- 1Department of Botany, Institute of Biology, Warsaw University of Life Sciences, Warsaw, Poland
- 2Department of Plant Genetics, Breeding and Biotechnology, Institute of Biology, Warsaw University of Life Sciences, Warsaw, Poland
- 3Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
- 4Center of Plant Systems Biology, VIB, Ghent, Belgium
Regulation of light absorption under variable light conditions is essential to optimize photosynthetic and acclimatory processes in plants. Light energy absorbed in excess has a damaging effect on chloroplasts and can lead to cell death. Therefore, plants have evolved protective mechanisms against excess excitation energy that include chloroplast accumulation and avoidance responses. One of the proteins involved in facilitating chloroplast movements in Arabidopsis thaliana is the J domain-containing protein required for chloroplast accumulation response 1 (JAC1). The function of JAC1 relates to the chloroplast actin filaments appearance and disappearance. So far, the role of JAC1 was studied mainly in terms of chloroplasts photorelocation. Here, we demonstrate that the function of JAC1 is more complex, since it influences the composition of photosynthetic pigments, the efficiency of photosynthesis, and the CO2 uptake rate. JAC1 has positive effect on water use efficiency (WUE) by reducing stomatal aperture and water vapor conductance. Importantly, we show that the stomatal aperture regulation is genetically coupled with JAC1 activity. In addition, our data demonstrate that JAC1 is involved in the fine-tuning of H2O2 foliar levels, antioxidant enzymes activities and cell death after UV-C photooxidative stress. This work uncovers a novel function for JAC1 in affecting photosynthesis, CO2 uptake, and photooxidative stress responses.
Introduction
Plants regulate the light absorption under naturally variable conditions, which is essential for photosynthesis optimization and acclimation. Light energy absorbed in excess (excess excitation energy, EEE) leads to photoinhibition and chloroplast retrograde-signalling and may cause chloroplast damage and ultimately cell death (Mühlenbock et al., 2008; Wituszyńska and Karpiński, 2013; Wituszyńska et al., 2015; Czarnocka and Karpiński, 2018; Górecka et al., 2020). Therefore, plants have evolved EEE avoidance and dissipation mechanisms to ensure the proper functionality of the photosynthetic apparatus and photosynthesis. One of these mechanisms is chloroplast movement (Demarsy et al., 2018).
Chloroplast photorelocation is an essential mechanism to adapt to the fluctuating light conditions (Gabryś, 2004; Suetsugu et al., 2010a). In the dark, chloroplasts are located along the anticlinal walls and at the cells’ bottom. Upon transferring plants to weak or moderate light, chloroplasts reposition themselves along the periclinal walls in order to efficiently capture light energy, which is termed the accumulation response. However, after exposure to strong light intensities, chloroplasts demonstrate avoidance response, which manifests itself by positioning at anticlinal cell walls and is aimed at lowering the risk of photoinhibition and photodamage (Wada and Kong, 2018). So far, only a couple of proteins have been identified to facilitate light intensity perception and light-mediated chloroplasts movements. Well-characterized are blue and UV-A/B photoreceptors, phototropins. Arabidopsis thaliana (Arabidopsis) possesses two such receptors, phototropin 1 (phot1) and phototropin 2 (phot2), that redundantly mediate the chloroplast accumulation response (Sakai et al., 2001). However, the avoidance response is regulated only by phot2 and not by phot1 (Kagawa et al., 2001). In Arabidopsis, the movement of chloroplasts and their anchoring to the plasma membrane is dependent on short actin filaments located along the chloroplast periphery (cp-actin filaments). Under light induction cp-actin filaments relocalize to the leading edge of chloroplasts and cause chloroplast movement that is regulated by both phot1 and phot2 (Kadota et al., 2009), and additionally by a J-domain protein required for chloroplast accumulation response 1 (JAC1) (Ichikawa et al., 2011).
JAC1 was originally identified in an Arabidopsis mutant screen deficient in chloroplast accumulation responses (Suetsugu et al., 2005). In jac1 mutant, chloroplasts remain localized to the side walls of palisade cells (Suetsugu et al., 2005; Gotoh et al., 2018). Apart from the important role in chloroplast accumulation at the cell surface in weak light, JAC1 is also required for the chloroplast relocation to the cell’s bottom in the darkness (Suetsugu et al., 2005). In palisade cells, the chloroplast dark positioning in the jac1 mutant is different from wild-type plants, because chloroplasts gather at the anticlinal cell walls (Hermanowicz et al., 2019). In addition, it was proven that under excess light, JAC1, to some extent, is also needed in the regulation of the avoidance response as cp-actin filament reorganization is impaired in jac1 (Kodama et al., 2010; Ichikawa et al., 2011). A similar perturbed cp-actin reorganization was observed in two other mutants, weak chloroplast movement under blue light 1 (web1) and plastid movement impaired 2 (pmi2) (Kodama et al., 2010), which show weak chloroplast movement during accumulation and avoidance responses. Even though the physical interaction between phot1 and phot2 (Sztatelman et al., 2016), as well as between WEB1 and PMI2 has been documented (Kodama et al., 2010), JAC1 does not physically interact with any of them (Suetsugu et al., 2005; Kodama et al., 2010). Therefore, a yet unidentified signal from plasma membrane-localized phototropins is received by JAC1 and passed along to chloroplasts to regulate the direction of their movement (Suetsugu et al., 2005). Moreover, it was shown that JAC1 is dispensable for phot2-mediated chloroplast avoidance response induced by high-light irradiation (Suetsugu et al., 2005).
The Arabidopsis JAC1 protein is 651 amino acids long and contains a 70 amino acid long auxilin-like J-domain at the C-terminus, which is essential for JAC1-mediated chloroplast photorelocation (Takano et al., 2010). A sequence-based classification assigned JAC1 to the group of type III J-proteins, including cochaperones of the heat shock protein 70 (Hsp70) chaperones (Takano et al., 2010). The JAC1 J-domain is similar to the J-domain of auxilins, enzymes that uncoat clathrin from the mature vesicles to promote the recycling of clathrin during endocytosis (Suetsugu et al., 2005). The highly conserved His-Pro-Asp (HPD) tripeptide within J-domain has been shown to facilitate the interactions with Hsp70 chaperones (Jiang et al., 2003). Structural analysis of the JAC1 J-domain indicates that HPD motif is indispensable for JAC1-dependent chloroplast light-induced movement and suggests that JAC1 cooperation with heat shock protein cognate 70 (Hsc70) chaperone is needed in this process (Suetsugu et al., 2010b). Auxilin J-domain targets and activates the Hsc70 that mediates clathrin uncoating (Eisenberg and Greene, 2007). JAC1 overexpression was shown to inhibit endocytosis process in root hair cells, which indirectly supports its role in clathrin uncoating (Ezaki et al., 2007). However, so far there is no evidence on the involvement of Hsc70 and clathrin-mediated endocytosis in chloroplast movement (Adamowski et al., 2018). The localization of JAC1 is most probably cytoplasmic, but its presence in the nucleus (Suetsugu et al., 2005) or within cellular membranes, cannot be excluded (Suetsugu et al., 2015). Furthermore, neither the expression level of JAC1 gene nor the protein level are regulated by light or by the phototropins (Suetsugu et al., 2005).
Even though the role of JAC1 has been broadly studied in terms of chloroplast movements, so far little is known about the other molecular and physiological processes that involve JAC1. This study demonstrates that JAC1 influences photosynthetic reactions, stomatal aperture and thus water vapor conductance and CO2 uptake. This regulation, at least partially, involves JAC1-dependent gene expression changes. In addition, we prove that JAC1 takes part in the fine-tuning of photooxidative-stress induced accumulation of hydrogen peroxide (H2O2), regulation of antioxidant enzymes activities and cell death. Our results shed a new light on the role of JAC1 in Arabidopsis.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana jac1-1 mutant in Columbia-0 gl1 (Col-0 gl1) background together with wild-type Col-0 gl1 plants were used in this study. The seeds were a kind gift from Prof. Masamitsu Wada (Tokyo Metropolitan University, Tokyo, Japan). Seeds were sown on Jiffy pots, stratified for two days in 4°C and moved for germination to standard laboratory conditions (8 h photoperiod, PPFD: 80 µmol photons m-² s-¹), 50% relative air humidity and temperature day/night: 22/18°C).
Measurements of Morphological and Physiological Traits
All morphological and physiological parameters were measured for four-week-old plants. Rosette size was analyzed with a FluorCam (Photon System Instruments PSI, Brno, Czech Republic) for 15 plants per genotype in two independent experiments (n=15). Rosette dry weight was measured for 12 plants for each genotype (n=12) after desiccation in 105°C for three days. The determination of water use efficiency (WUE) was performed for 12 plants per genotype in two independent experiments (n=12) according to the previously published protocol (Wituszyńska et al., 2013b; Wituszynska and Karpiński, 2014).
Microscopy and Image Analysis
The calculation of stomata number per mm² of leaf area was performed using the transparent glue imprints of the abaxial leaf side and with the use of Olympus AX70 Provis microscope (Olympus, Tokio, Japan). Three leaves, 6th, 7th and 8th were analyzed for 9 individual plants for each genotypes in two independent experiments (n=27). The number of stomata per mm² of leaf area was calculated from three frames of each microscopic sample. Stomatal aperture was measured with Image J program (Version 1.52e, National Institutes of Health, Bethesda, MD, USA) for 6th, 7th, and 8th leaf, for 77 and 74 individual stomata in the wild type and jac1 mutant, respectively (n=74–77). Arabidopsis mesophyll cells were assayed for intracellular chloroplast location with Leica TCS Sp5 confocal microscope (Leica Camera AG, Wetzlar, Germany), using chlorophyll autofluorescence upon argon laser (488 nm) excitation. 6th, 7th or 8th leaf was analyzed for 5 plants per each genotype per treatment in two independent experiments (n=5).
Chlorophyll a Fluorescence and Gas Exchange Parameters Measurements
Chlorophyll a fluorescence parameters and OJIP test were measured with a FluorCam system (Photon System Instruments PSI, Brno, Czech Republic), including four super bright LED panels 130 x 130 mm, providing actinic light (300 µmol photons m-2 s-1) and saturation pulses (4,000 µmol photons m-2 s-1). Measurements were performed with standard “Quenching” protocol implemented in the FluorCam7 software for 11–21 plants per genotypes per treatment in two separate experiments (n=11–21). Prior the measurements, plants were dark-adapted for 30 min in order to determine F0 and Fm parameters. Chlorophyll fluorescence terminology has been previously described (Baker, 2008; Wituszyńska et al., 2013a). CO2 uptake and water vapor conductance (GH2O) in variable conditions of photosynthetic active radiation (PAR) ranging between 0 and 1,900 µmol m-2 s-1 were determined using Portable Gas Exchange System GFS-3000 (Heinz Walz GmbH, Effeltrich, Germany) for 8–10 plants per genotype in two independent experiments (n=8–10). In order to obtain the slope of the regression line for each tested genotype, all the values for CO2 uptake (or GH2O) for each tested PAR intensity were used. To compare the regression line slopes the procedure from Wonnacott and Wonnacott (1990) was used. The statistical analysis was performed in the “R” version 2.12.1 using stats packages.
