- 1Department of Medical Microbiology, Faculty of Biology, Institute of Microbiology, University of Warsaw, Warsaw, Poland
- 2DNA Sequencing and Oligonucleotides Synthesis Laboratory at the Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland
- 3Department of Systems Biology, University of Warsaw, Warsaw, Poland
- 4Institute of Biochemistry and Biophysics, Polish Academy of Sciences (PAS), Warsaw, Poland
- 5Museum and Institute of Zoology, Polish Academy of Sciences, Warsaw, Poland
- 6Institute of Evolutionary Biology, Faculty of Biology, Biological and Chemical Research Centre, University of Warsaw, Poland
Of the Prototheca genus, Prototheca wickerhamii has the highest clinical significance in humans. However, neither nuclear nor organellar genomes of this species were sequenced until now. The hitherto determined and analyzed mitochondrial and plastid genomes of the alleged P. wickerhamii species belong in fact to another species, recently named Prototheca xanthoriae. This study provides a first insight into the organellar genomes of a true P. wickerhamii (type strain ATCC 16529). The P. wickerhamii mitochondrion had a 53.8-kb genome, which was considerably larger than that of Prototheca ciferrii (formerly Prototheca zopfii gen. 1) and Prototheca bovis (formerly Prototheca zopfii gen. 2), yet similarly functional, with the differences in size attributable to a higher number of introns and the presence of extra unique putative genes. The 48-kb plastid genome of P. wickerhamii, compared to autotrophic Trebouxiophyceae, was highly reduced due to the elimination of the photosynthesis-related genes. The gene content of the plastid genome of P. wickerhamii was, however, very similar to other colorless Prototheca species. Plastid genome-based phylogeny reinforced the polyphyly of the genus Prototheca, with Helicosporidium and Auxenochlorella branching within clades of Prototheca species. Phylogenetic reconstruction also confirmed the close relationship of P. wickerhamii and P. xanthoriae, which is reflected in the synteny of their organellar genomes. Interestingly, the entire set of atp genes was lost in P. wickerhamii plastid genome while being preserved in P. xanthoriae.
Introduction
The genus Prototheca comprises unicellular, nonphotosynthetic, saprophytic microalgae, usually associated with humid and organic-rich environments. These organisms are unique in the Plantae kingdom in that they have consistently been implicated in human and animal infections, collectively referred to as protothecosis (Jagielski and Lagneau, 2007). The taxonomy of the Prototheca genus has recently been revised based on phylogenetic analysis of partial cytb gene sequences. In the light of this new classification, 14 Prototheca species are proposed, split into two major lineages, comprising either human- or cattle-associated species (Jagielski et al., 2019).
The genus Prototheca along with another nonphotosynthetic genus Helicosporidium belongs to the predominantly photosynthetic clade of Trebouxiophyceae green algae. All Prototheca species evolved from photosynthetic algae that had lost their ability to photosynthesize yet retained vestigial plastids with substantially reduced genome (Suzuki et al., 2018). It has been suggested that the loss of photosynthesis happened independently three times in this lineage (Suzuki et al., 2018), and that might be reflected in the gene order and gene complement of the vestigial plastid genomes.
Prototheca wickerhamii represents the predominant etiological agent of human protothecosis, affecting both immunocompetent and immunocompromised patients (Todd et al., 2018). Clinically, the disease most frequently involves the skin and subcutaneous tissue followed by articular and disseminated manifestations. Treatment of protothecal infections is often difficult due to resistance of the algae to multiple antimicrobial agents (Lass-Flörl and Mayr, 2007; Todd et al., 2018).
Despite the pathogenic potential of Prototheca spp., the scientific knowledge on this genus remains very limited. Even more scarce are the genetic-level data so far accumulated for the Prototheca algae. Genome-wide sequencing has been attempted only thrice for four Prototheca species, namely Prototheca ciferrii (formerly P. zopfii gen. 1; strains: SAG 2063; 18125; N71), Prototheca bovis (formerly P. zopfii gen. 2; strains: SAG 2021; 50779), Prototheca cutis (JCM 15793), and Prototheca stagnora (JCM 9641) (Severgnini et al., 2018; Suzuki et al., 2018; Zeng et al., 2019). Furthermore, the previously reported mitochondrial (Wolff et al., 1994; Wolff and Kück, 1996) and plastid (Yan et al., 2015) genomes of P. wickerhamii were from the alleged P. wickerhamii strain SAG 263-11, which, according to the current taxonomy, represents a different species—Prototheca xanthoriae (Jagielski et al., 2019). Overall, studies at the genetic level may disclose the acquisition and evolution of the pathogenicity in Prototheca spp. as well as species-specific differences in the infectivity, pathogenicity, and clinical course (severity) of Prototheca infections.
In this work, we report, for the first time, the complete organellar genomes of the true P. wickerhamii species.
Materials and Methods
The type strain of P. wickerhamii (ATCC 16529) was used in the study. Genomic DNA was extracted with a previously described protocol (Jagielski et al., 2017).
Whole genome sequencing (WGS) was performed with a combination of the Illumina MiSeq (Illumina, USA; paired-end, 2 × 300 bp) and PacBio (Pacific Biosciences, USA) platforms using the manufacturer’s standard protocols.
Sequencing of the organellar P. wickerhamii DNA yielded a total of 134,966 and 9,601 reads for Illumina and PacBio respectively. This accounted for 1.1% (Illumina, USA) and 3.3% (PacBio) of the total number of reads for the entire P. wickerhamii genome.
Sequence reads were quality filtered and trimmed using FASTX toolkit (Pearson et al., 1997) and Cutadapt (Martin, 2011), respectively. The PacBio read sets were assembled de novo using wtdbg2 software (Ruan and Heng, 2019) and then corrected using Illumina data with Pilon software (Walker et al., 2014). All bioinformatics manipulations were done using the SeqMan software (DNAStar, USA) and CLCBio Genomic Workbench NGS pipeline (CLCBio, Denmark).
