
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 July 2020
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01150
This article is part of the Research Topic Nitrogen Use Efficiency and Sustainable Nitrogen Management in Crop Plants View all 23 articles
Proper allocation of nitrogen (N) from source leaves to grains is essential step for high crop grain yield and N use efficiency. In rice (Oryza sativa) grown in flooding paddy field, amino acids are the major N compounds for N distribution and re-allocation. We have recently identified that Lysine-Histidine-type Transporter 1 (OsLHT1) is the major transporter for root uptake and root-to-shoot allocation of amino acids in rice. In this study, we planted knockout mutant lines of OsLHT1 together wild-type (WT) in paddy field for evaluating OsLHT1 function in N redistribution and grain production. OsLHT1 is expressed in vascular bundles of leaves, rachis, and flowering organs. Oslht1 plants showed lower panicle length and seed setting rate, especially lower grain number per panicle and total grain weight. The concentrations of both total N and free amino acids in the flag leaf were similar at anthesis between Oslht1 lines and WT while significantly higher in the mutants than WT at maturation. The Oslht1 seeds contained higher proteins and most of the essential free amino acids, similar total starch but less amylose with lower paste viscosity than WT seeds. The mutant seeds showed lower germination rate than WT. Knockout of OsLHT1 decreased N uptake efficiency and physiological utilization efficiency (kg-grains/kg-N) by about 55% and 72%, respectively. Taken together, we conclude that OsLHT1 plays critical role in the translocation of amino acids from vegetative to reproductive organs for grain yield and quality of nutrition and functionality.
In natural soil ecosystem, nitrogen (N) is presented in diversity forms including both inorganic and organic N (Rasmussen et al., 2010; Robertson and Groffman, 2015). Rice (Oryza sativa) is commonly grown in flooding paddy filed where ammonium is dominant form of inorganic N (Xu et al., 2012). Rice absorbs N rapidly at the vegetative stage, especially at the early stage of growth (Muhammad and Kumazawa, 1974; Masclaux-Daubresse et al., 2010). When the panicles emerge, the N is highly distributed to both the flag leaves and panicles (Muhammad and Kumazawa, 1974; Itoh et al., 2005). During grain-filling stage, the N supplied for panicles is mainly redistributed from leaves and about 70% of N can be transported into the panicle (Muhammad and Kumazawa, 1974). The leaf blades are the main organ of photosynthesis and the N in the flag leaf contributes significantly to the yield (Ma et al., 1989).
In flooding paddy soil, ammonium absorbed in rice roots is further assimilated in plastid into amino acids which are then transferred from roots to leaves via xylem. Amino acids in leaves are loaded into sieve elements and companion cells complexes and released to seeds (Tegeder, 2012; Tegeder, 2014; Tegeder and Masclaux-Daubresse, 2018). Import of amino acids into phloem and seeds is the key event in source to sink partitioning of N, and the sink development and grain yield rely on the amounts of amino acids translocation (Santiago and Tegeder, 2016; Tegeder and Hammes, 2018). Several membrane proteins for amino acid allocation have been characterized recently and manipulation of these transporters is able to regulate plant growth. In Arabidopsis, AtLHT1, with the strong expression in the surface of roots, young leaves, flowers, and siliques, is involved in amino acid uptake and import into the mesophyll cells (Chen and Bush, 1997; Hirner et al., 2006). Disruption of AtLHT1 exhibited decrease of shoot biomass and seed yield while AtLHT1 overexpression improved the N use efficiency under limiting N (Hirner et al., 2006). Furthermore, AtAAP2 is expressed in the phloem and functions in xylem-to-phloem transfer step. Increased amino acids allocation to leaves in aap2 mutants resulted in higher seed yields (Zhang et al., 2010; Perchlik and Tegeder, 2018). AtAAP8 is shown to be expressed in leaf phloem and siliques. It functions in amino acid loading into the phloem and import into seeds. Decreased amino acid loading into phloem and partitioning to sinks in aap8 mutants led to decreased siliques and seed numbers (Schmidt et al., 2007; Santiago and Tegeder, 2016). AtAAP1 is localized to Arabidopsis embryos and regulates amino acid distribution to the developing embryos. Amino acid loaded into embryo via AtAAP1 could also affect seed protein reserves and seed yield (Sanders et al., 2009). Similarly, in pea plants, overexpression of Amino Acid Permease1 (AAP1) in the phloem and embryos was able to improve seed yield and N use efficiency (Zhang et al., 2015; Perchlik and Tegeder, 2017).
In rice, 85 putative amino acid transporters were identified (Zhao et al., 2012). However, only a few of them have been associated with N translocation from source leaves to grains. OsAAP3 is localized to roots and leaves. Disruption of OsAAP3 decreased amino acid levels in various tissues, and promoted the tiller growth, and further improved the grain yield (Lu et al., 2018). OsAAP5 is expressed in leaves and panicles and regulates tiller growth by affecting cytokinin levels. Loss-of-function of OsAAP5 could improve grain yield (Wang J. et al., 2019). Further, storage proteins act as a N sink and N allocation affects the composition of grain storage compounds (Perez et al., 1996; Chen et al., 2012). The major organic compounds of rice grains are starch, protein and lipids (Fitzgerald et al., 2009; Li et al., 2018). Amylose and amylopectin are the two types of starch (Shewry et al., 1995; Fitzgerald et al., 2009; Chen et al., 2012). Notably, OsAAP6 was revealed to function in affecting amino acid allocation and regulating grain protein content in rice (Peng B. et al., 2014).
Recently, based on the genetic association analysis between root 15N-aspartate uptake rate of rice core accessions and single-nucleotide polymorphisms of putative amino acids transporter genes, we found that Lysine-Histidine-type Transporter 1 (OsLHT1) is a key transporter for root amino acids uptake in rice (Guo et al., 2020). Repression of OsLHT1 also decreased amino acids allocation from root to shoot and rice growth (Wang X. et al., 2019; Guo et al., 2020). Notably, the reduction in shoot biomass of Oslht1 versus wild-type (WT) plants was accelerated during development (Guo et al., 2020). In this study, we analyzed the effect of OsLHT1 knockout on N allocation from source leaves to developing grains and the components of storage proteins and starches in the grains. The results show that OsLHT1 contributes to forming grain yield and quality, and high N use efficiency in rice.
Field experiments were conducted at the research base of Nanjing Agricultural University in Ledong county, Hainan Province, China. Seeds of Oslht1 mutant lines with Nipponbare background (Guo et al., 2020) were germinated and planted according to Chen et al. (2016) and Fan et al. (2016). Fields were managed according to local agricultural practices and 180-kg N/ha was applied during the growth season. For molecular and physiological analysis, rice plants were harvested at both anthesis and maturity. Panicles, leaves, and culms were collected, dried at 60°C for 3 days and weighed. Panicles were considered as reproductive tissues while leaves and culms were considered as vegetative tissues.
WT plants were planted in the paddy field (see above). At the anthesis stage, spikelet, rachis, leaf blades, leaf sheaths, nodes, and internodes were collected. Total RNA isolation and first-strand cDNA synthesis were performed according to Guo et al. (2020). RT-qPCR was performed with SYBR Premix Ex Taq™ II (Perfect Real Time) Kit (TaKaRa Biotechnology, Dalian, China) on the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, MA, USA) using OsLHT1-specific primers (F: 5′;-GGACTCCGGCAGATCATCA; R: 5′;-CTGGTTTCATCATGTGTGCCTA). Relative expression levels were normalized to the rice housekeeping gene OsActin1 using specific primer pairs (F: 5′;-CAACACCCCTGCTATGTACG; R: 5′;-CATCACCAGAGTCCAACACAA) and presented as 2-△CT.
Seeds of OsLHT1 promoter-GUS lines (Guo et al., 2020) were grown in the field (see above). GUS assays were done as described previously (Ai et al., 2009).
Oslht1 mutant and WT plants were grown in the field (see above). At the anthesis or/and maturity stage, flag leaves and dry seeds were collected for amino acid analyses. Flag leaf free amino acid extraction was performed according to Peng B. et al. (2014). Amino acid contents in dry seeds were analyzed according to Zhou et al. (2009) and Wang et al. (2008) with some modifications. Rice grains were husked and ground to a flour consistency in a mill (JFS-13A, Hangzhou, China). Seed flour (0.100 g) was used for extraction with 5 ml of 6 M HCl in a sealed air-evacuated tube at 110°C for 22 hours. The hydrolysate was diluted to 100 ml. 1 ml dilution was dried in a Termovap Sample Concentrator MD200-1 (Allsheng, China) and re-dissolved in 1 ml water. Amino acid concentrations were determined using an L-8900 automatic amino acid analyzer (Hitachi, Tokyo, Japan). All steps were performed according to the manufacturer’s instructions. Most individual proteinous amino acid were analyzed, and results show the combined total amounts of Asn and Asp, and Gln and Glu, respectively (Peng B. et al., 2014; Guo et al., 2020).