Photosynthetic Pigments Measurements
The determination of photosynthetic pigment compositions was performed according to the procedure described previously (Rusaczonek et al., 2015; Wituszyńska et al., 2015) for 12 plants per genotypes in two independent experiments (n=12).
Hydrogen Peroxide Levels Determination
The hydrogen peroxide (H2O2) content was determined for nine plants per genotype per treatment from two independent experiments (n=9), as previously described (Rusaczonek et al., 2015; Wituszyńska et al., 2015).
Antioxidant Enzyme Activity Measurements
The activities of superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX) were determined for nine plants per genotype per treatment from two independent experiments (n=9), as previously described (Rusaczonek et al., 2015; Wituszyńska et al., 2015).
UV-C Treatment and Cell Death Analysis
UV-C radiation (100 mJ cm-²) was used as oxidative-stress inducing factor. UV-C radiation was applied with 500 Crosslinker (Hoefer Pharmacia Biotech, San Francisco, CA, USA), equipped with lamps emitting light in the wavelength ranging from 250 to 258 nm (type G8T5, 8W; Sankyo Denki, Hiratsuka, Japan). Cell death analysis was performed by the measurement of ion leakage for 9–12 plants per genotype per treatment in two independent experiments (n=9–12), as previously described (Rusaczonek et al., 2015; Wituszyńska et al., 2015). Cell death visualization was performed by staining five leaves per genotype per treatment with 1% (m/v) Evans blue and vacuum infiltration for 30 min (Mergemann and Sauter, 2000). After staining, the leaves were washed three times with deionized water and chlorophyll was removed by the incubation in chloral hydrate (2.5 mg/mL) for two days. The pictures of representative leaves are presented.
RNA Extraction and cDNA Synthesis
The RNA was extracted from whole rosettes in three biological replicates, each consisting of at least four rosettes. Total RNA extraction was performed with a use of GeneMATRIX Universal RNA Purification Kit (EURX, Gdańsk, Poland). Additional step of on-column DNaseI digestion was performed. RNA concentration and purity were analyzed spectrophotometrically with Eppendorf BioSpectrometer (Eppendorf, Hamburg, Germany). The RNA integrity was tested by electrophoretic separation in 1% agarose gel and the same amount of RNA for each sample was used for reverse transcription. cDNA synthesis was performed with a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA).
Relative Gene Expression Measurement by Real-Time qPCR
Real-time qPCR was performed for four-week-old non-treated and UV-C treated plants (3, 6, 12, and 24 h after UV-C exposure), using the 7500 Fast Real-Time PCR System and Power SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). Each of three biological repeats per treatment was tested in three technical repeats. The following cycling programme was used in qPCR: 95°C for 10 min, followed by 40 cycles of denaturation in 95°C for 15 s and annealing/extension in 60°C for 60 s. Primers for JAC1 (AT1G75100) were designed with Primer3 software (Primer3Plus, Free Software Foundation, Inc., Boston, MA, USA) and their sequence was as follows, forward 5’- gatggttctaatgccaaggaa -3’ and reverse 5’- tgaaggtttctgatcccgatt -3’. YELLOW-LEAF-SPECIFIC GENE 8 (YLS8, AT5G08290) was used as a reference, according to the RefGenes tool incorporated in Genevestigator (Hruz et al., 2008) and amplified with the following primers: forward 5’- ttactgtttcggttgttctccattt-3’ and reverse 5’- cactgaatcatgttcgaagcaagt-3’. The specificity of each primer pair was analyzed by melting curve. The efficiency of real-time qPCR was calculated using LinRegPCR tool (Ramakers et al., 2003). Calculation of relative gene expression levels and the significance of difference between tested samples was performed using REST2009 (Pfaffl et al., 2002).
RNA Sequencing and RNA-Seq Analysis
The RNA was extracted as described above from four-week-old non-treated and UV-C treated plants 30 min after exposure to 100 mJ cm-² of UV-C radiation. RNA was isolated from three biological replicates, each consisting of at least four rosettes. RNA concentration and purity were determined spectrophotometrically using the Nanodrop ND-1000 (Nanodrop Technologies) and RNA integrity was assessed using a BioAnalyzer 2100 (Agilent). An amount of 1,000 ng of total RNA per sample was used as input. Poly-A containing mRNA was purified from the total RNA, using poly-T oligo-attached magnetic beads and the Illumina TruSeq® Stranded mRNA Sample Prep Kit (protocol version: Document # 1000000040498 v00 - October 2017). RNA was transcribed to cDNA in a reverse transcription reaction using random primers, and subsequently converted into double-stranded cDNA using DNA Polymerase I and RNAse H. The cDNA fragments were extended with a single ‘A’ base at the 3’ ends of the blunt-ended cDNA fragments, after which multiple indexing adapters were ligated introducing different barcodes for each sample. Finally, PCR was performed to enrich DNA fragments that possess adapter molecules on both ends. Sequence-libraries of each sample were equimolarly pooled and sequenced on Illumina HiSeq 4000 (Paired-end kit, 76 cycles, Dual Index, 4 lanes) at the VIB Nucleomics Core (www.nucleomics.be). Reads were aligned to the Arabidopsis genome by STAR (v2.5.2b) (Dobin et al., 2013), using the Araport11 annotation (Cheng et al., 2017). The number of reads per gene was quantified with the featureCounts function as implemented in the Subread package v1.6.2 (Liao et al., 2014). Only protein-coding genes quantified by at least 5 reads in at least three samples (20,013 genes) were retained for downstream differential gene expression analysis using the software package edgeR (Robinson et al., 2010) in R (v3.4.1). TMM normalization (Robinson and Oshlack, 2010) was applied using the calcNormFactors function. Variability in the data set was assessed with a MDS plot, showing clear separation according to genotype and UV-C treatment. In order to test user-defined hypotheses, a no-intercept single-factor model was defined combining genotype and treatment factors, e.g. such as jac1_UV. Dispersions were estimated with the estimateGLMRobustDisp function. A negative binomial regression model was used to model the overdispersed counts for each gene separately with fixed values for the dispersion parameter as outlined (McCarthy et al., 2012) and as implemented in the function glmFit using the above described model. Hypothesis testing was based on likelihood ratio tests. Contrasts of interest were the response between different genotypes under control conditions, the effect of UV stress in each genotype, and the interaction effect of UV stress and genotype. False discovery rate adjustments of the P values were performed with the method described by Benjamini and Hochberg (1995). The gene expression data were deposited in Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE143762. Gene ontology enrichment analysis was performed using ThaleMine tool (v4.1.2-20200127) within the Araport portal (Krishnakumar et al., 2015). Functional analysis of differentially expressed genes was performed using the MapMan tool (Thimm et al., 2004).
Results
JAC1 Influences Photosynthetic Efficiency and the Level of Photosynthetic Pigments
Since JAC1 is involved in the regulation of chloroplast movements (Supplementary Figure 1), our first aim was to assess if it has an influence on photosynthetic efficiency. Chlorophyll a fluorescence measurements proved that the jac1 mutant has higher maximum quantum efficiency of photosystem II (PSII) (Fv/Fm) (Figure 1A), but lower non-photochemical (NPQ) (Figure 1B) and photochemical quenching (qP) (Figure 1C), as compared to the wild-type plants.
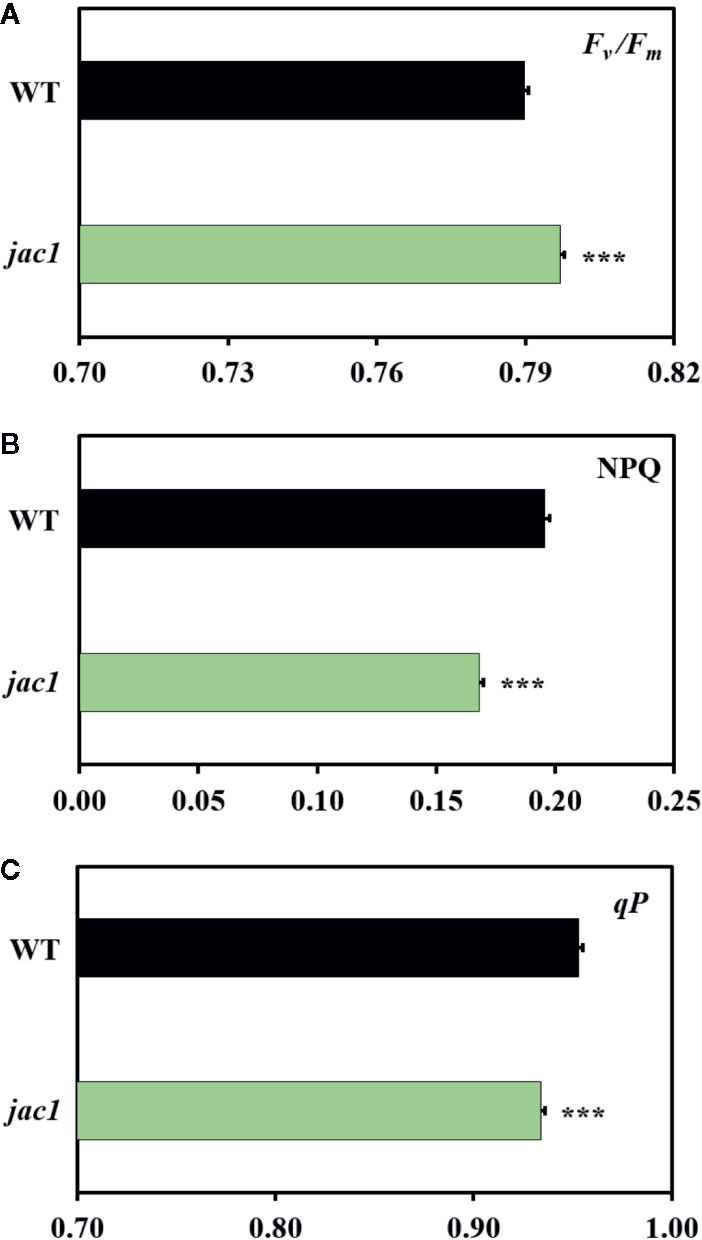
Figure 1 Chlorophyll a fluorescence parameters of 4-week-old Arabidopsis thaliana wild-type (WT) and jac1 mutant. (A) Fv/Fm, maximum quantum efficiency of PSII; (B) NPQ, non-photochemical quenching; (C) qP, photochemical quenching. Values are means (± SD) of 11–21 plants per genotype from two independent experiments (n=11–21). Asterisks indicate significant difference in comparison with the wild-type plants at the level P < 0.001 (***) according to the Tukey’s HSD multiple comparison test.
