- 1Department of Plant Sciences, North Dakota State University, Fargo, ND, United States
- 2Institute of Plant Protection, Postdoctoral Program, Heilongjiang Academy of Agricultural Sciences, Harbin, China
- 3USDA-ARS, Edward T. Schafer Agricultural Research Center, Fargo, ND, United States
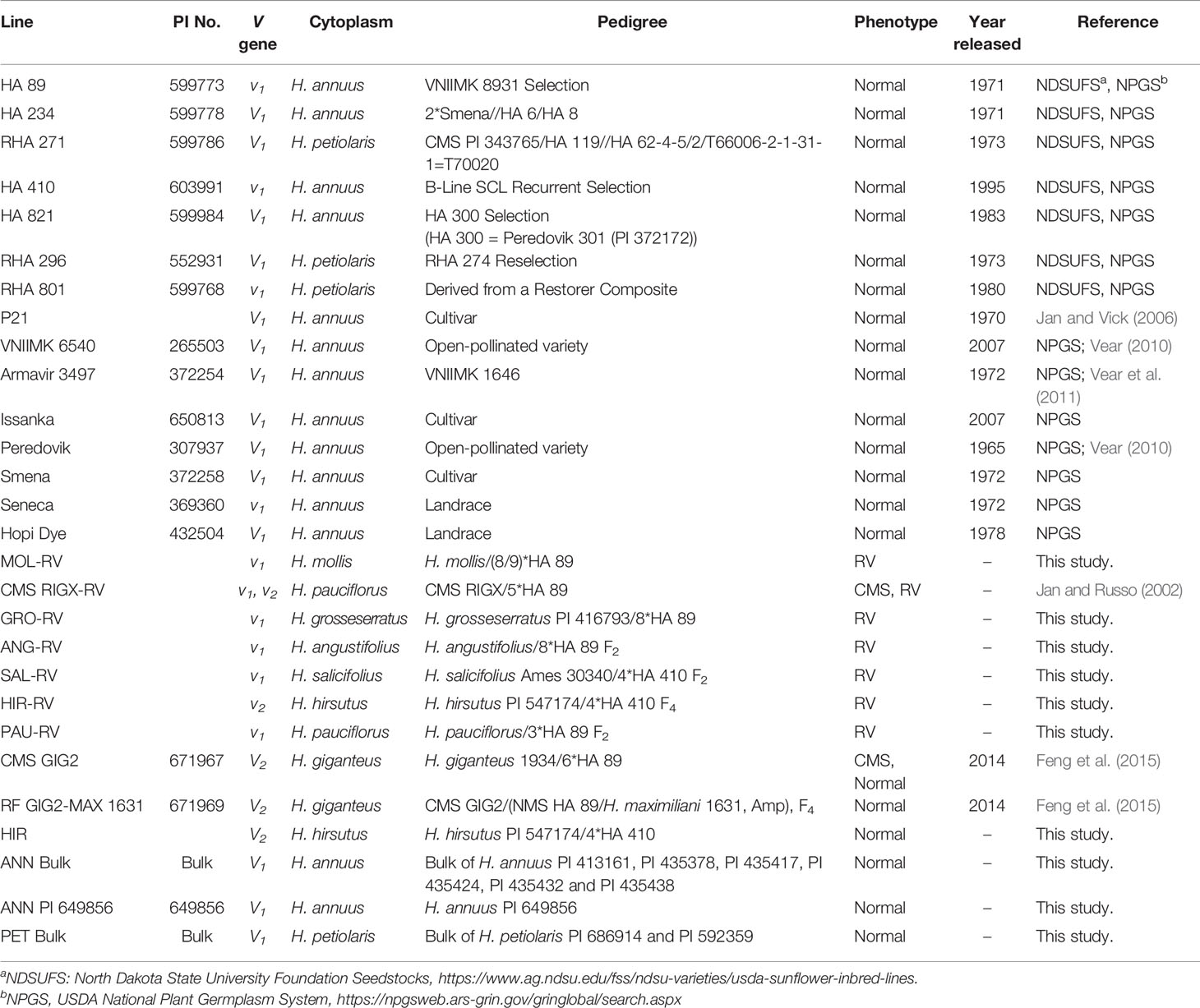
Wild Helianthus species are an important genetic resource for sunflower improvement, but sometimes there are adverse interactions between the wild and cultivated sunflowers. This study reports the inheritance of reduced vigor and its restoration resulting from an interaction of perennial Helianthus cytoplasms with nuclear genes of cultivated sunflower lines. The large number of vigor restoration (V) genes identified in cultivated lines are all located at the same locus, designated V1, suggesting a common origin of these genes. Additional V genes derived from the wild perennial species H. giganteus L. and H. hirsutus Raf. are located at a different locus than V1, designated V2. A major difference between the wild annual Helianthus cytoplasms and perennial cytoplasms is the lack of the vigor-reducing cytoplasms, but surprisingly V genes were observed in wild annual H. annuus L. and H. petiolaris Nutt. which were at the same locus as V1. A common vigor-reducing cytoplasmic effect of the perennial Helianthus species and the existence of a common vigor restoration V gene in most perennial Helianthus species could be explained as a result of vigor selection during Helianthus speciation. V1 was mapped on linkage group (LG) 7 of the sunflower genome, using an F2 population derived from MOL-RV/HA 821. V1 co-segregated with an InDel marker ZVG31, with three single-nucleotide polymorphism (SNP) markers, SFW01024, SFW07230, and SFW00604, located above it on the map at a genetic distance of 0.8 cM, and another SNP marker, SFW08671, below it at a distance of 0.4 cM. The physical distance between the two closest flanking SNP markers corresponds to 0.56 and 1.37 Mb on the HA 412-HO and XRQ assemblies, respectively. The tightly linked markers will help select normal vigor progenies when using perennial Helianthus cytoplasms in a breeding program, which will also provide a basis for studying the mechanism of the cytonuclear interaction, and the speciation of annual and perennial Helianthus species.
Introduction
Crop wild relatives (CWR) are an important genetic resource for crop improvement to biotic and abiotic stresses in many crops, such as wheat, rice, maize, barley, oat, cotton, and soybean (Yumurtaci, 2015; Mammadov et al., 2018). Cultivated sunflower (Helianthus annuus L., 2n = 2× = 34) is one of the few crops native to the United States. The Helianthus genus is well known for its taxonomic complexity, which includes 53 species (14 annual and 39 perennial) and 19 subspecies (Seiler et al., 2017; Anderson et al., 2019). The annual species are all diploid (including cultivated sunflower), and the perennial species include 26 diploid, three tetraploid, seven hexaploid, and three mixaploid species. As with other CWR, wild Helianthus species represent a large unexploited gene pool with genetic variation for different traits, such as resistance to Sclerotinia, Phomopsis, rust, and downy mildew diseases, and parasitic broomrape (Seiler et al., 2017). They are also a source of new cytoplasmic male sterility (CMS) for sunflower improvement. All the annual Helianthus species, except Helianthus agrestis Pollard, can be hybridized with cultivated sunflower using classical crossing methods (Seiler et al., 2017). However, utilization of the perennial diploid species which represent half of the Helianthus genus, is limited by poor crossability and F1 sterility in wild × cultivated interspecific hybrids. Development of a two-stage embryo rescue technique and a colchicine treatment of seedlings to double the chromosome number have minimized these problems and made it possible to produce interspecific amphiploids (Amp) (Jan, 1988; Sukno et al., 1999). These amphiploids have proven to be extremely valuable in transferring resistance to Orobanche cumana Wallr. (broomrape) race F, and in the introgression of fertility restoration genes into cultivated sunflower (Jan and Fernandez-Martinez, 2002; Jan et al., 2002; Feng and Jan, 2008; Liu et al., 2013).
Another limitation in the use of the perennial wild species is the existence of adverse cytonuclear interactions. Previously, reduced-vigor (RV) plants were observed in backcross progenies of an inbred line HA 89 in the cytoplasms of five perennial Helianthus species (H. mollis Lam., H. maximiliani Schrad., H. grosseserratus Mar., H. divaricatus L., and H. angustifolius L.) (Jan, 1992). The characteristics of RV plants included pale-green leaves, significantly reduced plant height, head diameter, seed weight, percent seed set, net photosynthesis, total leaf chlorophyll, and delayed flowering. The plant vigor reduction effects varied among the different cytoplasms. A cytoplasmic component of these effects has been confirmed by the occurrence of all-normal progenies in crosses of HA 89 with pollen from RV plants. Genetic studies suggested that each of the five species has a single dominant nuclear gene controlling plant vigor restoration (Jan, 1992). In addition, the segregation ratios of normal (N) to RV plants observed in the F2 progeny of diallel crosses among normal plants with heterozygous or homozygous vigor restoration genes derived from the above five interspecific crosses indicated a common perennial cytoplasmic deficiency in these wild perennial species, and that a common vigor restoration gene could restore the plant vigor (Jan, 1995).
