
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 August 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01192
Salt stress is one of the main factors that affect both growth and development of plants. Maintaining K+/Na+ balance in the cytoplasm is important for metabolism as well as salt resistance in plants. In the present study, we monitored the growth (height and diameter) of transgenic Populus alba × P. berolinensis trees (ABJ01) carrying JERF36s gene (a tomato jasmonic/ethylene responsive factors gene) over 4 years, which showed faster growth and significant salt tolerance compared with non-transgenic poplar trees (9#). The expression of NHX1 and SOS1 genes that encode Na+/H+ antiporters in the vacuole and plasma membranes was measured in leaves under NaCl stress. Non-invasive micro-test techniques (NMT) were used to analyse ion flux of Na+, K+, and H+ in the root tip of seedlings under treatment with100 mM NaCl for 7, 15, and 30 days. Results showed that the expression of NHX1 and SOS1 was much higher in ABJ01 compared with 9#, and the Na+ efflux and H+ influx fluxes of root were remarkable higher in ABJ01 than in 9#, but K+ efflux exhibited lower level. All above suggest that salt stress induces NHX1 and SOS1 to a greater expression level in ABJ01, resulting in the accumulation of Na+/H+ antiporter to better maintain K+/Na+ balance in the cytoplasm of this enhanced salt resistant variety. This may help us to better understand the mechanism of transgenic poplars with improving salt tolerance by overexpressing JERF36s and could provide a basis for future breeding programs aimed at improving salt resistance in transgenic poplar.
The total area of arid, semiarid and saline-alkali lands in China accounts for more than half of the total terrestrial with low underground water levels and serious salinization. Among them, soil salinization is a major abiotic stress, and approximately 7.6 million hm2 of farmland (almost 1/5 of the total) shows certain degrees of salinization (Song et al., 2016; Ke et al., 2017; Chen et al., 2019). Salt stress has various adverse effects in plant growth and development, including osmotic stress, ion toxicity, water shortage, and nutrient imbalances (Zhu, 2001; Kaleem et al., 2018; Liang et al., 2018; Yang and Guo, 2018; Chen et al., 2019). The prevalence and impact of high salinity on plant growth and productivity have drawn increasing attention (Widodo et al., 2009; Zhu et al., 2016; Yang and Guo, 2018). Populus is an economically and ecologically important perennial woody plant that is widely cultivated in the Northern Hemisphere because of their rapid growth and high biomass yields, with marked economic benefits and applications in the production of biofuels, pulp, paper, and other bio-based products, such as chemicals and adhesives (Sannigrahi et al., 2010; Meng and Wu, 2018; Zhang X. N. et al., 2019). However, soil salinization is an increasingly serious threat that limits poplar growth and afforestation program, so breeding salt-tolerant poplar has become very important and necessary.
The application of genetic engineering and transformation technology provides effective means of improving salt tolerance of poplar by introducing known salt-responsive genes into Populus (Jansson and Douglas, 2007; Li et al., 2009; Su et al., 2011; Ye et al., 2011; Zhou et al., 2014; Yao et al., 2017; Zhang X. N. et al., 2019). Many studies have shown that inroducing transcription factors (TFs) genes into plants is generally more efficient than structural genes, because TFs usually regulate the expression of many target genes in related pathways, leading to enhanced stress torlance in plants (Mcgrath et al., 2005; Li et al., 2009; Su et al., 2011; Licausi et al., 2013; Karim et al., 2015; Yao et al., 2016; Wei et al., 2017; Zhao H. et al., 2018; Charfeddine et al., 2019; Zhang C. H et al., 2019; Zhang X. N. et al., 2019; Chen et al., 2020; Jiang et al., 2020). Ethylene responsive factors (ERFs), which have a characteristic AP2/ERF DNA-binding domain, comprise one of the largest TF families in plants (Riechmann et al., 2000; Nakano et al., 2006; Licausi et al., 2013; Zhang X. N. et al., 2019; Chen et al., 2020). Overexpression of ERFs genes can effectively improve salt tolarence of plant under salt stress, such as ERF96 (in Arabidopsis thaliana) (Jiang et al., 2020), StERF94 (in potato) (Charfeddine et al., 2019), ERF76 (in di-haploid P. simonii × P. nigra) (Yao et al., 2016; Yao et al., 2017; Yao et al., 2019), PalERF109 (in P. alba var. pyramidalis) (Chen et al., 2020), and JERFs (in tomato) (Li et al., 2009; Su et al., 2011).
Populus alba × P. berolinensis exhibits fast growth and beautiful tree shape, and is widely used in urban greening and afforestation in northeast, northwest and north of China. However, P. alba × P. berolinensis is difficult to plant on salinity soil because of its susceptibility to salt stress, which seriously affects its popularizing application on Saline-alkali land. To improve salt resistance in this variety, a tomato jasmonic ethylene responsive factors (JERF36s) gene that encodes the ERF like transcription factor (Zhang and Huang, 2010) has been transferred to the genome of the P. alba × P. berolinensis in our previous study (Li et al., 2009). Overexpression of JERF36s resulted in the improved salt tolerance in transgenic poplar (ABJ01) compared with non-transgenic poplar (9#) (Li et al., 2009). However, how JERFs exactly improve salt resistance in transgenic poplar remains to be unravelled.
The maintenance of intracellular K+ and Na+ homeostasis is considered to be an essential component for plant growth or adaption in saline environments (Munns and Tester, 2008; Huang et al., 2018), which is a crucial factor affecting plant salinity tolerance (Chen et al., 2007; Huang et al., 2018; Zhang X. N. et al., 2019; Blumwald et al., 2000). Maintaining a cytosolic K+/Na+ homeostasis balance largely depends on preventing the accumulation of Na+ via Na+ expulsion from cell cytoplasm, sequestering Na+ into vacuoles, and increasing K+ absorption (Hasegawa, 2013; Bassil and Blumwald et al., 2014; Yang and Guo, 2018; Bassil et al., 2019; Su et al., 2019). The Na+, K+/H+ antiporters are the transmembrane protein locating in both plasma and vacuolar membranes (Guo et al., 2020). Na+, K+/H+ antiporter (NHX)-type exchangers are the critical regulators of vesicular tracking and cell volume, which catalyse the electroneutral exchange of K+ or Na+ for H+ and play vital roles in plant salt tolerance, as well as the Na+ and K+ homeostases of intracellular compartments (Bassil and Blumwald et al., 2014; Azhar et al., 2017). AtNHX1 and AtNHX2 are vacuolar-type NHXs and localized to the vacuolar membrane, which play diverse roles in maintaining Na+/K+ homeostasis under salt-stress (Leidi et al., 2010; Bassil et al., 2011), as well as in active K+ uptake in the tonoplast for stomatal function (Barragán et al., 2012). Another plasma membrane localized Na+/H+ exchange transporter SOS1 can be activated by SOS3-SOS2 protein kinase complex under high Na+ stress, resulting in the exclusion of excess Na+ from plant cells, which is essential for maintaining Na+ and K+ homeostasis in the cytoplasm and defense against salt stress (Zhu, 2002; Qiu et al., 2004; Hu et al., 2016; Zhu, 2016). SOS1, belongs to the NHX-type Na+(K+)/H+ exchange family genes (NHXs). Some studies have described that overexpression of NHXs, such as NHX1 and SOS1, can effectively maintain Na+/K+ homeostasis in the plant cells for the enhancement of salt tolerance (Wu, 1996; Shi et al., 2000; Barragán et al., 2012; Ma et al., 2014; Li et al., 2017; Fahmideh and Fooladvand, 2018; Huang et al., 2018; Liang et al., 2018; Meng and Wu, 2018; Wang et al., 2018; Acuña-Rodríguez et al., 2019; Liu et al., 2019; Mahi et al., 2019; Yao et al., 2019; Zhao et al., 2019). However, there are limited reports of the regulation of intracellular K+/Na+ balance by TFs in transgenic poplar with exogenous transcription factors.
In the present study, to explore the effects of JERFs on intracellular K+/Na+ balance, the transgenic poplar overexpressing JERF36s (ABJ01) and non-transgenic poplar (9#) as control were grown in field trials, and the expression of NHX1(Gene ID: 105132046) and SOS1(Gene ID: 7462643) genes, encoding Na+/H+ reverse transporters in the vacuole membrane and plasma membrane was measured. Non-invasive micro-test techniques (NMT) were used to analyse the ion flux of Na+, K+, and H+ in the root tip of seedlings stressed by 100 mM NaCl for 7, 15, and 30 days. These findings may provide a basis for future breeding programs aimed at improving salt resistance in transgenic poplar.
The hybrid poplar from Populus alba × P. berolinensis (9#) was used in this study. Transgenic poplar line (ABJ01) were produced as described previously (Li et al., 2009). Briefly, The transfected exogenous gene was the JERF36s gene, encoding AP2/EREBP plant transcription factors, which was under the control of the CaMV 35S promoter. The neomycin phosphotransferase II gene (NPT II) derived from Escherichia coli transposon Tn5 was used as a marker, which provided the plants with kanamycin resistance. The 4-year study plots of 9# and ABJ01 were established in 2004 in a forest located in Panjin, Liaoning Provence (41°170' N, 121°360' E) at an elevation of 14 m above sea level. The soil salt content was measured before the forest was established. Soil samples were taken from 0–10 cm, 10–30 cm, and 30–50 cm depths in three randomly chosen places. The average salt content and pH at the different depths were as follows; 0–10 cm = 0.38% and pH 7.30; 10–30 cm = 0.2% and pH 7.40; 30–50 cm = 0.18% and pH 7.50. The average total salt content was 0.25%. Established trees (1-year-old) from 9# and ABJ01 were planted in a randomized block design. The field trial consisted of four blocks, each containing three replicates for each line. The forest was planted in 2 m rows with 1 m between adjacent individuals within each row. Cuttings (15 cm) from annual seedlings of 9# and ABJ01 were used for ion flux measurements and qRT-PCR. Cuttings (15 cm) from annual seedlings of 9# and ABJ01 were placed in individual plastic pots of 10 cm in diameter and 10 cm in depth on the 15th of March 2014 and cultured in a greenhouse at the Chinese Academy of Forestry in Beijing, each line was 15 plants. The stroma in pots was a mixture of peat soil and perlite. On 21, April, 9 healthy and similar growth plants of each line were selected to transplant into larger pots (19 cm in height and 17 cm in diameter) in a mixture of sandy soil, peat soil, and pearlite at a ratio of 6:10:1 with 1.8 kg of soil per pot, which were used to measure the growth, ion flux and gene expression in the further. Seedlings were watered regularly, the temperature of the greenhouse was maintained between 25°C and 30°C, and the humidity was 55%.
