
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 June 2020
Sec. Plant Pathogen Interactions
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00919
 Ahmed S. M. Elnahal1,2
Ahmed S. M. Elnahal1,2 Jinyang Li1
Jinyang Li1 Xiaoxia Wang1
Xiaoxia Wang1 Chenyao Zhou1
Chenyao Zhou1 Guohong Wen3
Guohong Wen3 Jian Wang4
Jian Wang4 Hannele Lindqvist-Kreuze5
Hannele Lindqvist-Kreuze5 Yuling Meng6
Yuling Meng6 Weixing Shan6*
Weixing Shan6*Late blight is considered the most renowned devastating potato disease worldwide. Resistance gene (R)-based resistance to late blight is the most effective method to inhibit infection by the causal agent Phytophthora infestans. However, the limited availability of resistant potato varieties and the rapid loss of R resistance, caused by P. infestans virulence variability, make disease control rely on fungicide application. We employed an Agrobacterium tumefaciens-mediated transient gene expression assay and effector biology approach to understand late blight resistance of Chinese varieties that showed years of promising field performance. We are particularly interested in PiAvr3aEM, the most common virulent allele of PiAvr3aKI that triggers a R3a-mediated hypersensitive response (HR) and late blight resistance. Through our significantly improved A. tumefaciens-mediated transient gene expression assay in potato using cultured seedlings, we characterized two dominant potato varieties, Qingshu9 and Longshu7, in China by transient expression of P. infestans effector genes. Transient expression of 10 known avirulence genes showed that PiAvr4 and PiAvr8 (PiAvrsmira2) could induce HR in Qingshu9, and PiAvrvnt1.1 in Longshu7, respectively. Our study also indicated that PiAvr3aEM is recognized by these two potato varieties, and is likely involved in their significant field performance of late blight resistance. The identification of natural resistance mediated by PiAvr3aEM recognition in Qingshu9 and Longshu7 will facilitate breeding for improved potato resistance against P. infestans.
Potato (Solanum tuberosum L.) is regarded as the fourth-largest food crop and the main non-cereal crop worldwide which is influenced by the destructive and notorious late blight disease (Aguilera-Galvez et al., 2018). Phytophthora infestans is the causative agent that can destroy all potato parts, including leaves, stems and tubers (Fry, 2008), and is the main threat to potato production and responsible for 16% of yield losses globally (Haverkort et al., 2016). Similar to other crops, disease management using resistant varieties is one of the most effective strategies, environmentally and economically, to control late blight disease (Fry, 2016). Plant immunity is activated by detecting conserved microbial molecules, microbe (pathogen)-associated molecular patterns (MAMPs or PAMPs), known as pattern-triggered immunity (PTI), as well as by detecting the pathogen effectors, known as effector-triggered immunity (ETI) (Jones and Dangl, 2006). Plant pathogens can successfully colonize plant hosts by delivering effector proteins that repress host immunity and increase disease severity (Turnbull et al., 2017). In turn, few effectors might be recognized by the correspondent resistance (R) proteins, triggering a rapid immune response known as effector-triggered immunity (ETI), which often leads to an hypersensitive response (HR) cell death (Jones and Dangl, 2006; Turnbull et al., 2017).
To achieve effective control of late blight, potato breeders have to adopt novel techniques and strategies in various aspects such as detection and identification of new Rpi genes, their introgression and field application (Vleeshouwers and Oliver, 2014). Great efforts were made at the beginning of the last century to introgress Rpi genes into potato varieties from the wild Mexican species Solanum demissum to provide resistance to P. infestans in the cultivated potato S. tuberosum. This also led to the development of differential potato lines with 11 distinct recognition specificities, called R1-R11 (Black et al., 1953; Malcolmson and Black, 1966). Many R genes (Rpi) have been identified, cloned and some of them were introgressed into potato cultivars from wild Mexican Solanum species (Goodwin et al., 1992; Grunwald and Flier, 2005; Goss et al., 2014), including R1–R11, R3a, R3b, R9a and Rpi-demf1 from S. demissum (Ballvora et al., 2002; Huang et al., 2005; Lokossou et al., 2009; Jo et al., 2015), Rpi-blb1(RB), Rpi-blb2, Rpi-blb3, Rpi-abpt and Rpi-bt1 from S. bulbocastanum (Song et al., 2003; Van der Vossen et al., 2003; Park et al., 2005; Van der Vossen et al., 2005; Lokossou et al., 2009), Rpi-sto1,2, Rpi-pta1,2, and Rpi-plt1 from S. stoloniferum (Vleeshouwers et al., 2008; Wang et al., 2008; Champouret et al., 2009), Rpi-amr3 from S. americanum (Witek et al., 2016), Rpi-mch1 from S. michoacanum (Sliwka et al., 2012a), and Rpi1 from S. pinnatisectum (Kuhl et al., 2001; Lokossou et al., 2010). Additionally, further R genes have been identified in another center of genetic diversity of tuber-bearing Solanum, the Andean region in South America, such as Rpi-vnt1 from S. venturii (Foster et al., 2009; Pel et al., 2009), Rpi-mcq1 from S. mochiquense (Smilde et al., 2005), Rpi-ber from S. berthaultii (Rauscher et al., 2006), Rpi-mcd1 from S. microdontum (Tan et al., 2008), Rpi-pcs from S. paucissectum (Villamon et al., 2005), Rpi-cap1 from S. capsicibaccatum (Jacobs et al., 2010), Rpi-rzc1 from S. ruiz-ceballosii (syn. S. sparsipilum) (Śliwka et al., 2012b; Brylińska et al., 2015) and Rpi-chc1 from S. chacoense (Zhu et al., 2015).
Much attention has been paid to RXLR effectors since the cloned avirulence protein (Avr) of oomycete pathogens belong to this type of effector, such as P. sojae Avr1b (Shan et al., 2004), Hyaloperonospora arabidopsidis ATR13 (Allen et al., 2004) and ATR1 (Rehmany et al., 2005), and P. infestans PiAvr3a (Armstrong et al., 2005). So far, over 10 Avr genes, identified by 10 cognate-recognition Rpi genes, have been described in P. infestans, including PiAvr1 (Van der Lee et al., 2001; Ballvora et al., 2002), PiAvr2 (Gilroy et al., 2011), PiAvr3a (Armstrong et al., 2005), PiAvr3b (Rietman et al., 2012), PiAvr4 (Van Poppel et al., 2008), PiAvr8 (Vossen et al., 2016), PiAvrblb1 (Song et al., 2003; Vleeshouwers et al., 2008), PiAvrblb2 (Van der Vossen et al., 2005; Oh et al., 2009), PiAvrvnt1 (Foster et al., 2009; Pel et al., 2009), PiAvrSmira1 and PiAvrSmira2 (Rietman et al., 2012; Yoshida et al., 2013) as shown in (Supplementary Table S1). P. infestans is predicted to encode 563 RXLR effector genes which were mainly found located in repeat-rich or gene-sparse regions of the genome, meaning that more rapid evolution compared to other genes located in gene-dense regions (Haas et al., 2009; Yin et al., 2017). It might explain its ability to escape host defense mechanisms (Lenman et al., 2016). It has been well-documented that RXLR effectors play important roles in potato–P. infestans interactions. In rare cases of recognition by the cognate R genes, they mediate late blight resistance by triggering HR; in most cases, they are not recognized and function as typical virulence factors by interfering with host cell structure and function, resulting in enhancing plant susceptibility (Huang et al., 2019).
There are generally two strategies to improve late blight resistance. The first is the deployment of many different R genes to offer tentative durable resistance since changes of multiple effectors are predicted to increase the penalty. The second strategy is to identify genes that are capable of recognizing various effectors or core effectors. In fact, the identified R genes from varieties that showed durable disease resistance were confirmed to be able to recognize two or more effectors. For example, the potato Rpi-blb1, known as RB (Song et al., 2003) recognizes PiAvrblb1, ipiO1 and ipiO2 (Vleeshouwers et al., 2008; Chen et al., 2012); the Rps1k of soybean recognizes two P. sojae effectors, Avr1b and Avr1k (Shan et al., 2004; Dou et al., 2010).
Yet, P. infestans PiAvr3a is a well-characterized P. infestans RXLR effector that is highly expressed during the biotrophic phase of infection [2–3 days post in filtration (dpi)] (Haas et al., 2009; Chaparro-Garcia et al., 2015). PiAvr3a is essential for full virulence, pathogenicity and suppression of host immunity, including PTI and ETI, by suppressing the programmed cell death (PCD) triggered by the elicitin INF1, a secreted P. infestans protein with PAMP properties, by interacting with and stabilizing the host U-box E3 ligase CMPG1 (Bos et al., 2006; González-Lamothe et al., 2006; Bos et al., 2010; Gilroy et al., 2011), as well as targeting the receptor-mediated endocytosis dynamin-related protein 2B (DRP2B), clathrin-mediated endocytosis (CME) (Chaparro-Garcia et al., 2015). Two major allelic isoforms of PiAvr3a have been identified in P. infestans populations that have a difference in three amino acids in mature protein positions 19, 80 and 103 (Chapman et al., 2014). Avr3a (S19)E80M103 is known as PiAvr3aEM while Avr3a (C19)K80I103 is known as PiAvr3aKI (Vleeshouwers et al., 2011; Yang et al., 2018). Unlike PiAvr3aEM, PiAvr3aKI activates the potato R3a resistance protein to trigger ETI and confers avirulence to heterozygous or homozygous strains of the pathogen (Armstrong et al., 2005; Chaparro-Garcia et al., 2015). Therefore, P. infestans isolates, expressing only the PiAvr3aEM variant, can evade R3a recognition and do not trigger HR (Armstrong et al., 2005; Bos et al., 2006). Identification of natural R genes that can recognize PiAvr3aEM, are promising approaches to improve late blight resistance (Bos et al., 2010; Segretin et al., 2014).
