- 1BGPI, INRA, CIRAD, SupAgro, Univ. Montpellier, Montpellier, France
- 2Valle del Cauca, CIAT, Palmira, Colombia
- 3Institut Fédératif de Recherche 3450, Université de Toulouse, CNRS, UPS, Plateforme Imagerie TRI-Genotoul, Castanet-Tolosan, France
- 4LIPM, Université de Toulouse, INRA, CNRS, Castanet-Tolosan, France
- 5UMR Interactions Plantes-Microorganismes-Environnement (IPME), IRD-Cirad-Université Montpellier, Institut de Recherche pour le Développement, Montpellier, France
Background: Identifying new sources of disease resistance and the corresponding underlying resistance mechanisms remains very challenging, particularly in Monocots. Moreover, the modification of most disease resistance pathways made so far is detrimental to tolerance to abiotic stresses such as drought. This is largely due to negative cross-talks between disease resistance and abiotic stress tolerance signaling pathways. We have previously described the role of the rice ZBED protein containing three Zn-finger BED domains in disease resistance against the fungal pathogen Magnaporthe oryzae. The molecular and biological functions of such BED domains in plant proteins remain elusive.
Results: Using Nicotiana benthamiana as a heterologous system, we show that ZBED localizes in the nucleus, binds DNA, and triggers basal immunity. These activities require conserved cysteine residues of the Zn-finger BED domains that are involved in DNA binding. Interestingly, ZBED overexpressor rice lines show increased drought tolerance. More importantly, the disease resistance response conferred by ZBED is not compromised by drought-induced stress.
Conclusions: Together our data indicate that ZBED might represent a new type of transcriptional regulator playing simultaneously a positive role in both disease resistance and drought tolerance. We demonstrate that it is possible to provide disease resistance and drought resistance simultaneously.
Background
In agriculture, biotic and abiotic stresses cause important yield losses, making improvement in stress tolerance of crop plants a major aim for research. Rice (Oryza sativa) has a pivotal role for the food security of over half the world’s population (Calpe, 2006). Among biotic stresses, rice blast caused by the fungal pathogen Magnaporthe oryzae is arguably one of the most devastating diseases in rice because of its wide distribution and the potential of causing total losses under conducive conditions (Hyun-Khang and Valent, 2010). Additionally, under rainfed cultivation, rice is one of the most drought-susceptible crops because of its small root system, thin cuticular wax, and rapid stomata closure (Oladosu et al., 2019). Plants have developed regulatory networks allowing them to adapt, survive, and reproduce under several stress conditions (Kissoudis et al., 2014). Two major types of resistance against pathogens are often defined according to the molecular mechanism for pathogen recognition (Jones and Dangl, 2006). The first layer of active defense is triggered by highly conserved pathogen-associated molecular patterns (PAMPs). PAMP-triggered immunity (PTI), also called basal immunity, confers basal disease resistance following infection by a virulent pathogen. The other layer of defense, called effector-triggered immunity, is activated by direct or indirect recognition of effector molecules from the pathogen by resistance proteins of the plant and is leading to programmed cell death, preventing the spread of the pathogen in the host (Jones and Dangl, 2006). In both forms of resistance several physiological changes occur, including the generation of reactive oxygen species (ROS), ion fluxes, accumulation of defense hormones such as salicylic acid (SA), and induction of defense-related genes, such as pathogenesis-related (PR) genes (Corwin and Kliebenstein, 2017).
The main components of these regulatory networks are shared by abiotic stress as well, including ROS signaling, ion fluxes, and plant hormone signaling among others (Zhang and Sonnewald, 2017). However, negative cross-talks between disease resistance and abiotic tolerance pathways often render pathogen-resistant plants into abiotic-sensitive plants and the other way around (Kissoudis et al., 2014; Suzuki et al., 2014). For instance, the phytohormone abscisic acid (ABA), one of the main regulators of drought-stress response is also known to alter the plant response against pathogens by negatively regulating both salicylic acid (SA) and jasmonic acid (JA) biosynthesis, and signaling (Kissoudis et al., 2014; Ramegowda and Senthil-Kumar, 2015). Several studies of transgenic plants have shed light into some of the mechanisms by which rice copes with different stresses. For instance, the ERF family transcription factor OsLG3 rice protein increases tolerance against drought by scavenging reactive oxygen species (ROS) (Xiong et al., 2018). The stress response mitogen-activated protein kinase (MAPK) from rice OsMAPK5 positively regulates drought, salt, and cold tolerance but negatively modulates disease resistance against the rice blast fungus Magnaporthe oryzae and the bacterial pathogens Burkholderia glumae (Xiong and Yang, 2003) or Xanthomonas oryzae (Xoo) (Seo et al., 2011). Similarly, overexpression of NPR1, a gene acting as a positive regulator of disease resistance, leads to reduced tolerance to drought (Quilis et al., 2008). Out of 60 cases of transgenic rice plants showing enhanced disease resistance reviewed, 28 showed detrimental effects for other stress or developmental defects (Delteil et al., 2010). Conversely, there are few examples where the enhancement of disease resistance leads to an improved tolerance to abiotic stress. For instance, the calcium-dependent protein kinases OsCPK4 and OsCPK10 as well as the transcription factor OsNAC6 positively enhance tolerance against drought, salinity as well as resistance towards M. oryzae (Nakashima et al., 2007; Campo et al., 2014; Bundó and Coca, 2016; Bundó and Coca, 2017). Similarly, OsWRKY11 (Lee et al., 2018) and MAPK kinase 10.2 (Ma et al., 2017) enhance resistance against Xoo and drought. Additionally, the MADS-box transcription factor OsMADS26 negatively regulates resistance against M. oryzae, Xoo and drought in rice (Khong et al., 2015). These non-exhaustive examples demonstrate the complexity of the connections between plant response pathways to abiotic and biotic stresses.
The response of plants to combined stress, in particular combination of biotic and abiotic stresses, revealed to be even more complex than the response to single stresses (for review Zhang and Sonnewald, 2017). In the case of combined disease and drought stresses, the outcome of the interaction leading to either susceptibility or resistance is influenced by various factors including the nature of the pathogen, the developmental stage of the plant, as well as the severity and the duration of the drought stress (Ramegowda and Senthil-Kumar, 2015; Zhang and Sonnewald, 2017). For example, exposure to mild drought induces basal defenses (Ramegowda and Senthil-Kumar, 2015), while severe drought conditions reduce basal defenses and inhibit disease resistance mediated by resistance genes (Bidzinski et al., 2016). Thus, identifying genetic determinants conferring a disease resistance response that remains effective under abiotic stress like drought is an important challenge for research.
We previously identified a rice protein containing three zinc-finger BED domains called ZBED, which was shown to be a positive regulator of resistance against the rice blast fungus (Kroj et al., 2016). The BED-type zinc finger (zf-BED) domain is a protein domain that was named after the Drosophila BEAF (Boundary Element-associated factor) protein, thought to be involved in chromatin insulation and the Drosophila DREF protein, a transcriptional regulator for S-phase genes (Aravind, 2000). The BED finger is about 50 to 60 amino acid residues domain that contains a characteristic motif with two highly conserved aromatic positions, as well as a shared pattern of cysteines and histidines, predicted to form a zinc finger (zf). It has been suggested that DNA-binding is the general function of this domain. Some proteins known to contain BED domains are the Hobo-like transposases (Aravind, 2000); Caenorhabditis elegans Dpy-20 protein, a predicted cuticular gene transcriptional regulator (Inoue and Sternberg, 2010); and tobacco 3AF1 (Lam et al., 1990) and tomato E4/E8-BP1, light- and ethylene-regulated DNA-binding proteins (Coupe and Deikman, 1997). BED domains were also found to be integrated into plant resistance genes from different plant species like poplar (Populus trichocarpa) and in several monocot species such as Oryza sativa, Hordeum vulgare, Brachypodium distachyon and Triticum spp among others (Yoshimura et al., 1998; Kroj et al., 2016; Marchal et al., 2018; Read et al., 2020). However, the role of the zf-BED domain in plant resistance genes has not been elucidated. We proposed that such integrated BED domains could represent decoys of endogenous host BED proteins targeted by pathogen effectors (Read et al., 2019). However, the recent finding that the Yr7, Yr5, and YrSP resistance genes from wheat have identical zf-BED sequences as Xa1, CGS-Xo111, and Nb-xo15 from rice (Marchal et al., 2018; Read et al., 2019), suggests that this domain is not a specificity-determining decoy. Rather, it may have a role in downstream signaling, localization, or other. The way BED domains operate might include dimerization, recruitment of other interacting proteins, or DNA binding. Thus, the molecular and biological functions of BED-containing proteins in plants are still largely unknown. Given the multiple cases of negative cross-talks already mentioned, we wanted to better understand the potential role of ZBED in disease resistance by testing bacterial pathogens, and to assess whether ZBED has a role in abiotic stress response in rice. This led us to test if disease resistance conferred by ZBED is robust under drought.
Materials and Methods
Plant Genotypes
Rice plants (Oryzae sativa) Nipponbare background were used for Agrobacterium transformation to generate transgenic lines overexpressing the ZBED gene (LOC_Os01g36670) as described (Kroj et al., 2016). Briefly, the ZBED cDNA from rice cv Nipponbare (L.) was PCR amplified and cloned into the pBIOS2300OX transformation vector under the control of the constitutive maize (Zea mays) ubiquitin promoter and plants were transformed as described previously (Yuan et al., 2007). The number of T‐DNA insertions was estimated in the T1 and T2 families using PCR for the geneticyn/kanamycin selection marker as a diagnostic for the presence of T-DNA. Lines carrying single copy insertions (showing 3:1 PCR-positive; n > 20 plants analyzed) were conserved for further analysis. Siblings PCR-negative (not containing the transgene) were used as controls (azygous controls). Individual T2 plants (homozygous overexpressor and azygous) from three independent monolocus lines were selfed, and ZBED overexpression was confirmed in the T3 progeny (Grand et al., 2012). Three overexpressing ZBED lines (ZBED-OX1, ZBED-OX2, and ZBED-OX3) with their respective azygous lines (ZBED 1/0, ZBED 2/0, and ZBED 3/0) were used in this study.
Yeast Two-Hybrid Screening (Y2H)
For Y2H, we used the library (in the Co39 Indica background) and methods described in (Cesari et al., 2013). We also built a new library (RCL1) from japonica rice Nipponbare using RNA from leaves and shoot-apical meristem of flowering plants, after drought stress (root and leaves treated by Mannitol or salt) and disease-related stresses (leaves treated with blast fungus or chitin for one and two days). Both libraries were made in the Matchmaker system (Clontech) following the manufacturer’s instructions. Once identified, full-length cDNAs of ZBED interactors (KIP1, WRKY4, STE20 and STE11) were cloned and further used for BiFC analysis.
Bimolecular Fluorescence Complementation (BiFC)
Full-length cDNAs were cloned into the pDONR207 (Thermo Fischer Scientific). The N-terminal portion of yellow fluorescent protein (YFPn) was fused to the N-terminus of KIP1, STE20, STE11 and WRKY4. Each of those constructs was co-expressed in N. benthamiana leaves with YFPc-ZBED (carboxy-portion of YFP fused to the N-terminus of ZBED). Each construct was infiltrated in three N. benthamiana leaves (technical reps), and the experiment was repeated twice for a total of three biological reps. The YFP fluorescence complementation was analyzed by laser-scanning confocal microscope, laser power was set at 2% for all interactions, except WRKY4 which was set at 4%. The nuclei were DAPI-stained.
