- 1Department of Plant Science, Plant Genomics and Breeding Institute, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 2Division of Life Sciences, Incheon National University, Incheon, South Korea
- 3Graduate School of Agricultural and Life Sciences, Biotechnology Research Center, The University of Tokyo, Tokyo, Japan
- 4Department of Plant Molecular Systems Biotechnology, Crop Biotech Institute, Kyung Hee University, Yongin, South Korea
Leaf senescence is the final stage of leaf development and an important step that relocates nutrients for grain filling in cereal crops. Senescence occurs in an age-dependent manner and under unfavorable environmental conditions such as deep shade, water deficit, and high salinity stresses. Although many transcription factors that modulate leaf senescence have been identified, the mechanisms that regulate leaf senescence in response to environmental conditions remain elusive. Here, we show that rice (Oryza sativa) ETHYLENE RESPONSE FACTOR 101 (OsERF101) promotes the onset and progression of leaf senescence. OsERF101 encodes a predicted transcription factor and OsERF101 transcript levels rapidly increased in rice leaves during dark-induced senescence (DIS), indicating that OsERF101 is a senescence-associated transcription factor. Compared with wild type, the oserf101 T-DNA knockout mutant showed delayed leaf yellowing and higher chlorophyll contents during DIS and natural senescence. Consistent with its delayed-yellowing phenotype, the oserf101 mutant exhibited downregulation of genes involved in chlorophyll degradation, including rice NAM, ATAF1/2, and CUC2 (OsNAP), STAY-GREEN (SGR), NON-YELLOW COLORING 1 (NYC1), and NYC3 during DIS. After methyl jasmonate treatment to induce rapid leaf de-greening, the oserf101 leaves retained more chlorophyll compared with wild type, indicating that OsERF101 is involved in promoting jasmonic acid (JA)-induced leaf senescence. Consistent with the involvement of JA, the expression of the JA signaling genes OsMYC2/JA INSENSITIVE 1 (OsJAI1) and CORONATINE INSENSITIVE 1a (OsCOI1a), was downregulated in the oserf101 leaves during DIS. Transient transactivation and chromatin immunoprecipitation assays revealed that OsERF101 directly binds to the promoter regions of OsNAP and OsMYC2, which activate genes involved in chlorophyll degradation and JA signaling-mediated leaf senescence. These results demonstrate that OsERF101 promotes the onset and progression of leaf senescence through a JA-mediated signaling pathway.
Introduction
Leaf senescence is the final stage of leaf development and involves many molecular and physiological events, including chlorophyll degradation, the breakdown of other cellular components, and cell death, thus allowing plants to recycle nutrients from leaf tissues into seeds (Masclaux-Daubresse et al., 2008; Distelfeld et al., 2014; Yang et al., 2019). Environmental stresses, such as deep shade, water deficit, extreme temperatures, and high salinity, accelerate leaf senescence (Munné-Bosch and Alegre, 2004; Mickelbart et al., 2015). Premature leaf senescence shortens the vegetative growth period, inducing the precocious transition from the vegetative to the reproductive stage and reducing the plant’s nutritional capacity, thus negatively affecting crop productivity.
Age-dependent and stress-induced senescence share many regulatory networks and physiological, biochemical, and molecular mechanisms (Gepstein and Glick, 2013; Zhuang et al., 2019). Transcription factors (TFs) participate in the regulatory pathways for leaf senescence and abiotic stress tolerance. For instance, RNA interference (RNAi)-mediated suppression of rice (Oryza sativa) NAC2 (OsNAC2), a member of the NAC family, reduces the expression of chlorophyll-degradation and senescence-associated genes, leading to delayed leaf yellowing during dark-induced senescence (DIS) (Mao et al., 2017). Moreover, OsNAC2-RNAi transgenic plants display enhanced tolerance against drought and high salt stresses due to upregulation of stress-related and abscisic acid (ABA)-signaling genes (Shen et al., 2017). Overexpression of ONAC106 causes delayed leaf yellowing in rice, resulting in extended photosynthetic capacity at the reproductive stage in paddy fields and increased grain production. In addition to the biological function of ONAC106 in leaf senescence, transgenic rice overexpressing ONAC106 exhibit improved tolerance to salt stress (Sakuraba et al., 2015). The gain-of-function mutant prematurely senile 1-D (ps1-D) exhibits upregulated expression of OsNAP, leading to accelerated leaf yellowing during both natural senescence and DIS (Liang et al., 2014). In addition, overexpression of OsNAP confers tolerance to salt and drought stresses (Chen et al., 2014). These distinct phenomena reveal the role of OsNAP in regulating diverse genes involved in chlorophyll degradation and abiotic stress responses.
Plant hormones serve as signal molecules and are involved in the regulation of a variety of cellular processes, including leaf senescence and abiotic stress responses. Among them, jasmonic acid (JA) accumulates to high levels in plant cells when plants enter the senescence phase or in unfavorable conditions (He et al., 2002; Wasternack and Hause, 2013; Hou et al., 2016), JA biosynthesis is catalyzed by enzymes including lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and 12-oxo-PDA reductase (OPR) (He et al., 2002). Increased JA levels activate the downstream signal transduction pathways that are involved in senescence and abiotic stress tolerance. For instance, elevated JA levels are perceived by the F-box protein CORONATINE INSENSITIVE 1 (COI1), followed by ubiquitination and degradation of JA ZIM-domain (JAZ) proteins (Niu et al., 2011; Song et al., 2011; Qi et al., 2014). JAZ degradation causes the release of downstream TFs such as the basic helix-loop-helix (bHLH) TF family members MYC2, MYC3, and MYC4 (Fernández-Calvo et al., 2011). The oscoi1b mutant leaves retain their green color during DIS (Lee et al., 2015). Transgenic plants overexpressing a truncated OsJAZ8 protein that which lacks the ubiquitin-binding Jas domain also exhibit a delayed senescence phenotype (Uji et al., 2017). OsMYC2/OsJAI1 directly binds to the promoter of senescence-associated genes, and thus its overexpression leads to leaf yellowing during DIS (Uji et al., 2017). In Arabidopsis, MYC2/JAI1 and its homologous proteins MYC3 and MYC4 regulate the expression of the chlorophyll degradation genes NON-YELLOWING 1 (NYE1), NON-YELLOW COLORING 1 (NYC1), and PHEOPHYTINASE (PPH) by binding to their promoters (Zhu et al., 2015).
The APETALA2/Ethylene Response Factor (AP2/ERF) family can be divided into four subfamilies depending on the number of AP2/ERF domains and their amino acid similarity, such as APETALA2 (AP2), Related to ABI3/VP1 (RAV), Dehydration Responsive Element Binding protein (DREB), and Ethylene Response Factor (ERF) subfamilies (Sakuma et al., 2002; Nakano et al., 2006). DREB and ERF subfamilies have a single AP2/ERF domain that can bind to both the C-Repeat/Dehydration Responsive Element (CRT/DRE) and the Ethylene Response Element (ERE) in the promoter regions of their target genes. ERFs regulate the expression of abiotic stress-related genes in various plant species including rice (Fukao et al., 2011), Arabidopsis (Phukan et al., 2017), wheat (Triticum aestivum) (Gao et al., 2018), maize (Zea mays) (Liu et al., 2013), soybean (Glycine max) (Zhang et al., 2009), and tobacco (Nicotiana tabacum) (Park et al., 2001).
A recent study showed that the rice ETHYLENE RESPONSE FACTOR 101 (OsERF101) plays an important role in enhancing tolerance to drought stress in reproductive tissues (Jin et al., 2018). However, the regulatory functions of OsERF101 in leaf senescence have not been understood. In this study, our results substantially showed that OsERF101 positively regulates leaf senescence in rice, and directly activates the expression of OsNAP and OsMYC2 that play a central role in mediating chlorophyll degradation and JA-mediated leaf senescence. Thus, our findings provide a new molecular insight of OsERF101 function in leaf senescence.
Materials and Methods
Plant Materials, Growth Conditions, and Dark, Phytohormone, and Stress Treatments
The T-DNA insertion mutant oserf101 (PFG_2D-00368) was obtained from Crop Biotech Institute at Kyung Hee University, Republic of Korea (Jeong et al., 2002). The Oryza sativa japonica cultivar ’Dongjin’ (parental line) and the oserf101 mutant were cultivated in a paddy field under natural long day (NLD) conditions (>14 h sunlight/day, 37°N latitude) in Suwon, Republic of Korea. The germinated rice seedlings were transplanted in a paddy soil and grown in a growth chamber under long day (LD) conditions (14 h light/10 h dark, 37°N latitude) in Seoul, Republic of Korea. For the dark treatment, the detached leaves of rice plants grown in a growth chamber under LD conditions for 3 weeks were incubated on 3 mM MES (pH 5.8) buffer at 28°C in complete darkness. For phytohormone and stress treatments, the sterilized seeds were germinated on half-strength Murashige and Skoog (1/2 MS) solid medium under continuous light (90 µmol m-2 s-1) at 30°C. The 10-day-old seedlings were transferred to 1/2 MS liquid medium containing 100 µM 1-aminocyclo-propane-1-carboxylic acid (ACC), 100 µM MeJA, 100 µM ABA, 100 mM NaCl, and 20% polyethylene glycol (PEG) or were dehydrated by air drying. Rice seedlings incubated in 1/2 MS liquid medium without additional phytohormones were used as a mock control.