Agrobacterium tumefaciens-mediated transient gene expression technology is a rapid, widely and easily performed assay that is commonly sed in gene expression analysis and functional genomics studies in many plant species, including Arabidopsis thaliana, tobacco, tomato, soybean, citrus, grapevine and potato (Vleeshouwers et al., 2008). Typically, A. tumefaciens-mediated transient expression assays can be utilized for several purposes, such as i) functional genomics tools for transient overexpression of a gene in planta, ii) reverse genetic studies of a gene by virus-induced gene silencing (VIGS) or RNA interference (RNAi) technology, iii) rapid accessible production of recombinant proteins, iv) pathogen effector assays for the genetic components of the selected cultivars disease resistance.
In this study, we utilized the optimum conditions for A. tumefaciens-mediated transient assays in potato and performed analyses of two potato varieties for their capability to recognize a set of P. infestans known effectors, as part of our effort in understanding late blight resistance of potato varieties that showed promising field performance. This led to the identification of natural resistance, mediated by recognition of P. infestans Avr3aEM, which will facilitate potato breeding for improved late blight resistance.
Qingshu9 and Longshu7 are dominant potato varieties in Northwestern China. Qingshu9 was derived from crosses of two parents “387521.3 × APHRODITE”, while Longshu7 was derived from Fedori×Zhuangshu3. Potato cuttings have been cultured in a sterilized MS medium for four weeks (Murashige and Skoog, 1962). Next, the germinated seedlings were transferred for another four weeks into vermiculite, and then planted in pots that contain a mix of sterilized vermiculite and peat moss (V/V = 1:2). Also, some potato differentials, including R1, R2, R3a, R4 and R8, were used for the optimization of agroinfiltration assays. In addition, nine potato breeding lines were studied and agro-infiltrated with PiAvr3aEM. Progeny lines resulting from crossing Qingshu9 with Qingshu2, ND, NSS1-5, and Jizhang8, respectively, and Longshu7 with CIP01, CIP03, CIP16, CIP30, and CIPL06408, respectively, were evaluated. At least 20 progenies from each cross were evaluated. Potato plants were grown under standardized conditions in a greenhouse within a temperature range of 18–22°C and under a day/night regime of 16 h/8 h. Fully-expanded leaves of the 4-week old seedlings were used for infiltration with bacterial cell suspensions of A. tumefaciens strain AGL1 that carry a number of Avr genes to be evaluated.
All tested Avr genes were amplified from their plasmid DNA previously constructed into pK7WG2 vector, using TransStart® FastPfu DNA Polymerase (Applied Biosystems, USA) with Avr genes-specific primers containing the restriction enzyme recognition sites as shown in Supplementary Table S2. The PCR amplicons were purified using the TIANGEN Universal DNA Purification Kit (TianGen Biotech Co., Ltd., Beijing, China). The purified amplicons and the pART27 cloning vector were digested with the corresponding restriction enzymes and ligated together using T4 DNA ligase (Promega, USA). The ligation mixtures were transferred to E. coli DH5α competent cells by electroporation using standard protocols. Transformed colonies were cultured on LB medium supplemented with 100 µg ml−1 of spectinomycin and incubated at 37°C. Positive clones were confirmed by sequencing. The confirmed plasmid constructs were then transformed into A. tumefaciens strain AGL1 by the heat shock method. The transformed cell cultures were applied to LB plates containing antibiotics (100 µg ml−1 of spectinomycin, 20 µg ml−1 of rifampicin) and placed in a 28°C incubator for 2 days. A single colony was transferred using sterilized toothpicks to the liquid LB broth having the same antibiotics and incubated at 200 rpm in a shaker at 28°C for 2 days.
The optimized conditions of agroinfiltration-mediated transient expression assays were, 3–4 or 9–10 week-old potato seedlings, A. tumefaciens strain AGL1 and OD600 value of 0.4. A. tumefaciens cells were grown in LB medium (supplemented with 50 µg ml−1 of gentamicin, 20 µg m l−1 of rifampicin and 100 µg m l−1 of spectinomycin, 20 µg ml−1 of rifampicin, respectively) up to the log phase of development. The bacterial solution was then centrifuged at room temperature (20°C, 4,000g, 3 min), followed by resuspension in an inducing media (10 mM MES, 200 µM acetosyringone, 10 mM MgCl2, pH 5.6). The optical density of the A. tumefaciens suspensions was adjusted to OD600 value of 0.4 and incubated before infiltration for 1–3 h at room temperature. Agroinfiltration experiments were carried out at room temperature 20 ± 2°C (Dillen et al., 1997; Su et al., 2012) on potato seedling leaves 4-week-old and the results were scored from 3 dpi and typically photographed at 5–7 dpi.
To evaluate factors affecting the agro-infiltration assay, three parameters were assessed, including different A. tumefaciens strains (AGL1 and GV3101), bacterial cell densities (OD600 values of 0.2, 0.3, 0.4, 0.5, and 0.6), and different growth ages of cultured potato plants (3–4, 6–7 and 9–10 week-old). All experiments have been repeated three times with 20 replicates for each. In our study, potato seedling age refers to the time starting from tissue culture seedlings transferred to the soil matrix, after acclimatization in the vermiculite, to the time of experimentation.
Ten P. infestans Avr effectors were investigated including, PiAvr1, PiAvr2, PiAvr3aKI, PiAvr3b, PiAvr4, PiAvrblb1, PiAvrblb2, PiAvrsmira1, PiAvr8 (PiAvrsmira2) and PiAvrvnt1.1 (Supplementary Table S1). A. tumefaciens strain AGL1 carrying each of these effectors was used for infiltration in the two varieties with a concentration of OD600 value of 0.4. Further investigation was done for PiAvr3a alleles, PiAvr3aEM and PiAvr3aKI. Forty leaves were agro-infiltrated for each Avr effector in eight independent experiments with five replicates for each. All pictures were taken 5–7 days later of infiltration.
Genomic DNA was isolated from fresh leaf tissue of both potato varieties using the CTAB-based protocol. The genomic DNA was then subjected to PCR with primers specific to R8 and Rpi-vnt1 (Foster et al., 2009; Pel et al., 2009; Vossen et al., 2016), as listed in the Supplementary Table S3. PCR reactions were performed using FastPfu DNA polymerase (Applied Biosystems, USA). Each PCR reaction contained 30 μl PCR mix, including 6 μl 5× FastPfu Buffer, 2.4 μl dNTPs (0.2 mM), 1 μl total genomic DNA (100 ng), 0.2 μl MgSO4 (50mM), 0.6 μl each forward and reverse primers (0.2 mM), 0.6 μl FastPfu DNA polymerase (2.5 units) and 18.6 μl dH2O. The PCR amplification was carried out by denaturing at 95°C for 2 min, followed by 40 cycles of 94°C for 20 s, 55°C for 20 s and 72°C for 1 min, and a final extension time of 5 min at 72°C. PCR products were separated by gel electrophoresis on a 1% agarose gel and DNA bands were visualized under UV on the Quantum CX5 Imaging System.
P. infestans isolates were cultured and maintained on a rye sucrose agar (RSA) medium. All plates were then grown at 16°C in darkness for two weeks. The sporangial suspensions were prepared by washing and rubbing the culture with 5 ml distilled water. Then, the sporangial suspension concentration was adjusted to 4 × 104 sporangia/ml before cooled down for 2 h at 4°C to promote release of motile zoospores for inoculation (Tian et al., 2015). Leaflets of 6 –10 week-old potato plants were placed abaxially on plastic trays on a filter paper saturated with dH2O. All leaflets were drop-inoculated with 15 μl sporangial/zoospore suspension on the abaxial side. Six P. infestans isolates, PjY009, PjY048, PjY061, Pa21106, Pd21410 and F48, were used in the inoculation assays (Supplementary Figure S2). Inoculation with dH2O was considered as a control treatment. All plastic trays were covered by a plastic wrap and incubated in a growth chamber at 16–18°C with >75% relative humidity in the darkness in order to ensure infection. Results were recorded as a lesion diameter of the inoculated area were and pictures were taken five days after inoculation. Disease resistance or susceptibility were recorded by using a scale reported for disease severity (Sun, 2012).
The outcome of plant-Agrobacterium interactions is determined by the genetic background of both partners. In addition to the efficiency of transient gene expression, the frequent non-specific necrotic response is a major concern in the use of this assay. We therefore examined for suitable A. tumefaciens strains with reduced background necrotic reaction in potato. Six different A. tumefaciens strains, Agro-1D124g, GV3101, AGL1, 1100, LBA4404, and EHA105, were evaluated on ten different potato varieties (Data not shown). Even though the OD600 value was very low, strains 1100, LBA4404 and EHA105 induced a high rate of background necrosis on most of the potato cultivars. However, with a lower concentration of bacterial suspensions, GV3101 and AGL1 strains showed a significant reduction of background reaction on most potato varieties. Thus, strains GV3101 and AGL1 were employed to further investigate their transient expression efficiencies on the potato. The efficiency assay was examined by the HR symptoms that resulted from the co-infiltration of P. infestans Avr gene PiAvrblb1 and its cognate resistance gene RB. The results showed that the AGL1 strain was more efficient than GV3101 in terms of triggering specific HR mediated by co-expression of RB and PiAvrblb1 (Figure 1).