The cytoplasmic genome of plants contains 120–140 genes in the mitochondria and 95–100 genes in the chloroplast. Both chloroplasts and mitochondria require the import of nuclear-encoded proteins for organelle biogenesis (Levin, 2003). The results of above reciprocal crosses indicated that the phenotypes of RV plants in the progenies of the five perennial Helianthus species (Jan, 1992) may arise from cytonuclear interactions between the nuclear genome of the annual H. annuus and the cytoplasms of the perennials. Cytonuclear incompatibilities may play a role in establishing reproductive isolation among these species (Burton et al., 2013). The study of the reciprocal F1 hybrids and backcross families of H. annuus and H. petiolaris in xeric and mesic habitats of the parental species suggested that the parental species’ cytoplasms were strongly locally adapted and that cytonuclear interactions significantly affected the fitness and architecture of hybrid plants (Sambatti et al., 2008). Using a target enrichment approach, Stephens et al. (2015) studied phylogeny relationships across 37 diploid Helianthus species/subspecies with a total of 103 accessions using 170 nuclear genes and the chloroplast sequences. Their phylogeny analysis with nuclear genes supported three major clades including a large annual clade, a southeastern perennial clade, and another clade of primarily large-statured perennials. A rapid radiation and/or high levels of reticulate evolution among perennial Helianthus species was suggested in their study. Later, Lee-Yaw et al. (2019) analyzed the phylogenetic relationships among annual Helianthus species and individuals, using nuclear SNPs and chloroplast genomes sequences. The two perennial species H. nuttallii Torr. & A. Gray and H. maximiliani and one more distantly related genus Phoebanthus grandiflora Torr. & A. Gray were separated in a distinct clade when used as outgroups for the annual Helianthus species. These phylogenetic analyses indicated the clear distinction between the nuclear and chloroplast of annual and perennial Helianthus species (Stephens et al., 2015; Lee-Yaw et al., 2019).
Recently, RV and normal progenies have been observed in nine additional perennial Helianthus species when the wild species were used as maternal parents crossed with the cultivated sunflower inbred lines HA 89 or HA 410, with the goal of transferring Sclerotinia resistance and other useful genes from wild perennial Helianthus species into cultivated sunflower, including H. giganteus L., H. hirsutus Raf., H. salicifolius A. Dietr., H. pauciflorus Nutt., H. californicus DC., H. nuttallii T.& G., H. occidentalis Riddell subsp. plantagineus (T. & A. Gray) Heiser, H. schweinitzii T. & G., and H. tuberosus L. Therefore, reduced plant vigor and its restoration are commonly observed in utilizing CWR for sunflower improvement. The objectives of this study were to: 1) further examine reduced plant vigor and vigor restoration (V) genes for the vigor-reducing cytoplasms of wild perennial species with respect to cultivated sunflower, wild annual Helianthus species, and perennial H. giganteus and H. hirsutus; 2) examine the relationships among the V genes in cultivated sunflower, wild perennial and annual Helianthus species, and determine the inheritance of vigor restoration; and 3) map the common V gene from cultivated sunflower to a genetic map using an F2 population derived from the cross of MOL-RV/HA 821.
Materials and Methods
Plant Materials
Eight wild perennials plus two wild annual Helianthus species were used in this study. The wild perennials included H. mollis, H. giganteus, H. maximiliani, H. grosseserratus, H. angustifolius, H. salicifolius, H. hirsutus, and H. pauciflorus. The wild annuals included H. annuus and H. petiolaris. One alloplasmic line with H. mollis cytoplasm (MOL-RV), 14 cultivated sunflower lines (HA 89, HA 821, HA 234, RHA 271, RHA 296, RHA 801, P21, Peredovik, VNIIMK 6540, Smena, Seneca, Issanka, Armavir 3497, and Hopi Dye), and one RV CMS line derived from interspecific cross (CMS RIGX-RV) was used to study the inheritance of the V genes (Table 1). The normal and RV progeny plants derived from seven perennials (H. mollis, H. giganteus, H. grosseserratus, H. angustifolius, H. salicifolius, H. hirsutus, and H. pauciflorus) and two annuals (H. annuus and H. petiolaris) were used to determine the genetics and the relationships of the V genes. HA 89, HA 821 and HA 234 are oilseed maintainer lines, whereas RHA 271, RHA 296 and RHA 801 are oilseed restorer lines. These inbred lines were publicly released by USDA. P21 is a nuclear male-sterile line released by the USDA and the Texas Agricultural Experiment Station in 1970 (Jan and Vick, 2006). VNIIMK 6540, VNIIMK 8931 (for HA 89), Armavir 3497, Peredovik, and Smena were varieties developed by former Soviet Union. Issanka was developed in France. Seneca and Hopi Dye are Native American Indian Landraces. CMS RIGX-RV has perennial cytoplasm from H. pauciflorus.
The pedigree of the lines used in the study follow the nomenclature system of Purdy et al. (1968). Briefly, the symbol “/” indicated the primary cross and the backcrosses are indicated by numerals at the “/” symbol and placed on the same side of the symbol as the recurrent parent. Then numerals and the recurrent parent are separated by an asterisk “*”. The numerals indicate the number of times the recurrent parent was used, example, (H. giganteus/6*HA89). The “//” symbol indicates a secondary cross. Amphiploids were also used in the pedigree designated as i.e. NMS HA89/H. maximiliani 1631, Amp.
To study the relationship among V genes from different sources, six homozygous vigor restoration sources including HA 821; RF GIG2-MAX 1631 (pedigree: CMS GIG2/(NMS HA89/H. maximiliani 1631, Amp) F4, Normal); HIR (Pedigree: H. hirsutus PI 547174/4*HA 410, F4, Normal); two H. annuus sources, PI 649856 and a bulk of PI 413161, PI 435378, PI 435417, PI 435424, PI 435432 and PI 435438; plus a bulk of two H. petiolaris (PI 686914 and PI 592359) were pollinated onto five homozygous RV lines (GRO-RV, ANG-RV, SAL-RV, HIR-RV, and PAU-RV) with vigor-reducing cytoplasms of H. grosseserratus, H. angustifolius, H. salicifolius, H. hirsutus, and H. pauciflorus in 2015. CMS GIG2 is a normal-vigor progeny (pedigree: H. giganteus/6*HA89, CMS, Normal) (Feng et al., 2015). The pedigrees of the materials used in the study are listed in Table 1.
Progeny Test for Plant Vigor for Progenies Derived From MOL-RV and Cultivated Sunflower
Reduced-vigor progeny of H. mollis/8*HA 89 (MOL-RV) were grown in the greenhouse in 1998 and pollinated with 14 cultivated sunflower lines from diverse genetic backgrounds (Table 1). Vigor-restored normal (N) F1 plants were self-pollinated and F2 progeny evaluated in the greenhouse for plant vigor restoration under normal sunflower growth conditions in 1999. The F2 segregation ratios of N to RV plants were compared to hypothetical ratios using Chi-square analyses.
Eleven cultivated lines homogeneous or with a high frequency of vigor restoration genes (Jan and Ruso, 2000) were emasculated and pollinated with HA 89. All the F1s were self-pollinated to obtain F2 progenies. For each cross, 40 F2 progenies were planted in the greenhouse to observe the segregation of N and RV plants.
Half-Diallel Analysis of Vigor Restoration (V) Genes in Restoration Lines
To tentatively test the hypothesis that the V genes in cultivated lines originated from a common source, HA 271, HA 234, VNIIMK 6540, Armavir 3497, Issanka, and HA 821 were included in a half-diallel cross. Testcrosses were made by pollinating CMS RIGX-RV plants with a HA 89 background with 15 F1s (Jan and Ruso, 2000). The use of CMS RIGX-RV plants as the female parent assured cross-pollination. The testcross progenies were evaluated in the greenhouse for plant vigor segregation.
Molecular Mapping of the V1 Gene
An F2 population including 124 individuals of G99/501-625 derived from MOL-RV/HA 821 was used to map the V1 gene from HA 821. The N and RV segregation of the F2 progenies were examined in the greenhouse in 1999, and their genotypes was further confirmed by using F3 progeny grown in the greenhouse in 2013–2014, with 20–40 progeny seedlings each.
Genomic DNA was extracted according to the protocol of the Qiagen DNAeasy 96 Plant Kit (Qiagen, Valencia, CA, USA). The bulked segregant analysis (BSA) method was used for polymorphism screening (Michelmore et al., 1991). The two parents and the two bulks were used for screening. The two bulks included a homozygous normal bulk (Bulk-N) and a homozygous reduced vigor bulk (Bulk-RV), using equal quantities of DNA from 10 F2 plants for each bulk. The PCR amplification and genotyping for SSR markers followed Liu et al. (2012). PCR amplification was conducted following Tang et al. (2002) with minor modifications. The 15-µl PCR reaction mixture contained 1× PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.27 µM each of the forward and reverse primers, 40 ng DNA and 1-unit Taq DNA polymerase (Qiagen). PCR amplifications were performed using the “touchdown” profile (Lai et al., 2005; Feng and Jan, 2008) in an MJ Research (Watertown, MA, USA) single or Bio-Rad (Hercules, CA, USA) single or dual 96-well thermal cycler. The PCR products were separated on a 6.5% denaturing polyacrylamide gel after denaturation at 95°C for 5 min, at 60 W for 2.0 h (1× TBE) after pre-run for 1.0 h or on a 6.5% non-denaturing polyacrylamide gel at 60 W for 1.0 h (0.5× TBE), on a CBP Scientific gel electrophoresis system. The gels were analyzed after being stained with GelRed nucleic acid gel stain (Biotium Inc., CA, USA) and scanned with a Typhoon 9410 variable mode imager (Molecular Dynamics Inc., CA, USA).