Determination of Total Chlorophyll and Photosynthetic Capacity
To measure the total chlorophyll, pigments were extracted from an equal fresh weight of rice leaves grown in a paddy field, incubated in complete darkness, or treated with 100 µM MeJA using 80% ice-cold acetone. The extracts were centrifuged at 10,000 g for 10 min at 10°C and then the absorbance of the supernatants was measured at 647 and 663 nm using an UV/VIS spectrophotometer (BioTek). The concentration of chlorophyll was calculated as previously described (Porra et al., 1989). To determine the photosynthetic activity, the Fv/Fm ratio was measured using the OS-30p+ instrument (Opti-Sciences). The middle part of each flag leaf of plants grown in a paddy field under NLD conditions was adapted in the dark for 5 min and then the Fv/Fm ratio was determined in the flag leaves.
Determination of Phytohormone Sensitivity
To determine the sensitivity to phytohormones, detached leaves of 3-week-old plants grown in paddy soil were floated on 3 mM MES (pH 5.8) buffer containing 50 µM MeJA and 50 µM ABA and incubated in continuous light conditions (90 µmole·m−2s−1) at 30°C for 4 days. Detached leaves floated on 3 mM MES buffer (pH 5.8) without phytohormones were used as a control. To assess the ABA sensitivity of rice seedlings, the 10-day-old rice seedlings grown in 1/2 MS solid medium were transferred to 1/2 MS liquid medium containing 5 and 10 µM ABA. Seedlings incubated in 1/2 MS liquid medium without additional phytohormones were used as a mock control.
Quantitative Reverse Transcription PCR (qRT-PCR) and Semiquantitative RT-PCR
Total RNA was extracted from rice tissues using an RNA Extraction Kit (Macrogen, South Korea) according to manufacturer instructions. First-strand cDNA was synthesized with 2 µg of total RNA in a 25 µl volume using M-MLV reverse transcriptase and oligo(dT)15 primer (Promega), and diluted with 75 µl of water. The qPCR amplifications were conducted on the LightCycler 2.0 instrument (Roche Diagnostics). The 20 µl of qPCR mixture included 2 µl of the first-strand cDNA mixture, 10 µl of 2X GoTaq PCR Master Mix (Promega), and 1 µl of 10 pM primer pairs (Supplementary Table S1). The qPCR conditions were 95°C for 2 min, followed by 50 cycles at 95°C for 5 s, 59°C for 15 s, and 72°C for 10 s. The rice UBIQUITIN5 (OsUBQ5, AK061988) gene was used as an internal control for normalization. The semi-quantitative PCR was performed in a 20 µl volume containing 2 µl diluted cDNA, 1 unit Ex Taq polymerase (TaKaRa Biotechniques), and 1 µl of 10 pM primers (listed in Supplementary Table S1). The PCR program included initial denaturation at 94°C for 3 min, followed by specified cycles at 94°C for 30 sec, 55°C for 1 min, and 72°C for 40 sec, followed by a final extension at 72°C for 5 min. The PCR products were electrophoresed on a 1% agarose gel. OsUBQ5 was used as an equal loading control.
Plasmid Construction and Rice Transformation
To generate the overexpression construct, a full-length cDNA of OsERF101 was amplified using the primers listed in Supplementary Table S1. The amplified fragments were ligated into the pCR8/GW/TOPO TA cloning vector (Invitrogen) and then inserted into the pMDC32 Gateway-compatible binary vector through the LR recombination reaction. The pMDC32-OsERF101 plasmids were introduced into callus of Dongjin seeds by Agrobacterium tumefaciens (strain LBA4404)-mediated transformation (Jeon et al., 2000). Agrobacterium-infected calli were transferred to 1/2 MS solid medium containing cytokinin and auxin. Plantlets regenerated from the callus were grown under continuous light conditions (90 µmole·m−2 s−1) at 30°C.
Protoplast Transient Transactivation Assays
Reporter plasmids were constructed by the insertion of OsMYC2 (-1529 to -1 bp) and OsNAP (-1502 to -1 bp) promoters into the pJD301 vector (Luehrsen et al., 1992). To construct effector plasmids, OsERF101 cDNA from the WT cultivar was cloned upstream of a sequence encoding six copies of a MYC epitope tag in the pGA3697 vector. Protoplasts were isolated from young rice seedlings as previously described (Zhang et al., 2011). A reporter plasmid (4 µg) and an effector plasmid (8 µg) were co-transfected, together with 1 µg of an internal control plasmid (pUBQ10-GUS), into 5 × 104 protoplasts using the polyethylene glycol (PEG)-mediated transfection method (Yoo et al., 2007). Transfected protoplasts were incubated in protoplast culture solution (0.4 M mannitol, 15 mM MgCl2, 4 mM MES-KOH [pH 5.8]) in the dark at room temperature for 16 h. Luciferase (LUC) activity in each cell lysate was determined using the Luciferase Assay System Kit (Promega). LUC activity was normalized against β-glucuronidase (GUS) activity derived from the internal control plasmid.
Chromatin Immunoprecipitation (ChIP) Assays
For the ChIP assay, the Ubi : OsERF101-Myc and Ubi : Myc were transfected into rice protoplasts as previously described (Zhang et al., 2011). Protoplasts were then subjected to cross-linking with 1% formaldehyde for 30 min under vacuum. Then, nuclei were isolated and lysed, and chromatin complexes were isolated and sonicated, as described (Saleh et al., 2008). DNA was sonicated using a BIORUPTORII (COSMO BIO). Anti-Myc monoclonal antibody (Abcam, Cambridge, UK) and protein A agarose beads (Merck Millipore) were used for immunoprecipitation. DNA recovered from agarose beads was purified using the DNeasy Plant Mini Kit (Qiagen). Quantitative PCR was performed using the KAPA SYBR FAST qPCR Kit (KAPA Biosystems) and gene-specific primers (Supplementary Table S1).
Results
Expression of OsERF101 in Rice
Rice OsERF101 (LOC_Os04g32620) consists of 4366 bp of genomic DNA with an 807-bp open reading frame containing two exons and encoding a 268-amino acid protein belonging to the ERF transcription factor subfamily. Amino acid sequence alignments between OsERF101 and its putative orthologs demonstrated that they have a single AP2/ERF domain, which is highly conserved in plants (Supplementary Figure S1).
To investigate the expression of OsERF101, we measured OsERF101 transcript levels in various organs (root, leaf sheath, leaf blade, lamina joint, node, tiller base, panicle, stem, flag leaf, and internode). We used wild-type (WT; japonica rice cultivar ‘Dongjin’) plants grown in a growth chamber under long-day (LD) conditions (14.5 h light/9.5 h dark) at 21 days after germination (DAG, seedling stage) or in the paddy field under natural LD (NLD) conditions (≥14 h light/day in Suwon, South Korea, 37°N latitude) at 120 DAG (heading stage). Reverse transcription and quantitative real-time PCR (qRT-PCR) analysis revealed that the OsERF101 transcripts were most abundant in root, and then (in order of decreasing abundance), in lamina joint, node, leaf blade, and leaf sheath (Figure 1A).