Oslht1 mutant and WT plants were grown in the field (see above). At the anthesis or/and maturity stage, panicles, leaves, culms, and dry seeds were collected for total elemental N, protein, starch, and amylose analyses. Rice tissues were husked and ground to a flour consistency in a mill (JFS-13A, Hangzhou, China). Total N content was analyzed according to Kjeldahl method (Chen et al., 2016). Protein content was obtained by multiplying 5.95 as the protein conversion factor (GB5009.5-2016, National Food Safety Standards, China). Total starch was extracted using a starch assay kit (Megazyme, Wicklow, Ireland) according to the manufacturer’s instructions and determined with ultraviolet spectrophotometer U-1800 (Hitachi, Tokyo, Japan). Amylose content was analyzed according to Liu et al. (2009). Rapid Visco Analyser (RVA, Perton, Warriewood, NSW, Australia) were used for evaluating pasting properties of endosperm of both Oslht1 and WT seeds as described by Peng C. et al. (2014).
NUpE and NUtE were calculated according to Moll et al. (1982) and Xu et al. (2012). NUpE is the percentage ratio of total accumulated N in the above-ground shoot to total supplied fertilizer N. NUtE is the ratio of total grain yield to total accumulated N in the above-ground shoot.
A total of 100 respective fully filled seeds of Oslht1 mutant lines and Nipponbare WT were surface-sterilized for 30 min in 30% (v/v) NaClO solution, washed with sterilized water and then placed in water in Petri dishes (d=15 cm) with two sheets of filter paper. The seeds were incubated at 28°C in the dark for 2 days and then moved into a tissue culture chamber with 14-hour-days at a light intensity of 6000 lux, 40% humidity, and temperatures of 28°C. The water was exchanged every day. Seeds were photographed and plant root length, shoot length and germination rate were measured.
Data are generally presented as means ± SD (standard deviation) of at least 5 biological repetitions. Significant differences were analyzed using IBM SPSS Statistics 20 program at one-way ANOVA followed by Tukey’s test.
Sequence data from this article can be obtained from The Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/) under the following accession numbers: Oryza sativa japonica LOC_Os08g03350 (OsLHT1), LOC_Os03g50885 (OsActin1).
We have previously shown that OsLHT1 transcripts were detected in roots and leaf blades at the seedling stage (Guo et al., 2020). Here we show OsLHT1 expression highly in leaf blades, leaf sheaths and rachis, and relatively weak in spikelet, node and internode at the anthesis stage (Figures 1A, B). Since the leaf I (the youngest or flag leaf) and developing panicles are growing organs and need large amount of nitrogenous compounds transported via stem from leaves (Muhammad and Kumazawa, 1974), the abundant expression of OsLHT1 in leaves and rachis hints that OsLHT1 may play a role in leaf-to-panicle amino acid allocation. GUS staining was observed throughout all cell types of root-shoot junction, with the strong expression levels in the vascular bundles (Figure 1C). GUS activity was present in leaf sheaths and leaf blades, specifically in the cells of the major and minor veins (Figures 1D–F). In addition, OsLHT1 is strongly expressed in flowering organs, hulls and germinated seeds (Figures 1G, H), demonstrating its potential role in rice reproductive growth.

Figure 1 Expression levels of OsLHT1 in rice. Rice (cv. Nipponbare) was grown in paddy field (see materials and methods). (A) The individual organ of the plant at the anthesis stage to be sampled for extraction of total RNA. (B) The relative expression of OsLHT1 quantified by RT-qRCR. Rice housekeeping gene OsActin1 was used as an internal control. Values are means ± SD (n=5 biological replicates). (C–E) GUS staining in root-shoot junction (C), leaf sheath-leaf blade junction (D), and vascular bundles (VB) of mature leaf blades (E). (F) Cross-sections of a leaf blade showing a major (mVB) and minor (miVB) vascular bundles. Scale bar: 100 μm. (G) GUS staining in flower and lemma. (H) GUS staining in three-day-old germinated seeds.
To detect the contribution of OsLHT1 in rice grain production, we compared the performance of WT and Oslht1 mutants in the paddy field. At the maturation, in comparison to WT, Oslht1 plants showed shorter height, brown leaf and spikelet (Figure 2A), shorter panicle length (Figure 2B), lower grain number per panicle, seed setting rate, grain weight per panicle and total grain yield per plant (Figures 2C–F). The OsLHT1-mutation reduced 1000-grain weight about 10% (Figure 2G), which was relatively less affected in comparison to the other parameters.
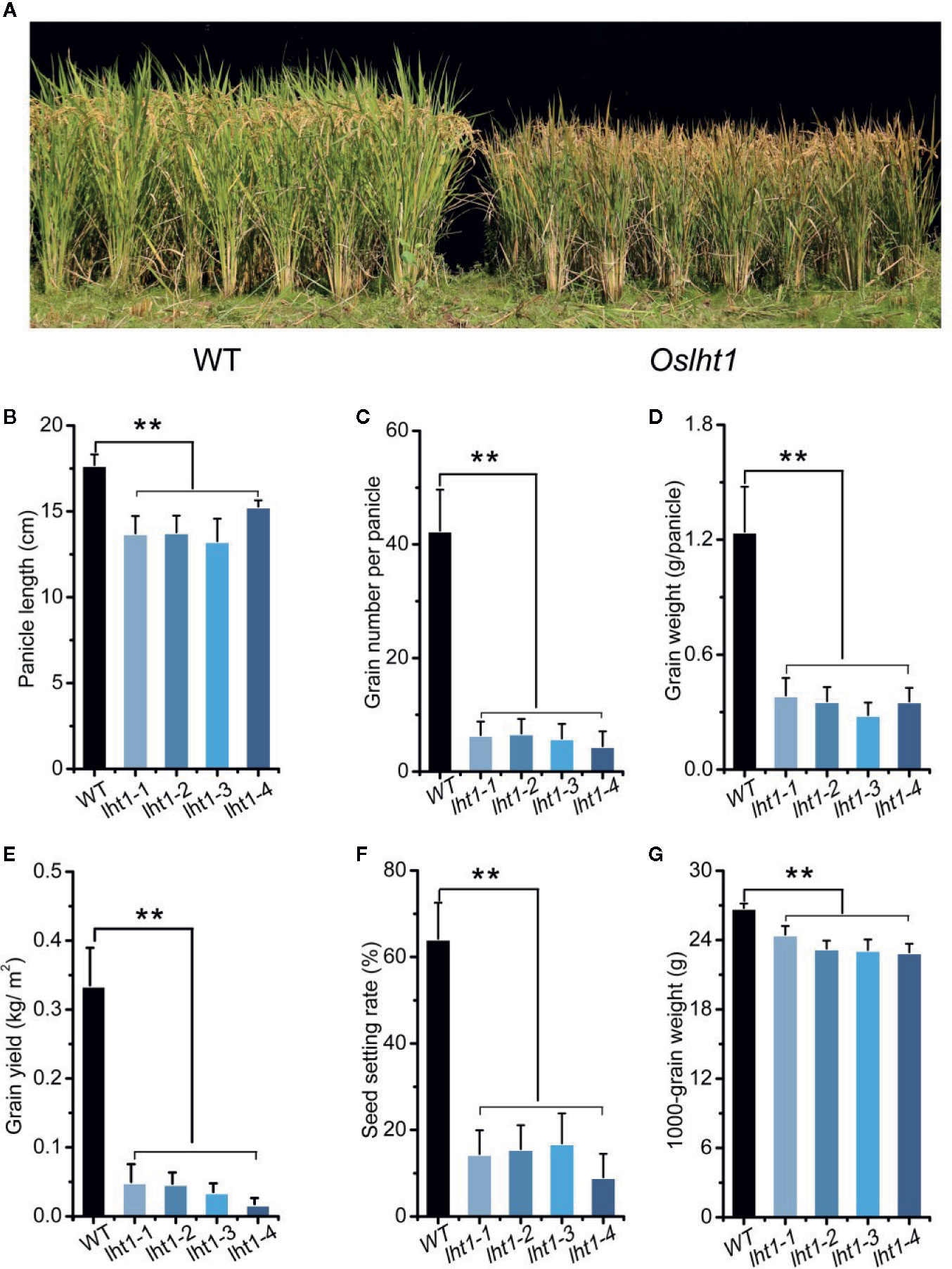
Figure 2 Effect of OsLHT1 mutation on rice growth and grain yield components. Both wild-type (WT, cv. Nipponbare) and Oslht1 mutants were grown in paddy field and harvested at the maturity stage for determining yield components (see Materials and Methods). (A) Image of Oslht1 and WT plants in the field. (B) Mean panicle length. (C) Total grain number per panicle. (D) Total grain weight per panicle. (E) Total grain yield per plant. (F) Mean seed setting rate. (G) Mean 1000-seeds weight. Values are means ± SD (n = 7). Asterisks indicate significant differences between each Oslht1 mutant line and WT detected by one-way ANOVA followed by Tukey’s test. **p < 0.01.