18 seedlings from each of the 9# and ABJ01 groups were selected when the height of the seedlings reached 40 cm. Nine seedlings from each group were set aside as controls, and nine were treated with 100 mM NaCl solution every 2 days for 7, 15, or 30 days with three triplicates, respectively. The growth was determined by measuring the height of each plant after 30 days of salt stress.
Growth measurements were completed in 2004, 2005, 2006, and 2007. The height (the distance from a point at ground level to the top of the tree) and diameter at breast height (DBH, 1.3 m above the ground level) of ABJ01 and 9# were measured each year after the growing season, each line was measured in 12 plants as replicates.
One or two leaves from the top of seedlings were harvested after treatment with 100 mM NaCl for 7, 15, or 30 days. Harvested leaves were frozen in liquid nitrogen immediately and stored at -80°C until needed for RNA extraction. Total RNA was isolated from leaves using a RNeasy Plant Mini Kit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized using the PrimerScript RT reagent Kit (TaKaRa, Dalian, China). Quantitative RT-PCR (qRT-PCR) was performed on a Roche LightCycle 480 Real-Time PCR system (Roche) with SYBR Green Realtime PCR Master Mix (TaKaRa) according to the manufacturer's instructions. Gene-specific primers were designed to amplify 120–130 bp fragments of JERFs, NHX1, and SOS1, and the relative gene expression were calculated by 2-ΔΔCt methods using a gene-specific primer pair for poplar ubiquitin (UBQ)-like gene (GenBank Accession BU871588) as a reference gene (Livak and Schmittgen, 2001). Primer sequences for the real-time PCR assay of the genes are listed in Supplementary Table S1. Six plants for each line were tested, and four PCR replicates were performed for each RNA sample.
The net Na+, K+, and H+ ion flux was measured using an NMT system (BIO-001A3, Younger USA Science & Technology Corporation, USA) (Xu et al., 2006; Xu et al., 2008; Sun et al., 2009a; Sun et al., 2009b; Yan et al., 2015a; Yan et al., 2015b; Liang et al., 2018; Zhao N. et al., 2018). The NMT system measured the concentration gradients of the target ions by moving the ion-selective microelectrode between two positions close to the plant material through a pre-set excursion distance (30 mm in our experiment) at a programmable frequency of 0.15 Hz (Sun et al., 2009a; Zhao N. et al., 2018). Ion-selective electrodes were made as described previously by Sun (2009a) and Yan (2015a; 2015b). Ion-selective electrodes were calibrated in calibration solutions (0.5 mM Na+, 5.0 mM NaCl; 0.1 mM K+, 1.0 mM KCl; H+, pH 5.5, and 6.5) prior to flux measurements (Sun et al., 2009a; Sun et al., 2009b; Yan et al., 2015a; Yan et al., 2015b; Zhao N. et al., 2018). Only electrodes with Nernstian slopes between 53 and 62 mV/[log-C (± z)] were used in our study (where z is the valence of the ion and C is the concentration of the ion) (Yan et al., 2015b). Ion flux was calculated by Fick's law of diffusion:
where J represents the ion flux, dc/dx is the ion concentration gradient, and D is the ion diffusion constant in a particular medium (Sun et al., 2009a; Sun et al., 2009b; Yan et al., 2015a; Yan et al., 2015b).
Fresh and tender roots from seedlings were treated with salt for 7, 15 or 30 days, and 20 mm from the root tip was removed from 2–3 seedlings in each group (performed in triplicate) for ion flux measurements. Washed root tips were placed in test solution for 25–30 min prior to measurement, then transferred to 10 mL of test solution in a transparent culture dish and fixed to the bottom of the dish. Ion-selective electrodes were passed through the root at 600 μm, 1,200 μm, and 1,800 μm from the root tip, and each measurement was made for at least 5 min to ensure a stable ion velocity. A positive value indicates external ion flux, and a negative value indicates internal ion flux.
One-way analysis of variance (ANOVA, = 0.05) was used to analysis the differences between the ABJ01 and 9# for growth, qRT-PCR and, Na+, K+, and H+ ion flux, and multiple comparison of means was done using Duncan's multiple range test. All analyses were done with Data Processing System (DPS) software (Tang and Feng, 2002).
Results showed that ABJ01 achieved a larger average height and diameter than 9#in 2004(1a), 2005(2a), 2006(3a), and 2007(4a), respectively (Figure 1). The plant height of ABJ01 was 35.60% and 22.94% remarkably higher than 9# in 2006 and 2007, respectively, and the diameter of ABJ01 was 39.31% and 36.32% significantly higher than 9# in 2005 and 2006, respectively, under salt stress conditions (Figure 1), which indicated that the salt tolerance of ABJ01 was enhanced compared with 9#. Moreover, the growth of ABJ01 and 9# in Panjin in 2005 and 2007 was shown in Figure 2.
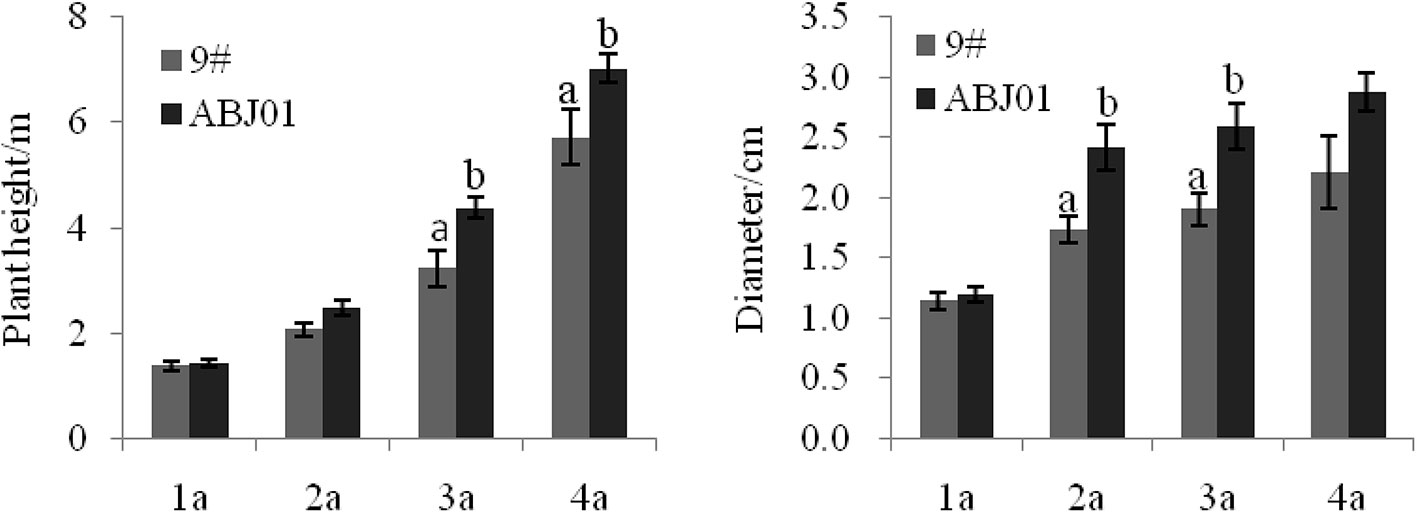
Figure 1 The growth (height and diameter) of 9# and ABJ01 over a 4-year period (2004–2007). The data shows mean ± S.E. of three blocks. Column with different letters indicate significant differences from control (9#) (p < 0.05). N = 12 plants.

Figure 2 The growth of ABJ01 and 9# in 2005 and 2007 in Panjin. The growth of ABJ01 was significantly faster than 9# in saline soil with 0.25% salt content at Panjin in 2005 and 2007.
For the greenhouse test, after the treatment with 100 mM NaCl for 7 days, some lower leaves of the 9# became yellow and begun to wilt. After 15 days treatment, some leaves had dropped off, the middle and uper leaves had black patches on the surface. After 30 days, more damaged leaves exisited in the middle. Compared to the control, the leaves of ABJ01 showed less wilting at the early stage of NaCl stress, and few lower and middle leaves had yellowed, and no leaves dropped by the end of the experiment (30 days) (Figure 3). Under the control condition (0 days) the height of ABJ01 (38.15 cm) was little greater than that of 9# (36.23 cm) with no significant difference. After 100 mM NaCl treatment for 30 days, ABJ01 was significantly higher than 9# with a 4.72% increase (Table 1).
To testify the stability of the exogenous gene expression, leaves of 4 years old transgenic popular was used for qRT-PCR analysis. As expected, the expression of JERF36s in ABJ01 was significantly higher than 9# (P<0.05), which suggested that it was stably expressed in transgenic line ABJ01 after 4 years (Figure 4).
