- 1Agricultural Environment and Resources Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, China
- 2Biotechnology Research Department, Ministry of Education, Mandalay, Myanmar
- 3Food Crops Research Institute/Yunnan Key Laboratory for Rice Genetic Improvement, Yunnan Academy of Agricultural Sciences, Kunming, China
- 4Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Kunming, China
- 5Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), UMR BGPI, TA A 54 K, Montpellier, France
- 6BGPI, Univ Montpellier, CIRAD, INRA, Montpellier, SupAgro, Montpellier, France
The discovery and deployment of new broad-spectrum resistance (R) genes from cultivated rice and its wild relatives is a strategy to broaden the genetic basis of modern rice cultivars to combat rice blast disease. Oryza glaberrima possessing many valuable traits for tolerance to biotic and abiotic stresses, is an elite gene pool for improvement of Asian cultivated rice. An introgression line IL106 derived from O. glaberrima (Acc. IRGC100137) confers complete resistance to Magnaporthe oryzae in blast nursery. Genetic analysis using 2185 BC6F2 progenies derived from a cross between IL106 and the recurrent parent Dianjingyou 1 showed that IL106 harbors a single dominant resistance gene against M. oryzae strain 09BSH-10-5A. This gene was preliminarily mapped on the long arm of chromosome 6 of rice in a region of ca. 0.9 cM delimited by two SSR markers (RM20650 and RM20701). In order to finely map this gene, 17,100 additional progenies were further analyzed. As a result, this gene was further narrowed down to a region flanked by two molecular markers STS69-15 and STS69-7, and co-segregated with 3 molecular markers, RM20676, STS69-21 and STS69-22 on the long arm of chromosome 6. Based on reference genome sequences, this R gene was mapped in silico in 76.1-Kb and 67.7-Kb physical intervals, and containing 4 and 3 NBS-LRR candidate genes in O. sativa cultivar Nipponbare and O. glaberrima cultivar CG14, respectively. Because no blast resistance gene was finely mapped in this physical interval before, this R gene was considered as not described yet and designated as Pi69(t), which is the first identified and finely mapped blast R gene from O. glaberrima, as far as we know. Evaluation of IL106 with 151 blast strains collected from 6 countries in Asia showed that 148 strains are avirulent on IL106, suggesting that Pi69(t) is a broad-spectrum blast R gene, and a promising resistant resource for improvement of Asian cultivated rice.
Introduction
The African cultivated rice, Oryza glaberrima Steud., is well adapted for cultivation in West Africa (Linares, 2002; Sarla and Swamy, 2005), and possesses many valuable traits for tolerance to abiotic stresses, such as salinity, drought and strong weed competitiveness (Sarla and Swamy, 2005). O. glaberrima is also reported to have high level of resistance against several diseases and insect pests, such as Rice yellow mottle virus (Ndjiondjop et al., 1999; Pidon et al., 2017), bacterial leaf blight (Djedatin et al., 2011), blast (Silué and Notteghem, 1991; Rama Devi et al., 2015), green rice leafhopper (Fujita et al., 2010), as well as rice gall midge (Ukwungwu et al., 1998). Although it contains a narrow genetic base compared with other Oryza species (Wang et al., 2014; Meyer et al., 2016; Ndjiondjop et al., 2017), O. glaberrima is considered as an excellent gene reservoir for improvement of Asian cultivated rice, due to its useful traits of agronomic importance (Linares, 2002; Sarla and Swamy, 2005).
Rice blast, caused by the ascomycete fungus Magnaporthe oryzae (syn., Pyricularia oryzae) (Couch and Kohn, 2002), is one of the most destructive diseases for rice, and is responsible for significant yield losses under favorable environmental conditions worldwide (Ou, 1980; Savary et al., 2019). Rice-M. oryzae interactions follow the gene-for-gene relationship (Silué et al., 1992; Jia et al., 2000). Utilization of resistance (R) genes is one of the most economical, effective and environment-friendly approaches for blast control. However, the R genes of rice cultivars are often overcome shortly after their release, due to the emergence of strains of the pathogen virulent on certain R genes (Zeigler et al., 1994). Thus, it is necessary to mine new genes with broad spectrum of resistance against M. oryzae from diversities of rice species and use them in appropriate management strategies for durable control of blast in rice production (Zhu et al., 2000; Raboin et al., 2012; Sester et al., 2014). To date, over 100 blast R genes have been identified and mapped on different chromosomal regions of rice, through broad genetic and linkage analysis in the past decades (Ballini et al., 2008; Ashkani et al., 2016). These R genes have mainly been identified from O. sativa, and only 5 R genes originated from wild species of genus Oryza, including Pi40 (O. australiensis), Pi54rh (O. rhizomatis), Pi54 (O. officinalis), Pi57(t) (O. longistaminata), and Pid3-A4 (O. rufipogon) (Jeung et al., 2007; Das et al., 2012; Lv et al., 2013; Devanna et al., 2014; Dong et al., 2017). O. glaberrima was domesticated from its wild progenitor O. barthii independently from O. sativa (Sweeney and McCouch, 2007). Although several studies previously reported that O. glaberrima expressed high level of resistance against rice blast (Silué and Notteghem, 1991; Rama Devi et al., 2015), no blast R gene locus was further identified and mapped yet. Whether blast R genes in O. glaberrima are different from R genes identified from other Oryza species so far remains unknown.
