- 1State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China
- 2Industrial Engineering Research Center of Mulberry, State Forestry and Grassland Administration, Chongqing, China
Terpenoids are considered to be the largest group of secondary metabolites and natural products. Studies have revealed 1-deoxy-D-xylulose 5-phosphate synthase (DXS) is the first and rate-limiting enzyme in the plastidial methylerythritol phosphate pathway, which produces isopentenyl diphosphate and its isoform dimethylallyl diphosphate as terpenoid biosynthesis precursors. Mulberry (Morus L.) is an economically and ecologically important perennial tree with diverse secondary metabolites, including terpenoids that protect plants against bacteria and insects and may be useful for treating human diseases. However, there has been relatively little research regarding DXS genes in mulberry and other woody plant species. In this study, we cloned and functionally characterized three Morus notabilis DXS genes (MnDXS1, MnDXS2A, and MnDXS2B). Bioinformatics analyses indicated MnDXS1 belongs to clade 1, whereas MnDXS2A and MnDXS2B are in clade 2. The three encoded MnDXS proteins are localized to chloroplasts. Additionally, substantial differences in MnDXS expression patterns were observed in diverse tissues and in response to insect feeding and methyl jasmonate treatment. Moreover, overexpression of MnDXS1 in Arabidopsis thaliana increased the gibberellic acid content and resulted in early flowering, whereas overexpression of MnDXS2A enhanced root growth and increased the chlorophyll and carotenoid content. Our findings indicate that MnDXS functions vary among the clades, which may be useful for further elucidation of the functions of the DXS genes in mulberry.
Introduction
Terpenoids, which are also known as terpenes, are the most diverse natural products that are widely distributed among various species, including higher plants, fungi, and bacteria. In plants, terpenoids are crucial for growth and development and have specialized functions related to responses to environmental stimuli (Zhang et al., 2019). Additionally, terpenoids have long been used in perfumes, pharmaceutical products, insecticides, and industrial compounds (Tholl, 2015). For example, artemisinin, which is a potent antimalarial drug, is a sesquiterpene endoperoxide isolated from Artemisia annua (Zheng et al., 2017; Zhang et al., 2018). The mechanisms underlying terpenoid biosynthesis need to be more comprehensively characterized to further elucidate terpenoid functions in plants and satisfy the demand for more sustainable terpenoid production methods.
Isopentenyl diphosphate (IPP) and its isoform dimethylallyl diphosphate (DMAPP), which are the common precursors of terpenoids, are produced via the mevalonate (MVA) and methylerythritol phosphate (MEP) pathways (Newman and Chappell, 1999; Querol et al., 2002). Although both pathways are thought to be compartmentally independent, there is some cross-talk between them (Laule et al., 2003; Dudareva et al., 2005). The MVA pathway occurs in the cytoplasm of most eukaryotes and is responsible for the synthesis of sesquiterpenes and triterpenes, whereas the MEP pathway occurs predominantly in eubacteria and the chloroplasts of photosynthetic eukaryotes to generate precursors for the synthesis of hormones, monoterpenes, and diterpenes (Rohdich et al., 2001; Dudareva et al., 2005). In the MEP pathway, 1-deoxy-D-xylulose 5-phosphate synthase (DXS) catalyzes the first reaction that converts pyruvate and glyceraldehyde-3-phosphate (GAP) to 1-deoxy-D-xylulose 5-phosphate (Lois et al., 1998; Querol et al., 2002). The MEP pathway subsequently produces IPP and DMAPP in a 5:1 ratio in six steps (Takahashi et al., 1998). There is experimental evidence that DXS is a rate-limiting enzyme and catalyzes an important step in the bottleneck of the pathway (Lange et al., 1998; Estevez et al., 2001). Unlike the other enzymes in the MEP pathway, which are mostly encoded by single-copy genes, DXS is usually encoded by multi-copy genes that are divided into three clades (Rodriguez-Concepcion and Boronat, 2002; Cordoba et al., 2009).
Previous studies on DXS focused on herbaceous plant species, including Arabidopsis thaliana, Solanum lycopersicum, Zea mays, and Oryza sativa. Of the three DXS genes identified in A. thaliana, only AtDXS1 in clade 1 encodes a functional DXS protein (Carretero-Paulet et al., 2013). A mutation in CLA1 (DXS1) disrupts normal chloroplast development and mesophyll tissue formation in A. thaliana (Estevez et al., 2000). Additionally, overexpression of DXS1 in transgenic A. thaliana affects the accumulation of various isoprenoids such as chlorophylls, tocopherols, carotenoids, abscisic acid, and gibberellins (Estevez et al., 2001). The ectopic expression of AtDXS1 in lavender increases the essential oil monoterpene content (Munoz-Bertomeu et al., 2006). Maize also carries three DXS genes, with each belonging to a different clade. Specifically, DXS1 is highly expressed in photosynthetic tissues during early developmental stages, whereas DXS2 is preferentially expressed in young seedling roots and may influence secondary metabolism (Cordoba et al., 2011). In contrast, the maize DXS3 gene belongs to clade 3, but it has not been functionally characterized. Research regarding S. lycopersicum DXS genes revealed that SlDXS1 is ubiquitously expressed, especially during the fruit-ripening stage, whereas SlDXS2 is highly expressed in only a few tissues (Garcia-Alcazar et al., 2017). Furthermore, the non-redundant function of SlDXS2 involves modulating isoprenoid metabolism and the plasticity in the allocation of the isoprenoid precursor (Paetzold et al., 2010). A recent study confirms that OsDXS2 plays an important role as a rate-limiting enzyme supplying IPP/DMAPPs for the seed-carotenoid accumulation. The carotenoid metabolism in rice seed could be largely enhanced without any significant transcriptional alteration of carotenogenic genes (You et al., 2020). These findings imply that the DXS genes in clade 1 likely have housekeeping functions that are essential for plant growth and development, whereas those in clade 2 contribute to secondary metabolism (Saladie et al., 2014).
Substantial progress has been made in clarifying the roles of DXS in herbaceous plant species, but there has been relatively little research regarding DXS in woody plants. One exception is Populus trichocarpa. Specifically, earlier investigations proved that overexpression of PtDXS in transgenic lines increases abscisic acid and gibberellic acid (GA) contents, enhances resistance to Septotis populiperda, and decreases feeding by Micromelalopha troglodyte (Wei et al., 2019; Xu et al., 2019).
Mulberry is widely planted in the Eurasian continent and an important tree used for rearing the domesticated silkworm. The mulberry tree is also attractive to farmers with its tasty fruit and multiple uses in traditional Chinese medicine (He et al., 2013). There are multiple active compounds in mulberry, including ursolic acid, which is a terpenoid isolated from Morus nigra with antibacterial and anticancer properties (Chen et al., 2017). Terpenoids in mulberry are required for physiological activities and defense systems. Therefore, clarifying the mechanisms underlying terpenoid functions in mulberry is warranted. In this study, we conducted a series of experiments to analyze mulberry DXS genes. First, we identified three MnDXS genes and determined that MnDXS1 belongs to clade 1, whereas MnDXS2A and MnDXS2B belong to clade 2. The encoded proteins were observed to localize in chloroplasts. Additionally, the expression patterns of the three MnDXS genes in various tissues and in response to silkworm feeding and methyl jasmonate (MeJA) treatment varied substantially. Transgenic lines overexpressing MnDXS1 exhibited early flowering and increased GA accumulation, whereas overexpression of MnDXS2A positively influenced root growth and increased the chlorophyll and carotenoid content. These findings revealed the functional differences among the MnDXS genes.
Materials and Methods
Plant Materials and Treatments
Leaves, roots, stems, male flowers, female flowers, and seeds were collected from Morus notabilis trees growing in Ya’an city, Sichuan province, China. The collected samples were immediately frozen in liquid nitrogen and then stored at −80°C until used. The seeds were peeled and then germinated in agar-solidified half-strength Murashige and Skoog (MS) medium prepared with tap water until they formed roots. The resulting seedlings were transplanted into pots filled with peat moss and perlite (5:1). The plantlets were grown in a PQX plant incubator (Ningbo Southeast Instrument Corporation, Ningbo, China) at 25°C with a 12-h light/12-h dark photoperiod until the aerial parts grew to approximately 25 cm.
