- State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai’an, China
SINA (Seven in absentia) proteins are a small family of ubiquitin ligases that play important roles in regulating plant growth and developmental processes as well as in responses to diverse types of biotic and abiotic stress. However, the characteristics of the apple SINA family have not been previously studied. Here, we identified 11 MdSINAs members in the apple genome based on their conserved, N‐terminal RING and C-terminal SINA domains. We also reconstructed a phylogeny of these genes; characterized their chromosomal location, structure, and motifs; and identified two major groups of MdSINA genes. Subsequent qRT-PCR analyses were used to characterize the expression of MdSINA genes in various tissues and organs, and levels of expression were highest in leaves. MdSINAs were significantly induced under ABA and carbon- and nitrate-starvation treatment. Except for MdSINA1 and MdSINA7, the other MdSINA proteins could interact with each other. Moreover, MdSINA2 was found to be localized in the nucleus using Agrobacterium-mediated transient expression. Western-blot analysis showed that MdSINA2 accumulated extensively under light, decreased under darkness, and became insensitive to light when the RING domain was disrupted. Finally, ABA-hypersensitive phenotypes were confirmed by transgenic calli and the ectopic expression of MdSINA2 in Arabidopsis. In conclusion, our results suggest that MdSINA genes participate in the responses to different types of stress, and that MdSINA2 might act as a negative regulator in the ABA stress response.
Introduction
Ubiquitination is one of the most important post-translational modifications in plant development and the responses of plants to environment (Devoto et al., 2003; Xu and Jaffrey, 2013; Stone, 2014; Zhang et al., 2015). The ubiquitin-proteasome system consists of ubiquitin (Ub), ubiquitin activating enzyme (E1), ubiquitin-binding enzyme (E2), ubiquitin ligase (E3), and the 26S proteasome, and functions in the degradation of target proteins in a three-enzyme cascade manner (Sadanandom et al., 2012; Jansen et al., 2014). Ubiquitin molecules are small spherical (76 residues) and highly conserved proteins that can covalently modify target proteins to alter their stability, function, and localization (Schimke, 1973; Callis et al., 1995; Dametto et al., 2015). Initially, E1 activates Ub to form the E1-Ub complex with ATP. The activated Ub is transferred to the cysteine residue of the E2 ubiquitin-binding enzyme, forming an intermediary. The E3 ubiquitin ligase then recognizes the E2-Ub intermediary and the substrate protein, and transfers the Ub from the E2-Ub intermediary to the target protein. Finally, the substrate protein labeled by ubiquitin can be transported to the 26S proteasome for degradation (Santner and Estelle, 2010).
Ubiquitin ligases bind to specific ubiquitin-binding enzymes and target proteins, and the E3 ligase determines the diversity and specificity of the target protein. In the plant kingdom, ubiquitin ligase can be classified into three types based on its reaction mechanism and special conserved domains: HECT (Homology to E6-Associated Carboxy-Terminus) ubiquitin ligases, RING (Really Interesting New Gene) ubiquitin ligases, and U-box ubiquitin ligases (Duplan and Rivas, 2014). Among ubiquitin ligases, RING type E3 ligases have been the most extensively studied. RING type E3 ligases contain a conserved RING domain that consists of histidine and cysteine and can bind to zinc ions (Stone et al., 2005; Metzger et al., 2014). RING ubiquitin ligases can be further divided into two groups: single subunit RING ubiquitin ligases and multi-subunit RING ubiquitin ligases. Multi-subunit RING ubiquitin ligases can be subdivided into four types: Skp1-Cullin-F-box, VHL-ELONGIN-CUL2/5, BTB-CUL3 and DDB-CUL4 (Hua and Vierstra, 2011; Sadanandom et al., 2012). In recent years, RING finger ubiquitin E3 ligases have been extensively studied and have been shown to play important roles in plant growth and development as well as in response to stress.
SINA (seven in absentia) proteins, RING-finger E3 ligases that were first identified in Drosophila (Carthew and Rubin, 1990), are involved in regulating the differentiation of light receptors (Li et al., 1997). Most SINA family members possess a highly conserved N-terminal RING finger domain and a C-terminal SINA domain. The N-terminal domain is for binding to E2, and the SINA domain recognizes the target protein which is subsequently degraded by the 26S proteasome (Hu and Fearon, 1999; Den Herder et al., 2012). In addition, studies have found that SINA homologs can regulate their target proteins to adapt to different developmental stages and environmental changes. For example, SINAT5Ler (Landsberg ecotype) can mediate the degradation of the transcriptional activator NAC1 which is involved in the auxin signaling pathway, thereby regulating lateral root development in Arabidopsis (Xie et al., 2002). Further study has shown that SINAT5Ler could interact with FLC (Flowering Locus C), LHY (LATE ELONGATEDHYPOCOTYL), and DET1 (DE-ETIOLATED1) to regulate flowering time in Arabidopsis by promoting the degradation of FLC and LHY (Park et al., 2007; Park et al., 2010). SINAT2 is involved in carotenogenesis by interacting with RAP2.2 (Welsch et al., 2007). All SINATs can interact with dephosphorylated BES1, which is one of the core transcription factors involved in BR signaling. However, only SINAT5Ler is known to be able to negatively regulate BR signaling by mediating the degradation of BES1.
Other major findings include the accumulation of SINATs proteins in light and their degradation in the dark (Yang et al., 2017) as well as the diverse synergistic and antagonistic functions of SINA members. For example, SINAT1 and SINAT2 can negatively regulate starvation-induced autophagy by ubiquitinating ATG6 (AUTOPHAGY PROTEIN6) or ATG13 (AUTOPHAGY PROTEIN13). Conversely, SINAT6 promotes autophagy by increasing the number of autophagic puncta under nutrient-rich or nutrient-poor conditions (Qi et al., 2017; Qi et al., 2020). A recent study has demonstrated that the ectopic expression of tomato SlSINA4 resulted in cell death in N. benthamiana leaves, but overexpression of any of the other five SlSINAs can suppress the hypersensitive response and cell death (Wang et al., 2018). Abscisic acid (ABA) plays a critical role in plant growth and stress adaptation (Knight and Knight, 2001). Overexpression of SINA2 (SEVEN IN ABSENTIA 2) increases tolerance to drought by inducing the closure of stomata in Arabidopsis (Bao et al., 2014). OsDIS1, a homologous protein of SINA in rice, is a negative regulator in the drought response (Ning et al., 2011). In addition, Siah1 and Siah2, human SINA homologs are involved in multiple processes such as synaptic transmission, apoptosis, tumor suppression, and stress response (Wheeler et al., 2002; Franck et al., 2006; Khurana et al., 2006; Fukuba et al., 2007).
Our knowledge of the SINA family in apple is limited. Here, we identified 11 MdSINA members in apple using bioinformatics analyses. We analyzed tissue expression patterns and the responses of these genes to abiotic stress. Additionally, we assessed the ability of these genes to form homodimers and heterodimers using yeast two-hybrid (Y2H) assays. Our results provide basic information on the function of SINA proteins in apple.
Materials and Methods
Identification of SINA Gene Family Members in Apple
Five SINA homologous proteins in Arabidopsis thaliana were obtained from TAIR (https://www.arabidopsis.org) (Lamesch et al., 2012). All protein sequences in apple (Malus × domestica) were downloaded from Apple Genome and Epigenome (https://iris.angers.inra.fr/gddh13/the-apple-genome- downloads.html) (Daccord et al., 2017). Local BLAST with an E-value of 1×10-5 was used to screen MdSINA genes. We confirmed the blast result by SMART (http://smart.embl-heidelberg.de/) and analyzed molecular weights and theoretical pI by ProtParam (http://web.expasy.org/protparam/). Subcellular localizations of MdSINAs were predicted by WoLF PSORT II (https://www.genscript.com/wolf-psort.html) (Horton et al., 2007).
Multiple Sequence Alignment and Phylogenetic Analysis
Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/), an online server, was used to perform multi-sequence alignment between the amino acid sequences of apple and A. thaliana, and Jalview (Jalview 2.10.5) was used to edit and visualize the comparison results.
MEGA: X was used to construct a neighbor-joining evolutionary tree with 1,000 bootstrap replications and a Poisson model (Kumar et al., 2018). Six SINA proteins in rice (Oryza sativa) were obtained from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html) (Goodstein et al., 2012). In MEGA X, ClustalW method was selected for sequence alignment, partial deletion and 95% site coverage cutoff were used gaps/missing data treatment.
Gene Structure, Chromosomal Location, and Conserved Motif Analysis
We used GSDS 2.0 (http://gsds.cbi.pku.edu.cn/) and MG2C (http://mg2c.iask.in/mg2c_v2.0/) to display the structure and chromosomal locations of MdSINA genes (Hu et al., 2015). Genome annotation information (gene_models_20170612.gff3.bz2) was downloaded from Apple Genome and Epigenome (https://iris.angers.inra.fr/gddh13/the-apple-genome-downloads.html). Conserved motifs were detected by MEME (http://memesuite.org/tools/meme). WebLogo (http://weblogo.berkeley.edu/logo.cgi) was used to redraw the motifs (Crooks et al., 2004; Bailey et al., 2009).
Synteny Analysis
The syntenic blocks information of the apple genome can be downloaded from the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). The genome sequence of the homologous SINA proteins in apples was evaluated using BLASTP. The MCScanX algorithm (Wang et al., 2012) was used to scan syntenic blocks containing the apple SINA genes.
Plant Materials and Treatments
Materials included various apple tissues, tissue-cultured apple seedlings (Malus × domestic “Royal Gala”), “Orin” apple calli, “Columbia” (WT) Arabidopsis seedlings (Col-0), and tobacco (Nicotiana benthamiana). The growth conditions of tissue-cultured apple seedlings, “Orin” apple calli, Arabidopsis seedlings and tobacco were the same as those described previously (An et al., 2019). Three-week-old shoot tissue culture seedlings were used for rooting with 0.2 mg/L IAA. We collected the roots, stems, leaves, flowers, and fruits of a 7-year-old “Gala” apple tree, which were immediately frozen in liquid nitrogen and stored at 80°C for tissue expression analysis.