Generation of Point Mutations in ZBED’s BED Domains
The ZBED (LOC_Os01g36670) cDNA, cloned into pDONR207 plasmid was used to generate point mutations in the three BED domains in order to determine whether BED domains were responsible of DNA binding. BED point mutations were done by substituting the two cysteines of the zinc finger to glycines in each of the three domains present in ZBED (Supplementary Figure 2). To this end, we used QuickChange lightning site-directed mutagenesis kit (Agilent Technologies) following the manufacturer’s instructions. Primers used are provided in Supplemental Table 1. Briefly, PCR conditions were initial denaturation: 95°C 2 min followed by 20 cycles of 95°C 15 s denaturation, 60°C 10 s annealing, and 68°C 4 min (30 s per kb) of extension step, with a final 68°C for 5 min. E. coli Top10 competent cells (Invitrogen™) were transformed with these plasmids and plated in LB agar with gentamicin selection and sent for sequencing to verify point mutation. Once the point mutation on each BED domain was confirmed, we proceeded to clone these constructs into a gateway plasmid 35S-eGFP-GWY (pB7FWG2 destination vector, https://gateway.psb.ugent.be) in order to transform Agrobacterium tumefaciens GV3101 cells for transient expression assays in Nicotiana benthamiana.
Plant Growth
Three-week-old rice plants were used for drought stress assays and inoculation with Magnaporthe oryzae Guy11 strain. Temperature was maintained between 29°C day and 21°C night with a 16 h light cycle. Plants were grown in Neuhaus S soil mixed with poudzolane (2 L/70 L). Standard fertilization solution containing nitrogen source (75% NO−3/25% NH+4; 40 mg/L) was supplied every Monday, and plants were inoculated one or two days after fertilization.
Four-week-old N. benthamiana plants were used for transient expression of ZBED using A. tumefaciens. Plants were grown in a growth chamber under 16 h light cycle and 22/20°C day/night temperatures. Plants were kept under these conditions during the course of the experiments.
Transient Expression Assays in Nicotiana benthamiana
Transient expression of recombinant proteins in N. benthamiana was performed as described previously with some modifications (Bos et al., 2006). Briefly, A. tumefaciens strains GV3101 were grown in LB medium at 28°C for 16 h and then centrifuged and resuspended in induction medium (10 mM MES (pH 5.6), 10 mM MgCl2, 150 mM acetosyringone) at an optical density at 600 nm (OD600) of 0.5 and incubated for 4 h at room temperature. Four-week-old N. benthamiana leaves (two per plant and five plants per biological rep, for a total of four biological reps) were infiltrated with Agrobacterium culture using a 1 ml needle-less syringe and evaluated for cell death phenotype at four and five days after infiltration (dai). For ZBED localization, samples were taken at two and three dai and observed under the confocal microscope.
Xanthomonas oryzae Inoculation and Scoring of Disease Symptoms
Four bacterial strains of X. oryzae pv oryzae (Xoo) PXO99A, MaiI, BAI3 and X11-5A(ptal6-TalC) and one strain of X. oryzae pv oryzicola (Xoc) BLS256, were kindly provided by Boris Szurek (IRD, Montpellier) for this study. All bacterial strains were grown in PSA medium (10 g of peptone, 10 g of sucrose, 1 g of glutamic acid, 16 g of agar per liter of H2O) for three days at 28°C. Bacterial cultures were suspended in sterile distilled H2O at an OD600 of 0.2 for Xoo leaf-clip inoculations (Kauffman et al., 1973) and 0.5 for infiltrations using a needle-less syringe for Xoc (Reimers and Leach, 1991). Three to four-week-old N-1 rice leaves from four plants per biological rep, with a total of four biological reps, were inoculated, and plants were kept in a phytotron (SANYO) under 12 h light cycle and 28/21°C day/night temperatures. Symptoms were evaluated two weeks after leaf-clip inoculation for Xoo by measuring lesion length and one week after Xoc infiltration by evaluating both water soaking phenotype and lesion length.
M. oryzae Inoculation and Scoring of Disease Symptoms
M. oryzae isolate Guy11 was cultured in rice agar for ten days (Berruyer et al., 2003). Spores were harvested in distilled water and filtered through two layers of nylon mesh. The spore concentration was adjusted to 30.000 spores/ml using a haemocytometer in 0.5% gelatin. Control plants were mock-inoculated with 0.5% gelatin. Inoculated and mock-inoculated plants (five per treatment) were transferred to an inoculation chamber at 25°C with 100% relative humidity for 16 h. Then plants were relocated in a phytotron 29°C day/21°C night in a 16 h light cycle. Disease symptoms were rated seven days after inoculation by measuring the percentage of susceptibility lesions, which are the number of susceptible lesions over the total number of lesions (susceptible plus resistant) multiplied by 100. The experiment was repeated four times.
DAB Staining to Detect Hydrogen Peroxide In Vivo in N. benthamiana
Four leaves of four-week-old N. benthamiana plants were agroinfiltrated with GFP-ZBED, GFP-ZBED mutants (BED point mutations) and control vector carrying GFP only. Leaves were cut 48 hai, and the petiole was dipped into diaminobenzidine (Sigma D-8001) 1 mg/ml of water and kept overnight in dark conditions at room temperature. Next day, tissue was cleared with ethanol/chloroform (4:1) overnight at room temperature. Once the tissue was cleared, the leaves were placed in 60% glycerol, and browning of cells was observed under the light microscope and quantified using automated phenotyping. Briefly, images are represented in multidimensional arrays containing pixel intensity values using EBImage (Pau et al., 2010), and with these results we then performed a T-test and considered significant differences for p-values < 0.05. The experiment was repeated three times for a total of four biological replications.
Preparation of Leaf Tissues for FRET-FLIM Analyses
A. tumefaciens-infiltrated N. benthamiana leaf disc samples (8-mm diameter, harvested 48 h post-infiltration) were vacuum-infiltrated in fixation solution (4% w/v) paraformaldehyde and 0.05 M (CH3)2AsO2Na and incubated for 20 min at 4°C. Samples were rinsed in TBS buffer (25 mM Tris-HCl (pH 7.5), 140 mM NaCl, 3 mM KCl) for 5 min. Permeabilization of fixed samples was performed by incubating in proteinase K buffer (50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1m MEDTA, 0.5% SDS, 200 mg/ml of proteinase K, Invitrogen) for 10 min at 37°C. Samples were rinsed in TBS buffer for 5 min. Nucleic acid staining was performed by vacuum infiltrating a 5 mM Sytox Orange (Invitrogen) solution (TBS buffer) and incubating samples for 30 min at room temperature in the dark. Fixed leaf discs were washed with and mounted on TBS buffer before observations on an inverted microscope (Eclipse TE2000E, Nikon). FRET-FLIM measurements were performed in at least 27 nuclei per construct (Table 1) as previously described (Le Roux et al., 2015).
RNA Extraction and RT-qPCR Analysis
RNA extraction was performed as described (Delteil et al., 2012). Quantitative RT-PCR was performed using LC 480 SYBR Green I Master Mix (Roche, Basel, Switzerland) and a LightCycler 480 instrument (Roche). The amount of rice RNA in each sample was normalized using actin (LOC_Os03g50890) as internal control. For N. benthamiana RNA in each sample was normalized using the housekeeping NbPP2A gene (Segonzac et al., 2011) as internal control. For each treatment we had four technical reps (each technical rep was the bulk of three leaf discs from three different plants); a total of four biological reps were done. Details of PCR primers are provided in Supplemental Table 1.
Drought-Stress Protocols
Greenhouse drought stress assay was done as previously described with small modifications. Rice genotypes were sown on day 1 in 60 g of soil per pot (for soil composition and greenhouse conditions, see above). Each replicate consisted of 10 plants grown in 7.5 cm × 7.5 cm × 6.5 cm (h × w × l) pots, and a total of five replicates per treatment per genotype. Plants were watered daily and fertilized on day 5, starting from the second week after sowing. At day 23 (10:00), drought stress was imposed by withholding water. Three days later (day 26; 10:00), drought-stressed plants were rehydrated prior to inoculation, and no fertilization was applied. The experiment was repeated twice for a total of three biological reps. This protocol was used to apply drought on greenhouse-grown plants shown in Figure 4B. Measurements in control plants indicated that this treatment significantly (T-test p<0,01) reduced Relative Water Content by ~15%, and published data indicate that drought-responsive genes were induced upon this treatment (Bidzinski et al., 2016).
For drought field experiments, a confined rain-out shelter field facility at CIAT (International Center for Tropical Agriculture, Cali Colombia) was used as previously described (Khong et al., 2015). Briefly, field trials were laid as a randomly complete block design with three replicates. Drought stress was imposed at panicle initiation (56 days after direct seeding) and continued until severe leaf rolling and wilting started in non-transgenic controls (three weeks approximately). Plants were re-watered until physiological maturity. Leaf rolling scores, plant height (centimeters), single-plant dry biomass (grams), and single-plant yield were recorded following the Standard Evaluation System for Rice (Gregorio et al., 1997). This protocol was used for drought measurements in field experiments shown in Figure 4B.
Salinity Stress Assay
Rice seeds were dehulled and surface-sterilized in 70% ethanol for 3 min. Ethanol was discarded, and seeds were submerged in 40% bleach for 30 min with shaking (discard bleach), then seeds were rinsed five times with sterile H2O under the hood. Ten surface-sterilized seeds per genotype were sown in one autoclaved magenta box containing 100 ml MS (Murashige & Skoog; Duchefa Biochemie) medium 4.3 g/l, with 8 g/l of agar amended with 120 mM NaCl (Electric conductivity 12dSm−1) for the salinity stress treatments or without NaCl for the controls (IRRI, 2014). Each genotype was sown under control MS and MS + NaCl in six independent experiments with similar results. The salt tolerant Pokkali accession which has the major QTL for salt tolerance Saltol was used as control (Thomson et al., 2010). Seed germination rates together with shoot and root length were measured three weeks after sowing. Root and shoot biomass were determined for three-week-old plants after drying in an oven at 65°C for one week.