Seedling leaves were treated with 2- or 3-d-old fifth instar silkworm larvae or were sprayed with 0.1 mM MeJA or with water. The seedling leaves without treatment were used as control check (CK). The seedlings were incubated for 2 h, after which the leaves were collected, immediately frozen in liquid nitrogen, and then stored at −80°C for a total RNA extraction. Untreated leaves were used as controls. All experiments were carried out in three independent biological replicates.
RNA Extraction and Quantitative Real-Time (qRT)-PCR Analysis
Total RNA was extracted from the collected mulberry roots, stems, male flowers, female flowers, and leaves as well as the stress-treated leaves with the RNAiso Plus Kit (Takara Bio., Shiga, Japan). Total RNA (1 μg) was used as the template to synthesize cDNA with the PrimeScript™ RT Reagent Kit (Perfect Real Time) (Takara Bio., Shiga, Japan). A qRT-PCR assay was completed with SYBR® Premix Ex Taq™ II (Takara Bio., Shiga, Japan). The CTR9 gene served as the reference control for normalizing CT values. Gene-specific qRT-PCR primers were designed with Primer Premier 5 (Table S1). The 2−ΔΔCT method was used to calculate the relative fold-changes in gene expression (Livak and Schmittgen, 2001). The qRT-PCR was carried out in three independent technical replicates.
Cloning of DXS Genes and Phylogenetic Analysis
The AtDXS1, AtDXS2, and AtDXS3 sequences were used as queries to screen the M. notabilis genome database with the BLAST algorithm to identify MnDXS genes (http://morus.swu.edu.cn/morusdb/). The cDNA prepared from M. notabilis leaves was used as the templates to clone genes with Ex Taq DNA Polymerase (Takara Bio., Shiga, Japan). The PCR primer pairs for amplifying specific coding sequences were designed based on the available MnDXS1, MnDXS2A, and MnDXS2B sequences (Table S1). The PCR conditions were as follows: 94°C for 4 min; 32 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2.5 min; 72°C for 7 min. The amplified DXS genes were cloned into the pMD19-T vector (TaKaRa, Dalian, China) and sequenced to verify accuracy. A phylogenetic tree based on the DXS protein sequences from plants, algae, and bacteria was constructed according to the neighbor-joining method of MEGA 7, with 1,000 bootstrap replicates. The Deinococcus radiodurans, Escherichia coli, and Aegilops variabilis sequences were used as references. The phylogenetic tree was drawn with iTOL (https://itol.embl.kde).
Subcellular Localization
The putative plastid transit peptides of DXS isoforms were predicted with TargetP 1.1. The fragments of three DXS genes amplified with specific primers were inserted into the BamHI site of the pTF486 expression vector, which includes an enhanced green fluorescent protein (EGFP) coding sequence (Table S1). For each recombinant plasmid, approximately 2 μg was introduced into A. thaliana mesophyll protoplasts via polyethylene glycol-mediated transformation (Yoo et al., 2007). The EGFP fluorescence and chlorophyll autofluorescence were observed with the FV1200 confocal laser scanning microscope (Olympus, Tokyo, Japan) at excitation wavelengths of 488 and 561 nm, respectively.
Construction of the MnDXS1, MnDXS2A and MnDXS2B Overexpression Plasmids
The full-length MnDXS1, MnDXS2A, and MnDXS2B coding sequences were amplified by PCR. They were then inserted between the BamHI and EcoRI sites of the pLGNL expression vector under the control of the cauliflower mosaic virus 35S promoter and followed at the 3′ end by the nopaline synthase gene terminator. The primers used for constructing the recombinant plasmids were listed in Table S1.
Transformation and Analysis of Transgenic Arabidopsisthaliana
The MnDXS1, MnDXS2A, and MnDXS2B overexpression plasmids were inserted in wild-type (WT) A. thaliana protoplasts (ecotype Columbia) via A. tumefaciens-mediated plant transformation involving the floral dipping method (Clough and Bent, 1998). Transformed seedlings identified on agar-solidified selective medium containing 50 mg/L kanamycin were transferred to soil. Plants were incubated in a growth chamber at 24°C under a 16-h light/8-h dark photoperiod. Finally, the homozygous transgenic A. thaliana lines from the T3 generation were analyzed further.
The WT and homozygous transgenic A. thaliana seedlings were grown on agar-solidified half-strength MS medium. The seedlings were incubated for 8 d in a growth chamber at 23°C, with a 16-h light/8-h dark photoperiod. The total root length of the seedlings was measured with the WinRHIZO Arabidopsis system. The root length measurement was performed on three independent biological replicates. After transferring the seedlings to soil, the time to flowering for six well-growing plants from each transgenic line was determined and calculated as the time from when the seedlings were transferred to soil until bolting to 1 cm. A qRT-PCR assay was carried out to assess the influence of MnDXS overexpression on the flowering time gene SOC1 (suppressor of overexpression of CO1). The AtActin2 gene was used as the internal control to normalize the expression levels. The leaves from the well-growing plants were used for RNA extraction and the qRT-PCR was carried out in three independent technical replicates. Total RNA isolation, reverse transcription, and qRT-PCR methods were completed as described previously.
The fresh leaves of WT plants and MnDXS1, MnDXS2A-overexpressing transgenic lines were collected for the measurement of chlorophyll and carotenoid using the Plant Chlorophyll and Carotenoid Content Kit (Cao Benyuan, Nangjing, China) by microplate method and the measurements were carried out in three independent biological replicates. The extracted solution of leaves was measured of absorbance at 470, 645, and 663 nm. The absorbance value of the extracting solution (acetone: ethyl alcohol = 2: 1) was used for CK. The corresponding pigment content was calculated according to the following formula: chlorophyll A (mg/g) = 0.02 × (12.7 × A663 − 2.69 × A645) × D ÷ M; chlorophyll B (mg/g) = 0.02 × (22.9 × A645 − 4.68 × A663) × D ÷ M; total chlorophyll (mg/g) = 0.02 × (20.21 × A645 + 8.02 × A663) × D ÷ M; carotenoid (mg/g) = 0.02 × ((1000 × A470 − 3.27Ca − 104Cb) ÷ 229) × D ÷ M. V stands for the volume of extracting solution, D stands for the dilution factor, and m stands for the weight of collected sample.
The stems and leaves of WT plants and MnDXS1 and MnDXS2A-overexpressing transgenic lines were collected at same position to measure the GA content using the Plant Gibberellic Acid ELISA Kit (Cao Benyuan, Nangjing, China). In addition, the whole plants were used for measuring abscisic acid (ABA) content with the Plant Hormone ABA ELISA Kit (Cao Benyuan, Nangjing, China). The measurements were carried out in three independent biological replicates. Double antibody sandwich method was used to determine the GA and ABA content in plants. Blank wells, standard wells, and sample wells were set for the determination of content. The blank wells were used for CK. In the experiment for determining the GA content, the concentration of standard solution was 0, 3, 6, 12, 24, and 48 nmol/L. According to the absorbance value at 450 nm of standard solution and its corresponding concentration, a standard curve (y = 0.0278x + 0.032, R2 = 0.9978) was produced. The GA content in tested samples was then calculated according to this formula. While in determining the ABA content, the standard solution in 0, 5, 10, 20, 40, and 80 ng/ml was used for producing the standard curve (y = 0.0142x − 0.0266, R2 = 0.9879). According to the formula, the ABA content was calculated.
qRT-PCR Analysis of Biosynthesis-Related Genes
The relative expression levels in A. thaliana of geranyl (geranyl) diphosphate synthase 11 (GGPPS11) for chlorophyll biosynthesis, phytoene synthase (PSY) for carotenoid biosynthesis, ent-copalyl diphosphate synthase (CPS), and entkaurene synthase (KS) for GA biosynthesis and nine-cis-epoxycarotenoid dioxygenase 2 (NCED2) for ABA biosynthesis of WT plants and MnDXS1, MnDXS2A-overexpressing transgenic lines were determined using qRT-PCR. Total RNA was extracted from the collected transgenic A. thaliana leaves, stems, and roots. The expression patterns of chlorophyll, carotenoid, GA, and ABA metabolism-related genes were analyzed using the RNAiso Plus Kit (Takara Bio., Shiga, Japan). All experiments were carried out in three independent biological replicates. Total RNA (1 μg) was used as the template to synthesize cDNA with the PrimeScript™ RT Reagent Kit (Perfect Real Time) (Takara Bio., Shiga, Japan). A qRT-PCR assay was completed with SYBR® Premix Ex Taq™ II (Takara Bio., Shiga, Japan). The AtActin2 gene was used as the internal control to normalize the expression levels. Gene-specific qRT-PCR primers were designed with Primer Premier 5 (Table S1). The 2−ΔΔCT method was used to calculate the relative fold-changes in gene expression (Livak and Schmittgen, 2001). The qRT-PCR was carried out in three independent technical replicates.