For ABA treatment, 3-week-old apple seedlings were treated with 100 μM ABA for 0, 1, 3, 6, 12, and 24 h following previously described methods (Zhao et al., 2019) and were then immediately frozen in liquid nitrogen and stored at −80°C for subsequent analysis. For the carbon-deprivation treatment, 3-week-old apple seedlings with roots grown in MS medium were treated without sucrose and in the dark for 0, 6, 12, 24, 36, and 48 h. For nitrate-deprivation treatment, 3-week-old seedlings grown on MS medium were transferred to nitrogen-deficient MS liquid medium and grown under normal growth conditions for 0, 6, 12, 24, 36, and 48 h.
RNA Extraction and qRT-PCR Analysis
Total RNAs of apple samples and calli was isolated using a RNA extraction kit (Tiangen, Beijing, China) and used for qRT-PCR analysis following previously described methods (Ren et al., 2019). Three technical and biological replicates were performed to detect the transcripts of MdSINA genes. Specific primers of the MdSINA genes were designed and used for qRT-PCR analysis (Supplementary Table 1).
Vector Construction and Genetic Transformation
The open reading frames of MdSINA2 was cloned into the pRI-GFP vector to construct the overexpression vector. The primer pairs MdSINA2-F (5′-GTCGAC ATGGACTTGGAAAGCATCGAGTGT-3′)/−R and (5′-GGATCC GCTACACAGGTTTGGTATGCA-3′) were used to amplify the full-length MdSINA2. “Orin” apple calli were infected by the recombinant plasmids 35S::GFP-MdSINA2 and WT (35S::GFP) via the Agrobacterium-mediated transformation method as previously described (An et al., 2016). The transgenic Arabidopsis were obtained by using the floral dip transformation method (Clough and Bent, 1998).
Yeast Two‐Hybrid (Y2H) Assays
Interactions among MdSINA proteins were characterized by Y2H assays. The full-length cDNA of MdSINAs was cloned and inserted into yeast vector pGBT9 and pGAD424. The recombinant plasmids of pGAD424-MdSINAs and pGBT9-MdSINAs were transformed into yeast “Y2H Gold.” The yeast was grown on selection medium as described previously (An et al., 2018); the primer pairs used are listed in Supplementary Table 2.
Subcellular Localization of MdSINA2
The recombinant plasmid 35S::GFP-MdSINA2 and the control vector 35S::GFP were injected into the epidermal cells of N. benthamiana leaves via Agrobacterium-mediated transient expression. The fluorescent signals were observed by a laser confocal microscope (Zeiss LSM 510 META, Jena, Germany).
Protein Extraction and Western-Blotting
The total protein of 2-week-old MdSINA2-GFP transgenic calli was isolated as described (Li et al., 2012). After the western blot assay, anti-GFP (Sigma-Aldrich) was used to detect protein abundance. Actin served as a protein loading control.
ABA-Sensitive Assays
Transgenic and WT calli were cultivated on MS medium containing 100 μM ABA. Fresh weight and superoxide anions content were measured after 18 days. The superoxide anions generation rates were determined by spectrophotometry using their corresponding assay kits (Keming, Suzhou, China).
Homozygous seeds of WT and transgenic Arabidopsis were plated on MS medium with or without either 1 or 3μM ABA and grown at 22°C under an 8/16 h dark/light photoperiod. Photographs were taken after 8 days.
Statistical Analysis
All experiments were performed in triplicate. Error bars showed the standard deviation of three biological replicates. GRAPHPAD PRISM 6.01 software was used to conduct a one-way ANOVA to detect significant differences (*, P < 0.05).
Results
Genome-Wide Identification of Apple SINA Genes
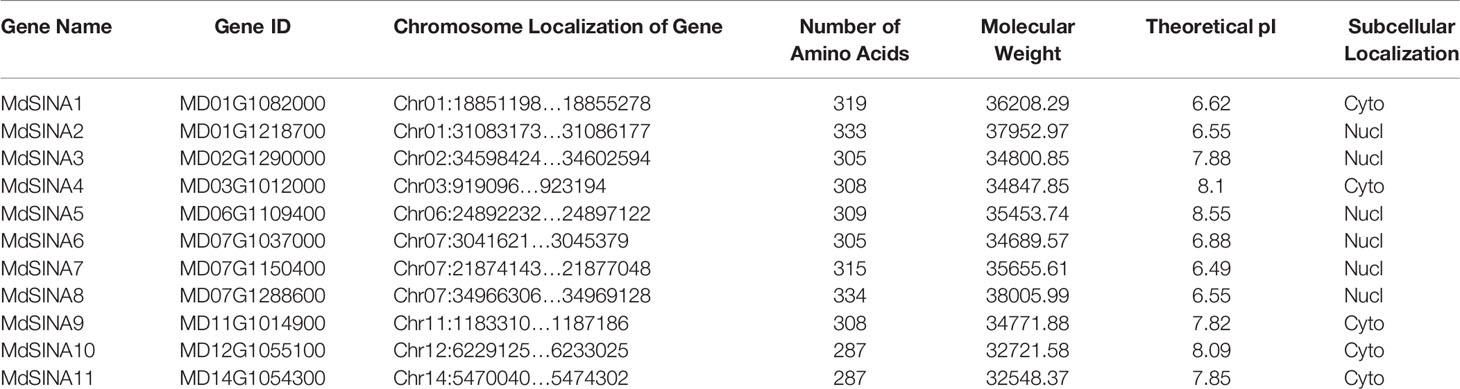
Five SINATs were previously identified in the A. thaliana genome, including AtSINA1 (SINAT1), AtSINA2 (SINAT2), AtSINA3 (SINAT3), AtSINA4 (SINAT4), and AtSINA5 (SINAT5Ler) (Xie et al., 2002; Wang et al., 2008). Five SINAT protein sequences were used to blastp in the apple genome database. After software screening and manual detection based on the conserved domains, 11 MdSINAs were obtained (Table 1). The MdSINA genes were named based on their chromosomal locations (MdSINA1–11). We found that the 11 MdSINA genes were unevenly distributed on eight chromosomes in the apple genome. Gene distribution density was highest on chromosome 7 (three genes), followed by chromosome 1 (two genes). The range of lengths of the MdSINA proteins was small, from 287 (MdSINA10/11) to 334 (MdSINA8) amino acids, and the molecular weights were predicted to be between 32,548–37,952 Da. The theoretical isoelectric point ranged from 6.49 (MdSINA7) to 8.55 (MdSINA5). In addition, the subcellular locations of the 11 MdSINA genes were predicted, suggesting that the MdSINA proteins were localized in either the cytoplasm or nucleus (Table 1).
Phylogenetic and Synteny Analyses of Apple SINA Genes
We performed a phylogenetic analysis of SINA proteins in apple, Arabidopsis and rice to gain additional insight into the evolutionary relationships of apple SINA proteins (Figure 1). The 22 SINA proteins used in the analysis were divided into two groups in these three species. Groups I contained five MdSINA proteins (MdSINA1, MdSINA2, MdSINA5, MdSINA7, and MdSINA8), and Group II contained six proteins (MdSINA3, MdSINA4, MdSINA6, MdSINA9, MdSINA10, and MdSINA11). We noticed that all genes except MdSINA5 appeared in pairs based on the phylogenetic tree of the apple SINA family, implying possible gene duplication have occurred during the evolution of the SINA gene family. Subsequently, we used MCScanX to detect gene duplication events in MdSINAs in the apple genome. Five pairs, MdSINA1/7, MdSINA2/8, MdSINA3/6, MdSINA4/9, and MdSINA10/11, were localized in duplicated genomic regions (Figure 2A). And those genes which are flanked the MdSINA genes are also present in the syntenic blocks (data not shown), indicating that segmental duplication events occurred. Moreover, 11 apple SINA genes distributed across eight chromosomes, including 1, 2, 3, 6, 7, 11, 12, 14 (Figure 2B), which have highlighted strong collinearity between large segments of chromosomes 3 and 11, and between shorter segments of chromosomes 1 and 7, 2 and 7, 12 and 14 (Velasco et al., 2010).
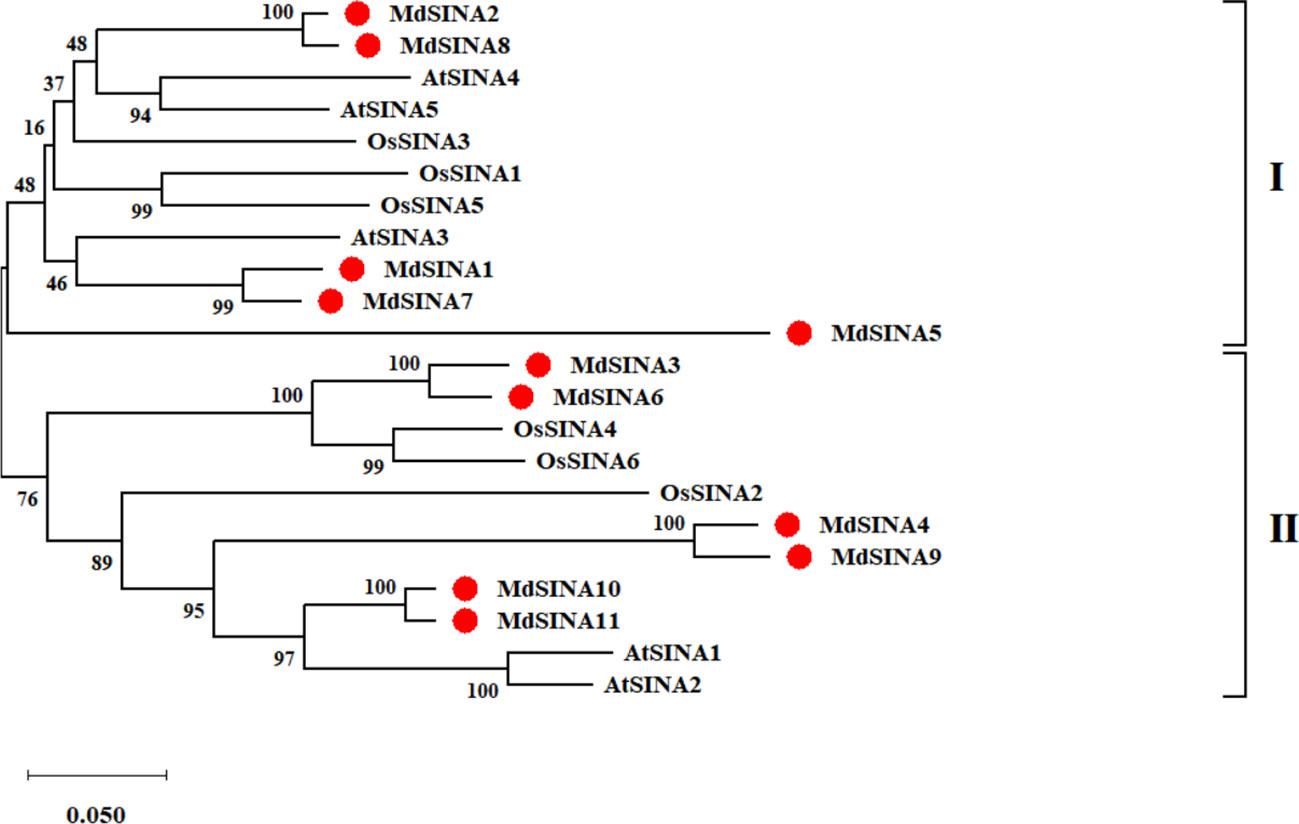
Figure 1 Phylogenetic analysis of SINA genes in apple, Arabidopsis, and rice. The phylogenetic tree was constructed using the NJ method and a bootstrap test with 1000 replicates.