In order to discover useful genes of agronomic importance from O. glaberrima, a set of BC5F4 introgression lines (ILs) was constructed through successive backcross strategy between IRGC100137, an accession of O. glaberrima and O. sativa cultivar Dianjingyou 1 (DJY1), an O. sativa subsp. japonica cultivar, used as male recurrent parent (Xu et al., 2014). The ILs were evaluated for blast resistance in blast nursery in the field and by artificial inoculation with M. oryzae isolates in the greenhouse. Twelve ILs showing complete resistance to M. oryzae compared with the susceptible recurrent parent DJY1 were obtained. In this study, we describe the identification and fine mapping of a new blast resistance locus Pi69(t) from O. glaberrima.
Materials and Methods
Rice Materials and Mapping Population Construction
Resistant introgression line IL106 derived from O. glaberrima (accession No. IRGC100137) was crossed with a susceptible recurrent parent Dianjingyou 1 (DJY1) to generate BC6F1 seeds, the BC6F1 seeds were further sown and grown in a greenhouse to generate BC6F2 population for linkage and genetic analysis for resistance to rice blast. Resistant donor IL106, 10 monogenic lines (IRBLZ-Fu (Piz), IRBLZ5-CA (Pi2), IRBLzt-T (Piz-t), IRBL9-W (Pi9), IRBL5-M (Pi5), IRBLKH-K3 (Pikh), IRBL1-CL (Pi1), IRBL7-M (Pi7), IRBL20-IR24 (Pi20), and IRBLTA2-PI (Pita2)), as well as susceptible control cultivar Lijiangxintuanheigu (LTH) were used to test resistant/susceptible phenotypes to 151 M. oryzae strains.
M. oryzae Cultivation and Spore Production
The M. oryzae isolate 09BSH-10-5A that is avirulent to IL106 and virulent to DJY1 was cultured on oatmeal medium (20 g of oatmeal, 15 g of agar, 10 g of sucrose, and 1 L of distilled water) for 7 days in the dark at 25°C. Then aerial mycelia were washed off by gentle rubbing with distilled water and paintbrush. The colony was then successively exposed to fluorescent light for 3 days to induce sporulation at 25°C. Conidia were harvested by softly scraping and flooding the medium surface with distilled water containing 0.01% Tween 20 detergent. The concentration of conidial suspension was adjusted to 50,000 conidia/ml for inoculation (Dong et al., 2017).
Plant Planting and Pathotesting
The BC6F2 population seeds derived from the cross between IL106 and DJY1 were sown in plastic trays of 20×12×5 cm filled with paddy soil, and each tray was sowed with 95 germinated seeds. Seedlings were inoculated with M. oryzae strain 09BSH-10-5A by spraying at 4-leaf stage with 20 ml conidial suspension per tray. The inoculated rice plants were incubated overnight in a dark chamber at 25°C for 24 h with over 95% relative humidity, and then transferred back to the greenhouse. Lesion types on rice leaves were observed 6–7 days after inoculation and scored according to a standard reference scale (Silué et al., 1992). Plants scored from 1 to 3 were considered to be resistant and plants scored from 4 to 6 were considered to be susceptible. Furthermore, 151 M. oryzae isolates from 6 countries were used to test the resistant spectrum of Pi69(t) gene carrying in IL106 and 10 known blast R genes carrying in monogenic lines.
Marker Development and Genetic Map Construction
Genomic DNA was extracted from fresh leaves of each plant following the method described by Edwards et al. (1991). A total of 229 SSR markers distributed evenly across all 12 rice chromosomes (McCouch et al., 2002) were used for identification of introgressed regions from O. glaberrima. Sequence-tagged site (STS) markers were developed within the critical region based on the sequence alignment of the genomic sequences of Nipponbare (O. sativa) and CG14 (O. glaberrima, http://plants.ensembl.org/Oryza_glaberrima/Info/Index).
PCR amplification conditions consisted of a denaturing step of 94°C/3 min, followed by 35 cycles of 94°C/30 s, annealing temperature 55°C/30 s, and 72°C/1 min, ending with an extension step of 72°C/7 min. Amplicons were separated by 8% polyacrylamide gel electrophoresis and detected by silver staining. Information of all primers used for gene mapping in this study is listed in Table 1. The genetic and linkage map of polymorphic markers was constructed using MAPMAKER/EXP 3.0 (Lander et al., 1987). The Kosambi mapping function was used to transform recombination frequency to genetic distance (cM).
Physical Map Construction In Silico and Candidate Gene Annotation
To construct physical map of Pi69(t) in silico based on the reference genome sequence of O. sativa subsp. japonica cultivar Nipponbare, all molecular markers were anchored on chromosome 6 of Os-Nipponbare-Reference-IRGSP-1.0 pseudomolecules by BLAST (https://blast.ncbi.nlm.nih.gov/). To annotate the candidate R genes, both the 76.1-Kb and 67.7-Kb target regions in Nipponbare and CG14 respectively were analyzed by using the FGENSH platform (http://www.softberry.com/).
Results
Genetic Analysis for Blast Resistance in IL106
The resistant donor IL106, recurrent parent DJY1, BC6F1 plants from IL106/DJY1 and BC6F2 population were inoculated with 09BSH-10-5A (Supplementary Figures S1A, B). The resistant donor IL106 and BC6F1 plants showed complete resistance, and recurrent parent DJY1 was susceptible to 09BSH-10-5A. The segregation of resistant and susceptible progenies among 2185 BC6F2 individuals fitted with an expected 3:1 ratio (resistant/susceptible: 1664/521, χ2 = 1.556, P = 0.212), indicating that a single dominant R gene from IL106 confers complete resistance to M. oryzae strain 09BSH-10-5A.