Results
Three Mulberry DXS Genes
The three MnDXS genes identified in the M. notabilis genome database, Morus008477, Morus022865, and Morus026786 were named as MnDXS1, MnDXS2A, and MnDXS2B, respectively. The MnDXS cDNA sequences were 2,289 (MnDXS1), 2,214 (MnDXS2A), and 2,145 bp (MnDXS2B) long. Additionally, the encoded amino acid sequences were more than 65% identical, with conserved GAP-binding and TPP-binding domains (Figure 1).
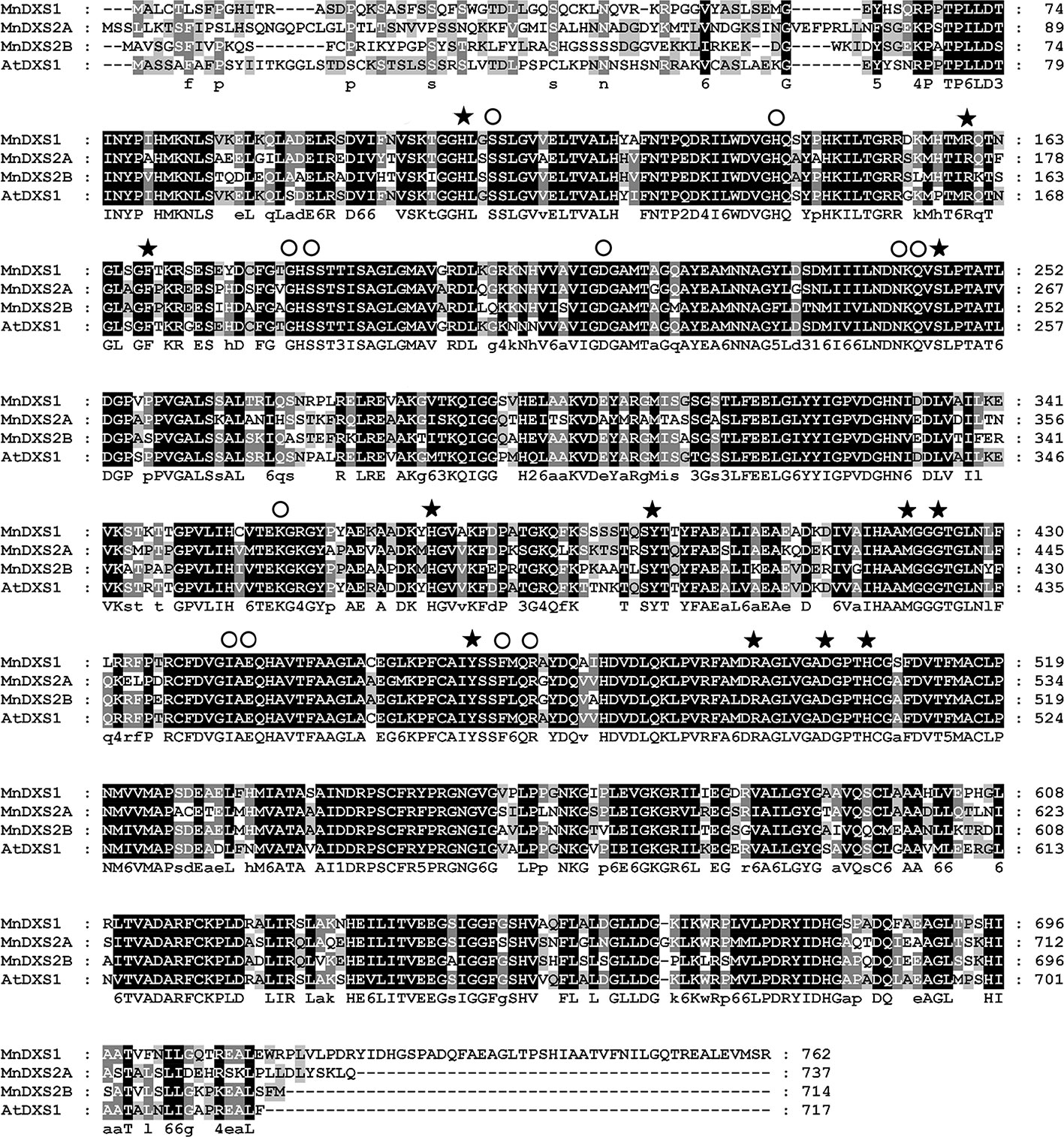
Figure 1 Alignment of the MnDXS1, MnDXS2A, and MnDXS2B amino acid sequences with the Arabidopsis thaliana DXS1 protein. Residues in the (GAP-binding and TPP-binding domains are marked with solid stars and circles, respectively.
The constructed phylogenetic tree revealed that MnDXS1 belongs to clade 1, which includes the well-characterized DXS1 proteins from A. thaliana, Z. mays, and other plant species (Figure 2). Clade 2 comprised MnDXS2A, MnDXS2B, and representatives from Medicago truncatula, Catharanthus roseus, and other species.
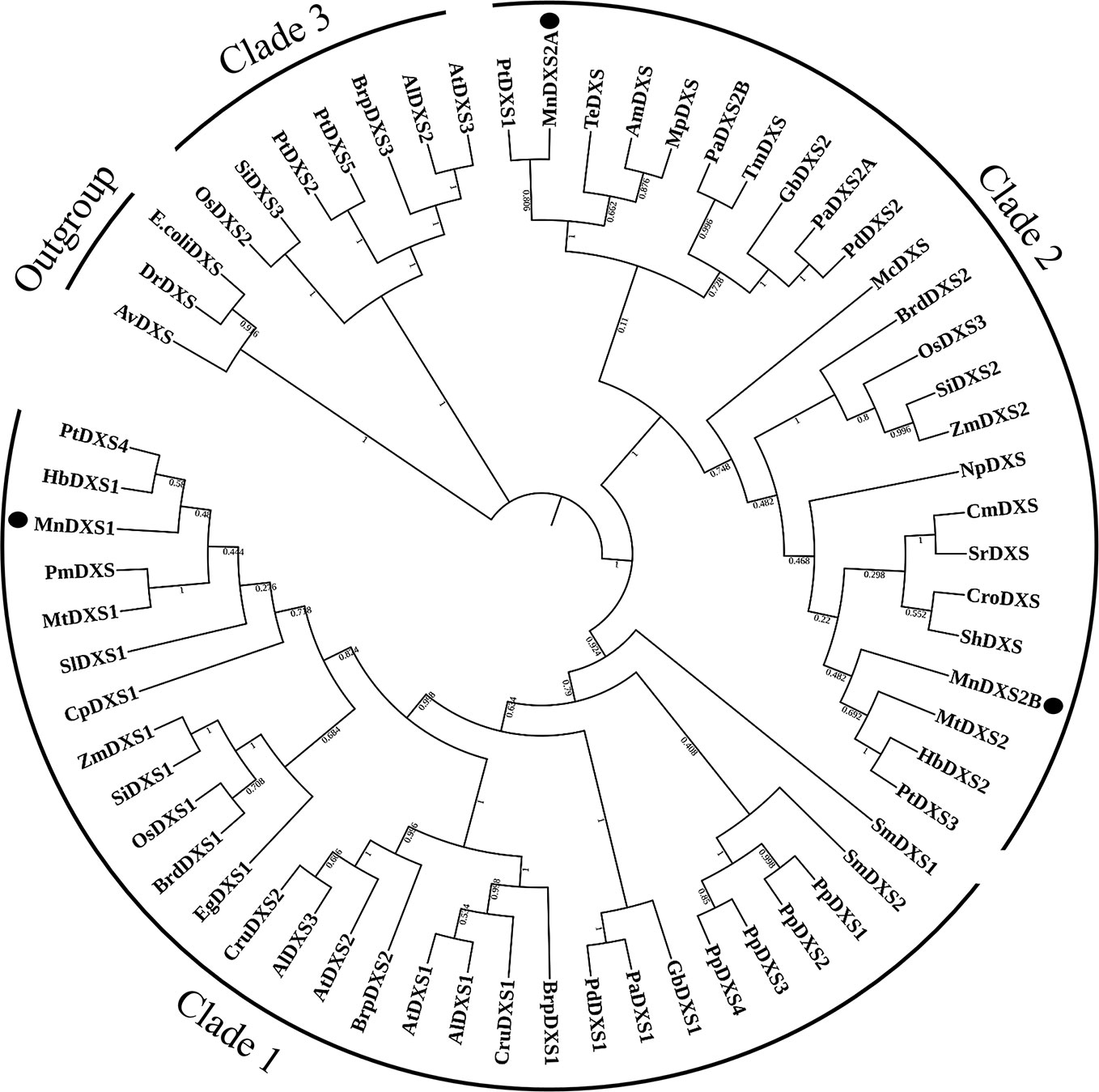
Figure 2 Phylogenetic relationships among DXS proteins. The National Center for Biotechnology Information (NCBI) accession numbers of the amino acid sequences included in the phylogenetic tree are listed in Table S2.