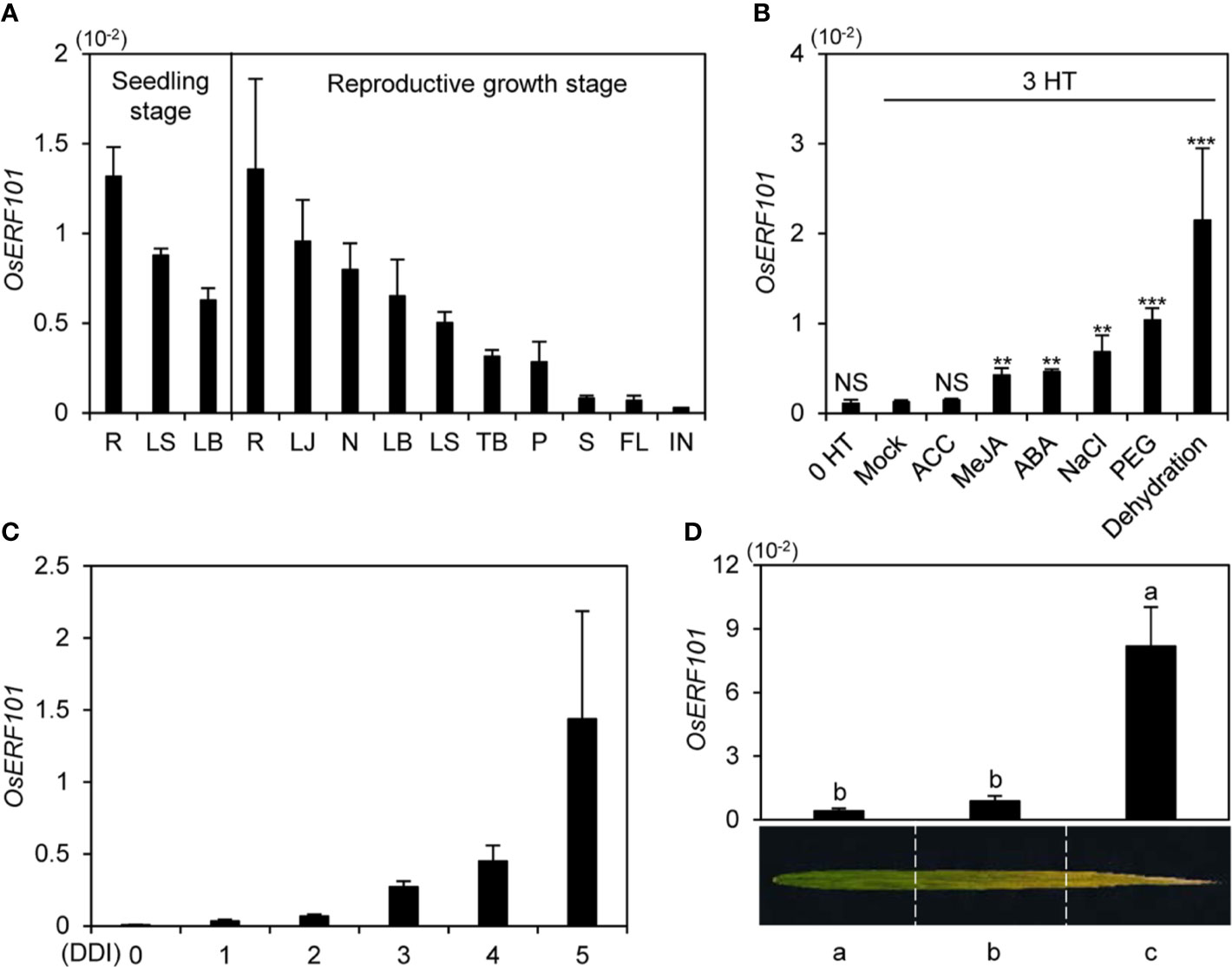
Figure 1 Expression profiles of OsERF101. (A) OsERF101 was differentially expressed in various japonica cultivar ’Dongjin’ (hereafter wild type; WT) tissues: root (R), leaf sheath (LS), leaf blade (LB), lamina joint (LJ), node (N), tiller base (TB), panicle (P), stem (S), flag leaf (FL), and internode (IN). WT seedlings were grown in a growth chamber for 3 weeks under long-day (LD) conditions (14 h light/10 h dark). WT plants were cultivated in a paddy field until the reproductive stage at 120 days after seeding (DAS) under natural long-day (NLD) conditions (>14 h light/day). (B) Expression patterns of OsERF101 in response to abiotic stresses and phytohormones. WT seedlings grown in half-strength Murashige and Skoog (1/2 MS) solid medium for 10 d under continuous light at 28°C were incubated in 1/2 MS liquid medium supplemented with 100 µM ACC, 100 µM MeJA, 100 µM ABA, 100 mM NaCl, or 20% PEG or dehydrated by air drying. Seedlings incubated in 1/2 MS liquid medium without treatment were used as a mock control. Total RNA was isolated from the leaves at 0 and 3 h of treatment (HT). Asterisks indicate statistically significant differences between treated samples and the mock control, as determined by Student’s t-test (**p < 0.01, ***p < 0.001). NS, not significant. (C) OsERF101 expression gradually increased in detached leaves of 3-week-old WT plants grown in a growth chamber under LD conditions. Detached leaves were subjected to complete darkness in 3 mM MES (pH 5.8) at 28°C. (D) Expression of OsERF101 was measured in flag leaves divided into three regions from the green sector (a) to the yellow sector (c) at the ripening stage (130 DAS). OsERF101 mRNA levels were determined by qRT-PCR analysis and normalized to that of OsUBQ5 (Os01g22490). Mean and standard deviations were obtained from at least three biological samples. Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (p < 0.05). Experiments were repeated twice with similar results.
To test whether phytohormones and stress treatments affect the expression of OsERF101, we examined the OsERF101 transcript levels in 10-day-old WT seedlings treated with 1-aminocyclo-propane-1-carboxylic acid (ACC), MeJA, ABA, NaCl, PEG, or under dehydration stress for 3 h. qRT-PCR analysis showed that OsERF101 expression increased significantly in response to the MeJA, ABA, NaCl, PEG, and water deficit treatments (Figure 1B). In addition, OsERF101 expression was sharply upregulated in the detached leaves of 2-week-old seedlings during DIS (Figure 1C). In the naturally senescing flag leaves, OsERF101 transcripts accumulated to higher levels in the yellowed tip (c) region than in the yellowing middle (b) or green bottom (a) regions (Figure 1D). These results strongly suggested that OsERF101 is involved in leaf senescence in rice.
OsERF101 Positively Regulates the Onset and Progression of Leaf Senescence
To examine the role of the OsERF101 TF in regulating leaf senescence, we obtained a loss-of-function mutant (PFG_2D-00368) from RiceGE (://signal.salk.edu/cgi-bin/RiceGE), in which a T-DNA fragment was integrated into the second exon of OsERF101 (Figure 2A). To verify the effect of the T-DNA insertion, we performed semi-quantitative RT-PCR analysis and found that in the detached leaves of 3-week-old plants, the OsERF101 transcript was completely absent in the PFG_2D-00368 line (Figure 2B) at 3 days of dark incubation (DDI), indicating that the PFG_2D-00368 line is a knockout mutant (hereafter termed oserf101). We next examined the progress of senescence in WT and oserf101 plants grown in the paddy field under NLD conditions. While there was no significant difference in leaf color between WT and oserf101 plants at 0 days after heading (DAH) (Figure 2C), the oserf101 leaves retained their green color during grain filling much more than the WT (Figures 2D, E). Consistent with the persistence of green leaf color, the chlorophyll levels remained much higher in the flag leaves of the oserf101 mutant compared to the WT (Figure 2F). In addition, the oserf101 mutant maintained a higher Fv/Fm ratio (efficiency of photosystem II) compared to the WT after 20 DAH (Figure 2G), indicating that the prolonged greenness of oserf101 leaves leads to improved photosynthetic activity during grain filling in the autumn fields.
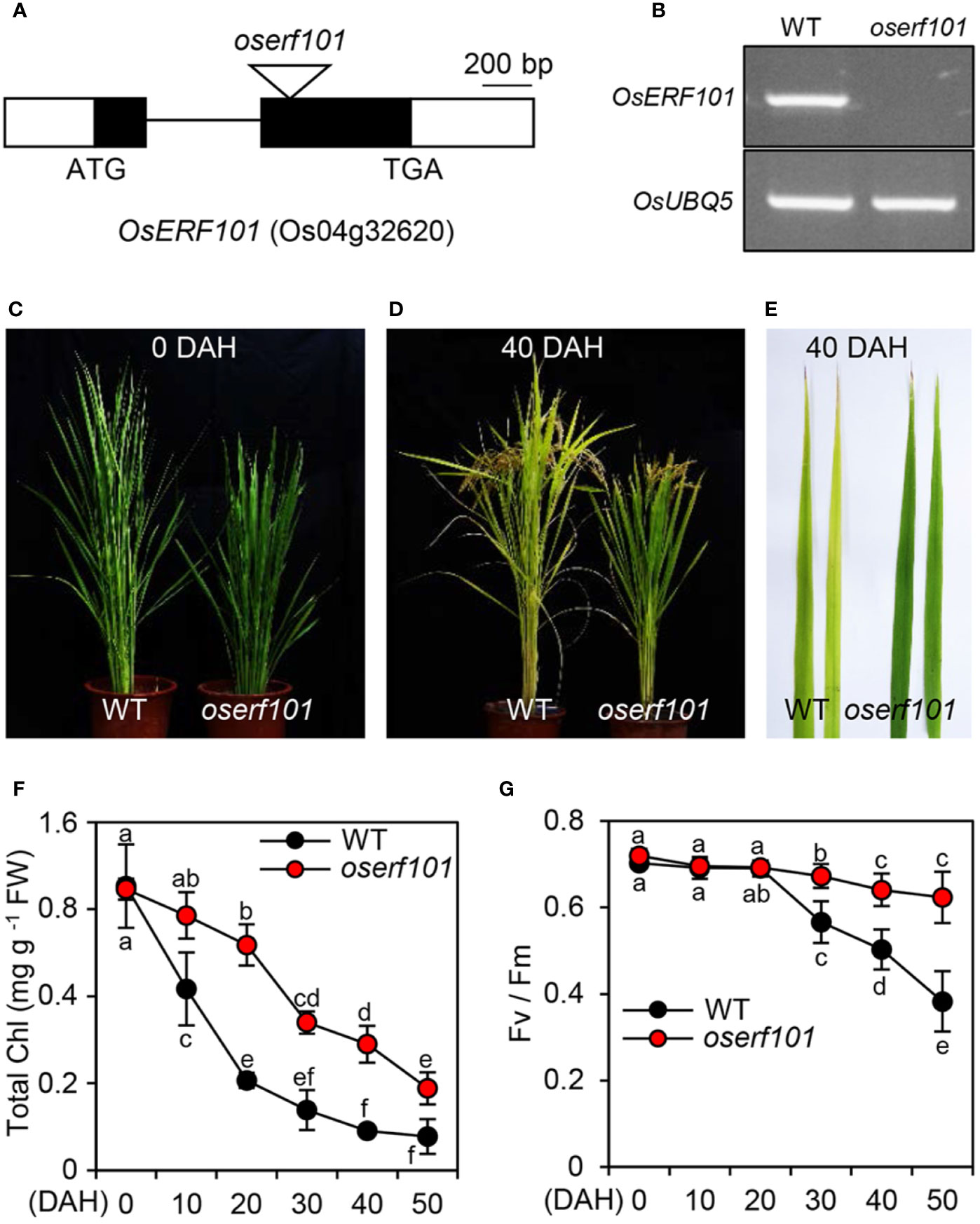
Figure 2 The oserf101 mutant exhibits delayed natural leaf senescence. (A) Position of the T-DNA insertion in the exon region of OsERF101 (LOC_Os04g32620). Black and white boxes represent exons and untranslated regions, respectively. The black line indicates an intron and the open triangle indicates the location of the T-DNA insertion (oserf101, PFG_2D-00368). (B) Mutation of OsERF101 was confirmed by semi-quantitative RT-PCR. Total RNA was isolated from detached second leaves of a 3-week-old WT (the parental japonica rice cultivar ’Dongjin’) plant and the oserf101 mutant in which senescence was induced for 3 days of dark incubation as shown in Figure 3A. OsUBQ5 (Os01g22490) was used as a loading control. (C, D) The comparison of senescence phenotypes of WT and oserf101 plants at 0 and 40 days after heading (DAH) under natural long-day conditions (>14 h light/day). (E) Senescing flag leaves of WT and oserf101 plants at 50 DAH. The images are representative of five independent experiments. (F, G) Changes in total chlorophyll (Chl) contents (F) and photosynthetic capacity (Fv/Fm) (G) of WT and oserf101 flag leaves after heading. Mean and standard deviations were obtained from more than 10 plants. Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (p < 0.05).