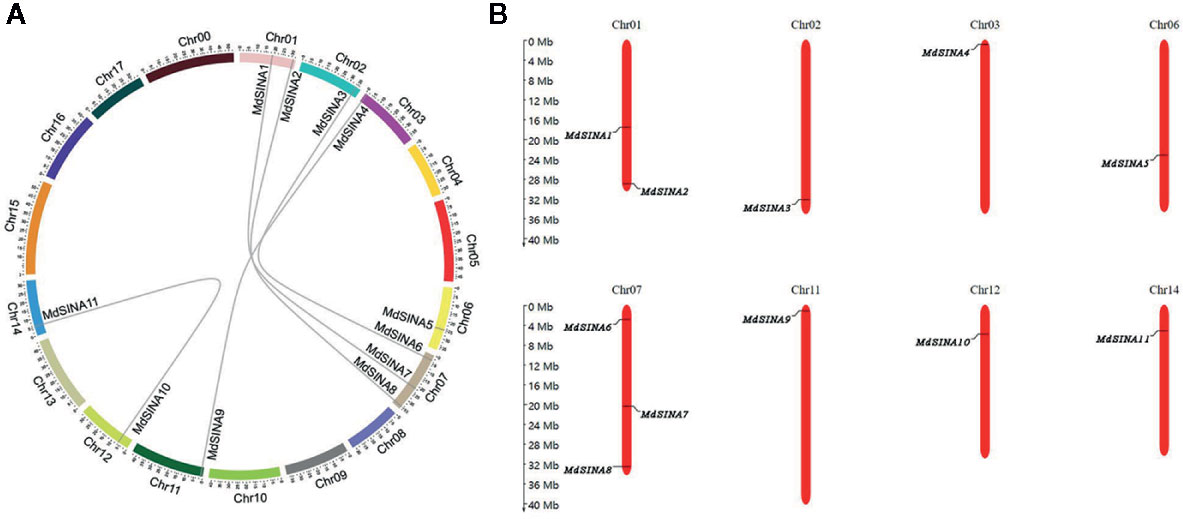
Figure 2 Evolutionary relationships among MdSINA gene family members and the chromosomal distribution of SINA genes. (A) Synteny analysis of SINA genes in apple. (B) Chromosomal location of SINA genes in apple.
Gene Structure and Multiple Sequences Alignment Analyses of MdSINA Genes
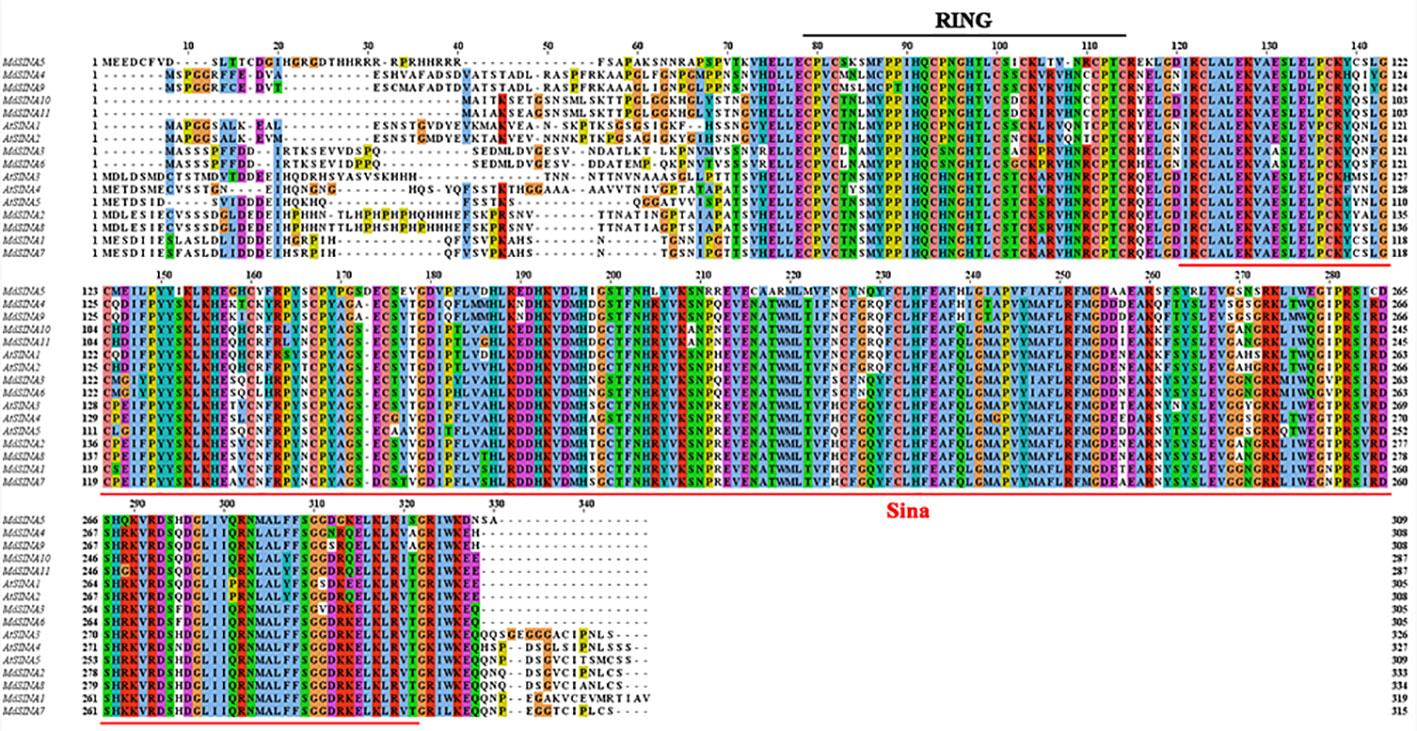
The exon-intron structure of the MdSINA genes was examined using the online server GSDS to provide more insight into the evolution of these genes (Figures 3A, B). Gene structures were highly similar within each of the two group in terms of exon number. Furthermore, a similar exon-intron structure was observed within the same group of MdSINA. For example, Group I genes contained two introns and three exons except for MdSINA1. However, Group II genes possessed three introns and three exons. Although the protein sequence similarity between MdSINA1 and MdSINA7 was over 95% (data not shown), the structures of their associated gene differed. These differences implied that these two genes possessed divergent functions during their evolution.
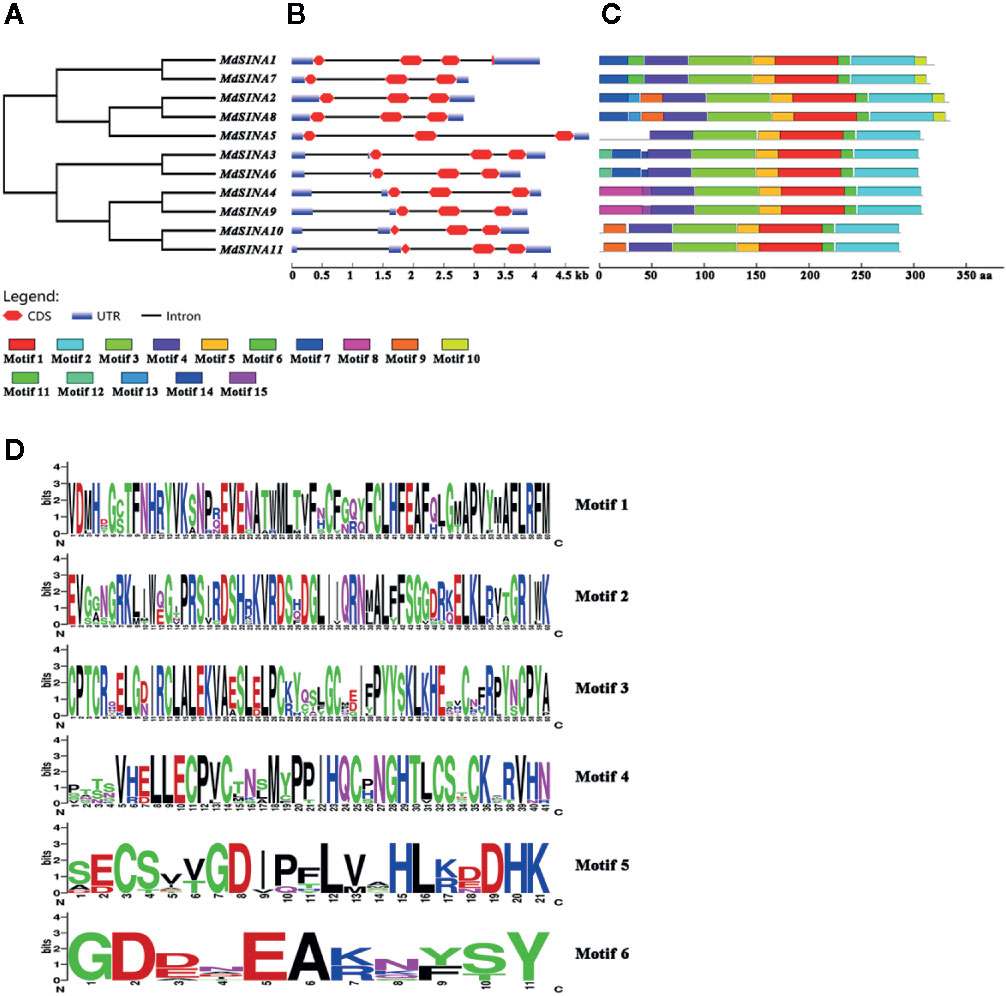
Figure 3 Gene characterization and structural analysis of MdSINA. (A) Phylogenetic tree of MdSINA genes. (B) Gene structure analysis of MdSINA genes was performed by GSDS. Red blocks and black lines represent exons and introns, respectively. (C) Conserved motifs of MdSINAs were detected by MEME. (D) Sequence logos of the conserved motifs. The degree of conservation was represented by the overall height of each position, while the height of each letter was proportional to its frequency.