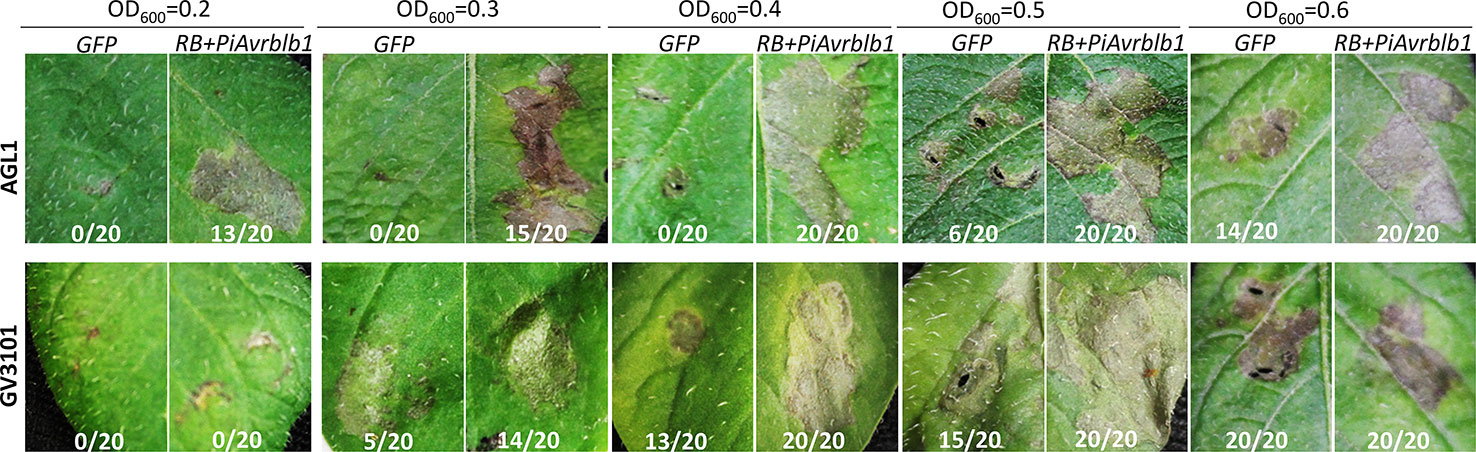
Figure 1 Specific hypersensitive response (HR) mediated by recognition of P. infestans Avr genes in potato is affected by strains used for A. tumefaciens infiltration. Strains AGL1 and GV3101 were examined with different optical densities, OD600 of 0.2, 0.3, 0.4, 0.5 and 0.6, in the potato differential genotype R3. Co-infiltration of agrobacteria carrying PiAvrblb1 and RB was used as a positive treatment, while an A. tumefaciens carrying Green Fluorescent Protein gene (GFP) was infiltrated as a negative control. The number of HR sites/total number of infiltration sites were indicated. The experiment was repeated three times with 20 replicates. Pictures were taken at 5–7 days dpi.
We also evaluated various potato seedling growth ages (3–4, 6–7 and 9–10 week-old) for the effect on the efficiency of the agro-infiltration assay. Five potato differential lines, including R1, R2, R3a, R4, and R8, were examined and agro-infiltrated with A. tumefaciens AGL1 suspensions (OD600 of 0.4) carrying the P. infestans Avr genes PiAvr1, PiAvr2, PiAvr3aKI, PiAvr4, and PiAvr8, respectively. The positive control was the co-infiltration of mixed agrobacteria carrying PiAvrblb1 and RB which would lead to HR while the negative control was the GFP. The most consistent and efficient infiltration was observed while using terminal leaflets from 3–4 and 9–10 week-old potato plants in all tested differential lines, while the 6–7 week potato leaves exhibited less efficient transient expression (Supplementary Figure S1). We speculated that the potato leaves were younger in 3–4 weeks when the leaves have just spread and the main veins were developed, but the lateral veins were not obvious, allowing easier infiltration in whole leaves. Meanwhile, the leaves of 9–10 week-old seedlings were fully developed, and the main and lateral veins were well developed, allowing efficient infiltration between the two lateral veins. However, the main veins and lateral veins of leaves of 6–7 week-old seedlings were all developed, still small interveinal spaces on the abaxial side hinder the infiltration process, making the bacterial solution restricted to a fixed grid, necessitating more infiltration sites. The optimum condition was utilized for further analysis which could be summarized as using the A. tumefaciens strain AGL1, with an OD600 value of 0.4 and leaves of the 3–4 or 9–10 week-old seedlings.
To further confirm our improved agroinfiltration assay, we examined known Avr effector genes for their capability in triggering genotype-specific HR. A. tumefaciens AGL1 bacterial suspensions carrying the P. infestans Avr genes PiAvr1, PiAvr2, PiAvr3aKI, PiAvr4, and PiAvr8 were infiltrated in potato differential lines carrying genotype-specific R genes R1, R2, R3a, R4 and R8, respectively. Each density (OD600 values of 0.2, 0.4 and 0.6) showed a different level of transient expression. The bacterial suspensions with OD600 value of 0.4 consistently displayed the highest efficiency in bacterial infiltration assays, as all tested Avr genes induced genotype-specific HR in all tested differential lines (Figure 2). While at a higher agrobacterial concentration of OD600 of 0.6, an increase of HR response for all tested Avr genes and significant background necrosis for the negative control of GFP expression were observed in all tested differentials, though at the lower agrobacterial concentration (OD600 of 0.2) the HR triggered by PiAvr3a and PiAvr8 in potato differential lines R3a and R8, respectively, were not visible.
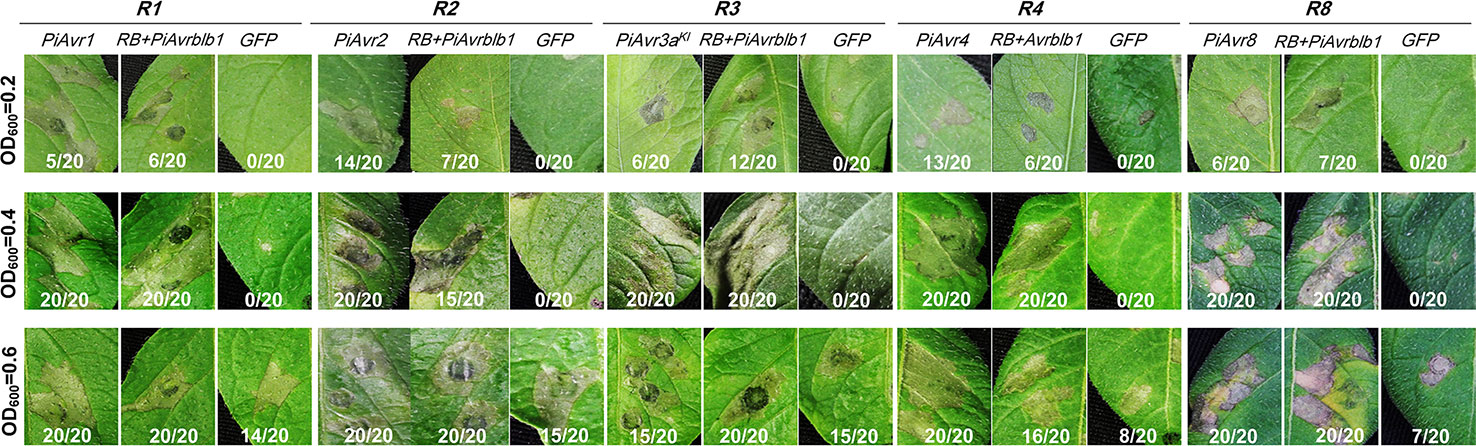
Figure 2 Effects of OD600 values of agrobacterial suspension on the HR response mediated by recognition of P. infestans Avr genes. Different OD600 values, 0.2, 0.4 and 0.6, were examined for genotype-specific HR triggered by P. infestans Avr genes on potato differential lines carrying the cognate R genes. A. tumefaciens AGL1 suspensions carrying P. infestans Avr genes PiAvr1, PiAvr2, PiAvr3aKI, PiAvr4, and PiAvr8, were infiltrated in potato differential genotypes carrying R1, R2, R3a, R4 and R8, respectively. Co-infiltration of agrobacteria carrying PiAvrblb1 and RB was used as a positive control, while GFP as a negative control. The number of HR sites/total number of infiltration sites were indicated. The experiment was repeated three times with 20 replicates. All images were taken at 5–7 dpi.