In order to obtain better insight into the specific photosynthetic reactions that are JAC1-dependent, we analyzed the fluorescence transient using the O–J–I–P test (Strasser et al., 2004). This demonstrated functional changes within light harvesting complexes (LHCs) and photosystems in jac1 mutant (Supplementary Figure 2). More specifically, a decreased ABS/RC ratio was apparent. This ratio represents the total number of photons absorbed by chlorophylls of all the reaction centres (RCs) divided by the total number of active RCs and thus suggests that jac1 mutant had more active RCs than wild-type plants. Besides measuring active RCs, the TRo/RC ratio is an estimate of maximal rate by which excitation is trapped by the RC resulting in the reduction of quinone A (QA). The decreased TRo/RC ratio in jac1 suggests that not the whole QA pool was reduced in this mutant, unlike in the wild-type. Furthermore, the jac1 mutant showed a significantly decreased reoxidation of reduced QA via electron transport in active RCs (ETo/RC). Because ETo/RC is represented only by the active centres, the lower ratio in the jac1 mutant implies again that there were more active centres than in the wild type. Finally, the effective dissipation of untrapped excitation energy from all RCs with respect to the number of active RCs (DIo/RC ratio) was also lower in jac1 mutant, indicating a better connectivity between the structurally heterogeneous units of PSII (Force et al., 2003; Wituszyńska et al., 2013a).
To study whether the observed JAC1-dependent changes in photosynthetic efficiency are caused by altered photosynthetic pigments levels, we analyzed their composition (Figure 2). The total chlorophyll level, concentration of chlorophyll b (but not chlorophyll a) were significantly elevated in jac1. We also observed that Chl a/b ratio was significantly decreased in jac1 mutant in relation to the wild type. Moreover, while β-carotene level was elevated in jac1 mutant, the content of violaxanthin, anteraxanthin and zeaxanthin were decreased in jac1 mutant, in comparison to the control plants.
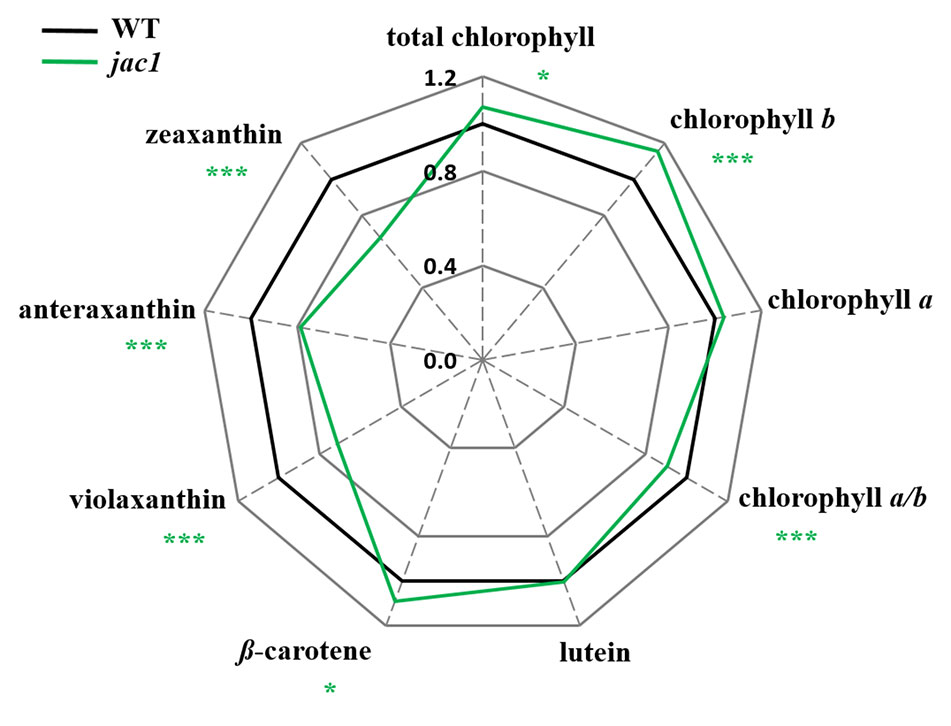
Figure 2 Photosynthetic pigment contents in 4-week-old Arabidopsis thaliana wild type and mutant plants. Total chlorophyll, chlorophyll a, chlorophyll b, chlorophyll a/b ratio; lutein; β-carotene; violaxanthin; antherxanthin; zeaxanthin. Values are means (± SD) of 12 plants per genotype from two independent experiments (n=12) expressed as the peak area (mg of dry weight-1). Asterisks indicate significant differences from the wild type according to the Tukey HSD test at the level of P < 0.05 (*), or P < 0.001 (***).
CO2 Uptake Rate and Water Vapor Conductance Are Negatively Affected by JAC1
Due to altered light-phase photosynthetic reactions in jac1 mutant, our next step was to assess whether JAC1 influences the carbon dioxide (CO2) uptake. In a range of photosynthetic active radiation (PAR) intensities that were tested, the CO2 uptake was consistently higher in jac1, reaching more than three times higher uptake in high light intensities (Figure 3A). The simultaneously measured water vapor conductance (GH2O) was elevated up to three times in low PAR intensities and, similarly to the CO2 uptake rate, a greater relative increase in jac1, compared to wild-type was apparent under high PAR (Figure 3B). Interestingly, the variation of CO2 uptake and GH2O parameters in a single measuring point was much greater in jac1 background, compared to the wild type.
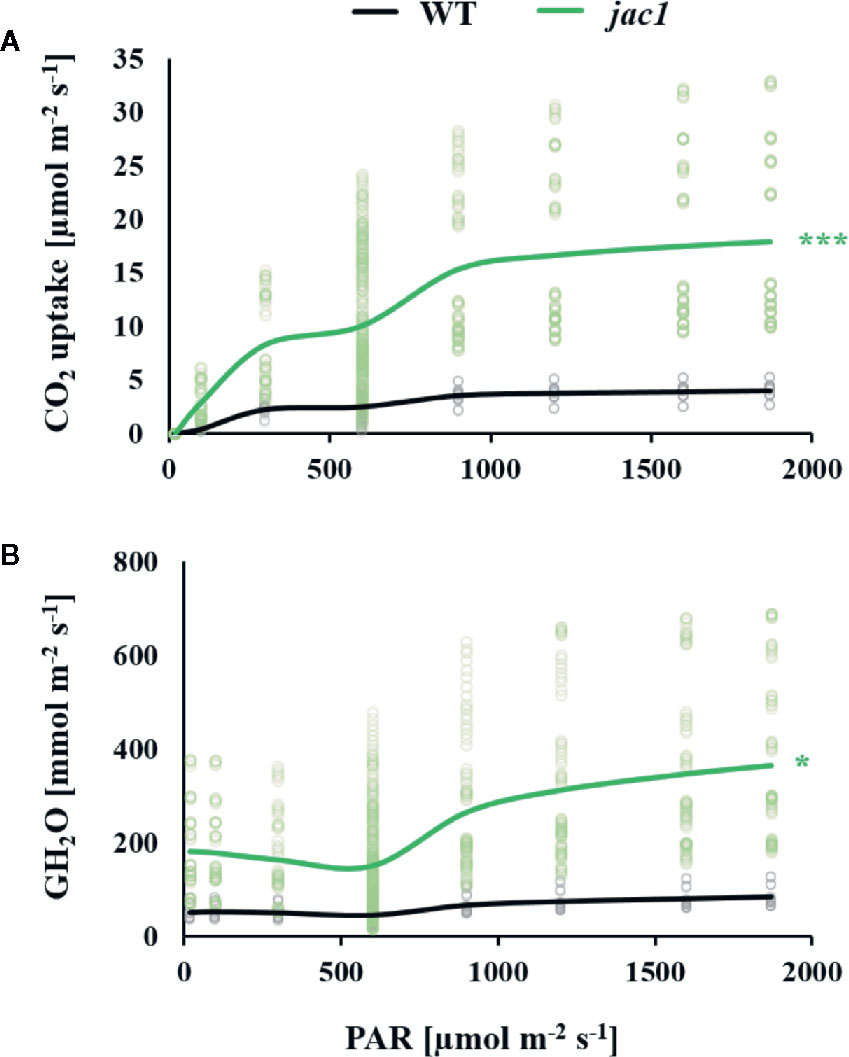
Figure 3 Gas exchange parameters in response to different light intensities (PAR) were measured for 4-week-old Arabidopsis thaliana wild type (WT) and jac1 mutant. Graphs represent relationships between light intensity and individual photosynthetic parameters. (A) CO2 uptake and (B) Water vapor conductance. Values are means of 8–10 plants per genotype from two independent experiments (n=8–10). Asterisks indicate significant difference of regression line slopes in comparison with the wild type at the level P < 0.05 (*) or P < 0.001 (***) according to the Tukey’s HSD test.
JAC1 Has an Impact on Water Use Efficiency Through the Reduction of Stomatal Conductance
Such high water vapor conductance and CO2 uptake rate in jac1 prompted us to analyze the impact of JAC1 activity on stomatal density and aperture. While the stomatal density was reduced in jac1 mutant, stomatal aperture was significantly elevated (Figures 4A–C). Moreover, water use efficiency (WUE), measured as dry weight per water used, proved to be significantly decreased in jac1 compared to wild-type plants (Figure 4D). Hence, jac1 consumed relatively more water for the production of dry weight, which is in agreement with the previously observed elevated water vapor conductance (Figure 3B), a measure of leaf transpiration efficiency. Importantly, our results showed that the rosette size and dry weight were not different between jac1 mutant and the wild type (Supplementary Figure 3, Supplementary Figures 4A, B).
