
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 July 2020
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01041
This article is part of the Research Topic Nitric Oxide in Plants View all 13 articles
 Rizwana Begum Syed Nabi1,2†
Rizwana Begum Syed Nabi1,2† Rupesh Tayade3†
Rupesh Tayade3† Qari Muhammad Imran1†
Qari Muhammad Imran1† Adil Hussain4
Adil Hussain4 Muhammad Shahid1
Muhammad Shahid1 Byung-Wook Yun1*
Byung-Wook Yun1*Advances in next-generation sequencing technologies facilitate the study of plant molecular functions in detail and with precision. Plant genome and proteome databases are continually being updated with large transcriptomic or genomic datasets. With the ever-increasing amount of sequencing data, several thousands of genes or proteins in public databases remain uncharacterized, and their domain functions are largely unknown. Such proteins contain domains of unknown function (DUF). In the present study, we identified 231 upregulated and 206 downregulated DUF genes from the available RNA-Seq-based transcriptome profiling datasets of Arabidopsis leaves exposed to a nitric oxide donor, S-nitroso-L-cysteine (CysNO). In addition, we performed extensive in silico and biological experiments to determine the potential functions of AtDUF569 and to elucidate its role in plant growth, development, and defense. We validated the expression pattern of the most upregulated and the most downregulated DUF genes from the transcriptomic data. In addition, a loss-of AtDUF569 function mutant was evaluated for growth, development, and defense against biotic and abiotic stresses. According to the results of the study, AtDUF569 negatively regulates biotic stress responses and differentially regulates plant growth under nitro-oxidative stress conditions.
Nitric oxide (NO) is a gaseous and highly reactive free radical involved as a signaling molecule in vital physiological processes in both animal and plant cells. Over the past few decades, various functional aspects of NO in animal and plant cell biology have been explored and described. NO in plants has increasingly emerged as an essential molecule involved in diverse plant functions, such as seed germination, growth, and development, plant defense, iron homeostasis, biotic and abiotic stresses, senescence, and cell death (reviewed by Domingos et al., 2015; Nabi et al., 2019) signaling pathways of indole acetic acid, abscisic acid (ABA), and other plant hormones (García-Mata and Lamattina, 2002; Pagnussat et al., 2003). Despite the importance and involvement of NO in diverse plant functions, its origin and mechanism of production in higher plants remain largely elusive.
One of the key roles of NO in the regulation of protein function is through posttranslational modifications such as S-nitrosation (or S-nitrosylation) and tyrosine nitration (Stamler et al., 1992; Greenacre and Ischiropoulos, 2001). Chief among them is S-nitrosation, the covalent attachment of NO to exposed cysteine residues of proteins to form S-nitrosothiols (Stamler et al., 1992). Such protein modification has been reported to play critical roles in cellular processes, enzyme pathways, protein–protein interactions, and protein localization (Tada, 2008; Wang et al., 2009; Yun et al., 2011).
In the current genomic era, advances in sequencing technologies are generating vast quantities of data at the transcriptomic and genomic levels. Although the genomes of several mammals, higher plants, and microbial species have been sequenced and annotated, several genes remain uncharacterized, and their biochemical and biological functions, in turn, remain unknown. Such uncharacterized proteins are deemed as proteins having domains of unknown function (DUF) and have been categorized in the Pfam database as a protein family with no functional annotation (Bateman et al., 2010; Finn et al., 2016; El-Gebali et al., 2019). Despite extensive research, DUF is one of the largest families in the Pfam database, representing more than 22% of the entire domains in the Pfam database, with approximately 4,000 DUFs that are poorly understood (Bateman et al., 2010; Finn et al., 2016).
Studies suggest that DUF domain-containing proteins play a vital role in plant stress responses. For instance, in wheat, the DUF622 domain-containing TaSRG (Triticum aestivum salt response gene) increased salt tolerance in overexpressed transgenic Arabidopsis and influenced gene expression levels under salt stress in rice (He et al., 2011). Similarly, in Arabidopsis, ABA and drought-induced ubiquitin ligase genes, AtRDUF1 and AtRDUF2 (RING-DUF1117E3), regulate ABA signaling and drought stress, while single and double knockout (KO) mutants of AtRDUF1 and AtRDUF2 exhibited reduced tolerance to ABA-mediated drought stress when compared with the wild type (WT) plants (Kim and Kim, 2013). Similarly, in another study on rice plant, Cui et al. (2016) reported that DUF1645 was upregulated in response to various stress factors and conferred improved drought tolerance when overexpressed.
Furthermore, negative regulation of DUF genes has also been reported. For example, in rice, the DUF966 domain-containing OsDSR2 encodes a protein that negatively regulates responses to salt and simulated drought stress, along with ABA signaling (Luo et al., 2014). Recently, the functions and expression levels of various members of the rice OsDUF866 family (OsDUF866.1–4) were analyzed (Li et al., 2017). Reports suggested that OsDUF866.1 positively regulated heat stress tolerance, while OsDUF866.2 expression decreased discernibly following exposure to drought conditions. OsDUF866.3 was highly expressed in response to drought and cold stress and decreased under heat stress, while OsDUF866.4 expression was upregulated under cold and heat stress and decreased under drought stress. In addition, the roles of other DUF genes in rice, such as OsDUF872, OsDUF829, and OsDUF946, have been described (Li et al., 2017; Li et al., 2018a; Li et al., 2018b).
Microarray and RNA-seq mediated transcriptome analyses have made it convenient to predict the global changes in gene expression in a particular genotype. A number of transcriptome analyses in response to various stress conditions have been conducted including H2O2-mediated oxidative stress (Desikan et al., 2001), ozone (Xu et al., 2015) cold, drought and oxidative stress (Sham et al., 2014). Nitrosative stress-induced changes in transcriptional regulation have been studied extensively. Microarray or RNA-Seq profiling has revealed several key genes with differential expression in response to 1 mM S-nitrosoglutathione (GSNO) and 0.1 and 1.0 mM sodium nitroprusside (SNP) in Arabidopsis (Parani et al., 2004; Begara-Morales et al., 2014). At the same time, such transcriptome data revealed several genes with unknown functions, which have exhibited differential expression in response to nitrosative stress. In our previous study involving Arabidopsis leaves, 1 mM S-nitroso-L-cysteine (CysNO)-mediated RNA-Seq-based transcriptome analysis revealed the differential expression of several thousand of genes (Hussain et al., 2016). In the present study, we identified approximately 440 DUF genes that exhibited differential responses to CysNO, including 235 upregulated and 205 downregulated genes in Arabidopsis thaliana, using high-throughput RNA-Seq data. We subsequently characterized them by in silico analysis to decipher any putative roles of the genes. In addition, we also studied the biological role of AtDUF569 in plant growth, development, and defense, using a functional genomics study. Our aim was to identify and characterize NO-induced genes encoding DUFs, with an emphasis on AtDUF569.
In a previous study, we reported approximately 6436 differentially expressed genes (DEGs), among which 440 were identified as containing DUFs (Hussain et al., 2016). The DUF name is assigned based on the functional annotation information. When the function of the protein has not been determined, it is deposited in the Pfam database (El-Gebali et al., 2019). Such proteins/genes are commonly termed DUFs (Bateman et al., 2010; Punta et al., 2012). All the 440 DUF domain containing DEGs exhibited significant (p ≤ 0.05) differential expression in response to 1 mM CysNO. The list of the DUF domain-containing DEGs was examined carefully for any duplicate values. To visualize the differential expression of the DUFs between the CysNO-treated and the untreated (control) samples, a heatmap with hierarchical clustering presented as a dendrogram was generated using R version 3.3.1.R (https://www.r-project.org/) using the fragments per kilobase of transcript per million mapped read (FPKM) values.
To validate the RNA-Seq data of the upregulated and downregulated DEGs qRT-PCR was performed as described previously (Imran et al., 2018a). Briefly, total RNA was extracted from A. thaliana leaf samples 6 h after 1 mM CysNO infiltration. The cDNA was synthesized using Biofact RT kit (BioFACT™, Korea) according to the manufacturer’s standard protocol. A two-step qRT-PCR reaction was performed using 2× Real-Time PCR Master mix (including SYBR Green I; BioFACT™) in an Illumina Eco Real-Time PCR system (Illumina™, USA). The PCR conditions were initial denaturation at 95°C for 15 min, followed by 40 cycles at 95°C for 20 s and 60°C for 40 s. The fold change in transcript accumulation of the selected genes was compared with that of the RNA-Seq data. Pearson’s correlation coefficient (R) for both data sets was calculated using MS Excel 2016 (Microsoft Corp., Redmond, WA, USA). The primers used in the present study are listed in Table S1.
To determine the NO-induced DUF genes, we used the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html) to find the corresponding pathways and biological information. The CysNO-induced DUF domain-containing DEGs were analyzed for associated Gene Ontology (GO) terms that describe biological processes, molecular functions, and cellular components through the PANTHER v14.1 (http://pantherdb.org) web interface, using the applicable search field and A. thaliana selected as the organism. The “statistical overrepresentation test” was selected, and the rest of the analyses were performed using default settings. Only the GO terms with significant (p < 0.05) fold enrichment were selected, and pie charts were constructed for visualization using MS Excel 2016.
To forecast the putative regulatory role of NO-responsive DUF genes, we obtained the promoter sequences 1 kb upstream of the transcription initiation site of the selected NO-responsive Arabidopsis DUF genes from The Arabidopsis Information Resource Center (TAIR) (https://www.arabidopsis.org/). The retrieved sequences were analyzed for potential cis-regulatory elements through the web-based MatInspector tool (https://omictools.com/matinspector-tool) with default settings. Only the transcription factor (TF) binding sites vital in plant regulatory responses to biotic and abiotic stresses were selected for visualization.
To examine the evolutionary relationship of the NO-responsive DUF DEGs, a list of 18 DUF DEGs including 10 upregulated and 8 downregulated (two downregulated genes were transposons) was used as query in Phytozome against rice (Oryza sativa), soybean (Glycine max), wheat (Triticum aestivum), maize (Zea mays) and tomato (Solanum lycopersicum). The protein sequences of these orthologs were retrieved from Phytozome web interface (https://phytozome.jgi.doe.gov/phytomine/begin.do) and aligned using ClustalW in Mega 7 (Kumar et al., 2016), the resultant alignment was used to make phylogenetic tree using maximum likelihood based on JTT matrix-based model (Jones et al., 1992) with bootstrap value of 1,000 replicates. All of the positions with less than 95% site coverage were eliminated; that is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. MEGA 7.0 (Kumar et al., 2016) was used for the analysis.
The seeds of WT A. thaliana (ecotype Col-0) and the mutant line atduf569 (AT1G69890) were obtained from the Nottingham Arabidopsis Stock Centre (NASC; http://arabidopsis.info/). In addition, atgsnor1-3 knockout mutant was used as disease susceptible control due to its established role in plant immunity (Feechan et al., 2005). Similarly, the atsid2 mutant deficient in the SA pathway was used as a control for SA-mediated defense pathway, and atcat2, a high H2O2-producing mutant deficient in AtCATALASE2 gene (AT1G74710; Hu et al., 2010) was used as susceptible controls for the nitro-oxidative stress assay and biotic stress-related experiments. Plants homozygous for T-DNA insertion were identified through genotyping using PCR with the T-DNA left border primers and gene-specific primers obtained from Signal Salk (http://signal.salk.edu/tdnaprimers.2.html; Figure S1, Table S1). The PCR conditions were an initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 58°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min. The confirmed homozygous mutant line was used for experimental biological evaluation after collecting the seeds in bulk. All the seeds were surface-sterilized using 50% bleach (commercial bleach) with 1% (v/v) Triton X-100 added, for about 1–2 min. After rinsing five times, the sterilized seeds were incubated in sterile water and stratified overnight at 4°C for uniform germination before sowing either on half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) or soil.
