- 1Department of Sustainable Crop Production, DI.PRO.VE.S., Università Cattolica del Sacro Cuore, Piacenza, Italy
- 2Dipartimento di Scienze Agrarie e Ambientali - Produzione, Territorio e Agroenergia (DiSAA), Università degli Studi di Milano, Milano, Italy
Grapevine downy mildew caused by Plasmopara viticola is one of the most important diseases in vineyards. Oospores that overwinter in the leaf litter above the soil are the sole source of inoculum for primary infections of P. viticola; in addition to triggering the first infections in the season, the oospores in leaf litter also contribute to disease development during the season. In the current study, a quantitative polymerase chain reaction (qPCR) method that was previously developed to detect P. viticola DNA in fresh grapevine leaves was assessed for its ability to quantify P. viticola oospores in diseased, senescent grapevine leaves. The qPCR assay was specific to P. viticola and sensitive to decreasing amounts of both genomic DNA and numbers of P. viticola oospores used to generate qPCR standard curves. When the qPCR assay was compared to microscope counts of oospores in leaves with different levels of P. viticola infestation, a strong linear relationship (R2 = 0.70) was obtained between the numbers of P. viticola oospores per gram of leaves as determined by qPCR vs. microscopic observation. Unlike microscopic observation, the qPCR assay was able to detect significant differences between leaf samples with a low level of oospore infestation (25% infested leaves and 75% non-infested leaves) vs. samples without infestation, and this ability was not influenced by the weight of the leaf sample. The results indicate that the qPCR method is sensitive and provides reliable estimates of the number of P. viticola oospores in grapevine leaves. Additional research is needed to determine whether the qPCR method is useful for quantifying P. viticola oospores in grapevine leaf litter.
Introduction
Diseases represent a constant threat to grapevine production and may cause considerable yield and economic losses. Grapevine downy mildew (DM), which is caused by the oomycete Plasmopara viticola (Berk et Curt.) Berlese and de Toni, is an important disease in vineyards located in areas with frequent rain and planted with susceptible cultivars of Vitis vinifera (Lafon and Clerjeau, 1988).
P. viticola has dimorphic reproductive forms, i.e., sexual and asexual forms, which are responsible for primary and secondary DM infections, respectively. Oospores represent the sexual stage of P. viticola and are formed after the fusion of an antheridium and an oogonium in the affected leaf tissue from mid-summer to autumn (Burruano, 2000; Rossi et al., 2013). The formation of oospores does not require particular temperatures but seems to be favoured by dry conditions (which impede asexual sporulation) or by leaf senescence (Gessler et al., 2011). Oospores overwinter in the leaf litter above the soil surface (Kennelly et al., 2007; Rossi et al., 2013). During winter, oospores reach morphological maturity, i.e., the oospore wall becomes thick, the nuclei fuse, an ooplast is formed, and large lipid globules separate into smaller ones (Vercesi et al., 1999). Germination of morphologically mature oospores is prevented by dormancy (Galet, 1977; Rossi and Caffi, 2007), a process regulated by the environment, nutrient permeability, and endogenous inhibitors. When the dormancy is broken, oospores are considered physiologically mature and are able to germinate under favorable environmental conditions (Rossi and Caffi, 2007). Oospore germination ends with the formation of a macrosporangium containing zoospores (Galbiati and Longhin, 1984). The germination of oospores requires a minimum temperature of 12–13°C (the optimum is between 20 and 24°C) (Laviola et al., 1986; Gessler et al., 2011), and also requires the moistening of the leaf litter by rainfall or water flux from the atmosphere (Arens, 1929; Rossi et al., 2008a; Rossi et al., 2013). Dry conditions prolong the dormant period and may damage oospores if protracted (Arens, 1929; Gessler et al., 2011). Some oospores remain dormant but viable for an entire season or even for several years (Kennelly et al., 2007; Caffi et al., 2011).
Oospores are the sole source of inoculum for primary infections of P. viticola, and were long considered to play a role only in triggering the epidemic in the early grapevine season; the subsequent explosive increase of the disease was attributed to asexual multiplication and secondary infections (Blaeser and Weltzien, 1979; Lalancette et al., 1988; Lafon and Clerjeau, 1988). The use of DNA microsatellites, which enables the identification of genotypes causing single DM leaf lesions, showed that new P. viticola genotypes enter into the epidemic during most of the grape-growing season, indicating that oospores continue to germinate throughout the season, and that the primary inoculum not only triggers epidemics but contributes to their progress (Kump et al., 1998; Gobbin et al., 2003; Rumbou and Gessler, 2004; Gobbin et al., 2005; Gobbin et al., 2006). Oospores constitute a very large and diverse inoculum pool, leading to a pathogen population with high genotypic diversity. This diversity allows the pathogen to adapt to several microclimates and various host varieties (Rossi et al., 2013).
The detection of P. viticola oospores in grape leaf tissue has been mainly based on two methods: i) direct microscopic observations after leaf clearing and staining (Burruano, 2000; Díez-Navajas et al., 2007; Taylor, 2018) or after filtration of the leaf tissue (Vercesi et al., 2000; Toffolatti et al., 2007); and ii) indirect estimation via a “floating grape leaf disc” bioassay (Hill, 1998). In the latter assay, leaf fragments containing oospores are immersed in water in the presence of non-infested floating leaf discs so that the zoospores originating from the oospores swim to the stomata of the discs and cause infection, the severity of which is proportional to the oospore number. These methods have been used to study oospore formation and germination processes (Lehoczky, 1965; Vercesi et al., 2002; Vercesi et al., 2010), mating types (Wong et al., 2001; Taylor, 2018), fungicide resistance in oospores (Bissbort et al., 1997; Wong and Wilcox, 2001; Gisi et al., 2007; Toffolatti et al., 2007; Toffolatti et al., 2011; Toffolatti et al., 2018), and oospore production in partially resistant grapevine cultivars (Brown et al., 1999; Delbac et al., 2018). The methods have also been used to assess the effect of environmental conditions on oospore maturation and germination dynamics in vineyards (Hill, 2001; Pertot and Zulini, 2003; Toffolatti et al., 2004; Rossi et al., 2008a; Caffi et al., 2009).
No studies have been conducted with the goal of enumerating the oospores in leaf litter in order to assess of the P. viticola inoculum potential in a vineyard. A possible reason is that both microscopy and leaf disc bioassays are too expensive and time-consuming for assessing the high numbers of oospores (as many as several hundred per g of leaf residue) that can be present in the leaf litter. Molecular methods that have been used for the quantification of the inoculum in other pathosystems (Pavón et al., 2008; Li et al., 2010; Hussain et al., 2014; Van der Heyden et al., 2019) could be potentially used for P. viticola. Although polymerase chain reaction (PCR) and real-time PCR (Valsesia et al., 2005; Gindro et al., 2014) have been developed for the detection and quantification of P. viticola in fresh grape leaves, molecular methods have not been developed for the detection and quantification of oospores.
The objective of this study was to determine whether a quantitative real-time PCR (qPCR) assay previously developed for the detection of P. viticola DNA in fresh leaves (Valsesia et al., 2005) could be used to quantify P. viticola oospores in diseased, senescent grapevine leaves.