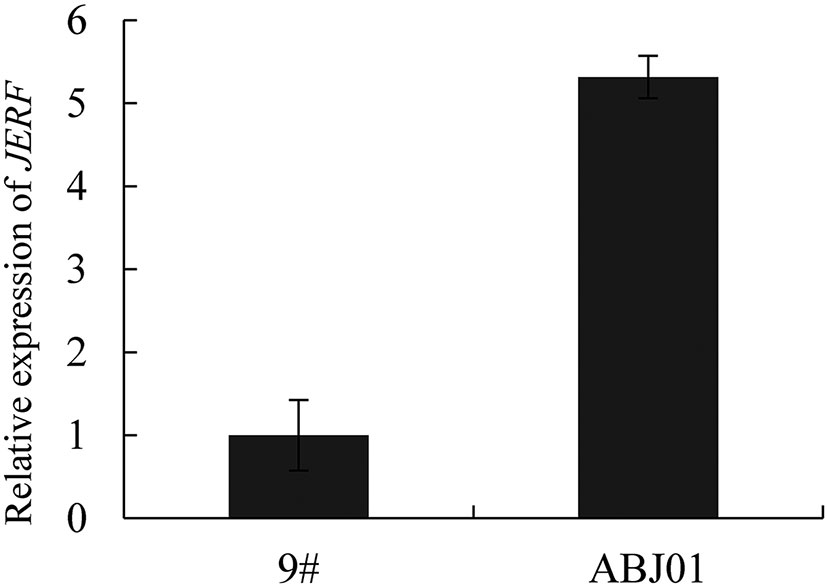
Figure 4 Relative expression of JERF36s in 4 years transgenic poplar (ABJ01). Data are given the means and standard errors (error bars) of four repetitions.
NHX1 encodes the Na+/H+ reverse transporter in the vacuole membrane that imports Na+ into the vacuole and decreases the Na+ concentration in the cytoplasm.
The expression of NHX1 in ABJ01 was much higher than in 9# under both 100mM NaCl stress and normal water condition (control). And compared with normal condition, ABJ01 and 9# displayed significantly higher expression of NHX1 under 15 days of salt stress, while the increased expression at 7 days showed no significance. After 30 days stress, the expression of NHX1 was reduced in 9# compared to control and that in 9# under 15 days treatment, while it was still increase in ABJ01. In addition, the expression of NHX1 was nearly 4-fold higher in ABJ01 than in 9# under NaCl stress for 30 days. These results suggested that, mild stress can induce the expression of NHX1 in both ABJ01 and 9#, and with the increase of time, the higher expression of NHX1 can be effectively maintained in ABJ01 (Figure 5).
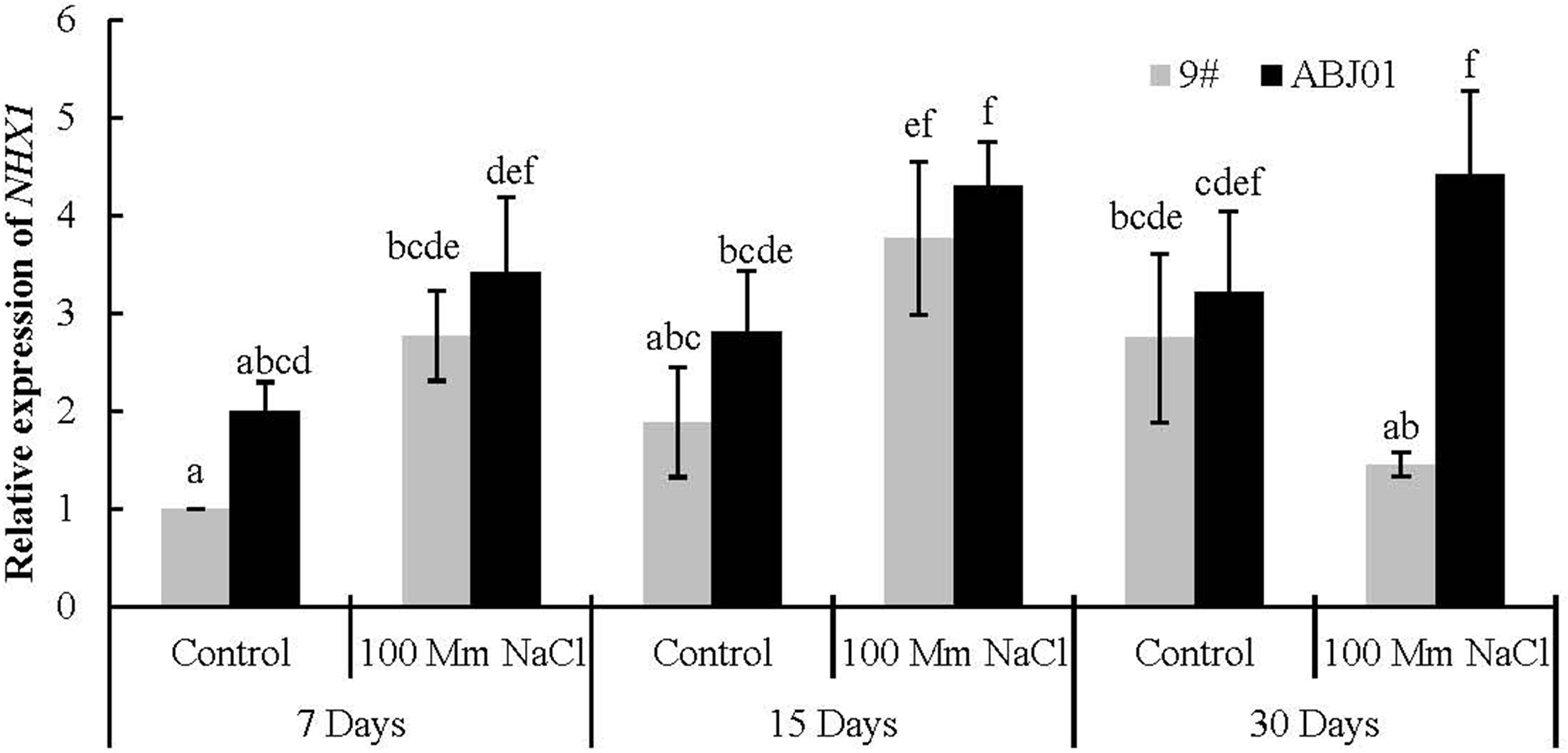
Figure 5 Expression of NHX1 in 9# and ABJ01 under 100 mM NaCl salt stress and normal water conditions (control) for 7 days, 15 days, and 30 days. The data shows mean ± S.E. of triplicate experiments. Column with different letters indicate significant differences at p < 0.05.
In normal water condition, the expression of SOS1 displayed non-significantly higher in ABJ01 than in 9#. The expression of SOS1 in ABJ01 changed significantly with first decreased and then increased trend from 7 days to 30 days under 100 mM NaCl salt stress, which was significant higher than in 9# under NaCl stress for 15 days and 30 days, while, the expression of SOS1 in 9# kept in a low level. In the other hand, the expression of SOS1 in ABJ01 also significant increased under 7 days and 30 days stressed compared with normal condition, but that was reduced in 9# at days 15 and 30 days. These results suggested that the expression of SOS1 can be induced in ABJ01 and inhibited in 9# under salt stress (Figure 6).
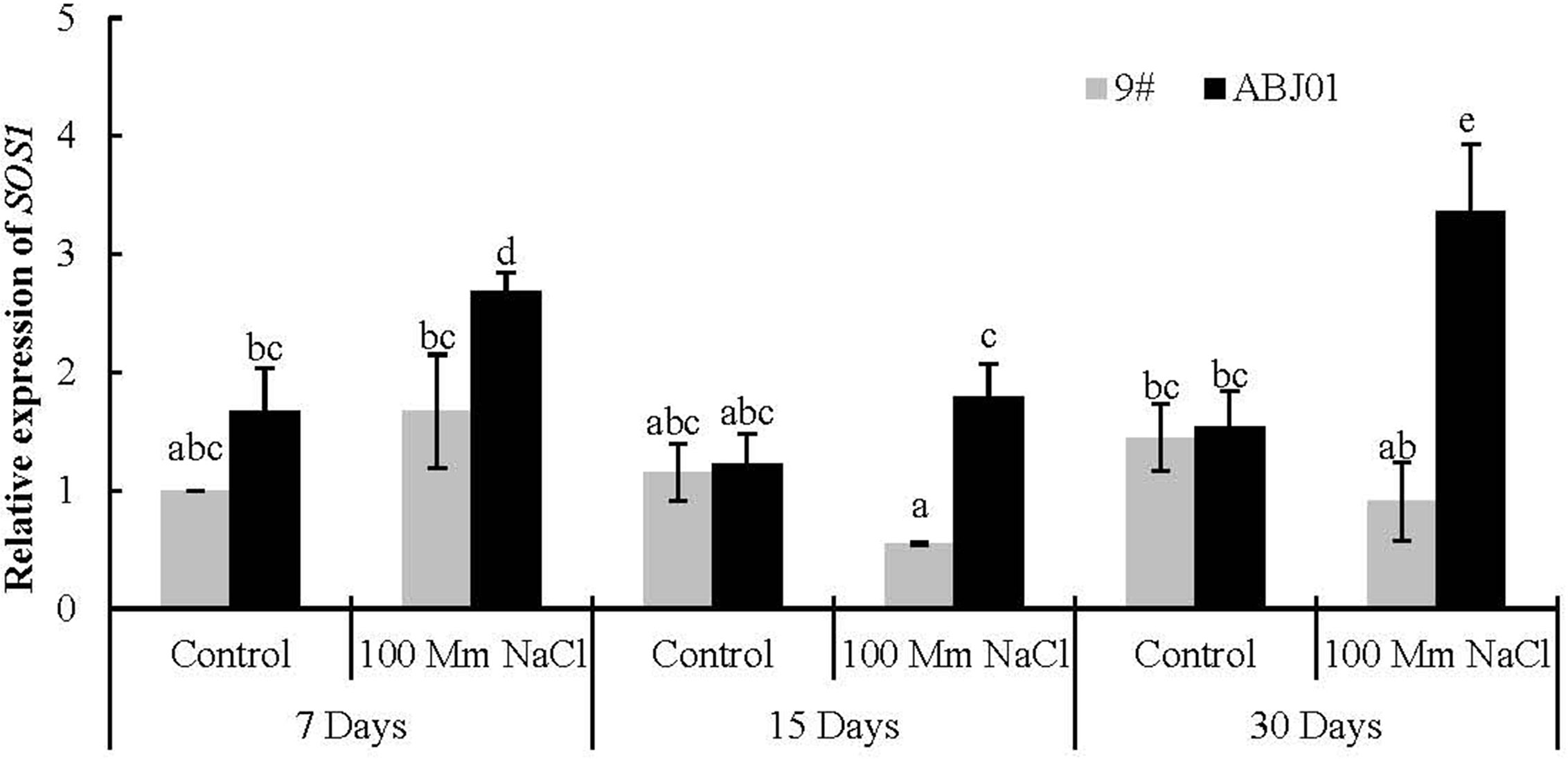
Figure 6 Expression of SOS1 in 9# and ABJ01 under 100 mM NaCl salt stress and normal water conditions (control) for 7 days, 15 days, and 30 days. The data shows mean ± S.E. of quadruplicate experiments. Column with different letters indicate significant differences at p < 0.05.