Subcellular Localization of MnDXS Proteins in Protoplasts
An analysis with TargetP 1.1 predicted the three MnDXS proteins are most likely localized in chloroplasts. The subcellular localization based on an examination of fluorescence by confocal laser scanning microscopy produced consistent results (Figure 3). Specifically, the EGFP signal overlapped with the chlorophyll autofluorescence signal. Because the MEP pathway is a plastid pathway, it is reasonable that these rate-limiting enzymes are localized in chloroplasts.
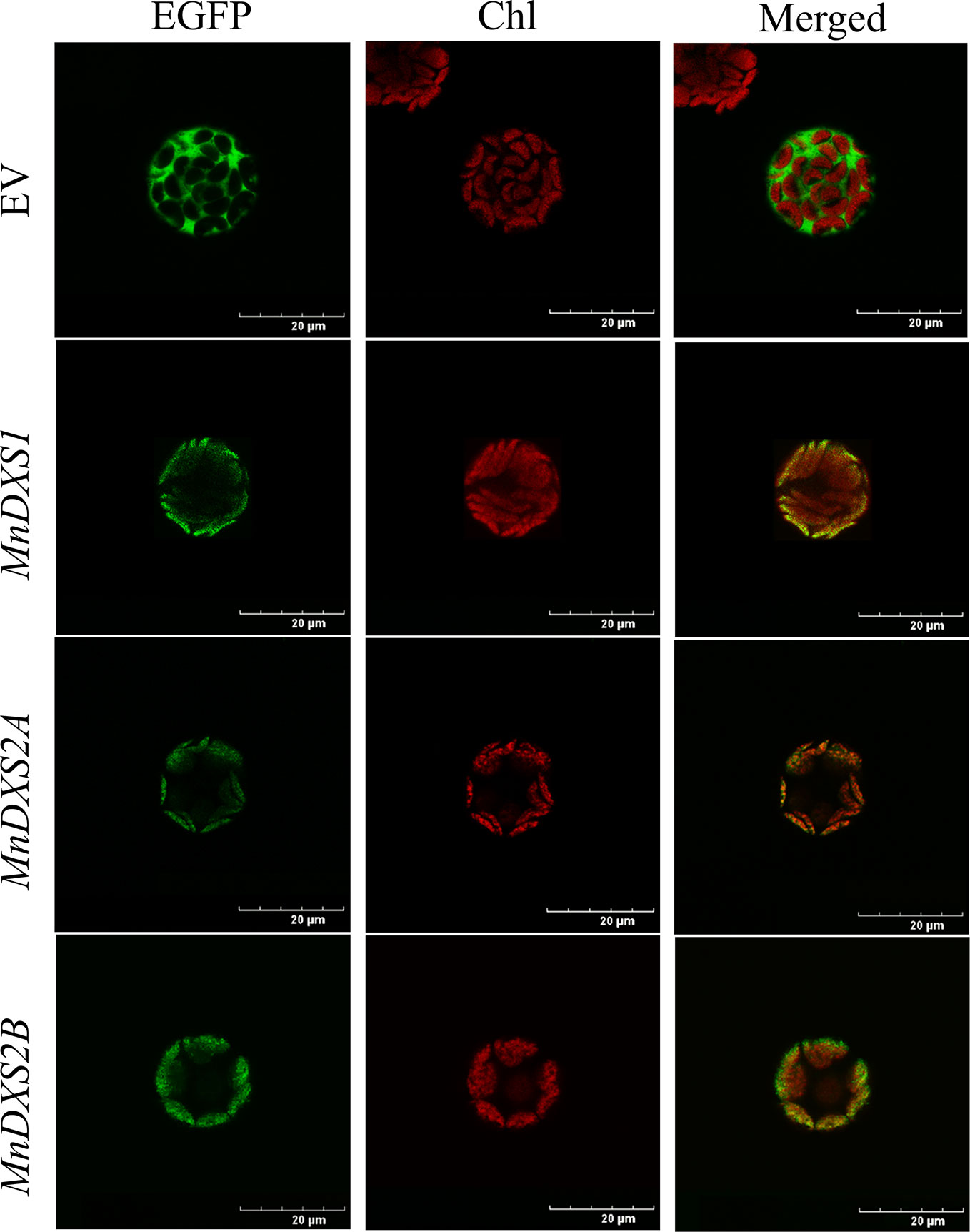
Figure 3 Images of Arabidopsis thaliana mesophyll protoplasts producing MnDXS-EGFP fusion proteins. The EGFP fluorescence and chlorophyll autofluorescence were detected by confocal laser scanning microscope.
Expression Analysis of the MnDXS Genes in Mulberry Tissues and in Response to Stress
The qRT-PCR assay revealed diverse MnDXS expression profiles among the examined tissues (Figure 4A). The MnDXS1 and MnDXS2A genes were similarly expressed at relatively high levels in all tissues. In contrast, although MnDXS2B was the most highly expressed gene in the roots, it was expressed at very low levels in the stems, leaves, and female flowers. The stress treatments affected the MnDXS1 expression level differently than the MnDXS2A and MnDXS2B expression levels (Figure 4B). The 2-h feeding by silkworm larvae and MeJA treatment tended to downregulate MnDXS1 expression, whereas these stresses had the opposite effect on MnDXS2A and MnDXS2B.
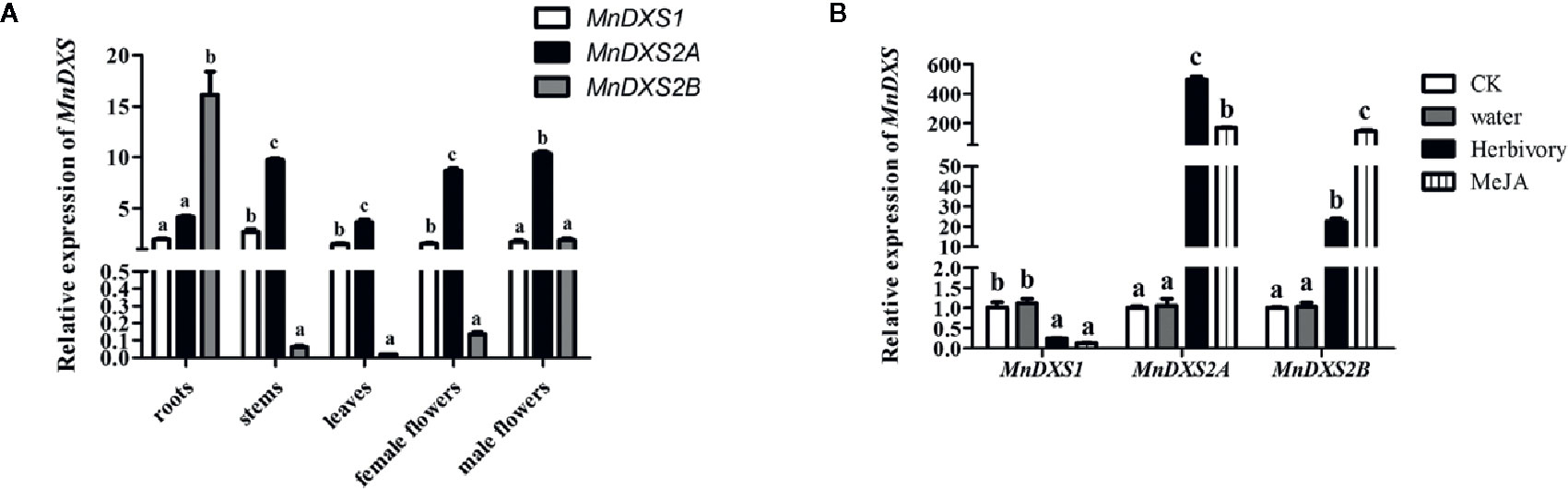
Figure 4 Relative MnDXS expression levels in various tissues and in response to stress treatments based on a qRT-PCR assay. The Morus notabilis CTR9 gene was used as the internal control to normalize the expression levels. (A) Relative MnDXS1, MnDXS2A, and MnDXS2B expression levels in the roots, stems, leaves, female flowers, and male flowers. (B) Relative MnDXS expression levels in leaves in response to silkworm feeding, MeJA and water. Data are presented as the mean ± standard error the mean of three replicates. The analysis of significant differences was conducted for each gene. CK stands for control without any treatment. Different letters above bars represent significant differences (p < 0.05) as determined with the one-way Duncan’s test.