To understand late blight resistance of two Chinese potato varieties, Qingshu9 and Longshu7, that showed excellent field performance with a low percentage of disease incidence and severity (Wang et al., 2018), we evaluated whether they contain known R genes by examining their capability to recognize corresponding 10 P. infestans Avr genes. Both Qingshu9 and Longshu7 showed typical genotype-specific HR phenotypes 5 days post infiltration with A. tumefaciens AGL1 cell suspensions with an OD600 of 0.4. Qingshu9 showed HR triggered by two P. infestans Avr genes, PiAvr4 and PiAvr8, suggesting the presence of R genes R4 and R8. Longshu7 showed HR triggered by PiAvrvnt1.1, indicating the existence of Rpi-vnt1 (Figure 3). Furthermore, the presence of R8 and Rpi-vnt1 in Qingshu9 and Longshu7, respectively, was preliminarily analyzed by PCR amplification using gene-specific primers (Foster et al., 2009; Pel et al., 2009; Vossen et al., 2016) (Supplementary Figure S3). However, whether they are functional R genes needs further validation. PCR amplification might provide possibility for their presence since it is highly dependent on the specific primers, while the potential presence of functional R gene homologs/alleles may lead to false negative results. The agroinfiltration assay using effector genes is an efficient method to detect the presence of functional R genes, such as R8 and Rpi-vnt1.1 in this research.
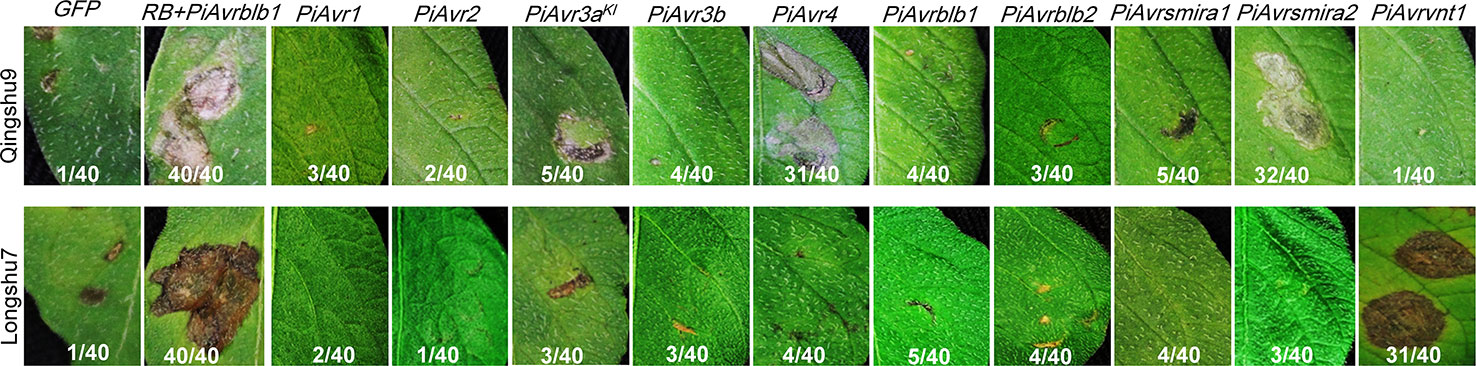
Figure 3 Specific HR induced by 10 known P. infestans Avr genes in two potato varieties, Qingshu9 and Longshu7. In Qingshu9, the typical HR was induced by PiAvr4 and PiAvrsmira2, whereas Longshu7 was responsive to PiAvrvnt1.1. A. tumefaciens AGL1 suspension with an OD600 value of 0.4 was used for infiltration of 4-week-old potato leaves. GFP was used as the negative control, shown at the left side, while the positive control was indicated by co-infiltration of agrobacteria carrying PiAvrblb1 and RB, respectively. The indicated are the number of HR-responsive leaves/total number of the infiltrated leaves. All pictures were taken at 5–7 dpi.
P. infestans Avr gene PiAvr3aKI can be specifically recognized by the cognate R3a. However, the number of its virulent alleles, that escaped recognition by R3a, is very limited and the virulent allele PiAvr3aEM is widely present around the world, suggesting the vital role of PiAvr3a in P. infestans pathogenesis. The identification of varieties with capable PiAvr3aEM recognition that make it possible for breeding new varieties with the capability to recognize both PiAvr3aKI and PiAvr3aEM, which is predicted to improve durable resistance against late blight. Both Qingshu9 and Longshu7 showed an HR upon PiAvr3aEM infiltration, but not upon PiAvr3aKI (Figure 4).
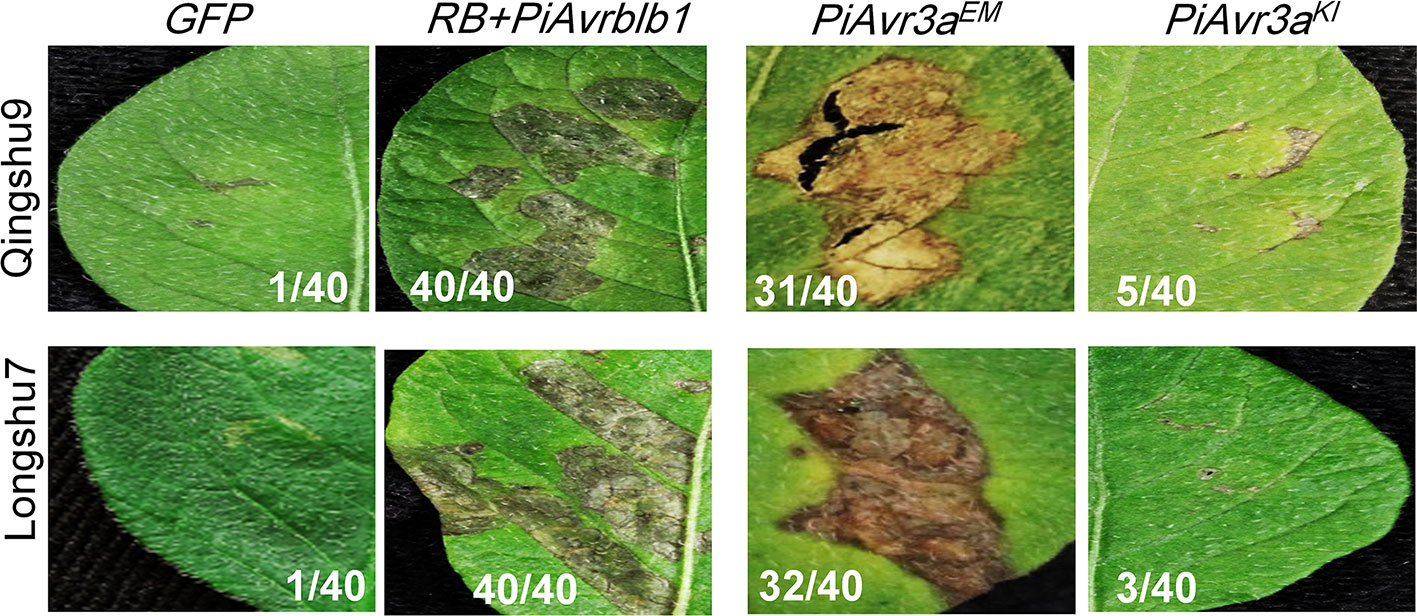
Figure 4 Potato varieties Qingshu9 and Longshu7 were capable of recognizing P. infestans PiAvr3aEM. Both varieties were triggered with HR when infiltrated with agrobacteria carrying PiAvr3aEM, a virulent allele of PiAvr3aKI, but not by PiAvr3aKI. The A. tumefaciens AGL1 bacterial suspension with an OD600 of 0.4 was used for infiltration of the 4-week-old potato leaves. The HR positive control was indicated by co-expression of PiAvrblb1 and RB, shown at the left side, and GFP was used as the negative control. The number of HR-responsive leaves/total number of the infiltrated leaves, were indicated. All pictures were taken at 5–7 dpi.
Given the fact that PiAvr3aKI (Armstrong et al., 2005) is recognized by R3a (Huang et al., 2005), a cloned and well-studied R gene, we predict that PiAvr3aEM is similarly recognized by a single R gene. We therefore employed several independent F1 segregation populations to ensure that the PiAvr3aEM recognition is conditioned by a single R gene, by examining whether the PiAvr3aEM recognition-triggered cell death segregates. We performed agroinfiltration assays for progenies derived from a total of nine crosses for the two responsive varieties, with five crosses using Longshu7 as a resistant parental with five non-responsive potato clones as the susceptible parental, including CIP01, CIP03, CIP16, CIP30, and CIPL06408. Qingshu9 as the resistant parental was crossed with four non-responsive potato clones as the susceptible parental, including Qingshu2, ND, NSS1-5, and Jizhang8. Twenty F1 progenies from each cross were tested for their response upon infiltration with PiAvr3aEM, with a total of 30 infiltration sites for each progenies. The results showed that progenies derived from two investigated Longshu7 crosses showed an HR response upon PiAvr3aEM infiltration at a rate of 1:1 for each cross, including Longshu7 X CIP01 and Longshu7 X CIP16 as shown in (Figure 5) and supplementary (Supplementary Table S4). While for Qingshu9 crosses, progenies from only one investigated cross (Qingshu9 X ND) showed an HR response with a rate of 1:1 (Figure 5, Supplementary Table S4). Although we did not perform comprehensive genetic analysis, the segregation of PiAvr3aEM recognition strongly suggests that PiAvr3aEM recognition by Qingshu9 and Longshu7 is conditioned by a single R gene. Also, the results suggest that the R genes for PiAvr3aEM recognition in Qingshu9 and Longshu7 were heterozygous, and most, if not all, parental lines crossed with Qingshu9 or Longshu7 were unable to recognize PiAvr3aEM. Another possibility is that a helper/sensor NLR might be required for R3a function to initiate the immune signaling, resulting in an HR response, similar to the case of NRC4, a helper NLR essential for immunity triggered by Rpi-blb2 (Wu et al., 2017).