Identification and Mapping of R Gene Locus in IL106
To identify and map the R locus in IL106, a total of 217 SSR markers distributed evenly across all 12 rice chromosomes were used to determine the polymorphism between resistance donor IL106 and recurrent parent DJY1. As expected, a large majority of markers were monomorphic between IL106 and its recurrent parent. Three SSR markers, RM345 on chromosome 6, RM6329 on chromosome 3, and RM3702 on chromosome 8, showed polymorphism between IL106 and DJY1, suggesting that three introgression fragments from O. glaberrima possessed in IL106. In order to verify the linkage relationship between these 3 SSR loci and R gene in IL106, 94 random susceptible individuals from BC6F2 population inoculated with 09BSH-10-5A were genotyped with these 3 SSR markers. The results showed that the severe segregating distortion (92 homozygotes of susceptible allele to 2 heterozygotes) was only detected for RM345, implying linkage between the R gene and this marker which located on chromosome 6.
To determine the O. glaberrima introgression length of the R gene region in IL106, 27 SSR markers located on the long arm of chromosome 6 were selected to survey the polymorphism between IL106 and DJY1. The result showed that the introgression fragment was located between SSR markers RM30 and RM345, and 4 SSR markers (RM20625, RM20650, RM20676, and RM20701) within this interval were also polymorphic. Subsequently, the mapping population consisting of 2,185 BC6F2 plants was genotyped with the two SSR markers RM30 and RM345, and the recombinants were further genotyped with 4 SSR markers (RM20625, RM20650, RM20676, and RM20701) to map the R gene location. Taken together, the R locus was mapped to a 0.9 cM region flanked by RM20650 and RM20701 on the long arm of chromosome 6, and co-segregated with RM20676 (Figure 1A).
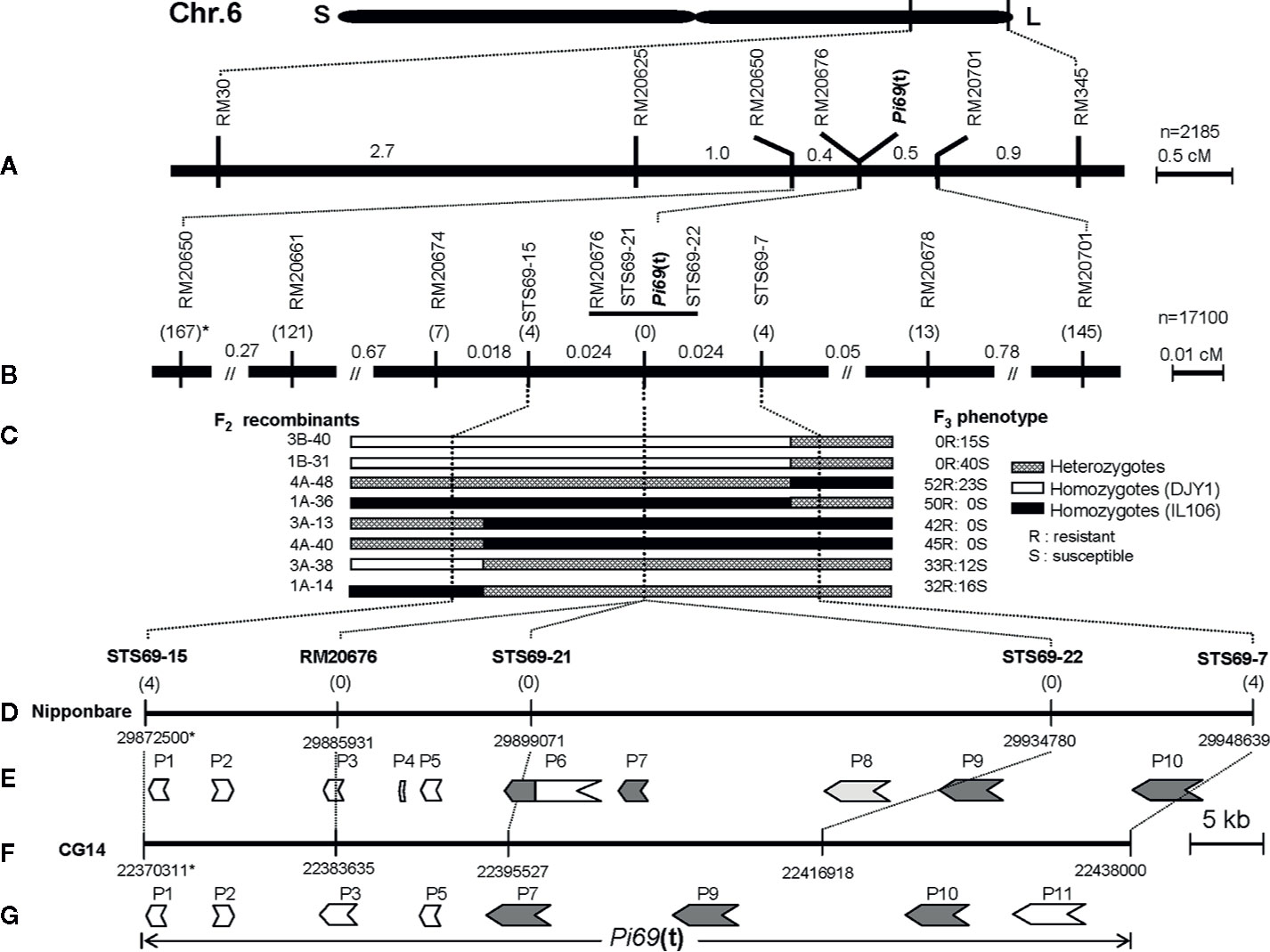
Figure 1 Genetic and physical maps of Pi69(t) locus on rice chromosome 6. (A) A genetic map of Pi69(t) locus. Map distances are in cM. (B) An integrated fine genetic map of Pi69(t) locus on chromosome 6, *: the numbers in parentheses under the markers present the number of recombinants between marker loci and Pi69(t); (C) The genotyping and phenotyping of key recombinants located between two markers STS69-15 and STS69-7. (D) Physical map of the Pi69(t) locus based on the reference genome sequence of O. savita cultivar Nipponbare. *: chromosomal position of markers on genomic sequence of chromosome 6 of Nipponbare; (E) Predicted candidate R genes for Pi69(t) in Nipponbare (O. sativa). (F) Physical map of the Pi69(t) locus based on the reference genome sequence of O. glaberrima accession CG14. *: chromosomal position of markers on genomic sequence of chromosome 6 of CG14; (G) Predicted candidate R genes for Pi69(t) in CG14 (O. glaberrima).