In cereal crops, there is a considerably higher distribution of N from flag leaf to the panicle after the emergence of panicles (Muhammad and Kumazawa, 1974; Zhang et al., 2007; Chen et al., 2016; Tegeder and Masclaux-Daubresse, 2018). As expected, the contents of both total N and free amino acids in the flag leaf was dramatically decreased from anthesis to mature stage in both WT and Oslht1 mutant lines of rice (Figures 3A, D), confirming that large portion of grain N was allocated from the flag leaf in addition of direct contribution of root acquired N from soil. Remarkably, there was no significant difference of the content of total N (mg/g), total free amino acids and most of individual free amino acids (mg/kg FW) in the flag leaf between WT and Oslht1 plants at the anthesis stage (Figures 3A–C). However, their contents were significantly higher except similar level of aspartate, asparagine and serine in the mutant lines than in WT at the maturity stage (Figures 3D–F). In comparison to the increase of total N in Oslht1 lines by about 15-30%, total free amino acids accumulated in the mutants were increased by about 60-120% at the maturation (Figures 3D, E). Since amino acids represent the main form of N that was transported over long distances to the reproductive tissues and OsLHT1 is an amino acid transporter (Guo et al., 2020), the results demonstrated that OsLHT1 greatly contributes N re-allocation from source leaves to developing grains.
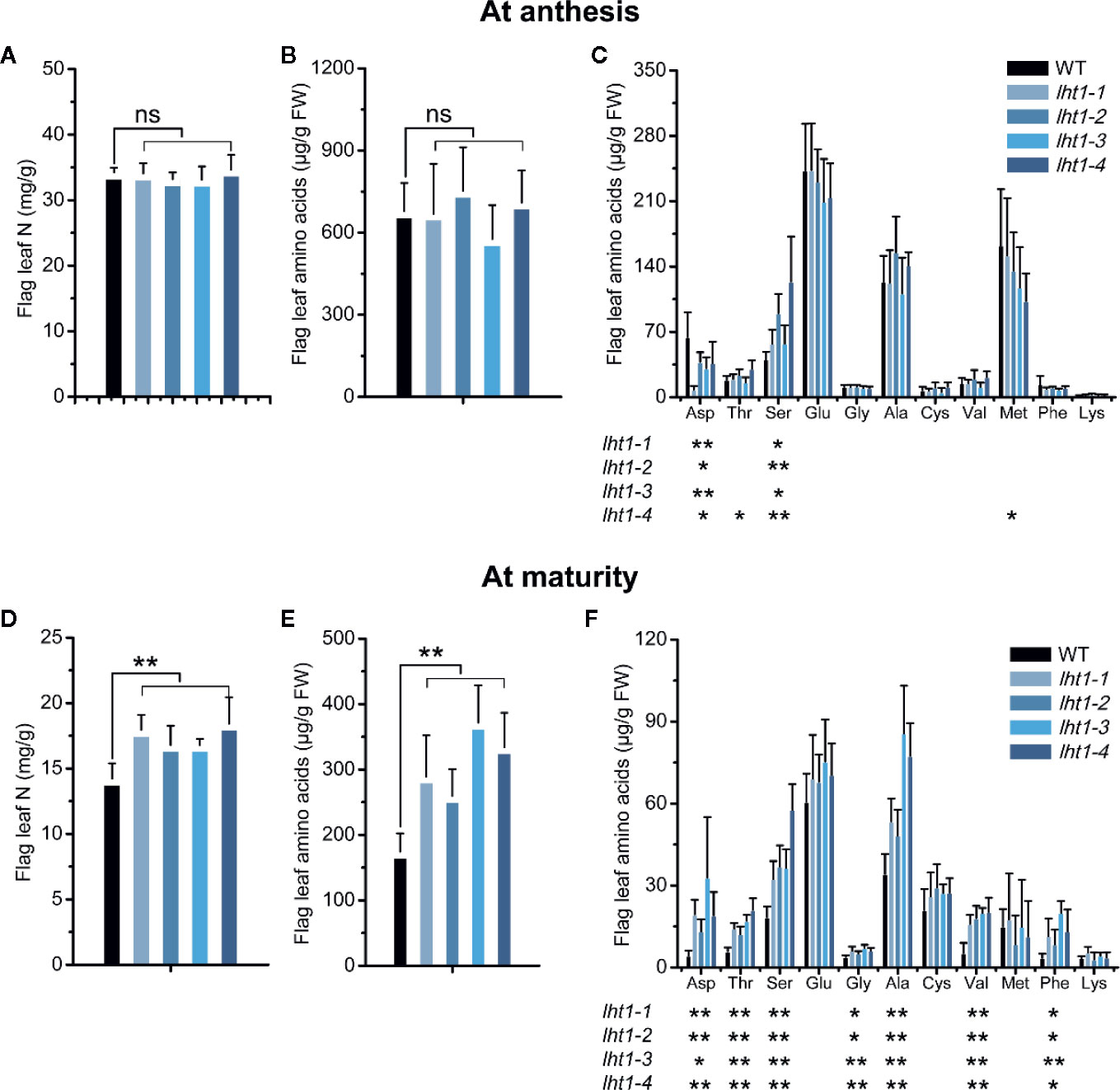
Figure 3 Effect of OsLHT1 mutation on content of total N, total, and individual free amino acid in rice leaf at anthesis and maturity. Both wild-type (WT, cv. Nipponbare) and Oslht1 mutants were grown in paddy field until mature. The flag leaf (fully expanded youngest leaf from top) of WT and Oslht1 plants was sampled for the analysis of total N (A, D), total free amino acids (B, E), and individual free amino acid (C, F). FW, fresh weight. Values are means ± SD (n = 7). Asterisks indicate significant differences between each Oslht1 mutant line and WT detected by one-way ANOVA followed by Tukey’s test. *p < 0.05; **p < 0.01; ns, not significant.
Since the mutation of OsLHT1 limited rice growth, as expected, total N per plant was significantly lower in the mutants than in WT at anthesis stage (Figure 4A), particularly at maturation stage (Figure 4C), while the difference of N in the leaves was relatively smaller than that in other organs (Figure 4C). At the anthesis stage, 67% of the total N was found in vegetative tissues and 33% in the reproductive tissues (Figure 4B). In contrast, in Oslht1 mutants, 77% of total N levels remained in the vegetative tissues while about 23% of the total plant N was detected in the reproductive tissues (Figure 4B). At the maturity stage, higher percentage of N was allocated to reproductive tissues compared to that at the anthesis stage. Oslht1 mutants only moved around 40% of total N to the reproductive tissues compared to 62% in WT plants (Figure 4D). These data indicated the great contribution of OsLHT1 to leaf-to-panicle allocation in the forms of amino acids.
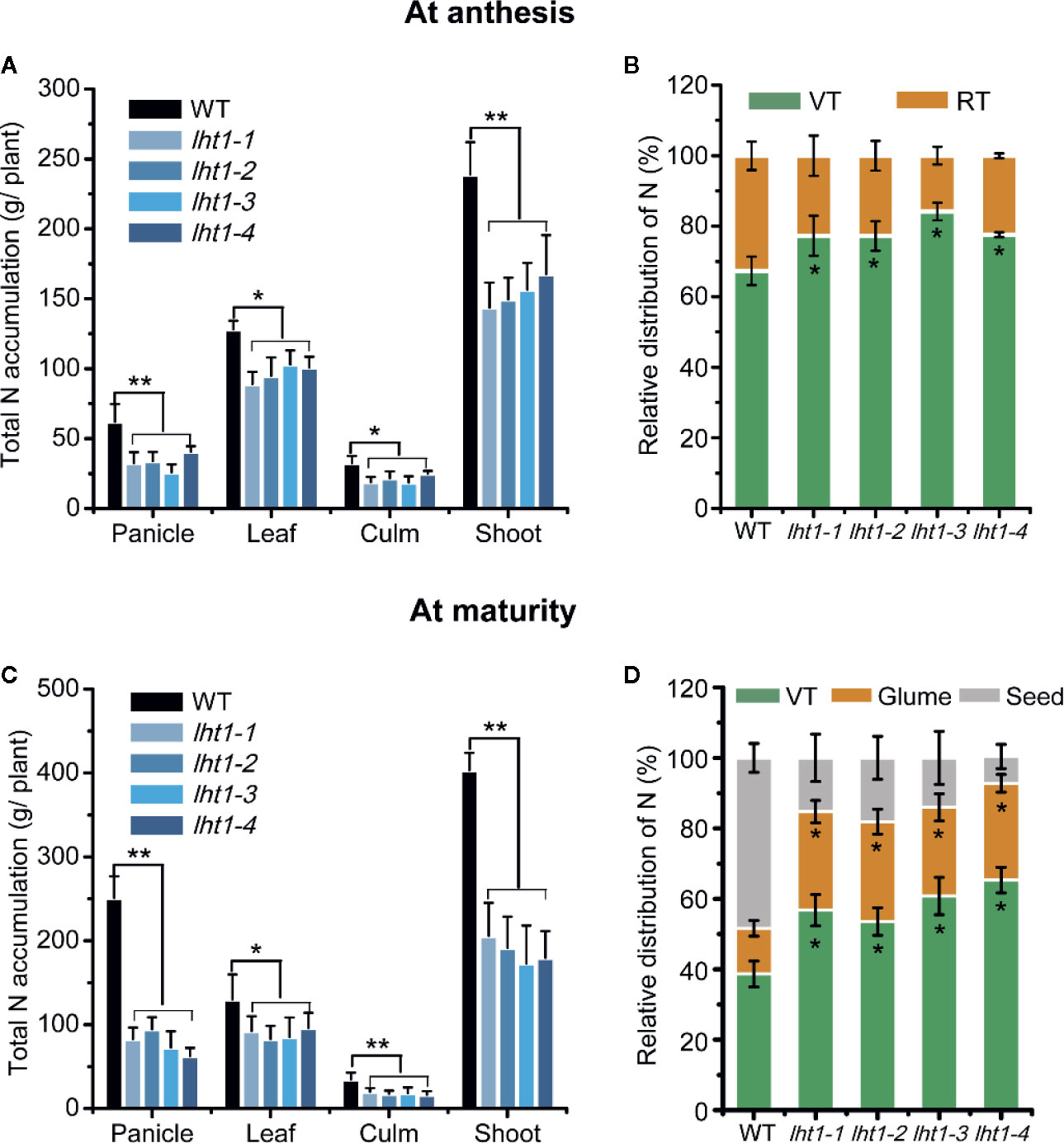
Figure 4 Effect of OsLHT1 mutation on total N content and relative distribution of N in different organs at anthesis and maturity. Both wild-type (WT, cv. Nipponbare) and Oslht1 mutants were grown in paddy field and sampled at anthesis and mature time. (A, C) Total N content in panicles, leaves, culms and total shoot. (B, D) Relative amount of N in vegetative tissues (VT) and reproductive tissues (RT). Leaves and culms were considered as vegetative tissues while panicles were considered as reproductive tissues. Values are means ± SD (n = 7). Asterisks indicate significant differences between each Oslht1 mutant line and wild-type (WT, Nipponbare) detected by one-way ANOVA followed by Tukey’s test. *p < 0.05; **p < 0.01.