To further examine the effects of OsERF101 on leaf de-greening in DIS, we monitored the color in sections of leaves detached from 3-week-old WT and oserf101 plants grown under LD conditions. The oserf101 leaves retained their green color much longer than the WT leaves (Figure 3A), consistent with their relatively higher chlorophyll contents at 3 and 4 DDI (Figure 3B). To confirm the positive role of OsERF101 in leaf senescence, we generated two independent transgenic rice plants overexpressing OsERF101 (OsERF101-OX1 and -OX2) (Figure 3C). The detached leaves of 3-week-old OsERF101-OX plants exhibited accelerated leaf yellowing compared to those of the WT at 3 DDI (Figure 3D). These results indicate that OsERF101 is a positive regulator of the onset and progression of leaf senescence.
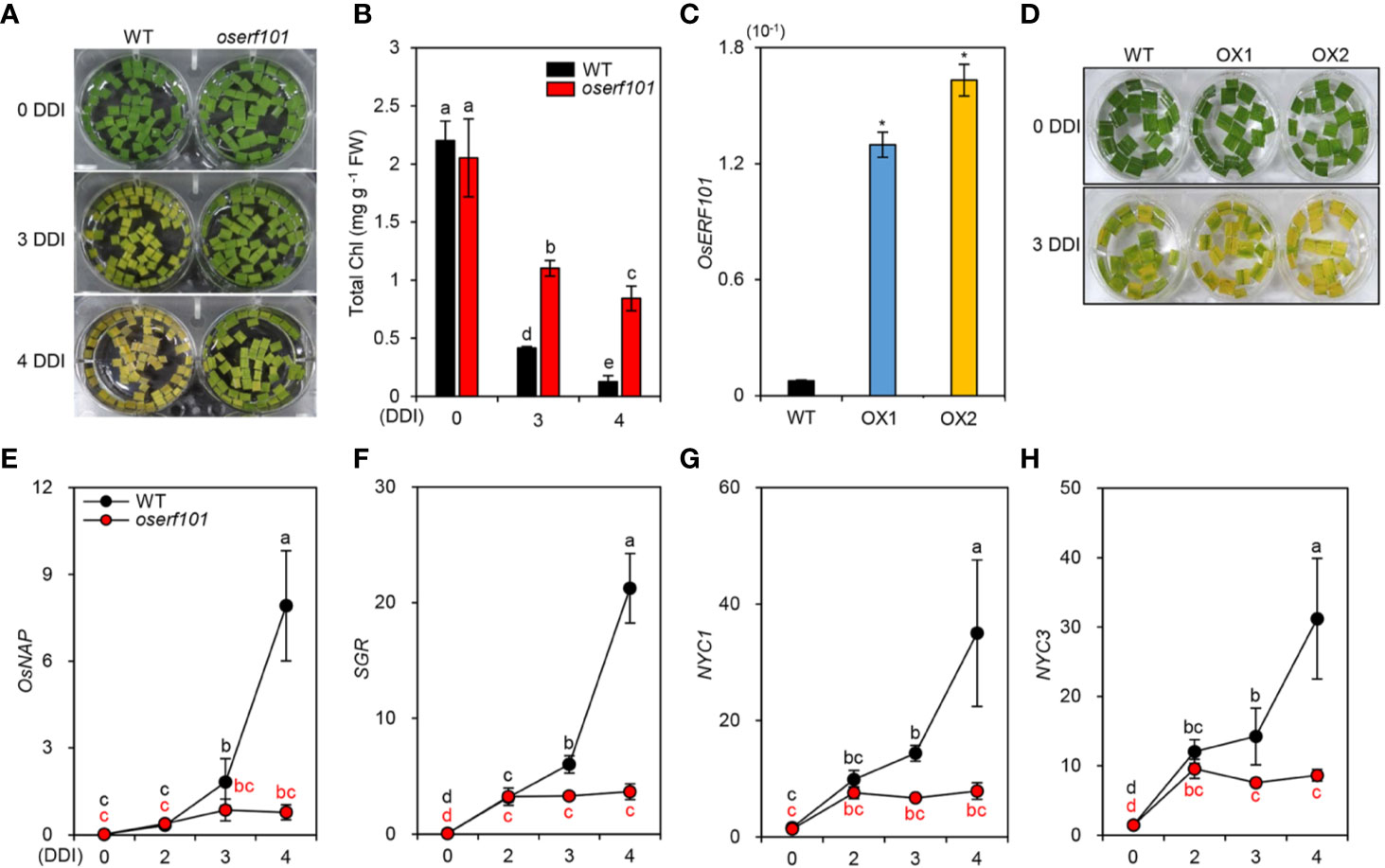
Figure 3 The oserf101 mutant exhibited delayed leaf yellowing during dark-induced senescence (DIS) conditions. (A, B) Detached leaves of 3-week-old WT and oserf101 plants grown in the greenhouse under natural long-day conditions (>14 h light/day) were incubated in 3 mM MES buffer (pH 5.8) with the abaxial side up at 28°C under complete darkness. The visual phenotypes (A) and total chlorophyll (Chl) contents (B) were observed after 0, 3, and 4 days of dark incubation (DDI). Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (p < 0.05). (C) Expression of OsERF101 measured in the leaves of WT and OsERF101-overexpressing (OsERF101-OX) transgenic plants (OX1, OX2) grown in the greenhouse for one month. (D) Detached leaves of 1-month-old OsERF101-OX plants exhibited accelerated leaf yellowing at 3 DDI. (E–H) Altered expression of OsNAP and chlorophyll degradation genes in detached leaves of 3-week-old WT and oserf101 seedlings during DIS. Total RNA was isolated from detached leaves at 0, 2, 3, and 4 DDI. The transcript levels of OsERF101 (C), OsNAP (E), and chlorophyll degradation genes (F–H) were determined by qRT-PCR and normalized to that of OsUBQ5 (Os01g22490). (C) Asterisks indicate statistically significant differences between WT and OsERF101-OX plants, as determined by Student’s t-test (*p < 0.05). (E–H) Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (p < 0.05). Mean and standard deviations were obtained from at least three biological samples. Experiments were repeated twice with similar results. FW, fresh weight.
Among the rice regulatory genes controlling leaf senescence, OsNAP expression is induced by senescence, and OsNAP directly activates the transcription of chlorophyll degradation genes (Liang et al., 2014) including STAY-GREEN (SGR, Park et al., 2007), NYC1 (Kusaba et al., 2007), and NYC3 (Morita et al., 2009). To investigate whether OsERF101 affects the expression of OsNAP and chlorophyll degradation genes, we measured their transcript levels in the detached leaves of 3-week-old WT and oserf101 seedlings during DIS. The qRT-PCR results showed that, while the transcript levels of OsNAP and chlorophyll degradation genes increased in the WT at 3 DDI, their expression was not altered in the oserf101 mutant during DIS (Figures 3E–G). These results indicate that OsERF101 acts as a positive regulator of leaf senescence by upregulating the expression of OsNAP and chlorophyll degradation genes.
OsERF101 Acts in JA-Mediated Leaf Senescence
In the WT, the expression of OsERF101 increased in response to treatment with ABA or MeJA (Figure 1B), implying that OsERF101 is positively involved in ABA- or JA-mediated leaf senescence. To assess the function of OsERF101 in JA signaling, we observed the progress of leaf yellowing in the detached leaves of 3-week-old WT and oserf101 seedlings incubated in 3 mM MES buffer (pH 5.8) containing 50 µM of ABA or MeJA under continuous light conditions. While there was no significant difference in leaf greenness and chlorophyll contents between the WT and oserf101 seedlings at 4 d of ABA treatment, the oserf101 leaves retained their green color more than WT leaves at 4 d of MeJA treatment and had higher total chlorophyll contents (Figures 4A, B). Consistent with these observations, chlorophyll contents were higher in the whole leaves of 3-week-old oserf101 seedlings than in those of the WT at 4 d of MeJA treatment (Supplementary Figure S2). In addition, we investigated the effects of MeJA on growth of WT and oserf101 seedlings grown on 1/2 MS solid medium for 10 days. MeJA treatment significantly retarded the growth of shoots and roots in WT and oserf101 seedlings (Figure 4C). However, the shoots and roots of the oserf101 mutant were much longer than those of the WT under MeJA treatment (Figures 4D, E). These results suggested that, compared with WT, the oserf101 mutant is less sensitive to MeJA.