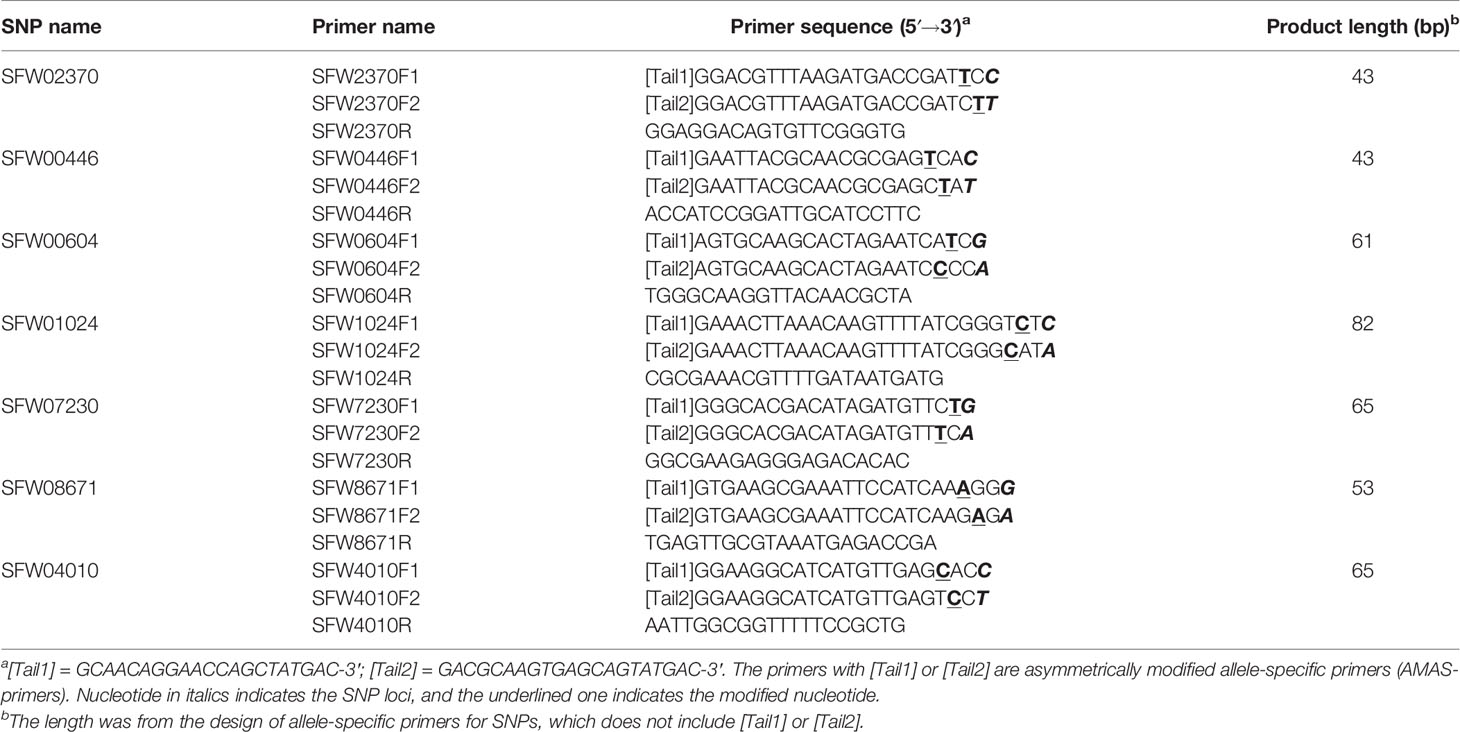
Bulked segregant analyses (Michelmore et al., 1991) were conducted for polymorphism screening using 550 SSR and expressed sequence tag (EST)-SSR primers on 17 LGs of sunflower, following the method of Liu et al. (2012). An additional 30 SSR/EST-SSR and InDel primers on the candidate LG 7 from 23 maps in the Sunflower CMap Database (http://sunflower.uga.edu/cgi-bin/cmap/map search) were used for polymorphism detection between the two parents. A total of 58 SSR/EST-SSR and InDel primers from LG 7 were used for polymorphism screening between parents and bulks. In addition, 30 sets of semi-thermal asymmetric reverse PCR (STARP) primers were designed according to the sequences of 30 SNP loci previously mapped on LG 7 (Bowers et al., 2012; Talukder et al., 2014; Hulke et al., 2015) following the method of Long et al. (2017). Each set of STARP primer included two universal priming element-adjustable primers (PEA-primers) and two asymmetrically modified allele-specific primers (AMAS-primers) in combination with one common reverse primer (Long et al., 2017). Polymorphism screening for the SNP markers was conducted among the two parents and Bulk-N. The PCR amplification system, program and product separation were performed as described in Liu et al. (2019). Briefly, the PCR program started with initial denaturation at 94°C for 3 min, followed by six cycles of 2-step touchdown PCR program, in which the Ta/e was decreased by 1°C per cycle starting at 94°C for 20 s and then 55°C for 2 min. PCR was continued with another 34 cycles of 2-step program at 94°C for 20 s and then 62°C for 2 min. The amplification was completed with a 2-min extension at 62°C. The PCR products for STARP markers were separated on a denaturing gel on IR2 4300/4200 DNA Analyzer (LI-COR, Lincoln, NE, USA). The polymorphic markers closely linked to the V1 gene were used for genotyping the mapping population.
The deviation analyses of the vigor trait and marker loci were compared with the expected Mendelian ratios in the F2 generation using the Chi-square test. The MAPMAKER/Exp version 3.0b program (Whitehead Institute, Cambridge, MA, USA) (Lander et al., 1987) was used for linkage analysis of the phenotypes and molecular genotypes, with a minimum LOD score of 4.0 and a maximum recombination frequency of 0.30. The Kosambi mapping function was used (Kosambi, 1944), with the “error detection on” command. The linkage map was generated using MapChart 2.3 (Voorrips, 2002).
Physical Location of V1 on LG 7
The analysis of the physical location of V1 and linked SNP and other markers on LG 7 was conducted by a BLASTn search against HA412.v1.1.bronze.20141015 on websites of the Sunflower Genome Database (https://sunflowergenome.org/) and the XRQ genome (https://www.heliagene.org/HanXRQ-SUNRISE/) on the INRA Sunflower Bioinformatics Resources (https://www.heliagene.org/) by using the sequences flanking the SNP markers and the sequences of other primers. The order of the linked markers on the genetic map of LG 7 was compared with those of Bowers et al. (2012) and Hulke et al. (2015).
Vigor Restoration of Cultivated Sunflower for Progenies With H. giganteus Cytoplasm
Helianthus giganteus 1934 was pollinated by HA 89 and the F1 plants were obtained via embryo rescue in 1995. One F1 plant was male-sterile and backcrossed with HA 89. Segregation of N and RV plants was observed in BC3F1. Three normal BC4F1 plants were pollinated with HA 89 pollen and the BC5F1 plants were evaluated for the segregation of N and RV plants. To study the genes controlling vigor restoration and fertility restoration, a normal-vigor and fertile F1 plant derived from the cross CMS GIG2//CMS GIG2/(NMS HA 89/H. maximiliani 1631, Amp) was selfed. The F2 progenies were phenotyped for both vigor and male fertility. Additionally, normal-vigor CMS GIG2 (pedigree: H. giganteus/6*HA89, CMS, Normal) plants were pollinated by HA 821 and testcrossed using HA 89 pollen. Progeny segregation patterns were used to determine the allelism of the vigor restoration genes derived from H. giganteus and in HA 821.
The F1 and Testcross Progeny Test for V Genes Derived From Different Sources
The F1s from crossing RV lines of HA 89 or HA 410 with five perennial Helianthus vigor-reducing cytoplasms of H. grosseserratus (GRO-RV), H. angustifolius (ANG-RV), H. salicifolius (SAL-RV), H. hirsutus (HIR-RV), and H. pauciflorus (PAU-RV) to six homozygous vigor restoration lines (HA 821, RF GIG2-MAX 1631, HIR, ANN PI 649856, ANN Bulk, and PET Bulk) were evaluated for segregation of plant vigor in 2016. Due to the discovery of homozygous V genes in both wild H. annuus (i.e. ANN Bulk and ANN PI 649856) and H. petiolaris (i.e. PET Bulk), partial half-diallel crosses among the six homozygous or heterozygous vigor restoration lines were established in the greenhouse in 2016, including three homozygous lines (HA 821, RF GIG2-MAX 1631, and HIR), and three heterozygous lines from ANN bulk, ANN PI 649856, and PET bulk. Then, six progeny plants from each cross were used to pollinate the CMS RIGX-RV with H. pauciflorus cytoplasm in 2016. Segregation of the N and RV F2s and testcross F1s was evaluated in the greenhouse in 2017. Progeny segregation of either 1N:1RV or no segregation suggests the same V gene for the two parents, and progeny segregation of either 1N:1RV or 3N:1RV suggests the two parents have different V genes. The data from progenies with segregation of 1N:1RV were not shown.
Results
Vigor Restoration of Cultivated Sunflower for Progenies With H. mollis Cytoplasm
Segregation patterns of the F1 progenies of the RV plants with H. mollis cytoplasm, MOL-RV, crossed with 14 cultivated sunflower lines in 1998 are shown in Table 2. Typical N and RV seedlings are shown in Figure 1. A high frequency of vigor restoration genes was found in the crosses involving 11 cultivated sunflower lines. The crosses of MOL-RV with HA 821, HA 234, and RHA 271 produced only normal progeny, suggesting the vigor restoration (V) genes in those lines were homozygous. The crosses of MOL-RV with HA 89, RHA 801, and Seneca produced only RV progeny, indicating that these lines did not have dominant vigor restoration genes. In addition, another inbred line, HA 410, doesn’t contain V genes (data not shown). Progeny from crosses with the remaining eight lines had a high frequency of normal plants, suggesting these lines contain V genes, although the V genes may not be homozygous. Since the MOL-RV plants were emasculated over several days, this low frequency of RV progeny could also be the result of accidental self-pollination.