Fine Mapping of R Gene Locus in IL106
To further map this R gene locus, 17,100 additional BC6F2 plants were genotyped with two flanking markers RM20650 and RM20701. All the 312 recombinants were then phenotyped for resistance to M. oryzae strain 09BSH-10-5A. As a result, 167 and 145 recombinants were detected between phenotypes (R or S) and RM20650 and RM20701 genotypes, respectively (Figure 1B). Meanwhile, new SSR markers located in RM20650-RM20701 interval were surveyed for polymorphism between IL106 and DJY1, and 3 polymorphic SSR markers RM20661, RM20674, and RM20678 were obtained. Then, the 312 recombinants were genotyped with these new SSR markers and RM20676. The recombination events btweeen the R locus and RM20661, RM20674, RM20676, and RM20678 were 121, 7, 0, and 13, respectively (Figure 1B), The R locus was linked to RM20661 and RM20674 by a genetic distance of ca. 0.71, and 0.041 cM, co-segregated with RM20676, and linked to RM20678 by 0.076 cM on the other side, respectively.
To further finely map the R locus, 24 STS markers were developed based on the genome sequences of Nipponbare and CG14. Five STS markers polymorphic between IL106 and DJY1 were used to genotype the 7 and 13 recombinants at RM20674 and RM20678 loci, respectively. As a result, 4 recombinants were detected at STS69-15 locus on RM20674 side, and 4 recombinants were detected at STS69-7 locus on RM20678 side. No recombinants were identified at RM20676, STS69-21, and STS69-22 loci. These results indicated that the R gene locus was narrowed down to the region flanked by STS69-15 and STS69-7, and co-segregated with 3 molecular markers RM20676, STS69-21, and STS69-22 (Figure 1B). The genotypes and phenotypes of 8 recombinants between STS69-15 and STS69-7 are shown in Figure 1C.
Differentiation Between R Gene Carrying in IL106 and Pi-tq1 in Teqing
The blast resistant gene Pi-tq1 from indica cultivar Teqing was previously mapped to a 4.24 Mb physical interval flanked by two RFLP markers, RZ682 and RZ508 (Tabien et al., 2000) that spanned over the R locus described here on the long arm of chromosome 6. To distinguish these two genes, IL106 and Teqing were inoculated with 3 M. oryzae strains (09BSH-10-5A, BS139, and HN09-1C-7) that are avirulent on IL106, but virulent on DJY1. The result showed that Teqing was resistant to both 09BSH-10-5A and BS139, but susceptible to HN09-1C-7 (Supplementary Figure S1C), suggesting that the R gene in IL106 could be different from Pi-tq1, because of their distinct reactions to HN09-1C-7. To demonstrate that the resistance of HN09-1C-7 is controlled by the same R gene in IL106, this strain was inoculated to 191 F2 progenies from the cross between IL106 and DJY1. The numbers of resistant and susceptible individuals were 149 and 42, respectively and fitted to a 3:1 ratio (P=0.337), confirming that IL106 possesses a single resistance gene to the strain HN09-1C-7. Genotyping of 42 susceptible and 4 resistant individuals with 4 molecular markers (RM30, RM345, STS69-7, and STS69-15) linked with Pi69(t), identified 4 and 3 recombinants at RM30 and RM345 loci, respectively, and no recombinants at both STS69-7 and STS69-15 loci (Supplementary Figure S2), indicating that the gene conferring resistance to HN09-1C-7 in IL106 was mapped to the same chromosomal region as that R gene conferring resistance to 09BSH-10-5A, and the resistance to both 09BSH-10-5A and HN09-1C-7 is controlled by the same R gene in IL106. Taken together, all these data confirmed that R locus in IL106 is different from Pi-tq1, due to their distinct reactions to M. oryzae strain HN09-1C-7. Because no blast R gene was finely mapped in chromosomal region flanked by STS69-15 and STS69-7 on chromosome 6 of rice to date, this major R gene carrying in IL106 from O. glaberrima was considered as a new gene and was tentatively designated as Pi69(t).