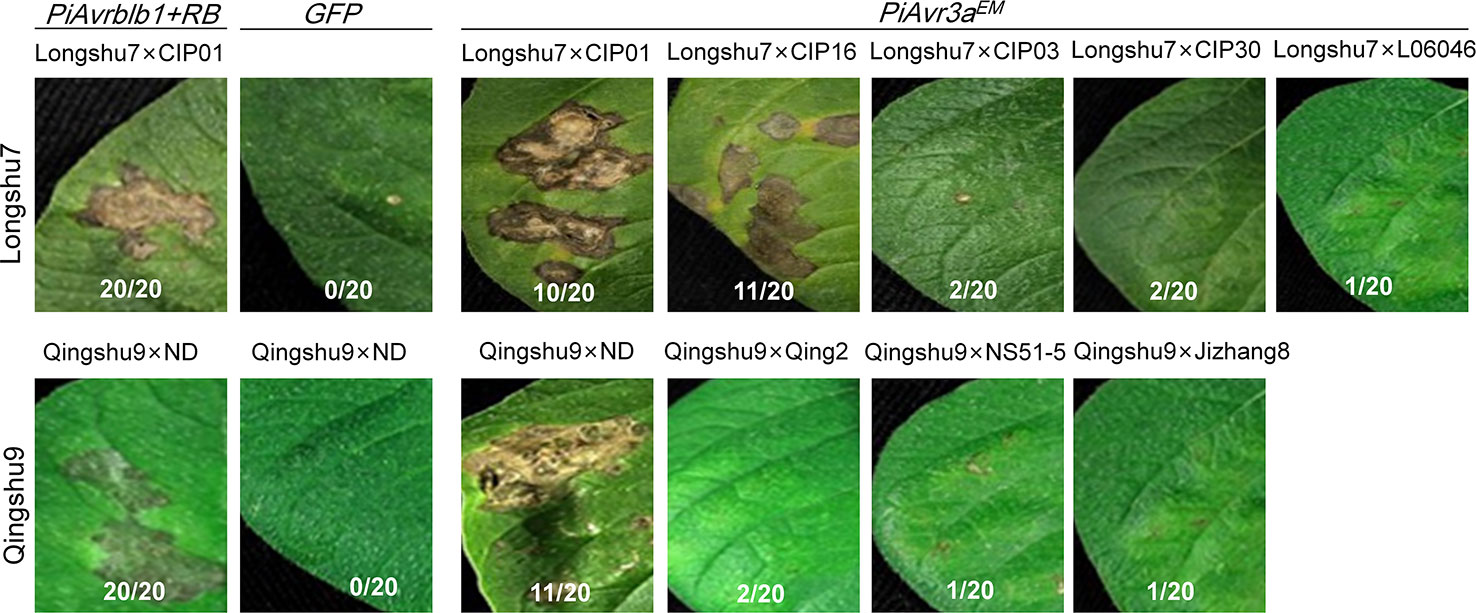
Figure 5 Segregation of HR induced by recognition of P. infestans PiAvr3aEM in F1 populations using Longshu7 or Qingshu9 as a parental. Progenies derived from crosses Longshu7 X CIP01 and Longshu7 X CIP16 showed typical HR as that in Longshu7, and progenies from cross Qingshu9 X ND showed typical HR as that in Qingshu9. GFP was used as negative control and co-expression of PiAvrblb1 and RB was used as a positive control. The number of HR-responsive progenies/total number of progenies, were indicated in each crosses, with each progenies examined with 30 infiltration sites. All pictures were taken at 5–7 dpi.
A critical step in the successful agroinfiltration-mediated transient expression is the establishment of harmonious interaction between the plant and A. tumefaciens. We also considered potential background non-specific necrotic reactions frequently caused by molecules from A. tumefaciens. We examined multiple A. tumefaciens strains (Agro-1D124g, GV3101, AGL1, 1100, LBA4404, and EHA105), potato genotypes, growth stages, and bacterial densities. This led to the conclusion that AGL1 was the most efficient strain with fewer background effects than GV3101, which is consistent with a report showing that strain AGL1 was preferred for potato whereas GV3101 was more suitable for Nicotiana benthamiana (Du et al., 2014).
Potato leaves at various growth stages were also a major concern and we found that the maximum level of bacterial infiltration was observed at the terminal leaflets from 3–4 to 9–10 week-old potato seedlings. The efficiency of infiltration became much lower when leaves were used from 6–7 week-old seedlings. Changes in the expression levels may be related to general changes in leaf physiology, especially soluble protein concentration during leaf aging (Halfhill et al., 2003; Wydro et al., 2006). Also leaf morphological characteristics including, leaf surface, the thickness of the cuticle layer and epidermis, stomatal size and frequency, veins, trichomes, the midrib structure, and interveinal distribution on the abaxial side leaves may affect infiltration efficiency (Abdullah and Halterman, 2018). Some developmental ages have a low density of trichomes on the abaxial side and the leaf veins on the surface are not prominent and as a result, facilitate infiltration (as 4-week-old leaves). Besides, the older leaves (9–10-week-old) where interveinal space has increased showed an increase in transient expression. Meanwhile, the 6–7-week-old leaves exhibited an irregular leaf surface with a high density of trichomes on the abaxial side, prominent veins and small interveinal spaces that can hinder the infiltration process.
Ten Avr genes had been described in P. infestans and their gain-of-virulence alleles were reported (Vleeshouwers and Oliver, 2014). In this study, we aimed at detecting and identifying R genes in potato varieties that showed excellent performance against late blight. Agroinfiltration analysis of 10 P. infestans Avr genes was done using A. tumefaciens strain AGL1 with OD600 value of 0.4 to evaluate two potato varieties, Qingshu9 and Longshu7, whether they encode R genes that may recognize these Avr genes. The agroinfiltration in Qingshu9 resulted in activating a typical HR towards three effector genes including, PiAvr4, PiAvr8 and PiAvr3aEM, which is a virulent allele of PiAvr3a that can be recognized by R3a, whereas Longshu7 exhibited a typical HR by two P. infestans effector genes, PiAvr3aEM and PiAvrvnt1. According to the gene-for-gene hypothesis, Avr genes are detected by its counterpart R genes (Anderson et al., 2015; Yin et al., 2017). These results suggest that Qingshu9 may carry at least three R genes, including R4, R8, and R3a*, whereas Longshu7 carries at least two R genes, Rpi-vnt1 and R3a*.
PiAvr3a was highlighted and has been extensively studied which is expected to be a useful target for potato breeders seeking durable resistance (Cooke et al., 2012). PiAvr3a appears to be a core effector of P. infestans since it's among few effectors that are conserved across several Phytophthora species and it is consistently induced within the early stages of P. infestans infection (Yin et al., 2017). Besides, it is involved in the suppression of PTI and ETI (Gilroy et al., 2011; Franco-Orozco et al., 2017). So far, there are only two detected PiAvr3a alleles among P. infestans populations (Bos et al., 2010). The avirulent allele, PiAvr3aKI, is recognized by R3a, while its virulent allele, PiAvr3aEM, evades recognition by R3a (Armstrong et al., 2005; Chapman et al., 2014).
Previous studies showed successful recognition of PiAvr3aEM by engineering potato resistance gene R3a (Chapman et al., 2014) and by screening the R3a variants library resulting from random mutagenesis of the full-length R3a coding sequence (Segretin et al., 2014). Remarkably, our results offer a new natural resistance gene that can recognize PiAvr3aEM in two potato varieties, suggesting that these two varieties are potentially undergoing R3a*-mediated recognition responses. Both varieties were derived from crosses that used different parents, for Longshu7 being derived from Fedori×Zhuangshu3, while Qingshu9 from 387521.3 × APHRODITE, suggesting that both varieties might contain a functional homolog of the R3a* resistance gene and both are very likely heterozygous. It's also possible that they might have two different forms of R3a* that mediate PiAvr3EM recognition. Both varieties didn't show any PiAvr3KI recognition, suggesting that they don't carry the known R3a.
Further work on the survey of PiAvr3EM -mediated HR on progenies derived from crosses using either Qingshu9 or Longshu7 as a parental indicated that progenies from two Longshu7 crosses were detected with PiAvr3aEM-mediated HR, while progenies from a single Qingshu9 cross were detected for inducing an HR response, suggesting that the recognition of PiAvr3aEM is most likely conditioned by a single R gene R3a* in both varieties. Most lines that were crossed with either Longshu7 or Qingshu9, if not all, do not carry R3a*. The lack of PiAvr3aEM response in some populations is likely resulted from the heterologous nature of PiAvr3aEM recognition in the resistant parents and short of PiAvr3aEM recognition in the other parents. It's also possible that we examined limited number of progenies. However, whether the PiAvr3aEM recognition in Longshu7 or Qingshu9 mediates late blight resistance needs additional pathogenicity tests. Under favorable infection conditions using detached leaves, our preliminary infection assays with diverse virulent P. infestans strains showed generally high levels of late blight resistance for Longshu7 and Qingshu9, though certain level of susceptibility was notable to several virulent strains (Supplementary Figure S2). There are potentially complicated interactions between effectors in suppression and triggering immune response. A promising efficient strategy to enhance late blight resistance is to integrate R3a that recognizes PiAvr3aKI and R3a* that mediates PiAvr3aEM response. However, whether such simple R gene combination is correlated with predicted enhanced durable late blight resistance needs confirmation by field assessments.