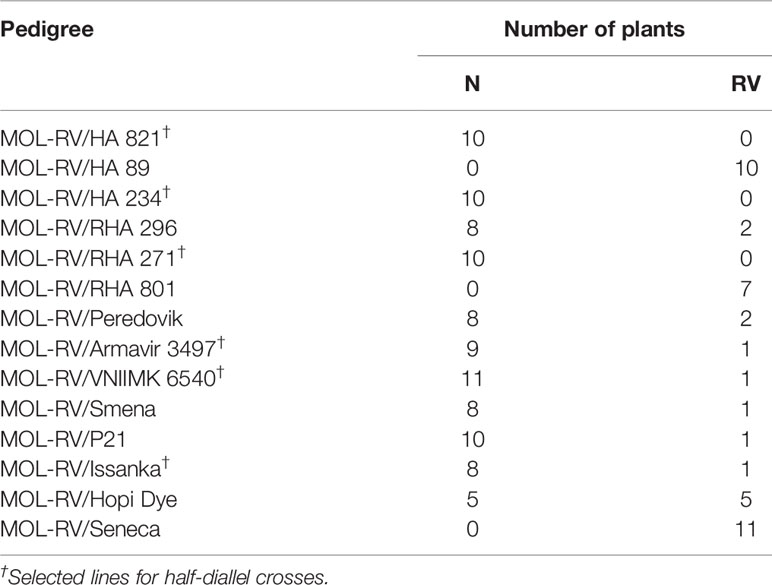
Table 2 Normal (N) and reduced-vigor (RV) plants in F1 progenies of MOL-RV crossed with 14 cultivated sunflower lines.

Figure 1 Representative reduced-vigor (RV) and normal (N) progeny plants of perennial Helianthus species crossed with cultivated sunflower lines at 35 days after planting. Left to right: RV progeny; normal progeny; HA 410; HA 89.
The F2 segregation ratios of N and RV plants of the MOL-RV line crossed with 10 cultivated sunflower lines are shown in Table 3. The segregation ratio of 3 N to 1 RV in nine of the 10 crosses was consistent with a single dominant gene hypothesis for control of vigor restoration. The segregation ratio of the cross MOL-RV/Armavir 3497 did not fit 3 N to 1 RV ratio (χ2 = 4.800, P = 0.028). The F2 plants of MOL-RV/Armavir 3497 were also tested for a segregation ratio of 15:1, which could not exclude two loci controlling the vigor restoration in Armavir 3497 (χ2 = 0.960, P = 0.327). However, the seeds used to produce reduced-vigor plants sometimes have lower germination rate compared to normal-vigor plants, so this maybe the most likely reason for the segregation ratio in this F2 population not fitting a 3:1 ratio. As a group, the homogeneity test with a probability of 0.241 also supports the single dominant gene hypothesis for vigor restoration.
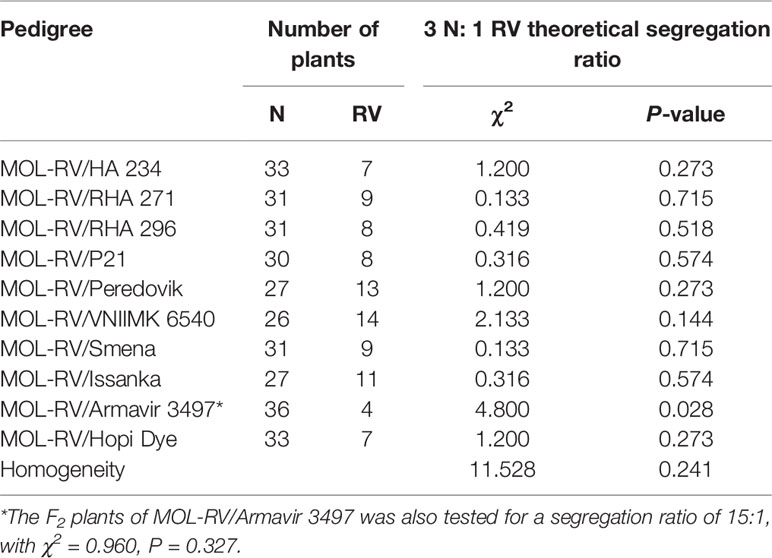
Table 3 Segregation of normal (N) and reduced-vigor (RV) F2 plants of MOL-RV crossed with 10 cultivated sunflower lines having vigor restoration (V) genes.
Half-Diallel Analysis of Vigor Restoration Genes in Cultivated Sunflower
The F1 hybrids of the 11 lines, including HA 271, HA 234, VNIIMK 6540, Armavir 3497, Issanka, HA 821, RHA 296, Peredovik, Smena, P21, and Hopi Dye, crossed with HA 89 were all N. With over 400 F2 progeny plants, 40 F2 progeny plants for each cross, there was not a single RV plant observed (data not shown). This suggested that there is no reduced vigor problem caused by cytonuclear interaction in these cultivated sunflower lines. The F1 progeny of the half-diallel crosses among six cultivated lines all had normal vigor, i.e., HA 271, HA 234, VNIIMK 6540, Armavir 3497, Issanka, and HA 821. The testcross progenies of the half-diallel crossed F1s among HA 271, HA 234, VNIIMK 6540, Armavir 3497, Issanka, and HA 821 onto the RV CMS RIGX were all normal, except for the testcross with VNIIMK 6540/Armavir 3497 F1, where a segregation ratio of 1 N to 1 RV plant was observed (Jan and Ruso, 2002). The 1 N to 1 RV segregation in the testcross using VNIIMK 6540/Armavir 3497 pollen could be due to a heterozygous F1 plant that resulted from a rare heterozygous parent. Therefore, the progeny test results suggested that all these lines possess the same V gene, designated V1.
Molecular Mapping of V1Gene on LG 7
The F2 population G99/501-625 derived from MOL-RV/HA 821 was used to map the V1 gene from HA 821. The 124 individuals in this population included 28 homozygous N, 59 heterozygous N, and 33 RV plants, confirmed by the progeny test, with four individuals not having enough seeds for progeny testing. Chi-square analysis indicated that the homozygous N: heterozygous N:RV phenotypes of the F2 population fit a 1:2:1 ratio (χ2 = 0.450, P = 0.799), suggesting a single dominant gene controlling the restoration of plant vigor.
BSA analysis using 550 SSR and EST-SSR primers from all 17 sunflower LGs among the two parents and the two bulks showed two polymorphic markers, ORS966 and ORS328, on LG 7. Further screening of 30 additional SSR markers on LG 7 identified three polymorphic markers, including two SSR markers, CRT136 and HT520, and one InDel marker ZVG31.
Of the 30 SNP markers tested on the LG 7 map, 18 showed polymorphisms between the two parents and Bulk-N (representative primers shown in Figure 2). The polymorphic SNPs were located at the 15.06–42.56 cM region on the scaffold-based genetic map of LG 7 of Hulke et al. (2015). The screening results showed that some markers were far from the V1 gene, with the Bulk-N containing the band from the RV parent. Therefore, the markers from the 26.94–35.55 cM region of the scaffold-based genetic map of LG 7 of Hulke et al. (2015) were used as the focal point to add more SNP markers around the V1 gene. Seven polymorphic PCR-based SNP markers from SFW02370 to SFW04010 were used to genotype the F2 mapping population, which were all co-dominant. The sequences and the product length of these STARP markers were shown in Table 4. As a result, a linkage map including 12 markers (four SSR, one InDel, and seven SNPs) and the V1 gene was constructed, covering a genetic distance of 36.7 cM (Figure 3B). The V1 gene co-segregated with the marker ZVG31, with three SNP markers, SFW01024, SFW07230, and SFW00604, located above V1 on the map at a genetic distance of 0.8 cM, and another SNP marker SFW08671, below it at a distance of 0.4 cM. Comparison of the order of markers on LG 7 containing V1 with those on the LG 7 of Hulke et al. (2015) (Figure 3A) and Bowers et al. (2012) (Figure 3C) showed the same order for most of the markers, except the order of ORS966, SFW00446, and ZVG31 between Figures 3B, C. The order of ORS966 and SFW00446 was reversed between Figures 3B, C. The distance between ORS966 and ZVG31 on Figure 3B was 3.2 cM, whereas they co-segregated on Figure 3C.