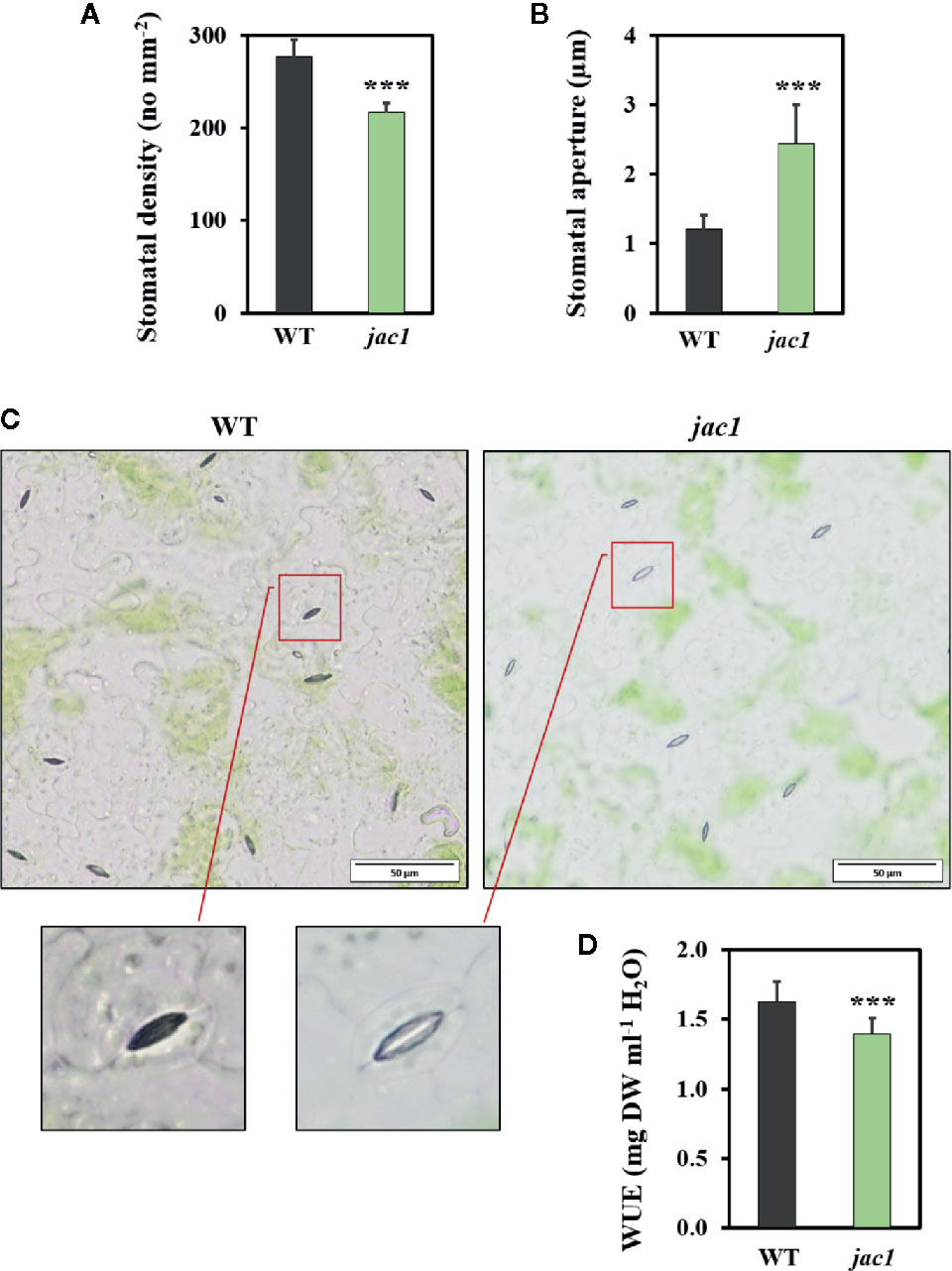
Figure 4 Morphological and physiological traits in 4-week-old Arabidopsis thaliana wild-type (WT) and jac1 mutant. (A) Stomatal density. Values are means (± SD) of 27 leaf fragments per genotype from two independent experiments (n=27); (B, C) Stomatal aperture. Values are means (± SD) of 74–77 stomata per genotype from two independent experiments (n=74–77); and (D) Water use efficiency (WUE) wild type and mutant plants grown under standard laboratory conditions. Values are means (± SD) of 12 plants per genotype from two independent experiments (n=12). Asterisks indicate significant difference in comparison with the wild-type plants at the level P < 0.001 (***) according to the Tukey’s HSD test.
Photooxidative Stress Causes Changes in JAC1 Expression Level and Shows the Influence of JAC1 on Photosynthetic Activity, Redox Homeostasis, and Cell Death Regulation
UV-C radiation has been shown to induce oxidative stress and photoinhibition. It was demonstrated that changes within chloroplasts are the first observed symptoms of UV-C induced cell death (Wituszyńska et al., 2015). Therefore, we used UV-C treatment to explore the influence of JAC1 in the photooxidative stress response and cell death
Firstly, we examined how the JAC1 expression changes after UV-C exposure (Figure 5). In the early hours (3 and 6 h) after UV-C treatment, JAC1 transcript abundance was slightly induced. However, between 6 and 12 h post UV-C stress, JAC1 expression dropped severely to reach 10% of the initial expression level after 12 h and 30% after 24 h.
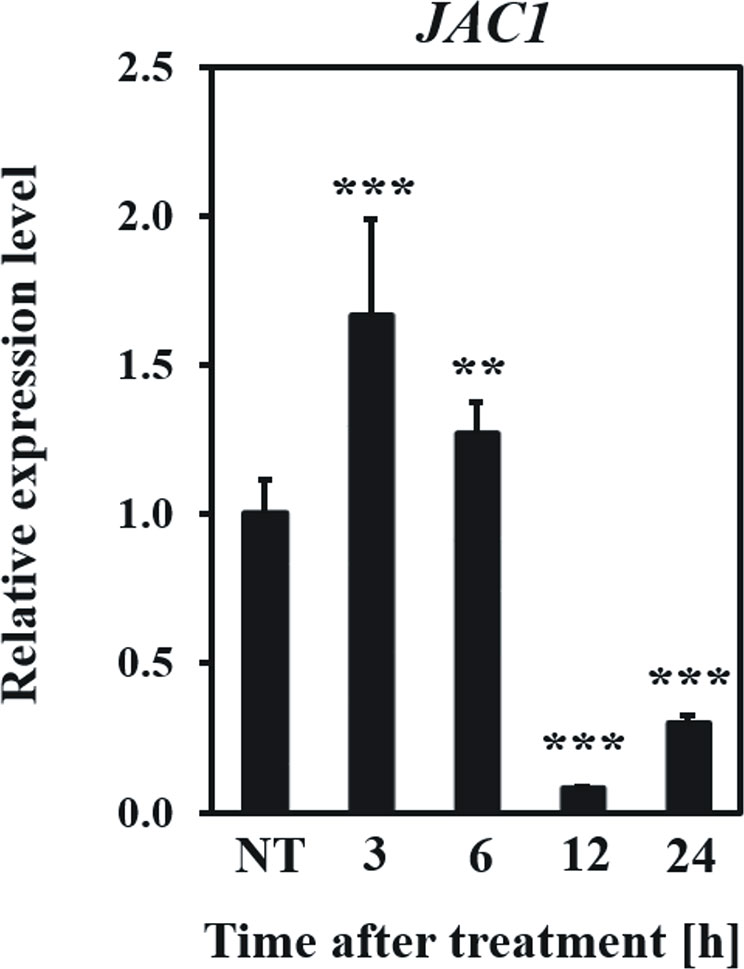
Figure 5 Relative expression level of JAC1 in the Arabidopsis thaliana wild-type plants in non-stress conditions (NT) and 3, 6, 12, 24 h after UV-C exposure. Values are means (± SD) from three biological replicates, for which three individual qPCR reactions were performed (n=9). Asterisks indicate statistically significant differences from non-treated plants, at the level P< 0.01 (**) or P< 0.001 (***).
In the next step, we explored the role of JAC1 during photosynthetic acclimation to oxidative stress. Our results demonstrated that both 48 and 96 h post UV-C exposure, jac1 had higher values of maximum and operational quantum efficiency of PSII (Fv/Fm and ΦPSII, respectively), as compared to the wild-type plants (Figures 6A, B). However, 96 h after UV-C incident, jac1 plants, compared to wild-type, exhibited decreased levels of the photochemical quenching (qP) parameter, signifying a lower proportion of PSII reaction centres to be open in jac1 (Figure 6C). In addition, the level of EEE dissipation through the NPQ reactions was decreased in jac1, compared to the wild type (Figure 6D). The overall leaf photosynthetic capacity, indicated by Rfd parameter, was elevated in jac1 mutant in relation to the control plants (Figure 6E). Taken together, this ensemble of parameters shows JAC1 to negatively influence the photosynthetic activity under photooxidative stress.
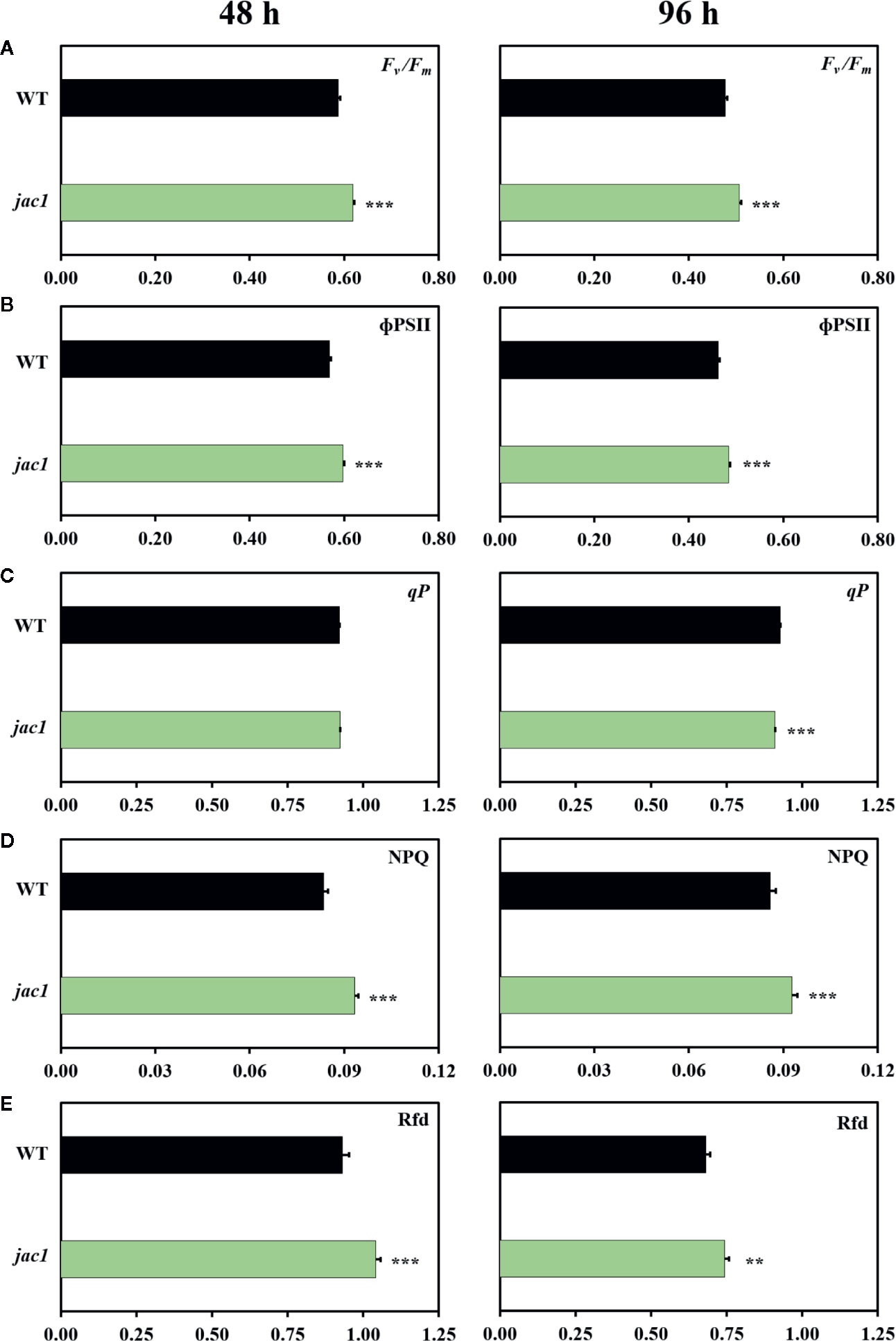
Figure 6 Chlorophyll a fluorescence parameters measured in 4-week-old Arabidopsis thaliana wild type and jac1 mutant 48 and 96 h after UV-C exposure (100 mJ cm-2). (A) Fv/Fm, maximum quantum efficiency of PSII; (B) ϕPSII, quantum yield of PSII; (C) qP, photochemical quenching; (D) NPQ, non-photochemical quenching; (E) Rfd, plant vitality parameter. Values are means (± SD) of 11–21 plants per genotype and time point from two independent experiments (n=11–21). Asterisks indicate significant difference in comparison with the wild-type plants at the level P < 0.005 (**), or P < 0.001 (***) according to the Tukey’s HSD multiple comparison test.