Resistance Spectrum of Pi69(t)
To determine the resistance spectrum of Pi69(t), IL106, and other 10 monogenic lines carrying broad-spectrum R genes were inoculated and assessed with 151 M. oryzae strains from Cambodia (16 strains), Laos (20 strains), Myanmar (4 strains), Thailand (20 strains), Vietnam (18 strains), and China (77 strains). IL106 was resistant to all strains from Cambodia and Laos, and susceptible to only 3 strains from China, Thailand and Vietnam (YX162, TH451, and VN4118; Supplementary Table S1). IL106 also showed broader resistant spectrum compared with nine monogenic lines carrying different known R genes, except for the line IRBL9-W carrying Pi9 that was resistant to all tested strains. These results suggest that Pi69(t) gene could confer broad-spectrum resistance against M. oryzae in IL106.
In Silico Physical Map Construction of Pi69(t) Gene Locus
To construct the physical map of Pi69(t) locus in silico, all the molecular markers closely linked to Pi69(t) were anchored to the genome sequences of both O. sativa subsp. japonica cultivar Nipponbare (IRGSP1.0) and O. glaberrima cultivar CG14 (AGI1.1) through BLAST analysis (http://plants.ensembl.org/). The two flanking markers and 3 co-segregating markers were anchored to the target region (Figures 1D, F). The physical distance between two closest flanking markers STS69-15 and STS69-24 was about 76.1 Kb (genomic position: 29872500-29948639) in Nipponbare, and 67.7 Kb (genomic position: 22370311- 22438000) in CG14.
Both the target genome sequences from Nipponbare and CG14 were annotated through the bioinformatics platform FGENSH (http://www.softberry.com). The annotation showed that 8 and 10 genes (named tentatively from P1 to P11) were predicted in Nipponbare and CG14, respectively (Figures 1E, G). Among all these annotated genes, P4 and P8 genes were absent in CG14, while P11 gene was absent in Nipponbare. Among these predicted genes, both P1(LOC_Os6g49300) and P3(LOC_Os6g49320) encode the putative genes homologous with glycosyltransferase; P2(LOC_Os6g49310) encodes a gene homologous to MATE efflux family protein; P4 (LOC_Os6g49330) annotated in Nipponbare only is an uncharacterized protein; P5 (LOC_Os6g49340) encodes a F-box and DUF domain containing protein; P6 (LOC_Os6g49350) and P11 encode a retrotransposon in Nipponabre and CG14 respectively; the remaining four genes (P7 (LOC_Os6g49360), P8 (LOC_Os6g49380), P9 (LOC_Os6g49390), and P10 (LOC_Os6g49420)) were predicted to be typical R genes encoding protein with the conserved structure of nucleotide-binding site and leucine-rich repeat (NBS-LRR; Figures 1E, G). In comparison with P7 in CG14, there are two genes (P6 and P7) in Nipponbare caused by an insertion of retrotransposon. Amino acids analysis among P7 in CG14, P6, and P7 in Nipponbare showed that the amino acid sequence on the 3’ side of P6 in Nipponbare has high similarity with those in P7 of CG14, and P7 in Nipponbare encoded only a truncated NBS-LRR protein compared with its P7 allele in CG14.
Evidence of O. glaberrima Genome Fragment Integration
To validate whether the fragment carrying Pi69(t) in IL106 was integrated from O. glaberrima, the introgression line IL106, the original O. glaberrima accession IRGC100137, the recurrent parent japonica cultivar DJY1, as well as two indica cultivars R498 and Teqing, were genotyped with 13 molecular markers used for mapping Pi69(t). The results showed that the size of all the DNA fragments amplified from IL106 were the same as those from IRGC100137 (Figure 2). Meanwhile, these molecular markers were polymorphic among line IL106, and all rice cultivars (DJY1, R498 and Teqing).
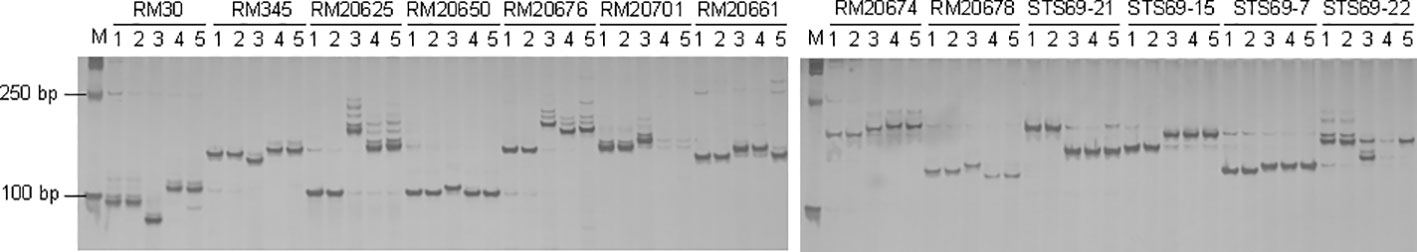
Figure 2 Confirmation of the origin of the introgression of the Pi69(t). Thirteen molecular markers linked with Pi69(t) were used to amplify the DNA fragments of the O. glaberrima (IRGC100137) donor parent (1), the introgression line IL106 (2), the recurrent parent DJY1 (3), and two indica cultivars: R498 (4), and Teqing (5). M, molecular weight marker DL2000. The PCR products were separated by 8% polyacrylamide gel.