The conserved motifs of apple SINA proteins were confirmed by MEME, and 15 distinct motifs were identified (Figure 3C). Motifs 1–6 were observed in all MdSINA proteins and contained conserved RING and SINA domains. Motif 4 was identified as the RING domain, while motifs 1, 2, 3, 5, and 6 were components of the SINA domain. Most members of Group I had motif 10 at the C‐terminus, while all Group II members lacked this motif. Five gene pairs had the similar, unique motif structure. For example, motifs 11 and13 were the unique motifs in MdSINA1/7 and MdSINA2/8, respectively. The conservation of SINA genes during evolution was substantiated by the distribution of motifs. The uniqueness of the motif distribution in different gene pairs reflects the conservation and differentiation of SINA genes function during evolution. Moreover, the consensus sequences of motifs is stacked, suggested that all MdSINA genes shared highly conserved or completely conserved sites (Figure 3D).
We performed multiple sequences alignment to detect the conserved domain of MdSINA proteins (Figure 4). In the BLASTP analysis, the highest percentage of amino acid sequence identity obtained was between pairs of MdSINA genes, and these genes were also closely grouped in the phylogenetic tree. Our results suggest that an N‐terminal C3HC4-type (Cys-X2-Cys-X-Cys-X-His-X-Cys-X2-Cys-X-Cys-X2-Cys) RING domain and a C-terminal SINA domain occurred in all members. The RING domain is generally composed of eight conserved cysteine (Cys) and histidine (His) residues, and these eight conserved amino acid residues can bind two zinc ions to form a zinc-finger structure.
Expression Patterns of MdSINAs in Different Tissues
SINAs, from both animals and plants, have been found to play vital roles in different physiological processes. In order to further investigate the roles of MdSINAs in apple growth and development, testing the expression level of MdSINAs in different tissues is necessary. We performed qRT-PCR analysis to examine the expression levels of MdSINAs in a variety of tissues, including root, stem, leaf, flower, and fruit. The five gene pairs exhibited similar spatial expression patterns (Figure 5). MdSINA1/7, MdSINA2/8, MdSINA4/9, and MdSINA10/11 showed relatively high expression levels in leaves and flowers, suggesting that these genes may be involved in leaf and flower development. In addition, MdSINA3/6 showed similar expression patterns with higher expression levels in stems, leaves, flowers, and fruit, indicating that they may play an important role in the growth and development of apples. MdSINA5 was only highly expressed in leaves. Overall the distinct spatial expression patterns imply that these genes play diverse roles in apple growth and development.
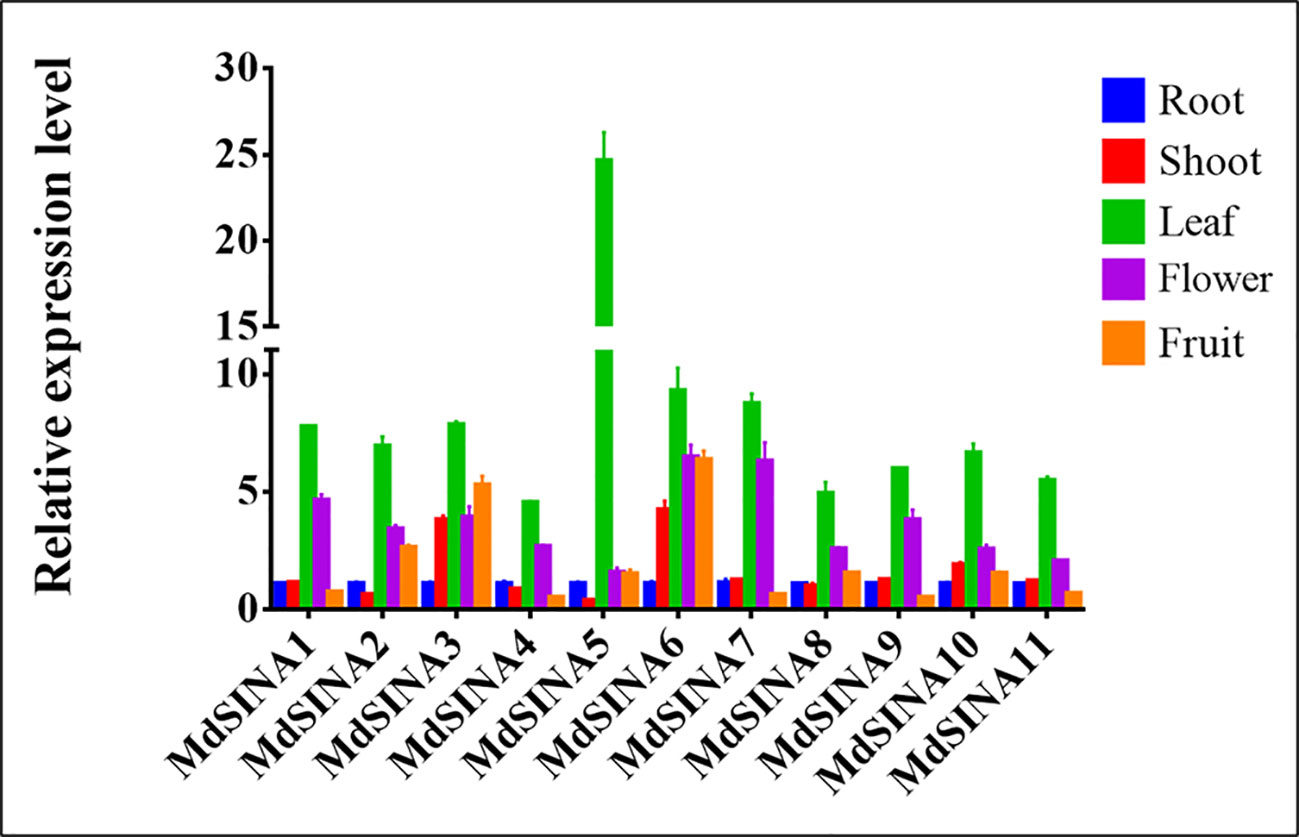
Figure 5 Expression patterns of MdSINA genes in different tissue. Transcripts of the 11 MdSINA genes in different tissues were determined by quantitative real-time PCR. Transcript values of the MdSINA genes determined in roots were set to 1. The apple Actin 18s gene was used as the reference transcript. Values are means ± SE of three biological replicates.
Expression Patterns of MdSINAs in Response to ABA and Nutrient Starvation
A previous study has shown that SINAT2 promotes drought tolerance in an ABA-dependent manner in Arabidopsis (Bao et al., 2014). To explore the potential response to ABA, the expression levels of MdSINAs were detected under ABA (100 μM) treatment. The expression levels of MdSINAs were highest at 3 h (approximately3–6-fold), and then returned to normal levels (Figure 6A). In addition, some of these genes had a higher expression level at 24 h, implying that MdSINAs were involved in ABA response and may act as important regulators of drought resistance as has been described previously.