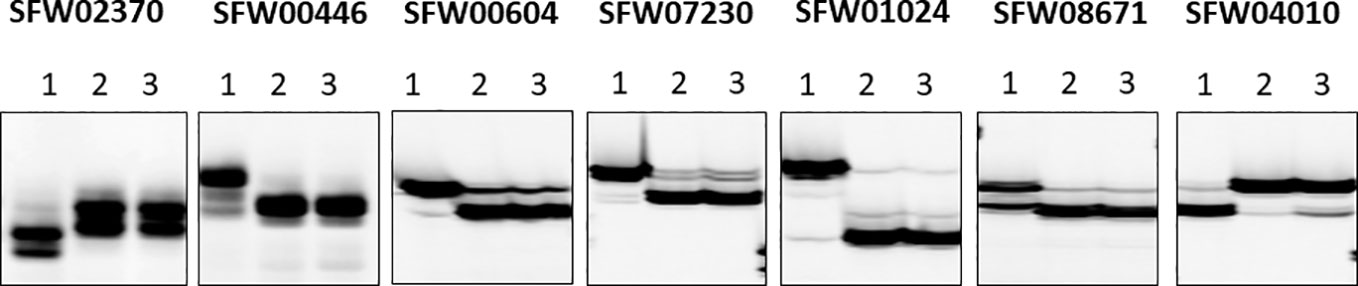
Figure 2 Representative image of seven semi-thermal asymmetric reverse PCR (STARP)-based SNP markers among parents and a bulk of normal F2 plants of the cross MOL-RV/HA 821 on a denaturing polyacrylamide gel. (Lane 1) Female parent MOL-RV; (Lane 2) Male parent HA 821; (Lane 3) Bulk-N.
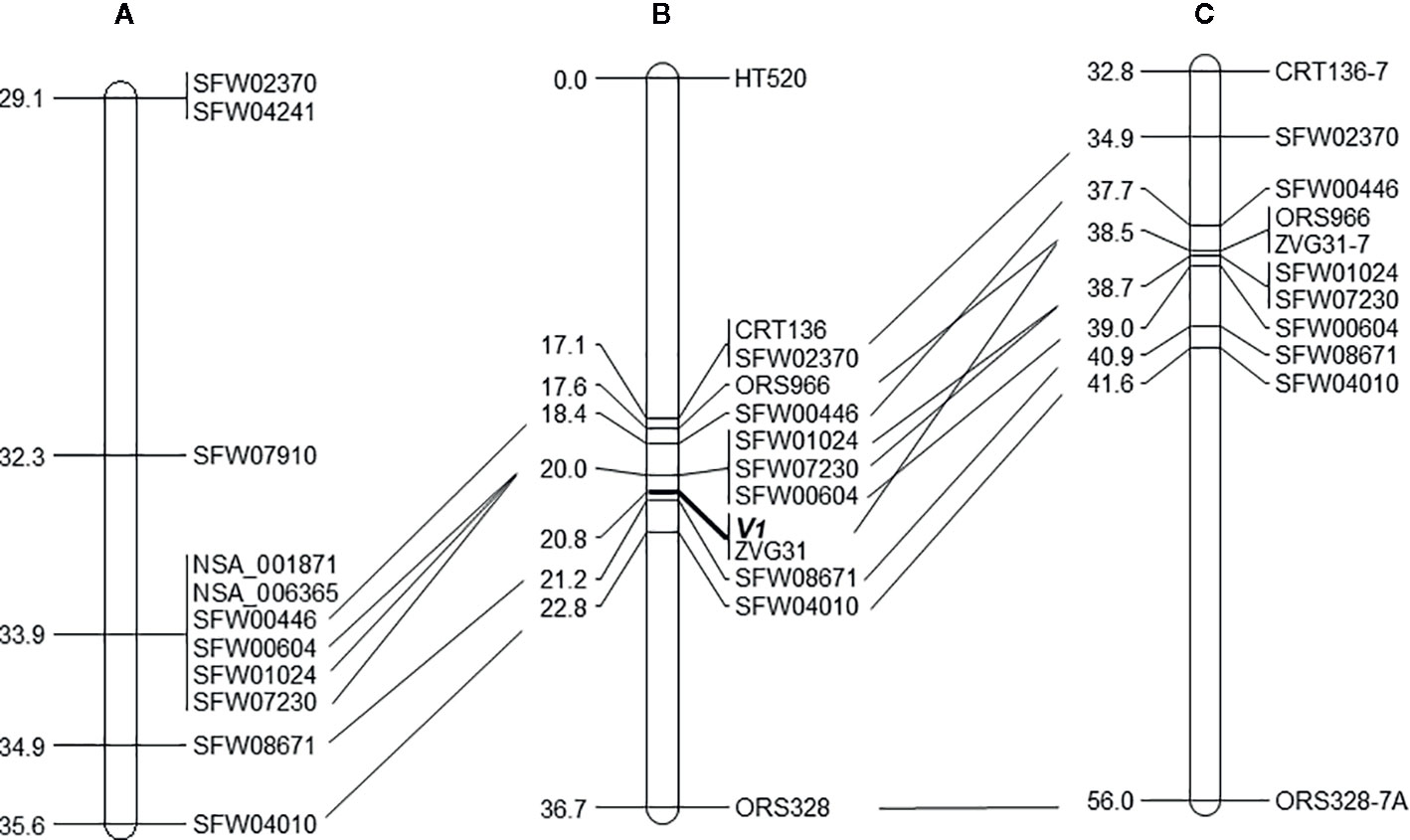
Figure 3 The position of the vigor restoration gene V1 on LG 7 of the sunflower map. (A) Scaffold-based genetic map of LG 7 with selected markers (Hulke et al., 2015); (B) Mapping result of the V1 gene on LG 7, with 12 linked markers, based on the analysis of 124 F2 plants derived from the cross of MOL-RV/HA 821; (C) Corresponding region of LG 7 of the genetic map of the sunflower genome based on multiple crosses (Bowers et al., 2012). The same markers are aligned by solid lines among the three maps.
Physical Location of V1 on LG 7
The sequences of the SNP markers and other markers closely linked to V1 were aligned to the reference genome sequences of HA 412-HO and XRQ, respectively (Table 5). The seven SFW SNP markers above and below the V1gene (from SFW02370 to SFW04010) on LG 7 span a 6.7-cM genetic distance on Figure 3C, which corresponds to a physical distance of 5.83 and 2.89 Mb on chromosome 7 of the HA 412-HO and XRQ assemblies, respectively. The order of the markers on the genetic maps is generally consistent with their physical order on the genome assemblies of HA 412-HO and XRQ, respectively, except for the order of the three co-segregated SNP markers SFW01024/SFW07230/SFW00604, and that of ZVG31 and SFW08671. SFW01024 was located above SFW00604 and SFW07230 on the HA 412-HO assembly, whereas it was below them on the XRQ assembly. The order of ZVG31 and SFW08671 was reversed compared between the HA 412-HO and XRQ assemblies. The V1gene was mapped between SNP markers SFW01024/SFW07230/SFW00604 and SFW08671 on LG 7, spanning a 1.2-cM genetic distance on Figure 3B. The physical distance between the two closest flanking SNP markers corresponds to 0.56 Mb at the 91,735,216–92,294,131 bp region (between SNP markers SFW07230 and SFW08671) on the HA 412-HO assembly, and 1.37 Mb at the 86,351,353–87,722,487 bp region (between SNP markers SFW01024 and SFW08671) on the XRQ assembly, respectively (Table 5). Interestingly, preliminary sequence analysis at the target region on the XRQ sunflower genome showed a chloroplastic NifU-like nitrogen fixation protein 3 gene, located at the 87,181,582–87,185,092 bp on LG 7, with a length of 3,511 bp. In addition, two other cytoplasmic-related genes were also detected at its nearby region of 87,185,542–87,205,817 bp. Previously, Yabe et al. (2004) reported a mutant Arabidopsis lacking a chloroplast-localized NifU-like nitrogen fixation protein AtCnfU-V exhibited a dwarf phenotype with faint pale-green leaves and had drastically impaired photosystem I accumulation.
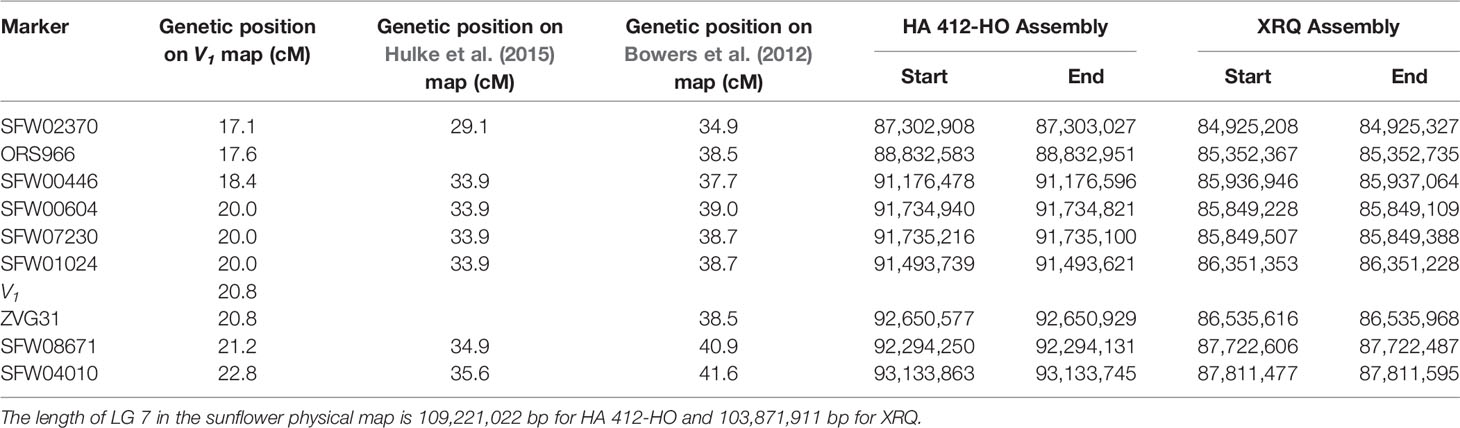
Table 5 Genetic and physical positions of the SNP and other markers linked to the V1 gene on linkage group (LG) 7.