Discussion
The discovery and deployment of broad-spectrum R genes from a large number of cultivated rice varieties and its wild relatives is an effective strategy to broaden the genetic basis of resistance of modern rice cultivars, to cope with the diversity and variability over time of pathogen population in rice production (Jeung et al., 2007; Su et al., 2015; Deng et al., 2017). Several incompatibility barriers such as pre- and post-fertilization barriers, hybrid sterility between Asian cultivated rice and its wild and cultivated relatives, however, have been hindering the utilization of favorable genes controlling important agronomic traits (Brar and Khush, 1997; Xu et al., 2014; Brar and Khush, 2018). Construction of introgression lines of these relatives with Asian cultivated rice cultivars has been proved to be one of the effective measures for further discovery and use of favorable genes from the wild species for rice breeding, and several genes conferring resistance to biotic stresses from wild species were identified in introgression lines (Brar and Khush, 1997; Gutierrez et al., 2010; Rama Devi et al., 2015; Brar and Khush, 2018). Extensive studies on exploiting and identification of blast resistance genes have been conducted, mainly focusing on the O. sativa and wild relatives. Relatively few investigations have been performed to identify new blast R genes in O. glaberrima (Silué and Notteghem, 1991; Rama Devi et al., 2015). In this study, we have successfully identified and finely mapped Pi69(t), the first blast R gene from O. glaberrima by using an introgression line IL106 derived from O. glaberrima. Pi69(t) confers a broad-spectrum resistance to M. oryzae diverse strains from 6 Asian countries, indicating that Pi69(t) is a promising resistance resource for improvement of Asian cultivated rice for resistance to rice blast.
Over 20 blast major R genes have been identified and mapped on rice chromosome 6, and the majority of them were mapped proximal to the centromere. The cloned R genes Piz-t, Pi2, Pi9, Pi50, and Pigm are members of the multigene family Pi2/Pi9 locus located on the short arm, while the Pid-2, Pid3/Pi25(t), and Pid3-A4 are located on the long arm (Chen et al., 2006; Shang et al., 2009; Chen et al., 2011; Lv et al., 2013). Using recombination inbred lines derived from a cross between Lemont and Teqing, Tabien et al. (2000) identified and mapped 3 blast resistance genes from Teqing (Pi-tq1, Pi-tq2, and Pi-tq3). Among them, Pi-tq1 was also mapped to chromosome 6 of rice but located proximal to telomeric side. Two flanking markers RZ682 and RZ508 defined a larger physical region of Pi-tq1 locus of around 4.24 Mb covering Pi69(t) locus. Although Pi69(t) could be differentiated from Pi-tq1 by using M. oryzae strain HN09-1C-7, due to their distinct reactions to this strain, whether Pi69(t) is allelic, or closely linked to Pi-tq1 remains to be determined through allelism test or fine mapping of Pi-tq1.
Pyramiding of R genes with different resistance specificity in the same cultivar is one effective measure to broaden the resistance spectrum against M. oryzae, and development of polymorphic molecular markers is the prerequisite to stack target genes into one cultivar with marker-assisted selection method (Hittalmani et al., 2000). The molecular markers developed in this study, tightly linked to Pi69(t), showed good polymorphisms among 5 tested rice lines/cultivars belonging to indica or japonica types. These markers are good candidates for pyramiding of Pi69(t) with other R genes for improvement of Asian cultivated cultivars in disease-resistant rice breeding program.
Most of cloned resistance genes from plants encode NBS-LRR like proteins that directly or indirectly recognize the pathogen effectors to trigger host defense responses (Dangl and Jones, 2001). Almost all the cloned rice R genes to blast encode NBS-LRR like proteins, except for Pid2, pi21, and Ptr genes, which encode a B-lectin receptor kinase, a proline-rich protein, and an atypical protein with amardillo repeat domain, respectively (Lv et al., 2013; Su et al., 2015; Ashkani et al., 2016; Deng et al., 2017; Zhao et al., 2018). O. glaberrima-derived Pi69(t) gene was located in a region containing a cluster of NBS-LRR like genes. These genes are potential or promising candidates for Pi69(t).
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material
Author Contributions
QY, DaT, LD, SL, PX, and DiT: Conceived idea and designed research. QY, DaT, and DiT wrote the manuscript. LD, SL, MK, QY, WD, XL, YB, LZ, JL, and JZ: Performed experiments and analyzed data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31860524) to LD, the Applied Basic Research Programs of Yunnan Academy of Agricultural Sciences (YJZ201803) to LD, the National Natural Science Foundation of China (31560493) to QY, the Applied Basic Research Programs of Yunnan Academy of Agricultural Sciences (YJM201707) to SL, the Key Research and Development Program of Yunnan Province (2019IB007), the Scientific Observing and Experimental Station of Crops Pests in Kunming, the Ministry of Agricultural, and the Rural Affairs of China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Khay S, Dr. Douangboupha B, Mr. Mya WA, Dr. Somrith A, Dr. Chung NH, and Dr. Zhang S for the provision of blast isolates, and to Dr. Zhu XY for providing seeds of the rice cultivar Teqing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01190/full#supplementary-material
References
Ashkani, S., Rafili, M. Y., Shabanimofrad, M., Ghasemzadeh, A., Ravanfar, S. A., Latif, M. A. (2016). Molecular progress on the mapping and cloning of functional genes for blast resistance in rice (Oryza sativa L.): current status and future considerations. Crit. Rev. Biotechnol. 36, 353–367. doi: 10.3109/07388551.2014.961403