To explore the effects of NHX1 and SOS1 expression on ion balance in ABJ01 and 9#, Na+, K+, and H+ ion flux in the apical root tip was measured by the NMT method after the seedlings were treated with 100 mM NaCl for 7, 15, and 30 days.
The results of Na+ flux in the apical root tip of ABJ01 and 9# under salt stress and normal condition showed that control groups (normal conditions) for ABJ01 and 9# both displayed lower Na+ efflux (<100 pmol cm-2 s-1) except after 15 days in ABJ01 roots at 1200 μm (Na+ efflux >100 pmol cm-2 s-1). Under salt stress, a higher Na+ efflux was observed in both lines. At 7 days, a significant efflux (430 pmol cm-2 s-1) was observed at the 600 μm root point in ABJ01, but 9# showed no significant differences in Na+ efflux. At 15 days of salt stress, a significant Na+ efflux (323.53 pmol cm-2 s-1) was observed at the 1,200 μm point in ABJ01, at the same time, a significant Na+ efflux was measured at 600 μm in 9#. At 30 days of salt treatment, Na+ efflux was significant at 600, 1200 and 1800 μm in ABJ01 roots. Compared with control, ABJ01 displayed a significantly higher Na+ efflux under salt stress, and 9# also exhibited a higher Na+ efflux than controls, but a lower efflux than ABJ01. The results therefore showed that Na+ efflux was much stronger in ABJ01 than 9# (Figure 7).
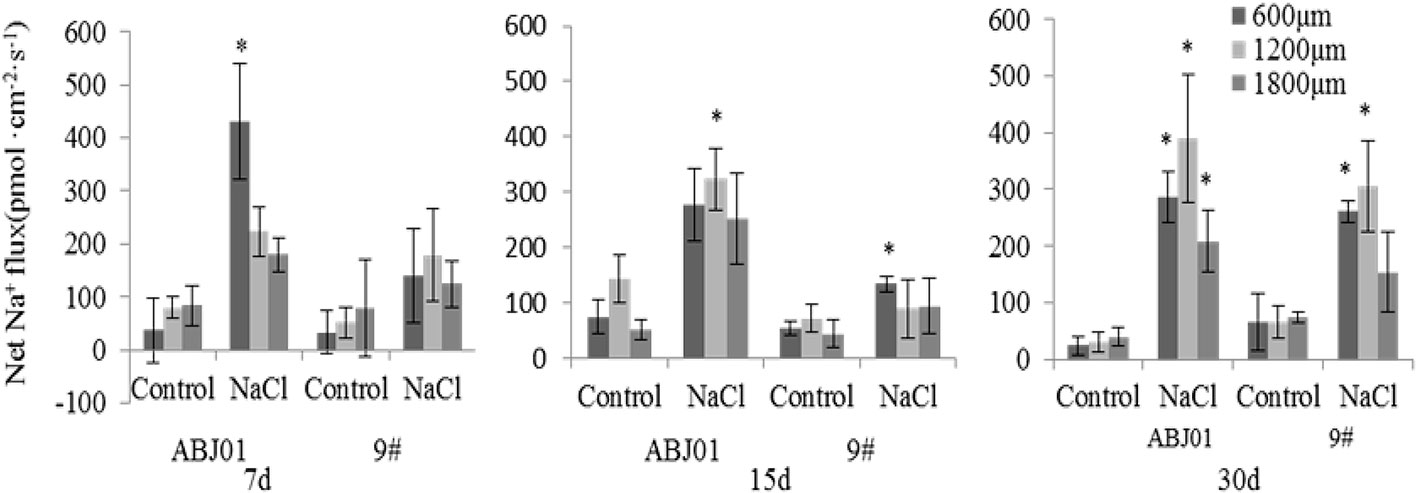
Figure 7 Effects of different salt stress on net Na+ flux in the root tip of transgenic (ABJ01) and non-transgenic lines (9#). Positive values indicate efflux, negative values indicate influx; *, significant differences from controls (p < 0.05). Data are given the means and standard errors (error bars) of six repetitions (n = 6).
Under normal condition, net K+ efflux showed weakly in ABJ01 at 7, 15, and 30 days, whereas it displayed influx weakly after 7 and 30 days and toward efflux at 15 days in 9#. Under 100mM NaCl salt stress, K+ efflux was significant in both ABJ01 and 9#, and the mean K+ efflux was highest after 15 days of salt stress in both varieties. After 7 days, ABJ01 showed a significant K+ efflux at 600 μm (82.69 pmol cm-2 s-1) and 1,800 μm (53.66 pmol cm-2 s-1), while K+ efflux in 9# was 115.40 pmol cm-2 s-1 at 600 μm 96.39 pmol cm-2 s-1 at 1,200 μm. After salt treatment for 15 days, K+ efflux was higher than after 7 days in both ABJ01 and 9#. ABJ01 displayed significant K+ efflux at 600 μm (170.53 pmol cm-2 s-1), and 9# also exhibited significant K+ efflux at 600 μm (396.30 pmol cm-2 s-1), but also at 1200 μm (346.27 pmol cm-2 s-1) and 1800 μm (209.46 pmol cm-2 s-1). K+ efflux was lower in both ABJ01 and 9# after salt treatment for 30 days, but K+ efflux remained significant in 9#. The results showed that K+ efflux is much lower in ABJ01 than in 9# under salt stress conditions, suggesting ABJ01 is more capable of maintain Na+/K+ balance in cells (Figure 8). Under normal condition, net H+ influx was observed for ABJ01, while H+ efflux was observed for 9# after 7 days. After 15 days, H+ efflux was observed in ABJ01 and influx was observed in 9#. Both ABJ01 and 9# displayed H+ efflux after 30 days. After 7 days salt stress treatment, ABJ01 (-9.73 pmol cm-2 s-1) and 9# (-5.58 pmol cm-2 s-1) exhibited significant H+ influx at 600 μm. After salt treatment for 15 days, H+ influx decreased in both genotypes. At 30 days of NaCl stress, both genotypes exhibited higher H+ influx. ABJ01 displayed significant H+ influx at 600 μm (-15.00 pmol cm-2 s-1), 1,200 μm (-9.04 pmol cm-2 s-1), and 1,800 μm (-6.18 pmol cm-2 s-1), while 9# displayed significant H+ influx at 1,200 μm (-6.89 pmol cm-2 s-1) and 1,800 μm (-2.26 pmol cm-2 s-1) (Figure 9). All above suggest that ABJ01 is more capable at moving H+ through roots than 9#.
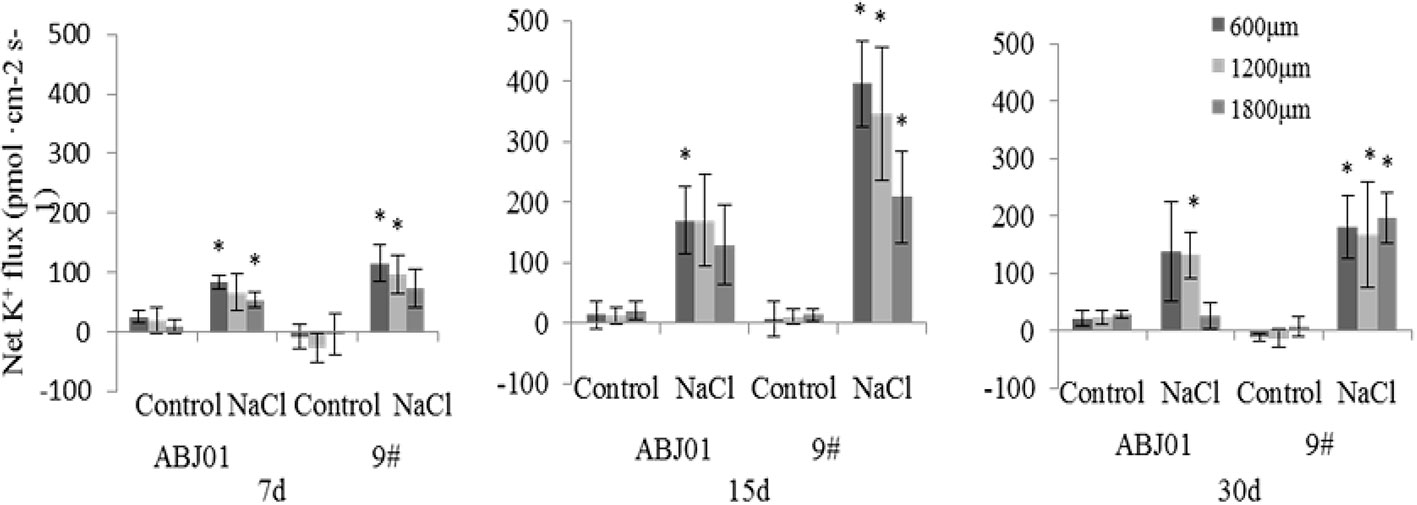
Figure 8 Effects of different salt stress on net K+ flux in the root tip of transgenic and non-transgenic lines. Positive values indicate efflux, negative values indicate influx; *, significant differences from controls (p < 0.05). Data are given the means and standard errors (error bars) of six repetition (n = 6).