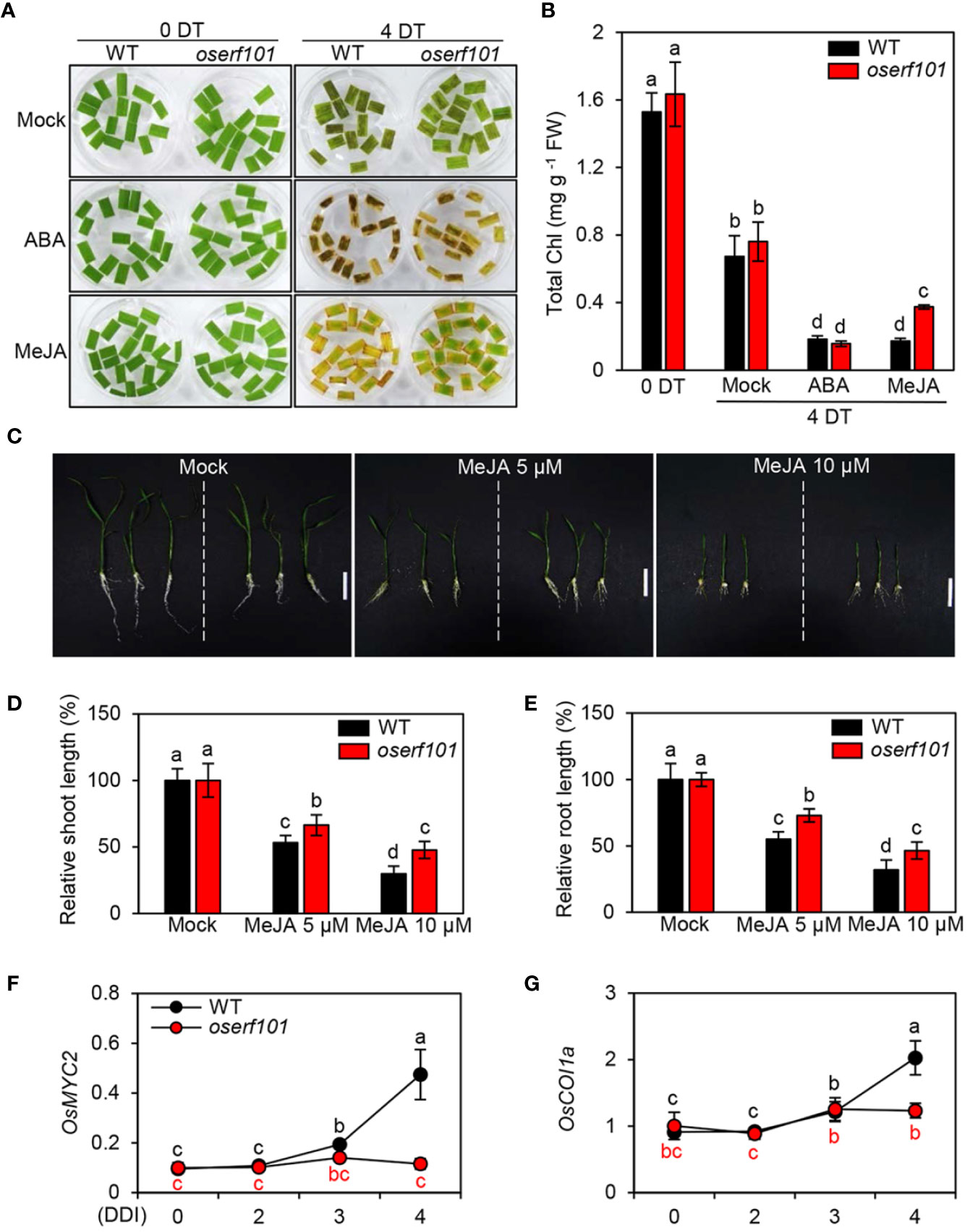
Figure 4 Jasmonic acid (JA) hyposensitivity of the oserf101 mutants. (A, B) Detached leaves of 3-week-old WT and oserf101 plants grown in the greenhouse under natural long-day conditions (>14 h light/day) were incubated in 3 mM MES buffer (pH 5.8) containing 50 µM abscisic acid (ABA) or 50 µM methyl jasmonate (MeJA) under continuous light at 28°C. Detached leaves floated on 3 mM MES buffer (pH 5.8) without phytohormones were used as a mock control. The MeJA hyposensitive phenotype (A) and total chlorophyll (Chl) contents (B) were observed at 0 and 4 days after treatment (DT). (C–E) The WT and oserf101 seeds germinated in half-strength Murashige and Skoog (1/2 MS) solid medium for 3 d were grown in 1/2 MS solid medium containing 5 or 10 µM MeJA for 10 d under long-day (LD) conditions (14 h light at 30°C/10 h dark at 25°C). Seedlings grown in 1/2 MS solid medium without MeJA were used as a mock control. The growth phenotype (C) and relative shoot (D) and root (E) length were observed (n = 10). (F, G) Altered expression of MeJA signaling genes OsMYC2 and OsCOI1a in detached leaves of 3-week-old WT and oserf101 seedlings during DIS. Total RNA was isolated from detached leaves at 0, 2, 3, and 4 DDI as shown in Figure 3A. The transcript levels of OsMYC2 (F) and OsCOI1a (G) were determined by qRT-PCR and normalized to that of OsUBQ5 (Os01g22490). Mean and standard deviations were obtained from at least three biological samples. Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (p < 0.05). FW, fresh weight.
To identify the effect of OsERF101 on JA signaling and biosynthesis during DIS, we investigated the transcript levels of JA related genes in the detached leaves of 3-week-old WT and oserf101 seedlings during DIS (Figure 3A). The expression of JA signaling genes (OsMYC2 and OsCOI1a) and JA biosynthesis genes (OsLOX2 and OsAOS1) was significantly upregulated in WT after 3 DDI, while the transcript levels were not altered in oserf101 mutant during DIS (Figures 4F, G and Supplementary Figure S3). Taken together, our results suggested that promotes the onset and progression of leaf senescence by upregulating JA signaling and biosynthesis pathways.
OsERF101 Binds to the OsNAP and OsMYC2 Promoter Regions
During DIS, the expression of OsNAP and OsMYC2 was downregulated in the oserf101 mutant compared to the WT (Figures 3E and 4F). Therefore, we used protoplast transient transactivation assays to determine if OsERF101 activates the transcription of OsNAP and OsMYC2 in planta. For the reporter construct, the promoter region of OsNAP (-1502 to -1) or OsMYC2 (-1529 to -1) was fused with the luciferase (LUC) reporter (Figure 5A). The LUC activities of the protoplasts transformed with the proOsNAP : LUC and proOsMYC2:LUC plasmids were significantly enhanced when each of them was co-transfected with the 35S:OsERF101:MYC effector plasmid (Figures 5B, C).