Results
Identification of ZBED Protein Interactors
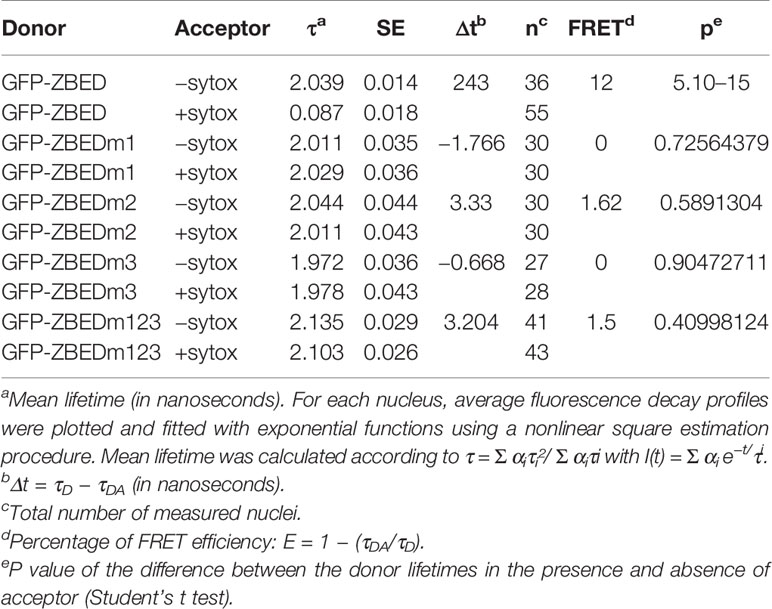
In order to gain a better understanding of ZBED’s molecular functions and predict the biological processes in which it could be involved, we performed a yeast two-hybrid (Y2H) screening to look for ZBED-interacting proteins in rice. We screened two rice libraries (Cesari et al., 2013; see Materials and Methods), using ZBED as bait. Among the various prey clones isolated, we identified KIP1 (LOC_Os01g07370), a kinase interacting protein predominantly expressed in pollen (Skirpan et al., 2001), STE11 (LOC_ Os01g50400), STE20/MAPK3 (LOC_Os12g0225; Jouannic et al., 1999) and WRKY4 (LOC_ Os03g55164), a transcriptional activator of rice defense responses against Rhizoctonia solani (Wang et al., 2015) (Figure 1A; ZBED auto-active controls Supplementary Figure 1). To confirm in planta the Y2H interacting data, we used a Bimolecular Fluorescent Complementation (BiFC) assay. A nuclear yellow-fluorescent protein (YFP) signal was detected when YFPc-ZBED was co-expressed either with YFPn-KIP1, YFPn-STE20 or YFPn-WRKY4 although it seems weaker than with KIP1 and STE20, but not with YFPn-STE11, confirming three of the four interactions detectable in yeast cells (Figure 1B).
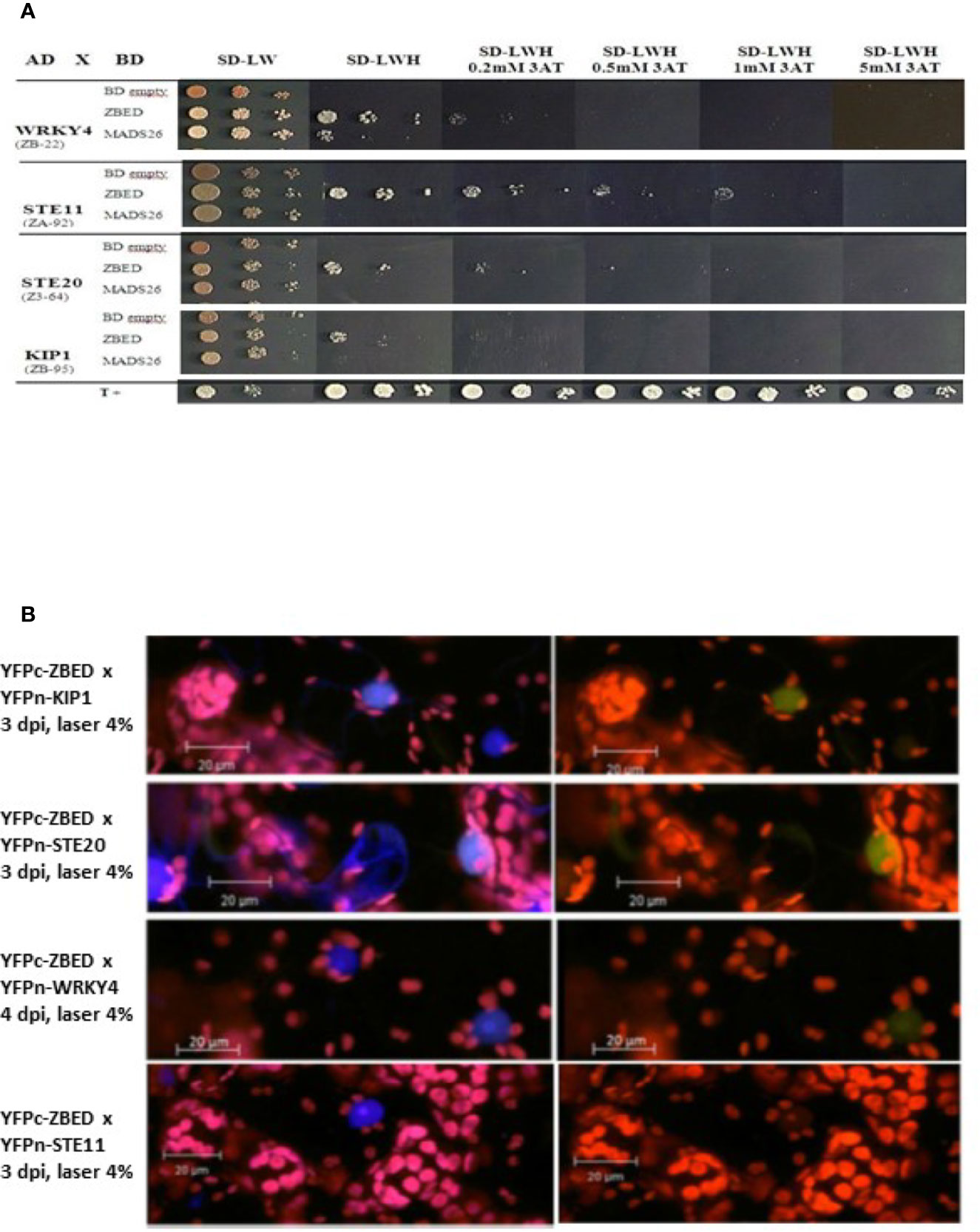
Figure 1 Identification of ZBED-interacting proteins. (A) Screening of two rice yeast two-hybrid cDNA libraries (CO39 and RCL1) with ZBED led to the identification of several putative interacting partners. WRKY4, STE11, STE20, and KIP1 were found to interact with ZBED in yeast cells. Yeasts co-expressing BD-ZBED with either AD-WRKY4, AD-STE11, AD-STE20, or AD-KIP1 were plated on selective media (SD-LWH) with different concentrations of 3AT. Empty pGBKT7-BD (BD-empty) was used as negative bait control. (B) Bimolecular Fluorescent Complementation (BiFC) assay was used to confirm yeast two-hybrid interactions (right panels). Transient co-expression of YFPc-ZBED either with YFPn-KIP1, YFPn-STE20, and YFPn-WRKY4, led to the detection of a nuclear fluorescent signal. Lack of YFP fluorescence upon co-expression of YFPc-ZBED with YFPn-STE11 suggests that these two proteins do not associate in planta. DAPI staining was used for nuclear staining (left panels). Observations were performed on N. benthamiana leaf samples collected three and four days post infiltration (dpi).
ZBED Localizes in the Plant Nucleus and Binds DNA
Since Zf-BED domains are hypothesized to bind DNA (Aravind, 2000), and considering our BiFC experiments (Figure 1B) indicating that ZBED is detected in the nucleus, we investigated the subcellular localization of ZBED and its putative DNA-binding ability. For this, ZBED was N-terminally fused to green fluorescent protein (GFP-ZBED) and transiently expressed in N. benthamiana. Laser-scanning confocal microscope analysis led to the detection of GFP fluorescence exclusively in plant nuclei indicating that ZBED is specifically targeted to this subcellular compartment (Figure 2A). A FRET-FLIM (fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy) assay (Le Roux et al., 2015) was performed to detect protein-nucleic acid interactions. GFP-ZBED was used as a donor fluorophore. A DNA-binding fluorescent dye (Sytox Orange) treatment performed on fixed plant material converts nuclear nucleic acids to FRET acceptors. In the absence of Sytox Orange treatment, an average GFP lifetime of 2.1 nanoseconds (ns) was measured (Figure 2B). A significant decrease of the GFP-ZBED lifetime was observed (1.8 ns), in Sytox orange-treated samples, indicating a close association of the GFP-ZBED partner (donor) and stained DNA (acceptor) due to FRET (Figure 2B). Because the cysteines and histidines are predicted to form a zinc-finger that binds DNA, each of the three BED domains were mutated separately by substituting their two conserved cysteines (which form zf) for two glycines to abolish DNA binding (Supplementary Figure 2). A single point mutation in any of the BED domains abolished the physical interaction between ZBED and DNA (Table 1), indicating that the integrity of each of ZBED’s zf-BED domains is strictly required for proper DNA-binding activity. The absence of DNA-binding in the zf-BED triple-mutant is depicted in Figure 2C. Together, our data indicate that ZBED localizes in the nucleus and binds DNA.
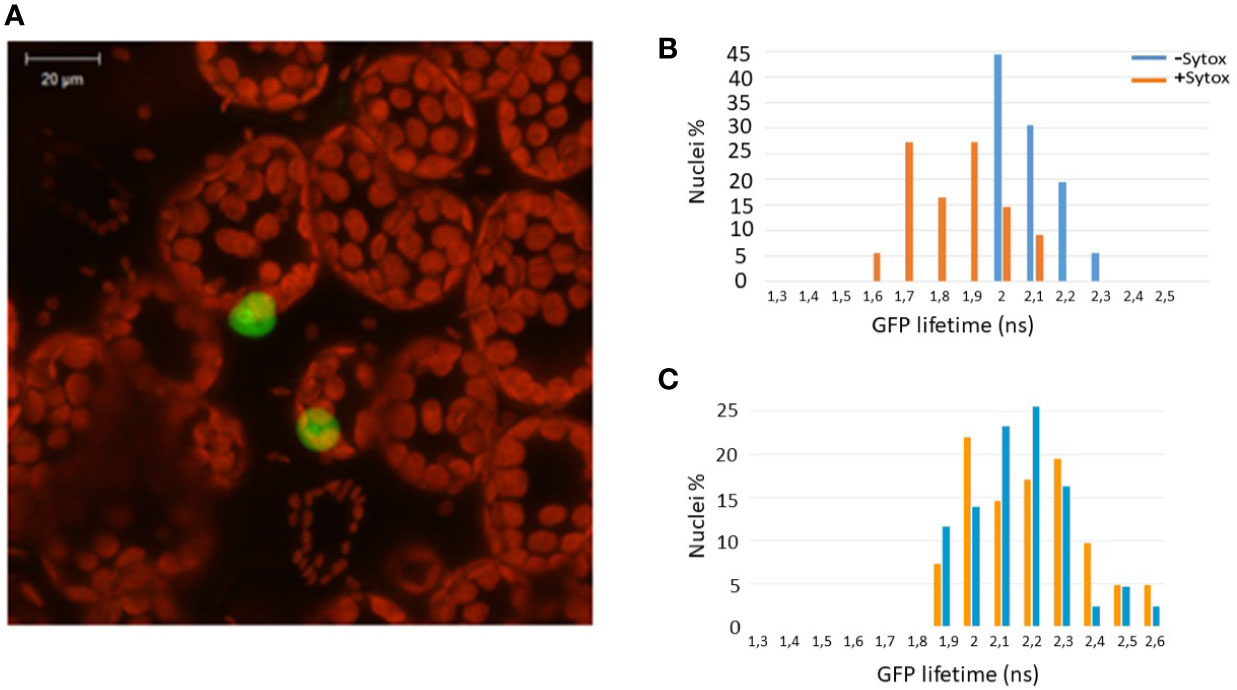
Figure 2 ZBED localizes to the nucleus and binds DNA. The GFP-ZBED fusion protein was transiently expressed in N. benthamiana. (A) GFP-ZBED accumulates in the plant nucleus; DNA binding using FRET/FLIM with or with Sytox (blue and orange bars respectively) with wild-type (B) and a mutated version of ZBED with point mutations of the cysteine to a glycine in each of three BED domains (C).