The responses of AtDUF569 (AT1G69890) to nitro-oxidative stress in plants were evaluated as described by Imran et al. (2016). Briefly, for nitro-oxidative stress conditions, Col-0 (WT), atgsnor1-3 (AT5G43940), atcat2 (AT1G74710), and atduf569 (AT1G69890) seeds were sterilized (See section Plant material and growth conditions) and grown on half-strength MS medium supplemented with 0.75 mM CysNO or GSNO as NO donors and 2 mM H2O2 or 1 µM MV as oxidative stress inducers, separately. The control and the treated plants were grown under 16/8 h light/dark conditions at 23 ± 2°C. The experiments were performed in triplicate and cotyledon development frequency (CDF) was measured after seven days, as described previously (Imran et al., 2018b). In addition, root and shoot lengths were measured 14 days after sowing.
Plants were subjected to pathogen inoculation as described previously (Yun et al., 2011). Briefly, Pseudomonas syringae pv. tomato virulent (Pst DC3000) and avirulent strains (Pst DC30000 avrB) were used for the pathogenicity assays. Bacterial cultures were grown and maintained as described previously (Whalen et al., 1991). Briefly, a single colony was cultured in Luria–Bertani (LB medium) broth with respective antibiotics added and incubated at 28°C overnight with shaking. The bacterial cultures (1 ml each) were centrifuged at 8,000 rpm (LABOGene 1736R) for 3 min. The bacterial pellet was resuspended in 1 ml of 10 mM MgCl2 solution. The prepared bacterial inoculum was then infiltrated into the abaxial side of the leaves via a needleless syringe at 5 × 105 CFU ml−1 for virulent Pst DC3000 and 0.002 at OD600 (1 × 106 CFU ml−1) for avirulent Pst DC30000 avrB. Mock plants were infiltrated with the 10 mM MgCl2 solution without bacteria. Leaf samples were collected over time for further gene expression analyses. In addition, leaf discs from Pst DC3000-inoculated leaves were crushed in 10 mM MgCl2 and diluted 10 times. To identify the responses of atduf569 and control genotypes to infection, the diluted samples were spread on LB agar plates containing the respective antibiotics. The development of symptoms was observed, and photos were captured using a digital camera (EOS 700D, Canon). Symptom development and pathogen growth in plant tissues were assessed 1, 2, and 3 d post-inoculation (dpi) by counting the bacterial CFU per leaf disc using the serial dilution method as described by Flors et al. (2008), and disease symptoms on the inoculated leaves were also observed.
Cell death induced by pathogen infection was assessed through electrolyte leakage (Arasimowicz-Jelonek et al., 2009). Briefly, triplicate leaf discs (about 1 cm2) with three leaf discs per replicate were collected from the plants inoculated with the avirulent bacterial pathogen (Pst DC3000 avrB) at an OD600 of 0.02 corresponding to 1 × 106 CFU ml−1. The leaf discs were washed three times with deionized water and maintained in approximately 300 µl of deionized water/well in a six-well plate (SPL Life Sciences, Korea). Electrolyte leakage of each sample was recorded over several time points using a portable conductivity meter (Huriba Twin Cond B-173).
The data were analyzed at p ≤ 0.05 or p ≤ 0.01 significance levels. A two-tailed t-test was performed to determine p-values using MS Excel 2016. The means and standard errors were calculated from at least three biological replicates and compared with the control.
To identify NO-responsive DUF domain-containing genes, we analyzed CysNO-induced DEGs reported previously (Hussain et al., 2016). All the RNA-seq data can be found in the public repository for Gene Expression Omnibus (GEO) and Short Read Achieve (SRA) under accession numbers GSE81361 and SRP074890, respectively. Notably, a considerable number (437 DEGs) of NO-responsive DUF domain-containing genes were differentially expressed in response to 1 mM CysNO (Supplementary Table S2). A list of top 10 up- and down-regulated DEGs is given in Table 1. We analyzed all the NO-responsive DUFs for associating Gene Ontology (GO) terms. GO terms for biological process suggested that NO responsive DUFs were associated with chloroplast organization, pattern specification process, cellular metabolic process and gene expression etc. (Figure 1A). In GO terms for molecular functions majority of DUFs were associated with Iron ion transmembrane transporter activity (Figure 1B). In GO terms of cellular component majority of DUFs were associated with Golgi apparatus, Golgi subcomponent, and Endosomes etc. (Figure 1C). A heatmap with hierarchical clustering between the control and the CysNO-treated samples was constructed from the FPKM values in triplicates and shows the expression patterns of the DUF domain-containing DEGs (Figure 2A). Samples 1 to 3 represent three replicates from control (buffer only) while samples 4 to 6 represent three samples from CySNO treated plants. The red color in the treated samples indicates upregulated genes, while the intensity of the color represents the intensity of expression. The more the red color, the more expression after CySNO treatment (Figure 2A). Some samples with black color show very low expression approaching to zero (Figure 2A). Among the 437 DUF domain-containing DEGs, 53% (236 DEGs) were upregulated, while 47% (205 DEGs) were downregulated, with at least a two-fold change in their expression relative to the control plants (Figure 2B, Supplementary Table S2). Furthermore, we evaluated the role of the NO-induced DUF569 (AT1G69890) in plant growth and defense against biotic and abiotic stresses. AtDUF569 had a fold change of 57.29 in the transcriptome profile.
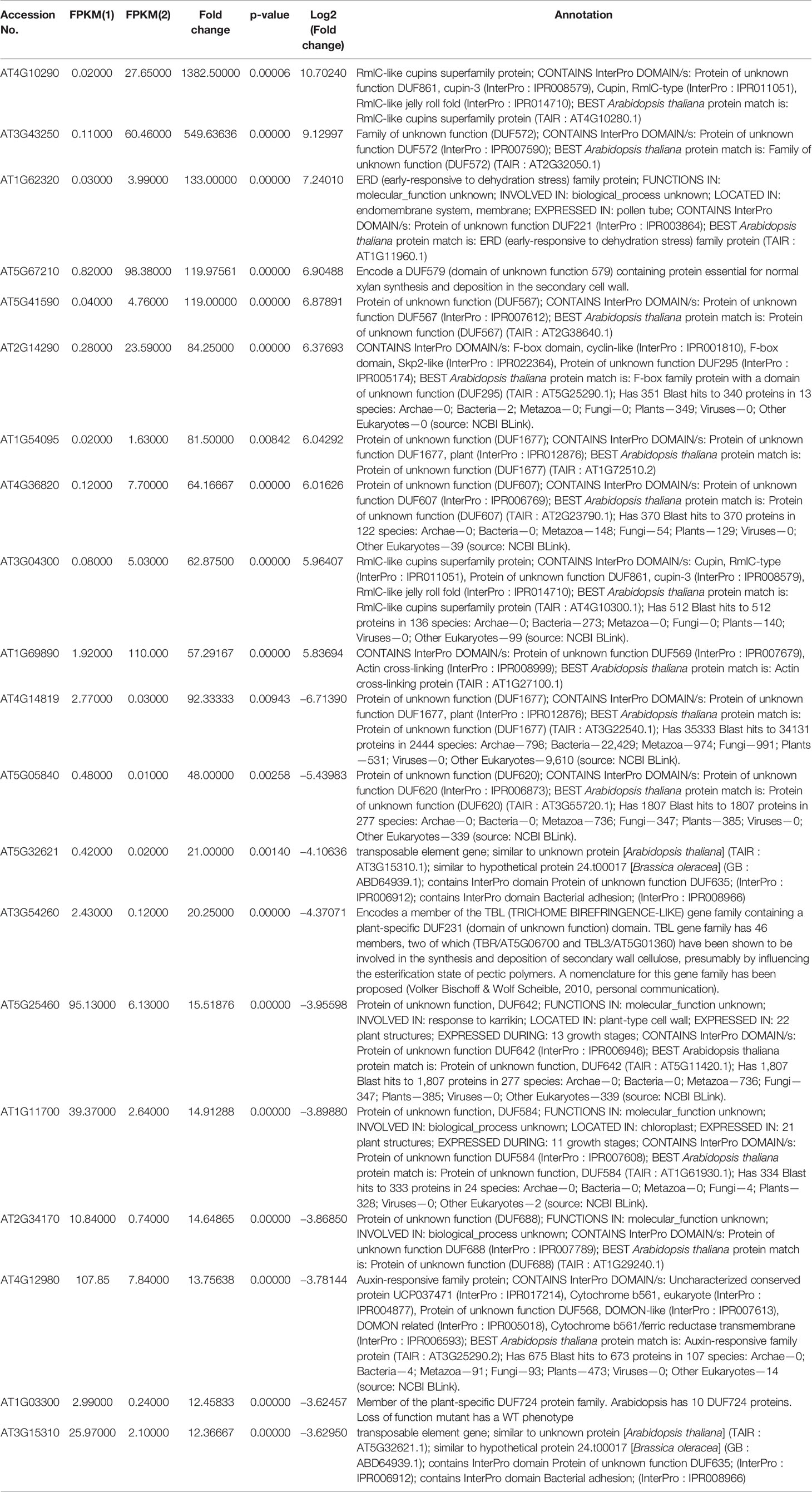
Table 1 List of top 10 up- and downregulated NO-responsive DUF domain containing genes with their fold change as per transcriptomic analysis.