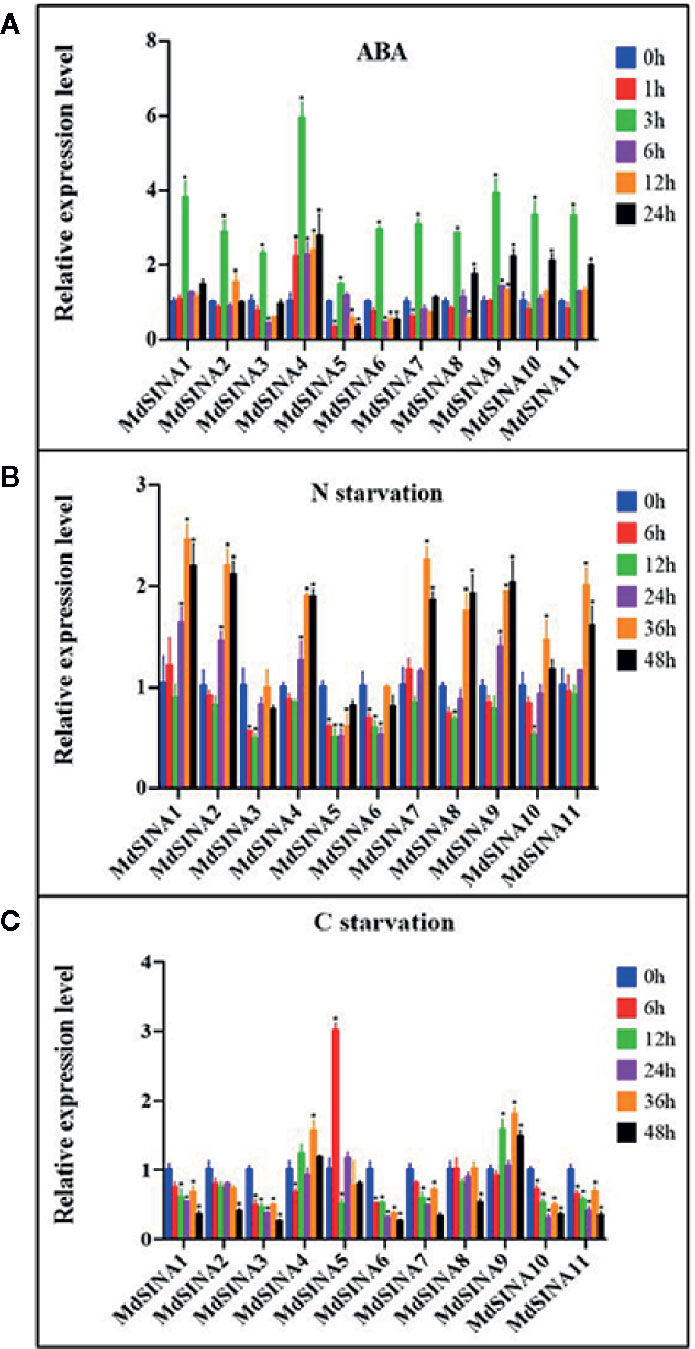
Figure 6 Expression profiles of MdSINAs in response to ABA (A), nitrate-starvation (B), and carbon-starvation (C). Transcript values of MdSINAs determined at 0 h were set to 1. The apple Actin 18s gene was used as the reference transcript. Values are means ± SE of three biological replicates. Significant difference are denoted by an asterisk (*P < 0.05).
Recently, studies have shown that SINAT1/2/6 are involved in starvation-induced autophagy in Arabidopsis (Nolan et al., 2017; Qi et al., 2017; Qi et al., 2020). To further characterize the potential roles of MdSINA genes in nutrient starvation, the expression levels of MdSINAs were examined under carbon- and nitrogen-starvation conditions. Under carbon starvation, MdSINA4/9 were slightly up-regulated, and MdSINA5 was significantly up-regulated (Figure 6B). Under nitrate-starvation conditions, the expression levels of MdSINA3/5/6 changed little; however, the expression of other MdSINAs was markedly up-regulated (Figure 6C). Thus, MdSINAs may be involved in the process of autophagy and function in different stress responses.
Interactions Among MdSINA Proteins
Previous studies have suggested that SINA proteins can form both homodimers and heterodimers to regulate their own stability via the proteasome degradation pathway and perform different biological functions (Hu and Fearon, 1999; Xie et al., 2002; Depaux et al., 2006; Wang et al., 2018). The homo- and hetero-dimerization of MdSINA proteins was confirmed by Y2H experiments. First, the full-length cDNA of each MdSINA gene was cloned into the bait vector pGBT9 and the prey vector pGAD424. The fusion plasmids were then co-transformed into yeast cells (the “Y2H Gold” strain) grown on selection medium. All MdSINA proteins except MdSINA1 and MdSINA7 can interact with each other to form homodimers and heterodimers, implying that MdSINA1/7 might function as a single E3 ligase (Figure 7). Specifically, MdSINA4/9-BD show weak autonomous activation. However, MdSINA4/9-AD can interact with other MdSINAs proteins except for MdSINA1 and MdSINA7. Thus MdSINAs can form homodimers and heterodimers.
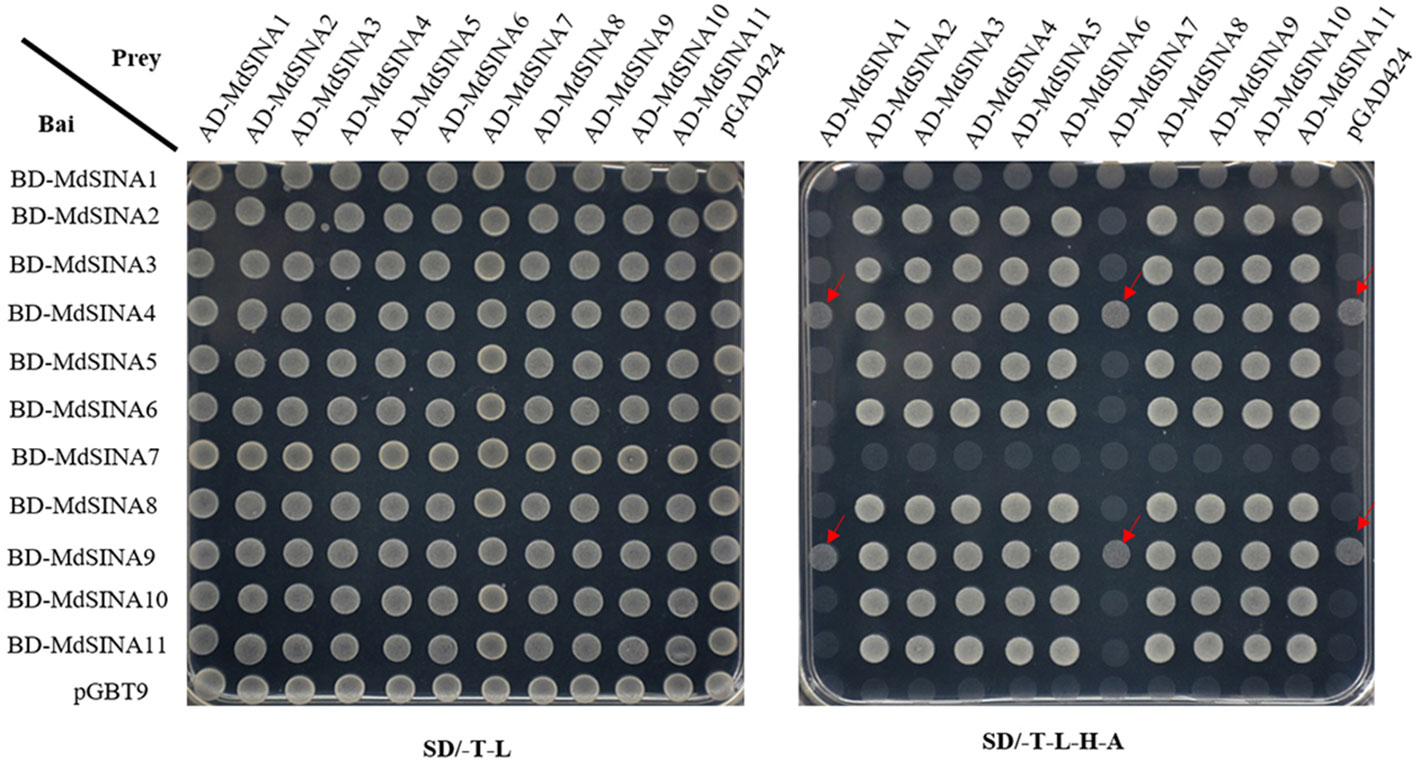
Figure 7 Y2H assay to test pair-wise interactions among MdSINA proteins. The yeast vector pGAD424 and pGBT9 were used as negative controls. The arrow indicates that BD-SINA4/9 showed weak autonomous activation.
Subcellular Localization of MdSINA2 Proteins
The predicted subcellular localization of MdSINA proteins is shown in Table 1, and MdSINA2 was selected for verification. Considering that ubiquitination can occur in the cytoplasm and nucleus to regulate the stability of cytoplasmic and nuclear proteins, the localization of ubiquitin ligase must localize in the cytoplasm, nucleus, or both (Xie et al., 2002; Heck et al., 2010; Yoo et al., 2013; Wang et al., 2018). To determine the subcellular localization of MdSINA2, we constructed the fusion plasmid 35S::MdSINA2-GFP and injected it into the epidermal cells of N. benthamiana leaves via Agrobacterium-mediated transient expression. After 48 h, we observed the GFP signal under confocal laser scanning microscopy. The fluorescent GFP signal was distributed throughout the cells or only in the nucleus of the epidermal cells of N. benthamiana leaves, which were injected with either 35S::GFP or 35S::GFP-MdSINA2, respectively (Figure S1). Thus MdSINA2 was localized in the nucleus, which was consistent with our initial prediction.
Light Signaling Mediates MdSINA2 Stability in a RING Domain-Dependent Manner
To gain further insight into the roles of MdSINA2, we cloned the full-length cDNA of MdSINA2 into plant expression vector with GFP label and transformed it into apple calli (Figure S2). SINA proteins are E3 ubiquitin ligases that have important functions in plants and animals, and understanding the factors that affect the stability of MdSINA proteins is important. In A. thaliana, levels of SINA proteins increase under light and decrease in darkness. However, SINAT5 protein without a RING domain, which represents the Columbia ecotype (Col-0), is insensitive to dark and light treatment (Yang et al., 2017). Therefore, we determined whether the effect of light on the stability of the SINA protein in apple was the same as that observed in Arabidopsis. Two-week-old MdSINA2 transgenic calli were grown on MS medium for 24 h under continuous light or dark conditions. We then transferred them to the dark or light for 1, 3, 6, 12, or 24 h. The protein level of MdSINA2 dramatically increased under light but decreased under dark conditions (Figure 8A).