Materials and Methods
Plant and P. viticola Material
In 2018, P. viticola sporangia and oospores were collected from vine plots that had not been treated with fungicides and that were located in an experimental vineyard (cv. Merlot) on the campus of Università Cattolica del Sacro Cuore (UCSC) in Piacenza (Italy).
In late May of 2018, sporangia of P. viticola were collected from leaves showing typical DM lesions (oil spots) with fresh sporulation on the abaxial surface of the leaf blade. A sterile cotton swab was used to gently remove the sporangia, which were suspended in sterile-distilled water and examined with a light microscope (40× magnification). After the sporangial suspension was centrifugated at 14,000 rpm for 10 min, the aqueous phase was discarded, and the sporangia in the pellet were stored at −20°C until they were used for extraction of genomic DNA.
In late September of 2018, leaves showing typical DM mosaic-like symptoms were collected from vines in the experimental vineyard (cv. Merlot) (Figure 1A). Although still attached to the vines at the time of collection, the leaves were diseased and senescent and would have soon fallen to the soil surface. The leaf petioles were removed and the leaf blades were wrapped in moistened blotting paper to prevent desiccation, placed in polyethylene bags, and stored at 4°C. To confirm the presence of P. viticola oospores in the freshly collected leaves, leaf pieces (1–2 cm2) with typical mosaic-like lesions were immersed in an acetic acid-ethanol (1/3 v/v) solution overnight at room temperature. The bleached leaf pieces were rinsed in distilled water and examined by light microscopy at 10–20× magnification (Figures 1B, C). Oospore suspensions were obtained as described by Vercesi et al. (2000), with a few modifications. Leaf pieces (approximately 1–2 cm2) were cut from the leaf blades with mosaic-like symptoms, weighed with an analytical balance (RADWAG PS45000/C2. Radom, Poland), and then finely ground in a mortar containing 20 mL of sterile-distilled water. The homogenate was passed through a series of three nylon filters with pore sizes of 100, 75, and 45 μm by thorough washing with sterile-distilled water. The material retained on the 45-μm mesh filter, which was presumed to contain the oospores based on oospore size, was resuspended in 15 mL of sterile-distilled water. The collected P. viticola oospores were counted with a hemacytometer at 40× (Figure 1D), and the counts were expressed as numbers of oospores per milliliter of suspension and per gram of leaf piece. The latter weight refers to leaves wrapped in blotting paper, for which 1 g of wrapped leaf = 1.3 g of fresh leaf = 0.34 g of dry leaf (i.e., after drying at 65°C for 24 h).
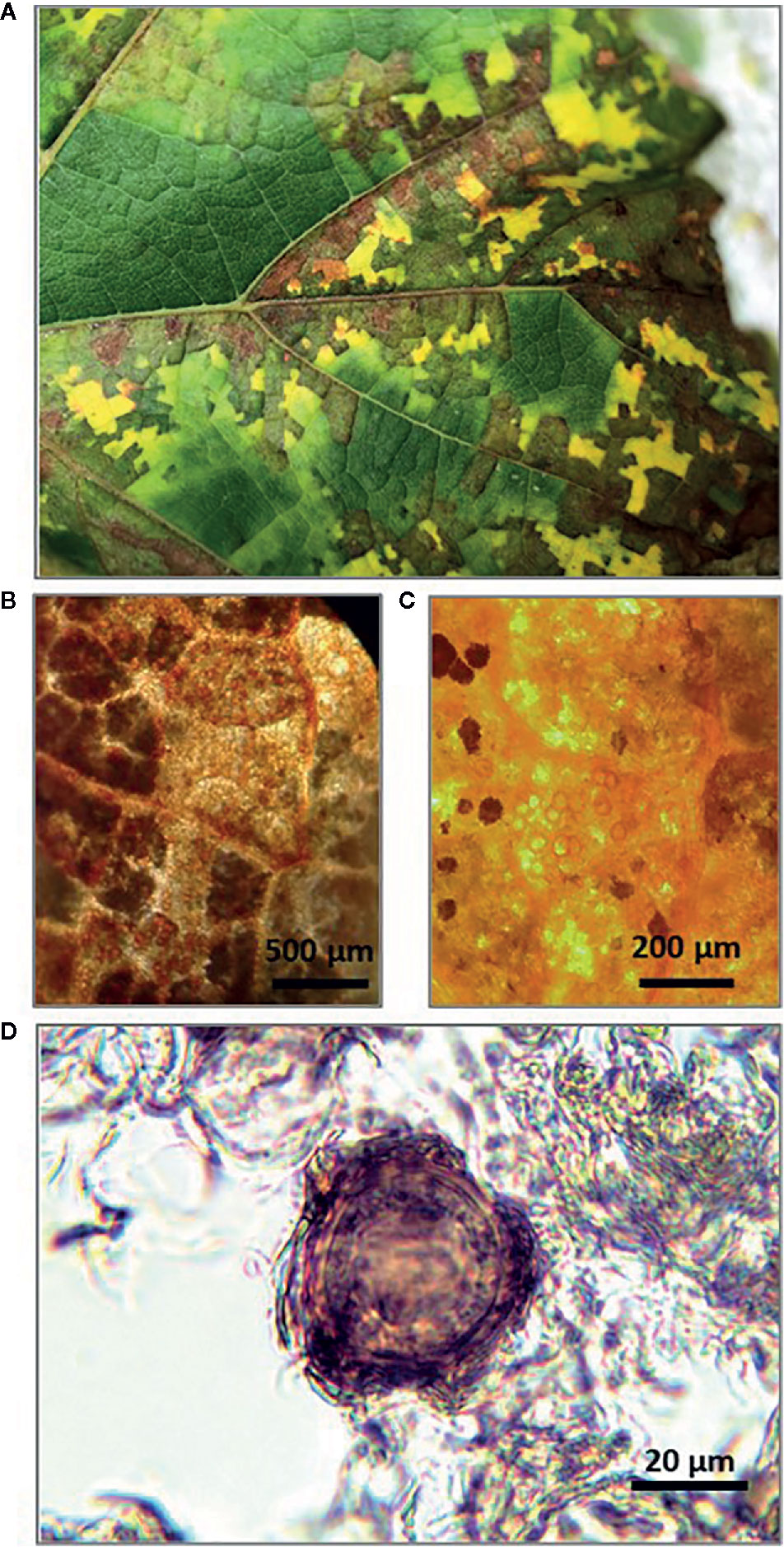
Figure 1 Senescent grape leaves showing typical downy mildew mosaic-like symptoms collected at the end of grapevine growing season (A); Stereomicroscope observation of Plasmopara viticola oospores inside bleached leaf pieces at 10× (B) and 20× (C); Light microscope observation of P. viticola oospore in suspension at 40× (D).
Non-infested, senescent leaves were also used, and these were collected from potted grape plants (cv. Merlot) that were growing in isolation in a greenhouse at the University campus.