Vigor Restoration of Cultivated Sunflower for Progenies With H. giganteus Cytoplasm
In the process of transferring Sclerotinia resistance and other useful genes from wild perennial Helianthus species into cultivated sunflower, we also observed the RV plants in the progenies derived from the crosses involving the perennial H. giganteus. Segregation of N and RV plants was observed starting with the BC3F1 generation of H. giganteus/HA 89 when the chromosome numbers ranged from 34 to 41. Approximately 50% of the BC4F1 plants had 2n = 34, while the remainder ranged from 2n = 35 to 36. Since HA 89 does not contain a vigor restoration gene for the vigor-reducing perennial species cytoplasm, normal BC4F1 plants with 2n = 34 must have obtained the vigor restoration gene from H. giganteus, and the BC5F1 progeny segregation of 33 N to 26 RV plants fit the 1 N to 1 RV ratio (χ2 = 0.831, P = 0.362), indicating a single dominant gene control of vigor restoration.
The amphiploid NMS HA 89/H. maximiliani 1631 provided the male-fertility restoration gene Rf4 to CMS GIG2 (Feng and Jan, 2008). Therefore, the F2 population derived from a normal-vigor and fertile F1 plant of cross CMS GIG2//CMS GIG2/(NMS HA 89/H. maximiliani 1631, Amp) segregated for both vigor and male-sterility. This F2 population included 82 N and 35 RV plants, which was consistent with the 3 N to 1 RV segregation ratio expected with single gene control of vigor restoration (χ2 = 1.507, P = 0.220). Meanwhile, the 79 male-fertile to 31 male-sterile progenies also fit a 3 MF to 1 MS ratio (χ2 = 0.594, P = 0.441), indicating a single gene control of fertility restoration. When the two traits, vigor and fertility restoration, were combined, the resulting 53, 26, 22, and nine plants of normal-vigor male-fertile, normal-vigor male-sterile, reduced vigor male-fertile, and reduced vigor male-sterile plants, respectively, fit a 9:3:3:1 ratio (χ2 = 0.748, P = 0.331), indicating the V and the Rf genes are not linked.
Progeny segregation of 14 normal plants of CMS GIG2/HA 821 crossed with HA 89 is shown in Table 6. Ten populations with segregation ratios of N to RV vigor plants fit the 3 N to 1 RV ratio, and four fit the 1 N to 1 RV ratio. CMS GIG2 was a normal plant, but its vigor restoration gene was heterozygous. The segregation of N and RV plants in all these progenies indicated that the vigor restoration gene derived from H. giganteus 1934 is different from the V1 gene commonly existing in cultivated lines, designated V2 here.
Table 6 Segregation of normal (N) and reduced-vigor (RV) plants in the progeny of cross CMS GIG2/HA 821//HA 89.
Other V Genes Derived From Wild Perennial Helianthus Species
The V genes derived from H. hirsutus and H. giganteus were compared to all other detected V genes. The F1 progenies of vigor-reducing lines with cytoplasms of H. grosseserratus (GRO-RV), H. angustifolius (ANG-RV), H. salicifolius (SAL-RV), H. hirsutus (HIR-RV), and H. pauciflorus (PAU-RV) substituted with the nuclear genomes of HA 89 or HA 410, pollinated by six normal lines having homozygous V genes (HA 821, RF GIG2-MAX 1631, HIR, ANN PI 649856, ANN Bulk and PET Bulk) were all normal (Table 7). The results of all normal progeny clearly indicated the common cytonuclear interaction defects in the progeny plant with perennial Helianthus cytoplasms and annual nuclear genomes, and the common function of vigor restoration genes from different sources of HA 821, H. giganteus, H. hirsutus, two H. annuus, and H. petiolaris.
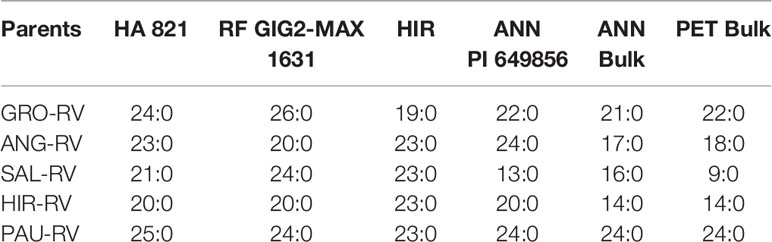
Table 7 Segregation of normal (N) and reduced-vigor (RV) progenies of F1s derived from RV lines of HA 89 or HA 410 with five perennial Helianthus vigor-reducing cytoplasms crossed with six homozygous vigor restoration lines.
The segregation of N and RV progenies of testcross progenies derived from the partial half-diallel crosses among the six homozygous or heterozygous vigor restoration lines are shown in Table 8. No segregation of plant vigor was observed in the progenies derived from the crosses involving four sources with normal vigor (HA 821, ANN Bulk, ANN PI 649856, and PET Bulk), indicating that they all contained the same V1gene for plant vigor restoration. Meanwhile, no segregation of plant vigor was observed in the progenies derived from the cross HIR/RF GIG2-MAX 1631 and the segregation of plant vigor was observed in the progenies derived from the crosses between HIR or RF GIG2-MAX 1631 and the four V1 sources, suggesting H. hirsutus PI 547174 and H. giganteus 1934 had the same V2 gene (Table 8).
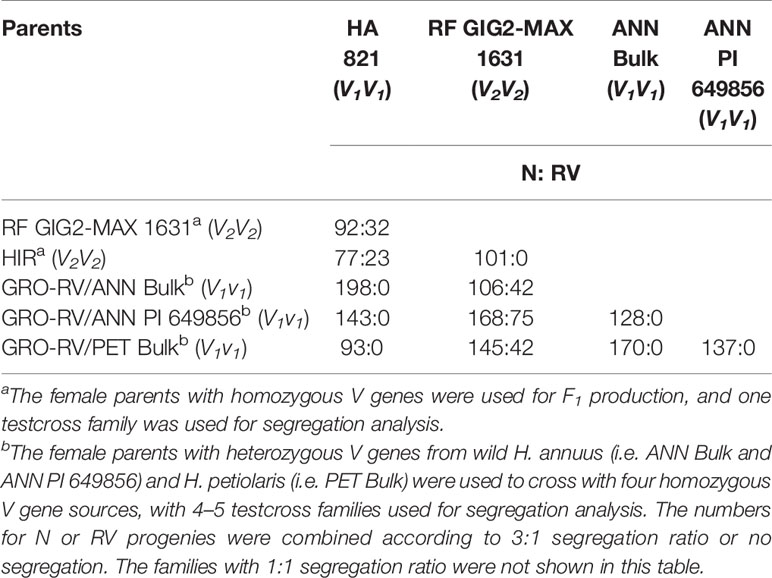
Table 8 Segregation of normal (N) and reduced-vigor (RV) progenies of testcross F1s derived from the partial half-diallel cross of females with homozygous or heterozygous V genes derived from six sources (HA 821, H. giganteus, H. hirsutus, H. annuus bulk, H. annuus PI 649856, and H. petiolaris bulk), pollinated onto CMS RIGX-RV (v1v1v2v2).
Discussion
The Existence of Vigor Restoration (V) Genes in Both Wild Helianthus Species and Cultivated Sunflower
Cytonuclear interactions could act as a source of variation for interspecific hybridization and may drive speciation (Levin, 2003). A study on wheat alloplasmic lines carrying the cytoplasm of Aegilops mutica showed that novel nuclear-cytoplasmic interactions can potentially trigger an epigenetic modification cascade in nuclear genes, which eventually change physiological traits, such as dry matter weight (Soltani et al., 2016). The research on Arabidopsis cytolines (each combining the nuclear genome of a natural variant with the cytoplasmic genomes of a different variant) indicated that genetic variation in organelle genomes could impact three seed physiological traits including dormancy, germination performance, and longevity (Boussardon et al., 2019). Their results also showed that natural parental accessions had contrasted contributions to the cytonuclear effect on germination phenotype depending whether they provided the nuclear or cytoplasmic genomes. In this study, we also detected a reduction of plant vigor with pale-green leaves and stunted growth in interspecific progenies involving different perennial Helianthus species, but only when using the wild perennial species as the maternal parent and cultivated sunflower as the paternal parent. No reduction of plant vigor was observed in the reciprocal crosses, or in progenies derived from the crosses involving wild annual Helianthus species. Therefore, a common cytoplasmic-nuclear interaction defect commonly exists in alloplasmic lines derived from wild perennial Helianthus species.
Since the vigor restoration gene was thought to exist in only the perennial Helianthus species, the high frequency of V in cultivated lines was not expected. Because the cytoplasms of other annual species do not cause adverse interaction with nuclear genes in cultivated sunflower, the CMS PET1 cytoplasm (derived from H. petiolaris, an annual species) (Leclercq, 1969) has been used successfully for hybrid sunflower production for about 50 years. However, if there is ever a need to use perennial Helianthus species cytoplasms, the abundance of V genes in cultivated germplasm lines should not hinder the utilization of perennial species cytoplasms in sunflower breeding programs. Our earlier work only demonstrated that wild annual Helianthus species did not produce RV plants, likely because they didn’t have vigor-reducing cytoplasms. For the tested materials in this study, the reduced-vigor trait was only observed in the progenies for the crosses using wild perennial species as the maternal parent, not for the two annual species. The current study has shown that the V genes in wild annual Helianthus species, including H. annuus and H. petiolaris, are the same as the V1 gene that commonly exists in the cultivated sunflower lines. Future research with diverse sources of annual and perennial species may be necessary to determine whether there is deficiency in their cytoplasms causing reduced vigor and the evolutionary role of V gene for the annual Helianthus species.