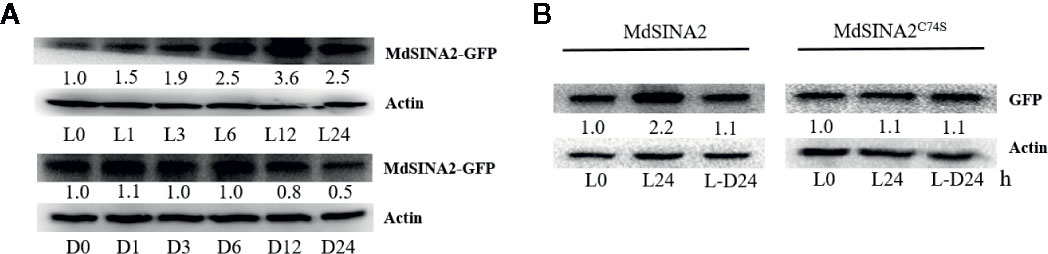
Figure 8 Light mediates the stability of MdSINA2. (A) MdSINA2 protein increased under light and decreased under dark condition. (B) MdSINA2C74S protein was more stable than MdSINA2 both under light and darkness. Two-week-old 35S::MdSINA2 and 35S::MdSINA2C74S apple calli were grown under continuous light (L) or dark (D) for the indicated time. L, continuous light; L-D, the continuous light-grown calli were transferred to the dark. MdSINA2-GFP was detected with anti-GFP antibodies. Actin was used for the loading control. The protein levels of MdSINA2-GFP determined at 0 h were set to 1. The assays were conducted in three biological replicates.
Disruption of the ubiquitination site of the SINA protein is known to result in the loss of self-ubiquitination activity (Xie et al., 2002; Wang et al., 2018). To test whether the degradation of MdSINA2 protein depends on the RING domain, we obtained a mutant protein, Cys74→Ser (C74S), in which the RING domain was disrupted following the methods of a previous study (Xie et al., 2002). Although the MdSINA2 protein level in MdSINA2-GFP calli increased and decreased under light and darkness, respectively, there were no other noticeable differences between MdSINA2C74S-GFP calli (Figure 8B). Thus, the stability of light-mediated MdSINA2 protein is dependent on its RING domain.
MdSINA2 Improves Sensitivity to ABA Treatment in Transgenic Apple Calli and Seed Germination of Arabidopsis
MdSINA2 transcripts were significantly induced by ABA treatment. To evaluate the role of MdSINA2 in the ABA response, 2-week-old MdSINA2 transgenic calli and WT were grown on MS medium with 100 µM ABA. MdSINA2-OX calli grew much slower than WT (Figures 9A, B). Moreover, MdSINA2-OX calli had lower fresh weight and accumulated more superoxide anions than WT (Figures 9C, D).
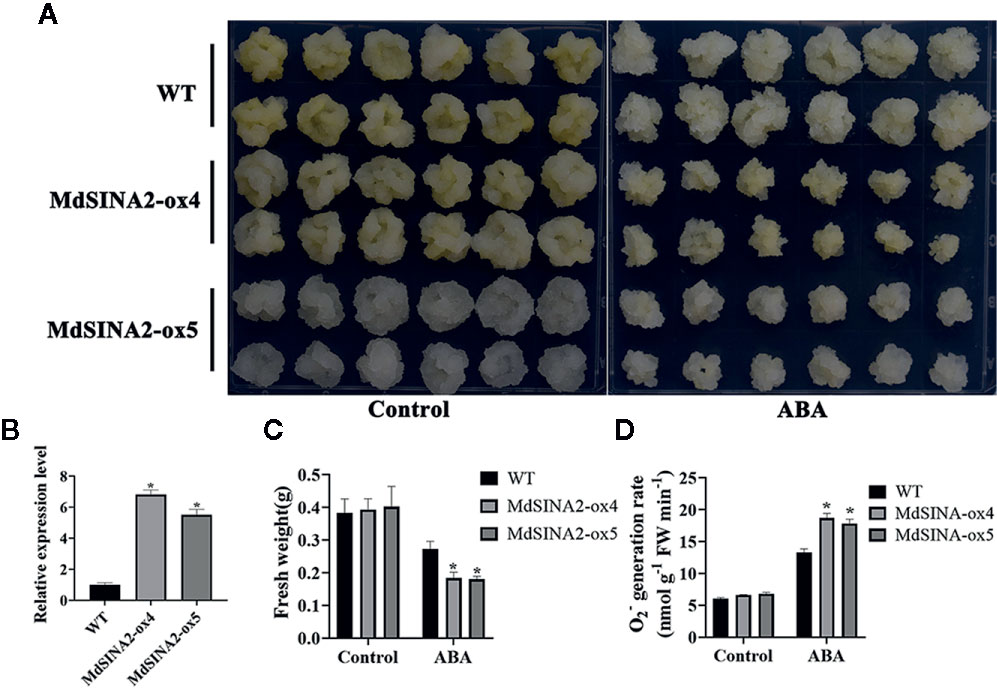
Figure 9 Overexpression of MdSINA2 results in sensitivity to ABA treatment in transgenic apple calli. (A) Phenotype of transgenic and WT calli grown in MS medium supplemented with 100 μM ABA. (B) Transcripts analysis of MdSINA2 in transgenic and WT calli. (C) Fresh weight statistics of transgenic and WT calli in (A). (D) O2- generation rates of transgenic and WT calli in (A). Values are means ± SE of three biological replicates. Significant differences between groups are indicated by an asterisk (*P < 0.05).
In addition, MdSINA2 was successfully transformed into wild-type Arabidopsis (Figure S2). Three independent lines were selected for seed germination experiments. The transgenic Arabidopsis seeds were sown on MS medium supplemented with different concentrations of ABA. The germination rate was negatively correlated with ABA concentration (Figure 10). There was no significant difference in germination rates between WT and transgenic seeds on MS medium. However, the germination rate of transgenic seeds was significantly lower on MS medium supplemented with 1 μM or 3 μM ABA compared with WT. Moreover, the transgenic Arabidopsis seeds hardly germinated on medium with 3 μM ABA even after 8 days. Therefore, overexpression of MdSINA2 results in an ABA-hypersensitive phenotype.
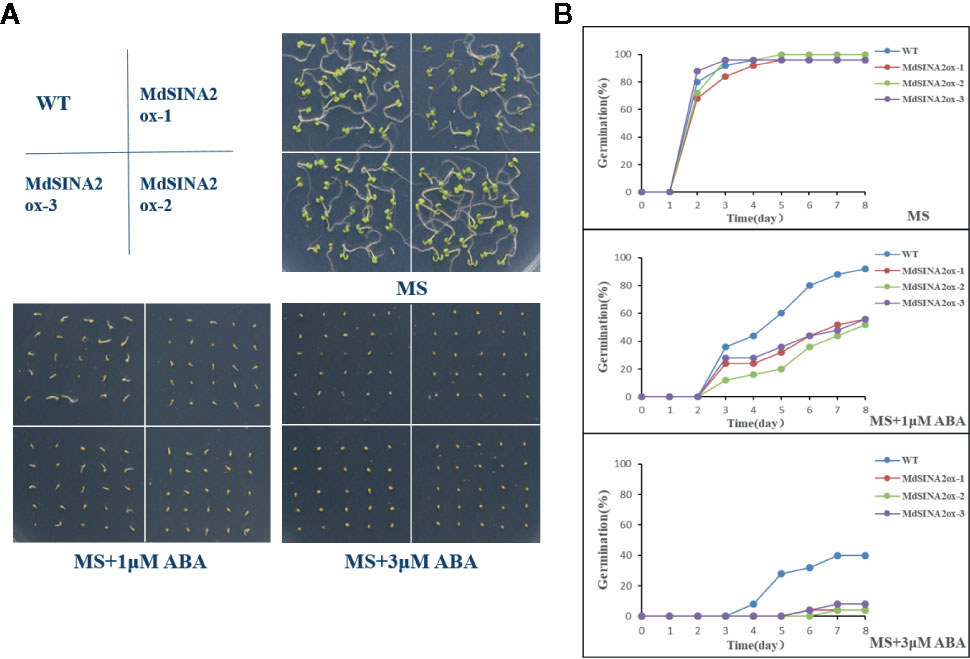
Figure 10 Overexpression of MdSINA2 in Arabidopsis results in a lower germination rate under different ABA concentration treatments. (A) Seed germination of WT and MdSINA2-OX Arabidopsis on MS medium supplement with 1 or 3 μM ABA. (B) Seed germination rate of WT and MdSINA2-OX Arabidopsis within 8 days.
Discussion
The function of SINA (Seven in absentia) homologous proteins have been extensively studied in animals, especially in the medical field, and these studies have revealed that SINA proteins are associated with resistance to various diseases such as cancer (Liu et al., 2008; Cai et al., 2015; Siswanto et al., 2018). Recently, an increasing number of studies investigating the function of SINA homologs have been conducted in plants (Zhang et al., 2019). SINA E3 ligase regulates plant growth and the response to different stresses via degradation of multiple targeted proteins. In this study, 11 MdSINA genes were identified in apple by genome-wide analysis (Table 1). MdSINA genes were found to be unevenly distributed across eight chromosomes through chromosomal localization. Five SINA homologous proteins are known in A. thaliana (SINAT1–5), 10 homologous proteins in Populus trichocarpa, six homologous proteins in O. sativa, at least six homologous proteins in Zea mays, and six homologous proteins in both Medicago truncatula and Solanum lycopersicum (Den Herder et al., 2008; Wang et al., 2008; Wang et al., 2018). However, the 11 SINA genes in apple may reflect an evolutionary expansion of the SINA family.
Multiple sequence alignments indicated that all MdSINA proteins contained two conserved domains: a RING domain and a SINA domain. Phylogenetic analysis was conducted to characterize the evolutionary relationships of SINA genes in apple, Arabidopsis, and rice (Figure 1). The results showed that SINA genes could be divided into two groups (Groups I and II) that differed in gene structure and motif composition between Group I and II. For instance, genes in the same group, such as MdSINA2 and MdSINA8 or MdSINA4 and MdSINA9, exhibited similar exon-intron structures and motif compositions. The main mechanism driving the evolution of SINA genes appears to be gene duplication followed by divergence (Ohno et al., 1968; Flagel and Wendel, 2009). A previous study has shown multiple segmental duplication events in Arabidopsis, poplar, and rice (Wang et al., 2008). Our results suggest that segmental duplication events have also occurred in apple. Five gene pairs showing similar gene structures and protein sequences imply that these proteins show some degree of functional redundancy, such as MdSINA2/8.