ZBED Induces Cell Death and Activates Basal Defense Responses in N. benthamiana
While using N. benthamiana as a heterologous system to determine ZBED localization, we noticed that after 4–5 days, there was a cell death phenotype caused by ZBED transient overexpression (Figure 3A), although this phenotype was not as strong as with the rice resistance protein RGA4 used as a positive control. This weak cell death was abolished with all ZBED-mutants, even when a single BED domain was mutated (Figure 3A shows phenotype with ZBED-triple mutant), suggesting that their DNA-binding activities are important for the cell death phenotype. To characterize the mechanisms by which ZBED was inducing cell death in N. benthamiana, we first checked whether ZBED was able to induce basal immunity, using four PTI-related marker genes: NbACRE31, NbGRAS2, NbPTI5, and NbWIPK (Hao et al., 2014). We measured their expression by RT-qPCR at three different time points (72, 96, and 120 h) after ZBED expression in N. benthamiana leaves. As shown in Figure 3B, ZBED overexpression enhanced the expression level of all four marker genes suggesting that it activates PTI, although to lower levels than with the RGA4 positive control. However, PTI induction was no longer detected with the ZBED-mutants, suggesting that this induction depends on the ability of ZBED to bind DNA (Figure 3B). Additionally, we explored whether ZBED was able to activate the production of reactive oxygen species (ROS), a well described readout of basal immunity. For this, N. benthamiana leaf samples expressing either GFP-ZBED, GFP-ZBED-mutant, or GFP alone were collected 48 h after Agrobacterium infiltrations and subjected to a DAB staining procedure. Transient expression of GFP-ZBED induced ROS while both the GFP-ZBED mutant and GFP did not (Figure 3C).
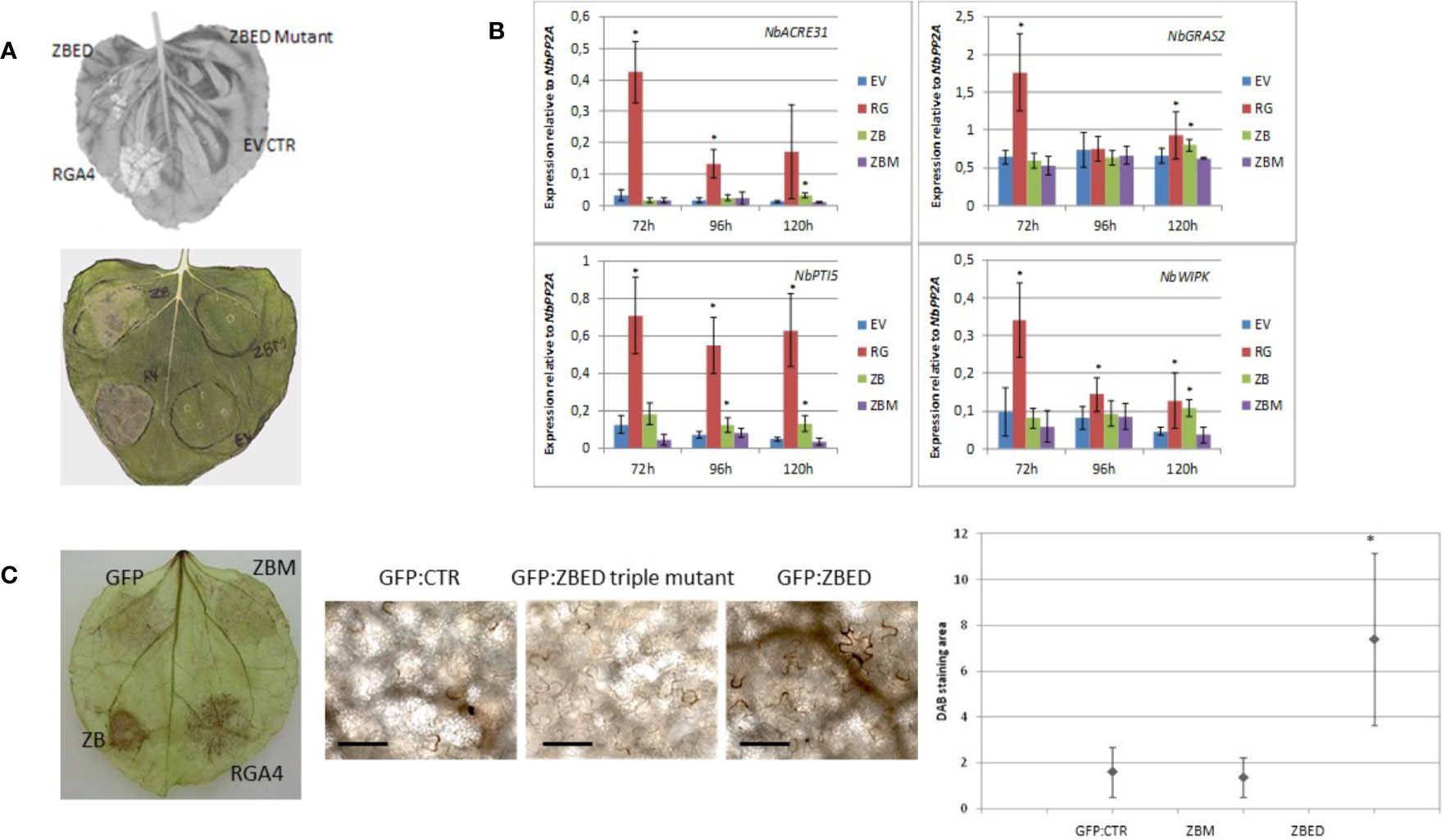
Figure 3 ZBED phenotypes related to basal immunity in N. benthamiana. (A) Transient expression of ZBED in N. benthamiana induces cell death. Transient expression of ZBED caused cell death four to five days after infiltration. ZBED mutants (ZBM) or GFP empty vector (EV) did not cause cell death. Positive control autoactive rice resistance gene RGA4 (R4-RGA4). (B) ZBED induces PAMP Triggered Immunity (PTI) in N. benthamiana as measured by RT-qPCR with four different PTI markers: NbACRE31, NbGRAS2, NbPTI5, and NbWIPK. ZBED triple mutant (ZBM) behaved like the GFP empty vector (EV) negative control at all time points and with all markers. Positive control autoactive rice resistance gene RGA4 (RG) (T-test *p < 0.05). (C) ZBED induced reactive oxygen species (ROS) measured with DAB staining, while ZBED mutant (ZBM) and GFP empty vector (EV) control did not. Quantification was done using EBImage (Pau et al., 2010; T-test *p < 0.05); scale bar is 100 µm.
Role of ZBED in Resistance to the Bacterial Pathogen Xanthomonas oryzae
Individual T2 plants (homozygous overexpressor and non-overexpressor–azygous) from three independent lines carrying single copy insertions were selfed, and ZBED overexpression was confirmed in the T3 progeny (see Material and Methods; Grand et al., 2012). ZBED overexpressor lines (ZBED-OX1, ZBED-OX2, and ZBED-OX3) were more resistant to M. oryzae (Kroj et al., 2016) than the non-overexpressor azygous lines (ZBED 1/0, ZBED 2/0 and ZBED 3/0 respectively). Our initial hypothesis postulated that ZBED could be involved in X. oryzae resistance (Zuluaga et al., 2017) functioning as a decoy for pathogen effectors, because the rice gene Xa1 has an integrated BED domain (Yoshimura et al., 1998; Zuluaga et al., 2017). We used the strongest ZBED overexpressor (ZBED-OX1) and its respective azygous line (ZBED 1/0) (Kroj et al., 2016) to test whether ZBED conferred resistance against different strains of this bacterial pathogen. Four different strains of the vascular pathogen X. oryzae pv oryzae (Xoo) (PXO99A, MaiI, BAI3, X11-5A (ptal6-TalC)) and one strain of X. oryzae pv oryzicola (Xoc) BLS256 were used. However, there was no effect on pathogenicity after inoculation of the ZBED-OX1 plants, compared to ZBED 1/0 plants, suggesting that ZBED-overexpressing lines do not show resistance to these bacterial pathogens (Supplementary Figure 3).
Rice ZBED Overexpressor Lines Are Resistant to Drought but Not to Salt Stress
Because of the frequent negative cross-talk between disease resistance and abiotic stress responses, we hypothesized that ZBED overexpressor (ZBED-OX) plants with enhanced resistance against M. oryzae (Kroj et al., 2016), might be less tolerant to abiotic stresses. Thus, we tested whether ZBED conferred susceptibility to drought and salinity stresses. Drought experiments were done in both the greenhouse and field (Figure 4A). Three independent transformation lines (ZBED-OX1, ZBED-OX2, and ZBED-OX3, collectively called ZBED-OX), together with their respective azygous lines (ZBED 1/0, ZBED 2/0, ZBED 3/0, collectively called AZY), were tested under controlled field conditions using a confined rain-out shelter field facility at CIAT (Centro Internacional de Agricultura Tropical, Colombia). ZBED-OX lines were significantly more resistant to drought-induced stress than the corresponding AZY lines as measured by different agronomical traits (Figure 4B). Single plant yield measured as grams of seeds produced per plant, was significantly higher in ZBED-OX compared to AZY lines under drought stress, although it must be noticed that drought-stressed plants had ten times less yield than well-watered plants, regardless of the genotype (Figure 4B). Under control conditions, single plant biomass measured as foliage dry weight, was higher for ZBED-1OX and 2OX compared to the azygous controls, except for 3OX plants. Drought had a more negative impact on AZY lines biomass thane on that of ZBED-OX lines, especially ZBED-3OX. As a result, biomass was significantly higher in all ZBED-OX lines compared to all AZY lines (Figure 4B) even though all genotypes (OX and AZY) showed an important decline (around ten times), in plant biomass after drought stress. Moreover, drought stress did not seem to have an impact on panicle length or number of productive tillers (t-test > 0.05; Supplementary Figure 4). To gain a better understanding on how ZBED was contributing to drought resistance, we further challenged ZBED-OX plants using a well-established drought protocol under controlled greenhouse conditions (Bidzinski et al., 2016). Greenhouse experiments showed that leaf rolling and wilting during the drought-stress peak were lower in ZBED-OX lines than in their corresponding AZY lines (Figure 4A). Because increased resistance to drought is often associated with salinity resistance due to the overlap of plant responses mechanisms to both stresses (Roychoudhury et al., 2013), we tested whether ZBED-OX lines conferred enhanced tolerance to salinity. To this end, we performed in-vitro assays by sowing seeds on MS medium supplemented with 120mM NaCl (corresponding to an electrical conductivity of dSm−1 12.2) which has been reported to cause a decrease in rice germination and seedling survival (Senguttuvel et al., 2013). Our data indicate that ZBED overexpression did not increase tolerance to salinity stress at the seedling stage (Supplementary Figure 5). Thus, plants overexpressing ZBED showed elevated resistance against M. oryzae, enhanced tolerance to a mild drought episode and did not develop any phenotype in response to salt at the seedling stage.