Real-Time qPCR
DNA Extraction Method
Genomic DNA was obtained from P. viticola sporangia suspensions, oospore suspensions, and oospore-containing leaf pieces. The DNA was extracted as described by Toffolatti et al. (2007) with minor modifications in sample preparation; a 1-mL volume of each sporangia and oospore suspension, prepared as described above, was centrifuged at 14000 rpm for 10 min, and the aqueous phase was discarded. The pellets of sporangia and oospores as well as leaf pieces containing oospores (100 mg) were placed in 2-mL microcentrifuge tubes containing 500 µL of cetyl trimethylammonium bromide (CTAB) extraction buffer (2% CTAB, 100 mM Tris-HCl pH 8.0, 20 mM ethylenediaminetetraacetic acid [EDTA], 1.4 M NaCl, and 1% polyvinylpyrrolidone [PVP]), 100 mg of glass sand (425–600 µm diameter), and two glass beads (5 mm diameter). The sporangia, oospores, or leaf pieces in the tubes were then ground for 1 min at 30 cycles/s with a TissueLyser II (Qiagen, Milano). The tubes were subsequently placed in a heat block at 65°C for 90 min. DNA was purified with chloroform-isoamyl alcohol (24:1) (v:v), precipitated with isopropanol, and resuspended in 40 µL of sterile ultrapure water. The yield and purity of the extracted DNA were determined using a NanoPhotometer® N60 (Implen GmbH, München).
qPCR Assay
The qPCR assay was based on two specific primers and a hydrolysis probe (Giop) designed to target the internal transcribed spacer 1 (ITS 1) -5.8S rDNA of P. viticola (Valsesia et al., 2005), with the fluorescent reporter FAM (6-carboxyfluorescein) as a substitute for VIC reporter dye. The primer sequences were as follows: Giop F: 5′-TCC TGC AAT TCG CAT TAC GT-3′; Giop R: 5′-GGT TGC AGC TAA TGG ATT CCT A-3′; Giop P: 5′-6-FAM-TCG CAG TTC GCA GCG TTC TTC A-BHQ-1-3′. Reaction mixtures contained 1× Luna Universal Probe qPCR Master Mix (New England Biolabs, Ipswich), 250 nM of probe GiopP, 700 nM of each primer (GiopF/R), and 2 µL of DNA template in a final volume of 10 µL. An Applied Biosystems StepOnePlus™ System (Thermo Fisher Scientific Inc., Waltham) was used, and reaction conditions consisted of an initial incubation at 95°C for 1 min followed by 40 cycles of 95°C for 15 s and 60°C for 30 s.
Specificity
The specificity of the qPCR assay for P. viticola was determined in a test that included fungi and other oomycetes frequently found in grapevines (Table 1); these species were not previously assessed by Valsesia et al. (2005). The fungal strains were obtained from the culture collection of the Department of Sustainable Crop Production of the UCSC, Piacenza (Italy). Erysiphe necator Schwein. isolates were collected in the field in 2017 and 2018 and were maintained in the greenhouse on inoculated grape plants (cv. Merlot). Except in the case of P. viticola and E. necator, genomic DNA was obtained from 100 mg of fresh mycelium (obtained by scrapping the surface of 10-day-old colonies growing on potato dextrose agar, PDA). Genomic DNA was obtained from sporangial suspensions in the case of P. viticola and from leaf discs with sporulating powdery mildew colonies (100 mg of leaf material) in the case of E. necator. The DNA extraction method was previously described by Si Ammour et al. (2019).
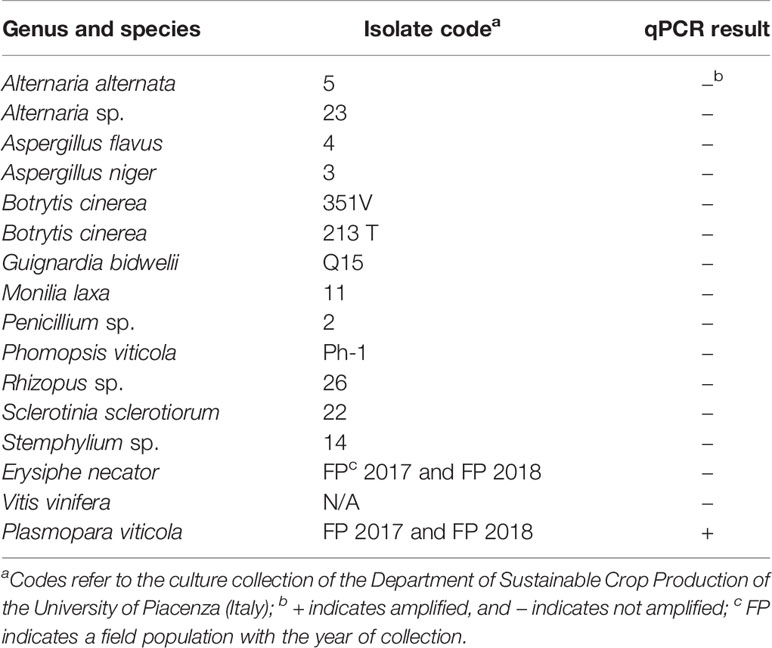
Table 1 List of fungi, oomycetes, and plants used for testing the specificity of a real-time qPCR assay targeting Plasmopara viticola DNA, and the corresponding results.
qPCR Standard Curves
The analytical sensitivity of the assay was assessed following an absolute quantification approach. Standard curves of the Giop assay were obtained using genomic DNA of P. viticola as template in a 10-fold dilution series (from 1 to 10–5 ng), and DNA from oospores suspensions as template in a 10-fold dilution series (from 12,000 to 1 oospore/mL, equivalent to 6,000 to 0.5 oospores/g of leaf sample). Real-time qPCR assays were carried out twice, and for each DNA template, each dilution was replicated three times. A water control was included in triplicate in each assay. Standard curves of the qPCR assay were produced by linear regression, and the coefficient of determination (R2) was calculated. The amplification efficiency (E) was determined from the slope of the standard curves (Bustin et al., 2009).
Microscope Counts vs. qPCR Assay
In experiment 1, five leaf samples with different levels of P. viticola infestation were prepared by mixing DM-infested and non-infested leaf pieces, described above, in the following proportions: 0 g infested/100 g non-infested (infestation 0%), 25/75 (25%), 50/50 (50%), 75/25 (75%), and 100/0. In experiment 2, leaf samples were prepared that consisted of 1, 2, or 3 g of DM-infested leaves.
Samples of both experiments were managed as described earlier, and numbers of oospores were determined by the Giop qPCR assay in three biological replicates in each assay and by microscope counts of oospore suspensions in four replicate samples for each infestation level in experiment 1 or in two replicate samples for each sample weight in experiment 2. Six replicate microscopic counts were carried out for each biological replicate sample in each experiment. The two experiments were performed twice.
Data Analysis
All statistical analyses were carried out using SPSS (version 24; IBM SPSS Statistics, IBM Corp., USA). An analysis of variance (ANOVA) was used to test differences among leaf samples in the two experiments. Before ANOVA, numbers of oospores per g of leaf were transformed using the decimal logarithm function to make variances homogeneous. A post-hoc comparison of means was conducted by using the Student-Newman-Keuls test at P<0.05.