As reported, the dry seeds of both Oslht1 mutants and WT plants contained a high amount of total and individual free amino acids in addition to rich starch (Figure 5) (Fitzgerald et al., 2009; Li et al., 2018). Compared with WT, Oslht1 seeds accumulated higher total N and proteins (Figures 5A, D), total and individual free amino acids (Figures 5B, C) with the increase by up to 30-35%. In contrast, amylose content in Oslht1 seeds was reduced by about 31% (Figure 5F) even though total starch content in the mutant and WT seeds was kept at the same level (Figure 5E).
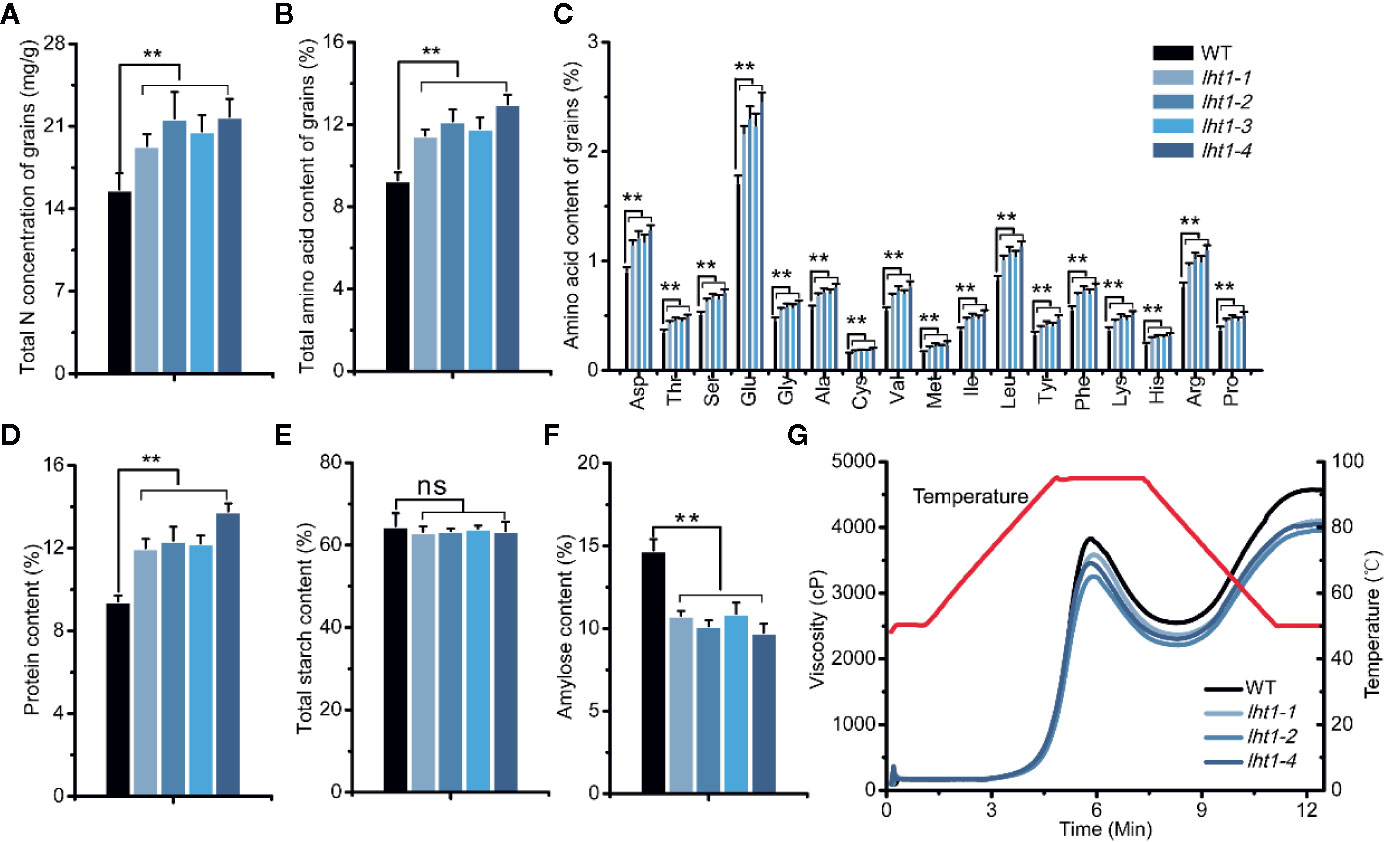
Figure 5 Effect of OsLHT1 mutation on content of total N, total and individual amino acid, total crude protein, total starch and amylose, and pasting properties of endosperm starch in harvested grains. Both wild-type (WT, cv. Nipponbare) and Oslht1 mutants were grown in paddy field until mature. (A) Total N concentration. (B, C) Total and individual amino acid content. (D) Total crude protein content. (E) Total starch content. (F) Amylose content. Values in a-f are means ± SD (n = 7). Asterisks indicate significant differences between each Oslht1 mutant line and WT detected by one-way ANOVA followed by Tukey’s test. **p < 0.01; ns, not significant. (G) Pasting properties of endosperm starch. The viscosity value at each temperature is the mean of three replicates. The red line indicates the temperature changes during the measurements.
The altered storage compounds in Oslht1 grains affected cooked texture and pasting properties of the endosperm starch. Both WT and Oslht1 seed starches showed the similar patterns of pasting with peak viscosity and breakdown when the temperature was increased. However, compared with WT, the peak and cool paste viscosity of the Oslht1 starch were all decreased (Figure 5G).
Increasing both the grain and N harvest indexes to drive N acquisition and utilization is an important approach for breeding future high N use efficient cultivars (Xu et al., 2012). In this study, we also analyzed both N uptake efficiency (NUpE) and N utilization efficiency (NUtE) (Xu et al., 2012) of Oslht1 plants and WT grown in the paddy field with the moderate application level of N fertilizer (180-kg/ha). Knockout of OsLHT1 resulted in decrease of NUpE by about 50% while it decreased NUtE by about 70% to 90% (Figure 6). The extreme low NUtE of Oslht1 was caused mainly by low grain harvest index (the grain to straw ratio) due to limited N re-allocation.
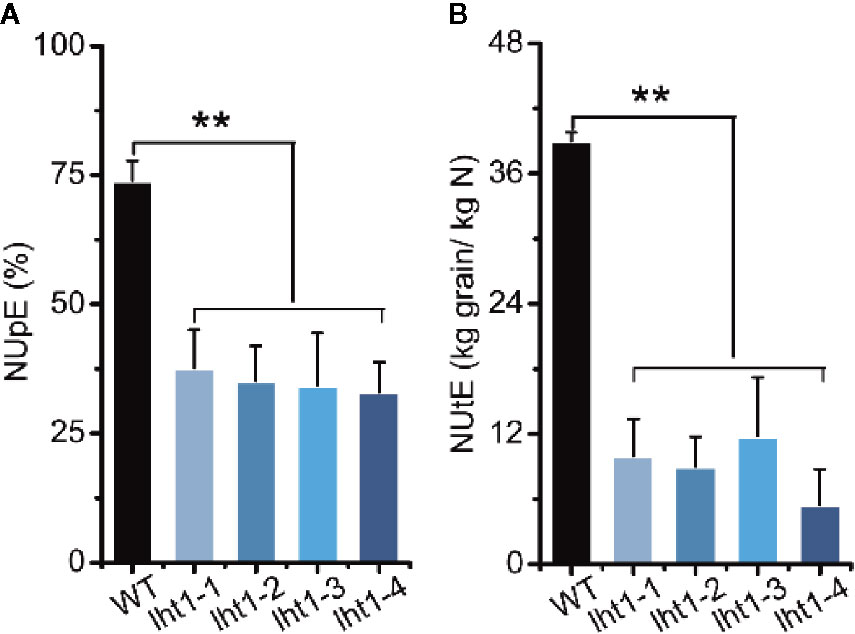
Figure 6 Effect of OsLHT1 mutation on nitrogen uptake, utilization, and total use efficiency. Both wild-type (WT, cv. Nipponbare) and Oslht1 mutants were grown in paddy field until mature. (A) NUpE: nitrogen uptake efficiency = (total accumulated N/total supplied fertilizer N)*100%. (B) NUtE: nitrogen utilization efficiency = total grain yield/total accumulated N. Values are means ± SD (n = 7). Asterisks indicate significant differences between each Oslht1 mutant line and WT detected by one-way ANOVA followed by Tukey’s test. **p < 0.01.