RNA was isolated using Total RNA kit (A&A Biotechnology, Poland) with RNase-free DNase (A&A Biotechnology, Poland) treatment step. Libraries were generated and sequenced according to the producer’s protocol on a MiSeq instrument (Illumina, USA).
Gene prediction and annotation of organellar DNA were performed using the GeSeq ver. 1.76 (Tillich et al., 2017). To predict tRNA genes, online tools tRNAscan v2.0.3 (Chan and Lowe, 2019), MFannot (https://megasun.bch.umontreal.ca/cgi-bin/dev_mfa/mfannotInterface.pl) and RNAweasel (https://megasun.bch.umontreal.ca/cgi-bin/RNAweasel/RNAweaselInterface.pl) were used. Automatically generated gene models have been validated manually using Artemis v.18.0.3 genome browser (Carver et al., 2012). Additionally, localization of rRNA genes was confirmed by manually comparing with RNAseq data. Mitochondrial and plastid genomes of P. wickerhamii were compared with the reference genomes listed in Table 1, which had been annotated previously (Severgnini et al., 2018). Apart from the Prototheca spp., the comparative analysis included other Trebouxiophyceae green algae, Chlorella variabilis, and the closest Prototheca relatives, i.e. the photosynthetic, mixotrophic alga Auxenochlorella protothecoides, and nonphotosynthetic obligate entomoparasite Helicosporidium sp.
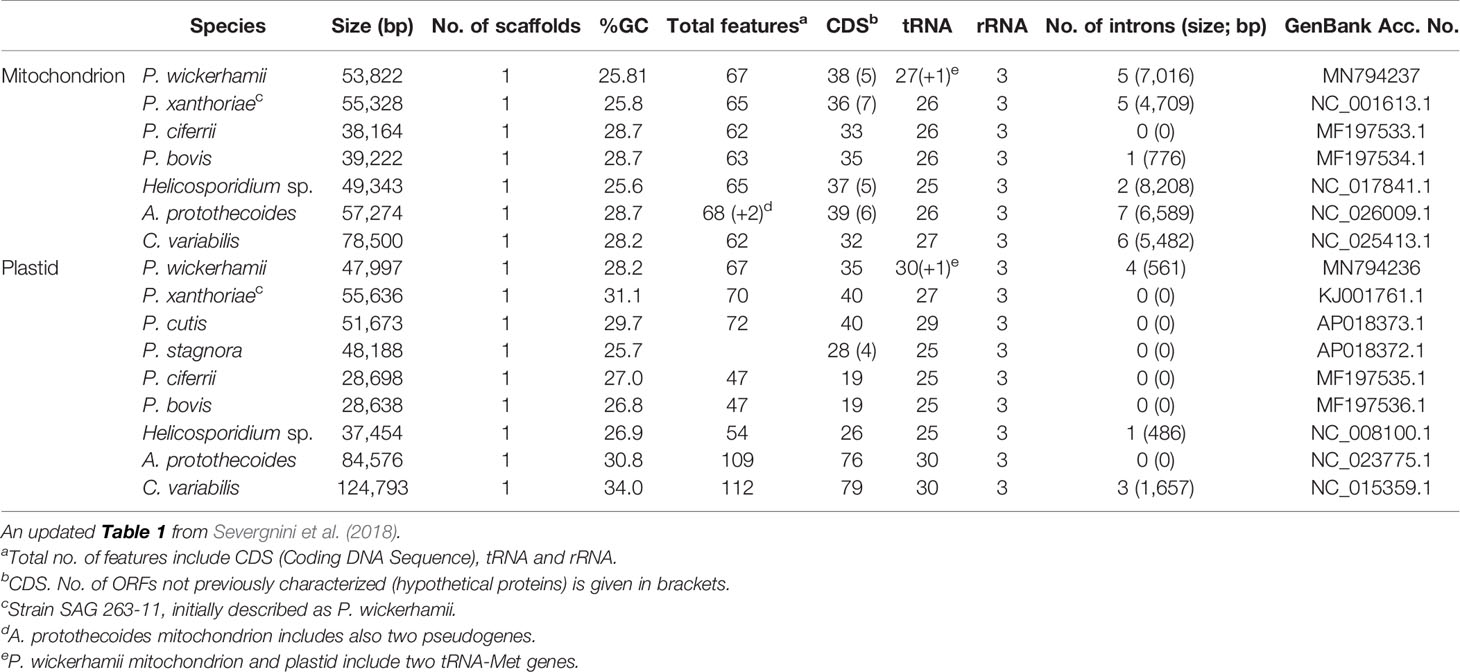
Table 1 General features of the mitochondrial and plastid genomes of Prototheca spp., and closely related species, i.e. C. variabilis, A. protothecoides, and Helicosporidium sp.
Plastid-based maximum likelihood phylogenomic analysis was performed using IQ-TREE v1.6.12 (Nguyen et al., 2015; Chernomor et al., 2016) with 1,000 bootstrap replicates on all 79 nonhypothetical, protein-coding genes present in the plastid genomes of the 24 investigated taxa. All genes were translated into amino acid sequences, aligned with MAFFT v7.271 (Katoh and Standley, 2013), and concatenated in Geneious 10.2.6 (Kearse et al., 2012) to produce a single alignment with total length of 37,109 amino acids, which was subsequently used as the input dataset for tree reconstruction. The sequence evolution model was selected automatically by IQ-TREE (-m TEST parameter; Kalyaanamoorthy et al., 2017) for every partition (gene), and the selected models are shown in Supplementary Table 1.