Next, to examine the role of JAC1 in UV-C-triggered photooxidative stress response, we analyzed the concentration of H2O2 and activities of antioxidant enzymes in jac1 mutant and wild-type plants. Before the UV-C exposure, the H2O2 content, as well as superoxide dismutase (SOD) and catalase (CAT) activities were similar in jac1 and wild-type plants (Figures 7A–C). In contrast, non-stressed jac1 plants showed significantly higher activity of ascorbate peroxidase (APX) (Figure 7D).
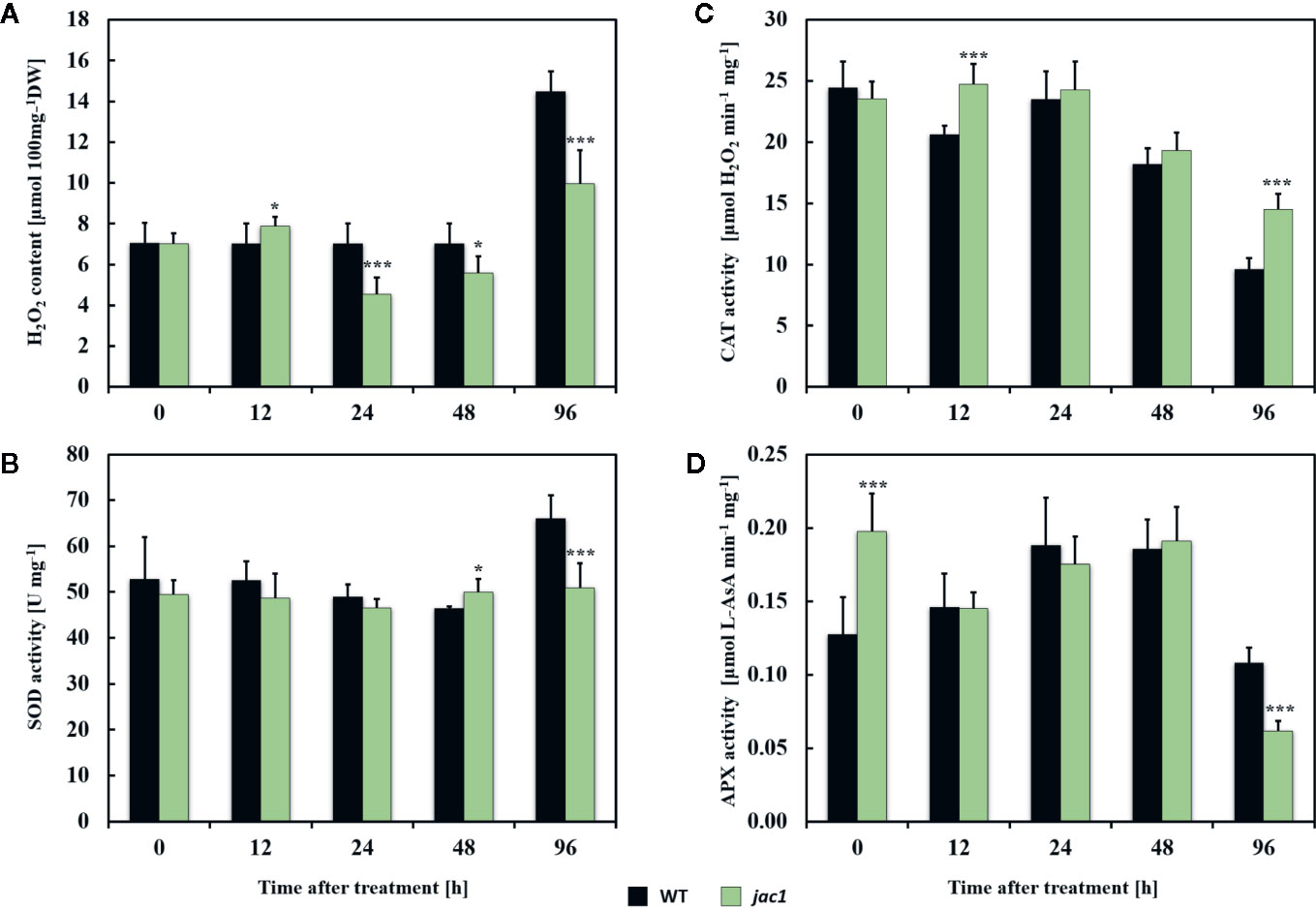
Figure 7 Hydrogen peroxide content and activities of selected antioxidant enzymes in 4-week-old Arabidopsis thaliana wild type and jac1 mutant plants measured for untreated plants and 12, 24, 48, and 96 h after UV-C exposure (100 mJ cm-2). (A) H2O2, hydrogen peroxide content; (B) SOD, superoxide dismutase activity; (C) CAT, catalase activity; (D) APX, ascorbate peroxidase activity. Values are means (± SD) of 9 plants per genotype per time point from two independent experiments (n=9). Asterisks indicate significant difference in comparison with the wild-type plants at the level P<0.05 (*) or P<0.001 (***) according to the Tukey’s HSD test.
Twelve hours after UV-C exposure, jac1 mutant demonstrated higher levels of H2O2 (Figure 7A), which corresponded with elevated CAT activity (Figure 7C). In contrast, samples analyzed in all time points after 24 h post UV-C stress showed lower content of H2O2 (Figure 7A). The pattern of SOD and APX activities in jac1 mutant was similar to the wild-type, except 96 h after UV-C exposure, when activities of those antioxidant enzymes were significantly decreased in relation to the controls (Figures 7B, D).
In order to assess the influence of JAC1 activity on UV-C-triggered cell death, we performed the analysis of cellular leakage and Evans blue staining of dead cells. Our results proved that 48 h after UV-C treatment, jac1 mutant demonstrated significantly higher ion leakage (Figure 8A). Higher cell death level was also confirmed by Evans blue staining (Figure 8B), which was visibly more intense at all time points starting from 12 h after UV-C stress.
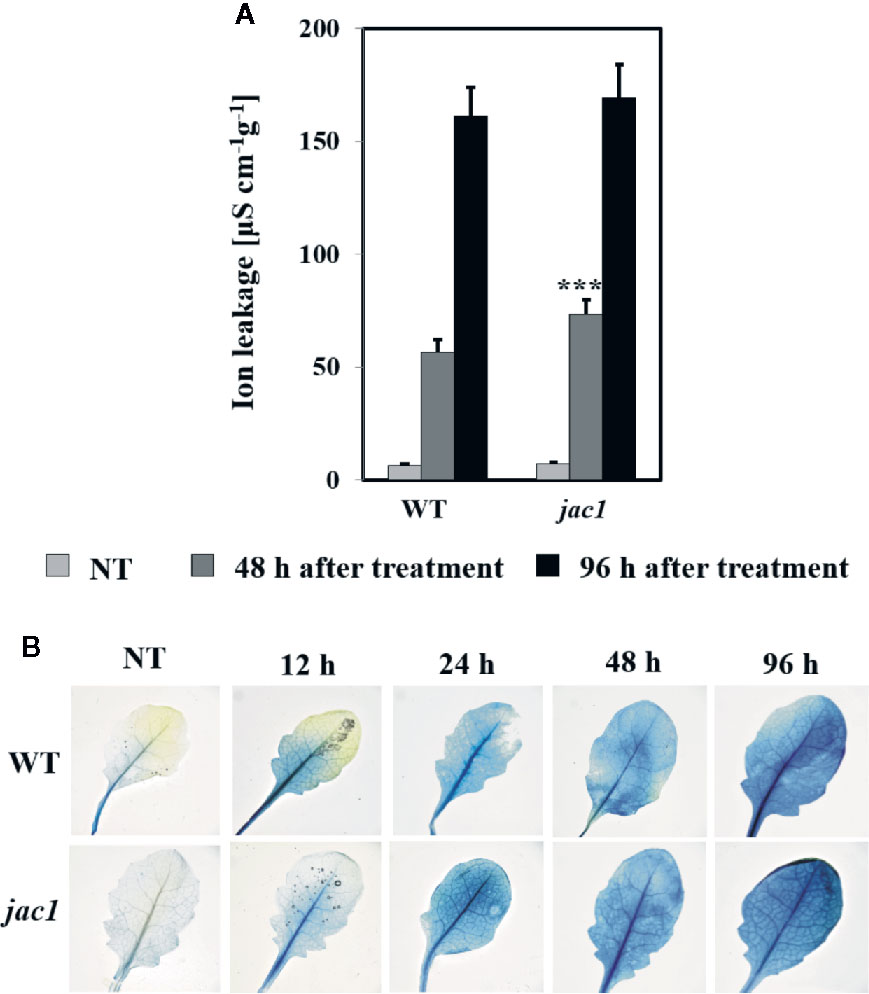
Figure 8 Cell death in 4-week-old Arabidopsis thaliana wild type and jac1 mutant measured for untreated plants (NT) and 48 and 96 h after UV-C exposure (100 mJ cm-2). Cell death (A) quantified by cellular electrolyte leakage; and (B) visualized using Evans blue staining. Values are means (± SD) of 9–12 plants per genotype per time point from two independent experiments (n=9–12). Asterisks indicate significant difference in comparison with the wild-type plants at the level P < 0.001 (***) according to the Tukey’s HSD test.