Rpi-Smira2 (R8) confers quantitative resistance under field conditions and associates with PiAvrSmira2 (PiAvr8) (Rietman et al., 2012; Hajianfar et al., 2014). In our study, Qingshu9 exhibited an HR response upon PiAvr8/PiAvrSmira2 infiltration, suggesting the presence of R8/Rpi-smira2 in Qingshu9. The PiAvr8/PiAvrSmira2-triggered HR in Qingshu9 was consistent with a previous report in which R8 is correlated with quantitative resistance and PiAvr8/PiAvrSmira2 triggered R8-mediated resistance (Rietman et al., 2012). Our findings are also consistent with a report in which genotype-specific HR was induced after R8-PiAvr8 co-infiltration as well as R8-like co-infiltration with PiAvr8 (Jiang et al., 2018). Notably, the NB-LRR gene R8 has been cloned and was thought to provide broad-spectrum and durable field resistance against P. infestans (Vossen et al., 2016; Jiang et al., 2018). It has been reported that Rpi-Smira2 co-localized with the R8 locus and both loci conferred similar resistance levels (Jo, 2013; Stefańczyk et al., 2017). Hence, it was suggested that Rpi-Smira2 and R8 are identical or functional homologs (Jo et al., 2011). In addition, many P. infestans isolates carry PiAvr8 that was reported to trigger an HR response of the R8 gene in disease resistant potato varieties and lines, such as Sarpo Mira from Europe, PB-06, S-60, and QTL dPI09c from China, and Jacqueline Lee from USA (Vossen et al., 2016; Jiang et al., 2018), suggesting its vital role in the pathogen and the effectiveness of R8.
PiAvrvnt1 is recognized by the potato resistance gene Rpi-phu1/Rpi-vnt1 (Foster et al., 2009). Because of its polymorphism, it is associated with a response to a diversified target protein or recognition avoidance (Pel et al., 2009; Pais et al., 2017). Longshu7 showed HR toward Avrvnt1.1, suggesting that it carries the functional Rpi-vnt1 gene which may provide a high level and wide-spectrum late blight resistance (Stefańczyk et al., 2018).
In summary, we developed and used the optimized A. tumefaciens-mediated transient expression assays to evaluate two potato varieties Qingshu9 and Longshu7 that showed years of promising field late blight resistance for R genes they might carry, by detecting the presence of HR triggered by 10 known P. infestans Avr genes. This led to the identification of natural resistance mediated by recognition of PiAvr3aEM, a globally present virulence allele of PiAvr3aKI that plays vital roles in potato-P. infestans interactions. Interestingly, cloning and analysis of R3a* that mediates PiAvr3aEM recognition and other detected R genes in Qingshu9 and Longshu7 will be interesting to make both good use of late blight resistance and improved understanding of disease resistance in future. Together with the identification of additional complementary R genes in the two varieties, these findings will facilitate the development of potato lines with a high level of late blight resistance, by pyramiding these promising R genes.
The datasets generated for this study are available on request to the corresponding author.
WS and YM designed the experiments. AE, JL, XW, CZ, and YM performed the experiments. AE, JL, YM, GW, JW, HL-K, and WS analyzed the data. AE, YM, and WS wrote the manuscript with contribution from all authors.
This work was supported by National Natural Science Foundation of China (31561143007), China Agriculture Research System (CARS-09), Potato Breeding Program from Department of Science and Technology of Ningxia (#2019NYYZ01), and the Programme of Introducing Talents of Innovative Discipline to Universities (project 111) from the State Administration of Foreign Experts Affairs (#B18042).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Vivianne Vleeshouwers of Wageningen University for providing the initially used Avr gene constructs.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00919/full#supplementary-material
Supplementary Table S1 | List of Avr genes of late blight pathogen P. infestans with their corresponding R genes.
Supplementary Table S2 | List of all specific primers used for cloning P. infestans Avr genes.
Supplementary Table S3 | List of specific primers used for detection of R8 and Rpi-vnt1 by PCR amplification.
Supplementary Table S4 | Segregation for PiAvr3aEM response in the F1 populations of crosses Longshu7 X CIP01, Longshu7 X CIP16, and Qingshu9 X ND.
Supplementary Figure S1 | Effect of various growth ages of potato seedlings on the efficiency of Agrobacterium tumefaciens-mediated transient expression. Leaves of three different growth stages, 3-4, 6-7 and 9-10 week-old seedlings, were investigated. Five P. infestans Avr genes PiAvr1, PiAvr2, PiAvr3aKI, PiAvr4 and PiAvr8 were expressed by A. tumefaciens AGL1, OD600 of 0.4, in five potato differential genotypes carrying cognate R genes R1, R2, R3a, R4 and R8, respectively. Co-infiltration of PiAvrblb1 and RB was used as a positive control, whereas GFP used as a negative control. The number of HR sites/total number of infiltration sites were indicated in each treatment. All photographs were taken 5 dpi.
Supplementary Figure S2 | Infection of Qingshu9 and Longshu7 to different P. infestans strains. Detached leaflets of 9-10 week-old plants were drop-inoculated with 4×104 sporangia/mL of six P. infestans strains with known virulence spectrum, including PjY009, PjY048, PjY061 (Yunnan, Southwestern China), Pa21106, Pd21410 (Ningxia, Northwestern China) and F48 (Fujian, Southeastern China), with sterilized dH2O as the control treatment. Qingshu9 was highly resistant (R) to Pa21106, Pd21410 and F48, but susceptible (S) to PjY009, PjY048 and PjY061. Longshu7 was resistant to PjY061, Pa21106, Pd21410 and F48, but susceptible to PjY009 and PjY048. Disease index (DI) were scored and photographs were taken 5 dpi. The experiment was repeated four timed with five replicates.
Supplementary Figure S3 | Detection of R8 and Rpi-vnt1 in Qingshu9 and Longshu7 by PCR amplification using gene-specific primers. Two sets of gene-specific primers for R8 and Rpi-vnt1 each were used for their detection.
Abdullah, S., Halterman, D. (2018). “Methods for transient gene expression in wild relatives of potato,” in Plant Pathogenic Fungi and Oomycetes (New York: Humana Press), 131–138. doi: 10.1007/978-1-4939-8724-5_11
Aguilera-Galvez, C., Champouret, N., Rietman, H., Lin, X., Wouters, D., Chu, Z., et al. (2018). Two different R gene loci co-evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Stud. Mycol. 89, 105–115. doi: 10.1016/j.simyco.2018.01.002
Allen, R. L., Bittner-Eddy, P. D., Grenville-Briggs, L. J., Meitz, J. C., Rehmany, A. P., Rose, L. E., et al. (2004). Host-parasite coevolutionary conflict between Arabidopsis and downy mildew. Science 306, 1957–1960. doi: 10.1126/science.1104022
Anderson, R. G., Deb, D., Fedkenheuer, K., McDowell, J. M. (2015). Recent progress in RXLR effector research. Mol. Plant-Microbe Interact. 28, 1063–1072. doi: 10.1094/MPMI-01-15-0022-CR
Armstrong, M. R., Whisson, S. C., Pritchard, L., Bos, J. I. B., Venter, E., Avrova, A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102, 7766–7771. doi: 10.1073/pnas.0500113102
Ballvora, A., Ercolano, M. R., Weiß, J., Meksem, K., Bormann, C. A., Oberhagemann, P., et al. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30, 361–371. doi: 10.1046/j.1365-313X.2001.01292.x
Black, W., Mastenbroek, C., Mills, W. R., Peterson, L. C. (1953). A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivates. Euphytica 2, 173–179. doi: 10.1007/BF00053724
Bos, J. I., Kanneganti, T. D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Bos, J. I. B., Armstrong, M. R., Gilroy, E. M., Boevink, P. C., Hein, I., Taylor, R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107, 9909–9914. doi: 10.1073/pnas.0914408107
Brylińska, M., Tomczyńska, I., Jakuczun, H., Wasilewicz-Flis, I., Witek, K., Jones, J. D., et al. (2015). Fine mapping of the Rpi-rzc1 gene conferring broad-spectrum resistance to potato late blight. Eur. J. Plant Pathol. 143, 193–198. doi: 10.1007/s10658-015-0663-2
Champouret, N., Bouwmeester, K., Rietman, H., van der Lee, T., Maliepaard, C., Heupink, A., et al. (2009). Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant-Microbe Interact. 22, 1535–1545. doi: 10.1094/MPMI-22-12-1535