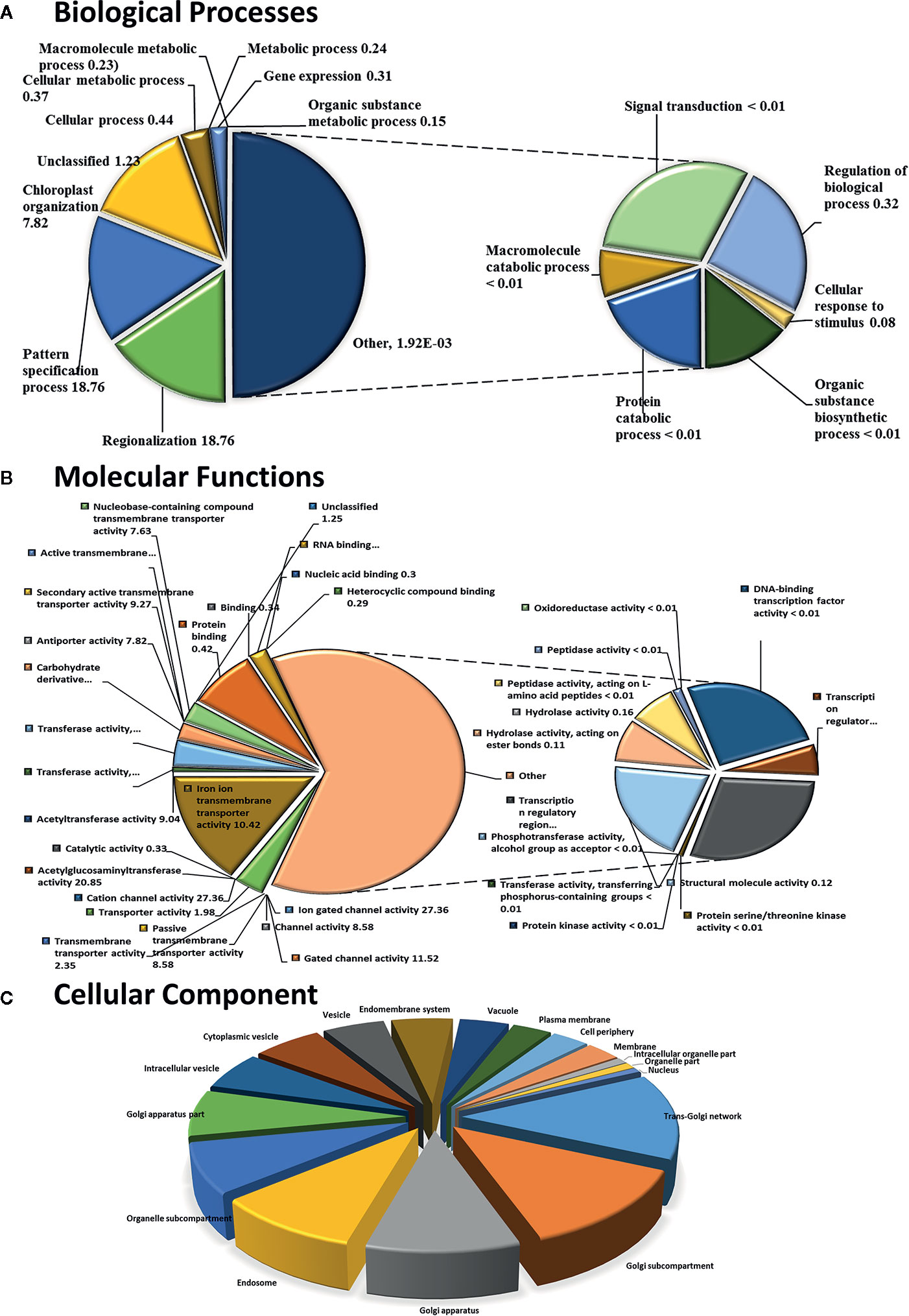
Figure 1 Functional GO term classification of CysNO-responsive DUF genes of Arabidopsis leaf transcripts. Gene ontologies for (A) Biological Processes (B) Molecular Functions and (C) Cellular Process were determined using Arabidopsis thaliana as a reference genome. Out of the 443 CySNO responsive genes, 440 were mapped with the reference genome, PANTHER version 14.1 (http://pantherdb.org) at P < 0.05 for the PANTHER GO-slim and the fold enrichment-values shown.
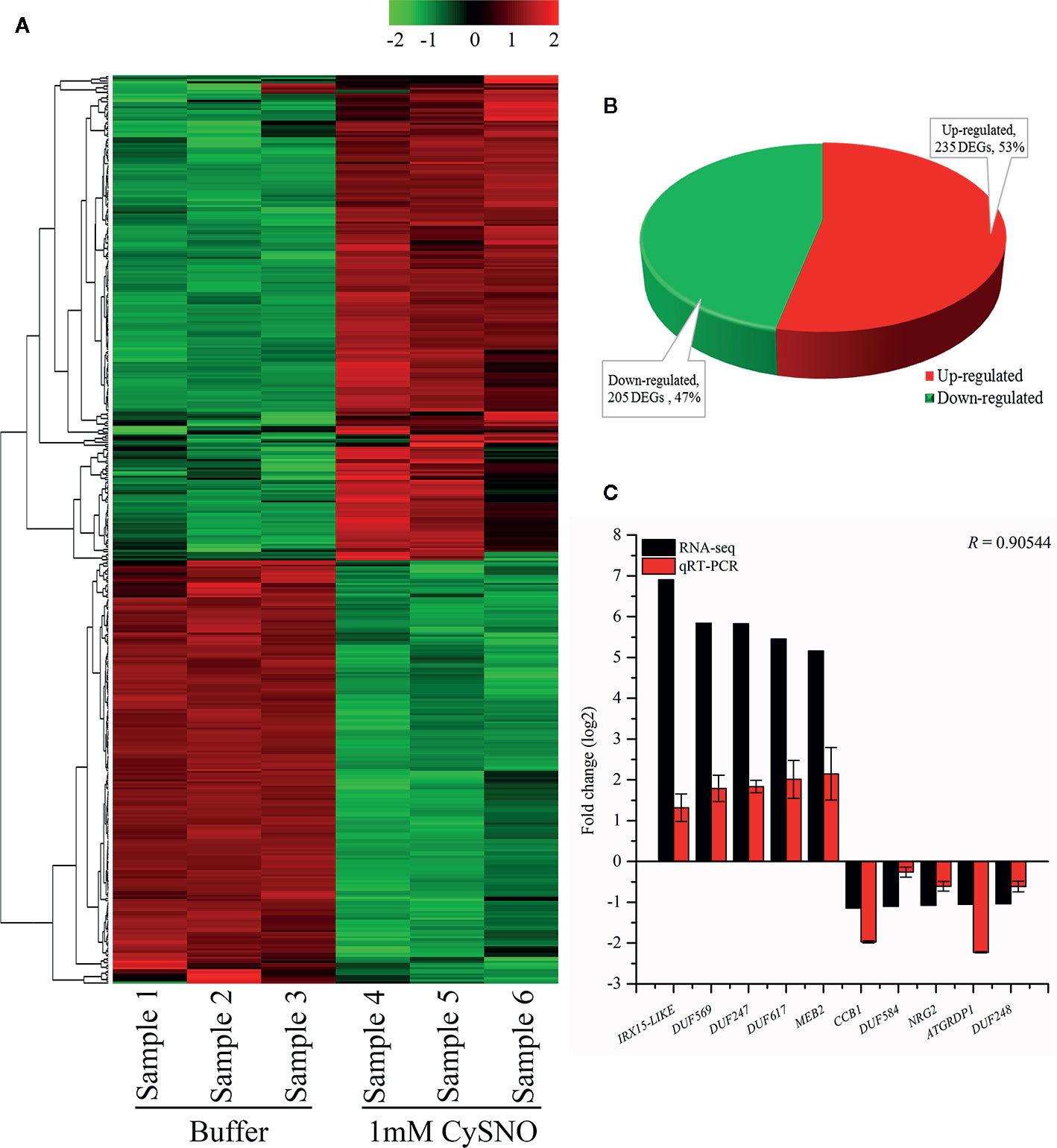
Figure 2 Identification and analysis of NO-responsive DUF domain-containing genes. (A) Heatmap revealing the expression pattern of transcriptome-wide, DEGs in response to S-nitroso L cysteine (CySNO) and hierarchical clustering generated from the FPKM values of CysNO-responsive DUF domain-containing genes. Red and green colors indicate the expression values in the respective sample. Samples 1to 3 are buffer-treated while 4 to 6 are CySNO infiltrated. A color key representing the intensities of the expression values is also given. (B) Pie-chart illustrating the total 440 number of up- and downregulated NO-responsive DUF domain-containing DEGs; these gene were identified at a Q-value < 0.05 in the transcriptome. (C) qRT-PCR validation of RNA-Seq data. About 10 DUF domain-containing genes (five each from up- and downregulated DEGs) were selected based on their fold change for validation through qRT-PCR. The R value represents Pearson’s correlation coefficient and was calculated among RNA-Seq and qRT-PCR datasets. The data points in C are the mean of three replicates while error bars represent standard error.
The transcriptional changes in the expression of DUF domain-containing genes were validated by analyzing the 10 representative DEGs (five upregulated and five downregulated) through qRT-PCR. The genes were selected based on the highest fold change from both upregulated and downregulated genes in response to CysNO infiltration. The expression patterns in qRT-PCR corroborated the RNA-Seq results for all of the 10 analyzed DUF domain-containing DEGs. The RNA-Seq and qRT-PCR datasets were compared based on the Pearson’s correlation coefficient (r = 0.905), revealing a high similarity between the datasets (Figure 2C).
We analyzed the promoter region of selected NO-induced DUF genes 1 Kb upstream of the transcription initiation site and the cis-elements or TF binding sites that were involved in plant stress were subsequently mapped (Figure S2). A detailed list of these TF binding sites is given in Supplementary Table S3. We identified the critical binding sites, including sites for the prolamin box (P-box), which was observed in 100% of the analyzed sequences. Similarly, two other elements, the W-box (WRKY TF family) and MAD-box (RIN), were found in 100% of the analyzed sequences. The AP2L ethylene-responsive TF was found in 54.1%, ETTIN (Auxin Response Factor 3) in 70.8%, ethylene-responsive TF ERF017 in 83.3%, ATMYB77-binding site in 83.3%, GT-box elements (GT2) in 70.8%, ANT (Arabidopsis protein AINTEGUMENTA) in 29.1%, SP8BF in 62.5%, HSFA1A in 95.8%, CDM1 in 29.6%, TATA-box in 91.6%, ANAC087 in 95.8%, and HBP1B in 91.6% of the analyzed sequences, respectively (Figure S2; Table S3). Among the above major elements, AP2L recognizes motifs within pathogenesis-related promoters, which may mediate the regulation of gene expression under biotic stress and components of stress signal transduction pathways (Xie et al., 2019). Similarly, other elements, such as ATMYB77 (an R2R3-type MYB TF), WRKY20, and ANAC087 (a NAC domain-containing protein), have been reported to play important roles in biotic and abiotic stresses. Therefore, the identification of regulatory elements and their modules is a prerequisite step for the understanding of gene expression and regulation (Nuruzzaman et al., 2013).
To observe the ancestral evolutionary relationship of Arabidopsis DUF genes Top 10 upregulated and downregulated DEGs were used as query to identify their orthologs in agronomically important species such as rice, soybean, wheat, maize, and tomato. About 151 protein sequences from all the species were used to generate a phylogenic tree. From the analysis, we can predict that Arabidopsis DUFs are distributed almost in every clade. All the peptides can be divided into four major clades and subclades. From an evolutionary perspective, rice and Arabidopsis were distributed throughout the tree, interestingly, majority of the wheat proteins were grouped together with tomato and were evolutionarily distant from Arabidopsis, rice, and soybean (Figure 3) Together these results suggest that these species may share common ancestors and that NO-responsive DUF protein functions could be conserved in these species.