Gene synteny analysis was performed with MAUVE v2.3.1 plugin (Darling et al., 2010), integrated into Geneious 10.2.6 software (Kearse et al., 2012).
The mitochondrial and plastid genomes of P. wickerhamii were deposited in the GenBank under MN794237 and MN794236 accession numbers, respectively.
All raw sequence data produced in this study were deposited in the NCBI Short Reads Archive (SRA) under project numbers PRJNA646401 (mitochondrion genome) and PRJNA646400 (plastid genome).
Results and Discussion
The P. wickerhamii mitochondrial DNA (mtDNA), comprising 53,878 and 3,861 reads for Illumina and PacBio, respectively and a 475-fold coverage, was AT-rich, circular mapping molecule (Figure 1A, Table 1). It was characterized with 42.4% of noncoding DNA including introns (36.1% excluding introns) and an average intergenic space of 340.6 bp. Overall, the P. wickerhamii mtDNA architecture mirrored P. xanthoriae mtDNA (Wolff et al., 1994; Severgnini et al., 2018; Jagielski et al., 2019). The P. wickerhamii mitochondrial genome was significantly larger than that of P. ciferrii and P. bovis (Table 1). These differences might be explained by putative rearrangements and/or reduction events. Exemplarily, in P. wickerhamii and P. xanthoriae, the cox1 gene contains three (4,975 bp) and four exons (5,376 bp), respectively, whereas in P. bovis and P. ciferrii, it is a single exon gene of 1,574 bp. Noteworthy, in the former two species, the cox1 gene contains two additional genes (in P. wickerhamii designated as DBVPGmt_008 and DBVPGmt_009), which are missing in the mitochondrial genomes of P. bovis and P. ciferrii (Figure 1A).
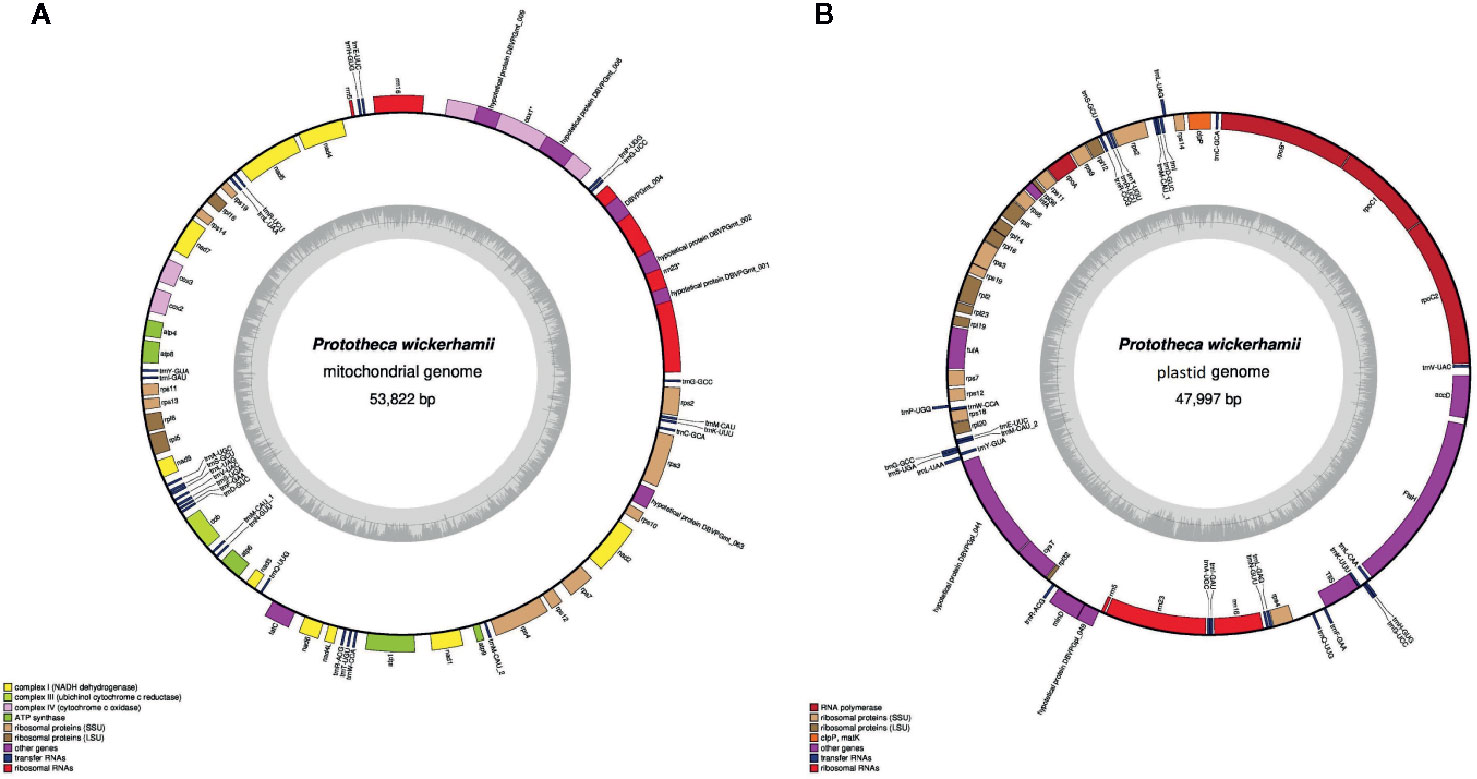
Figure 1 P. wickerhamii mitochondrion (A) and plastid (B) circular plot. Genes (filled boxes) located outside/inside the map are transcribed clockwise/counterclockwise. tRNA genes are indicated by the “trn” followed by one-letter amino acid code and anticodon given behind the dash. Innermost circle represents GC content.