Chaparro-Garcia, A., Schwizer, S., Sklenar, J., Yoshida, K., Petre, B., Bos, J. I., et al. (2015). Phytophthora infestans RXLR-WY effector AVR3a associates with dynamin-related protein 2 required for endocytosis of the plant pattern recognition receptor FLS2. PloS One 10, e0137071. doi: 10.1371/journal.pone.0137071
Chapman, S., Stevens, L. J., Boevink, P. C., Engelhardt, S., Alexander, C. J., Harrower, B., et al. (2014). Detection of the virulent form of AVR3a from Phytophthora infestans following artificial evolution of potato resistance gene R3a. PloS One 9, e110158. doi: 10.1371/journal.pone.0110158
Chen, Y., Liu, Z., Halterman, D. A. (2012). Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PloS Pathog. 8, e1002595. doi: 10.1371/journal.ppat.1002595
Cooke, D. E. L., Cano, L. M., Raffaele, S., Bain, R. A., Cooke, L. R., Etherington, G. J., et al. (2012). Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PloS Pathog. 8, e1002940. doi: 10.1007/s00299-014-1656-x
Dillen, W., De Clercq, J., Kapila, J., Zambre, M., Van Montagu, M., Angenon, G. (1997). The effect of temperature on Agrobacterium tumefaciens-mediated gene transfer to plants. Plant J. 12, 1459–1463. doi: 10.1046/j.1365-313x.1997.12061459.x
Dou, D., Kale, S. D., Liu, T., Tang, Q., Wang, X., Arredondo, F. D., et al. (2010). Different domains of Phytophthora sojae effector Avr4/6 are recognized by soybean resistance genes Rps4 and Rps6. Mol. Plant-Microbe Interact. 23, 425–435. doi: 10.1094/MPMI-23-4-0425
Du, J., Rietman, H., Vleeshouwers, V. G. A. (2014). Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. J. Vis. Exp. 83, e50971. doi: 10.3791/50971
Foster, S. J., Park, T. H., Pel, M., Brigneti, G., Sliwka, J., Jagger, L., et al. (2009). Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol. Plant-Microbe Interact. 22, 589–600. doi: 10.1094/MPMI-22-5-0589
Franco-Orozco, B., Berepiki, A., Ruiz, O., Gamble, L., Griffe, L. L., Wang, S., et al. (2017). A new proteinaceous pathogen-associated molecular pattern (PAMP) identified in Ascomycete fungi induces cell death in Solanaceae. New Phytol. 214, 1657–1672. doi: 10.1111/nph.14542
Fry, W. E. (2008). Phytophthora infestans: the plant (and R gene) destroyer. Mol. Plant Pathol. 9, 385–402. doi: 10.1111/j.1364-3703.2007.00465.x
Fry, W. E. (2016). Phytophthora infestans: New tools (and old ones) lead to new understanding and precision management. Annu. Rev. Phytopathol. 54, 529–547. doi: 10.1146/annurev-phyto-080615-095951
Gilroy, E. M., Breen, S., Whisson, S. C., Squires, J., Hein, I., Kaczmarek, M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. doi: 10.1111/j.1469-8137.2011.03736.x
González-Lamothe, R., Tsitsigiannis, D. I., Ludwig, A. A., Panicot, M., Shirasu, K., Jones, J. D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083. doi: 10.1105/tpc.106.040998
Goodwin, S. B., Spielman, L. J., Matuszak, J. M., Bergeron, S. N., Fry, W. E. (1992). Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology 82, 955–961. doi: 10.1094/Phyto-82-955
Goss, E. M., Tabima, J. F., Cooke, D. E. L., Restrepo, S., Fry, W. E., Forbes, G. A., et al. (2014). The Irish potato famine pathogen Phytophthora infestans originated in central Mexico rather than the Andes. Proc. Natl. Acad. Sci. U.S.A. 111, 8791–8796. doi: 10.1073/pnas.1401884111
Grunwald, N. J., Flier, W. G. (2005). The biology of Phytophthora infestans at its center of origin. Annu. Rev. Phytopathol. 43, 171–190. doi: 10.1094/PHYTO.2001.91.9.882
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H. Y., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398. doi: 10.1038/nature08358
Hajianfar, R., Polgar, Z. S., Wolf, I., Takacs, A., Cernák, I., Taller, J. (2014). Complexity of late blight resistance in potato and its potential in cultivar improvement. Acta Phytopathol. Entomol. Hung 49, 141–161. doi: 10.1556/APhyt.49.2014.2.2
Halfhill, M. D., Millwood, R. J., Rufty, T. W., Weissinger, A. K., Stewart, C. N. (2003). Spatial and temporal patterns of green fluorescent protein (GFP) fluorescence during leaf canopy development in transgenic oilseed rape, Brassica napus L. Plant Cell Rep. 22, 338–343. doi: 10.1007/s00299-003-0696-4
Haverkort, A. J., Boonekamp, P. M., Hutten, R., Jacobsen, E., Lotz, L. A. P., Kessel, G. J. T., et al. (2016). Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Res. 59, 35–66. doi: 10.1007/s11540-015-9312-6
Huang, S. W., van der Vossen, E. A., Kuang, H., Vleeshouwers, V. G. A., Zhang, N., Borm, T. J., et al. (2005). Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251–261. doi: 10.1111/j.1365-313X.2005.02365.x
Huang, G., Liu, Z., Gu, B., Zhao, H., Jia, J., Fan, G., et al. (2019). An RXLR effector secreted by Phytophthora parasitica is a virulence factor and triggers cell death in various plants. Mol. Plant Pathol. 20, 356–371. doi: 10.1111/mpp.12760
Jacobs, M. M. J., Vosman, B., Vleeshouwers, V. G. A., Visser, R. G. F., Henken, B., van den Berg, R. G. (2010). A novel approach to locate Phytophthora infestans resistance genes on the potato genetic map. Theor. Appl. Genet. 120, 785–796. doi: 10.1007/s00122-009-1199-7
Jiang, R., Li, J., Tian, Z., Du, J., Armstrong, M., Baker, K., et al. (2018). Potato late blight field resistance from QTL dPI09c is conferred by the NB-LRR gene R8. J. Exp. Bot. 69, 1545–1555. doi: 10.1093/jxb/ery021
Jo, K. R., Arens, M., Kim, T. Y., Jongsma, M., Visser, R., Jacobsen, E., et al. (2011). Mapping of the S. demissum late blight resistance gene R8 to a new locus on chromosome IX. Theor. Appl. Genet. 123, 1331–1340. doi: 10.1007/s00122-011-1670-0
Jo, K. R., Visser, R. G., Jacobsen, E., Vossen, J. (2015). Characterization of the late blight resistance in potato differential MaR9 reveals a qualitative resistance gene, R9a, residing in a cluster of Tm-2 2 homologs on chromosome IX. Theor. Appl. Genet. 128, 931–941. doi: 10.1007/s00122-015-2480-6
Jo, K. R. (2013). Unveiling and deploying durability of late blight resistance in potato: from natural stacking to cisgenic stacking. PhD thesis (Wageningen, The Netherlands: Wageningen University).
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kuhl, J. C., Hanneman, R. E. J., Havey, M. J. (2001). Characterization and mapping of Rpi1, a late blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol. Genet. Genomics 265, 977–985. doi: 10.1007/s004380100490
Lenman, M., Ali, A., Mühlenbock, P., Carlson-Nilsson, U., Liljeroth, E., Champouret, N., et al. (2016). Effector-driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93-1015. Theor. Appl. Genet. 129, 105–115. doi: 10.1007/s00122-015-2613-y
Lokossou, A. A., Park, T. H., van Arkel, G., Arens, M., Ruyter-Spira, C., Morales, J., et al. (2009). Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBSLRR family of late blight resistance genes from potato linkage group IV. Mol. Plant-Microbe Interact. 22, 630–641. doi: 10.1094/MPMI-22-6-0630
Lokossou, A. A., Rietman, H., Wang, M., Krenek, P., van der Schoot, H., Henken, B., et al. (2010). Diversity, distribution, and evolution of Solanum bulbocastanumlate blight resistance genes. Mol. Plant-Microbe Interact. 23, 1206–1216. doi: 10.1094/MPMI-23-9-1206
Malcolmson, J. F., Black, W. (1966). New R genes in Solanum demissum Lindl. and their complementary races of Phytophthora infestans (Mont.) de Bary. Euphytica 15, 199–203. doi: 10.1007/BF00022324
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plantarum 15, 437–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Oh, S. K., Young, C., Lee, M., Oliva, R., Bozkurt, T. O., Cano, L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21, 2928–2947. doi: 10.1105/tpc.109.068247
Pais, M., Yoshida, K., Giannakopoulou, A., Pel, M. A., Cano, L. M., Oliva, R. F., et al. (2017). Gene expression polymorphism underpins evasion of host immunity in an asexual lineage of the Irish potato famine pathogen. BMC Evol. Biol. 18, 93. doi: 10.1186/s12862-018-1201-6
Park, T. H., Gros, J., Sikkema, A., Vleeshouwers, V. G. A., Muskens, M., Allefs, S., et al. (2005). The late blight resistance locus Rpi-blb3 from Solanum bulbocastanum belongs to a major late blight R gene cluster on chromosome 4 of potato. Mol. Plant-Microbe Interact. 18, 722–729. doi: 10.1094/MPMI-18-0722
Pel, M. A., Foster, S. J., Park, T. H., Rietman, H., Van Arkel, G., Jones, J. D. G., et al. (2009). Mapping and cloning of late blight resistance genes from Solanum venturiiusing an interspecific candidate gene approach. Mol. Plant-Microbe Interact. 22, 601–615. doi: 10.1094/MPMI-22-5-0601