Results
qPCR Specificity and Standard Curves
In the specificity test for P. viticola, the Giop probe/primer set did not amplify the purified DNA of non-target organisms but did amplify the purified DNA of P. viticola (Table 1). The Giop standard curve generated by a 10-fold serial dilution of P. viticola purified DNA obtained from sporangia suspensions was linear, with an efficiency of 110% and an R2 (the coefficient of determination) of 0.99 (Table 2, eq. 1). Similarly, the oospore standard curves generated by five-fold serial dilutions of extracted oospore DNA were linear, with an efficiency of 121% and an R2 of 0.99 (Table 2, eq. 2). The Giop assay was able to amplify the lowest tested concentration of P. viticola purified DNA, which was 10−5 ng, and a P. viticola oospore concentration as low as 1 oospore/mL (not shown). A third linear regression derived from the superposition of the two previous regressions was calculated for the relationship between DNA quantity (ng) and the number of oospores in a Cq range from 18 to 37, with an R2 of 0.99 (Table 2, eq. 3). Based on this equation, the oospore number increased linearly with the quantity of DNA in the leaf sample.
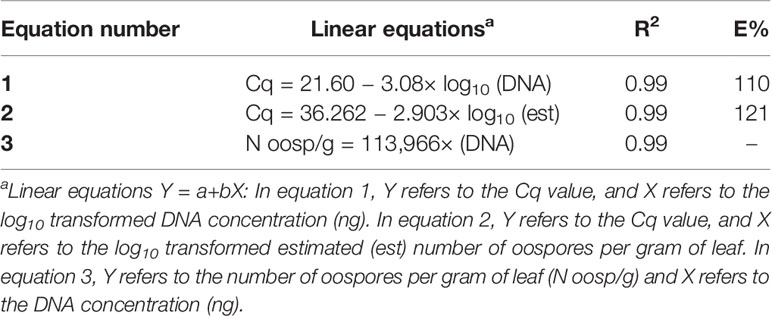
Table 2 Linear regressions, coefficients of determination (R2), and reaction efficiencies (E, %) for the qPCR standard curves used for the quantification of Plasmopara viticola oospores; Cq is the qPCR quantification cycle.
Microscope Counts vs. qPCR Assay
In experiment 1, oospore numbers were estimated by microscope counts and by the qPCR assay in samples with the following percentages of P. viticola-infested leaves: 0, 25, 50, 75, and 100%. The ANOVA revealed significant differences among infestation levels, but the qPCR assay provided more consistent differences than the microscope counts, as indicated by the post-hoc test (Table 3). For instance, the post-hoc test for microscope counts revealed no significant difference between the number of oospores in the 25% and 0% infection level, which was not the case for the qPCR assay. The standard errors for the oospore estimation were greater with microscope counts than with qPCR estimation, especially for the 25 and 50% infestation levels, i.e., the variability among replicates was greater for the microscope counts than for the qPCR estimates (Table 3).
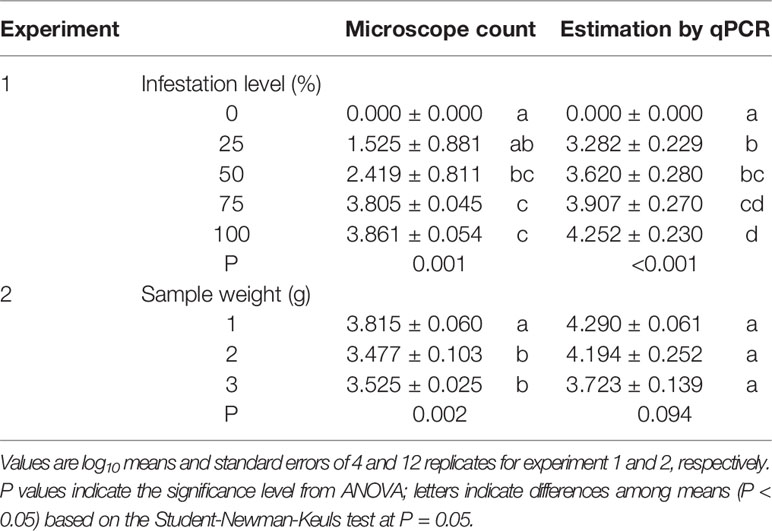
Table 3 Numbers of oospores per gram of leaf estimated using two methods (microscope count and qPCR) in grape leaves with different levels of infestation by Plasmopara viticola (experiment 1) and with different sample weights of infested leaves (experiment 2).
In experiment 2, the number of oospores per g of leaf was determined by microscope counts and by the qPCR assay in three sub-samples of different weights (1, 2, and 3 g); the sub-samples were obtained from the same infested leaf sample, with the expectation that the sub-sample weight would not influence the number of oospores detected per g of leaf. The ANOVA revealed significant differences between sample weights for microscope counts but not for qPCR (Table 3), showing that the former method was affected by the leaf sample weight while the latter was not.
There was a linear relationship between the number of P. viticola oospores per gram of leaf (log10 transformed) estimated by qPCR and enumerated with the microscope, with R2 = 0.70 (Figure 2, Table 4 eq. 1). In this linear equation, the intercept (a=−0.147 ± 0.362) and slope (b=1.157 ± 0.100) were not significantly different from zero (P=0.686) and one (P=0.134), respectively (Figure 2), indicating that the qPCR did not estimate the presence of oospores when there were no oospores in the leaf sample, and that the number of oospores detected by qPCR was equal to the number detected by microscopic observation. The relationship between the number of P. viticola oospores per gram of leaf (log10 transformed) estimated by qPCR and the Cq value is described by a logistic equation with a R2 of 0.87 (Table 4 eq. 2).