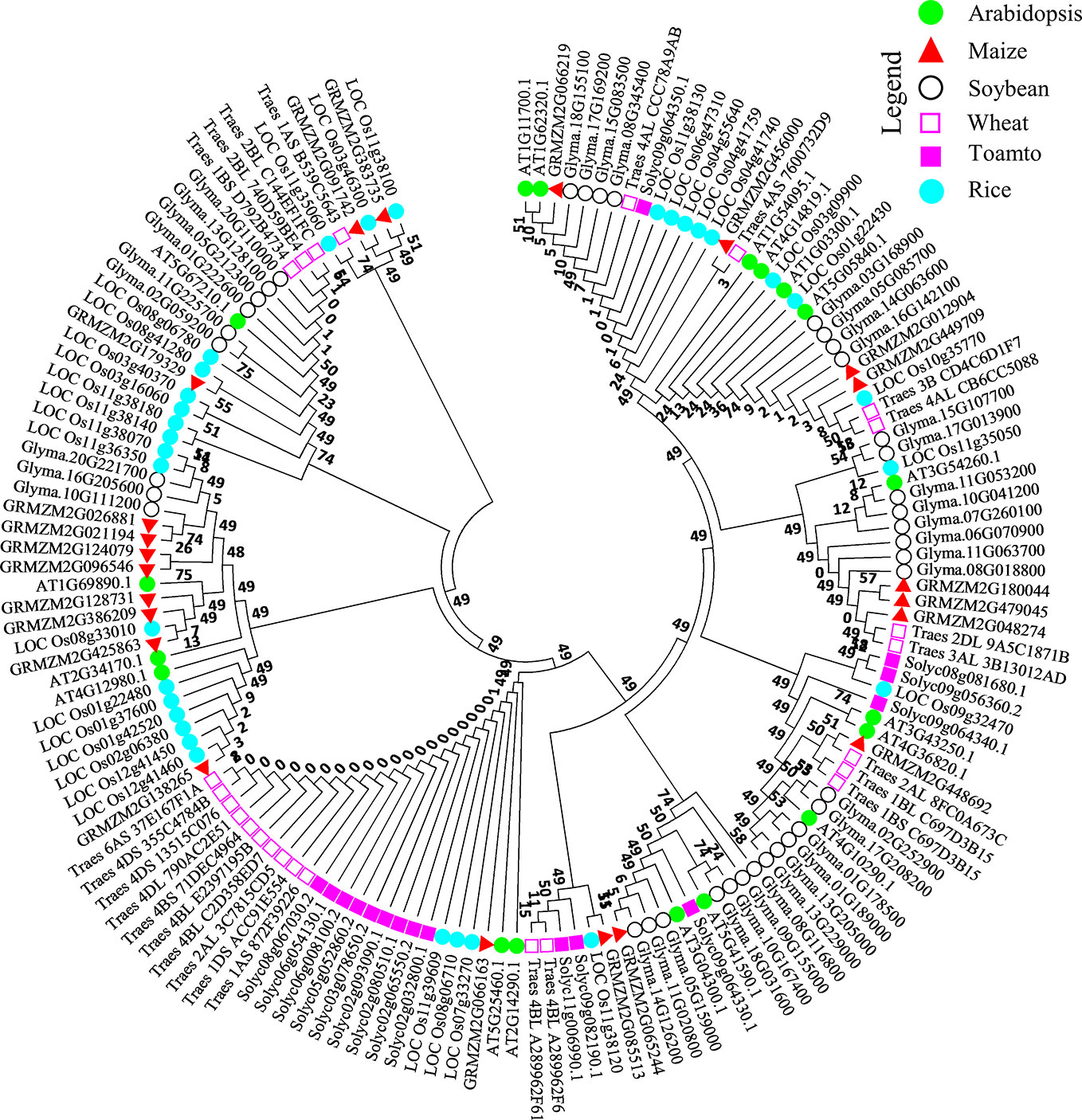
Figure 3 Molecular phylogenetic analysis of the AtDUF569 protein with homologs from other plant species. About 18 NO-responsive DUF DEGs were used as query against rice, soybean, wheat, maize, and tomato databases in Phytozome. The best-hit orthologs were identified and sequences were retrieved and aligned using ClustalW in Mega 7. The resultant alignment was used to make phylogenetic tree using maximum likelihood method on JTT matrix-based model through which evolutionary history was inferred. The bootstrap value was 1,000 replicates and branches corresponding to partitions reproduced in less than 50% of bootstrap replicates were eliminated. About 151 peptide sequences from Arabidopsis and various other crops mentioned above were used for this analysis. The positions with less than 95% site coverage were collapsed. The phylogenetic tree was constructed using Mega 7 software. Different species are labeled with different shapes and colors and can be found in the legend in the up-right corner of the figure.
We have searched for interactions between the CysNO-induced AtDUF569 (AT1G69890) and other proteins using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/). We observed some interesting interactions between the DUF569 and other proteins, and we identified 10 predicted functional partners, which included the uncharacterized protein, AT3G49790, known as a carbohydrate-binding protein. Its function is described as ATP-binding, but the underlying biological mechanism remains unknown. The PHLOEM PROTEIN 2-LIKE A10 is another carbohydrate-binding protein located in the mitochondria and found in several plant species and at different growth stages. The CYB-1 and ACYB-2 proteins are potentially transmembrane ascorbate ferrireductase 2, which contains two-heme-cytochrome (1 and 2) and is involved in the catalyzation of ascorbate-dependent transmembrane ferric-chelate reduction. Such interactions of DUF569 protein with other important proteins are illustrated in Figure 4 and Table S4.
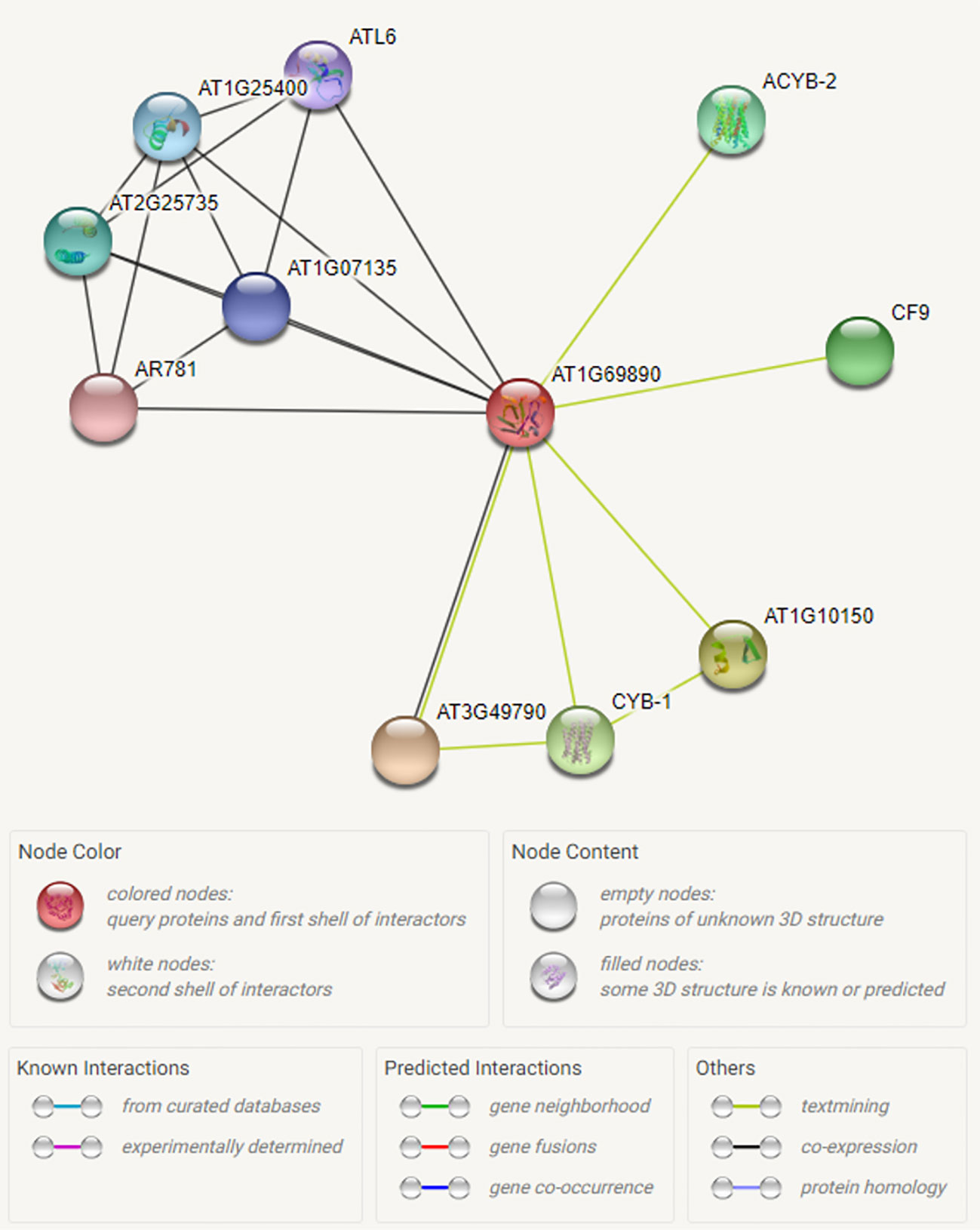
Figure 4 Interactome of the CysNO-induced AtDUF569. To predict possible interaction of ATDUF569 with other proteins, the accession number of DUF569 was used as “Protein name” in the search option in online Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/) with default parameters. A list of interacting proteins with DUF569 is given in Table S2 with relevant information. Different colors represent proteins or various types of interactions.
The in silico and experimental observations suggested that the AtDUF569 protein interacts with various proteins involved in cellular trafficking machinery and carbohydrate-binding and with glycine-rich proteins that participate in cellular stress responses and signaling (Czolpinska and Rurek, 2018).
We explored and applied the CDF as a development or fitness score for each plant. In the data recorded after seven days of treatment, our KO mutant atduf569 exhibited a noticeably higher CDF compared with Col-0 during exposure to CysNO- and GSNO-induced nitrosative stress, while all plants shared similar CDF result 14 days post-treatment. However, regarding MV-induced oxidative stress, KO mutant exhibited markedly reduced CDF at both seven- and 14-days post-stress treatment. On the contrary, under H2O2 exposure, the CDF was markedly higher for the KO mutant than for the Col-0 plants at both seven and 14 days after stress treatment (Figures 5 and 6). The results suggested that the atduf569 mutant was tolerant to CysNO- and GSNO-mediated nitrosative stress but was considerably tolerant and sensitive to H2O2 and MV, respectively. We also measured other plant attributes, such as shoot and root length, of the mutant in comparison to the WT (Col-0) and other designated control mutants. The KO mutant atduf569 exhibited higher shoot length under oxidative stress (induced by H2O2 and MV) 14 days post stress treatment relative to the other plants. However, shoot and root length displayed differential patterns under different nitrosative stress conditions. In addition, shoot and root lengths of KO mutant atduf569 were higher under CysNO, while the KO mutant under GSNO stress presented reduced length and increased shoot and root lengths, respectively (Figures 5 and 6). Overall, the results revealed that AtDUF659 (AT1G69890) differentially regulates plant growth and development under nitro-oxidative stress conditions.