Overexpression of MnDXS Genes Altered Arabidopsis thaliana Phenotypes
A comparison of the root lengths of WT and MnDXS-overexpressing transgenic lines revealed the genes differentially affected root growth (Figure S2). In agar-solidified half-strength MS medium, the roots of WT and MnDXS1-overexpressing lines were 32–35 mm long, whereas the roots of MnDXS2A-overexpressing plants were 40–56 mm long.
The MnDXS1-overexpressing transgenic lines flowered earlier and produced fewer rosette leaves than the WT plants (Figures 5A–D). Moreover, the SOC1 expression level was higher in the MnDXS1-overexpressing transgenic lines than those in the WT plants and other transgenic lines (Figure 5E). In MnDXS2A-overexpressing lines, greener leaves and better growth were observed (Figures 5A, B).
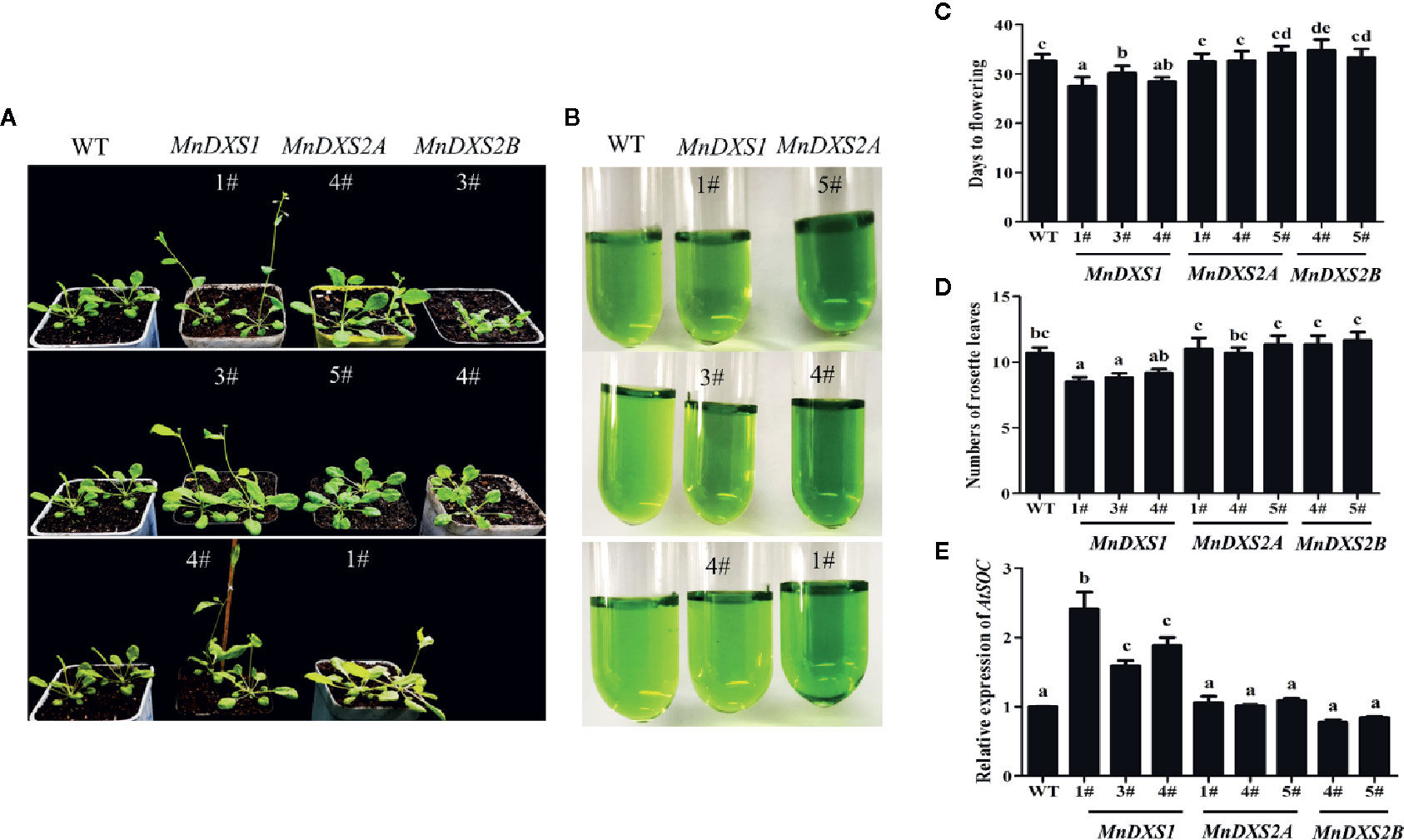
Figure 5 Days to flowering, the number of rosette leaves, and the pigment content for the WT and transgenic A. thaliana plants. (A) Phenotypes of 30-d-old wild-type and MnDXS1-, MnDXS2A-, and MnDXS2B-overexpressing transgenic A. thaliana plants. (B) The pigment extracted solution of WT and transgenic A. thaliana plants. (C, D) Days to flowering, the number of rosette leaves for the WT and transgenic A. thaliana plants. (E) qRT-PCR analyses of relative AtSOC1 expression levels in WT and transgenic A. thaliana plants based on a qRT-PCR assay, with Actin2 used as the internal control. Data are presented as the mean ± standard error of the mean of three replicates. Different letters above bars represent significant differences (p < 0.05) as determined with the one-way Duncan’s test.
As shown in Figures 6A–D, the total chlorophyll (including chlorophyll A and B) and carotenoid content were higher in MnDXS2A transgenic lines than those of WT plants and MnDXS1-overexpressing lines. The GA content of MnDXS1-overexpressing transgenic lines was significantly higher than that of WT plants, while the ABA content was decreased (Figures 6E, F). However, the GA content in different MnDXS2A transgenic lines displayed divergent trends compared with WT plants and the ABA content was slightly decreased (Figures 6E, F).
