent of the CLV3 expression domain is determined by both WUS and HAM (Zhou et al., 2018). One of the key themes in this model is that the more basally localized HAM proteins are responsible for preventing WUS induction of CLV3 expression (Zhou et al., 2018). Here we found overlapping and distinct roles of HAM family members in control of CLV3 patterning, closely related to their protein expression domains in the deeper cell layers of SAMs. This work supports the previously proposed WUS-HAM-CLV3 regulatory circuit (Zhou et al., 2015; Zhou et al., 2018) and further defines the cell layer-specific roles of HAM, both in established and in initiating meristems.
It has been reported that the expression patterns of the epidermis (L1)-specific marker, ATML1 and the rib meristem (L3)-specific marker ATHB23 (HOMEOBOX PROTEIN23) remain unaltered in a ham123 triple mutant, comparable to that in wild type (Schulze et al., 2010). These results suggest that the ectopic expression of CLV3 in the L3 in a ham123 mutant is not due to a respecification of clonally distinct cell layers (from L1 to corpus/L3) in the SAMs. In line with these findings, we showed here that HAM1 and HAM2 proteins have the overlapping expression patterns in corpus/L3 (Figures 1 and 2) and they both are responsible for CLV3 repression (Figure 4). In contrast, the endogenous HAM3 protein is dispensable for CLV3 patterning (Figure 4), although it plays a role in other aspects of HAM family-mediated developmental processes (Figure S5). Furthermore, when the HAM3 protein is expressed in a broader region comprising L3 and RM by use of a HAM2 promoter, it complements to a large extent the defective CLV3 expression pattern in a ham123 mutant (Figure 5). These results suggest that the specific protein expression domain is crucial for the function of HAM family members. The confocal imaging of all the pHAM::YPET-HAM translational reporters was performed using inflorescence SAMs. In the future, it will be important to quantitatively determine the expression patterns of these translational reporters in vegetative SAMs and in the developing AMs, to get a more comprehensive view of HAM protein localization and function during meristem development.
Differently from WUS and CLV3 that are specifically expressed in a few cells, HAM1 and HAM2 are expressed in a much broader domain of the SAM (Figures 1 and 2) (Zhou et al., 2015), suggesting the possibility HAM1/2 have additional roles that are independent of CLV3 and/or WUS, such as regulating STM (Schulze et al., 2010). Future experiments will determine whether HAM1/2 can regulate the apical-basal polarity of the expression of genes other than CLV3 and whether HAM1/2 also shape shoot architecture through CLV3-independent pathways.
All datasets generated for this study are included in the article/Supplementary Material.
HH, XL, AY, and YZ conceived the research direction. HH, YG, XL, and YZ performed the experiments. LG contributed research reagents. AY and EM discussed and commented on the experimental results. HH, XL, and YZ wrote the manuscript. AY and EM revised the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by Purdue University start-up and funds from Purdue Center for Plant Biology to YZ. The work in EM group was funded by the Howard Hughes Medical Institute.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the Purdue Bindley Bioscience Imaging Facility for access to the ZEISS LSM880 Laser scanning confocal microscope.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.541968/full#supplementary-material
Figure S1 | Control for the RNA in situ experiment shown in Figure 4D. (A–B) RNA in situ hybridization of CLV3 in the SAMs of Ler wild type (A) and ham123 (B) at the same developmental stage (27 DAG), which were grown in the identical conditions and analyzed with identical procedures together with the SAM of pHAM2::YPET-HAM2 in ham123 (shown in Figure 4D). Scale bar (A–B): 50 µm.
Figure S2 | Control for the RNA in situ experiment shown in Figure 4E. (A–B) RNA in situ hybridization of CLV3 in the SAMs of Ler wild type (A) and ham123 (B) at the same developmental stage (27 DAG). These samples were grown in the identical conditions and analyzed with identical procedures together with the SAM of pHAM3::YPET-HAM3 in ham123 (shown in Figure 4E). Scale bar (A–B): 50 µm.
Figure S3 | Roles of HAM1 and HAM2 in control of CLV3 patterning in SAMs. (A–E) RNA in situ hybridization of CLV3 in the SAMs of ham123 (A) and four additional ham12 plants (samples 2-5, B–E) at the same developmental stage (27 DAG). These samples were grown in the identical conditions and analyzed with identical procedures together with the SAM of ham12 (sample 1, shown in Figure 4F). Scale bar (A–E): 50 µm.
Figure S4 | Partial loss-of function of HAM genes leads to the misregulation of CLV3 in the developing AMs. (A–D) RNA in situ hybridization of CLV3 in the AMs from wild type (Ler) (A–B) and MIR171OE (C–D) at both early and late stages. Arrows indicate CLV3 expressing cells. Scale bar: 100 µm.
Figure S5 | Roles of different HAM genes in control of vegetative growth and leaf development. Plants with indicated genotypes, Ler wild type (A), ham123 (B), pHAM1::YPET-HAM1 in ham123 (C), pHAM2::YPET-HAM2 in ham123 (D), and pHAM3::YPET-HAM3 in ham123 were grown in the same conditions (short days) and imaged at the same age (23 DAG). Scale bar: 0.5 cm.