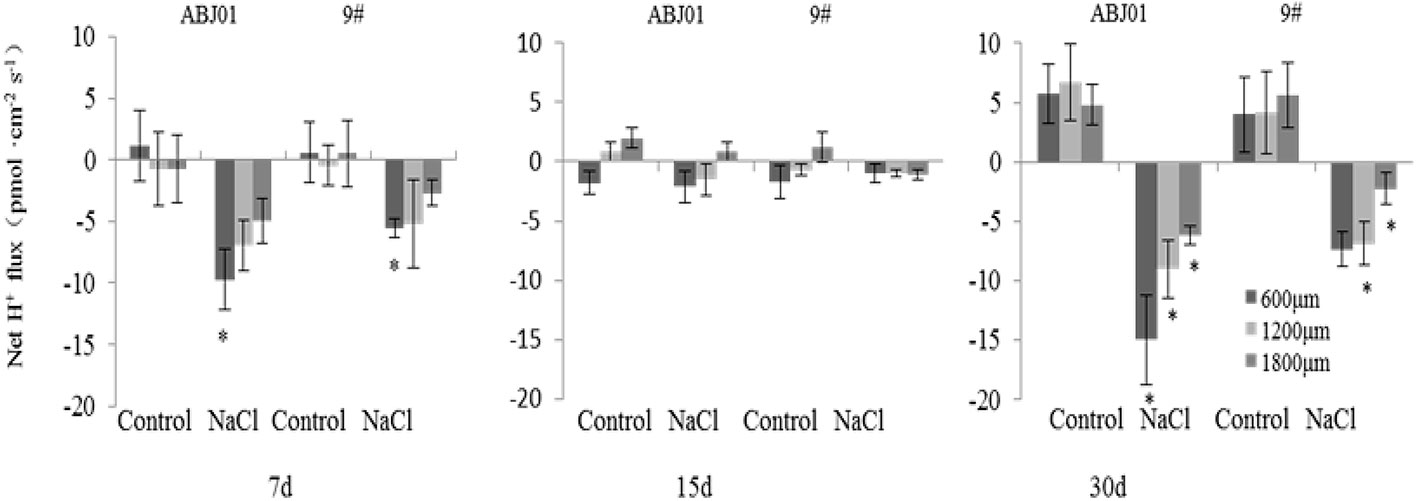
Figure 9 Effects of salt stress on net H+ flux in the apical root tip of transgenic and non-transgenic lines. Positive values indicate efflux, negative values indicate influx; *, significant differences from controls (p < 0.05). Data are given the means and standard errors (error bars) of six repetition (n = 6).
It is well known that, maintaining the cellular ion homeostasis by restricting the accumulation of toxic sodium (Na+) is the one key response to salt stress, such as a suitable K+:Na+ ratio to maintain K+/Na+ balance in the cell, which prevents cellular damage and nutrient deficiency (Clarkson and Hanson, 1980; Tester and Davenport, 2003; Li et al., 2017; Liang et al., 2018; Yang and Guo, 2018; Zhang X. N. et al., 2019). The SOS pathway (exports Na+ ions out of cells and is highly conserved in plants) plays an important role in maintaining ion homeostasis, and salt tolerance at cellular in plants (Zhu, 2000; Ji et al., 2013; Yang and Guo, 2018; Cheng et al., 2019; Liu et al., 2019; Mahi et al., 2019; Zhang X. N. et al., 2019). Previous studies have shown that SOS1, SOS2 and SOS3 are the key components of the SOS signalling pathway involved in Na+ extrusion (Zhu et al., 1998; Zhu, 2003; Quintero et al., 2011; Cheng et al., 2019; Foster and Miklavcic, 2019; Zhang X. N. et al., 2019). The Na+/H+ antiporter responsible for Na+ active efflux is encoded by SOS1, and proteins encoded by SOS2 and SOS3 mainly modulate the activity of SOS1 (Zhu, 2003; Quintero et al., 2011; Fahmideh and Fooladvand, 2018; Foster and Miklavcic, 2019; Yin et al., 2020). And the SOS1 is the most sensitive gene to different levels of hypersensitivity to NaCl (Wu, 1996; Shi et al., 2000; Yang and Guo, 2018; Yin et al., 2020). Salt tolerance in plants is mediated by Na+ extrusion from the cytosol by the plasma membrane Na+/H+ antiporter SOS1, which transports excess Na+ from the cytoplasm to the extracellular region (Shi et al., 2000; Cheng et al., 2019; Zhang X. N. et al., 2019; Yin et al., 2020). Overexpression of genes in the SOS pathway, such as AtSOS1, can increase salt tolerance in plant, including Arabidopsis, Festuca arundinacea, Nicotiana tabacum, and Agrostis grass (Shi et al., 2003; Yue et al., 2012; Ma et al., 2014; Liu et al., 2019). As a primary adaptive mechanism for reducing cytoplasmic ion toxicity in plants grown under high salt concentrations, vacuolar Na+ compartmentalization is mediated by the vacuolar Na+/H+ antiporters (NHX), which are ubiquitous and catalyze the electroneutral exchange of H+ for Na+ or K+ (Barragán et al., 2012; Yang and Guo, 2018; Bassil et al., 2019; Zhang X. N. et al., 2019). The K+(Na+)/H+ antiporter located in the vacuole membrane is encoded by NHX1 (Venema et al., 2002; Barragán et al., 2012; Zhang X. N. et al., 2019). Plants overexpressing AtNHX1 have higher vacuolar K+ content than Na+ even under moderate NaCl stress (Leidi et al., 2010; Li et al., 2017; Yang et al., 2017; Liu et al., 2019). The NHX1 protein can transport Na+ into the vacuole and decreases the Na+ concentration in the cytoplasm (Mahajan et al., 2008; Ding et al., 2010; Tang et al., 2010; Yamaguchi et al., 2013; Fahmideh and Fooladvand, 2018; Zhang X. N. et al., 2019). Overexpression of NHX1 in salt-stressed Arabidopsis, Brassica napus, Zea mays, Triticum aestivum, and A. grass can improve salt resistance owing to altering the distribution of Na+ and K+ (Apse et al., 1999; Zhang et al., 2001; Xue et al., 2004; Li et al., 2017; Yang et al., 2017; Liu et al., 2019). In our study, the expression of NHX1 and SOS1 were significant higher in ABJ01 than in 9# after 15 days or 30 days salt treatment, and was up-regulated significantly in ABJ01 compared with normal water condition and down-regulated in 9# after 30 days. These results may suggest that the ABJ01 have potential high Na+, K+/H+ antiporter activity and better maintaining intracellular ion homeostasis in the cytoplasm effectively, which greatly reduces the ion toxicity and nutrient deficiency.
Plants minimize K+ loss through active efflux of Na+ and separation of Na+ in the vacuole to maintain K+/Na+ balance (Zhu, 2003; Chen et al., 2007; Li et al., 2017; Liang et al., 2018; Yang and Guo, 2018; Zhang X. N. et al., 2019). The ability of exporting Na+ from cytoplasm to apoplast and partitioning vacuolar Na+ into the vacuole becomes a primary adaptive mechanism for reducing cytoplasmic ion toxicity in plants grown under high salt concentration (Hasegawa, 2013; Yang and Guo, 2018; Bassil et al., 2019; Su et al., 2019; Zhang X. N. et al., 2019). And the capability to retain more intracellular K+ than Na+ helps plants to withstand stress from Na+ shock. Electrogenic proton (H+) pumps depend on counter-ion fluxes to establish transmembrane pH gradients at the plasma membrane and endomembranes, alkali cation/H+ antiporters can alter pH and/or cation homeostasis locally and transientl (Li et al., 2017; Liang et al., 2018; Sze and Chanroj, 2018; Yang and Guo, 2018; Zhao N. et al., 2018; Bassil et al., 2019; Su et al., 2019; Zhang X. N. et al., 2019). Thus, H+ is important to maintain the vacuole ion balance for plant under salt stress. Our ion flux results showed that, Na+ efflux and H+ influx were higher in ABJ01 than in 9# under NaCl stress, consistent with a previous study on P. euphratica (Sun et al., 2009a). Other researchers also reported a higher H+ influx in wild Arabidopsis grown under salt stress (Shabala, 2000; Shabala et al., 2005). Na+ can compete with K+ and inhibit K+ influx, resulting in an imbalance of the K+/Na+ ratio that eventually causes ion toxicity in plants (Zhang X. N. et al., 2019). Therefore, restricting the K+ efflux is important for the salt resistance of Populus. Our results showed that K+ efflux was lower in ABJ01 than in 9# under salt stress, indicating less K+ loss in ABJ01. This was also the case for Populus (Sun et al., 2009b; Zhao N. et al., 2018), Hordeum vulgare (Chen et al., 2007), and T. aestivum (Cuin et al., 2008). In H. vulgare (Chen et al., 2007) and T. aestivum (Cuin et al., 2008), the root maturation region was measured at 10,000 μm from the root tip, whereas we measured 600, 1,200, and 1,800 μm from the root tip, which corresponds to the meristematic and elongation zones of the root. P. euphratica (Sun et al., 2009b) revealed a change on K+ flux in the root tip area (200–2,000 μm) and a smaller change in the meristematic zone (10,000-12,000 μm). Na+ efflux and K+ influx in roots of two poplar species (P. euphratica, P. popularis) were recorded in theapical region (ca. 300 μm from the root tip) (Zhao N. et al., 2018). Exogenous expression of Vacuolar membrane-localized GmNHX1 enhances plant salt tolerance through increasing the efflux rate of Na+ in roots and maintaining a high K+/Na+ ratio along with inducing the expression of Na+/H+ exchanger SKOR, SOS1, and AKT1 (Sun et al., 2019a). All above suggest that higher net Na+ efflux, H+ influx, and a greater capacity to inhibit K+ efflux in ABJ01 can effectively improve the salt tolerance, which may be caused by the higher expression of NHX1 and SOS1 in ABJ01.