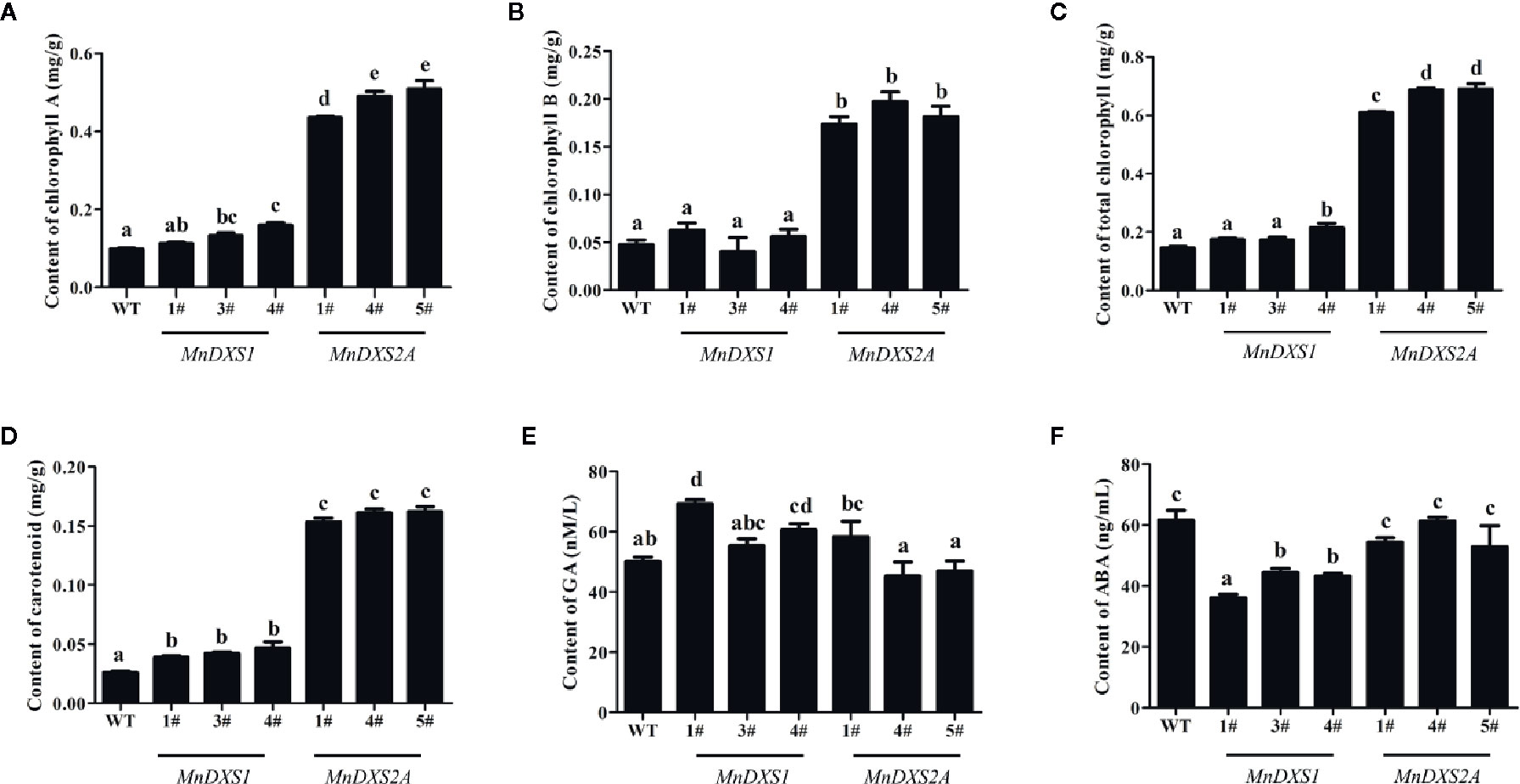
Figure 6 Chlorophyll (A–C), carotenoid (D), GA (E), and ABA (F) content in WT plants and MnDXS1- MnDXS2A-overexpressing transgenic lines. Data are presented as the mean ± standard error of the mean of three replicates. Different letters above bars represent significant differences (p < 0.05) as determined with the one-way Duncan’s test.
Overexpression of MnDXS1 and MnDXS2A Changed Relative Expression Levels of Biosynthesis-Related Genes
As shown in Figure 7, in the MnDXS1-overexpressing lines, the expression levels of AtKS and AtNCED2 were downregulated to 76% and 57% compared with the WT plants. The relative expression levels of AtGGPPS11 and AtPSY were kept at the same levels as the WT plants, while those of AtCPS was upregulated to 220%. In the MnDXS2A-overexpressing lines, the expression levels of AtGGPPS11 were upregulated to 300%, while those of AtPSY were downregulated to 42%. The relative expression levels of AtCPS, AtKS, and AtNCED2 were maintained at the same levels as the WT plants.
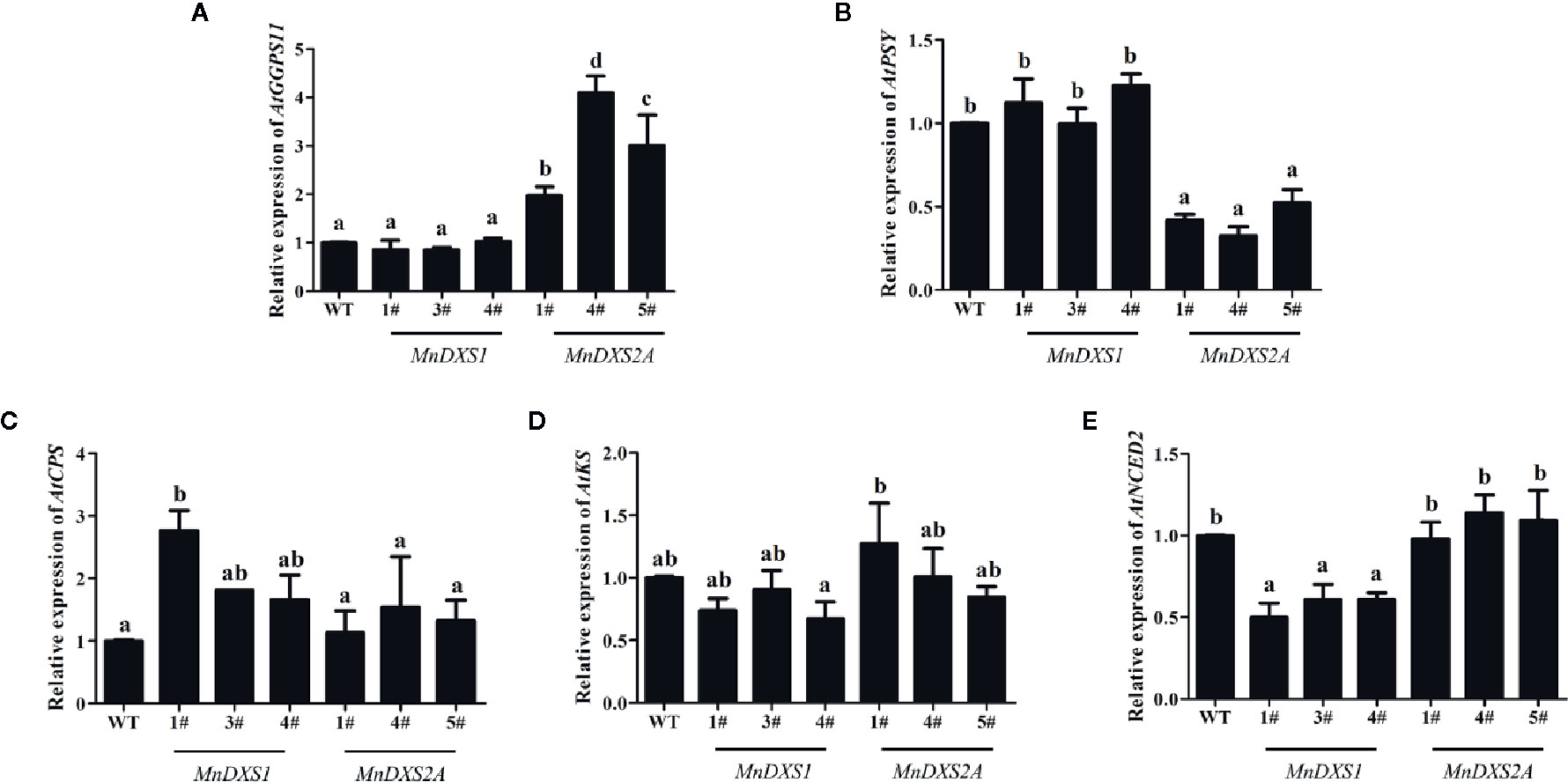
Figure 7 Relative expression levels of biosynthesis-related genes- geranyl (geranyl) diphosphate synthase 11 (GGPPS11) for chlorophyll synthesis (A), phytoene synthase (PSY) for carotenoid synthesis (B), ent-copalyl diphosphate synthase (CPS) and entkaurene synthase (KS) for GA synthesis (C, D), and nine-cis-epoxycarotenoid dioxygenase 2 (NCED2) for ABA synthesis (E) in WT plants and MnDXS1-, MnDXS2A-overexpressing transgenic lines. The Actin2 gene was used as the internal control to normalize the expression levels. Data are presented as the mean ± standard error of the mean of three replicates. Different letters above bars represent significant differences (p < 0.05) as determined with the one-way Duncan’s test.