Ballini, E., Morel, J. B., Droc, G., Price, A., Courtois, B., Notteghem, J. L., et al. (2008). A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 21, 859–868. doi: 10.1094/MPMI-21-7-0859
Brar, D. S., Khush, G. S. (1997). Alien introgression in rice. Plant Mol. Biol. 35, 35–47. doi: 10.1023/A:1005825519998
Brar, D. S., Khush, G. S. (2018). “Wild relatives of rice: a valuable genetic resource for genomics and breeding research,” in The Wild Oryza Genomes, Compendium of Plant Genomes. Eds. Mondal, T. K., Henry, R. J. (Cham: Springer), 1–25. doi: 10.1007/978-3-319-71997-9_1
Chen, X. W., Shang, J. J., Chen, D. X., Lei, C. L., Zou, Y., Zhai, W. X., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. doi: 10.1111/j.1365-313X.2006.02739.x
Chen, J., Shi, Y. F., Liu, W. Z., Chai, R. Y., Fu, Y. P., Zhuang, J. Y., et al. (2011). A Pid3 allele from rice cultivar Gumei 2 confers resistance to Magnaporthe oryzae. J. Genet. Genomics 38, 209–216. doi: 10.1016/j.jgg.2011.03.010
Couch, B. C., Kohn, L. M. (2002). A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94, 683–693. doi: 10.1080/15572536.2003.11833196
Dangl, J. L., Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Das, A., Soubam, D., Singh, P., Thakur, S., Singh, N., Sharma, T. (2012). A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genomic. 12, 215–228. doi: 10.1007/s10142-012-0284-1
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Devanna, N. B., Vijayan, J., Sharma, T. R. (2014). The blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PloS One 9 (8), e104840. doi: 10.137/journal.pone.0104840
Djedatin, G., Ndjiondjop, M. N., Mathieu, T., Vera Cruz, C. M., Sanni, A., Ghesquière, A., et al. (2011). Evaluation of African cultivated rice Oryza glaberrima for resistance to bacterial blight. Plant Dis. 95, 441–447. doi: 10.1094/PDIS-08-10-0558
Dong, L., Liu, S., Xu, P., Deng, W., Li, X., Tharreau, D., et al. (2017). Fine mapping of Pi57(t) conferring broad spectrum resistance against Magnaporthe oryzae in introgression line IL-E1454 derived from Oryza longistaminata. PloS One 12 (10), e0186201. doi: 10.1371/journal.pone.0186201
Edwards, K., Johnstone, C., Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic. Acids Res. 19, 1349. doi: 10.1093/nar/19.6.1349
Fujita, D., Doi, K., Yoshimura, A., Yasui, H. (2010). A major QTL for resistance to green rice leafhopper (Nephotettix cincticeps Uhler) derived from African rice (Oryza glaberrima Steud.). Breed. Sci. 60, 336–341. doi: 10.1270/jsbbs.60.336
Gutierrez, A., Carabali, S., Giraldo, O. X., Martinez, C. P., Correa, F., Prado, G., et al. (2010). Identification of a Rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines. BMC Plant Biol. 10, 6. doi: 10.1186/1471-2229-10-6
Hittalmani, S., Parco, A., Mew, T. V., Zeigler, R. S., Huang, N. (2000). Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 100, 1121–1128. doi: 10.1007/s001220051395
Jeung, J. U., Kim, B. R., Cho, Y. C., Han, S. S., Moon, H. P., Lee, Y. T., et al. (2007). A novel gene, Pi40(t), linked to the DNA markers derived from NBS-LRR motifs confers broad spectrum of blast resistance in rice. Theor. Appl. Genet. 115, 1163–1177. doi: 10.1007/s00122-007-0642-x
Jia, Y. L., McAdams, S. A., Bryan, G. T., Hershey, H. P., Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. doi: 10.1093/emboj/19.15.4004
Lander, E. S., Green, P., Abrahanson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkages of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Linares, O. F. (2002). African rice (Oryza glaberrima): History and future potential. Proc. Natl. Acad. Sci. U.S.A. 99, 16360–16365. doi: 10.1073/pnas.252604599
Lv, Q., Xu, X., Shang, J., Jiang, G., Pang, Z., Zhou, Z., et al. (2013). Functional analysis of Pid3-A4, an ortholog of rice blast resistance gene Pid3 revealed by allele mining in common wild rice. Phytopathology 103, 594–599. doi: 10.1094/PHYTO-10-12-0260-R
McCouch, S., Teytelman, L., Xu, Y., Lobos, K., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Meyer, R. S., Choi, J. Y., Sanches, M., Plessis, A., Flowers, J. M., Amas, J., et al. (2016). Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat. Genet. 48, 1083–1088. doi: 10.1038/ng.3633
Ndjiondjop, M. N., Albar, L., Fargette, D., Fauquet, C., Ghesquière, A. (1999). The genetic basis of high resistance to rice yellow mottle virus (RYMV) in cultivars of two cultivated rice species. Plant Dis. 83, 931–935. doi: 10.1094/PDIS.1999.83.10.931
Ndjiondjop, M. N., Semagn, K., Gouda, A. C., Kpeki, S. B., Dro Tia, D., Sow, M., et al. (2017). Genetic variation and population structure of Oryza glaberrima and development of a mini-core collection using DArTseq. Front. Plant Sci. 8, 1748. doi: 10.3389/fpls.2017.01748
Ou, S. H. (1980). Pathogen variability and host resistance in rice blast disease. Annu. Rev. Phytopathol. 18, 167–187. doi: 10.1146/annurev.py.18.090180.001123
Pidon, H., Ghesquière, A., Chéron, S., Issaka, S., Hébrard, E., Sabot, F., et al. (2017). Fine mapping of RYMV3: a new resistance gene to rice yellow mottle virus from Oryza glaberrima. Theor. Appl. Genet. 130, 807–818. doi: 10.1007/s00122-017-2853-0
Raboin, L. M., Ramanantsoanirina, A., Dusserre, J., Razasolofonanahary, F., Tharreau, D., Lannou, C., et al. (2012). Two-component cultivar mixtures reduce rice blast epidemics in an upland agrosystem. Plant Pathol. 61, 1103–1111. doi: 10.1111/j.1365-3059.2012.02602.x
Rama Devi, S. J. S., Singh, K., Umakanth, B., Vishalakshi, B., Renuka, P., Vijay Sudhakar, K., et al. (2015). Development and identification of novel rice blast resistant sources and their characterization using molecular markers. Rice Sci. 22, 300–308. doi: 10.1016/j.rsci.2015.11.002
Sarla, N., Swamy, B. P. M. (2005). Oryza glaberrima: A source for the improvement of Oryza sativa. Curr. Sci. 89, 955–963.