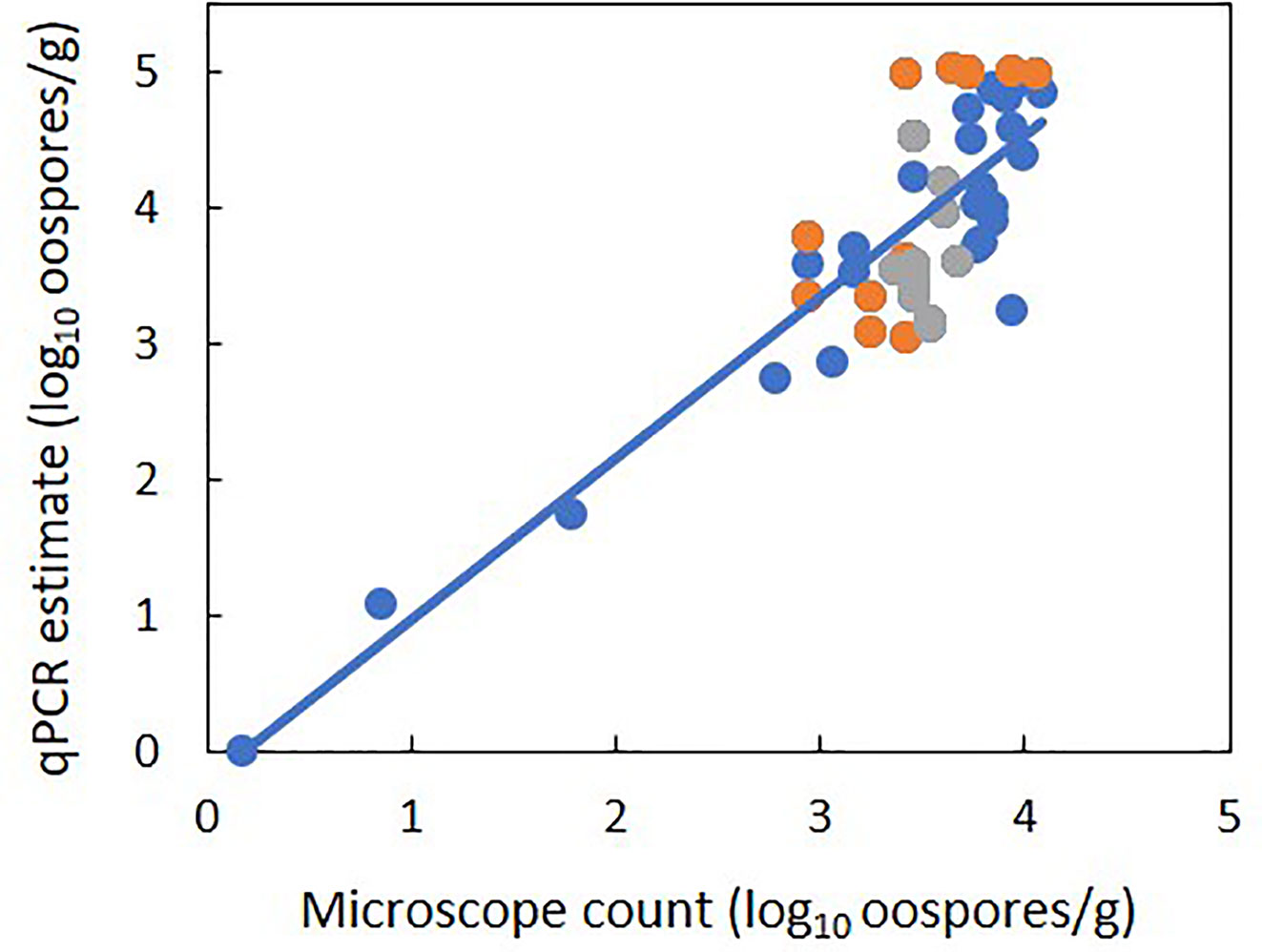
Figure 2 Relationship between estimates of the number of Plasmopara viticola oospores per gram of leaf (log10 transformed) based on the real-time qPCR assay and on microscope counts. Blue and orange dots represent mean values from experiment 1 and 2, respectively. Experiment 1 had five levels of P. viticola infested leaves (0, 25, 50, 75, and 100%) with a constant sample weight of 1 g. Experiment 2 had three samples weights of infested leaves: 1, 2, and 3 g. The line represents linear regression 1 in Table 4.
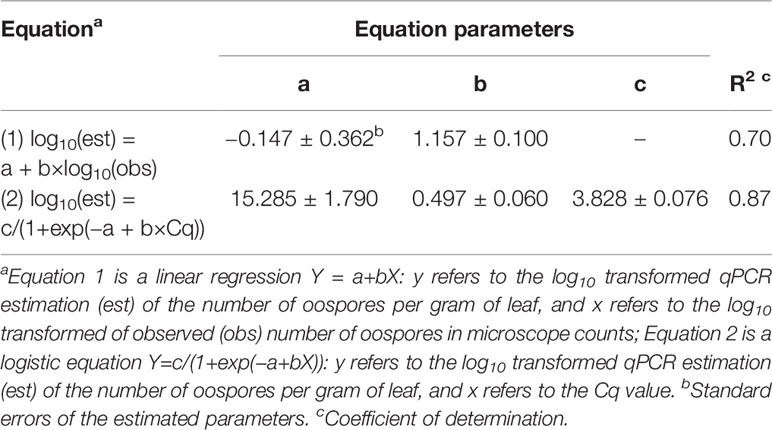
Table 4 Parameters and statistics of the regression equations used for fitting the relationships between the qPCR estimation of the number of Plasmopara viticola oospores per gram of leaf (dependent variable Y) and 1) the microscope counts of oospores (independent variable X) and 2) quantification cycle (Cq, independent variable X).
Discussion
In 2005, Valsesia et al. (2005) described a qPCR assay for the detection of P. viticola in fresh grape leaves. The current study assessed the suitability of that assay (with minor modifications) for the quantification of pathogen’s overwintering inoculum, i.e., oospores in senescent grapevine leaves. One expectation in developing a detection method is that the new method performs better than or at least as well as an existing one. In our experiments, the qPCR assay was therefore compared with microscope counts of oospores. Our results showed that the two methods are comparable in that there was a strong linear relationship between the number of P. viticola oospores per gram of grapevine leaves estimated by qPCR and enumerated with the microscope. Furthermore, unlike microscope counts, the qPCR assay could detect significant differences among samples with low vs. no oospore infestation (25 vs. 0% infestation level), and was not influenced by the leaf sample weight, indicating that the qPCR method provides a more sensitive and reliable estimate of the number of oospores than microscope counts. In the comparison of the two methods (qPCR and microscope counts), it is also important to note that the microscopic counting of oospores in leaf tissue or in suspensions requires careful observation by experts. In our experiments, for example, the P. viticola oospores in suspensions could be easily confused with similar structures visible at the same magnification (e.g., spores of soilborne pathogens and saprophytes). Moreover, despite the thorough and time-consuming filtration process used to prepare oospore suspensions, oospores could remain trapped in microscopic fragments of leaf tissue, making observation and identification difficult. The qPCR assay, in contrast, provides less subjective estimates of P. viticola oospore numbers. These results, which need to be confirmed with a range of field samples, show that the qPCR assay provides accurate and consistent results.
It can be argued that the qPCR estimation is based on the quantification of total genomic DNA of P. viticola in leaf samples, which may include the oospores but also the endophytic mycelium that generated the oospores (Lafon and Clerjeau, 1988; Gobbin et al., 2006; Rumbou and Gessler, 2006), which would result in an overestimation of the number of oospores. The amount of mycelium in the leaf tissue, however, should be proportional to the number of oospores because the oospores form following crosses of antheridia and oogonia produced by the mycelium itself (Wong et al., 2001; Scherer and Gisi, 2006); therefore, if an overestimation exists, it should be proportional to the oospore numbers. Consistent with the latter assumption, the results of the present work revealed a direct, linear relationship between the quantity of P. viticola DNA detected by qPCR and microscope counts in senescent grapevine leaves that would soon drop to the soil surface and become leaf litter.