Barrell, P. J., Conner, A. J. (2006). Minimal T-DNA vectors suitable for agricultural deployment of transgenic plants. BioTechniques 41, 708–710. doi: 10.2144/000112306
Biedermann, S., Laux, T. (2018). Plant Development: Adding HAM to Stem Cell Control. Curr. Biol. 28, R1261–R1263. doi: 10.1016/j.cub.2018.09.039
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Brand, U., Grünewald, M., Hobe, M., Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. doi: 10.1104/pp.001867
Clark, S. E., Williams, R. W., Meyerowitz, E. M. (1997). The CLAVATA1gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/S0092-8674(00)80239-1
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Daum, G., Medzihradszky, A., Suzaki, T., Lohmann, J. U. (2014). A mechanistic framework for noncell autonomous stem cell induction in Arabidopsis. Proc. Natl. Acad. Sci. 111, 14619–14624. doi: 10.1073/pnas.1406446111
Engstrom, E. M., Andersen, C. M., Gumulak-Smith, J., Hu, J., Orlova, E., Sozzani, R., et al. (2011). Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 155, 735–750. doi: 10.1104/pp.110.168757
Fletcher, J. C., Brand, U., Running, M. P., Simon, R., Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Geng, Y., Zhou, Y. (2019). Confocal live imaging of shoot apical meristems from different plant species. J. Visual. Exp. 145, e59369. doi: 10.3791/59369
Graf, P., Dolzblasz, A., Würschum, T., Lenhard, M., Pfreundt, U., Laux, T. (2010). MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell 22, 716–728. doi: 10.1105/tpc.109.068296
Gruel, J., Deichmann, J., Landrein, B., Hitchcock, T., Jönsson, H. (2018). The interaction of transcription factors controls the spatial layout of plant aerial stem cell niches. NPJ Syst. Biol. Appl. 4, 36. doi: 10.1038/s41540-018-0072-1
Han, H., Liu, X., Zhou, Y. (2020a). Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 53, 50–56. doi: 10.1016/j.pbi.2019.10.004
Han, H., Yan, A., Li, L., Zhu, Y., Feng, B., Liu, X., et al. (2020b). A signal cascade originated from epidermis defines apical-basal patterning of Arabidopsis shoot apical meristems. Nat. Commun. 11, 1214. doi: 10.1038/s41467-020-14989-4
Heckman, K. L., Pease, L. R. (2007). Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932. doi: 10.1038/nprot.2007.132
Kinoshita, A., Betsuyaku, S., Osakabe, Y., Mizuno, S., Nagawa, S., Stahl, Y., et al. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920. doi: 10.1242/dev.048199
Krizek, B. A. (1999). Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236. doi: 10.1002/(SICI)1520-6408(1999)25:3<224::AID-DVG5>3.0.CO;2-Y
Laux, T., Mayer, K., Berger, J., Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Li, W., Zhou, Y., Liu, X., Yu, P., Cohen, J. D., Meyerowitz, E. M. (2013). LEAFY controls auxin response pathways in floral primordium formation. Sci. Signaling 6, ra23–ra23. doi: 10.1126/scisignal.2003937
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/S0092-8674(00)81703-1
Meyerowitz, E. M. (1997). Genetic control of cell division patterns in developing plants. Cell 88, 299–308. doi: 10.1016/S0092-8674(00)81868-1
Nimchuk, Z. L., Tarr, P. T., Ohno, C., Qu, X., Meyerowitz, E. M. (2011). Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr. Biol. 21, 345–352. doi: 10.1016/j.cub.2011.01.039
Nimchuk, Z. L., Zhou, Y., Tarr, P. T., Peterson, B. A., Meyerowitz, E. M. (2015). Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142, 1043–1049. doi: 10.1242/dev.119677
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G., Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-X
Schulze, S., Schäfer, B. N., Parizotto, E. A., Voinnet, O., Theres, K. (2010). LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 64, 668–678. doi: 10.1111/j.1365-313X.2010.04359.x
Truernit, E., Bauby, H., Dubreucq, B., Grandjean, O., Runions, J., Barthélémy, J., et al. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20, 1494–1503. doi: 10.1105/tpc.107.056069
Yadav, R. K., Perales, M., Gruel, J., Girke, T., Jönsson, H., Reddy, G. V. (2011). WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 25, 2025–2030. doi: 10.1101/gad.17258511
Zhou, Y., Liu, X., Engstrom, E. M., Nimchuk, Z. L., Pruneda-Paz, J. L., Tarr, P. T., et al. (2015). Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517, 377. doi: 10.1038/nature13853
Keywords: shoot development, Arabidopsis, HAIRY MERISTEM, stem cells, confocal imaging, shoot apical meristems
Citation: Han H, Geng Y, Guo L, Yan A, Meyerowitz EM, Liu X and Zhou Y (2020) The Overlapping and Distinct Roles of HAM Family Genes in Arabidopsis Shoot Meristems. Front. Plant Sci. 11:541968. doi: 10.3389/fpls.2020.541968
Received: 11 March 2020; Accepted: 19 August 2020;
Published: 04 September 2020.
Edited by:
Patrick Laufs, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
David Jackson, Cold Spring Harbor Laboratory, United StatesCopyright © 2020 Han, Geng, Guo, Yan, Meyerowitz, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Zhou, emhvdXl1bkBwdXJkdWUuZWR1
†Present address: Lei Guo, Department of Cell Biology and Molecular Genetics, University of Maryland, College Park, MD, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.