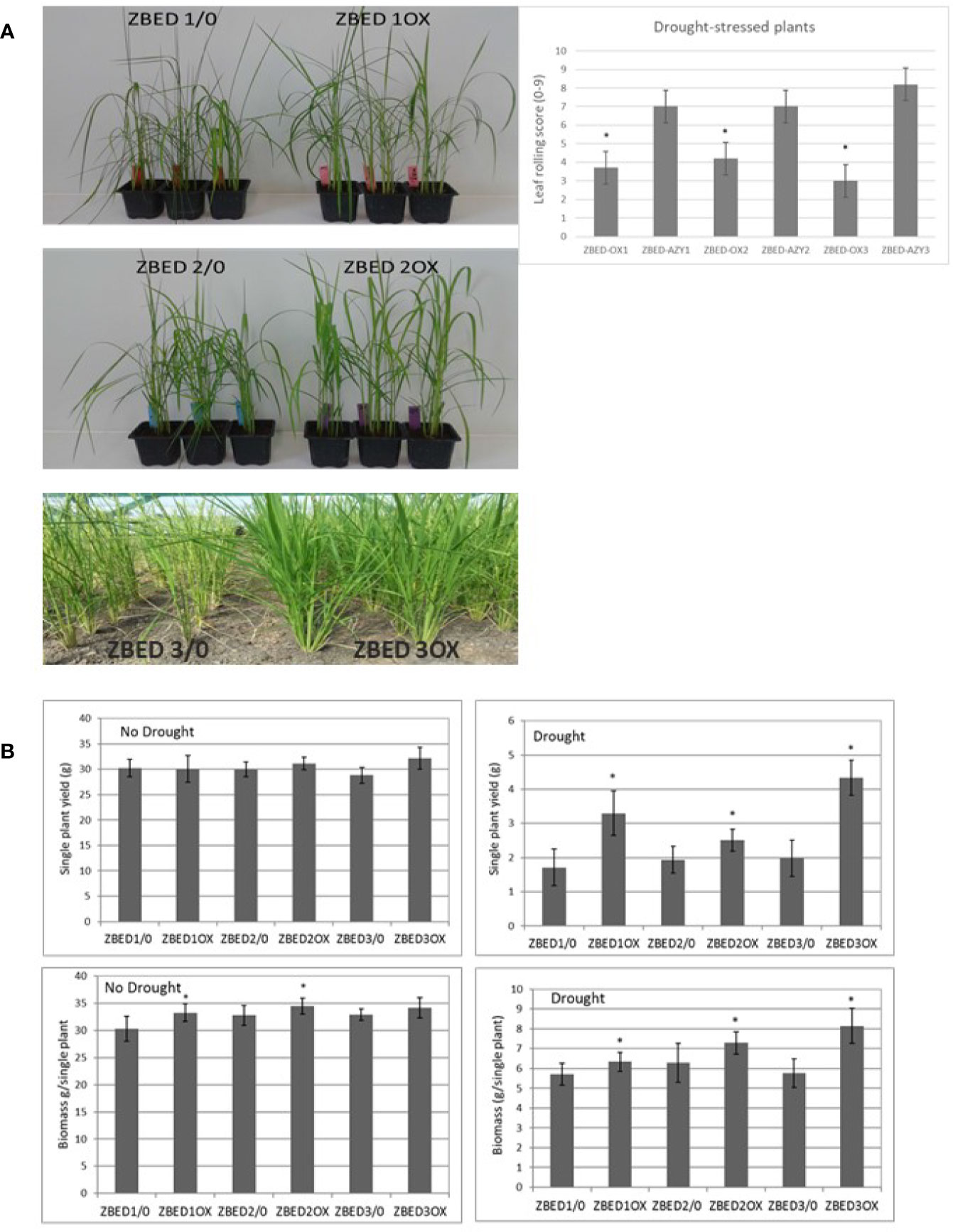
Figure 4 ZBED over-expression enhances drought tolerance. (A) Rice ZBED overexpressor lines are resistant to drought stress both in controlled (greenhouse) and non-controlled (Field, CIAT, Colombia) conditions; leaf-rolling score graph shows that ZBED-OX lines have less wilting symptoms than the azygous controls three days after water was withheld, using a scale from 0: non leaf rolling to 9: leaf completely wilted (n = 3; T-test *p < 0.05). (B) Agronomical traits in three independent ZBED overexpressor lines and their respective azygous lines under optimal water conditions and drought in the confined field rainout shelter facility (CIAT, Colombia). Single plant yield and single plant biomass are significantly higher in ZBED overexpressor lines under drought stress than in their respective azygous controls (T-test *p < 0.05).
ZBED Positively Regulates Stress-Responsive Genes
To gain a better understanding of the mechanism by which ZBED was conferring drought tolerance and disease resistance, we measured constitutive expression of stress-related markers under normal growth conditions (no inoculation and no drought stress). To this end, we performed RT-qPCR of six key regulatory genes of abiotic stress: OsDhn1, Oshox22, OsDREB2A, OsMAPK5, OsMYB4 and OsNAC6 (Xiong and Yang, 2003; Nakashima et al., 2007; Agarwal and Jha, 2010; Cui et al., 2011; Zhang et al., 2012; Kumar et al., 2014). We found that contrary to the AZY lines, ZBED-OX lines had significantly higher levels of dehydrin OsDhn1 and OsMAPK5 expression under control conditions (t-test p < 0.05; Figure 5A). Next, we quantified the expression level of five central regulators and marker genes of disease resistance: OsNPR1, OsPAL, OsPBZ1, OsPR1, and OsWRKY45 (Jwa et al., 2001; Yuan et al., 2007; Mitsuhara et al., 2008; Qiu and Yu, 2009; Tonnessen et al., 2014). We observed differential expression of components of the salicylic acid (SA) pathway; both the master regulator OsNPR1 (Yuan et al., 2007) and the OsPBZ1 gene had significantly higher expression in overexpressor plants compared to azygous controls (t-test p < 0.05; Figure 5B). Additionally, the OsPAL gene a key component of the phenylpropanoid pathway was strongly expressed in overexpressor plants compared to azygous controls. Thus, ZBED-OX plants showed elevated expression of both abiotic and biotic key regulatory genes expressed under control conditions, indicating that ZBED positively regulates some stress-responsive genes which might prime plants against both episodes of mild drought and M. oryzae.
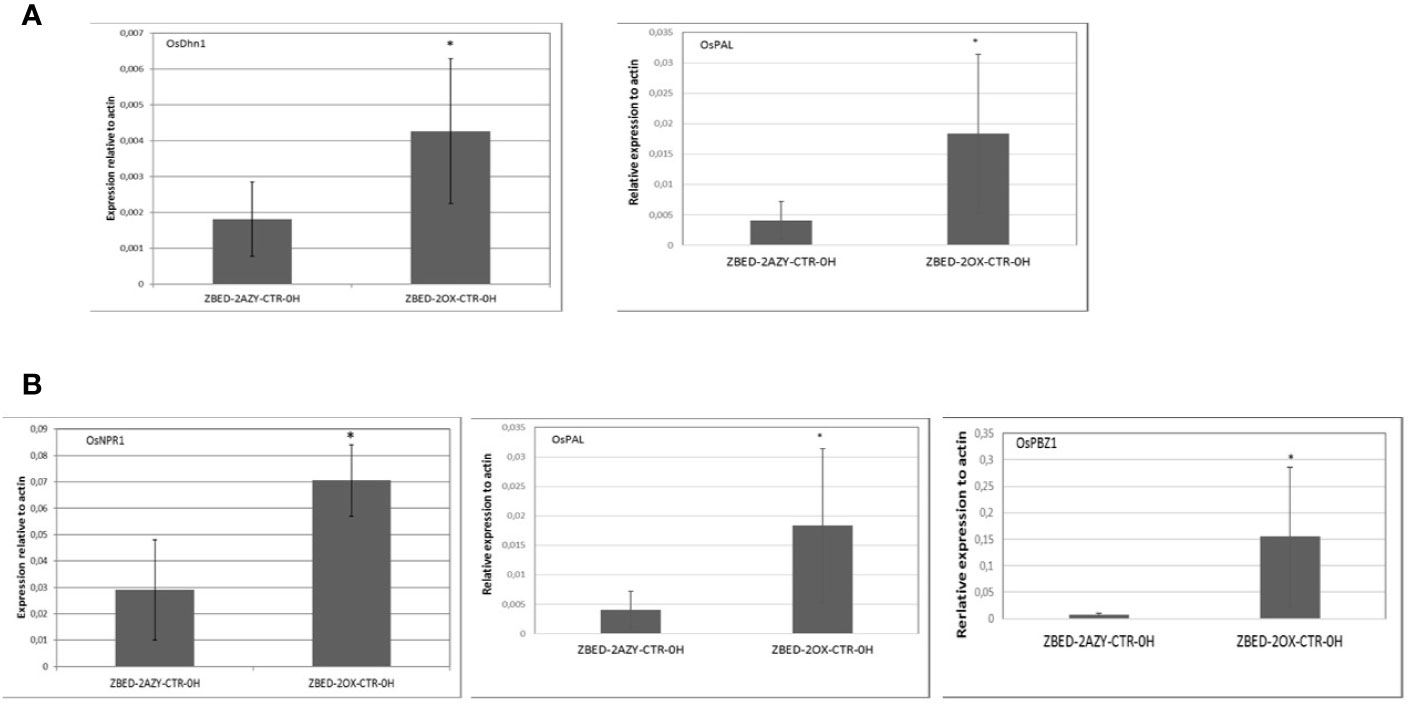
Figure 5 Constitutive expression of drought marker genes and disease-related marker genes in ZBED over-expressor. (A) Quantification of abiotic stress and (B) disease-related markers in one overexpressor line and its respective azygous, under control conditions (no inoculation/no drought). ZBED overexpressor (ZBED-OX) line showed a higher expression of OsDhn1, OsMAPK5, OsNPR1, OsPAL, and OsPBZ1 (T-test *p < 0.05; bars represent SD for four biological reps) than its azygous control (AZY).
Characterization of Resistance to M. oryzae After Drought Stress
We and others have demonstrated that rice plants are more susceptible to M. oryzae after a drought-stress episode, a phenomenon called drought-induced susceptibility (DIS) (Bonman, 1992; Bidzinski et al., 2016). Because ZBED-OX lines showed increased resistance against M. oryzae and tolerance to mild drought stresses occurring separately, we wanted to test whether ZBED-OX lines were able to confer resistance against the rice blast fungus after a mild drought-stress episode. For this, 22-day-old plants were submitted to a mild drought stress for three days. On the third day, plants were re-watered 4 h prior to inoculation at which point they no longer showed a drought-stress phenotype (leaf rolling or wilting). Plants were inoculated with M. oryzae and symptoms were scored seven days after inoculation. We observed that ZBED-OX genotypes remained significantly more resistant to M. oryzae than the corresponding AZY lines (ANOVA, Tukey HSD p < 0.05) despite the drought stress prior to inoculation, suggesting that mild drought did not interfere with ZBED-triggered resistance (Figures 6A, B).