The mitochondrion of P. wickerhamii encoded 38 proteins, a number that almost equaled that of A. protothecoides (39), yet being higher than in other analyzed species, where it ranged from 32 (C. variabilis) to 37 (Helicosporidium sp.) (Table 1).
Mitochondria and plastids originated from a primary endosymbiotic event, yet the subsequent evolution of the two organelles differ. Whereas mitochondria have evolved in a vertical inheritance, plastid evolution has involved both vertical and horizontal spread (Archibald, 2015; Martin et al., 2015; de Vries and Archibald, 2018).
A standard set of 32 mitochondrial protein-coding genes was present in P. wickerhamii, namely ribosomal proteins, apocytochrome b, subunits of the ATPase, cytochrome oxidase, NADH dehydrogenase complexes, and Twin-arginine translocation protein. Almost all of them were found among the analyzed species (Supplementary Table 2). One exception was atp8 coding for ATP synthase F0 subunit 8, which was demonstrated in all algal species but P. xanthoriae (Supplementary Table 2). Furthermore, the rpl10 gene coding for a ribosomal protein was found only in P. ciferrii and P. bovis. Transfer of the atp8 gene from the mitochondrial genome to the nuclear genome had already been reported in various eukaryotic lineages, including ciliates (Burger et al., 2000), apicomplexans, dinoflagellates (Slamovits et al., 2007), and Chlorophyceae (Martínez-Alberola et al., 2019). The rpl10 gene was lost several times in Chlorophytes, including Chlorophyceae, Ulvophyceae, perhaps Prasinophyceae, and some Trebouxiophyceae (Martínez-Alberola et al., 2019). Whether the atp8 and rpl10 were lost entirely or transferred to the protothecal nuclear genomes remains to be answered. Overall, the mitochondrial gene content appears to be highly conserved among the analyzed species.
A total of five introns in two genes (2/67; 3%) were characterized in P. wickerhamii mtDNA, with a total length of 7,016 bp (Table 1). Those introns split the cox1 and rrn23, into three and four exons, respectively. A more complex intron structure, with up to seven introns and the total length reaching 8,200 bp was observed in P. xanthoriae, C. variabilis, A. protothecoides, and Helicosporidium sp. Interestingly, P. bovis showed only a single intron in the rrn23 gene (Table 1). In P. wickerhamii, four introns were annotated either as LAGLIDADG endonuclease (DBVPGmt_004) or had LAGLIDADG motifs (DBVPGmt_001, DBVPGmt_002, DBVPGmt_009), which indicates a putative endonuclease function for the protein (Pombert and Keeling, 2010). LAGLIDADG motifs, commonly found in group I introns (Haugen and Bhattacharya, 2004), had previously been described in P. bovis and P. xanthoriae, but not in P. ciferrii (Supplementary Table 2).
The P. wickerhamii plastid DNA (ptDNA), comprised 81,088 and 5,740 reads for Illumina and PacBio, respectively, had 730-fold coverage (Figure 1B, Table 1). It was similar in size to the plastid of P. stagnora and almost double in size compared to plastids of P. ciferrii and P. bovis. Structurally, the P. wickerhamii plastid was compact, with about 25% of noncoding DNA including introns (23.9% excluding introns) and an average intergenic space of 171.2 bp.
The P. wickerhamii ptDNA was predicted to contain 35 protein-coding genes, which was somewhat lower compared with P. xanthoriae and P. cutis (40), yet clearly higher as compared with P. ciferrii and P. bovis (19). More than twice as much proteins as in P. wickerhamii were encoded by the plastid genomes of the two photosynthetic species (Table 1).
The so far sequenced plastid genomes of colorless Prototheca spp. were shown to be highly reduced due to the elimination of photosynthesis-related genes. Moreover, comparative analyses of the ptDNAs revealed that the gene content for plastid functions was highly conserved among these nonphotosynthetic lineages (Severgnini et al., 2018; Suzuki et al., 2018; Maciszewski and Karnkowska, 2019; Zeng et al., 2019;). The plastid genomes of Prototheca spp. lacked cytochrome complex, photosystem I and II proteins, and genes involved in chlorophyll biosynthesis when compared with their photosynthetic relatives, i.e. A. protothecoides and C. variabilis. The gene content differed also between the Prototheca species (Supplementary Table 3). Only P. wickerhamii and P. xanthoriae retained all large ribosomal subunits. Only P. wickerhamii, P. xanthoriae, and P. cutis preserved all small ribosomal subunits, two protein quality control genes—ftsH and clpP, two translation mediating genes—tufA and infA, and cell division gene—minD. Algae that contain minD in their plastid genome are exclusively monoplastidic (de Vries and Gould, 2018). However, the number of plastids in P. wickerhamii remains unknown (Nadakavukaren and McCracken, 1977; Kiyohara et al., 2006). P. wickerhamii, P. xanthoriae, P. cutis, and P. stagnora retained also all rpo subunits, which are absent from the plastid genomes of P. ciferrii and P. bovis (Supplementary Table 3).
As it was previously hypothesized, changes in ycf1 occur concomitantly with changes in FtsH (de Vries et al., 2017). Unexpectedly, P. wickerhamii did encode FtsH but not ycf1 (Supplementary Table 3). Although FtsH is a plastid-encoded component of the photosystem II maintenance machinery, ycf1 function is still debatable.