Both methods (qPCR and microscope counts) fail to distinguish between living and dead oospores, and that could bias the assessment of the inoculum potential in a vineyard. Long-term survival of oospores is influenced by several factors. For instance, exposure to high temperatures (40 to 53°C) for 1 to 24 h resulted in the death of oospores of Phytophthora capsici (Etxeberria et al., 2011) and the inhibition of the germination of oospores of P. kernoviae and P. infestans (Fay and Fry, 1997; Widmer, 2011). Trichoderma asperellum, a common soil-borne fungus, can penetrate oospores of P. capsici, develop hyphae, and produce conidia leading to the disintegration of oospores (Jiang et al., 2016). Other microorganisms, including the biocontrol agents Bacillus subtilis and Trichoderma hartianum T39, can prevent the germination of Plasmopara viticola oospores (Vecchione et al., 2005; Dagostin et al., 2006). Glucosinolates and their degradation products, which are generated when Brassica plants are incorporated into soil as green manure, also prevented the germination of Pythium irregulare oospores (Manici et al., 2000) and significantly reduced the viability of oospores of P. capsici when used in combination with solarization (Lacasa et al., 2015). Therefore, the quantification of total oospores (viable and non-viable) may overestimate the inoculum potential in a vineyard. Methods to differentiate between dead and viable cells by quantifying only DNA from viable cells have been developed (Fittipaldi et al., 2012); when propidium monoazide (PMA) is used with qPCR, for example, the PMA enters non-viable cells, binds to DNA, and inhibits DNA amplification during PCR (Nocker et al., 2007). A method that used PMA-qPCR to quantify the viable resting spores of Plasmodiophora brassicae in soil was developed by Al-Daoud et al. (2017). This method was subsequently used to demonstrate that a large proportion of the DNA of P. brassicae detected in soil was derived from non-viable or immature resting spores (Gossen et al., 2019). Adaptation of the method of Al-Daoud et al. (2017) for P. viticola would probably result in a more accurate and reliable estimation of the inoculum density, and this warrants further study.
In this work, the qPCR assay was used to detect the oospores in grapevine leaves, but oospores in grapevine leaves represent only part of the total oospores present in a vineyard. Once the leaf litter decomposes, the oospores are incorporated into the soil. Although these gradually die, some remain viable for at least 65 months (Caffi et al., 2011). The oospore population in a vineyard is therefore composed of oospores in the leaf litter and in the soil. There is no information about the epidemiological role of the oospores in the soil and their ability to produce zoospores that can be splash-transported from soil to grape leaves, but it would be useful to determine whether the qPCR assay can be used to estimate oospore numbers in vineyard soil samples. In this regard, researchers have recently developed techniques that overcome some of the problems in using qPCR to quantify pathogen inoculum in soil (Pavón et al., 2008; Schena et al., 2013; Hussain et al., 2014; Gossen et al., 2019; Van der Heyden et al., 2019). Another limitation of this research is that it was conducted with diseased senescent leaves that would have soon fallen to the soil surface and become litter rather than with leaf litter. It follows that the qPCR method and microscopic counts should now be compared for determination of oospore numbers in grapevine leaf litter.
The qPCR assay for P. viticola oospores could be useful for managing DM in vineyards. Such management is currently based on the assumption that the potential for severe disease outbreak is always present, even when the disease was not severe in the previous season. With this assumption, vineyard managers tend to apply fungicides whether they are needed or not. Managers would be less likely to apply unneeded fungicides if they knew the oospore inoculum levels (as indicated by the qPCR assay) in their vineyards and how those levels related to DM severity. The modification of disease management according to inoculum level has occurred with other pathosystems. Apple scab, for example, can be effectively controlled by delaying fungicide applications based on estimates of the potential ascospore numbers of Venturia inaequalis in an orchard (MacHardy et al., 1993). Information on the P. viticola oospore numbers in vineyards could also be used for adapting the mathematical models for predicting P. viticola oospore dynamics and primary infections (Caffi et al., 2007; Rossi et al., 2008b; Caffi et al., 2009). Further study could be performed in order to establish the relationship between oospore density in vineyard and DM severity.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
VR, MS, and ST mainly contributed to the conception and the design of the study. MS and FB carried out the experiments. VR, MS, and FB contributed to the analysis of results. MS and FB wrote the first draft of the manuscript. VR and ST contributed to the critical analysis of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Financial support for carrying out this research was partially provided by the Project BIOVINE supported by transnational funding Bodies, being partners of the H2020 ERA-net project "CORE Organic Cofund", and the cofund from the European Commission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Luca Languasco for technical assistance. FB was supported by the Ph.D. in Agro-Food System (Agrisystem) of the Università Cattolica del Sacro Cuore (Italy).
References
Al-Daoud, F., Gossen, B. D., Robson, J., McDonald, M. R. (2017). Propidium monoazide improves quantification of resting spores of Plasmodiophora brassicae with qPCR. Plant Dis. 101 (3), 442–447. doi: 10.1094/PDIS-05-16-0715-RE
Arens, K. (1929). Untersuchungen über Keimung und Zytologie der oosporen von Plasmopara viticola (Berl, et de Toni). Jahrbuch für Wissenschaftliche Botanik 70, 57–92.
Bissbort, S., Albert, G., Schlösser, E. (1997). Effects of dimethomorph on the oospore formation of Plasmopara viticola. J. Plant Dis. Prot. 104 (2), 126–132.
Blaeser, M., Weltzien, H. C. (1979). Epidemiologische Studien an Plasmopara viticola zur Verbesserung der Spritzterminbestimmung/Epidemiological studies to improve the control of grapevine downy mildew (Plasmopara viticola). Z. für Pflanzenkrankheiten und Pflanzenschutz/Journal Plant Dis. Prot. 86 (8), 489–498.
Brown, M. V., Moore, J. N., Fenn, P., McNew, R. W. (1999). Comparison of leaf disk, greenhouse, and field screening procedures for evaluation of grape seedlings for downy mildew resistance. HortScience 34 (2), 331–333. doi: 10.21273/HORTSCI.34.2.331
Burruano, S. (2000). The life-cycle of Plasmopara viticola, cause of downy mildew of vine. Mycologist 14 (4), 179–182. doi: 10.1016/S0269-915X(00)80040-3
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi: 10.1373/clinchem.2008.112797
Caffi, T., Rossi, V., Cossu, A., Fronteddu, F. (2007). Empirical vs. mechanistic models for primary infections of Plasmopara viticola Vol. 37 (Oxford, UK: Blackwell Publishing Ltd), 261–271.
Caffi, T., Rossi, V., Bugiani, R., Spanna, F., Flamini, L., Cossu, A., et al. (2009). A model predicting primary infections of Plasmopara viticola in different grapevine-growing areas of Italy. J. Plant Pathol. 91 (3), 535–548.
Caffi, T., Rossi, V., Lusitani, M. (2011). Long-term survival of Plasmopara viticola oospores. Long-term survival of Plasmopara viticola oospores Vol. 67 (Dijon, France: IOBC-bulletin), 111–114.
Dagostin, S., Vecchione, A., Zulini, L., Ferrari, A., Gobbin, D., Pertot, I. (2006). Potential use of biocontrol agents to prevent Plasmopara viticola oospore germination. IOBC WPRS Bull. 29 (11), 43.
Delbac, L., Delière, L., Schneider, C., Delmotte, F. (2018). Evidence for sexual reproduction and fertile oospore production by Plasmopara viticola on the leaves of partially resistant grapevine cultivars. In XII Int. Conf. Grapevine Breed. Genet. 1248, 607–620. doi: 10.17660/ActaHortic.2019.1248.82
Díez-Navajas, A. M., Greif, C., Poutaraud, A., Merdinoglu, D. (2007). Two simplified fluorescent staining techniques to observe infection structures of the oomycete Plasmopara viticola in grapevine leaf tissues. Micron 38, 680–683. doi: 10.1016/j.micron.2006.09.009
Etxeberria, A., Mendarte, S., Larregla, S. (2011). Thermal inactivation of Phytophthora capsici oospores. Rev. Iberoamericana micologia 28 (2), 83–90. doi: 10.1016/j.riam.2011.01.004
Fay, J. C., Fry, W. E. (1997). Effects of hot and cold temperatures on the survival of oospores produced by United States strains of Phytophthora infestans. Am. Potato J. 74 (5), 315–323. doi: 10.1007/BF02851575
Fittipaldi, M., Nocker, A., Codony, F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 91 (2), 276–289. doi: 10.1016/j.mimet.2012.08.007
Galbiati, C., Longhin, G. (1984). Indagini sulla formazione e sulla germinazione delle oospore di Plasmopara viticola. Rivista di Patol. Vegetale 20 (2), 66–80.