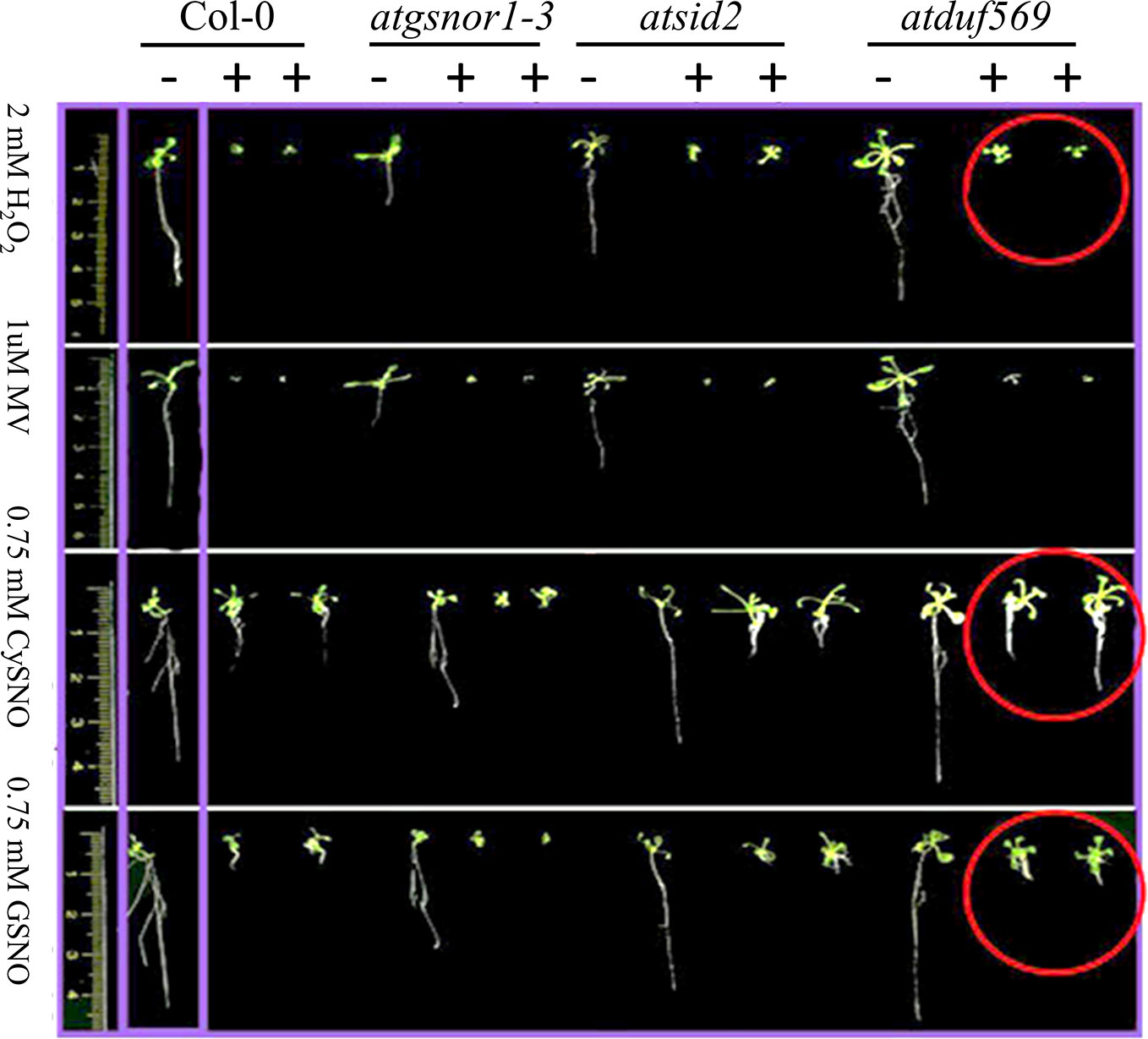
Figure 5 Differential role of AtDUF569 in plant development. Phenotypic evaluation of the genotypes under the control (−) and following exposure to various oxidative and nitrosative stresses (+). For the control genotype, a single plant is presented, whereas two plants are illustrated for the treatment.
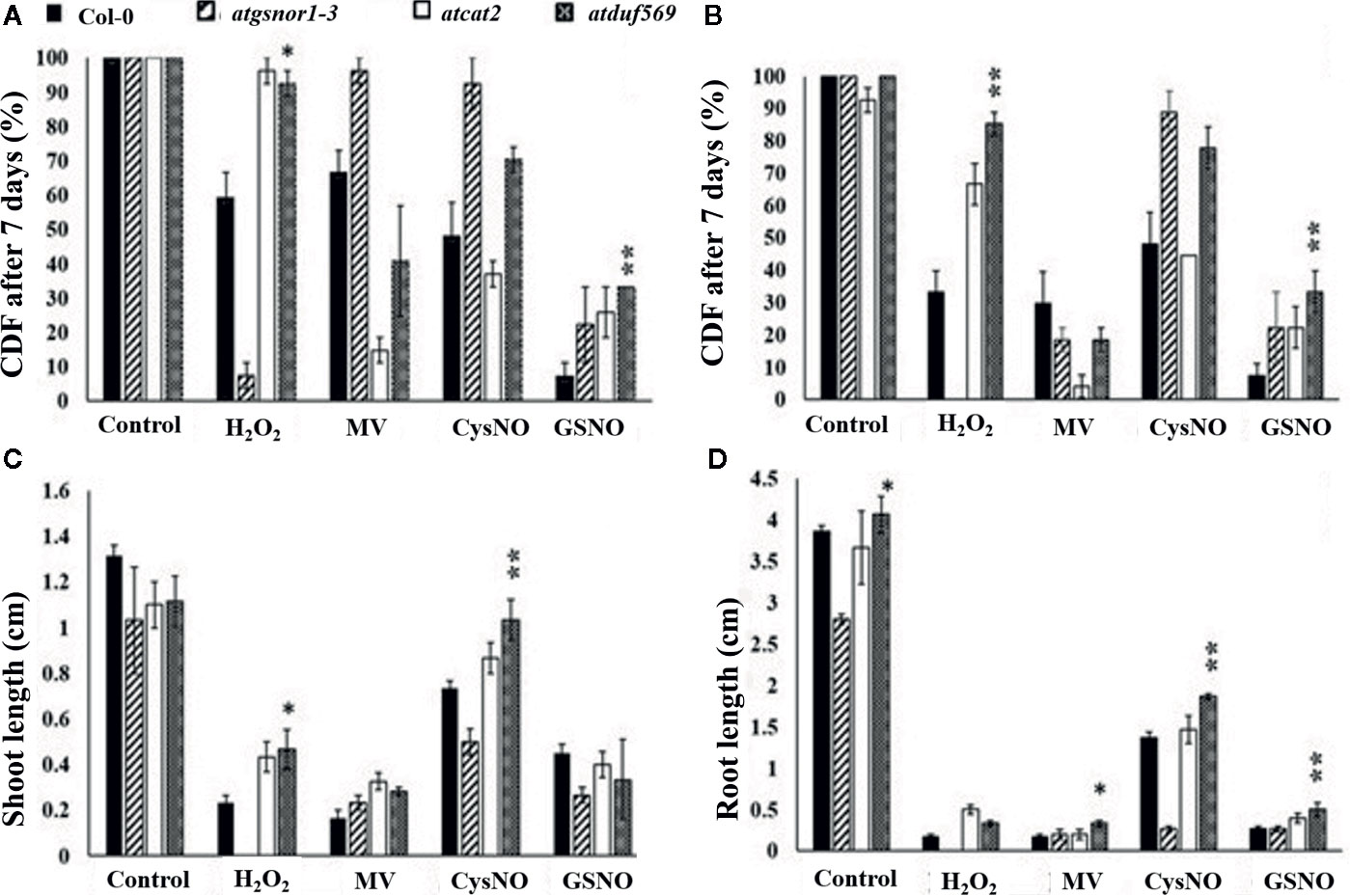
Figure 6 Exogenous application of CysNO, GSNO, or H2O2 and MV mediated nitro-oxidative stress. Designated genotypes Col-0, atgsnor1–3, atcat2, and atdfu569, were grown on half-strength Murashige and Skoog (MS) medium, with and without supplementation with either H2O2 and methyl viologen (Medina-Rivera et al., 2015) or CysNO and S-nitrosoglutathione (GSNO) for oxidative and nitrosative stress conditions, respectively. (A) Cotyledon development frequency (CDF) of the designated genotypes seven days after sowing under indicated stress. (B) CDF of the designated genotypes 14 days after sowing under indicated stress. (C) Shoot lengths of the designated genotypes 14 days after indicated stress. (D) Root lengths of the designated genotypes 14 days after indicated stress. All data points represent the means of triplicates. The experiment was repeated three times, with similar results. Error bars represent the standard error. Significant interactions are indicated by an asterisk (Student’s t-test with a 95% confidence level).
To examine the role of AtDUF569 (AT1G69890) in plant basal defense, we observed the phenotypic response of the atduf569 KO mutant line under the control conditions, as well as post pathogen inoculation as described elsewhere (Koornneef et al., 1991). The atduf569 KO mutant line was in the Col-0 genetic background; therefore, WT Col-0 plants were used as a control, while atgsnor1–3 KO mutant was used as a disease-susceptible control (Feechan et al., 2005). The loss-of-function mutant for Salicylic acid induction-deficient 2 (SID2) inhibits pathogen-induced salicylic acid (SA) production and makes the plants highly susceptible to infection by pathogens. Therefore, atsid2 was used as a second control for pathogenicity assessment. The pathogenicity assay revealed that the atduf569 KO line exhibited disease symptoms at 1 dpi, but in the later assessment, it was completely resistant toward the Pst DC3000 infection and exhibited no disease symptoms compared with Col-0 (WT), atgsnor1–3, and atsid2 plants (Figure 7A). Similar results were observed for the bacterial CFU counts at 3 dpi (Figure 7B). Based on the results, we hypothesized that the enhanced resistance to pathogens in the atduf569 line could have been due to the upregulation of the SA signaling pathway. Therefore, we performed an expression analysis for Pathogenesis-related 1 (PR1) and PR2 genes, which are regulators of SA-dependent gene expression. The qRT-PCR analysis revealed that the transcript accumulations of PR1 and PR2 were significantly (P < 0.05) lower in atduf569 than in the WT (Col-0) but higher than those of recognized sensitive genotypes, atgsnor1–3 and atsid2, 24 h post-inoculation (hpi) and 48 hpi, at which it displayed the highest expression. At 72 dpi, however, the expression level was lower in comparison to the expression level in WT, although the level was higher than in recognized sensitive genotypes atgsnor1–3 and atsid2 (Figures 8A, B).
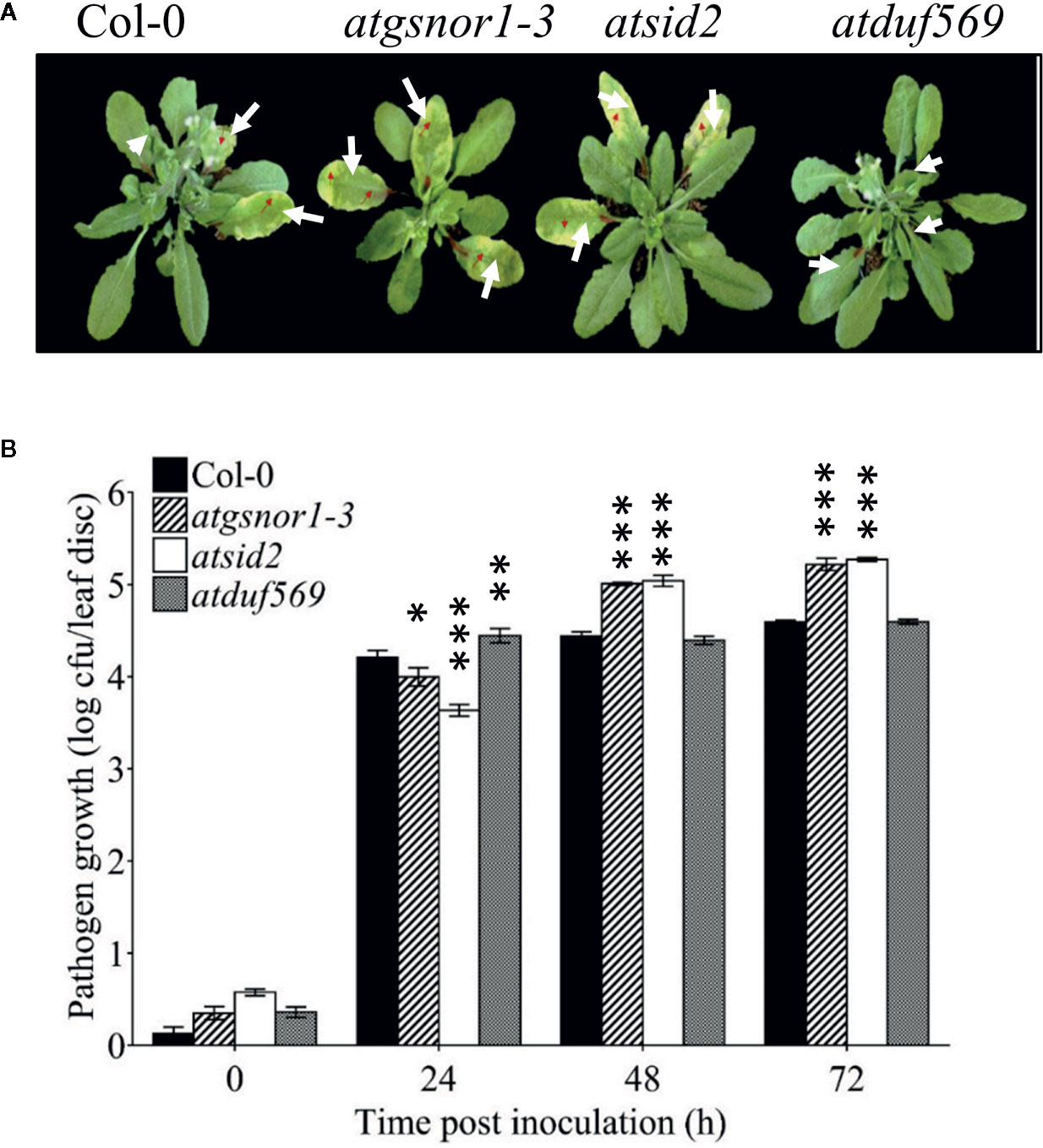
Figure 7 AtDUF569 negatively regulates basal defense. (A) Development of symptoms post-inoculation with virulent Pst DC3000, (B) Growth of the virulent Pst DC3000 in plants.