A more comprehensive characterization of the MdSINA gene family is necessary given the high functional diversity of SINA homologs that has been documented in different species. Therefore, qRT-PCR analyses of the expression patterns of MdSINA genes were conducted. These 11 MdSINA genes were differentially regulated at the transcriptional level in different apple tissues. The transcript levels of most MdSINA family members were significantly higher in leaves and lower in roots (Figure 5), which is in contrast with the transcriptional patterns of these genes documented in tomato (Wang et al., 2018), implying that they might play an important role in leaf physiology in apple. Given that ABA signaling plays a vital role in regulating the response to different types of stress (Cutler et al., 2010), such as drought, cold, and salt, we analyzed the expression profiles of MdSINA genes under ABA treatment. The expression levels of MdSINA genes were altered under different degrees of ABA induction, suggesting that MdSINAs might participate in the stress response as a functional component of the ABA signaling pathway.
Furthermore, increasing evidence has demonstrated that autophagy can be induced by nutrient starvation and transport needless proteins or damaged organelles into vacuoles for degradation and circulation (Lee et al., 2010; Avin-Wittenberg et al., 2015; Chen et al., 2017; Islam et al., 2019). A large number of autophagy-related proteins (ATGs) have been documented in plants; these ATGs play an important role in regulating core autophagy mechanisms (Soto-Burgos et al., 2018; Zhuang et al., 2018). To date, a few studies have shown that SINA homologs are involved in the autophagy process in Arabidopsis, and SINAT-RNAi plants exhibit a starvation hypersensitivity phenotype. SINAT1/2 were negative regulators in autophagy process under nutrient starvation condition by mediating the ubiquitylation and degradation of ATG6/13. Here, we observed different expression patterns of MdSINAs under carbon- and nitrogen-starvation treatments (Figures 6B, C), implying that MdSINAs may play different functions in different periods and under different starvation treatments.
The stability and activity of SINA ligases in vivo depend on their dimerization (Hu and Fearon, 1999; Polekhina et al., 2002; Depaux et al., 2006). Hence, we detected the dimerization of MdSINA proteins. The SINA proteins in apple are surprisingly different from those in tomato and M. truncatula (Den Herder et al., 2008; Wang et al., 2018), all of which can interact with each other. We found that MdSINA1/7 neither formed homodimers nor heterodimers with any of the other MdSINA proteins. Thus, MdSINA proteins may function via the formation of homodimers and heterodimers or in another manner.
SINA ubiquitin ligases have a wide range of functions in plants and animals. They can regulate the stability of many important proteins in plants and animals. Previous studies have shown that the ABA response factor can regulate the transcription level of SINA2, but it remains unclear how the protein levels of SINATs are regulated (Bao et al., 2014). The identification of key factors regulating the stability of SINA proteins could enhance our understanding of the function of SINA proteins. We found that the MdSINA2 degraded in the dark, and it became insensitive to light after its RING domain had been destroyed. Based on this result and those of previous studies (Qi et al., 2017; Qi et al., 2020), we speculate that the degradation of MdSINA2 in the dark may stem from the lack of energy available under dark conditions.
ABA plays an important biological role in stomatal closure, stress resistance, seed germination, and senescence (Finkelstein et al., 2002; Assmann, 2003; Zhu, 2016; Sussmilch et al., 2017). The observation that MdSINA2 is significantly induced by ABA (Figure 5) and is highly expressed in leaves suggests that MdSINA2 may be a functional component of ABA signaling and may be involved in the tolerance of different types of stress by regulating leaf development. Indeed, when we compared the germination rate of wild type (Col-0) and 35S::MdSINA2 on MS media supplemented with different concentrations of ABA, significant differences were observed in seed germination (Figure 10). Our results differed from those documented for SINA2 in transgenic Arabidopsis (Bao et al., 2014) under different ABA concentration treatments, where no significant differences were observed among wild-type and experimental groups. Although the specific mechanism of MdSINA2 in the responses to ABA and different types of stress remains unclear, our results suggest that altering MdSINA2 expression can significantly improve sensitivity to ABA in transgenic calli and Arabidopsis. Additional screening of possible targets with which MdSINA2 could interact in ABA signaling pathways will help elucidate the role that it plays in ABA signaling.
In conclusion, 11 MdSINA genes were identified in the apple genome. Bioinformatics analyses and analyses of gene expression were conducted to characterize their roles of these genes in apple growth and development. The functional characterization suggested that apple SINA members may function by responding to ABA signaling. These findings provided new insight into the ability of plants to resist abiotic stress.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
H-LL and Y-JH conceived the experiments. H-LL, XW, X-LJ, and Z-WQ performed the research. H-LL, XW, and X-LJ analyzed the data. H-LL and XW wrote the manuscript. X-LJ and Y-JH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The National Key Research and Development Program of China (2018YFD1000200), the National Natural Science Foundation of China (U1706202 and 31772288), the Ministry of Agriculture of China (CARS-27).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Xutong Wang in Purdue University for his suggestions and analyses on bioinformatics in this article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01109/full#supplementary-material
References
An, J. P., Li, H. H., Song, L. Q., Su, L., Liu, X., You, C. X., et al. (2016). The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple. Plant Physiol. Biochem. 108, 24–31. doi: 10.1016/j.plaphy.2016.06.032
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., Hao, Y. J. (2018). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 41 (11), 2678–2692. doi: 10.1111/pce.13393
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., Hao, Y. J. (2019). MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 222 (2), 735–751. doi: 10.1111/nph.15628
Assmann, S. M. (2003). OPEN STOMATA1 opens the door to ABA signaling in Arabidopsis guard cells. Trends Plant Sci. 8 (4), 151–153. doi: 10.1016/S1360-1385(03)00052-9
Avin-Wittenberg, T., Bajdzienko, K., Wittenberg, G., Alseekh, S., Tohge, T., Bock, R., et al. (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27 (2), 306–322. doi: 10.1105/tpc.114.134205
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Web Server issue), W202–W208. doi: 10.1093/nar/gkp335
Bao, Y., Wang, C., Jiang, C., Pan, J., Zhang, G., Liu, H., et al. (2014). The tumor necrosis factor receptor-associated factor (TRAF)-like family protein SEVEN IN ABSENTIA 2 (SINA2) promotes drought tolerance in an ABA-dependent manner in Arabidopsis. New Phytol. 202 (1), 174–187. doi: 10.1111/nph.12644
Cai, Z. L., Xu, J., Xue, S. R., Liu, Y. Y., Zhang, Y. J., Zhang, X. Z., et al. (2015). The E3 ubiquitin ligase seven in absentia homolog 1 may be a potential new therapeutic target for Parkinson’s disease. Neural Regener. Res. 10 (8), 1286–1291. doi: 10.4103/1673-5374.162763
Callis, J., Carpenter, T., Sun, C. W., Vierstra, R. D. (1995). Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 139 (2), 921–939. doi: 10.1101/gad.9.3.370
Carthew, R. W., Rubin, G. M. (1990). seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63 (3), 561–577. doi: 10.1016/0092-8674(90)90452-k
Chen, L., Su, Z. Z., Huang, L., Xia, F. N., Qi, H., Xie, L. J., et al. (2017). The AMP-Activated Protein Kinase KIN10 Is Involved in the Regulation of Autophagy in Arabidopsis. Front. Plant Sci. 8:1201:1201. doi: 10.3389/fpls.2017.01201
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Crooks, G. E., Hon, G., Chandonia, J. M., Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14 (6), 1188–1190. doi: 10.1101/gr.849004
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Daccord, N., Celton, J. M., Linsmith, G., Becker, C., Choisne, N., Schijlen, E., et al. (2017). High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet. 49 (7), 1099–1106. doi: 10.1038/ng.3886
Dametto, A., Buffon, G., Dos Reis Blasi, E. A., Sperotto, R. A. (2015). Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal Behav. 10 (9), e1057369. doi: 10.1080/15592324.2015.1057369
Den Herder, G., De Keyser, A., De Rycke, R., Rombauts, S., Van de Velde, W., Clemente, M. R., et al. (2008). Seven in absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol. 148 (1), 369–382. doi: 10.1104/pp.108.119453
Den Herder, G., Yoshida, S., Antolin-Llovera, M., Ried, M. K., Parniske, M. (2012). Lotus japonicus E3 ligase SEVEN IN ABSENTIA4 destabilizes the symbiosis receptor-like kinase SYMRK and negatively regulates rhizobial infection. Plant Cell 24 (4), 1691–1707. doi: 10.1105/tpc.110.082248
Depaux, A., Regnier-Ricard, F., Germani, A., Varin-Blank, N. (2006). Dimerization of hSiah proteins regulates their stability. Biochem. Biophys. Res. Commun. 348 (3), 857–863. doi: 10.1016/j.bbrc.2006.07.092
Devoto, A., Muskett, P. R., Shirasu, K. (2003). Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 6 (4), 307–311. doi: 10.1016/s1369-5266(03)00060-8
Duplan, V., Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5:42:42. doi: 10.3389/fpls.2014.00042