JERF36s belong to the ERF transcription factor class, which are involved in signal transduction, hormone synthesis, and stress responses (Li et al., 2009; Zhang and Huang, 2010; Su et al., 2011). Previous researches have demonstrated that ERF transcription factors could enhance stress resistance in A. thaliana, N. tabacum, Lilium longifloruma, and Populus (Gilmour et al., 2000; Wang et al., 2004; Li et al., 2006; Li et al., 2009; Su et al., 2011; Yao et al., 2016; Yao et al., 2017; Charfeddine et al., 2019; Chen et al., 2020; Jiang et al., 2020). Expression of JERFs activates the expression of resistance-associated genes and promotes the ability of plants to adapt to environmental stress. And these induced genes in turn may participate in the generation of hormones like ABA (abscisic acid), salicylic acid, and ethylene. In the other hand, these gene products may amplify the initial signal and initiate a second round of signalling that may follow the same pathway or altogether different component of signalling pathway (Basu and Roychoudhury, 2014). For example, ERF76 (a highly salt-inducible ERF gene) was proved to activate the expression of other transcription factors in the transgenic poplar lines overexpressing ERF76 under multiple abiotic stresses (Yao et al., 2019). Overexpression of ERF76 in transgenic poplar upregulated the expression of 16 transcription factor genes and 45 stress-related genes and also increased the ability of ABA and GA (gibberellic acid) biosynthesis, which resulted in stronger tolerance to salt stress (Yao et al., 2016). Overexpression of PalERF109 increased the transcript abundance of salt stress-related genes, enhanced the ABA signaling pathway, and induced expression of salt many ion channel proteins as well as transporters in transgenic poplars with higher salt tolerance (Chen et al., 2020). Plant hormones and plant hormone signaling pathways regulate complex signaling networks related to plant development and responses to environmental stresses (Bari and Jones, 2009; Zhu, 2016). Under the exogenous ABA treatment, the expression of SOS-3 and NHX-1 genes was upregulated in the all rice varieties (Basu and Roychoudhury, 2014). Transgenic lines revealed significantly increased transcript levels of BjSOS2 gene after ABA treatment (Kaur et al., 2015). In our study, ABJ01 stably overexpressing JERF36s grew significantly higher (by more than 20%–30%) and reached a larger diameter than 9# in 2005 and 2006 at Panjin with 0.25% salt content. The higher expression of NHX1 and SOS1 in ABJ01 than 9# both was found under both 100 mM NaCl stress and normal condition, especially at 15 days. This may be explained that the overexpression of JERF36s in ABJ01 has regulate some signaling networks such as ABA signaling pathway and salt-overly-sensitive (SOS) pathway for responses against environmental stresses, and lead to the increased content of plant growth regulators, such as ABA, gibberellin 3 (GA3), salicylic acid (SA), and jasmonic acid (JA), and then up-regulated the expression of downstream target genes (NHX1 and SOS1) in plant for improving the salt tolerance. Consider of this, up-regulating the expression of genes encoding Na+, K+/H+ antiporters, such as of NHX1 and SOS1, by overexpression of exogenous JERF36s may be one of the reason for the improved salt tolerance of transgenic poplars under salt stress.
In conclusion, ABJ01 exhibited stable expression of JERF36s, higher expression of SOS1 and NHX1, stronger Na+ efflux and H+ influx, and smaller K+ loss than 9# in this study, suggesting that exogenous JA/ET responsive transcription factors could regulates Na+/H+ exchanger SOS1 and NHX1 expression, which improve the ability maintain K+/Na+ homeostasis in root cells for the improved tolerance to salt stress in plants. However, the mechanism by which JERFs modulate the expression of SOS1 and NHX1 remains to be elucidated and requires further research.
All datasets generated for this study are included in the article/Supplementary Material.
Conceptualization: XS and QH. Methodology: WZ, DL, and CD. Formal analysis: WZ and DL. Data curation: DL and CD. Investigation: YD and JL. Writing—original draft preparation: DL and CD. Writing—review and editing: CD and WZ. Supervision: QH. Project administration: CD and WZ. Funding acquisition: XS and CD. All authors contributed to the article and approved the submitted version.
This research was funded by the National Major Project of GMO New Species Cultivation (Grant No. 2018ZX08020002) and the National High-Tech R&D Program of China (863 Program) (Grant No. 2011AA100201).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Wenxu Zhu and Yanbo Wang for reading and commenting on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01192/full#supplementary-material
Acuña-Rodríguez, I. S., Hansen, H., Gallardo-Cerda, J., Atala, C., Molina-Montenegro, M. A. (2019). Antarctic Extremophiles: Biotechnological alternative to crop productivity in saline soils. Front. Bioeng. Biotech. 7, 22. doi: 10.3389/fbioe.2019.00022
Apse, M. P., Aharon, G. S., Snedden, W. A., Blumwald, E. (1999). Salt tolerance conferred by overexpression of a vacuolar antiporter in Arabidopsis thaliana. Science 285 (5431), 1256–1258. doi: 10.1126/science.285.5431.1256
Azhar, N., Su, N., Shabala, L., Shabala, S. (2017). Exogenously applied 24-epibrassinolide (EBL) ameliorates detrimental effects of salinity by reducing K+ efflux via depolarization-activated K+ channels. Plant Cell Physiol. 58, 802–810. doi: 10.1093/pcp/pcx026
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 (4), 473–488. doi: 10.1007/s11103-008-9435-0
Barragán, V., Leidi, E. O., Andrés, Z., Rubio, L., De Luca, A., Fernández, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24 (3), 1127–1142. doi: 10.1105/tpc.111.095273
Bassil, E., Blumwald, E. (2014). The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 22, 1–6. doi: 10.1016/j.pbi.2014.08.002
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M. A., Ushijima, K., Nakano, R., et al. (2011). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Bassil, E., Zhang, S. Q., Gong, H. J., Tajima, H., Blumwald, E. (2019). Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant Physiol. 179, 616–629. doi: 10.1104/pp.18.01103
Basu, S., Roychoudhury, A. (2014). Expression profiling of abiotic stress-inducible genes in response to multiple stresses in rice (Oryza sativa L.) varieties with contrasting level of stress tolerance. BioMed. Res. Int. 2014 (4), 706890. doi: 10.1155/2014/706890
Blumwald, E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12 (4), 431–434. doi: 10.1016/S0955-0674(00)00112-5
Charfeddine, M., Charfeddine, S., Ghazala, I., Bouaziz, D., Bouzid, R. G. (2019). Investigation of the response to salinity of transgenic potato plants overexpressing the transcription factor StERF94. J. Biosci. 44, 141. doi: 10.1007/s12038-019-9959-2
Chen, Z. H., Pottosin, I. I., Cuin, T. A., Fuglsang, A. T., Tester, M., Jha, D., et al. (2007). Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 145 (4), 1714–1725. doi: 10.1104/pp.107.110262
Chen, Z. X., Zhao, X. Q., Hu, Z. H., Leng, P. S. (2019). Nnitric oxide modulating ion balance in Hylotelephium erythrostictum roots subjected to Nacl stress based on the analysis of transcriptome, fluorescence, and ion fluxes. Sci. Rep. 9, 18317. doi: 10.1038/s41598-019-54611-2
Chen, N. N., Tong, S. F., Tang, H., Zhang, Z. Y., Liu, B., Lou, S. L., et al. (2020). The PalERF109 transcription factor positively regulates salt tolerance via PalHKT1; 2 in Populus alba var. pyramidalis. Tree Physiol. 40 (6), 717–730. doi: 10.1093/treephys/tpaa018
Cheng, C. H., Zhong, Y. M., Wang, Q., Cai, Z. M., Wang, D. D., Li, C. M. (2019). Genome-wide identification and gene expression analysis of SOS family genes in tuber mustard (Brassica juncea var. tumida). PloS One 14 (11), e0224672. doi: 10.1371/journal.pone.0224672
Clarkson, D. T., Hanson, J. B. (1980). The mineral nutrition of higher plants. Annu. Rev. Plant Physiol. 31, 239–298. doi: 10.1146/annurev.pp.31.060180.001323
Cuin, T. A., Betts, S. A., Chalmandrier, R., Shabala, S. (2008). A root's ability to retain K correlates with salt tolerance in wheat. J. Exp. Bot. 59 (10), 2697–2706. doi: 10.1093/jxb/ern128
Ding, M. Q., Hou, P. C., Shen, X., Wang, M. J., Deng, S. R., Sun, J., et al. (2010). Salt-induced expression of genes related to Na and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol. Biol. 73 (3), 251–269. doi: 10.1007/s11103-010-9612-9