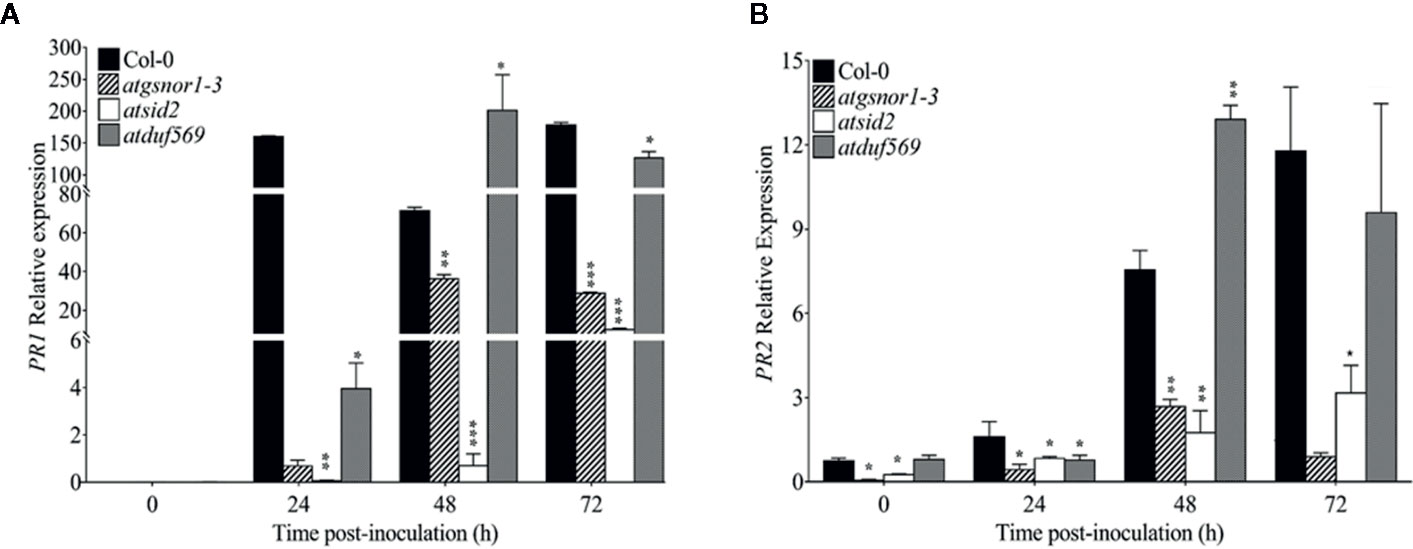
Figure 8 Relative expression levels of PR1 and PR2 in basal defense. (A) Relative expression of PR1 gene after Pst DC3000 (virulent) inoculation, (B) Relative expression of PR2 gene after Pst DC3000 (virulent) inoculation. The experiments were conducted with three replicates and repeated four times, with similar results. Data were analyzed using the Student’s t-test: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
To examine whether the AtDUF569 function is required for R-gene mediated resistance, we inoculated plants with Pst DC3000 avrB. Our results suggested early, high transcript accumulation of the in atduf569 when compared to the WT plants (Figure 9A) at 0, 3, 6, and 12 hpi. On the contrary, PR2 expression was significantly (P < 0.05) lower relative to the WT at 3 hpi with a gradual increase in expression at 6, 12, 24, and 48 hpi (Figure 9B). The observation indicated that PR2 transcript accumulation might have a higher contribution to the control of pathogen growth at later time points. Furthermore, the electrolyte leakage assay, performed after Pst DC3000 avrB inoculation, revealed higher electrolyte leakage in the atduf569 plants over time relative to the other genotypes except for atgsnor1-3 (Figure 9C).
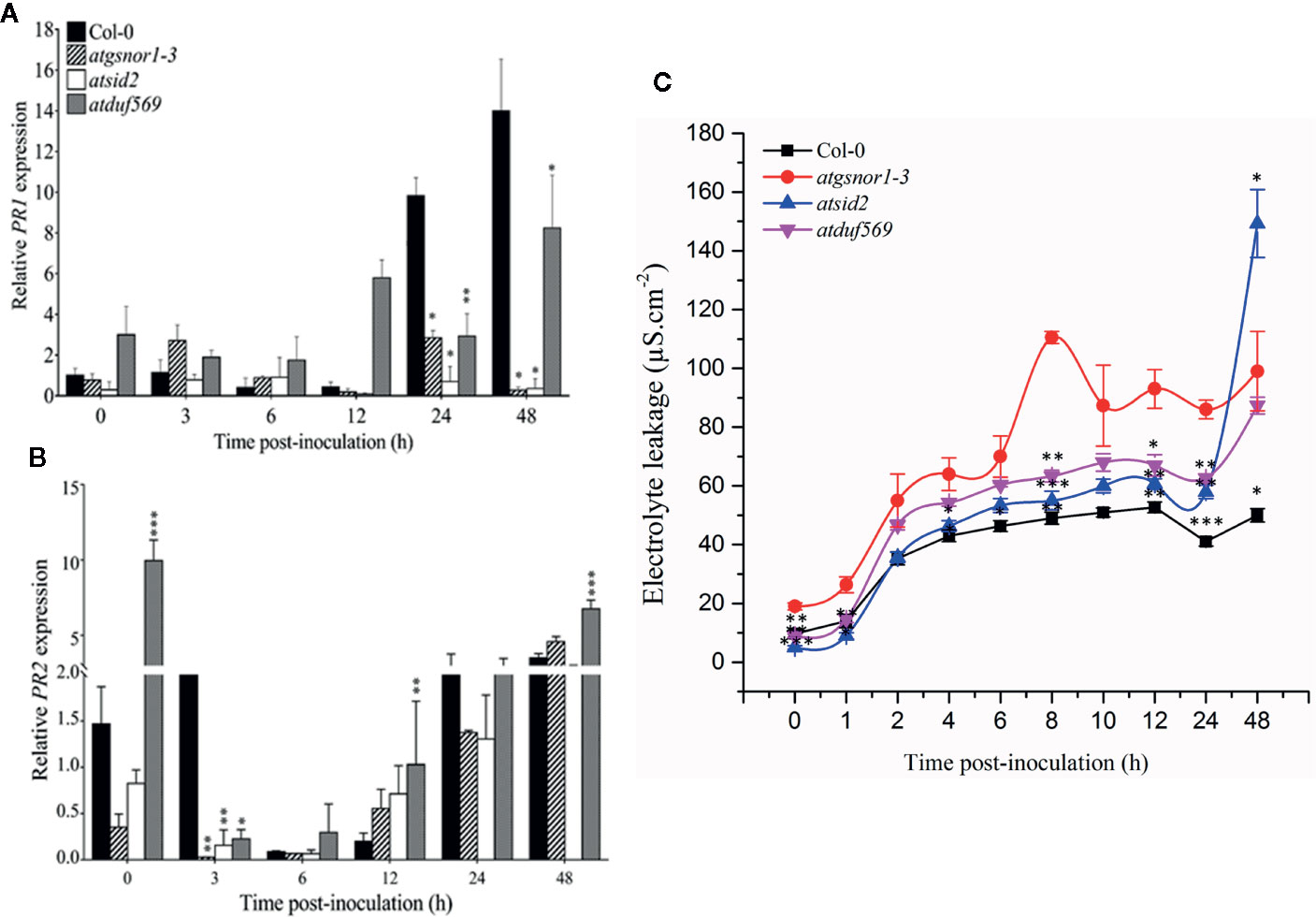
Figure 9 Relative expression of PR1 and PR2 during evaluation of the genotypes for R-gene-mediated resistance. Plants were inoculated with the Pst DC3000 avrB, and symptoms were recorded for analyzing the gene expression of the pathogenesis-related PRs genes. (A) PR1, (B) PR2 expression, (C) Electrolyte leakage over time. All the data points are the mean of three replicates. Data were analyzed using the Student’s t-test: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
In plants, signaling molecules, such as azelaic acid, glycerol-3-phosphate, and SA-PR1 and -PR2, are involved in mediating SAR in plants. The expression patterns of PR1 and PR2 revealed a significantly (P < 0.05) higher level of transcript accumulation in atduf569 at 0, 6, and 12 hpi but a drastically decreased level at 24 and 48 hpi in comparison to WT, atgsnor1–3, and atsid2 plants (Figures 10A, B). In addition, the expression levels of AZI were significantly (P ≤ 0.05) higher at 12 and 24 hpi and G3pdh at 0, 6, 12, 14, and 48 hpi (Figures 10C, D). Based on the expression pattern, the initial upregulation of PR1 and PR2 genes at 0, 6, and 12 hpi and the differential upregulation of AZI and G3pdh potentially contributed to the atduf569 mutant plants’ resistance to the Pst DC3000 infection. The results provide further evidence that the atduf569-resistant phenotype responded via the upregulation of SA-dependent PRs at the initial stage of pathogenicity, which controlled the effects of the pathogen and protected the mutant phenotype from later-stage disease development.