Discussion
The MEP pathway generates IPP and DMAPP, which are precursors for the biosynthesis of hormones, monoterpenes, and diterpenes (Chappell, 2002). Terpenoids are necessary for plant growth and development and responses to environmental factors. They are also useful for humans. Therefore, the mechanisms underlying plant terpenoid metabolism and regulation will need to be more thoroughly characterized. In the MEP pathway, DXS catalyzes the first and the rate-limiting reaction, and mutations to the DXS-encoding genes in A. thaliana reportedly result in an albino phenotype (Estevez et al., 2000). A recent report suggested DXS may be an important target in Nicotiana benthamiana for manipulating the biosynthesis of taxanes, which are diterpenoids that have anticancer properties (Li et al., 2019). Previous studies on DXS confirmed that it is an essential enzyme in plants and a key target for manipulating certain tepenoids. Mulberry is a woody plant species abundant in metabolites that may be useful for treating diseases. In this study, we demonstrated that the MnDXS genes from different clades have non-redundant functions. Additionally, overexpression of MnDXS influences plant development and metabolism.
An analysis of three M. notabilis DXS genes uncovered sequences encoding conserved TPP-binding and GAP-binding domains, which are necessary for the biosynthesis of thiamine (vitamin B1) and pyridoxol (vitamin B6) (Xiang et al., 2007). Although the three MnDXS proteins are more than 65% similar regarding their amino acid sequences and are localized in chloroplasts, the corresponding genes belong to different clades. Specifically, MnDXS1 is in clade 1, whereas MnDXS2A and MnDXS2B belong to clade 2. These genes were differentially expressed in various mulberry tissues. More importantly, stresses imposed by silkworm feeding and MeJA treatments decreased MnDXS1 expression levels, but considerably increased MnDXS2A and MnDXS2B expression levels. We hypothesized that the upregulated expression of MnDXS2A and MnDXS2B might be related to certain metabolites involved in the plant defense system against herbivores. Additionally, MnDXS1 likely has a housekeeping function. Analyses of MnDXS-overexpressing lines supported our hypothesis that different MnDXS genes from different clades had distinct function. Surprisingly, the MnDXS1-overexpressing transgenic lines unexpectedly flowered earlier than the WT plants. Moreover, overexpression of MnDXS2A induced root growth. We speculated that overexpression of MnDXS2A may increase the abundance of specific terpenoids secreted from the roots to soil and further promote the absorption of nutrients by the roots.
It has been reported that the carotenoid, chlorophyll, ABA, and GA are derived from the common precursor geranylgeranyl diphosphate (GGPP) (Moreno et al., 2016). Overexpression of MnDXS genes belonging to different clades contributed to distinct content of these substances. The total chlorophyll content (including chlorophyll A and B) and carotenoid content were both significantly higher in MnDXS2A-overexpressing transgenic lines than those in WT and MnDXS1-overexpressing plants. The highest levels of GA content were observed in MnDXS1-overexpressing lines while the ABA content was obviously decreased. Although GGPP is the common precursor, it seemed that MnDXS2A was involved in the synthesis of chlorophyll and carotenoid, and overexpression of MnDXS1 contributed to the GA accumulation. The functions of MnDXS genes were differentiated.
The high levels of GA content were observed in the MnDXS1-overexpressing lines. The increased GA content may result in early flowering. A previous study on A. thaliana identified the following five flowering-time pathways: age, autonomous, gibberellin, photoperiod, and vernalization (Amasino and Michaels, 2010). GA is a tetracyclic diterpene growth factor with an essential role in many aspects of plants development, including seed germination, stem and petiole elongation, floral induction, and floral organ development. Experimental evidence also suggests that the GA biosynthesis is correlated with the MEP pathway (Yamaguchi and Kamiya, 2000). The fact that the GA content was highest in the MnDXS1-overexpressing transgenic lines is consistent with our hypothesis that higher GA content could lead to earlier flowering. A flowering-related function for DXS was detected in an earlier investigation involving rose petals, in which DXS gene expression levels increased from the bud to blooming stages (Feng et al., 2014). Additionally, in the gibberellin synthesis pathway, the CPS and KS catalyze GGDP to ent-kaurene, and CPS is a rate-limiting enzyme that determines the level of GGDP converted into gibberellins (Huang et al., 2012). It was reported that overexpression of AtCPS in A. thaliana results in high levels of ent-kaurene, but overexpression of AtKS does not increase the ent-kaurene content, indicating that they have different regulation mechanisms (Fleet et al., 2003). In the present study, AtCPS was higher expressed in MnDXS1 transgenic lines with higher GA content. The relative expression levels of GGPPS11 involved in chlorophyll synthesis were upregulated in MnDXS2A transgenic lines and those of AtNCED2 involved in ABA synthesis were downregulated in MnDXS1 lines. However, the expression profile of the gene involved in carotenoid synthesis was not consistent to the content variation. It is reasonable that the transcriptional expression of the intrinsic metabolism gene (AtPSY) might not be proportional to the expression levels of their encoded-proteins. And the protein stability of the intermediate biosynthetic enzymes might be equally as important as the enhanced activity of rate-limiting enzymes for the content enhancement (You et al., 2020).
The data presented herein prove that the MnDXS genes in different clades are functionally diverse and have non-redundant roles. The clade 1 gene MnDXS1 influences the GA content and the flowering process. Moreover, to the best of our knowledge, this is the first study to observe an early-flowering phenotype during a functional analysis of DXS. The clade 2 MnDXS genes participate in defense mechanisms and are related to root development, which are associated with secondary metabolism (Floss et al., 2008). And the chlorophyll and carotenoid content significantly increase in MnDXS2A-overexpressing lines. Although our findings are important for the molecular characterization of MnDXS genes, there remain unresolved issues. For example, we determined that MnDXS genes encoding similar amino acid sequences have distinct functions, but the underlying mechanisms remain unclear. Also the heterologous expression of MnDXS genes in A. thaliana was not sufficient enough to elucidate the function of MnDXS genes. Nevertheless, our results may be useful for future research regarding DXS genes in mulberry and related species.
Conclusion
We cloned three MnDXS genes and revealed the functional diversity among these genes. Overexpression of MnDXS1 in A. thaliana increased the GA content and resulted in early flowering. The MnDXS genes from clade 2 might mediate the synthesis of terpenoids related to plant defenses against herbivores. Additionally, overexpression of MnDXS2A resulted in higher chlorophyll and carotenoid content and promoted root growth. In conclusion, diverse DXS types influence various physiological activities in plants. These results may provide new perspectives for further research on DXS in M. notabilis and related plant species.
Data Availability Statement
The NCBI accession numbers for the three MnDXS genes are XM_024171193.1, XM_010104684.2, and XM_010115150.2.