To resolve if the change in Oslht1 grain storage compounds affects seed viability, we performed germination test of the seeds. The seeds of both WT and mutants began to germinate after soaking in water for 24 h and peaked at 72 h. After 72 h, the germination rate of WT seeds reached 95%, while Oslht1 seeds germinated only between 80-86% and no further increase after that (Figure 7A). Furthermore, the significant differences of seedling growth were also observed between Oslht1 and WT (Figure 7B). After 12-day growth in nutrient-free water, in comparison to WT, Oslht1 root and shoot length showed incredibly significant decrease (Figures 7C, D). The results clearly showed an inhibitory effect on the germination efficacy of the seeds in the loss-of-function of OsLHT1.
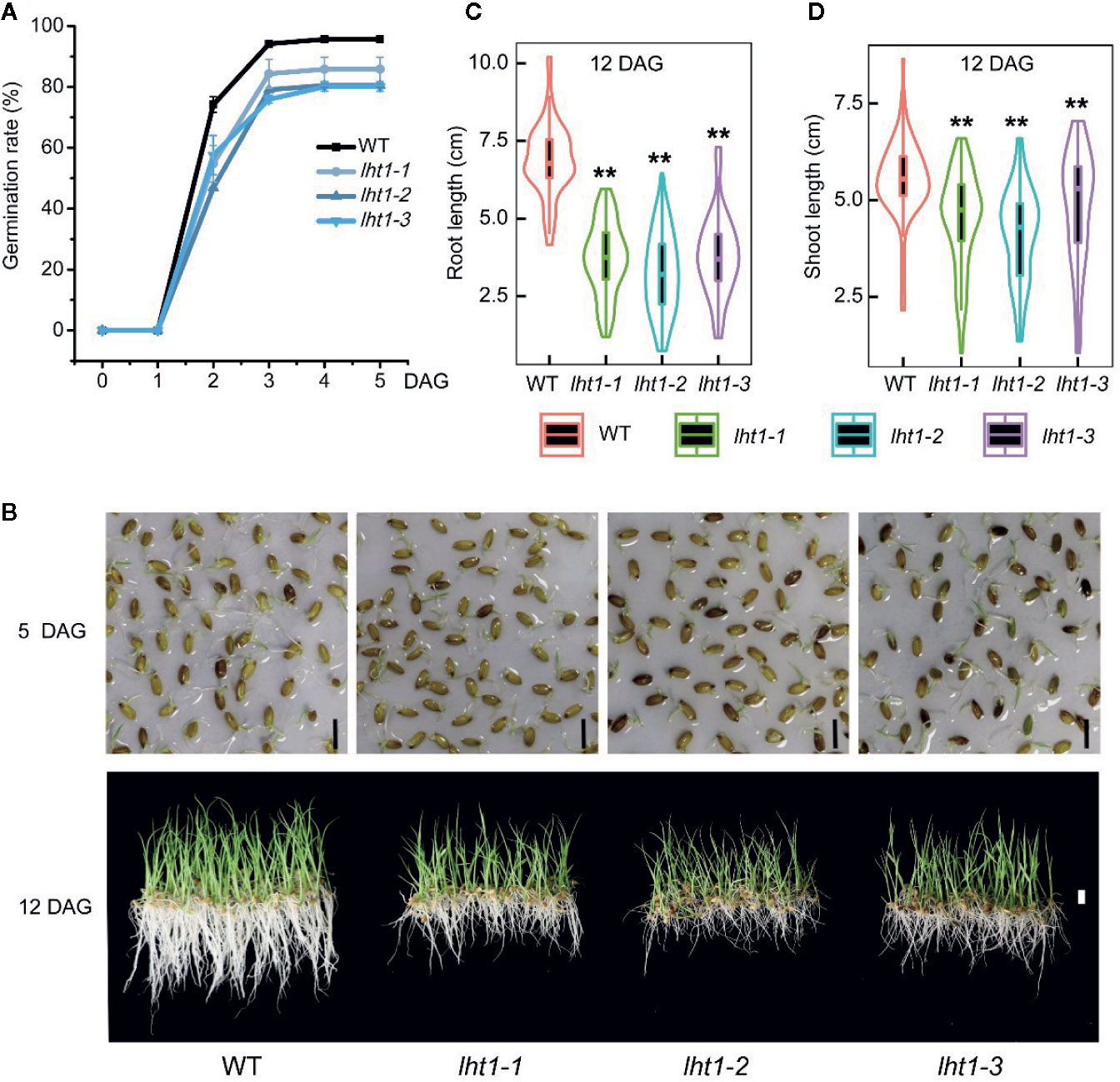
Figure 7 Effect of OsLHT1 mutation on seed germination, root and shoot growth rate. 100-seeds of each homozygote T4 Oslht1 mutant line and wild-type (WT, Nipponbare) were placed in nutrient-free tap-water. (A) Seed germination rate. DAG, day after germination. (B) Images of seedlings on nutrient-free water on 5 DAG and 12 DAG. Scale bar= 1 cm. (C, D) Root and shoot length of each plant grown in the nutrient-free water for 12 days. Asterisks indicate significant differences between each Oslht1 mutant line and WT detected by the Student’s t-test, **p < 0.01.
Amino acids in mature leaves are transported via phloem of minor veins to sink tissues (Noctor et al., 2002; Tegeder, 2014). In our previous study, the promoter-GUS assay in rice indicated that OsLHT1 is localized in the leaf blade, especially in the cells of the major and minor veins (Guo et al., 2020). Here we further present high expression of OsLHT1 in multiple tissues including leaf blades, sheaths, nodes, rachis and spikelets at the anthesis stage, especially in the major and minor veins of leaf blade (Figure 1), supporting the role of OsLHT1 in the phloem loading of leaf or root-synthesized amino acids.
Next, OsLHT1 functions in N allocation most probably in the broad spectrum of amino acids as shown in the seedling stage (Guo et al., 2020). In this study, N translocation from source leaves to panicles was clearly established with analysis of four independent Oslht1 mutant lines. Inactivation of OsLHT1 resulted in increased accumulation of total N and amino acids in the flag leaves of Oslht1 mutants from anthesis to maturity stage (Figure 3). Considering that OsLHT1 is involved in root-to-shoot allocation and loss function of OsLHT1 reduced root-synthesized amino acid levels in leaves (Guo et al., 2020), the large amounts of amino acids accumulated in the mutant leaves suggested that the process of amino acids synthesized in source leaves might be stimulated, or less amino acid amounts were allocated into phloem. Since lower percentage of N was contained in the reproductive tissues of Oslht1 mutants (Figure 4), it provides strong support that lack of OsLHT1 function reduced the amino acids allocation from source leaves to panicles. Since OsLHT1 is localized in the major and minor veins of leaf blade and serves as an amino acid importer (Figure 1F; Guo et al., 2020), this decrease might result from the reduced amino acid import into phloem. In Arabidopsis, many members of AAP family are expressed in the phloem of major and minor veins and contribute to source to sink allocation at the reproductive stage (Sanders et al., 2009; Zhang et al., 2010; Santiago and Tegeder, 2016; Perchlik and Tegeder, 2018). Although AAP and LHT transporters are two distinct groups, they usually have overlapping functions in amino acid translocation (Tegeder and Ward, 2012; Perchlik et al., 2014). Therefore, OsLHT1, a member of LHT family, is one of the major transporters responsible for the transfer process of amino acids in rice.
Amino acids move from the location of the synthesis to the sites of usage to satisfy the demand of plant growth (Tegeder, 2014). At the seedling stage, amino acids are synthesized in the root and exported to supply the developing leaves. In our previous studies, compared with WT, total amounts of free amino acids were reduced in leave blades of Oslht1 mutants (Guo et al., 2020). At the reproductive stage, amino acids could move from roots to flag leaves and, at the same time, flag leaf could also serve as the source organs to supply amino acids to panicles (Muhammad and Kumazawa, 1974). Since no difference of the free amino acid content in the flag leaf between WT and Oslht1 lines was observed at the anthesis stage, amino acids imported into flag leaves and exported from flag leaves might be dynamically balanced before anthesis (Figures 3A, B). At the maturity stage, much higher percentage of amino acids needs to be allocated to the panicles for meeting the demand of seed production. The higher free leaf amino acids and total N in Oslht1 mutants than in WT (Figures 3D–F) indirectly supports the conclusion that OsLHT1 functions in amino acid allocation to panicles and grains.