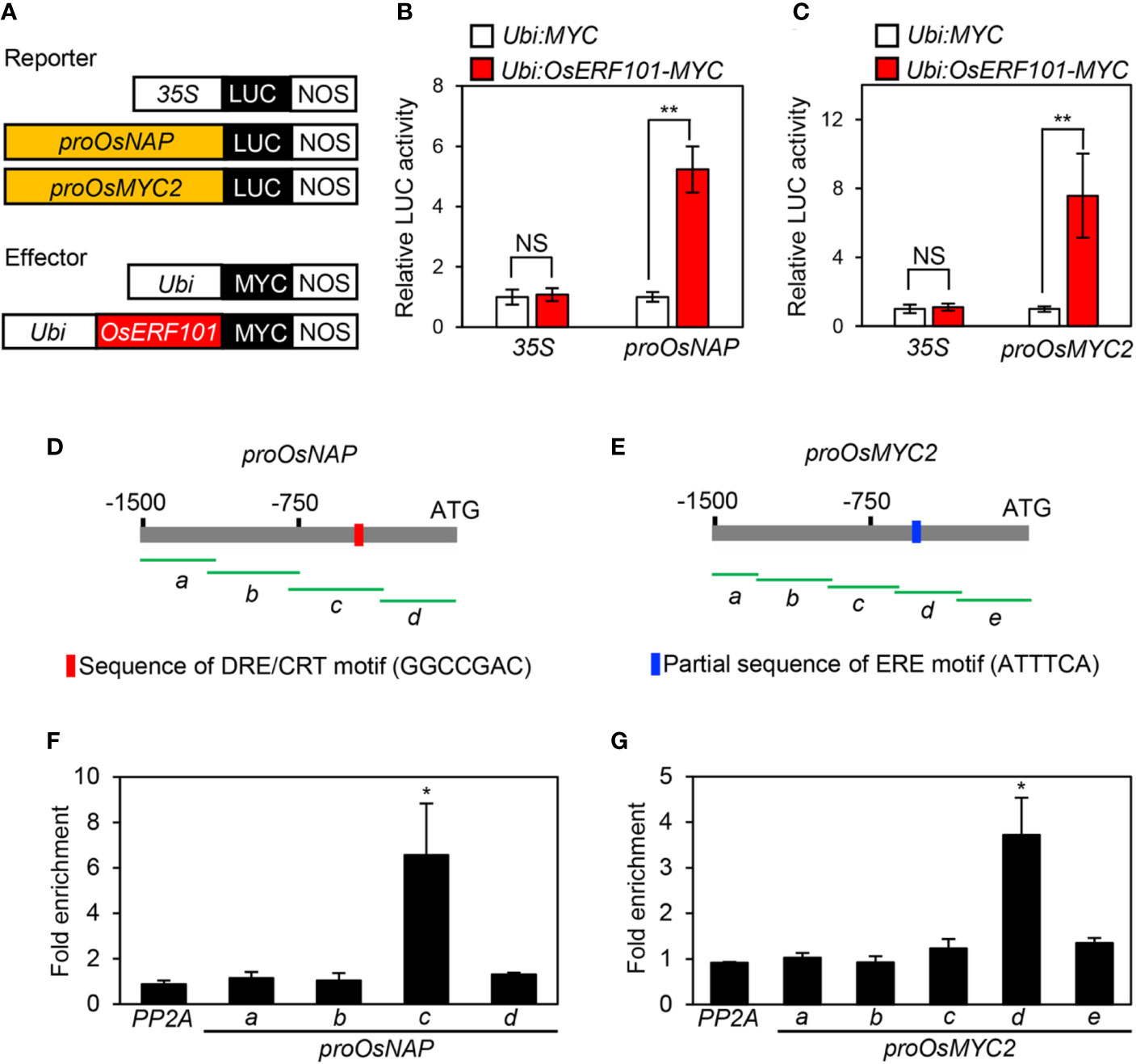
Figure 5 OsERF101 directly activates the transcription of OsNAP and OsMYC2. (A) Reporter and effector constructs used in the transactivation assay. (B, C) The activation of the OsNAP and OsMYC2 promoter (proOsNAP and proOsMYC2) by OsERF101-MYC in the protoplast transient assay. The 35S promoter was used as a negative control. (D, E) Positions of fragments used for the ChIP assay (green horizontal lines) in the promoter regions of OsNAP and OsMYC2. Red and blue bars indicate the position of the DRE/CRT and partial ERE binding motifs, respectively. (F, G) OsERF101 binding affinity assays to the promoter regions of OsNAP and OsMYC2 in planta examined by ChIP assays. OsERF101-Myc was transiently expressed in protoplasts isolated from 10-day-old WT seedlings. Fold-enrichment of the promoter fragments was measured by immunoprecipitation with an anti-Myc antibody (see Methods). The SERINE/THREONINE PROTEIN PHOSPHATASE 2A (PP2A) gene was used as a negative control. Mean and standard deviations were obtained from more than five biological repeats. Asterisks indicate a significant difference compared with the negative control (Student’s t-test, *p < 0.05, **p < 0.01).
We conducted chromatin immunoprecipitation (ChIP) assays to examine whether OsERF101 directly binds to the promoter regions of OsNAP and/or OsMYC2. The 1500-bp region upstream of the transcription start site of OsNAP or OsMYC2 was divided into four or five fragments with 50-bp overlap (Figures 5D, E). OsERF101 strongly bound to the amplicon-c region of proOsNAP, which includes the DRE/CRT motif (GGCCGAC, -447 to -440), and to the amplicon-d region of proOsMYC2, which includes a partial ERE motif (ATTTCA, -483 to -477) (Figures 5F, G). We further checked the expression levels of OsNAP and OsMYC2 in the developing leaves of WT and OsERF101-OX plants grown for one month in the paddy field under NLD conditions. The qRT-PCR analysis revealed that transcripts of OsNAP and OsMYC2 were significantly abundant in two OsERF101-OX lines than the transcript levels in the WT (Supplementary Figure S4). These results demonstrated that OsERF101 functions as an upstream activator of OsNAP and OsMYC2 by directly binding to their promoter regions.
Loss of Function of OsERF101 Negatively Affects Panicle Development and Grain Yield
In addition to exploring the molecular genetic function of OsERF101 in leaf senescence, we examined several agronomic traits in the oserf101 mutant plants grown in the paddy field under NLD conditions. We evaluated the number of panicles per plant, panicle length, number of grains per panicle, fertility, and 500-grain weight. Interestingly, although the panicle length was shorter in the oserf101 mutant than in the WT (Figure 6A), WT and oserf101 plants produced a similar number of spikelets per panicle (Figure 6B), implying that the mutation of OsERF101 led to higher spikelet density (Figure 6H). However, the number of panicles per plant, spikelet fertility, and 500-grain weight were lower in the oserf101 mutant (Figures 6C–E), resulting in reduced grain yield per plant in the mutant (Figures 6F, G). Thus, the defect of panicle development and low spikelet fertility in the oserf101 mutant appear to attenuate the advantage of high spikelet density on improving grain yield.
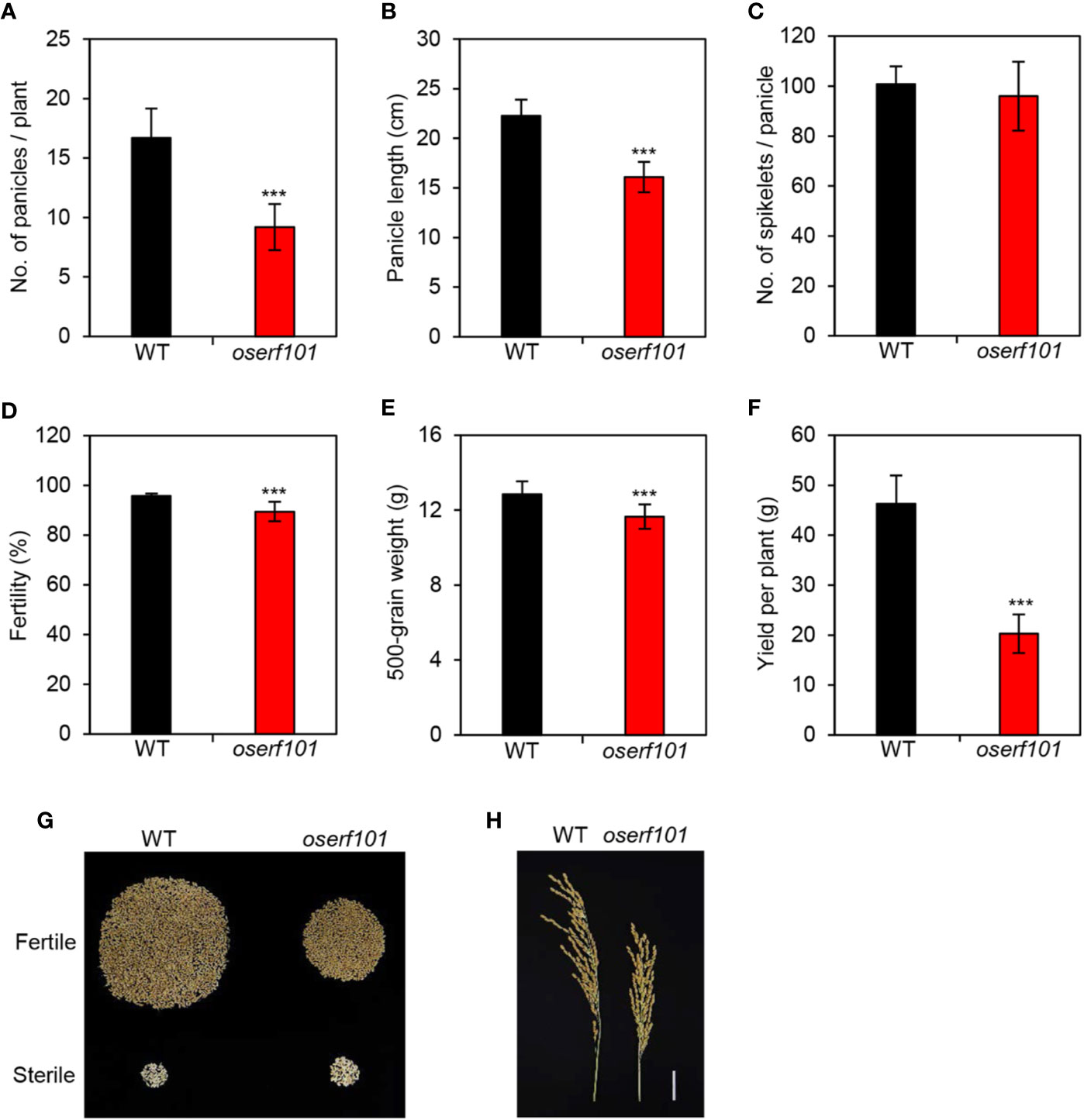
Figure 6 Agronomic traits measured in the oserf101 mutant. Agronomic traits were compared between the WT and oserf101 plants after harvest in the autumn. (A) Number of panicles per plant. (B) Panicle length. (C) Number of spikelets per panicle. (D) Fertility. (E) 500-grain weight. (F) Grain yield per plant. (G) The relative abundances of fertile and sterile spikelets in the WT and oserf101 plants. (H) Phenotype of panicles. Mean and standard deviations were obtained from 10 rice plants. Asterisks indicate a significant difference between the WT and oserf101 mutants (Student’s t-test, ***p < 0.001).