Similarly, an explanation for the high frequency of a V gene in cultivated lines without any obvious selective advantage is not clear. Since H. tuberosus has been used extensively in the improvement of cultivated sunflower (Fick and Miller, 1997), it is also possible that the V gene is tightly linked with genes controlling desirable agronomic traits and was simultaneously selected and maintained in those lines. As more sunflower genes are mapped, the prevalence of the V gene in cultivated lines may eventually be more clearly explained.
H. hirsutus PI 547174 and H. giganteus 1934 Had a Different V Gene Than Other Helianthus Species
The crosses involving five perennial Helianthus vigor-reducing cytoplasms with six homozygous vigor restoration lines have assessed the commonality of the cytoplasmic-nuclear interaction defect of RV cytoplasms from different perennial Helianthus species, as well as the vigor restoration genes. The segregation patterns of the progenies of the partial half-diallel crosses among the six homozygous/heterozygous vigor restoration lines indicated that H. hirsutus PI 547174 and H. giganteus 1934 had a different V gene than other Helianthus species. For the convenience of future research, we have designated the vigor restoration gene identified in 1992 (Jan, 1992) and those identified in cultivated lines as V1, and the V from H. giganteus and H. hirsutus as V2, respectively. Although the two V genes are located on different loci, they both can restore the plant vigor of the progeny containing different cytoplasms, which suggests that they can compensate for a common cytonuclear interaction defect causing reduced plant vigor.
The segregation of normal plants in the F1 and F2 progeny of MOL-RV/P21 indicated that P21 has the V gene to restore the reduced vigor trait (Tables 2 and 3). P21 has been used to produce several amphiploids for sunflower improvement (Liu et al., 2013; North Dakota State University Foundation Seedstocks (NDSUFS), website: https://www.ag.ndsu.edu/fss/ndsu-varieties/usda-sunflower-inbred-lines). Therefore, in the study of V genes in sunflower, one needs to avoid adding more V genes by carefully checking their pedigree not involving P21 in amphiploids. The V gene discovered in other sources will need to be compared with the V1 and V2 genes for allelism, as well as their effectiveness for restoring other perennial RV cytoplasms. Further study of the RV and its restoration caused by the cytoponuclear gene interaction in multi-species may help to elucidate the speciation of annual and perennial Helianthus species.
The V1 Gene Was Mapped on LG 7 Using SNP and Other Markers
According to BSA screening results between Bulk-N and Bulk-RV using the already mapped SSR/InDel markers, the V1 gene was mapped to LG 7. However, only five markers were linked to V1. With the aim of adding more markers close to V1, according to the linkage maps with high-density of SNPs, 30 SNP markers were selected to design PCR-based SNP markers. The markers in a focused region from 26.94 to 35.55 cM on the scaffold-based genetic map of LG 7 of Hulke et al. (2015) were used for further genotyping of the F2 population. Seven co-dominant SNP markers were successfully added to the map on both sides of V1. Using the flanking sequences of the SNP markers and primer sequences of other markers, the V1 gene has been located onto the physical map of chromosome 7 of the sunflower genome, i.e., a 0.56 Mb region on the HA 412-HO assembly, and a 1.37 Mb region on the XRQ assembly. The tightly linked molecular markers identified in this study will facilitate the marker-assisted selection for the lines with vigor restoration genes at the early stages of sunflower breeding, especially utilizing the CWR.
The pattern of cytonuclear interactions is the result of a long-term coevolution between nuclear and organellar genomes under selection pressure, which is essential for the proper function of plant cells (Postel and Touzet, 2020). When using organellar markers to evaluate phylogenetic relationships for characterizing genetic diversity, mitochondrial and chloroplast genes often show markedly different phylogenetic patterns from nuclear markers, which is called “cytonuclear discordance” (Lee-Yaw et al., 2019). Phylogenetic analyses using whole-chloroplast sequence data in combination with over 1000 nuclear SNPs in wild annual Helianthus indicate that cytonuclear discordance is widespread both among species and among individuals within species. Since mitochondria and chloroplasts affect key physiological processes, selection may have played a role in driving patterns of plastid variation (Lee-Yaw et al., 2019). In this study, the existence of V genes to different Helianthus cytoplasms and the vigor restoration ability across the Helianthus species provides another piece of evidence for cytonuclear discordance in Helianthus genus. Since there is clear distinction between the nuclear and chloroplast of annual and perennial Helianthus species (Stephens et al., 2015; Lee-Yaw et al., 2019), the V genes contained in the nuclear genomes of both annual and perennial species suggest that the V genes may have evolved before the speciation of annual and perennial Helianthus species.
Many cytonuclear incompatibilities are caused by plastid-nuclear incompatibilities, which has been reported in many flowering plants, such as in Passiflora, Oenothera and Pisum. Such incompatibilities often produce lutescent, chlorosis/virescence, or variegation, because of a decreased photosynthetic function of plastid complex malfunction in the plants (Greiner et al., 2011; Postel and Touzet, 2020). On the other hand, the incompatibility between mitochondrial and the nuclear genomes will often cause CMS (Postel and Touzet, 2020). As a result, several corresponding nuclear Rf gene have been identified and molecularly mapped in sunflower for different CMS sources, such as Rf1 for CMS PET-1, Rf4 for CMS GIG2, and Rf6 for CMS 514A (Horn et al., 2003; Feng and Jan, 2008; Liu et al., 2013).
In this study, we have mapped the vigor restoration gene V1 to the LG 7 of HA 412-HO and XRQ sunflower assemblies. With these targeted regions containing the V1 gene, identification of more molecular markers, such as SNPs or SSR, according to the genomic DNA sequences of the sunflower genome will facilitate fine mapping of the V1 gene, as well as future map-based cloning of the V1 gene. Further fine-mapping, detailed analysis of the genes contained in the corresponding regions of the two assemblies, plus using microarray and RNA-Seq (RNA sequencing) techniques (Wang et al., 2009) will help to identify candidate genes for plant vigor restoration. Using other methods such as RT-PCR, gene knock-out, or gene-editing will confirm the function of the gene, and thus will reveal the mechanism for vigor reduction and restoration in sunflower and will help to understand the interaction between the cytoplasm and nuclear genes. Therefore, the inheritance study and molecular mapping of vigor restoration gene in this study will also provide evidence for the speciation of annual and perennial Helianthus species. Finally, the results of these studies will provide the basis for better and more efficient utilization of sunflower CWR in crop improvement.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
C-CJ and ZL conceived and designed the research. ZL, GU, C-CJ performed the experiments, ZL and C-CJ analyzed the data and ZL, C-CJ and GS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The project was supported by the USDA-ARS National Sclerotinia Initiative, Grant No. 3060-21220-028-00D, the USDA-ARS CRIS Project No. 3060-21000-043-00D, and the Heilongjiang Postdoctoral Fund of China, Grant No. LBH-Z14190. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity lender, provider, and employer.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Lisa A. Brown, Cheryl A. Huckle, and Drs. Yunming Long and Zahirul Talukder for technical assistance, and Dr. Lili Qi for kindly providing wild H. annuus pollen and several important primers in this study.
References
Anderson, J., Kantar, M., Bock, D., Grubbs, K. C., Schilling, E., Rieseberg, L. (2019). Skim-sequencing reveals the likely origin of the enigmatic endangered sunflower Helianthus schweinitzii. Genes (Basel) 10 (12), pii: E1040. doi: 10.3390/genes10121040
Boussardon, C., Martin-Magniette, M. L., Godin, B., Benamar, A., Vittrant, B., Citerne, S., et al. (2019). Novel cytonuclear combinations modify Arabidopsis thaliana seed physiology and vigor. Front. Plant Sci. 10, 32. doi: 10.3389/fpls.2019.00032
Bowers, J. E., Bachlava, E., Brunick, R. L., Rieseberg, L. H., Knapp, S. J., Burke, J. M. (2012). Development of a 10,000-locus genetic map of the sunflower genome based on multiple crosses. Genes Genomes Genet. 2, 721–729. doi: 10.1534/g3.112.002659
Burton, R. S., Pereira, R. J., Barreto, F. S. (2013). Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44, 281–302. doi: 10.1146/annurev-ecolsys-110512-135758
Feng, J., Jan, C. C. (2008). Introgression and molecular tagging of Rf4, a new male fertility restoration gene from wild sunflower Helianthus maximiliani L. Theor. Appl. Genet. 117, 241–249. doi: 10.1007/s00122-008-0769-4
Feng, J., Liu, Z., Seiler, G. J., Jan, C. C. (2015). Registration of cytoplasmic male-sterile oilseed sunflower genetic stocks CMS GIG2 and CMS GIG2-RV, and fertility restoration lines RF GIG2-MAX 1631 and RF GIG2-MAX 1631-RV. J. Plant Regist. 9, 125–127. doi: 10.3198/jpr2014.05.0029crgs
Fick, G. N., Miller, J. F. (1997). “Sunflower breeding,” in Sunflower Technology and Production. Ed. Schneiter, A. A. (Madison, WI: ASA, CSSA, and SSSA, Press), 395–440.