Fahmideh, L., Fooladvand, Z. (2018). Isolation and semi quantitative PCR of Na+/H+ antiporter (SOS1 and NHX) genes under salinity stress in. Kochia scoparia. Biol. Proced. Online 20, 11. doi: 10.1186/s12575-018-0076-7
Foster, K. J., Miklavcic, S. J. (2019). A comprehensive biophysical model of ion and water transport in plant roots. II. Clarifying the roles of SOS1 in the salt stress response in Arabidopsis. Front. Plant Sci. 10, 1121. doi: 10.3389/fpls.2017.01326
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., Thomashow, M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124 (4), 1854–1865. doi: 10.1104/pp.124.4.1854
Guo, W. F., Li, G. Q., Wang, N., Yang, C. F., Zhao, Y. N., Peng, H. K., et al. (2020). A Na+/H+ antiporter, K2−NhaD, improves salt and drought tolerance in cotton (Gossypium hirsutum L.). Plant Mol. Biol. 102, 553–567. doi: 10.1007/s11103-020-00969-1
Hasegawa, P. M. (2013). Sodium (Na+) homeostasis and salt tolerance of plants. Environ. Exp. Bot. 92 (8), 19–31. doi: 10.1016/j.envexpbot.2013.03.001
Hu, D. G., Ma, Q. J., Sun, C. H., Sun, M. H., You, C. X., Hao, Y. J. (2016). Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plantarum. 156, 201–214. doi: 10.1111/ppl.12354
Huang, Y. H., Cui, X., Cen, H. F., Wang, K. H., Zhang, Y. W. (2018). Transcriptomic analysis reveals vacuolar Na+ (K+)/H+ antiporter gene contributing to growth, development, and defense in switchgrass (Panicum virgatum L.). BMC Plant Biol. 18, 57. doi: 10.1186/s12870-018-1278-5
Jansson, S., Douglas, C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Boil. 58, 435–458. doi: 10.1146/annurev.arplant.58.032806.103956
Ji, H. T., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., Li, X. (2013). The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6 (2), 275–286. doi: 10.1093/mp/sst017
Jiang, L., Yang, J., Liu, C. X., Chen, Z. P., Yao, Z. C., Cao, S. Q. (2020). Overexpression of ethylene response factor ERF96 gene enhances selenium tolerance in Arabidopsis. Plant Physiol. Bioch. 149, 294–300. doi: 10.1016/j.plaphy.2020.02.024
Kaleem, F., Shabir, G., Aslam, K., Rasul, S., Manzoor, H., Shah, S. M., et al. (2018). An overview of the genetics of plant response to salt stress: Present status and the way forward. Appl. Biochem. Biotech. 186, 306–334. doi: 10.1007/s12010-018-2738-y
Karim, A., Jiang, Y. Z., Guo, L., Ling, Z. Y., Ye, S. L., Duan, Y. J., et al. (2015). Isolation and characterization of a subgroup IIa WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 35 (10), 1129–1139. doi: 10.1093/treephys/tpv084
Kaur, C., Kumar, G., Kaur, S., Ansari, M. W., Pareek, A., Sopory, S. K., et al. (2015). Molecular cloning and characterization of salt overly sensitive gene promoter from Brassica juncea (BjSOS2). Mol. Biol. Rep. 42, 1139–1148. doi: 10.1007/s11033-015-3851-4
Ke, Q., Kim, H. S., Wang, Z., Ji, C. Y., Jeong, J. C., Lee, H. S., et al. (2017). Down-regulation of GIGANTEA-like genes increases plant growth and salt stress tolerance in poplar. Plant Biotech. J. 15 (3), 331–343. doi: 10.1111/pbi.12628
Leidi, E. O., Barragán, V., Rubio, L., El-Hamdaoui, A., Ruiz, M. T., Cubero, B., et al. (2010). The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61, 495–506. doi: 10.1111/j.1365-313X.2009.04073.x
Li, W. Z., Zhang, H. W., Wang, J. Y., Huang, R. F. (2006). Ethylene responsive factors and their application in improving tobacco tolerance to biotic and abiotic stresses. Biotechnol. Bull. 4, 30–34. doi: 10.13560/j.cnki.biotech.bull.1985.2006.04.007
Li, Y. L., Su, X. H., Zhang, B. Y., Huang, Q. J., Zhang, X. H., Huang, R. F. (2009). Expression of jasmonic ethylene responsive factor gene in transgenic polar tree leads to increased salt tolerance. Tree Physiol. 29 (2), 273–279. doi: 10.1093/treephys/tpn025
Li, N. N., Wang, X., Ma, B. J., Du, C., Zheng, L. L., Wang, Y. C. (2017). Expression of a Na+/H+ antiporter RtNHX1 from a recretohalophyte Reaumuria trigyna improved salt tolerance of transgenic. Arabidopsis thaliana. J. Plant Physiol. 218, 109–120. doi: 10.1016/j.jplph.2017.07.015
Liang, W. J., Ma, X. L., Wan, P., Liu, L. Y. (2018). Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 495 (1), 286–291. doi: 10.1016/j.bbrc.2017.11.043
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA2/Ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, T. Z., Zhuang, L. L., Huang, B. R. (2019). Metabolic adjustment and gene expression for root sodium transport and calcium signaling contribute to salt tolerance in Agrostis grass species. Plant Soil 443, 219–232. doi: 10.1007/s11104-019-04140-8
Livak, K. J., Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCt Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, D. M., Xu, W. R., Li, H. W., Jin, F. X., Guo, L. N., Wang, J., et al. (2014). Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 251, 219–231. doi: 10.1007/s00709-013-0540-9
Mahajan, S., Pandey, G. K., Tuteja, N. (2008). Calcium and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471 (2), 146–158. doi: 10.1016/j.abb.2008.01.010
Mahi, H. E., Pérez-Hormaeche, J., Luca, A. D., Villalta, I., Espartero, J., Gámez-Arjona, F., et al. (2019). A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180 (2), 1046–1065. doi: 10.1104/pp.19.00324
Mcgrath, K. C., Dombrecht, B., Manners, J. M., Schenk, P. M., Edgar, C. I., Maclean, D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. doi: 10.1104/pp.105.068544
Meng, K. B., Wu, Y. X. (2018). Footprints of divergent evolution in two Na+/H+ type antiporter gene families (NHX and SOS1) in the genus. Populus. Tree Physiol. 38 (6), 813–824. doi: 10.1093/treephys/tpx173
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 (1), 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 (2), 411–432. doi: 10.1104/pp.105.073783
Qiu, Q. S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., Zhu, J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279, 207–215. doi: 10.1074/jbc.M307982200
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X. Y., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter salt-overlysensitive1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108 (6), 2611–2616. doi: 10.1073/pnas.1018921108
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 (5499), 2105–2110. doi: 10.1126/science.290.5499.2105
Sannigrahi, P., Ragauskas, A. J., Tuskan, G. A. (2010). Poplar as a feedstock for biofuels: a review of compositional characteristics. Biofuel. Bioprod. Bior. 4, 209–226. doi: 10.1002/bbb.206
Shabala, L., Cuin, T. A., Newman, L. A., Shabala, S. (2005). Salinity-induced ion flux patterns from the excised roots of Arabidopsis SOS mutants. Planta 222 (6), 1041–1050. doi: 10.2307/23389100
Shabala, S. (2000). Ionic and osmotic components of salt stress specifically modulate net ion fluxes from bean leaf mesophyll. Plant Cell Environ. 23 (8), 825–837. doi: 10.1046/j.1365-3040.2000.00606.x
Shi, H. Z., Ishitani, M., Kim, C., Zhu, J. K. (2000). The arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97 (12), 6896–6901. doi: 10.1073/pnas.120170197
Shi, H. Z., Lee, B. H., Wu, S. J., Zhu, J. K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21 (1), 81–85. doi: 10.1038/nbt766
Song, X. Q., Yu, X., Hori, C., Demura, T., Ohtani, M., Zhuge, Q. (2016). Heterologous overexpression of poplar SnRK2 genes enhanced salt stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 7, 612. doi: 10.3389/fpls.2016.00612
Su, X. H., Chu, Y. G., Li, H., Hou, Y. J., Zhang, B. Y., Huang, Q. J., et al. (2011). Expression of multiple resistance genes enhances tolerance to environmental stressors in transgenic poplar (Populus euramericana ‘Guariento'). PloS One 6 (9), e24614. doi: 10.1371/journal.pone.0024614
Su, N. N., Wu, Q., Chen, J. H., Shabala, L., Mithöfer, A., Wang, H. Y., et al. (2019). GABA operates upstream of H+-ATPase and improves salinity tolerance in Arabidopsis by enabling cytosolic K+ retention and Na+ exclusion. J. Exp. Bot. 70 (21), 6349–6361. doi: 10.1093/jxb/erz367
Sun, J., Chen, S. L., Dai, S. X., Wang, R. G., Li, N. Y., Shen, X., et al. (2009a). NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 149 (2), 1141–1153. doi: 10.1104/pp.108.129494
Sun, J., Dai, S. X., Wang, R. G., Chen, S. L., Li, N. Y., Zhou, X. Y., et al. (2009b). Calcium mediates root K+/Na+ homeostasis in poplar species differing in salt tolerance. Tree Physiol. 29 (9), 1175–1186. doi: 10.1093/treephys/tpp048
Sze, H., Chanroj, S. (2018). Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 177 (3), 875–895. doi: 10.1104/pp.18.00142
Tang, Q. Y., Feng, M. G. (2002). DPS data processing system for practical statistics (Beijing: Science Press).