Discussion
Leaf senescence largely occurs in an age-dependent manner, but can also be triggered by abiotic stresses. The onset and progression of leaf senescence are controlled by multiple regulatory networks in response to environmental cues. Leaf senescence is accompanied by chlorophyll degradation through phytohormone signaling pathways that are driven by many plant-specific TFs (Buchanan-Wollaston, 1997; Lee et al., 2001; Kim et al., 2016). OsERF101 transcript levels were upregulated by treatment with phytohormones (ABA, MeJA), and by salt (NaCl), osmotic (PEG), and dehydration stresses (Figures 1B–D), implying that OsERF101 is involved in senescence and abiotic stress responses through mediating ABA and MeJA signaling. Overexpression of OsERF101 confers enhanced tolerance to osmotic and drought stresses at vegetative and reproductive stages, respectively (Jin et al., 2018). The finding that ABA-responsive genes such as LATE EMBRYOGENESIS ABUNDANT 3 (LEA3), PEROXIDASE 2 (POD2), and RESPONSIVE TO DEHYDRATION 22 (RD22) are upregulated in OsERF101-overexpressed transgenic rice seedlings in response to osmotic stress treatment (20% PEG) suggesting that OsERF101 participates in the ABA signaling pathway under osmotic stress conditions (Jin et al., 2018). In the present study, we found that overexpression of OsERF101 led to early leaf yellowing during DIS and natural senescence (Figure 3). The detached leaves of the oserf101 mutant were less sensitive to MeJA, but were sensitive to ABA in continuous light (Figure 4 and Supplementary Figure S2), indicating that OsERF101 mediates leaf senescence via the JA signaling pathway, but not the ABA signaling pathway. Indeed, the expression of the JA signaling-associated genes OsCOI1b and OsMYC2 was suppressed in the oserf101 mutant during DIS (Figures 4F, G). Moreover, the ChIP assay showed that OsERF101 directly binds to a partial ERF motif in the promoter region of OsMYC2 (Figures 5E, G). OsMYC2 directly regulates the expression of senescence-associated genes (SIMILAR TO SAG and OsSAG12), leading to rapid leaf yellowing in OsMYC2-OX plants (Uji et al., 2017). Together, these results support the idea that OsERF101 acts as a positive regulator of leaf senescence in response to JA.
In addition to the involvement of OsERF101 in regulation of leaf senescence via the JA signaling pathway, OsERF101 is involved in two independent senescence pathways mediated by OsNAP (Figure 7). The first is the OsNAP-mediated positive feedback loop modulating JA biosynthesis. OsERF101 activates OsNAP transcription by directly binding to the DRE/CRT motif of the OsNAP promoter region (Figures 5D, F). OsNAP is involved in various regulatory pathways regulating abiotic stress responses and leaf senescence. Here, we discussed the regulatory function of OsNAP in JA-regulated leaf senescence. Overexpression of OsNAP led to an increased level of expression of JA biosynthesis genes including OsLOX2 and OsAOC, thereby upregulating endogenous JA levels in the transgenic plants (Zhou et al., 2013). Thus, the downregulation of JA biosynthesis genes (OsLOX2 and OsAOS1) in the oserf101 mutant during DIS suggests that OsERF101 participates in JA biosynthesis (Supplementary Figure S3). Finally, increased levels of JA can enhance the transcription of OsERF101, and consequently activate the JA–OsERF101–OsNAP positive feedback regulatory loop to promote leaf senescence. The second regulatory function of OsNAP is the OsNAP-dependent chlorophyll degradation pathway. In addition, OsNAP is required for the induction of chlorophyll degradation genes including SGR, NYC1, and NYC3 (Liang et al., 2014). Our observations indicated that the oserf101 leaves retained their green color much longer than the WT due to the lower expression of OsNAP and chlorophyll degradation genes during DIS, indicating that OsERF101 upregulates OsNAP-induced chlorophyll degradation.
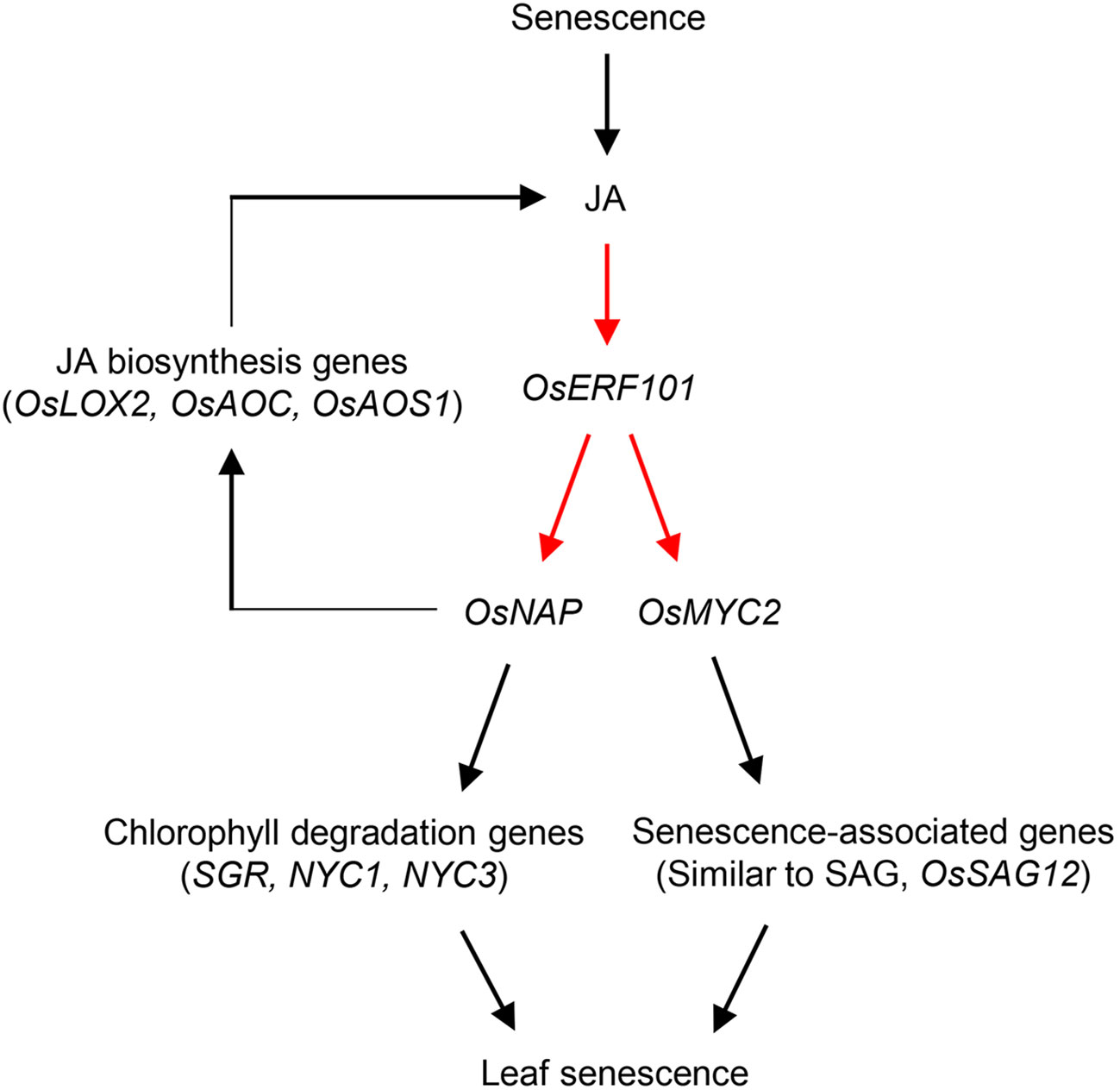
Figure 7 Proposed model of the role of OsERF101 in the regulation in leaf senescence through JA signaling pathways involving OsNAP and OsMYC2. Red arrows indicate our findings in this study, and black arrows represent pathways identified in previous studies (Zhou et al., 2013; Liang et al., 2014; Uji et al., 2017).
In higher plants, endogenous JA levels and aqrJA signal transduction play important roles in shoot growth, lateral and adventitious root formation, seed germination, and embryo and pollen development (Ueda and Kato, 1980; McConn et al., 1997; Kang et al., 2010; Dave et al., 2011; Valenzuela et al., 2016). For instance, inhibition of JA signaling by knockdown of OsCOI1a and OsCOI1b expression led to increases in plant height, internode length, and cell length (Yang et al., 2012). Mutation of OsMADS1 causes a defect in spikelet development due to a lack of glume formation (Chen et al., 2006). OsJAZ1 interacts with the OsMYC2 TF to suppress its activity, which leads to downregulation of the expression of OsMADS1 (Cai et al., 2014). Genetic studies have indicated that the gain-of-function mutant of OsJAZ1/EG2, eg2-1D, exhibits downregulated expression of OsMADS1, resulting in low spikelet fertility. Thus, the poor spikelet fertility of the oserf101 mutant appears to be caused by inhibition of JA signaling due to downregulation of OsCOI1b and OsMYC2, ultimately leading to lower grain yield (Figures 6D, F).