A number of studies indicated that rice sink production is positively related to root N uptake and the amounts of N supplied by vegetative tissues (Fan et al., 2016; Wang et al., 2018). In Arabidopsis, LHT1 is crucial for organic N uptake from soil (Ganeteg et al., 2017). Atlht1 T-DNA insertion mutants displayed dramatic growth inhibition when grown in soil (Hirner et al., 2006). In Oslht1 mutants, less N distributed into shoot at early stage and panicle sinks at late stage decreased shoot biomass and grain yield production (Figure 2; Wang X. et al., 2019; Guo et al., 2020). We have discussed the mechanism of OsLHT1 function in dramatically alteration of rice growth (Guo et al., 2020). We proposed that OsLHT1 mutation could decrease both xylem loading and leaf amino acid import, resulting in N deficiency and growth impairment as observed in Oslht1 mutant (Hermans et al., 2006; Grechi et al., 2007; Guo et al., 2020). The altered leaf structure and size of Oslht1 mutants can affect transpiration, water transport and photosynthesis which further limits root N uptake, allocation and re-distribution, as well as downstream carbon-N interaction (Oliveira and Coruzzi, 1999; Lejay et al., 2003; Palenchar et al., 2004; Gutiérrez et al., 2007). In addition, the reduced import of amino acids into mesophyll cells by Oslht1 mutation could trigger a stress response which in turn restricts plant growth (Pilot et al., 2004; Hirner et al., 2006; Liu et al., 2010; Yang et al., 2014; Sonawala et al., 2018). When an amino acid is not allocated at a sufficient step, it could also serve as the limiting step for rice growth and development (Mo et al., 2006; Muñoz-Bertomeu et al., 2009; Xia et al., 2014). The decreased grain yield was partly due to the extremely low seed setting as well as reduction in grain weight (Figures 2F, G). Specific C-, N-regulatory and C/N interaction pathways play crucial roles in seed germination (Osuna et al., 2015). OsLHT1-mutation resulted in significant increase of total N and most of individual amino acids, and decrease of amylose concentration in rice grains (Figure 5). Internal C/N status could affect seed germination by moderating gibberellin acid and abscisic acid metabolism (Koornneef et al., 2002; Yamaguchi and Kamiya, 2002; Osuna et al., 2015). In addition, seed size is commonly showing positive correlation with seed germination (Ambika et al., 2014). Therefore, the relatively smaller seed size, lower amylose and higher amino acids of Oslht1 mutants could contribute to the delay of seed germination (Figure 2G). Furthermore, decreased grain weight was partially resulted from less seed storage amylose content (Figure 5F) while few seed number was caused by decreased amino acid allocation (Figures 3 and 4) (Tan et al., 2010; Santiago and Tegeder, 2016). Both root-to-shoot and leaf-to-panicle amino acid allocation were reduced, and eventually led to low grain number (Figure 2) (Guo et al., 2020). However, total N and protein levels per grains were still higher in Oslht1 mutants (Figures 5A, D). The results may hint that the trade-off occurs between producing more seed number and keeping high N per seed during the grain filling phase (Seiffert et al., 2004; Drechsler et al., 2015). Overexpressing AAP1 in pea plants could allocate more N to seeds and produce high seed number (Perchlik and Tegeder, 2017). However, in Arabidopsis, decreased amino acid partitioning to sinks led to decreased seed numbers while seed protein levels were unchanged (Schmidt et al., 2007; Santiago and Tegeder, 2016). In rice, amino acid transporter OsAAP6 functions as an important regulator of grain protein content and nutritional quality. OsAAP6 showed no effect on grain yield. However, transgenic plants with higher OsAAP6 expression levels produced much more grain storage protein (Peng B. et al., 2014). The variation in seed numbers and seed N pools may be caused by the species or other unknown complex mechanism (Fageria and Baligar, 2005; Hirel et al., 2007; Sanders et al., 2009).
The OsLHT1 mutation induced incredibly low yield and alteration of storage compounds in the kernels. In comparison to WT, Oslht1 brown rice kernel accumulated significant higher protein and lower amylose (Figures 5D, F), which may cause the change of pasting properties of endosperm starch (Figure 5G). It has been shown that the formation of protein–starch complexes affects rice starch gelatinization properties, tending to increase flour peak viscosity (Saleh and Meullenet, 2015). The decrease in flour breakdown viscosity of Oslht1 kernels (Figure 5G) suggests a greater protection of the amylose granule integrity. This study demonstrated that changes in rice protein impacted rice flour pasting properties.
Plant N use efficiency (NUE) is inherently complex and it is the combination of N uptake efficiency (NUpE) and N utilization efficiency (NUtE) (Xu et al., 2012). NUE is governed by multiple interacting genetic, environmental and management factors (Balasubramanian et al., 2004; Xu et al., 2012). Many genes involved in N uptake, assimilation, translocation and regulation have been identified to make great contributions to the NUE, such as amino acid transporter AtAAP2 in Arabidopsis, nitrate transporter OsNRT1.1b, OsNRT2.3b and ammonium transporter in rice (Zhang et al., 2010; Ranathunge et al., 2014; Bao et al., 2015; Hu et al., 2015; Fan et al., 2016). Rice plants mainly acquire ammonium and transport of N to above ground parts in the forms of amino acids (Xu et al., 2012). Since OsLHT1 plays the key role in the allocation of amino acids from root to shoot (Guo et al., 2020) and N re-distribution from source leaves to developing grains (Figures 3–5), the inhibitory effect of Oslht1 mutation on shoot growth acted the feedback role in preventing root N acquisition, which resulted in low NUpE (Figure 6). The confined panicle development in Oslht1 plants decreased grain to straw ratio, which in turn reduced NUtE for root acquired N to produce grains (Figure 6).
In summary, as shown in Figure 8, OsLHT1 is not only for root acquisition and long-distance transport of amino acids, it also plays the major role in N allocation from source leaves to developing panicle and grains for reaching the potential grain yield and quality of nutrition and functionality.
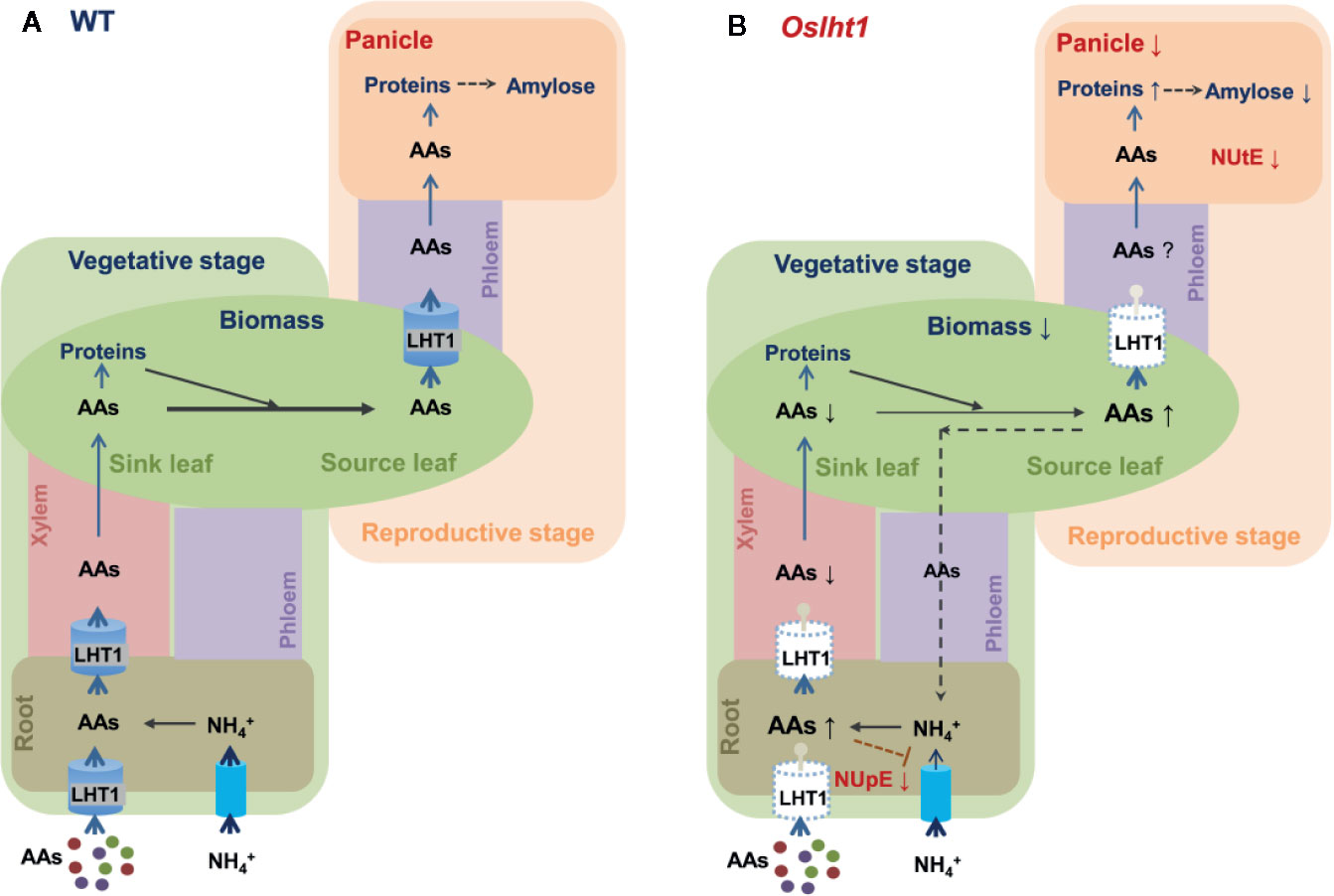
Figure 8 Overview model illustrating the OsLHT1 functions in maintaining both N uptake efficiency and N utilization efficiency in rice. In wild type (A), rice roots directly acquire ammonium and amino acids (AAs) from soil. Ammonium in root cells is rapidly assimilated into AAs that are delivered to shoot (sink leaves) in xylem at vegetative stage. Our previous study (Guo et al., 2020) has shown that OsLHT1 (for simplicity, it is indicated by LHT1 in the figure) directly contributes root acquisition and root to shoot transport of a broad spectrum of amino acids. At reproductive stage, large portion of N is re-allocated mainly in the form of amino acids from source leaves to panicles occurring in the phloem. OsLHT1 plays critical role in this process and functions in grain yield and nutrition quality. (B) Knockout of OsLHT1 dramatically reduces N supply from source leaves to developing panicles which are accompanied by reduced shoot biomass and grain yield with higher storage proteins and lower amylose. The higher levels of N in roots or leaves might negatively affect root ammonium uptake in oslht1 plants, probably by a feedback regulatory mechanism, which results in lower N uptake efficiency (NUpE). The limited grain yield by loss of OsLHT1 function results in low N utilization efficiency (NUtE). The sizes of the arrows located to the right of the features analyzed indicate the significant changes in Oslht1 mutants compared with wild-type plants (up, increase; down, decrease).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
NG and HQ conceived the research and analyzed the data. NG, JH, and MG performed the experiments. NG, HQ, and GX wrote the article. All authors contributed to the article and approved the submitted version.