Greiner, S., Rauwolf, U., Meurer, J., Herrmann, R. G. (2011). The role of plastids in plant speciation. Mol. Ecol. 20, 671–691. doi: 10.1111/j.1365-294X.2010.04984.x
Horn, R., Kusterer, B., Lazarescu, E., Prüfe, M., Friedt, W. (2003). Molecular mapping of the Rf1 gene restoring pollen fertility in PET1-based F1 hybrids in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 106, 599–606. doi: 10.1007/s00122-002-1078-y
Hulke, B. S., Grassa, C. J., Bowers, J. E., Burke, J. M., Qi, L. L., Talukder, Z. I., et al. (2015). A unified SNP map of sunflower (Helianthus annuus L.) derived from the sunflower genome project resources. Crop Sci. 55, 1696–1702. doi: 10.2135/cropsci2014.11.0752
Jan, C. C., Fernandez-Martinez, J. M. (2002). Interspecific hybridization, gene transfer, and the development of resistance to broomrape race F in Spain. Helia 25, 123–136. doi: 10.2298/HEL0236123J
Jan, C. C., Ruso, J. (2000). Vigor reducing cytoplasms of perennial Helianthus species and their nuclear fertility restoration genes in cultivated sunflower lines, in: Proceedings of the 22nd Sunflower Research Workshop, Fargo, ND, January 18-19, 2000, (Bismarck, ND: National Sunflower Assoc.) p. 51–53.
Jan, C. C., Ruso, J. (2002). Nuclear vigor restoration genes in cultivated sunflower that restore the vigor reducing cytoplasms of perennial Helianthus species, in: Proceedings of the 24th Sunflower Research Workshop, Fargo, ND, January 17-18, 2002, (Bismarck, ND: National Sunflower Assoc.) p. 159–161.
Jan, C. C., Vick, B. A. (2006). Registration of seven cytoplasmic male-sterile and four fertility restoration sunflower germplasms. Crop Sci. 46, 1829–1830. doi: 10.2135/cropsci2005.12-0510
Jan, C. C., Fernandez-Martinez, J. M., Ruso, J., Munoz-Ruz, J. (2002). Registration of four sunflower germplasms with resistance to Orobanche cumana Race F. Crop Sci. 42, 2217–2218. doi: 10.2135/cropsci2002.2217
Jan, C. C. (1988). “Chromosome doubling of wild x cultivated sunflower interspecific hybrids and its direct effect on backcross success,” in Proceedings of the 12th International Sunflower Conference, Novi Sad, Yugoslavia, July 25-29, 1988, (Paris, France: International Sunflower Assoc.), 287–292.
Jan, C. C. (1992). Cytoplasmic-nuclear gene interaction for plant vigor in Helianthus species. Crop Sci. 32, 320–323. doi: 10.2135/cropsci1992.0011183X003200020007x
Jan, C. C. (1995). Allelic relationship among vigor restoration genes derived from five perennial Helianthus species. Agronomy Abstr. (Madison, WI: ASA Press), 81.
Kosambi, D. D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Lai, Z., Livingstone, K., Zou, Y., Church, S. A., Knapp, S. J., Andrews, J., et al. (2005). Identification and mapping of SNPs from ESTs in sunflower. Theor. Appl. Genet. 111, 1532–1544. doi: 10.1007/s00122-005-0082-4
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Stephen, E. L., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Leclercq, P. (1969). Une sterilite male cytoplasmique chez le tournesol. Ann. Amelior. Plant 19, 99–106.
Lee-Yaw, J. A., Grassa, C. J., Joly, S., Andrew, R. L., Rieseberg, L. H. (2019). An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytol. 221, 515–526. doi: 10.1111/nph.15386
Levin, D. A. (2003). The cytoplasmic factor in plant speciation. Syst. Bot. 28, 5–11. doi: 10.1043/0363-6445-28.1.5
Liu, Z., Gulya, T. J., Seiler, G. J., Vick, B. A., Jan, C. C. (2012). Molecular mapping of the Pl16 downy mildew resistance gene from HA- R4 to facilitate marker-assisted selection in sunflower. Theor. Appl. Genet. 125, 121–131. doi: 10.1007/s00122-012-1820-z
Liu, Z., Wang, D., Feng, J., Seiler, G. J., Cai, X., Jan, C. C. (2013). Diversifying sunflower germplasm by integration and mapping of a novel male fertility restoration gene. Genetics 193, 727–737. doi: 10.1534/genetics.112.146092
Liu, Z., Zhang, L., Ma, G. J., Seiler, G. J., Jan, C. C., Qi, L. L. (2019). Molecular mapping of the downy mildew and rust resistance genes in a sunflower germplasm line TX16R. Mol. Breed. 39, 19. doi: 10.1007/s11032-018-0921-z
Long, Y. M., Chao, W. S., Ma, G. J., Xu, S. S., Qi, L. L. (2017). An innovative SNP genotyping method adapting multiple platforms and throughputs. Theor. Appl. Genet. 130, 597–607. doi: 10.1007/s00122-016-2838-4
Mammadov, J., Buyyarapu, R., Guttikonda, S. K., Parliament, K., Abdurakhmonov, I. Y., Kumpatla, S. P. (2018). Wild relatives of maize, rice, cotton, and soybean: Treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 9, 886. doi: 10.3389/fpls.2018.00886
Michelmore, R. W., Paran, I., Kesseli, R. V. (1991). Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Postel, Z., Touzet, P. (2020). Cytonuclear genetic incompatibilities in plant speciation. Plants (Basel) 9 (4), pii: E487. doi: 10.3390/plants9040487
Purdy, L. H., Loegering, W. Q., Konzak, C. F., Peterson, C. J., Allan, R. E. (1968). A proposed standard method for illustrating pedigrees of small grain varieties. Crop Sci. 8, 405–406. doi: 10.2135/cropsci1968.0011183X000800040002x
Sambatti, J. B., Ortiz-Barrientos, D., Baack, E. J., Rieseberg, L. H. (2008). Ecological selection maintains cytonuclear incompatibilities in hybridizing sunflowers. Ecol. Lett. 11, 1082–1091. doi: 10.1111/j.1461-0248.2008.01224.x
Seiler, G. J., Qi, L. L., Marek, L. F. (2017). Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 57, 1083–1101. doi: 10.2135/cropsci2016.10.0856
Soltani, A., Kumar, A., Mergoum, M., Pirseyedi, S. M., Hegstad, J. B., Mazaheri, M., et al. (2016). Novel nuclear-cytoplasmic interaction in wheat (Triticum aestivum) induces vigorous plants. Funct. Integr. Genomics 16, 171–182. doi: 10.1007/s10142-016-0475-2
Stephens, J. D., Rogers, W. L., Mason, C. M., Donovan, L. A., Malmberg, R. L. (2015). Species tree estimation of diploid Helianthus (Asteraceae) using target enrichment. Am. J. Bot. 102, 910–920. doi: 10.3732/ajb.1500031
Sukno, S., Ruso, J., Jan, C. C., Melero-Vara, J. M., Fernandez-Martinez, J. M. (1999). Interspecific hybridization between sunflower and wild perennial Helianthus species via embryo rescue. Euphytica 106, 69–78. doi: 10.1023/A:1003524822284
Talukder, Z. I., Gong, L., Hulke, B. S., Pegadaraju, V., Song, Q. J., Schultz, Q., et al. (2014). A high-density SNP map of sunflower derived from RAD-sequencing facilitating fine-mapping of the rust resistance gene R12. PloS One 9 (7), e98628. doi: 10.1371/journal.pone.0098628
Tang, S., Yu, J. K., Slabaugh, B., Shintani, K., Knapp, J. (2002). Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105, 1124–1136. doi: 10.1007/s00122-002-0989-y
Vear, F., Cadic, E., Vincourt, P. (2011). Diversity among cultivated sunflower resources and use in breeding. Helia 34, 21–30. doi: 10.2298/HEL1155021V
Vear, F. (2010). “Classic Genetics and Breeding,” in Genetics, Genomics and Breeding of Sunflower. Eds. Hu, J., Seiler, G., Kole, C. (Science Publisher: Enfield, NH), 51–78.
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Z., Gerstein, M., Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Yabe, T., Morimoto, K., Kikuchi, S., Nishio, K., Terashima, I., Nakai, M. (2004). The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16, 993–1007. doi: 10.1105/tpc.020511
Keywords: Helianthus, vigor-reducing cytoplasms, vigor restoration genes, SNP, gene mapping
Citation: Liu Z, Gu W, Seiler GJ and Jan C-C (2020) A Unique Cytoplasmic–Nuclear Interaction in Sunflower (Helianthus annuus L.) Causing Reduced-Vigor Plants and the Genetics of Vigor Restoration. Front. Plant Sci. 11:1010. doi: 10.3389/fpls.2020.01010
Received: 02 March 2020; Accepted: 19 June 2020;
Published: 10 July 2020.
Edited by:
Petr Smýkal, Palacký University, CzechiaReviewed by:
Françoise Budar, INRA UMR1318 Institut Jean Pierre Bourgin, FranceAlejandro Presotto, Universidad Nacional del Sur, Argentina
Copyright © 2020 Liu, Gu, Seiler and Jan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerald J. Seiler, gerald.seiler@usda.gov
 Zhao Liu1
Zhao Liu1 Wei Gu
Wei Gu Gerald J. Seiler
Gerald J. Seiler