UV-C Stress Reshapes the Arabidopsis Transcriptome
In a next phase, we profiled gene expression changes under UV-C stress using high-throughput mRNA sequencing (RNA-seq). In the wild type, UV-C treatment resulted in 2,094 differentially expressed genes (DEGs) (FDR < 0.01, absolute log2 fold change (FC) > 1) (Supplementary Data Sheet 2). In total, 1,579 of these DEGs (75.4%) showed higher expression level after UV-C treatment. Gene set enrichment demonstrated that many DEGs encode membrane, cell wall and chloroplast proteins, especially those integral to thylakoids (Supplementary Data Sheet 3). Interestingly, 73 genes encoded by chloroplast genome were highly upregulated after UV-C treatment (Supplementary Table 1). In addition, amongst the DEGs, there were many genes encoding stress-responsive proteins, including transcription factors and proteins possessing oxidoreductase activity (Supplementary Data Sheet 3). There was also an enrichment of genes encoding tetrapyrrole (including chlorophyll)-binding proteins, light-harvesting complex proteins and photosystems subunits (Supplementary Figure 5), as well as biotic- and abiotic-stress responsive proteins (Supplementary Figure 6), mitochondrial electron transport chain proteins (Supplementary Figure 7), cell wall modification proteins, proteins involved in hormone and redox homeostasis, and protein modifying/degrading enzymes (Supplementary Figure 8). These results indicate that UV-C treatment in wild-type plants has pronounced effects on gene expression level.
Changes in jac1 Transcriptome in Non-Stress and After Oxidative Stress Conditions
Under control conditions, 558 DEGs were apparent in jac1 in comparison to the wild type (FDR < 0.01, |log2 FC| > 1) (Supplementary Data Sheet 2). Enrichment analysis showed that relatively high proportion of DEGs encoded extracellular (16%) and cell wall (6%) proteins (Supplementary Data Sheet 4). In terms of biological processes, jac1 mutant, in relation to the wild type, demonstrated changes in expression level of genes encoding proteins involved in abiotic stresses (mainly light-induced stress), circadian rhythm and transcriptional regulation (Supplementary Data Sheet 4). Given the observed redox- and photosynthetic-related changes in non-stressed jac1, we inspected the gene expression of several photosynthetic components and redox homeostasis enzymes. We observed induced expression of both LHCI (LHCA1, LHCA4) and LHCII (LHCB1.1, LHCB1.2, LHCB1.4, LHCB2.1, LHCB2.2, LHCB2.3, LHCB3) components in jac1, in comparison to the wild type (Supplementary Data Sheet 4). Furthermore, in non-stress conditions we found five DEGs involved in redox regulation, encoding thioredoxins, glutaredoxins, and enzymes involved in ascorbate and glutathione metabolism. For instance, monodehydroascorbate reductase (ATMDAR, AT3G09940) was highly induced in jac1 background. This enzyme takes part in ascorbate recovery, which provides reducing power to APX. In addition, we observed a relatively high number of DEGs encoding cell wall modifying enzymes in non-treated jac1, compared to the wild type. Also, genes encoding proteins involved in proteolysis and signalling, especially light- and calcium-induced signal transduction demonstrated changed expression in jac1, in relation to wild-type plants. Interestingly, seven genes involved in calcium signalling expressed in stomata guard cells were significantly induced in jac1 when compared to the wild type (Supplementary Data Sheet 4): MULTICOPY SUPPRESSORS OF SNF4 DEFICIENCY IN YEAST 3 (AT2G43290), calmodulin binding SAR DEFICIENT 1 (AT1G73805), CALMODULIN LIKE 23 (AT1G66400), CALMODULIN 9 (AT3G51920), TOUCH 2 (AT5G37770), TOUCH 3 (AT2G41100) and a calcium-binding endonuclease/exonuclease/phosphatase family gene (AT5G54130) (Obulareddy et al., 2013). Moreover, a gene encoding calcium-binding EF hand family protein (AT4G27280), engaged in stomata movements (Cominelli et al., 2005) was up-regulated in jac1, compared to the wild type. CHLORIDE CHANNEL B (AT3G27170), and two water channels, PLASMA MEMBRANE INTRINSIC PROTEIN 2 (AT3G53420) and RESPONSIVE TO DESICCATION 28 (AT2G37180), all expressed in guard cells (Obulareddy et al., 2013; Wang et al., 2016), also demonstrated significantly higher expression level in jac1 mutant, compared to wild-type counterparts.
After UV-C stress, jac1 displayed 1,017 DEGs in comparison to wild-type plants (Supplementary Data Sheet 2). Within the “cellular component” category we observed enrichment in genes encoding components of cell wall, plasma membrane and photosystems (Supplementary Data Sheet 5). DEGs in UV-treated jac1 versus UV-treated wild type were over-represented by genes encoding proteins engaged in response to stimulus (especially abiotic stress), and response to hormones. Among stress-related genes, we identified genes encoding proteins involved in oxidative stress signalling and response, such as catalase 3 (CAT3, AT1G20620), galactinol synthase 2 (GolS2, AT1G56600), and GolS3 (AT1G09350) to be up-regulated, in comparison to the wild type (Sun et al., 2013; Song et al., 2016; Selvaraj et al., 2017). Moreover, significantly changed expression level in UV-C treated jac1 background, in relation to the wild type, included genes encoding transcription factors (10% of all the DEGs), enzymes with oxidoreductase activity (8% of all the DEGs), tetrapyrole-binding proteins (3% of all the DEGs). Furthermore, MapMan categorization proved that after UV-C stress in jac1 mutant, compared to wild-type plants, there were a lot of genes (both up- and down-regulated) encoding proteins involved in signal transduction (i.e. receptor kinases, proteins involved in light-, calcium-dependent signalling), intra- and inter-cellular transport, protein degradation and development (i.e. senescence-associated genes) (Supplementary Data Sheet 5). After UV-C treatment, similarly to non-treated samples, jac1 demonstrated higher expression level of LHCI (LHCA1, LHCA3, LHCA4) and LHCII (LHCB1.1, LHCB1.2, LHCB1.4, LHCB2.2, LHCB2.4, LHCB3, LHCB4.2) components.
Discussion
Under variable natural conditions plants need to optimize the absorption of light in order to efficiently perform photosynthetic reactions. EEE, which is the energy absorbed in excess, may lead to photoinhibition, photosystem damage, chloroplasts disruption, and finally to cell death. Thus, plants are equipped in various avoidance and dissipation mechanisms that enable them to protect photosynthetic apparatus from EEE and perform optimal, condition-dependent photosynthesis.
Among the EEE protective mechanisms we can distinguish chloroplast movements, NPQ, and state-transitions (Karpiński et al., 2013; Gawroński et al., 2014; Gilroy et al., 2016; Czarnocka and Karpiński, 2018; Górecka et al., 2020). Chloroplast accumulation and avoidance response are dependent on blue light and UV-A/B radiation receptors, phototropins, and a non-receptor protein, JAC1 (Suetsugu et al., 2005; Kodama et al., 2010; Ichikawa et al., 2011). Even though the JAC1 function was well described in terms of chloroplast movements, so far little was known about its involvement in photosynthetic reactions, response to EEE and cell death.
The analysis of chlorophyll a fluorescence performed in this study proved that jac1 mutant has elevated value of maximum quantum efficiency of PSII (Fv/Fm), more active centres, better connectivity between the structurally heterogeneous PSII units and lower QA pool reduction, compared to the wild type. The results indicate that even though the mutation in JAC1 positively influences PSII maximum quantum efficiency (Fv/Fm), it has negative impact on the conversion of the absorbed photon energy into photochemistry (qP). Higher Fv/Fm in jac1, compared to the wild type, may be connected with the lack of chloroplast accumulation ability and thus the need of induced potential photosynthetic efficiency and suggests that the jac1 mutant has elevated capacity of acclimation to variable light conditions, in relation to the wild type. Moreover, higher Fv/Fm in jac1 versus wild type was in agreement with elevated level of total chlorophyll and up-regulation of genes encoding LHC subunits.
We also observed decreased values of NPQ in jac1 mutant, compared to the wild type, which well corresponded with decreased xanthophyll pigments level. Xanthophyll pigments are involved in light energy dissipation during violaxanthin-antheraxanthin-zeaxanthin (VAZ) cycle (Demmig-Adams and Adams, 1992) and thus protect plants against photodamage (Havaux and Niyogi, 1999). Lower NPQ value and reduced content of xanthophyll pigments in jac1, in relation to the wild type, may be caused by the defects in jac1 chloroplasts photorelocation. Because the jac1 chloroplasts are constantly located close to the anticlinal cell walls, low light-acclimated mutant has less necessity in energy quenching. These results indicate that JAC1-dependent chloroplast movement positively influences non-photochemical reaction rates and light energy dissipation.
Most importantly, we proved that apart from positively affecting photosynthetic electron transport, JAC1 negatively influences plant CO2 uptake. The jac1 mutant demonstrated significantly elevated CO2 uptake rate, compared to the wild type, especially in higher light intensities. This observation was most probably caused by the chloroplasts inhibited mobility. It was shown that chloroplast surface area exposed to intercellular airspaces positively correlates with the CO2 conductance, and thus it was suggested that chloroplast movement control the CO2 uptake by modulating the CO2 diffusion path length from intercellular airspaces to the chloroplast stroma (Terashima et al., 2006). It was also proven that the chloroplast avoidance response decreases the internal conductance to CO2 diffusion in Arabidopsis leaves (Tholen et al., 2008). Taking into consideration that jac1 mutant is to some extent disturbed in avoidance response (Kodama et al., 2010; Ichikawa et al., 2011), relatively higher chloroplast surface area should be adjacent to intercellular airspaces, in comparison to wild-type plants, especially in high light intensities. Therefore, the diffusion path length of CO2 to the chloroplast stroma may be lower in jac1, in relation to control plants, which can be one of the factors increasing the CO2 uptake rate in jac1 mutant. Furthermore, higher variation of CO2 uptake and GH2O parameters in a single measuring point in jac1 mutant, compared to the wild type, suggests that the lack of functional JAC1 affects the ability of whole-leaf stomatal aperture stabilization, most likely due to changes in the guard cells functioning.