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Sester, M., Raveloson, H., Tharreau, D., Dusserre, J. (2014). Conservation agriculture cropping system to limit blast disease in upland rainfed rice. Plant Pathol. 63, 373–381. doi: 10.1111/ppa.12099
Shang, J., Tao, Y., Chen, X., Zou, Y., Lei, C., Wang, J., et al. (2009). Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site-leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182, 1303–1311. doi: 10.1534/genetics.109.102871
Silué, D., Notteghem, J. (1991). Resistance of 99 Oryza glaberrima varieties to blast. Int. Rice Res. Newsl. 16, 13–14.
Silué, D., Notteghem, J. L., Tharreau, D. (1992). Evidence of a gene-for-gene relationship in the Oryza sativa-Magnaporthe grisea pathosystem. Phytopathology 82, 577–580. doi: 10.1094/Phyto-82-577
Su, J., Wang, W., Han, J., Chen, S., Wang, C., Zeng, L., et al. (2015). Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theor. Appl. Genet. 128, 2213–2225. doi: 10.1007/s00122-015-2579-9
Sweeney, M., McCouch, S. (2007). The complex history of the domestication of rice. Ann. Bot. 100, 951–957. doi: 10.1093/aob/mcm128
Tabien, R. E., Li, Z., Paterson, A. H., Marchetti, M. A., Stansel, J. W., Pinson, S. R. M. (2000). Mapping of four major rice blast resistance genes from ‘Lemont’ and ‘Teqing’ and evaluation of their combinatorial effect for field resistance. Theor. Appl. Genet. 101, 1215–1225. doi: 10.1007/s001220051600
Ukwungwu, M. N., Williams, C. T., Okhidievbie, O. (1998). Screening of African rice Oryza glaberrima Steud, for resistance to the African rice gall midge Orseolia oryzivora Harris and Gagné. Insect Sci. Applic. 18, 167–170. doi: 10.1017/S1742758400007827
Wang, M., Yu, Y., Haberer, G., Marri, P. R., Fan, C., Goicoechea, J. L., et al. (2014). The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46, 982–991. doi: 10.1038/ng.3044
Xu, P., Zhou, J., Li, J., Hu, F., Deng, X., Feng, S., et al. (2014). Mapping three new interspecific hybrid sterile loci between Oryza sativa and O. glaberrima. Breed. Sci. 63, 476–482. doi: 10.1270/jsbbs.63.476
Zeigler, R. S., Tohme, J., Nelson, J., Levy, M., Correa, F. (1994). “Linking blast population analysis to resistance breeding (1994) A proposed strategy for durable resistance,” in Rice Blast Disease. Eds. Zeigler, R. S., Leong, S. A., Teng, P. S. (Wallingford, United Kindom: CAB International and IRRI), 16–26.
Zhao, H., Wang, X., Jia, Y., Minkenberg, B., Wheatley, M., Fan, J., et al. (2018). The rice blast resistance Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9, 2039. doi: 10.1038/s41467-018-04369-4
Keywords: Oryza glaberrima, introgression line, Magnaporthe oryzae, Pyricularia oryzae, resistance gene, fine mapping
Citation: Dong L, Liu S, Kyaing MS, Xu P, Tharreau D, Deng W, Li X, Bi Y, Zeng L, Li J, Zhou J, Tao D and Yang Q (2020) Identification and Fine Mapping of Pi69(t), a New Gene Conferring Broad-Spectrum Resistance Against Magnaporthe oryzae From Oryza glaberrima Steud. Front. Plant Sci. 11:1190. doi: 10.3389/fpls.2020.01190
Received: 14 April 2020; Accepted: 22 July 2020;
Published: 07 August 2020.
Edited by:
Hanwei Mei, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Yan Li, Sichuan Agricultural University, ChinaMingfu Zhao, Fujian Academy of Agricultural Sciences, China
Copyright © 2020 Dong, Liu, Kyaing, Xu, Tharreau, Deng, Li, Bi, Zeng, Li, Zhou, Tao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayun Tao, dGFvZHkxMkBhbGl5dW4uY29t; Qinzhong Yang, cXpoeWFuZ0AxNjMuY29t
†These authors have contributed equal to this work
 Liying Dong
Liying Dong Shufang Liu
Shufang Liu May Sandar Kyaing
May Sandar Kyaing Peng Xu
Peng Xu Didier Tharreau
Didier Tharreau Wei Deng3
Wei Deng3 Xundong Li
Xundong Li Yunqing Bi
Yunqing Bi Li Zeng
Li Zeng Jing Li
Jing Li Jiawu Zhou
Jiawu Zhou Dayun Tao
Dayun Tao Qinzhong Yang
Qinzhong Yang