Tang, R. J., Liu, H., Bao, Y., Lv, Q. D., Yang, L., Zhang, H. X. (2010). The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 74 (4-5), 367–380. doi: 10.1007/s11103-010-9680-x
Tester, M., Davenport, R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91 (5), 503–527. doi: 10.1093/aob/mcg058
Venema, K., Quintero, F. J., Pardo, J. M., Donaire, J. P. (2002). The Arabidopsis Na+/H+ exchanger AtNHX1 catalyzes low affinity Na+ and K+ transport in reconstituted liposomes. J. Boil. Chem. 277 (4), 2413–2418. doi: 10.1074/jbc.M105043200
Wang, H., Huang, Z. J., Chen, Q., Zhang, Z. J., Zhang, H. B., Wu, Y. M., et al. (2004). Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 55 (2), 183–192. doi: 10.1007/s11103-004-0113-6
Wang, H. Y., Ding, Q., Wang, H. L. (2018). A new Na+/H+ antiporter gene KvNHX1 isolated from the halophyte Kosteletzkya virginica improves salt tolerance in transgenic tobacco. Biotechnol. Biotec. Eq. 32 (6), 1378–1386. doi: 10.1080/13102818.2018.1522972
Wei, Q. H., Luo, Q. C., Wang, R. B., Zhang, F., He, Y., Zhang, Y., et al. (2017). A wheat R2R3-type MYB transcription factor TaODORANT1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front. Plant Sci. 8, 1374. doi: 10.3389/fpls.2017.01374
Widodo, P., H, J., Newbigin, E., Tester, M., Bacic, A., Roessner, U. (2009). Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 60 (14), 4089–4103. doi: 10.1093/jxb/erp243
Wu, S. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 (4), 617. doi: 10.1105/tpc.8.4.617
Xu, Y., Sun, T., Yin, L. P. (2006). Application of non-invasive microsensing system to simultaneously measure both H+ and O2 fluxes around the pollen tube. J. Integr. Plant Biol. 48 (7), 823–831. doi: 10.1111/j.1744-7909.2006.00281.x
Xu, H. X., Jiang, X. Y., Zhan, K. H., Cheng, X. Y., Chen, X. Z., Pardo, J. M., et al. (2008). Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473 (1), 8–15. doi: 10.1016/j.abb.2008.02.018
Xue, Z. Y., Zhi, D. Y., Xue, G. P., Zhang, H., Zhao, Y. X., Xia, G. M. (2004). Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci. 167 (4), 849–859. doi: 10.1016/j.plantsci.2004.05.034
Yamaguchi, T., Hamamoto, S., Uozumi, N. (2013). Sodium transport system in plant cells. Front. Plant Sci. 4, 410. doi: 10.3389/fpls.2013.00410
Yan, S. L., Luo, S. T., Dong, S. S., Zhang, T., Sun, J. R., Wang, N. N., et al. (2015a). Heterotrimeric G-proteins involved in the MeJA regulated ion flux and stomatal closure in Arabidopsis thaliana. Funct. Plant Biol. 42 (2), 126–135. doi: 10.1071/FP14162
Yan, S. L., McLamore, E. S., Dong, S. S., Gao, H. B., Taguchi, M., Wang, N. N., et al. (2015b). The role of plasma membrane H+-ATPase in jasmonate-induced ion fluxes and stomatal closure in Arabidopsis thaliana. Plant J. 83 (4), 638–649. doi: 10.1111/tpj.12915
Yang, Y. Q., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217 (2), 523. doi: 10.1111/nph.14920
Yang, L., Liu, H., Fu, S. M., Ge, H. M., Tang, R. J., Yang, Y., et al. (2017). Na+/H+ and K+/H+ antiporters AtNHX1 and AtNHX3 from Arabidopsis improve salt and drought tolerance in transgenic poplar. Biol. Plantarum 61, 641–650. doi: 10.1007/s10535-017-0724-9
Yao, W. J., Wang, S. J., Zhou, B., Jiang, T. B. (2016). Transgenic poplar overexpressing the endogenous transcription factor ERF76 gene improves salinity tolerance. Tree Physiol. 36 (7), 896–908. doi: 10.1093/treephys/tpw004
Yao, W. J., Zhang, X. M., Zhou, B., Zhao, K., Li, R. H., Jiang, T. B. (2017). Expression pattern of ERF gene family under multiple abiotic stresses in Populus simonii ×. P. nigra. Front. Plant Sci. 8, 181. doi: 10.3389/fpls.2017.00181
Yao, W. J., Zhou, B. R., Zhang, X. M., Zhao, K., Cheng, Z. H., Jiang, T. B. (2019). Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii ×. P. nigra. Gene 707, 189–197. doi: 10.1016/j.gene.2019.04.071
Ye, X., Busov, V., Zhao, N., Meilan, R., Mcdonnell, L., Coleman, H. D., et al. (2011). Transgenic Populus trees for forest products, bioenergy, and functional genomics. Crit. Rev. Plant Sci. 30, 415–434. doi: 10.1080/07352689.2011.605737
Yin, X. C., Xia, Y. Q., Xie, Q., Cao, Y. X., Wang, Z. Y., Hao, G. P., et al. (2020). The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J. Exp. Bot. 71 (6), 1801–1814. doi: 10.1093/jxb/erz549
Yue, Y. S., Zhang, M. C., Zhang, J. C., Duan, L. S., Li, Z. H. (2012). SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 169 (3), 255–261. doi: 10.1016/j.jplph.2011.10.007
Zhang, Z. J., Huang, R. F. (2010). Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Boil. 73, 241–249. doi: 10.1007/s11103-010-9609-4
Zhang, H. X., Hodson, J. N., Williams, J. P., Blumwald, E. (2001). Engineering salt tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 98 (22), 12832–12836. doi: 10.1073/pnas.231476498
Zhang, C. H., Luo, W. C., Li, Y. D., Qin, Z. H., Xiao, Y. F., Zhang, Y., et al. (2019). Transcriptomic Analysis of Seed Germination Under Salt Stress in Two Desert Sister Species (Populus euphratica and P. pruinosa). Front. Genet. 10, 231. doi: 10.3389/fgene.2019.00231
Zhang, X. N., Liu, L. J., Chen, B. W., Qin, Z. H., Xiao, Y. F., Zhang, Y., et al. (2019). Progress in understanding the physiological and molecular responses of Populus to salt stress. Int. J. Mol. Sci. 20 (6), 1312. doi: 10.3390/ijms20061312
Zhao, H., Zhao, X. Y., Li, M. Y., Jiang, Y., Xu, J. Q., Jin, J. J., et al. (2018). Ectopic expression of Limonium bicolor (Bag.) Kuntze DREB (LbDREB) results in enhanced salt stress tolerance of transgenic Populus ussuriensis Kom. Plant Cell Tiss. Org. 132 (1), 123–136. doi: 10.1007/s11240-017-1317-1
Zhao, N., Zhu, H. P., Zhang, H. L., Sun, J., Zhou, J. C., Deng, C., et al. (2018). Hydrogen sulfide mediates K+ and Na+ homeostasis in the roots of salt-resistant and salt-sensitive poplar species subjected to NaCl stress. Front. Plant Sci. 9, 1366. doi: 10.3389/fpls.2018.01366
Zhao, G., Zhao, Y. Y., Lou, W., Su, J. C., Wei, S. Q., Yang, X. M., et al. (2019). Nitrate reductase-dependent nitric oxide is crucial for multi-walled carbon nanotubes-induced plant tolerance against salinity. Nanoscale 11, 10511–10523. doi: 10.1039/C8NR10514F
Zhou, J., Wang, J. Q., Bi, Y. F., Wang, L. K., Tang, L. Z., Yu, X., et al. (2014). Overexpression of PtSOS2 Enhances Salt Tolerance in Transgenic Poplars. Plant Mol. Biol. Rep. 32, 185–197. doi: 10.1007/s11105-013-0640-x
Zhu, J. K., Liu, J. P., Xiong, L. M. (1998). Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10 (7), 1181–1191. doi: 10.2307/3870720
Zhu, J. K. (2000). Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124 (3), 941–948. doi: 10.1104/pp.124.3.941
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6 (2), 66–71. doi: 10.1016/S1360-1385(00)01838-0
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Boil. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 (5), 441–445. doi: 10.1016/S1369-5266(03)00085-2
Keywords: poplar, JERF36s, ion balance, NHX1, SOS1, salt resistance
Citation: Ding C, Zhang W, Li D, Dong Y, Liu J, Huang Q and Su X (2020) Effect of Overexpression of JERFs on Intracellular K+/Na+ Balance in Transgenic Poplar (Populus alba × P. berolinensis) Under Salt Stress. Front. Plant Sci. 11:1192. doi: 10.3389/fpls.2020.01192
Received: 12 March 2020; Accepted: 22 July 2020;
Published: 14 August 2020.
Edited by:
Marcello Mastrorilli, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Shaoliang Chen, Beijing Forestry University, ChinaCopyright © 2020 Ding, Zhang, Li, Dong, Liu, Huang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinjun Huang, aHVhbmdxakBjYWYuYWMuY24=; Xiaohua Su, c3V4aEBjYWYuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.