Increased CO2 uptake rate, correlated with JAC1 absence, was connected with almost three-times elevated water vapor conductance and two-times higher stomatal aperture. Performed transcriptomic profiling showed that many genes, expressed in stomata guard cells and involved in stomata movements, were all up-regulated in jac1, compared to wild-type counterparts. These were genes encoding proteins involved in calcium binding (Cominelli et al., 2005; Obulareddy et al., 2013) as well as chloride and water channel proteins (Obulareddy et al., 2013; Wang et al., 2016). Therefore, it indicates that JAC1 activity is genetically coupled with the negative regulation of stomatal conductance, and thus, affects water vapor conductance and CO2 uptake rate. We cannot exclude that JAC1 regulates the gene expression directly, because of its possible presence in the nucleus (Suetsugu et al., 2005). Higher stomatal aperture and water vapor conductance in jac1 mutant were most probably the reason for higher transpiration and water loss. Therefore, JAC1 activity positively influenced water use efficiency (WUE). Despite these changes, we did not observe any statistically important changes in jac1 mutant size or dry weight, as compared to the wild-type plants. Interestingly, elevated CO2 uptake in jac1 mutant did not correlate with increased biomass production. Our observations are contrary to the study by Gotoh and co-workers (2018), where reduced photosynthesis, CO2 uptake, smaller rosette size and lesser biomass production were observed for jac1 mutant. However, this research was performed in moderate light conditions (120 µmol photons m-² s-¹), while the experiments described in the current work were all performed for relatively low light conditions (80 µmol photons m-² s-¹). This may indicate that, similarly to our previous studies on the regulators of EEE responses, such as LSD1 (Wituszyńska et al., 2013b; Wituszyńska et al., 2015), the role of JAC1 may be condition-dependent. Therefore, we postulate that JAC1 may participate in the condition-dependent optimization of the photon energy use in photosynthetic light phase reaction and the CO2 uptake.
We have shown before that UV-C treatment induces chloroplast retrograde signalling, causes photoinhibition, photooxidative stress and finally cell death (Rusaczonek et al., 2015; Wituszyńska et al., 2015). Since chloroplast movement is an important mechanism of EEE avoidance, we wanted to define the role of JAC1 protein in the activation of UV-C triggered cellular response. In a very recent study, it was shown that UV-B treatment did not trigger chloroplast rearrangements in the upper parts of palisade cells in the jac1 mutant, which means that upon UV-B, the jac1 chloroplasts were stuck and did not demonstrate avoidance response (Hermanowicz et al., 2019). Despite the greater exposure of chloroplasts to UV-C, we demonstrated that the jac1 mutant had higher values of maximum and operational quantum efficiency of PSII, and the overall leaf photosynthetic capacity (Rfd), when compared to the wild-type counterparts, thus indicating that JAC1 activity negatively affects the PSII protection under UV-C stress. These results suggest that JAC1 influences acclimatory processes, leading to LHCs and PS photo-protection.
When assessing differences in H2O2 levels and antioxidant enzyme activities in jac1 mutant and wild type plants after UV-C exposure, jac1 mutant showed higher level of H2O2 12 h after treatment. In contrast, in later time points, H2O2 levels were decreased, when compared to the wild type. The H2O2 content 12 h after UV-C exposure corresponded well with the CAT activity, which was higher in jac1, when compared to the wild type, and thus processed H2O2 more efficiently. In fact, transcriptomic profiling proved that the expression of CAT3 (AT1G20620), located in peroxisomes (Li et al., 2015), was up-regulated in jac1 background after UV-C treatment. Additionally, in non-treated jac1 we observed higher APX activity, which corresponded with higher expression of monodehydroascorbate reductase (ATMDAR), engaged in ascorbate recovery.
Moreover, UV-induced cell death was more pronounced in jac1 mutant after 48 h, but not after 96 h post UV-C exposure, which suggests that JAC1 negatively influences the fine-tuning of cell death signalling and progression. The only work so far studying the impact of JAC1 on stress response showed that Arabidopsis line with significantly higher expression of JAC1 demonstrated lower level of aluminium-induced oxidative stress and higher resistance towards Al stress, than the wild-type plants (Ezaki et al., 2007). This study is convergent with our conclusion on the role of JAC1 in the increasing of resistance towards cell death. Interestingly, the expression level of JAC1 demonstrated dynamic pattern, as it was up-regulated 3 and 6 h after UV-C exposure and strongly down-regulated afterwards. It seems that Arabidopsis wild-type plants tend to diminish JAC1 expression to reduce its negative impact on photosynthetic activity and lower the spread of cell death under oxidative stress. All these results indicate that JAC1 is most important at the early stages of photooxidative stress as it is engaged in the fine-tuning of the cellular redox homeostasis and cell death.
In the present work, we also identified genes that are transcriptionally affected by the absence of JAC1 activity. Interestingly, in jac1 mutant, when compared to the wild type, we found deregulation of many genes encoding proteins engaged in calcium signalling, such as calmodulins (CaMs), CaM-like and CaM-binding proteins. It was previously shown that JAC1 binds calmodulin 1 (CaM1) and CaM-like CML10 (Popescu et al., 2007). Here, we prove that the regulation of CaMs is not only at the protein-protein interaction level, but also at the transcriptional stage. Importantly, two times more genes showed significant change in the expression level in UV-C treated jac1, compared to the UV-C treated wild type. It may be caused by altered chloroplast positioning in jac1, that due to impaired avoidance response capture more UV-C and hence provoke more plastid signals affecting nuclear gene expression. Although we have identified many genes that are influenced by the lack of JAC1 activity, the identification of JAC1-interacting proteins may further clarify the role of JAC1 in chloroplast movement, photosynthetic reactions, stomatal conductance, and stress responses.
In conclusion, we have identified a new role of JAC1 in the photosynthetic apparatus efficiency and plant stomatal conductance and thus CO2 uptake. Our results prove that the movements of stomata are genetically coupled with JAC1 activity. From a broader perspective, we demonstrate that chloroplast altered positioning, dependent on JAC1, influences plant photosynthetic performance, H2O2 foliar levels, antioxidant enzymes activities, and cell death after UV-C photooxidative stress.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WC, AR, and SK planned the experiments and postulated the hypotheses tested in this paper. WC measured hydrogen peroxide content and activities of antioxidant enzymes, prepared the RNA for NGS sequencing, performed the qPCRs, and analyzed functionally the RNAseq results. AR performed morphological analyses (measured plant size, dry weight, stomatal density), analyzed water use efficiency, chlorophyll a fluorescence, and ion leakage. PW did the annotation and statistical analysis of the RNAseq data. MS-R performed analysis of stomatal aperture and Evans Blue staining. WC wrote the manuscript and together with AR prepared the figures. SK and FB reviewed and approved its final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Maestro 6 project (2014/14/A/NZ1/00218) granted to SK by the National Science Centre. PW is supported by the FWO grant 3G006916.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01124/full#supplementary-material
References
Adamowski, M., Narasimhan, M., Kania, U., Glanc, M., De Jaeger, G., Friml, J. (2018). A Functional Study of AUXILIN-LIKE1 and 2, Two Putative Clathrin Uncoating Factors in Arabidopsis. Plant Cell 30, 700–716. doi: 10.1105/tpc.17.00785
Baker, N. R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59, 89–113. doi: 10.1146/annurev.arplant.59.032607.092759
Benjamini, Y., Hochberg, Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Cheng, C.-Y., Krishnakumar, V., Chan, A. P., Thibaud-Nissen, F., Schobel, S., Town, C. D. (2017). Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. Cell Mol. Biol. 89, 789–804. doi: 10.1111/tpj.13415
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A Guard-Cell-Specific MYB Transcription Factor Regulates Stomatal Movements and Plant Drought Tolerance. Curr. Biol. 15, 1196–1200. doi: 10.1016/j.cub.2005.05.048
Czarnocka, W., Karpiński, S. (2018). Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 122, 4–20. doi: 10.1016/j.freeradbiomed.2018.01.011
Demarsy, E., Goldschmidt-Clermont, M., Ulm, R. (2018). Coping with “Dark Sides of the Sun” through Photoreceptor Signaling. Trends Plant Sci. 23, 260–271. doi: 10.1016/j.tplants.2017.11.007
Demmig-Adams, B., Adams, W. W. (1992). Photoprotection and Other Responses of Plants to High Light Stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 599–626. doi: 10.1146/annurev.pp.43.060192.003123
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl. 29, 15–21. doi: 10.1093/bioinformatics/bts635
Eisenberg, E., Greene, L. E. (2007). Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic Cph. Den. 8, 640–646. doi: 10.1111/j.1600-0854.2007.00568.x
Ezaki, B., Kiyohara, H., Matsumoto, H., Nakashima, S. (2007). Overexpression of an auxilin-like gene (F9E10.5) can suppress Al uptake in roots of Arabidopsis. J. Exp. Bot. 58, 497–506. doi: 10.1093/jxb/erl221
Force, L., Critchley, C., van Rensen, J. J. S. (2003). New fluorescence parameters for monitoring photosynthesis in plants. Photosynth. Res. 78, 17. doi: 10.1023/A:1026012116709
Gabryś, H. (2004). Blue light-induced orientation movements of chloroplasts in higher plants: Recent progress in the study of their mechanisms. Acta Physiol. Plant 26, 473–479. doi: 10.1007/s11738-004-0038-3
Gawroński, P., Witoń, D., Vashutina, K., Bederska, M., Betliński, B., Rusaczonek, A., et al. (2014). Mitogen-activated protein kinase 4 is a salicylic acid-independent regulator of growth but not of photosynthesis in Arabidopsis. Mol. Plant 7, 1151–1166. doi: 10.1093/mp/ssu060