Interestingly, P. wickerhamii, P. stagnora, P. bovis, and P. ciferrii, in contrast to P. xanthoriae and P. cutis, lacked six genes for the ATP synthase subunits, typically involved in the photosynthesis (Supplementary Table 3). Genes of the ATP synthase/hydrolysis complex were also detected in nonphotosynthetic plastids of a diatom Nitzschia despite the lack of genes for photosynthesis, carbon fixation, and chlorophyll production. It has been hypothesized that ATP synthase subunits present in Nitzschia may produce a proton gradient between the thylakoids and stroma, which is involved in protein translocation (Kamikawa et al., 2015). Moreover, reconstruction of plastid metabolism of this diatom suggested that the ATP synthase complex might function to regulate activities of plastid proteins involved in amino acid biosynthesis, reductive pentose phosphate pathway, and glycolysis/gluconeogenesis by pumping protons between the stroma and thylakoid lumen (Kamikawa et al., 2017). The function of the ATP synthase in Prototheca spp. might be similar. However, it appeared not indispensable in this lineage since most of the known Prototheca ptDNAs completely lacked all genes required for the plastid ATP synthase. In the absence of ATP synthase, some ATP might be imported from the cytosol by plastid ATP transporters as it was shown in diatoms (Ast et al., 2009). Still, whether or not that is the case in Prototheca cannot be answered without plastid proteome reconstruction. At this place, it is worth to note that P. wickerhamii was demonstrated, upon electron microscopy studies, to contain double-membraned plastids with starch grains and rudimentary lamellar-like structures (Nadakavukaren and McCracken, 1977; Kiyohara et al., 2006).
The observed differences in the gene content among Prototheca spp. may reflect an independent loss of photosynthesis in several protothecal lineages. Therefore, various lineages and species might be at a different stage of the reductive evolution of plastid functions after the loss of photosynthesis.
Plastid genome-based phylogeny resolved Prototheca as a polyphyletic genus, composed of two major clades (Figure 2). The first clade contains two pairs of sister species: P. cutis and P. wickerhamii, and P. xanthoriae and Auxenochlorella protothecoides. The second clade contains P. stagnora, P. bovis, and P. ciferrii, with Helicosporidium sp. situated on a long branch at its base. All nodes within the aforementioned clades have absolute bootstrap support. Despite limited taxon sampling, our results are fully concordant with cytb-based single-gene phylogeny of Prototheca and their relatives by Jagielski et al., 2019. Although no plastid genome sequence is available for the Prototheca lectotype strain, which is P. zopfii ATCC 16533, the recent work of Jagielski et al., 2019 resolves the phylogenetic position of this species as sister to P. ciferrii. Therefore, the lectotype would certainly be within the second of the aforementioned clades in our phylogeny, suggesting that only this clade should be recognized as the genus Prototheca. Except A. protothecoides, the rest of the analyzed species, including Prototheca spp. and Helicosporidium sp., are secondary nonphotosynthetic Trebouxiophyceae. The tree topology suggests that photosynthesis has been lost at least three times in this lineage, first in P. xanthoriae, the second time in the common ancestor of P. wickerhamii and P. cutis, and the third time in the second clade encompassing Helicosporidium, P. stagnora, P. bovis, and P. ciferrii. Three independent losses of photosynthesis in this group are in agreement with earlier reconstructions (Suzuki et al., 2018), and are an excellent example of convergent reductive evolution, reflected in the nearly identical plastid-genome complement in all the Prototheca species.
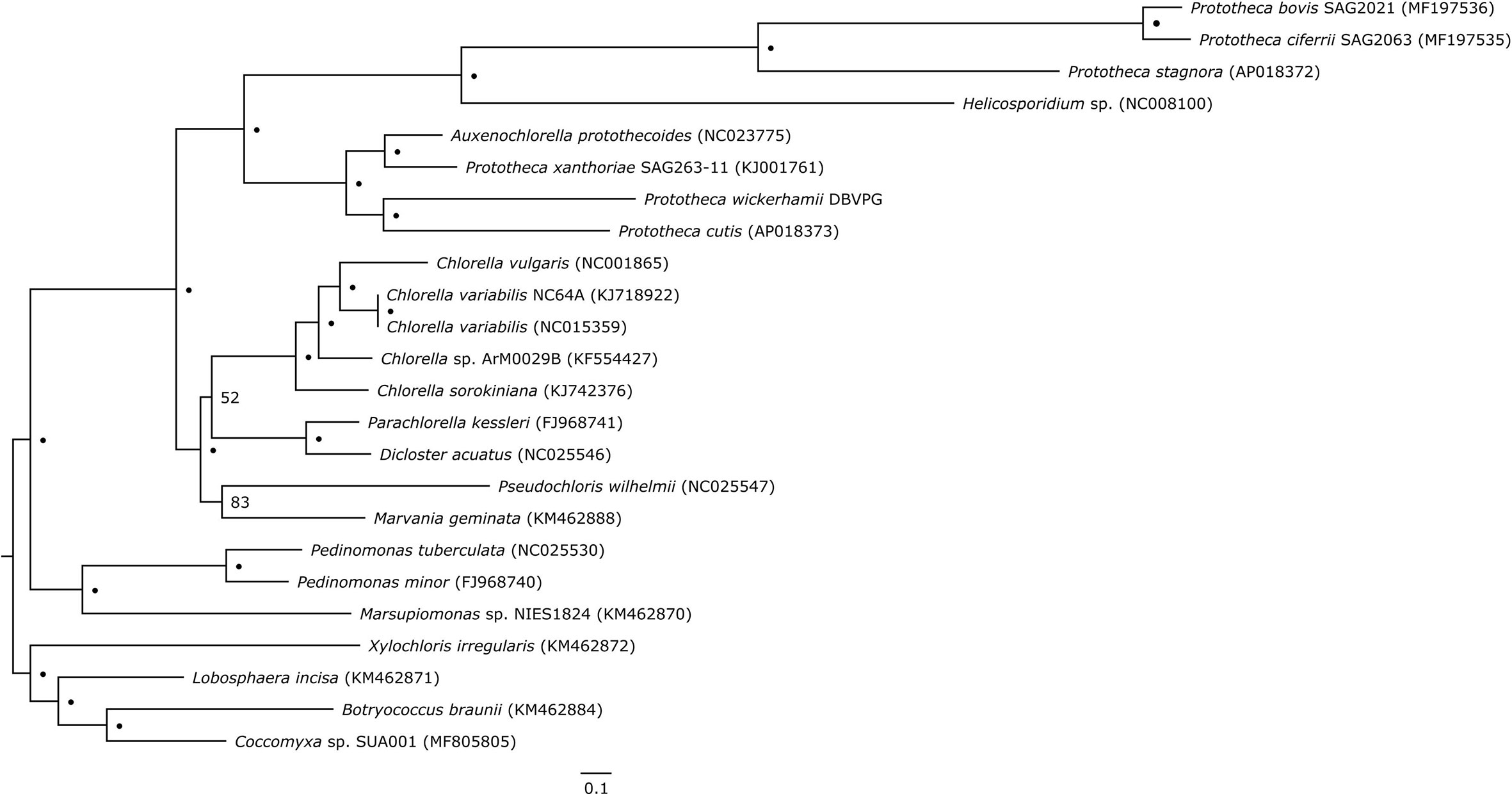
Figure 2 Plastid genome-based phylogenetic tree of Prototheca spp. and related genera. Dots (•) represent absolute bootstrap support (100); for nodes without absolute support, numerical bootstrap values are provided. Scale bar at the bottom indicates 0.1 substitution per 10 amino acid positions.