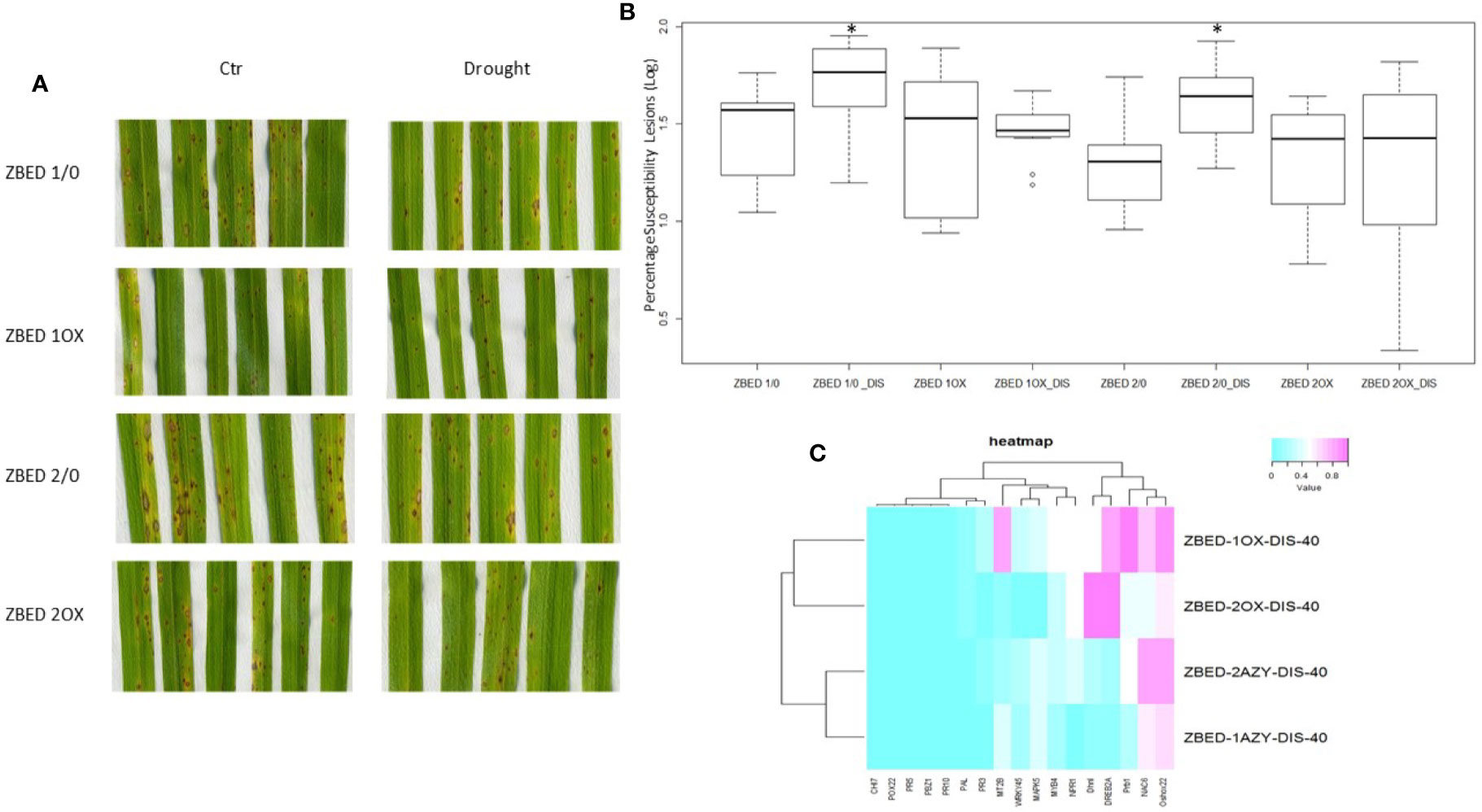
Figure 6 Disease resistance in ZBED-OX after drought stress. (A) Phenotypic evaluation of ZBED plants inoculated with M. oryzae after drought stress. ZBED-OX lines have smaller and less susceptible lesions after drought stress than the AZY lines. (B) ZBED overexpressor lines subjected to drought stress are significantly more resistant to M. oryzae than the azygous controls (ANOVA, Tukey HSD *p < 0.05). (C) Heatmap of marker genes for both abiotic and biotic stresses occurring simultaneously at 40 h after inoculation (hai). Except for OsNPR1, all the other biotic stress marker genes tested were not differentially expressed in ZBED overexpressor lines 1OX and 2OX compared to their respective azygous lines 1/0 and 2/0 under both drought and M. oryzae infection. However, the abiotic stress markers DREB2A and Dhn1 show a higher expression in ZBED-OX plants compared to AZY.
Leaf tissue was collected at 40, 64, and 88 h after inoculation (hai), and RT-qPCR analysis for both abiotic and biotic marker genes were done in order to test if the differential expression of these genes could explain pathogen resistance under drought in ZBED-OX plants. We observed that both ZBED-OX lines tested showed higher expression than azygous plants of the stress marker genes OsDhn1, OsDREB2A and OsNPR1 after a drought stress episode at early stages of the interaction (40 hai; Figure 6C). Additionally, there was a change in gene expression throughout infection, with the drought marker genes OsMAPK5, OsMYB4, and OsNAC6 expressed higher in OX lines compared to AZY lines at 64 and 88 hai (Supplementary Figures 6A–C). Conversely, the biotic stress marker genes that we studied were not differentially expressed between ZBED-OX and AZY lines throughout the interaction, suggesting that they do not contribute to the observed resistance phenotype (Supplementary Figures 6D–F).
Discussion
ZBED Molecular Function
The molecular function of BED domain remains elusive in plants. Only one report indicates that they bind DNA (Coupe and Deikman, 1997), and they were recently shown to direct proteins into the plasma membrane (Khong et al., 2015). Here we demonstrate that ZBED localizes to the nucleus and binds DNA in a heterologous system. ZBED DNA-binding activity seems to require the integrity of each of the three cysteine residues in the zinc-fingers of the BED domains of ZBED (Table 1). This observation is consistent with previously published data showing that BED domains bind DNA in Arabidopsis (Bundock and Hooykaas, 2005). Whether the ability of ZBED to bind DNA requires zinc ions (Zn2+), like for other Zn-Finger proteins, remains to be determined. In animals, BED proteins display various molecular functions, including transcriptional activation (Wang et al., 2013). In support of such function in rice, transient expression of wild-type ZBED in N. benthamiana induced the expression of several stress-related genes, whereas the triple-mutated ZBED did not (Figure 5). Transcriptional regulation activity of ZBED is consistent with the observation that ZBED interacts in the nucleus with WRKY4 (Figure 2) previously described as a transcriptional activator (Wang et al., 2015).
In animals, only a few reports identified protein partners of BED proteins (Chen et al., 2009). To date in plants, no protein partner has been described for a BED-containing protein. Using Y2H and BiFC assays, we found that ZBED interacts in the nucleus with the MAP kinase STE20 (Jouannic et al., 1999), the transcriptional regulator WRKY4 (Wang et al., 2015) and the kinase interacting protein KIP1. Interestingly, two thirds of all MAPK localize to both the cytosol and the nucleus where most of their substrates identified so far are transcription factors (Bigeard and Hirt, 2018). For instance, in rice the OsBWMK1 localizes predominantly to the cytosol, but is translocated into the nucleus within 1 to 12 h after treatment with H2O2 and SA (Koo et al., 2007). This ZBED-interacting protein network shades some light into its possible molecular function. One can speculate that putative ZBED transcriptional activity could be modulated by kinase activities and coordinated through interactions with additional partners including transcription factors like WRKY4.
Additionally, we determined that ZBED, when transiently expressed in N. benthamiana, induces components of basal defense such as PTI, ROS and cell death. Remarkably, ZBED overexpression in rice plants did not trigger cell death. Different explanations can account for this result. On the one hand, it suggests that rice, not N. benthamiana, has a negative regulator suppressing the cell death-inducing activities of ZBED. This regulator could be similar to the product of the rice disease resistance gene RGA4, used here as a positive control, that causes severe cell death when transiently expressed in N. benthamiana but induces no phenotype when overexpressed in rice (Cesari et al., 2014a). On the other hand, it might be an artefact of expressing ZBED in a heterologous system, where high amounts of protein could cause the cell death phenotype.
Role of ZBED in Resistance to Xanthomonas oryzae
Recently, it has been demonstrated that decoy proteins with a role in effector recognition can be integrated into plant immune receptors, giving rise to the integrated decoy model (Cesari et al., 2014b). In rice, a BED domain was found integrated with the Xa1 resistance protein, which confers resistance against Xanthomonas oryzae (Yoshimura et al., 1998). Thus, it was tempting to hypothesize that the BED domain of Xa1 acts as a decoy whose targeting by X. oryzae effectors would trigger resistance response activation against this pathogen in rice (Zuluaga et al., 2017). According to this model, ZBED with its BED domains could represent a virulence target for X. oryzae effectors. However, we did not observe any difference in pathogenicity after inoculation with Xoc or Xoo isolates between ZBED-OX and AZY lines (Supplementary Figure 3). Our results suggest that ZBED-OX lines do not show resistance to these bacterial pathogens, invalidating the model we recently proposed (Zuluaga et al., 2017). Consistent with this observation, three resistance genes necessary to confer resistance against a wide range of the yellow rust fungus, which is unrelated to Xanthomonas, have recently been shown to carry a BED domain in their N-terminus (Marchal et al., 2018). Thus, the BED domain found in resistance proteins may not act as a decoy but could rather function as a downstream signaling or localization module required for some resistance proteins to activate immune responses.
Rice ZBED-OX Lines Are Resistant to Rice Blast Even After Drought Stress
ZBED-OX plants showed increased drought tolerance after a mild drought as well as disease resistance. Thus, the ZBED gene represents one of the few cases where disease resistance and drought tolerance are simultaneously improved, similar to OsMADS26, MAPKK10.2, OsNAC6, OsCPK4, OsCPK10, WRKY11, or OsHAP2E (Nakashima et al., 2007; Campo et al., 2014; Alam et al., 2015; Khong et al., 2015; Bundó and Coca, 2016; Bundó and Coca, 2017; Ma et al., 2017; Lee et al., 2018). The mechanisms by which ZBED-OX increases disease resistance and drought tolerance were explored in this study. Several disease-related marker genes were highly induced in ZBED-OX plants under normal growth and in the absence of infection (Figure 5). For instance, expression of OsNPR1 encoding for a key regulator of SA-mediated resistance in rice (Yuan et al., 2007) was significantly increased in ZBED-OX. Likewise, OsPAL that encodes for a key enzyme in the SA biosynthesis and the phenylpropanoid pathway (Tonnessen et al., 2014) was significantly induced. Additionally, two executors of defense, OsPBZ1 and PRb1, were induced as well (Figure 5). Although gene expression was not measured in the field, it is noteworthy that yield was not impacted in ZBED-OX plants (Figure 4B) despite a possible constitutive overexpression of these stress-related genes.