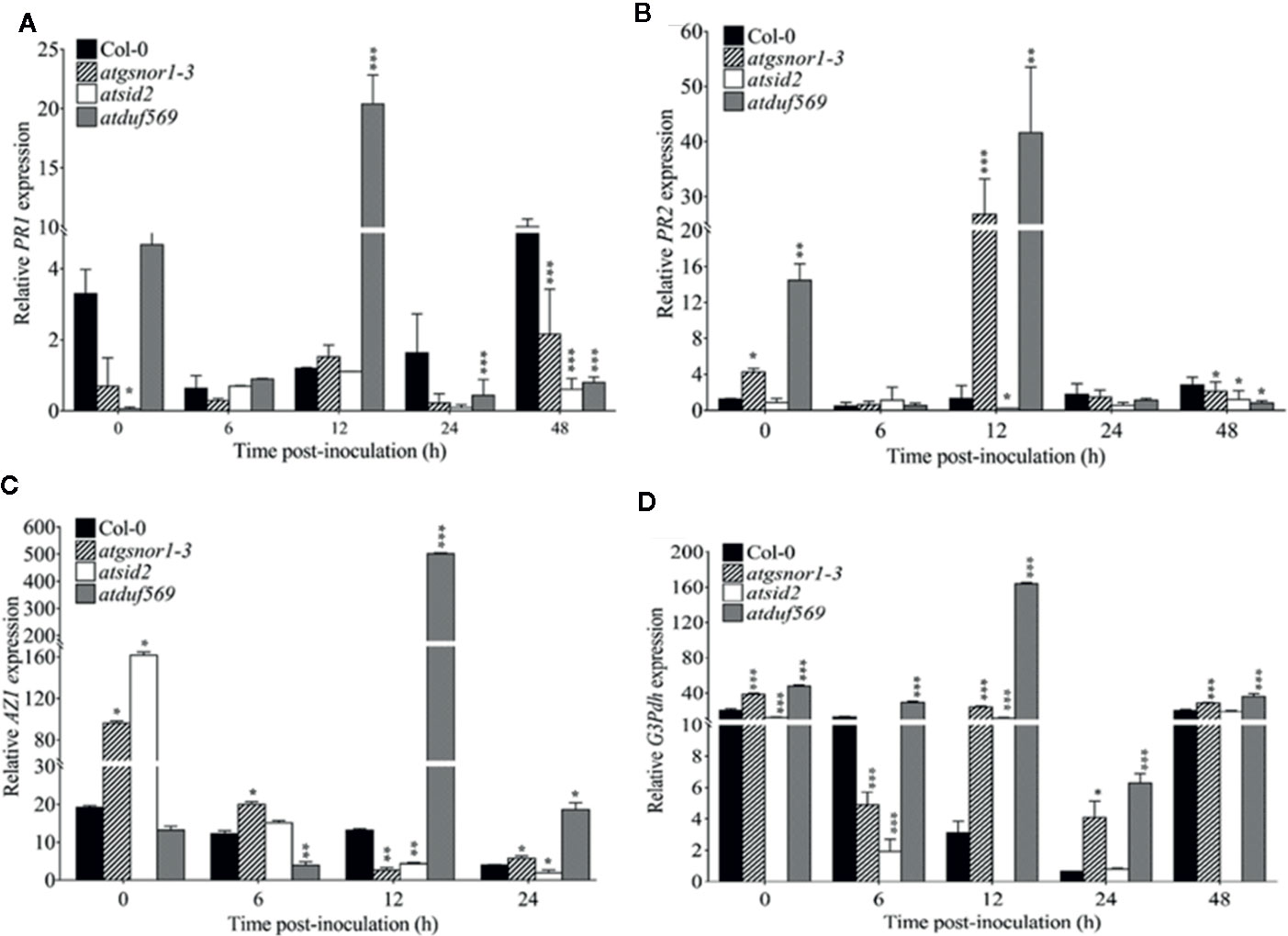
Figure 10 Relative expression of PR1, PR2, AZI, and G3DPH. AtDUF569 negatively regulates systemic acquired resistance. Plants were inoculated with the Pst DC3000 avrB, and symptoms were recorded to analyze the gene expression dynamics of the pathogenesis-related genes. All the data points are the mean of three replicates (A) PR1, (B) PR2, (C) AZI, and (D) G3DPH. Data were analyzed using the Student’s t-test: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
NO is a small, diatomic, and highly reactive gaseous molecule that orchestrates a plethora of physiological functions both in plants and animals. Although, its role has been well-explored in the mammalian system (Kerwin et al., 1995), NO opened a new pandora-box in plant science when it was reported as a signaling molecule during plant defense (Delledonne et al., 1998; Durner et al., 1998). In plants, contrary to other signaling cascades, NO transfer its bio-activity via posttranslational modifications (PTMs) such as S-nitrosation (Stamler et al., 2001), tyrosine nitration (Ischiropoulos et al., 1992), and metal nitrosylation (Gupta et al., 2011). These PTMs are known to regulate protein function under various stress conditions; however, it used to be very difficult to predict that a particular protein may undergo NO-mediated PTMs. The evolution of in silico tools such as GPS-SNO (Xue et al., 2010) made it quite easy to predict if a protein may undergo S-nitrosation or not. Similar tools are available for other PTMs too. Global gene expression such as RNA-seq has made it easy to identify candidate genes that are responsive to a stimulus. In the last couple of decades, a number of transcriptomes have revealed a bulk of data that generated interesting information about NO-mediated transcriptional changes. For example, transcriptome analysis of Arabidopsis leaves and roots in response to 1 mM GSNO exogenous application differentially expressed about 3,263 genes after 3 h of treatment (Begara-Morales et al., 2014). Similarly, Parani et al. (2004) showed 422 DEGs in response to 0.1 mM sodium nitroprusside (SNP). Recently in RNA-seq mediated transcriptome analysis we reported more than six thousand genes (Hussain et al., 2016) and transcription factors (Imran et al., 2018a) that showed differential expression in response to 1 mM CySNO. In the present study, using a transcriptome-wide search, we identified a total of 440 NO-responsive DUF genes that showed differential expression in response to 1 mM CysNO. Among the 437 DUF domain-containing DEGs, 231 (53%) were upregulated, while 206 (47%) were downregulated, with at least a two-fold change in their expression, implying that the genes have a key role in transcriptional regulation of various physiological processes. The differential expression of such huge number of DUF genes suggests their involvement in NO biology and possibly in NO-mediated post-translational modifications. In silico analyses of DUF569 for targeting potential sites for phosphorylation suggest at least five different sites in DUF569 that could undergo phosphorylation (Figure S3B). Similarly, the 3D protein structure and simulation by GPS SNO for identification of potential sites for S-nitrosylation also suggested the presence of exposed Cysteine residues that could be potential target for S-nitrosylation (Figures S3B, C). DUF genes have also been identified in response to other stimuli. In Arabidopsis transcriptomic profiling in response to 0.1 and 1.0 mM SNP, 126 genes (98 upregulated and 28 downregulated) with unknown functions were reported (Parani et al., 2004). Similarly, a transcriptomic study of Arabidopsis root exposed to 250 µM SNP revealed the largest category of genes with unknown and unclassified proteins known to date (Badri et al., 2008). More recently, in other plant species, such as upland cotton, transcriptome analysis based on Taq sequencing found 265 genes with uncharacterized proteins (Huang et al., 2018). Individual DUF genes have also been characterized. In a transcriptomic study of Arabidopsis plants exposed to SNP and O3, there was a more than two-fold change in expression and the upregulation of AtDUF569 (AT1G69890) (Ahlfors et al., 2009), suggesting that the AtDUF569 participates in O3‐induced cell death and NO production.
We observed a significant increase in the expression of AtDUF569 (57-fold) in response to NO donor application, which indicates relatively upstream and direct involvement of AtDUF569 in NO-related responses in A. thaliana. The presence of the P-box binding site for prolamin box-binding factor 1 (pbf1) and W-box sequences in the promoters of all the DUF genes was particularly interesting because W-box sequences have been recognized as positive regulators of senescence in the Minghui 63 rice variety, in which they are bound by leaf senescence-specific proteins (Liu et al., 2016). Furthermore, the W-box is the cognate cis-element for WRKY proteins, which regulates essential cellular functions (Imran et al., 2018b). The presence of other cis-elements, such as RIN, the GT-box, ANT, HSF, and TATA, also indicates the involvement of DUF genes in key physiological processes. TFs are involved in plant development and defense, inducing or repressing the transcription of specific genes in signaling pathways that ultimately control the differential responses toward plant growth and defense (Jin et al., 2017). Reports have suggested that NO-responsive genes contain a significantly higher number of certain transcription factor binding sites (TFBS) in their promoter regions (Palmieri et al., 2008). Palmieri et al. (2008) using in silico approach analyzed 28447 Arabidopsis genes and suggested that several TFBS were found at least 15% more often in NO-induced genes. Furthermore, promoter analysis of other non-NO-responsive DUF genes may help scientists understand the full potential of the gene family. The functions of DUF genes in plants may well be highly similar and evolutionarily conserved, as indicated by the high homology observed during phylogenetic analysis of DUF genes from different plant species, which could be one of the reasons why the exact functions of the genes remain elusive, as one would expect different functions in different plant species; simple/model or complex.
Through the predicted interactome analysis, we observed that AtDUF569 could interact with potentially important proteins. For example, out of ten proteins, DUF569 interacted with AT3G49790 which helps in ATP binding, therefore AtDUF569 is suggested to have important role during carbohydrate metabolism. Similarly, another interacting protein ATL6 E3 ubiquitin protein ligase is reported to be involved in regulating early steps of plant defense signaling, suggesting that AtDUF569 may regulate plant defense functions directly or by interacting with other proteins. Another interesting interacting protein is AR781 which is a pheromone receptor-like protein (DUF1645); if characterized fully, this gene might be a key in entomological research to attract a particular type of insects such as the honeybee. Thus, the interaction of AtDUF569 yields interesting and important information that can be further explored by an in-depth study.
We further sought to determine the role of DUF genes in the biological system using functional genomics approach. We, therefore, selected DUF569 which was among the top 10 DUFs that had the highest fold change in response to CysNO. Our results suggested that DUF569 negatively regulates shoot and root growth under nitro-oxidative stress, while in the case of pathogenicity, duf569 showed sensitive phenotype at an early time point (24 h) followed by more pathogen growth while it showed a resistant phenotype afterward (Figures 7A, B). This was in accordance with the PR gene expression which showed low expression at 24 h while significantly higher expression at 48 h.
The trend indicates that the atduf569-resistant phenotype could be due to the upregulation of SA-dependent PR genes at the initial stage of pathogenicity, which influences the effect of the pathogen effect and, in turn, protects the mutant phenotype from later-stage toxicity and disease symptoms.
Collectively, our results suggested the possible involvement of DUF genes in plant metabolism and energy generation and/or turnover involving active trafficking between cellular organelles. Furthermore, biological data on atduf569 KO plants suggested the involvement of the gene in regulating plant shoot and root growth besides plant responses to oxidative as well as nitro-oxidative stress conditions.
The current study illustrates the role of AtDUF569 in plant biology using a functional genomics approach. To the best of our knowledge, this is the first study attempting to functionally characterize NO-induced DUF genes in Arabidopsis. To date, no study has attempted to characterize NO-induced DUF genes in plant systems. Our study presents an exemplary model for future transcriptomic studies on similar DUF genes or proteins to elucidate molecular and functional aspects in detail in different plant species. This study motivates further studies to aid the comprehensive characterization of AtDUF569 about its cellular functioning.
The datasets generated for this study can be found in the the public repository for Gene Expression Omnibus (GEO) and Short Read Achieve (SRA) under accession numbers GSE81361 and SRP074890.