The mtDNA gene order analysis revealed two lineages among the Prototheca spp. investigated: the first allocated P. wickerhamii and P. xanthoriae, the second contained P. ciferrii and P. bovis (Figure 3A). Not surprisingly, highly syntenic pairs of genomes represented closely related taxa (Jagielski et al., 2019).
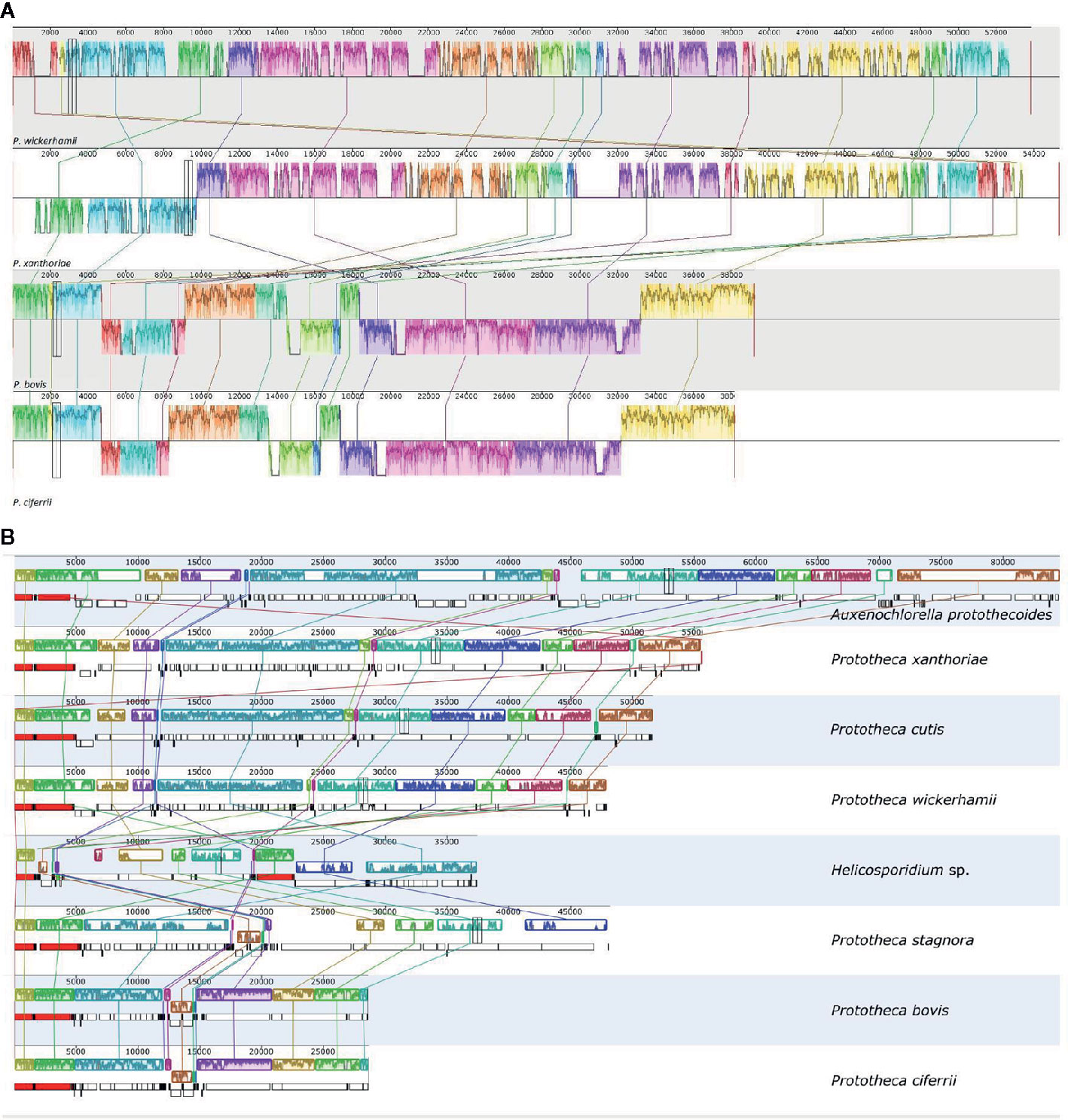
Figure 3 Comparison of the gene order in the mitochondrial (A) and plastid (B) genomes of the analyzed species. Syntenic regions are marked with matching colors and connected by lines. Numbers indicate nucleotide positions in the mt- and ptDNA.