This work was supported by National Key Research and Development Program of China (2016YFD0100700), Jiangsu Collaborative Innovation Center for Solid Organic Waste Resource Utilization and the Innovative Research Team Development Plan of the Ministry of Education of China (IRT17R56, KYT201802).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ms. Xiaoli Dai and Ms. Kaiyun Qian from College of Resources and Environmental Sciences, Nanjing Agriculture University for their technique support in conducting the chemical analyses.
Ai, P., Sun, S., Zhao, J., Fan, X., Xin, W., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation. Plant J. 57 (5), 798–809. doi: 10.1111/j.1365-313X.2008.03726.x
Ambika, S., Manonmani, V., Somasundar, G. (2014). Review on effect of seed size on seedling vigour and seed yield. Res. J. Seed Sci. 7 (2), 31–38. doi: 10.3923/rjss.2014.31.38
Balasubramanian, V., Alves, B., Aulakh, M., Bekunda, M., Cai, Z., Drinkwater, L., et al. (2004). Crop, environmental, and management factors affecting nitrogen use efficiency. Agriculture and the Nitrogen Cycle: Assessing the impacts of fertilizer use on food production and the environment. SCOPE (65), 19–33.
Bao, A., Liang, Z., Zhao, Z., Cai, H. (2015). Overexpressing of OsAMT1-3, a high affinity ammonium transporter gene, modifies rice growth and carbon-nitrogen metabolic status. Int. J. Mol. Sci. 16 (5), 9037–9063. doi: 10.3390/ijms16059037
Chen, L., Bush, D. R. (1997). LHT1, A Lysine- and Histidine-Specific amino acid transporter in Arabidopsis. Plant Physiol. 115 (3):1127. doi: 10.1104/pp.115.3.1127
Chen, Y., Wang, M., Ouwerkerk, P. B. (2012). Molecular and environmental factors determining grain quality in rice. Food Energy Secur. 1 (2), 111–132. doi: 10.1002/fes3.11
Chen, J., Zhang, Y., Tan, Y., Zhang, M., Zhu, L., Xu, G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving OsNRT2.1 expression with the OsNAR2.1 promoter. Plant Biotechnol. J. 14 (8), 1705–1715. doi: 10.1111/pbi.12531
Drechsler, N., Zheng, Y., Bohner, A., Nobmann, B., von Wirén, N., Kunze, R., et al. (2015). Nitrate-dependent control of shoot K homeostasis by the nitrate transporter1/peptide transporter family member NPF7. 3/NRT1. 5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol. 169 (4), 2832–2847. doi: 10.1104/pp.15.01152
Fageria, N., Baligar, V. (2005). Enhancing nitrogen use efficiency in crop plants. Adv. Agron. 88, 97–185. doi: 10.1016/S0065-2113(05)88004-6
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. U.S.A. 113 (26), 7118–7123. doi: 10.1073/pnas.1525184113
Fitzgerald, M. A., McCouch, S. R., Hall, R. D. (2009). Not just a grain of rice: the quest for quality. Trends Plant Sci. 14 (3), 133–139. doi: 10.1016/j.tplants.2008.12.004
Ganeteg, U., Ahmad, I., Jamtgard, S., Aguetoni-Cambui, C., Inselsbacher, E., Svennerstam, H., et al. (2017). Amino acid transporter mutants of Arabidopsis provides evidence that a non-mycorrhizal plant acquires organic nitrogen from agricultural soil. Plant Cell Environ. 40 (3), 413–423. doi: 10.1111/pce.12881
Grechi, I., Vivin, P., Hilbert, G., Milin, S., Robert, T., Gaudillère, J.-P. (2007). Effect of light and nitrogen supply on internal C: N balance and control of root-to-shoot biomass allocation in grapevine. Environ. Exp. Bot. 59 (2), 139–149. doi: 10.1016/j.envexpbot.2005.11.002
Guo, N., Hu, J., Yan, M., Luo, L., Qu, H., Tegeder, M., et al. (2020). Oryza sativa Lysine-Histidine-type Transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. 103 (1), 395–411. doi: 10.1111/tpj.14742
Gutiérrez, R. A., Lejay, L. V., Dean, A., Chiaromonte, F., Shasha, D. E., Coruzzi, G. M. (2007). Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 8 (1), R7. doi: 10.1186/gb-2007-8-1-r7
Hermans, C., Hammond, J. P., White, P. J., Verbruggen, N. (2006). How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci. 11 (12), 610–617. doi: 10.1016/j.tplants.2006.10.007
Hirel, B., Le Gouis, J., Ney, B., Gallais, A. (2007). The challenge of improving nitrogen use efficiency in crop plants: towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 58 (9), 2369–2387. doi: 10.1093/jxb/erm097
Hirner, A., Ladwig, F., Stransky, H., Okumoto, S., Keinath, M., Harms, A., et al. (2006). Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 18 (8), 1931–1946. doi: 10.1105/tpc.106.041012
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47 (7), 834–838. doi: 10.1038/ng.3337
Itoh, J.-I., Nonomura, K.-I., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., et al. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46 (1), 23–47. doi: 10.1093/pcp/pci501
Koornneef, M., Bentsink, L., Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5 (1), 33–36. doi: 10.1016/S1369-5266(01)00219-9
Lejay, L., Gansel, X., Cerezo, M., Tillard, P., Müller, C., Krapp, A., et al. (2003). Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15 (9), 2218–2232. doi: 10.1105/tpc.013516
Li, Y., Xiao, J., Chen, L., Huang, X., Cheng, Z., Han, B., et al. (2018). Rice functional genomics research: past decade and future. Mol. Plant 11 (3), 359–380. doi: 10.1016/j.molp.2018.01.007
Liu, L., Ma, X., Liu, S., Zhu, C., Jiang, L., Wang, Y., et al. (2009). Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71 (6), 609–626. doi: 10.1007/s11103-009-9544-4
Liu, G., Ji, Y., Bhuiyan, N. H., Pilot, G., Selvaraj, G., Zou, J., et al. (2010). Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22 (11), 3845–3863. doi: 10.1105/tpc.110.079392
Lu, K., Wu, B., Wang, J., Zhu, W., Nie, H., Qian, J., et al. (2018). Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 16 (10), 1710–1722. doi: 10.1111/pbi.12907
Ma, J., Nishimura, K., Takahashi, E. (1989). Effect of silicon on the growth of rice plant at different growth stages. Soil Sci. Plant Nutr. 35 (3), 347–356. doi: 10.1080/00380768.1989.10434768
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105 (7), 1141–1157. doi: 10.1093/aob/mcq028
Mo, X., Zhu, Q., Li, X., Li, J., Zeng, Q., Rong, H., et al. (2006). The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol. 141 (4), 1425–1435. doi: 10.1104/pp.106.084178
Moll, R., Kamprath, E., Jackson, W. (1982). Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron. J. 74 (3), 562–564. doi: 10.2134/agronj1982.00021962007400030037x
Muhammad, S., Kumazawa, K. (1974). The absorption, distribution, and redistribution of 15N-labelled ammonium and nitrate nitrogen administered at different growth stages of rice. Soil Sci. Plant Nut 20 (1), 47–55. doi: 10.1080/00380768.1974.10433227
Muñoz-Bertomeu, J., Cascales-Miñana, B., Mulet, J. M., Baroja-Fernández, E., Pozueta-Romero, J., Kuhn, J. M., et al. (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 151 (2), 541–558. doi: 10.1104/pp.109.143701
Noctor, G., Novitskaya, L., Lea, P. J., Foyer, C. H. (2002). Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J. Exp. Bot. 53 (370), 939–945. doi: 10.1093/jexbot/53.370.939
Oliveira, I. C., Coruzzi, G. M. (1999). Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 121 (1), 301–310. doi: 10.1104/pp.121.1.301
Osuna, D., Prieto, P., Aguilar, M. (2015). Control of seed germination and plant development by carbon and nitrogen availability. Front. Plant Sci. 6:1023:1023. doi: 10.3389/fpls.2015.01023
Palenchar, P. M., Kouranov, A., Lejay, L. V., Coruzzi, G. M. (2004). Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biol. 5 (11), R91. doi: 10.1186/gb-2004-5-11-r91