RN performed the experiments and analyzed the data. RN, RT, and QI drafted the manuscript. AH and MS helped with the conceptualization of the study. QI, MS, and RN prepared illustrations, figures, tables, and references. B-WY, AH, and QI edited the manuscript. B-WY contributed critical comments to the draft and approved the manuscript. All authors contributed to the article and approved the submitted version
This work was supported by Next-Generation Bio Green21 Program (SSAC, Grant No. PJ01342501) Rural Development Administration, South Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01041/full#supplementary-material
Ahlfors, R., Brosché, M., Kollist, H., Jaakko, K. (2009). Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 58, 1–12. doi: 10.1111/j.1365-313X.2008.03756.x
Arasimowicz-Jelonek, M., Floryszak-Wieczorek, J., Kubiś, J. (2009). Involvement of nitric oxide in water stress-induced responses of cucumber roots. Plant Sci. 177, 682–690. doi: 10.1016/j.plantsci.2009.09.007
Badri, D. V., Loyola-Vargas, V. M., Du, J., Stermitz, F. R., Broeckling, C. D., Iglesias-Andreu, L., et al. (2008). Transcriptome analysis of Arabidopsis roots treated with signaling compounds: A focus on signal transduction, metabolic regulation and secretion. New Phytol. 179, 209–223. doi: 10.1111/j.1469-8137.2008.02458.x
Bateman, A., Coggill, P., Finn, R. D. (2010). DUFs: Families in search of function. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 66, 1148–1152. doi: 10.1107/S1744309110001685
Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., Valderrama, R., Mata-Pérez, C., López-Jaramillo, J., et al. (2014). Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 65, 527–538. doi: 10.1093/jxb/ert396
Cui, Y., Wang, M., Zhou, H., Li, M., Huang, L., Yin, X., et al. (2016). OsSGL, a novel DUF1645 domain-containing protein, confers enhanced drought tolerance in transgenic rice and arabidopsis. Front. Plant Sci. 7, 1–13. doi: 10.3389/fpls.2016.02001
Czolpinska, M., Rurek, M. (2018). Plant glycine-rich proteins in stress response: An emerging, still prospective story. Front. Plant Sci. 9, 1–13. doi: 10.3389/fpls.2018.00302
Delledonne, M., Xia, Y., Dixon, R. A., Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394 (6693), 585–588. doi: 10.1038/29087
Desikan, R., Mackerness, S. A. H., Hancock, J. T., Neill, S. J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127 (1), 159–172. doi: 10.1104/pp.127.1.159
Domingos, P., Prado, A. M., Wong, A., Gehring, C., Feijo, J. A. (2015). Nitric oxide: A multitasked signaling gas in plants. Mol. Plant 8, 506–520. doi: 10.1016/j.molp.2014.12.010
Durner, J., Wendehenne, D., Klessig, D. F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. 95 (17), 10328–10333. doi: 10.1073/pnas.95.17.10328
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Feechan, A., Kwon, E., Yun, B.-W., Wang, Y., Pallas, J., Loake, G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U. S. A. 102, 8054–8059. doi: 10.1073/pnas.0501456102
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Flors, V., Ton, J., Van Doorn, R., Jakab, G., García-Agustín, P., Mauch-Mani, B. (2008). Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 54, 81–92. doi: 10.1111/j.1365-313X.2007.03397.x
García-Mata, C., Lamattina, L. (2002). Nitric oxide and abscisic acid cross talk in guard cells. Plant Physiol. 128, 790–792. doi: 10.1104/pp.011020
Greenacre, S. A. B., Ischiropoulos, H. (2001). Tyrsosine nitration: localization, quantification, consequences for protein function and signal transduction. Free Rad. Res. 34, 541–581. doi: 10.1080/10715760100300471
Gupta, K. J., Hebelstrup, K. H., Mur, L. A. J., Igamberdiev, A. U. (2011). Plant hemoglobins: Important players at the crossroads between oxygen and nitric oxide. FEBS Lett. 585 (24), 3843–3849. doi: 10.1016/j.febslet.2011.10.036
He, X., Hou, X., Shen, Y., Huang, Z. (2011). TaSRG, a wheat transcription factor, significantly affects salt tolerance in transgenic rice and Arabidopsis. FEBS Lett. 585, 1231–1237. doi: 10.1016/j.febslet.2011.03.055
Hu, Y. Q., Liu, S., Yuan, H. M., Li, J., Yan, D. W., Zhang, J. F., et al. (2010). Functional comparison of catalase genes in the elimination of photorespiratory H2O2 using promoter- and 3’-untranslated region exchange experiments in the Arabidopsis cat2 photorespiratory mutant. Plant Cell Environ. 33, 1656–1670. doi: 10.1111/j.1365-3040.2010.02171.x
Huang, J., Wei, H., Li, L., Yu, S. (2018). Transcriptome analysis of nitric oxide-responsive genes in upland cotton (Gossypium hirsutum). PloS One 13, 1–18. doi: 10.1371/journal.pone.0192367
Hussain, A., Mun, B.-G., Imran, Q. M., Lee, S.-U., Adamu, T. A., Shahid, M., et al. (2016). Nitric Oxide Mediated Transcriptome Profiling Reveals Activation of Multiple Regulatory Pathways in Arabidopsis thaliana. Front. Plant Sci. 7, 975. doi: 10.3389/fpls.2016.00975
Imran, Q. M., Falak, N., Hussain, A., Mun, B.-G., Sharma, A., Lee, S.-U., et al. (2016). Nitric oxide responsive heavy metal-associated gene AtHMAD1 contributes to development and disease resistance in Arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01712
Imran, Q. M., Hussain, A., Lee, S. U., Mun, B. G., Falak, N., Loake, G. J., et al. (2018a). Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci. Rep. 8, 1–14. doi: 10.1038/s41598-017-18850-5
Imran, Q. M., Hussain, A., Mun, B. G., Lee, S. U., Asaf, S., Ali, M. A., et al. (2018b). Transcriptome wide identification and characterization of NO-responsive WRKY transcription factors in Arabidopsis thaliana L. Environ. Exp. Bot. 148, 128–143. doi: 10.1016/j.envexpbot.2018.01.010
Ischiropoulos, H., Zhu, L., Chen, J., Tsai, M., Martin, J. C., Smith, C. D., et al. (1992). Peroxynitrite-Mediated Tyrosine Nitration Catalyzed by Superoxide-Dismutase. Arch. Biochem. Biophysics 298 (2), 431–437. doi: 10.1016/0003-9861(92)90431-U
Jin, Y., Eser, U., Struhl, K., Churchman, L. S. (2017). The Ground State and Evolution of Promoter Region Directionality. Cell 170, 889–898.e10. doi: 10.1016/j.cell.2017.07.006
Jones, D. T., Taylor, W. R., Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kerwin, J. F., Jr., Lancaster, J. R., Jr., Feldman, P. L. (1995). Nitric oxide: a new paradigm for second messengers. J. Med. Chem. 38 (22), 4343–4362. doi: 10.1021/jm00022a001
Kim, J. H., Kim, W. T. (2013). The arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 162, 1733–1749. doi: 10.1104/pp.113.220103
Koornneef, M., Hanhart, C. J., van der Veen, J. H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. MGG Mol. Gen. Genet. 229, 57–66. doi: 10.1007/BF00264213
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, L., Xie, C., Ye, T., Xu, J., Chen, R., Gao, X., et al. (2017). Molecular characterization, expression pattern and function analysis of the rice OsDUF866 family. Biotechnol. Biotechnol. Equip. 31, 243–249. doi: 10.1080/13102818.2016.1268932
Li, L. H., Lv, M. M., Li, X., Ye, T. Z., He, X., Rong, S. H., et al. (2018a). The Rice OsDUF810 Family: OsDUF810.7 May be Involved in the Tolerance to Salt and Drought. Mol. Biol. (Mosk). 52, 567–575. doi: 10.1134/S0026898418040122
Li, L., Lv, M., Ye, T., He, X., Dong, Y., Rong, S., et al. (2018b). Genome-wide identification and analyses of the rice OsDUF639 family. Int. J. Agric. Biol. 20, 1117–1122. doi: 10.17957/IJAB/15.0611
Liu, L., Xu, W., Hu, X., Liu, H., Lin, Y. (2016). W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 6, 1–9. doi: 10.1038/srep20881
Luo, C., Guo, C., Wang, W., Wang, L., Chen, L. (2014). Overexpression of a new stress-repressive gene OsDSR2 encoding a protein with a DUF966 domain increases salt and simulated drought stress sensitivities and reduces ABA sensitivity in rice. Plant Cell Rep. 33, 323–336. doi: 10.1007/s00299-013-1532-0
Medina-Rivera, A., Defrance, M., Sand, O., Herrmann, C., Castro-Mondragon, J. A., Delerce, J., et al. (2015). RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 43, W50–W56. doi: 10.1093/nar/gkv362
Murashige, T., Skoog, F. (1962). A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nabi, R. B. S., Tayade, R., Hussain, A., Kulkarni, K. P., Imran, Q. M., Mun, B. G., et al. (2019). Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ. Exp. Bot. 161, 120–133. doi: 10.1016/j.envexpbot.2019.02.003
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 1–16. doi: 10.3389/fmicb.2013.00248
Pagnussat, G. C., Lanteri, M. L., Lamattina, L. (2003). Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 132, 1241–1248. doi: 10.1104/pp.103.022228
Palmieri, M. C., Sell, S., Huang, X., Scherf, M., Werner, T., Durner, J., et al. (2008). Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J. Exp. Bot. 59 (2), 177–186. doi: 10.1093/jxb/erm345
Parani, M., Rudrabhatla, S., Myers, R., Weirich, H., Smith, B., Leaman, D. W., et al. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2, 359–366. doi: 10.1111/j.1467-7652.2004.00085.x
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, 290–301. doi: 10.1093/nar/gkr1065
Sham, A., Al-Azzawi, A., Al-Ameri, S., Al-Mahmoud, B., Awwad, F., Al-Rawashdeh, A., et al. (2014). Transcriptome Analysis Reveals Genes Commonly Induced by Botrytis cinerea Infection, Cold, Drought and Oxidative Stresses in Arabidopsis. PloS One 9 (11). doi: 10.1371/journal.pone.0113718
Stamler, J. S., Simon, D. I., Osborne, J. A., Mullins, M. E., Jaraki, O., Michel, T., et al. (1992). S-Nitrosylation of proteins with nitric oxide: Synthesis and characterization of biologically active compounds. Proc. Natl. Acad. Sci. U. S. A. 89, 444–448. doi: 10.1073/pnas.89.1.444
Stamler, J. S., Lamas, S., Fang, F. C. (2001). Nitrosylation: the prototypic redox-based signaling mechanism. Cell 106 (6), 675–683.
Tada (2008). Plant Immunity Requires Conformational Charges of NPR1 via S- Nitrosylation and Thioredoxins. Bone 23, 1–7. doi: 10.1161/CIRCULATIONAHA.110.956839
Wang, Y. Q., Feechan, A., Yun, B. W., Shafiei, R., Hofmann, A., Taylor, P., et al. (2009). S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137. doi: 10.1074/jbc.M806782200
Whalen, M. C., Innes, R. W., Bent, A. F., Staskawicz, B. J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. doi: 10.1105/tpc.3.1.49
Xie, Z., Nolan, T. M., Jiang, H., Yin, Y. (2019). AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 10, 1–17. doi: 10.3389/fpls.2019.00228
Xu, E. J., Vaahtera, L., Brosche, M. (2015). Roles of Defense Hormones in the Regulation of Ozone-Induced Changes in Gene Expression and Cell Death. Mol. Plant 8 (12), 1776–1794. doi: 10.1016/j.molp.2015.08.008
Xue, Y., Liu, Z. X., Gao, X. J., Jin, C. J., Wen, L. P., Yao, X. B., et al. (2010). GPS-SNO: Computational Prediction of Protein S-Nitrosylation Sites with a Modified GPS Algorithm. PloS One 5 (6), ARTN e11290. doi: 10.1371/journal.pone.0011290
Keywords: Arabidopsis, transcriptome, DUF569, differentially expressed genes, nitro-oxidative stress
Citation: Nabi RBS, Tayade R, Imran QM, Hussain A, Shahid M and Yun B-W (2020) Functional Insight of Nitric-Oxide Induced DUF Genes in Arabidopsis thaliana. Front. Plant Sci. 11:1041. doi: 10.3389/fpls.2020.01041
Received: 24 January 2020; Accepted: 24 June 2020;
Published: 14 July 2020.
Edited by:
Mounira Chaki, University of Jaén, SpainReviewed by:
Pedro Carrasco, University of Valencia, SpainCopyright © 2020 Nabi, Tayade, Imran, Hussain, Shahid and Yun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Wook Yun, Ynd5dW5Aa251LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.