After fungal infection, the expression of disease-related marker genes was not strongly different between ZBED-OX and AZY. This suggests that ZBED is modifying plant’s immunity before and not after infection, leading to a priming effect which could be sufficient to increase resistance (Vergne et al., 2010). Another possibility is that other disease-resistant pathways are induced after pathogen infection, but that we did not use the correct markers to detect them. With respect to drought-related markers measured, we observed that OsDhn1 and OsMAPK5 were constitutively expressed in well-watered, non-inoculated ZBED-OX plants (Figure 5). The OsDhn1 gene has been shown to protect rice plants against drought and salinity stress by scavenging ROS (Kumar et al., 2014). Quite surprisingly, the OsMAPK5 which overexpression induces salt, drought, and cold resistance but negatively modulates disease resistance against M. oryzae (Xiong and Yang, 2003), was also constitutively expressed in ZBED-OX lines, without an evident trade-off in disease resistance against rice blast. Thus, more gene expression studies will be required to fully understand how ZBED overexpression enhances both resistance to the fungal pathogen (but not bacterial) and tolerance to drought (but not salt or osmotic stress).
One last important result of our work is that we show that the disease resistance conferred by ZBED is still functional after drought stress, demonstrating that it is possible to build such robust resistance. This was unexpected because it was previously shown that drought stress slightly reduces the constitutive expression of defense genes and strongly diminishes its induction under infection (Bidzinski et al., 2016). It is important to highlight that this is one of the few studies where the outcome of two stresses applied sequentially was analyzed. Indeed, most of the effects of one given transgene on resistance/tolerance to different stresses have been done by applying and evaluating the outcome of each stress separately. Evaluating rice plants overexpressing OsMADS26, OsNAC6, OsCPK4, OsCPK10 among others, under both drought and pathogen infection occurring simultaneously or sequentially, will indicate whether ZBED behaviour is unique of its kind.
Conclusions
Using different approaches, we broaden the knowledge on the molecular function of BED domains in plants. Since these domains are now frequently found in plant resistance genes, our work will be of interest to groups working on the molecular function of resistance genes. Additionally, we demonstrate that overexpression of ZBED gene in rice, represents one of the few cases where disease resistance to the rice blast fungus is maintained even after a drought episode. This provides the first example to our knowledge of transgenic plants that display robust disease resistance under abiotic stress occurring simultaneously.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AZ performed experiments, analyzed data, and wrote the paper. PB, AD, EC, BC, MG, MI, LD, and CM performed experiments. TK, BS, and RK provided conceptual advice. J-BM conceived the project, analyzed data, and wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This publication has been written with the support of the AgreenSkills+ fellowship programme which has received funding from the EU’s Seventh Framework Programme under grant agreement N° FP7-609398 (AgreenSkills+) for PZ. This work was supported by an INRA project from the SPE division (Decoy) as well as the Agropolis and Cariplo Foundations under the reference « Rice Connections » 1201- 001 which is supported by the French ANR program “Investissement d’Avenir” ANR-10-LABX-0001-01.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to L. Fontaine for taking care of the plants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01265/full#supplementary-material
Supplementary Figure 1 | Yeast two-hybrid ZBED auto-activation tests. Yeasts co-expressing BD-ZBED with AD-empty were plated on selective media (SD-LWH) with different concentrations of 3AT to test for auto-activation. A SAP9 protein from another project was added as an auto-activation positive control.
Supplementary Figure 2 | Point mutations in BED domains. Two point-mutations were done in each of the BED domain to substitute the cysteine for glycine, to determine whether the BED domains where responsible for DNA-binding.
Supplementary Figure 3 | Determining if ZBED is a target for Xanthomonas oryzae effectors. ZBED azygous (ZBED 1/0) and ZBED overexpressor (ZBED 1OX) inoculated with Xanthomonas oryzae pv oryzae (Xoo) PXO99A (A), MaiI (B) and X. oryzae oryzicola (Xoc) BLS256 (D). There is no significant difference (T test p > 0.05) in pathogenicity between ZBED overexpressor lines vs azygous controls with any of the Xoo or Xoc tested.
Supplementary Figure 4 | Evaluation of agronomical traits in rice ZBED overexpressor vs ZBED azygous lines in the field (CIAT, Colombia). Number of productive tillers and panicle length are not significantly different ZBED overexpressor lines under drought stress than in their respective azygous controls (T test p > 0.05).
Supplementary Figure 5 | ZBEDOX and AZY seeds were germinated in MS media, supplemented with 120 mM NaCl to induce salt stress. Pokkali seedlings were used as the resistant control. Relative weight of salt stressed plants over control plants (MS media no-salt added) is shown for root biomass (left) and shoot biomass (right). There is no significant difference between overexpressors or azygous ZBED lines in salt resistance at seedling stage (T test p > 0.05).
Supplementary Figure 6 | Heatmap of marker genes for abiotic stress at 40 (A), 64 (B) and 88 (C) h after inoculation (hai) under drought stress (DIS) or no drought (CTR). At 40 hai, abiotic stress markers DREB2A and DhnI show a higher expression in OX plants compared to AZY, while at 64 hai MYB4 and MAPK5 are more expressed in ZBED-OX than ZBED-AZY independently of whether they are inoculated or not. Finally, at 88 hai the expression of MAPK5, NPR1, NAC6, and MYB4 is higher in ZBED-OX plants in both drought and control plants. Heatmap of marker genes for biotic stress at 40, 64, and 88 h after inoculation (hai) (D, E, and F respectively) under drought stress (DIS) or no drought (CTR). Biotic stress markers tested are not differentially expressed in ZBED overexpressor lines 1OX and 2OX compared to their respective azygous lines 1/0 and 2/0 under both drought and M. oryzae infection.
Abbreviations
PAMPs, pathogen-associated molecular patterns; PTI, PAMP-triggered immunity; ROS, reactive oxygen species; SA, salicylic acid; PR, pathogenesis-related genes; ABA, abscisic acid; JA, jasmonic acid; MAPK, mitogen-activated protein kinase; Xoo, Xanthomonas oryzae; OsCPK, calcium-dependent protein kinases; Y2H, yeast two-hybrid (Y2H); KIP, kinase interacting protein; BiFC, Bimolecular Fluorescent Complementation; YFP, yellow-fluorescent protein; GFP, green-fluorescent protein; FRET-FLIM, fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy; OX, Overexpressor; AZY, Azygous; DAI, days after infiltration; ZF, zinc-finger.
References
Agarwal, P. K., Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol. Plant 54, 201. doi: 10.1007/s10535-010-0038-7
Alam, M. M., Tanaka, T., Nakamura, H., Ichikawa, H., Kobayashi, K., Yaeno, T., et al. (2015). Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol. J. 13 (1), 85–96. doi: 10.1111/pbi.12239
Aravind, L. (2000). The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25, 421–423. doi: 10.1016/S0968-0004(00)01620-0
Berruyer, R., Adreit, H., Milazzo, J., Gaillard, S., Berger, A., Dioh, W., et al. (2003). Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 107, 1139–1147. doi: 10.1007/s00122-003-1349-2
Bidzinski, P., Ballini, E., Ducasse, A., Michel, C., Zuluaga, P., Genga, A., et al. (2016). Transcriptional Basis of Drought-Induced Susceptibility to the Rice Blast Fungus Magnaporthe oryzae. Front. Plant Sci. 7, 1558. doi: 10.3389/fpls.2016.01558
Bigeard, J., Hirt, H. (2018). Nuclear Signaling of Plant MAPKs. Front. Plant Sci. 9, 469. doi: 10.3389/fpls.2018.00469
Bonman, J. M. (1992). Durable resistance to rice blast disease-environmental influences. Euphytica. 63, 115–123. doi: 10.1007/BF00023917
Bos, J. I. B., Kanneganti, T. D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48 (2), 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Bundó, M., Coca, M. (2016). Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 14 (6), 1357–1367. doi: 10.1111/pbi.12500
Bundó, M., Coca, M. (2017). Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 68 (11), 2963–2975. doi: 10.1093/jxb/erx145
Bundock, P., Hooykaas, P. (2005). An Arabidopsis hAT-like transposase is essential for plant development. Nature. 436 (7048), 282–284. doi: 10.1038/nature03667
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., San Segundo, B. (2014). Overexpression of a Calcium-Dependent Protein Kinase Confers Salt and Drought Tolerance in Rice by Preventing Membrane Lipid Peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The Rice Resistance Protein Pair RGA4/RGA5 Recognizes the Magnaporthe oryzae Effectors AVR-Pia and AVR1-CO39 by Direct Binding. Plant Cell. 25 (4), 1463–1481. doi: 10.1105/tpc.112.107201
Cesari, S., Kanzaki, H., Fujiwara, T., Bernoux, M., Chalvon, V., Kawano, Y., et al. (2014a). The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33 (17), 1941–1959. doi: 10.15252/embj.201487923
Cesari, S., Bernoux, M., Moncuquet, P., Kroj, T., Dodds, P. N. (2014b). A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 25 (5), 606. doi: 10.3389/fpls.2014.00606
Chen, T., Li, M., Ding, Y., Zhang, L. S., Xi, Y., Pan, W. J., et al. (2009). Identification of zinc-finger BED domain-containing 3 (Zbed3) as a novel axin-interacting protein that activates Wnt/β-catenin signaling. J. Biol. Chem. 284 (11), 6683–6689. doi: 10.1074/jbc.M807753200
Corwin, J. A., Kliebenstein, D. J. (2017). Quantitative Resistance: More Than Just Perception of a Pathogen. Plant Cell. 29, 655–665. doi: 10.1105/tpc.16.00915
Coupe, S. A., Deikman, J. (1997). Characterization of a DNA-binding protein that interacts with 5’ flanking regions of two fruit-ripening genes. Plant J. 11, 1207–1218. doi: 10.1046/j.1365-313X.1997.11061207.x
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. doi: 10.1016/j.plaphy.2011.09.012
Delteil, A., Zhang, J., Lessard, P., Morel, J. B. (2010). Potential candidate genes for improving rice disease resistance. Rice. 3, 56–71. doi: 10.1007/s12284-009-9035-x
Delteil, A., Blein, M., Faivre-Rampant, O., Guellim, A., Estevan, J., Hirsch, J., et al. (2012). Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defence. Mol. Plant Pathol. 13 (1), 72–82. doi: 10.1111/j.1364-3703.2011.00731.x
Grand, X., Espinoza, R., Michel, C., Cros, S., Chalvon, V., Jacobs, J., et al. (2012). Identification of positive and negative regulators of disease resistance to rice blast fungus using constitutive gene expression patterns. Plant Biotechnol. J. 10, 840–850. doi: 10.1111/j.1467-7652.2012.00703.x
Gregorio, G. B., Senadhira, D., Mendoza, R. D. (1997). Screening rice for salinity tolerance. International Rice Research Institute Discussion paper series Number 22.