Rauscher, G. M., Smart, C. D., Simko, I., Bonierbale, M., Mayton, H., Greenland, A., et al. (2006). Characterization and mapping of Rpi-ber, a novel potato late blight resistance gene from Solanum berthaultii. Theor. Appl. Genet. 112, 674–687. doi: 10.1007/s00122-005-0171-4
Rehmany, A. P., Gordon, A., Rose, L. E., Allen, R. L., Armstrong, M. R., Whisson, S. C., et al. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17, 1839–1850. doi: 10.1105/tpc.105.031807
Rietman, H., Bijsterbosch, G., Cano, L. M., Lee, H. R., Vossen, J. H., Jacobsen, E., et al. (2012). Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol. Plant-Microbe Interact. 25, 910–919. doi: 10.1094/MPMI-01-12-0010-R
Segretin, M. E., Pais, M., Franceschetti, M., Chaparro-Garcia, A., Bos, J. I., Banfield, M. J., et al. (2014). Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol. Plant-Microbe Interact. 27, 624–637. doi: 10.1094/MPMI-02-14-0040-R
Shan, W., Cao, M., Leung, D., Tyler, B. M. (2004). The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol. Plant-Microbe Interact. 17, 394–403. doi: 10.1094/mpmi.2004.17.4.394
Sliwka, J., Jakuczun, H., Chmielarz, M., Agnieszka, H., Iga, T., Andrzej, K., et al. (2012a). A resistance gene against potato late blight originating from Solanum michoacanum maps to potato chromosome VII. Theor. Appl. Genet. 124, 397–406. doi: 10.1007/s00122-011-1715-4
Śliwka, J., Jakuczun, H., Chmielarz, M., Hara-Skrzypiec, A., Tomczyńska, I., Kilian, A., et al. (2012b). Late blight resistance gene from Solanum ruiz-ceballosii is located on potato chromosome X and linked to violet flower colour. BMC Genet. 13, 11. doi: 10.1186/1471-2156-13-11
Smilde, W. D., Brigneti, G., Jagger, L., Perkins, S., Jones, J. D. (2005). Solanum mochiquense chromosome IX carries a novel late blight resistance gene Rpi-moc1. Theor. Appl. Genet. 110, 252–258. doi: 10.1007/s00122-004-1820-8
Song, J., Bradeen, J. M., Naess, S. K., Raasch, J. A., Wielgus, S. M., Haberlach, G. T., et al. (2003). Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. U.S.A. 100, 9128–9133. doi: 10.1073/pnas.1533501100
Stefańczyk, E., Sobkowiak, S., Brylińska, M., Śliwka, J. (2017). Expression of the potato late blight resistance gene Rpi-phu1 and Phytophthora infestans effectors in the compatible and incompatible interactions in potato. Phytopathology 107, 740–748. doi: 10.1094/PHYTO-09-16-0328-R
Stefańczyk, E., Brylińska, M., Brurberg, M. B., Naerstad, R., Elameen, A., Sobkowiak, S., et al. (2018). Diversity of Avr-vnt1 and AvrSmira1 effector genes in Polish and Norwegian populations of Phytophthora infestans. Plant Pathol. 67, 1792–1802. doi: 10.1111/ppa.12875
Su, G., Park, S., Lee, S., Murai, N. (2012). Low co-cultivation temperature at 20°C resulted in the reproducible maximum increase in both the fresh weight yield and stable expression of GUS activity after Agrobacterium tumefaciens-mediated transformation of tobacco leaf disks. Am. J. Plant Sci. 3, 537. doi: 10.4236/ajps.2012.34064
Sun, J. (2012). Virulence diversity of Phytophthora infestans on potato in northwest of China. Master thesis (Yangling, China: Northwest A&F Univ.).
Tan, M. Y., Hutten, R. C. B., Celis, C., Park, T. H., Niks, R. E., Visser, R. G. F., et al. (2008). The RPi-mcd1 locus from Solanum microdontum involved in resistance to Phytophthora infestans, causing a delay in infection, maps on potato chromosome 4 in a cluster of NBS-LRR genes. Mol. Plant-Microbe Interact. 21, 909–918. doi: 10.1094/MPMI-21-7-0909
Tian, Y., Sun, J., Li, H., Wang, G., Ma, Y., Liu, D., et al. (2015). Dominance of a single clonal lineage in the Phytophthora infestans population from northern Shaanxi, China revealed by genetic and phenotypic diversity analysis. Plant Pathol. 64, 200–206. doi: 10.1111/ppa.12251
Turnbull, D., Yang, L., Naqvi, S., Breen, S., Welsh, L., Stephens, J., et al. (2017). RXLR effector AVR2 up-regulates a brassinosteroid responsive bHLH transcription factor to suppress immunity. Plant Physiol. 174, 356–369. doi: 10.1104/pp.16.01804
Van der Lee, T., Robold, A., Testa, A., van't Klooster, J. W., Govers, F. (2001). Mapping of avirulence genes in Phytophthora infestans with amplified fragment length polymorphism markers selected by bulked segregant analysis. Genetics 157, 949–956.
Van der Vossen, E., Sikkema, A., Hekkert, B. T. L., Gros, J., Stevens, P., Muskens, M., et al. (2003). An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. doi: 10.1046/j.1365-313X.2003.01934.x
Van der Vossen, E. A., Gros, J., Sikkema, A., Muskens, M., Wouters, D., Wolters, P., et al. (2005). The Rpi-blb2 gene from Solanum bulbocastanum is a Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44, 208–222. doi: 10.1111/j.1365-313X.2005.02527.x
Van Poppel, P. M. J. A., Guo, J., van de Vondervoort, P. J. I., Jung, M. W., Birch, P. R., Whisson, S. C., et al. (2008). The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol. Plant-Microbe Interact. 21, 1460–1470. doi: 10.1094/MPMI-21-11-1460
Villamon, F. G., Spooner, D. M., Orrillo, M., Mihovilovich, E., Perez, W., Bonierbale, M. (2005). Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana). Theor. Appl. Genet. 111, 1201–1214. doi: 10.1007/s00122-005-0053-9
Vleeshouwers, V. G. A., Oliver, R. P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant- Microbe Interact. 27, 196–206. doi: 10.1094/MPMI-10-13-0313-IA
Vleeshouwers, V. G. A., Rietman, H., Krenek, P., Champouret, N., Young, C., Oh, S. K., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PloS One 3, e2875. doi: 10.1371/journal.pone.0002875
Vleeshouwers, V. G. A., Raffaele, S., Vossen, J. H., Champouret, N., Oliver, R., Segretin, M. E., et al. (2011). Understanding and exploiting late blight resistance in the age of effectors. Ann. Rev. Phytopathol. 49, 507–531. doi: 10.1146/annurev-phyto-072910-095326
Vossen, J. H., van Arkel, G., Bergervoet, M., Jo, K., Jacobsen, E., Visser, R. G. F. (2016). The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor. Appl. Genet. 129, 1785–1796. doi: 10.1007/s00122-016-2740-0
Wang, M., Allefs, S., van den Berg, R. G., Vleeshouwers, V. G. A., van der Vossen, E. A., Vosman, B. (2008). Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor. Appl. Genet. 116, 933–943. doi: 10.1007/s00122-008-0725-3
Wang, P., Li, F., Guo, T., Dou, J., Qi, X., Jie, W., et al. (2018). Identification and evaluation of late blight resistance and yield of potato varieties. Chin. J. Potato 32 (4), 199–204. doi: 10.3969/j.issn.1672-3635.2018.04.002
Witek, K., Jupe, F., Witek, A. I., Baker, D., Clark, M. D., Jones, J. D. (2016). Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotech. 34, 656–660. doi: 10.1038/nbt.3540
Wu, C. H., Abd-El-Haliem, A., Bozkurt, T. O., Belhaj, K., Terauchi, R., Vossen, J. H., et al. (2017). NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U.S.A. 114, 8113–8118. doi: 10.1073/pnas.1702041114
Wydro, M., Kozubek, E., Lehmann, P. (2006). Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 53, 289. doi: 10.18388/abp.2006_3341
Yang, L., Ouyang, H. B., Fang, Z. G., Zhu, W., Wu, E. J., Luo, G. H., et al. (2018). Evidence for intragenic recombination and selective sweep in an effector gene of Phytophthora infestans. Evol. Appl. 11, 1342–1353. doi: 10.1111/eva.12629
Yin, J., Gu, B., Huang, G., Tian, Y., Quan, J., Lindqvist-Kreuze, H., et al. (2017). Conserved RXLR effector genes of Phytophthora infestans expressed at the early stage of potato infection are suppressive to host defense. Front. Plant Sci. 8, 2155. doi: 10.3389/fpls.2017.02155
Yoshida, K., Schuenemann, V. J., Cano, L. M., Pais, M., Mishra, B., Sharma, et al. (2013). The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. eLife 2, e00731. doi: 10.7554/eLife.00731
Keywords: potato late blight, Phytophthora infestans, PiAvr3aEM, Qingshu9, Longshu7, Agrobacterium tumefaciens, RXLR effectors, hypersensitive response
Citation: Elnahal ASM, Li J, Wang X, Zhou C, Wen G, Wang J, Lindqvist-Kreuze H, Meng Y and Shan W (2020) Identification of Natural Resistance Mediated by Recognition of Phytophthora infestans Effector Gene Avr3aEM in Potato. Front. Plant Sci. 11:919. doi: 10.3389/fpls.2020.00919
Received: 16 February 2020; Accepted: 05 June 2020;
Published: 19 June 2020.
Edited by:
Zuhua He, Chinese Academy of Sciences, ChinaReviewed by:
Jack Vossen, Wageningen University and Research, NetherlandsCopyright © 2020 Elnahal, Li, Wang, Zhou, Wen, Wang, Lindqvist-Kreuze, Meng and Shan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weixing Shan, d3hzaGFuQG53YWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.