Galet, P. (1977). Les maladies et les parasites de la vigne (Montpellier, France: Les imprimeries du Midi) No. 634.81, G3.
Gessler, C., Pertot, I., Perazzolli, M. (2011). Plasmopara viticola: A review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol. Mediterr. 50, 3–44.
Gindro, K., Lecoultre, N., Molino, L., de Joffrey, J.-P., Schnee, S., Voinesco, F., et al. (2014). Development of rapid direct PCR assays to identify downy and powdery mildew and grey mould in Vitis vinifera tissues. J. Int. Des. Sci. la Vigne du Vin 48 (4), 261–268. doi: 10.20870/oeno-one.2014.48.4.1697
Gisi, U., Waldner, M., Kraus, N., Dubuis, P. H., Sierotzki, H. (2007). Inheritance of resistance to carboxylic acid amide (CAA) fungicides in Plasmopara viticola. Plant Pathol. 56 (2), 199–208. doi: 10.1111/j.1365-3059.2006.01512.x
Gobbin, D., Pertot, I., Gessler, C. (2003). Genetic structure of a Plasmopara viticola population in an isolated Italian mountain vineyard. J. Phytopathol. 151 (11-12), 636–646. doi: 10.1046/j.0931-1785.2003.00779.x
Gobbin, D., Jermini, M., Loskill, B., Pertot, I., Raynal, M., Gessler, C. (2005). Importance of secondary inoculum of Plasmopara viticola to epidemics of grapevine downy mildew. Plant Pathol. 54 (4), 522–534. doi: 10.1111/j.1365-3059.2005.01208.x
Gobbin, D., Rumbou, A., Linde, C. C., Gessler, C. (2006). Population genetic structure of Plasmopara viticola after 125 years of colonization in European vineyards. Mol. Plant Pathol. 7 (6), 519–531. doi: 10.1111/j.1364-3703.2006.00357.x
Gossen, B. D., Al-Daoud, F., Dumonceaux, T., Dalton, J. A., Peng, G., Pageau, D., et al. (2019). Comparison of techniques for estimation of resting spores of Plasmodiophora brassicae in soil. Plant Pathol. 68 (5), 954–961. doi: 10.1111/ppa.13007
Hill, G. K. (1998). Studies on the germination of Plasmopara viticola oospores with a floating disc test. IOBC WPRS Bull. 21, 1–2.
Hill, G. K. (2001). The survival of Plasmopara viticola macrosporangia under various moisture conditions. IOBC WPRS Bull. 24 (7), 33–36.
Hussain, T., Singh, B. P., Anwar, F. (2014). A quantitative Real Time PCR-based method for the detection of Phytophthora infestans causing Late blight of potato, in infested soil. Saudi J. Biol. Sci. 21 (4), 380–386. doi: 10.1016/j.sjbs.2013.09.012
Jiang, H., Zhang, L., Zhang, J. Z., Ojaghian, M. R., Hyde, K. D. (2016). Antagonistic interaction between Trichoderma asperellum and Phytophthora capsici in vitro. J. Zhejiang University-SCIENCE B 17 (4), 271–281. doi: 10.1631/jzus.B1500243
Kennelly, M. M., Gadoury, D. M., Wilcox, W. F., Magarey, P. A., Seem, R. C. (2007). Primary infection, lesion productivity, and survival of sporangia in the grapevine downy mildew pathogen Plasmopara viticola. Phytopathology 97 (4), 512–522. doi: 10.1094/PHYTO-97-4-0512
Kump, I., Blaise, P., Gessler, C. (1998). The use of RAPD-markers to estimate genetic diversity of Plasmopara viticola in a single vineyard. – In: Magarey et al. (eds.), Third Int. Workshop on Grapevine Downy and Powdery Mildew. Adelaide, Australia: SARDI Research Report Series, 50 (176 pp.), 72–73.
Lacasa, C. M., Martínez, V., Hernández, A., Ros, C., Lacasa, A., del Mar Guerrero, M., et al. (2015). Survival reduction of Phytophthora capsici oospores and P.nicotianae chlamydospores with Brassica green manures combined with solarization. Scientia Hortic. 197, 607–618. doi: 10.1016/j.scienta.2015.10.024
Lafon, R., Clerjeau, M. (1988). Downy mildew Pages 11-13 in: Compendium of grape diseases. Eds. Pearson, R. C., Goheen, A. C. (St. Paul, MN, USA: APS Press).
Lalancette, N., Ellis, M. A., Madden, L. V. (1988). Development of an infection efficiency model for Plasmopara viticola on American grape based on temperature and duration of leaf wetness. Phytopathology 78 (6), 794–800. doi: 10.1094/Phyto-78-794
Laviola, C., Burruano, S., Strazzeri, S. (1986). Influenza della temperatura sulla germinazione delle oospore di Plasmopara viticola (Berk. Curt.) Berl. Toni. Phytopathol. Mediterr. 25, 80–84.
Lehoczky, J. (1965). Observations on oospore production by Plasmopara viticola in floating leaf discs in artificial culture. Vitis 5, 17–19.
Li, M., Senda, M., Komatsu, T., Suga, H., Kageyama, K. (2010). Development of real-time PCR technique for the estimation of population density of Pythium intermedium in forest soils. Microbiol. Res. 165 (8), 695–705. doi: 10.1016/j.micres.2009.11.010
MacHardy, W. E., Gadoury, D. M., Rosenberger, D. A. (1993). Delaying the onset of fungicide programs for control of apple scab in orchards with low potential ascospore dose of Venturia inaequalis. Plant Dis. 77, 372–372. doi: 10.1094/PD-77-0372
Manici, L. M., Lazzeri, L., Baruzzi, G., Leoni, O., Galletti, S., Palmieri, S. (2000). Suppressive activity of some glucosinolate enzyme degradation products on Pythium irregulare and Rhizoctonia solani in sterile soil. Pest Manage. Sci.: Formerly Pesticide Sci. 56 (10), 921–926. doi: 10.1002/1526-4998(200010)56:10<921::AID-PS232>3.0.CO;2-L
Nocker, A., Sossa-Fernandez, P., Burr, M. D., Camper, A. K. (2007). Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73 (16), 5111–5117. doi: 10.1128/AEM.02987-06
Pavón, C. F., Babadoost, M., Lambert, K. N. (2008). Quantification of Phytophthora capsici oospores in soil by sieving-centrifugation and real-time polymerase chain reaction. Plant Dis. 92 (1), 143–149. doi: 10.1094/PDIS-92-1-0143
Pertot, I., Zulini, L. (2003). Studies on Plasmopara viticola oospore germination in Trentino, Italy. IOBC WPRS Bull. 26 (8), 43–46.