Hao, G., Pitino, M., Ding, F., Lin, H., Stover, E., Duan, Y. (2014). Induction of innate immune responses by flagellin from the intracellular bacterium, “Candidatus Liberibacter solanacearum.” BMC Plant Biol. 14:211. doi: 10.1186/s12870-014-0211-9
Hyun-Khang, C., Valent, B. (2010). “Magnaporthe oryzae and Rice Blast Disease,” in Cellular and Molecular Biology of Filamentous Fungi. Eds. Borkovich, K., Ebbole, D. (Washington, DC: ASM Press), p 593–p 606. doi: 10.1128/9781555816636.ch37
Inoue, T., Sternberg, P. W. (2010). C. elegans BED domain transcription factor BED-3 controls lineage-specific cell proliferation during organogenesis. Dev. Biol. 338 (2), 226–236. doi: 10.1016/j.ydbio.2009.12.005
IRRI (2014). Standard evaluation system for rice (SES). 5th edn (Los Banos, Philippines: International Rice Research Institute).
Jouannic, S., Hamal, A., Leprince, A. S., Tregear, J. W., Kreis, M., Henry, Y. (1999). Characterisation of novel plant genes encoding MEKK/STE11 and RAF-related protein kinases k. Gene 229 (1-2), 171–181. doi: 10.1016/s0378-1119(99)00012-8
Jwa, N. S., Kumar Agrawal, G., Rakwal, R., Park, C. H., Prasad Agrawal, V. (2001). Molecular cloning and characterization of a novel jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativa L.) seedling leaves. Biochem. Biophys. Res. Commun. 286 (5), 973–983. doi: 10.1006/bbrc.2001.5507
Kauffman, H., Reddy, A., Hsieh, S., Merca, S. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Khong, G. N., Pati, P. K., Richaud, F., Parizot, B., Bidzinski, P., Mai, C. D., et al. (2015). OsMADS26 Negatively Regulates Resistance to Pathogens and Drought Tolerance in Rice. Plant Physiol. 169 (4), 2935–2949. doi: 10.1104/pp.15.01192
Kissoudis, C., van de Wiel, C., Visser, R. G. F., van der Linden, G. (2014). Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular cross-talk. Front. Plant Sci. 5, 207. doi: 10.3389/fpls.2014.00207
Koo, S. C., Yoon, H. W., Kim, C. Y., Moon, B. C., Cheong, Y. H., Han, H. J., et al. (2007). Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochem. Biophys. Res. Commun. 360 (1), 188–193. doi: 10.1016/j.bbrc.2007.06.052
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X., Morel, J. B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210 (6), 618–626. doi: 10.1111/nph.13869
Kumar, M., Lee, S. C., Kim, J. Y., Kim, S. J., Aye, S. S., Kim, S. R. (2014). Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 57, 383–393. doi: 10.1007/s12374-014-0487-1
Lam, E., Kano-Murakami, Y., Gilmartin, P., Niner, B., Chua, N. (1990). A metal-dependent DNA-binding protein interacts with a constitutive element of a light-responsive promoter. Plant Cell 2, 857–866. doi: 10.1105/tpc.2.9.857
Le Roux, C., Huet, G., Jauneau, A., Camborde, L., Trémousaygue, D., Kraut, A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 165 (5), 1074–1088. doi: 10.1016/j.cell.2015.04.025
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H. S., Park, S. R., et al. (2018). Rice WRKY11 Plays a Role in Pathogen Defense and Drought Tolerance. Rice. 11, 5. doi: 10.1186/s12284-018-0199-0
Ma, H., Chen, J., Zhang, Z., Ma, L., Yang, Z., Zhang, Q., et al. (2017). MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 92 (4), 557–570. doi: 10.1111/tpj.13674
Marchal, C., Zhang, J., Zhang, P., Fenwick, P., Steuernagel, B., Adamski, N. M., et al. (2018). BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat. Plants. 4, 662–668. doi: 10.1038/s41477-018-0236-4
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics. 279 (4), 415–427. doi: 10.1007/s00438-008-0322-9
Nakashima, K., Tran, L. S. M., Nguyen, D. V., Fujita, M., Maruyama, M., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51 (4), 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Oladosu, Y., Rafii, M. Y., Samuel, C., Fatai, A., Magaji, U., Kareem, I., et al. (2019). Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 20 (14), 3519. doi: 10.3390/ijms20143519
Pau, G., Fuchs, F., Sklyar, O., Boutros, M., Huber, W. (2010). EBImage-an R package for image processing with applications to cellular phenotypes. Bioinformatics. 26 (7), 979–981. doi: 10.1093/bioinformatics/btq046
Qiu, Y., Yu, D. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65 (1), 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Quilis, J., Peñas, G., Messeguer, J., Brugidou, C., San Segundo, B. (2008). The Arabidopsis AtNPR1 Inversely Modulates Defense Responses Against Fungal, Bacterial, or Viral Pathogens While Conferring Hypersensitivity to Abiotic Stresses in Transgenic Rice. MPMI 21, 1215–1231. doi: 10.1094/MPMI-21-9-1215
Ramegowda, V., Senthil-Kumar, M. (2015). The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 176, 47–54. doi: 10.1016/j.jplph.2014.11.008
Read, A. C., Moscou, M. J., Zimin, A. V., Pertea, G., Meyer, R. S., Purugganan, M. D., et al. (2020). Genome assembly and characterization of a complex 1 zfBED-NLR gene-containing disease resistance locus in Carolina Gold Select rice with Nanopore sequencing. PLoS Genet. 16 (1), e1008571. doi: 10.1371/journal.pgen.1008571
Reimers, P. J., Leach, J. E. (1991). Race-specific resistance to Xanthomonas oryzae pv. oryzae conferred by bacterial blight resistance gene Xa-10 in rice (Oryza sativa) involves accumulation of a lignin-like substance in host tissues. Physiol. Mol. Plant Pathol. 38 (1), 39–55. doi: 10.1016/S0885-5765(05)80141-9
Roychoudhury, A., Paul, S., Basu, S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32 (7), 985–1006. doi: 10.1007/s00299-013-1414-5
Segonzac, C., Feike, D., Gimenez-Ibanez, S., Hann, D. R., Zipfel, C., Rathjen, J. P. (2011). Hierarchy and Roles of Pathogen-Associated Molecular Pattern-Induced Responses in Nicotiana benthamiana. Plant Physiol. 156 (2), 687–699. doi: 10.1104/pp.110.171249
Senguttuvel, P., Vijayalakshmi, C., Thiyagarajan, K., Sritharan, R., Geetha, S., KannanBapu, J. R., et al. (2013). Differential response of rice seedlings to salt stress in relation to antioxidant enzyme activity and membrane stability index. Arch. Agron. Soil Sci. 59 (10), 1359–1371. doi: 10.1080/03650340.2012.724170
Seo, Y. S., Chern, M., Bartley, L. E., Han, M., Jung, K. H., Lee, I., et al. (2011). Towards establishment of a rice stress response interactome. PloS Genet. 7 (4), e1002020. doi: 10.1371/journal.pgen.1002020
Skirpan, A. L., McCubbin, A. G., Ishimizu, T., Wang, X., Hu, Y., Dowd, P. E., et al. (2001). Isolation and Characterization of Kinase Interacting Protein 1, a Pollen Protein That Interacts with the Kinase Domain of PRK1, a Receptor-Like Kinase of Petunia 1. Plant Physiol. 126 (4), 1480–1492. doi: 10.1104/pp.126.4.1480
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203 (1), 32–34. doi: 10.1111/nph.12797
Thomson, M. J., de Ocampo, M., Egdane, J., Rahman, M. A., Godwin-Sajise, A., Adorada, D. L., et al. (2010). Characterizing the Saltol Quantitative Trait Locus for Salinity Tolerance in Rice. Rice 2010) 3, 148–160. doi: 10.1007/s12284-010-9053-8
Tonnessen, B. W., Manosalva, P., Lang, J. M., Baraoidan, M., Bordeos, A., Mauleon, R., et al. (2014). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9
Vergne, E., Grand, X., Ballini, E., Chalvon, V., Saindrenan, P., Tharreau, D., et al. (2010). Preformed expression of defense is a hallmark of partial resistance to rice blast fungal pathogen Magnaporthe oryzae. BMC Plant Biol. 10, 206. doi: 10.1186/1471-2229-10-206
Wang, X., Jiang, L., Wallerman, O., Engström, U., Ameur, A., Gupta, R. K., et al. (2013). Transcription factor ZBED6 affects gene expression, proliferation, and cell death in pancreatic beta cells. Proc. Natl. Acad. Sci. 110 (40), 15997–16002. doi: 10.1073/pnas.1303625110
Wang, H., Meng, J., Peng, X., Tang, X., Zhou, P., Xiang, J., et al. (2015). Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. doi: 10.1007/s11103-015-0360-8
Xiong, L., Yang, Y. (2003). Disease Resistance and Abiotic Stress Tolerance in Rice Are Inversely Modulated by an Abscisic Acid-Inducible Mitogen-Activated Protein Kinase. Plant Cell Online. 15 (3), 745–759. doi: 10.1105/tpc.008714
Xiong, H., Yu, J., Miao, J., Li, J., Zhang, H., Wang, X., et al. (2018). Natural Variation in OsLG3 Increases Drought Tolerance in Rice by Inducing ROS Scavenging. Plant Physiol. 178, 451–467. doi: 10.1104/pp.17.01492
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z. X., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. PNAS 95 (4), 1663–1668. doi: 10.1073/pnas.95.4.1663
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5 (2), 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zhang, H., Sonnewald, U. (2017). Differences and commonalities of plant responses to single and combined stresses. Plant J. 90 (5), 839–355. doi: 10.1111/tpj.13557
Zhang, S., Haider, I., Kohlen, W., Jiang, L., Bouwmeester, H., Meijer, A. H., et al. (2012). Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80 (6), 571–585. doi: 10.1007/s11103-012-9967-1
Keywords: Zn-finger BED domains, Magnaporthe oryzae, disease resistance, drought resistance, plant–pathogen interactions
Citation: Zuluaga AP, Bidzinski P, Chanclud E, Ducasse A, Cayrol B, Gomez Selvaraj M, Ishitani M, Jauneau A, Deslandes L, Kroj T, Michel C, Szurek B, Koebnik R and Morel J-B (2020) The Rice DNA-Binding Protein ZBED Controls Stress Regulators and Maintains Disease Resistance After a Mild Drought. Front. Plant Sci. 11:1265. doi: 10.3389/fpls.2020.01265
Received: 26 November 2019; Accepted: 31 July 2020;
Published: 18 August 2020.
Edited by:
Eric Ruelland, UMR7618 Institut d’écologie et des sciences de l’environnement de Paris (IEES), FranceReviewed by:
Tetiana Kalachova, Institute of Experimental Botany (ASCR), CzechiaAnne Repellin, Université Paris-Est Créteil Val de Marne, France
Copyright © 2020 Zuluaga, Bidzinski, Chanclud, Ducasse, Cayrol, Gomez Selvaraj, Ishitani, Jauneau, Deslandes, Kroj, Michel, Szurek, Koebnik and Morel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Benoit Morel, amVhbi1iZW5vaXQubW9yZWxAaW5yYS5mcg==
 A. Paola Zuluaga
A. Paola Zuluaga Przemyslaw Bidzinski
Przemyslaw Bidzinski Emilie Chanclud
Emilie Chanclud Aurelie Ducasse1
Aurelie Ducasse1 Michael Gomez Selvaraj
Michael Gomez Selvaraj Boris Szurek
Boris Szurek Ralf Koebnik
Ralf Koebnik