Finkelstein, R. R., Gampala, S. S., Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 Suppl, S15–S45. doi: 10.1105/tpc.010441
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 (3), 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Franck, T., Krueger, R., Woitalla, D., Muller, T., Engelender, S., Riess, O. (2006). Mutation analysis of the seven in absentia homolog 1 (SIAH1) gene in Parkinson’s disease. J. Neural Transm. (Vienna) 113 (12), 1903–1908. doi: 10.1007/s00702-006-0480-z
Fukuba, H., Yamashita, H., Nagano, Y., Jin, H. G., Hiji, M., Ohtsuki, T., et al. (2007). Siah-1 facilitates ubiquitination and degradation of factor inhibiting HIF-1alpha (FIH). Biochem. Biophys. Res. Commun. 353 (2), 324–329. doi: 10.1016/j.bbrc.2006.12.051
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Heck, J. W., Cheung, S. K., Hampton, R. Y. (2010). Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. U.S.A. 107 (3), 1106–1111. doi: 10.1073/pnas.0910591107
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 (Web Server issue), W585–W587. doi: 10.1093/nar/gkm259
Hu, G., Fearon, E. R. (1999). Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell Biol. 19 (1), 724–732. doi: 10.1128/mcb.19.1.724
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene features visualization server. Bioinformatics 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Hua, Z., Vierstra, R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334. doi: 10.1146/annurev-arplant-042809-112256
Islam, M. S., Proshad, R., Kormoker, T., Tusher, T. R. (2019). Autophagy-mediated Nutrient Recycling and Regulation in Plants: A Molecular View. J. Plant Biol. 62 (5), 307–319. doi: 10.1007/s12374-019-0213-0
Jansen, A. H., Reits, E. A., Hol, E. M. (2014). The ubiquitin proteasome system in glia and its role in neurodegenerative diseases. Front. Mol. Neurosci. 7:73:73. doi: 10.3389/fnmol.2014.00073
Khurana, A., Nakayama, K., Williams, S., Davis, R. J., Mustelin, T., Ronai, Z. (2006). Regulation of the ring finger E3 ligase Siah2 by p38 MAPK. J. Biol. Chem. 281 (46), 35316–35326. doi: 10.1074/jbc.M606568200
Knight, H., Knight, M. R. (2001). Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 6 (6), 262–267. doi: 10.1016/s1360-1385(01)01946-x
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40 (Database issue), D1202–D1210. doi: 10.1093/nar/gkr1090
Lee, Y. R., Yuan, W. C., Ho, H. C., Chen, C. H., Shih, H. M., Chen, R. H. (2010). The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 29 (10), 1748–1761. doi: 10.1038/emboj.2010.62
Li, S., Li, Y., Carthew, R. W., Lai, Z. C. (1997). Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell 90 (3), 469–478. doi: 10.1016/s0092-8674(00)80507-3
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, H. L., Shu, H. R., et al. (2012). MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 160 (2), 1011–1022. doi: 10.1104/pp.112.199703
Liu, M., Aneja, R., Wang, H., Sun, L., Dong, X., Huo, L., et al. (2008). Modulation of multidrug resistance in cancer cells by the E3 ubiquitin ligase seven-in-absentia homologue 1. J. Pathol. 214 (4), 508–514. doi: 10.1002/path.2312
Metzger, M. B., Pruneda, J. N., Klevit, R. E., Weissman, A. M. (2014). RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843 (1), 47–60. doi: 10.1016/j.bbamcr.2013.05.026
Ning, Y., Jantasuriyarat, C., Zhao, Q., Zhang, H., Chen, S., Liu, J., et al. (2011). The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 157 (1), 242–255. doi: 10.1104/pp.111.180893
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell 41 (1), 33–46 e37. doi: 10.1016/j.devcel.2017.03.013
Ohno, S., Wolf, U., Atkin, N. B. (1968). Evolution from fish to mammals by gene duplication. Hereditas 59 (1), 169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x
Park, B. S., Sang, W. G., Yeu, S. Y., Choi, Y. D., Paek, N.-C., Kim, M. C., et al. (2007). Post-translational regulation of FLC is mediated by an E3 ubiquitin ligase activity of SINAT5 in Arabidopsis. Plant Sci. 173 (2), 269–275. doi: 10.1016/j.plantsci.2007.06.001
Park, B. S., Eo, H. J., Jang, I. C., Kang, H. G., Song, J. T., Seo, H. S. (2010). Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 398 (2), 242–246. doi: 10.1016/j.bbrc.2010.06.067
Polekhina, G., House, C. M., Traficante, N., Mackay, J. P., Relaix, F., Sassoon, D. A., et al. (2002). Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat. Struct. Biol. 9 (1), 68–75. doi: 10.1038/nsb743
Qi, H., Xia, F. N., Xie, L. J., Yu, L. J., Chen, Q. F., Zhuang, X. H., et al. (2017). TRAF Family Proteins Regulate Autophagy Dynamics by Modulating AUTOPHAGY PROTEIN6 Stability in Arabidopsis. Plant Cell 29 (4), 890–911. doi: 10.1105/tpc.17.00056
Qi, H., Li, J., Xia, F. N., Chen, J. Y., Lei, X., Han, M. Q., et al. (2020). Arabidopsis SINAT Proteins Control Autophagy by Mediating Ubiquitylation and Degradation of ATG13. Plant Cell 32 (1), 263–284. doi: 10.1105/tpc.19.00413
Ren, Y. R., Yang, Y. Y., Zhang, R., You, C. X., Zhao, Q., Hao, Y. J. (2019). MdGRF11, an apple 14-3-3 protein, acts as a positive regulator of drought and salt tolerance. Plant Sci. 288:110219. doi: 10.1016/j.plantsci.2019.110219
Sadanandom, A., Bailey, M., Ewan, R., Lee, J., Nelis, S. (2012). The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 196 (1), 13–28. doi: 10.1111/j.1469-8137.2012.04266.x
Santner, A., Estelle, M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61 (6), 1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x
Schimke, R. T. (1973). Control of enzyme levels in mammalian tissues. Adv. Enzymol. Relat. Areas Mol. Biol. 37, 135–187. doi: 10.1002/9780470122822.ch3
Siswanto, F. M., Jawi, I. M., Kartiko, B. H. (2018). The role of E3 ubiquitin ligase seven in absentia homolog in the innate immune system: An overview. Vet. World 11 (11), 1551–1557. doi: 10.14202/vetworld.2018.1551-1557
Soto-Burgos, J., Zhuang, X., Jiang, L., Bassham, D. C. (2018). Dynamics of Autophagosome Formation. Plant Physiol. 176 (1), 219–229. doi: 10.1104/pp.17.01236
Stone, S. L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E., Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 (1), 13–30. doi: 10.1104/pp.104.052423
Stone, S. L. (2014). The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 5:135:135. doi: 10.3389/fpls.2014.00135
Sussmilch, F. C., Brodribb, T. J., McAdam, S. A. (2017). What are the evolutionary origins of stomatal responses to abscisic acid in land plants? J. Integr. Plant Biol. 59 (4), 240–260. doi: 10.1111/jipb.12523
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42 (10), 833–839. doi: 10.1038/ng.654
Wang, M., Jin, Y., Fu, J., Zhu, Y., Zheng, J., Hu, J., et al. (2008). Genome-wide analysis of SINA family in plants and their phylogenetic relationships. DNA Seq. 19 (3), 206–216. doi: 10.1080/10425170701517317
Wang, Y., Tang, H., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wang, W., Fan, Y., Niu, X., Miao, M., Kud, J., Zhou, B., et al. (2018). Functional analysis of the seven in absentia ubiquitin ligase family in tomato. Plant Cell Environ. 41 (3), 689–703. doi: 10.1111/pce.13140
Welsch, R., Maass, D., Voegel, T., Dellapenna, D., Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145 (3), 1073–1085. doi: 10.1104/pp.107.104828
Wheeler, T. C., Chin, L. S., Li, Y., Roudabush, F. L., Li, L. (2002). Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J. Biol. Chem. 277 (12), 10273–10282. doi: 10.1074/jbc.M107857200
Xie, Q., Guo, H. S., Dallman, G., Fang, S., Weissman, A. M., Chua, N. H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 (6903), 167–170. doi: 10.1038/nature00998
Xu, G., Jaffrey, S. R. (2013). Proteomic identification of protein ubiquitination events. Biotechnol. Genet. Eng. Rev. 29, 73–109. doi: 10.1080/02648725.2013.801232
Yang, M., Li, C., Cai, Z., Hu, Y., Nolan, T., Yu, F., et al. (2017). SINAT E3 Ligases Control the Light-Mediated Stability of the Brassinosteroid-Activated Transcription Factor BES1 in Arabidopsis. Dev. Cell 41 (1), 47–58 e44. doi: 10.1016/j.devcel.2017.03.014
Yoo, S. H., Mohawk, J. A., Siepka, S. M., Shan, Y., Huh, S. K., Hong, H. K., et al. (2013). Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152 (5), 1091–1105. doi: 10.1016/j.cell.2013.01.055
Zhang, Z., Li, J., Liu, H., Chong, K., Xu, Y. (2015). Roles of ubiquitination-mediated protein degradation in plant responses to abiotic stresses. Environ. Exp. Bot. 114, 92–103. doi: 10.1016/j.envexpbot.2014.07.005
Zhang, C., Hao, Z., Ning, Y., Wang, G. L. (2019). SINA E3 Ubiquitin Ligases: Versatile Moderators of Plant Growth and Stress Response. Mol. Plant 12 (5), 610–612. doi: 10.1016/j.molp.2019.03.013
Zhao, X. Y., Qi, C. H., Jiang, H., You, C. X., Guan, Q. M., Ma, F. W., et al. (2019). The MdWRKY31 transcription factor binds to the MdRAV1 promoter to mediate ABA sensitivity. Hortic. Res. 6, 66. doi: 10.1038/s41438-019-0147-1
Zhu, J. K. (2016). Abiotic Stress Signaling and Responses in Plants. Cell 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: apple, bioinformatics analysis, expression analysis, MdSINA2, ABA sensitivity
Citation: Li H-L, Wang X, Ji X-L, Qiao Z-W, You C-X and Hao Y-J (2020) Genome-Wide Identification of Apple Ubiquitin SINA E3 Ligase and Functional Characterization of MdSINA2. Front. Plant Sci. 11:1109. doi: 10.3389/fpls.2020.01109
Received: 29 April 2020; Accepted: 06 July 2020;
Published: 24 July 2020.
Edited by:
Agnieszka Ludwików, Adam Mickiewicz University, PolandReviewed by:
Zhaoqing Chu, Shanghai Chenshan Plant Science Research Center (CAS), ChinaDerek Gingerich, University of Wisconsin–Eau Claire, United States
Copyright © 2020 Li, Wang, Ji, Qiao, You and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Jin Hao, aGFveXVqaW5Ac2RhdS5lZHUuY24=
 Hong-Liang Li
Hong-Liang Li Yu-Jin Hao
Yu-Jin Hao