Taken together, we concluded that OsERF101 affects multiple aspects of plant development, including leaf senescence and grain yield through regulating OsNAP and JA signaling. These results provide insight into the multiple regulatory mechanisms of OsERF101 in leaf senescence and spikelet development in rice.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
KK and N-CP designed experiments. CL, YSh, and YSa performed experiments and analyzed data. GA developed plant materials. CL, KK, YSh, and N-CP wrote the article.
Funding
This work was carried out with the support of the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (NRF-2017R1A2B3003310 to N-CP and NRF-2019R1I1A1A01060494 to KK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01096/full#supplementary-material
References
Buchanan-Wollaston, V. (1997). The molecular biology of leaf senescence. J. Exp. Bot. 48, 181–199. doi: 10.1093/jxb/48.2.181
Cai, Q., Yuan, Z., Chen, M., Yin, C., Luo, Z., Zhao, X., et al. (2014). Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 3476. doi: 10.1038/ncomms4476
Chen, Z.-X., Wu, J.-G., Ding, W.-N., Chen, H.-M., Wu, P., Shi, C.-H. (2006). Morphogenesis and molecular basis on naked seed rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223, 882–890. doi: 10.1007/s00425-005-0141-8
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55, 604–619. doi: 10.1093/pcp/pct204
Dave, A., Hernández, M. L., He, Z., Andriotis, V. M. E., Vaistij, F. E., Larson, T. R., et al. (2011). 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23, 583–599. doi: 10.1105/tpc.110.081489
Distelfeld, A., Avni, R., Fischer, A. M. (2014). Senescence, nutrient remobilization, and yield in wheat and barley. J. Exp. Bot. 65, 3783–3798. doi: 10.1093/jxb/ert477
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J.-M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Fukao, T., Yeung, E., Bailey-Serres, J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 23, 412–427. doi: 10.1105/tpc.110.080325
Gao, T., Li, G.-Z., Wang, C.-R., Dong, J., Yuan, S.-S., Wang, Y.-H., et al. (2018). Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency. Int. J. Mol. Sci. 19:1465. doi: 10.3390/ijms19051465
Gepstein, S., Glick, B. R. (2013). Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 82, 623–633. doi: 10.1007/s11103-013-0038-z
He, Y., Fukushige, H., Hildebrand, D. F., Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. doi: 10.1104/pp.010843
Hou, Q., Ufer, G., Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress: Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 39, 1029–1048. doi: 10.1111/pce.12666
Jeon, J.-S., Lee, S., Jung, K.-H., Jun, S.-H., Jeong, D.-H., Lee, J., et al. (2000). T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570. doi: 10.1046/j.1365-313x.2000.00767.x
Jeong, D.-H., An, S., Kang, H.-G., Moon, S., Han, J.-J., Park, S., et al. (2002). T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 130, 1636–1644. doi: 10.1104/pp.014357
Jin, Y., Pan, W., Zheng, X., Cheng, X., Liu, M., Ma, H., et al. (2018). OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol. Biol. 98, 51–65. doi: 10.1007/s11103-018-0762-5
Kang, J.-H., Liu, G., Shi, F., Jones, A. D., Beaudry, R. M., Howe, G. A. (2010). The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 154, 262–272. doi: 10.1104/pp.110.160192
Kim, H. J., Nam, H. G., Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., Fujimoto, M., et al. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Lee, R., Wang, C., Huang, L., Chen, S. G. (2001). Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 52, 1117–1121. doi: 10.1093/jexbot/52.358.1117
Lee, S.-H., Sakuraba, Y., Lee, T., Kim, K.-W., An, G., Lee, H. Y., et al. (2015). Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b ) delays leaf senescence: OsCOI1b in leaf senescence. J. Integr. Plant Biol. 57, 562–576. doi: 10.1111/jipb.12276
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U. S. A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9, e1003790. doi: 10.1371/journal.pgen.1003790
Luehrsen, K. R., de Wet, J. R., Walbot, V. (1992). Transient expression analysis in plants using firefly luciferase reporter gene. Methods Enzymol. 216, 397–414. doi: 10.1016/0076-6879(92)16037-k
Mao, C., Lu, S., Lv, B., Zhang, B., Shen, J., He, J., et al. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174, 1747–1763. doi: 10.1104/pp.17.00542
Masclaux-Daubresse, C., Reisdorf-Cren, M., Orsel, M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. 10, 23–36. doi: 10.1111/j.1438-8677.2008.00097.x
McConn, M., Creelman, R. A., Bell, E., Mullet, J. E., Browse, J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 94, 5473–5477. doi: 10.1073/pnas.94.10.5473
Mickelbart, M. V., Hasegawa, P. M., Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Morita, R., Sato, Y., Masuda, Y., Nishimura, M., Kusaba, M. (2009). Defect in Non-Yellow Coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 59, 940–952. doi: 10.1111/j.1365-313X.2009.03919.x
Munné-Bosch, S., Alegre, L. (2004). Die and let live: leaf senescence contributes to plant survival under drought stress. Funct. Plant Biol. 31, 203. doi: 10.1071/FP03236
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Niu, Y., Figueroa, P., Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154. doi: 10.1093/jxb/erq408
Park, J. M., Park, C.-J., Lee, S.-B., Ham, B.-K., Shin, R., Paek, K.-H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Park, S.-Y., Yu, J.-W., Park, J.-S., Li, J., Yoo, S.-C., Lee, N.-Y., et al. (2007). The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19, 1649–1664. doi: 10.1105/tpc.106.044891
Phukan, U. J., Jeena, G. S., Tripathi, V., Shukla, R. K. (2017). Regulation of Apetala2/Ethylene Response Factors in plants. Front. Plant Sci. 8:150. doi: 10.3389/fpls.2017.00150
Porra, R. J., Thompson, W. A., Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta (BBA) - Bioenerg. 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Qi, T., Huang, H., Wu, D., Yan, J., Qi, Y., Song, S., et al. (2014). Arabidopsis DELLA and JAZ proteins bind the WD-Repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26, 1118–1133. doi: 10.1105/tpc.113.121731
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sakuraba, Y., Piao, W., Lim, J.-H., Han, S.-H., Kim, Y.-S., An, G., et al. (2015). Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56, 2325–2339. doi: 10.1093/pcp/pcv144
Saleh, A., Alvarez-Venegas, R., Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Shen, J., Lv, B., Luo, L., He, J., Mao, C., Xi, D., et al. (2017). The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 7:40641. doi: 10.1038/srep40641
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. doi: 10.1105/tpc.111.083089
Ueda, J., Kato, J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66, 246–249. doi: 10.1104/pp.66.2.246
Uji, Y., Akimitsu, K., Gomi, K. (2017). Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 245, 1241–1246. doi: 10.1007/s00425-017-2697-5
Valenzuela, C. E., Acevedo-Acevedo, O., Miranda, G. S., Vergara-Barros, P., Holuigue, L., Figueroa, C. R., et al. (2016). Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 67, 4209–4220. doi: 10.1093/jxb/erw202
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Yang, D.-L., Yao, J., Mei, C.-S., Tong, X.-H., Zeng, L.-J., Li, Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U. S. A. 109, E1192–E1200. doi: 10.1073/pnas.1201616109
Yang, Y. N., Safarova, R. B., Park, S.-Y., Sakuraba, Y., Oh, M.-H., Zulfugarov, I. S., et al. (2019). Chlorophyll Degradation and Light-harvesting Complex II Aggregate Formation During Dark-induced Leaf Senescence in Arabidopsis Pheophytinase Mutants. J. Plant Biol. 62, 27–38. doi: 10.1007/s12374-018-0242-0
Yoo, S.-D., Cho, Y.-H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Zhou, Y., Huang, W., Liu, L., Chen, T., Zhou, F., Lin, Y. (2013). Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 13:132. doi: 10.1186/1471-2229-13-132
Zhu, X., Chen, J., Xie, Z., Gao, J., Ren, G., Gao, S., et al. (2015). Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84, 597–610. doi: 10.1111/tpj.13030
Keywords: rice, leaf senescence, chlorophyll degradation, jasmonic acid signaling, OsERF101, OsNAP, OsMYC2
Citation: Lim C, Kang K, Shim Y, Sakuraba Y, An G and Paek N-C (2020) Rice ETHYLENE RESPONSE FACTOR 101 Promotes Leaf Senescence Through Jasmonic Acid-Mediated Regulation of OsNAP and OsMYC2. Front. Plant Sci. 11:1096. doi: 10.3389/fpls.2020.01096
Received: 05 May 2020; Accepted: 03 July 2020;
Published: 16 July 2020.
Edited by:
Randy D. Allen, Oklahoma State University, United StatesCopyright © 2020 Lim, Kang, Shim, Sakuraba, An and Paek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nam-Chon Paek, bmNwYWVrQHNudS5hYy5rcg==
†ORCID: Nam-Chon Paek orcid.org/0000-0002-9827-2287
‡These authors have contributed equally to this work
 Chaemyeong Lim
Chaemyeong Lim Kiyoon Kang
Kiyoon Kang Yejin Shim
Yejin Shim Yasuhito Sakuraba3
Yasuhito Sakuraba3 Gynheung An
Gynheung An Nam-Chon Paek
Nam-Chon Paek