Rossi, V., Caffi, T. (2007). Effect of water on germination of Plasmopara viticola oospores. Plant Pathol. 56 (6), 957–966. doi: 10.1111/j.1365-3059.2007.01685.x
Rossi, V., Caffi, T., Bugiani, R., Spanna, F., Valle, D. D. (2008a). Estimating the germination dynamics of Plasmopara viticola oospores using hydro-thermal time. Plant Pathol. 57 (2), 216–226. doi: 10.1111/j.1365-3059.2007.01738.x
Rossi, V., Caffi, T., Giosuè, S., Bugiani, R. (2008b). A mechanistic model simulating primary infections of downy mildew in grapevine. Ecol. Modeling 212 (3–4), 480–491. doi: 10.1016/j.ecolmodel.2007.10.046
Rossi, V., Caffi, T., Gobbin, D. (2013). Contribution of molecular studies to botanical epidemiology and disease modelling: Grapevine downy mildew as a case-study. Eur. J. Plant Pathol. 135, 641–654. doi: 10.1007/s10658-012-0114-2
Rumbou, A., Gessler, C. (2006). Particular structure of Plasmopara viticola populations evolved under Greek island conditions. Phytopathology 96 (5), 501–509. doi: 10.1094/PHYTO-96-0501
Rumbou, A., Gessler, C. (2004). Genetic dissection of Plasmopara viticola population from a Greek vineyard in two consecutive years. Eur. J. Plant Pathol. 110 (4), 379–392. doi: 10.1023/B:EJPP.0000021061.38154.22
Schena, L., Li Destri Nicosia, M. G., Sanzani, S. M., Faedda, R., Ippolito, A., Cacciola, S. O. (2013). Development of quantitative PCR detection methods for phytopathogenic fungi and oomycetes. J. Plant Pathol. 95 (1), 7–24.
Scherer, E., Gisi, U. (2006). Characterization of genotype and mating type in European isolates of Plasmopara viticola. J. Phytopathol. 154, 489–495. doi: 10.1111/j.1439-0434.2006.01136.x
Si Ammour, M., Fedele, G., Morcia, C., Terzi, V., Rossi, V. (2019). Quantification of Botrytis cinerea in Grapevine Bunch Trash by Real-Time PCR. Phytopathology 109 (7), 1312–1319. doi: 10.1094/PHYTO-11-18-0441-R
Taylor, A. (2018). Genetic structure, survival mechanisms and spread of downy mildew in Western Australian and Australian vineyards (Doctoral dissertation, Murdoch University).
Toffolatti, S., Vavassori, A., Prandato, M., Vercesi, A. (2004). Effect of different overwintering conditions on the germination dynamics of Plasmopara viticola oospores. J. Plant Pathol. 86 (4, Special issue), 336–336.
Toffolatti, S. L., Serrati, L., Sierotzki, H., Gisi, U., Vercesi, A. (2007). Assessment of QoI resistance in Plasmopara viticola oospores. Pest Manage. Sci.: Formerly Pesticide Sci. 63 (2), 194–201. doi: 10.1002/ps.1327
Toffolatti, S. L., Prandato, M., Serrati, L., Sierotzki, H., Gisi, U., Vercesi, A. (2011). Evolution of Qol resistance in Plasmopara viticola oospores. Eur. J. Plant Pathol. 129 (2), 331–338. doi: 10.1007/s10658-010-9677-y
Toffolatti, S. L., Russo, G., Campia, P., Bianco, P. A., Borsa, P., Coatti, M., et al. (2018). A time-course investigation of resistance to the carboxylic acid amide mandipropamid in field populations of Plasmopara viticola treated with anti-resistance strategies. Pest Manage. Sci. 74 (12), 2822–2834. doi: 10.1002/ps.5072
Valsesia, G., Gobbin, D., Patocchi, A., Vecchione, A., Pertot, I., Gessler, C. (2005). Development of a high-throughput method for quantification of Plasmopara viticola DNA in grapevine leaves by means of quantitative real-time polymerase chain reaction. Phytopathology 95, 672–678. doi: 10.1094/PHYTO-95-0672
Van der Heyden, H., Wallon, T., Lévesque, C. A., Carisse, O. (2019). Detection and quantification of Pythium tracheiphilum in soil by multiplex real-time qPCR. Plant Dis. 103 (3), 475–483. doi: 10.1094/PDIS-03-18-0419-RE
Vecchione, A., Zulini, L., Pertot, I., Musetti, R. (2005). Biological control of Plasmopara viticola: a multisite approach. In Int. Workshop Adv. Grapevine Wine Res. 754, 361–366.
Vercesi, A., Tornaghi, R., Sant, S., Burruano, S., Faoro, F. (1999). A cytological and ultrastructural study on the maturation and germination of oospores of Plasmopara viticola from overwintering vine leaves. Mycological Res. 103 (2), 193–202. doi: 10.1017/S095375629800700X
Vercesi, A., Sirtori, C., Vavassori, A., Setti, E., Liberati, D. (2000). Estimating germinability of Plasmopara viticola oospores by means of neural networks. Med. Biol. Eng. Comput. 38 (1), 109–112. doi: 10.1007/BF02344698
Vercesi, A., Vavassori, A., Faoro, F., Bisiach, M. (2002). Effect of azoxystrobin on the oospores of Plasmopara viticola. In Advances in downy mildew research. (Dordrecht: Springer), 195–199.
Vercesi, A., Toffolatti, S. L., Zocchi, G., Guglielmann, R., Ironi, L. (2010). A new approach to modelling the dynamics of oospore germination in Plasmopara viticola. Eur. J. Plant Pathol. 128 (1), 113–126. doi: 10.1007/s10658-010-9635-8
Widmer, T. (2011). Effect of temperature on survival of Phytophthora kernoviae oospores, sporangia, and mycelium. New Z. J. Forestry Sci. 41S, S15–S23.
Wong, F. P., Wilcox, W. F. (2001). Comparative physical modes of action of azoxystrobin, mancozeb, and metalaxyl against Plasmopara viticola (grapevine downy mildew). Plant Dis. 85 (6), 649–656. doi: 10.1094/PDIS.2001.85.6.649
Keywords: grapevine downy mildew, oospore density, qPCR, infestation level, overwintering inoculum, microscope counts
Citation: Si Ammour M, Bove F, Toffolatti SL and Rossi V (2020) A Real-Time PCR Assay for the Quantification of Plasmopara viticola Oospores in Grapevine Leaves. Front. Plant Sci. 11:1202. doi: 10.3389/fpls.2020.01202
Received: 04 June 2020; Accepted: 24 July 2020;
Published: 07 August 2020.
Edited by:
Andreia Figueiredo, University of Lisbon, PortugalReviewed by:
Xiao Yin, Northwest A and F University, ChinaYuejin Wang, Northwest A and F University, China
Annalisa Polverari, University of Verona, Italy
Copyright © 2020 Si Ammour, Bove, Toffolatti and Rossi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vittorio Rossi, dml0dG9yaW8ucm9zc2lAdW5pY2F0dC5pdA==
 Melissa Si Ammour
Melissa Si Ammour Federica Bove
Federica Bove Silvia Laura Toffolatti
Silvia Laura Toffolatti Vittorio Rossi
Vittorio Rossi