Peng, B., Kong, H., Li, Y., Wang, L., Zhong, M., Sun, L., et al. (2014). OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 5, 4847. doi: 10.1038/ncomms5847
Peng, C., Wang, Y., Liu, F., Ren, Y., Zhou, K., Lv, J., et al. (2014). FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant J. 77 (6), 917–930. doi: 10.1111/tpj.12444
Perchlik, M., Tegeder, M. (2017). Improving plant nitrogen use efficiency through alteration of amino acid transport processes. Plant Physiol. 175 (1), 235–247. doi: 10.1104/pp.17.00608
Perchlik, M., Tegeder, M. (2018). Leaf amino acid supply affects photosynthetic and plant nitrogen use efficiency under nitrogen stress. Plant Physiol. 178 (1), 174–188. doi: 10.1104/pp.18.00597
Perchlik, M., Foster, J., Tegeder, M. (2014). Different and overlapping functions of Arabidopsis LHT6 and AAP1 transporters in root amino acid uptake. J. Exp. Bot. 65 (18), 5193–5204. doi: 10.1093/jxb/eru278
Perez, C. M., Juliano, B O., Liboon, S. P., Alcantara, J. M., Cassman, K. G. (1996). Effects of late nitrogen fertilizer application on head rice yield, protein content, and grain quality of rice. Cereal Chem. 73 (5), 556–560. doi: 10.1021/bp960226z
Pilot, G., Stransky, H., Bushey, D. F., Pratelli, R., Ludewig, U., Wingate, V. P., et al. (2004). Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16 (7), 1827–1840. doi: 10.1105/tpc.021642
Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y. M., Rothstein, S. J. (2014). AMT1; 1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J. Exp. Bot. 65 (4), 965–979. doi: 10.1093/jxb/ert458
Rasmussen, J., Sauheitl, L., Eriksen, J., Kuzyakov, Y. (2010). Plant uptake of dual-labeled organic N biased by inorganic C uptake: Results of a triple labeling study. Soil Biol. Biochem. 42 (3), 524–527. doi: 10.1016/j.soilbio.2009.11.032
Robertson, G. P., Groffman, P. M. (2015). “Nitrogen transformations,” in Soil Microbiology, Ecology and Biochemistry, 4th ed. Ed. Paul, E. A. (Burlington, MA: Academic Press), 421–446. doi: 10.1016/b978-0-12-415955-6.00014-1
Saleh, M., Meullenet, J. (2015). Cooked rice texture and rice flour pasting properties; impacted by rice temperature during milling. J. Food Sci. Tech. Mys 52 (3), 1602–1609. doi: 10.1007/s13197-013-1180-y
Sanders, A., Collier, R., Trethewy, A., Gould, G., Sieker, R., Tegeder, M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 59 (4), 540–552. doi: 10.1111/j.1365-313X.2009.03890.x
Santiago, J. P., Tegeder, M. (2016). Connecting source with sink: the role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol. 171 (1), 508–521. doi: 10.1104/pp.16.00244
Schmidt, R., Stransky, H., Koch, W. (2007). The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226 (4), 805–813. doi: 10.1007/s00425-007-0527-x
Seiffert, B., Zhou, Z., Wallbraun, M., Lohaus, G., Möllers, C. (2004). Expression of a bacterial asparagine synthetase gene in oilseed rape (Brassica napus) and its effect on traits related to nitrogen efficiency. Physiol. Plantarum 121 (4), 656–665. doi: 10.1111/j.1399-3054.2004.00361.x
Shewry, P. R., Napier, J. A., Tatham, A. S. (1995). Seed storage proteins: structures and biosynthesis. Plant Cell 7 (7):945. doi: 10.1105/tpc.7.7.945
Sonawala, U., Dinkeloo, K., Danna, C. H., McDowell, J. M., Pilot, G. (2018). Functional linkages between amino acid transporters and plant responses to pathogens. Plant Sci. 277, 79–88. doi: 10.1016/j.plantsci.2018.09.009
Tan, Q., Zhang, L., Grant, J., Cooper, P., Tegeder, M. (2010). Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiol. 154 (4), 1886–1896. doi: 10.1104/pp.110.166389
Tegeder, M., Hammes, U. Z. (2018). The way out and in: phloem loading and unloading of amino acids. Curr. Opin. Plant Biol. 43, 16–21. doi: 10.1016/j.pbi.2017.12.002
Tegeder, M., Masclaux-Daubresse, C. (2018). Source and sink mechanisms of nitrogen transport and use. New Phytol. 217 (1), 35–53. doi: 10.1111/nph.14876
Tegeder, M., Ward, J. M. (2012). Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 3:21:21. doi: 10.3389/fpls.2012.00021
Tegeder, M. (2012). Transporters for amino acids in plant cells: some functions and many unknowns. Curr. Opin. Plant Biol. 15 (3), 315–321. doi: 10.1016/j.pbi.2012.02.001
Tegeder, M. (2014). Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J. Exp. Bot. 65 (7), 1865–1878. doi: 10.1093/jxb/eru012
Wang, L., Zhong, M., Li, X., Yuan, D., Xu, Y., Liu, H., et al. (2008). The QTL controlling amino acid content in grains of rice (Oryza sativa) are co-localized with the regions involved in the amino acid metabolism pathway. Mol. Breed. 21 (1), 127–137. doi: 10.1007/s11032-007-9141-7
Wang, W., Hu, B., Yuan, D., Liu, Y., Che, R., Hu, Y., et al. (2018). Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30 (3), 638–651. doi: 10.1105/tpc.17.00809
Wang, J., Wu, B., Lu, K., Wei, Q., Qian, J., Chen, Y., et al. (2019). The Amino Acid Permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 180 (2), 1031–1045. doi: 10.1104/pp.19.00034
Wang, X., Yang, G., Shi, M., Hao, D., Wei, Q., Wang, Z., et al. (2019). Disruption of an amino acid transporter LHT1 leads to growth inhibition and low yields in rice. BMC Plant Biol. 19 (1), 268. doi: 10.1186/s12870-019-1885-9
Xia, J., Yamaji, N., Che, J., Shen, R. F., Ma, J. F. (2014). Normal root elongation requires arginine produced by argininosuccinate lyase in rice. Plant J. 78 (2), 215–226. doi: 10.1111/tpj.12476
Xu, G., Fan, X., Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yamaguchi, S., Kamiya, Y. (2002). Gibberellins and light-stimulated seed germination. J. Plant Growth Regul. 20 (4), 369–376. doi: 10.1007/s003440010035
Yang, H., Postel, S., Kemmerling, B., Ludewig, U. (2014). Altered growth and improved resistance of Arabidopsis against Pseudomonas syringae by overexpression of the basic amino acid transporter AtCAT1. Plant Cell Environ. 37 (6), 1404–1414. doi: 10.1111/pce.12244
Zhang, W., Zhou, Y., Dibley, K. E., Tyerman, S. D., Furbank, R. T., Patrick, J. W. (2007). Review: Nutrient loading of developing seeds. Funct. Plant Biol. 34 (4), 314–331. doi: 10.1071/FP06271
Zhang, L., Tan, Q., Lee, R., Trethewy, A., Lee, Y. H., Tegeder, M. (2010). Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22 (11), 3603–3620. doi: 10.1105/tpc.110.073833
Zhang, L., Garneau, M. G., Majumdar, R., Grant, J., Tegeder, M. (2015). Improvement of pea biomass and seed productivity by simultaneous increase of phloem and embryo loading with amino acids. Plant J. 81 (1), 134–146. doi: 10.1111/tpj.12716
Zhao, H., Ma, H., Yu, L., Wang, X., Zhao, J. (2012). Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PloS One 7 (11), e49210. doi: 10.1371/journal.pone.0049210
Keywords: rice, amino acids, transporter, OsLHT1, nitrogen allocation, grain yield, grain quality
Citation: Guo N, Gu M, Hu J, Qu H and Xu G (2020) Rice OsLHT1 Functions in Leaf-to-Panicle Nitrogen Allocation for Grain Yield and Quality. Front. Plant Sci. 11:1150. doi: 10.3389/fpls.2020.01150
Received: 14 May 2020; Accepted: 15 July 2020;
Published: 29 July 2020.
Edited by:
Nandula Raghuram, Guru Gobind Singh Indraprastha University, IndiaReviewed by:
Daniel R. Bush, Colorado State University, United StatesCopyright © 2020 Guo, Gu, Hu, Qu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongye Qu, aG9uZ3llcXVAbmphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.