Author Contributions
SZ, GD, and NH conceived and designed the study. GD performed the bioinformatics analysis. KL and JT did the RNA extraction and qRT-PCR analysis. SZ and WH performed the plant transformation and subcellular localization. SZ and WH managed Arabidopsis. SZ and GD analyzed the data. YL provided the technical assistance. SZ wrote the manuscript and NH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the National Key R&D Program of China (No. 2019YFD1001200), Natural Science Foundation of China (No. 31572323), and Fundamental Research Funds for the Central Universities (XDJK2019C012 and XDJK2019C088).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01142/full#supplementary-material
References
Amasino, R. M., Michaels, S. D. (2010). The timing of flowering. Plant Physiol. 154, 516–520. doi: 10.1104/pp.110.161653
Carretero-Paulet, L., Cairo, A., Talavera, D., Saura, A., Imperial, S., Rodríguez-Concepción, M., et al. (2013). Functional and evolutionary analysis of DXL1, a non-essential gene encoding a 1-deoxy-D-xylulose 5-phosphate synthase like protein in Arabidopsis thaliana. Gene 524, 40–53. doi: 10.1016/j.gene.2012.10.071
Chappell, J. (2002). The genetics and molecular genetics of terpene and sterol origami. Curr. Opin. Plant Biol. 5, 151–157. doi: 10.1016/s1369-5266(02)00241-8
Chen, H., Yu, W., Chen, G., Meng, S., Xiang, Z., He, N., et al. (2017). Antinociceptive and Antibacterial Properties of Anthocyanins and Flavonols from Fruits of Black and Non-Black Mulberries. Molecules 23. doi: 10.3390/molecules23010004
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cordoba, E., Salmi, M., Leon, P. (2009). Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60, 2933–2943. doi: 10.1093/jxb/erp190
Cordoba, E., Porta, H., Arroyo, A., San Roman, C., Medina, L., Rodríguez-Concepción, M., et al. (2011). Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 62, 2023–2038. doi: 10.1093/jxb/erq393
Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., David, R., et al. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. U. S. A. 102, 933–938. doi: 10.1073/pnas.0407360102
Estevez, J. M., Cantero, A., Romero, C., Kawaide, H., Jimenez, L. F., Kuzuyama, T., et al. (2000). Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 124, 95–104. doi: 10.1104/pp.124.1.95
Estevez, J. M., Cantero, A., Reindl, A., Reichler, S., Leon, P. (2001). 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276, 22901–22909. doi: 10.1074/jbc.M100854200
Feng, L., Chen, C., Li, T., Wang, M., Tao, J., Zhao, D., et al. (2014). Flowery odor formation revealed by differential expression of monoterpene biosynthetic genes and monoterpene accumulation in rose (Rosa rugosa Thunb.). Plant Physiol. Biochem. 75, 80–88. doi: 10.1016/j.plaphy.2013.12.006
Fleet, C. M., Yamaguchi, S., Hanada, A., Kawaide, H., David, C. J., Kamiya, Y., et al. (2003). Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 132, 830–839. doi: 10.1104/pp.103.021725
Floss, D. S., Hause, B., Lange, P. R., Kuster, H., Strack, D., Michael, H. W., et al. (2008). Knock-down of the MEP pathway isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 inhibits formation of arbuscular mycorrhiza-induced apocarotenoids, and abolishes normal expression of mycorrhiza-specific plant marker genes. Plant J. 56, 86–100. doi: 10.1111/j.1365-313X.2008.03575.x
Garcia-Alcazar, M., Gimenez, E., Pineda, B., Capel, C., Garcia-Sogo, B., Sibilla, S., et al. (2017). Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci. Rep. 7:45333. doi: 10.1038/srep45333
He, N., Zhang, C., Qi, X., Zhao, S., Tao, Y., Yang, G., et al. (2013). Draft genome sequence of the mulberry tree Morus notabilis. Nat. Commun. 4, 2445. doi: 10.1038/ncomms3445
Huang, Y., Yang, W., Pei, Z., Guo, X., Liu, D., Sun, J., et al. (2012). The genes for gibberellin biosynthesis in wheat. Funct. Integr. Genomics 12, 199–206. doi: 10.1007/s10142-011-0243-2
Lange, B. M., Wildung, M. R., Mccaskill, D., Croteau, R. (1998). A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. U. S. A. 95, 2100–2104. doi: 10.1073/pnas.95.5.2100
Laule, O., Furholz, A., Chang, H. S., Zhu, T., Wang, X., Peter, B., et al. (2003). Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 100, 6866–6871. doi: 10.1073/pnas.1031755100
Li, J., Mutanda, I., Wang, K., Yang, L., Wang, J., Wang, Y., et al. (2019). Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 10, 4850. doi: 10.1038/s41467-019-12879-y
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lois, L. M., Campos, N., Putra, S. R., Danielsen, K., Rohmer, M., Boronat, A., et al. (1998). Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 95, 2105–2110. doi: 10.1073/pnas.95.5.2105
Moreno, J. C., Cerda, A., Simpson, K., Lopez-Diaz, I., Carrera, E., Handford, M., et al. (2016). Increased Nicotiana tabacum fitness through positive regulation of carotenoid, gibberellin and chlorophyll pathways promoted by Daucus carota lycopene β-cyclase (Dclcyb1) expression. J. Exp. Bot. 67, 2325–2338. doi: 10.1093/jxb/erw037
Munoz-Bertomeu, J., Arrillaga, I., Ros, R., Segura, J. (2006). Up-regulation of 1-deoxy-D-xylulose-5-phosphate synthase enhances production of essential oils in transgenic spike lavender. Plant Physiol. 142, 890–900. doi: 10.1104/pp.106.086355
Newman, J. D., Chappell, J. (1999). Isoprenoid biosynthesis in plants: carbon partitioning within the cytoplasmic pathway. Crit. Rev. Biochem. Mol. Biol. 34, 95–106. doi: 10.1080/10409239991209228
Paetzold, H., Garms, S., Bartram, S., Wieczorek, J., Uros-Gracia, E. M., Rodríguez-Concepción, M., et al. (2010). The isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 controls isoprenoid profiles, precursor pathway allocation, and density of tomato trichomes. Mol. Plant 3, 904–916. doi: 10.1093/mp/ssq032
Querol, J., Grosdemange-Billiard, C., Rohmer, M., Boronat, A., Imperial, S. (2002). Enzymatic synthesis of 1-deoxysugar-phosphates using E. coli 1-deoxy-d-xylulose 5-phosphate synthase. Tetrahedron Lett. 43, 8265–8268. doi: 10.1016/S0040-4039(02)02018-X
Rodriguez-Concepcion, M., Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089. doi: 10.1104/pp.007138
Rohdich, F., Kis, K., Bacher, A., Eisenreich, W. (2001). The non-mevalonate pathway of isoprenoids: genes, enzymes and intermediates. Curr. Opin. Chem. Biol. 5, 535–540. doi: 10.1016/s1367-5931(00)00240-4
Saladie, M., Wright, L. P., Garcia-Mas, J., Rodriguez-Concepcion, M., Phillips, M. A. (2014). The 2-C-methylerythritol 4-phosphate pathway in melon is regulated by specialized isoforms for the first and last steps. J. Exp. Bot. 65, 5077–5092. doi: 10.1093/jxb/eru275
Takahashi, S., Kuzuyama, T., Watanabe, H., Seto, H. (1998). A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 95, 9879–9884. doi: 10.1073/pnas.95.17.9879
Tholl, D. (2015). Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 148, 63–106. doi: 10.1007/10_2014_295
Wei, H., Movahedi, A., Xu, C., Sun, W., Almasi Zadeh Yaghuti, A., Wang, P., et al. (2019). Overexpression of PtDXS Enhances Stress Resistance in Poplars. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20071669
Xiang, S., Usunow, G., Lange, G., Busch, M., Tong, L. (2007). Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 282, 2676–2682. doi: 10.1074/jbc.M610235200
Xu, C., Wei, H., Movahedi, A., Sun, W., Ma, X., Li, D., et al. (2019). Evaluation, characterization, expression profiling, and functional analysis of DXS and DXR genes of Populus trichocarpa. Plant Physiol. Biochem. 142, 94–105. doi: 10.1016/j.plaphy.2019.05.034
Yamaguchi, S., Kamiya, Y. (2000). Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 41, 251–257. doi: 10.1093/pcp/41.3.251
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
You, M. K., Lee, Y. J., Kim, J. K., Baek, S. A., Jeon, Y. A., Lim, S. H., et al. (2020). The organ-specific differential roles of rice DXS and DXR, the first two enzymes of the MEP pathway, in carotenoid metabolism in Oryza sativa leaves and seeds. BMC Plant Biol. 20, 167. doi: 10.1186/s12870-020-02357-9
Zhang, F., Liu, W., Xia, J., Zeng, J., Xiang, L., Zhu, S., et al. (2018). Molecular Characterization of the 1-Deoxy-D-Xylulose 5-Phosphate Synthase Gene Family in Artemisia annua. Front. Plant Sci. 9:952. doi: 10.3389/fpls.2018.00952
Zhang, L., Zhang, X., Wang, X., Xu, J., Wang, M., Li, L., et al. (2019). SEED CAROTENOID DEFICIENT Functions in Isoprenoid Biosynthesis via the Plastid MEP Pathway. Plant Physiol. 179, 1723–1738. doi: 10.1104/pp.18.01148
Keywords: terpenoids, 1-deoxy-D-xylulose 5-phosphate synthase, Morus notabilis, functional differentiation, early flowering
Citation: Zhang S, Ding G, He W, Liu K, Luo Y, Tang J and He N (2020) Functional Characterization of the 1-Deoxy-D-Xylulose 5-Phosphate Synthase Genes in Morus notabilis. Front. Plant Sci. 11:1142. doi: 10.3389/fpls.2020.01142
Received: 19 December 2019; Accepted: 14 July 2020;
Published: 24 July 2020.
Edited by:
Dorothea Tholl, Virginia Tech, United StatesReviewed by:
Axel Schmidt, Max Planck Institute for Chemical Ecology, GermanyTianhu Sun, Cornell University, United States
Copyright © 2020 Zhang, Ding, He, Liu, Luo, Tang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ningjia He, aGVqaWFAc3d1LmVkdS5jbg==
 Shaoyu Zhang
Shaoyu Zhang Guangyu Ding
Guangyu Ding Wenmin He
Wenmin He Kai Liu
Kai Liu Yiwei Luo
Yiwei Luo Jiaqi Tang
Jiaqi Tang Ningjia He
Ningjia He