The ptDNA protein-coding gene order was exactly identical in the entire clade containing A. protothecoides, P. wickerhamii, P. cutis, and P. xanthoriae—a sole rearrangement was observed in P. cutis, where a small block of three tRNA-coding genes (trnG, trnH, and trnL) was inverted. An overall similar, yet definitely separate ptDNA synteny type was observed in the other clade of Prototheca, composed of P. stagnora, P. bovis, and P. ciferrii (Figure 3B). In this group, four locally collinear ptDNA blocks are translocated in comparison to the first Prototheca clade, with a fifth (tilS-rps4) block being additionally inverted, which is a unique case of protein-coding gene inversion in protothecan ptDNA since their diversification from the last common ancestor of the entire genus.
The presence of two Prototheca lineages, evidenced by pt- and mtDNA structure raises a question, if the human- and cattle-associated clades, represented by P. wickerhamii and P. bovis, respectively, have acquired pathogenic features independently and in parallel rather than from a common ancestor. The WGS data of Prototheca spp. will give a better understanding of the pathobiology and evolution of this genus.
Conclusions
In conclusion, this study provides a first, brief insight into the organellar genomes of P. wickerhamii. The mtDNA of P. wickerhamii preserved its functionality similar to other related organisms, with its size extension, mostly due to a higher number of introns (five in both P. wickerhamii and P. xanthoriae), as well as some unique putative genes unseen in other species (P. bovis and P. ciferrii). Compact and simplified structure was observed in the P. wickerhamii plastid genome, driven by the lack of photosynthesis-related genes. The architecture of the P. wickerhamii mitochondrial and plastid genomes resembles more that of closely related saprophytic P. xanthoriae than of pathogenic P. ciferrii and P. bovis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
ZB performed culturing, analyzed the data, and wrote the article. JG performed DNA isolation and genome sequencing (Illumina). RG provided genome annotation. PS performed synteny analysis. JP performed genome sequencing (PacBio). KM performed phylogenetic analysis, reviewed synteny analysis and genome annotation. AG performed RNA-seq analysis. AK performed phylogenetic analysis and reviewed the manuscript. TJ conceptualized and supervised the study, provided the funding, critical revision. All authors contributed to the article and approved the submitted version.
Funding
The study was financed by the National Science Centre grants «PRELUDIUM» (2013/09/N/NZ2/00248) and «SONATA» (2014/15/D/NZ7/01797). Additionally, AK and KM were supported by the National Science Centre grant (“SONATA” 2016/21/D/NZ8/01288) and EMBO Installation Grant (to AK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01296/full#supplementary-material
References
Archibald, J. M. (2015). Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 25, 911–921. doi: 10.1016/j.cub.2015.07.055
Ast, M., Gruber, A., Schmitz-Esser, A., Neuhaus, H. E., Kroth, P. G., Horn, M., et al. (2009). Diatom plastids depend on nucleotide import from the cytosol. PNAS 106, 3621–3626. doi: 10.1073/pnas.0808862106
Burger, G., Zhu, Y., Littlejohn, T. G., Greenwood, S. J., Schnare, M. N., Lang, B. F., et al. (2000). Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J. Mol. Biol. 297, 365–380. doi: 10.1006/jmbi.2000.3529
Carver, T., Harris, S. R., Berriman, M., Parkhill, J., McQuillan, J. A. (2012). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. doi: 10.1093/bioinformatics/btr703
Chan, P. P., Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chernomor, O., von Haeseler, A., Minh, B. Q. (2016). Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 65, 997–1008. doi: 10.1093/sysbio/syw037
de Vries, J., Gould, S. B. (2018). The monoplastidic bottleneck in algae and plant evolution. J. Cell Sci. 13, jcs203414.
Darling, A. E., Mau, B., Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5, e11147. doi: 10.1371/journal.pone.0011147
de Vries, J., Archibald, J. M. (2018). Plastid genomes. Curr. Biol. 28, 336–337. doi: 10.1016/j.cub.2018.01.027
de Vries, J., Archibald, J. M., Gould, S. B. (2017). The carboxy terminus of ycf1 contains a motif conserved throughout >500 myr of streptophyte evolution. Genome Biol. Evol. 9, 473–479. doi: 10.1093/gbe/evx013
Haugen, P., Bhattacharya, D. (2004). The spread of LAGLIDADG homing endonuclease genes in rDNA. Nucleic Acids Res. 32, 2049–2057. doi: 10.1093/nar/gkh520
Jagielski, T., Lagneau, P. E. (2007). Protothecosis. A pseudofungal infection. J. Mycol. Méd 17, 261–270. doi: 10.1016/j.mycmed.2007.08.003
Jagielski, T., Gawor, J., Bakuła, Z., Zuchniewicz, K., Gromadka, R. (2017). Żak IAn optimized method for high quality DNA extraction from microalga Prototheca wickerhamii for genome sequencing. Plant Methods 13, 105. doi: 10.1186/s13007-017-0228-9
Jagielski, T., Bakula, Z., Gawor, J., Maciszewski, K., Dyląg, M., Nowakowska, J., et al. (2019). The genus Prototheca (Trebouxiophyceae, Chlorophyta) revisited: implications from molecular taxonomic studies. Alg. Res. 43, 101639. doi: 10.1016/j.algal.2019.101639