Gilroy, S., Białasek, M., Suzuki, N., Górecka, M., Devireddy, A. R., Karpiński, S., et al. (2016). ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants1[OPEN]. Plant Physiol. 171, 1606–1615. doi: 10.1104/pp.16.00434
Górecka, M., Lewandowska, M., Dąbrowska-Bronk, J., Białasek, M., Barczak-Brzyżek, A., Kulasek, M., et al. (2020). Photosystem II 22kDa protein level - a prerequisite for excess light-inducible memory, cross-tolerance to UV-C and regulation of electrical signalling. Plant Cell Environ. 43, 649–661. doi: 10.1111/pce.13686
Gotoh, E., Suetsugu, N., Yamori, W., Ishishita, K., Kiyabu, R., Fukuda, M., et al. (2018). Chloroplast Accumulation Response Enhances Leaf Photosynthesis and Plant Biomass Production. Plant Physiol. 178, 1358–1369. doi: 10.1104/pp.18.00484
Havaux, M., Niyogi, K. K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. 96, 8762–8767. doi: 10.1073/pnas.96.15.8762
Hermanowicz, P., Banaś, A. K., Sztatelman, O., Gabryś, H., Łabuz, J. (2019). UV-B Induces Chloroplast Movements in a Phototropin-Dependent Manner. Front. Plant Sci. 10:1279. doi: 10.3389/fpls.2019.01279
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008:420747. doi: 10.1155/2008/420747
Ichikawa, S., Yamada, N., Suetsugu, N., Wada, M., Kadota, A. (2011). Red light, Phot1 and JAC1 modulate Phot2-dependent reorganization of chloroplast actin filaments and chloroplast avoidance movement. Plant Cell Physiol. 52, 1422–1432. doi: 10.1093/pcp/pcr087
Jiang, J., Taylor, A. B., Prasad, K., Ishikawa-Brush, Y., Hart, P. J., Lafer, E. M., et al. (2003). Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry 42, 5748–5753. doi: 10.1021/bi034270g
Kadota, A., Yamada, N., Suetsugu, N., Hirose, M., Saito, C., Shoda, K., et al. (2009). Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 13106–13111. doi: 10.1073/pnas.0906250106
Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., et al. (2001). Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. doi: 10.1126/science.291.5511.2138
Karpiński, S., Szechyńska-Hebda, M., Wituszyńska, W., Burdiak, P. (2013). Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 36, 736–744. doi: 10.1111/pce.12018
Kodama, Y., Suetsugu, N., Kong, S.-G., Wada, M. (2010). Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 107, 19591–19596. doi: 10.1073/pnas.1007836107
Krishnakumar, V., Hanlon, M. R., Contrino, S., Ferlanti, E. S., Karamycheva, S., Kim, M., et al. (2015). Araport: the Arabidopsis Information Portal. Nucleic Acids Res. 43, D1003–D1009. doi: 10.1093/nar/gku1200
Li, J., Liu, J., Wang, G., Cha, J.-Y., Li, G., Chen, S., et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27, 908–925. doi: 10.1105/tpc.114.135095
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma. Oxf. Engl. 30, 923–930. doi: 10.1093/bioinformatics/btt656
McCarthy, D. J., Chen, Y., Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Mergemann, H., Sauter, M. (2000). Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 124, 609–614. doi: 10.1104/pp.124.2.609
Mühlenbock, P., Szechynska-Hebda, M., Plaszczyca, M., Baudo, M., Mateo, A., Mullineaux, P. M., et al. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356. doi: 10.1105/tpc.108.059618
Obulareddy, N., Panchal, S., Melotto, M. (2013). Guard Cell Purification and RNA Isolation Suitable for High-Throughput Transcriptional Analysis of Cell-Type Responses to Biotic Stresses. Mol. Plant-Microbe Interact.® 26, 844–849. doi: 10.1094/MPMI-03-13-0081-TA
Pfaffl, M. W., Horgan, G. W., Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. doi: 10.1093/nar/30.9.e36
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Seay, M., Gerstein, M., et al. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. 104, 4730–4735. doi: 10.1073/pnas.0611615104
Ramakers, C., Ruijter, J. M., Deprez, R. H. L., Moorman, A. F. M. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/s0304-3940(02)01423-4
Robinson, M. D., Oshlack, A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. doi: 10.1186/gb-2010-11-3-r25
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl. 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rusaczonek, A., Czarnocka, W., Kacprzak, S., Witoń, D., Ślesak, I., Szechyńska-Hebda, M., et al. (2015). Role of phytochromes A and B in the regulation of cell death and acclimatory responses to UV stress in Arabidopsis thaliana. J. Exp. Bot. 66, 6679–6695. doi: 10.1093/jxb/erv375
Sakai, T., Kagawa, T., Kasahara, M., Swartz, T. E., Christie, J. M., Briggs, W. R., et al. (2001). Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U. S. A. 98, 6969–6974. doi: 10.1073/pnas.101137598
Selvaraj, M. G., Ishizaki, T., Valencia, M., Ogawa, S., Dedicova, B., Ogata, T., et al. (2017). Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 15, 1465–1477. doi: 10.1111/pbi.12731
Song, C., Chung, W. S., Lim, C. O. (2016). Overexpression of Heat Shock Factor Gene HsfA3 Increases Galactinol Levels and Oxidative Stress Tolerance in Arabidopsis. Mol. Cells 39, 477–483. doi: 10.14348/molcells.2016.0027
Strasser, R. J., Tsimilli-Michael, M., Srivastava, A. (2004). “Analysis of the Chlorophyll a Fluorescence Transient,” in Chlorophyll a Fluorescence: A Signature of Photosynthesis Advances in Photosynthesis and Respiration. Eds. Papageorgiou, G. C., Govindjee (Dordrecht: Springer Netherlands), 321–362. doi: 10.1007/978-1-4020-3218-9_12
Suetsugu, N., Kagawa, T., Wada, M. (2005). An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 139, 151–162. doi: 10.1104/pp.105.067371
Suetsugu, N., Dolja, V. V., Wada, M. (2010a). Why have chloroplasts developed a unique motility system? Plant Signal. Behav. 5, 1190–1196. doi: 10.4161/psb.5.10.12802
Suetsugu, N., Takano, A., Kohda, D., Wada, M. (2010b). Structure and activity of JAC1 J-domain implicate the involvement of the cochaperone activity with HSC70 in chloroplast photorelocation movement. Plant Signal. Behav. 5, 1602–1606. doi: 10.4161/psb.5.12.13915
Suetsugu, N., Higa, T., Kong, S.-G., Wada, M. (2015). Plastid Movement Impaired1 and Plastid Movement Impaired1-Related1 Mediate Photorelocation Movements of Both Chloroplasts and Nuclei. Plant Physiol. 169, 1155–1167. doi: 10.1104/pp.15.00214
Sun, Z., Qi, X., Wang, Z., Li, P., Wu, C., Zhang, H., et al. (2013). Overexpression of TsGOLS2, a galactinol synthase, in Arabidopsis thaliana enhances tolerance to high salinity and osmotic stresses. Plant Physiol. Biochem. 69, 82–89. doi: 10.1016/j.plaphy.2013.04.009
Sztatelman, O., Łabuz, J., Hermanowicz, P., Banaś, A. K., Bażant, A., Zgłobicki, P., et al. (2016). Fine tuning chloroplast movements through physical interactions between phototropins. J. Exp. Bot. 67, 4963–4978. doi: 10.1093/jxb/erw265
Takano, A., Suetsugu, N., Wada, M., Kohda, D. (2010). Crystallographic and functional analyses of J-domain of JAC1 essential for chloroplast photorelocation movement in Arabidopsis thaliana. Plant Cell Physiol. 51, 1372–1376. doi: 10.1093/pcp/pcq089
Terashima, I., Hanba, Y. T., Tazoe, Y., Vyas, P., Yano, S. (2006). Irradiance and phenotype: comparative eco-development of sun and shade leaves in relation to photosynthetic CO2 diffusion. J. Exp. Bot. 57, 343–354. doi: 10.1093/jxb/erj014
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. Cell Mol. Biol. 37, 914–939. doi: 10.1111/j.1365-313x.2004.02016.x
Tholen, D., Boom, C., Noguchi, K., Ueda, S., Katase, T., Terashima, I. (2008). The chloroplast avoidance response decreases internal conductance to CO2 diffusion in Arabidopsis thaliana leaves. Plant Cell Environ. 31, 1688–1700. doi: 10.1111/j.1365-3040.2008.01875.x
Wada, M., Kong, S.-G. (2018). Actin-mediated movement of chloroplasts. J. Cell Sci. 131, 1–8. doi: 10.1242/jcs.210310
Wang, C., Hu, H., Qin, X., Zeise, B., Xu, D., Rappel, W.-J., et al. (2016). Reconstitution of CO2 Regulation of SLAC1 Anion Channel and Function of CO2-Permeable PIP2;1 Aquaporin as CARBONIC ANHYDRASE4 Interactor. Plant Cell 28, 568–582. doi: 10.1105/tpc.15.00637
Wituszyńska, W., Karpiński, S. (2013). Programmed Cell Death as a Response to High Light, UV and Drought Stress in Plants. Abiotic Stress - Plant Responses Appl. Agric. 207–246. doi: 10.5772/53127
Wituszynska, W., Karpiński, S. (2014). Determination of Water Use Efficiency for Arabidopsis thaliana. BIO-Protoc. 4 (3), e1041. doi: 10.21769/BioProtoc.1041
Wituszyńska, W., Gałązka, K., Rusaczonek, A., Vanderauwera, S., Van Breusegem, F., Karpiński, S. (2013a). Multivariable environmental conditions promote photosynthetic adaptation potential in Arabidopsis thaliana. J. Plant Physiol. 170, 548–559. doi: 10.1016/j.jplph.2012.11.016
Wituszyńska, W., Ślesak, I., Vanderauwera, S., Szechyńska-Hebda, M., Kornaś, A., Kelen, K. V. D., et al. (2013b). Lesion Simulating Disease1, Enhanced Disease Susceptibility1, And Phytoalexin Deficient4 Conditionally Regulate Cellular Signaling Homeostasis, Photosynthesis, Water Use Efficiency, and Seed Yield in Arabidopsis. Plant Physiol. 161, 1795–1805. doi: 10.1104/pp.112.208116
Wituszyńska, W., Szechyńska-Hebda, M., Sobczak, M., Rusaczonek, A., Kozłowska-Makulska, A., Witoń, D., et al. (2015). Lesion simulating disease 1 and enhanced disease susceptibility 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 38, 315–330. doi: 10.1111/pce.12288
Keywords: chloroplast movement, stomata, photosynthesis, oxidative stress, transcriptome
Citation: Czarnocka W, Rusaczonek A, Willems P, Sujkowska-Rybkowska M, Van Breusegem F and Karpiński S (2020) Novel Role of JAC1 in Influencing Photosynthesis, Stomatal Conductance, and Photooxidative Stress Signalling Pathway in Arabidopsis thaliana. Front. Plant Sci. 11:1124. doi: 10.3389/fpls.2020.01124
Received: 22 March 2020; Accepted: 08 July 2020;
Published: 29 July 2020.
Edited by:
Joerg Durner, Helmholtz Zentrum München, GermanyReviewed by:
Peter J. Gollan, University of Turku, FinlandVivek Dogra, Institute of Himalayan Bioresource Technology (CSIR), India
Copyright © 2020 Czarnocka, Rusaczonek, Willems, Sujkowska-Rybkowska, Van Breusegem and Karpiński. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weronika Czarnocka, d2Vyb25pa2FfY3phcm5vY2thQHNnZ3cuZWR1LnBs
 Weronika Czarnocka
Weronika Czarnocka Anna Rusaczonek
Anna Rusaczonek Patrick Willems
Patrick Willems Marzena Sujkowska-Rybkowska
Marzena Sujkowska-Rybkowska Frank Van Breusegem
Frank Van Breusegem Stanisław Karpiński
Stanisław Karpiński