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kamikawa, R., Tanifuji, G., Ishikawa, S. A., Ishii, K., Matsuno, Y., Onodera, N. T., et al. (2015). Proposal of a twin arginine translocator system-mediated constraint against loss of ATP synthase genes from nonphotosynthetic plastid genomes. Mol. Biol. Evol. 32, 2598–2604. doi: 10.1093/molbev/msv134
Kamikawa, R., Moog, D., Zauner, S., Tanifuji, G., Ishida, K. I., Miyashita, H., et al. (2017). A non-photosynthetic diatom reveals early steps of reductive evolution in plastids. Mol. Biol. Evol. 34, 2355–2366. doi: 10.1093/molbev/msx172
Katoh, K., Standley, D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kiyohara, N., Osafune, T., Ehara, T., Hiroshi, H., Ito-Kuwa, S., Aoki, S. (2006). Immuno-electron microscopic studies on plastid DNA and photosynthetic proteins in Prototheca wickerhamii. Cytol. - CYTOL. TOKYO 71, 309–314. doi: 10.1508/cytologia.71.309
Lass-Flörl, C., Mayr, A. (2007). Human protothecosis. Clin. Microbiol. Rev. 20, 230–242. doi: 10.1128/CMR.00032-06
Maciszewski, K., Karnkowska, A. (2019). Should I stay or should I go?Retention and loss of components in vestigial endosymbiotic organelles. Curr. Opin. Genet. Dev. 58-59, 33–39. doi: 10.1016/j.gde.2019.07.013
Martin, W. F., Garg, S., Zimorski, V. (2015). Endosymbiotic theories for eukaryote origin. Phil. Trans. R Soc. B 370, 20140330. doi: 10.1098/rstb.2014.0330
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EmbNET J. 17, 10–12. doi: 10.14806/ej.17.1.200
Martínez-Alberola, F., Barreno, E., Casano, L. M., Gasulla, F., Molins, A., del Campo, E. M. (2019). Dynamic evolution of mitochondrial genomes in Trebouxiophyceae, including the first completely assembled mtDNA from a lichen-symbiont microalga (Trebouxia sp. TR9). Sci. Rep. 9, 8209. doi: 10.1038/s41598-019-44700-7
Nadakavukaren, M. J., McCracken, D. A. (1977). An ultrastructural survey of the genus Prototheca with special reference to plastids. Mycopathologia 61, 117–119. doi: 10.1007/BF00443840
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Pearson, W. R., Wood, T., Zhang, Z., Miller, W. (1997). Comparison of DNA sequences with protein sequences. Genomics 46, 24–36. doi: 10.1006/geno.1997.4995
Pombert, J. F., Keeling, P. J. (2010). The mitochondrial genome of the entomoparasitic green alga Helicosporidium. PLoS One 5, e8954. doi: 10.1371/journal.pone.0008954
Ruan, J., Heng, L. (2020). Fast and accurate long-read assembly with wtdbg2. Nat. Methods, 17, 155–158.
Severgnini, M., Lazzari, B., Capra, E., Chessa, S., Luini, M., Bordoni, R., et al. (2018). Genome sequencing of Prototheca zopfii genotypes 1 and 2 provides evidence of a severe reduction in organellar genomes. Sci. Rep. 8, 14637. doi: 10.1038/s41598-018-32992-0
Slamovits, C. H., Saldarriaga, J. F., Larocque, A., Keeling, P. J. (2007). The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J. Mol. Biol. 372, 356–368. doi: 10.1016/j.jmb.2007.06.085
Suzuki, S., Endoh, R., Manabe, R. I., Ohkuma, M., Hirakawa, Y. (2018). Multiple losses of photosynthesis and convergent reductive genome evolution in the colourless green algae Prototheca. Sci. Rep. 17, 940. doi: 10.1038/s41598-017-18378-8
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, 6–11. doi: 10.1093/nar/gkx391
Todd, J. R., Matsumoto, T., Ueno, R., Murugaiyan, J., Britten, A., King, J. W., et al. (2018). Medical phycology 2017. Med. Mycol. 56, 188–204. doi: 10.1093/mmy/myx162
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wolff, G., Kück, U. (1996). Transcript mapping and processing of mitochondrial RNA in the chlorophyte alga Prototheca wickerhamii. Plant Mol. Biol. 30, 577–595. doi: 10.1007/BF00049333
Wolff, G., Plante, I., Lang, B. F., Kück, U., Burger, G. (1994). Complete sequence of the mitochondrial DNA of the chlorophyte alga Prototheca wickerhamii. Gene content and genome organization. J. Mol. Biol. 18, 75–86. doi: 10.1006/jmbi.1994.1210
Yan, D., Wang, Y., Murakami, T., Shen, Y., Gong, J., Jiang, H., et al. (2015). Auxenochlorella protothecoides and Prototheca wickerhamii plastid genome sequences give insight into the origins of non-photosynthetic algae. Sci. Rep. 25, 14465. doi: 10.1038/srep14465
Keywords: Prototheca wickerhamii, protothecosis, colorless algae, mitochondrial genome, plastid genome
Citation: Bakuła Z, Gromadka R, Gawor J, Siedlecki P, Pomorski JJ, Maciszewski K, Gromadka A, Karnkowska A and Jagielski T (2020) Sequencing and Analysis of the Complete Organellar Genomes of Prototheca wickerhamii. Front. Plant Sci. 11:1296. doi: 10.3389/fpls.2020.01296
Received: 09 June 2020; Accepted: 07 August 2020;
Published: 01 September 2020.
Edited by:
Miroslav Obornik, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Jan de Vries, University of Göttingen, GermanyJan Janouskovec, University of Oslo, Norway
Copyright © 2020 Bakuła, Gromadka, Gawor, Siedlecki, Pomorski, Maciszewski, Gromadka, Karnkowska and Jagielski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomasz Jagielski, t.jagielski@biol.uw.edu.pl
 Zofia Bakuła
Zofia Bakuła